The receptor-like kinase IOS1 is critical for priming of Arabidopsis thaliana innate immunity and is required for optimal function of BAK1-dependent and BAK1-independent cell surface immune receptors.
Abstract
Plasma membrane-localized pattern recognition receptors (PRRs) such as FLAGELLIN SENSING2 (FLS2), EF-TU RECEPTOR (EFR), and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) recognize microbe-associated molecular patterns (MAMPs) to activate pattern-triggered immunity (PTI). A reverse genetics approach on genes responsive to the priming agent β-aminobutyric acid (BABA) revealed IMPAIRED OOMYCETE SUSCEPTIBILITY1 (IOS1) as a critical PTI player. Arabidopsis thaliana ios1 mutants were hypersusceptible to Pseudomonas syringae bacteria. Accordingly, ios1 mutants showed defective PTI responses, notably delayed upregulation of the PTI marker gene FLG22-INDUCED RECEPTOR-LIKE KINASE1, reduced callose deposition, and mitogen-activated protein kinase activation upon MAMP treatment. Moreover, Arabidopsis lines overexpressing IOS1 were more resistant to bacteria and showed a primed PTI response. In vitro pull-down, bimolecular fluorescence complementation, coimmunoprecipitation, and mass spectrometry analyses supported the existence of complexes between the membrane-localized IOS1 and BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 (BAK1)-dependent PRRs FLS2 and EFR, as well as with the BAK1-independent PRR CERK1. IOS1 also associated with BAK1 in a ligand-independent manner and positively regulated FLS2-BAK1 complex formation upon MAMP treatment. In addition, IOS1 was critical for chitin-mediated PTI. Finally, ios1 mutants were defective in BABA-induced resistance and priming. This work reveals IOS1 as a novel regulatory protein of FLS2-, EFR-, and CERK1-mediated signaling pathways that primes PTI activation.
INTRODUCTION
Plants possess multilayered recognition systems that detect pathogens at various stages of infection and proliferation. Recognition of microbial invasion is essentially based upon the host’s ability to distinguish between self and non-self components. Early microbial pathogens detection is performed by cell surface-localized pattern recognition receptors (PRRs) that sense pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs) (Monaghan and Zipfel, 2012). Major examples of MAMPs are lipopolysaccharides present in the envelope of Gram-negative bacteria, eubacterial flagellin, eubacterial elongation factor Tu (EF-Tu), peptidoglycans from Gram-positive bacteria, methylated bacterial DNA fragments, and fungal cell wall-derived chitins (Girardin et al., 2002; Cook et al., 2004; Boller and Felix, 2009). MAMP recognition promptly triggers the activation of pattern-triggered immunity (PTI) (Tsuda and Katagiri, 2010). Early PTI responses, such as calcium influx, production of reactive oxygen species (ROS), and activation of mitogen-activated protein (MAP) kinases, induce transcriptional reprogramming mediated by plant WRKY transcription factors as well as calmodulin binding proteins (Boller and Felix, 2009; Tena et al., 2011). In addition, Arabidopsis thaliana plants close stomata in a MAMP-dependent manner when in contact with bacteria (Melotto et al., 2006; Singh et al., 2012). Callose deposition and PTI marker gene upregulation are usually observed later (Zipfel and Robatzek, 2010). Activation of PTI leads to broad resistance to pathogens (Nicaise et al., 2009; Tsuda and Katagiri, 2010; Zeng et al., 2010; Desclos-Theveniau et al., 2012). Virulent bacterial pathogens inject proteins, some of which suppress PTI (Deslandes and Rivas, 2012; Feng and Zhou, 2012). Often, recognition of microbial effectors by plant intracellular nucleotide binding site and leucine-rich repeat proteins activates effector-triggered immunity (ETI). ETI is a rapid and robust response, usually associated with a hypersensitive reaction (Maekawa et al., 2011; Gassmann and Bhattacharjee, 2012).
In Arabidopsis, the most extensively studied PRRs are the leucine-rich repeat receptor-like kinases (LRR-RLKs) FLAGELLIN SENSING2 (FLS2) and EF-Tu receptor (EFR). FLS2 and EFR recognize bacterial flagellin (or the derived peptide flg22) and EF-Tu (or the derived peptides elf18/elf26), respectively (Gómez-Gómez and Boller, 2000; Zipfel et al., 2006). Upon ligand binding, FLS2 and EFR rapidly associate with another LRR-RLK, BRI1-ASSOCIATED RECEPTOR-LIKE KINASE/SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE3 (BAK1/SERK3), forming a ligand-inducible complex that triggers downstream PTI responses (Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011). In addition to associating with FLS2, BAK1 recognizes the C terminus of the FLS2-bound flg22, thus acting as a coreceptor (Sun et al., 2013). BAK1-LIKE1/SERK4 also cooperates with BAK1 to regulate PRR-mediated signaling (Roux et al., 2011). Interestingly, the BAK1-INTERACTING RECEPTOR KINASE2 (BIR2) prevents BAK1 interaction with FLS2 before elicitation. Importantly, BIR2 is released from BAK1 upon MAMP perception, allowing FLS2-BAK1 association and PTI activation (Halter et al., 2014). While BAK1 and other SERKs are the primary regulators downstream of FLS2 and EFR, the perception of the fungal MAMP chitin and signaling through CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) does not require BAK1 (Shan et al., 2008; Kemmerling et al., 2011; Ranf et al., 2011). Although CERK1 was considered as the major PRR for chitin (Miya et al., 2007; Wan et al., 2008, 2012), recent data suggest that the LYSIN MOTIF RECEPTOR KINASE5 (LYK5) is the primary receptor for chitin (Cao et al., 2014). Upon chitin elicitation, CERK1 and LYK5 form a complex to activate plant innate immunity (Cao et al., 2014). CERK1 is also involved in the recognition of peptidoglycans (Willmann et al., 2011). Other proteins downstream of PRRs modulate the PTI response. Typically, the receptor-like cytoplasmic kinase BOTRYTIS-INDUCED KINASE1 (BIK1) plays a critical role in mediating early flagellin signaling from the FLS2/BAK1 receptor complex and regulates responses induced by elf18, Pep1, and chitin and thus acts as a convergent point downstream of multiple PRRs (Lu et al., 2010a; Zhang et al., 2010). Other receptor-like cytoplasmic kinase, such as PTI COMPROMISED RECEPTOR-LIKE CYTOPLASMIC KINASE1 (PCRK1) and PCRK2, function downstream of multiple PRRs (Sreekanta et al., 2015; Kong et al., 2016). In addition, BRASSINOSTEROID-SIGNALING KINASE1 (BSK1) associates with unstimulated FLS2 (Shi et al., 2013). The DENN (Differentially Expressed in Normal and Neoplastic cells) domain protein STOMATAL CYTOKINESIS-DEFECTIVE1 (SCD1) is also necessary for some FLS2- and EFR-mediated responses and associates in a ligand-independent manner with FLS2 in vivo (Korasick et al., 2010). Furthermore, lectin receptor kinases (LecRKs) such as LecRK-VI.2 and LecRK-V.5 modulate early PTI signaling (Desclos-Theveniau et al., 2012; Singh et al., 2012; Singh and Zimmerli, 2013; Huang et al., 2014).
In addition to PTI and ETI, other resistance responses, such as systemic acquired resistance and induced systemic resistance, are activated after pathogen challenges (Durrant and Dong, 2004; Van Wees et al., 2008). Organic and inorganic compounds can also induce systemic resistance in plants. The non-protein amino acid β-aminobutyric acid (BABA) is a potent inducer of resistance against abiotic stress (Jakab et al., 2005; Zimmerli et al., 2008), nematodes (Oka et al., 1999), insects (Hodge et al., 2005), and microbial pathogens (Jakab et al., 2001; Zimmerli et al., 2001; Cohen, 2002; Ton and Mauch-Mani, 2004; Po-Wen et al., 2013). BABA-induced resistance is associated with a faster activation of defense mechanisms upon stress perception, a phenomenon known as priming (Conrath et al., 2006; Návarová et al., 2012). Although accumulation of defense signaling components before stress exposure (Beckers et al., 2009; Singh et al., 2012) and epigenetic modifications (Jaskiewicz et al., 2011; Luna et al., 2012; Rasmann et al., 2012; Slaughter et al., 2012; Po-Wen et al., 2013) are suggested to be critical for priming, the identity of signaling components involved in priming is still largely unknown.
In an effort to identify novel critical players in Arabidopsis immunity and priming, we used a reverse genetic approach by testing mutants of genes whose expression levels are induced by the priming agent BABA (Tsai et al., 2011). Three independent insertion lines in the malectin-like/LRR-RLK IMPAIRED OOMYCETE SUSCEPTIBILITY1 (IOS1) (Hok et al., 2011) were found to be hypersusceptible to bacterial pathogens. IOS1 is known to contribute to disease caused by filamentous (hemi)biotrophs and to attenuate abscisic acid (ABA) responses in Arabidopsis (Hok et al., 2011, 2014). Through loss- and gain-of-function analyses and biochemical approaches, we show that IOS1 is an important modulator of Arabidopsis PTI that associates with the LRR-RLKs FLS2, EFR, and BAK1 in a ligand-independent manner, notably controlling the complex formation between FLS2 and BAK1. IOS1 also associates with the LysM-domain RLK CERK1 and controls chitin-mediated PTI.
RESULTS
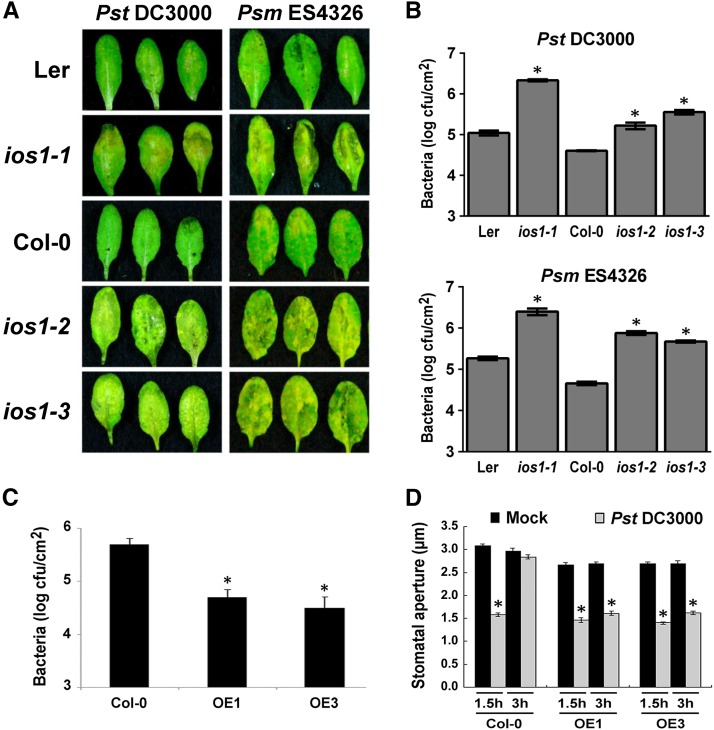
IOS1 Is Required for Resistance to Hemibiotrophic Bacteria
To identify Arabidopsis genes involved in immunity to bacteria, we followed a reverse genetic analysis of genes upregulated by the priming agent BABA (Tsai et al., 2011). One of these genes is the Malectin-like/LRR-RLK IOS1 (At1g51800) (Hok et al., 2011, 2014). For our analyses, we used ios1-1, a transcriptional knockout Ds transposon insertion line in Ler-0 background (GT_5_22250) recently isolated (Hok et al., 2011), and ios1-2 (Salk_137388) and ios1-3 (SAIL_343_B11), two independent T-DNA insertion lines in Col-0 background still producing truncated IOS1 transcripts (Supplemental Figure 1). To test whether IOS1 is required for antibacterial immunity, the three insertion lines were dip-inoculated with the virulent hemibiotrophic bacteria Pseudomonas syringae pv tomato DC3000 (Pst DC3000) or Pseudomonas syringae pv maculicola ES4326 (Psm ES4326). At 3 d postinoculation (dpi), ios1-1, ios1-2, and ios1-3 developed stronger symptoms than wild-type plants, as illustrated by increased chlorosis and necrosis formation (Figure 1A). This phenotype was associated with significantly higher bacterial titers (Figure 1B). Typically, the susceptibility phenotype of ios1-2 to Pst DC3000 is similar to the mutant bak1-5 (Supplemental Figure 2). We also evaluated the susceptibility of ios1 mutants to the Pst DC3000 hrcC- mutant, a strain defective in delivering type III effectors that cannot repress the PTI response, and consequently is mostly nonvirulent on Arabidopsis (Brooks et al., 2004). All three ios1 mutants tested allowed more growth of Pst DC3000 hrcC- than wild-type plants upon syringe infiltration (Supplemental Figure 3), suggesting a defective PTI response in ios1 mutants.
Figure 1.
A Critical Role for IOS1 in Arabidopsis Resistance to Hemibiotrophic Bacteria.
(A) Disease symptoms in ios1 mutants. Five-week-old Arabidopsis were dip-inoculated in a bacterial solution of 106 cfu/mL Pst DC3000 or 5 × 105 cfu/mL Psm ES4326. Symptoms were evaluated at 3 dpi. These experiments were repeated at least twice with similar results.
(B) Bacterial growth in ios1 mutants. Five-week-old Arabidopsis were dip-inoculated as in (A) and bacterial titers were evaluated at 2 dpi. Values are means ± se of two independent experiments each consisting of three plants (n = 6). Asterisks indicate a significant difference to the respective wild-type control based on a paired two-tailed t test (P < 0.01).
(C) Growth of Pst DC3000 in lines overexpressing IOS1. Bacterial titers in 5-week-old Col-0 and IOS1 overexpression lines OE1 and OE3 were determined at 3 dpi with 106 cfu/mL Pst DC3000. Values are means ± se of three independent biological replicates each with three plants (n = 9). Asterisks indicate a significant difference to Col-0 wild-type based on a paired two-tailed t test (*P < 0.01).
(D) Stomatal innate immunity in lines overexpressing IOS1. Stomatal apertures in leaf epidermal peels from 5-week-old Col-0 and IOS1 overexpression lines OE1 and OE3 were analyzed after 1.5 or 3 h exposure to MgSO4 (Mock) or 108 cfu/mL Pst DC3000. Values are shown as means ± se of three independent experiments each consisting of at least 60 stomata (n > 180). Asterisks indicate a significant difference to respective mock controls based on a paired two-tailed t test analysis (P < 0.001).
To test whether IOS1 is also required for immunity to pathogens other than bacteria, susceptibility of ios1 mutants to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola was evaluated by droplet inoculation. The ios1-1, ios1-2, and ios1-3 mutants were as susceptible as wild-type plants to both pathogens (Supplemental Figures 4A and 4B), suggesting that IOS1 is critical for immunity to virulent hemibiotrophic bacteria, but not to necrotrophic fungi such as B. cinerea and A. brassicicola.
The role of IOS1 in antibacterial immunity was further evaluated by analyzing the susceptibility to Pst DC3000 of transgenic Arabidopsis lines overexpressing IOS1 mRNA (OE1 and OE3) (Supplemental Figure 5). Both IOS1-OE lines were significantly less susceptible to Pst DC3000 (Figure 1C). Notably, although ios1 mutants did not show any defect in stomatal innate immunity (Supplemental Figure 6), overexpression of IOS1 inhibited the bacteria-mediated reopening of stomata (Figure 1D). Together, these data are consistent with a positive role of IOS1 in antibacterial immunity.
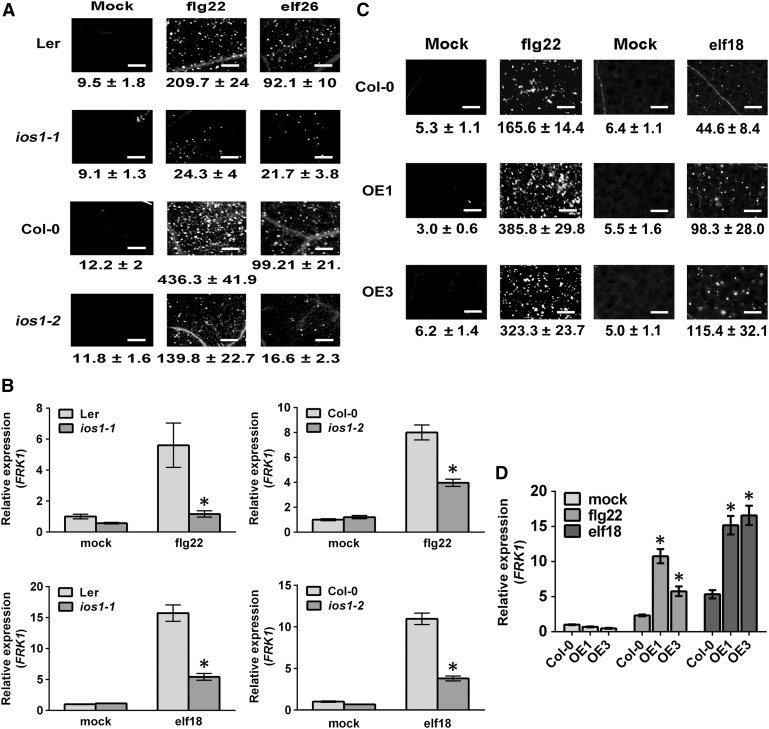
IOS1 Is Critical for Late PTI Responses
To analyze whether IOS1 is involved in PTI responses, we first monitored IOS1 mRNA expression levels by RT-qPCR after treatment of seedlings with 100 nM flg22 or elf18. Both MAMPs induced IOS1 transcripts accumulation at 1 h after treatment (Supplemental Figure 7). To evaluate the role of IOS1 in late PTI responses, we measured callose deposition in ios1 mutants after infiltration with the bacterial MAMPs flg22 or elf26. Aniline blue staining and image analysis indicated lower levels of callose deposition in ios1-1 and ios1-2 than in wild-type leaves (Figure 2A). These results suggest that IOS1 is critical for PTI-induced callose deposition. To further evaluate late PTI responses, we monitored expression levels of the PTI marker gene FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1) (Asai et al., 2002; Xiao et al., 2007; Boudsocq et al., 2010) after treatment with flg22 or elf18. At 1 h after MAMP treatment, both ios1-1 and ios1-2 mutants demonstrated lower FRK1 upregulation levels (Figure 2B). Furthermore, we analyzed late PTI responses in the IOS1-OE lines. Interestingly, these lines did not exhibit constitutive callose deposition, while more callose deposits were observed in OE1 and OE3 upon elicitation with the MAMPs flg22 or elf18 (Figure 2C). Similarly, constitutive upregulation of FRK1 was not observed in IOS1-OE lines, but FRK1 expression levels were potentiated in the OE1 and OE3 lines upon flg22 or elf18 treatments (Figure 2D). These data suggest that overexpression of IOS1 primes late PTI responses and that IOS1 positively regulates several late PTI responses.
Figure 2.
Altered Late PTI Responses in ios1 Mutants and IOS1-OE Lines.
(A) and (C) Callose deposition. Leaves of 5-week-old ios1-1 and ios1-2 (A) were syringe infiltrated with 1 µM flg22 or elf26 and samples were collected 9 h (flg22) or 24 h (elf26) later for aniline blue staining. For IOS1-OE lines (C), leaves of 5-week-old Arabidopsis or 10-d-old seedlings were respectively syringe infiltrated with 1 µM flg22 or treated with 100 nM elf18 and samples were collected 6 h (flg22) or 16 h (elf18) later for aniline blue staining. Mock samples were infiltrated with MgSO4 (for flg22 and elf26) or water (for elf18). Numbers under the pictures are average ± sd of the number of callose deposits per square millimeter from at least two independent experiments each consisting of 6 plants (n = 12). Bar = 200 µm.
(B) and (D) PTI-responsive gene FRK1 upregulation. Relative FRK1 expression levels were evaluated at 30 min posttreatment with 100 nM flg22 or elf18 in ios1-1 and ios1-2 mutants (B) or at 45 min posttreatment with 50 nM flg22 or elf18 in IOS1-OE lines (D). UBQ10 was used for normalization. Relative gene expression levels were compared with wild-type mock (Ler-0 or Col-0) (defined value of 1) by RT-qPCR analyses. The values are means ± sd of two independent experiments each with three batches of 20 plantlets (n = 6). Asterisks indicate a significant difference to wild-type controls based on a paired two-tailed t test (P < 0.01).
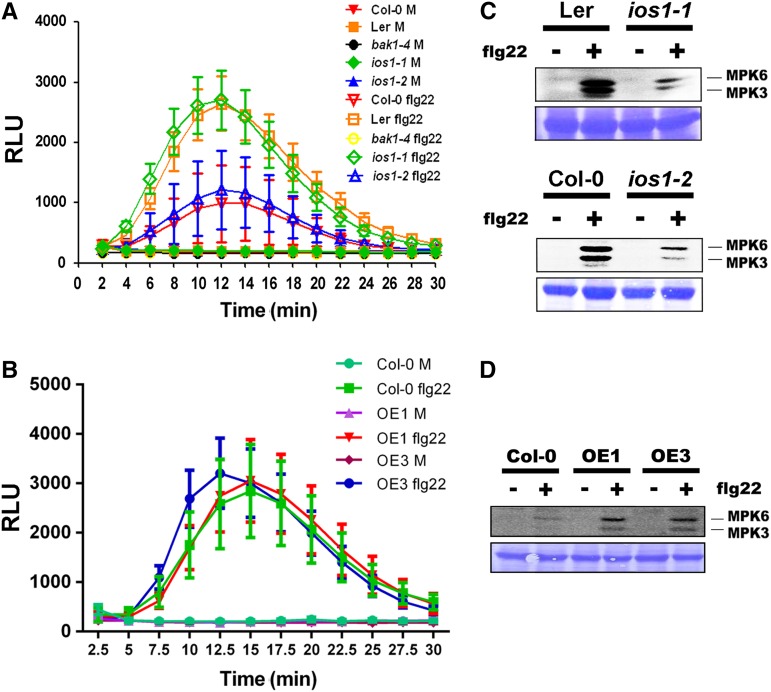
IOS1 Modulates Several Early PTI Responses
To test whether IOS1 is required for early PTI events, we analyzed ROS production in response to 10 nM flg22 or elf26 for 30 min in wild-type, ios1-1, ios1-2, and IOS1-OE leaves. Both mutants and OE lines displayed wild-type levels of ROS production, while MAMP-mediated ROS production was strongly reduced in bak1-4 (Figures 3A and 3B; Supplemental Figures 8A and 8B). Treatments with MAMPs rapidly activate Arabidopsis MAP kinases MPK3 and MPK6 (Nühse et al., 2000). Notably, both ios1-1 and ios1-2 mutants demonstrated a weaker activation of MPK3 and MPK6 than the wild type following treatment with flg22 or elf18 (Figure 3C; Supplemental Figures 8C and 8D). On the other hand, MPK3 and MPK6 activation was stronger than the wild type in the OE1 and OE3 transgenic lines (Figure 3D; Supplemental Figure 8E). Together, these results suggest that IOS1 is required for full MPK activation, but not for the ROS burst after MAMP perception. This observation is consistent with these responses being uncoupled (Segonzac et al., 2011; Xu et al., 2014a). However, since we do not provide a kinetic analysis, we cannot exclude a slower or faster MPK response in ios1 mutants or OE lines, respectively.
Figure 3.
Early PTI Responses.
(A) ROS production in ios1 mutants. Responsiveness of 5-week-old Ler-0 and Col-0 wild-type controls and respective mutants ios1-1 and ios1-2 to 10 nM flg22. bak1-4 was used as a negative control. Production of ROS in Arabidopsis leaf discs is expressed as relative light units (RLU) for a period of 30 min after elicitation. Values are means ± se of three independent experiments each with six leaf discs (n = 18). Differences between ios1 mutants and the wild type were not statistically significant based on a paired two-tailed t test (P < 0.01).
(B) ROS production in IOS1-OE lines. Responsiveness of 5-week-old overexpression lines OE1 and OE3 and Col-0 wild-type control to 10 nM flg22. Production of ROS in Arabidopsis leaf discs is expressed as relative light units for a period of 30 min after elicitation. Values are means ± se of three independent experiments each with six leaf discs (n = 18). Differences between OE lines and the wild type were not statistically significant based on a paired two-tailed t test (P < 0.01).
(C) MPK activation in ios1 mutants. Ten-day-old Ler-0 and ios1-1 or Col-0 and ios1-2 were treated with 100 nM flg22 for 5 min. Immunoblot analysis using phospho-p44/42 MPK antibody is shown in top panel. Lines indicate the positions of MPK3 and MPK6. Coomassie blue staining is used to estimate equal loading in each lane (bottom panel). Similar results were observed in another independent repeat.
(D) MAPK activation in IOS1-OE lines. Ten-day-old Col-0 and IOS1 overexpression lines OE1 and OE3 were treated with 50 nM flg22 for 5 min. Immunoblot analysis using phospho-p44/42 MAP kinase antibody is shown in the top panel. Lines indicate the positions of MPK3 and MPK6. Coomassie blue staining is used to estimate equal loading in each lane (bottom panel). Similar results were observed in another independent repeat.
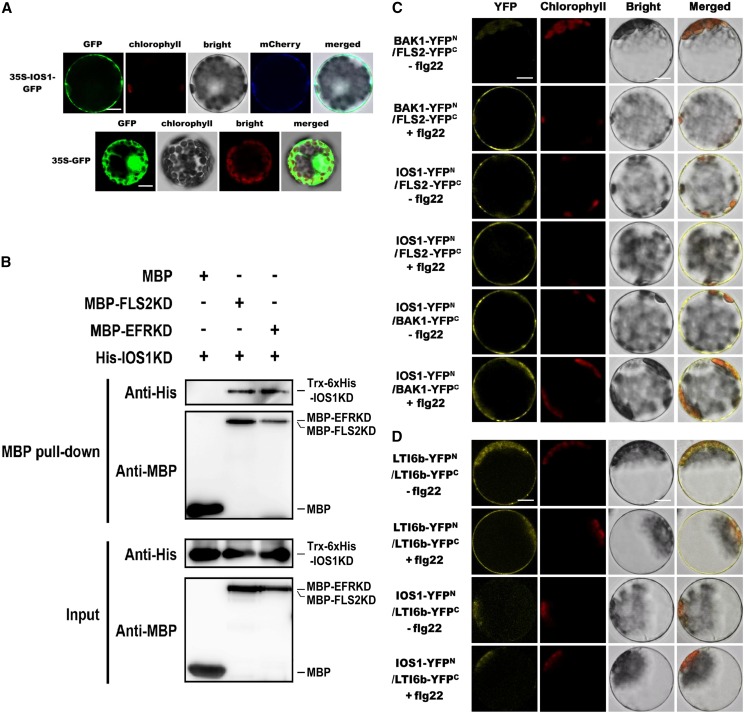
IOS1 Likely Localizes to the Plasma Membrane
IOS1 is a predicted transmembrane RLK (Hok et al., 2011). We analyzed IOS1 subcellular localization by transiently expressing IOS1-GFP fusion protein driven by the cauliflower mosaic virus 35S promoter in Arabidopsis mesophyll protoplasts. The fluorescence signal was mainly confined to the cell surface with a pattern similar to the plasma membrane marker pm-rk CD3-1007 (Nelson et al., 2007), while the control protoplasts expressing GFP alone showed a nuclear/cytoplasmic localization (Figure 4A). These data suggest that similarly to the PRRs FLS2 and EFR (Robatzek et al., 2006; Häweker et al., 2010), IOS1 is likely localized at the plasma membrane.
Figure 4.
IOS1 Localization, Pull-Down, and BiFC Analyses of IOS1 Interaction with PRRs.
(A) Subcellular localization of IOS1-GFP fusion protein in Arabidopsis mesophyll protoplasts. IOS1-GFP expression was driven by the cauliflower mosaic virus 35S promoter and transiently expressed in Arabidopsis mesophyll protoplasts. The images of the GFP fluorescence (GFP), the chlorophyll autofluorescence (chlorophyll), the bright-field image (bright), the plasma membrane marker (pm-rk CD3-1007)-mCherry fluorescence localization, and the combined images (merged) are shown. Similar observations were made in another independent repeat. Bars = 10 µm.
(B) In vitro MBP pull-down assay of IOS1 interaction with FLS2 and EFR. E. coli expressed MBP (negative control), MBP-FLS2KD, or MBP-EFRKD were incubated with Trx-6xHis-IOS1KD and pulled down with amylose resin beads. Input and bead-bound proteins were analyzed by immunoblotting with specific antibodies. Experiments were repeated three times with similar results.
(C) BiFC analyses of IOS1 interactions with FLS2 and BAK1. Arabidopsis protoplasts were cotransfected with BAK1-YFPN + FLS2-YFPC, IOS1-YFPN + FLS2-YFPC, and IOS1-YFPN + BAK1-YFPC and treated with (+) or without (–) 100 nM flg22 for 10 min. The YFP fluorescence (yellow), chlorophyll autofluorescence (red), bright-field, and the combined images were visualized under a confocal microscope 16 h after transfection. Images are representative of multiple protoplasts. Experiments were repeated at least twice with similar results. Bars = 10 µm.
(D) BiFC of LTI6b and IOS1 interaction. Arabidopsis protoplasts were cotransfected with LTI6b-YFPN + LTI6b-YFPC or IOS1-YFPN + LTI6b-YFPC and treated with (+) or without (−) 100 nM flg22 for 10 min. The YFP fluorescence (yellow), chlorophyll autofluorescence (red), bright-field, and the combined images were visualized under a confocal microscope 16 h after transfection. Images are representative of multiple protoplasts. Experiments were repeated twice with similar results. Bars = 10 µm.
IOS1 Associates with FLS2, EFR, and BAK1 in a Ligand-Independent Manner
IOS1 acts upstream of MPK in flg22- and elf26- or elf18-triggered PTI signaling cascades. We thus evaluated whether IOS1 associates with PRRs such as FLS2 or EFR. We first used pull-down analysis to show that a Trx-6xHis-tagged IOS1 kinase domain interacted with MBP-tagged FLS2 and EFR in vitro (Figure 4B). Next, interactions were evaluated by bimolecular fluorescence complementation (BiFC) assays (Walter et al., 2004) in Arabidopsis protoplasts. To test whether our experimental conditions were appropriate, we first analyzed the interactions between BAK1 and FLS2 or EFR that occur only upon elicitation (Chinchilla et al., 2007; Heese et al., 2007; Roux et al., 2011). As expected, the YFP signal was clearly observed after flg22 or elf18 treatment (Figure 4C; Supplemental Figure 9A). YFP fluorescence was detected before and after elicitation with flg22 or elf18 when testing IOS1 interaction with FLS2 or EFR, respectively (Figure 4C; Supplemental Figure 9A). Similarly, IOS1 interacted with BAK1 in a ligand-independent manner (Figure 4C; Supplemental Figure 9A). The low temperature and salt-responsive protein 6B (LTI6b/RCI2B) fused to GFP, which is known to localize at the plasma membrane (Cutler et al., 2000), was used as a negative control. LTI6b is known to dimerize (Huang et al., 2014), and YFP fluorescence was indeed observed when Arabidopsis protoplasts were transfected with LTI6b-YFPN and LTI6b-YFPC (Figure 4D; Supplemental Figure 9B), indicating that both constructs were functional. Importantly, no YFP fluorescence at the plasma membrane was observed when testing IOS1 interaction with LTI6b, even after elicitation with flg22 or elf18 (Figure 4D; Supplemental Figure 9B). Although we cannot exclude artifacts inherent to overexpression in protoplasts, these data suggest that IOS1 interacts at the plasma membrane with the PRRs FLS2 and EFR and the coreceptor BAK1 in a ligand-independent manner.
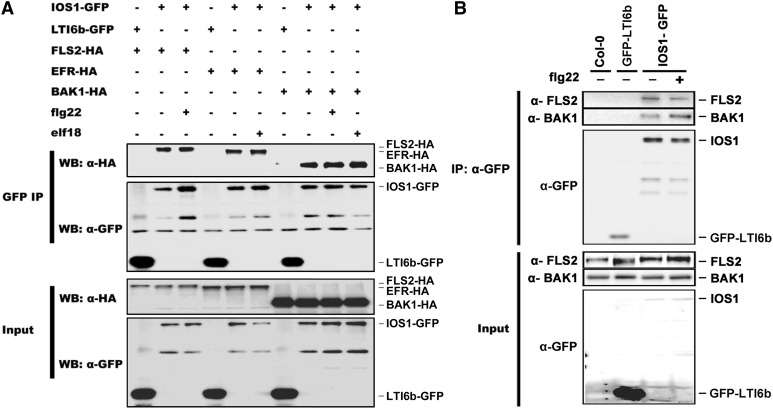
To test whether IOS1 associates with FLS2 in vivo, we transiently coexpressed FLS2-HA3 with GFP epitope-tagged IOS1 in Arabidopsis protoplasts. Equal amounts of IOS1 were immunoprecipitated with GFP-Trap beads and analyzed for presence of FLS2-HA3 using anti-HA immunoblotting. FLS2 could be detected in mock- and flg22-treated samples (Figure 5A). Similarly, we analyzed the possible association of IOS1 with EFR and BAK1 before and after elicitation with elf18 (for EFR) or flg22 and elf18 (for BAK1). For that purpose, IOS1-GFP was transiently coexpressed with EFR-HA3 or BAK1-HA3 in Arabidopsis protoplasts, and IOS1 was immunoprecipitated with GFP-Trap beads. EFR and BAK1 could also be detected in the IOS1 immunoprecipitate before and after MAMP treatment (Figure 5A). As a negative control, we tested the association of IOS1 with LTI6b by immunoprecipitating equal amounts of LTI6b with GFP-trap beads and by analyzing the presence of FLS2-HA3, EFR-HA3, and BAK1-HA3 using anti-HA immunoblotting. FLS2, EFR, and BAK1 could not be detected, suggesting that they do not associate with GFP at the plasma membrane (Figure 5A). These observations suggest that IOS1 associates with FLS2, EFR, and BAK1 in a ligand-independent manner. Of note, IOS1 homodimerized independently of flg22 treatment (Supplemental Figure 10), as previously reported for FLS2 (Sun et al., 2012). In addition, the GFP fusion of IOS1 does not affect its function as it can complement the defective MPK3 and MPK6 activation observed in ios1-1 and ios1-2 mutants (Supplemental Figure 11).
Figure 5.
IOS1 Associates with Unstimulated and Stimulated FLS2, EFR, and BAK1.
(A) Coimmunoprecipitation of IOS1, FLS2, EFR, and BAK1 proteins. Arabidopsis protoplasts expressing IOS1-GFP and FLS2-HA3 (lanes 2 and 3), IOS1-GFP and EFR-HA3 (lanes 5 and 6), or IOS1-GFP and BAK1-HA3 (lanes 8 to 10) constructs were treated (+) or not (−) with 100 nM flg22 or elf18 for 10 min. LTI6b-GFP, a known plasma membrane protein, was used as a control to illustrate that FLS2-HA3, EFR-HA3, and BAK1-HA3 do not associate with GFP at the plasma membrane (lanes 1, 4, and 7). Total proteins (input) were subjected to immunoprecipitation with GFP trap beads followed by immunoblot analysis with anti-HA antibodies to detect FLS2-HA3, EFR-HA3, and BAK1-HA3. Anti-GFP antibodies detect IOS1-GFP and LTI6b-GFP. Experiments were repeated twice with similar results.
(B) Coimmunoprecipitation of FLS2, BAK1, and IOS1 proteins in Arabidopsis. Transgenic Arabidopsis seedlings overexpressing IOS1-GFP (OE3) were treated (+) or not (−) with 100 nM flg22 for 10 min. Total proteins (input) were subjected to immunoprecipitation with anti-GFP magnetic beads followed by immunoblot analysis with anti-FLS2 antibodies, anti-BAK1 antibodies, or anti-GFP antibodies to detect FLS2, BAK1, and IOS1-GFP. Untransformed Col-0 Arabidopsis tissue was used as a control to show that FLS2 and BAK1 do not adhere nonspecifically to anti-GFP magnetic beads (lane 1). LTI6b, a known plasma membrane protein was used as a control to illustrate that FLS2 and BAK1 do not associate with GFP at the plasma membrane (lane 2). This experiment is one of two independent replicates.
To test whether IOS1-GFP associates with FLS2 in Arabidopsis as well, we performed coimmunoprecipitation experiments using transgenic lines overexpressing IOS1-GFP. IOS1-GFP was immunoprecipitated with anti-GFP magnetic beads and analyzed for the presence of endogenous BAK1 and FLS2 using anti-BAK1 and anti-FLS2 immunoblotting. As negative controls, anti-GFP magnetic beads were incubated with protein extracts of untransformed Col-0 and transgenic plants expressing LTI6b fused to GFP. Signals for FLS2 and BAK1 upon LTI6b-GFP immunoprecipitation were largely weaker than those observed upon IOS1-GFP, suggesting that FLS2 and BAK1 do not nonspecifically bind to anti-GFP magnetic beads, nor do they interact with GFP itself at the plasma membrane (Figure 5B). In contrast, we could detect a clear association of IOS1-GFP with native FLS2 and BAK1 (Figure 5B). Treatment with flg22 did not affect significantly or reproducibly the associations of IOS-GFP with FLS2 and BAK1 (Figure 5B). Moreover, we found IOS1 as part of the in vivo EFR complex in an unbiased manner while searching for EFR-associated proteins in planta by proteomics analysis. In these experiments, anti-GFP immunoprecipitates were prepared from untreated and elf18-treated transgenic efr-1/EFRp:EFR-eGFP seedlings, as well as from untreated efr-1 null mutant or Col-0 seedlings, in order to reveal proteins that nonspecifically bind to GFP beads. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis identified eight different peptides matching IOS1 in the EFR-eGFP immunoprecipitates, but none in the negative controls (Supplemental Table 1). The IOS1 peptides were found in both untreated and elf18-treated samples, corroborating the fact that IOS1 associates with the PRRs FLS2 and EFR in a ligand-independent manner in vivo.
IOS1 Is Required for Optimal flg22-Induced FLS2-BAK1 Association, but Functions Independently of BIK1
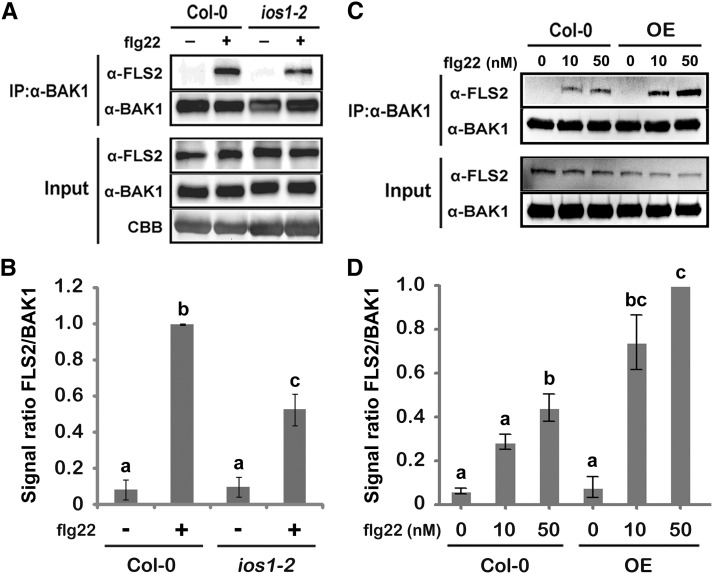
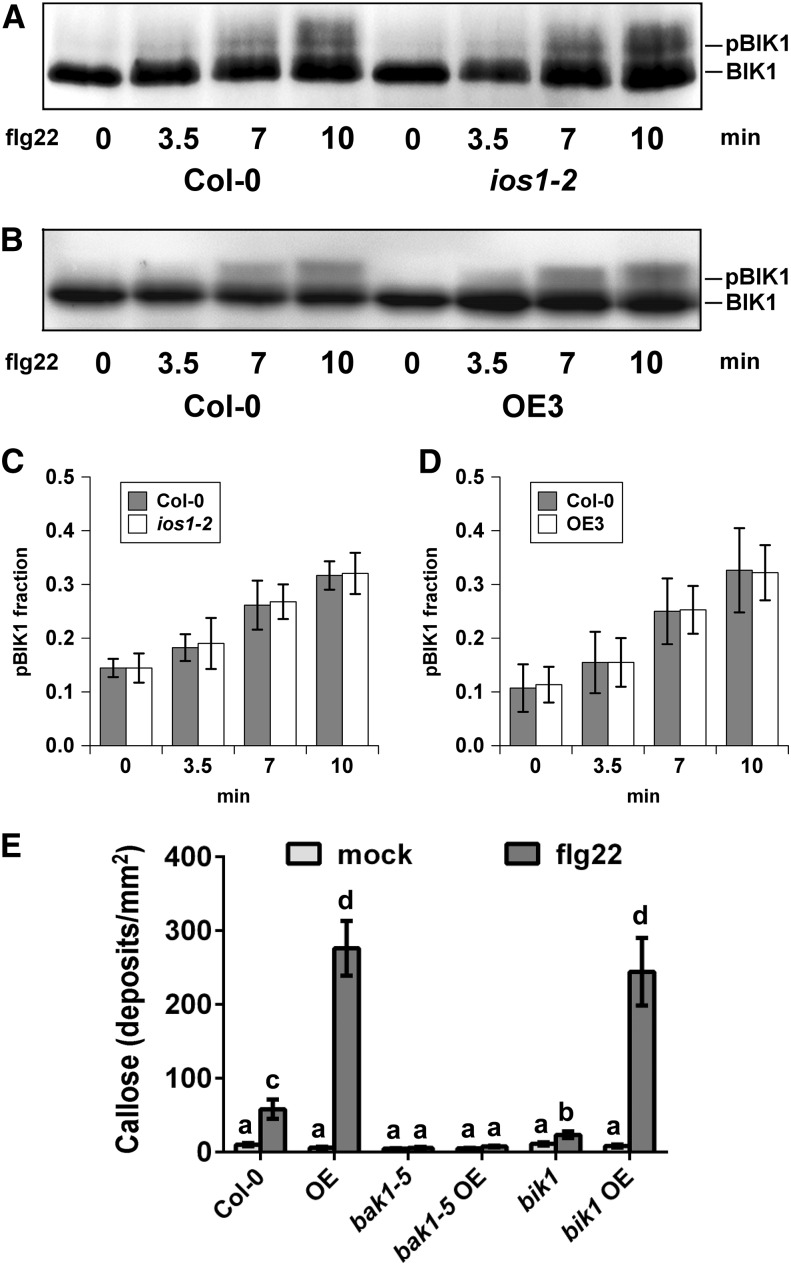
To test whether associations between IOS1 and both FLS2 and BAK1 impact other biochemical events within the FLS2 complex, we analyzed ligand-induced FLS2-BAK1 association (Chinchilla et al., 2007; Heese et al., 2007). Toward this goal, BAK1 was first immunoprecipitated from ios1-2 plants treated or not with 100 nM flg22 and associated FLS2 was revealed by anti-FLS2 immunoblotting. After flg22 treatment, the mutant ios1-2 displayed significantly less FLS2 coimmunoprecipitated with BAK1 than the wild-type control (Figures 6A and 6B). By contrast, a significant increase in coimmunoprecipitated FLS2 was observed in Arabidopsis overexpressing IOS1 treated with two different concentrations of flg22 (Figures 6C and 6D). These data show that the active kinase IOS1 (Supplemental Figure 12) positively regulates the association of BAK1 with FLS2. However, flg22-mediated phosphorylation of BIK1, which is a direct substrate of FLS2 (Lu et al., 2010a; Zhang et al., 2010), was not affected in ios1-2 and the OE3 line (Figures 7A to 7D). Together, these results indicate that IOS1 modulates FLS2-BAK1 association upon elicitation but is not critical for BIK1 phosphorylation. We further evaluated IOS1 dependency to BAK1 and BIK1 by analyzing callose deposition in lines overexpressing IOS1 in bak1-5 and bik1 mutant backgrounds. The bak1-5 and bik1 mutants are largely defective in flg22-mediated callose deposition (Figure 7E; Zhang et al., 2010). While overexpression of IOS1 strongly primed callose deposition in the Col-0 wild-type control, bak1-5 mutation completely abolished IOS1-mediated priming of callose deposition (Figure 7E). However, lines overexpressing IOS1 in the bik1 mutant background still displayed a large increase in callose deposits after flg22 treatment (Figure 7E). Collectively, these results suggest that IOS1 functions in a BAK1-dependent, but BIK1-independent, manner in the FLS2 complex.
Figure 6.
IOS1 Regulates Ligand-Induced FLS2/BAK1 Association.
(A) and (B) Ligand-dependent association of FLS2 to BAK1 is reduced in the ios1-2 mutant. Col-0 or ios1-2 seedlings were treated (+) or not (−) with 100 nM flg22 for 10 min. Total proteins (input) were subjected to immunoprecipitation (IP) with anti-BAK1 antibodies and IgG beads followed by immunoblot analysis using anti-FLS2 and anti-BAK1 antibodies. For (A), Coomassie blue (CBB) is used to estimate equal loading (bottom panel). The experiment shown in (A) is one of three independent replicates pooled together in (B).
(C) and D) Ligand-dependent association of FLS2 to BAK1 is augmented in the IOS1-OE3 line. Col-0 or OE3 seedlings were treated with MgSO4 (0) or 10 or 50 nM flg22 for 10 min. Total proteins (input) were subjected to immunoprecipitation with anti-BAK1 antibodies and IgG beads followed by immunoblot analysis using anti-FLS2 and anti-BAK1 antibodies. The experiment shown in (C) is one of three independent replicates pooled together in (D). For both (B) and (D), signals were evaluated with the ImageJ software. Values are means ± sd of three independent biological replicates (n = 3). Different letters denote significant difference based on a one-way ANOVA with post-hoc Tukey HSD (P < 0.05).
Figure 7.
IOS1 Functions in a BAK1-Dependent but BIK1-Independent Manner in the FLS2 Complex.
(A) to (D) Immunoblot analysis of BIK1 phosphorylation revealed by gel mobility shift. Nonphosphorylated (BIK1) and phosphorylated (pBIK1) BIK1 signals are indicated. Protoplasts from Col-0 leaves and ios1-2 ([A] and [C]) or OE3 ([B] and [D]) were treated 4 h after transfection using 0.75 µM flg22 for 3.5, 7, and 10 min. The reaction was stopped by immersion in liquid nitrogen following concentration by low speed centrifugation. Experiments were repeated at least five times with similar results. For (C) and (D), phosphorylated over nonphosphorylated BIK1 fractions were calculated by measuring digital signals with the ImageJ software. Values are means ± sd of five independent biological replicates (n = 5). For each time point, differences between the wild type and the ios1-2 mutant or the OE3 line were not statistically significant based on a paired two-tailed t test (P < 0.01).
(E) Callose deposition upon elicitation with flg22. Fourteen-day-old Col-0 wild-type, IOS1-OE3 (OE), bak1-5 or bik1 mutants, and IOS1-OE in bak1-5 or bik1 mutant background were treated with 100 nM flg22 and samples were collected 16 h later for aniline blue staining. Each bar represents average ± se of callose deposits per square millimeter from two independent experiments each with six plants (n = 12). For IOS1-OE lines in the bak1-5 and bik1 backgrounds, data represent two independent transformation events for each genotype. Different letters denote significant differences among different lines based on a one-way ANOVA with post-hoc Tukey HSD (P < 0.01).
Defective Chitin Responses in ios1 Mutants
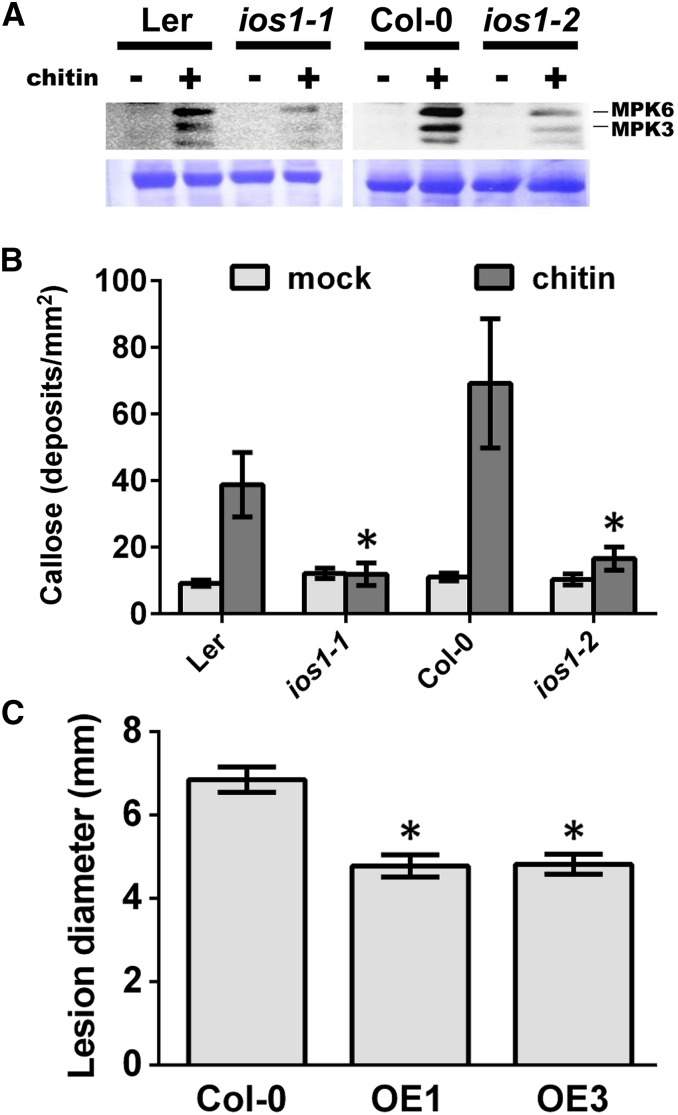
To evaluate whether IOS1 function is uniquely linked with BAK1, we analyzed MPK3/6 activities upon elicitation with the MAMP chitin. Fungal chitin recognition is mediated by LysM-domain RLKs such as CERK1 (Miya et al., 2007; Wan et al., 2008, 2012), and BAK1 is not required for chitin perception and signaling (Shan et al., 2008; Kemmerling et al., 2011; Ranf et al., 2011). Reduced MPK3/6 activities were observed after chitin treatment in the ios1-1 and ios1-2 mutants (Figure 8A), suggesting that IOS1 also plays a role in PRR complexes that do not recruit BAK1. To further investigate whether IOS1 is necessary for the chitin-mediated PTI response, callose deposition in ios1-1 and ios1-2 mutants was analyzed. Both mutants accumulated less callose than wild-type Arabidopsis at 16 h after chitin treatment (Figure 8B), indicating that IOS1 is necessary for chitin-mediated callose deposition. The fungal pathogen B. cinerea produces the MAMP chitin (Windram et al., 2012) and A. brassicicola is commonly used in chitin perception studies (Miya et al., 2007), but ios1 mutants demonstrated wild-type resistance to both pathogens (Supplemental Figure 4). We thus tested the resistance response of the OE1 and OE3 lines toward these necrotrophic pathogens. Both OE lines harbored smaller B. cinerea-mediated disease lesions (Figure 8C), while they did not show increased resistance toward A. brassicicola (Supplemental Figure 13). Taken together, these data suggest that IOS1 is critical for chitin-mediated PTI and plays a positive role in Arabidopsis resistance to some, but not all, pathogens that produce the MAMP chitin.
Figure 8.
A Role for IOS1 in the Chitin Response.
(A) MPK activation upon elicitation with chitin. Fourteen-day-old seedlings from Ler-0 or Col-0 wild type, ios1-1, or ios1-2 were syringe-infiltrated with 0.2 mg/mL chitin for 5 min. Immunoblot analysis using phospho-p44/42 MPK antibody is shown in top panel. Lines indicate the positions of MPK3 and MPK6. Coomassie blue staining is used to estimate equal loading in each lane (bottom panel). An independent experiment showed similar results.
(B) Callose deposition upon elicitation with chitin. Fourteen-day-old seedlings from Ler-0 and ios1-1 or Col-0 and ios1-2 were treated with 0.2 mg/mL chitin and samples were collected 16 h later for aniline blue staining. Numbers are averages ± se of callose deposits per square millimeters from two independent experiments each including six seedlings (n = 12). Asterisks indicate a significant difference to wild-type controls based on a paired two-tailed t test (P < 0.01).
(C) B. cinerea-mediated lesions. Arabidopsis leaves of Col-0 and IOS1 overexpression lines were droplet-inoculated (10 μL) with 105 B. cinerea spores/mL and lesion diameters were evaluated at 3 dpi. Data are average ± se of lesion diameters from two independent experiments each with six plants (n = 12). Asterisks indicate a significant difference to wild-type controls based on a paired two-tailed t test (P < 0.01).
IOS1 Associates with CERK1
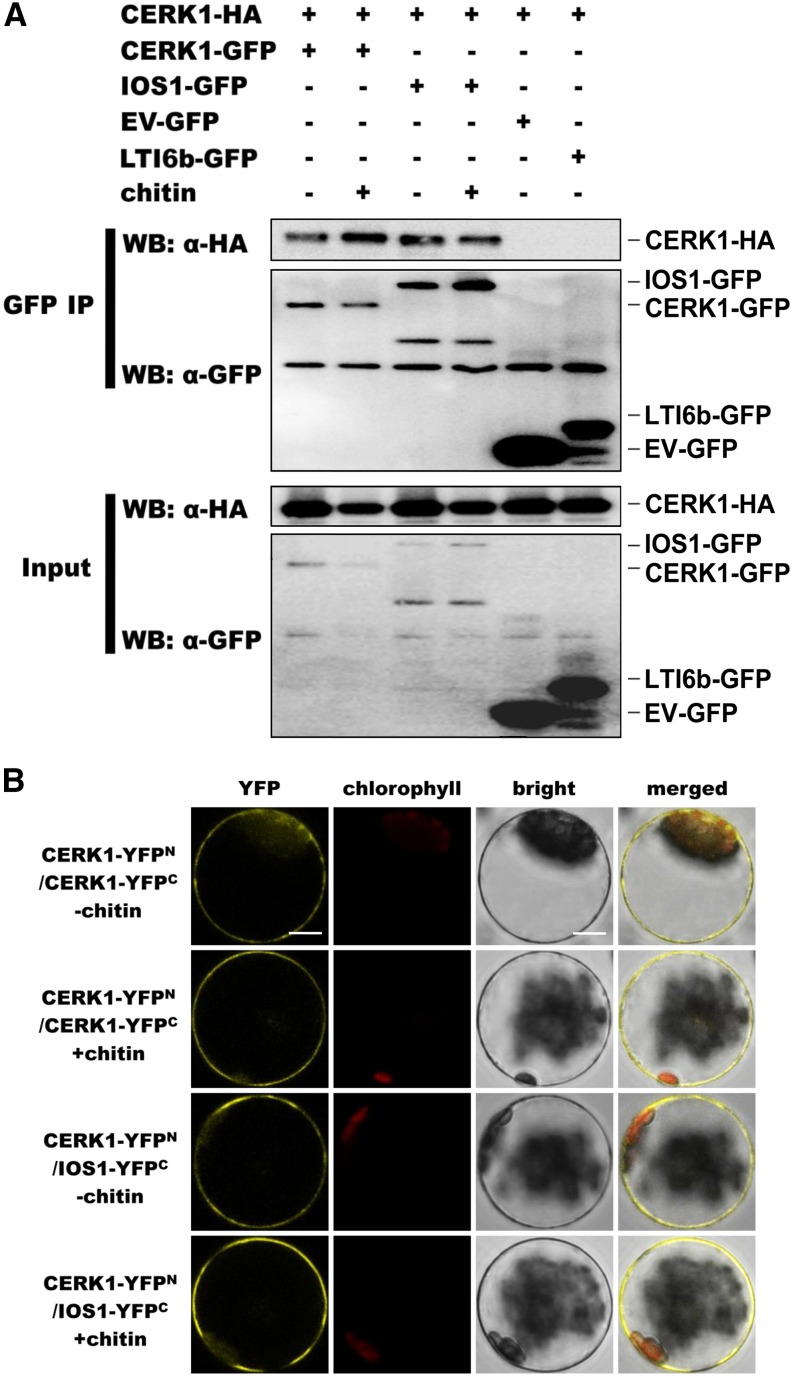
Possible association of IOS1 with CERK1 was tested by coimmunoprecipitation in Arabidopsis protoplasts. The previously described CERK1 dimerization (Liu et al., 2012) was observed, indicating that the CERK1-GFP and CERK1-HA3 constructs were functional (Figure 9A). Association of IOS1 with CERK1 was then performed by immunoprecipitating equal amounts of IOS1 with GFP-Trap beads and by analyzing CERK1-HA3 presence using anti-HA immunoblotting. CERK1 could clearly be detected before and after elicitation with chitin (Figure 9A), suggesting ligand-independent association. We also coexpressed CERK1-HA3 with empty vector (EV)-GFP. No signal was observed in this negative control, suggesting that the association between CERK1-HA3 and IOS1-GFP is not due to a direct binding of CERK1-HA3 with GFP proteins or GFP-Trap beads (Figure 9A). As an additional negative control, we tested the association of CERK1 with LTI6b after elicitation with chitin by immunoprecipitating an equal amount of LTI6b with GFP-trap beads and by analyzing CERK1-HA3 presence using anti-HA immunoblotting. CERK1-HA3 could not be detected or at very low levels (Figure 9A). Together, these data suggest specific association of IOS1 with CERK1 at the plasma membrane.
Figure 9.
IOS1 Associates with CERK1.
(A) Coimmunoprecipitation of IOS1 with CERK1 proteins in Arabidopsis protoplasts. Arabidopsis protoplasts expressing CERK1-GFP and CERK1-HA3 (lanes 1 and 2), IOS1-GFP and CERK1-HA3 (lanes 3 and 4), EV-GFP and CERK1-HA3 (lane 5), or LTI6b-GFP and CERK1-HA3 (lane 6) constructs were treated with (+) or without (−) 0.2 mg/mL chitin for 10 min. Total proteins (input) were subjected to immunoprecipitation (IP) with GFP trap beads followed by immunoblot analysis with anti-HA antibodies to detect CERK1-HA3. EV-GFP and LTI6b-GFP, a known plasma membrane protein, were used as controls to illustrate that CERK1-HA3 does not stick to GFP beads or associate with GFP at the plasma membrane, respectively. This experiment was repeated twice with similar results.
(B) BiFC analyses of IOS1 interactions with CERK1. Arabidopsis protoplasts were cotransfected with CERK1-YFPN + CERK1-YFPC and CERK1-YFPN + IOS1-YFPC, and treated with (+) or without (−) 0.2 mg/mL chitin for 10 min. The YFP fluorescence (yellow), chlorophyll autofluorescence (red), bright-field, and the combined images were visualized under a confocal microscope 16 h after transfection. Images are representative of multiple protoplasts. At least two independent experiments were performed with similar results. Bars = 10 μm.
These observations suggest that IOS1 is part of the CERK1 receptor complex. To further verify the IOS1-CERK1 complex, we used BiFC in Arabidopsis protoplasts to examine the direct interaction of IOS1 with CERK1. The dimerization of CERK1 was first used to demonstrate that CERK1-YFPN and CERK1-YFPC constructs are functional (Liu et al., 2012). As expected, the YFP signal was observed when both CERK1-YFPN and CERK1-YFPC were cotransfected in Arabidopsis protoplasts before and after chitin treatment indicating dimerization of CERK1 (Figure 9B). Similarly, a clear YFP signal was visible independently of chitin treatment with the CERK1-YFPN and IOS1-YFPC constructs (Figure 9B), suggesting that both proteins can directly interact in Arabidopsis protoplasts. Collectively, these results suggest that in addition to FLS2 and EFR, IOS1 also interacts with CERK1.
IOS1 Is Necessary for BABA-Induced Resistance and Priming
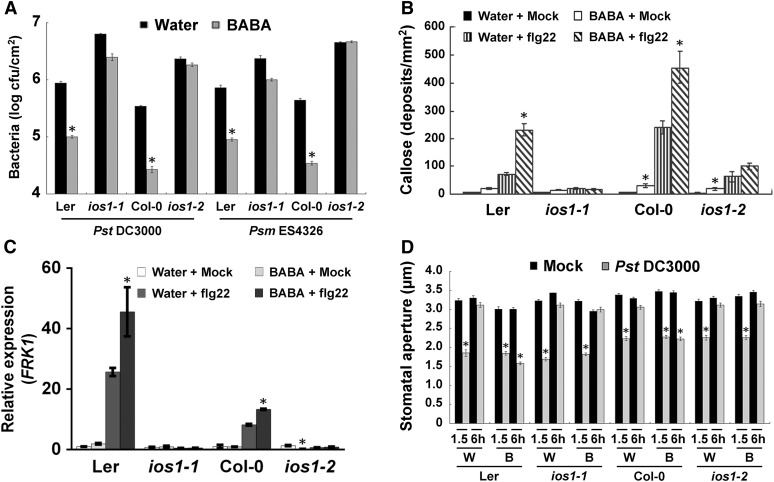
Since overexpression of the BABA-responsive IOS1 primes Arabidopsis PTI (Figures 2C, 2D, and 3D), we tested whether IOS1 is required for induced resistance to Pst DC3000 and Psm ES4326 triggered by the priming agent BABA (Zimmerli et al., 2000; Tsai et al., 2011). While BABA treatments protected both Col-0 and Ler-0 wild type against Pst DC3000 and Psm ES4326 infection, ios1-1 and ios1-2 mutants demonstrated a defective BABA-induced resistance toward these hemibiotrophic bacteria (Figure 10A). BABA is known to prime the PTI response in Arabidopsis (Singh et al., 2012; Po-Wen et al., 2013). We thus tested the role of IOS1 in BABA-induced priming of PTI responses. Notably, the priming effect of BABA on flg22-induced callose deposition and FRK1 expression was largely abolished in ios1-1 and ios1-2, in comparison to the wild type (Figures 10B and 10C). IOS1 positively modulates flg22-mediated FLS2-BAK1 association (Figure 6) and BABA treatment upregulates IOS1 expression (Tsai et al., 2011). We thus asked whether BABA affects FLS2-BAK1 association upon flg22 elicitation. No clear increase in FLS2-BAK1 association upon flg22 treatment was observed in BABA-treated Col-0 plants (Supplemental Figure 14).
Figure 10.
BABA Action Is Defective in ios1 Mutants.
(A) BABA-induced resistance. Bacterial titers in 5-week-old Ler-0, ios1-1, Col-0, and ios1-2 were determined at 2 dpi with 106 cfu/mL Pst DC3000 or 5 × 105 cfu/mL Psm ES4326. Two days before bacterial inoculation, plants were soil-drenched with water as a control or 225 µM BABA. Values are means ± se of three independent experiments each with three plants (n = 9). Asterisks indicate a significant difference to respective water-treated control based on a paired two-tailed t test (P < 0.01).
(B) BABA priming of PTI-mediated callose deposition. Leaves of water- or BABA-pretreated (225 µM) Ler-0 and ios1-1 or Col-0 and ios1-2 were syringe-infiltrated with 1 µM flg22 and samples were collected 6 h later for aniline blue staining. Values are average ± sd from three independent experiments each consisting of nine plants (n = 27). Asterisks indicate a significant difference to water-treated respective controls based on a paired two-tailed t test (P < 0.01).
(C) BABA priming of PTI-mediated FRK1 expression. Ten-day-old Ler-0 and ios1-1 or Col-0 and ios1-2 seedlings grown on 0.5× MS medium supplemented with 30 µM BABA (BABA) or not (Water) were submerged with water (Mock) or 1 µM flg22, and FRK1 expression levels were analyzed 60 min later by RT-qPCR. UBQ10 was used for normalization. Relative gene expression levels were compared with respective water + mock-treated wild type (defined value of 1). Values are means ± sd of two independent experiments each with three plants (n = 6). Asterisks indicate a significant difference to water-treated respective controls based on a paired two-tailed t test (P < 0.01).
(D) BABA inhibition of bacteria-mediated stomatal reopening. Stomatal apertures in epidermal peels from water- (W) or BABA-treated (225 µM) (B) Ler-0 and ios1-1 or Col-0 and ios1-2 were analyzed after 1.5 and 6 h exposure to MgSO4 (Mock) or 108 Pst DC3000. Results are shown as mean ± se of three independent experiments each consisting of at least 60 stomata (n > 180). Asterisks indicate a significant difference to respective mock controls based on a paired two-tailed t test analysis (P < 0.001).
Since BABA inhibits bacteria-mediated stomatal reopening (Tsai et al., 2011), we tested whether IOS1 is involved in this phenomenon. While BABA inhibited bacteria-mediated stomatal reopenings in Col-0 and Ler-0 wild type, it did not in ios1-1 and ios1-2 mutants (Figure 10D). Taken together, these data suggest a positive role for IOS1 in BABA-induced resistance and BABA-mediated priming of PTI, including strengthening of stomatal innate immunity.
DISCUSSION
PRRs are critical to elicit PTI responses and to restrict pathogen ingress (Boller and Felix, 2009; Nicaise et al., 2009; Zhang and Zhou, 2010; Huang and Zimmerli, 2014). To date, all known plant PRRs are modular transmembrane proteins containing ligand binding ectodomains that function as part of multiprotein complexes (Böhm et al., 2014; Zipfel, 2014). In this work, we analyzed the role of the Arabidopsis malectin-like/LRR-RLK IOS1 in innate immunity and priming with genetic and biochemical approaches. The results support the following conclusions.
IOS1 Is Necessary for Full Activation of PTI in Arabidopsis
Our reverse genetic approach identified three independent IOS1 insertion mutants with hypersusceptibility to virulent hemibiotrophic Pst DC3000 and Psm ES4326 bacteria, but with wild-type sensitivity to the necrotrophic fungal pathogens B. cinerea and A. brassicicola. These observations suggest that IOS1 is critical for resistance to hemibiotrophic bacteria, but not to necrotrophic fungi. However, lines overexpressing IOS1 were more resistant to B. cinerea, suggesting a role for IOS1 in Arabidopsis resistance to this necrotrophic pathogen. A partially defective PTI response in ios1 mutants may not be sufficient to produce a visible increased susceptibility phenotype upon infection by B. cinerea. Necrotrophic fungal pathogens such as B. cinerea produce toxins, cell wall degrading enzymes, and ROS to promote disease and macerate plant tissue (Prins et al., 2000), possibly hiding the effect of a defective PTI. By contrast, a stronger PTI in IOS1 OE lines may restrict early B. cinerea infection at least partially independently of toxins, cell wall degrading enzymes, and ROS produced by B. cinerea, leading to increased resistance. Redundancy may also explain the lack of increased sensitivity in ios1 loss-of-function mutants. Lines overexpressing IOS1 demonstrated a wild-type resistance to A. brassicicola, suggesting that Arabidopsis resistance to this necrotrophic fungus occurs independently of IOS1. The mutant ios1-1 is known to be more resistant to the filamentous oomycete pathogens Hyaloperonospora arabidopsidis and Phytophthora parasitica (Hok et al., 2011, 2014). IOS1 could be a direct or indirect target of oomycete effectors necessary for pathogen virulence. IOS1 absence in ios1-1 would not allow H. arabidopsidis or P. parasitica to fully repress Arabidopsis PTI (Hok et al., 2014). Taken together, these observations suggest that IOS1 is involved in Arabidopsis immunity to various microbial pathogens.
Increased susceptibility of ios1 mutants to virulent bacteria was associated with a defective PTI response. Typically, bacteria- and MAMP-induced callose depositions were dramatically reduced in ios1 mutants. In addition, upregulation of the PTI-responsive gene FRK1 was delayed in plants with a defective IOS1. Consistent with this, Arabidopsis overexpressing IOS1 demonstrated increased accumulation of callose and potentiated expression levels of FRK1 upon MAMP elicitation. By contrast, both FRK1 expression and callose deposition were not affected in the ios1-1 mutant after inoculation with filamentous pathogens (Hok et al., 2014). These discrepancies may be explained by early (this work) versus late time point analyses (Hok et al., 2014). MPK3/6 activation was reduced in ios1 mutants and augmented in IOS1 overexpression lines, suggesting that IOS1 acts upstream of MPK3/6 in PTI signaling. These observations point to the fact that IOS1 is necessary for full activation of both early and late PTI responses. Similarly, LecRK-VI.2 is necessary for full activation of some early and late PTI responses (Singh et al., 2012). However, while Arabidopsis overexpressing LecRK-VI.2 demonstrate a constitutive PTI response, IOS1 overexpression lines showed a strengthened PTI only upon elicitation by bacteria or MAMPs, suggesting a different mechanism of action for these two positive regulators of PTI. Importantly, PTI-mediated ROS production was at wild-type levels in ios1 mutants and in IOS1-OE lines, suggesting that IOS1 may not regulate all aspects of the PTI response. The PRR-associated kinase BIK1 directly regulates apoplastic ROS production during PTI (Kadota et al., 2014, 2015; Li et al., 2014). So, the apparent absence of IOS1 regulation of ROS production could be explained by the fact that IOS1 acts largely in a BIK1-independent manner (Figures 7). The ios1 mutants demonstrated wild-type bacteria-mediated stomatal closure, while IOS1-OE lines harbored a strengthened stomatal immunity. Redundancy may explain stomatal innate immunity discrepancy between ios1 mutants and IOS1-OE lines (compared with Figure 1D and Supplemental Figure 6). Other malectin-like/LRR-RLKs may indeed play a redundant role in stomatal closure (Hok et al., 2011), thus masking the possible function of IOS1 in this early PTI response. Stomata of the ios1-1 mutant are hyperresponsive to ABA (Hok et al., 2014), and ABA signaling is critical for stomatal immunity (Melotto et al., 2006; Desclos-Theveniau et al., 2012), but we observed wild-type stomatal closure in ios1-1 after Pst DC3000 inoculation. As suggested by Hok et al. (2014), IOS1 may use different signaling pathways for the activation of PTI in response to bacteria and in the downregulation of ABA upon infection with filamentous pathogens. Taken together, these data reveal IOS1 as a major positive regulator of Arabidopsis PTI against bacteria, acting upstream of MPK3/6 in FLS2- and EFR-dependent defense signaling pathways. Noteworthy, typical concentrations of MAMPs were used for each PTI assay resulting in the use of various concentrations of MAMPs in different experiments. We thus cannot fully exclude dose-dependent effects.
IOS1 Associates with FLS2, EFR, and BAK1 in a Ligand-Independent Manner
Having genetically demonstrated the importance of IOS1 in bacteria-, flg22-, and elf26/elf18-triggered PTI upstream of MPK3/6, and also considering that IOS1 is a LRR-RLK with two LRR motifs, a transmembrane domain, and a complete extracellular malectin-like domain (Hok et al., 2011), we further investigated whether IOS1 is part of PRR complexes recognizing bacterial MAMPs. We first showed that the kinase domain of IOS1 associates in vitro with the KDs of FLS2 and EFR using a pull-down approach. In addition, in vivo association of IOS1 with FLS2, EFR, and/or the regulatory LRR-RLK BAK1 were evaluated by BiFC and coimmunoprecipitation analyses. We performed BiFC assays and coimmunoprecipitation experiments in Arabidopsis protoplasts and found that IOS1 constitutively associates with FLS2 and EFR and that elicitation with flg22 or elf18 does not significantly affect the association. The constitutive IOS1 and FLS2 association was further confirmed in Arabidopsis using transgenic lines overexpressing IOS1-GFP, while IOS1 was also found to be part of unstimulated and stimulated EFR complexes by in planta proteomics analysis on EFR-associated proteins (Supplemental Table 1). In addition to associating with PRRs, IOS1 interacts with the BSK3 in Arabidopsis (Xu et al., 2014b). In Arabidopsis, only few proteins are known to be present in PRR complexes before elicitation by MAMPs. Notably, the cytoplasmic kinases BIK1/PBLs and BSK1 interact constitutively with FLS2 and are released upon elicitation (Lu et al., 2010a; Zhang et al., 2010; Shi et al., 2013). Both PCRK1 and PCRK2 associate with FLS2 (Kong et al., 2016), and heterotrimeric G proteins directly interact with FLS2 to regulate PTI (Liang et al., 2016). Additionally, the DENN domain protein SCD1 that negatively regulates innate immunity associates in a ligand-independent manner with FLS2 in vivo (Korasick et al., 2010). Furthermore, the ubiquitin E3 ligases PUB12/13 interact with BAK1 prior elicitation and ubiquitinate FLS2 upon flg22-induced FLS2/BAK1 complex formation, leading to FLS2 degradation (Lu et al., 2011), and BIR2 negatively regulates Arabidopsis PTI by association before elicitation with BAK1 (Halter et al., 2014). This work thus identifies a component of the FLS2 and EFR protein complexes.
IOS1 Positively Regulates FLS2-BAK1 Complex Formation
Since the malectin-like LRR-RLK IOS1 constitutively associates with the PRRs FLS2 and EFR and the regulatory LRR-RLK BAK1, and since ios1 mutants demonstrate a defective PTI, we hypothesized that IOS1 affects early events at PRR complexes. The flg22-mediated association of FLS2 and BAK1 was indeed reduced in ios1-2 and increased in the OE3 line when compared with wild-type Col-0 controls. By contrast, the positive regulator of PTI LecRK-VI.2 does not modulate flg22-mediated association of FLS2 and BAK1 (Singh et al., 2012). The heterodimerization between FLS2 and BAK1 occurs within seconds (Schulze et al., 2010) with BAK1 acting as coreceptor for flg22 (Sun et al., 2013), indicating that both LRR-RLKs most likely exist in close proximity at the plasma membrane, as recently suggested in the case of BAK1 and BRI1 (Bücherl et al., 2013). We thus propose that the plasma membrane-localized IOS1 is required for promoting rapid FLS2-BAK1 complex formation upon flg22 binding. Importantly, the flg22-mediated association between FLS2 and BAK1 was not completely abolished in ios1-2. Other players such as other malectin-like LRR-RLKs may generate the partial FLS2-BAK1 association observed upon flg22 elicitation in ios1-2. IOS1 constitutively interacts with both FLS2 and BAK1; however, FLS2 and BAK1 complex formation only occurs after flg22 treatment. In addition, FLS2-FLS2 and IOS1-IOS1 homodimerization could be observed independently of elicitation (Sun et al., 2012; Supplemental Figure 10). IOS1 monomers or dimers may thus bind both FLS2 and BAK1 in different complexes before PTI elicitation. Upon flg22 treatment, IOS1 may participate in the formation of a new complex that integrates both FLS2 and BAK1. Contrarily to IOS1, BIR2 negatively regulates FLS2-BAK1 complex formation (Halter et al., 2014). Thus, BIR2 may directly or indirectly antagonize IOS1.
Treatments with flg22 induce rapid phosphorylation of BIK1, which further increases phosphorylation of FLS2 and BAK1 (Lu et al., 2010a; Zhang et al., 2010). BIK1 phosphorylation occurs within minutes after flg22 treatment and is thus considered a good marker of PRRs activities. Surprisingly, BIK1 phosphorylation was at wild-type levels in flg22-treated ios1-2 mutant and in the OE3 line. Since MPK3/6 activities were altered in ios1 mutants and in IOS1-OE lines, a BIK1-independent signaling cascade that affects MPK3/6 activities must be present in ios1-2 and in IOS1-OE lines. This observation is in agreement with Zhou and colleagues, who demonstrated that BIK1 and the closely related PBL1 are not required for flg22-induced MAPK activation (Feng et al., 2012). Therefore, other receptor-like cytoplasmic kinases could play a role in regulating different branches of PTI signaling (Lu et al., 2010b). In addition, Arabidopsis overexpressing IOS1 in the bik1 mutant background still demonstrated a strong priming of callose deposition. By contrast, augmented callose deposition after flg22 treatment was strongly abolished in lines overexpressing IOS1 in the bak1-5 background. Taken together, these data suggest that an altered FLS2-BAK1 association in ios1-2 impacts MPK3/6 activation independently of BIK1 phosphorylation.
IOS1 Is Necessary for Chitin-Mediated PTI Responses and Associates with CERK1
IOS1 plays a critical role in BAK1-dependent PRR complexes such as FLS2 and EFR in a BIK1-independent manner. We next asked whether IOS1 is also necessary for a full PTI response activated by PRR complexes functioning in a BAK1-independent manner. Contrary to FLS2 that associates with the BAK1 coreceptor to sense flagellin (Sun et al., 2013), the LysM-domain RLK CERK1 that is part of PRR complexes that recognize chitin (Miya et al., 2007; Wan et al., 2008, 2012; Cao et al., 2014) and peptidoglycans (Willmann et al., 2011) functions in a BAK1-independent manner. Notably, BAK1 is not required for chitin perception and signaling (Shan et al., 2008; Kemmerling et al., 2011; Ranf et al., 2011). Both ios1-1 and ios1-2 mutants demonstrated defective MPK3/6 activation and reduced callose deposition after chitin treatment. In addition, coimmunoprecipitation and BiFC analyses suggested direct association of IOS1 with CERK1. We thus propose that IOS1 acts at the CERK1 receptor complex to positively regulate chitin-mediated PTI responses. IOS1 thus regulates the PTI response at both BAK1-dependent and BAK1-independent PRR complexes. Similarly, BIK1 is involved in both FLS2/EFR and CERK1 complexes to activate downstream PTI responses (Lu et al., 2010a; Zhang et al., 2010). Notably, BIK1 regulates CERK1-mediated chitin responses, including the accumulation of ROS and the induction of defense genes (Zhang et al., 2010). Since CERK1 does not associate with BAK1 upon elicitation, IOS1 should modulate the chitin-mediated PTI response through other regulatory mechanisms than PRR association with BAK1 as observed for FLS2 (Figure 6). CERK1 is also involved in bacterial resistance (Gimenez-Ibanez et al., 2009) and is a critical member of the PRR complex that recognizes bacterial peptidoglycans (Willmann et al., 2011), suggesting that IOS1 plays a role in at least three PRR complexes recognizing bacterial MAMPs. This may explain the strong phenotypes of ios1 mutants and IOS1-OE lines observed upon bacterial infection. By contrast, the role of IOS1 in Arabidopsis resistance against necrotrophic fungi such as B. cinerea and A. brassicicola was rather weak. In addition, the ios1-1 mutant is more resistant to the biotrophic fungus Erysiphe cruciferarum (Hok et al., 2014). These observations suggest that IOS1 role in the Arabidopsis resistance against pathogens producing the MAMP chitin is not critical. As suggested for oomycete pathogens (Hok et al., 2011), fungal pathogens may produce effectors that target IOS1, and the absence of IOS1 may result in wild-type or enhanced Arabidopsis resistance levels even though IOS1 is critical for a full chitin-mediated defense response.
IOS1 Plays a Critical Role in Priming of PTI
Accumulation of positive regulators of defense such as MPK3/6 or LecRK-VI.2 prior to stress challenge is critical for priming (Beckers et al., 2009; Singh et al., 2012). Plants overexpressing IOS1 demonstrated potentiated expression of PTI-responsive genes, primed callose deposition, and increased MPK3/6 activities upon PTI elicitation. These observations further suggest that increased accumulation of positive regulators of PTI before elicitation is sufficient to prime PTI and consequently to increase resistance to pathogens. BABA-mediated accumulation of IOS1 mRNA (Tsai et al., 2011) may thus be critical for BABA-mediated priming of PTI (Singh et al., 2012; Po-Wen et al., 2013). We therefore investigated whether ios1 mutants are defective in BABA-mediated priming. While the lecrk-VI.2-1 mutant is only partially defective in BABA priming (Singh et al., 2012), ios1-1 and ios1-2 mutants were largely deficient in BABA-induced resistance to bacteria, priming of FRK1 expression and callose deposition, and in BABA-mediated strengthening of stomatal innate immunity. These results suggest that IOS1 plays a predominant role during priming of PTI by the non-protein amino acid BABA. Surprisingly, BABA had no effect on flg22-mediated FLS2-BAK1 association, suggesting that the reported role of IOS1 in BABA-triggered priming involves another regulatory mechanism.
LRR-RLKs such as FLS2, EFR, and CERK1 or PEPR1/2 are receptors for MAMPs or DAMPs, respectively (Huffaker et al., 2006; Yamaguchi et al., 2006, 2010; Miya et al., 2007; Ryan et al., 2007; Wan et al., 2008; Krol et al., 2010). Another LRR-RLK, BAK1, functions in several PRR complexes as a coreceptor (Chinchilla et al., 2007; Heese et al., 2007; Boller and Felix, 2009; Schulze et al., 2010; Sun et al., 2013). Our data identified the malectin-like LRR-RLK IOS1 as a novel member of FLS2 and EFR PRR complexes that also associates in a ligand-independent manner with BAK1. In addition, IOS1 regulates CERK1-dependent PTI responses that are BAK1-independent (Shan et al., 2008; Kemmerling et al., 2011; Ranf et al., 2011). This work identifies a novel LRR-RLK regulating BAK1-dependent and -independent PTI responses and further reveals the intricate regulation of the PRR complex dynamics needed for transmitting and regulating PTI signaling, which requires additional components beyond the ligand binding receptor and coreceptor.
METHODS
Biological Materials and Growth Conditions
Arabidopsis thaliana ecotypes Col-0 and Landsberg erecta (Ler-0) were grown in commercial potting soil/perlite (3:2) at 22 to 24°C day and 17 to 19°C night temperature under a 9-h-light/15-h-dark photoperiod. The lighting was supplied at an intensity of ∼100 µE m–2 s–1 by fluorescence tubes (TOA, model FL40D/38). The Ds transposon insertion line (Ler-0) ios1-1 (GT_5_22250) and the T-DNA insertion mutants (Col-0) ios1-2 (Salk_137388) and ios1-3 (SAIL_343_B11) were obtained from the ABRC. The mutant bak1-4 (Salk_116202) and bik-1 have been described elsewhere (Chinchilla et al., 2007; Lu et al., 2010a; Zhang et al., 2010). Bacterial strains Pst DC3000 and the Pst DC3000 hrcC- mutant (CB200) were provided by B.N. Kunkel (Washington University, St. Louis, MO), while Psm ES4326 was a gift from J. Glazebrook (Minnesota University, St. Paul, MN). All bacteria were cultivated at 28°C and 340 rpm in King’s B medium with 50 mg/L rifampicin (Pst DC3000), 50 mg/L rifampicin and kanamycin (CB200), or 50 mg/L streptomycin (Psm ES4326). The fungi Botrytis cinerea and Alternaria brassicicola were obtained from C.Y. Chen (National Taiwan University, Taipei, Taiwan) and grown at room temperature (18°C to ∼25°C) on PDA-agar plates (Zimmerli et al., 2001).
Pathogen Infection Assays
Five-week-old Arabidopsis were dipped in 106 cfu/mL Pst DC3000 or 5 × 105 cfu/mL Psm ES4326 in 10 mM MgSO4 containing 0.01% Silwet L-77 (Lehle Seeds) for 15 min. After inoculation, plants were kept at 100% relative humidity, and symptoms were evaluated 3 d later. Bacterial titers were determined as previously described (Zimmerli et al., 2000). For B. cinerea and A. brassicicola infection, spores were diluted to105 and 5 × 105 spores/mL in 0.5× potato dextrose broth medium, respectively. Droplets of 10 μL 0.5× potato dextrose broth with B. cinerea or A. brassicicola spores were deposited on leaf surfaces of 5-week-old plants (three leaves per plant). Leaves of same age were used for droplet inoculation. Disease symptoms and lesion diameters were determined at 3 dpi. At least 18 lesion diameters were evaluated for each independent experiment (six plants).
IOS1 Overexpression Plants
The DNA plasmids (pH35GWG) expressing IOS1 protein fused with GFP at the C terminus under the control of the cauliflower mosaic virus 35S promoter were obtained from the ABRC (Gou et al., 2010; ABRC stock S1G51800HGF). Agrobacterium tumefaciens strain GV3101 was used for the transformation of Col-0 plants. Successful transformations were determined by selecting on 0.6% Murashige and Skoog (MS) agar plates containing 50 mg/mL hygromycin B and raised to homozygous T3 lines. For the generation of IOS1-OE lines in the bak1-5 or bik1 mutant background, mutant plants were dip-inoculated with Agrobacterium strains GV3101 carrying Pro35S-IOS1-GFP (pFAST-R05) using OLE1-TagRFP as a screenable marker (Shimada et al., 2010) and raised to T2 for analyses.
BABA and MAMP Treatments
For bacteria titer, callose deposition, and stomatal aperture evaluations, 5-week-old Arabidopsis was soil drenched with BABA (Fluka) at a final concentration of 225 µM 2 d before bacteria inoculation or MAMP treatments. BABA was dissolved in water and controls were soil drenched with water only. For FRK1 expression and ligand-induced FLS2-BAK1 association, seedlings grown on 0.5× MS plates with or without 30 µM BABA for 10 d were submerged in a 1 µM or 100 nM flg22 solution, respectively, for 60 min before sample collection.
The flg22 and elf26 or elf18 peptides were purchased from Biomer Technology and dissolved in 10 mM MgSO4, MgSO4 only, or water for seedling treatment was used as control. Chitin from shrimp shells (Sigma-Aldrich) was dissolved in water. Water-only treatments were used as controls. MAMPs were syringe-infiltrated into leaves and samples were harvest at indicated time points.
Callose Deposition
Five-week-old Arabidopsis leaves were syringe infiltrated with 1 µM flg22 in 10 mM MgSO4. Control plants were infiltrated with 10 mM MgSO4 only. Nine leaf discs from three different plants were selected for analyses at the indicated time points. Callose deposition evaluation on seedlings was performed on 14-d-old Arabidopsis grown on 0.5× MS plates that were transferred to 0.5× MS liquid medium one night before treatment with 100 nM flg22, 100 nM elf18, or 0.2 mg/mL chitin for 16 h. Six seedlings were selected for analyses for each sample. Callose deposits were visualized as described (Singh et al., 2012).
RT-qPCR
For RT-qPCR, Arabidopsis seedlings grown on 0.5× MS plates for 10 d were transferred to liquid 0.5× MS one night before treatments with 100 nM flg22 or elf26 for ios1 mutants and with 50 nM flg22 or elf18 for the OE lines, and samples were collected at the indicated time points. Total RNA isolation, cDNA biosynthesis, and real-time PCR analyses were performed as described (Wu et al., 2010). Normalization of gene expression was conducted with At4g05320 (UBQ10). For RT-PCR, one microliter of cDNA was used as template and standard PCR conditions were applied as described (Singh et al., 2012). At4g05320 (UBQ10) was used as a loading control. Primers used are in Supplemental Table 2.
MAP Kinase Assay
Twenty 10-d-old plants were incubated in 0.5× MS supplemented with 100 nM (ios1 mutants) or 50 nM (OE lines) flg22 or elf18 dissolved in water or with water only (control), 0.2 mg/mL chitin, or water (control) for 5 min before being pooled for harvest. For complementation assays, protoplasts from 5-week-old Arabidopsis were transfected with plasmids (pH35GWG) carrying Pro35S-IOS1-GFP or the vector only by polyethylene glycol (Sigma-Aldrich), and samples were collected 5 min after flg22 or water treatment. MAP kinase assays were performed as described (Singh et al., 2012).
ROS Burst
ROS assays were performed as previously described (Huang et al., 2013). Briefly, six leaf discs (10 mm diameter) from three 5-week-old Arabidopsis (two discs/plant) were incubated in double-distilled water in 96-well plates overnight. The following day, the water was replaced by 10 nM flg22 or elf26 in 10 mM MgSO4 buffer or by 10 mM MgSO4 buffer only for the mock controls containing 2 µM luminol (Sigma-Aldrich) and 10 µg/mL peroxidase (Sigma-Aldrich). The plates were analyzed every 2 min for mutants or every 2.5 min for OE lines after addition of MAMPs for a period of 30 min using a CentroLIApc LB 692 plate luminometer (Berthold Technologies).
Stomatal Assay
Five-week-old plants were kept under light (100 µE m–2 s–1) for at least 3 h to open stomata before the beginning of the experiments. For each biological replicate, stomatal apertures were evaluated from 12 epidermal peels from four plants (three epidermal peels/plant) as described (Tsai et al., 2011).
Subcellular Localization in Protoplasts
For transient expression of the GFP fusion proteins, constructs expressing 35S-IOS1-GFP (plasmid pH35GWG, ABRC stock S1G51800HGF) or vector alone were transfected into Arabidopsis mesophyll protoplasts according to He et al. (2007). The GFP-fusion constructs were cotransfected with the plasma membrane marker pm-rkCD3-1007 (Nelson et al., 2007). Transfected protoplasts were visualized using a confocal laser scanning microscope (Zeiss LSM 780 Confocal; Carl Zeiss) with excitation at 488 nm and emission at 490 to 515 nm; autofluorescence was observed at 650 to 700 nm. The plasma membrane marker was detected with excitation at 594 nm and emission at 595 to 650 nm.
BIK1 Phosphorylation
Mesophyll protoplasts were obtained as described by He et al. (2007). BIK1 phosphorylation assays on protoplasts treated with 0.75 µM flg22 for 3.5, 7, and 10 min were performed as described (Singh et al., 2013).
Cloning, Expression, and Purification of Recombinant Proteins
In order to generate a Trx-6xHis N-terminal fusion of the IOS1 kinase domain, the sequence coding for the IOS1 cytosolic domain was amplified from the pH35GWG vector expressing the IOS1-GFP fusion using primers carrying BamHI and XhoI restriction sites (Supplemental Table 2) and introduced into the polylinker of the pET-32a(+) expression vector (Novagen). To produce an inactive kinase fusion protein, a point mutation at the kinase active site (D710N) was introduced into the expression vector by primer extension (Supplemental Table 2) using the Phusion polymerase (New England Biolabs), followed by DpnI (New England Biolabs) digestion according to manufacturer’s instructions. The Trx-6xHis-IOS1KD and Trx-6xHis-IOS1KDm (kinase dead) fusion proteins were expressed in the Escherichia coli strain Rosetta (DE3) pLysS (Novagen). After overnight induction at 16°C with 0.4 mM IPTG, the bacteria were pelleted by centrifugation, resuspended in 100 mL binding buffer (20 mM sodium phosphate, 0.5 M NaCl, 20 mM imidazole, and 0.04% 2-mercaptoethanol), and sonicated. The soluble His-tagged proteins were affinity purified using a HisTrap FF column (GE Healthcare) according to the manufacturer's instructions. All constructs were confirmed by Sanger sequencing and purified Trx-6xHis-IOS1KD and Trx-6xHis-IOS1KDm were analyzed by LC-MS/MS.
The production of MBP-tagged FLS2 and EFR kinase domain constructs was performed as described by Schwessinger et al. (2011). MBP was expressed from pMAL-c5X (New England Biolabs). MBP and the two MBP-tagged proteins were expressed as described above, but using the E. coli strain BL21 (DE3) pLysS (Novagen) and purified using amylose resins (MBPTrap HP; GE Healthcare) following the manufacturer's instructions. Finally, MBP and the two MBP-tagged proteins were dialyzed against dialysis buffer (20 mM Tris-HCl and 100 mM NaCl, pH 7.4).
In Vitro Pull-Down Assay
One microgram of MBP, MBP-FLS2KD, or MBP-EFRKD was incubated with 2 µg of Trx-6xHis-IOS1KD in a binding buffer (20 mM Tris-HCl, 200 mM NaCl, and 1 mM EDTA, pH 7.4) under agitation at 4°C. After 2 h, 50 μL of amylose resin beads (GE Healthcare) were added, and the incubation continued for another 2 h. The beads were then washed five times with washing buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA, and 0.6% Triton X-100, pH 7.4). Input and pulled-down proteins were resolved by 8% SDS-PAGE and detected by immunoblotting using appropriate antibodies.
BiFC Assay
Full-length coding sequences of FLS2, EFR, BAK1, CERK1, LTI6b, and IOS1 without stop codon amplified from cDNA of Arabidopsis Col-0 were inserted into the entry vector pCR8/GW/TOPO and subcloned into YN (pEarleyGate201-YN) or YC (pEarleyGate202-YC) vectors (Lu et al., 2010c) through LR reaction (Invitrogen). The constructs were transfected into Arabidopsis protoplasts by polyethylene glycol (Sigma-Aldrich) for transient expression (Yoo et al., 2007). Sixteen hours later, transfected cells were treated with or without 100 nM flg22, 100 nM elf26, or 0.2 mg/mL chitin for 10 min before being imaged using a confocal laser scanning microscope (Zeiss LSM 780 Confocal).
Transient Expression in Arabidopsis Protoplasts
For the coimmunoprecipitation, TOPO plasmid containing full-length coding sequences of FLS2, EFR, BAK1, CERK1, LTI6b, or IOS1 without stop codon were recombined into GFP (pEarlyGate103) or HA (modified pEarleyGate100 with a AvrII-3xHA-SpeI fragment introduced after the attR2 recombination site) vectors. Amplification of the coding sequences was performed using the primers described in Supplemental Table 2. All constructs were confirmed by DNA sequencing. The 35S-FLS2-GFP-His and 35S-EFR-GFP-His constructs were as described (Schwessinger et al., 2011).
Protein Extraction and Immunoprecipitation in Arabidopsis Protoplasts
Protein extraction and immunoprecipitation were performed as described (Yeh et al., 2015). Briefly, plasmids containing HA3 or GFP tag constructs were cotransfected into Arabidopsis protoplasts by polyethylene glycol (Sigma-Aldrich) for transient expression (Yoo et al., 2007). Total proteins were extracted with 0.5 mL protein extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10% glycerol, 10 mM DTT, 10 mM EDTA, 1 mM NaF, 1 mM Na2MoO4·2H2O, 1% [v/v] IGEPAL CA-630 [Sigma-Aldrich], and 1% [v/v] Roche protease inhibitor cocktail) and incubated with gentle shaking at 4°C for 1 h. Samples were then centrifuged at 14,000 rpm for 15 min at 4°C. Supernatants (1.5 mL) were adjusted to 2 mg/mL protein and incubated for 2 h at 4°C with 20 mL GFP Trap-A beads (Chromotek). Following incubation, beads were washed four times with TBS containing 0.5% (v/v) IGEPALCA-630. Total proteins (input) or immunoprecipitated proteins were separated by 10% SDS-PAGE and then transferred to a polyvinylidene fluoride membrane (Immobilon-P; Millipore). GFP and HA3 fusion proteins were detected by immunoblotting with anti-GFP and anti-HA primary antibodies, respectively
Protein Extraction and Immunoprecipitation in Arabidopsis
The protocol for protein extraction was described by Roux et al. (2011). Arabidopsis seedlings grown in liquid 0.5× MS medium were used for immunoprecipitation assays. For immunoprecipitation of endogenous BAK1, supernatants were incubated with 25 μL true-blot anti-rabbit Ig beads (Ebioscience) and 20 μL anti-BAK1 antibody (Schulze et al., 2010) for 4 h at 4°C. For immunoprecipitation of IOS1-GFP, supernatants were incubated with 50 to 200 μL of anti-GFP magnetic beads (Miltenyi Biotec) for 2 h at 4°C (Kadota et al., 2014, 2016). Following incubation, beads were washed three to five times with extraction buffer, before adding SDS loading buffer (Schwessinger et al., 2011).
SDS-PAGE and Immunoblotting
For immunoblotting, 8 to 10% SDS-PAGE gels were run at 80 to 140 V for 2 h before electroblotting on PVDF membrane (Millipore) at 100 V for 1 h at 4°C. Membranes were rinsed in TBS and blocked in 5% (w/v) nonfat milk powder in TBS-Tween 0.1% (v/v) for 2 h. Primary antibodies were diluted in TBS-Tween solution to the following ratios: mouse anti-His (Santa Cruz Biotechnology sc-8036) 1:1000; rabbit anti-MBP 1:4000 (Sigma-Aldrich SAB2108749); rabbit anti-GFP (Santa Cruz Biotechnology sc-9996) 1:3000; mouse anti-HA (Santa Cruz Biotechnology sc-7392) 1:3000; anti-BAK1 1:500 and anti-FLS2 1:1000 (Schwessinger et al., 2011), and incubated overnight. Membranes were washed three times in TBS-Tween before 1-h incubation with secondary antibodies anti-mouse-HRP (Santa Cruz Biotechnology sc-2005; 1:3000) or anti-rabbit-HRP (Santa Cruz Biotechnology sc-2004; 1:3000). Signals were visualized using an enhanced chemiluminescence system (Immobilon Western; Millipore) and a LAS-3000 (Fujifilm) scanner following the manufacturer’s instructions.
Mass Spectrometry
Proteins were separated by SDS-PAGE (Nupage precast gel system; Invitrogen) and after staining with Coomassie Brilliant Blue (SimplyBlue SafeStain; Invitrogen), the proteins were cut out and were digested by trypsin as described previously (Ntoukakis et al., 2009). LC-MS/MS analysis was performed using a LTQ-Orbitrap mass spectrometer (Thermo Scientific) and a nanoflow-HPLC system (nanoAcquity; Waters) as described previously (Ntoukakis et al., 2009). The entire TAIR10 (www.arabidopsis.org) and E. coli O157 databases were searched using Mascot (with the inclusion of sequences of common contaminants, such as keratins and trypsin). Parameters were set for 65 ppm peptide mass tolerance and allowing for Met oxidation and two missed tryptic cleavages. Carbamidomethylation of Cys residues was specified as a fixed modification, and oxidized Met and phosphorylation of Ser or Thr residues were allowed as variable modifications. Scaffold (v2_06_01; Proteome Software) was used to validate MS/MS-based peptide and protein identifications.
In Vitro Kinase Assay
The in vitro kinase assay was performed as described previously (Singh et al., 2013). Briefly, 2 µg of purified Trx-6xHis-IOS1KD and Trx-6xHis-IOS1KDm was incubated for 30 min at 28°C in 30 μL kinase buffer (50 mM Tris-Cl, pH 7.5, 50 mM KCl, 2 mM DTT, 10% [v/v] glycerol, 5 mM MnCl2, and 5 mM MgCl2). Phosphorylation was initiated with the addition of 10 mM ATP and terminated by adding 30 μL of 2× SDS page loading buffer. Of these, 30 μL were separated on a 8% polyacrylamide gel and the phosphorylation level of the proteins was detected using the Pro-Q Diamond Phosphoprotein Gel Stain (Invitrogen) according to the manufacturer's instructions. The fluorescent signal was imaged using a Typhoon 9400 scanner (Amersham Biosciences), and the same gel was subsequently stained for total protein with Coomassie Brilliant Blue.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: IOS1 (At1g51800), FRK1 (At2g19190), and UBQ10 (At4g05320).
Supplemental Data
Supplemental Figure 1. The mutant ios1-1 is a knockout but ios1-2 and ios1-3 produce some IOS1 transcripts.
Supplemental Figure 2. Susceptibility phenotypes of ios1-2 and bak1-5 to Pst DC3000.
Supplemental Figure 3. ios1 mutants demonstrate increased susceptibility to Pst DC3000 hrcC-.
Supplemental Figure 4. Resistance of ios1 mutants to necrotrophic fungal pathogens.
Supplemental Figure 5. IOS1 mRNA expression levels in two independent IOS1 overexpression lines.
Supplemental Figure 6. Stomatal innate immunity in ios1 mutants.
Supplemental Figure 7. Expression of IOS1 is upregulated by bacterial MAMPs.
Supplemental Figure 8. Early PTI responses.
Supplemental Figure 9. Bimolecular fluorescence complementation analyses of IOS1 interactions with EFR and BAK1.
Supplemental Figure 10. FLS2-FLS2 and IOS1-IOS1 dimerization.
Supplemental Figure 11. Complementation of defective MAPK activation in ios1-1 and ios1-2 mutants by IOS1-GFP.
Supplemental Figure 12. IOS1 in vitro autophosphorylation.
Supplemental Figure 13. A. brassicicola-mediated lesions in lines overexpressing IOS1.
Supplemental Figure 14. BABA does not regulate ligand-induced FLS2-BAK1 association.
Supplemental Table 1. Identification of IOS1 tryptic peptides by HPLC-ESI-MS/MS analysis of EFR immunoprecipitates.
Supplemental Table 2. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank the ABRC for providing seeds and constructs. We thank B.N. Kunkel, J. Glazebrook, and C.Y. Chen for the pathogens. We also thank P. He for providing the BIK1 construct and bik1 mutant seeds and D. Chinchilla for FLS2 antibody. We appreciate the help from the staff of Technology Commons, College of Life Science, National Taiwan University, in microscopy and for the RT-qPCR equipment. We thank the Proteomics Core Laboratory sponsored by the Institute of Plant and Microbial Biology and the Agricultural Biotechnology Research Center, Academia Sinica, for mass spectrometric protein identifications and analyses. We also thank A. Jones and J. Sklenar from TSL Proteomics for their excellent service. We acknowledge Y.S. Cheng and I.F. Chang for their technical support in protein expression and analyses as well as B. Schulze, M. Desclos-Theveniau, and members of Zimmerli’s laboratory for critical comments. This work was supported by the National Science Council of Taiwan Grants 99-2628-B-002-053-MY3 and 102-2628-B-002-011-MY3 (to L.Z.), a Frontier and Innovative Research grant of the National Taiwan University (code number 99R70436; to L.Z.), The European Research Council, and The Gatsby Charitable Foundation (to C.Z.). Y.K. was supported by fellowships from KAKENHI (#23580068), the Excellent Young Researcher Overseas Visit Program, and the Uehara memorial foundation. M.R. was part of the John Innes Centre/The Sainsbury Laboratory PhD Rotation Program.
AUTHOR CONTRIBUTIONS
Y.-H.Y., D.P., Y.K., C.Z., and L.Z. designed the research. Y.-H.Y., D.P., Y.K., Y.-C.H., P.-Y.H., C.-N.T., M.R., H.-C.C., T.-C.C., and P.-W.C performed research. Y.-H.Y., D.P., Y.K., P.-Y.H., M.R., C.Z., and L.Z. analyzed data. L.Z. wrote the article with contributions from Y.-H.Y., D.P., Y.K., and C.Z.
This article is a resubmission after the retraction of the Chen et al. (2014) article: Chen, C.W., Panzeri, D., Yeh, Y.H., Kadota, Y., Huang, P.Y., Tao, C.N., Roux, M., Chien, S.C., Chin, T.C., Chu, P.W., Zipfel, C., and Zimmerli, L. (2014). The Arabidopsis malectin-like leucine-rich repeat receptor-like kinase IOS1 associates with the pattern recognition receptors FLS2 and EFR and is critical for priming of pattern-triggered immunity. Plant Cell 26: 3201–3219. Retraction comments are available at in Plant Cell 27: 1563. Briefly, experiments performed by C.W. Chen, the first author of the 2014 article, had to be redone. Other figures were retained from the 2014 article as noted below.
For this new version of the manuscript, Y.-H.Y performed the experiments for the new Figures 1A, 1B, 3C, 3D, 4A, 4C, 4D, 5A, 8A, 8C, 9A, and 9B. Y.-C.H. performed experiments for the new Figures 2C (elf18), 7E, and 10C, while new Figures 2B, 2D, and 8B were done by Y.-H.Y and Y.-C.H. Experiments for the retained Figures 1C and 1D were performed by Y.-H.Y. and C.-N.T., respectively. Retained Figure 2A was done by H.-C.C. Experiments for retained Figures 2C (flg22), 3A, and 3B were performed by Y.-H.Y. Figure 4B was provided by P.-Y.H., while retained Figure 5B was done by Y.K. Retained Figures 6A and 6C were provided by Y.K., and experiments for Figures 6B and 6D were performed by D.P. Retained Figures 7A to 7D are from D.P. Retained Figure 10A was provided by Y.-H.Y. and C.-N.T., and retained Figure 10B was provided by H.-C.C. and T.-C.C. Experiments for retained Figure 10D were performed by C.-N.T. The new Supplemental Figures 2, 4, 5, 7, 8C to 8E, 9 to 11, 13, and 14 were provided by Y.-H.Y. Data provided by P.-W.C. were used for retained Supplemental Figure 1. Retained Supplemental Figures 3 and 6 were done by C.-N.T. Retained Supplemental Figures 8A and 8B are from Y.-H.Y. Experiments for retained Supplemental Figure 12 were performed by D.P. Data for retained Supplemental Table 1 are from M.R.
Glossary
- PRR
pattern recognition receptor
- MAMP
microbe-associated molecular pattern
- PTI
pattern-triggered immunity
- ROS
reactive oxygen species
- MAP
mitogen-activated protein
- ETI
effector-triggered immunity
- BABA
β-aminobutyric acid
- ABA
abscisic acid
- dpi
days postinoculation
- BiFC
bimolecular fluorescence complementation
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
References
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Beckers G.J.M., Jaskiewicz M., Liu Y., Underwood W.R., He S.Y., Zhang S., Conrath U. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H., Albert I., Fan L., Reinhard A., Nürnberger T. (2014). Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 20: 47–54. [DOI] [PubMed] [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.H., Sheen J. (2010). Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D.M., Hernández-Guzmán G., Kloek A.P., Alarcón-Chaidez F., Sreedharan A., Rangaswamy V., Peñaloza-Vázquez A., Bender C.L., Kunkel B.N. (2004). Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 17: 162–174. [DOI] [PubMed] [Google Scholar]
- Bücherl C.A., van Esse G.W., Kruis A., Luchtenberg J., Westphal A.H., Aker J., van Hoek A., Albrecht C., Borst J.W., de Vries S.C. (2013). Visualization of BRI1 and BAK1(SERK3) membrane receptor heterooligomers during brassinosteroid signaling. Plant Physiol. 162: 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Liang Y., Tanaka K., Nguyen C.T., Jedrzejczak R.P., Joachimiak A., Stacey G. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3: 03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500. [DOI] [PubMed] [Google Scholar]
- Cohen Y.R. (2002). beta-aminobutyric acid-induced resistance against plant pathogens. Plant Dis. 86: 448–457. [DOI] [PubMed] [Google Scholar]
- Conrath U., et al. ; Prime-A-Plant Group (2006). Priming: getting ready for battle. Mol. Plant Microbe Interact. 19: 1062–1071. [DOI] [PubMed] [Google Scholar]
- Cook D.N., Pisetsky D.S., Schwartz D.A. (2004). Toll-like receptors in the pathogenesis of human disease. Nat. Immunol. 5: 975–979. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desclos-Theveniau M., Arnaud D., Huang T.Y., Lin G.J., Chen W.Y., Lin Y.C., Zimmerli L. (2012). The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 8: e1002513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L., Rivas S. (2012). Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17: 644–655. [DOI] [PubMed] [Google Scholar]
- Durrant W.E., Dong X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42: 185–209. [DOI] [PubMed] [Google Scholar]
- Feng F., Zhou J.M. (2012). Plant-bacterial pathogen interactions mediated by type III effectors. Curr. Opin. Plant Biol. 15: 469–476. [DOI] [PubMed] [Google Scholar]
- Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C., Zhou J.M. (2012). A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485: 114–118. [DOI] [PubMed] [Google Scholar]
- Gassmann W., Bhattacharjee S. (2012). Effector-triggered immunity signaling: from gene-for-gene pathways to protein-protein interaction networks. Mol. Plant Microbe Interact. 25: 862–868. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Hann D.R., Ntoukakis V., Petutschnig E., Lipka V., Rathjen J.P. (2009). AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr. Biol. 19: 423–429. [DOI] [PubMed] [Google Scholar]
- Girardin S.E., Sansonetti P.J., Philpott D.J. (2002). Intracellular vs extracellular recognition of pathogens--common concepts in mammals and flies. Trends Microbiol. 10: 193–199. [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gou X., He K., Yang H., Yuan T., Lin H., Clouse S.D., Li J. (2010). Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter T., et al. (2014). The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24: 134–143. [DOI] [PubMed] [Google Scholar]
- Häweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., Robatzek S., von Schaewen A. (2010). Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285: 4629–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Shan L., Sheen J. (2007). The use of protoplasts to study innate immune responses. Methods Mol. Biol. 354: 1–9. [DOI] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M.E., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge S., Thompson G.A., Powell G. (2005). Application of DL-beta-aminobutyric acid (BABA) as a root drench to legumes inhibits the growth and reproduction of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphididae). Bull. Entomol. Res. 95: 449–455. [DOI] [PubMed] [Google Scholar]
- Hok S., et al. (2014). The receptor kinase IMPAIRED OOMYCETE SUSCEPTIBILITY1 attenuates abscisic acid responses in Arabidopsis. Plant Physiol. 166: 1506–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hok S., Danchin E.G.J., Allasia V., Panabières F., Attard A., Keller H. (2011). An Arabidopsis (malectin-like) leucine-rich repeat receptor-like kinase contributes to downy mildew disease. Plant Cell Environ. 34: 1944–1957. [DOI] [PubMed] [Google Scholar]
- Huang P.Y., Yeh Y.H., Liu A.C., Cheng C.P., Zimmerli L. (2014). The Arabidopsis LecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity. Plant J. 79: 243–255. [DOI] [PubMed] [Google Scholar]
- Huang P.Y., Zimmerli L. (2014). Enhancing crop innate immunity: new promising trends. Front. Plant Sci. 5: 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.Y., Desclos-Theveniau M., Chien C.T., Zimmerli L. (2013). Arabidopsis thaliana transgenics overexpressing IBR3 show enhanced susceptibility to the bacterium Pseudomonas syringae. Plant Biol (Stuttg) 15: 832–840. [DOI] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C.A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 103: 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab G., Cottier V., Toquin V., Rigoli G., Zimmerli L., Metraux J.P., Mauch-Mani B. (2001). beta-Aminobutyric acid-induced resistance in plants. Eur. J. Plant Pathol. 107: 29–37. [Google Scholar]
- Jakab G., Ton J., Flors V., Zimmerli L., Métraux J.P., Mauch-Mani B. (2005). Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 139: 267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz M., Conrath U., Peterhänsel C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12: 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Macho A.P., Zipfel C. (2016). Immunoprecipitation of plasma membrane receptor-like kinases for identification of phosphorylation sites and associated proteins. Methods Mol. Biol. 1363: 133–144. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Zipfel C. (2015). Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56: 1472–1480. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D.G., Shirasu K., Menke F., Jones A., Zipfel C. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54: 43–55. [DOI] [PubMed] [Google Scholar]
- Kemmerling B., Halter T., Mazzotta S., Mosher S., Nürnberger T. (2011). A genome-wide survey for Arabidopsis leucine-rich repeat receptor kinases implicated in plant immunity. Front. Plant Sci. 2: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q., Sun T., Qu N., Ma J., Li M., Cheng Y.T., Zhang Q., Wu D., Zhang Z., Zhang Y. (2016). Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis in Arabidopsis. Plant Physiol. 171: 1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick D.A., McMichael C., Walker K.A., Anderson J.C., Bednarek S.Y., Heese A. (2010). Novel functions of Stomatal Cytokinesis-Defective 1 (SCD1) in innate immune responses against bacteria. J. Biol. Chem. 285: 23342–23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E., Mentzel T., Chinchilla D., Boller T., Felix G., Kemmerling B., Postel S., Arents M., Jeworutzki E., Al-Rasheid K.A., Becker D., Hedrich R. (2010). Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285: 13471–13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li M., Yu L., Zhou Z., Liang X., Liu Z., Cai G., Gao L., Zhang X., Wang Y., Chen S., Zhou J.M. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15: 329–338. [DOI] [PubMed] [Google Scholar]
- Liang X., Ding P., Lian K., Wang J., Ma M., Li L., Li L., Li M., Zhang X., Chen S., Zhang Y., Zhou J.M. (2016). Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife 5: e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Liu Z., Song C., Hu Y., Han Z., She J., Fan F., Wang J., Jin C., Chang J., Zhou J.M., Chai J. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336: 1160–1164. [DOI] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010a). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., He P., Shan L. (2010b). Phosphorylation of receptor-like cytoplasmic kinases by bacterial flagellin. Plant Signal. Behav. 5: 598–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S.E., Keller W.A., Tsang E.W., Harada J.J., Rothstein S.J., Cui Y. (2010c). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61: 259–270. [DOI] [PubMed] [Google Scholar]
- Luna E., Bruce T.J., Roberts M.R., Flors V., Ton J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158: 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Kufer T.A., Schulze-Lefert P. (2011). NLR functions in plant and animal immune systems: so far and yet so close. Nat. Immunol. 12: 817–826. [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980. [DOI] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J., Zipfel C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15: 349–357. [DOI] [PubMed] [Google Scholar]
- Návarová H., Bernsdorff F., Döring A.C., Zeier J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nicaise V., Roux M., Zipfel C. (2009). Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol. 150: 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis V., Mucyn T.S., Gimenez-Ibanez S., Chapman H.C., Gutierrez J.R., Balmuth A.L., Jones A.M., Rathjen J.P. (2009). Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 324: 784–787. [DOI] [PubMed] [Google Scholar]
- Nühse T.S., Peck S.C., Hirt H., Boller T. (2000). Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 275: 7521–7526. [DOI] [PubMed] [Google Scholar]
- Oka Y., Cohen Y., Spiegel Y. (1999). Local and systemic induced resistance to the root-knot nematode in tomato by DL-beta-amino-n-butyric acid. Phytopathology 89: 1138–1143. [DOI] [PubMed] [Google Scholar]
- Po-Wen C., Singh P., Zimmerli L. (2013). Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol. Plant Pathol. 14: 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins T.W., Tudzynski P., Tiedemann A.V., Tudzynski B., Have A.T., Hansen M.E., Tenberge K., van Kan J.A.L. (2000). Infection strategies of Botrytis cinerea and related necrotrophic pathogens. In Fungal Pathology, Kronstad J., ed (Kluwer Academic Publishers; ), pp. 33–64. [Google Scholar]
- Ranf S., Eschen-Lippold L., Pecher P., Lee J., Scheel D. (2011). Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 68: 100–113. [DOI] [PubMed] [Google Scholar]
- Rasmann S., De Vos M., Casteel C.L., Tian D., Halitschke R., Sun J.Y., Agrawal A.A., Felton G.W., Jander G. (2012). Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 158: 854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Chinchilla D., Boller T. (2006). Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 20: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux M., Schwessinger B., Albrecht C., Chinchilla D., Jones A., Holton N., Malinovsky F.G., Tör M., de Vries S., Zipfel C. (2011). The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C.A., Huffaker A., Yamaguchi Y. (2007). New insights into innate immunity in Arabidopsis. Cell. Microbiol. 9: 1902–1908. [DOI] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A.K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285: 9444–9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B., Roux M., Kadota Y., Ntoukakis V., Sklenar J., Jones A., Zipfel C. (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C., Feike D., Gimenez-Ibanez S., Hann D.R., Zipfel C., Rathjen J.P. (2011). Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol. 156: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L., He P., Li J., Heese A., Peck S.C., Nürnberger T., Martin G.B., Sheen J. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Shen Q., Qi Y., Yan H., Nie H., Chen Y., Zhao T., Katagiri F., Tang D. (2013). BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 25: 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T.L., Shimada T., Hara-Nishimura I. (2010). A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61: 519–528. [DOI] [PubMed] [Google Scholar]
- Singh P., Zimmerli L. (2013). Lectin receptor kinases in plant innate immunity. Front. Plant Sci. 4: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Kuo Y.C., Mishra S., Tsai C.H., Chien C.C., Chen C.W., Desclos-Theveniau M., Chu P.W., Schulze B., Chinchilla D., Boller T., Zimmerli L. (2012). The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 24: 1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Chien C.C., Mishra S., Tsai C.H., Zimmerli L. (2013). The Arabidopsis LECTIN RECEPTOR KINASE-VI.2 is a functional protein kinase and is dispensable for basal resistance to Botrytis cinerea. Plant Signal. Behav. 8: e22611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter A., Daniel X., Flors V., Luna E., Hohn B., Mauch-Mani B. (2012). Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 158: 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekanta S., Bethke G., Hatsugai N., Tsuda K., Thao A., Wang L., Katagiri F., Glazebrook J. (2015). The receptor-like cytoplasmic kinase PCRK1 contributes to pattern-triggered immunity against Pseudomonas syringae in Arabidopsis thaliana. New Phytol. 207: 78–90. [DOI] [PubMed] [Google Scholar]
- Sun W., Cao Y., Jansen Labby K., Bittel P., Boller T., Bent A.F. (2012). Probing the Arabidopsis flagellin receptor: FLS2-FLS2 association and the contributions of specific domains to signaling function. Plant Cell 24: 1096–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li L., Macho A.P., Han Z., Hu Z., Zipfel C., Zhou J.M., Chai J. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342: 624–628. [DOI] [PubMed] [Google Scholar]
- Tena G., Boudsocq M., Sheen J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14: 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J., Mauch-Mani B. (2004). Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38: 119–130. [DOI] [PubMed] [Google Scholar]
- Tsai C.H., Singh P., Chen C.W., Thomas J., Weber J., Mauch-Mani B., Zimmerli L. (2011). Priming for enhanced defence responses by specific inhibition of the Arabidopsis response to coronatine. Plant J. 65: 469–479. [DOI] [PubMed] [Google Scholar]
- Tsuda K., Katagiri F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13: 459–465. [DOI] [PubMed] [Google Scholar]
- Van Wees S.C.M., Van der Ent S., Pieterse C.M.J. (2008). Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11: 443–448. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wan J., Tanaka K., Zhang X.C., Son G.H., Brechenmacher L., Nguyen T.H., Stacey G. (2012). LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in Arabidopsis. Plant Physiol. 160: 396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Zhang X.C., Neece D., Ramonell K.M., Clough S., Kim S.Y., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann R., et al. (2011). Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 108: 19824–19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windram O., et al. (2012). Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 24: 3530–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.C., Singh P., Chen M.C., Zimmerli L. (2010). L-Glutamine inhibits beta-aminobutyric acid-induced stress resistance and priming in Arabidopsis. J. Exp. Bot. 61: 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., He P., Abramovitch R.B., Dawson J.E., Nicholson L.K., Sheen J., Martin G.B. (2007). The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J. 52: 595–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Xie J., Yan C., Zou X., Ren D., Zhang S. (2014a). A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77: 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Xu S.L., Li Z.J., Tang W., Burlingame A.L., Wang Z.Y. (2014b). A brassinosteroid-signaling kinase interacts with multiple receptor-like kinases in Arabidopsis. Mol. Plant 7: 441–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A., Bryan A.C., Tax F.E., Ryan C.A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Pearce G., Ryan C.A. (2006). The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 103: 10104–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.H., Chang Y.H., Huang P.Y., Huang J.B., Zimmerli L. (2015). Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 6: 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zeng W., Melotto M., He S.Y. (2010). Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr. Opin. Biotechnol. 21: 599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhou J.M. (2010). Plant immunity triggered by microbial molecular signatures. Mol. Plant 3: 783–793. [DOI] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301. [DOI] [PubMed] [Google Scholar]
- Zimmerli L., Hou B.H., Tsai C.H., Jakab G., Mauch-Mani B., Somerville S. (2008). The xenobiotic beta-aminobutyric acid enhances Arabidopsis thermotolerance. Plant J. 53: 144–156. [DOI] [PubMed] [Google Scholar]
- Zimmerli L., Jakab G., Metraux J.P., Mauch-Mani B. (2000). Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta-aminobutyric acid. Proc. Natl. Acad. Sci. USA 97: 12920–12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli L., Métraux J.P., Mauch-Mani B. (2001). beta-Aminobutyric acid-induced protection of Arabidopsis against the necrotrophic fungus Botrytis cinerea. Plant Physiol. 126: 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2014). Plant pattern-recognition receptors. Trends Immunol. 35: 345–351. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S. (2010). Pathogen-associated molecular pattern-triggered immunity: veni, vidi...? Plant Physiol. 154: 551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D.G., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.