Abstract
The large and increasing volume of genomic data analyzed by comparative methods provides information about transcription factors and their binding sites that, in turn, enables statistical analysis of correlations between factors and sites, uncovering mechanisms and evolution of specific protein-DNA recognition. Here we present an online tool, Prot-DNA-Korr, designed to identify and analyze crucial protein-DNA pairs of positions in a family of transcription factors. Correlations are identified by analysis of mutual information between columns of protein and DNA alignments. The algorithm reduces the effects of common phylogenetic history and of abundance of closely related proteins and binding sites. We apply it to five closely related subfamilies of the MerR family of bacterial transcription factors that regulate heavy metal resistance systems. We validate the approach using known 3D structures of MerR-family proteins in complexes with their cognate DNA binding sites and demonstrate that a significant fraction of correlated positions indeed form specific side-chain-to-base contacts. The joint distribution of amino acids and nucleotides hence may be used to predict changes of specificity for point mutations in transcription factors.
Introduction
Specific binding of transcription factors to DNA is a major mechanism of regulation of gene expression, hence boosting interest to the problem of the protein-DNA recognition code. Initial hopes stemmed from the observations that single amino acid substitutions can drastically change the protein affinity to its DNA sites. On the other hand, the structure of the DNA double helix is relatively rigid. An early (mid-70s) paper suggested that specific recognition depends on hydrogen bonds between side chains of amino acid residues and nucleotides bases, demonstrated that this recognition is easier in the major groove of the double helix than in the minor one, and discussed the role of the guanidine group of arginine in the recognition of the GC base pair [1].
The substantial progress in the 80s and 90s was based on the analysis of X-ray structures of protein-DNA complexes. It has been established that the recognition depends not only on hydrogen bonds, but on other types of weak interactions, and some empirical rules of the protein-DNA recognition have been suggested. Analysis of twenty structures demonstrated that the most common contacts between amino acid residues and nucleotide bases may be explained by the physical and chemical properties of the residues—the hydrophobic methyl group of alanine often interacts with the methyl group of thymine; arginine forms two hydrogen bonds with guanine; asparagine forms two hydrogen with adenine; etc. [2]. Moreover, while the orientation of DNA-binding protein structural elements varies in different protein families, within a family the binding is defined by a fixed, limited set of positions. For example, in the helix-turn-helix (HTH) domains, the binding element is the second α-helix, with residues 1, 2, 6 recognizing four successive bases in the major groove [2].
These rules were subsequently confirmed in a larger study that analyzed 129 protein-DNA complexes with close homologs filtered out [3]. About one third of ‘residue side chain—base’ hydrogen bonds are involved in complex interactions where one residue interacts with two consecutive nucleotides in DNA. In addition to universal contacts, there exist context-dependent contacts contributing to the recognition specificity, but unique for a given complex.
Currently it is widely accepted that, unlike protein-protein contacts, the regions of protein-DNA contacts are rich in polar residues (Arg, Ser, Tyr, Thr, Asn) [4]. The most positively charged patch on the protein surface often coincides with the DNA-binding site. Purines are more selective in their contacts than pyrimidines [4]. Aromatic amino acids have different specificities, e.g. phenylalanine prefers adenine and thymine, and histidine prefers thymine and guanine [5].
At the same time, there are as many exceptions as there are rules [6]. There is no simple relationship between the amino acid sequence of a protein and the nucleotide sequence of its binding DNA site, and the protein-DNA code is degenerate on both sides [7]. This is not surprising, given the existence of complex contacts [2, 8] and diversity of contact geometries and docking surfaces [9], even for structurally similar proteins [10]. Moreover, the protein’s interaction with its sites is not an all-or-nothing, but rather a quantitative parameter [11], not limited to the chemical identity of the interacting residues and bases, but involving changes in the protein and/or DNA conformation upon interaction known as indirect readout [6]. This shifted the focus of attention from identification of empirical rules to creation of statistical functions based on structural data [6] using neural networks [12, 13], support vector machines [13], or Bayesian classifiers [14] trained on known structures and then applied to protein sequences.
High-throughput experimental techniques such as SELEX [15], ChIP-chip [16, 17], DIP-ChIP [18], ChIP-Seq [19] and PBMs [20], as well as comparative genomic analyses [21, 22] provide large number of binding sites for a given TF. Available data on binding sites of transcription factors, collected in databases such as TRANSFAC [23], JASPAR [24], Factorbook [25], RegTransBase [26], and RegPrecise [27], exceed by orders of magnitude the number of solved structures of protein-DNA complexes and even transcription factors without DNA. Hence statistical analysis of correlations between transcription factors and their sites becomes both a possibility and a necessity.
Transcription factors (TFs) from one structural family tend to recognize similar DNA motifs [8, 28, 29] and that allows one to construct family-specific motifs that may be used both for the identification of candidate binding sites (BS) and for the classification of transcription factors [30]. The correlation between the level of conservation of specific residues in DNA-binding proteins and that of DNA sites has been demonstrated for 21 protein families [31]. The residues contacting the sugar-phosphate backbone are conserved, whereas the residues contacting nucleotide bases are conserved if binding motifs are similar for all proteins from a family, and variable otherwise. Within a genome, there is a correlation between the degree of conservation of a consensus nucleotide and the number of contacts it forms with DNA [32]. In the TAL-effector family of Xanthomonas TFs, injected into plant cells during infection, there exists a recognition code linking pairs of amino acid residues, so-called repeat-variable diresidues, and base pairs in the recognized site [33, 34], and this code may be used to predict TAL-effector targets [35, 36]. A similar code was suggested for the Cro family of phage TFs [37].
These and similar observations formed a base for the identification of specificity-determining positions in aligned, homologous protein sequences divided into groups by specificity towards ligands, cofactors or DNA motifs [38]. For each alignment column, the mutual information is calculated as a measure of correlation between the positional amino acid distribution and the division into specificity groups. This method was applied to identification of specificity-determining positions in prokaryotic [38, 39] and eukaryotic [40] transcription factors, and the predictions were in good agreement with the structural and mutagenesis data. The main drawback of the method, the need to define specificity groups in advance, may be partially offset by automated clustering of protein sequences [40, 41].
Similar methods based on measuring the mutual information are widely used for the identification of protein-protein interactions (e.g. [42, 43]) or even prediction of the protein three-dimensional structure [44]. They do not require structural or phylogenetic information. Such methods were applied to identify a fraction of functionally important contacts in several families of eukaryotic TFs [45, 46] and the LacI family of bacterial TFs [28]. A caveat is that this method requires large training samples and an estimate of expected mutual information. It also, by construction, underestimates the importance of conserved positions. One more problem is that it is sensitive to shared evolutionary history of the analyzed factors (phylogenetic trace), and special techniques need to be developed to get rid of the latter [38, 43]. A related approach, applied to the EGR subfamily of eukaryotic zinc finger TFs [47] and to bacterial LacI and TetR families [48], is assigning interaction energies to contacting pairs of residues and bases, and it may suffer from similar drawbacks. Direct analysis of available structures supplemented with calculation of a physical energy function was used to redefine binding motifs for 67 yeast TFs [49, 50]. Binding specificity predictions derived from 3D structures are systemized in the 3D-footprint database [51].
Predicted specific interactions were used to construct mutant TFs with new specificities for a variety of families, both eukaryotic, e.g. zinc fingers [52, 53] and bHLH [54], and prokaryotic, such as TAL effectors [55], LacI [28], and CRP/FNR [56]. On the other hand, extensive experimental screens sometimes produced discouraging results: randomization of DNA-interacting residues of a zinc-finger protein Zif268 [57] and LacI-family TFs [58] did not yield consistent, family-specific protein-DNA interaction codes. Most residues, including non-contacting ones, were shown to influence binding of LacI-family TFs [59, 60]. Contacting residues are not sufficient to explain binding specificity of eukaryotic FOX (forkhead box) TFs [61].
Previously we adapted a number of techniques used to identify specificity determining positions [39] to the identification of correlated protein and nucleotide positions, likely important for protein-DNA recognition. In addition to simple computation of mutual information, our algorithm assesses statistical significance correcting for (possible) overrepresentation of closely related TFs and common ancestry (phylogenetic trace) of some subgroups in a dataset. An objective threshold is set based on probabilistic calculations (the so-called Bernoulli threshold). The algorithm was implemented as a web server Prot-DNA-Korr (http://bioinf.fbb.msu.ru/Prot-DNA-Korr) and applied to study co-evolution of TFs and binding motifs in the NrtR [62] and Rex [63] families of TFs. Here we describe it in detail and apply to the MerR family of bacterial TFs.
MerR family
TFs from the MerR family regulate response to various stresses: antibiotics, heavy metals, oxidative stress [64], nitrosative stress [65, 66], heat shock [67, 68], carbonyl stress [66, 69, 70], as well as polyamine degradation [71], nitrogen metabolism [72], carotenoid biosynthesis [73], curli and biofilm formation [74], degradation of isoprenoids [75] and branched-chain amino acids [76]. In particular, the family contains a group of TFs that act as transcriptional activators of heavy metal resistance (HMR) systems. These HMR regulators form a distinct cluster within the MerR family (GenBank CDD accession number cl02600). The spectrum of toxic metals includes mercury, copper, zinc, cadmium, lead, silver, and gold.
Experimentally studied proteins, MerR, HmrR, CueR, ZntR, CadR, PbrR, GolS use mono- and divalent metal ions as ligands [64, 77, 78]. In addition, several heavy-metal resistance regulons (sets of operons regulated by particular TFs) were subject for a comparative-genomics study [79]. The binding sites of these TFs are located between the promoter −35 and −10 boxes of the regulated operons, an arrangement being typical for MerR-family transcriptional activators. Moreover, the distance between the promoter boxes in such promoters equals 19–20 bp instead of usual 16–17 bp [64, 69, 70, 79, 80]. The mechanism of transcriptional activation is known from structural and mutational studies [81]. DNA untwisting and base pair distortion decrease the distance between the promoter boxes and set them in a conformation capable of binding by the RNA polymerase. This distance change approximately equals 2 bp. Deletion of 2 bp from the promoter spacer has the same effect on transcription.
The crystal structures in complexes with DNA are known for six MerR-family proteins: BmrR [81–84], MtaN [82] and GlnR [85] from Bacillus subtilis, TnrA from Bacillus megaterium [85], SoxR from Escherichia coli [86, 87], and TipAL from Streptomyces lividans (PDB ID 2VZ4). None of them are involved in heavy metal resistance. DNA-free structures are available for BmrR [88] and Mta [89] from B. subtilis, CueR and ZntR from E. coli [90], NmlR from Bacillus thuringiensis (PDB ID 3GPV), BC_0953 from Bacillus cereus (PDB ID 3HH0), LMOf2365_2715 (PDB ID 3GP4) and lmo0526 (PDB ID 3QAO) from Listeria monocytogenes, and SCO5550 from Streptomyces coelicolor [91]. These structures show that TFs from the MerR family have very similar spatial conformations, GlnR, TnrA and SCO5550 being exceptions. The DNA-binding winged helix-turn-helix (WHTH) domain is located in the N-terminus followed by the antiparallel coiled coil providing dimerization. The ligand-binding domains located in the C-terminus may differ in length, sequence and structure. SCO5550 has a different dimerization domain resulting in a different overall structure. GlnR and TnrA have a dimerization domain located in the N-terminus that results in a different mode of interaction between monomers also yielding a different overall dimer architecture. Similar crystal structures and promoter organization suggest that the mechanism of transcriptional activation is the same for all MerR-family activators sharing this structural organization.
Methods
Here we describe an outline of the algorithm for the identification of correlated pairs of positions. The details for each step are presented in the Results section. The program takes TFs and TFBSs alignments as an input. For each pair of alignment positions we calculate the frequencies of ‘nucleotide—amino acid’ (NT-AA) pairs. From the observed and expected (under hypothesis of independence) frequences we derive a measure of correlation between pair of columns, mutual information. Applying the above steps for randomly generated pairs of columns, we obtain the expected mutual information values, which are then corrected by linear transformation to take into account shared ancestry of sequences as described in [38]. From the observed and expected mutual information values, a measure of statistical significance, Z-score, is then derived. Pairs with top Z-scores are designated as statistically significantly correlated. The actual number of pairs is determined by the Bernoulli cutoff procedure [39].
Study of MerR-family regulators of heavy-metal resistance
Genomic and protein sequences were taken from GenBank RefSeq database (release 55) [92]. Three-dimensional structures of proteins were taken from the PDB database [93]. The GenBank CDD database [94] was used for classification of transcription factor (TF) into subfamilies. Protein-DNA molecular contacts were taken from the NPIDB database [95]. Van der Waals contacts were taken from the articles in which the structures were published. In the NrtR, Rex, MerR cross-family study, Van der Waals contacts were obtained using the HBPLUS utility [96]. Structure-based multiple protein sequence alignments were built using the PROMALS3D program [97]. Phylogenetic trees were constructed using the MEGA5 package [98]. The GenomeExplorer package [99] was used to build positional weighted matrices (PWMs) and to search genomic sequences for transcription factor binding sites (TFBSs) and promoters. TFBS and operon data were submitted to the RegPrecise database [27]. Ancestral protein and DNA sequences were reconstructed using the PAML package [100]. Sequence logos were generated using the WebLogo program [101].
Results
Algorithm for the identification of correlated pairs of positions
The correlation between the residues A in an amino acid alignment column i and the bases N in a nucleotide alignment column j is measured using the mutual information:
| (1) |
where fi,j(a, n) is the observed weighted frequency of a pair (amino acid a in the TF alignment column i, nucleotide n in the site alignment column j) and is the expected weighted frequency of this pair computed as a product of fi(a), the weighted frequency of the amino acid a at the column i, and fj(n), the weighted frequency of the nucleotide n at the column j.
To estimate the statistical significance of the observed mutual information values, one needs the distribution of mutual information for a random pair of columns . In order to obtain it, TF-site pairs are randomly reconnected 10,000 times. Further, a linear transformation is applied to take into account shared ancestries (the phylogenetic trace), as described in [38]. Finally the Z-score, a measure of statistical significance, is calculated as:
| (2) |
where and are the mean and the standard deviation, respectively.
The pairs are ranked by calculated Z-scores, and the top k pairs are selected, where k is determined by the Bernoulli cutoff procedure [39]. In a nutshell it minimizes the probability (reported as p-value) to observe k given Z-scores from the Gaussian distribution.
Weighting
To avoid overrepresentation of similar, and closely related sequences, we introduce weights of pairs TF–site as products of weights of individual TF and site sequences: w(rs) = w(r) × w(s).
Here, the number of pairs residue a—nucleotide n (further denoted by [a − n]) in column [i, j] is calculated as the sum of weights of TF–site pairs:
| (3) |
where is the set of TF–site pairs with the pair [a − n] in the columns [i, j].
Similarly, residues a in the column i are counted as:
| (4) |
where is the set of TF–site pairs with the residue a at position i in TF.
Weights of TFs are determined using the Gerstein-Sonnhammer-Chothia algorithm [102]. To do that, the phylogenetic tree of TFs was constructed using the neighbor-joining method implemented in Clustal [103] and rooted in the middle of the longest path between leaves.
Pseudocounts
To account for non-observed data and to avoid null frequencies, we introduced pseudocounts supplementing the set of N observed sequences by random sequences with amino acid and nucleotide frequencies drawn from the respective alignment columns. At that, the amino acid pseudocounts reflected the amino acid substitution matrix, as in the SDPPred algorithm [39], and the normalized frequency of the amino acid a in the alignment column i was defined as:
| (5) |
where Ni(a) is the weighted count of amino acid a in column i, N is the total number of residues in the alignment column, P(b → a) is the probability of substitution b → a computed by the BLOSUM [104] matrix at identity level 30–40%, κ = 0.5 is a parameter regulating the contribution of pseudocounts.
The nucleotide pseudocounts are introduced in the same way with substitution probabilities P(m → n) = 1/4 for each pair m, n.
Finally, the frequency of a pair [a − n] in columns [i, j] is computed as:
| (6) |
By our null hypothesis nucleotide and residue substitutions are independent, thus P(b, m → a, n) = P(b → a) × 1/4 and:
| (7) |
Implementation
The algorithm is implemented in the Java language and thus can be executed on any computer provided a Java virtual machine is installed. The program and the source code may be accessed from the web at http://bioinf.fbb.msu.ru/Prot-DNA-Korr. Calculated Z-scores are graphically represented via an interactive heatmap plot (TFs vs TFBSs). A detailed NT-AA contingency table for a requested pair of positions can be drawn for in-depth analysis. Under- and overrepresented NT-AA pairs in the table are emphasized by coloring based on an arbitrary χ2-score summand cutoff (50 by default). The contingency tables, along with the tables of χ2 and mutual information summands, as well as list of Z-scores may be exported as a plain text.
Analysis of heavy-metal resistance regulators from the MerR family
Identification of transcription factors and construction of motifs
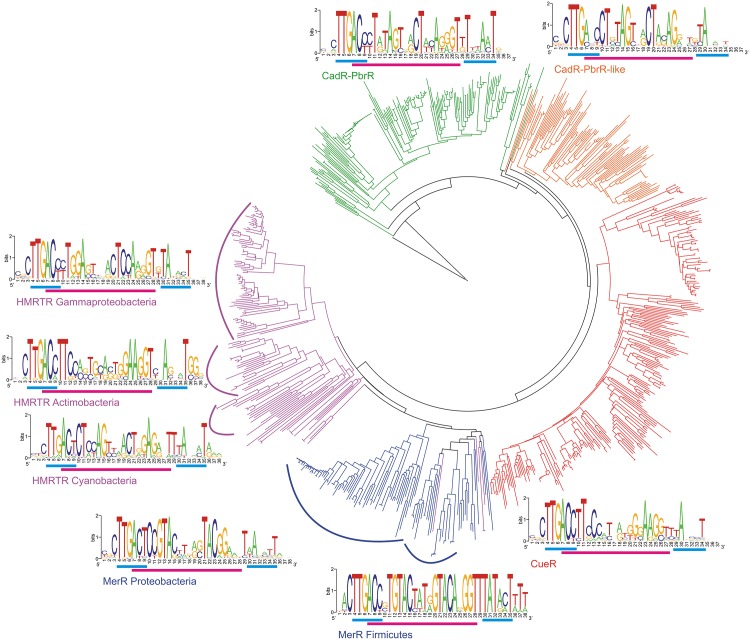
Proteins containing HTH_CueR, HTH_MerR1, HTH_CadR-PbrR, HTH_CadR-PbrR-like and HTH_HMRTR conserved domains (Specific Protein option in GenBank CDD) were downloaded from the GenBank RefSeq database. Further in this study they are referred to as TFs from the CueR, MerR, CadR-PbrR, CadR-PbrR-like and HMRTR subfamilies, respectively. Only proteins encoded in completely assembled genomes were retained for the analysis. In total they contained 1516 TFs (see Table 1 for details). TFs with sequences longer than 190 bp and shorter than 110 bp were excluded from the study, given that typical proteins of these subfamilies have the length of 130–140 bp [79, 90]. Structure-based multiple sequence alignments were constructed using structural information from CueR (PDB ID 1Q05, 1Q06, 1Q07) and ZntR (PDB ID 1Q08, 1Q09, 1Q0A) from Escherichia coli [90]. Phylogenetic trees for each subfamily were built by the neighbor-joining method with pairwise gap deletion option that keeps the information from gap-containing columns. BmrR from Bacillus subtilis (GI 50812267) was used as an outgroup. Only one of each group of nearly identical proteins (distance between the leaves on the tree less than 0.02) encoded in genomes of different strains of the same species was retained for further study. Following the application of these procedures, 906 TFs remained in the studied set (Table 1). Most of them (783 TFs) are encoded in genomes of Proteobacteria. Other phyla represented in this set include Actinobacteria (52 TFs), Cyanobacteria (22 TFs), and Firmicutes (18 TFs).
Table 1. TFs and TFBSs statistics.
| Subfamily | TFs (counts) | TFs after filtering | TFs with identified TFBSs | TFBSs |
|---|---|---|---|---|
| CueR | 511 | 260 | 238 | 324 |
| MerR | 205 | 123 | 105 | 106 |
| CadR-PbrR | 253 | 193 | 172 | 174 |
| CadR-PbrR-like | 189 | 147 | 100 | 110 |
| HMRTR | 358 | 183 | 148 | 170 |
| Total | 1516 | 906 | 763 | 884 |
We built selective PWMs for searching the genomes for putative TFBSs using sites from [79] as a starting point. One PWM per subfamily was built with the exception for MerR and HMRTR where a single PWM did not provide desired sensitivity. Hence, two PWMs were constructed for the MerR subfamily and three for the HMRTR subfamily, each corresponding to a separate smaller branch on the phylogenetic tree of studied TFs. The PWMs are presented in S1 File. The length of CueR, CadR-PbrR and CadR-PbrR-like motifs was 21 bp, whereas the length of MerR and HMRTR motifs was 22 bp. The selected genomes were searched for TFBSs in regions from −400 to +50 bp relative to the gene translation start sites annotated in GenBank. The threshold for TFBS search was set to 3.5.
Exclusion of false positive TFBS
Numerous experimental and computational studies of promoters regulated by transcriptional activators from the MerR family show that these TFs bind specific sites located between the promoter boxes of the regulated operons [64, 69, 70, 79, 80]. Moreover, the distance between the promoter boxes in such promoters equals 19–20 bp instead of usual 16–17 bp. Previous studies [105] demonstrated that the distance between the center of the TFBS and the 3’-end of the −35 promoter box is fixed within a subfamily. Putative promoters were found using the E. coli σ70 promoter consensus TTGACA-()-TATAAT. The distance between the TFBS centers and the −35 promoter boxes equals 7 bp for CueR, CadR-PbrR and CadR-PbrR-like sites (21-bp long) and 8 bp for MerR and HMRTR sites (22-bp long). TFBSs scoring above the threshold were considered false positives if they did not overlap with candidate promoters having 19–20 bp spacers or the distance between the center of the site and the 3’-end of the −35 promoter box is other than 7 bp for 21-bp sites and 8 bp for 22-bp sites. Further, only sites co-localized with the TF gene and/or located upstream of genes with relevant function (heavy metal resistance) were retained.
Using this procedure, 884 TFBSs were identified for 763 TFs (Table 1, S2 File). We tested how the usage of site and promoter overlap affects the number of found sites. We did this for weak sites (with scores from 3.5 to 5.0) and strong sites (with scores above 5.0). For strong sites, the number of candidates grows only slightly when the promoter information is omitted. In contrast, for weak sites this number grows tremendously. On average, 97% of candidate sites in a genome are weak sites without promoter support (S3 File). Therefore we used the information about putative promoters.
Sequence Logos for the sites of each studied subfamily are presented in Fig 1. Each Logo includes the binding motif as well as the −35 and −10 promoter boxes and three flanking positions. Then we built the phylogenetic tree for TFs with identified sites (Fig 1). TFs from different subfamilies form distinct branches on the tree with only several exceptions, in agreement with manually curated conserved-domain classification of HMR TFs from the MerR family provided in GenBank CDD (GenBank CDD accession number cl02600). The identified TFBSs were then aligned: one central position was deleted from the 21-bp long sites, and two, from the 22-bp long sites. For the computation of correlations, the alignment block containing 74 columns was taken from the alignment of TFs of all studied subfamilies. This block completely covers the N-terminal DNA-binding winged helix-turn-helix (WHTH) domain of these proteins. A set of corresponding pairs of protein and DNA sequences was formed by this block and the alignment of TFBSs. After deleting duplicate pairs, we obtained a set containing 776 unique pairs of corresponding protein and DNA sequences.
Fig 1. Phylogenetic tree of TFs from studied subfamilies.
Subfamily branches are colored: CueR—red, MerR—blue, CadR-PbrR—green, CadR-PbrR-like—orange, HMRTR—purple. Sequence Logos represent binding motifs (magenta bars) with −10 and −35 promoter boxes (cyan bars) and 3 flanking positions.
Identification and analysis of correlated positions
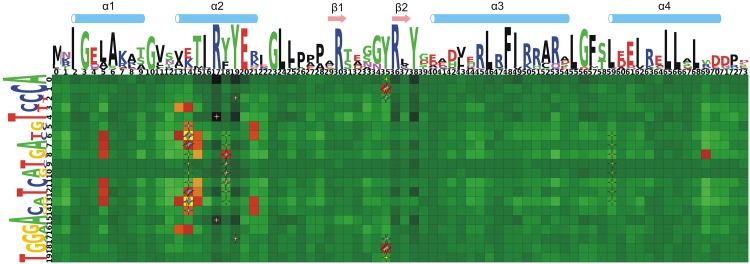
At the B-cutoff step (S1 Fig), Prot-DNA-Korr suggested 32 correlated pairs corresponding to the global minimum of the p-value. Correlation Z-scores are listed in S4 File. The heatmap showing the correlated positions is presented in Fig 2. This heatmap shows imperfect symmetry due to imperfect symmetry of TF binding sites and respective binding motifs. We searched the literature and the NPIDB database [95] for the contacts of TFs from the MerR family with DNA (Fig 2). All protein-DNA contacts (side chains to bases, side chains to DNA backbone and protein backbone to DNA backbone) are presented in S2 Fig overlaid with the same heatmap. A pair of positions was marked as interacting if the interaction was reported at least once. Since CueR and ZntR structures were resolved in the DNA-free form [90], the experimental contacts come from crystal structures of proteins from subfamilies not included in the present study [81–85, 87]. However, these experimental data are relevant, as the structures of WHTH DNA-binding domains of the TFs from the MerR family are conserved [82, 85, 87, 91]. These crystal structures of dimeric TFs (except MtaN and GlnR from B. subtilis) consist of one monomer and one DNA strand. The GlnR structure includes one monomer and one double-stranded half-site and MtaN structure includes both monomers and complete double-stranded site. Therefore we performed a mirror reflection of the contacts to cover both half-sites. This results in a strictly symmetrical map of contacts (Fig 2, S2 Fig).
Fig 2. Heatmap of protein-DNA correlations.
TF positions are along the horizontal axis and at the Logo above. Site positions are along the vertical axis and at Logo on the left. The color denotes the Z-score for a pair of positions with the color palette for significantly correlated pairs in the yellow to red interval, while black through light green colors denoting positions below the significance threshold. Protein side chain—DNA base interactions are shown as stars: blue—hydrogen bonds; red—Van der Waals contacts; yellow—water bridges; green—hydrophobic contacts. Interactions observed in the structures of complexes at least once are shown. Elements of protein secondary structure (from the crystal structure of E. coli CueR—PDB ID 1Q05) are shown at the top.
Overall, 36 experimentally identified interacting pairs (side chain to base) were found. Nine pairs appear as both correlated and forming side-chain-to-base contacts (Fisher’s exact test p-value of 1.96 × 10−8). This proves the relevance of the applied procedure and cutoff selection. Of 32 correlated pairs, 23 are located in the recognition α-helix of the HTH domain of MerR-family proteins (positions 13–21 in the protein alignment in Fig 2). Other correlated pairs correspond to the α1-helix of the WHTH domain and the β-hairpin between the β1 and β2 strands that constitutes the first wing of the WHTH domain. The most significantly correlated pairs of positions are symmetrical (6,14) and (13,14). Other correlations are much less significant.
Hereinafter pairs of positions are referred to as (j, i), where the TFBS position comes before the comma and the TF position, after. Symmetrical TFBS positions give 19 when summed. The ‘nucleotide—amino acid’ pairs for the respective pairs of positions are denoted as NT-AA.
Over- and underrepresented NT-AA pairs along with subfamilies where they preferably occur are listed in Table 2.
Table 2. Correlated pairs of positions with over- and underrepresented pairs ‘nucleotide—amino acid’.
| Positions | Residues | Count | Type | Note |
|---|---|---|---|---|
| (6,14) | T-E | 200 | + | MerR, CadR-PbrR, CadR-PbrR-like |
| C-K | 191 | + | CueR, HMRTR (Actinobacteria) | |
| G-D | 76 | + | HMRTR (Gammaproteobacteria) | |
| A-A | 7 | + | ||
| T-K | 7 | - | ||
| C-E | 3 | - | ||
| (13,14) | A-E | 184 | + | CadR-PbrR, CadR-PbrR-like, HMRTR |
| G-K | 180 | + | CueR, HMRTR (Actinobacteria) | |
| C-D | 79 | + | HMRTR (Gammaproteobacteria) | |
| T-R | 9 | + | ||
| A-K | 10 | - | ||
| G-E | 1 | - | ||
| (8,15) | A-M | 104 | + | CueR |
| G-M | 3 | - | ||
| A-T | 6 | - | ||
| (11,15) | T-M | 102 | + | CueR |
| C-M | 1 | - | ||
| T-T | 4 | - | ||
| (3,13) | C-V | 208 | + | MerR, CadR-PbrR, CadR-PbrR-like |
| T-A | 146 | + | CueR, HMRTR (Gammaproteobacteria) | |
| G-K | 3 | + | ||
| C-A | 12 | - | ||
| T-V | 9 | - | ||
| (16,13) | A-A | 136 | + | CueR, HMRTR (Gammaproteobacteria) |
| A-V | 6 | - | ||
| (12,15) | G-M | 93 | + | CueR |
| T-M | 13 | - | ||
| G-T | 13 | - | ||
| (7,15) | C-M | 86 | + | CueR |
| A-M | 18 | - | ||
| C-T | 16 | - | ||
| (5,14) | G-E | 190 | + | MerR, CadR-PbrR, CadR-PbrR-like |
| C-K | 152 | + | CueR, HMRTR (Actinobacteria and Cyanobacteria) | |
| A-Q | 38 | + | CadR-PbrR-like | |
| G-K | 24 | - | ||
| C-E | 1 | - | ||
| (14,14) | C-E | 211 | + | MerR, CadR-PbrR, CadR-PbrR-like |
| G-K | 143 | + | CueR, HMRTR (Actinobacteria) | |
| T-Q | 27 | + | ||
| T-V | 13 | + | ||
| G-E | 4 | - | ||
| (6,15) | C-M | 106 | + | CueR |
| A-Q | 6 | + | ||
| (13,15) | G-M | 96 | + | CueR |
| (6,21) | T-R | 159 | + | MerR, CadR-PbrR, CadR-PbrR-like |
| C-S | 64 | + | CueR | |
| C-E | 56 | + | CueR | |
| C-R | 7 | - | ||
| (13,21) | A-R | 149 | + | MerR, CadR-PbrR, CadR-PbrR-like |
| G-E | 53 | + | CueR | |
| C-K | 61 | + | HMRTR (Gammaproteobacteria) | |
| (7,5) | C-A | 87 | + | CueR |
| C-L | 5 | - | ||
| (12,5) | G-A | 95 | + | CueR |
| G-L | 6 | - | ||
| (14,21) | G-R | 3 | - | |
| (5,21) | C-R | 4 | - | |
| (3,14) | T-K | 191 | + | CueR, HMRTR (Actinobacteria) |
| A-H | 2 | + | ||
| C-K | 18 | - | ||
| T-E | 23 | - | ||
| (6,14) | A-K | 186 | + | CueR, HMRTR (Actinobacteria) |
| C-H | 2 | + | ||
| G-K | 22 | - | ||
| A-E | 16 | - | ||
| (12,14) | G-K | 124 | + | CueR, HMRTR (Actinobacteria) |
| (7,14) | C-K | 127 | + | CueR, HMRTR (Actinobacteria) |
| A-K | 67 | - | ||
| (8,5) | A-A | 97 | + | CueR |
| A-L | 6 | - | ||
| (11,5) | T-A | 93 | + | CueR |
| T-L | 6 | - | ||
| (13,5) | G-A | 110 | + | CueR |
| G-L | 35 | - | ||
| (6,5) | C-A | 117 | + | CueR |
| (1,35) | A-Q | 26 | + | |
| T-R | 22 | + | ||
| (18,35) | T-H | 11 | + | |
| A-I | 17 | + | ||
| A-R | 15 | + | ||
| (6,13) | C-A | 123 | + | CueR |
| (13,13) | A-V | 163 | + | MerR, CadR-PbrR, CadR-PbrR-like, HMRTR (Cyanobacteria) |
| G-A | 108 | + | ||
| (8,18) | A-H | 47 | + | |
| (8,69) | A-W | 106 | + | CueR |
Pairs of positions are ordered by decrease of statistical significance. ‘Residues’ column shows pairs of residues. In ‘Type’ column ‘+’ stands for overrepresented pair, ‘-’ stands for underrepresented pair. ‘Notes’ column shows preferred occurrence of the ‘nucleotide—amino acid’ pair.
We mapped correlated pairs on the phylogenetic tree of the studied TFs (Fig 1), using only pairs where several overrepresented pairs of residues had large (over 50) counts: (3, 13)—S3 Fig, (5,14)—S4 Fig, (6,14)—S5 Fig and (6,21)—S6 Fig. These data show that the same overrepresented pairs NT-AA appeared several times independently in course of evolution. We tested whether mutations in the TF DNA-binding domains lead to subsequent changes in binding motifs. At that, we reconstructed ancestral sequences of studied TFs and their binding sites in internal nodes of the phylogenetic tree of the TFs (data not shown). We used the Jones-Taylor-Thornton (JTT) substitution model for amino acids and general time-reversible (REV/GTR) model for nucleotides. However, we could not observe a prevalence of either protein–DNA or DNA–protein order of mutations leading to the formation of overrepresented pairs.
Algorithm performance analysis
Input data bootstraping
We studied to what extent our method tolerates inadequate data in the input. For that, we progressively shuffled residues in 10%, 20%, etc. of aligned protein sequences, simulating misalignment and wrong input data. Each progressive step was performed 100 times independently. For each pair we calculated the number of its occurrences in the top 32 correlated pairs, which corresponds to the previously established significance threshold.
Bootstrap Table 3 shows that half of 32 significantly correlated pairs remain in the list even if 50% of the data is scrambled. Moreover, top two correlated pairs remain in the list with only 30% of the valid input data. On the other hand, the weaker 1/3 of the list fall below the threshold with only 10% of scrambled data. While the ranks of the said pairs usually drop only slightly below the 32 rank threshold (S5 File), this happens in a consistent manner. For instance, the (18,35) pair originally having rank 31 never gets to the top 32 pairs with 10% of the data scrambled.
Table 3. Occurrence of the pair in the top 32 pairs of the list with fraction of the input being scrambled over 100 iterations.
| pair | 0 | 0.1 | 0.2 | 0.3 | 0.4 | 0.5 | 0.6 | 0.7 | 0.8 | 0.9 | 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| (6,14) | 100 | 100 | 100 | 100 | 100 | 100 | 99 | 90 | 46 | 12 | 3 |
| (13,14) | 100 | 100 | 100 | 100 | 100 | 100 | 98 | 85 | 40 | 5 | 2 |
| (3,13) | 100 | 100 | 100 | 100 | 100 | 99 | 87 | 67 | 35 | 14 | 5 |
| (11,15) | 100 | 100 | 100 | 100 | 99 | 100 | 91 | 88 | 63 | 37 | 7 |
| (8,15) | 100 | 100 | 100 | 100 | 98 | 97 | 90 | 81 | 52 | 29 | 3 |
| (5,14) | 100 | 100 | 100 | 99 | 98 | 86 | 60 | 40 | 18 | 5 | 1 |
| (16,13) | 100 | 100 | 100 | 97 | 87 | 83 | 53 | 45 | 13 | 12 | 2 |
| (3,14) | 100 | 100 | 99 | 99 | 95 | 89 | 72 | 55 | 26 | 6 | 7 |
| (8,69) | 100 | 100 | 99 | 96 | 94 | 95 | 85 | 72 | 56 | 31 | 10 |
| (16,14) | 100 | 100 | 98 | 98 | 93 | 82 | 70 | 37 | 27 | 7 | 2 |
| (14,14) | 100 | 100 | 97 | 94 | 83 | 71 | 43 | 25 | 7 | 2 | 1 |
| (12,15) | 100 | 99 | 100 | 100 | 98 | 98 | 93 | 81 | 59 | 28 | 10 |
| (7,15) | 100 | 99 | 100 | 99 | 97 | 94 | 82 | 66 | 46 | 14 | 2 |
| (12,14) | 100 | 98 | 91 | 86 | 74 | 73 | 41 | 36 | 14 | 8 | 4 |
| (7,14) | 100 | 96 | 90 | 87 | 62 | 62 | 39 | 29 | 19 | 10 | 2 |
| (6,21) | 100 | 95 | 74 | 68 | 52 | 42 | 32 | 15 | 11 | 7 | 5 |
| (13,21) | 100 | 87 | 67 | 50 | 41 | 26 | 29 | 10 | 9 | 4 | 3 |
| (14,21) | 100 | 87 | 56 | 38 | 20 | 21 | 11 | 9 | 2 | 1 | 0 |
| (5,21) | 100 | 84 | 61 | 41 | 31 | 24 | 12 | 6 | 3 | 1 | 1 |
| (6,15) | 100 | 73 | 78 | 77 | 75 | 67 | 59 | 33 | 27 | 7 | 2 |
| (6,13) | 100 | 55 | 57 | 40 | 33 | 15 | 20 | 16 | 13 | 6 | 0 |
| (13,13) | 100 | 49 | 34 | 33 | 29 | 13 | 15 | 12 | 9 | 6 | 1 |
| (12,5) | 100 | 48 | 36 | 28 | 40 | 29 | 18 | 19 | 13 | 5 | 0 |
| (7,5) | 100 | 41 | 39 | 26 | 31 | 24 | 17 | 18 | 13 | 1 | 3 |
| (1,35) | 100 | 35 | 33 | 24 | 19 | 12 | 5 | 2 | 2 | 0 | 1 |
| (13,15) | 100 | 32 | 39 | 45 | 44 | 40 | 28 | 28 | 20 | 6 | 4 |
| (8,5) | 100 | 11 | 17 | 24 | 29 | 27 | 24 | 23 | 16 | 10 | 3 |
| (11,5) | 100 | 6 | 12 | 11 | 26 | 24 | 12 | 24 | 22 | 11 | 3 |
| (13,5) | 100 | 3 | 4 | 9 | 18 | 12 | 13 | 14 | 12 | 5 | 5 |
| (8,18) | 100 | 2 | 11 | 12 | 11 | 22 | 18 | 10 | 18 | 14 | 2 |
| (18,35) | 100 | 0 | 3 | 3 | 2 | 4 | 5 | 6 | 2 | 1 | 0 |
| (6,5) | 99 | 1 | 4 | 8 | 12 | 17 | 12 | 12 | 8 | 4 | 2 |
| (11,69) | 1 | 100 | 100 | 92 | 96 | 91 | 84 | 70 | 63 | 38 | 5 |
| (12,69) | 0 | 88 | 80 | 74 | 75 | 55 | 53 | 44 | 30 | 18 | 5 |
| (6,69) | 0 | 78 | 66 | 45 | 52 | 33 | 41 | 23 | 17 | 10 | 4 |
| (11,12) | 0 | 72 | 60 | 56 | 45 | 41 | 47 | 20 | 28 | 11 | 5 |
| (13,1) | 0 | 64 | 79 | 70 | 61 | 49 | 32 | 30 | 7 | 7 | 1 |
| (6,1) | 0 | 60 | 72 | 72 | 51 | 56 | 36 | 27 | 20 | 4 | 5 |
| (7,1) | 0 | 54 | 49 | 48 | 44 | 40 | 32 | 28 | 15 | 12 | 3 |
| (7,69) | 0 | 53 | 47 | 37 | 20 | 23 | 20 | 19 | 12 | 10 | 5 |
| (13,69) | 0 | 52 | 37 | 26 | 31 | 17 | 23 | 13 | 14 | 10 | 4 |
The bootstrap table suggests that the bottom 1/3 of the correlated list are sensitive to the input data quality and, together with some pairs falling just below the significance threshold may be considered as the “grey area”.
We also performed negative control of our method by providing shuffled regulator-site pairs from the initial input data. The shuffling was performed similarly to shuffling for expected mutual information required for Z-scores calculation. The B-cutoff global minimum log(p − value) = −14 (S7 Fig) obtained is negligible compared to −1250 yielded by the original data (S1 Fig).
Conservation and correlation
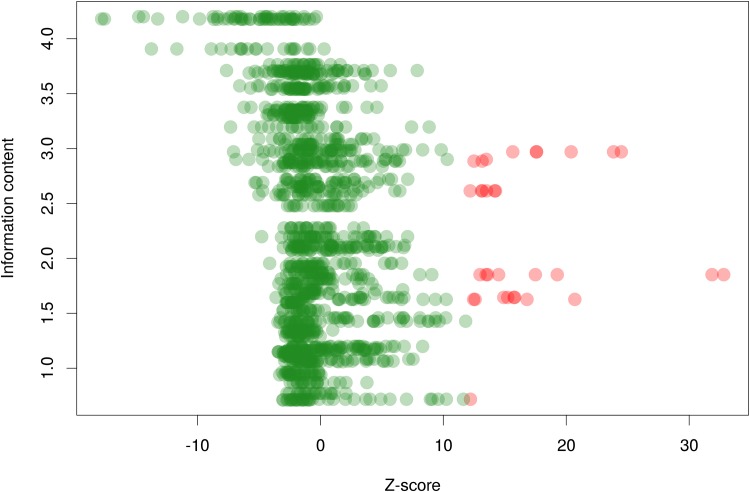
Protein positional information content used in the logo generation as a measure of conservation was compared with Z-scores for corresponding pairs of columns. The correlated protein positions appear to be moderately conserved. Fig 3 suggests that overall highly conserved residues tend to have lower z-scores.
Fig 3. Z-score vs. protein column conservation.
Red—significantly correlated pairs. Green—other pairs. Y-axis is the protein positional information content for corresponding pair of columns after weighting and adding pseudocounts. X-axis is the Z-score of a pair.
Conserved correlated residues that form contacts are very rare. In three PDB (3ikt, 3gz6, 1r8e) structures from the Rex, NrtR and MerR families, respectively, we found 43 contacts with either of the partners being conserved (S6 File). Only two such contacts in the NrtR structure appeared to be correlated.
Discussion
We developed and implemented an algorithm for the identification of pairs of positions that are important for the protein-DNA recognition. Our method requires multiple alignments of DNA-binding proteins and of their respective sites. The method does not rely on known 3D structures of protein-DNA complexes, here we rather use them to validate our results. It should be noted that of necessity the contacts and correlations were identified on different sets of TFs belonging to different subfamilies.
The comparison with structural data shows good agreement both in quantitative and qualitative terms. The sets of correlated and contacting pairs strongly overlap (Fisher’s test p-value 1.96 × 10−8). The recognition helix of the HTH domain contains a large cluster of correlated pairs. According to classic Suzuki studies of spatial structures [2], residues 1, 2, 6 of the recognition helix that face the DNA major groove are most important for the protein-DNA recognition as they form hydrogen bonds with DNA bases hence allowing the protein to read the DNA sequence. Here these residues participated in correlated pairs, with residue 2 being the most correlated. The MerR and previously studied Rex [63] and NrtR [62] families provide correlation data on three families HTH binding domains. Residue 6 participates in correlations in all three families and residues 1, 2 in two familes each.
Among hydrogen bonds, Van der Waals interactions, and hydrophobic contacts in the three families we do not see any preference for either type to correspond to correlations (S5 File). However, the data are not sufficient to form a solid conclusion.
Although significantly correlated pairs are likely to be contacting ones, our algorithm is not merely a substitute for a 3D structures analysis. Conserved interactions will not demonstrate correlations due to the lack of sequence variation [45]. On the other hand, some residues may affect specificity indirectly, and it would be difficult to identify them in 3D structures. The correlation analysis identifies all coevolving pairs of positions and along with overrepresented NT-AA pairs thus providing hints for future experimental investigations [56].
In most correlated pairs of positions, overrepresented NT-AA pairs appear independently multiple times in course of evolution of the studied TFs. It has been shown that binding sites for existing TFs can emerge rather rapidly from sequences that resemble weak sites [106, 107]. This model implies that changes in a TF sequence, decreasing its affinity to pre-existing sites, yield changes in the sites, hence restoring the effective binding. The binding motif (in the simplest form, the consensus of the sites) changes accordingly. We reconstructed ancestral sequences of TFs and the respective DNA motifs, but failed to confirm the hypothesis about the leading role of substitutions in TFs yielding subsequent substitutions in recognized sites and hence motifs.
We used several crystal structures of related TFs in the DNA-bound form to demonstrate high level of coincidence between correlated pairs and contacting positions. At that, it is plausible that the conserved positions provide for the initial DNA binding, whereas correlated positions fine-tune interactions with specific sites. A proof of concept was provided by an experimental study of CRP, that demonstrated lack of specific binding after individual mutations in either the TF or the site, but partially reconstituted binding after dual TF-site mutations substituting one preferred NT-AA pair to another pair preferred at the given positions [56]. While existing computational methods may not predict DNA motif given only TF sequence and 3D structure, some progress has been already made. For example, it is possible to match each TF from a given family, present in a genome, to the respective motif from a given set of motifs recognized by these TFs in the same genome [108]. The latter situation arises in comparative-genomic prediction of transcriptional networks.
TFBS prediction and regulon reconstruction in multiple related genomes using comparative genomic approaches has become a major source of information about regulatory networks. Combined with identification of correlations between the sequences of TFs and their binding sites, they may become powerful tools for studying the evolution of TF families and coevolution of interacting protein and DNA sequences using sequence data alone.
Supporting Information
(PDF)
Data on different subfamilies are presented on separate sheets. Only the first members of regulated operons are shown. TFBS positions are given relative to translation starts of regulated genes annotated in the genomes.
(XLS)
Horizontal axis shows the percentage of sites from a given category from all sites found in genome. The vertical axis shows the number of such genomes.
(PDF)
(PDF)
(XLS)
(XLS)
Global minimum p-value corresponds to 32 pairs.
(PDF)
TF positions are along the horizontal axis and at the Logo above. Site positions are along the vertical axis and at Logo on the left. The color denotes the Z-score for a pair of positions with the color palette for significantly correlated pairs in the yellow to red interval, while black through light green colors denote positions below the significance threshold. Protein-DNA interactions are shown as stars. Interactions observed in the structures of complexes at least once are shown. Elements of protein secondary structure (from the crystal structure of E. coli CueR – PDB ID 1Q05) are shown at the top.
(PDF)
Colors of branches show overrepresented pairs of residues in positions (3,13) (see color codein the picture). Background colors show TF subfamilies: red—CueR, blue – MerR, green—CadR-PbrR, beige—CadR-PbrR-like, pink—HMRTR.
(PDF)
Colors of branches show overrepresented pairs of residues in positions (5,14) (see color code in the picture). Background colors show TF subfamilies: red—CueR, blue – MerR, green—CadR-PbrR, beige—CadR-PbrR-like, pink—HMRTR.
(PDF)
Colors of branches show overrepresented pairs of residues in positions (6,14) (see color code in the picture). Background colors show TF subfamilies: red—CueR, blue – MerR, green—CadR-PbrR, beige—CadR-PbrR-like, pink—HMRTR.
(PDF)
Colors of branches show overrepresented pairs of residues in positions (6,21) (see color code in the picture). Background colors show TF subfamilies: red—CueR, blue – MerR, green—CadR-PbrR, beige—CadR-PbrR-like, pink—HMRTR.
(PDF)
(PDF)
Acknowledgments
We are grateful to Georgii A. Bazykin for useful discussions. We also thank Prof. A.S. Kondrashov for sharing computational facilities provided under Russian Ministry of Science and Education grant [11.G34.31.0008].
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Russian Science Foundation (http://www.rscf.ru/en/), grant No 14-24-00155 (received by YDK, IAZ, MSG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976. March;73(3):804–808. 10.1073/pnas.73.3.804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Suzuki M, Brenner SE, Gerstein M, Yagi N. DNA recognition code of transcription factors. Protein Eng. 1995. April;8(4):319–328. 10.1093/protein/8.4.319 [DOI] [PubMed] [Google Scholar]
- 3. Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001. July;29(13):2860–2874. 10.1093/nar/29.13.2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones S, Shanahan HP, Berman HM, Thornton JM. Using electrostatic potentials to predict DNA-binding sites on DNA-binding proteins. Nucleic Acids Res. 2003. December;31(24):7189–7198. 10.1093/nar/gfg922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baker CM, Grant GH. Role of aromatic amino acids in protein-nucleic acid recognition. Biopolymers. 2007;85(5-6):456–470. Available from: 10.1002/bip.20682 [DOI] [PubMed] [Google Scholar]
- 6. Sarai A, Kono H. Protein-DNA recognition patterns and predictions. Annu Rev Biophys Biomol Struct. 2005;34:379–398. Available from: 10.1146/annurev.biophys.34.040204.144537 [DOI] [PubMed] [Google Scholar]
- 7. Benos PV, Lapedes AS, Stormo GD. Is there a code for protein-DNA recognition? Probab(ilistical)ly… Bioessays. 2002. May;24(5):466–475. Available from: 10.1002/bies.10073 [DOI] [PubMed] [Google Scholar]
- 8. Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1(1):REVIEWS001 10.1186/gb-2000-1-1-reviews001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pabo CO, Nekludova L. Geometric analysis and comparison of protein-DNA interfaces: why is there no simple code for recognition? J Mol Biol. 2000. August;301(3):597–624. Available from: 10.1006/jmbi.2000.3918 [DOI] [PubMed] [Google Scholar]
- 10. Siggers TW, Silkov A, Honig B. Structural alignment of protein–DNA interfaces: insights into the determinants of binding specificity. J Mol Biol. 2005. February;345(5):1027–1045. Available from: 10.1016/j.jmb.2004.11.010 [DOI] [PubMed] [Google Scholar]
- 11. Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981. November;20(24):6929–6948. 10.1021/bi00527a028 [DOI] [PubMed] [Google Scholar]
- 12. Ofran Y, Mysore V, Rost B. Prediction of DNA-binding residues from sequence. Bioinformatics. 2007. July;23(13):i347–i353. Available from: 10.1093/bioinformatics/btm174 [DOI] [PubMed] [Google Scholar]
- 13. Ahmad S, Sarai A. PSSM-based prediction of DNA binding sites in proteins. BMC Bioinformatics. 2005;6:33 Available from: 10.1186/1471-2105-6-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yan C, Terribilini M, Wu F, Jernigan RL, Dobbs D, Honavar V. Predicting DNA-binding sites of proteins from amino acid sequence. BMC Bioinformatics. 2006;7:262 Available from: 10.1186/1471-2105-7-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oliphant AR, Brandl CJ, Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol Cell Biol. 1989. July;9(7):2944–2949. 10.1128/MCB.9.7.2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Bakel H, van Werven FJ, Radonjic M, Brok MO, van Leenen D, Holstege FCP, et al. Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification. Nucleic Acids Res. 2008. March;36(4):e21 Available from: 10.1093/nar/gkm1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004. September;431(7004):99–104. Available from: 10.1038/nature02800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu X, Noll DM, Lieb JD, Clarke ND. DIP-chip: rapid and accurate determination of DNA-binding specificity. Genome Res. 2005. March;15(3):421–427. Available from: 10.1101/gr.3256505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014. March;42(5):2976–2987. Available from: 10.1093/nar/gkt1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berger MF, Bulyk ML. Protein binding microarrays (PBMs) for rapid, high-throughput characterization of the sequence specificities of DNA binding proteins. Methods Mol Biol. 2006;338:245–260. Available from: 10.1385/1-59745-097-9:245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rodionov DA. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem Rev. 2007. August;107(8):3467–3497. Available from: 10.1021/cr068309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113 Available from: 10.1186/1471-2105-7-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006. January;34(Database issue):D108–D110. Available from: 10.1093/nar/gkj143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014. January;42(Database issue):D142–D147. Available from: 10.1093/nar/gkt997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang J, Zhuang J, Iyer S, Lin XY, Greven MC, Kim BH, et al. Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res. 2013. January;41(Database issue):D171–D176. Available from: 10.1093/nar/gks1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kazakov AE, Cipriano MJ, Novichkov PS, Minovitsky S, Vinogradov DV, Arkin A, et al. RegTransBase–a database of regulatory sequences and interactions in a wide range of prokaryotic genomes. Nucleic Acids Res. 2007. January;35(Database issue):D407–D412. Available from: 10.1093/nar/gkl865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, et al. RegPrecise 3.0–a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics. 2013;14:745 Available from: 10.1186/1471-2164-14-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Camas FM, Alm EJ, Poyatos JF. Local gene regulation details a recognition code within the LacI transcriptional factor family. PLoS Comput Biol. 2010;6(11):e1000989 10.1371/journal.pcbi.1000989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rigali S, Schlicht M, Hoskisson P, Nothaft H, Merzbacher M, Joris B, et al. Extending the classification of bacterial transcription factors beyond the helix-turn-helix motif as an alternative approach to discover new cis/trans relationships. Nucleic Acids Res. 2004;32(11):3418–3426. Available from: 10.1093/nar/gkh673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sandelin A, Wasserman WW. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J Mol Biol. 2004. April;338(2):207–215. Available from: 10.1016/j.jmb.2004.02.048 [DOI] [PubMed] [Google Scholar]
- 31. Luscombe NM, Thornton JM. Protein-DNA interactions: amino acid conservation and the effects of mutations on binding specificity. J Mol Biol. 2002. July;320(5):991–1009. 10.1016/S0022-2836(02)00571-5 [DOI] [PubMed] [Google Scholar]
- 32. Mirny LA, Gelfand MS. Structural analysis of conserved base pairs in protein-DNA complexes. Nucleic Acids Res. 2002. April;30(7):1704–1711. 10.1093/nar/30.7.1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009. December;326(5959):1501 Available from: 10.1126/science.1178817 [DOI] [PubMed] [Google Scholar]
- 34. Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009. December;326(5959):1509–1512. Available from: 10.1126/science.1178811 [DOI] [PubMed] [Google Scholar]
- 35. Grau J, Wolf A, Reschke M, Bonas U, Posch S, Boch J. Computational predictions provide insights into the biology of TAL effector target sites. PLoS Comput Biol. 2013;9(3):e1002962 Available from: 10.1371/journal.pcbi.1002962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pérez-Quintero AL, Rodriguez-R LM, Dereeper A, López C, Koebnik R, Szurek B, et al. An improved method for TAL effectors DNA-binding sites prediction reveals functional convergence in TAL repertoires of Xanthomonas oryzae strains. PLoS One. 2013;8(7):e68464 Available from: 10.1371/journal.pone.0068464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hall BM, Lefevre KR, Cordes MHJ. Sequence correlations between Cro recognition helices and cognate O(R) consensus half-sites suggest conserved rules of protein-DNA recognition. J Mol Biol. 2005. July;350(4):667–681. Available from: 10.1016/j.jmb.2005.05.025 [DOI] [PubMed] [Google Scholar]
- 38. Mirny LA, Gelfand MS. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J Mol Biol. 2002. August;321(1):7–20. 10.1016/S0022-2836(02)00587-9 [DOI] [PubMed] [Google Scholar]
- 39. Kalinina OV, Mironov AA, Gelfand MS, Rakhmaninova AB. Automated selection of positions determining functional specificity of proteins by comparative analysis of orthologous groups in protein families. Protein Sci. 2004. February;13(2):443–456. Available from: 10.1110/ps.03191704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Donald JE, Shakhnovich EI. Predicting specificity-determining residues in two large eukaryotic transcription factor families. Nucleic Acids Res. 2005;33(14):4455–4465. Available from: 10.1093/nar/gki755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mazin PV, Gelfand MS, Mironov AA, Rakhmaninova AB, Rubinov AR, Russell RB, et al. An automated stochastic approach to the identification of the protein specificity determinants and functional subfamilies. Algorithms Mol Biol. 2010;5:29 Available from: 10.1186/1748-7188-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gloor GB, Martin LC, Wahl LM, Dunn SD. Mutual information in protein multiple sequence alignments reveals two classes of coevolving positions. Biochemistry. 2005. May;44(19):7156–7165. Available from: 10.1021/bi050293e [DOI] [PubMed] [Google Scholar]
- 43. Dunn SD, Wahl LM, Gloor GB. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008. February;24(3):333–340. Available from: 10.1093/bioinformatics/btm604 [DOI] [PubMed] [Google Scholar]
- 44. Shackelford G, Karplus K. Contact prediction using mutual information and neural nets. Proteins. 2007;69 Suppl 8:159–164. Available from: 10.1002/prot.21791 [DOI] [PubMed] [Google Scholar]
- 45. Mahony S, Auron PE, Benos PV. Inferring protein-DNA dependencies using motif alignments and mutual information. Bioinformatics. 2007. July;23(13):i297–i304. Available from: 10.1093/bioinformatics/btm215 [DOI] [PubMed] [Google Scholar]
- 46. Yang S, Yalamanchili HK, Li X, Yao KM, Sham PC, Zhang MQ, et al. Correlated evolution of transcription factors and their binding sites. Bioinformatics. 2011. November;27(21):2972–2978. Available from: 10.1093/bioinformatics/btr503 [DOI] [PubMed] [Google Scholar]
- 47. Benos PV, Lapedes AS, Stormo GD. Probabilistic code for DNA recognition by proteins of the EGR family. J Mol Biol. 2002. November;323(4):701–727. 10.1016/S0022-2836(02)00917-8 [DOI] [PubMed] [Google Scholar]
- 48. Sahota G, Stormo GD. Novel sequence-based method for identifying transcription factor binding sites in prokaryotic genomes. Bioinformatics. 2010. November;26(21):2672–2677. Available from: 10.1093/bioinformatics/btq501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morozov AV, Havranek JJ, Baker D, Siggia ED. Protein-DNA binding specificity predictions with structural models. Nucleic Acids Res. 2005;33(18):5781–5798. Available from: 10.1093/nar/gki875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morozov AV, Siggia ED. Connecting protein structure with predictions of regulatory sites. Proc Natl Acad Sci U S A. 2007. April;104(17):7068–7073. Available from: 10.1073/pnas.0701356104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Contreras-Moreira B. 3D-footprint: a database for the structural analysis of protein-DNA complexes. Nucleic Acids Res. 2010. January;38(Database issue):D91–D97. Available from: 10.1093/nar/gkp781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Desjarlais JR, Berg JM. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc Natl Acad Sci U S A. 1993. March;90(6):2256–2260. 10.1073/pnas.90.6.2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sera T. Zinc-finger-based artificial transcription factors and their applications. Adv Drug Deliv Rev. 2009. July;61(7-8):513–526. Available from: 10.1016/j.addr.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 54. De Masi F, Grove CA, Vedenko A, Alibés A, Gisselbrecht SS, Serrano L, et al. Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res. 2011. June;39(11):4553–4563. Available from: 10.1093/nar/gkr070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Geissler R, Scholze H, Hahn S, Streubel J, Bonas U, Behrens SE, et al. Transcriptional activators of human genes with programmable DNA-specificity. PLoS One. 2011;6(5):e19509 Available from: 10.1371/journal.pone.0019509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Desai TA, Rodionov DA, Gelfand MS, Alm EJ, Rao CV. Engineering transcription factors with novel DNA-binding specificity using comparative genomics. Nucleic Acids Res. 2009. May;37(8):2493–2503. Available from: 10.1093/nar/gkp079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu H, Yang WP, Barbas C 3rd. Building zinc fingers by selection: toward a therapeutic application. Proc Natl Acad Sci U S A. 1995. January;92(2):344–348. 10.1073/pnas.92.2.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Milk L, Daber R, Lewis M. Functional rules for lac repressor-operator associations and implications for protein-DNA interactions. Protein Sci. 2010. June;19(6):1162–1172. Available from: 10.1002/pro.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tungtur S, Meinhardt S, Swint-Kruse L. Comparing the functional roles of nonconserved sequence positions in homologous transcription repressors: implications for sequence/function analyses. J Mol Biol. 2010. January;395(4):785–802. Available from: 10.1016/j.jmb.2009.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tungtur S, Parente DJ, Swint-Kruse L. Functionally important positions can comprise the majority of a protein’s architecture. Proteins. 2011. May;79(5):1589–1608. Available from: 10.1002/prot.22985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc Natl Acad Sci U S A. 2013. July;110(30):12349–12354. Available from: 10.1073/pnas.1310430110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang N, Ingeniis JD, Galeazzi L, Mancini C, Korostelev YD, Rakhmaninova AB, et al. Structure and function of an ADP-ribose-dependent transcriptional regulator of NAD metabolism. Structure. 2009. July;17(7):939–951. Available from: 10.1016/j.str.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ravcheev DA, Li X, Latif H, Zengler K, Leyn SA, Korostelev YD, et al. Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex. J Bacteriol. 2012. March;194(5):1145–1157. Available from: 10.1128/JB.06412-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003. June;27(2-3):145–163. 10.1016/S0168-6445(03)00051-2 [DOI] [PubMed] [Google Scholar]
- 65. Spiro S. Regulators of bacterial responses to nitric oxide. FEMS microbiology reviews. 2007;31(2):193–211. 10.1111/j.1574-6976.2006.00061.x [DOI] [PubMed] [Google Scholar]
- 66. McEwan AG, Djoko KY, Chen NH, Couñago RLM, Kidd SP, Potter AJ, et al. Novel bacterial MerR-like regulators their role in the response to carbonyl and nitrosative stress. Adv Microb Physiol. 2011;58:1–22. Available from: 10.1016/B978-0-12-381043-4.00001-5 [DOI] [PubMed] [Google Scholar]
- 67. Bucca G, Ferina G, Puglia AM, Smith CP. The dnaK operon of Streptomyces coelicolor encodes a novel heat-shock protein which binds to the promoter region of the operon. Mol Microbiol. 1995. August;17(4):663–674. 10.1111/j.1365-2958.1995.mmi_17040663.x [DOI] [PubMed] [Google Scholar]
- 68. Zomer A, Fernandez M, Kearney B, Fitzgerald GF, Ventura M, van Sinderen D. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J Bacteriol. 2009. November;191(22):7039–7049. Available from: 10.1128/JB.00897-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kidd SP, Potter AJ, Apicella MA, Jennings MP, McEwan AG. NmlR of Neisseria gonorrhoeae: a novel redox responsive transcription factor from the MerR family. Mol Microbiol. 2005. September;57(6):1676–1689. Available from: 10.1111/j.1365-2958.2005.04773.x [DOI] [PubMed] [Google Scholar]
- 70. Nguyen TTH, Eiamphungporn W, Mäder U, Liebeke M, Lalk M, Hecker M, et al. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR). Mol Microbiol. 2009. February;71(4):876–894. Available from: 10.1111/j.1365-2958.2008.06568.x [DOI] [PubMed] [Google Scholar]
- 71. Woolridge DP, Martinez JD, Stringer DE, Gerner EW. Characterization of a novel spermidine/spermine acetyltransferase, BltD, from Bacillus subtilis. Biochem J. 1999. June;340 (Pt 3):753–758. 10.1042/bj3400753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fisher SH. Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence! Mol Microbiol. 1999. April;32(2):223–232. 10.1046/j.1365-2958.1999.01333.x [DOI] [PubMed] [Google Scholar]
- 73. Pérez-Marín MC, Padmanabhan S, Polanco MC, Murillo FJ, Elías-Arnanz M. Vitamin B12 partners the CarH repressor to downregulate a photoinducible promoter in Myxococcus xanthus. Mol Microbiol. 2008. February;67(4):804–819. Available from: 10.1111/j.1365-2958.2007.06086.x [DOI] [PubMed] [Google Scholar]
- 74. Ogasawara H, Yamamoto K, Ishihama A. Regulatory role of MlrA in transcription activation of csgD, the master regulator of biofilm formation in Escherichia coli. FEMS Microbiol Lett. 2010. November;312(2):160–168. Available from: 10.1111/j.1574-6968.2010.02112.x [DOI] [PubMed] [Google Scholar]
- 75. Díaz-Pérez AL, Zavala-Hernández AN, Cervantes C, Campos-García J. The gnyRDBHAL cluster is involved in acyclic isoprenoid degradation in Pseudomonas aeruginosa. Appl Environ Microbiol. 2004. September;70(9):5102–5110. Available from: 10.1128/AEM.70.9.5102-5110.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kazakov AE, Rodionov DA, Alm E, Arkin AP, Dubchak I, Gelfand MS. Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in proteobacteria. J Bacteriol. 2009. January;191(1):52–64. Available from: 10.1128/JB.01175-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen PR, He C. Selective recognition of metal ions by metalloregulatory proteins. Curr Opin Chem Biol. 2008. April;12(2):214–221. Available from: 10.1016/j.cbpa.2007.12.010 [DOI] [PubMed] [Google Scholar]
- 78. Summers AO. Damage control: regulating defenses against toxic metals and metalloids. Curr Opin Microbiol. 2009. April;12(2):138–144. Available from: 10.1016/j.mib.2009.02.003 [DOI] [PubMed] [Google Scholar]
- 79. Permina EA, Kazakov AE, Kalinina OV, Gelfand MS. Comparative genomics of regulation of heavy metal resistance in Eubacteria. BMC Microbiol. 2006;6:49 Available from: 10.1186/1471-2180-6-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ahmed M, Lyass L, Markham PN, Taylor SS, Vázquez-Laslop N, Neyfakh AA. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J Bacteriol. 1995. July;177(14):3904–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Heldwein EE, Brennan RG. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature. 2001. January;409(6818):378–382. Available from: 10.1038/35053138 [DOI] [PubMed] [Google Scholar]
- 82. Newberry KJ, Brennan RG. The structural mechanism for transcription activation by MerR family member multidrug transporter activation, N terminus. J Biol Chem. 2004. May;279(19):20356–20362. Available from: 10.1074/jbc.M400960200 [DOI] [PubMed] [Google Scholar]
- 83. Newberry KJ, Huffman JL, Miller MC, Vazquez-Laslop N, Neyfakh AA, Brennan RG. Structures of BmrR-drug complexes reveal a rigid multidrug binding pocket and transcription activation through tyrosine expulsion. J Biol Chem. 2008. September;283(39):26795–26804. Available from: 10.1074/jbc.M804191200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bachas S, Eginton C, Gunio D, Wade H. Structural contributions to multidrug recognition in the multidrug resistance (MDR) gene regulator, BmrR. Proc Natl Acad Sci U S A. 2011. July;108(27):11046–11051. Available from: 10.1073/pnas.1104850108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Schumacher MA, Chinnam NB, Cuthbert B, Tonthat NK, Whitfill T. Structures of regulatory machinery reveal novel molecular mechanisms controlling B. subtilis nitrogen homeostasis. Genes Dev. 2015. February;29(4):451–464. Available from: 10.1101/gad.254714.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Watanabe S, Kita A, Kobayashi K, Takahashi Y, Miki K. Crystallization and preliminary X-ray crystallographic studies of the oxidative-stress sensor SoxR and its complex with DNA. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006. December;62(Pt 12):1275–1277. Available from: 10.1107/S1744309106048482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Watanabe S, Kita A, Kobayashi K, Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Natl Acad Sci U S A. 2008. March;105(11):4121–4126. Available from: 10.1073/pnas.0709188105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kumaraswami M, Newberry KJ, Brennan RG. Conformational plasticity of the coiled-coil domain of BmrR is required for bmr operator binding: the structure of unliganded BmrR. J Mol Biol. 2010. April;398(2):264–275. Available from: 10.1016/j.jmb.2010.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Godsey MH, Baranova NN, Neyfakh AA, Brennan RG. Crystal structure of MtaN, a global multidrug transporter gene activator. J Biol Chem. 2001. December;276(50):47178–47184. Available from: 10.1074/jbc.M105819200 [DOI] [PubMed] [Google Scholar]
- 90. Changela A, Chen K, Xue Y, Holschen J, Outten CE, O’Halloran TV, et al. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003. September;301(5638):1383–1387. Available from: 10.1126/science.1085950 [DOI] [PubMed] [Google Scholar]
- 91. Hayashi T, Tanaka Y, Sakai N, Watanabe N, Tamura T, Tanaka I, et al. Structural and genomic DNA analysis of a putative transcription factor SCO5550 from Streptomyces coelicolor A3(2): regulating the expression of gene sco5551 as a transcriptional activator with a novel dimer shape. Biochem Biophys Res Commun. 2013. May;435(1):28–33. Available from: 10.1016/j.bbrc.2013.04.017 [DOI] [PubMed] [Google Scholar]
- 92. Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009. January;37(Database issue):D32–D36. Available from: 10.1093/nar/gkn721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, et al. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002. June;58(Pt 6 No 1):899–907. 10.1107/S0907444902003451 [DOI] [PubMed] [Google Scholar]
- 94. Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007. January;35(Database issue):D237–D240. Available from: 10.1093/nar/gkl951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kirsanov DD, Zanegina ON, Aksianov EA, Spirin SA, Karyagina AS, Alexeevski AV. NPIDB: Nucleic acid-Protein Interaction DataBase. Nucleic Acids Res. 2013. January;41(Database issue):D517–D523. Available from: 10.1093/nar/gks1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994. May;238(5):777–793. Available from: 10.1006/jmbi.1994.1334 [DOI] [PubMed] [Google Scholar]
- 97. Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008. April;36(7):2295–2300. Available from: 10.1093/nar/gkn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011. October;28(10):2731–2739. Available from: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Mironov AA, Vinokurova NP, Gelfand MS. Software for analysis of bacterial genomes. Molecular Biology. 2000;34(2):222–231. Available from: 10.1007/BF02759643 [DOI] [Google Scholar]
- 100. Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007. August;24(8):1586–1591. Available from: 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- 101. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004. June;14(6):1188–1190. Available from: 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gerstein M, Sonnhammer EL, Chothia C. Volume changes in protein evolution. J Mol Biol. 1994. March;236(4):1067–1078. 10.1016/0022-2836(94)90012-4 [DOI] [PubMed] [Google Scholar]
- 103. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003. July;31(13):3497–3500. Available from: http://dx.doi.org/10.1093%2Fbioinformatics%2Fbtm404 10.1093/nar/gkg500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Henikoff JG, Henikoff S. Blocks database and its applications. Methods Enzymol. 1996;266:88–105. 10.1016/S0076-6879(96)66008-X [DOI] [PubMed] [Google Scholar]
- 105. Zharov IA, Gelfand MS, Kazakov AE. Regulation of multidrug resistance genes by transcription factors of the BltR subfamily. Molecular Biology. 2011;45(4):658–666. Available from: 10.1134/S002689331103023X [DOI] [PubMed] [Google Scholar]
- 106. Berg J, Willmann S, Lässig M. Adaptive evolution of transcription factor binding sites. BMC Evol Biol. 2004. October;4:42 Available from: 10.1186/1471-2148-4-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. MacArthur S, Brookfield JFY. Expected rates and modes of evolution of enhancer sequences. Mol Biol Evol. 2004. June;21(6):1064–1073. Available from: 10.1093/molbev/msh105 [DOI] [PubMed] [Google Scholar]
- 108. Fedonin GG, Rakhmaninova AB, Korostelev YD, Laikova ON, Gelfand MS. Machine learning study of DNA binding by transcription factors from the LacI family. Molecular Biology. 2011;45(4):667–679. Available from: 10.1134/S0026893311040054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data on different subfamilies are presented on separate sheets. Only the first members of regulated operons are shown. TFBS positions are given relative to translation starts of regulated genes annotated in the genomes.
(XLS)
Horizontal axis shows the percentage of sites from a given category from all sites found in genome. The vertical axis shows the number of such genomes.
(PDF)
(PDF)
(XLS)
(XLS)
Global minimum p-value corresponds to 32 pairs.
(PDF)
TF positions are along the horizontal axis and at the Logo above. Site positions are along the vertical axis and at Logo on the left. The color denotes the Z-score for a pair of positions with the color palette for significantly correlated pairs in the yellow to red interval, while black through light green colors denote positions below the significance threshold. Protein-DNA interactions are shown as stars. Interactions observed in the structures of complexes at least once are shown. Elements of protein secondary structure (from the crystal structure of E. coli CueR – PDB ID 1Q05) are shown at the top.
(PDF)
Colors of branches show overrepresented pairs of residues in positions (3,13) (see color codein the picture). Background colors show TF subfamilies: red—CueR, blue – MerR, green—CadR-PbrR, beige—CadR-PbrR-like, pink—HMRTR.
(PDF)
Colors of branches show overrepresented pairs of residues in positions (5,14) (see color code in the picture). Background colors show TF subfamilies: red—CueR, blue – MerR, green—CadR-PbrR, beige—CadR-PbrR-like, pink—HMRTR.
(PDF)
Colors of branches show overrepresented pairs of residues in positions (6,14) (see color code in the picture). Background colors show TF subfamilies: red—CueR, blue – MerR, green—CadR-PbrR, beige—CadR-PbrR-like, pink—HMRTR.
(PDF)
Colors of branches show overrepresented pairs of residues in positions (6,21) (see color code in the picture). Background colors show TF subfamilies: red—CueR, blue – MerR, green—CadR-PbrR, beige—CadR-PbrR-like, pink—HMRTR.
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.