Abstract
Sterol regulatory element-binding protein (SREBP) transcription factors are master regulators of cellular lipid homeostasis in mammals and oxygen-responsive regulators of hypoxic adaptation in fungi. SREBP C-terminus binds to the WD40 domain of SREBP cleavage-activating protein (SCAP), which confers sterol regulation by controlling the ER-to-Golgi transport of the SREBP-SCAP complex and access to the activating proteases in the Golgi. Here, we biochemically and structurally show that the carboxyl terminal domains (CTD) of Sre1 and Scp1, the fission yeast SREBP and SCAP, form a functional 4:4 oligomer and Sre1-CTD forms a dimer of dimers. The crystal structure of Sre1-CTD at 3.5 Å and cryo-EM structure of the complex at 5.4 Å together with in vitro biochemical evidence elucidate three distinct regions in Sre1-CTD required for Scp1 binding, Sre1-CTD dimerization and tetrameric formation. Finally, these structurally identified domains are validated in a cellular context, demonstrating that the proper 4:4 oligomeric complex formation is required for Sre1 activation.
Keywords: lipid homeostasis, SREBP, SCAP
Introduction
The sterol regulatory element-binding protein (SREBP) transcription factors are master regulators of cellular lipid homeostasis1. Three SREBP isoforms SREBP1a, SREBP1c and SREBP2 control the supply of phospholipids, triglycerides and cholesterol in cells2. SREBPs are synthesized as membrane-bound inactive precursors in the endoplasmic reticulum (ER) where SREBP cleavage-activating protein (SCAP) binds and stabilizes the SREBP precursor3. SREBP is a hairpin-like membrane-bound protein with its N- and C-termini both facing the cytosol4. The N-terminal domain is a basic helix-loop-helix leucine zipper (bHLH-LZ) transcription factor that functions only after proteolytic release from the membrane and binds to DNA regulatory elements as a dimer5. The C-terminal domain contains a regulatory domain required for SCAP binding3. SCAP contains an N-terminal domain with eight transmembrane segments (TMs) that function as a sterol sensor and a cytosol C-terminal domain that binds SREBP6.
Under sterol-replete conditions, the SREBP-SCAP complex binds to the ER-resident protein insulin-induced gene (INSIG), and the complex is retained in the ER. Under sterol-depleted conditions, the SREBP-SCAP complex disassociates from INSIG, and SCAP escorts SREBP from the ER to the Golgi, where SREBP is sequentially cleaved by the Site-1 and Site-2 proteases, producing an active N-terminal transcription factor fragment (SREBP-N)7,8.
A homologous pathway consisting of the SREBP Sre1 and SCAP Scp1 was identified in the fission yeast Schizosaccharomyces pombe and has a critical role in hypoxic adaptation9,10. Despite modest overall sequence identities with their mammalian homologs, Sre1 and Scp1 are regulated by sterols and have a conserved domain organization9. Scp1 binds and stabilizes Sre1 precursor in the ER9,11. Under conditions of low sterols or low oxygen, Scp1 escorts Sre1 from the ER to the Golgi where it is proteolytically liberated from the membrane12. Upon inhibition of Sre1 cleavage in the Golgi, Scp1 is degraded as seen for SCAP in mammalian cells13. Thus, fission yeast Sre1 and Scp1 serve as a simplified model to understand SREBP-SCAP function and regulation.
The SREBP pathway has been of tremendous interest to both basic researchers and pharmaceutical industry owing to its central role in the regulation of lipid homeostasis14. Accumulating studies suggest that the SREBP pathway is also involved in cancer, diabetes and the immune response15. Structural information of the key players in this pathway has been sought for mechanistic understanding and potential drug development. However, due to technical challenges concerning eukaryotic membrane proteins, progress in the structural and biochemical studies has been disproportionally slow compared with studies of the pathway's physiological functions. Thus far, homologs in SREBP pathway from simpler organisms have served as surrogates for structural characterizations of the integral membrane proteins, exemplified by the structural resolution of an archaeal homolog of S2P and a mycobacterial homologue of INSIG16,17. Notwithstanding these achievements, the structure of the complex between SREBP and SCAP remains elusive.
Considering the fact that the SREBP C-terminus and SCAP C-terminus are sufficient for SREBP-SCAP binding3, we focused on the structural characterization of these domains from the fission yeast homologs: the C-terminal regulatory domain of Sre1 (Sre1-CTD) and the C-terminal WD40 domain of Scp1 (Scp1-WD40). Previously, we obtained well-behaved recombinant proteins of Sre1-CTD and Scp1-WD40, reconstituted the complex formation in vitro, and resolved the crystal structure of Scp1-WD4018. Structure-guided mutagenesis analysis showed that an Arg/Lys-enriched surface area (designated the RK-patch) of Scp1-WD40 is required for binding to Sre118.
In this study, we present the 3.5 Å crystal structure of the Sre1-CTD and the 5.4 Å structure of the oligomeric complex of Sre1-CTD and Scp1-WD40 by single-particle cryo-EM. Supported by in vitro biochemical and in vivo functional experiments, our studies revealed that the Sre1-CTD, which has a helical fold, forms a dimer of dimers and Sre1-CTD and Scp1-WD40 form a 4:4 oligomeric complex that is essential for Sre1 activation.
Results
Sre1-CTD and Scp1-WD40 form a 4:4 oligomer
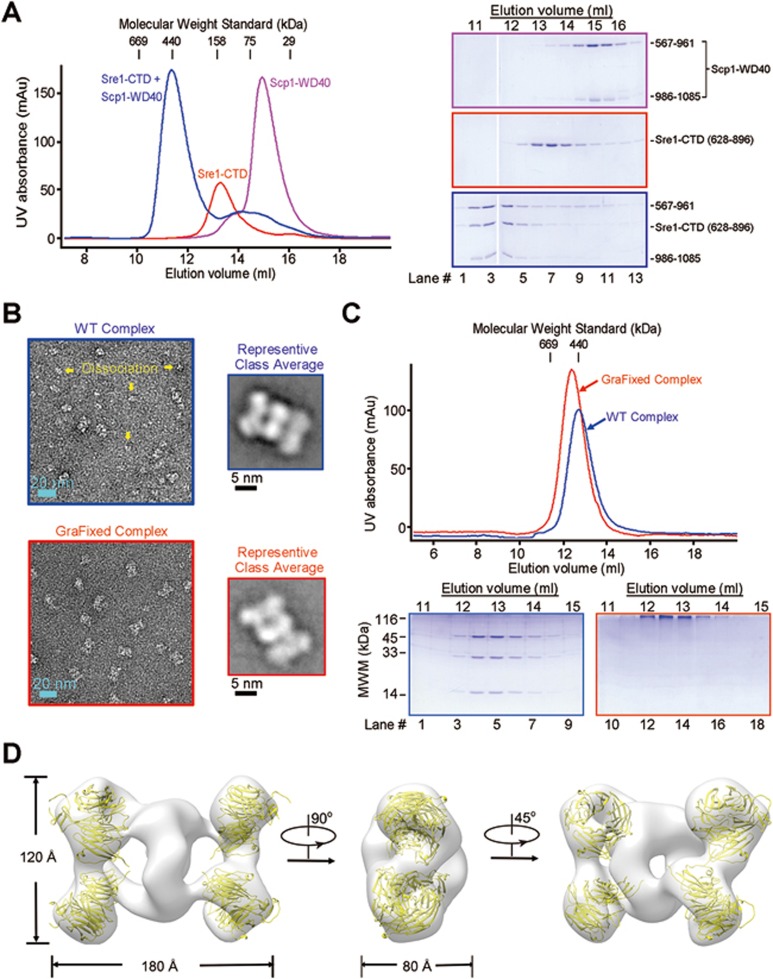
When the complex of Sre1-CTD and Scp1-WD40 was applied to size exclusion chromatography (SEC), the elution volume corresponded to a molecular weight of ∼440 kDa, whereas the calculated molecular weights of Sre1-CTD (residues 628-896) and Scp1-WD40 (co-expression of two segments consisting of residues 567-961 and 986-1 085) are ∼30 and 56 kDa, respectively. In addition, Sre1-CTD was eluted at a volume close to the molecular weight standard of 158 kDa, considerably bigger than the monomeric Scp1-WD40, whose elution volume was consistent with its calculated molecular weight. Further characterization of the proteins with static light scattering (SLS) and analytical ultracentrifugation sedimentation velocity (AUC-SV) yielded similar but varied molecular weights, all supporting that Sre1-CTD may form oligomers and that the Sre1-Scp1 complex may exhibit a higher order of oligomerization (Figure 1A, Supplementary information, Figure S1A and S1B).
Figure 1.
Sre1-CTD and Scp1-WD4 can form a 4:4 complex. (A) The Sre1-CTD and Scp1-WD40 form a high-order oligomer on the size exclusion chromatography (SEC). For better solution behavior, the Scp1-WD40 protein was obtained by co-expressing segments containing residues 567-961 and residues 986-1 085 as reported previously18. (B) The negative staining EM analysis of the unprocessed (WT) and GraFixed complex of Sre1-CTD and Scp1-WD40. The WT proteins display considerable degree of sample heterogeneity, indicating complex dissociation. In contrast, the GraFixed samples were more homogeneous, sharing similar structural feature with a subgroup of WT particles. (C) GraFixed Sre1-CTD and Scp1-WD40 complex gave rise to well-behaved proteins. The experimental details of GraFix cross-linking can be found in Materials and Methods. The GraFixed samples were eluted from SEC at a slightly advanced volume, suggesting that the WT complex may undergo dissociation at the tested concentration. (D) Reconstruction of the negative staining EM structure of the GraFixed Sre1-CTD and Scp1-WD40 complex. The overall structure resembles a flattened dumbbell with a seemingly four-fold symmetry related by two perpendicular axes. Each of the four corners of the EM map exhibits a distinct donut shape that can be docked with the WD40 domain of Scp1. All structure figures and the EM maps were prepared with PyMol41 and Chimera40, respectively. See Supplementary information, Figures S1 and S2 for additional characterizations.
To reveal the oligomerization status, in particular, the stoichiometric ratio of the Sre1-Scp1 complex, we took electron microscopic (EM) images of the negative-stained complex samples (Figure 1B, top). The complex of wild-type (WT) proteins exhibited extreme size heterogeneity. After careful particle picking and 2D classification, a subgroup of particles exhibited a flattened dumbbell shape (Figure 1B, top). However, the heterogeneity of the protein samples impeded further data processing. To overcome this hurdle, we made several attempts, including screening of different buffer conditions, engineering of proteins with distinct boundaries and designed mutations, and cross-linking. Among all the endeavors, cross-linking of the protein complex through the GraFix approach19 gave rise to well-behaved proteins (Figure 1C).
The GraFixed products appeared homogeneous when imaged with negative staining EM (Figure 1B, bottom). The majority of 2D classifications gave rise to the same shape as the aforementioned unprocessed protein complex (Figure 1B). When applied to SEC, the GraFixed Sre1-Scp1 complex was eluted as a mono-dispersed peak at a position slightly earlier than the unprocessed protein complex (hereafter referred to as the WT complex) (Figure 1C). Consistently, AUC-SV and SLS analyses both gave rise to higher molecular weights of the GraFixed complex than those of the WT complex (Supplementary information, Figure S1B and S1C). One possible explanation is that the WT complex may dissociate. Supporting this notion, SLS measurements showed reduced molecular weight upon serial dilution of the protein concentration (Supplementary information, Figure S1D).
As the GraFixed complex does resemble one class of the WT proteins but with substantially improved data quality of negative-staining EM, we focused on the GraFixed sample for EM analysis. A 26 Å EM map was reconstructed out of 17 000 negative-stained particles (Figure 1D and Supplementary information, Figure S2). At this resolution, the “dumbbell” appears to be composed of several twisted rings. The handle of the dumbbell resembles an oval ring with the two axes of ∼100 and 80 Å long. Each end of the dumbbell is made of two small donuts, each characteristic of a WD40 protein. In fact, the structure of a monomeric Scp1-WD40 can be docked perfectly into each donut, leaving Sre1-CTD at the central region in the reconstruction (Figure 1D). As the SLS and AUC-SV measured molecular weights of GraFixed complex were ∼352 and 322 kDa, respectively, the complex should contain four Sre1-CTDs (Supplementary information, Figure S1B and S1C).
Sre1-CTD exists as a dimer of dimers
The lack of structural information of Sre1-CTD or mammalian SREBPs prevented model building from the low-resolution negative-staining EM map. We, therefore, sought to determine the structure of Sre1-CTD. Prior to that, we systematically examined the oligomerization state of Sre1-CTD employing cross-linking, SLS and AUC analyses.
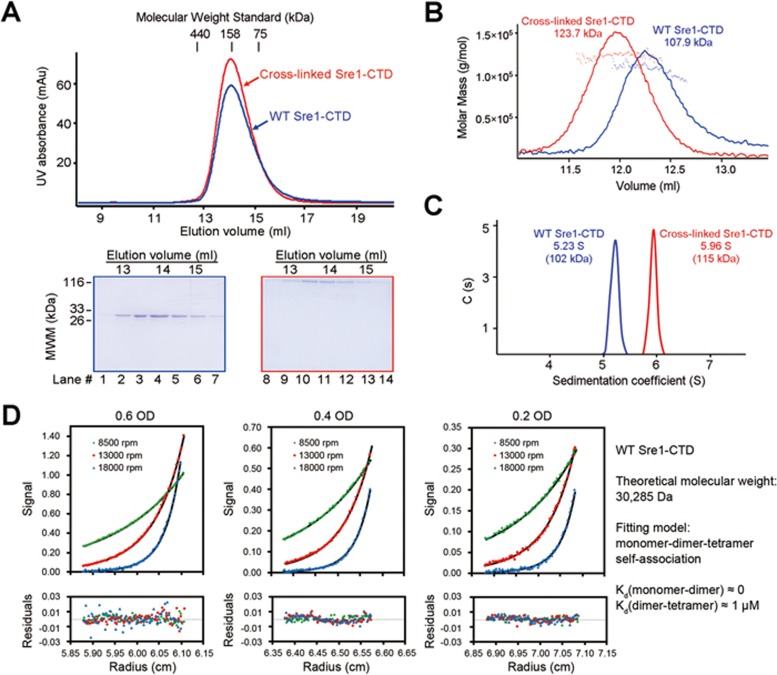
Glutaraldehyde-mediated cross-linking resulted in the formation of dimers and tetramers (Supplementary information, Figure S3A). When the proteins were cross-linked with 0.64% (w/v) glutaraldehyde for 2 h, the majority became tetramer (Supplementary information, Figure S3A, right panel). When the cross-linked Sre1-CTD was applied to SEC, it migrated to the similar position to the unprocessed WT protein, supporting the oligomeric formation of Sre1-CTD (Figure 2A). Interestingly, despite similar elution volumes on SEC, cross-linked Sre1-CTD was measured as a tetramer with SLS and AUC-SV, whereas the unprocessed protein appeared smaller by ∼15 kDa in both measurements (Figure 2B and 2C).
Figure 2.
Sre1-CTD exists as a dimer of dimer. (A) The Sre1-CTD may exist as an oligomer. The Sre1-CTD (residues 628-896) was eluted from SEC at a position corresponding to the molecular weight of a tetramer. The cross-linked Sre1-CTD, which showed a molecular weight of ∼120 kDa on SDS-PAGE, was eluted at a similar position to the WT protein on SEC. (B) Static light scattering (SLS) analysis of the Sre1-CTD. SLS analysis of the WT and cross-linked samples suggested that the Sre1-CTD may form a less stable tetramer as the cross-linked protein was measured with a molecular weight of a tetramer, whereas the WT protein exhibited a slightly reduced molecular weight. (C) Analytical ultracentrifugation analysis (AUC) of the Sre1-CTD. Consistent with the SLS analysis, the sedimentation velocity AUC (AUC-SV) analysis of the WT and cross-linked Sre1-CTD supported the formation of an unstable tetramer of Sre1-CTD. (D) Sedimentation equilibrium AUC analysis of the Sre1-CTD. The AUC-SE analysis yielded a dissociation coefficient (Kd) of 1 μM between tetramer and dimer, suggesting a dimer of dimer organization of the Sre1-CTD. See also Supplementary information, Figure S3.
On the basis of the slightly smaller molecular weight of unprocessed Sre1-CTD and the presence of dimer, but not trimer, of the cross-linked products, we reasoned that Sre1-CTD may exist in equilibrium between tetramer and dimer (Supplementary information, Figure S3A). To test this proposition, we applied the sample to sedimentation equilibrium AUC (AUC-SE) analysis (Figure 2D). Data fitting supported a dimer-tetramer equilibrium with a dissociation coefficient (Kd) of ∼1 μM (Figure 2D). The cross-linked Sre1-CTD oligomer fit into a single species model with a measured molecular weight of ∼114 kDa (Supplementary information, Figure S3B).
Crystal structure of dimeric Sre1-CTD
To obtain the crystal structure of Sre1-CTD, we screened a variety of boundaries. Eventually, protein lacking the C-terminal 24 residues and larboring two Cys-to-Ser substitutions (designated the ΔC-tail variant, residues 628-876, C644S/C672S) crystallized in the space group of P3121. The structure was determined by selenium-based single-wavelength anomalous dispersion and refined to 3.5 Å resolution (Supplementary information, Figure S4 and Table S1).
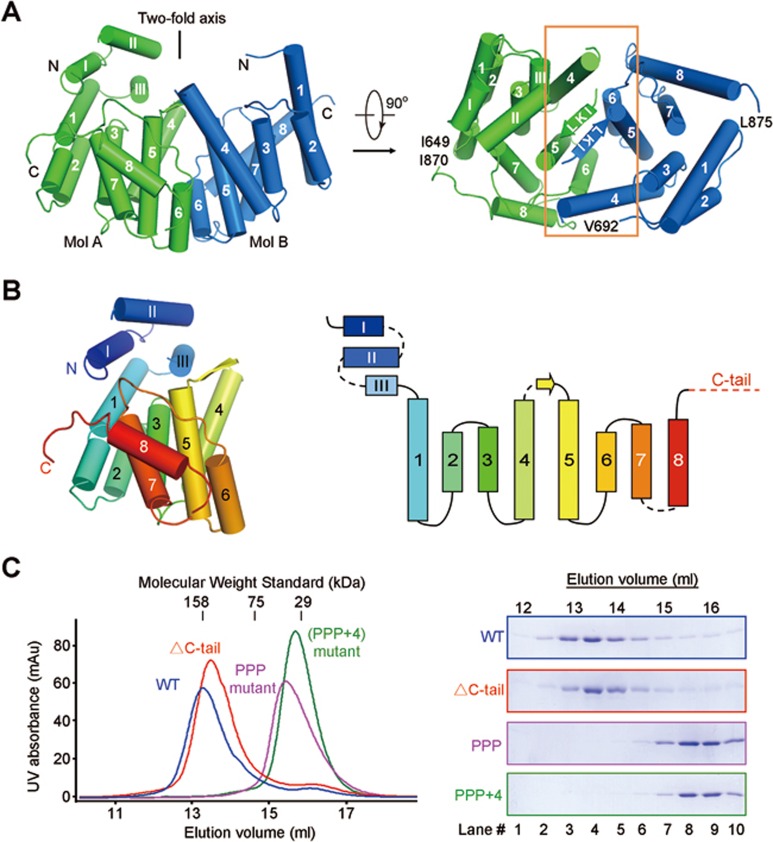
There are two Sre1-CTD molecules in each asymmetric unit (Figure 3A). The two molecules, named Mol A and Mol B, were resolved for residues 649-870 and residues 692-875, respectively. The N-terminal segment of Mol A, which is invisible in Mol B, form three short helices, designated αI-αIII. The C-terminal segment consisting of residues 863-870 unfolds to a flexible loop in Mol A. Although Mol B lacks the N-terminal helices, its last 23 amino acids form a consecutive helix (Supplementary information, Figure S5A). Other than the deviations of the two termini, the structures of the two protomers can be superimposed with a root mean square deviation value of 0.63 Å over 133 Cα atoms (Supplementary information, Figure S5A). The core of each Sre1-CTD protomer comprises eight α helices that form a tilted bundle with the additional helices αI-αIII in Mol A capped on top (Figure 3B).
Figure 3.
Crystal structure of Sre1-CTD dimer at 3.5 Å resolution. (A) The Sre1-CTD forms a dimer in the crystal structure. The two protomers within each asymmetric unit exhibits a two-fold pseudo-symmetry relative by an axis that traverses the interface constituted by the anti-parallel β-strands and the α4-α6 helices of the two protomers. (B) The overall structure of a Sre1-CTD protomer. Shown here is Mol A in each asymmetric unit. There are three short helices on the amino termini of Mol A that are invisible in Mol B. The shared structure core of the two protomers comprises 8 α helices and a short β strand that connects α4 and α5. Right panel: The topological structure of Sre1-CTD. (C) Biochemical validation of the structurally revealed interface. PPP mutant that was designed to disrupt the anti-parallel β strands led to disruption of the oligomer. Mutations of four additional amino acids (K766E/V809D/R812E/F818D) on helices α4 and α6 completely disrupted of the oligomeric interface (designated PPP+4 mutant). The C terminus-deleted (designated ΔC-tail) protein remained as an oligomer. See also Supplementary information, Figures S4 and S5.
The connecting segments between α4 and α5 from the two protomers form anti-parallel β-strands, constituting a major dimer interface (Figure 3A). Helices α4 and α6 from the two protomers also contribute to dimer formation (Supplementary information, Figure S5B). The two protomers are related to each other by 180° rotation around an axis that is approximately perpendicular to the anti-parallel β-strands and penetrates the dimeric interface. The side view of the Sre1-CTD dimer resembles an arch with the N-termini of both protomers on the two sides of the concave side, whereas the top view appears to be an oval with the long and short axes of 70 Å and 30 Å, respectively (Figure 3A).
To biochemically verify the dimeric interface observed in the crystal structure, we replaced the β-strand residues with three consecutive Pro (designated the PPP mutant, residues 628-896 and L783P/K784P/I785P), which may disrupt β-strand formation. Indeed, at low concentration, the PPP mutant appeared to be a monomer (Figure 3C). As a control, deletion of the C-tail (residues 877-900) had no effect on oligomer formation (Figure 3C). Notably, the SEC elution volume of the PPP mutant was concentration-dependent, suggesting incomplete disruption of the oligomeric interface (Supplementary information, Figure S5C, middle panel). When four additional amino acids (K766E/V809D/R812E/F818D) on helices α4 and α6 were mutated (named the PPP+4 variant), the protein was a monomer at any tested concentration (Figure 3C, Supplementary information, Figure S5B and S5C).
Although the biochemical characterization suggested that Sre1-CTD may form a dimer of dimers in isolation and in complex with Scp1-WD40 (Figures 1D and 2D), none of the two dimers from adjacent asymmetric units in the crystal structure could be fit into the EM reconstruction (Figure 1D, Supplementary information, Figure S4C), suggesting potential disruption of the tetramer during crystallization.
Cryo-EM structure of the 4:4 complex between Sre1-CTD and Scp1-WD40
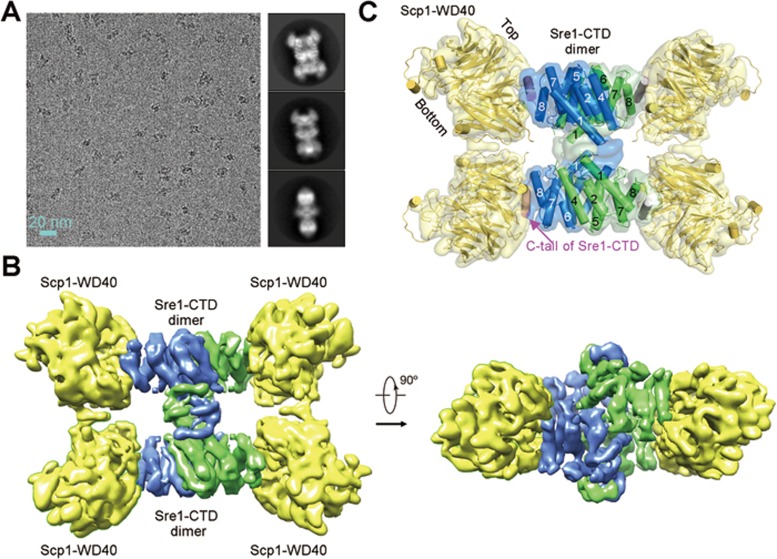
The Sre1-Scp1 complex defied crystallization despite numerous attempts. The low-resolution negative-stained EM reconstruction did not support docking of the Sre1-CTD dimer with confidence. Single-particle cryo-EM was employed and a reconstruction of the GraFixed Sre1-CTD (residues 628-896) and Scp1-WD40 (residues 567-961 and 986-1 085) complex was generated out of 157 243 selected particles to an overall resolution of 5.4 Å according to the gold standard Fourier shell correlation (FSC) 0.143 criterion (Figure 4A, 4B, and Supplementary information, Figure S6).
Figure 4.
Cryo-EM structure of the Sre1-CTD/Scp1-WD40 complex. (A) Cryo-EM imaging of the GraFixed Sre1-CTD/Scp1-WD40 complex. Shown on the left and right are representative micrographs as well as 2D classifications. (B) The overall EM map of the Sre1-CTD and Scp1-WD40 complex. (C) Docking of the crystal structures of Sre1-CTD dimer and Scp1-WD40 into the EM map. The crystal structures of the individual components can be easily docked into the EM map in Chimera. The two protomers in each Sre1-CTD dimer are colored green and blue for visual clarity. The modest resolution does not allow discrimination of any conformational differences between the two protomers within each dimer. See also Supplementary information, Figure S6.
At this resolution, most secondary structural elements of the complex were well resolved, allowing faithful docking of the crystal structures of Scp1-WD40 (PDB ID: 4YHC) and the Sre1-CTD dimer (Figure 4C). In particular, the asymmetric feature of Scp1-WD40 resulting from the extended loop in propeller #6 facilitated structural docking with correct orientation18. Two Sre1-CTD dimers were modeled into the map. After docking of all the known structural elements, additional densities remain to be resolved. Four patches may correspond to a predicted α helix in the C-tail of Sre1-CTD (Supplementary information, Figure S7). We modeled a poly-Ala α helix into these densities. Two patches are involved in the interface between Sre1-CTD dimers that will be discussed in a later session (Figure 4C).
Interface between Sre1-CTD and Scp1-WD40
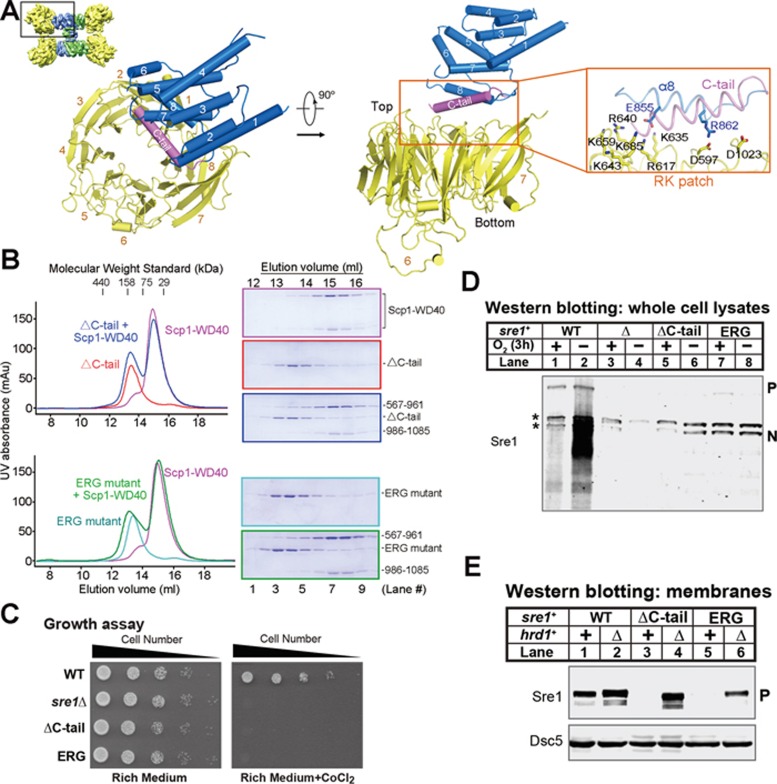
The cryo-EM structure of the Sre1-CTD and Scp1-WD40 complex reveals an unambiguous 4:4 stoichiometric ratio of the two domains and allows examination of the specific contacts between Sre1 and Scp1. Each Sre1-CTD binds to one Scp1-WD40 through a well-defined interface (Figure 5A). Consistent with our previous structure-guided mutational analysis, the RK patch of Scp1-WD40 mediates the interaction with α8 and C-tail helices of Sre1 (Figure 5A, right panel)18. The 5.4 Å resolution did not permit assignment of side chains of the C-tail helix of Sre1-CTD. Nevertheless, docking of the crystal structure of the preceding segments suggested that conserved residues Glu855 and Arg862 on the α8 helix of Sre1 may form electrostatic interactions with charged residues on Scp1 (Supplementary information, Figure S7). Indeed, either deletion of the C-tail (designated ΔC-tail mutant) or mutation of the conserved residues in the α8 helix (designated ERG mutant, E855K/R862E/G866D), disrupted complex formation between Sre1-CTD and Scp1-WD40 in vitro (Figure 5B).
Figure 5.
Interface between Sre1-CTD and Scp1-WD40. (A) The interface of the Sre1-CTD and Scp1-WD40 involves helix α8 and the succeeding C-tail helix, which is missing in the crystal structure, and the RK patch of Scp1, which was biochemically identified previously18. (B) Biochemical verification of the interface between Sre1-CTD and Scp1-WD40. Deletion of the C-tail or mutation of the conserved residues on helix α8 resulted in compromised complex formation. (C) WT, sre1Δ and the indicated sre1 mutants (5 000, 1 000, 200, 40 and 8 cells) were grown on rich medium (3 days) or rich medium containing cobalt chloride (1.6 mM; 6 days). (D) Western blot of whole-cell lysates from WT, sre1Δ and ΔC-tail and ERG sre1 mutants grown in the presence or absence of oxygen for 3 h with anti-Sre1 serum. P and N denote Sre1 precursor and nuclear forms, respectively. Asterisks indicate cross-reacting proteins. (E) Western blot of membrane fractions from WT and sre1 mutant strains (ΔC-tail and ERG) with or without hrd1+ probed with anti-Sre1 serum or anti-Dsc5 serum.
To characterize the Sre1-Scp1 interaction in a cellular context, we generated fission yeast strains expressing full-length WT sre1 or mutants (ΔC-tail and ERG) from the endogenous sre1+ promoter. Our previous studies indicated that both stabilization of ER-localized Sre1 precursor and activation of Sre1 processing required Scp1 binding. scp1Δ cells or cells expressing Scp1 mutants that do not bind Sre1 fail to grow on cobalt chloride-containing medium and fail to activate Sre1 under low oxygen9,18. In addition, Sre1 precursor in these strains is rapidly degraded in the ER through a pathway that requires the E3 ubiquitin ligase Hrd111,18. On the basis of these findings, we expected that Sre1 mutants defective for Scp1 binding would: (i) fail to grow in the presence of cobalt, (ii) fail to accumulate Sre1N under hypoxia and (iii) show reduced Sre1 precursor due to Hrd1-dependent degradation. As expected, both the ΔC-tail and ERG mutants failed to grow in the presence of cobalt (Figure 5C) or activate Sre1 under hypoxia (Figure 5D, lanes 5-8). In addition, the steady-state Sre1 precursor levels in ΔC-tail or ERG mutants decreased dramatically compared with WT cells (Figure 5D, lanes 1, 5 and 7). Deletion of hrd1+ restored Sre1 precursor levels in these strains (Figure 5E), indicating that low Sre1 precursor levels resulted from Hrd1-dependent degradation in the ER. These cellular data demonstrate that both the C-tail helix and ERG residues in α8 helix of Sre1 have essential roles in Scp1 binding.
Tetrameric formation of Sre1-CTD is required for Sre1 activation
The EM structure of Sre1-CTD and Scp1-WD40 complex allowed examination of the tetrameric organization of Sre1-CTD. For simplicity, we refer to the interface between Sre1-CTD dimers as the tetrameric interface (Figure 6A). The two Sre1-CTD dimers, each reminiscent of an arch from the side view, together form the closed oval ring seen in middle of the low-resolution negative-staining EM reconstruction (Figure 1D). The two arches contact each other through the α1 helices and unresolved density immediately preceding α1, likely contributed by the N-terminal segments of Sre1-CTD consisting of residues 628–692. However, the low resolution prevented modeling of any segment to this density (Figure 6A).
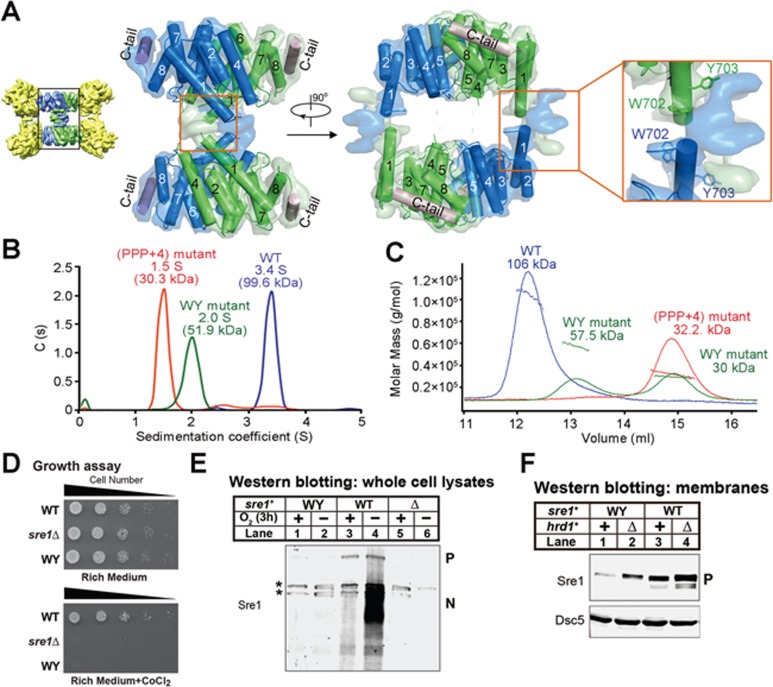
Figure 6.
Tetrameric formation of Sre1-CTD. (A) The N-terminus of the α1 helix contains two aromatic residues Trp702/Tyr703 that are highly conserved in related species and are shown as sticks. (B) AUC-SV analysis of the Sre1-CTD oligomeric mutants. The AUC-SV analysis of the WT, PPP+4 mutant and WY mutant of Sre1-CTD supported that the conserved WY motif contributed to the tetrameric formation. (C) SLS analysis of the Sre1-CTD oligomeric mutants. SLS characterization of WY mutant produced two peaks corresponding to 57.5 kDa and 30 kDa, respectively, suggesting less stable dimer formation. (D) WT, sre1Δ and sre1 WY mutant cells (5 000, 1 000, 200, 40 and 8 cells) were grown on rich medium (3 days) or rich medium containing cobalt chloride (1.6 mM; 6 days). (E) Western blot of whole-cell lysates from WT, sre1Δ and sre1 WY cells grown in the presence or absence of oxygen for 3 h with anti-Sre1 serum. P and N denote Sre1 precursor and nuclear forms, respectively. Asterisks indicate cross-reacting proteins. (F) Western blot of membrane fractions from WT and WY mutant cells with or without hrd1+ probed with anti-Sre1 serum or anti-Dsc5 serum.
The N-terminus of the α1 helices features two aromatic residues Trp702/Tyr703 that are highly conserved in related species and project into the unresolved EM map (Supplementary information, Figure S7). We conjectured that these two residues (designated WY motif) may contribute to the tetrameric organization. To test this, we introduced two point mutations W702D/Y703D to Sre1-CTD (residues 628-896). Indeed, the WY mutant appeared as a dimer with a molecular weight of 51.9 kDa measured by AUC-SV, whereas molecular weights of dimers of the (PPP+4) mutant and WT protein were 30.3 and 99.6 kDa, respectively (Figure 6B). However, WY variant was eluted from SEC in two peaks (Supplementary information, Figure S8A). Similarly, SLS characterization produced two peaks corresponding to 57.5 and 30 kDa, respectively, suggesting less stable dimer formation (Figure 6C). When the WY variant was mixed with Scp1-WD40, the two proteins co-migrated on SEC in two peaks, both smaller than the 4:4 complex of the WT proteins (Supplementary information, Figure S8A). These biochemical characterizations are consistent with the structural observation that the WY motif and the tetrameric interface of Sre1-CTD are away from the Scp1-WD40, and thus unlikely to be involved in Scp1-WD40 binding (Figure 6A).
To characterize the physiological relevance of the tetrameric organization of Sre1-CTD, we examined the WY mutations in the cellular context. Fission yeast strains expressing full-length WT sre1 or the WY mutant from the endogenous sre1+ promoter were generated. The WY mutant failed to grow in the presence of cobalt (Figure 6D) or activate Sre1 under low oxygen (Figure 6E, lanes 1-2). In addition, the WY mutant showed reduced Sre1 precursor level compared with the WT strain (Figure 6F, lanes 1 and 3). Deletion of hrd1+ in WY mutant cells restored Sre1 precursor level (Figure 6F), indicating that the Sre1 WY precursor was degraded in a Hrd1-dependent manner. These data suggest that the tetrameric formation of Sre1-CTD has a critical role in Sre1 activation under low oxygen.
Discussion
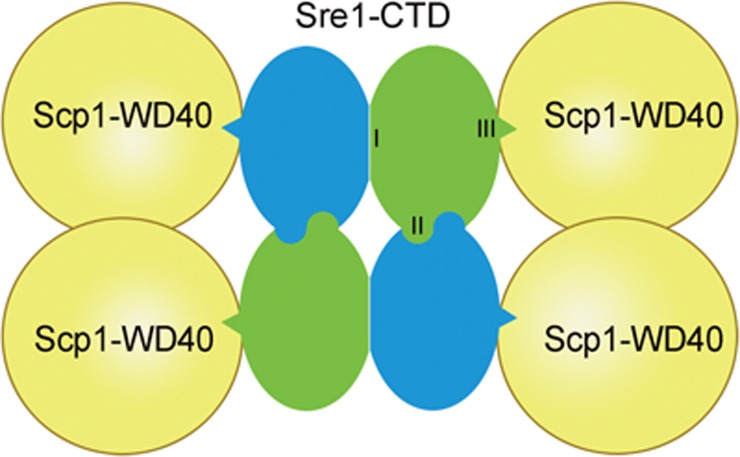
The SREBP pathway controls cellular lipid homeostasis, and the interaction between SREBP and SCAP is key to understanding this important pathway. In this study, we identified a biochemically tractable Sre1-Scp1 system to study SREBP-SCAP interaction, marking a major step forward in this field. Using combined biochemical and structural approaches, we characterized the SREBP-SCAP cytosolic complex, which reveals an intriguing 4:4 stoichiometric ratio between Sre1-CTD and Scp1-WD40. Our combined structural, biochemical and in vivo characterizations demonstrate that Sre1-CTD contains three distinct functional domains. The C-tail and helix α8 of Sre1-CTD are essential for binding to the RK patch of Scp1-WD40 (Figure 7, Domain I); Helix α1 and the N-terminal segments mediate dimerization of Sre1-CTD dimers (Domain II); and Helices α4 and α6 plus β strand 4-5 are required for dimerization of Sre1-CTD monomers (Domain III).
Figure 7.
Three distinct regions in the Sre1-CTD required for Scp1 binding, Sre1-CTD dimerization and tetrameric formation. The simplified cartoon highlights the three distinct surface areas of Sre1-CTD for homo- and heterotypic interactions. (I) the dimer interface, (II) the interface between dimers, (III) the interface with Scp1-WD40. The green and blue colors of the two protomers in each Sre1-CTD are for visual clarity only.
Notwithstanding this progress, there are several intriguing questions to be further addressed. Despite the retained interaction between the WY mutant of the Sre1-CTD and Scp1-WD40 (Supplementary information, Figure S8A), full-length Sre1 containing W702D/Y703D mutations was unstable (Figure 6F), indicating that Scp1 binding is not sufficient to stabilize Sre1 precursor. Rather, dimerization of Sre1-CTD dimers has an additional stabilizing role. These observations suggest that correct assembly of the Sre1-CTD oligomeric complex has a central role in Sre1 function. Intriguingly, mutations introduced into Sre1-CTD helices α4 to α6 and β strand 4-5 disrupted dimerization of Sre1-CTD monomers in vitro. The same mutations in full-length Sre1 reduced function but did not prevent Sre1 activation (Supplementary information, Figure S8B and S8C), suggesting that either Sre1-CTD dimer formation is not essential for proper Sre1 function or more likely that these mutations failed to disrupt full-length Sre1 complex formation in vivo.
Although the SREBP pathway has been studied in great detail, the stoichiometry of the SREBP-SCAP complex was unknown. The cryo-EM structure of Sre1-CTD and Scp1-WD40 provides the first experimental evidence that the SREBP-SCAP complex consists of four SREBP molecules and four SCAP molecules. This 4:4 model is consistent with the previous observation that purified recombinant hamster SCAP (TM1-8) forms a tetramer in detergent20. Importantly, the 4:4 structure answers several open questions in the SREBP field. The N-terminal transcription factor domains of SREBPs belong to the bHLH-LZ family, which bind to DNA regulatory elements as a dimer5. In the human genome, there are multiple bHLH-LZ transcription factors such as MYC, MAD, TFE and USF21, and they can form functional heterodimers executing distinct biological functions22. However, whether dimerization of SREBP-N happens before or after proteolytic release from the membrane is unknown. If dimerization happens after cleavage, SREBP-N may form heterodimers with other soluble bHLH-LZ transcription factors like Myc or Max. The fact that Sre1-CTD forms a dimer of dimers in complex with Scp1 indicates that the Sre1 bHLH-LZ domain forms a dimer before cleavage. Dimer formation immediately following synthesis and prior to proteolytic release would prevent inappropriate heterodimer formation with other soluble bHLH-LZ transcription factors. We observed that disruption of Sre1-CTD dimerization had no effect on Sre1 activation (Supplementary information, Figure S8). In the context of full-length Sre1, it is possible that the N-terminal bHLH-LZ domain stabilizes 4:4 complex formation, suppressing the in vitro binding defects of the monomeric mutants.
Mammalian cells express three SREBP isoforms: SREBP1a, SREBP1c and SREBP2. SREBP1a and 1c share the identical CTD, and SREBP1-CTD is highly similar to SREBP2-CTD. Dimerization of the Sre1-CTD provides a mechanism for formation of SREBP isoform heterodimers. For example, SREBP1a and SREBP1c could form heterodimers through their CTD. SREBP1c is a weaker transcriptional activator than SREBP1a, and this could provide a mechanism for modulating SREBP1 activity. Indeed, soluble SREBP NTDs from all three isoforms can cooperate as heterodimers in cells to modulate gene expression23. In addition, assuming that the 4:4 complex exits the ER as a unit, a single complex could contain a combination of SREBP1 and SREBP2 dimers or heterodimers.
It is known that the SREBP N-terminal transcription factor domain is liberated from the membrane through sequential proteolytic cleavage24. Recent evidence suggests that Sre1 activation also requires sequential proteolytic cleavage, first by a rhomboid protease and then a signal peptide peptidase-like enzyme25,26,27,28. Why the second proteolytic event requires an initiating cleavage is unknown. The 4:4 complex model provides a possible structural explanation. In addition to the soluble CTD domains, SREBP and SCAP contain two and eight N-terminal TMs, respectively, for a total of 40 TMs per complex. The 4:4 structure reveals that SREBP sits at the core of the complex surrounded by four SCAP molecules. It is possible that this configuration blocks access of the second protease to full-length SREBP, and that cleavage requires protease-dependent dissociation of SREBP from the 4:4 complex. With regard to mammalian cells, the minimal element of the Site-2 protease crystal contained two protease molecules16. On the basis of data presented here, mammalian SREBP bHLH-LZ likely dimerizes prior to Site-1 cleavage, and a dimeric configuration of Site-2 protease may facilitate recognition and processing of this dimeric SREBP substrate.
In sum, our studies of fission yeast SREBP and SCAP provide a framework for exploring whether a parallel organization exists in mammalian cells. A complete understanding of sterol regulation of this system awaits the structure of the SREBP-SCAP full-length proteins as the SCAP N-terminal domain mediates cholesterol binding.
Materials and Methods
In vitro reconstitution of the Sre1-CTD and Scp1-WD40 complex
The Sre1-CTD and Scp1-WD40 were overexpressed and purified following the previously reported protocol18. The complex of Sre1-CTD and Scp1-WD40 was reconstituted by incubating the purified Sre1-CTD and Scp1-WD40 at a molar ratio of ∼1:1.2 on ice for 30 min and was further purified through size-exclusion chromatography (Superdex 200, GE Healthcare) in the buffer containing 25 mM Tris (pH 8.0), 50 mM NaCl and 1 mM TCEP.
GraFix of the Sre1-CTD/Scp1-WD40 complex
The GraFix preparation for the Sre1-CTD/Scp1-WD40 complex was carried out as described with minor modifications19. Briefly, the purified complex was applied to a 10%-40% (v/v) glycerol gradient combined with a 0%-0.1% glutaraldehyde gradient in the buffer containing 25 mM HEPES (pH 7.5), 50 mM NaCl, 1 mM TECP. Samples were subjected to ultracentrifugation for 20 h at 35 000 rpm in a SW41Ti rotor (Beckman) and recovered by gradient fractionation. The recovered peak was quenched by the addition of Tris (pH 8.0) to a final concentration of 50 mM and then concentrated. The GraFixed complex was further applied to size-exclusion chromatography in buffer containing 25 mM Tris (pH 8.0), 50 mM NaCl and 1 mM TCEP. The peak fractions of target protein complex were pooled and concentrated for biochemical analysis and EM analysis.
Gel filtration assay
The gel filtration assay was performed with a SD200 (Superdex 200 HR10/300, GE Healthcare) in the buffer containing 25 mM Tris (pH 8.0) and 150 mM NaCl. To examine the interactions between the indicated proteins or the oligomeric state of the indicated proteins, 500 μl proteins or protein mixtures were applied to SD200.
Analytical ultracentrifugation analyses
Analytical ultracentrifugation (sedimentation velocity and sedimentation equilibrium) experiments were carried out using a Beckman XL-I analytical ultracentrifuge (Beckman Coulter) equipped with an eight-cell An-50 Ti rotor. For sedimentation velocity, absorbance at 280 nm versus radial location was recorded during centrifugation at 50 000 rpm at 20 °C. For sedimentation equilibrium, data were collected by UV detector at 4 °C for three protein concentrations (OD280 = 0.2, 0.4 and 0.6) at three rotor speeds (8 500, 13 000 and 18 000 rpm). The data of sedimentation velocity and sedimentation equilibrium were analyzed using Sedfit and Sedphat29.
SLS measurements
An 18-angle light scattering detector (DAWN HELEOS, Wyatt Technology) was connected immediately downstream of Superdex 200 10/300 or Superose 6 10/300 size exclusion column (GE Healthcare) to collect static light scattering data. Protein in 100 μl solution was injected into the system equilibrated with buffer containing 25 mM Tris (pH 8.0) and 150 mM NaCl. The data were analyzed with the manufacturer's recommended software (ASTRA) and the recommended procedure was followed.
Glutaraldehyde cross-linking assay
About 0.3 mg/ml Sre1-CTD was mixed with glutaraldehyde at the indicated concentrations and incubated at 4 °C for the indicated time in the buffer containing 25 mM HEPES and 150 mM NaCl. Then the reaction was quenched by the addition of Tris (pH 8.0) to a final concentration of 50 mM and cross-linking result was analyzed by SDS-PAGE. The Sre1-CTD cross-linked by 0.4% glutaraldehyde at 4 °C for 3 h was further purified by gel filtration. The peak fractions were pooled and concentrated for biochemical analysis.
Crystallization and structure determination of Sre1-CTD
Crystals of Sre1-CTD (residues 628-876, C644S/C672S) were grown at 18 °C by the hanging drop vapor diffusion method mixing 1 μl protein (at a concentration of ∼30 mg/ml) with 1 μl reservoir solution containing 0.1 M BTP (pH 7.0), 1.5 M NaNO3. Crystals appeared in 1-2 days and grew to full size in 1 week. Se-Met-labeled protein was crystallized similarly. Both native and Se-Met crystals were soaked in mother liquor containing 20% ethylene glycol before flash-freezing in a cold nitrogen stream at 100 K.
Both the native- and SeMet-derived data were collected at SSRF beamline BL17U. All data sets were integrated and scaled using HKL200030. Further processing was carried out using programs from CCP4 suite31. Data collection statistics are summarized in Supplementary information, Table S1. The phase was solved by single anomalous diffraction (SAD) of Se-Met. Then a crude helical model was manually built in the program Coot32. Using this partial model as input, the identified Se atom positions were refined and phases were recalculated using the SAD experimental phasing module of the program Phaser33. With the improved map, the molecular boundary was unambiguously defined and two molecules were found in an asymmetry unit. Sequence assignment was aided with the selenium sites in the anomalous difference Fourier map. The model was further rebuilt with Coot and refined with Phenix34.
Negative staining electron microscopy
Samples of WT or GraFixed Sre1-CTD/Scp1-CTD complex (4 μl) were applied to glow-discharged continuous carbon-coated grids (Zhongjingkeyi Technology) and were stained with 2% uranyl acetate for 1 min. The negative-stained grids were imaged on a FEI Tecnai T12 microscope at 120 kV with a 4k × 4k charge-coupled device camera (UltraScan 4000, Gatan). For each sample, ∼100 images (for the GraFixed sample, untilted and 40° tilt-pair micrographs were collected) were collected and about 20 000 particles were semi-automatically picked to perform 2D classification with EMAN235. Then a cylinder model (60 pixels × 30 pixels × 30 pixels) was built and used as an initial model to refine the bin2 particles from the GraFixed sample with Spider36. After several iterative refinements, the model was convergent and was used as an initial model for the cryo-EM data processing.
Cryo-EM data acquisition
Aliquots (4 μl) of Grafixed Sre1-CTD/Scp1-WD40 complex at a concentration of ∼20 mg/ml, which had been supplemented with 0.01% DDM were placed onto glow-discharged holey carbon grids (Cu R 1.2/1.3 200 mesh, Quantifoil). Grids were blotted for 2.5 s and flash frozen in liquid ethane using an FEI Vitrobot. Images were taken on FEI Titan Krios electron microscope operating at 300 kV with a nominal magnification of 22 500×. Images were recorded by a K2 Summit detector (Gatan) with the super-resolution mode, and binned to a pixel size of 1.32 Å. Defocus values varied from 1.5 to 2.5 μm. Each image was dose-fractionated to 32 frames with a dose rate of ∼8.2 counts/sec/physical-pixel (∼4.7 e−/sec/Å2), total exposure time of 8 and 0.25 s per frame. UCSFImage4 was used for all data collection (developed by XL).
Image processing
A total of 1 064 cryo-EM micrographs were collected. All images were aligned and summed using whole-image motion correction37. The defocus value of each image was determined by CTFFIND338. Auto-picking was performed using the reference-based particle picking subroutine in RELION and the templates for particle auto-picking were obtained from the 2D class averages calculated from ∼5 000 manually picked particles. After manual check of the auto-picking results, 306 675 particles were extracted. Reference-free 2D classification was applied to remove ice and contaminants yielding 274 341 good particles. After two rounds of 3D classification, 157 243 particles were selected and 3D reconstruction with D2 symmetry was preformed resulting in a resolution of 7.3 Å. Then soft mask was performed and the reported resolution was increased to 5.4 Å. All reported resolutions are based on the gold-standard FSC = 0.143. All processions were performed in RELION39.
Docking and modeling into the cryo-EM map
Given the availability of a high-quality crystal structure of Scp1-WD40 (PDB ID: 4YHC), which fitted very cleanly into the correct hand of the 5.4 Å density map, we docked four Scp1-WD40 molecules into the peripheral donuts first. Residues 1 055-1 086 of the Scp1-WD40, which were not resolved in the crystal structure, were also not modeled into the map. Then, two Sre1-CTD dimeric crystal structures were docked into the central moiety. The αI-αIII helices of Sre1-CTD were removed in COOT as there was no matched density in the cryo-EM map. Besides, a poly-A helices was built to the C-terminus of Sre1-CTD in COOT as clear density of a helix was seen in the cryo-EM map. Initial rigid-body fitting of all models was carried out using UCSF Chimera40. To correct the geometry of the final models and to remove steric clashes, the model was refined in PHENIX by using phenix.real_space_refine and manually checked in COOT34.
S. pombe strains, plasmids and cell culture
We obtained WT haploid S. pombe KGY425 (h-) and KGY461 (h+) from ATCC. Strains sre1Δ, scp1-13×Myc and hrd1Δ have been described previously9,11,18. sre1Δ strain PEY1765 with natMX marker was generated by transforming linearized pFA6a-NatMX into PEY522 and selecting on medium containing 100 μg/l clonNAT. To generate sre1+ WT and mutants (ΔC-tail, ERG, PPP, PPP+4 and WY), sre1+ integrating plasmids were linearized, transformed into sre1Δ strain PEY1765 and selected on medium containing 100 μg/l Geneticin. To construct sre1+ integrating plasmids for strain generation, pFA6a-13×Myc-kanMX was digested by BglII and NcoI to remove TEF promoter upstream of kanMX, and a fragment containing 500 bp ura4+ promoter flanked by BglII and NcoI restriction sites was inserted. We then inserted a 2 710 bp fragment starting 457 bp upstream of the sre1+ start codon to the sequence coding for Pro751 to generate the intermediate plasmid pWS571. Fragments coding for WT or mutated sre1+ (PPP and PPP+4) were inserted between sequences coding for P751 and adh1 terminator by Gibson cloning to generate sre1+ integrating plasmids, respectively. Mutated sre1+ (ΔC-tail, ERG and WY) integrating plasmids were generated by mutagenesis on WT sre1+ integrating plasmid. To further delete hrd1+from sre1+ WT and mutants (ΔC-tail, ERG and WY), hrd1Δ strain PEY1756 was mated to sre1+ WT and mutants (ΔC-tail, ERG and WY), respectively, and selected on medium containing both clonNAT (100 μg/l) and Geneticin (100 μg/l). To generate scp1-13×Myc, hrd1Δ, sre1+ WT or WY mutant triple strains, scp1-13×Myc strain PEY1777 was mated to hrd1Δsre1+ WT or WY mutant, respectively, and selected on medium containing both clonNAT (100 μg/l) and Geneticin (100 μg/l). Supplementary information, Table S2 contains a complete list of strains used in this study. Standard S. pombe cell culture, growth assay and protein extraction were described previously18. Co-immunoprecipitation of Sre1 precursor by Scp1-13×Myc was described previously9.
Accession codes
The atomic coordinates for the crystal structure of Sre1-CTD and the cryo-EM structure of Sre1-CTD/Scp1-WD40 complex have been deposited in the Protein Data Bank (PDB) with the accession codes 5GPD and 5GRS, respectively. The cryo-EM maps of Sre1-CTD/Scp1-WD40 complex have been deposited in the Electron Microscopy Data Bank (EMDB) with the accession code EMD-9537.
Author Contributions
NY conceived the project. XG, HQ, WS, PE and NY designed all the experiments. XG, HQ and JL performed all the in vitro biochemical experiments. WS performed the in vivo experiments. JW solved the crystal structure. WL helped with the data analysis of AUC and SLS results. J-JL and H-WW helped with the negative staining EM reconstruction. All the authors contributed to data analysis. XG, HQ, WS, PE and NY contributed to the manuscript preparation.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Jianlin Lei, Yanji Xu and Xiaomei Li for the technical support. We thank the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for providing the facility support. The computation was completed on the “Explorer 100” cluster system of Tsinghua National Laboratory for Information Science and Technology. This work was supported by funds from the Ministry of Science and Technology of China (2015CB9101012014, ZX09507003006), the National Natural Science Foundation of China (31321062 and 81590761) and National Institute of Health (HL 077588 to PE). The research of NY was supported in part by an International Early Career Scientist grant from the Howard Hughes Medical Institute and an endowed professorship from Bayer Healthcare.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
SLS and AUC-SV analyses of the Sre1-CTD, Scp1-WD40, and their complex.
Ab initio negative staining EM reconstruction of the Sre1-CTD/Scp1-WD40 complex.
Verification of the tetrameric organization of Sre1-CTD.
Structural determination ofthe Sre1-CTD.
The dimer interface of Sre1-CTD.
Cryo-EM analysis of the Sre1-CTD/Scp1-WD40 complex.
Sequence alignment of the Sre1-CTD from eight closely-related species.
In vitro and in vivo characterization of Sre1-CTD dimerization mutants.
Crystallographic data collection and refinement statistics
S. pombes train list
References
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997; 89:331–340. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 2002; 109:1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, Goldstein JL. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J Biol Chem 1997; 272:20213–20221. [DOI] [PubMed] [Google Scholar]
- Hua XX, Sakai J, Ho YK, Goldstein JL, Brown MS. Hairpin orientation of sterol regulatory element-binding protein-2 in cell-membranes as determined by protease protection. J Biol Chem 1995; 270:29422–29427. [DOI] [PubMed] [Google Scholar]
- Parraga A, Bellsolell L, Ferre-D'Amare AR, Burley SK. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 angstrom resolution. Structure 1998; 6:661–672. [DOI] [PubMed] [Google Scholar]
- Motamed M, Zhang YX, Wang ML, et al. Identification of luminal loop 1 of scap protein as the sterol sensor that maintains cholesterol homeostasis. J Biol Chem 2011; 286:18002–18012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade PJ, Hughes AL. Regulation of sterol synthesis in eukaryotes. Annu Rev Genet 2007; 41:401–427. [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 1999; 99:703–712. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 2005; 120:831–842. [DOI] [PubMed] [Google Scholar]
- Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol Cell Biol 2006; 26:2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes BT, Nwosu CC, Espenshade PJ. Degradation of sterol regulatory element-binding protein precursor requires the endoplasmic reticulum-associated degradation components Ubc7 and Hrd1 in fission yeast. J Biol Chem 2009; 284:20512–20521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EV, Nwosu CC, Tong ZT, et al. Yeast SREBP cleavage activation requires the Golgi Dsc E3 ligase complex. Mol Cell 2011; 42:160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Espenshade PJ. Sterol regulatory element-binding protein (SREBP) cleavage regulates Golgi-to-endoplasmic reticulum recycling of SREBP cleavage-activating protein (SCAP). J Biol Chem 2014; 289:7547–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. A century of cholesterol and coronaries: from plaques to genes to statins. Cell 2015; 161:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab 2012; 16:414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Yan H, Wu Z, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science 2007; 318:1608–1612. [DOI] [PubMed] [Google Scholar]
- Ren R, Zhou X, He Y, et al. PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels. Science 2015; 349:187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Li J, Shao W, et al. Structure of the WD40 domain of SCAP from fission yeast reveals the molecular basis for SREBP recognition. Cell Res 2015; 25:401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Fischer N, Golas MM, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods 2008; 5:53–55. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol Cell 2004; 15:259–268. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci USA 1997; 94:5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol 2000; 16:653–699. [DOI] [PubMed] [Google Scholar]
- Datta S, Osborne TF. Activation domains from both monomers contribute to transcriptional stimulation by sterol regulatory element-binding protein dimers. J Biol Chem 2005; 280:3338–3345. [DOI] [PubMed] [Google Scholar]
- Sakai J, Rawson RB, Espenshade PJ, et al. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell 1998; 2:505–514. [DOI] [PubMed] [Google Scholar]
- Kim J, Ha HJ, Kim S, et al. Identification of Rbd2 as a candidate protease for sterol regulatory element binding protein (SREBP) cleavage in fission yeast. Biochem Biophys Res Commun 2015; 468:606–610. [DOI] [PubMed] [Google Scholar]
- Vaknin Y, Hillmann F, Iannitti R, et al. Identification and characterization of a novel Aspergillus fumigatus rhomboid family putative protease RbdA involved in hypoxia sensing and virulence. Infect Immun 2016; 84:1866–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bat-Ochir C, Kwak JY, Koh SK, et al. The signal peptide peptidase SppA is involved in sterol regulatory element-binding protein cleavage and hypoxia adaptation in Aspergillus nidulans. Mol Microbiol 2016; 100:635–655. [DOI] [PubMed] [Google Scholar]
- Dhingra S, Kowlaski CH, Thammahong A, Beattie SR, Bultman KM, Cramer RA. RbdB, a rhomboid protease critical for SREBP activation and virulence in Aspergillus fumigatus. mSphere 2016; 1:pii: e00035-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem 2003; 320:104–124. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The Ccp4 Suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 1994; 50:760–763. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 1999; 128:82–97. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 2008; 3:1941–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XM, Mooney P, Zheng S, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 2013; 10:584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 2003; 142:334–347. [DOI] [PubMed] [Google Scholar]
- Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system on world wide web. Available from: URL: www.pymol.org. 2002.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SLS and AUC-SV analyses of the Sre1-CTD, Scp1-WD40, and their complex.
Ab initio negative staining EM reconstruction of the Sre1-CTD/Scp1-WD40 complex.
Verification of the tetrameric organization of Sre1-CTD.
Structural determination ofthe Sre1-CTD.
The dimer interface of Sre1-CTD.
Cryo-EM analysis of the Sre1-CTD/Scp1-WD40 complex.
Sequence alignment of the Sre1-CTD from eight closely-related species.
In vitro and in vivo characterization of Sre1-CTD dimerization mutants.
Crystallographic data collection and refinement statistics
S. pombes train list