Abstract
DNA polymerases in family B are workhorses of DNA replication that carry out the bulk of the job at a high speed with high accuracy. A polymerase in this family relies on a built-in exonuclease for proofreading. It has not been observed at the atomic resolution how the polymerase advances one nucleotide space on the DNA template strand after a correct nucleotide is incorporated, that is, a process known as translocation. It is even more puzzling how translocation is avoided after the primer strand is excised by the exonuclease and returned back to the polymerase active site once an error occurs. The structural events along the bifurcate pathways of translocation and proofreading have been unwittingly captured by hundreds of structures in Protein Data Bank. This study analyzes all available structures of a representative member in family B and reveals the orchestrated event sequence during translocation and proofreading.
INTRODUCTION
Eukaryotic replicative DNA polymerases δ and ϵ belong to the polymerase family B. They are workhorses of genome replication during cell division (1,2). In bacteriophage T4, and a phylogenic variant RB69 in the T4 family, the gene product 43 (gp43) is a DNA polymerase in family B. RB69 DNA polymerase carries built-in 3′-exonuclease activity in a single polypeptide chain of 903 amino acid residues (3). This DNA polymerase incorporates hundreds of nucleotides to a primer strand in each second under instructions of a template strand (4–6). At the most fundamental level of the polymerase mechanism, this enzyme transfers a phosphoryl bond on Pα of an incoming deoxyribonucleoside triphosphate (dNTP) to the 3′ hydroxyl on the terminal ribose of the primer so that the primer strand becomes extended by one more nucleotide (7). It is well known that the phosphoryl transfer reaction utilizes two or three divalent metal ions, often Mg2+ (8,9). This chemical reaction is highly regulated. First, a correct dNTP that satisfies Watson–Crick base pairing preferentially binds in the polymerase active site prior to the reaction. Second, the enzyme translocates by one nucleotide space along the single-stranded DNA template after a correct incorporation. Third, if an incorrect nucleotide is accidentally incorporated into the primer, the 3′ terminus of the primer is switched to the exonuclease active site more than 30 Å away for an excision. When the corrected primer returns back to the polymerase active site, the translocation step is either bypassed or undone. This is equivalent to the function of the backspace key and termed proofreading (10). It is remarkable that RB69 DNA polymerase of 903 residues is fully capable of accomplishing such complex tasks. Good processivity in DNA synthesis requires additional accessory proteins to work jointly with the polymerase (3). The conformational changes associated with the catalytic and regulatory events are not yet clear due to the lack of suitable biophysical methods to directly observe the process at the atomic resolution. Therefore, the structural events associated with translocation and active site switching for processive proofreading (11) are largely unknown except for the chemical reaction of phosphoryl transfer.
This work employs a strategy of joint structural analysis to examine all known structures of RB69 DNA polymerase, and to reverse engineer the working mechanics of this polymerase. Nearly 170 independently observed structures of this polymerase are available in the Protein Data Bank (PDB). More than 40 other polymerase structures in family B are also available. This joint analysis is able to extract a conformational pathway that clearly shows detailed structural changes during translocation along a DNA template. A further extension of the translocation pathway leads to processive switching between the polymerase and exonuclease active sites. This joint analysis of more than 200 polymerase structures suggests a unified driving mechanism for translocation and proofreading. Extensive structural and functional data from mutants of this polymerase and DNA templates containing various lesion sites are discussed in light of the proposed mechanism.
MATERIALS AND METHODS
Structural meta-analysis based on singular value decomposition (SVD) of interatomic distance matrices has enabled a large-scale joint analysis of related protein structures (12–14). This computational method of structural analysis takes advantage of a mathematical, yet practical, definition of conformational space with limited dimensionality (12). Each experimentally determined static structure is a snapshot of the protein structure. See (15,16) for examples within the context of polymerase structures. A large number of such snapshots taken under a variety of experimental conditions would collectively provide a survey of the accessible conformational space. Such joint analytical strategy would not be effective in early years when far fewer protein structures were determined to atomic resolution. Recent rapid growth in protein crystallography and structural genomics (17,18) has supplied the necessarily wide sampling of protein structures for a joint analytical strategy to come of age. The vacancies or gaps in a conformational space between well-populated conformational clusters often correspond to less stable transient states whose conformations are difficult, if not impossible, to capture by static crystallography. These conformations are often key to mechanistic understanding and could be explored by a back calculation based on molecular distance geometry (12), the chief computational algorithm in nucleic magnetic resonance spectroscopy. These computed structures at atomic resolution may reveal short-lived intermediate conformation hard to be captured in crystal lattices.
This structural meta-analysis has been applied to tetrameric human hemoglobin (12,13) and other macromolecular systems. Extensive conformational changes have been observed along the continuous trajectories of the functional pathways of these proteins. The discrete structural states separated by large taboo gaps in their conformational spaces are required to accomplish their physiological functions. Many RB69 DNA polymerase structures were previously determined to study how the enzyme and its mutants would handle various types of lesions. In effect, these experiments developed a variety of chemical means to trap the enzyme in different points along its reaction trajectory. The structural meta-analysis presented here reveals structural events embedded in a large collection of structural data already available in PDB. The algebraic protocols employed in meta-analysis are briefly highlighted in this section (12,13).
A distance matrix contains M pairwise inter-atomic distances of a structure that is archived in PDB in the form of Cartesian coordinates of all observed atoms. An everyday example of distance matrix is an intercity mileage chart on the traveler's map. Due to its symmetry, only the lower triangle is necessary. N such lower triangles calculated from a collection of N related structures are assembled into a large data matrix A. Each column of A contains a distance matrix of one structure. It is worthwhile to note that differences in the molecular orientation, choice of origin and crystal lattice among all N experimentally determined structures have no contribution to the distance matrices even without any structural alignment protocol. If a particular structure has missing residues, the distance values involving these residues will be filled with the average values of the corresponding distances so that the missing parts would have a minimal impact to the results of the analysis (see Supplementary Figure S1 for the effect of missing residues). SVD of the data matrix A results in A = U W VT. The matrix U contains N left singular vectors Uk of M elements, which can be restructured to form decomposed lower triangles. Since each vector Uk has a unit length, all left singular vectors have the same average value <u> = 1/√M. The square matrix W contains all zeros except for N positive values on its major diagonal, known as the singular values wk. The magnitude of wk can be considered as a weight or significance of its corresponding Uk. Each column of V or row of its transpose VT, known as a right singular vector, contains the relative compositions of Uk. A singular triplet denotes (i) a decomposed lower triangle Uk, (ii) its singular value wk and (iii) the composition function Vk. Singular triplets are often sorted in a descending order of their singular values wk. Only a small number of n significant lower triangles identified by the greatest singular values w1 through wn can be used in a linear combination of Uk to reproduce a distance matrix that closely resembles the original one calculated from a structure, where n < N. As a result, the structural information evenly distributed in all distance matrices is effectively concentrated into the n significant lower triangles after the SVD procedure. The coefficient set of the linear combination is wkVk. The average value <u> of all elements in Uk can be used to scale the coefficient sets back to distances in Å, that is, ck = <u>wkVk = wkVk/√M.
ck in the first a few dimensions where k = 1, …, n as judged by the magnitudes of the singular values wk form a conformational space of the N structures that constitute the data matrix A (12). When N is large enough, it is said that the collection of N structures samples the entire conformational space of the protein under investigation. With this SVD analysis of distance matrices, each protein structure is mapped to a point in its conformational space, such as those in Figures 2 and 5. Conversely, any given point in the conformational space leads to a unique structure in real space as a solution of molecular distance geometry. Since a distance in the conformational space is equivalent to an rmsd value between two structures, two similar structures are mapped to two points close to each other in the conformational space. Two points far apart in the conformational space indicate two distinct structures. More importantly, vectors with different orientations in the conformational space represent structural changes in different aspects. As a result, a one-on-one correspondence is established between three-dimensional (3D) structures and their conformational space up to the first n most significant dimensions.
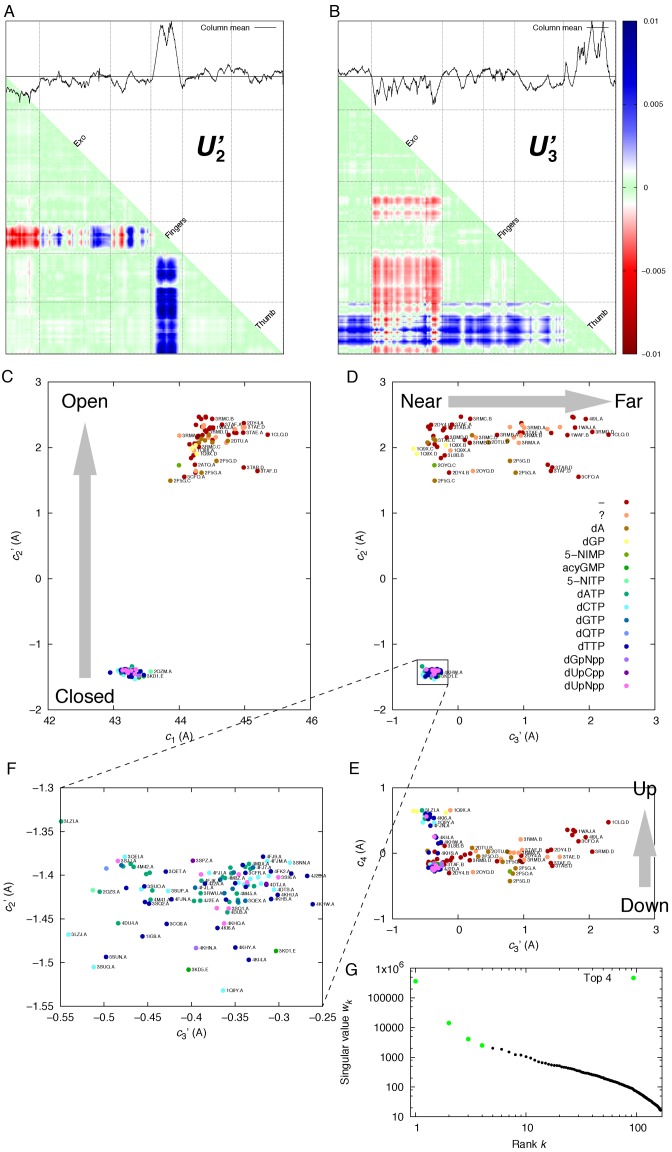
Figure 2.
SVD analysis of RB69 DNA polymerase structures. (A and B), the second and third components U’2 and U’3, also known as left singular vectors (‘Materials and Methods’ section). See also Supplementary Figure S1 for more details on SVD results. The motion of the fingers is decoupled from those of exo and thumb domains. These motions are separated into two independent components U’2 and U’3. The domains are divided by grids and marked on the matrix diagonals. The motions of exo and thumb are highly concerted, since the red and blue bands of these domains are equally strong in a same component (B). (C, D, E and F), the coefficient sets in the first four dimensions, also known as right singular vectors. Each dot represents a polymerase structure. Its distance matrix could be approximately reconstructed as a linear combination c1U1 + c2U2 + c3U3 + c4U4. That is to say, two dots close to each other in the conformational space indicate two similar linear combinations thus represent two similar structures. Two dots far apart represent two distinct structures. The distance between two dots is equivalent to the rmsd value between two structures. The down→up arrow indicates that β-hairpin movement is possible among all structures (Supplementary Figure S1d). The color of each dot indicates the type of ligand bound opposite the template base in the polymerase active site. A dash (-) indicates no ligand; and a question mark (?) indicates that a ligand could be bound but not observed in the structure. See Supplementary Table S1 for all types of ligands. Sixty-two open structures stretch out horizontally at c′2 ≈ 2, but 96 closed structures share constant values of c′2 ≈ −1.4 and c′3 ≈ −0.4. The tight cluster of closed structures is also magnified in (F). The other 11 structures with missing parts in the fingers domain are omitted for clarity. The closed→open and near→far arrows indicate discrete and continuous conformational changes, respectively. See also Supplementary Movie S1 for the continuous motion from near to far conformation. (G) Singular values with the top four highlighted in green.
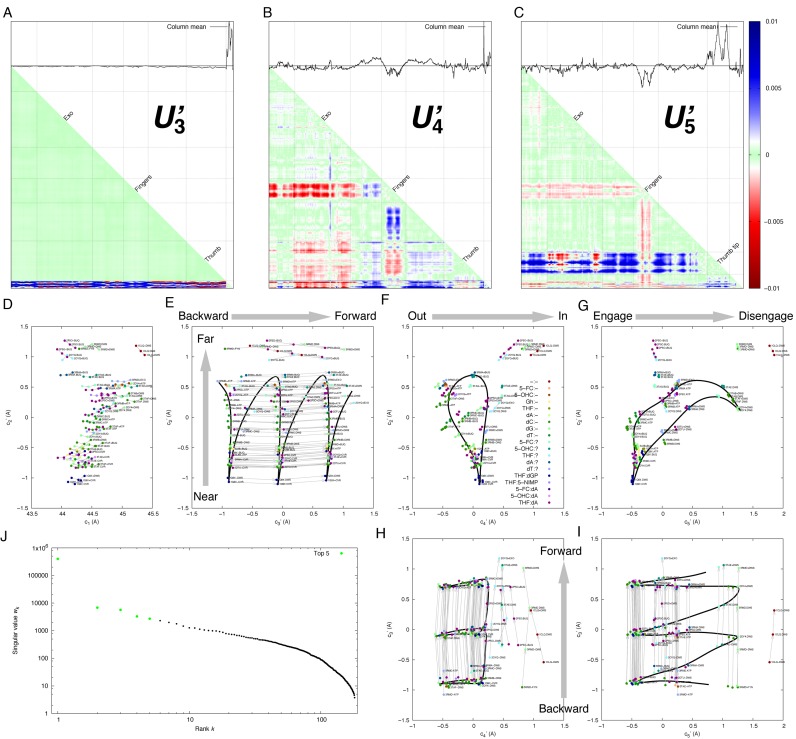
Figure 5.
SVD analysis of open structures with template and primer strands. Sixty-three open structures complexed with DNA template and primer strands are duplicated twice to generate structures on the circular reaction coordinate that are equivalent to the original structures except with backward and forward translocation (Supplementary Text). The first and second decomposed component, or left singular vectors, produced by SVD analysis of these open structures are very similar to the first and third components from SVD of all structures (Supplementary Figure S1a and Figure 2B). The component showing the large swing of the fingers (Figure 2A) is no longer present because only open structures are analyzed. (A) The third component U3′ is artificially inserted by permutation of nucleotide numbering (Supplementary Text). The small bottom portion shows the interactions between the DNA and polymerase, which is the only non-zero portion of U3′ indicating the permutation of nucleotide numbering. (B) The fourth component U4’ shows that the fingers are approaching the N-terminal, exo and thumb domains, but leaving the palm domain, that is, moving toward in-position (Supplementary Figure S2). (C) The fifth component U5′ mainly features the departure of the thumb tip from the rest of the structure including DNA. (D–I), the coefficient sets proportional to the right singular vectors in the first five dimensions. Each dot represents a complex structure. A thin gray arrow connects from a structure with backward translocation to the original structure and to that with forward translocation. The color of each dot indicates the pair of nucleotides in the polymerase active site (F). The nucleotide in the template is before the colon (:), and that in the primer is after the colon. A dash (-) indicates no nucleotide; and a question mark (?) indicates that no nucleotide is observed in the structure but there could be one present. See Supplementary Table S1 for all abbreviations. The solid curves in E–I mark a plausible trajectory of the translocation pathway for more than two consecutive cycles. Each cycle begins and ends at the extreme near ends (E) and traverses along backward→forward direction (E, H and I). The loops in F and G start from the lower end and traverse counterclockwise for forward translocation. See Movie S2 for conformational changes along the pathway. (J) singular values with the top five highlighted in green.
Since U1 is essentially the average of all distance matrices in A except that its length is scaled to a unit, each element in the coefficient vector c1 can be considered as an average of all inter-atomic or intra-molecular distances in a structure, thus a good measure of the particle size. For example, RB69 DNA polymerase in the open state is about 1 Å larger than that in the closed state judging by the difference in c1 for these two states (Figure 2C).
Due to the non-uniqueness of SVD, the conformational space could rotate in some or all dimensions to transform into an equivalent conformational space. The purpose of rotation may vary, but often it would be desirable to align a certain feature of structural changes along a principal dimension such as the motion of the fingers along the second dimension and the motions of the exonuclease and thumb domains along the third dimension (Figure 2). Rotation in a subspace of some dimensions would not affect other orthonormal dimensions. For example, a rotation in X-Y subspace does not change the Z coordinate in a 3D Cartesian space. See reference 19 for example of rotation after SVD analysis of spectroscopic data. Rotation in a subspace is frequently used in this study (Figures 2, 5 and Supplementary Figure S1).
This structural meta-analysis technique places each structure from a large collection into a conformational space with respect to all other structures in the collection. The relationship among these structures is revealed by this large-scale structural comparison that results in the distribution of structures in their conformational space. Therefore, an order could be assigned to each structure based on an assumed smoothness of conformational changes along a reaction trajectory. This dot-to-dot order is important new information that is not associated with each individual structure but could be retrieved from this joint structural analysis. Causation and consequence of structural motions are revealed from the order of the structures in a series, which further lead to structural mechanism.
RESULTS
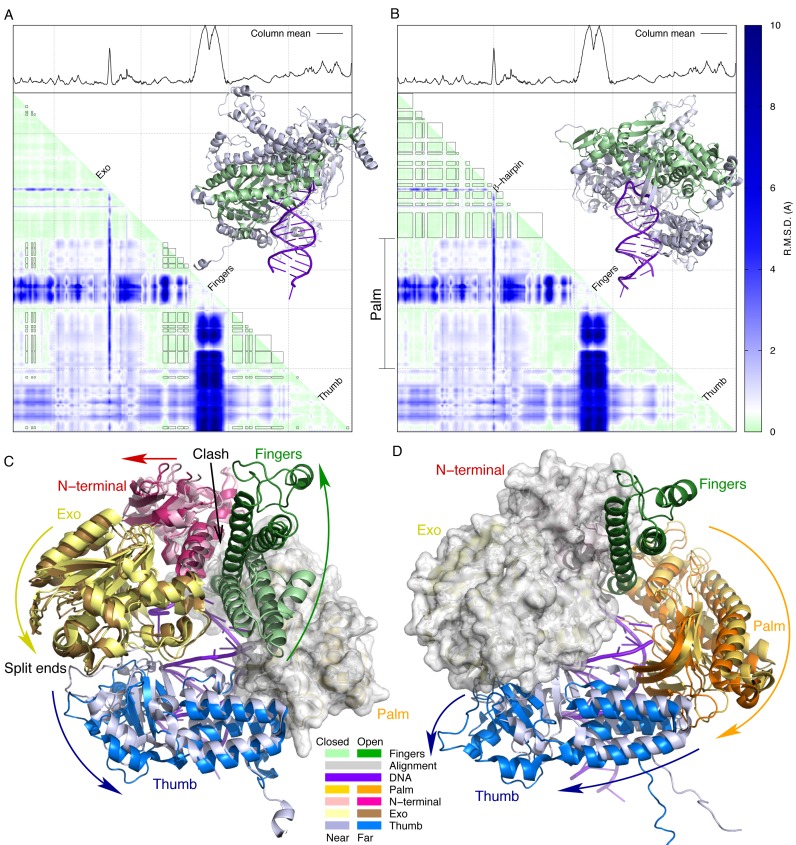
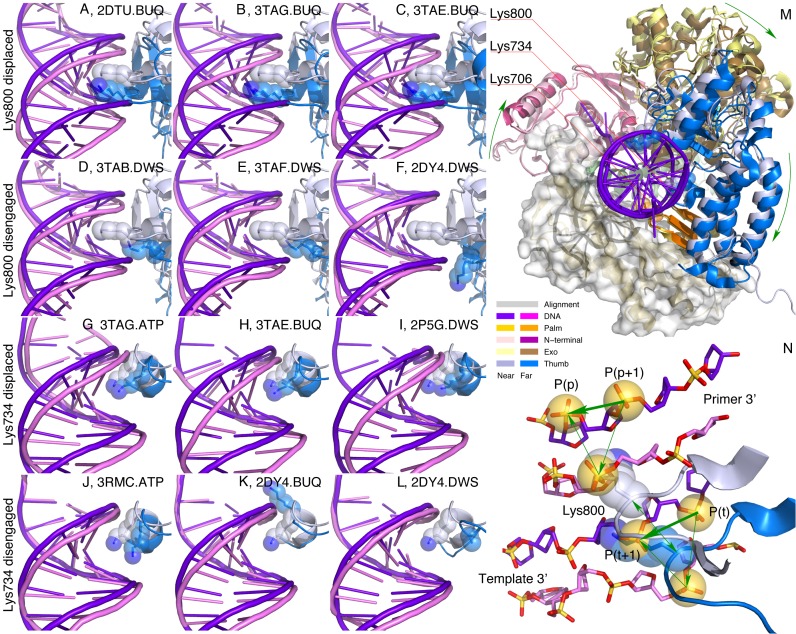
DNA polymerase structures in family B are represented by those from bacteriophages, herpesvirus (20), bacteria (1Q8I), archaea (21–23) and eukaryotes (24,25). They share a common architecture. The N-terminal (residues 1–380 in RB69 numbering), palm (404–729) and thumb (730–903) domains are arranged to form a split ring structure similar to a lock washer (Figure 1A and C). The exonuclease domain (109–339; exo hereafter) located at one of the split ends extends from and returns to the N-terminal domain, thus dividing the N-terminal domain into two separate segments. Other major structural features include a β-hairpin (248–265) in the exo domain, the most mobile fingers domain (469–571) consisting of two antiparallel long helices extending from the palm domain and the thumb domain that can be divided further into a three-helix bundle subdomain (738–774 and 868–888) and a thumb tip (775–867) at the other split end opposite the exo domain (Figure 1A and C).
Figure 1.
A cartoon representation of the main findings. DNA double helix is drawn as intertwined arcs as viewed along the major and minor grooves on one side of the DNA duplex. α helices in the fingers domain are shown as cylinders. (A and C) viewed from a similar perspective show that the overall shape of RB69 DNA polymerase resembles a split ring such as the lock washer. It transforms its shape between the shaded and outlined rings in the open state, denoted as near and far conformations, respectively. (B and D) are viewed from another direction orthogonal to (A and C) that shows the fingers domain swings between the open and closed states. The key structural events involved in translocation and active site switching are the displacement and disengagement of thumb domain exemplified by Lys734 and 800.
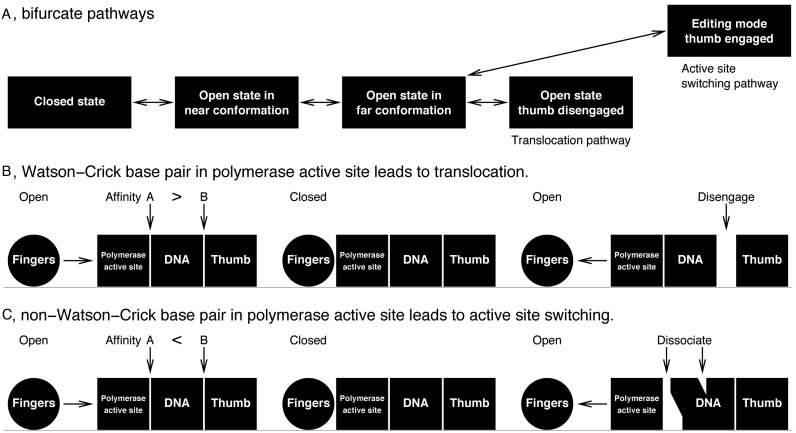
I first present an overview of the main findings and hypotheses of this work (Figure 1). The detailed evidences that support these conclusions are analytically deduced from a collection of experimentally observed structures of RB69 DNA polymerase. This enzyme catalyzes incorporation of hundreds of nucleotides each second in host cells. That is to say, its fingers domain is wagging between the open and closed positions at a frequency of several hundred Hz during normal elongation (300 Hz is used in calculations throughout this paper). The fingers domain acts as an oscillator just like one in a mechanical clock and is responsible for power management of both translocation and processive active site switching. Once this oscillation is established, each leaving pyrophosphate (PPi) bound to the fingers in the closed conformation replenishes energy to maintain the oscillation. With each push from a newly cleaved PPi resulting from dNTP hydrolysis, the closed fingers accelerate to adopt an open conformation. At the end of a large swing, the fingers slam into the N-terminal domain. When the motion of the fingers is completely halted at the open position, their momentum is transferred to power the translocation. The motion of the thumb drives a back-and-forth displacement of the product duplex during translocation. The structural events during translocation and active site switching share much of the same conformational pathway until they branch out at an advanced stage. The association affinity of a nascent base pair, correct or incorrect, in its binding pocket is the determining factor that allocates energy to choose one from the bifurcate pathways. However, this is not to say that the choice is always appropriate. Along the translocation branch more likely following a Watson–Crick base pairing, the thumb is momentarily disengaged from the minor groove that facilitates the relative sliding between protein and DNA, a key step for translocation. Alternatively, the thumb is constantly holding the duplex in the minor groove to avoid translocation while the primer shuttles to and from the exonuclease active site in the event of an accidental mismatch. The rest of this paper presents detailed evidences resulting from a joint analysis of the abundant structural and functional data in support of the mechanisms of translocation and proofreading outlined above. Non-processive switching of active sites based on dissociation and re-association of polymerase and DNA is not discussed here.
Conformational distribution of open and closed states
Nearly 170 structures of RB69 DNA polymerase can be unambiguously divided into open and closed states depending on two discrete positions of the fingers. Apparently, such discreteness of states is distinct from the behavior of polymerases in family A (26). The swing of the fingers as wide as 35° is estimated to have an average angular speed of at least 3500 rpm, given that the enzyme incorporates 300 nt per second (4–6). Such large motion between two discrete states is also evidenced in the structural meta-analysis based on SVD (‘Materials and Methods’ section) by a sizable gap between two groups of structures compared to the deviation within each group (Figure 2 and Supplementary Figure S2). Only the closed state is catalytically capable of the phosphoryl transfer reaction. It is postulated that the highly discrete states of the closed and open conformations are associated with an energy minimum and an entropy maximum, respectively (27). Interestingly, no intermediate structure between them has ever been observed by static crystallography (Figure 2C and D; Supplementary Figure S1f). The discreteness of states has also been detected in ionic current traces when a single molecule of ϕ29 DNA polymerase, another member in family B, performs translocation on an α-hemolysin nanopore in an electric field (28,29). Concerted and continuous motions of the exo and thumb are observed only when the polymerase is in the open state as many open structures scatter along c3′ dimension, suggesting a rather flat, low energy valley. The two end conformations along this dimension are denoted as near and far conformations, respectively (Figure 2D). In contrast, similar motions are absent in the closed state. That is to say, the large swing of the fingers is not in concert with motions of the exo and thumb as indicated by two orthogonal arrows closed→open and near→far (Figure 2C and D). The open conformation of the fingers is apparently a prerequisite for the concerted motions of the exo and thumb domains.
Tight clustering of structures in the conformational space indicates high conformational similarity in the closed state where the only significant motion is at the β-hairpin as c4 varies slightly (Figure 2E and Supplementary Figure S1d). Binding of any type of triphosphate ligand, such as dNTP or non-hydrolyzable analogs (30–32), at the polymerase active site seems to be sufficient to induce the closed conformation. On the other hand, all 69 structures adopt the open state in the absence of bound triphosphate. The only exceptions are the chimeric structures 3KD1 and 3KD5 with extensively altered amino acid sequences in the fingers. A key substitute V478W among many other changes introduces a clash in the open state, which significantly shifts the equilibrium toward the closed state regardless whether a triphosphate is bound (33). Both chimeric structures with and without phosphonoformic acid bound in the active site are located at the extreme of the closed conformation where c′2 ≈ −1.5 (Figure 2F).
The largest conformational changes between the closed and open states and between the near and far conformations are summarized in Figure 3, based on structural alignment according to rigid frameworks identified by the rmsd matrix (12,13) of RB69 DNA polymerase (Supplementary Text). The concerted near→far motions of the exo and thumb are shown in Movie S1. The exo and thumb located at the split ends of the ring structure rub against each other on two surfaces consisting of main chain and rigid side chains such as Pro, Ala and Gly, but devoid of flexible side chains. The flexible side chains are largely hidden in the concave grooves of both surfaces, which smoothens the relative motions between the exo and thumb (Figure 4). It can be postulated that changing this property of the interface at the split ends will have significant impact, although the interface is away from both active sites.
Figure 3.
Domain movements of RB69 DNA polymerase. (A and B) The rmsd matrix calculated from all polymerase structures (Supplementary Text) contains two sub-matrices of small values outlined by solid rectangles. These sub-matrices of average values of 0.22 and 0.23 Å respectively indicate very small internal movements within each of these two blocks in the polymerase structure. These two well-separate invariant structural frameworks are highlighted in green in the insets. Both open and closed fingers are shown. The first framework (A) consists of large parts of the palm domain (residues 397–431, 436–464, 581–609, 614–633 and 641–715) excluding the fingers, small parts of the N-terminal domain (34-41, 49–62 and 361–367), and a few residues in the thumb (749–755 and 883–889). The second framework (B) includes most of the N-terminal and exo domains (residues 2–42, 49–72, 80–163, 180–250, 262–281, 290–300 and 318–384). (C and D) Structures are aligned according to the rigid frameworks identified in panels A and B, respectively. The alignment segments are rendered in white surfaces. The colored arrows indicate domain movements from closed to open fingers or from near to far conformations (Figure 2). (C) The opening fingers clash into the second segment of the N-terminal domain (residues 340–380), and move the N-terminal domain to the left. Another motion component of the N-terminal domain is along the opening direction of the fingers, which is hardly visible from this view but more obvious in Figure 9A and Supplementary Movie S4. The motion of the N-terminal domain is transmitted to the exo domain, as these domains are relatively invariant (B). The exo and thumb domains move in concert because they interact at the two split ends of the lock washer structure (Figure 1). (D) The palm domain is a major component in the first rigid framework (A). It moves with respect to the N-terminal and exo domains, the second rigid framework (B). The motion of the palm is transmitted to two subdomains of the thumb. The thumb tip at one of the split ends moves in a different direction from the rest of the thumb, because of its interaction with the exo domain at the other split end.
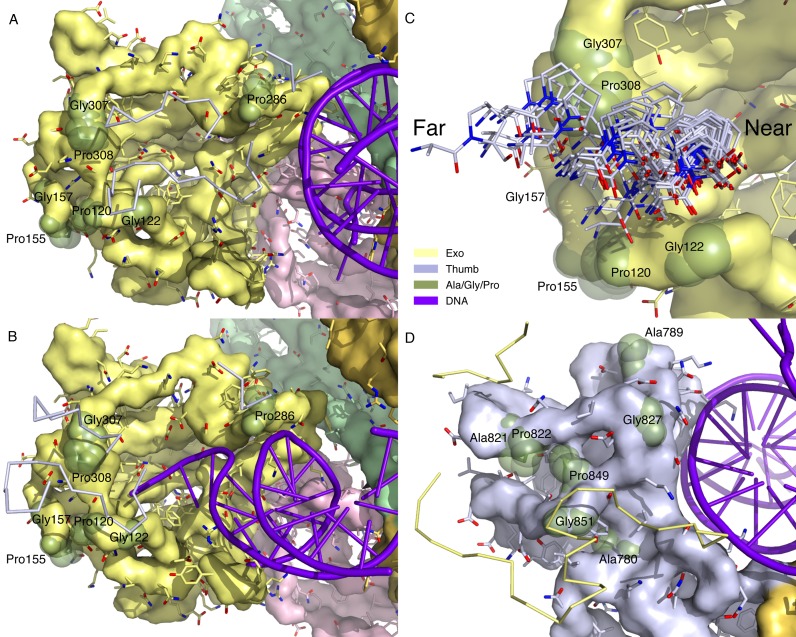
Figure 4.
Relative motions between two split ends. The exo and thumb domains are located at the split ends of the lock washer structure of RB69 DNA polymerase (Figure 1). The protein backbone is depicted in surface models. Side chains are shown as stick models protruded from the backbone surface. Gly, Ala and Pro residues are highlighted with transparent green spheres. The convex surfaces of both sides of the split ends interface are largely composed of the protein main chain and rigid side chains including Gly, Ala and Pro, which facilitates relative rubbing and sliding between two split ends, for example, Gly122, 157, 307, 827, 851, Ala780, 789, 821, Pro120, 155, 286, 308, 822 and 849. The other flexible side chains are mostly hidden in the concave grooves thus not in contact with the opposite sides. The relative sliding between these split ends are important to near-far conformational changes. All open structures are aligned together according to the second rigid framework of the N-terminal and exo domains (Figure 3B and D). (A) Exo end interface in near conformation. The thumb domain is removed to expose the interface of the exo end except a few residues rendered as thin sticks in the foreground. (B) Exo end interface in far conformation. Compared to the near conformation (A), the thumb domain in the foreground indicated by a few residues in sticks moves to the left passing several convex knobs on the exo end interface. (C) Ala821 and Pro822 on the thumb interface transit over Pro308 from near to far conformations. These residues from 69 structures in the open state are rendered in stick models. The entire range of the thumb motion spans 15 Å. (D) Thumb end interface in near conformation viewed from the opposite direction of (A). A few residues on the exo interface are rendered as yellow thin sticks in the foreground.
In summary, distribution of the experimentally determined structures in the conformational space defined by SVD (Supplementary Text and Figure 2) enables a comprehensive survey and joint analysis of a large number of structures. The density of structural distribution also offers a glimpse of the energy landscape in the conformational space. The exo and thumb domains move in concert continuously along a rather leveled path in an energy valley connecting the near conformation to the far in the editing mode. Such motions of the exo and thumb are largely decoupled from the highly discrete positions of the fingers. The open and closed states strictly depend on whether or not a triphosphate is bound in the polymerase active site. And the closed structures are highly uniform in conformation and clustered in a small and deep energy well distinct from the open structures.
Translocation pathway
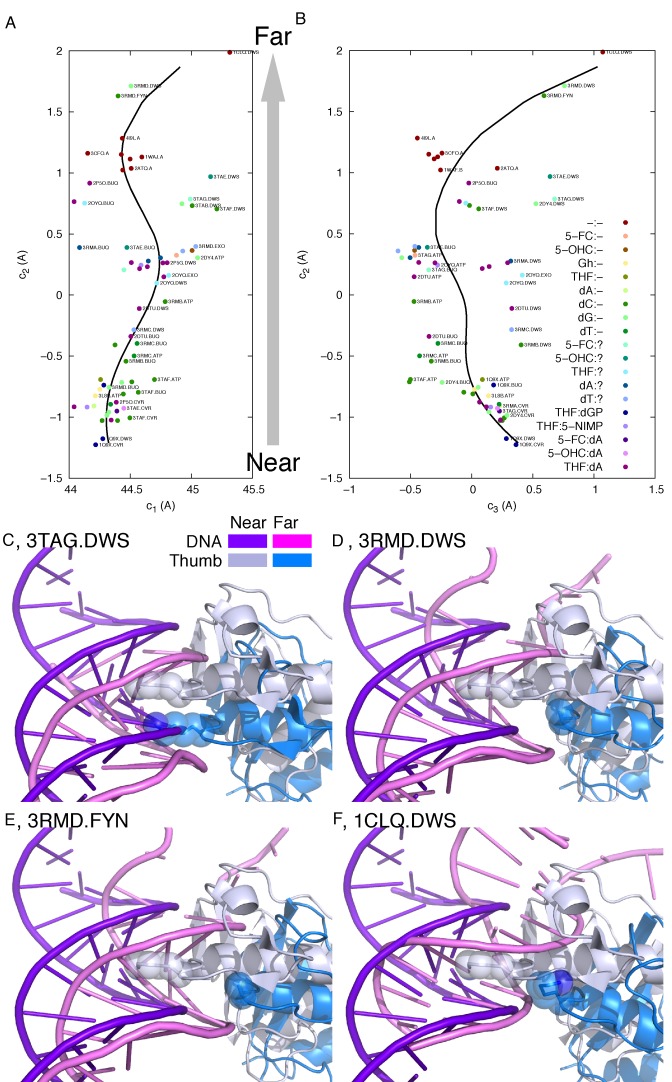
Translocation is a key step in the functional cycle of a polymerase that follows a circular reaction coordinate. In other words, an experimentally observed structure represents a conformational snapshot of the polymerase between two consecutive translocation events. Some open structures represent a conformation immediately prior to the step of translocation, in which dAMP (2DTU, 2P5G, 2P5O, 3TAE and 3TAG) or 5-NIMP (2OYQ) has been incorporated to the 3′-end of the primer (34–36). Some others, as in 2DY4, 3L8B, 3RMA, 3RMB, 3RMC, 3RMD, 3TAB and 3TAF, correspond to a conformation immediately after translocation, in which the space opposite the template nucleotide in the active site is vacant (36–38). Others are uncertain. Nevertheless, each structure represents both pre- and post-translocation conformations with respect to two consecutive translocation steps regardless whether a nascent nucleotide has been added to the primer or not. In other words, both pre- and post-translocation structures adopt the same conformation along the circular reaction coordinate but in two consecutive cycles.
Each open structure is therefore duplicated with a permutation of the nucleotide numbering to produce the structures after a step of forward and backward translocation (Supplementary Figure S3 and Supplementary Text). A joint SVD analysis of these duplicated open structures maps out the translocation pathway both before and after the translocation step and identifies those intermediate structures during process of translocation (Figure 5). The open structures do not seem to correlate with what is situated in the polymerase active site as evidenced by the random order of the colored dots in Figure 5. The permutation of nucleotide numbering introduces an additional dimension c3′ to the SVD conformational space (Figure 5E). Most of the open structures including their duplicates are located in three separate columns. The original structure 1Q9X.CVR and its duplicates, 1Q9X−CVR and 1Q9X+CVR, (see Supplementary Text for notation) are located at three near ends of the columns corresponding to an extreme conformation (Figure 5E). This protein conformation is shared by two different moments in the reaction coordinate when the fingers have just opened before the translocation process begins and the fingers are poised to return to the closed position after the completion of translocation. The gaps between the columns are wide and clear at the near conformation but are gradually narrowing down toward the far conformation, which suggests possible crossover from one column to the next, that is, one step of translocation. In other words, translocation occurs while near→far conformational changes are developing. The conformational pathway of translocation traverses toward the far end in one column, crosses over the gap as evidenced by several experimental structures protruded into the gaps (Figure 5E), and then returns to the near end of the next column. That is to say, translocation does not occur directly between two extreme dispositions. Rather, the entire translocation pathway traverses through each observed open structure in the near section twice, once on the way out and the second time on the way back. The round-trip of protein conformational changes couples with one complete translocation process of the DNA duplex (Supplementary Movie S2).
The translocation pathway marked as the black curves in Figure 5E–I traverses through most open structures but does not visit those of extreme far conformations that are in the editing mode. The U-turn pathway also spans the fourth (Figure 5B, F and H) and fifth (Figure 5C, G and I) dimensions of the SVD analysis. Along the fourth dimension, the fingers are at the ‘in’ position during near→far development, but move to the ‘out’ position momentarily before returning to the near conformation (see in and out positions in Supplementary Figure S2). The fifth dimension reveals disengagement of the thumb tip from the DNA duplex (see below for detail).
This translocation pathway prompts a closer examination of a number of open structures in real space, especially for those that deviate from the columns and protrude into the gaps. Three types of structural events during translocation are identified: (i) displacement of the DNA duplex away from the polymerase active site (Figure 6A–C and G–I), (ii) disengagement of the thumb from the minor groove of the duplex (Figure 6D–F and J–L) and (iii) an overall rotation of the N-terminal and thumb domains around the DNA duplex (Figure 6M). These structural events embedded in the experimentally determined crystal structures support the translocation pathway derived from the SVD analysis, and provides key insights into the translocation mechanism of this polymerase.
Figure 6.
Translocation events captured by experimental observations. 1Q9X.CVR is located at the very near end of the translocation pathway (Figure 5E–G), thus serves as a reference for the open state. Each open structure is aligned with the reference according to the first rigid framework that spans the palm domain (Figure 3A and C). Several Lys residues interacting with the DNA are rendered in stick and transparent sphere models. (A–C) The product duplex and Lys800 are displaced in concert toward the direction of outgoing DNA when Lys800 remains engaged in the minor groove. (D–F) The product duplex is not significantly displaced if Lys800 is disengaged from the minor groove when sufficiently advanced toward the far conformation. (G–I) Various extents of Lys734 displacement have also been captured together with DNA displacements. (J–L) Lys734 may also disengage from the minor groove resulting little displacement of the DNA. (M) Viewed along the outgoing product duplex, near→far conformational changes appear to be a clockwise rotation of 15° when an advanced far conformation is reached. This observed rotation is here interpreted as the polymerase threading on the double helix toward the growing direction of the primer. See Supplementary Movie S1 for this rotation. (N) Atomic details of translocation. DNA structures in both pre- and post-translocation positions are in purple except that a nucleotide t in the template is translocated to t + 1 and a nucleotide p + 1 in the primer is translocated to p. The thick green arrows mark the outcome of a complete translocation step. However, no motion following the green arrows has been observed. Instead, many intermediate structures represented by the far conformation are displaced from both the pre- and post-translocation structures. The thin green arrows show the motions of intermediate steps. Lys800 from a loop in the thumb tip drives the displacement and returns to its original position (double green arrow), but the DNA duplex translocates one nucleotide space indirectly. Lys734 at a different position in the minor groove plays a similar role. N, O, and P atoms are in blue, red and gold, respectively.
First, the thumb motion from the conformational changes of near→far is perpendicular to the minor grove (Supplementary Movie S1), which is most effective for Lys734 and 800 bound in the minor grove to displace the product duplex. Since the largest displacement of 5 Å is greater than a typical stacking distance of ∼3.3 Å between two adjacent base pairs, it is more than sufficient to drive translocation by one nucleotide space. The back-and-forth motions of the thumb give away some of the displacement; and their vectorial sum accomplishes a complete translocation of one nucleotide space (Figure 6N). Second, at the far end of the translocation pathway, Lys734 and 800 disengage from the minor grove momentarily. Loosened DNA–protein contact even for a brief period allows a chance for these Lys residues to disengage from the minor groove, slide relative to the DNA duplex, and reengage into the minor grove at the next nucleotide position. This disengagement has also been captured in structures such as 2DY4, 3TAB, 3TAF and 3RMC. If such disengagement occurs in a structure, the DNA duplex usually exhibits little displacement (Figure 6D–F and J–L). In other words, a significant displacement of DNA duplex requires the fully engaged Lys734 and 800 in the minor groove. Third, when viewed along the axis of the DNA product duplex (Figure 6M and Supplementary Movie S1, lower panel), the exo and thumb domains move clockwise for approximately 15° around the DNA double helix from the near to far conformation, which accounts for >40% of a complete step (35°) of translocation in the anticipated direction of forward translocation. This rotation of the thumb ensures that Lys734 and 800 re-engage with the minor groove one step toward the 5′-end of the template. The remaining 60% rotation of a complete translocation step has not been captured by static crystallography and can be illustrated by computed structures (Supplementary Movie S2).
The reconstructed movie of translocation shows a brief period of the thumb disengagement from the DNA duplex, while the disengaged DNA duplex moves back toward its original position momentarily (right tip in Figure 5G and Supplementary Movie S2). This duplex motion originates mainly from several disengaged static structures that support the thumb disengagement (Figure 6). A duplex disengaged from the thumb could not stay at a displaced position in a static structure, therefore returned duplexes have been observed in 2DY4, 3TAB, 3TAF and 3RMC, which is unlikely to occur during a dynamic process in real time. It can be imagined that a more realistic transient structure should feature maximum duplex displacement even during the brief disengagement of the thumb. Although the captured conformation by several static structures may not exactly reflect the short-lived reality, these static structures are very important to demonstrate, first, that the thumb indeed disengages from the minor groove when a structure approaches the far end of the conformational space, and second, that the thumb powers the displacement of the DNA as evidenced by the returned duplex in its original position without an engaged thumb.
In summary, the polymerase conformation in the open state does not seem to depend on whether the polymerase active site is empty, half occupied by a nucleotide from the template, or fully occupied by a base pair. No correlation with the phosphoryl transfer reaction is obvious either. Any given open conformation could be found in both forward and backward pathways of the translocation loop. The thumb holds the DNA duplex in its minor groove with Lys734 and 800 and drives the translocation of the product duplex. These Lys residues alternate between a mode engaged in the minor groove and a brief period disengaged from the duplex. Cycling between these two modes during the thumb movement propels the polymerase to advance one nucleotide space at a time along the template. The SVD procedure applied to the original and duplicated sets of open structures identifies the key structural events that have been collectively captured in many structures. These structural events only become apparent when the joint structural analysis is able to isolate the consistent motion components from random fluctuations and systematic differences among a large number of experimental structures.
Switching between polymerase and exonuclease active sites
The built-in 3′→5′ exonuclease activity provides proofreading capability to this polymerase in the event of a misincorporation. In the meantime, the exonuclease active site in the same protein threatens to remove the nucleotide, even if it is a correct one, that the polymerase has just incorporated to the 3′-end of the primer. Therefore, the precise mechanism of active site switching is key to the function of this polymerase. In the SVD-defined conformational space, the observed structures in the editing mode are located further toward the far conformation away from those structures undergoing translocation (Figure 5). Therefore, the event of active site switching shares many conformational changes with translocation, but branches out from the translocation pathway to take a much longer detour. The long excursion of conformational changes to and from the editing mode could very likely follow a loop, that is, the conformational changes along the forward and reverse paths are not necessarily the same. Unfortunately, there are currently insufficient structures to define the loop of conformational trajectory with high confidence. Only a linear pathway to and from the editing mode is computed here (Figure 7 and Supplementary Movie S3). Interestingly, several apo polymerase structures adopt similar conformations en route to the editing mode, of course, judged solely by the protein conformations. This observation implies that the polymerase conformation is somewhat strained in order to bind DNA and the strained conformation of the complex relaxes while approaching the editing mode. Supplementary Movie S3 displays the dramatic motions along the extended pathway to and from the farthest conformation captured in 1CLQ. It is noteworthy that the thumb tip accompanies the DNA duplex throughout the entire excursion without disengagement (39), even at the very far end of the pathway as shown by 1CLQ and 3RMD. But Lys706 of the palm domain used to bind in the minor groove has to disengage from the DNA in the editing mode. Both DNA strands depart from the polymerase active site. The primer strand partially separates from the template strand (38,40,41) and passes behind the β-hairpin to reach the exonuclease active site (34,42) (see Discussion for more). The exo domain also moves toward the upcoming 3′-end of the primer strand. These coordinated motions together accomplish active site switching.
Figure 7.
Active site switching pathway. (A and B) SVD analysis of all open structures including the structures of apo polymerase. The solid spline curves in the first three dimensions of SVD indicate the active site switching pathway from the polymerase active site at the near end to the exonuclease active site at the far end. The return path follows the exact same trajectory, which may be an oversimplification. See Supplementary Movie S3 for conformational changes along the pathway. (C–F) Four structures in the extreme far conformations are aligned with the reference 1Q9X.CVR according to the first rigid framework that spans the palm domain (Figure 3A and C). Lys800 is rendered in stick and transparent sphere models. Judging by the relative positions of the DNA and the thumb tip, Lys800 remains engaged in the minor groove, although its side chain is not observed in some structures.
Three outstanding questions remain to be answered for a better understanding of the structural mechanism of this and other polymerases in family B. First, how does a successful Watson–Crick base pairing signal the system to take a shortcut pathway for translocation; and why would an incorrect base pair switch the system into a long excursion to the editing mode? Second, the switching between the active sites has been hypothesized as an equilibrium of DNA binding in two active sites depending on their binding affinities to correct and incorrect base pairs. How is such equilibrium achieved given that two active sites are partitioned by the β-hairpin in two separate binding pockets (41)? Third, how does the polymerase advance one nucleotide space during translocation while it stays at the same location on the template after proofreading? Specific structural events with atomic details involved in these regulatory functions are not yet clear. Here a detailed illustration of structural events is presented to address these questions based on the results of structural meta-analysis.
As proposed above, the rapidly swinging fingers are halted once it collides with the N-terminal domain at the open position (Figure 3C). Their kinetic energy is transferred to drive the system through either translocation or active site switching pathway. That is to say, each cycle of translocation or active site switching begins with a constant amount of momentum determined by the angular speed of the oscillating fingers. Disassociation of the DNA duplex from the polymerase active site and separation of the two DNA strands cost less energy, if a non-Watson–Crick base pair has just been mistakenly formed (43). As a result, there is sufficient energy left to power a long excursion that sends the 3′-end of the primer into the exonuclease active site (Figure 8C). In contrast, it would take more energy to break a Watson–Crick base pair and to have them dissociate from the binding pocket (Figure 8B), which in effect dampens the impact of the oscillating fingers so that the remaining energy would be just enough to complete translocation via the relatively short pathway (Figure 8). The decision to take which of the bifurcated pathways is a minor factor contributing to the overall replication fidelity compared to selection of the right dNTP to incorporate (2). It is therefore understandable that small difference in free energy cost of only ∼0.3 kcal/mol between right and wrong incorporations does not seem to account for the overall fidelity (43). The hypothesis that the DNA duplex undergoes thumb-assisted one-dimensional diffusion driven by an equilibrium between polymerizing and proofreading (39) implies abundant thermodynamic events. The main addition from this work is a sequence of specific structural events inferred from a computed order of structures in the conformational space in which the motions of the DNA strands, the exo and thumb domains are powered by the oscillating fingers.
Figure 8.
Bifurcate pathways for translocation and active site switching. (A) The translocation and active site switching pathways share common conformational changes and bifurcate at an advanced stage. (B and C) Schematic presentations differentiating a Watson–Crick base pairing and a mismatch. When a Watson–Crick base pairing occurs, the collision of the fingers domain, here represented as a rolling ball, leads to disengagement of the thumb domain from the DNA, since the affinity of a Watson–Crick base pair in the polymerase active site is relatively higher. On the other hand, when a non-Watson–Crick base pairing occurs, the mismatched base pair separates from the polymerase active site due to its poor affinity, which maintains the thumb binding in the minor groove and avoids translocation. The point of separation is largely located between two DNA strands, and to a lesser extent between the template and the protein. A separation at one of these points with lower affinity prevents dissociation at the other of higher affinity. Even if the difference in free energy cost is small, this mechanism of pathway branching amplifies the difference and results in decisive choices. On the other hand, if the affinities were too close to differentiate, both pathways would be possible.
The large conformational changes of near→far generate significant structural strain. Disengagement of Lys734 and 800 of the thumb from the minor groove alleviates the strain in the event of a newly formed Watson–Crick base pair, so that the DNA duplex is expected to stay intact during translocation (Figures 5 and 6; Supplementary Movie S2). In the meantime, necessary sliding between the protein and DNA can take place. Weakened association between a non-Watson–Crick base pair and in the polymerase active site are an alternative release of the structural strain, which maintains the relatively stronger association between the duplex and the thumb throughout the long shuttling to and from the exonuclease active site (Figure 7). As a result, translocation is avoided. The mechanism of this pathway branching essentially depends on the relative association affinities between the nascent base pair in the polymerase active site and the interaction of the product duplex to the thumb (Figure 8). A correct or an incorrect base pairing would tip the balance and set off a sequence of structural events along a proper pathway. Relative small difference in free energy cost is successfully translated to sharply varying probability to choose the appropriate structural events and to avoid the harmful ones. This hypothesis is fully consistent with the view of correlated and balanced activities between two active sites (10). This study provides atomic details to illustrate these principles in ‘Discussion’ section below.
DISCUSSION
Single-stranded DNA template
The single-stranded DNA template is prepared by a helicase at the branching point of the replication fork and temporarily protected by single strand binding proteins (3,44). The available structures of RB69 DNA polymerase are usually complexed with template strands with only 2–4 nt prior to the polymerase active site. It has been proposed that the single-stranded template enters the active site from the groove between the N-terminal and exo domains and interacts with the β-hairpin (34,39). Without the β-hairpin, the polymerase is prone to disassociate from the DNA during active site switching (34). In contrast to the translocation of the product duplex, the anticipated motion of the single-stranded template has not been clearly observed in the available static structures, largely due to more specific DNA interactions in the polymerase active site compared to the interactions between the thumb and the DNA duplex. Nevertheless, large motions of the N-terminal and exo domains are clearly present between the closed and open states (Figures 3 and 9; Supplementary Movie S4). One of the motion components is along a direction similar to that of the opening fingers, thus could be attributed to the impact of the oscillating fingers (Figures 3C and 9). The groove between the N-terminal domain and the β-hairpin is lined with many positively charged Arg and Lys residues (Arg18, 25, 27, 29, 66, 74, 109, 145, 241, 246, 249, Lys2, 73, 89, 102, 107, 185, 208, 240, 247 and 251). All their side chains move along the same direction with the N-terminal domain (Figure 9A and Supplementary Movie S4). It is highly plausible that this Arg-Lys-rich groove is the binding track for the negatively charged, single-stranded template. These long-stretching side chains move toward the polymerase active site and signify the translocation of the single-stranded template. Unfortunately, no structure reported so far explicitly demonstrates DNA binding to this track, as the previous template constructs were not long enough to achieve stable binding for crystallization. The interaction between this long single-stranded template and the Arg-Lys-rich groove provides part of the driving force for translocation that works together with the motion of the thumb to power the movement of the product duplex. While the thumb disengages from the minor groove momentarily (Figure 6 and Supplementary Movie S2), the translocating single-stranded template pushes the product duplex to slide relative to the thumb, which ensures that the thumb would reengage to the next position in the minor groove. Without the motions of the N-terminal and exo domains feeding the template single strand into the polymerase active site, the thumb may reinsert back to its previous position in the product duplex. Such effect of power loss during replication has been observed as an increased difficulty to replicate at the end of the template (45), which is an indication of the diminishing interaction between the Arg-Lys-rich groove and the ending single-stranded template.
Figure 9.
Motions related to the single stranded DNA template. (A) Concerted domain motions between the open and closed states. Open and closed structures are aligned according to the rigid framework of the palm domain (Figure 3A and C). The exo domain is rendered in Cα trace to reveal more of the fingers in the background. Arg and Lys residues in the N-terminal and exo domains are shown as stick models. This view shows the motion components that are not clear in Figure 3C. See also Supplementary Movie S4 for these motions. (B and C) β-hairpin motion. The β-hairpin is believed to associate with the incoming single-stranded DNA template and its motion is captured as one of the major motion components in all RB69 DNA polymerase structures (Figure 2E and Supplementary Figure S1d). The palm domain is removed to expose the tunnel connecting the polymerase and exonuclease active sites. The 3′-end nucleotide of the primer in the polymerase active site is rendered in spheres. Several side chains in the exonuclease active site are also in spheres at the end of the tunnel. (B) The β-hairpin is down separating two active sites. (C) The β-hairpin is lifted up to open the tunnel so that the 3′ end of the primer can reach the exonuclease active site in the white circle.
In case of a mismatch, the thumb retains its interaction with the product duplex to avoid translocation (Figure 8). If the Arg-Lys-rich groove advances the single-stranded template toward the polymerase active site (Figure 9), but the product duplex is withheld by the thumb not yielding more room (Figure 7), the template strand would have to bulge upward due to the temporary molecular congestion. Since several Arg and Lys residues from the β-hairpin could bind to the template (Figure 9A), the β-hairpin would be lifted up by the bulging template (Figure 9B and C). The β-hairpin is more extensive in yeast DNA polymerase δ (25), but shorter in ϵ (24). The available structures in both open and closed states have shown that the β-hairpin is able to lift, which is one of the major motion components (Figure 2E and Supplementary Figure S1d) (34). The uplifted β-hairpin would create a passage behind it to allow the primer strand to shuttle between two active sites (Figure 9B and C). However, I conjecture that the β-hairpin would not move upward during normal translocation, but keeps the passage shut to retain the primer strand in the polymerase active site, since no congestion is caused by simultaneous translocation of the single-stranded template and the product duplex. It is apparently a safeguard to have the β-hairpin to partition two active sites (41) and to open only when necessary in case of differential translocation caused by a mismatch. This is an important feature of this duo-enzyme as the potent 3′→5′ exonuclease nearby is a direct threat of degradation to the newly synthesized DNA primer (46). Such safeguard is however unnecessary for DNA polymerase I, e.g. the Klenow fragment from Escherichia coli, as its exonuclease is three to four orders of magnitude less active (10).
Coupling between polymerase and exonuclease active sites
Two distinct, but not mutually exclusive trends have emerged from abundant mutational data in vivo and in vitro from mostly T4 and RB69 DNA polymerases. First, the polymerase and exonuclease activities are largely correlated among a variety of mutants, however with important outliers (47,48). At one end of the correlation, both activities are vibrant. At the other end, severe loss of exonuclease activity often significantly impairs polymerase activity, for example, E191A/D324G (49) and Y320A/Q/F (48) of T4 DNA polymerase. These observations clearly suggest the coupling between two active sites. The second trend is the balance between these activities (10). The mechanisms of translocation and active site switching found in this study shed light into the structural basis of the coupling between two activities (Figure 8).
Since translocation is avoided after switching back from the exo domain, the untrimmed 3′-end of the primer would reoccupy the polymerase active site, if the exonuclease were severely deficient. Thus no new dNTP could enter the binding pocket, which leads to no new phosphoryl transfer reaction and no additional energy to replenish the fingers oscillation. If there were sufficient momentum left in the oscillator to propel one more cycle of shortcut pathway, a successful translocation would revive the reaction as a new dNTP refills the system; such as D222A/D327A of RB69 polymerase, an exonuclease deficient double mutant, retains a large fraction of the polymerase activity. If the oscillator stalls, the polymerase activity diminishes with the exonuclease activity—positive correlation.
This mechanism also applies to how the polymerase treats an abasic lesion. If an abasic site in the template is encountered, any nucleotide incorporated into the primer strand is likely to be shuttled into the exo domain, since an unpaired nucleotide has poor affinity in the binding pocket (Supplementary Figure S4g–i). If a functional exonuclease removes the 3′ nucleotide and sends the primer back, the system is refilled by new hydrolysis of dNTP and the process would repeat again and again as an idle reaction without extension beyond the abasic lesion (45). On the other hand, a deficient exonuclease would return the untrimmed 3′ nucleotide back into the polymerase active site. The base pair binding pocket has the same content before and after the excursion to the deficient exonuclease—one unpaired nucleotide. The remaining momentum is only sufficient for the short pathway of translocation without a new dNTP hydrolysis. This explains why the wild-type T4 DNA polymerase stops at an abasic site on both linear and minicircle DNA templates, but D219A and D112A/E114A exonuclease deficient mutants perform translesion replication on the same templates—anti-correlation (50,51).
Similarly, an oxidative thymine lesion, where a non-planar thymine glycol in the template fits poorly in the binding pocket, blocks the wild-type T4 (52,53) and RB69 (54) DNA polymerases, but pauses the procession of an exonuclease deficient mutant of RB69 DNA polymerase (38,54). The ability to bypass a lesion, presumably a character of the polymerase active site, is apparently regulated by the exonulease activity. Here the same mechanism of energy consumption and replenishment during translocation and active site switching simultaneously explains both positive and negative correlations between the polymerase and exonuclease activities.
The A-rule
These bifurcate pathways (Figure 8) also elucidate several phenomena related to the A-rule. (i) dA is preferentially incorporated into the primer opposite an abasic site in the template. This is because an unpaired dA has the smallest dissociation constant among unpaired nucleotides (55) due to its large size and the best ability to stack with other bases (56,57), so that dA has the greatest probability to escape from the infinite loop of active site switching and to divert onto the translocation pathway. (ii) An additional dA could also be preferentially incorporated at the 3′ end of the primer after the template has been completely paired (58). This indicates the runoff of momentum in the oscillating fingers before they come to a full stop. Nucleotides other than dA would have greater probability to be trimmed by the exonulease, which makes dA as the favored overhang beyond the end of the template. (iii) If a dA has been incorporated opposite an abasic site, it is more efficient to extend further than other scenarios in which a nucleotide other than dA has been previously incorporated opposite an abasic site (57). A nascent base pair would have poor affinity in the binding pocket, if its floor formed by the penultimate base pair were incomplete (Supplementary Figure S4j–l). Therefore, the nascent base pair would be shuttled into the exo domain, even if it were a correct match. An unpaired dA is the least incomplete floor, thus the next base pair would have the greatest probability to take the translocation pathway and to extend successfully beyond the abasic site. (iv) A short deletion also tends to occur opposite an abasic lesion in the template (51,59). This can be explained by the overall poor affinity for unpaired nucleotides in the base pair binding pocket. An unpaired nucleotide would lead to a short-term energy upsurge that drives the fingers to oscillate extra vigorously. Consecutive trips on the long excursion to the exonucease active site may trim the 3′-end multiple times without a new addition. (v) It is not surprising that no specific context in the template sequence around an abasic lesion was found correlated to a deletion (51). Based on the mechanism of the bifurcated pathways, I can foresee that a deletion opposite an abasic site would mostly depend on what nucleotides are incorporated to the primer and would have little correlation to the sequence context except repetition in the template (60). It can be predicted that a short deletion would mostly occur after two back-to-back incorporations of dT opposite the abasic site, since a dTTP replenishes the same amount of energy to the polymerase machinery, but an unpaired dT has the least affinity in the binding pocket. Not only both newly incorporated dTs would be removed, but also the previous nucleotide(s) due to the sudden energy surplus.
Balance between polymerase and exonuclease active sites
The balance between polymerase and exonulease activities has been well studied in detail using three variants of T4 DNA polymerase with ethylation on a phosphodiester linkage of a template: I417V mutant (V) with reduced DNA binding affinity in the polymerase active site, wild-type T4 DNA polymerase (W) and D112A/E114A exonuclease deficient mutant (X) (45). To illustrate the balance between polymerase and exonulease activities, these three variants V, W and X are sorted in the order of rising polymerase activity and declining exonuclease activity. The wild-type W is partially blocked by an ethyl phosphotriester on the template but is able to pass the blockage in sufficiently high concentration of dNTP. The exo- mutant X is only deterred temporarily at an ethyl phosphotriester even in low concentration of dNTP. In contrast, I417V mutant V is completely blocked by an ethyl phosphotriester and may only pass it in high concentration of dNTP for a long time. This is to say, weaker exonuclease activity seems to boost the translesion ability of the polymerase active site. It is apparent that a newly formed base pair immediately beyond the ethyl phosphotriester is treated as a mismatch. X, W and V are more and more capable of trimming the 3′-end nucleotide on the primer, incorporating a new dNTP, running into the idle reaction loop, depleting dNTP at low concentration and thus more and more likely to be blocked by an ethyl phosphotriester (X < W < V being blocked). On the other hand, V, W and X would have increasing probability to switch into the translocation pathway and pass beyond the ethylation site (V < W < X passing). However, in high concentration of dNTP for a long time, first the supply could not be easily depleted, and second each loop of the idle reaction carries a small chance to sidetrack into the translocation shortcut. Over a long time, all of the polymerase molecules may have escaped from the infinite loop and run to the completion even for V.
Mutational impact to fidelity
Numerous mutants of RB69 DNA polymerase have been studied to test hypotheses of fidelity, some structures of which have been determined. Here the most important mutants are discussed in light of the mechanisms proposed in this study for translocation and active site switching, which involve Tyr567 located at the center of the back wall between Tyr416 and Gly568 in the base pair binding pocket (Supplementary Figure S4d–f). Substitution for this residue with Ala substantially increases the error rate and also leads to other puzzling features (61,62). The available structures of Y567A mutant (e.g. 3NDK) show that a deep hole is created in the back wall. However, other residues surrounding this empty space partially collapse into it (Supplementary Figure S4a and f). Therefore the overall structural effect of Y567A is a smaller, rather than larger (63), binding pocket except that the back wall is no longer intact. Compared to the wild-type, Y567A mutant has reduced affinity to bind normal DNA duplex but elevated affinity to bind DNA duplex with mismatched base pair (62). Similar results also show that this substitution increases binding affinity of dGTP by 45-fold opposite a template G compared to the wild-type (64). The reduced size of the binding pocket resulting from the removal of the large site chain of Tyr567 seems counterintuitive. The translocation mechanism revealed in this study explains the reduced fidelity from a perspective of statistical choice of conformational pathways (Figure 8). The wild-type discriminates the correct base pairs from the incorrect ones with 5.5-fold difference in dissociation constants, while Y567A mutant cannot decisively discriminate them with <2-fold difference in dissociation constants, therefore both conformational pathways are almost equally probable. Compared to an exonuclease deficient double mutant (D222A/D327A), additional mutation Y567A slows down incorporation of correctly matched nucleotides, but speeds up incorporation of mismatched nucleotides by about 10 times (61). First, the increased error rate of Y567A mutant could be a result of some mismatches that are not sent into the exo domain for editing. Because of the higher affinity, the mismatched DNA is somehow tolerated and takes the translocation shortcut, as evidenced by the 10-fold speedup in incorporation of mismatches. Secondly, some Watson–Crick base pairs are mistakenly sent into the exonuclease active site and slow down the overall reaction since the affinity for correct base pairs is reduced (10). These experimental measurements of turnover rates and binding affinities from Y567A mutant support one another under the mechanism of translocation proposed in this study. These specific examples of wild-type and mutant functions illustrate the bifurcated pathways of translocation and active site switching at work.
To a limited extent, the structural mechanisms of translocation and active site switching extracted here from 200 polymerase structures in family B may offer some hints for understanding polymerases in family A, as some structural domains are in common between these families. Important differences must exist originating from the distinct overall architectures (65). A similar meta-analysis of family A is expected to be more challenging due to fewer structures available from the same organism.
Supplementary Material
Acknowledgments
I thank X. Yang and P. Rice for critical reading and discussion of the manuscript. The following database and software are used in this work: CCP4 (ccp4.ac.uk), CNS (cns-online.org), Coot (www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot), dynamiX™ (Renz Research, Inc., Westmont, IL, USA), gnuplot (gnuplot.info), PDB (rcsb.org), PyMOL (pymol.org), Python (python.org) and SciPy (scipy.org).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: NIH [R01EY024363].
Conflict of interest statement. None declared.
REFERENCES
- 1.Garcia-Diaz M., Bebenek K. Multiple functions of DNA polymerases. Crit. Rev. Plant Sci. 2007;26:105–122. doi: 10.1080/07352680701252817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCulloch S.D., Kunkel T.A. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benkovic S.J., Valentine A.M., Salinas F. Replisome-mediated DNA replication. Annu. Rev. Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 4.Capson T.L., Peliska J.A., Kaboord B.F., Frey M.W., Lively C., Dahlberg M., Benkovic S.J. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry. 1992;31:10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- 5.Yang G., Franklin M., Li J., Lin T.-C., Konigsberg W. Correlation of the kinetics of finger domain mutants in RB69 DNA polymerase with its structure. Biochemistry. 2002;41:2526–2534. doi: 10.1021/bi0119924. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H., Rhee C., Bebenek A., Drake J.W., Wang J., Konigsberg W. The L561A substitution in the nascent base-pair binding pocket of RB69 DNA polymerase reduces base discrimination. Biochemistry. 2006;45:2211–2220. doi: 10.1021/bi052099y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steitz T.A. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 8.Freudenthal B.D., Beard W.A., Shock D.D., Wilson S.H. Observing a DNA polymerase choose right from wrong. Cell. 2013;154:157–168. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura T., Zhao Y., Yamagata Y., Hua Y., Yang W. Watching DNA polymerase η make a phosphodiester bond. Nature. 2012;487:196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reha-Krantz L.J. DNA polymerase proofreading: multiple roles maintain genome stability. Biochim. Biophys. Acta. 2010;1804:1049–1063. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Reddy M.K., Weitzel S.E., von Hippel P.H. Processive proofreading is intrinsic to T4 DNA polymerase. J. Biol. Chem. 1992;267:14157–14166. [PubMed] [Google Scholar]
- 12.Ren Z. Reaction trajectory revealed by a joint analysis of Protein Data Bank. PLoS One. 2013;8:e77141. doi: 10.1371/journal.pone.0077141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren Z. Reverse engineering the cooperative machinery of human hemoglobin. PLoS One. 2013;8:e77363. doi: 10.1371/journal.pone.0077363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren Z., Šrajer V., Knapp J.E., Royer W.E. Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures. Proc. Natl. Acad. Sci. U.S.A. 2012;109:107–112. doi: 10.1073/pnas.1109213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung A.C.M., Cramer P. A movie of RNA polymerase II transcription. Cell. 2012;149:1431–1437. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Hogg M., Wallace S.S., Doublié S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman H.M., Kleywegt G.J., Nakamura H., Markley J.L. The Protein Data Bank at 40: reflecting on the past to prepare for the future. Structure. 2012;20:391–396. doi: 10.1016/j.str.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandonia J.-M., Brenner S.E. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 19.Henry E.R., Hofrichter J. Singular value decomposition: application to analysis of experimental data. In: Brand L, Johnson ML, editors. Numerical Computer Methods. Vol. 210. Massachusetts: Academic Press; 1992. pp. 129–192. [Google Scholar]
- 20.Liu S., Knafels J.D., Chang J.S., Waszak G.A., Baldwin E.T., Deibel M.R., Thomsen D.R., Homa F.L., Wells P.A., Tory M.C., et al. Crystal structure of the herpes simplex virus 1 DNA polymerase. J. Biol. Chem. 2006;281:18193–18200. doi: 10.1074/jbc.M602414200. [DOI] [PubMed] [Google Scholar]
- 21.Bergen K., Betz K., Welte W., Diederichs K., Marx A. Structures of KOD and 9°N DNA polymerases complexed with primer template duplex. Chembiochem. 2013;14:1058–1062. doi: 10.1002/cbic.201300175. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto H., Nishioka M., Fujiwara S., Takagi M., Imanaka T., Inoue T., Kai Y. Crystal structure of DNA polymerase from hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. J. Mol. Biol. 2001;306:469–477. doi: 10.1006/jmbi.2000.4403. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y., Jeruzalmi D., Moarefi I., Leighton L., Lasken R., Kuriyan J. Crystal structure of an archaebacterial DNA polymerase. Structure. 1999;7:1189–1199. doi: 10.1016/s0969-2126(00)80053-2. [DOI] [PubMed] [Google Scholar]
- 24.Hogg M., Osterman P., Bylund G.O., Ganai R.A., Lundström E.-B., Sauer-Eriksson A.E., Johansson E. Structural basis for processive DNA synthesis by yeast DNA polymerase ϵ. Nat. Struct. Mol. Biol. 2013;21:49–55. doi: 10.1038/nsmb.2712. [DOI] [PubMed] [Google Scholar]
- 25.Swan M.K., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat. Struct. Mol. Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu E.Y., Beese L.S. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an ‘ajar’ intermediate conformation in the nucleotide selection mechanism. J. Biol. Chem. 2011;286:19758–19767. doi: 10.1074/jbc.M110.191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai Y.-C., Johnson K.A. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahl J.M., Mai A.H., Cherf G.M., Jetha N.N., Garalde D.R., Marziali A., Akeson M., Wang H., Lieberman K.R. Direct observation of translocation in individual DNA polymerase complexes. J. Biol. Chem. 2012;287:13407–13421. doi: 10.1074/jbc.M111.338418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lieberman K.R., Dahl J.M., Mai A.H., Cox A., Akeson M., Wang H. Kinetic mechanism of translocation and dNTP binding in individual DNA polymerase complexes. J. Am. Chem. Soc. 2013;135:9149–9155. doi: 10.1021/ja403640b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacewicz A., Trzemecka A., Guja K.E., Plochocka D., Yakubovskaya E., Bebenek A., Garcia-Diaz M. A remote palm domain residue of RB69 DNA polymerase is critical for enzyme activity and influences the conformation of the active site. PLoS One. 2013;8:e76700. doi: 10.1371/journal.pone.0076700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia S., Wang M., Blaha G., Konigsberg W.H., Wang J. Structural insights into complete metal ion coordination from ternary complexes of B family RB69 DNA polymerase. Biochemistry. 2011;50:9114–9124. doi: 10.1021/bi201260h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia S., Eom S.H., Konigsberg W.H., Wang J. Bidentate and tridentate metal-ion coordination states within ternary complexes of RB69 DNA polymerase. Protein Sci. 2012;21:447–451. doi: 10.1002/pro.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zahn K.E., Tchesnokov E.P., Gotte M., Doublie S. Phosphonoformic acid inhibits viral replication by trapping the closed form of the DNA polymerase. J. Biol. Chem. 2011;286:25246–25255. doi: 10.1074/jbc.M111.248864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hogg M., Aller P., Konigsberg W., Wallace S.S., Doublie S. Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J. Biol. Chem. 2006;282:1432–1444. doi: 10.1074/jbc.M605675200. [DOI] [PubMed] [Google Scholar]
- 35.Zahn K.E., Belrhali H., Wallace S.S., Doublié S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- 36.Zahn K.E., Averill A., Wallace S.S., Doublié S. The miscoding potential of 5-hydroxycytosine arises due to template instability in the replicative polymerase active site. Biochemistry. 2011;50:10350–10358. doi: 10.1021/bi201219s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aller P., Ye Y., Wallace S.S., Burrows C.J., Doublié S. Crystal structure of a replicative DNA polymerase bound to the oxidized guanine lesion guanidinohydantoin. Biochemistry. 2010;49:2502–2509. doi: 10.1021/bi902195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aller P., Duclos S., Wallace S.S., Doublié S. A crystallographic study of the role of sequence context in thymine glycol bypass by a replicative DNA polymerase serendipitously sheds light on the exonuclease complex. J. Mol. Biol. 2011;412:22–34. doi: 10.1016/j.jmb.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franklin M.C., Wang J., Steitz T.A. Structure of the replicating complex of a Pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 40.Marquez L.A., Reha-Krantz L.J. Using 2-aminopurine fluorescence and mutational analysis to demonstrate an active role of bacteriophage T4 DNA polymerase in strand separation required for 3′ right-arrow 5′-exonuclease activity. J. Biol. Chem. 1996;271:28903–28911. doi: 10.1074/jbc.271.46.28903. [DOI] [PubMed] [Google Scholar]
- 41.Shamoo Y., Steitz T.A. Building a replisome from interacting pieces. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 42.Stocki S.A., Nonay R.L., Reha-Krantz L.J. Dynamics of bacteriophage T4 DNA polymerase function: Identification of amino acid residues that affect switching between polymerase and 3′ → 5′ exonuclease activities. J. Mol. Biol. 1995;254:15–28. doi: 10.1006/jmbi.1995.0595. [DOI] [PubMed] [Google Scholar]
- 43.Petruska J., Goodman M.F., Boosalis M.S., Sowers L.C., Cheong C., Tinoco I.J. Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pomerantz R.T., O'Donnell M. Replisome mechanics: insights into a twin DNA polymerase machine. Trends Microbiol. 2007;15:156–164. doi: 10.1016/j.tim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Tsujikawa L., Weinfield M., Reha-Krantz L.J. Differences in replication of a DNA template containing an ethyl phosphotriester by T4 DNA polymerase and Escherichia coli DNA polymerase I. Nucleic Acids Res. 2003;31:4965–4972. doi: 10.1093/nar/gkg722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reha-Krantz L.J. Regulation of DNA polymerase exonucleolytic proofreading activity: studies of bacteriophage T4 ‘antimutator’ DNA polymerases. Genetics. 1998;148:1551–1557. doi: 10.1093/genetics/148.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reha-Krantz L.J., Nonay R.L. Genetic and biochemical studies of bacteriophage T4 DNA polymerase 3′->5′-exonuclease activity. J. Biol. Chem. 1993;268:27100–27108. [PubMed] [Google Scholar]
- 48.Sattar A.K.M.A., Lin T.-C., Jones C., Konigsberg W.H. Functional consequences and exonuclease kinetic parameters of point mutations in bacteriophage T4 DNA polymerase. Biochemistry. 1996;35:16621–16629. doi: 10.1021/bi961552q. [DOI] [PubMed] [Google Scholar]
- 49.Reha-Krantz L.J., Stocki S., Nonay R.L., Dimayuga E., Goodrich L.D., Konigsberg W.H., Spicer E.K. DNA polymerization in the absence of exonucleolytic proofreading: in vivo and in vitro studies. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2417–2421. doi: 10.1073/pnas.88.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Gac N.T., Delagoutte E., Germain M., Villani G. Inactivation of the 3′-5′ exonuclease of the replicative T4 DNA polymerase allows translesion DNA synthesis at an abasic site. J. Mol. Biol. 2004;336:1023–1034. doi: 10.1016/j.jmb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 51.Hatahet Z., Zhou M., Reha-Krantz L.J., Ide H., Morrical S.W., Wallace S.S. In Vitro selection of sequence contexts which enhance bypass of abasic sites and tetrahydrofuran by T4 DNA polymerase holoenzyme. J. Mol. Biol. 1999;286:1045–1057. doi: 10.1006/jmbi.1998.2520. [DOI] [PubMed] [Google Scholar]
- 52.Clark J.M., Beardsley G.P. Functional effects of cis-thymine glycol lesions on DNA synthesis in vitro. Biochemistry. 1987;26:5398–5403. doi: 10.1021/bi00391a027. [DOI] [PubMed] [Google Scholar]
- 53.Clark J.M., Beardsley G.P. Template length, sequence context, and 3′-5′ exonuclease activity modulate replicative bypass of thymine glycol lesions in vitro. Biochemistry. 1989;28:775–779. doi: 10.1021/bi00428a054. [DOI] [PubMed] [Google Scholar]
- 54.Aller P., Rould M.A., Hogg M., Wallace S.S., Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl. Acad. Sci. U.S.A. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berdis A.J. Dynamics of translesion DNA synthesis catalyzed by the bacteriophage T4 exonuclease-deficient DNA polymerase. Biochemistry. 2001;40:7180–7191. doi: 10.1021/bi0101594. [DOI] [PubMed] [Google Scholar]
- 56.Kool E.T. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 57.Xia S., Vashishtha A., Bulkley D., Eom S.H., Wang J., Konigsberg W.H. Contribution of partial charge interactions and base stacking to the efficiency of primer extension at and beyond abasic sites in DNA. Biochemistry. 2012;51:4922–4931. doi: 10.1021/bi300296q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clark J.M. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shibutani S., Takeshita M., Grollman A.P. Translesional synthesis on DNA templates containing a single abasic site. J. Biol. Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 60.Kunkel T.A. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- 61.Bebenek A., Dressman H.K., Carver G.T., Ng S., Petrov V., Yang G., Konigsberg W.H., Karam J.D., Drake J.W. Interacting fidelity defects in the replicative DNA polymerase of bacteriophage RB69. J. Biol. Chem. 2001;276:10387–10397. doi: 10.1074/jbc.M007707200. [DOI] [PubMed] [Google Scholar]
- 62.Trzemecka A., Jacewicz A., Carver G.T., Drake J.W., Bebenek A. Reversal of a mutator activity by a nearby fidelity-neutral substitution in the RB69 DNA polymerase binding pocket. J. Mol. Biol. 2010;404:778–793. doi: 10.1016/j.jmb.2010.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang H., Beckman J., Wang J., Konigsberg W. RB69 DNA polymerase mutants with expanded nascent base-pair-binding pockets are highly efficient but have reduced base selectivity. Biochemistry. 2009;48:6940–6950. doi: 10.1021/bi900422b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang G., Wang J., Konigsberg W. Base selectivity is impaired by mutants that perturb hydrogen bonding networks in the RB69 DNA polymerase active site. Biochemistry. 2005;44:3338–3346. doi: 10.1021/bi047921x. [DOI] [PubMed] [Google Scholar]
- 65.Wang J., Sattar A.K.M.A., Wang C.C., Karam J.D., Konigsberg W.H., Steitz T.A. Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.