Abstract
Benzylisoquinoline alkaloids are a large group of plant-specialized metabolites displaying an array of biological and pharmacological properties associated with numerous structural scaffolds and diverse functional group modification. N-Methylation is one of the most common tailoring reactions, yielding tertiary and quaternary pathway intermediates and products. Two N-methyltransferases accepting (i) early 1-benzylisoquinoline intermediates possessing a secondary amine and leading to the key branch-point intermediate (S)-reticuline and (ii) downstream protoberberines containing a tertiary amine and forming quaternary intermediates destined for phthalideisoquinolines and antimicrobial benzo[c]phenanthridines were previously characterized. We report the isolation and characterization of a phylogenetically related yet functionally distinct N-methyltransferase (NMT) from opium poppy (Papaver somniferum) that primarily accepts 1-benzylisoquinoline and aporphine substrates possessing a tertiary amine. The preferred substrates were the R and S conformers of reticuline and the aporphine (S)-corytuberine, which are proposed intermediates in the biosynthesis of magnoflorine, a quaternary aporphine alkaloid common in plants. Suppression of the gene encoding reticuline N-methyltransferase (RNMT) using virus-induced gene silencing in opium poppy resulted in a significant decrease in magnoflorine accumulation and a concomitant increase in corytuberine levels in roots. RNMT transcript levels were also most abundant in roots, in contrast to the distribution of transcripts encoding other NMTs, which occur predominantly in aerial plant organs. The characterization of a third functionally unique NMT involved in benzylisoquinoline alkaloid metabolism will facilitate the establishment of structure-function relationships among a large group of related enzymes.
Keywords: biosynthesis, enzyme catalysis, functional genomics, plant biochemistry, secondary metabolism, N-methyltransferase, aporphine alkaloid, benzylisoquinoline alkaloid, biochemical genomics, magnoflorine
Introduction
Magnoflorine is a quaternary benzylisoquinoline alkaloid (BIA)2 occurring as a specialized metabolite in several angiosperm orders including the Ranunculales, Laurales, Sapindales, Piperales, Rosales, Malpighiales, and Magnoliales (1). Key physiological and ecological roles for magnoflorine in plants can be predicted on the basis of its widespread taxonomic distribution, potent enzyme inhibitory, and cytotoxic effects and diverse pharmacological properties that include potential anti-diabetic, anti-inflammatory, antimicrobial, antitumor, sedative, and anxiolytic applications (2–6). In the Ranunculaceae family, magnoflorine is typically found in subterranean organs where it has been purported to function as an allelopathic agent or in the defense against pathogens (7, 8). The ecological functions of magnoflorine also appear to include plant-insect interactions. The larvae of the Battus polydamus butterfly were shown to accumulate magnoflorine after feeding on Aristolochia in manner similar to the defensive sequestration of anti-feedant compounds, such as the aristolochic acids (9).
As with all BIAs, the biosynthesis of magnoflorine begins with the condensation of two tyrosine derivatives, dopamine and 4-hydroxyphenylacetaldehyde, which yields the first committed pathway intermediate (S)-norcoclaurine (Fig. 1). Subsequent O-methylations, N-methylation, and aromatic ring hydroxylation yield (S)-reticuline, which serves as a key branch point intermediate to several BIA subclasses with structurally diverse scaffolds. Notable BIAs (and their structural subclasses) include the narcotic analgesic morphine (morphinan), the antitussive and potential anticancer drug noscapine (phthalideisoquinoline), the vasodilator papaverine (1-benzylisoquinoline), and the antimicrobials berberine (protoberberine) and sanguinarine (benzo[c]phenanthridine) (10).
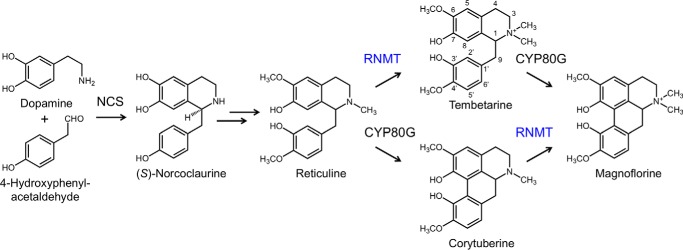
FIGURE 1.
Biosynthesis of the quaternary alkaloid magnoflorine in opium poppy. The condensation of dopamine and 4-HPAA by norcoclaurine synthase (NCS) yields the central BIA intermediate, (S)-norcoclaurine, which is converted to (S)-reticuline via two O-methylations, an N-methylation, and an aromatic ring hydroxylation. (S)-Reticuline can subsequently undergo stereochemical inversion yielding (R)-reticuline. Carbon-carbon phenol coupling of reticuline by a cytochrome P450 related to C. japonica CYP80G2 (11) putatively establishes the aporphine scaffold in corytuberine, which can be N-methylated to magnoflorine. Alternatively, the N-methylation of reticuline yields the quaternary alkaloid tembetarine, which may also serve as a substrate for a CYP80G2-like enzyme forming magnoflorine via an alternative route. Reticuline N-methyltransferase (RNMT) is shown herein to efficiently catalyze the N-methylation of (S)-reticuline, (R)-reticuline, and (S)-corytuberine in support of the pathways shown.
Magnoflorine is a member of the aporphine subclass of BIAs, which are formed from reticuline via carbon-carbon phenol coupling involving C8 of the isoquinoline moiety and either C2′ (e.g. magnoflorine) or C6′ (e.g. glaucine) of the benzyl group (Fig. 1). Formation of the aporphine scaffold is catalyzed by the cytochrome P450 corytuberine synthase (CYP80G2), which has been isolated and characterized from Coptis japonica (Ranunculaceae) (11). CYP80G2 efficiently converted (S)-reticuline to (S)-corytuberine, which was assumed to undergo N-methylation yielding magnoflorine. Alternatively, an initial N-methylation of reticuline would yield tembetarine, which hypothetically could also serve as a substrate for C8-C2′ coupling. Early tracer experiments using Cocculus laurifolius (Menispermaceae) showed the incorporation of both radiolabeled reticuline and tembetarine into magnoflorine, suggesting that either route is possible or that C8-C2′ bond formation occurs subsequent to N-methylation (12). However, CYP80G2 did not accept tembetarine as a substrate supporting a role only for corytuberine as an intermediate in magnoflorine biosynthesis, at least in C. japonica (11). Moreover, corytuberine, but not tembetarine, was detected in Podophyllum spp., which also accumulate magnoflorine (Ranunculaceae) (8). Although C. japonica coclaurine N-methyltransferase (CNMT) was reported to accept (S)-corytuberine as a minor substrate (13), an N-methyltransferase (NMT) specifically involved in magnoflorine biosynthesis has not been isolated. Magnoflorine contains a chiral center, although the stereochemistry of the compound has not been determined in most plants, including opium poppy.
Three related, yet functionally distinct, NMT isoforms involved in BIA metabolism have been isolated and characterized: CNMT (13, 14), tetrahydroprotoberberine cis-N-methyltransferase (TNMT) (15), and pavine N-methyltransferase (PavNMT) (16). Enzymes classified as a CNMT display a relatively broad substrate range but predominantly target the secondary amine of the 1-benzylisoquinoline (S)-coclaurine yielding (S)-N-methylcoclaurine, which is an intermediate in the formation of (S)-reticuline (13, 14). In contrast, TNMT enzymes show greater specificity in accepting various protoberberine substrates containing tertiary amines and yielding products with quaternary nitrogen atoms (15, 16). PavNMT from Thalictrum flavum (Ranunculaceae) is the only partially characterized member of an isoform group distinct from CNMT and TNMT because of its predominant activity on the secondary amine of (+/−)-pavine, which possesses a different scaffold structure compared with benzylisoquinolines and protoberberines (16). The biosynthesis of magnoflorine and related quaternary apophines (Fig. 1) requires an NMT that efficiently targets the tertiary amines in 1-benzylisoquinoline (e.g. (S)-reticuline) or aporphine (e.g. corytuberine) substrates. Such an NMT is not represented among the characterized enzymes. In this paper, we report the isolation and characterization of a cDNA encoding reticuline N-methyltransferase (RNMT) from opium poppy (Papaver somniferum), and we demonstrate a physiological role for the enzyme in the biosynthesis of magnoflorine in the plant.
Results
Isolation and Phylogenetic Analysis
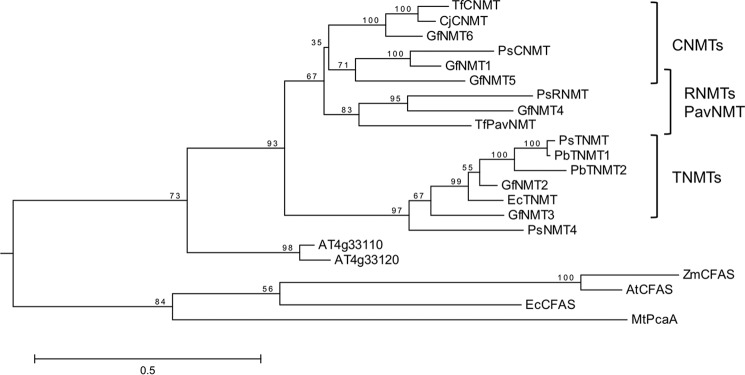
Opium poppy transcripts encoding two putative NMTs were identified in stem and root transcriptome databases based on their predicted amino acid sequence identity with CNMT and TNMT (51 and 61%, respectively). The isolated NMT3 (RNMT) cDNA perfectly matched the nucleotide sequence identity of assembled contigs discovered in the transcriptome analysis, whereas the NMT4 cDNA contained 12 mismatched nucleotides resulting in four amino acid substitutions. The predicted RNMT and NMT4 translation products featured molecular masses of 41.9 and 40.7 kDa and predicted pI values of 5.9 and 5.2, respectively. RNMT showed 51, 44, and 46% amino acid identity with respect to opium poppy CNMT, TNMT, and NMT4, whereas NMT4 displayed 49 and 60% amino acid identity with respect to CNMT and TNMT, respectively. Phylogenetic relationships among functionally characterized NMTs implicated in BIA metabolism from opium poppy and other plant species, and a selection of proteins with relevant amino acid identity, placed RNMT in a subclade containing PavNMT from T. flavum and a partially characterized NMT from Glaucium flavum (GfNMT4), which preferentially N-methylate (S)-coclaurine and (+/−)-pavine, respectively (Fig. 2). In contrast, opium poppy NMT4 emerged as the most distal member of a subclade containing only TNMTs.
FIGURE 2.
Rooted maximum likelihood phylogenetic tree for characterized NMTs involved in BIA metabolism and related enzymes. Bootstrap frequencies displayed at each node are percentages of 500 bootstrap replicates. The analysis involved 22 amino acid sequences and a total of 222 positions following the elimination of gaps and missing data. Protein names and accession numbers are provided under “Experimental Procedures.” The scale bar represents the length corresponding to 0.5 substitution/site.
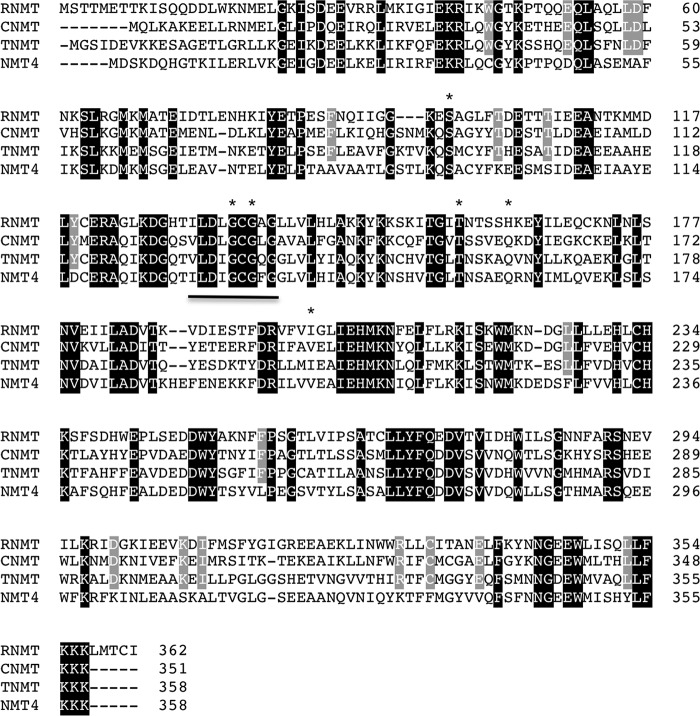
Amino acid sequence alignment of all four characterized and candidate NMTs from opium poppy showed the conservation of several residues, including a canonical GXGXG motif and four of six additional residues reportedly involved in S-(5′-adenosyl)-l-methionine iodide (AdoMet) binding (Fig. 3) (17, 18). Fifteen conserved residues in CNMT, TNMT, and RNMT were substituted in NMT4. The TargetP program (19) did not detect a putative cellular targeting sequence associated with the deduced amino acid sequences, suggesting the cytoplasmic localization of all four enzymes. Interestingly, CNMT, TNMT, and NMT4 terminate in a potential tri-Lys (KKK) ER retention signal (20), whereas RNMT extends by five additional residues.
FIGURE 3.
Alignment of opium poppy RNMT, CNMT, TNMT, and NMT4. Residues conserved across the four NMT sequences are shaded in black, whereas residues differing only in NMT4 are shaded in gray. Residues implicated in AdoMet binding are indicated with asterisks (17). The conserved AdoMet-dependent methyltransferase region, motif I (18), is underlined.
Functional Characterization
Recombinant His6-tagged RNMT and NMT4 produced in Escherichia coli were purified using cobalt affinity chromatography (supplemental Fig. S1). The purified proteins showed apparent molecular masses of 45 and 43 kDa, respectively, which are consistent with the masses of the predicted translation products. Both proteins were screened for enzymatic activity using 32 potential BIA substrates displaying a wide range of structural scaffolds and functional group modifications (Table 1). Reaction products consistent with N-methylation (i.e. a gain of 14 atomic mass units with respect to the substrate) were detected for RNMT. In contrast, NMT4 failed to show conversion of any of the tested BIA at both pH 7.0 and 8.5. RNMT accepted a variety of BIAs with three distinctly different structural scaffolds: 1-benzylisoquinoline, aporphine, and pthalideisoquinoline. Within each structural subclass, RNMT showed a strong preference for substrates with a tertiary amine, although alkaloids containing a secondary amine (e.g. (R,S)-tetrahydropapaverine and, to a lesser extent, (S)-coclaurine) were also accepted as substrates. In the case of substrates with secondary amines, RNMT catalyzed two sequential N-methylations yielding detectable products showing an increase in 14 and 28 atomic mass units. The substrate showing the highest relative conversion rate was (R)-reticuline, with the turnover rate for (S)-reticuline reduced by ∼50% (Fig. 4). The aporphine alkaloid (S)-corytuberine displayed the second highest turnover rate after (R)-reticuline, and other aporphines (i.e. (+)-glaucine and (+)-bulbocapnine) showed substantial conversion. The phthalideisoquinoine noscapine was accepted at relatively lower levels. NMR spectroscopy confirmed the identification of (S)-tembetarine as the RNMT reaction product obtained from (S)-reticuline (supplemental Table S1 and supplemental Data File S1).
TABLE 1.
Substrate range for RNMT
Substrate conversion values represent the means of the percentages of maximum activity ± S.D. for three independent replicates, with the single replicate showing the highest substrate consumption overall set to 100%. The values for (S)-coclaurine and (S)-tetrahydropapaverine include reaction products with increases of both 14 and 28 atomic mass units. nd, not detected. Indicated variable groups on core structures correspond to the specific alkaloids tested: 1-benzylisoquinoline, R1 = H or CH3, R2 and R3 = OH or OCH3; protoberberine, R1, R2, R3 and R4 = OH, OCH3 or OCH2O; protopine, R1 and R2 = OCH3 or OCH2O; aporphine, R1 and R2 = OH, OCH3, or OCH2O; R3, R4, and R5 = H, OH, or OCH3; morphinan, R1, and R2 = OH or OCH3 (with corresponding double bond adjustment in thebaine); secoberbine, R1 = OH or CHO, R2 = H, OH, or OAc, R3 = H, OH, or OCH3; phthalideisoquinoline, R1 = H, OH or OCH3, R2 = H, OH or OCH3, R3 and R4 = OCH3 or OCH2O.
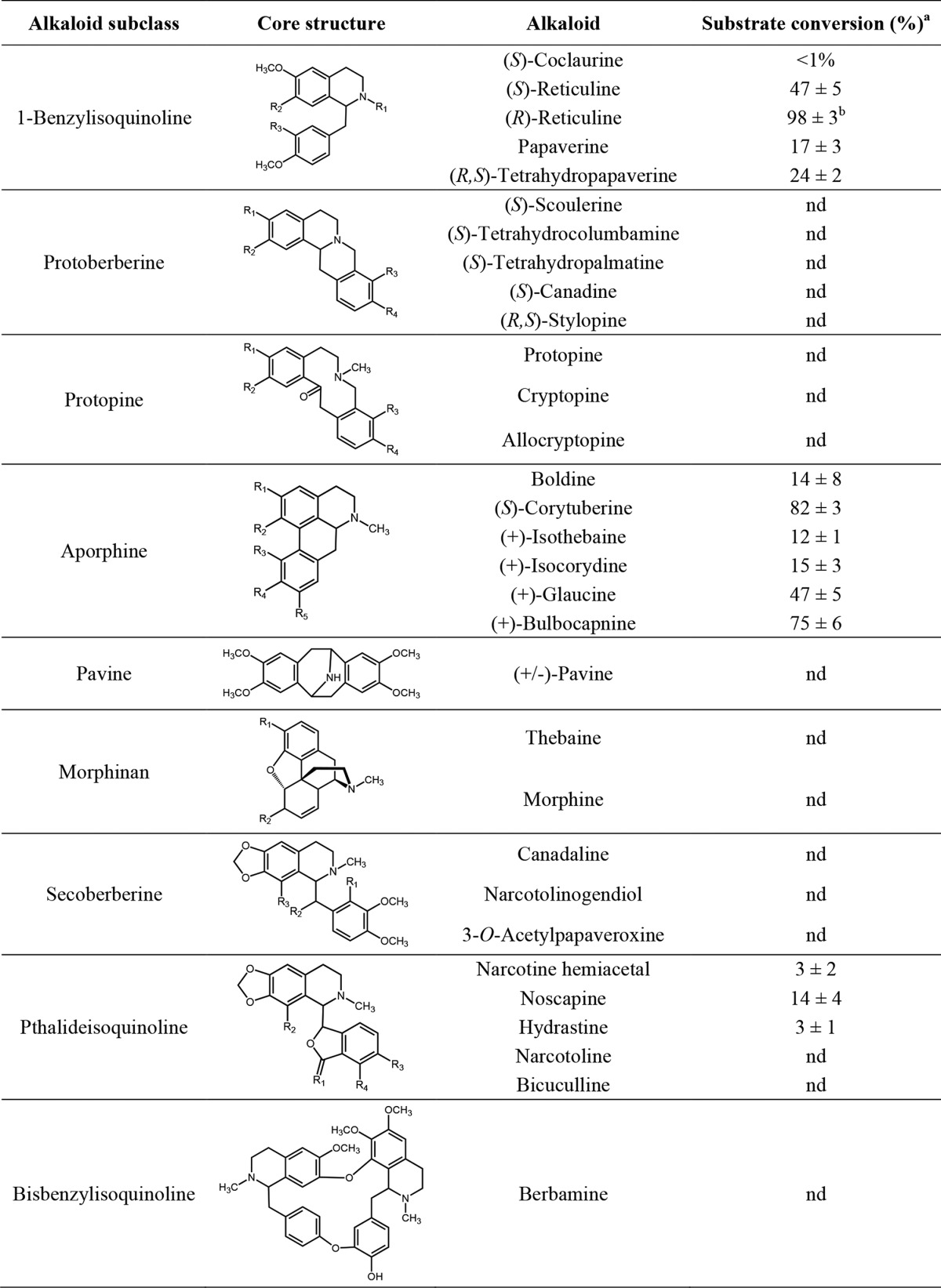
FIGURE 4.
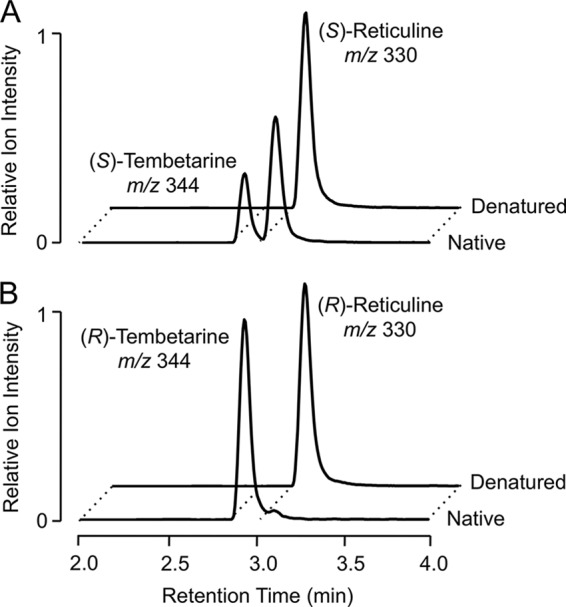
Extracted ion chromatograms of enzyme assays showing the N-methylation of (S)-reticuline and (R)-reticuline by RNMT. A, assays performed using (S)-reticuline (m/z 330) as the substrate and yielding (S)-tembetarine (m/z 344) as the product. B, assays performed using (R)-reticuline (m/z 330) as the substrate and yielding (R)-tembetarine (m/z 344) as the product. Boiled enzyme extracts were used as the negative controls. The reaction products were identified as tembetarine by NMR (supplemental Table S1).
RNMT showed a pH optimum of 8.5 using (R)-reticuline as the substrate, but a relatively broader optimum between pH 7.0 and 9.0 for (S)-reticuline (supplemental Fig. S2). With both substrates, maximum RNMT activity was detected at 30 °C. Consequently, the determination of kinetic parameters was performed at pH 7.0 and 30 °C. RNMT exhibited Km values of 42 and 85 μm for (S)-reticuline and (R)-reticuline, respectively (Table 2 and supplemental Fig. S2). The maximum reaction rates for each substrate at saturation were estimated at 39.6 and 74.8 pmol min−1 μg−1, with (S)-reticuline showing modest inhibition at concentrations ≥500 μm. RNMT displayed an apparent Km of 168 μm for AdoMet with no indication of inhibition at higher concentrations. Under saturating conditions, the number of (S)-reticuline and (R)-reticuline molecules turned over per second (kcat) was calculated as 0.028 and 0.053, respectively (Table 2). Overall, catalytic efficiencies (kcat/Km) were similar for (S)-reticuline and (R)-reticuline at 655 and 618 s−1 m−1, respectively.
TABLE 2.
Michaelis-menten enzyme kinetic parameters for RNMT with respect to (S)-reticuline, (R)-reticuline, and AdoMet
Parameters for AdoMet were determined at a fixed, saturating concentration of (R)-reticuline.
| Substrate | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|
| μm | pmol min−1 μg−1 protein | s−1 | s−1 m−1 | |
| (S)-Reticuline | 42 ± 3 | 39.6 ± 0.9 | 0.028 | 655 |
| (R)-Reticuline | 85 ± 11 | 74.8 ± 3.6 | 0.053 | 618 |
| AdoMet | 168 ± 8 | 66.5 ± 1.4 | 0.047 | 280 |
Virus-induced Gene Silencing
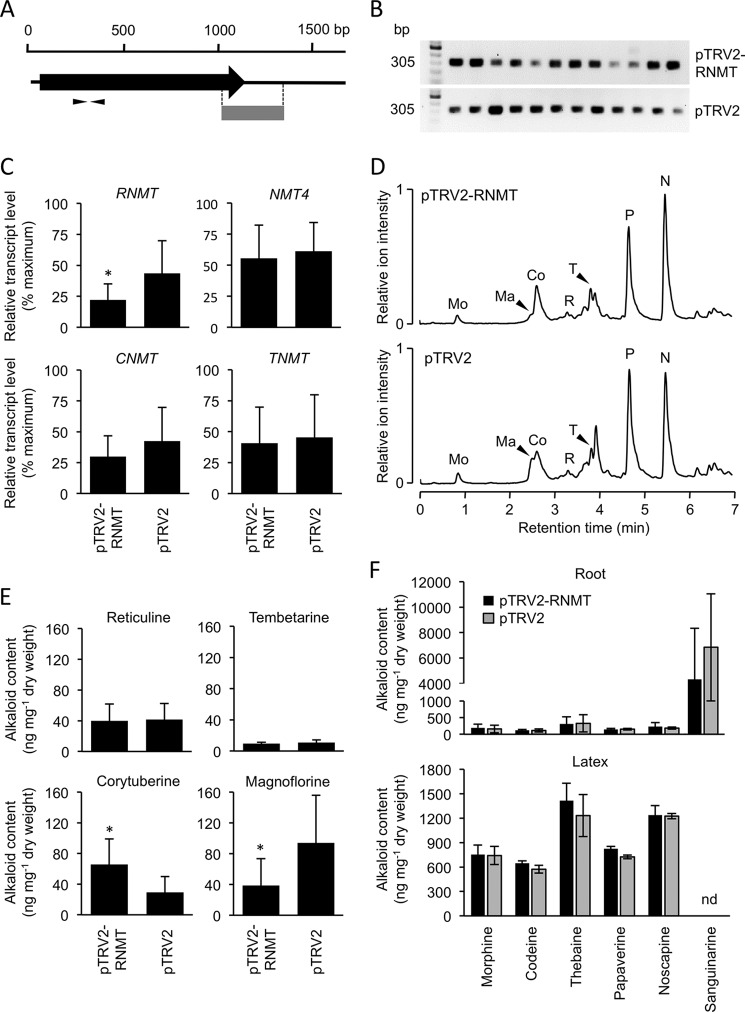
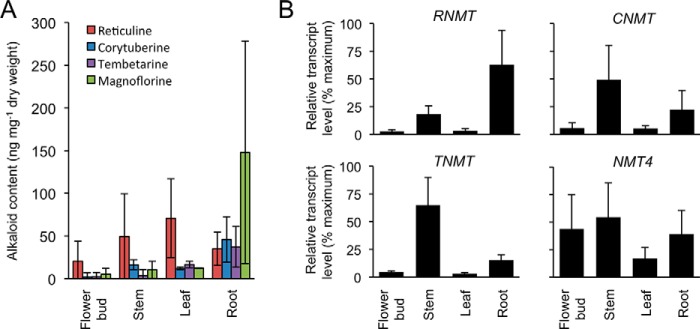
The physiological significance of RNMT in opium poppy BIA biosynthesis was investigated by knockdown of the cognate gene transcript levels using VIGS (Fig. 5A). Screening mature opium poppy plants for coat protein transcripts by RT-PCR revealed 12 TRV-infected individuals previously exposed to Agrobacterium tumefaciens containing either the empty pTRV2 vector or the pTRV2-RNMT construct (Fig. 5B). Analysis by qRT-PCR showed that RNMT transcript levels were significantly (p ≤ 0.0340) reduced in plants treated with pTRV2-RNMT compared with those treated with the empty pTRV2 vector (Fig. 5C). RT-PCR confirmed the suppression of RNMT transcript levels (supplemental Fig. S3 A). Transcript levels for paralogous genes (i.e. CNMT, TNMT, and NMT4) were not significantly affected. Plants with suppressed RNMT transcript levels showed specific changes in root alkaloid profile compared with controls (Fig. 5D). However, the relative abundance of magnoflorine was significantly (p ≤ 0.0038) reduced in RNMT-silenced plants compared with controls (Fig. 5E). In contrast, the relative abundance of corytuberine was significantly (p ≤ 0.0138) increased, whereas levels of tembetarine and reticuline were not affected. Major alkaloids in both root and latex were not altered (Fig. 5F). Relative RNMT transcript levels in individual RNMT-silenced and control plants showed a strong correlation with the accumulation of magnoflorine, whereas the relationship with corytuberine abundance was more variable (supplemental Fig. S3B).
FIGURE 5.
Effect of RNMT suppression on the accumulation of magnoflorine in opium poppy roots. A, position of the region targeted for virus-induced gene silencing (gray block) and the sites of the qRT-PCR primers on the RNMT transcript. B, detection of TRV2 coat protein transcripts by RT-PCR showing the infection of 12 RNMT-silenced (pTRV2-RNMT) and 12 control (pTRV2) plants. C, qRT-PCR analysis of infected plants showing significant (p < 0.05) and specific knockdown of transcript levels encoding RNMT, NMT4, CNMT, and TNMT in RNMT-silenced versus control plants. D, representative total ion chromatograms of root alkaloid extracts for RNMT-silenced and control plants. The major alkaloid peaks are indicated: Mo, morphine; Ma, magnoflorine; Co, codeine; R, reticuline; T, thebaine; P, papaverine; N, noscapine. E, abundance of reticuline, tembetarine, corytuberine, and magnoflorine in roots of RNMT-silenced and control plants. F, abundance of major alkaloids in roots and latex of RNMT-silenced and control plants. All values represent the means ± S.D. of 12 individuals. Statistical significance was calculated using Welch's t test.
Organ-specific Transcript and Alkaloid Levels
In mature opium poppy plants, magnoflorine accumulation was highest in root and was also detected in flower bud, stem, and leaf (Fig. 6A). Corytuberine and tembetarine were also most abundant in root but accumulated to lower levels than magnoflorine. Reticuline accumulation showed the most variation among individual plants and occurred at more consistent levels in different organs. RNMT transcript levels were also highest in root, followed by stem, and were detected at low relative abundance in flower bud and leaf (Fig. 6B). In contrast, CNMT and TNMT transcripts were most abundant in stem and showed relatively higher accumulation in root compared with flower bud and leaf. NMT4 transcripts were more uniformly distributed in different organs but were lowest in leaf.
FIGURE 6.
Relative abundance of magnoflorine pathway alkaloids and NMT transcripts in different opium poppy organs. LC-MS analysis (A) and qRT-PCR (B) were performed using alkaloids and total RNA isolated from five individual plants. For qRT-PCR analysis, the geometric mean of actin and ubiquitin was used as internal reference, and the sample with the highest ΔCt served as calibrator for each transcript. Relative transcript abundance (2−ΔΔCt) was converted to the percentage of maximum.
Discussion
We have shown that an uncharacterized cDNA from opium poppy with homology to known NMTs involved in BIA metabolism efficiently catalyzes the previously unreported N-methylation of both S and R conformers of the 1-benzylisoquinoline alkaloid reticuline yielding the corresponding enantiomers of tembetarine, a quaternary ammonium compound and putative intermediate in the biosynthesis of magnoflorine. RNMT also displayed substantial activity on the aporphine alkaloid (S)-corytuberine, formed via the C8-C2′ phenol coupling of (S)-reticuline, yielding magnoflorine, which also contains a quaternary amine. Gene-specific suppression of RNMT transcript levels in opium poppy plants confirmed the physiological role of RNMT in magnoflorine biosynthesis. Previously, formation of the quaternary amine in magnoflorine was assigned to CNMT because of a low efficiency activity on (S)-corytuberine (11, 16). We also show that RNMT is preferentially expressed in opium poppy root, which accumulates magnoflorine to the highest levels compared with aerial plant organs. In contrast, transcripts encoding CNMT and TNMT, which are primarily involved in the formation of the central branch point intermediate (S)-reticuline and the major latex alkaloid noscapine, occur most abundantly in aerial organs. RNMT also accepted a variety of aporphine and phthalideisoquinoline alkaloids, including noscapine (Table 1), and appears responsible for the formation of an array of N,N-dimethylated alkaloids including N,N-dimethylnarcotine, which has been detected in opium poppy latex (21). RNMT also N-methylated the opium poppy latex alkaloid papaverine, yielding the quaternary alkaloid densiberine (N-methylpapaverine), which has been reported in Berberis densiflora (22). In contract to RNMT, NMT4 showed no detectable catalytic activity on any tested substrate, and a gene expression profile inconsistent with general alkaloid accumulation profiles in major opium poppy organs, suggesting that the enzyme does not participate in BIA metabolism.
Unlike O-methyltransferases, which are considered monophyletic in origin (23), plant NMTs were clearly recruited independently from genes encoding several unrelated ancestral enzymes. Major and distinct clades are represented in plants by (i) putrescine NMTs (24), (ii) phosphoethanolamine NMTs (25), (iii) xanthine NMTs (26, 27), (iv) enzymes related to tocopherol C-methyltransferases and involved in monoterpenoid indole alkaloid biosynthesis (28, 29), and (v) NMTs involved in BIA metabolism (13, 15, 16). Most plant NMTs carry out a single methylation of the acceptor nitrogen atom; however, sequential methylation is a common feature of phosphoethanolamine NMTs, which yield the quaternary ammonium betaines used as osmolytes under certain stress conditions. The only other known NMTs that target a tertiary amine are TNMTs, which almost exclusively accept protoberberine alkaloids as substrates. The characterization of RNMT extends the list of enzymes that preferentially methylate a tertiary nitrogen atom. However, in contrast to TNMT, substrates possessing 1-benzylisoquinoline, aporphine and pthalideisoquinoline structural scaffolds and containing a tertiary amine bound in a single ring system were preferred. RNMT also accepted (R,S)-tetrahydropapaverine, a 1-benzylisoquinoline with a secondary amine, yielding both tertiary and quaternary alkaloids as reaction products. Overall, RNMT displays catalytic properties combining those of CNMT (i.e. a preference for 1-benzylisoquinoline substrates) and TNMT (i.e. efficient acceptance of tertiary amines). However, unlike CNMT and TNMT, RNMT did not accept (S)-coclaurine or protoberberines, respectively.
The overall topology of the presented NMT phylogeny (Fig. 2) is in agreement with previous analyses (15, 30). Functionally characterized and putative BIA NMTs cluster in several subclades with a strong correlation with reported catalytic properties. Opium poppy TNMT and functionally related enzymes from Papaver bracteatum (PbTNMT), Eschscholzia californica (EcTNMT), and G. flavum (GfNMT2 and GfNMT3), all of which preferentially accept protoberberine substrates, occur in a distinct subclade. NMT4, which did not exhibit enzymatic activity on any tested BIA, was also associated with this clade in a more distant position, suggesting a potentially common ancestral function with respect to TNMTs. More divergent NMT-like proteins from Arabidopsis have also been reported to lack activity on BIA substrates (15). Opium poppy RNMT belongs to a subclade with PavNMT and GfNMT4 from T. flavum and G. flavum, respectively, which forms a larger clade with characterized CNMTs from T. flavum (TfCNMT), C. japonica (CjCNMT), opium poppy (PsCNMT), and G. flavum (GfNMT1 and GfNMT5). GflNMT4 has been shown to catalyze the N-methylation of 1-benzylisoquinonlines and to accept (S)-reticuline as a substrate (30). In contrast, PavNMT was reported to prefer pavine rather than protoberberines as a substrate and did not appear to accept 1-benzylisoquinolines. The widespread occurrence of RNMT orthologs in species belonging to four different plant families, most of which accumulate moderate to high levels of magnoflorine (31, 32), suggests a generally ubiquitous occurrence of a potentially dedicated NMT involved in magnoflorine or potentially broader quaternary aporphine alkaloid biosynthesis.
All BIA NMTs are predicted to have a Rossmann-like fold producing a highly conserved AdoMet-binding domain (33). The hallmark binding motif I (GXGXG) is represented in CNMT, TNMT, and RNMT (Fig. 3). All but two key residues (i.e. Gln-99 and Ile-136) involved in AdoMet binding to bacterial cyclopropane fatty acid synthase (MtPcaA) are mostly conserved in BIA NMTs. Replacement of Gln-99 with Val and Ile-136 with His could maintain an interaction with the nitrogen atom of the amine group and preserve an ability to hydrogen bond with ribosyl hydroxyl moieties, respectively. Although no subcellular localization signals were detected using standard prediction servers, the C terminus of all three opium poppy BIA NMTs contain a tri-Lys (KKK) motif reminiscent of a canonical ER retrieval signal (KKXX). The C-terminal tri-Lys motif is highly represented in eukaryotic species and is proposed to reflect a loose consensus sequence for interaction with an unknown class of subcellular targeting chaperones (34). The effect of the additional C-terminal amino acids in RNMT is not known.
RNMT showed a modest preference for (R)-reticuline over (S)-reticuline as a substrate (Table 1). Although RNMT displayed an ∼2-fold higher Vmax for (R)-reticuline compared with (S)-reticuline, catalytic efficiency (Kcat/Km) calculations indicated that the turnover rate was essentially equivalent at low substrate concentrations. Moreover, opium poppy latex has been reported to accumulate S and R enantiomers of reticuline in an ∼3:1 ratio, potentially minimizing the physiological relevance of the detected in vitro preference for the R conformer. Furthermore, differences in the turnover of reticuline enantiomers were relatively small at a cytosolic pH of 7.0 and only became more substantial at pH of >8.0 (supplemental Fig. S2). The stereo-specificity of NMTs involved in BIA metabolism appears generally species-specific with no apparent phylogenetic influence. CNMTs from Dicentra spacabilis (Fumarioideae), Argenome mexicana (Papaveraceae), and Tinospora cordifolia (Menispermaceae) were reported to N-methylate only the S conformer of coclaurine (35). In contrast, CNMTs from Fumaria capreolata (Fumarioideae), Chondrodendron tomentosum (Menispermaceae) P. somniferum (Papaveraceae), Berberis stolonifera (Berberidaceae), and Berberis juliana (Berberidaceae) accepted both S and R enantiomers of coclaurine as substrates (35). Similarly, purified CNMT from C. japonica (Ranunculaceae) accepted both enantiomers, with a reported moderate preference for (R)-coclaurine (14). Substrate range also exhibits similar diversity among characterized CNMTs. Tinospora cordifolia CNMT was reported to not accept (R,S)-tetrahydropapaverine, which was a substrate for RNMT (Table 1), whereas CNMT from Berberis vulgaris N-methylated tetrahydropapaverine with a substantial preference for the R conformer (36). Various CNMTs also displayed differential acceptance of several O-substituted 1-benzylisoquinoline derivatives (13, 14, 35, 36). The diversity in substrate specificity among apparent CNMTs isoforms from even a single plant was suggested from the purification of three distinct enzymes from Berberis koetineae (37). Two isoforms showed a preference for (R)-coclaurine and a substantial acceptance of the S conformer, whereas a third isoform preferred (R)-tetrahydropapaverine and also converted the S enantiomer.
TNMTs generally show strict preference for protoberberine as opposed to 1-benzylisoquinoline substrates and an absolute specificity for S conformers (15, 16, 30, 38). Variations in the specific protoberberine substrate range are also apparent. For example, TNMT from P. somniferum accepted both (S)-stylopine and (S)-canadine, whereas the ortholog from P. bracteatum was not active on (S)-canadine. This is potentially notable because P. somniferum but not P. bracteatum accumulates noscapine, which is derived from (S)-canadine (16). A thorough screen of related NMTs from G. flavum (Papaveraceae) revealed six active enzymes, two of which displayed the substrate specificities for (S)-coclaurine of a CNMT and one of which showed the specificity for protoberberine substrates of a TNMT and exhibited an absolute preference for (S)-reticuline (30). The G. flavum enzymes also presented phylogenetic relationships with other NMTs consistent these functions (Fig. 2). Interestingly, two additional enzymes showed substrate ranges that crossed the general boundaries between TMNT and RNMT and between CNMT and RNMT, with the former efficiently N-methylating (S)-stylopine and (S)-reticuline and the latter preferring (S)-coclaurine and to a lesser extent (S)-reticuline. Overall, the specific functional parameters of NMTs involved in BIA metabolism appear relatively sensitive to mutations in amino acid sequence. However, the occurrence of dedicated CNMT, TNMT, and RNMT variants across plant species suggests the evolutionary selection and maintenance of dedicated enzymes responsible for the formation of primarily (S)-reticuline, benzophenanthridines (e.g. sanguinarine), phthalideisoquinolines (e.g. noscapine), and aporphines (e.g. magnoflorine).
The Km values of RNMT for the S and R enantiomers of reticuline (Table 2) are in agreement with those reported for the binding of (S)-coclaurine (36 μm) and (S)-norcoclaurine (52 μm) to CNMT. In contrast, the reported Km of CNMT for AdoMet (44 μm) was substantially lower than the measured value for RNMT (35). The lower affinity of RNMT for the methyl donor could result from the observed sequential methylation activity, which would be enhanced by facile release and replacement of the AdoMet substrate. For comparison, the Km values (0.6 and 40 μm) of TNMTs for protoberberine substrates are generally lower than those reported for the 1-benzylisoquinoline substrates of RNMT and CNMTs (15, 16, 38).
The stereochemistry of magnoflorine has not been determined in most plants, including opium poppy (1). The C8-C2′ phenol-coupling corytuberine synthase (CYP80G2) yielding the aporphine scaffold was reported to display strict stereo-specificity for (S)-reticuline, suggesting that only (S)-magnoflorine occurs in C. japonica (11). However, a CYP80G2 in opium poppy has not been characterized. Recently, a unique fusion protein, reticuline epimerase, consisting of cytochrome P450 and aldo-keto reductase domains, was identified as the enzyme responsible for the stereochemical inversion of (S)-reticuline (39, 40). Reticuline epimerase is important in opium poppy because the gateway enzyme, salutaridine synthase, to morphine biosynthesis accepts only (R)-reticuline. A lack of stereo-specificity in some plants among NMTs and potentially other biosynthetic enzymes involved in the formation of (S)-reticuline belies the absolute stereoselectivity of norcoclaurine synthase (Fig. 1), all known variants of which yield only (S)-norcoclaurine (10); thus de novo (R)-reticuline production is precluded, and stereochemical inversion is required. Nevertheless, the ∼3:1 ratio of S and R reticuline conformers, coupled with the lack of RNMT stereospecificity, suggests proportional levels of tembetarine enantiomers in opium poppy. A prediction of whether both S and R conformers of magnoflorine occur in the plant will require characterization of a CYP80G2 ortholog or direct enantiomer analysis in opium poppy.
Gene-specific suppression of RNMT caused a significant reduction in the accumulation of magnoflorine in roots, with a concomitant and significant increase in corytuberine levels, compared with control plants (Fig. 5). The relative abundance of major alkaloids, along with the levels of reticuline and the putative pathway intermediate tembetarine, were not affected by RNMT suppression, suggesting that the major route to magnoflorine is via corytuberine rather than tembetarine. The generally stoichiometric shift in the relative abundance of corytuberine and magnoflorine in response to a reduction in RNMT transcript levels confirms the occurrence of a largely root-specific NMT variant (Fig. 6) in opium poppy dedicated to magnoflorine biosynthesis. The widespread occurrence of magnoflorine (31, 32) and RNMT orthologs (30) in plants belonging to four different families suggests an important physiological function for this quaternary aporphine alkaloid, most likely related to its allelopathic or antimicrobial properties. RNMT extends the range of distinct N-methylating enzymes involved in BIA metabolism to those acting primarily on 1-benzylisoquinoline, protoberberine, and aporphine substrates, and containing primarily tertiary amines. RNMT accounts for the biosynthesis of a broad array of BIA pathway intermediates and quaternary ammonium products. The availability of a set of functionally divergent NMTs involved in BIA metabolism will facilitate the establishment of structure-function relationships for this large and diverse group of AdoMet-dependent enzymes.
Experimental Procedures
Plant Material
Opium poppy (P. somniferum L. cultivar Bea's Choice) cultivation was performed as previously described (21). Plant materials for analysis were harvested 1–2 days before anthesis, corresponding to ∼60–80 days after seed germination.
Chemicals and Reagents
(S)-Coclaurine, thebaine, canadaline, narcotolinogendiol, 3-O-acetylpapaveroxine, and narcotine hemiacetal was purchased from Toronto Research Chemicals, Mississauga, Canada). (S)-Reticuline oxalate was a gift from Tasmanian Alkaloids (Westbury, Australia), and morphine and codeine were gifts from Sanofi-Aventis (Paris, France). (R)-Reticuline was purchased from Santa Cruz Biotechnology (Dallas, TX). (S)-Canadine, hydrastine, and bicuculline were obtained from Latoxan (Valence, France), and protopine was purchased from Indofine (Hillsborough, NJ). (S)-Tetrahydrocolumbamine (41), (R,S)-stylopine (15), and (S)-scoulerine (42) were prepared as previously described. Cryptopine and allocryptopine were purchased from ChromaDex (Irvine, CA) and MP Biomedicals (Santa Ana, CA), respectively. (+)-Bulbocapnine, (+)-isocorydine, (S)-corytuberine, (+)-glaucine, (+)-isothebaine, and berbamine were purchased from Sequoia Research Products (St. James Close, UK). (S)-Tetrahydropapaverine and (+/−)-pavine were isolated from commercial pavine (Sigma-Aldrich), and narcotoline was isolated as previously described (21). Noscapine, papaverine, sanguinarine, and AdoMet were purchased from Sigma-Aldrich. Restriction endonucleases, T4 DNA ligase, and Q5 High Fidelity DNA polymerase were obtained from New England Biolabs (Ipswitch, MA), and Taq polymerase was from Bioshop Canada (Burlington, Canada). SuperScript III reverse transcriptase was purchased from Thermo Fisher. SYBR FAST ABI PRISM master mix was purchased from KAPA Biosystems (Wilmington, MA). All other chemicals were from Sigma-Aldrich or Bioshop Canada.
Preparation of (S)-Tembetarine
(S)-Reticuline (3 mm) was incubated for 5 h at 30 °C with 400 μg of purified of recombinant RNMT and 5 mm AdoMet in a total volume of 2 ml. The reaction was terminated by adding an equal volume of methanol followed by centrifugation at 18,000 × g to remove debris. The supernatant was transferred to a new tube, reduced to dryness, and redissolved in 0.5 ml of 100 mm ammonium acetate, pH 6.0, for separation using a Strata X-CW column (Phenomenex, Golden, CO). The column was conditioned with 0.5 ml of acetonitrile and equilibrated with 0.5 ml of 100 mm ammonium acetate, pH 6.0. After loading the reaction mixture and successive wash steps of 0.5 ml of 5% (w/v) ammonium hydroxide, pH 12.0, and 1 ml of methanol, elution was achieved using 1 ml of formic acid:acetonitrile (5:95). The separation of (S)-reticuline in the methanol wash and (S)-tembetarine in the elution was confirmed by LC-MS. The (S)-tembetarine fraction was subjected to semi-preparative HPLC using a mobile phase of 0.08% (v/v) acetic acid and 5% (v/v) acetonitrile in water (Solvent A) and 100% acetonitrile (Solvent B), and a Luna C18(2) 100A column (5 μm; 250 × 10 mm; Phenomenex) on a 1260 HPLC system (Agilent, Santa Clara, CA). Fractions corresponding to (S)-tembetarine were pooled and reduced to dryness, and 2 mg was subjected to NMR analysis. The concentration of (S)-tembetarine was estimated using the molar extinction coefficient of (S)-reticuline. 1H and 13C NMR spectra and gc2hsqcse, gCOSY, and gHMBCAD correlation spectroscopy confirmed the identity of (S)-tembetarine (Table 1).
Identification of Candidates
Previously reported opium poppy stem and root transcriptomes (42) were searched to identify sequences encoding potential N-methyltransferase candidates involved in BIA biosynthesis, using the tblastn algorithm and known BIA NMTs as queries (TNMT, Q108P1; CNMT, AAP45316).
Phylogenetic Analysis
Amino acid alignments of select plant NMTs were performed with the MUSCLE algorithm (43) in MEGA7 (44) using default parameters. Evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix-based model assuming equal substitution rates, followed by a bootstrap test of phylogeny. The tree with the highest log likelihood was drawn with branch lengths proportional to the number of substitutions per site and supported by calculating the percentage of 500 bootstrapped trees where the associated taxa clustered together (45). GenBankTMaccession numbers were: T. flavum TfCNMT (AAU20766), C. japonica CjCNMT (BAB71802), P. somniferum PsCNMT (AAP45316), T. flavum TfPavNMT (EU883010), P. somniferum PsTNMT (AAY79177), P. bracteatum PbTNMT1 (C3SBU5) and PbTNMT2 (C3SBU4), E. californica EcTNMT (EU882977), Arabidopsis thaliana CNMT-like AT4G33110 (AAM97074) and AT4G33120 (BAE99366), Zea mays cyclopropane fatty-acid synthase ZmCFAS (NP_001147782), Aegilops tauschii AtCFAS (EMT06798), E. coli EcCFAS (WP_044067789), and Mycobacterium tuberculosis MtCFAS pcaA (EAY58886). P. somniferum PsRNMT (NMT3) and PsNMT4 were deposited in NCBI under accession numbers KX369612 and KX369613, respectively. The six G. flavum sequences (GfNMT1-GfNMT6) were previously reported (30).
Isolation and Expression Vector Construction
Plant tissues were ground to a power under liquid N2 using a Tissue Lyser II (Qiagen), and total RNA was extracted using the cetyl trimethyl ammonium bromide method (46). First strand cDNA synthesis was performed on 2 μg of RNA using SuperScript III reverse transcriptase and oligo(dT)20 according to the manufacturer's instructions (Thermo Fisher). Opium poppy RNMT and NMT4 open reading frames were amplified from cDNA using Q5 high fidelity DNA polymerase (New England Biolabs) and sequence-specific primers incorporating attB sites for Gateway cloning (RNMT, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGGTCGACAACAATGGAGACAACTAAAAT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTATCAGATGCAAGTCATTAATTTCTTT-3′; and NMT4, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGGGATTCTAAAGATCAACAAGG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTATCACTTTTTCTTGAAGAGAAAATG-3′). PCR products were purified using a Genejet gel extraction kit (Thermo Fisher). Recombination reactions using Gateway BP clonase II were used to construct entry plasmids pDONR221-PsNMT3 and pDONR221-PsNMT4. Following isolation, a second recombination reaction using the entry plasmids and Gateway LR clonase II yielded the destination/expression plasmids pHGWA-PsNMT3 and pHGWA-PsNMT4. Expression plasmids were used to transform E. coli expression strain ArcticExpress (DE3) (Agilent) grown on lysogenic broth containing 100 μg/ml ampicillin. The pHGWA expression plasmid encoding a recombinant C-terminal His6 tag protein was previously described (47).
Recombinant Gene Expression and Purification
Single colonies of E. coli ArcticExpress (DE3) transformed with pHGWA-PsNMT3 and pHGWA-PsNMT4 were used to inoculate 50 ml of lysogenic broth supplemented with 100 μg/ml ampicillin and 10 μg/ml gentamycin. Cultures were grown for 16 h at 30 °C with shaking at 200 rpm, and cells were harvested by centrifugation at 2000 × g. The cells were resuspended in fresh lysogenic broth and used to inoculate 1 liter of Studier's autoinduction media (ZYP-5052) supplemented with 100 μg/ml ampicillin to a starting A600 of ∼0.2. Autoinduction cultures were grown at 30 °C to A600 of ∼1.0 and then transferred to 16 °C for an additional 24 h. The cells were harvested by centrifugation at 14,000 × g for 20 min, after which the pellet was flash frozen in liquid N2 and stored at −80 °C until purification.
Cell pellets were resuspended in protein extraction buffer (50 mm sodium phosphate, pH 7.0, 300 mm NaCl, 10% (w/v) glycerol) supplemented with 1 mg/ml lysozyme, followed by sonication on ice for 6 min. The crude lysate was centrifuged at 4 °C at 18,000 × g for 20 min to remove cellular debris, and the supernatant was transferred to a fresh tube containing 1 ml of equilibrated TALON resin (Clontech), which was then incubated on ice for 30 min on a gyratory shaker at 50 rpm. The resin was washed with 40 ml of protein extraction buffer, followed by 20 ml of extraction buffer containing 20 mm imidazole. Purified protein was eluted using 4 ml of extraction buffer containing 200 mm imidazole. The imidazole concentration was reduced to less than 1 mm by repeated ultrafiltration on Amicon Ultra 30K columns (EMD Millipore, Billerica, MA) using extraction buffer. The purified protein concentration was determined using the Bradford reagent according to the manufacturer's instructions (Thermo Fisher).
Enzyme Assays
Initial substrate acceptance screens were conducted in 100 mm Tris-HCl, pH 7.0 and 8.5, using 10 μm alkaloid, 20 μm AdoMet, and 6 μg of purified, desalted recombinant protein in a reaction volume of 50 μl. Assays were incubated for 16 h at 30 °C and quenched with the addition of 150 μl of stop solution (methanol containing 1% (v/v) acetic acid). Quenched assays were diluted 1:1 with Solvent A (10 mm ammonium acetate, pH 5.5, 5% (v/v) acetonitrile) and analyzed by LC-MS. Relative conversion rates for substrates yielding a reaction product showing an increased m/z of 14 and/or 28 were performed under identical assays conditions, except that incubation times of 10 min and 1 h were used, both of which were within the linear assay range. The substrate showing the highest turnover (measured as a decrease in substrate peak area) was set to 100% activity, and other substrate conversion rates were scaled proportionally. For the determination of pH optima, assays were performed in 100 mm MES, pH 5.0–7.0, or in Tris-HCl, pH 7.0–9.0, using 500 μm (S)-reticuline or (R)-reticuline, 500 μm AdoMet, and 1.5 μg of purified recombinant protein in a reaction volume of 50 μl. Assays were incubated for 1 h at 30 °C and quenched with the addition of 200 μl of stop solution. Assays to determine temperature optima were performed under identical conditions except that the buffer was 100 mm Tris-HCl, pH 7.0, and incubation temperatures between 4 and 50 °C were used. Kinetic parameters were measured for 1 h under the same conditions using concentrations of (S)-reticuline and (R)-reticuline between 0.5 to 500 μm, and at a fixed AdoMet concentration of 500 μm. AdoMet concentrations between 0.5 and 500 μm were tested at a fixed (R)-reticuline concentration of 500 μm. Negative control assays were performed using purified recombinant protein denatured in boiling water for 15 min. Product formation was quantified by comparison to a five-point curve for authentic (S)-reticuline between 80 nm and 50 μm. Saturation curves and kinetic constants were determined based on Michaelis-Menten kinetics using the enzyme kinetics module in SigmaPlot 13.0.
Virus-induced Gene Silencing
Gene-specific silencing constructs were based on the pYL156/TRV2 system. The target nucleotide sequence for RNMT was designed to straddle the 3′ coding and untranslated regions because of the relatively high sequence identity among Rossmann fold/AdoMet-binding domains near the 5′ end. The target sequence was amplified from opium poppy cDNA using Q5 high fidelity DNA polymerase (New England Biolabs) and PCR primers incorporating 5′ BamHI and 3′ EcoRI restriction sites. The PCR product was ligated into the corresponding restriction sites of pTRV2. The resulting pTRV2-NMT3 construct was transformed into A. tumefaciens strain GV3101 by electroporation. A. tumefaciens strains harboring pTRV2-PsNMT3 or the empty pTRV2 vector were combined in equal amounts with a strain containing pTRV1 and used to infiltrate 2-week-old opium poppy seedlings as previously described (48). Latex, stem, and root samples were collected from each plant 1–2 days prior to anthesis and ground to fine powder under liquid N2 using a TissueLyzer II (Qiagen). Total RNA was extracted from ground stem tissue using the cetyl trimethyl ammonium bromide method (46), and cDNA synthesis was performed as described above. Plants were screened for infection by reverse transcription-PCR to identify individuals containing TRV coat protein transcripts using specific primers (TRV2-CP-F, 5′-CTGACTTGATGGACGATTC-3′; and TRV2-CP-R, 5′-TGTGTTTGGATTCGCAG-3′). Twelve infected plants for each pTRV2 construct were analyzed by (i) qRT-PCR to determine relative transcript abundance of NMT genes and (ii) LC-MS to determine the abundance of selected BIAs. Briefly, latex and ground root samples were freeze-dried for 24 h, after which the tissue samples were suspended in 100 ml/g acetonitrile, sonicated for 10 min on ice, and extracted at −20 °C for 24 h. Extracts were diluted 100-fold in Solvent A and subjected to LC-MS analysis.
Gene Expression Analysis
Plant organs were collected 1–2 days prior to anthesis from five individual opium poppy plants and flash frozen in liquid N2. Tissue samples were ground to a fine powder under liquid N2 using a TissueLyzer II (Qiagen), and aliquots were used for RNA and alkaloid extraction, followed by RT-qPCR and LC-MS analysis.
Quantitative Real Time PCR
qRT-PCR was performed to determine the relative abundance of NMT transcripts using SYBR Green detection in an ABI 7300 (Applied Biosystems, Foster City, CA). RNA extraction and cDNA synthesis were performed as described above. Each 10-μl reaction contained 2 μl of cDNA, 2 pmol of each primer (CNMT, 5′-CGTTGGACGAAGCTGAGATAG-3′ and 5′-CACCCAATCCACACCCTAAA-3′; TNMT, 5′-CTTCCTCCAAGTCCAGGTAATAG-3′ and 5′-CACATGGCTCGTTCAGTAGA-3′; RNMT, 5′-CAGTAAACAAACCCGCACTTTC-3′ and 5′-GGCAACAGAGATAGACACGTTAG-3′; and NMT4, 5′-GACATTGGTTGCGGCTTTG-3′ and 5′-TCTTTGCTCGGCGGAATTAG-3′) and KAPA SYBR FAST qPCR Master Mix used as recommended by the manufacturer (Kapa Biosystems, Wilmington, MA). Transcripts encoding actin (5′-TCTCAACCCAAAGGCTAATCG-3′ and 5′-CCCCAGAATCCAAGACAATACC-3′) and ubiquitin-10 (5′-GGGAACACAAACGACACCAAA-3′ and 5′-TCGTCTTCGTGGTGGTAACTAGAG-3′) were used as the constitutive reference. PCR was performed as follows: 3 min at 95 °C, followed by 40 cycles of 3 s at 95 °C and 20 s at 60 °C. Amplification specificity was verified by melt curve analysis and agarose gel electrophoresis of reaction products. Primer efficiency was verified using LinRegPCR (49). The 2−ΔΔCt method was used to calculate relative transcript abundance (50), whereby the geometric mean of actin and ubiquitin-10 used as reference genes and the results for each transcript are normalized to the plant showing the highest expression level.
Liquid Chromatography-Mass Spectrometry
LC-MS was performed using a 1200 HPLC coupled with an 6410 triple-quadrupole MS (Agilent). Samples (2 μl) were injected onto a Poroshell C18 HPLC column (Agilent), and analytes were eluted in a gradient of solvent A (10 mm ammonium acetate, pH 5.5, 5% (v/v) acetonitrile) and solvent B (100% acetonitrile) at a flow rate of 400 μl/min. The gradient began at 0% B, reached 80% B by 6 min, and then increased to 100% at 7 min. Subsequently, the mixture returned to 0% B for a 3-min re-equilibration period. Analytes were applied to the mass analyzer using an electrospray ionization probe operating in positive mode with the following conditions: capillary voltage, 4000 V; fragmentor voltage, 100 V; source temperature, 350 °C; nebulizer pressure, 50 PSI; and gas flow, 10 liters/min. For full-scan analysis, quadrupole 1 and 2 were set to radio frequency only, whereas the third quadrupole scanned from 200 to 700 m/z (for alkaloid extracts and substrate range assays). Selected ion mode (m/z 330 and 344) was used for pH and temperature optima and kinetic parameter analyses. Positive mode electrospray ionization, collision-induced dissociation spectra were analyzed, the precursor m/z was selected in quadrupole 1, and collision energy of 25 eV was applied in quadrupole 2 and an argon collision gas pressure of 1.8 × 10−3 Torr. The resulting MS2 fragments were resolved by quadrupole 3 scanning from 40 to 5 m/z greater than the precursor ion m/z. Compounds were identified in extracted ion chromatograms (supplemental Fig. S4) based on retention times and positive mode electrospray ionization collision-induced dissociation spectra compared with authentic standards (supplemental Table 2). Alkaloids were quantified by comparison to five-point (80 nm to 50 μm) curves of authentic standards.
Author Contributions
J. S. M. performed all experimental work and wrote the first draft of the manuscript. P. J. F. conceived of the project, supervised the research, and edited the final draft of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Darcy Burns (Department of Chemistry, University of Toronto) for assistance with the assignment of NMR spectra.
Note Added in Proof
Another article published in this issue (51) reports the structure of pavine N-methyltransferase, a related enzyme that shares some functional properties and substantial amino acid sequence identity with reticuline N-methyltransferase.
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (to P. J. F.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental data file, Tables S1 and S2, and Figs. S1–S4.
- BIA
- benzylisoquinoline alkaloid
- AdoMet
- S-adenosyl-l-methionine
- NMT
- N-methyltransferase
- CNMT
- coclaurine NMT
- TNMT
- tetrahydroprotoberberine NMT
- RNMT
- reticuline NMT
- PavNMT
- pavine NMT
- contig
- group of overlapping clones
- PbTNMT
- P. bracteatum NMT
- GfNMT
- G. flavum NMT
- TfCNMT
- T. flavum CNMT
- CjCNMT
- C. japonica CNMT
- PsCNMT
- P. somniferum CNMT
- qRT-PCR
- quantitative real time PCR.
References
- 1. Shulgin A. T., and Perry W. E. (2002) The Simple Plant Isoquinolines, Transform Press, Berkeley, CA [Google Scholar]
- 2. Hung T. M., Lee J. P., Min B. S., Choi J. S., Na M., Zhang X., Ngoc T. M., Lee I., and Bae K. (2007) Magnoflorine from Coptidis rhizoma protects high density lipoprotein during oxidant stress. Biol. Pharm. Bull. 30, 1157–1160 [DOI] [PubMed] [Google Scholar]
- 3. Hung T. M., Na M., Min B. S., Zhang X., Lee I., Ngoc T. M., Thuong P. T., Sok D. E., and Bae K. (2007) Protective effect of magnoflorine isolated from Coptidis rhizoma on Cu2+-induced oxidation of human low density lipoprotein. Planta Med. 73, 1281–1284 [DOI] [PubMed] [Google Scholar]
- 4. Patel M. B., and Mishra S. (2012) Isoquinoline alkaloids from Tinospora cordifolia inhibit rat lens aldose reductase. Phytother. Res. 26, 1342–1347 [DOI] [PubMed] [Google Scholar]
- 5. Chen J. H., Du Z. Z., Shen Y. M., and Yang Y. P. (2009) Aporphine alkaloids from Clematis parviloba and their antifungal activity. Arch. Pharm. Res. 32, 3–5 [DOI] [PubMed] [Google Scholar]
- 6. de la Peña J. B., Lee H. L., Yoon S. Y., Kim G. H., Lee Y. S., and Cheong J. H. (2013) The involvement of magnoflorine in the sedative and anxiolytic effects of Sinomeni Caulis et Rhizoma in mice. J. Nat. Med. 67, 814–821 [DOI] [PubMed] [Google Scholar]
- 7. Willaman J. J., and Schubert B. G. (1961) Alkaloid-bearing plants and their contained alkaloids, U.S. Department of Agriculture, Government Printing Office, Washington, DC [Google Scholar]
- 8. Marques J. V., Dalisay D. S., Yang H., Lee C., Davin L. B., and Lewis N. G. (2014) A multi-omics strategy resolves the elusive nature of alkaloids in Podophyllum species. Mol. Biosyst. 10, 2838–2849 [DOI] [PubMed] [Google Scholar]
- 9. Priestap H. A., Velandia A. E., Johnson J. V., and Barbieri M. A. (2012) Secondary metabolite uptake by the Aristolochia-feeding papilionoid butterfly Battus polydamas. Biochem. Syst. Ecol. 40, 126–137 [Google Scholar]
- 10. Hagel J. M., and Facchini P. J. (2013) Benzylisoquinoline alkaloid metabolism: A century of discovery and a brave new world. Plant Cell Physiol. 54, 647–672 [DOI] [PubMed] [Google Scholar]
- 11. Ikezawa N., Iwasa K., and Sato F. (2008) Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C-C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 283, 8810–8821 [DOI] [PubMed] [Google Scholar]
- 12. Bhakuni D. S., Jain S., and Singh R. S. (1980) Biosynthesis of magnoflorine and laurifoline. Tetrahedron 36, 2525–2528 [Google Scholar]
- 13. Choi K.-B., Morishige T., Shitan N., Yazaki K., and Sato F. (2002) Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J. Biol. Chem. 277, 830–835 [DOI] [PubMed] [Google Scholar]
- 14. Choi K.-B., Morishige T., and Sato F. (2001) Purification and characterization of coclaurine N-methyltransferase from cultured Coptis japonica cells. Phytochemistry 56, 649–655 [DOI] [PubMed] [Google Scholar]
- 15. Liscombe D. K., and Facchini P. J. (2007) Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J. Biol. Chem. 282, 14741–14751 [DOI] [PubMed] [Google Scholar]
- 16. Liscombe D. K., Ziegler J., Schmidt J., Ammer C., and Facchini P. J. (2009) Targeted metabolite and transcript profiling for elucidating enzyme function: isolation of novel N-methyltransferases from three benzylisoquinoline alkaloid-producing species. Plant J. 60, 729–743 [DOI] [PubMed] [Google Scholar]
- 17. Huang C. C., Smith C. V., Glickman M. S., Jacobs W. R. Jr., and Sacchettini J. C. (2002) Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis. J. Biol. Chem. 277, 11559–11569 [DOI] [PubMed] [Google Scholar]
- 18. Martin J. L., and McMillan F. M. (2002) SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12, 783–793 [DOI] [PubMed] [Google Scholar]
- 19. Emanuelsson O., Brunak S., von Heijne G., and Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protocols 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 20. Gidda S. K., Shockey J. M., Rothstein S. J., Dyer J. M., and Mullen R. T. (2009) Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: Functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol. Biochem. 47, 867–879 [DOI] [PubMed] [Google Scholar]
- 21. Desgagné-Penix I., Farrow S. C., Cram D., Nowak J., and Facchini P. J. (2012) Integration of deep transcript and targeted metabolite profiles for eight cultivars of opium poppy. Plant Mol. Biol. 79, 295–313 [DOI] [PubMed] [Google Scholar]
- 22. Khamidov I. I., Aripova S. F., Telezhenetskaya M. V., Karimov A., and Dzhenberov I. (1997) Berberis alkaloids: XXXIX. New alkaloids from B. densiflora. Chem. Nat. Comp. 33, 323–325 [Google Scholar]
- 23. Lam K. C., Ibrahim R. K., Behdad B., and Dayanandan S. (2007) Structure, function, and evolution of plant O-methyltransferases. Genome 50, 1001–1013 [DOI] [PubMed] [Google Scholar]
- 24. Hibi N., Higashiguchi S., Hashimoto T., and Yamada Y. (1994) Gene expression in tobacco low-nicotine mutants. Plant Cell 6, 723–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nuccio M. L., Ziemak M. J., Henry S. A., Weretilnyk E. A., and Hanson A. D. (2000) cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J. Biol. Chem. 275, 14095–14101 [DOI] [PubMed] [Google Scholar]
- 26. Kato M., Mizuno K., Crozier A., Fujimura T., and Ashihara H. (2000) Caffeine synthase gene from tea leaves. Nature 406, 956–957 [DOI] [PubMed] [Google Scholar]
- 27. McCarthy A. A., and McCarthy J. G. (2007) The structure of two N-methyltransferases from the caffeine biosynthetic pathway. Plant Physiol. 144, 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liscombe D. K., Usera A. R., and O'Connor S. E. (2010) Homolog of tocopherol C-methyltransferases catalyzes N-methylation in anticancer alkaloid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 18793–18798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levac D., Cázares P., Yu F., and De Luca V. (2016) A picrinine N-methyltransferase belongs to a new family of γ-tocopherol-like methyltransferases found in medicinal plants that make biologically active monoterpenoid indole alkaloids. Plant Physiol. 170, 1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hagel J. M., Morris J. S., Lee E.-J., Desgagné-Penix I., Bross C. D., Chang L., Chen X., Farrow S. C., Zhang Y., Soh J., Sensen C. W., and Facchini P. J. (2015) Transcriptome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol. 15, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Farrow S. C., Hagel J. M., and Facchini P. J. (2012) Transcript and metabolite profiling in cell cultures of 18 plant species that produce benzylisoquinoline alkaloids. Phytochemistry 77, 79–88 [DOI] [PubMed] [Google Scholar]
- 32. Hagel J. M., Mandal R., Han B., Han J., Dinsmore D. R., Borchers C. H., Wishart D. S., and Facchini P. J. (2015) Metabolome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol. 15, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kozbial P. Z., and Mushegian A. R. (2005) Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Austin R. S., Provart N. J., and Cutler S. R. (2007) C-terminal motif prediction in eukaryotic proteomes using comparative genomics and statistical over-representation across protein families. BMC Genomics 8, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Loeffler S., Deus-Nuemann B., and Zenk M. H. (1995) S-adenosyl-l-methionine: (S)-coclaurine-N-methyltransferase from Tinospora cordifolia. Phytochemistry 38, 1387–1395 [Google Scholar]
- 36. Wat C.-K., Steffens P., and Zenk M. H. (1986) Partial purification and characterization of S-adenosyl-l-methionine: norreticuline N-methyltransferases from Berberis cell suspension cultures. Z. Naturforsch. 41c, 126–134 [Google Scholar]
- 37. Frenzel T., and Zenk M. H. (1990) Purification and characterization of three isoforms of S-adenosyl-l-methionine: (R,S)-tetrahydrobenzylisoquinoline-N-methyltransferase from Berberis koetineana cultures. Phytochemistry 29, 3491–3497 [Google Scholar]
- 38. Rueffer M., Zumstein G., and Zenk M. H. (1990) Partial purification and properties of S-adenosyl-l-methionine: (S)-tetrahydroprotoberberine-cis-N-methyltransferase from suspension-cultured cells of Eschscholzia and Corydalis. Phytochemistry 29, 3727–3733 [Google Scholar]
- 39. Farrow S. C., Hagel J. M., Beaudoin G. A., Burns D. C., and Facchini P. J. (2015) Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy. Nat. Chem. Biol. 11, 728–732 [DOI] [PubMed] [Google Scholar]
- 40. Winzer T., Kern M., King A. J., Larson T. R., Teodor R. I., Donninger S. L., Li Y., Dowle A. A., Cartwright J., Bates R., Ashford D., Thomas J., Walker C., Bowser T. A., and Graham I. A. (2015) Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science 349, 309–312 [DOI] [PubMed] [Google Scholar]
- 41. Dang T. T., and Facchini P. J. (2012) Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol. 159, 618–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hagel J. M., Beaudoin G. A., Fossati E., Ekins A., Martin V. J., and Facchini P. J. (2012) Characterization of a flavoprotein oxidase from opium poppy catalyzing the final steps in sanguinarine and papaverine biosynthesis. J. Biol. Chem. 287, 42972–42983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kumar S., Stecher G., and Tamura K. (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Felsenstein J. (1985) Confidence limits on phylogenetics: an approach using the bootstrap. Evolution 39, 783–791 [DOI] [PubMed] [Google Scholar]
- 46. Meisel L., Fonseca B., González S., Baeza-Yates R., Cambiazo V., Campos R., Gonźalez M., Orellana A., Retamales J., and Silva H. (2005) A rapid and efficient method for purifying high quality total RNA from peaches (Prunus persica) for functional genomics analyses. Biol. Res. 38, 83–88 [DOI] [PubMed] [Google Scholar]
- 47. Busso D., Delagoutte-Busso B., and Moras D. (2005) Construction of a set Gateway-based destination vectors for high-throughput cloning and expression screening in Escherichia coli. Anal. Biochem. 343, 313–321 [DOI] [PubMed] [Google Scholar]
- 48. Hagel J. M., and Facchini P. J. (2010) Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat. Chem. Biol. 6, 273–275 [DOI] [PubMed] [Google Scholar]
- 49. Ramakers C., Ruijter J. M., Deprez R. H., and Moorman A. F. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66 [DOI] [PubMed] [Google Scholar]
- 50. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 51. Torres M. A., Hoffarth E., Eugenio L., Savtchouk J., Chen X., Morris J., Facchini P. J., and Ng K. K.-S. (2016) Structural and functional studies of pavine N-methyltransferase from Thalictrum flavum reveal novel insights into substrate recognition and catalytic mechanism. J. Biol. Chem. 291, 23403–23415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.