Abstract
The present review focuses on microbial type I fatty acid synthases (FASs), demonstrating their structural and functional diversity. Depending on their origin and biochemical function, multifunctional type I FAS proteins form dimers or hexamers with characteristic organization of their catalytic domains. A single polypeptide may contain one or more sets of the eight FAS component functions. Alternatively, these functions may split up into two different and mutually complementing subunits. Targeted inactivation of the individual yeast FAS acylation sites allowed us to define their roles during the overall catalytic process. In particular, their pronounced negative cooperativity is presumed to coordinate the FAS initiation and chain elongation reactions. Expression of the unlinked genes, FAS1 and FAS2, is in part constitutive and in part subject to repression by the phospholipid precursors inositol and choline. The interplay of the involved regulatory proteins, Rap1, Reb1, Abf1, Ino2/Ino4, Opi1, Sin3 and TFIIB, has been elucidated in considerable detail. Balanced levels of subunits α and β are ensured by an autoregulatory effect of FAS1 on FAS2 expression and by posttranslational degradation of excess FAS subunits. The functional specificity of type I FAS multienzymes usually requires the presence of multiple FAS systems within the same cell. De novo synthesis of long-chain fatty acids, mitochondrial fatty acid synthesis, acylation of certain secondary metabolites and coenzymes, fatty acid elongation, and the vast diversity of mycobacterial lipids each result from specific FAS activities. The microcompartmentalization of FAS activities in type I multienzymes may thus allow for both the controlled and concerted action of multiple FAS systems within the same cell.
INTRODUCTION
In contrast to their structural simplicity, the biological functions of fatty acids are impressively diverse. As constituents of neutral and polar lipids, as side chains in some coenzymes and secondary metabolites, as covalent attachments to distinct eucaryotic proteins, and as parts of eucaryotic second-messenger molecules, they fulfill central roles not only in biological energy storage or in the integrity and dynamics of biological membranes but also in the control of cellular metabolism and cell physiology. In accordance with this diversity of biological functions, the ability to synthesize fatty acids de novo is an elementary capacity of most cells. Even among archebacteria, which contain exclusively diphytanylglycerol diethers rather than diacylglycerides as membrane lipids, the synthesis of myristic acid for membrane protein acylation was discussed (26, 109). Thus, the enzyme system involved in de novo fatty acid synthesis, fatty acid synthase (FAS), is one of the household enzymes of the cell. Catalyzing a well-defined multistep reaction pathway, it soon became one of the paradigm multienzymes of enzyme biochemistry. Even though there is considerable variation in the molecular structures of FASs from different sources, the reaction mechanism of de novo fatty acid synthesis is essentially the same in all biological systems. The basic features of this pathway, i.e., its constituent chemical reactions and the identity of the respective component enzymes, were elucidated more than three decades ago in the laboratories of Lynen (81), Vagelos (167, 168), and Wakil (163), working with bacterial and yeast systems. According to the textbook reaction mechanism, fatty acid synthase is an “iterative” multienzyme, performing several successive cycles of a distinct reaction sequence in combination with a specific initiation and termination reaction at the beginning and end of the overall process, respectively. The individual FAS component enzymes are ac(et)yltransferase (AC), malonyl/acetyl- or malonyl/palmitoyl-transacylase (AT, MPT), ketoacyl synthase (KS) ketoacyl reductase (KR), dehydratase (DH), enoyl reductase (ER), acyl carrier protein (ACP), and thioesterase (TE). As discussed below, the occurrence of AT and TE activities, on the one hand, and of MPT, on the other, is mutually exclusive and depends on the particular FAS system.
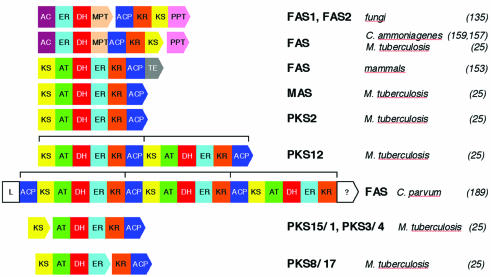
During evolution, a number of functionally differentiated FAS variants as well as a large family of FAS-related enzymes have developed which produce, by slight variation of the FAS pathway, a broad spectrum of natural compounds. FAS structural variants may be assigned to three general classes. The first class is represented by the dissociated type II FAS systems, which occur in most bacteria as well as in the organelles of procaryotic descent, i.e., mitochondria and chloroplasts. The component enzymes of type II fatty acid synthases are independent proteins which are encoded by a series of separate genes. These enzymes are contrasted by the highly integrated type I FAS multienzymes, which contain the various catalytic activities of the reaction sequence as discrete functional domains, either on a single polypeptide chain or, in some cases, on two different multifunctional proteins of comparable size. Type I FAS multienzymes are characteristically found in the eucaryotic cytoplasm (82, 111, 135) and, as a remarkable procaryotic exception, also among the mycolic acid producing subgroup of the Actinomycetales (13, 144). The type I systems may be further subdivided according to the domain organization of the multifunctional proteins and, concomitantly, according to their subunit stoichiometry. Microbial type I FASs are hexamers with a domain sequence of AC-ER-DH-MPT/ACP-KR-KS forming either α6β6 (fungi) or α6 (bacteria) oligomers (type Ia). In contrast, animal FASs are α2 dimers with the domain sequence KS-AT-DH-ER-KR-ACP-TE (type Ib). Occasionally, more than one set of FAS domains may be fused to a multimodular synthase. For instance, the pks12 gene of Mycobacterium tuberculosis (25, 149) has two complete sets of FAS domains, and the FAS gene of the parasitic protist Cryptosporidium parvum (189) has three (Fig. 1). Apart from these structural differences, type I FASs may also be functionally differentiated on the basis of to various parameters which are discussed in more detail, below Furthermore, individual FASs may also differ by their specific cellular compartmentation being localized not only in the cytoplasm but also in organelles (16, 145) and microsomal membranes (45, 114). Occasionally, they are also found associated with subcellular structures, which subsequently serve as acceptors of their reaction products (39, 40, 42, 166, 174).
FIG. 1.
Domain organization of known type I FASs and related multienzymes. Arrows indicate open reading frames. Their subdivision into functional domains is not shown to scale. With the exception of FAS1 and FAS2, the indicated gene pairs are chromosomally linked in tandem orientation. The indicated PKS combinations encode putative heteromeric multienzymes comprising a complete set of FAS domains. Brackets indicate intramolecular FAS modules. L, acyl-CoA ligase. Adapted from references 25 and 93.
FASs may be considered the forebears of most members of the large family of polyketide synthases (PKSs). PKS and FAS systems contain basically the same set of component enzymes. However, in contrast to FASs, the typical PKS pathways are not “iterative” reaction sequences. Instead, they catalyze one or several rounds of FAS-like reaction sequences, each specifically missing one or even more of the canonical FAS reactions. Usually, different enzyme combinations are used in successive PKS cycles ranging from complete to more or less incomplete FAS sequences (51). Thus, polyketides retain, at distinct positions, the characteristic functional groups of certain FAS intermediates such as keto groups, hydroxyl groups, or double bonds. The distinction between FAS and PKS systems, in this review, therefore refers to the ability of an enzyme to produce long-chain saturated or monounsaturated fatty acids. Furthermore, this review is restricted to microbial type I FAS systems, focusing on their molecular structure and function, the redundancy of cellular FAS systems, the regulation of FAS biosynthesis, and the specific physiological roles of some FAS products. For the discussion of related systems such as animal type I FAS (152, 174), bacterial (104, 113) and plant (49, 104) type II FAS, or type I and II PKS-(51) systems, the reader is referred to several excellent and comprehensive reviews which have recently appeared elsewhere.
GENERAL PRINCIPLES AND FUNCTIONAL DIVERSITY IN BIOLOGICAL FATTY ACID SYNTHESIS
The detailed reaction mechanisms of paradigm FASs from bacterial (112), fungal (82), plant (49, 104), or animal (111, 152) sources have been published elsewhere and are not described here. Instead, a short overview focusing on the functional diversity of naturally occurring FAS systems emphasizes some specific facets of type I FASs which are discussed in more detail later in this review.
FAS Reaction Mechanism
Fatty acid biosynthesis is initiated by the FAS component enzyme acetyltransferase, loading the acyl primer, usually acetate, from coenzyme A (CoA) to a specific binding site on FAS. At the end of the process, termination of chain elongation occurs by removing the product from FAS either by transesterification to an appropriate acceptor or by hydrolysis. The respective enzymes are usually palmitoyl transferase and thioesterase. The reaction sequence between initiation and termination involves the elongation of enzyme-bound intermediates by several iterative cycles of a distinct set of reaction steps. Each cycle includes malonyl-transacylation from CoA to the enzyme by malonyl transferase condensation of acyl-enzyme with enzyme-bound malonate to 3-ketoacyl-enzyme by 3-ketoacyl synthase, reduction of the 3-keto- to the 3-hydroxyacyl intermediate by ketoacyl reductase, dehydration of 3-hydroxyacyl enzyme to 2,3-trans-enoate by dehydratase, and, finally, reduction of the enoate to the saturated acyl-enzyme by enoyl reductase. A central role in substrate binding, processing of intermediates, and communicating of intermediates between the various catalytic centers of FAS is played by the prosthetic group, 4′-phosphopantetheine. This cofactor is covalently bound to a specific serine hydroxyl group of the ACP domain or, depending on the FAS system, to the ACP component of FAS. In some bacteria, the iterative sequence of elongation cycles may be interrupted, at a chain length of 10 carbons, by one cycle involving an intrinsic isomerase converting the 2-trans- into the 3-cis-decenoyl intermediate, which is subsequently not reduced but further elongated to long-chain monounsaturated fatty acids (86, 112).
Primer and Chain Extender Specificities
Even though in most organisms acetyl-CoA represents the preferred primer and C16 to C18 fatty acids are the main products of de novo fatty acid synthesis, the FAS initiation and termination reactions nevertheless may vary considerably, depending on the particular organism or tissue systems. For instance, several bacterial species synthesize terminally branched iso-, anteiso- or omega-alicyclic fatty acids from branched, short-chain carboxylic acid primers (57, 58). Furthermore, Corynebacterium ammoniagenes type I FAS is capable to utilize, besides acetyl-CoA, longer-chain acyl-CoAs up to a length of 10 carbons as primers (61). Conversely, for yeast FAS, nonanoyl-CoA and its higher homologous are also used, with low efficiency and within a limited concentration range, as FAS primers (107). For mammalian FAS, butyryl-CoA represents even a more efficient primer than acetyl-CoA (13, 79). The mycobacterial type II FAS exclusively uses long-chain (C16 to C24) primers for synthesizing the extraordinarily long (C50 to C60) meromycolic acids (120, 151). In the same class of bacteria, the FAS-like mycerosic and phthioceranic acid synthases elongate C12 to C20 fatty acyl primers to C22 to C34 carbon chains (110, 148). Similarly, the FAS-like elongation systems of yeast, and probably also those of other eucaryotes, elongate C12 to C18 acyl-CoA primers to C18 to C26 very-long-chain fatty acids (VLCFAs) (114). A rather unusual primer, benzoyl-CoA, is presumed to start phenolphthiocerol biosynthesis by a FAS-like multienzyme in pathogenic mycobacteria (8). In vitro, fatty acid synthesis is often initiated even in the absence of any externally added primer. Although primerless fatty acid synthesis is slow with most FAS preparations, it is very efficient with C. ammoniagenes FAS (3, 144). This capacity is attributed to the inherent malonyl-decarboxylase activity of FAS being part of the ketoacyl synthase component (71). Malonyl decarboxylation generates the highly reactive acetyl thioester carbanion, which subsequently undergoes Claisen condensation with the saturated acyl-enzyme thioester (5). In contrast to the high spontaneous activity of C. ammoniagenes FAS, the malonyl decarboxylase of most other FASs is stimulated by several orders of magnitude only on acylation of the catalytically reactive cysteine of ketoacyl synthase. Carboxymethylation of this cysteine in yeast FAS (71) or its replacement by glutamine in the animal enzyme (153) were reported to drastically stimulate spontaneous malonyl-decarboxylation too. These modifications were presumed to structurally mimic the acylated cysteine within the KS catalytic center.
Apart from the diversity of the eventually used primers, variation of the extender substrate is also possible. For instance, several FASs also accept methylmalonyl-CoA instead of malonyl-CoA as a substrate. Thereby, (multi)methyl-branched fatty acids are produced. For instance, mycobacterial mycocerosic and phthioceranic acid synthases are FAS-like enzymes which use exclusively methylmalonyl-CoA for chain extension (110, 148). In contrast, animal FAS uses methylmalonyl-CoA only in certain tissues, such as the uropygial glands of birds, as an alternative substrate under conditions of malonyl-CoA limitation (19, 64). Here, an organ- and substrate-specific and FAS-independent malonyl-CoA decarboxylase selectively lowers the malonyl-CoA level and thereby restricts fatty acid synthesis to the available methylmalonyl-CoA. In other tissues of the same organisms, animal FAS synthesizes straight-chain fatty acids according to its usual preference for malonyl-CoA (19).
Product Release and Acyl Acceptors
An important characteristic of every FAS is the specificity of its chain termination reaction, which determines both the chain length and the acceptor of the FAS product. In Escherichia coli, long-chain fatty acids are transacylated by specific glycerol phosphate transacylases from acyl-ACP directly to the membrane phospholipids. Alternatively, certain shorter-chain intermediates may be diverted from the elongation process for other reactions such as lipopolysaccharide or coenzyme biosyntheses (for a review, see reference 112). The type I FASs of yeast, mycobacteria, corynebacteria, and Euglena use an integral palmitoyl transferase activity for transacylation of palmitate from the enzyme to CoA (13, 82,144). Animal FAS, on the other hand, releases its products as free fatty acids after hydrolysis by an intrinsic thioesterase (111, 153). During sterigmatocystin or aflatoxin biosynthesis in Aspergillus species, a specific fatty acid synthase (sFAS) is presumed to produce enzyme-bound hexanoic acid, which is subsequently directly transferred to an associated poyketide synthase, NorS, to start the remaining part of the toxin biosynthetic pathway (174, 175). Similarly, mycocerosic acid synthesized by mycobacterial mycocerosic acid synthase appears to be directly transesterified to its final acceptor, phthiocerol, without intermediate release of acyl-CoA (174). As reported by Ueno (166), a putative FAS preparation from Bombyx mori, as well as a homologous, embryonic mouse FAS, exhibited distinct protein palmitoylation activites. As discussed previously by Sumper et al. (161) and by Bloch and Vance (13), the enzymes involved in the chain-terminating transacylation reactions compete with β-ketoacyl synthase for enzyme-bound acyl-chains as substrates at the end of the elongation cycle. The relative activities and/or substrate affinities of the competing enzymes are presumed to alter on chain elongation in opposite directions. Thus, deacylation will finally prevail and overcome elongation.
Chain Length Determination
The chain length of its products is probably an inherent property of every FAS, even though the structural basis for this characteristic continues to be elusive. Depending on the particular organism and FAS system, the chain lengths of FAS products may vary over a wide range. For instance, hexanoic and octanoic acids are specifically provided for the pathways of aflatoxin (174, 175) and lipoic acid (16,97, 169) synthesis, respectively. In African trypanosomes, massive amounts of 14:0 are synthesized by a type II system and used for remodeling the glycosylphosphatidylinositol anchors of their surface glycoproteins (96). The vast majority of FAS products being incorporated into the phospholipids of biological membranes comprise 16- to 18-carbon fatty acids together with a small proportion (<5%) of 26- to 30-carbon VLCFAs in eucaryotes (21, 92, 176). Mycobacterial meromycolic acid, on the other hand, extend to lengths of 50 or more carbon atoms. In mycobacteria (13) and in a strain of Vibrio (95), the respective type I and II FAS systems exhibit bimodal product patterns with chain length maxima of 16 to 18 and 24 to 30 carbon atoms. Apart from intrinsic determinants of the FAS proteins, external factors may interfere with the elongation process too. For instance, FAS-independent and short-chain-specific thioesterases are responsible for pre-early-chain termination and medium-chain-length fatty acid production in some specialized animal tissues such as the lactating mammary gland, several sebaceous glands (for a review, see reference 111), and the oil seeds of certain plants (78, 108). In vitro, acyl-CoA binding substances such as bovine serum albunim (BSA) or certain mycobacterial polysaccharides were capable of dramatically shifting the FAS product pattern of mycobacterial type I FAS toward shorter-chain acids (13). With yeast FAS, a similar effect was observed on addition of acyl-CoA binding protein (ACBP) to the assay mixture, causing a dramatic decrease in the chain length of acyl-CoA reaction products (121). If BSA is omitted from the assay mixture, the in vitro products of yeast FAS are mainly 18- to 20-carbon fatty acids, while in the presence of BSA, 14- to 18-carbon chains are produced (82). The synthesis of 14- to 18-carbon fatty acids in vivo is therefore presumed to result from interaction of FAS with ACBP or another functionally related factor.
MOLECULAR STRUCTURE AND REACTIVITY OF YEAST FATTY ACID SYNTHASES
The pathway of de novo fatty acid synthesis in yeast, together with the enzyme system involved, has been under extensive investigation for more than three decades. Today, FASs have been isolated from a variety of yeasts and fungi (35, 50, 56, 59, 60, 81, 84, 88, 90, 100, 105), but Saccharomyces cerevisiae FAS still represents the archetype of this class of enzymes and, hence, is used here to demonstrate the properties of fungal FASs in general. The general topics of fatty acid biosynthesis are well established and have been repeatedly reviewed (49, 82, 104, 111, 112, 167, 172). Therefore, they are not treated in detail here. In a review specializing in type I FASs, it may be appropriate to address primarily the aspects which are specifically related to the integrated structure of these enzymes. Other than in E. coli, where ACP is an abundant cellular protein (112), only a single ACP domain is available, in type I FASs, to the various component enzymes of the FAS elongation cycle. Thus, distinct structural constraints must ensure not only the accessibility of ACP-bound intermediates to each of these activities but also the coordinate interaction of chain extension and acyl modification reactions. Besides this, the integrated structure of type I FAS multienzymes provides an opportunity of long- or short-range conformational changes induced by the more than 30 different substrates, intermediates, and products which are covalently bound to the enzyme in the course of the synthetic process. Conformational changes may in fact be considered one of the crucial parameters controlling the dynamics of type I FAS reactivity. To understand the topology and interaction of catalytic sites within the FAS multienzyme at the molecular level, the tertiary structure of yeast FAS and, hence, its molecular and functional architecture have always been of primary interest. Apart from an early report by Oesterhelt et al. (102), however, on the crystallization of yeast FAS, which unfortunately proved unsuited for crystallographic analysis, yeast FAS has been refractory to crystallization ever since. There is no satisfactory explanation for this technical problem. Possibly, the multitude of acyl binding sites and their acylation by a wealth of possible intermediates, even in purified FAS, introduces a structural microheterogeneity which impedes crystallization.
FAS Catalytic Centers
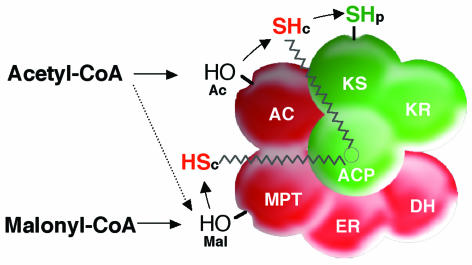
Our present knowledge about the various substrate binding sites and their interaction in yeast FAS is still based on results originally obtained in Lynen's laboratory (7, 37, 70, 82, 124, 137, 190), characterizing acylated peptides which had been obtained on incubation of FAS with radioactively labeled acetyl-, palmitoyl- or malonyl-CoA. Originally, kinetic studies using N-ethylmaleimide and iodoacetamide as specific inhibitors of FAS activity, together with substrate competition experiments, had led to the identification of two chemically different reactive thiols in yeast FAS. These were defined as “central” SH-group (SHc), represented by ACP-bound phosphopantetheine, and “peripheral” SH-group (SHp), which belongs to a specific cysteine within the ketoacyl synthase catalytic domain (70, 101, 190). In addition, two specific serine residues in the acetyl- and malonyl-/palmitoyl-transferase domains, respectively, represent nonthiol acylation sites which transiently bind substrates or products during their transacylation between CoA and the reactive thiol groups, SHc and SHp. From acyl binding and FAS inhibition studies, Lynen and coworkers developed a hypothetical scheme of FAS acylation, intraenzyme transacylation, and subsequent condensation reactions involving the two thiol and the two nonthiol acylation sites on yeast FAS (Fig. 2), (82). According to this scheme, acetate is transferred from CoA to SHp in a three-step process with the acetyl-transacylase hydroxyl-group and the ACP thiol group (SHc) as transient acetylation sites. Other than acetate, malonate is excluded from SHp and directed exclusively to SHc. Subsequently, acetyl-Sp and malonyl-Sc thioesters condense to enzyme-bound acetoacetate, thereby initiating the elongation cycle. Isolation and sequencing of the two S. cerevisiae FAS genes, FAS1 and FAS2, subsequently confirmed the chemical structure and location of the two thiol and two hydroxyl acylation sites in yeast FAS (23, 67, 94, 138, 141, 142). SHp was identified as Cys1305 in the KS domain of FAS2. The SHc-bearing phosphopantetheine is bound to Ser180 of FAS2. The acetyl- and malonyl-/palmitoyl-transacylation sites correspond to Ser819 and Ser5421 of the respective transferases in FAS1 (158). By deletion mapping, five of the catalytic FAS domains were assigned to FAS1-encoded subunit β in the following order. AC, ER, DH, and MPT. Accordingly, the FAS2-encoded subunit α contained the remaining domains in the order ACP, KR, and KS. As discussed below, the reading frame of the apoFAS activating phosphopantetheine transferase (PPT) was recently localized, as C-terminal domain, within the FAS2 reading frame (41).
FIG. 2.
Specificity of acyl binding sites and interdomain transacylation reactions in yeast FAS. Subunits α (green) and β (red) are marked by different colors. Functional domains are arranged according to the reaction sequence. The zigzag line indicates the “swinging arm” of ACP-bound phosphopantetheine. Abbreviations are defined in the text.
Specificity and Interaction of Substrate Binding Sites
Based on the above-described knowledge of FAS acylation sites, we replaced, in separate experiments, the four substrate binding amino acids of yeast FAS by the functionally inert amino acids glycine, alanine, and glutamine, respectively. The mutated FAS proteins were isolated and analyzed for their substrate binding capacities on incubation with the radioactively labeled substrate acetyl-CoA or malonyl-CoA. Protein-bound acyl groups were differentiated by performic acid oxidation as O-ester or thioester linkages. The results indicated that the SHc of ACP-bound pantetheine is in fact acylated by both acetate and malonate, as was suggested from the work of Lynen and coworkers (82, 131, 190). Similarly, the SHp proved as a specific acetyl binding site. Surprisingly and in contrast to the model of Lynen and coworkers (190), however, only Ser819 proved as specific and reacted exclusively with acetate, while Ser5421 was acylated by both acetate and malonate. Thus, the access of malonate to the enzyme depends strictly on Ser 5421 but acetate may enter the enzyme by both the malonyl and acctyl transacylation domains on subunit β (131). These findings are in accordance with the characteristics of AT mutants, which were consistently leaky and exhibited, in contrast to mutants with mutations of other FAS functions, a considerable amount of residual FAS activity (10 to 20%) (140). According to its acylation characteristics, the MPT domain of yeast FAS is actually a malonyl-/palmityl/-acetyl transacylation site and thus resembles, to some extent, the acetyl-/malonyl transacylation domain of animal FAS (111). Nevertheless, the yeast MPT domain substitutes only in part, not completely, for the AT function. These data support earlier results obtained by Pirson et al. (107), reporting on the competition of decanoyl and malonyl residues for the same binding site. This competition increased with the chain length of the decanoyl homologues and finally limited chain growth by displacing malonate from the enzyme. The various possible transacylation routes of yeast FAS are summarized in Fig. 2.
The observed specificities of yeast FAS acylation ensure that the priming substrate, acetate, enters the enzyme only at the beginning of the process. During chain elongation, however, the access of acetate to SHp is prevented by the occupation of SHc by either malonate or fatty acyl intermediates. Since exclusively long-chain saturated fatty acids are produced by yeast FAS, it is obvious that new malonate is excluded from SHc during a particular elongation cycle, i.e., as long as SHc-bound acyl intermediates are not yet fully reduced. Thus, both acylation of SHc by FAS intermediates and inhibition of malonyl transacylation by long-chain acyl products control FAS malonylation and thus the progress of chain elongation. For animal FAS, inhibition of FAS internal transacylation between SHc and SHp by longer-chain products has been demonstrated by Rangan and Smith (111). In contrast to the animal enzyme, however, release of end products from the FAS protein is not a chain-length-specific reaction in yeast. In vitro, the palmitoyl transferase of purified yeast FAS exhibited comparable specific activities to those of acyl-CoA substrates between 6 and 18 carbon atoms long (139). Even though chain elongation is, in a complete system, not reinitiated before a particular elongation cycle is finished, Yalpani et al. (184) observed that under conditions of NADPH limitation, the ketoacid intermediate does in fact condense with additional malonate and thus is obviously relocated from SHc to SHp. Thereby, the triketide triacetolactone, rather than a saturated fatty acid, is produced.
Negative Cooperativity
Using wild-type FAS as well as distinct acylation site-defective FAS mutants, the kinetics of binding of acetate and malonate to yeast FAS were investigated quantitatively. Determining the molar amounts of substrate bound per mole of enzyme, it was found that neither acetate nor malonate binds stoichiometrically, even under saturation conditions, to any of the four acylation sites of yeast FAS. Instead, only 2 to 3 rather than 12 mol of malonate and 6 to 7 rather than 24 mol of acetate were covalently bound by 1 mol of hexameric FAS (131). In accordance with the relative stabilities of O and thioester linkages, 20 to 30% of the malonyl enzyme and 35 to 50% of the acetyl enzyme occurred as performic acid-labile thioesters. Complete acylation of yeast FAS was achieved when the equilibrium of the reaction
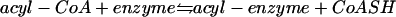 |
was shifted to the product side on addition of N-ethylmaleimide as a CoASH-trapping agent (131). Thus, all substrate binding sites of FAS, rather than only some of them, are actually available to acylation. These data are at variance with those of Wakil and coworkers (156), who reported on the binding of 3 mol of acetate per αβ FAS protomer when using p-nitrothiophenylacetate as a model substrate. According to Rangan and Smith (111), however, substoichiometric substrate binding is also observed with animal FAS. Possibly, negative cooperativity represents a general characteristic of type I FASs and is essential for their particular reaction mechanism. The capacity to exert negative cooperativety may in fact be one of the reasons for the persistently oligomeric structure of all known type I FAS proteins. Originally, the dimerization of identical subunits in an antiparallel side-by-side arrangement was considered a structural prerequisite for typeI FAS activity (134, 153, 155). It was presumed that only the oppositely oriented dimers allow the interaction, in trans, of catalytic domains which are located at distal ends of the same subunit and which, therefore, cannot interact directly. The finding by Smith et al. (153) that dissociation renders the animal FAS dimer enzymatically inactive supported this conclusion. However, recent results by these authors showing that heterodimeric animal FASs bear different mutations on each subunit led to a revision of this model (see references 53, 76, and 153 for reviews). It was observed that heterodimeric animal FAS consisting of one wild-type subunit and a second subunit compromised in all seven of its catalytic domains was nevertheless capable of palmitate synthesis. Thus, functional interaction of all catalytic domains within the same subunit is in fact possible. Therefore, oligomerization may be required primarily for stabilization of the catalytically active conformation of animal FASs rather than for a half-of-the-sites reaction mechanism. The negative cooperativity observed with both yeast and animal FASs ensures that the majority of acyl binding sites in the oligomer remain empty and thus available to the intermediates of chain elongation rather than being blocked by the substrates acetate or malonate. Other than with acetyl-CoA or malonyl-CoA alone, the various binding sites are exhausively acylated on incubation of yeast FAS with the complete reaction mixture under steady-state conditions of fatty acid synthesis (131, 147). This demonstrates, for example, that all binding sites are in fact available for acylation.
Even though the individual αβ monomer of yeast FAS is possibly capable of palmitic acid synthesis, cooperation between both identical and nonidentical subunits within the α6β6 oligomer has in fact been demonstrated. Genetic complementation studies with yeast mutants which were specifically defective in one of the various FAS domains revealed that overall FAS activity was always restored, both in vitro and in vivo, whenever two mutations which affected two different catalytic activities were combined (74, 140, 178). For complementation, it was irrelevant whether the affected domains were located on the same or on two different subunits (140, 178). Thus, every catalytic site of yeast FAS is obviously capable of interacting with any other site, at least within an α2β2 FAS subcomplex. The various phosphopantetheine “arms” within the oligomer may be instrumental for this cooperation. It should be mentioned that in contrast to these characteristics of yeast FAS, interallelic complementation within dimeric animal FAS is not generally observed but is subjected to certain restrictions (153).
Phosphopantetheinylation of apoFAS
In all FASs, the ACP domain requires posttranslational attachment of the prosthetic group, 4′-phosphopantetheine. By the action of a distinct enzyme, PPT, phosphopantetheine is transferred from CoA to a specific serine hydroxyl group of ACP (36). Depending on their origin, the degree of substrate specificity varies considerably among different PPTs (54, 75). In yeast, each of the three known phosphopantetheine-containing proteins, i.e., cytoplasmic FAS, mitochondrial ACP, and α-aminoadipate semialdehyde dehydrogenase, is activated by its own specific PPT (41, 160). Recently, several lines of evidence, such as sequence alignment studies, isolation of pantetheine-deficient FAS mutants, and in vitro autoactivation of recombinant apoFAS, indicated that the cytoplasmic FAS protein of yeast is pantetheinylated by a PPT activity which is integrated in the FAS complex rather than being an independent cellular protein (41, 136). On the basis of to its significant sequence similarity to several known PPT enzymes and the specific loss of FAS pantetheinylation in the respective mutants, the PPT domain of yeast FAS was localized at the C terminus of FAS2. For efficient autoactivation of recombinant yeast apoFAS in vitro, the presence of Mg2+ ions and CoA was both necessary and sufficient (75). Even though the genes of PPT and its cognate apoproteins are often closely linked, their fusion is observed only with the FAS2 genes of yeast and all other fungi so far investigated.
MULTIPLE FATTY ACID SYNTHASES IN YEAST AND OTHER FUNGI
Mitochondrial Fatty Acid Synthesis
In organisms other than green plants, the FAS responsible for bulk fatty acid synthesis is a soluble cytoplasmic enzyme. In addition to this main source of cellular fatty acids, other FAS systems occur in eucaryotes, serving a variety of specialized functions. For instance, mitochondria from various sources such as fungi (16, 91), plants (46), and animals (188) contain their own organellar FAS, which is structurally and functionally independent of the cytoplasmic system. Mitochondrial fatty acid synthesis and the involvement of a type II ACP in this process were originally observed by Brody and coworkers in Neurospora crassa (15, 91). Subsequently, it was demonstrated by several groups (116, 117, 187) that this typical component of a dissociated, bacterial type of FAS represents, in Neurospora and mammalian mitochondria, one of the subunits of respiratory chain complex I. Independently, an S. cerevisiae nuclear gene, CEM1, with significant sequence similarity at the amino acid level to bacterial Ketoacyl synthase and, hence, another component of a hypothetical mitochondrial type II FAS, had been identified by Slonimski and coworkers (48). The screening of the S. cerevisiae genome by different groups finally led to the identification of essentially all the components of a bacterial type II FAS encoded by nuclear genes but obviously located in the mitochondrial compartment (16, 122, 164). Discovering that in Neurospora and mammals, one of the central components of prokaryotic FAS, i.e., ACP, was part of the respiratory chain was rather puzzling at first. However, in yeast which is void of respiratory complex I, ACP is an idependent mitochondrial protein. Hence, its association with the respiratory chain in some organisms appears to be more accidental than functionally relevant.
A cell-free system prepared from null mutants of yeast cytoplasmic FAS incorporates radioactively labeled malonyl-CoA into long-chain fatty acids at about 2% of the wild-type rate (114). This activity is sensitive to cerulenin inhibition and is abolished on deletion of the nuclear gene, ACP1, which encodes mitochondrial ACP (114). Hence, this FAS I-independent fatty acid synthesis in yeast was interpreted as mitochondrial FAS activity. The products of mitochondrial FAS in yeast, Neurospora, and plants were reported to be medium and long-chain fatty acids of 8 to 18 carbon atoms (46, 91, 114). Therefore, the mitochondrial and cytoplasmic FAS systems appear to be partially redundant functionally. Nevertheless, the organellar system is obviously unable to compensate for the loss of cytoplasmic FAS in fas1 or fas2 yeast mutants since the phenotype of the mutant is not leaky. Either the low efficiency or the organellar compartmentation of mitochondrial FAS may be responsible for this failure. At present, convincing evidence for an eventual function of mitochondrially made long-chain fatty acids is still missing. A possible role in remodeling of mitochondrial membrane lipids has been proposed (122). Apart from the long-chain products, however, it became obvious that in yeast (16) and plant (46) systems, mitochondrially made octanoic acid serves as a precursor of mitochondrial lipoic acid synthesis. ACP1-defective yeast mutants contained 10 to 20 times less lipoic acid than did wild-type cells (16). The importance of lipoate-dependent α-ketoacid dehydrogenases for the functioning of the tricarboxylic acid cycle is in agreement with the inability of yeast mutants defective in whatever component of mitochondrial FAS to grow on the nonfermentable carbon sources lactate and glycerol (16, 116, 122, 160). In contrast to bacteria, yeast cells are not able to effectively incorporate external lipoic acid, nor does cytoplasmic FAS produce sufficient amounts of octanoic acid to fulfill the need for cellular lipoate synthesis. For this reason, the mitochondrial FAS exhibiting appropriate product spectra and intracellular localization may have been conserved during evolution of the endosymbiont.
Plant mitochondria were shown to contain both malonyl-CoA synthetase and malonyl-CoA:ACP transacylase (46). Furthermore, mitochondrial malonate was thought to originate from cytoplasmic acetyl-CoA carboxylation and subsequent importation by the mitochondrial dicarboxylic acid carrier into the organelle (46). Thus, the joint activities of malonyl-CoA synthetase and malonyl-CoA:ACP transacylase may provide the malonyl-ACP required for mitochondrial fatty acid synthesis. In yeast mitochondria, however, malonyl-CoA appears to be synthesized by an alternative route, using a specific, organellar acetyl-CoA carboxylase. According to recent data from our laboratory, mitochondrial acetyl-CoA carboxylase appears to be encoded by the nuclear gene, HFA1, and closely resembles, in both its molecular mass and amino acid sequence, the cytoplasmic acetyl-CoA carboxylase, Accl. HFA1 mutants are lactate negative and lipoic acid deficient and thus exhibit the same phenotype as mitochondrial FAS mutants. The suggested role of HFA1 is further supported by the finding that the Hfa1 protein exhibits acetyl-CoA carboxylase activity in vitro (E. Schweizer, unpublished data).
Fatty Acid Elongases
The vast majority of cellular fatty acids have chain lengths between 14 and 18 carbon atoms. As well as this, a small proportion (0.5 to 3% in wild-type yeast) of VLCFA with 20 or more carbon atoms are characteristic components of all eucaryotic membranes (21, 30, 92, 176). Yeast VLCFAs have chain lengths of 24 to 26 carbon atoms and are usually attached by amide likages to the sphingosine backbone of sphingolipids (for a review, see reference 27). In spite of their very low cellular concentrations, VLCFAs are, for reasons which are not fully understood, essential for cell viability (65, 123). The VLCFA-synthesizing enzyme systems are designated fatty acid elongases rather than as FASs. Nevertheless, mechanistically they may represent FASs that use long-chain fatty acyl-CoA rather than acetyl-CoA as a primer. Similar to FASs, elongases use malonyl-CoA and NADPH as extender and reducing substrates, respectively (30, 114). FASs and elongases are considered to catalyze homologous reaction sequences utilizing a functionally homologous set of component enzymes. As a possible exception and mechanistic pecularity, however, it remains to be demonstrated whether, like FASs, elongases use enzyme-bound phosphopantetheine for the malonyl-CoA dependent condensation reaction. So far, evidence is missing for the presence of an additional functional yet unassigned pantetheinylated protein in yeast. As is known from the chalcon and stilben synthases of plants, pantetheine-independent malonyl-CoA condensations have evolved in some systems independently of the usual FAS and PKS systems (72). In agreement with this view and with the characteristics of chalcon synthesis, yeast elongase is insensitive to the FAS ketoacyl synthase inhibitor cerulenin (114). As a further difference from most FAS systems, elongases are not soluble cytoplasmic enzymes but are localized in the microsomal membrane fraction. Even though the purification and molecular characterization of a distinct elongase system has not been achieved so far, available data suggest that elongases have a nonaggregated molecular structure consisting of physically independent enzyme entities. By genetic inactivation and subsequent isolation, two putative yeast elongase genes, YBR159w and TSC13, have recently been identified. They encode, respectively, an elongase-specific β-ketoacyl reductase and an enoyl reductase (10, 47, 65, 114). Using a specific in vitro elongase assay, at least three different elongase systems have been identified in yeast. They may be differentiated according to their differential primer usage and product specificities: -elongase I extends C12 to C16 primers to C16 to C18 fatty acids, elongase II converts C16 to C18 acyl-CoAs to C22 fatty acids, and elongase III synthesizes C24 to C26 fatty acids from C18-CoA (114). Elongase I has no function in VLCFA synthesis but probably serves to extend medium-chain-length fatty acids which eventually result from pre-early-chain termination of cytoplasmic FASs. For each elongation system, one of the three closely related yeast genes, ELO1, ELO2, and ELO3, fulfills an essential although presently still elusive function (30, 103, 114, 162). Consequently, elo1, elo2, and elo3 mutants are specifically defective in one of the elongase I to III. Elongases II and III share some of their components, such as β-ketoacyl reductase and enoyl reductase (114, 47). The presence of redundant elongases or, alternatively, the lethality of their functional loss prevented the isolation of yeast elongase mutants for a long time. In contrast to the fatty acid requirement of yeast FAS mutants, elongase-defective mutants cannot be supplemented by the elongase reaction products, i.e., external VLCFAs. Recently, however, elo1 mutants have been isolated based on the failure of elo1/fas double mutants to elongate and, consequently, to use 12:0 as a fatty acid supplement (30, 162). In contrast, ELO1-positive fas mutants readily convert 12:0 to the essential C14 to C18 acids. The enoyl reductase and ketoacyl reductase components of elongases II and III were identified on the basis of the abnormal sphingolipid composition of a respective mutant (TSC13) (65) and by an extensive database search with subsequent biochemical characterization of candidate mutants (YBR159w) (47, 114). Elongase II and III mutants are viable even though they are VLCFA negative in vitro (114). In contrast to these in vitro characteristics, VLCFA synthesis is reduced but not absent in vivo (47, 114). The VLCFA level in the ybr159wΔ mutant was 20 to 30% of that in the wild type. Thus, an additional elongation system appears to be functional in intact yeast cells. This system is obviously inactivated by addition of the FAS inhibitor cerulenin to the growth medium or, alternatively, by mutational inactivation of cytoplasmic FAS. Both methods induce synthetic lethality in the elongase mutants (114). This data may indicate that yeast FAS participates not only in de novo fatty acid synthesis but also in fatty acid elongation. In contrast to the structurally related FAS of mycobacteria, which synthesizes both C16 to C18 and C24 to C26 fatty acids, a bimodal product pattern of yeast FAS is not evident in vitro. It may nevertheless be speculated that in vivo, a fraction of cellular FAS is associated with the microsomal membrane and thereby engages in VLCFA synthesis. The general potential of yeast FAS to synthesize VLCFAs has been demonstrated in Schizosaccharomyces pombe. Here, certain FAS2 mutants unaffected in de novo FAS activity are reported to produce a considerable amount of C30 fatty acid (185). As a consequence, these mutants exhibited distinct alterations in cell shape and physiology (118). Remarkably, a membrane-bound FAS variant was identified in Drosophila melanogaster that was distinctly different from the homologous cytoplasmic FAS (45). The possible involvement of this variant in VLCFA synthesis remains to be demonstrated.
In the yeast cell extract, the relative activities of FAS and elongase differ by a factor of more than 20, even under conditions of malonyl-CoA saturation (114). Due to this difference, excessive VLCFA synthesis and the interference of VLCFAs with the physicochemical properties of membrane lipids are precluded. The moderate affinity of elongase to malonyl-CoA, which is about 17 timer lower than that of FAS, together with the marked inhibition of elongase by acyl-CoAs of increasing chain lengths (30), represent additional control mechanisms keeping the rate of cellular VLCFA synthesis low.
FASs of Secondary Metabolism
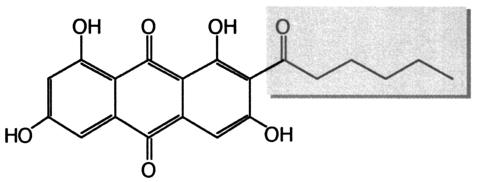
At least 20 different species of Aspergillus produce the secondary metabolite sterigmatocystin, while only few sythesize the related compound, aflatoxin (180). Both metabolites are derived from a common precursor, norsorolinic acid, which is composed of an anthrachinone-like polyketide to which a hexanoic acid side chain is attached (Fig. 3). Studies of the pathway of norsorolinic acid biosynthesis in several laboratories revealed the existence of two structurally related although functionally differentiated FASs in these Aspergillus strains (references 17, 175, and 180 and references therein). One of them (FAS) is responsible for primary metabolism and normal long-chain fatty acid synthesis, while the other (sFAS) synthesizes the fatty acyl side chain of the secondary metabolite. Consequently, primary FAS, although not sFAS mutants, requires long-chain fatty acids for growth, while biosynthesis of the secondary metabolites is specifically abolished in the sFAS mutants (18). Sterigmatocystin biosynthesis may be restored by adding hexanoic acid to the growth medium (18, 180). The sFAS genes, sFASα/HexA and sFASβ/HexB closely resemble, in their size and deduced amino acid sequences, the yeast genes FAS2 and FAS1 respectively (17, 83, 180). Nevertheless, the isolated sFAS genes were unable to complement Aspergillus mutants defective in the homologous primary FAS genes, fasA and fasB (18). Available data on norsorolinic acid biosynthesis suggest that the process starts by the sFAS-dependent formation of hexanoic acid. This fatty acid subsequently serves as a primer for the PKS complex synthesizing the tricyclic polyketide. According to Watanabe and Townsend and (174), the enzymes of norsorolinic acid synthesis, sFAS and PKS, are physically associated with an α2β2γ2 complex. The hexanoic acid may thus be transferred directly from sFAS (α2β2) to PKS (γ2) without release of an acyl-CoA intermediate. Accordingly, model substrates such as hexanoyl-CoA or S-hexanoyl-N-acetyl cysteamine were remarkably inefficient in starting norsorolinic acid synthesis (174, 175). It is possible that the interaction of PKS and sFAS limits fatty acid biosynthesis by sFAS to only two elongation cycles.
FIG. 3.
Norsorolinic acid. The chemical structure and hexanoic acid side chain (boxed) are shown.
Nonribosomally synthesized peptides of bacterial or fungal secondary metabolism sometimes contain an aliphatic acid in addition to uncommon amino acids. For instance, the maize pathogenic fungus Cochlibolus carbonum produces the phytotoxic and cytostatic HC-toxin containing 2-amino-9,10-epoxy-8-oxodecanoic acid as part of a cyclic tetrapeptide (173). Biosynthesis of the decanoic acid backbone in HC-toxin was correlated with the FAS1-like gene Tox-C, encoding a protein with significant sequence similarity to subunit β of yeast FAS (1). Tox-C is present in three copies within the fungal genome (2). Since fatty acid synthesis requires both FAS1 and FAS2 genes, it is speculated that the putative FAS2 gene involved in HC-toxin production remains to be identified or, alternatively, that the primary and secondary FAS systems in this fungus have the same α subunit (1, 173).
REGULATION OF FATTY ACID SYNTHASE BIOSYNTHESIS IN YEAST
In most cells, FAS belongs to the housekeeping enzymes fulfilling elementary functions in cellular metabolism and cell proliferation. Accordingly, FAS expression occurs mostly at an intermediate, constitutive level; however, it may be modulated by distinct metabolic conditions (33, 86, 143). For instance, synthesis of unsaturated fatty acids by bacterial FAS is regulated according to the needs of membrane fluidity (86). In mammalian tissues, regulation of FAS biosynthesis is complex and depends on tissue-specific, hormone- and cell cycle-responsive determinants (33, 143). With the exception of special tissues such as the mammary gland, FAS is down-regulated in most human cells. In contrast, dysregulation of FAS is observed in certain human tumors, leading to FAS overexpression preferentially in the aggressive varieties of these cancers (73). In the yeast S. cerevisiae, FAS biosynthesis is basically constitutive but may eventually be increased by a factor of 2 according to the needs of cellular phospholipid synthesis. In our hands, FAS biosynthesis was not affected by external free fatty acids (90). In contrast, a fatty acid-induced two- to threefold reduction of FAS mRNA levels was reported by Chirala et al. (22). Possibly, these characteristics were related to the particular yeast strain used by these authors. In fact, we found FAS biosynthesis fully repressed in the fatty acid-degrading yeast Yarrowia lipolytica when it was grown growth on fatty acid-supplemented media (90).
FAS Gene Organization
Today, our knowledge about regulation of microbiological type I FAS expression is most advanced for the fungal system of the yeast, S. cerevisiae. Biosynthesis of the heteromultimeric fungal type I FASs requires the coordinate expression of two genes, FAS1 and FAS2, which encode subunits β (FAS1) and α (FAS2) of the α6β6 FAS complex. The balanced production of the two subunits is suggested by the absence of nonaggregated FAS proteins from the yeast cell extract (28, 128). In fungi other than yeast, such as Neurospora crassa and Aspergillus nidulans, the FAS1 and FAS2 genes are closely linked and arranged, in opposite orientation, around a putative common promoter. Coordinate expression in these systems may thus be achieved by divergent transcription of FAS1 and FAS2 from the intergenic region (18). Similarly, the two FAS genes involved in Aspergillus secondary metabolism, sFASα/HexA and sFASβ/HexB, are located adjacent and in opposite orientation within the sterigmatocystin and aflatoxin gene clusters and therefore are likely to be coordinately transcribed (17, 18, 180). In contrast, the FAS1 and FAS2 genes of S. cerevisiae are unlinked and map to two different chromosomes (20, 146). The coordinate synthesis of subunits α and β in yeast was therefore expected to be controlled by considerably more elaborate mechanisms.
General and Metabolic Control of FAS Expression
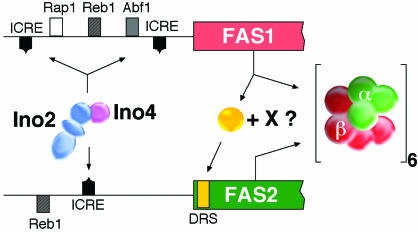
Studying the coregulation of FAS expression and phospholipid synthesis in yeast, Schüller and coworkers demonstrated that transcription of FAS1 and FAS2 is activated by a distinct cis-acting element present in two copies and one copy in the FAS1 and FAS2 promoters, respectively (126). This element is commonly observed in the promoters of S. cerevisiae structural genes which are involved in phospholipid biosynthesis (reviewed in reference 132). The element mediates transcriptional activation under conditions of inositol/choline (IC) limitation, whereas transcription is suppressed by an excess of these phospholipid precursors (132). Consequently, the element was designated ICRE (inositol/choline-responsive element) (126) or UASINO (80). Transcriptional activation of ICRE-dependent genes requires the regulatory genes, INO2 and INO4, which encode proteins with a basic helix-loop-helix (bHLH) structural motif (52, 99). Like other members of the large group of eucaryotic bHLH regulatory proteins, Ino2 and Ino4 bind, as an Ino2-Ino4 heterodimer, to the consensus sequence WYTTCAYRTG. This sequence is present in the ICRE motifs of both FAS genes (129). Schwank et al. (133) demonstrated that transcriptional activation of target genes by the Ino2-Ino4 heterodimer is mediated exclusively by the Ino2 subunit, which contains two separate activation domains in its N-terminal section. Remarkably, overexpression of INO2, but not of INO4, counteracts IC repression, suggesting that Ino2 is the primary target of IC repression (132). In contrast, deletion of INO4, abolishing Ino2-Ino4-mediated gene activation, reduced the transcriptional potential of FAS1 and FAS2 promoters to 36 and 51%, respectively, of the original values (127). Likewise, the cellular FAS level in ino4Δ mutants was decreased to about 50% of the wild-type level (130). From these results, it was evident that besides ICRE, additional transcriptional control elements must exist in the promoters of both FAS genes. Subsequent screening of the FAS1 and FAS2 promoters by serial deletion analyses and in vitro DNase footprint studies revealed binding sites for the general yeast transcription factors Rap1, Abf1, and Reb1 in the FAS1 upstream DNA and one site for Reb1 in the upstream region of FAS2 (127). Successive elimination of these sites in an ICRE-defective FAS1 promoter led to a gradual decrease of its transcriptional potency from about 50 to 2-10% of the wild-type level (127). Thus, transcription of yeast FAS genes is obviously subjected to both the pathway-specific regulation of ICRE-dependent phospholipid synthesis genes and the activation by general transcription factors. The latter elements allow the constitutive expression of FAS at a level which satisfies the needs of phospholipid-independent fatty acylation reactions of the cell.
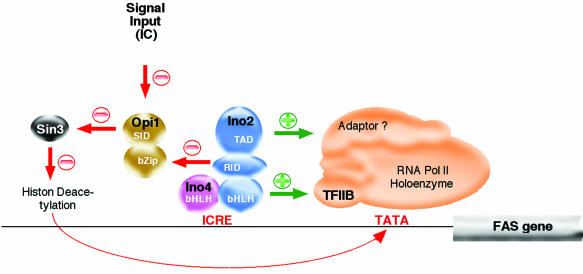
Transcriptional inactivation of ICRE-dependent genes occurs, in the presence of excess IC, by interaction of the Ino2-Ino4 activator with the negative regulator, Opil. On overexpression of OPI1, inactivation is observed even in the absence of IC (170). Conversely, constitutive expression of ICRE-dependent genes, i.e., abolition of IC repression, is observed in opi1-defective strains (44). Using in vivo and in vitro interaction assays, Wagner et al. (171) characterized the functional role of Opi1, which obviously serves as a link between the Ino2-Ino4 transcriptional activator and the pleiotropic yeast repressor, Sin3. Accordingly, these authors observed deregulation of an ICRE-controlled reporter gene on Sin3 inactivation in vivo. Furthermore, it was found that Ino2 contains separate functional domains for dimerization with Ino4, DNA binding, transcriptional activation, and interaction with Sin3, respectively. This functional diversity pointed to the central role of Ino2 in ICRE-mediated gene regulation. Wagner et al. (171) suggested that Opi1 may serve as the most upstream recipient of the IC-regulatory signal, thereby acting as a regulatory switch between the positively acting Ino2-Ino4 and the negatively acting Sin3 transcription factors. Even though important details of the signal transduction process still remain to be elucidated, the existence of an ICRE-bound ternary complex between Ino2, Ino4, and Opi1, or even a higher aggregate including Sin3, may be implicated (171). Since Sin3 is presumed to be associated with histone deacetylase activity, its repressor function may be correlated with local alterations of the chromatin structure and consequently with the promoter inactivation of target genes (55). Recently, it was suggested by Dietz et al. (29) that not only the N terminus but also the C terminus of Ino2 is involved in transcriptional activation. These authors found that part of the HLH dimerization domain of INO2 interacted with the basal transcription factor, TFIIB, of yeast. This factor is considered an important link in the signal transduction chain between transcriptional activators and the RNA polymerase II holoenzyme. Thus, regulation of ICRE-dependent genes implies complex interactions between a variety of regulatory proteins including Ino2, Ino4, Opi1, Sin3, and TFIIB. A hypothetical scheme based on the available data for these interactions is depicted in Fig. 4.
FIG. 4.
Regulatory interactions of yeast transcription factors involved in ICRE-dependent FAS expression. Interactions with activating (green) or repressing (red) effects on FAS transcription are marked by different colors. TAD, transcription activation domain; RID, repressor interaction domain; SID, Sin3-interaction domain; RNA Pol II, RNA polymerase II. Adapted from reference 17.
Autoregulation and Posttranslational Control
The presence of similar control elements in the promoters of FAS1 and FAS2 may explain their basically coordinate expression in yeast. Nevertheless, additional mechanisms are required to ensure an exactly balanced ratio of the two FAS subunits as observed in vivo. Studies by Wenz et al. (177) of the transcription of FAS2 in a fas1Δ deletion strain and of FAS1 in a fas2Δ deletion strain suggested that the two genes were in fact not expressed independently of each other. While transcription of FAS1 was unaffected in the fas2Δ mutant, deletion of FAS1 caused a dramatic reduction of FAS2 transcription. Compared to the fas1Δ null mutant, FAS2 transcript levels increased about 10-fold in the presence of multiple FAS1 genes copies. Using appropriate FAS2-lacZ reporter constructs, Wenz et al. (177) demonstrated that a “downstream repression site” (DRS) within the first 66 nucleotides of the FAS2 reading frame was responsible fort this effect. On deletion of the DRS sequence, FAS2 expression became derepressed, even in the absence of FAS1. Thus, FAS1 obviously interferes with DRS-dependent retardation of FAS2 transcription. The molecular mechanism of the DRS-dependent retardation of FAS2 transcription is still elusive. According to the hypothetical scheme shown in Fig. 5, excess FAS1-encoded subunit β may relieve DRS function either by direct interaction or in association with an unknown factor X. Thereby, synthesis of subunit α is stimulated to a level where the two subunits are present in balanced cellular concentrations. Its role as a primary target of both constitutive and IC-dependent transcriptional regulation makes FAS1 a key determinant of this autoregulation of FAS biosynthesis.
FIG. 5.
Autoregulation of FAS expression (127, 177). General (Rap1, Rep1, Abf1) and ICRE transcriptional activation sites in the FAS1 and FAS2 upstream regions are indicated. The DRS in the FAS2 coding sequence is presumed to interact with Fas1 either directly or in combination with an unknown factor (X).
Besides transcriptional and translational regulation, cellular FAS levels are subject to posttranslational control. Selective degradation of nonassembled FAS subunits by vacuolar (subunit β) and cytoplasmic (subunit α) proteases has been demonstrated. While the intact α6β6 FAS oligomer is proteolytically stable, its individual subunits are rapidly degraded both in vivo and in vitro (28, 34, 128). In conclusion, the hierarchy of regulatory mechanisms controlling FAS biosynthesis in yeast may be summarized as follows: (i) the pleiotropic transcriptional activators Rap1, Abf1, and Reb1 activate the constitutive part of FAS1 and FAS2 expression at a rate which ensures the housekeeping functions of cellular fatty acid synthesis; (ii) ICRE-dependent transcriptional control of FAS1 and FAS2 by the heterodimeric Ino2-Ino4 activator modulates FAS biosynthesis according to the needs of phospholipid production; (iii) Fas1-dependent activation of FAS2 transcription adjusts the production of subunit α to that of subunit β; and (iv) proteolytic degradation of an eventual excess of nonassembled FAS subunits represents a fine-tuning device supporting the regulation of FAS biosynthesis under certain conditions.
FATTY ACID SYNTHESIS IN MYCOLIC ACID-PRODUCING BACTERIA
Mycobacterial Fatty Acids
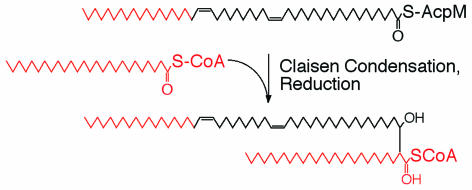
Mycolic acids are the predominant and characteristic lipid components of the cell envelopes of mycobacteria, corynebacteria, rhodococci, and nocardiae (12, 14, 154). They are high-molecular-weight α-alkyl, β-hydroxy fatty acids with a remarkable chemical structure, containing a species-specific “short” arm of 22 to 26 carbon atoms and a, “long” meromycolic acid arm of 50 to 60 carbon atoms (Fig. 6) (12, 14, 66, 77, 93, 125). As an exception, the corynemycolates of corynebacteria contain, instead of the long meronycolic acid branch, an additional short arm of 12 to 18 carbon atoms (77). Being covalently linked to the arabinogalactan-peptidoglycan matrix of the cell wall, mycolic acids form, as well as the conventional plasma membrane, part of a second and complex outer membrane bilayer (66, 98). The extremely low fluidity and, consequently, low permeability of this particular cell envelope contribute to the pathogenicity and extraordinary infectivity of mycobacteria. Even though the mechanism of mycolic acid biosynthesis remains to be elucidated in its details, formation of the mycolic acid backbone is thought to result from a remarkable Claisen-type condensation of two long-chain fatty acyl thioesters with subsequent reduction of the keto group (Fig. 6) (6, 77). A transient α-carbon carboxylation of one of the reactants, comparable to malonyl CoA formation in de novo fatty acid synthesis, is not supported experimentally (77). Regarding the enzymology of fatty acid synthesis, the mycolic acid-producing branch of the Actimomycetales represents a remarkable exception within the kingdom of procaryotes since these organisms use, like eucaryotes, a structurally integrated type 1 FAS rather than the usual procaryotic type II system for de novo long-chain fatty acid synthesis (13, 38, 62, 63, 144, 183). All catalytic domains of this unique bacterial type I FAS are contained within a single protein chain. Nevertheless, mycobacteria and related strains which produce the exceptionally long meromycolic acid side chains contain, in addition to the type I FAS, an ACP-dependent dissociated type II system (12, 66). In contrast to the type II synthases of other bacteria, the mycobacterial type II FAS is incapable of de novo fatty acid synthesis from acetyl-CoA. Instead, it elongates medium-chain-length C12 to C16 fatty acids to the very-long-chain meromycolic acids. The in vitro observed preference of some of the type II FAS components for longer-chain acyl thioester substrates supports this functional differentiation (24, 69, 85, 119, 120, 181). The preference for long-chain acyl substrates was correlated in these cases, by nuclear magnetic resonance or X-ray diffraction studies, to distinct structural features of the respective proteins. As discussed below in more detail, the long-chain acyl-CoA products of mycobacterial type I FAS in pathogenic mycobacteria not only function in meromycolic acid synthesis but also function as primers for the synthesis of multimethyl-branched fatty acids such as mycocerosic and phthioceranic acids. Apart from these specific pathways of fatty acid metabolism, the acyl-CoA products of mycobacterial type I FAS are also incorporated, as in all other organisms, into the conventional phospholipids of the plasma membrane.
FIG. 6.
Proposed pathway of mycolic acid biosynthesis (186). Carbon atoms in the meromycolic and fatty acyl chains originating from either type I (red) or type II (black) FAS are marked.
Bacterial Type I FAS
The bacterial type I FAS multienzyme has been isolated from several mycobacterial strains (13, 38, 63, 183) and from C. ammoniagenes (62, 144). In all cases, the purified enzymes were hexamers of identical subunits combining the entire set of catalytic FAS domains within a single polypeptide chain. Isolation and sequencing of the respective genes revealed a domain organization of these multifunctional FAS proteins which was comparable to a head-to-tail fusion of the yeast FAS subunits β und α (38, 89, 157). Thus, the microbial type I FASs, on the one hand, and animal FAS, on the other, represent different patterns of intramolecular and supramolecular organization. The size of the bacterial FAS subunits, comprising about 3,000 amino acids, was in between those of animal FAS (about 2,500 amino acids) and the αβ dimer of yeast FAS (3,940 amino acids). In contrast to yeast FAS, the bacterial apoFAS activating phosphopantetheine transferase is, in the bacterial system, encoded by a separate gene rather than being integrated into the FAS protein (25, 157). Even though this PPT gene is closely linked to the FAS-B reading frame in C. ammoniagenes, its product obviously functions as an independent enzyme activating both cellular FAS proteins, FAS-A and FAS-B (159). Similarly, it appears that a single PPT coding sequence in the M. tuberculosis genome is sufficient for the activation of all pantetheinylated cellular proteins. The bacterial FAS multienzyme differs from other known type I FASs by using different reductants, NADPH and NADH, for its ketoacyl and enoyl reduction steps, respectively (13, 144). Accordingly, two distinct nucleotide binding sites were identified in the bacterial FAS sequence, in contrast to only one site in other type I synthases (93).
The type I FAS multienzymes of mycobacteria and C. ammoniagenes are structurally very similar but nevertheless functionally slightly differentiated. A unique feature of mycobacterial type I FAS which is not observed with the related enzyme from corynebacteria was first reported by Bloch and Vance for M. smegmatis FAS (13) and subsequently also by the group of Kolattukudy (38, 63) for that of M. tuberculosis. According to these authors, the purified enzymes produced two classes of long-chain fatty acids with chain lengths of 16 to 18 and 24 to 26 carbon atoms, respectively, in vitro The relative amounts of these products were variable within a wide range, but the pattern remained persistently bimodal. Compared to the corynebacterial enzyme (144), no additional domains are evident from the mycobacterial sequence (25) to which this unique elongating capacity could be assigned. The two enzymes are very similar in size and amino acid sequence, comprising 3,063 and 3,069 amino acids, respectively.
According to Bloch and Vance (13), the rate-limiting step of M. smegmatis FAS activity occurs during termination of the synthetic cycle, i.e., the acyl transfer from the enzyme to CoA or the release of acyl-CoA from the enzyme. Distinct mycobacterial polysaccharides were found to increase both the production of shorter-chain fatty acids and the overall rate of fatty acid synthesis (9, 13, 106, 165, 182). The promoting effect of these polysaccharides was attributed in particular to the facilitated release of long-chain acyl-CoA from the enzyme (13). In pathogenic mycobacteria, which probably derive most of their conventional fatty acids from the host, the type I FAS system may function primarily as an elongase converting the host fatty acids into C24 or C26 products (179). These very-long-chain type I FAS products subsequently participate, by Claisen condensation with meromycolic acid, in mycolic acid biosynthesis. From the early work by Kawaguchi and coworkers (62, 144), it was known that purified C. ammoniagenes FAS synthesizes both saturated (16:0 and 18:0) and unsaturated (18:1) fatty acids. The subsequent isolation and characterization of the C. ammoniagenes FAS DNA by Meurer et al. (89) and by Stuible et al. (157, 158) revealed that this organism in fact contained two independent type I FAS genes. The encoded proteins, FAS-B and FAS-A, were individually purified on heterologous expression in E. coli and found to synthesize saturated (FAS-B) and a mixture of saturated and unsaturated (FAS-A) fatty acids (157). The FAS-B content is 5 to 10% of the total cellular FAS protein. According to Seyama and Kawaguchi (144), oleic synthesis by FAS-A is oxygen independent and sensitive to the inhibitor 3-decynoyl-N-acetyl cysteine (4). Therefore, the involvement of an additional component, β-hydroxydecanoyl thioester dehydratase, as is known from bacterial type II FAS, was suggested, even though the respective catalytic domain is not clearly evident from the comparison of FAS-B and FAS-A amino acid sequences. Obviously, FAS-B is a unique type I FAS since it combines the structural characteristics of eucaryotic type I FASs with one of the functional characteristics of procaryotic type II FASs. The β,γ-dehydratase of FAS-B, responsible for the synthesis of oleic acid, obviously has a remarkably high degree of chain length specificity. Using propionyl-CoA instead of acetyl-CoA as a primer, Arai et al. (3) obtained only 17:0 (no 17:1) products. Due to the redundancy of the two cellular palmitic acid-synthesizing activities, FAS-A and FAS-B, in C. ammoniagenes, only oleic acid-dependent (no saturated fatty acid-requiring) mutants are isolated on conventional mutagenesis. Even among corynebacteria, the occurrence of an oleate-synthesizing type I FAS appears to be an exception rather than the rule. Among six different strains of corynebacteria investigated, the FAS enzymes of only two, C. ammoniagenes and C. glutamicum, exhibited these characteristics (4). The other strains produced exclusively saturated fatty acids and thus are likely to lack the FAS-A variant.
Mycolic Acid Synthesis
Apart from its preference for long-chain acyl-CoA primers, the type II FAS of mycobacteria is likely to be functionally homologous to the dissociated type II systems of other procaryotic species. As in E. coli, three distinct ketoacyl synthases, KasA, KasB, and mtFabH, operate in the mycobacterial pathway, with mtFabH obviously functioning as a link between the mycobacterial type I and II FAS systems (24, 120, 151). Using palmitoyl-CoA rather than acetyl-CoA as a substrate, mtFabH is presumed to catalyze the starting reaction of meromycolic acid synthesis. Like the initiating ketoacyl synthase of E. coli, but in contrast to KasA and KasB, it prefers CoA over ACP thioesters as substrates (24). According to in vitro data obtained by Schaeffer et al. (120) and by Slayden and Barry (151), the KasA and KasB ketoacyl synthases are involved, with distinct chain length specificities (68, 119), in the subsequent elongation of AcpM acyl thioesters to C40 and C54 carbon chains, respectively. Extension of palmitoyl-CoA to 50 to 60 carbon acyl chains by more than 20 successive elongation cycles represents, for a single enzyme, a most remarkable and unique property. Even though our knowledge of the details of meromycolic acid synthesis and its subsequent condensation with palmitoyl-CoA is still fragmentary, this view has been widely accepted for a long time. Nevertheless, an alternative mechanism has occasionally been discussed. Since meromycolic acids are usually composed of three distinct polymethylenic sequences that are linked by C—C double bonds (Fig. 6), Asselineau et al. (6) suggested that successive condensations of four shorter-chain fatty acids with subsequent reduction and dehydration of the condensation products may also represent a possible mechanism.
FAS-Like Enzyme Systems in Mycobacteria
Only a few organisms produce a comparable amount and diversity of lipids to those of pathogenic mycobacteria. Up to 60% of the dry mass of mycobacterial cell walls is composed of lipids. Accordingly, a considerable portion of the M. tuberculosis genome (about 250 genes, compared to only 50 in E. coli) are devoted to lipid metabolism (25). Apart from mycolic acids and conventional phospholipids, pathogenic mycobacteria produce an impressive diversity of very long and methyl-branched fatty acids (12, 66, 93). Biosynthesis of these acids is initiated by straight-chain acyl-CoA primers, and subsequent chain elongation is catalyzed by methylmalonyl-CoA-specific FAS-like synthases. The resulting products are mono- to octamethyl-branched fatty acids, depending on the particular enzyme involved. The tetramethyl-branched mycocerosic acids and the hepta- and octamethyl-branched phthioceranic and hydroxyphthioceranic acids are prominent members of this class of lipids. The genetics and enzymology of the biosynthetic pathways involved in the production of this remarkable variety of different fatty acids are only beginning to be understood. So far, only two of these enzymes, the mycocerosic acid synthases MAS and SMAS, have been purified and biochemically characterized in greater detail (43, 110, 186). For several others, the biochemical functions were deduced from the lipid patterns of respective mutants (8, 115, 148-150). In vitro, MAS and SMAS synthesize tetramethyl C28 to C32 and C22 to C26 acids from straight-chain acyl-CoA primers of 16 to 20 (MAS) or 10 to 14 (SMAS) carbon atoms, respectively. Enzymes like MAS, which are often referred to as PKSs, may be termed FASs as well. They comprise complete sets of FAS component enzymes and synthesize fully reduced and saturated fatty acids. MAS is a homodimeric type I synthase composed of identical 280-kDa subunits. Cloning and sequencing of the mas gene revealed a domain organization similar to that of animal FAS and differing from that of the homologous, mycobacterial FAS (Fig. 1) (139). On heterologous expression in E. coli, the AT and KS domains of MAS were shown to be responsible for the methylmalonyl-CoA specificity of MAS. They directed the bacterial FAS system to incorporate methylmalonyl-CoA into methyl-branched fatty acids (39). Inspection of the M. tuberculosis genome revealed a number of MAS-like genes, all of them containing a complete set of FAS domains (25). Several other, MAS-related genes encode putative type I synthases lacking one or more of the canonical FAS domains. These enzymes either may synthesize distinct poyketides or may aggregate, in appropriate combinations, to heteromeric multienzymes with a complete set of FAS domains, as is observed with subunits α and β of yeast FAS (Fig. 1). According to Dubey et al. (32), the combination of pks8 and pks15 gene products appears to be responsible for the synthesis of 2-methyl-branched unsaturated C18 fatty acids. Cooperation of several FAS- and PKS-like multienzymes in phthiocerol biosynthesis was suggested by Kolattukudy's group (148-150). In contrast to conventional FAS, but similar to sFAS of Aspergillus (174, 175), the products of MAS are not released from the enzyme but seem to be directly transferred to the long-chain diols phthiocerol and phenolphthiocerol (66, 149). It is suggested that an open reading frame which is located in close proximity to the mas gene encodes an acyl-CoA ligase-like enzyme that may be implicated in this transesterification (42, 66). Similar open reading frame adjacent to several other pks- and mas-like genes in the M. tuberculosis genome may exhibit corresponding functions. This channeling of products to their cognate acceptors may explain why individual classes of mycobacterial lipids usually contain specific varieties of methyl-branched fatty acids.
CONCLUDING REMARKS
The reasons why fatty acid synthesis, in some organisms, is performed by dissociated type II systems while others contain integrated type I FAS proteins, will of course remain a matter of speculation. The frontier delimiting these two classes of organisms is obviously not the same as that separating the kingdoms of procaryotes and eucaryotes. It may be reasoned that multienzymes and aggregated enzyme complexes are kinetically more efficient than dissociated systems and also that the coordinated synthesis of multienzymes is readily controlled. However, most bacteria obviously overcome the kinetic disadvantage of type II FASs by having high cellular concentrations of ACP, while the regulatory aspect is probably not decisive for a housekeeping enzyme. On the other hand, it becomes particularly evident from the variety of FAS and PKS systems found in mycobacteria that only a multiplicity of discrete and functionally differentiated multienzymes allows for the controlled synthesis of a correspondingly diverse spectrum of different products. This control refers not only to the biosynthesis and activity of each system but also to its cellular localization and to the eventual product channeling in some cases. By this diversification, a minimum of mutual interference and, simultaneously, qualitatively and quantitatively controlled product patterns are ensured. The functional inflexibility of integrated multienzymes is particularly suited for iterative systems, such as FAS, which periodically repeat the same sequence of reactions. The programming of type I multienzymes for performing nonidentical reaction sequences in subsequent cycles appears to be difficult. With the possible exception of methylsalicylic acid synthase (11) from Penicillium patulum and also of FAS-B from C. ammoniagenes (157), where a double bond is introduced during one of the elongation cycles, a set of different multienzyme modules is normally used for these noniterative biosyntheses. This principle becomes most evident from the biosynthesis of polyketides such as erythonolid B in Saccharopolyspora erythraea (31) or phthiocerol in M. tuberculosis (8). In both cases, several multifunctional type I PKS modules, each catalyzing one cycle of specific reaction steps, cooperate in the overall biosynthetic pathway. In fatty acid biosynthesis, different FAS modules within the same cell allow the spatially or functionally differentiated production of fatty acids. In this way, targeting of fatty acids to the cytoplasm, to mitochondria and chloroplasts, to the microsomal membranes, or to distinct PKS systems is achieved. At the same time, biosynthesis of distinct and structurally defined fatty acids as well as their differential utilization in specific acylation reactions may thus be ensured.
REFERENCES
- 1.Ahn, J.-H., and J. D. Walton. 1996. A fatty acid synthase gene in Cochliobolus carbonum required for production of HC-toxin, cyclo(d-prolyl-l-alanyl-d-alanyl-l-2-amino-9,10-epoxi-8-oxodecanoyl). Mol. Plant-Microbe Interact. 10:207-214. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J.-H., and J. D. Walton. 1996. Chromosomal organization of TOX2, a complex locus controlling host-selective toxin biosynthesis in Cochliobolus carbonum. Plant Cell 8:887-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai, K., A. Kawaguchi, Y. Saito, N. Koike, Y. Seyama, T. Yamakawa, and S. Okuda. 1982. Propionyl-CoA induced synthesis of even-chain-length fatty acids by fatty acid synthetase from Brevibacterium ammoniagenes. J. Biochem. 91:11-18. [DOI] [PubMed] [Google Scholar]
- 4.Ariga, N., K. Maruyama, and A. Kawaguchi. 1984. Comparative studies of fatty acid synthases of corynebacteria. J. Gen. Appl. Microbiol. 30:87-95. [Google Scholar]
- 5.Arnstadt, K.-I., G. Schindlbeck, and F. Lynen. 1975. Zum Mechanismus der Kondensationsreaktion der Fettsäurebiosynthese. Eur. J. Biochem. 55:561-571. [DOI] [PubMed] [Google Scholar]
- 6.Asselineau, C., J. Asselineau, G. Lanéelle, and M.-A. Lanéelle. 2002. The biosynthesis of mycolic acids by mycobacteria: current and alternative hypotheses. Prog. Lipid Res. 41:501-523. [DOI] [PubMed] [Google Scholar]
- 7.Ayling, J., R. Pirson, and F. Lynen. 1972. Participation of covalently linked fatty acyl coenzyme A products in the action of yeast fatty acid synthetase. Biochemistry 11:526-532. [DOI] [PubMed] [Google Scholar]
- 8.Azad, A. K., T. D. Sirakova, N. D. Fernandes, and P. E. Kolattukudy. 1997. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J. Biol. Chem. 272:16741-16745. [DOI] [PubMed] [Google Scholar]
- 9.Banis, R. J., D. O. Peterson, and K. Bloch. 1977. Mycobacterium smegmatis fatty acid synthetase. Polysaccharide stimulation of the rate-limiting step. J. Biol. Chem. 252:5740-5744. [PubMed] [Google Scholar]
- 10.Beaudoin, F., K. Gable, O. Sayanova, T. Dunn, and J. A. Napier. 2002. A Saccharomyces cerevisisae gene required for heterologous fatty acid elongase activitiy encodes a microsomal β-keto-reductase. J. Biol. Chem. 277:11481-11488. [DOI] [PubMed] [Google Scholar]
- 11.Beck, J., S. Ripka, A. Siegner, E. Schiltz, and E. Schweizer. 1990. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur. J. Biochem. 192:487-498. [DOI] [PubMed] [Google Scholar]
- 12.Besra, G. S., and D. Chatterjee. 1994. Lipids and carbohydrates of Mycobacterium tuberculosis, p. 285-306. In B. R. Bloom (ed.), Tuberculosis, pathogenesis, protection and control. American Society for Microbiology, Washington, D.C.
- 13.Bloch, K., and D. Vance. 1977. Control mechanisms in the synthesis of saturated fatty acids. Annu. Rev. Biochem. 46:263-298. [DOI] [PubMed] [Google Scholar]
- 14.Brennan, P. J., and H. Nikaido. 1995. The envelope of Mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 15.Brody, S., and S. Mikolajczyk. 1988. Neurospora mitochondria contain an acyl-carrier protein. Eur. J. Biochem. 173:353-359. [DOI] [PubMed] [Google Scholar]
- 16.Brody, S., C. Oh, U. Hoja, and E. Schweizer. 1997. Mitochondrial acyl carrier protein is involved in lipoic acid synthesis in Saccharomyces cerevisiae. FEBS Lett. 408:217-220. [DOI] [PubMed] [Google Scholar]
- 17.Brown, D. W., J.-H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown, D. W., T. H. Adams, and N. P. Keller. 1996. Aspergillus has distinct fatty acid synthases for primary and secondary metabolism. Proc. Natl. Acad. Sci. USA 93:14873-14877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buckner, J. S., P. E. Kolattukudy, and L. Rogers. 1978. Synthesis of multimethyl-branched fatty acids by avian and mammalian fatty acid synthetase and its regulation by malonyl-CoA decarboxylase in the uropygial gland. Arch. Biochem. Biophys. 186:152-163. [DOI] [PubMed] [Google Scholar]
- 20.Burkl, G., H. Castorph, and E. Schweizer. 1972. Mapping of a complex gene locus coding for part of the Saccharomyces cerevisiae fatty acid synthetase multienzyme complex. Mol. Gen. Genet. 119:315-322. [DOI] [PubMed] [Google Scholar]
- 21.Cassagne, C., D. Darriet, and J. M. Bourre. 1978. Biosynthesis of very long chain fatty acids by the sciatic nerve of the rabbit. FEBS Lett. 90:336-340. [DOI] [PubMed] [Google Scholar]
- 22.Chirala, S. S. 1992. Coordinated regulation and inositol-mediated and fatty acid-mediated repression of fatty acid synthase genes in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89:10232-10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chirala, S. S., M. A. Kuziora, D. M. Spector, and S. J. Wakil. 1987. Complementation of mutations and nucleotide sequence of FAS1 gene encoding β-subunit of yeast fatty acid synthase. J. Biol. Chem. 262:4231-4240. [PubMed] [Google Scholar]
- 24.Choi, K. H., L. Kremer, G. S. Besra, and C. O. Rock. 2000. Identification and substrate specificity of beta-ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis. J. Biol. Chem. 275:28201-28207. [DOI] [PubMed] [Google Scholar]
- 25.Cole, S. T., R. Brosch, J. Parkhill, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 26.De Rosa, M., and A. Gambacorta. 1988. The lipids of Archaebacteria. Prog. Lipid Res. 27:153-175. [DOI] [PubMed] [Google Scholar]
- 27.Dickson, R. C. 1998. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu. Rev. Biochem. 67:27-48. [DOI] [PubMed] [Google Scholar]
- 28.Dietlein, G., and E. Schweizer. 1975. Control of fatty-acid-synthetase biosynthesis in Saccharomyces cerevisiae. Eur. J. Biochem. 58:177-184. [DOI] [PubMed] [Google Scholar]
- 29.Dietz, M., W.-T. Heyken, J. Hoppen, S. Geburtig, and H.-J. Schüller. 2003. TFIIB and subunits of the SAGA complex are involved in transcriptional activation of phospholipid biosynthetic genes by the regulatory protein Ino2 in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 48:1119-1130. [DOI] [PubMed] [Google Scholar]
- 30.Dittrich, F., D. Zajonc, K. Hühne, U. Hoja, A. Ekici, E. Greiner, H. Klein, J. Hofmann, J.-J. Bessoule, P. Sperling, and E. Schweizer. 1998. Fatty acid elongation in yeast. Biochemical characteristics of the enzyme system and isolation of elongation-defective mutants. Eur. J. Biochem. 252:477-485. [DOI] [PubMed] [Google Scholar]
- 31.Donadio, S., M. J. Staver, J. B. McAlpine, S. J. Swanson, and L. Katz. 1991. Modular organization of genes required for complex polyketide biosynthesis. Science 252:675-679. [DOI] [PubMed] [Google Scholar]
- 32.Dubey, V. S., T. D. Sirakova, M. H. Cynamon, and P. E. Kollatukudy. 2003. Biochemical Function of msl5 (pks8 plus pks17) in Mycobacterium tuberculosis H37Rv: biosynthesis of monomethyl branched unsaturated fatty acids. J. Bacteriol. 185:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duplus, E., and C. Forest. 2002. Is there a single mechanism for fatty acid regulation of gene transcription? Biochem. Pharmacol. 64:893-901. [DOI] [PubMed] [Google Scholar]
- 34.Egner, R., M. Thumm, M. Straub, A. Simeon, H.-J. Schüller, and D. H. Wolf. 1993. Tracing intracellular proteolytic pathways. Proteolysis of fatty acid synthase and other cytoplasmic proteins in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268:27269-27276. [PubMed] [Google Scholar]
- 35.Elovson, J. 1975. Purification and properties of the fatty acid synthetase complex from Neurospora crassa, and the nature of the fas mutation. J. Bacteriol. 124:534-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elovson, J., and P. R. Vagelos. 1968. Acyl carrier protein. X. Acyl carrier protein synthetase. J. Biol. Chem. 243:3603-3611. [PubMed] [Google Scholar]
- 37.Engeser, J., K. Hübner, J. Straub, and F. Lynen. 1979. Identity of malonyl and palmitoyl transferase of fatty acid synthetase from yeast. 2. A comparison of active-site peptides. Eur. J. Biochem. 101:413-422. [DOI] [PubMed] [Google Scholar]
- 38.Fernandes, N. D., and P. E. Kolattukudy. 1996. Cloning, sequencing and characterization of a fatty acid synthase-encoding gene from Mycobacterium tuberculosis var. bovis BCG. Gene 170:95-99. [DOI] [PubMed] [Google Scholar]
- 39.Fernandes, N. D., and P. E. Kolattukudy. 1997. Methylmalonyl coenzyme A selectivity of cloned and expressed acyltransferase and beta-ketoacyl synthase domains of mycocerosic acid synthase from Mycobacterium bovis BCG. J. Bacteriol. 179:7538-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernandes, N. D., and P. E. Kolattukudy. 1998. A newly identified methyl- branched chain fatty acid synthesizing enzyme from Mycobacterium tuberculosis var. bovis BCG. J. Biol. Chem. 273:2823-2828. [DOI] [PubMed] [Google Scholar]
- 41.Fichtlscherer, F., C. Wellein, M. Mittag, and E. Schweizer. 2000. A novel function of yeast fatty acid synthase. Subunit α is capable of self-pantetheinylation. Eur. J. Biochem. 267:2666-2671. [DOI] [PubMed] [Google Scholar]
- 42.Fitzmaurice, A. M., and P. E. Kolattukudy. 1998. An acyl-CoA synthase (acoas) gene adjacent to the mycocerosic acid synthase (mas) locus is necessary for mycocerosyl lipid synthesis in Mycobacterium tuberculosis var. bovis BCG. J. Biol. Chem. 273:8033-8039. [DOI] [PubMed] [Google Scholar]
- 43.Goodrich-Tanrikulu, M., D. H. Jacobson, A. E. Stafford, J. T. Lin, and T. A. McKeon. 1999. Characterization of Neurospora crassa mutants isolated following repeat-induced point mutation of the beta subunit of fatty acid synthase. Curr. Genet. 36:147-152. [DOI] [PubMed] [Google Scholar]
- 44.Greenberg, M. L., P. Goldwasser, and S. A. Henry. 1982. Characterization of yeast regulatory mutant constitutive for synthesis of inositol-1-phosphate synthase. Mol. Gen. Genet. 186:157-163. [DOI] [PubMed] [Google Scholar]
- 45.Gu, P., W. H. Welch, L. Guo, K. M. Schegg, and G. J. Blomquist. 1997. Characterization of a novel microsomal fatty acid synthetase (FAS) compared to a cytosolic FAS in the housefly, Musca domestica. Comp. Biochem. Physiol. 118:447-456. [DOI] [PubMed] [Google Scholar]
- 46.Gueguen, V., D. Macherel, M. Jaquinod, R. Douce, and J. Bourguignon. 2000. Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J. Biol. Chem. 275:5016-5025. [DOI] [PubMed] [Google Scholar]
- 47.Han, G., K. Gable, S. D. Kohlwein, F. Beaudoin, J. A. Napier, and T. M. Dunn. 2002. The Saccharomyces cerevisiae YBR159w gene encodes the 3- ketoreductase of the microsomal fatty acid elongase. J. Biol. Chem. 277:35440-35449. [DOI] [PubMed] [Google Scholar]
- 48.Harrington, A., C. H. Herbert, B. Tung, G. S. Getz, and P. P. Slonimski. 1993. Identification of a new nuclear gene (CEM1) encoding a protein homologous to a β-keto-acyl synthase which is essential for mitochondrial respiration in Saccharomyces cerevisiae. Mol. Microbiol. 9:545-555. [DOI] [PubMed] [Google Scholar]
- 49.Harwood, J. L. 1996. Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophys. Acta 1301:7-56. [DOI] [PubMed] [Google Scholar]
- 50.Holtermüller, K. H., E. Ringelmann, and F. Lynen. 1970. Reinigung und Charakterisierung der Fettsäure-Synthetase aus Penicillium patulum. Hoppe-Seyler's Z:Physiol. Chem. 351:1411-1422. [PubMed] [Google Scholar]
- 51.Hopwood, D. A., and D. H. Sherman. 1990. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu. Rev. Genet. 24:36-66. [DOI] [PubMed] [Google Scholar]
- 52.Hoskizaki, D. K., J. E. Hill, and S. A. Henry. 1990. The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J. Biol. Chem. 265:4736-4745. [PubMed] [Google Scholar]
- 53.Joshi, A. K., V. S. Rangan, A. Witkowski, and S. Smith. 2003. Engineering of an active animal fatty acid synthase dimer with only one competent subunit. Chem. Biol. 10:169-173. [DOI] [PubMed] [Google Scholar]
- 54.Joshi, A. K., L. Zhang, V. S. Rangan, and S. Smith. 2003. Cloning, expression, and characterization of a human 4′-phosphopantetheinyl transferase with broad substrate specificity. J. Biol. Chem. 278:33142-33149. [DOI] [PubMed] [Google Scholar]
- 55.Kadosh, D., and K. Struhl. 1998. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell. Biol. 18:5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kajiwara, S., T. Oura, and K. Shishido. 2001. Cloning of a fatty acid synthase component FAS1 gene from Saccharomyces kluyveri and its functional complementation of S. cerevisiae fas1 mutant. Yeast 18:1339-1345. [DOI] [PubMed] [Google Scholar]
- 57.Kaneda, T. 1977. Fatty acids of the genus Bacillus: an example of branched- chain preference. Bacteriol. Rev. 41:391-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaneda, T. 1991. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol. Rev. 55:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaneda, T., and E. J. Smith. 1980. Relationship of primer specificity of fatty acid de novo synthetase to fatty acid composition in 10 species of bacteria and yeasts. Can. J. Microbiol. 8:893-898. [DOI] [PubMed] [Google Scholar]
- 60.Kawaguchi, A., H. Tomoda, S. Okuda, and S. Omura. 1981. Fatty acid synthase from Cephalosporium caerulens. Methods Enzymol. 71:117-120. [DOI] [PubMed] [Google Scholar]
- 61.Kawaguchi, A., Y. Seyama, T. Yamakawa, and S. Okuda. 1981. Fatty acid synthase from Brevibacterium ammoniagenes. Methods Enzymol. 71:120-127. [Google Scholar]
- 62.Kawaguchi, K., and S. Okuda. 1977. Fatty acid synthetase from Brevibacterium ammoniagenes: formation of monounsaturated fatty acids by a multienzyme complex. Proc. Natl. Acad. Sci. USA 74:3180-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kikuchi, S., D. L. Rainwater, and P. E. Kollatukudy. 1992. Purification and characterization of an unusually large fatty acid synthase from Mycobacterium tuberculosis var. bovis BCG. Arch. Biochem. Biophys. 295:318-326. [DOI] [PubMed] [Google Scholar]
- 64.Kim, Y. S., and P. E. Kolattukudy. 1978. Malonyl-CoA decarboxylase from the uropygial gland of waterfowl: purification, properties, immunological comparison, and role in regulating the synthesis of multi-methyl-branched fatty acids. Arch. Biochem. Biophys. 190:585-597. [DOI] [PubMed] [Google Scholar]
- 65.Kohlwein, S. D., S. Eder, C.-S. Oh, C. E. Martin, K. Gable, D. Bacikova, and T. Dunn. 2001. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:109-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolattukudy, P. E., N. D. Fernandes, A. K. Azad, A. M. Fitzmaurice, and T. D. Sirakova. 1997. Biochemistry and molecular genetics of cell-wall lipid biosynthesis in Mycobacteria. Mol. Microbiol. 24:263-270. [DOI] [PubMed] [Google Scholar]
- 67.Köttig, H., G. Rottner, K.-F. Beck, M. Schweizer, and E. Schweizer. 1991. The pentafunctional FAS1 genes of Saccharomyces cerevisiae and Yarrowia lipolytica are co-linear and considerably longer than previously estimated. Mol. Gen. Genet. 226:310-314. [DOI] [PubMed] [Google Scholar]
- 68.Kremer, L., L. G. Dover, S. Carrere, K. M. Nampoothiri, S. Lesjean, A. K. Brown, P. J. Brennan, D. E. Minnikin, C. Locht, and G. S. Besra. 2002. Mycolic acid biosynthesis and enzymic characterization of the beta-ketoacyl-ACP synthase A-condensing enzyme from Mycobacterium tuberculosis. Biochem. J. 364:423-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kremer, L., K. M. Nampoothiri, S. Lesjean, L. G. Dover, S. Graham, J. Betts, P. J. Brennan, D. E. Minnikin, C. Locht, and G. S. Besra. 2001. Biochemical characterization of acyl carrier protein (AcpM) and malonyl-CoA: AcpM transacylase (mtFabD), two major components of Mycobacterium tuberculosis fatty acid syntase II. J. Biol. Chem. 276:27967-27974. [DOI] [PubMed] [Google Scholar]
- 70.Kresze, G.-B., L. Steber, D. Oesterhelt, and F. Lynen. 1977. Reaction of yeast fatty acid synthetase with iodoacetamide. 3. Malonyl-coenzyme A decarboxylase as product of the reaction of fatty acid synthetase with iodoacetamide. Eur. J. Biochem. 79:191-199. [DOI] [PubMed] [Google Scholar]
- 71.Kresze, G.-B., L. Steber, D. Oesterhelt, and F. Lynen. 1977. Reaction of yeast fatty acid synthetase with iodoacetamide. 2. Identification of the amino acid residues reacting with iodoacetamide and primary structure of a peptide containing the peripheral sulfhydryl group. Eur. J. Biochem. 79:181-190. [DOI] [PubMed] [Google Scholar]
- 72.Kreuzaler, F., H. Ragg, W. Heller, R. Tesch, I. Witt, D. Hammer, and K. Hahlbrock. 1979. Flavanone synthase from Petroselinum hortense: molecular weight, subunit, composition, size of messenger RNA, and absence of pantetheinyl residue. Eur. J. Biochem. 99:89-96. [DOI] [PubMed] [Google Scholar]
- 73.Kuhajda, F. P. 2000. Fatty acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition 16:202-208. [DOI] [PubMed] [Google Scholar]
- 74.Kühn, L., H. Castorph, and E. Schweizer. 1972. Gene linkage and gene- enzyme relations in the fatty-acid-synthetase system of Saccharomyces cerevisiae. Eur. J. Biochem. 24:492-497. [DOI] [PubMed] [Google Scholar]
- 75.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. Lacelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 76.Leadley, P., and A. Baerga-Otiz. 2003. Mammalian fatty acid synthase: Closure on a textbook mechanism? Chem. Biol. 10:101-106. [DOI] [PubMed] [Google Scholar]
- 77.Lee, R. E., J. W. Armour, K. Takkayama, P. J. Brennan, and G. S. Besra. 1997. Mycolic acid biosynthesis: definition and targeting of the Claisen condensation step. Biochim. Biophys. Acta 1346:275-284. [DOI] [PubMed] [Google Scholar]
- 78.Leonard, J. M., S. J. Knapp, and M. B. Slabaugh. 1998. A Cuphea beta- ketoacyl-ACP synthase shifts the synthesis of fatty acids towards shorter chains in Arabidopsis seeds expressing Cuphea FatB thioesterases. Plant J. 13:621-628. [DOI] [PubMed] [Google Scholar]
- 79.Lin, C. Y., and S. Kumar. 1972. Pathway for the synthesis of fatty acids in mammalian tissues. J. Biol. Chem. 247:604-606. [PubMed] [Google Scholar]
- 80.Lopes, J. M., J. P. Hirsch, P. A. Chorgo, K. L. Schulze, and S. A. Henry. 1991. Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipids precursors. Nucleic Acids Res. 19:1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lynen, F. 1969. Yeast fatty acid synthase. Methods Enzymol. 14:17-32. [Google Scholar]
- 82.Lynen, F. 1980. On the structure of fatty acid synthetase of yeast. Eur. J. Biochem. 112:431-442. [DOI] [PubMed] [Google Scholar]
- 83.Mahanti, N., D. Bhatnagar, J. W. Cary, J. Joubran, and J. E. Linz. 1996. Structure and function of fas-1A, a gene encoding a putative fatty acid synthetase directly involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 62:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mahmoud, Y. A., S. M. Abu el Souod, and W. G. Niehaus. 1996-1997. Purifiicaton and characterization of fatty acid synthetase from Cryptococcus neoformans. Mycopathologia 136:75-84. [DOI] [PubMed] [Google Scholar]
- 85.Marrakchi, H., S. Ducasse, G. Labesse, H. Montrozier, E. Margeat, L. Emorine, X. Charpentier, M. Daffe, and A. Quemard. 2002. MabA (FabG1), a Mycobacterium tuberculosis protein involved in the long-chain fatty acid elongation system FAS-II. Microbiology 148:951-960. [DOI] [PubMed] [Google Scholar]
- 86.Marrakchi, H., Y. M. Zhang, and C. O. Rock. 2001. Mechanistic diversity and regulation of type II fatty acid synthesis. Biochem. Soc. Trans. 30:1050-1055. [DOI] [PubMed] [Google Scholar]
- 87.Mathur, M., and P. E. Kolattukudy. 1992. Molecular cloning and sequencing of the gene for mycocerosic acid synthase, a novel fatty acid elongating multifunctional enzyme, from Mycobacterium tuberculosis var. bovis Bacillus Calmette-Guerin. J. Biol. Chem. 267:19388-19395. [PubMed] [Google Scholar]
- 88.McElhaney-Feser, G. E., and C. L. Cihlar. 1994. Purification and characterization of fatty acid synthase from Candida albicans strain 4918 and two derived spontaneous cerulenin-resistant mutants. J. Med. Vet. Mycol. 32:13-20. [PubMed] [Google Scholar]
- 89.Meurer, G., G. Biermann, A. Schütz, S. Harth, and E. Schweizer. 1991. Molecular structure of the multifunctional fatty acid synthetase genes of Brevibacterium ammoniagenes: its sequence of catalytic domains is formally consistent with a head to tail fusion of the two yeast genes FAS1 and FAS2. Mol. Gen. Genet. 232:106-116. [DOI] [PubMed] [Google Scholar]
- 90.Meyer, K.-H., and E. Schweizer. 1976. Control of fatty-acid synthetase levels by exogeneous long-chain fatty acids in the yeasts Candida lipolytica and Saccharomyces cerevisiae. Eur. J. Biochem. 65:317-324. [DOI] [PubMed] [Google Scholar]
- 91.Mikolajczyk, S., and S. Brody. 1990. De novo fatty acid synthesis mediated by acyl-carrier protein in Neurospora crassa mitochondria. Eur. J. Biochem. 187:431-437. [DOI] [PubMed] [Google Scholar]
- 92.Millar, A. A., and L. Kunst. 1997. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12:121-131. [DOI] [PubMed] [Google Scholar]
- 93.Minnikin, D. E., L. Kremer, L. G. Dover, and G. S. Besra. 2002. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem. Biol. 9:545-553. [DOI] [PubMed] [Google Scholar]
- 94.Mohamed, A. H., S. S. Chirala, N. H. Mody, W.-Y. Huang, and S. J. Wakil. 1988. Primary structure of the multifunctional α subunit protein of yeast fatty acid synthase derived from FAS2 gene sequence. J. Biol. Chem. 263:12315-12325. [PubMed] [Google Scholar]
- 95.Morita, N., N. Okajima, M. Gotoh, H. Hayashi, H. Okuyama, and S. Sasaki. 1992. Synthesis in vitro of very long chain fatty acids in Vibrio sp. strain ABE-1. Arch. Microbiol. 157:223-228. [DOI] [PubMed] [Google Scholar]
- 96.Morita, Y. S., K. S. Paul, and P. T. Englund. 2000. Specialized fatty acid synthesis in African trypanosomes: myristate for GPI anchors. Science 288:140-143. [DOI] [PubMed] [Google Scholar]
- 97.Morris, T. W., K. E. Read, and J. E. Cronan. 1995. Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J. Bacteriol. 177:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nikaido, H., S.-H. Kim, and E. Y. Rosenberg. 1993. Physical organization of lipids in the cell wall of Mycobacterium chelonae. Mol. Microbiol. 8:1025-1030. [DOI] [PubMed] [Google Scholar]
- 99.Nikoloff, D. M., P. McGraw, and S. A. Henry. 1992. The INO2 gene of Saccharomyces cerevisiae encodes a helix-loop-helix protein that is required for activation of phospholipid synthesis. Nucleic Acids Res. 20:3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Niwa, H., E. Katayama, M. Yanagida, and K. Morikawa. 1998. Cloning of the fatty acid synthetase beta subunit from fission yeast, coexpression with the alpha subunit, and purification of the intact multifunctional enzyme complex. Protein Expression Purif. 13:403-413. [DOI] [PubMed] [Google Scholar]
- 101.Oesterhelt D., H. Bauer, G.-B. Kresze, L. Steber, and F. Lynen. 1977. Reaction of yeast fatty acid synthetase with iodoacetamide. 1. Kinetics of inactivation and extent of carboxyamidomethylation. Eur. J. Biochem. 79:173-180. [DOI] [PubMed] [Google Scholar]
- 102.Oesterhelt, D., H. Bauer, and F. Lynen. 1969. Crystallization of a multienzyme complex: fatty acid synthetase from yeast. Proc. Natl. Acad. Sci. USA 63:1377-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oh, C.-S., D. A. Toke, S. Mandala, and C. E. Martin. 1997. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272:17376-17384. [DOI] [PubMed] [Google Scholar]
- 104.Ohlrogge, J. B., and J. G. Jaworski. 1997. Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:109-136. [DOI] [PubMed] [Google Scholar]
- 105.Packter, N. M., and A. Alam. 1980. Characterization of subspecies from a fungal fatty acid synthetase. Biochim. Biophys. Acta 615:497-508. [DOI] [PubMed] [Google Scholar]
- 106.Peterson, D. O., and K. Bloch. 1977. Mycobacterium smegmatis fatty acid synthetase. Long chain transacylase chain length specificity. J. Biol. Chem. 252:5735-5739. [PubMed] [Google Scholar]
- 107.Pirson, W., L. Schuhmann, and F. Lynen. 1973. The specificity of yeast fatty-acid-synthetase with respect to the “priming” substrate. Decanoyl-CoA and derivatives as “primers” of fatty acid synthesis in vitro. Eur. J. Biochem. 36:16-24. [DOI] [PubMed] [Google Scholar]
- 108.Pollard, M. R., L. Anderson, C. Fan, D. J. Hawkins, and H. M. Davies. 1991. A specific acyl-ACP thioesterase implicated in medium-chain fatty acid production in immature cotyledons of Umbellularia californica. Arch. Biochem. Biophys. 284:306-312. [DOI] [PubMed] [Google Scholar]
- 109.Pugh, E. L., and M. Kates. 1994. Acylation of proteins of the archebacteria Halobacterium cutirubrum and Methanobacterium thermoautotrophicum. Biochim. Biophys. Acta 1196:38-44. [DOI] [PubMed] [Google Scholar]
- 110.Rainwater, D. L., and P. E. Kolattukudy. 1985. Fatty acid biosynthesis in Mycobacterium tuberculosis var. bovis Bacillus Calmette-Guérin. Purification and characterization of a novel fatty acid synthase, mycocerosic acid synthase, which elongates n-fatty acyl-CoA with methylmalonyl-CoA. J. Biol. Chem. 260:616-623. [PubMed] [Google Scholar]
- 111.Rangan, V. S., and S. Smith. 2003. Fatty acid synthesis in eukaryotes, p. 151-179. In D. E. Vance and J. E. Vance (ed.), Biochemistry of lipids, lipoproteins and membranes. Elsevier, Amsterdam, The Netherlands.
- 112.Rock, C. O., and J. E. Cronan. 1996. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochim. Biophys. Acta 1302:1-16. [DOI] [PubMed] [Google Scholar]
- 113.Rock, C. O., and S. Jackowski. 2002. Forty years of bacterial fatty acid synthesis. Biochem. Biophys. Res. Commun. 292:1155-1166. [DOI] [PubMed] [Google Scholar]
- 114.Rössler, H., C. Rieck, T. Delong, U. Hoja, and E. Schweizer. 2003. Functional differentiation and selective inactivation of multiple Saccharomyces cerevisiae genes involved in very-long-chain fatty acid synthesis. Mol. Gen. Genet. 269:290-298. [DOI] [PubMed] [Google Scholar]
- 115.Rousseau, C., T. D. Sirakkova, V. S. Dubey, Y. Bordat, P. E. Kolattukudy, B. Gicquel, and M. Jackson. 2003. Virulence attenuation of two Mas-like polyketide synthase mutants of Mycobacterium tuberculosis. Microbiology 149:1837-1847. [DOI] [PubMed] [Google Scholar]
- 116.Runswick, M. J., I. M. Fearnley, J. M. Skehel, and J. E. Walker. 1991. Presence of an acyl carrier protein in NADH: ubiquinone oxireductase from bovine heart mitochondria. FEBS Lett. 286:121-124. [DOI] [PubMed] [Google Scholar]
- 117.Sackmann, U., R. Zensen, D. Röhlen, U. Jahnke, and H. Weiss. 1991. The acyl-carrier protein in Neurospora crassa mitochondria is a subunit of NADH: ubiquinone reductase (complex I). Eur. J. Biochem. 200:463-469. [DOI] [PubMed] [Google Scholar]
- 118.Saitoh, S., K. Takahashi, K. Nabeshima, Y. Yamashita, Y. Nakaseko, A. Hirata, and M. Yanagida. 1996. Aberrant mitosis in fission yeast mutants defective in fatty acid synthetase and acetyl CoA carboxylase. J. Cell Biol. 134:949-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Scarsdale, J. N., G. Kazanina, X. He, K. A. Reynolds, and H. T. Wright. 2001. Crystal structure of the Mycobacterium tuberculosis β-ketoacyl-acyl carrier protein synthase III. J. Biol. Chem. 276:20516-20522. [DOI] [PubMed] [Google Scholar]
- 120.Schaeffer, M. L., G. Agnihotri, C. Volker, H. Kallender, P. J. Brennan, and J. T. Lonsdale. 2001. Purificaton and biochemical characterization of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthases KasA and KasB. J. Biol. Chem. 276:47029-46037. [DOI] [PubMed] [Google Scholar]
- 121.Schjerling, C. K., R. Hummel, J. K. Hansen, C. Borsting, J. M. Mikkelsen, K. Kristiansen, and J. Knudsen. 1996. Disruption of the gene encoding the acyl-CoA-binding protein (ACB1) perturbs acyl-CoA metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 271:22514-22521. [DOI] [PubMed] [Google Scholar]
- 122.Schneider, R., B. Brors, F. Bürger, S. Camrath, and H. Weiss. 1997. Two genes of the putative mitochondrial fatty acid synthase in the genome of Saccharomyces cerevisiae. Curr. Genet. 32:384-388. [DOI] [PubMed] [Google Scholar]
- 123.Schneiter, R., C. E. Guerra, M. Lampl, G. Gogg, S. Kohlwein, and H. L. Klein. 1999. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Δ is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 19:3415-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schreckenbach, T., H. Wobser, and F. Lynen. 1977. The palmityl binding sites of fatty acid synthetase from yeast. Eur. J. Biochem. 80:13-23. [DOI] [PubMed] [Google Scholar]
- 125.Schroeder, E. K., N. Souza, D. S. Santos, J. S. Blachard and L. A. Basso. 2002. Drugs that inhibit mycolic acid biosynthesis in Mycobacterium tuberculosis. Curr. Pharm. Biotechnol. 3:197-225. [DOI] [PubMed] [Google Scholar]
- 126.Schüller, H.-J., A. Hahn., F. Tröster, A. Schütz, and E. Schweizer. 1992. Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 11:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schüller, H.-J., A. Schütz, S. Knab, B. Hoffmann, and E. Schweizer. 1994. Importance of general regulatory factors Rap1p, Abf1p and Reb1p for the activation of yeast fatty acid synthase genes FAS1 and FAS2. Eur. J. Biochem. 225:213-222. [DOI] [PubMed] [Google Scholar]
- 128.Schüller, H.-J., B. Förtsch, B. Rautenstrauss, D. H. Wolf, and E. Schweizer. 1992. Differential proteolytic sensitivity of yeast fatty acid synthetase subunits α and β contributing to a balanced ratio of both fatty acid synthetase components. Eur. J. Biochem. 203:607-614. [DOI] [PubMed] [Google Scholar]
- 129.Schüller, H.-J., K. Richter, B. Hoffmann, R. Ebbert, and E. Schweizer. 1995. DNA binding site of the yeast heteromeric Ino2p/Ino4p basic helix-loop-helix transcription factor: structural requirements as defined by saturation mutagenesis. FEBS Lett. 370:149-152. [DOI] [PubMed] [Google Scholar]
- 130.Schüller, H.-J., R. Schorr, B. Hoffmann, and E. Schweizer. 1992. Regulatory gene INO4 of yeast phospholipid biosynthesis is positively autoregulated and functions as a transactivator of fatty acid synthase genes FAS1 and FAS2 from Saccharomyces cerevisiae. Nucleic Acids Res. 20:5955-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schuster, H., B. Rautenstrauss, M. Mittag, D. Stratmann, and E. Schweizer. 1995. Substrate and product binding sites of yeast fatty acid synthase. Stoichiometry and binding kinetics of wild type and in vitro mutated enzymes. Eur. J. Biochem. 228:417-424. [PubMed] [Google Scholar]
- 132.Schwank, S., B. Hoffmann, and H.-J. Schüller. 1997. Influence of gene dosage and autoregulation of the regulatory genes INO2 and INO4 on inositol/choline-repressible gene transcription in the yeast Saccharomyces cerevisiae. Curr. Genet. 31:462-468. [DOI] [PubMed] [Google Scholar]
- 133.Schwank, S., R. Ebbert, K. Rautenstrauβ, E. Schweizer, and H.-J. Schüller. 1995. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix- loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res. 23:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schweizer, E. 1989. Biosynthesis of fatty acids and related compounds, p. 3-50. In C. Ratledge and S. G. Wilkinson (ed.), Microbial lipids, vol. 2. Academic Press Ltd., London, United Kingdom.
- 135.Schweizer, E. 1996. Fettsäuresynthasen-Funktionsstrategien eines Multienzyms. Naturwissenschaften 83:347-358. [PubMed] [Google Scholar]
- 136.Schweizer, E., B. Kniep, H. Castorph, and U. Holzner. 1973. Pantetheine- free mutants of the yeast fatty-acid-synthetase complex. Eur. J. Biochem. 39:353-362. [DOI] [PubMed] [Google Scholar]
- 137.Schweizer, E., F. Piccinini, C. Duba, S. Gunther, E. Ritter, and F. Lynen. 1970. Die Malonyl-Bindungsstellen des Fettsäuresynthetase-Komplexes aus Hefe. Eur. J. Biochem. 15:483-499. [DOI] [PubMed] [Google Scholar]
- 138.Schweizer, E., G. Müller, L. M. Roberts, M. Schweizer, J. Rösch, P. Wiesner, J. Beck, D. Stratmann, and I. Zauner. 1987. Genetic control of fatty acid synthetase biosynthesis and structure in lower fungi. Fat Sci. Technol. 89:570-577. [Google Scholar]
- 139.Schweizer, E., I. Lerch, L. Kroeplin-Rueff, and F. Lynen. 1970. Fatty acyl transferase. Characterization of the enzyme as part of the yeast fatty acid synthetase complex by the use of radioactively labeled coenzyme A. Eur. J. Biochem. 15:472-482. [DOI] [PubMed] [Google Scholar]
- 140.Schweizer, E., K. K. Werkmeister, and M. K. Jain. 1978. Fatty acid biosynthesis in yeast. Mol. Cell. Biochem. 21:95-107. [DOI] [PubMed] [Google Scholar]
- 141.Schweizer, M., C. Lebert, J. Höltke, L. M. Roberts, and E. Schweizer. 1984. Molecular cloning of the yeast fatty acid synthetase genes, FAS1 and FAS2: illustrating the structure of the FAS1 cluster gene by transcript mapping and transformation studies. Mol. Gen. Genet. 194:457-465. [DOI] [PubMed] [Google Scholar]
- 142.Schweizer, M., L. M. Roberts, H.-J. Höltke, K. Takabayashi, E. Höllerer, B. Hoffmann, G. Müller, H. Köttig, and E. Schweizer. 1986. The pentafunctional FAS1 gene of yeast: its nucleotide sequence and order of the catalytic domains. Mol. Gen. Genet. 203:479-486. [DOI] [PubMed] [Google Scholar]
- 143.Semenkovich, C. F. 1997. Regulation of fatty acid synthase (FAS). Prog. Lipid Res. 36:43-53. [DOI] [PubMed] [Google Scholar]
- 144.Seyama, Y., and A. Kawaguchi. 1987. Fatty acid synthesis and the role of pyridine nucleotides, p. 381-431. In D. Dolphin, R. Poulson, and O. Avramovic, (ed.), Pyridine nucleotide coenzymes: chemical, biochemical, and medical aspects, vol. 2B. John Wiley & Sons, Inc., New York, N.Y.
- 145.Shintani, D. K. and J. B. Ohlrogge. 1994. The characterization of a mitochondrial acyl carrier protein isoform isolated from Arabidopsis thaliana. Plant Physiol. 104:1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Siebenlist, U., J. Nix, M. Schweizer, D. Jäger, and E. Schweizer. 1990. Mapping of the trifunctional fatty acid synthetase gene FAS2 on chromosome XVI of Saccharomyces cerevisiae. Yeast 6:411-415. [DOI] [PubMed] [Google Scholar]
- 147.Singh, N., S. J. Wakil, and J. K. Stoops. 1985. Yeast fatty acid synthase: Structure to function relationship. Biochemistry 24:6598-6602. [DOI] [PubMed] [Google Scholar]
- 148.Sirakova, T. D., A. K. Thirumala, V. S. Dubey, H. Sprecher, and P. E. Kolattukudy. 2001. The Mycobacterium tuberculosis pks2 gene encodes the synthase for the hepta- and octamethyl-branched fatty acids required for sulfolipid synthesis. J. Biol. Chem. 276:16833-16839. [DOI] [PubMed] [Google Scholar]
- 149.Sirakova, T. D., V. S. Dubey, H.-J. Kim, M. H. Cynamon, and P. E. Kolattukudy. 2003. The Largest open reading frame (pks12) in the Mycobacterium tuberculosis genome is involved in pathogenesis and dimycocerosyl phthiocerol synthesis. Infect. Immun. 71:3794-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sirakova, T. D., V. S. Dubey, M. H. Cynamon, and P. E. Kolattukudy. 2003. Attenuation of Mycobacterium tuberculosis by disruption of a mas-like gene or a chalcone synthase-like gene, which causes deficiency in dimycocerosyl phthiocerol synthesis. J. Bacteriol. 185:2999-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Slayden, R. A., and C. E. Barry. 2002. The role of KasA and KasB in the biosynthesis of meromycolic acids and isoniazid resistance in Mycobacterium tuberculosis. Science 82:149-160. [DOI] [PubMed] [Google Scholar]
- 152.Smith, S. 1994. The animal fatty acid synthase: one gene, one polypeptide, seven enzymes. FASEB J. 8:1248-1259. [PubMed] [Google Scholar]
- 153.Smith, S., A. Witkowski, and A. K. Joshi. 2003. Structural and functional organization of the animal fatty acid synthase. Prog. Lipid Res. 42:289-317. [DOI] [PubMed] [Google Scholar]
- 154.Stackebrandt, E., F. A. Rainey, and N. L. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 155.Stoops, J. K., and S. J. Wakil. 1981. The yeast fatty acid synthetase. Structure- function relationship and the role of the active cysteine-SH and pantetheine-SH. J. Biol. Chem. 256:8364-8370. [PubMed] [Google Scholar]
- 156.Stoops, J. K., N. Singh, and S. J. Wakil. 1990. The yeast fatty acid synthase. J. Biol. Chem. 265:16971-16977. [PubMed] [Google Scholar]
- 157.Stuible, H.-P., C. Wagner, I. Andreou, G. Huter, J. Haselmann, and E. Schweizer. 1996. Identification and functional differentiation of two type I fatty acid synthases in Brevibacterium ammoniagenes. J. Bacteriol. 178:4787-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stuible, H.-P., G. Meurer, and E. Schweizer. 1997. Heterologous expression and biochemical characterization of two functionally different type I fatty acid synthases from Brevibacterium ammoniagenes. Eur. J. Biochem. 247:268-273. [DOI] [PubMed] [Google Scholar]
- 159.Stuible, H.-P., S. Meier, and E. Schweizer. 1997. Identification, isolation and biochemical characterization of a phosphopantetheine: protein transferase that activates the two type-I fatty acid synthases of Brevibacterium ammoniagenes. Eur. J. Biochem. 248:481-487. [DOI] [PubMed] [Google Scholar]
- 160.Stuible, H.-P., S. Meier, C. Wagner, E. Hannappel, and E. Schweizer. 1998. A novel phosphopantetheine: Protein transferase activating yeast mitochondrial acyl carrier protein. J. Biol. Chem. 273:22334-22339. [DOI] [PubMed] [Google Scholar]
- 161.Sumper, M., D. Oesterhelt, C. Riepertinger, and F. Lynen. 1969. Die Synthese verschiedener Carbonsäuren durch den Multienzymkomplex der Fettsäuresynthese aus Hefe und die Erklärung ihrer Bildung. Eur. J. Biochem. 10:377-387. [DOI] [PubMed] [Google Scholar]
- 162.Toke, D. A., and C. E. Martin. 1996. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J. Biol. Chem. 271:18413-18422. [DOI] [PubMed] [Google Scholar]
- 163.Toomey, R. F., and S. J. Wakil. 1966. Studies on the mechanism of fatty acid synthesis. XVI. Preparation and general properties of acyl-malonyl-acyl carrier protein condensing enzyme from Escherichia coli. J. Biol. Chem. 241:1159-1165. [PubMed] [Google Scholar]
- 164.Torkko, J. M., K. T. Koivuranta, I. J. Minalainen, A. I. Yagi, W. Schmitz, A. J. Kastaniotis, T. T. Airenne, A. Gurvitz, and K. J. Hiltunen. 2001. Candida tropicalis Etr1p and Saccharomyces cerevisiae Ybr 026p (Mrf1p), 2-enoyl thioester reductases essential for mitochondrial respiratory competence. Mol. Cell. Biol. 21:6243-6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tuffal, G., C. Ponthus, C. Picard, M. Rivière, and G. Puzo. 1995. Structural elucidation of novel methylglucose-containing polysaccharides from Mycobacterium xenopi. Eur. J. Biochem. 233:377-383. [DOI] [PubMed] [Google Scholar]
- 166.Ueno, K. 2000. Involvement of fatty acid synthase in axonal development in mouse embryos. Genes Cells 5:859-869. [DOI] [PubMed] [Google Scholar]
- 167.Vagelos, P. R. 1971. Regulation of fatty acid biosynthesis. Curr. Top. Cell. Regul. 4:119-166. [Google Scholar]
- 168.Vagelos, P. R., A. W. Alberts, and P. W. Majerus. 1969. Mechanism of saturated fatty acid biosynthesis in Escherichia coli. Methods Enzymol. 14:39-42. [Google Scholar]
- 169.Wada, H., D. Shintari, and J. Ohlrogge. 1997. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc. Natl. Acad. Sci. USA 94:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wagner, C., M. Blank, B. Strohmann, and H.-J. Schüller. 1999. Overproduction of the Opil repressor inhibits transcriptional activation of structural genes required for phospholipids biosynthesis in the yeast Saccharomyces cerevisiae. Yeast 15:843-854. [DOI] [PubMed] [Google Scholar]
- 171.Wagner, C., M. Dietz, J. Wittmann, A. Albrecht, and H.-J. Schüller. 2001. The negative regulator Opil of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol. 41:155-166. [DOI] [PubMed] [Google Scholar]
- 172.Wakil, S. J. 1991. Fatty acid synthase, a proficient multifunctional enzyme, p. 141-148. In H. Neurath (ed.), Perspectives in biochemistry, vol. 2. American Chemical Society, Washington, D.C. [DOI] [PubMed]
- 173.Walton, J. D. 1996. Host-selective toxins: Agents of compatibility. Plant Cell 8:1723-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Watanabe, C. M., and C. A. Townsend. 2002. Initial characterization of a type I fatty acid synthase and polyketide synthase multienzyme complex NorS in the biosynthesis of aflatoxin B1. Chem. Biol. 9:981-988. [DOI] [PubMed] [Google Scholar]
- 175.Watanabe, C. M., D. Wilson, J. E. Linz, and C. A. Townsend. 1996. Demonstration of the catalytic roles and evidence for the physical association of type I fatty acid synthases and a polyketide synthase in the biosynthesis of aflatoxin B1. Chem. Biol. 3:463-469. [DOI] [PubMed] [Google Scholar]
- 176.Welch, J. W., and A. L. Burlingame. 1973. Very long chain fatty acids in yeast. J. Bacteriol. 115:464-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wenz, P., S. Schwank, U. Hoja, and H. J. Schüller. 2001. A downstream regulatory element located within the coding sequence mediates autoregulated expression of the yeast fatty acid synthase gene FAS2 by the FAS1 gene product. Nucleic Acids Res. 29:4625-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Werkmeister, K., R. B. Johnston, and E. Schweizer. 1981. Complementation in vitro between purified mutant fatty acid synthetase complexes of yeast. Eur. J. Biochem. 116:303-309. [DOI] [PubMed] [Google Scholar]
- 179.Wheeler, P. R., K. Bulmer, and C. Ratledge. 1990. Enzymes for biosynthesis de novo and elongation of fatty acids in mycobacteria grown in host cells: is Mycobacterium leprae competent in fatty acid biosynthesis? J. Gen. Microbiol. 136:211-217. [DOI] [PubMed] [Google Scholar]
- 180.Woloshuk, C. P., and R. Prieto. 1998. Genetic organization and function of the aflatoxin B1 biosynthetic genes. FEMS Microbiol. Lett. 160:169-176. [DOI] [PubMed] [Google Scholar]
- 181.Wong, H. C., G. Liu, Y.-M. Zhang, C. O. Rock, and J. Zheng. 2002. The solution structure of acyl carrier protein from Mycobacterium tuberculosis. J. Biol. Chem. 277:15874-15880. [DOI] [PubMed] [Google Scholar]
- 182.Wood, W. I., D. O. Peterson, and K. Bloch. 1977. Mycobacterium smegmatis fatty acid synthetase. A mechanism based on steady state rates and product distributions. J. Biol. Chem. 252:5745-5749. [PubMed] [Google Scholar]
- 183.Wood, W. I., D. O. Peterson, and K. Bloch. 1978. Subunit structure of Mycobacterium smegmatis fatty acid synthetase. Evidence for identical multifunctional polypeptide chains. J. Biol. Chem. 253:2650-2656. [PubMed] [Google Scholar]
- 184.Yalpani, M., K. Willecke, and F. Lynen. 1968. Triacetic acid lactone, a derailment product of fatty acid biosynthesis. Eur. J. Biochem. 8:495-502. [DOI] [PubMed] [Google Scholar]
- 185.Yokoyama, K., S. Saitoh, M. Ishida, Y. Yamakawa, K. Nakamura, K. Inoue, R. Taguchi, A. Tokumura, M. Nishijima, M. Yanagida, and M. Setaka. 2001. Very-long-chain fatty acid-containing phospholipids accumulate in fatty acid synthase temperature-sensitive mutant strains of the fission yeast Schizosaccharomyces pombe fas2/lsd1. Biochim. Biophys. Acta 1532:223-233. [DOI] [PubMed] [Google Scholar]
- 186.Yuan, Y., R. E. Lee, G. S. Besra, J. T. Belisle, and C. E. Barry. 1995. Identification of a gene involved in the biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 92:6630-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zensen, R., H. Husmann, R. Schneider, T. Peine, and H. Weiss. 1992. De novo synthesis and desaturation of fatty acids at the mitochondrial acyl-carrier protein, a subunit of NADH:ubiquinone oxidoreductase in Neurospora crassa. FEBS Lett. 310:179-181. [DOI] [PubMed] [Google Scholar]
- 188.Zhang, L., A. K. Joshi, and S. Smith. 2003. Cloning, expression, characterization, and interaction of two components of a human mitochondrial fatty acid synthase. Malonyltransferase and acyl carrier protein. J. Biol. Chem. 278:40067-40074. [DOI] [PubMed] [Google Scholar]
- 189.Zhu, G., M. J. Marchewka, K. M. Woods, S. J. Upton, and J. S. Keithly. 2000. Molecular analysis of a type I fatty acid synthase in Cryptosporidium parvum. Mol. Biochem. Parasitol. 105:253-260. [DOI] [PubMed] [Google Scholar]
- 190.Ziegenhorn, J., R. Niedermeier, C. Nüssler, and F. Lynen. 1972. Charakterisierung der Acetyltransferase in der Fettsäuresynthetase aus Hefe. Eur. J. Biochem. 30:285-300. [DOI] [PubMed] [Google Scholar]