Abstract
Burkitt’s Lymphomas (BLs) acquire consistent point mutations in a conserved domain of Myc, Myc Box I. We report that the enhanced transforming activity of BL-associated Myc mutants can be uncoupled from loss of phosphorylation and increased protein stability. Furthermore, two different BL-associated Myc mutations induced similar gene expression profiles independently of T58 phosphorylation, and these profiles are dramatically different from MycWT. Nol5a/Nop56, which is required for rRNA methylation, was identified as a gene hyperactivated by the BL-associated Myc mutants. We show that Nol5a is necessary for Myc-induced cell transformation, enhances MycWT-induced cell transformation, and increases the size of MycWT induced tumors. Thus, Nol5a expands the link between Myc-induced regulation of nucleolar target genes which are rate-limiting for cell transformation and tumor growth.
INTRODUCTION
Burkitt’s Lymphoma (BL) is an aggressive mature B cell lymphoma1, in which the Myc proto-oncogene is translocated to the immunoglobulin loci.2 In addition to the consistent chromosomal translocations, a majority of Burkitt’s lymphomas have mutations in the coding region of Myc which cluster in a conserved region known as Myc Box I (MBI), including E39D, A44V, P57S, T58A, and T58I.3-9 The repeated identification of these mutations in tumors suggests that they are selected for during Myc-driven tumorigenesis. The majority of investigations into the activity of the Burkitt’s lymphoma Myc mutants have focused on MycT58A.10-12 Myc is phosphorylated on T58, and this phosphorylation is reported to be required for Myc degradation.13 MycT58A has been found to have an increased half-life and expression level, an increased transforming activity, and/or an impairment in Myc induced apoptosis.10,14-18 Studies in systems in which Myc does not induce apoptosis still show increases in transformation by MycT58A expressing cells10,14,15, suggesting that changes in apoptosis are not solely responsible for changes in Myc oncogenic activity.
Myc likely drives cell proliferation through its broad influence on gene expression which controls a number of essential functions; metabolism, protein synthesis, mRNA cap methylation, and promoting the activity of RNA pol I and III.19-21,22,23-25 The question arises whether the transformed phenotype induced by Myc overexpression is the result of a general increase in all Myc functions or due to altered regulation of specific genes. In cancers of several origins, it has been demonstrated that deregulation of specific Myc targets genes plays an integral role in promoting the transformed phenotype.19,20 Studies on the effect of the T58A mutation on target gene regulation vary widely, but some specific examples of genes and proteins differentially expressed in response to MycWT and MycT58A expression have been found.17,26,27 To date there has been scant investigation of the more common BL-associated Myc mutants, MycE39D and MycT58I.10,28 In this study, we applied global gene expression analysis to examine how the BL-associated Myc Box I mutants modulate Myc function and to identify the target genes that these mutants regulate to enhance Myc-induced transforming activity. We find that different mutations converge on remarkably similar and substantial changes in Myc-responsive genes compared to MycWT and that Nol5a/Nop56, a gene involved in ribosome biogenesis, may be a critical gene involved in Myc-mediated oncogenic transformation.
RESULTS
BL-associated Myc mutations increase the oncogenic potential of Myc, independent of T58 phosphorylation
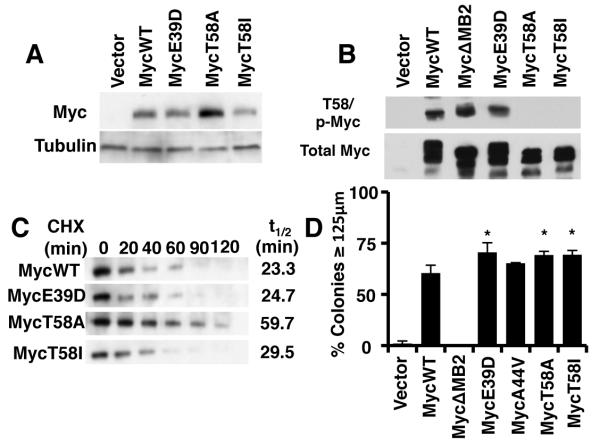
To investigate the role of BL-associated Myc mutants in Myc-mediated tumorigenesis, we created a panel of mutants corresponding to the most common mutations; MycE39D, MycT58I, and MycT58A.10 Upon stable overexpression of these mutants, we observed variable levels of Myc expression (Figure 1A). The mutants MycE39D and MycT58I expressed similarly to Myc wildtype, whereas MycT8A showed a substantial increase (2.6 fold) in expression compared to Myc wildtype. To determine if the increase in expression was due to changes in protein stability, we treated the cells with cycloheximide and observed protein levels at varying times (Figure 1B). The half-life of MycE39D was equivalent to that of MycWT (23 ±6.2 minutes), similar to previously reported half-lives for Myc, whereas MycT58A had an increased half-live of more than twice that of MycWT (59.7 ±10.4 min). This result is consistent with previous studies of MycT58A half-life.29 Surprisingly we did not observe any appreciable increase in MycT58I stability compared to that of MycWT, consistent with the equivalent steady state level. We observed that MycWT and MycE39D retained T58 phosphorylation, whereas both the MycT58A and MycT58I mutation where no longer phosphorylated in both HEK293 and myc−/− fibroblasts (Fig 1C and data not shown). One explanation for the differential stability of MycT58A versus MycT58I could be previously observed differences in S62 phosphorylation10, which contribute- to changes in stability.13
Figure 1. Burkitt’s Lymphoma Myc mutants enhance Myc-induced cell transformation independently of loss of T58 phosphorylation and increased expression.
A) Rat1a fibroblasts were engineered to express MycWT, the indicated Burkitt’s Lymphoma (BL) Myc mutants or vector control. Cell extracts were analyzed by Western blot using anti-Myc or anti-Tubulin antibodies. B) MycWT and the BL-associated Myc mutants were transiently expressed in 293 cells. Cell extracts were analyzed by parallel western blots using anti-phospho-Myc (p58/p62) or anti-Myc (phospho-independent). C) The half-life of MycWT and BL-associated mutants was determined by engineering myc−/− fibroblast to express the indicated proteins, and cells were treated with 50 μg/mL cycloheximide for the up to 120 minutes. Cell extracts were analyzed by western blot using anti-Myc antibodies D) Log phase Rat1a fibroblasts expressing MycWT or BL-associated Myc mutants were used in a soft agar transformation assay. The chart depicts the diameter of colonies from each cell line after 1 week of growth. Error bars indicate the standard deviation (SD) for three biological replicates using two independently engineered cell line. Significance was determined using student’s t-test; *p≤0.05.
Since the E39D and T58I mutations do not increase protein stability, we wanted to determine if these mutations were still able to enhance Myc-mediated transformation (Figure 1D). Although we found no change in the proliferation rates (Supplemental Figure 1), all three mutants (MycE39D, MycT58A, and MycT58I) enhanced anchorage-independent growth compared to MycWT or the transformation-defective MycΔMB2 mutant. We observed a 35-40% increase in Myc-mediated transformation on average. Thus, Myc-induced transformation is enhanced with BL-associated Myc mutants, independent of protein stability and T58 phosphorylation status. A previous study failed to show similar increases in these Myc mutant alleles.10 However these studies used an oncogene cooperation assay to determine oncogenic potential which may lack the sensitivity to observe these more subtle qualitative difference from the more robust quantitative difference between c-Myc overexpression.
MycE39D and MycT58I regulate target gene expression similarly but significantly different from MycWT
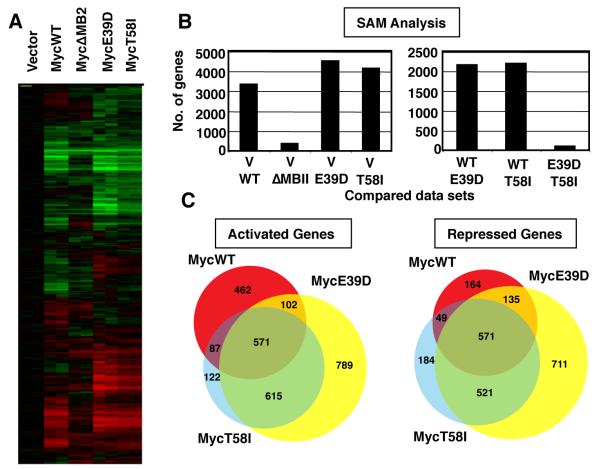
We investigated whether the enhanced transforming activity of the BL-associated Myc mutants could be correlated with differential regulation of target genes. To avoid contributions of differential protein stability, we transduced MycE39D, MycT58I, and MycΔMB2 (a non-transforming Myc mutant) into myc−/− fibroblasts.30 Myc protein expression in this polyclonal cell system is relatively low, and MycE39D and MycT58I were expressed equivalently to MycWT and at physiological levels similar to parental myc+/+ fibroblasts. (Supplemental Figure 2A). Proliferation rates and cell morphology were also similar to parental fibroblasts. RNA was harvested from two independent log-phase pools for each Myc variant or vector control and analyzed for gene expression by whole genome microarray. A graphic comparison of gene expression profiles can be visualized by hierarchical clustering of the complete data set (Figure 2A). After filtering and normalizing the data, the number of genes up-regulated and down-regulated by 1.5-, 1.75- and 2-fold in response to each Myc protein was compared to vector control (Supplemental Figure 2B). Consistent with microarray data from this and other cell systems, expression of MycWT was found to regulate a large number of genes in a MB2-dependent manner, and there were generally equivalent numbers of genes up-regulated and down-regulated.
Figure 2. The BL-associated mutants, MycE39D and MycT58I, have a highly similar set of target genes which is significantly different from MycWT target genes.
A) myc−/− fibroblasts were engineered to express MycWT, a transactivation defective mutant MycΔMB2, or the indicated BL-associated Myc mutants. RNA was extracted from two independent polyclonal cell populations expressing the indicated Myc protein or vector control. Expression microarrays were performed using Agilent 4X44K Whole Rat genome microarrays. The figure shows the clustered heat map of all genes with signals 1.5 fold above background and normalized to vector control. B) Significance Analysis of Microarray (SAM) analysis was performed to compare the number of genes that were differentially regulated between the cell lines expressing the Myc proteins and vector control (left panel), and between the cell lines expressing the different Myc proteins (right panel). C) Venn diagram representing target gene overlap of MycWT and BL-associated mutants for genes up-regulated or down-regulated 1.5-fold over vector.
A key question was whether MycWT and the BL-associated mutants regulated the same or different genes. We carried out a “Significance Analysis of Microarrays” or SAM31 which analyzes the differences in gene expression profiles between groups of cell lines (Figure 2B). At a false discovery rate of 1% in a 2-class analysis, SAM determined that there were 3326 genes differentially expressed between MycWT expressing and vector control myc−/− cells. Similarly, 4494 genes were differentially expressed between MycE39D expressing and vector control cells, and 4116 genes were differentially expressed between MycT58I expressing and vector control cells. The most significant question is whether MycWT and the BL-associated Myc mutants activate the same or different genes. We made the unexpected finding that a large number of genes are differentially regulated by MycWT and the Myc mutants. Using the same 1% false discovery rate, there were 2150 genes differentially regulated between cells expressing MycWT and MycE39D, and 2186 genes between cells expressing MycWT and MycT58I (Supplemental Table S1). Most strikingly, there was little difference between the gene sets expressed in MycE39D and MycT58I. SAM determined that only 100 genes were expressed at a significantly different level between these BL-associated Myc mutants. Since MycE39D is phosphorylated at both T58/S62 whereas MycT58I is not, this substantial shift in Myc-responsive genes is independent of phosphorylation status. Thus, two independent mutations skew the Myc response in a remarkably similar way from the MycWT profile, correlating with an enhanced oncogenic potential for both mutants.
We next examined the overlap of genes up-regulated and down-regulated 1.5-fold or more over vector in MycWT and BL associated mutants gene sets (Figure 2C, Supplemental Table 2 & 3). Of the 1222 genes activated by MycWT, 462 (38%) were activated by MycWT alone, whereas a nearly equivalent number (615; 50%) were jointly activated by the BL-associated mutants alone. Similarly, 164 of 919 (18%) genes were repressed by MycWT alone, whereas 521 (56%) were jointly repressed by BL-associated mutants alone. We noted that the total number of genes activated and repressed by MycE39D was greater (approximately 50%) than MycT58I and MycWT. However, 1186 of 1345 (89%) were up-regulated and 1092 of 1329 (82%) were down-regulated similarly between MycT58I and MycE39D. We performed a GO term analysis on these subsets of genes (MycWT only, BL-associated mutant only, and commonly regulated) to determine if any particular pathway changes coud account for the observed increase in tumorigenesis. While there were slight changes in gene numbers in a variety of pathways, we did not observe any significant changes in important pathways for the BL-associated mutants.
It is apparent that broad groups of genes are differentially regulated by the Myc mutants versus MycWT, and gene expression sometimes reversed from activated to repressed (or visa versa) between MycWT and mutants. This demonstrates that, although all the Myc proteins regulate a subset of the same genes, there are large groups of genes that are regulated differently by the Myc mutants and MycWT. We also noted that the MycE39D responsive genes have generally more robust signals than MycT58I, consistent with the larger number of genes that pass each threshold.
Differential target gene regulation results in changes in gene expression and promoter histone modifications associated with Myc co-factor binding
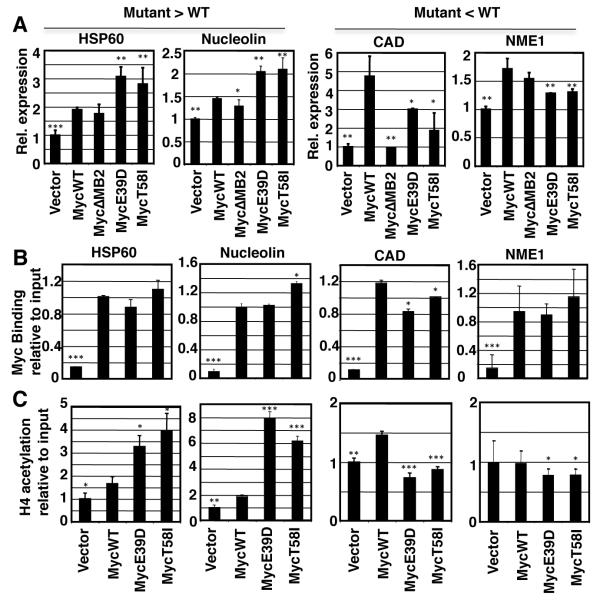
We wanted to investigate how BL-associated Myc mutations could alter the Myc target gene expression profile. Myc promotes transcription by binding to target genes and recruiting histone acetyltransferase complexes.12,33,34 We investigated Myc recruitment and histone acetylation on two genes, HSP60 and Nucleolin, that are hyperactivated by the Myc mutants compared to MycWT. These were compared to two genes, CAD and NME1, which had reduced activation by the Myc mutants compared to MycWT. We used independent RNA samples to confirm differential expression by the Myc mutants compared to MycWT (Figure 3A). CAD and NME1 were induced more by MycWT than the Myc mutants, whereas HSP60 and Nucleolin were induced more by the Myc mutants. For all four genes, we used chromatin-immunoprecipitation (ChIP) to investigate Myc recruitment to the E-box closest to the transcription initiation site (Figure 3B). We found that MycWT, MycE39D and MycT58I were all recruited equivalently to the E-boxes of all four Myc target genes. As a negative control we found that the Myc proteins were not significantly recruited to the ß-globin promoter (data not shown). We next investigated histone acetylation in response to MycWT and the Myc mutants at the same genes. We found Myc-dependent histone H4 acetylation at all four target genes (Figure 3C). In correlation with the increased HSP60 and Nucleolin expression, we observed increased H4 acetylation at the HSP60 and NUCL genes in response to expression of the Myc mutants compared to that induced by MycWT. Conversely, in correlation with the reduced CAD and NME1 expression observed in response to MycE39D and MycT58I, we observed reduced histone acetylation at the CAD and NME1 genes compared to that induced by MycWT. Thus the Myc mutations affect local chromatin modifications but do not affect promoter occupancy, consistent with their location in the Myc transactivation domain and not the DNA binding domain.
Figure 3. Changes in target gene expression between MycWT and BL associated mutants correspond with changes in H4 acetylation, but not Myc promoter occupancy.
A) Validation of expression microarrays was performed by RT-PCR on Rat1a engineered cell lines expressing indicated form of Myc or vector, to confirm expression levels of known Myc target genes HSP60, Nucleolin, CAD, NME1. The differential regulation of these target genes is consistent with relative expression levels determined by expression microarray. B) ChIP assays using anti-Myc antibodies were performed to determine Myc binding to E boxes in the promoters of indicated target gene C) ChIP assay using anti-acetylated H4 antibodies were performed at the same sites to analyze histone acetylation by the indicated Myc protein. Error bars indicate SD over three biological replicates. Asterisk indicates significant difference compared to MycWT; student’s t-test, *p≤0.05, **p≤0.01,***p≤0.001.
Myc mutations differentially regulate Nol5a, a gene required for ribosome biogenesis
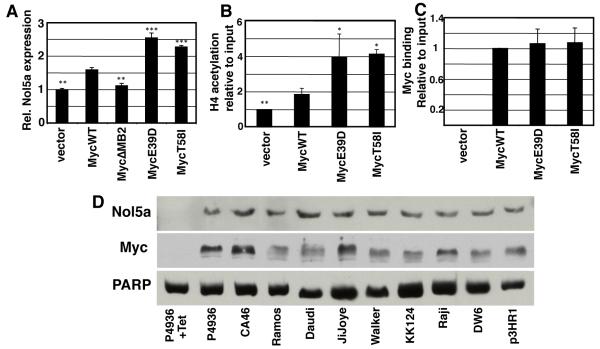
The BL-associated mutations result in increased Myc-induced cell transformation and a qualitative change in Myc responsive genes. We wanted to test the hypothesis that the increase in cell transformation was a result of specific changes in gene expression. Myc regulates all aspects of protein synthesis and ribosome biogenesis35,36, and recent evidence suggests that ribosome biogenesis may be a primordial function of Myc, conserved throughout evolution in even the most primitive organisms that harbor Myc and Max orthologs.37 Furthermore, mice that overexpress Myc but express only one functional copy of ribosomal protein L24 have significantly reduced translation rates and impaired Myc mediated oncogenesis.38 We therefore focused our studies on an essential ribosome biogenesis target gene that was differentially activated in Myc mutants and is used as a molecular diagnostic for Burkitt’s Lymphoma.39 Nol5a (nucleolar protein 5a/Nop56) encodes a component of an rRNA methylation complex that also regulates rRNA processing.40-42 We first established that Nol5a expression was also increased in Rat1a fibroblasts in response to MycWT (Figure 4A). Nol5a expression was further elevated in cells expressing MycE39D and MycT58I. Histone acetylation on the Nol5a gene was higher in cells expressing the Myc mutants compared to those expressing MycWT, correlating with Nol5a expression levels (Figure 4B). In accord with the earlier examples of Myc target genes, the Myc mutants and MycWT exhibit equivalent recruitment to the E-box closest to the Nol5a transcription initiation site (Figure 4C). Nol5a expression was also elevated in response to the activation of Myc expression in P493-6 cells and in BL cell lines (Figure 4D).
Figure 4. The nucleolar protein gene, Nol5a, is differentially induced and acetylated by MycE39D and MycT58I, and highly expressed in Burkitt’s Lymphoma cell lines.
A) The relative expression level of Nol5a in Rat1A fibroblasts engineered to express MycWT or the indicated BL associated mutations was determined by qPCR. B) ChIP assays were performed at the same site in the Nol5a promoter using anti-acetylated H4 antibodies to determine relative acetylation by the indicated myc proteins. C) ChIP assays were performed using anti-Myc antibodies to determine Myc binding at a conical E box in the Nol5a promoter. D) P493-6 human B cells express high levels of Myc which can be repressed by tetracycline. After 72 hour treatment with 0.1 μg/mL tetracycline or vehicle, nuclear lysates for P493-6 cells as well as indicated Burkitt’s Lymphoma cell lines were prepared and analyzed by western blots using an anti-c-myc and anti-Nop56 antibodies. Error bars (SD) represent three biological replicates. Asterisk indicates significant difference compared to MycWT; student’s t-test, *p≤0.05, **p≤0.01,***p≤0.001.
Nol5a overexpression is required for Myc mediated transformation
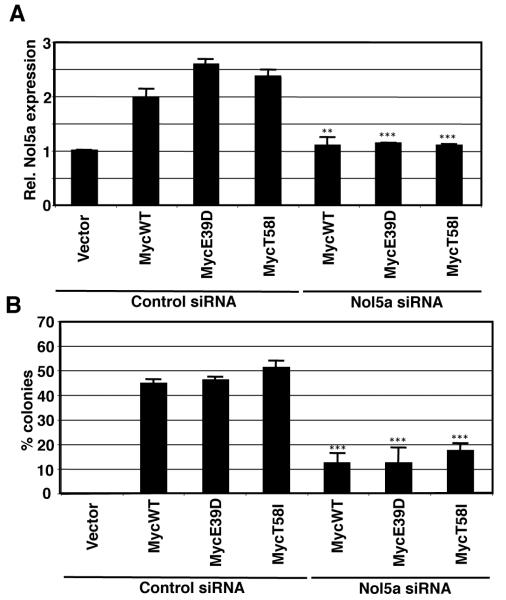
To explore a functional role for Nol5a in Myc transformation, Nol5a expression was carefully titrated using siRNAs. As expected, nearly complete depletion of Nol5a expression resulted in severely reduced proliferation rates and increased cell death (not shown), consistent with its essential role in ribosome biogenesis.40-42 However, when levels of Nol5a were reduced by 50% in Rat1a/Myc cells, equivalent to levels in cells not overexpressing Myc, proliferation rates were unaffected and cells remained viable (Figure 5A and Supplemental Figure 3). We next examined how the reduction of Nol5a expression would effect the transformation potential of MycWT, MycE39D, and MycT58I. Soft agar colony growth confirmed a substantial increase in transformation in cells overexpressing MycWT, MycE39D, and MycT58I. Upon depletion of Nol5a, there was significant loss in transformation (Figure 5B). In summary, cell transformation by Myc is dependent on Myc-induced Nol5a expression, whereas a maximal cell proliferation rate only requires vector control levels of Nol5a expression.
Figure 5. Myc induced Nol5a overexpression is necessary for MycWT and BL mutant cellular transformation.
A) Rat1A fibroblast cells expressing the indicated proteins were treated with control or Nol5a siRNA. After 48 hrs, RNA was obtained and qPCR was performed to determine relative levels of Myc induced Nol5a. Nol5a levels were titrated to levels equivalent to cells containing control vector. B) Cells which expressed Nol5a at equivalent levels as vector cells were replated and assessed for their ability to grow in soft agar. Error bars (SD) represent three biological replicates. Asterisk indicates significant difference between control and Nol5a siRNA treated cells; student’s t-test, *p≤0.05, **p≤0.01,***p≤0.001.
Nol5a enhances MycWT-induced transformation and tumor growth
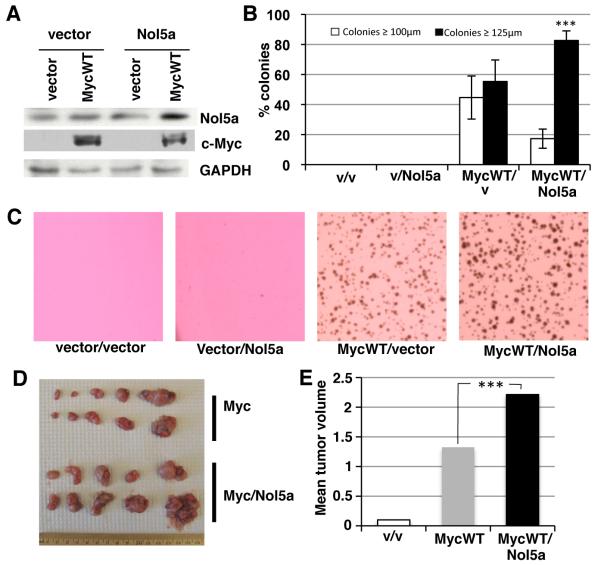
Expression of the Myc mutants increases Nol5a expression to a level above that induced by MycWT. We wanted to investigate whether this hyperactivation had functional significance. To this end, we experimentally increased Nol5a levels above those found in MycWT expressing cells by ectopically expressing Nol5a in MycWT/Rat1a cells. We confirmed that ectopic expression of Nol5a did not enhance endogenous or exogenous MycWT expression (Figure 6A). Cells were plated in soft agar, and colonies were counted and measured. (Figure 6B). As described earlier, expression of MycWT allowed cells to grow into robust colonies in soft agar. We found that further increasing the expression of Nol5a in MycWT expressing cells enhanced cell transformation, producing significantly larger colonies, as depicted in colony images and diameter measurements (Figures 6B and 6C). Increased Nol5a expression alone was not sufficient to permit cell growth in soft agar (Figure 6B). The increased colony size was not due to any increase in the cell proliferation rate of cells expressing MycWT and elevated Nol5a (Supplemental Figure 3B) or in rRNA processing (Supplemental Figure 4). In summary, expression of Nol5a at levels above those induced by MycWT, as found in response to the BL-associated Myc mutants, is functionally significant and can enhance MycWT-induced cell transformation.
Figure 6. Nol5a synergizes with Myc WT to increase cellular transformation and tumor size.
A) Rat1A fibroblast were engineered to express control vector, Nol5a, and/or MycWT. Lysates were assayed by western blot using anti-Nol5a (Sigma), anti-Myc, and anti-GAPDH. B) Cells expressing the indicated vectors were plated in soft agar and cellular transformation was measured by colonies formation and scored for size after 7 days. Colonies were scored as ≤ 100 μm or ≥ 125 μm divisions using a reticule; student’s t-test, p≤0.001 C) Representative soft agar plates expressing indicated vectors 7 days after plating. D) Rat1A cells engineered to express vector (n=10), MycWT (n=10), or MycWT/Nol5a (n=10) were injected subcutaneously into the flanks of nude mice and allowed to form tumors. The tumors were extracted after 3 wks of growth. E) Individual tumors were weighed and mean mass for each tumor type was calculated. The mass of tumors induced by MycWT, and MycWT/Nol5a were determined to be significantly different; student’s t-test, p≤0.001.
Since Nol5a expression enhances MycWT-induced cell transformation in the soft agar assay, we wanted to expand upon these assays and measure in vivo tumor growth. We injected Rat1a cells expressing MycWT, MycWT plus Nol5a, or vector control subcutaneously into the flanks of nude mice (Figures 6D and 6E). Vector control injections resulted in no tumors or nearly undetectable tumors (0.1 g or below), while all mice injected with cells expressing MycWT or MycWT and Nol5a produced sizeable tumors. We compared the mean mass of each of the groups, 1.20g and 2.14g respectively, and determined that the mass of MycWT/Nol5a tumors was significantly higher than those produced from MycWT (P value > 0.001). Therefore, Nol5a enhances Myc-induced tumor formation similarly to oncogenic transformation assays in cell culture.
DISCUSSION
We have been interested in a detailed understanding of how Burkitt’s lymphoma-associated Myc mutations contribute to cancer. The most frequently studied mutation has been MycT58A, which can have a modest effect on Myc protein levels through reduced turnover. However, we show that BL-associated mutations enhance cell transformation independently of loss of T58 phosphorylation or increases in Myc expression. All Myc mutants investigated have increased soft agar colony size. Thus, the effect of T58 mutations appear to be allele-specific, with some leading to increased half-life and others unchanged from MycWT. These observations are consistent with previous findings that Myc mutations do not alter Myc protein levels in a murine B-lymphoma model.17 Thus the mutations contribute to qualitative rather than quantitative changes in Myc protein biochemistry and activity.
To understand how the BL-associated Myc mutants increase cell transformation, we used expression microarrays to investigate the endogenous gene response to these mutants. We focused our study on MycE39D, which retains T58 phosphorylation, and on MycT58I, which lacks the T58 phosphorylation site, to determine if this phosphorylation status qualitatively effected target gene expression. We found that the MycE39D mutation increases the number of Myc target genes by approximately 50%, but the T58I mutation had no effect on the absolute number of activated or repressed genes at each threshold. Therefore a global increase or decrease in gene expression is not necessary for increased cell transformation. However, analysis of the transcriptome of MycE39D and MycT58I expressing cells revealed that these point mutants had a significant effect on the profile of responsive genes compared to MycWT. From gene clustering and SAM analysis, MycWT, MycE39D and MycT58I were found to regulate a subset of the same genes, but a large proportion of the BL-associated Myc mutant responsive genes are significantly different from those regulated by MycWT. The surprising finding was that MycE39D and MycT58I converge to regulate an almost identical set of target genes that is distinct from MycWT. Thus, in this system, subtle mutations in the Myc transactivation domain substantially alter the responsive genes, and blocking T58 phosphorylation is not responsible for altering the Myc transcriptome. Recent publications suggest a model for Myc function as a ‘transcriptional amplifier’ of all active genes in a cell 43,44. While this model can account for the broad effect that Myc has on genome wide transcription, it does not account for the allele-specific differences in individual gene expression quantitated in our study.
Myc promotes increased transcription by increasing histone acetylation at target gene promoters.12,33,34 To gain insight into the mechanism by which the BL-associated point mutations were altering the Myc transcriptome, we investigated Myc recruitment and histone acetylation at four well-established Myc target genes. The BL-associated mutations did not influence Myc recruitment to the genes tested, but we found significant changes in histone acetylation that paralleled changes in mRNA levels. We suggest that the BL-associated mutations induce similar conformational changes in the Myc transactivation domain from that of MycWT which results in altered recruitment of chromatin modifying and transcription promoting complexes.
We found that over 2000 genes were differentially regulated between MycWT and the Myc mutants. While it is possible that many of these genes are collectively responsible for the increased transforming activity of the mutants, we focused our attention on a single differentially regulated gene, Nol5a/Nop56. Nol5a is a nucleolar protein that is a component of a ribonuclear protein complex which methylates rRNA and regulates rRNA processing, although it may also have other functions.40-42 Our attention was drawn to Nol5a because its expression has been found to be a highly associated with Burkitt’s lymphoma and serves as a molecular marker for diagnosis39, suggesting a potentially biological role for its differential expression in these tumors. In addition Nol5a has been shown to be upregulated in two additional B cell lymphomas, diffuse B cell lymphoma and chronic lymphocytic leukemia.45,46 Nol5a has previously been found to be a Myc target gene by several global analyses of gene expression, including in B cells.47-50 We show that Nol5a expression is also high in Burkitt’s Lymphoma cell lines and induced in response to expression of MycWT in B cells and in fibroblasts. Nol5a is further increased by the MycE39D and MycT58I mutations. The higher levels of Nol5a expression induced by MycE39D and MycT58I, above that induced by MycWT, is functionally significant because it enhanced MycWT-induced transformation in two different assays. This demonstrates that increased Nol5a expression by the BL-associated Myc mutants is critical for enhanced oncogenicity.
Nol5a is an essential gene for cell viability, and it has been demonstrated to be rate-limiting for cell proliferation in yeast and mammalian cells.40,51 Notably, a screen for genes that promote a shortened G1 phase of the cell cycle in yeast identified Nop56/Sik1 as an important mediator of cell cycle progression.52 We also found that severe Nol5a depletion inhibited cell proliferation. However when we titrated the levels of Nol5a in Myc expressing cells to levels found in vector control cells, cell transformation was inhibited but there was no effect on cell proliferation. Therefore Nol5a is rate-limiting for biological effects over a broad range of expression. A basal level of Nol5a is necessary and sufficient for cell proliferation, however the MycWT-induced increase in Nol5a is necessary for cell transformation, and further Nol5a increases enhances cell transformation.
Nol5a joins the emerging group of nucleolar proteins which are essential for ribosome biogenesis and which have also been found to promote cell transformation and tumorigenesis.53,54 Myc proteins are recognized to regulate expression of a large number of genes involved in ribosome biogenesis48,55-57 including NPM158, and DKC159, as well as rRNA synthesis itself.19 Furthermore, ribosome biogenesis is the only cellular process in which the genes harbor conserved Myc/Max binding sites from primitive metazoans to humans.37 Although the only known function of Nol5a is in ribosome biogenesis, it may also have an extraribosomal function that remains to be discovered.60 Nucleophosmin, which is frequently mutated in human cancer, offers a precedent for a novel non-nucleolar function since it is essential for ribosome biogenesis but also has a number of additional roles including regulating the activity and stability of Myc, p53 and ARF, and in centrosome duplication.58,61 Nevertheless, the activity of Nol5a in oncogenic transformation strengthens the link between Myc overexpression and ribosome biogenesis and suggests that the protein synthetic capacity of cancer cells may be crucial for Myc-induced tumor formation.
MATERIAL AND METHODS
Cell Culture and Stable Transfections
Rat1a fibroblast, myc−/− fibroblast (HO15.19)30, and retroviral packaging Phoenix cell lines were maintained in DMEM supplemented with 10% FBS. Burkitt’s Lymphoma lines were maintained in Iscove’s modified MEM/10%F FBS. c-Myc repressible P493-6 cells were maintained in RPMI/10% FBS and exogenous c-Myc was repressed by treatment with 0.1 μg/mL Tetracycline for 72 hrs. Retroviral expression of HO15.19 and Rat1a cells was performed using Phoenix retroviral transduction system.
Western Blots and Immunoprecipitation
Rat1a fibroblast, myc−/− fibroblast, and BL cell lines were lysed in F-buffer (10 mM Tris ph 7.05, 50 mM NaCl, 30mM Na Pyrophosphate, 5 mM ZnCl, 10% Glycerol, 0.1% Triton). Cell extracts were normalized for protein content and either immunoprecipitated or whole cell lysates run for western blotting. Myc was immunoprecipitated in HO cells using either monoclonal anti-Myc (C33, Santa Cruz) or anti-pyo (AFC-115P, Covance), and western blots performed with polyclonal anti-Myc (N262, Santa Cruz) for total Myc. To determine the T58 phosphorylation status. 293 cells were transiently transfected, lysed in RIPA buffer (Pierce) with phosphatase inhibitors (Sigma), and western blots performed using anti-phospho-Myc (T58/S62, Santa Cruz) which recognizes both phosphorylated residues. Whole cell lysates were analyzed using polyclonal anti-Myc (N262), anti-Nop56 (BL lines; C19, Santa Cruz) and anti-Nol5a (Rat1A lines; AV40645, Sigma) and anti-tubulin or anti-GAPDH controls (Santa Cruz).
siRNA Knockdowns
Nol5a or scrambled siRNA (Ambion) was transfected in a 6 well plate according to manufacturers instructions. Cells were harvested and replated for soft agar 48 hours post transfection. RNA and protein was harvested for analysis 48 hours post transfection. Error bars (SD) represent three biological replicates and significance was determined using student t-test.
Protein Stability
Treated c-Myc stably expressing myc−/− cells with 50 μg/ml cycloheximide for the up to 120 min. Cells were then lysed using F-buffer, immunoprecipitated and blotted as described above. Band intensity was determined by ImageJ image processing software. Results are from two independent experiments.
Real Time PCR
RT-PCR was performed using iScript cDNA Synthesis Kit (BioRad) according to manufacturers instructions. For each experiment RNA from two independently established cell lines was analyzed. Quantitative PCR was performed by mixing cDNA, appropriate primers, and iQ SYBR Green Supermix (BioRad) and analyzed on a C1000 Thermal Cycler (BioRad). Data was analyzed using Bio-Rad CFX Manager 2.0 software. Error bars (SD) represent three biological replicates and significance was determined using student t-test.
Chromatin Immunoprecipitation
Chromatin Immunoprecipitations were performed using the Upstate Chip kit. The following antibodies were used, polyclonal anti-Myc (N-262; Santa Cruz), anti-Acetyl-H4 (Upstate), and control IgG (Santa Cruz). Error bars (SD) represent three biological replicates and significance was determined using student t-test.
Transformation assays
Soft agar assays were performed as described in.62 Seven days after plating, the longest diameter of 100 colonies from duplicate wells was measured using a reticule. After pretreated with Nol5a or control siRNA, 100 colonies/well from duplicate wells were measured 3 days after plating, and the size (μm) of colonies over 3 reticule divisions (75 μm) in diameter were recorded. Differences in colony size are represented by percentage of colonies greater than 125 μm. Assays were repeated in duplicate wells over three independent experiments with similar results. Significance was determined using student’s t-test.
Microarrays
RNA was harvested on 2 independent occasions from 2 independent cultures of myc-null fibroblasts expressing vector control, MycWT, MycΔMBII, MycE39D and MycT58I using Trizol, followed by phenol chloroform extraction. RNA integrity and concentration was verified by gel electrophoresis. RNA was analyzed on Agilent 4x44K Whole Rat genome microarrays as described in.62 RNA was labeled using the oligo-dT primed linear-amplification protocol according to manufacturers instructions. Only genes with a signal 50% above background and with at least 70% good data across the arrays were considered for further analysis. The data were centered to the average of the two independent vector-only arrays and filtered for average changes in signal for either MycWT or Myc mutants that were above the threshold indicated.
Supervised SAM analysis, GO term analysis, and student’s t-tests
Significance Analysis of Microarrays31, was performed using MeV 4.0 software. To identify the genes that are most significantly different between the different cell types, we employed 2-class SAM analysis with a false discovery rate of 1%. Only those clones in which both replicate spots were found significant were selected. GO term analysis (David, NIAID) was performed on genes up-regulated 1.5fold over vector for subsets representing MycWT only, BL-associated mutants only, or commonly regulated genes. All other statistical significance was determined using non-paired students t-test with Graphpad Prism 6.0b, (La Jolla, CA).
Tumorigenicity assay
1×106 cells in 100 μL PBS were injected subcutaneously in the flanks of 6 week old female nude mice (Charles River) N=10 for all cell types injected. Tumors were excised after 3 weeks and weighed and measured. Significance in mean tumor volume was determined using student’s t-test.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Steve Hann and Chi Dang for providing cell lines. This project was supported by a grant from the National Cancer Institute (CA055248) (MDC) and by Award Number T32GM008704 from the National Institute of General Medical Sciences (SAT). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
REFERENCES
- 1.Yustein JT, Dang CV. Biology and treatment of Burkitt's lymphoma. Curr Opin Hematol. 2007;14:375–381. doi: 10.1097/MOH.0b013e3281bccdee. [DOI] [PubMed] [Google Scholar]
- 2.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 3.Albert T, Urlbauer B, Kohlhuber F, Hammersen B, Eick D. Ongoing mutations in the N-terminal domain of c-Myc affect transactivation in Burkitt's lymphoma cell lines. Oncogene. 1994;9:759–763. [PubMed] [Google Scholar]
- 4.Bhatia K, et al. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nature Genetics. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- 5.Bhatia K, et al. Mutations in the coding region of c-myc occur frequently in acquired immunodeficiency syndrome-associated lymphomas. Blood. 1994;84:883–888. [PubMed] [Google Scholar]
- 6.Brennscheidt U, et al. Burkitt-like mutations in the c-myc gene locus in prolymphocytic leukemia. Leukemia. 1994;8:897–902. [PubMed] [Google Scholar]
- 7.Clark HM, et al. Mutations in the coding region of c-MYC in AIDS-associated and other aggressive lymphomas. Cancer Res. 1994;54:3383–3386. [PubMed] [Google Scholar]
- 8.Johnston JM, Yu MT, Carroll WL. c-myc hypermutation is ongoing in endemic, but not all Burkitt's lymphoma. Blood. 1991;78:2419–2425. [PubMed] [Google Scholar]
- 9.Yano T, et al. Clustered mutations in the second exon of the MYC gene in sporadic Burkitt's lymphoma. Oncogene. 1993;8:2741–2748. [PubMed] [Google Scholar]
- 10.Chang DW, Claassen GF, Hann SR, Cole MD. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol Cell Biol. 2000;20:4309–4319. doi: 10.1128/mcb.20.12.4309-4319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hann SR. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol. 2006;16:288–302. doi: 10.1016/j.semcancer.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Vervoorts J, et al. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sears RC. The life cycle of C-myc: from synthesis to degradation. Cell Cycle. 2004;3:1133–1137. [PubMed] [Google Scholar]
- 14.Henriksson M, Bakardjiev A, Klein G, Luscher B. Phosphorylation sites mapping in the N-terminal domain of c-myc modulate its transforming potential. Oncogene. 1993;8:3199–3209. [PubMed] [Google Scholar]
- 15.Pulverer BJ, et al. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 16.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 17.Hemann MT, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, et al. Phosphorylation regulates c-Myc's oncogenic activity in the mammary gland. Cancer Res. 2011;71:925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 20.Dang CV, et al. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Arabi A, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 22.Cowling VH, Cole MD. The Myc Transactivation Domain Promotes Global Phosphorylation of the RNA pol II Carboxy-terminal Domain Independently of Direct DNA Binding. Mol Cell Biol. 2007;6:2059–2073. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 24.Grandori C, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 25.Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol. 2005;7:295–302. doi: 10.1038/ncb1223. [DOI] [PubMed] [Google Scholar]
- 26.Benassi B, et al. c-Myc phosphorylation is required for cellular response to oxidative stress. Mol Cell. 2006;21:509–519. doi: 10.1016/j.molcel.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Watnick RS, Cheng YN, Rangarajan A, Ince TA, Weinberg RA. Ras modulates Myc activity to repress thrombospondin-1 expression and increase tumor angiogenesis. Cancer Cell. 2003;3:219–231. doi: 10.1016/s1535-6108(03)00030-8. [DOI] [PubMed] [Google Scholar]
- 28.Hoang AT, et al. A link between increased transforming activity of lymphoma-derived MYC mutant alleles, their defective regulation by p107, and altered phosphorylation of the c-Myc transactivation domain. Mol Cell Biol. 1995;15:4031–4042. doi: 10.1128/mcb.15.8.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sears R, et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 31.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez PC, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 36.Ruggero D. Revisiting the nucleolus: from marker to dynamic integrator of cancer signaling. Sci Signal. 2012;5:pe38. doi: 10.1126/scisignal.2003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown SJ, Cole MD, Erives AJ. Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics. 2008;9:442. doi: 10.1186/1471-2164-9-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barna M, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dave SS, et al. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 40.Gautier T, Berges T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayano T, et al. Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J Biol Chem. 2003;278:34309–34319. doi: 10.1074/jbc.M304304200. [DOI] [PubMed] [Google Scholar]
- 42.Newman DR, Kuhn JF, Shanab GM, Maxwell ES. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. Rna. 2000;6:861–879. doi: 10.1017/s1355838200992446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goy A, et al. The feasibility of gene expression profiling generated in fine-needle aspiration specimens from patients with follicular lymphoma and diffuse large B-cell lymphoma. Cancer. 2006;108:10–20. doi: 10.1002/cncr.21500. [DOI] [PubMed] [Google Scholar]
- 46.Vallat LD, Park Y, Li C, Gribben JG. Temporal genetic program following B-cell receptor cross-linking: altered balance between proliferation and death in healthy and malignant B cells. Blood. 2007;109:3989–3997. doi: 10.1182/blood-2006-09-045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci U S A. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlosser I, et al. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003;31:6148–6156. doi: 10.1093/nar/gkg794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaffer AL, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 50.Watson JD, Oster SK, Shago M, Khosravi F, Penn LZ. Identifying genes regulated in a Myc-dependent manner. J Biol Chem. 2002;277:36921–36930. doi: 10.1074/jbc.M201493200. [DOI] [PubMed] [Google Scholar]
- 51.Machida YJ, et al. Targeted comparative RNA interference analysis reveals differential requirement of genes essential for cell proliferation. Mol Biol Cell. 2006;17:4837–4845. doi: 10.1091/mbc.E06-04-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bogomolnaya LM, et al. A new enrichment approach identifies genes that alter cell cycle progression in Saccharomyces cerevisiae. Curr Genet. 2004;45:350–359. doi: 10.1007/s00294-004-0497-5. [DOI] [PubMed] [Google Scholar]
- 53.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 54.Pandolfi PP. Aberrant mRNA translation in cancer pathogenesis: an old concept revisited comes finally of age. Oncogene. 2004;23:3134–3137. doi: 10.1038/sj.onc.1207618. [DOI] [PubMed] [Google Scholar]
- 55.Boon K, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. Embo J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S, Li Q, Dang CV, Lee LA. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc Natl Acad Sci U S A. 2000;97:11198–11202. doi: 10.1073/pnas.200372597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perna D, et al. Genome-wide mapping of Myc binding and gene regulation in serum-stimulated fibroblasts. Oncogene. 2012;31:1695–1709. doi: 10.1038/onc.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Z, Hann SR. Nucleophosmin is essential for c-Myc nucleolar localization and c-Myc-mediated rDNA transcription. Oncogene. 2012 doi: 10.1038/onc.2012.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alawi F, Lee MN. DKC1 is a direct and conserved transcriptional target of c-MYC. Biochem Biophys Res Commun. 2007;362:893–898. doi: 10.1016/j.bbrc.2007.08.071. [DOI] [PubMed] [Google Scholar]
- 60.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6:493–505. doi: 10.1038/nrc1885. [DOI] [PubMed] [Google Scholar]
- 62.Cowling VH, Chandriani S, Whitfield ML, Cole MD. A conserved Myc protein domain, MBIV, regulates DNA binding, apoptosis, transformation, and G2 arrest. Mol Cell Biol. 2006;26:4226–4239. doi: 10.1128/MCB.01959-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.