Abstract
The Helicobacter pylori energy sensor TlpD determines tactic behaviour under low energy conditions and is important in vivo. We explored protein-protein interactions of TlpD and their impact on TlpD localisation and function. Pull-down of tagged TlpD identified protein interaction partners of TlpD, which included the chemotaxis histidine kinase CheAY2, the central metabolic enzyme aconitase (AcnB) and the detoxifying enzyme catalase (KatA). We confirmed that KatA and AcnB physically interact with TlpD. While the TlpD-dependent behavioural response appeared not influenced in the interactor mutants katA and acnB in steady-state behavioural assays, acetone carboxylase subunit (acxC) mutant behaviour was altered. TlpD was localised in a bipolar subcellular pattern in media of high energy. We observed a significant change in TlpD localisation towards the cell body in cheAY2-, catalase- or aconitase-deficient bacteria or in bacteria incubated under low energy conditions, including oxidative stress or respiratory inhibition. Inactivation of tlpD resulted in an increased sensitivity to iron limitation and oxidative stress and influenced the H. pylori transcriptome. Oxidative stress, iron limitation and overexpressing the iron-sulfur repair system nifSU altered TlpD-dependent behaviour. We propose that TlpD localisation is instructed by metabolic activity and protein interactions, and its sensory activity is linked to iron-sulfur cluster integrity.
Helicobacter pylori colonises about 50% of the world’s population. Chronic colonisation of the human stomach, usually starting in early childhood, results in chronic gastritis. Persistent H. pylori infection can also progress to gastric and duodenal ulcer, adenocarcinoma and MALT lymphoma, accounting for more than 500,000 deaths per year worldwide1. H. pylori is adapted to survive in a challenging niche, the human stomach. Particular challenges of this environment are an acidic luminal pH of about 22, high pepsin activity, which readily inactivates the bacteria3, constant mucus turn-over, scarceness of nutrients and metal ions, and prominent host defence mechanisms, for instance oxidative stress4,5,6. These obstacles constantly threaten the survival and persistence of the bacteria. Under these challenging and variable conditions, the majority of H. pylori cells is found in close proximity to the epithelial cell layer, within 15 μm above the mucosal surface7,8, and deep in the stomach glands9. Targeted motility and adherence of H. pylori to gastric epithelial cells are major factors in colonisation9,10,11,12,13.
Several in vivo studies showed that motility and taxis are of particular importance for H. pylori in its niche to navigate to conditions of optimal growth and survival. Non-motile mutants or mutants without a functional chemotaxis system displayed severe colonisation defects or did not colonise at all, which was demonstrated in piglet, mouse and gerbil models10,11,13,14. The H. pylori chemotaxis system comprises one set of core chemotaxis components15,16 and four chemotaxis receptors/transducer-like proteins (Tlps)16,17,18,19,20, TlpA, TlpB, TlpC, TlpD. Cues described so far for chemotactic sensing in H. pylori include zinc, nickel21, urea22,23, low pH18, bicarbonate, and the amino acid arginine24. As in other bacteria, signals sensed by taxis receptors result in a flagellar switch from counterclockwise to clockwise rotation and thereby induce stops, direction changes and backward movements (reversals) of the bacteria, allowing directed motility25,26. Two of the four H. pylori chemotaxis receptors, TlpB and TlpD, have been reported to be linked to energy taxis. Energy taxis can mediate a metabolism-dependent response to intrabacterial energy levels, presumably helping to navigate the bacterium to metabolically favourable environments13,18,26. The H. pylori transmembrane receptor TlpB mediates a repellent response to low pH (<pH 3)18; low extracellular pH directly influences the proton motive force of the bacteria27,28, suggesting a connection between TlpB and energy metabolism. TlpB has also been reported to be involved in repellent taxis against the autoinducer metabolite AI-229. We have previously shown that the soluble taxis sensor TlpD, which does not contain transmembrane domains, mediates chemotactic behaviour in response to intrabacterial energy levels in H. pylori13,26. We concluded that TlpD promotes a repellent response to low energy conditions and to inhibitors of the respiratory chain in H. pylori. Energy-dependent behaviour was clearly attributed to TlpD in a triple transducer knock-out mutant, and TlpD exerted a dominant effect over the other three sensors under low energy conditions in vitro26. In vivo colonisation experiments in mouse and gerbil models demonstrated that the role of TlpB in vivo is variable in mice and relatively minor in the gerbil model11,12,18, while TlpD was very important for the colonisation of the gerbil stomach13, and was required for high-level colonisation in the antrum of mice12. Our previous inhibitor studies on the function of TlpD suggested that a functional electron transport chain is important for TlpD-dependent sensing and taxis26, but more research is clearly required to define the sensing mechanism.
Energy taxis is suggested to play a decisive role for the survival of different bacteria in their niches30,31. Tlps providing energy-dependent taxis have been described in several species, albeit for most receptors, little is known about the energy sensing mechanisms. Known modes of sensing related to the intrabacterial energy state are diverse30. The dedicated E. coli energy sensor Aer detects the cellular redox state using a PAS domain and involving FAD cofactor binding, although its precise mode of sensing remains unresolved27. A direct function of protein-protein interactions in chemotaxis or energy taxis might merit further investigations, since a direct impact of NADH dehydrogenase on Aer-mediated sensing in E. coli was discussed27. For the E. coli metabolic enzyme ATP synthase, a colocalisation with the flagellar basal body and a direct influence on flagellar rotation was previously reported32. Protein-protein interactions involving conformational change might permit quick sensory responses, since shifts of protein conformation can occur rapidly with changing protein activities.
In the present study, we have investigated protein-protein interactions of TlpD and their potential role in TlpD localisation and TlpD-dependent energy sensing of H. pylori. We initially identified novel TlpD protein interaction partners using a pull-down approach in H. pylori. In addition, we confirmed the direct interaction of TlpD with the enzymes catalase and aconitase. The localisation of the soluble sensor TlpD, which was determined here for the first time, was altered in media of different energy levels and in cheAY2, acnB and katA mutants. Functional assays using a TlpD wild type strain, overexpression strains, and interactor mutants suggested a role of iron depletion, oxidative stress and iron-sulfur cluster proteins in the TlpD sensing mechanism. The absence of TlpD induced a shift in global transcript activities. These findings indicate a functional connection between the soluble receptor/transducer-like sensor TlpD, its protein-protein interactions and metabolic homeostasis.
Results
Protein-protein interactions of H. pylori TlpD uncovered by pull-down assay and mass spectrometry
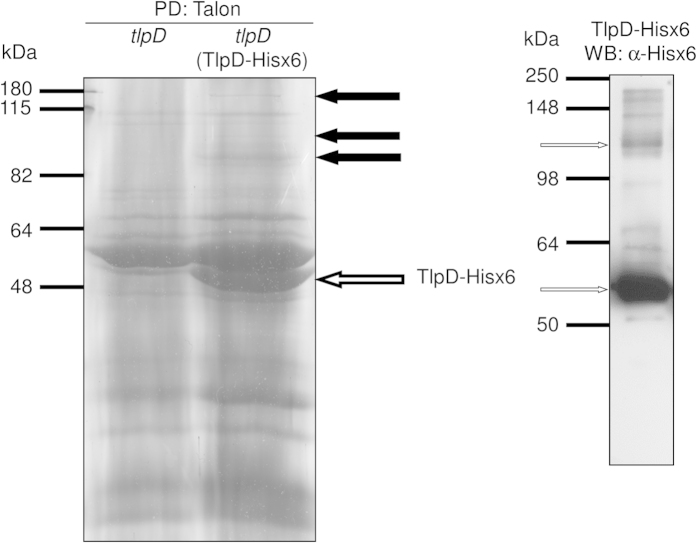
The transducer-like protein TlpD of H. pylori mediates energy tactic behaviour, but its mode of action is still unknown. Its C-terminus is homologous to the signal transducing domain of canonical Tlps26, and contains an additional C-terminal zinc-binding domain33. The N-terminal domain of TlpD, which is potentially involved in sensing, contains no sequence homology to any known sensing domain in databases (26, and own recent comparison with other proteins in databases). TlpD does not bear strong similarities to other known soluble sensors, e.g. TlpT of Rhodobacter sphaeroides34,35. In order to test the hypothesis that TlpD interacts with other proteins, which may be relevant to its function, we used a pull-down approach to identify potential protein interaction partners of TlpD. We expressed histidine-tagged TlpD (TlpD-Hisx6), which is fully functional in H. pylori (Methods), in a H. pylori N6 tlpD::aphA3 insertion mutant from an expression plasmid (pHel2::tlpD-hisx6). We then performed pull-down experiments from H. pylori grown under standard high energy conditions (blood plates) using Talon affinity beads and visualised TlpD-bound proteins on Coomassie-stained SDS gels. Several protein bands were exclusively detected in the TlpD-Hisx6-expressing strain in comparison to the tlpD mutant control (Fig. 1). A prominent TlpD-Hisx6 band and a potential TlpD dimer were detected by anti-His-tag Western blot (Fig. 1). We reproducibly recovered several TlpD-associated protein bands at higher molecular masses (>82 kDa) in three independent pull-down experiments (see black arrows, Fig. 1), which were separately excised and subjected to mass spectrometry analysis. In these analyses (for full results see Table 1), we identified various peptides in each excised band, corresponding to overall six different proteins in addition to TlpD. Peptides for five of the six proteins (chemotaxis histidine kinase CheAY2, aconitate hydratase AcnB, catalase KatA, isoleucyl-tRNA synthetase IleS, general protein secretion translocase SecA, bifunctional RNA polymerase subunit RpoBC), were detected in two independent experimental repeats of the pull-down, respectively. Notably, the only chemotaxis histidine kinase present in H. pylori, CheAY210,36, which is expected to be interacting with Tlp-orthologous taxis sensors, as previously shown in E. coli37,38,39, was identified in both experimental repeats (Table 1). The enzymes AcnB and KatA were also identified in both TlpD pull-down experiments. Due to their direct or indirect importance in metabolic processes, we selected KatA and AcnB for further characterisation in the context of TlpD. We also included the H. pylori acetone carboxylase subunit gene (acxC) in further functional studies, since AcxC had been identified as a protein that potentially interacts with TlpD in a previous high-throughput Yeast-Two-Hybrid screen40 testing for H. pylori protein-protein interactions.
Figure 1. Pull-down of TlpD-Hisx6 with potential protein interaction partners from H. pylori N6 cleared cell lysates.
Pull-down (PD) precipitates of TlpD-Hisx6 from N6 tlpD (TlpD-Hisx6) and N6 tlpD (negative control not expressing any TlpD; precipitated using Talon (Cobalt2+–coupled matrix)) were separated in 12% SDS gels and stained with Coomassie blue (left panel). Black arrows point at specific higher molecular mass protein bands that were detected only in the pull-down material of H. pylori N6 tlpD (TlpD-Hisx6) but not in the N6 tlpD mutant (control strain). These gel sections, containing differentially detected bands which are numbered 1, 2, 3 from top to bottom (corresponding to numbers in Table 1), were subsequently cut from the gels separately and analysed using mass spectrometry (MS/MS; see Supplementary Methods). The white arrow points at TlpD-Hisx6 monomer. The right panel shows a corresponding Western blot (WB) of the purified TlpD-Hisx6 fraction which was developed using α-Hisx6 antibody. The arrows in the right panel point at TlpD-Hisx6 monomer (lower arrow) and TlpD-Hisx6 dimer (upper arrow).
Table 1. H. pylori proteins of which peptides were identified by mass spectrometry in two independent pull-down assays of TlpD-Hisx6 from H. pylori tlpD (TlpD-Hisx6) cleared cell lysates.
| Protein | Number of identified peptidesa |
|---|---|
| Histidine kinase (CheAY2) | 2;1 (band 1) |
| peptides: | |
| Expt. 1: IYGDVEER; MDGYTFASEVR | |
| Expt. 2: ISQDEIYTVDGK | |
| Aconitate hydratase (AcnB) | 5;8 (band 1) |
| peptides: | |
| Expt. 1: GVPVAYVGDVVGTGSSR; IPLIIGR; | |
| VTTVGSQDTTGAMTR; ILEIEGLPDIK; | |
| MEQAFELSDASAER | |
| Expt. 2: AEFLAQLSQK; DIPFVPNKR; IPLIIGR; | |
| VTTVGSQDTTGAMTRox;MEQAFELSDASAERox; LWVVPPSK | |
| Isoleucyl-tRNA synthetase (IleS) | 3;2 (band 1) |
| peptides: | |
| Expt. 1: TSLENPTLFR; SAFNEELDR; LSETPAFTLFK | |
| Expt. 2: TSLENPTLFR; VEFVPSSGK | |
| Catalase (KatA) | 1;1 (bands 2, 3) |
| peptides: | |
| Expt. 1: SLPADEKER | |
| Expt. 2: FSTVAGER | |
| Protein translocase (SecA) | 1;1 (band 1) |
| peptides: | |
| Expt. 1: TPLIISGPVDR | |
| Expt. 2: TPLIISGPVDR | |
| DNA-dependent RNA polymerase (RpoBC) | -;7 (band 3) |
| peptides: | |
| Expt. 2: DSYDSFLYSK; VLSAYEEEK; KPETINYR; | |
| FAVSDVNELYR; VLDQGNIIATSAGR; NASILVVEPK; | |
| VSELFEAR | |
| Methyl-accepting protein TlpD (HylB)b | 38;21 (bands 1, 2, 3) |
Distinct protein bands of corresponding mass were excised for both experiments. Bands are indicated with three black arrows in Fig. 1, from top (1) to bottom (3). Only proteins which were detected in both replicate experiments, or which were identified with high score (>8; ProteinLynx) by more than 5 peptides in one single experiment, are listed. Only identification scores for proteins of equal or above 7 (ProteinLynx) were considered. The names of the identified proteins in the table correspond to the database designations. aExperiment 1; Experiment 2: bands 1,2, 3 designate bands that were excised from SDS gels after TlpD pull-down for mass spectrometry. For details of mass spectrometry analyses see Supplementary Methods.
bpull-down target (single peptides not listed).
oxrespective peptide was identified twice - as an unmodified and as a methyl-oxidated variant.
Confirmation of direct H. pylori TlpD-KatA and TlpD-AcnB interactions by biochemical interaction studies
Our next goal was to verify and characterise the direct interactions of TlpD with KatA and AcnB. We investigated purified recombinant TlpD (see Methods) for binding to recombinant active purified KatA41 or purified native AcnB (V5-tagged) from H. pylori26 by biolayer interferometry (BLI), an optical biosensing method that yields kinetic and affinity information in a manner similar to surface plasmon resonance (SPR)42. We tested sensor-bound Hisx6-TlpD with different AcnB-V5 concentrations as analyte (Fig. 2A). The data indicated binding and was fit to a two-state parallel binding model which yielded equilibrium dissociation constants KD1 of 1*10−8 M (fast on, slow off) and KD2 of 1.21*10−7 M (slow on, fast off) (R2 = 0.998). The slow association constant kon1 was 2.3*104 M−1*s−1 and a faster kon2 was 2.1*105 M−1*s−1. Dissociation rate constants koff1 and koff2 were 2.4*10−4 s−1 and 2.5*10−2 s−1, respectively. We also obtained interaction signals for the binding between Hisx6-TlpD (ligand) and KatA (analyte) in the same setting, with a KD1 of 3.6 *10−8 M and a KD2 of 7.8 *10−9 M, with a confidence interval R2 of 0.96 (data not shown). For direct KatA-TlpD interactions, a two-state parallel binding model also provided the best fit. The KatA-TlpD interaction was also analysed in an inverse experimental setting with KatA as ligand and TlpD (V5-tagged) as analyte (Fig. 2B). We determined KD values of 2.75*10−7 M (fast on, slow off) and 2*10−6 M (slow on, fast off) in this inverse setting (R2 = 0.95). Values of the corresponding rate constants for the association were kon1 4.7*103 M−1*s−1 for the fast and kon2 6.1*102 M−1*s−1 for the slow component (KatA at sensor). For the KatA-TlpD interaction, koff for the two-state model could not be determined due to the very slow off rate. Taken together, we detected in vitro reproducible direct, specific interactions of TlpD with AcnB or with KatA, respectively.
Figure 2. Biochemical analysis of direct interactions of purified H. pylori TlpD and AcnB or TlpD and KatA proteins using biolayer interferometry.
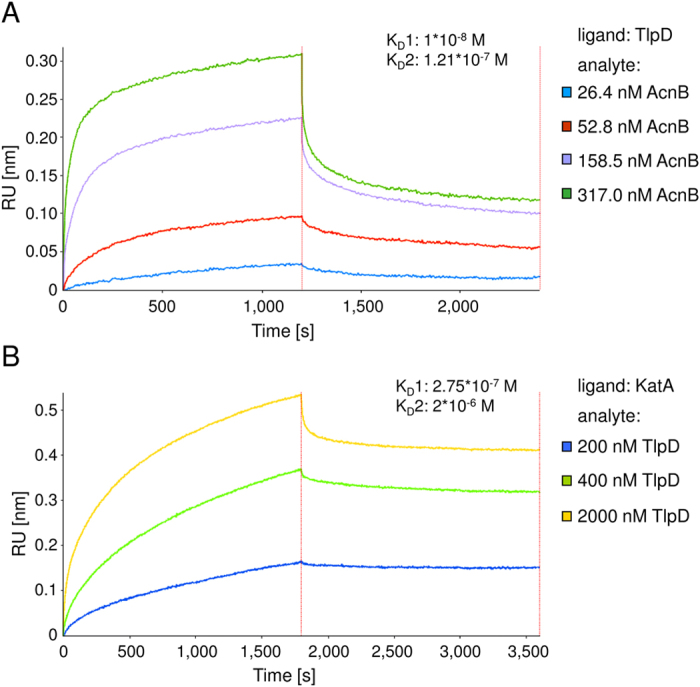
Recombinantly purified TlpD was tested for direct interactions with purified H. pylori AcnB or KatA using biolayer interferometry. While one purified protein (ligand) was coupled to the sensor surface via a hexa-histidine tag or free amine groups (see Supplementary Methods), the second protein (analyte) was applied as a solute in assay buffer. Sensors were dipped into different analyte solutions for 1,200 sec (A) or 1,800 sec (B), before dissociation was monitored in assay buffer. (A) TlpD-AcnB interaction: ligand Hisx6-TlpD (purified from E. coli) was immobilised to the sensor surface, while analyte H. pylori AcnB (AcnB-V5, purified from H. pylori) was applied in assay buffer (Methods and Supplementary Methods) at four different concentrations (26.4 nM, 52.8 nM, 158.5 nM and 317.0 nM). (B) TlpD-KatA interaction: immobilised H. pylori KatA (recombinant, tag-free, purified from E. coli) was coupled on the sensor surface via free amine groups, while analyte TlpD-V5 (purified from H. pylori) was supplied diluted in assay buffer at 200 nM, 400 nM and 2,000 nM. Values derived from a ligand-coupled sensor that was dipped in assay buffer only over the full time course of the interaction assay were subtracted as the baseline from the interaction curves.
Alterations of subcellular localisation of TlpD in H. pylori in the presence or absence of protein interaction partners
We had previously found that the predicted soluble receptor TlpD is partially membrane-associated26. The subcellular localisation and potential differential distribution patterns of TlpD, which may be interconnected with its function, were unresolved. To allow for specific localisation studies using immunofluorescence (IF; Supplementary Materials and Methods), we expressed a C-terminally V5-tagged fully functional version of TlpD26 in the chromosomal rdxA locus (Methods) in the H. pylori N6 parental strain.
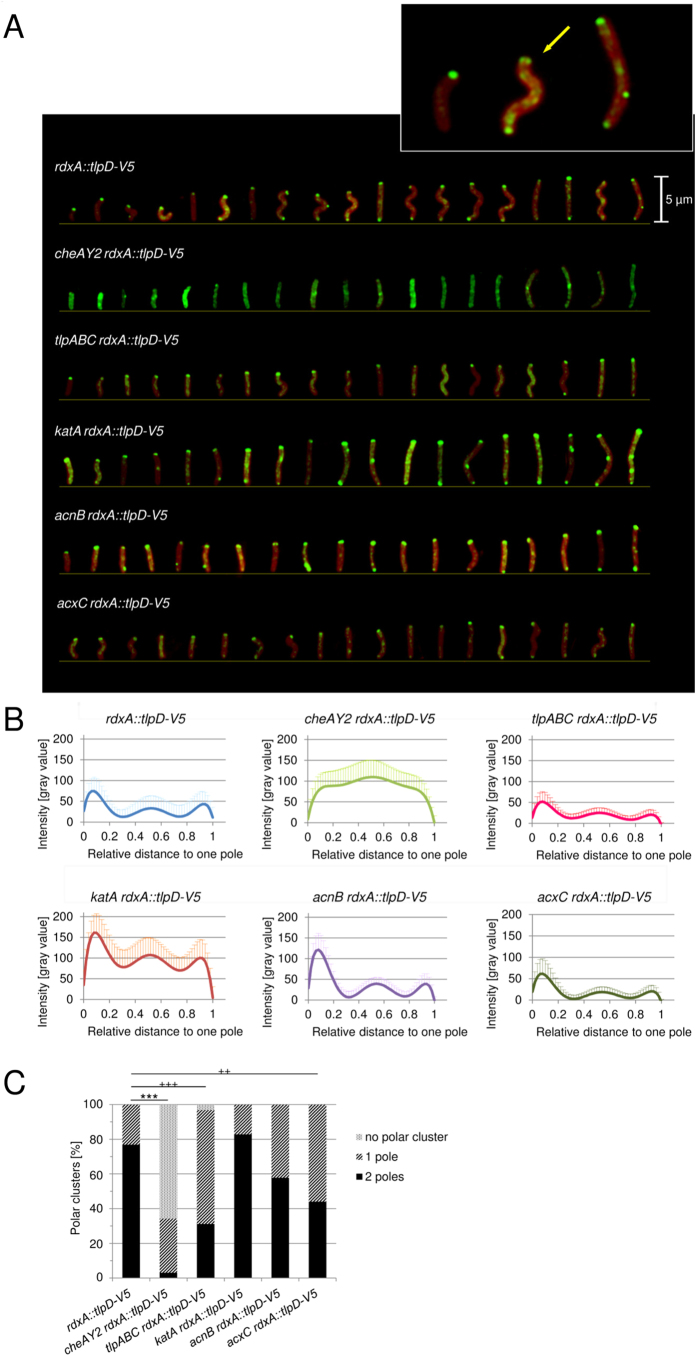
We initially used IF microscopy to determine the localisation of TlpD in plate-grown bacteria (mid-log phase; growth-dependent high-energy conditions). Immediately upon harvest, bacteria were resuspended in fixing agent to maintain the current energetic conditions. In the reference strain (N6 rdxA::tlpD-V5), TlpD was detectable at clearly defined dots at both poles (Fig. 3; Supplementary Fig. S1). In some bacteria, two separate sub-terminal clusters were noted at both sides, close to one pole, corresponding to previous cryotomography results for taxis sensor arrays accumulating on both sides of the flagellar bundle in the related species Helicobacter hepaticus43. Furthermore, we detected a subfraction of TlpD in more diffuse patches along the whole cell body. Less frequently, dot-like clusters were also present in the bacterial mid-cell region (Fig. 3A). The same distribution of TlpD-V5 was observed in intact cells of strains N6 rdxA::tlpD-V5 (reference) and N6 tlpD rdxA::tlpD-V5 which both served as IF positive controls for the expression of functional TlpD26 (Methods) (Fig. 3; Supplementary Fig. S1).
Figure 3. Subcellular localisation of TlpD in H. pylori in the presence and absence of potential interaction partners.
(A) Subcellular localisation of TlpD in intact, permeabilised H. pylori cells grown on plates for 20 h (standard high-energy conditions) was imaged by immunofluorescence microscopy (IF; for details of sample preparation see Supplementary Methods). Cells were immediately immersed in fixing agent after harvest. Green: TlpD-V5 was detected using primary mouse α-V5 antibody (1:1,000), combined with secondary α-mouse IgG coupled to Alexa-488 (diluted 1:5,000); Red: SynaptoRed FM4-64 (1:5,000) membrane marker. Specimens were visualised in a Zeiss Apotome fluorescence microscope at a 63-fold lens magnification. A constant exposure time and the same digital image lighting correction functions were used and adjusted equally for all specimens; in each panel, 19 representative cells from one experiment are arranged according to cell length. Arrows point at polar TlpD localisation foci. Inset: three representative images of the N6 rdxA::tlpD-V5 reference strain were magnified by an additional 4.5 fold. The size bar represents 5 μm. (B) Subcellular distribution of TlpD-V5-derived fluorescence intensities (α-V5 MAB) in the H. pylori parental strain and corresponding mutant strains deficient in TlpD-interacting proteins AcnB, KatA or AcxC. A tlpABC triple transducer mutant, lacking three of the four H. pylori Tlps, and a cheAY2 (chemotaxis null) mutant were used as additional control strains. Growth conditions and sample preparation were the same as in panel A. The fluorescence intensity distribution patterns were quantitated by averaging the pixel intensities in transversal sections (of one pixel length and 8 pixel width) along the longitudinal (x) axes for 15 bacteria from each strain using ImageJ, and then calculating average distribution patterns for all bacteria of each strain (Table 3). The polar and non-polar distributions of TlpD were compared statistically (for details see Supplementary Methods).(C) Quantification of polar TlpD clusters per cell for ≥30 bacteria per strain. Clusters at one and both poles were counted visually, and levels of significance were calculated by Fisher’s exact test, for the two conditions polar vs. non-polar clusters (***p ≤ 0.001), and for cluster distribution at 1 vs. 2 poles (+++p ≤ 0.001, ++p ≤ 0.01).
Since we hypothesised that interactions of TlpD with other proteins may have consequences for TlpD localisation and/or function, we constructed isogenic allelic exchange mutants in H. pylori N6 (wild-type strain) and in the H. pylori N6 rdxA::tlpD-V5 parental strain (reference to test TlpD localisation), each deficient in one of the genes encoding the H. pylori TlpD interaction partners, cheAY2, acnB, katA, and the potential interaction partner acxC40 (Table 2). Except for the acnB mutants, which had a severe growth defect, none of the other mutants showed an obvious growth deficiency in vitro or other gross phenotypical alterations in comparison to the parental strains. All the mutants also displayed normal morphology in transmission electron microscopy (not shown).
Table 2. Helicobacter pylori strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| H. pylori strains and short designations | Strain characteristics and full designations | |
| N6 | parental strain; human patient isolate | 79 |
| N6 tlpD | H. pylori N6 tlpD::aphA3 | 26 |
| HP87P7 | wild type, gerbil-adapted | 13 |
| HP87P7 tlpD | H. pylori HP87 P7 tlpD::aphA3 | 13 |
| N6 tlpD (TlpD-Hisx6) | H. pylori N6 tlpD::aphA3 (pHel2::tlpD-hisx6) | this study |
| N6 tlpD (pHel2::tlpD-V5) | H. pylori N6 tlpD::aphA3 (pHel2::tlpD-V5) | 26 |
| N6 rdxA::tlpD-V5 | H. pylori N6 rdxA::tlpD-V5 CmR (chromosomal complementation) | this study |
| N6 tlpD rdxA::tlpD-V5 | H. pylori N6 tlpD::aphA3 rdxA::tlpD-V5 CmR | this study |
| N6 cheAY2 rdxA::tlpD-V5 | H. pylori N6 cheAY2::aphA3 rdxA::tlpD-V5 CmR | this study |
| N6 tlpABC rdxA::tlpD-V5 | H. pylori N6 tlpA::aphA3 tlpBC (unmarked partial deletions) rdxA::tlpD-V5 CmR | this study |
| N6 katA rdxA::tlpD-V5 | H. pylori N6 katA::aphA3 rdxA::tlpD-V5 CmR | this study |
| N6 acnB rdxA::tlpD-V5 | H. pylori N6 acnB::aphA3 rdxA::tlpD-V5 CmR | this study |
| N6 acxC rdxA::tlpD-V5 | H. pylori N6 acxC::aphA3 rdxA::tlpD-V5 CmR | this study |
| N6 (AcnB-V5) | H. pylori N6 (pHel2::acnB-V5) CmR | this study |
| N6 acnB | H. pylori N6 acnB::aphA3 | this study |
| N6 katA | H. pylori N6 katA::aphA3 | this study |
| N6 acxC | H. pylori N6 acxC::aphA3 | this study |
| N6 cheAY2 | H. pylori N6 cheAY2::aphA3 | 26 |
| N6 (NifSU+) | H. pylori N6 (pHel2::nifSU) | this study |
| Plasmids | Plasmid characteristics | |
| pUC18 | AmpR, RepEc; high-copy-number vector | 86 |
| pUT18 | AmpR; RepEc; pBR322 derivative; cloning vector | 87 |
| pILL570 | SpecR; RepEc; cloning vector | 88 |
| pEF6-V5 | AmpR, BlastR; RepEc; source of V5-tag sequence | Invitrogen Life Technologies, Darmstadt, Germany |
| pET28a | KmR; RepEc; high-copy-number expression vector, includes hexa-histidine tag sequence | Novagen |
| pHel2 | CmR; RepEc, RepHp; multi-copy-number vector; H. pylori expression plasmid | 89 |
| pKSF | KmR; RepEc; source of the kan-sacB construct | 90 |
| pCJ522 | CmR; tlpD-V5 fusion gene including the potential tlpD promoter region in pHel2 shuttle vector | 26 |
| pCJ545 | CmR; tlpD-hisx6 fusion gene including the potential tlpD promoter region in pHel2 shuttle vector | this study |
| pCJ1341 | KmR; pET28a derivative; tlpD fused N terminally with Hisx6 | this study |
| pCJ542 | AmpR, CmR; pUC18 derivative; tlpD-V5 expression construct and CAT cassette inserted in rdxA flanking regions (for chromosomal complementation) | 13 |
| pCJ1306 | AmpR, KmR; pUT18 derivative; acnB disrupted by aphA3 cassette | this study |
| pCJ1308 | AmpR, KmR; pUT18 derivative; katA disrupted by aphA3 cassette | this study |
| pCJ537 | AmpR, KmR; pUC18 derivative; acxC disrupted by aphA3 cassette | this study |
| pCJ513 | SpecR, KmR; pILL570 derivative; tlpB disrupted by kan-sacB construct (kanamycin resistant, sucrose sensitive) | this study |
| pCJ514 | SpecR; pILL570 derivative; tlpB partially deleted (unmarked) | this study |
| pCJ1396 | AmpR, KmR; pUC18 derivative; tlpA disrupted by aphA3 cassette | this study |
| pCJ1314 | CmR; acnB-V5 construct and potential acnB promoter region in pHel2 shuttle vector | this study |
| pCJ1350 | CmR; nifSU gene cluster including the potential promoter region in pHel2 shuttle vector | this study |
AmpR ampicillin resistance, KmR kanamycin resistance, CmR chloramphenicol resistance, BlastR blasticidin resistance, SpecR spectinomycin resistance, RepEc replication in E. coli, RepHp replication in H. pylori.
We then used IF microscopy to determine the localisation of TlpD in these strains (plate-grown, same conditions as for the N6 rdxA::tlpD-V5 parental strain). We noted obvious differences between the parental strain and its cheAY2 and katA mutants. In the former, TlpD appeared to be almost lost at the polar localisation, while, in the latter, relatively more TlpD was observed throughout the cell body (Fig. 3). For the more precise comparison of TlpD localisation between the different interactor mutants, we quantitated signal intensities in IF (Fig. 3; Supplementary Figs S1 and S2) and Western blots (Supplementary Fig. S3), using densitometry. First of all, we verified that TlpD-V5 was present at similar levels in all tested strains, which was indeed the case (Supplementary Fig. S3). We subsequently quantitated the polar TlpD clusters and the average fluorescence intensities of TlpD at different subcellular locations (polar, non-polar; Methods). We compared the localisation patterns for each cell by enumerating the polar clusters, and by averaging multiple cells of each strain followed by statistical evaluation (uni- versus bipolar or no polar clusters; Fig. 3C; Methods) between the reference strains and all interactor mutants. Furthermore, we compared the fluorescence intensities in the cell body versus the polar regions (Fig. 3B; Table 3). The mutant in the central chemotaxis histidine kinase gene cheAY2, a taxis null mutant, exhibited a significant delocalisation of TlpD away from both poles (Fig. 3A–C; Table 3) and a loss of polar TlpD in almost all cells (see Table 3 for percent values, Fig. 3A–C). Even in the virtual absence of polar clusters in the cheAY2 mutant, TlpD was still detectable in diffuse clusters along the entire cell body (Fig. 3A; Supplementary Figure S2). The triple transducer knockout strain tlpABC that we used as an additional control strain, which contains TlpD as the only Tlp sensor, partially lost polar TlpD. In the latter strain, we detected TlpD predominantly at one pole in contrast to the bipolar distribution in the parental strain (Fig. 3A–C). Remarkably, the katA mutant displayed a significantly increased TlpD signal in the cell body over the polar regions in comparison to the parental strain (Fig. 3A–C; Table 3). In contrast, the acxC mutant showed a less obvious but still significant shift of relative TlpD intensity towards polarly localised signal, with a preferentially unipolar distribution (Fig. 3A–C). In contrast, the acnB mutant did not exhibit a significant change in subcellular TlpD localisation under these standard incubation conditions (20 h plate-grown bacteria, directly resupended in fixing agent; Fig. 3A–C; Table 3).
Table 3. Distribution of TlpD-V5 between cell poles and cell body in H. pylori strains lacking potential interaction partners.
| Genotype | Mean fraction of intensity at polesa/cell bodyb [%] ± SDc | Significance (p value)d | Maximum amplitude as fraction of maximum absolute intensity |
|---|---|---|---|
| N6 rdxA::tlpD-V5 | 50.7/49.3 ± 21.7 | – | 83.5% |
| N6 cheAY2 rdxA::tlpD-V5 | 20.7/79.3 ± 7.1 | 2.2*10–5 | not determined |
| N6 tlpABC rdxA::tlpD-V5 | 44.0/56.0 ± 17.2 | ns | 82.7% |
| N6 katA rdxA::tlpD-V5 | 27.6/72.4 ± 7.3 | 5.7*10–4 | 56.9% |
| N6 acnB rdxA::tlpD-V5 | 53.0/47.0 ± 9.7 | ns | 94.4% |
| N6 acxC rdxA::tlpD-V5 | 59.8/40.2 ± 12.2 | ns | 95.6% |
apolar regions were defined uniformly as six pixel lengths of highest fluorescence intensity from both poles inward.
bcell body was defined as the rest of pixels along the longitudinal axis of each bacterium, after subtracting polar regions defined in a.
cmean and standard deviation derived from 15 bacteria.
dStudent’s t test for significance of difference (p value) between N6 rdxA::tlpD-V5 and each of the mutants.
SD: standard deviation; ns: not significant.
To refine the in situ characterisation of TlpD subcellular localisation, we used Western blots of whole cell lysates of the katA, acnB, and acxC mutant strains and control strains fractionated into soluble and insoluble (membrane-enriched) fractions. TlpD was quantitated in both fractions of each strain by densitometry (Supplementary Fig. S3). In all tested mutants, we detected TlpD in both the insoluble and soluble protein fractions without any significant differences in TlpD amounts or distribution between the strains (Supplementary Fig. S3; approximately 5% TlpD in insoluble, 95% in soluble fraction). Interestingly, higher molecular mass (MM) protein bands containing TlpD were similarly present in wild type and mutants even with denaturing SDS gels in the presence of reducing agent (Supplementary Fig. S3; Methods). In control strains, harbouring single knock-out alleles in acnB, katA or acxC, which did not contain an additional tlpD-V5 allele in the rdxA locus, a similar distribution of TlpD between the soluble and insoluble bacterial fractions was also detected in all strains (not shown).
Inactivation of TlpD interaction partners KatA, AcnB and AcxC maintains TlpD-dependent steady-state behaviour
We hypothesised that potential TlpD interaction partners can be, either directly or indirectly, involved in energy taxis, and, in this context, might influence TlpD-mediated energy-dependent behaviour. In order to test this hypothesis, katA, acnB, acxC, and cheAY2 isogenic mutant strains expressing wild type TlpD were subjected to steady-state behavioural assays (single cell tracking assays) in liquid media under conditions of low intrabacterial energy, under which TlpD is functionally dominant and the TlpD-dependent phenotype well characterised26. Under these conditions, TlpD mediates a high number of stops and direction changes13,26.
katA and acnB mutants did not show a significantly altered swimming speed or motile behaviour in comparison to the parental strain, indicating that the respective proteins do not play a role in TlpD-mediated behaviour in these assay settings (Fig. 4). The acxC mutant showed a slight but statistically significant upshift in TlpD-mediated steady-state behaviour (increased stops). The cheAY2 mutant, as expected, was uniformly smooth-swimming under all incubation conditions, which is a known chemotaxis null phenotype as described before26. We had already previously assayed tlpABC mutants26, which exhibited a reduced TlpD-dependent stopping behaviour in comparison to the parental strain.
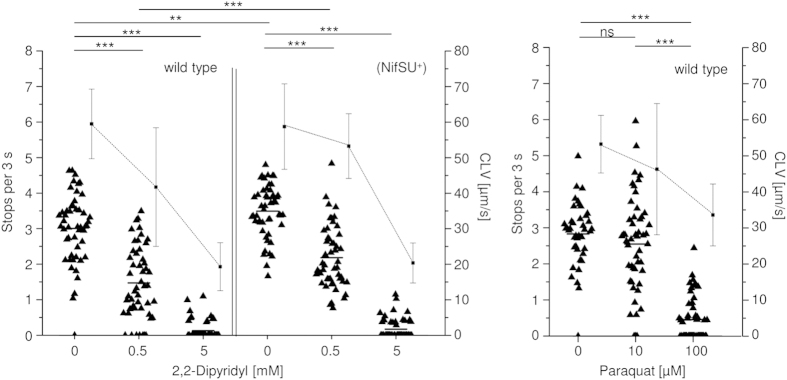
Figure 4. Characterisation of behavioural patterns for different H. pylori mutants in genes coding for TlpD-interacting proteins (swim assay).
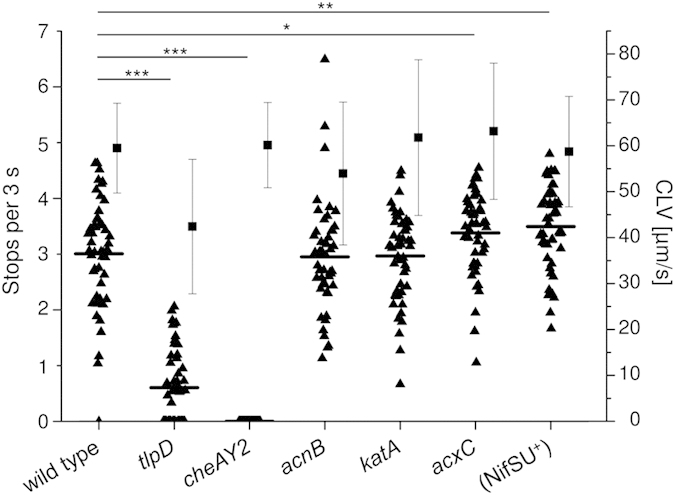
Behavioural patterns of H. pylori were determined by tracking assays using 50 bacteria per strain under previously determined steady state conditions of low bacterial energy yield13,26 in RPMI 1640 + 3% FCS at 37 °C and ambient air enriched with 5% CO2. Bacteria were collected from blood agar plates after 20 h of growth and resuspended in the liquid medium of low energy yield. Under these conditions, stopping frequency is dominated by TlpD26. Stops per three seconds (black triangles with mean) and curvilinear velocity (CLV; average with standard deviation as black squares with whiskers) were quantified for the different mutants. Significance values for differences between the strains were calculated by Student’s t test.
H. pylori tlpD mutants are more susceptible towards iron depletion and oxidative stress and show an altered transcriptome
Iron-limiting conditions and oxidative stress are two major factors characteristic of the H. pylori in vivo environment13. The host mucosal milieu is poor in iron, due to strong iron-sequestering factors of mammalian cells, and the proinflammatory host innate immune response provides strong oxidative stressors. Previous functional assays13,26 had suggested that intrabacterial energy and oxidative stress, which impacts on central energy metabolism via central iron-sulfur proteins44,45, both may play a role in the TlpD-mediated sensing process. Taken together with our previous, unexpected observation that the tlpD mutant has an increased susceptibility to oxidative stress13 these results strengthened a link between TlpD and iron acquisition or iron-sulfur homeostasis.
Supplementing our previous results of paraquat treatment13, we specifically investigated the impact of iron depletion on the growth of H. pylori N6 wild type, tlpD mutant and the complementation strain, using 2,2-dipyridyl (2,2-DP) in agar diffusion assays. tlpD mutants in two H. pylori strains, N6 and HP87P7, were more sensitive to iron depletion by 2,2-DP (significantly increased zone of growth inhibition) than the respective wild type (Table 4), similarly as previously found in the presence of paraquat13. The phenotype was reconstituted back to wild type by tlpD complementation in the chromosomal rdxA locus (Table 4), which confirmed that the phenotype was indeed mediated by the lack of TlpD.
Table 4. Growth inhibition of H. pylori N6 and HP87 P7 wild type, tlpD mutant and tlpD complemented strains in agar diffusion assays with different concentrations of the iron chelator 2,2-dipyridyl (DP).
| Diameter of zone with growth inhibition [mm]a,b | ||||
|---|---|---|---|---|
| Strain | 10 mM 2,2-DP | Significance (p value) | 20 mM 2,2-DP | Significance (p value) |
| N6 wild type | 9.6 ± 0.5 | 15.8 ± 1.0 | ||
| N6 tlpD | 11.9 ± 0.9 | 0.017 | 17.8 ± 0.6 | 0.015 |
| N6 tlpD rdxA::tlpD-V5 | 10.0 ± 0.0 | 0.205 (ns) | 16.9 ± 0.3 | 0.118 (ns) |
| HP87P7 wild type | 8.2 ± 0.2 | 16.1 ± 0.4 | ||
| HP87P7 tlpD | 9.9 ± 0.7 | 0.029 | 18.3 ± 1.0 | 0.010 |
| HP87P7 tlpD rdxA::tlpD-V5 | 10.1 ± 1.3 | 0.138 (ns) | 17.7 ± 1.5 | 0.094 (ns) |
amean diameter of growth inhibition combined from duplicate experiments and standard deviation.
bp values for significance of difference calculated by Student’s t test in comparison to respective wild type.
ns: not significant.
To investigate whether a change in gene regulation may be one cause of different susceptibility of the tlpD mutant to these conditions, we performed genome-wide microarray transcriptome analysis of the tlpD mutant in comparison to the H. pylori N6 parental strain. Overall, 68 genes were found either at >2-fold higher (20) or <2-fold lower transcript levels (48) in the tlpD mutant in comparison to the wild type strain (Supplementary Tables S1 and S2). Several transcripts coding for iron-provisioning proteins or iron-sulfur proteins were detected at increased amounts in the tlpD mutant, including the iron-sulfur biosynthesis protein operon NifSU (HP0220-HP0221)46,47. An increase in nifU transcript was confirmed by qRT-PCR in tlpD mutants of two strains (Supplementary Fig. S4A,B).
TlpD-dependent steady-state behaviour of H. pylori is altered under conditions of low iron or high oxidative stress, and by NifSU supplementation
We performed behavioural assays in the presence of paraquat (10 μM and 100 μM) or iron depletion by 2,2-DP (0.5 mM and 5 mM) for N6 wild type, acnB and katA mutants. The assays dose-dependently revealed a reduced TlpD-mediated stopping frequency for all tested strains, similarly with paraquat or 2,2-DP (Fig. 5, results for the parental strain; mutants not shown). Almost no stops were observed at the higher concentrations of 100 μM paraquat or 5 mM 2,2-DP (Fig. 5), respectively, suggesting a comparable effect of both conditions on behaviour.
Figure 5. Characterisation of H. pylori motility patterns under conditions of iron depletion or oxidative stress.
Tracking assays were performed under steady state conditions as described before (see also legend to Fig. 4; low-energy liquid medium RPMI 1640 with 3% FCS). The iron chelator 2,2-dipyridyl or the oxidative stress-inducer paraquat were added to the medium at indicated concentrations, and motile steady-state behaviour was analysed by single cell tracking and quantitated by stop frequencies and curvilinear velocity (CLV) (black squares with whiskers). For each condition, at least 50 cells were traced (black triangles with mean). Significance of differences was calculated using Student’s t test. ***p ≤ 0.001; **p ≤ 0.01 ns is non-significant. N6 (NifSU+) is a strain expressing the H. pylori nifSU genes from a plasmid (Results and Supplementary Methods).
The tlpD mutant, as shown above, was more sensitive than the wild type to oxidative and iron depletion-related stress, although the transcriptome analyses revealed an increase in transcript amounts of some genes, including the nifSU operon, which potentially counteract these environmental stressors (Supplementary Fig. S4). In order to test the hypothesis that iron-sulfur cluster damage or repair may exert an influence on energy-dependent sensing by TlpD, we targeted the essential nifSU genes (HP0220-HP0221). We introduced an expression plasmid for NifSU into H. pylori N6, providing additional gene copies under the control of the intrinsic promoter. This strain, N6 (pHel2::nifSU) (N6 (pCJ1350), in short: N6 (NifSU+)) was viable and showed no growth changes, but had an elevated cellular ATP content (not shown; quantitated as one marker for metabolic activity as described in26) compared to the parental strain. When we tested the N6 (NifSU+) strain for swimming behaviour, TlpD-mediated behaviour was reproducibly altered in comparison to the parental strain (Figs. 4 and 5). Bacterial stopping frequencies were significantly increased in comparison to the parental strain, suggesting that a reactivity threshold for TlpD-dependent responses was shifted in this strain. As expected, the resistance of the N6 (NifSU+) strain towards low-iron stress (iron depletion by 2,2-DP supplementation) was enhanced in comparison to the parental strain (Supplementary Fig. S5).
TlpD shows altered subcellular localisation in H. pylori under incubation conditions of different energy yield, respiratory inhibition and oxidative stress
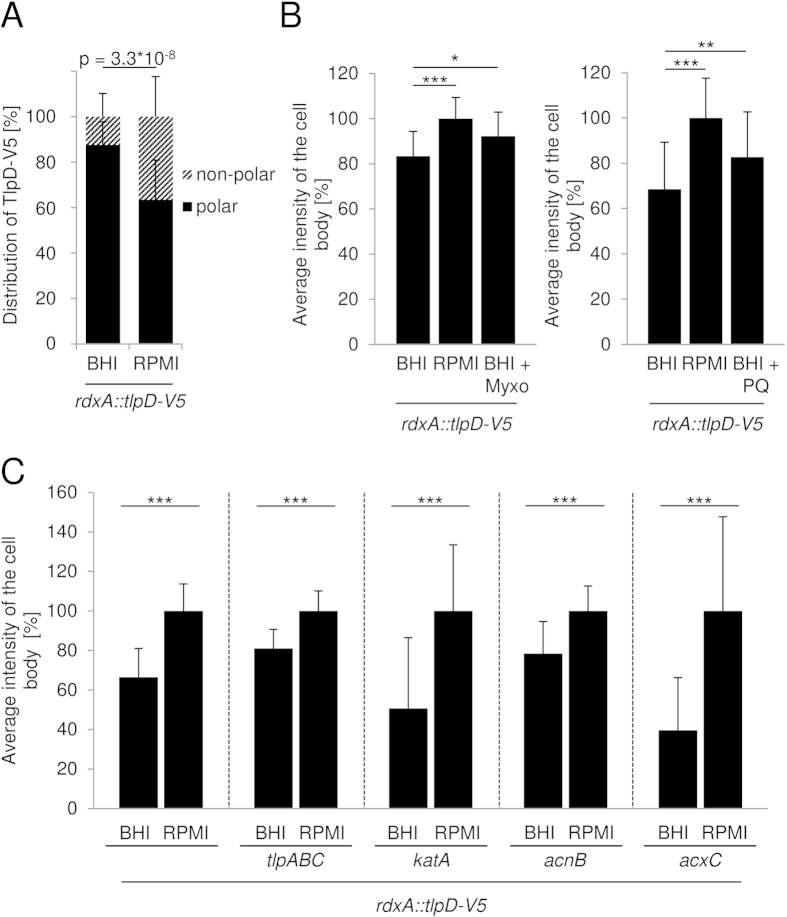
Previously, we were able to match the differences in ATP production by H. pylori in different media to the TlpD-mediated behavioural responses: for instance, BHI containing fetal calf serum (FCS) provided higher ATP levels to H. pylori, while ATP levels were markedly lower in the defined medium RPMI 1640 containing the same amount of serum26; concomitantly, TlpD-mediated responses were prominent in the low-energy medium, while TlpD-mediated responses were minor in the higher energy medium26. In addition, a metabolic inhibitor of the electron transport chain, myxothiazol, induced a TlpD-mediated repellent response and a concentration-dependent change in steady-state swimming behaviour26. In order to verify whether the subcellular distribution of TlpD was associated with the energy content of the growth medium, we shifted H. pylori from plate-grown conditions to the two different liquid media of higher (BHI-FCS) and lower (RPMI-FCS) energy yield26. We incubated the bacteria for a short period of 20 min in these liquid media, pelleted them at low temperature, and immediately resuspended them in fixing agent to preserve their energetic state. We also included additional conditions, inducing alterations in the bacterial energy status in energy-rich liquid medium (BHI-FCS) by direct (myxothiazol added at 10 μM; 26) or indirect metabolic inhibition (oxidative stress by paraquat supplementation, at 10 μM concentration). Under these different energy repletion or depletion conditions, we compared the relative localisation of TlpD by the quantitation of localised fluorescence intensities in the above strains expressing TlpD-V5 from the chromosomal rdxA locus.
Remarkably, in the reference strain (H. pylori N6 rdxA::tlpD-V5) incubated in liquid media of different energy yield, we observed a differential subcellular distribution of IF-labelled TlpD between high energy and low energy medium. In fresh, high-nutrient growth medium in the parental strain, TlpD was predominantly polar (87.6% polar intensity versus 12.4% non-polar, standard error 10.2%; Fig. 6A; Supplementary Fig. S6A). In energy-limited medium at the same time point post incubation (20 min), this relative distribution was significantly shifted towards the cell body (63.5% polar versus 36.5% non-polar TlpD intensity, standard error 17.4%; Fig. 6A, Supplementary Fig. S6A). Likewise, when oxidative stress was applied in high-energy medium using paraquat, or metabolic stress in the presence of the respiratory inhibitor myxothiazol, a significantly larger fraction of total TlpD in the reference strain was non-polar as compared to the energy-rich medium. This was reflected by significantly higher relative TlpD intensities in the cell body under these conditions of metabolic stress (Fig. 6B; Supplementary Fig. S6A). When we tested the TlpD interactor mutants (acnB, katA, acxC) under the same conditions in two different media, they also showed a statistically significant enrichment of TlpD in the cell body in energy-limited medium versus high-energy liquid medium (Fig. 6C). The effect of short-term incubation in liquid media was particularly pronounced for the acnB mutant, which, in comparison to the parental strain, showed a significant redistribution of TlpD towards the cell body even in the high-energy liquid medium (Supplementary Fig. S6C). The katA mutant, which already differed from the parental strain under standard growth conditions (relative TlpD intensity higher in the cell body), also showed a significant difference in relative TlpD intensity in the cell body between high-energy and low-energy liquid medium (Figs. 3 and 6C).
Figure 6. Subcellular localisation of H. pylori TlpD under different energetic conditions.
(A) TlpD-V5 localisation at bacterial poles and cell body was quantitated by immunofluorescence (IF) and compared for N6 rdxA::tlpD-V5 (reference strain) in BHI-yeast, 3% FCS (BHI, high energy yield26) and RPMI 1640, 3% FCS (RPMI, low energy yield26). For IF quantitation, 20 h plate-grown bacteria were incubated in media for 20 min, then immediately fixed and immunolabelled (Supplementary Methods). Total cell intensities were set to 100%; polar and cell body pixel intensities are depicted in percent of total intensities. p-value indicates significant difference between conditions for poles or cell body. (B) TlpD-derived intensities for the cell body only were determined for the reference strain to detect energy-dependent differences in non-polar TlpD. Incubation conditions were: BHI, RPMI, BHI and myxothiazol13 (Myxo, 10 μM for 10 min; left panel, for respiratory inhibition), or BHI and paraquat (PQ, 10 μM for 30 min; right panel for oxidative stress). Myxothiazol or PQ was added after 20 min equilibration in BHI. The fluorescence intensity of the sum of all pixels in the cell body without the poles was first calculated in percent of the total cell pixel intensities added up for each strain; subsequently, cell body-associated intensities in RPMI were set to 100%. Fluorescence intensities of the bacterial cell body omitting the poles were averaged for 30 bacteria under each condition (Supplementary Methods). (C) TlpD localisation in the cell body in the reference strain and the corresponding acnB, katA, tlpABC, acxC mutants was determined under two energetic conditions. 20 h plate-grown bacteria were resuspended in liquid media RPMI or BHI for 20 min. Cell body intensities in RPMI were calculated as in B and set to 100% (right bar for each strain), with normalised relative values in percent for the BHI condition (Supplementary Methods). For all panels and settings, mean and standard deviations of TlpD-derived intensities averaged for 30 bacteria are depicted. IF images were taken at 63-fold magnification. Poles and cell body regions were defined as in Table 3. Significance of differences was determined using Student’s t test (***p ≤ 0.001, **p ≤ 0.01, *p<= 0.05).
Discussion
Energy sensor TlpD interacts with several non-chemotaxis proteins, including catalase and aconitase
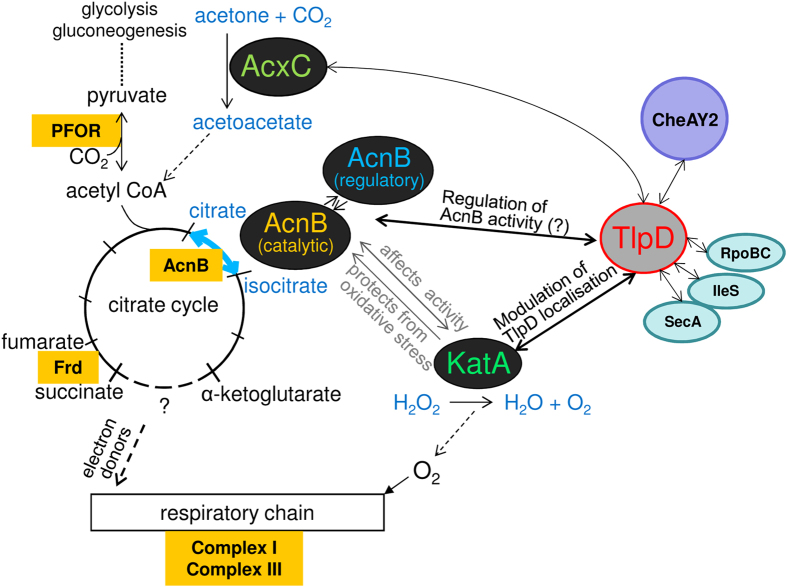
A Yeast-Two-Hybrid protein-protein interaction study of H. pylori proteins had suggested before that H. pylori TlpD is able to interact with other bacterial proteins40, providing one motivation for the current study. Protein-protein interactions involving conformational change may be a means to modulate TlpD-related functions, which might be a key to understand mechanisms of TlpD function. Interactions of chemoreceptors with other proteins may be involved in the mechanism of sensing and also in soluble taxis sensor localisation, although few examples have been reported. For instance, interaction of the soluble cytoplasmic MCP of Rhodobacter sphaeroides, TlpT, with a novel type of partitioning protein, PpfA, determines the subcellular positioning and cluster segregation of the cytoplasmic sensor upon cell division34. Using a pull-down approach against Hisx6-tagged TlpD combined with mass spectrometry of specifically enriched protein bands from H. pylori lysates, we identified six novel potential protein interaction partners of TlpD, CheAY2, AcnB, KatA, IleS, SecA, and RpoBC. The mass spectrometry analyses did not reveal a dedicated partitioning protein interacting with TlpD. Since the analysis was restricted to the fraction of TlpD that became soluble during the lysis procedure, we might have missed yet other potential interactors. Among the novel TlpD-bound proteins was CheAY2, the histidine kinase propagating signals from Tlp sensors towards the flagellar basal body10. In E. coli, the current concept for MCP interactions with the chemotaxis core proteins is that MCPs can dock with CheW25 or with CheA directly, using the CheA P5 domain37,39,48. Although we have not experimentally followed up specific binding between these two proteins, TlpD and CheAY2, in the current study, the detection of the intrabacterial TlpD-CheAY2 interaction in H. pylori is therefore highly plausible and indicates that the pull-down approach was able to reveal true in-vivo interactions. Interestingly, two proteins directly involved in bacterial metabolic functions were also identified as TlpD interactors, aconitate hydratase (AcnB49, a key enzyme in the central metabolism and tricarboxylic acid [TCA] cycle, containing one iron-sulfur cluster per subunit) and catalase (KatA41,50, crucial in cellular detoxification of hydrogen peroxide and oxygen radicals). KatA is the only catalase in H. pylori50. The bidirectional catalytic reaction mediated by AcnB in the TCA cycle (Fig. 7) is considered to be central to two major functions of the capnophilic H. pylori TCA cycle, first by providing substrates for CO2 fixation and glycogenic pyruvate assimilation by the pyruvate-ferredoxin oxidoreductase (PFOR) enzyme, upstream of citrate51,52; and second, by generating electron donors (NADPH+) for the respiratory chain through the TCA enzymes downstream of isocitrate53 (see Fig. 7 for AcnB function and integration into metabolic circuitry). H. pylori acnB49 and katA50 mutants as well as another mutant in the acx operon (acxB; 54) were described before. All three genes were non-essential in vitro. acnB, katA and acx(B) mutants displayed a severe colonisation-deficient phenotype in mice54,55,49, underlining their important role in vivo.
Figure 7. Model of protein-protein interactions of the H. pylori energy sensor TlpD and their connection to metabolism and gene regulation.
Several novel TlpD protein-protein interactions were identified in the present study (KatA, AcnB, CheAY2, IleS, RpoBC, SecA). This model depicts how some of these proteins, jointly with TlpD, integrate into the network of H. pylori metabolism and energy-related functions, which may feed back into TlpD-mediated energy sensing. AcnB is a bifunctional enzyme of the TCA cycle, converting citrate into isocitrate and vice versa. In H. pylori, microaerobic metabolism is thought to favour the generation of citrate by AcnB, feeding into CO2 fixation and gluconeogenesis53. KatA80 detoxifies H2O2 and oxygen radicals, and might thereby protect metabolic enzymes containing iron-sulfur clusters such as AcnB. The putative interactor AcxC is a subunit of acetone carboxylase54, which synthesises acetoacetate, an important precursor of the gluconeogenesis and TCA cofactor acetyl-CoA. Pyruvate-ferredoxin oxidoreductase (PFOR), an iron-sulfur enzyme essential for the microaerobic, capnophilic metabolism of H. pylori51,52, can also govern the direction of metabolic flow in H. pylori from the citrate cycle to gluconeogenesis by CO2 fixation and pyruvate generation. By repleting acetyl-CoA, acetone carboxylase also supplements the PFOR reaction and gluconeogenesis. Under conditions of oxidative stress, AcnB switches between a catalytic and a regulatory function75. TlpD may interfere with AcnB activity and influence regulatory functions of AcnB under stress conditions. KatA possibly protects AcnB from oxidative stress to maintain AcnB catalytic function under oxidative conditions. Proteins containing iron-sulfur clusters46 which are integrated into this part of the metabolic network and may influence TlpD sensing under oxidative or metabolic stress (AcnB, fumarate reductase Frd, PFOR, and subunits of the respiratory complexes I and III) are highlighted by orange colour. Verified interactions between proteins are indicated by bold arrows. Additional potential interactors of TlpD whose direct interaction has not yet been clarified (SecA, IleS, RpoBC) are depicted for completeness.
We verified that KatA and AcnB can act as direct physical binding partners for TlpD in vitro using biolayer interferometry. Both proteins interacted with TlpD with a KD in the nanomolar range. A two-state parallel binding model was best fitting the data for both interactions (TlpD-KatA and TlpD-AcnB). The determined complexity of the binding curves may reflect particularities of the BLI method such as presentation of the ligand to the analyte, but may also be suggestive of the so far unknown mechanisms of the biological interactions. Interestingly, an earlier study which used a Strep-tag pull-down approach to identify large protein complexes in H. pylori identified both AcnB and KatA as components of a large complex involving urease subunits28. This earlier publication already provided an indication that both AcnB and KatA jointly can be part of one single large protein complex, involved in metabolic homeostasis. Three other intrabacterial protein interactions of TlpD identified by the pull-down were to isoleucyl-tRNA synthetase IleS, to the bifunctional RNA polymerase subunit RpoBC, and to the protein translocase-ATPase SecA. These three additional interactions remain to be characterised with respect to direct or indirect binding to TlpD, regulatory function, TlpD activity, or TlpD subcellular positioning.
For the functional characterisation of TlpD, we included a third metabolism-related protein, AcxC, in addition to AcnB and KatA. AcxC (HP0697, encoding an acetone carboxylase subunit54), was identified earlier as a potentially TlpD-interacting protein in a large-scale H. pylori Yeast-Two-Hybrid interaction study40. However, we did not perform direct binding analyses for AcxC to confirm this proposed interaction, and it is therefore not excluded that this candidate was a false positive.
TlpD localisation in H. pylori is influenced by protein interactions, cellular energy status and oxidative stress
The subcellular localisation and distribution patterns of TlpD were previously unresolved. In order to gain better insight into TlpD localisation, function, or potential mechanisms of TlpD energy sensing, we investigated the subcellular localisation of the soluble sensor TlpD under different conditions. While we had previously obtained a preliminary result of membrane-associated localisation of TlpD using an expression plasmid which introduced a gene copy effect26, we were now able by chromosomal integration to obtain more accurate data on the intracellular TlpD localisation in situ which also permitted to dissect subtle differences. Under standard culture conditions in situ, we found TlpD to be localised both focally at the cell poles and in diffuse clusters along the cell body. Although TlpD was predicted to be a soluble sensor by sequence, it was invariably present in similar proportions in the insoluble as well as soluble fractions in all fractionated lysates of mutants and parental strain.
We hypothesised that TlpD localisation might be connected to its function. Therefore, we addressed TlpD localisation in H. pylori isogenic mutants lacking TlpD-interacting proteins and in the context of metabolic and oxidative stress. Some of the newly identified protein interaction partners had an influence on TlpD subcellular localisation. Inactivation of the H. pylori core chemotaxis kinase CheAY210 (taxis null phenotype) had the strongest impact and led to an almost complete loss of TlpD at the polar localisation foci. Both, TlpD polar localisation and a loss of polar accumulation in the absence of CheAY2, were not expected, since TlpD would not necessarily have to interact at the poles with CheAY2 to be active. However, we interpret this localisation as a result of the interaction of TlpD with a fraction of CheAY2 at the poles, which is supposed to be maintained there, for instance via direct interaction with the canonical membrane-bound chemoreceptors37,38,48,56. Very recently, CheAY2 was indeed reported in H. pylori to localise to the bacterial poles, mediated by the presence of the Tlps57, which is consistent with our interpretation. In the same study, the authors also detected a predominantly bipolar localisation of the H. pylori Tlp family, with some signal distant from the poles. TlpD localisation on its own in situ was not addressed in the other study; however, the results match our findings that both bipolar and non-polar TlpD clusters exist in the H. pylori cell.
Although the three transmembrane Tlp sensors of H. pylori are not essential for TlpD function, TlpD-mediated behaviour was significantly diminished in tlpABC triple mutants which express TlpD as the only taxis sensor, as shown before26. When testing the tlpABC mutants for TlpD localisation, we demonstrated that they retained TlpD at one pole, but virtually lost the bipolar localisation characteristic of the parental strain. In accordance with other recent data on the subcellular localisation of chemotaxis components in H. pylori57, it is plausible to speculate that TlpD is retained preferentially at one pole by associating with the polarly localised transmembrane Tlps57, while it may be kept at the other pole by so far unknown interactions possibly involving CheAY2. With regard to a role in TlpD localisation of the additional novel TlpD interactors identified by pull-down, we also detected a clear alteration in bacterial subcellular localisation of TlpD in katA mutants, where a significant shift of TlpD towards the cell body occurred under standard incubation conditions. In acxC mutants, TlpD showed a shift towards a preferentially unipolar localisation, similar to the triple-transducer mutant. We currently have no mechanistic explanation for the latter result.
With respect to conditions that impact on metabolic activities, we have shown here that in the parental strain TlpD resided primarily at both poles (more than 80% of total TlpD-derived fluorescence signal) under defined conditions of high nutrient availability, under which TlpD net activity is minor26. Under nutrient-limited conditions, which favour a dominance of TlpD function over the other H. pylori Tlps26, a significant proportion of TlpD was delocalised towards the cell body. The same effect occurred under the influence of metabolic inhibitors or under conditions of oxidative stress. This raised the question whether this was an effect of protein redistribution, indicating protein dynamics, or of de novo synthesis of TlpD, possibly starting at non-polar sites. This open question will need to be addressed in detail, for example by conducting the experiments in the presence of protein synthesis inhibitors. Interestingly, a metabolism-dependent localisation of a soluble energy taxis receptor was described previously in a seminal study for AerC, a PAS-domain-containing taxis sensor in Azospirillum brasilense58. AerC distributed differentially, either polar or disperse, in different states of bacterial nitrogen fixing activity58. A more recent study concluded that metabolic conditions can guide the dynamic positioning of a respiratory membrane complex (nitrate reductase) in E. coli to shift between the poles (anoxic) and the total circumference of the bacteria (oxic conditions)59. The latter study also revealed that protein de novo synthesis was involved in the dynamics of the respiratory complex when the bacteria were shifted from oxic to anoxic conditions, but not vice versa59. It was not clarified whether the contribution of protein synthesis was direct or indirect in the process.
What can be the mechanisms responsible for TlpD localisation differences in certain mutants? For one, protein-protein interactions can directly influence localisation. We assume that in cheAY2 and triple transducer mutants, shifts in TlpD localisation away from one or both poles are most likely mediated by direct interactions within heterogeneous protein complexes, as explained above. However, we have no unambiguous explanation for the significant alterations in TlpD localisation in katA and acnB mutants versus the parental strain. While a role of the direct protein interactions is possible, an alternative scenario which could explain some changes in TlpD localisation is a general shift of metabolic activities in the mutant bacteria. A strong contribution of the latter scenario appears most probable for the acnB mutant, which showed a severe growth defect and a reduced metabolic activity in several assays (own unpublished data). In accordance, the loss of AcnB appeared to mediate a significant difference in subcellular distribution of TlpD towards the cell body only in high-energy liquid medium with respect to the parental strain. However, in the katA mutant, while we did not observe a change in growth, ATP content, respiratory activity, or altered TlpD-mediated behavioural response, TlpD was significantly enriched in the cell body versus the poles. Therefore, we currently assume a direct influence of KatA on TlpD localisation. The localisation differences suggest that KatA, of which a significant insoluble portion exists in H. pylori according to our present results (Fig. S3), may contribute to retaining TlpD at the poles under conditions of high energy, in which the contribution of TlpD to the net bacterial behaviour was minor26. We still have to clarify whether TlpD performs its function at any particular subcellular localisation, i.e. whether it might be more active at the non-polar location versus the poles.
In other bacterial species, dedicated energy harvesting complexes of the respiratory chain were located in close proximity to the basal body of the flagellar apparatus32. Whether TlpD incorporates into similar complexes will need to be further investigated. As AcnB catalytic activity was reduced in H. pylori katA mutants (own unpublished data), KatA might provide a functional connection between TlpD and AcnB, which needs to be further explored (see also model in Fig. 7). Taken together, our data hint toward a functional and possibly dynamic, physical connection between metabolically relevant enzymes such as KatA, AcnB, and other energy-generating complexes in H. pylori with components of the chemotaxis and motility machineries.
TlpD-related behaviour remains unchanged by deficiency in the interactors AcnB or KatA, but is modulated by oxidative stress, iron depletion and iron-sulfur homeostasis via NifSU
In addition to localisation, we hypothesised that AcnB, KatA or other potentially interacting proteins, by catalytic activity status or conformational alterations, might influence the energy-dependent taxis function of TlpD. We phenotypically characterised mutants in the potential interaction partners AcnB, KatA and AcxC for changes in their behavioural repertoire linked to the TlpD sensor. Since our microarray analyses revealed compensatory regulation in the absence of functional TlpD, for instance a transcript increase of the iron-sulfur cluster maintenance gene nifU46, we also included the characterisation of NifSU overexpression with respect to TlpD function.
We did not observe a significant modulation of H. pylori steady-state motile behaviour when analysing the interactor mutants katA or acnB, despite the fact that TlpD localisation, in particular in the katA mutant, was changed. Differential influences of mutants on TlpD localisation versus function are not excluded by prior data, and future investigation will have to take other interacting proteins, protein complexes and alternative incubation conditions into account. The tlpABC triple mutant had a reduced TlpD response activity as noted previously26. Interestingly, AcxC deficiency and overexpression of the NifSU iron-sulfur cluster biogenesis proteins also led to a significant modulation in TlpD-mediated steady-state behaviour. Since the NifSU gene products in H. pylori are involved in the maintenance and repair of iron-sulfur cluster proteins such as aconitase, PFOR, fumarate reductase, hydrogenase, ferredoxins and other enzymes feeding into or directly involved in the H. pylori respiratory chain46,47, they could provide one missing functional link between intrabacterial energy generation, redox homeostasis and energy taxis. Such a link was previously suggested by experiments conducted with inhibitors of the respiratory chain which hampered TlpD function and induced a repellent response26. TlpD-dependent stopping frequency was augmented by NifSU. This result was correlated to the inverse effect of iron depletion (2,2-dipyridyl) and increased oxidative stress (paraquat) which both diminished the behavioural response by TlpD. It is a possible scenario that NifSU supplementation mediates a general metabolic shift in the bacteria by acting on a host of important metabolic iron-sulfur-cluster enzymes simultaneously and thus might influence TlpD-dependent sensing indirectly.
Although the tlpD-deficient strain was more sensitive than its parent against iron depletion or oxidative stress, H. pylori in general showed a relatively weak sensitivity. This can be explained by the technical limitations of the used test system (agar diffusion) and by strong redundancies in the bacterial detoxifying systems in H. pylori which counteract iron and oxidative stresses60,61.
The TlpD-dependent behavioural phenotype13,26 was very robust in different H. pylori mutants. Except for the chemotaxis null mutants cheAY210,26, cheW62,63, cheY10,62 and mutants in tlpD itself, no mutant is yet known which has lost TlpD function. In vivo, H. pylori tactic abilities are involved in modulating or avoiding the inflammatory host response against H. pylori which generates oxidative stress11,13,19,64. Oxidative stress and iron depletion both can act on the same cellular targets by affecting iron-sulfur cluster integrity and thereby the activity of essential metabolic dehydratase enzymes by damaging their active centres44,45. Among the most important iron-sulfur proteins in H. pylori are aconitase, fumarate reductase, hydrogenase, PFOR and the NifSU proteins themselves46,65. H. pylori nifU was an essential gene in vitro46,66 and hydrogenase was required for survival in a mouse model67. The modulatory effect of NifSU on TlpD behavioural activity corresponds to a major role of oxidative or iron stress with regard to TlpD functionality and requirement in vivo, as previously reported by us13 and others12. This mutuality implies a connection of TlpD function to iron-sulfur protein homeostasis, which, however, could not yet be pinpointed to any direct protein interaction in the present study. Future studies will have to clarify if interactions with other non-chemotactic proteins, possibly containing iron-sulfur clusters, are required for the sensing mechanism of TlpD.
TlpD contributes to gene regulation in H. pylori by influencing multiple transcripts
An involvement of dedicated chemotaxis receptors in bacterial gene regulation has rarely been reported, and a role for TlpD in gene regulation has not been determined either. Direct or indirect functions of transducer-like proteins in gene regulation are conceivable, especially when the overall number of regulators is low, as in H. pylori. Prüss et al.68 described an effect of the E. coli aerotaxis sensor Aer on transcript amounts which may be linked to an important role in maintaining intrabacterial energy levels. On the basis of the assumption that gene regulation events can be helpful in deducing the TlpD sensing mechanism or protein interactions relevant to understanding its function, or finding causes of functional deficiencies of the tlpD mutant, we have tested how the transcriptome is affected in H. pylori tlpD mutants. In addition, the possibility of a regulatory compensation for the loss of energy taxis in tlpD mutants needed to be explored. In the tlpD mutant in comparison with the wild type strain, numerous differentially expressed transcripts in diverse functional categories were identified. The effects on transcript amounts were moderate rather than drastic which suggested an indirect role of TlpD. This raised the question of how the genes might be differentially regulated in the absence of TlpD. nifU and katA transcripts were positively affected by the loss of TlpD. Both nifU and katA were previously found positively regulated by the HP1021 orphan response regulator, whose sensory input is unknown69. Some of the changed transcripts in the tlpD mutant also appeared to overlap with the Fur or other metal-dependent regulons70,71. nifSU transcripts were earlier reported to be upregulated under conditions of oxidative stress, excess iron and by the ferric uptake regulator Fur70. Only a fraction of the transcript changes in the tlpD mutant can be linked to the canonical regulators HP1021 or Fur, which calls for other possible explanations. One way how TlpD might be linked to regulation is via its identified interaction with AcnB. As a basis, previous studies have indeed demonstrated that aconitase B in other bacteria (E. coli)72,73, mammals74, and recently also in H. pylori49, performs a central “moonlighting” role as a posttranscriptional gene regulator, in particular under conditions of iron depletion and oxidative stress. This moonlighting function was inversely correlated with AcnB catalytic activity, which is dependent on the integrity of its iron-sulfur complexes and determines its catalytic or regulatory protein conformation72. Austin et al.49 noted that the iron chelator 2,2-DP that we also observed to impact on TlpD sensing, can inactivate he catalytic activity H. pylori aconitase and enhance the regulatory function of aconitase. Some transcripts that we found higher expressed in tlpD mutants in microarrays or real-time PCR overlapped with results of a recent proteome analysis of an acnB mutant75, among those ureA (functional urease subunit gene), tpx (thiol peroxidase gene), ahpC (alkylhydroperoxidase subunit gene), porA, (pyruvate-ferredoxin oxidoreductase subunit gene A), trx2 (thioredoxin 2 gene), trxR2 (thioredoxin reductase gene). Thus it is plausible that the regulatory potential of AcnB in H. pylori might be harnessed by the TlpD interaction, which needs to be clarified. In addition, an intriguing finding of the latter study with regard to our results was that CheAY2, SecA and IleS, which we recovered in the TlpD pull-down, were all enriched in membrane fractions of an acnB mutant75. Thus, the finding of TlpD-dependent transcript changes also underlines a second hypothesis regarding the regulatory effect in the tlpD mutant and functions of TlpD in the context of confirmed protein-protein interactions: namely, that binding to TlpD might influence the catalytic status and regulatory activity of interacting proteins, for instance of AcnB (Fig. 7). In tune with this hypothesis, a slight reduction in AcnB catalytic activity was noted in H. pylori lysate of a TlpD overexpressing strain in contrast to the parent (own unpublished data).
Thus, in summary, a compensatory gene regulation in tlpD mutants was identified, possibly connected to AcnB-mediated posttranscriptional regulation, which strengthens a link of TlpD to metabolism, oxidative stress and iron provisioning.
Conclusions
With TlpD and the acid sensor TlpB, two of four H. pylori behavioural receptors may observe a function possibly associated with intrabacterial energy status, indicating a prominent role of energy taxis for this bacterium in its challenging environment. Since major oxidative stress by the host defence and metal stress by varying metal ion availability in vivo are assumed to play a crucial role in the stomach niche of H. pylori, energy-associated behavioural functions are anticipated to exert a major influence on the short-term environmental flexibility of the organism in vivo. TlpD interacted with several other bacterial cytoplasmic proteins in addition to its expected interaction with the CheAY2 signalling complex. At present, combined data on TlpD localisation and behavioural activity in the tested mutants do not provide conclusive evidence as to where in the cell TlpD is active. Fine in situ localisation of CheAY2 and other interaction partners under various conditions will be required to interpret TlpD localisation in the context of TlpD function. A current model of TlpD interactions and metabolic crosstalk is presented in Fig. 7: confirmed TlpD interactors aconitase B and catalase both play an important role in metabolic functions and regulation or protect against iron and oxidative stress which threaten the bacterial central metabolism and respiratory chain (Fig. 7). Although we could not link any of the interactions, including KatA and AcnB, directly to the TlpD sensing and signalling mechanisms so far, TlpD affected multiple transcripts, possibly in part via AcnB-dependent regulation (Fig. 7). By fine-tuning posttranscriptional gene regulation, TlpD might act in a flexible manner on cellular functions which feed back into behaviour via expression changes in metabolism, redox functions and sensory proteins. Thereby, TlpD may provide a short to medium-term global adaptation process to metabolic threats, in case TlpD energy taxis functions are not sufficient to quickly evade these threats. In support of this, functional data demonstrated that sensing and the repellent mechanism initiated by the TlpD sensor are strongly connected to cellular functions depending on iron and damaged by oxidative stress. TlpD sensing is probably connected to at least one of the iron-sulfur cluster proteins in the central metabolism45,46,65 (Fig. 7), which can act as sensors for redox and iron76 and are intimately linked to the energetic status of the cell. Conditional depletion of specific essential iron-sulfur proteins and the structural investigation of TlpD will permit to clarify remaining questions concerning protein-protein interactions or functional implications of these proteins for energy-dependent behaviour.
Methods
See Supplementary Methods for details about DNA and protein methods, protein-based pull-down-assays, construction of H. pylori allelic exchange mutants (Table 2, Supplementary Table S3), RNA preparation from H. pylori and cDNA synthesis, immunofluorescence labelling and microscopy26,77, and statistics.
Mass spectrometry after TlpD pull-down
For tandem mass spectrometry, peptides recovered after TlpD-Hisx6 based pull-down and trypsin digest78 were dissolved in 2% ACN, 0.1% formic acid, and reverse phase chromatography using acetonitrile as an eluent was performed on a nanoACQUITY UPLC system (Waters) equipped with an analytical column (Waters, BEH130C18, 100 μm × 100 mm, 1.7 μm particle size), coupled online to an ESI Q-TOF (Q-TOF MS/MS; Ultima, Waters, Milford, MA, USA). Spectra were recorded in positive reflection mode, and peptides were automatically subjected to fragmentation (MS/MS). Protein and peptide identification were performed using the program ProteinLynx™ Global Server (Version 2.1, Waters) and the MASCOT search engine (Matrix Science). A more detailed description of peptide identification parameters, database, mass spectrometry settings and scoring is provided in the Supplementary Methods.
Bacterial strains and growth conditions
Helicobacter pylori strains N679 and HP87P713 and their isogenic mutants are listed in Table 2. For details of growth conditions see Supplementary Methods.
Purification of H. pylori TlpD-V5, AcnB-V5, Hisx6-TlpD and catalase using affinity chromatography
We purified recombinant Hisx6-TlpD (N-terminally fused Hisx6 Tag) from E. coli BL21(DE3) upon expression from a pET28a based plasmid (pCJ1341). TlpD-V5 and AcnB-V5 were affinity-purified from cleared lysates of H. pylori expressing TlpD-V5 or AcnB-V5 from plasmids. Catalase was purified after recombinant expression in E. coli as previously described80. For details of purification procedures, see Supplementary Methods.
Testing of direct interactions of TlpD and interaction partners using biolayer interferometry
To test for direct protein-protein interaction, biolayer interferometry (BLI) was performed on the Octet RED96 System (ForteBio, Menlo Park, USA). BLI is an optical biosensing method that yields kinetic and affinity information in a manner similar to surface plasmon resonance (SPR)42. Purified recombinant H. pylori catalase (KatA)41,80 and V5-purified native H. pylori aconitase (AcnB-V5) were used as interaction partners and analytes for sensor-coupled Hisx6-TlpD (ligand). Alternatively, recombinant KatA (ligand) was tested with native TlpD-V5 (analyte) purified from H. pylori. Analyses of the binding curves were performed using the Octet Data Analysis Software 6.4.. For a detailed description, see Supplementary Methods.
Single cell tracking and analysis of steady-state behaviour of H. pylori
The behaviour of H. pylori N6 and its derivatives in liquid medium was determined using established cell tracking protocols26. Briefly, freshly grown bacteria (grown about 20 h on blood agar plates) were resuspended in pre-warmed defined media at an OD600 of 0.03. Bacteria were visualised using a live cell microscope (Olympus IX-80 inverted Cell-R microscope) equipped with a climate chamber26. Before recording movies, bacteria were pre-incubated and equilibrated at 37 °C and ambient air enriched with 5% CO2 for 15 min. Thus, we determined for each condition a steady-state behaviour under equilibrated conditions. We did not observe an overt change of behaviour within the first 15 min of equilibration before the measurements were started. One might suspect that adaptation can occur during these 15 min. However, behavioural adaptation by sensor methylation is not known to occur in H. pylori, and cheB/cheR genes for methylating and demethylating Tlp sensors are not present in the H. pylori genome16. For quantification of bacterial traces, 3 to 4 movies (250 frames, 19 frames s−1), were recorded for each bacterial strain, and the swimming patterns of at least 50 single cells per strain (see figure legends) were analysed using the Cell-R system as described before13,26. We counted each single stop, and each stop in conjunction with a direction change, as one stop. In addition, we counted as one stop a full reversal of the bacteria (including one back–up movement, and one immediately following forward-run), if there was no direction change involved. Bacteria on the bottom of the field of view, which are bacteria in close contact with the solid support and could be sticking to the surface, were excluded from the counts and analyses. For the motility analysis of the strain N6 (pHel2::nifSU), we also tested an empty pHel2 plasmid as a control, which showed no behavioural difference in comparison to the parental N6 strain (data not shown).
Testing growth and behaviour of H. pylori under iron-depleted conditions
H. pylori N6 wild type and tlpD mutant were tested for growth defects and behaviour in agar diffusion assays and steady-state tracking assays under iron-limited conditions by adding the iron-chelator 2,2-dipyridyl (Sigma-Aldrich, St. Louis, USA). For agar diffusion assays, bacteria grown on blood agar plates for about 20 h were resuspended in BHI medium to an OD600 of 0.07. 100 μl of the bacterial suspension were plated on Blood Agar Base No. 2 plates (Oxoid) supplemented with 5% horse serum. 10 μl of 10 mM, 20 mM and 40 mM 2,2-dipyridyl (dissolved in ethanol) as iron chelator or 10 μl of appropriate dilutions of ethanol in PBS (negative control) were applied to the plates on paper discs (diameter 5 mm). The diameter of the growth inhibition zone was measured upon incubation of bacteria at standard conditions for 48 to 72 h. Tracking assays under iron-limited conditions were performed as described above. After equilibrating the bacteria in liquid medium (see above), 0.5 mM or 5 mM 2,2-dipyridyl was added to the medium or the bacteria were left untreated. Movies were recorded 15 min and 30 min after the addition of 2,2-dipyridyl and steady-state behaviour was evaluated as described above. Steady-state assays with paraquat (10 μM, 100 μM) were carried out according to the same procedure.
Transcript analysis using whole genome microarray
Comparative analyses of transcript levels in N6 and HP87 P7 and their respective tlpD mutants were performed using whole genome DNA microarrays as previously described81. Microarrays were generated and quality-controlled as previously described81,82. The custom-made whole-genome microarray consists of 1,649 gene-specific PCR products generated with specific primer pairs derived from the genome sequences of H. pylori strains 2669516 and J9915, which were all spotted in duplicates. Microarray hybridisation, raw data generation, and data analysis including normalisation and quantitation were performed as previously reported81,83. Raw data of the competitive hybridisations were analysed using ImaGene v5.0 (BioDiscovery, USA) and MAVI v2.5 (MWG Biotech AG, Ebersberg, Germany) using previously described settings83. Four microarray slides in total were evaluated in dual-colour mode for strain N6, and dyes were swapped between the parental strain and tlpD mutant in one of the four experiments. Finally, the mean values of four separate arrays, including three biological experiments and one dye swap (each array comprising in addition two technical replicates for each gene), of H. pylori N6 and its isogenic tlpD mutant were summarised and evaluated jointly for the final dataset (Supplementary Tables S1 and S2). Significance Analysis of Microarray (SAM84, Version 5.0) analysis was performed for the dataset with the following parameters: Data Type: One class; Arrays centered: true; delta: 0.32022; number of permutations: 100; input percentile for exchangeability factor s0: automatic; number of neighbours for KNN: 10; seed for random number generator: 1234567. These settings calculate a number of significantly regulated transcripts of 81, a probability of correct assignments of 95% and a false discovery rate (FDR) of 5.3%. Statistically significant changes are indicated for single transcripts in Supplementary Tables S1 and S2. Stringent criteria for the evaluation of regulated transcripts for the final dataset were set as lower and upper limits for the transcript ratios of 0.5- and 2-fold changed, respectively. Changes in transcript levels of selected regulated transcripts in microarray results of two H. pylori strains were additionally verified using semi-quantitative and quantitative RT PCR (Supplementary Fig. S4). Microarray data shown in the manuscript have been deposited in the GEO omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE79364.
Quantitation of fluorescence intensities for single bacterial cells and statistics
Subcellular fluorescence distribution in H. pylori (visualised with fluorescently labelled anti-V5 by α-V5 antibody) was determined using ImageJ densitometry85of TIFF images, which were obtained using the same exposure and lighting settings for each strain. Measurements were then used to generate intensity histograms of TlpD for each cell along the longitudinal axis of at least 15 or 30 separate bacterial cells per strain and evaluated statistically. Details of quantitation methodology and statistics are described in the Supplementary Methods.
Additional Information
How to cite this article: Behrens, W. et al. Localisation and protein-protein interactions of the Helicobacter pylori taxis sensor TlpD and their connection to metabolic functions. Sci. Rep. 6, 23582; doi: 10.1038/srep23582 (2016).
Supplementary Material
Acknowledgments
We acknowledge the international graduate program IRTG1273 (German Research Foundation) for support to C.J. and W.B., and the graduate school HBRS of Hannover Medical School for additional funding. The support through grants Jo 344/3-1 by the German Research Foundation and HeldivPAT (grant no. 0315905E) by the German Ministry for Education and Research provided to C.J. is also gratefully acknowledged. C.J. was also supported in the framework of the German Center of Infection Research (DZIF). We thank Sarah A. Kuehne for experimental contributions to the microarray experiments. Work in the lab of P.C.L. was supported by the Discovery Grant 9600 from the Natural Sciences and Engineering Council (NSERC) of Canada and the Canada Research Chair Program.
Footnotes
Author Contributions W.B., T.S. and C.J. designed the experiments, analysed the data, wrote the manuscript and prepared the figures and tables; W.B., T.S., F.B., S.M. and C.J. performed the experiments; J.M. provided support for Octet analyses and KD determination. P.L. provided material and contributed to experimental design, T.S., C.J. and F.B. performed and analysed mass spectrometry experiments, and all authors contributed to editing and revising the manuscript.
References
- Suerbaum S. & Michetti P. Helicobacter pylori infection. N. Engl. J. Med. 347, 1175–1186 (2002). [DOI] [PubMed] [Google Scholar]
- Annibale B. et al. Concomitant alterations in intragastric pH and ascorbic acid concentration in patients with Helicobacter pylori gastritis and associated iron deficiency anaemia. Gut 52, 496–501 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S. et al. Rapid loss of motility of Helicobacter pylori in the gastric lumen in vivo. Infect. Immun. 73, 1584–1589 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suerbaum S. & Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat. Rev. Microbiol. 5, 441–452 (2007). [DOI] [PubMed] [Google Scholar]
- Salama N. R., Hartung M. L. & Muller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Microbiol. 11, 385–399 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I., Nell S. & Suerbaum S. Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol. Rev. 37, 736–761 (2013). [DOI] [PubMed] [Google Scholar]
- Schreiber S. et al. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl. Acad. Sci. USA 101, 5024–5029 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücker R. et al. Helicobacter pylori colonization critically depends on postprandial gastric conditions. Sci. Rep. 2, 994, doi: 10.1038/srep00994 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt M. R. et al. ChePep controls Helicobacter pylori Infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. MBio 2, e00098–11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foynes S. et al. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68, 2016–2023 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee D. J. et al. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 73, 1820–1827 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolig A. S., Shanks J., Carter J. E. & Ottemann K. M. Helicobacter pylori requires TlpD-driven chemotaxis to proliferate in the antrum. Infect. Immun. 80, 3713–3720 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens W. et al. Role of energy sensor TlpD of Helicobacter pylori in gerbil colonization and genome analyses after adaptation in the gerbil. Infect. Immun. 81, 3534–3551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton K. A., Suerbaum S., Josenhans C. & Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64, 2445–2448 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm R. A. et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397, 176–180 (1999). [DOI] [PubMed] [Google Scholar]
- Tomb J. F. et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388, 539–547 (1997). [DOI] [PubMed] [Google Scholar]
- Andermann T. M., Chen Y. T. & Ottemann K. M. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect. Immun. 70, 5877–5881 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen M. A., Sisson G., Melano R. & Hoffman P. S. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 188, 2656–2665 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. M. et al. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 75, 3747–3757 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertsethtakarn P., Ottemann K. M. & Hendrixson D. R. Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 65, 389–410 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders L. & Andermann T. M. & Ottemann,K.M. A supplemented-soft agar chemotaxis assay demonstrates Helicobacter pylori chemotactic response to zinc and nickel. Microbiology 159, 46–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizote T., Yoshiyama H. & Nakazawa T. Urease-independent chemotactic responses of Helicobacter pylori to urea, urease inhibitors, and sodium bicarbonate. Infect. Immun. 65, 1519–1521 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Y. et al. Chemodetection and Destruction of Host Urea Allows Helicobacter pylori to Locate the Epithelium. Cell Host Microbe 18, 147–156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda O., Rivas A. & Toledo H. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol. Lett. 224, 175–181 (2003). [DOI] [PubMed] [Google Scholar]
- Wadhams G. H. & Armitage J. P. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5, 1024–1037 (2004). [DOI] [PubMed] [Google Scholar]
- Schweinitzer T. et al. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J. Bacteriol. 190, 3244–3255 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. C., Johnson M. S. & Taylor B. L. Differentiation between electron transport sensing and proton motive force sensing by the Aer and Tsr receptors for aerotaxis. Mol. Microbiol. 62, 823–837 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl K., Uhlemann E. M., Schmid R., Altendorf K. & Bakker E. P. Energetics of Helicobacter pylori and its implications for the mechanism of urease-dependent acid tolerance at pH 1. J. Bacteriol. 184, 3053–3060 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader B. A. et al. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology 157, 2445–2455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinitzer T. & Josenhans C. Bacterial energy taxis: a global strategy? Arch. Microbiol. 192, 507–520 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre G. Coupling metabolism and chemotaxis-dependent behaviours by energy taxis receptors. Microbiology 156, 2283–2293 (2010). [DOI] [PubMed] [Google Scholar]
- Zarbiv G. et al. Energy complexes are apparently associated with the switch-motor complex of bacterial flagella. J. Mol. Biol. 416, 192–207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper J., Karplus K. & Ottemann K. M. Identification of a chemoreceptor zinc-binding domain common to cytoplasmic bacterial chemoreceptors. J. Bacteriol. 193, 4338–4345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. A., Wadhams G. H., Hadfield K. A., Tickner S. & Armitage J. P. ParA-like protein uses nonspecific chromosomal DNA binding to partition protein complexes. Proc. Natl. Acad. Sci. USA 109, 6698–6703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A. et al. Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. Elife 3, e02151, doi: 10.7554/eLife.02151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Pearson M. A., Delany I., Scarlato V. & Beier D. Phosphate flow in the chemotactic response system of Helicobacter pylori. Microbiology 151, 3299–3311 (2005). [DOI] [PubMed] [Google Scholar]
- Li X. et al. The 3.2 A resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays. Biochemistry 52, 3852–3865 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasta K. N., Ulliman C. J., Slivka P. F., Crane B. R. & Falke J. J. Defining a key receptor-CheA kinase contact and elucidating its function in the membrane-bound bacterial chemosensory array: a disulfide mapping and TAM-IDS Study. Biochemistry 52, 3866–3880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Vu A., Lee K. & Dahlquist F. W. CheA-receptor interaction sites in bacterial chemotaxis. J. Mol. Biol. 422, 282–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain J. C. et al. The protein-protein interaction map of Helicobacter pylori. Nature 409, 211–215 (2001). [DOI] [PubMed] [Google Scholar]
- Switala J. & Loewen P. C. Diversity of properties among catalases. Arch. Biochem. Biophys. 401, 145–154 (2002). [DOI] [PubMed] [Google Scholar]
- Abdiche Y., Malashock D., Pinkerton A. & Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 377, 209–217 (2008). [DOI] [PubMed] [Google Scholar]
- Briegel A. et al. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. USA 106, 17181–17186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche B. et al. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim. Biophys. Acta. 1827, 455–469 ( 2013). [DOI] [PubMed] [Google Scholar]
- Imlay J. A. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson J. W., Agar J. N., Johnson M. K. & Maier R. J. Characterization of the NifU and NifS Fe-S cluster formation proteins essential for viability in Helicobacter pylori. Biochemistry 39, 16213–16219 (2000). [DOI] [PubMed] [Google Scholar]
- Tokumoto U., Kitamura S., Fukuyama K. & Takahashi Y. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J. Biochem. 136, 199–209 (2004). [DOI] [PubMed] [Google Scholar]
- Briegel A. et al. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. USA 109, 3766–3771 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin C. M. & Maier R. J. Aconitase-mediated posttranscriptional regulation of Helicobacter pylori peptidoglycan deacetylase. J. Bacteriol. 195, 5316–5322 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenbreit S., Wieland B. & Haas R. Cloning and genetic characterization of Helicobacter pylori catalase and construction of a catalase-deficient mutant strain. J Bacteriol. 178, 6960–6967 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes N. J., Chalk P. A., Clayton C. L. & Kelly D. J. Identification of carboxylation enzymes and characterization of a novel four-subunit pyruvate:flavodoxin oxidoreductase from Helicobacter pylori. J. Bacteriol. 177, 3953–3959 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Maurice M. et al. Flavodoxin:quinone reductase (FqrB): a redox partner of pyruvate:ferredoxin oxidoreductase that reversibly couples pyruvate oxidation to NADPH production in Helicobacter pylori and Campylobacter jejuni. J. Bacteriol. 189, 4764–4773 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitson S. M., Mendz G. L., Srinivasan S. & Hazell S. L. The tricarboxylic acid cycle of Helicobacter pylori. Eur. J. Biochem. 260, 258–267 (1999). [DOI] [PubMed] [Google Scholar]
- Brahmachary P. et al. The human gastric pathogen Helicobacter pylori has a potential acetone carboxylase that enhances its ability to colonize mice. BMC Microbiol. 8(4), 14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A. G. et al. Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology 149, 665–672 (2003). [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Falke J. J. & Parkinson J. S. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33, 9–19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lertsethtakarn P., Howitt M. R., Castellon J., Amieva M. R. & Ottemann K. M. Helicobacter pylori CheZ and ChePep form a novel chemotaxis-regulatory complex distinct from the core chemotaxis signaling proteins and the flagellar motor. Mol. Microbiol. 6, 1063–78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Ulrich L. E., Zhulin I. B. & Alexandre G. PAS domain containing chemoreceptor couples dynamic changes in metabolism with chemotaxis. Proc. Natl. Acad. Sci. USA 107, 2235–2240 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberge F. et al. Dynamic subcellular localization of a respiratory complex controls bacterial respiration. Elife 4, e05357; doi: 10.7554/eLife.05357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelhalder C., Gerstenecker B., Kersten A., Schiltz E. & Kist M. Purification of Helicobacter pylori superoxide dismutase and cloning and sequencing of the gene. Infect. Immun. 61, 5315–5325 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit S. L., Bayyareddy K., Mahawar M., Sharp J. S. & Maier R. J. Alkyl hydroperoxide reductase repair by Helicobacter pylori methionine sulfoxide reductase. J. Bacteriol. 195, 5396–5401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry K., Williams S. M., Connolly L. & Ottemann K. M. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect. Immun. 73, 803–811 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman M. S., Goodwin M. & Kelly D. J. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147, 2493–2504 (2001). [DOI] [PubMed] [Google Scholar]
- Rolig A. S., Carter J. E. & Ottemann K. M. Bacterial chemotaxis modulates host cell apoptosis to establish a T-helper cell, type 17 (Th17)-dominant immune response in Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 108, 19749–19754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamuri P., Mehta N., Burk A. & Maier R. J. Regulation of the Helicobacter pylori Fe-S cluster synthesis protein NifS by iron, oxidative stress conditions, and Fur. J. Bacteriol. 188, 5325–5330 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama N. R., Shepherd B. & Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186, 7926–7935 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson J. W. & Maier R. J. Molecular hydrogen as an energy source for Helicobacter pylori. Science 298, 1788–1790 (2002). [DOI] [PubMed] [Google Scholar]
- Prüss B. M. et al. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185, 534–543 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflock M. et al. The orphan response regulator HP1021 of Helicobacter pylori regulates transcription of a gene cluster presumably involved in acetone metabolism. J. Bacteriol. 189, 2339–2349 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst F. D. et al. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151, 533–546 (2005). [DOI] [PubMed] [Google Scholar]
- Danielli A. & Scarlato V. Regulatory circuits in Helicobacter pylori: network motifs and regulators involved in metal-dependent responses. FEMS Microbiol. Rev. 34, 738–752 (2010). [DOI] [PubMed] [Google Scholar]
- Tang Y., Guest J. R., Artymiuk P. J. & Green J. Switching aconitase B between catalytic and regulatory modes involves iron-dependent dimer formation. Mol. Microbiol. 56, 1149–1158 (2005). [DOI] [PubMed] [Google Scholar]
- Kiley P. J. & Beinert H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6, 181–185 (2003). [DOI] [PubMed] [Google Scholar]
- Lushchak O. V., Piroddi M., Galli F. & Lushchak V. I. Aconitase post-translational modification as a key in linkage between Krebs cycle, iron homeostasis, redox signaling, and metabolism of reactive oxygen species. Redox. Rep. 19, 8–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin C. M., Wang G. & Maier R. J. Aconitase functions as a pleiotropic posttranscriptional regulator in Helicobacter pylori. J. Bacteriol. 197, 3076–3086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T. A. & Klausner R. D. Iron-sulfur clusters as biosensors of oxidants and iron. Trends Biochem. Sci. 21, 174–177 (1996). [PubMed] [Google Scholar]
- Rust M. et al. The Helicobacter pylori anti-sigma factor FlgM is predominantly cytoplasmic and cooperates with the flagellar basal body protein FlhA. J Bacteriol. 191, 4824–4834 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J. V. & Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006). [DOI] [PubMed] [Google Scholar]
- Behrens W., Bönig T., Suerbaum S. & Josenhans C. Genome sequence of Helicobacter pylori hpEurope strain N6. J. Bacteriol. 194, 3725–3726 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen P. C. et al. Structure of Helicobacter pylori catalase, with and without formic acid bound, at 1.6 A resolution. Biochemistry 43, 3089–3103 (2004). [DOI] [PubMed] [Google Scholar]
- Niehus E. et al. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52, 947–961 (2004). [DOI] [PubMed] [Google Scholar]
- Gressmann H. et al. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS. Genet. 1, e43 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterzenbach T. et al. Role of the Helicobacter hepaticus flagellar sigma factor FliA in gene regulation and murine colonization. J. Bacteriol. 190, 6398–6408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R. & Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S. & Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J. & Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19, 259–268 (1982). [DOI] [PubMed] [Google Scholar]
- Omori K., Akatsuka H. & Komatsubara S. Versatile Escherichia coli expression vectors for production of truncated proteins. Plasmid 31, 297–299 (1994). [DOI] [PubMed] [Google Scholar]
- Josenhans C., Labigne A. & Suerbaum S. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 177, 3010–3020 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuermann D. & Haas,R. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257, 519–528 (1998). [DOI] [PubMed] [Google Scholar]
- Copass M., Grandi G. & Rappuoli R. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infect. Immun. 65, 1949–1952 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.