ABSTRACT
VirF, an AraC-like activator, is required to trigger a regulatory cascade that initiates the invasive program of Shigella spp., the etiological agents of bacillary dysentery in humans. VirF expression is activated upon entry into the host and depends on many environmental signals. Here, we show that the virF mRNA is translated into two proteins, the major form, VirF30 (30 kDa), and the shorter VirF21 (21 kDa), lacking the N-terminal segment. By site-specific mutagenesis and toeprint analysis, we identified the translation start sites of VirF30 and VirF21 and showed that the two different forms of VirF arise from differential translation. Interestingly, in vitro and in vivo translation experiments showed that VirF21 is also translated from a leaderless mRNA (llmRNA) whose 5′ end is at position +309/+310, only 1 or 2 nucleotides upstream of the ATG84 start codon of VirF21. The llmRNA is transcribed from a gene-internal promoter, which we identified here. Functional analysis revealed that while VirF30 is responsible for activation of the virulence system, VirF21 negatively autoregulates virF expression itself. Since VirF21 modulates the intracellular VirF levels, this suggests that transcription of the llmRNA might occur when the onset of the virulence program is not required. We speculate that environmental cues, like stress conditions, may promote changes in virF mRNA transcription and preferential translation of llmRNA.
IMPORTANCE
Shigella spp. are a major cause of dysentery in humans. In bacteria of this genus, the activation of the invasive program involves a multitude of signals that act on all layers of the gene regulatory hierarchy. By controlling the essential genes for host cell invasion, VirF is the key regulator of the switch from the noninvasive to the invasive phenotype. Here, we show that the Shigella virF gene encodes two proteins of different sizes, VirF30 and VirF21, that are functionally distinct. The major form, VirF30, activates the genes necessary for virulence, whereas the minor VirF21, which shares the C-terminal two-thirds of VirF30, negatively autoregulates virF expression itself. VirF21 is transcribed from a newly identified gene-internal promoter and, moreover, is translated from an unusual leaderless mRNA. The identification of a new player in regulation adds complexity to the regulation of the Shigella invasive process and may help development of new therapies for shigellosis.
INTRODUCTION
Shigella spp. are highly adapted human pathogens that cause bacillary dysentery (1). The sophisticated infectious strategy of Shigella depends on the capacity to invade, disrupt, and cause inflammatory destruction of the intestinal epithelial barrier (2, 3). Activation of the invasive program is exceptionally complex and involves many signals affecting gene regulation at different levels. A key factor is VirF, an AraC-like transcription factor (TF) whose expression is activated as Shigella bacteria sense entry into the host environment (4, 5). In a cascade model, VirF triggers activation of the virB and icsA genes. IcsA affects bacterial intracellular spreading, and VirB promotes expression of several virulence genes, including those encoding a type III secretion system (T3SS), its effectors, and the last regulator of the cascade, MxiE (6, 7). Interestingly, MxiE, another AraC-like TF, appears to rely on high-level transcriptional slippage to generate its reading frame from two separate open reading frames (ORFs) (8). The genes virF, icsA, virB, and those controlled by VirB are located on a virulence plasmid (pINV) and are silenced outside the human host (9). At low temperatures, the nucleoid-associated protein H-NS represses transcription of the virulence genes (5). In a temperature-dependent manner, H-NS interacts with two sites within the virF promoter, spaced by an intrinsically curved DNA region, to prevent access of RNA polymerase (5, 10, 11). At a permissive temperature (37°C), reduced DNA curvature counteracts H-NS binding (4, 12) and unmasks a binding site for the nucleoid protein FIS to activate virF transcription (13). VirF then relieves H-NS-mediated repression of virB and icsA and directly stimulates transcription (14, 15). By binding upstream of the virF promoter between two H-NS sites, VirB also counteracts H-NS-dependent repression of virF transcription (16). Expression of virF is further modulated by integration host factor (IHF) (17) and other environmental factors, such as pH and osmolarity (7), and is affected by tRNA modifications (18).
The relevance of virF activation for the invasive program is further supported by posttranscriptional regulation of icsA. RnaG is a cis-encoded antisense RNA that promotes premature termination of the icsA mRNA (19). VirF binds the RnaG promoter and decreases rnaG expression (14). Thus, VirF plays a dual role: (i) it relieves H-NS-mediated repression to activate icsA transcription, and (ii) it represses RnaG transcription, thus increasing the level of icsA mRNA (14). VirF also globally activates the expression of chromosomal genes in both Shigella and Escherichia coli. In particular, VirF appears to play a role in shaping the Shigella transcriptional program to better match the requirements of an effective intracellular life (20–22).
Like other members of the AraC family of transcriptional regulators, VirF has a conserved, carboxy-terminal DNA-binding domain with two helix-turn-helix (HTH) motifs. AraC-like proteins are typically insoluble and, accordingly, problems with VirF purification have hampered biochemical studies (23). Mutagenesis experiments indicated that the N-terminal domain of VirF promotes dimerization while C-terminal HTH2 motif mutants are nonfunctional (24).
While attempting a thorough characterization of VirF, we found that the virF mRNA (R1) is subject to differential translation, giving rise to two forms of VirF. VirF30 activates the virulence system and some chromosomal genes, whereas VirF21 exerts negative feedback control on virF expression itself.
Moreover, we identified a second virF mRNA species (R2) with a 5′ end at position nucleotide (nt) +309/+310. This leaderless yet translation-competent mRNA is transcribed from a gene-internal promoter. Possible implications in an interplay between environmental sensing and virulence gene expression are discussed.
RESULTS
The virF gene encodes two independently translated proteins, VirF30 and VirF21.
Earlier experiments on E. coli minicells carrying the virF gene on recombinant plasmids from Shigella flexneri and Shigella sonnei indicated two main VirF protein forms of about 30 and 21 kDa and a minor form of 27 kDa (25, 26). The significance of the 27- and 21-kDa forms remained unclear, and it seemed possible that they were degradation products of full-length VirF (27). To analyze which VirF forms are present in Shigella, a 3×FLAG tag sequence was inserted at the 3′ end of the S. flexneri M90T virF ORF. Western blot analysis (Fig. 1A) confirmed that two VirF proteins, VirF30 (30 kDa) and VirF21 (21 kDa), are expressed by S. flexneri. The 27-kDa form was not detected.
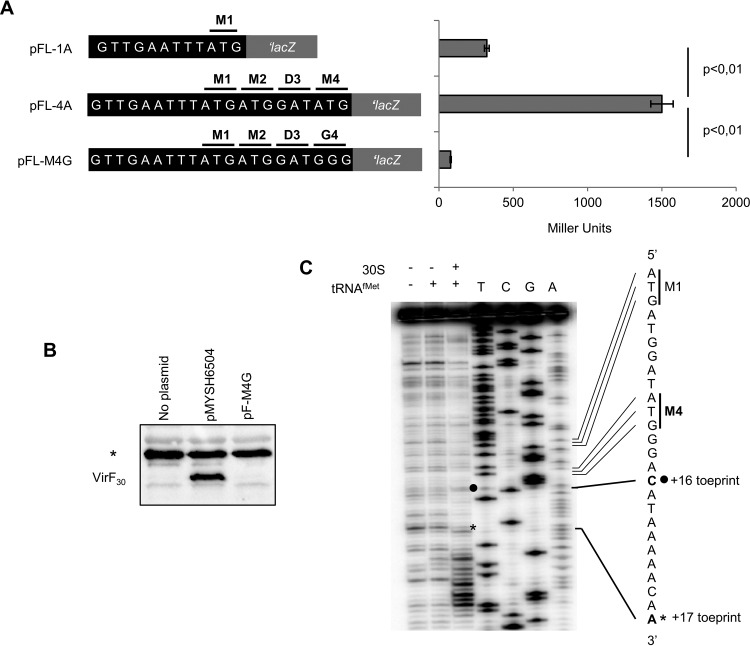
FIG 1 .
Detection of two VirF protein variants. (A) Western blot analysis with anti-FLAG antibody of whole-cell extracts of S. flexneri M90T carrying virF-3×FLAG. Two forms are indicated, VirF30 and VirF21. (B) Schematic organization of the virF gene of Shigella. The methionine (M) codons relevant for this study are highlighted. The transcription start site (+1) was identified previously (5).
The sequence of the virF gene contains three putative start codons, all in the same frame, for VirF30 and an internal ATG codon, consistent with independent translation of VirF21 (25). Thus, we determined at which ATG codons VirF30 and VirF21 translation initiates. In the absence of a recognizable Shine-Dalgarno (SD) sequence upstream, prediction of the ATG encoding the N-terminal methionine of VirF30 was difficult. Thus, each of the ATG codons (codons 1, 2, and 4; codon 3 encodes Asp) (Fig. 1B) was tested for translation initiation activity by using plasmids carrying the virF promoter followed by a virF-lacZ translational fusion. Plasmid pFL-4A is fused in frame after the fourth virF codon (third Met codon), and pFL-1A is fused after the first ATG (Fig. 2A). β-Galactosidase activities indicated that the construct with all three ATGs has ≈5-fold-higher activity than the one fused after ATG1. Thus, ATG2 and/or ATG4 appear to be required for high translation of VirF30, and ATG1 gives a minor contribution. ATG4, which has a short upstream SD-like (GAA) sequence, was tested by introducing an ATG4 → GGG (Gly) mutation into pFL-4A. This plasmid, pFL-M4G, in which ATG1 and ATG2 are still present, gave very low reporter gene activity (Fig. 2A), suggesting ATG4 as the main VirF30 start codon. Western blot analysis supported this. VirF30 was produced only from the wild-type (wt) virF gene, but not when ATG4 had mutated (Fig. 2B, cf. pMYSH6504 and pF-M4G). To corroborate this finding in vitro, we used a toeprinting assay to analyze the formation of ribosomal initiation complexes on virF mRNA (28). A predominant toeprint was seen 17 nt downstream of AUG4 and a minor one 16 nt downstream of AUG1 (Fig. 2C), in line with our in vivo results (Fig. 2A and B). Additional bands downstream of position +17 of AUG4 implicated possible 30S binding-driven structure changes resulting in reverse transcription pauses. In conclusion, translation of VirF30 initiates predominantly at ATG4. Throughout the remainder of this paper, codon positions are accordingly renumbered, with ATG4 as codon 1.
FIG 2 .
Identification of the translation start codon of VirF30. (A, left) Schematic of the pFL plasmids used. Plasmids pFL-1A and pFL-4A carry a translational fusion between the 5′-UTR of virF mRNA after Met1 (pFL-1A) or Met4 (pFL-4A), in frame with lacZ. (Right) pFL-M4G, the Met4 codon, was replaced by Gly (ATG to GGG). β-Galactosidase activities of E. coli strain DH10b carrying the same virF-lacZ plasmids are shown. Strains harboring the pRS414 vector showed very low β-galactosidase activity levels (2 to 4 Miller units) relative to the values obtained. Values are averages of three experiments, and standard deviations are indicated. (B) Western blot with VirF antibodies on extracts from MG1655 harboring pMYSH6504, a plasmid carrying the wt virF gene of S. flexneri, or pF-M4G (pMYSH6504 with the M4G substitution). The asterisk indicates unspecific cross-hybridization with a protein in the extract. (C) Toeprint assay results on the +1 virF mRNA (see Materials and Methods). The mRNA was incubated alone (lane 1), with 30S (lane 2), and with 30S and tRNAfMet (lane 3). Toeprints at position +16 from ATG1 and at +17 from ATG4 are indicated by a black circle and asterisk, respectively. Sequencing ladders were generated with the same 5′-end-labeled primer.
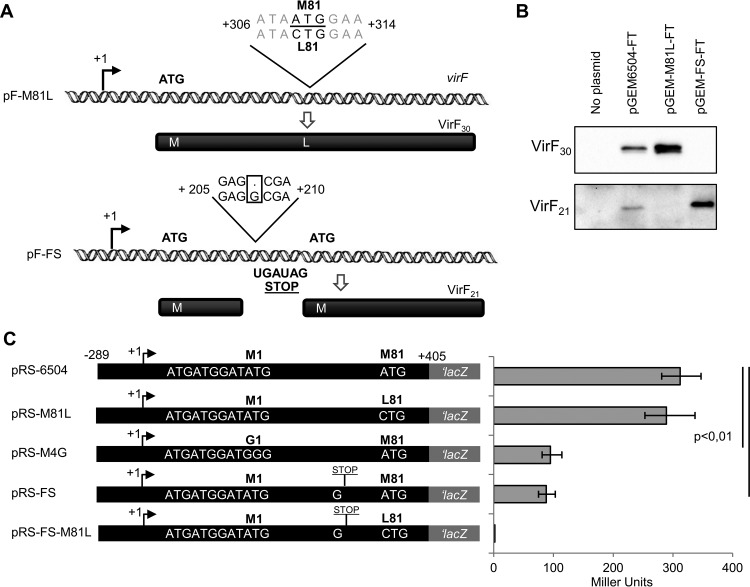
While searching for a VirF21 translation start site, we noticed an in-frame ATG codon within virF at position 311 to 313 (relative to +1 of virF) (Fig. 1B), consistent with translation of the minor form of VirF. To validate ATG81 (formerly ATG84) as the start codon for VirF21, two mutations were introduced into virF, generating a codon change and a frameshift, respectively. To mutate ATG81 to a different codon that would retain VirF30 function, we changed the ATG (mRNA position 311 to 313) to CTG (Met to Leu; pF-M81L) (Fig. 3A) or to ATC (Met to Ile; pF-M81I). Neither mutation should affect VirF30 translation but should abolish independent translation of VirF21. Both mutant VirF30 proteins were tested for activated expression of virB in a virF-defective S. flexneri strain (M90TFd) (see Table S1 in the supplemental material) carrying plasmids expressing wt VirF, VirFM81L, or VirFM81I. VirFM81L but not VirFM81I activated virB to a level comparable to wt (see Fig. S1 in the supplemental material). Thus, the substitution in VirFM81I impairs VirF30 functionality, and therefore only pF-M81L was used in subsequent experiments. Moreover, the exclusive expression of VirF30 upon Met → Leu substitution (Fig. 3B) identified ATG81 as the start codon for VirF21.
FIG 3 .
Differential translation of VirF30 and VirF21. (A) Schematic representation of the constructs used to exclusively produce VirF30 or VirF21. Sequences relevant for the construction of plasmid pF-M81L (M81L substitution) or plasmid pF-FS (insertion of G at position +208) are highlighted. Plasmids are derivatives of pMYSH6504. (B) Western blot analysis (with anti-FLAG antibody) of extracts of E. coli DH10b carrying pGEM-6504-FT (VirF30 and VirF21), pGEM-M81L-FT (only VirF30), or pGEM-FS-FT (only VirF21). (C) β-Galactosidase activity levels of the virF-lacZ plasmids obtained by fusing a fragment (−289 to +405) of the virF gene of pMYSH6504 (pRS-6504), pF-M81L (pRS-M81L), pF-M4G (pRS-M4G), pF-FS (pRS-FS), or pF-FS-M81L (pRS-FS-M81L) as the control, to the promoterless lacZ gene of pRS414. Values are averages of three experiments, and standard deviations are indicated.
To uncouple the translation of VirF30 and VirF21, we inserted a single G between positions +207 and +208 of virF to create a frameshift (pF-FS) into two stop codons (UGA and UAG, +243 to +249), upstream of ATG81 of VirF21 (Fig. 3A). The wt virF gene and its M81L (substitution) and frameshift mutant variants were FLAG tagged, resulting in plasmids pGEM-6504-FT, pGEM-M81L-FT, and pGEM-FS-FT. Western blot analysis of total protein from E. coli cells showed that only VirF30 is translated from pGEM-M81L-FT and only VirF21 is translated from pGEM-FS-FT (Fig. 3B). Western blotting assays with cells with untagged plasmids confirmed this result (see Fig. S2 in the supplemental material). Since premature termination of frameshifted virF only abolished the synthesis of VirF30 and not that of VirF21, both proteins are independently translated.
The relative expression of the two VirF forms was further analyzed by translational lacZ fusions. Four virF-lacZ fusions enabled us to monitor translation of VirF21 (pRS-M4G and pRS-FS) and VirF30 (pRS-M81L), in comparison to total wt virF mRNA translation (pRS-6504). The β-galactosidase levels from pRS-M4G and pRS-FS were about 3-fold lower than those from pRS-6504 and pRS-M81L, which is congruent with the Western blot results shown in Fig. 3B; under these experimental conditions, VirF21 is a minor fraction of the total VirF protein pool.
VirF21 negatively autoregulates the virF gene.
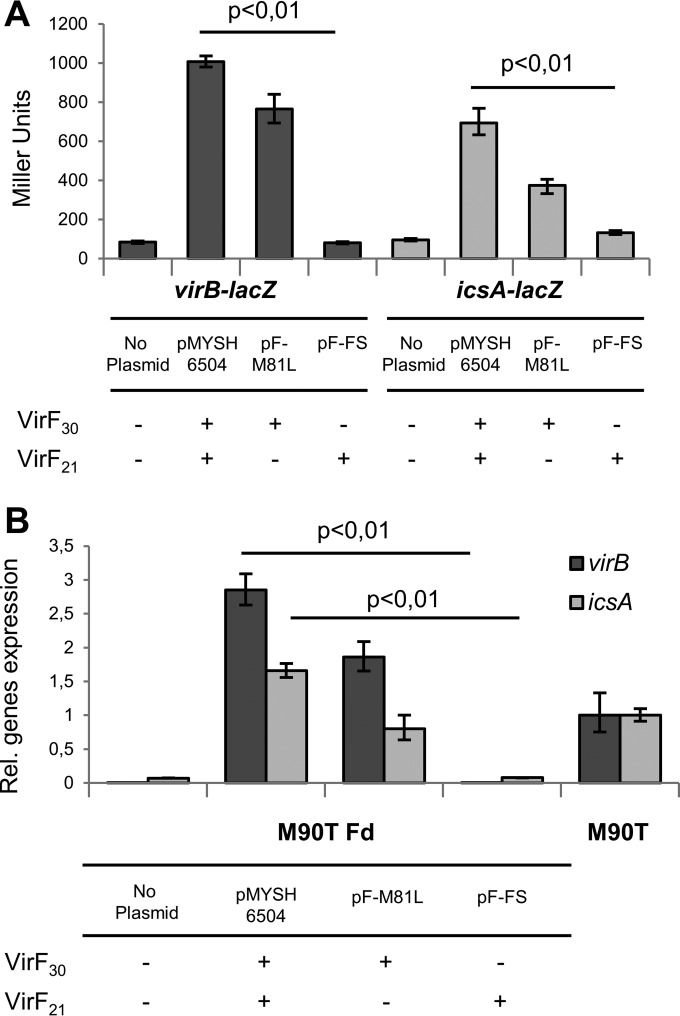
What is the function of the independently translated VirF21? To test whether VirF21 can activate virulence genes, the promoters of virB and icsA were transcriptionally fused to lacZ and transferred to the chromosome of the E. coli K-12 strain P90C, generating P90CλB and P90CλA, respectively (see Materials and Methods). Activation by wt VirF30 and VirF21 (pMYSH6504), VirF30 (pF-M81L), and VirF21 (pF-FS) was monitored in strain P90C λB or P90C λA.
Figure 4A shows that VirF30 alone (pF-M81L) induced the expression of both lacZ fusions to a level similar to that in the presence of both VirF30 and VirF21 (pMYSH6504). VirF21 alone (pF-FS) failed to activate (Fig. 4A). Quantitative reverse transcription-PCR (qRT-PCR) results with the S. flexneri strain M90T Fd (virF defective) carrying the same three plasmids supported this conclusion (Fig. 4B). Thus, a role for VirF21 in the activation of the virulence cascade of Shigella is ruled out. A qRT-PCR experiment also confirmed that the previously shown VirF-dependent activation of some chromosomal heat shock genes (20) cannot be carried out by VirF21 (see Fig. S3 in the supplemental material).
FIG 4 .
Functional analysis of VirF30 and VirF21. (A) β-Galactosidase activity of E. coli P90C carrying virB-lacZ and icsA-lacZ transcriptional fusions, transformed with a plasmid expressing VirF30 and VirF21 (pMYSH6504), only VirF30 (pF-M81L), or only VirF21 (pF-FS). Values are averages of three experiments, with standard deviations indicated. (B) In vivo levels of virB and icsA mRNA as a function of the two VirF forms, monitored by qRT-PCR in a ΔvirF S. flexneri strain (M90T Fd) transformed with pMYSH6504 (VirF30 and VirF21), pF-M81L (VirF30), or pF-FS (VirF21). Expression levels were monitored in M90T or M90T Fd as controls. Samples were run in triplicate, and error bars show the calculated maximum (RQMax) and minimum (RQMin) levels that represent the standard error of the mean expression level (RQ value).
To address putative functions of VirF21, we investigated its role in positive or negative autoregulation of the virF gene. An E. coli K-12 strain harboring a PvirF-lacZ fusion (DH10b pvirF-lacZ) was transformed with plasmids that expressed either Ptac promoter-driven VirF30 (pAC-30) or VirF21 (pAC-21). Figure 5A clearly shows that VirF21, but not VirF30, strongly repressed virF expression, and qRT-PCR on the same samples showed corresponding decreases in lacZ mRNA levels in the presence of VirF21 (Fig. 5B). To validate VirF21-mediated repression of virF transcription in Shigella, we asked whether increased VirF21 levels would reduce the expression of the VirF-activated virB gene. qRT-PCR experiments in the virF-defective strain M90T Fd expressing VirF30 from pF-M81L confirmed severely reduced virB transcription upon induction (isopropyl-β-d-thiogalactopyranoside [IPTG]) of VirF21 (pAC-21) (Fig. 5C). To monitor the VirF21 induction-dependent effect on the VirF protein level, we introduced pAC-21 in the S. flexneri strain that contained the 3×FLAG virF gene (M90T F3xFT; see above). This setup permitted us to assess the levels of VirF21 and VirF30 encoded by pINV by FLAG-tagged antibodies as a function of increasing levels of untagged VirF21 expressed from pAC-21 (monitored via a halon anti-VirF antibody). Figure 5D shows that increasing the VirF21 concentration resulted in a decrease in VirF30, confirming that VirF21 negatively autoregulates virF expression.
FIG 5 .
VirF21 autoregulates virF expression. (A) β-Galactosidase activity of virF-lacZ fusions in response to increased levels of VirF21 or VirF30. Ectopic expression of VirF21 or VirF30 was obtained in E. coli pvirF-lacZ strains carrying pAC-21 or pAC-30, respectively. pGIP7, empty vector. Values are averages of three experiments, and standard deviations indicated. (B) In vivo levels of lacZ mRNA were monitored in the same samples used in the β-galactosidase assay summarized in panel A. Triplicate samples were evaluated, and error bars indicate standard errors of the mean expression levels (RQ values). (C) In vivo levels of virB mRNA were monitored in the ΔvirF S. flexneri strain (M90T Fd) carrying pF-M81L (VirF30) or ectopically expressing VirF21 under IPTG control (pAC-21). Triplicate samples were evaluated; error bars show standard errors of the mean expression levels (RQ values). (D) Western blot analysis of cell extracts of M90T F3xFT carrying pAC-21, with or without ectopic induction of VirF21. The level of expression of VirF30 was monitored with an anti-FLAG antibody. VirF21 induction was monitored with an anti-VirF antibody. Asterisks indicate unspecific cross-hybridization with an unknown protein in the extract. (E) Identification of the VirF21 binding site on the virF promoter based on DNase I footprinting results. Plasmid pMYSH6504 DNA (41) was incubated with 0, 10 or 20 µl of in vitro-translated VirF21. The samples were DNase I treated and subsequently analyzed as described in Materials and Methods, using ML-U30 and ML-U29 as primers. Sequencing ladders were generated with the same 5′-end-labeled plus- or minus-strand-specific primers. The VirF21-protected site is indicated by vertical black lines and shown by shading on both strands of the virF promoter sequence.
In addition, we performed DNase I footprinting by in vitro-translated VirF21 on both strands of the virF promoter region. VirF21 was translated in an in vitro system (PureSystem) (see Materials and Methods), using a PCR-generated DNA template for virF21-only transcription and translation. VirF21 translation was verified by Western blotting (see Fig. S4 in the supplemental material). Figure 5E indicates that VirF21 binding conferred protection of the virF promoter region between positions −90 and −20 on the plus strand and approximate positions −60 to −10 on the minus strand and enhanced minus-strand cleavage from about positions −70 to −90. This result, together with data from in vivo experiments (Fig. 5A and B), strongly suggests that the transcriptional repression of virF by VirF21 depends on its direct binding to the consensus virF promoter elements.
Identification of a VirF21-encoding leaderless mRNA.
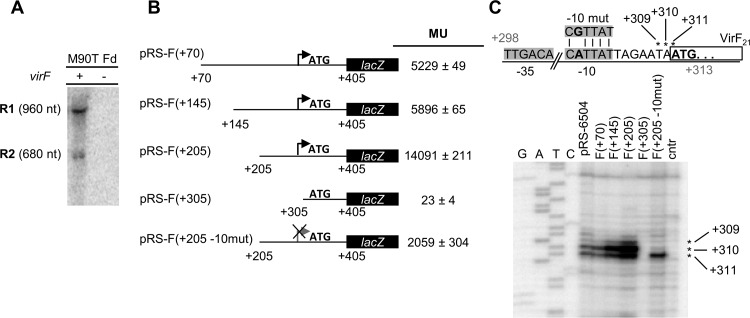
The above results showed that two VirF proteins are independently translated. Whether both are translated from the same mRNA, or different versions of virF transcripts, was unknown. The possibility of different mRNAs was suggested by two virF mRNA variants detected in a Northern blot assay performed with total RNA from strain M90T Fd complemented with the virF-encoding pMYSH6504 and with plasmid-free M90T Fd (Fig. 6A). An ≈960-nt band (full-length virF mRNA; R1) and an ≈680-nt mRNA that might support translation of VirF21 (R2) were visible. To test whether R2 virF mRNA is transcribed from a virF internal promoter or generated by processing, virF-lacZ transcriptional fusions and primer extension (PE) analyses were used. We constructed four virF-lacZ fusions starting at positions +70, +145, +205, and +305; all were fused at +405. The β-galactosidase activities clearly indicated the presence of a promoter between +205 and +305; truncation up to position +305 produced background values (Fig. 6B). A promoter was indeed predicted by PromoterHunt (29), with consensus −10 (CATTAT; +298 to +303) and −35 elements (TTGACA; +276 to +289) (Fig. 6C). After mutagenesis of the −10 box [CATTAT to CGTTAT; pRS-F(+205 −10mut)], we observed a severe reduction (≈7-fold) in the β-galactosidase level. This new promoter was further delineated by PE analysis on RNA extracted from E. coli cells harboring the different virF-lacZ plasmids. This showed 5′ ends at positions +309, +310 (major band), and +311. All three bands were absent in the PE on pRS-F(+305), while with pRS-F(+205 −10mut) the +309/310 bands were not detected. The weaker band at +311 is consistent with a shifted −10 box (data not shown). Thus, the R2 virF mRNA is transcribed from a second virF promoter, with a transcription start site at position +309/+310.
FIG 6 .
In vivo analysis of virF transcripts. (A) Northern hybridization of 10 µg of total RNA from S. flexneri strain M90TFd, with or without pmysh6504 (virF wt) and a virF-specific 32P-labeled probe indicated two major mRNA variants. (B) Schematic representation of virF-lacZ transcriptional fusions carrying truncations of the region upstream of the translational start site of VirF21. The ATG for VirF21 is indicated as a reference, and the putative promoter is depicted by an arrow. The β-galactosidase activities of the virF-lacZ fusion strains were determined. Values reported are in Miller units and represent the averages ± standard deviations of five independent experiments. (C) Schematic representation of the new virF promoter. The positions of the −35 and −10 elements are indicated, and the mutated −10 box (−10mut) is shown above. PE analysis results are shown for total RNA extracted from E. coli cells carrying the indicated plasmids. Three 5′ ends at position +309, +310, and +311 are indicated by asterisks. The −10 mutation shows only a 5′ end at +311.
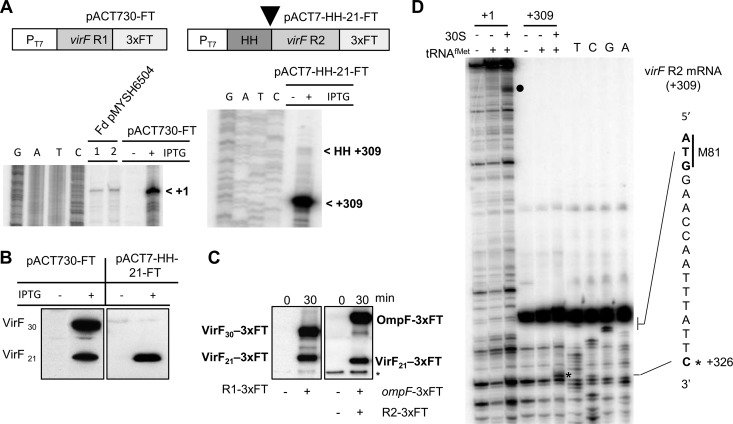
The 5′ ends at +309 to 311 and the start codon at +311 to 313 imply that the R2 mRNA is leaderless (Fig. 6C). To test whether the llmRNA is VirF21 translation competent, we cloned the sequence corresponding to R2 mRNA, and also the entire R1 mRNA as a control, downstream of a T7 promoter. To ensure a correct 5′ end of the R2 mRNA in vivo (5′ U+309 as +1) (Fig. 6C), a hammerhead ribozyme sequence downstream of the T7 promoter (see Fig. S5 in the supplemental material) was introduced to generate an R2 mRNA starting at position +309. The plasmids carrying the R1 or R2 transcripts, pAC-T730-FT (R1; virF +1 to +888) and pAC-T7-HH-21-FT (R2; virF +309 to +888) also harbored 3′ FLAG tags in virF. Upon IPTG induction, virF mRNA transcription from the T7 promoter was induced in E. coli BL21(D3) harboring either plasmid. PE analysis verified the expected 5′ ends of both transcripts (Fig. 7A).
FIG 7 .
Analysis of VirF21 translation from the virF R2 transcript. (A) Primer extension analysis of total RNA from BL21(DE3) cells carrying pAC-T730FT or pACT7-HH-21-FT with or without induction using IPTG. The arrowhead indicates the position of hammerhead cleavage. (B) Western blot analysis of total protein extracts from strain BL21(DE3) cells carrying pAC-T730FT or pACT7-HH-21-FT, with or without induction by IPTG. Shown is translation of both VirF30 and VirF21 in the presence of pACT7-HH-21-FT or of only VirF21 in the presence of pACT7-HH-21-FT. (C) In vitro translation of virF R1-3XFT (start, + 1) and R2-3XFT (start, + 309) mRNAs. Both VirF30 and VirF21 were translated from virF R1-3XFT, but only VirF21 was translated from virF R2-3XFT mRNA. Asterisk, unspecific cross-hybridization with a protein in the extract. In the blot on the right, we included ompF mRNA as an internal canonical, SD-dependent translation control. (D) Toeprint assay results with +1 (full-length) and +309 (leaderless) virF mRNAs. The mRNAs were incubated alone (control; lanes 1 and 4), with 30S (lanes 2 and 5), or with 30S and tRNA−fMet (lanes 3 and 6). A specific toeprint was observed on full-length mRNA (+1) near the 5′ end (black circle) (compare with Fig. 2C). A second toeprint, specific to the llmRNA, is at position +326 (black asterisk). Sequencing ladders were generated with the same 5′-end-labeled primer.
VirF21 translation from the leaderless R2 mRNA was tested by immunoblot analysis on protein extracts after induction. VirF21 was detected in cells carrying pACT7-HH-21-FT, confirming that R2 is a leaderless translation-proficient mRNA (Fig. 7B, right panel). Translation of both VirF forms was observed in cells harboring pAC-T730-FT (Fig. 7B, left panel). In vitro translation in the PureSystem (30) was tested on R1 and R2 mRNAs carrying FLAG tag sequences. Translation products were analyzed with anti-FLAG antibodies. In agreement with the in vivo results, R1 mRNA supported translation of both VirF forms, whereas the leaderless R2 transcript only produced VirF21 (Fig. 7C). Furthermore, toeprint experiments on in vitro-transcribed virF R1 mRNA (start, +1) showed a strong RT stop near the 5′ end, indicating initiation complex formation at AUG4 (compare with Fig. 2C). In contrast, a specific toeprint was observed at position +326 for the llmRNA R2 (start, +309) (Fig. 7D). This toeprint was absent on R1 mRNA, indicating a strong preference for VirF30 translation from the full-length mRNA. Together, these results suggest that a new virF promoter generates a llmRNA variant (R2 mRNA) dedicated to the exclusive translation of VirF21.
DISCUSSION
The complex regulatory cascade for activation of the Shigella virulence genes depends on the VirF protein (7). VirF is at the heart of the switch from the noninvasive to the invasive phenotype. Thus, it is not surprising that its expression is triggered by many environmental signals and that it is controlled at several levels (2, 4, 10, 17). Since its discovery, VirF was known to be present in three forms that differ in size: 30, 27, and 21 kDa (25). The smaller forms were ignored as likely degradation products. Here, we report that the VirF 21-kDa form is translated as an independent polypeptide. Our results address how the VirF21 variant is produced and suggest an autoregulatory role in virF expression. As a first step, we identified the translation start sites of VirF30 and VirF21. Of the three Met codons among the first four codons of the predicted virF ORF, only ATG4 was essential for VirF30 translation (Fig. 1 and 2). Replacement with GGG drastically reduced VirF, as monitored by Western blotting or β-galactosidase activity of virF-lacZ translational fusions (Fig. 2A and B). The identification of ATG4 as a start codon was further supported by toeprint analysis (Fig. 2C). The start codon consistent with the size of VirF21 is ATG81; accordingly, replacement with CTG blocks VirF21 production (Fig. 3B).
Interestingly, while the wt virF mRNA is translated into both VirF30 and VirF21 in vivo, a frameshift mutation upstream of ATG81 affects only the production of VirF30, and not that of VirF21. Thus, the two forms are independently translated (Fig. 3B); consequently, a derivative with either the FS mutation or the M81L substitution gives only VirF21 or only VirF30, respectively. β-Galactosidase fusion and immunoblot analyses (Fig. 3C) showed that the expression level of VirF21 under our experimental conditions is generally lower than that of VirF30.
VirF21 is clearly not functionally redundant with VirF30. Unlike VirF30, it does not restore the expression of VirF-regulated genes in a virF-defective S. flexneri mutant (Fig. 4). Instead, overexpression of VirF21 negatively autoregulates virF expression, reducing intracellular levels of VirF30 and causing reduced virB expression (Fig. 5B). This negative autoregulation is likely due to VirF21 binding to the virF promoter, as indicated by the position of a DNase I footprint (Fig. 5E) which is predicted to interfere with RNA polymerase access.
An arrangement based on a smaller protein controlling a larger one, with both of them encoded by the same gene, applies to Tn5 transposase (31). The form of Tn5 transposase lacking the first 55 amino acids posttranslationally forms nonproductive complexes with transposase, thus blocking its activity at IS50 inverted repeats (31). Superficially similar in setup, the shorter VirF21 also lacks a large N-terminal portion of the longer VirF30 protein, but here, the shorter form alone is sufficient to exert control at the level of virF transcription (Fig. 5A). Though the known C-terminal DNA-binding domain is present in both VirF variants, our data suggest different DNA recognition preferences. Further work will test whether N-terminal sequences affect binding properties of VirF30 and whether protein folding differences in the shared domain can account for the observed specificity differences.
Since VirF30 and VirF21 originate from differential translation, we investigated the virF transcripts in more detail. Long (R1) and shorter (R2) virF mRNAs of lengths compatible with VirF30 and VirF21 were detected (Fig. 6A). Evaluation of deletions in the region upstream of the VirF21 ORF, along with PE analyses, identified a new gene-internal virF promoter that drives the transcription of the virF R2 mRNA. In vivo and in vitro data support that the leaderless R2 is translated into VirF21; plasmid vectors encoding R2 (start site, +309) support in vivo translation of VirF21 (Fig. 7B). Moreover, leaderless translation of VirF21 by R2 also occurs in vitro (Fig. 7C), and initiation complex formation occurs at the appropriate position (Fig. 7D).
In recent years, noncanonical translation initiation mechanisms have been reported, including so-called leaderless transcripts, i.e., those lacking a 5′-untranslated region (UTR) and an SD sequence (32–34). Most leaderless genes identified so far in E. coli reside in mobile DNA, including λ, P2, and Tn1721. The virF gene is also located within an IS-rich region on an extrachromosomal element, the large Shigella/EIEC invasive plasmid (9). Sequencing data for bacteria and archaea suggest that a leaderless model may not be uncommon (35, 36).
The mechanisms underlying synthesis and translation of llmRNAs are not yet fully understood. Vesper et al. (37) showed that induction of the MazEF toxin-antitoxin (TA) system in E. coli produces a leaderless mRNA population and, simultaneously, specialized “stress” ribosomes with a preference to translate proteins from llmRNAs. The endoribonuclease MazF cleaves single-stranded mRNAs, sometimes at ACA sequences upstream of AUG start codons, generating llmRNA. MazF also cleaves 16S rRNA, removing the anti-SD sequence required for translation on canonical mRNAs. Thereby, a subpopulation of ribosomes is generated for selective translation on llmRNA (37). It is well established that Shigella bacteria sense and respond to environmental conditions within and outside the host, with corresponding reprogramming of transcription. Since VirF21 modulates the intracellular level of VirF, this suggests that the transcription of the leaderless R2 mRNA could occur under conditions where the activation of the virulence program is undesirable. A possible coupling between stress conditions that might promote changes in R2 virF mRNA transcription and/or preferential translation of leaderless R2 mRNA and effects on virulence gene regulation is an exciting possibility that we intend to pursue. In particular, the environmental cues that may regulate transcription of the shorter virF mRNA, and the translation of VirF21 from the llmRNA under stress and infection-relevant conditions, will be addressed. In summary, this study has added new, entirely unexpected elements to the complex regulation of the Shigella virulence system and of its major regulator, the VirF protein.
MATERIALS AND METHODS
Oligodeoxyribonucleotides.
Oligodeoxynucleotides used in this study (see Table S1 in the supplemental material) were purchased from Metabion.
Bacterial strains and general methods.
Strains used in this study are listed in Table S2 in the supplemental material. Cloning was performed wtih strain DH10b. E. coli strain P90CλB was obtained by transferring a PvirB-lacZ fusion from plasmid pRS415 via homologous recombination to the lac transducing phage λRS45 and then integration (38) into the the λ att site of E. coli P90C. P90CλA was previously described (see Table S2). Strains M90T-F3xFT and M90T Fd(ΔvirF) were previously constructed (21).
Bacteria were grown aerobically in LB medium at 37°C. Antibiotics and chemicals were used at the following concentrations: ampicillin, 50 µg/ml; chloramphenicol, 25 µg/ml; kanamycin, 30 µg/ml; streptomycin, 10 µg/ml; tetracycline, 5 µg/ml; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, 20 mg/ml. β-Galactosidase assays were performed as described elsewhere (39). Reported values represent the means of at least three separate measurements. DNA isolation, PCR, restriction digests, cloning, and other DNA manipulation methods were performed as described previously (39). Plasmids are listed in Table S3 in the supplemental material. In addition, plasmid constructions are detailed in Text S1 in the supplemental material.
Analysis of virF mRNA.
S. flexneri M90T Fd (ΔvirF) (Table S2) cells with or without pMYSH6504 were grown in LB broth at 37°C to an optical density at 600 nm of 0.4 to 0.5. Total RNA extraction and Northern blot assays with an α-32P-labeled virF-specific probe were performed as described elsewhere (21). Loading controls entailed rRNA staining. Radioactivity was quantified using a Typhoon 9200 instrument (GE Healthcare).
qRT-PCR was performed using Power SYBR green PCR master mix on a 7300 real-time PCR system (Applied Biosystems) as described previously (19). The levels of virB, icsA, and lacZ transcripts were analyzed using the 2−ΔΔCT (cycle threshhold [CT]) method (40), and results are reported that the fold increase relative to the reference. Primers for mdh (endogenous control) and for virB, icsA, and lacZ transcripts were designed by using Primer Express software v2.0 and validated. The following oligonucleotides were used (see Table S1 in the supplemental material): mdhQF/mdhQR, virBQF/virBQR, icsAQL/icsAQR, and lacZQF/lacZQR.
Primer extensions.
Total RNA from exponentially growing plasmid-carrying E. coli strains was extracted (41). Total RNA (10 to 20 µg) was hybridized with 5′-32P-labeled ML-512 and ML-1314 primers. Reverse transcription experiments were done at 42°C using the reverse transcriptase ImProm-II (Promega). Reaction products were analyzed on an 8% polyacrylamide gel in parallel with sequencing reaction products obtained using the same primers.
DNase I footprinting.
Supercoiled plasmid pMYSH6504 (42) (200 ng/sample) was preincubated for 20 min at room temperature with the indicated volumes of the translation mixture, which contained VirF21 or control (no-template) PureSystem reagent in 30 µl of binding buffer (40 mM Tris-HCl [pH 8.0], 50 mM KCl, 10 mM Mg-acetate, and 0.5 mM dithiothreitol). The DNA-protein complex was incubated with 1 U of DNase I for 40 s. After stopping the reaction, the DNA was precipitated and separately analyzed by primer extension on either DNA strand with 3 pmol of 5′-end-labeled primers ML-U30 or ML-U29 as described previously (14). The extension products and corresponding sequencing reactions were run on 7% sequencing gels and then fixed for 5 min (10% ethanol–6% acetic acid) and dried. Signals were detected using a phosphorimager screen.
Immunodetection of VirF protein.
Western blot assays were carried out as described in reference 21. Incubation with primary antibodies (polyclonal halon anti-VirF, anti-FLAG [Sigma F1804]) was at 4°C in phosphate-buffered saline–Tween (PBS-T) containing 2% dried skim milk. Membranes were washed and incubated at room temperature for 1 h with a secondary anti-rabbit (1:10,000) or anti-mouse (1:5,000) horseradish peroxidase-conjugated antibody in PBS-T. After washing with PBS-T, membranes were developed for 5 min for enhanced chemiluminescence and visualized on a ChemiDoc XRS+ system.
RNA in vitro transcription.
The virF-3XFT mRNAs R1 and R2 were transcribed for in vitro translation and toeprint assays. For virF mRNA R1-3XFT (start, +1), DNA templates contained a T7 promoter (PCR with primers ML-U1/ML-982). For virF mRNA R2-3XFT (start, +309), a fragment with a T7 promoter and a hammerhead ribozyme sequence in front of the virF sequence was produced by PCR (primers ML-U20/ML-982) on pAC-T7-HH-21-FT as the template (for the hammerhead sequence, Fig. S5 in the supplemental material). DNA templates were in vitro transcribed as described in reference 43. To obtain virF R2-3XFT, an additional ribozyme self-cleavage step was performed after in vitro transcription according methods described previously (44).
Toeprint assay.
Toeprint assays were performed as in reference 45. Aliquots of 0.2 pmol of unlabeled virF-3xft mRNAs R1 and R2 were annealed with 0.5 pmol 5′-end-labeled ML-U25 or ML-U26 primer in water at 95°C for 1 min and chilled on ice for 2 min. After addition of renaturing buffer (20 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 100 mM NH4Cl) and incubation for 10 min at 37°C, 2 pmol of 30S ribosomal subunits was added. After 15 min, 4 pmol of tRNA-fMet was added, and incubation continued for 20 min before cDNA synthesis with avian myeloblastosis virus reverse transcriptase (7.5 U; Invitrogen) and deoxynucleoside triphosphates (100 nM). Reactions were stopped by phenol-chloroform-isoamyl alcohol extraction followed by ethanol precipitation. The cDNAs and sequencing reactions were run on 8% denaturing polyacrylamide gels that were fixed for 5 min (10% ethanol–6% acetic acid) and dried for 1 h at 80°C. Signals were detected using a phoshorimager screen.
In vitro translation.
To generate VirF21 for DNase I footprinting, 500 ng of a PCR product containing a T7 promoter and the virF21 coding sequence was used as the template in the PureSystem Express (New England BioLabs [NEB]) transcription-translation system at 37°C for 4 h. VirF21 translation was analyzed by immunoblotting using anti-VirF antibodies (see Fig. S4 in the supplemental material). For the in vitro translation of different virF mRNAs (Fig. 7C), each purified transcript was denatured for 2 min at 95°C, chilled for 1 min on ice, diluted in TMN (20 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 150 mM NaCl), and incubated for 15 min at room temperature. In vitro translation (mRNA at 50 nM) was performed with the PureSystem Express (NEB) translation system at 37°C. Translation products were analyzed by immunoblotting with anti-FLAG antibodies.
SUPPLEMENTAL MATERIAL
In vivo levels of virB expression as a function of VirF30 were monitored by qRT-PCR in a virF-defective S. flexneri strain (M90TFd) transformed with pMYSH6504, pF-M81L, or pF-M81I. Expression levels in M90TFd were used as the control. At least three wells were run for each sample, and error bars display the calculated maximum (RQMax) and minimum (RQMin) levels of the standard error of the mean expression level (RQ value). Download
Western blot analysis (with anti-VirF halon serum) of whole extracts of DH10b carrying pMYSH6504 (expressing both VirF30 and VirF21), pF-M81L (expressing only VirF30), or pF-FS (expressing only VirF21). The asterisks indicate unspecific cross-hybridization with other proteins in the extracts. Download
In vivo levels of groEL and htpG mRNAs as a function of VirF30 or VirF21 were monitored by real-time PCR in a virF-defective S. flexneri strain (M90TFd) transformed with pMYSH6504, pF-M81L, or pF-FS. Expression levels monitored in M90T were used as a control. At least three wells were run for each sample, and error bars display the calculated maximum (RQMax) and minimum (RQMin) levels, which represent standard errors of the mean expression levels (RQ values). The following oligonucleotides were used (see Table S1): groELQL/groELQR and htpGQL/htpGQR. Download
In vitro translation of VirF21 protein used for the DNase I footprinting experiment (Fig. 5E). In vitro transcription-translation was done in the PureExpress system using a PCR-generated DNA template with a T7 promoter immediately followed by the virF21 ORF sequence. A 2.5-µl aliquot of the reaction was loaded and the protein was detected with a polyclonal anti-VirF halon antibody. In parallel, 2.5 µl of extract minus template was analyzed. Asterisk, unspecific cross-hybridization with proteins in the reaction mixture. Download
Secondary structure of the hammerhead ribozyme used to generate the virF R2 leaderless mRNA (start, +309). The ribozyme sequence was designed according to methods described elsewhere (J. M. Avis, G. L. Conn, S. C. Walker SC, Methods Mol Biol 941:83–98, 2012, http://dx.doi.org/10.1007/978-1-62703-113-4_7). Download
Oligodeoxyribonucleotides used in this study.
Strains used in this study.
Plasmids used in this study.
Plasmid constructions and related references. Download
ACKNOWLEDGMENTS
We thank Davide Roncarati for advice on DNaseI footprinting. We also thank Gioacchino Micheli and Mikael Sellin for critical reading of the manuscript. This work was supported by grants from Sapienza University, PRIN2012-WWJSX8K and PTR 24-2016 (B.C. and G.P.) and from the Swedish Research Council (E.G.H.W.).
Funding Statement
This work was funded by grants from Sapienza University, from PRIN2012-WWJSX8K and from Institut Pasteur PTR-24-2016 to B.C. and G.P., and from the Swedish Research Council to E.G.H.W. This work, including the efforts of Maria Letizia Di Martino, was supported by grants from Sapienza University, from PRIN2012-WWJSX8K, from Istituto Pasteur Italia, and from the Swedish Research Council. This work, and the efforts of Cédric Romilly, was supported by the Swedish Research Council.
Footnotes
Citation Di Martino ML, Romilly C, Wagner EGH, Colonna B, Prosseda G. 2016. One gene and two proteins: a leaderless mRNA supports the translation of a shorter form of the Shigella VirF regulator. mBio 7(6):e01860-16. doi:10.1128/mBio.01860-16.
REFERENCES
- 1.The HC, Thanh DP, Holt KE, Thomson NR, Baker S. 2016. The genomic signatures of Shigella evolution, adaptation and geographical spread. Nat Rev Microbiol 14:235–250. doi: 10.1038/nrmicro.2016.10. [DOI] [PubMed] [Google Scholar]
- 2.Schroeder GN, Hilbi H. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev 21:134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ray K, Marteyn B, Sansonetti PJ, Tang CM. 2009. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol 7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 4.Prosseda G, Falconi M, Giangrossi M, Gualerzi CO, Micheli G, Colonna B. 2004. The virF promoter in Shigella: more than just a curved DNA stretch. Mol Microbiol 51:523–537. doi: 10.1046/j.1365-2958.2003.03848.x. [DOI] [PubMed] [Google Scholar]
- 5.Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi CO. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J 17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogawa M, Handa Y, Ashida H, Suzuki M, Sasakawa C. 2008. The versatility of Shigella effectors. Nat Rev Microbiol 6:11–16. doi: 10.1038/nrmicro1814. [DOI] [PubMed] [Google Scholar]
- 7.Di Martino ML, Falconi M, Micheli G, Colonna B, Prosseda G. 2016. The multifaceted activity of the VirF regulatory protein in the Shigella lifestyle. Front Mol Biosci 3:61. doi: 10.3389/fmolb.2016.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penno C, Sansonetti P, Parsot C. 2005. Frameshifting by transcriptional slippage is involved in production of MxiE, the transcription activator regulated by the activity of the type III secretion apparatus in Shigella flexneri. Mol Microbiol 56:204–214. doi: 10.1111/j.1365-2958.2004.04530.x. [DOI] [PubMed] [Google Scholar]
- 9.Buchrieser C, Glaser P, Rusniok C, Nedjari H, D’Hauteville H, Kunst F, Sansonetti P, Parsot C. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol 38:760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 10.Dorman CJ. 2004. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol 2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 11.Dorman CJ. 2007. H-NS, the genome sentinel. Nat Rev Microbiol 5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- 12.Prosseda G, Mazzola A, Di Martino ML, Tielker D, Micheli G, Colonna B. 2010. A temperature-induced narrow DNA curvature range sustains the maximum activity of a bacterial promoter in vitro. Biochemistry 49:2778–2785. doi: 10.1021/bi902003g. [DOI] [PubMed] [Google Scholar]
- 13.Falconi M, Prosseda G, Giangrossi M, Beghetto E, Colonna B. 2001. Involvement of FIS in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Mol Microbiol 42:439–452. doi: 10.1046/j.1365-2958.2001.02646.x. [DOI] [PubMed] [Google Scholar]
- 14.Tran CN, Giangrossi M, Prosseda G, Brandi A, Di Martino ML, Colonna B, Falconi M. 2011. A multifactor regulatory circuit involving H-NS, VirF and an antisense RNA modulates transcription of the virulence gene icsA of Shigella flexneri. Nucleic Acids Res 39:8122–8134. doi: 10.1093/nar/gkr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. 1993. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by virF and repression by H-NS. J Bacteriol 175:6142–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kane KA, Dorman CJ. 2012. VirB-mediated positive feedback control of the virulence gene regulatory cascade of Shigella flexneri. J Bacteriol 194:5264–5273. doi: 10.1128/JB.00800-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porter ME, Dorman CJ. 1997. Positive regulation of Shigella flexneri virulence genes by integration host factor. J Bacteriol 179:6537–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurt JK, Olgen S, Garcia GA. 2007. Site-specific modification of Shigella flexneri virF mRNA by tRNA-guanine transglycosylase in vitro. Nucleic Acids Res 35:4905–4913. doi: 10.1093/nar/gkm473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giangrossi M, Prosseda G, Tran CN, Brandi A, Colonna B, Falconi M. 2010. A novel antisense RNA regulates at transcriptional level the virulence gene icsA of Shigella flexneri. Nucleic Acids Res 38:3362–3375. doi: 10.1093/nar/gkq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbagallo M, Di Martino ML, Marcocci L, Pietrangeli P, De Carolis E, Casalino M, Colonna B, Prosseda G. 2011. A new piece of the Shigella pathogenicity puzzle: spermidine accumulation by silencing of the speG gene. PLoS One 6:e27226. doi: 10.1371/journal.pone.0027226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leuzzi A, Di Martino ML, Campilongo R, Falconi M, Barbagallo M, Marcocci L, Pietrangeli P, Casalino M, Grossi M, Micheli G, Colonna B, Prosseda G. 2015. Multifactor regulation of the MdtJI polyamine transporter in Shigella. PLoS One 10:e0136744. doi: 10.1371/journal.pone.0136744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prosseda G, Di Martino ML, Campilongo R, Fioravanti R, Micheli G, Casalino M, Colonna B. 2012. Shedding of genes that interfere with the pathogenic lifestyle: the Shigella model. Res Microbiol 163:399–406. doi: 10.1016/j.resmic.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Emanuele AA, Garcia GA. 2015. Mechanism of action and initial, in vitro SAR of an inhibitor of the Shigella flexneri virulence regulator VirF. PLoS One 10:e0137410. doi: 10.1371/journal.pone.0137410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter ME, Dorman CJ. 2002. In vivo DNA-binding and oligomerization properties of the Shigella flexneri AraC-like transcriptional regulator VirF as identified by random and site-specific mutagenesis. J Bacteriol 184:531–539. doi: 10.1128/JB.184.2.531-539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai T, Sasakawa C, Makino S, Yoshikawa M. 1986. DNA sequence and product analysis of the virF locus responsible for Congo red binding and cell invasion in Shigella flexneri 2a. Infect Immun 54:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato J, Ito K, Nakamura A, Watanabe H. 1989. Cloning of regions required for contact hemolysis and entry into LLC-MK2 cells from Shigella sonnei form I plasmid: virF is a positive regulator gene for these phenotypes. Infect Immun 57:1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter ME, Mitchell P, Roe AJ, Free A, Smith DG, Gally DL. 2004. Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol Microbiol 54:1117–1133. doi: 10.1111/j.1365-2958.2004.04333.x. [DOI] [PubMed] [Google Scholar]
- 28.Fechter P, Chevalier C, Yusupova G, Yusupov M, Romby P, Marzi S. 2009. Ribosomal initiation complexes probed by toeprinting and effect of trans-acting translational regulators in bacteria. Methods Mol Biol 540:247–263. doi: 10.1007/978-1-59745-558-9_18. [DOI] [PubMed] [Google Scholar]
- 29.Klucar L, Stano M, Hajduk M. 2010. phiSITE: database of gene regulation in bacteriophages. Nucleic Acids Res 38:D366–D370. doi: 10.1093/nar/gkp911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohashi H, Kanamori T, Shimizu Y, Ueda T. 2010. A highly controllable reconstituted cell-free system—a breakthrough in protein synthesis research. Curr Pharm Biotechnol 11:267–271. doi: 10.2174/138920110791111889. [DOI] [PubMed] [Google Scholar]
- 31.Mahnke Braam LA, Goryshin IY, Reznikoff WS. 1999. A mechanism for Tn5 inhibition. Carboxyl-terminal dimerization. J Biol Chem 274:86–92. doi: 10.1074/jbc.274.1.86. [DOI] [PubMed] [Google Scholar]
- 32.Moll I, Grill S, Gualerzi CO, Bläsi U. 2002. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol Microbiol 43:239–246. doi: 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- 33.Malys N, McCarthy JE. 2011. Translation initiation: variations in the mechanism can be anticipated. Cell Mol Life Sci 68:991–1003. doi: 10.1007/s00018-010-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moll I, Engelberg-Kulka H. 2012. Selective translation during stress in Escherichia coli. Trends Biochem Sci 37:493–498. doi: 10.1016/j.tibs.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer P, Gäbel K, Pfeiffer F, Soppa J. 2014. Haloferax volcanii, a prokaryotic species that does not use the Shine Dalgarno mechanism for translation initiation at 5′-UTRs. PLoS One 9:e94979. doi: 10.1371/journal.pone.0094979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT, Gray TA. 2015. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet 11:e1005641. doi: 10.1371/journal.pgen.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. 2011. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147:147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 39.Prosseda G, Latella MC, Casalino M, Nicoletti M, Michienzi S, Colonna B. 2006. Plasticity of the P junc promoter of ISEc11, a new insertion sequence of the IS1111 family. J Bacteriol 188:4681–4689. doi: 10.1128/JB.00332-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Campilongo R, Di Martino ML, Marcocci L, Pietrangeli P, Leuzzi A, Grossi M, Casalino M, Nicoletti M, Micheli G, Colonna B, Prosseda G. 2014. Molecular and functional profiling of the polyamine content in enteroinvasive E. coli: looking into the gap between commensal E. coli and harmful Shigella. PLoS One 9:e106589. doi: 10.1371/journal.pone.0106589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakai T, Sasakawa C, Makino S, Kamata K, Yoshikawa M. 1986. Molecular cloning of a genetic determinant for Congo red binding ability which is essential for the virulence of Shigella flexneri. Infect Immun 51:476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romilly C, Lays C, Tomasini A, Caldelari I, Benito Y, Hammann P, Geissmann T, Boisset S, Romby P, Vandenesch F. 2014. A non-coding RNA Promotes bacterial persistence and decreases virulence by regulating a regulator in Staphylococcus aureus. PLoS Pathog 10:e1003979. doi: 10.1371/journal.ppat.1003979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fechter P, Rudinger J, Giegé R, Théobald-Dietrich A. 1998. Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett 436:99–103. doi: 10.1016/S0014-5793(98)01096-5. [DOI] [PubMed] [Google Scholar]
- 45.Holmqvist E, Unoson C, Reimegård J, Wagner EG. 2012. A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp. Mol Microbiol 84:414–427. doi: 10.1111/j.1365-2958.2012.07994.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vivo levels of virB expression as a function of VirF30 were monitored by qRT-PCR in a virF-defective S. flexneri strain (M90TFd) transformed with pMYSH6504, pF-M81L, or pF-M81I. Expression levels in M90TFd were used as the control. At least three wells were run for each sample, and error bars display the calculated maximum (RQMax) and minimum (RQMin) levels of the standard error of the mean expression level (RQ value). Download
Western blot analysis (with anti-VirF halon serum) of whole extracts of DH10b carrying pMYSH6504 (expressing both VirF30 and VirF21), pF-M81L (expressing only VirF30), or pF-FS (expressing only VirF21). The asterisks indicate unspecific cross-hybridization with other proteins in the extracts. Download
In vivo levels of groEL and htpG mRNAs as a function of VirF30 or VirF21 were monitored by real-time PCR in a virF-defective S. flexneri strain (M90TFd) transformed with pMYSH6504, pF-M81L, or pF-FS. Expression levels monitored in M90T were used as a control. At least three wells were run for each sample, and error bars display the calculated maximum (RQMax) and minimum (RQMin) levels, which represent standard errors of the mean expression levels (RQ values). The following oligonucleotides were used (see Table S1): groELQL/groELQR and htpGQL/htpGQR. Download
In vitro translation of VirF21 protein used for the DNase I footprinting experiment (Fig. 5E). In vitro transcription-translation was done in the PureExpress system using a PCR-generated DNA template with a T7 promoter immediately followed by the virF21 ORF sequence. A 2.5-µl aliquot of the reaction was loaded and the protein was detected with a polyclonal anti-VirF halon antibody. In parallel, 2.5 µl of extract minus template was analyzed. Asterisk, unspecific cross-hybridization with proteins in the reaction mixture. Download
Secondary structure of the hammerhead ribozyme used to generate the virF R2 leaderless mRNA (start, +309). The ribozyme sequence was designed according to methods described elsewhere (J. M. Avis, G. L. Conn, S. C. Walker SC, Methods Mol Biol 941:83–98, 2012, http://dx.doi.org/10.1007/978-1-62703-113-4_7). Download
Oligodeoxyribonucleotides used in this study.
Strains used in this study.
Plasmids used in this study.
Plasmid constructions and related references. Download