Abstract
Recently, a number of advances have been implemented into the core ChIP-seq (chromatin immunoprecipitation coupled with next-generation sequencing) methodology to streamline the process, reduce costs or improve data resolution. Several of these emerging ChIP-based methods perform additional chemical steps on bead-bound immunoprecipitated chromatin, posing a challenge for generating similarly treated input controls required for artifact removal during bioinformatics analyses. Here we present a versatile method for producing technique-specific input controls for ChIP-based methods that utilize additional bead-bound processing steps. This reported method, termed protein attached chromatin capture (PAtCh-Cap), relies on the non-specific capture of chromatin-bound proteins via their carboxylate groups, leaving the DNA accessible for subsequent chemical treatments in parallel with chromatin separately immunoprecipitated for the target protein. Application of this input strategy not only significantly enhanced artifact removal from ChIP-exo data, increasing confidence in peak identification and allowing for de novo motif searching, but also afforded discovery of a novel CTCF binding motif.
INTRODUCTION
ChIP-seq (chromatin immunoprecipitation coupled with next-generation sequencing) has emerged as a powerful and widely used methodology for defining the site-specific localization of transcription factors and histone marks in the context of the cellular genome (1–3). Since inception of this core technique nearly a decade ago, several improvements have been implemented to expand the capabilities (4–11), reduce costs (12,13) or maximize resolution (14–16). In many of the recently enhanced ChIP-based methods including ChIP-exo, ChIP-nexus, lobChIP and ChIPmentation (12–15), several steps of sample and/or library preparation are performed on bead-bound immunoprecipitated chromatin, posing a challenge in generating a similarly treated input control required for downstream bioinformatic analysis and data quality assessments.
Numerous reports have focused on the sources and methods for removal of artifacts in ChIP-based data sets, many of which highlight the necessity and critical importance of having a proper input control (2,17–22). For ChIP-seq, input controls are normally derived from isolated cellular DNA that has been cross-linked, sheared and ideally chemically treated in an analogous manner to DNA immunoprecipitated for the protein of interest. When subjected to high-throughput sequencing in parallel with the ChIP sample, the input control informs on the genomic locations of technique-specific artifact peaks that exist in the ChIP-seq data set. As such, the use of an input control has become a core component of the communally agreed upon standards and guidelines for ChIP-seq experiments (20). In lieu of having a comparable input, less ideal methods for artifact removal must be implemented such as: (i) utilizing an IgG control, which typically pulls-down comparably less DNA resulting in lower library complexity and significant sequencing biasing relative to the ChIP sample (19,20); (ii) relying on only filtering ChIP-seq derived blacklisted peaks, which is unable to eliminate technique-specific false positives; and/or (iii) applying alternative peak caller strategies in which P-values may provide less reliable false discovery rates (FDRs), reducing confidence in the statistical significance of identified peaks (22). Thus, a general procedure for producing a matched input control that can facilitate technique-specific artifact removal would greatly increase the quality and confidence of information gained from the above listed modified ChIP-based methodologies that perform additional bead-bound processing steps (12–15).
Indeed, bioinformatics treatments commonly utilized in ChIP-seq data analysis, such as blacklist filtering and duplicate read removal, have proven to be inappropriate for eliminating artifacts in ChIP-exo data (17,23). It is understood that due to the narrow peak distributions observed in high-resolution methods such as ChIP-exo, removal of duplicate reads would discard essential peak information (17,23), whereas the failure of blacklisting to improve these data sets is likely a direct result of technique-specific variances in artifact generation. Similarly, it has been well established that ChIP-chIP and ChIP-seq data sets have variable artifact signatures (24). While methods like ChIP-exo (15) and ChIP-nexus (14) have undoubtedly increased resolution over standard ChIP-seq, the high number of washing and digestion steps in conjunction with decreased library complexity (23) result in significant false positive peaks that can considerably impact downstream data analysis. This may be of minimal concern for histone proteins, high occupancy transcription factors, or transcription factors for which the consensus binding motif is well characterized. However, for analysis of transcription factors where the consensus sequence has yet to be identified, or for proteins that have a globally low genomic occupancy, the persistence of false positives in these data present a significant barrier in reliably identifying peaks with high confidence and discerning de novo binding motifs.
Here, we report a method for non-specifically capturing cross-linked chromatin complexes via protein carboxylate groups that allows for the DNA to be subjected to all downstream chemical treatments in parallel with bead-bound chromatin separately immunoprecipitated for the target of interest. This input control method, termed protein attached chromatin capture (PAtCh-Cap), is designed to be facile and universally applicable to any of the current (12–15) and future ChIP-based techniques that perform additional chemical and library preparation steps on bead-bound chromatin. Applying our input control method to the analysis of CTCF ChIP-exo data demonstrated that we were able to selectively remove artifacts in both pericentromeric and gene proximal regions, significantly increasing confidence in peak identification, revealing previously unidentifiable peaks and affording the capability of performing a de novo motif search analysis. This improved analysis capability within a high-resolution ChIP-exo data set was essential for the identification of a novel CTCF motif that appears to have an independent cellular function.
MATERIALS AND METHODS
Cell culturing
HeLa cells (from the laboratory of Prof. C.J. Burrows; University of Utah) were cultured in Dulbecco's modified Eagle medium supplemented with 4.5 g/l glucose, 10% fetal bovine serum and 2 mM glutamine and maintained in a humidified incubator at 37°C and 5% CO2. Cell counting and viability analysis was performed on a Countess Automated Cell Counter (Thermo Fisher Scientific). Cell line authentication to confirm lack of cross-contamination was routinely verified by short tandem repeat (STR) DNA profiling.
ChIP-exo
For each of the two ChIP-exo replicates, 20 × 106 HeLa cells were fixed with 1% formaldehyde for 15 min to cross-link protein:DNA complexes, followed by a quench with 125 mM glycine. The IP, exonuclease digestions and library generation procedures were all performed using a commercially available ChIP-exo Kit (Active Motif) following the manufacturer's instructions with the few noted modifications. A Diagenode Bioruptor Standard sonication device (run at max amplitude for 5 × 15 min in ice water) was used to shear the cross-linked DNA to 100–400 bp fragments. Cell debris was removed by centrifugation and the supernatant containing the solubilized chromatin DNA:protein complexes was isolated. Prior to further treatment, 10% of the sheared chromatin sample volume was removed from each replicate for input sample preparation (see PAtCh-Cap section below). For the IP step, protein G coated magnetic beads were pre-functionalized with CTCF antibody (Millipore) prior to incubation with the sheared chromatin sample. DNA purification after reverse cross-linking was performed with the MinElute PCR Purification Kit (Qiagen). It should be noted that the library preparations performed with the Active Motif ChIP-exo Kit are designed to be compatible with the Illumina sequencing platform (25). Final purified DNA libraries were sequenced by the High-Throughput Genomics Core within the University of Utah Huntsman Cancer Institute using the Illumina HiSeq 2000 platform.
Protein attached chromatin capture (PAtCh-Cap) for ChIP-exo
Following two series of washes with 0.01 M PBS (pH 7.4), 50 μg of M-280 streptavidin coated Dynabeads (Thermo Fisher Scientific; equivalent to the number of beads utilized per reaction in the Active Motif ChIP-exo Kit) were conjugated with 10 μl of 50 nM EZ-link amine-PEG3-Biotin (Thermo Fisher Scientific; PEG = polyethylene glycol) in 0.01 M PBS (pH 7.4) at room temperature for 20 min. These beads were selected as they have the same size and core material composition as the protein G coated magnetic beads utilized in the Active Motif ChIP-exo Kit. The biotinylated beads were then washed twice with 0.01 M PBS (pH 7.4) and once with 0.1 mM MES (pH 5.0) to remove any non-conjugated material. For each of the two replicates, the input sample (obtained as discussed above) was combined with 300 μl of 0.1 M EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) along with the pre-functionalized biotinylated magnetics beads and incubated in 10 ml 0.1 mM MES buffer (pH 5.0) for 3 h at room temperature on a mechanical rotator. Once covalently bound to the magnetic beads, the input samples were treated identically as described above for the CTCF ChIP-exo samples utilizing reagents and materials from the Active Motif ChIP-exo Kit.
Preparation of input DNA for ChIP-seq
For each of the three replicates, 20 × 106 HeLa cells were fixed with 1% formaldehyde for 15 min to cross-link protein:DNA complexes, followed by a quench with 125 mM glycine. Cells were washed with cold 0.01 M PBS (pH 7.4) and lysed for 10 min at 40°C in cell lysis buffer (50 mM HEPES (pH 8.0), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100). Cellular nuclei were then washed (10 mM Tris–HCl (pH 8.0), 200 mM EDTA), centrifuged and re-suspended in nuclear lysis buffer (50 mM Tris–HCl (pH 8.0), 100 mM NaCl, 10 mM EDTA, 1% SDS). A Diagenode Bioruptor Standard sonication device (run at max amplitude for 5 × 15 min in ice water) was used to shear the cross-linked DNA to 100–400 bp fragments. Proteinase K digestion was performed overnight at 55°C. DNA was purified with the MinElute PCR Purification Kit (Qiagen). DNA quantification, library construction and sequencing were all performed by the High-Throughput Genomics Core within the University of Utah Huntsman Cancer Institute. The input DNA libraries were sequenced using the Illumina HiSeq 2000 platform.
RNA interference and RNA-seq
HeLa cells were transfected with either a scrambled siRNA or one of two CTCF siRNAs (Thermo Fisher Scientific) in triplicate for each siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific) for 24 hrs. Cells were washed with 0.01 M PBS (pH 7.4) and resuspended in TRIzol (Thermo Fisher Scientific) prior to RNA extraction with the Direct-zol RNA Kit (Zymo Research). Prior to submission for high-throughput sequencing analysis, an aliquot of the RNA from each sample was reversed transcribed using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). The amount of CTCF was then determined in each sample by quantitative real-time PCR (qRT-PCR), utilizing HPRT1 as a normalization control, to ensure that sufficient CTCF knock-down was achieved. In parallel, protein was extracted from HeLa cells after siRNA transfection utilizing NP-40 buffer (50 mM Tris (pH 8.0), 150 mM NaCl, 1.0% NP-40) supplemented with protease inhibitors (Roche) and separated by gel electrophoresis. Proteins were then transferred to nitrocellulose membranes and immunoblotted using a CTCF antibody (Millipore) following standard procedures, to ensure sufficient knock-down was also achieved at the protein level. For RNA-seq, RNA quality control measurements, purification, library construction and sequencing were all performed by the High-Throughput Genomics Core within the University of Utah Huntsman Cancer Institute. In short, RNA quality was measured on a Bioanalyzer RNA 6000 Nano Chip. Total RNA was then further purified with the RiboMinus Eukaryote Kit for RNA-seq (Thermo Fisher Scientific). Small and long directional RNA-seq libraries were then constructed using Illumina TruSeq Stranded mRNA Sample Prep with poly(A) selection and sequenced with a 50 bp single-end run on the Illumina HiSeq 2000 platform.
Sequencing data analyses
ChIP-exo fastq files were aligned to the human genome (hg19) using Novoalign (Novocraft, Inc.) and the following parameters: -0 SAM –r. Sam files were then sorted and indexed with SAMtools (26). Peak calling was performed with and without the input control by MACS2 (27) using the following standard parameters: model fold settings of 5–50 q-values with a cutoff of 0.05. Bedgraph outputs from MACS2 were viewed on the Integrated Genome Browser (IGB) (28). The MACS2 bdgcmp command was further performed to obtain the read coverage tracks after ChIP-exo normalization to the PAtCh-Cap derived input control. RNA-seq fastq files were similarly aligned to the human genome (hg19) using Novoalign (Novocraft, Inc.) and peak called with the USeq suite (21). RNA-seq reads were aligned with all known and theoretical splice junctions using the following parameters: -r All 50 -t 40 -o SAM 90 –k. The USeq NovoalignParser application was then used to parse the alignment files into binary point data by setting the posterior probability to 0 and alignment score threshold to 60. The MultipleReplicaDefinedRegionScanseqs USeq application, which utilizes the DESeq R package (29), identified statistically significant differentially expressed genes between cells treated with the scrambled siRNA and CTCF depleted cells.
Bioinformatics analyses
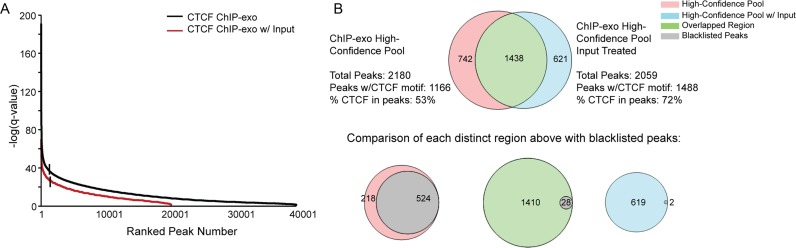
To identify the high-confidence ChIP-exo peaks, the peak confidence level was plotted against the ranked peak number (refer to Figure 2A). Upon doing this, a clearly observable inflection point separated a sub-set of peaks with the highest intensity from the overall peaks analyzed. Therefore, in this analysis high-confidence CTCF peaks were selected from enriched regions characterized by a –log(q-value) higher than the inflection point and a line with a slope of –tan 1 to the curve. Further, ChIP-seq input identified blacklisted genomic regions from the DAC, DER and UHS lists (30) were intersected with the above determined high-confidence pools for both the CTCF ChIP-exo data with and without input treatment. The DAC and DER blacklisted regions were downloaded from the UCSC table browser (http://hgwdev.cse.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeMapability) (31) whereas the UHS regions were extracted from the following site: https://sites.google.com/site/anshulkundaje/projects/blacklists.
Figure 2.
Application of PAtCh-Cap to CTCF ChIP-exo data allowed for significant artifact removal and improved confidence in peak identification. (A) To identify high-confidence CTCF peaks, all peaks called with a 0.05 q-value threshold from the ChIP-exo data with (red) and without (black) input treatment were plotted as the –log(q-value) versus ranked peak number. High confidence peaks were determined to be those characterized by a –log(q-value) higher than the inflection point and a line with a slope of –tan 1 to the curve (denoted by vertical lines). (B) Venn diagram demonstrating the overlap of high-confidence peaks identified from data sets with and without input treatment (top). The number of CTCF motifs found within each pool is denoted and clearly shows that the percentage of CTCF containing peaks relative to the total increases substantially after input treatment. Venn diagrams for the overlap of blacklisted peaks with each of the above regions (bottom).
High confidence peaks were then subjected to de novo motif discovery analysis using RSAT (32–34) with the following parameters: peak -motifs -v 1 -title bedfile -i $RSAT -max_seq_len 200 -markov auto -disco oligos,positions,local_words -nmotifs 8 -minol 6 -maxol 6 -no_merge_lengths -2str -origin center -motif_db jaspar_core_vertebrates. In addition to the expected CTCF consensus sequence, two additional motifs were identified from the de novo motif analysis in the input treated ChIP-exo data. FIMO (35) was then used to identify matches for all of these motifs in all peak regions (‘fimo -bgfile flanking.bg–motif 1 motif.meme.txt) with a default P-value threshold of 10−4. To determine the preferential spacing and co-localization of the additionally identified motifs relative to the CTCF consensus sequence, the CTCF core motif was extended by ±30 nucleotides and analyzed by SpaMo (spamo -png -bgfile -dumpseqs -inc 1 meme.motif.txt) (36). Average logos representing all the extended motifs around the centralized CTCF consensus were created using MEME (37). The read tag and nucleotide base heatmaps as well as aggregate plots were generated using in-house python scripts. Each pool of sequences containing the motifs of interest were trimmed to the same size and centered at their CTCF core motifs using the USeq scoreSequences application. All instances in which the identified CTCF motifs were localized within gene promoters (defined as ±1000 bps around the transcription start site (TSS)) were intersected with the genes identified to be differentially expressed from the above RNA-seq analysis (0.05 adjusted Benjamini–Hochberg P-value). For each set of filtered differentially expressed genes, Ingenuity Pathway Analysis (IPA, www.ingenuity.com) was performed to identify uniquely significant biological pathways correlated with each motif.
RESULTS AND DISCUSSION
Development of a method for non-specific protein mediated chromatin capture
Development of a general method to generate matched input controls for ChIP-based techniques that perform additional preparation steps on bead-bound immunoprecipitated chromatin complexes required that two major challenges be addressed. First, it was necessary to identify a way to non-specifically pull-down a random sampling of cross-linked chromatin complexes that could be affixed to magnetic beads with a sufficient affinity to remain associated throughout the following treatment steps. Second, it was ideal to determine a strategy for affixing these chromatin complexes to the magnetic beads via the proteins, leaving the chromatin DNA accessible for further processing. These criteria eliminated the possibility of utilizing an antibody based approach as no single protein-specific antibody would be capable of immunoprecipitating a completely unbiased background representation of the chromatin complexes in a given sample with adequate affinity.
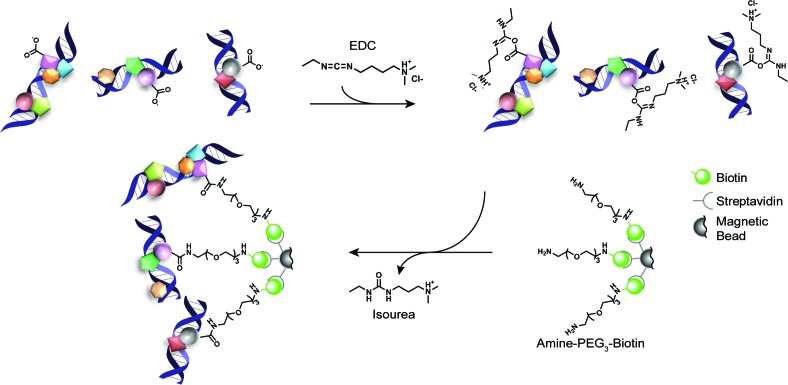
Thus, to facilitate protein mediated bead-bound capture of cross-linked chromatin complexes, we took advantage of the readily available amine functionalized pegylated biotin reagents commonly utilized for the conjugation of EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) activated protein carboxylates. In short, after standard cross-linking, chromatin isolation and DNA shearing procedures, an aliquot of the sample (typically 10%) is removed to generate the input control. In parallel, streptavidin coated magnetic beads are conjugated with an amine functionalized pegylated biotin (amine-PEG3-biotin). These pre-conjugated beads are then incubated with the isolated cross-linked chromatin complexes in the presence of EDC producing bead-bound chromatin complexes covalently linked via protein carboxylate groups (Figure 1). These pulled-down complexes can then be subjected to additional chemical processing steps in parallel with bead-bound chromatin separately immunoprecipitated for the target of interest. Compared to other chromatin capture methods that pull-down on modified DNA (38,39), which may result in disruption of protein:DNA interactions prior to cross-linking and/or mask accessibility of the chromatin fragments, our PAtCh-Cap method achieves the goal of non-specifically pulling-down chromatin bound proteins, leaving the cross-linked DNA freely accessible for additional treatments.
Figure 1.
Schematic overview for the protein attached chromatin capture (PAtCh-Cap) method. Streptavidin coated magnetic beads are first conjugated with amine-PEG3-biotin (where PEG is polyethylene glycol). After standard cross-linking, chromatin isolation and DNA shearing procedures, 10% of the sample volume is removed and incubated with the pre-conjugated beads in the presence of EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride). EDC reacts with protein carboxylate groups, forming unstable o-acylisourea activated esters that can undergo nucleophilic attack by the primary amines on the amine-PEG3-biotin prosthetics. This forms a covalent amide linkage with proteins in the chromatin complexes and releases an isourea by-product. Once bead-bound, these chromatin complexes can be subjected to additional chemical processing steps in parallel with bead-bound chromatin that was separately immunoprecipitated for the target of interest.
Application of PAtCh-Cap to generate an input control for technique-specific artifact removal in ChIP-exo data
As a proof-of-principle, we applied our PAtCh-Cap strategy to produce a ChIP-exo input control that could be utilized in downstream bioinformatics analysis of CTCF genomic occupations in HeLa cells. After cross-linking and shearing, 10% of the chromatin complex sample volume was removed for the input control and treated with the PAtCh-Cap method, generating the bead-bound input control as described above. In parallel, protein G coated magnetic beads were conjugated with CTCF antibody and utilized to selectively immunoprecipitate CTCF-bound chromatin fragments from the remaining cross-linked chromatin complex pool. At this stage, both the bead-bound input control and isolated CTCF-containing chromatin complexes were simultaneously and identically subjected to all subsequent exonuclease digestions, library preparation steps, reverse cross-linking, purification procedures, quality assessments and high-throughput sequencing.
To assess whether the use of sequencing data from a similarly treated input control was able to sufficiently remove artifacts and improve peak identification within our CTCF ChIP-exo data set, we first defined pools of high-confidence peaks from this data with and without input treatment. These high-confidence pools were defined from enriched regions by plotting the -log(q-value) versus the ranked peak number and only considering peaks above the inflection point and a line with the slope of –tan 1 to the curve (Figure 2A). Once identified, the high-confidence peaks from the pools with and without input treatment were intersected and found to have a high degree of overlap, though in each case there were also a significant number of peaks that were individual to each pool (Figure 2B). To discern whether artifact peaks were eliminated from the ChIP-exo data set, we first performed a weight matrix search (21) for the core CTCF motif within each set of high-confidence peaks. This search revealed an approximate 20% increase in peaks containing the CTCF motif out of total high-confidence peaks called for the input treated data relative to the non-treated data set (Figure 2B). The majority of remaining peaks that do not harbor the core CTCF motif in the cleaned ChIP-exo data likely represent genomic occupations for the numerous CTCF protein interacting partners (40). Together, these findings indicate that bioinformatic treatment of the CTCF ChIP-exo data with the input control not only removes a significant number of artifact peaks but also allowed for identification of additional CTCF-containing peaks that were otherwise masked.
As further confirmation, we separately intersected known ChIP-seq blacklisted peaks with the above determined high-confidence pools for the CTCF ChIP-exo data with and without input treatment. Blacklisted peaks constitute known genomic artifact regions systematically observed in input controls from standard ChIP-seq data and it has become an acceptable practice to exclude these reads out of ChIP-seq data sets (30,41–43). As can be seen in Figure 2B, nearly all of the blacklisted peaks are present in the pool of ChIP-exo peaks excluded by the input control treatment. This clearly demonstrates that the majority of these blacklisted peaks are captured by the ChIP-exo input control and that as previously observed, the removal of blacklisted peaks alone is not sufficient to remove all ChIP-exo specific artifacts (17).
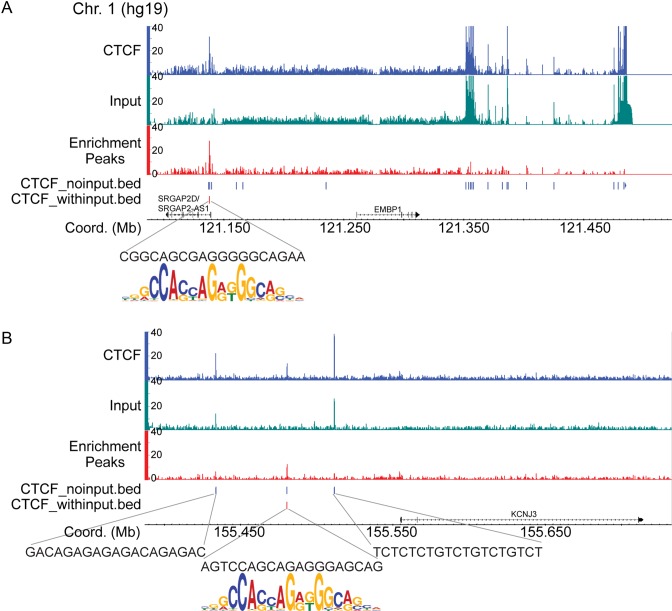
Furthermore, we prepared a ChIP-seq input control from our HeLa cells and compared this with our ChIP-exo input. Comparisons of enrichment profiles and genome-wide correlation analysis between these input controls demonstrates that there are a number of artifact peaks that are common between the two methods, though there are many more ChIP-exo specific false positives (Supplementary Figure S1A (green loci) and Supplementary Figure S1B). Indeed, intersection of the identified peaks in the ChIP-seq and ChIP-exo input controls results in nearly all of the ChIP-seq input peaks being captured by the ChIP-exo input control (Supplementary Figure S1C). Additionally, example read coverage tracks of CTCF ChIP-exo with and without input treatment indicate that use of the input control dramatically cleans up artifacts not only within pericentromeric, but also gene proximal regions (Figure 3). Analysis of genomic sequences underneath the peaks removed by the ChIP-exo input determined that these sites do not contain the CTCF motif, whereas remaining peaks do.
Figure 3.
Representative CTCF ChIP-exo read coverage tracks for the pericentromeric region of chromosome 1 (A) and the promoter of the KCNJ3 gene (B). The CTCF reads (blue) were normalized to the reads from the input control (green) using MACS2 (27) to generate the enrichment read coverage tracks (red). Peaks identified by the MACS2 peak caller (represented in the .bed tracks) are denoted as red or blue vertical lines for the CTCF ChIP-exo data sets with and without input treatment, respectively. Analysis of the genomic sequences underneath the remaining peaks after input treatment (vertical red lines) definitively showed that these sites contain the core CTCF motif as evidenced by alignment of the CTCF sequence logo beneath.
Finally, we sought to determine whether the increased incidence of artifact peaks in the ChIP-exo data was a consequence of biasing from the exonuclease digestion or GC content during sequencing. Comparative analysis of the nucleotide frequency and GC content plots from the ChIP-exo input controls and CTCF ChIP-exo sequencing data relative to standard ChIP-seq input controls indicates that there is a minor biasing common between all of the ChIP-exo data sets that likely results from the exonucleases used in the processing of these samples (Supplementary Figure S2). Nonetheless, this observed exonuclease biasing in the ChIP-exo samples is not sufficient to account for the significant increase in generated artifacts relative to ChIP-seq data sets. Thus, it is likely that the remainder of these artifacts result from non-specific interactions that occur during the additional processing steps performed on the bead-bound samples. This further suggests that other bead-bound ChIP-based methods (12–14) may also generate an increase in technique-specific artifacts relative to standard ChIP-seq and that a strategy such as PAtCh-Cap to produce a matched input is needed. Together all of the above findings clearly demonstrate that ChIP-exo data sets harbor significant technique-specific false positive peaks and that our PAtCh-Cap method was not only able to remove these artifacts, but also improved confidence in peak identification.
It should be noted that while we clearly demonstrated that use of an appropriate input control can dramatically improve artifact removal in ChIP-exo data, new bioinformatics tools will need to be developed that are capable of implementing input treatments while maintaining full resolution of ChIP-exo data sets. For the analysis presented here, we utilized the MACS2 peak caller (27) as it has the built-in capability of determining peak enrichment over input controls and using local statistics to reduce biasing and calculate empirical FDRs. However, peak calling is administered by performing read-shifting to account for the offset in forward and reverse strand reads. In contrast, peak callers specifically designed for analysis of ChIP-exo data sets, such as GeneTrack (44) or MACE (45), rely on retention of strand information by using 5′ cross-link borders on each strand to define ‘peak-pairs’ that result in identification of high-resolution protein footprints (23). Currently, neither of these ChIP-exo specific peak callers are capable of readily normalizing these data sets to input controls. Thus, it would be ideal to have a peak caller suite that can normalize the data relative to an input control without compromising resolution by eliminating strand information.
Improved ChIP-exo data analysis identified a novel CTCF binding site
Once peaks within the high-confidence pools for the ChIP-exo data with and without input treatment were identified as described above (Figure 2A), we subjected each pool to a de novo motif search analysis (32–34). This search returned not only the core CTCF motif, but also its previously characterized flanking 5′-site (15,46) for both the untreated and input treated ChIP-exo CTCF data sets (Supplementary Figure S3A). It has been shown previously that depending on the genomic occupation context, various combinations of the 11 CTCF zinc finger subsets recognize the CTCF core alone or in conjunction with conserved 5′- and 3′-flanking regions (15,46). This modularity in CTCF motif recognition is believed to allow for tunability in strength of the DNA binding interaction and subsequently chromatin residence time (46). Surprisingly, we also identified a novel consensus site in the de novo motif search, but only for the CTCF data set in which the input control was utilized (Supplementary Figure S3A). This observation was confirmed by the fact that the peaks containing this new motif were nearly all localized within the high-confidence peak population that was only identified after input treatment of the ChIP-exo data (Supplementary Figure S3B). Thus, identification of this novel motif was only possible when the ChIP-exo specific artifact peaks were removed prior to de novo motif analysis.
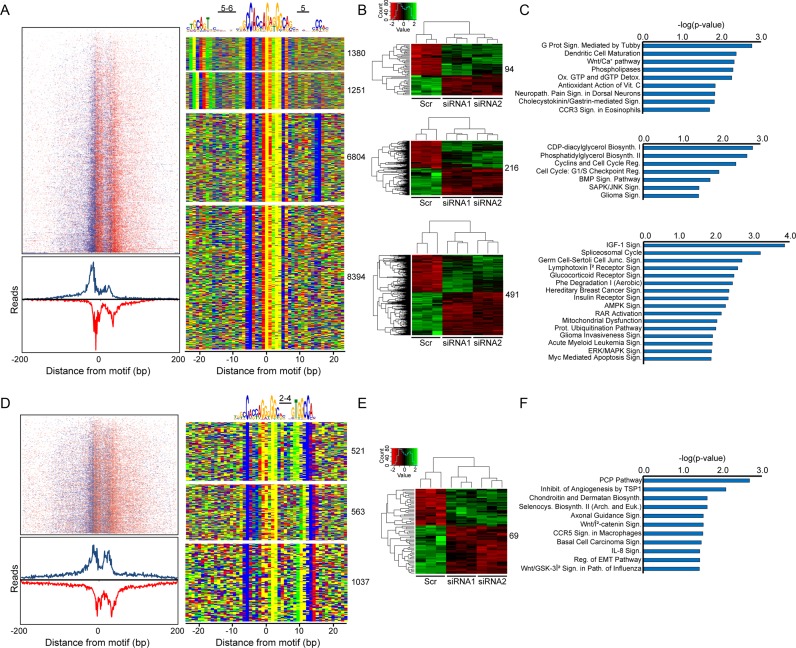
Next, a comprehensive weight matrix analysis was performed to identify all peaks containing the core CTCF sequence within the complete input treated CTCF ChIP-exo data set, which resulted in 19,950 hits. To validate that all sequences found in the de novo motif search (Supplementary Figure S3A) were localized proximal to the core CTCF site, a spaced motif analysis (SpaMo) was used (36). This search demonstrated that there was a significantly enriched spacing between the CTCF motif and the other two conserved sequences identified from the de novo search, including the new consensus site which was found to be localized 2–4 base pairs 3′ of the CTCF core (Figure 4A and D). In addition, a previously characterized flanking 5′-site to the CTCF core sequence was also found (15,46). Enrichment profiles depicting CTCF genomic localizations represented by both aggregate plots and heat maps (left panels in Figure 4A and D) also confirmed CTCF occupancy at all sites identified within the input treated ChIP-exo data set.
Figure 4.
From the input treated CTCF ChIP-exo data set, (A) read tag distributions around all genomic CTCF-bound sites shown in the four binned motif combinations (right panel) were centered on the midpoint of the CTCF consensus to generate a heat map (top left) which is summed below as an aggregate plot. Denoted in blue and red are the sense and antisense strand read enrichments around the core CTCF motif, respectively. The centralized CTCF core sequence and adjacent motifs are depicted above a color map representation of 50 bp DNA stretches containing the various motif combinations (right panel). (B) Heat maps from RNA-seq data depicting gene transcripts exhibiting a two-fold up- (green) or down-regulation (red) after CTCF depletion relative to the scrambled siRNA control (Scr). For each motif group, CTCF promoter occupation sites (defined as ±1000 bps around the transcription start site (TSS)) were intersected with the RNA-seq data and resulting altered gene sets were binned as individual heat maps. (C) Each gene set from (B) was subjected to Ingenuity Pathway Analysis (IPA, www.ingenuity.com) to identify biological pathways uniquely modulated by each of the CTCF motif combinations. (D–F) The same analyses in (A–C) were performed separately on the core CTCF consensus with the newly identified 3′-CTCF motif.
To determine the biological relevance of this newly identified 3′-flanking sequence, we analyzed RNA-seq data from CTCF depleted HeLa cells. CTCF depletion was confirmed by both immunoblot and qRT-PCR (Supplementary Figure S4). In the absence of CTCF, a substantial number of genes showed a significant transcriptional alteration relative to the control (Supplementary Figure S5). Independent intersection of these altered genes with the CTCF ChIP-exo data with and without input treatment showed that many more promoter proximal peaks overlapped within the untreated ChIP-exo data set (Supplementary Table S1). However, the number of peaks actually containing the core CTCF motif was essentially the same as the input treated data set. Thus, the percentage of real peaks containing the CTCF core motif localized within promoters of regulated genes was improved >30% after input treatment. This analysis highlights the importance and necessity for being able to remove technique-specific artifacts from ChIP-exo data sets and further demonstrates that our PAtCh-Cap derived ChIP-exo input control is able to perform this task remarkably well.
In the context of the input treated CTCF ChIP-exo data, analysis of the altered genes binned by the various motif combinations denoted in Figure 4A and D showed that there was a clear set of transcriptional alterations that were specific to each of these bins, including the core CTCF consensus in conjunction with the newly identified 3′-site (Figure 4B and E). As further evidence, analysis of the biological pathways associated with each of the binned gene sets demonstrated that several pathways were uniquely regulated by this newly identified extended CTCF motif (Figure 4C and F). While this analysis is not able to delineate direct versus indirect transcriptional regulation by CTCF, it does establish that CTCF occupation at sites containing the CTCF core in conjunction with the newly identified 3′-flanking sequence independently modulates the transcriptional outcome of certain genes.
Here, we have provided a simple and convenient method for generating technique-specific input controls that affords increased confidence in peak identification and the ability to perform de novo motif searches for ChIP-based methods that utilize additional bead-bound processing steps. We anticipate that the presented PAtCh-Cap input control strategy will remove a barrier preventing advanced bead-bound ChIP-based techniques such as ChIP-exo from becoming mainstream, allowing them to be accessible to a broader range of proteins and increasing the level at which we can interrogate interesting biological questions. Indeed, using our approach afforded identification of a novel binding motif for the very well characterized protein CTCF that appears to have an independent physiological function; the significance of which will require further investigation.
AVAILABILITY
Sequencing data are available within NCBI Gene Expression Omnibus under accession number GSE79565.
Supplementary Material
Acknowledgments
We thank B. Dalley for his assistance with the high-throughput sequencing, B. Milash, T. Mosbruger and D. Nix for their bioinformatics expertise. HeLa cells were a generous gift from Prof. C.J. Burrows (University of Utah).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Chemistry at the University of Utah; [RSG-14-185-01-DMC] from the American Cancer Society; National Cancer Institute for the Huntsman Cancer Institute core facilities [P30CA042014]. Funding for open access charge: University of Utah internal funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 2.Park P.J. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberstson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 4.Adli M., Bernstein B.E. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat. Protoc. 2011;6:1656–1668. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balakrishnan L., Milavetz B. Dual agarose magnetic (DAM) ChIP. BMC Res. Notes. 2009;2:250. doi: 10.1186/1756-0500-2-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakobsen J.S., Bagger F.O., Hasemann M.S., Schuster M.B., frank A.-K., Waage J., Vitting-Seerup K., Porse B.T. Amplification of pico-scale DNA mediated by bacerial carrier DNA for small-cell-number transcription factor ChIP-seq. BMC Genetics. 2014;16:46. doi: 10.1186/s12864-014-1195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S., et al. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachs M., Onodera C., Blaschke K., Ebata K.T., Song J.S., Ramalho-Santos M. Bivalent chromatin marks devlopment regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 2013;3:1777–1784. doi: 10.1016/j.celrep.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shankaranarayanan P., Mendoza-Parra M.-A., Walia M., Wang L., Li N., Trindade L.M., Gronemeyer H. Single-tube linear DNA amplification (LinDa) for robust ChIP-seq. Nat. Methods. 2011;8:565–567. doi: 10.1038/nmeth.1626. [DOI] [PubMed] [Google Scholar]
- 10.Shen J., Jiang D., Fu Y., Wu X., Guo H., Feng B., Pang Y., Streets A.M., Tang F., Huang Y. H3K4me3 epigenomic landscape derived from ChIP-seq of 1000 mouse early embryonic cells. Cell Res. 2015;25:143–147. doi: 10.1038/cr.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwart W., Koornstra R., Wesseling J., Rutgers E., Linn S., Carroll J.S. A carrier-assisted ChIP-seq method for estrogen receptor-chromatin interactions from breast cancer core needle biopsy samples. BMC Genomics. 2013;14:232. doi: 10.1186/1471-2164-14-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidl C., Renderio A.F., Sheffield N.C., Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods. 2015;12:963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallerman O., Nord H., Bysani M., Borghini L., Wadelius C. lobChIP: from cells to sequencing ready ChIP libraries in a single day. Epigenet. Chromatin. 2015;8:25. doi: 10.1186/s13072-015-0017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Q., Johnston J., Zeitlinger J. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat. Biotechnol. 2015;33:395–401. doi: 10.1038/nbt.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhee H.S., Pugh B.J. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skene P.J., Henikoff S. A simple method for generating high-resolution maps of genome-wide protein binding. eLife. 2015;4:e09225. doi: 10.7554/eLife.09225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll T.S., Liang Z., Salama R., Stark R., de Santiago I. Impact of artifact removal on ChIP quality metrics in ChIP-seq and ChIP-exo data. Front. Genet. 2014;5:1–11. doi: 10.3389/fgene.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., Negre N., Li Q., Mieczkowska J.O., Slattery M., Liu T., Zhang Y., Kim T.-K., He H.H., Zieba J., et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat. Methods. 2012;9:609–614. doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kidder B.l., Hu G., Zhao K. ChIP-Seq: technical considerations for obtaining high quality data. Nat. Immunol. 2013;12:918–922. doi: 10.1038/ni.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P., et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Gen. Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nix D.A., Courdy S.J., Boucher K.M. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523–531. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pepke S., Wold B., Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahony S., Pugh B.F. Protein-DNA binding in high-resolution. Crit. Rev. Biochem. Mol. Biol. 2015;50:269–283. doi: 10.3109/10409238.2015.1051505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho J.W.K., Bishop E., Karchenko P.V., Negre N., White K.P., Park P.J. ChIP-chIP versus ChIP-seq: Lessons for experimental design and data analysis. BMC Genomics. 2011;12:134. doi: 10.1186/1471-2164-12-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serandour A.A., Brown G.D., Cohen J.D., Carroll J.S. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol. 2013;14:R147. doi: 10.1186/gb-2013-14-12-r147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137.131–R137.139. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicol J.W., Helt G.A., Blanchard S.G., Jr, Raja A., Loraine A.E. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dreszer T.R., Karolchik D., Zweig A.S., Hinrichs A.S., Raney B.J., Kuhn R.M., Meyer L.R., Wong M., Sloan C.A., Rosenbloom K.R., et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medina-Rivera A., Defrance M., Sand O., Herrmann C., Castro-Mondragon J., Delerce J., Jaeger S., Blanchet C., Vincens P., Caron C., et al. RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 2015;43:W50–W56. doi: 10.1093/nar/gkv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas-Chollier M., Defrance M., Medina-Rivera A., Sand O., Herrmann C., Thieffry D., van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39:86–91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas-Chollier M., Sand O., Turatsinze J.V., Janky R., Defrance M., Vervisch E., Brohee S., van Helden J. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant C.E., Bailey T.L., Noble W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitington T., Frith M.C., Johnson J., Bailey T.L. Inferring transcription factor complexes from ChIP-seq data. Nucleic Acids Res. 2011;39:e98. doi: 10.1093/nar/gkr341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Machanick P., Bailey T.L. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kliszczak A.E., Rainey M.D., Harhen B., Boisvert F.M., Santocanale C. DNA mediated chromatin pull-down for the study of chromatin replication. Sci. Rep. 2011;1:95. doi: 10.1038/srep00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sirbu B., Couch F.B., Feigerle J.T., Bhaskara S., Hiebert S.W., Cortez D. Analysis of protein dynanmics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zlatanova J., Caiafa P. CTCF and its protein partners: divide and rule. J. Cell Sci. 2009;122:1275–1284. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]
- 41.Bailey T., Krajewski P., Ladunga I., Lefebvre C., Li Q., Liu T., Madrigal P., Taslim C., Zhang J. Practical guidelines for the comprehensive analysis of ChIP-seq data. PLoS Comput. Biol. 2013;9:e1003326. doi: 10.1371/journal.pcbi.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kharchenko P.V., Tolstorukov M.Y., Park P.J. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffman M.M., Ernst J., Wilder S.P., Kundaje A., Harris R.S., Libbrecht M., Giardine B., Ellenbogen P.M., Bilmes J.A., Birney E., et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albert I., Wachi S., Jiang C., Pugh F.B. GeneTrack — a genomic data processing and visualization framework. Bioinformatics. 2008;24:1305–1306. doi: 10.1093/bioinformatics/btn119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L., Chen J., Wang C., Uuskula-Reimand L., Chen K., Medina-Rivera A., Young E.J., Zimmermann M.T., Yan H., Sun Z., et al. MACE: model based analysis of ChIP-exo. Nucleic Acids Res. 2014;42:e156. doi: 10.1093/nar/gku846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakahashi H., Kwon K.-R.K., Resch W., Vian L., Dose M., Stavreva D., Hakim O., Pruett N., Nelson S., Yamane A., et al. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013;3:1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.