Abstract
Background
Hydroxyproline-rich glycoproteins (HRGPs) constitute a plant cell wall protein superfamily that functions in diverse aspects of growth and development. This superfamily contains three members: the highly glycosylated arabinogalactan-proteins (AGPs), the moderately glycosylated extensins (EXTs), and the lightly glycosylated proline-rich proteins (PRPs). Chimeric and hybrid HRGPs, however, also exist. A bioinformatics approach is employed here to identify and classify AGPs, EXTs, PRPs, chimeric HRGPs, and hybrid HRGPs from the proteins predicted by the completed genome sequence of poplar (Populus trichocarpa). This bioinformatics approach is based on searching for biased amino acid compositions and for particular protein motifs associated with known HRGPs with a newly revised and improved BIO OHIO 2.0 program. Proteins detected by the program are subsequently analyzed to identify the following: 1) repeating amino acid sequences, 2) signal peptide sequences, 3) glycosylphosphatidylinositol lipid anchor addition sequences, and 4) similar HRGPs using the Basic Local Alignment Search Tool (BLAST).
Results
The program was used to identify and classify 271 HRGPs from poplar including 162 AGPs, 60 EXTs, and 49 PRPs, which are each divided into various classes. This is in contrast to a previous analysis of the Arabidopsis proteome which identified 162 HRGPs consisting of 85 AGPs, 59 EXTs, and 18 PRPs. Poplar was observed to have fewer classical EXTs, to have more fasciclin-like AGPs, plastocyanin AGPs and AG peptides, and to contain a novel class of PRPs referred to as the proline-rich peptides.
Conclusions
The newly revised and improved BIO OHIO 2.0 bioinformatics program was used to identify and classify the inventory of HRGPs in poplar in order to facilitate and guide basic and applied research on plant cell walls. The newly identified poplar HRGPs can now be examined to determine their respective structural and functional roles, including their possible applications in the areas plant biofuel and natural products for medicinal or industrial uses. Additionally, other plants whose genomes are sequenced can now be examined in a similar way using this bioinformatics program which will provide insight to the evolution of the HRGP family in the plant kingdom.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0912-3) contains supplementary material, which is available to authorized users.
Keywords: Arabinogalactan-protein, Bioinformatics, Extensin, Hydroxyproline-rich glycoprotein, Plant cell wall, Poplar, Populus trichocarpa, Proline-rich protein
Background
The hydroxyproline-rich glycoproteins (HRGPs) constitute a diverse superfamily of glycoproteins found throughout the plant kingdom [1–6]. Based on their patterns of proline hydroxylation and subsequent glycosylation, HRGPs are separated into three families: arabinogalactan-proteins (AGPs), extensins (EXTs), and proline-rich proteins (PRPs). These differences in proline hydroxylation and glycosylation are ultimately determined by the primary amino acid sequence, particularly with respect to the location and distribution of proline residues. Specifically, AGPs typically contain non-contiguous proline residues (e.g., APAPAP) which are hydroxylated and glycosylated with arabinogalactan (AG) polysaccharides [7–9]. In contrast, EXTs typically contain contiguous prolines (e.g., SPPPP) that are hydroxylated and subsequently glycosylated with arabinose oligosaccharides [2, 10]. The PRPs typically contain stretches of contiguous proline residues which are shorter than those found in EXTs; these proline residues may be hydroxylated and subsequently glycosylated with arabinose oligosaccharides. Thus, AGPs are extensively glycosylated, EXTs are moderately glycosylated, and PRPs are lightly glycosylated, if at all. In addition, most HRGPs have an N-terminal signal peptide that results in their insertion into the endomembrane system and delivery to the plasma membrane/cell wall. Certain families of HRGPs, particularly the AGPs, are also modified with a C-terminal glycosylphosphatidylinositol (GPI) membrane anchor, which tethers the protein to the outer leaflet of plasma membrane and allows the rest of the glycoprotein to extend toward the cell wall in the periplasm [11–13]. These characteristic amino acid sequences and sequence features allow for the effective identification and classification of HRGPs from proteomic databases by bioinformatic approaches involving biased amino acid composition searches and/or HRGP amino acid motif searches [14–17]. In addition, Newman and Cooper [18] utilized another bioinformatic approach involving searching for proline-rich tandem repeats to identify numerous HRGPs as well as other proteins in a variety of plant species.
The AGP family can be divided into the classical AGPs, which include a subset of lysine-rich classical AGPs, and the AG peptides. In addition, chimeric AGPs exist, most notably the fasciclin-like AGPs (FLAs) and the plastocyanin AGPs (PAGs), but also other proteins which have AGP-like regions along with non-HRGP sequences. Classical AGPs are identified using a search for proteins whose amino acid composition consists of at least 50 % proline (P), alanine (A), serine (S), and theronine (T), or more simply, 50 % PAST [14, 16]. Similarly, AG peptides are identified with a search of 35 % PAST, but are size limited to be between 50 and 90 amino acids in length. EXTs contain characteristic SPPP and SPPPP repeats. As such, EXTs are identified by searching for proteins that contain at least two SPPP repeats. Finally, PRPs are identified by searching for proteins that contain at least 45 % PVKCYT or contain two or more repeated motifs (PPVX[KT] or KKPCPP). Similar to AGPs, chimeric versions of EXTs and PRPs also exist. Each HRGP identified here in this poplar study can then be subjected to BLAST searches against both the Arabidopsis and poplar databases for several purposes: 1) to ensure that the protein identified is similar in sequence to some known HRGPs in Arabidopsis, 2) to identify if the protein is similar to other proteins in poplar which were identified as HRGPs by using the BIO OHIO 2.O program, and 3) to identify similar proteins that may be HRGPs, but which do not meet the search criteria.
Although the numbers and types of HRGPs in Arabidopsis are well established [14, 16], much less is known in other plant species. As more plant genome sequencing projects are completed, comprehensive identification and analysis of HRGPs in these species can be completed. This knowledge can be used to facilitate and guide basic and applied research on these cell wall proteins, potentially with respect to plant biofuel research that utilizes cell wall components for energy production. In fact, a paper was recently published linking poplar EXTs to recalcitrance [19]. Moreover, comparisons can be made with what is already known in Arabidopsis, which will potentially provide further insight into the roles that these particular classes of HRGPs play in the plant as well as their evolution. A comprehensive inventory of HRGPs in poplar, or trees in general, is lacking, although a search for proline-rich tandem repeat proteins in poplar recently identified several HRGP sequences [18]. Additionally, 15 fasciclin-like AGPs (FLAs) were identified in Populus tremula × P. alba, a hybrid related to Populus trichocarpa, and found to be highly expressed in tension wood [20].
Here, the completed genome sequence, or more precisely the encoded proteome, of Populus trichocarpa was utilized to successfully conduct a comprehensive bioinformatics based approach for the identification of HRGPs in this species (Fig. 1). This approach utilizes a newly revised and improved BIO OHIO 2.0 program. Since Arabidopsis and poplar are both dicots, they are expected to have a similar inventory of HRGPs, as opposed to the monocots, which may prove to be considerably different. Nevertheless, Arabidopsis and poplar are morphologically different from one another with Arabidopsis being a small annual herbaceous plant and with poplar being a large woody deciduous tree. Distinct differences were reflected in their inventories of HRGPs, which can now be used to guide further research on the functional roles, commercial applications, and evolution of these ubiquitous and highly modified plant glycoproteins.
Fig. 1.
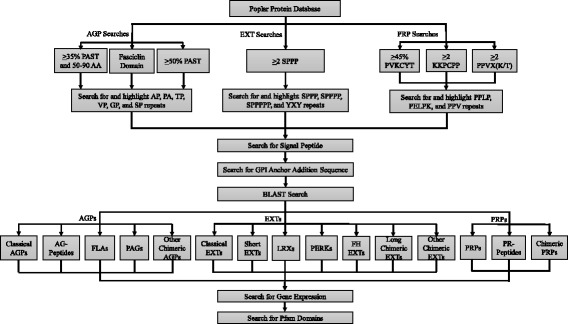
Workflow diagram for the identification, classification, and analysis of HRGPs (AGPs, EXTs, and PRPs) in poplar using a newly revised and improved BIO OHIO 2.0. Classical AGPs were characterized as containing greater than 50 % PAST. AG peptides were characterized to be 50 to 90 amino acids in length and containing greater than 35 % PAST. FLAs were characterized as having a fasciclin domain. Chimeric AGPs were characterized as containing greater than 50 % PAST coupled with one or more domain(s) not known in HRGPs. All AGPs feature the presence of AP, PA, TP, VP, GP, and SP repeats distributed throughout the protein. EXTs were defined as containing two or more SPPP repeats coupled with the distribution of such repeats throughout the protein; chimeric extensins, including LRXs, PERKs, FH EXTs, long chimeric EXTs (>2000 aa), and other chimeric EXTs, were similarly identified but were distinguished from the classical EXTs by the localized distribution of such repeats in the protein and the presence of non-HRGP sequences/domains, many of which were identified by the Pfam analysis; and short extensins were defined to be less than 200 amino acids in length coupled with the EXT definition. PRPs were identified to contain greater than 45 % PVKCYT or two or more KKPCPP or PVX(K/T) repeats coupled with the distribution of such repeats and/or PPV throughout the protein. Chimeric PRPs were similarly identified but were distinguished from PRPs by the localized distribution of such repeats in the protein. Other integrated functional modules include searching for the presence of a signal peptide to provide added support for the identification of an HRGP; the presence of a GPI anchor addition sequence for added support for the identification of AGPs, and BLAST searches to provide some support to the classification. Tissue/organ-specific expression data were also obtained for identified HRGPs to guide for future research
Methods
Identification of AGPs, EXTs, and PRPs using BIO OHIO 2.0
The Populus trichocarpa protein database (Ptrichocarpa_210_v3.0.protein.fa.gz) was downloaded from the Phytozome v11.0 website (www.phytozome.org) [21]. The protein database was searched for AGPs, EXTs, and PRPs using the newly revised and improved BIO OHIO 2.0 software [16, 22]. Compared to the previous version, this new version integrated more functional modules that include searching for the presence of a signal peptide at the SignalP server (www.cbs.dtu.dk/services/SignalP/) [23], searching for the presence of GPI anchor addition sequences using the big-PI plant predictor (mendel.imp.ac.at/gpi/plant_server.html) [24], as well as an automated BLAST search against Arabidopsis proteome. In cases where no signal peptide was identified using the default parameters for a sequence, the sensitive mode was then used which lowered the D-cutoff values to 0.34 [23]. These improvements make the program an ideal bioinformatic tool to study cell wall proteins/glycoproteins within any sequenced plant species. The program is freely available upon request. Briefly, classical AGPs were identified as proteins of any length that consisted of 50 % or greater of the amino acids P, A, S, and T (PAST). AG peptides were identified as proteins of 50–90 amino acids in length consisting of 35 % or greater PAST. FLAs were designated as proteins containing the following consensus motif:
EXTs were identified by searching with a regular expression for the occurrence of two or more SPPP repeats in the protein. Hits were examined for the location and distribution of SP3 and SP4 repeats as well as for the occurrence of other repeating sequences, including YXY. PRPs were identified by searching for a biased amino acid composition of greater than 45 % PVKCYT or for sequences containing two or more repeated motifs (PPVX[KT] or KKPCPP) [25].
BLAST Analysis against Arabidopsis and poplar proteomes
All proteins identified by the BIO OHIO 2.0 searches were subjected to protein-protein BLAST (blastp) analysis. BLAST analysis against Arabidopsis HRGPs was conducted as an integrated module within BIO OHIO 2.0. BLAST analysis against the poplar database (Ptrichocarpa_210_v3.0.protein.fa) was conducted using NCBI BLAST+ (2.2.30) downloaded from the NCBI website. BLAST searches were conducted with the “filter query” option both on and off.
Pfam database and poplar HRGP Gene Expression Database
All proteins identified in this study were subjected to a sequence search using Pfam database 30.0 (http://pfam.xfam.org/) to identify Pfam matches within the protein sequences [26], and the Poplar eFP Browser (http://bar.utoronto.ca/efppop/cgi-bin/efpWeb.cgi) for organ/tissue-specific expression data [27]. Specifically, protein sequences of poplar v3.0 were entered into the Pfam database, while poplar v2.0 identifiers were entered into the Poplar eFP Browser since the eFP browser currently does not recognize poplar v3.0 identifiers.
Results
Arabinogalactan-proteins (AGPs)
Among the 73,013 proteins in the poplar database, 86 proteins were found to have at least 50 % PAST, while 194 peptides have at least 35 % PAST, and are between 50 and 90 amino acids in length (Table 1). Several chimeric AGPs were identified in the 50 % PAST search, but the FLAs in particular required a unique test as they typically do not meet the 50 % PAST threshold. Previously in Arabidopsis, a consensus sequence for the fasciclin H1 domain was utilized to search for these proteins, and this consensus sequence was again utilized here [16]. A total of 43 proteins were found to contain this sequence.
Table 1.
AGPs, EXTs, and PRPs identified from the Populus trichocarpa protein database based on biased amino acid compositions, size, and repeat units
| Search Criteria | Total | Classical AGPs | Lys-Rich AGPs | AG Peptides | FLAs | PAGs | Other Chimeric AGPs | EXTs | Short EXTs | LRXs | PERKs | FH EXTs | Other Chimeric EXTs | PRPs | PR Peptides | Chimeric PRPs | Others |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≥50 % PAST | 86 | 10 | 5 | 0 | 1 | 5 | 0 | 7 | 4 | 0 | 0 | 0 | 0 | 1 | 16 | 0 | 37 |
| ≥35 % PAST and 50-90 AA | 194 | 0 | 0 | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 163 |
| Fasciclin domain | 43 | 0 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 |
| ≥2 SPPP | 162 | 1 | 1 | 0 | 0 | 2 | 0 | 8 | 21 | 10 | 12 | 5 | 3 | 0 | 0 | 0 | 99 |
| ≥2 KKPCPP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ≥2 PPV.[KT] | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 25 |
| ≥45 % PVKCYT | 240 | 4 | 5 | 0 | 0 | 0 | 1 | 8 | 8 | 0 | 0 | 0 | 0 | 10 | 10 | 0 | 194 |
In addition to meeting one of the search criteria, several other factors were considered in determining if the proteins were classified as HRGPs. All proteins were examined for signal peptides and for GPI membrane anchor addition sequences, as these are known to occur in AGPs. In addition, sequences were examined for certain dipeptide repeats which are characteristic of AGPs, including AP, PA, SP, TP, VP, and GP [3, 28]. The presence of these repeats was used to determine if a protein identified by the search was classified as an AGP. The various searches for AGPs combined with BLAST searches identified a total of 162 poplar proteins that were determined to be AGPs (Table 2). In total, 27 classical AGPs (which include six lysine-rich AGPs) and 35 AG peptides were identified. In terms of chimeric AGPs, FLAs were particularly abundant in poplar with 50 being identified. Using the consensus sequence that identifies all 21 of the Arabidopsis FLAs, a total of 24 FLAs were identified in poplar. However, because a single amino acid change in the consensus sequence would result in a particular FLA not being identified, the additional 26 FLAs were identified with BLAST searches. Another particularly common class of chimeric AGPs identified in Arabidopsis was the plastocyanin AGPs, or PAGs. Only five PAGs were identified with the 50 % PAST search, but 34 others were identified that fall below the 50 % PAST threshold with BLAST searches. Finally, 11 other chimeric AGPs were also identified. Representative AGP sequences from each class are shown in Fig. 2, while sequences from all 162 AGPs identified are available in Additional file 1: Figure S1.
Table 2.
Identification and analysis of AGP genes in Populus trichocarpa
| Locus Identifier 3.0 (ID 2.0) a | Name | Class | AP/PA/SP/TP/GP/VP Repeats | % PAST | Amino Acids | Pfamb | SPc | GPI | Organ/tissue-specific Expressiond | Arabidopsis HRGP BLAST Hits | Poplar HRGP BLAST Hitse |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Potri.017G050200 | PtAGP1C | Classical | 3/3/12/2/1/1 | 66 % | 137 | Y | Y | AtAGP1C, AtAGP17K, AtAGP18K, AtAGP7C | PtAGP2C, PtAGP7C, PtAGP9C, PtAGP5C, Potri.005G077100 | ||
| Potri.017G050300 (POPTR_0017s07700) | PtAGP2C | Classical | 5/5/9/2/1/1 | 64 % | 133 | Y | Y | Female catkins | AtAGP1C, AtAGP10C, AtAGP3C, AtPAG11 | PtAGP9C, PtAGP1C, Potri.004G161700, Potri.001G376400, Potri.009G009600 | |
| Potri.005G161100 (POPTR_0005s17440) | PtAGP3C | Classical | 11/9/8/5/0/2 | 59 % | 161 | Y | N | Roots | AtAGP10C, AtAGP3C, AtAGP5C, AtAGP18K, AtPERK13 | Potri.013G119700, Potri.009G124200, Potri.004G162500, Potri.001G376400, Potri.013G112500 | |
| Potri.014G135100 (POPTR_0014s12960) | PtAGP4C | Classical | 4/4/6/1/2/0 | 54 % | 140 | Y | Y | Dark etiolated seedlings, light-grown seedling, young leaf | AtAGP26C, AtAGP27C, AtAGP25C | PtAGP47C, PtAGP48C, PtAGP49K, Potri.013G119700, Potri.004G196400 | |
| Potri.001G339700 (POPTR_0001s35940) | PtAGP5C | Classical | 9/8/4/3/4/0 | 59 % | 144 | Y | Y | Male catkins | AtAGP6C, AtAGP11C, AtAGP17K | PtAGP50C, Potri.003G031800, PtAGP51C, PtAGP52C, Potri.003G143000 | |
| Potri.001G259700 | PtAGP6C | Classical | 1/3/20/3/0/1 | 57 % | 197 | Y | N | None | PtAGP43P, PtPtEXT7, PtPtEXT4 | ||
| Potri.001G310300 (POPTR_0001s31780) | PtAGP7C | Classical | 6/7/8/5/0/2 | 63 % | 126 | Y | Y | Young leaf | AtAGP6C | PtAGP1C, PtAGP9C, Potri.002G256200, Potri.002G235500, Potri.005G049100 | |
| Potri.001G367600 | PtAGP8C | Classical | 7/8/29/4/1/1 | 68 % | 265 | Y | Y | None | Potri.004G145800 | ||
| Potri.001G310400 (POPTR_0001s31790) | PtAGP9C | Classical | 6/7/9/3/0/2 | 62 % | 137 | Y | Y | Young leaf | AtAGP18K, AtAGP1C, AtPEX4, AtAGP10C | PtAGP2C, Potri.009G085400, Potri.013G119700, PtAGP7C, Potri.005G043900 | |
| Potri.017G047500 (POPTR_0017s07480) | PtAGP10C | Classical | 0/2/4/5/1/3 | 50 % | 207 | Y | Y | Female catkins | None | Potri.011G046900, Potri.010G094700, PtPRP23, Potri.004G038300, PtPRP28 | |
| Potri.002G207500 (POPTR_0020s00250) | PtAGP47C | Classical | 4/4/6/1/2/0 | 49 % | 141 | Y | N | Xylem | AtAGP26C, AtAGP27C | PtAGP4C, PtAGP48C, PtAGP49K, Potri.013G119700, Potri.003G164300 | |
| Potri.010G031700 (POPTR_0010s03290) | PtAGP48C | Classical | 2/2/9/2/1/2 | 44 % | 169 | Y* | N | Xylem | AtAGP26C, AtAGP25C, AtAGP27C | PtAGP49K, PtAGP4C, PtAGP47C, Potri.008G153000, Potri.008G147100 | |
| Potri.008G182400 (POPTR_0008s18270) | PtAGP50C | Classical | 3/2/1/0/3/1 | 47 % | 101 | Y | Y | Male catkins | AtAGP50C, AtAGP6C, AtAGP5C | PtAGP52C, PtAGP51C, PtAGP5C, Potri.013G011700, Potri.018G128000 | |
| Potri.015G093700 (POPTR_0015s10580) | PtAGP51C | Classical | 6/3/0/0/2/1 | 49 % | 115 | Y | Y | Male catkins | AtAGP50C, AtAGP6C, AtAGP15P | PtAGP52C, PtAGP50C, PtAGP5C, Potri.014G159300, Potri.009G065300 | |
| Potri.012G095900 (POPTR_0012s09790) | PtAGP52C | Classical | 6/5/0/0/2/1 | 49 % | 115 | Y | Y | Male catkins | AtAGP50C, AtAGP6C, AtAGP3C | PtAGP51C, PtAGP50C, PtAGP5C, Potri.014G159300, Potri.019G095800 | |
| Potri.005G169000 | PtAGP64C | Classical | 10/9/4/1/0/3 | 48 % | 216 | PF14368.4 | Y | N | AtAGP29I | PtAGP60I, PtAGP57I, PtAGP58I, Potri.001G210100, PtAGP69C | |
| Potri.008G155200 (POPTR_0008s15500) | PtAGP65C | Classical | 4/4/3/4/0/7 | 45 % | 219 | PF14368.4 | Y* | Y | Xylem, male catkins, female catkins | AtAGP29I | Potri.010G085200, PtAGP66C, PtAGP67C, PtAGP68C, PtAGP69C |
| Potri.005G212000 (POPTR_0005s23360) | PtAGP66C | Classical | 4/4/5/4/2/2 | 45 % | 207 | PF14368.4 | Y | Y | Roots | AtAGP29I | PtAGP67C, Potri.010G085200, PtAGP65C, PtAGP69C, PtAGP68C |
| Potri.002G050200 (POPTR_0002s05110) | PtAGP67C | Classical | 4/5/5/4/2/2 | 46 % | 205 | PF14368.4 | Y | N | AtAGP29I | PtAGP66C, Potri.010G085200, PtAGP65C, PtAGP68C, PtAGP69C | |
| Potri.010G085400 (POPTR_0010s09550) | PtAGP68C | Classical | 0/2/4/4/0/1 | 44 % | 170 | PF14368.4 | Y | Y | Male catkins | AtAGP29I | PtAGP69C, Potri.005G211800, Potri.002G050500, Potri.002G050300, Potri.005G211900 |
|
Potri.008G155100
(POPTR_0008s15490) |
PtAGP69C | Classical | 1/2/5/2/0/1 | 44 % | 170 | PF14368.4 | Y | Y | Male catkins | AtAGP29I | PtAGP68C, Potri.005G211800, Potri.002G050500, Potri.010G085300, Potri.002G050300 |
| Potri.009G092300 (POPTR_0009s09530) | PtAGP11K | Lysine-rich | 11/19/8/11/1/2 | 69 % | 196 | Y | Y | Xylem | AtAGP17K, AtAGP18K, AtPRP1 | PtAGP14K, Potri.004G181200, Potri.001G310900, PtAGP71I | |
| Potri.010G132500 (POPTR_0010s14250) | PtAGP12K | Lysine-rich | 18/24/10/12/0/4 | 65 % | 241 | Y | N | Xylem | AtAGP19K | PtAGP15K, Potri.013G003500, Potri.007G013600 | |
| Potri.007G051600 (POPTR_0007s10230) | PtAGP13K | Lysine-rich | 12/12/9/11/2/5 | 60 % | 204 | Y | Y | Dark etiolated seedlings, young leaf | AtAGP17K, AtAGP18K | PtAGP14K, Potri.013G003500, PtAGP72I, Potri.018G122900 | |
| Potri.005G144900 (POPTR_0005s18840) | PtAGP14K | Lysine-rich | 11/12/9/10/3/4 | 62 % | 208 | Y | Y | Female catkins | AtAGP18K, AtAGP17K, AtPRP1 | PtAGP13K, Potri.002G008600, Potri.005G049100, Potri.006G234100 | |
| Potri.008G111000 (POPTR_0008s11040) | PtAGP15K | Lysine-rich | 23/33/14/12/0/2 | 66 % | 276 | Y | Y | None | PtAGP12K, PtPtPAG5 | ||
| Potri.008G195700 (POPTR_0008s20030) | PtAGP49K | Lysine-rich | 2/2/9/1/1/4 | 45 % | 194 | Y | N | Female catkins | AtAGP25C, AtAGP27C, AtAGP26C | PtAGP48C, PtAGP4C, PtAGP47C, Potri.008G147100, Potri.010G094700 | |
| Potri.009G063600 (POPTR_0006s05460) | PtAGP16P | AG peptide | 2/2/1/0/0/0 | 48 % | 60 | Y | Y | AtAGP43P, AtAGP23P, AtAGP40P, AtAGP14P, AtAGP15P | PtAGP41P, PtAGP24P, Potri.016G052000, PtAGP29P, PtAGP28P | ||
| Potri.009G062700 | PtAGP17P | AG peptide | 2/2/0/0/0/0 | 36 % | 68 | Y | Y | AtAGP22P, AtAGP16P | PtAGP38P, PtAGP29P, PtAGP22P, PtAGP28P, PtAGP25P | ||
| Potri.009G063200 | PtAGP18P | AG peptide | 3/2/0/0/0/0 | 40 % | 69 | Y | Y | AtAGP43P | PtAGP39P, PtAGP19P, PtAGP29P, PtAGP38P, PtAGP53P | ||
| Potri.009G063000 | PtAGP19P | AG peptide | 3/2/0/0/0/0 | 41 % | 70 | Y | Y | None | PtAGP18P, PtAGP39P, PtAGP29P, PtAGP53P, PtAGP38P | ||
| Potri.013G057500 (POPTR_0013s05400) | PtAGP20P | AG peptide | 2/2/1/0/0/1 | 41 % | 60 | Y | Y | Male catkins | AtAGP14P, AtAGP12P, AtAGP13P, AtAGP21P, AtAGP15P | PtAGP54P, PtAGP33P, PtAGP44P, PtAGP41P, PtAGP30P | |
| Potri.003G136600 (POPTR_0003s13640) | PtAGP21P | AG peptide | 3/2/0/0/0/0 | 39 % | 69 | PF06376.10 | Y | Y | Female catkins, male catkins | AtAGP20P, AtAGP16P, AtAGP22P, AtAGP41P, AtAGP15P | PtAGP40P, PtAGP30P, PtAGP45P, PtAGP35P, PtAGP54P |
| Potri.006G056000 (POPTR_0831s00200) | PtAGP22P | AG peptide | 3/2/0/0/0/0 | 36 % | 68 | Y | Y | Xylem | AtAGP40P, AtAGP43P | PtAGP53P, PtAGP28P, PtAGP29P, PtAGP27P, PtAGP25P | |
| Potri.006G055700 (POPTR_0006s05460) | PtAGP23P | AG peptide | 4/3/0/0/0/0 | 42 % | 66 | Y | Y | male catkins, dark etiolated seedlings | AtAGP16P, AtAGP43P | PtAGP29P, PtAGP27P, PtAGP22P, PtAGP25P, PtAGP28P | |
| Potri.006G056200 (POPTR_0006s05490) | PtAGP24P | AG peptide | 2/1/1/0/0/0 | 47 % | 61 | Y | Y | Male catkins | AtAGP43P, AtAGP23P, AtAGP40P, AtAGP13P, AtAGP14P | Potri.016G052000, PtAGP16P, PtAGP41P, PtAGP29P, PtAGP23P | |
| Potri.006G055900 | PtAGP25P | AG peptide | 3/2/0/0/0/0 | 37 % | 67 | Y | Y | AtAGP43P, AtPAG2 | PtAGP27P, PtAGP28P, PtAGP22P, PtAGP29P, PtAGP53P | ||
| Potri.006G055500 (POPTR_0006s05440) | PtAGP26P | AG peptide | 4/3/1/0/0/0 | 39 % | 69 | Y | Y | Dark etiolated seedlings | AtAGP12P, AtAGP43P, AtAGP15P | PtAGP23P, PtAGP29P, PtAGP28P, PtAGP22P, PtAGP27P | |
| Potri.006G055800 | PtAGP27P | AG peptide | 3/2/0/0/0/0 | 37 % | 67 | Y | Y | AtAGP43P, AtPAG2 | PtAGP25P, PtAGP28P, PtAGP22P, PtAGP29P, PtAGP53P | ||
| Potri.016G052400 (POPTR_0016s05280) | PtAGP28P | AG peptide | 3/2/0/0/0/0 | 37 % | 67 | Y | Y | Dark etiolated seedlings | AtAGP40P, AtAGP15P | PtAGP27P, PtAGP22P, PtAGP25P, PtAGP53P, PtAGP29P | |
| Potri.016G052200 (POPTR_0016s05270) | PtAGP29P | AG peptide | 3/2/1/0/0/1 | 38 % | 67 | Y | Y | Male catkins | AtAGP40P, AtAGP28I AtAGP43P, AtAGP12P | PtAGP22P, PtAGP27P, PtAGP25P, PtAGP28P, PtAGP53P | |
| Potri.015G022600 (POPTR_0015s06130) | PtAGP30P | AG peptide | 2/1/1/0/0/0 | 37 % | 64 | PF06376.10 | Y | Y | AtAGP20P, AtAGP22P, AtAGP16P, AtAGP41P, AtAGP15P | PtAGP45P, PtAGP35P, PtAGP40P, PtAGP21P, Potri.001G070600 | |
| Potri.015G139200 | PtAGP31P | AG peptide | 2/0/0/1/0/0 | 35 % | 57 | Y | N | None | Potri.015G139100, Potri.012G137400, Potri.006G150100, Potri.008G094200, Potri.007G131100 | ||
| Potri.002G226300 (POPTR_0002s21530) | PtAGP32P | AG peptide | 1/1/4/0/1/1 | 37 % | 74 | Y | N | None | PtAGP34P, Potri.012G138200, Potri.001G274200, Potri.002G121800, Potri.015G140000 | ||
| Potri.019G035500 (POPTR_0019s05110) | PtAGP33P | AG peptide | 2/2/1/0/0/1 | 44 % | 59 | Y | Y | AtAGP14P, AtAGP12P, AtAGP13P, AtAGP21P, AtAGP22P | PtAGP20P, PtAGP54P, PtAGP44P, PtAGP41P, PtAGP30P | ||
| Potri.014G156600 (POPTR_0014s15480) | PtAGP34P | AG peptide | 1/0/2/1/0/1 | 37 % | 74 | Y | N | None | PtAGP32P, Potri.001G274200, Potri.012G138200, Potri.015G140000, Potri.010G111200 | ||
| Potri.014G094800 (POPTR_0014s09050) | PtAGP35P | AG peptide | 3/3/2/0/0/0 | 42 % | 76 | PF06376.10 | Y | N | Male catkins | AtAGP20P, AtAGP16P, AtAGP22P, AtAGP41P, AtAGP15P | PtAGP30P, PtAGP45P, PtAGP40P, PtAGP21P, PtAGP17P |
| Potri.T142100 | PtAGP36P | AG peptide | 1/2/2/1/0/0 | 36 % | 90 | Y | N | None | Potri.004G234800, Potri.014G034500, Potri.005G136800, Potri.007G041500, Potri.007G041400 | ||
| Potri.001G387800 (POPTR_0001s39620) | PtAGP37P | AG peptide | 1/0/3/0/0/0 | 37 % | 78 | Y | N | Female catkins, male catkins, young leaf | None | Potri.004G061300, Potri.011G070500, Potri.003G125800, Potri.008G019500, Potri.002G195300 | |
| Potri.001G268400 (POPTR_0001s27530) | PtAGP38P | AG peptide | 3/2/0/0/0/0 | 39 % | 68 | Y | Y | AtAGP22P, AtPAG1 | PtAGP17P, PtAGP29P, PtAGP22P, PtAGP28P, PtAGP27P | ||
| Potri.001G268500 (POPTR_0001s27540) | PtAGP39P | AG peptide | 3/3/0/0/0/0 | 40 % | 69 | Y | Y | AtAGP15P, AtAGP14P, AtAGP28I AtAGP13P, AtPAG1 | PtAGP18P, PtAGP19P, PtAGP29P, PtAGP53P, PtAGP38P | ||
| Potri.001G094700 (POPTR_0001s10310) | PtAGP40P | AG peptide | 3/2/0/0/0/0 | 42 % | 69 | PF06376.10 | Y | Y | AtAGP20P, AtAGP16P, AtAGP22P, AtAGP41P, AtAGP12P | PtAGP21P, PtAGP30P, PtAGP45P, PtAGP35P, Potri.016G086300 | |
| Potri.001G268800 | PtAGP41P | AG peptide | 2/1/1/0/0/0 | 46 % | 60 | Y | Y | AtAGP43P, AtAGP23P, AtAGP40P, AtAGP12P, AtAGP15P | PtAGP16P, PtAGP24P, Potri.016G052000, PtAGP29P, PtAGP28P | ||
| Potri.001G268900 (POPTR_0001s27570) | PtAGP42P | AG peptide | 1/1/0/0/0/0 | 36 % | 66 | Y | Y | None | PtAGP29P, PtAGP56P, Potri.010G100200, Potri.011G126900, PtAGP23P | ||
| Potri.001G259500 | PtAGP43P | AG peptide | 0/0/3/1/0/0 | 37 % | 67 | Y | N | None | PtAGP6C, PtEXT7, PtEXT4, Potri.018G145800, Potri.007G096600 | ||
| Potri.001G004100 (POPTR_0001s04130) | PtAGP44P | AG peptide | 2/1/1/0/0/1 | 40 % | 59 | Y | Y | AtAGP14P, AtAGP12P, AtAGP13P, AtAGP21P, AtAGP15P | PtAGP54P, PtAGP20P, PtAGP33P, PtAGP41P, PtAGP60I | ||
| Potri.012G032000 (POPTR_0012s01350) | PtAGP45P | AG peptide | 2/1/1/0/0/0 | 39 % | 64 | PF06376.10 | Y | Y | Male catkins | AtAGP20P, AtAGP16P, AtAGP22P, AtAGP41P, AtAGP15P | PtAGP30P, PtAGP35P, PtAGP40P, PtAGP21P, PtAGP54P |
| Potri.012G144100 | PtAGP46P | AG peptide | 1/1/1/2/0/1 | 41 % | 89 | Y | N | None | Potri.002G258000, Potri.007G124600, Potri.003G086400, Potri.001G148100, Potri.013G051400 | ||
| Potri.016G052300 | PtAGP53P | AG peptide | 3/2/1/0/0/0 | 32 % | 110 | Y* | Y | AtAGP15P, AtAGP40P, AtPAG11, AtAGP43P, AtPERK3 | PtAGP22P, PtAGP28P, PtAGP27P, PtAGP25P, PtAGP29P | ||
| Potri.003G220900 (POPTR_0003s21020) | PtAGP54P | AG peptide | 3/1/1/1/0/1 | 37 % | 139 | Y* | Y | AtAGP14P, AtAGP12P, AtAGP13P, AtAGP21P, AtAGP22P | PtAGP44P, PtAGP20P, PtAGP33P, PtAGP41P, Potri.004G067400 | ||
| Potri.006G056100 (POPTR_0006s05480) | PtAGP55P | AG peptide | 1/1/0/1/0/0 | 33 % | 66 | Y | N | None | PtAGP56P, PtAGP28P, PtAGP29P, PtAGP22P, PtAGP25P | ||
| Potri.016G052100 (POPTR_0016s05260) | PtAGP56P | AG peptide | 1/1/0/1/0/0 | 31 % | 66 | Y | N | Xylem | None | PtAGP55P, PtAGP29P, PtAGP25P, PtAGP27P, PtAGP22P | |
| Potri.010G244900 (POPTR_0010s25110) | PtFLA1 | Chimeric | 10/4/0/0/3/1 | 26 % | 459 | PF02469.20 | Y | N | AtFLA17, AtFLA16, AtFLA18, AtFLA15, AtFLA12 | PtFLA19, PtFLA6, PtFLA8, PtFLA41, Potri.012G006200 | |
| Potri.009G012200 (POPTR_0009s01740) | PtFLA2 | Chimeric | 8/7/3/2/2/0 | 39 % | 254 | PF02469.20 | Y | N | AtFLA11, AtFLA12, AtFLA13, AtFLA9, AtFLA6 | PtFLA34, PtFLA10, PtFLA23, PtFLA40, PtFLA48 | |
| Potri.013G120600 (POPTR_0013s12490) | PtFLA3 | Chimeric | 4/2/2/3/1/1 | 34 % | 238 | PF02469.20 | Y | Y | Dark etiolated seedlings, roots, female catkins | AtFLA6, AtFLA9, AtFLA13, AtFLA11, AtFLA12 | PtFLA15, PtFLA9, PtFLA7, PtFLA10, PtFLA23 |
| Potri.013G152200 (POPTR_0013s14840) | PtFLA4 | Chimeric | 5/0/5/0/1/0 | 31 % | 353 | PF02469.20 | N | N | Female catkins | AtFLA21, AtFLA19, AtFLA20, AtFLA15, AtFLA16 | Potri.019G125200, PtFLA36, PtFLA42, PtFLA44, Potri.T118500 |
| Potri.011G093500 (POPTR_0011s09590) | PtFLA5 | Chimeric | 7/4/2/2/1/2 | 32 % | 408 | PF02469.20 | Y | Y | AtFLA1, AtFLA2, AtFLA8, AtFLA10, AtFLA14 | PtFLA22, PtFLA16, PtFLA17, PtFLA21, PtFLA37 | |
| Potri.006G200300 (POPTR_0006s21460) | PtFLA6 | Chimeric | 8/2/1/0/3/1 | 27 % | 466 | PF02469.20 | Y | N | AtFLA17, AtFLA18, AtFLA16, AtFLA15, AtFLA11 | PtFLA8, PtFLA1, PtFLA19, PtFLA41, Potri.012G006200 | |
| Potri.006G129200 (POPTR_0006s13120) | PtFLA7 | Chimeric | 6/5/2/1/1/2 | 36 % | 227 | PF02469.20 | Y | N | AtFLA11, AtFLA12, AtFLA6, AtFLA13, AtFLA9 | PtFLA9, PtFLA10, PtFLA23, PtFLA32, PtFLA49 | |
| Potri.016G066500 (POPTR_0016s06680) | PtFLA8 | Chimeric | 7/2/2/1/3/1 | 27 % | 466 | PF02469.20 | Y | N | Male catkins, and light etiolated seedlings, light grown seedling | AtFLA17, AtFLA18, AtFLA16, AtFLA15, AtFLA11 | PtFLA6, PtFLA1, PtFLA19, PtFLA41, Potri.012G006200 |
| Potri.016G088700 (POPTR_0016s09010) | PtFLA9 | Chimeric | 7/6/2/1/1/2 | 37 % | 239 | PF02469.20 | Y | Y | Xylem | AtFLA11, AtFLA12, AtFLA6, AtFLA13, AtFLA9 | PtFLA7, PtFLA10, PtFLA23, PtFLA32, PtFLA49 |
| Potri.015G129400 (POPTR_0015s14570) | PtFLA10 | Chimeric | 5/5/3/2/1/1 | 37 % | 240 | PF02469.20 | Y | Y | Xylem | AtFLA11, AtFLA12, AtFLA6, AtFLA13, AtFLA9 | PtFLA23, PtFLA34, PtFLA2, PtFLA20, PtFLA28 |
| Potri.T130300 (POPTR_0018s03790) | PtFLA11 | Chimeric | 8/3/3/1/2/2 | 40 % | 271 | Y | Y | Male catkins | AtFLA3, AtFLA5, AtFLA14, AtFLA8, AtFLA10 | PtFLA25, PtFLA26, PtFLA21, PtFLA17, PtFLA16 | |
| Potri.002G223300 (POPTR_0002s22020) | PtFLA12 | Chimeric | 8/7/5/4/1/1 | 41 % | 263 | PF02469.20 | Y | Y | Xylem | AtFLA7, AtFLA6, AtFLA11, AtFLA9, AtFLA12 | PtFLA18, PtFLA3, PtFLA9, PtFLA7, PtFLA23 |
| Potri.019G122600 (POPTR_0019s14350) | PtFLA13 | Chimeric | 7/5/1/0/0/2 | 39 % | 215 | PF02469.20 | N | N | AtFLA12, AtFLA11, AtFLA13, AtFLA9, AtFLA6 | PtFLA45, PtFLA35, PtFLA39, PtFLA29, PtFLA47 | |
| Potri.019G120800 (POPTR_0019s14320) | PtFLA14 | Chimeric | 10/10/2/1/0/1 | 43 % | 214 | PF02469.20 | N | N | AtFLA12, AtFLA11, AtFLA9, AtFLA13, AtFLA6 | PtFLA39, PtFLA28, 7PtFLA13, PtFLA45, PtFLA35 | |
| Potri.019G093300 (POPTR_0019s12310) | PtFLA15 | Chimeric | 6/5/3/0/1/1 | 34 % | 245 | PF02469.20 | Y | Y | Dark etiolated seedlings | AtFLA6, AtFLA9, AtFLA13, AtFLA11, AtFLA12 | PtFLA3, PtFLA9, PtFLA7, PtFLA10, PtFLA23 |
| Potri.014G168100 (POPTR_0014s16610) | PtFLA16 | Chimeric | 9/1/0/0/1/0 | 30 % | 397 | PF02469.20 | Y | Y | Roots | AtFLA2, AtFLA1, AtFLA8, AtFLA10, AtFLA4 | PtFLA22, PtFLA5, PtFLA17, PtFLA21, PtFLA37 |
| Potri.014G071700 (POPTR_0014s06740) | PtFLA17 | Chimeric | 13/7/7/4/1/3 | 42 % | 421 | PF02469.20 | Y | Y | Xylem | AtFLA10, AtFLA8, AtFLA2, AtFLA1, AtFLA14 | PtFLA16, PtFLA22, PtFLA5, PtFLA21, PtFLA25 |
| Potri.014G162900 (POPTR_0014s16100) | PtFLA18 | Chimeric | 7/6/7/4/1/1 | 40 % | 262 | PF02469.20 | Y | Y | Xylem | AtFLA7, AtFLA6, AtFLA9, AtFLA11, AtFLA12 | PtFLA12, PtFLA3, PtFLA9, PtFLA7, PtFLA23 |
| Potri.008G012400 (POPTR_0008s01310) | PtFLA19 | Chimeric | 11/4/1/0/3/1 | 27 % | 463 | PF02469.20 | Y | N | Xylem | AtFLA17, AtFLA16, AtFLA18, AtFLA15, AtFLA12 | PtFLA1, PtFLA6, PtFLA8, PtFLA41, Potri.012G006200 |
| Potri.001G320800 (POPTR_0001s32800) | PtFLA20 | Chimeric | 7/6/3/1/1/1 | 37 % | 243 | PF02469.20 | Y | Y | Xylem | AtFLA11, AtFLA12, AtFLA6, AtFLA13, AtFLA9 | PtFLA10, PtFLA23, PtFLA39, PtFLA34, PtFLA13 |
| Potri.001G037800 (POPTR_0001s07490) | PtFLA21 | Chimeric | 2/5/7/2/4/2 | 43 % | 281 | PF02469.20 | Y | Y | Male catkins | AtFLA14, AtFLA8, AtFLA10, AtFLA3, AtFLA2 | PtFLA26, PtFLA25, PtFLA11, PtFLA17, PtFLA5 |
| Potri.001G367900 (POPTR_0001s37650) | PtFLA22 | Chimeric | 7/4/2/2/1/1 | 33 % | 406 | PF02469.20 | Y | Y | Dark etiolated seedlings, young leaf | AtFLA1, AtFLA2, AtFLA8, AtFLA10, AtFLA14 | PtFLA5, PtFLA16, PtFLA17, PtFLA21, PtFLA37 |
| Potri.012G127900 (POPTR_0012s14510) | PtFLA23 | Chimeric | 5/3/2/2/2/1 | 35 % | 240 | PF02469.20 | Y | Y | Xylem | AtFLA11, AtFLA12, AtFLA6, AtFLA9, AtFLA13 | PtFLA10, PtFLA22, PtFLA34, PtFLA2, PtFLA20 |
| Potri.001G440800 (POPTR_0001s43130) | PtFLA24 | Chimeric | 8/5/8/16/3/2 | 50 % | 399 | Y | Y | Male catkins | AtFLA20, AtFLA19, AtFLA21, AtFLA15, AtFLA17 | Potri.T118500, PtFLA44, PtFLA36, Potri.019G125200, PtFLA19 | |
| Potri.018G005100 | PtFLA25 | Chimeric | 8/3/3/1/2/2 | 40 % | 271 | Y | Y | AtFLA3, AtFLA5, AtFLA14, AtFLA8, AtFLA10 | PtFLA11, PtFLA26, PtFLA21, PtFLA17, PtFLA16 | ||
| Potri.006G276200 (POPTR_0006s29110) | PtFLA26 | Chimeric | 11/11/4/4/4/2 | 38 % | 393 | Y* | Y | Male catkins | AtFLA3, AtFLA14, AtFLA5, AtFLA8, AtFLA10 | PtFLA11, PtFLA25, PtFLA21, PtFLA17, PtFLA16 | |
| Potri.012G015000 (POPTR_0012s02210) | PtFLA27 | Chimeric | 8/6/2/1/1/2 | 38 % | 269 | PF02469.20 | Y | Y | AtFLA11, AtFLA12, AtFLA13, AtFLA6, AtFLA9 | PtFLA48, PtFLA10, PtFLA23, PtFLA39, PtFLA28 | |
| Potri.013G014200 (POPTR_0013s01570) | PtFLA28 | Chimeric | 8/8/2/2/0/2 | 42 % | 266 | PF02469.20 | Y | Y | AtFLA12, AtFLA11, AtFLA13, AtFLA9, AtFLA6 | PtFLA39, PtFLA47, PtFLA50, PtFLA32, PtFLA49 | |
| Potri.019G121200 (POPTR_0019s14420) | PtFLA29 | Chimeric | 8/8/3/1/0/2 | 42 % | 263 | PF02469.20 | Y | Y | Xylem | AtFLA11, AtFLA12, AtFLA13, AtFLA9, AtFLA6 | PtFLA50, PtFLA32, PtFLA49, PtFLA28, PtFLA39 |
| Potri.006G174900 (POPTR_0006s18920) | PtFLA30 | Chimeric | 1/4/5/3/0/2 | 38 % | 426 | PF02469.20 | Y* | Y | Xylem | AtFLA4, AtFLA8, AtFLA10, AtFLA1, AtFLA2 | PtFLA37, PtFLA17, PtFLA16, PtFLA5, PtFLA22 |
| Potri.008G127500 (POPTR_0008s12640) | PtFLA31 | Chimeric | 1/0/3/1/0/1 | 29 % | 292 | PF02469.20 | Y | N | Male catkins | AtFLA20, AtFLA19, AtFLA21, AtFLA10, AtFLA12 | PtFLA36, PtFLA42, Potri.019G125200, PtFLA44, PtFLA4 |
| Potri.019G123200 (POPTR_0019s14430) | PtFLA32 | Chimeric | 10/9/1/1/0/2 | 42 % | 263 | PF02469.20 | Y | Y | AtFLA11, AtFLA12, AtFLA9, AtFLA13, AtFLA6, | PtFLA49, PtFLA50, PtFLA28, PtFLA39, PtFLA29 | |
| Potri.019G120900 (POPTR_0019s14330) | PtFLA33 | Chimeric | 8/8/3/1/0/2 | 42 % | 227 | PF02469.20 | Y | Y | Xylem | AtFLA11, AtFLA12, AtFLA13, AtFLA9, AtFLA6 | PtFLA43, PtFLA50, PtFLA32, PtFLA49, PtFLA29 |
| Potri.004G210600 (POPTR_0004s22030) | PtFLA34 | Chimeric | 10/5/3/3/2/0 | 40 % | 268 | PF02469.20 | Y | N | Xylem | AtFLA11, AtFLA12, AtFLA9, AtFLA13, AtFLA6 | PtFLA2, PtFLA10, PtFLA23, PtFLA39, PtFLA40 |
| Potri.019G123000 (POPTR_0019s14410) | PtFLA35 | Chimeric | 11/9/2/1/0/1 | 39 % | 269 | PF02469.20 | Y | Y | AtFLA12, AtFLA11, AtFLA13, AtFLA9, AtFLA6 | PtFLA45, PtFLA39, PtFLA28, PtFLA47, PtFLA13 | |
| Potri.008G128200 (POPTR_0008s12720) | PtFLA36 | Chimeric | 1/0/1/1/0/2 | 28 % | 344 | PF02469.20 | Y | Y | Female catkins, male catkins | AtFLA20, AtFLA21, AtFLA19, AtFLA12, AtFLA6 | PtFLA31, PtFLA42, PtFLA44, PtFLA4, Potri.T118500 |
| Potri.019G002300 (POPTR_0019s01620) | PtFLA37 | Chimeric | 1/2/3/0/0/2 | 29 % | 283 | Y | N | Female catkins, young leaf | AtFLA19, AtFLA21, AtFLA20, AtFLA17, AtFLA16 | Potri.001G306800, PtFLA4, Potri.T118500, PtFLA24, Potri.019G049600 | |
| Potri.018G097000 (POPTR_0018s10600) | PtFLA38 | Chimeric | 2/2/5/2/0/3 | 38 % | 427 | PF02469.20 | Y* | N | Xylem | AtFLA4, AtFLA8, AtFLA10, AtFLA1, AtFLA2, | PtFLA30, PtFLA17, PtFLA16, PtFLA5, PtFLA22 |
| Potri.013G151300 (POPTR_0013s14760) | PtFLA39 | Chimeric | 9/5/2/1/0/2 | 39 % | 269 | PF02469.20 | Y | Y | Xylem | AtFLA12, AtFLA11, AtFLA13, AtFLA6, AtFLA9 | PtFLA40, PtFLA28, PtFLA47, PtFLA45, PtFLA50 |
| Potri.013G151400 (POPTR_0013s14780) | PtFLA40 | Chimeric | 9/9/2/1/0/2 | 40 % | 269 | PF02469.20 | Y | Y | Xylem | AtFLA11, AtFLA12, AtFLA13, AtFLA9, AtFLA6 | PtFLA39, PtFLA28, PtFLA47, PtFLA50, PtFLA32 |
| Potri.019G008400 (POPTR_0073s00210) | PtFLA41 | Chimeric | 9/4/0/0/3/1 | 27 % | 361 | PF02469.20 | N | N | Xylem | AtFLA17, AtFLA16, AtFLA18, AtFLA15, AtFLA7 | PtFLA1, Potri.012G006200, PtFLA19, PtFLA6, PtFLA8 |
| Potri.017G111600 (POPTR_0017s14020) | PtFLA42 | Chimeric | 5/2/4/2/0/2 | 30 % | 352 | PF02469.20 | Y | N | Male catkins | AtFLA20, AtFLA21, AtFLA19, AtFLA10, AtFLA6 | PtFLA36, PtFLA31, PtFLA44, PtFLA4, Potri.019G125200 |
| Potri.019G122800 (POPTR_0019s14390) | PtFLA43 | Chimeric | 9/8/3/0/0/2 | 41 % | 252 | PF02469.20 | Y | Y | Xylem | AtFLA11, AtFLA12, AtFLA9, AtFLA13, AtFLA6 | PtFLA50, PtFLA32, PtFLA49, PtFLA29, PtFLA28 |
| Potri.005G079500 (POPTR_0005s08130) | PtFLA44 | Chimeric | 3/3/5/2/1/6 | 33 % | 442 | Y | N | Male catkins | AtFLA21, AtFLA20, AtFLA19, AtFLA15 | PtFLA36, PtFLA42, Potri.T118500, PtFLA24, PtFLA4 | |
| Potri.019G121100 (POPTR_0019s14370) | PtFLA45 | Chimeric | 10/9/2/1/0/1 | 41 % | 262 | PF02469.20 | Y | N | AtFLA11, AtFLA12, AtFLA13, AtFLA9, AtFLA6 | PtFLA35, PtFLA39, PtFLA13, PtFLA28, PtFLA47 | |
| Potri.009G012100 (POPTR_0009s01730) | PtFLA46 | Chimeric | 6/7/2/0/1/2 | 36 % | 263 | PF02469.20 | Y | N | Xylem | AtFLA11, AtFLA12, AtFLA9, AtFLA13, AtFLA6 | PtFLA2, PtFLA48, PtFLA27, PtFLA28, PtFLA10 |
| Potri.013G151500 (POPTR_0013s14790) | PtFLA47 | Chimeric | 8/9/2/2/0/2 | 42 % | 264 | PF02469.20 | Y | N | Xylem | AtFLA12, AtFLA11, AtFLA13, AtFLA9, AtFLA6, | PtFLA28, PtFLA39, PtFLA40, PtFLA50, PtFLA32 |
| Potri.015G013300 (POPTR_0015s01560) | PtFLA48 | Chimeric | 7/5/2/0/1/3 | 36 % | 267 | PF02469.20 | Y | Y | Xylem | AtFLA11, AtFLA12, AtFLA13, AtFLA9, AtFLA6 | PtFLA27, PtFLA23, PtFLA10, PtFLA2, PtFLA34 |
| Potri.019G121300 | PtFLA49 | Chimeric | 10/9/1/1/0/2 | 42 % | 263 | PF02469.20 | Y | Y | AtFLA11, AtFLA12, AtFLA9, AtFLA13, AtFLA6 | PtFLA32, PtFLA50, PtFLA28, PtFLA39, PtFLA29 | |
| Potri.019G123100 | PtFLA50 | Chimeric | 8/8/3/1/0/2 | 42 % | 263 | PF02469.20 | Y | Y | AtFLA11, AtFLA12, AtFLA13, AtFLA9, AtFLA6 | PtFLA29, PtFLA32, PtFLA49, PtFLA28, PtFLA39 | |
| Potri.011G117800 (POPTR_0011s11860) | PtPAG1 | Chimeric | 10/10/22/9/4/3 | 52 % | 343 | PF02298.15 | Y | Y | Roots | AtPAG17, AtPAG11, AtPAG10, AtPAG14, AtPAG7 | PtPAG5, PtPAG6, PtPAG7, PtPAG8, PtPAG9 |
| Potri.006G067300 (POPTR_0006s06640) | PtPAG2 | Chimeric | 9/13/13/13/1/0 | 54 % | 322 | PF02298.15 | Y* | Y | Male catkins | AtPAG4, AtPAG3, AtPAG5, AtPAG16, AtPAG7 | PtPAG3, PtPAG10, PtPAG11, PtPAG4, PtPAG12 |
| Potri.018G129200 (POPTR_0018s12930) | PtPAG3 | Chimeric | 4/7/14/12/0/0 | 60 % | 250 | PF02298.15 | Y | Y | Roots | AtPAG5, AtPAG4, AtPAG7, AtPAG17, AtPAG3 | PtPAG2, PtPAG10, PtPAG11, PtPAG4, PtPAG12 |
| Potri.018G129400 (POPTR_0018s12950) | PtPAG4 | Chimeric | 1/1/3/4/1/0 | 50 % | 183 | PF02298.15 | Y | Y | AtPAG16, AtPAG5, AtPAG7, AtPAG3, AtPAG8 | PtPAG11, PtPAG10, PtPAG13, PtPAG2, PtPAG3 | |
| Potri.001G398800 (POPTR_0001s40940) | PtPAG5 | Chimeric | 15/11/23/8/5/3 | 51 % | 377 | PF02298.15 | Y | Y | Light-grown seedling, young leaf | AtPAG17, AtPAG11, AtPAG10, AtPAG14, AtPAG7 | PtPAG1, PtPAG6, PtPAG7, PtPAG9, PtPAG14 |
| Potri.017G011200 (POPTR_0017s04390) | PtPAG6 | Chimeric | 1/3/5/2/2/0 | 33 % | 212 | PF02298.15 | Y | Y | AtPAG11, AtPAG14, AtPAG17, AtPAG10, AtPAG7 | PtPAG7, PtPAG1, PtPAG5, PtPAG16, PtPAG14 | |
| Potri.017G012300 (POPTR_0017s00580) | PtPAG7 | Chimeric | 1/3/5/2/2/0 | 33 % | 212 | PF02298.15 | Y | Y | AtPAG11, AtPAG14, AtPAG17, AtPAG10, AtPAG7 | PtPAG6, PtPAG1, PtPAG5, PtPAG16, PtPAG14 | |
| Potri.011G135400 (POPTR_0011s13870) | PtPAG8 | Chimeric | 2/2/3/2/2/2 | 35 % | 208 | PF02298.15 | Y | Y | Roots, young leaf | AtPAG7, AtPAG13, AtPAG2, AtPAG12, AtPAG17 | PtPAG14, PtPAG16, PtPAG1, PtPAG5, PtPAG15 |
| Potri.018G018200 (POPTR_0018s02630) | PtPAG9 | Chimeric | 1/2/2/0/2/0 | 26 % | 178 | PF02298.15 | Y | Y | Young leaf | AtPAG13, AtPAG2, AtPAG15, AtPAG12, AtPAG1 | PtPAG16, PtPAG15, PtPAG1, PtPAG5, PtPAG6 |
| Potri.001G192100 (POPTR_0001s19280) | PtPAG10 | Chimeric | 2/1/5/3/1/1 | 41 % | 210 | PF02298.15 | Y | Y | Male catkins | AtPAG2, AtPAG4, AtPAG3, AtPAG16, AtPAG7 | PtPAG2, PtPAG3, PtPAG4, PtPAG11, PtPAG17 |
| Potri.006G067400 (POPTR_0006s06650) | PtPAG11 | Chimeric | 0/1/3/0/1/0 | 39 % | 163 | PF02298.15 | Y | Y | Light-grown seedling | AtPAG16, AtPAG5, AtPAG8, AtPAG3, AtPAG13 | PtPAG4, PtPAG2, PtPAG3, PtPAG10, PtPAG13 |
| Potri.003G047300 (POPTR_0003s04580) | PtPAG12 | Chimeric | 1/0/4/2/1/2 | 35 % | 217 | PF02298.15 | Y | Y | Female catkins | AtPAG16, AtPAG4, AtPAG5, AtPAG3, AtPAG8 | PtPAG18, PtPAG19, Potri.006G259100, PtPAG20, Potri.006G259000 |
| Potri.014G049600 (POPTR_0014s04850) | PtPAG13 | Chimeric | 2/1/1/5/1/1 | 48 % | 192 | PF02298.15 | Y | Y | Dark etiolated seedlings | AtPAG9, AtPAG8, AtPAG6, AtPAG3, AtPAG5 | PtPAG21, PtPAG22, PtPAG290, PtPAG23, PtPAG12 |
| Potri.001G419200 (POPTR_0001s44510) | PtPAG14 | Chimeric | 4/5/2/3/0/2 | 35 % | 221 | PF02298.15 | Y | Y | Roots | AtPAG7, AtPAG17, AtPAG15, AtPAG11, AtPAG12 | PtPAG8, PtPAG15, PtPAG6, PtPAG1, PtPAG7 |
| Potri.006G184100 (POPTR_0006s19770) | PtPAG15 | Chimeric | 2/2/3/0/2/0 | 29 % | 178 | PF02298.15 | Y | Y | AtPAG13, AtPAG2, AtPAG15, AtPAG12, AtPAG1 | PtPAG16, PtPAG9, PtPAG8, PtPAG14, PtPAG1 | |
| Potri.006G264600 (POPTR_0006s28040) | PtPAG16 | Chimeric | 2/3/3/0/2/0 | 28 % | 179 | PF02298.15 | Y | Y | Young leaf | AtPAG13, AtPAG2, AtPAG15, AtPAG1, AtPAG12 | PtPAG9, PtPAG15, PtPAG8, PtPAG1, PtPAG6 |
| Potri.013G061300 (POPTR_0013s05800) | PtPAG17 | Chimeric | 2/2/3/1/0/1 | 29 % | 155 | PF02298.15 | Y | N | Female catkins, male catkins | AtPAG5, AtPAG4, AtPAG3, AtPAG16, AtPAG13 | PtPAG39, PtPAG24, PtPAG25, PtPAG26, PtPAG27 |
| Potri.002G161300 (POPTR_0002s16270) | PtPAG18 | Chimeric | 2/2/2/0/1/0 | 31 % | 169 | PF02298.15 | Y | Y | Male catkins | AtPAG16, AtPAG4, AtPAG3, AtPAG5, AtPAG13 | PtPAG19, Potri.002G156100, Potri.002G156400, Potri.006G259000, Potri.006G259100 |
| Potri.001G268700 (POPTR_0001s27560) | PtPAG19 | Chimeric | 1/2/4/0/0/0 | 31 % | 165 | PF02298.15 | Y | Y | Male catkins | AtPAG16, AtPAG4, AtPAG3, AtPAG5, AtPAG13 | PtPAG18, Potri.002G156100, Potri.002G156400, Potri.006G259000, PtPAG20 |
| Potri.002G052500 (POPTR_0002s05340) | PtPAG20 | Chimeric | 0/1/2/0/1/0 | 28 % | 169 | PF02298.15 | Y | Y | Young leaf | AtPAG16, AtPAG4, AtPAG3, AtPAG5, AtPAG13 | PtPAG18, PtPAG19, Potri.002G156100, Potri.002G156400, Potri.006G259000 |
| Potri.001G080700 (POPTR_0001s11680) | PtPAG21 | Chimeric | 1/2/0/0/0/1 | 30 % | 184 | PF02298.15 | Y | Y | AtPAG5, AtPAG8, AtPAG9, AtPAG16, AtPAG3 | PtPAG22, PtPAG13, PtPAG28, PtPAG23, PtPAG290 | |
| Potri.003G150300 (POPTR_0003s15000) | PtPAG22 | Chimeric | 1/1/1/0/0/0 | 31 % | 183 | PF02298.15 | Y | Y | AtPAG5, AtPAG16, AtPAG8, AtPAG3, AtPAG4 | PtPAG21, PtPAG13, PtPAG28, PtPAG23, PtPAG290 | |
| Potri.002G101300 (POPTR_0002s10170) | PtPAG23 | Chimeric | 0/1/3/1/0/4 | 42 % | 188 | PF02298.15 | Y | Y | Xylem | AtPAG5, AtPAG8, AtPAG6, AtPAG3, AtPAG9 | PtPAG290, PtPAG13, PtPAG12, PtPAG22, PtPAG24 |
| Potri.013G030000 (POPTR_0013s03090) | PtPAG24 | Chimeric | 0/1/3/2/1/3 | 31 % | 168 | PF02298.15 | Y | Y | Male catkins | AtPAG5, AtPAG4, AtPAG3, AtPAG16, AtPAG13 | PtPAG25, PtPAG30, PtPAG26, PtPAG27, Potri.001G114200 |
| Potri.013G030200 (POPTR_0986s00200) | PtPAG25 | Chimeric | 0/1/3/2/1/3 | 31 % | 168 | PF02298.15 | Y | Y | Male catkins | AtPAG5, AtPAG4, AtPAG3, AtPAG16, AtPAG13 | PtPAG24, PtPAG30, PtPAG26, PtPAG27, Potri.001G114200 |
| Potri.019G037800 | PtPAG26 | Chimeric | 1/1/1/2/0/0 | 32 % | 155 | PF02298.15 | Y | Y | AtPAG5, AtPAG16, AtPAG4, AtPAG9, AtPAG3 | PtPAG27, PtPAG39, PtPAG24, PtPAG25, PtPAG30 | |
| Potri.T070900 (POPTR_0019s05370) | PtPAG27 | Chimeric | 1/1/1/2/0/0 | 32 % | 155 | PF02298.15 | Y | Y | Male catkins | AtPAG5, AtPAG16, AtPAG4, AtPAG9, AtPAG3 | PtPAG26, PtPAG39, PtPAG24, PtPAG25, PtPAG30 |
| Potri.007G120200 (POPTR_0007s02750) | PtPAG28 | Chimeric | 2/6/13/7/1/0 | 49 % | 247 | PF02298.15 | Y | Y | Dark etiolated seedlings | AtPAG5, AtPAG17, AtPAG4, AtPAG3, AtPAG8 | PtPAG21, PtPAG22, PtPAG13, PtPAG12, PtPAG31 |
| Potri.002G101200 (POPTR_1040s00200) | PtPAG29 | Chimeric | 0/1/4/3/0/4 | 37 % | 249 | PF02298.15 | Y* | Y | AtPAG5, AtPAG8, AtPAG3, AtPAG6, AtPAG9 | PtPAG23, PtPAG13, PtPAG12, PtPAG22, PtPAG21 | |
| Potri.003G117900 (POPTR_0003s11780) | PtPAG30 | Chimeric | 0/0/6/1/0/2 | 33 % | 167 | PF02298.15 | Y | Y | Male catkins, female catkins | AtPAG5, AtPAG4, AtPAG3, AtPAG16, AtPAG9 | PtPAG24, PtPAG25, PtPAG26, PtPAG27, PtPAG17 |
| Potri.001G332200 (POPTR_0001s33960) | PtPAG31 | Chimeric | 1/1/2/1/0/0 | 33 % | 168 | PF02298.15 | Y | Y | Xylem | AtPAG5, AtPAG4, AtPAG3, AtPAG13, AtPAG16 | PtPAG24, PtPAG25, Potri.009G136200, PtPAG28, PtPAG23 |
| Potri.008G151000 (POPTR_0008s15040) | PtPAG32 | Chimeric | 3/1/2/0/1/3 | 35 % | 185 | PF02298.15 | Y | N | Xylem | AtPAG16, AtPAG3, AtPAG4, AtPAG5, AtPAG13 | PtPAG38, PtPAG18, Potri.006G259000, Potri.006G259100, PtPAG19 |
| Potri.017G088500 (POPTR_0017s12450) | PtPAG33 | Chimeric | 2/2/1/1/0/0 | 23 % | 175 | PF02298.15 | Y* | Y | Roots | AtPAG16, AtPAG9, AtPAG1, AtPAG5, AtPAG2, | Potri.001G219900, Potri.001G219800, Potri.017G088600, Potri.003G183300, Potri.001G043600 |
| Potri.015G114300 (POPTR_0015s12570) | PtPAG34 | Chimeric | 0/2/0/0/0/1 | 20 % | 131 | PF02298.15 | Y | N | AtPAG11, AtPAG7, AtPAG13, AtPAG2, AtPAG14 | Potri.015G114700, Potri.015G113300, Potri.015G115600, Potri.015G117100, Potri.015G114600 | |
| Potri.010G243600 (POPTR_0010s24980) | PtPAG35 | Chimeric | 3/3/6/0/1/2 | 34 % | 214 | PF02298.15 | Y | Y | Male catkins | AtPAG11, AtPAG5, AtPAG17, AtPAG2, AtPAG4, | PtPAG2, PtPAG4, PtPAG3, PtPAG18, PtPAG12 |
| Potri.001G187700 (POPTR_0001s18820) | PtPAG36 | Chimeric | 1/1/2/2/1/0 | 27 % | 181 | PF02298.15 | Y | Y | Male catkins, female catkins | AtPAG11, AtPAG7, AtPAG2, AtPAG17, AtPAG14 | PtPAG37, Potri.015G052000, PtPAG8, PtPAG1, Potri.001G338800 |
| Potri.003G050500 (POPTR_0003s04900) | PtPAG37 | Chimeric | 2/0/2/1/0/0 | 26 % | 180 | PF02298.15 | Y | Y | AtPAG17, AtPAG2, AtPAG13, AtPAG7, AtPAG15 | PtPAG36, Potri.015G052000, PtPAG15, Potri.001G338800, PtPAG1 | |
| Potri.010G089900 (POPTR_0010s10020) | PtPAG38 | Chimeric | 1/2/2/1/1/2 | 34 % | 185 | PF02298.15 | Y | N | Xylem | AtPAG16, AtPAG3, AtPAG4, AtPAG5, AtPAG13 | PtPAG32, PtPAG18, Potri.006G259000, Potri.006G259100, Potri.002G156100 |
| Potri.013G054500 (POPTR_0013s05140) | PtPAG39 | Chimeric | 2/1/0/1/0/0 | 29 % | 156 | PF02298.15 | Y | N | Female catkins | AtPAG5, AtPAG16, AtPAG4, AtPAG3, AtPAG9 | PtPAG26, PtPAG27, PtPAG24, PtPAG25, PtPAG17 |
| Potri.002G092800 (POPTR_0002s09340) | PtAGP57I | Chimeric | 10/7/3/0/0/1 | 46 % | 193 | PF14368.4 | Y | N | AtAGP29I | PtAGP60I, PtAGP64C, PtAGP58I, PtAGP61I, PtAGP69C | |
| Potri.003G020200 (POPTR_0003s01440) | PtAGP58I | Chimeric | 6/5/2/1/1/0 | 43 % | 179 | PF14368.4 | Y | Y | Xylem, young leaf | AtAGP29I | PtAGP61I, PtAGP60I, PtAGP64C, PtAGP57I, PtAGP68C |
| Potri.006G261800 (POPTR_0006s27770) | PtAGP59I | Chimeric | 3/11/9/5/2/4 | 36 % | 484 | PF00704.26 | Y | N | Male catkins | None | Potri.018G112100, Potri.006G188400, Potri.006G188300, Potri.018G111600, Potri.006G262000 |
| Potri.005G167500 (POPTR_0005s16550) | PtAGP60I | Chimeric | 10/9/4/1/0/3 | 48 % | 216 | PF14368.4 | Y | N | Male catkins, female catkins | AtAGP29I | PtAGP64C, PtAGP57I, PtAGP58I, PtAGP61I, PtAGP69C |
| Potri.001G210100 (POPTR_0001s21750) | PtAGP61I | Chimeric | 8/5/3/0/0/0 | 41 % | 178 | PF14368.4 | Y | Y | Young leaf | AtAGP29I, AtAGP3C | PtAGP58I, PtAGP60I, PtAGP64C, PtAGP57I, Potri.001G231400 |
| Potri.010G085200 (POPTR_0010s09530) | PtAGP62I | Chimeric | 4/1/6/5/2/4 | 47 % | 216 | PF14368.4 | Y | Y | Male catkins | AtAGP29I | PtAGP65C, PtAGP66C, PtAGP67C, PtAGP68C, PtAGP69C |
| Potri.005G003500 (POPTR_0005s00550) | PtAGP63I | Chimeric | 7/15/6/9/0/5 | 41 % | 624 | PF07983.11 | Y | Y | AtPRP13, AtPEX4 | Potri.013G003500, PtAGP70I, PtAGP71I, PtAGP72I, PtAGP73I | |
| Potri.002G059600 (POPTR_0002s06050) | PtAGP70I | Chimeric | 0/1/4/7/0/3 | 47 % | 255 | PF07983.11 | Y | N | AtPRP13 | PtAGP73I, PtAGP71I, PtAGP72I, PtAGP63I, Potri.011G094400 | |
| Potri.001G353400 (POPTR_0001s34420) | PtAGP71I | Chimeric | 1/7/5/9/1/5 | 49 % | 286 | PF07983.11 | Y | N | AtPRP13 | PtAGP72I, PtAGP70I, PtAGP73I, PtAGP63I, Potri.013G003500 | |
| Potri.011G078500 (POPTR_0011s02870) | PtAGP72I | Chimeric | 1/7/5/10/1/1 | 46 % | 304 | PF07983.11 | Y | Y | AtPRP13 | PtAGP71I, PtAGP70I, Potri.013G003500, PtAGP63I, PtAGP73I | |
| Potri.005G202400 | PtAGP73I | Chimeric | 1/2/4/5/0/3 | 44 % | 261 | PF07983.11 | Y | N | AtPRP13 | PtAGP70I, PtAGP71I, PtAGP72I, PtAGP63I, Potri.013G003500 |
a Protein identifiers of the version 2.0 are shown in the parenthesis. Italics indicates a protein that was identified only by a BLAST search
b The domains indicated by the Pfam number are: PF14368.4, LTP_2 domain (Probable lipid transfer); PF06376.10, AGP domain (Arabinogalactan peptide); PF02469.20, Fasciclin domain (Fasciclin domain); PF02298.15, Cu_bind_like domain (Plastocyanin-like domain); PF00704.26, Glyco_hydro_18 domain (Glycoside hydrolase family 18); PF07983.11, X8 domain (X8 domain)
c Asterisk indicates a protein that is predicted to have a signal peptide either using the sensitive mode in the SignalP website or only if amino acids at the N terminus are discarded
d Expression data are shown only when available at http://bar.utoronto.ca/efppop/cgi-bin/efpWeb.cgi
e A locus ID indicates that it is not identified as an HRGP
Fig. 2.
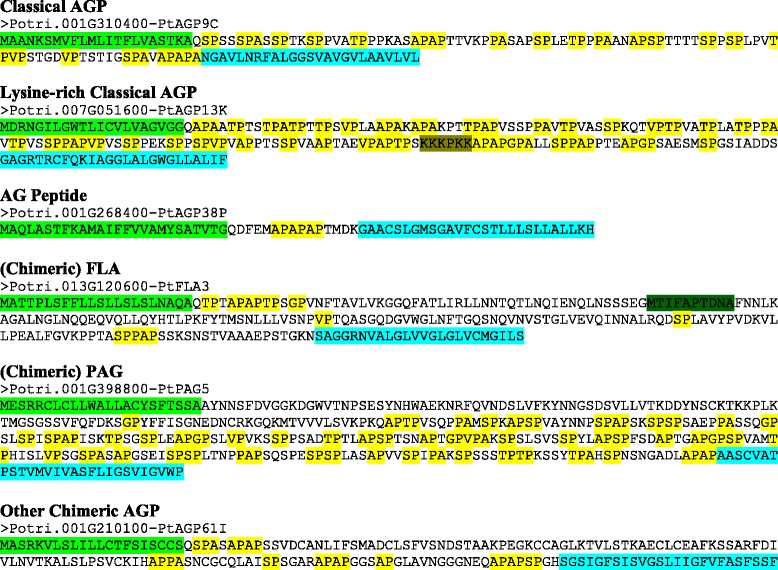
Protein sequences encoded by the representative AGP gene classes in Populus trichocarpa. The colored sequences at the N and C terminus indicate predicted signal peptides (green) and GPI anchor addition sequences (light blue) if present. AP, PA, SP, TP, VP, and GP repeats (yellow), lysine-rich regions (olive) and core fasciclin motif (dark green) are also indicated
The vast majority (97 %) of the identified AGPs were predicted to have a signal peptide and many (70 %) were predicted to have a GPI anchor, both of which are characteristic features of the AGP family. Of the 162 AGPs identified, only four FLAs were predicted to lack a signal peptide. A total of 114 of the 162 AGPs (70 %) were predicted to have a GPI anchor addition sequence. BLAST searches against the Arabidopsis protein database found that all but 21 of the putative AGPs were similar to at least one known Arabidopsis AGP, providing further evidence that these proteins are likely AGPs.
Extensins (EXTs)
Poplar had a smaller number of the classical EXTs containing large numbers of SPPPP repeats compared to Arabidopsis. For instance, a search for proteins with at least 15 SPPPP repeats in Arabidopsis found 21 “hits” while a similar search in poplar yielded only six, two of which are chimeric EXTs. The largest number of SPPPP repeats found in a single protein in poplar is 25, while in Arabidopsis one EXT contains 70 SPPPP repeats. Interestingly, although the abundance of these classical EXTs is decreased, many chimeric EXTs found in Arabidopsis were also in poplar in similar numbers, including the leucine-rich repeat extensins (LRXs) and proline-rich extensin-like receptor protein kinases (PERKs). By searching for proteins that contain at least two SPPP repeats, 162 poplar proteins were identified (Table 1). In all, 59 proteins identified in the search criteria were determined to be EXTs (Table 3). The only exception is a short EXT (i.e., Potri.T139000 or PtEXT33) identified by a BLAST search with one SPPPP that is homologous to several other short EXTs. These 60 proteins included 8 classical EXTs, 22 Short EXTs, 10 LRXs, 12 PERKs, 5 Formin Homology proteins (FHs), and 3 other chimeric EXTs (Fig. 3 and Additional file 2: Figure S2). YXY repeats were observed in 45 % of the EXT sequences; such sequences are involved in cross-linking EXTs [29–33]. Twenty-seven of the 60 EXTs identified contained YXY sequences in which X is quite variable. In contrast, 40 of the 59 EXTs in Arabidopsis (i.e., 68 %) contained YXY sequences in which X was often V [16]. Many of the classical EXTs and some of the LRXs also contained a SPPPP or SPPPPP sequence and Y residue at the C-terminus of their sequences as previously observed in Arabidopsis EXTs [33].
Table 3.
Identification and analysis of EXT genes in Populus trichocarpa
| Locus Identifier 3.0 (ID 2.0)a | Name | Class | SP3/SP4/SP5/YXY Repeats | Amino Acids | Pfamb | SPc | GPI | Organ/issue-specific Expression11 | Arabidopsis HRGP BLAST Hits | Poplar HRGP BLAST Hitse |
|---|---|---|---|---|---|---|---|---|---|---|
| Potri.018G050100 (POPTR 0018 s05480) | PtEXT1 | Classical EXT | 1/6/4/5 | 190 | PF04554.11 | Y | N | Young leaf | AtEXT22, AtEXT21 | Potri.001G201800 |
| Potri.001G019700 (POPTR 0001 s05720) | PtEXT2 | Classical EXT | 1/21/0/11 | 213 | Y | N | AtEXT3/5 | PtEXT8 | ||
| Potri.001G122100 (POPTR_0001 s00420) | PtEXT3 | Classical EXT | 2/5/6/0 | 238 | PF14547.4 | Y | N | Male catkins | AtPRP16, AtPRP15, AtPRP14, AtHAE4 | Potri.013G128800, Potri.002G200100, Potri.018G025900, Potri.001G158400, Potri.014G059800 |
| Potri.001G259600 (POPTR 0001 s26690) | PtEXT4 | Classical EXT | 2/8/2/0 | 500 | Y | N | AtAGP51C | PtEXT7, AGP6C, AGP43P | ||
| Potri.001G020100 (POPTR 0001 s05740) | PtEXT5 | Classical EXT | 1/22/0/13 | 257 | Y | N | None | PtEXT6, PtEXT8 | ||
| Potri.001G019900 | PtEXT6 | Classical EXT | 1/25/0/14 | 259 | Y* | N | None | PtEXT8, PtEXT5 | ||
| Potri.001G260200 (POPTR_0001 s26680) | PtEXT7 | Classical EXT | 4/6/1/0 | 222 | Y | N | None | AGP43P, AGP6C, PtEXT4, Potri.003G074200 | ||
| Potri.001G020000 | PtEXT8 | Classical EXT | 1/23/0/16 | 267 | Y* | N | AtEXT3/5 | PtEXT6, PtEXT5 | ||
| Potri.010G001200 (POPTR_0010s003 50) | PtEXT9 | Short EXT | 1/6/0/3 | 174 | Y | Y | AtEXT37, AtEXT41 | PtEXT24, Potri.008G129200, Potri.010G128900, Potri.008G117500, FLA21 | ||
| Potri.010G113300 (POPTR_0010s12360) | PtEXT10 | Short EXT | 0/2/0/0 | 131 | Y | N | AtEXT31, AtEXT33 | PtEXT23, Potri.006G106800, Potri.005G033000, Potri.001G371600, PossiblePtEXT5 | ||
| Potri.T091000 | PtEXT11 | Short EXT | 1/1/0/0 | 106 | Y | N | None | PtEXT12, PtEXT19, Potri.005G079400 | ||
| Potri.013G045700 (POPTR 0013 s04290) | PtEXT12 | Short EXT | 1/1/0/0 | 111 | Y | N | None | PtEXT11, PtEXT19 | ||
| Potri.003G064900 (POPTR_0003 s063 50) | PtEXT13 | Short EXT | 1/1/3/0 | 167 | Y | N | AtEXT32, AtAGP57C, AtPERK5 | PtEXT26, Potri.009G013500, Potri.006G276200 | ||
| Potri.006G225400 (POPTR_0006s24190) | PtEXT14 | Short EXT | 2/0/1/3 | 186 | Y | Y | Male catkins, roots | AtEXT38, AtEXT7 | Potri.015G147200, Potri.008G168300, Potri.010G094700, Potri.012G144400, PtFH2 | |
| Potri.002G070100 | PtEXT15 | Short EXT | 0/1/2/2 | 102 | Y | N | AtEXT3/5, AtEXT1/4, AtEXT22 | PtEXT20, Potri.017G110900, PtEXT1, PtLRX3 | ||
| Potri.019G015900 (POPTR_0019s03210) | PtEXT16 | Short EXT | 0/2/0/0 | 108 | Y | N | None | PtEXT18, PtEXT33, PtEXT17, Potri.019G015700, Potri.T139100 | ||
| Potri.019G015800 (POPTR_0019s03200) | PtEXT17 | Short EXT | 0/2/0/0 | 107 | Y | N | Male catkins | None | PtEXT33, PtEXT18, PtEXT16, Potri.T139100, Potri.019G015700 | |
| Potri.019G016000 | PtEXT18 | Short EXT | 0/2/0/0 | 116 | Y | N | None | PtEXT16, PtEXT33, PtEXT17, Potri.019G015700, Potri.T139100 | ||
| Potri.019G017300 (POPTR_0019s03400) | PtEXT19 | Short EXT | 0/2/0/0 | 110 | Y* | N | Dark etiolated seedlings | AtPERK6, AtAGP45P | PtEXT11, PtEXT12, Potri.005G257000, Potri.010G244800, Potri.006G136900 | |
| Potri.005G190100 | PtEXT20 | Short EXT | 1/2/0/2 | 115 | Y | N | AtEXT3/5, AtEXT1/4, AtPRP3, AtPRP1 | Potri.019G083200, Potri.013G112500, PtLRX3, Potri.007G090300, Potri.005G077700 | ||
| Potri.014G124700 | PtEXT21 | Short EXT | 0/2/0/0 | 168 | Y | N | AtEXT34, AtEXT41, AtPERK3, AtPERK5 | Potri.015G147200, Potri.012G144400, Potri.001G371600, Potri.004G143700, PtFH2 | ||
| Potri.T082000 | PtEXT22 | Short EXT | 1/1/1/0 | 177 | Y* | N | None | PtAEH4, PtEXT28, PtEXT27, Potri.001G042100, Potri.008G043900 | ||
| Potri.008G129100 (POPTR_0008s12800) | PtEXT23 | Short EXT | 0/3/0/0 | 155 | Y | Y | Female catkins, xylem | AtEXT31, AtEXT33, AtPAG10 | PtEXT10, Potri.010G094700, Potri.015G147200, Potri.006G163700, Potri.018G086100 | |
| Potri.008G213600 (POPTR_0008s22980) | PtEXT24 | Short EXT | 0/1/1/2 | 172 | Y | Y | Male catkins | AtEXT37, AtPERK6, AtEXT41 | PtEXT9, Potri.008G129200, PossiblePtEXT15, Potri.010G094700, Potri.004G143700 | |
| Potri.008G125400 (POPTR_0008s12430) | PtEXT25 | Short EXT | 2/0/0/0 | 80 | Y* | N | None | Potri.005G239200, Potri.010G094700, Potri.010G006800, Potri.002G189300, Potri .005G239200 | ||
| Potri.001G169200 (POPTR 0001 s16930) | PtEXT26 | Short EXT | 0/0/2/0 | 147 | Y | N | None | PtEXT13, Potri.010G006800 | ||
| Potri.001G042200 (POPTR 0001 s03370) | PtEXT27 | Short EXT | 2/2/0/1 | 177 | Y | N | None | PtEXT28, PtEXT22, PtAEH4, Potri.001G042100, Potri.001G316500 | ||
| Potri.T179500 (POPTR_0523s00220) | PtEXT28 | Short EXT | 1/0/1/0 | 176 | Y* | N | None | PtAEH4, PtEXT22, PtEXT27, Potri.001G042100, Potri.005G030300 | ||
| Potri.T101300 (POPTR_0017 s06820) | PtEXT29 | Short EXT | 0/2/0/0 | 151 | Y* | N | AtAGP56C | Potri.007G120100, Potri.002G054100, Potri.001G371600, Potri.015G147200, Potri.002G235500 | ||
| Potri.T139000 | PtEXT33 | Short EXT | 0/1/0/0 | 107 | Y | N | None | PtEXT17, PtEXT18, PtEXT16, Potri.019G015700, Potri.T139100 | ||
| Potri.009G108100 (POPTR_0009s 11130) | PtLRX1 | Chimeric | 5/16/6/1 | 982 | PF13855.4 | Y | N | Female catkins | AtPEX3, AtPEX1, AtPEX4, AtPEX2, AtLRX4 | PtLRX2, PtLRX10, PtLRX3, PtLRX6, PtLRX7 |
| Potri.004G146400 (POPTR_0004s15360) | PtLRX2 | Chimeric | 2/19/1/1 | 603 | PF13855.4 | Y | N | Male catkins | AtPEX3, AtPEX4, AtPEX1, AtPEX2, AtLRX4 | PtLRX1, PtLRX10, PtLRX3, PtLRX4, PtLRX7 |
| Potri.006G081200 | PtLRX3 | Chimeric | 2/1/3/0 | 584 | PF13855.4 PF08263.10 | Y* | N | AtLRX2, AtLRX1, AtLRX4, AtLRX3, AtLRX5 | PtLRX7, PtLRX6, PtLRX4, PtLRX2, PtLRX10 | |
| Potri.006G245600 (POPTR_0006s26190) | PtLRX4 | Chimeric | 2/2/5/1 | 549 | PF08263.10 | Y | N | Xylem | AtLRX4, AtLRX3, AtLRX5, AtLRX7, AtLRX6 | PtLRX8, PtLRX5, PtLRX9, PtLRX6, PtLRX3 |
| Potri.006G162300 (POPTR_0024s00730) | PtLRX5 | Chimeric | 2/3/3/0 | 569 | PF13855.4 | Y | N | Male catkins | AtLRX4, AtLRX3, AtLRX2, AtLRX1, AtPEX4 | PtLRX9, PtLRX6, PtLRX4, PtLRX8, PtLRX3 |
| Potri.018G075900 (POPTR_0018s06150) | PtLRX6 | Chimeric | 1/2/5/0 | 509 | PF13855.4 | Y | N | Male catkins, young leaf, xylem | AtLRX3, AtLRX5, AtLRX2, AtLRX7, AtLRX1 | PtLRX5, PtLRX9, PtLRX4, PtLRX8, PtLRX3 |
| Potri.018G151000 (POPTR_0018s14790) | PtLRX7 | Chimeric | 1/6/1/0 | 481 | PF08263.10 PF13855.4 | Y | N | Male catkins | AtLRX2, AtLRX1, AtLRX4, AtLRX3, AtLRX5 | PtLRX3, PtLRX6, PtLRX5, PtLRX9, PtLRX4 |
| Potri.018G035100 (POPTR_0018s01010) | PtLRX8 | Chimeric | 0/3/2/1 | 496 | PF08263.10 | Y | N | Male catkins | AtLRX4, AtLRX3, AtLRX5, AtLRX7, AtLRX6 | PtLRX4, PtLRX6, Potri.010G083000, PtLRX3, PtLRX7 |
| Potri.T016600 (POPTR_0028s00200) | PtLRX9 | Chimeric | 2/3/4/0 | 573 | PF13855.4 | Y | N | Male catkins | AtLRX4, AtLRX3, AtLRX2, AtLRX1, AtPEX4 | PtLRX5, PtLRX6, PtLRX8, PtLRX3, PtLRX7 |
| Potri.014G036700 (POPTR_0014s03600) | PtLRX10 | Chimeric | 1/5/1/1 | 474 | PF13855.4 | Y | N | Male catkins | AtPEX3, AtPEX1, AtPEX4, AtPEX2, AtLRX4 | PtLRX2, PtLRX1, PtLRX3, PtLRX7, Potri.007G139200 |
| Potri.010G041400 (POPTR_0010s05110) | PtPERK1 | Chimeric | 5/0/2/1 | 700 | PF07714.15 | N | N | AtPERK13, AtPERK12, AtPERK11, AtPERK10, AtPERK8 | PtPERK11,PtPERK3, PtPERK6, PtPERK3, PtPERK12 | |
| Potri.010G132900 (POPTR_0010s14290) | PtPERK2 | Chimeric | 5/4/2/1 | 765 | PF00069.23 | N | N | AtPERK8, AtPERK13, AtPERK1, AtPERK15, AtPERK4 | PtPERK12, PtPERK11, PtPERK1, PtPERK8, PtPERK10 | |
| Potri.017G110400 (POPTR_0017s14140) | PtPERK3 | Chimeric | 5/5/0/1 | 724 | PF07714.15 | N | N | Dark etiolated and light-grown seedlings | AtPERK8, AtPERK10, AtPERK13, AtPERK12, AtPERK3 | PtPERK6, PtPERK12, PtPERK2, PtPERK1, PtPERK11 |
| Potri.009G115200 (POPTR_0009s 11810) | PtPERK4 | Chimeric | 1/6/2/1 | 649 | PF07714.15 | N | N | Male catkins | AtPERK5, AtPERK4, AtPERK15, AtPERK3, AtPERK13 | PtPERK10, PtPERK9, PtPERK8, Potri.001G183000, Potri.T140000 |
| Potri.004G153600 (POPTR_0004s16100) | PtPERK5 | Chimeric | 3/3/3/1 | 656 | PF07714.15 | N | N | AtPERK5, AtPERK7, AtPERK4, AtPERK6, AtPERK15 | PtPERK4, PtPERK10, PtPERK9, PtPERK8, Potri.001G183000 | |
| Potri.004G105200 (POPTR_0004s10490) | PtPERK6 | Chimeric | 6/4/0/2 | 724 | PF07714.15 | N | N | Dark etiolated seedlings | AtPERK10, AtPERK12, AtPERK13, AtPERK3, AtPERK15 | PtPERK3, PtPERK2, PtPERK1, PtPERK11, PtPERK10 |
| Potri.006G242800 | PtPERK7 | Chimeric | 2/0/0/1 | 706 | PF07714.15 | N | N | AtPERK1, AtPERK5, AtPERK14, AtPERK15, AtPERK3 | PtPERK10, PtPERK9, Potri.001G183000, Potri.003G053300, Potri.T140000 | |
| Potri.018G081300 (POPTR_0018s08800) | PtPERK8 | Chimeric | 0/2/2/0 | 672 | PF07714.15 | N | N | Xylem | AtPERK1, AtPERK4, AtPERK5, AtPERK15, AtPERK6 | Potri.001G183000, PtPERK10, PtPERK9, Potri.003G053300, PtPERK5 |
| Potri.007G027000 (POPTR_0007s12680) | PtPERK9 | Chimeric | 2/3/5/1 | 639 | PF07714.15 | N | N | AtPERK5, AtPERK7, AtPERK6, AtPERK15, AtPERK13 | PtPERK10, PtPERK8, PtPERK5, Potri.003G053300, Potri.T140000 | |
| Potri.005G124400 (POPTR_0005s12590) | PtPERK10 | Chimeric | 2/1/5/0 | 592 | PF07714.15 | N | N | Female catkins, male catkins | AtPERK4, AtPERK5, AtPERK7, AtPERK6, AtPERK1 | PtPERK9, PtPERK8, PtPERK5, PtPERK4, Potri.001G183000 |
| Potri.008G189700 (POPTR_0008s19400) | PtPERK11 | Chimeric | 5/3/1/1 | 733 | PF07714.15 | N | N | Male catkins | AtPERK13, AtPERK11, AtPERK8, AtPERK10, AtPERK15 | PtPERK1, PtPERK3, PtPERK6, PtPERK12, PtPERK2 |
| Potri.008G111600 (POPTR_0008s11080) | PtPERK12 | Chimeric | 0/6/2/1 | 728 | PF07714.15 | N | N | AtPERK13, AtPERK1, AtPERK5, AtPERK15, AtPERK3 | PtPERK2, PtPERK1, PtPERK8, PtPERK11, Potri.001G183000 | |
| Potri.003G103800 (POPTR_0003 s10280) | PtFH1 | Chimeric | 1/0/2/0 | 1226 | PF02181.21 PF10409.7 | N | N | Female catkins, male catkins | None | Potri.018G019600, PtFH5, Potri.018G108000, Potri.006G263700, Potri.015G061000 |
| Potri.011G131700 (POPTR 0011 s13510) | PtFH2 | Chimeric | 1/0/2/0 | 987 | PF02181.21 | Y | N | Roots | None | Potri.001G416100, Potri.007G119900, Potri.007G054900, PtFH4, Potri.017G009900 |
| Potri.002G240200 (POPTR_0002s24130) | PtFH3 | Chimeric | 1/0/1/0 | 1066 | PF02181.21 | Y | N | Young leaf, male catkins | None | PtFH4, Potri.007G140200, Potri.017G009900, Potri.007G054900, Potri.013G017900 |
| Potri.014G174700 (POPTR_0014s17310) | PtFH4 | Chimeric | 0/0/2/0 | 1071 | PF02181.21 | Y | N | Roots, light-grown seedling | AtPERK5 | PtFH3, Potri.007G140200, Potri.017G009900, Potri.007G054900, Potri.013G017900 |
| Potri.012G067900 (POPTR_0012s06980) | PtFH5 | Chimeric | 0/0/2/0 | 1400 | PF10409.7 PF02181.21 | N | N | Xylem, male catkins | None | Potri.015G061000, Potri.018G019600, Potri.006G185500, Potri.018G108000, PtFH1 |
| Potri.009G145700 (POPTR_0009s14810) | PtEXT30 | Chimeric | 5/0/0/0 | 467 | PF06830.9 | Y | N | Male catkins, roots | AtEXT51 | Potri.009G097400, Potri.012G145400, Potri.011G127900, Potri.009G012600, Potri.009G012500 |
| Potri.014G115700 (POPTR_0014s11110) | PtEXT31 | Chimeric | 8/0/0/0 | 526 | PF00295.15 | Y* | N | Roots | None | Potri.002G190600, Potri.005G005500, Potri.013G005000, Potri.010G152000, Potri.008G100500 |
| Potri.011G066900 (POPTR_0011s07300) | PtEXT32 | Chimeric | 0/1/2/2 | 498 | PF00112.21 PF00396.16 PF08246.10 | Y | N | Female catkins, male catkins | AtAGP4C | Potri.011G066800, Potri.004G057700, Potri.005G232900, Potri.014G024100, Potri.001G302100 |
| Potri.004G024500 | PtAEH1 | AGP EXT Hybrid | 0/1/1/1 | 673 | PF01657.15 PF07714.15 | Y | N | None | Potri.004G024600, PtAEH2, Potri.004G025800, Potri.011G028400, Potri.004G025900 | |
| Potri.004G024800 | PtAEH2 | AGP EXT Hybrid | 0/1/1/1 | 678 | PF01657.15 PF07714.15 | Y | N | None | Potri.004G024600, Potri.004G025800, PtAEH1, Potri.011G028400, Potri.004G025900 | |
| Potri.003G082300 (POPTR_0003 s08030) | PtAEH3 | AGP EXT Hybrid | 2/0/0/0 | 188 | Y* | Y | Dark and light-grown seedlings, young leaf | AtPRP1 | Potri.005G191900, Potri.016G025300, Potri.004G162500, PossibleHybrid2, Potri.015G147200 | |
| Potri.003G184500 | PtAEH4 | AGP EXT Hybrid | 1/1/1/0 | 177 | Y* | N | None | PtEXT22, PtEXT28, PtEXT27, Potri.001G042100, Potri.019G047600 |
a Protein identifiers of the version 2.0 are shown in the parenthesis. Italics indicates a protein that was identified only by a BLAST search
b The domains indicated by the Pfam number are: PF04554.11, Extensin_2 domain (Extensin-like region); PF14547.4, Hydrophob_seed domain (Hydrophobic seed protein); PF13855.4, LRR_8 domain (Leucine rich repeat); PF08263.10, LRRNT_2 domain (Leucine rich repeat N-terminal domain); PF07714.15, Pkinase_Tyr domain (Protein tyrosine kinase); PF00069.23, Pkinase domain (Protein kinase domain); PF02181.21, FH2 domain (Formin Homology 2 Domain); PF10409.7, PTEN_C2 domain (C2 domain of PTEN tumour-suppressor protein); PF06830.9, Root_cap domain (Root cap); PF00295.15, Glyco_hydro_28 domain (Glycoside hydrolase family 28); PF00112.21, Peptidase_C1 domain (Papain family cysteine protease); PF00396.16, Granulin domain (Granulin); PF08246.10, Inhibitor_I29 domain (Cathepsin propeptide inhibitor domain); PF01657.15, Stress-antifung domain (Salt stress response/antifungal); PF07714.15, Pkinase_Tyr domain (Protein tyrosine kinase)
c Asterisk indicates a protein that is predicted to have a signal peptide either using the sensitive mode in the SignalP website or only if amino acids at the N terminus are discarded
d Expression data are shown only when available at http://bar.utoronto.ca/efppop/cgi-bin/efpWeb.cgi
e A locus ID indicates that it is not identified as an HRGP
Fig. 3.
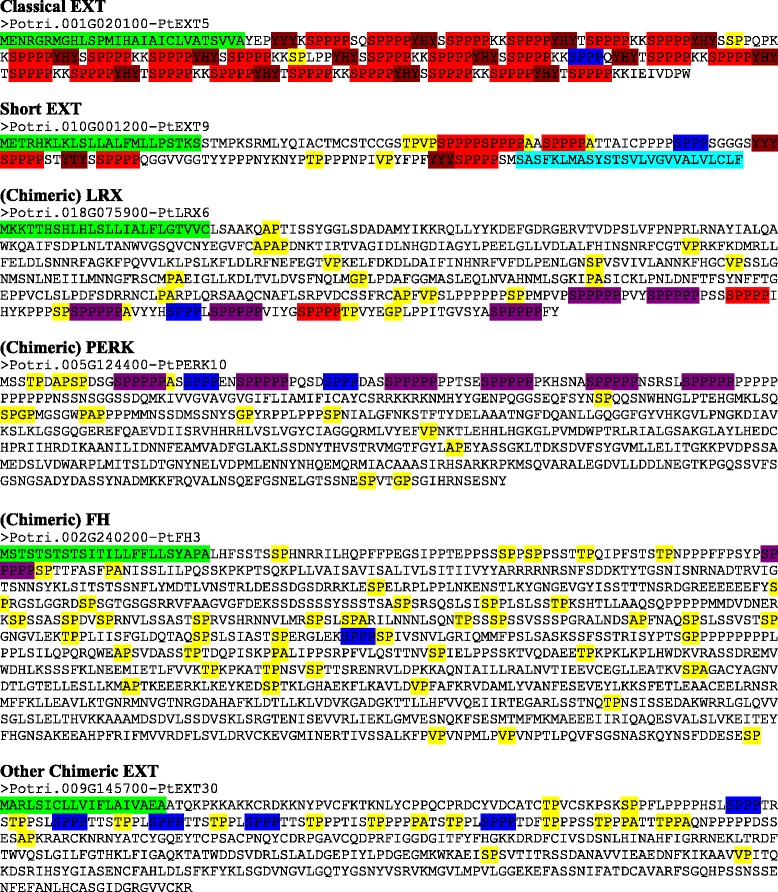
Protein sequences encoded by the representative EXT gene classes in Populus trichocarpa. The colored sequences at the N and C terminus indicate predicted signal peptides (green) and GPI anchor addition sequences (light blue) if present in the sequences. The SP3 (blue), SP4 (red), SP5 (purple), and YXY (dark red) repeats are also indicated in the sequences. The sequences typical of AGPs, specifically AP, PA, SP, TP, VP, and GP repeats, are also indicated (yellow)
In addition to the presence of SPPP and SPPPP repeats, the presence of a signal peptide was another factor in determining if a protein was considered an EXT. As with the AGPs, all the potential EXTs identified by the search were examined for signal peptides and GPI anchors. Signal peptides are known to occur in EXTs, but certain chimeric EXTs, notably the PERKs, lack a signal peptide [34]. In total, 46 of the 60 EXTs (77 %) identified have a signal peptide. Only four EXTs with GPI anchor addition sequences were identified, all of which were classified as short EXTs. This novel class of short EXTs with GPI anchor addition sequences was also observed in Arabidopsis [16].
Because EXTs were identified by searching for proteins with at least two SPPP sequences, many proteins were identified that contain only a few SPPP or SPPPP repeats among a much larger protein sequence. Many of these potential chimeric EXTs are not included in Table 3, but the sequences are available in Additional file 3: Figure S3 for further review. These may in fact be chimeric EXTs, but many lack a signal peptide and have only a few SPPP or SPPPP repeats among a much larger protein that does not belong to a class of previously characterized chimeric EXTs, such as PERKs, LRXs, or FHs.
Proline-rich Proteins (PRPs)
PRPs were identified by searching for proteins that contain at least 45 % PVKCYT or contain two or more repeated motifs (PPVX[KT] or KKPCPP) (Table 1). Although this search generates a large number of false positives and proteins identified as AGPs and EXTs by other searches as described above, it was effective in the identification of PRPs in Arabidopsis [16]. Of the 240 poplar proteins meeting the 45 % PVKCYT criteria, 20 of the proteins were determined to be PRPs based on sequence analysis, the presence of a signal peptide, and BLAST analysis. The PPVX[KT] motif search returned 29 candidate proteins of which four were determined to be PRPs, while the other motif (KKPCPP) search returned no candidate protein despite its effectiveness in Arabidopsis (Table 4 and Additional file 4: Figure S4). Additional proteins were identified by BLAST searches that fall below the 45 % threshold. Some of these proteins were also determined to be PRPs based on a spectrum of information, including the presence of a signal peptide and Pfam domains, the number of motif repeats, and BLAST hits against Arabidopsis HRGPs. BLAST searches against the Arabidopsis database were particularly beneficial in determining if a protein was a PRP. In total, 49 proteins were determined as PRPs, including 16 PRPs, 30 PR-peptides, and three chimeric PRPs (Fig. 4 and Additional file 4: Figure S4). Indeed, each of the 49 putative PRPs identified here is similar to at least one PRP previously identified in Arabidopsis [16].
Table 4.
Identification and analysis of PRP genes in Populus trichocarpa
| Locus Identifier 3.0 (ID 2.0)a | Name | Class | % PVKCYT | PPV/PPLP/PELPK Repeats | Amino Acids | Pfamb | SPc | GPI | Organ/issue - Specific Expressiond | Arabidopsis HRGP BLAST Hits | Poplar HRGP BLAST Hitse |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Potri.004G168600 (POPTR 0004 s17590) | PtPRP1 | PRP | 64 % | 24/8/0 | 554 | PF01190.15 | Y | N | Dark etiolated seedlings | AtPRP2, AtPRP1, AtPRP11 | PtPRP6, PtPRP32, PtPRP33, PtPRP143, Potri.016G006200 |
| Potri.016G015500 (POPTR_0016s01720) | PtPRP2 | PRP | 70 % | 13/0/0 | 449 | PF14547.4 | Y | N | Dark and +3 h light etiolated seedlings | AtPRP18, AtPEX4 | Potri.012G076700, Potri.015G071500, Potri.019G083900, Potri.T155100, Potri.005G239100 |
| Potri.014G126200 (POPTR 0014 s12100) | PtPRP3 | PRP | 51 % | 0/0/0 | 372 | PF01190.15 | Y | N | AtPRP9, AtPRP10 | PtPRP24, PtPRP22, PtPRP28, PtPRP26, PtPRP21 | |
| Potri.014G126500 (POPTR_0014s12120) | PtPRP4 | PRP | 52 % | 0/0/0 | 366 | PF01190.15 | Y | N | AtPRP7, AtPRP3, AtPRP1, AtAGP30I, AtAGP31I | PtPRP35, PtPRP3, PtPRP4, Potri.014G126300, PtPRP39 | |
| Potri.018G126000 (POPTR 0018 s12630) | PtPRP5 | PRP | 62 % | 15/9/0 | 310 | PF14547.4 | Y* | N | AtPRP9, AtPRP10, AtPERK15 | PtPRP44, PtPRP42, PtPRP41, PtPRP43, Potri.011G060200 | |
| Potri.009G129900 (POPTR 0009 s13250) | PtPRP6 | PRP | 48 % | 2/1/0 | 283 | PF01190.15 | Y* | N | AtPRP9, AtPRP10, AtPRP1 | Potri.019G082700, PtPRP21, PtPRP26, PtPRP18, PtPRP28 | |
| Potri.003G111300 (POPTR 0003 s11060) | PtPRP7 | PRP | 46 % | 4/1/0 | 234 | PF14547.4 | Y* | N | Male catkins | AtPRP9, AtPRP10, AtPRP15 | PtPRP27, PtPRP30, PtPRP21, PtPRP26, PtPRP22 |
| Potri.006G008300 | PtPRP8 | PRP | 59 % | 8/0/0 | 234 | PF14547.4 | Y | N | AtPRP9, AtPRP10 | PtPRP49, PtPRP26, PtPRP22, PtPRP23, PtPRP24 | |
| Potri.T162800 (POPTR 0006 s01030) | PtPRP9 | PRP | 50 % | 2/0/0 | 216 | PF14547.4 | Y | N | AtPRP9, AtPRP10 | PtPRP48, PtPRP26, PtPRP22, PtPRP28, PtPRP23 | |
| Potri.006G008600 | PtPRP10 | PRP | 53 % | 4/0/0 | 214 | PF14547.4 | Y | N | Young leaf | AtPRP16, AtPRP14, AtPRP17, AtPRP15, AtHAE4 | PtPRP15, PtPRP13, PtPRP5, PtPRP11, Potri.018G025900 |
| Potri. 002G201800 (POPTR 0002 s20290) | PtPRP34 | PRP | 37 % | 0/0/0 | 213 | PF01190.15 | Y | N | Young leaf, male catkins | AtPRP9, AtPRP10 | PtPRP22, PtPRP23, PtPRP26, PtPRP24, PtPRP29 |
| Potri. 017G145800 (POPTR 0017 s01230) | PtPRP35 | PRP | 42 % | 0/0/0 | 272 | PF01190.15 | Y | N | AtPRP9, AtPRP10 | PtPRP22, PtPRP26, PtPRP21, PtPRP23, PtPRP24 | |
| Potri. 001G060500 (POPTR_0001s13450) | PtPRP38 | PRP | 39 % | 0/7/0 | 332 | PF01190.15 | Y | N | Dark and +3 h light etiolated seedlings | AtPRP11, AtAGP31I, AtPRP1 | PtPRP33, PtPRP36, Potri.001G326200, Potri.017G068400, PtPRP38 |
| Potri. 003G167100 (POPTR_0003s16550) | PtPRP40 | PRP | 39 % | 0/2/0 | 299 | PF01190.15 | Y | N | Female catkins | AtPRP7, AtPRP1, AtPRP3, AtAGP30I, AtAGP31I | PtPRP34, PtPRP4, PtPRP3, Potri.014G126300, PtPRP39 |
| Potri.007G114400 | PtPRP44 | PRP | 43 % | 0/1/10 | 275 | Y | N | Roots | AtPRP7, AtPRP3, AtPRP1, AtAGP30I, AtAGP31I | PtPRP34, PtPRP35, PtPRP4, PtPRP3, Potri.014G126300 | |
| Potri. 013 G111600 (POPTR 0013 s11600) | PtPRP46 | PRP | 39 % | 0/4/0 | 216 | Y | N | AtPRP9, AtPRP10, AtPERK5 | PtPRP45, PtPRP44, PtPRP42, PtPRP43, PtPRP28 | ||
| Potri.006G065500 (POPTR 0006 s06430) | PtPRP11 | PR Peptide | 56 % | 5/2/0 | 198 | PF14547.4 | Y | N | Dark and +3 h light etiolated seedlings | AtPRP7, AtPRP3, AtPRP1, AtAGP30I, AtPRP9 | PtPRP4, PossiblePtPRP6, Potri.002G201700, PtPRP34, PtPRP35 |
| Potri.001G350600 (POPTR_0001s34750) | PtPRP12 | PR Peptide | 63 % | 6/0/0 | 191 | PF02704.12 | Y | N | AtPRP7, AtPRP3, AtPRP1, AtPRP9, AtAGP30I | PtPRP3, PossiblePtPRP6, Potri.002G201700, PtPRP34, PtPRP35 | |
| Potri.T162900 (POPTR_0006s01020) | PtPRP13 | PR Peptide | 52 % | 4/0/0 | 184 | PF14547.4 | Y | N | Young leaf | AtPRP15, AtPRP14, AtPRP17, AtPRP2, AtPRP1 | PtPRP11, PtPRP7, PtPRP13, PtPRP15, PtPRP8 |
| Potri.010G072200 (POPTR 0010 s08290) | PtPRP14 | PR Peptide | 50 % | 6/0/0 | 179 | PF02095.13 | Y | N | Mature leaf | AtPRP2, AtPRP4, AtPRP11 | PtPRP1.8, PtPRP32, PtPRP33, PtPRP36, Potri.005G041400 |
| Potri.006G008500 | PtPRP15 | PR Peptide | 53 % | 4/0/0 | 179 | PF14547.4 | Y | N | Roots | AtPRP14, AtPRP15, AtPRP16, AtPRP17 | PtPRP11, PtPRP5, PtPRP2, PtPRP13, PtPRP15 |
| Potri.007G113900 (POPTR_0007s03420) | PtPRP16 | PR Peptide | 47 % | 0/4/0 | 130 | Y | N | AtPRP16, AtPRP17, AtPRP15, AtPRP14, AtHAE4 | PtPRP15, PtPRP13, PtPRP9, PtPRP2, PtPRP11 | ||
| Potri.007G114100 (POPTR_0007s03400) | PtPRP17 | PR Peptide | 46 % | 0/3/0 | 119 | Y | N | AtPRP16, AtPRP17, AtPRP14, AtPRP15, AtHAE4 | PtPRP10, PtPRP13, PtPRP8, PtPRP2, PtPRP11 | ||
| Potri.007G113700 (POPTR_0007s03440) | PtPRP18 | PR Peptide | 47 % | 0/4/0 | 119 | Y | N | AtPRP16, AtPRP17, AtPRP14, AtPRP15, AtAGP30I | PtPRP9, PtPRP13, PtPRP8, PtPRP2, PtPRP15 | ||
| Potri.017G047400 (POPTR_0017s07470) | PtPRP19 | PR Peptide | 46 % | 0/3/0 | 113 | Y | N | Dark etiolated seedlings, light-grown seedling | AtPRP15, AtPRP14, AtPRP17, AtPRP2 | PtPRP5, PtPRP7, PtPRP13, PtPRP15, PtPRP8 | |
| Potri.019G082600 (POPTR_0019s11220) | PtPRP20 | PR Peptide | 45 % | 0/4/0 | 112 | Y | N | light-grown seedling | AtPRP16, AtPRP17, AtPRP14, AtPRP15, AtHAE4, | PtPRP15, PtPRP8, PtPRP10, PtPRP9, PtPRP11 | |
| Potri.017G047200 (POPTR_0017s07450) | PtPRP21 | PR Peptide | 43 % | 0/3/0 | 130 | Y | N | Young leaf, male catkins | AtPRP1, AtPRP2, AtPEX4 | Potri.004G110100, Potri.010G211100, Potri.004G109000, Potri.T018900, Potri.004G109900 | |
| Potri.017G045800 (POPTR_0017 s07310) | PtPRP22 | PR Peptide | 43 % | 0/3/0 | 116 | Y | N | AtPRP16, AtPRP17, AtPRP14, AtPRP15, AtHAE4, AtPERK5 | PtPRP13, PtPRP10, PtPRP2, PtPRP9, PtPRP11 | ||
| Potri.017G046700 (POPTR 0017 s07400) | PtPRP23 | PR Peptide | 40 % | 0/3/0 | 116 | Y | N | AtPRP9, AtPRP10, AtPRP15 | PtPRP21, PtPRP26, PtPRP31, Potri.017G046800, PtPRP27 | ||
| Potri.017G046400 (POPTR 0017 s07370) | PtPRP24 | PR Peptide | 43 % | 0/3/0 | 116 | Y | N | Roots | AtPRP9, AtPRP10 | PtPRP21, PtPRP30, PtPRP27, Potri.017G046800, PtPRP18 | |
| Potri.017G045900 (POPTR 0017 s07320) | PtPRP25 | PR Peptide | 43 % | 0/3/0 | 116 | Y | N | AtPRP9, AtPRP10, AtPRP15 | PtPRP19, PtPRP21, PtPRP27, PtPRP30, Potri.017G046800 | ||
| Potri.017G047000 (POPTR_0017 s07430) | PtPRP26 | PR Peptide | 42 % | 0/3/0 | 116 | Y | N | AtPRP9, AtPRP10 | PtPRP18, PtPRP21, Potri.017G046800, PtPRP27, PtPRP30 | ||
| Potri.017G047100 | PtPRP27 | PR Peptide | 44 % | 0/4/0 | 134 | Y | N | Female catkins | AtPRP9, AtPRP10, AtPRP15 | PtPRP21, PtPRP18, PtPRP26, PtPRP37, PtPRP19 | |
| Potri.017G045600 (POPTR 0017 s07290) | PtPRP28 | PR Peptide | 44 % | 0/3/0 | 126 | Y | N | Roots | AtPRP9, AtPRP10 | PtPRP30, Potri.017G046800, PtPRP27, PtPRP18, PtPRP17 | |
| Potri.017G046100 (POPTR 0017 s07340) | PtPRP29 | PR Peptide | 42 % | 0/3/0 | 116 | Y | N | AtPRP9, AtPRP10 | PtPRP26, PtPRP25, PtPRP24, PtPRP23, PtPRP29 | ||
| Potri.T178800 (POPTR 2000 s00200) | PtPRP30 | PR Peptide | 42 % | 0/4/0 | 135 | Y | N | Xylem | AtPRP9, AtPRP10 | PtPRP22, PtPRP23, PtPRP26, PtPRP21, PtPRP28 | |
| Potri.007G114200 (POPTR 0007 s03390) | PtPRP31 | PR Peptide | 44 % | 0/4/0 | 121 | Y | N | AtPRP9, AtPRP10 | PtPRP22, PtPRP26, PtPRP21, PtPRP23, PtPRP28 | ||
| Potri. 017G045000 | PtPRP37 | PR Peptide | 40 % | 0/3/0 | 105 | Y | N | Roots | AtPRP9, AtPRP10, AtPRP15 | PtPRP16, PtPRP21, PtPRP26, Potri.017G046800, PtPRP27 | |
| Potri. 002G201900 (POPTR_0002s20300) | PtPRP39 | PR Peptide | 33 % | 0/0/0 | 179 | PF01190.15 | Y | N | AtPRP11, AtAGP31I, AtPRP1 | PtPRP32, PtPRP36, Potri.001G326200, Potri.017G068400, PtPRP38 | |
| Potri. 017G044800 (POPTR_0017s07230) | PtPRP41 | PR Peptide | 34 % | 0/1/3 | 112 | Y | N | Young leaf, male catkins | AtPRP11, AtPRP1, AtAGP31I, AtPRP2 | PtPRP32, Potri .001G326200, Potri.017G068400, PtPRP38, PtPRP40 | |
| Potri. 017G044900 | PtPRP42 | PR Peptide | 39 % | 0/0/5 | 109 | Y | N | AtPRP9, AtPRP10 | PtPRP26, PtPRP21, PtPRP22, PtPRP28, PtPRP23 | ||
| Potri. 018G146200 | PtPRP43 | PR Peptide | 42 % | 0/1/2 | 114 | Y | N | Young leaf | AtPRP9 | PtPRP40, Potri.017G068400, Potri.001G326200, PtPRP32, PtPRP33 | |
| Potri.007G114700 (P0PTR_0007s03340) | PtPRP45 | PR Peptide | 38 % | 0/0/4 | 107 | Y | N | AtPRP11 | PtPRP38, Potri.017G068400, Potri.001G326200, PtPRP33, PtPRP32 | ||
| Potri. 017G046800 (POPTR 0017 s07440) | PtPRP47 | PR Peptide | 41 % | 0/5/0 | 174 | Y* | N | AtPRP9, AtPRP10, AtPEX2 | PtPRP45, PtPRP44, PtPRP43, PtPRP41, PtPRP18 | ||
| Potri. 017G045700 (POPTR 0017 s07300) | PtPRP48 | PR Peptide | 38 % | 0/2/0 | 97 | Y | N | AtPRP9, AtPRP10 | PtPRP44, PtPRP45, PtPRP42, PtPRP41, PtPRP37 | ||
| Potri. 017G046500 (POPTR 0017 s07380) | PtPRP49 | PR Peptide | 38 % | 0/3/0 | 97 | Y* | N | AtPRP10, AtPRP9, AtPEX2 | PtPRP45, PtPRP43, PtPRP42, PtPRP41, Potri.017G052100 | ||
| Potri. 004G114300 (POPTR 0004 s11300) | PtPRP32I | Chimeric | 41 % | 2/5/0 | 319 | PF01190.15 | Y | N | AtPRP9, AtPRP10 | PtPRP22, PtPRP21, PtPRP23, PtPRP28, PtPRP24 | |
| Potri. 004G114400 | PtPRP33I | Chimeric | 41 % | 0/6/0 | 365 | PF01190.15 | Y | N | AtPRP9, AtPRP10 | PtPRP30, Potri.017G046800, PtPRP21, PtPRP17, PtPRP18 | |
| Potri. 017G100600 (POPTR_0017s13490) | PtPRP36I | Chimeric | 43 % | 0/5/0 | 410 | PF01190.15 | Y | N | AtPRP9, AtPRP10 | PtPRP27, PtPRP21, Potri.017G046800, PtPRP17, PtPRP18 |
a Protein identifiers of the version 2.0 are shown in the parenthesis. Italics indicates a protein that was identified only by a BLAST search
b The domains indicated by the Pfam number are: PF01190.15, Pollen_Ole_e_I domain (Pollen proteins Ole e I like); PF14547.4, Hydrophob_seed domain (Hydrophobic seed protein); PF02704.12, GASA domain (Gibberellin regulated protein); PF02095.13, Extensin_1 domain (Extensin-like protein repeat)
c Asterisk indicates a protein that is predicted to have a signal peptide either using the sensitive mode in the SignalP website or only if amino acids at the N terminus are discarded
d Expression data are shown only when available at http://bar.utoronto.ca/efppop/cgi-bin/efpWeb.cgi
e A locus ID indicates that it is not identified as an HRGP
Fig. 4.

Protein sequences encoded by the representative PRP gene classes in Populus trichocarpa. The colored sequences at the N terminus indicate predicted signal peptides (green). PPV (pink) repeats typical of PRPs are indicated. The sequences typical of AGPs, specifically AP, PA, SP, TP, VP, and GP repeats, are also indicated (yellow) if present
Interestingly, 30 short PRPs were identified in poplar, most of which contain a single SPPP repeat at the C-terminus. Nearly all of the 30 proteins show similarity to AtPRP9 and AtPRP10 based on BLAST searches. These novel 30 proteins were grouped into a new class known as the proline-rich peptides (PR peptides) due to their much shorter amino acid length compared to the typical PRPs identified. These PR peptides can be further subdivided based on the presence of two pentapeptide repeat sequences, PPLP and PELPK. The PPLP repeat is present in 23 of these PR peptides and in a few other PRPs and chimeric PRPs, while the PELPK repeat is found only in one PRP and four PR peptides including two that contain PPLP repeats. It is also interesting to note that the 23 genes encoding the PPLP-containing PR peptides are clustered on chromosome 17, while the genes encoding only the PELPK-containing PR peptides are clustered on chromosome 7. All of the 49 PRPs had a predicted signal peptide, while none had a GPI anchor predicted.
Discussion
A Bioinformatics Approach for Identifying HRGPs
As more plant genome sequencing projects are completed, vast amounts of biological data are being generated. Bioinformatics and in particular the BIO OHIO 2.0 program, which was recently revised and improved to provide a more rapid, reliable, and efficient method to identify proteins with biased amino acid compositions and known repetitive motifs [16, 22]. For instance, the BIO OHIO/Prot-Class program can search through over 73,000 proteins in the poplar proteomic database and identify those containing at least 50 % PAST in one minute. Using the various search criteria, we have predicted 271 HRGPs in poplar, including 162 AGPs, 60 EXTs, and 49 PRPs.
Although HRGPs were identified primarily through searching for biased amino acid compositions and repetitive motifs, the possibility that other HRGPs could be found in the poplar genome exists. Not all AGPs meet the 50 % PAST threshold, for instance, one classical AGP, PtAGP51C, contains only 49 % PAST. Similar problems exist for identifying chimeric AGPs. Because these proteins may contain only a small AGP region within a much larger sequence, they are likely to contain less than 50 % PAST. The possibility remains that other classes of chimeric AGPs or individual proteins that contain AGP-like regions exist and were not identified by the search parameters used in this study. A similar problem could exist for AG peptides that fall below the 35 % PAST cut-off or for PRPs that fall below 45 % PVKCYT.
One possible solution is to simply lower the thresholds and continue to search, but the number of false positives increases markedly as thresholds are lowered, making such searches less feasible. For instance, lowering the threshold for the AG peptide search to 30 % would identify 877 proteins compared to the 194 identified with a 35 % threshold.
In such a scenario, BLAST provides an alternative means to find additional candidate proteins. When using identified proteins as queries, BLAST is effective in finding a few related family members. For example, when using identified FLAs as queries, BLAST is capable of finding additional FLAs that don’t meet the criteria of the BIO OHIO 2.0 program. However, it is not particularly effective in finding other members of HRGP superfamily and thus could not be utilized in a comprehensive manner.
Indeed, a bioinformatics search that identifies HRGPs, especially chimeric HRGPs without also identifying a very large number of false positives remains difficult. Nevertheless, the search parameters and BLAST searches used here provide an efficient means to identify HRGPs and distinguish them from a limited number of false positive sequences. Of course, future molecular and biochemical analysis of the HRGPs predicted from this study will be necessary to validate these predictions more completely and elucidate their biological functions. Only when such work is completed will it become possible to conclusively distinguish HRGPs from false positive sequences.
HRGPs exist as a spectrum of proteins
Although HRGPs are divided into AGPs, EXTs, and PRPs, the distinction between these categories is not always clear, since many HRGPs appear to exist as members of a spectrum of proteins rather than distinct categories. Indeed, several HRGPs identified here as well as some previously identified in Arabidopsis have characteristics of multiple families and can be considered hybrid HRGPs. For instance, many of the PRPs identified here, particularly some chimeric PRPs, also contain dipeptide repeats that are characteristic of AGPs. As such, it is difficult to determine if these should be considered as AGPs, PRPs, or classified as a hybrid HRGP. Determining whether these are actually AGPs or PRPs would depend on whether the proline residues are hydroxylated and subsequently glycosylated with arabinogalactan polysaccharides, which are characteristic of AGPs. Similarly, PtEXT4 also contains large numbers of characteristic AGP repeats (Additional file 2: Figure S2). In addition, BLAST searches revealed that it is similar in sequence to AtAGP51. Given that it contains many SPPP and SPPPP repeats, it was classified as an EXT. However, there is a possibility that this protein may also be glycosylated with the addition of AG polysaccharides, in which case it could potentially be grouped as a hybrid HRGP. Another example is the novel class identified here as the PR peptides (Table 4). Although grouped here as PRPs, these short sequences (i.e., PtPRP16-31 and PtPRP37) also contain a SPPP sequence characteristic of an EXT as well as the dipeptide repeats characteristic of AGPs, particularly AP, PA, and VP (Additional file 4: Figure S4).
Other difficulties arise when chimeric HRGPs are considered. For instance, the plastocyanins range from those that contain a majority of AGP repeats and easily pass the 50 % PAST test to those that contain only a few AP, PA, SP, VP, and GP repeats to those that contain no characteristic AGP repeats. The exact cutoff between proteins that are considered chimeric AGPs and those that are simply plastocyanin proteins is difficult to determine. Again, biochemical studies would be required to examine which of the proteins are actually glycosylated to make a final determination for classification. However, all those proteins annotated here as PAGs have at least a few characteristic AGP repeats, contain a signal peptide, and most have predicted GPI membrane anchor addition sequences, all of which is consistent with the chimeric AGP designation (Additional file 1: Figure S1).
A similar situation also exists for the chimeric EXTs, such as the PERKs and LRXs. How many SPPP or SPPPP repeats are required for a protein to be considered a LRX and not simply a leucine-rich repeat (LRR) protein? Here the cutoff was arbitrarily set to at least two repeats. As such, there may be LRR proteins that contain one SPPP that are not considered here as LRXs. Another example which illustrates this classification difficulty concerns the four proteins (PtAGP70I, PtAGP71I, PtAGP72I, and PtAGP73I) which are similar to AtPRP13 based on BLAST searches. However, these four proteins also contain numerous SP and AP repeats that would be more characteristic of an AGP. Exactly how proteins such as these should be classified is certainly debatable. Indeed it is human nature to group and classify items to facilitate understanding, while Mother Nature operates without such regard.
Comparisons with previously identified poplar HRGPs
This study identified 271 poplar HRGPs (162 AGPs, 60 EXT, and 49 PRPs) in contrast to the 24 HRGPs (3 AGPs, 10 EXT, and 11 PRPs) identified by Newman and Cooper [18]. The more stringent search criteria for proline-rich tandem repeats and a less comprehensive poplar proteomic database based on EST and NCBI Non-Redundant protein sequences data from10/04/09 likely account for the fewer poplar HRGPs identified in this earlier study. In addition, homologs of the 15 FLA AGPs reported by Lafarguette et al. [20] in a Populus tremula × P. alba hybrid related to Populus trichocarpa were also identified in addition to 35 other FLAs. Thus, the present study represents the most comprehensive and detailed picture of the HRGP inventory in poplar to date.
Comparisons with Arabidopsis
Findings here allow for a comparison of the HRGPs identified in Arabidopsis to those in poplar (Table 5). For AGPs, the classical AGPs identified in poplar showed a similar number as in Arabidopsis. Specifically, 27 classical AGPs including six lysine-rich AGPs were identified in poplar, while 25 classical AGPs including three lysine-rich AGPs were identified in Arabidopsis. Among other AGPs, particularly notable is the large increase the number of FLAs, PAGs, and AG peptides in poplar compared to Arabidopsis. While 21 FLAs, 17 PAGs and 16 AG peptides were identified in Arabidopsis, 50 FLAs, 39 PAGs and 35 AG peptides are identified here in poplar. There is also a noticeable increase in the number of other chimeric AGPs in poplar compared to Arabidopsis. Here, 11 other chimeric AGPs were identified in poplar, while only 6 were found in Arabidopsis.
Table 5.
Comparison of HRGPs identified in Populus trichocarpa and Arabidopsis thaliana
| HRGP family | HRGP subfamily | Poplar | Arabidopsisa |
|---|---|---|---|
| AGPs | Classical AGPs | 21 | 22 |
| Lysine-Rich Classical AGPs | 6 | 3 | |
| AG-Peptides | 35 | 16 | |
| (Chimeric) FLAs | 50 | 21 | |
| (Chimeric) PAGs | 39 | 17 | |
| Other Chimeric AGPs | 11 | 6 | |
| All AGP subfamilies | 162 | 85 | |
| EXTs | Classical EXTs | 8 | 20 |
| Short EXTs | 22 | 12 | |
| (Chimeric) LRXs | 10 | 11 | |
| (Chimeric) FHs | 5 | 6 | |
| (Chimeric) PERKs | 12 | 13 | |
| Other Chimeric EXTs | 3 | 3 | |
| All EXT subfamilies | 60 | 59 | |
| PRPs | PRPs | 16 | 11 |
| PR Peptides | 30 | 1 | |
| Chimeric PRPs | 3 | 6 | |
| All PRP subfamilies | 49 | 18 | |
| Total | 271 | 168 |
a The Arabidopsis HRGP data shown here are from Showalter et al. [16] with the exceptions that 6 chimeric FH EXTs were added and that one PR-peptide was found out of originally identified 12 PRPs as part of this study
Among EXTs, the classical EXTs with large numbers of SPPPP repeats are markedly decreased in poplar, while similar numbers of the chimeric EXTs exist in both species. The reduction in the number of classical EXTs in poplar is dramatic and likely indicates that many EXT genes or EXT functions are dispensable in poplar, and therefore not conserved in evolution. A similar loss of EXTs has also been observed in analysis of certain monocot species [unpublished data,18]. Moreover, far fewer poplar EXTs contain putative cross-linking YXY sequences compared to Arabidopsis, and this can be largely explained by the reduced number of classic EXT sequences, which typically contain such cross linking sequences. The various chimeric EXTs, namely the LRXs/PEXs, PERKs, and FHs, are conserved in both species. Although FHs were not reported in Showalter et al. [16], a reexamination of the Arabidopsis proteome shows 6 FH sequences (AtFH1-At3g2550, AtFH5-At5g54650, AtFH8-At1g70140, AtFH13-At5g58160, AtFH16-At5g07770, and AtFH20-At5g07740) contain two or more SPPP sequences. These 6 formins are included in Table 5 and are a subset of the 21 reported formins in Arabidopsis [35]. Similar to the chimeric EXTs, the short EXTs are also conserved in Arabidopsis and poplar. The short EXTs are a particularly interesting class because EXTs are not known to have GPI membrane anchors, a feature commonly found in many AGPs and associated with proteins found in lipid rafts [36]. The finding that several of these short EXTs encode a predicted GPI-anchor sequence are conserved in poplar and Arabidopsis certainly prompts the question of what role these proteins are playing in the plant. Currently, no publications verifying their biochemical existence or examining their roles exist, but this class stands out in terms of having interesting candidates for further investigation, particularly with respect to confirming their plasma membrane localization, hydroxylation, and glycosylation.
PRPs are similar in both species with the notable exception of the PR-peptides, which is a much expanded class in poplar compared to Arabidopsis, which is now recognized to have only one PR-peptide following a reexamination prompted by this study. All of the PR-peptides in poplar are similar in sequence with most containing LPPLP repeats and having a single SPPP repeat at the C terminus, although some contained PELPK repeats. In addition, most of these PR-peptides are similar to AtPRP9 and AtPRP10 based on BLAST analysis; both of these Arabidopsis proteins contain PELPK repeats as well. Indeed, AtPRP9 is quite short and similar in sequence to the PR peptides found in poplar but lacks the C terminal SPPP repeat. However, this is the only such protein found in Arabidopsis, while 30 were observed in poplar. AtPRP10 contains some similarity in sequence but is much longer than the poplar PR-peptides. Indeed, the large number of LPPLP- and PELPK- containing PR-peptides in poplar clustered respectively in two chromosomal locations indicates that these two gene subfamilies likely result from tandem gene duplication events, analogous to a unique, clustered set of PEHK-containing PRP genes in the grape family [18].
Although most sub-families of HRGPs exist in both the Arabidopsis and poplar inventories, certain species-specific differences do exist, which is reflected in the difference of number of certain groups and the total number of HRGPs (271 in poplar versus 168 in Arabidopsis). Precisely why certain classes of HRGPs are increased or decreased in abundance in a particular species remains to be determined, but these results lay the groundwork for future experimentation in this area.
Poplar HRGPs genome 2.0 release and expression analysis
The study revealed that the poplar genome 3.0 release is quite different from 2.0 release in terms of HRGPs. Only 33 % of HRGPs identified in 3.0 are the same as counterparts in 2.0, others may differ from a few amino acids in sequence to a distinct start and/or stop position. For several such cases, a green highlight indicated a likely signal sequence placed internally, either because these signal sequences were at the N terminus in the 2.0 release or they should be at N terminus based on analysis of sequences in this study.
In addition, tissue/organ-specific HRGP expression data were obtained from the poplar eFP browser. However, this database does not contain all HRGP data, and it only accepts query IDs in poplar genome version 2.0 format. Judging from the available information, one could observe that HRGPs in general have high expression in seedlings, leaves, and reproductive tissues (Tables 2, 3, and 4). In particular, a number of FLAs were specifically expressed in xylem, while some PAGs were found to be highly expressed in male catkins. Many PRPs have high expression in seedlings and leaves. Interestingly, several LRXs are found to be uniquely expressed in male catkins; this finding is consistent with previous research in Arabidopsis and rice that a group of LRXs are pollen-specific LRXs, or PEXs [37].
Pfam analysis of poplar HRGPs
All 271 poplar HRGPs identified in this study were subjected to Pfam analysis to identify specific domains within them. Pfam domains were found in 160 of the 271 proteins (59 %). More specifically, Pfam domains were identified in 105 of the 162 AGPs, 32 of the 62 EXTs, and 23 of the 49 PRPs. In particular, Pfam analysis exceled at finding domains within chimeric HRGPs, such as FLAs, PAGs, LRXs, PERKs, and FH EXTs. In contrast, such analysis often failed to find domains in classical AGPs or EXTs, possibly due to the variable sequences and numbers of sequence repeats associated with many of the HRGPs. Interestingly, many of the PRPs were found to contain Pollen Ole domains and Hydrophob seed domains. Pfam analysis also has merit in identifying domains in the chimeric HRGPs identified in the study. Indeed, while Pfam analysis alone is not sufficient for identifying HRGPs in a comprehensive manner, it can add valuable information to identified HRGPs, and thus a Pfam analysis module will likely be incorporated into future versions of the BIO OHIO program.
Conclusions
The new and improved BIO OHIO 2.0 bioinformatics program was used to identify and classify the current inventory of HRGPs in poplar. This information will allow researchers to determine the structure and function of individual HRGPs and to explore potential industrial applications of these proteins in such areas as plant biofuel production, food additives, lubricants, and medicine. Other plant proteomes/genomes can also be examined with the program to provide their respective HRGP inventories and facilitate comparative evolutionary analysis of the HRGP family in the plant kingdom [16, 38]. Finally, while this program was specifically developed for HRGP identification, it can also be used to examine other plant or non-plant genomes/proteomes in order to identify proteins or protein families with any particular amino acid bias and/or amino acid sequence motif, making it useful throughout the tree domains and six kingdoms of life.
Acknowledgments
The authors thank Carol Morris Showalter for reading this manuscript and providing valuable comments and suggestions.
Funding
No funding was obtained for this study.
Availability of data and materials
All relevant data are within the paper and its Additional files 1, 2, 3 and 4.
Authors’ contributions
Conceived and designed the experiments: AMS BDK XL. Performed the experiments: BDK XL. Analyzed the data: AMS BDK XL. Contributed reagents/materials/analysis tools: JL LW. Wrote the paper: AMS. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- AGPs
Arabinogalactan-proteins
- EXTs
Extensins
- FHs
Formin homology proteins
- FLAs
Fasciclin-like AGPs
- GPI
Glycosylphosphatidylinositol
- HRGPs
Hydroxyproline-rich glycoproteins
- LRXs
Leucine-rich repeat extensins
- PAGs
Plastocyanin AGPs
- PERKs
Proline-rich extensin-like receptor protein kinases
- PRPs
Proline-rich proteins
Additional files
Protein sequences encoded by the predicted AGP genes in Populus trichocarpa. The colored sequences at the N and C terminus indicate predicted signal peptides (green) and GPI anchor addition sequences (light blue) if present in the sequences. AP, PA, SP, TP, VP, and GP repeats (yellow) and lysine-rich regions (olive) are also indicated. Additionally, EXT SP3 (blue), SP4 (red), SP5 (purple) repeats and sequences typical of PRPs, PPV repeats, are indicated (pink) if present. Note that green font indicates a predicted signal peptide using the sensitive mode from the SignalP website. Internal green highlights indicate the presence of a predicted signal peptide only if amino acids at the N terminus are discarded. (PDF 69 kb)
Protein sequences encoded by the predicted EXT genes in Populus trichocarpa. The colored sequences at the N and C terminus indicate predicted signal peptides (green) and GPI anchor addition sequences (light blue) if present in the sequences. The SP3 (blue), SP4 (red), SP5 (purple), and YXY (dark red) repeats are also indicated in the sequences. The sequences typical of AGPs, specifically AP, PA, SP, TP, VP, and GP repeats, are also indicated (yellow) in the sequences. Note that green font indicates a predicted signal peptide using the sensitive mode from the SignalP website. Internal green highlights indicate the presence of a predicted signal peptide only if amino acids at the N terminus are discarded. (PDF 72 kb)
Protein sequences encoded by the potential chimeric EXT genes in Populus trichocarpa. The colored sequences at the N and C terminus indicate the predicted signal peptides (green) and GPI anchor addition sequences (light blue) if present in the sequences. The SP3 (blue), SP4 (red), SP5 (purple), and YXY (dark red) repeats are also indicated in the sequences. The sequences typical of AGPs, specifically AP, PA, SP, TP, VP, and GP repeats, are also indicated (yellow) in the sequences. (PDF 23 kb)
Protein sequences encoded by the predicted PRP genes in Populus trichocarpa. The colored sequences at the N terminus indicate the predicted signal peptides (green). PPV (pink) repeats typical of PRPs are indicated. Repetitive motifs PPLP (teal) and PELPK (dark yellow) are also indicated. Additionally, EXT SP3 (blue) repeats, YXY (dark red) and sequences typical of AGPs, specifically AP, PA, SP, TP, VP, and GP repeats, are indicated (yellow) if present. Note that green font indicates a predicted signal peptide using the sensitive mode from the SignalP website. Internal green highlights indicate the presence of a predicted signal peptide only if amino acids at the N terminus are discarded. (PDF 47 kb)
Contributor Information
Allan M. Showalter, Phone: 740-593-1135, Email: showalte@ohio.edu
Brian D. Keppler, Email: bdkeppler@wisc.edu
Xiao Liu, Email: xl224810@ohio.edu.
Jens Lichtenberg, Email: jl476805@ohio.edu.
Lonnie R. Welch, Email: welch@ohio.edu
References
- 1.Showalter AM. Structure and function of plant cell wall proteins. Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kieliszewski MJ, Lamport DTA. Extensin: Repetitive motifs, functional sites, posttranslational codes and phylogeny. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313X.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- 3.Nothnagel EA. Proteoglycans and related components in plant cells. Int Rev Cytol. 1997;174:195–291. doi: 10.1016/S0074-7696(08)62118-X. [DOI] [PubMed] [Google Scholar]
- 4.Cassab GI. Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:281–309. doi: 10.1146/annurev.arplant.49.1.281. [DOI] [PubMed] [Google Scholar]
- 5.Jose-Estanyol M, Puigdomenech P. Plant cell wall glycoproteins and their genes. Plant Physiol Biochem (Paris) 2000;38:97–108. doi: 10.1016/S0981-9428(00)00165-0. [DOI] [Google Scholar]
- 6.Seifert GJ, Roberts K. The biology of arabinogalactan proteins. Annu Rev Plant Biol. 2007;58:137–161. doi: 10.1146/annurev.arplant.58.032806.103801. [DOI] [PubMed] [Google Scholar]
- 7.Tan L, Leykam JF, Kieliszewski MJ. Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiol. 2003;132:1362–1369. doi: 10.1104/pp.103.021766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan L, Qiu F, Lamport DTA, Kieliszewski MJ. Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J Biol Chem. 2004;279:13156–13165. doi: 10.1074/jbc.M311864200. [DOI] [PubMed] [Google Scholar]
- 9.Tan L, Showalter AM, Egelund J, Hernandez-Sanchez A, Doblin MS, Bacic A. Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front Plant Sci. 2012;3:1–10. doi: 10.3389/fpls.2012.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shpak E, Barbar E, Leykam JF, Kieliszewski MJ. Contiguous Hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J Biol Chem. 2001;276:11272–11278. doi: 10.1074/jbc.M011323200. [DOI] [PubMed] [Google Scholar]
- 11.Youl JJ, Bacic A, Oxley D. Arabinogalactan-proteins from Nicotiana alata and Pyrus communis contain glycosylphosphatidylinositol membrane anchors. Proc Natl Acad Sci U S A. 1998;95:7921–7926. doi: 10.1073/pnas.95.14.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherrier DJ, Prime TA, Dupree P. Glycosylphosphatidylinositol-anchored cell surface proteins from Arabidopsis. Electrophoresis. 1999;20:2027–2035. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2027::AID-ELPS2027>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 13.Svetek J, Yadav MP, Nothnagel EA. Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J Biol Chem. 1999;274:14724–14733. doi: 10.1074/jbc.274.21.14724. [DOI] [PubMed] [Google Scholar]
- 14.Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A. Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol. 2002;129:1448–1463. doi: 10.1104/pp.003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham MA, Silverstein KAT, Cannon SB, VandenBosch KA. Computational identification and characterization of novel genes from legumes. Plant Physiol. 2004;135:1179–1197. doi: 10.1104/pp.104.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 2010;153:485–513. doi: 10.1104/pp.110.156554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma H, Zhao J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.) J Exp Bot. 2010;61:2647–2668. doi: 10.1093/jxb/erq104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman AM, Cooper JB. Global analysis of proline-rich tandem repeat proteins reveals broad phylogenetic diversity in plant secretomes. PLoS One. 2011;doi:10.1371/journal.pone.0023167 [DOI] [PMC free article] [PubMed]
- 19.Fleming MB, Decker SR, Bedinger PA. Investigating the role of extensin proteins in poplar biomass recalcitrance. BioResources. 2016;11:4727–4744. doi: 10.15376/biores.11.2.4727-4744. [DOI] [Google Scholar]
- 20.Lafarguette F, Leplé J-C, Déjardin A, Laurans F, Costa G, Lesage-Descauses M-C, et al. Poplar genes encoding fasciclin-like arabinogalactan proteins are highly expressed in tension wood. New Phytol. 2004;164:107–121. doi: 10.1111/j.1469-8137.2004.01175.x. [DOI] [PubMed] [Google Scholar]
- 21.Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 22.Lichtenberg J, Keppler BD, Conley T, Gu D, Burns P, Welch LR, et al. Prot-Class: a bioinformatics tool for protein classification based on amino acid signatures. Nat Sci. 2012;4:1161–1164. [Google Scholar]
- 23.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 24.Eisenhaber B, Wildpaner M, Schultz CJ, Borner GHH, Dupree P, Eisenhaber F. Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol. 2003;133:1691–1701. doi: 10.1104/pp.103.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fowler TJ, Bernhardt C, Tierney ML. Characterization and expression of four proline-rich cell wall protein genes in Arabidopsis encoding two distinct subsets of multiple domain proteins. Plant Physiol. 1999;121:1081–1091. doi: 10.1104/pp.121.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009;149:981–993. doi: 10.1104/pp.108.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schultz CJ, Ferguson KL, Lahnstein J, Bacic A. Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana. Endoplasmic reticulum and glycosylphosphatidylinositol-anchor signal cleavage sites and hydroxylation of proline. J Biol Chem. 2004;279:45503–45511. doi: 10.1074/jbc.M407594200. [DOI] [PubMed] [Google Scholar]
- 29.Brady JD, Sadler IH, Fry SC. Di-isodityrosine, a novel tetrameric derivative of tyrosine in plant cell wall proteins: a new potential cross-link. Biochem J. 1996;315:323–327. doi: 10.1042/bj3150323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnabelrauch LS, Kieliszewski MJ, Upham BL, Alizedeh H, Lamport DTA. Isolation of pI 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link site. Plant J. 1996;9:477–489. doi: 10.1046/j.1365-313X.1996.09040477.x. [DOI] [PubMed] [Google Scholar]
- 31.Brady JD, Sadler IH, Fry SC. Pulcherosine, an oxidatively coupled trimer of tyrosine in plant cell walls: Its role in cross-link formation. Phytochemistry. 1998;47:349–353. doi: 10.1016/S0031-9422(97)00592-X. [DOI] [PubMed] [Google Scholar]
- 32.Held MA, Tan L, Kamyab A, Hare M, Shpak E, Kieliszewski MJ. Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross linked in vitro. J Biol Chem. 2004;279:55474–55482. doi: 10.1074/jbc.M408396200. [DOI] [PubMed] [Google Scholar]
- 33.Cannon MC, Terneus K, Hall Q, Tan L, Wang Y, Wegenhart BL, et al. Self-assembly of the plant cell wall requires an extension scaffold. Proc Natl Acad Sci U S A. 2008;105:2226–2231. doi: 10.1073/pnas.0711980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakhamchik A, Zhao Z, Provart NJ, Shiu SH, Keatley SK, Cameron RK, et al. A comprehensive expression analysis of the Arabidopsis proline-rich extensin-like receptor kinase gene family using bioinformatic and experimental approaches. Plant Cell Physiol. 2004;45:1875–1881. doi: 10.1093/pcp/pch206. [DOI] [PubMed] [Google Scholar]
- 35.Cvrčková F, Grunt M, Žárský V. Expression of GFP-mTalin reveals an actin related role for the Arabidopsis Class II formin AtFH12. Biol Plant. 2012;56:431–440. doi: 10.1007/s10535-012-0071-9. [DOI] [Google Scholar]
- 36.Borner GHH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, MacAskill A, et al. Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 2005;137:104–116. doi: 10.1104/pp.104.053041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumberger N, Doesseger B, Guyot R, Diet A, Parsons RL, Clark MA, et al. Whole-genome comparison of leucine rich repeat extensins in Arabidopsis and rice: a conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 2003;131:1313–1326. doi: 10.1104/pp.102.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Wolfe R, Welch LR, Domozych DS, Popper ZA, Showalter AM. Bioinformatic identification and analysis of extensins in the plant kingdom. PLoS One. 2016;doi:10.1371/journal.pone.0150177 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Additional files 1, 2, 3 and 4.


