Abstract
Yersinia pestis, the causative agent of plague, is a highly uniform clone that diverged recently from the enteric pathogen Yersinia pseudotuberculosis. Despite their close genetic relationship, they differ radically in their pathogenicity and transmission. Here, we report the complete genomic sequence of Y. pseudotuberculosis IP32953 and its use for detailed genome comparisons with available Y. pestis sequences. Analyses of identified differences across a panel of Yersinia isolates from around the world reveal 32 Y. pestis chromosomal genes that, together with the two Y. pestis-specific plasmids, to our knowledge, represent the only new genetic material in Y. pestis acquired since the the divergence from Y. pseudotuberculosis. In contrast, 149 other pseudogenes (doubling the previous estimate) and 317 genes absent from Y. pestis were detected, indicating that as many as 13% of Y. pseudotuberculosis genes no longer function in Y. pestis. Extensive insertion sequence-mediated genome rearrangements and reductive evolution through massive gene loss, resulting in elimination and modification of preexisting gene expression pathways, appear to be more important than acquisition of genes in the evolution of Y. pestis. These results provide a sobering example of how a highly virulent epidemic clone can suddenly emerge from a less virulent, closely related progenitor.
Strong molecular evidence supports the fact that Yersinia pseudotuberculosis, responsible for yersiniosis in animals and humans, is the recent ancestor to Yersinia pestis, the etiologic agent of bubonic and pneumonic plague (1–3). However, whereas Y. pseudotuberculosis is a soil- and water-borne enteropathogen, Y. pestis is much more dangerous and is of current interest due to its potential use in bioterrorism and as a biological weapon. Present-day Y. pestis strains, although all similarly pathogenic, can be classified into three biovars [Antiqua (A), Medievalis (M), and Orientalis (O)] on the basis of their ability to use glycerol and to reduce nitrate. These phenotypic differences and molecular typing methods in conjunction with strain geographical origins have served to correlate these biovars with the three recorded plague pandemics.
Of special importance to the pathogenic process of both Y. pseudotuberculosis and Y. pestis is the shared requirement of a virulence plasmid pCD1 (pYV in enteropathogenic Yersinia) that encodes a type III secretion system (4), which is responsible for injecting into host cells a number of cytotoxins and effectors (Yersinia outer proteins) that inhibit bacterial phagocytosis and processes of innate immunity (5, 6). Two additional plasmids unique to Y. pestis, termed pPCP1 (9.6 kb) and pMT1 (102 kb), play roles in tissue invasion (7, 8) and capsule formation (9), as well as infection of the plague flea vector (10, 11), respectively. However, the presence of these plasmids by themselves cannot account for the remarkable increase in virulence observed in Y. pestis (12–15). Despite many extensive studies of the plasmid-encoded virulence determinants induced during the infectious process, and the recent availability of the genome sequences of a Y. pestis O. strain, CO92 (16), and an M strain, KIM10+ (17), the mechanism(s) underlying the strikingly different clinical manifestations of Y. pseudotuberculosis and Y. pestis have remained elusive. Although a microarray-based comparison of these two Yersinia species has been reported recently (18), the detailed comparison between the completed genomes of Y. pestis and that of Y. pseudotuberculosis IP32953 (serotype I) presented here provides the first opportunity, to our knowledge, to examine all differences in genome structure and at the nucleotide level. These comparisons reveal many of the molecular details that were involved in the speciation and emergence of Y. pestis and may hold the key to the exceptional virulence of the plague bacillus.
Materials and Methods
Whole-genome shotgun libraries were obtained and were sequenced as described (19). The whole-genome sequence of Y. pseudotuberculosis IP32953 was obtained from 85,000 end sequences (8.8-fold redundancy), and was assembled by using the program phrap (P. Green, University of Washington, Seattle). All gaps were closed by primer walking on gap-spanning clones or PCR products and a large insert scaffold was used to verify proper genome assembly. Gene modeling and genome annotation was performed as described (19). Genome comparisons between the Yersinia sequences were viewed by using the artemis comparison tool, which can be accessed at www.sanger.ac.uk/software/ACT.
The Yersinia strains studied came from the collection at the Institut Pasteur. Analysis of these strains was performed by screening PCR results, and, if necessary, sequencing the resulting products. Specifically, for the strain- or species-specific genes, primers were designed to amplify an ≈500-bp region within the gene (or gene portion) that was found to be missing from the other strains or species. For the insertion sequence (IS)-interrupted genes, primers were designed to amplify a 300- to 600-bp region of the WT gene or a 1- to 2.5-kb fragment that includes the interrupting IS element. Due to homologous recombination between IS elements, the alternative and sometimes expected result was a negative one (e.g., no PCR product when the IS in question underwent recombination, or in the event of a deletion removing at least one of the priming sites). For the other pseudogenes, sequencing each PCR product was followed by multiple alignments of the sequences to identify wild-type versus mutant loci. In all cases, experiments yielding negative results were repeated under the same conditions and also by using a lower annealing temperature in the event that the region in question had undergone divergence.
The Y. pestis and Y. pseudotuberculosis genomic DNAs that were used in panel-screens were isolated from the following strains of Y. pestis (biovars A, M, and O) Harbin (Former Soviet Union, A), Japan (Japan, A), Margaret (Kenya, A), 343 (Belgium Congo, A), PKH-4 (Kurdistan, M), PKR292 (Kurdistan, M), PAR13 (Iran, M), 297RR (Vietnam, O), Exu184 (Brazil, O), Hambourg10 (Germany, O), and 6/69 (Madagascar, O); and Y. pseudotuberculosis, IP33134 (Russia, serotype I), IP32790 (Italy, serotype I), IP32950 (France, serotype I), IP30215 (Denmark, serotype II), IP32802 (Italy, serotype III), IP32889 (Spain, serotype III), IP31833 (England, serotype IV), and IP32952 (France, serotype V). The controls used were Y. pestis CO92 (United States, O) and Y. pseudotuberculosis IP32953 (France, serotype I). Results for Y. pestis KIM10+ were predicted by using the available genome sequence.
Results and Discussion
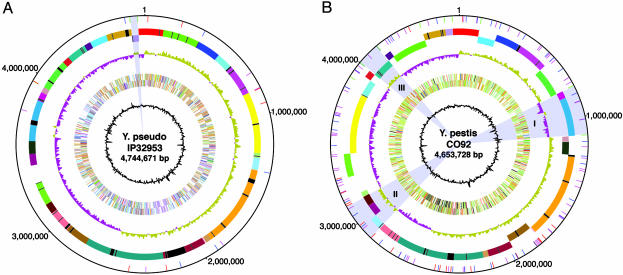
Genome Organization of Y. pseudotuberculosis IP32953. The genome of strain IP32953, a fully virulent clinical isolate from a human patient, consists of a single circular chromosome (4,744,671 bp), the pYV virulence plasmid (68,526 bp), and an atypical 27,702-bp cryptic plasmid, designated pYptb32953. The general features of the IP32953 genome are listed in Table 1 and the chromosome is represented in Fig. 1. Comparisons of pYV to the previously sequenced pCD1 plasmids from Y. pestis KIM5 (20, 21) and Y. pestis CO92 (16) revealed an essentially conserved colinear backbone, differing by the presence in pCD1 of an IS100 element, a coding sequence encoding a 68-aa hypothetical protein of unknown function, and an apparent internal in-frame 12-aa insertion in the middle of the yopM gene, which is consistent with the known heterogeneity found among Yersinia outer protein M in yersiniae (22).
Table 1. General features of Y. pseudotuberculosis IP32953 and chromosomal comparison with Y. pestis KIM10+ and CO92.
|
Y. pseudotuberculosis IP32953
|
Y. pestis KIM10+
|
Y. pestis CO92
|
|||
|---|---|---|---|---|---|
| Property | pYV32953 | pYptb32953 | Chromosome | Chromosome | Chromosome |
| Size, bp | 68,526 | 27,702 | 4,744,671 | 4,600,755 | 4,653,728 |
| G+C, % | 44.60 | 44.59 | 47.61 | 47.64 | 47.64 |
| CDS, total* | 99 | 43 | 3,974 | 4,090 | 4,016 |
| CDS, % | 80.7 | 86.5 | 83.6 | 83.4 | 83.9 |
| RNA operons | 0 | 0 | 7 | 7 | 6 |
| tRNAs | 0 | 0 | 85 | 73 | 70 |
| Total IS | 0 | 0 | 20 | 117 | 138 |
| IS100 | 5 | 35 | 44 | ||
| IS1541 | 5 | 55 | 65 | ||
| IS1661 | 3 | 8 | 8 | ||
| IS285 | 7 | 19 | 21 | ||
| Pseudogenes† | |||||
| Total | 4 | 1 | 62 | 54 (+202) | 149 (+149) |
| Role unknown | 0 | 0 | 10 | 8 (+60) | 34 (+58) |
| Unique regions‡ | |||||
| Total | 0 | 0 | 35 | 21 | 21 (+1) |
| Phage regions | 0 | 0 | 5 | 3 | 3 (+1) |
| Unique CDS‡ | |||||
| Total | 0 | 0 | 304 | 112 | 112 (+12) |
| Phage-related | 183 | 59 | 59 (+12) | ||
| Assigned role | 56 | 11 | 11 | ||
| Role unknown | 65 | 42 | 42 | ||
| Pseudogenes | 6 | 5 | 5 | ||
CDS, coding regions.
Based on published values for KIM10+ and CO92; discrepancies between chromosome size versus CDS can be attributed to differences in annotation.
For CO92 and KIM10+, numbers in parentheses are pseudogenes identified in this study.
In comparisons between IP32953 and either KIM10+ or CO92, the additional CO92-specific filamentous phage is presented in parentheses.
Fig. 1.
Circular genome map of IP32953 and comparison with Y. pestis CO92. (A) Genome of IP32953. (B) Genome of CO92. (A and B) Circle 1 (from center outward), G+C content; circles 2 and 3, all genes coded by function (forward and reverse strand); circle 4, GC skew (G–C/G+C); circles 5 and 6, genome divided into locally colinear blocks (when IP32953 and CO92 are compared with one another); each block is distinguished by a unique color (black segments within colored blocks represent regions specific to that genome in the comparison), and the orientation of each block is indicated by strand; [circle 5, –ve strand; circle 6, +ve strand); circle 7, locations of IS elements (IS100 is blue, IS285 is red, IS1661 is green, and IS1541 is magenta)]. In A, the gray highlighted region near the 12 o'clock position indicates the proposed IP32953 inversion (see text), whereas the remainder of the genome denotes the stable “ancestral” arrangement that has prevailed through the present. B illustrates the complexity of the molecular events that gave rise to the inversions or translocations in the Y. pestis genome first proposed (16) solely on the basis of the dramatic shifts in G/C skew (gray highlights serotypes I, II, and III), but now extended through whole-genome comparison. For example, gray highlight II is composed of three distinct blocks, two that are derived from distinct places within the same replichore (origin to terminus half), whereas the third block originated from the other replichore (light blue block).
The plasmid pYptb32953 is likely a conjugative cryptic plasmid and bears similarity to the recently described cryptic plasmid of Yersinia enterocolitica strain 29930 (23). The similarity (≈60–65%) extends to the plasmid mobilization machinery involving TraE and MobB/C homologues, and the entire cluster of type IV conjugation genes involved in plasmid transfer, suggesting that pYptb32953 may be self-transmissible. The latter operon also displays similarity to the conjugation genes of the IncX plasmid R6K (24) and to the Brucella spp. virB operon (25). Examination of the presence of this plasmid across a large number of isolates belonging to the three pathogenic Yersinia species indicated that its distribution is quite narrow (present in 3 of 81 strains; see Table 2, which is published as supporting information on the PNAS web site). Thus, this plasmid cannot account for any important virulence-associated characteristic of Y. pseudotuberculosis.
Few chromosomal features have been known to distinguish Y. pestis from Y. pseudotuberculosis strains (26, 27). However, comparisons between the chromosome of IP32953 and the Y. pestis chromosomes of CO92 and KIM10+ revealed several major differences (Table 1 and Fig. 1). The IP32953 chromosome encodes 3,974 predicted genes of which 2,976 (75%) have greater than or equal to 97% identity to their homologues in Y. pestis. Likewise, the synteny of the Yersinia genomes is readily discernable, and the breaks in colinearity have been mapped precisely (Fig. 1).
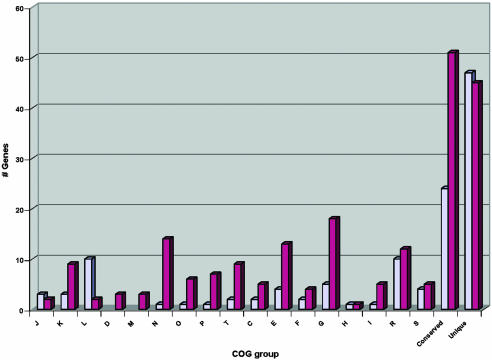
Unique Chromosomal Regions in Y. pseudotuberculosis. Thirty-six IP32953-specific regions, ranging in size between 500 bp and 122 kb, are scattered throughout the chromosome and contain a total of 317 putative genes that are not found in either CO92 or KIM10+. A list of putative proteins encoded in these regions and their gene locations can be found in Table 3, which is published as supporting information on the PNAS web site. More than one-half (188) of the genes in these unique regions are distributed in five clusters composed of phage-like products, the largest of which is a 122-kb region consisting of a 60-kb core of unknown function flanked by two parallel, but different, ≈30-kb P2-like mosaic phage regions. Another seven nonphage clusters encode 49 genes involved primarily in transposition and restriction modification, and together with the phage-associated regions, are also likely to have been horizontally acquired. It appears that the great majority of the remaining clusters encoding 80 genes have been deleted from the Y. pestis genome as demonstrated by the presence of partial gene remnants, IS elements, etc. The distribution of these genes (other than phage regions) into functional categories is shown in Fig. 2. Approximately one-third of all of the IP32953-specific genes in this group are hypothetical or conserved hypothetical genes, whereas others include genes that encode general metabolic functions that appear to have been lost in Y. pestis.
Fig. 2.
Functional classification of genes missing or inactivated in Y. pestis. Distribution of Y. pestis-specific lost functions by gene region deletion (light blue) or by gene inactivation (i.e., pseudogene, dark purple) in COG functional groups: C, energy production; D, cell division and/or chromosome partitioning; E, amino acid metabolism; F, nucleotide metabolism; G, carbohydrate metabolism; H, coenzyme metabolism; I, lipid metabolism; J, translation; K, transcription; L, DNA replication and/or repair; M, cell envelope biogenesis; N, cell motility, secretion; O, posttranslational modification; P, inorganic ion metabolism; R, general function prediction only; S, function unknown; T, signal transduction; conserved, conserved hypothetical genes with no significant COG hits; and unique, hypothetical genes with no significant COG hit.
Because species-specific regions and other species-specific genomic features should be conserved across a broad section of strains, a panel of 19 geographically and phenotypically diverse strains of Y. pestis and Y. pseudotuberculosis was selected and were screened for the presence or absence of these features. Of 85 IP32953-specific genes tested by PCR, 11 genes were found to be specific for the Y. pseudotuberculosis species (i.e., present in all Y. pseudotuberculosis and absent from all Y. pestis isolates; see Table 4, which is published as supporting information on the PNAS web site). Only one of these genes, YPTB0537, lies within one of the 12 above-described regions of putative foreign origin. Four of these 11 genes encode hypothetical proteins, whereas four others encode a putative restriction modification system component (YPTB0537) and proteins that are involved in glucan biosynthesis (YPTB2493 and YPTB2494) and uracil transport (YPTB2793). The last three encode metabolism-related functions: aspartate aminotransferase, enolase-phosphatase E1, and 5-methylthioribose kinase, respectively. These differences in metabolic enzymes may reflect the differences in Y. pestis and Y. pseudotuberculosis host ranges. In addition, the Y. pseudotuberculosis-specific regions may account for important virulence factors uniquely required in Y. pseudotuberculosis, such as the opg operon (YPTB2493–YPTB2495), which is required for the synthesis of periplasmic branched glucans that serve in other organisms as an osmoprotectant (28), but may not be needed in Y. pestis, an obligate parasite of eukaryotes, which is unlikely to experience wide fluctuations in environmental osmotic conditions.
Unique Chromosomal Regions in Y. pestis. We also identified 112 KIM10+- and CO92-specific (i.e., not found in IP32953) genes distributed in 21 clusters of 300 bp to 41.7 kb scattered throughout the genome (Table 5, which is published as supporting information on the PNAS web site). Approximately three categories of genes were identified in these 21 regions: (i) 39 genes (35% of the total) are hypothetical or are conserved hypothetical, (ii) 59 genes (53%) are phage or transposon-related, and (iii) 14 genes (12%) can be attributed a putative function. Among those genes with an ascribed function are membrane proteins, lipoproteins, a putative esterase, a DNA-binding protein, and a methyltransferase. Our studies indicate that a CO92 9-kb filamentous prophage region, previously believed to be O biovar-specific (18, 27), is in fact also present in some members of the A biovar (ref. 29, and Table 6, which is published as supporting information on the PNAS web site), and is absent from IP32953.
Of the 112 genes uniquely associated with the two Y. pestis genomes, 105 were tested for their presence or absence in our panel of 19 Yersinia strains (Table 6). Only 32 genes, located in six clusters, were present in all Y. pestis and were absent from all Y. pseudotuberculosis strains that were examined. Four of these clusters have been recently identified by using microarray analysis (18). However, genome sequence comparison coupled with PCR has identified two additional regions not found by hybridization and has eliminated the five other regions previously determined as unique to Y. pestis (Table 6) by that method.
Four of the Y. pestis-specific gene clusters encode predominantly putative proteins with little, if any, similarity to known or predicted proteins (with the exception of a methylase). Another cluster consists of bacteriophage-related genes (YPO2084-103, YPO2114 in CO92; and y2227-y2211, y2201 in KIM10+); whereas the last cluster (YPO1668-71 in CO92; y1829-y1832) encodes putative membrane proteins, a translation initiation inhibitor, and conserved hypothetical proteins. Although there were no obvious virulence factors encoded in these regions, their role in pathogenicity deserves further study.
Inactivated Genes. Sixty-two pseudogenes are found in IP32953, 43 of which are also pseudogenes in one or both sequenced Y. pestis strains (Table 7, which is published as supporting information on the PNAS web site). The remaining 19 genes likely represent recent Y. pseudotuberculosis-acquired mutations that have arisen since their divergence. Of these genes, the functions most frequently affected included outer membrane transport and exported proteins, perhaps reflecting the organism's interaction with its environment. Two of the 19 genes were integrases with substantial similarity to one another: a P4-like integrase (YPTB0534) and the previously described (30) high-pathogenicity island integrase (YPTB1602). Although the significance of the other Y. pseudotuberculosis-specific inactivated P4-like integrase is not known, the intact counterpart in Y. pestis, may be involved in the increased frequency of IS transposition in the latter.
Of the 149 originally reported CO92 pseudogenes (16), only 84 are pseudogenes in the KIM10+ strain and yet are intact genes in IP32953. Three-way gene-by-gene comparisons among the Yersinia strains enabled us to identify 149 additional putative pseudogenes in CO92 (Table 8, which is published as supporting information on the PNAS web site), of which 124 are also pseudogenes in the KIM10+ genome, yet only two are pseudogenes in IP32953. Thus, a closer approximation to the factual number of potentially lost functions by this evolutionary mechanism in Y. pestis is 208 (84 plus 124), so that as much as 5% of the gene complement may have been selectively inactivated in Y. pestis. A summary of this subset of inactivated genes and their distribution by the Clusters of Orthologous Groups (COG) database functional classes is shown in Fig. 2.
By using the same panel of 19 strains, we also examined the distribution of 52 randomly selected CO92 pseudogenes. Forty-six genes could be grouped into five discernable categories, the largest of which comprises 28 pseudogenes specific to Y. pestis (Table 9, which is published as supporting information on the PNAS web site). Members of this group are potentially the most interesting because they affect traits that are unique to Y. pestis strains, and thus, may represent good targets for studying their novel pathogenic properties and for quick identification in clinical settings. Genes disrupted in this group range from conserved hypothetical, to genes of general metabolism such as metB (responsible for the observed methionine requirement of Y. pestis), to regulatory genes (e.g., putative two-component sensor kinase, etc.), and potential virulence-associated genes (invasin, toxin transporter, etc.).
A second group of seven pseudogenes was only found in members of the biovar O, and includes the arginine-binding periplasmic protein 2 precursor (argJ), the N-terminal region of Escherichia coli prepilin peptidase-dependent protein (ppdA), the exonuclease encoded by sbcC, and the aerobic glycerol phosphate dehydrogenase (glpD), which is likely responsible for the glycerol-minus phenotype of the biovar O (31).
Six IS-interrupted pseudogenes comprise a third category, including aroG, penicillin-binding protein 1C (pbpC), and setA, a sugar efflux transporter. These pseudogenes are in all members of the O biovar and are in one or both of the African A strains (from Kenya and Congo), and are intact genes in Y. pseudotuberculosis, the M lineage, and the non-African A strains. This finding, in addition to the previously alluded to filamentous phage distribution pattern, supports the notion that the O and M lineages arose independent from the A biovar.
A fourth category of two other pseudogenes, a putative surface protein (YPO0902 in CO92 and y3288 in KIM10+) and a pectin-degradation protein (YPO1726 in CO92 and y1888 in KIM10+) are found in all Y. pestis strains and are also present in several Y. pseudotuberculosis strains. These pseudogenes may represent mutations acquired before the emergence of Y. pestis because they are unlikely to have been independently acquired by each species (one is a partial deletion and the other is interrupted by an IS285).
The fifth and last category comprises a single IS100-interrupted acetylornithine aminotransferase, argD, a CO92-specific pseudogene, which is likely the result of a very recent IS mobility that supports the idea of a continuously fluid genome.
Metabolism. Because Y. pseudotuberculosis is a chemoheterotroph, a full complement of biosynthetic and intermediary metabolic pathways was expected and has been verified. As already indicated, several of the IP32953-specific regions encode general metabolic functions, and thus, may account for some of the observed physiological differences between the two species. Noteworthy among this group are genes of purine and aspartate metabolism as well as genes of the methionine salvage pathway (32–34). Gene inactivations that may account for the Y. pestis-specific biochemical phenotypes include a cysteine synthase (cysM) frameshift (the cysteine requirement of Y. pestis), a missense point mutation affecting amino acid 363 in the aspartate ammonia lyase (aspA) of Y. pestis likely accounting for the stimulatory effect of CO2 on growth (35), and a proline substitution present in amino acid 161 of glucose 6 P-dehydrogenase (zwf) that likely prevents utilization of hexose via the pentose-phosphate pathway (36). The significance of these last two types of mutations will require further functional analyses.
Pathogenicity. Genomic differences that may play a role in the unique pathogenic characteristics of these two species include alterations in lipid A biosynthesis that is exemplified by the absence in Y. pestis of lipid A acyltransferase gene htrB (YPTB2490), which adds an acyl group to lipid A, and may account for the differences in lipid A between the two species. Because lipid A acylation changes are known to alter endotoxic properties and interactions with the innate immune system, this difference could be of significance for pathogenesis.
Several hemolysin/hemagglutinin homologues of different pathogens are present in the yersiniae. In IP32953, a cluster of nine coding sequences (YPTB3450–YPTB3459) encodes several hemolysin homologues in a region absent from Y. pestis. A hemolysin activator is a pseudogene in both IP32953 (YPTB3651) and KIM10+ (y0002) but is wild-type in CO92 (YPO3720). However, because this mutation occurs at a homopolymeric tract of C's (11 in IP32953 and KIM10+, and only 7 in CO92), it may simply represent a spontaneous reversion similar to that shown to occur in ureD, in which silencing and reactivation of urease in Y. pestis is determined by a spontaneous addition/excision of a single G residue in the ureD gene (37). Another hemolysin gene that is inactivated by partial deletion in IP32953 (YPTB2524) and all other Y. pseudotuberculosis strains is found intact in Y. pestis (Table 6; gene YPO2486 in CO92 and y1701 in KIM10+). Although the role of hemolysins in Yersinia virulence remains unclear, their conserved nature and clear differences among the species suggest the need for further studies to investigate their possible function.
The insecticidal toxin homologues found either in complete or inactivated form in the Y. pestis genomes have been implicated in the adaptation of this organism to the flea life cycle (16, 18, 38). Thus, it has been suggested that the observed inactivation of tcaB, encoding an insecticidal toxin protein, is required for flea life cycle but this argument can now be refuted because this gene is complete and normal in several M and A Y. pestis strains (Table 9). Similarly, the in-frame deletion of tcaC in Y. pestis cannot alone account for its ability to colonize the flea midgut because this gene is even shorter in Y. pseudotuberculosis; neither can the same function be attributed to the viral enhancing protein previously described in CO92 (16) because it is also present in IP32953. Thus, the precise role of insecticidal toxin homologues in flea midgut colonization remains largely unresolved.
Two loci (srfA and srfB) encoding putative virulence factors, along with the gene for the Cu-Zn superoxide dismutase sodC, have in-frame insertion/deletions in KIM10+ and CO92, but are wild-type in IP32953. If these mutations affect protein function, they could play a role in species-specific virulence. Similarly, an IS1541 neighboring csrB, a small noncoding RNA that antagonizes CsrA, an S-layer protein involved in adherence to cells, may modify the transcription and/or stability of this RNA and thus may have an effect on virulence in Y. pestis. Another region that could have a role in virulence in these organisms is a high pathogenicity island-like region, HPI-2 (noted in the CO92 genome, GenBank accession no. AL590842). This region is wild-type in Y. pseudotuberculosis but is defective in Y. pestis, in which the siderophore synthesis protein (YPO0778 and YPO1012 in CO92; y3406 and y3410 in KIM10+) is inactivated by an IS100 insertion.
Regulatory Genes. At least nine regulatory genes that are inactivated in Y. pestis could have effects on its phenotype, including virulence. A frameshift in Y. pestis homologues of YPTB0553 affects a gene similar to sorC, a transcriptional regulator required for sorbose use, whereas a frameshift in the Y. pestis homologues of YPTB1259 may affect the regulation of the synthesis of polysaccharide colanic acid. This capsular polysaccharide has been implicated in blocking the specific binding between uropathogenic E. coli and inert substrates (39). These inactivations in Y. pestis may be consistent with the general loss of adhesins that are unnecessary for its lifestyle. The gene flhD may be one of many genes inactivated in Y. pestis responsible for altered motility in this organism. Its absence may have a positive impact on Yersinia outer protein expression (40), and a possible pleiotropic effect on virulence and metabolism, as demonstrated in other enterobacteria (41). Also inactivated in Y. pestis is the rhafR homologue (YPO1728 in CO92 and y2579 in KIM10+), which may lead to derepression of the rhaffinose utilization pathway in this organism.
A frameshift in the transcriptional regulator iclR, carried by Y. pestis, leads to constitutive glyoxylate bypass in this organism, explaining an already known phenomenon (42). Furthermore, because the glyoxylate bypass has been shown to be necessary for virulence in other bacterial pathogens and fungi (43, 44), constitutive expression may also enhance Y. pestis virulence.
UhpB (YPTB3846), a transcriptional activator of genes involved in the uptake and metabolism of hexose phosphates, is inactivated in many Y. pestis strains. Finally, the gene encoding sigma N modulating factor (YPTB3527) possesses a stop codon in position 36 in Y. pestis, which could lead to modified expression of sigma 54-dependent genes.
IS Elements, Genome Rearrangements, and Evolution. Only 20 IS elements were found in the IP32953 chromosome, which is in stark contrast to the 117 in KIM10+ and 138 in CO92 (Table 1). Twelve of the 20 IS elements in IP32953 share integration locations with those in the two Y. pestis strains, suggesting that only eight recent transposition events have occurred in IP32953, whereas an extraordinary expansion of each IS family took place in Y. pestis strains since their divergence. Examination of the shared IS locations within CO92 and KIM10+ suggests that their most recent common ancestor carried 109 IS elements and that since the divergence of this ancestral representative and the present-day KIM10+ and CO92 strains, 8 and 28 new insertions occurred, respectively. What remains unclear is whether the rate of transposition in Yersinia is periodically stimulated or whether these events occurred in a punctuated fashion on some as-yet-unknown induction.
Despite the dense distribution of IS elements in Y. pestis and their potential for generating homologous recombination-mediated deletions, there are surprisingly few (only five) IP32953-specific regions that can be the result of excision of intervening sequence by means of recombination at flanking direct IS elements in Y. pestis.
Deng et al. (17) first alluded to the important role played by repeat elements (namely IS elements) in explaining the unique genome arrangement displayed by the two sequenced Y. pestis strains. Analyses by using the structural organization of IP32953 for comparison further support the role played by IS elements in genome evolution and confirms the ancestral character of Y. pseudotuberculosis because IP32953 most often has no “equivalent” IS element when compared with Y. pestis. This finding implies that most rearrangements have occurred only recently in the Y. pestis lineage and that the genome structural organization of IP32953 more closely reflects that of the ancestral type.
In a manner analogous to that used in the KIM10+/CO92 comparisons (17), we can identify some 32 syntenic colinear blocks conserved between IP32953 and CO92 (Fig. 1) and 25 between IP32953 and KIM10+. The genome organization of the last common ancestral genome of the two Y. pestis strains, as well as the ancestral genome of both species, could be deduced by investigating the precise locations of these rearrangements. Thus, IP32953 has undergone at least one and likely no more than three intrachromosomal recombinations since the split from the last common ancestor. A probable recombination in IP32953 that generated a large inversion between two IS1661 is supported by the distinct shift in the GC skew associated with this region (Fig. 1). Two other putative IP32953 rearrangements are exemplified by the mobile pathogenicity island region HPI, which typically integrates at one of three asn-tRNAs in Yersinia spp. (45), and a recombination at a P4-like integrase (YPTB0534) that is common to all three sequenced strains. All other rearrangements appeared to have occurred in the Y. pestis lineage.
Allowing for the three possible rearrangements proposed during the evolution of IP32953, the progenitor of both CO92 and KIM10+, must have undergone at least 11 recombination/rearrangement events (undoubtedly influenced by the 97 additional IS elements gained since diverging from Y. pseudotuberculosis). KIM10+ and CO92 have since undergone an additional 10 and 18 rearrangements, respectively, which is commensurate to their respective increased levels of IS transposition. It is thus quite likely that the insertion elements themselves and/or the subsequent rearrangements they have generated have played an important role in the emergence of Y. pestis from its Y. pseudotuberculosis ancestor.
Implications in Virulence and Pathogen Evolution. The genome sequence of Y. pseudotuberculosis IP32953 and its comparison with Y. pestis reveal aspects of the evolutionary processes that evidently transformed a common enteropathogenic ancestor, and later gave rise to two present-day pathogens of vastly distinct clinical manifestations. Molecular events that likely operated during the evolution of alternatively free-living Y. pseudotuberculosis (capable of causing localized chronic disease) contrast with those involved in the evolution of Y. pestis (capable of causing vector-dependent acute disease). The extensive chromosomal rearrangements that occurred during the emergence of Y. pestis undoubtedly are indicative of the mechanisms that drove the evolution of this pathogen. IS element expansion and its corollary, the increased fluidity of the genome, together with massive gene inactivation, almost surely have played a role in this process. A direct comparison between the work presented in this study and the calculated evolutionary distances between these two Yersinia species presented by Achtman et al. (1) is difficult to make without a reliable molecular clock to measure the rates of genome rearrangement, IS transposition, and gene inactivation. Because the mechanism(s) that account for IS element expansion and increased gene inactivation in Y. pestis is unknown, we can only surmise that these processes were driven by selection for lethality as well as evolutionary pressures that further enabled colonization of the flea. In this scenario, gene inactivation or IS-mediated rearrangements (either before or after the lateral transfer of pPCP1 and pMT1) might have led to changes that increased virulence (high septicemia) and that facilitated fleaborne transmission. The concomitant and dramatic change in lifestyle undergone by Y. pestis, ensuing from its continuous association with the host and dependency on the flea vector for survival, would have been sufficient to provide the selective pressure that resulted in wholesale inactivation of as much as 13% of its genome that we observe today. This result may represent an intermediate stage in genome compaction, a process that has been proposed in the evolution of other pathogens closely associated with their hosts such as Salmonella typhi (46) and Mycobacterium lepae (47). Finally, the significance of horizontal gene transfer into the chromosome of Y. pestis is uncertain. It may be hypothesized that the acquisition of at least some of the six chromosomal regions uniquely conserved in Y. pestis strains, in conjunction with the high degree of gene inactivation has been responsible for the increased pathogenicity of this species. Whole-genome comparisons of pathogen nearneighbors of distinct characteristics, such as those described in this study, lay the foundation for future mutational, functional, and animal studies that will ultimately help elucidate the mechanisms underlying the emergence of new pathogens.
Supplementary Material
Acknowledgments
We thank S. Falkow, D. Monack, P. Agron, M. Chu, and C. Kim for helpful discussions and review of the manuscript. This work was supported by the U.S. Department of Energy and the Lawrence Livermore National Laboratory under Contract W-7405-ENG-48, the Oak Ridge National Laboratory under Contract DE-AC05-00OR22725, and the French Délégation Générale à l'Armement under Contract 99 01 110 00 470 94 50.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IS, insertion sequence; A, Antiqua; M, Medievalis; O, Orientalis; COG, Clusters of Orthologous Groups.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. BX936398 (chromosome), BX936399 (pYV), and BX936400 (pYptb32953)].
References
- 1.Achtman, M., Zurth, K., Morelli, G., Torrea, G., Guiyoule, A. & Carniel, E. (1999) Proc. Natl. Acad. Sci. USA 96, 14043–14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner, D. J., Steigerwalt, A. G., Falcao, D. P., Weaver, R. E. & Fanning, G. R. (1976) Int. J. Syst. Bacteriol. 26, 180–194. [Google Scholar]
- 3.Moore, R. L. & Brubaker, R. R. (1975) Int. J. Syst. Bacteriol. 25, 336–339. [Google Scholar]
- 4.Cornelis, G. R. & Van Gijsegem, F. (2000) Annu. Rev. Microbiol. 54, 735–774. [DOI] [PubMed] [Google Scholar]
- 5.Brubaker, R. R. (2003) Infect. Immun. 71, 3673–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornelis, G. R. (2002) Nat. Rev. Mol. Cell Biol. 3, 742–752. [DOI] [PubMed] [Google Scholar]
- 7.Brubaker, R. R., Beesley, E. D. & Surgalla, M. J. (1965) Science 149, 422–424. [DOI] [PubMed] [Google Scholar]
- 8.Lahteenmaki, K., Virkola, R., Saren, A., Emody, L. & Korhonen, T. K. (1998) Infect. Immun. 66, 5755–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kutyrev, V. V., Popov, IuA. & Protsenko, O. A. (1986) Mol. Gen. Mikrobiol. Virusol. 6, 3–11. [PubMed] [Google Scholar]
- 10.Hinnebusch, B. J., Rudolph, A. E., Cherepanov, P., Dixon, J. E., Schwan, T. G. & Forsberg, A. (2002) Science 296, 733–735. [DOI] [PubMed] [Google Scholar]
- 11.Hinnebusch, B. J. (2003) Ad. Exp. Med. Biol. 529, 55–62. [DOI] [PubMed] [Google Scholar]
- 12.Filippov, A. A., Solodovnikov, N. S., Kookleva, L. M. & Protsenko, O. A. (1990) FEMS Microbiol. Lett. 55, 45–48. [DOI] [PubMed] [Google Scholar]
- 13.Friedlander, A. M., Welkos, S. L., Worsham, P. L., Andrews, G. P., Heath, D. G., Anderson, G. W., Jr., Pitt, M. L., Estep, J. & Davis, K. (1995) Clin. Infect. Dis. 21, Suppl. 2, S178–S181. [DOI] [PubMed] [Google Scholar]
- 14.Kutyrev, V., Mehigh, R. J., Motin, V. L., Pokrovskaya, M. S., Smirnov, G. B. & Brubaker, R. R. (1999) Infect. Immun. 67, 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welkos, S. L., Andrews, G. P., Lindler, L. E., Snellings, N. J. & Strachan, S. D. (2004) Plasmid 51, 1–11. [DOI] [PubMed] [Google Scholar]
- 16.Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T., Prentice, M. B., Sebaihia, M., James, K. D., Churcher, C., Mungall, K. L., et al. (2001) Nature 413, 523–527. [DOI] [PubMed] [Google Scholar]
- 17.Deng, W., Burland, V., Plunkett, G., III, Boutin, A., Mayhew, G. F., Liss, P., Perna, N. T., Rose, D. J., Mau, B., Zhou, S., et al. (2002) J. Bacteriol. 184, 4601–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinchliffe, S. J., Isherwood, K. E., Stabler, R. A., Prentice, M. B., Rakin, A., Nichols, R. A., Oyston, P. C., Hinds, J., Titball, R. W. & Wren, B. W. (2003) Genome Res. 13, 2018–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chain, P., Lamerdin, J., Larimer, F., Regala, W., Lao, V., Land, M., Hauser, L., Hooper, A., Klotz, M., Norton, J., et al. (2003) J. Bacteriol. 185, 2759–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, P., Elliott, J., McCready, P., Skowronski, E., Garnes, J., Kobayashi, A., Brubaker, R. R. & Garcia, E. (1998) J. Bacteriol. 180, 5192–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry, R. D., Straley, S. C., Fetherston, J. D., Rose, D. J., Gregor, J. & Blattner, F. R. (1998) Infect. Immun. 66, 4611–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boland, A., Havaux, S. & Cornelis, G. R. (1998) Microb. Pathog. 25, 343–348. [DOI] [PubMed] [Google Scholar]
- 23.Strauch, E., Goelz, G., Knabner, D., Konietzny, A., Lanka, E. & Appel, B. (2003) Microbiology 149, 2829–2845. [DOI] [PubMed] [Google Scholar]
- 24.Nunez, B., Avila, P. & de la Cruz, F. (1997) Mol. Microbiol. 24, 1157–1168. [DOI] [PubMed] [Google Scholar]
- 25.Boschiroli, M. L., Ouahrani-Bettache, S., Foulongne, V., Michaux-Charachon, S., Bourg, G., Allardet-Servent, A., Cazevieille, C., Lavigne, J. P., Liautard, J. P., Ramuz, M. & O'Callaghan, D. (2002) Vet. Microbiol. 90, 341–348. [DOI] [PubMed] [Google Scholar]
- 26.Brubaker, R. R. (2000) The Prokaryotes, an Evolving Electronic Resource for the Microbiological Community, ed. Stackelbrandt, E. (Springer, New York), Vol. 2000.
- 27.Radnedge, L., Agron, P. G., Worsham, P. L. & Andersen, G. L. (2002) Microbiology 148, 1687–1698. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy, E. P. (1996) in Escherichia coli and Salmonella, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. I, pp. 1064–1071. [Google Scholar]
- 29.Gonzalez, M. D., Lichtensteiger, C. A., Caughlan, R. & Vimr, E. R. (2002) J. Bacteriol. 184, 6050–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesic, B., Bach, S., Ghigo, J. M., Dobrindt, U., Hacker, J. & Carniel, E. (2004) Mol. Microbiol. 52, 1337–1348. [DOI] [PubMed] [Google Scholar]
- 31.Motin, V. L., Georgescu, A. M., Elliott, J. M., Hu, P., Worsham, P. L., Ott, L. L., Slezak, T. R., Sokhansanj, B. A., Regala, W. M., Brubaker, R. R. & Garcia, E. (2002) J. Bacteriol. 184, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortlock, R. P. (1962) J. Bacteriol. 84, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brubaker, R. R. (1970) Infect. Immun. 1, 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dreyfus, L. A. & Brubaker, R. R. (1978) J. Bacteriol. 136, 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baugh, C. L., Lanham, J. W. & Surgalla, M. J. (1964) J. Bacteriol. 88, 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortlock, R. P. & Brubaker, R. R. (1962) J. Bacteriol. 84, 1122–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sebbane, F., Devalckenaere, A., Foulon, J., Carniel, E. & Simonet, M. (2001) Infect. Immun. 69, 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wren, B. W. (2003) Nat. Rev. Microbiol. 1, 55–64. [DOI] [PubMed] [Google Scholar]
- 39.Hanna, A., Berg, M., Stout, V. & Razatos, A. (2003) Appl. Environ. Microbiol. 69, 4474–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bleves, S., Marenne, M. N., Detry, G. & Cornelis, G. R. (2002) J. Bacteriol. 184, 3214–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruss, B. M., Campbell, J. W., Van Dyk, T. K., Zhu, C., Kogan, Y. & Matsumura, P. (2003) J. Bacteriol. 185, 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hillier, S. & Charnetzky, W. T. (1981) J. Bacteriol. 145, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorenz, M. C. & Fink, G. R. (2002) Eukaryot. Cell 1, 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lorenz, M. C. & Fink, G. R. (2001) Nature 412, 83–86. [DOI] [PubMed] [Google Scholar]
- 45.Buchrieser, C., Brosch, R., Bach, S., Guiyoule, A. & Carniel, E. (1998) Mol. Microbiol. 30, 965–978. [DOI] [PubMed] [Google Scholar]
- 46.Parkhill, J., Dougan, G., James, K. D., Thomson, N. R., Pickard, D., Wain, J., Churcher, C., Mungall, K. L., Bentley, S. D., Holden, M. T., et al. (2001) Nature 413, 848–852. [DOI] [PubMed] [Google Scholar]
- 47.Cole, S. T., Eiglmeier, K., Parkhill, J., James, K. D., Thomson, N. R., Wheeler, P. R., Honore, N., Garnier, T., Churcher, C., Harris, D., et al. (2001) Nature 409, 1007–1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.