Abstract
Transcriptional silencing in Saccharomyces cerevisiae occurs at several genomic sites including the silent mating-type loci, telomeres, and the ribosomal DNA (rDNA) tandem array. Epigenetic silencing at each of these domains is characterized by the absence of nearly all histone modifications, including most prominently the lack of histone H4 lysine 16 acetylation. In all cases, silencing requires Sir2, a highly-conserved NAD+-dependent histone deacetylase. At locations other than the rDNA, silencing also requires additional Sir proteins, Sir1, Sir3, and Sir4 that together form a repressive heterochromatin-like structure termed silent chromatin. The mechanisms of silent chromatin establishment, maintenance, and inheritance have been investigated extensively over the last 25 years, and these studies have revealed numerous paradigms for transcriptional repression, chromatin organization, and epigenetic gene regulation. Studies of Sir2-dependent silencing at the rDNA have also contributed to understanding the mechanisms for maintaining the stability of repetitive DNA and regulating replicative cell aging. The goal of this comprehensive review is to distill a wide array of biochemical, molecular genetic, cell biological, and genomics studies down to the “nuts and bolts” of silent chromatin and the processes that yield transcriptional silencing.
Keywords: chromatin, silencing, Sir2, yeast, histone deacetylation
AMONG yeast molecular biologists, the term transcriptional silencing refers to a form of transcriptional repression that is generally independent of the identity of the gene and one that involves inactivation of chromosomal domains the size of kilobases rather than individual gene promoters. Transcriptional silencing involves a specialized chromatin structure often referred to as silent chromatin, or sometimes yeast heterochromatin in deference to functional similarities to heterochromatin in higher eukaryotes. In general, heterochromatin yields repression that is both variegated and heritable, meaning that genes within heterochromatic domains are silenced in many cells of a population but not in all cells and that these expression states are propagated through successive cell divisions. Silent chromatin domains of Saccharomyces cerevisiae are epigenetically inherited and exhibit variegated expression under certain circumstances. In contrast to heterochromatin in Schizosaccharomyces pombe and higher eukaryotes, budding yeast silent chromatin lacks a number of hallmark characteristics including methylation of histones, heterochromatin protein 1 (HP1), and the participation of RNA interference (RNAi) (Martienssen and Moazed 2015). In essence, S. cerevisiae silent chromatin is a stripped-down version of heterochromatin found in other organisms. Despite these differences, the lessons learned from the study of silent chromatin have been instructive in understanding how large, repressive chromatin domains assemble and impose transcriptional regulation.
Silent chromatin of S. cerevisiae contains histone octamers that lack most post-translational modifications. Silent chromatin binds a set of nonhistone proteins called the Silent-information regulators, or Sir proteins (Sir1, Sir2, Sir3, and Sir4). Most of the current data suggests that these factors incur silencing by blocking access to DNA. Silent chromatin also embodies structural features that extend beyond a simple beads-on-a-string model of chromatin. Silent chromatin domains (1) fold into three-dimensional structures, (2) engage in long-range chromatin-chromatin interactions, and (3) compartmentalize into subvolumes of the nucleus. Here we begin by describing features and functions of silent chromatin, leaving mechanistic discussion of how silent chromatin assembles to later sections.
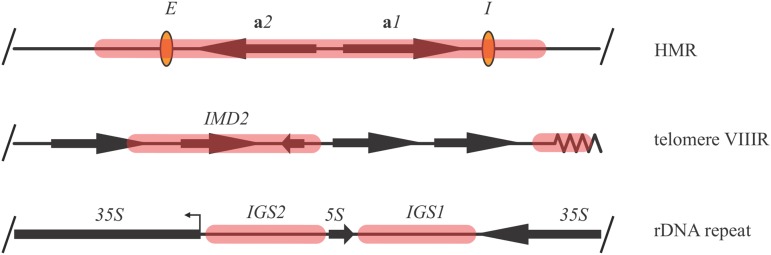
Similar, if not identical, forms of silent chromatin assemble at the HM mating-type loci and telomeres. The only significant differences are how structures at the two locations are nucleated and the size of the domains (Figure 1). At the HM loci, silent chromatin spans several kilobases and represses pairs of genes, the a genes at HMR and the α genes at HML, required for haploid mating-type identity. At telomeric sites, the size of the domains varies substantially from one telomere to the next with many telomeres possessing only small and often discontinuous silent chromatin domains (Fourel et al. 1999; Pryde and Louis 1999; Takahashi et al. 2011; Ellahi et al. 2015). Only a fraction of subtelomeric genes are subject to transcriptional silencing by the Sir proteins. The common misconception that silent chromatin domains extend continuously and extensively from all chromosome ends is largely based on experiments with model telomeres that are unusually adept at silent chromatin assembly. Nevertheless, studies with these telomeres have yielded significant information about the nature of silent chromatin.
Figure 1.
Representative silent chromatin domains of budding yeast. (A) The HMR locus. Silent chromatin is shown in pink and cis-acting silencer elements, E and I, are shown in orange. At HMR, silent chromatin silences the a mating-type genes. (B) Discontinuous silent chromatin domains at telomere VIIIR silence the subtelomeric IMD2 gene. (C) A ribosomal rDNA repeat element. Silent chromatin domains spanning the intergenic sequences, IGS1 and IGS2, suppress Pol II transcription. rDNA silencing requires Sir2 but not Sir1, Sir3, or Sir4. Loci are not drawn to scale.
In contrast to the HM loci and telomeres, transcriptional silencing in the rDNA array requires Sir2 but not the other Sir proteins. This indicates that silent chromatin at the rDNA is structurally distinct even if the domain-style of repression is similar.
General Features and Functions of Silent Chromatin
Silent chromatin is best known for its role in transcriptional silencing of the mating-type genes at the HM mating loci. Silent chromatin at these sites also blocks the action of Ho, the endonuclease that cuts DNA selectively at the MAT locus to initiate switching between mating types (Haber 2012). Thus, a more apt picture of silent chromatin is that of a repressive structure that hinders natural transactions of DNA. Obligatory DNA-based events, like the firing of replication origins, are delayed or blocked entirely at some silent chromatin locations (Stevenson and Gottschling 1999; Zappulla et al. 2002). Similarly, silent chromatin impedes the repair of UV lesions in DNA (Livingstone-Zatchej et al. 2003). Limitations on DNA repair, whether transcription-coupled or otherwise, may account for the rapid evolution of DNA sequences flanking HM loci (Teytelman et al. 2008).
How does silent chromatin inhibit such diverse genome functions? A simple and prevalent model is based on steric hindrance: silent chromatin adopts a structure that hinders access to the underlying DNA. Supporting this model, silent chromatin was found to limit modification of unbiased probes of DNA accessibility, like bacterial DNA methyltransferases and restriction endonucleases (Gottschling 1992; Singh and Klar 1992; Loo and Rine 1994; Ansari and Gartenberg 1999). Importantly, the property of limited access extends to the proteins that mediate transcription. A variety of studies have shown that RNA polymerase (Pol) II and basal transcription factors fail to bind promoters in silenced regions (Chen and Widom 2005; Lynch and Rusche 2009; Kitada et al. 2012; note that exceptions have been reported by Sekinger and Gross 2001; Gao and Gross 2008). Upstream transcriptional activators also fail to find their targets within silent chromatin (Chen and Widom 2005; Kitada et al. 2012). When the activators are overexpressed or provided before de novo silent chromatin assembly, however, they promote full expression of reporter genes within silenced domains (Aparicio and Gottschling 1994; Kitada et al. 2012). These results suggest that silencing factors compete with transcription factors for DNA access. Under typical physiological conditions silent chromatin at the HM loci prevails, permitting the rare production of a functional transcript only once per thousand cell divisions (Dodson and Rine 2015).
Silent chromatin is not universally refractory to intervention. Mating-type switching requires that the HM mating-type loci serve as genetic donors during directed homologous recombination. Similarly, the domains must eventually be replicated by DNA polymerases. On these occasions, specific chromatin remodelers may facilitate DNA access to DNA synthesis and processing enzymes. Sir3 does in fact associate with the SWI/SNF chromatin-remodeling complex. While the interaction is important for telomeric silencing, it is, however, not required for mating-type switching (Sinha et al. 2009; Manning and Peterson 2014).
Distinctive molecular features of silent chromatin
Several distinguishing molecular features of silent chromatin have been elucidated from studies with live cells and reconstituted systems. A broad overview of the gross structural features is presented here. The most important distinguishing feature of silent chromatin is the presence of the Sir proteins that bind silenced loci (Figure 2A) (Hecht et al. 1996; Strahl-Bolsinger et al. 1997; Lieb et al. 2001; Zhang et al. 2002; Sperling and Grunstein 2009). A protein complex containing Sir2, Sir3, and Sir4 is recruited to specific sites, known as silencers, by DNA-bound factors and Sir1. The Sir2/3/4 complex interacts with histones and DNA adjacent to silencers, thereby imposing greater nucleosome occupancy and more precise nucleosome positioning across silenced domains (Weiss and Simpson 1998; Ravindra et al. 1999; Wang et al. 2015). When Sir proteins are reconstituted with chromatin templates in vitro, nucleosomes aggregate; sometimes forming distinctive 20-nm fibers visible by electron microscopy (EM) (Onishi et al. 2007; Johnson et al. 2009).
Figure 2.
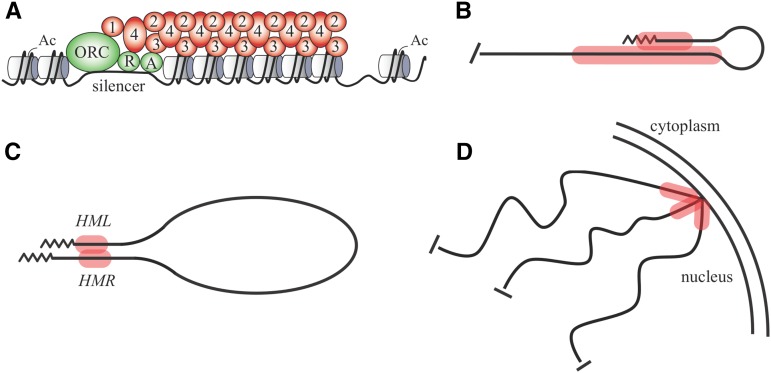
Local and long-range interactions of silent chromatin. (A) The arrangements of components within a typical locus of silent chromatin. Sir2 (2), Sir3 (3) and Sir4 (4) form the Sir2/3/4 complex that binds histones throughout the silenced domain. Histones within the domain lack post-translational modifications with the exception of H2AS129 phosphorylation and some H4K12 acetylation. ORC, Rap1 (R), Abf1 (A) and Sir1 (1) bind to cis-acting silencer elements and interact with proteins of the Sir2/3/4 complex. Specific interactions between these components are documented in subsequent figures. Ac, acetylation. (B) The folded-back structure of silent chromatin at a telomere. (C) The long-range interactions between the silent chromatin domains at HML and HMR cause chromosome III to fold back upon itself. (D) Interactions between the silent chromatin domains of different telomeres and interactions between silent chromatin and docking sites at the nuclear membrane cause chromosome ends to cluster at the nuclear periphery.
A second distinguishing feature of silent chromatin domains is the depletion of nearly all post-transcriptional histone modifications typically found in bulk active chromatin (for examples, see Braunstein et al. 1993; Suka et al. 2001). Early genetic studies identified residues in the histone H4 N-terminal tail that were critical for silencing (Kayne et al. 1988; Johnson et al. 1990; Megee et al. 1990; Park and Szostak 1990). Subsequent studies identified residues on the histone octamer core that were equally important (Ng et al. 2002; van Leeuwen et al. 2002). Acetylation of K16 on the H4 tail and methylation of K79 on the H3 globular domain stood out as being particularly important because mutations that mimicked the modifications were strikingly detrimental to silencing. It is now understood that H4K16 and H3K79 lie at the nucleosomal docking site for Sir3 and that modification of either residue interferes with Sir3 binding (Onishi et al. 2007; Armache et al. 2011). While other histone modifications might impose similar constraints, still others may be missing from silent chromatin domains simply due to steric occlusion of the enzymes that create them. One exception is acetylation of H4K12, which is reduced but not absent in silent chromatin (Braunstein et al. 1996; de Bruin et al. 2000; Zhou et al. 2011). Another exception is the phosphorylation of S129 on H2A, which is typically associated with sites of DNA damage and replication stress (Szilard et al. 2010; Kitada et al. 2011; Kirkland and Kamakaka 2013).
Higher-order structures within silenced chromosomal domains
A growing number of studies provide compelling evidence that the Sir proteins fold chromatin into a higher-order structure. The exact nature of the folded structure is not known, but it likely involves Sir-mediated contact of nonadjacent chromatin sites. The first evidence came from studies of telomeres where it was found that Rap1, which binds directly to terminal telomeric repeat sequences, also cross-linked to subtelomeric sites. The results were taken as evidence that silent chromatin in subtelomeric regions folds back upon itself (Figure 2B) (Strahl-Bolsinger et al. 1997; de Bruin et al. 2000). Standing alone, the results could alternatively suggest that Rap1 arrives on subtelomeric DNA as a passenger of Sir proteins without binding to DNA at all. However, the notion of a silent chromatin-mediated fold back was supported by the use of novel, transcriptional-reporter constructs that detected long-range interactions within silenced subtelomeric regions (de Bruin et al. 2001). Later studies suggested that the HM loci also fold back upon themselves, consistent with earlier findings that the DNA supercoiling of the silenced domains was altered (Bi and Broach 1997; Cheng et al. 1998; Valenzuela et al. 2008). Molecular genetic studies showed that distant silencers synergize one another, as if they interact physically (Boscheron et al. 1996; Fourel et al. 1999; Pryde and Louis 1999; Cheng and Gartenberg 2000; Oki et al. 2004; Valenzuela et al. 2008). Even silent chromatin domains separated by great distances, like the HM loci at the ends of chromosome III, have been shown to interact (Figure 2C); in essence by folding back upon one another (Lebrun et al. 2003; Miele et al. 2009; Kirkland and Kamakaka 2013). The specific interactions that promote folding back in each of these situations are not entirely understood. Kamakaka and colleagues suggested that DNA-repair proteins and DNA homology contribute to long-range interactions between the HM loci (Kirkland and Kamakaka 2013). More generally, interactions between the subtelomeric silent chromatin of different chromosomes are driven by self-association of Sir3 and are independent of DNA homology (Ruault et al. 2011; Guidi et al. 2015). At high concentrations, pure Sir3 causes nucleosome arrays to condense and aggregate (McBryant et al. 2008; Swygert et al. 2014). Irrespective of mechanism, the interactions that form higher-order structures appear to mask epitopes of some of the resident Sir proteins and histones in silenced regions (Thurtle and Rine 2014).
Dynamic nuclear compartmentalization of silent chromatin
The interactions between Sir3 at different telomeres cause chromosome ends to cluster into a small number of foci in exponentially-growing cells (Figure 2D) (Klein et al. 1992; Palladino et al. 1993; Gotta et al. 1996). Clustering is a dynamic process where individual telomeres split from clusters and rejoin on a time scale of minutes (Schober et al. 2008). Telomeres at the ends of chromosome arms of similar length associate with one another more frequently, owing to the comigration of centromeres at anaphase (Schober et al. 2008; Therizols et al. 2010). Interestingly, long-lived stationary-phase cells group all telomeres into a single Sir3-dependent cluster, a genomic restructuring event that extends chronological life span (Guidi et al. 2015).
In addition to clustering, the ends of chromosomes also associate reversibly with the inner nuclear membrane. The proteins that mediate membrane anchoring of telomeres fall into two primary pathways (Hediger et al. 2002; Taddei et al. 2004). The first is defined by Ku, a protein complex that binds directly to telomeric ends, as well as to double-stranded DNA breaks. The second pathway is defined by Sir4 of subtelomeric silent chromatin. The silent chromatin of HMR, although not contiguous with subtelomeric silent chromatin, is similarly anchored (Gartenberg et al. 2004). The contributions of both the Ku and Sir4 pathways vary from one telomere to the next with additional variation imparted by the stage of the cell cycle (Hediger et al. 2002; Taddei et al. 2004). To achieve anchoring, Ku and Sir4 associate with a network of docking sites on the inner nuclear membrane, as described later. Sir4 and Ku also interact directly (Tsukamoto et al. 1997; Roy et al. 2004; Hass and Zappulla 2015). Thus, the anchoring pathways defined by the two proteins may not be entirely independent.
Why has budding yeast evolved mechanisms to sequester telomeres at the nuclear membrane? Current data points to a role in chromosome-end protection. Disruption of anchoring can lead to amplification of the subtelomeric Y′ repeat elements, inappropriate lengthening of terminal repeat sequences by telomerase, as well as a senescence phenotype that is reminiscent of telomerase loss in strains lacking the ataxia telangiectasia mutated-kinase homolog Tel1 (Schober et al. 2009; Ferreira et al. 2011). The extent to which silent chromatin contributes directly to telomere protection is difficult to unravel because the Ku- and Sir4-anchoring pathways are so interwoven. Nevertheless, it should be noted that simply eliminating the Sir proteins causes changes in telomere length (Palladino et al. 1993).
There are likely other inherent advantages to maintaining silent chromatin at the nuclear periphery. The supply of Sir proteins is limiting for transcriptional silencing. Increased concentration of Sir proteins at one site diminishes the pool of available proteins for silencing at another (Buck and Shore 1995; Maillet et al. 1996; Marcand et al. 1996; Larin et al. 2015). By sequestering individual silent chromatin domains at the periphery, the concentration of perinuclear Sir proteins is raised for all other silent chromatin domains anchored at the periphery. Indeed, tethering a suboptimal silencer to the Sir-enriched nuclear membrane results in silencing of the tethered chromosomal domain (Andrulis et al. 1998; Taddei et al. 2009). In essence, telomere anchoring and clustering creates a nonequilibrium enrichment of Sir proteins at the edge of the nucleus (Gasser et al. 2004). A corollary of this so-called “Circe effect” is that Sir proteins are sequestered from the rest of the genome that occupies the bulk of the nucleus. In this way, sequestration offers a level of specificity for a set of general chromatin repressors like the Sir proteins that might cause promiscuous silencing. In this paradigm, controlled release of the Sir proteins from telomeres can be used to rewire the cell’s transcriptional program in response to environmental cues. For example, in response to environmental stressors Sir3 is phosphorylated, causing derepression of some stress-response genes near telomeres and a shortened replicative life span (RLS) (Stone and Pillus 1996; Ai et al. 2002; Ray et al. 2003). Globally, dispersion of telomeric Sir proteins causes downregulation of genes involved in ribosome biogenesis (Taddei et al. 2009). Thus, sequestration of Sir proteins at telomeres may maintain a pool of transcriptional repressors, readily available for gene reprogramming in response to changes in the environment.
Silencers
Silencers were first defined genetically as discrete DNA sequences of the silenced HM mating-type loci that were required for transcriptional repression of the endogenous genes and heterologous reporters (Abraham et al. 1984; Feldman et al. 1984; Brand et al. 1985). Silencers act in a relatively distance-independent manner up to several kilobases. Silencers also function on adjacent genes when moved to ectopic locations, albeit not as efficiently (Lee and Gross 1993; Thompson et al. 1994; Shei and Broach 1995; Maillet et al. 1996). While some silencers operate bidirectionally, nucleosome gaps adjacent to other silencers impose unidirectional function (Zou et al. 2006).
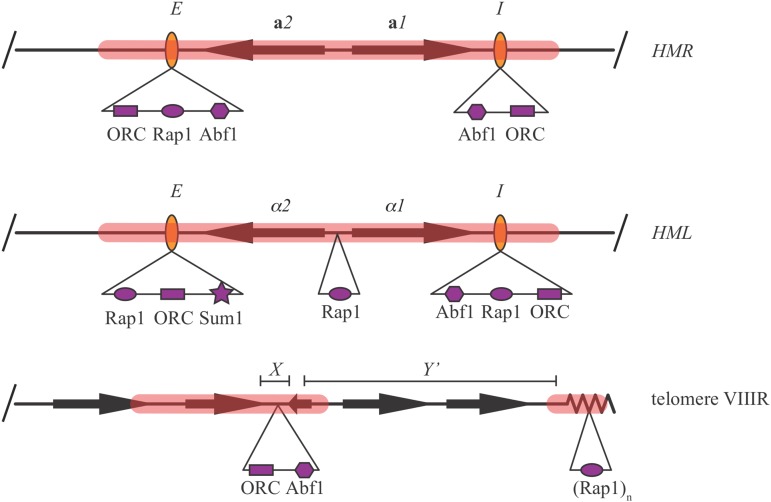
Silencers of the HM loci are compact elements spanning <150 bp. Each contains binding sites for at least two of three essential factors: origin recognition complex (ORC), Rap1, and Abf1 (Figure 3) (Brand et al. 1987; Buchman et al. 1988; Kimmerly et al. 1988; Mahoney et al. 1991; Boscheron et al. 1996). The multi-subunit ORC complex recognizes DNA replication origins in all eukaryotes (Bell and Stillman 1992). Abf1 is a transcription factor that binds the promoters of a diverse set of genes (Rhode et al. 1992, and references therein). The protein also frequently binds near origins of DNA replication where it facilitates origin firing. Rap1 is a transcription factor that binds hundreds of genes involved in protein synthesis and glycolysis (Lieb et al. 2001). Densely-packed Rap1 binding sites reside within the terminal TG1–3 repeat sequences of telomeres and act as silencers at chromosomal ends (Kurtz and Shore 1991; Gilson et al. 1993; Kyrion et al. 1993; Cockell et al. 1995; Hecht et al. 1996).
Figure 3.
DNA binding sites within silencers and proto-silencers. Direct DNA binding by Sum1 contributes to the function of the HML-E silencer. The X and Y′ subtelomeric repeat elements of telomere VIIIR are shown.
The silencers of HMR and HML were given the names “E” or “I” based on their relative contributions to silencing in early assays (Abraham et al. 1984; Feldman et al. 1984). In a chromosomal context, HMR-E is sufficient for silencing whereas the contribution of HMR-I can only be detected with sensitized assays that weaken silencing (Brand et al. 1985; Rivier et al. 1999; Lynch and Rusche 2010). The E and I silencers of HML are both sufficient for complete silencing of the locus in the genome (Mahoney and Broach 1989). Thus, under normal laboratory conditions, the HM silencers appear to be functionally redundant for their role in repression.
Each of the silencer binding proteins contributes to silencer function (Sussel and Shore 1991; Foss et al. 1993; Liu et al. 1994; Loo et al. 1995). Nevertheless, redundancy can be found within individual silencers, as exemplified by the study of HMR-E. Elimination of single binding sites within the element caused only a partial loss of activity or no loss at all (Brand et al. 1987). This probably owes to the complexity of natural silencers. When a synthetic silencer was reconstituted from just oligonucleotide binding sites for each of the factors, each binding site became essential (McNally and Rine 1991).
Mechanism of silencer action
Silencers function by recruiting Sir proteins to chromatin, as described in more detail later. In support of a simple recruitment mechanism, individual silencer binding proteins can be replaced by tethering Sir proteins directly to DNA with protein fusions (Chien et al. 1993; Marcand et al. 1996; Cuperus et al. 2000). It is likely that proximity and perhaps positioning of these factors at silencers achieves a combinatorial affinity necessary for Sir protein nucleation. ORC, Abf1, and Rap1 each bind hundreds of sites throughout the yeast genome yet the vast majority of sites do not nucleate silencing. Evolutionary forces likely shaped the relative affinity of the silencer-bound factors for Sir proteins, as well as the intracellular level and distribution of Sir proteins to avoid promiscuous binding.
Proto-silencers
Curiously, individual binding sites for ORC, Abf1, and Rap1 that make no contribution to silencing in isolation can augment the function of a bona fide silencer when situated nearby. Such sites are termed proto-silencers (Boscheron et al. 1996). Proto-silencers can act at distances of up to several kilobases. One illustrative example is the Rap1 binding site within the divergent promoters of the α mating-type genes (Figure 3). When these genes are present at MAT, the site contributes to gene expression (Giesman et al. 1991). When the genes are present at HML, the site augments the action of the HML silencers (Cheng and Gartenberg 2000). Proto-silencers within subtelomeric DNA elements extend silent chromatin domains specified by terminal telomeric silencers (Fourel et al. 1999; Pryde and Louis 1999). These proto-silencers include ORC binding sites within repetitive subtelomeric X elements (Figure 3). Unexpectedly, Ume6 binding sites in the promoters of some subtelomeric seripauperin genes (PAU genes) also act as proto-silencers (Radman-Livaja et al. 2011; Ellahi et al. 2015).
Proto-silencers do not typically recruit Sir proteins on their own (Rusche et al. 2002). Thus, it is not clear how the elements contribute to silencing. One possibility is that proto-silencers favor extension of heterochromatin nucleated at nearby silencers by providing localized sites of enhanced Sir affinity. Another possibility is that proto-silencers interact directly with nearby silencers to synergistically enhance Sir recruitment. Chromosome conformation capture (3C) studies indicated that the silenced HM loci fold back upon themselves, placing the flanking silencers in close proximity (Valenzuela et al. 2008). Although these interactions required Sir proteins, earlier work showed that the silencer binding factor Rap1 can associate with distal sites and loop out intervening DNA (Hofmann et al. 1989).
The Sir Proteins
Genes encoding the Sir proteins were first identified as mutations unlinked to HML and HMR that caused sterility yet restored sporulation in certain yeast strains (Haber and George 1979; Klar et al. 1979; Rine 1979; Rine et al. 1979). The genes were appropriately understood to derepress the silenced mating-type loci. Sir2, Sir3, and Sir4 are now known to form a complex with 1:1:1 stoichiometry that is recruited to chromatin by Sir1. Each of the Sir proteins contributes one or more unique and critical activities to the assembly and function of silent chromatin. In the following sections, the four proteins are discussed in detail.
Sir1
The curious behavior of sir1 mutants distinguished the gene from the other SIRs, each of which caused total derepression when mutated. Loss of SIR1 yielded mixed populations of cells with one fraction bearing HM loci that were derepressed and other fractions in which one or both of the loci were silenced (Pillus and Rine 1989; Xu et al. 2006). The transcriptional states were heritable, lasting tens of generations before switching events converted one expression state to the other. Initially, the silencing phenotype was attributed solely to a defect in establishing silencing because derepressed cells did not immediately restore transcriptional repression. This notion was further supported by studies in which reintroduction of SIR1, either the wild-type gene or elaborate synthetic alleles, led to rapid restoration of silencing (Fox et al. 1997; Enomoto and Berman 1998; Kirchmaier and Rine 2001; Li et al. 2001). Only recently has it become apparent that Sir1, like the silencers to which it binds, is also required for perpetuating the silent state (Dodson and Rine 2015).
At a molecular level, the Sir1 protein differs from the other Sir proteins by acting primarily in nucleation of silent chromatin. The Orc1 subunit of the ORC complex recruits Sir1 to the HM silencers and to ORC-based proto-silencers within subtelomeric X repeats (Triolo and Sternglanz 1996; Fourel et al. 1999). An α-helical domain in the Orc1 N-terminal end interacts with a small region of the Sir1 C-terminal end named the ORC interacting region (OIR) (Hou et al. 2005; Hsu et al. 2005). Disruption of the interface by mutation blocks Sir1 recruitment and phenocopies a sir1 null mutation (Gardner et al. 1999; Zhang et al. 2002; Bose et al. 2004). A second OIR motif in the Sir1 N-terminal end also contributes to silencing, presumably through binding a second protein partner (Connelly et al. 2006; Hou et al. 2009).
The C-terminal end of Sir1 also associates with Sir4, which in turn binds the other Sir proteins (Triolo and Sternglanz 1996; Bose et al. 2004). Thus, Sir1 acts as a molecular adaptor between silencer-bound factors and the Sir2/3/4 complex. All cells would lose silencing in the absence of Sir1 if not for the additional contacts between Sir3 and Sir4, and factors bound directly to silencers (Moretti et al. 1994; Moretti and Shore 2001).
Given the long-held view of Sir1 as a silent chromatin nucleator, it might be expected that the protein acts only at silencers. Here the experimental record has been equivocal. Some studies have found Sir1 limited to silencers (Zhang et al. 2002), whereas others have found the protein and ORC subunits distributed throughout the HM loci (Gardner and Fox 2001; Rusche et al. 2002; Ozaydin and Rine 2010). Whether Sir1 and ORC function throughout silent chromatin domains or whether they simply hitchhike onto silent chromatin as passengers of Sir4 is not yet clear.
Sir2
Evolutionary considerations of Sir2
Sir2 is the founding member of a protein family of NAD+-dependent histone/protein deacetylases called the sirtuins, which are highly conserved from bacteria to humans (Brachmann et al. 1995; Frye 1999). Yeast S. cerevisiae possesses five sirtuins: Hst1, Hst2, Hst3, and Hst4, as well as Sir2 (Brachmann et al. 1995; Derbyshire et al. 1996). Sir2 and Hst1, paralogs derived from an ancient genome duplication, have functionally diverged but retain a level of redundancy (Kellis et al. 2004; Hickman and Rusche 2007). Mammals possess seven sirtuins (SIRT1 through SIRT7). SIRT1, the closest mammalian homolog to yeast Sir2, can partially restore silencing function when overexpressed in a sir2∆ mutant (Gaglio et al. 2013).
Sir2 as an NAD+-dependent histone deacetylase
The first indication that Sir2 could be a histone deacetylase (HDAC) came from collaborative work between the Broach and Allis laboratories. In an early application of chromatin immunoprecipitation (ChIP), these investigators found that the H3 and H4 N-terminal tails at HML and HMR were hypoacetylated (Braunstein et al. 1993). Importantly, H3 and H4 at HML and HMR acquired acetylation in a sir2∆ mutant, and bulk histone acetylation decreased when SIR2 was overexpressed (Braunstein et al. 1993). These experiments suggested that Sir2 was either a histone deacetylase or that the protein somehow indirectly regulated histone acetylation. At the time, attempts to demonstrate HDAC activity of Sir2 biochemically failed because the requirement for an NAD+ cofactor was not yet known.
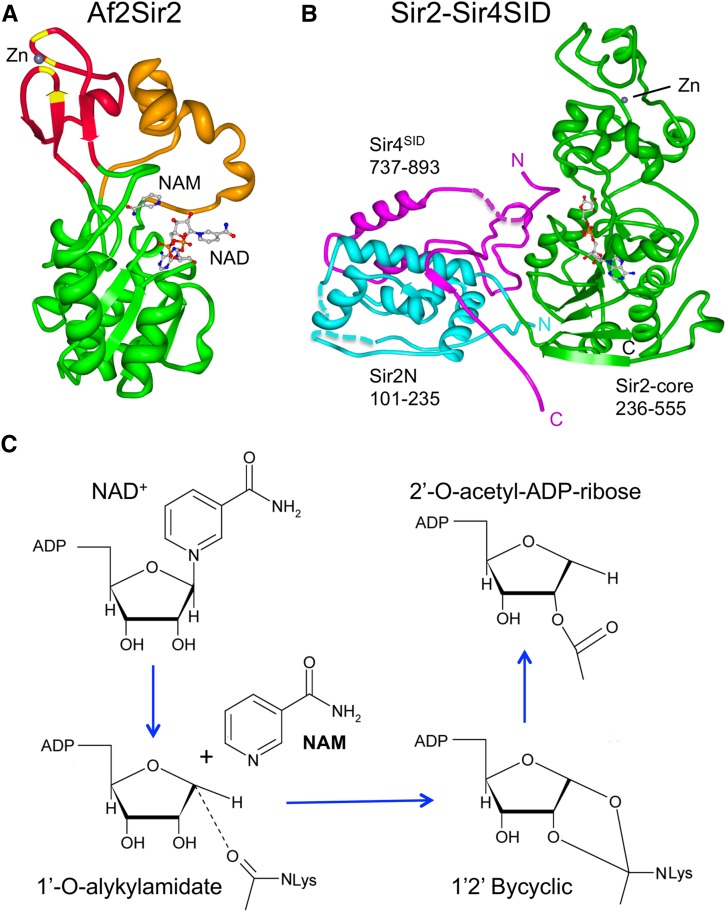
A key turning point came when the sequence of Sir2 was found to resemble Salmonella CobB, a protein of the cobalamin synthesis pathway. CobB was thought to be a nicotinic acid mononucleotide (NaMN) phosphoribosyltransferase (Tsang and Escalante-Semerena 1998). Direct testing of Sir2 and human SIRT2 showed that these proteins did indeed possess some mono-ADP-ribosyltransferase activity (Frye 1999; Tanny et al. 1999). However, the predominant enzymatic activity of sirtuins was uncovered later while studying ADP ribosylation in reactions that contained acetylated histone substrates (Imai et al. 2000; Landry et al. 2000b). In studies with Sir2 and SIRT1, the acetyl groups were removed in the presence of NAD+. The results indicated that sirtuins are primarily NAD-dependent histone deacetylases. The deacetylase activity is conserved for sirtuins from Archaea and Eubacteria species to humans (Smith et al. 2000). In fact, some of the original sirtuin X-ray crystallographic structures were generated with sirtuins from Archaeogloblus fulgidus (Sir2Af1 and Sir2Af2, see Figure 4A) (Min et al. 2001; Avalos et al. 2002; Chang et al. 2002).
Figure 4.
Sir2 structure and enzymatic activity. (A) Crystal structure of the Archaeoglobus fulgidus homolog Sir2Af2. The nucleotide-binding Rossmann fold is shown in green, zinc-binding (ribbon) in red, and small helical domain in orange. Zn2+ ion is shown coordinated with four cysteine residues (yellow). The positions of NAM and NAD+ binding are indicated. Protein Data Bank (PDB) accession number 1S7G. (B) Crystal structure of S. cerevisiae Sir2 core (green) and N-terminally extended (cyan) domains are shown in contact with the SID of Sir4 (magenta). PDB accession number 4IAO. (C) Chemical reaction of lysine deacetylation by Sir2 and other sirtuins.
Sir2 forms a stable homotrimeric complex, but the histone deacetylation activity of purified recombinant protein is relatively weak (Cubizolles et al. 2006). In yeast cells, Sir2 associates with partner proteins to form either the Sir2/3/4 complex or the regulator of nucleolar silencing and telophase exit (RENT) complex, as discussed below (Ghidelli et al. 2001; Tanny et al. 2004). In contrast to pure Sir2, the complexes possess strong H4K16 deacetylation activities on purified histones, indicating that the Sir2 partner proteins stimulate Sir2 enzymatic activity. Surprisingly, neither complex displays significant deacetylation activity on purified nucleosomes, strongly suggesting that additional factors contribute to the robust activity on chromatin in cells (Tanny et al. 2004).
Sir2 protein structure
Each of the sirtuins shares a conserved catalytic core that consists of a large Rossmann-fold domain and a smaller domain with four conserved cysteine residues that coordinate zinc, usually within a zinc-ribbon structure (Figure 4A) (Finnin et al. 2001; Min et al. 2001). Acetylated substrate peptides and NAD+ bind in a cleft between the Rossmann fold and zinc-containing domain. Thus, substrate lysines must reside within a relatively flexible region of the target protein (Figure 4A) (Min et al. 2001; Avalos et al. 2002). The core of S. cerevisiae Sir2 differs from other sirtuins by a 30 amino acid insertion following the four cysteines that changes the zinc ribbon into a motif that more closely resembles a plant homeodomain finger (Figure 4B) (Hsu et al. 2013). The function of this insertion remains unclear.
Some sirtuins, including Sir2, Hst1, and SIRT1, possess long N- and/or C-terminal domains that extend beyond the catalytic core. These flanking domains associate with secondary proteins that confer substrate specificity to the sirtuins. The extended N-terminal domain of Sir2 interacts with a Sir2-interacting domain (SID) within Sir4 (amino acids 737–893), which stabilizes the Sir2 core structure and allosterically stimulates the histone deacetylase activity (Figure 4B) (Hsu et al. 2013).
Sirtuin biochemistry and NAD+ homeostasis
Sir2 and other sirtuins consume one NAD+ molecule for each deacetylation of a lysine (Landry et al. 2000a; Tanny and Moazed 2001). The cleavage of NAD+ into nicotinamide (NAM) and ADP-ribose is coupled to transfer of the acetyl group from a target lysine onto the ADP-ribose moiety, yielding one molecule each of 2′-O-acetyl-ADP-ribose (OAADPr), NAM, and the deacetylated protein (Figure 4C) (Tanner et al. 2000; Sauve et al. 2001; Tanny and Moazed 2001; Jackson and Denu 2002). In principle, the NAD+-dependent, catalytic activity of sirtuin-mediated deacetylation could deplete cellular NAD+ pools. However, cellular NAD+ levels do not change in most yeast sirtuin mutants, with the exception of hst1∆, which has elevated NAD+ due to derepression of NAD+ biosynthesis genes (Bedalov et al. 2003). Reductions in cellular NAD+, on the other hand, strongly affect the function of sirtuins. For example, silencing and replicative aging defects emerge in an npt1∆ mutant, where NAD+ levels are reduced by approximately two- to threefold (Lin et al. 2000; Smith et al. 2000). Npt1 is a nicotinic acid phosphoribosyltransferase that converts nicotinic acid (NA) into NaMN, the rate-limiting step of the Preiss–Handler NAD+ salvage pathway (Rajavel et al. 1998). Npt1 and other enzymes of the salvage pathway are enriched in the nucleus (Anderson et al. 2002; Sandmeier et al. 2002a), suggesting that there could be a nuclear flux or pool of NAD+ that is critical for maintaining nuclear Sir2 functions (see below).
Sir2 is strongly inhibited by NAM in vitro (IC50 50 µM), and in vivo by the addition of NAM to the growth medium (Bitterman et al. 2002). Feedback inhibition of Sir2 by NAM is normally prevented by the conversion of NAM to NA via the nicotinamidase Pnc1 (Anderson et al. 2003a; Gallo et al. 2004). In the absence of Pnc1, intracellular NAM levels rise ∼10-fold (Sauve et al. 2005), and silencing becomes hypersensitive to exogenous NAM in the media (Gallo et al. 2004).
Other small molecule regulators of sirtuin function have been identified, and these drugs have often been useful in investigating sirtuin functions (Bedalov et al. 2001; Grozinger et al. 2001; Howitz et al. 2003). For example, splitomicin was identified in a cell-based study as an inhibitor of Sir2 enzymatic activity. When added to cells arrested for growth, splitomicin caused loss of silencing, indicating that continual deacetylation by Sir2 is required to maintain the silent state (Bedalov et al. 2001). Splitomicin and another inhibitor, sirtinol, are commonly used to inhibit mammalian sirtuins (Yeung et al. 2004; Ota et al. 2006).
The functional silencing relationship between Sir2 and its paralog, Hst1
A screen for mutants that could restore mating in a yeast strain lacking Sir2 isolated a dominant missense mutation in the SUM1 gene (Klar et al. 1985; Livi et al. 1990; Laurenson and Rine 1991; Chi and Shore 1996). The mutant, SUM1-1, suppressed the mating defect by restoring silencing of the HM loci not only in a sir2 mutant but also in mutants lacking Sir3 or Sir4. How a point mutation in a seemingly-unrelated protein could bypass the entire silencing apparatus at HM loci has been a source of fascination to scientists in the field. It is now understood that Sum1 forms a complex with Hst1 and Rfm1 that normally represses the promoters of genes involved in sporulation, de novo NAD+ biosynthesis, and thiamine biosynthesis (Xie et al. 1999; Bedalov et al. 2003; Li et al. 2010). Remarkably, the Sum1-1 protein gains the ability to self-associate and interact with ORC while reducing an affinity for direct DNA binding (Safi et al. 2008). These features convert the Sum1-1/Rfm1/Hst1 complex from a short-range repressor of specific promoters to a complex that can bind silencers and spread long-range repression to the promoters of the mating-type genes at the HM loci. Spreading is in many ways analogous to the behavior of the Sir2/3/4 complex (e.g., H4K16 is deacetylated) yet in other ways distinct (e.g., unstable and nonheritable repression) (Rusche and Rine 2001; Sutton et al. 2001; Lynch et al. 2005; Valenzuela et al. 2006; Prescott et al. 2011). In this context, it should also be mentioned that Sum1 normally associates with the HML-E silencer at a 10-bp site called D2, where it is required for silencing under weakened conditions (see Figure 3; Irlbacher et al. 2005). Hst1 is not required for HML silencing. It is possible that Sir2 substitutes as a Sum1 binding partner at this location, since Sir2 is known to associate with Sum1 when HST1 is deleted (Hickman and Rusche 2007).
Sir2 post-translational modifications
Given that Sir2 and other sirtuins govern numerous physiological pathways, it would not be surprising if the enzymatic activities and targeting specificities of the proteins were regulated by reversible post-translational events. Indeed, Sir2 was recently reported to be modified and regulated by sumoylation and phosphorylation (Hannan et al. 2015; Kang et al. 2015). Sir2 is sumoylated by the SUMO ligase Siz2 on three lysines (K106, K132, and K215) within the N-terminal extension domain that associates with Sir4. Modification of K215 appears to be important in regulating Sir2 distribution between telomeres and the rDNA (Hannan et al. 2015), perhaps by disrupting the Sir2-Sir4 interaction.
Sir2 is phosphorylated on a highly-conserved Ser473 residue located within the catalytic core domain (Kang et al. 2015). The modification alters the acetylation state and expression level of the PMA1 gene, where Sir2 acts to control life span. Whether the modification controls the enzymatic activity of Sir2 has not yet been tested, and whether the modification affects silencing of the classical targets of Sir2 (e.g., the HM loci) has not yet been reported.
Lastly, yeast Sir2 was reported to be a substrate for self-modification by mono-ADP ribosylation (Tanny et al. 1999). Whether Sir2-mediated ADP ribosylation of Sir2 (and Sir2-mediated ADP ribosylation of histones) is physiologically relevant has never been proven. We suspect that additional modifications of Sir2 will eventually be identified.
Sir3
Overview
Sir3 plays a central structural role in silent chromatin by binding nucleosomes. The association is attenuated by post-translational histone modifications that are known to interfere with transcriptional silencing. Investigations of the Sir3 interaction with nucleosomes have provided fundamental insight into how transcriptionally silent chromatin domains assemble.
Evolutionary considerations of Sir3
SIR3 arose from the gene encoding the largest subunit of ORC, ORC1, during an ancient whole-genome duplication of budding yeast (Kellis et al. 2004; Byrne and Wolfe 2005). Silencing was not a new function to the emergent SIR3 gene. ORC1 in the yeast Kluyveromyces lactis, which diverged from S. cerevisiae before genome duplication, functions in both DNA replication and transcriptional silencing (Hickman and Rusche 2010). The subfunctionalization of SIR3 and ORC1 has been substantial: S. cerevisiae Sir3 cannot replace Orc1 in DNA replication and Orc1 cannot replace Sir3 in silencing (Bell et al. 1995).
Sir3 protein structure
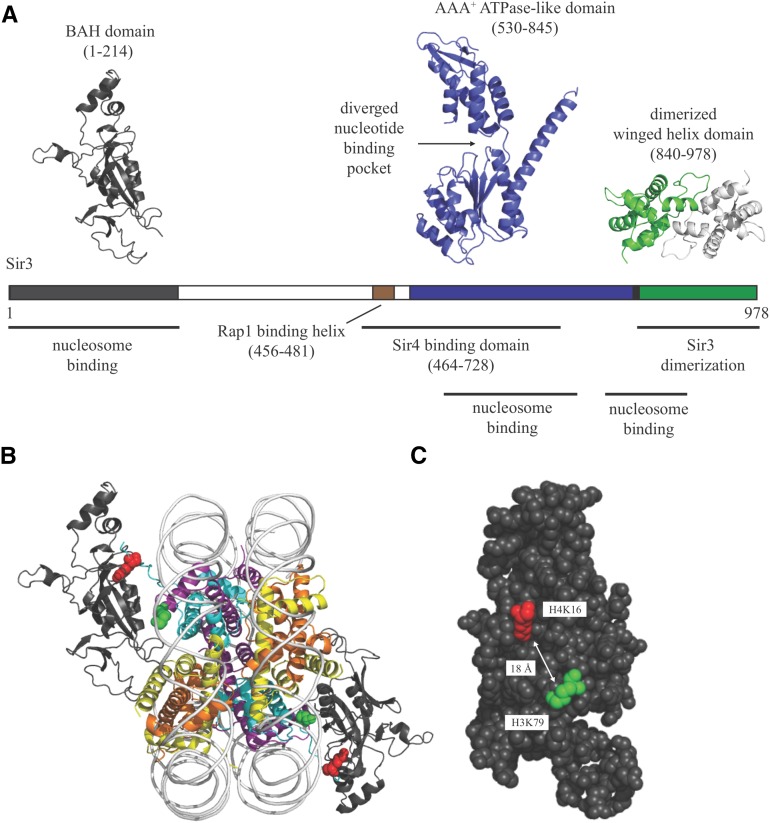
Sequence comparisons of Sir3 with Orc1 and related proteins have identified three conserved domains: an N-terminal bromo-adjacent homogy (BAH) domain, a central AAA+ ATPase-like domain, and a C-terminal winged-helix domain (Figure 5A). The structures of each of the Sir3 domains have been determined at atomic resolution.
Figure 5.
Domain structure and nucleosome binding of Sir3. (A) The structural and functional domains of Sir3. The BAH domain from PDB accession number 3TU4, the AAA+ ATPase-like domain from 3TE6, and the winged-helix domain from 3ZCO. (B) Crystal structure of the Sir3 BAH domain bound to the nucleosome core particle. The BAH domain is shown with a dark gray ribbon. The H4K16 and H3K79 histone residues critical for Sir3 binding are shown in red and green space-filling spheres, respectively. PDB accession number 3TU4. (C) The positions of H4K16 and H3K79 on the nucleosome-binding surface of the Sir3 BAH domain.
The BAH domain:
The N-terminal 214 amino acids of Sir3 contains a BAH domain (Zhang et al. 2002; Connelly et al. 2006; Hou et al. 2006). Genetic suppressor studies over the course of many years suggested that the domain mediates binding of nucleosomes. Mutations that mapped to the BAH domain suppressed mutations of either the H4 tail or a patch on the nucleosome surface known as the LRS domain (Johnson et al. 1990; Thompson et al. 2003; Norris et al. 2008; Sampath et al. 2009). The BAH domain was sufficient for partial silencing when forced to dimerize, strengthening the notion that the BAH domain was sufficient for nucleosome recognition (Connelly et al. 2006). From intensive biochemical and biophysical study, the nucleosome-binding activity of the Sir3 BAH domain is now understood at atomic resolution, as described below.
The AAA+ ATPase domain:
The center span of Sir3 (amino acids 530–845) forms an AAA+ ATPase-like domain (Ehrentraut et al. 2011). AAA+ domains are found in a large number of proteins that use ATP hydrolysis (Neuwald et al. 1999). In Sir3, noncanonical residues in the ATP binding pocket likely occlude binding of nucleotide triphosphates (Ehrentraut et al. 2011). Nevertheless, this central portion of the protein contributes to silencing by interacting with Sir4 (Chang et al. 2003; King et al. 2006). Mutation of Sir3 at the protein-protein interface disrupts Sir4 binding and silent chromatin assembly (Ehrentraut et al. 2011). In addition to Sir4 binding, a small region upstream of the AAA+ ATPase-like domain (amino acids 456–481) binds Rap1 at silencers to aid nucleation of the Sir2/3/4 complex on chromatin (Moretti and Shore 2001; Chen et al. 2011).
The winged-helix domain:
The C-terminal 138 amino acids of Sir3 fold into a winged helix (Oppikofer et al. 2013). Although winged helices often function as DNA binding modules, the winged helix in Sir3 contains an unusual 30-amino acid insertion that mediates homodimerization (Gajiwala and Burley 2000; Liaw and Lustig 2006; Oppikofer et al. 2013). Dimerization may in fact be the sole function of this winged helix because an unrelated self-associating peptide can substitute for the domain in silencing (Oppikofer et al. 2013). Whether the winged helix dimerizes pairs of Sir3 proteins that bind the same nucleosome or separate nucleosomes is not yet known.
Histone binding by Sir3 BAH domain
When overexpressed, Sir3 extends silent chromatin domains without additional binding by either Sir2 or Sir4 (Renauld et al. 1993; Strahl-Bolsinger et al. 1997). This feature, together with genetic suppressor linkage of histone H4 and Sir3, suggested that the silencing factor associated with chromatin (Johnson et al. 1990). Direct association of Sir3 with nucleosomes was first shown with recombinant protein and later with native Sir3 from yeast (Georgel et al. 2001; Onishi et al. 2007). Binding affinity was diminished when critical residues like H4K16 or H3K79 were altered, or if the histone tails were removed (Liou et al. 2005; Onishi et al. 2007; McBryant et al. 2008; Johnson et al. 2009; Martino et al. 2009). Remarkably, the BAH domain alone recapitulated the binding properties of full-length Sir3, suggesting that the domain represents an independent nucleosome-binding module (Connelly et al. 2006; Onishi et al. 2007).
Crystallographic determination of the Sir3 BAH domain in complex with the nucleosome core particle was a major step forward in understanding silent chromatin (Armache et al. 2011). Obtaining cocrystals required the use of a SIR3 hypermorphic allele D205N, which suppresses silencing defects caused by mutations in histones and other silencing factors (Johnson et al. 1990; Liu and Lustig 1996; Norris et al. 2008). The mutation increases the affinity of Sir3 for nucleosomes in vitro (Connelly et al. 2006). In the structure, the BAH domain contacts all four histones with a protein-protein interface spanning an astonishing 1750 Å2 (Figure 5B). The points of physical contact between the BAH domain and nucleosome correspond extensively to residues identified by earlier genetic analyses. The H4 N-terminal tail, which is unstructured in the free nucleosome, folds onto the nucleosome surface with the critical H4K16 joining H3K79 of the LRS domain at the BAH interface. These two critical residues for silencing are separated by 18 Å, thereby highlighting the expanse of the protein-protein interface (Figure 5C). Acetylation of H4K16 or methylation of H3K79 would break some of the extended contact with Sir3 in this region and reduce binding affinity. Two additional relevant residues of the H4 tail, R17 and R19, contact phosphates of nucleosomal DNA in a cocrystal, forming a clamp that may favor silencing by increasing nucleosome stability (Wang et al. 2013). Otherwise, the nucleosomal DNA makes a surprisingly minimal contribution to the structure.
In crystals, a pair of BAH domains interact with the two symmetry-related faces of the nucleosome in a 2:1 stoichiometry (Armache et al. 2011). Stringent biophysical measurements confirmed a 2:1, Sir3-nucleosome stoichiometry in solution (Swygert et al. 2014). Moreover, a similar 2:1 stoichiometry was found with Sir2/3/4 complexes and nucleosomes when reconstitutions were performed with elevated Sir protein levels (Martino et al. 2009). It is not known whether Sir3 (or Sir2/3/4) binds nucleosomes as a dimer or two monomers. However, an attractive model holds that Sir3 dimers bridge pairs of nucleosomes. Bridging could occur between adjacent nucleosomes, or distant nucleosomes, to create higher-order structures.
Histone binding by other Sir3 domains
Histone binding activity has also been attributed to other parts of Sir3. A C-terminal fragment of the protein lacking the BAH domain binds nucleosomes, as well as peptides corresponding to just the H4 and H3 tails and the LRS domain (Hecht et al. 1995; Carmen et al. 2002; Santos-Rosa et al. 2004; Altaf et al. 2007; Ehrentraut et al. 2011). Mutational analyses indicate that this C-terminal domain (actually two domains in close proximity) contributes to silencing (Figure 5A) (Hecht et al. 1995; Stone et al. 2000; Buchberger et al. 2008; Ehrentraut et al. 2011). Importantly, acetylation or methylation of the critical lysines in histone peptides blocks binding by the Sir3 C-terminal. It is both intriguing and perplexing that both the N- and C- termini of Sir3 exhibit similar specificities for nucleosomal features. How might both domains operate within silent chromatin? One possibility is that the different Sir3 domains are used sequentially during a step-wise assembly of Sir3-nucleosome complexes. A second possibility is that the BAH domain and C-terminal of an individual Sir3 protein bind simultaneously to either different nucleosomes or to the opposite faces of the same nucleosome. Further study of the Sir3 C-terminal is required to understand its full contribution to nucleosome recognition.
Post- and cotranslational modification of Sir3
The N-terminal ends of Sir3 and Orc1 are processed during translation. After cleavage of the initiator methionine, the penultimate alanine is acetylated by the Nα acetyltransferase NatA (Geissenhoner et al. 2004; Wang et al. 2004). Mutations that block Nα acetylation of Sir3 abolish silencing and impede assembly of extended silent chromatin domains (Whiteway et al. 1987; Mullen et al. 1989; Ruault et al. 2011). Genetic studies suggested and biochemical studies proved that Sir3 Nα acetylation increases the affinity of Sir3 for nucleosomes (Connelly et al. 2006; Onishi et al. 2007; van Welsem et al. 2008; Sampath et al. 2009). Crystallographic studies showed that the Sir3 modification stabilizes the surface of the Sir3 BAH domain at the nucleosome-binding interface (Arnaudo et al. 2013; Yang et al. 2013).
As described earlier, Sir3 is also phosphorylated by the Slt2 mitogen-activated protein kinase on a patch of serines between residues 275 and 295 (Stone and Pillus 1996; Ai et al. 2002; Ray et al. 2003). A variety of environmental stresses trigger phosphorylation, which causes partial release of Sir proteins from telomeres and a commensurate increase in silencing at HM loci and the rDNA, as well as a shortened RLS. In this way, silent chromatin can respond to changes in the environment.
Sir4
Sir4 as a scaffold for Sir2/3/4 complex assembly
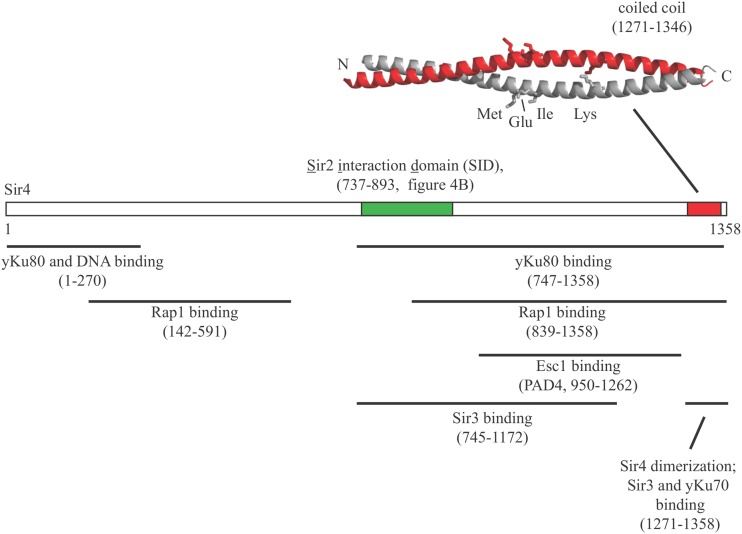
Sir4 binds each of the other Sir proteins directly, thus providing a scaffold for Sir2/3/4 complex assembly (Figure 6) (Hecht et al. 1996; Triolo and Sternglanz 1996; Moazed et al. 1997; Strahl-Bolsinger et al. 1997; Chang et al. 2003; Rudner et al. 2005; Cubizolles et al. 2006). The protein contains operationally-defined N- and C-terminal domains that complement one another in trans (Marshall et al. 1987; Kueng et al. 2012). The extreme C-terminal end contains the only recognizable structural motif to date: an α-helical domain that dimerizes via formation of a coiled coil (amino acids 1271–1346) (Chang et al. 2003; Murphy et al. 2003). The coiled-coil domain also interacts with Sir3 (Moretti et al. 1994; Moazed et al. 1997; Park et al. 1998). Point mutations that disrupt either dimerization or Sir3 binding abolish silencing (Chang et al. 2003; Rudner et al. 2005). A second Sir3 interaction module is located within amino acids 745–1172 (Liou et al. 2005).
Figure 6.
The structural and functional domains of Sir4. Each of the amino acids highlighted in the coiled-coil structure, M1307, E1310, I1311, and K1324 disrupts Sir3 binding when mutated. PDB accession number 1NYH.
A more central domain of Sir4 (amino acids 737–893) makes extensive contacts with Sir2 (Moazed et al. 1997; Cockell et al. 2000; Ghidelli et al. 2001; Hoppe et al. 2002; Hsu et al. 2013). Mutations at the protein-protein interface disrupt silencing at the HM loci but not at the rDNA where Sir2 acts as a subunit of RENT (Cuperus et al. 2000). Sir4 binding stimulates the deacetylase activity of Sir2 (Tanny et al. 2004; Cubizolles et al. 2006; Hsu et al. 2013). Allosteric regulation of this kind may limit Sir2 activity until a targeting factor, like Sir4, brings the enzyme to nucleation sites on chromatin.
The N-terminal half of Sir4 (amino acids 1–746) is dispensable under normal laboratory conditions (Kueng et al. 2012). However, under conditions where silencing is suboptimal the domain is essential. Biochemical studies indicate that the N-terminal 270 amino acids associate with linker DNA and increase the nucleosome-binding affinity of the Sir2/3/4 complex (Martino et al. 2009; Kueng et al. 2012). This N-terminal domain of Sir4 also contains cyclin-dependent kinase consensus sites, some of which are phosphorylated disproportionately in M phase when Sir proteins partially disperse from telomeres and silent chromatin is more susceptible to derepression (Aparicio and Gottschling 1994; Laroche et al. 2000). Mutation of the consensus sites alters silencing in a variety of assays, suggesting that Sir4 phosphorylation may regulate silent chromatin function, perhaps by modulating the DNA binding activity of the protein.
Sir4 protein levels might also be regulated to modulate silencing. Sir4 ubiquitylation is specified by Dia2, an F-box protein that is a component of the SCFDia2 E3 ubiquitin ligase (Burgess et al. 2012). In the absence of Dia2, Sir proteins mislocalize and silencing is disrupted. Sir4 levels also change in response to environmental stimuli. Levels of the protein drop precipitously in cells subjected to extended arrest by the α-factor mating pheromone (Larin et al. 2015).
Sir4 in recruitment of the Sir2/3/4 complex to chromatin
In addition to providing a scaffold for assembly of the Sir2/3/4 complex, Sir4 targets the complex to chromatin by associating with a variety of other proteins. As cited earlier, the protein makes numerous contacts with factors bound at silencers, including Sir1 (Triolo and Sternglanz 1996). The N- and C-termini of Sir4 associate with Rap1 (Moretti et al. 1994; Luo et al. 2002). Sir3 reinforces contacts between the Sir2/3/4 complex and silencers but Sir4 appears to be the linchpin. The protein persists at silencers in mutants lacking the other Sir proteins (Hoppe et al. 2002; Luo et al. 2002; Rusche et al. 2002). These findings suggest that recruitment of Sir4 is a key initial step in silent chromatin assembly
At telomeres, Sir4 is recruited by densely-packed Rap1 proteins that bind terminal telomeric TG1–3 sequences. The telomere end-binding protein Ku also contributes in recruitment. Ku subunits, yKu70 and yKu80, interact with both the N- and C-terminal ends of Sir4 (Tsukamoto et al. 1997; Laroche et al. 1998; Mishra and Shore 1999; Luo et al. 2002; Roy et al. 2004; Taddei et al. 2004; Hass and Zappulla 2015).
Sir4 has also been reported to bind the tails of histones H3 and H4 (Hecht et al. 1995). An interaction of this kind would contribute to formation of extended silent chromatin domains. To fairly judge the relevance of this activity, further work is required to identify the histone binding domain and to determine the specificity of the interaction.
Sir4 and heterochromatin anchoring at the nuclear membrane
In addition to determining where Sir2/3/4 complexes assemble on chromatin, Sir4 also specifies the localization of silent chromatin at the nuclear periphery. Anchoring to inner nuclear membrane is achieved through interactions between Sir4 and three different docking partners. The first, Esc1, is a membrane-associated factor that interacts with a C-terminal domain of Sir4 named partitioning and anchoring domain of Sir4 (PAD4; amino acids 950–1262) (Ansari and Gartenberg 1997; Andrulis et al. 2002; Taddei et al. 2004). Esc1 is still somewhat obscure. Recent work has shown that the protein associates with the Mlp proteins that form the nuclear basket of nuclear pore complexes, as well as a protein network on the inner nuclear membrane (Niepel et al. 2013, and references therein). A second anchor is created by interaction of Sir4 with a nucleoporin, Nup170 (Van de Vosse et al. 2013). The protein resides at the core of the nuclear pore complex, closely opposed to other nucleoporins that span the nuclear membrane. A third anchor is created by interaction of a central domain of Sir4 (amino acids 839–950) with Mps3, a SUN-domain protein initially known for its role in spindle pole body duplication (Bupp et al. 2007). The protein is now known to play a significant role in organizing chromosome ends at the edge of the nucleus during mitotic and meiotic growth. Sir4 also interacts with Ku, which in turn interacts with proteins at the nuclear membrane (Tsukamoto et al. 1997; Roy et al. 2004; Taddei et al. 2004; Schober et al. 2009). Anchoring of silent chromatin to each of these docking sites is cell cycle regulated and likely to be highly dynamic; residence times at the nuclear membrane span no more than minutes (Hediger et al. 2002).
Evolutionary considerations of SIR4
SIR4 is the least conserved of the SIR genes, having been found in only the Saccharomycetaceae family of yeasts (Fabre et al. 2005; Zill et al. 2010). In K. lactis, Candida galbrata, and S. bayanus, the gene maintains a conserved role in transcriptional silencing (Åström and Rine 1998; Iraqui et al. 2005; Gallagher et al. 2009). SIR4 has undergone a rapid rate of evolution that has been matched by, or perhaps driven by, rapid evolutionary changes in silencers and the Sir1 proteins that associate with them (Zill et al. 2010). This coevolutionary trait may highlight the shared role of Sir1 and Sir4 as adaptors that link highly-conserved silencer binding proteins, like ORC, to other more conserved components of silent chromatin, like Sir2 and Sir3. Interestingly, S. cerevisiae contains a distantly-related SIR4 paralog named ASF2 with no known physiological function. Overexpression of ASF2 disrupts silencing, possibly because the gene product can compete with Sir4 for binding Sir3 (Le et al. 1997; Buchberger et al. 2008).
Molecular Basis of Silent Chromatin Assembly
“Establishment” and “maintenance” of silent chromatin
Long before silent chromatin assembly could be described in molecular terms, silencing factors were assigned operationally-defined roles on the basis of the behavior of mutants. Whereas some factors were required continuously to maintain silencing, other factors seemed to participate primarily in establishing the silent state. Operational definitions like establishment and maintenance have been useful in framing the subsequent genetic and biochemical data that ultimately fleshed out the true molecular roles of silencing factors. SIR3, for example, was initially known as a gene required continuously to maintain silent chromatin (Miller and Nasmyth 1984). The gene product is now known to possess a central nucleosome-binding motif (Armache et al. 2011). The terms establishment and maintenance are still useful today, but they are now used in parallel with terminology like nucleation, spreading, and maturation that better describe the molecular details of silent chromatin.
Nucleation and spreading in silent chromatin assembly
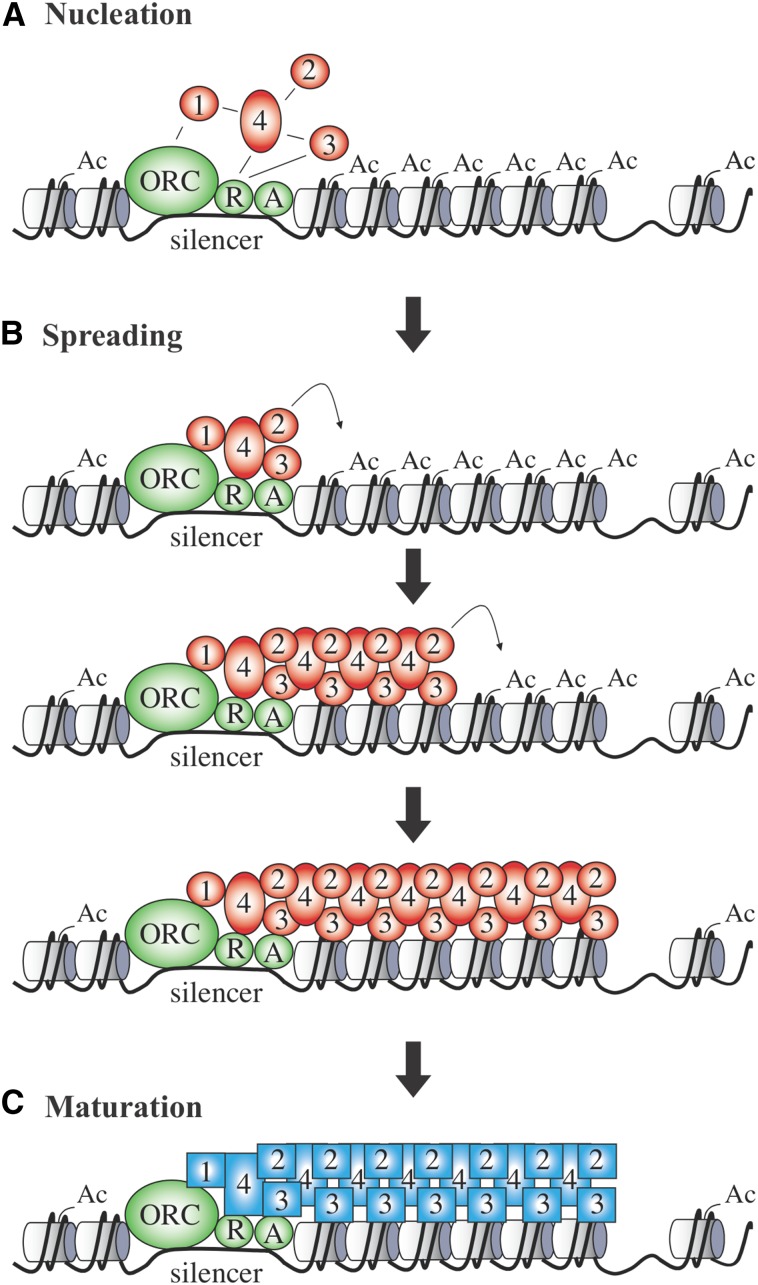
At the molecular level, assembly of silent chromatin occurs in at least two steps: nucleation and spreading. Nucleation describes the initial step when the Sir2/3/4 complex is recruited to silencers. The much-less-understood spreading step describes the subsequent process by which the complex assembles an extended domain of silent chromatin. Nucleation and spreading steps are coupled because both arise from the intrinsic properties of the Sir proteins. Mutations and other experimental manipulations are required to study nucleation without spreading and spreading without nucleation.
Nucleation is a relatively straightforward process. A network of interactions recruits the Sir2/3/4 complex to proteins bound at silencers (Figure 7A). Sir3 and Sir4 associate with Rap1 and Sir4 associates with ORC-bound Sir1 (Moretti et al. 1994; Triolo and Sternglanz 1996; Moretti and Shore 2001; Chen et al. 2011). In catalytic mutants of Sir2, the Sir2/3/4 complex is restricted to silencers (Hoppe et al. 2002; Rusche et al. 2002; Ellahi et al. 2015). Thus, the action of Sir2 on histone substrates triggers the transition from nucleation to spreading. The original sequential model of spreading posits that Sir2 acts first on nucleosomes adjacent to silencers, creating additional recruitment sites for Sir2/3/4 complexes (Hoppe et al. 2002; Luo et al. 2002; Rusche et al. 2002). The deacetylated H4 tail, amino acid H4K16 in particular, is a preferred binding site for Sir3 (Hecht et al. 1995; Liou et al. 2005; Johnson et al. 2009). A more elaborate view, based on an unexpected affinity of the Sir2/3/4 complex for acetylated H4K16, holds that the complex binds acetylated nucleosomes first and then acquires additional stability by H4K16 deacetylation and docking of Sir3 to the deacetylated tails (Oppikofer et al. 2011). Either way, the sequential-spreading model holds that rounds of Sir2/3/4 binding, histone deacetylation, and interactions between Sir2/3/4 complexes expand the growing silent chromatin domain until a barrier is reached or the pool of free Sir proteins falls below a threshold necessary for efficient binding (Figure 7B). According to this view, sequential spreading of Sir2/3/4 complexes is analogous to a linear polymerization reaction.
Figure 7.
Nucleation, spreading, and maturation of silent chromatin. (A) Nucleation. The known network of interactions between Sir proteins and silencer-bound factors is shown. (B) Spreading. Deacetylation of neighboring histones by Sir2 creates additional binding sites for Sir3 and Sir4. Successive rounds of histone deacetylation and Sir2/3/4 binding expands the silent chromatin domain until a barrier is reached. (C) Maturation. Numerous conditions have been found where Sir2/3/4 spreading does not produce transcriptional repression. These circumstances suggest that nascent silent chromatin may undergo a final maturation step (e.g., removal of H3K79 methylation) to yield transcriptional silencing.
The sequential-spreading model predicts that silent chromatin assembles uninterrupted structures, emanating from silencers to span across the entire silenced domain. At HMR and HML, this must be inferred because some regions of the loci are refractory to analysis by traditional ChIP methods (Thurtle and Rine 2014). At telomeres, silent chromatin domains also challenge the simple model because blocks of silent chromatin are small and interspersed with active chromatin segments (Fourel et al. 1999; Pryde and Louis 1999; Zill et al. 2010; Takahashi et al. 2011; Ellahi et al. 2015). A sequential-spreading model cannot account for discontinuities in silent chromatin domains. Lastly, sequential spreading of silent chromatin would likely be complicated by the highly-dynamic nature of Sir proteins, which are thought to equilibrate on and off silent chromatin rapidly (Cheng and Gartenberg 2000). Constant exchange of the proteins might create an unstable platform upon which to sequentially expand silent chromatin.
Attempts to visualize sequential spreading of the Sir2/3/4 complex have been met with mixed success. In various studies, de novo assembly has been triggered by reintroduction of a Sir protein that was experimentally withheld (usually Sir3). At model telomeres and sites distant from strong silencers, time-dependent expansion of silent chromatin domains was detected (Katan-Khaykovich and Struhl 2005; Lynch and Rusche 2009; Radman-Livaja et al. 2011). Even in these best-case scenarios, however, true processivity of the spreading reaction (i.e., template commitment) was not demonstrated. At the better-characterized HMR locus, Sir proteins were found to increase evenly across the entire domain over time. A simple explanation holds that the experimental methods used lacked the temporal resolution and/or synchronicity to observe spreading. An alternative explanation is that spreading occurs in a nonsequential fashion (Lynch and Rusche 2009). If a silencer were to contact adjacent nucleosomes as well as distant nucleosomes, say through looping out the intervening DNA, then small domains of silent chromatin could assemble in a piecemeal fashion. Such a nonlinear model for spreading could also help explain the discontinuities in silent chromatin at telomeres.
Irrespective of whether silent chromatin assembles in a linear or nonlinear fashion, transcriptional silencing occurs rapidly. Following induction of Sir3 in population-based assays, messenger RNAs (mRNAs) from genes in newly-silenced regions diminished significantly within a single cell cycle (Katan-Khaykovich and Struhl 2005; Lynch and Rusche 2009). Assays based on single cell measurements showed an even greater speed of silencing onset. Using restoration of mating competence as a functional criterion, de novo silencing occurred within 1–2 generations in most cells expressing native Sir proteins (Osborne et al. 2009). Taken together, these results indicate that a silent domain can form over extended domains to repress resident genes rapidly and efficiently.
Lessons learned from silent chromatin reconstitution
The interactions between Sir proteins and nucleosomes that underlie spreading have been reproduced with purified components. Recombinant Sir2, Sir3, and Sir4 form a 1:1:1 stoichiometric complex (Liou et al. 2005; Cubizolles et al. 2006). The complex binds both nucleosome core particles and reconstituted nucleosome arrays. The stringent specificity for deacetylated H4K16 seen with Sir3 alone is muted with Sir2/3/4, perhaps owing to additional nucleosome contacts provided by the other Sir proteins (Johnson et al. 2009; Martino et al. 2009). Nevertheless, NAD-dependent deacetylation of H4K16 by Sir2 increased the affinity of Sir2/3/4 for nucleosomes (Oppikofer et al. 2011). Curiously, addition of OAADPr, the small molecule byproduct of the NAD-dependent reaction, increased the affinity of Sir2/3/4 for nucleosomes (Martino et al. 2009). OAADPr also induced conformational and stoichiometric changes in the Sir2/3/4 complex in the absence of nucleosomes (Liou et al. 2005). These findings suggest that OAADPr might contribute to silent chromatin assembly as an allosteric effector. A binding site for the metabolite has yet to be defined within the Sir2/3/4 complex. To test whether OAADPr was essential for silencing and whether other deacetylases could substitute for Sir2, silent chromatin was assembled in vivo with Hos3, an Rpd3-family deacetylase that neither consumes NAD nor produces OAADPr (Chou et al. 2008). Hos3 was targeted to assembling silent chromatin domains by creating a Sir3-Hos3 fusion protein. The chimera yielded robust transcriptional silencing, even in strains that lacked all of the NAD-dependent deacetylases. Thus, if OAADPr contributes to silencing, it is not likely to make an essential contribution.
Reconstitution of Sir2/3/4 with oligonucleosome arrays created higher-order structures, visualized as compact clusters and intriguing fibers on EM grids (Onishi et al. 2007; Johnson et al. 2009). The reconstituted material blocked digestion by nucleases and transcription by RNA polymerases, like silent chromatin in vivo (Johnson et al. 2009; Oppikofer et al. 2011; Kitada et al. 2012). More detailed studies of transcription by Pol II showed that binding of an upstream transcriptional activator was not impeded. However, interactions between the activator and coactivators were disrupted and elongation by initiated polymerases was hindered (Johnson et al. 2013). Thus, it appears that silent chromatin has the capacity to block latter stages in transcription in cases where transcription activators evade steric occlusion.
Does silencing require a maturation step after Sir protein binding?
The simplest form of the nucleation and spreading model predicts that silencing should occur once Sir proteins assemble on chromatin. Several studies, however, have described experimental situations where association of Sir2/3/4 with chromatin is not sufficient (for examples, see Lau et al. 2002; Kirchmaier and Rine 2006; Yang and Kirchmaier 2006; Xu et al. 2007). Results of this nature suggest that at least one additional maturation step is involved in creating the silent state (Figure 7C). Conceptually, maturation could involve conformational changes of bound components, the acquisition or removal of additional chromatin modifications, and/or the binding of small molecule regulators, like OAADPr. In kinetic studies of de novo silencing establishment, loss of H3K4 and H3K79 methylation were among the last events observed in silent chromatin assembly (Katan-Khaykovich and Struhl 2005). Moreover, elimination of the enzymes that methylate these residues accelerated the rate of silencing onset (Katan-Khaykovich and Struhl 2005; Osborne et al. 2009; Osborne et al. 2011). Additionally, in a wild-type population of cells with variegated expression of a telomeric reporter gene, histone methylation was found to be the only chromatin feature to distinguish the cells that still permitted transcription of the reporter from those that did not (Kitada et al. 2012). Sir proteins bound to the reporter gene equivalently in both silent and nonsilent cells. Coupled with the observation that H3K79 methylation prevents transcriptional repression of reconstituted silent chromatin, these studies suggest that demethylation of H3K79 may represent a late or final maturation step during silent chromatin assembly.
At least three different pathways attenuate histone H3 methylation in silent chromatin domains. First, Sir4 recruits Ubp10, a protease that removes ubiquitin from K123 of H2B (Gardner et al. 2005). Ubiquitylation of H2B is a prerequisite for methylation of H3K4 and H3K79. In ubp10 mutants, silencing was compromised and methylation of telomeric histone H3 increased. Second, the Ino80 chromatin-remodeling complex is recruited to silent chromatin (Xue et al. 2015). Ino80 suppresses transcription-associated H3K79 methylation. In mutants lacking Ino80 subunits, silencing was compromised and methylation of telomeric H3K79 increased. Third, H3K79 methylation might also be diminished passively by reassembly of chromatin with unmodified histones following replication fork passage (Katan-Khaykovich and Struhl 2005). In assays of de novo silencing establishment, replicating chromatin templates lost H3K79 methylation more rapidly than nonreplicating DNA circles.
Barriers and Antisilencing
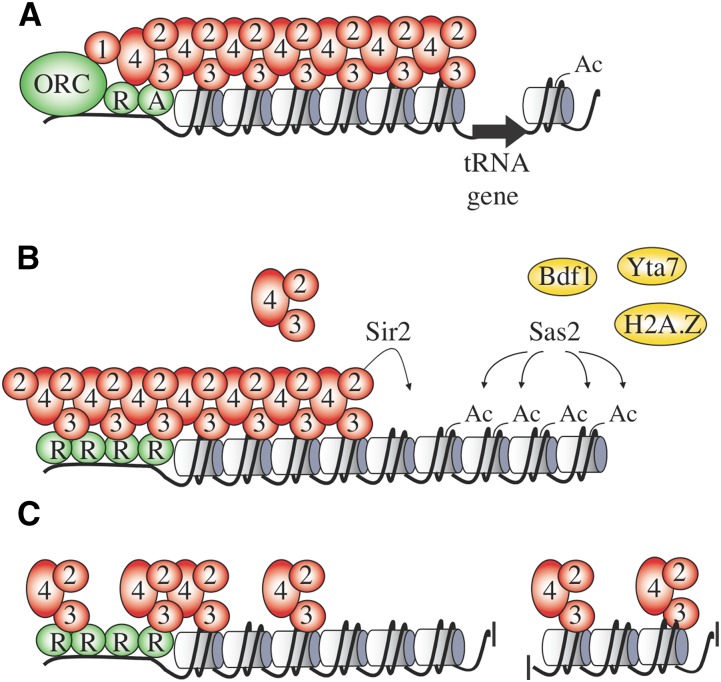
Evolutionary forces likely restricted the affinities of the Sir proteins toward one another and toward chromatin to prevent promiscuous creation and undesirable expansion of silent chromatin domains (Rusche and Lynch 2009). In addition to these intrinsic limitations, two types of physical barriers to silent chromatin spreading have been described. The first consists of discrete boundary elements that coincide with strong promoters or DNA-bound transcription factors (Figure 8A). The best-characterized boundary element is the threonine transfer RNA (tRNA) gene on the telomere-proximal side of HMR (Donze et al. 1999; Donze and Kamakaka 2001). Pol III transcription factors TFIIIB and TFIIIC are required for boundary function, but transcription by Pol III is dispensable (Simms et al. 2008; Valenzuela et al. 2009). How the transcription factors block the spread of silent chromatin is uncertain, but it is likely related to the discontinuity in the nucleosomal template caused by missing or rapidly-exchanging histones. Chromatin modifiers that both create a nucleosome-free region around the tRNA gene and facilitate recruitment of transcription factors, like RSC and Isw2 complexes, promote barrier activity by the gene, possibly in conjunction with the actions of DNA polymerase ε and the Rtt109 histone acetyltransferase (Tackett et al. 2005; Dhillon et al. 2009). Contact of chromatin with nuclear pore complexes was initially thought to also be an underlying feature of silent chromatin barriers (Ishii et al. 2002). While the tRNA boundary element at HMR does indeed contact nucleoporins of nuclear pore complexes, it is now clear that the contact does not contribute to the barrier function of the gene (Ruben et al. 2011).
Figure 8.
Antisilencing and barriers to silent chromatin spreading. (A) Discontinuities in chromatin created by highly-dynamic or displaced nucleosomes disfavor silent chromatin spreading. A barrier created by the nucleosome-depleted tRNA gene next to HMR is shown. (B) At silent chromatin borders, the Sas2 acetyltransferase and Sir2 histone deacetylase compete to determine the acetylation state of H4K16. The histone tail is then bound by effector proteins that demarcate the silent chromatin boundary. In the case of deacetylation, the Sir2/3/4 complex binds to extend the silent chromatin domain and in the case of acetylation, barrier proteins Bdf1 and Yta7 bind to create a boundary. H4K16ac also favors incorporation of histone variant H2A.Z, another barrier to silent chromatin spreading. (C) Loss of genome-wide antisilencing. Global histone modifications that disfavor Sir2/3/4 complex binding increase the available pool of Sir proteins available for binding at telomeres. When these histone modifications are lost, as shown in the figure, Sir proteins are titrated from bona fide silent chromatin sites by nonspecific binding elsewhere.
Other discrete barriers include the upstream activation sequence of the CHA1 gene near HML (Donze and Kamakaka 2001) and binding sites for transcription factors Reb1 and Tbf1 within subtelomeric repeat sequences [generically referred to as subtelomeric antisilencing regions (STARs)] (Fourel et al. 1999). A unifying feature of these barriers may be that they too favor the formation of nucleosome-free regions (Moreira and Holmberg 1998; Hartley and Madhani 2009). Indeed, synthetic constructs that disfavor nucleosome formation also create discrete barriers to silent chromatin spreading (Bi and Broach 1999; Bi et al. 2004).
In addition to discrete boundary elements, like those described above, a second class of barrier is defined by active chromatin states that impede Sir protein spreading (Figure 8B). These two classes of barriers need not be mutually exclusive if chromatin that disfavors silent chromatin assembly abuts a nucleosome-free region (Oki and Kamakaka 2005). One such active chromatin barrier is created by Sas2, a histone acetyltransferase that accounts for the bulk of genomic H4K16 acetylation (H4K16ac) (Meijsing and Ehrenhofer-Murray 2001; Osada et al. 2001). Sas2 was initially identified as a paradoxical factor that seemed to hinder silencing at one HM locus but favor silencing at the other (Reifsnyder et al. 1996; Ehrenhofer-Murray et al. 1997). Its role in barrier formation became clear with the study of silencing at telomeres. Silent chromatin domains were found to expand outward from telomeric ends when Sas2 was eliminated whereas the domains shrunk when Sas2 was overexpressed (Kimura et al. 2002; Suka et al. 2002). Thus, a barrier to spreading is created by the competition of Sas2 and Sir2 enzymes over the fate of H4K16ac. Sir2 deacetylation dominates at sites near the telomeric nucleation of silent chromatin whereas Sas2 acetylation dominates at sites more distal from the telomere.
If Sir3 and Sir4 are considered effectors of silent chromatin by binding the deacetylated lysines created by Sir2, then effectors of the H4K16ac created by Sas2 should also exist. Indeed, bromodomain-containing proteins Bdf1 and Yta7 bind H4K16ac and block expansion of silent chromatin domains (Ladurner et al. 2003; Jambunathan et al. 2005; Tackett et al. 2005). In addition, H4 acetylation by Sas2 facilitates the incorporation of the histone variant H2A.Z, which acts as another inhibitor to silent chromatin spreading (Meneghini et al. 2003; Shia et al. 2006). H2A.Z is enriched at the borders of nucleosome-free regions of gene promoters, including those of telomere-proximal genes where barrier activity is observed (Raisner et al. 2005). H2A.Z is also acetylated, and according to one study the modification is required for barrier function by the histone variant (Babiarz et al. 2006).
One caveat to the Sas2/Sir2 competition model emerges from high-resolution maps of Sir protein binding (Thurtle and Rine 2014; Ellahi et al. 2015). The competition model predicts that silent chromatin domains abut domains with high levels of H4K16ac. Surprisingly, domains lacking H4K16ac extend far beyond where the Sir proteins can be detected. One possible explanation is that Sir protein binding is more transient in the transition zones, leaving deacetylated nucleosomes without long-lived binding partners. A second possibility is that Sir2 within silent chromatin acts far beyond the nucleosomes to which it is bound, thereby creating zones of potential Sir3 binding sites. A final possibility is that additional deacetylases, such as Rpd3, participate in creating transition zones (Ehrentraut et al. 2010).
While the consequences of Sas2 on silencing are evident near telomeres, it is important to recognize that Sas2 acts genome wide. Therefore, histone acetylation by the enzyme might also block silencing at distal sites where Sir proteins would otherwise assemble promiscuously. The term antisilencing is often used to describe such processes where chromatin modifications prevent inappropriate binding of Sir proteins. By reducing the loss of Sir proteins to off-target genome-wide binding, antisilencing increases the concentration of Sir proteins available for bona fide silent chromatin assembly (Figure 8C). Thus, antisilencing can be viewed as a mechanism that promotes efficiency and specificity of silencing factors that are limiting in the nucleus. As a general rule of thumb, any process that promotes genome-wide incorporation of histone variants or histone modifications that hinder Sir2/3/4 binding should be considered as a mediator of antisilencing. Two particular histone modifications, both methylations of lysines in histone H3, illustrate the antisilencing concept in the following paragraphs.
The Set1 enzyme methylates lysine 4 on the N-terminal tail of histone H3 in a transcription-coupled process (Briggs et al. 2001; Roguev et al. 2001; Krogan et al. 2003). Silenced loci are devoid of the modification, and methylation of H3K4 in vitro blocks Sir3 binding (Santos-Rosa et al. 2004). Mutants lacking Set1 exhibit transcriptional silencing defects (Nislow et al. 1997). Moreover, inactivation of the methyltransferase causes dispersal of Sir3 from typical silent chromatin locations and corresponding increases in Sir3 binding at subtelomeric sites and euchromatic sites far from telomeres (Santos-Rosa et al. 2004; Venkatasubrahmanyam et al. 2007).
A second histone methyltransferase, Dot1, modifies lysine H3K79 on the core of the histone octamer in a reaction stimulated by prior H4K16ac (Lacoste et al. 2002; Ng et al. 2002; van Leeuwen et al. 2002; Altaf et al. 2007). The majority of the genome carries the mono-, di-, and trimethylated H3K79 but the modifications are missing from silenced chromatin domains (Ng et al. 2002, 2003). H3K79 methylation interferes with Sir3 binding in most in vitro assays and blocks silencing by Sir-bound nucleosomes in vivo (Altaf et al. 2007; Onishi et al. 2007; Martino et al. 2009; Kitada et al. 2012). In dot1 mutants, Sir3 redistributes from telomeric regions, although the magnitude of the redistribution may be limited (San-Segundo and Roeder 2000; Takahashi et al. 2011). At a minimum, a prominent shift of Sir3 from one telomeric end to subtelomeric domains was reported (van Leeuwen et al. 2002).
Silent Chromatin Establishment and the Cell Cycle
A requirement for cell cycle progression in silent chromatin establishment
Early work from the Nasmyth laboratory was the first to suggest that establishment of transcriptional silencing was subject to cell cycle-specific regulation. These first experiments used strains with temperature-sensitive alleles of the SIR genes. Upon restoration to permissive temperature, silencing only reemerged if the cells progressed through a portion of the cell cycle that contained S phase (Miller and Nasmyth 1984). Thus, it was concluded that de novo establishment of silencing requires some event during S phase. Wild-type cells already possess silent chromatin and never establish silencing de novo. However, the rationale behind investigations like these was that regulatory events uncovered in such controlled situations would correspond to normal processes during the heritable propagation of silencing.
Obligatory remodeling of chromatin during replication-fork passage presented a logical explanation for the S-phase requirement in establishment. That silencers curiously possessed DNA replication origins lent support to this view. However, an obvious role of DNA replication in triggering silent chromatin assembly was challenged with three types of experiments. First, genetic studies identified separation-of-function mutants in ORC that supported replication but not silencing (Bell et al. 1995; Dillin and Rine 1997). Second, the DNA replication origins of silencers were found to be dispensable for silencing if Sir1 was tethered directly to DNA (Fox et al. 1997). Third, silencing could be established on DNA rings that did not replicate at all (Kirchmaier and Rine 2001; Li et al. 2001). Taken together, these results indicated that neither origins of replication nor passage of replication forks were needed to remodel chromatin for silencing during S phase.
Further work has sought to identify a single elusive cell cycle event required for establishment of silencing. Several studies point to the erasure of euchromatic features, like the removal of H3K79 methylation described above (Katan-Khaykovich and Struhl 2005; Osborne et al. 2009; Osborne et al. 2011). Complementary to this view were reports that the timing of silencing establishment was promoter-specific and not strictly S-phase dependent. Specifically, while the a1 mating-type gene at HMR was largely repressed during S phase, additional silencing occurred only with passage from M phase to G1 (Lau et al. 2002). Similarly, silencing of a telomeric URA3 reporter occurred only during passage from M to G1 phases (Martins-Taylor et al. 2004; Martins-Taylor et al. 2011). Finally, the α1 mating-type gene at HML was silenced in cells that were not cycling at all (Ren et al. 2010; Lazarus and Holmes 2011). That the different reporter genes shut off in such different ways during establishment assays may indicate that the timing of silencing onset is regulated predominantly by the factors that perpetuate transcription of the genes, rather than the action of a silent chromatin-specific trigger.
Potential cell cycle regulators of silent chromatin establishment
Other support for DNA replication in silencing establishment:
Over the years of searching for cell cycle-dependent triggers of silent chromatin establishment, focus has recurrently fallen on two processes. Foremost, DNA replication has continued to be a strong candidate despite evidence that a particular locus need not be replicated to establish silencing. Repeatedly, mutants in replication fork-associated factors [DNA helicase, Polε, proliferating cell nuclear antigen (PCNA), and RF-C] and replication-coupled chromatin assembly factors (CAF-1, Rtt106, and Asf1) were found to compromise silencing (Kaufman et al. 1997; Singer et al. 1998; Ehrenhofer-Murray et al. 1999; Smith et al. 1999; Zhang et al. 2000; Iida and Araki 2004; Huang et al. 2005; Miller et al. 2010). Many of these factors can be linked to genome-wide replication deposition of histones with acetylation patterns that inhibit Sir2/3/4 binding (summarized in Young and Kirchmaier 2013). Global suppression of Sir2/3/4 binding by an antisilencing mechanism, as discussed earlier, can favor silencing at designated locations by increasing the pool of free Sir proteins. Thus, an apparent role for DNA replication in silencing establishment inferred from the behavior of replication mutants may in reality be an indirect consequence of misassembling chromatin genome wide. The common use of the nucleotide biosynthetic gene URA3 as a reporter in silencing assays has further clouded the issue of DNA replication in silencing establishment. The Stillman laboratory recently showed that mutations in PCNA, and possibly other genes involved in replication-coupled chromatin assembly, sensitize cells to 5-fluoroorotic acid (5FOA), the drug used to monitor URA3 expression (Rossmann et al. 2011). Hypersensitivity to 5FOA can easily be mistaken as derepression in a URA3-based silencing assay.
The notion of DNA replication contributing to silencing establishment should not be discounted entirely. Intriguingly, tight protein-DNA interactions, like those formed by the Pol III machinery at tRNA genes, promote recruitment of Sir2/3/4 complexes to chromatin in certain contexts (Dubarry et al. 2011). Typically, this activity is suppressed by the Rrm3 helicase that facilitates replication-fork passage through nonhistone DNA complexes. The SWI/SNF chromatin remodeler might further attenuate accumulation of the Sir proteins at sites of replication stress (Manning and Peterson 2014). SWI/SNF interacts with Sir3 and displaces the repressor from chromatin (Sinha et al. 2009). These results raise the possibility that replication-fork pausing at the tightly-bound proteins of silencers might influence establishment of silencing during S phase (Nikolov and Taddei 2015).
Cohesin in silencing establishment:
Sister chromatid cohesion is the second cell cycle-dependent process that has often been linked to cell cycle control of silencing. Cohesion is mediated by cohesin, a protein complex that holds sister chromatids together from the time they are replicated during S phase until anaphase onset. Cohesin is also a negative regulator of silencing establishment, blocking complete silent chromatin assembly and transcriptional repression until after cohesin is destroyed in M phase (Lau et al. 2002; Martins-Taylor et al. 2011). The complex appears to act directly because it binds at HMR to mediate cohesion of the locus (Chang et al. 2005). Cohesin action at silent loci is regulated by the extreme C-terminal of Sir2 (Wu et al. 2011). Interestingly, silencing persists in a sir2 mutant that blocks cohesion but still recruits cohesin to chromatin (Chen et al. 2016). This result indicates that cohesion is dispensable for silencing even if the cohesin complex is not.
In budding yeast, cohesin loads onto chromatin at discrete sites (usually heavily-transcribed genes) and then migrates to distal sites where it accumulates (Lengronne et al. 2004; Lopez-Serra et al. 2014). In the vicinity of HMR, cohesin loading is mediated by the tRNA gene that demarcates the telomere-proximal, silent chromatin boundary (Dubey and Gartenberg 2007). Intriguingly, cohesin subunits are required for boundary function of the tRNA gene and tethering cohesin subunits directly to DNA creates a synthetic boundary element (Donze et al. 1999; Martins-Taylor et al. 2011). The tRNA gene (and presumably the cohesin that it loads) appears to be a modular regulatory element of silencing: when the gene was deleted from HMR, silencing could be established in all phases of the cell cycle; when the gene was transferred to HML it imposed cell cycle constraints on silencing establishment (Lazarus and Holmes 2011). Histone variant H2A.Z, which flanks tRNA genes and other nucleosome-free regions genome wide, has also been implicated in cell cycle control of silencing and cohesin function (Martins-Taylor et al. 2011; Sharma et al. 2013). These results suggest that the cohesin complex not only impedes establishment of silencing in a cell cycle-dependent manner but also limits the expansion of silent chromatin domains by blocking spreading. The molecular basis for regulation of these events by cohesin has not been elucidated.
Variegated Expression and Inheritance
How silent chromatin is propagated from one generation to the next may be the most-fascinating and least-understood aspect of the silencing phenomenon. The terms establishment and maintenance figure prominently in the discussion. Faithful inheritance requires that silent chromatin domains be maintained stably once they have assembled, and that they be reestablished quickly following perturbations imposed by DNA replication and other restrictive cell cycle events. Inheritance of silent chromatin can be a remarkably efficient process. The mating-type genes at the core of the HM loci derepress exceedingly rarely (Dodson and Rine 2015). Even then, derepression events are transient, yielding only low levels of mRNA. It is precisely this high efficiency that makes it important to distinguish faithful inheritance from efficient de novo establishment each cell cycle; a situation where inheritance would not be required at all.
The behavior of reporter genes at the edges of silent chromatin domains has offered a window into the nature of inheritance. When reporters were placed at an artificial telomere or beyond the silencers at HMR expression, their expression was variegated (Gottschling et al. 1990; Simms et al. 2008; Mano et al. 2013). In the case of the ADE2 reporter, silenced cells acquired a red pigment whereas genetically identical, nonsilent cells remained white. Importantly, the silenced and nonsilenced expression states were heritably propagated in lineages of cells, yielding colonies with both red and white sectors (Figure 9). Switches occurred between expression states but the switched states were also heritable, as evidenced by sectors within sectors. These experiments showed that silencing could be maintained and inherited in a subset of cells. Switching events that yielded nonsilent cells indicated that maintenance and/or inheritance occasionally failed. Finally, the persistence of a nonsilent subset of cells showed that establishment was not guaranteed. The situation was highly reminiscent of position-effect variegation of reporter genes at the edges of heterochromatin domains in Drosophila. In yeast, the phenomenon was first observed with reporter genes in telomeric silent chromatin, and thus it was named telomere position effect or TPE.
Figure 9.
Epigenetic inheritance of the silent state. Genetically identical colonies with a telomeric ADE2 reporter were plated on low adenine media. The red sectors contain lineages of cells with ADE2 in the silenced state. The white sectors contain cell lineages with the ADE2 gene in the nonsilent state. Red and white sectors intermingle within a given colony, indicating that cells periodically switch between silent and nonsilent states.
Similar patterns of variegated expression and epigenetic inheritance were elicited from silenced genes at the core of the HM loci. However, to see the phenomenon the loci had to be sensitized by partial disruption of silencers, mutation of Rap1 or complete loss of Sir1 (Pillus and Rine 1989; Mahoney et al. 1991; Sussel et al. 1993; Xu et al. 2006). The existence of a stable subset of nonsilent cells cemented the notion that silencers and the proteins that bind them promote establishment of silencing. These factors also suppressed switches from the silent to nonsilent state, suggesting that they also promoted maintenance and inheritance. Direct examination showed that silencers do indeed act continuously to maintain the silent state even after silent chromatin has assembled. When the elements were removed from HM loci by site-specific recombination, silent chromatin disassembled rapidly in actively growing cells, and even in cells arrested for growth if no proto-silencers were present (Holmes and Broach 1996; Cheng and Gartenberg 2000). Sir1, originally thought to be solely a mediator of establishment, was also shown to act continuously. HM loci derepressed 35 times more frequently in strains depleted of the gene by outcrossing (Dodson and Rine 2015).
If inheritance was determined solely by events at silencers then fluctuations in silencing at one silent domain might occur independently of fluctuations at another. This question has been addressed with pairs of fluorescent reporter genes integrated at separate silenced locations. In one study, correlated switches in expression state were observed in cell lineages when the genes were placed at the edges of the silent chromatin domains at HMR and HML (Mano et al. 2013). These results indicate that inheritance of silent chromatin is influenced by a property that is generic to the cell and not specific to the locus. For example, stochastic changes in levels of a limiting silencing factor, like Sir3 or Sir4, could impart cell-wide effects. Additionally, physical interactions between the distant silent chromatin domains could help coordinate the response to limiting silencing factors (Valenzuela et al. 2008; Miele et al. 2009). While these findings agree with earlier studies that relied on genetic criteria (Pillus and Rine 1989; Sussel et al. 1993), it should be noted that a study using sir1 strains and similar reporter gene constructs arrived at the opposite conclusion. In that case, the variegated silencing patterns of HMR and HML were not correlated, even when the reporter genes were placed at homologous HML loci of a diploid (Xu et al. 2006). Perhaps residual silencer activity becomes a dominant cis-acting determinant of inheritance in the absence of Sir1.
Demonstration that inheritance occurs, even if in only a subset of cells, indicates that silent chromatin is self-templating during cell duplication. How might this occur? Parental nucleosomes distribute randomly between sister chromatids in the wake of the replication fork. If Sir2/3/4 complexes remain associated with parental nucleosomes, then interactions between Sir complexes and further recruitment by silencers could foster replacement Sir complexes for newly-assembled nucleosomes. Even if some Sir complexes are stripped from nucleosomes during replication, the absence of histone modifications in parental nucleosomes and the absence of transcription-associated modifications in newly-assembled histone octamers may favor rapid reassembly directed by silencers. According to this view, inheritance of the silent state is a facilitated-reassembly process following replication-induced perturbations of silent chromatin.
Heterochromatin Distinctions Between S. cerevisiae and S. pombe
The contributions of both transcriptional and post-transcriptional mechanisms to silencing are consistent with the mechanism of heterochromatin formation in S. pombe, where an active RNAi system plays a major role in the establishment of silencing (Hall et al. 2002; Volpe et al. 2002). RNAi-dependent silencing in S. pombe involves methylation of H3K9 by the histone methyltransferase Clr4 (Volpe et al. 2002). Methylated H3K9 is recognized and bound by Swi6, the HP1 homolog of S. pombe (Bannister et al. 2001; Hall et al. 2002). Importantly, SpSir2 is also critical for heterochromatin formation at telomeres, centromeres, silent mating-type loci, and the rDNA. In this context H3K9 must be deacetylated prior to methylation, and deacetylation of H4K16 is also required for silencing in S. pombe (Shankaranarayana et al. 2003; Wiren et al. 2005). S. cerevisiae relies primarily on histone deacetylation and lacks other typical features of eukaryotic heterochromatin, like H3K9 methylation, an HP1/Swi6 homolog, and an RNAi system. Structural contributions from the Sir proteins likely functionally substitute for HP1 and other H3K9-associated factors such as the RNA-induced transcriptional silencing complex (Verdel et al. 2004). The addition of H3K9 methylation appears to add a layer of more stable trans-generational epigenetic inheritance (see section on Variegated Expression and Inheritance).
rDNA Silencing
Overview
The genes encoding ribosomal RNA (rRNA) are transcribed at an extraordinarily-high level, accounting for ∼60% of the total cellular transcription in yeast (Warner 1999). Given this extreme activity, the discovery that the chromosomal domain containing the rRNA genes (rDNA) hinders transcription by Pol II came as an unexpected surprise (Bryk et al. 1997; Smith and Boeke 1997). The phenomenon resembled silencing at the HM loci and telomeres but was different in important ways. Most prominently, SIR2 was required but the other SIR genes were not. In this section we describe the initial characterization of rDNA silencing, more recent mechanistic analyses, and the interesting links between rDNA silencing, rDNA stability, and aging.
The yeast rDNA locus consists of 150–200 tandem copies of a 9.1-kb repeat on chromosome XII that packs into the distinct crescent-shaped nucleolus at the edge of the nucleus (Figure 10A) (Petes 1979). The repeat units encode alternating copies of the Pol I-transcribed 35S precursor rRNA and the Pol III-transcribed 5S RNA, which are separated from one another by spacers, termed nontranscribed spacer 1 (NTS1) and NTS2 [but sometimes referred to as intergenic spacers (IGS), and intervening sequences (IVS) in the literature] (Granneman and Baserga 2005). These intergenic regions were originally thought to be devoid of transcription. NTS2 contains the Pol I promoter and an autonomous replication element (ARS), while NTS1 contains Pol I-termination sequences and a replication fork block (RFB) site. The spacers accumulate high levels of Sir2 and it is at these sites where rDNA silencing is strongest (Smith and Boeke 1997; Huang and Moazed 2003).
Figure 10.
Sir2-dependent silencing in the rDNA locus. (A) Schematic representation of the rDNA repeat organization on chromosome XII. The array consists of ∼150 repeats of 9.1 kb each. Purple arrows represent individual Pol I transcription units of the 35S precursor rRNA. In between are the IGS consisting of NTS1 and NTS2, divided by the Pol III-transcribed 5S gene. Pol I sits at the rDNA promoter region in NTS2 and Fob1 on the RFB site in NTS1. The RENT complex is recruited to NTS2 and NTS1 via interactions with Pol I and Fob1, respectively. (B) Example of Sir2-dependent rDNA silencing phenotype using the mURA3 reporter gene integrated at NTS1. (C) Example of rDNA silencing using MET15 at NTS2 as a colorimetric reporter on lead nitrate-containing plates. White color indicates loss of silencing. Dark brown sectors indicate loss of the marker due to rDNA recombination. CEN, centromere; Chr, chromosome; TEL, telomere; WT, wild type.
Sir2-mediated rDNA stabilization and silencing
The first indication that Sir2 was involved in regulating rDNA chromatin came from the Esposito laboratory, which showed that deleting SIR2, but not the other SIR genes, increased meiotic and mitotic recombination at the rDNA array by an order of magnitude (Gottlieb and Esposito 1989). Given the size and repetitive nature of the array, the remarkably-low intrinsic recombination rate of the locus had been something of a mystery. Recombination at the rDNA, when it can be measured, occurs by unequal sister chromatid exchange and gene conversion (Szostak and Wu 1980; Gangloff et al. 1996). Sir2 suppresses these recombination pathways by funneling strand exchange events into an equal sister chromatid exchange pathway (Gottlieb and Esposito 1989; Kobayashi et al. 2004). In this way Sir2 maintains the size and stability of the rDNA.
The role of Sir2 in silencing Pol II transcription within the rDNA array was discovered later through the study of Ty1 retrotransposition (Bryk et al. 1997; Smith and Boeke 1997). The Boeke and Curcio laboratories found that transposons integrated into the NTS elements were transcriptionally quiescent. Repression was dependent on Sir2 as well as other chromatin factors (histones H2A/H2B) and chromatin modifiers (Top1 and Ubc2). Like rDNA stability, the other Sir proteins were not required. Repression also occurred for a wide variety of Pol II-transcribed reporters engineered directly into the locus (examples shown in Figure 10, B and C), although the extent of silencing depended on the strength of the promoter and the assay used (Fritze et al. 1997; Smith and Boeke 1997). This Sir2-dependent position effect on Pol II transcription was coined “rDNA silencing”.
Why does a mechanism to silence Pol II transcription exist in a locus devoted to rRNA synthesis by Pol I and Pol III? While initially characterized using reporter genes, rDNA silencing was eventually found to regulate TAR1, a mitochondrial protein gene encoded on the antisense strand of the 35S transcribed region (Coelho et al. 2002). RENT also represses noncoding RNAs expressed from NTS1 and NTS2 (Li et al. 2006; Vasiljeva et al. 2008). Transcription from NTS1 actually destabilizes the rDNA (Kobayashi and Ganley 2005). Thus, Sir2-mediated rDNA silencing controls rDNA stability by regulating Pol II transcription within the NTS regions. A more detailed description of the process is provided below.
The RENT complex
Sir2 in the nucleolus associates with Net1 and Cdc14 to form a protein complex named RENT (Shou et al. 1999; Straight et al. 1999). The name derives from two functions first attributed to the complex: rDNA silencing and regulation of mitotic exit complex. As shown in this section, the subunits of this complex each carry out distinct functions in addition to those originally identified.
One role of the Net1 subunit is to serve as a scaffold for recruitment and targeting of the other RENT components (Figure 10A). Net1 tethers RENT to the 35S promoter in NTS2 by interacting with Pol I (Straight et al. 1999; Shou et al. 2001). At this location, Net1 also plays a critical role in 35S transcription (Shou et al. 2001). Mutants lacking NET1 grow very slowly, while mutants lacking SIR2 grow normally (Straight et al. 1999). Thus, in addition to sequestering Sir2 and Cdc14 in the RENT complex at the rDNA, Net1 facilitates rRNA synthesis. Net1 independently targets RENT to the RFB site in NTS1 through a direct physical interaction with Fob1 (Figure 10A), a replication fork blocking factor that prevents collision of replication forks with oncoming Pol I transcription (Kobayashi and Horiuchi 1996; Huang and Moazed 2003; Zaman et al. 2016). RENT recruitment also requires phosphorylation of the Fob1 C-terminus (Zaman et al. 2016). At both NTS1 and NTS2, recruitment of RENT delivers Sir2 to chromatin.
Within the RENT complex Sir2 functions as a histone H3 and H4 deacetylase (Ghidelli et al. 2001; Tanny et al. 2004), though additional unidentified nonhistone targets are certainly possible. In vitro, purified RENT primarily deacetylates K16 of histone H4, with little activity on the H3 N-terminal tail. In vivo, H3K9 and H3K14 are also deacetylated suggesting that these residues may also be Sir2 targets (Buck et al. 2002). Indeed, a recent study demonstrated that H3K14 is uniquely important for silencing at the rDNA, but not telomeres (Xu et al. 2016). The Sir2 deacetylase activity hinders Pol II activity within NTS1 and NTS2 but it does not regulate Pol I transcription (Armstrong et al. 2002; Sandmeier et al. 2002b).
The Cdc14 subunit of RENT is an essential protein phosphatase that inactivates mitotic cyclins, thereby promoting mitotic exit (Visintin et al. 1998). Cdc14 function is regulated by spatial constraint. The phosphatase is sequestered as part of RENT in the nucleolus during most of the cell cycle. In telophase, Cdc14 is released to modify its cyclin targets throughout the nucleus and cytoplasm (Shou et al. 1999). Curiously, Sir2 is also released from RENT at telophase, although the reason is not clear (Straight et al. 1999). Net1, on the other hand, always remains associated with the rDNA.
While retained in the nucleolus, Cdc14 carries out additional functions. The phosphatase inhibits Pol I transcription during anaphase, which is required for proper chromosome condensation (Clemente-Blanco et al. 2009). The phosphatase also contributes to Pol II repression within the IGS in a Sir2-independent manner (Clemente-Blanco et al. 2011). In summary, the multiple subunits of the RENT complex affect many cellular processes, including rDNA silencing, rDNA stabilization, rRNA synthesis, and cell cycle control.
Intracellular competition for limiting amounts of Sir2
Modest overexpression of SIR2 enhances silencing at both the rDNA and telomeres, indicating that Sir2 is limiting in the nucleus (Fritze et al. 1997; Smith et al. 1998; Cockell et al. 2000). Various studies have shown that these sites of Sir2 action compete for the silencing factor. For example, disrupting the Sir2/3/4 complex by deleting all or just part of Sir4, causes Sir2 to dissociate from the HM loci and telomeres and strengthen rDNA silencing (Gotta et al. 1997; Kennedy et al. 1997; Smith et al. 1998). Sir3 also redistributes to the rDNA in the absence of Sir4 (Gotta et al. 1997), perhaps as a passenger of Sir2; but Sir3 is not required for the enhanced rDNA silencing (Smith et al. 1998).
Competition between the rDNA and telomeres for Sir2 goes both ways. Spontaneous shrinkage of the rDNA array, for example, releases sufficient Sir2 to strengthen silencing at telomeres (Michel et al. 2005). Similarly, overexpression of an N-terminal Sir3 fragment displaces Sir2 from the rDNA such that telomeric silencing increases (Gotta et al. 1998). Collectively, these studies show that Sir2 distribution is dynamic, and could readily respond to physiological changes that cause release from one locus and recruitment at another.
Nucleation and the spread of rDNA silencing
rDNA silencing is strongest within NTS1 and NTS2 where the RENT complex is recruited (Gotta et al. 1997; Smith and Boeke 1997; Buck et al. 2002; Huang and Moazed 2003). The recruitment sites are akin to the silencers that nucleate silent chromatin at the HM loci and telomeres. Recruitment of RENT by Pol I at NTS2 is dynamic, as might be expected for a translocating enzyme. ChIP assays suggest that RENT tracks with the polymerase for a short distance after initiation, spreading into the 5′ externally-transcribed spacer (5′-ETS) region (Huang and Moazed 2003; Li et al. 2013). Only ∼50% of the rDNA genes in a growing cell are transcribed by Pol I at a given time (Dammann et al. 1993), so it is possible that RENT recruitment to NTS2 is restricted to actively-transcribed rDNA genes. Supporting this idea, rDNA silencing of Pol II transcription actually requires Pol I transcription. Mutants lacking Pol I activity are defective for rDNA silencing, regardless of whether the reporter or endogenous noncoding genes are located in NTS1, NTS2, or 35S regions (Buck et al. 2002; Cioci et al. 2003; Cesarini et al. 2010). Therefore, transcription of the rDNA genes by Pol I also has a much broader and undefined role in rDNA silencing outside its function in recruiting RENT.
Spreading of rDNA silencing has been best characterized at the centromere-proximal (left) flank of the rDNA array where the unique non-rDNA sequences enable a simple and direct readout of spreading (Buck et al. 2002). Spreading does not occur on the telomere-proximal (right) flank. The left edge of the tandem array terminates precisely within the RFB of the terminal NTS1 element (Figure 11) (Bairwa et al. 2010). Fob1 typically binds two cis-acting sequences within the RFB, known as Ter1 and Ter2 (Kobayashi 2003; Mohanty and Bastia 2004). Only Ter2 remains at the left flank, resulting in attenuated recruitment of RENT and suboptimal silencing (Bairwa et al. 2010; Buck et al. 2016). Nevertheless, binding of RENT nucleates a Sir2-dependent structure that spreads into the adjoining chromatin in a fashion analogous to spreading by the Sir2/3/4 complex at HM loci and telomeres (Buck et al. 2002). The other Sir proteins are not required. When Sir2 is overexpressed, the size of the silenced domain expands nearly fivefold (∼2700 bp) until a tRNA gene [tQ(UUG)L] blocks further spreading (Figure 11) (Biswas et al. 2009). This boundary element, coupled with the weakened terminal RFB and the relatively large distance from the rDNA, likely insulates downstream Pol II-transcribed genes from fluctuations in Sir2 availability and the unusual nucleolar chromatin structure. At the HM loci and telomeres, Sir3 and Sir4 associate with deacetylated H4K16 to facilitate spreading of the Sir2/3/4 complex. The analogous “readers” of deacetylated chromatin that operate with RENT within and adjacent to the rDNA are not known.
Figure 11.
Spreading of rDNA silencing. Schematic representation of chromosome XII organization at the interface between the leftmost (centromere-proximal) rDNA gene and unique sequence. NTS1 sequence of the leftmost repeat (thick purple arrow) is truncated at the middle of the Ter1 RFB site. Sir2 overexpression results in spreading of silencing ∼2.7 kb, up to tRNAGln gene, tQ(UUG)L, which acts as a boundary element.
Why does silencing occur downstream of rDNA genes if RENT is recruited to the Pol I promoter in NTS2? One theory holds that the highly-transcribed Pol I transcription units fold the rDNA into a higher-order chromatin structure, like the gene loops that hold the 5′ and 3′ ends of Pol II genes together (Hampsey et al. 2011). Along these lines, 3C approaches have shown that 35S genes are extruded into loops that are brought together at their bases by Fob1 bound to RFBs (Choudhury et al. 2015). Structures like these may facilitate silencing at locations seemingly separated by great distances (Figure 7B and Figure 11). A second model is based on transcription-driven supercoiling (Liu and Wang 1987). Transcription produces positive DNA supercoiling ahead of the polymerase. Positive supercoils generated by heavy Pol I transcription might accumulate in NTS1 where they could facilitate silencing by RENT at the RFB. Indeed, topoisomerase I is required for rDNA silencing, and a lack of TOP1 results in accumulation of negative supercoiling in the rDNA genes, presumably due to the selective relaxation of positive supercoils by another topoisomerase (Bryk et al. 1997; French et al. 2011). Also consistent with a supercoiling model, transcriptional elongation by Pol I was shown to be necessary for downstream silencing (Buck et al. 2016).
rDNA stability, cohibin, and perinuclear anchoring of the rDNA
In addition to RENT, Fob1 also recruits the “cohibin” complex to the RFB (Huang et al. 2006). Cohibin contains a Csm1 homodimer bound to two loss of rDNA silencing 4 (Lrs4) proteins (Huang et al. 2006; Mekhail et al. 2008; Corbett et al. 2010) (Figure 12A). LRS4 was originally identified through a genetic screen for factors that function in rDNA silencing (Smith et al. 1999), and CSM1 through a genetic screen for meiosis defects (Rabitsch et al. 2001). Deletion of either gene causes loss of rDNA silencing and increased DNA recombination frequency. During mitotic growth, cohibin subunits remain in the nucleolus and associate with Fob1 via a Net1 paralog Tof2 (Huang et al. 2006). During meiosis, however, cohibin relocalizes to kinetochores of meiotic chromosomes where it associates with the meiosis-specific protein Mam1 to form the “monopolin” complex (Rabitsch et al. 2003). Monopolin (Lrs4/Csm1/Mam1) mediates monopolar attachment of sister chromatids to facilitate proper homolog separation at meiosis I (Rabitsch et al. 2003). Therefore, like the components of RENT, the components of cohibin form a multifunctional complex that acts in silencing as well as additional life cycle events.
Figure 12.
Silencing and endogenous noncoding RNAs and cohibin function in the rDNA. (A) X-ray crystal structure of the cohibin complex consisting of Csm1 (full length 190 amino acids) and an N-terminal portion of Lrs4 (amino acids 1–102), although only a small N-terminal α-helical portion of Lrs4 (purple, amino acids 3–33) is visible in structure. PDB accession number 3N7M. (B) Model for cohibin function at the rDNA where it bridges an interaction between Fob1, RENT, and Tof2 bound to the RFB sites, with the inner nuclear membrane CLIP complex. Cohesin and condensin then associate to align rDNA repeats and stabilize rDNA array structure. (C) Schematic diagram indicating sites of noncoding RNA transcription emanating from NTS1 and NTS2 (green dashed arrows), including bidirectional transcription from E-pro. CAR indicates a cohesin-associated region in NTS2. The rDNA ARS in NTS2 is also indicated.
Cohibin also helps anchor the rDNA to the nuclear periphery through interaction with Heh1 and Nur1, two proteins that span the inner nuclear membrane (Figure 12B) (Mekhail et al. 2008). The proteins are collectively referred to as chromosome linkage inner membrane proteins (CLIP). Disruption of CLIP-mediated rDNA anchoring to the inner nuclear membrane destabilizes the rDNA by allowing it to release from the nucleolar mass and become more accessible to recombination machinery (Mekhail et al. 2008). Although the rDNA becomes destabilized in CLIP mutants, Sir2-dependent silencing remains normal, indicating that silencing is not sufficient to maintain rDNA stability. Anchorage points are also required. Similarly, silencing of the HM loci can occur independently of subnuclear chromatin organization (Gartenberg et al. 2004).
rDNA stability and the silencing of Pol II transcription in the rDNA
While rDNA silencing was initially discovered fortuitously with reporter genes, study of the phenomenon ultimately led to the realization that Pol II naturally transcribes the NTS1 and NTS2 elements that are repressed by Sir2 (Figure 12C) (Kobayashi and Ganley 2005; Li et al. 2006; Vasiljeva et al. 2008; Cesarini et al. 2010). Such noncoding RNAs are derived from NTS1 and NTS2, and range in size from ∼1000 to 1700 nt. Foremost among these noncoding transcription units is the 520-bp EXP (expansion of rDNA repeats) region located with NTS1 (Ganley et al. 2005). Strong transcription from the bidirectional EXP promoter, named E-pro (Figure 12C), causes rDNA instability. Sir2 normally silences E-pro transcription. Therefore, the rDNA instability phenotype of a sir2∆ mutant can be traced in part to E-pro derepression.
How does E-pro transcription cause rDNA instability? The predominant model centers on the cohesin complex and cohesion of the rDNA. Cohesin binds avidly to the NTS regions, perhaps through recruitment by cohibin (Huang et al. 2006). In mediating cohesion of the rDNA array, cohesin is thought to maintain register of the rDNA repeats, and thus promote equal sister chromatid exchange should DNA recombination between the sister chromatids occur (Kobayashi et al. 2004). The need for DNA repair in the rDNA array should not be underestimated: Fob1-dependent pausing of the DNA replication fork at the RFB yields double-strand DNA breaks that induce mitotic rDNA recombination (Defossez et al. 1999; Weitao et al. 2003). Kobayashi and coworkers showed that transcription from E-pro, or from heterologous promoters engineered into the E-pro site, displaced cohesin, which in turn caused destabilization of the rDNA (Kobayashi and Ganley 2005). Thus, in this framework, Sir2 preserves cohesion of the rDNA by preventing E-pro transcription. Further stabilization likely comes from Sir2 directly guiding cohesin to chromatin (Wu et al. 2011).
Like cohesin, condensin also contributes to the maintenance of proper nucleolar structure and rDNA array stability. The complex associates with both NTS elements but is particularly enriched at the RFB through cooperative recruitment by Fob1, Tof2, and cohibin (Johzuka and Horiuchi 2009). At NTS1, the complex facilitates proper segregation of the rDNA during mitosis, probably through formation of higher-ordered chromatin looping and folding (Johzuka and Horiuchi 2009).
Surprisingly little is known about the roles of cohesin and condensin in Sir2-mediated rDNA silencing. With regard to cohesin, one study showed that mutations in the cohesin loading complex did not affect repression of reporter genes within the NTS elements (Gard et al. 2009). In another study, separation-of-function mutations of SIR2 that abolished cohesion of the mating-type loci had no impact on silencing there or at the rDNA (Chen et al. 2016). Thus, the evidence suggests that while Sir2 is required for cohesion; cohesion is not required for silencing. With regard to condensin, one study showed that condensin mutations actually strengthen rDNA silencing by causing Sir2 to relocalize from telomeres to the nucleolus (Machin et al. 2004). A more detailed study found that condensin mediates a position effect within the rDNA array whereby Pol II reporter genes in the middle of the array were silenced during starvation, while genes toward the outer edges of the array were derepressed (Wang et al. 2016). Condensin likely contributes to a chromatin architecture that facilitates such a long-range silencing gradient.
Sir2-independent rDNA silencing
Silencing in yeast is typically defined as a Sir2-dependent process, but there are clear instances of “silencing” in the rDNA that are Sir2-independent. As introduced above, the Cdc14 subunit of RENT silences noncoding RNAs (ncRNAs) from NTS1 and NTS2 independently of Net1 and Sir2 (Clemente-Blanco et al. 2011). In this case, Cdc14 directly represses Pol II transcription within the rDNA by dephosphorylating serines 2 and 5 on the C-terminal repeat domain of the second largest Pol II subunit (Clemente-Blanco et al. 2011). This is yet another example of the multifunctional nature of the RENT complex.
Cryptic nascent transcripts that escape Sir2-dependent silencing are limited by a Sir2-independent mechanism. The transcripts are terminated by the Nrd1/Sen1/Nab3 complex and then degraded by the exosome with assistance from TRAMP4 (Houseley et al. 2007; Vasiljeva et al. 2008).TRAMP4 is an exosome cofactor that contains the Trf4 poly(A) polymerase (Houseley et al. 2007). Interestingly, disruption of the Nrd1/Sen1/Nab3 complex also causes an increase in histone acetylation within NTS1 and even derepresses the mURA3 reporter positioned at NTS2 (Vasiljeva et al. 2008). How loss of proper termination and ncRNA degradation changes rDNA chromatin structure remains uncharacterized, but it is important to note that such mutants also cause silencing defects at telomeres (Houseley et al. 2007; Vasiljeva et al. 2008). Pol II transcription of cryptic noncoding RNAs in the rDNA is therefore silenced by both transcriptional and post-transcriptional mechanisms.
The fascinating connection between Sir2 and rDNA silencing to replicative aging
Sir2 is best known as a transcriptional silencing factor but has gained almost equal attention as a longevity factor. Linking Sir2 to longevity jump-started a modern wave of research on the molecular genetics of aging. The first indication that silencing was linked to aging came from a screen for stress-resistant mutants that also extended RLS, defined as the number of times a mother cell divides before dying (Figure 13A) (Mortimer and Johnston 1959; Muller et al. 1980; Kennedy et al. 1995). One of the isolated long-lived mutants harbored a dominant mutation in SIR4 called SIR4-42. This mutation truncates Sir4, blocking the ability of Rap1 to recruit the Sir2/3/4 complex at the HM loci and telomeres, thereby liberating Sir2 to accumulate at the rDNA and enhance silencing (Gotta et al. 1997; Kennedy et al. 1997; Smith et al. 1998). These studies established that the rDNA array was central to the replicative aging process.
Figure 13.
Replicative aging in S. cerevisiae. (A) Mother cells (M) produce daughter cells (D) that leave behind chitinous bud scars (blue circles). Older mother cells therefore harbor more bud scars, which is commonly used as an indicator of average replicative age of a population. (B) Example of a typical survival curve showing short life span of a sir2∆ strain and extended life span when Sir2 is modestly overexpressed (Sir2oe).
During normal yeast replicative aging, old mother cells become sterile due to loss of silencing at HML and HMR (Smeal et al. 1996), and also acquire fragmented nucleoli due to rDNA instability (Sinclair et al. 1997). The fragmented nucleoli are a symptom of elevated rDNA recombination reactions, one of which produces extrachromosomal rDNA circle (ERC) byproducts from the repetitive rDNA repeats (Sinclair and Guarente 1997). Each repeat contains an ARS that allows the ERCs to replicate during S phase (Miller and Kowalski 1993). Mother cells preferentially retain ERCs by a septin-based lateral diffusion barrier at the bud neck (Shcheprova et al. 2008). In an ERC-centric model for aging, the accumulation of ERCs in old mothers promotes senescence by titrating replication and transcription factors from nuclear genes (Sinclair and Guarente 1997). According to the model, Fob1 shortens RLS by instigating DNA recombination events at the RFB that produce ERCs (Defossez et al. 1999). Sir2, on the other hand, lengthens RLS by suppressing rDNA recombination and thus, reducing the production of ERCs (Figure 13B) (Kaeberlein et al. 1999). However, the short RLS of a sir2∆ mutant is only partially suppressed by simultaneously deleting FOB1, suggesting there are additional mechanisms by which Sir2 controls aging. Among these, Sir2-dependent H4K16 deacetylation at subtelomeric chromatin domains has been implicated in promoting RLS, though the mechanism remains unclear (Dang et al. 2009).
Several experimental observations are inconsistent with an ERC-centric model for replicative aging. For example, more recent studies demonstrated that ERCs accumulate in aging fob1∆ cells significantly more than originally reported (Lindstrom et al. 2011). Thus, ERCs are present even when aging is not apparent. Furthermore, the level of rDNA instability, as measured by marker loss and pulsed-field gel analysis, tracks more closely with RLS than does the ERC level when the rDNA-ARS activity is manipulated (Ganley et al. 2009). This suggests that rDNA instability, and not the ERCs themselves, is important for aging. Based on these findings and others related to the ARS with the rDNA, an alternative model for replicative aging emerged. Specifically, QTL analysis of long- and short-lived yeast isolates identified the rDNA, and specifically the ARS, as a major RLS determinant (Stumpferl et al. 2012; Kwan et al. 2013). The rDNA-ARS element is relatively inefficient (Miller et al. 1999). Polymorphisms in the ARS that increase its initiation activity shorten RLS, perhaps due to genome-wide replication stress caused by titration of factors away from non-rDNA origins (Kwan et al. 2013).
Taken together, the current general replicative aging model is that Sir2-dependent silencing of E-pro within NTS1 is important to maintain cohesin recruitment and prevent unequal sister chromatid exchange (rDNA instability) in younger cells (Kobayashi and Ganley 2005; Ganley et al. 2009). Sir2 protein levels naturally decrease in replicatively old mother cells (Dang et al. 2009), so deleting SIR2 accelerates rDNA destabilization and aging (Figure 13B) (Kobayashi and Ganley 2005; Ganley et al. 2009; Saka et al. 2013). Increased DNA replication stress in older cells also leads to rDNA instability through a mechanism independent of Sir2 (Lindstrom et al. 2011). Therefore, in both premature and normal aging, the key appears to be rDNA instability or replication, leading to an “rDNA theory” of aging that continues to evolve (Ganley and Kobayashi 2014).
The above model raises the critical question of how rDNA instability leads to replicative aging. Evidence to date indicates that dramatically-reduced rDNA copy number leads to most of the remaining rDNA genes being highly transcribed by Pol I. This makes the cells sensitive to DNA damaging agents such as UV light or MMS, and prone to double-strand breaks; thus indicating a more general effect of the rDNA array on genome-wide stability (Ide et al. 2010). Low rDNA copy number also releases Sir2 from the tandem array, and the cell compensates by somehow downregulating overall Sir2 protein levels, perhaps though autoregulation (Michel et al. 2005). This reduction in Sir2 could potentially impact the regulation of other Sir2 targets, including numerous nonhistone proteins involved in metabolism and other cellular processes.
Caloric restriction and silencing
Since the identification of Sir2 as an NAD+-dependent histone deacetylase (Imai et al. 2000; Landry et al. 2000b), there has been tremendous interest in Sir2 as a possible link between metabolism, chromatin regulation, and aging. This interest largely stems from findings that Sir2 activity is regulated in vivo by changes in cellular NAD+ concentration (Lin et al. 2000; Smith et al. 2000). Caloric restriction (CR) for yeast consists of reducing the media glucose concentration from 2 to 0.5% or lower, and this consistently extends RLS (Jiang et al. 2000; Lin et al. 2000). Early studies implicated Sir2 as being required for CR-mediated RLS extension, presumably because CR was somehow inducing the HDAC activity of Sir2 (Lin et al. 2000). The NAD+/NADH ratio is elevated by CR growth conditions (Lin et al. 2004), but NADH is a very weak Sir2 inhibitor (Anderson et al. 2003b; Schmidt et al. 2004). Furthermore, CR has little effect on rDNA silencing strength (Riesen and Morgan 2009; Smith et al. 2009), which is normally sensitive to changes in Sir2 dosage or activity (Smith et al. 1998). Therefore, in yeast, the idea of CR activating Sir2 globally via changes in NAD+ has recently been downplayed, though it remains a popular notion in mammalian studies of sirtuins and disease.
Sir2-dependent repression of specific genes such as PMA1, which encodes an H+-ATPase, has been associated with longevity. In this case, phosphorylation of Sir2 via the cyclic AMP-PKA pathway was shown to inhibit Sir2 activity (Kang et al. 2015). CR reduces PKA activity independently of NAD+ or NADH. If PKA were targeted to specific promoters, like that of PMA1, then Sir2 could potentially be regulated on a locus-by-locus basis, rather than globally. Further work will be required to show if CR-induced modification of Sir2 such as phosphorylation has any additional function at other gene promoters related to age-associated pathways.
Other factors that function in rDNA silencing
Sir2/RENT and other proteins discussed in this review are not the only factors that impact rDNA silencing. For example, numerous chromatin-modifying enzymes impact rDNA silencing using silencing reporter gene assays such as Ty1, mURA3, MET15, or ADE2 (Bryk et al. 1997; Fritze et al. 1997; Smith et al. 1999). Many of these also regulate silencing at HM loci and/or telomeres. Space constraints do not allow for extended discussion of all rDNA silencing regulatory factors, but those that were not discussed in the text have been compiled into Supplemental Material, Table S1 for easy reference. Mutants that show increased rDNA silencing tend to be defective in telomeric silencing, most likely reflecting redistribution of Sir2 from telomeres to the rDNA. In summary, Sir2-dependent silencing in the rDNA is highly complex and our mechanistic understanding of the phenomenon remains behind that of the HM loci and telomeres. More research is clearly needed to catch up.
Acknowledgments
This work was funded by grants from the National Institutes of Health (GM-51402 to M.R.G. and GM-075240 to J.S.S.).
Footnotes
Communicating editor: J. Boeke
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.112.145243/-/DC1.
Literature Cited
- Abraham J., Nasmyth K., Strathern J. N., Klar A. J. S., Hicks J. B., 1984. Regulation of mating-type information in yeast. J. Mol. Biol. 176: 307–331. [DOI] [PubMed] [Google Scholar]
- Ai W., Bertram P. G., Tsang C. K., Chan T.-F., Zheng X. F. S., 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10: 1295–1305. [DOI] [PubMed] [Google Scholar]
- Altaf M., Utley R. T., Lacoste N., Tan S., Briggs S. D., et al. , 2007. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28: 1002–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Cohen H., et al. , 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277: 18881–18890. [DOI] [PubMed] [Google Scholar]
- Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A., 2003a Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423: 181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. M., Latorre-Esteves M., Neves A. R., Lavu S., Medvedik O., et al. , 2003b Yeast life-span extension by calorie restriction is independent of NAD fluctuation. Science 302: 2124–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E., Neiman A. M., Zappulla D. C., Sternglanz R., 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592–595. [DOI] [PubMed] [Google Scholar]
- Andrulis E. D., Zappulla D. C., Ansari A., Perrod S., Laiosia C. V., et al. , 2002. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol. Cell. Biol. 22: 8292–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A., Gartenberg M. R., 1997. The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol. Cell. Biol. 17: 7061–7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A., Gartenberg M. R., 1999. Persistence of an alternate chromatin structure at silenced loci in vitro. Proc. Natl. Acad. Sci. USA 96: 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O. M., Gottschling D. E., 1994. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell-cycle dependent way. Genes Dev. 8: 1133–1146. [DOI] [PubMed] [Google Scholar]
- Armache K. J., Garlick J. D., Canzio D., Narlikar G. J., Kingston R. E., 2011. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334: 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C. M., Kaeberlein M., Imai S. I., Guarente L., 2002. Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity. Mol. Biol. Cell 13: 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaudo N., Fernandez I. S., McLaughlin S. H., Peak-Chew S. Y., Rhodes D., et al. , 2013. The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle. Nat. Struct. Mol. Biol. 20: 1119–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åström S. U., Rine J., 1998. Theme and variation among silencing proteins in Saccharomyces cerevisiae and Kluyveromyces lactis. Genetics 148: 1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos J. L., Celic I., Muhammad S., Cosgrove M. S., Boeke J. D., et al. , 2002. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell 10: 523–535. [DOI] [PubMed] [Google Scholar]
- Babiarz J. E., Halley J. E., Rine J., 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20: 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa N. K., Zzaman S., Mohanty B. K., Bastia D., 2010. Replication fork arrest and rDNA silencing are two independent and separable functions of the replication terminator protein Fob1 of Saccharomyces cerevisiae. J. Biol. Chem. 285: 12612–12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., et al. , 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. [DOI] [PubMed] [Google Scholar]
- Bedalov A., Gatbonton T., Irvine W. P., Gottschling D. E., Simon J. A., 2001. Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. USA 98: 15113–15118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedalov A., Hirao M., Posakony J., Nelson M., Simon J. A., 2003. NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 7044–7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P., Stillman B., 1992. ATP-dependent recognition of eukaryotic origins of replication by a multisubunit complex. Nature 357: 128–134. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Mitchell J., Leber J., Kobayashi R., Stillman B., 1995. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell 83: 563–568. [DOI] [PubMed] [Google Scholar]
- Bi X., Broach J. R., 1997. DNA in transcriptionally silent chromatin assumes a distinct topology that is sensitive to cell cycle progression. Mol. Cell. Biol. 17: 7077–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Broach J. R., 1999. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 13: 1089–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Yu Q., Sandmeier J. J., Zou Y., 2004. Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 24: 2118–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas M., Maqani N., Rai R., Kumaran S. P., Iyer K. R., et al. , 2009. Limiting the extent of the RDN1 heterochromatin domain by a silencing barrier and Sir2 protein levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 29: 2889–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman K. J., Anderson R. M., Cohen H. Y., Latorre-Esteves M., Sinclair D. A., 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SirT1. J. Biol. Chem. 277: 45099–45107. [DOI] [PubMed] [Google Scholar]
- Boscheron C., Maillet L., Marcand S., Tsai-Pflugfelder M., Gasser S. M., et al. , 1996. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 15: 2184–2195. [PMC free article] [PubMed] [Google Scholar]
- Bose M. E., McConnell K. H., Gardner-Aukema K. A., Muller U., Weinreich M., et al. , 2004. The origin recognition complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Mol. Cell. Biol. 24: 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Sherman J. M., Devine S. E., Cameron E. E., Pillus L., et al. , 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9: 2888–2902. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Breeden L., Abraham J., Sternglanz R., Nasmyth K., 1985. Characterization of a ‘silencer’ in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 41: 41–48. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K., 1987. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell 51: 709–719. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose A. B., Holmes S. G., Allis C. D., Broach J. R., 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7: 592–604. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Sobel R. E., Allis C. D., Turner B. M., Broach J. R., 1996. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol. 16: 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., et al. , 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15: 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M., Banerjee M., Murphy M., Knudsen K. E., Garfinkel D. J., et al. , 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11: 255–269. [DOI] [PubMed] [Google Scholar]
- Buchberger J. R., Onishi M., Li G., Seebacher J., Rudner A. D., et al. , 2008. Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 28: 6903–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D., 1988. Two DNA binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 210–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck S. W., Shore D., 1995. Action of a Rap1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 9: 370–384. [DOI] [PubMed] [Google Scholar]
- Buck S. W., Sandmeier J. J., Smith J. S., 2002. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111: 1003–1014. [DOI] [PubMed] [Google Scholar]
- Buck S. W., Maqani N., Matecic M., Hontz R. D., Fine R. D., et al. , 2016. RNA Polymerase I and Fob1 contributions to transcriptional silencing at the yeast rDNA locus. Nucleic Acids Res. (in press). 10.1093/nar/gkw212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bupp J., Martin A. E., Stensrud E. S., Jaspersen S. L., 2007. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J. Cell Biol. 179: 845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R. J., Zhou H., Han J., Li Q., Zhang Z., 2012. The SCFDia2 ubiquitin E3 ligase ubiquitylates Sir4 and functions in transcriptional silencing. PLoS Genet. 8: e1002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne K. P., Wolfe K. H., 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15: 1456–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen A. A., Milne L., Grunstein M., 2002. Acetylation of the yeast histone H4 N-terminus regulates its binding to heterochromatin protein Sir3. J. Biol. Chem. 277: 4778–4781. [DOI] [PubMed] [Google Scholar]
- Cesarini E., Mariotti F. R., Cioci F., Camilloni G., 2010. RNA polymerase I transcription silences noncoding RNAs at the ribosomal DNA locus in Saccharomyces cerevisiae. Eukaryot. Cell 9: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. R., Wu C. S., Hom Y., Gartenberg M. R., 2005. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 19: 3031–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. F., Hall B. E., Tanny J. C., Moazed D., Filman D., et al. , 2003. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure 11: 637–649. [DOI] [PubMed] [Google Scholar]
- Chang J. H., Kim H. C., Hwang K. Y., Lee J. W., Jackson S. P., et al. , 2002. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J. Biol. Chem. 277: 34489–34498. [DOI] [PubMed] [Google Scholar]
- Chen L., Widom J., 2005. Mechanism of transcriptional silencing in yeast. Cell 120: 37–48. [DOI] [PubMed] [Google Scholar]
- Chen Y., Rai R., Zhou Z. R., Kanoh J., Ribeyre C., et al. , 2011. A conserved motif within Rap1 has diversified roles in telomere protection and regulation in different organisms. Nat. Struct. Mol. Biol. 18: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-F., Chou C.-C., Gartenberg M. R., 2016. Determinants of Sir2-mediated, silent chromatin cohesion. Mol. Cell. Biol. DOI: 10.1128/mcb.00057-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T.-H., Gartenberg M. R., 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14: 452–463. [PMC free article] [PubMed] [Google Scholar]
- Cheng T.-H., Li Y.-C., Gartenberg M. R., 1998. Persistence of an alternate chromatin structure at silenced loci in the absence of silencers. Proc. Natl. Acad. Sci. USA 95: 5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi M.-H., Shore D., 1996. SUM1–1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol. Cell. Biol. 16: 4281–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C.-T., Buck S., Sternglanz R., Shore D., 1993. Targeting of Sir1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75: 531–541. [DOI] [PubMed] [Google Scholar]
- Chou C. C., Li Y. C., Gartenberg M. R., 2008. Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing. Mol. Cell 31: 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury M., Zaman S., Jiang J. C., Jazwinski S. M., Bastia D., 2015. Mechanism of regulation of ‘chromosome kissing’ induced by Fob1 and its physiological significance. Genes Dev. 29: 1188–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioci F., Vu L., Eliason K., Oakes M., Siddiqi I. N., et al. , 2003. Silencing in yeast rDNA chromatin: reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell 12: 135–145. [DOI] [PubMed] [Google Scholar]
- Clemente-Blanco A., Mayan-Santos M., Schneider D. A., Machin F., Jarmuz A., et al. , 2009. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 458: 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Blanco A., Sen N., Mayan-Santos M., Sacristan M. P., Graham B., et al. , 2011. Cdc14 phosphatase promotes segregation of telomeres through repression of RNA polymerase II transcription. Nat. Cell Biol. 13: 1450–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M., Palladino F., Laroche T., Kyrion G., Liu C., et al. , 1995. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: Evidence for a multicomponent complex required for yeast telomeric silencing. J. Cell Biol. 129: 909–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M. M., Perrod S., Gasser S. M., 2000. Analysis of Sir2p domains required for rDNA and telomeric silencing in Saccharomyces cerevisiae. Genetics 154: 1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho P. S., Bryan A. C., Kumar A., Shadel G. S., Snyder M., 2002. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA. Genes Dev. 16: 2755–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J. J., Yuan P., Hsu H. C., Li Z., Xu R. M., et al. , 2006. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol. Cell. Biol. 26: 3256–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K. D., Yip C. K., Ee L. S., Walz T., Amon A., et al. , 2010. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell 142: 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubizolles F., Martino F., Perrod S., Gasser S. M., 2006. A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol. Cell 21: 825–836. [DOI] [PubMed] [Google Scholar]
- Cuperus G., Shafaatian R., Shore D., 2000. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 19: 2641–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R., Lucchini R., Koller T., Sogo J. M., 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21: 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W., Steffen K. K., Perry R., Dorsey J. A., Johnson F. B., et al. , 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin D., Kantrow S. M., Liberatore R. A., Zakian V. A., 2000. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol. Cell. Biol. 20: 7991–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin D., Zaman Z., Liberatore R. A., Ptashne M., 2001. Telomere looping permits gene activation by a downstream UAS in yeast. Nature 409: 109–113. [DOI] [PubMed] [Google Scholar]
- Defossez P.-A., Prusty R., Kaeberlein M., Lin S.-J., Ferrigno P., et al. , 1999. Elimination of replication block protein Fob1 extends the lifespan of yeast mother cells. Mol. Cell 3: 447–455. [DOI] [PubMed] [Google Scholar]
- Derbyshire M. K., Weinstock K. G., Strathern J. N., 1996. HST1, a new member of the SIR2 family of genes. Yeast 12: 631–640. [DOI] [PubMed] [Google Scholar]
- Dhillon N., Raab J., Guzzo J., Szyjka S. J., Gangadharan S., et al. , 2009. DNA polymerase ε, acetylases and remodellers cooperate to form a specialized chromatin structure at a tRNA insulator. EMBO J. 28: 2583–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A., Rine J., 1997. Separable functions of ORC5 in replication initiation and silencing in Saccharomyces cerevisiae. Genetics 147: 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson A. E., Rine J., 2015. Heritable capture of heterochromatin dynamics in Saccharomyces cerevisiae. eLife 4: e05007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., Kamakaka R. T., 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20: 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., Adams C. R., Rine J., Kamakaka R. T., 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13: 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry M., Loiodice I., Chen C. L., Thermes C., Taddei A., 2011. Tight protein-DNA interactions favor gene silencing. Genes Dev. 25: 1365–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey R. N., Gartenberg M. R., 2007. A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev. 21: 2150–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenhofer-Murray A. E., Rivier D. H., Rine J., 1997. The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics 145: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenhofer-Murray A. E., Kamakaka R., Rine J., 1999. A role for replication proteins PCNA, RF-C, polymerase ε and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics 153: 1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrentraut S., Weber J. M., Dybowski J. N., Hoffmann D., Ehrenhofer-Murray A. E., 2010. Rpd3-dependent boundary formation at telomeres by removal of Sir2 substrate. Proc. Natl. Acad. Sci. USA 107: 5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrentraut S., Hassler M., Oppikofer M., Kueng S., Weber J. M., et al. , 2011. Structural basis for the role of the Sir3 AAA+ domain in silencing: interaction with Sir4 and unmethylated histone H3K79. Genes Dev. 25: 1835–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellahi A., Thurtle D. M., Rine J., 2015. The chromatin and transcriptional landscape of native Saccharomyces cerevisiae telomeres and subtelomeric domains. Genetics 200: 505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S., Berman J., 1998. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 12: 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E., Muller H., Therizols P., Lafontaine I., Dujon B., et al. , 2005. Comparative genomics in hemiascomycete yeasts: evolution of sex, silencing, and subtelomeres. Mol. Biol. Evol. 22: 856–873. [DOI] [PubMed] [Google Scholar]
- Feldman J. B., Hicks J. B., Broach J. R., 1984. Identification of the sites required for repression of a silent mating-type locus in yeast. J. Mol. Biol. 178: 815–834. [DOI] [PubMed] [Google Scholar]
- Ferreira H. C., Luke B., Schober H., Kalck V., Lingner J., et al. , 2011. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat. Cell Biol. 13: 867–874. [DOI] [PubMed] [Google Scholar]
- Finnin M. S., Donigian J. R., Pavletich N. P., 2001. Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 8: 621–625. [DOI] [PubMed] [Google Scholar]
- Foss M., McNally F. J., Laurenson P., Rine J., 1993. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in Saccharomyces cerevisiae. Science 262: 1838–1843. [DOI] [PubMed] [Google Scholar]
- Fourel G., Revardel E., Koering C. E., Gilson E., 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18: 2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C. A., Ehrenhofer-Murray A. E., Loo S., Rine J., 1997. The origin of recognition complex, SIR1, and the S phase requirement for silencing. Science 276: 1547–1551. [DOI] [PubMed] [Google Scholar]
- French S. L., Sikes M. L., Hontz R. D., Osheim Y. N., Lambert T. E., et al. , 2011. Distinguishing the roles of Topoisomerases I and II in relief of transcription-induced torsional stress in yeast rRNA genes. Mol. Cell. Biol. 31: 482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritze C. E., Verschueren K., Strich R., Easton Esposito R., 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16: 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R. A., 1999. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260: 273–279. [DOI] [PubMed] [Google Scholar]
- Gaglio D., D’Alfonso A., Camilloni G., 2013. Functional complementation of sir2Δ yeast mutation by the human orthologous gene SIRT1. PLoS One 8: e83114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajiwala K. S., Burley S. K., 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10: 110–116. [DOI] [PubMed] [Google Scholar]
- Gallagher J. E., Babiarz J. E., Teytelman L., Wolfe K. H., Rine J., 2009. Elaboration, diversification and regulation of the Sir1 family of silencing proteins in Saccharomyces. Genetics 181: 1477–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo C. M., Smith D. L., Jr, Smith J. S., 2004. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 24: 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., Zou H., Rothstein R., 1996. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 15: 1715–1725. [PMC free article] [PubMed] [Google Scholar]
- Ganley A. R., Kobayashi T., 2014. Ribosomal DNA and cellular senescence: new evidence supporting the connection between rDNA and aging. FEMS Yeast Res. 14: 49–59. [DOI] [PubMed] [Google Scholar]
- Ganley A. R., Hayashi K., Horiuchi T., Kobayashi T., 2005. Identifying gene-independent noncoding functional elements in the yeast ribosomal DNA by phylogenetic footprinting. Proc. Natl. Acad. Sci. USA 102: 11787–11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley A. R., Ide S., Saka K., Kobayashi T., 2009. The effect of replication initiation on gene amplification in the rDNA and its relationship to aging. Mol. Cell 35: 683–693. [DOI] [PubMed] [Google Scholar]
- Gao L., Gross D. S., 2008. Sir2 silences gene transcription by targeting the transition between RNA polymerase II initiation and elongation. Mol. Cell. Biol. 28: 3979–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard S., Light W., Xiong B., Bose T., McNairn A. J., et al. , 2009. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J. Cell Biol. 187: 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K. A., Fox C. A., 2001. The Sir1 protein’s association with a silenced chromosome domain. Genes Dev. 15: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K. A., Rine J., Fox C. A., 1999. A region of Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics 151: 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. G., Nelson Z. W., Gottschling D. E., 2005. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Mol. Cell. Biol. 25: 6123–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg M. R., Neumann F. N., Laroche T., Blaszczyk M., Gasser S. M., 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119: 955–967. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Hediger F., Taddei A., Neumann F. R., Gartenberg M. R., 2004. The function of telomere clustering in yeast: the Circe effect. Cold Spring Harb. Symp. Quant. Biol. 69: 327–337. [DOI] [PubMed] [Google Scholar]
- Geissenhoner A., Weise C., Ehrenhofer-Murray A. E., 2004. Dependence of ORC silencing function on NatA-mediated Nα acetylation in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 10300–10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgel P. T., Palacios DeBeer M. A., Pietz G., Fox C. A., Hansen J. C., 2001. Sir3-dependent assembly of supramolecular chromatin structures in vitro. Proc. Natl. Acad. Sci. USA 98: 8584–8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidelli S., Donze D., Dhillon N., Kamakaka R. T., 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 20: 4522–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesman D., Best L., Tatchell K., 1991. The role of RAP1 in the regulation of the MATa locus. Mol. Cell. Biol. 11: 1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Roberge M., Giraldo R., Rhodes D., Gasser S. M., 1993. Distortion of the DNA double helix by Rap1 at silencers and multiple telomeric binding sites. J. Mol. Biol. 231: 293–310. [DOI] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., et al. , 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134: 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Strahl-Bolsinger S., Renauld H., Laroche T., Kennedy B. K., et al. , 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16: 3243–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Palladino F., Gasser S. M., 1998. Functional characterization of the N terminus of Sir3p. Mol. Cell. Biol. 18: 6110–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S., Esposito R. E., 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56: 771–776. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., 1992. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc. Natl. Acad. Sci. USA 89: 4062–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A., 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Granneman S., Baserga S. J., 2005. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr. Opin. Cell Biol. 17: 281–286. [DOI] [PubMed] [Google Scholar]
- Grozinger C. M., Chao E. D., Blackwell H. E., Moazed D., Schreiber S. L., 2001. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 276: 38837–38843. [DOI] [PubMed] [Google Scholar]
- Guidi M., Ruault M., Marbouty M., Loiodice I., Cournac A., et al. , 2015. Spatial reorganization of telomeres in long-lived quiescent cells. Genome Biol. 16: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., 2012. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191: 33–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E., George J. P., 1979. A mutation that permits the expression of normally silent copies of mating-type information in Saccharomyces cerevisiae. Genetics 93: 13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall I. M., Shankaranarayana G. D., Noma K., Ayoub N., Cohen A., et al. , 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237. [DOI] [PubMed] [Google Scholar]
- Hampsey M., Singh B. N., Ansari A., Laine J. P., Krishnamurthy S., 2011. Control of eukaryotic gene expression: Gene loops and transcriptional memory. Adv. Enzyme Regul. 15: 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan A., Abraham N. M., Goyal S., Jamir I., Priyakumar U. D., et al. , 2015. Sumoylation of Sir2 differentially regulates transcriptional silencing in yeast. Nucleic Acids Res. 43: 10213–10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley P. D., Madhani H. D., 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass E. P., Zappulla D. C., 2015. The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae. eLife 4: e07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A., Laroche T., Strahl-Bolsinger S., Gasser S. M., Grunstein M., 1995. Histone H3 and H4 N-termini interact with Sir3 and Sir4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell 80: 583–592. [DOI] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger S., Grunstein M., 1996. Spreading of transcriptional repressor Sir3 from telomeric heterochromatin. Nature 383: 92–95. [DOI] [PubMed] [Google Scholar]
- Hediger F., Neumann F. R., Van Houwe G., Dubrana K., Gasser S. M., 2002. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12: 2076–2089. [DOI] [PubMed] [Google Scholar]
- Hickman M. A., Rusche L. N., 2007. Substitution as a mechanism for genetic robustness: the duplicated deacetylases Hst1p and Sir2p in Saccharomyces cerevisiae. PLoS Genet. 3: e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M. A., Rusche L. N., 2010. Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proc. Natl. Acad. Sci. USA 107: 19384–19389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J. F.-X., Laroche T., Brand A. H., Gasser S. M., 1989. Rap1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell 57: 725–737. [DOI] [PubMed] [Google Scholar]
- Holmes S. G., Broach J. R., 1996. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 10: 1021–1032. [DOI] [PubMed] [Google Scholar]
- Hoppe G. J., Tanny J. C., Rudner A. D., Gerber S. A., Danaie S., et al. , 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22: 4167–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Bernstein D. A., Fox C. A., Keck J. L., 2005. Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proc. Natl. Acad. Sci. USA 102: 8489–8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Danzer J. R., Fox C. A., Keck J. L., 2006. Structure of the Sir3 protein bromo adjacent homology (BAH) domain from S. cerevisiae at 1.95 Å resolution. Protein Sci. 15: 1182–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Danzer J. R., Mendoza L., Bose M. E., Muller U., et al. , 2009. Phylogenetic conservation and homology modeling help reveal a novel domain within the budding yeast heterochromatin protein Sir1. Mol. Cell. Biol. 29: 687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J., Kotovic K., El Hage A., Tollervey D., 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 26: 4996–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz K. T., Bitterman K. J., Cohen H. Y., Lamming D. W., Lavu S., et al. , 2003. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425: 191–196. [DOI] [PubMed] [Google Scholar]
- Hsu H. C., Stillman B., Xu R. M., 2005. Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing. Proc. Natl. Acad. Sci. USA 102: 8519–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H. C., Wang C. L., Wang M., Yang N., Chen Z., et al. , 2013. Structural basis for allosteric stimulation of Sir2 activity by Sir4 binding. Genes Dev. 27: 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Moazed D., 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 17: 2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Brito I. L., Villen J., Gygi S. P., Amon A., et al. , 2006. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 20: 2887–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Zhou H., Katzmann D., Hochstrasser M., Atanasova E., et al. , 2005. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc. Natl. Acad. Sci. USA 102: 13410–13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S., Miyazaki T., Maki H., Kobayashi T., 2010. Abundance of ribosomal RNA gene copies maintains genome integrity. Science 327: 693–696. [DOI] [PubMed] [Google Scholar]
- Iida T., Araki H., 2004. Noncompetitive counteractions of DNA polymerase ε and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S.-I., Armstrong C. M., Kaeberlein M., Guarente L., 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800. [DOI] [PubMed] [Google Scholar]
- Iraqui I., Garcia-Sanchez S., Aubert S., Dromer F., Ghigo J. M., et al. , 2005. The Yak1p kinase controls expression of adhesins and biofilm formation in Candida glabrata in a Sir4p-dependent pathway. Mol. Microbiol. 55: 1259–1271. [DOI] [PubMed] [Google Scholar]
- Irlbacher H., Franke J., Manke T., Vingron M., Ehrenhofer-Murray A. E., 2005. Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes Dev. 19: 1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Arib G., Lin C., Van Houwe G., Laemmli U. K., 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109: 551–562. [DOI] [PubMed] [Google Scholar]
- Jackson M. D., Denu J. M., 2002. Structural identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of β-NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 277: 18535–18544. [DOI] [PubMed] [Google Scholar]
- Jambunathan N., Martinez A. W., Robert E. C., Agochukwu N. B., Ibos M. E., et al. , 2005. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics 171: 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J. C., Jaruga E., Repnevskaya M. V., Jazwinski S. M., 2000. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 14: 2135–2137. [DOI] [PubMed] [Google Scholar]
- Johnson A., Li G., Sikorski T. W., Buratowski S., Woodcock C. L., et al. , 2009. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol. Cell 35: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A., Wu R., Peetz M., Gygi S. P., Moazed D., 2013. Heterochromatic gene silencing by activator interference and a transcription elongation barrier. J. Biol. Chem. 288: 28771–28782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. M., Kayne P. S., Kahn E. S., Grunstein M., 1990. Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87: 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johzuka K., Horiuchi T., 2009. The cis element and factors required for condensin recruitment to chromosomes. Mol. Cell 34: 26–35. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M., McVey M., Guarente L., 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13: 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W. K., Kim Y. H., Kang H. A., Kwon K. S., Kim J. Y., 2015. Sir2 phosphorylation through cAMP-PKA and CK2 signaling inhibits the lifespan extension activity of Sir2 in yeast. eLife 4: e09709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y., Struhl K., 2005. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 24: 2138–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman P. D., Kobayashi R., Stillman B., 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11: 345–357. [DOI] [PubMed] [Google Scholar]
- Kayne P. S., Kim U., Han M., Mullen J. R., Yoshizaki F., et al. , 1988. Extremely conserved histone H4 N-terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell 55: 27–39. [DOI] [PubMed] [Google Scholar]
- Kellis M., Birren B. W., Lander E. S., 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624. [DOI] [PubMed] [Google Scholar]
- Kennedy B. K., Austriaco N. R., Jr, Zhang J., Guarente L., 1995. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 80: 485–496. [DOI] [PubMed] [Google Scholar]
- Kennedy B. K., Gotta M., Sinclair D. A., Mills K., McNabb D. S., et al. , 1997. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in Saccharomyces cerevisiae. Cell 89: 381–391. [DOI] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J., 1988. Roles of two DNA binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 7: 2241–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Umehara T., Horikoshi M., 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32: 370–377. [DOI] [PubMed] [Google Scholar]
- King D. A., Hall B. E., Iwamoto M. A., Win K. Z., Chang J. F., et al. , 2006. Domain structure and protein interactions of the silent information regulator Sir3 revealed by screening a nested deletion library of protein fragments. J. Biol. Chem. 281: 20107–20119. [DOI] [PubMed] [Google Scholar]
- Kirchmaier A. L., Rine J., 2001. DNA replication-independent silencing in S. cerevisiae. Science 291: 646–650. [DOI] [PubMed] [Google Scholar]
- Kirchmaier A. L., Rine J., 2006. Cell cycle requirements in assembling silent chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland J. G., Kamakaka R. T., 2013. Long-range heterochromatin association is mediated by silencing and double-strand DNA break repair proteins. J. Cell Biol. 201: 809–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T., Schleker T., Sperling A. S., Xie W., Gasser S. M., et al. , 2011. γH2A is a component of yeast heterochromatin required for telomere elongation. Cell Cycle 10: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T., Kuryan B. G., Tran N. N., Song C., Xue Y., et al. , 2012. Mechanism for epigenetic variegation of gene expression at yeast telomeric heterochromatin. Genes Dev. 26: 2443–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Fogel S., Macleod K., 1979. MAR1—a regulator of the HMa and HMα loci in Saccharomyces cerevisiae. Genetics 93: 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., Kakar S. N., Ivy J. M., Hicks J. B., Livi G. P., et al. , 1985. SUM1, an apparent positive regulator of the cryptic mating-type loci in Saccharomyces cerevisiae. Genetics 111: 745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Laroche T., Cardenas M. E., Hofmann J. F.-X., Schweizer D., et al. , 1992. Localization of Rap1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J. Cell Biol. 117: 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., 2003. The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol. Cell. Biol. 23: 9178–9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Ganley A. R., 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Horiuchi T., 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1: 465–474. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Horiuchi T., Tongaonkar P., Vu L., Nomura M., 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441–453. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., et al. , 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11: 721–729. [DOI] [PubMed] [Google Scholar]
- Kueng S., Tsai-Pflugfelder M., Oppikofer M., Ferreira H. C., Roberts E., et al. , 2012. Regulating repression: roles for the Sir4 N-terminus in linker DNA protection and stabilization of epigenetic states. PLoS Genet. 8: e1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Shore D., 1991. Rap1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 5: 616–628. [DOI] [PubMed] [Google Scholar]
- Kwan E. X., Foss E. J., Tsuchiyama S., Alvino G. M., Kruglyak L., et al. , 2013. A natural polymorphism in rDNA replication origins links origin activation with calorie restriction and lifespan. PLoS Genet. 9: e1003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G., Liu K., Liu C., Lustig A. J., 1993. Rap1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 7: 1146–1159. [DOI] [PubMed] [Google Scholar]
- Lacoste N., Utley R. T., Hunter J. M., Poirier G. G., Cote J., 2002. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 277: 30421–30424. [DOI] [PubMed] [Google Scholar]
- Ladurner A. G., Inouye C., Jain R., Tjian R., 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 11: 365–376. [DOI] [PubMed] [Google Scholar]
- Landry J., Slama J. T., Sternglanz R., 2000a Role of NAD+ in the deacetylase activity of the Sir2-like proteins. Biochem. Biophys. Res. Commun. 278: 685–690. [DOI] [PubMed] [Google Scholar]
- Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., et al. , 2000b The silencing protein Sir2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97: 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larin M. L., Harding K., Williams E. C., Lianga N., Dore C., et al. , 2015. Competition between heterochromatic loci allows the abundance of the silencing protein, Sir4, to regulate de novo assembly of heterochromatin. PLoS Genet. 11: e1005425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T., Martin S. G., Gotta M., Gorham H. C., Pryde F. E., et al. , 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8: 653–656. [DOI] [PubMed] [Google Scholar]
- Laroche T., Martin S. G., Tsai-Pflugfelder M., Gasser S. M., 2000. The dynamics of yeast telomeres and silencing proteins through the cell cycle. J. Struct. Biol. 129: 159–174. [DOI] [PubMed] [Google Scholar]
- Lau A., Blitzblau H., Bell S. P., 2002. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 16: 2935–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenson P., Rine J., 1991. SUM1–1: a suppressor of silencing defects in Saccharomyces cerevisiae. Genetics 129: 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus A. G., Holmes S. G., 2011. A cis-acting tRNA gene imposes the cell cycle progression requirement for establishing silencing at the HMR locus in yeast. Genetics 187: 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S., Davis C., Konopka J. B., Sternglanz R., 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13: 1029–1042. [DOI] [PubMed] [Google Scholar]
- Lebrun E., Fourel G., Defossez P. A., Gilson E., 2003. A methyltransferase targeting assay reveals silencer-telomere interactions in budding yeast. Mol. Cell. Biol. 23: 1498–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Gross D. S., 1993. Conditional silencing: the HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol. Cell. Biol. 13: 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G. P., et al. , 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Mueller J. E., Bryk M., 2006. Sir2 represses endogenous polymerase II transcription units in the ribosomal DNA nontranscribed spacer. Mol. Biol. Cell 17: 3848–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Petteys B. J., McClure J. M., Valsakumar V., Bekiranov S., et al. , 2010. Thiamine biosynthesis in Saccharomyces cerevisiae is regulated by the NAD+-dependent histone deacetylase Hst1. Mol. Cell. Biol. 30: 3329–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Valsakumar V., Poorey K., Bekiranov S., Smith J. S., 2013. Genome-wide analysis of functional sirtuin chromatin targets in yeast. Genome Biol. 14: R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-C., Cheng T.-H., Gartenberg M. R., 2001. Establishment of transcriptional silencing in the absence of DNA replication. Science 291: 650–653. [DOI] [PubMed] [Google Scholar]
- Liaw H., Lustig A. J., 2006. Sir3 C-terminal domain involvement in the initiation and spreading of heterochromatin. Mol. Cell. Biol. 26: 7616–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb J. D., Liu X., Botstein D., Brown P. O., 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28: 327–334. [DOI] [PubMed] [Google Scholar]
- Lin S. J., Defossez P. A., Guarente L., 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289: 2126–2128. [DOI] [PubMed] [Google Scholar]
- Lin S. J., Ford E., Haigis M., Liszt G., Guarente L., 2004. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 18: 12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom D. L., Leverich C. K., Henderson K. A., Gottschling D. E., 2011. Replicative age induces mitotic recombination in the ribosomal RNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 7: e1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou G. G., Tanny J. C., Kruger R. G., Walz T., Moazed D., 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121: 515–527. [DOI] [PubMed] [Google Scholar]
- Liu C., Lustig A. J., 1996. Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics 143: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Mao X., Lustig A. J., 1994. Mutational analysis defines a C-terminal tail domain of Rap1 essential for telomeric silencing. Genetics 138: 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C., 1987. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 84: 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livi G. P., Hicks J. B., Klar A. J., 1990. The SUM1–1 mutation affects silent mating-type gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone-Zatchej M., Marcionelli R., Moller K., de Pril R., Thoma F., 2003. Repair of UV lesions in silenced chromatin provides in vivo evidence for a compact chromatin structure. J. Biol. Chem. 278: 37471–37479. [DOI] [PubMed] [Google Scholar]
- Loo S., Rine J., 1994. Silencers and domains of generalized repression. Science 264: 1768–1771. [DOI] [PubMed] [Google Scholar]
- Loo S., Fox C. A., Rine J., Kobayashi R., Stillman B., et al. , 1995. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol. Biol. Cell 6: 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Serra L., Kelly G., Patel H., Stewart A., Uhlmann F., 2014. The Scc2-Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat. Genet. 46: 1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Vega-Palas M. A., Grunstein M., 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16: 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch P. J., Rusche L. N., 2009. A silencer promotes the assembly of silenced chromatin independently of recruitment. Mol. Cell. Biol. 29: 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch P. J., Rusche L. N., 2010. An auxiliary silencer and a boundary element maintain high levels of silencing proteins at HMR in Saccharomyces cerevisiae. Genetics 185: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch P. J., Fraser H. B., Sevastopoulos E., Rine J., Rusche L. N., 2005. Sum1p, the origin recognition complex, and the spreading of a promoter-specific repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 25: 5920–5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machin F., Paschos K., Jarmuz A., Torres-Rosell J., Pade C., et al. , 2004. Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr. Biol. 14: 125–130. [PubMed] [Google Scholar]
- Mahoney D. J., Broach J. R., 1989. The HML mating-type cassette of Saccharomyces cerevisiae is regulated by two separate but functionally equivalent silencers. Mol. Cell. Biol. 9: 4621–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney D. J., Marquardt R., Shei G.-J., Rose A. B., Broach J. R., 1991. Mutations in HML E silencer of Saccharomyces cerevisiae yield metastable inheritance of transcriptional repression. Genes Dev. 5: 605–615. [DOI] [PubMed] [Google Scholar]
- Maillet L., Boscheron C., Gotta M., Marcand S., Gilson E., et al. , 1996. Evidence of silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev. 10: 1796–1811. [DOI] [PubMed] [Google Scholar]
- Manning B. J., Peterson C. L., 2014. Direct interactions promote eviction of the Sir3 heterochromatin protein by the SWI/SNF chromatin remodeling enzyme. Proc. Natl. Acad. Sci. USA 111: 17827–17832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano Y., Kobayashi T. J., Nakayama J., Uchida H., Oki M., 2013. Single cell visualization of yeast gene expression shows correlation of epigenetic switching between multiple heterochromatic regions through multiple generations. PLoS Biol. 11: e1001601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Buck S. W., Moretti P., Gilson E., Shore D., 1996. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap1 protein. Genes Dev. 10: 1297–1309. [DOI] [PubMed] [Google Scholar]
- Marshall M., Mahoney D., Rose A., Hicks J. B., Broach J. B., 1987. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 4441–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R., Moazed D., 2015. RNAi and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 7: a019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino F., Kueng S., Robinson P., Tsai-Pflugfelder M., van Leeuwen F., et al. , 2009. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol. Cell 33: 323–334. [DOI] [PubMed] [Google Scholar]
- Martins-Taylor K., Dula M. L., Holmes S. G., 2004. Heterochromatin spreading at yeast telomeres occurs in M phase. Genetics 168: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Taylor K., Sharma U., Rozario T., Holmes S. G., 2011. H2A.Z (Htz1) controls the cell-cycle-dependent establishment of transcriptional silencing at Saccharomyces cerevisiae telomeres. Genetics 187: 89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBryant S. J., Krause C., Woodcock C. L., Hansen J. C., 2008. The silent information regulator 3 protein, SIR3p, binds to chromatin fibers and assembles a hypercondensed chromatin architecture in the presence of salt. Mol. Cell. Biol. 28: 3563–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F. J., Rine J., 1991. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 5648–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee P. C., Morgan B. A., Mittman B. A., Smith M. M., 1990. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science 247: 841–845. [DOI] [PubMed] [Google Scholar]
- Meijsing S. H., Ehrenhofer-Murray A. E., 2001. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 15: 3169–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K., Seebacher J., Gygi S. P., Moazed D., 2008. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456: 667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini M. D., Wu M., Madhani H. D., 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736. [DOI] [PubMed] [Google Scholar]
- Michel A. H., Kornmann B., Dubrana K., Shore D., 2005. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 19: 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A., Bystricky K., Dekker J., 2009. Yeast silent mating-type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet. 5: e1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A., Chen J., Takasuka T. E., Jacobi J. L., Kaufman P. D., et al. , 2010. Proliferating cell nuclear antigen (PCNA) is required for cell cycle-regulated silent chromatin on replicated and nonreplicated genes. J. Biol. Chem. 285: 35142–35154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. M., Nasmyth K. A., 1984. Role of DNA replication in the repression of silent mating-type loci in yeast. Nature 312: 247–251. [DOI] [PubMed] [Google Scholar]
- Miller C. A., Kowalski D., 1993. cis-acting components in the replication origin from ribosomal DNA of Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 5360–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. A., Umek R. M., Kowalski D., 1999. The inefficient replication origin from yeast ribosomal DNA is naturally impaired in the ARS consensus sequence and in DNA unwinding. Nucleic Acids Res. 27: 3921–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Landry J., Sternglanz R., Xu R. M., 2001. Crystal structure of a Sir2 homolog-NAD complex. Cell 105: 269–279. [DOI] [PubMed] [Google Scholar]
- Mishra K., Shore D., 1999. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by Rif proteins. Curr. Biol. 9: 1123–1126. [DOI] [PubMed] [Google Scholar]
- Moazed D., Kistler A., Axelrod A., Rine J., Johnson A. D., 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: A Sir2/Sir4 complex and evidence for a regulatory domain in Sir4 that inhibits its interaction with Sir3. Proc. Natl. Acad. Sci. USA 94: 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B. K., Bastia D., 2004. Binding of the replication terminator protein Fob1p to the Ter sites of yeast causes polar fork arrest. J. Biol. Chem. 279: 1932–1941. [DOI] [PubMed] [Google Scholar]
- Moreira J. M. A., Holmberg S., 1998. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA polII mutants defective in vivo in response to acidic activators. EMBO J. 17: 6028–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P., Shore D., 2001. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol. Cell. Biol. 21: 8082–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P., Freeman K., Coodly L., Shore D., 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8: 2257–2269. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Johnston J. R., 1959. Life span of individual yeast cells. Nature 183: 1751–1752. [DOI] [PubMed] [Google Scholar]
- Mullen J. R., Kayne P. S., Moerschell R. P., Tsunasawa S., Gribskov M., et al. , 1989. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 8: 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller I., Zimmermann M., Becker D., Flomer M., 1980. Calendar life span vs. budding life span of Saccharomyces cerevisiae. Mech. Ageing Dev. 12: 47–52. [DOI] [PubMed] [Google Scholar]
- Murphy G. A., Spedale E. J., Powell S. T., Pillus L., Schultz S. C., et al. , 2003. The Sir4 C-terminal coiled coil is required for telomeric and mating-type silencing in Saccharomyces cerevisiae. J. Mol. Biol. 334: 769–780. [DOI] [PubMed] [Google Scholar]
- Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V., 1999. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9: 27–43. [PubMed] [Google Scholar]
- Ng H. H., Robert F., Young R. A., Struhl K., 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16: 806–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng H. H., Ciccone D. N., Morshead K. B., Oettinger M. A., Struhl K., 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100: 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M., Molloy K. R., Williams R., Farr J. C., Meinema A. C., et al. , 2013. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell 24: 3920–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov I., Taddei A., 2016. Linking replication stress with heterochromatin formation. Chromosoma 125: 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C., Ray E., Pillus L., 1997. a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 8: 2421–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A., Bianchet M. A., Boeke J. D., 2008. Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction. PLoS Genet. 4: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki M., Kamakaka R. T., 2005. Barrier function at HMR. Mol. Cell 19: 707–716. [DOI] [PubMed] [Google Scholar]
- Oki M., Valenzuela L., Chiba T., Ito T., Kamakaka R. T., 2004. Barrier proteins remodel and modify chromatin to restrict silenced domains. Mol. Cell. Biol. 24: 1956–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M., Liou G. G., Buchberger J. R., Walz T., Moazed D., 2007. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 28: 1015–1028. [DOI] [PubMed] [Google Scholar]
- Oppikofer M., Kueng S., Martino F., Soeroes S., Hancock S. M., et al. , 2011. A dual role of H4K16 acetylation in the establishment of yeast silent chromatin. EMBO J. 30: 2610–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppikofer M., Kueng S., Keusch J. J., Hassler M., Ladurner A. G., et al. , 2013. Dimerization of Sir3 via its C-terminal winged helix domain is essential for yeast heterochromatin formation. EMBO J. 32: 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada S., Sutton A., Muster N., Brown C. E., Yates J. R., 3rd, et al. , 2001. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev. 15: 3155–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne E. A., Dudoit S., Rine J., 2009. The establishment of gene silencing at single-cell resolution. Nat. Genet. 41: 800–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne E. A., Hiraoka Y., Rine J., 2011. Symmetry, asymmetry, and kinetics of silencing establishment in Saccharomyces cerevisiae revealed by single-cell optical assays. Proc. Natl. Acad. Sci. USA 108: 1209–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H., Tokunaga E., Chang K., Hikasa M., Iijima K., et al. , 2006. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene 25: 176–185. [DOI] [PubMed] [Google Scholar]
- Ozaydin B., Rine J., 2010. Expanded roles of the origin recognition complex in the architecture and function of silenced chromatin in Saccharomyces cerevisiae. Mol. Cell. Biol. 30: 626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F., Laroche T., Gilson E., Axelrod A., Pillus L., et al. , 1993. Sir3 and Sir4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75: 543–555. [DOI] [PubMed] [Google Scholar]
- Park E. C., Szostak J. W., 1990. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating-type locus HML. Mol. Cell. Biol. 10: 4932–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y., Hanish J., Lustig A. J., 1998. Sir3p domains involved in the initiation of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., 1979. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. USA 76: 410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus L., Rine J., 1989. Epigenetic inheritance of transcriptional states in Saccharomyces cerevisiae. Cell 59: 637–647. [DOI] [PubMed] [Google Scholar]
- Prescott E. T., Safi A., Rusche L. N., 2011. A region of the nucleosome required for multiple types of transcriptional silencing in Saccharomyces cerevisiae. Genetics 188: 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F. E., Louis E. J., 1999. Limitations of silencing at native telomeres. EMBO J. 18: 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch K. P., Toth A., Galova M., Schleiffer A., Schaffner G., et al. , 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11: 1001–1009. [DOI] [PubMed] [Google Scholar]
- Rabitsch K. P., Petronczki M., Javerzat J. P., Genier S., Chwalla B., et al. , 2003. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev. Cell 4: 535–548. [DOI] [PubMed] [Google Scholar]
- Radman-Livaja M., Ruben G., Weiner A., Friedman N., Kamakaka R., et al. , 2011. Dynamics of Sir3 spreading in budding yeast: secondary recruitment sites and euchromatic localization. EMBO J. 30: 1012–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner R. M., Hartley P. D., Meneghini M. D., Bao M. Z., Liu C. L., et al. , 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajavel M., Lalo D., Gross J. W., Grubmeyer C., 1998. Conversion of a cosubstrate to an inhibitor: phosphorylation mutants of nicotinic acid phosphoribosyltransferase. Biochem. 37: 4181–4188. [DOI] [PubMed] [Google Scholar]
- Ravindra A., Weiss K., Simpson R. T., 1999. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol. Cell. Biol. 19: 7944–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Hector R. E., Roy N., Song J. H., Berkner K. L., et al. , 2003. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat. Genet. 33: 522–526. [DOI] [PubMed] [Google Scholar]
- Reifsnyder C., Lowell J., Clarke A., Pillus L., 1996. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat. Genet. 14: 42–49. [DOI] [PubMed] [Google Scholar]
- Ren J., Wang C. L., Sternglanz R., 2010. Promoter strength influences the S phase requirement for establishment of silencing at the Saccharomyces cerevisiae silent mating-type loci. Genetics 186: 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H., Aparicio O. M., Zierath P. D., Billington B. L., Chhablani S. K., et al. , 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7: 1133–1145. [DOI] [PubMed] [Google Scholar]
- Rhode P. R., Elsasser S., Campbell J. L., 1992. Role of multifunctional autonomously replicating sequence binding factor 1 in the initiation of DNA replication and transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 1064–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesen M., Morgan A., 2009. Calorie restriction reduces rDNA recombination independently of rDNA silencing. Aging Cell 8: 624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine, J., 1979 Regulation and Transposition of Cryptic Mating Type Genes in Saccharomyces cerevisiae Ph.D. Thesis, University of Oregon, Eugene. [Google Scholar]
- Rine J., Strathern J. N., Hicks J. B., Herskowitz I., 1979. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics 93: 877–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier D. H., Ekena J. L., Rine J., 1999. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics 151: 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A., Schaft D., Shevchenko A., Pijnappel W. W., Wilm M., et al. , 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20: 7137–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. P., Luo W., Tsaponina O., Chabes A., Stillman B., 2011. A common telomeric gene silencing assay is affected by nucleotide metabolism. Mol. Cell 42: 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R., Meier B., McAinsh A. D., Feldmann H. M., Jackson S. P., 2004. Separation-of-function mutants of yeast Ku80 reveal a Yku80p-Sir4p interaction involved in telomeric silencing. J. Biol. Chem. 279: 86–94. [DOI] [PubMed] [Google Scholar]
- Ruault M., De Meyer A., Loiodice I., Taddei A., 2011. Clustering heterochromatin: Sir3 promotes telomere clustering independently of silencing in yeast. J. Cell Biol. 192: 418–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben G. J., Kirkland J. G., MacDonough T., Chen M., Dubey R. N., et al. , 2011. Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS One 6: e21923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A. D., Hall B. E., Ellenberger T., Moazed D., 2005. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell. Biol. 25: 4514–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Lynch P. J., 2009. Assembling heterochromatin in the appropriate places: A boost is needed. J. Cell. Physiol. 219: 525–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Rine J., 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15: 955–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safi A., Wallace K. A., Rusche L. N., 2008. Evolution of new function through a single amino acid change in the yeast repressor Sum1p. Mol. Cell. Biol. 28: 2567–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka K., Ide S., Ganley A. R., Kobayashi T., 2013. Cellular senescence in yeast is regulated by rDNA noncoding transcription. Curr. Biol. 23: 1794–1798. [DOI] [PubMed] [Google Scholar]
- Sampath V., Yuan P., Wang I. X., Prugar E., van Leeuwen F., et al. , 2009. Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes. Mol. Cell. Biol. 29: 2532–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo P. A., Roeder G. S., 2000. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell 11: 3601–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier J. J., Celic I., Boeke J. D., Smith J. S., 2002a Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD+ salvage pathway. Genetics 160: 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier J. J., French S., Osheim Y., Cheung W. L., Gallo C. M., et al. , 2002b RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21: 4959–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Bannister A. J., Dehe P. M., Geli V., Kouzarides T., 2004. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 279: 47506–47512. [DOI] [PubMed] [Google Scholar]
- Sauve A. A., Celic I., Avalos J., Deng H., Boeke J. D., et al. , 2001. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochem. 40: 15456–15463. [DOI] [PubMed] [Google Scholar]
- Sauve A. A., Moir R. D., Schramm V. L., Willis I. M., 2005. Chemical activation of Sir2-dependent silencing by relief of nicotinamide inhibition. Mol. Cell 17: 595–601. [DOI] [PubMed] [Google Scholar]
- Schmidt M. T., Smith B. C., Jackson M. D., Denu J. M., 2004. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J. Biol. Chem. 279: 40122–40129. [DOI] [PubMed] [Google Scholar]
- Schober H., Kalck V., Vega-Palas M. A., Van Houwe G., Sage D., et al. , 2008. Controlled exchange of chromosomal arms reveals principles driving telomere interactions in yeast. Genome Res. 18: 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H., Ferreira H., Kalck V., Gehlen L. R., Gasser S. M., 2009. Telomerase and SUN-domain protein Mps3 anchor and protect telomeres in budding yeast. Genes Dev. 23: 928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger E. A., Gross D. S., 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105: 403–414. [DOI] [PubMed] [Google Scholar]
- Shankaranarayana G. D., Motamedi M. R., Moazed D., Grewal S. I., 2003. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 13: 1240–1246. [DOI] [PubMed] [Google Scholar]
- Sharma U., Stefanova D., Holmes S. G., 2013. Histone variant H2A.Z functions in sister chromatid cohesion in Saccharomyces cerevisiae. Mol. Cell. Biol. 33: 3473–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcheprova Z., Baldi S., Frei S. B., Gonnet G., Barral Y., 2008. A mechanism for asymmetric segregation of age during yeast budding. Nature 454: 728–734. [DOI] [PubMed] [Google Scholar]
- Shei G.-J., Broach J. R., 1995. Yeast silencers can act as orientation-dependent gene inactivation centers that respond to environmental signals. Mol. Cell. Biol. 15: 3496–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia W. J., Li B., Workman J. L., 2006. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 20: 2507–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W., Seol J. H., Shevchenko A., Baskerville C., Moazed D., et al. , 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233–244. [DOI] [PubMed] [Google Scholar]
- Shou W., Sakamoto K. M., Keener J., Morimoto K. W., Traverso E. E., et al. , 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell 8: 45–55. [DOI] [PubMed] [Google Scholar]
- Simms T. A., Dugas S. L., Gremillion J. C., Ibos M. E., Dandurand M. N., et al. , 2008. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot. Cell 7: 2078–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D. A., Guarente L., 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Sinclair D. A., Mills K., Guarente L., 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277: 1313–1316. [DOI] [PubMed] [Google Scholar]
- Singer M. S., Kahana A., Wolf A. J., Meisinger L. L., Peterson S. E., et al. , 1998. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 150: 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Klar A. J. S., 1992. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 6: 186–196. [DOI] [PubMed] [Google Scholar]
- Sinha M., Watanabe S., Johnson A., Moazed D., Peterson C. L., 2009. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell 138: 1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeal T., Claus J., Kennedy B., Cole F., Guarente L., 1996. Loss of transcriptional silencing causes sterility in old mother cells of Saccharomyces cerevisiae. Cell 84: 633–642. [DOI] [PubMed] [Google Scholar]
- Smith D. L., Jr, Li C., Matecic M., Maqani N., Bryk M., et al. , 2009. Calorie restriction effects on silencing and recombination at the yeast rDNA. Aging Cell 8: 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. S., Boeke J. D., 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11: 241–254. [DOI] [PubMed] [Google Scholar]
- Smith J. S., Brachmann C. B., Pillus L., Boeke J. D., 1998. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149: 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. S., Caputo E., Boeke J. D., 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19: 3184–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. S., Brachmann C. B., Celic I., Kenna M. A., Muhammad S., et al. , 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97: 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling A. S., Grunstein M., 2009. Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc. Natl. Acad. Sci. USA 106: 13153–13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson J. B., Gottschling D. E., 1999. Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev. 13: 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E. M., Pillus L., 1996. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J. Cell Biol. 135: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E. M., Heun P., Laroche T., Pillus L., Gasser S. M., 2000. MAP kinase signaling induces nuclear reorganization in budding yeast. Curr. Biol. 10: 373–382. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M., 1997. Sir2 and Sir4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11: 83–93. [DOI] [PubMed] [Google Scholar]
- Straight A. F., Shou W., Dowd G. J., Turck C. W., Deshaies R. J., et al. , 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97: 245–256. [DOI] [PubMed] [Google Scholar]
- Stumpferl S. W., Brand S. E., Jiang J. C., Korona B., Tiwari A., et al. , 2012. Natural genetic variation in yeast longevity. Genome Res. 22: 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N., Suka Y., Carmen A. A., Wu J., Grunstein M., 2001. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell 8: 473–479. [DOI] [PubMed] [Google Scholar]
- Suka N., Luo K., Grunstein M., 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32: 378–383. [DOI] [PubMed] [Google Scholar]
- Sussel L., Shore D., 1991. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc. Natl. Acad. Sci. USA 88: 7749–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L., Vannier D., Shore D., 1993. Epigenetic switching of transcriptional states: cis- and trans- acting factors affecting establishment of silencing at the HMR locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 3919–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A., Heller R. C., Landry J., Choy J. S., Sirko A., et al. , 2001. A novel form of transcriptional silencing by Sum1–1 requires Hst1 and the origin recognition complex. Mol. Cell. Biol. 21: 3514–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swygert S. G., Manning B. J., Senapati S., Kaur P., Lindsay S., et al. , 2014. Solution-state conformation and stoichiometry of yeast Sir3 heterochromatin fibres. Nat. Commun. 5: 4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard R. K., Jacques P. E., Laramee L., Cheng B., Galicia S., et al. , 2010. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat. Struct. Mol. Biol. 17: 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Wu R., 1980. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature 284: 426–430. [DOI] [PubMed] [Google Scholar]
- Tackett A. J., Dilworth D. J., Davey M. J., O’Donnell M., Aitchison J. D., et al. , 2005. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 169: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Hediger F., Neumann F. R., Bauer C., Gasser S. M., 2004. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 23: 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., Van Houwe G., Nagai S., Erb I., van Nimwegen E., et al. , 2009. The functional importance of telomere clustering: global changes in gene expression result from SIR factor dispersion. Genome Res. 19: 611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y. H., Schulze J. M., Jackson J., Hentrich T., Seidel C., et al. , 2011. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol. Cell 42: 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner K. G., Landry J., Sternglanz R., Denu J. M., 2000. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 97: 14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J. C., Moazed D., 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98: 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J. C., Dowd G. J., Huang J., Hilz H., Moazed D., 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99: 735–745. [DOI] [PubMed] [Google Scholar]
- Tanny J. C., Kirkpatrick D. S., Gerber S. A., Gygi S. P., Moazed D., 2004. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell. Biol. 24: 6931–6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L., Eisen M. B., Rine J., 2008. Silent but not static: accelerated base-pair substitution in silenced chromatin of budding yeasts. PLoS Genet. 4: e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therizols P., Duong T., Dujon B., Zimmer C., Fabre E., 2010. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc. Natl. Acad. Sci. USA 107: 2025–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Johnson L. M., Grunstein M., 1994. Specific repression of the yeast silent mating locus HMR by an adjacent telomere. Mol. Cell. Biol. 14: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Snow M. L., Giles S., McPherson L. E., Grunstein M., 2003. Identification of a functional domain within the essential core of histone H3 that is required for telomeric and HM silencing in Saccharomyces cerevisiae. Genetics 163: 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurtle D. M., Rine J., 2014. The molecular topography of silenced chromatin in Saccharomyces cerevisiae. Genes Dev. 28: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triolo T., Sternglanz R., 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381: 251–253. [DOI] [PubMed] [Google Scholar]
- Tsang A. W., Escalante-Semerena J. C., 1998. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem. 273: 31788–31794. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y., Kato J.-I., Ikeda H., 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388: 900–903. [DOI] [PubMed] [Google Scholar]
- Valenzuela L., Gangadharan S., Kamakaka R. T., 2006. Analyses of SUM1–1-mediated long-range repression. Genetics 172: 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L., Dhillon N., Dubey R. N., Gartenberg M. R., Kamakaka R. T., 2008. Long-range communication between the silencers of HMR. Mol. Cell. Biol. 28: 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L., Dhillon N., Kamakaka R. T., 2009. Transcription independent insulation at TFIIIC-dependent insulators. Genetics 183: 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse D. W., Wan Y., Lapetina D. L., Chen W. M., Chiang J. H., et al. , 2013. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 152: 969–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F., Gafken P. R., Gottschling D. E., 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756. [DOI] [PubMed] [Google Scholar]
- van Welsem T., Frederiks F., Verzijlbergen K. F., Faber A. W., Nelson Z. W., et al. , 2008. Synthetic lethal screens identify gene silencing processes in yeast and implicate the acetylated amino terminus of Sir3 in recognition of the nucleosome core. Mol. Cell. Biol. 28: 3861–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L., Kim M., Terzi N., Soares L. M., Buratowski S., 2008. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell 29: 313–323. [DOI] [PubMed] [Google Scholar]
- Venkatasubrahmanyam S., Hwang W. W., Meneghini M. D., Tong A. H., Madhani H. D., 2007. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A. Z. Proc. Natl. Acad. Sci. USA 104: 16609–16614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., et al. , 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., et al. , 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2: 709–718. [DOI] [PubMed] [Google Scholar]
- Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., et al. , 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Wang D., Mansisidor A., Prabhakar G., Hochwagen A., 2016. Condensin and Hmo1 mediate a starvation-induced transcriptional position effect within the ribosomal DNA array. Cell Reports 14: 1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Li G., Altaf M., Lu C., Currie M. A., et al. , 2013. Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA. Proc. Natl. Acad. Sci. USA 110: 8495–8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Connelly J. J., Wang C. L., Sternglanz R., 2004. Importance of the Sir3 N terminus and its acetylation for yeast transcriptional silencing. Genetics 168: 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Bryant G., Zhao A., Ptashne M., 2015. Nucleosome avidities and transcriptional silencing in yeast. Curr. Biol. 25: 1215–1220. [DOI] [PubMed] [Google Scholar]
- Warner J. R., 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Weiss K., Simpson R. T., 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMLα. Mol. Cell. Biol. 18: 5392–5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitao T., Budd M., Hoopes L. L., Campbell J. L., 2003. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem. 278: 22513–22522. [DOI] [PubMed] [Google Scholar]
- Whiteway M., Freedman R., Van Arsdell S., Szostak J. W., Thorner J., 1987. The yeast ARD1 gene product is required for repression of cryptic mating-type information at the HML locus. Mol. Cell. Biol. 7: 3713–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiren M., Silverstein R. A., Sinha I., Walfridsson J., Lee H. M., et al. , 2005. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 24: 2906–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. S., Chen Y. F., Gartenberg M. R., 2011. Targeted sister chromatid cohesion by Sir2. PLoS Genet. 7: e1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Pierce M., Gailus-Durner V., Wagner M., Winter E., et al. , 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18: 6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu E. Y., Zawadzki K. A., Broach J. R., 2006. Single-cell observations reveal intermediate transcriptional silencing states. Mol. Cell 23: 219–229. [DOI] [PubMed] [Google Scholar]
- Xu F., Zhang Q., Zhang K., Xie W., Grunstein M., 2007. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell 27: 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. H., Su T., Xue Y., 2016. Histone H3 N-terminal acetylation sites especially K14 are important for rDNA silencing and aging. Sci. Rep. 6: 21900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Van C., Pradhan S. K., Su T., Gehrke J., et al. , 2015. The Ino80 complex prevents invasion of euchromatin into silent chromatin. Genes Dev. 29: 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Kirchmaier A. L., 2006. Bypassing the catalytic activity of SIR2 for SIR protein spreading in Saccharomyces cerevisiae. Mol. Biol. Cell 17: 5287–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Fang Q., Wang M., Ren R., Wang H., et al. , 2013. Nα-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain. Nat. Struct. Mol. Biol. 20: 1116–1118. [DOI] [PubMed] [Google Scholar]
- Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., et al. , 2004. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23: 2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T. J., Kirchmaier A. L., 2013. Cell cycle regulation of silent chromatin formation. Biochim. Biophys. Acta 1819: 303–312. [DOI] [PubMed] [Google Scholar]
- Zaman S., Choudhury M., Jiang J. C., Srivastava P., Mohanty B. K., et al. , 2016. Mechanism of regulation of intrachromatid recombination and long-range chromosome interactions in Saccharomyces cerevisiae. Mol. Cell. Biol. 36: 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla D. C., Sternglanz R., Leatherwood J., 2002. Control of replication timing by a transcriptional silencer. Curr. Biol. 12: 869–875. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Shibahara K.-I., Stillman B., 2000. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408: 221–225. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Hayashi M. K., Merkel O., Stillman B., Xu R. M., 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21: 4600–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B. O., Wang S. S., Zhang Y., Fu X. H., Dang W., et al. , 2011. Histone H4 lysine 12 acetylation regulates telomeric heterochromatin plasticity in Saccharomyces cerevisiae. PLoS Genet. 7: e1001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zill O. A., Scannell D., Teytelman L., Rine J., 2010. Co-evolution of transcriptional silencing proteins and the DNA elements specifying their assembly. PLoS Biol. 8: e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Yu Q., Bi X., 2006. Asymmetric positioning of nucleosomes and directional establishment of transcriptionally silent chromatin by Saccharomyces cerevisiae silencers. Mol. Cell. Biol. 26: 7806–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]