Abstract
Fetal exposure to essential and toxic metals can influence life-long health trajectories. The placenta regulates chemical transmission from maternal circulation to the fetus and itself exhibits a complex response to environmental stressors. The placenta can thus be a useful matrix to monitor metal exposures and stress responses in utero, but strategies to explore the biologic effects of metal mixtures in this organ are not well-developed. In this proof-of-concept study, we used laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS) to measure the distributions of multiple metals in placental tissue from a low-birth-weight pregnancy, and we developed an approach to identify the components of metal mixtures that colocalized with biological response markers. Our novel workflow, which includes custom-developed software tools and algorithms for spatial outlier identification and background subtraction in multidimensional elemental image stacks, enables rapid image processing and seamless integration of data from elemental imaging and immunohistochemistry. Using quantitative spatial statistics, we identified distinct patterns of metal accumulation at sites of inflammation. Broadly, our multiplexed approach can be used to explore the mechanisms mediating complex metal exposures and biologic responses within placentae and other tissue types. Our LA-ICP-MS image processing workflow can be accessed through our interactive R Shiny application ‘shinyImaging’, which is available at https://mniedz.shinyapps.io/shinyImaging/.
Keywords: laser ablation-inductively coupled plasma-mass spectrometry, mass spectrometry imaging, image processing, colocalization, prenatal exposures, metallomics, metal mixtures, biological response, placenta, humans, exposome
Graphical abstract
Introduction
The adverse health effects of metal exposures—including toxic metal exposures and micronutrient deficiencies and excesses—may be exacerbated or ameliorated when exposure occurs in the context of a mixture.1 This is of particular importance in utero, when critical windows of susceptibility exist for fetal programming events that may trigger negative health outcomes later in life.2 In exposure biology, an emerging paradigm is the concept of the pregnancy exposome, which seeks to understand the interactions among all environmental exposures that pass through or are stopped at the placenta,3 as well as simultaneously evaluate the physiological responses to these exposures.4,5 An ‘exposomic’ approach has the potential to enable new discoveries about the role of mixed-metal exposures in the fetal origins of disease; however, the successful application of this paradigm is predicated upon the development of novel biomarkers and analytic approaches for assessing multiple exposures simultaneously.
The exposome is a complex concept that involves not only how exposures change over time, but also how a given chemical distributes in the body or even within a tissue. In theory, two people with an identical level of exposure (i.e. dose) may have different biological responses. Inherent in this concept are genetic and epigenetic susceptibility; while genetics and epigenetics no doubt play a role in chemical distribution, many other factors, some of which may be stochastic, also play a role. Thus, how a chemical distributes in a tissue, which is a reflection of where it is binding, may be more informative than only measuring epigenetic or genetic factors that partially drive that distribution difference. Measuring all these factors together is one of the long-term goals of the exposome.
Mass spectrometry imaging (MSI) is a powerful tool for the direct quantitative visualization of the distributions of multiple analytes in tissues.6 One common MSI technique, laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS), allows the sensitive, high-resolution spatial mapping of element concentrations.7,8 MSI holds great promise for advancing the study of the human exposome, and we propose that multidimensional elemental imaging of placental tissues is a powerful approach to uncover the biologic effects of metal mixtures during pregnancy.9 We recognized that the widespread integration of elemental imaging in exposomic research, particularly in large-scale human studies, necessitates a workflow involving the (i) rapid and objective pre-processing of elemental imaging datasets, (ii) integration of data from elemental imaging with other imaging approaches, and (iii) rigorous statistical analysis to quantify the relationships among analytes measured across different imaging modalities.
With the goal to create a workflow enabling a more nuanced assessment of the biologic impacts of metal mixture exposures in the human placenta, we combined LA-ICP-MS elemental imaging10 with multiplexed immunohistochemistry (IHC) as a novel approach to measure element and protein distributions within tissues. Here, we present a proof-of-principle example showing how our workflow can be used to assess the spatial associations between metals and markers of inflammation in the placenta.
Results
Overview of elemental imaging workflow
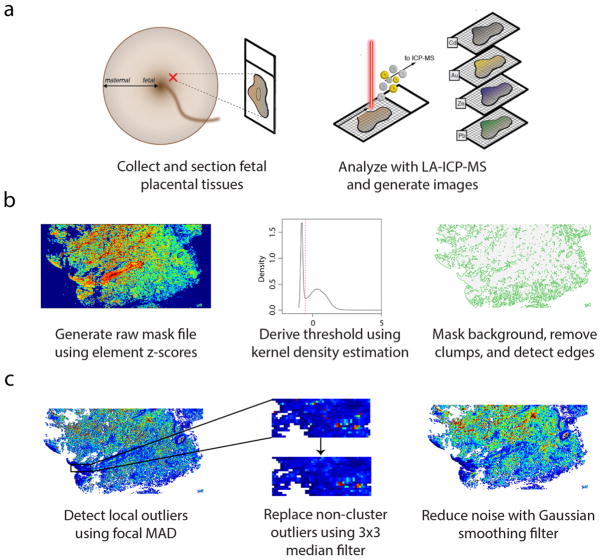
The sample collection, preparation and imaging protocol is summarized in Figure 1; a video displaying the implementation of our workflow using our interactive software tool shinyImaging (https://mniedz.shinyapps.io/shinyImaging/) can be found in Supplementary Movie 1 and https://vimeo.com/156307893. Briefly, tissue was collected from the fetal side of the placenta, fixed in formalin, and embedded in paraffin. Tissue sections (5-μm thick) were affixed to glass microscope slides and imaged using a LA-ICP-MS system, and signals for the major (or sole) isotopes for elements of interest were collected and assembled into multidimensional grid stacks using our R script, which was written to accompany the R package ‘raster’.11 Our data reduction approach combines all individual .csv data files (or ablation lines) into a single file structured as a stack of matrices, with one element per matrix (Fig. 1). Following pre-processing (see Materials and Methods), the matrix stack is converted to the RasterStack file format, in which matrices are RasterLayers with identical grid structures (spatial extent and resolution). The element RasterStack can be further analyzed in R or written to a file in numerous single and multiband formats, including but not limited to ascii (.asc), ENVI (.envi), and GeoTiff (.tif).
Fig. 1. Overview of LA-ICP-MS imaging workflow.
(a) Formalin-fixed, paraffin-embedded placental tissues, sampled from the fetal aspect, were sectioned (5-μm thick) and affixed to glass slides. Spatial distribution of multiple elements was determined using LA-ICP-MS, and data was read into R as a grid stack for image processing. (b) Background pixels were masked using a global thresholding algorithm. A raw mask file was created by converting the elements in the grid stack to z-scores, removing elements from the stack with high background noise, and finding the mean pixel z-scores across all elements in the stack, weighted on the squared gas-blank z-score means for each element. A threshold cutoff value was determined from the first-occurring local minimum value from the kernel density estimate of the weighted mean grid, and pixels with values below the cutoff were set to null values. Then, the algorithm removed small clusters of remaining background pixels and identified tissue edges. (c) Extreme outliers were detected using a focal median absolute deviation algorithm that ignored outliers in biologically-relevant clusters, and these outliers were replaced using a 3×3 median filter. After outlier replacement, images were smoothed using a Gaussian filter (σ = 2/3, r = 2).
In quantitative image analysis, the presence of background pixels can lead to biased results and should be removed prior to statistical analyses.12 Here, we devised a global thresholding algorithm to classify tissue and background pixels uniformly across all layers of a multidimensional element image stack (see Materials and Methods for an in-depth description of the algorithm). Briefly, our algorithm calculates the variance in element intensities in the overall image and within the gas-blank region to assess the ability of each element to differentiate between tissue and background pixels, and a composite ‘mask’ layer is created by calculating the mean intensity for each pixel across all elements in the stack, with each element weighted on its pixel differentiation ability. Next, kernel density estimation, a non-parametric smoothing technique, is used to estimate the probability density function for the mask layer. Since the distribution is bimodal (a mixture of two component distributions, target and background), the algorithm determines the threshold value by finding the value at the first-occurring local minimum in this distribution. Then, the algorithm sets all pixels in the mask layer with values below the threshold to null (NA). (If necessary, remaining background pixels can be removed using a clump identification algorithm that assigns null values to small ‘clumps’ [i.e., patches of connected pixels] and/or manually removed using a region-of-interest selection tool.) Finally, an edge detection algorithm delineates tissue boundaries to create a final mask layer with edge, non-edge, and background pixels assigned values of 1, 0, and NA respectively (Fig. 1b). Masked element stacks are generated by assigning null values to all corresponding pixels classified as “NA” in the mask layer, consistently across each layer in the stack.
Grids then undergo a robust cleaning and smoothing procedure (Fig. 1c). We designed an approach for the spatial assessment of local extreme outliers that are likely to result from shot noise or other instrumentation-related issues, since clusters of high values identified as potential outliers by a global assessment may instead represent biologically-relevant areas of metal accumulation. As such, a moving window MAD assessment identifies pixels in masked grids with values that exceed the local median by a specific magnitude (here, 5 times MAD), while ignoring outliers that are located in clusters that are likely to be biologically relevant (see Materials and Methods). Outliers are replaced using a focal filter that replaces values for outliers only. To further reduce noise and remove imaging artefact, grids are smoothed with a Gaussian filter, which is performed after outlier removal to prevent inadvertent integration of extreme outliers into the final images.
To examine the performance and speed of our workflow, we analyzed randomly-selected regions from the fetal and maternal sides from 5 placental samples (10 sections in total; 90 ablation lines each). Our masking algorithm was successful for all 10 sections and performed excellently despite the presence of shot noise and complex tissue architectures (Supplementary Figures 1–10). Using our shinyImaging application, we were able to process all 10 samples (raw ablation file upload, background correction, masking, outlier removal, smoothing, and ‘cleaned’ file download) in approximately 30 minutes.
Proof-of-concept: metals, inflammation and low birth weight
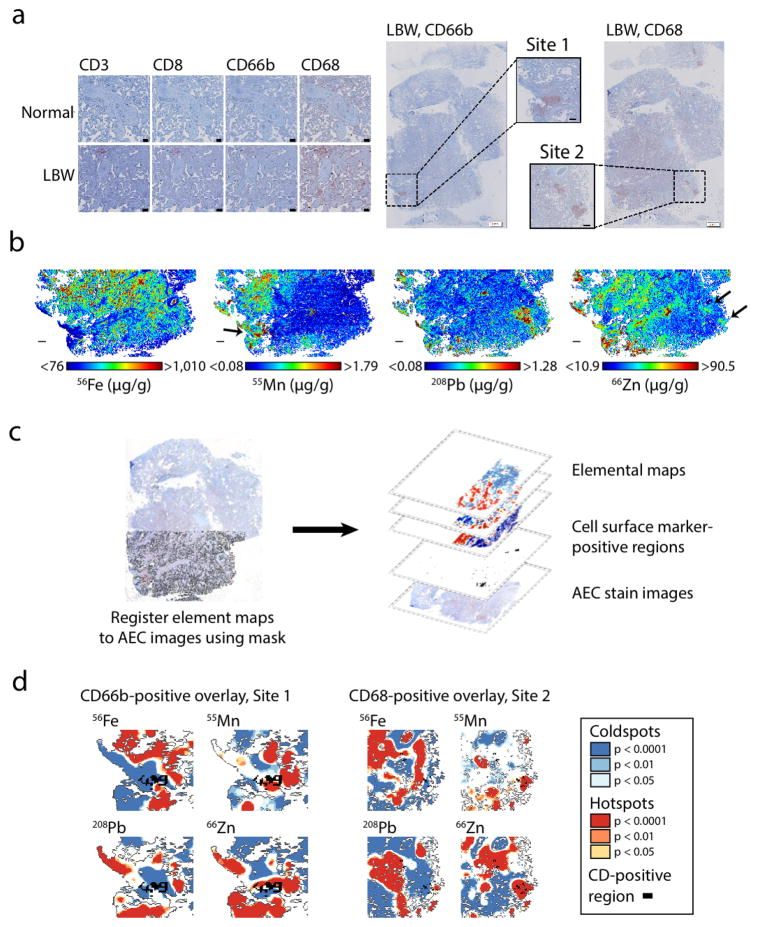
Low birth weight (LBW; birth weight < 2500 g, regardless of gestational age) is a significant predictor of infant morbidity and mortality.13,14 Inflammation in the placenta is believed to be a major pathway of LBW in preterm pregnancies,15 but the source of this inflammation is not always known. As a proof-of-concept to demonstrate how our workflow could be used to explore the relationships between trace metals and placental inflammation in LBW—and to demonstrate how elemental imaging can be integrated with other imaging techniques—we stained for CD3, CD8, CD68, and CD66b using 3-amino-9-ethylcarbazole (AEC)-based multiplexed immunohistochemistry in a single tissue section sampled from the fetal side of the placenta from a LBW birth (2,185 g; born at 35 weeks gestation) and normal weight birth (2,828 g; born at 34 weeks gestation); both infants were male and delivered via Cesarean section (Figure 2). Expression of inflammatory markers CD68 (marker of macrophages) and CD66b (marker of neutrophils) was markedly increased in the chorionic villi of the LBW placenta (Fig. 2a).
Fig. 2. Metals and inflammation in a low-birth-weight placenta.
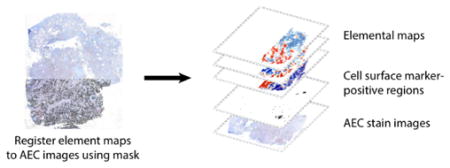
(a) Left: immunohistochemical stains for inflammatory cell surface markers (CD3, CD8, CD66b, and CD68) in placentas from normal-weight and low-birth-weight pregnancies (scale bar = 100 μm). Right: low-resolution stains in low-birth-weight placenta for CD66b and CD68 (scale bar = 2 mm) with higher-resolution images from sites of CD66b- and CD68-positive staining (Site 1 and Site 2, respectively; scale bar = 500 μm). (b) Element maps for Fe, Mn, Pb, and Zn from adjacent serial section in low-birth-weight placenta. Scale bar = 1 mm. (c) Immunohistochemical stains and element maps were overlaid by registering the tissue mask to the stain image, then creating a multilayered map with stain images, stain-positive pixels, and element maps registered to the same spatial coordinate system. (d) Overlay of CD66b- and CD68-positive regions with Getis Ord Gi* hotspot and coldspots for Fe, Mn, Pb, and Zn.
Visual inspection of the low-resolution stains for the LBW section identified one site of CD66b accumulation (Site 1) and another site of CD68 accumulation (Site 2; Fig. 2a). In an adjacent section in the LBW placenta, we performed LA-ICP-MS imaging (40-μm spot size; pixel area = 1600 μm2) for Fe, Zn, Mn, Pb, and Cd (Fig. 2b). Although Cd was below detection limits, we found unique distributions for the other elements that appeared to coincide with these inflammation sites: e.g., Mn and CD66b seemed to be found together at Site 1, and Zn and CD68 appeared to coincide at Site 2 (Fig. 2b).
To explore the relationships between the inflammatory markers and metals in a more quantitative manner, we constructed a multilayered map in which the AEC stain images and elemental maps were registered to the same spatial coordinate system (Fig. 2c). We aligned the AEC stain images using automated 2D rigid body image registration, then registered the element maps to the histologically stained images by manually selecting tie points (corresponding features) between the stains and the tissue mask layer using QGIS 2.10.1,16 an open-source geographic information systems (GIS) program. We classified the AEC stains by separating the hematoxylin and AEC stains using color deconvolution and applying an intensity threshold algorithm to identify positive and negative pixels. Our final GIS map structure consisted of the AEC base images with spatially-registered elemental maps and AEC-positive areas for each cell surface marker that could be easily overlaid (Fig. 2c).
Based on the image overlays at Site 2, CD68-positive areas appeared to coincide with Zn hotspots (Fig. 2d). We identified element hotspots at Site 1 and Site 2 using site-specific Getis-Ord Gi* analyses, which objectively identifies areas of high and low element accumulation (details in Materials and Methods). We quantified the overlap between the CD68-positive regions and element hotspots (Getis Ord Gi* z-scores > 1.96) using the ‘cell surface marker fractional’ (fCDx: fraction of cell surface marker-positive pixels that were also classified as element hotspots) and confirmed that all CD68-positive pixels were also Zn hotspots (fCD68 = 1.00) at Site 2. While the CD66b-positive regions at Site 1 appeared to possibly colocalize with hotspots for both Mn and Zn, the fCD66b scores for the Mn and Zn hotspots showed less evidence of strong colocalization (Mn, fCD66b = 0.33; Zn, fCD66b = 0.39).
Discussion
The placenta is critical in maintaining the health of the fetus, and biomarkers of placenta-mediated changes to the intrauterine environment may serve as predictors of future health disorders.17,18 Recognizing the central role of the placenta in fetal development, major initiatives, including the Human Placenta Project of the US National Institutes of Health, have been instigated to develop novel methods for better understanding of how placental function is affected by a variety of external stressors.19,20 To uncover the role of environmental chemical exposures in fetal programming, it is important to characterize the pathways connecting environmental exposures, often occurring as complex mixtures of chemicals, with the biologic responses elicited within the placenta and to the effects on the developing fetal systems.18 Traditional approaches to analyzing placentae have not considered its complex architecture but have instead analyzed bulk chemical concentrations in tissue segments,21 which does not uncover regional variations in placental chemical processing that is likely cell-specific.
To overcome this barrier—and as a step toward the integration of MSI in human exposomic research—we developed a rapid and automated elemental image processing workflow for the quantitative assessment of the biologic impacts of complex metal mixtures. Our overarching goal was to develop an approach to consider the spatial distributions of metal exposures with cellular response proteins in human placental tissues. By doing so, we believe additional information on chemical effects can be deciphered when compared to standard methods that use whole tissues to measure chemical exposures or protein expression.
To demonstrate the potential of our method in health outcome research, we investigated the associations between metals and inflammatory markers in placental sections from a LBW case and a typical birth weight control. We also display the versatility of our workflow, which readily integrates multielemental MSI imaging with data generated from other modalities (chromogen-based stains in this case). Using quantitative spatial statistics, we observed that sites of inflammation coincided with variations in Fe, Mn and Zn. While we intended for this sample to serve as a proof-of-concept of our workflow and thus do not draw any biological conclusions from the metal/protein associations, we note that previous reports have observed complex relationships of Zn and Mn with neutrophil and macrophage activities.22,23 Although no inferences can be drawn from the current study, we propose that the integration of elemental imaging with immune marker detection has the potential to provide insight into the roles of these metals in the immune response in future studies. We used formalin-fixed, paraffin-embedded tissues to test the applicability of this method for banked human samples. However, our workflow can be readily applied to tissues prepared with other methods, such as frozen sections, when there are concerns that fixation may perturb elemental signatures.24–26
Conclusions
Broadly, the workflow we present here can be used to quantitatively explore the relationships between metal mixture exposures and biologic response mediators in placenta and other tissues. The multiplexed nature of these assays make them amenable to data-driven computational approaches to identify disease-relevant interactions between metals and physiologic pathways, and are a crucial step towards the application of MSI in uncovering the role of the placental exposome in fetal programming of life-long health trajectories.
Materials and Methods
Sample collection and processing
Samples were collected from mother-child dyads enrolled from hospitals in Mexico City, Mexico. All subjects provided informed consent, and study protocols were approved by the Institutional Review Boards of all participating institutions. Placental tissues were sampled immediately after birth and placed in 4% neutral formaldehyde for transport. After arrival at the laboratory, the maternal, maternal decidua, and fetal zones were identified, and a small fragment from each zone was collected and fixed in 4% neutral formaldehyde overnight. The fragments were embedded in paraffin, cut to 5-um sections, and affixed to glass microscope slides, which were preserved at 4°C until use.
Cell surface marker immunohistochemistry
Placental sections were dewaxed in xylene and rehydrated in decreased concentrations of ethanol (EtOH) to pure water. Sections were incubated in Target Retrieval Solution pH 9.0 (Dako, S2367) for antigen retrieval (95°C, 30 minutes). Sections were incubated with 3% hydrogen peroxide and protein block serum-free (Dako, X0909) before adding primary antibody (CD3, clone 2GV6) followed by secondary antibody coupled with biotin. The binding of biotinylated antibodies was revealed by streptavidin-horseradish peroxidase, and peroxidase activity was revealed using 3-amino-9-ethylcarbazole (AEC, Vector, SK-4200). Tissue sections were then counterstained with hematoxylin Harris modified (Sigma, HHS16) and the slides mounted with aqueous mounting medium. Images were acquired using an Olympus whole-slide scanner operated with Olyvia software. After scanning, the CD3 staining was bleached and the slides subjected to another cycle of tagging (MICSSS method, Remark et al., submitted). Thus, the same slides were stained for CD8 (clone C8/144b) using Target Retrieval Solution pH 9, for antigen retrieval, scanned and de-stained. By following the same workflow, the same slides were then sequentially stained for CD68 (clone KP1) and CD66b (clone G10F5) using 10 mM citrate buffer pH 6 and Target Retrieval Solution pH 9 for antigen retrieval, respectively.
LA-ICP-MS analysis
A New Wave Research NWR-193 laser ablation system was connected to an Agilent Technologies 8800 ICP-MS by Tygon® tubing. A flow of 0.8 L min−1 He gas carried ablated material from the ablation chamber and was mixed with 0.65 L min−1 Ar via a y-piece prior to the ICP-MS. A 40 μm diameter laser beam was rastered at a speed of 100 μm s−1. ICP-MS isotope dwell times for manganese (55Mn), iron (56Fe), zinc (66Zn), cadmium (111Cd) and lead (208Pb) were adjusted to maintain sample dimensions27 (data points that correspond to a pixel size of approximately 40 × 40 μm). Laser fluence and repetition rate was optimized for ablation of the tissue whilst preventing ablation of the underlying glass slide (0.5 J cm−2, 40 Hz). A 3.0 mL min−1 flow of H2 in the ICP-MS octopole reaction system (ORS®) was used to minimize interferences and improve signal stability.28 In the absence of suitable tissue standards, the certified reference material NIST612 glass was used to correct signal variation between analyses and provide indicative concentration values. LA-ICP-MS quantification is dependent on sample matrix;29 therefore, the concentration values provided in this study are considered semi-quantitative.
LA-ICP-MS data processing
Following LA-ICP-MS imaging, ablation line-specific .csv files generated by the MassHunter Workstation software (Agilent) were read into RStudio (version 0.98.1102),30 and CPS values for each element were coerced into a list of m by n element-specific matrices, where m = number of ablation lines and n = number of ablation spots per ablation line. For each matrix, row-specific gas blank CPS medians—which here corresponded to the median values in pixels (ablation spots) collected during the first 10 s of each ablation line—were subtracted from each pixel in the corresponding row. Gas-blank corrected matrices were converted to the RasterStack format using the R package ‘raster’,11 where each Xm x n matrix was converted to a RasterLayer with spatial extent (xmin = 0, xmax = n, ymin = 0, ymax = m).
Image segmentation and background removal
Image segmentation into target and background pixels is often achieved using histogram-based thresholding of grayscale images, such as Otsu’s method.31 Here, we designed a histogram-based global thresholding algorithm that could be applied to LA-ICP-MS multidimensional imaging data. In an elemental imaging stack, element CPS values follow a finite mixture distribution with two component distributions,
| (1) |
where ptissue(x) and pback(x) represent the probability density functions for tissue and background pixels, respectively, and w = weights where wi ≥ 0 and wtissue + wback = 1. Additionally, there is a varying level of shot (background) noise, which follows a Poisson distribution that can be approximated by a Gaussian distribution,
| (2) |
Our algorithm derives a global threshold cutoff value from a single composite image created from an element RasterStack, with a method designed to (i) identify and remove elements with high background noise and (ii) place more weight on elements with higher tissue to background signals (xtissue/xback). First, CPS values for each element were scaled and centered to generate standardized scores (z-scores), and elements with high background noise (high z-score variability in gas-blank regions based on a user-defined cutoff; here, standard deviation (s) > 0.15) were removed from the element stack. Next, a single composite layer (CL) was created by calculating the mean z-score for each pixel among all remaining element layers in the stack, with elements weighted by the squared z-score mean in pixels located in their gas-blank region, such that
| (3) |
where n = number of RasterLayers in the stack, (xi j)k = z-score for pixel (i,j) in RasterLayer Xk, and wk = squared z-score mean in the gas-blank region of Xk, since elements with a better ability to differentiate between tissue and background will have z-scores in gas-blank pixels that deviate further below the overall image z-score mean of 0. From the vectorized values in this composite layer, the kernel density function was estimated using the R function density. The basic kernel density estimator can be expressed as follows,
| (4) |
where K = kernel and h = bandwidth; here, our algorithm uses a Gaussian kernel and the “nrd0” bandwidth selector, which defaults to Silverman’s “rule of thumb”,32
| (5) |
where R = the interquartile range X[0.75n] − X[0.25n]. The cutoff value for background masking (ncutoff) was calculated by finding the first-occurring local minimum value from the n = 512 equally-spaced points at which the density was estimated. Background pixels (BP) were classified by thresholding the weighted mean z-scores in the composite mask layer based on the criterion
| (6) |
All corresponding BP pixels in the mask layer were set to null (NA), while values in non-BP pixels were not changed.
After background thresholding, remaining pixels in the background region were removed using an automated clump deletion algorithm. First, unique ‘clumps’, or patches of connected pixels, were detected and assigned unique IDs using the clump function in the ‘raster’ package, with cells considered adjacent based on the Rook’s case contiguity rule. Next, all unique clumps in the mask layer that were comprised of less than a user-defined number of pixels (here, all clumps of ≤3 pixels) were assigned null (NA) values. A small group of remaining background clumps in the LBW sample mask was removed by manually drawing a polygon around the region using a region-of-interest selection tool (the drawPoly function in the ‘raster’ package) and setting pixels within this polygon region to NA. In the final step of the creation of the mask layer, tissue edges were identified based on Rook’s case contiguity using the boundary function in the ‘raster’ package. Pixels in the final mask (FM) layer were represented by values
| (7) |
Masked element stacks were created by assigning NA values to all pixels identified as background (NA) pixels in the mask layer, uniformly across each layer in the stack.
Outlier identification and spatial smoothing
We designed a local outlier detection algorithm to identify and replace local extreme, right-tail outliers that were likely to result from random shot noise and/or other instrumentation-related issues. Since LA-ICP-MS is a scanning technique, adjacent pixels in the y-axis are not collected sequentially, but are instead separated by a time overhead as the laser is rastered across the sample in x-axis rows. Thus, outliers that cluster along both the x- and y-axes are less likely to be due to random instrumentation fluctuation and more likely to reflect true biological variance. First, outliers were identified in masked element grids using a moving-window (user-defined dimensions; here, 7 × 7 pixels) method, where the median absolute deviation
| (8) |
was calculated for 7 × 7 moving windows using the focal function in the ‘raster’ package, and pixels with values deviating from the local median at a user-defined magnitude greater than the MAD (here, 5 times) were classified as outliers. Next, a cluster identification algorithm identified outliers that were present in clusters occurring in ≥ 2 rows, which were reclassified as non-outlier pixels. Remaining outliers were assigned null values and replaced using a user-defined filter (here, a 3 × 3 median filter), which only replaced values in tissue-associated (non-background) pixels with null values. Finally, all non-null pixels in the masked images were spatially smoothed using a Gaussian smoothing operator,
| (9) |
where σ = 2/3; the default radius in the focalWeight function in the ‘raster’ package, which was used here for spatial smoothing, is 3σ. The masked and smoothed elemental images were the final images used for all subsequent statistical analyses. (Note: prior to spatial smoothing, the shinyImaging application replaces outliers using a weighted mean 7 × 7 filter [Gaussian filter: σ=1, r=3] rather than a median filter).
Integration of data from elemental imaging and immunohistochemistry
Cell surface marker (AEC) images (.vsi files) were imported into ImageJ 1.49v and converted to RGB color images. The images were background subtracted, compiled into a stack, and aligned using 2D rigid body image registration with the StackReg plugin.33 After unstacking the images, positive AEC pixels were identified by separating the hematoxylin and AEC stains with the “H AEC” vector in the Color Deconvolution 1.7 plugin, then applying a global threshold (using Tsai’s method34) to the 8-bit AEC-separated images. The registered original images and segmented AEC-positive images were saved as .tif files, which were each imported into QGIS 2.10.116 as raster layers within a project file. To register the element maps to the cell surface marker images, the tissue mask (.asc file) was opened in the QGIS Georeferencer, tie points (corresponding features) were manually selected between the tissue mask and the stain images, and the mask was aligned to the stains using a thin plate spline transformation. Tie point coordinates were saved and used to register the element .asc files, which were saved as raster layers in the project file.
Statistical analysis of analyte colocalization
We used a Getis-Ord Gi* hotspot analysis35 for an objective method to identify statistically-significant areas of high and low element accumulation. Here, the Getis-Ord Gi* statistic was calculated for each element raster using the lisa function in the ‘usdm’ R package36, using a local neighborhood size corresponding to 175 μm, and pixels with z-scores ≥ 1.96 or ≤ −1.96 were classified as significant ‘hotspots’ or ‘coldspots,’ respectively. To assess the degree of overlap between element hotspots and immune marker-positive regions (AEC-positive regions) using a method similar to the Manders overlap coefficient37, we calculated three scores originally proposed by Nawaz et al.38:
Cell surface marker fractional (fCDx): fraction of AEC-positive regions that were also element hotspots;
Element hotspot fractional (felement): fraction of element hotspots that were also AEC-positive regions; and
Overlap fractional (foverlap): fraction of AEC-positive regions and/or element hotspots that were both AEC-positive regions and element hotspots (different from the colocalization statistic proposed by Nawaz et al.35).
Supplementary Material
Acknowledgments
We thank Joseph Gilbert for his help with the collection and preparation of the placental tissues. M.A. is supported by DP2ES025453 (New Innovator Award) and R00ES019597 from the National Institutes of Health. R.O.W. is supported by P30ES023515, R01ES013744, and R01ES021357 from the National Institutes of Health.
Footnotes
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Cui Y, Zhu YG, Zhai R, Huang Y, Qiu Y, Liang J. Exposure to metal mixtures and human health impacts in a contaminated area in Nanning, China. Environment International. 2005;31:784–790. doi: 10.1016/j.envint.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 2.Hu H, Shine J, Wright RO. The challenge posed to children’s health by mixtures of toxic waste: The Tar Creek Superfund site as a case-study. Pediatric Clinics of North America. 2007;54:155–175. doi: 10.1016/j.pcl.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson O, Vrijheid M. The pregnancy exposome. Current Environmental Health Reports. 2015;2:204–213. doi: 10.1007/s40572-015-0043-2. [DOI] [PubMed] [Google Scholar]
- 4.De Felice A, Ricceri L, Venerosi A, Chiarotti F, Calamandrei G. Multifactorial origin of neurodevelopmental disorders: approaches to understanding complex etiologies. Toxics. 2015;3:89–129. doi: 10.3390/toxics3010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wild CP. The exposome: From concept to utility. International Journal of Epidemiology. 2012;41:24–32. doi: 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- 6.McDonnell LA, Heeren RM. Imaging mass spectrometry. Mass Spectrometry Reviews. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 7.Hare DJ, New EJ, de Jonge MD, McColl G. Imaging metals in biology: Balancing sensitivity, selectivity and spatial resolution. Chemical Society Reviews. 2015;44:5941–58. doi: 10.1039/c5cs00055f. [DOI] [PubMed] [Google Scholar]
- 8.Pozebon D, Scheffler GL, Dressler VL, Nunes MAG. Review of the applications of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) to the analysis of biological samples. Journal of Analytical Atomic Spectrometry. 2014;29:2204–2228. [Google Scholar]
- 9.Austin C, Niedzwiecki M, Arora M. Multielemental bioimaging of tissues in children’s environmental health research. Current Opinion in Pediatrics. 2016 doi: 10.1097/MOP.0000000000000328. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hare DJ, Lei P, Ayton S, Roberts BR, Grimm R, George JL, et al. An iron–dopamine index predicts risk of parkinsonian neurodegeneration in the substantia nigra pars compacta. Chemical Science. 2014;5:2160. [Google Scholar]
- 11.Hijmans RJ. Raster: Geographic data analysis and modeling. R package version 2.3–24 2015 [Google Scholar]
- 12.Chen TW, Lin BJ, Brunner E, Schild D. In situ background estimation in quantitative fluorescence imaging. Biophysical Journal. 2006;90(7):2534–4. doi: 10.1529/biophysj.105.070854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIntire DD, Bloom SL, Casey BM, Leveno KJ. Birth weight in relation to morbidity and mortality among newborn infants. New England Journal of Medicine. 1999;340:1234–1238. doi: 10.1056/NEJM199904223401603. [DOI] [PubMed] [Google Scholar]
- 14.McCormick MC. The contribution of low birth weight to infant mortality and childhood morbidity. The New England Journal of Medicine. 1985;312:82–90. doi: 10.1056/NEJM198501103120204. [DOI] [PubMed] [Google Scholar]
- 15.Mwanyumba F, Inion I, Gaillard P, Mandaliya K, Praet M, Temmerman M. Placental inflammation and perinatal outcome. European Journal of Obstetrics, Gynecology, and Reproductive Biology. 2003;108:164–170. doi: 10.1016/s0301-2115(02)00438-4. [DOI] [PubMed] [Google Scholar]
- 16.QGIS Team. QGIS geographic information system. Open Source Geospatial Foundation Project 2015 [Google Scholar]
- 17.Mairesse J, Lesage J, Breton C, Bréant B, Hahn T, Darnaudéry M, et al. Maternal stress alters endocrine function of the feto-placental unit in rats. American Journal of Physiology Endocrinology and Metabolism. 2007;292:E1526–33. doi: 10.1152/ajpendo.00574.2006. [DOI] [PubMed] [Google Scholar]
- 18.Prouillac C, Lecoeur S. The role of the placenta in fetal exposure to xenobiotics: Importance of membrane transporters and human models for transfer studies. Drug Metabolism and Disposition. 2010;38:1623–1635. doi: 10.1124/dmd.110.033571. [DOI] [PubMed] [Google Scholar]
- 19.Guttmacher AE, Maddox YT, Spong CY. The human placenta project: Placental structure, development, and function in real time. Placenta. 2014;35:303–304. doi: 10.1016/j.placenta.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser J. Gearing up for a closer look at the human placenta. Science. 2014;344:1073. doi: 10.1126/science.344.6188.1073. [DOI] [PubMed] [Google Scholar]
- 21.Esteban-Vasallo MD, Aragones N, Pollan M, Lopez-Abente G, Perez-Gomez B. Mercury, cadmium, and lead levels in human placenta: A systematic review. Environmental Health Perspectives. 2012;120:1369–1377. doi: 10.1289/ehp.1204952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Current Opinion in Chemical Biology. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibs K-H, Rink L. Zinc-altered immune function. The Journal of Nutrition. 2003;133:1452S–1456S. doi: 10.1093/jn/133.5.1452S. [DOI] [PubMed] [Google Scholar]
- 24.Hackett MJ, McQuillan JA, El-Assaad F, Aitken JB, Levina A, Cohen DD, et al. Chemical alterations to murine brain tissue induced by formalin fixation: Implications for biospectroscopic imaging and mapping studies of disease pathogenesis. The Analyst. 2011;136:2941–2952. doi: 10.1039/c0an00269k. [DOI] [PubMed] [Google Scholar]
- 25.Sarafanov AG, Todorov TI, Kajdacsy-Balla A, Gray MA, Macias V, Centeno JA. Analysis of iron, zinc, selenium and cadmium in paraffin-embedded prostate tissue specimens using inductively coupled plasma mass-spectrometry. Journal of Trace Elements in Medicine and Biology: Organ of the Society for Minerals and Trace Elements (GMS) 2008;22:305–314. doi: 10.1016/j.jtemb.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Punshon T, Chen S, Finney L, Howard L, Jackson BP, Karagas MR, Ornvold K. High-resolution elemental mapping of human placental chorionic villi using synchrotron X-ray fluorescence spectroscopy. Analytical and Bioanalytical Chemistry. 2015;407(22):6839–50. doi: 10.1007/s00216-015-8861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lear J, Hare D, Adlard P, Finkelstein D, Doble P. Improving acquisition times of elemental bio-imaging for quadrupole-based LA-ICP-MS. Journal of Analytical Atomic Spectrometry. 2012a;27:159–164. [Google Scholar]
- 28.Lear J, Hare DJ, Fryer F, Adlard PA, Finkelstein DI, Doble PA. High-resolution elemental bioimaging of Ca, Mn, Fe, Co, Cu, and Zn employing LA-ICP-MS and hydrogen reaction gas. Analytical Chemistry. 2012b;84:6707–6714. doi: 10.1021/ac301156f. [DOI] [PubMed] [Google Scholar]
- 29.Craig C-A, Jarvis KE, Clarke LJ. An assessment of calibration strategies for the quantitative and semi-quantitative analysis of calcium carbonate matrices by laser ablation-inductively coupled plasma-mass spectrometry (LA-ICP-MS) Journal of Analytical Atomic Spectrometry. 2000;15:1001–1008. [Google Scholar]
- 30.R Team. Rstudio: Integrated development for R. Boston, MA: RStudio, Inc; 2015. [Google Scholar]
- 31.Otsu N. A threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man, and Cybernetics. 1979;9:62–66. [Google Scholar]
- 32.Silverman BW. Monographs on Statistics and Applied Probability. London: Chapman and Hall; 1986. Density estimation for statistics and data analysis. [Google Scholar]
- 33.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Transactions on Image Processing. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 34.Tsai W-H. Moment-preserving thresolding: A new approach. Computer Vision, Graphics, and Image Processing. 1985;29:377–393. [Google Scholar]
- 35.Getis A, Ord JK. The analysis of spatial association by use of distance statistics. Geographical Analysis. 1992;24:189–206. [Google Scholar]
- 36.Naimi B. R package version 1.1–12. 2013. Usdm: Uncertainty analysis for species distribution models. [Google Scholar]
- 37.Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. Journal of Microscopy. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 38.Nawaz S, Heindl A, Koelble K, Yuan Y. Beyond immune density: Critical role of spatial heterogeneity in estrogen receptor-negative breast cancer. Mod Pathol. 2015;28:766–777. doi: 10.1038/modpathol.2015.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.