Abstract
Typically disease-causing CAG/CTG repeats expand, but rare affected families can display high levels of contraction of the expanded repeat amongst offspring. Understanding instability is important since arresting expansions or enhancing contractions could be clinically beneficial. The MutSβ mismatch repair complex is required for CAG/CTG expansions in mice and patients. Oddly, by unknown mechanisms MutSβ-deficient mice incur contractions instead of expansions. Replication using CTG or CAG as the lagging strand template is known to cause contractions or expansions respectively; however, the interplay between replication and repair leading to this instability remains unclear. Towards understanding how repeat contractions may arise, we performed in vitro SV40-mediated replication of repeat-containing plasmids in the presence or absence of mismatch repair. Specifically, we separated repair from replication: Replication mediated by MutSβ- and MutSα-deficient human cells or cell extracts produced slipped-DNA heteroduplexes in the contraction- but not expansion-biased replication direction. Replication in the presence of MutSβ disfavoured the retention of replication products harbouring slipped-DNA heteroduplexes. Post-replication repair of slipped-DNAs by MutSβ-proficient extracts eliminated slipped-DNAs. Thus, a MutSβ-deficiency likely enhances repeat contractions because MutSβ protects against contractions by repairing template strand slip-outs. Replication deficient in LigaseI or PCNA-interaction mutant LigaseI revealed slipped-DNA formation at lagging strands. Our results reveal that distinct mechanisms lead to expansions or contractions and support inhibition of MutSβ as a therapeutic strategy to enhance the contraction of expanded repeats.
Introduction
At least 14 neurological, neurodegenerative, and neuromuscular diseases, including myotonic dystrophy type 1 (DM1) and Huntington’s disease (HD), are caused by CAG/CTG repeat expansions in specific genes. In healthy individuals, repeat tract lengths typically range from 5–24 repeats, which are genetically stable. Expanded tracts of >35 repeats are unstable and tend to expand further in transmissions and in tissues, leading to disease. Typically CAG/CTG repeats expand, but rare DM1, HD, SCA1 and SBMA families can display high levels of contraction of the expanded repeat [1–17]. Furthermore, while most somatic tissues show ongoing expansions as the patient ages, some tissues, such as the male germline of spinocerebellar ataxia type 8, Friedreich’s ataxia, and fragile × men, show clear signs of contractions of the inherited expansion (reviewed in [18]). Most studies of the role of mammalian mismatch repair (MMR) on CAG/CTG instability have focused upon the mechanism of expansion; little is known regarding the mechanism of mammalian MMR involvement in repeat contractions. Understanding the process of repeat contractions is important since harnessing this process may be clinically beneficial. It is thought that repeat expansions and contractions occur due to the formation of slipped-DNA structures, which may form during DNA replication, damage, or repair; however, the process through which slipped-DNAs actually form has not been revealed.
While repeat expansions clearly arise in non-proliferating tissues, such as patients’ brains [19], DNA replication is likely to be involved under some contexts. Several studies have demonstrated links between DNA replication and trinucleotide repeat (TNR) instability, including those using cell lines derived from myotonic dystrophy patients, where spontaneous CTG expansions occur under proliferating but not non-proliferating conditions [20–22]. In bacteria, yeast, an SV40 replication system, HeLa, and DM1 patient cell lines, the direction of replication through an expanded repeat tract was found to affect repeat instability [23–26]. In all systems, repeat contractions were predominant when the CTG strand was the lagging strand template. Biases for repeat expansions were evident specifically when CAG was the lagging strand template in a primate replication system, DM1 patient cell lines, and tissues of DM1 transgenic mice [26–30]. In agreement with this, CAG serves as the lagging strand template in the replication directions observed at the mutant HD, SCA7, and DM1 loci in cell lines derived from HD, SCA7, and DM1 patients [28, 31]. Evidence explaining the replication-direction sensitive tendencies towards expansions or contractions has not been provided, but it has been hypothesized to be due to the differential propensity of CAG versus CTG repeats to form slipped-DNA heteroduplexes, or their differential processing by repair machinery at replication forks.
A key function of the mismatch repair pathway is to maintain genome stability by repairing mispaired DNA heteroduplexes. There are two complexes involved in mismatch recognition – MutSα (MSH2 + MSH6) and MutSβ (MSH2 + MSH3). MutSα acts in the repair of some base-base mispairs and short insertion/deletions (IDLs), while MutSβ acts in repair of selected base-base mispairs and both short and long IDLs [32–37]. Both MutSα and MutSβ are part of replication fork complexes [38–40], and recent evidence indicates that MMR protein expression correlates with tissue proliferation status in mice [41]. Despite their role in genome maintenance, MMR proteins have been found to be required for trinucleotide repeat expansion mutations (reviewed in [42]). In many, but not all transgenic mice harboring unstable CAG/CTG repeats which show a repeat expansion bias both in somatic tissues and in transmissions to offspring, deficiencies in MSH2 or MSH3, but not MSH6, led to a striking switch from an expansion bias to a contraction bias [43–48]. Not all transgenic mice display this switch to a contraction bias, but some have repeats that are stabilized [47–50] – suggesting a link between MMR and instability. In DM1 mice with >300 CTG repeats, a contraction bias was observed on a defective Ligase I background (46BRLigIm/m) in the maternal germline, a phenomenon explained by the unique replication pattern in these cells [30]. In human embryonic stem cells derived from germ cells of patients with DM1 (expanded CAG/CTG repeats), it was shown that the DM1 repeat was stabilized upon cell differentiation – coinciding with a decrease in MMR protein expression [51]. Similar requirements of human MSH2-MSH3 proteins for CAG/CTG expansions were found in iPS DM1 patient cells [52]. Thus, in the case of CAG/CTG repeats, human and murine MMR seems to be required for expansion mutations instead of protecting against them – yet the processes through which this occurs are unknown.
Previously we studied the role of MMR proteins in the in vitro repair of slipped-CAG/CTG repeats. The involvement of MMR in slipped-DNA repair is dependent upon the length and number of slip-outs. Repair of large slip-outs with 20 or 25 excess repeats does not require MMR [53, 54], while repair of shorter slip-outs (1–3 repeats) is dependent upon MMR – specifically MutSβ [55]. Repair of intermediate lengths (5–10 excess repeats) is moderately improved by the presence of MutSβ [55, 56]. Surprisingly, the clustering of multiple short slip-outs interferes with repair activity (attempts to repair several closely situated slip-outs can lead to repeat expansions) [55]. We recently identified clustered slipped-DNAs at the unstable trinucleotide repeats (CTG)n•(CAG)n of the myotonic dystrophy disease locus in non-mitotic tissues (brain, muscle, heart, etc.) of DM1 patients [57]. The amounts of slipped-DNA molecules were greater in tissues having greater levels of CTG expansions — supporting the formation of clustered slipped-DNAs as persistent mutation products of repeat expansions in non-mitotic states, and not merely as transient mutagenic intermediates. While repair of pre-formed slipped-DNAs has been observed, the role of human MutSα or MutSβ during replication of repeat tracts has not been reported.
To assess the effect of MMR proteins upon the replication of CAG/CTG repeat tracts, we replicated repeat-containing templates using human cell extracts in the presence or absence of MMR-proteins. This SV40 replication fork model reflects many, but not all aspects of chromosomal replication forks. The same replication-direction sensitive repeat expansions and contractions evident in human, murine, and yeast cells occur [23–26]. Importantly, MMR proteins track with the SV40 replication complex, as it does with chromosomal replication forks [40]. In this system both replication and repair could occur simultaneously or can be separated from each other. In the presence of MMR, templates displayed either an expansion- or contraction-bias, determined by the direction of replication through the repeat [26, 27]. Unexpectedly, in the absence of MMR we detected the formation of slipped-DNA heteroduplexes during replication in one direction but not the other. The results suggest that slipped heteroduplexes form and then are processed following replication fork passage by a MutSβ-dependent mechanism.
Results
Slipped-DNA formation is MMR- and replication direction-dependent
To assess the role of MMR proteins in the replication of CAG/CTG repeats, we used a well-established in vitro SV40 replication assay. We previously mapped the initiation of replication to the SV40 origin sequence, regardless of the presence of the CAG/CTG repeat [26, 27]. The site of initiation was unique and identical to that reported by many independent studies ([58–61], reviewed in [62–64]).
Circular SV40-replication templates containing 79 CTG repeats (Figure 1) were replicated using either HeLa (MMR-proficient) or LoVo (MMR-deficient; genetically MSH2−/− but also deficient in MSH3 and MSH6 proteins [55]) extracts in the presence of SV40 large T-antigen. MMR is known to be active in these replication assays, as the same extracts and conditions (with the exception of SV40 large T-antigen) are used for in vitro mismatch repair assays [54]. Alterations to the repeat following in vitro replication were measured using the STRIP assay [26, 27]. Briefly, fully-replicated products of in vitro replication were obtained by eliminating unreplicated or partially replicated material using DpnI digestion, DpnI-resistant material was transformed into bacteria, and then plasmid DNA was isolated from individual colonies (Figure S1). Repeat tracts in the replicated DNA were analyzed by releasing the repeat-containing fragment by restriction digestion followed by polyacrylamide gel electrophoresis. It is expected that each individual colony is representative of one product of replication due to the small ratio of DNA:bacteria during transformation [65]. Since the absence of MSH2 or MSH3 in mice led to high levels of repeat contractions, we focused upon a replication template which yields a contraction-bias when replicated by HeLa, in which the SV40-ori is placed 98 base pairs upstream of the CTG repeat (pDM79E – CTG strand as the lagging strand template) (Figure 1) [26, 27].
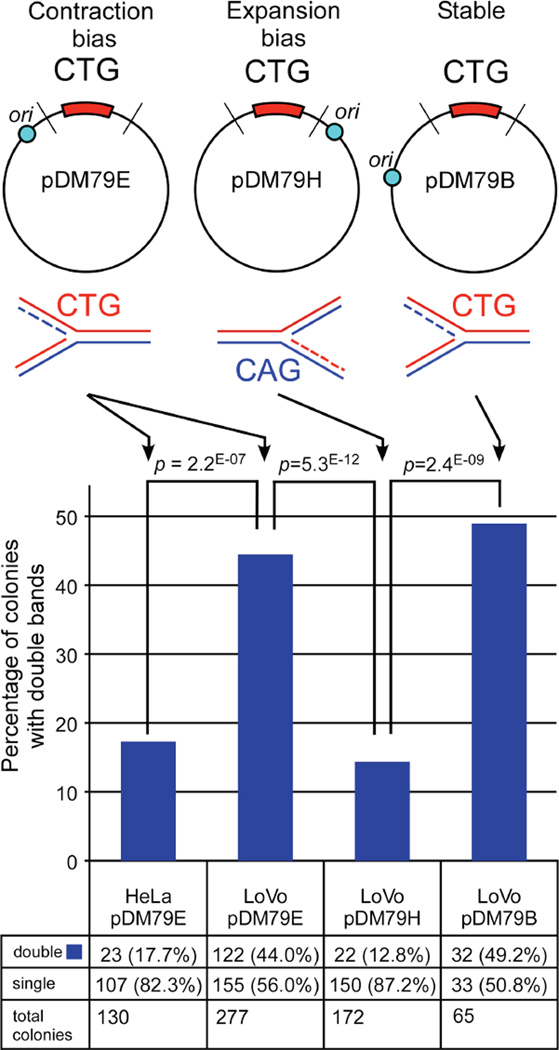
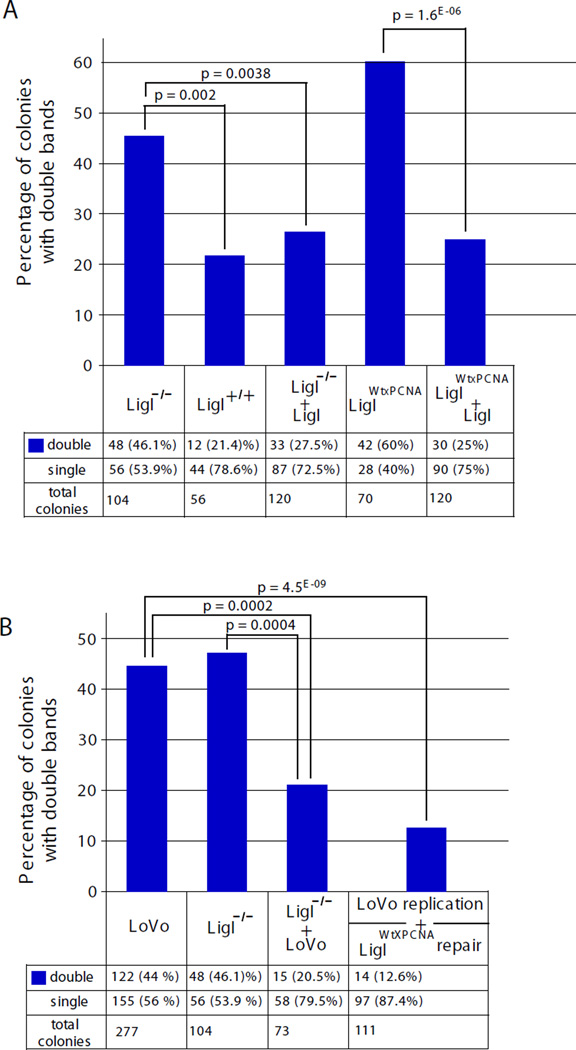
Figure 1. Replication-direction sensitive production of two repeat-containing fragments per colony by LoVo extracts.
SV40 replication templates used in in vitro replication assays, all with 79 CTG/CAG repeats. Locations of the SV40 replication origin (blue circle) relative to the repeat tract are: pDM79E-98 bp upstream; pDM79H-103 bp downstream; pDM79B-332 bp upstream. Lines intersecting the plasmids indicate restriction digestion sites to release the repeat containing fragment. Schematics below plasmids indicate the sequence of the lagging strand template. Templates were replicated with human cell extracts and assessed by bacterial STRIP analysis (Methods). Graph shows percentage of bacterial colonies from STRIP containing two (double) repeat-containing fragments. Actual numbers are shown at bottom and significance level indicated above graphs (χ2 test). NB, while both pDM79E and pDM79B are equally susceptible to slippage events, differences in either the site of Okazaki initiation relative to the repeat [66] and/or post-replication repair permits pDM79E and pDM79B to be contraction-biased and stable respectively following replication by HeLa extracts.
To assess the effect of MMR upon replication of the contraction-biased plasmid pDM79E, we performed replication using MMR-deficient LoVo cell extracts. Unexpectedly, analysis of individual replication products from these reactions yielded many mixed bacterial colonies that contained two repeat-containing fragments (Figure S2A, Figure 1). The high number of mixed colonies was specific for replication mediated by LoVo cell extracts, as there was a significant increase in the number of mixed colonies compared to the same template replicated by HeLa extracts (Figure 1: 44.0% versus 17.7%, χ2 test: p=2.2 * 10−7). The number of double repeat-containing colonies in HeLa replicated material (17.7%) was similar to that present using only the starting template (DNA not exposed to human cellular extract), 25.4%. Incubation of pDM79E with LoVo in the absence of T-antigen (no replication; no DpnI digestion) also yielded a similar result as the starting material: 25.7% mixed colonies (19 of 74 colonies). Thus, an increased number of mixed colonies depended upon LoVo-mediated replication.
The large number of mixed colonies also depended upon the direction of replication (CAG versus CTG as the lagging strand template), as LoVo replication of an expansion-biased template pDM79H did not lead to a high level of mixed colonies (12.8%, Figure 1). The presence of mixed colonies appeared to depend upon replication direction, but may also be due to a predisposition towards contractions versus expansions. To test this possibility, we assessed the levels of mixed colonies produced from replication template pDM79B (Figure 1) that is replicated in the same direction as the contraction-biased pDM79E, but initiates replication from a more distal origin yielding a stable repeat tract following replication [27]. Replication of pDM79B by LoVo yielded 49.2% double repeat fragments, similar to what was seen for pDM79E (44.0%) and significantly greater than what was seen for pDM79H (12.8%) (Figure 1; p = 2*10−5). Thus, the large number of replicated products producing mixed colonies was specific to replication direction as well as the MMR-deficiency.
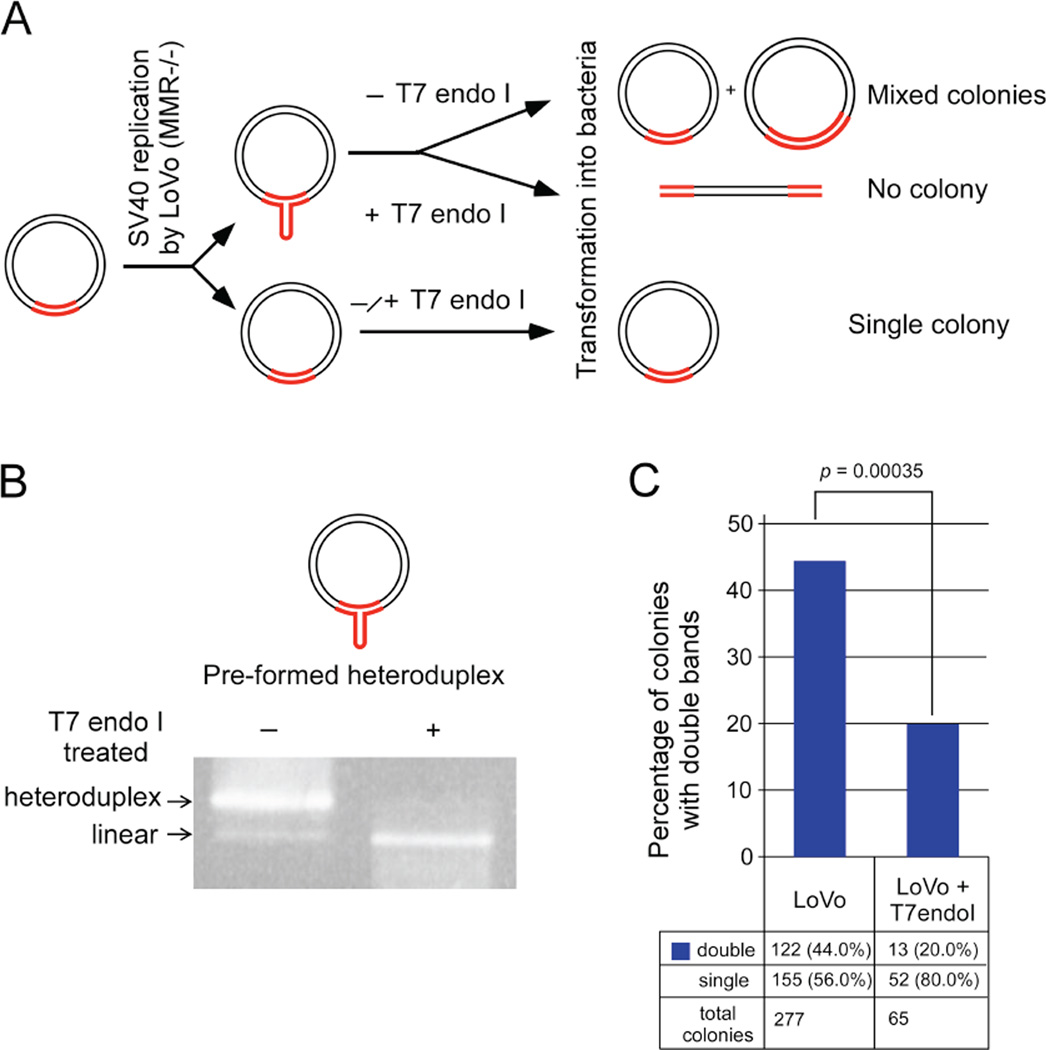
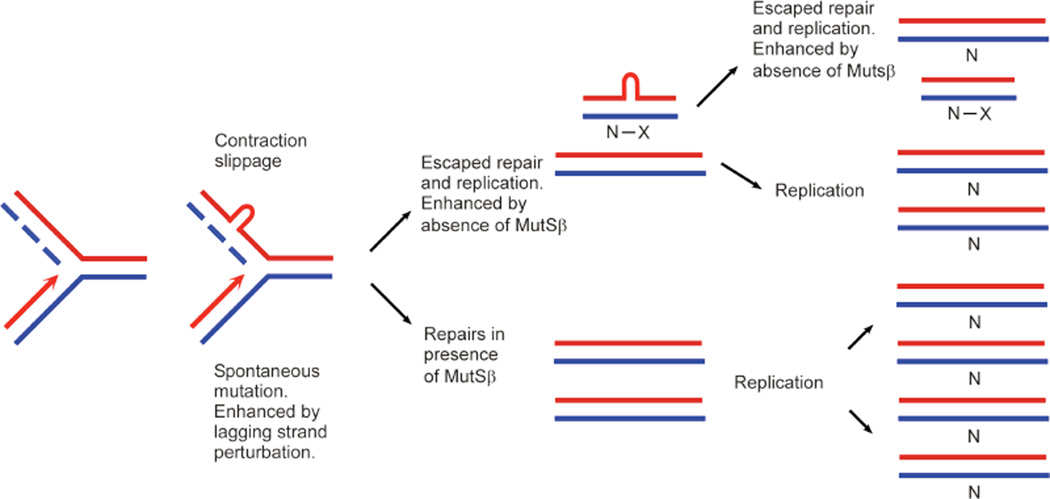
Since each bacterial colony represents one replication product, only one repeat–containing fragment is expected per colony. The presence of double repeat-containing fragments within a single colony might be due to any of four scenarios: 1) each colony may have a single plasmid that recombined to harbor two tandem repeat-containing fragments; 2) the persistence of linked newly-replicated circular daughter molecules with different repeat lengths; 3) unprocessed recombination intermediates between two plasmids containing different repeat lengths; or 4) SV40 replication products contain slipped-DNA heteroduplexes. A series of experiments tested and ruled-out the first three options (Figure S3–5), leading us to focus upon the possibility that slipped-DNAs had formed at the repeats. Specifically, during in vitro SV40 replication, DNA slippage may lead to a differential number of repeat units between the nascent and template strands – essentially yielding a heteroduplexed replication product, which may escape repair. Upon transformation into bacteria, the subsequent replication of the heteroduplex plasmid would result in mixed colonies containing two plasmids with different repeat lengths, each derived from the parental strands of the transformed heteroduplex. Transformation of plasmids harbouring heteroduplexes into bacteria has previously been reported to yield mixed colonies [67–72]. To test whether MMR-deficient replication indeed produced heteroduplex plasmids, replication products from LoVo extracts were digested with T7 endonuclease I (T7endoI), which cleaves across DNA-junctions including slipped-DNAs [73]. This enzyme cuts 5’ to the slip-out in heteroduplex DNA, resulting in linearization of the DNA at the slip-out (Figure 2A and 2B) which would eliminate any colonies with two repeat-containing fragments that were caused by heteroduplexes (Figure 2A). E. coli is unable to re-circularize linearized plasmids [74], a phenomenon that we confirmed with repeat-containing plasmids [75]. Thus, T7endoI digestion should leave only fully-duplexed circular DNAs with the same number of repeats on both strands to be transformed into bacteria and fewer double fragments should appear. Conditions for T7endoI cleavage of slipped DNAs were optimized using a pre-formed circular slipped-DNA (Figure 2B). LoVo replicated DNAs isolated by DpnI digestion were subjected to T7endoI cleavage or mock treated and assessed in bacteria. T7endoI cleavage significantly reduced the levels of double repeat-containing colonies from 44% down to 20% (p = 0.00035; Figure 2C). While T7endoI can also cleave DNA junctions other than slipped-DNAs, such as recombination intermediates, 2D-gel analyses of replication products ruled-out the possibility that the double repeat colonies were the result of recombination intermediates (Figure S5). Thus, the elimination of the doublet-colonies by T7endoI digestion demonstrated that the double repeat fragments were likely caused by the presence of slipped-DNA heteroduplexes in the SV40 replicated material.
Figure 2. Mechanism of double fragment formation.
A: Schematic showing likely scenario leading to double repeat-containing fragment formation, tested by T7endoI digestion. B: Gel showing digestion of pre-formed circular slipped-heteroduplex to linear plasmid by T7endoI digestion at optimized conditions. C: Graph shows percentage of colonies with double repeat-containing fragments in LoVo replicated pDM79E material with and without T7endoI digestion prior to bacterial transformation. Actual numbers are shown at bottom and significance level indicated above graphs (χ2 test). For other possible avenues see Figure S2–5.
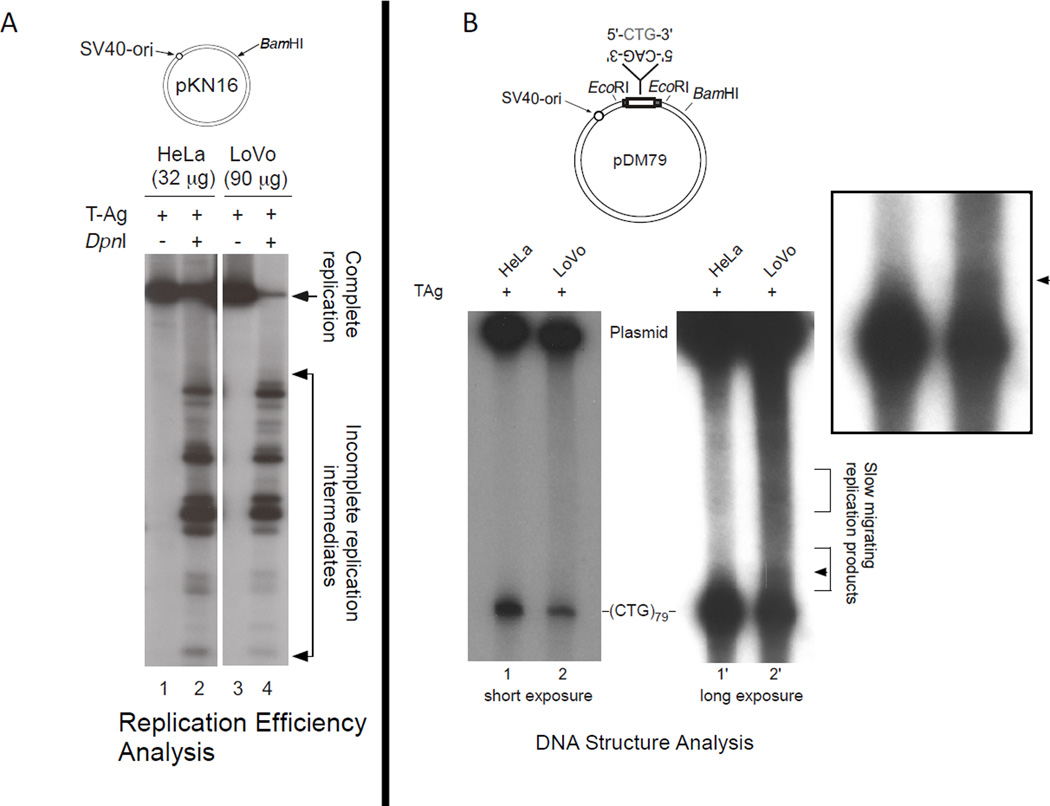
We attempted to independently detect the slipped DNAs using various approaches. Electron microscopic analysis failed to reveal any molecules with slip-outs, possibly due to the poor efficiency of replication and/or the small slip-out size. We assessed the relative replication efficiencies of LoVo and HeLa extracts (Figure 3A, see arrow). The amount of fully-replicated material produced by LoVo extracts was considerably less than the amount produced by HeLa extracts. Southern blotting, which in theory might be expected to yield a signal of slow-migrating slipped-DNA species also failed to detect a singular slipped-species. This was unsuccessful likely due to the heterogenous nature of the slip-outs yielding a smear of different species where there was not sufficient amounts of any one species to be detectable by Southern. We previously observed error-prone repair products during repair of pre-formed slipped–DNAs [54]. Error-prone-repair products could only be detected by radio-incorporation of all four radionucleotides; neither Southern blotting, nor electron microscopy were able to detect these heteroduplexes. Using a similar approach, we assessed whether the electrophoretic migration pattern of radio-incorporated replication products produced by LoVo differed from those produced by HeLa extracts. There was a considerable smear of electrophoretic species migrating slower than the starting repeat length in the LoVo replicated material compared to the HeLa replicated material (Figure 3B, see arrow). Slow-migration might be caused by the presence of slipped-heteroduplex DNA at the repeat region. Within this slow-migrating smear is an enrichment of a species evident as a band, more evident in darker exposures (Figure S5). These results, coupled with the T7endoI results above, support the presence of rare, short slip-out species produced by LoVo replication.
Figure 3. LoVo extracts have reduced SV40 replication efficiency, and yield more slow-migrating replication products.
A) The efficiency of replication by LoVo extracts was considerably reduced compared to the MMR-proficient HeLa cell extracts: Compare the amounts of fully-replicated products (linearized, see arrow) to the unreplicated or partially replicated material digested by DpnI (DpnI cleaves bi-methylated adenosine residues at GATC sites of bacterial but not primate replication). B) Replication products produced in the presence of all four radionucleotides were assessed for their electrophoretic migration following release of the repeat-containing fragment and resolved on 4% PAGE. Slipped-DNA heteroduplexes migrate slower than fully-duplexed species [76]. A slow-migrating smear representing a high-proportion of the repeat-containing fragment was evident for the LoVo replicated material compared to the HeLa replicated material. Moreover, a distinct species was just evident in the LoVo but not HeLa replicated material, see arrow.
Slipped-DNAs formed during contraction-biased replication are eliminated via MutSβ-mediated post-replication repair
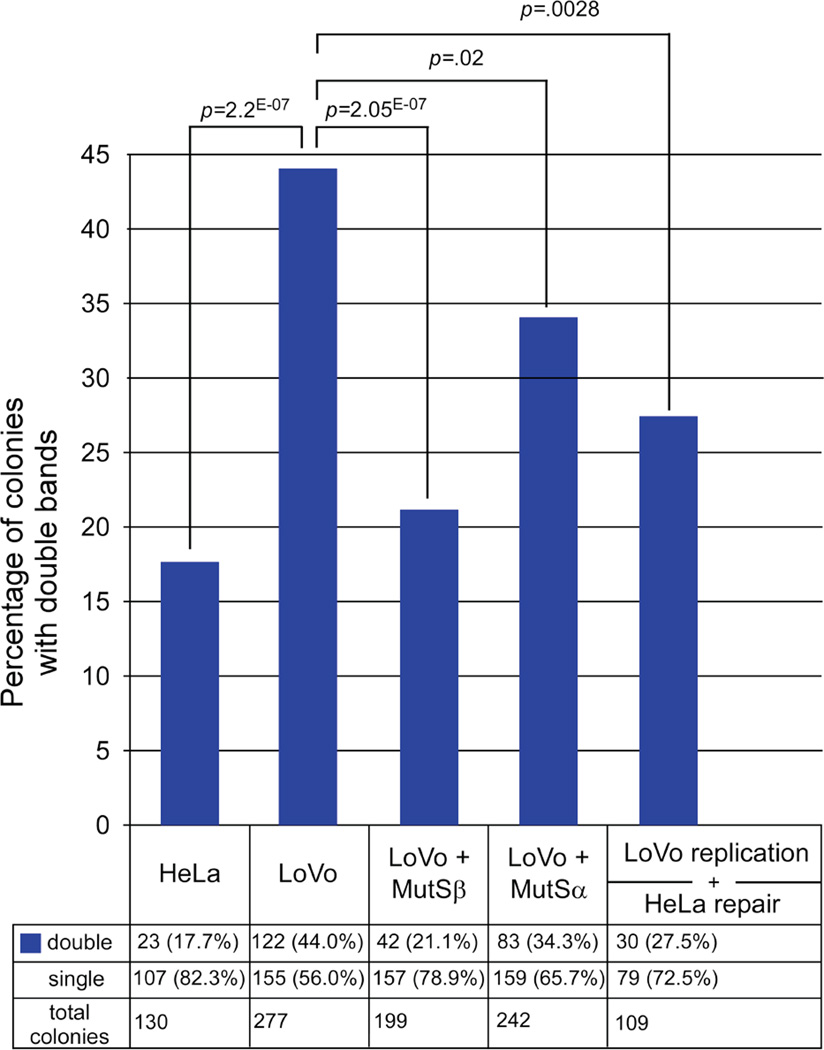
If slip-outs formed during replication by the MMR-deficient LoVo extracts escaped repair due to the absence of MMR proteins (MSH2, MSH3, and MSH6), their inclusion during replication should eliminate the presence of slip-outs in the replicated products. To test this in an isogenic manner, recombinant purified proteins were added to the LoVo extracts during replication and the presence of slipped-heteroduplexes in the replicated material was assessed. Addition of MutSβ to the LoVo replication reaction led to a highly significant decrease in heteroduplex formation (p = 2*10−7 compared with LoVo), reducing the number of double fragment colonies to levels present in the starting material and present in HeLa replicated material (Figure 4, 21%; p = 0.37 compared to starting material, p = 0.45 compared to HeLa). Addition of MutSα to the LoVo replication reaction led to a partial decrease in slipped heteroduplex formation (34.3% double fragments, Figure 4), which was significantly lower than in uncomplemented LoVo (p = 0.02), although still significantly more than found in HeLa (p = 0.0007). The complete rescue by addition of MutSβ indicates that it is involved in eliminating slipped-DNAs formed during replication. Since human MMR proteins are in complex with the replication fork machinery [38–40], it is likely that this repair occurs coincident with, or immediately following replication. The partial elimination of slipped-DNAs with MutSα and their complete elimination with MutSβ indicates that the heteroduplexes which form during replication are likely short slip-outs (≤3 repeat units), since MMR proteins are not involved in the repair of long slip-outs but are absolutely required to repair short slip-outs, a process preferentially mediated by MutSβ [55]. Thus, the inclusion of MutSβ or to a lesser degree MutSα, during replication of a contraction-biased template disfavours the retention of replication products harbouring slipped-DNA heteroduplexes.
Figure 4. MutSβ is required for repair of slipped-DNAs formed during replication.
Graph shows percentage of colonies with double repeat-containing fragments in HeLa- or LoVo-replicated pDM79E material. Levels of double fragments significantly decreased with the inclusion of MutSβ or MutSα proteins to LoVo extracts during replication or when LoVo-replicated material was de-proteinized, purified and subjected to post-replication repair with MMR-proficient HeLa extract. Actual numbers are shown at bottom and significance level indicated above graphs (χ2 test).
The retention of slipped-DNAs following replication by MMR-deficient LoVo extracts might be due to their inability to be repaired; it should be possible to repair these errors through in vitro repair of the replication products using MMR-proficient HeLa extracts. To test this, replication products produced by LoVo extracts were purified and then treated with HeLa extracts in the absence of SV40 large T-antigen (no replication can occur, allowing only repair). Following this post-replication repair, the number of slipped-DNA/mixed colonies was significantly decreased from 44% to 27.5% (p = 0.0028); levels similar to that present in the starting material (27.5% double fragments, p = 0.71 compared to starting material; Figure 4). Thus, slipped-DNAs formed during the passage of the MMR-deficient replication fork could be eliminated by post-replication repair using MMR-proficient HeLa extracts.
Slipped-DNAs form during lagging strand synthesis
In our system, changing replication direction led to a significant difference in slipped-DNA structure formation. It is likely that slipped-DNA formation was increased preferentially in the contraction-biased replication direction because the most stable structure forming CTG repeats were in the lagging strand template, and structures are more prone to forming in the lagging strand template as it is left single-stranded prior to the synthesis of Okazaki fragments. To test this, we assessed the effect of ligase I deficiency on slipped-DNA formation. Ligase 1 (LigI) is required for efficient joining of Okazaki fragments on the lagging strand, so a LigI deficiency would have a greater impact on lagging strand synthesis than on leading strand synthesis [77–81]. We used a human cell line, 46BR.1G1, which expresses a mutant DNA LigI, which maintains only 3–5% activity compared with wild type hLigI [77–81]. Replication of the contraction-biased pDM79E template by 46BRLigIm/m extracts led to a high level of slipped-DNAs/mixed colonies, which was significantly reduced in similar assays with extracts from a derivative that stably expresses wild type LigI 46BRLigIm/m;wt or through the addition of purified wild type hLigI protein to the replication reaction (Figure 5A). Similar results were obtained with extracts of a 46BR.1G1 derivative that stably expresses a mutant version of hLigI with a defective ability to interact with PCNA (46BRLigIm/m;wt-PCNA) (Figure 5A). Together these results support the idea that slipped-DNAs are formed during lagging strand synthesis and suggest that these structures are formed more readily when ligation of Okazaki fragments is delayed due to LigI-deficiency.
Figure 5. Human LigI–deficiency enhances slipped-DNA formation in pDM79E replicated DNAs.
Replication was performed using extracts derived from a human cell line, 46BR.1G1 (46BRLigIm/m), that expresses a mutant DNA LigI which maintains only 3–5% of ligase activity compared with the non-mutant hLigI, as well as a series of cell lines derived from 46BRLigIm/m: The Lig-deficient line (LigI−/−) was complemented to stably express wildtype LigI (46BRLigIm/m;wt = LigI+/+), or complemented to stably express a wildtype hLigI with a defective ability to interact with PCNA (46BRLigIm/m;wt-PCNA = LigIWtXPCNA). A: Graph shows percentage of colonies with double repeat-containing fragments in replicated pDM79E material mediated by extracts of cell derivatives of hLigI-deficient cells, in some reactions purified wiltype human ligase I protein (LigI) was added. Actual numbers are shown at bottom and significance level indicated above graphs (χ2 test). B: LigI defective and LoVo cell extracts complement each other and reduce levels of colonies with double repeat-containing fragments in replicated pDM79E material. LoVo-replicated material was de-proteinized, purified and subjected to post-replication repair with MMR-proficient, but replication-deficient LigIWtXPCNA extract. Actual numbers are shown at bottom and significance level indicated above graphs (χ2 test).
Slipped-DNAs formed during replication in the absence of MMR are eliminated by post-replication repair
The above results suggest that slipped-DNAs formed during replication fork passage may escape efficient post-replication repair. Replication by a mixture of MMR-deficient LoVo extracts with LigI deficient 46BRLigIm/m extracts did not lead to increased slipped-DNAs, indicating that these cell extracts complemented each other for both defective replication and repair activities in the formation/repair of slipped-DNAs (Figure 5B). To further delineate the timing of the formation and repair of slipped-DNAs we replicated the contraction-biased template in the absence of MMR (LoVo) and the replication products were then repaired in the absence of T-antigen, by the PCNA interaction-defective hLigI (46BRLigIm/m;wt-PCNA) extract. The product of this post-replication repair did not contain high levels of slipped-DNAs, consistent with the ability of this LigI-defective extract to perform repair (short path BER and slipped DNA repair) [81, 82] (Figure 5B). The fact that slip-outs produced during replication by MMR-deficient LoVo extracts can be repaired in the absence of replication, by MMR-competent extracts (LoVo+MutSβ, HeLa, or 46BRLigIm/m;wt-PCNA, Figures 4 & 5) indicates that post-replication repair processes following the passage of the replication fork are responsible for repairing slipped-DNAs formed in a contraction biased direction. This finding is consistent with the ability of HeLa extracts to repair slipped-DNAs formed by LoVo-mediated replication (Figure 4). Thus, the formation of slipped-DNAs most likely arises during Okazaki fragment synthesis and their repair likely follows replication fork passage. Towards addressing whether slipped-DNAs could form on chromatinized DNAs, we transfected the contraction templates into human cells, where templates are compacted into chromatin before DNA replication [83], in contrast to in vitro replication. Replication products isolated from HeLa or LoVo cells were assessed for repeat lengths in bacteria, as above. Replication in HeLa cells yielded 6.25% double repeat-containing fragments/slipped-DNAs (6 out of 96 colonies). In contrast, significantly more slipped-DNAs were present in DNAs replicated in LoVo cells 25% (24 out of 96, p = 0.00035). These results support the formation of slipped DNAs in chromatinized DNA.
Bacteria do not faithfully process slipped-heteroduplexes
Our initial aim was to assess repeat instability following MMR-deficient replication; however, the presence of multiple repeat-containing fragments per colony hindered this analysis. Since in vitro replication only permits a single-round of replication [59], at least one of the heteroduplex strands should contain the template starting length (79 repeats). Provided bacteria faithfully replicate each of the parental strands of the slipped-heteroduplex, we would expect mixed colonies where one of the plasmids contained the starting length and the other would have a mutation (expansion or contraction). The majority of the mixed colonies that we observed had two repeat lengths that both differed from the template, indicating that repeat length was being altered by the bacteria. This was confirmed by transformation of a pre-formed circular slipped heteroduplex into bacteria. While all of the colonies had two repeat lengths, the majority (>76%) of these mixed colonies had lengths distinct from either parental strand, indicating that bacterial processing of repeat-containing slipped-heteroduplexes is mutagenic – however faithfully represents the fact that a slipped-structure was used for transformation (Figure S6) (transformation of a fully-duplexed plasmid did not yield significant numbers of mixed colonies (<20%, p = 2.7 × 10−8)). Similar results were obtained using bacterial strains deficient for mismatch repair. Since bacteria process the slipped-DNAs to lengths that are distinct from the input heteroduplex strands, it is not possible to accurately assess instability of the LoVo-replicated products: if neither of the two repeat-fragments in a mixed colony is the same length of the starting material, it is impossible to know which of the two fragments was derived from the expansion or contraction event and which is from the template strand. However, our results can confirm that many, but not all of the mixed colonies arising from LoVo-replicated material are generated as a result of slipped-DNAs, and reveal the utility of bacterial transformation as a means to identify the presence of slipped-heteroduplexes. (A portion of the mixed colonies are of unknown origin, but likely arise following replication of the plasmids in bacteria as DNAs that have not been exposed to primate cell extracts yield some mixed colonies.) Despite our inability to report changes in instability, the existence of the double repeat-containing fragments/slipped-DNAs could provide clues as to how repeats are processed at replication forks in the absence of MMR proteins.
Discussion
A variety of mechanisms and metabolic processes seem to be involved in the expansion and contraction of trinucleotide repeats. Whether instability occurs during DNA replication, repair, or by another mechanism, the mutation process is invariably thought to involve the formation of slipped-DNAs. Recent evidence revealed that DM1 patient tissues harboured slipped-DNAs at the mutant DM1 locus in patient tissues [57]. Leffak and colleagues showed hairpin formation in cells harbouring an expanded CTG/CAG tract [25]. However, data revealing the process through which slipped-DNAs are formed and retained has been limited. The idea of slipped-DNAs forming during replication and then being repaired inaccurately is an idea that has long-persisted despite a lack of data supporting interdependence between replication and repair in CAG/CTG instability (Figure 6).
Figure 6. Slipped-DNA structures formed during replication that are intermediates of contraction events are repaired by MutSβ.
See Discussion.
The existence of rare DM1, HD and SBMA families that consistently display contractions of expanded repeats rather than expansions, suggests that processes leading to contractions may be distinct from processes leading to expansions [1–17]. Further evidence indicating differences in mechanisms leading to repeat contractions versus expansions is the switch from expansion-biased mutations to contraction-biased mutations in numerous CAG/CTG transgenic mouse models upon loss of MMR factors [43, 45, 48–50]. Moreover, the high level of CTG contractions in the female germline only of DM1 mice in a LigI-deficient background clearly indicates that there are distinct expansion and contraction processes [30].
In affected families and in transgenic mouse models of CAG/CTG instability, the repeats show expansions both upon transmission to offspring as well as in somatic tissues during ageing (reviewed in [42]). In many of these mouse models, deficiencies of MSH2 or MSH3 eliminated CAG/CTG repeat expansions and led to a repeat contraction bias [43, 45, 48–50]. In contrast to the consistent effect of MMR on CAG/CTG expansions in transgenic mice, mammalian and patient cell models, the role of MMR on CAG/CTG instability in non-mammalian systems has varied widely, with reports in yeast, bacteria, and flies claiming either no effect [84–86], stabilization [87–90], or a destabilizing effect [88, 91–93]. In all non-mammalian models, contractions predominated and expansions were rare. Where evident, the effect of MMR seemed to depend upon the direction of replication in some [91, 93] but not all [89] bacterial and yeast systems. However, the link between replication and repair in each study has been unclear.
In this study, we used an in vitro replication assay that permitted inclusion, exclusion, or separation of repair to investigate a potential link between DNA replication and mismatch repair in CAG/CTG repeat instability. Interestingly, we found that slipped-DNAs are formed mainly in the replication-direction that can lead to contractions, and these slipped-DNAs are repaired using MMR proteins. In contrast, any mutagenic intermediates formed on the expansion-biased replication template are processed in the presence or absence of MMR. Our results suggest that the role of MMR is different for contraction versus expansion events.
In all systems tested thus far for effects of replication on repeat instability, replication direction is a crucial factor [66], albeit replication is not the only contributor to instability. The direction of replication through a repeat strand will be determined by the location of initiation relative to the repeat. Previously, we revealed that while there were two replication origins at the human DM1 locus, upstream and downstream of the repeat, the replication direction for expansions on the mutant allele used the CAG strand as the lagging strand template [28], the same as what we observe for expansions in the SV40 system, and the same as has been mapped for the HD and SCA7 loci in patient cells [31]. We proposed that the switch from CAG/CTG expansions to contractions due to a LigI-deficiency in the maternal germline of the DM1 mice might be due to the maternal germline-specific activation of the upstream DM1 origin of replication coupled with de-activation of the downstream origin [30]. This change in replication direction would then make the CTG strand the lagging strand template, which in the presence of perturbed LigI, would enhance contractions. Such contractions might be mediated by slipped-DNA formation, as observed herein with the SV40 system. While the SV40 system has fundamental differences from chromosomal replication, the similar replication polarity effects for expansions between it and the replication direction at disease loci in DM1, HD, and SCA7 patient cells (all use CAG as lagging strand template) [28, 31] support SV40 as a relevant model for instability. Furthermore, we were able to observe increased slipped-DNA formation in chromatinized plasmids, further supporting the SV40 system as an informative model of repeat instability.
In our system, changing replication direction led to a significant difference in slipped-DNA structure formation. It is likely that slipped-heteroduplex formation was increased when CTG repeats were in the lagging strand template because CTG repeats form biophysically more stable structures than CAG repeats. Slipped-DNA structures are likely more prone to form in the lagging strand template as it is left single-stranded for stretches of ~300 nucleotides prior to the synthesis of Okazaki fragments [58, 94]. We show that a deficiency of human ligase I, either as result of reduced catalytic activity or a defect in binding to PCNA, also increased the formation of slipped-DNAs, indicating that slipped-DNA formation occurs on the lagging strand, as LigI is critically required for efficient joining of Okazaki fragments [77–81]. Perturbation of lagging strand synthesis may enhance the propensity of slippage. Similarly, changing MMR status may also exacerbate structure formation at the lagging strand template because MMR is more active on the lagging strand than the leading strand [95, 96].
Recently Romanova & Crouse reported that MutSα has a strong bias toward repair of insertion loops, while MutSβ has an even stronger bias toward repair of deletion loops [97]. These authors suggested that this bias in repair could be due to the different interactions of the MutS complexes with the various MutL complexes, initially demonstrated by the labs of Jinks-Robertson, Liskay and colleagues [98]. Our findings suggest that human MutSβ protects against CTG/CAG contraction events, which is consistent with a preference of MutSβ for deletion loops [97], explaining the repeat contraction bias in many CTG/CAG transgenic mice deficient in either MSH2 or MSH3 [43–47, 49, 50].
Repeated attempts to directly visualize slipped-DNAs by electron microscopy were unsuccessful, possibly due to their small size. Since repair of short slip-outs (≤3 repeat units) is MutSβ-dependent [55], the slip-outs we detected were probably short because their presence was decreased to background levels with the addition of exogenous MutSβ. Slipped-DNAs may also form during HeLa replication, but short slip-outs would be repaired by the MMR machinery and hence would be undetectable. Further data supporting that slip-outs formed were short is the relative slow-electrophoretic migration of the replicated material, much like short slip-outs (Figure 3) [76]. Our data suggest that slipped-DNAs formed during LoVo replication persisted by escaping processing due to the lack of MMR proteins, as we were able to reduce the incidence of these structures by the addition of MutSβ. Other factors driving the formation of slipped-DNAs may exist – as revealed by a LigI-deficiency. Taken all together, the greater propensity for lagging strand CTG repeat structure formation and the persistence of these slip-outs in the absence of MMR, might explain why slipped-DNAs arise only in the contraction-biased replication template, and may indicate that contractions occur by a different mechanism than expansions.
There are large differences between repeat lengths in various tissues in DM1 patients due to somatic instability [19]. The ongoing somatic expansions of trinucleotide repeats are thought to contribute to disease progression and severity. Notably, individuals with smaller expansion sizes have later age-of-onset, less severe disease and slower progression [1–17]. Due to differences in gene expression profiles between tissues [41, 99], or differences in replication origin usage [27], certain tissues could have a greater propensity to form slipped-DNA structures, which could alter trinucleotide repeat instability. If these structures require MMR for their repair, then when MMR is lacking (genetically, or due to low expression levels) these DNA structures may persist leading to contractions. Therapeutically, increasing contractions of expanded repeats would be advantageous – even if expansions were still occurring, if contraction frequencies were greatly enhanced then the net result would either be a decrease in repeat length, or shorter length increases. Since the MMR protein MutSβ is needed to repair contraction intermediates, disrupting MutSβ function would most likely increase repeat contractions and thus mitigate disease progression and severity.
Materials and Methods
Cell Lines and Cell Culture
the repair proficient HeLa cells and the MMR-deficient LoVo cells (genetically deficient in MSH2, and deficient in MSH3 and MSH6 proteins), have previously been described and characterized [55]. Three different ligase I-defective variant cell lines derived from 46BR.1G1 (46BRLigI; hemizygous or homozygous for R771W with only 3–5% of wt ligase activity) have been described [81, 82]. Briefly, these were created by stably transfecting pRC/RSV plasmids (Invitrogen) into the original patient cell line [81]; they are (i) 46BRLigIm/m carrying an empty vector, (ii) 46BRLigIm/m,wt expressing a wild type hLigI, and (iii) 46BRLigIm/m,wt-PCNA expressing a hLigI cDNA mutant in PCNA binding. 46BRLigIm/m;wt and 46BRLigIm/m;wt-PCNA are complemented hLigI cell lines but not truly corrected because the endogenous hLigI mutation is still present in all derivative cell lines. Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, 1% penicillin/streptomycin, and 300 ug/ml Geneticin as described [81].
Replication Templates / Heteroduplexed DNA
Replication template design has been previously described [26]. Briefly, the pDM79 circular plasmid contains a fragment of a genomic DM1 clone with 79 CTG repeats. pDM79E, pDM79H, and pDM79B have an SV40-ori containing fragment inserted into the EcoRI, HindIII, and BglI sites of the pBluescript KSII+ plasmid, respectively. The EcoRI site is 98 nucleotides upstream of the CTG repeat, the HindIII site is 103 nts downstream of the CTG repeat, and the BglI site is 332 nts upstream of the CTG repeat. pDM79E-2 and pDM79H-2 each have an insert between the SV40-ori and the repeat such that they are ~800 nucleotides apart for improved 2D gel resolution (Figure S5). Heteroduplexed DNA with 48 CTG repeats paired with 47 CAG repeats was made as previously described [54, 55].
In Vitro Replication/Repair
Cell extracts of HeLa/LoVo were made and replication and repair reactions were performed as described [26, 54, 55]. Briefly, plasmid DNA containing 79 CAG/CTG repeats (150 ng) was replicated in a reaction containing: 100 µM of each dNTP, 200 µM of GTP, UTP and CTP, 4 mM ATP, 40 mM creatine phosphatase (Roche), 100 µg/mL creatine kinase (Roche), 1 µg of SV40 T-antigen (Chimerx), and 30 µL of whole cell extracts (concentration of ~4 mg/mL). Reactions were incubated at 37 °C for 4 hours, and stopped for 1 hour (2× stop solution: 2 mg/mL proteinase K, 2% SDS, 50 mM EDTA pH 8.0). This system does not permit re-initiation, hence products have undergone only one round of replication [59]. DNA was purified by phenol/chloroform extraction followed by ethanol precipitation and resuspension in water. DpnI digestion reaction conditions ensured that digestion eliminated only unreplicated and partially replicated DNAs [65, 100].
Post-replication repair reactions were identical to replication reactions except that SV40 T-antigen was not included, and the reaction time was only 30 minutes.
Recombinant hMutSα, hMutSβ and LigI Expression
Expression and purification of recombinant proteins using baculoviruses expressing hMSH2, hMSH3, and hMSH6 was as previously described [55]. Human LigI protein was expressed and purified as described [81].
Analysis of Replication Products
Replication products were analyzed as previously described [26]. Briefly, purified replication products were digested with DpnI to remove un-replicated material, and then transformed into DH5α at DNA:bacteria ratios that permit less than one plasmid per bacterial cell [65]. In the absence of T-antigen during replication reactions, all DNA was sensitive to DpnI digestion and no bacterial colonies were obtained, indicating that DpnI digestion was effective. Bacterial colonies were cultured for ~6 hours, and plasmids were purified. Mini-prepped DNA was digested with HindIII or PstI to release the repeat containing fragment and run on 4% acrylamide gels to assess the products of replication.
TopoII treatment
TopoII alpha (TopoGEN, TG2000H) treatment in the presence of extracts (Figure S4) was carried out by adding the enzyme directly to the extract during incubation. As a control for activity, 0.1 µg of kDNA was treated with TopoII in reaction buffer (50 mM Tris-HCl pH 8, 120 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 0.5 mM ATP, 30 µg BSA/mL) for 15 min at 37 °C. TopoII is active on kDNA in both TopoII reaction buffer and in cell extracts (Figure S4).
T7endoI treatment
Heteroduplexed DNA [54], and LoVo replicated material before transformation, was treated with T7endoI (New England Biolabs, M0302S) in NEBuffer 2 for 15 minutes at 37 °C. T7endoI treated replication material was re-purified by phenol/chloroform extraction before transformation.
2D gel electrophoresis
HeLa and LoVo cells were co-transfected with 1093 the SV40 large T-antigen expression construct ([101], kindly provided by Ellen Fanning) and either pDM79E-2 or pDM79H-2 using Fugene 6 (Roche). After 24 hours, plasmid DNA was collected via Hirt’s lysis. pDM79E-2 was digested with AvrII/XmnI and pDM79H-2 was digested with AlwNI/AvrII, then DNA was purified by phenol/chloroform extraction. First dimension was run on a 0.4% agarose gel in the absence of ethidium bromide (EtBr) for 22 hours at 25V at room temperature. Second dimension was run on a 1% agarose gel in the presence of EtBr for 6 hours at 150V. Southern blotting was performed after the second dimension, probing for the XmnI/AlwNI fragment (bold in Figure S5A). Some of the Hirt’s material was treated with DpnI to eliminate un-replicated material, and 1093 was linearized using BbsI, which uniquely cuts it to eliminate its contribution to the colonies. These DNAs were transformed and assessed by STRIP analysis.
Supplementary Material
Highlights.
-
-
slipped DNAs persist following replication of a contraction biased template in the absence of mismatch repair
-
-
repair of contraction intermediate slipped DNAs is performed by MutSβ
-
-
role of MMR proteins is distinct for repeat expansion versus contraction events
Acknowledgments
We thank Ellen Fanning for her kind gift of the SV40 Large T-antigen expression construct. We thank Sergei Mirkin, Massimo Lopes, and Katharina Zwicky for helpful discussions regarding our 2D gel analysis. We thank Yuh-Hwa Wang for her efforts to visualize our slipped-DNAs via electron microscopy. The Pearson Lab was supported by the Muscular Dystrophy Association Canada, the Canadian Institutes of Health Research (MOP-97896), the Paul Wellstone Muscular Dystrophy Cooperative Research Center, and by Grant U54NS48843 from the National Institutes of Health, Tribute Communities, the Kazman Family Fund, and the Marigold Foundation. M.M.S. was supported by studentships from The Hospital for Sick Children Research Training Competition and the Canadian Institutes of Health Research Collaborative Graduate Training Program in Molecular Medicine. A.L.C. was supported by a RESTRACOMP fellowship. A.B.P. was supported by The Sick Kids SSURE program. The Tomkinson Lab was supported by the National Institutes of Health (NIH R01 GM57479).
Bibliography
- 1.Abeliovich D, Lerer I, Pashut-Lavon I, Shmueli E, Raas-Rothschild A, Frydman M. Negative expansion of the myotonic dystrophy unstable sequence. American journal of human genetics. 1993;52:1175–1181. [PMC free article] [PubMed] [Google Scholar]
- 2.Achiron A, Magal N, Shem-Tov N, Noy S, Shohat M, Gadoth N. Myotonic dystrophy gene analysis in affected Israeli families. Israel journal of medical sciences. 1994;30:622–625. [PubMed] [Google Scholar]
- 3.Amiel J, Raclin V, Jouannic JM, Morichon N, Hoffman-Radvanyi H, Dommergues M, Feingold J, Munnich A, Bonnefont JP. Trinucleotide repeat contraction: a pitfall in prenatal diagnosis of myotonic dystrophy. Journal of medical genetics. 2001;38:850–852. doi: 10.1136/jmg.38.12.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashizawa T, Anvret M, Baiget M, Barcelo JM, Brunner H, Cobo AM, Dallapiccola B, Fenwick RG, Jr, Grandell U, Harley H, et al. Characteristics of intergenerational contractions of the CTG repeat in myotonic dystrophy. American journal of human genetics. 1994;54:414–423. [PMC free article] [PubMed] [Google Scholar]
- 5.Aziz NA, van Belzen MJ, Coops ID, Belfroid RD, Roos RA. Parent-of-origin differences of mutant HTT CAG repeat instability in Huntington's disease. European journal of medical genetics. 2011;54:e413–e418. doi: 10.1016/j.ejmg.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Brunner HG, Jansen G, Nillesen W, Nelen MR, de Die CE, Howeler CJ, van Oost BA, Wieringa B, Ropers HH, Smeets HJ. Brief report: reverse mutation in myotonic dystrophy. The New England journal of medicine. 1993;328:476–480. doi: 10.1056/NEJM199302183280705. [DOI] [PubMed] [Google Scholar]
- 7.Chung MY, Ranum LP, Duvick LA, Servadio A, Zoghbi HY, Orr HT. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nature genetics. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- 8.Giordano M, De Angelis MS, Mutani R, Richiardi PM. Origin of a regressed myotonic dystrophy allele. Journal of medical genetics. 1994;31:130–132. doi: 10.1136/jmg.31.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez de Munain A, Cobo AM, Saenz A, Blanco A, Poza JJ, Martorell L, Marti-Masso JF, Baiget M. Frequency of intergenerational contractions of the CTG repeats in myotonic dystrophy. Genetic epidemiology. 1996;13:483–487. doi: 10.1002/(SICI)1098-2272(1996)13:5<483::AID-GEPI4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 10.Matsumura R, Namikawa T, Miki T, Kihira T, Yamagata H, Mano Y, Takayanagi T. An intergenerational contraction of the CTG repeat in Japanese myotonic dystrophy. Journal of the neurological sciences. 1996;139:48–51. [PubMed] [Google Scholar]
- 11.Nahhas FA, Garbern J, Krajewski KM, Roa BB, Feldman GL. Juvenile onset Huntington disease resulting from a very large maternal expansion. American journal of medical genetics. Part A. 2005;137A:328–331. doi: 10.1002/ajmg.a.30891. [DOI] [PubMed] [Google Scholar]
- 12.Norremolle A, Hasholt L, Petersen CB, Eiberg H, Hasselbalch SG, Gideon P, Nielsen JE, Sorensen SA. Mosaicism of the CAG repeat sequence in the Huntington disease gene in a pair of monozygotic twins. American journal of medical genetics. Part A. 2004;130A:154–159. doi: 10.1002/ajmg.a.30128. [DOI] [PubMed] [Google Scholar]
- 13.O'Hoy KL, Tsilfidis C, Mahadevan MS, Neville CE, Barcelo J, Hunter AG, Korneluk RG. Reduction in size of the myotonic dystrophy trinucleotide repeat mutation during transmission. Science. 1993;259:809–812. doi: 10.1126/science.8094260. [DOI] [PubMed] [Google Scholar]
- 14.Puymirat J, Giguere Y, Mathieu J, Bouchard JP. Intergenerational contraction of the CTG repeats in 2 families with myotonic dystrophy type 1. Neurology. 2009;73:2126–2127. doi: 10.1212/WNL.0b013e3181c677e1. [DOI] [PubMed] [Google Scholar]
- 15.Sharp A, Hurst J. Somatic instability of the androgen receptor CAG repeat in a normal female. American journal of medical genetics. Part A. 2003;117A:161–163. doi: 10.1002/ajmg.a.10897. [DOI] [PubMed] [Google Scholar]
- 16.Shelbourne P, Winqvist R, Kunert E, Davies J, Leisti J, Thiele H, Bachmann H, Buxton J, Williamson B, Johnson K. Unstable DNA may be responsible for the incomplete penetrance of the myotonic dystrophy phenotype. Human molecular genetics. 1992;1:467–473. doi: 10.1093/hmg/1.7.467. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y, Wang Y, Yang P, Liu Y, Wang B, Podolsky R, McIndoe R, Wang CY. Intergeneration CAG expansion and contraction in a Chinese HD family. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2006;141B:242–244. doi: 10.1002/ajmg.b.30261. [DOI] [PubMed] [Google Scholar]
- 18.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nature reviews. Genetics. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 19.Castel A. Lopez, Nakamori M, Tome S, Chitayat D, Gourdon G, Thornton CA, Pearson CE. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Human molecular genetics. 2011;20:1–15. doi: 10.1093/hmg/ddq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrell BT, Lahue RS. CAG*CTG repeat instability in cultured human astrocytes. Nucleic acids research. 2006;34:4495–4505. doi: 10.1093/nar/gkl614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martorell L, Monckton DG, Gamez J, Johnson KJ, Gich I, Lopez de Munain A, Baiget M. Progression of somatic CTG repeat length heterogeneity in the blood cells of myotonic dystrophy patients. Human molecular genetics. 1998;7:307–312. doi: 10.1093/hmg/7.2.307. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Lau R, Marcadier JL, Chitayat D, Pearson CE. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. American journal of human genetics. 2003;73:1092–1105. doi: 10.1086/379523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freudenreich CH, Stavenhagen JB, Zakian VA. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Molecular and cellular biology. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nature genetics. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 25.Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG) × (CAG) repeat hairpins in human cells. Nature chemical biology. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panigrahi GB, Cleary JD, Pearson CE. In vitro (CTG)*(CAG) expansions and deletions by human cell extracts. The Journal of biological chemistry. 2002;277:13926–13934. doi: 10.1074/jbc.M109761200. [DOI] [PubMed] [Google Scholar]
- 27.Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nature genetics. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 28.Cleary JD, Tome S, Castel A. Lopez, Panigrahi GB, Foiry L, Hagerman KA, Sroka H, Chitayat D, Gourdon G, Pearson CE. Tissue- and age-specific DNA replication patterns at the CTG/CAG-expanded human myotonic dystrophy type 1 locus. Nature structural & molecular biology. 2010;17:1079–1087. doi: 10.1038/nsmb.1876. [DOI] [PubMed] [Google Scholar]
- 29.Panigrahi GB, Slean MM, Simard JP, Pearson CE. Human mismatch repair protein hMutLalpha is required to repair short slipped-DNAs of trinucleotide repeats. The Journal of biological chemistry. 2012;287:41844–41850. doi: 10.1074/jbc.M112.420398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tome S, Panigrahi GB, Castel A. Lopez, Foiry L, Melton DW, Gourdon G, Pearson CE. Maternal germline-specific effect of DNA ligase I on CTG/CAG instability. Human molecular genetics. 2011;20:2131–2143. doi: 10.1093/hmg/ddr099. [DOI] [PubMed] [Google Scholar]
- 31.Nenguke T, Aladjem MI, Gusella JF, Wexler NS, Arnheim N, Venezuela HDP. Candidate DNA replication initiation regions at human trinucleotide repeat disease loci. Human molecular genetics. 2003;12:1021–1028. doi: 10.1093/hmg/ddg111. [DOI] [PubMed] [Google Scholar]
- 32.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edelmann W, Umar A, Yang K, Heyer J, Kucherlapati M, Lia M, Kneitz B, Avdievich E, Fan K, Wong E, Crouse G, Kunkel T, Lipkin M, Kolodner RD, Kucherlapati R. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer research. 2000;60:803–807. [PubMed] [Google Scholar]
- 34.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. The Journal of biological chemistry. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 35.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Current biology : CB. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 36.Risinger JI, Umar A, Boyd J, Berchuck A, Kunkel TA, Barrett JC. Mutation of MSH3 in endometrial cancer and evidence for its functional role in heteroduplex repair. Nature genetics. 1996;14:102–105. doi: 10.1038/ng0996-102. [DOI] [PubMed] [Google Scholar]
- 37.Srivatsan A, Bowen N, Kolodner RD. Mispair-specific recruitment of the Mlh1-Pms1 complex identifies repair substrates of the Saccharomyces cerevisiae Msh2-Msh3 complex. The Journal of biological chemistry. 2014;289:9352–9364. doi: 10.1074/jbc.M114.552190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. The Journal of biological chemistry. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 39.Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes & development. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masih PJ, Kunnev D, Melendy T. Mismatch Repair proteins are recruited to replicating DNA through interaction with Proliferating Cell Nuclear Antigen (PCNA) Nucleic acids research. 2008;36:67–75. doi: 10.1093/nar/gkm943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tome S, Simard JP, Slean MM, Holt I, Morris GE, Wojciechowicz K, te Riele H, Pearson CE. Tissue-specific mismatch repair protein expression: MSH3 is higher than MSH6 in multiple mouse tissues. DNA repair. 2013;12:46–52. doi: 10.1016/j.dnarep.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Slean MM, Panigrahi GB, Ranum LP, Pearson CE. Mutagenic roles of DNA "repair" proteins in antibody diversity and disease-associated trinucleotide repeat instability. DNA repair. 2008;7:1135–1154. doi: 10.1016/j.dnarep.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Foiry L, Dong L, Savouret C, Hubert L, te Riele H, Junien C, Gourdon G. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Human genetics. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- 44.Kovtun IV, Thornhill AR, McMurray CT. Somatic deletion events occur during early embryonic development and modify the extent of CAG expansion in subsequent generations. Human molecular genetics. 2004;13:3057–3068. doi: 10.1093/hmg/ddh325. [DOI] [PubMed] [Google Scholar]
- 45.Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, te Riele H, Junien C, Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. The EMBO journal. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savouret C, Garcia-Cordier C, Megret J, te Riele H, Junien C, Gourdon G. MSH2-dependent germinal CTG repeat expansions are produced continuously in spermatogonia from DM1 transgenic mice. Molecular and cellular biology. 2004;24:629–637. doi: 10.1128/MCB.24.2.629-637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wheeler VC, Lebel LA, Vrbanac V, Teed A, te Riele H, MacDonald ME. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Human molecular genetics. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 48.Dragileva E, Hendricks A, Teed A, Gillis T, Lopez ET, Friedberg EC, Kucherlapati R, Edelmann W, Lunetta KL, MacDonald ME, Wheeler VC. Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiology of disease. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nature genetics. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 50.van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, Groenen PJ, Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Human molecular genetics. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 51.Seriola A, Spits C, Simard JP, Hilven P, Haentjens P, Pearson CE, Sermon K. Huntington's and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Human molecular genetics. 2011;20:176–185. doi: 10.1093/hmg/ddq456. [DOI] [PubMed] [Google Scholar]
- 52.Du J, Campau E, Soragni E, Jespersen C, Gottesfeld JM. Length-dependent CTG.CAG triplet-repeat expansion in myotonic dystrophy patient-derived induced pluripotent stem cells. Human molecular genetics. 2013;22:5276–5287. doi: 10.1093/hmg/ddt386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou C, Chan NL, Gu L, Li GM. Incision-dependent and error-free repair of (CAG)(n)/(CTG)(n) hairpins in human cell extracts. Nature structural & molecular biology. 2009;16:869–875. doi: 10.1038/nsmb.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nature structural & molecular biology. 2005;12:654–662. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 55.Panigrahi GB, Slean MM, Simard JP, Gileadi O, Pearson CE. Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutSbeta, but clustered slip-outs are poorly repaired. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:12593–12598. doi: 10.1073/pnas.0909087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang T, Huang J, Gu L, Li GM. In vitro repair of DNA hairpins containing various numbers of CAG/CTG trinucleotide repeats. DNA repair. 2012;11:201–209. doi: 10.1016/j.dnarep.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Axford MM, Wang YH, Nakamori M, Zannis-Hadjopoulos M, Thornton CA, Pearson CE. Detection of slipped-DNAs at the trinucleotide repeats of the myotonic dystrophy type I disease locus in patient tissues. PLoS genetics. 2013;9:e1003866. doi: 10.1371/journal.pgen.1003866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hay RT, DePamphilis ML. Initiation of SV40 DNA replication in vivo: location and structure of 5' ends of DNA synthesized in the ori region. Cell. 1982;28:767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- 59.Li JJ, Kelly TJ. Simian virus 40 DNA replication in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stillman B, Gerard RD, Guggenheimer RA, Gluzman Y. T antigen and template requirements for SV40 DNA replication in vitro. The EMBO journal. 1985;4:2933–2939. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dean FB, Borowiec JA, Ishimi Y, Deb S, Tegtmeyer P, Hurwitz J. Simian virus 40 large tumor antigen requires three core replication origin domains for DNA unwinding and replication in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:8267–8271. doi: 10.1073/pnas.84.23.8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erdile LF, Collins KL, Russo A, Simancek P, Small D, Umbricht C, Virshup D, Cheng L, Randall S, Weinberg D, et al. Initiation of SV40 DNA replication: mechanism and control. Cold Spring Harbor symposia on quantitative biology. 1991;56:303–313. doi: 10.1101/sqb.1991.056.01.037. [DOI] [PubMed] [Google Scholar]
- 63.Stillman B. Initiation of eukaryotic DNA replication in vitro. BioEssays : news and reviews in molecular, cellular and developmental biology. 1988;9:56–60. doi: 10.1002/bies.950090205. [DOI] [PubMed] [Google Scholar]
- 64.Stillman B. Initiation of eukaryotic DNA replication in vitro. Annual review of cell biology. 1989;5:197–245. doi: 10.1146/annurev.cb.05.110189.001213. [DOI] [PubMed] [Google Scholar]
- 65.Vassilev L, Johnson EM. Evaluation of autonomous plasmid replication in transfected mammalian cells. Nucleic acids research. 1988;16:7742. doi: 10.1093/nar/16.15.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cleary JD, Pearson CE. Replication fork dynamics and dynamic mutations: the fork-shift model of repeat instability. Trends in genetics : TIG. 2005;21:272–280. doi: 10.1016/j.tig.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Abastado JP, Cami B, Dinh TH, Igolen J, Kourilsky P. Processing of complex heteroduplexes in Escherichia coli and Cos-1 monkey cells. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:5792–5796. doi: 10.1073/pnas.81.18.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cami B, Chambon P, Kourilsky P. Correction of complex heteroduplexes made of mouse H-2 gene sequences in Escherichia coli K-12. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:503–507. doi: 10.1073/pnas.81.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Campbell CR, Ayares D, Watkins K, Wolski R, Kucherlapati R. Single-stranded DNA gaps, tails and loops are repaired in Escherichia coli. Mutation research. 1989;211:181–188. doi: 10.1016/0027-5107(89)90118-8. [DOI] [PubMed] [Google Scholar]
- 70.Carraway M, Marinus MG. Repair of heteroduplex DNA molecules with multibase loops in Escherichia coli. Journal of bacteriology. 1993;175:3972–3980. doi: 10.1128/jb.175.13.3972-3980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fishel RA, Siegel EC, Kolodner R. Gene conversion in Escherichia coli. Resolution of heteroallelic mismatched nucleotides by co-repair. Journal of molecular biology. 1986;188:147–157. doi: 10.1016/0022-2836(86)90300-1. [DOI] [PubMed] [Google Scholar]
- 72.Westmoreland J, Porter G, Radman M, Resnick MA. Highly mismatched molecules resembling recombination intermediates efficiently transform mismatch repair proficient Escherichia coli. Genetics. 1997;145:29–38. doi: 10.1093/genetics/145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pearson CE, Wang YH, Griffith JD, Sinden RR. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n. (CAG)n repeats from the myotonic dystrophy locus. Nucleic acids research. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Conley EC, Saunders VA, Jackson V, Saunders JR. Mechanism of intramolecular recyclization and deletion formation following transformation of Escherichia coli with linearized plasmid DNA. Nucleic acids research. 1986;14:8919–8932. doi: 10.1093/nar/14.22.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marcadier JL, Pearson CE. Fidelity of primate cell repair of a double-strand break within a (CTG).(CAG) tract. Effect of slipped DNA structures. The Journal of biological chemistry. 2003;278:33848–33856. doi: 10.1074/jbc.M304284200. [DOI] [PubMed] [Google Scholar]
- 76.Tam M, Erin Montgomery S, Kekis M, Stollar BD, Price GB, Pearson CE. Slipped (CTG).(CAG) repeats of the myotonic dystrophy locus: surface probing with anti-DNA antibodies. Journal of molecular biology. 2003;332:585–600. doi: 10.1016/s0022-2836(03)00880-5. [DOI] [PubMed] [Google Scholar]
- 77.Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69:495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 78.Mackenney VJ, Barnes DE, Lindahl T. Specific function of DNA ligase I in simian virus 40 DNA replication by human cell-free extracts is mediated by the amino-terminal non-catalytic domain. The Journal of biological chemistry. 1997;272:11550–11556. doi: 10.1074/jbc.272.17.11550. [DOI] [PubMed] [Google Scholar]
- 79.Prigent C, Satoh MS, Daly G, Barnes DE, Lindahl T. Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Molecular and cellular biology. 1994;14:310–317. doi: 10.1128/mcb.14.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goetz JD, Motycka TA, Han M, Jasin M, Tomkinson AE. Reduced repair of DNA double-strand breaks by homologous recombination in a DNA ligase I-deficient human cell line. DNA repair. 2005;4:649–654. doi: 10.1016/j.dnarep.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 81.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Current biology : CB. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 82.Castel A. Lopez, Tomkinson AE, Pearson CE. CTG/CAG repeat instability is modulated by the levels of human DNA ligase I and its interaction with proliferating cell nuclear antigen: a distinction between replication and slipped-DNA repair. The Journal of biological chemistry. 2009;284:26631–26645. doi: 10.1074/jbc.M109.034405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cereghini S, Yaniv M. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. The EMBO journal. 1984;3:1243–1253. doi: 10.1002/j.1460-2075.1984.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jackson SM, Whitworth AJ, Greene JC, Libby RT, Baccam SL, Pallanck LJ, La Spada AR. A SCA7 CAG/CTG repeat expansion is stable in Drosophila melanogaster despite modulation of genomic context and gene dosage. Gene. 2005;347:35–41. doi: 10.1016/j.gene.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 85.Miret JJ, Pessoa-Brandao L, Lahue RS. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miret JJ, Pessoa-Brandao L, Lahue RS. Instability of CAG and CTG trinucleotide repeats in Saccharomyces cerevisiae. Molecular and cellular biology. 1997;17:3382–3387. doi: 10.1128/mcb.17.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hashem VI, Rosche WA, Sinden RR. Genetic assays for measuring rates of (CAG).(CTG) repeat instability in Escherichia coli. Mutation research. 2002;502:25–37. doi: 10.1016/s0027-5107(02)00026-x. [DOI] [PubMed] [Google Scholar]
- 88.Schmidt KH, Abbott CM, Leach DR. Two opposing effects of mismatch repair on CTG repeat instability in Escherichia coli. Molecular microbiology. 2000;35:463–471. doi: 10.1046/j.1365-2958.2000.01727.x. [DOI] [PubMed] [Google Scholar]
- 89.Schumacher S, Fuchs RP, Bichara M. Expansion of CTG repeats from human disease genes is dependent upon replication mechanisms in Escherichia coli: the effect of long patch mismatch repair revisited. Journal of molecular biology. 1998;279:1101–1110. doi: 10.1006/jmbi.1998.1827. [DOI] [PubMed] [Google Scholar]
- 90.Schweitzer JK, Livingston DM. Destabilization of CAG trinucleotide repeat tracts by mismatch repair mutations in yeast. Human molecular genetics. 1997;6:349–355. doi: 10.1093/hmg/6.3.349. [DOI] [PubMed] [Google Scholar]
- 91.Jaworski A, Rosche WA, Gellibolian R, Kang S, Shimizu M, Bowater RP, Sinden RR, Wells RD. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:11019–11023. doi: 10.1073/pnas.92.24.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kantartzis A, Williams GM, Balakrishnan L, Roberts RL, Surtees JA, Bambara RA. Msh2-Msh3 interferes with Okazaki fragment processing to promote trinucleotide repeat expansions. Cell reports. 2012;2:216–222. doi: 10.1016/j.celrep.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parniewski P, Jaworski A, Wells RD, Bowater RP. Length of CTG.CAG repeats determines the influence of mismatch repair on genetic instability. Journal of molecular biology. 2000;299:865–874. doi: 10.1006/jmbi.2000.3796. [DOI] [PubMed] [Google Scholar]
- 94.Anderson S, DePamphilis ML. Metabolism of Okazaki fragments during simian virus 40 DNA replication. The Journal of biological chemistry. 1979;254:11495–11504. [PubMed] [Google Scholar]
- 95.Kow YW, Bao G, Reeves JW, Jinks-Robertson S, Crouse GF. Oligonucleotide transformation of yeast reveals mismatch repair complexes to be differentially active on DNA replication strands. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:11352–11357. doi: 10.1073/pnas.0704695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Current biology : CB. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- 97.Romanova NV, Crouse GF. Different roles of eukaryotic MutS and MutL complexes in repair of small insertion and deletion loops in yeast. PLoS genetics. 2013;9:e1003920. doi: 10.1371/journal.pgen.1003920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Erdeniz N, Dudley S, Gealy R, Jinks-Robertson S, Liskay RM. Novel PMS1 alleles preferentially affect the repair of primer strand loops during DNA replication. Molecular and cellular biology. 2005;25:9221–9231. doi: 10.1128/MCB.25.21.9221-9231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mason AG, Tome S, Simard JP, Libby RT, Bammler TK, Beyer RP, Morton AJ, Pearson CE, La Spada AR. Expression levels of DNA replication and repair genes predict regional somatic repeat instability in the brain but are not altered by polyglutamine disease protein expression or age. Human molecular genetics. 2014;23:1606–1618. doi: 10.1093/hmg/ddt551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu L, Patel H, Bissler JJ. Optimizing DpnI digestion conditions to detect replicated DNA. BioTechniques. 2002;33:316–318. doi: 10.2144/02332st03. [DOI] [PubMed] [Google Scholar]
- 101.Schneider J, Fanning E. Mutations in the phosphorylation sites of simian virus 40 (SV40) T antigen alter its origin DNA-binding specificity for sites I or II and affect SV40 DNA replication activity. Journal of virology. 1988;62:1598–1605. doi: 10.1128/jvi.62.5.1598-1605.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.