Identification of LATE BLOOMER2 in pea as a CYCLING DOF FACTOR gene sheds light on the mechanism of long-day photoperiodism, a crucial factor for the spread and cultivation of many crop species.
Abstract
The molecular pathways responsible for the flowering response to photoperiod have been extensively studied in Arabidopsis thaliana and cereals but remain poorly understood in other major plant groups. Here, we describe a dominant mutant at the LATE BLOOMER2 (LATE2) locus in pea (Pisum sativum) that is late-flowering with a reduced response to photoperiod. LATE2 acts downstream of light signaling and the circadian clock to control expression of the main photoperiod-regulated FT gene, FTb2, implying that it plays a primary role in photoperiod measurement. Mapping identified the CYCLING DOF FACTOR gene CDFc1 as a strong candidate for LATE2, and the late2-1D mutant was found to carry a missense mutation in CDFc1 that impairs its capacity to bind to the blue-light photoreceptor FKF1 in yeast two-hybrid assays and delays flowering in Arabidopsis when overexpressed. Arabidopsis CDF genes are important negative regulators of CONSTANS (CO) transcription, but we found no effect of LATE2 on the transcription of pea CO-LIKE genes, nor on genes in any other families previously implicated in the activation of FT in Arabidopsis. Our results reveal an important component of the pea photoperiod response pathway and support the view that regulation of FTb2 expression by photoperiod occurs via a CO-independent mechanism.
INTRODUCTION
In plants, a key factor in the transition from vegetative to reproductive development is the ability to detect and respond to changes in photoperiod. This capacity enables plants to synchronize flowering time with seasonal conditions that are conducive to maximum reproductive fitness. For instance, wild progenitors and primitive cultivars of rice (Oryza sativa) originating from tropical regions are typically short-day (SD) plants with a long basic vegetative growth phase that maximizes accumulation of resources before flowering in response to lengthening nights at the end of summer (Wu et al., 2013). By contrast, the progenitors of many important crop species known to have originated in the Fertile Crescent of southwest Asia, such as wheat (Triticum spp), barley (Hordeum vulgare), pea (Pisum sativum), lentil (Lens culinaris), and flax (Linum usitatissimum), are long-day (LD) plants, flowering in response to the lengthening days of spring and enabling seed maturation and harvest before the arrival of late summer droughts (Nakamichi, 2015).
The relaxation of such photoperiod requirements for flowering is considered to have been essential to the spread and cultivation of numerous crop species (Nakamichi, 2015). For example, it is thought that reduced function at LD repressor loci enabled cultivation of rice at higher latitudes, where the growing season is restricted to spring and summer, and long photoperiods would otherwise be noninductive (Takahashi and Shimamoto, 2011; Gómez-Ariza et al., 2015). Similarly, molecular evidence suggests that it was a loss of function in genes conferring a strong LD requirement that first permitted ancestral forms of wheat, barley, pea, and lentil to flower early and be cultivated as spring crops at higher latitudes in Europe where summers are short (Turner et al., 2005; Comadran et al., 2012; Weller et al., 2012).
Because of the relevance of photoperiod responsiveness to plant adaptation in new cropping environments, the underlying molecular processes have received considerable attention, particularly in the model species Arabidopsis thaliana and rice (Shrestha et al., 2014). In both species, studies of photoperiod response variants have identified genes in the FT family as the ultimate targets of photoperiod regulation and key factors in communication between photoperiod sensing in leaves and meristem identity gene networks at the shoot apex (Suárez-López et al., 2001; Ishikawa et al., 2005).
In Arabidopsis, the B-box transcription factor CONSTANS (CO) plays an important role in the LD-specific induction of FT expression through direct interaction with the FT promoter (Tiwari et al., 2010). CO expression is diurnally regulated by the circadian clock, with peak expression occurring at night during SD, but in the afternoon under LD (Suárez-López et al., 2001). During peak CO expression in LD, the blue light photoreceptor FLAVIN BINDING KELCH REPEAT F-BOX1 (FKF1) is activated by light and forms a complex with GIGANTEA (GI; Sawa et al., 2007; Song et al., 2012). This complex binds to and directs the degradation of CYCLING DOF FACTOR (CDF) proteins, which are transcriptional repressors of CO and FT, allowing CO expression and flowering to occur (Imaizumi et al., 2005). Light-activated FKF1 also binds directly to CO and contributes to its stabilization (Song et al., 2012). In contrast to LD conditions, CO expression under SD occurs in the dark period, when FKF1 is not active. As a result, FKF1-dependent release of CO expression and stabilization of CO protein do not occur, resulting in persistent repression of FT (Sawa et al., 2007; Song et al., 2012).
Evidence from several other photoperiodic species also indicates an important role for CO homologs in photoperiod responsiveness. In rice, the CO-like gene Heading date 1 (Hd1) is involved in photoperiod measurement and photoperiod-specific regulation of genes in the FT family (Brambilla and Fornara, 2013), but unlike Arabidopsis CO, it serves a dual function as both a promoter of FT under SD and an inhibitor under LD (Izawa et al., 2002; Kojima et al., 2002). Functional analyses in potato (Solanum tuberosum) and sugar beet (Beta vulgaris) have also demonstrated a role for CO-like genes in photoperiod responses (Chia et al., 2008; González-Schain et al., 2012). Evidence for conserved CO function is less clear in other species such as barley and poplar (Campoli et al., 2012; Hsu et al., 2012).
In legumes, the molecular basis of photoperiod measurement has also attracted much interest. However, most research has focused specifically on light perception, circadian clock function, or genes from the FT family (e.g., Hecht et al., 2007, 2011; Liew et al., 2009; Kong et al., 2010; Watanabe et al., 2012), and comparatively little is known about the mechanism by which clock and photoreceptor inputs are integrated to provide photoperiod-specific regulation of FT genes. Recent analysis of CONSTANS-LIKE (COL) genes in the temperate LD legume Medicago truncatula suggests that the central role of CO in photoperiod measurement is not conserved in temperate legumes (Wong et al., 2014), and it seems likely that CO-independent pathways may play a greater role in regulating photoperiod sensing in legumes generally.
We previously conducted a mutant screen to isolate late-flowering photoperiod-insensitive mutants that might define genes encoding components of the photoperiod sensing mechanism in pea and identified one such gene, LATE BLOOMER1 (LATE1), as the pea ortholog of Arabidopsis GI (Hecht et al., 2007). In this study, we describe a second locus that contributes to photoperiod responsiveness, LATE2. We present genetic and molecular evidence that LATE2 is a CDF homolog that can regulate FT expression and flowering time without affecting the expression of CO-like genes. Together with results from previous studies, this supports the idea that CO-like genes do not have a significant role in the photoperiod response of pea and other temperate legumes.
RESULTS
A New Photoperiod Response Locus, LATE2, Defined by a Dominant Late-Flowering Mutant
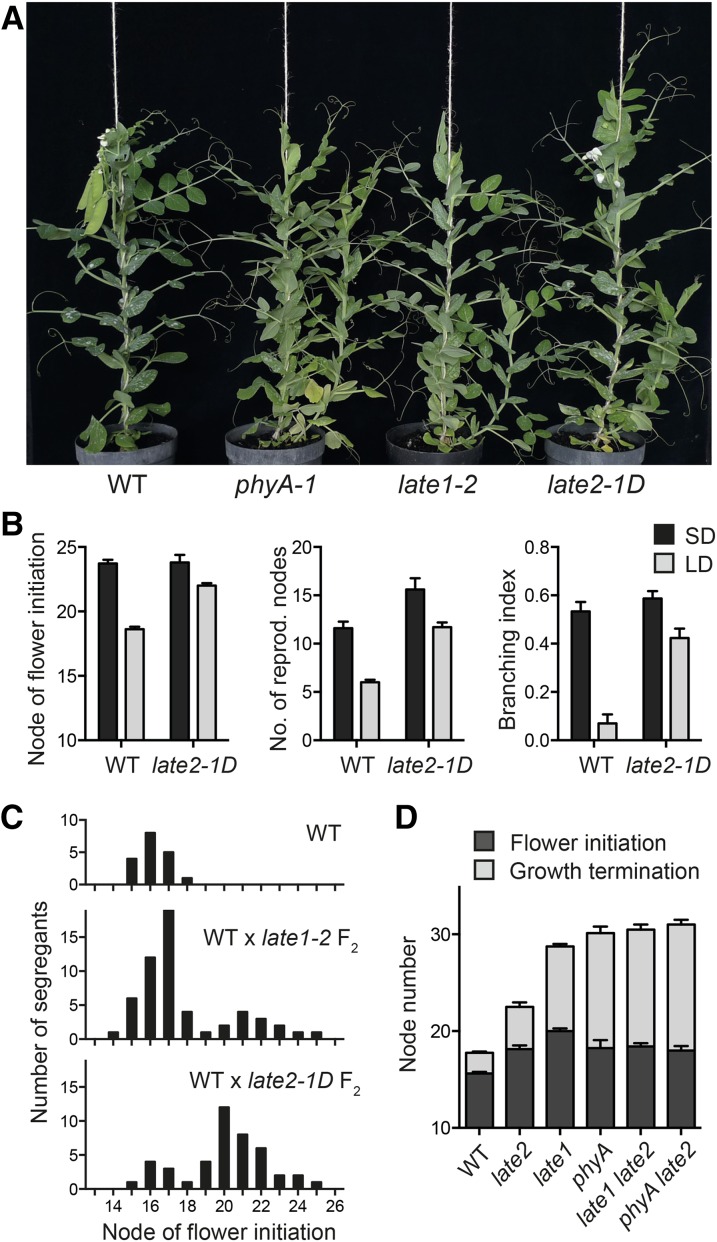
Screening of an EMS-mutagenized population of the dwarf pea line NGB5839 under 18-h LD conditions identified a line with several characteristics of a photoperiod response mutant, including delayed flowering, an extended flowering phase, and an increased tendency for basal branching (Hecht et al., 2007; Figures 1A and 1B). These phenotypes cosegregated and showed dominant inheritance in the F2 population of a cross to the NGB5839 progenitor (Figure 1C). Crosses to other late-flowering photoperiod response mutants phyA-1 and late1-2 returned wild-type progeny in the F2, indicating that the mutant defined a new locus, which we termed LATE2, with the single mutant allele designated late2-1D. A comparison of phenotypes under LD and SD conditions showed that, like the phyA-1 and late1-2 mutants, the late2-1D mutant flowered later than the wild type under LD conditions and showed a reduced response to photoperiod (Figure 1B). However, the late2-1D phenotype under LD was clearly weaker than that of late1 and phyA null mutants (Weller et al., 1997; Hecht et al., 2007; Figures 1A and 1D). To test the genetic relationship of LATE2 to LATE1 and PHYA, we generated double mutants for late2-1D with the null late1-2 and phyA-1 alleles (Weller et al., 2004; Hecht et al., 2007). Figure 1D shows that neither the late2 late1 nor the late2 phyA double mutants initiated flowering later or produced more reproductive nodes than the respective single mutants, consistent with all three mutations affecting the same pathway. The marginally earlier flowering in the late1 late2 double mutant relative to late1 mutant controls (P = 0.03) shown in Figure 1D was not consistently observed in segregating populations.
Figure 1.
The Dominant late2-1D Mutant Shows Reduced Responsiveness to Photoperiod.
(A) Comparison of late2-1D with photoperiod response mutants phyA-1 and late1-2 and their wild-type progenitor line NGB5839 (WT) grown under LD conditions.
(B) Effect of photoperiod on flowering and other traits in wild-type (NGB5839) and late2-1D plants. Values represent mean ± se for n = 6 to 8 replicates.
(C) Distribution of flowering time in F2 progeny of the cross NGB5839 (WT) × late2-1D. The distribution in an F2 progeny of the cross wild type × late1-2 is shown for comparison.
(D) Genetic interaction of late2-1D with phyA-1 and late1-2 in control of flower initiation and growth termination under LD conditions. Values represent mean ± se for n = 6 to 8 replicates.
The late2-1D Mutation Impairs Graft-Transmissible Promotion of Flowering and Disrupts Expression of FT Genes
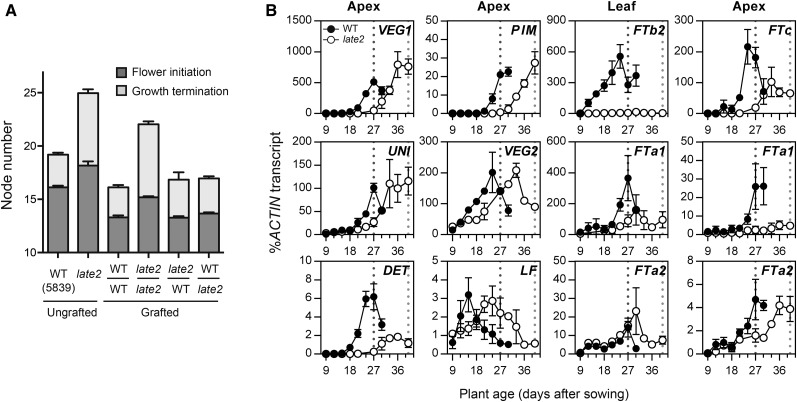
We previously showed that the late-flowering LD phenotypes of the photoperiod response mutants phyA and late1 can be rescued by grafting to wild-type stocks (Weller et al., 1997; Hecht et al., 2007). A similar grafting experiment was performed to examine whether the same might be true for late2-1D. Figure 2A shows that self-grafted late2 scions flowered approximately two nodes later than self-grafted wild-type scions and produced more than twice the number of reproductive nodes at maturity (P < 0.0001 in both cases). However, grafting late2 scions to wild-type stocks fully restored the node of flower initiation (13.28 ± 0.14 versus 13.31 ± 0.18 in wild-type self-grafts; P = 0.87) and significantly reduced the number of reproductive nodes (3.56 ± 0.20 versus 6.85 ± 0.27 in late2 self-grafts; P < 0.0001). These results show that like phyA and late1, the late2-1D mutation impairs the production of a graft-transmissible signal.
Figure 2.
The late2-1D Mutation Affects Graft-Transmissible Flowering Signals and Induction of Flowering Genes.
(A) Node of flower initiation and growth termination for graft combinations of wild-type NGB5839 (WT) and the late2-1D mutant, in comparison to intact and self-grafted controls. For each graft combination, the genotypes of scion (top) and stock (bottom) are shown, separated by a horizontal line. Six-day-old shoots (scions) were excised above the cotyledons and the epicotyl grafted onto the uppermost internode of 3-week-old stocks. Values represent mean ± se for n = 10 to 20 plants grown in LD.
(B) Gene expression in the wild type (NGB5839) and the late2-1D mutant during development under continuous light. Relative transcript levels were determined in dissected shoot apices or the second-uppermost fully expanded leaf. Mean values ± se are shown for n = 2 to 3 biological replicates, each consisting of pooled material from two plants. Developing floral buds were first visible in the wild type 27 d after sowing and in late2-1D 40 d after sowing (broken lines).
Previous work distinguished two different graft-transmissible signals in pea that are correlated to the expression of different FT genes (Hecht et al., 2011). The graft-transmissible promotion of flowering by LD is closely associated with the induction of the FTb2 gene, which does not occur in wild-type plants grown under SD or in late1 mutants (Hecht et al., 2011). We therefore examined whether late2-1D might also be defective in FTb2 regulation. Under constant light, the late2-1D mutant flowered four nodes later than the wild type (20.4 ± 0.24 nodes versus 16.2 ± 0.13), and Figure 2B shows this was reflected in a delay in induction of VEGETATIVE1 (VEG1), a developmental marker for legume inflorescence development (Berbel et al., 2012), which was first detectable above background at 18 d after sowing in the wild type and 27 d in late2-1D. Other flowering genes expressed at the shoot apex also showed a delay in induction or in the timing of peak expression levels in late2-1D, including APETALA1 homolog PROLIFERATING INFLORESCENCE MERISTEM (PIM), LEAFY ortholog UNIFOLIATA (UNI), FD homolog VEGETATIVE2(VEG2), and TERMINAL FLOWER1 homologs DETERMINATE (DET) and LATE FLOWERING (LF), which is generally consistent with the delay in flowering. The late2-1D mutant also completely failed to induce expression of FTb2 in leaves throughout the entire time course examined, even though flower initiation occurred within the time frame of the experiment, ∼40 d after sowing. Expression of other FT genes was also altered in late2-1D, but to a lesser extent, with weaker induction of FTa1 in leaves and delayed induction of both FTa1 and FTc in shoot apical tissue. Overall, this pattern of FT gene misregulation is very similar to that described for the late1 mutant (Hecht et al., 2011).
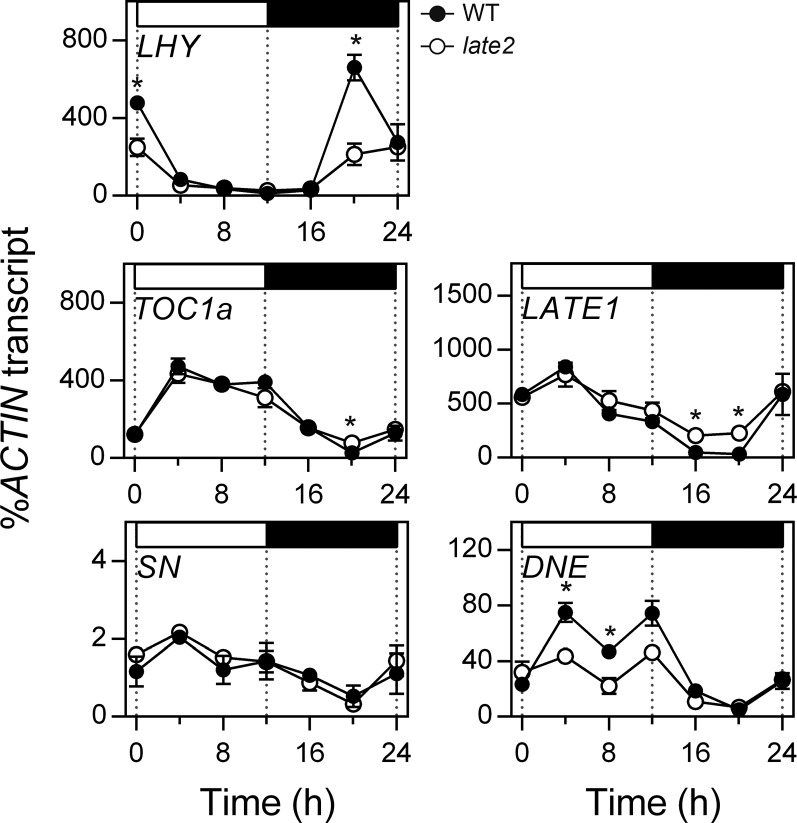
The late2-1D Mutant Does Not Affect Photomorphogenesis or the Rhythmic Expression of Circadian Clock Genes
As in Arabidopsis and other species, the photoperiod response in pea depends both on photoreceptor signaling and the circadian clock (Weller et al., 2004, 2012; Hecht et al., 2007; Liew et al., 2009, 2014), and we considered that the late2-1D phenotype could therefore result from a defect in either of these systems. In order to test whether the mutation influenced clock function, we examined the rhythmic expression of several central circadian clock genes under light/dark cycles. Figure 3 shows that under a 12-h-light/12-h-dark diurnal cycle, the expression rhythms of clock genes LATE ELONGATED HYPOCOTYL (LHY), TIMING OF CAB EXPRESSION 1a (TOC1a), and STERILE NODES (SN) were unaffected by the late2-1D mutation. The rhythmic phase of other clock-related genes LATE1 and DIE NEUTRALIS (DNE) was also unaffected by late2-1D, although there were significant differences in expression level of both genes at several points in the diurnal cycle, suggesting that LATE2 could potentially influence the expression of these genes in a manner unrelated to their rhythmic regulation. Similar patterns and effects of late2-1D were also seen under LD conditions (Supplemental Figure 1).
Figure 3.
The late2-1D Mutation Does Not Affect the Rhythmic Expression Patterns of Circadian Clock Genes under SD Cycles.
Transcript levels were determined in the uppermost fully expanded leaf of 3-week-old wild-type (NGB5839) and late2-1D mutant plants grown under a 12-h photoperiod at 20°C. Mean values ± se are shown for n = 2 to 6 biological replicates, each consisting of pooled material from two plants. Asterisks indicate significantly different values (P ≤ 0.05). Day and night periods are respectively indicated by white and black bars above the graph. Note that both genotypes carry the hr mutation (Weller et al., 2012).
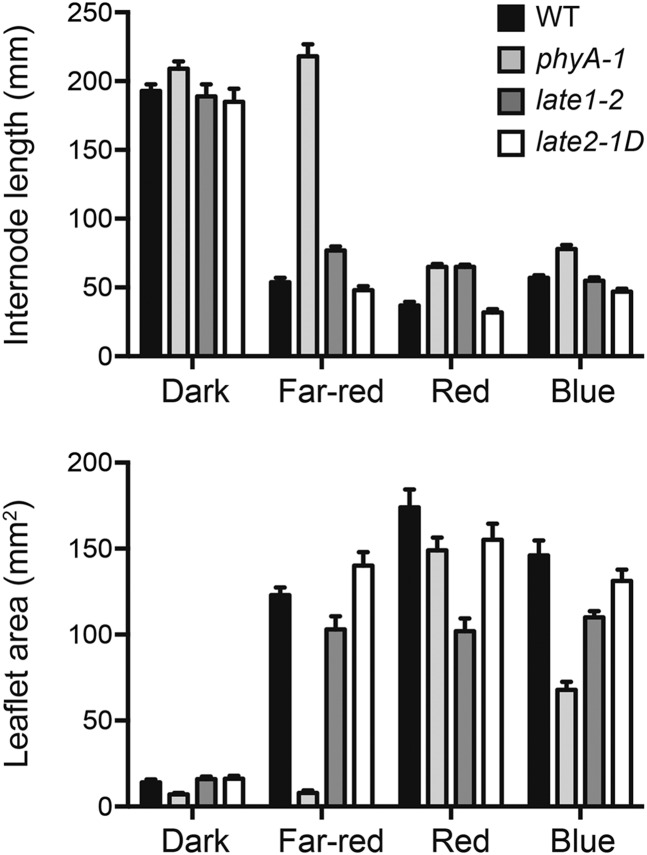
We also examined whether the late2-1D mutation affected seedling responses to light. Figure 4 shows that both phyA and late1 mutations disrupt normal photomorphogenesis, consistent with previous reports (Weller et al., 1997; Hecht et al., 2007). In contrast, late2-1D mutants did not differ from the wild type for either stem elongation or leaf expansion under monochromatic red, blue, or far-red light conditions (P > 0.05 for each comparison). This indicates that the late2-1D flowering phenotype is unlikely to be the result of a primary defect in light perception or signaling and suggests that LATE2 may participate specifically in the photoperiod response mechanism.
Figure 4.
The late2-1D Mutation Does Not Affect Photomorphogenesis.
Internode elongation (upper panel) and leaflet area (lower panel) of wild-type, phyA-1, late1-2, and late2-1D seedlings grown from sowing in darkness or under monochromatic red, blue, or far-red light (15 µmol m−2 s−1). Internode length was measured as the length between nodes 1 and 3, and leaflet area was estimated as the product of the length and width of the larger of the leaflets from leaf 3. Values represent mean ± se for n = 4 to 14 plants.
The late2-1D Mutant Carries a Substitution of a Highly Conserved C-Terminal Residue in a CYCLING DOF FACTOR Homolog
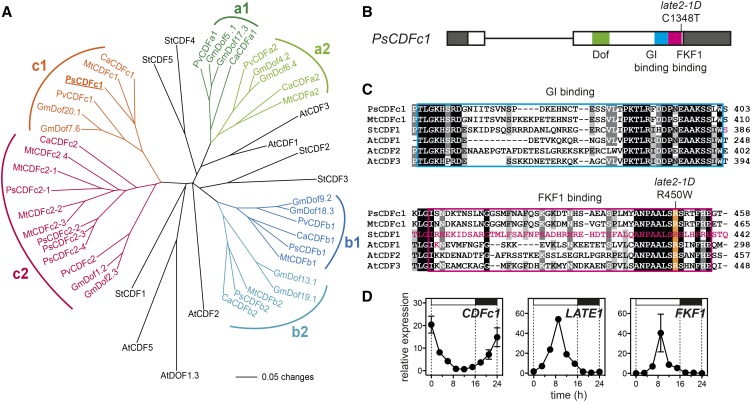
In order to narrow the range of potential candidate genes, we mapped LATE2 in the F2 progeny of a cross between late2-1D and cv Térèse (n = 219). Initial results placed LATE2 in linkage group VII, in a region syntenic with the middle of M. truncatula chromosome 4 (Supplemental Figure 2). Several genes potentially related to flowering were identified within this region, including homologs of the MADS domain gene SHORT VEGETATIVE PHASE (SVP), HUA2, and a CYCLING DOF FACTOR (CDF) gene. Fine-mapping of LATE2 in relation to these genes identified recombination with SVP and HUA2 but not with the CDF gene, indicating a distance of <0.3 cM between LATE2 and this CDF, and defining an interval comprising 226 annotated genes in the corresponding region of the M. truncatula genome. Apart from the CDF gene, no other genes with reported effects on flowering were present in this interval.
In Arabidopsis, the CDF genes comprise a small subgroup of DOF transcription factor genes distinguished by their rhythmic expression and ability to delay flowering when overexpressed (Imaizumi et al., 2005; Fornara et al., 2009). Phylogenetic analysis of legume DOF genes identified three distinct subclades, designated here as CDFa-CDFc, within the group II DOF clade (Yanagisawa, 2002). This clade includes Arabidopsis CDF1-CDF3 and CDF5, which share redundant roles and a similar expression pattern (Fornara et al., 2009), and potato CDF1-CDF5 (Figure 5A; Supplemental Figure 3). The CDF gene identified as a positional candidate for LATE2 in our mapping population corresponded to CDFc1 in this analysis. Other legume DOF proteins fall outside this core clade and are more closely related to Arabidopsis CDF4 and related proteins (Supplemental Figure 3). Overall, the phylogenetic relationships of the legume clades to specific CDF genes in Arabidopsis and potato are not well resolved, but the clades appear to predate legumes, as a more extensive analysis shows that they include genes from non-legume taxa (Supplemental Figure 3). A duplication is apparent within each of the legume CDFa, CDFb, and CDFc clades, such that the basic complement of genes appears to be six, as seen in chickpea (Cicer arietinum), or 12 in tetraploid soybean (Glycine max; Figure 5A). Other legume taxa may have suffered the loss of one or more genes, as bean (Phaseolus vulgaris) CDFb2, M. truncatula CDFa1, and pea CDFa1 and CDFa2 do not appear in any sequence databases. In addition, the CDFc2 gene has undergone multiple duplications in both pea and M. truncatula, with the multiple copies in M. truncatula arranged in tandem (Supplemental Table 1).
Figure 5.
The late2-1D Mutant Carries a Mutation in a CDF Homolog.
(A) Phylogram of the legume CDF family with reference to CDF genes from Arabidopsis and potato. Branches with bootstrap values <55% obtained from 1000 trees have been collapsed. The analysis is based on the sequence alignment shown in Supplemental File 1. Sequence details are given in Supplemental Table 1.
(B) Diagram of the pea CDFc1 gene showing the nature and location of the mutation in late2-1D in the FKF1 binding domain. Exons are shown as boxes, with coding sequence in white and untranslated regions in dark gray. Functional domains are indicated.
(C) Alignment of pea CDFc1 and other CDF homologs showing conserved residues in the GI binding (blue box) and FKF1 binding (pink box) domains. The arginine affected by the late2-1D mutation is highlighted in orange. The residues deleted in the naturally occurring potato StCDF1.2 and 1.3 variants, and altered in the Arabidopsis AtCDF1 K253A variant, all of which affect FKF1 binding (Imaizumi et al., 2005; Kloosterman et al., 2013), are indicated in pink. Shading indicates degree of conservation (black = 100%, dark gray = 80%, and light gray = 60%).
(D) Comparison of rhythmic expression patterns for CDFc1, LATE1, and PsFKF1 under LD conditions. Transcript levels were determined in the uppermost fully expanded leaf of 3-week-old wild-type (NGB5839) plants grown under a 16-h photoperiod at 20°C. Mean values ± se are shown for n = 2 biological replicates, each consisting of pooled material from two plants. Day and night periods are indicated by white and black bars, respectively, above the graph.
At, Arabidopsis thaliana; Ca, Cicer arietinum; Gm, Glycine max; Mt, Medicago truncatula; Ps, Pisum sativum; Pv, Phaseolus vulgaris; St, Solanum tuberosum.
Sequencing of CDFc1 cDNA in the wild type and late2 revealed a C1348T transition resulting in a substitution of arginine by tryptophan at residue 450 (Figure 5B). As expected, a marker for this mutation also cosegregated perfectly with the late2-1D phenotype (Supplemental Figure 2). The affected amino acid is located within the FKF1 binding domain defined in Arabidopsis and potato (Imaizumi et al., 2005; Kloosterman et al., 2013) and is one of several highly conserved basic residues within this C-terminal region suggested to potentially influence the CDF-FKF1 interaction (Figure 5C), strongly implicating this mutation as the likely cause of the late2-1D phenotype.
In Arabidopsis, the three genes most closely related to PsCDFc1 (AtCDF1-3) all show rhythmic expression patterns, with a morning peak under LD (Imaizumi et al., 2005; Fornara et al., 2009), and we found this also to be true for PsCDFc1 (Figure 5D). This expression pattern contrasted with those of LATE1 and FKF1, which both showed peaks in the middle of the day (Figure 5D). We also consulted the pea gene expression atlas (Alves-Carvalho et al., 2015) for information about tissue specificity of PsCDFc1 expression and found it to be expressed most strongly in leaf tissue, similar to AtCDF1 (Imaizumi et al., 2005). In addition, among the six other CDF genes shown in Figure 5A, only two (PsCDFb1 and PsCDFc2-1) showed significant expression in the gene atlas. These genes also showed a similar pattern of expression to PsCDFc1 but were expressed at levels ∼4-fold and 12-fold lower than PsCDFc1, respectively.
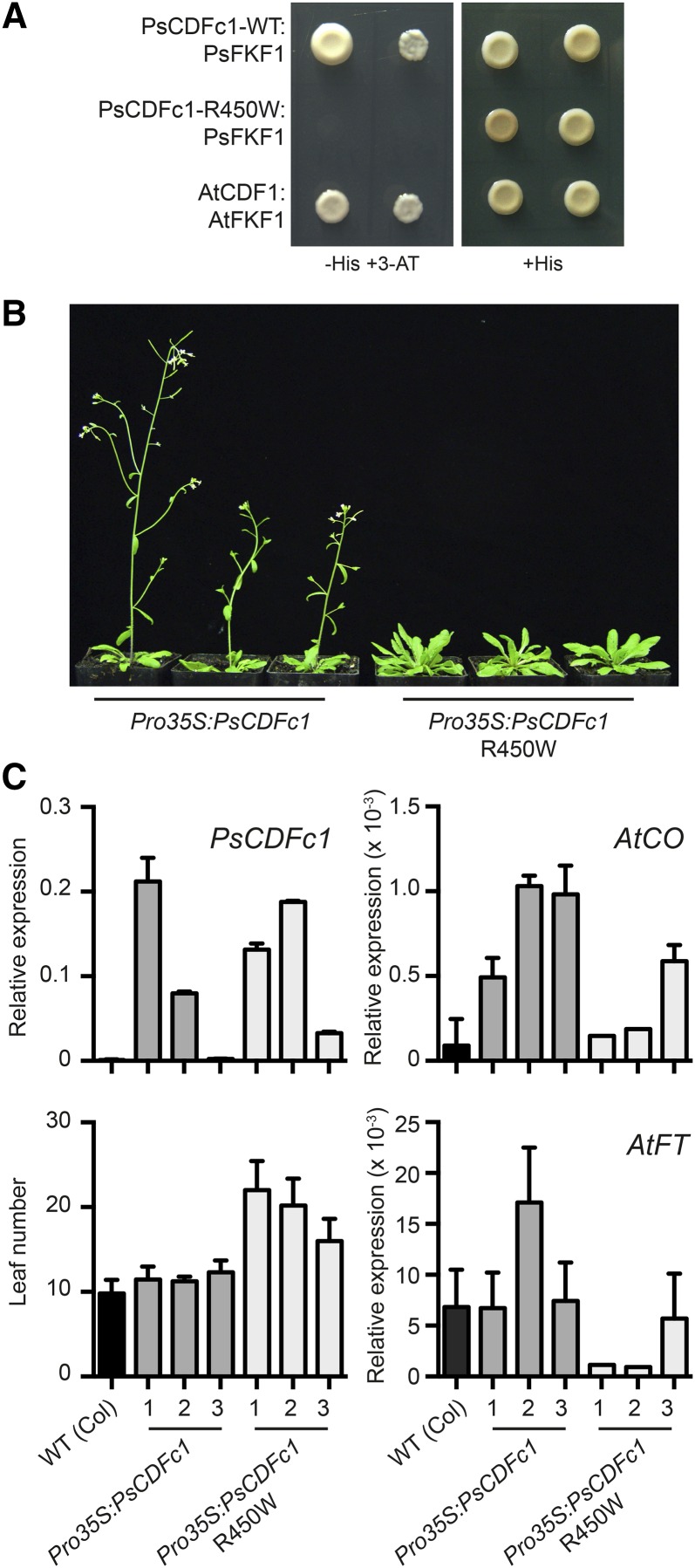
The Function of the CDFc1 Protein Is Altered by the R450W Mutation
The nature of the R450W mutation suggested that the late2-1D flowering phenotype might be due to an inability of PsCDFc1 to bind to PsFKF1, resulting in impaired light-dependent degradation of PsCDFc1 and persistent transcriptional repression of FTb2. To test this idea, we used a yeast two-hybrid assay system to examine the ability of both wild-type and R450W mutant forms of PsCDFc1 to interact with the C-terminal half of PsFKF1 (Supplemental Figure 4; Alves-Carvalho et al., 2015), which contains several KELCH repeats involved in protein-protein interactions (Nelson et al., 2000; Imaizumi et al., 2005). We first confirmed that none of the bait or prey constructs showed autoactivation (Supplemental Figure 5A). Yeast two-hybrid assay results in Figure 6A show that the wild-type form of CDFc1 interacts strongly with FKF1, whereas the late2-1D R450W variant does not. This provides a clear demonstration that the regulation of CDFc1 is likely to be impaired by the R450W mutation and supports the idea that this mutation is the cause of the late flowering phenotype in the late2-1D mutant.
Figure 6.
The R450W Mutation in late2-1D Affects the Function of the CDFc1 Protein.
(A) Yeast two-hybrid analysis of the interaction between pea FKF1 and the wild-type and R450W mutant form of CDFc1. The previously characterized interaction between Arabidopsis FKF1 and CDF1 proteins (Imaizumi et al., 2005) was included as a positive control. Each panel shows bait/prey interactions in both orientations, either FKF1/CDF (left) or CDF/FKF1 (right). Each clone is shown after 4 d of growth at 30°C on selective medium (with 3-amino-1,2,4-triazole added; -His+3-AT) and nonselective medium (+His). Negative controls are shown in Supplemental Figure 5.
(B) Representative plants from three independent Arabidopsis lines (Columbia accession) transformed either with wild-type or R450W mutant forms of pea CDFc1, grown under LD conditions.
(C) Relative expression levels at ZT 15.45 of PsCDFc1, AtCO, and AtFT and leaf number at flowering in transgenic Arabidopsis lines. Labels 1 to 3 denote independent transformants. Values represent mean ± se for n = 2 biological replicates (expression data) and n = 6 to 10 plants (leaf number).
To further test this idea, we confirmed that the R450W variant also impaired the ability of PsCDFc1 to interact physically with Arabidopsis FKF1 (Supplemental Figure 5B), and we next compared the ability of wild-type and mutant forms of PsCDFc1 to influence flowering when expressed in Arabidopsis. Figures 6B and 6C show that expression of PsCDFc1 from the 35S promoter had no effect on the flowering time of wild-type Arabidopsis plants grown under LD, whereas expression of the late2-1D R450W variant significantly delayed flowering in multiple independent transformed lines. This delayed flowering was also reflected in reduced levels of endogenous CO and FT transcript (Figure 6C), showing that the mutant form of PsCDFc1 is more effective than the wild-type form in repressing flowering through the CO/FT pathway. Together, these results strengthen the evidence that the R450W mutation is indeed the cause of the late2-1D late-flowering phenotype and show that when expressed in transgenic Arabidopsis, the mutant PsCDFc1 is capable of repressing flowering via CO in a manner similar to the endogenous Arabidopsis CDF1. However, the lack of effect of Pro35S:PsCDFc1 was in contrast to previous reports that overexpression of AtCDF1 from the 35S promoter causes late flowering (Imaizumi et al., 2005). This could reflect differences in the relative expression levels or location of the two proteins, or in the efficiency with which they are processed by the Arabidopsis FKF1/GI degradation mechanism. Consistent with this interpretation, we found that when PsCDFc1 was instead expressed from the Arabidopsis SUC2 promoter, which drives expression specifically in phloem companion cells, a small delay in flowering was observed (Supplemental Figure 6).
LATE2 Does Not Affect the Expression of COL Genes
In Arabidopsis, overexpression of CDF1 results in strong repression of CO expression in the afternoon and during the night under LD conditions (Imaizumi et al., 2005). A similar effect was seen in lines expressing a mutant form of the CDF1 protein that can no longer bind to FKF1 (Imaizumi et al., 2005). This regulatory relationship is conserved in potato, where lines carrying a truncated, degradation-resistant form of the potato CDF1 protein show increased repression of CO expression in comparison to the wild type (Kloosterman et al., 2013). Previously, we described 11 members of the CO-like gene family in legumes and showed that the temperate LD legume M. truncatula has only a single CO co-ortholog (COLa), which does not affect flowering (Wong et al., 2014). In addition, both M. truncatula COLa and its pea ortholog exhibit a morning phased diurnal expression rhythm that lacks the afternoon peak characteristic of CO and is more similar to that of Arabidopsis COL1 and COL2 (Hecht et al., 2007; Wong et al., 2014).
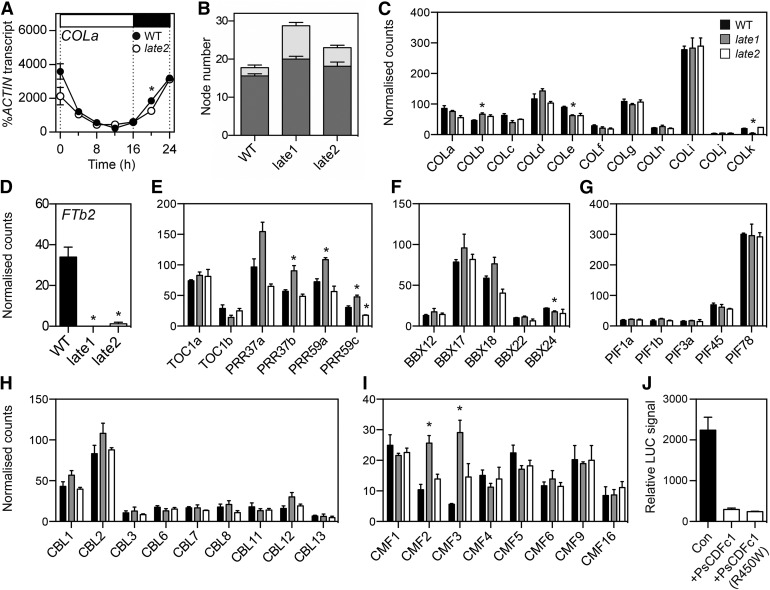
In view of the apparent conservation in CDF function between Arabidopsis and potato, we first examined whether the late2-1D mutation might also affect expression of the pea COLa ortholog. Figure 7A shows that the diurnal rhythm of COLa expression is very similar in the wild type and late2-1D and thus provides no evidence that late2-1D may affect flowering through regulation of COLa transcript level. This is consistent with the previous finding that a null late1 mutation also does not appear to influence the COLa expression rhythm (Hecht et al., 2007).
Figure 7.
Effects of LATE2 on FTb2 Expression Are Not Mediated by Expression Changes in Key Transcription Factor Families and May Result from Direct Transcriptional Repression.
(A) Expression of COLa in wild-type NGB5839 (WT) and late2-1D mutant plants under a 16-h LD photoperiod. Transcript levels were determined in the second uppermost fully expanded leaf of 3-week-old plants grown at 20°C. Mean values ± se are shown for n = 2 biological replicates, each consisting of pooled material from two plants. Day and night periods are indicated by white and black bars, respectively, above the graph.
(B) Flowering phenotypes of late1-2 and late2-1D mutants grown from sowing under continuous white light. Values represent mean ± se for n = 8 replicates. Dark gray, node of flower initiation; light gray, node of growth termination.
(C) to (I) Transcript levels of FTb2 (D) and genes in the COL (C), PRR (E), BBX (F), PIF (G), CBL (H), and CMF (I) families in expanded leaf tissue from wild-type, late1-2, and late2-1D mutants grown from sowing under continuous white light. Values represent mean ± se for n = 3 biological replicates, each consisting of pooled material from two plants. Asterisks indicate values significantly different from the wild type: P ≤ 0.05 for (A) and P ≤ 0.025 for (C) to (I).
(J) Luciferase activity driven from the M. truncatula FTb1 promoter:LUC construct coinfiltrated with Pro35S:GUS (control, denoted as “Con”), Pro35S:PsCDFc1 (wild type), or Pro 35S:PsCDFc1 (mutant R450W) and transiently expressed in N. benthamiana leaves. Data represents mean ± se for n = 8, 3, and 10 independently infiltrated plants, for control, PsCDFc1 wild-type, and PsCDFc1 mutant respectively.
Using a combination of database searches and PCR-based approaches, we isolated 10 further pea COL genes orthologous to the previously described M. truncatula COLb-k genes (Wong et al., 2014; Supplemental Figures 7 and 8). We then examined whether the expression of any of these genes was altered in the late2-1D mutant by performing RNA-sequencing with RNA from leaf tissue of 3-week-old seedlings. We also included the late1 mutant in this experiment, with the reasoning that as the Arabidopsis ortholog GI regulates CO through an effect on CDF proteins, any gene necessary for LATE2 regulation of FTb2 should show similar misregulation in both late1-2 and late2-1D mutants in pea. In an attempt to minimize any light-driven rhythmic component to the differential regulation, plants were grown under conditions of continuous light from sowing. Under these conditions, both late1-2 and late2-1D mutants showed a characteristic late-flowering phenotype similar to that seen under a standard 16-h photoperiod (Figure 7B). In order to verify the absence of any expression rhythms under these continuous light conditions, we used RT-qPCR to examine expression of clock and COL genes in leaf tissue of 3-week-old wild-type plants over a 24-h time course. Supplemental Figure 9 confirms that rhythmic regulation was effectively absent for key clock genes and the 11 members of the pea COL family. RNA-seq analysis of a single time point showed that, as expected, the late2-1D mutant had a negligible level of FTb2 transcript, similar to late1-2, and much lower than the wild type (Figure 7D). Figure 7C confirmed that COLa transcript levels did not differ significantly between wild-type and late2-1D mutants. In the late1-2 mutant, COLb showed a small but statistically significant increase in expression relative to the wild type, while COLe and COLk transcript levels were slightly reduced; however, there was no corresponding misregulation of any of the COL genes in late2-1D mutants (Figure 7C).
LATE2 Does Not Affect the Expression of Other Genes Implicated in the Activation of FT in Arabidopsis
While CO homologs have been implicated in photoperiod-specific FT expression across a range of diverse species (Kojima et al., 2002; Navarro et al., 2011; Yang et al., 2014), recent evidence from species within the Brassicaceae suggests this may reflect convergent evolution (Simon et al., 2015) and raises the possibility of CO-independent mechanisms for photoperiod regulation of flowering in other species. One possible mechanism is suggested by recent results from sugar beet, where the CCT-domain pseudo-response regulator gene BTC1 and the B-box (BBX) gene B2 have similar roles in the regulation of FT genes and are proposed to act together to confer a CO-like activity (Pin et al., 2012; Dally et al., 2014). In addition, in Arabidopsis, a number of other transcription factors have been shown to directly activate FT, including members of the CIB (CRYPTOCHROME-INTERACTING BASIC-helix-loop-helix) and PIF (PHYTOCHROME INTERACTING FACTOR) families of bHLH (basic helix-loop-helix) proteins (Liu et al., 2008; Kumar et al., 2012; Liu et al., 2013). Members of the AP2 family of transcription factors have also been implicated as regulators of photoperiodic flowering and FT expression (Jung et al., 2007).
To look for evidence that LATE2 might act through one or more similar genes in pea, we next annotated all CCT, BBX, CIB/BEE-like (CBL), PIF, and AP2 family genes in the recently released pea transcriptome and other publicly available pea transcript resources (Franssen et al., 2011; Kaur et al., 2012; Alves-Carvalho et al., 2015). We identified a total of 7 PRR genes, 14 CBL genes, 12 additional BBX genes, 9 PIF genes, 5 AP2 family genes, and 16 additional CCT MOTIF FAMILY (CMF) genes in pea (Supplemental Figures 7, 8, and 10 to 12). Transcript levels for expressed genes in these families did not differ significantly between the wild type and late2-1D, with the exception of PRR59c, which showed a small reduction (Figures 7E to 7I; Supplemental Figure 13). In the late1 mutant, expression of PRR59c was significantly elevated relative to the wild type, along with PRR37b, PRR59a, CMF2, and CMF3, while BBX24 showed a small but significant reduction in expression. However, overall, none of these genes in any of the families examined showed comparable expression changes in both late2-1D and late1-2, suggesting that the absence of FTb2 induction in these mutants is unlikely to result from misregulation of these genes at the transcriptional level. In addition, consistent with the data in Supplemental Figure 1, we found no significant misregulation of LATE1/GI, FKF1, or other clock-related genes in the late2-1D mutant (Supplemental Figure 13), with the possible exception of LHY, which showed a reduction in expression at the margin of significance (P = 0.025).
In the absence of any candidate transcription factors showing clear coregulation with FTb2, we considered the possibility that CDFc1 might be able to repress the transcription of FTb2 through direct interaction with its promoter. In addition to transcriptional repression of CO, Arabidopsis CDF1 has been reported to associate with the FT promoter (Sawa and Kay, 2011; Song et al., 2012), and it has been suggested that FKF1-dependent removal of CDF1 may contribute to FT regulation in a CO-independent manner (Song et al., 2012). As the available pea genome sequence is incomplete in the region of PsFTb2, we were unable to isolate the PsFTb2 promoter, and as an alternative we used the promoter of the M. truncatula FTb1 gene, which is known to show qualitative induction under LD, similar to PsFTb2 (Laurie et al., 2011). Figure 7J shows that in the Nicotiana benthamiana transient assay system, constructs expressing either the wild-type or the late2-1D variant of PsCDFc1 from the 35S promoter were able to significantly repress expression of the LUCIFERASE reporter driven from the MtFTb1 promoter, relative to control. This result provides evidence that the PsCDFc1 protein is capable of directly repressing the expression of MtFTb1 and suggests that the same is likely to be true for PsFTb2.
DISCUSSION
In comparison to Arabidopsis and cereal systems, our understanding of the genetic mechanisms conferring photoperiod responsiveness in legumes is less advanced. Although several previous studies in pea and soybean have characterized components of the photoperiod response pathway and confirmed the conserved importance of light signaling and the circadian clock (Weller et al., 2001; Hecht et al., 2007; Liew et al., 2009; Xia et al. 2012; Watanabe et al., 2012), the mechanism by which light and clock signals are integrated to confer photoperiod-specific induction of FT genes is still not clear. In this study, we characterized a component of the pea photoperiod response pathway, LATE2, and identified it as a CDF gene that appears to act downstream of light signaling and the circadian clock to repress the expression of FTb2, the main photoperiod-regulated FT gene in pea.
A Conserved Role for the LATE2/CDFc1 Gene in the Pea Photoperiod Response
In the Arabidopsis photoperiod response pathway, CDF proteins appear to have a primary role as indirect repressors of FT expression, acting via transcriptional repression of the direct FT activator, CO. Light-dependent binding of the FKF1-GI complex to a region near the C terminus of CDF proteins targets them for degradation in the afternoon, which releases CO from repression and allows the induction of FT and flowering (Imaizumi et al., 2005; Sawa et al., 2007; Fornara et al., 2009). A similar interaction is seen between potato homologs of these proteins (Kloosterman et al., 2013). In both species, CDF variants with disrupted FKF1 binding domains are no longer subject to posttranslational regulation by the FKF1-GI complex, and this leads to accumulation of CDF protein and persistent repression of CO and FT transcription (Imaizumi et al., 2005; Kloosterman et al., 2013). The late2-1D mutant is a late flowering, dominantly inherited photoperiod response mutant that carries a mutation in a CDF gene, CDFc1, which is tightly linked to the LATE2 locus (Figures 1 and 5; Supplemental Figure 2). The mutation directs the substitution of a highly conserved residue (R450W) within the FKF1 binding domain of the CDFc1 protein and impairs its physical interaction with PsFKF1 (Figures 5 and 6), suggesting that LD-specific CDFc1 protein degradation may also be impaired. Consistent with this interpretation, overexpression of this mutant form in transgenic Arabidopsis delays flowering (Figure 6). Together, these findings support the R450W substitution as the cause of the late2-1D late-flowering phenotype and indicate an endogenous role for CDFc1 in repression of flowering under noninductive conditions.
Like LATE2, the pea GI ortholog LATE1 also participates in photoperiod-specific induction of FTb2 (Hecht et al., 2007, 2011), and the similarity of late-flowering phenotypes in late2-1D, late1, and the double mutant is consistent with these two genes acting in the same pathway (Figure 1). In addition, the patterns of rhythmic expression for all three genes (LATE2, FKF1, and LATE1) are similar to their Arabidopsis equivalents (Figure 5D; Supplemental Figure 1; Imaizumi et al., 2005; Fornara et al., 2009), suggesting a similar temporal pattern of interaction within the daily cycle. These data are consistent with the conservation in pea of a mechanism in which orthologs of GI and FKF1 interact to regulate CDF protein stability. The conservation of this mechanism within legumes is also supported by a study showing interaction of FKF1, GI, and CDF homologs in soybean (Li et al., 2013). In the future, it will be of interest to test other facets of this mechanism directly, including FKF1 function and regulation of CDF protein abundance.
Other aspects of the late2-1D phenotype raise questions about whether LATE2 might influence flowering through other mechanisms. We also observed small effects of the late2-1D mutation on the expression levels of several genes related to circadian clock function. These differences seem unlikely to represent a significant change to clock function, as they occur at different points in the daily cycle and do not appear to constitute a change in rhythmic phase, nor are they reflected in changes to expression levels or rhythmic phases of other clock-related or clock-regulated genes such as SN/LUX or TOC1a. In this respect, the effects of LATE2 are different from those previously reported for LATE1 (Hecht et al., 2007; Liew et al., 2009). In addition, the observed differences are difficult to reconcile with the known functions of these genes in flowering time control (as, for example, DNE is known to inhibit flowering in SD, but its expression is lower in the late-flowering late2-1D mutant; Figure 3; Supplemental Figure 1). We conclude that these differences are unlikely to be important for the observed effects of LATE2 on flowering, but further work may be needed to establish this definitively.
The Regulatory Relationship between CDF and CO Homologs Is Not Conserved in Pea
In contrast to the apparent conservation of the FKF1/GI/CDF mechanism in pea and its role in the regulation of photoperiod-specific FT expression, there is no evidence for a conserved role of CO-like genes as flowering time regulators. Diurnal expression dynamics of the pea CO co-ortholog COLa do not conform to those of Arabidopsis CO and are not altered in photoperiod response mutants such as late1 and dne, which influence the circadian clock (Hecht et al., 2007; Liew et al., 2009). The observation that expression of COLa under LD conditions was also unaffected by the late2-1D mutation (Figure 7A) further indicates that the regulatory relationship between CDF and CO seen in Arabidopsis and potato is not present in pea and that COLa is unlikely to mediate the effects of LATE2 on FTb2. Further support for this conclusion is provided by recent observations that a null mutant for M. truncatula COLa does not have a flowering phenotype under LD and that MtCOLa does not promote flowering when overexpressed in Arabidopsis (Wong et al., 2014). Interestingly, recent evidence suggests that Group Ia COL genes in soybean may contribute to delay of flowering under noninductive conditions (Cao et al., 2015), although the significance of this for the overall response to photoperiod has yet to be explored in detail.
Demonstration that a close homolog of CO contributes to flowering time control and the photoperiod response in rice (Hayama et al., 2003) has generated speculation that a “CO/FT” module could be widely conserved as a central feature of the photoperiod response mechanism in angiosperms (Hayama and Coupland, 2004; Izawa, 2007; Valverde, 2011). However, this assumption has increasingly been challenged by observations inconsistent with such a model (Ballerini and Kramer, 2011). The recent study of Simon et al. (2015) is particularly definitive, showing that the characteristic regulation and function of Arabidopsis CO arose from a tandem duplication within the Brassicaceae, which placed the CO promoter adjacent to multiple DOF binding sites and thereby under the control of CDF proteins. This implies that CO-like genes are unlikely to be important for flowering time control in all species, but that where they are, this role has evolved independently from the one well known in Arabidopsis and may involve a different regulatory mechanism or represent convergent evolution. Consistent with these conclusions, COLa genes in both pea and M. truncatula show a morning-phased expression pattern similar to that of the Arabidopsis COL1 and COL2 genes (Figure 7; Ledger et al., 2001; Hecht et al., 2007), which is likely to represent an ancestral, CDF-independent pattern of regulation for this class of COL genes (Simon et al., 2015).
LATE2 Does Not Regulate Genes Homologous to Direct Transcriptional Activators of Arabidopsis FT
If the pea CO ortholog does not mediate the effect of LATE2 on FTb2 expression and flowering, then how else might this be achieved? One possibility is that CO function could be assumed by another protein in the wider COL family, or by association of two different proteins, each containing one of the two domains (BBX and CCT) characteristic of COL proteins, as has recently been proposed for FT-dependent bolting control in sugar beet (Pin et al., 2012; Dally et al., 2014). Another possibility is that this role may be performed by an unrelated protein that acts in a manner analogous to CO: as an activator of FTb2 transcription that is itself transcriptionally repressed by LATE2 under SD. In Arabidopsis, several proteins other than CO have been demonstrated to directly bind to the FT promoter and activate transcription, including members of the PIF and CIB families of bHLH transcription factors (Liu et al., 2008, 2013; Kumar et al., 2012). However, in a comprehensive analysis of genes in the COL, CCT, BBX, PIF, and CIB families, we did not identify any with expression that was dependent on LATE1 and LATE2 and correlated with expression of FTb2 or flowering time (Figure 7). This implies that none of these genes serves as an intermediate in the regulation of FTb2 expression by LATE2.
Other Possible Mechanisms for the CO-Independent Response to Photoperiod in Pea
Another explanation for our findings is that a function analogous to that of CO may not exist in pea, and the mechanism for regulation of FTb2 by FKF1/GI/CDF may be more direct or occur via alternative mechanism(s). Evidence in Arabidopsis suggests that CDF1 protein, in addition to binding to the CO promoter (Imaizumi et al., 2005), can also bind directly to the FT promoter to repress transcription under noninductive conditions (Song et al., 2012), and the FKF1/GI complex might therefore relieve this direct repression by promoting CDF degradation. Other observations in Arabidopsis indicate that GI can induce FT expression independently of CO, and this is suggested to operate through interaction with direct repressors of FT including SVP and TEM (Sawa and Kay, 2011) and regulation of miR172 abundance (Jung et al., 2007). Whether or not CDF proteins participate in either of these mechanisms has not been thoroughly examined, but one recent study reports a regulatory link between AtCDF2 and miR172 levels (Sun et al., 2015). However, as none of the AP2 family of miRNA172 target genes showed differential expression in our RNA-seq experiment (Supplemental Figure 13), it seems the latter mechanism is unlikely to operate in pea. In Arabidopsis, both FKF1 and GI have also been reported to associate with the FT promoter (Imaizumi et al., 2005; Sawa and Kay, 2011), suggesting that they could have a more direct role in activation of FT and that interaction between GI, FKF1, and CDF proteins at the FT promoter may be significant. The ability of PsCDFc1/LATE2 to repress transcription from the photoperiod-regulated MtFTb1 promoter (Figure 7J) provides an indication that direct regulation of FT genes may indeed be an important mechanism through which CDF proteins regulate flowering in temperate LD legumes and might even form a central feature of the photoperiod response mechanism in this group. Whether PsGI/LATE1 and PsFKF1 are able to activate target FT genes directly and how levels of LATE1, LATE2, and FKF1 proteins may be regulated by photoperiod are intriguing questions for future research.
CO-independent mechanisms for FT upregulation have been reported in several other species. In rice, the main pathway for photoperiod-specific flowering acts in parallel to Hd1/CO and involves the action of EARLY HEADING DATE1, which encodes a B-type response regulator, with no clear ortholog in Arabidopsis. This gene is in turn suppressed by Ghd7, a CCT domain protein with similarity to wheat VERNALIZATION2 (Trevaskis et al., 2006; Xue et al., 2008). There is also evidence in rice that the Dof transcription factor OsDOF12 can regulate the expression of the FT homolog Hd3a without effect on Hd1/CO expression (Li et al., 2009). In soybean, the B3 transcription factor-like gene E1 is rhythmically regulated and acts downstream of light signaling to repress expression of the FT genes FT2a and FT5a under noninductive LD (Xia et al., 2012). Interestingly, E1 also acts by promoting expression of the FTb-class gene FT4, an inhibitor of flowering (Zhai et al., 2014), suggesting that it may have the capacity to act as a transcriptional activator. E1-like genes exist in temperate legumes, including pea, but a transcript from the E1 ortholog was not detected in our RNA-seq data set. Whether these genes may participate in the long-day photoperiod response mechanism is not yet known.
Any explanation for the CO-independent integration of light and clock signals for photoperiod responsiveness in pea must also take into account the role of PHYA, another important component, which has a similar mutant phenotype to those of late1 and late2-1D (Figure 1). In Arabidopsis, PHYA promotes FT expression by regulating CO activity, opposing the COP1-dependent degradation of CO protein (Yanovsky et al., 2001; Valverde et al., 2004; Laubinger et al., 2006; Jang et al., 2008). However, it also mediates light input to the circadian clock (Somers et al., 1998), suggesting the potential to influence the rhythmic expression of CO transcript. In addition to its effects on CO, COP1 also acts together with ELF3 to promote degradation of GI (Yu et al., 2008), representing a third potential mechanism through which light could affect FT expression. Pea orthologs of all of these genes have been functionally characterized, and it will be interesting in the future to explore the effects of PHYA, the COP1 ortholog LIP1 and the ELF3 ortholog HR on the expression and/or protein stability of LATE1 and LATE2.
Increasing evidence that the role of CO in photoperiodic regulation of flowering time is not widely conserved raises fundamental questions about the evolution of photoperiod sensing mechanisms in plants. At present, our understanding of these mechanisms in species where there is no clear role of CO-like genes is in its infancy. In this study, we established an important role for a CDF gene in a major crop group where there is mounting evidence against a conserved regulatory role for CO-like genes in flowering time. Our results provide evidence that a FKF1/GI/CDF module in pea controls FT expression and contributes to photoperiod measurement. However, they also suggest that CO homologs are not targets of this module and point to the existence of an alternative mechanism, possibly one in which it is more directly involved in FT regulation. In the future, clarification of the precise means by which LATE2 regulates flowering time will be a key step in understanding photoperiod regulation in legumes and the basis for diversification of photoperiod response mechanisms. One plausible hypothesis is that direct regulation of FT genes by a GI/FKF1/CDF module could be an ancestral mechanism for flowering time control that has been reinforced in various taxa by other, diverse mechanisms. It will thus be of interest to determine whether this is a widespread phenomenon.
METHODS
Plant Material, Growth Conditions, and Grafting
As with the previously described phyA-1 and late1-2 mutants (Weller et al., 1997; Hecht et al., 2007), the late2-1D mutant was derived from EMS mutagenesis of pea (Pisum sativum) line NGB5839, a dwarf derivative of ‘Torsdag’ carrying the le-3 and hr mutations (Lester et al., 1999; Weller et al., 2012) using the mutagenesis protocol described by Weller et al. (1997). Plants used for both the photomorphogenesis experiment (Figure 4) and expression experiments (Figures 2B, 3, and 7; Supplemental Figures 1, 9, and 13) were conducted in growth cabinets at 20°C. All other experiments were conducted in the Hobart phytotron using previously described growth media, light sources, phytotron conditions, and grafting protocols (Hecht et al., 2007). Standard phytotron SD conditions consisted of an 8-h photoperiod of natural light, which was extended with white light from fluorescent tubes and incandescent globes at an irradiance of 10 µmol m−2 s−1 to provide a LD photoperiod. Branching index (Figure 1B) represents the ratio of the total length of lateral branches to the total height of the main shoot axis.
Mapping
The late2-1D × Térèse F2 population consisted of 219 plants. F3 seed from plants displaying the mutant phenotype were screened for segregation of the late2-1D phenotype in order to determine the genotype of F2 plants. Markers used in the linkage analysis were designed to target the introns of genes identified in the relevant interval of the Medicago truncatula genome (v4.0; www.jcvi.org/medicago) that were also present in pea sequence databases in GenBank (www.ncbi.nlm.nih.gov). Details of these markers and their method of detection are provided in Supplemental Table 2.
Phylogenetic Analysis
For phylogenetic analyses, genes were identified by performing BLAST searches at NCBI (www.ncbi.nlm.nih.gov), Phytozome (www.phytozome.jgi.doe.gov), and the pea RNA-seq gene atlas (bios.dijon.inra.fr/FATAL/cgi/pscam.cgi), with reciprocal BLAST searches performed against Arabidopsis at TAIR (www.arabidopsis.org) to confirm gene identity. For each alignment (Supplemental Files 1 to 8), full-length amino acid sequences were aligned using ClustalX (Thompson et al., 1997) or the MAFFT algorithm in Geneious (v8.1.7; Biomatters) and adjusted manually, where necessary, using GeneDoc (version 2.7.000; Nicholas and Nicholas, 1997; http://www.psc.edu/biomed/genedoc). Using these alignments, distance-based methods were used for phylogenetic analyses in PAUP* 4.0b10 (http://paup.csit.fsu.edu/), using either 1000 or 10,000 bootstrap replications, as indicated in the figure legends.
Gene Expression Studies
Harvested tissue used in RT-qPCR experiments (Figures 2B, 3, and 7A) consisted of both leaflets from a fully expanded leaf or dissected apical buds (∼2 mm) from two plants. Samples were frozen in liquid nitrogen and total RNA extracted using the SV Total RNA Isolation System (Promega). RNA concentrations were determined using a NanoDrop 8000 (Thermo Scientific). Reverse transcription was performed in 20 μL with 1 µg of total RNA using the Tetro cDNA synthesis kit (Bioline). RT-negative (no enzyme) controls were included to monitor for genomic DNA contamination. First-strand cDNA was diluted five times and 2 μL was used in each RT-qPCR. RT-qPCR reactions using SYBR green chemistry (SensiFast; Bioline) were set up with a CAS-1200N robotic liquid handling system (Corbett Research) and run for 50 cycles in a Rotor-Gene Q (Qiagen). Two technical replicates and at least two biological replicates were performed for each sample. Primer details are included in Supplemental Table 3.
Yeast Two-Hybrid Assay
For the yeast two-hybrid assay (Figure 6A), full-length coding sequences of PsCDFc1 and AtCDF1 and the KELCH repeats region of PsFKF1 and AtFKF1 (Imaizumi et al., 2005; amino acids 296 to 636 and 284 to 619, respectively) were amplified from wild-type pea cv NGB5839 and Arabidopsis thaliana (Col-0) cDNA. In addition, the R450W allele of PsCDFc1, comprising the full-length coding sequence, was isolated from the late2-1D mutant. These sequences were then cloned in-frame into the pCR8/GW/TOPO entry vector (Invitrogen) and transferred by Gateway LR reaction (Invitrogen) into both pDEST32 (bait; BD) and pDEST22 (prey; AD) vectors (Invitrogen). Primers used to amplify these fragments are provided in Supplemental Table 3. Using the method described by de Folter and Immink (2011), the haploid yeast strains PJ69-4α and PJ69-4A were transformed with bait and prey constructs, respectively, and mated to create diploid colonies containing both interaction partners. Clones containing empty pDEST32 and pDEST22 vectors were also created as controls to test autoactivation. Diploid colonies were subcultured onto Synthetic Complete (SC) medium lacking leucine (Leu) and tryptophan (Trp). Individual colonies were suspended in 200 μL and were directly spotted onto nonselective medium (SC -Leu -Trp) and selective medium also lacking histidine (SC -Leu -Trp -His) with 35 mM 3-amino-1,2,4-triazole added and grown at 30°C for 4 d to test interactions. Negative controls and controls confirming interaction between AtFKF1 and PsCDFc1 are shown in Supplemental Figure 5.
Construct Preparation and Plant Transformation
For the Arabidopsis expression studies (Figure 6; Supplemental Figure 6), pCR8/GW/TOPO entry clones were generated for the wild-type (NGB5839) and R450W (late2-1D) alleles of PsCDF1. Entry clones were then linearized with either XbaI or XhoI (New England Biolabs) and transferred by LR reaction (Invitrogen) into the pB2GW7 vector, which contains the CaMV 35S promoter. The PsCDF1 entry clones were also transferred by LR reaction into a version of the pB2GW7 vector where the CaMV 35S promoter was replaced with the SUC2 promoter (Lee et al., 2013). For the transient assays (Figure 7J), the M. truncatula FTb1 (Medtr7g006630.1) promoter:LUC construct was generated by PCR amplifying from genomic DNA a 453-bp promoter fragment (from an upstream HindIII site to the FTb1 ATG translation initiation site, which was converted to a NcoI site; GenBank ID NC_016413.2, chr7:777018-776566) and cloning it into the pGreen 0800-5-LUC binary vector (Hellens et al., 2005). The GUS (Escherichia coli uidA) gene was also cloned into the pB2GW7 vector and used as a control in Figure 7J. The binary vectors were transformed into Agrobacterium tumefaciens GV3101 by electroporation. Arabidopsis (Col-0) transformation was conducted using the method described by Martinez-Trujillo et al. (2004). For transient expression in Nicotiana benthamiana leaves (Figure 7J), infiltrations were performed as described by Hellens et al. (2005), and expression was assayed 5 d after infiltration.
RNA-Seq Experiment and Data Analysis
Plants were grown in constant light for 3 weeks, and three leaflets (one each from leaves 6, 7, and 8) were harvested and pooled from two plants per replicate. As for gene expression studies, samples were frozen in liquid nitrogen and total RNA extracted using the SV Total RNA Isolation System (Promega). RNA concentration and RNA quality were determined using a Fragment Analyzer (Advanced Analytical). Sequencing library preparations were conducted with 1 µg of total RNA using the Truseq Stranded Total RNA library preparation kit with Ribozero Plant (Illumina). Three replicates per genotype were prepared separately. Each replicate was sequenced on a Miseq sequencer using a Miseq Reagent v3 150 cycles kit (Illumina). Quality checking of sequenced reads was performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Mapping of sequenced reads was performed using STAR (Spliced Transcript Alignment to a Reference; Dobin et al., 2013) on the pea transcriptome (PsUniLowCopy; Alves-Carvalho et al., 2015), and mapping results are presented in Supplemental Table 4. Raw read counts were obtained using the R package Rsubread (Liao et al., 2013) and normalized using the DESeq method (Love et al., 2014).
Statistical Analysis
For statistical analysis of the data presented in Figures 2, 3, and 7A, a two-tailed unpaired t test (with Welch’s correction for Figure 3) was performed, while data in Figure 4 were analyzed using a two-way ANOVA with Dunnett’s multiple comparison test. Differences between wild-type and mutant normalized count data in Figures 7C to 7I were also subjected to a two-tailed unpaired t test, with an adjusted threshold P value of 0.025 to account for the two comparisons performed for each gene. All tests were performed using GraphPad Prism (v6.05; GraphPad Software).
Accession Numbers
The accession numbers for genes newly reported in this study can be found in Supplemental Table 1 and in the supplemental figures.
Supplemental Data
Supplemental Figure 1. Rhythmic expression patterns of circadian clock genes under LD cycles.
Supplemental Figure 2. Position of LATE2 in pea linkage group VII.
Supplemental Figure 3. Phylogram of the legume CDF family and related DOF transcription factors.
Supplemental Figure 4. Phylogram of the legume FKF1 protein family.
Supplemental Figure 5. Yeast two-hybrid controls.
Supplemental Figure 6. Leaf number at flowering in both wild-type and transgenic Arabidopsis lines expressing wild-type and R450W forms of PsCDFc1 from the SUC2 promoter.
Supplemental Figure 7. Phylogram of the legume BBX protein family.
Supplemental Figure 8. Phylogram of the legume CCT protein family.
Supplemental Figure 9. Expression patterns of clock-related and COL family genes under constant light.
Supplemental Figure 10. Phylogram of legume CIB/BEE-like (CBL) proteins within the bHLH subfamily XII.
Supplemental Figure 11. Phylogram of legume PIF-like proteins within the bHLH subfamily VII(a+b).
Supplemental Figure 12. Phylogram of the legume AP2 protein family.
Supplemental Figure 13. Transcript levels in expanded leaf tissue from wild-type, late1-2, and late2-1D mutants grown from sowing under continuous white light.
Supplemental Table 1. Details of sequences for DOF transcription factors used in phylogenetic analyses and alignments.
Supplemental Table 2. Mapping marker details.
Supplemental Table 3. Primer details.
Supplemental Table 4. RNA-seq mapping results.
Supplemental File 1. Alignment for Figure 5A.
Supplemental File 2. Alignment for Supplemental Figure 3.
Supplemental File 3. Alignment for Supplemental Figure 4.
Supplemental File 4. Alignment for Supplemental Figure 7.
Supplemental File 5. Alignment for Supplemental Figure 8.
Supplemental File 6. Alignment for Supplemental Figure 10.
Supplemental File 7. Alignment for Supplemental Figure 11.
Supplemental File 8. Alignment for Supplemental Figure 12.
Supplementary Material
Acknowledgments
We thank Tracey Winterbottom and Michelle Lang for help with plant maintenance, Albert Wong for assistance with initial mapping and characterization of the late2 mutant, and Lim Chee Liew for assistance with expression analyses of COL genes. We acknowledge the use of facilities administered by the UTAS Central Science Laboratory. This work was supported by the Australian Research Council (J.L.W.) and the New Zealand Marsden Fund (R.C.M.).
AUTHOR CONTRIBUTIONS
J.L.W., S.R., R.C.M., V.H., and F.C.S conceived and designed the experiments. S.R., F.C.S., V.H., J.K.V.S., R.L., and J.L.W. performed the experiments. J.B. and G.A. contributed new analytical tools. S.R., F.C.S., V.H., J.K.V.S., J.L.W., R.L., and R.C.M. analyzed the data. S.R., J.L.W., and F.C.S. wrote the article.
Glossary
- SD
short-day
- LD
long-day
References
- Alves-Carvalho S., et al. (2015). Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J. 84: 1–19. [DOI] [PubMed] [Google Scholar]
- Ballerini E.S., Kramer E.M. (2011). In the light of evolution: a reevaluation of conservation in the CO-FT regulon and its role in ohotoperiodic regulation of flowering time. Front. Plant Sci. 2: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbel A., Ferrándiz C., Hecht V., Dalmais M., Lund O.S., Sussmilch F.C., Taylor S.A., Bendahmane A., Ellis T.H., Beltrán J.P., Weller J.L., Madueño F. (2012). VEGETATIVE1 is essential for development of the compound inflorescence in pea. Nat. Commun. 3: 797. [DOI] [PubMed] [Google Scholar]
- Brambilla V., Fornara F. (2013). Molecular control of flowering in response to day length in rice. J. Integr. Plant Biol. 55: 410–418. [DOI] [PubMed] [Google Scholar]
- Campoli C., Drosse B., Searle I., Coupland G., von Korff M. (2012). Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. Plant J. 69: 868–880. [DOI] [PubMed] [Google Scholar]
- Cao D., et al. (2015). GmCOL1a and GmCOL1b function as flowering repressors in soybean under long-day conditions. Plant Cell Physiol. 56: 2409–2422. [DOI] [PubMed] [Google Scholar]
- Chia T.Y., Müller A., Jung C., Mutasa-Göttgens E.S. (2008). Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J. Exp. Bot. 59: 2735–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comadran J., et al. (2012). Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 44: 1388–1392. [DOI] [PubMed] [Google Scholar]
- Dally N., Xiao K., Holtgräwe D., Jung C. (2014). The B2 flowering time locus of beet encodes a zinc finger transcription factor. Proc. Natl. Acad. Sci. USA 111: 10365–10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S., Immink R.G. (2011). Yeast protein-protein interaction assays and screens. Methods Mol. Biol. 754: 145–165. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K.C., Gissot L., Sauerbrunn N., Rühl M., Jarillo J.A., Coupland G. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17: 75–86. [DOI] [PubMed] [Google Scholar]
- Franssen S.U., Shrestha R.P., Bräutigam A., Bornberg-Bauer E., Weber A.P.M. (2011). Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics 12: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Ariza J., Galbiati F., Goretti D., Brambilla V., Shrestha R., Pappolla A., Courtois B., Fornara F. (2015). Loss of floral repressor function adapts rice to higher latitudes in Europe. J. Exp. Bot. 66: 2027–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Schain N.D., Díaz-Mendoza M., Zurczak M., Suárez-López P. (2012). Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. 70: 678–690. [DOI] [PubMed] [Google Scholar]
- Hayama R., Coupland G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M., Shimamoto K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722. [DOI] [PubMed] [Google Scholar]
- Hecht V., Knowles C.L., Vander Schoor J.K., Liew L.C., Jones S.E., Lambert M.J., Weller J.L. (2007). Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144: 648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Laurie R.E., Vander Schoor J.K., Ridge S., Knowles C.L., Liew L.C., Sussmilch F.C., Murfet I.C., Macknight R.C., Weller J.L. (2011). The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23: 147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.Y., Adams J.P., No K., Liang H., Meilan R., Pechanova O., Barakat A., Carlson J.E., Page G.P., Yuceer C. (2012). Overexpression of CONSTANS homologs CO1 and CO2 fails to alter normal reproductive onset and fall bud set in woody perennial poplar. PLoS One 7: e45448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297. [DOI] [PubMed] [Google Scholar]
- Ishikawa R., Tamaki S., Yokoi S., Inagaki N., Shinomura T., Takano M., Shimamoto K. (2005). Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice. Plant Cell 17: 3326–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T. (2007). Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58: 3091–3097. [DOI] [PubMed] [Google Scholar]
- Izawa T., Oikawa T., Sugiyama N., Tanisaka T., Yano M., Shimamoto K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C.S., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Pembleton L.W., Cogan N.O.I., Savin K.W., Leonforte T., Paull J., Materne M., Forster J.W. (2012). Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genomics 13: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman B., Abelenda J.A., Gomez Mdel.M., Oortwijn M., de Boer J.M., Kowitwanich K., Horvath B.M., van Eck H.J., Smaczniak C., Prat S., Visser R.G., Bachem C.W.B. (2013). Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495: 246–250. [DOI] [PubMed] [Google Scholar]
- Kojima S., Takahashi Y., Kobayashi Y., Monna L., Sasaki T., Araki T., Yano M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43: 1096–1105. [DOI] [PubMed] [Google Scholar]
- Kong F., Liu B., Xia Z., Sato S., Kim B.M., Watanabe S., Yamada T., Tabata S., Kanazawa A., Harada K., Abe J. (2010). Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol. 154: 1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.V., Lucyshyn D., Jaeger K.E., Alós E., Alvey E., Harberd N.P., Wigge P.A. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Marchal V., Le Gourrierec J., Wenkel S., Adrian J., Jang S., Kulajta C., Braun H., Coupland G., Hoecker U. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133: 3213–3222. [DOI] [PubMed] [Google Scholar]
- Laurie R.E., Diwadkar P., Jaudal M., Zhang L., Hecht V., Wen J., Tadege M., Mysore K.S., Putterill J., Weller J.L., Macknight R.C. (2011). The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156: 2207–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledger S., Strayer C., Ashton F., Kay S.A., Putterill J. (2001). Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 26: 15–22. [DOI] [PubMed] [Google Scholar]
- Lee R., Baldwin S., Kenel F., McCallum J., Macknight R. (2013). FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat. Commun. 4: 2884. [DOI] [PubMed] [Google Scholar]
- Lester D., Mackenzie-Hose A., Davies P., Ross J., Reid J. (1999). The influence of the null le-2 mutation on gibberellin levels in developing pea seeds. Plant Growth Regul. 27: 83–89. [Google Scholar]
- Li D., Yang C., Li X., Gan Q., Zhao X., Zhu L. (2009). Functional characterization of rice OsDof12. Planta 229: 1159–1169. [DOI] [PubMed] [Google Scholar]
- Li F., Zhang X., Hu R., Wu F., Ma J., Meng Y., Fu Y. (2013). Identification and molecular characterization of FKF1 and GI homologous genes in soybean. PLoS One 8: e79036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew L.C., Hecht V., Sussmilch F.C., Weller J.L. (2014). The pea photoperiod response gene STERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol. 165: 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew L.C., Hecht V., Laurie R.E., Knowles C.L., Vander Schoor J.K., Macknight R.C., Weller J.L. (2009). DIE NEUTRALIS and LATE BLOOMER 1 contribute to regulation of the pea circadian clock. Plant Cell 21: 3198–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., Lin C. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539. [DOI] [PubMed] [Google Scholar]
- Liu Y., Li X., Li K., Liu H., Lin C. (2013). Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 9: e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Trujillo M., Limones-Briones V., Cabrera-Ponce J., Herrera-Estrella L. (2004). Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol. Biol. Report. 22: 63–70. [Google Scholar]
- Nakamichi N. (2015). Adaptation to the local environment by modifications of the photoperiod response in crops. Plant Cell Physiol. 56: 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C., Abelenda J.A., Cruz-Oró E., Cuéllar C.A., Tamaki S., Silva J., Shimamoto K., Prat S. (2011). Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478: 119–122. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Lasswell J., Rogg L.E., Cohen M.A., Bartel B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340. [DOI] [PubMed] [Google Scholar]
- Nicholas K.B., Nicholas H.B. (1997). GeneDoc: a tool for editing and annotating multiple sequence alignments (distributed by the author). http://www.psc.edu/biomed/genedoc.
- Pin P.A., et al. (2012). The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr. Biol. 22: 1095–1101. [DOI] [PubMed] [Google Scholar]
- Sawa M., Kay S.A. (2011). GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 11698–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R., Gómez-Ariza J., Brambilla V., Fornara F. (2014). Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. (Lond.) 114: 1445–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S., Rühl M., de Montaigu A., Wötzel S., Coupland G. (2015). Evolution of CONSTANS regulation and function after gene duplication produced a photoperiodic flowering switch in the Brassicaceae. Mol. Biol. Evol. 32: 2284–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Devlin P.F., Kay S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490. [DOI] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120. [DOI] [PubMed] [Google Scholar]
- Sun Z., Guo T., Liu Y., Liu Q., Fang Y. (2015). The roles of Arabidopsis CDF2 in rranscriptional and posttranscriptional regulation of primary microRNAs. PLoS Genet. 11: e1005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Shimamoto K. (2011). Heading date 1 (Hd1), an ortholog of Arabidopsis CONSTANS, is a possible target of human selection during domestication to diversify flowering times of cultivated rice. Genes Genet. Syst. 86: 175–182. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187: 57–66. [DOI] [PubMed] [Google Scholar]
- Trevaskis B., Hemming M.N., Peacock W.J., Dennis E.S. (2006). HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol. 140: 1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R.P., Laurie D.A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034. [DOI] [PubMed] [Google Scholar]
- Valverde F. (2011). CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 62: 2453–2463. [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006. [DOI] [PubMed] [Google Scholar]
- Watanabe S., Harada K., Abe J. (2012). Genetic and molecular bases of photoperiod responses of flowering in soybean. Breed. Sci. 61: 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J.L., Murfet I.C., Reid J.B. (1997). Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol. 114: 1225–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J.L., Beauchamp N., Kerckhoffs L.H., Platten J.D., Reid J.B. (2001). Interaction of phytochromes A and B in the control of de-etiolation and flowering in pea. Plant J. 26: 283–294. [DOI] [PubMed] [Google Scholar]
- Weller J.L., Batge S.L., Smith J.J., Kerckhoffs L.H., Sineshchekov V.A., Murfet I.C., Reid J.B. (2004). A dominant mutation in the pea PHYA gene confers enhanced responses to light and impairs the light-dependent degradation of phytochrome A. Plant Physiol. 135: 2186–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J.L., et al. (2012). A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. USA 109: 21158–21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.C.S., Hecht V.F., Picard K., Diwadkar P., Laurie R.E., Wen J., Mysore K., Macknight R.C., Weller J.L. (2014). Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 5: 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., et al. (2013). Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. USA 110: 2775–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z., et al. (2012). Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 109: E2155–E2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., Zhang Q. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. (2002). The Dof family of plant transcription factors. Trends Plant Sci. 7: 555–560. [DOI] [PubMed] [Google Scholar]
- Yang S., Weers B.D., Morishige D.T., Mullet J.E. (2014). CONSTANS is a photoperiod regulated activator of flowering in sorghum. BMC Plant Biol. 14: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M.J., Mazzella M.A., Whitelam G.C., Casal J.J. (2001). Resetting of the circadian clock by phytochromes and cryptochromes in Arabidopsis. J. Biol. Rhythms 16: 523–530. [DOI] [PubMed] [Google Scholar]
- Yu J.W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai H., Lü S., Liang S., Wu H., Zhang X., Liu B., Kong F., Yuan X., Li J., Xia Z. (2014). GmFT4, a homolog of FLOWERING LOCUS T, is positively regulated by E1 and functions as a flowering repressor in soybean. PLoS One 9: e89030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.