Abstract
Natural selection is crucial for the adaptation of populations to their environments. Here, we present the first global study of natural selection in the Hominidae (humans and great apes) based on genome-wide information from population samples representing all extant species (including most subspecies). Combining several neutrality tests we create a multi-species map of signatures of natural selection covering all major types of natural selection. We find that the estimated efficiency of both purifying and positive selection varies between species and is significantly correlated with their long-term effective population size. Thus, even the modest differences in population size among the closely related Hominidae lineages have resulted in differences in their ability to remove deleterious alleles and to adapt to changing environments. Most signatures of balancing and positive selection are species-specific, with signatures of balancing selection more often being shared among species. We also identify loci with evidence of positive selection across several lineages. Notably, we detect signatures of positive selection in several genes related to brain function, anatomy, diet and immune processes. Our results contribute to a better understanding of human evolution by putting the evidence of natural selection in humans within its larger evolutionary context. The global map of natural selection in our closest living relatives is available as an interactive browser at http://tinyurl.com/nf8qmzh.
Keywords: evolution, adaptation, comparative genomics, primates.
Introduction
Understanding the adaptive genetic changes that led to the emergence of modern humans continues to be a major focus of modern genomics (Pritchard et al. 2010; Enard et al. 2014). However, despite much work in this field, many central questions remain unanswered. For example, it is still unclear what percentage of the human genome has been shaped by natural selection, which genetic variants are responsible for the phenotypes that make humans unique, and to what extent demographic factors have influenced the rate of adaptive evolution through human history. These questions can only be answered through a deeper understanding of the evolution both of the human genome and also of other closely related species. While laboratory studies on adaptation in organisms such as Drosophila have furthered our understanding of adaptive evolution (Lee et al. 2014), the usefulness of these model organisms for understanding adaptation in humans is limited by the wide disparities that exist between them and humans, in both physiology and demography. Investigation of the molecular basis of adaptation is also hindered by differences in the structure and content of the genomes of more distantly related organisms. Studying our closest living relatives, the great apes, is therefore crucial for furthering our understanding of human evolution.
The Hominidae (humans and great apes) share several traits that make them particularly interesting. Relative to their ancestors they have evolved larger brains, more complex social systems and, arguably, the ability to create and maintain cultural traditions (McGrew 2004). Furthermore, the Hominidae species differ from one another in important ways (including their morphology, physiology, behavior and life history traits) that may result from their independent adaptation to particular environments. Evolutionary genomic information can help us to understand the origin and molecular bases of both shared and species-specific traits in the Hominidae.
The Hominidae also provide an excellent system for comparative studies. This is because although the species are very closely related (with all lineages diverging over the last 12 My) they differ substantially in relevant features such as the effective size of their populations (Ne) (Prado-Martinez et al. 2013). This makes them well-suited for addressing longstanding theoretical questions in evolutionary biology. A central principle of population genetics is that the effective size of a population influences the efficacy of selection (Charlesworth 2009). Populations with a large Ne are expected to be more efficient at both fixing beneficial alleles and removing deleterious ones, when compared with populations with small long-term Ne or that have experienced severe bottlenecks. Empirical attempts to quantify this effect have been limited, with exceptions that include work in yeast (Elyashiv et al. 2010), Drosophila (Jensen and Bachtrog 2011) and eukaryotes (Grossman et al. 2013), as well as comparisons of very divergent lineages (Corbett-Detig et al. 2015). It remains unclear to what extent differences in Ne between closely related mammalian species impact the process of natural selection (Ellegren and Galtier 2016). Full genome sequences of humans and great apes provide a unique opportunity to investigate this question over a relatively short evolutionary timescale.
The signatures of natural selection have been extensively studied in humans (Bustamante et al. 2005; Sabeti et al. 2006; Nielsen et al. 2009; Andrés et al. 2010) and some of the apes (Mikkelsen et al. 2005; Locke et al. 2011; Prüfer et al. 2012; Scally et al. 2012; Bataillon et al. 2015; McManus et al. 2015). However, no study has comprehensively investigated the evidence for natural selection across the Hominidae lineages. We analyzed whole-genome sequence data from multiple individuals from lineages covering all major Hominidae species and subspecies (except Gorilla beringei beringei) (Prado-Martinez et al. 2013) and present the first investigation of the impact of natural selection using this dataset. We focus on attributes of the data that allow us to detect the different types of selection across evolutionary timescales. We then integrate these results to investigate the influence of Ne on the efficacy of natural selection, the targeted functional elements, the genes and biological processes targeted by each type of selection, and the conservation of selective pressures across Hominidae lineages.
Results
Sample Processing
In order to assess the influence of natural selection, we use a dataset of 54 non-human great ape and nine human genomes sequenced to an average of 25-fold coverage (supplementary table S1, Supplementary Material online). Because of differences in demography and selective pressures on autosomes and sex chromosomes, we focus exclusively on the autosomes.
We take particular care to minimize the influence of errors and biases in genomic data and ensure that our data is of the highest possible quality—something particularly important when comparing species. All reads were mapped to the same reference genome (human hg18). We built on the extensive data filtering strategy of Prado-Martinez et al. (2013) (see “Dataset” in “Methods” section). This conservative filtering strategy resulted in the exclusion of 726 Mb (∼23%) of the autosomal genome. This includes tandem repeats (∼38 Mb), segmental duplications (∼154 Mb) and structural variants annotated in at least one species (∼334 Mb) (see supplementary fig. S1, Supplementary Material online), all identified by unusual read-depth, so alternative methods (Gokcumen et al. 2013; Sudmant et al. 2015) may identify nonidentical regions. While certain genomic regions and gene families may be enriched in structural variation and be disproportionately affected by this filtering step, their removal is essential to minimize artifacts. We also excluded genomic gaps (∼226 Mb) and base pairs that were not covered by a minimum of five reads in all individuals per species. The resulting dataset includes on an average 2,099 Mb of analyzable genome sequence per species (see supplementary fig. S1, Supplementary Material online). Although every filtering strategy has limitations and putative biases, we aim for a conservative approach that minimizes the presence of artifacts. The result is a high-quality comparative genomic dataset that allows us to investigate the signatures of natural selection and compare them across species (see supplementary materials Sample Processing, Supplementary Material online).
Neutrality Tests
We selected a set of neutrality tests that explore different aspects of the patterns of polymorphism and in combination allow us to detect the signatures of different types of natural selection across different time depths, from the emergence of the Hominidae ∼12 Ma to recent and ongoing species-specific selective sweeps (fig. 1). Many neutrality tests exist; among them we chose those that utilize the type of information that we have (i.e., that do not require phased genomes), that have been shown to have high power to detect selection (Zhai et al. 2009), that explore relatively independent signatures and that provide information on different timescales. To keep the analyses manageable, we focus on four tests (see fig. 2):
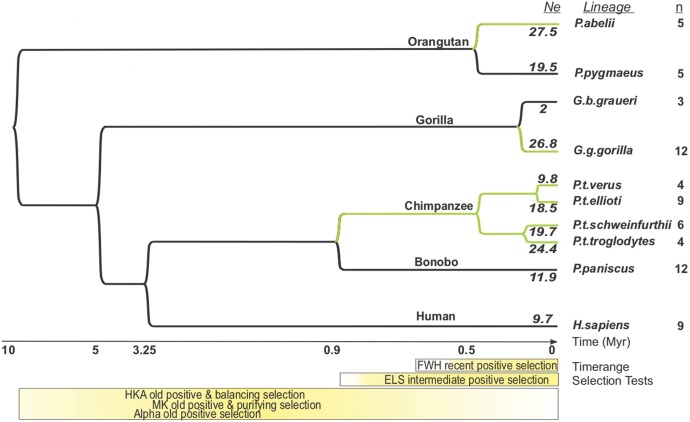
Fig. 1.
Timescale of neutrality tests. Hominidae phylogeny with the approximate time ranges where each neutrality test has power to detect signatures of natural selection. (a) The lineages with the number of genomes used in this study are shown on the right. The X-axis shows the timescale, in units of millions of years. Split times of lineages from Prado-Martinez et al. (2013). For FWH, MK and HKA, the approximate time range where the tests are inferred to have most power to detect selection are represented by color intensity. For ELS, we label in green the branches where the test has power to detect selection. n: number of individuals in each lineage. Ne: estimates of effective population size in units of thousands of individuals according to Watterson’s estimator, taken from Prado-Martinez et al. (2013).
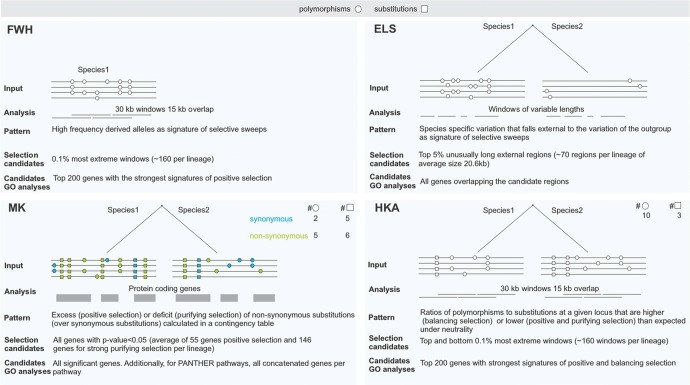
Fig. 2.
Summary of the neutrality tests used. Each box presents the input (the information used), the analysis strategy (how each test was applied on the genome-wide data), the pattern (the signatures of selection explored), the criteria to select selection candidates (the top candidates for each test) and the criteria to select candidates for GO analyses (the candidate used for gene ontology analyses).
To detect signals of purifying and positive selection on the coding sequences of proteins, we applied the McDonald–Kreitman test (MK test; McDonald and Kreitman 1991). The MK test is run on a protein-coding gene-by-gene basis (supplementary materials MK 1, Supplementary Material online). By using information on sequence divergence, it has power to detect signatures of positive and purifying selection along the entire branch lengths of the Hominidae.
To detect long-term balancing selection and positive selection that could have occurred at a deep evolutionary time, we applied a statistic based on the Hudson–Kreitman–Aguadé test (HKA; Hudson et al. 1987), which has been found to be a highly powerful method to detect positive selection (Zhai et al. 2009). The HKA statistic was calculated across the genome in 30-kb windows with a 15-kb overlap between windows (Methods and supplementary materials HKA 1, Supplementary Material online). As it uses both divergence and diversity data, the HKA statistic has power to detect positive selection over broad timescales as well as long-term balancing selection, including persistent balancing selection that predates the emergence of the Hominidae, such as on the MHC region (Hedrick 1999).
To detect lineage-specific positive selection that occurred after the divergence of an ancestral population into two species, we applied the Extended Lineage Sorting test (ELS; SOM 13 in Green et al. 2010; Supplementary Information 7 in Prüfer et al. 2012; Supplementary Information 19a in Prüfer et al. 2014). The test is run across the genome and identifies regions without a predefined size (Methods and supplementary materials ELS 1, Supplementary Material online).
To detect recent selective sweeps, we applied Fay and Wu’s H statistic (FWH; Fay and Wu 2000). The FWH statistic was calculated across the genome in 30-kb windows with 15-kb overlap between windows (Methods and supplementary materials FWH 1, Supplementary Material online).
Together, these tests detect the signatures of purifying, balancing and positive selection, old and recent (we refer to events in the order of millions of years for old and of hundreds of thousands of years up to present day for recent selection), in each lineage (fig. 1). Integrating all results provides an unprecedentedly broad picture of the targets of natural selection in the genomes of humans and great apes.
Ne and the Strength of Natural Selection in the Great Apes
As discussed earlier, empirical data is limited regarding the effect of long-term Ne on the efficacy of natural selection over short evolutionary timescales in vertebrates. The relationship between population size, selection and levels of neutral diversity in populations continues to be a matter of considerable debate (Ellegren and Galtier 2016). A recent study (Corbett-Detig et al. 2015) proposed that the effects of linked selection can explain Lewontin’s paradox (1974), namely that neutral diversity does not scale as expected with population size. Though a recent reanalysis of this data suggests that while linked selection influences diversity along genomes, fluctuations in Ne are the major driver of levels of diversity between species (Coop 2016). The debate has so far been hampered by the limited availability of population-level genome sequence data across species (Ellegren and Galtier 2016). Our dataset therefore provides an ideal opportunity to investigate the relationship between Ne and selection in closely related species.
We find that the ratio of nonsynonymous to synonymous substitutions negatively correlates with long-term Ne in this dataset (Prado-Martinez et al. 2013), as expected with more efficient purifying selection in populations with higher Ne. Here we aim to: (1) infer the distribution of fitness effects (DFE) in each species, (2) quantify the magnitude of the influence of Ne on the DFE, (3) compare the influence of long-term versus short-term Ne, and (4) investigate its influence not only on purifying, but also on positive selection.
Ne and the Strength of Purifying Selection
We first inferred the DFE of deleterious mutations for 3,859 one-to-one orthologous protein-coding genes with DFE-alpha (Eyre-Walker and Keightley 2009), which is based on the MK test (see Methods and MK supplementary materials, Supplementary Material online). The method fits a demographic model to the SFS of neutral sites, and, simultaneously, estimates the gamma-distributed DFE of new nonneutral mutations and the fraction of adaptive substitutions (α) (supplementary fig. S16, Supplementary Material online). For all lineages, the shape parameter of the gamma distribution is <1, indicative of highly leptokurtic (L shaped) DFEs and most nonsynonymous mutations being strongly deleterious. Indeed, in all lineages the proportion of nonsynonymous mutations with a NeS > 10 (S being the mean homozygous effect of a deleterious variant) is >65% (supplementary table S104 and fig. S18, Supplementary Material online), similar to estimates for humans (Eyre-Walker and Keightley 2009) and gorillas (McManus et al. 2015). We observe that the proportion of predicted neutral or nearly neutral mutations correlates negatively with long-term Ne (correlation of −0.64, P value= 0.04 after accounting for phylogenetic nonindependence using BayesTraitsV2 random walk/maximum likelihood method; Pagel and Meade 2013). This correlation reflects stronger purifying selection in great ape species with a larger long-term Ne.
Efficient purifying selection reduces also the accumulation of linked genetic variation due to background selection. Within the bins in the middle range of the HKA distribution (see “Methods” section), which are particularly sensitive to purifying selection, lower HKA scores associate with stronger background selection (lower B scores, supplementary fig. S6, Supplementary Material online) and a higher proportion of protein-coding exons (fig. 3). This is expected if background selection reduces diversity around protein-coding and other functional regions. Across lineages, and in agreement with the DFE-alpha results, the effects of purifying selection increase with larger Ne both when considering the proportion of protein-coding exons and the B scores (supplementary materials HKA 3 and supplementary table S5, Supplementary Material online) (McVicker et al. 2009). Incidentally, the effect is much weaker for nonprotein coding exons (supplementary table S5 and supplementary fig. 1E, Supplementary Material online). These results are virtually unchanged if we use only lineages with less than ten individuals or only lineages with more than five individuals, suggesting that sample size differences between lineages do not affect our observations (supplementary materials subsampling analysis 1.4 and supplementary table S106 and supplementary fig. S30, Supplementary Material online).
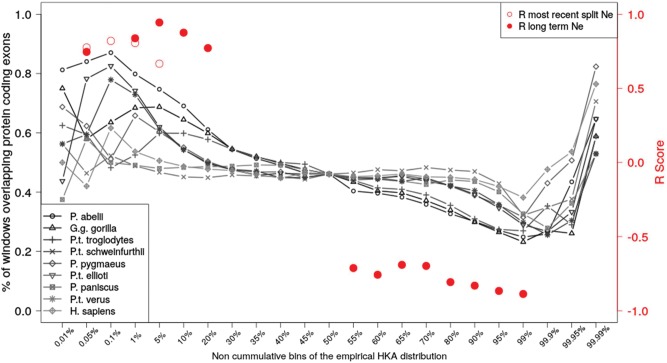
Fig. 3.
Percentage of windows overlapping protein coding exons. Percentage of windows overlapping protein coding exons for noncumulative bins of the HKA empirical distribution (X-axis). Each lineage is plotted as a shaded line. The Pearson’s correlation (R) between the percentage of windows overlapping protein coding exons and Ne within each HKA bin and across all lineages is shown on the right Y-axis. Pearson’s correlation coefficient was computed both with an estimate of short- and long-term Ne (from Prado-Martinez et al. 2013). Only R values with significant P values (P <0.05) are shown.
The correlations with Ne above are almost always stronger with long-term Ne than with recent Ne (for 21 of the 23 HKA bins; supplementary table S3, Supplementary Material online), indicating that we detect the effects of long-term evolutionary history rather than only differences in power due to overall levels of diversity (although differences in the accuracy of the Ne estimates may affect this comparison). Therefore, despite the recent and ongoing population declines experienced by many of these species, their long-term Ne appears to be a better predictor than recent Ne of the past efficacy of purifying selection.
Ne and Adaptive Evolution
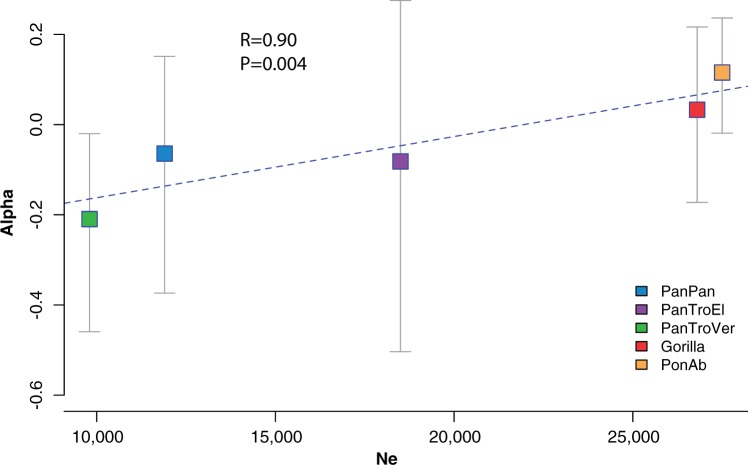
With the MK-based DFE-alpha, it is possible to estimate the proportion (α) of nonsynonymous substitutions driven by positive selection, as well as the ratio of adaptive to neutral divergence (ω(α)) (supplementary table S103, Supplementary Material online). With the exceptions of Pongo pygmaeus (with poor bootstrap support), and Pan t. schweinfurthii (where two inbred individuals (Prado-Martinez et al. 2013) dramatically increase the estimates) (supplementary table S103, Supplementary Material online), both the estimated proportion (α) and the estimated rate of adaptive evolution are low (0–12% and 0–2%, respectively) in agreement with previous estimates (Eyre-Walker 2006; McManus et al. 2015). We observe that in nonhuman great apes both the proportion and the rate of adaptive substitutions are positively correlated with long-term Ne, after phylogenetic nonindependence is accounted for using a generalized least square approach (supplementary fig. S17 and supplementary materials MK test 2.3, Supplementary Material online). The correlation is high and significant when all nonhuman species, except the problematic Pongo pygmaeus and Pan troglodytes schweinfurthii, are considered (Pearson’s R = 0.9, P value = 0.004) (see fig. 4).
Fig. 4.
Correlation between rate of adaptive substitutions (α) and effective population size (Ne). The X-axis shows the effective population size. On the Y-axis, the rate of adaptive substitutions is plotted as α. Correlations were calculated while controlling for the phylogenetic nonindependence using a generalized least square approach and a random walk/maximum likelihood method (see Supplementary Materials MK 2.3, Supplementary Material online).
The effect of positive selection on linked variation also increases with long-term Ne. In the bottom bins of the HKA empirical distribution, which are enriched in targets of positive selection, the percentage of protein-coding windows correlates positively with Ne (0.05–1% bins, P values = 0.0001–0.02). Only the lowest 0.01% HKA bin is not significant, potentially due to the spatial clustering of windows as a result of selective sweeps (supplementary materials HKA 4 and supplementary table S12, Supplementary Material online).
Thus, our results indicate that long-term Ne of populations significantly affects the efficacy of both purifying and positive selection. These correlations are remarkable because these species are very closely related and their long-term Ne varies by a maximum difference of 3-fold.
The Candidate Targets of Natural Selection
As most genomic sites evolve neutrally or nearly neutrally (Kimura 1979; Kelley et al. 2006), we expect an enrichment of targets of natural selection in the extreme tails of the genome-wide distributions of neutrality test statistics. Therefore, we can identify candidate targets of natural selection without relying on a simulated neutral expectation, which is vulnerable to parameter misspecification, an important problem given the complex evolutionary history of the Hominidae lineages. Given the little we know about the strongest targets of purifying, positive and balancing selection in nonhuman apes, this catalog is highly relevant. In addition, these loci allow us to investigate the tempo, conservation, and biological function of natural selection in the great apes. The nature of each of the tests considered means that their implementation in the genome varies and the criteria to define candidate targets of selection necessarily varies too (see fig. 2).
Sample Size and the Candidate Targets of Natural Selection
Sample size, which varies among lineages, may influence the power to detect signatures of natural selection. We assess how differences in sample size might influence our results with down-sampling analyses. We randomly down-sample, 100 times, four or eight individuals from the two lineages with the largest sample size (Pan paniscus and Gorilla gorilla gorilla) and run all neutrality tests for chromosome 1 (except the ELS test for Pan paniscus because this test is not appropriate for this lineage). We then measure the overlap between the candidates from the down-samples (0.1% or 1% tail of the empirical distribution) with the equivalent candidates from the original results (supplementary materials subsampling analysis 1, Supplementary Material online).
The impact of sample size differs between selection tests. HKA appears very robust to sample size variation for signatures of positive selection, showing a mean overlap between the original and down-sampled results of at least 86% (supplementary materials subsampling analysis 1.1 and supplementary figs. S21–24, Supplementary Material online). This is likely to be because the HKA is not strongly affected by the allele frequency of polymorphisms.
In contrast, FWH and ELS appear more sensitive to sample size (supplementary materials subsampling analysis 1.2–1.3 and supplementary figs. S25–28, Supplementary Material online). This may be due to the influence of sample size on the estimates of allele frequency. Therefore, we find that the HKA test is better suited for comparative analyses where sample sizes are low or unequal between populations.
An Available Genome-Wide Map of Natural Selection in Hominidae
The genome-wide map of signatures of natural selection includes information about different types of selection over varying time frames. As such, it provides a broad picture of the influence of natural selection in each genomic region and Hominidae species. All the information is available as an interactive browser at webpage: http://tinyurl.com/nf8qmzh following the criteria and configuration of a recently published human dataset (Pybus et al. 2014, 2015). The UCSC-style format facilitates the integration with the rich UCSC browser tracks, a search mask allows easy access to results for specific genes or genomic regions, and the raw scores (test statistic value and rank score/empirical P value) can be conveniently downloaded using the UCSC Table function. We expect this to be a valuable resource for a wide range of analyses.
The Functional Targets of Natural Selection
The relative contributions of variants in regulatory versus protein coding regions of the genome to adaptive evolution remains a matter of controversy (Halligan et al. 2013). Since King and Wilson (1975) the relative importance of coding and regulatory variation to adaptive evolution has been contentious. Protein-coding DNA constitutes ∼1.5% of the genome but 10–15% appears to be functionally constrained (Ponting and Hardison 2011). The role of nonprotein-coding genes and other nongenic elements in genome function and evolution remains debated (Encode Project Consortium 2011; Doolittle 2013) with several lines of work suggesting that nongenic regions (including some gene desserts) can play an important role in phenotype and adaptation (Bejerano et al. 2006; Libioulle et al. 2007; McPherson et al. 2007; Hubisz and Pollard 2014). Although the stringent filtering of the data and the imperfect annotation of nonprotein-coding functional elements hampers the comparison of protein-coding versus nonprotein regions, we investigated the functional annotations of our candidates.
Except for MK, the neutrality tests we used are agnostic about functional annotation. Still, most of our candidate targets of positive selection contain functional annotations: mean values across species are 72% for HKA, 71% for ELS and 80% for FWH. This is significantly greater than genome-wide expectations based on random sampling of the callable genome (P values < 0.05 in all lineages except Pan paniscus, P value = 0.2, and Pongo pygmaeus, P value = 0.11) (supplementary materials HKA 7 and supplementary figs. S7–15, Supplementary Material online). Among these annotations, the overlap with protein-coding exons (HKA = 62%, ELS = 59%, FWH = 45%) is significantly enriched in all lineages except Pan paniscus (P value = 0.15) and Pan troglodytes verus (P value = 0.06). In contrast, the mean overlap with exons from nonprotein coding genes (HKA = 18%, ELS = 2%, FWH = 20%), is not significantly elevated relative to genome-wide levels (supplementary figs. S7–15, Supplementary Material online, lowest P value = 0.16 in Gorilla gorilla gorilla).
Candidate targets of balancing selection are also highly enriched in protein-coding exons (e.g., 64% in the top 0.01% bin) and in the top HKA bins this proportion sharply increases with the HKA score (fig. 3). The increased levels of diversity in these windows cannot be explained by technical artifacts, as these regions are not unusual in terms of coverage or mapping quality (supplementary materials HKA 1.3–1.4, Supplementary Material online) or by current models of neutral evolution or purifying selection, and are instead best explained by long-term balancing selection acting on or near these protein-coding exons.
The Biological Pathways Targeted by Natural Selection
According to our results above, protein-coding genes appear to be a key target of natural selection in the Hominidae. We thus investigated the biological functions that these genes are involved in. For each neutrality test and lineage, we identified the genes in candidate regions of positive or balancing selection (see fig. 2 and “Methods” section for details) and performed gene enrichment analyses using WebGestalt (Zhang et al. 2005). Our necessarily strict data filtering may disproportionally affect certain Gene Ontology (GO) categories (e.g., olfactory receptors), but we discuss below the categories that retain the strongest signatures for each type of natural selection.
Pathways Targeted by Balancing Selection
The top genes for balancing selection include a number of well-established targets, such as the major histocompatibility complex (MHC) genes (Hughes and Yeager 1998; Hedrick 1999). Indeed, windows containing MHC genes appear among those with the strongest signals of balancing selection in all lineages (supplementary tables S6 and S13, Supplementary Material online). In addition, in all lineages there is a significant enrichment of immunity-related categories such as the GO “Antigen processing and presentation” category (closely related to the MHC) (supplementary tables S16–40, Supplementary Material online). This provides evidence that balancing selection has a strong influence on immunological pathways in all lineages.
To test whether there were strong signatures of balancing selection beyond the MHC complex, we re-ran the GO enrichment analysis excluding all genes in the MHC region on chromosome 6 (supplementary tables S57–65, Supplementary Material online). Doing so removes the enrichment for the GO category “Antigen processing and presentation” in all lineages. Interestingly, in three of the four Pan troglodytes lineages (excluding Pan troglodytes schweinfurthii) there is significant enrichment for the GO category “Cornified envelope” (supplementary tables S360, S62, and S63, Supplementary Material online), driven by the three genes SCEL, SPRR2B and SPRR2G. The cornified envelope is the most exterior layer of the skin and consists of dead cells. Related to this, we note that in Pan troglodytes verus the GO category “keratinocyte differentiation”, involved in the development of the most common cell type in the epidermis is also significantly enriched (P value = 0.0026). This is interesting because keratins and proteins similarly involved in epithelial barrier formation have been proposed as targets of balancing selection (see “Discussion” section).
Pathways Targeted by Strong Purifying Selection
Since Hominidae are closely related, we would expect that similar regions are evolving under purifying selection. We therefore tested whether the pathways showing the strongest signatures of constraint are consistent among lineages. From the MK test, 53 of the 152 evaluated pathways showed signatures of strong purifying selection in more than one lineage. In particular, the “Integrin signaling” pathway and “Wnt signaling” pathway, which regulate basic cellular and developmental processes and the “Alzheimer’s disease-presenilin” pathway are significantly constrained across all lineages (supplementary table S102, Supplementary Material online).
Pathways Targeted by Positive Selection
For the HKA, several lineages show evidence of positive selection targets being enriched for GO categories related to immune function. For example, the GO category “Complement activation” (genes that activate the innate immune system) is significantly enriched in Pan paniscus (P value = 0.042; all P values adjusted for multiple testing), whereas the related pathway “Complement receptor activity” is enriched in Pongo abelii (P value = 0.011) and the GO category “Viral receptor activity” in Gorilla gorilla gorilla (P value = 0.0004) (supplementary tables S41, S46, and S54, Supplementary Material online).
We find that the FWH candidate targets of positive selection show enrichment in several GO categories related to brain development and function, exclusively within the African Hominidae lineages. This includes for example the GO categories “Dendrite” (Homo sapiens P value = 0.040; Pan troglodytes troglodytes P value = 0.010; Gorilla gorilla P value = 0.0024) and “Neuron spine” (Pan troglodytes troglodytes P value = 0.001; Gorilla gorilla gorilla P value = 0.006). Several additional neurological categories are enriched in single lineages (supplementary tables S75–86, Supplementary Material online). For example, Homo sapiens is the only lineage with significant enrichment of the GO category “Glutamate receptor activity” (P value = 0.002); glutamate is the main excitatory neurotransmitter in the brain.
For the MK, the set of genes with an excess of divergence is small (supplementary table S96, Supplementary Material online) but we found a significant enrichment in genes involved in, for instance, “Ion channel activity” in Pan paniscus (P value =0.034), and in “Glycosaminoglycan biosynthesis” in Gorilla gorilla gorilla (P value = 0.020), among other pathways (supplementary tables S98–101, Supplementary Material online).
Overlap betweenTargets of Positive and Balancing Selection across Lineages
The Hominidae lineages have shared, along their evolutionary history, similar physiologies and environments. As such, they have likely been subject to common selective pressures even after their lineages split. To investigate this possibility, we identified genes that show similar signals of natural selection in multiple lineages. Since we use an empirical approach to identify candidate targets of natural selection (as the demographic models for these species are not well established), we use the same 0.1% cut-off to identify outliers from both tails of the HKA empirical distribution and one tail of the FWH distribution. Therefore, we cannot make general claims about the relative frequency of positive and balancing selection in primate species. We can though explore the level of sharing across lineages of these candidate targets. To be conservative, we only consider genes to be shared targets of selection if they appear as candidates in at least three lineages.
Overlap between Selection Targets
We find no signals of positive selection that are shared across all lineages. In fact, there is modest sharing across lineages, a possible indication of the lineage-specific nature of the adaptive process (although we note that we are highly conservative in our selection of candidate genes and the power of FWH is reduced with lower sample sizes) (supplementary table S15, Supplementary Material online). We observe that the HKA candidates show lower sharing across lineages than those from the FWH (supplementary tables S14 and S73, Supplementary Material online). For the HKA, only 27 genes (of the 200 candidates per lineage) are shared in at least three lineages compared with 67 for the FWH, which detects more recent selective events. We note that shared signals among the Pan troglodytes sub-species may not reflect truly independent signatures of selection as signals may predate their divergence into separate lineages and the possibility of admixture between sub-species. However, of these 67 genes, only a minority (8) are shared exclusively among the Pan troglodytes subspecies, potentially reflecting their recently shared ancestry. The majority (59) show evidence of recent positive selection across a range of lineages (supplementary table S73, Supplementary Material online) suggesting putative parallel adaptive events.
Turning to the biological function of these genes, we find limited enrichment among HKA targets (the only significant GO category is “Structural molecule activity”, P value = 0.009; supplementary table S3AAB, Supplementary Material online). However, the 67 shared FWH candidate genes are significantly enriched in multiple functional categories (supplementary tables S87–89, Supplementary Material online), including several neuronal pathways, suggesting that these are a common target of recent positive selection across the Hominidae.
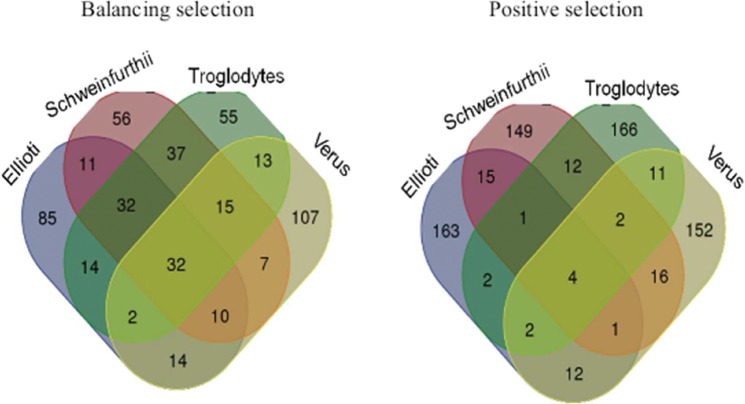
Genes targeted by balancing selection show much greater sharing across lineages, with 156 genes showing signatures of balancing selection across at least three lineages (supplementary table S13, Supplementary Material online, fig. 5 for an example across Pan troglodytes lineages). Nine genes, primarily from the MHC region, are shared across all lineages (supplementary table S13, Supplementary Material online). This likely reflects the long-term and persistent nature of balancing selection on immunity-related genes, in particular in the MHC.
Fig. 5.
Venn Diagram of shared targets of balancing and positive selection among Pan troglodytes lineages. Overlap of the number of putative target genes of balancing and positive selection as inferred by the HKA test for all P. troglodytes lineages.
Discussion
We present a global investigation of the signatures of purifying, positive, and balancing selection at different time scales and across the Hominidae lineages. We observe strong evidence for the signatures of each of these types of natural selection on patterns of genomic variation. By carefully avoiding technical differences, we can compare, for the first time, the patterns of different types of natural selection across the great ape species. All genomic analyses of signatures of selection are complicated to some extent by demographic processes which can result in patterns of genomic variation that obscure signatures of selection or produce false-positives. We tried to mitigate this by utilizing tests that identified putatively selected regions as outliers based on the entire distribution of patterns of genomic variation, under the assumption that the majority of the genome is evolving neutrally.
We find that even with the relatively similar Ne of the great apes (with a maximum difference of 3-fold), Ne has a significant effect on the efficacy of natural selection. This appears to be true for both purifying and positive selection. The evidence for adaptive evolution is stronger in protein-coding than in nonprotein coding genes, and it is overrepresented not only on loci with relevance to immune function, but also on loci involved in the development and maintenance of the brain. Long-term balancing selection, which most clearly affects the evolution of immune and skin-related loci, is more often shared across lineages than positive selection. In what follows, we briefly discuss these observations, as well as some of the biological insights from the loci identified.
Effective Population Size Significantly Influences the Efficacy of Natural Selection in the Hominidae
We estimate that at least 65% of mutations are deleterious (NeS > 10) in all lineages. This estimate agrees very well with results in humans (Eyre-Walker and Keightley 2009). Our results also agree with a recent study in gorillas (McManus et al. 2015) where the DFE-alpha method provided a very similar gamma shape parameter and proportion of strongly deleterious and neutral alleles (66% and 23.8%, compared with our 65% and 22%). Regarding the prevalence of positive selection, our estimates in Gorilla gorilla gorilla (3%) also overlap with previous estimates (McManus et al. 2015). Our results thus confirm the limited information that exists for the Hominidae and greatly expand upon it.
Having information for many great apes enables us to start to compare the different species. The strength of purifying selection on nonsynonymous sites, and its effects on linked variation, correlate with the long-term Ne of the populations. Similar correlations have been observed among other, more distant, species (Leffler et al. 2012; Corbett-Detig et al. 2015). However, our results indicate that even the modest Ne differences that exist among the Hominidae have also affected the efficacy of positive selection, which is likely to be less prevalent and more dependent on environmental changes than purifying selection. Therefore, the Hominidae lineages with the largest long-term effective population sizes, such as Gorilla gorilla gorilla and Pan troglodytes troglodytes, are better able to both remove deleterious alleles and fix adaptive alleles than lineages such as Pan paniscus and Homo sapiens. Even the relatively recent differences in long-term Ne between Pan troglodytes sub-species seem to have resulted in differences in the efficacy of natural selection. This may be extremely important because the long-term survival of these species, which live in small populations and are endangered, may depend on their ability to adapt to changes in their local environments.
Biological Interpretation of Candidate Genes
Our data indicates that adaptive evolution (positive and balancing selection) often targets protein-coding regions (be that the protein-coding sequence or the surrounding regulatory elements). This suggests that in these species variants that affect proteins have been important drivers of novel adaptations. It also supports the comparison of protein-coding versus nonprotein coding regions to establish patterns of positive selection, as long as additional confounding factors are accounted for (Coop et al. 2009; Key et al. 2016).
The candidate genes targeted by positive selection show only moderate overlap between species. This is not surprising, as different populations likely adapt differently at the genetic level even to similar environmental pressures. Nevertheless, there are certain genes, and even gene categories, that show evidence of positive selection in several lineages, which could reflect recurrent evolution at the genomic level. We note that the sharing across lineages is substantially higher when we turn to balancing selection. This is expected because we target only long-term balancing selection, which may predate the divergence of the great ape lineages.
There are several possible interpretations of shared signals. When the signature is shared across closely related lineages these most likely reflect shared events. When the signature is shared across distant lineages, this may reflect independent adaptive evolution. In these cases, selection may be favoring independent phenotypes in each species, for example if it affects different, neighboring functional elements in each species or, due to pleiotropy, the same functional element for a different phenotype in each species. Alternatively, these regions may represent cases of convergent evolution, where the same phenotype is selected for across species. For example, many genes involved in brain development have shared evidence for positive selection across different species. We speculate that there has been ongoing positive selection for neurological phenotypes across the great apes and that although this was likely to be highly polygenic, some of the same genes may have been involved across species.
The detection of specific genes that have been under adaptive evolution is of great interest, especially when dealing with lineages closely related to humans. Here we discuss some of the most interesting findings. For an extended discussion of putatively selected genes, see supplementary materials section 7, Supplementary Material online.
Immunity
Balancing Selection on the MHC
Host–pathogen co-evolution can result in strong selective pressures (Anderson and May 1982). In agreement with this we find, in all lineages, evidence of balancing selection maintaining adaptive diversity on immunity-related genes. As expected with long-term balancing selection, where the time to the most recent common ancestor may predate the species split, many cases are shared among closely related lineages. These results provide further evidence that advantageous diversity is extremely important for the immune system, as has been shown in humans (Ferrer-Admetlla et al. 2008; reviewed in Key et al. 2014). Not surprisingly, the MHC genes are among the top candidate targets of balancing selection in all lineages.
Balancing Selection on the Skin Barrier
The cornified envelope is a layer of dead keratinocytes (corneocytes) that are linked to structural proteins. They form a protective barrier in the outermost layer of the epidermis, known as the stratum corneum, which acts as an external wall that protects the body from physical injury and bacterial invasion. We find that the candidate targets of balancing selection are enriched in genes involved in keratinocyte differentiation in Pan troglodytes verus. They are also enriched in the related biological process “cornified envelope development” in Pan t. ellioti, Pan troglodytes troglodytes and Pan troglodytes verus (the genes involved are SCEL, SPRR2B and SPRR2G) and include two additional cornified envelope genes (late cornified envelope genes 3D and 3E, LCE3D and LCE3E). In LCE3D and LCE3E, the signatures are in flanking regions (the protein-coding portions of the genes are filtered out by the segmental duplication filter). We also identify CDSN, which encodes corneodesmosin, an adhesive protein involved in skin barrier integrity and has previously been shown to have signatures of balancing selection in humans (Andrés et al. 2009; Cagliani et al. 2011), as a putative target of balancing selection in three lineages (Homo sapiens, Pan paniscus, Pan troglodytes verus) (supplementary table S13, Supplementary Material online).
A hypothesis for why genes involved in epidermal differentiation may evolve under balancing selection has been proposed in humans in relation to the filaggrin (FLG) gene (Irvine and McLean 2006), which is essential for the formation of the cornified envelope yet it has two common loss-of-functions alleles (5% frequency each in Europeans) that cause icthyosis vulgaris and strongly predispose to atopic dermatitis (Irvine and McLean 2006; Smith et al. 2006). It has been proposed that the loss-of-function alleles might result in a leaky skin barrier through which low levels of pathogens can penetrate, promoting innate immunity through a process of “natural vaccination” (Irvine and McLean 2006).
Humans homozygous for loss-of-function CDSN alleles frequently show skin barrier defects and are susceptible to Staphylococcus aureus superinfections early in life, suggesting variation in the gene influences the ability of pathogens to penetrate the skin barrier (Oji et al. 2010). Interestingly, heterozygote carriers of this loss-of-function allele do not present these phenotypes, suggesting that heterozygotes may obtain benefits without the deleterious costs of homozygous carriers. Therefore, a leaky skin barrier that promotes “natural vaccination” may be a hitherto under-appreciated mechanism driving balancing selection on a variety of genes involved in development of the stratum corneum across species. We hypothesize that this mechanism may underlie the strong signatures of balancing selection we detect in CDSN (corneodesmosin) and other genes involved in the development of the cornified envelope. The presence of advantageous variation on genes involved in the formation of the epithelial barrier may therefore be more widespread than previously recognized.
Positive Selection on HIV/SIV-Related Genes
We also find evidence for pervasive positive selection on immune-related processes, as seen in H. sapiens and other Hominidae before (Mikkelsen et al. 2005; Cagliani et al. 2010; Casals et al. 2011). MK, HKA and FWH candidate targets of positive selection all are significantly enriched in genes related to immune response (supplementary tables S16–55, S75–86 and S99, Supplementary Material online). The particular genes vary between lineages, although some are shared across lineages.
Immunity-related genes with signals of selection in multiple lineages may reveal convergent adaptive response to pathogens, or adaptive introgression. The gene IDO2 is identified as a FWH candidate of recent positive selection in all four Pan troglodytes lineages and Pan paniscus. IDO2 encodes the enzyme indoleamine 2, 3-dioxygenase 2, which is involved in T-cell regulation and the Tryptophan oxidation pathway (Metz et al. 2014). This pathway is activated after HIV infection and causes chronic inflammation (Murray 2010), likely underlying HIV-1 immunopathogenesis (Boasso and Shearer 2008). Interestingly, blocking expression of the IDO genes in rhesus macaques infected with SIV/HIV improves health outcomes (Boasso et al. 2009). Therefore, selection on this functional pathway may contribute to the ability of some Pan troglodytes individuals to be resistant to AIDS progression after HIV infection, which has been attributed to a lack of HIV induced T-cell dysfunction (Heeney et al. 1993). The MK test also identifies HIVEP1 as a target of positive selection in Pan troglodytes schweinfurthii and Pan paniscus. The transcription factor encoded by HIVEP1 binds enhancer elements of several promoters of viruses, including HIV-1. Investigation of these selection signals may be of relevance to treating HIV infections in humans.
In summary, we find strong evidence of shared signals of both balancing and, less frequently, positive selection on genes involved in immunity in the Hominidae. This likely reflects the strong and continuous selective pressure that infection and disease exerts on these populations, and the close evolutionary history of the Hominidae, which results in exposure to similar pathogens and similar genetic responses.
Neurological Functions
All Hominidae lineages are known to possess sophisticated cognitive abilities (Tomasello and Call 1997; McGrew 2004) related to their increased brain size and changes in brain organization relative to other primates (Semendeferi et al. 2002). We find some of the categories with the strongest evidence of purifying selection (with MK) are involved in brain function. It is intriguing that the candidate targets of recent positive selection are also enriched in neurological functional categories, with some genes involved in brain development and function showing signatures across multiple lineages.
The gene with signatures of positive selection across the highest number of species and timescales is NRXN3, which codes for neurexin 3. The gene is mainly expressed in the brain and encodes for a protein involved in synaptic transmission and plasticity; it belongs to a gene family associated with several cognitive diseases (Südhof 2008). NRXN3 shows signatures of positive selection in six lineages, with a FWH signal of recent positive selection in Pan troglodytes ellioti, Pan troglodytes schweinfurthii, Pan troglodytes troglodytes and Pongo pygmaeus, an HKA signal of positive selection in Homo sapiens, and an ELS signature in all lineages where it was performed (Pan troglodytes, Gorilla gorilla gorilla and Pongo abelii). Therefore, this gene may have been involved in the cognitive evolution of multiple Hominidae lineages, including our own.
Several additional prominent candidates of positive selection are involved in cognitive and neurodevelopmental phenotypes. This includes AUTS2, identified by ELS in G. g. gorilla (second highest rank) and Pan troglodytes troglodytes (fourth highest rank), and implicated in neuronal development and autism in humans (Oksenberg and Ahituv 2013). In addition, CSMD1, the gene with FWH signatures in the most lineages (Homo sapiens, Pan paniscus, Pan troglodytes ellioti, Pan troglodytes schweinfurthii, Gorilla gorilla gorilla and Pongo pygmaeus) (supplementary table S73, Supplementary Material online), whose function is unknown but that is highly expressed in the central nervous system (Kraus et al. 2006) and harbors variants associated with schizophrenia (Håvik et al. 2011). Further, of the four genes with FWH signatures in five lineages, two are associated with neuronal phenotypes: KCNIP4, which encodes an A-type potassium channel modulatory protein, and harbors variants associated with attention deficit hyperactive disorder (Weißflog et al. 2013) and NRG3 (Neuregulin 3), which is crucial in the development of the nervous system and whose variants are associated to schizophrenia (Chen et al. 2009).
In addition, 12 genes detected as positively selected by MK are related to neurodevelopmental disorders in humans (supplementary table S98, Supplementary Material online). Five (MCPH1, CASC5, PHGDH, FTO and NBN) can display a phenotype of microcephaly when mutated (Faheem et al. 2015), with mutations in MCPH1 and CASC5 being responsible for autosomal recessive primary microcephaly (MCPH) (Woods et al. 2005; Genin et al. 2012). MCPH1 (identified here in H. sapiens) has been described as a target of positive selection in primate evolution (Wang and Su 2004; Shi et al. 2013); CASC5 (identified here in Pan troglodytes ellioti and Pan paniscus) contains, in Homo sapiens, a nonsynonymous mutation that reached fixation since the split with Neandertals (Prüfer et al. 2014), suggesting recent positive selection also in our lineage. MCPH1. CENPJ, another MCPH gene, shows marginally nonsignificant evidence of positive selection (P value = 0.055 in P.t. verus). Together these results show putative adaptive evolution in genes that may have contributed to changes in brain size and function during primate evolution.
Conclusion
We present a comparative population genomic analysis that investigates the influence of natural selection across the Hominidae. This information sheds light on the past adaptations of each of these populations. As expected, immune function was a strong selective force in all species. Given the close evolutionary relationship, similar physiology and shared pathogens of humans with the other Hominidae lineages, further functional study of these immunity-related genes may be of medical relevance. In addition, the evidence of positive selection in neuronal pathways of several lineages suggests differential adaptations in phenotypes that distinguish the Hominidae species from one another. For example, genes that show strong signals of positive selection solely on the human lineage constitute the best candidates to explain human-specific neurological phenotypes. Similarly, genes with evidence of positive selection in species that differ from one another in phenotypes including size, locomotion, morphology or diet help us to understand the genetic basis of these adaptations.
The fact that even the modest differences in long-term Ne between Hominidae lineages has had discernible impacts on the efficacy of natural selection, both to remove deleterious alleles and to favor adaptive ones, has additional implications. The different great ape species, all of which (except for humans) are currently endangered, may thus differ significantly in their ability to adapt to environmental change. This may affect their ability to adapt not only to constantly changing pathogens, but also to the often human-induced changes to their habitats.
Methods
Dataset
The dataset we analyzed consists of whole-genome autosomal sequences from 83 individuals across all the major lineages of the Hominidae (with the exception of Gorilla beringei beringei) (fig. 1 and supplementary table S1, Supplementary Material online). The dataset was originally presented in Prado-Martinez et al. (2013; SOM), where the SNP calling pipeline and filtering criteria are described in detail. All reads are mapped to the human reference genome (hg18). This approach has three main advantages. First, we take advantage of the extensive data-quality exploration and filtering performed in the original publication. Second, mapping to the human genome ensures that all species are mapped to a high-quality genome, avoiding the (hard to account for) biases that would result from mapping to genomes of low and varying qualities. Third, the human genome has the most comprehensive annotation of gene coding regions, which is very important in this study.
To avoid errors introduced by miss-mapping due to paralogous variants, we also restricted all analyses to a set of sites with a unique mapping to the human genome. To address the possible influence of unknown copy number variants (which would result in collapsing several genomic regions during mapping and produce false SNP calls), we took several steps (supplementary fig. S1, Supplementary Material online). Using UCSC tracks we excluded from analysis all repetitive regions (∼248 Mb), segmental duplications (∼154 Mb), genomic gaps (∼226 Mb) and tandem repeats (∼38 Mb). We also excluded structural variants detected in any of the great ape lineages (∼334 Mb) based on the most comprehensive catalogue available which was itself generated using this dataset and read-depth methods (Sudmant et al. 2013). Furthermore, sites with depth of coverage (DP) < (mean read depth/8.0) and DP > (mean read depth×3), were also removed. To maximize the number of sites to be analyzed, we excluded multiple individuals with low coverage (supplementary tables S1 and S2, Supplementary Material online). Additionally, we also required positions to have at least 5× coverage in all individuals per species. Only the resulting set of sites, which we termed “callable sites”, were used in further analyses; this minimizes, as much as possible, the effects of filtering in all enrichment analyses. This resulted in a mean of 2,099 Mb of analyzable genome sequence per species (supplementary fig. S1, Supplementary Material online). We caution that despite our many efforts, which include multiple stringent filtering steps and the manual curation of targets presented in the main text, we cannot discard the presence of some artifact in our data (e.g., undetected structural variants in the candidate targets of balancing selection) although we expect that them to have a weak influence in our overall results.
Tests
Hudson–Kreitman–Aguadé Test (HKA)
To detect long-term balancing selection and positive selection that could have occurred at a deep evolutionary timescale, we used a statistic based on the HKA test (Hudson et al. 1987). Here, the HKA statistic is simply the ratio of polymorphic (SNPs) to divergent (substitutions) sites in a window. We consider as a polymorphism a genomic position that was identified as a single nucleotide variant (SNV) in Prado-Martinez et al. (2013). We consider a substitution (a divergent site) a genomic position that is identified as a fixed difference between the tested and the outgroup lineage. For consistency, Homo sapiens was used as an outgroup for all lineages. When performing the test for Homo sapiens, we used the combined Pan troglodytes lineages as the outgroup.
For each lineage, the genome was divided into 30-kb genomic windows with 15-kb overlap and the HKA statistic was calculated. We consider only windows that contain at least 300 callable and 6 informative sites, where an informative site is a SNV or substitution (see supplementary materials HKA, Supplementary Material online). Each window in the genome was ranked according to its HKA score, and the rank was considered the window’s empirical P value (see supplementary fig. S4, Supplementary Material online, for an example of the distribution of polymorphic sites and substitutions across the HKA empirical distribution). To ensure that our results were not influenced by variation in data quality across the genome, we tested whether extreme HKA scores are biased in terms of coverage or mapping quality. We find no evidence for such artifacts influencing our results (see supplementary materials HKA and supplementary figs. S2 and S3, Supplementary Material online).
Fay and Wu H Test (FWH)
To detect complete or nearly complete positive selective sweeps caused by recent or ongoing positive selection, which result in an excess of high-frequency derived alleles, we used the FWH statistic (Fay and Wu 2000). We confirmed our implementation of the FWH statistic was capable of detecting recent selective sweeps using simulations (see supplementary materials FWH 1 and supplementary fig. S19, Supplementary Material online). For each lineage, the genome was divided into 30-kb windows with 15-kb overlap using the same strategy as the HKA test (see above and supplementary materials FWH 1, Supplementary Material online). Windows with less than 300 callable sites were removed. Each window in the genome was ranked according to its FWH score, and the rank was considered the window’s empirical P value.
McDonald–Kreitman Test (MK)
To detect positive and purifying selection on protein coding genes, we used the McDonald–Kreitman test (McDonald and Kreitman 1991). The MK test was calculated for all lineages with at least five individuals, as this was considered the minimum sample size for sufficient polymorphism data (supplementary table S93, Supplementary Material online). Only Pan troglodytes troglodytes did not meet these criteria. Pan t. verus met the criterion only by including Donald, an individual excluded in all other analyses because of evidence of admixture between P.t. verus and Pan troglodytes troglodytes (Prado-Martinez et al. 2013). Coordinates for coding regions of all autosomal transcript unique identifiers were taken from RefSeq hg18 and intersected with the callable sites in our data (Pruitt et al. 2012). This resulted in ∼15.1 Mb of coding sequence available for analysis. For each lineage, we count all polymorphisms and substitutions that are predicted to have appeared after the most recent common ancestor with an outgroup (Homo sapiens was used for all lineages, except when performing the test for Homo sapiens, in which case Pan troglodytes was used) (supplementary table S94, Supplementary Material online). The total number of transcripts tested for each species can be seen in supplementary table S6C, Supplementary Material online, and the significant transcripts for either positive or purifying selection in supplementary tables S96 and S97, Supplementary Material online. Variants were annotated as either synonymous or nonsynonymous using ANNOVAR (Wang et al. 2010). Multiallelic sites were excluded (see supplementary materials MK 1.4, Supplementary Material online).
Extended Lineage Sorting Test (ELS)
To detect lineage-specific positive selection that occurred after the divergence of two closely related lineages, we scan the genome for a signal of extended lineage sorting (see SOM 13 in Green et al. 2010; see Supplementary Information 7 in Prüfer et al. 2012; see Supplementary Information 19a in Prüfer et al. 2014), i.e., genomic regions where the lineage of a closely related outgroup falls basal to the lineages of a test-group of individuals. The test requires a particular relationship between the test-group and the closely related outgroup individual where the outgroup individual is sufficiently close and the test-group is sufficiently diverse so that the outgroup often falls within the diversity of the test-group. To determine which population pairs are suitable for ELS, we performed neutral coalescent simulations with ms (Hudson 2002) (supplementary table S66, Supplementary Material online). The fraction of derived sites in the simulations was compared with the fraction in the data, which closely matched the simulations in most cases (supplementary fig. S20 and supplementary tables S66 and S67, Supplementary Material online). Three lineage pairs showed a sufficiently close relationship and were used for the ELS test: Pan troglodytes—Pan paniscus, Gorilla g. gorilla—Gorilla b. graueri, Pongo abelii—Pongo pygmaeus.
We use an implementation of the ELS test that is based on a hidden Markov model (HMM) that analyses SNPs in individuals from one population and the genotype from a single individual from the outgroup population (Prüfer et al. 2014, SOM). The HMM then infers the posterior probability for the hidden states internal (the outgroup falls within the diversity of the test group) and external (the outgroup falls basal to the lineages of the test group) at all SNP positions.
Following Prüfer et al. (2012, Supplementary Information 7), external regions were defined as a run of SNPs with a probability of >0.8 for being external that is not interrupted by SNPs with a probability of >0.8 for being internal, and scored by their genetic length using the 1-Mb average human recombination rate from Kong et al. (2002).
For each population, the HMM was run repeatedly with each “outgroup” individual. To combine these multiple outputs, we disregarded any external region that was not in the top 5% of the empirical distribution in all runs (as truly external regions are shared among all outgroup individuals) and the remaining external regions were then assigned a final rank based on their cumulative rank score from the multiple runs (supplementary materials ELS 1.3 and supplementary tables S68–70, Supplementary Material online).
Region Annotation and GO Category Enrichment
Regions were annotated as genic (protein-coding and nonprotein coding) if at least 1 bp of the region overlapped with a gene using GENCODE hg18 gene coordinates (Harrow et al. 2012).
To test for evidence of functional enrichment among the genes that we detect as putative targets of natural selection, we performed biological category enrichment analysis using the software WebGestalt (Zhang et al. 2005). For the HKA and FWH tests, we selected the 200 genes with the strongest signatures of selection as our test set of candidate genes. For the ELS test, we considered all genes in the 5% longest external regions. For the MK test, we selected by species all genes with at least one transcript presenting a nominal P value of ≤0.05.
For each test and lineage, we tested for functional enrichment using several databases of biological pathway and functional information: GO categories (Harris et al. 2004) “biological processes”, “molecular functions” and “cellular components”; the Kyoto encyclopedia of genes and genomes (KEGG) pathway database (Kanehisa et al. 2004) based on mammalian and human phenotype ontology; and the PheWas database, which is based on the human PheWas ontology (Denny et al. 2010). We set a significance threshold of 0.05 and used the Bonferroni correction for multiple hypothesis testing. Significant categories driven by only one gene were discarded due to the high potential for spurious signals in such cases. For HKA results, see supplementary tables S3N–S3AAA, Supplementary Material online, for ELS see supplementary tables S71 and S72, Supplementary Material online, for FWH see supplementary tables S5C–S5N, Supplementary Material online, for MK test see supplementary table S101, Supplementary Material online.
We note that all gene pathways used were annotated for humans. While this is not ideal for pathway enrichment analyses of nonhuman species, the putative biases should be minor. This is because these functional elements are evolutionarily constrained and these species are extremely closely related (all within only 12 My). For example, there have only been 96 gene-deletion events in the great apes (Prado-Martinez et al. 2013), which should have a minimal impact on an enrichment analyses that uses thousands of genes. Furthermore, any putative annotation errors between species should be random with respect to biological pathways and not systematically biasing gene enrichment results.
We tested the potential effect of gene length bias on the results by repeating the enrichment analyses after randomly selecting equal numbers of windows and exploring the overlap of these (random) categories with our results (see supplementary materials HKA 7, Supplementary Material online).
Data Access
An interactive browser with the signatures of natural selection for each species is available at http://tinyurl.com/nf8qmzh (last accessed October 10, 2016).
Supplementary Material
Supplementary figures S1–S22, tables S1–S107 and supplementary materials are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Matthias Ongyerth for assistance with data preparation, Michael Lachmann and Mark Stoneking for discussions and valuable comments, and Michael Lachmann for help with the ELS test. We thank the members of the Great Ape Genome Diversity Consortium for support throughout this work. This work was supported by funding from the Max Planck Society to K.P. and A.M.A.; by grants from the Ministerio de Economía y Competitividad in Spain (grant BFU2013-43726-P) and the Secretaria d’Universitats i Recerca de la Generalitat de Catalunya (grant GRC 2014 SGR 866) to J.B.; by an European Research Council Advanced Grant (233297) to S.Pääbo and European Research Council Starting Grant (260372) to T.M.B.; and by European Molecular Biology Organization Young Investigator Award and Ministerio de Ciencia e Innovación in Spain (BFU2014-55090-P) to T.M.B.
References
- Anderson RM, May RM. 1982. Coevolution of hosts and parasites. Parasitology 85(02):411–426. [DOI] [PubMed] [Google Scholar]
- Andrés AM, Dennis MY, Kretzschmar WW, Cannons JL, Lee-Lin S-Q, Hurle B, Schwartzberg PL, Williamson SH, Bustamante CD, Nielsen R, et al. 2010. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 6:e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés AM, Hubisz MJ, Indap A, Torgerson DG, Degenhardt JD, Boyko AR, Gutenkunst RN, White TJ, Green ED, Bustamante CD, Clark AG. 2009. Targets of balancing selection in the human genome. Mol Biol Evol. 26(12):2755–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataillon T, Duan J, Hvilsom C, Jin X, Li Y, Skov L, Glemin S, Munch K, Jiang T, Qian Y, Hobolth A. 2015. Inference of purifying and positive selection in three subspecies of chimpanzees (Pan troglodytes) from exome sequencing. Genome Biol Evol. 7(4):1122–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. 2006. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441(7089):87–90. [DOI] [PubMed] [Google Scholar]
- Boasso A, Shearer GM. 2008. Chronic innate immune activation as a cause of HIV-1 immunopathogenesis. Clin Immunol. 126:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boasso A, Vaccari M, Fuchs D, Hardy AW, Tsai W-P, Tryniszewska E, Shearer GM, Franchini G. 2009. Combined effect of antiretroviral therapy and blockade of IDO in SIV-infected rhesus macaques. J Immunol. 182:4313–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, Civello D. 2005. Natural selection on protein-coding genes in the human genome. Nature 437(7062):1153–1157. 20 [DOI] [PubMed] [Google Scholar]
- Cagliani R, Riva S, Biasin M, Fumagalli M, Pozzoli U, Lo Caputo S, Mazzotta F, Piacentini L, Bresolin N, Clerici M, et al. 2010. Genetic diversity at endoplasmic reticulum aminopeptidases is maintained by balancing selection and is associated with natural resistance to HIV-1 infection. Hum Mol Genet. 19:4705–4714. [DOI] [PubMed] [Google Scholar]
- Cagliani R, Riva S, Pozzoli U, Fumagalli M, Comi GP, Bresolin N, Clerici M, Sironi M. 2011. Balancing selection is common in the extended MHC region but most alleles with opposite risk profile for autoimmune diseases are neutrally evolving. BMC Evol Biol. 11(1):171.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casals F, Sikora M, Laayouni H, Montanucci L, Muntasell A, Lazarus R, Calafell F, Awadalla P, Netea MG, Bertranpetit J. 2011. Genetic adaptation of the antibacterial human innate immunity network. BMC Evol Biol. 11:202.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 2009. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 10:195–205. [DOI] [PubMed] [Google Scholar]
- Chen PL, Avramopoulos D, Lasseter VK, McGrath JA, Fallin MD, Liang KY, Nestadt G, Feng N, Steel G, Cutting AS, Wolyniec P. 2009. Fine mapping on chromosome 10q22-q23 implicates Neuregulin 3 in schizophrenia. Am J Hum Genet. 84:21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G. 2016. Does linked selection explain the narrow range of genetic diversity across species? bioRxiv 1:042598. [Google Scholar]
- Coop G, Pickrell JK, Novembre J, Kudaravalli S, Li J, Absher D, Myers RM, Cavalli-Sforza LL, Feldman MW, Pritchard JK. 2009. “The role of geography in human adaptation.”. PLoS Genet. 5:e1000500.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Hartl DL, Sackton TB. 2015. Natural selection constrains neutral diversity across a wide range of species. PLoS Biol. 13:e1002112.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. 2010. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 26:1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. 2013. Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci. 110(14):5294–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Galtier G. 2016. Determinants of genetic diversity. Nat Rev Genet 17:422–433. [DOI] [PubMed] [Google Scholar]
- Elyashiv E, Bullaughey K, Sattath S, Rinott Y, Przeworski M, Sella G. 2010. Shifts in the intensity of purifying selection: an analysis of genome-wide polymorphism data from two closely related yeast species. Genome Res. 20:1558–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard D, Messer PW, Petrov DA. 2014. Genome-wide signals of positive selection in human evolution. Genome Res. 24:885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2011. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9(4):e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. 2006. The genomic rate of adaptive evolution. Trends Ecol Evol. 21:569–575. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. 2009. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 26:2097–2108. [DOI] [PubMed] [Google Scholar]
- Faheem M, Naseer MI, Rasool M, Chaudhary AG, Kumosani TA, Ilyas AM, Pushparaj P, Ahmed F, Algahtani HA, Al-Qahtani MH, et al. 2015. Molecular genetics of human primary microcephaly: an overview. BMC Med Genomics 8(Suppl 1):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. 2000. Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A, Bosch E, Sikora M, Marquès-Bonet T, Ramírez-Soriano A, Muntasell A, Navarro A, Lazarus R, Calafell F, Bertranpetit J, et al. 2008. Balancing selection is the main force shaping the evolution of innate immunity genes. J Immunol. 181:1315–1322. [DOI] [PubMed] [Google Scholar]
- Genin A, Desir J, Lambert N, Biervliet M, Van der Aa N, Pierquin G, Killian A, Tosi M, Urbina M, Lefort A, et al. 2012. Kinetochore KMN network gene CASC5 mutated in primary microcephaly. Hum Mol Genet. 21:5306–5317. [DOI] [PubMed] [Google Scholar]
- Gokcumen O, Tischler V, Tica J, Zhu Q, Iskow RC, Lee E, Fritz MH, Langdon A, Stütz AM, Pavlidis P, Benes V. 2013. Primate genome architecture influences structural variation mechanisms and functional consequences. Proc Natl Acad Sci. 110(39):15764–15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, et al. 2013. Identifying recent adaptations in large-scale genomic data. Cell 152:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halligan DL, Kousathanas A, Ness RW, Harr B, Eöry L, Keane TM, Adams DJ, Keightley PD. 2013. Contributions of protein-coding and regulatory change to adaptive molecular evolution in murid rodents. PLoS Genet. 5:e1003995.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32:D258–D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvik B, Le Hellard S, Rietschel M, Lybæk H, Djurovic S, Mattheisen M, Mhleisen TW, Degenhardt F, Priebe L, Maier W, et al. 2011. The complement control-related genes CSMD1 and CSMD2 associate to schizophrenia. Biol Psychiatry. 70:35–42. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. 1999. Balancing selection and MHC. Genetica 104:207–214. [DOI] [PubMed] [Google Scholar]
- Heeney J, Jonker R, Koornstra W, Dubbes R, Niphuis H, Di Rienzo AM, Gougeon ML, Montagnier L. 1993. The resistance of HIV-infected chimpanzees to progression to AIDS correlates with absence of HIV-related T-cell dysfunction. J Med Primatol. 22:194–200. [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS. 2014. Exploring the genesis and functions of human accelerated regions sheds light on their role in human evolution. Curr Opin Genet Dev. 29:15–21. [DOI] [PubMed] [Google Scholar]
- Hudson RR. 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18:337–338. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Yeager M. 1998. Natural selection at major histocompatibility complex loci of vertebrates. Annu Rev Genet. 32:415–435. [DOI] [PubMed] [Google Scholar]
- Irvine AD, McLean WHI. 2006. Breaking the (un)sound barrier: filaggrin is a major gene for atopic dermatitis. J Investig Dermatol. 126:1200–1202. [DOI] [PubMed] [Google Scholar]
- Jensen JD, Bachtrog D. 2011. Characterizing the influence of effective population size on the rate of adaptation: Gillespie’s Darwin domain. Genome Biol Evol. 3:687–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32:D277–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. 2006. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 16:980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key FM, Fu Q, Romagné F, Lachmann M, Andrés AM. 2016. Human adaptation and population differentiation in the light of ancient genomes. Nat Commun. 18:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key FM, Teixeira JC, Filippo C, de, Andrés AM. 2014. Advantageous diversity maintained by balancing selection in humans. Curr Opin Genet Dev. 29:45–51. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1979. The neutral theory of molecular evolution. Sci Am. 241:98–126. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188:107–116. [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. 2002. A high-resolution recombination map of the human genome. Nat Genet. 31:241–247. [DOI] [PubMed] [Google Scholar]
- Kraus DM, Elliott GS, Chute H, Horan T, Pfenninger KH, Sanford SD, Foster S, Scully S, Welcher AA, Holers VM. 2006. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. J Immunol. 176:4419–4430. [DOI] [PubMed] [Google Scholar]
- Lee Y, Langley C, Begun D. 2014. Differential strengths of positive selection revealed by hitchhiking effects at small physical scales in Drosophila melanogaster. Mol Biol Evol 31(4):804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, Bullaughey K, Matute DR, Meyer WK, Ségurel L, Venkat A, Andolfatto P, Przeworski M. 2012. Revisiting an old riddle: what determines genetic diversity levels within species?. PLoS Biol. 10(9):e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. 1974. The genetic basis of evolutionary change. New York: Columbia University Press. [Google Scholar]
- Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, De Vos M, Dixon A, Demarche B. 2007. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13. 1 and modulates expression of PTGER4. PLoS Genet. 3(4):e58.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, Mitreva M. 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469(7331):529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654. [DOI] [PubMed] [Google Scholar]
- McGrew WC. 2004. The cultured chimpanzee Reflections on cultural primatology. Cambridge University Press. [Google Scholar]
- McManus KF, Kelley JL, Song S, Veeramah KR, Woerner AE, Stevison LS, Ryder OA, Ape Genome Project G, Kidd JM, Wall JD, et al. 2015. Inference of gorilla demographic and selective history from whole-genome sequence data. Mol Biol Evol. 32:600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E. 2007. A common allele on chromosome 9 associated with coronary heart disease. Science 316(5830):1488–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker G, Gordon D, Davis C, Green P. 2009. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5(5):e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz R, Smith C, DuHadaway JB, Chandler P, Baban B, Merlo LMF, Pigott E, Keough MP, Rust S, Mellor AL, et al. 2014. IDO2 is critical for IDO1-mediated T-cell regulation and exerts a non-redundant function in inflammation. Int Immunol. 26:357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hillier LW, Eichler EE, Zody MC, Jaffe DB, Yang S-P, Enard W, Hellmann I, Lindblad-Toh K, Altheide TK, et al. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Murray MF. 2010. Insights into therapy: tryptophan oxidation and HIV infection. Sci Transl Med. 2:32ps23.. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Hubisz MJ, Hellmann I, Torgerson D, Andrés AM, Albrechtsen A, Gutenkunst R, Adams MD, Cargill M, Boyko A, Indap A. 2009. Darwinian and demographic forces affecting human protein coding genes. Genome Res. 19(5):838–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oji V, Eckl KM, Aufenvenne K, Natebus M, Tarinski T, Ackermann K, Seller N, Metze D, Nurnberg G, Folster-Holst R, et al. 2010. Loss of corneodesmosin leads to severe skin barrier defect, pruritus, and atopy: unraveling the peeling skin disease. Am J Hum Genet. 87(2):274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg N, Ahituv N. 2013. The role of AUTS2 in neurodevelopment and human evolution. Trends Genet. 29:600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M, Meade A. (2013). BayesTraits V2. Software and manual. Reading: University of Reading. http://www.evolution.rdg.ac.uk/BayesTraitsV2Beta.html.
- Ponting CP, Hardison RC. 2011. What fraction of the human genome is functional? Genome Res. 21(11):1769–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, et al. 2013. Great ape genetic diversity and population history. Nature 499:471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK, Coop G. 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 20(4):R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B, Koren S, Sutton G, Kodira C, Winer R, et al. 2012. The bonobo genome compared with the chimpanzee and human genomes. Nature 486:527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Brown GR, Maglott DR. 2012. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 2(40):D130–D135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus M, Dall’Olio GM, Luisi P, Uzkudun M, Carreño-Torres A, Pavlidis P, Laayouni H, Bertranpetit J, Engelken J. 2014. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 42 (Database issue):D903–D909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus M, Luisi P, Dall’Olio G, Uzkudun M, Laayouni H, Bertranpetit J, Engelken J. 2015. Hierarchical boosting: a machine-learning framework to detect and classify hard selective sweeps in human populations. Bioinformatics 31(24):3946–3952. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. 2006. Positive natural selection in the human lineage. Science 312 (5780):1614–1620. [DOI] [PubMed] [Google Scholar]
- Scally A, Dutheil J, Hillier L. 2012. Insights into hominid evolution from the gorilla genome sequence. Nature 483:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semendeferi K, Lu A, Schenker N, Damasio H. 2002. Humans and great apes share a large frontal cortex. Nat Neurosci. 5:272–276. [DOI] [PubMed] [Google Scholar]
- Shi L, Li M, Lin Q, Qi X, Su B. 2013. Functional divergence of the brain-size regulating gene MCPH1 during primate evolution and the origin of humans. BMC Biol. 11:62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FJD, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, et al. 2006. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 38:337–342. [DOI] [PubMed] [Google Scholar]
- Südhof TC. 2008. Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455:903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Huddleston J, Catacchio CR, Malig M, Hillier LW, Baker C, Mohajeri K, Kondova I, Bontrop RE, Persengiev S, Antonacci F. 2013. Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 23(9):1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, Konkel MK. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasello M, Call J. 1997. Primate cognition. USA: Oxford University Press [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Su B. 2004. Molecular evolution of microcephalin, a gene determining human brain size. Hum Mol Genet. 13:1131–1137. [DOI] [PubMed] [Google Scholar]
- Weißflog L, Scholz CJ, Jacob CP, Nguyen TT, Zamzow K, Groß-Lesch S, Renner TJ, Romanos M, Rujescu D, Walitza S, et al. 2013. KCNIP4 as a candidate gene for personality disorders and adult ADHD. Eur Neuropsychopharmacol. 23:436–447. [DOI] [PubMed] [Google Scholar]
- Woods CG, Bond J, Enard W. 2005. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet. 76:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Nielsen R, Slatkin M. 2009. An investigation of the statistical power of neutrality tests based on comparative and population genetic data. Mol Biol Evol. 26(2):273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kirov S, Snoddy J. 2005. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 1(33):W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.