Abstract
The 1,4-naphthoquinones (1,4-NQs) are a diverse group of natural products found in every kingdom of life. Plants, including many horticultural species, collectively synthesize hundreds of specialized 1,4-NQs with ecological roles in plant–plant (allelopathy), plant–insect and plant–microbe interactions. Numerous horticultural plants producing 1,4-NQs have also served as sources of traditional medicines for hundreds of years. As a result, horticultural species have been at the forefront of many basic studies conducted to understand the metabolism and function of specialized plant 1,4-NQs. Several 1,4-NQ natural products derived from horticultural plants have also emerged as promising scaffolds for developing new drugs. In this review, the current understanding of the core metabolic pathways leading to plant 1,4-NQs is provided with additional emphasis on downstream natural products originating from horticultural species. An overview on the biochemical mechanisms of action, both from an ecological and pharmacological perspective, of 1,4-NQs derived from horticultural plants is also provided. In addition, future directions for improving basic knowledge about plant 1,4-NQ metabolism are discussed.
Introduction
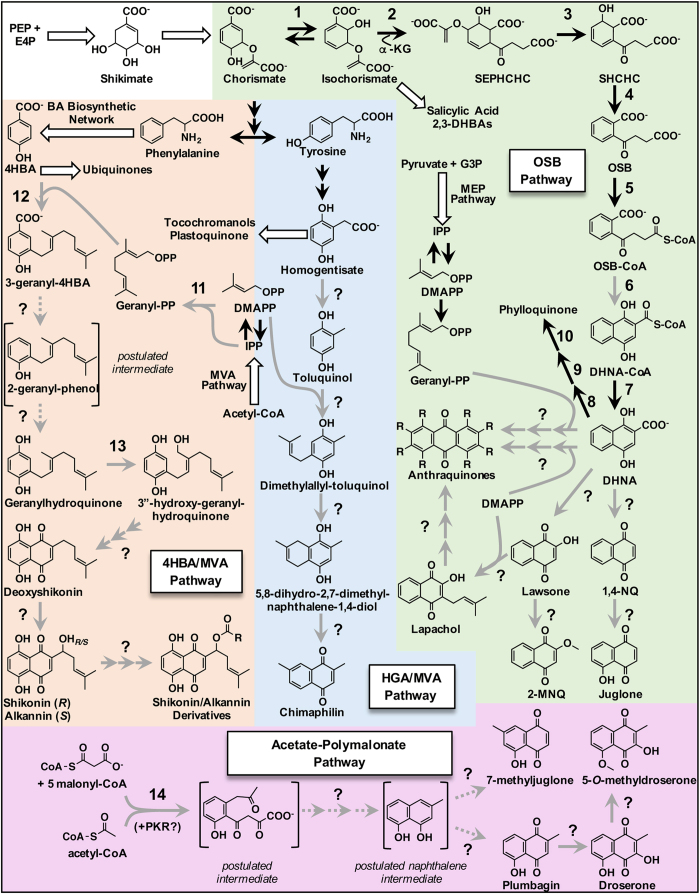
The 1,4-naphthoquinones (1,4-NQs) are redox active compounds structurally related to naphthalene that are comprised of a benzene moiety (ring A) linearly fused with a fully conjugated cyclic diketone (ring B) in which the carbonyl groups are arranged in the para orientation (Figure 1a). In living organisms, 1,4-NQs encompass a class of natural products containing a 1,4-naphthalenoid ring, often bearing one or more methyl, hydroxyl and/or methoxy substitutions, and, in some molecules, a liposoluble side chain.
Figure 1.
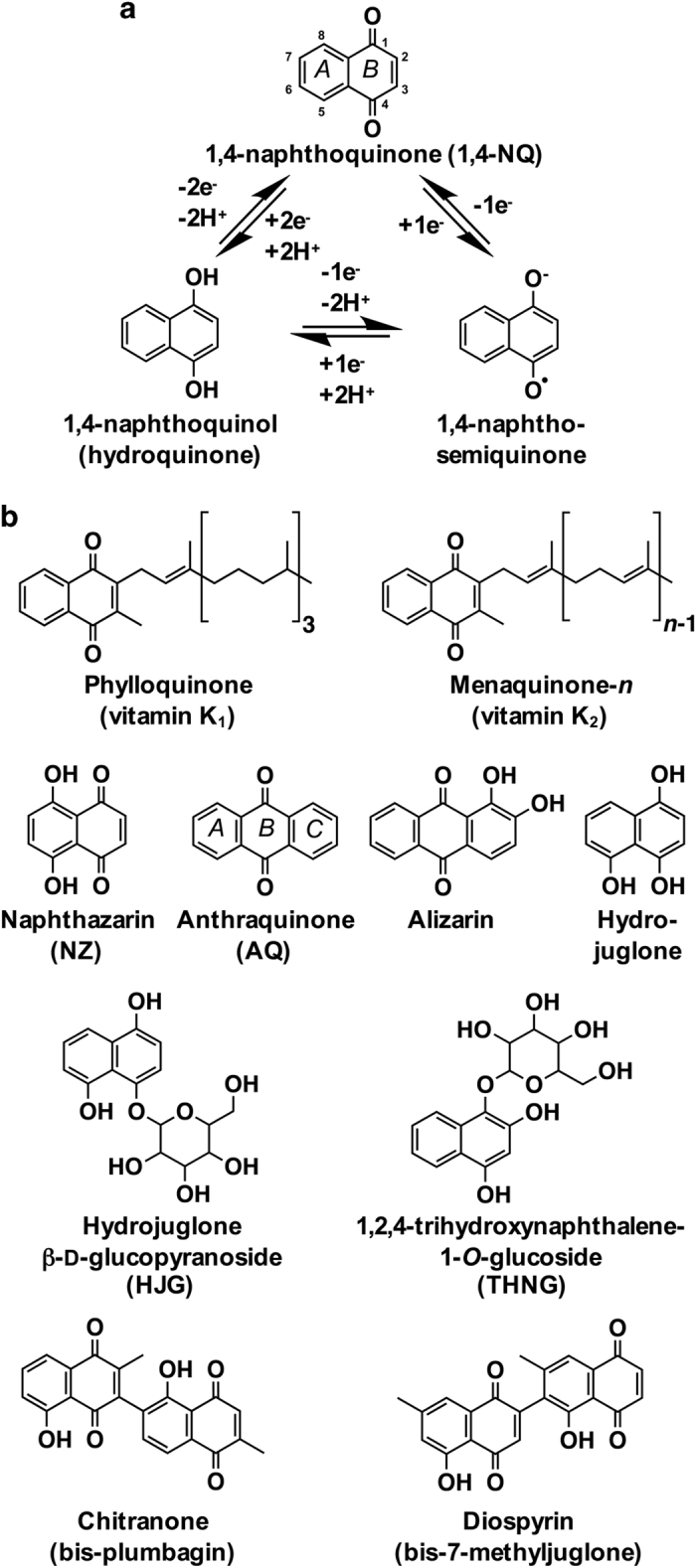
(a) Basic structure and redox forms of 1,4-NQs and (b) examples of 1,4-NQ natural products referenced in the text.
The 1,4-NQs are synthesized by organisms throughout all kingdoms of life (described below) and are involved in vital metabolic processes and/or contribute toward adaptation to ecological niches. Filamentous fungi synthesize dozens of 1,4-NQ-based compounds,1 some of which are reported to be responsible for coloring of sexual fruiting bodies and thought to confer protection against ultraviolet, desiccation and insects.2 Although restricted to only a handful of lineages, several animals also produce 1,4-NQs, such as those found in secretions of a few tenebrionid beetles3 and in the scent-producing glands of certain arachnids.4 Moreover, the sea urchin, Strongylocentrotus purpuratus, is reported to make a red-colored 1,4-NQ called echinochrome in its pigment-producing cells.5,6 Within bacteria, the Actinomycetes produce numerous 1,4-NQs,7 as well as substituted 5,8-dihydroxy-1,4-NQs called naphthazarins (NZs; Figure 1b) that form core moieties in the antimicrobial rubromycins.8 Many extant archaea and bacteria have retained the ability to synthesize menaquinone (vitamin K2; Figure 1b), a prenylated 1,4-NQ suggested to be the ancestral quinone involved in anaerobic respiratory electron transport chains.9 In some cyanobacteria, rhodophytes (red algae)10 and most diatoms (protists),11 menaquinone fulfills the role of phylloquinone (vitamin K1; Figure 1b), which is the 1,4-NQ involved in photosynthesis in plants,12 green algae,13 many cyanobacteria9 and some euglenoids (for example, Euglena gracilis14).
Perhaps the greatest diversity of 1,4-NQs is found amongst the specialized natural products synthesized by plants, particularly those by horticultural species (see refs 7,15,16,17,18,19,20 for further information on the occurrence of plant 1,4-NQs). Collectively, using several different metabolic pathways, plants produce hundreds of specialized 1,4-NQs, NZs and derived metabolites, including certain anthraquinones (AQs; Figure 1b). Together, these natural products possess a multitude of biochemical properties modulating numerous ecological and pharmacological roles, offering new targets for addressing challenges in modern horticulture and providing scaffolds for developing novel drugs.
This review summarizes the current knowledge on the different plant biosynthetic pathways involved in forming simple 1,4-naphthalenoid rings and on the metabolism of downstream 1,4-NQs derived from horticultural species. Advances made in uncovering the molecular mechanisms of action, ecological functions and pharmacological activities of select specialized horticultural plant 1,4-NQs are also highlighted. Table 1 summarizes the horticultural species and 1,4-NQ natural products covered in this review. As the production of some 1,4-NQ natural products involves intermediates shared in phylloquinone biosynthesis, relevant discoveries that have improved the understanding of this primary metabolic pathway in Arabidopsis thaliana will also be described. However, more comprehensive reviews on this pathway have recently become available,21,22 as have reviews concerning the metabolism of precursors for each of the 1,4-NQ biosynthetic pathways (for example, for the shikimate pathway,23 benzoic acids,24 isoprenoids25 and polyketides26). Finally, this report will cover future directions for addressing gaps still remaining in understanding specialized plant 1,4-NQ metabolism.
Table 1. Major 1,4-NQ natural products produced by horticultural species highlighted in this review.
| Common name | Scientific name | Major 1,4-NQ natural product(s) present |
|---|---|---|
|
Medicinal or ethnobotanical
| ||
| Henna | Lawsonia inermis | Lawsone |
| Pau d’arco tree | Tabebuia impetiginosa | Lapachol |
| Madder | Rubia tinctorum | Alizarin |
| Purple gromwell (Zi cao) Arnebia | Lithospermum erythrorhizon Arnebia euchroma | Shikonins |
| Alkanet Arizona popcorn flower | Alkanna tinctoria Plagiobothrys arizonicus | Alkannins |
| Pipsissewa One-flowered wintergreen | Chimaphila umbellate Moneses uniflora | Chimaphilins |
| Indian leadwort | Plumbago indica | Plumbagin |
|
Ornamental
| ||
| Garden balsam | Impatiens balsamina | Lawsone |
| Himalayan balsam | Impatiens glandulifera | Lawsone, 2-MNQ |
| Venus fly trap | Dionaea muscipula | Plumbagin, droserone |
| Pitcher plants | Nepenthes sp. | Plumbagin, droserone, 7-Methyljuglone |
|
Nuts and seeds
| ||
| Black walnut English walnut | Juglans nigra Juglans regia | Juglone |
| Pecan | Carya illinoensis | Juglone |
| Sesame | Sesamum indicum | Anthrasesamones |
Abbreviations: 1,4-NQ, 1,4-naphthoquinone; 2-MNQ, 2-methoxy-1,4-NQ.
Plants have evolved several pathways to synthesize 1,4-naphthalenoid rings
In nature, 1,4-NQs are known to be derived from several metabolic pathways: the o-succinylbenzoate (OSB; Figure 2) pathway; the 4-hydroxybenzoic acid (4HBA; Figure 2)/geranyl diphosphate (GPP; Figure 2) pathway; the acetate-polymalonate pathway; the homogentisate (HGA; Figure 2)/mevalonic acid (MVA) pathway; and the futalosine pathway. Except for the futalosine pathway, which was recently discovered to be an alternative route toward menaquinone in some bacteria,27 all of these pathways are present in the plant kingdom. In the OSB, 4HBA/MVA and HGA/MVA pathways, chorismate, the product of the shikimate pathway,23 ultimately provides one of the rings in the core 1,4-naphthalenoid structure, although the chorismate product from which each pathway starts is different (Figure 2). The precursor for the second ring is another feature that differentiates these three pathways (Figure 2). Finally, specialized plant 1,4-NQs synthesized via the acetate-polymalonate pathway, as the name implies, are derived from the condensation of acetyl-CoA with multiple malonyl-CoA molecules (Figure 2).
Figure 2.
The plant 1,4-NQ biosynthetic network. Presented is the current understanding of the enzymes and intermediates involved in the core metabolic pathways for synthesizing 1,4-naphthalenoid rings in plants and for producing some of the major horticultural 1,4-NQs. Subcellular architecture is not depicted but is discussed in the text. Black arrows indicate the existence of genetic evidence to support biosynthetic reactions, while gray arrows signify a lack of genetic evidence. Tandem triplicate arrows indicate an unknown number of multiple steps to go from a given intermediate to the next metabolite. Dotted arrows are used to represent steps to and from postulated intermediates. White block arrows represent entire metabolic pathways to relevant or noteworthy metabolites not addressed in this review. Question marks next to arrows indicate that enzymatic activities for those steps have not been described. Numbers next to arrows represent characterized enzymes or detected enzymatic activities: 1, isochorismate synthase; 2, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-2-carboxylate (SEPHCHC) synthase; 3, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-2-carboxylate (SHCHC) synthase; 4, o-succinylbenzoate (OSB) synthase; 5, OSB-CoA ligase; 6, Dihydroxynaphthoyl-CoA (DHNA-CoA) synthase; 7, DHNA-CoA thioesterase; 8, Dihydroxynaphthoic acid (DHNA) phytyl transferase; 9, NAD(P)H dehydrogenase C1 (NDC1); 10, Demethylphylloquinone methyltransferase; 11, cytosolic geranyl diphosphate synthase (GPPS); 12, p-hydroxybenzoate:geranyltransferase (PGT); 13, geranylhydroquinone (GHQ) 3″-hydroxylase; 14, polyketide synthase (PKS). BA, benzoic acid; DHBA, dihydroxybenzoic acid; DMAPP; dimethylallyl diphosphate; E4P, D-erythrose 4-phosphate; G3P, glyceraldehyde 3-phosphate; IPP, isopentenyl diphosphate; MEP, methylerythritol 4-phosphate; MVA, mevalonic acid; PEP, phosphoenolpyruvate; PKR, polyketide reductase; PP, diphosphate.
The OSB pathway
The OSB pathway consists of a core set of seven reactions that convert chorismate to 1,4-dihydroxy-2-naphthoate (DHNA; Figure 2), which supplies the 1,4-naphthalenoid ring for menaquinone in most bacteria and for phylloquinone in all plants. In some plants, DHNA is also the precursor for specialized 1,4-NQs, such as lawsone (2-hydroxy-1,4-NQ; Figure 2) and juglone (5-hydroxy-1,4-NQ; Figure 2). The first indication for the existence of the OSB pathway came in the 1960s when it was shown that [U-14C]-shikimate fed to Escherichia coli and to etiolated maize shoots labeled menaquinone28 and phylloquinone,29 respectively. Experiments demonstrating that labeling from [U-14C]-shikimate could also be retrieved in the benzene moiety (ring A) of lawsone30,31 and juglone32 soon followed. First evidence for the origin of the quinone moiety (ring B) in OSB-derived 1,4-NQs came from tracer studies in Impatiens balsamina (Garden balsam) showing that [2-14C]-glutamate33 and [U-14C]-α-ketoglutarate34 labeled lawsone in a specific pattern. Extension of this finding led to further investigations establishing that OSB is an intermediate and that DHNA is the product from which the OSB pathway branches toward production of various 1,4-NQs.35–39
Nearly all the plant OSB pathway genes have been identified and functionally characterized from biochemical and genetic studies investigating phylloquinone biosynthesis in Arabidopsis.40–44 The OSB route begins with the isomerization of chorismate to isochorismate by isochorismate synthase41,42 (ICS; reaction 1, Figure 2), an enzyme that is shared with plant pathways for salicylic acid40,41,45 and 2,3-dihydroxybenozic acid46,47 biosynthesis. Isochorismate is then the substrate for PHYLLO,42 a trifunctional enzyme with 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-2-carboxylate (SEPHCHC) synthase, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-2-carboxylate (SHCHC) synthase and OSB synthase domains (reactions 2–4, Figure 2). On the basis of biochemical characterization of their bacterial orthologs, these enzymes, respectively, are known to sequentially catalyze the addition of α-ketoglutarate48 (ultimately providing the succinyl side chain of OSB) to form SEPHCHC, the 2,5-elimination of the pyruvyl side chain to form SHCHC,49 and dehydration to produce OSB,50,51 the aromatized ring of which serves as the benzene moiety (ring A) in the eventual 1,4-naphthalenoid structure of DHNA (Figure 2). Next, the succinyl side chain of OSB is activated to its corresponding CoA-ester by OSB-CoA ligase (reaction 5, Figure 2)43,52 and cyclized by DHNA-CoA synthase (reaction 6, Figure 2)53 (formerly misnamed as DHNA synthase) to produce the quinone moiety (ring B) in the resulting product, DHNA-CoA (Figure 2). Although no plant DHNA-CoA synthase has been functionally characterized, a predicted ortholog of the E. coli DHNA-CoA synthase gene, menB, is present in Arabidopsis and co-expresses with other known phylloquinone biosynthetic genes.43 Phylogenetic reconstruction has revealed that plant DHNA-CoA synthases belong to the type I class, which rely on bound bicarbonate as the catalytic base,54 suggesting the OSB pathway may be regulated by cellular bicarbonate levels. The final step of the core OSB pathway is hydrolysis of DHNA-CoA to DHNA (Figure 2), a reaction that was previously assigned to DHNA-CoA synthase, then to SHCHC synthase and finally thought to occur spontaneously.55,56 Only recently was it demonstrated that cyanobacteria,57 E. coli58 and plants44 contain thioesterases catalyzing the hydrolysis of DHNA-CoA to DHNA (reaction 7, Figure 2). Once formed, DHNA is then used to synthesize phylloquinone in all plants. First, DHNA is phytylated by DHNA phytyl transferase (reaction 8, Figure 2),59 a reaction that is accompanied by decarboxylation and spontaneous oxidation of the 1,4-naphthalenoid ring.60 A demethylnaphthoquinone oxidoreductase then reduces the resulting demethylphylloquinone product to demethylphylloquinol (reaction 9, Figure 2),61 which is promptly transmethylated by demethylphylloquinone methyltransferase to phylloquinol (reaction 10, Figure 2),61,62 the reduced form of phylloquinone.
One interesting, yet poorly understood, aspect of the plant OSB pathway is its split between plastids and peroxisomes. Fluorescent protein fusion experiments revealed that the conversion of chorismate to OSB by ICS and PHYLLO occurs in plastids.41,42 The site(s) of OSB-CoA formation, however, remains enigmatic as fluorescent protein fusion experiments showed OSB-CoA ligase is dual localized in plastids43 and peroxisomes.63 Peptide fragments of the spinach MenB ortholog, a priori catalyzing the formation of DHNA-CoA from OSB-CoA, were retrieved in proteomes obtained from leaf peroxisomes, and the Arabidopsis ortholog was demonstrated through fluorescent protein fusion experiments to localize to peroxisomes.63 In Arabidopsis, DHNA-CoA thioesterase activity was detected in purified peroxisomes, and found to be absent in plastids.44 Moreover, based on fluorescent protein fusion experiments and proteomics evidence, the cognate enzymes were established to localize to peroxisomes.44,64 Together, these data suggest that OSB and/or OSB-CoA is exported from plastids and converted to DHNA in peroxisomes. The final three enzymes in phylloquinone biosynthesis are localized in plastids,59,61,62 definitively indicating that DHNA must be transported from peroxisomes to plastids. It is also likely that DHNA is needed in plastids to synthesize AQs derived from the OSB pathway (Figure 1). Labeling studies with Rubia tinctorum65 and Cinchona ‘Robusta’66 cell cultures showed that the methylerythritol 4-phosphate (MEP) pathway, which is localized in plastids, is overwhelmingly the dominant source of isopentenyl diphosphate/dimethylallyl diphosphate (DMAPP) used to synthesize ring C of their respective AQs (Figure 2). Similarly, labeling patterns retrieved in the anthrasesamone type AQs produced by sesame (Sesamum indicum) hairy root cultures fed with [1-13C]-glucose revealed DHNA from the OSB pathway and GPP produced by the MEP pathway as the sources of rings A and B, and ring C, respectively.67 These studies are in agreement with those performed by Leistner showing that [2-14C]- and [5-14C]-mevalonic acid are negligibly incorporated into ring C of alizarin (Figure 1b), a red pigment produced in roots of madder (R. tinctorum).68,69 It still remains an open question if there are additional subcellular destinations for DHNA in plants as, as described below, none of the specialized 1,4-NQ biosynthetic enzymes downstream of DHNA have been identified.
In many members of the Juglandaceae, including black walnut (Juglans nigra) and English walnut (Juglans regia), DHNA is an intermediate in the synthesis of juglone and several other related 1,4-NQs.70 Chemical degradation of juglone isolated from J. regia leaves fed with radiolabeled precursor revealed that the carboxyl group of shikimate is equally distributed between the keto groups (C1 and C4) in the quinone moiety of juglone, leading to the hypothesis that a symmetrical intermediate like 1,4-naphthoquinone (Figure 2) must be an intermediate in the pathway.32 Indeed, 1,4-naphthoquinone was found to be present in J. regia leaves and to be labeled by radiolabeled OSB.39,71 This suggests the existence of an enzyme that decarboxylates DHNA to 1,4-naphthoquinone. The subsequent conversion of 1,4-naphthoquinone to juglone is likely to be carried out by a hydroxylase, perhaps belonging to the cytochrome P45072 or 2-oxoglutarate/Fe(II)-dependent dioxygenase (2-ODD)73 families.
Phenolic compounds are often glycosylated to increase their solubility and stability, to aide in transport and sequestration, and to render the compounds physiologically inactive in plants.74 It should come as no surprise then that in several species, such as black and English walnut,75 J. major, J. microcarpa76 and a number of pecan (Carya illinoensis) cultivars,77,78 juglone has been found to accumulate in many tissues in its glycosylated form, hydrojuglone glucoside (HJG; 1,5-dihydroxy-4-naphthalenyl-β-D-glucopyranoside; Figure 1b). This modification may allow juglone to be stored in large quantities and to reduce the potential for autotoxicity. In English walnut leaves, glycosyltransferase activity with a benzoquinone substrate has been detected,76 though the responsible enzyme is unknown and activity with HJG has yet to be demonstrated. Moreover, glycosylation depends on the quinone substrate being in its reduced form,76 thus implicating the existence of an oxidoreductase capable of first reducing juglone to hydrojuglone (1,4,5-trihydroxynaphthalene; Figure 1b). Although there have been no reports on the presence of such an enzyme in Juglans species, a quinone oxidoreductase that uses NADH or NADPH as electron donors to reduce a variety of quinones, including juglone, has been identified in roots of the parasitic plant Triphysaria versicolor.79 Just as reduction followed by glycosylation inactivates juglone, deglycosylation of HJG by an unknown β-glucosidase purified from English walnut husks releases hydrojuglone aglycone, which then spontaneously oxidizes to generate active juglone.80
Another major specialized 1,4-NQ derived from DHNA is lawsone, the molecule responsible for the reddish-orange dyestuff extracted from Henna (Lawsonia inermis) leaves. Lawsone is also found in the flowering aerial parts and roots of several Impatiens species.81–83 Similar to juglone, lawsone is a simple hydroxylated 1,4-NQ, though its biosynthesis is quite different. This idea first emerged after feeding studies with I. balsamina showed that labeled shikimate incorporated into juglone and lawsone in different patterns.31,32 Later, using the same model species, Chung et al.71 demonstrated with stable-isotopically labeled [1-13C]-OSB that the C1 keto group of lawsone was substantially more highly labeled compared with the C4 keto group, indicating that OSB is asymmetrically incorporated into lawsone.71 Therefore, in contrast to juglone, the biosynthesis of lawsone does not proceed through a symmetrical 1,4-naphthoquinone intermediate and is instead likely formed via oxidative decarboxylation of DHNA by an unknown enzyme (Figure 2). The glucosylated form of reduced lawsone, 1,2,4-trihydroxynaphthalene-1-O-glucoside (Figure 1b), has been reported in Impatiens glandulifera (Himalayan balsam), thus pointing to the presence of an oxidoreductase and a glycosyltransferase analogous to those involved in metabolizing juglone.
Lawsone is also precursor to other 1,4-NQ natural products. An allelopathic methylated lawsone derivative, 2-methoxy-1,4-NQ (2-MNQ; Figure 2), is found in many tissues of several Impatiens species,81,83–85 and is almost certainly formed via an S-adenosylmethionine-dependent O-methyltransferase.86 In several native Central and South American trees, including the Pau d’arco tree (Red Lapacho; Tabebuia impetiginosa, syn. Tabebuia avellanedae)87 and Tabebuia guayacan,15 lawsone is proposed to provide the hydroxylated naphthalenoid structure of the prenylated 1,4-NQ lapachol (Figure 2).16 The identity of the responsible prenyltransferase is unknown and it is still unclear if the DMAPP moiety of lapachol is predominantly derived from the MEP or MVA pathway. Lapachol is also thought to be a precursor for other 1,4-NQ, 1,2-NQ (for example, β-lapachone) and AQ derivatives (Figure 2) contributing to the resistance of Tabebuia trees to marine borers88,89 and to their wide range of medicinal properties.16,87
The 4HBA/MVA pathway
Many boraginaceous species utilize the 4HBA/MVA pathway to synthesize a subclass of 1,4-NQs called isohexenylnaphthazarins (IHNs). These compounds are comprised of a NZ ring (Figure 1b) conjugated with a lipophilic side chain on the quinone moiety. The IHNs encompass the red-pigmented compounds shikonin, alkannin and at least 40 other acylated derivatives (Figure 2) synthesized in roots of medicinal species like Lithospermum erythrorhizon and Alkanna tinctoria.90 Early tracer experiments demonstrated that phenylalanine (via cinnamic acid and 4HBA) and mevalonic acid are precursors for the benzene and quinone rings, respectively, of alkannin produced in Plagiobothrys arizonicus.91 This finding, in combination with isolation of 3-geranyl-4HBA and geranylhydroquinone from cell cultures of L. erythrorhizon,92,93 led to the hypothesis that alkannin and shikonin are likely synthesized via a pathway analogous to ubiquinone biosynthesis with subsequent ring closure reactions (Figure 2).
Biosynthesis of benzoic acids from phenylalanine in plants involves a complex network of metabolic routes branching off the core phenylpropanoid pathway.24 Administration of the phenylalanine-ammonia lyase (PAL) inhibitor aminoindan-2-phosphonic acid to L. erythrorhizon cell cultures effectively blocked shikonin formation.94 Yazaki et al.95 demonstrated that the enzymatic formation of 4HBA from 4-coumaric acid (4CA) in L. erythrorhizon cell cultures partially proceeds through the ‘non-oxidative route’ based on the presence of a 4-hydroxybenzaldehyde intermediate, dependence on NAD and the lack of an ATP or CoA requirement.95 It was later discovered that L. erythrorhizon cell cultures are also capable of converting 4CA-CoA to 4HBA via the ‘β-oxidative route’,96 which was recently shown to provide the 4HBA ring for ubiquinone biosynthesis in Arabidopsis.97 Genes encoding the core phenylpropanoid pathway enzymes PAL, cinnamic acid 4-hydroxylase (C4H) and 4CA-CoA ligase (4CL) have been cloned and studied from L. erythrorhizon98,99 and Arnebia euchroma,100 but those involved in the ‘non-oxidative’ and ‘β-oxidative’ routes have not. Benzoic acid ‘β-oxidative route’ genes have been identified in other species, while all but one of the ‘non-oxidative route’ genes remain unknown across all plants.24 Once synthesized, 4HBA can be glucosylated by a cytosolic glucosyltransferase and stored in the vacuole until released to its free form by a cytosolic β-glucosidase upon stimulation of shikonin production.101
In plants, GPP is predominantly synthesized from isopentenyl diphosphate and DMAPP derived from the MEP pathway using a GPP synthase (GPPS) localized in plastids.25 It is therefore noteworthy that the GPP precursor ultimately providing ring B of shikonin- and alkannin-type 1,4-NQs was shown to be derived from the MVA pathway (based on labeling91 and inhibitor studies94,100) and to originate via the only known cytosolically localized GPPS (reaction 11, Figure 2).102,103 Expression of multiple MVA pathway genes in A. euchroma,100 and the activity and cognate gene expression of the key MVA pathway enzyme 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) in L. erythrorhizon,104 correlate with shikonin production. Generally, MVA pathway genes are more highly expressed in non-photosynthetic tissues, like roots, whereas MEP pathway genes are primarily active in green tissues.25
The committed step of shikonin and alkannin biosynthesis begins with the addition of GPP to 4HBA by a GPP:4HBA 3-geranyltransferase (p-hydroxybenzoate:geranyltransferase, PGT; reaction 12, Figure 2). Activity of this enzyme was first reported in L. erythrorhizon extracts.105 Later, PGT was shown to be localized to the endoplasmic reticulum106 and to have a high affinity for GPP (Km=18.4 μM) and 4HBA (Km=13.8 μM).107 Two cDNAs encoding PGTs with 93% identity were isolated from L. erythrorhizon cell cultures.108 Subsequent biochemical characterization of one isoform, LePGT1, revealed that the N-terminal 130 amino acids are responsible for its specificity for GPP109 and that it is inhibited by aromatic substrates with two phenolic hydroxyl groups.110
After the PGT-catalyzed reaction, very little is known about the biosynthesis of shikonin- and alkannin-type 1,4-NQs. It is likely the next steps entail decarboxylation and hydroxylation of the C1 position of the 3-geranyl-4HBA product of PGT. In bacterial ubiquinone biosynthesis, the C1 position of the polyprenylated 4HBA product is non-oxidatively decarboxylated using a prenylated flavin cofactor to produce a prenylphenol intermediate,111,112 which is hydroxylated further down the pathway.113 It is possible that 3-gernanyl-4HBA is decarboxylated and hydroxylated in a similar fashion by discrete enzymes to produce geranylhydroquinone (GHQ), which has been detected in planta,92,93 via a 2-geranyl-phenol intermediate (Figure 2). Alternatively, 3-geranyl-4HBA may be directly converted to GHQ by oxidative decarboxylation. The next reaction in the pathway is hydroxylation of the GHQ isoprenoid side chain to produce 3″-hydroxy-GHQ by a GHQ 3″-hydroxylase (reaction 13, Figure 2). A GHQ 3″-hydroxylase was partially purified from the microsomal fraction of L. erythrorhizon cell cultures and shown to require NADPH and molecular oxygen as cofactors, suggesting it is a cytochrome P450-dependent monooxygenase.114 Moreover, the purified GHQ 3″-hydroxylase was found to have a Km for GHQ of 1.5 μM and to be inhibited by the shikonin derivative acetylshikonin at a concentration as low as 10 μM.114 It has been proposed that cyclization of 3″-hydroxy-GHQ to form the quinone moiety of the 1,4-NQ skeleton occurs via oxidation of the C3″ position to generate an aldehyde capable of forming the aromatic nucleus via an electrophilic reaction.114 As deoxyshikonin has been detected in shikonin-producing species,115,116 it is probable that it is the final intermediate in the pathway and is converted to shikonin (or alkannin in alkannin-producing species) by GHQ 3″-hydroxylase or a similar enzyme. To date no enzymes downstream of GHQ have been identified.
Production of shikonin and its derivatives is influenced by many external factors, as has been previously summarized,117–120 and is in large part modulated by transcriptional regulation of metabolic genes.100,121,122 Biosynthesis was also shown to be controlled by auxin,123 methyl jasmonate124 and ethylene.125–127 Overexpression of the L. erythrorhizon MYB1 (LeMYB1) transcription factor gene, an ortholog of the Nicotiana tabacum MYB involved in regulating phenylpropanoid metabolism,128 led to increased expression of PAL, HMGR and PGT.129 Non-biosynthetic regulators of shikonin production have also been identified in L. erythrorhizon, including an unknown cell wall protein, LePS-2, perhaps involved in deploying shikonin,130 and LeDI-2, a small hydrophobic dark-inducible protein of unknown function.131 Downregulation of LeDI-2 reduced the shikonin pool size, though had no affect on the expression of PAL or activity of PGT.131 In addition to increasing expression of biosynthetic genes, LeMYB1 overexpression also increased expression of the shikonin regulators LeDI-2 and LePS-2.129
The HGA/MVA pathway
The HGA/MVA pathway (also referred to as the toluhydroquinone or toluquinol pathway) is widely distributed throughout, although limited to, plants within the Pyroloideae subfamily of the Ericaceae. Very little progress has been made toward understanding the metabolism of this pathway as labeling experiments in Chimaphila umbellata (pipsissewa) established that tyrosine132 and DMAPP derived from the MVA pathway133 provide precursors for chimaphilin (2,7-dimethyl-1,4-NQ; Figure 2). One unique feature of the HGA/MVA pathway is that shikimate (via tyrosine) ultimately provides the quinone moiety of the 1,4-naphthalenoid ring, compared with the OSB and 4HBA/MVA pathways in which shikimate (via isochorismate and phenylalanine, respectively) provides the benzene moiety (Figure 2).
Bolkart and Zenk showed that the β-carbon atom of tyrosine is exclusively incorporated into the 2-methyl position of chimaphilin via homogentisate and toluquinol intermediates,132,134 and that the C7 methyl group arises from the C2 of mevalonic acid.133 It can thus be envisioned that upon decarboxylation of homogentisate, which is also precursor for tocochromanols and plastoquinone synthesized in plastids (recently reviewed135), DMAPP is attached to the toluquinol product (Figure 2). The resulting prenylated intermediate, dimethylallyl-toluquinol, would then be cyclized to 5,8-dihydro-2,7-dimethylnaphthalene-1,4-diol, which would subsequently be aromatized to produce chimaphilin (Figure 2). Support for such a biosynthetic pathway architecture comes from the isolation of toluquinol, its glucoside (homoarbutin) and the glucoside of 5,8-dihydro-2,7-dimethylnaphthalene-1,4-diol (renifolin) from Pyrola media and Pyrola incarnata.136–138 Beyond supporting the postulated pathway leading to chimaphilin, these findings raise questions about the role of glycosylation in the HGA/MVA pathway. Similarly, Moneses uniflora produces chimaphilin derivatives, including 8-chlorochimaphilin, 8-hydroxychimaphilin and 3-hydroxychimaphilin, that also appear to be subject to glycoslylation in their reduced forms.139
The acetate-polymalonate pathway
A fourth route to synthesize plant 1,4-NQs, the acetate-polymalonate pathway (also referred to as the polyketide pathway), relies on CoA-linked acetate and malonate substrates, occurs throughout at least half a dozen unrelated families,7 and is most notably responsible for the production of plumbagin (5-hydroxy-2-methyl-1,4-NQ), droserone (3,5-dihydroxy-2-methyl-1,4-NQ), 5-O-methyldroserone and 7-methyljuglone (Figure 2), as well as bis-1,4-NQs, such as chitranone and diospyrin (Figure 1b).140 Support for the acetate-polymalonate pathway surfaced from experiments conducted by Durand and Zenk showing that labeled acetate precursors were incorporated into plumbagin in young Plumbago europaea shoots141 and Drosophyllum lusitanicum leaves.142 Moreover, it was revealed that neither shikimate- nor methionine-labeled plumbagin strongly indicating that its synthesis (and by extension, the synthesis of droserone, 5-O-methyldroserone and 7-methyljuglone) does not proceed through a juglone intermediate derived from the OSB pathway (Figure 2).142
On the basis of stable-isotope feeding experiments in Triphyophyllum peltatum callus cultures, which confirmed the acetogenic origin of plumbagin and droserone, Bringmann et al. proposed the involvement of polyketide synthases (PKSs) in the acetate-polymalonate pathway.143,144 In plants, PKSs belong to the type III class, which catalyze C–C bond formation in a single active site through a series of decarboxylation, condensation and cyclization reactions using a CoA-ester substrate (for example, acetyl-CoA) and CoA-ester extenders (for example, malonyl-CoA).26 Recently, cDNAs encoding type III PKSs (reaction 14, Figure 2) from Plumbago indica roots and D. lusitanicum calluses have been isolated.145,146 Biochemical characterization of the cognate recombinant enzymes revealed that both were capable of accepting acetyl-CoA (Km=31 μM for D. lusitanicum PKS) as starter and catalyzing multiple sequential decarboxylative condensations with malonyl-CoA (Km=83 μM for D. lusitanicum PKS).145,146 However, under the tested in vitro conditions, neither formed expected naphthalene products and instead produced α-pyrones, which may have been an artifact given the absence of pyrone metabolites in the tissues from which the cDNAs were isolated.145,146 Together with the lack of other PKS candidates and the high plumbagin content in the tissues examined, it is likely the identified PKSs provide the 1,4-NQ backbone via the postulated intermediates depicted in Figure 2.145,146 The acetate-polymalonate pathway is also likely to rely on a polyketide reductase to remove the oxygen atom of the third acetate unit before the initial cyclization.146
That plumbagin, droserone and 5-O-methyl droserone have been described to co-occur in Nepenthes species147,148 suggests these compounds may be part of a linear biosynthetic pathway as depicted in Figure 2. The postulated naphthalene intermediate is likely oxidized to generate either 7-methyljuglone or plumbagin, an idea consistent with the fact that many Droseraceae species exclusively contain one or the other of these 1,4-NQs.149,150 Perhaps using enzymes belonging to the same classes describuned above for juglone, lawsone and 2-MNQ synthesis, plumbagin may be further oxidized to produce droserone, and then methylated at the C5 hydroxyl group to generate 5-O-methyldroserone (Figure 2). Similar to other aforementioned 1,4-NQs, acetate-polymalonate-derived 1,4-NQs are subject to modifications, including glycosylation.151,152 Certain types of AQs, such as emodin produced by rhubarb and related species,153,154 are also synthesized via PKSs with acetyl-CoA and malonyl-CoA precursors. However, their biosynthesis does not proceed through a 1,4-NQ like those derived from the OSB pathway, so they will not be covered in this review. Generally, AQs synthesized via PKSs contain modifications in both rings A and C, while those derived from the OSB pathway (for example, alizarin) only contain functional groups on ring C, although exceptions exist.66
Biochemical perspectives on the functions of specialized 1,4-NQs produced by horticultural species
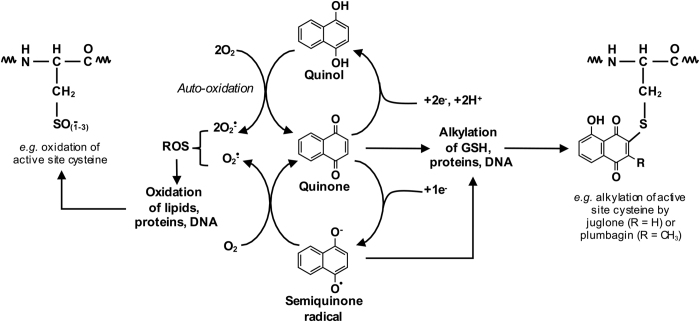
An understanding of the ecological significance of 1,4-NQ production in horticultural plants requires recognition of the biochemical properties of quinones in general and 1,4-NQs in particular. Quinones undergo redox cycling and alkylation reactions, generating reactive oxygen species (ROS) and adduct formation with proteins and DNA.155 Alkylation (also termed arylation) of reduced glutathione (GSH) or cysteine residues of proteins is particularly common, leading to depletion of GSH levels and/or protein cysteine chemical modifications.155,156 The one- or two-electon reduction of quinones to the semiquinone radical or quinol, respectively, leads to their autoxidation by molecular oxygen to the superoxide anion radical (O2−), which subsequently disproportionates into O2 and H2O2, promoting oxidation of lipids, proteins and DNA (Figure 3).155
Figure 3.
Potential mechanisms of action for 1,4-NQs. Plant 1,4-NQ redox cycling may lead to the generation of reactive oxygen species (ROS), which can oxidize certain cellular macromolecules. The quinone and/or semiquinone forms of plant 1,4-NQs can react with nucleophiles to form adducts.
In the case of 1,4-NQs, the cellular reduction of the quinone moieties in mammalian cells can be mediated by cytochrome P450 reductase, forming the semiquinone, or by NAD(P)H: quinone oxidoreductase-1 (NQO-1, DT-diaphorase), forming the quinol. Thus, both reactions may occur at the expense of NADH or NADPH. Quinols may undergo detoxification via coupling of the hydroxyl moieties to water-soluble molecules,157 such as glycosylation as described above for juglone,80 lawsone,83 7-methyljuglone and droserone.152 It is tempting to speculate that such quinol conjugates are the predominant forms in planta and afford protection against autotoxicity. However, naphthoquinone glycosides in the genus Drosera usually appear only as minor components of the total naphthoquinone pool.18 Moreover, the 1,4-NQ glucosides rossoliside and plumbaside A, isolated from Nepenthes, showed no incorporation after feeding of either [U-13C2]-sodium acetate or [U-13C3,15N]-alanine, suggesting these glycosides are storage forms with very low turnover rates.158
Depending on ring modifications, some 1,4-NQs are also good electrophiles that react with nucleophiles, including cysteine residue thiol groups in some proteins, to form adducts in cells (Figure 3).156,157 Such alkylation reactions can occur when a free C3 position is present in the 1,4-naphthalenoid structure, though the C2 substituent must also allow access and sufficient electrophilicity.157 For example, lawsone, which has a free C3 position, but a hydroxyl group at C2 (Figure 2), is considered a weak alkylating agent compared with juglone (free C2 and C3 positions; Figure 3) and plumbagin (free C3 position and methyl group at C2; Figure 3).157 The type and placement of functional groups in the benzene moiety also influences the bioreactivity and cytotoxicity of 1,4-NQs. Juglone, plumbagin, NZs and other 1,4-NQs with at least one hydroxyl group in the benzene moiety are more potent topoisomerase inhibitors compared with unsubstituted 1,4-NQs and 1,4-NQs hydroxylated on the quinone moiety (for example, lawsone).159,160
Given the aforementioned reactivity of 1,4-NQs in biological systems, it should not be surprising that 1,4-NQs have frequently been observed to induce marked perturbations of metabolism, including ROS production, thiol depletion, alkylation/arylation of numerous target proteins, DNA damage and genotoxicity in diverse biological systems. Specific examples of these effects, induced by 1,4-NQ natural products derived from horticultural species, in plants, microorganisms, insects and mammals are described below, with particular emphasis on potential alkylation/arylation target sites.
Plant–plant interactions (allelopathy)
Allelopathy is the term used to describe the harmful effect one plant exerts on another via the release of natural products into the environment.161 This is the definition that will be adopted in the remainder of this review, though it is recognized that allelopathy is also used to generally describe any direct or indirect effect, beneficial or harmful, one plant has another plant.162
The classic example of allelopathy is the release of the phytotoxic 1,4-NQ, juglone, from black walnut.163,164 Other 1,4-NQ-producing plants reported to exhibit allelopathy and notable as noxious invaders include Echium plantaginerum (Paterson’s curse; produces shikonin and its derivatives)165 and I. glandulifera (producer of lawsone and 2-MNQ).85,166 Undoubtedly, secretion of 1,4-NQs from the roots and/or leaching of 1,4-NQs from leaves and leaf litter may have contributed to the ecological success of these invading species via their phytotoxic effects on native species by the general mechanisms described above (that is, ROS production, GSH depletion and/or alkylation of proteins and DNA of neighboring plants). The best studied plant-derived phytotoxic 1,4-NQs are plumbagin and juglone, which induce ROS production in tobacco BY-2 cells, ultimately resulting in programmed cell death.167 In lettuce, juglone induces oxidative damage to the root apical meristem via ROS and a cascade of cellular changes, including decreased mitochondrial potential, chromatin condensation and DNA fragmentation.168 Juglone also triggers a large number of changes in gene transcription associated with cell growth, cell wall formation, chemical detoxification and abiotic stress responses via rapid induction of ROS in rice roots.169
Juglone’s specific cellular alkylation/arylation protein targets have been less intensively investigated in plants than in mammalian systems (see Pharmacology section below), but a number of potential targets can be postulated from the literature. Rapid irreversible growth inhibition of maize coleoptiles and inhibition of auxin-induced growth in maize coleoptile segments strongly suggests that the plasma membrane H+-ATPase may be a juglone alkylation/arylation target.170 Direct arylation of cysteine residues of jack bean urease by juglone (but not by lawsone) has been reported.171 Juglone is also a potent inhibitor of Malus domestica MdPin1, a homolog of a phosphorylation-specific peptidyl prolyl cis/trans isomerase (PPIase) in humans called Pin1 that has an important role in cell cycle regulation.172
The molecular mechanisms governing the large interspecies variability within the plant kingdom with respect to the toxic effects of juglone summarized by Willis164 are virtually unexplored. It is still unclear whether species unaffected by juglone are equipped with enzymes that facilitate detoxification and/or proteins that regulate juglone exclusion, transport and/or sequestration/compartmentation.
Plant–microbe interactions
The toxicity of plant-derived 1,4-NQs toward various bacteria, fungi and other microorganisms is widely recognized. Examples of the growth inhibitory effects that plant 1,4-NQs have on microorganisms documented to be associated with oxidative stress and/or disrupted thiol metabolism in the target organisms include lawsone on E. coli;173 juglone on Staphylococcus aureus174 and Acanthamoeba castellanii;175 plumbagin on Candida albicans and S. aureus;176 7-methyljuglone on Mycobacterium tuberculosis;177 and shikonin on Candida albicans.178
Reddy et al.179 investigated the antimicrobial effects of plumbagin in Bacillus subtilis by identifying differentially expressed proteins, and found evidence suggesting the 1,4-NQ represses the tricarboxylic acid cycle, the electron transport chain and the fatty acid synthesis; however, specific arylation targets have yet to be defined in this organism. Juglone inactivates the E. coli PPIase by covalent modification of cysteine residues.180 In yeast, juglone may not only inhibit the PPIase homolog, ESS1 but also RNA polymerase II, most likely by modification of sulfhydryl groups.181
The toxicity of plant 1,4-NQs to human pathogens (for example M. tuberculosis) is of particular interest and has obvious overlaps with the Pharmacology section below. In M. tuberculosis, 7-methyljuglone is a subversive substrate for mycothiol disulfide reductase.177 Moreover, 7-methyljuglone and its bis form, diospyrin (Figure 1b), produced by Diospyros montana,182 are potent inhibitors of DNA gyrases of M. tuberculosis, E. coli and S. aureus.183 DNA gyrase is a DNA topoisomerase that is present in bacteria and plants, but not in animals, and has been widely exploited as a target for antimicrobial chemotherapy.183 Whereas animal DNA topoisomerase of the type II (topo II) class are thought to be inhibited by formation of cysteine adducts with quinones in the N-terminal (ATPase) domain, the inhibition of DNA gyrase may be different. The naphthoquinone-binding site is within the N-terminal domain of GyrB, but as one of the enzymes examined (S. aureus gyrase) lacks Cys residues in the ATPase domain, it is unlikely that covalent adducts with Cys residues form part of the mode of binding.183 Moreover, there was no evidence of adduct formation with M. tuberculosis gyrase.183 Microarray analysis of M. tuberculosis in response to plumbagin challenge identified 103 and 171 up- and downregulated genes, respectively, but it is presently unknown whether these transcriptional responses were the sole consequence of DNA gyrase inhibition.184
Being that plants are susceptible to pathogens, it is tempting to speculate that the production of 1,4-NQs by plants may not only serve a role in allelopathy (above) but also in plant disease defense. Supporting this notion, juglone has been shown to be a potent and specific inhibitor of the growth of the fire blight pathogen, Erwinia amylovara.185 Similarly, it was reported that plumbagin is a potent growth inhibitor of a number of phytopathogenic fungi.186
Certain soil bacteria, such as Pseudomonas putida,187 are capable of degrading juglone, although little is understood about the role juglone has in shaping the soil microflora. It has been proposed that arbuscular mycorrhizal fungal hyphae may have a key role in transporting juglone in the rhizosphere.188,189 Hook et al. argue that because the production of antimicrobial 1,4-NQs is often restricted to specific root cells and elicited by soil-borne microbes, this suggests their role is in plant defense at the cellular level in the rhizosphere.20
The production of 1,4-NQs by many carnivorous plants (especially in the family Droseraceae)150 may have a key role in maintaining sterility of the digestive fluids secreted by these organisms, an idea that has been speculated for Nepenthes sp. (pitcher plants).148,190 Plumbagin secreted by the Venus fly trap (Dionaea muscipula) also has antimicrobial activity against food-related pathogenic and putrefactive bacteria.191 Perhaps more significantly, the chemical composition of the Nepenthes pitcher fluid may promote a specific microbiome dedicated to chitinolytic, proteolytic, amylolytic, and cellulolytic and xylanolytic activities for digesting pitcher-captured insects.192 Resistance mechanisms used by pitcher-associated microbes to pitcher plant 1,4-NQs remain unexplored. Clues to the molecular mechanism of bacterial plumbagin resistance have come from studies with E. coli demonstrating that two plumbagin-responsive genes ygfZ and sodA are required for counteracting toxicity.193 Furthermore, it was found that Cys228 in YgfZ is needed for the degradation of plumbagin, which may be excreted in a methylated and less-toxic form.193
Plant–insect interactions
The toxicity of plant-derived 1,4-NQs toward insects has also been documented. It appears that plumbagin, juglone and 2-MNQ may specifically target ecdysone 20-monooxygenase, an enzyme responsible for converting the molting hormone ecdysone to its more physiologically active metabolite, 20-hydroxyecdysone.194,195 Plumbagin is also an inhibitor of chitin synthetase194 and, similar to other 1,4-NQs, is an effective anti-feedant defensive agents against insects.195–197 It is also plausible that certain insects that have gained the ability to feed on 1,4-NQ-producing plants may utilize these molecules for their own defense; however, the precise mechanisms of tolerance of these organisms has been underexplored.198 Piskorski et al.199,200 propose that the larvae of the codling moth, Cydia pomonella, are able to survive on walnut trees by excreting hydrojuglone in their frass.199,200 However, this hypothesis is difficult to reconcile with the observation that hydrojuglone rapidly autoxidizes to juglone in air.80
Insectivorous plants face a dilemma in both attracting insects for pollination and as prey. The sundew, Drosera auriculata, maintains a distinct profile of volatiles emitted from flowers in comparison with their sticky traps, with plumbagin restricted to the trap, suggesting a possible role for plumbagin in attracting insect prey with volatile preferences that are distinct from insect pollinators.201 It has been suggested that plumbagin may contribute to oxidative protein modification as a predigestive mechanism in Venus fly trap.202 Plumbagin production has also been observed to be induced in response to chitin in Drosophyllum lusitanicum suspension cultures.203 In Nepenthes khasiana, chitin specifically induces production of droserone and 5-O-methyl droserone to protect pitcher fluid against microbes brought by visiting prey and perhaps to act as molecular triggers in prey capture and digestion.147
Pharmacology (plant–human interactions)
Humans have exploited plants producing 1,4-NQs for centuries as wound-healing agents. In part this may be a function of the antibacterial and antifungal properties of 1,4-NQs discussed above, preventing opportunistic wound infections. Thomson204 and Hook et al.20 have reviewed the numerous bioactive properties of plant-derived NQs and discuss their cytotoxic, anticancer, antibacterial, antifungal, anti-inflammatory and anti-parasitic activities. These and other reviews on the pharmacological properties of individual 1,4-NQ-producing species, including Henna,205–208 Plumbago seylanica209 and Venus fly trap,210 or individual 1,4-NQs of plant origin, such as the alkannins and shikonins,90,211–214 plumbagin and its analogs,215 7-methyljuglone,216 and laphachol,16 demonstrate the considerable pharmacological interest in this class of molecules in recent years.
As described above for other biological systems, plant-derived 1,4-NQs can elicit oxidative stress and disrupt thiol metabolism in mammalian cell systems, including juglone in melanoma (B16F1) tumor cells;217 juglone, shikonin and plumbagin in rat liver microsomes;218 juglone and plumbagin in HaCaT human keratinocytes;219,220 plumbagin in human prostate cancer cells;221 and shikonin in human glioma cells,222 thyroid cancer cells223 and leukemia HL-60 cells.224,225 Beyond these effects, it is also evident that plant 1,4-NQs have specific effects on downregulating central mediators of mammalian inflammation.156 Plant-derived 1,4-NQs inhibit Kelch-like enoyl-CoA-hydratase (ECH) associated protein 1 (Keap1) most likely by alkylation/arylation of key cysteine residues in the protein.156,157 In addition, it has been proposed that 1,4-NQs promote glutathionylation of Keap1.226 This may be due in part to accumulation of oxidized glutathione (GSSG),227 by GSH-mediated S-transarylation,228 and/or via upregulation of glutathione S-transferase pi, which potentiates S-glutathionylation of Keap1.229 Regardless of the precise mechanism of Keap1 cysteine modifications, it is well established that these modifications collectively alter the binding of Keap1 to transcription factor Nrf2 (NF-E2-related factor-2). Normally, Keap1 sequesters Nrf2 in the cytoplasm and bridges it to a ubiquitin ligase, cullin 3 (Cul3), to facilitate proteasomal Nrf2 degradation.157 Disruption of the Keap1–Nrf2 interaction by 1,4-NQs results in Nrf2 accumulation, leading to Nrf2 nuclear transport and induction of genes containing ‘antioxidant response elements’ (AREs).157 Consistent with this, plumbagin and shikonin strongly activate Nrf2–ARE signaling, and were found to activate genes encoding heat shock proteins.230,231 The Keap1–Nrf2–ARE pathway has now emerged as a promising target to develop drugs that upregulate expression of ARE-controlled cytoprotective oxidative stress response enzymes to treat a number of diseases and conditions.232
Nrf2 not only regulates oxidative/xenobiotic stress response but also represses inflammation by opposing transcriptional upregulation of a number of pro-inflammatory cytokine genes.233 Specific anti-inflammatory effects of plumbagin appear consistent with inhibiting the activation of the transcription factor nuclear factor-κB (NF-κB)234 in lymphocytes,235 macrophages and liver cells.236 The underlying mechanism of plumbagin and shikonin inhibition of NF-κB activation appears to entail suppression of an inhibitor of κBα (IκBα) phosphorylation and degradation, thus precluding the phosphorylation of the p65 subunit of NF-κB.237–241 However, it is still unclear whether the inhibition of NF-κB activation by plumbagin, shikonin and 1,4-naphthoquinone (see ref. 226) is solely mediated via Keap1-dependent activation of Nrf2/ARE signaling or by other mechanisms, especially considering Keap1-independent mechanisms of regulating Nrf2 have been well documented227 Furthermore, it is well established that 1,4-NQs are inhibitors of topoisomerases.242–249 Topoisomerase 1 (Top 1) inhibition is known to suppresses inflammatory genes, including tumor necrosis factor-α (TNF-α), and to protect against lethal inflammation in vivo.250 Thus, it seems plausible that anti-inflammatory effects of 1,4-NQs may also be mediated by Top 1 inhibition.
Similar to the M. domestica, yeast and E. coli PPIases described above, human Pin1 is inhibited by juglone.180,251 Pin1 is now recognized as a molecular switch for TNF-α-induced priming of the NADPH oxidase in human neutrophils,252 as a modulator of the type 1 immune response of T cells253 and as an enhancer of the oncogenic activity of the Rel proteins in the NF-κB family.254 Therefore, inhibition of Pin1 by juglone may contribute to the anti-inflammatory and anticancer actions of this molecule. Additional pharmacological mechanisms of action for plant-derived 1,4-NQs are listed in Table 2.
Table 2. Additional examples of pharmacological mechanisms of action for plant-derived 1,4-NQs.
| Plant-derived 1,4-NQ(s) | Pharmacological mechanism of action | Reference(s) |
|---|---|---|
| Shikonin | Protection of brain against ischemic stroke damage by attenuated TLR4, p-p38MAPK, NF-κB, TNF-α and MMP-9 expression, and upregulated claudin-5 expression | 255 |
| Suppression of epithelial–mesenchymal transition and downregulation of expression of Slug and MMP-2, -9 and -14 in thyroid cancer cells | 223 | |
| Management of inflammatory bowel disease by inhibiting activation of NF-κB and STAT3 | 256 | |
| Promotion of intestinal wound healing via induction of TGF-β release | 257 | |
| Inhibition of expression of the pro-inflammatory cytokine TNF-α through selective blockade of pre-mRNA splicing | 258 | |
| Inhibition of IFN-γ induced K17 overexpression by interfering with STAT3 signaling in psoriasis pathogenesis | 259 | |
| Inhibition of lipopolysaccharide-induced release of HMGB1 via IFN-β and NF-κB signaling pathways in inflammation | 260 | |
| Inhibition of STAT3-, FAK- and Src-mediated signaling in breast cancer | 261 | |
| Suppression of IL-17-induced VEGF expression via blockage of the JAK2/STAT3 pathway in psoriasis pathogenesis | 262 | |
| Inhibition of c-MYC expression with involvement of ERK/JNK/MAPK and AKT pathways in leukemia cells | 263 | |
| Suppression of orphan nuclear receptor Nr4a family gene expression for treating allergic diseases | 264 | |
| Shikonin derivatives | Inhibition of the transcriptional activation of the human TNF-α promoter in treating inflammatory diseases | 265 |
| Inhibition of tumor angiogenesis via inhibition of VEGFRs | 266 | |
| Shikonin and alkannin | Inhibition of cancer cell glycolysis via inhibition of tumor-specific PKM2 | 267 |
| Plumbagin | Decreased expression of TNF-α, IFN-γ and IL-17 in murine ulcerative colitis | 268 |
| Amelioration of autoimmune encephalomyelitis via downregulation of JAK–STAT and NF-κB signaling pathways | 269 | |
| Antiproliferative activity against lung epithelium carcinoma cells by disruption of the microtubule network through tubulin binding | 270 | |
| Inhibition of cytochrome P450s | 271,272 | |
| Inhibition of telomerase and induction of cell death in human brain tumor cells | 273 | |
| Induction of cell cycle arrest and autophagy; suppression of epithelial to mesenchymal transition involving the PI3K/Akt/mTOR-mediated pathway in human pancreatic cancer cells | 274 | |
| Induction of G2/M arrest, apoptosis, and autophagy via p38 MAPK- and PI3K/Akt/mTOR-mediated pathways in human tongue carcinoma cells | 275 | |
| Binding to and inhibition of five cancer signaling proteins (PI3Kc, AKT1/PKBa, Bcl-2, NF-κB and Stat3) | 276 | |
| Interference with the binding of ER-alpha to ERE and antagonism at the death receptor complex in BRCA1 breast cancer cells | 277 | |
| 5-O-Acyl plumbagins | Inhibition of mammalian DNA polymerase and suppression of inflammatory response | 278 |
| 7-Methyljuglone | Suppression of PI3K/Akt signaling in breast cancer cells | 279 |
| Juglone | Inactivation of cysteine-rich proteins required for progression through mitosis | 251 |
| Prevention of metabolic endotoxemia-induced hepatitis and neuroinflammation via suppression of the TLR4/NF-κB signaling pathway | 280 | |
| Juglone and plumbagin | Inhibition of protein tyrosine phosphatases, leading to increased phosphorylation and activation of epidermal growth factor receptor in HaCaT keratinocytes | 219 |
| Acetylshikonin, shikonin, juglone, lawsone, plumbagin and lapachol | Inhibition of monoamine oxidases regulating neurotransmitter levels and cell signaling, growth and differentiation | 281–283 |
Abbreviations: ERE, estrogen responsive elements; IFN, interferon; IL-17, interleukin-17; MMP, matrix metallopeptidase; NF-κB, nuclear factor κB; 1,4-NQ, 1,4-naphthoquinone; PKM2, pyruvate kinase-M2; TGF-β, transforming growth factor-β; TLR4, Toll-like receptor 4; TNF-α, tumor-necrosis factor-α; VEGF, vascular endothelial growth factor; VEGFR, VEGF receptor.
The bewildering array of actions elicited by plant 1,4-NQs listed in Table 2 are likely just the tip of the iceberg. Proteomics studies by Lame et al.284 with 14C-labeled 1,4-naphthoquinone indicate that this molecule targets a number of different proteins in human bronchial epithelial cells, including nucleophosmin, galectin-1, protein disulfide isomerase (PDI) and probable PDI, 60 kDa heat shock protein, mitochondrial stress-70 protein, epithelial cell marker protein and S100-type calcium-binding protein A14. A quantitative proteomic study using stable-isotope labeling by amino acids in cell culture revealed that there were at least 1225 and 267 proteins interacting with plumbagin and 341 and 107 signaling pathways and cellular functions potentially regulated by plumbagin in human PC-3 and DU145 prostate cancer cells, respectively.285 These proteins and pathways have critical roles in the regulation of cell cycle, apoptosis, autophagy, epithelial to mesenchymal transition and ROS generation.285
The diversity of targets and mechanisms of action of plant 1,4-NQs have stimulated great pharmacological interest, particularly in the area of ROS initiation and signaling, cancer therapeutic strategies and as anti-inflammatory agents.214,286 Moreover, numerous analogs of 1,4-NQs have been designed and synthesized to enhance their toxicity toward specific human cancer cell lines,277,287–289 specific proteins (for example, Hsp90),290 selected pathogenic organisms (for example, Trypanosoma sp)291–293 and insects.294
Conclusions and future prospectives
Plant 1,4-NQs are a diverse class of metabolites possessing a wide range of ecological functions contributing to plant fitness, particularly in the horticultural species highlighted in this review (Table 1). At least four different metabolic pathways to synthesize 1,4-NQs exist in the plant kingdom. The OSB route is present in all plants to produce phylloquinone, though some species are capable of synthesizing additional 1,4-naphthalenoid natural products branching off this pathway. The 4HBA/MVA, HGA/MVA and acetate-polymalonate pathways are each restricted to certain families. Regardless of the metabolic origin, however, 1,4-NQs have key roles in the interactions certain plants have with their biotic environment. New evidence has also emerged demonstrating that menadione, a synthetic 1,4-NQ, which is also present in Juglans,70 is capable of priming crops against abiotic stress.295 This raises the prospect that other plant 1,4-NQ natural products may also have similar roles in planta.
Medicinal plants synthesizing 1,4-NQs have been used for centuries based on the numerous pharmacological applications of these compounds.7 Today, molecular studies are beginning to validate these claims (see text and Table 2), making 1,4-NQs strong candidates for developing novel drugs (for example, shikonin against breast cancer296). The 1,4-NQs are also targets for synthetic strategies to develop more potent therapeutics, though there is undoubtedly a number of plant 1,4-NQs still undiscovered in nature that may already offer such drugs.
Beyond the core metabolic pathways (for example, phenylpropanoid, benzenoid and terpenoid) providing precursors for synthesizing specialized 1,4-NQs, there are only a couple identified 1,4-NQ biosynthetic genes. Although there is solid biochemical support for the involvement of these genes, there is a lack of genetic evidence to corroborate these results. This is in part due to the lack of genetically amenable systems in which to study these pathways. Therefore, there is a critical need to develop methods for generating transgenics in 1,4-NQ-synthesizing species, thus allowing for functional screening of candidates identified, for example, by comparative transcriptomic approaches. With the decreased costs and increased capabilities of sequencing technologies, forward genetic screens in existing horticultural models are also a favorable strategy to consider.
Indications are that biosynthesis of plant 1,4-NQs is highly compartmentalized and spread across multiple subcellular locations. This implicates the contribution of undefined transporters and other protein-mediated trafficking steps to the movement of metabolic intermediates and 1,4-NQs throughout the cell. That plant 1,4-NQs are often secreted into the environment further suggests the involvement of unknown plasma membrane-localized transporters and/or vesicular exocytosis for deployment.165,297–299
Despite extensive work over the last several decades, specialized plant 1,4-NQs are clearly an understudied, yet extremely promising class of metabolites for developing novel drugs and innovative strategies to address horticultural challenges, especially in pest management. To harness these metabolites for practical applications, however, will require a monumental improvement in the basic knowledge encompassing 1,4-NQ synthesis, transport and the molecular mechanisms behind their modes of action and release into the environment.
Acknowledgments
This work was supported by start-up funds from Purdue University to JRW.
The authors declare no conflict of interest.
References
- Medentsev AG, Akimenko VK. Naphthoquinone metabolites of the fungi. Phytochemistry 1998; 47: 935–959. [DOI] [PubMed] [Google Scholar]
- Studt L, Wiemann P, Kleigrewe K, Humpf HU, Tudzynski B. Biosynthesis of fusarubins accounts for pigmentation of fusarium Fujikuroi perithecia. Appl Environ Microbiol 2012; 78: 4468–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankewitz F, Hilker M. Polyketides in insects: ecological role of these widespread chemicals and evolutionary aspects of their biogenesis. Biol Rev 2008; 83: 209–226. [DOI] [PubMed] [Google Scholar]
- Raspotnig G, Fauler G, Leis M, Leis HJ. Chemical profiles of scent gland secretions in the cyphophthalmid opilionid harvestmen, Siro duricorius and S. exilis. J Chem Ecol 2005; 31: 1353–1368. [DOI] [PubMed] [Google Scholar]
- Calestani C, Rast JP, Davidson EH. Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development 2003; 130: 4587–4596. [DOI] [PubMed] [Google Scholar]
- Castoe TA, Stephens T, Noonan BP, Calestani C. A novel group of type I polyketide synthases (PKS) in animals and the complex phylogenomics of PKSs. Gene 2007; 392: 47–58. [DOI] [PubMed] [Google Scholar]
- Babula P, Adam V, Havel L, Kizek R. Noteworthy secondary metabolites naphthoquinones—their occurrence, pharmacological properties and analysis. Curr Pharm Anal 2009; 5: 47–68. [Google Scholar]
- Atkinson DJ, Brimble MA. Isolation, biological activity, biosynthesis and synthetic studies towards the rubromycin family of natural products. Nat Prod Rep 2015; 32: 811–840. [DOI] [PubMed] [Google Scholar]
- Collins MD, Jones D. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 1981; 45: 316–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida E, Nakamura A, Watanabe T. Reversed-phase HPLC determination of chlorophyll a’ and naphthoquinones in photosystem I of red algae: existence of two menaquinone-4 molecules in photosystem I of Cyanidium caldarium. Anal Sci 2003; 19: 1001–1005. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Komura M, Watanabe M, Minami C, Koike H, Itoh S et al. Photosystem I complexes associated with fucoxanthin-chlorophyll-binding proteins from a marine centric diatom, Chaetoceros gracilis. Biochim Biophys Acta 2008; 1777: 351–361. [DOI] [PubMed] [Google Scholar]
- Brettel K, Sétif P, Mathis P. Flash-induced absorption changes in photosystem I at low temperature: evidence that the electron acceptor A1 is vitamin K1. FEBS Lett 1986; 203: 220–224. [Google Scholar]
- Lefebvre-Legendre L, Rappaport F, Finazzi G, Ceol M, Grivet C, Hopfgartner G et al. Loss of phylloquinone in Chlamydomonas affects plastoquinone pool size and photosystem II synthesis. J Biol Chem 2007; 282: 13250–13263. [DOI] [PubMed] [Google Scholar]
- Seeger JW, Bentley R. Phylloquinone (vitamin K1) biosynthesis in Euglena gracilis strain Z. Phytochemistry 1991; 30: 3585–3589. [Google Scholar]
- Seigler DS. Benzoquinones, naphthoquinones, and anthraquinones. In: Seigler DS (ed.) Plant Secondary Metabolism. Boston: Springer, 1998: 76–93. [Google Scholar]
- Hussain H, Krohn K, Ahmad VU, Miana GA, Green IR. Lapachol: an overview. Arkivoc 2007; 2007: 145. [Google Scholar]
- Thomson RH. Naphthoquinones. Naturally Occurring Quinones IV. Dordrecht: Springer, 1997: 112–308. [Google Scholar]
- Egan PA, Van Der Kooy F. Phytochemistry of the carnivorous sundew genus Drosera (Droseraceae)—future perspectives and ethnopharmacological relevance. Chem Biodivers 2013; 10: 1774–1790. [DOI] [PubMed] [Google Scholar]
- Malik S, Bhushan S, Sharma M, Ahuja PS. Biotechnological approaches to the production of shikonins: a critical review with recent updates. Crit Rev Biotechnol 2014; 8551: 1–14. [DOI] [PubMed] [Google Scholar]
- Hook I, Mills C, Sheridan H. Bioactive naphthoquinones from higher plants. In: Atta-ur-Rahman (ed.) Studies in Natural Products Chemistry. Netherlands: Elsevier, 2014: 119–160. [Google Scholar]
- van Oostende C, Widhalm JR, Furt F, Ducluzeau A-L, Basset GJ. Vitamin K1 (Phylloquinone): function, enzymes and genes. In: Fabrice Rébeillé and Roland Douce (ed.) Advances in Botanical Research. Amsterdam: Elsevier, 2011: 229–261. [Google Scholar]
- Fitzpatrick TB, Basset GJC, Borel P, Carrari F, DellaPenna D, Fraser PD et al. Vitamin deficiencies in humans: can plant science help? Plant Cell 2012; 24: 395–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Dudareva N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 2012; 63: 73–105. [DOI] [PubMed] [Google Scholar]
- Widhalm JR, Dudareva N. A familiar ring to it: biosynthesis of plant benzoic acids. Mol Plant 2015; 8: 83–97. [DOI] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 2013; 64: 665–700. [DOI] [PubMed] [Google Scholar]
- Flores-Sanchez IJ, Verpoorte R. Plant polyketide synthases: a fascinating group of enzymes. Plant Physiol Biochem 2009; 47: 167–174. [DOI] [PubMed] [Google Scholar]
- Hiratsuka T, Furihata K, Ishikawa J, Yamashita H, Itoh N, Seto H et al. An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 2008; 321: 1670–1673. [DOI] [PubMed] [Google Scholar]
- Cox GB, Gibson F. Biosynthesis of vitamin K and ubiquinone relation to the shikimic acid pathway in Escherichia coli. Biochim Biophys Acta 1964; 93: 204–206. [DOI] [PubMed] [Google Scholar]
- Whistance GR, Threlfall DR, Goodwin TW. Incorporation of [G-14C]shikimate and [U-14C]para-hydroxybenzoate into phytoquinones and chromanols. Biochem Biophys Res Commun 1966; 23: 849–853. [DOI] [PubMed] [Google Scholar]
- Chen D, Bohm BA. Naphthoquinone biosynthesis in higher plants: I. Studies on 2-hydroxy-l,4-naphthoquinone in Impatiens balsamina L. Can J Biochem 1966; 44: 1389–1395. [DOI] [PubMed] [Google Scholar]
- Zenk MH, Leistner E. [On the mode of incorporation of shikimic acid into 2-hydroxy-1,4-naphthoquinone (lawsone)]. Z Naturforsch B 1967; 22: 460. [DOI] [PubMed] [Google Scholar]
- Leistner E, Zenk MH. Zur Biogenese von 5-Hydroxy-1,4-naphthochinon (Juglon) in Juglans regia L.. Z Naturforsch B 1968; 23: 259–268. [PubMed] [Google Scholar]
- Campbell IM. The roles of alanine, aspartate and glutamate in lawsone biosynthesis in Impatiens balsamina. Tetrahedron Lett 1969; 10: 4777–4780. [Google Scholar]
- Grotzinger E, Campbell IM. The role of 2-ketoglutarate in lawsone biosynthesis in Impatiens balsamina. Tetrahedron Lett 1972; 13: 4685–4686. [Google Scholar]
- Dansette P, Azerad R. A new intermediate in naphthoquinone and menaquinone biosynthesis. Biochem Biophys Res Commun 1970; 40: 1090–1095. [DOI] [PubMed] [Google Scholar]
- Robins DJ, Yee RB, Bentley R. Biosynthetic precursors of vitamin K as growth promoters for Bacteroides melaninogenicus. J Bacteriol 1973; 116: 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley R. Biosynthesis of vitamin K and other natural naphthoquinones. Pure Appl Chem 1975; 41: 47–68. [Google Scholar]
- Thomas G, Threlfall DR. Incorporation of shikimate and 4-(2′-carboxyphenyl)-4-oxobutyrate into phylloquinone. Phytochemistry 1974; 13: 807–813. [Google Scholar]
- Müller W, Leistner E. 1,4-Naphthoquinone, an intermediate in juglone (5-hydroxy-1,4-naphthoquinone) biosynthesis. Phytochemistry 1976; 15: 407–410. [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001; 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Garcion C, Lohmann A, Lamodière E, Catinot J, Buchala A, Doermann P et al. Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol 2008; 147: 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J, Won KC, Lezhneva L, Falk J, Krupinska K, Shinozaki K et al. A plant locus essential for phylloquinone (vitamin K1) biosynthesis originated from a fusion of four eubacterial genes. J Biol Chem 2006; 281: 17189–17196. [DOI] [PubMed] [Google Scholar]
- Kim HU, Van Oostende C, Basset GJC, Browse J. The AAE14 gene encodes the Arabidopsis o-succinylbenzoyl-CoA ligase that is essential for phylloquinone synthesis and photosystem-I function. Plant J 2008; 54: 272–283. [DOI] [PubMed] [Google Scholar]
- Widhalm JR, Ducluzeau AL, Buller NE, Elowsky CG, Olsen LJ, Basset GJC. Phylloquinone (vitamin K1) biosynthesis in plants: two peroxisomal thioesterases of lactobacillales origin hydrolyze 1,4-dihydroxy-2-naphthoyl-coa. Plant J 2012; 71: 205–215. [DOI] [PubMed] [Google Scholar]
- Mustafa NR, Kim HK, Choi YH, Erkelens C, Lefeber AWM, Spijksma G et al. Biosynthesis of salicylic acid in fungus elicited Catharanthus roseus cells. Phytochemistry 2009; 70: 532–539. [DOI] [PubMed] [Google Scholar]
- Muljono RAB, Scheffer JJC, Verpoorte R. Isochorismate is an intermediate in 2,3-dihydroxybenzoic acid biosynthesis in Catharanthus roseus cell cultures. Plant Physiol Biochem 2002; 40: 231–234. [Google Scholar]
- Bartsch M, Bednarek P, Vivancos PD, Schneider B, Von Roepenack-Lahaye E, Foyer CH et al. Accumulation of isochorismate-derived 2,3-dihydroxybenzoic 3-O-β-D-xyloside in Arabidopsis resistance to pathogens and ageing of leaves. J Biol Chem 2010; 285: 25654–25665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Cao Y, Guo ZF, Chen M, Chen X, Guo Z. Menaquinone biosynthesis in Escherichia coli: identification of 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate as a novel intermediate and re-evaluation of MenD activity. Biochemistry 2007; 46: 10979–10989. [DOI] [PubMed] [Google Scholar]
- Jiang M, Chen X, Guo ZF, Cao Y, Chen M, Guo Z. Identification and characterization of (1R,6R)-2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase in the menaquinone biosynthesis of Escherichia coli. Biochemistry 2008; 47: 3426–3434. [DOI] [PubMed] [Google Scholar]
- Meganathan R, Bentley R. Thiamine pyrophosphate requirement for o-succinylbenzoic acid synthesis in Escherichia coli and evidence for an intermediate. J Bacteriol 1983; 153: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DRJ, Garrett JB, Sharma V, Meganathan R, Babbitt PC, Gerlt JA. Unexpected divergence of enzyme function and sequence: ‘N-acylamino acid racemase’ is o-succinylbenzoate synthase. Biochemistry 1999; 38: 4252–4258. [DOI] [PubMed] [Google Scholar]
- Kwon O, Bhattacharyya DK, Meganathan R. Menaquinone (vitamin K2) biosynthesis: overexpression, purification, and properties of o-succinylbenzoyl-coenzyme A synthetase from Escherichia coli. J Bacteriol 1996; 178: 6778–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truglio JJ, Theis K, Feng Y, Gajda R, Machutta C, Tonge PJ et al. Crystal Structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis. J Biol Chem 2003; 278: 42352–42360. [DOI] [PubMed] [Google Scholar]
- Jiang M, Chen M, Guo ZF, Guo Z. A bicarbonate cofactor modulates 1,4-dihydroxy-2-naphthoyl-coenzyme a synthase in menaquinone biosynthesis of Escherichia coli. J Biol Chem 2010; 285: 30159–30169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meganathan R. Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q): a perspective on enzymatic mechanisms. Vitam Horm 2001; 61: 173–218. [DOI] [PubMed] [Google Scholar]
- Sakuragi Y, Bryant DA. Genetic manipulation of quinone biosynthesis in cyanobacteria. In: John H. Golbeck (ed.) Photosystem I. Dordrecht: Springer, 2006: 205–222. [Google Scholar]
- Widhalm JR, van Oostende C, Furt F, Basset GJC. A dedicated thioesterase of the Hotdog-fold family is required for the biosynthesis of the naphthoquinone ring of vitamin K1. Proc Natl Acad Sci USA 2009; 106: 5599–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Ma X, Chen X, Jiang M, Song H, Guo Z. Identification of a hotdog fold thioesterase involved in the biosynthesis of menaquinone in Escherichia coli. J Bacteriol 2013; 195: 2768–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Ohno R, Shibata M, Ikegami I, Onai K, Ohto MA et al. Inactivation and deficiency of core proteins of photosystems I and II caused by genetical phylloquinone and plastoquinone deficiency but retained lamellar structure in a T-DNA mutant of Arabidopsis. Plant J 2005; 41: 627–637. [DOI] [PubMed] [Google Scholar]
- Meganathan R, Kwon O. Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q). Ecosal Plus 2009; 3: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatihi A, Latimer S, Schmollinger S, Block A, Dussault PH, Vermaas WF et al. A dedicated type II NADPH dehydrogenase performs the penultimate step in the biosynthesis of vitamin K1 in Synechocystis and Arabidopsis. Plant Cell 2015; 27: 1730–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann A, Schöttler MA, Bréhélin C, Kessler F, Bock R, Cahoon EB et al. Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J Biol Chem 2006; 281: 40461–40472. [DOI] [PubMed] [Google Scholar]
- Babujee L, Wurtz V, Ma C, Lueder F, Soni P, Van Dorsselaer A et al. The proteome map of spinach leaf peroxisomes indicates partial compartmentalization of phylloquinone (vitamin K1) biosynthesis in plant peroxisomes. J Exp Bot 2010; 61: 1441–1453. [DOI] [PubMed] [Google Scholar]
- Reumann S, Quan S, Aung K, Yang P, Manandhar-Shrestha K, Holbrook D et al. In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol 2009; 150: 125–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger D, Bacher A, Zenk MH, Eisenreich W. Quantitative assessment of metabolic flux by C-13 NMR analysis. Biosynthesis of anthraquinones in Rubia tinctorum. J Am Chem Soc 1999; 121: 7469–7475. [Google Scholar]
- Han YS, Van der Heijden R, Lefeber AWM, Erkelens C, Verpoorte R. Biosynthesis of anthraquinones in cell cultures of Cinchona ‘Robusta’ proceeds via the methylerythritol 4-phosphate pathway. Phytochemistry 2002; 59: 45–55. [DOI] [PubMed] [Google Scholar]
- Furumoto T, Hoshikuma A. Biosynthetic origin of 2-geranyl-1,4-naphthoquinone and its related anthraquinone in a Sesamum indicum hairy root culture. Phytochemistry 2011; 72: 871–874. [DOI] [PubMed] [Google Scholar]
- Leistner E. Biosynthesis of morindone and alizarin in intact plants and cell suspension cultures of Morinda citrifolia. Phytochemistry 1973; 12: 1669–1674. [Google Scholar]
- Leistner E. Mode of incorporation of precursors into alizarin (1,2-dihydroxy-9,10-anthraquinone). Phytochemistry 1973; 12: 337–345. [Google Scholar]
- Binder RG, Benson ME, Flath RA. Eight 1,4-naphthoquinones from Juglans. Phytochemistry 1989; 28: 2799–2801. [Google Scholar]
- Chung D, Maier UH, Inouye H, Zenk MH. Different mode of incorporation of o-succinylbenzoic acid into the naphthoquinones juglone and lawsone in higher plants. Z Naturforsch C 1994; 49: 885–887. [Google Scholar]
- Mizutani M, Ohta D. Diversification of P450 genes during land plant evolution. Annu Rev Plant Biol 2010; 61: 291–315. [DOI] [PubMed] [Google Scholar]
- Farrow SC, Facchini PJ. Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front Plant Sci 2014; 5: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy J, Huss B, Creach A, Hawkins S, Neutelings G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front Plant Sci 2016; 7: 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W-U, Leistner E. Metabolic relation between naphthalene derivatives in Juglans. Phytochemistry 1978; 17: 1735–1738. [Google Scholar]
- Müller W-U, Leistner E. Aglycones and glycosides of oxygenated naphthalenes and a glycosyltransferase from Juglans. Phytochemistry 1978; 17: 1739–1742. [Google Scholar]
- Gueldner RC, Yates IE, Smith MT. Levels of a hydrojuglone glucoside in developing pecan leaves in relation to scab susceptibility. J. Am. Soc. Hort. Sci. 1994; 119: 498–504. [Google Scholar]
- Hedin PA, Collum DH, Langhans VE, Graves CH. Distribution of juglone and related compounds in pecan and their effect on Fusicladium effusum. J Agric Food Chem 1980; 28: 340–342. [Google Scholar]
- Wrobel RL, Matvienko M, Yoder JI. Heterologous expression and biochemical characterization of an NAD(P)H:quinone oxidoreductase from the hemiparasitic plant Triphysaria versicolor. Plant Physiol Biochem 2002; 40: 265–272. [Google Scholar]
- Duroux L, Delmotte FM, Lancelin JM, Keravis G, Jay-Allemand C. Insight into naphthoquinone metabolism: beta-glucosidase-catalysed hydrolysis of hydrojuglone beta-D-glucopyranoside. Biochem J 1998; 333275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobstein A, Brenne X, Feist E, Metz N, Weniger B, Anton R. Quantitative determination of naphthoquinones of Impatiens species. Phytochem Anal 2001; 12: 202–205. [DOI] [PubMed] [Google Scholar]
- Sakunphueak A, Panichayupakaranant P. Increased production of naphthoquinones in Impatiens balsamina root cultures by elicitation with methyl jasmonate. Bioresour Technol 2010; 101: 8777–8783. [DOI] [PubMed] [Google Scholar]
- Tříska J, Vrchotová N, Sýkora J, Moos M. Separation and Identification of 1,2,4-Trihydroxynaphthalene-1-O-glucoside in Impatiens glandulifera Royle. Molecules 2013; 18: 8429–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapelle J-P. 2-Methoxy-1, 4-naphthoquinone in Impatiens glandulifera and related species. Phytochemistry 1974; 13: 662. [Google Scholar]
- Ruckli R, Hesse K, Glauser G, Rusterholz H-P, Baur B. Inhibitory potential of naphthoquinones leached from leaves and exuded from roots of the invasive plant Impatiens glandulifera. J Chem Ecol 2014; 40: 371–378. [DOI] [PubMed] [Google Scholar]
- Liscombe DK, Louie GV, Noel JP. Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat Prod Rep 2012; 29: 1238–1250. [DOI] [PubMed] [Google Scholar]
- Gómez Castellanos JR, Prieto JM, Heinrich M. Red Lapacho (Tabebuia impetiginosa)—a global ethnopharmacological commodity? J Ethnopharmacol 2009; 121: 1–13. [DOI] [PubMed] [Google Scholar]
- Manners GD, Jurd L, Wong R, Palmer K. Constituents of Tabebuia guayacan. Tetrahedron 1975; 31: 3019–3024. [Google Scholar]
- Manners GD, Jurd L, Wong R, Palmer K. Constituents of Tabebuia guayacan-II. Tetrahedron 1976; 32: 543–547. [Google Scholar]
- Papageorgiou V, Assimopoulou A, Samanidou V, Papadoyannis I. Recent advances in chemistry, biology and biotechnology of alkannins and shikonins. Curr Org Chem 2006; 10: 2123–2142. [Google Scholar]
- Schmid HV, Zenk MH. p-hydroxybenzoic acid and mevalonic acid as precursors of the plant naphthoquinone alkannin. Tetrahedron Lett 1971; 12: 4151–4155. [Google Scholar]
- Inouye H, Ueda S, Inoue K, Matsumura H. Biosynthesis of shikonin in callus cultures of Lithospermum erythrorhizon. Phytochemistry 1979; 18: 1301–1308. [Google Scholar]
- Yazaki K, Fukui H, Tabata M. Isolation of the intermediates and related metabolites of shikonin biosynthesis from Lithospermum erythrorhizon cell cultures. Chem Pharm Bull (Tokyo) 1986; 34: 2290–2293. [Google Scholar]
- Gaisser S, Heide L. Inhibition and regulation of shikonin biosynthesis in suspension cultures of Lithospermum. Phytochemistry 1996; 41: 1065–1072. [Google Scholar]
- Yazaki K, Heide L, Tabata M. Formation of p-hydroxybenzoic acid from p-coumaric acid by cell free extract of Lithospermum erythrorhizon cell cultures. Phytochemistry 1991; 30: 2233–2236. [Google Scholar]
- Löscher R, Heide L. Biosynthesis of p-Hydroxybenzoate from p-Coumarate and p-Coumaroyl-Coenzyme A in Cell-Free Extracts of Lithospermum erythrorhizon Cell Cultures. Plant Physiol 1994; 106: 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Widhalm JR, Fatihi A, Cahoon RE, Wamboldt Y, Elowsky C et al. The origin and biosynthesis of the benzenoid moiety of ubiquinone (coenzyme Q) in Arabidopsis. Plant Cell 2014; 26: 1938–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K, Kataoka M, Honda G, Severin K, Heide L. cDNA Cloning and gene expression of phenylalanine ammonia-lyase in Lithospermum erythrorhizon. Biosci Biotechnol Biochem 1997; 61: 1995–2003. [DOI] [PubMed] [Google Scholar]
- Yamamura Y, Ogihara Y, Mizukami H. Cinnamic acid 4-hydroxylase from Lithospermum erythrorhizon: cDNA cloning and gene expression. Plant Cell Rep 2001; 20: 655–662. [Google Scholar]
- Singh RS, Gara RK, Bhardwaj PK, Kaachra A, Malik S, Kumar R et al. Expression of 3-hydroxy-3-methylglutaryl-CoA reductase, p-hydroxybenzoate-m-geranyltransferase and genes of phenylpropanoid pathway exhibits positive correlation with shikonins content in Arnebia [Arnebia euchroma (Royle) Johnston]. BMC Mol Biol 2010; 11: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K. Intracellular localization of UDPG: p-hydroxybenzoate glucosyltransferase and its reaction product in Lithospermum cell cultures. Phytochemistry 1995; 38: 1127–1130. [Google Scholar]
- Sommer S, Severin K, Camara B, Heide L. Intracellular localization of geranylpyrophosphate synthase from cell cultures of Lithospermum erythrorhizon. Phytochemistry 1995; 38: 623–627. [Google Scholar]
- Li S-M, Hennig S, Heide L. Shikonin: a geranyl diphosphate-derived plant hemiterpenoid formed via the mevalonate pathway. Tetrahedron Lett 1998; 39: 2721–2724. [Google Scholar]
- Markus Lange B, Severin K, Bechthold A, Heide L. Regulatory role of microsomal 3-hydroxy-3-methylglutaryl-coenzyme A reductase for shikonin biosynthesis in Lithospermum erythrorhizon cell suspension cultures. Planta 1998; 204: 234–241. [DOI] [PubMed] [Google Scholar]
- Heide L, Tabata M. Geranylpyrophosphate: p-hydroxybenzoate geranyltransferase activity in extracts of Lithospermum erythrorhizon cell cultures. Phytochemistry 1987; 26: 1651–1655. [Google Scholar]
- Yamaga Y, Nakanishi K, Fukui H, Tabata M. Intracellular localization of p-hydroxybenzoate geranyltransferase, a key enzyme involved in shikonin biosynthesis. Phytochemistry 1993; 32: 633–636. [Google Scholar]
- Mühlenweg A, Melzer M, Li SM, Heide L. 4-Hydroxybenzoate 3-geranyltransferase from Lithospermum erythrorhizon: purification of a plant membrane-bound prenyltransferase. Planta 1998; 205: 407–413. [DOI] [PubMed] [Google Scholar]
- Yazaki K, Kunihisa M, Fujisaki T, Sato F. Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon: cloning and characterization of a key enzyme in shikonin biosynthesis. J Biol Chem 2002; 277: 6240–6246. [DOI] [PubMed] [Google Scholar]
- Ohara K, Muroya A, Fukushima N, Yazaki K. Functional characterization of LePGT1, a membrane-bound prenyltransferase involved in the geranylation of p-hydroxybenzoic acid. Biochem J 2009; 421: 231–241. [DOI] [PubMed] [Google Scholar]
- Ohara K, Mito K, Yazaki K. Homogeneous purification and characterization of LePGT1—a membrane-bound aromatic substrate prenyltransferase involved in secondary metabolism of Lithospermum erythrorhizon. FEBS J 2013; 280: 2572–2580. [DOI] [PubMed] [Google Scholar]
- White MD, Payne KAP, Fisher K, Marshall SA, Parker D, Rattray NJW et al. UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 2015; 522: 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne KaP, White MD, Fisher K, Khara B, Bailey SS, Parker D et al. New cofactor supports α,β-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature 2015; 522: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aussel L, Pierrel F, Loiseau L, Lombard M, Fontecave M, Barras F. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta 2014; 1837: 1004–1011. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Inoue K, Li S, Heide L. Geranylhydroquinone 3′′-hydroxylase, a cytochrome P-450 monooxygenase from Lithospermum erythrorhizon cell suspension cultures. Planta 2000; 210: 312–317. [DOI] [PubMed] [Google Scholar]
- Weston PA, Weston LA, Hildebrand S. Metabolic profiling in Echium plantagineum: presence of bioactive pyrrolizidine alkaloids and napthoquinones from accessions across southeastern Australia. Phytochem Rev 2013; 12: 831–837. [Google Scholar]
- Fujita Y, Maeda Y, Suga C, Morimoto T. Production of shikonin derivatives by cell suspension cultures of Lithospermum erythrorhizon. III. Comparison of shikonin derivatives of cultured cells and ko-shikon. Plant Cell Rep 1983; 2: 192–193. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Zhao P, Yazaki K, Inoue K. Regulation of lithospermic acid B and shikonin production in Lithospermum erythrorhizon cell suspension cultures. Chem Pharm Bull (Tokyo) 2002; 50: 1086–1090. [DOI] [PubMed] [Google Scholar]
- Brigham LA, Michaels PJ, Flores HE. Cell-specific production and antimicrobial activity of naphthoquinones in roots of Lithospermum erythrorhizon 1. Plant Physiol 1999; 119: 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Bhushan S, Sharma M, Singh Ahuja P. Physico-chemical factors influencing the shikonin derivatives production in cell suspension cultures of Arnebia euchroma (Royle) Johnston, a medicinally important plant species. Cell Biol Int 2011; 35: 153–158. [DOI] [PubMed] [Google Scholar]
- Liu Z, Qi J-L, Chen L, Zhang M-S, Wang X-Q, Pang Y-J et al. Effect of light on gene expression and shikonin formation in cultured Onosma paniculatum cells. Plant Cell Tissue Organ Cult 2005; 84: 38–48. [Google Scholar]
- Yazaki K, Matsuoka H, Ujihara T, Sato F. Shikonin biosynthesis in Lithospermum erythrorhizon. Light-induced negative regulation of secondary metabolism. Plant Biotechnol 1999; 16: 335–342. [Google Scholar]
- Wu SJ, Qi JL, Zhang WJ, Liu SH, Xiao FH, Zhang MS et al. Nitric oxide regulates shikonin formation in suspension-cultured onosma paniculatum cells. Plant Cell Physiol 2009; 50: 118–128. [DOI] [PubMed] [Google Scholar]
- Tabata M, Mizukami H, Hiraoka N, Konoshima M. Pigment formation in callus cultures of Lithospermum erythrorhizon. Phytochemistry 1974; 13: 927–932. [Google Scholar]
- Yazaki K, Takeda K, Tabata M. Effects of methyl jasmonate on shikonin and dihydroechinofuran production in Lithospermum cell cultures. Plant Cell Physiol 1997; 38: 776–782. [Google Scholar]
- Fang R, Wu F, Zou A, Zhu Y, Zhao H, Zhao H et al. Transgenic analysis reveals LeACS-1 as a positive regulator of ethylene-induced shikonin biosynthesis in Lithospermum erythrorhizon hairy roots. Plant Mol Biol 2016; 90: 345–358. [DOI] [PubMed] [Google Scholar]
- Fang R, Zou A, Zhao H, Wu F, Zhu Y, Zhao H et al. Transgenic studies reveal the positive role of LeEIL-1 in regulating shikonin biosynthesis in Lithospermum erythrorhizon hairy roots. BMC Plant Biol 2016; 16: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zou A, Miao J, Yin Y, Tian R, Pang Y et al. LeERF-1, a novel AP2/ERF family gene within the B3 subcluster, is down-regulated by light signals in Lithospermum erythrorhizon. Plant Biol 2011; 13: 343–348. [DOI] [PubMed] [Google Scholar]
- Gális I, Šimek P, Narisawa T, Sasaki M, Horiguchi T, Fukuda H et al. A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant J 2006; 46: 573–592. [DOI] [PubMed] [Google Scholar]
- Zhao H, Chang QS, Zhang DX, Fang RJ, Zhao H, Wu FY et al. Overexpression of LeMYB1 enhances shikonin formation by up-regulating key shikonin biosynthesis-related genes in Lithospermum erythrorhizon. Biol Plant 2015; 59: 429–435. [Google Scholar]
- Yamamura Y, Sahin FP, Nagatsu A, Mizukami H. Molecular cloning and characterization of a cDNA encoding a novel apoplastic protein preferentially expressed in a shikonin-producing callus strain of Lithospermum erythrorhizon. Plant Cell Physiol 2003; 44: 437–446. [DOI] [PubMed] [Google Scholar]
- Yazaki K, Matsuoka H, Shimomura K, Bechthold A, Sato F. A novel dark-inducible protein, LeDI-2, and its involvement in root-specific secondary metabolism in Lithospermum erythrorhizon. Plant Physiol 2001; 125: 1831–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolkart KH, Zenk MH. [Tyrosine, a precursor of the quinone ring of 2,7-dimethyl-naphthoquinone (Chimaphilin)]. Naturwissenschaften 1968; 55: 444–445. [DOI] [PubMed] [Google Scholar]
- Bolkart KH, Knobloch M, Zenk MH. Mevalonic acid, the precursor of the substituted benzenoid ring of chimaphilin. Naturwissenschaften 1968; 55: 445. [DOI] [PubMed] [Google Scholar]
- Bolkart KH, Zenk MH. The homogentisate pathway in the biosynthesis of 2,7-dimethyl-1,4-naphthoquinone (chimaphilin). Z Pflanzenphysiol 1969; 61: 356–359. [Google Scholar]
- DellaPenna D, Mène-Saffrané L. Vitamin E. In: Fabrice Rébeillé and Roland Douce (ed.) Current Trends in Eye Tracking Research. Amsterdam: Elsevier, 2011: 179–227. [Google Scholar]
- Burnett AR, Thomson RH. Naturally occurring quinones. Part XIV. The quinonoid constituents of Pyrola media Sw. (Pyrolaceae). J Chem Soc C Org 1968; 3: 857. [Google Scholar]
- Inouye H. Ein Naphthochinon-Farbstoff aus Pirola incarnata FISCH. Pharm Soc Japan 1956; 76: 976–977. [Google Scholar]
- Inouye H, Arai T. Uber die Bestandteile der Pyrolazeen. XI. Die Bestandteile der Pyrola renifolia MAXIM. (1). Chem Pharm Bull (Tokyo) 1964; 12: 533–539. [DOI] [PubMed] [Google Scholar]
- Saxena G, Farmer SW, Hancock REW, Towers GHN. Chlorochimaphilin: a new antibiotic from Moneses uniflora. J Nat Prod 1996; 59: 62–65. [DOI] [PubMed] [Google Scholar]
- Sankaram AVB, Srinivasarao A, Sidhu GS. Chitranone—a new Binaphthaquinone from Plumbago zeylanica. Phytochemistry 1976; 15: 237–238. [Google Scholar]
- Durand R, Zenk MH. Biosynthesis of plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone) via the acetate pathway in higher plants. Tetrahedron Lett 1971; 12: 3009–3012. [Google Scholar]
- Durand R, Zenk MH. The homogentisate ring-cleavage pathway in the biosynthesis of acetate-derived naphthoquinones of the droseraceae. Phytochemistry 1974; 13: 1483–1492. [Google Scholar]
- Bringmann G, Rischer H, Wohlfarth M, Schlauer J, Assi LA. Droserone from cell cultures of Triphyophyllum peltatum (Dioncophyllaceae) and its biosynthetic origin. Phytochemistry 2000; 53: 339–343. [DOI] [PubMed] [Google Scholar]
- Bringmann G, Feineis D. Stress-related polyketide metabolism of Dioncophyllaceae and Ancistrocladaceae. J Exp Bot 2001; 52: 2015–2022. [DOI] [PubMed] [Google Scholar]
- Springob K, Samappito S, Jindaprasert A, Schmidt J, Page JE, De-Eknamkul W et al. A polyketide synthase of Plumbago indica that catalyzes the formation of hexaketide pyrones. FEBS J 2007; 274: 406–417. [DOI] [PubMed] [Google Scholar]
- Jindaprasert A, Springob K, Schmidt J, De-Eknamkul W, Kutchan TM. Pyrone polyketides synthesized by a type III polyketide synthase from Drosophyllum lusitanicum. Phytochemistry 2008; 69: 3043–3053. [DOI] [PubMed] [Google Scholar]
- Raj G, Kurup R, Hussain AA, Baby S. Distribution of naphthoquinones, plumbagin, droserone, and 5-O-methyl droserone in chitin-induced and uninduced Nepenthes khasiana: molecular events in prey capture. J Exp Bot 2011; 62: 5429–5436. [DOI] [PubMed] [Google Scholar]
- Eilenberg H, Pnini-Cohen S, Rahamim Y, Sionov E, Segal E, Carmeli S et al. Induced production of antifungal naphthoquinones in the pitchers of the carnivorous plant Nepenthes khasiana. J Exp Bot 2010; 61: 911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenk MH, Fürbringer M, Steglich W. Occurrence and distribution of 7-methyljuglone and plumbagin in the droseraceae. Phytochemistry 1969; 8: 2199–2200. [Google Scholar]
- Culham A, Gornall RJ. The taxonomic significance of naphthoquinones in the Droseraceae. Biochem Syst Ecol 1994; 22: 507–515. [Google Scholar]
- Lin LC, Yang LL, Chou CJ. Cytotoxic naphthoquinones and plumbagic acid glucosides from Plumbago zeylanica. Phytochemistry 2003; 62: 619–622. [DOI] [PubMed] [Google Scholar]
- Budzianowski J. Naphthohydroquinone glucosides of Drosera rotundifolia and D. intermedia from in vitro cultures. Phytochemistry 1996; 42: 1145–1147. [Google Scholar]
- Izhaki I. Emodin—a secondary metabolite with multiple ecological functions in higher plants. New Phytol 2002; 155: 205–217. [Google Scholar]
- Abe I, Utsumi Y, Oguro S, Noguchi H. The first plant type III polyketide synthase that catalyzes formation of aromatic heptaketide. FEBS Lett 2004; 562: 171–176. [DOI] [PubMed] [Google Scholar]
- Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ. Role of quinones in toxicology. Chem Res Toxicol 2000; 13: 135–160. [DOI] [PubMed] [Google Scholar]
- Kumagai Y, Shinkai Y, Miura T, Cho AK. The chemical biology of naphthoquinones and its environmental implications. Annu Rev Pharmacol Toxicol 2012; 52: 221–247. [DOI] [PubMed] [Google Scholar]
- Klotz LO, Hou X, Jacob C. 1,4-naphthoquinones: from oxidative damage to cellular and inter-cellular signaling. Molecules 2014; 19: 14902–14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rischer H, Hamm A, Bringmann G. Nepenthes insignis uses a C2-portion of the carbon skeleton of L-alanine acquired via its carnivorous organs, to build up the allelochemical plumbagin. Phytochemistry 2002; 59: 603–609. [DOI] [PubMed] [Google Scholar]
- Hernández-Muñoz LS, Goméz M, González FJ, González I, Frontana C. Towards a molecular-level understanding of the reactivity differences for radical anions of juglone and plumbagin: an electrochemical and spectroelectrochemical approach. Org Biomol Chem 2009; 7: 1896–1903. [DOI] [PubMed] [Google Scholar]
- Babich H, Stern A. In vitro cytotoxicities of 1,4-naphthoquinone and hydroxylated 1,4-naphthoquinones to replicating cells. J Appl Toxicol 1993; 13: 353–358. [DOI] [PubMed] [Google Scholar]
- Willis RJ. The History of Allelopathy. Springer: Dordrecht, Netherlands. 2007. [Google Scholar]
- Rice EL. Allelopathy2nd ednAcademic Press: New York, NY, USA. 1984. [Google Scholar]
- Soderquist CJ. Juglone and allelopathy. J Chem Educ 1973; 50: 782–783. [DOI] [PubMed] [Google Scholar]
- Willis RJ. Juglans spp., juglone and allelopathy. Allelopath J 2000; 7: 1–55. [Google Scholar]
- Zhu X, Skoneczny D, Weidenhamer JD, Mwendwa JM, Weston PA, Gurr GM et al. Identification and localization of bioactive naphthoquinones in the roots and rhizosphere of Paterson’s curse (Echium plantagineum), a noxious invader. J Exp Bot 2016; 67: 3777–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrchotová N, Šerá B, Krejčová J. Allelopathic activity of extracts from Impatiens species. Plant Soil Environ 2011; 57: 57–60. [Google Scholar]
- Babula P, Adam V, Kizek R, Sladký Z, Havel L. Naphthoquinones as allelochemical triggers of programmed cell death. Environ Exp Bot 2009; 65: 330–337. [Google Scholar]
- Babula P, Vaverkova V, Poborilova Z, Ballova L, Masarik M, Provaznik I. Phytotoxic action of naphthoquinone juglone demonstrated on lettuce seedling roots. Plant Physiol Biochem 2014; 84: 78–86. [DOI] [PubMed] [Google Scholar]
- Chi WC, Fu SF, Huang TL, Chen YA, Chen CC, Huang HJ. Identification of transcriptome profiles and signaling pathways for the allelochemical juglone in rice roots. Plant Mol Biol 2011; 77: 591–607. [DOI] [PubMed] [Google Scholar]
- Rudnicka M, Polak M, Karcz W. Cellular responses to naphthoquinones: Juglone as a case study. Plant Growth Regul 2014; 72: 239–248. [Google Scholar]
- Kot M, Karcz W, Zaborska W. 5-Hydroxy-1,4-naphthoquinone (juglone) and 2-hydroxy-1,4-naphthoquinone (lawsone) influence on jack bean urease activity: elucidation of the difference in inhibition activity. Bioorg Chem 2010; 38: 132–137. [DOI] [PubMed] [Google Scholar]
- Yao JL, Kops O, Lu PJ, Lu KP. Functional conservation of phosphorylation-specific prolyl isomerases in plants. J Biol Chem 2001; 276: 13517–13523. [DOI] [PubMed] [Google Scholar]
- Sauriasari R, Wang DH, Takemura Y, Tsutsui K, Masuoka N, Sano K et al. Cytotoxicity of lawsone and cytoprotective activity of antioxidants in catalase mutant Escherichia coli. Toxicology 2007; 235: 103–111. [DOI] [PubMed] [Google Scholar]
- Wang J, Cheng Y, Wu R, Jiang D, Bai B, Tan D et al. Antibacterial activity of juglone against Staphylococcus aureus: from apparent to proteomic. Int J Mol Sci 2016; 17: 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha BK, Jung H, Seo I, Suh S-I, Suh M, Baek W. Juglone induces cell death of Acanthamoeba through increased production of reactive oxygen species. Exp Parasitol 2015; 159: 100–106. [DOI] [PubMed] [Google Scholar]
- Nair SV, Baranwal G, Chatterjee M, Sachu A, Vasudevan AK, Bose C et al. Antimicrobial activity of plumbagin, a naturally occurring naphthoquinone from Plumbago rosea, against Staphylococcus aureus and Candida albicans. Int J Med Microbiol 2016; 306: 237–248. [DOI] [PubMed] [Google Scholar]
- Mahapatra A, Mativandlela SPN, Binneman B, Fourie PB, Hamilton CJ, Meyer JJM et al. Activity of 7-methyljuglone derivatives against Mycobacterium tuberculosis and as subversive substrates for mycothiol disulfide reductase. Bioorganic Med Chem 2007; 15: 7638–7646. [DOI] [PubMed] [Google Scholar]
- Miao H, Zhao L, Li C, Shang Q, Lu H, Fu Z et al. Inhibitory effect of shikonin on Candida albicans growth. Biol Pharm Bull 2012; 35: 1956–1963. [DOI] [PubMed] [Google Scholar]
- Reddy PJ, Ray S, Sathe GJ, Prasad TSK, Rapole S, Panda D et al. Proteomics analyses of Bacillus subtilis after treatment with plumbagin, a plant-derived naphthoquinone. Omi A J Integr Biol 2015; 19: 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Christner C, Kipping M, Schelbert B, Rücknagel KP, Grabley S et al. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry 1998; 37: 5953–5960. [DOI] [PubMed] [Google Scholar]
- Chao SH, Greenleaf AL, Price DH. Juglone, an inhibitor of the peptidyl-prolyl isomerase Pin1, also directly blocks transcription. Nucleic Acids Res 2001; 29: 767–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie TJ, Musgrave OC, Skoyles D. Ebenaceae extractives. Part V. New diospyrin derivatives from Diospyros montana Roxb. J Chem Soc Perkin Trans 1976; 1: 2155–2161. [Google Scholar]
- Karkare S, Chung TTH, Collin F, Mitchenall LA, McKay AR, Greive SJ et al. The naphthoquinone diospyrin is an inhibitor of DNA gyrase with a novel mechanism of action. J Biol Chem 2013; 288: 5149–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Liu Z, Guo A, Liang J, Guo N, Zeng F et al. Global transcriptional profiles of Mycobacterium tuberculosis treated with plumbagin. World J Microbiol Biotechnol 2011; 27: 2261–2269. [Google Scholar]
- Fischer TC, Gosch C, Mirbeth B, Gselmann M, Thallmair V, Stich K. Potent and specific bactericidal effect of juglone (5-Hydroxy-1,4- naphthoquinone) on the fire blight pathogen Erwinia amylovora. J Agric Food Chem 2012; 60: 12074–12081. [DOI] [PubMed] [Google Scholar]
- Shin K-S, Lee S-K, Cha B-J. Antifungal Activity of Plumbagin purified from leaves of Nepenthes ventricosa x maxima against phytopathogenic fungi. Plant Pathol J 2007; 23: 113–115. [Google Scholar]
- Rettenmaier H, Kupas U, Lingens F. Degradation of juglone by Pseudomonas putida J 1. FEMS Microbiol Lett 1983; 19: 193–195. [Google Scholar]
- Achatz M, Morris EK, Müller F, Hilker M, Rillig MC. Soil hypha-mediated movement of allelochemicals: Arbuscular mycorrhizae extend the bioactive zone of juglone. Funct Ecol 2014; 28: 1020–1029. [Google Scholar]
- Achatz M, Rillig MC. Arbuscular mycorrhizal fungal hyphae enhance transport of the allelochemical juglone in the field. Soil Biol Biochem 2014; 78: 76–82. [Google Scholar]
- Buch F, Rott M, Rottloff S, Paetz C, Hilke I, Raessler M et al. Secreted pitfall-trap fluid of carnivorous Nepenthes plants is unsuitable for microbial growth. Ann Bot 2013; 111: 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara H, Endou F, Furukawa S, Matsufuji H, Suzuki K, Anzai H. Antimicrobial activity of the carnivorous plant. Biocontrol Sci 2013; 18: 151–155. [DOI] [PubMed] [Google Scholar]
- Chan X-Y, Hong K-W, Yin W-F, Chan K-G. Microbiome and biocatalytic bacteria in monkey cup (Nepenthes pitcher) Digestive Fluid. Sci Rep 2016; 6: 20016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-N, Syu W-J, Sun W-SW, Chen J-W, Chen T-H, Don M-J et al. A role of ygfZ in the Escherichia coli response to plumbagin challenge. J Biomed Sci 2010; 17: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MJ, Smith SL. Effects of the chitin synthetase inhibitor plumbagin and its 2-demethyl derivative juglone on insect ecdysone 20-monooxygenase activity. Experientia 1988; 44: 990–991. [DOI] [PubMed] [Google Scholar]
- Mitchell MJ, Brescia AI, Smith SL, Morgan ED. Effects of the compounds 2-methoxynaphthoquinone, 2-propoxynaphthoquinone, and 2-isopropoxynaphthoquinone on ecdysone 20-monooxygenase activity. Arch Insect Biochem Physiol 2007; 66: 45–52. [DOI] [PubMed] [Google Scholar]
- Tokunaga T, Dohmura A, Takada N, Ueda M. Cytotoxic antifeedant from Dionaea muscipula Ellis: a defensive mechanism of carnivorous plants against predators. Bull Chem Soc Jpn 2004; 77: 537–541. [Google Scholar]
- Tokunaga T, Takada N, Ueda M. Mechanism of antifeedant activity of plumbagin, a compound concerning the chemical defense in carnivorous plant. Tetrahedron Lett 2004; 45: 7115–7119. [Google Scholar]
- Thiboldeaux RL, Lindroth RL, Tracy JW. Differential toxicity of juglone (5-hydroxy-1,4-naphthoquinone) and related naphthoquinones to saturniid moths. J Chem Ecol 1994; 20: 1631–1641. [DOI] [PubMed] [Google Scholar]
- Piskorski R, Dorn S. How the oligophage codling moth Cydia pomonella survives on walnut despite its secondary metabolite juglone. J Insect Physiol 2011; 57: 744–750. [DOI] [PubMed] [Google Scholar]
- Piskorski R, Ineichen S, Dorn S. Ability of the oriental fruit moth Grapholita molesta (Lepidoptera: Tortricidae) to detoxify juglone, the main secondary metabolite of the non-host plant walnut. J Chem Ecol 2011; 37: 1110–1116. [DOI] [PubMed] [Google Scholar]
- El-Sayed AM, Byers JA, Suckling DM. Pollinator-prey conflicts in carnivorous plants: when flower and trap properties mean life or death. Sci Rep 2016; 6: 21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galek H, Osswald WF, Elstner EF. Oxidative protein modification as predigestive mechanism of the carnivorous plant Dionaea muscipula: an hypothesis based on in vitro experiments. Free Radic Biol Med 1990; 9: 427–434. [DOI] [PubMed] [Google Scholar]
- Nahálka J, Nahálková J, Gemeiner P, Blanárik P. Elicitation of plumbagin by chitin and its release into the medium in Drosophyllum lusitanicum Link. suspension cultures. Biotechnol Lett 1998; 20: 841–845. [Google Scholar]
- Thomson RH. Naphthaquinones. In: Thomson RH (ed.) Naturally Occurring Quinones. Netherlands: Elsevier, 1971: 198–366. [Google Scholar]
- Chaudhary G, Goyal S, Poonia P, Linn L. Lawsonia inermis Linnaeus: a phytopharmacological review. Int J Pharm Sci Drug Res 2010; 2: 91–98. [Google Scholar]
- Singh DK, Luqman S. Lawsonia inermis (L.): a perspective on anticancer potential of mehndi/henna. Biomed Res Ther 2014; 1: 112–120. [Google Scholar]
- Singh DK, Luqman S, Mathur AK. Lawsonia inermis L.—a commercially important primaeval dying and medicinal plant with diverse pharmacological activity: a review. Ind Crops Prod 2015; 65: 269–286. [Google Scholar]
- Badoni Semwal R, Semwal DK, Combrinck S, Cartwright-Jones C, Viljoen A. Lawsonia inermis L. (henna): ethnobotanical, phytochemical and pharmacological aspects. J Ethnopharmacol 2014; 155: 80–103. [DOI] [PubMed] [Google Scholar]
- Chauhan MA. A review on morphology, phytochemistry and pharmacological activities of medicinal herb Plumbago Zeylanica Linn. J Pharmacogn Phytochem 2014; 3: 95–118. [Google Scholar]
- Gaascht F, Dicato M, Diederich M. Venus flytrap (Dionaea muscipula Solander ex Ellis) contains powerful compounds that prevent and cure cancer. Front Oncol 2013; 3: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Yang L, Oppenheim JJ, Howard OMZ. Cellular pharmacology studies of shikonin derivatives. Phyther Res 2002; 16: 199–209. [DOI] [PubMed] [Google Scholar]
- Papageorgiou VP, Assimopoulou AN, Couladouros Ea, Hepworth D, Nicolaou KC. The chemistry and biology of alkannin, shikonin, and related naphthazarin natural products. Angew Chem Int Ed 1999; 38: 270–300. [DOI] [PubMed] [Google Scholar]
- Papageorgiou VP, Assimopoulou AN, Ballis AC. Alkannins and shikonins: a new class of wound healing agents. Curr Med Chem 2008; 15: 3248–3267. [DOI] [PubMed] [Google Scholar]
- Andújar I, Ríos JL, Giner RM, Recio MC. Pharmacological properties of shikonin—a review of literature since 2002. Planta Med 2013; 79: 1685–1697. [DOI] [PubMed] [Google Scholar]
- Padhye S, Dandawate P, Yusufi M, Ahmad A, Sarkar FH. Perspectives on medicinal properties of plumbagin and its analogs. Med Res Rev 2012; 32: 1131–1158. [DOI] [PubMed] [Google Scholar]
- Mbaveng AT, Kuete V. Review of the chemistry and pharmacology of 7-Methyljugulone. Afr Health Sci 2014; 14: 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiran Aithal B, Sunil Kumar MR, Nageshwar Rao B, Udupa N, Satish Rao BS. Juglone, a naphthoquinone from walnut, exerts cytotoxic and genotoxic effects against cultured melanoma tumor cells. Cell Biol Int 2009; 33: 1039–1049. [DOI] [PubMed] [Google Scholar]
- Murakami K, Haneda M, Iwata S, Yoshino M. Effect of hydroxy substituent on the prooxidant action of naphthoquinone compounds. Toxicol Vitr 2010; 24: 905–909. [DOI] [PubMed] [Google Scholar]
- Klaus V, Hartmann T, Gambini J, Graf P, Stahl W, Hartwig A et al. 1,4-Naphthoquinones as inducers of oxidative damage and stress signaling in HaCaT human keratinocytes. Arch Biochem Biophys 2010; 496: 93–100. [DOI] [PubMed] [Google Scholar]
- Inbaraj JJ, Chignell CF. Cytotoxic action of juglone and plumbagin: a mechanistic study using HaCaT keratinocytes. Chem Res Toxicol 2004; 17: 55–62. [DOI] [PubMed] [Google Scholar]
- Powolny AA, Singh SV. Plumbagin-induced apoptosis in human prostate cancer cells is associated with modulation of cellular redox status and generation of reactive oxygen species. Pharm Res 2008; 25: 2171–2180. [DOI] [PubMed] [Google Scholar]
- Chen C-H, Lin M-L, Ong P-L, Yang J-T. Novel multiple apoptotic mechanism of shikonin in human glioma cells. Ann Surg Oncol 2012; 19: 3097–3106. [DOI] [PubMed] [Google Scholar]
- Yang Q, Ji M, Guan H, Shi B, Hou P. Shikonin inhibits thyroid cancer cell growth and invasiveness through targeting major signaling pathways. J Clin Endocrinol Metab 2013; 98: 1909–1917. [DOI] [PubMed] [Google Scholar]
- Gao D, Hiromura M, Yasui H, Sakurai H. Direct reaction between shikonin and thiols induces apoptosis in HL60 cells. Biol Pharm Bull 2002; 25: 827–832. [DOI] [PubMed] [Google Scholar]
- Zhang B, Chen N, Chen H, Wang Z, Zheng Q. The critical role of redox homeostasis in shikonin-induced HL-60 cell differentiation via unique modulation of the Nrf2/ARE pathway. Oxid Med Cell Longev 2012; 2012: 781516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambhir L, Checker R, Thoh M, Patwardhan RS, Sharma D, Kumar M et al. 1,4-naphthoquinone, a pro-oxidant, suppresses immune responses via KEAP-1 glutathionylation. Biochem Pharmacol 2014; 88: 95–105. [DOI] [PubMed] [Google Scholar]
- Bryan HK, Olayanju A, Goldring CE, Park BK. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 2013; 85: 705–717. [DOI] [PubMed] [Google Scholar]
- Abiko Y, Kumagai Y. Interaction of Keap1 modified by 2-tert-Butyl-1,4-benzoquinone with GSH: evidence for S-transarylation. Chem Res Toxicol 2013; 26: 1080–1087. [DOI] [PubMed] [Google Scholar]
- Carvalho AN, Marques C, Guedes RC, Castro-Caldas M, Rodrigues E, van Horssen J et al. S-Glutathionylation of Keap1: a new role for glutathione S-transferase pi in neuronal protection. FEBS Lett 2016; 590: 1455–1466. [DOI] [PubMed] [Google Scholar]
- Son TG, Camandola S, Arumugam TV, Cutler RG, Telljohann RS, Mughal MR et al. Plumbagin, a novel Nrf2/ARE activator, protects against cerebral ischemia. J Neurochem 2010; 112: 1316–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K, Furusawa Y, Tabuchi Y, Emam HF, Piao J-L, Hassan MA et al. Chemical inducers of heat shock proteins derived from medicinal plants and cytoprotective genes response. Int J Hyperthermia 2012; 28: 1–8. [DOI] [PubMed] [Google Scholar]
- Abed DA, Goldstein M, Albanyan H, Jin H, Hu L. Discovery of direct inhibitors of Keap1-Nrf2 protein-protein interaction as potential therapeutic and preventive agents. Acta Pharm Sin B 2015; 5: 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi EH, Suzuki T, Funayama R, Nagashima T, Hayashi M, Sekine H et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun 2016; 7: 11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS. Signaling to NF- B. Genes Dev 2004; 18: 2195–2224. [DOI] [PubMed] [Google Scholar]
- Checker R, Sharma D, Sandur SK, Khanam S, Poduval TB. Anti-inflammatory effects of plumbagin are mediated by inhibition of NF-κB activation in lymphocytes. Int Immunopharmacol 2009; 9: 949–958. [DOI] [PubMed] [Google Scholar]
- Checker R, Patwardhan RS, Sharma D, Menon J, Thoh M, Sandur SK et al. Plumbagin, a vitamin K3 analogue, abrogates lipopolysaccharide-induced oxidative stress, inflammation and endotoxic shock via NF-κB suppression. Inflammation 2014; 37: 542–554. [DOI] [PubMed] [Google Scholar]
- Sandur SK, Ichikawa H, Sethi G, Ahn KS, Aggarwal BB. Plumbagin (5-Hydroxy-2-methyl-1,4-naphthoquinone) suppresses NF-κB activation and NF-κB-regulated gene products through modulation of p65 and IκBa kinase activation, leading to potentiation of apoptosis induced by cytokine and chemotherapeutic agents. J Biol Chem 2006; 281: 17023–17033. [DOI] [PubMed] [Google Scholar]
- Luo P, Wong YF, Ge L, Zhang ZF, Liu Y, Liu L et al. Anti-inflammatory and analgesic effect of plumbagin through inhibition of nuclear factor-κB activation. J Pharmacol Exp Ther 2010; 335: 735–742. [DOI] [PubMed] [Google Scholar]
- Li J, Shen L, Lu F, Qin Y, Chen R, Li J et al. Plumbagin inhibits cell growth and potentiates apoptosis in human gastric cancer cells in vitro through the NF-κB signaling pathway. Acta Pharmacol Sin 2012; 33: 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Furumura M, Gouya T, Iwanaga A, Teye K, Numata S et al. Shikonin promotes skin cell proliferation and inhibits nuclear factor-κB translocation via proteasome inhibition in vitro. Chin Med J (Engl) 2015; 128: 2228–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andújar I, Recio M, Bacelli T, Giner R, Ríos J. Shikonin reduces oedema induced by phorbol ester by interfering with IκBα degradation thus inhibiting translocation of NF-κB to the nucleus. Br J Pharmacol 2010; 160: 376–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii N, Yamashita Y, Arima Y, Nagashima M, Nakano H. Induction of topoisomerase II-mediated DNA cleavage by the plant naphthoquinones plumbagin and shikonin. Antimicrob Agents Chemother 1992; 36: 2589–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman B, Marton LJ, Sun JS, Neder K, Witiak DT, Liu AA et al. Induction of DNA topoisomerase II-mediated DNA cleavage by beta-lapachone and related naphthoquinones. Cancer Res 1997; 57: 620–627. [PubMed] [Google Scholar]
- Ray S, Hazra B, Mittra B, Das A, Majumder HK. Diospyrin, a bisnaphthoquinone: a novel inhibitor of type I DNA topoisomerase of Leishmania donovani. Mol Pharmacol 1998; 54: 994–999. [DOI] [PubMed] [Google Scholar]
- Plyta ZF, Li T, Papageorgiou VP, Mellidis AS, Assimopoulou AN, Pitsinos EN et al. Inhibition of topoisomerase I by naphthoquinone derivatives. Bioorg Med Chem Lett 1998; 8: 3385–3390. [DOI] [PubMed] [Google Scholar]
- Kennedy S, DiCesare JC, Sheaff RJ. Topoisomerase I inactivation by a novel thiol reactive naphthoquinone. Biochem Biophys Res Commun 2011; 410: 152–158. [DOI] [PubMed] [Google Scholar]
- Gurbani D, Kukshal V, Laubenthal J, Kumar A, Pandey A, Tripathi S et al. Mechanism of inhibition of the ATpase domain of human topoisomerase IIα by 1,4-benzoquinone, 1,2-naphthoquinone, 1,4-naphthoquinone, and 9,10-phenanthroquinone. Toxicol Sci 2012; 126: 372–390. [DOI] [PubMed] [Google Scholar]
- Virupaksha B, Alpana G, Prashant K, Uday D, Alessandro D. Analysis of naphthoquinone derivatives as topoisomerase I inhibitors using fragment based QSAR. J Cheminform 2013; 5: P22. [Google Scholar]
- Zhang FL, Wang P, Liu YH, Liu LB, Liu XB, Li Z et al. Topoisomerase I inhibitors, shikonin and topotecan, inhibit growth and induce apoptosis of glioma cells and glioma stem cells. PLoS One 2013; 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rialdi A, Campisi L, Zhao N, Lagda AC, Pietzsch C, Ho JSY et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science 2016; 352: aad7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fila C, Metz C, Van Der Sluijs P. Juglone inactivates cysteine-rich proteins required for progression through mitosis. J Biol Chem 2008; 283: 21714–21724. [DOI] [PubMed] [Google Scholar]
- Boussetta T, Gougerot-Pocidalo MA, Hayem G, Ciappelloni S, Raad H, Derkawi RA et al. The prolyl isomerase Pin1 acts as a novel molecular switch for TNF-a-induced priming of the NADPH oxidase in human neutrophils. Blood 2010; 116: 5795–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault S, Braun RK, Shen ZJ, Xiang Z, Heninger E, Love RB et al. Pin1 modulates the type 1 immune response. PLoS One 2007; 2: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Fan Y, Gupta N, Matsuura I, Liu F, Xiao ZZ et al. Peptidyl-prolyl isomerase Pin1 markedly enhances the oncogenic activity of the Rel proteins in the nuclear factor-κB family. Cancer Res 2009; 69: 4589–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li Z, Zhang X, Wang S, Zhu C, Miao J et al. Protective effect of shikonin in experimental ischemic stroke: Attenuated TLR4, p-p38MAPK, NF-κB, TNF-α and MMP-9 expression, up-regulated claudin-5 expression, ameliorated BBB permeability. Neurochem Res 2014; 39: 97–106. [DOI] [PubMed] [Google Scholar]
- Andújar I, Ríos JL, Giner RM, Miguel Cerdá J, Recio MDC. Beneficial effect of shikonin on experimental colitis induced by dextran sulfate sodium in Balb/C mice. Evid Based Complement Alternat Med 2012; 2012: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andújar I, Ríos JL, Giner RM, Recio MC. Shikonin promotes intestinal wound healing in vitro via induction of TGF-β release in IEC-18 cells. Eur J Pharm Sci 2013; 49: 637–641. [DOI] [PubMed] [Google Scholar]
- Chiu S-C, Yang N-S. Inhibition of tumor necrosis factor-a through selective blockade of pre-mRNA splicing by shikonin. Mol Pharmacol 2007; 71: 1640–1645. [DOI] [PubMed] [Google Scholar]
- Liu L, Wu Y, Cao K, Xu Y-Y, Gao X-H, Chen H-D et al. Shikonin inhibits IFN-γ-induced K17 over-expression of HaCaT cells by interfering with STAT3 signaling. Int J Clin Exp Pathol 2015; 8: 9202–9207. [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang J, Yang Q, Wu S, Yang Z, Zhu H et al. Shikonin inhibits the lipopolysaccharide-induced release of HMGB1 in RAW264.7 cells via IFN and NF-κB signaling pathways. Int Immunopharmacol 2014; 19: 81–87. [DOI] [PubMed] [Google Scholar]
- Thakur R, Trivedi R, Rastogi N, Singh M, Mishra DP. Inhibition of STAT3, FAK and Src mediated signaling reduces cancer stem cell load, tumorigenic potential and metastasis in breast cancer. Sci Rep 2015; 5: 10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu X, Gao X, Chen H, Geng L. Shikonin suppresses IL-17-induced VEGF expression via blockage of JAK2/STAT3 pathway. Int Immunopharmacol 2014; 19: 327–333. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Assimopoulou AN, Klauck SM, Damianakos H, Chinou I, Kretschmer N et al. Inhibition of c-MYC with involvement of ERK/JNK/MAPK and AKT pathways as a novel mechanism for shikonin and its derivatives in killing leukemia cells. Oncotarget 2015; 6: 38934–38951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hayashi S, Umezaki M, Yamamoto T, Kageyama-Yahara N, Kondo T et al. Shikonin, a constituent of Lithospermum erythrorhizon exhibits anti-allergic effects by suppressing orphan nuclear receptor Nr4a family gene expression as a new prototype of calcineurin inhibitors in mast cells. Chem Biol Interact 2014; 224: 117–127. [DOI] [PubMed] [Google Scholar]
- Staniforth V, Wang SY, Shyur LF, Yang NS. Shikonins, phytocompounds from Lithospermum erythrorhizon, inhibit the transcriptional activation of human tumor necrosis factor α promoter in vivo. J Biol Chem 2004; 279: 5877–5885. [DOI] [PubMed] [Google Scholar]
- Komi Y, Suzuki Y, Shimamura M, Kajimoto S, Nakajo S, Masuda M et al. Mechanism of inhibition of tumor angiogenesis by beta-hydroxyisovalerylshikonin. Cancer Sci 2009; 100: 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xie J, Jiang Z, Wang B, Wang Y, Hu X. Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2. Oncogene 2011; 30: 4297–4306. [DOI] [PubMed] [Google Scholar]
- Pile JE, Navalta JW, Davis CD, Sharma NC. Interventional effects of plumbagin on experimental ulcerative colitis in mice. J Nat Prod 2013; 76: 1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Jing J, Bai Y, Li Z, Liu L, Luo J et al. Amelioration of experimental autoimmune encephalomyelitis by plumbagin through down-regulation of JAK-STAT and NF-κB signaling pathways. PLoS One 2011; 6: e27006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya BR, Bhattacharyya B, Chakrabarti G. The natural naphthoquinone plumbagin exhibits antiproliferative activity and disrupts the microtubule network through tubulin binding. Biochemistry 2008; 47: 7838–7845. [DOI] [PubMed] [Google Scholar]
- Chen A, Zhou X, Tang S, Liu M, Wang X. Evaluation of the inhibition potential of plumbagin against cytochrome P450 using LC-MS/MS and cocktail approach. Sci Rep 2016; 6: 28482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumsakul W, Chaijaroenkul W, Na-Bangchang K. In vitro inhibitory effects of plumbagin, the promising antimalarial candidate, on human cytochrome P450 enzymes. Asian Pac J Trop Med 2015; 8: 914–918. [DOI] [PubMed] [Google Scholar]
- Khaw AK, Sameni S, Venkatesan S, Kalthur G, Hande MP. Plumbagin alters telomere dynamics, induces DNA damage and cell death in human brain tumour cells. Mutat Res Toxicol Environ Mutagen 2015; 793: 86–95. [DOI] [PubMed] [Google Scholar]
- Wang F, Wang Q, Zhou Z-W, Yu S-N, Pan S-T, He Z-X et al. Plumbagin induces cell cycle arrest and autophagy and suppresses epithelial to mesenchymal transition involving PI3K/Akt/mTOR-mediated pathway in human pancreatic cancer cells. Drug Des Devel Ther 2015; 9: 537–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S-T, Qin Y, Zhou Z-W, He Z, Zhang X, Yang T et al. Plumbagin induces G2/M arrest, apoptosis, and autophagy via p38 MAPK- and PI3K/Akt/mTOR-mediated pathways in human tongue squamous cell carcinoma cells. Drug Des Devel Ther 2015; 9: 1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal MS, Parveen S, Beg MA, Suhail M, Chaudhary AGA, Damanhouri GA et al. Anticancer compound plumbagin and its molecular targets: a structural insight into the inhibitory mechanisms using computational approaches. PLoS One 2014; 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thasni KA, Ratheeshkumar T, Rojini G, Sivakumar KC, Nair RS, Srinivas G et al. Structure activity relationship of plumbagin in BRCA1 related cancer cells. Mol Carcinog 2013; 52: 392–403. [DOI] [PubMed] [Google Scholar]
- Onodera T, Kuriyama I, Sakamoto Y, Kawamura M, Kuramochi K, Tsubaki K et al. 5-O-Acyl plumbagins inhibit DNA polymerase activity and suppress the inflammatory response. Arch Biochem Biophys 2015; 573: 100–110. [DOI] [PubMed] [Google Scholar]
- Kawiak A, Lojkowska E. Ramentaceone, a naphthoquinone derived from Drosera sp., induces apoptosis by suppressing PI3K/Akt signaling in breast cancer cells. PLoS One 2016; 11: e0147718. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Peng X, Nie Y, Wu J, Huang Q, Cheng Y. Juglone prevents metabolic endotoxemia-induced hepatitis and neuroinflammation via suppressing TLR4/NF-κB signaling pathway in high-fat diet rats. Biochem Biophys Res Commun 2015; 462: 1–6. [DOI] [PubMed] [Google Scholar]
- Choi WH, Hong SS, Lee SA, Han XH, Lee KS, Lee MK et al. Monoamine oxidase inhibitory naphthoquinones from the roots of Lithospermum erythrorhizon. Arch Pharm Res 2005; 28: 400–404. [DOI] [PubMed] [Google Scholar]
- Coelho-Cerqueira E, Netz PA, Do Canto VP, Pinto AC, Follmer C. Beyond topoisomerase inhibition: antitumor 1,4-naphthoquinones as potential inhibitors of human monoamine oxidase. Chem Biol Drug Des 2014; 83: 401–410. [DOI] [PubMed] [Google Scholar]
- Mostert S, Petzer A, Petzer JP. Evaluation of natural and synthetic 1,4-naphthoquinones as inhibitors of monoamine oxidase. Chem Biol Drug Des 2016; 87: 737–746. [DOI] [PubMed] [Google Scholar]
- Lamé MW, Jones AD, Wilson DW, Segall HJ. Protein targets of 1,4-benzoquinone and 1,4-naphthoquinone in human bronchial epithelial cells. Proteomics 2003; 3: 479–495. [DOI] [PubMed] [Google Scholar]
- Qiu JX, Zhou ZW, He ZX, Zhao RJ, Zhang X, Yang L et al. Plumbagin elicits differential proteomic responses mainly involving cell cycle, apoptosis, autophagy, and epithelial-to-mesenchymal transition pathways in human prostate cancer PC-3 and DU145 cells. Drug Des Devel Ther 2015; 9: 349–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz L-O. Reactive oxygen species as initiators and mediators of cellular signaling processes. In: Roberts SM et al (ed.) Studies on Experimental Toxicology and Pharmacology. Switzerland: Springer International Publishing, 2015: 149–171. [Google Scholar]
- Kishore N, Binneman B, Mahapatra A, Van De Venter M, Du Plessis-Stoman D, Boukes G et al. Cytotoxicity of synthesized 1,4-naphthoquinone analogues on selected human cancer cell lines. Bioorganic Med Chem 2014; 22: 5013–5019. [DOI] [PubMed] [Google Scholar]
- Wang S-H, Lo C-Y, Gwo Z-H, Lin H-J, Chen L-G, Kuo C-D et al. Synthesis and biological evaluation of lipophilic 1,4-naphthoquinone derivatives against human cancer cell lines. Molecules 2015; 20: 11994–12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Chen XF, Liu J, Lin HY, Han HW, Liu HC et al. Novel shikonin derivatives targeting tubulin as anticancer agents. Chem Biol Drug Des 2014; 84: 603–615. [DOI] [PubMed] [Google Scholar]
- Kyle Hadden M, Hill SA, Davenport J, Matts RL, Blagg BSJ. Synthesis and evaluation of Hsp90 inhibitors that contain the 1,4-naphthoquinone scaffold. Bioorganic Med Chem 2009; 17: 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura Pinto A, Lisboa de Castro S. The trypanocidal activity of naphthoquinones: a review. Molecules 2009; 14: 4570–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogindo CO, Khraiwesh MH, George M, Brandy Y, Brandy N, Gugssa A et al. Novel drug design for Chagas disease via targeting Trypanosoma cruzi tubulin: homology modeling and binding pocket prediction on Trypanosoma cruzi tubulin polymerization inhibition by naphthoquinone derivatives. Bioorg Med Chem 2016; 24: 3849–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieretti S, Haanstra JR, Mazet M, Perozzo R, Bergamini C, Prati F et al. Naphthoquinone derivatives exert their antitrypanosomal activity via a multi-target mechanism. PLoS Negl Trop Dis 2013; 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad KR, Babu KS, Rao RR, Suresh G, Rekha K, Murthy JM et al. Synthesis and insect antifeedant activity of plumbagin derivatives. Med Chem Res 2012; 21: 578–583. [Google Scholar]
- Borges AA, Jiménez-Arias D, Expósito-Rodríguez M, Sandalio LM, Pérez JA. Priming crops against biotic and abiotic stresses: MSB as a tool for studying mechanisms. Front Plant Sci 2014; 5: 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duru N, Gernapudi R, Zhou Q. Chemopreventive activities of shikonin in breast cancer. Biochem Pharmacol Open Access 2014; 03: 3–4. [Google Scholar]
- Tabata M. The mechanism of shikonin biosynthesis in Lithospermum cell cultures. Plant Tissue Cult Lett 1996; 13: 117–125. [Google Scholar]
- Weston LA, Ryan PR, Watt M. Mechanisms for cellular transport and release of allelochemicals from plant roots into the rhizosphere. J Exp Bot 2012; 63: 3445–3454. [DOI] [PubMed] [Google Scholar]
- Tatsumi K, Yano M, Kaminade K, Sugiyama A, Sato M, Toyooka K et al. Characterization of shikonin derivative secretion in Lithospermum erythrorhizon hairy roots as a model of lipid-soluble metabolite secretion from plants. Front Plant Sci 2016; 7: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]