Whole-plant glutamine mobilization aids local tissue, pathway-specific auxin biosynthesis in maize roots as a way to stimulate root regrowth after herbivory by the western corn rootworm.
Abstract
The western corn rootworm (WCR; Diabrotica virgifera virgifera LeConte) is a major pest of maize (Zea mays) that is well adapted to most crop management strategies. Breeding for tolerance is a promising alternative to combat WCR but is currently constrained by a lack of physiological understanding and phenotyping tools. We developed dynamic precision phenotyping approaches using 11C with positron emission tomography, root autoradiography, and radiometabolite flux analysis to understand maize tolerance to WCR. Our results reveal that WCR attack induces specific patterns of lateral root growth that are associated with a shift in auxin biosynthesis from indole-3-pyruvic acid to indole-3-acetonitrile. WCR attack also increases transport of newly synthesized amino acids to the roots, including the accumulation of Gln. Finally, the regrowth zones of WCR-attacked roots show an increase in Gln turnover, which strongly correlates with the induction of indole-3-acetonitrile-dependent auxin biosynthesis. In summary, our findings identify local changes in the auxin biosynthesis flux network as a promising marker for induced WCR tolerance.
The western corn rootworm (WCR; Diabrotica virgifera virgifera LeConte; Supplemental Fig. S1) is a voracious pest of maize (Zea mays). Larvae hatch in the soil during late spring and immediately begin feeding on the crop’s root system. Over time, active feeding can result in substantial root damage with significant loss of water and/or nutrient uptake, thus weakening plants (Flint-Garcia et al., 2009). Plants also become highly susceptible to lodging when major damage is inflicted upon the anchoring root system. Taken together, these effects can result in significant corn yield losses and management costs totaling between $650 million to $1 billion in the U.S. annually (Flint-Garcia et al., 2009; Gray et al., 2009).
History reveals the enormous resilience and adaptability of this pest and just how quickly it can evolve to overcome management strategies. For example, resistance to application of chemical pesticides, including cyclodienes (benzene hexachloride, aldrin) and organophosphates (methyl parathion), was seen over just a 10-year period of their use in Nebraska’s cornfields during the 1950s and 1990s, respectively (Ball and Weekman, 1963; Meinke et al., 1998). Alternate management practices, including rotation of corn with other crops on a seasonal basis, was generally considered the best choice for management since 1909 (Levine et al., 2002). In east/central Illinois, 95% to 98% of cropland had adopted a management strategy using only soybean as the rotation crop. Unfortunately, the enthusiastic adoption of this strategy over a broad area combined with the efficacy of the technique created a strong selection that favored a less common D. v. virgifera phenotype with reduced egg laying fidelity to cornfields. Over time, natural selection afforded a strong reproductive advantage to females laying their eggs in soybean fields. Since the late 1990s, a strain of the western corn rootworm with resistance to crop rotation can be found in parts of Illinois, Indiana, and parts of bordering states (Gray et al., 2009; Levine et al., 2002).
More recently, D. v. virgifera resistance to deployed genetically modified organisms has been reported. First introduced into the market to target this pest back in 2003, genetically altered Bt-maize expressing one or more proteins from the soil bacteria Bacillus thuringiensis provided enhanced plant defenses to larval feeding. When a vulnerable insect ate the Bt-containing plant, the protein became activated in its gut, forming a toxin that paralyzed the digestive system and caused it to stop feeding. Unfortunately, resistance began to show within three generations of selection (Meihls et al., 2008).
An alternative strategy to reduce the negative impact of D. v. virgifera attack without triggering counter adaptations in the pest is plant tolerance, which relies on a plant’s capacity to maintain growth and yield even in the presence of substantial damage. While D. v. virgifera-tolerant maize germplasms exhibiting slight to moderate tolerances to D. v. virgifera have been reported (Flint-Garcia et al., 2009), more effective lines are needed. Unfortunately, we know very little about the underlying mechanisms for crop tolerance. Over the years, one resounding message has been that the physiological processes affected by herbivory should be better characterized before breeding tools can be leveraged in a rational way to generate improved varieties that maintain high yields under herbivore pressure (Riedell, 1990). Rational decision making in the breeding selection process requires rigorous phenotyping; however, present phenotyping tools tell us little about the plasticity of root systems, especially when it comes to understanding mechanisms for crop tolerance to attack belowground. It was recently suggested that the timing for allocation of newly fixed carbon resources as soluble sugars between leaves, stalks, and root systems, and their coordination with mobilization of other resources including amino acids, may play significant roles in determining the ability of maize plants to survive an attack by D. v. virgifera (Orians et al., 2011; Robert et al., 2014).
In this work, our systematic evaluation of the physiological, metabolic, and genetic basis for root regrowth as a tolerance trait sheds new light on the regulation of the growth hormone auxin (indole-3-acetic acid [IAA]) and its role in this process. Radioactive decay of 11C (β+ emitter, t1/2 = 20.4 min), dynamic whole-plant positron emission tomography, root autoradiography, and radiometabolite flux analyses allowed us to map the transport, allocation and metabolism of carbon and nitrogen resources against genetic and radiolabeled biochemical markers including [11C]IAA, [11C]indole, [11C]indole-3-acetonitrile ([11C]IAN), [11C]indole-3-acetamide ([11C]IAM), and l-[5-11C]Gln (Supplemental Fig. S2). Taken together, these tools enabled us for the first time, to our knowledge, to rigorously map out the auxin biosynthesis flux network at regional tissue levels and in turn provide new insights on auxin regulation and its coordination with the availability of a key amino acid, l-Gln. The developed phenotyping tools can now be employed for the rapid identification and selection of D. v. virgifera-tolerant maize germplasm.
RESULTS
Root Herbivore Attack Induces Asymmetric Formation of Lateral Root Primordia
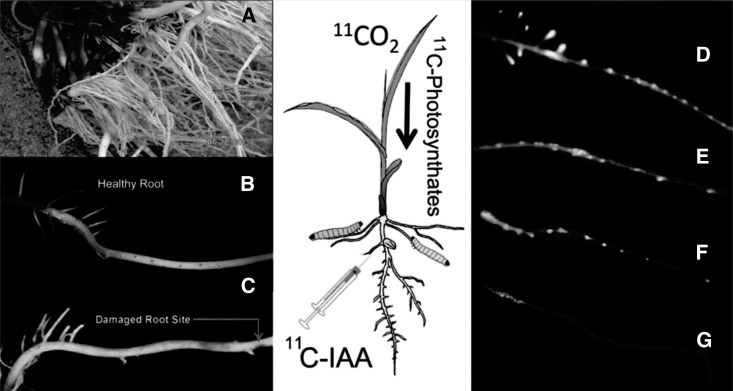
In the field, D. v. virgifera-tolerant maize plants often display a pronounced increase in lateral root growth (Fig. 1A). Initial imaging studies set out to determine whether this trait can be characterized in a laboratory setting using 3-week-old maize seedlings at their V2 stage. Seedlings at this stage of development have mature nodal or crown roots showing developing lateral root primordia (LRP) and lateral root structures (Fig. 1B). The high degree of LRP spatial symmetry (Fig. 1B) found in healthy nodal roots is lost upon larva feeding (Fig. 1C). After leaf administration of 11CO2, autoradiographic imaging of the gross [11C]photosynthate distribution reveals that in healthy undamaged roots, the lateral root meristems and LRP have a high accumulation of 11C radioactivity, indicating that these sites are strong sinks for resources (Fig. 1D). Furthermore, the spatial patterning of radioactivity in the healthy root image was highly symmetrical. In contrast to this, roots mildly damaged by herbivore feeding show a nonsymmetrical patterning of [11C]photosynthate in the LRP (Fig. 1E). These LRP sites of high sink strength (and presumably high metabolic activity) coincide with high specific binding of [11C]IAA both in healthy roots (Fig. 1F) and in mildly damaged roots (Fig. 1G) suggesting a strong correlation between auxin signaling, cellular metabolism, and compensatory growth as a form of herbivore tolerance.
Figure 1.
A, Ectopic branch root patterning of maize plant observed in the University of Missouri planting fields after infestation by D. v. virgifera. B and C, Photographs of healthy and damaged nodal roots. Loss of lateral root spatial symmetry is noted when roots were damaged by herbivore feeding. D and E, Root radiographic images showing [11C]photosynthate distributions in healthy (D) and damaged (E) nodal roots. Images were acquired 90 min after administration of a dose of 11CO2 to a source leaf. 11C activity shows good spatial correlation with sites of LRP, suggesting these are sites are strong sinks with high metabolic activity. F, Root radiographic image acquired after a subcortical injection of [11C]IAA into a healthy undamaged root—a similar symmetrical spatial patterning of 11C tracer binding is seen between this image and the healthy undamaged root image presented in D for [11C]photosynthate. G, Root radiographic image acquired after a subcortical injection of [11C]IAA into a WCR-damaged root—a similar nonsymmetrical spatial patterning of 11C tracer binding is seen between this image and the damaged root image presented in E for [11C]photosynthate. The spatial correlations of 11C radioactivity seen between [11C]IAA and [11C]photosynthate images suggests that LRP are sites of high auxin receptor density.
Compensatory Growth Is Associated with Distinct Shifts in Auxin Biosynthesis and Transport
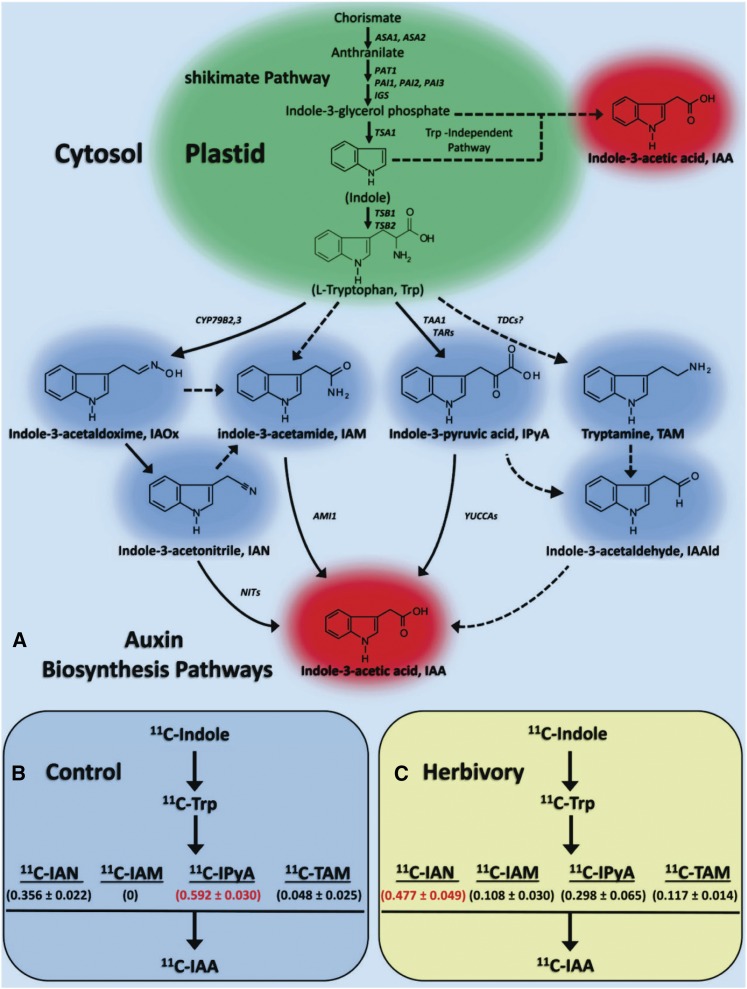
Root growth is in part determined by the hormone auxin. We therefore conducted a series of experiments to understand whether the observed regrowth phenotype is accompanied by changes in auxin biosynthesis and its distribution. Endogenous auxin (IAA) concentrations in nodal roots were seen to increase significantly (by 45%) as a function of root herbivore attack reaching levels of 0.051 ± 0.012 ng mg FW−1 (Supplemental Fig. S3). Even so, this result says little about auxin biosynthesis and its regulation. IAA can be traced back to chorismic acid, which is biosynthesized via the shikimate pathway within the cell chloroplast (Fig. 2A). Chorismic acid undergoes rapid amination to anthranilic acid via the action of l-Gln. Anthranilic acid is then transformed through several steps into different indole compounds and eventually into key aromatic amino acids including l-Trp, Phe, and Tyr that are essential for primary and secondary metabolism in plants (Maeda and Dudareva, 2012).
Figure 2.
A, The biosynthesis of IAA precursors, such as indole-3-glycerol phosphate and l-Trp, takes place in plastids. l-Trp, the major IAA precursor, is generated via the shikimate pathway. The subsequent l-Trp-dependent IAA biosynthesis pathways are believed to occur in the cytosol. Four putative pathways for l-Trp-dependent IAA biosynthesis in higher plants are shown: the IAOx, IAM, IPyA, and TAM pathways. The enzymes known to operate in each pathway are shown in italics. Solid pathway arrows reflect presumably active processes. Dashed pathway arrows are suggested to exist, but have yet to be proven. Modifications to past published pathways (McSteen, 2010; Normanly, 2010; Ljung, 2013) are shown in A. Abbreviations not defined in the text: IAAld, indole-3-acetaldehyde; TDCs, Trp decarboxylases. B and C, The IAA biosynthesis flux network across the four l-Trp-dependent pathways was quantified using [11C]indole. Measurements were taken 1 h after incubation with tracer. In unstressed control plants (B), the IPyA pathway was dominant, the IAOx pathway through IAN was second in importance, the TAM pathway was minor, and the IAM pathway was not active. In herbivore-stressed plants (C), the IAOx pathway through IAN became the dominant pathway, and the IAM pathway was now active.
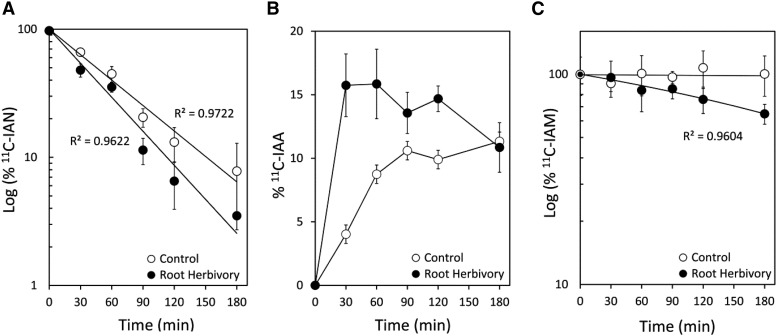
Two major pathways for IAA biosynthesis have been postulated in higher plants (Fig. 2A; McSteen, 2010; Normanly, 2010; Mashiguchi et al., 2011; Won et al., 2011; Ljung, 2013; Wang et al., 2015): a Trp-dependent route including (1) the indole-3-acetaldoxime (IAOx) pathway, (2) the IAM pathway, (3) the indole-3-pyruvic acid (IPyA) pathway, and (4) the tryptamine (TAM) pathway, as well as a Trp-independent route branching either from indole-3-glycerol phosphate or indole has also been suggested (Wright et al., 1991). Because the scientific community has lacked a strong set of biochemical markers enabling rigorous mapping of the IAA biosynthesis flux network, little is known about the importance of individual pathways as a function of environmental stress. We applied several [11C]indole-containing tracers to rigorously map key branch points of this complex network (Reid et al., 2011; Lee et al., 2015a, 2015b). Using [11C]indole, we mapped the Trp-dependent IAA biosynthesis pathways occurring within the upper nodal root tissues 1 h after incubation with tracer for two experimental conditions including (1) unstressed (control) conditions (Fig. 2B) and (2) biotic stress conditions elicited by WCR larvae feeding (Fig. 2C). We observed the following: (1) IAA biosynthesis in unstressed control roots is dominated by the IPyA pathway with no detectable contributions from the IAM pathway; and (2) IAA biosynthesis in herbivore stressed roots is dominated by the IAOx pathway (leading through IAN), and the IAM pathway is now active. These results prompted our further investigation into the kinetics of some of these pathways. Using [11C]IAN, we mapped the temporal profile for the disappearance of this substrate in upper nodal roots with and without WCR attack (Fig. 3A). The kinetic loss of substrate is first order, and the rate of metabolism increases significantly upon treatment. We also mapped the growth of the [11C]IAA product derived from [11C]IAN metabolism (Fig. 3B) and found that the rate of product formation increases significantly with treatment but that the rate of free [11C]IAA utilization (either through conjugation, metabolism, or protein binding) increases significantly as well. Finally, using the [11C]IAM tracer (Fig. 3C), we verified that the IAM pathway is not active in unstressed control roots but becomes active with treatment exhibiting first-order kinetics for substrate metabolism.
Figure 3.
A, Logarithmic plot of [11C]IAN levels after subcortical injection of tracer into the upper proximal zone of a nodal root (n = 5, control; n = 7, root herbivory). Results reflect first-order kinetics for [11C]IAN metabolism and a strong root herbivore treatment effect with increased rate of substrate metabolism. B, Relative percent of [11C]IAA product derived from [11C]IAN metabolism shown in A. Herbivory increases the rate of [11C]IAA formation, as well as increases loss of free [11C]IAA from metabolism, conjugation, and/or receptor binding. C, Logarithmic plot of [11C]IAM levels after subcortical injection of tracer into the upper proximal zone of a nodal root (n = 4, control; n = 4, root herbivory). Results show that [11C]IAM is not metabolized in unstressed control roots but is actively metabolized in first-order kinetics after root herbivore treatment.
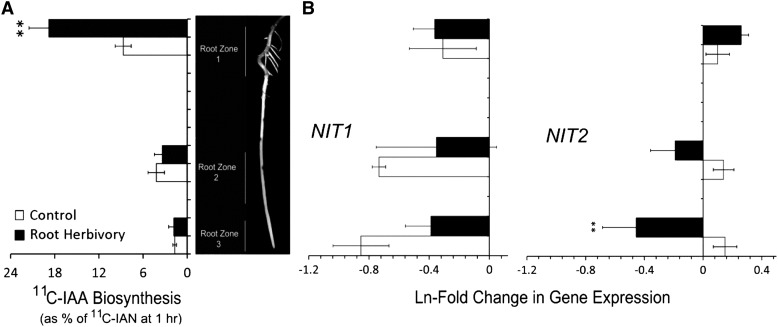
Next, we spatially mapped the amount of [11C]IAA that was formed from [11C]IAN after 1 h incubation with tracer (Fig. 4A) across three regions of the nodal root length including the upper proximal portion where lateral roots are visibly growing (zone 1), the midroot section extending down to the elongation zone (zone 2), and the root tip (zone 3). Overall, our findings show a significantly higher rate of metabolic turnover of [11C]IAN in the upper proximal zone than in lower zones, as well as a significant treatment effect in this zone.
Figure 4.
A, Spatial mapping of [11C]IAA biosynthesis from [11C]IAN across three nodal root zones, including the upper proximal region (zone 1), the midroot region including the elongation zone (zone 2), and the root tip (zone 3; n = 5, control; n = 6, root herbivory). Higher levels of [11C]IAN metabolism are indicated in zone 1. Root herbivore treatment significantly increased tracer metabolism in this zone but had no effect in zones 2 and 3. B, Ln-fold change in NIT1 and NIT2 gene expression measured across the same three root zones as a function of root herbivore treatment. No change in NIT1 gene expression was noted across any of the zones or as a function of treatment (n = 3). On the other hand, NIT2 gene expression was significantly down-regulated with herbivore treatment in zones 2 and 3 but exhibited a slight up-regulation of expression in zone 1 (n = 4), though not statistically significant (P = 0.062). Where appropriate, statistical significance was denoted as *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Because enzymes catalyzing the conversion of IAOx to IAN are not known, we decided to target the nitrilase (NIT) genes that have been characterized in maize and may play an important role in the conversion of IAN to IAA. NIT1 and NIT2 gene expression patterns were quantified by reverse transcriptase real-time polymerase chain reactions and mapped across the same three root zones as described above (Fig. 4B). NIT1 gene expression does not show a statistical difference across the three root zones, nor does root herbivore treatment impose any effect on this gene’s expression level. NIT2 also doesn’t exhibit spatial patterning in gene expression across the root zones for the unstressed controls. However, WCR attack significantly lowers NIT2 expression in zones 2 and 3 relative to controls while possibly slightly increasing expression in zone 1, though this was not considered statistically significant (P = 0.062).
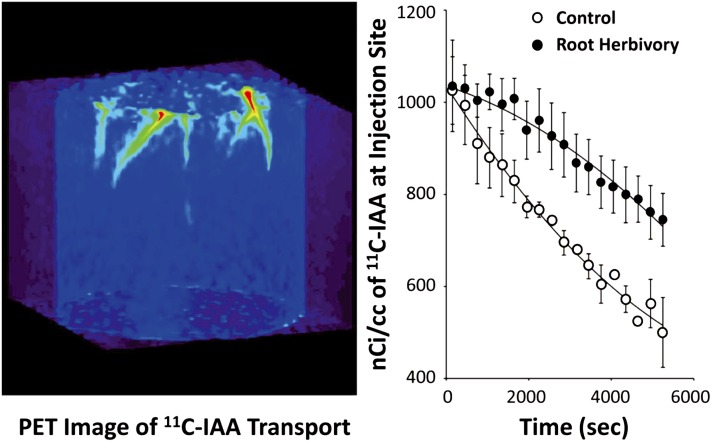
Finally, redistribution of [11C]IAA via physical transport was measured using dynamic positron emission tomography (PET) imaging (Fig. 5A). Using subcortical microliter injections of tracer in the upper root zone 1, PET imaging enabled us to visualize the movement of [11C]IAA over time, as well as enabled us to quantify its transport speed (Fig. 5B). By measuring the time of arrival of tracer across two distinct ROIs, an auxin transport speed of 17.0 ± 5.7 mm h−1 is observed for the unstressed control state. WCR attack significantly lowers transport by a factor of 2.6. Taken together, these results show that WCR attack leads to a pronounced reorganization of auxin biosynthesis and transport.
Figure 5.
A, Projection PET image showing [11C]IAA movement from the injection sites in the upper proximal zone of undamaged (control) nodal roots. B, Time-activity plots reflecting levels of decay corrected [11C]IAA activity at the injection site over time in seconds (n = 4, control; n = 5, root herbivory). Root herbivory decreased [11C]IAA transport 2.6-fold from an average speed of 17.0 ± 5.7 mm h−1 as measured from time of arrival of the activity from across two distinct ROIs along an individual root.
Root Herbivore Attack Increases Amino Acid Transport to the Roots
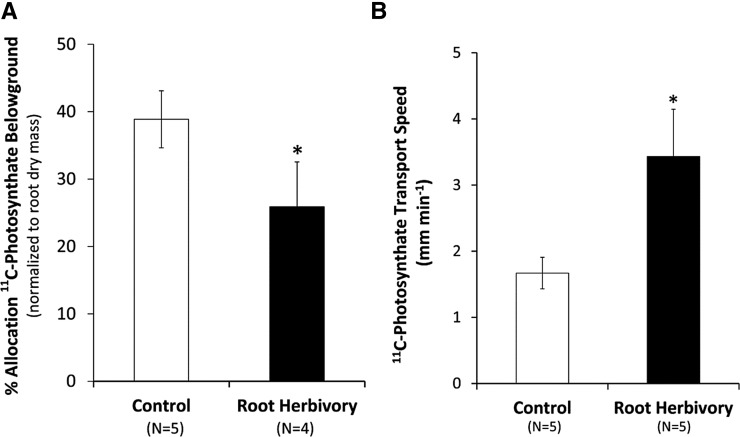
The regulation of key hormones like auxin that are important for controlling root growth requires a steady supply of essential carbon and nitrogen resources. To understand how WCR influences resource allocation, we used 11CO2 administered to source leaves in combination with dynamic PET imaging to trace the physiological and biochemical fate of “new carbon” (as 11C) at the whole-plant level. We observed that allocation of gross [11C]photosynthates belowground decreases significantly with WCR attack from 38.87% ± 4.23% of fixed 11CO2 to 25.91% ± 6.64% (Fig. 6A). By contrast, transport speeds of gross [11C]photosynthates more than doubled with herbivore treatment from 1.67 ± 0.24 to 3.43 ± 0.71 mm min−1 (Fig. 6B), suggesting that the turnover of photosynthates increases under WCR attack.
Figure 6.
A, Allocation belowground of gross [11C]photosynthates derived from fixed 11CO2. Root herbivore treatment significantly decreased belowground allocation of these resources. B, The speed of [11C]photosynthate transport (mm min−1) shown in B increased significantly with treatment. Where appropriate, statistical significance was denoted as *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
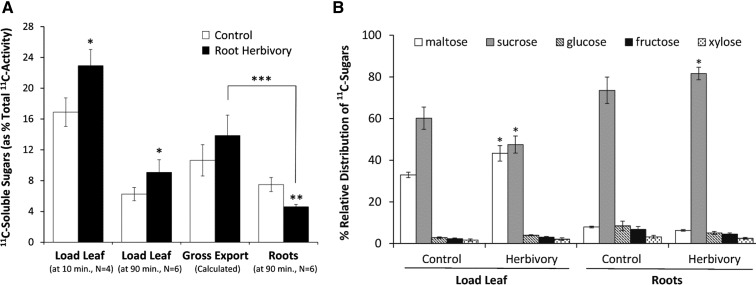
Absolute amounts of 11C-soluble sugars were measured both in the load (source) leaf at 10 and 90 min postadministration of 11CO2, as well as in the roots at 90 min, for control plants and plants subjected to root herbivore treatment (Fig. 7A). Gross export of 11C-labeled sugars was calculated from the difference between the load leaf levels of 11C-labeled sugars measured at the 10 and 90 min time points (ses propagated). Three general observations are worth noting on the effects of root herbivore treatment on whole-plant sugar status: (1) it increases the flux of new carbon into source-leaf-soluble sugar pools; (2) it increases the export of these sugar resources from those leaves (though not statistically significant); and (3) it decreases their transport belowground to sites of attack. Specifically, gross sugar export from the source load leaf increases from 10.64% ± 2.04% to 13.85% ± 2.66% (based on total 11CO2 fixed) as a function of treatment, but roots receive less of these sugars, showing a significant decrease from 7.49% ± 0.91% to 4.60% ± 0.32%. The relative profile of the individual sugars is also revealing. Here, we captured profile information for both the load leaf and the roots at 90 min (Fig. 7B). At the source leaf supply side, root herbivore treatment significantly increases the metabolic partitioning of new carbon into [11C]maltose and [11C]Suc, relative to controls, while generating a slightly higher proportion of [11C]Suc in the roots.
Figure 7.
A, Levels of 11C-labeled soluble sugars (presented as % total 11C activity fixed as 11CO2) were plotted for the following tissues: load leaf at 10 min postadministration of 11CO2, load leaf at 90 min postadministration of 11CO2, and roots at 90 min postadministration of 11CO2 to leaves. The gross export of 11C-labeled soluble sugars was calculated from the difference in load leaf activities at 10 and 90 min. Root herbivore treatment manifested in a significant reduction in root allocation of 11C-labeled soluble sugars. B, Relative distributions of 11C-labeled soluble sugars are presented for the load leaf and roots at 90 min postadministration of 11CO2 and as a function of root herbivore treatment.
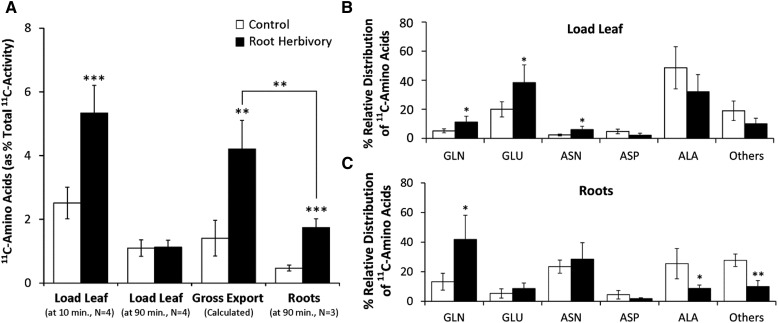
Similarly, absolute amounts of 11C-labeled amino acids were measured in the load (source) leaf at 10 and 90 min postadministration of 11CO2, as well as in the roots at 90 min, for control plants and plants subjected root herbivore treatment (Fig. 8A). Like before, the gross export of 11C-labeled amino acids was calculated from the difference between the levels of 11C-labeled amino acids measured at the 10 and 90 min time points. Three general observations are worth noting on the effects of WCR attack on whole-plant amino acid status: (1) it significantly increases the flux of new carbon into source leaf amino acid pools; (2) it significantly increases the export of these resources from those source leaves; and (3) it significantly increases their transport belowground to sites of attack. Specifically, gross amino acid export from the source load leaf increases from 1.41% ± 56% to 4.21% ± 0.89% as a function of treatment. Roots receive more of these amino acids where allocations significantly increase from 0.47% ± 0.09% to 1.74% ± 0.28%. Furthermore, studies using the l-[5-11C]Gln tracer, applied to an abraded source leaf tip, show that root herbivore treatment doubles the phloem transport speed of this substrate (Supplemental Fig. S4). Like before, we captured amino acid profile information for the load leaf (Fig. 8B) and the roots (Fig. 8C) at 90 min. On the source leaf supply side, root herbivore treatment significantly increases the metabolic partitioning of new carbon into [11C]Gln, [11C]Glu, and [11C]Asn relative to controls. Roots also show a significantly higher proportion of [11C]Gln with treatment but lower proportions of [11C]Ala and other [11C] amino acids. These results show that WCR attack triggers strong and opposing changes in carbohydrate and amino acid allocation to the roots, which reflects the reorganization of root metabolism and growth upon herbivore infestation.
Figure 8.
A, Levels of 11C-labeled amino acids (presented as % total 11C activity fixed as 11CO2) were plotted for the following tissues: load leaf at 10 min postadministration of 11CO2, load leaf at 90 min postadministration of 11CO2, and roots at 90 min postadministration of 11CO2 to leaves. The gross export of 11C-labeled amino acids was calculated from the difference in load leaf activities at 10 and 90 min. Root herbivore treatment manifested in a significant increase in gross export of amino acids from source leaves, as well as in a significant increase in their allocation to roots. B and C, Relative distributions of 11C-labeled amino acids are presented for the load leaf (B) and roots (C) at 90 min postadministration of 11CO2 and as a function of root herbivore treatment. Treatment increased source leaf levels of [11C]Gln, [11C]Glu, and [11C]Asn. Treated roots showed significantly higher levels of [11C]Gln, as well as lower levels of [11C]Ala and other less-abundant radiolabeled amino acids that were summed together. Where appropriate, statistical significance was denoted as *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Changes in Gln and Auxin Metabolism Are Strongly Correlated
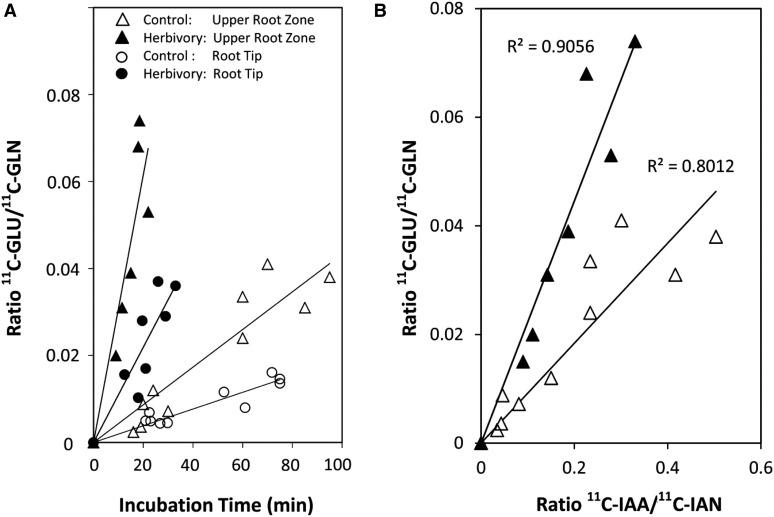
Because Gln is important to maintaining auxin homeostasis (Ljung, 2013) through its involvement in the shikimate pathway (Maeda and Dudareva, 2012) providing indole-like substrates in support of auxin biosynthesis, we further investigated the dynamics of its metabolism in roots. Using the l-[5-11C]Gln tracer, we mapped its metabolism to [11C]Glu as a function of root tissue type (upper nodal root versus lower root tip) and root herbivore treatment (Fig. 9A). Results show that Gln metabolism is significantly faster in upper root tissues than in tips. Furthermore, WCR attack increases metabolic turnover of l-[5-11C]Gln, for both of these tissue types. Lastly, by mapping l-[5-11C]Gln metabolism in upper root tissues (Fig. 9B) against [11C]IAA biosynthesis (derived from the [11C]IAN precursor) we see a strong correlation between the two processes and their response to WCR attack, suggesting that Gln accumulation and IAA signaling are tightly coupled in maize roots.
Figure 9.
A, [11C]Gln metabolism, represented as the ratio of [11C]Glu to [11C]Gln, was plotted against tracer incubation for upper root tissues and root tips as a function of root herbivore treatment. Two key observations include (1) Gln metabolism is higher in upper root tissues than in tips, and (2) root herbivore treatment increases Gln metabolism across all spatial zones of the root. B, [11C]Gln metabolism, represented as the ratio of [11C]Glu to [11C]Gln was plotted against [11C]IAN metabolism, represented as the ratio of [11C]IAA to [11C]IAN, as a function of root herbivore treatment. Regression analysis reveals a strong correlation between the two biochemical processes as well as a significant positive herbivore treatment effect.
DISCUSSION
Influence of Biotic Stress on Auxin Regulation
In an environment where higher plants are continuously subjected to a broad array of biotic and abiotic stimuli, their adaptability to such conditions requires a high degree of plasticity across scales of the whole plant. Plasticity, best reflected by rapid changes in a plant’s physiology and/or central metabolism, can give rise to significant changes in growth and morphology, as well as in changes secondary metabolism. Often, active defensive strategies of a plant against would-be “attackers” involves changes to secondary metabolism, producing toxins that can repel, deter, or kill their attacker (Howe and Jander, 2008). Defense can also involve tolerance mechanisms that allow the plant to regrow or reproduce after significant damage and/or tissue loss (Strauss and Agrawal, 1999). Underlying mechanisms for triggering such responses are traced to long-range signaling involving specialized hormones that help coordinate allocation of resources to targeted distal tissues and organs, as well as the metabolic partitioning of these resources (Thorpe et al., 2007).
Hormones like auxin are known classically for their roles in regulating plant development including axis formation and patterning during postembryogenesis, vascular elongation, leaf expansion, inflorescence, fruit development, tropism, and apical dominance (Woodward and Bartel, 2005; Kazan and Manners, 2009). Most particularly, auxin can stimulate “reshaping” of root system architecture by triggering branch root patterning and root hair formation (McSteen, 2010).
The supply of IAA to targeted tissues and organs of the plant is thought to rely on a fine balance between its biosynthesis and its physical transport both in and out of cells, as well as across tissues. Through a combination of efflux carrier proteins, including the PIN family and ABCB family (Zazímalová et al., 2010) and influx carrier proteins, including the AUX1/LAX family (Swarup and Péret, 2012), polar auxin transport can be established at a cellular level, providing the plant with a mechanism for effectively transporting this polar substrate across longer distances. Often, gradients in auxin concentration are found in targeted tissues that are thought to provide developmental cues for the plant (Blilou et al., 2005; Benková et al., 2009). For example, auxin is typically found at its highest concentration at the root tip (Agtuca et al., 2013).
As described earlier (Fig. 2A), several pathways can play a role in producing auxin. Of these, we were primarily interested in the IAOx and IAM pathways. IAN is a putative IAA precursor that is synthesized from IAOx. NITs are important in converting IAN-to-IAA. ZmNIT1 and ZmNIT2, which are encoded in the maize genome and are expressed in developing kernels and seedlings (Park et al., 2003), have been identified. Prior research using a Zmnit2 knockout maize mutant showed a significantly lower root IAA concentration than wild type, and it exhibited a strong root phenotype, suggesting that a pathway through IAN is important (Kriechbaumer et al., 2007). Indeed, our metabolic flux assays show that this pathway is the second most dominant pathway, next to the IPyA pathway, contributing to auxin in unstressed maize roots, and is the most dominant pathway when roots come under herbivore attack. Furthermore, the regional specificity exhibited by this pathway, as evidenced by higher NIT2 gene expression and faster [11C]IAN kinetics in the upper root zone where lateral root growth is evident, directly links this biochemical response to regrowth and crop tolerance. We note that past studies that spatially mapped both the IAA biosynthesis rate (Ljung et al., 2005) and the expression level of key genes linked with auxin biosynthesis (Birnbaum et al., 2003) across similar root zones in Arabidopsis (Arabidopsis thaliana) found higher rates of auxin biosynthesis and higher levels of NIT expression in the upper zone where lateral root growth occurs. Hence, the significant down-regulation that we see in NIT2 expression in the lower maize root zones where no lateral root growth occurs suggests that the IAOx pathway may be uniquely tied to a plant stress response mechanism that aligns with the development lateral roots.
The IAM pathway, though considered an important pathway in bacteria, manifests in only very minute amounts of IAM in certain plants (Novák et al., 2012). The IAM hydrolase (AtAMI1) enzyme, responsible for converting IAM-to-IAA, has also been observed in some plant species (Pollmann et al., 2006; Nemoto et al., 2009). Even so, little is known about the importance of the IAM pathway across the plant kingdom, or of its connectivity with other pathway intermediates such as IAN in making IAA (see Fig. 2A). Through two different tracer experiments, our results clearly show that the IAM pathway is not active in unstressed plants, but activates under biotic stress. Even so, this pathway only contributes a minor portion of substrate to the overall auxin biosynthesis flux network. Furthermore, the first-order kinetics for metabolism of both [11C]IAN and [11C]IAM substrates clearly indicates that pathway interconnectivity does not exist for this system. As noted in Figure 2, there has been some speculation on the interconnectivity of these pathways based on identification of key enzymes. However, we note that these precursors can also play important roles in plant secondary defense chemistry, leading to defense compounds such as indole glucosinolates and camalexin, and perhaps the existence of these enzymes in plants is to serve this purpose and not auxin biosynthesis (Ljung, 2013).
Influence of Biotic Stress on Central Metabolism and Resource Allocation
While resistance mechanisms have been extensively studied over the years, much less is known about the mechanistic basis of tolerance (Stowe et al., 2000). Resource reallocation following real or simulated leaf attack aboveground has been well documented across numerous plant species, including tomato, tobacco, maize, barley, and poplar, where the response has been increased export of leaf photosynthates to the stem and roots (Babst et al., 2005, 2008; Gómez et al., 2010; Hanik et al., 2010a, 2010b; Henkes et al., 2008; Schwachtje et al., 2006). Similarly, nitrogen reallocation from leaves-to-roots was found to increase in tomato plants that were subjected to simulated herbivory aboveground (Gómez et al., 2010). More recently, our work using real root herbivore treatments showed decreased allocation of gross [11C]photosynthates from source leaves to nodal roots in maize, as well as decreased root meristematic activity (Robert et al., 2012, 2014). However, we have shown that both biotic (Hanik et al., 2010a) and abiotic stresses (Pankievicz et al., 2015) can drastically and rapidly alter new carbon fluxes into key metabolite pools that make up plant photosynthates. Most particularly, increased partitioning of new carbon into 11C-labeled amino acid pools was noted, which coincided with compensatory reduction of the 11C-labeled sugar pools. In this work, we also observe a 2.6-fold increase in new carbon partitioning into source leaf 11C-labeled amino acid pools as a function of root herbivory.
Unlike before, there was no compensatory decrease in the 11C-labeled sugar pool, but rather a slight increase following WCR attack. This may be due to the fact that 11CO2 fixation (i.e. input of new carbon) actually increases when roots come under attack (Robert et al., 2014)—a feature that was not evident in prior aboveground herbivore studies. Upon closer inspection of our data, we begin to see an interesting story unfold where an increase in metabolic partitioning of new carbon into source leaf amino acids (specifically into highly transportable amino acids like Gln) enables the plant to quickly translocate more nitrogen resources to the roots when they come under attack. This statement is substantiated by key observations, including: (1) increases in allocation of 11C-labeled amino acids to roots, (2) increases in relative amounts of [11C]Gln in the root amino acid pool, and (3) increases in the rate of transport of the l-[5-11C]Gln tracer. We suspect this strong physiological response is being driven by the increase in [11C]Gln metabolism we see in roots when they come under attack. In concert with this, there is significant inhibition in the flow of sugars to roots. Because the profile of 11C-labeled sugars in these sink tissues reflect higher levels of [11C]Suc, we suspect that this strong physiological response is driven by decreased sink demand for sugars. Indeed, our prior studies revealed a significant reduction in root invertase enzyme activity with root herbivore treatment (Robert et al., 2014).
CONCLUSION
In summary, this work has revealed several new insights about root regrowth in crop tolerance to root herbivore attack as it relates to auxin biosynthesis and its regulation, including the following: (1) auxin biosynthesis is tightly regulated at the local root tissue level with minimal hormone translocation away from site of biosynthesis; (2) auxin biosynthesis is highly pathway specific, with the IAOx pathway through IAN dominating under stress; and (3) auxin biosynthesis is tightly coordinated, with Gln mobilization to roots, and with increased Gln metabolism at sites of regrowth.
Given the redundant nature through which auxin is biosynthesized in higher plants, it is unlikely that up-regulating a specific pathway using genetic engineering will satisfy the local tissue demands for this hormone. Other pathways will likely just down-regulate, enabling the auxin flux network to maintain status quo based on the supply of substrates needed to support the overall process. A more viable approach is to target increasing the supply of these essential substrates. Our observations suggest that improving upon whole plant Gln utilization through increased Gln biosynthesis and/or transport would be a good place to start for crop breeding programs or for engineering new genetically modified organisms.
MATERIALS AND METHODS
Materials
All chemicals used in these studies were obtained from Sigma-Aldrich and used without any further purification.
Plant Growth and Treatment
Corn (Zea mays) kernels of the B73 line (U.S. Department of Agriculture Agricultural Research Service-Germplasm Resource Network) were germinated in darkness for 48 h on wetted KimWipe tissues placed in 7 × 7 cm petri dishes. The dishes were wrapped with aluminum foil and kept at room temperature. After 48 h, the petri dishes were unwrapped and placed under Agro lights giving 100 µmol m−2 s−1 of light intensity during a 16 h photoperiod. Once primary roots of germinating seedlings reached 1 to 2 cm in length, they were transplanted into 600 mL glass growth cells filled with Hoagland’s fortified agar gel.
Agar gels were prepared out of 3 L of deionized water, 4.9 g Hoagland modified basal salt mixture (PhytoTechnology Laboratories), and 1.66 g MES hydrate. The pH of the solution was adjusted to 5.9 by adding 1 n sodium hydroxide solution. While stirring, 8.4 g Gelzan CM (Sigma-Aldrich) were added. The solution was autoclaved (Harvey SterileMax; Thermo Fisher Scientific) for 15 min at 121°C and mixed at high speed to enable aeration of the viscous solution before it had a chance to set as a gel. Sterilized plastic inserts were placed within each cell before the gel set. This allowed us to position the germinated kernel into a notch in the top portion of the cell. Following this, the tops of the cells were sealed with Parafilm and completely wrapped with aluminum foil to block out light. A small hole at the top of the cell enabled the developing plant to grow through. The cells were cultivated in growth chambers (Conviron) at 23°C using 320 µmol m−2 s−1 of light intensity on a 12-h photoperiod. Three-week-old plants that measured ∼35 to 40 cm in height were used for studies.
For root herbivore studies involving attack by Diabrotica virgifera virgifera, 3-week-old plants were subjected to feeding by twelve D. v. virgifera larvae at their second-instar stage. Larvae were allowed to free-feed for 4 d prior to screening plant physiological and biochemical responses using 11C.
11CO2 Production and Administration
11CO2 was produced via the 14N(p, α)11C nuclear transformation from a 20 mL target filled with high-purity nitrogen gas containing 1.13 × 103 mg m−3 oxygen (400 mL @ STP) using 18 MeV protons from the TR-19 (Ebco Industries) cyclotron at Brookhaven National Laboratory and captured on a molecular sieve (4 Å). The 11CO2 that was trapped on the molecular sieve was desorbed and quickly released into an air stream at 200 mL/min as a discrete pulse for labeling a leaf affixed within a 5 × 10 cm lighted (320 µmol m−2 s−1) leaf cell at 21°C to ensure a steady level of fixation. The load leaf affixed within the cell was pulse-fed 11CO2 for 1 min, then chased with normal air for the duration of exposure. A PIN diode radiation detector (Carroll Ramsey Associates) affixed to the bottom of the leaf cell enabled continuous measurement of radioactivity levels within the cell during the initial pulse and in the minutes right after the pulse giving information on 11CO2 fixation (Ferrieri et al., 2005).
Radiosyntheses of Complex 11C-Labeled Substrates
The radiosynthesis of 11C-IAA and its biosynthetic precursors 11C-IAN and 11C-IAM, using nucleophilic 11C-cyanations on gramine, was previously reported by us as a way to examine hormone signaling pathways in plants noninvasively (Reid et al., 2011). In the past year, we further refined this original synthesis method to include a two-step, one-pot process with integrated solid-phase extractions that enabled reliable and remotely controllable production of these radiotracers in less than 1 h yielding 0.22 to 0.46 GBq of product at specific activities of 47.4 ± 12.5 GBq µmol−1 and >98% radiochemical purity (Reid et al., 2011; Lee et al., 2015a). The improvements to radiochemistry allowed for final product formulation in extremely small volumes (0.15–0.3 mL of deionized water at pH 6–7) that were suitable for in vivo PET plant imaging and/or metabolic flux assays.
The radiosynthesis of [2-11C]indole was recently reported by us (Lee et al., 2015b), which leveraged the nucleophilic 11C-cyanation reaction on 2-nitrobenzyl bromide generating a highly reactive 2-(2-nitrophenyl)-[1-11C]acetonitrile intermediate that was subjected to nickel-catalyzed reductive cyclization to yield the desired product in less than 1 h at >98% radiochemical purity and with a specific activity of 176 ± 24.8 GBq μmol−1.
A previously reported [11C]cyanide end-capping labeling method (Qu et al., 2012) was recently refined by us (Gleede et al., 2015) to create a milder, more reliable, two-step method for radiosynthesizing 185 to 370 MBq batches of l-[5-11C]-Gln in less than 50 min at >93% radiochemical purity and possessing a L/D enantiomeric ratio of 94% ± 2.5/2.2% ± 1.7, and specific activity of 7.0 ± 1.5 GBq µmol−1.
Complex organic radiotracers were introduced into targeted root tissues using subcortical injections (< 1 μL). We compared this method for tracer administration to that of grinding fresh tissue with tracer and obtained similar results on radiometabolite profiles.
Plant Imaging
We used microPET imaging (Concorde MicroSystems) to carry out 90 min dynamic scanning on roots. The system was used in its default configuration as described previously for all dynamic imaging experiments (Knoess et al., 2003; Alexoff et al., 2003). Images reconstructed by filtered back projection had a volumetric resolution of ∼5 µL at the center of the field of view increasing to ∼30 µL at a radius of 4 cm (Knoess et al., 2003). Data corrections for random coincidences and dead-time losses were provided by the manufacturer. Scattered coincidences were subtracted using a tail-fitting method described previously (Alexoff et al., 2003). ASIPro VM software (RSI Research Systems) was used to create regions of interest (ROI) on reconstructed images giving information on radioactivity time-of-arrival between those ROIs. Distances between ROIs were also measured in pixels using the same software and related to mm spatial scales for calculating velocities (mm min−1). After the microPET imaging, plants were removed from their growth cells and selected tissues (roots and leaves) were imaged using autoradiography (Typhoon 7000; GE Healthcare). Image data were quantified using ImageQuant TL 7.0 software (GE Healthcare Bio-Sciences AB).
[11C]Sugar and [11C]Amino Acid Analyses
Targeted tissues were extracted in 4× (w/v) methanol, briefly vortexed (VWR analog vortex mixer; Sigma-Aldrich), and then sonicated (Branson Bransonic 32; Sigma-Aldrich) in an iced water bath for 10 min with intermittent vortexing to ensure complete mixing. The tubes were centrifuged (Eppendorf Centrifuge 5424) for 2 min at 15,000 rpm, and the supernatant was filtered through 0.2 µm Acrodiscs (Gelman Sciences). The pellet contained all insoluble components that comprised mostly cell wall polymers and starch. The filtrate contained small soluble compounds, including soluble sugars and amino acids. Sugars were separated and analyzed by thin-layer chromatography (TLC; Babst et al., 2013). Glass backed NH2-silica HPTLC plates (200 µm, w/UV254) were used for the sugar separation (Sorbent Technologies). Plates were prespotted with sugar standards of maltose, Suc, Fru, Glc, and Xyl. Aliquots of radioactive tissue extract were applied to TLC plates using a semiautomatic Linomat 5 sample applicator (Camag Scientific) for high precision of spot size and sample volume. TLC plates were developed using a mobile phase consisting of 75:25 acetonitrile:water (v/v). Developed plates were imaged using autoradiography to determine the fraction of each radiolabeled sugar. The plates were then heat treated (200°C for 10 min) to initiate chemical reaction of individual sugars with the amino functionalized silica support that gave fluorescence under long-wavelength (365 nm) UV light providing coregistration of the 12C-sugar standards with individual 11C-labeled components. ImageQuant TL software 7.0 (GE Healthcare Bio-Sciences AB) was used to analyze the radiographic image data to determine the amount of 11C within each sugar.
An additional 150 µL of tissue extract was also delivered into 2 mL brown-glass vials (Fisher Scientific) where equal volumes of O-phthaldialdehyde amino acid derivatizing reagent containing 0.1% (v/v) mercaptoethanol (Sigma Life Science) were delivered. Mixtures were vortexed and then allowed to react at ambient temperature for 3 min. Fifty microliters of the derivatized samples were injected onto a reversed-phase analytical HPLC column (Phenomenex; Ultramex C18, 10 µm particle size, 250 mm × 4.6 mm inner diameter) using a precolumn gradient mixer (Sonntek) and a mobile phase composed of A (deionized water), B (0.01 m NaH2PO4 buffered at pH 6.8 using trifluoroacetic acid), and C (methanol). At injection, the mobile phase (1.5 mL min−1) was sustained at 75% A; 25% B for 5 min, and then programmed to attain 20% B; and 80% C by 30 min. Retention times of O-phthaldialdehyde-derivatized amino acid were measured against authentic standards using a fluorescence detector (280 nm excitation, 350 nm emission; Hitachi LaChrom Elite l-2485; Sonntek). The outlet of the fluorescence detector was connected in series to a NaI gamma radiation detector (Ortec) that enabled direct measurement of the amount of radioactivity associated with each substrate eluting the column.
[11C]IAA and Related [11C]Indole Compound Analyses
Root tissues were subjected to the same methanol extraction procedure as described above. However, the alcoholic extracts were then subjected to a phase separation using equal volumes of ethyl acetate to remove undesirable sugars and amino acids from the subsequent analysis. Ethyl acetate fractions were reduced to dryness in a vacuum centrifuge (LabConco) and then reconstituted in 50 µL of methanol, where aliquots measured on a gamma counter for 11C activity as well as injected onto a radio HPLC. The radio HPLC gradient system was identical to that described above, with the exception of the column and mobile phase used (Phenomenex; Luna PFP (2), 5 µm particle size, 250 × 4.6 mm inner diameter). The mobile phase was composed of A (2.5% aqueous acetic acid adjusted to pH 3.8 using 1 n KOH) and B (80:20 acetonitrile:water). At injection, the mobile phase (1.0 mL min−1) was programmed to adjust from 80% A; 20% B, to 50% A; and 50% B over 25 min.
Two radio-TLC methods were also deployed to enable rapid assay of pathway-specific auxin biosynthesis kinetic fluxes using [11C]IAN and [11C]IAM radiotracers. In the first method, the silica TLC plate was developed using 1.5:1, hexane:ethyl acetate (0.1% formic acid [v/v]) as the solvent, which allowed rapid separation of [11C]IAN and [11C]IAA and leaving [11C]IAM at the origin. In the second method, the silica TLC plates were pretreated using NH3(aq) (29%) and developed using pure ethyl acetate solvent to enable a different Rf elution profile of the three components with [11C]IAN carried with the solvent front. Activity distributions on the plates were made visible by autoradiography and quantified using ImageQuant TL 7.0 software (GE Healthcare Bio-Sciences AB).
Endogenous Auxin Hormone Analysis
Root tissues were placed in Eppendorf tubes and ground to a fine powder at liquid nitrogen temperature using a Retsch ball mill grinder (Verder Scientific). Five hundred microliters of 0.05 m sodium phosphate buffer, pH 7.0, containing 0.02% (w/v) sodium diethyldithiocarbamate as antioxidant and 250 ng of anthranilic acid (Sigma-Aldrich) as an interna1 standard were added to the Eppendorf tube, and the sample was extracted under continuous shaking for 1 h at 4°C. After extraction, the pH was adjusted to approximately 2.6 with 1 m HC1, and the sample was slurried with 35 mg of Amberlite XAD-7HP (weakly polar polymeric resin, 20–60 mesh; Sigma-Aldrich) for 30 min. After removal of the buffer, the XAD-7 was washed twice using 500 µL of 1% acetic acid before being slurried twice more using 500 µL of dichloromethane for 30 min. The combined dichloromethane fractions were reduced to dryness in a vacuum centrifuge (LabConco) and then trimethylsilylated with 25 µL of acetonitrile and 25 µL of N,O-bis(trimethylsilyl)-trifluoroacetamide with 1% (v/v) trimethylchlorosilane (Sigma-Aldrich) at 70°C for 15 min. After evaporation to dryness (LabConco), samples were dissolved in hexane. One microliter aliquots were split injected (100:1 [v/v]) onto a Hewlett-Packard 5890A capillary gas chromatograph equipped with a 30 m HP-5 column (0.25 mm diameter, 0.25 µm film thickness: Agilent Technologies) and a nitrogen-phosphorous detector.
Quantitative RT-PCR Analysis
NIT1 and NIT2 gene expression patterns along three root zones (including upper proximal, midroot, and root tip) were quantified by performing a quantitative reverse transcriptase real-time polymerase chain reactions as previously described (Erb et al., 2010). The gene-specific primers were used for NIT1 and NIT2 as previously described (Park et al., 2003). The final Ct values were corrected for the housekeeping genes Actin and GapC and normalized to controls.
Statistical Analysis
Data were subjected to the Student’s t test for unpaired samples assuming an unequal variance. Statistical significance levels were assigned to the following rating scale: *P < 0.05; **P < 0.01; ***P < 0.001.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. A, Adult beetle of D. v. virgifera, LeConte; B, mature larva of D. v. virgifera.
Supplemental Figure S2. Complex organic 11C-labeled radiotracers used in this study.
Supplemental Figure S3. Endogeneous nodal root IAA concentrations presented as ng mg FW−1 and plotted as a function of root herbivore treatment.
Supplemental Figure S4. Transport speed (mm min−1) of l-[5-11C]Gln plotted as a function of root herbivore treatment.
Supplementary Material
Acknowledgments
This article has been authored by Brookhaven Science Associates, LLC, under contract number DE-AC02-98CH10886 with the United States Department of Energy, Office of Biological and Environmental Research. Additional support was provided by the German Academic Exchange Service (Deutscher Akademischer Austauschdienst, DAAD) Bonn, the Marie Curie Intra-European Fellowship program, and the Swiss National Foundation Fellowship program. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.
Glossary
- WCR
western corn rootworm
- LRP
lateral root primordia
Footnotes
Articles can be viewed without a subscription.
References
- Agtuca B, Rieger E, Hilger K, Song L, Robert CAM, Erb M, Karve A, Ferrieri RA (2013) Carbon-11 reveals opposing roles of auxin and salicylic acid in regulating leaf physiology, leaf metabolism and resource allocation patterns that impact root growth in Zea mays. J Plant Growth Regul 33: 328–339 [Google Scholar]
- Alexoff DL, Vaska P, Marsteller D, Gerasimov T, Li J, Logan J, Fowler JS, Taintor NB, Thanos PK, Volkow ND (2003) Reproducibility of 11C-raclopride binding in the rat brain measured with the microPET R4: effects of scatter correction and tracer specific activity. J Nucl Med 44: 815–822 [PubMed] [Google Scholar]
- Babst BA, Ferrieri RA, Gray DW, Lerdau M, Schlyer DJ, Schueller M, Thorpe MR, Orians CM (2005) Jasmonic acid induces rapid changes in carbon transport and partitioning in Populus. New Phytol 167: 63–72 [DOI] [PubMed] [Google Scholar]
- Babst BA, Ferrieri RA, Thorpe MR, Orians CM (2008) Lymantria dispar herbivory induces rapid changes in carbon transport and partitioning in Populus nigra. Entomol Exp Appl 128: 117–125 [Google Scholar]
- Babst BA, Karve AA, Judt T (2013) Radio-metabolite analysis of carbon-11 biochemical partitioning to non-structural carbohydrates for integrated metabolism and transport studies. Plant Cell Physiol 54: 1016–1025 [DOI] [PubMed] [Google Scholar]
- Ball HJ, Weekman GT (1963) Differential resistance of corn rootworms to insecticides in Nebraska and adjoining states. J Econ Entomol 56: 553–555 [Google Scholar]
- Benková E, Ivanchenko MG, Friml J, Shishkova S, Dubrovsky JGA (2009) A morphogenetic trigger: is there an emerging concept in plant developmental biology? Trends Plant Sci 14: 189–193 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Erb M, Foresti N, Turlings TCJ (2010) A tritrophic signal that attracts parasitoids to host-damaged plants withstands disruption by non-host herbivores. BMC Plant Biol 10: 247–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrieri RA, Gray DW, Babst BA, Schueller MJ, Schlyer DJ, Thorpe MR, Orians CM, Lerdau M (2005) Use of carbon-11 in Populus shows that exogenous jasmonic acid increases biosynthesis of isoprene from recently fixed carbon. Plant Cell Environ 25: 591–602 [Google Scholar]
- Flint-Garcia SA, Dashiell KE, Prischmann DA, Bohn MO, Hibbard BE (2009) Conventional screening overlooks resistance sources: rootworm damage of diverse inbred lines and their B73 hybrids is unrelated. J Econ Entomol 102: 1317–1324 [DOI] [PubMed] [Google Scholar]
- Gleede T, Riehl B, Shea C, Kersting L, Cankaya AS, Alexoff D, Schueller M, Fowler JS, Qu W (2015) Investigation of SN2 [11C]cyanation for base-sensitive substrates: an improved radiosynthesis of L-[5-11C]-glutamine. Amino Acids 47: 525–533 [DOI] [PubMed] [Google Scholar]
- Gómez S, Ferrieri RA, Schueller M, Orians CM (2010) Methyl jasmonate elicits rapid changes in carbon and nitrogen dynamics in tomato. New Phytol 188: 835–844 [DOI] [PubMed] [Google Scholar]
- Gray ME, Sappington TW, Miller NJ, Moeser J, Bohn MO (2009) Adaptation and invasiveness of western corn rootworm: intensifying research on a worsening pest. Annu Rev Entomol 54: 303–321 [DOI] [PubMed] [Google Scholar]
- Hanik N, Gómez S, Best M, Schueller M, Orians CM, Ferrieri RA (2010a) Partitioning of new carbon as ¹¹C in Nicotiana tabacum reveals insight into methyl jasmonate induced changes in metabolism. J Chem Ecol 36: 1058–1067 [DOI] [PubMed] [Google Scholar]
- Hanik N, Gómez S, Schueller M, Orians CM, Ferrieri RA (2010b) Use of gaseous 13NH3 administered to intact leaves of Nicotiana tabacum to study changes in nitrogen utilization during defence induction. Plant Cell Environ 33: 2173–2179 [DOI] [PubMed] [Google Scholar]
- Henkes GJ, Thorpe MR, Minchin PEH, Schurr U, Röse USR (2008) Jasmonic acid treatment to part of the root system is consistent with simulated leaf herbivory, diverting recently assimilated carbon towards untreated roots within an hour. Plant Cell Environ 31: 1229–1236 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM (2009) Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci 14: 373–382 [DOI] [PubMed] [Google Scholar]
- Knoess C, Siegel S, Smith A, Newport D, Richerzhagen N, Winkeler A, Jacobs A, Goble RN, Graf R, Wienhard K, et al. (2003) Performance evaluation of the microPET R4 PET scanner for rodents. Eur J Nucl Med Mol Imaging 30: 737–747 [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V, Park WJ, Piotrowski M, Meeley RB, Gierl A, Glawischnig E (2007) Maize nitrilases have a dual role in auxin homeostasis and β-cyanoalanine hydrolysis. J Exp Bot 58: 4225–4233 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Alexoff DL, Shea C, Kim D, Schueller MJ, Fowler JS, Qu W (2015a) Tetraethylene glycol promoted two-step, one-pot rapid synthesis of indole-3-[1-11C]acetic acid. Tetrahedron Lett 56: 517–520 [Google Scholar]
- Lee SJ, Fowler JS, Alexoff D, Schueller M, Kim D, Nauth A, Weber C, Kim SW, Hooker JM, Ma L, Qu W (2015b) An efficient and practical synthesis of [2-(11)C]indole via superfast nucleophilic [(11)C]cyanation and RANEY® Nickel catalyzed reductive cyclization. Org Biomol Chem 13: 11235–11243 [DOI] [PubMed] [Google Scholar]
- Levine E, Spencer JL, Isard SA, Onstad DW, Gray ME (2002) Adaptation of the western corn rootworm, Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae), to crop rotation: Evolution of a new strain in response to a cultural management practice. Amer Entomol 48: 94–107 [Google Scholar]
- Ljung K. (2013) Auxin metabolism and homeostasis during plant development. Development 140: 943–950 [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G (2005) Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17: 1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Dudareva N (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci USA 108: 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P. (2010) Auxin and monocot development. Cold Spring Harb Perspect Biol 2: a001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke LJ, Siegfried BD, Wright RJ, Chandler LD (1998) Adult susceptibility of Nebraska western corn rootworm (Coleoptera: Chrysomelidae) populations to selected insecticides. J Econ Entomol 91: 594–600 [Google Scholar]
- Meihls LN, Higdon ML, Siegfried BD, Miller NJ, Sappington TW, Ellersieck MR, Spencer TA, Hibbard BE (2008) Increased survival of western corn rootworm on transgenic corn within three generations of on-plant greenhouse selection. Proc Natl Acad Sci USA 105: 19177–19182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto K, Hara M, Suzuki M, Seki H, Muranaka T, Mano Y (2009) The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Lett 583: 487–492 [DOI] [PubMed] [Google Scholar]
- Normanly J. (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2: a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K (2012) Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J 72: 523–536 [DOI] [PubMed] [Google Scholar]
- Orians CM, Thorn A, Gómez S (2011) Herbivore-induced resource sequestration in plants: why bother? Oecologia 167: 1–9 [DOI] [PubMed] [Google Scholar]
- Pankievicz VCS, do Amaral FP, Santos KFDN, Agtuca B, Xu Y, Schueller MJ, Arisi ACM, Steffens MBR, de Souza EM, Pedrosa FO, et al. (2015) Robust biological nitrogen fixation in a model grass-bacterial association. Plant J 81: 907–919 [DOI] [PubMed] [Google Scholar]
- Park WJ, Kriechbaumer V, Möller A, Piotrowski M, Meeley RB, Gierl A, Glawischnig E (2003) The Nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiol 133: 794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann S, Neu D, Lehmann T, Berkowitz O, Schäfer T, Weiler EW (2006) Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta 224: 1241–1253 [DOI] [PubMed] [Google Scholar]
- Qu W, Oya S, Lieberman BP, Ploessl K, Wang L, Wise DR, Divgi CR, Chodosh LA, Thompson CB, Kung HF (2012) Preparation and characterization of L-[5-11C]-glutamine for metabolic imaging of tumors. J Nucl Med 53: 98–105 [DOI] [PubMed] [Google Scholar]
- Riedell WE. (1990) Rootworm and mechanical damage effects on root morphology and water relations in maize. Crop Sci 30: 628–631 [Google Scholar]
- Reid AE, Kim SW, Seiner B, Fowler FW, Hooker J, Ferrieri RA, Babst BA, Fowler JS (2011) Radiosynthesis of C-11 labeled auxin (3-indolyl[1-11C]acetic acid) and its derivatives from gramine. J Labelled Comp Radiopharm 54: 433–437 [Google Scholar]
- Robert CAM, Ferrieri RA, Schirmer S, Babst BA, Schueller MJ, Machado RAR, Arce CCM, Hibbard BE, Gershenzon J, Turlings TCJ, et al. (2014) Induced carbon reallocation and compensatory growth as root herbivore tolerance mechanisms. Plant Cell Environ 37: 2613–2622 [DOI] [PubMed] [Google Scholar]
- Robert CAM, Veyrat N, Glauser G, Marti G, Doyen GR, Villard N, Gaillard MDP, Köllner TG, Giron D, Body M, et al. (2012) A specialist root herbivore exploits defensive metabolites to locate nutritious tissues. Ecol Lett 15: 55–64 [DOI] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PEH, Jahnke S, van Dongen JT, Schittko U, Baldwin IT (2006) SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc Natl Acad Sci USA 103: 12935–12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe KA, Marquis RJ, Hochwender CG, Simms EL (2000) The evolutionary ecology of tolerance to consumer damage. Annu Rev Ecol Syst 31: 565–595 [Google Scholar]
- Strauss SY, Agrawal AA (1999) The ecology and evolution of plant tolerance to herbivory. Trends Ecol Evol 14: 179–185 [DOI] [PubMed] [Google Scholar]
- Swarup R, Péret B (2012) AUX/LAX family of auxin influx carriers-an overview. Front Plant Sci 3: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe MR, Ferrieri AP, Herth MM, Ferrieri RA (2007) 11C-imaging: methyl jasmonate moves in both phloem and xylem, promotes transport of jasmonate, and of photoassimilate even after proton transport is decoupled. Planta 226: 541–551 [DOI] [PubMed] [Google Scholar]
- Wang B, Chu J, Yu T, Xu Q, Sun X, Yuan J, Xiong G, Wang G, Wang Y, Li J (2015) Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc Natl Acad Sci USA 112: 4821–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci USA 108: 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AD, Sampson MB, Neuffer MG, Michalczuk L, Slovin JP, Cohen JD (1991) Indole-3-acetic-acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science 254: 998–1000 [DOI] [PubMed] [Google Scholar]
- Zazímalová E, Murphy AS, Yang H, Hoyerová K, Hošek P (2010) Auxin transporters—why so many? Cold Spring Harb Perspect Biol 2: a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.