Abstract
Studying the molecular consequences of rare genetic variants has the potential to identify novel and hitherto uncharacterized pathways causally contributing to phenotypic variation. Here, we characterize the functional consequences of a rare coding variant of TAO3, previously reported to contribute significantly to sporulation efficiency variation in Saccharomyces cerevisiae. During mitosis, the common TAO3 allele interacts with CBK1—a conserved NDR kinase. Both TAO3 and CBK1 are components of the RAM signaling network that regulates cell separation and polarization during mitosis. We demonstrate that the role of the rare allele TAO3(4477C) in meiosis is distinct from its role in mitosis by being independent of ACE2—a RAM network target gene. By quantitatively measuring cell morphological dynamics, and expressing the TAO3(4477C) allele conditionally during sporulation, we show that TAO3 has an early role in meiosis. This early role of TAO3 coincides with entry of cells into meiotic division. Time-resolved transcriptome analyses during early sporulation identified regulators of carbon and lipid metabolic pathways as candidate mediators. We show experimentally that, during sporulation, the TAO3(4477C) allele interacts genetically with ERT1 and PIP2, regulators of the tricarboxylic acid cycle and gluconeogenesis metabolic pathways, respectively. We thus uncover a meiotic functional role for TAO3, and identify ERT1 and PIP2 as novel regulators of sporulation efficiency. Our results demonstrate that studying the causal effects of genetic variation on the underlying molecular network has the potential to provide a more extensive understanding of the pathways driving a complex trait.
Keywords: rare variant, TAO3, meiosis, transcriptome profiling, allelic variant
The ‘common disease, common variant’ rationale of genome-wide association studies (GWAS) is being challenged owing to the limited fraction of disease heritability explained by mapped common variants (Manolio et al. 2009; Zuk et al. 2014). Not considering the potential effects of rare variants has been suggested as one of the potential contributors to this ‘missing’ heritability (Saint Pierre and Génin 2014). This view has been substantiated by identification of rare variants carrying a considerable risk for autism, schizophrenia, and epilepsy (Stankiewicz and Lupski 2010). Thus, characterizing the functional role of rare variants associated with complex diseases has the potential to reveal new biology, and to provide opportunities for treatment (Cirulli and Goldstein 2010; Zuk et al. 2014). Although multiple variants for various diseases have been mapped, they have not been able to provide targets for treatment. This is because many variants have been mapped in noncoding regions of the genome, and we do not understand their functional role in disease development. Moreover, detailed characterization is required even for causal coding variants to fully understand their role in phenotypic variation. This necessitates the need to identify the mediating molecular pathways connecting a variant to the phenotype, which has the potential to greatly expand the set of possible targets for molecular intervention (Gagneur et al. 2013).
Yeast sporulation efficiency is a complex trait, and many polymorphisms contributing to this trait variation have been mapped in yeast strains from diverse ecological niches. These include sporulation genes such as IME1, an initiator of meiosis (Gerke et al. 2009), and RIM15, a glucose-sensing regulator of meiosis (Lorenz and Cohen 2014). In addition to several sporulation genes, coding polymorphisms were also identified in two genes, MKT1, a putative RNA-binding protein, and TAO3, a putative scaffolding protein (Deutschbauer and Davis 2005), both of which were uncharacterized for their role in meiosis. The high sporulating SK1 strain contains the causative nonsynonymous polymorphisms MKT1 (89G) and TAO3 (4477C), while the low sporulating S288c strain contains MKT1(89A) and TAO3(4477G) (Deutschbauer and Davis 2005). In our previous work, we showed that the MKT1(89G) variant increased sporulation efficiency by interacting with regulators of mitochondrial retrograde signaling and nitrogen starvation during sporulation (Gupta et al. 2015). TAO3 encodes a highly conserved scaffolding protein that is a component of the RAM (Regulation of Ace2p activity and cellular Morphogenesis) signaling network. In addition, Tao3 activates another RAM network protein, Cbk1—a NDR protein kinase (Du and Novick 2002; Hergovich et al. 2006). The RAM network, which consists of Cbk1, Hym1, Kic1, Mob2, Sog2, and Tao3 proteins, is involved in an Ace2-dependent cell separation and cellular progression during mitotic division (Nelson et al. 2003). Ace2, a transcription factor, peaks early in mitosis and is involved in G1/S transition (Spellman et al. 1998). The RAM network regulates cellular progression in a Ace2-independent manner as well (Bogomolnaya et al. 2006). While components of the RAM network interact with TAO3 during mitosis, none of these interactions provide clues as to its role in the developmental processes of meiosis and sporulation.
Here, we characterized the functional role of TAO3(4477C) in sporulation efficiency variation by elucidating the molecular pathways linking this mitotic gene to meiosis. We compared phenotypes of a pair of S288c-background strains differing only in the causal TAO3 polymorphism. By studying the genome-wide transcriptional dynamics of these strains during sporulation, we predicted TAO3(4477C)-associated candidate mediator genes. A genetic interaction assay between these candidate genes and TAO3 alleles identified regulators of tricarboxylic acid cycle and gluconeogenic enzymes as causal and novel regulators of sporulation efficiency.
Materials and Methods
Yeast strains and media
The yeast strains were grown in standard conditions at 30° in YPD (1% yeast extract, 2% bacto peptone, 2% dextrose). Allele replacement strain YAD331 (Deutschbauer and Davis 2005) was a S288c-background diploid strain containing the homozygous causative sporulation polymorphism TAO3(4477C). Whole-genome resequencing of YAD331 with S288c strain as the reference strain identified two additional polymorphisms (Supplemental Material, Figure S6 and Table S7). Three consecutive backcrosses were performed between the haploid derivative of YAD331 and the haploid reference strain (S288c) to remove these secondary polymorphisms. After the backcrosses, the sole genetic difference between the reference S288c strain and the backcrossed allele replacement strain was at the TAO3(G4477C) position, which was confirmed by performing PCR-based sequencing 650 bp up and downstream around the two secondary polymorphisms and the TAO3 polymorphic nucleotide. This backcrossed strain was diplodized to make it homozygous at the TAO3(4477C) position, and was called “T strain” in this study; the diploid parental strain S288c was called “S strain”. All gene deletions in the study were made in haploids of the T and S strains, except for those made in strain SK1 (Table S8). Deletions were performed and verified as described previously (Goldstein and McCusker 1999; Gietz and Woods 2002). The haploid strains were diplodized using pHS2 plasmid (containing a functional HO) and mating types were confirmed by performing MAT PCR (Huxley et al. 1990). All experiments in this study were performed using the diplodized parent strains and their diploid derivatives. To replace the endogenous TAO3 promoter (–150 to –1 bp upstream of the start site) in the T strain with a tetracycline-responsive promoter, a tetO7-based promoter substitution cassette containing kanMX4 was amplified from the plasmid pCM225 (Bellí et al. 1998b). The diploid T strain with this tetO7-based cassette is termed PTet-TAO3(4477C) strain. The primers for sequencing, deletions, and their confirmations are listed in Table S9.
Phenotyping
Sporulation efficiency estimation at 48 hr, progression through meiotic landmark events meiosis I (MI) and meiosis II (MII), and its quantitation was done as described previously (Gupta et al. 2015). For quantitation of meiotic landmarks in the T strain, parametric curves assuming delayed and 1st order kinetics were fitted to DAPI-stained meiotic progression time-course data, and fitting uncertainties were estimated by bootstrapping (File S1). Cell cycle progression data for S288c and SK1 strains was taken from Gupta et al. (2015) (Figure 1, D and E). Conditional expression of TAO3(4477C) was performed by constructing strain PTet-TAO3 (details in File S1), which was responsive to the tetracycline analog doxycycline (Bellí et al. 1998a, 1998b). Doxycycline (2 µg/ml) was added to growth and sporulation media to decrease expression of the TAO3 gene. For each strain, a minimum of three biological replicates was used, and the experiment was carried out a minimum of two times; ∼300 cells were counted per replicate. Fold difference was calculated as the ratio of mean sporulation efficiencies of the two strains A and B when the sporulation efficiency of A is greater than that of B. Growth curve analysis was performed for individual strains grown in YPD in 96-well plates. Cells were grown overnight in YPD to saturation, reinoculated in YPD in transparent 96-well plates with a starting OD600 of 0.01, and grown with shaking at 30° for 24 hr in a Tecan Infinite M200 microplate reader. Doubling times were calculated from OD measurements of liquid cultures at a wavelength of 600 nm in the Tecan reader. For each strain, four technical replicates for each of the three biological replicates were used. Raw sporulation efficiency values are given in Table S10.
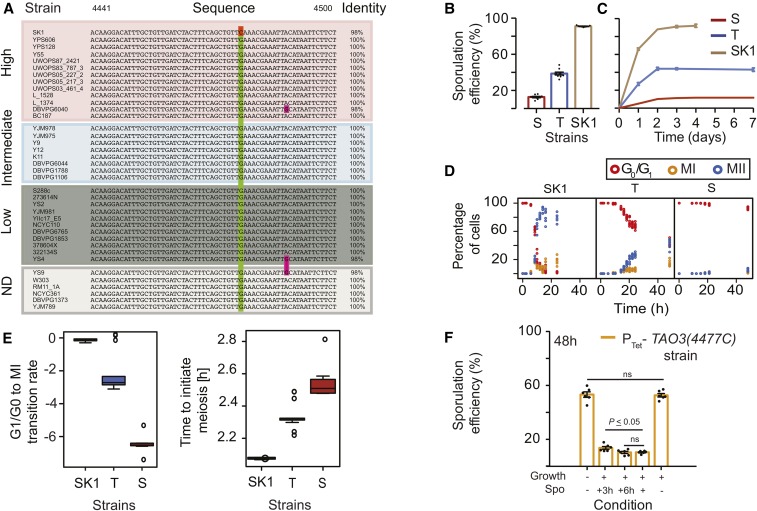
Figure 1.
Role of TAO3 in sporulation efficiency. (A) Comparison of genomic sequences of TAO3 (4441–4500) across the SGRP collection (Liti et al. 2009). The 4477th position of TAO3 consists of the sporulation causative variant where identical nucleotides are indicated by the same color. Identity indicates the percentage match between the nucleotides in the shown region of the gene. The strains are ordered according to their mean sporulation efficiency (Tomar et al. 2013): high (60–100%), intermediate (10–60%), low (0–10%) and ND (not determined). (B) Bar plots represents the mean sporulation efficiency after 48 hr of the SK1, T and S strains. The sporulation efficiency data are indicated as circles. (C) Line graphs represent the mean sporulation efficiency of the S, T, and SK1 strains measured until saturation, i.e., until sporulation efficiency did not vary for three consecutive days. (D) Percentage of one-, two- and four-nuclei states of the T strain (y-axis) vs. time in sporulation medium (x-axis). One-nucleus stage is indicated as red circles (G0/G1 phase), two-nuclei state as yellow circles (completion of Meiosis I, MI phase), and blue circles is four-nuclei stage (completion of Meiosis II, MII phase). (E) Bootstrap distribution of the time to initiate meiosis and the rate of transition from G1/G0 into MI, estimated from time courses in (D). See Materials and Methods for details. (F) Conditional expression of TAO3(4477C) during sporulation in PTet-TAO3(4477C) strain. The y-axis is the mean sporulation efficiency in 48 hr. No doxycycline in growth (YPD) or spo (YPA + sporulation) medium is depicted as “–” condition on the x-axis, and addition of doxycycline is depicted as “+” under those conditions. The “+3h” condition in Spo indicates that doxycycline was present throughout in the growth medium, and in the sporulation medium until 3 hr, after which cells were sporulated in the absence of doxycycline. The “+6h” condition indicates that doxycycline was present throughout in both the growth and sporulation medium until 6 hr, after which cells were sporulated in the absence of doxycycline. P values were calculated by an unpaired t-test. Error bars are SEM.
Statistical test for calculating sporulation efficiency
To compare sporulation efficiency, two statistical tests were used: the pair test and the interaction test. The pair test tests the null hypothesis that the two given strains (S and T) have the same sporulation efficiency.
The number, , of sporulated cells (four-nuclei count) among the total number of cells, , of strain i in replicate experiment k was modeled with a quasi-binomial generalized linear model using the logit link function, and subject to a common log-odds ratio,, between replicates, i.e.,:
for all k, where .
The pair test tests the null hypothesis of equality of log-odds ratios for two strains, i and j, i.e., .
In the case of the S and T strains, the interaction test tests the null hypothesis that the effect of mutation A is independent of the effect of mutation B, taking strain T as the reference background. This test thus compares four strains: mutation A only, mutation B only, both A and B, and neither A nor B (T strain). Here, the S strain was considered as a T strain mutated for TAO3(4477). For every interaction test, we considered the dataset of the four strains of interest, and fitted a quasi-binomial generalized linear model using the logit link function and subject to:
for all k, where Ai and Bi are indicator variables of the mutations A and B in strain i, respectively. The interaction test tested the null hypothesis that the odds ratio of sporulation in the double mutant equals the product of the odds ratios of each mutation, i.e., .
Both the pair test and the interaction test were implemented in the statistical language R with the function glm() assuming a constant variance function fitted by maximizing the quasi-likelihood, and using the t-test on the tested parameters (Gupta et al. 2015).
Whole genome gene-expression profiling
Sporulating yeast cell collection at 0 hr, 30 min, 45 min, 1 hr and 10 min, 1 hr and 40 min, 2 hr and 30 min, 3 hr and 50 m, 5 hr and 40 min, and 8 h and 30 min (logarithmic time-series), RNA isolation, and cDNA preparation were performed as previously described (Xu et al. 2009). Samples were hybridized to S. cerevisiae yeast tiling array (Affymetrix, Cat# 520055). Arrays at each time point for both strains were normalized to each other using the vsn normalization method (Huber et al. 2002). For qPCR, aliquots of cDNA were used in real-time PCR analyses with reagents from Kapa SYBR fast Universal qPCR master mix (Kapa Biosystems) in the Eppendorf Real-time PCR system according to manufacturer’s protocol. For each strain, four technical replicates for each of the three biological replicates were used. The primers used are given in Table S9.
Whole genome gene-expression analysis
Within each strain, the log2 expression values obtained were smoothed using locfit at optimized bandwidth parameter h = 1.2 (Figure S7), and base transformed for each transcript by subtracting the expression value at each time point from the baseline value at time point t = 0 hr (t0, Table S11). This log2 fold change value with respect to t0 is described as “expression” throughout the manuscript. To identify genes showing temporal differential expression between the T and S strains (Table S1), the method implemented in EDGE software was used to calculate statistically significant changes in expression between the T and S strains over time (Storey et al. 2005). The differentially expressed genes were clustered according to their temporal expression patterns using the time abstraction clustering algorithm implemented in the TimeClust software (Magni et al. 2008, see File S1). Four major clusters were identified in each strain: Cluster I (early trend), Cluster II (increasing trend), Cluster III (late trend), Cluster IV (repressing trend) (Table S2). The transcription factors regulating a cluster of genes were extracted using the YEASTRACT database (Teixeira et al. 2013). Only those transcription factors whose target genes were significantly enriched in the corresponding cluster were considered as candidate genes (P ≤ 0.05, odds ratio ≥ 1.5). The YEASTRACT database was also used in this study to obtain the regulation matrix of yeast for identifying target genes of regulators such as UME6. Target genes for ACE2 were obtained from Nelson et al. (2003). Significantly enriched gene ontology (GO) terms by biological process (Bonferroni corrected P < 0.05, Table 1) were obtained from SGD Yeastmine (Balakrishnan et al. 2012).
Table 1. Functional GO categories of clusters in the T and S strains.
| Cluster | Functional GO Category | Genes |
|---|---|---|
| Early in T strain (Cluster I) | Carbohydrate metabolic process | DOG1, YPI1 |
| Ion transport | AVT4, DAL5 | |
| Mitochondrion organization | PPE1, UPS3 | |
| Cellular respiration | COX5B | |
| Early in T strain (Cluster I) repressed in S strain (Cluster IV) | Carbohydrate metabolic process | ALG6, DEP1, DOG1, TPS3, YPI1 |
| Mitochondrial organization | ATG33, COX20, PPE1, UPS3 |
Comparison of functional GO categories of differentially expressed genes in the T strain clusters with the S strain. See Table S2 for the full list of genes in each cluster.
Data availability
The array data for the T strain has been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) with accession number E-MTAB-3889. The entire genome sequence data for the T strain has been deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) with the accession number PRJEB8698. The array data and the whole genome sequence data for the S strain were downloaded from Gupta et al. (2015). TAO3 gene sequence data for Saccharomyces Genome Resequencing Project (SGRP) strains (Liti et al. 2009) was downloaded from (http://www.moseslab.csb.utoronto.ca/sgrp/). An additional 24 TAO3 sequences were downloaded from the Saccharomyces Genome Database (SGD, http://www.yeastgenome.org/cgi-bin/FUNGI/alignment.pl?locus=YIL129C, date accessed: March 1, 2016). Detailed methods are described in File S1. Table S1 lists all differentially expressed genes between the T and S strains with their P and Q values calculated using EDGE. Table S2 lists genes in each cluster using TimeClust. Table S3 lists transcription factors regulating unique early (Cluster I) genes of the T strain. Table S4 lists transcription factors regulating unique increasing (Cluster II) genes of the T strain. Table S5 lists differentially expressed target genes of regulators of candidate genes mediating the affect of TAO3. Table S6 lists transcription factors regulating unique repressing (Cluster IV) genes of the S strain. Whole genome resequencing results for the TAO3 allele replacement strain are described in Table S7. All the strains used in the study are listed in Table S8, which are available upon request. All the primers used in the study are listed in Table S9. The raw sporulation efficiency values of the strains are given in Table S10. Table S11 contains smoothed expression data, base transformed with respect to t0 for the T and S strains. Figure S1 shows mathematical modeling to identify stages of meiosis affected by TAO3 causal allele. Figure S2 shows growth phenotype and TAO3 expression in PTet-TAO3(4477C) strain. Figure S3 shows comparison of global gene expression between the T and S strains at t = 0h. Figure S4 shows comparison of genes showing early (Cluster I) and increasing trend (Cluster II) between the T and S strains. Figure S5 shows genes having early expression in the T strain show expression at later time points or repressed in the S strain. Figure S6 shows whole genome resequencing of TAO3 allele replacement strain (YAD331, (Deutschbauer and Davis 2005) in comparison to the S288c reference strain. Figure S7 shows smoothing of normalized temporal data using locfit.
Results
Role of causative allele of TAO3 in sporulation efficiency variation
Analysis of TAO3 nucleotide sequence of 38 S. cerevisiae strains in the SGRP database (Liti et al. 2009), and 24 strains in SGD, showed that the TAO3(4477C) allele of strain SK1 was a rare variant (minor allele frequency = 1.6%, Figure 1A). Deutschbauer and Davis (2005) mapped TAO3(4477C) as one of the causal alleles contributing to high sporulation efficiency while studying the genetic basis of phenotypic variation between S288c (low sporulating) and SK1 (high sporulating) strains. By introducing TAO3(4477G) in the S288c background, they constructed the allele replacement strain YAD331, which differed from the parental S288c strain [TAO3(4477G)] only for this variant. They also showed that YAD331 showed significantly higher sporulation efficiency than S288c in 48 hr. For this study, we confirmed by sequencing the presence of the TAO3(4477G) allele in the YAD331 strain, but also identified and removed several background mutations present in this strain (Materials and Methods). This cleaned version of the YAD331 allele replacement strain was called “T strain”. Sporulation analysis of the T strain reconfirmed that it sporulated threefold more efficiently at 48 hr in comparison to the S288c strain (“S strain”, P = 1.8 × 10−10, pair test in Materials and Methods, Figure 1B). Moreover, this fold-difference between the two strains remained constant even after a week in sporulation medium (Figure 1C). Studying the progression of meiotic phases showed that the T strain initiated meiosis within 12 hr (Figure 1, D and E). Quantitative comparison of the ‘time to initiate meiosis’, and the ‘rate of transition from G1/G0 into Meiosis I stage’, showed significant differences between the T and S strains (Figure 1, D and E, and Figure S1). This suggested that TAO3(4477C) affected entry of the T strain cells into initiating meiosis within 12 hr during sporulation. To further resolve when, during these 12 hr in the cells entering meiosis, TAO3(4477C) plays a functional role, this allele was placed under a tetracycline-responsive promoter [PTet-TAO3(4477C) strain, see Materials and Methods]. In the absence of the tetracycline analog doxycycline, higher expression of TAO3(4477C) was observed in strain PTet-TAO3(4477C) compared to its expression in the S strain (Materials and Methods, Figure S2). Addition of doxycycline significantly reduced TAO3 expression, making it equivalent to the S strain (Materials and Methods, Figure S2). Concomitantly, sporulation efficiency of the PTet-TAO3(4477C) strain was high in the absence of doxycycline, being equivalent to the S strain in the presence of doxycycline (Figure 1F). This suggested that high TAO3(4477C) expression was required for the high sporulation efficiency phenotype. We next reduced TAO3(4477C) expression only for 3 hr and 6 hr in sporulation medium by sporulating the PTet-TAO3(4477C) strain in the presence of doxycycline for these specific time-periods. Sporulation efficiency of the PTet-TAO3(4477C) strain was equivalent to the S strain whether TAO3 expression was reduced for 48 hr or only for the first 6 hr during sporulation (Figure 1F). Reduction of TAO3 expression for the first 3 hr only did reduce the sporulation efficiency of PTet-TAO3(4477C) strain, but it was not equivalent to S strain (P = 0.02, Figure 1F). This showed that the TAO3(4477C) allele played a functional role in sporulation within the first 6 hr.
Role of TAO3 in meiosis is distinct from its role during mitosis
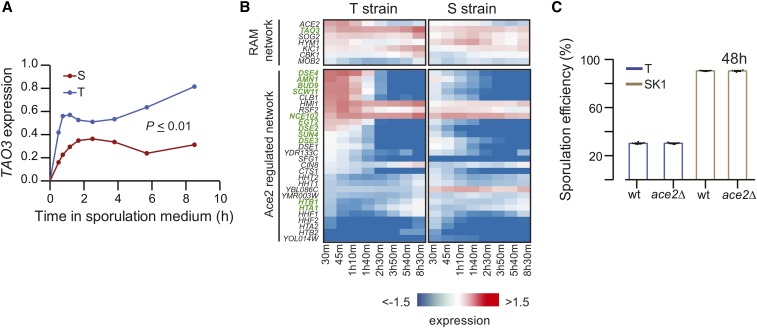
Varying the gene expression of TAO3(4477C) affected the sporulation efficiency phenotype. Hence, to identify the molecular pathways affected by this causative allele, we studied the global gene expression dynamics during sporulation in the allele replacement strains. Time-resolved transcriptomes of the T and S strains were compared from 0 hr to 8 hr, 30 min in sporulation medium (Materials and Methods). At the initial time point (t = 0 hr), only 190 out of 6960 transcripts (∼3%) showed differential expression, with an enrichment for a single GO term, iron ion homeostasis (P = 0.04, post Holm-Bonferroni corrected, Figure S3). In contrast 1122 transcripts (including noncoding stable unannotated transcripts, Table S1) showed statistically significant differences in gene expression dynamics as a function of time between the two strains (false discovery rate cut-off 10%, when controlling for expression at t = 0 hr) . While TAO3 was among the transcripts showing differential expression dynamics during sporulation (P = 0.004), none of its mitotic interactors were differentially expressed (Figure 2, A and B). Instead, we identified 11 ACE2-regulated genes (shown in green in Figure 2B) showing differential expression, and so we studied the effect of ace2∆ in the T strain and high sporulating SK1 strain. ACE2 is known to regulate the budding phenotype (Voth et al. 2005), and we recapitulated the clumping phenotype of ace2∆ in both the T and SK1 strains. However ace2∆ did not affect sporulation efficiency of either the T or the SK1 strain (Figure 2C). An Ace2-independent effect of the RAM network on cellular polarization has been observed previously (Nelson et al. 2003); therefore, it is possible that this network could still be involved in meiosis.
Figure 2.
The role of TAO3 in meiosis is distinct from its role during mitosis. (A) Expression profile (log2 fold change t0) of TAO3 is given in the y-axis for the T (purple) and S strains (red), and the x-axis denotes the time in sporulation medium (data in Table S1 and Table S11). (B) Heatmap showing gene expression of RAM network genes and Ace2-regulated genes in the T and S strains. Gene names in green show differential expression (data in Table S1 and Table S11). (C) Bar plots represent the mean sporulation efficiency after 48 hr of the SK1 and T wild type (wt) and ace2∆ deletion strains. Pair and interaction tests (Materials and Methods) were performed to test significance.
To determine whether the mitotic interactors of TAO3 could be distinct from its meiotic interactors, we again used PTet-TAO3(4477C) strain and reduced TAO3 expression only during the mitotic growth phase, i.e., in glucose rich (YPD) medium. We observed no growth difference between PTet-TAO3(4477C) strain with or without doxycycline and the T strain (Figure S2). Moreover, reduction of TAO3 expression during growth had no effect on the high sporulation efficiency of PTet-TAO3(4477C) strain (Figure 1F). These results suggested that TAO3(4477C) allele could have distinct meiosis-associated interactors that could explain its functional role during sporulation.
Temporal gene expression profiling predicts TAO3(4477C)-specific interactors during sporulation
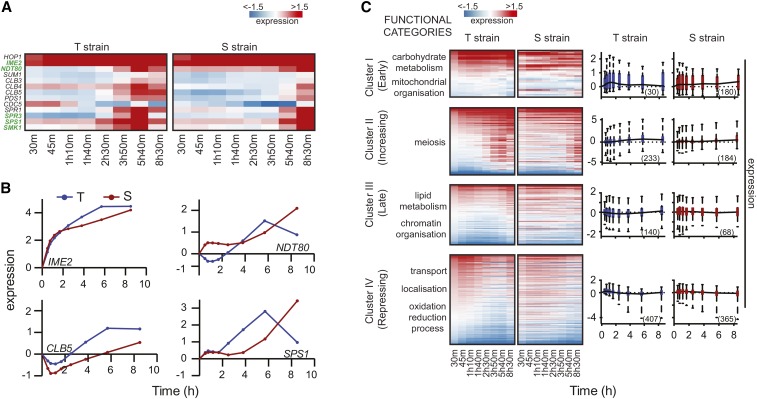
Since we observed the functional role of TAO3(4477C) within the first 6 hr of sporulation (Figure 1F), we investigated the differentially expressed genes showing an early and increasing trend in their expression profiles uniquely in the T strain. These genes were identified by comparing the clustered differentially expressed genes separately for T and S strains (Materials and Methods). Various sporulation genes, including crucial regulators of meiosis, namely IME1, IME2, DMC1, and NDT80, were enriched (P = 5.5 × 10−12) in a cluster showing increasing expression trend (Cluster II) during sporulation in the T strain (Figure 3B, Materials and Methods); ∼50% of Cluster II genes of the T strain showed a similar increasing trend in the S strain, including IME1, IME2, DMC1, ECM11, and NDT80 (Figure S4 and Table S2). Interestingly, very few early-expressing genes (Cluster I) of the T strain overlapped with the S strain (7%, Figure S4). These genes belonged to biological processes that regulated entry into sporulation, such as carbohydrate metabolic process, ion transport, mitochondrial organization, and cellular respiration (Table 1). Furthermore genes involved in biological processes like carbohydrate metabolism and mitochondrial organization showed repression in the S strain (Table 1 and Figure S5). Therefore, to study the early effects of the causal TAO3 allele, we identified regulators of only those differentially expressed genes that showed early and increasing expression uniquely in the T strain (Table S3 and Table S4).
Figure 3.
Global gene expression variation in presence of causative TAO3 allele. (A) Temporal heatmap of meiotic genes in the T and S strains. The gene names shown in green are differentially expressed in the presence of TAO3(4477C). (B) The expression profile (log2 fold change t0) for the meiotic landmark genes is given in the y-axis, and the x-axis denotes the time in sporulation medium. The red line represents the expression profile of the respective gene in the S strain, and the blue line is the same in the T strain. (C) Heatmap of the T and S strains showing differentially expressed genes across time, clustered according to expression profiles in T strain. Each row represents a single gene, and columns are time points of each strain (for gene list in each cluster, see Table S2). The order of genes is based on clustering of the T strain. Both the heatmap and boxplot consists of genes in each cluster of the T strain, and represent their expression profiles in both T and S strains. Functional GO categories of genes in each cluster of T strain are shown on left. The boxplots on the right represent the average expression profile of each cluster in the T, and the same genes in S strains. The number of genes in each cluster in a strain is indicated in brackets.
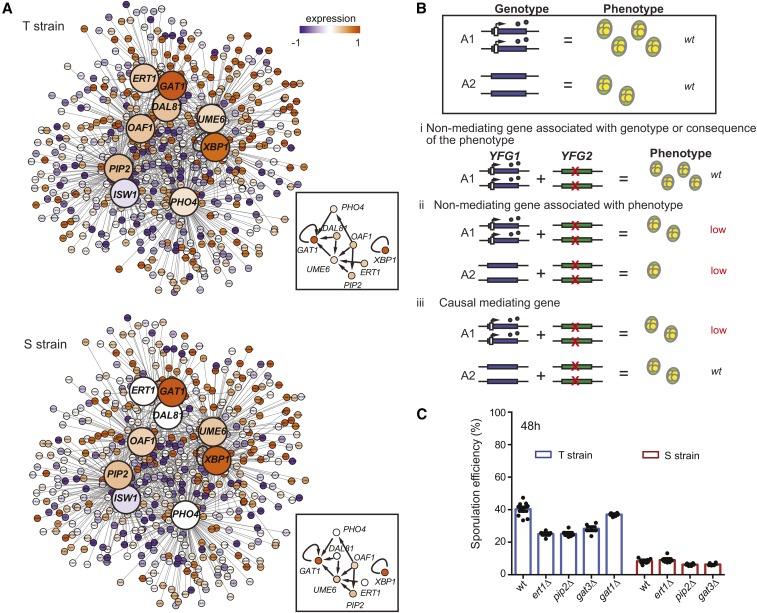
Regulators of unique genes in Cluster I and II of the T strain were enriched in nutrient metabolism and chromatin modification. These biological processes are important for initiation of meiosis (Neiman 2011). A core sporulation gene, UME6, which, together with IME1, induces expression of early meiotic genes (Kassir et al. 2003), thereby regulating pathways that initiate meiosis (Table 2; Lardenois et al. 2015), was also identified. Interestingly, along with UME6, we also identified 25 upstream regulators of UME6 (Figure 4A, Table S3, Table S4, and Table S5), such as ERT1, OAF1-PIP2, and DAL81. ERT1, a regulator of carbon source utilization (Turcotte et al. 2010), is involved in the switch from fermentation to respiration in glucose-limiting conditions (Gasmi et al. 2014). OAF1-PIP2 is a protein complex regulating lipid metabolism (Karpichev and Small 1998). DAL81 is a regulator of the nitrogen-degradation pathway (Marzluf 1997). Interestingly, like UME6, OAF1 target genes were repressed in the S strain (Cluster IV, Table S6). Earlier work in S288c and SK1 strains had shown upregulation of ERT1, PIP2, and DAL81 in SK1 strain during sporulation (Primig et al. 2000). However, their deletion in S288c strain had no effect on sporulation efficiency (Deutschbauer et al. 2002). A few other interesting regulators that we identified, not upstream UME6 (Table S3 and Table S4), included GAT1, a regulator of nitrogen metabolism (Ljungdahl and Daignan-Fornier 2012) and GAT3, a regulator of spore wall assembly (Lin et al. 2013). We next tested if these metabolic regulators interacted genetically with TAO3(4477C) during sporulation.
Table 2. Functional GO categories of regulators of clusters in the T and S strains.
| Functional GO Category | Regulators | P Value |
|---|---|---|
| Carbon metabolism | ERT1, OAF1, PIP2, MIG1, MIG2 | 1.9 × 10−6 |
| Nitrogen catabolite regulation | DAL81, DAL82, GAT1, UME6 | 1.7 × 10−5 |
| Chromatin modification | ISW1, PHO2, PHO4, UME6, OAF1, XBP1, SIF2, RSC2 | 1.4 × 10−5 |
Figure 4.
Identifying candidate genes mediating the allele-specific effects of TAO3 during sporulation using the temporal gene expression data. (A) Regulatory network of candidate genes predicted to mediate the effects of TAO3(4477C) in sporulation. The candidate mediating genes are shown as bigger nodes (large circles), with their target genes (small circles) connected to them as straight lines. The box contains the protein network interactions of the candidate genes with the core sporulation gene UME6, obtained from YEASTRACT (Materials and Methods). Colors inside the nodes were calculated as an average of the first six time points in sporulation (early phase). For a complete list of interacting genes and their expression values, see Table S5 and Table S11, respectively. (B) Genetic model for functional validation of allele-specific interactors mediating sporulation efficiency variation. The wild type effect comparison of the two alleles A1 and A2 of the YFG1 gene is shown inside the box. A1 is associated with high sporulation efficiency (wild type genotype and phenotype shown), and A2 is associated with low sporulation efficiency (wild type genotype and phenotype shown). Genetic interaction of these YFG1 alleles with candidate mediating genes (YFG2) is represented: (i) representation of nonmediating gene associated with genotype only, or is a consequence of the phenotype, since yfg2∆ in the presence of A1 does not affect the wild type phenotype of A1; (ii) representation of nonmediating gene associated with the phenotype independent of the allele, since yfg2∆ in the presence of both A1 and A2 lowers (low) the phenotype; (iii) representation of causal mediating gene since yfg2∆ only in presence of allele A1 lowers the phenotype, and in the presence of allele A2 does not change the wild type phenotype of A2. (C) Bar plots represent the mean sporulation efficiency after 48 hr of the T and S wild type (wt), and the ert1∆, pip2∆, and gat3∆ strains. Pair and interaction tests (Materials and Methods) were performed to test significance. For the T strain, gat1∆ was nonsignificant, but ert1∆ (P = 2.1 × 10−12), pip2∆ (P = 6.1 × 10−13), and gat3∆ (P = 9.6 × 10−10) significantly reduced the mean sporulation efficiency. Significant interaction terms were obtained between the genetic backgrounds (S and T) and ert1∆ (P = 2.3 × 10−4) and pip2∆ (P = 0.04).
Allele-specific functional validation identifies TAO3(4477C)-specific genetic interactors during sporulation
The candidate genes predicted from the above analysis could be either causal, mediating genes interacting with TAO3(4477C) during sporulation, or nonmediating consequential genes associated with only the genotype or only the phenotype. To identify the causal mediating genes, we used a genetic model described previously (Figure 4B; Gupta et al. 2015). According to this model, if a gene is associated only with the TAO3 genotype, and not with the sporulation phenotype, or is expressed as a consequence of the phenotype, its deletion would not affect the T strain phenotype. On the contrary, if a gene had an independent role in the sporulation phenotype, irrespective of the TAO3 genotype, its deletion will result in both a reduction in phenotype, and an additive effect, irrespective of the genetic background. Any significant deviation from this expectation would imply dependence on the genotype, with epistasis being an extreme case. In this scenario, deleting the candidate gene in T strain would affect the phenotype, while deleting the same gene in the S strain would not have an effect on the phenotype, making it a causal mediating gene. We used this model on the regulators we had identified as candidate genes. While gat1∆ had no effect on sporulation efficiency of the T strain, ert1∆, pip2∆, and gat3∆ significantly reduced the mean sporulation efficiency of the T strain by about 1.5-fold (P = 2.1 × 10−12, P = 6.1 × 10−13, P = 9.6 × 10−10, respectively, pair test in Materials and Methods, Figure 4C). Significant interaction terms were obtained between the genetic backgrounds (S and T), and ert1∆ and pip2∆ (P = 2.3 × 10−4, P = 0.04, Materials and Methods) but not for gat3∆. This showed that the effect of ert1∆ and pip2∆ on sporulation efficiency was specific to TAO3(4477C), making them causal mediating genes. GAT1 and GAT3 were nonmediating genes, the former could be associated with genotype only, or could be a sporulation-consequential gene, and the latter affected sporulation independently of the genotype. Therefore, the genetic model aided identification of true causal genes, namely ERT1 and PIP2, which mediate the effect of the TAO3 allelic variant on sporulation efficiency.
Discussion
Strong effects on phenotypic variation have been observed as a consequence of rare coding variants (Cohen et al. 2004, 2005). Tao3 is conserved from yeast to humans (Hergovich et al. 2006), and its common allele has been functionally annotated solely for mitotic cell division (Du and Novick 2002; Nelson et al. 2003). Hence, it was surprising when the rare TAO3 variant was mapped for sporulation efficiency variation (Deutschbauer and Davis 2005). In this study, using time-resolved transcriptome analysis, and an allele-specific genetic interaction assay, we identified ERT1 and PIP2 as the TAO3(4477C)-dependent mediators contributing to efficient meiosis. These genetic interactors of TAO3(4477C) are distinct from the mitotic interactors of TAO3(4477G). In this study, we identified their novel regulatory role in sporulation efficiency.
During sporulation, the sole nonfermentable carbon source, such as acetate, becomes internalized into the tricarboxylic acid (TCA) and glyoxylate cycles. Gluconeogenesis utilizes TCA cycle intermediates, and synthesizes storage carbohydrates like trehalose that are utilized during late sporulation processes (Ray and Ye 2013). Hence, TCA, glyoxylate, and gluconeogenesis are the metabolic processes that are crucial for sporulation to progress. Reduced flux through any of these metabolic pathways is capable of reducing yeast sporulation efficiency (Aon et al. 1996). Genes encoding the crucial enzymes of these metabolic processes, such as PFK1, CIT1, and CIT2, are essential for sporulation (Deutschbauer et al. 2002). ERT1 and PIP2 are transcription factors that regulate these metabolic enzymes (Baumgartner et al. 1999; Gasmi et al. 2014). Taken together, our results show that the TAO3(4477C) allele interacts genetically with regulators of the TCA cycle and gluconeogenic enzymes during sporulation. The presence of the sporulation-associated polymorphism could modulate this interaction, thereby modulating the metabolic flux during early sporulation that could result in sporulation efficiency variation.
IME1 acts as a bottleneck for the sporulation decision pathway. Lorenz and Cohen (2014) observed many natural sporulation-associated polymorphisms in genes that interacted with this input–output gene IME1, such as RIM15, a nutrient-sensing regulator of IME2. While TAO3 and MKT1 (Gupta et al. 2015) do not directly regulate IME1, in this study we show that variants in these two genes regulate early upstream metabolic processes that impinge on IME1. This provides support for the hypothesis that genes surrounding the signal transduction bottlenecks are reservoirs for accumulating causal genetic variants.
Tao3 localizes to polarized bud sites during mitosis (Nelson et al. 2003). Further determination of colocalization of TAO3(4477C) with membrane-associated ERT1 and beta-oxidation regulators OAF1-PIP2 can provide interesting clues of its function during sporulation. Similar to other scaffolding proteins like Fry (Drosophila) and SAX-2 (Caenorhabditis elegans), Tao3 has multiple conserved Armadillo-like repeats (Hergovich et al. 2006), and the causal sporulation variant resides in one of them. Tao3(1493E) physically interacts with the RAM network proteins in rich growth conditions. It would be interesting to determine binding partners of causal Tao3(1493Q) during sporulation, and to study how the polymorphism affects the binding of this putative scaffolding protein. Additionally, a few iron metabolism genes were differentially expressed during growth phase prior to incubation in the sporulation medium (t = 0 hr). It would be interesting to study whether this metabolic effect of TAO3 also plays a role in sporulation.
Even if the basic cellular network of an organism is known, it is crucial to understand how natural genetic variation and stress conditions modulate the molecular interactions within this network, resulting in differences in phenotypic outcomes (Gasch et al. 2016). This study highlights how the molecular interaction of TAO3 variant with metabolic genes causes different phenotypic outcomes. Performing such functional studies following GWAS and linkage analysis could provide a deeper understanding of how causal genetic variants function at a molecular level. This understanding is crucial, especially in the field of personalized medicine, to make more reliable predictions regarding the functional consequences of an individual’s genotype on disease predisposition and treatment (Burga and Lehner 2013).
Supplementary Material
Acknowledgments
We thank Manu Tekkedil for help with whole genome sequencing sample preparation, and Allan Jones and Gyan Bhanot for helpful comments. This research was supported by Tata Institute of Fundamental Research intramural funds 12P-0120, and Department of Biotechnology grant BT/PR14842/BRB/10/881/2010 (H.S.); Bavarian Research Center for Molecular Biosystems and Bundesministerium für Bildung und Forschung through the Juniorverbund in der Systemmedizin “mitOmics” grant FKZ 01ZX1405A (J.G.); the National Institutes of Health, Deutsche Forschungsgemeinschaft and a European Research Council Advanced Investigator Grant (L.M.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.029900/-/DC1
Communicating editor: C. Boone
Literature Cited
- Aon J. C., Rapisarda V. A., Cortassa S., 1996. Metabolic fluxes regulate the success of sporulation in Saccharomyces cerevisiae. Exp. Cell Res. 222: 157–162. [DOI] [PubMed] [Google Scholar]
- Balakrishnan R., Park J., Karra K., Hitz B. C., Binkley G., et al. , 2012. YeastMine—an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford) 2012: bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner U., Hamilton B., Piskacek M., Ruis H., Rottensteiner H., 1999. Functional analysis of the Zn2Cys6 transcription factors Oaf1p and Pip2p—different roles in fatty acid induction of β-oxidation in Saccharomyces cerevisiae. J. Biol. Chem. 274: 22208–22216. [DOI] [PubMed] [Google Scholar]
- Bellí G., Garí E., Aldea M., Herrero E., 1998a Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 14: 1127–1138. [DOI] [PubMed] [Google Scholar]
- Bellí G., Garí E., Piedrafita L., Aldea M., Herrero E., 1998b An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 26: 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolnaya L. M., Pathak R., Guo J., Polymenis M., 2006. Roles of the RAM signaling network in cell cycle progression in Saccharomyces cerevisiae. Curr. Genet. 49: 384–392. [DOI] [PubMed] [Google Scholar]
- Burga A., Lehner B., 2013. Predicting phenotypic variation from genotypes, phenotypes and a combination of the two. Curr. Opin. Biotechnol. 24: 803–809. [DOI] [PubMed] [Google Scholar]
- Cirulli E. T., Goldstein D. B., 2010. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 11: 415–425. [DOI] [PubMed] [Google Scholar]
- Cohen J. C., Kiss R. S., Pertsemlidis A., Marcel Y. L., McPherson R., et al. , 2004. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 305: 869–872. [DOI] [PubMed] [Google Scholar]
- Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., et al. , 2005. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37: 161–165. [DOI] [PubMed] [Google Scholar]
- Deutschbauer A. M., Davis R. W., 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 37: 1333–1340. [DOI] [PubMed] [Google Scholar]
- Deutschbauer A. M., Williams R. M., Chu A. M., Davis R. W., 2002. Parallel phenotypic analysis of sporulation and postgermination growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99: 15530–15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L. L., Novick P., 2002. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneur J., Stegle O., Zhu C., Jakob P., Tekkedil M. M., et al. , 2013. Genotype-environment interactions reveal causal pathways that mediate genetic effects on phenotype. PLoS Genet. 9: e1003803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Payseur B. A., Pool J. E., 2016. The power of natural variation for model organism biology. Trends Genet. 32: 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasmi N., Jacques P. E., Klimova N., Guo X., Ricciardi A., et al. , 2014. The switch from fermentation to respiration in Saccharomyces cerevisiae is regulated by the Ert1 transcriptional activator/repressor. Genetics 198: 547–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J., Lorenz K., Cohen B., 2009. Genetic interactions between transcription factors cause natural variation in yeast. Science 323: 498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D. R., Woods R. A., 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gupta S., Radhakrishnan A., Raharja-Liu P., Lin G., Steinmetz L. M., et al. , 2015. Temporal expression profiling identifies pathways mediating effect of causal variant on phenotype. PLoS Genet. 11: e1005195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Stegert M. R., Schmitz D., Hemmings B. A., 2006. NDR kinases regulate essential cell processes from yeast to humans. Nat. Rev. Mol. Cell Biol. 7: 253–264. [DOI] [PubMed] [Google Scholar]
- Huber W., von Heydebreck A., Sültmann H., Poustka A., Vingron M., 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18: S96–S104. [DOI] [PubMed] [Google Scholar]
- Huxley C., Green E. D., Dunham I., 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 6: 236. [DOI] [PubMed] [Google Scholar]
- Karpichev I. V., Small G. M., 1998. Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 6560–6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y., Adir N., Boger-Nadjar E., Raviv N. G., Rubin-Bejerano I., et al. , 2003. Transcriptional regulation of meiosis in budding yeast. Int. Rev. Cytol. 224: 111–171. [DOI] [PubMed] [Google Scholar]
- Lardenois A., Becker E., Walther T., Law M. J., Xie B., et al. , 2015. Global alterations of the transcriptional landscape during yeast growth and development in the absence of Ume6-dependent chromatin modification. Mol. Genet. Genomics 290: 2031–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. P., Kim C., Smith S. O., Neiman A. M., 2013. A highly redundant gene network controls assembly of the outer spore wall in S. cerevisiae. PLoS Genet. 9: e1003700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl P. O., Daignan-Fornier B., 2012. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 190: 885–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz K., Cohen B. A., 2014. Causal variation in yeast sporulation tends to reside in a pathway bottleneck. PLoS Genet. 10: e1004634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magni P., Ferrazzi F., Sacchi L., Bellazzi R., 2008. TimeClust: a clustering tool for gene expression time series. Bioinformatics 24: 430–432. [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf G. A., 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61: 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M., 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189: 737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B., Kurischko C., Horecka J., Mody M., Nair P., et al. , 2003. RAM: A conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14: 3782–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M., Williams R. M., Winzeler E. A., Tevzadze G. G., Conway A. R., et al. , 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26: 415–423. [DOI] [PubMed] [Google Scholar]
- Ray D., Ye P., 2013. Characterization of the metabolic requirements in yeast meiosis. PLoS One 8: e63707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint Pierre A., Génin E., 2014. How important are rare variants in common disease? Brief. Funct. Genomics 13: 353–361. [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., et al. , 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J. R., 2010. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61: 437–455. [DOI] [PubMed] [Google Scholar]
- Storey J. D., Xiao W., Leek J. T., Tompkins R. G., Davis R. W., 2005. Significance analysis of time course microarray experiments. Proc. Natl. Acad. Sci. USA 102: 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M. C., Monteiro P. T., Guerreiro J. F., Gonçalves J. P., Mira N. P., et al. , 2013. The YEASTRACT database: an upgraded information system for the analysis of gene and genomic transcription regulation in Saccharomyces cerevisiae. Nucleic Acids Res. 42: D161–D166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar P., Bhatia A., Ramdas S., Diao L., Bhanot G., et al. , 2013. Sporulation genes associated with sporulation efficiency in natural isolates of yeast. PLoS One 8: e69765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B., Liang X. B., Robert F., Soontorngun N., 2010. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 10: 2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth W. P., Olsen A. E., Sbia M., Freedman K. H., Stillman D. J., 2005. ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot. Cell 4: 1018–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., et al. , 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O., Schaffner S. F., Samocha K., Do R., Hechter E., et al. , 2014. Searching for missing heritability: designing rare variant association studies. Proc. Natl. Acad. Sci. USA 111: E455–E464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The array data for the T strain has been deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) with accession number E-MTAB-3889. The entire genome sequence data for the T strain has been deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) with the accession number PRJEB8698. The array data and the whole genome sequence data for the S strain were downloaded from Gupta et al. (2015). TAO3 gene sequence data for Saccharomyces Genome Resequencing Project (SGRP) strains (Liti et al. 2009) was downloaded from (http://www.moseslab.csb.utoronto.ca/sgrp/). An additional 24 TAO3 sequences were downloaded from the Saccharomyces Genome Database (SGD, http://www.yeastgenome.org/cgi-bin/FUNGI/alignment.pl?locus=YIL129C, date accessed: March 1, 2016). Detailed methods are described in File S1. Table S1 lists all differentially expressed genes between the T and S strains with their P and Q values calculated using EDGE. Table S2 lists genes in each cluster using TimeClust. Table S3 lists transcription factors regulating unique early (Cluster I) genes of the T strain. Table S4 lists transcription factors regulating unique increasing (Cluster II) genes of the T strain. Table S5 lists differentially expressed target genes of regulators of candidate genes mediating the affect of TAO3. Table S6 lists transcription factors regulating unique repressing (Cluster IV) genes of the S strain. Whole genome resequencing results for the TAO3 allele replacement strain are described in Table S7. All the strains used in the study are listed in Table S8, which are available upon request. All the primers used in the study are listed in Table S9. The raw sporulation efficiency values of the strains are given in Table S10. Table S11 contains smoothed expression data, base transformed with respect to t0 for the T and S strains. Figure S1 shows mathematical modeling to identify stages of meiosis affected by TAO3 causal allele. Figure S2 shows growth phenotype and TAO3 expression in PTet-TAO3(4477C) strain. Figure S3 shows comparison of global gene expression between the T and S strains at t = 0h. Figure S4 shows comparison of genes showing early (Cluster I) and increasing trend (Cluster II) between the T and S strains. Figure S5 shows genes having early expression in the T strain show expression at later time points or repressed in the S strain. Figure S6 shows whole genome resequencing of TAO3 allele replacement strain (YAD331, (Deutschbauer and Davis 2005) in comparison to the S288c reference strain. Figure S7 shows smoothing of normalized temporal data using locfit.