Abstract
Over the past decade, protein-protein interactions have gone from being neglected as “undruggable” to being considered attractive targets for the development of therapeutics. Recent advances in computational analysis, fragment-based screening and molecular design have revealed promising strategies to address the basic molecular recognition challenge: how to target large protein surfaces with specificity. Several systematic and complementary workflows have been developed to yield successful inhibitors of protein-protein interactions. Herein we review the major contemporary approaches utilized for the discovery of inhibitors and focus on a structure-based workflow, from the selection of a biological target through design.
Keywords: inhibitors, protein-protein interactions, rational design, screening, fragment-based design, protein domain mimics
Approaches to Targeting Protein-Protein Interactions
Selective recognition of one protein by another – protein-protein interactions, or PPIs – governs the three key dimensions of cellular life: growth, survival, and differentiation. Modulators of these interactions are critical both for understanding cellular networks governing biological functions and for developing new therapeutics. Despite their fundamental role, PPIs are often considered unattractive targets for drug discovery as illustrated by the fact that less than 0.01% of PPIs composing the interactome have been targeted with an inhibitor [1]. However, recent advances in proteomics, computational chemistry, and ligand design provide new road maps for manipulating these recalcitrant targets.
In this perspective, we focus on structure-guided approaches to develop PPI inhibitors. We begin with a brief discussion of workflows for phenotypic and target-guided screens as well as structure-based design. Within structure-based design, we highlight two complementary approaches rooted in fragment-based design and protein domain mimicry for rational design of PPI inhibitors. These methods rely on the mimicry of interfacial residues that contribute most significantly to binding. Identification of these critical contacts, termed “hot spot” residues, is facilitated by computational assessment of protein-protein complexes.
Phenotypic Screens
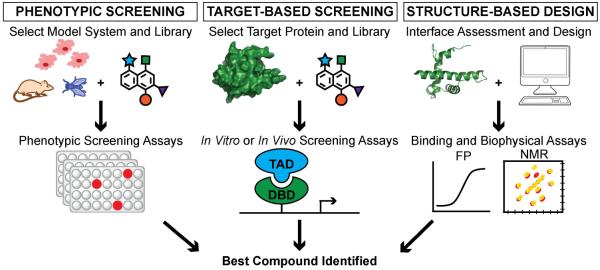
Approaches to inhibitor design can be categorized into (a) phenotypic screening, (b) target-based screening and (c) structure-based design (Figure 1) [2, 3]. In phenotypic screens, also referred to as “forward chemical genetics,” the goal is to find a hit from a collection of compounds that leads to a desired and specific biological result such as inhibition of mitosis, modulation of transcription of a particular gene, or inhibition of specific kinase signaling[4]. Phenotypic screens are often performed with libraries of drug-like molecules, and compounds that emerge from these screens become attractive leads for drug discovery [3]. A key benefit of phenotypic screens is that it provides impetus for finding new targets that drive the desired biological activity[4]. Several compounds that gave the field of chemical genetics its initial appeal have been discovered through phenotypic screens. Monastrol, an inhibitor of mitotic spindle formation, was found in a small molecule library during a search for compounds that induced changes in spindle formation without perturbing tubulin polymerization[5, 6]. Discovery of monastrol also led to the discovery of its target, motor protein Eg5, establishing the elegance and potential of forward chemical genetics[6]. Similarly, the anticancer drug lenalidomide, which has been approved by the USA FDA, was discovered from a phenotypic screen, but the elucidation of its target, an E3 ligase protein cereblon, did not occur until years after its approval in 2012[3, 7]. Not surprisingly, target identification and determination of mechanism of action of advanced lead compounds remain significant bottlenecks[2]. Several high-throughput mass spectrometry strategies have been implemented to mitigate this challenge. They include, but are not limited to, classic affinity pull-down assays, activity-based protein profiling (ABPP), chemical capture compound assays, stable isotope labeling by amino acids in cell culture (SILAC), isotope-coded affinity tag (ICAT), isobaric tags for relative and absolute quantification (iTRAQ), drug affinity responsive target stability (DARTS), and stability of protein from rates of oxidation (SPROX)[4, 8, 9].
Figure 1.
Approaches to inhibitor design can be categorized into phenotypic screening, target-based screening and structure-based design. Left: Phenotypic Screening. A compound library is screened in a model system (i.e. cells, mice, flies) and analyzed for a specific phenotype. Center: Target-Based Screening. A library is screened against a particular protein target of interest in cell free or cell culture assays. Right: Structure-Based Design. A protein of interest is computationally assessed to design a modulator. Binding and biophysical assays are then performed on designed modulators to determine the best compound.
Target-based Screens
In target-based drug screening, also referred to as “reverse chemical genetics,” specific compounds are screened to modulate a particular target or protein of interest[4]. This approach requires a biologically validated target or pathway; however, a high-resolution structure of the target is not needed. Target-based drug discovery has gained prominence with growing understanding of cellular networks and molecular targets from genome sequencing[3, 10]. Several methods, including ELISA-based screens, split luciferase, and yeast two-hybrid assays, are widely used to screen compounds against a desired protein of interest both in vitro and in vivo[11]. These approaches do not require an intimate knowledge of the molecular details of targeted protein interfaces. Nutlins, which are small molecule ligands of Mdm2 and potent inhibitors of the p53/Mdm2 interaction, were discovered from a target-based high-throughput screen[12, 13]. While high-throughput screening has become relatively low cost and efficient, replication of the protein-protein interaction within the assay often remains problematic. For example, only part of the protein target may be able to be expressed and amenable to an assay format, or multi-protein complexes and other co-factors play a more substantial role in vivo as compared to what is replicated in the assay[14]. Another general challenge of screening approaches for PPI targeting is that often the compound libraries are not structurally diverse enough to target large and diffuse interfaces[15]. To address this challenge, several groups are developing strategies for the synthesis of complex natural product-like libraries[16-18].
Structure-based Design
In contrast to the screening techniques, structure-based design relies on the use of structural models to rationally design small molecules or peptidomimics for targeting a PPI. Homology models may be utilized in the absence of high-resolution X-ray crystal or NMR structures[19]; however, the availability of high-resolution structures enables in silico evaluation of the target complex, thereby significantly streamlining identification of PPI modulators[20, 21]. Several structure-based design approaches have been developed including fragment-based design and mimicry of folded protein domains that display the important binding functionality.
Critical steps in the rational design process begin with the selection of the target. The target must be both biologically relevant and the PPI interfaces must suggest that the complex is amenable to disruption by a synthetic modulator. Numerous biochemical and biophysical assays, as well as computational prediction algorithms, have been developed and utilized to identify both binary PPIs and multi-protein complexes[22, 23]. The recent explosion of information emanating from the “omics” fields has produced a wealth of data resulting in over 300 pathway and interaction databases [22, 24]. Gene knockdown strategies such as RNAi or CRISPR-Cas9 screens, offer efficient methods for experimentally defining the biological relevance of an interaction in a pathway[25, 26]. Additionally, synthetic lethality assays have aided the elucidation of genes and proteins linked with disease states[27]. Combined, these strategies have greatly contributed to the understanding of PPIs associated with cancer and other disorders and revealed attractive PPI targets.
AbbVie’s venetroclax, which was approved by the FDA in April 2016, provides a landmark example of structure-based drug design[28]. Generation of lead compounds for its target protein, apoptotic regulator Bcl2, was achieved by a novel NMR-based approach[29, 30]. Although the redesign of analogs and clinical trials culminating in the approval of the drug took over 20 years, the biochemical tools and methods implemented for the discovery of this PPI inhibitor provide a roadmap for future success.
Mode of Modulation
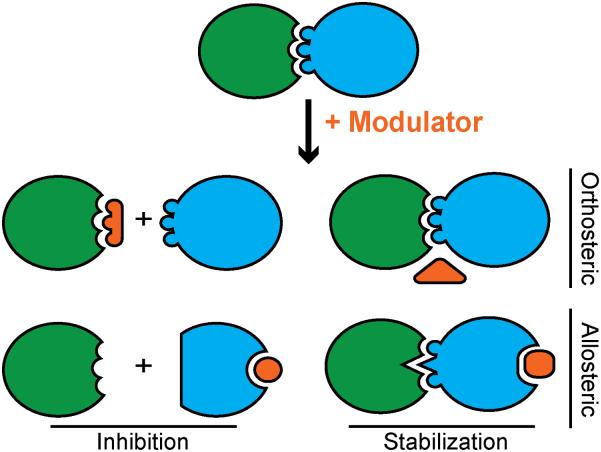
Both orthosteric and allosteric PPI inhibitors have been described (Figure 2)[31, 32]. Modulation of PPIs by either mode of binding can lead to complex inhibition or stabilization. Similarly, both complex inhibition and stabilization can lead to either inhibition or activation of biological function. In regards to the mode of modulator binding, allosteric modulators are attractive because large molecules may not be required to morph the protein-binding surface by altering protein conformation[33, 34]. However, prediction of protein motion and dynamics in response to ligand binding remains non-trivial, thus rational design approaches often seek to develop orthosteric modulators that mimic critical features of the binding interface.
Figure 2.
Modulators for PPIs may function using orthosteric and allosteric mechanisms to lead to PPI inhibition or stabilization.
PPI stabilization represents a promising mode of modulation because binding to a pre-existing complex is more energetically favorable relative to inhibition of complex formation[31, 35, 36]. Roche has described synthetic stabilizers, RO-2443 and RO-5963, which activate p53 signaling and induce apoptosis in breast cancer cells by stabilizing MdmX dimers. MdmX regulates p53 activity, and its complex with p53 has been a long-standing target for inhibitor design[13, 37]. The Roche compounds illustrate that the same desired biological results may be obtained, for MdmX-overexpressing cancers, through an alternative mode of action[38]. This elegant result highlights the considerable potential of PPI stabilizers.
Computational Analysis of Protein Complexes
The central hypothesis guiding rational design of PPI inhibitors is that while the interfaces are large and diffuse, some local regions are more critical for binding interactions than others[39, 40]. These binding regions often feature a small subset of residues that contribute significantly to the free energy of binding[41]. Several computational methods have been developed to quantify the influence each residue has on the overall binding of a protein-protein complex.
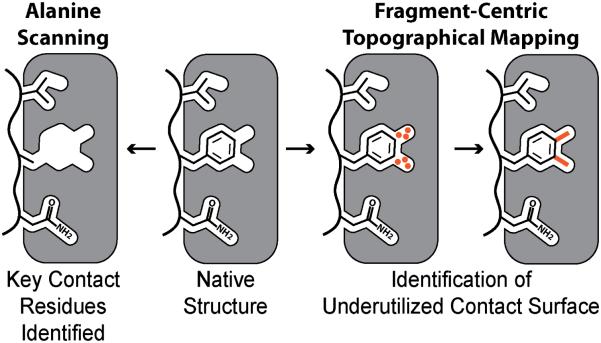
Alanine scanning mutagenesis offers an effective approach for identifying hot spot residues - residues whose substitution with alanine leads to a decrease in binding energy by ΔΔG ≥1 kcal/mol are considered important contributors (Figure 3)[42-44]. Identification of hot spot residues at protein-protein interfaces provides a powerful starting point for rational design[45, 46]. Small molecules or peptidomimetics that reproduce the functionality of these hot spot residues have been shown to be potent inhibitors of PPIs. For example, in the well-studied p53/Mdm2 interaction, three residues (Phe19, Trp23, and Leu26) from the p53 activation domain are known to be strong contributors to binding as shown alanine scanning[47, 48]. Several compounds that mimic Phe19, Trp23 and Leu26 with either small-molecules or peptide-based backbone scaffolds have been shown to successfully inhibit the p53/Mdm2 interaction[13, 37, 49-51]. Additionally, analysis of changes in solvent-accessible surface area (ΔSASA) upon binding also offers a valuable tool for judging the relative importance of interfacial residues[52]. Both alanine scanning and ΔSASA measurements reveal native residues that make important contacts with the target surface using different metrics.
Figure 3.
Computational analysis of PPIs. Starting from the native structure, alanine scanning mutagenesis (left) can be performed on the “ligand” to quantify how much each contact residue contributes to the overall binding of the complex. The example shows a phenylalanine residue mutated to alanine to analyze the contribution of Phe to binding. While, fragment-centric topographical mapping of the “receptor’s” surface (right) reveals underutilized contact surface area. The native residue may not be optimal and a nonnatural amino acid may provide added contacts. Surface mapping allows judicious exploration of nonnatural residues.
Emerging in silico approaches are also exploring a complementary question: which native interfacial residues can be optimized to make increased contacts to the target protein? This question is pertinent because nature has not designed all protein-protein interactions to have the highest possible affinity, thus not all native residues make the best possible contacts. Computational approaches that systematically reveal underutilized contact surfaces provide a powerful tool for rational design (Figure 3)[53-55]. Both natural and non-natural residues have been used to aid in the design of potent inhibitors to optimize native hydrophobic and electrostatic contacts with the protein surface[56]. The inherent structural plasticity of protein-protein interactions provides a major challenge for structure-based efforts that often utilize static models for inhibitor design. Recent efforts with molecular dynamic simulations suggest the exciting possibility of revealing cryptic surface pockets that may be suitable for modulation by allosteric ligands[57-60].
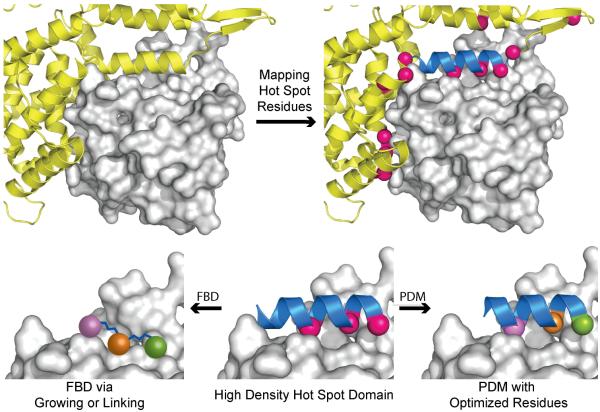
Fundamentally, inhibitor design may be approached from engagement of the target “receptor” surface or mimicry of the “ligand,” i.e., hot spot residues. Following these ideas, the strategies that utilize hot spot information to develop potent inhibitors may be subdivided into fragment-based design to target pockets on the “receptor” or direct mimicry of protein contact residues on the “ligand.” Both strategies capitalize on the identification of key contact residues and pockets found in the native PPI but often differ in their approach to targeting these regions. Fragment-based design searches for potential “anchors” for further elaboration with less of a focus on the connection between these “anchors,” while protein secondary and tertiary structure mimicry efforts consider the folded protein backbones as scaffolds that display critical side chains[52]. Figure 4 provides a schematic illustrating this decision point for a protein complex. After computational analysis of the PPI, an inhibitor can be designed to engage and fill the receptor’s pockets (a fragment-centric approach) or mimic the backbone geometry of the ligand protein chain displaying hot spot residues (a protein domain mimic approach). It should be noted that both fragment-based design and protein domain mimicry can be achieved with protein backbone scaffolds or small molecules[61]. Both techniques are plausible as long as the pockets on the receptor are targeted, or the hot spots on the ligand are mimicked, respectively.
Figure 4.
Structure-guided design of PPI inhibitors. Hot spot contact residues (hot pink spheres) are identified through experimental or computational alanine scanning, mapped on the ligand protein (yellow ribbon), and analyzed to understand how they interact with the receptor protein surface (grey surface). Protein domain mimetics (PDMs) with high hot spot density (blue ribbon) can be chosen as starting points for inhibitor design. Nonnative residues may be utilized to optimize binding interactions. In a fragment-based design (FBD) approach, the consecutive hot spots on the high-density domain may be utilized as initial fragments, which can be optimized via fragment linking or growing.
Fragment-based Design
Successful PPI modulators are generally larger than traditional drugs, typically double or triple the molecular weight range preferred for enzyme inhibitors[62]. Drug-like libraries, developed for traditional drug targets, often lack the characteristics needed to engage a protein’s surface[62, 63]. Thus, screening of drug-like compound libraries against PPI surfaces often leads to nonspecific and low affinity hits. To address these limitations, fragment-based screening techniques have been developed[63, 64]. These techniques work well since modulators of PPIs may be viewed as assemblies of multiple drug-like molecules or fragments stitched together.
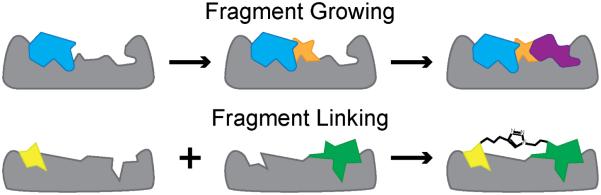
Many screening and validation techniques as applied to PPIs are the same when applied to traditional drug targets but have required modifications to interpret weaker affinities between fragment and target. Due to the reduced contact area and increased solvent exposure of a protein binding interface, the affinities of initial fragment hits for a PPI (KD 0.1-5 mM) are often lower than those for a traditional drug target[64, 65]. To address this problem, pioneering work by Wells et al. developed a covalent ‘tethering’ approach in which fragments containing thiol moieties are utilized to form disulfide bonds with strategically located native or engineered cysteine residues[66-68]. Optimization of a fragment-based modulator can be performed through: (i) linking small fragment moieties after concurrently optimizing each piece for a particular pocket, known as fragment linking, or (ii) progressively growing the fragment chain with successive optimization after each step of chain elongation, known as fragment growing (Figure 5)[69, 70].
Figure 5.
Fragment-based design. Top: Fragment growing. A single fragment is progressively grown to optimize contacts with the target protein. Bottom: Fragment linking. Multiple fragments that bind in nearby sites are individually optimized and subsequently linked together.
Complementary innovative efforts have established the potential of NMR to find fragment binding and elaboration sites[71]. Fragment-based lead discovery and determination of structure-activity relationships by NMR have now become established methods for the development of PPI inhibitors[29, 30, 72-76]. Other methods including X-ray crystallography, differential scanning fluorimetry (DSF), surface plasmon resonance (SPR), isothermal titration calorimetry (ITC), and fluorescence spectroscopy assays have also been adapted to address the challenge of finding lead fragments[64, 65, 77-79].
Protein Domain Mimics
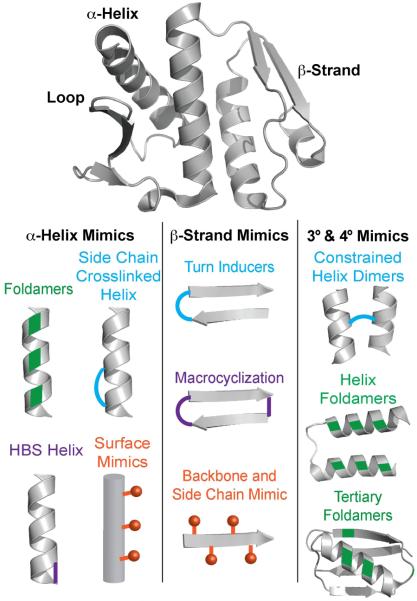
Proteins often utilize secondary and tertiary structures to display hot spot residues[80]. These folded domains are attractive targets for mimicry and inhibitor development since they naturally reproduce side chains in a favorable orientation to engage the target protein’s surface. The synthesis of protein domain mimics (PDMs) has a rich history in bioorganic chemistry with a focus on increasing cellular uptake and proteolytic stability. Recently, both peptidic and nonpeptidic molecules have been shown to be potent leads for the development of therapeutics targeting PPIs[15, 37, 81]. The motivation for the development of PDMs is straightforward: if nature uses these domains to bring proteins together, the same strategy would allow competitive inhibition of complex formation. Below, we discuss the different classes of PDMs, ranging from small molecule secondary structure mimetics to peptide and protein-derived tertiary structure analogs (Figure 6).
Figure 6.
Surface-exposed protein secondary structures often mediate binding of one protein with another (Top). Mimics of these folded domains can lead to potent PPI inhibitors. Several methods for mimicking protein motifs (Bottom). For example, mimicking an α-helix can be achieved by using side chain crosslinks, nonnatural oligomers that adopt helical conformations, hydrogen bond surrogates (HBS) and topographical/surface mimics that reproduce the side chain disposition. β-strands may be mimicked by nonnatural backbones, and turn inducers or macrocycles that hold peptide stands in β-sheet conformations. Emerging methods allow tertiary (3°) and quaternary (4°) structures to be stabilized by crosslinks or nonnatural residues. Key: Green denotes backbone analogs or nonnatural residues (i.e. β-amino acid residues). Blue and purple indicate constraints or cross-linkings. Orange indicates side chain residues placed to reproduce the orientation of the side chains coming from the canonical secondary structures. Protein (PDB: 3QKR), Helix Foldamer (PDB: 3S1K), Tertiary Foldamer (PDB: 2QMT).
We and others have comprehensively surveyed the PDB using computational alanine scanning mutagenesis[43] analysis to define complexes mediated by helices[45, 82], strands[83], loops[84], and helix dimer interfaces[85] to provide a systematic starting point for the exploration of PPIs using PDMs.
Helix Mimics
An analysis of high-resolution structures deposited in the Protein Data Bank (PDB) revealed that roughly 60% of multiprotein complexes feature a helix at the interface [45]. Further analysis revealed that a majority of these helices contain hot spot residues on one helical face, with the rest displaying critical functionality on two or all three faces for recognition. Helix mimicry has become a fertile avenue for discovery of potent PPI inhibitors. Two general strategies for helix mimicry have been described: (a) topographical helix mimics or helix surface mimics and (b) stabilized helices or foldamers.
Topographical helix mimics contain a non-peptidic scaffold to orient protein-like side chains into proper vectors to mimic the display of side chains on α-helices. This class of mimics includes generally low molecular weight compounds that mimic a single helix face[12, 61, 82]. Since the report of the first example of a topographical helix mimetic, which was based on an aromatic scaffold[86], many different scaffolds have been described to afford compounds that are less hydrophobic than the original designs and can target more than one face of a helix[12, 56, 61, 82].
Alternatively, stabilized helices or foldamers are often able to mimic up to 2-3 faces of the helix depending on the stabilization technique[87]. For example, helix side chain staples – involving lactams, thiols, triazole linkages, and hydrocarbons – require one face for stabilization and allow two faces for recognition [12, 61, 82]. Additionally, hydrogen bond surrogate (HBS) helices with stabilization through the peptide backbone, as well as foldamers comprised of judiciously placed α- and β-amino acids, can mimic proteins that require three faces for recognition[12, 61, 82, 88]. These mimics have been shown to modulate PPIs in cell culture and animal models[37, 81, 89, 90]. Recent advances in helix mimicry have been extensively reviewed, and we refer the reader to these excellent reports[12, 61, 91].
β-Strand and β-Sheet Mimics
Despite the fundamental role of strands and sheets at protein-protein interfaces, application of β-strand or β-sheet mimics as modulators of PPIs is limited. Strand designs are challenging because mimics with appropriately placed hydrogen-bonding groups tend to aggregate. An analysis of the PDB for β-strands found at PPI interfaces reveals that β-strands interact with protein partners in multiple ways: as a lone strand or a sheet, side chain recognition, and with or without engagement of backbone hydrogen bonding[83]. A number of scaffolds for each type of these structures have been designed[92-100].
Macrocycles
Relative to α-helix and β-strand mimics, non-regular secondary structure (loop, turn, etc.) mimicry as a means to modulate PPIs is vastly unexplored. Analysis of the PDB identifies non-regular structures appearing at the interface of 50% of heterocomplexes and 41% of homodimers[80]. Loops have become a non-regular secondary structure of particular interest due to the potential to mimic these structures with macrocycles. Macrocycles have also seen a resurgence because they exhibit higher proteolytic and conformational stability and increased cellular uptake compared to linear peptides[101]. The attractiveness of macrocycles has grown with the emergence of in vitro selection[102], as well as on-bead screening strategies[103, 104] for isolation of compounds that bind desired targets. Systematic analysis of the PDB for loops at protein interfaces has provided a compelling impetus for rational design of macrocycles as PPI inhibitors[84].
Tertiary and Quaternary Structure Mimics
The impressive success in the design of protein secondary structure mimics has been paralleled by substantive progress in the development of protein tertiary and quaternary structures where hot spot residues required for recognition lie on more than one secondary structure. Miniature proteins consisting of helical bundles, β-sheet barrels, and loops, along with synthetic antibodies[105-109] are now routinely used to enrich ligands for protein targets, especially for extracellular receptors. Significantly, many of these tertiary and quaternary protein structure scaffolds have been engineered for phage display, allowing rapid selection of potent sequences for desired targets. Synthetic protein tertiary structures with non-natural backbones have also been described[110-112].
Gellman et al. have demonstrated that protease-resistant oligomers consisting of both α- and β-amino acid residues can mimic peptides derived from the three-helix bundle “Z-domain” scaffold and target cell surface receptors with high affinity[110]. Arora and coworkers have described a strategy for constraining short helix dimers for inhibition of protein-protein interactions that are mediated by such motifs[85, 113]. Successes in mimicking tertiary and quaternary motifs through design or recombinant technologies suggest the tractability of targeting PPIs with larger domains.
Concluding Remarks
Inhibition of protein-protein interactions is rapidly becoming an attractive approach for generation of new therapeutics. The progress in the field is aided by the establishment of systematic approaches for target assessment, screening, and structure-based design. While it remains non-trivial to discover new inhibitors for protein complexes, computational tools now provide reliable assessments of hot spots and pockets found in PPIs, which offer reasonable starting points for inhibitor design. The optimism in the field stands in stark contrast to the rejection of PPIs as largely “undruggable” less than a decade ago.
It is now recognized that molecules larger than traditional drug compounds will be required for specific disruption of PPIs. While small molecules remain attractive as therapeutics, especially for oral bioavailablity, peptides and proteins are increasingly considered viable candidates. The successes in modulating proteins with peptides and peptidomimetics has been reflected in the number of FDA-approved peptide therapeutics. An unprecedented number of peptides have been recently approved or are currrently in clinical trials[114]. The success of peptides and proteins in the clinic promises to lead to a larger number of PPI therapeutics.
A critical challenge for structure-based design of ligands for protein surfaces – by small molecules, peptidomimetics, or designer proteins – is that natural PPIs are not always optimized for affinity or specificity. Typical drugs are potent molecules that exhibit high therapeutic index, i.e. selectivity for the target over a non-selective response. PPIs are often dynamic and transient in nature with partner proteins exhibiting weak affinity for each other[1, 115]. The challenge of developing potent selective binders is particularly difficult for strategies that seek to mimic surface residues. Since synthetic inhibitors only mimic a portion of the native “ligand” and bind to a limited surface area on the “receptor”, direct mimicry leads to weaker affinity than observed for the native interaction. This inherent limitation in protein surface mimicry may be overcome by use of nonnatural residues that access hidden pockets not accessible to native residues. Critical advances in computation are needed to create high-resolution surface maps and to define cryptic, transient pockets on protein surfaces thereby enabling design of exquisitely selective synthetic inhibitors.
We predict that in the near future, compounds will be designed for more challenging PPI targets such as intrinsically disordered proteins, which form a unique structure upon complex formation (see Outstanding Questions)[116-118]. Additionally, bi-specific PPI modulators that selectively target multiple proteins remain an unexplored area of research but will likely lead to exciting classes of compounds from both fundamental molecular recognition and drug design perspectives.
Outstanding Questions.
Protein-protein interactions are considered challenging targets for drug design because drug-like small molecules often cannot inhibit these complexes. Would the larger compounds be orally bioavailable?
How can we add stability, uptake and solubility properties to peptide-based protein epitope mimics?
Fragment-based screens and protein domain mimetics seek to develop inhibitors using seemingly different hypothesis. Will we see a convergence of fragment-based screening and protein domain mimics?
Can intrinsically disordered proteins, that do not adopt a stable conformation until in complex with a partner, be targeted with synthetic ligands?
Trends Box.
Natural protein complexes are not always optimized for affinity. Computational approaches to locate underutilized and cryptic pockets are leading to new classes of potent inhibitors.
Traditional modulators of PPIs consist of orthosteric inhibitors; however, we are now seeing a rise in allosteric modulators as well as stabilizers of PPIs
Protein secondary mimics have been well developed over the past decade, and now there is a push for the development of protein tertiary and quaternary mimics.
Advances in proteomics are paving the way to identify direct protein targets of modulators.
ACKNOWLEDGEMENTS
We thank the National Institutes of Health (GM073943) for financial support. A.E.M. is grateful for the Margaret and Herman Sokol Predoctoral Fellowship and the Margaret Strauss Kramer Graduate Fellowship from the NYU Chemistry Department.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Thompson AD, et al. Fine-Tuning Multiprotein Complexes Using Small Molecules. ACS Chem. Biol. 2012;7:1311–1320. doi: 10.1021/cb300255p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swinney DC, Anthony J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011:10. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 3.Moffat JG, Rudolph J, Bailey D. Phenotypic screening in cancer drug discovery – past, present and future. Nat. Rev. Drug Discov. 2014;13:588–402. doi: 10.1038/nrd4366. [DOI] [PubMed] [Google Scholar]
- 4.Schenone M, Dancik V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013;9:232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 6.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing Spindle Assembly Mechanisms with Monastrol, a Small Molecule Inhibitor of the Mitotic Kinesin, Eg5. J. Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Girona A.e.a. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Futamura YM, Osada H. Target identification of small molecules based on chemical biology approaches. Mol. Biosyst. 2013;9:897–914. doi: 10.1039/c2mb25468a. M. [DOI] [PubMed] [Google Scholar]
- 9.Lenz T, Fischer JJ, Dregar M. Probing small molecule–protein interactions: A new perspective for functional proteomics. Journal Of Proteomics. 2011;75:100–115. doi: 10.1016/j.jprot.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 10.Craik DJF, Liras S, Price D. The Future of Peptide-based Drugs. Chem Biol Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. D. P. [DOI] [PubMed] [Google Scholar]
- 11.Hennemann HW, Carl C. Cell-based peptide screening to access the undruggable target space. Eur. J. Med. Chem. 2015;94:489–496. doi: 10.1016/j.ejmech.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 12.Azzarito V, et al. Inhibition of alpha-helix-mediated protein-protein interactions using designed molecules. Nature Chemistry. 2013;5:161–173. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- 13.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 14.Nero TL, et al. Oncogenic protein interfaces: small molecules, big challenges. Nat. Rev. Cancer. 2014;14:248–262. doi: 10.1038/nrc3690. [DOI] [PubMed] [Google Scholar]
- 15.Tsomaia N. Peptide therapeutics: Targeting the undruggable space. Eur. J. Med. Chem. 2015;94:459–470. doi: 10.1016/j.ejmech.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Koes D, Khoury K, Huang Y, Wang W, Bista M, Popowicz GM, Wold W, Holak TA, Domling A, Camacho CJ. Enabling Large-Scale Design, Synthesis and Validation of Small Molecule Protein-Protein Antagonists. PLoS One. 2012;7:e32839. doi: 10.1371/journal.pone.0032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cragg GMN. Natural Products: A Continuing Source of Novel Drug Leads. Biochim Biophys Acta. 2013;1830:3670–3695. doi: 10.1016/j.bbagen.2013.02.008. D. J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan DS. Diversity-oriented synthesis: exploring the intersections between chemistry and biology. Nat. Chem. Biol. 2005;1:74–84. doi: 10.1038/nchembio0705-74. [DOI] [PubMed] [Google Scholar]
- 19.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.London NR, Schueler-Furman O. Peptide docking and structure-based characterization of peptide binding: from knowledge to know-how. Curr. Opin. Struct. Biol. 2013;23:894–902. doi: 10.1016/j.sbi.2013.07.006. B. [DOI] [PubMed] [Google Scholar]
- 21.Tanrikulu Y, Kruger B, Proschak E. The holistic integration of virtual screening in drug discovery. Drug Discov. Today. 2013;18:358–364. doi: 10.1016/j.drudis.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Garcia J, et al. Networks of Protein-Protein Interactions: From Uncertainty to Molecular Details. Molecular Informatics. 2012;31:342–362. doi: 10.1002/minf.201200005. [DOI] [PubMed] [Google Scholar]
- 23.Westermarck J, et al. Identification of protein interactions involved in cellular signaling. Mol Cell Proteomics. 2013;12:1752–1763. doi: 10.1074/mcp.R113.027771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Las Rivas J, Fontanillo C. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput. Biol. 2010;6:e1000807. doi: 10.1371/journal.pcbi.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohr SE, et al. RNAi screening comes of age: improved techniques and complementary approaches. Nat. Rev. Mol. Cell Biol. 2014;15:591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shalem O, et al. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan DA, Giaccia AJ. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat. Rev. Drug Discov. 2011;10:351–364. doi: 10.1038/nrd3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullard A. Pioneering apoptosis-targeted cancer drug poised for FDA approval. Nat. Rev. Drug Discov. 2016;15:147–149. doi: 10.1038/nrd.2016.23. [DOI] [PubMed] [Google Scholar]
- 29.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 30.Shuker SB, et al. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 31.Fischer G, et al. Alternative modulation of protein–protein interactions by small molecules. Curr. Opin. Biotechnol. 2015;35:78–85. doi: 10.1016/j.copbio.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arkin MRT, Wells JA. Small-Molecule Inhibitors of Protein-Protein Interactions: Progressing toward the Reality. Chem. Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostrem JM, et al. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang N, et al. Dissecting allosteric effects of activator–coactivator complexes using a covalent small molecule ligand. Proc. Natl. Acad. Sci. USA. 2014;111:12061–12066. doi: 10.1073/pnas.1406033111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarzycka B, et al. Stabilization of protein–protein interaction complexes through small molecules. Drug Discov. Today. 2016;21:48–57. doi: 10.1016/j.drudis.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Thiel P, et al. Small-Molecule Stabilization of Protein–Protein Interactions: An Underestimated Concept in Drug Discovery? Angew. Chem. Int. Ed. 2012;51:2012–2018. doi: 10.1002/anie.201107616. [DOI] [PubMed] [Google Scholar]
- 37.Chang YS, et al. Stapled α−helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. USA. 2013 doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graves B, et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc. Natl. Acad. Sci. USA. 2012;109:11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 40.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 41.London N, et al. Druggable protein-protein interactions--from hot spots to hot segments. Curr. Opin. Chem. Biol. 2013;17:952–959. doi: 10.1016/j.cbpa.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 42.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 43.Kortemme T, et al. Computational alanine scanning of protein-protein interfaces. Sci. Signal. 2004;2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 44.Massova I, Kollman PA. Computational alanine scanning to probe protein-protein interactions: A novel approach to evaluate binding free energies. J. Am. Chem. Soc. 1999;121:8133–8143. [Google Scholar]
- 45.Jochim AL, Arora PS. Systematic Analysis of Helical Protein Interfaces Reveals Targets for Synthetic Inhibitors. ACS Chem. Biol. 2010;5:919–923. doi: 10.1021/cb1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall DR, et al. Lessons from Hot Spot Analysis for Fragment-Based Drug Discovery. Trends in Pharmacological Sciences. 2015;36:724–736. doi: 10.1016/j.tips.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 48.Picksley SM, et al. Immunochemical analysis of the interaction of p53 with MDM2;--fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- 49.Yu S, et al. Potent and orally active small-molecule inhibitors of the MDM2-p53 interaction. J. Med. Chem. 2009;52:7970–7973. doi: 10.1021/jm901400z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin H, et al. Terphenyl-Based Helical Mimetics That Disrupt the p53/HDM2 Interaction. Angew. Chem. Int. Ed. 2005;44:2704–2707. doi: 10.1002/anie.200462316. [DOI] [PubMed] [Google Scholar]
- 51.Kritzer JA, et al. Helical β-peptide inhibitors of the p53-hDM2 interaction. J. Am. Chem. Soc. 2004;126:9468–9469. doi: 10.1021/ja031625a. [DOI] [PubMed] [Google Scholar]
- 52.Rajamani D, et al. Anchor residues in protein–protein interactions. Proc. Natl. Acad. Sci. U S A. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rooklin D, et al. AlphaSpace: Fragment-Centric Topographical Mapping To Target Protein–Protein Interaction Interfaces. J. Chem. Inf. Model. 2015;55:1585–1599. doi: 10.1021/acs.jcim.5b00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laskowski RA. SURFNET: A program for visualizing molecular surfaces, cavities, and intermolecular interactions. Journal of Molecular Graphics. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 55.Ngan CH, et al. FTMAP: extended protein mapping with user-selected probe molecules. Nucleic Acids Res. 2012;40:W271–W275. doi: 10.1093/nar/gks441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lao BB, et al. Rational Design of Topographical Helix Mimics as Potent Inhibitors of Protein-Protein Interactions. J. Am. Chem. Soc. 2014;136:7877–7888. doi: 10.1021/ja502310r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eyrisch S, Helms V. Transient Pockets on Protein Surfaces Involved in Protein−Protein Interaction. J. Med. Chem. 2007;50:3457–3464. doi: 10.1021/jm070095g. [DOI] [PubMed] [Google Scholar]
- 58.Johnson DK, Karanicolas J. Druggable Protein Interaction Sites Are More Predisposed to Surface Pocket Formation than the Rest of the Protein Surface. PLoS Comput. Biol. 2013;9:e1002951. doi: 10.1371/journal.pcbi.1002951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rakers C, et al. Computational close up on protein–protein interactions: how to unravel the invisible using molecular dynamics simulations? Wiley Interdisciplinary Reviews: Computational Molecular Science. 2015;5:345–359. [Google Scholar]
- 60.Bowman GR, Geissler PL. Equilibrium fluctuations of a single folded protein reveal a multitude of potential cryptic allosteric sites. Proc. Natl. Acad. Sci. USA. 2012;109:11681–11686. doi: 10.1073/pnas.1209309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pelay-Gimeno M, et al. Structure-Based Design of Inhibitors of Protein–Protein Interactions: Mimicking Peptide Binding Epitopes. Angew. Chem. Int. Ed. 2015;54:8896–8927. doi: 10.1002/anie.201412070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sperandio O, et al. Rationalizing the chemical space of protein-protein interaction inhibitors. Drug Discov Today. 2010;15:220–229. doi: 10.1016/j.drudis.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 63.Turnbull AP, et al. Fragment-based drug discovery and protein-protein interactions. Research and Reports in Biochemistry. 2014;4:13–26. [Google Scholar]
- 64.Valkov E, et al. Targeting protein-protein interactions and fragment-based drug discovery. Top Curr Chem. 2012;317:145–179. doi: 10.1007/128_2011_265. [DOI] [PubMed] [Google Scholar]
- 65.Magee TV. Progress in discovery of small-molecule modulators of protein-protein interactions via fragment screening. Bioorg. Med. Chem. Lett. 2015;25:2461–2468. doi: 10.1016/j.bmcl.2015.04.089. [DOI] [PubMed] [Google Scholar]
- 66.Erlanson DA, et al. Site-directed ligand discovery. Proc. Natl. Acad. Sci. USA. 2000;97:9367–9372. doi: 10.1073/pnas.97.17.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Erlanson DA, et al. In situ assembly of enzyme inhibitors using extended tethering. Nat Biotechnol. 2003;21:308–314. doi: 10.1038/nbt786. [DOI] [PubMed] [Google Scholar]
- 68.Lodge JM, et al. FP Tethering: a screening technique to rapidly identify compounds that disrupt protein-protein interactions. Medchemcomm. 2014;5:370–375. doi: 10.1039/C3MD00356F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arkin MR, et al. Binding of small molecules to an adaptive protein-protein interface. Proc. Natl. Acad. Sci. USA. 2003;100:1603–1608. doi: 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erlanson DA. Introduction to fragment-based drug discovery. Top Curr Chem. 2012;317:1–32. doi: 10.1007/128_2011_180. [DOI] [PubMed] [Google Scholar]
- 71.Barile E, Pellecchia M. NMR-Based Approaches for the Identification and Optimization of Inhibitors of Protein–Protein Interactions. Chem. Rev. 2014;114:4749–4763. doi: 10.1021/cr500043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chu S, Gochin M. Identification of fragments targeting an alternative pocket on HIV-1 gp41 by NMR screening and similarity searching. Bioorg. Med. Chem. Lett. 2013;23:5114–5118. doi: 10.1016/j.bmcl.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rega MF, et al. SAR by interligand nuclear overhauser effects (ILOEs) based discovery of acylsulfonamide compounds active against Bcl-x(L) and Mcl-1. J. Med. Chem. 2011;54:6000–6013. doi: 10.1021/jm200826s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun Q, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Takeuchi K, Wagner G. NMR studies of protein interactions. Curr Opin Struct Biol. 2006;16:109–117. doi: 10.1016/j.sbi.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 76.Wu B, et al. HTS by NMR of combinatorial libraries: a fragment-based approach to ligand discovery. Chem. Biol. 2013;20:19–33. doi: 10.1016/j.chembiol.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scott DE, et al. Using a fragment-based approach to target protein-protein interactions. ChemBiochem. 2013;14:332–342. doi: 10.1002/cbic.201200521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Molle I, et al. Dissecting fragment-based lead discovery at the von Hippel-Lindau protein:hypoxia inducible factor 1alpha protein-protein interface. Chem. Biol. 2012;19:1300–1312. doi: 10.1016/j.chembiol.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Winter A, et al. Biophysical and computational fragment-based approaches to targeting protein-protein interactions: applications in structure-guided drug discovery. Q Rev Biophys. 2012;45:383–426. doi: 10.1017/S0033583512000108. [DOI] [PubMed] [Google Scholar]
- 80.Guharoy M, Chakrabarti P. Secondary structure based analysis and classification of biological interfaces: identification of binding motifs in protein-protein interactions. Bioinformatics. 2007;23:1909–1918. doi: 10.1093/bioinformatics/btm274. [DOI] [PubMed] [Google Scholar]
- 81.Johnson LM, et al. A Potent α/β-Peptide Analogue of GLP-1 with Prolonged Action in Vivo. J. Am. Chem. Soc. 2014;136:12848–12851. doi: 10.1021/ja507168t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bullock BN, et al. Assessing Helical Protein Interfaces for Inhibitor Design. J. Am. Chem. Soc. 2011;133:14220–14223. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watkins AM, Arora PS. Anatomy of beta-strands at protein-protein interfaces. ACS Chem. Biol. 2014;9:1747–1754. doi: 10.1021/cb500241y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gavenonis J, et al. Comprehensive analysis of loops at protein-protein interfaces for macrocycle design. Nat. Chem. Biol. 2014;10:716–722. doi: 10.1038/nchembio.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watkins AM, et al. Protein-Protein Interactions Mediated by Helical Tertiary Structure Motifs. J. Am. Chem. Soc. 2015;137:11622–11630. doi: 10.1021/jacs.5b05527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Orner BP, et al. Toward proteomimetics: Terphenyl derivatives as structural and functional mimics of extended regions of an alpha-helix. J. Am. Chem. Soc. 2001;123:5382–5383. doi: 10.1021/ja0025548. [DOI] [PubMed] [Google Scholar]
- 87.Henchey LK, et al. Contemporary strategies for the stabilization of peptides in the alpha-helical conformation. Curr. Opin. Chem. Biol. 2008;12:692–697. doi: 10.1016/j.cbpa.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sawada T, Gellman SH. Structural mimicry of the alpha-helix in aqueous solution with an isoatomic alpha/beta/gamma-peptide backbone. J. Am. Chem. Soc. 2011;133:7336–7339. doi: 10.1021/ja202175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kushal S, et al. Protein domain mimetics as in vivo modulators of hypoxia-inducible factor signaling. Proc. Natl. Acad. Sci. USA. 2013;110:15602–15607. doi: 10.1073/pnas.1312473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Walensky LD, Bird GH. Hydrocarbon-Stapled Peptides: Principles, Practice, and Progress. J. Med. Chem. 2014;57:6275–6288. doi: 10.1021/jm4011675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Milroy L-G, et al. Modulators of Protein–Protein Interactions. Chem. Rev. 2014;114:4695–4748. doi: 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- 92.Angelo NG, Arora PS. Nonpeptidic foldamers from amino acids: synthesis and characterization of 1,3-substituted triazole oligomers. J. Am. Chem. Soc. 2005;127:17134–17135. doi: 10.1021/ja056406z. [DOI] [PubMed] [Google Scholar]
- 93.Bag SS, et al. Triazolo-beta-aza-epsilon-amino acid and its aromatic analogue as novel scaffolds for beta-turn peptidomimetics. Chem Commun (Camb) 2015;51:5242–5245. doi: 10.1039/c4cc08414d. [DOI] [PubMed] [Google Scholar]
- 94.Freire F, Gellman SH. Macrocyclic design strategies for small, stable parallel beta-sheet scaffolds. J. Am. Chem. Soc. 2009;131:7970–7972. doi: 10.1021/ja902210f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fuller AA, et al. Evaluating beta-turn mimics as beta-sheet folding nucleators. Proc. Natl. Acad. Sci. USA. 2009;106:11067–11072. doi: 10.1073/pnas.0813012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kang CW, et al. beta-Strand mimics based on tetrahydropyridazinedione (tpd) peptide stitching. Chem Commun (Camb) 2015;51:16259–16262. doi: 10.1039/c5cc07189e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kang CW, et al. Substituted imidazo[1,2-a]pyridines as beta-strand peptidomimetics. Org. Lett. 2012;14:6162–6165. doi: 10.1021/ol302850n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lingard H, et al. Diphenylacetylene-linked peptide strands induce bidirectional beta-sheet formation. Angew Chem Int Ed Engl. 2014;53:3650–3653. doi: 10.1002/anie.201309353. [DOI] [PubMed] [Google Scholar]
- 99.Loughlin WA, et al. Update 1 of: Beta-strand mimetics. Chemical reviews. 2010;110:PR32–69. doi: 10.1021/cr900395y. [DOI] [PubMed] [Google Scholar]
- 100.Robinson JA. Beta-hairpin peptidomimetics: design, structures and biological activities. Acc Chem Res. 2008;41:1278–1288. doi: 10.1021/ar700259k. [DOI] [PubMed] [Google Scholar]
- 101.Villar EA, et al. How proteins bind macrocycles. Nat. Chem. Biol. 2014;10:723–731. doi: 10.1038/nchembio.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Passioura T, et al. Selection-Based Discovery of Druglike Macrocyclic Peptides. Annu. Rev. Biochem. 2014;83:727–752. doi: 10.1146/annurev-biochem-060713-035456. [DOI] [PubMed] [Google Scholar]
- 103.White CJ, Yudin AK. Contemporary strategies for peptide macrocyclization. Nat. Chem. 2011;3:509–524. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- 104.Lian W, et al. Screening Bicyclic Peptide Libraries for Protein–Protein Interaction Inhibitors: Discovery of a Tumor Necrosis Factor-α Antagonist. J. Am. Chem. Soc. 2013;135:11990–11995. doi: 10.1021/ja405106u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Adams JJ, Sidhu SS. Synthetic antibody technologies. Curr. Opin. Struct. Biol. 2014;24:1–9. doi: 10.1016/j.sbi.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 106.Gilbreth RN, Koide S. Structural insights for engineering binding proteins based on non-antibody scaffolds. Curr. Opin. Struct. Biol. 2012;22:413–420. doi: 10.1016/j.sbi.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lewis RJ, Garcia ML. Therapeutic potential of venom peptides. Nat. Rev. Drug Discov. 2003;2:790–802. doi: 10.1038/nrd1197. [DOI] [PubMed] [Google Scholar]
- 108.Kariolis MS, et al. Beyond antibodies: using biological principles to guide the development of next-generation protein therapeutics. Curr. Opin. Biotechnol. 2013;24:1072–1077. doi: 10.1016/j.copbio.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 109.Hosse RJ, et al. A new generation of protein display scaffolds for molecular recognition. Protein Sci. 2006;15:14–27. doi: 10.1110/ps.051817606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Checco JW, et al. Targeting diverse protein–protein interaction interfaces with α/β-peptides derived from the Z-domain scaffold. Proc. Natl. Acad. Sci. USA. 2015;112:4552–4557. doi: 10.1073/pnas.1420380112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Daniels DS, et al. High-resolution structure of a beta-peptide bundle. J. Am. Chem. Soc. 2007;129:1532–1533. doi: 10.1021/ja068678n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tavenor NA, et al. Comparison of design strategies for alpha-helix backbone modification in a protein tertiary fold. Chem Commun (Camb) 2016 doi: 10.1039/c6cc00273k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wuo MG, et al. An Effective Strategy for Stabilizing Minimal Coiled Coil Mimetics. J. Am. Chem. Soc. 2015;137:11618–11621. doi: 10.1021/jacs.5b05525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kaspar AA, Reichert JM. Future directions for peptide therapeutics development. Drug Discov Today. 2013;18:807–817. doi: 10.1016/j.drudis.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 115.Mapp AK, et al. Targeting transcription is no longer a quixotic quest. Nat. Chem. Biol. 2015;11:891–894. doi: 10.1038/nchembio.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dunker AK, et al. Intrinsic Disorder and Protein Function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 117.Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr. Opin. Chem. Biol. 2010;14:481–488. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]