Abstract
Aberrant expression and activation of FGFR3 is associated with disease states including bone dysplasia and malignancies of bladder, cervix, and bone marrow. MS analysis of protein-phosphotyrosine in multiple myeloma cells revealed a prevalent phosphorylated motif, D/EYYR/K, derived from the kinase domain activation loops of tyrosine kinases including FGFR3 corresponding to a recognition sequence of phosphotyrosine phosphatases PTPN1. Knockdown of PTPN1 or the related enzyme PTPN2 by RNAi resulted in ligand-independent activation of FGFR3. Modulation of FGFR3 activation loop phosphorylation by both PTPN1 and PTPN2 was a function of receptor trafficking and PTP compartmentalization. The FGFR3 activation loop motif DYYKK650 is altered to DYYKE650 in the oncogenic variant FGFR3K650E, and consequently it is constitutively fully activated and unaffected by activation loop phosphorylation. FGFR3K650E was nevertheless remarkably sensitive to negative regulation by PTPN1 and PTPN2. This suggests that in addition to modulating FGFR3 phosphorylation, PTPN1 and PTPN2 constrain the kinase domain by fostering an inactive-state. Loss of this constraint in response to ligand or impaired PTPN1/N2 may initiate FGFR3 activation. These results suggest a model wherein PTP expression levels may define conditions that select for ectopic FGFR3 expression and activation during tumorigenesis.
Keywords: activation loop, endocytosis, glycosylation, multiple myeloma, phosphorylation, trafficking, tyrosine kinase
1 Introduction
Fibroblast growth factor receptor 3 (FGFR3) is one of four members of the FGFR family of receptor tyrosine kinases (RTKs). Approximately 15 % of multiple myeloma (MM) are associated with a t(4;14) translocation, generally resulting in aberrant expression of FGFR3. In some cases FGFR3 point mutations render it to some extent constitutively active and ligand-independent [1, 2]. In this context, FGFR3 acts as an oncogene involved in tumor progression and is considered a therapeutic target in MM [3-5], which despite therapeutic advances remains an incurable, fatal disease.
FGFRs are temporally and spatially regulated, which involves intracellular trafficking, protein-protein interactions, and post-translational modifications. Nascent FGF receptors are processed through the endoplasmic reticulum (ER) and Golgi apparatus in order to guide their N-glycosylation and localization to the plasma membrane. FGFR3 activation and signaling, similar to general models for RTKs, is normally initiated by dimerization as a consequence of ligand binding. This relieves inhibitory constraints on kinase activity, which have not been fully defined, resulting in an ordered series of trans-auto-phosphorylations that stimulate enzymatic activity and generate binding sites for signaling proteins containing phosphotyrosine (pY) binding domains (e.g. SH2 and PTB) [6]. The resulting pY-dependent protein-protein interactions trigger cellular responses by stimulating effector pathways involving RAS-ERK, PLCγ, and PI3-K [5, 7-9]. From the cell surface, FGFRs are subject to ligand-stimulated endocytosis, ubiquitination-dependent endosome sorting, and then recycling to the cell surface or lysosome-mediated degradation [10]. Perturbations in the down-regulation of activated RTKs by endosome-lysosome trafficking can lead to inappropriate, oncogenic signaling [see 11, 12].
Strict control of FGFR3 phosphorylation is critical for its signaling functions. Ligand-activated FGFR dimers are associated with phosphorylated tandem tyrosine residues in the kinase domain activation loop (A-loop; pY647pY648 in FGFR3), which stabilizes an active-state conformation that is approximately 500-fold activated relative to the non-phosphorylated kinase domain, and 50-fold activated compared to a singly-phosphorylated kinase domain [7, 13]. The FGFR3 kinase domain adopts a virtually identical activate-state structure in the absence of A loop phosphorylation as a consequence of a K-to-E substitution adjacent to the tandem tyrosines, which changes the A-loop sequence from DYYKK650 to DYYKE650 [14]. The FGFR3K650E kinase domain is constitutively fully activated, and does not require A-loop phosphorylation [15, 16]. K650E results from an oncogenic somatic event in MM and other malignancies and causes developmental skeletal dysplasia including the neonatal lethal syndrome Thanatophoric Dysplasia Type II, in germline contexts [1, 2, 17-20]. RTK kinase activity is normally tempered during nascent processing and during endocytosis by the ER-associated protein-phosphotyrosine (pY) phosphatase PTPN1 (PTP-1B) [21-23]. PTPN1 is anchored to the ER through a C-terminal hydrophobic tail [21]. The structurally-related enzyme PTPN2 (TC-PTP) expresses a 48 kDa splice variant (TC48) anchored to the ER, and a 45-kDA variant (TC45) that translocates from the nucleus to the cytoplasm in response to various extracellular stimuli [24-26]. PTPN1 and PTPN2 interact with common substrates such as RTKs, including EGFR [26-28], insulin receptor (IR) [29, 30], MET [31], PDGFR [32-34], VEGFR [35, 36], and CSF1R [37, 38]. However, PTPN1 and PTPN2 fulfill non-redundant roles, and in some instances this is manifest through shared substrates [reviewed in ref. 39]. PTPN1, along with the SH2-containing PTPs (PTN6, PTPN11) and PTPN7 were identified as major determinants of protein-pY patterns in a set of MM cell lines [40]. Therefore, the regulation of RTKs by PTPs is pervasive and dependent on spatial and temporal contexts [41].
A comprehensive analysis of protein-pY in MM models defined a network of proteins modulated by FGFR3 and PTPs including subsets involved in trafficking and signaling [42]. Herein we report that FGFR3 is subject to context-dependent regulation by PTPN1 and PTPN2. Activated tyrosine kinases in FGFR3-expressing MM cells were found to share a tandem tyrosine motif corresponding to a PTPN1 and PTPN2 recognition sequence. Loss of either PTP resulted in ligand-independent activation of FGFR3 in a manner reflecting their co-localization. However, even a cytosolic, constitutively activated FGFR3 variant remained highly sensitive to negative regulation by PTPN1 and especially PTPN2. These data provide insight into how the cellular protein-pY profile arises as a function of both activated tyrosine kinase and PTP activities, and how dysregulation of PTP-mediated FGFR3 regulation may contribute to FGFR3-associated pathophysiology, including t(4;14) MM.
2 Materials and Methods
2.1 Cell culture and reagents
Human MM cells were cultured in IMDM (Gibco,), while HEK293 cells were maintained in DMEM (Gibco,) and both media types were supplemented with 10 % FBS (Hyclone Laboratories, Thermo Scientific, South Logan) and penicillin (10,000 U/ml)/streptomycin (10,000 mg/ml; Bioshop, Burlington). All cells were maintained in 5 % CO2 incubators at 37 °C. PD173074 (Pfizer, Ann Arbor, MI) treatment was performed at a final concentration of 100 nM for 4 h, essentially as described previously [3]. FGF1 (Cedarlane, R&D Systems) was used at 80 ng/ml with sodium heparin salt (30 μg/ml; Sigma) to activate FGFR3. Pervanadate (50 μM) was generated by oxidation of sodium orthovanadate (Calbiochem) with H2O2. Tunicamycin (Sigma-Aldrich) was used for 16 h at 1 μg/ml.
2.2 Generation of FGFR3 constructs
Carboxy-terminal FLAG-tagged FGFR3 constructs were generated from PCR (Herculase II Fusion, Agilent) of cDNA generated by reverse transcription (Superscript III ,Invitrogen) of RNA (Trizol, Invitrogen) extracted from murine B9 cells expressing WT and K650E FGFR3-IIIc. PCR products were then inserted into the mammalian expression vector pCDNA3.1(−)A (Invitrogen). C-terminal-truncated WT and K650E FGFR3 constructs with WT or mutant myristylation sequence were provided by Dr. Daniel Donoghue (University of California, San Diego). Selection for stable cell lines was performed by using G418 (800 μg/ml).
2.3 Immunofluorescence confocal microscopy
Cells were grown on poly-L-lysine-coated coverslips 24-well vessels, fixed with 3.7 % paraformaldehyde in PBS at room temperature for 20 minutes, and permeabilized with 0.2 % Triton X-100 and 5 % goat serum-containing PBS for 10 min. Coverslips were incubated with primary (FGFR3 (C15), 1:100, 1 h at 23 °C) and Alexa fluorescent secondary antibodies (1:100, 1 h at 23 °C) in 0.1 % Triton X-100 and PBS containing 5 % goat serum. Data acquisition was performed on a Zeiss LSM 510 META confocal microscope (Carl Zeiss, Inc., Thornwood, NY) with a 63x objective. Iterative restoration (95 % confidence with maximum of 20 iterations) of convoluted images was performed by using Volocity image analysis software (version 6; PerkinElmer).
2.4 Lentiviral production and cell infection for shRNA-mediated gene knockdown
Lentiviral particles containing target shRNA of interest were produced by transfecting 1 μg of hairpin-pLKO.1 plasmid, 100 ng of envelope plasmid pCMV-VSV-G and 900 ng of packaging plasmid pCMV-R8.74psPAX2 in the HEK293T cell helper system using FuGene6 (Roche) reagent. Two sequential viral harvests, 40- and 64-h post-transfection, of 6 ml each were performed. For each PTPN1 (GCTGCTCTGCTATATGCCTTA and TGCGACAGCTAGAATTGGAAA) and PTPN2 (GAAGATGTGAAGTCGTATTAT and GTGCAGTAGAATAGACATCAA) knockdown, 2 different hairpin sequences were tested and the hairpin displaying the most efficient knockdown was used for the study (PTPN1=GCTGCTCTGCTATATGCCTTA; PTPN2=GAAGATGTGAAGTCGTATTAT; see Supplementary Information, Figure S1). For infection, HEK293 cells were seeded in 6-well plates in 2 ml DMEM (FBS, P/S) supplemented with 8 μg/ml hexadimethrine bromide (Sigma). Lentiviral supernatant (200 μl from 12 ml preparation) was added and cells were incubated at 37 °C (5 % CO2) for 16 h. Media was then replaced with puromycin-containing (2 μg/ml) DMEM for selection of stably infected cells. Myeloma cells were infected essentially as previously described [reviewed in ref. 43]. Briefly, cells were plated in 2 ml (2×106 cells/ml) in 8 μg/ml hexadimethrine bromide (Sigma) in 6-well vessels. Lentiviral supernatant (200 μl of 12 ml preparation) was added to cells prior to centrifugation of culture vessels at 930 g for 90 min at 30 °C. Cells were then incubated at 37 °C (5 % CO2) for 16 h. Media was then replaced with puromycin-containing (2 μg/ml) IMDM. Knockdown efficiency was verified by Western blot.
2.5 Western blotting and immunoprecipitation sample preparations
Protein lysates for Western analysis were prepared by lysis of cells in Laemmli sample buffer. Following sonication to reduce viscosity, protein lysates were quantified using the RC DC Protein Assay (Bio-Rad). For protein immunoprecipitation, cell lysis was performed in buffer containing 1 % Triton X-100, 50 mM Hepes (pH 7.5), 150 mM NaCl, 10 % glycerol, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 1 mM sodium pyrophosphate, 1 mM sodium vanadate, 10 μg/ml leupeptin, 10 μg/ml aprotinin and 1 mM AEBSF. FGFR3 antibodies C-15 and B9 were from Santa Cruz Biotechnology, anti-phosphotyrosine 4G10 antibodies were purified from ascites fluid, anti-PTPN1 and anti-Flag M2 antibody beads were from Sigma, anti-PTPN2 was obtained from Calbiochem, and antibodies to IGF1R and insulin receptor (IR) were from Cell Signaling Technology (CST; Danvers, MA). Deglycosylation of FGFR3 with PNGase F (500 units) and/or Endo H (500 units; New-England Biolabs) was performed for 16 h according to the manufacturer’s protocol.
2.6 Affinity enrichment of peptides and liquid chromatography-tandem mass spectrometry
Enrichment of pY-modified peptides was performed by using PhosphoScan reagents (CST) essentially following the manufacturer recommendations [44]. Inline reversed-phase chromatography was performed using an integrated nano-LC system (Easy-nLC, Proxeon Biosystems A/S, Odense, Denmark). In the initial profile of protein-pY with MM cells, the LC system was coupled to a linear ion trap-Orbitrap MS system (LTQ-Orbitrap, Thermo, San Jose, CA). MS spectra were acquired at a resolution of 60,000 full-width half-maximum (FWHM) in profile mode in the Orbitrap whereas MS/MS spectra were acquired in the LTQ linear ion trap. Subsequent analyses of pY in PTPN1/PTPN2 knockdown cells was performed on a quadrupole-Orbitrap instrument (Q-Exactive, Thermo) with 70,000 FWHM in MS mode and 17,500 FWHM in MS/MS mode.
2.7 Database Searches
MS/MS samples analyzed on the LTQ-Orbitrap platform were searched using Mascot (Matrix Science). Mascot was set up to search the IPI human v3.87 database (91464 entries) assuming tryptic digest. Fragment ion mass tolerance of 0.50 Da and a parent ion tolerance of 10.0 ppm were used. For samples analyzed on the Q-Exactive, SEQUEST (Thermo Scientific) was used for database searching of the IPI human v3.83 database (93290 entries) with a parent ion tolerance of 15.0 ppm and 0.02 Da fragment ion mass tolerance. Carbamidomethylation of cysteine was specified as a fixed modification. Methionine oxidation and phosphorylation of serine, threonine and tyrosine were specified as variable modifications.
2.8 Peptide and protein identification
Scaffold software (v3.6.5, Proteome Software Inc., Portland, OR) was used to display and validate MS/MS-based peptide and protein identifications. The Peptide Prophet cut off was set at 95 % [45] and additionally, MS/MS spectra of all pY-peptides were individually inspected as previously described [46] to ensure high quality spectrum filtering. In addition, each phospho-peptide was assigned an Ascore [47] by using Scaffold PTM (v2.1.0, Proteome Software Inc., Portland, OR) as a measure of phosphorylation site assignment confidence. Motif analysis of identified tyrosine kinase pY-peptides was performed using the Motif Analysis Generator from PhosphoSitePlus® (www.phosphosite.org)[48].
3 Results
3.1 PTP regulation of protein-pY and tyrosine kinases in multiple myeloma cells
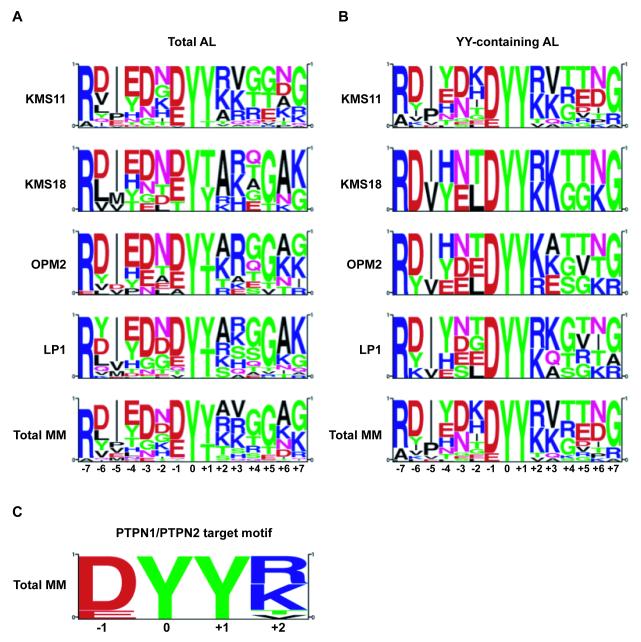
In order to reveal latent tyrosine kinase and PTP substrates, pY profiles of MM cells treated with the PTP inhibitor pervanadate were generated. This approach of global PTP inhibition has been applied with numerous cells types and technical platforms as a method to broadly potentiate proteome tyrosine phosphorylations [49-51], which quantitative MS analysis verified as being consistently naturally occurring modifications [52]. MS analysis of trypsin-digested lysates from MM cell lines KMS11 (Y373C), OPM2 (K650E), LP1 (WT) and KMS18 (G384D), which express various FGFR3 alleles (as indicated), was completed following enrichment for pY-containing peptides essentially as described previously [45, 47]. Fourteen pY-containing peptides derived from kinase domain A-loop regions corresponding to different protein tyrosine kinases were observed (Table 1). Phosphorylated A-loop peptides from FGFR3, IR/IGF1R and YES/LCK/FYN were observed in all 4 cell lines, and accounted for nearly 70 % of all MS/MS spectral counts of identified tyrosine kinase A-loop peptides. FGFR3 alone represented 34 % of all tyrosine kinase A-loop MS/MS events. Motif analysis revealed a tandem tyrosine motif in 8 of the 14 A-loop peptides corresponding to a perfect (FGFR3, IR/IGF1R, FAK2/PTK2B, MERTK/TYRO3, TYK2, FGFR2) or near-perfect (JAK1 and JAK3) D/EYYR/K consensus PTPN1 substrate sequence [53] (Fig. 1). Of these, the regulation of IR signaling by both PTPN1 and PTPN2 has been extensively characterized [reviewed in ref. 39], while targeting of IGF1-R and TYK2 by PTPN1, as well as interactions between PTPN2 and JAK1/JAK3 have also been reviewed [41]. Phospho-proteomic and PTPome analyses in the MM cell model KMS11 revealed a network of proteins subject to tyrosine phosphorylation downstream of constitutively dimerized, activated FGFR3Y373C [47], and implicated PTPN1 in the regulation of protein-pY in these cells [40].
Table 1. Protein tyrosine kinase A-loop pY peptides identified by LC-MS/MS in MM cells.
Phosphotyrosine-containing tryptic peptides were generated from the indicated multiple myeloma (MM) cell lines, and characterized by LC-MS/MS. Dots indicate the detection of the indicated A-loop peptide in a given cell type. As a rough estimation of relative abundance, The total number of MS/MS events (spectrum counts) for each phospho-peptide, summed across all the samples, is indicated in the last column.
| Protein Kinase1 | A-Loop Peptide2 | MM Cells: FGFR3: |
KMS11 Y373C |
OPM2 K650E |
LP1 WT |
KMS18 G384D |
MS/MS3 |
|---|---|---|---|---|---|---|---|
| FGFR3 | DVHNLDyyK | • | • | • | • | 82 | |
| IR/IGF1R | DIYETDyYRK | • | • | • | • | 40 | |
| YES/LCK/FYN | LIEDNEyTAR | • | • | • | • | 38 | |
| HCK/LYN | VIEDNEyTAR | • | • | • | 14 | ||
| JAK3 | LLPLDKDyYVVR | • | • | 12 | |||
| FAK2 | YIEDEDyYKASVTR | • | • | • | 12 | ||
| MER/TYRO3 | KIYSGDyYR | • | • | 10 | |||
| TYK2 | AVPEGHEyYR | • | 8 | ||||
| FER | QEDGGVySSSGLK | • | • | 5 | |||
| JAK1 | AIETDKEyYTVK | • | 3 | ||||
| TEC | YVLDDQyTSSSGAK | • | 3 | ||||
| FGFR2 | DINNIDyYKK | • | 2 | ||||
| ABL | LMTGDTyTAHAGAK | • | • | 2 | |||
| EphR | VLEDDPEAAyTTR | • | 1 |
Multiple protein names are given in cases of shared, identical peptides
Lowercase y denotes phosphotyrosine
Sum of total MS/MS spectral counts across all the indicated cell lines
Figure 1. Global PTP inhibition of MM cells reveals PTPN1/PPN2 consensus target sequence in the activation loop of activated protein tyrosine kinases.
Motif analysis, using the PSP algorithm (Cell Signaling Technology), was performed using (A) the protein tyrosine kinase-associated activation loop pY-peptides or (B) the subset of these same peptides that display a YY motif in their activation loop identified by LC-MS/MS in pervanadate-treated LP1 (WT), KMS11 (Y373C), KMS18 (G384D) and OPM2 (K650E) MM cell lines, (C) revealing a PTPN1/PTPN2 recognition motif around the YY activation loop residues.
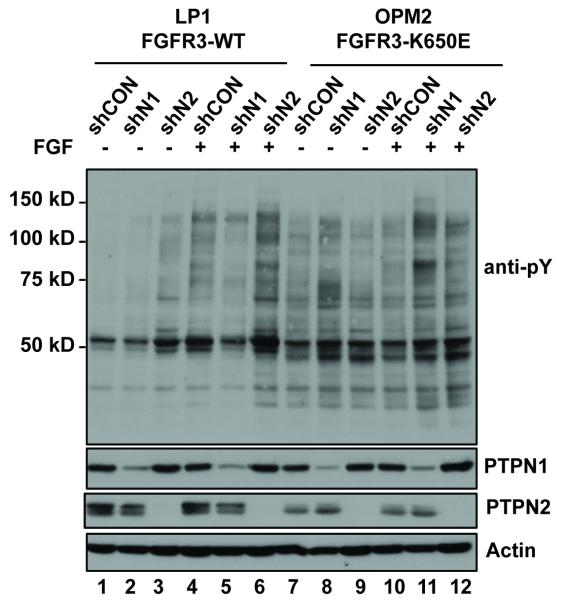
The results captured in Fig. 1 and Table 1 implicate functional interactions between PTPN1 and activated tyrosine kinases including especially FGFR3 and its variants in KMS11 and other FGFR-expressing MM cells. Therefore, we sought to address the role of PTPN1, and the structurally-related enzyme PTPN2 in FGFR3 regulation by using shRNA-mediated knockdown of PTPN1 and PTPN2 in MM tumor-derived cell lines that express FGFR3 as a product of the 4(4;14) translocation. The line LP1 expresses wild type FGFR3 (containing a functionally silent F384L polymorphism), while OPM2 expresses the activated K650E variant [54]. The two cell types were infected with lentivirus encoding either control non-silencing shRNA, or constructs directed at PTPN1 or PTPN2 (Fig. 2). In each case two independent shRNA constructs were tested with equivalent results (Fig. S1). The level of knockdown of PTPN1 was approximately 90%, whereas knockdown of the 45 kDa and 48 kDa PTPN2 isoforms appeared complete (Fig. 2). The infected cells were maintained in serum-containing medium and then treated with or without FGF. Western blot analysis showed that the level of cellular protein-pY is relatively low in MM cells ectopically expressing FGFR3, which is consistent with our previous observations [42]. In LP1, knockdown of PTPN1 caused a very slight increase in total cellular protein-pY, whereas a stronger response was associated with PTPN2 knockdown, and all instances the total protein-pY signal was further augmented when the serum-deprived cells were stimulated with FGF ligand (Fig. 2, lanes 1-6). OPM2, expressing the constitutively active FGFR3K650E variant, had a higher basal level of total cellular protein-pY. This was further increased when PTPN1 was knocked down, and to a much lesser extent when PTPN2 was knocked down. FGF ligand stimulation was ineffective with OPM2 control cells (shCON), but with PTPN2 and especially with PTPN1 knockdown, protein-pY signals were increased in response to FGF (Fig. 2, lanes 7-12).
Figure 2. PTPN1/PTPN2 knockdown in MM cells.
Shown are western blots of whole cell lysates from MM cell lines LP1 (FGFR3WT) and OPM2 (FGFR3K650E) with and without shRNA-mediated knockdown of PTPN1 (shN1) or PTPN2 (shN2), or control cells expressing a non-silencing control shRNA (shCON). Cells were cultured without serum and then treated with or without FGF1/Heparin for 10 min (B). Whole cell protein aliquots were resolved by SDS-PAGE and then subjected to western immuno-blotting with the indicated antibodies.
3.2 Context-dependent regulation of FGFR3 activity by PTPN1 and PTPN2
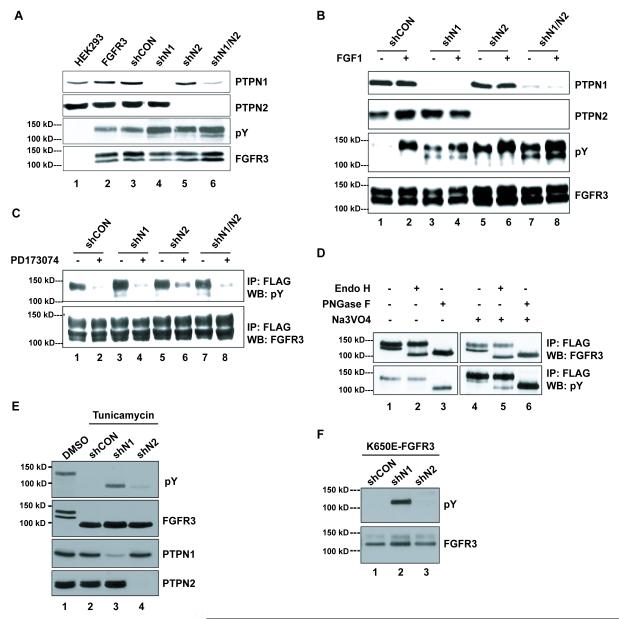
In order to further explore the ability of PTPN1 and PTPN2 to modulate FGFR3 the HEK293 cell system was used, which in contrast to typical MM cell lines is readily transfected, amenable to the establishment of stable cell lines, and is an established model system for the analysis of tyrosine kinase signaling and trafficking [e.g. 55]. Also, we showed previously that compared with various MM cell lines, including those used in this study, HEK293 express a similar set (i.e. >80%) of PTPs including PTPN1 and PTPN2 [40]. As shown in Fig. 3A, HEK293 stably expressing wild-type FGFR3-FLAG were infected with lentivirus expressing small hairpin RNA (shRNA) to knockdown PTPN1 and/or PTPN2 protein expression, or a control, non-silencing shRNA (shCON; Fig. 3A). Little effect was observed on FGFR3 pY in cells maintained in serum-containing medium (Figure 3A). However, under serum starvation conditions and in the absence of FGFR3-specific stimulation, knockdown of either PTPN1 or PTPN2 resulted in a considerable increase in FGFR3 pY levels (Figure 3B, lanes 3 and 5). To determine if the observed FGFR3 phosphorylation required FGFR3 kinase activity, FGFR3 was immunoprecipitated from PTP knockdown cells treated with PD173074 [3], and assessed by anti-pY blotting as a measure of activation. PD173074 is an ATP-competitive inhibitor of FGFR [56], and results in loss of the doubly phosphorylated (i.e. pY647pY648) isoform of FGFR3 in treated cells [42]. Figure 3C shows that PD173074 treatment countered most of the effect of the PTP knockdown, thus suggesting that the increased phosphorylation following the loss of PTPN1 or PTPN2 is dependent upon the intrinsic kinase activity of FGFR3.
Figure 3. PTPN1 and PTPN2 regulate FGFR3 tyrosine phosphorylation by targeting different glycosylated species of the receptor.
(A) Western blot of whole cell lysates from HEK293 cells with (FGFR3) or without (HEK293) stable WT-FGFR3-FLAG expression and stably expressing either scrambled shRNA (shCON), PTPN1 shRNA (shN1), PTPN2 shRNA (shN2) or both PTPN1 and PTPN2 shRNAs (shN1/N2). Cells were grown in serum-containing media. (B) Western blot of whole cell lysates from HEK293 stably expressing WT-FGFR3-FLAG expression and either scrambled shRNA (shCON), PTPN1 shRNA (shN1), PTPN2 shRNA (shN2) or both PTPN1 and PTPN2 shRNAs (shN1/N2). Cells were starved for 16 hours prior to a 10-minute stimulation with a combination of FGF1 (80 ng/ml) and Heparin (10 μg/ml). (C) Western blot of anti-FLAG immunoprecipitates from lysates of cells described in (B). Cells were starved for 12 hours in media without serum prior to a 4-hour treatment with the FGFR3 inhibitor PD173074 (100 nM). (D) Anti-FLAG immunoprecipitates from WT-FGFR3-FLAG-expressing HEK293 cell lysates with or without pervanadate treatment (Na3VO4)50 μM, 10 minutes). Immunoprecipitates were subjected to 1-hour glycosidase treatments with 500 units of either Endo H or PNGase F prior to Western analysis. (E) Western of lysates from WT-FGFR3-expressing HEK293 cells +/− shPTPN1 or PTPN2 treated with tunicamycin (1 μg/ml, 16 hours). (F) Western of lysates from HEK293 cells expressing K650E-FGFR3 +/− shPTPN1 or PTPN2. Arrows are aligned with the 125 kDa bands.
Knockdown of PTPN1 resulted in the activation of FGFR3 species migrating at both 125 kDa and 135 kDa (Fig. 3B, lanes 3, 4), while the PTPN2 knockdown activated predominantly the 135 kDa isoform (Fig. 3B, lanes 5, 6). To determine if these two differently migrating forms of FGFR3 represented differentially glycosylated isoforms, FGFR3 was isolated by IP and then subjected to in vitro treatment with glycosidase. Treatment with Endo H caused an apparent conversion of the 125 kDa isoform to an approximately 100 kDa species (Fig. 3D, lane 2). Treatment with PNGase F reduced both the 125 kDa and 135 kDa isoforms to the faster migrating, approx. 100 kDa form (Fig. 3D, lane 3). This indicates that the bands at 125 kDa and 135 kDa correspond to the mannose-rich, immature form of the receptor, and the fully processed species, respectively. Both of the FGFR3 glyco-isoforms became tyrosine-phosphorylated when cells were treated with pervanadate (Fig. 3D, lanes 4-6).
Interestingly, while loss of either PTPN1 or PTPN2 resulted in increased FGFR3 phosphorylation, there were qualitative differences that suggested the knockdowns were affecting different FGFR3 glyco-isoforms. Loss of PTPN1 resulted in a major increase in the phosphorylation of both the mannose-rich 125 kDa and mature 135 kDa forms of FGFR3, whereas loss of PTPN2 caused increased phosphorylation of only the fully processed, 135 kDa species (Fig. 3B). Simultaneous knockdown of both phosphatases also increased the pY levels of both forms of the receptor (Fig. 3B, lanes 7, 8). Treatment of cells with FGF1 did not change the pattern of FGFR3 glyco-isoform expression, but did cause an increase in tyrosine phosphorylation of the fully processed receptor species (Figure 3B). These findings suggested a role for the PTPs in modulating FGFR3 activity during the various stages of receptor maturation and processing. This was further supported by treatment of cells with tunicamycin in order to inhibit protein glycosylation. Following tunicamycin treatment of cells, the 125 kDa and 135 kDa species of FGFR3 were replaced with a ~100 kDa form, which is the expected size of nascent, un-modified FGFR3 (Fig. 3E). Figure 2E shows that cells lacking PTPN1 accumulated tyrosine-phosphorylated, non-glycosylated FGFR3 (lane 3), while loss of PTPN2 produced only a trace amount of pY-containing FGFR3 (lane 4).
The K650E variant of FGFR3 is known to be impaired for ER-Golgi processing and maturation [57]. To examine if PTPN1 and PTPN2 expression levels affect this phenomenon, the PTP knockdowns were replicated in cells expressing the K650E variant of FGFR3 (Fig. 3F). As shown in Figure 3F, in both control and cells lacking PTPN1 or PTPN2, FGFR3K650E was mostly the 125 kDa high-mannose form, and with a much lesser amount of the fully processed 135 kDa form. Probing for pY revealed that in the absence of PTPN1, but not PTPN2, the K650E variant became tyrosine phosphorylated (Figure 3F). These data suggested that FGFR3 auto-phosphorylation was modulated as a function of FGFR3-PTPN1/N2 co-localization, as reflected in the state of receptor glycosylation.
3.3 FGFR3 regulation by PTPN1 and PTPN2 depend on FGFR3 localization and A-loop sequence
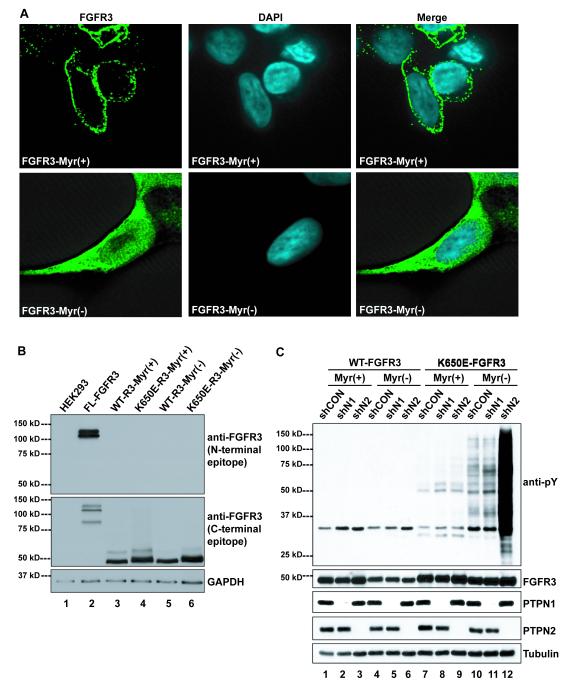
Next, an approach was taken to further examine the ability of the PTPs to modulate FGFR3 phosphorylation and activity as a function of subcellular localization, but independent of glycosylation processing, and ligand-stimulated activation. This involved characterization of FGFR3 variants lacking the transmembrane and extracellular domains. These amino-truncated variants were comprised of the intracellular region and modified to include, or not, an amino-terminal myristylation signal sequence, as described previously [58]. Immunofluorescence microscopy of HEK293 cells expressing either the myristylated FGFR3-Myr(+) or non-myristylated FGFR3-Myr(−) truncated variant revealed clear differences between their subcellular localization. FGFR3-Myr(+) displayed staining mostly restricted to the cell periphery, while FGFR3-Myr(−) was distributed throughout the cell (Fig. 4A). This suggested the proteins were, respectively, membrane-anchored and cytosolic as anticipated. As a further confirmation of their structure the truncated variants were detected by Western blot when probed with antibodies directed against the FGFR3 carboxyl-terminal region, but not the amino-terminal region (Figure 4B). As shown in Figure 4C, the truncated variant proteins including both wild type and K650E versions with or without the myristylation tag were efficiently expressed, and unaffected in terms of abundance by the knockdown of PTPN1 or PTPN2 (Fig. 4C, lower 4 panels).
Figure 4. Truncation of the extracellular domain and the K650E mutation in the activation loop of FGFR3 affect its pY regulation by PTPN1 and PTPN2.
(A) Immunofluorescence microscopy of FGFR3-Myr(+) and FGFR3-Myr(−) HEK293 cells. (B) Western blot of whole cell lysates from HEK293 cells stably expressing either full-length WT-FGFR3-FLAG (FL-FGFR3), or Myr(+) wild-type (WT-R3-Myr(+)), K650E mutant (K650E-R3-Myr(+)), or Myr(−) wild-type (WT-R3-Myr(−)), K650E mutant (K650E-R3-Myr(−)) FGFR3 lacking the complete extracellular domain. FGFR3 blots were probed with an antibody raised against the amino- (N-terminal epitope) or carboxy- (C-terminal epitope) terminal region of FGFR3. (C) Western blot of whole cell lysates or immune-precipitated cell lysates from HEK293 cells stably expressing wild-type (WT-FGFR3) or K650E mutant (K650E-FGFR3), myristylated (Myr(+)) or non-myristylated (Myr(−)) FGFR3 +/− shPTPN1 or shPTPN2.
To gauge the state of activation of the truncated, intracellular FGFR3 variants, profiles of whole cell protein-pY were measured by Western blot (Fig. 4C). The level of cellular protein-pY was very low in cells expressing wild type FGFR3-Myr(+) or FGFR3-Myr(−), and knockdown of PTPN1 or PTPN2 had no effect (Figure 4C, lanes 1-6). This is consistent with our previous analysis of protein-pY in t(4;14) MM cells expressing ectopic FGFR3, which generally display only low amounts of protein-pY relative to cells such as A431 that over-express wild type EGFR as a consequence of gene amplification [47]. By comparison, a minor increase in total protein-pY was observed in cells expressing FGFR3K650E-Myr(+) following knockdown of either PTPN1 or PTPN2 (Fig. 3C, lanes 7-9). Interestingly, total cellular protein-pY was higher in cells expressing the cytosolic variant FGFRK650E-Myr(−), and while this was slightly augmented by knockdown of PTPN1 (lanes 10, 11), a pronounced increase was associated with PTPN2 knockdown (Fig. 4C lane 12).
3.4 MS analysis indicates that loss of PTPN1 or PTPN2 increases FGFR3 A-loop phosphorylation
Following the observations of increased tyrosine phosphorylation upon loss of PTPN1 or PTPN2 in cells expressing the truncated FGFRK650E variants (Fig. 4C), a phospho-proteomic assessment was completed. Immuno-affinity purified pY-containing peptides were analyzed by high-resolution MS in an Orbitrap, thus allowing for high confidence identification of pY-containing peptides, and with additional verification based on the Ascore algorithm [59]. From 1 mg (as protein) starting material per sample more than 900 unique proteins containing over 1500 unique pY sites were identified across the set of samples (Table 2; Table S2). As shown in Table 2, the number of unique sites identified in each of the samples was consistent with the pY patterns observed by Western blot analysis (Fig. 4C), including the observation of increased pY associated with PTPN1 and PTPN2 knockdown in cells expressing the membrane-attached FGFRK650E-Myr(+) protein. The level of cellular protein-pY was higher in cells expressing the cytosolic variant FGFRK650E-Myr(−). Consistent the with the results shown in Fig. 4C, a dramatic increase in tyrosine phosphorylation was associated with knockdown of PTPN2 in cells expressing the cytosolic variant FGFRK650E-Myr(−). As shown in Table 3, 4 FGFR3 pY sites were identified (599, 657, 648, and 724), with most of the MS/MS events (81/88) matching pY-peptides containing A-loop residues (Y647 or Y648) contained on either singly (64/81) or doubly (17/81) phosphorylated peptides. Doubly phosphorylated FGFR3 A-loop peptides were only identified in the FGFRK650E-Myr(−) samples. Indeed most (80%) of all FGFR3 activation loop MS/MS events (65/81) were identified in this background (Table 3).
Table 2. Total cell phospho-proteome (pY) analysis of FGFR3 variants as a function of PTPN1/N2 expression.
Numbers indicate the non-redundant number of unique pY-containing proteins and peptides measured in HEK293 cells expressing truncated FGFR3 variants lacking the transmembrane and extracellular domains and with (+) or without (−) a myristate localization tag as indicated. Cells were infected with lentivirus encoding a non-silencing control shRNA (shCON) or shRNA directed at PTPN1 (shPTPN1) or PTPN2 (shPTPN2). Total indicates the sum of non-redundant pY sites and protein detected across the set of samples.
| K650E-FGFR3-Myr(+) |
K650E-FGFR3-Myr(−) |
Total | |||||
|---|---|---|---|---|---|---|---|
| shCON | shPTPN1 | shPTPN2 | shCON | shPTPN1 | shPTPN2 | ||
| pY proteins1 | 21 | 67 | 108 | 339 | 464 | 686 | 910 |
| pY sites1 | 22 | 71 | 122 | 438 | 610 | 1154 | 1558 |
Numbers indicate unique pY proteins or pY sites identified
Table 3. Spectral counts of identified FGFR3 pY peptides.
Various FGFR3-derived pY-containing peptides are shown along with the indicated sites of phosphorylation and their relative abundance as reflected in the indicated numbers of MS/MS events.
| Peptide1 | Site | FGFR3-K650E-Myr(+)2 |
FGFR3-K650E-Myr(−)2 |
Total | ||||
|---|---|---|---|---|---|---|---|---|
| shCON | shPTPN1 | shPTPN2 | shCON | shPTPN1 | shPTPN2 | |||
| DVHNLDyYK | Y647 | 1 | 4 | 5 | 6 | 7 | 7 | 30 |
| DVHNLDyYKETTNGR | Y647 | 0 | 2 | 1 | 2 | 5 | 5 | 15 |
| DVHNLDyYKETTNGRLPVK | Y647 | 0 | 1 | 2 | 2 | 5 | 5 | 15 |
| DVHNLDyyKETTNGRLPVK | Y647, Y648 | 0 | 0 | 0 | 2 | 3 | 5 | 10 |
| DVHNLDyyKETTNGR | Y647, Y648 | 0 | 0 | 0 | 1 | 2 | 4 | 7 |
| DVHNLDYyKETTNGRLPVK | Y648 | 0 | 0 | 0 | 0 | 1 | 2 | 3 |
| DVHNLDYyKETTNGR | Y648 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| DLVSCAyQVAR | Y599 | 0 | 0 | 0 | 2 | 2 | 2 | 6 |
| MDKPANCTHDLyMIMR | Y724 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
|
|
||||||||
| Sum: | 1 | 7 | 8 | 15 | 26 | 31 | 88 | |
Lowercase y denotes phosphotyrosine
Numbers indicate all MS/MS events associated with listed pY-peptides
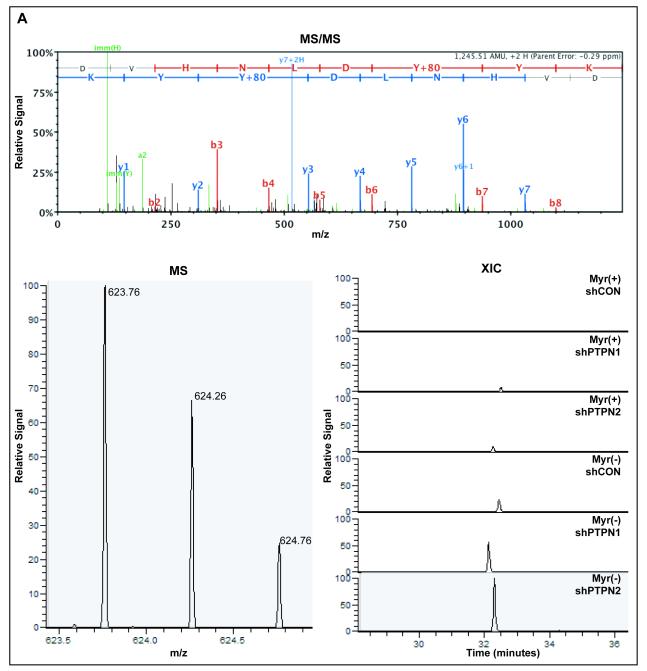
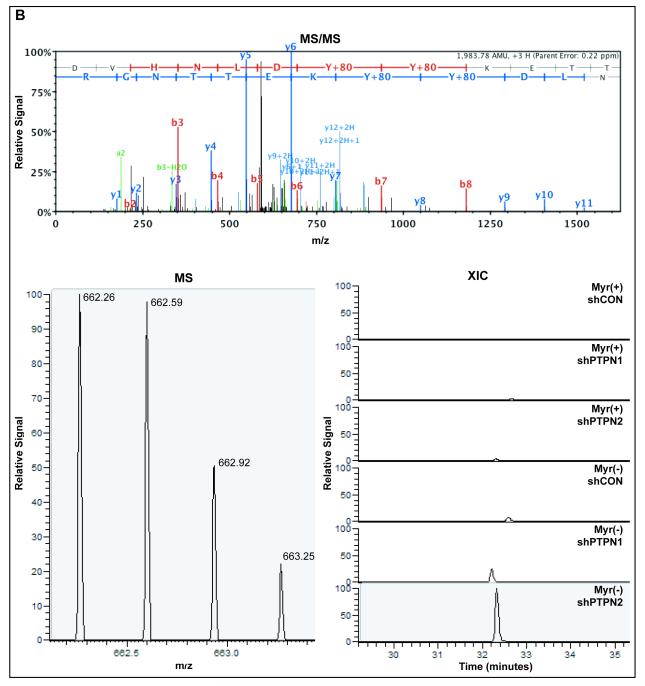
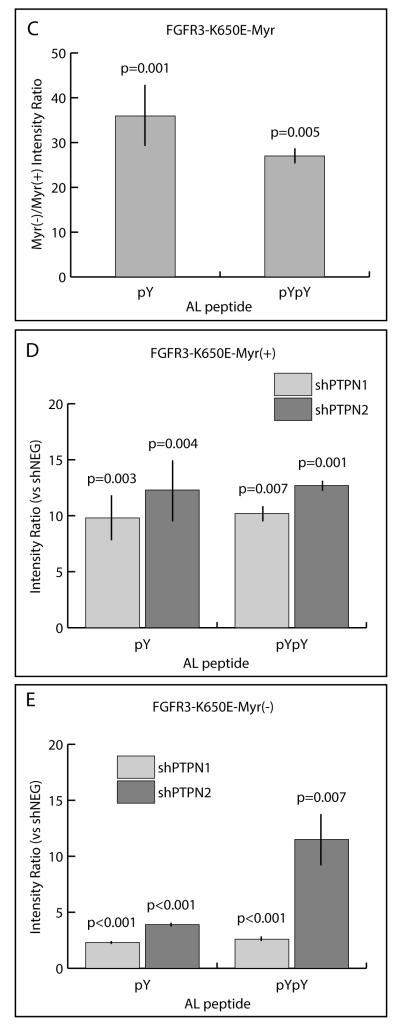
In order to obtain more quantitative information about FGFR3 A-loop peptides, MS ion currents of the singly (Fig. 5A) and doubly (Fig. 5B) phosphorylated A-loop peptides were extracted across FGFR3-Myr(+) and FGFR3-Myr(−) samples with or without loss of PTPN1 or PTPN2. Considerable increases were observed in Myr(−) compared to Myr(+) in shCON backgrounds for both the singly (18-fold) and doubly (29-fold) phosphorylated A-loop peptides (Fig. 5C). In both the Myr(+) (Fig. 5D) and Myr(−) (Fig. 5E) backgrounds, loss of PTPN1 or PTPN2 resulted in increases in both singly and doubly phosphorylated A-loop peptides. Moreover, increased FGFR3 A-loop phosphorylation associated with knockdown of PTPN1 or PTPN2 were similar, except for the doubly phosphorylated A-loop peptide in the Myr(−) background, for which the level was increased to a greater extent following loss of PTPN2 (9.9-fold over shCON) compared to PTPN1 (2.7-fold over shCON).
Figure 5. MS analysis of FGFR3 A-loop phosphorylation as a function of PTPN1/PTPN2 expression.
(A) Top panel; Representative MS/MS fragmentation spectrum of the singly phosphorylated (pY647) FGFR3 activation loop peptide. Bottom left panel; MS spectrum of singly phosphorylated (pY647) FGFR3 activation loop peptide isotopes. Bottom right panel; Extracted ion current of pY647 peptide from Myr (+) and Myr(−) of FGFR3K650E cells ±PTPN1 or PTPN2 knockdown. (B) Top panel; Representative MS/MS fragmentation spectrum of the doubly phosphorylated (pY647/pY648) FGFR3 activation loop peptide. Bottom left panel; MS spectrum of pY647/pY648 peptide isotopes. Bottom right panel; Extracted ion current of pY647pY648 peptide from Myr (+) and Myr(−) of FGFR3K650E cells ±PTPN1 or PTPN2 knockdown. (C) Quantification of integrated extracted ion current (XIC) for pY647 and pY647/pY648 in cells expressing FGFR3K650E–Myr(−). Abscissa represents ratio of integrated XICs, Myr(−):Myr(+). (D) Quantification of integrated XIC for pY647 and pY647/pY648 in cells expressing FGFR3K650E-Myr(+) ±PTPN1 or PTPN2 knockdown. Abscissa represents ratio of integrated XICs, shPTP:shNEG. (E) Quantification of integrated XIC for pY647 and pY647/pY648 in cells expressing FGFR3K650E-Myr(−) ±PTPN1 or PTPN2 knockdown. Abscissa represent ratio of integrated XICs, shPTP:shNEG. Error bars represent standard deviations; n ≥4. P values were generated from paired, 2-tailed Student's t-tests.
4 Discussion
Little is known of substrate sequences that confer specificity to PTPs. Indeed, despite several strategies developed to identify such motifs [reviewed in ref. 60], the identification of pY-proximal sequences that render sites exclusively or optimally susceptible to one or a given set of PTPs has remained elusive. Our pY screen of MM cell lines under conditions of global PTP inhibition identified pY A-loop peptides from several tyrosine kinases with tandem tyrosine motifs, several of which corresponded to the D/EYYR/K recognition motif of PTPN1/PTPN2 (Fig. 1, Table 1). Of these, the FGFR3 pY A-loop peptides were the most abundantly identified, indicating that FGFR3 in MM is effectively constrained by cellular PTP activities. Peptides with tandem pY residues N-terminally flanked by acidic amino acids are preferred PTPN1 substrates [53, 61-63], and structural and catalytic data of the IR A-loop peptide containing tandem tyrosine residues pY1162pY1163 within a DYYR context indicated that the C-terminal R residue is involved in several surface interactions with the catalytic domain of PTPN1 [53]. PTPN2 has not been as extensively characterized, but based on its structural similarity to PTPN1 and our findings, it is expected that substrates containing the D/EYYR/K motif may be common targets of PTPN1 and PTPN2 [53]. Indeed we found that knockdown of either PTPN1 or PTPN2 caused increased FGFR3 A-loop phosphorylation. While this does not confirm a preference of PTPN1/PTPN2 for the D/EYYR/K recognition motif, it supports the body of evidence that these PTPs affect the phosphorylation of D/EYYR/K-containing tyrosine kinase A-loops.
FGFR3 is not consistently activated by mutation in MM [5, 64]. Our observation of FGFR3 activation as a consequence of PTPN1/N2 knockdown supports a model wherein t(4;14) translocation, as an early event in MM, could result in the production of activated wild type FGFR3 as a function of low PTPN1 and PTPN2 expression or activity. Therefore, we speculate the t(4;14) translocation that causes FGFR3 ectopic over-expression in MM may arise through selective pressure for FGFR3 expression in pre-MM cells whose PTPome (i.e. their complement of expressed PTPs) [ 40] is permissive for the accumulation of phosphorylated D/EYYR/K sequence motifs, such as the FGFR3 A-loop. Circumstantial support for this model comes from our characterization of MM PTPomes [40], which indicated a relatively low expression of PTPN1 and PTPN2 in t(4;14) MM cell lines and much higher expression of PTPN2 in particular, in the MM cell line KMS12, which is not 4(4;14), and therefore does not express ectopic FGFR3. A more thorough investigation of PTP expression in MM is required to test this model.
In contrast to FGFR3, the increase in Met phosphorylation that accompanies loss of PTPN1 or PTPN2 remains ligand (HGF) dependent. Since Met migrates as a single band during electrophoresis, no differences were observed between PTPN1 and PTPN2 in regards to their preference for fully processed or immature receptor species [31]. Nevertheless, loss of both PTPs results in a greater increase in Met phosphorylation than loss of either PTP alone, thus suggesting that PTPN1 and PTPN2 non-redundantly regulate Met [31]. Similarly, A-loop residues Y1162/Y1163 of IR are differently regulated by PTPN1 and PTPN2 [65]. Thus, PTPN1 and PTPN2 exert non-overlapping roles in the regulation of various RTKs, and the mechanisms that govern this specificity may vary across different receptors, and receptor alleles as discussed below.
Our results indicate that FGFR3 is constrained in vivo, in terms of A-loop phosphorylation and cognate kinase domain activity, by PTPN1 and PTPN2. This modulation is clearly spatially and temporally regulated as evidenced by the findings relating FGFR3 glycosylation with PTPN1/N2 susceptibility (Fig. 3). Loss of PTPN1, but not PTPN2 affected the phosphorylation of nascent FGFR3 (Fig. 3B), indicating that during ER-Golgi processing PTPN1 rather than PTPN2 controls FGFR3 phosphorylation. This is consistent with the ER-localization of PTPN1 and agrees with the demonstrated context (i.e. localization)–dependent susceptibilities of various RTKs to PTPN1 and PTPN2 [66]. The K650E substitution prevents FGFR3 maturation resulting in ER retention [57]. It has been deduced this is a consequence of a failure to restrain K650E kinase activity, since a related variant, murine FGFR3 K644R (corresponding to human K650R), which is not catalytically activated, is properly processed and trafficked to the plasma membrane [67]. Indeed the requirement to quell nascent Y-kinase activity applies to other RTKs including FLT-3, PDFGRβ, Kit, as well as a TrkA-Ros chimera [68]. Loss of PTPN1 also increased the phosphorylation of the mature, fully processed form of FGFR3, indicating that PTPN1 also targets FGFR3 beyond the ER. Indeed ER-anchored PTPN1 has substrates located at the plasma membrane [69] as well as endosomes and multi-vesicular body through points of contact between these subcellular compartments and the ER [22, 70, 71]. Consistent with this, we previously identified numerous tyrosine phosphorylated markers of endocytosis as part of the FGF-stimulated FGFR3 pY network in a MM model [47]. Hence, our findings that PTPN1 levels affect the phosphorylation of both nascent and mature forms of FGFR3 support the model wherein PTPN1-FGFR3 interactions are inextricably linked to FGFR3 trafficking.
Our data reveal that that the A-loop sequence of FGFR3 functions as a determinant of PTPN1/N2 susceptibility. The K650E kinase domain favors an active-state conformation essentially superimposable with that of the doubly phosphorylated activated kinase domains of FGFR1 and FGFR2 rendering it constitutively activated [16]. Indeed, Y-to-F substitution at Y647 and/or Y648 did not impair K650E intrinsic kinase activity [15, 16], and the constitutive kinase activity of K650E was similar to that attained by the fully auto-phosphorylated wild type FGFR3 kinase domain [16]. These data might suggest that K650E would not be regulated by PTPN1/N2, to the extent that this normally occurs through modulation of A-loop phosphorylation. However, we found that K650E, especially when stripped of the trafficking constraints conferred by its transmembrane sequence, was in fact extremely sensitive to the expression of PTPN2. A model to explain this is that PTPN2, possibly through direct interaction with the FGFR3 kinase domain, shifts the K650E equilibrium towards the inactive-state conformation. This model respects the ligand-independent activation of wild type FGFR3 that accompanied PTPN2 knockdown. In this scenario, in the absence of ligand, PTPN2 stabilizes the inactive-state of the kinase domain; ligand binding to the extracellular domain transduces structural rearrangements that disrupt this constraint, allowing the initiation of the ordered series of auto-phosphorylations that results in full kinase activation and accumulation of pY moieties as signaling protein binding sites [9, 13]. By analogy, the model predicts PTPN1 plays a similar role in constraining the intrinsic activity of nascent and internalized FGFR3. The model described above is currently under investigation.
In conclusion, this study highlights that several regulatory mechanisms including kinase A-loop sequence, compartmentalization, ligand and substrate availability, and PTPN1 and PTPN2 expression levels profoundly affect FGFR3 activity. This further illustrates the impact of the PTPome on the kinome, and hence cellular protein-pY profiles, which are of critical importance in normal and cancer cells.
Supplementary Material
Acknowledgements
This research was supported by funding provided to MFM by the Canadian Cancer Society Research Institute and Canada Research Chairs program. We thank Prof. Daniel Donoghue for generously providing the truncated FGFR3 expression constructs.
Abbreviations
- MM
multiple myeloma
- FGFR3
fibroblast growth factor receptor 3
- pY
phosphotyrosine
- PTP
phosphotyrosine phosphatase
- RTK
receptor tyrosine kinase
- ER
endoplasmic reticulum
- XIC
extracted ion current
- shRNA
short hairpin RNA
Footnotes
The authors have declared no conflict of interest.
5 References
- [1].Chesi M, Nardini E, Brents LA, Schrock E, et al. Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet. 1997;16:260–264. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Richelda R, Ronchetti D, Baldini L, Cro L, et al. A novel chromosomal translocation t(4; 14)(p16.3; q32) in multiple myeloma involves the fibroblast growth-factor receptor 3 gene. Blood. 1997;90:4062–4070. [PubMed] [Google Scholar]
- [3].Trudel S, Ely S, Farooqi Y, Affer M, et al. Inhibition of fibroblast growth factor receptor 3 induces differentiation and apoptosis in t(4;14) myeloma. Blood. 2004;103:3521–3528. doi: 10.1182/blood-2003-10-3650. [DOI] [PubMed] [Google Scholar]
- [4].Paterson JL, Li Z, Wen XY, Masih-Khan E, et al. Preclinical studies of fibroblast growth factor receptor 3 as a therapeutic target in multiple myeloma. Br J Haematol. 2004;124:595–603. doi: 10.1111/j.1365-2141.2004.04814.x. [DOI] [PubMed] [Google Scholar]
- [5].Chesi M, Brents LA, Ely SA, Bais C, et al. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood. 2001;97:729–736. doi: 10.1182/blood.v97.3.729. [DOI] [PubMed] [Google Scholar]
- [6].Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- [8].Kanai M, Goke M, Tsunekawa S, Podolsky DK. Signal transduction pathway of human fibroblast growth factor receptor 3. Identification of a novel 66-kDa phosphoprotein. J Biol Chem. 1997;272:6621–6628. doi: 10.1074/jbc.272.10.6621. [DOI] [PubMed] [Google Scholar]
- [9].Mohammadi M, Schlessinger J, Hubbard SR. Structure of the FGF receptor tyrosine kinase domain reveals a novel autoinhibitory mechanism. Cell. 1996;86:577–587. doi: 10.1016/s0092-8674(00)80131-2. [DOI] [PubMed] [Google Scholar]
- [10].Haugsten EM, Malecki J, Bjørklund SMS, Olsnes S, Wesche J. Ubiquitination of fibroblast growth factor receptor 1 is required for its intracellular sorting but not for its endocytosis. Mol Biol Cell. 2008;19:3390–3403. doi: 10.1091/mbc.E07-12-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Abella JV, Park M. Breakdown of endocytosis in the oncogenic activation of receptor tyrosine kinases. Am J Physiol Endocrinol Metab. 2009;296:E973–984. doi: 10.1152/ajpendo.90857.2008. [DOI] [PubMed] [Google Scholar]
- [12].Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- [13].Lew ED, Furdui CM, Anderson KS, Schlessinger J. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci Signal. 2009;2:ra6. doi: 10.1126/scisignal.2000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang Z, Chen H, Blais S, Neubert TA, et al. Structural Mimicry of A-Loop Tyrosine Phosphorylation by a Pathogenic FGF Receptor 3 Mutation. Structure. 2013 doi: 10.1016/j.str.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Webster MK, D'Avis PY, Robertson SC, Donoghue DJ. Profound ligand-independent kinase activation of fibroblast growth factor receptor 3 by the activation loop mutation responsible for a lethal skeletal dysplasia, thanatophoric dysplasia type II. Mol Cell Biol. 1996;16:4081–4087. doi: 10.1128/mcb.16.8.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Huang Z, Chen H, Blais S, Neubert TA, et al. Structural mimicry of a-loop tyrosine phosphorylation by a pathogenic FGF receptor 3 mutation. Structure. 2013;21:1889–1896. doi: 10.1016/j.str.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, et al. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- [18].Shiang R, Thompson LM, Zhu YZ, Church DM, et al. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78:335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- [19].Tavormina PL, Shiang R, Thompson LM, Zhu YZ, et al. Thanatophoric dysplasia (types I and II) caused by distinct mutations in fibroblast growth factor receptor 3. Nat Genet. 1995;9:321–328. doi: 10.1038/ng0395-321. [DOI] [PubMed] [Google Scholar]
- [20].Cappellen D, De Oliveira C, Ricol D, de Medina S, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- [21].Haj FG, Verveer PJ, Squire A, Neel BG, Bastiaens PI. Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science. 2002;295:1708–1711. doi: 10.1126/science.1067566. [DOI] [PubMed] [Google Scholar]
- [22].Haj FG, Sabet O, Kinkhabwala A, Wimmer-Kleikamp S, et al. Regulation of signaling at regions of cell-cell contact by endoplasmic reticulum-bound protein-tyrosine phosphatase 1B. PLoS One. 2012;7:e36633. doi: 10.1371/journal.pone.0036633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Stuible M, Tremblay ML. In control at the ER: PTP1B and the down-regulation of RTKs by dephosphorylation and endocytosis. Trends in cell biology. 2010;20:672–679. doi: 10.1016/j.tcb.2010.08.013. [DOI] [PubMed] [Google Scholar]
- [24].Lorenzen JA, Dadabay CY, Fischer EH. COOH-terminal sequence motifs target the T cell protein tyrosine phosphatase to the ER and nucleus. J Cell Biol. 1995;131:631–643. doi: 10.1083/jcb.131.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tiganis T, Bennett AM, Ravichandran KS, Tonks NK. Epidermal growth factor receptor and the adaptor protein p52Shc are specific substrates of T-cell protein tyrosine phosphatase. Mol Cell Biol. 1998;18:1622–1634. doi: 10.1128/mcb.18.3.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tiganis T, Kemp BE, Tonks NK. The protein-tyrosine phosphatase TCPTP regulates epidermal growth factor receptor-mediated and phosphatidylinositol 3-kinase-dependent signaling. J Biol Chem. 1999;274:27768–27775. doi: 10.1074/jbc.274.39.27768. [DOI] [PubMed] [Google Scholar]
- [27].Flint AJ, Tiganis T, Barford D, Tonks NK. Development of "substrate-trapping" mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sangwan V, Abella J, Lai A, Bertos N, et al. Protein-tyrosine phosphatase 1B modulates early endosome fusion and trafficking of Met and epidermal growth factor receptors. J Biol Chem. 2011;286:45000–45013. doi: 10.1074/jbc.M111.270934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Galic S, Klingler-Hoffmann M, Fodero-Tavoletti MT, Puryer MA, et al. Regulation of insulin receptor signaling by the protein tyrosine phosphatase TCPTP. Mol Cell Biol. 2003;23:2096–2108. doi: 10.1128/MCB.23.6.2096-2108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Seely BL, Staubs PA, Reichart DR, Berhanu P, et al. Protein tyrosine phosphatase 1B interacts with the activated insulin receptor. Diabetes. 1996;45:1379–1385. doi: 10.2337/diab.45.10.1379. [DOI] [PubMed] [Google Scholar]
- [31].Sangwan V, Paliouras GN, Abella JV, Dube N, et al. Regulation of the Met receptor tyrosine kinase by the protein tyrosine phosphatases PTP1B and TCPTP. J Biol Chem. 2008 doi: 10.1074/jbc.M805916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu F, Chernoff J. Protein tyrosine phosphatase 1B interacts with and is tyrosine phosphorylated by the epidermal growth factor receptor. Biochem J. 1997;327(Pt 1):139–145. doi: 10.1042/bj3270139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Markova B, Herrlich P, Ronnstrand L, Bohmer FD. Identification of protein tyrosine phosphatases associating with the PDGF receptor. Biochemistry. 2003;42:2691–2699. doi: 10.1021/bi0265574. [DOI] [PubMed] [Google Scholar]
- [34].Persson C, Savenhed C, Bourdeau A, Tremblay ML, et al. Site-selective regulation of platelet-derived growth factor beta receptor tyrosine phosphorylation by T-cell protein tyrosine phosphatase. Mol Cell Biol. 2004;24:2190–2201. doi: 10.1128/MCB.24.5.2190-2201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mattila E, Auvinen K, Salmi M, Ivaska J. The protein tyrosine phosphatase TCPTP controls VEGFR2 signalling. J Cell Sci. 2008;121:3570–3580. doi: 10.1242/jcs.031898. [DOI] [PubMed] [Google Scholar]
- [36].Nakamura Y, Patrushev N, Inomata H, Mehta D, et al. Role of protein tyrosine phosphatase 1B in vascular endothelial growth factor signaling and cell-cell adhesions in endothelial cells. Circ Res. 2008;102:1182–1191. doi: 10.1161/CIRCRESAHA.107.167080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Heinonen KM, Dube N, Bourdeau A, Lapp WS, Tremblay ML. Protein tyrosine phosphatase 1B negatively regulates macrophage development through CSF-1 signaling. Proc Natl Acad Sci U S A. 2006;103:2776–2781. doi: 10.1073/pnas.0508563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Simoncic PD, Bourdeau A, Lee-Loy A, Rohrschneider LR, et al. T-cell protein tyrosine phosphatase (Tcptp) is a negative regulator of colony-stimulating factor 1 signaling and macrophage differentiation. Mol Cell Biol. 2006;26:4149–4160. doi: 10.1128/MCB.01932-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tiganis T. PTP1B and TCPTP - nonredundant phosphatases in insulin signaling and glucose homeostasis. FEBS J. 2012 doi: 10.1111/j.1742-4658.2012.08563.x. [DOI] [PubMed] [Google Scholar]
- [40].Karisch R, Fernandez M, Taylor P, Virtanen C, et al. Global proteomic assessment of the classical protein-tyrosine phosphatome and “redoxome”. Cell. 2011;146:826–840. doi: 10.1016/j.cell.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bourdeau A, Dube N, Tremblay ML. Cytoplasmic protein tyrosine phosphatases, regulation and function: the roles of PTP1B and TC-PTP. Curr Opin Cell Biol. 2005;17:203–209. doi: 10.1016/j.ceb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- [42].St-Germain JR, Taylor P, Tong J, Jin LL, et al. Multiple myeloma phosphotyrosine proteomic profile associated with FGFR3 expression, ligand activation, and drug inhibition. Proc Natl Acad Sci U S A. 2009;106:20127–20132. doi: 10.1073/pnas.0910957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Barlow AL, Macleod A, Noppen S, Sanderson J, Guerin CJ. Colocalization analysis in fluorescence micrographs: verification of a more accurate calculation of pearson's correlation coefficient. Microsc Microanal. 16:710–724. doi: 10.1017/S143192761009389X. [DOI] [PubMed] [Google Scholar]
- [44].Luo B, Cheung HW, Subramanian A, Sharifnia T, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rush J, Moritz A, Lee KA, Guo A, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- [46].Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- [47].St-Germain JR, Taylor P, Tong J, Jin LL, et al. Multiple Myeloma Phosphotyrosine Proteomic Profile Associated with FGFR3 expression, ligand activation, and drug inhibition. Proc Natl Acad Sci, USA. 2009;106:20127–20132. doi: 10.1073/pnas.0910957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic acids research. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Boeri Erba E, Matthiesen R, Bunkenborg J, Schulze WX, et al. Quantitation of multisite EGF receptor phosphorylation using mass spectrometry and a novel normalization approach. J Proteome Res. 2007;6:2768–2785. doi: 10.1021/pr060675m. [DOI] [PubMed] [Google Scholar]
- [50].Kamps MP, Sefton BM. Most of the substrates of oncogenic viral tyrosine protein kinases can be phosphorylated by cellular tyrosine protein kinases in normal cells. Oncogene Res. 1988;3:105–115. [PubMed] [Google Scholar]
- [51].Irish JM, Hovland R, Krutzik PO, Perez OD, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- [52].Pan C, Gnad F, Olsen JV, Mann M. Quantitative phosphoproteome analysis of a mouse liver cell line reveals specificity of phosphatase inhibitors. Proteomics. 2008;8:4534–4546. doi: 10.1002/pmic.200800105. [DOI] [PubMed] [Google Scholar]
- [53].Salmeen A, Andersen JN, Myers MP, Tonks NK, Barford D. Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol Cell. 2000;6:1401–1412. doi: 10.1016/s1097-2765(00)00137-4. [DOI] [PubMed] [Google Scholar]
- [54].Ronchetti D, Greco A, Compasso S, Colombo G, et al. Deregulated FGFR3 mutants in multiple myeloma cell lines with t(4;14): comparative analysis of Y373C, K650E and the novel G384D mutations. Oncogene. 2001;20:3553–3562. doi: 10.1038/sj.onc.1204465. [DOI] [PubMed] [Google Scholar]
- [55].Tong J, Taylor P, Moran MF. Proteomic analysis of the EGFR interactome and post-translational modifications associated with receptor endocytosis in response to EGF and stress. Mol Cell Proteomics. 2014;13:1644–1658. doi: 10.1074/mcp.M114.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mohammadi M, Froum S, Hamby JM, Schroeder MC, et al. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17:5896–5904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lievens PM, Liboi E. The thanatophoric dysplasia type II mutation hampers complete maturation of fibroblast growth factor receptor 3 (FGFR3), which activates signal transducer and activator of transcription 1 (STAT1) from the endoplasmic reticulum. The Journal of Biological Chemistry. 2003;278:17344–17349. doi: 10.1074/jbc.M212710200. [DOI] [PubMed] [Google Scholar]
- [58].Webster MK, Donoghue DJ. Enhanced signaling and morphological transformation by amebrane-localized derivative of the fibroblast growth factor receptor 3 kinase domain. Molecular and Cellular Biology. 1997;17:5739–5747. doi: 10.1128/mcb.17.10.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- [60].Zhang H, Zha X, Tan Y, Hornbeck PV, et al. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- [61].Zhang ZY, Thieme-Sefler AM, Maclean D, McNamara DJ, et al. Substrate specificity of the protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1993;90:4446–4450. doi: 10.1073/pnas.90.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jia Z, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- [63].Ren L, Chen X, Luechapanichkul R, Selner NG, et al. Substrate specificity of protein tyrosine phosphatases 1B, RPTPalpha, SHP-1, and SHP-2. Biochemistry. 2011;50:2339–2356. doi: 10.1021/bi1014453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chesi M, Bergsagel PL, Kuehl WM. The enigma of ectopic expression of FGFR3 in multiple myeloma: a critical initiating event or just a target for mutational activation during tumor progression. Curr Opin Hematol. 2002;9:288–293. doi: 10.1097/00062752-200207000-00005. [DOI] [PubMed] [Google Scholar]
- [65].Galic S, Hauser C, Kahn BB, Haj FG, et al. Coordinated regulation of insulin signaling by the protein tyrosine phosphatases PTP1B and TCPTP. Mol Cell Biol. 2005;25:819–829. doi: 10.1128/MCB.25.2.819-829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lammers R, Bossenmaier B, Cool DE, Tonks NK, et al. Differential activities of protein tyrosine phosphatases in intact cells. J Biol Chem. 1993;268:22456–22462. [PubMed] [Google Scholar]
- [67].Lievens PM, Mutinelli C, Baynes D, Liboi E. The kinase activity of fibroblast growth factor receptor 3 with activation loop mutations affects receptor trafficking and signaling. The Journal of Biological Chemistry. 2004;279:43254–43260. doi: 10.1074/jbc.M405247200. [DOI] [PubMed] [Google Scholar]
- [68].Schmidt-Arras DE, Bohmer A, Markova B, Choudhary C, et al. Tyrosine phosphorylation regulates maturation of receptor tyrosine kinases. Mol Cell Biol. 2005;25:3690–3703. doi: 10.1128/MCB.25.9.3690-3703.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Anderie I, Schulz I, Schmid A. Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell Signal. 2007;19:582–592. doi: 10.1016/j.cellsig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- [70].Faure R, Baquiran G, Bergeron JJ, Posner BI. The dephosphorylation of insulin and epidermal growth factor receptors. Role of endosome-associated phosphotyrosine phosphatase(s) J Biol Chem. 1992;267:11215–11221. [PubMed] [Google Scholar]
- [71].Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nature cell biology. 2010;12:267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.