Abstract
Depolymerization of the actin cytoskeleton induces nuclear trafficking of regulatory proteins and global effects on gene transcription. We here show that in mesenchymal stem cells (MSCs), cytochalasin D treatment causes rapid cofilin-/importin-9-dependent transfer of G-actin into the nucleus. The continued presence of intranuclear actin, which forms rod-like structures that stain with phalloidin, is associated with induction of robust expression of the osteogenic genes osterix and osteocalcin in a Runx2-dependent manner, and leads to acquisition of osteogenic phenotype. Adipogenic differentiation also occurs, but to a lesser degree. Intranuclear actin leads to nuclear export of Yes-associated protein (YAP); maintenance of nuclear YAP inhibits Runx2 initiation of osteogenesis. Injection of cytochalasin into the tibial marrow space of live mice results in abundant bone formation within the space of 1 week. In sum, increased intranuclear actin forces MSC into osteogenic lineage through controlling Runx2 activity; this process may be useful for clinical objectives of forming bone.
Keywords: Mesenchymal stem cells, Cytoskeleton, Bone, Cofilin, Importin 9, Runx2, Yes-associated protein
Introduction
The cell cytoskeleton, which participates in regulation of signal transduction, protein transport, and signal compartmentalization, can undergo reorganization in response to its microenvironment. Cytoskeletal reorganization in response to static physical cues from substrate attachment has been shown to influence lineage allocation of mesenchymal stem cells (MSCs) [1, 2]. Dynamic physical forces induce rearrangement of focal adhesions and interconnecting F-actin struts, leading to enhanced signaling [3, 4] and force transfer [5]. Competition for G-actin can limit development of F-actin networks in discrete cellular locations [6]. In particular, interest has increased in understanding the processes regulating G-actin monomer transport to the nucleus [7], where actin is known to support gene transcription [8, 9] as well as affect nuclear stiffness [10]. Whether intranuclear actin in monomeric or polymerized forms serves specific regulatory roles is unknown [11].
Nuclear actin has both global and specific effects on gene expression [12], affecting chromatin remodeling as well as transcript elongation [13], and its depletion is associated with cell quiescence [14]. Availability of monomeric or polymeric actin in the cytoplasm and nucleus may serve to regulate cell fate through differential binding affinities to multiple transcription factors. myocardin-like protein 1 (MAL/MKL1) affinity for actin monomers has led to its calponin domains being used to generate monomeric actin probes [15]; increased cytoplasmic actin monomers have recently been shown to sequester MKL1 outside of the nucleus [9], relieving suppression of the adipocyte transcription factor PPARγ and thus promoting adipogenesis of MSCs [16]. In a similar fashion, binding of two related transcriptional co-activators, Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ), to polymeric actin promotes their localization outside of the nucleus [17]. This suggests that actin disposition may regulate the localization of transcription factors involved in stem cell lineage decisions.
Actin transport into the nucleus is dependent on the co-regulatory functions of importin-9 and the actin binding protein cofilin, which limits actin polymerization [18]. Nuclear steady state actin levels are also determined by the amount of monomeric actin substrate available for transport, as well as by an export machinery consisting of paired profilin and exportin 6 [19]. Once intranuclear, actin can be found in filamentous forms [15, 20], as well as in a formation defined by the presence of actin-cofilin rods [21]. The role of intranuclear actin formation in controlling gene transcription is poorly understood.
In the case of MSC differentiation, it has been suggested that greater cytoskeletal structure due to increased F-actin stress fibers will enhance differentiation towards an osteoblastic lineage and prevent adipocyte differentiation [1, 22]. The cytoskeletal response to attachment on hard surfaces is thought to be an in vitro representation of osteogenic differentiation occurring along the mineralized surface of skeletal tissue. Dynamic physical force in the form of exercise also reinforces the skeleton [23, 24] and its withdrawal promotes bone resorption [25], effects subtended under Wolff’s Law that form is adapted to function [26]. To better understand cytoskeletal effects on MSC differentiation, we here consider that actin turnover, where F-actin and G-actin dynamically cycle in response to a changing mechanical environment [27, 28], might have a regulatory role in MSC osteogenesis.
In this work, we show that influx of actin to the nucleus, and its maintenance there, regulates osteogenesis via activation of the transcription factor, Runx2, in part by relieving Runx2 from its repressive interaction with YAP. Cytochalasin D (CytoD) or latrunculin directed cytoplasmic F-actin disassembly induces the importin 9/cofilin dependent transport of monomeric G-actin into the nucleus. Formation of intranuclear actin filaments (or actin–cofilin rods) is coincident with osteogenic gene expression, resulting in a robust initiation and acceleration of MSC entry into the osteogenic lineage. Disruption of the cytoplasmic actin cytoskeleton also serves to induce osteogenesis in vivo, where injection of cytochalasin into the tibial marrow compartment, which is replete with bone marrow MSCs, generates abundant trabecular bone formation within 1 week. These data demonstrate that intranuclear actin can exert a potent pro-osteogenic stimulus to MSC differentiation.
Materials and Methods
Reagents
Fetal bovine serum (FBS) was from Atlanta Biologicals (Atlanta, GA). Culture media, trypsin-EDTA reagent, antibiotics, CytoD, and latrunculin B were from Sigma-Aldrich (sigmaaldrich.com). Wnt10b was from R&D. Leptomycin B was from Santa Cruz (scbt.com). YAP expression construct was a gift from Dr. H Zhang (Chongqing Medical University, China). siRNAs were as follows: Importin 9: 5′-CCCAGCUCUUCAACCUGCUUAUGGA and control (nucleotide change within same sequence) 5′-CCCTCTCCTAACCGTTCATTGAGGA; Cofilin 1: 5′-AAACTAGGTGGCAGCGCCGTCATTT and the control 5′-TCATTTCCCTGGAGGGCAAGCCTTT; for Runx2: 5′-CCAGGTTCAACGATCTGAGATTTGT and control 5′-CCATTACCAAGCTGTGATATGGTGT; for Exportin 6: 5′-CAGCAAGTAGGAGCTTGGAGATTCT and control 5′-CAGTGAGGACGAGTTGAGTACATCT; for β-catenin: 5′-CCCTCAGATGGTGTCTGCCATTGTA and control 5′-CCCGATAGGGTCTGTCCTATCTGTA.
Cells and Culture Conditions
Mouse marrow-derived MSC (mdMSC) cells were harvested from murine marrow using a published protocol [29] and human marrow-derived MSC (hMSC) were purchased from Texas A&M hMSC Cell Inventory (Texas). mdMSC were maintained in minimal essential medium (MEM) containing 10% FBS, 100 μg/ml penicillin/streptomycin. hMSC were maintained in MEM containing 15% FBS, 2 mM glutamine, and 100 μg/ml penicillin/streptomycin. For experiments, the cells were plated at a density of 10,000 cells per square centimeter in six-well culture plates (Fisher, fishersci.com) and cultured for 1 day before application of treatments. Osteogenic medium consisted of 50 μg/ml ascorbic acid and 10 μM β-glycerophosphate. Adipogenic medium included 0.5 mM 3-isobutyl-1-methyl xanthine (IBMX), 5 μg/ml insulin, and 1 μM dexamethasone.
Cell Staining
Cultures were fixed in 10% formalin. Alkaline phosphatase staining was performed with a kit (Sigma-Aldrich) as described previously [30]. Alizarin red S staining performed as manufacturer’s instruction to detect calcified matrix.
Immunofluorescence
For microscopy, cells were fixed with 4% paraformaldehyde for 10 minutes, permeabilized in 0.1% Triton-X 100 for 5 minutes, blocked sequentially with 0.2 M glycine for 10 minutes separated by 3× 10-minute phosphate-buffered saline (PBS) washes between steps. Silicone membranes were cut from plates and transferred to six-well plate surface. Actin stress fibers were visualized with Alexa Fluor 488-conjugated phalloidin (Invitrogen, lifetechnologies.com). After 3× 10-minute washes, membranes were sealed with mounting medium on glass. Cells were imaged on an Olympus BX61 inverted microscope system using filters: Alexa Fluor 488 Phalloidin: Semroc 3540B. For 3D pictures, cells were imaged on Olympus IX70 confocal microscope.
Live Cell Imaging
hMSC were plated in six-well plate at 3 × 104 cells per well and labeled with CellLight Actin-RFP (Invitrogen) for 48 hours. The live cells were imaged at every 2 minutes with Zeiss LSM700 CO2 microscope. CytoD (0.1 μg/ml) was added in the culture at 30 minutes after imaging.
Real Time Reverse Transcriptase Polymerase Chain Reaction
Total RNA was isolated with the RNeasy mini kit (Qiagen, Qiagen.com) and treated with DNase I. Reverse transcription of 1 μg of RNA in a total volume of 20 μl was performed before real time polymerase chain reaction PCR (Bio-Rad iCycler). The 25-μl amplification reactions contained primers (0.5 μM), dNTPs (0.2 mM each), 0.03 units Taq polymerase, and SYBR-green (Molecular Probes, Eugene, OR) at 1:150,000. Aliquots of cDNA were diluted 5- to 5,000-fold to generate relative standard curves to which sample cDNA was compared. Apn, β-catenin, Osx, Runx2, Ocn, and 18S primers were as in ref. 31–33. Alp forward primer: 5′-AACCCAGACACAAGCATTCC-3′ and reverse primer: 5′-GCCTTTGAGGTTTTTGGTCA-3′; aP2 forward primer: 5′-CATCAGCGTAAATGGGGATT-3′ and reverse primer: 5′-TCGACTTTCCATCCCACTTC-3′; Axin2 forward primer: 5′-TAGGCGGAATGAAGATGGAC-3′ and reverse primer: 5′-CTGGTCACCCAACAAGGAGT-3′. Standards and samples were run in triplicate. PCR products were normalized to 18S amplicons in the RT sample, and standardized on a dilution curve from RT sample.
Nuclear and Cytoplasmic Protein Fractionation
Cells were washed with 1× PBS, and the cell pellets were resuspended in 0.33 M sucrose, 10 mM Hepes, pH 7.4, 1 mM MgCl2, 0.1% Triton X-100 (pellet vs. buffer, 1:5), and placed on ice for 15 minutes. After 3,000 rpm for 5 minutes, the supernatant was collected (cytoplasmic fraction). The pellet was resuspended in 0.45 M NaCl and 10mM Hepes, pH 7.4, and placed on ice for 15 minutes. After centrifugation at 12,000 rpm for 5 minutes, the nuclear fraction supernatant was collected.
Immunoblot
Whole cell lysates were prepared with lysis buffer (150 mM NaCl, 50 mM Tris HCl, 1 mM EGTA, 0.24% sodium deoxycholate, 1% Igepal, pH 7.5) containing 25 mM NaF and 2 mM Na3VO4; aprotinin, leupeptin, pepstatin, and phenylmethylsulfonyl fluoride were added before each lysis. An amount of 5–20 μg of fractionated or whole lysate proteins were loaded onto a 7%–10% polyacrylamide gel for chromatography and transferred to polyvinylidene difluoride membrane. After blocking, primary antibody was applied overnight at 4°C including antibodies against active β-catenin (clone 8E7; Upstate, Temecula, CA), osteocalcin, PARP, LDH, actin, tubulin, YAP (Santa Cruz), and cofilin1 (Cell Signaling, CellSignal.com). Secondary antibody conjugated with horseradish peroxidase was detected with ECL plus chemiluminescence kit (Amersham Biosciences, gelifesciences.com). The images were acquired with aHPScanjet and densitometry determined using NIH ImageJ, 1.37v.
Intra-Tibial Injection
C57BL/6 wild type mice, 6 weeks old, were used for the CytoD injection experiment. 12 mice were divided into two groups to receive CytoD (0.8 μg in 10 μl) or 10 μl MEM medium control injected into the right tibial marrow space. (IACUC protocol approved).
Histochemical Staining
One week after CytoD injection, the right tibia were excised and fixed in 4% paraformaldehyde solution for 48 hours. The samples were then decalcified in 14% EDTA for 7 days, dehydrated in serial dilutions of ethanol, and embedded in paraffin. After hardening, the samples were sectioned along the long axis into 5 μm thickness, transferred onto 3-aminopropyltriethoxysilane (APES)-coated glass slides, and stained with H&E for histological analysis.
Bone Microarchitecture
The 4% paraformaldehyde fixed tibia was subjected to microcomputed tomography (μCT) analysis. Bone morphology parameters of the proximal tibial metaphysis and mid diaphysis were quantified ex vivo using high-resolution X-ray μCT (Scanco Medical; Wayne, PA). These included bone volume (BV), BV fraction (BV/TV), trabecular number, thickness and spacing, and measures of cortical area and thickness. Beginning 200 μm distal to the growth plate of the metaphysis was evaluated for trabecular bone; from 300 μm to 200 μm for cortical bone, at 12 μm resolution and 55 keV intensity settings. A threshold for each slice was set exclusively for cortical and trabecular bone using an automated script [34]. The reconstructed solid 3D images were then used to quantify bone microarchitecture.
Statistical Analysis
Results are expressed as mean ± SEM. Statistical significance was evaluated by one-way analysis of variance or t-test as appropriate (GraphPad Prism). All experiments were replicated at least three times to assure reproducibility.
Results
Increasing G-Actin Availability Initiates and Enhances Osteogenesis
Increased cytoskeletal tension is thought to contribute to the commitment of MSC to osteogenic lineage [22]. On stiff extracellular matrix, mimicking the natural bone environment, MSCs differentiate more rapidly into osteoblasts when treated with an osteogenic media containing ascorbate and β-glycerophosphate, preceded by an upregulation of the osteogenic transcription factor Runx2 [1]. To understand whether the polymerized state of actin affected MSC lineage, we induced F-actin depolymerization in mdMSC, maintaining the depolymerized state continuously over several days. CytoD (0.1 μg/ml applied daily, Fig. 1A), induced an osteogenic gene program by day 3 even in the absence of an osteogenic medium (Fig. 1B); osterix (Osx) and osteocalcin (Ocn) mRNA expression increased five- and twofold, respectively, in non-osteogenic growth medium. When mdMSC were grown in osteogenic medium, 3 days of CytoD both accelerated and enhanced the induction of the osteogenic gene program (Fig. 1C); Osx and Ocn mRNA were upregulated 40- and 20-fold, respectively. If the CytoD treatment was limited to the first day only, and then removed, MSC did not undergo osteogenesis, indicating that continuous repression of actin depolymerization was required (Supporting Information Fig. S1A). Alkaline phosphatase mRNA also rose, with protein reflected by increased enzymatic activity and stain for alkaline phosphatase activity (Fig. 1D).
Figure 1.
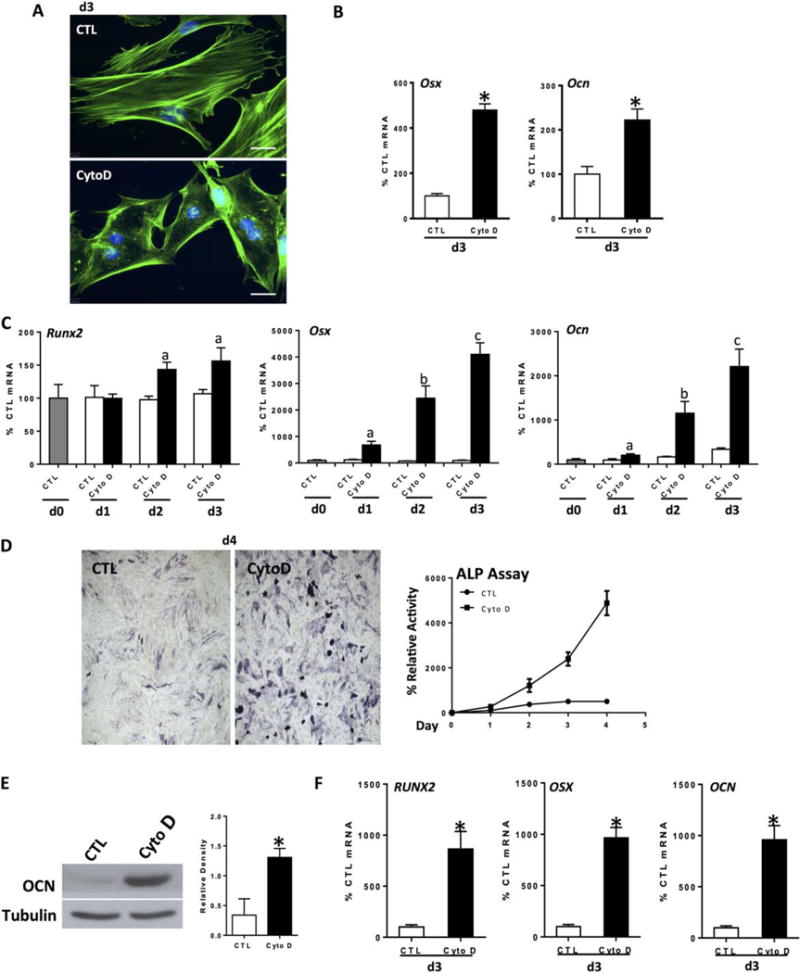
Actin depolymerization initiates and enhances osteogenesis. Mouse marrow-derived mesenchymal stem cell (mdMSC) or human marrow-derived MSC (hMSC) were treated with CytoD (0.1 μg/ml) for indicated times. Except for panel (B), cells were cultured in osteogenic medium. (A): Control and CytoD-treated mdMSC stained with phalloidin, day 3. Scale bars = 25 μm. (B): mdMSC cultured in MEM; Osx and Ocn reverse transcriptase polymerase chain reaction. (C): mdMSC response to continuous CytoD, day 3; notations a, b, c ≠ control and differ from each other with p < 0.05. (D): ALP assay (at d1, control without CytoD = 5.1 nmol nNP/μg total protein per minutes) and ALP stain, mdMSC. (E): Osteocalcin (OCN) protein at 5 d, mdMSC. 3 experiments assessed for densitometry of Ocn, shown in graphs to the right, confirm a significant increase; *, p< 0.05. (F): hMSC response to CytoD, 3 days; *, p < 0.01. Abbreviations: ALP, alkaline phosphatase; CTL, control; CytoD, cytochalasin D; OCN, osteocalcin.
As a corollary, culture with latrunculin B, which inhibits actin filament polymerization by binding to actin monomers [35], also enhanced osteogenic gene expression (Supporting Information Fig. S1B). These results suggest that free G-actin monomers are of significant regulatory importance for osteogenesis.
Runx2 rose in response to continuous actin depolymerization, lagging behind the appearance of specific bone mRNA expression. Both Osx and Ocn, downstream targets of Runx2, rose 24 hours after the application of CytoD (Fig. 1C), with osteocalcin protein rising to threefold more than that in osteogenic medium alone (Fig. 1E). This suggests that Runx2 activity is enhanced during actin depolymerization. This effect was reproduced in hMSC, where CytoD induced a nearly 10-fold increase in osteogenic genes, Osx and Ocn after 3 days (Fig. 1F). Runx2 expression was earlier and perhaps of greater magnitude in the human MSC.
Staining for hydroxyapatite with Alizarin red S to confirm osteoblast function showed that both control and CytoD treated cells generated a mineralized matrix (Supporting Information Fig. S1E). Bone nodules formed in the control plate were typically observed as dense packets amid confluent cells. The number of nodules formed in a culture correlates with the total number of cells initially plated with each bone nodule developing from the progeny of an osteoprogenitor cell type present in the original cultures at a frequency of approximately one in approximately 250 cells [36]. In contrast, although the dose of CytoD used limited cell proliferation (Supporting Information Fig. S1F) as actin filaments are necessary during the process of cell division [37], a large proportion of plated cells entered the osteogenic lineage and achieved capacity to generate a mineralized matrix. This is supported by the increased proportion of cells attaining alkaline phosphatase staining after CytoD treatment, shown in Supporting Information Figure S1C. Actin depolymerization thus appears to induce osteogenic maturation of MSCs.
It has been demonstrated previously that cytoskeletal disruption can promote adipogenesis [16, 38], and we have confirmed that here. CytoD treatment of mdMSC cultured in growth medium at 5 days induces aP2 and adiponectin gene expression at the same time as increasing osteogenic markers (Supporting Information Fig. S2A). In osteogenic medium, however, actin depolymerization enhances osteogenic genes by 10-fold (osterix) and 20-fold (Ocn) while inducing a smaller increase in adipogenic genes (fourfold for aP2 and 10-fold for adiponectin), suggesting that in these conditions the effect to enhance osteogenic genes was more pronounced (Supporting Information Fig. S2B).
Actin Transport into the Nucleus Is Required for Enhanced Osteogenic Gene Expression due to CytoD
Actin stress fiber depolymerization by CytoD generates free monomeric G-actin. G-actin can shuttle between the nucleus and cytoplasm in a highly regulated process [13]. We, therefore, considered that the monomeric G-actin generated by CytoD treatment might regulate osteogenic gene expression after transport into the nucleus. We tracked the location of actin between the cytoplasm and nucleus after CytoD treatment of mdMSCs. Nuclear actin increased by nearly threefold, measured after 3 days treatment with CytoD, with a concurrent decrease in cytoplasmic actin (Fig. 2A). To monitor the actin transport into the nucleus of live cells, RFP-labeled actin was transfected into human MSC: after CytoD treatment, stress fibers disassembled quickly (Supporting Information Movie S1) and accumulated in the nucleus within 30 minutes (Fig. 2B). The 3D images clearly confirmed the nuclear accumulation of actin at 8 hours in CytoD treated cells compared with control cells (Supporting Information Movies S2, S3).
Figure 2.
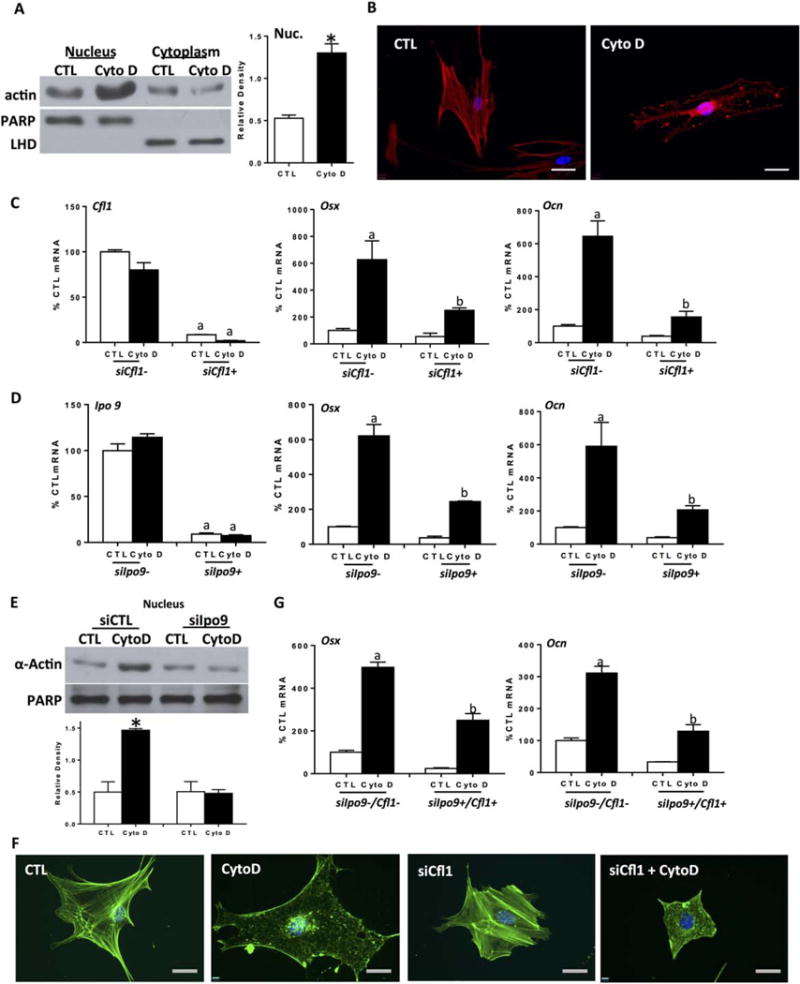
Actin transport into the nucleus is required for enhanced osteogenic gene expression after CytoD. (A): Nuclear and cytoplasmic IB, mouse marrow-derived mesenchymal stem cell (mdMSC) ± CytoD, 3 days. Densitometry of nuclear actin, shown in graphs to the right, confirms a significant increase compared with control, n = 3, *, p< 0.05. (B): CellLight Actin-RFP transfected human marrow-derived MSC imaged 8 hours after ± CytoD. Scale bars = 25μM. (C, D, G). Reverse transcriptase polymerase chain reaction analysis after siRNA treatment for cofilin1 (C), importin 9 (D) and importin 9 + cofilin 1 (G) ±CytoD for 3 days. For Panels (C), (D), and Alkaline phosphatase a, b ≠ control and differ from each other, p < 0.01. (E): Nuclear and cytoplasmic IB. Densitometry for nuclear actin blots reflect n = 3, *, p < 0.05. (F): Phalloidin stain. Scale bars = 25 μM. Abbreviations: CTL, control; CytoD, cytochalasin D; LDH, lactate dehydrogenase; PARP, polyribose polymerase.
Cofilin and importin 9 cooperate to transport actin monomers and dimers into nucleus [39]. Depletion of cofilin 1, the major isoform of cofilin in these cells, with siRNA limited CytoD induced osteogenesis (Fig. 2C). As well, siRNA targeting importin 9 inhibited stimulation of osteogenesis measured at 3 days (Fig. 2D). It is important to note that knockdown of either actin co-transporter also decreased osteogenic gene expression in the absence of CytoD (Osx and Ocn, white bars in Fig. 2C, 2D), suggesting that nuclear actin transport is supportive of osteogenic processes.
To verify that actin was not transported into the nucleus when the actin transport system was interrupted, we confirmed that importin 9 deficient cells failed to show a rise in nuclear actin after CytoD treatment (Fig. 2E). Actin transport was also inhibited when cofilin 1 was specifically targeted by siRNA (Supporting Information Fig. S3A). Phalloidin staining after CytoD treatment revealed nuclear filamentous actin, while staining was absent in the nucleus in cells depleted of either cofilin (Fig. 2F) or importin 9 (Supporting Information Fig. S3B), despite clear depolymerization of the cytoplasmic F-actin cytoskeleton. Simultaneous knockdown of cofilin 1 and importin 9 did not produce an additive inhibitory effect on osteogenesis (Fig. 2G), supporting that these factors work in concert to transport actin, and that the process of actin transport into the nucleus promotes osteogenic gene transcription.
An active export process involving exportin 6 and profilin is required transport G-actin out of the nucleus [18, 40]. To ascertain whether nuclear actin export was involved in CytoD induced osteogenesis, exportin 6 was decreased by RNA interference. Knock down of exportin 6 neither induced osteogenesis nor prevented the increase of osteogenic gene expression due to CytoD (Supporting Information Fig. S3C). That eliminating exportin 6 failed to enhance osteogenesis suggests that sustained actin nuclear influx through a cofilin/importin 9 transport mechanism is critical for this process.
Osteogenesis Resulting from Increased Nuclear Actin Requires Runx2
As Runx2 is the master regulator of osteogenic differentiation of MSC [41] we hypothesized that actin depolymerization might activate Runx2. To investigate this, Runx2 was knocked down using siRNA. Depletion of Runx2 prevented CytoD induction of osteogenic gene expression (Fig. 3A), consistent with a requirement of Runx2 for deployment of the osteogenic gene program. Interestingly, the CytoD induced increase in alkaline phosphatase expression was unaffected by Runx2 knockdown. Alkaline phosphatase (ALP) is a ubiquitous cellular protein [42], even though it can serve as an early marker of osteogenic differentiation. Here, CytoD induced ALP expression was independent of Runx2, and thus dependent on other effects mediated via cytoskeletal remodeling.
Figure 3.
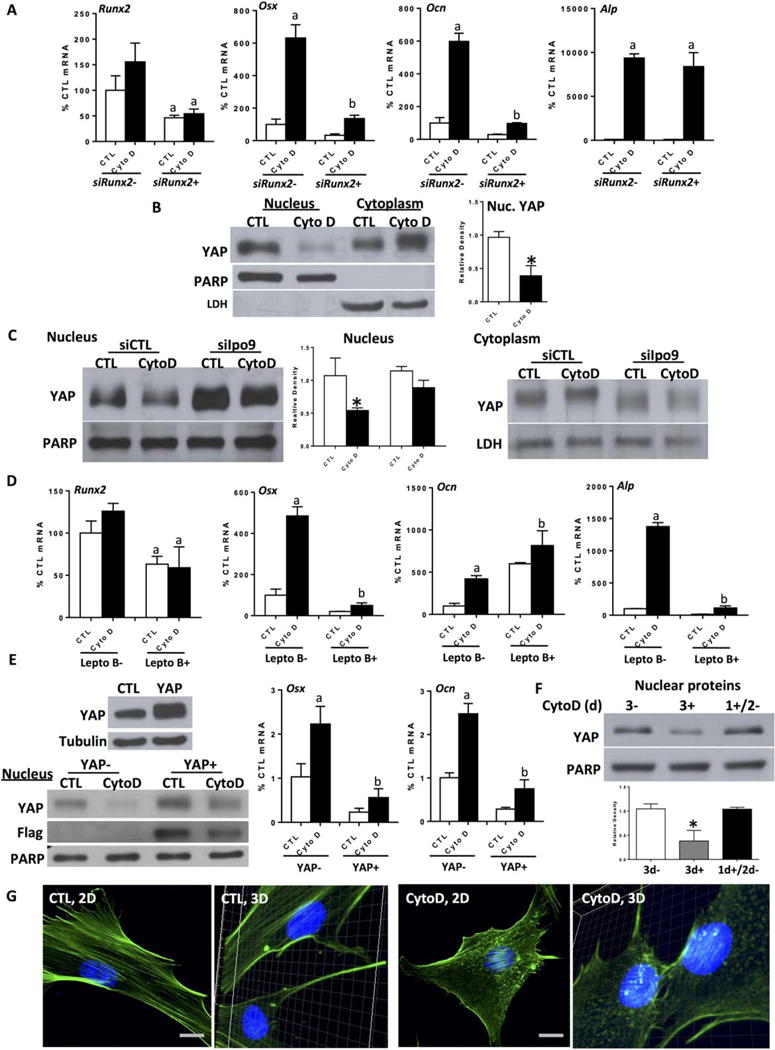
Osteogenesis resulting from increased nuclear actin requires Runx2 activity. (A): Mouse marrow-derived mesenchymal stem cell (mdMSC), ± CytoD for 3 days at 24 hours after Runx2 siRNA treatment; a, b ≠ control and differ from each other, p < 0.01. (B, C): Nuclear and cytoplasmic IB, 3-day treatment of cytochalasin D after importin 9 knock-down. Densitometry of nuclear Yes-associated protein (YAP) is shown in (B, C), n = 3, *, p < 0.05. (D): Reverse transcriptase polymerase chain reaction (RT-PCR) analysis ± leptomycin B (5 ng/ml) and ± CytoD for 3 days; a, b ≠ control and differ from each other with p < 0.01. (E): mdMSC transfected with YAP construct or empty vector. IB, left panel. RT-PCR ± CytoD for 3 days. a, b ≠ control and differ from each other, p <0.01. (F): mdMSC treated with CytoD for 0 (3−), 3d (3+), or on the first day only (1+/2−). IB for nuclear YAP. Densitometry of nuclear YAP bands is shown, n = 3, *, p< 0.05. (G): 2D and 3D images. mdMSC ± CytoD for 3 days, phalloidin stain. Scale bars = 25 μm. Abbreviations: CTL, control; CytoD, cytochalasin D; LDH, lactate dehydrogenase; PARP, polyribose polymerase; YAP, Yes-associated protein.
The activity of Runx2 is inhibited by YAP, which interacts directly with the Runx2 protein to suppress Runx2 transcriptional activity in a dose-dependent manner [43]. As YAP is known to translocate from the nucleus to the cytoplasm during actin depolymerization [44, 45], we considered that YAP trafficking might be involved in Runx2 de-repression. First we showed that after 3 days treatment with CytoD, when osteogenesis is in effect, nuclear YAP was decreased by more than half, and accompanied by increases in the cytoplasmic compartment (Fig. 3B). Immunofluorescence staining showed that intense nuclear YAP stain was diminished and found in the cytoplasm after actin depolymerization (Supporting Information Fig. S4B). We next asked whether the nuclear import of free G-actin was involved in the YAP nuclear egress. siRNA depletion of importin 9, which prevents nuclear actin import, prevented YAP export from the nucleus after CytoD (Fig. 3C). This indicates that G-actin nuclear transfer contributes to the effects of actin depolymerization to induce YAP translocation to the cytoplasm.
Leptomycin B inhibits exportin 1 (CRM1), an evolutionarily conserved receptor for nuclear protein export [46]. The fact that blockade of nuclear export with leptomycin B preserved nuclear localization of YAP in MSC treated with cytoskeletal inhibitors [44] suggested that this inhibitor might restrain osteogenic induction by maintaining nuclear YAP. Indeed, shown in Figure 3D, pretreatment of mdMSC with leptomycin B prevented CytoD stimulation of Osx and Runx2 expression. These data support that maintenance of nuclear YAP contributes regulation of Runx2 activity. Ocn was surprisingly unaffected by leptomycin B pretreatment (Fig. 3D); however, we noted that even in the absence of CytoD, Ocn expression was induced by the presence of leptomycin B. This may be consistent with a direct effect of leptomycin B on osteocalcin transcription.
To further verify an effect of YAP to regulate osteogenic gene expression, YAP protein was overexpressed. Excess YAP protein interfered with CytoD induced osteogenesis in MSC, with retention of some FLAG-YAP expression in the nucleus after CytoD treatment (Fig. 3E). We further showed that YAP localization to the cytoplasm required persistent actin fiber depolymerization: in Supporting Information Figure S4A removing CytoD after 24 hours allowed the actin cytoskeleton to reassemble. Replacing CytoD containing medium with fresh medium after 1 day caused YAP to relocalize from the cytoplasm to the nucleus when measured 24 hours later (Fig. 3F). As such, persistence of nuclear actin is requisite for the nuclear exclusion of YAP, as well as for the induction of osteogenesis (Supporting Information Fig. S1A).
Forcing actin into the nucleus by tagging with a nuclear localization sequence when cofilin is silenced does not rescue actin responsive RNA polymerase II activity [18]. As noted, G-actin nuclear transfer is dependent on nonmuscle cofilin-1, which is also found in complex with actin and phosphorylated RNA polymerase II. Depleting cofilin affects active transcription [47], which might implicate cofilin as a partner in nuclear actin’s effect to modulate gene transcription. Indeed, after actin enters the nucleus, it can form rod like structures with cofilin; these structures are inhibited in the presence of leptomycin B [21]. We examined formation of such actin–cofilin rod structures within nuclei, finding them only in nuclei of CytoD treated cells: Figures 2D, 3D, and 3G show many rod-like phalloidin-stained structures appearing after CytoD treatment. Furthermore, these rods are clearly visible within the nucleus as shown in Supporting Information Movies S4 and S5. Taken together, these results indicate that osteogenic differentiation due to cytoskeletal actin depolymerization depends on Runx2 activation, a process that involves YAP translocation from the nucleus. YAP translocation and potentially gene activation may require the formation of actin/cofilin rods which form when G-actin and cofilin reach appropriate concentrations within the nucleus.
Bone Formation Is Induced by Intra-Tibial Injection of CytoD
Prior investigations have suggested that greater cytoskeletal structure due to increased F-actin stress fibers will enhance osteogenic differentiation of MSC. We, instead, found that rapid and sustained loss of actin cytoskeleton can also lead to osteogenesis in vitro. To test the effect of actin depolymerization in a site where endogenous marrow MSC are readily accessible, we injected CytoD (0.8 μg in 10 μl), or 10 μl MEM medium control, into the right tibial marrow space marrow of 6-week-old mice (IACUC protocol approved). One week after injection, tibiae were excised and analyzed with μCT. The vertical sections revealed that the CytoD-injected tibiae contained more trabeculae in the marrow cavity, with trabecular extension into the diaphysis (Fig. 4A; Supporting Information Fig. S5A). Trabecular parameters measured by μCT showed that trabecular BV/TV, trabecular thickness (Tb.Th), and trabecular number (Tb.N) significantly increased and trabecular spacing (Tb.S) significantly decreased, consistent with new bone formation (Fig. 4B, p > 0.01). In cross-sections, we found that cortical thickness in the CytoD group was increased compared with the control group (Fig. 4A; Supporting Information Fig. S4B, p < 0.01). Cortical bone showed significant increase in cortical bone area (Ct.Ar), total area (Tt.Ar), and in thickness (Ct.Th) in the CytoD treated group (Fig. 4C). H&E staining confirmed the abundant increase in trabecular bone formation in CytoD treated tibiae (Fig. 4D). As such, this indicates that CytoD not only strongly induces formation in trabecular bone but also enhances cortical bone area.
Figure 4.
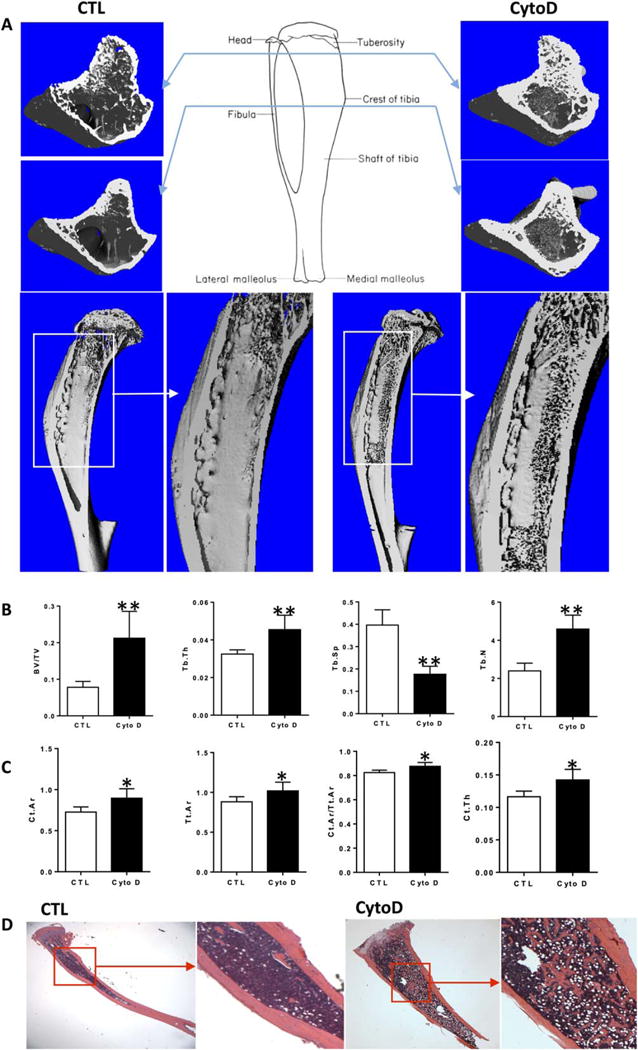
Bone formation is induced by intra-tibial injection of cytochalasin D. (A): 3D images of cross and vertical sections of right tibia reconstructed from μCT. (B, C): Trabecular and cortical quantitative measurement for the same tibia as (A). (D): H&E staining for the right tibia with/without CytoD treatment. Asterisks indicate significant difference, *, p< 0.05; **, p < 0.01. Abbreviations: CTL, control; CytoD, cytochalasin D.
Diminishing cellular tension can promote adipogenic differentiation [2, 22], and as we found with ALP, intracellular actin can generally increase gene activity [8, 12]. Histology of tibial bones (Fig. 4D) suggested that CytoD treated marrow had more white spaces, consistent with marrow adipocytes, than did control mouse tibiae. This is consistent with reverse transcriptase PCR (RT-PCR) results showed that gene expression level of adipogenic markers were increased in mdMSC treated with CytoD (Supporting Information Fig. S2). Importantly, in a bone environment, these levels were less than those reflecting osteogenic gene expression (Supporting Information Fig. S2C). Thus, although depolymerization of actin stress fiber caused adipogenesis in vitro and in vivo, the induction of osteogenesis in MSC in a bone environment predominates over that of adipogenesis.
Discussion
In this study, we provide direct evidence that cofilin-/importin 9-dependent nuclear translocation of actin initiates a robust osteogenic differentiation program in marrow derived MSC. The sustained presence of nuclear actin results in osteoblastogenesis in vitro, an effect recapitulated in vivo as intramarrow injection of CytoD causes robust local bone formation. It has been shown that structural dynamics of nuclear actin filaments can modulate gene transcription [20, 48, 49]. Nuclear entry of G-actin affects the expression profile of many genes either through sequestration of inhibitors and activators [9], or potentially the binding of such regulators to chromatin [48]. Examples include the interaction of MAL, a cofactor of serum response factor (SRF), with monomeric actin in the regulation of SRF induced gene transcription [9] and the reactivation of the pluripotency gene Oct4 by nuclear actin [49]. Here, we show that increased nuclear actin causes YAP relocalization to the cytoplasm resulting in the de-repression of the specific osteogenic transcription factor, Runx2.
Actin, lacking a nuclear localization sequence, requires cofilin to gain access to the nucleus. Depletion of cofilin, or importin 9, its transporter partner, prevented actin induced osteogenesis (Fig. 2). Furthermore, the depolymerization constraint had to be maintained, as removal of CytoD after 24 hours is followed by reformation of F-actin fibers (Supporting Information Fig. S4A), YAP re-entry (Fig. 3F) and failure to induce osteogenesis (Supporting Information Fig. S1A). Either very high intranuclear actin is necessary, or there may be a specific nontransport role for cofilin in Runx2 activation. Besides its key role in nuclear translocation of actin, cofilin participates in the formation of actin/cofilin rods [18, 21]. The formation of actin/cofilin rods during cell stress decreases transcription of most steady-state proteins while increasing transcription and translation of chaperones and other stress related proteins [50]. Here, we show that F-actin structures, perhaps representing actin/cofilin rods, are present within the nucleus (Fig. 3G) after CytoD induced cytoplasmic actin depolymerization.
The increase in the osteogenic gene program exemplified by expression of osterix and osteocalcin followed by mineralization of MSC was found to require the participation of Runx2. The Runx2 binding profile, or its “cistrome,” is altered during osteoblastogenesis [51]. The PY motif of Runx2 has been previously shown to recruit YAP to Runx2 binding sites at heterochromatin, where its presence represses Runx2 activity [43]. Our data suggest that Runx2 activation, consistent with the report by Zaidi, may be regulated through nuclear availability of YAP. Indeed we showed that overexpression of YAP and its nuclear retention almost entirely prevented the osteogenic response to CytoD (Fig. 3E). Recently it has become apparent that the nuclear/cytoplasmic location of YAP and its affiliated protein TAZ are dependent on mechanical and cytoskeletal cues [52]. A highly structured cytoplasmic F-actin network promotes YAP localization within the nucleus [53, 54]. The obverse state, actin depolymerization, results in YAP nuclear egress [55], and is consistent with our data showing that increased nuclear actin promotes YAP nuclear exit. Although extra-nuclear YAP is commonly associated with a decrease in proliferation [56], it may have contextually dependent effects on differentiation: in mechanically sensitive cardiac progenitor cells silencing YAP/TAZ not only decreases proliferation but also promotes endothelial commitment [57]. In the bone marrow MSC studied here, maintenance of YAP exclusion from the nucleus supports osteogenic differentiation.
Cell tension stimulated by internal or external force induces actin polymerization, which is broadly thought to enhance commitment of MSC to osteogenic lineage [58]. However, the fact that actin depolymerization induced by CytoD strongly enhanced osteogenic differentiation both in vitro and in vivo challenges the simplicity of this theory, and suggests that temporal (during MSC differentiation) and contextual (environmental) effects strongly pertain. The turnover of actin polymerization and depolymerization is a common process during cell migration [59], a process that may be crucial to increasing periosteal circumference of loaded bones. Furthermore, osteoblast differentiation in vitro takes place within confluent cultures; here, cells are less spread than in subconfluent dishes, consistent with extra-nuclear localization of YAP. In support of the concept that the cellular fraction of G-actin will enhance osteogenesis, work has shown that inhibition of actin bundling with the ROCK inhibitor, Y27632, augments BMP induced ectopic bone formation in vivo [60]. The ROCK inhibitor also increased osteocalcin production and enhanced bone nodule formation in primary cultures of neonatal murine calvarial cells [60].
In MSC, along with the predominant stimulation of Runx2 dependent osteogenesis, nuclear actin has a Runx2 independent effect to increase alkaline phosphatase mRNA and protein (Fig. 3A) as well as factors associated with adipogenesis. Monomeric G-actin generated by depolymerization of F-actin interacts with (MAL/MKL1) to prevent nuclear access of MKL1, allowing expression of the adipocyte transcription factor PPARγ [16]. We also demonstrated adipogenesis, confirming these findings (Supporting Information Fig. S2), and further showed histological evidence of increased adipocytes after CytoD injection in vivo (Fig. 4D). Quantitative measures showed significant increases in bone formation 1 week after CytoD injection into the in vivo bone environment containing MSC. Nuclear actin may also underlie the periosteal bone formation seen in a study where CytoD was injected into the periosteum [61], this study did not evaluate the mechanism or the target cell. In the case of marrow MSC, nuclear actin’s effect on Runx2 activity to cause osteogenesis dominates the differentiation response under osteogenic conditions both in vivo and in vitro. It is known that actin participates in gene expression through multiple mechanisms, including chromatin remodeling, RNA processing, and is required for transcription by all RNA polymerases [62]; it is likely that the high degree of cofilin-dependent influx of actin in our experiments affects the differentiation state of MSC dependent on multiple mechanisms which are temporally and spatially controlled by cues in the microenvironment.
Finally, delivery of a chemical disruptor of F-actin within mouse tibiae where MSC reside induces abundant trabecular and even cortical bone formation. Whether this effect is sustained, and whether biochemical control of actin turnover could offer a clinical strategy for building bone, remains to be explored. There are limitations to our in vivo experiment. First, we did not follow for longer than 7 days to measure continued bone apposition, or whether the new bone was maintained. Second, the mice used were young, presumably with abundant multipotential MSC, which may not be present in older animals. Furthermore, it is possible that the effect of cytochalasin within the marrow space might be indirect, with an injury response driving resident MSC, and indeed that actin transposition to the nucleus is not involved at all. These questions will need to be addressed before we can propose actin regulation of osteogenesis in the clinic.
Conclusion
In conclusion, depolymerization of the MSC actin cytoskeleton induces a robust osteogenic gene program in marrow derived MSC that leads to bone formation. Such osteogenesis is dependent on actin nuclear transport, where it forms fibrillar intranuclear structures that are associated with YAP relocalization to the cytoplasm and thus de-repression of Runx2 activity. As such regulation of the MSC actin cytoskeleton, and nuclear actin content, should have implications for understanding differentiation of stem cells.
Supplementary Material
Significance Statement.
Actin is a multi-functional protein that forms F-actin. Cytochalasin D binds to actin filaments and blocks polymerization which results in rapid cofilin/importin-9 dependent transfer of free-actin into nucleus. Intranuclear actin induces Runx2-dependent expression of osteogenic genes, and leads to acquisition of osteogenic cell phenotype. Intranuclear actin causes nuclear export of YAP which inhibits Runx2 initiation of osteogenesis in nucleus. In live mice, injection of cytochalasin into MSC-rich tibial marrow results in abundant bone formation. In sum, increased intranuclear actin forces MSC into osteogenic lineage through enhancing Runx2 activity, a mechanism which could be harnessed for clinical objectives of forming bone.
Acknowledgments
This work was supported by NIH grants AR056655 and AR042360.
Footnotes
Author Contributions
B.S.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript; Z.X.: collection and/or assembly of data, data analysis and interpretation, and final approval of manuscript; G.U., W.T., M.S., and X.W.: data analysis and interpretation, and final approval of manuscript; J.R.: conception and design, financial support, data analysis and interpretation, manuscript writing, and final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Engler AJ, Sen S, Sweeney HL, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 2.Swift J, Ivanovska IL, Buxboim A, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen B, Xie Z, Case N, et al. mTORC2 regulates mechanically induced cytoskeletal reorganization and lineage selection in marrow derived mesenchymal stem cells. J Bone Miner Res. 2014;29:78–89. doi: 10.1002/jbmr.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson WR, Guilluy C, Xie Z, et al. Mechanically activated Fyn utilizes mTORC2 to regulate RhoA and adipogenesis in mesenchymal stem cells. Stem Cells. 2013;31:2528–2537. doi: 10.1002/stem.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu S, Chen J, Butler JP, et al. Prestress mediates force propagation into the nucleus. Biochem Biophys Res Commun. 2005;329:423–428. doi: 10.1016/j.bbrc.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 6.Burke TA, Christensen JR, Barone E, et al. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol. 2014;24:579–585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald D, Carrero G, Andrin C, et al. Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–552. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapoor P, Chen M, Winkler DD, et al. Evidence for monomeric actin function in INO80 chromatin remodeling. Nat Struct Mol Biol. 2013;20:426–432. doi: 10.1038/nsmb.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vartiainen MK, Guettler S, Larijani B, et al. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 10.Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baarlink C, Grosse R. Formin’ actin in the nucleus. Nucleus. 2014;5:15–20. doi: 10.4161/nucl.28066. 10.4161/nucl.28066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Percipalle P. Co-transcriptional nuclear actin dynamics. Nucleus. 2013;4:43–52. doi: 10.4161/nucl.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosse R, Vartiainen MK. To be or not to be assembled: Progressing into nuclear actin filaments. Nat Rev Mol Cell Biol. 2013;14:693–697. doi: 10.1038/nrm3681. [DOI] [PubMed] [Google Scholar]
- 14.Spencer VA, Costes S, Inman JL, et al. Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J Cell Sci. 2011;124:123–132. doi: 10.1242/jcs.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belin BJ, Cimini BA, Blackburn EH, et al. Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell. 2013;24:982–994. doi: 10.1091/mbc.E12-09-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nobusue H, Onishi N, Shimizu T, et al. Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. Nat Commun. 2014;5:3368. doi: 10.1038/ncomms4368. [DOI] [PubMed] [Google Scholar]
- 17.Aragona M, Panciera T, Manfrin A, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 18.Dopie J, Skarp KP, Rajakyla EK, et al. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci USA. 2012;109:E544–E552. doi: 10.1073/pnas.1118880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skarp KP, Huet G, Vartiainen MK. Steady-state nuclear actin levels are determined by export competent actin pool. Cytoskeleton. 2013;70:623–634. doi: 10.1002/cm.21116. [DOI] [PubMed] [Google Scholar]
- 20.Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340:864–867. doi: 10.1126/science.1235038. [DOI] [PubMed] [Google Scholar]
- 21.Munsie LN, Desmond CR, Truant R. Cofilin nuclear-cytoplasmic shuttling affects cofilin-actin rod formation during stress. J Cell Sci. 2012;125:3977–3988. doi: 10.1242/jcs.097667. [DOI] [PubMed] [Google Scholar]
- 22.McBeath R, Pirone DM, Nelson CM, et al. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 23.Gross TS, Srinivasan S, Liu CC, et al. Noninvasive loading of the murine tibia: An in vivo model for the study of mechanotransduction. J Bone Miner Res. 2002;17:493–501. doi: 10.1359/jbmr.2002.17.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warden SJ, Mantila Roosa SM, Kersh ME, et al. Physical activity when young provides lifelong benefits to cortical bone size and strength in men. Proc Natl Acad Sci USA. 2014;111:5337–5342. doi: 10.1073/pnas.1321605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross TS, Rubin CT. Uniformity of resorptive bone loss induced by disuse. J Orthop Res. 1995;13:708–714. doi: 10.1002/jor.1100130510. [DOI] [PubMed] [Google Scholar]
- 26.Wolff J. Das Gesetz Der Transformation Der Knochen. Berlin, Germany: Verlag August Hirschwald; 1892. [Google Scholar]
- 27.Bershadsky AD, Balaban NQ, Geiger B. Adhesion-dependent cell mechanosensitivity. Annu Rev Cell Dev Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 28.Lessey EC, Guilluy C, Burridge K. From mechanical force to RhoA activation. Biochemistry. 2012;51:7420–7432. doi: 10.1021/bi300758e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peister A, Mellad JA, Larson BL, et al. Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood. 2004;103:1662–1668. doi: 10.1182/blood-2003-09-3070. [DOI] [PubMed] [Google Scholar]
- 30.Fan X, Biskobing DM, Fan D, et al. Macrophage colony stimulating factor down-regulates MCSF-receptor expression and entry of progenitors into the osteoclast lineage. J Bone Miner Res. 1997;12:1387–1395. doi: 10.1359/jbmr.1997.12.9.1387. [DOI] [PubMed] [Google Scholar]
- 31.Sen B, Guilluy C, Xie Z, et al. Mechanically induced focal adhesion assembly amplifies anti-adipogenic pathways in mesenchymal stem cells. Stem Cells. 2011;29:1829–1836. doi: 10.1002/stem.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sen B, Styner M, Xie Z, et al. Mechanical loading regulates NFATc1 and beta-catenin signaling through a GSK3beta control node. J Biol Chem. 2009;284:34607–34617. doi: 10.1074/jbc.M109.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sen B, Xie Z, Case N, et al. Mechanical strain inhibits adipogenesis in mesenchymal stem cells by stimulating a durable beta-catenin signal. Endocrinology. 2008;149:6065–6075. doi: 10.1210/en.2008-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pagnotti GM, Adler BJ, Green DE, et al. Low magnitude mechanical signals mitigate osteopenia without compromising longevity in an aged murine model of spontaneous granulosa cell ovarian cancer. Bone. 2012;51:570–577. doi: 10.1016/j.bone.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pendleton A, Pope B, Weeds A, et al. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem. 2003;278:14394–14400. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- 36.Bellows CG, Heersche JN, Aubin JE. Determination of the capacity for proliferation and differentiation of osteoprogenitor cells in the presence and absence of dexamethasone. Dev Biol. 1990;140:132–138. doi: 10.1016/0012-1606(90)90060-v. [DOI] [PubMed] [Google Scholar]
- 37.Lancaster OM, Baum B. Shaping up to divide: Coordinating actin and microtubule cytoskeletal remodelling during mitosis. Semin Cell Dev Biol. 2014;34:109–115. doi: 10.1016/j.semcdb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Meyers VE, Zayzafoon M, Douglas JT, et al. RhoA and cytoskeletal disruption mediate reduced osteoblastogenesis and enhanced adipogenesis of human mesenchymal stem cells in modeled microgravity. J Bone Miner Res. 2005;20:1858–1866. doi: 10.1359/JBMR.050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huet G, Skarp KP, Vartiainen MK. Nuclear actin levels as an important transcriptional switch. Transcription. 2012;3:226–230. doi: 10.4161/trns.21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuven T, Hartmann E, Gorlich D. Exportin 6: A novel nuclear export receptor that is specific for profilin. actin complexes. EMBO J. 2003;22:5928–5940. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu JC, Lengner CJ, Gaur T, et al. Runx2 protein expression utilizes the Runx2 P1 promoter to establish osteoprogenitor cell number for normal bone formation. J Biol Chem. 2011;286:30057–30070. doi: 10.1074/jbc.M111.241505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchet R, Millan JL, Magne D. Multisystemic functions of alkaline phosphatases. Meth Mol Biol. 2013;1053:27–51. doi: 10.1007/978-1-62703-562-0_3. [DOI] [PubMed] [Google Scholar]
- 43.Zaidi SK, Sullivan AJ, Medina R, et al. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–799. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupont S, Morsut L, Aragona M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 45.Yu FX, Guan KL. The Hippo pathway: Regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishi K, Yoshida M, Fujiwara D, et al. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 47.Obrdlik A, Percipalle P. The F-actin severing protein cofilin-1 is required for RNA polymerase II transcription elongation. Nucleus. 2011;2:72–79. doi: 10.4161/nucl.2.1.14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang W, Ghisletti S, Saijo K, et al. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470:414–418. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyamoto K, Pasque V, Jullien J, et al. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev. 2011;25:946–958. doi: 10.1101/gad.615211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morimoto RI. The heat shock response: Systems biology of proteotoxic stress in aging and disease. Cold Spring Harbor Symp Quantit Biol. 2011;76:91–99. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 51.Meyer MB, Benkusky NA, Pike JW. The RUNX2 cistrome in osteoblasts: Characterization, downregulation following differentiation and relationship to gene expression. J Biol Chem. 2014;289:16016–16031. doi: 10.1074/jbc.M114.552216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 53.Posern G, Sotiropoulos A, Treisman R. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol Biol Cell. 2002;13:4167–4178. doi: 10.1091/mbc.02-05-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Posern G, Miralles F, Guettler S, et al. Mutant actins that stabilise F-actin use distinct mechanisms to activate the SRF coactivator MAL. EMBO J. 2004;23:3973–3983. doi: 10.1038/sj.emboj.7600404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Low BC, Pan CQ, Shivashankar GV, et al. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett. 2014;588:2663–2670. doi: 10.1016/j.febslet.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 56.Wada K, Itoga K, Okano T, et al. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 57.Mosqueira D, Pagliari S, Uto K, et al. Hippo pathway effectors control cardiac progenitor cell fate by acting as dynamic sensors of substrate mechanics and nanostructure. ACS Nano. 2014;8:2033–2047. doi: 10.1021/nn4058984. [DOI] [PubMed] [Google Scholar]
- 58.Chen JC, Jacobs CR. Mechanically induced osteogenic lineage commitment of stem cells. Stem Cell Res Ther. 2013;4:107. doi: 10.1186/scrt318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tojkander S, Gateva G, Lappalainen P. Actin stress fibers—Assembly, dynamics and biological roles. J Cell Sci. 2012;125:1855–1864. doi: 10.1242/jcs.098087. [DOI] [PubMed] [Google Scholar]
- 60.Yoshikawa H, Yoshioka K, Nakase T, et al. Stimulation of ectopic bone formation in response to BMP-2 by Rho kinase inhibitor: A pilot study. Clin Orthop Relat Res. 2009;467:3087–3095. doi: 10.1007/s11999-009-0976-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakai D, Kii I, Nakagawa K, et al. Remodeling of actin cytoskeleton in mouse periosteal cells under mechanical loading induces periosteal cell proliferation during bone formation. PloS One. 2011;6:e24847. doi: 10.1371/journal.pone.0024847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ’network of networks’. Nat Rev Mol Cell Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


