Abstract
Alternative splicing increases the diversity of transcriptomes and proteomes in metazoans. The extent to which alternative splicing is active and functional in unicellular organisms is less understood. Here, we exploit a single-molecule long-read sequencing technique and develop an open-source software program called SpliceHunter to characterize the transcriptome in the meiosis of fission yeast. We reveal 14,353 alternative splicing events in 17,669 novel isoforms at different stages of meiosis, including antisense and read-through transcripts. Intron retention is the major type of alternative splicing, followed by alternate “intron in exon.” Seven hundred seventy novel transcription units are detected; 53 of the predicted proteins show homology in other species and form theoretical stable structures. We report the complexity of alternative splicing along isoforms, including 683 intra-molecularly co-associated intron pairs. We compare the dynamics of novel isoforms based on the number of supporting full-length reads with those of annotated isoforms and explore the translational capacity and quality of novel isoforms. The evaluation of these factors indicates that the majority of novel isoforms are unlikely to be both condition-specific and translatable but consistent with the possibility of biologically functional novel isoforms. Moreover, the co-option of these unusual transcripts into newly born genes seems likely. Together, the results of this study highlight the diversity and dynamics at the isoform level in the sexual development of fission yeast.
Splicing is a fundamental process which removes intragenic noncoding regions (introns) and forms mature mRNAs for proper translation (Wang and Burge 2008; Lee and Rio 2015). It provides an important checkpoint and a posttranscriptional layer of gene expression control (Le Hir et al. 2003; McGlincy and Smith 2008; Braunschweig et al. 2013; Bentley 2014). One of the regulatory mechanisms is alternative splicing (AS), which leads to multiple transcripts from the same gene (Wang et al. 2008). AS is achieved mainly by different combinations of exons and introns or AS sites, producing a vast expansion of transcriptome diversity and, potentially, protein diversity (Keren et al. 2010; Nilsen and Graveley 2010). AS is considered a potent regulator of gene expression in multicellular organisms given the rationale that isoforms generated by AS are differentially regulated in different tissues or conditions (Wang et al. 2008). However, AS in unicellular organisms is less extensively explored. Important questions are: What is the complexity of single cell transcriptomes? How is AS differentially regulated across different conditions? Is AS even functionally relevant to eukaryotic microorganisms? Understanding these questions in unicellular organisms will extend our knowledge of AS and, in particular, the origin of AS.
Saccharomyces cerevisiae and Schizosaccharomyces pombe are the two most documented unicellular model organisms. S. cerevisiae has ∼280 intron-containing genes, and the vast majority of them have a single intron. On the contrary, >2200 genes in S. pombe are currently known to contain introns, and half of these genes have multiple introns. Therefore, S. pombe is a more suitable unicellular model to study AS and transcriptome diversity than S. cerevisiae. Thousands of AS events have been identified recently under various conditions in WT and mutant cells via different techniques such as short read RNA-seq and lariat sequencing (Awan et al. 2013; Bitton et al. 2015; Stepankiw et al. 2015). RNA metabolism kinetics, including synthesis, splicing, and decay rates have also been determined in vegetative fission yeast (Eser et al. 2016). A broad spectrum of AS types is observed, similar to multicellular organisms. These studies suggest prevalent AS in this unicellular organism. However, these findings are limited in defining isoforms, which represent the complete structure and sequence of transcripts. Characterizing isoforms is critical to predicting the effects on the encoded proteome. Whether these AS events are functionally relevant or condition-specific also remains poorly explored.
To address these questions, we exploited a single-molecule real-time (SMRT) sequencing technique based on the Pacific Biosciences (PacBio) platform (Eid et al. 2009) and developed SpliceHunter, a novel open-source software program to systematically explore the transcriptome in S. pombe. PacBio sequencing features very long reads, which are suitable for full-length cDNA sequencing. Recently, PacBio sequencing has been used to examine the transcriptome in multiple metazoan organisms, such as chicken and human, and multiple plants, and identified thousands of annotated and novel isoforms (Au et al. 2013; Sharon et al. 2013; Martin et al. 2014; Thomas et al. 2014; Tilgner et al. 2014; Dong et al. 2015; Minoche et al. 2015). To systematically characterize isoforms and explore their differential regulations in fission yeast, we performed time-course isoform-level profiling during the meiosis of S. pombe. The transcriptome is dramatically reshaped by meiosis, and multiple meiosis-specific AS events have been observed (Mata et al. 2002; Wilhelm et al. 2008). In this study, we summarize the diversity and dynamics of the transcriptome at the isoform level during S. pombe meiosis and explore the conservation and translational potential of AS isoforms.
Results
Experimental design, workflow and definition of AS types
To explore the diversity and dynamics of alternative splicing in S. pombe, we collected six time points of WT cells during meiosis from 0 to 10 h at 2-h intervals (Supplemental Fig. S1). Upon nitrogen starvation, S. pombe is synchronized and enters the premeiotic S phase. After ∼4 h, meiotic division begins and is completed by ∼6 h. Spore maturation follows, and mature spores are usually observed after 8–10 h. Poly(A)+ RNA was purified, reverse-transcribed to cDNA, and sequenced with a PacBio sequencer. Five SMRT cells were used for two RNA replicates of each time point. Raw reads were clustered and polished using the Iso-Seq pipeline, and high-quality (HQ), full-length (FL), polished consensus sequences (referred to here as Iso-Seq reads) were output (Rhoads and Au 2015). Replicates were pooled for downstream analysis. We developed SpliceHunter, a novel software program that detects, quantifies, and compares complex splicing patterns along novel isoforms inferred from Iso-Seq reads. SpliceHunter reports the number of supporting reads of isoforms per sample or time point and predicts protein sequences. Based on the Iso-Seq algorithm, each Iso-Seq read is associated with multiple FL CCS (circular-consensus) reads and non-FL CCS reads. FL CCS reads are defined by co-existence of 5′ and 3′ adaptors and poly(A)+ tail. Therefore, we use the number of FL CCS reads as a proxy for abundance and dynamics analysis.
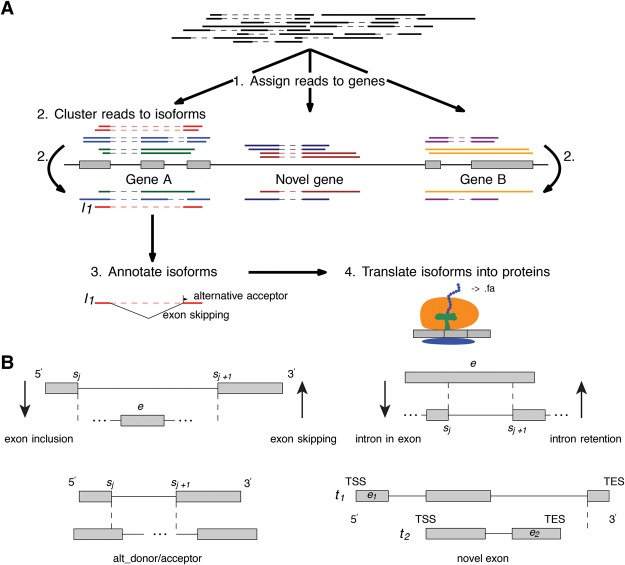
After mapping reads to a reference genome, SpliceHunter assigns reads to annotated or novel transcription units (TUs) (Fig. 1A, step 1) according to well-defined and adjustable criteria. After collapsing compatible reads to isoforms, each isoform is compared to the previously annotated exon-intron structure to detect AS events (Fig. 1A, steps 2, 3), antisense, and read-through transcripts. Finally, the effect of AS events on the encoded protein sequence is analyzed (Fig. 1A, step 4), including their combined shift in the open reading frame (ORF). We consider the following types of AS events (Fig. 1B): exon skipping, exon inclusion, intron retention, intron in exon, alternative acceptors and donors, and novel exons. See Methods for a formal definition of AS types and a detailed description of software tool SpliceHunter.
Figure 1.
Summary of the transcriptome analysis using PacBio sequencing. (A) A pipeline of SpliceHunter analysis. (B) Eight AS types are defined and examined in this study. (Upper left) Exon e is strictly contained in intron (sj,sj+1) and both primary RNA sequences contain sites sj and sj+1 if TSS and TES lie beyond the dashed vertical lines. The skipping and inclusion of exon e is a symmetric event with respect to the roles of reference and novel transcript. (Upper right) Exon e retains intron (sj,sj+1). Intron retention and intron in exon are symmetric events with respect to the roles of reference and novel transcripts. (Lower left) sj and sj+1 form an alternative donor/acceptor pair. They are contained in both primary RNA sequences if TSS and TES lie beyond the dashed vertical lines. (Lower right) Exons e1 and e2 are novel in transcripts t1 and t2, respectively. Although e2 lies within the primary RNA sequence of t1, it does not constitute an inclusion even according to Definition 2 as long as the TES of t2 lies to the left of the dashed vertical line.
Characterization of PacBio reads
In total, we obtained 2,266,791 CCS reads, and 1,311,840 of those were FL CCS reads (Fig. 2A; Supplemental Table S2). Through the Iso-Seq pipeline, 424,511 Iso-Seq reads were generated. The average length of CCS reads was 1285 bp, which was slightly longer than previous reports (Fig. 2B; Sharon et al. 2013; Thomas et al. 2014; Tilgner et al. 2014). The average length of FL CCS reads and Iso-Seq reads were 1094 and 1178 bp, respectively, and the length distributions did not vary across time points (Supplemental Fig. S2A). Six thousand one hundred ninety-nine S. pombe genes (∼90% of all genes) were recovered from PacBio sequencing. Median coverage of FL CCS reads per gene was 71, and many genes had coverage > 100, which enabled deep discovery of novel isoforms (Fig. 2C). Pearson correlation coefficients of read counts between replicates ranged from 0.93 to 0.96 at different time points (Supplemental Fig. S2B), suggesting high technical reproducibility in our study. Additionally, FL CCS read counts showed strong correlation with expression levels inferred from Illumina-based short-read RNA-seq data despite variations in strains, conditions, and techniques (Supplemental Fig. S2C). This high correlation supports the reliability of our time course analysis, which is based on large differences between PacBio read counts at different time points and smooth trends. With respect to PomBase v2.29 definitions of (non-)coding RNA and pseudogenes, 4993 (97.1%) protein-coding genes, 1121 (73%) noncoding RNA (ncRNA) genes, and 18 (62.1%) pseudogenes were recovered. Of the FL CCS reads, 97.4% were assigned to protein-coding genes which represented 80% of all captured genes, indicating higher coverage for protein-coding genes (Fig. 2D). Interestingly, the ratio of RNA molecules corresponding to protein-coding genes versus ncRNA decreased in the middle of meiosis and increased at the late stage (Fig. 2E). We further explored the complexity of PacBio reads by examining the number of introns per read. Although the majority of reads contained only 0 or 1 intron, there were still 190,101 reads with multiple introns, together providing substantial data to explore alternative splicing (Fig. 2F). Lastly, we explored the distances from the 5′ or 3′ end of reads to the corresponding gene transcription start site (TSS) or transcription end site (TES). We focused on reads which extended beyond the annotated TSS or TES because the SMARTer cDNA preparation method does not select for 7-methylguanosine (7mG) which marks intact 5′ end mRNA molecules. On average, reads extend 232 bp upstream of 5′ ends (Fig. 2G) and 188 bp downstream from 3′ ends (Fig. 2H) of annotated genes, suggesting alternative 5′ and 3′ UTRs.
Figure 2.
General properties of PacBio reads. (A) Numbers of CCS reads, full-length CCS reads (CCS-FL), and Iso-Seq reads from two replicates. The second batch has 4× size of the first batch. (B) Length distribution of CCS, CCS-FL, and Iso-Seq reads. (C) Box plot showing the 25th, 50th, and 75th percentiles of the numbers of FL CCS reads for each gene. Whiskers represent 1.5 inter-quartile range. (D) Pie plot showing the percentage of FL CCS reads corresponding to protein-coding genes, noncoding RNA genes, and pseudogenes. (E) Overall trends of FL CCS reads matching protein-coding genes, noncoding RNA genes, and pseudogenes. (F) Counts of FL CCS reads with different numbers of introns. (G) Distribution of distances between the 5′ end of reads to the annotated TSS sites (aTSSs). Reads only outside the annotated TSS sites were counted. (H) Distribution of distances between the 3′ end of reads to the annotated TES sites (aTESs). Reads only outside the annotated TES sites were counted.
Various types of alternative splicing detected in S. pombe meiosis
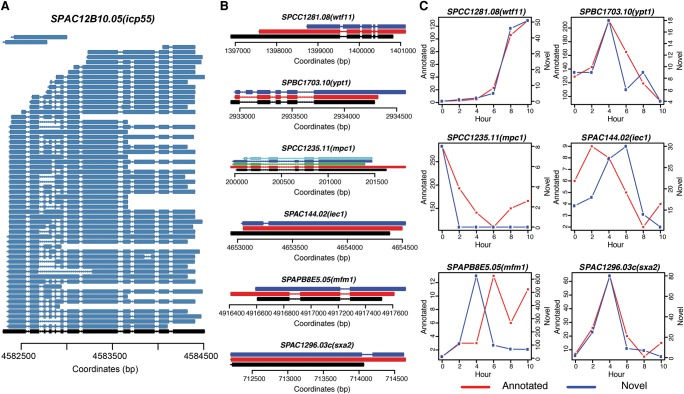
Next, we explored AS isoforms in S. pombe. Although multiple studies have revealed different AS events, this is to our knowledge the first study to explore AS at the isoform level. Not surprisingly, we unveiled complex cases of alternative splicing. For example, we observed 59 distinct polyadenylated mRNA isoforms for SPAC12B10.05(icp55), containing different types of AS events either in the same or different isoforms (Fig. 3A). The capacity to detect multiple AS events in the same molecule by PacBio sequencing avoids underestimating isoform complexity.
Figure 3.
Examples of alternative splicing during S. pombe meiosis. (A) Fifty-nine different isoforms were detected for SPAC12B10.05(icp55). Black represents the annotation and dark blue represents detected isoforms. Aligned nucleotides are denoted by solid lines and alignment gaps indicating introns are marked by thin lines with arrows indicating the strand. (B) Six different types of alternative splicing structures. SPCC1281.08(wtf11) (novel splicing donor or acceptor); SPBC1703.10(ypt1) (exon inclusion); SPCC1235.11(mpc1) (exon skipping); SPAC144.02(iec1) (intron in exon); SPAPB8E5.05(mfm1) (intron retention); SPAC1296.03c(sxa2) (novel exon). Black marks the annotated structure; red depicts isoforms that match the internal annotated structure; all other colors (dark blue, light blue, green) denote structurally different AS isoforms. (C) Dynamics of annotated isoform and novel isoform for each of the six examples shown in B. Red lines represent numbers of FL CCS reads for annotated isoforms and blue lines represent numbers of FL CCS reads for novel isoforms during meiosis.
Alternative splicing comes in many potential forms, and we realized that a formal nomenclature/classification scheme was needed to unambiguously discuss the many events we observed. Thus, we developed a rigorous classification scheme and nomenclature that embraces commonly known instances such as exon skipping, intron inclusion, etc. Based on the definitions implemented in our algorithm (see Methods), we observed examples for all types of AS in S. pombe: an alternative splicing acceptor in SPCC1281.08(wtf11), an exon inclusion in SPBC1703.10(ypt1), multiple exon skipping events in SPCC1235.11(mpc1), an intron in exon in SPAC144.02(iec1), an intron retention in SPAPB8E5.05(mfm1), and a novel exon in SPAC1296.03c(sxa2) (Fig. 3B). Interestingly, some of the novel isoforms had comparable or even more numerous supporting reads than the corresponding annotated isoforms (Fig. 3C). This implies that these isoforms were unlikely to represent rare erroneous byproducts that escaped mRNA surveillance. Although many novel isoforms showed splicing patterns well-correlated with the corresponding annotated isoforms, some novel isoforms exhibited distinct temporal patterns compared to the corresponding annotated isoforms (Fig. 3C), suggesting that the novel isoforms could be temporally differentially regulated. More examples are shown in Supplemental Figure S3.
Landscape of alternative splicing
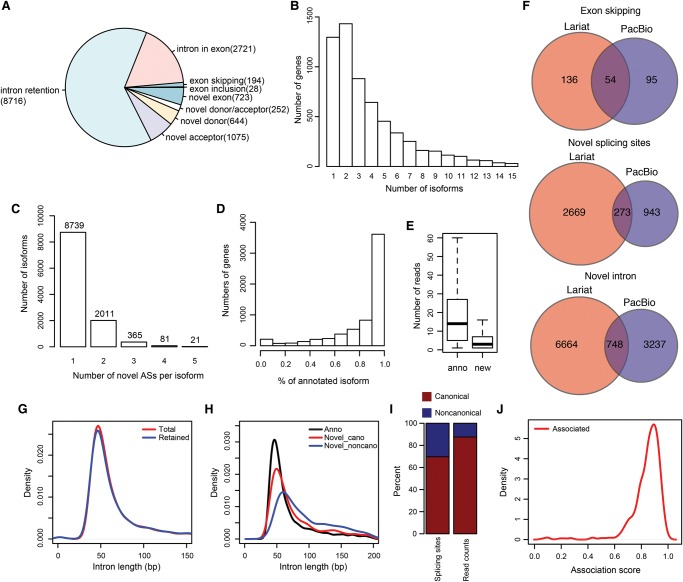
After examining individual examples, we next sought to explore the landscape of AS and isoforms (Supplemental Table S1). The dominant type of AS event observed in S. pombe was intron retention, probably caused by a bypass of individual splicing sites (Fig. 4A; Supplemental Table S3). The retained introns are distributed similarly in length to the total of all annotated introns (Fig. 4G). The next dominant type was “intron in exon,” which can be considered the opposite of intron retention. Other types of AS events were observed but less abundant, including novel splicing sites, novel exon, and exon skipping/inclusion. Next, we asked how many genes have multiple isoforms. Interestingly, only ∼1300 genes have a single detected isoform in this study and 1432 genes have two isoforms with a minimum of one FL CCS read per isoform (Fig. 4B). More than 3000 genes have >2 isoforms, suggesting pervasive complexity of the S. pombe transcriptome and potential AS-mediated regulation. Isoform counts per gene were further analyzed by increasing the minimum number of FL CCS reads required to support an isoform (Supplemental Table S4). Additionally, for the first time we are able to distinguish isoforms with single AS events (8739 or 77.8%) and mRNA isoforms with multiple AS events (22.2%) (Fig. 4C). To examine the molecular association of AS events, we examined 1708 pairs of alternatively retained introns (see Methods), the most abundant type in this study. We found 683 significantly dependent intron pairs at an FDR of 0.05 using Fisher's exact test. To quantify the association, we adopt the intragenic molecular association score previously defined (Tilgner et al. 2015), which is the ratio of the number of reads that retained both introns or skipped both introns to the total number of reads spanning both introns. Associated intron pairs show higher scores, indicating that their retention is co-associated rather than mutually exclusive (Fig. 4J). Only two pairs of the reverse type of event, novel introns in exons, were alternatively spliced in the same molecule, both independently. Similarly, none of 124 mixed pairs of these two types of events were identified to be dependent. Although AS was prevalent in S. pombe, the annotated isoform was the dominant form in the majority of genes (3677 genes had >90% of reads assigned to the annotated isoform) (Supplemental Table S5). However, there were also 648 genes for which the annotated isoforms accounted for less than the sum of the number of reads supporting alternative isoforms (Fig. 4D). In general, FL CCS reads matching annotated isoforms were approximately eightfold more abundant than the reads matching novel isoforms. Besides typical AS isoforms, we also discovered mRNAs apparently encoding 770 new TUs (Supplemental Table S1) and ∼3800 antisense isoforms (Supplemental Table S6) supported by at least one read. Six hundred twenty-five of the new TUs had no intron, with a maximum of 118 supporting reads, and 145 new TUs had introns with a maximum of 60 supporting reads. Five hundred fifty-six of 635 annotated antisense RNA genes were detected from our data, and we additionally detected antisense isoforms targeting 3178 other genes, consistent with a previous study showing the prevalence of antisense meiotic transcripts (Ni et al. 2010). However, the number of reads supporting novel antisense isoforms was generally substantially lower than the number of reads matching annotated antisense isoforms (Fig. 4E). Therefore, there was potentially a mix of real antisense isoforms and transcriptional noise. Furthermore, we examined the conserved intronic dinucleotides at the novel splicing sites. At annotated splicing sites, 99.94% of the dinucleotides are GU-AG, with only three exceptions. However, GU-AG only appeared in 69.67% of novel splicing sites or in 87.5% of reads with novel introns (Fig. 4I) after filtering for high-confidence alignments (see Methods). Neither the dinucleotides nor the full 5′ hexamers from noncanonical novel introns show any consensus sequence (Supplemental Fig. S4F), implying that many of these noncanonical introns might be aberrantly spliced (Supplemental Table S7). Intriguingly, novel introns with canonical splicing sites have similar lengths as annotated introns, but novel introns with noncanonical splicing sites are generally longer (Fig. 4H). Pairs of dinucleotides with small hamming distances to GU-AG occur more often, such as 75 pairs of GU-UG, 55 pairs of GU-GG, etc.
Figure 4.
Landscape of alternative splicing events during S. pombe meiosis. (A) Pie chart showing the fraction of different types of alternative splicing events in S. pombe. Numbers of alternative splicing for each type are listed after the names. (B) Count of genes with different number of isoforms. (C) Isoforms broken up by the number of novel alternative splicing events discovered in individual isoforms. (D) Genes broken up by the percentage of FL CCS reads corresponding to the annotated isoforms. (E) Box plot showing the numbers of FL CCS reads supporting annotated and novel antisense transcripts. (F) Venn graphs showing the comparison between AS events detected by PacBio sequencing and lariat sequencing. (G) Length distribution of all annotated introns and of retained introns. (H) Length distribution of annotated introns, novel introns with canonical splicing sites, and novel introns with noncanonical splicing sites. (I) Fractions of canonical and noncanonical splicing sites (left) and of their supporting read counts (right). (J) Distribution of intra-genic association score for alternative intron pairs at FDR = 0.05.
Interestingly, we discovered some read-through transcripts that spanned two consecutive genes on the same strand, 57 of which contained annotated introns from both genes (Supplemental Tables S1, S8). Among read-through transcripts with introns, 14 had supporting reads more abundant than at least one of the individual transcripts, suggesting that these were less likely to represent chimeric reads. This is consistent with a previous finding of widespread “polycistronic” transcripts in fungi (Gordon et al. 2015). Supplemental Figure S4 shows four examples of read-through transcripts, which are further discussed in the Supplemental Material.
Different methods have been applied in S. pombe to identify alternative splicing, including short-read RNA-seq, ribosomal profiling, and lariat sequencing (Awan et al. 2013; Duncan and Mata 2014; Bitton et al. 2015; Stepankiw et al. 2015). Here, we compared our results with corresponding events predicted from short-read RNA-seq, lariat sequencing, and ribosomal profiling. Fifty-five percent of skipped exons and 38% of novel introns/splice sites reported in this study were also identified by short-read RNA-seq (Supplemental Fig. S4E; Bitton et al. 2015). Ribosomal profiling was performed in meiosis, too (Duncan and Mata 2014), and with the exception of novel start and stop codons, which were not predicted by our study, the remaining eight new isoforms detected were all validated in our study. Lariat sequencing was done in a dbr1Δ mutant strain from log phase, diauxic shift, and heat shock conditions (Stepankiw et al. 2015). The occurrences of three types of AS events were compared, including exon skipping, novel splicing sites, and novel intron. Surprisingly, all three types of AS events showed little overlap between the two data sets (Fig. 4F), presumably because the conditions in the two studies were distinct and the techniques captured different RNA molecules. This comparison suggests that the true complexity of AS in S. pombe is even higher than thus far reported.
Dynamics of AS during meiosis
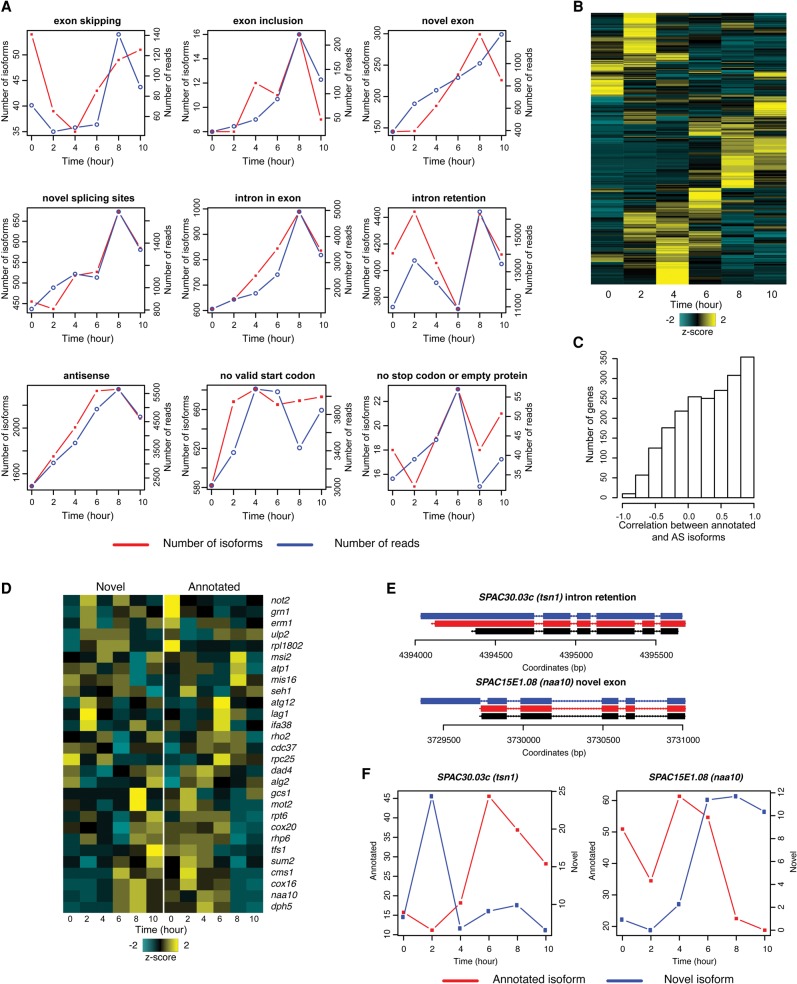
When we examined AS events, we noticed that many alternative isoforms exhibited unexpected temporal patterns compared to annotated isoforms. To explore the dynamics of AS isoforms, we first summarized the general dynamic patterns of different types of AS events by examining the number of isoforms and number of reads supporting each type of AS. Most types of AS increased in meiosis (Fig. 5A), which was likely to be at least partially related to decreased RNA surveillance. However, exon-skipping events decreased at early stages of meiosis and increased at the late stage; intron retention was relatively unchanged. The dynamics of exon skipping suggest a condition-driven alternative usage of different exons between mitosis and meiosis.
Figure 5.
Dynamics of alternative splicing during meiosis. (A) Trends of different types of alternative splicing during meiosis. Red lines represent numbers of isoforms and blue lines represent numbers of FL CCS reads. (B) Heat map showing the temporal pattern of novel isoforms. (C) Histogram showing the correlation between annotated and alternative isoforms during meiosis. (D) Heat map showing the temporal pattern of novel and annotated isoforms. Genes with correlation smaller than −0.4, FL CCS read count > 100, and percentage of annotated isoform < 90% were chosen for hierarchical clustering. (E,F) Examples of anti-correlated isoforms in meiosis. Panel E shows the exon-intron structures of the examples; panel F shows the temporal patterns of the annotated and alternative isoforms.
Next, we examined isoform dynamics at the gene level. The abundance of the majority of individual AS isoforms was also increased during meiosis, consistent with the general patterns (Fig. 5B). We hypothesized that if AS is functionally associated with different conditions, the two isoforms for the same gene are more likely to be uncorrelated or anti-correlated. Therefore, Pearson's correlation coefficients were calculated between the number of reads supporting annotated and alternative isoforms for genes with comparable abundance of each isoform. A left-skewed histogram of coefficients (Fig. 5C; Supplemental Table S9) suggests that the majority of alternative isoforms were correlated in abundance with annotated isoforms. However, there were still 519 genes with anti-correlated isoforms and 104 genes with correlation ≤ −0.5. To explore the anti-correlated isoforms, we selected 28 genes that were supported by more than 100 FL CCS reads for further analysis. The temporal patterns of novel and annotated isoforms were compared in a heat map (Fig. 5D). Some genes had alternative isoforms expressed in mitosis and annotated isoforms expressed in meiosis, whereas a second cluster of genes had annotated isoforms expressed in mitosis and switched to alternative isoforms in late meiosis.
SPAC30.03c (tsn1) encodes a translin protein (Laufman et al. 2005) and is up-regulated in middle and late meiosis (Mata et al. 2002; Martín-Castellanos et al. 2005). Interestingly, besides an increment of annotated isoform expression in middle to late meiosis, we also observed a 48-bp intron retention isoform expressed in early meiosis (Fig. 5E,F). SPAC15E1.08 (naa10) encodes an N-acetyltransferase catalytic subunit, which is essential for survival, and mutations in the S. cerevisiae homolog ARD1 cause sporulation defects (Mullen et al. 1989). The abundance of the novel isoform, which has a novel exon and an intron upstream of the annotated start codon, increased in late meiosis. It is possible that the new 5′ UTR is involved in the gene's expression regulation or provides new start sites. A few examples were further examined in Supplemental Figure S5.
Prediction of novel proteins
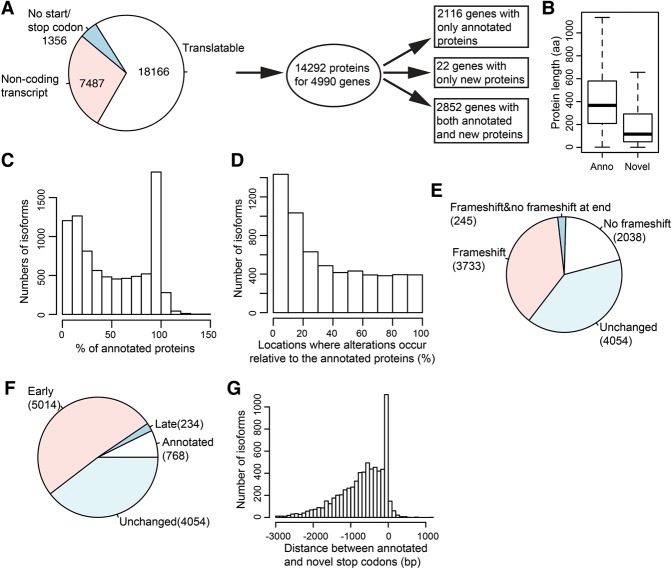
One major role of alternative splicing is to generate distinct functional proteins from the same gene. On the contrary, a product of aberrant splicing is not responsible for generating functional proteins. Therefore, we performed systematic prediction of protein sequences from all isoforms detected in our study (Supplemental Table S10). Translation prediction always started from the annotated start codon if available and stopped at the first stop codon. By this definition, 18,166 isoforms were predicted to be translatable, while the remaining isoforms either belonged to genes that do not encode proteins or lacked the stop codon or annotated start codon (Fig. 6A). About 1200 of these lack the annotated start codon, and all but one of these contain at least one alternate ATG and a stop codon in the same frame and thus might in principle also encode functional proteins, but these isoforms were not studied further.
Figure 6.
Predicting translational products from detected isoforms. (A) Summary of protein prediction. The left pie plot shows the number of translatable isoforms. From 4990 genes, 14,292 protein sequences were predicted, and these genes were further broken up by the status of annotated and new protein sequences. (B) Box plot indicating the length of annotated and novel protein sequences. (C) For isoforms encoding novel protein sequences, the histogram shows the ratio of the length of novel protein sequences relative to the length of the corresponding annotated protein sequence. (D) Histogram showing where alteration occurs in the predicted protein relative to the corresponding annotated protein. (E) Fractions of translatable isoforms with novel AS events which contain frameshift or not. Unchanged refers to AS isoforms with the same protein sequence as the annotated one. (F) Fractions of translatable isoforms with novel AS events which have early, late, or annotated stop codons. (G) Histogram showing the distances between annotated and novel stop codons for isoforms with novel predicted proteins.
From 4990 genes, 14,292 distinct translatable sequences were predicted. Among these genes, 2116 genes were predicted to generate exclusively annotated proteins and 2852 genes were predicted to encode both annotated and novel protein sequences. Interestingly, for 22 genes, we observed a substantial number of reads supporting AS isoforms that encode protein sequences different from the annotated sequence, while the annotated isoform was not supported by any read (Supplemental Table S11). If not misannotated, these isoforms are potentially expressed at a very low level.
A predicted novel protein may not actually be produced. We sought to theoretically address this question by quantifying the impact of alternative splicing on the protein level with respect to the annotated sequence. We examined important protein sequence properties, including its length, relative position of protein sequence alteration caused by AS, how it affects the reading frame, and where the hypothetical translation of novel proteins terminates compared to the corresponding annotated proteins. In general, novel protein sequences were shorter than annotated protein sequences (Fig. 6B). We observed a major mode at isoforms which encode novel protein sequences with lengths similar to annotated proteins and a minor mode at isoforms which encode novel proteins with <20% of annotated protein length (Fig. 6C; Supplemental Fig. S6D). Furthermore, the alteration in predicted protein sequences due to alternative splicing usually occurs at the beginning of a coding sequence (Fig. 6D; Supplemental Fig. S6E). Overall, 40.3% of translatable isoforms with AS events encoded the same proteins as annotated isoforms. More than half of the remaining isoforms which encode novel protein sequences are at least partially not in the same reading frame as the corresponding annotated isoform (Fig. 6E). Furthermore, ∼33.9% of isoforms encoding novel protein sequences were in the same reading frame as annotated isoforms and another ∼4.07% of isoforms had alterations that shifted the reading frame followed by a second alteration that restored the frame. Furthermore, 47.9% of AS isoforms had translation ending at the annotated stop codons (Fig. 6F). Only for 2.32% of AS isoforms, translation ended downstream from, but close to, annotated stop codons (Fig. 6G). Most isoforms with altered protein sequences (49.8%) showed early translation termination with varying distances between annotated and novel stop codons. We call a stop codon novel if it terminates the predicted translation of AS isoforms at a different location than the annotated stop codon.
In addition, we attempted to predict protein-coding regions for the 770 new TUs. The protein length was generally very short with a median length of ∼30 aa; the longest was >200 aa (Supplemental Fig. S6A–C). The majority of novel TUs had no ORF longer than 25% of their full-length sequence. Previous work has hypothesized that some or most ncRNAs represent transcriptional noise (Hüttenhofer et al. 2005; Costa 2007) but may also represent fodder for the birth of new genes (Carvunis et al. 2012). This is supported by the observation that only 13% of the novel TUs we identified were shown to bind to ribosomes in a previous ribosomal profiling study (Duncan and Mata 2014).
Protein sequence conservation and secondary structure
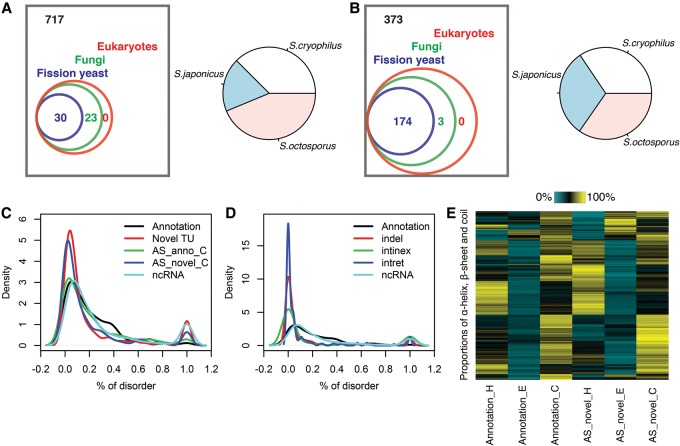
Following the protein prediction analysis, we next sought to check the quality of these novel protein sequences by evaluating their conservation and secondary structure formation. Two major classes of novel protein sequences were examined. Hypothetical proteins predicted from novel TUs and novel C-terminal amino acid sequences starting at the first alteration in AS isoforms. Conservation of novel sequences was assessed by searching for sequence-similar proteins in fission yeasts, fungi, and eukaryotes using BLAST (Gish and States 1993). Secondary and tertiary structures and their properties were predicted by RaptorX (Källberg et al. 2012). For 53 novel TUs, we found evidence for homology in other species with less than 100 matching amino acids (Supplemental Fig. S7C), indicating their potential translation. The majority of them had homologs in other related fission yeasts without significant preference (Fig. 7A; Supplemental Table S12). For example, the predicted protein of NG_554 highly resembles a membrane transporter such as allantoate permease in other fission yeasts, and it could form a structure with 64% α-helix (Supplemental Fig. S7A). The longest protein sequence predicted from a novel TU (NG_569) is similar to retrotransposons in different fission yeasts (Supplemental Fig. S7B). Additionally, we examined conservation of C terminus novel amino acid sequences from 550 AS isoforms which have >2 CCS FL supporting reads and are at least 19 aa long. One hundred seventy-seven AS isoforms have homologs in other species (Fig. 7B; Supplemental Table S12).
Figure 7.
Conservation and secondary structure analysis. (A) Left panel shows the protein BLAST results when using the 770 novel TUs as query sequences. Numbers of novel TUs with alignment hits in different taxa are shown, where the same TU potentially has hits in multiple taxa. Right panel shows the proportion of hits in different fission yeasts. (B) Left panel shows protein BLAST results using as query sequence only the modified part of the 550 protein sequences encoded by AS isoforms. Numbers of AS isoforms with alignment hits in different taxa are shown, where the same AS isoform potentially has hits in multiple taxa. Right panel shows the proportion of hits in different fission yeasts. (C) Distribution of percent of disorder predicted by RaptorX (Källberg et al. 2012). Five classes of protein sequences were examined: full length annotated proteins, longest proteins predicted from novel TUs, C terminus amino acid sequence starting at the first alteration in AS isoforms and its annotated counterpart, and longest proteins predicted from annotated ncRNA. (D) Distributions of percent of disorder predicted for annotated proteins, amino acids affected by insertions and deletions (indel), intron retentions (intret) and introns in exons (intinex) from AS isoforms, and annotated ncRNA. (E) Proportions of α-helix (H), β-sheet (E) and coil (C) predicted from C terminus amino acid sequence starting at the first alteration in AS isoforms and its annotated counterpart.
Next, we systematically evaluated secondary structure formation of novel proteins. Annotated proteins uniformly exhibit low levels of disorder while hypothetical proteins corresponding to the longest ORFs of annotated ncRNA would have a minor mode with complete disorder (Fig. 7C). A similar bimodal pattern was observed for novel TUs and novel C terminus proteins from AS isoforms, as opposed to annotated C terminus proteins. We further explored this pattern of disordered residues among inserted (by intron retentions) and deleted (by introns in exons) amino acid sequences. We consistently observed a major mode of low disorder and a minor mode of complete disorder (Fig. 7D). Then, we asked how alternative C-terminal amino acids in AS isoforms affect secondary structures. Figure 7E and Supplemental Figure S7, D and E suggest that the majority of altered C termini maintain the proportion of α-helix (H), β-sheet (E). and coil (C).
Discussion
In summary, we systematically characterized the transcriptome diversity and dynamics at the isoform level during fission yeast meiosis. We identified thousands of novel isoforms and AS events, including unexpected findings such as novel TUs and read-through transcripts. We further carefully evaluated the functional relevance of AS isoforms by scrutinizing three factors—the abundance of AS isoforms, the differential regulation of annotated and AS isoforms during meiosis, and the translational capacity and quality of AS isoforms. The majority of AS isoforms show similar temporal patterns compared to the annotated isoforms, and the majority of AS isoforms also have shifted reading frames and short predicted protein sequences. However, a few genes show anti-correlated abundance of annotated versus AS isoforms, and these AS isoforms have similar length of translatable sequences, suggesting the possibility of AS-mediated regulation of gene activity in the unicellular organism. We developed SpliceHunter, a novel computational tool for the discovery of AS events and isoform-based sequence analysis from PacBio long sequencing reads. SpliceHunter can process single or multiple samples (e.g., time series). Our results show that the analysis of isoform dynamics is feasible using PacBio sequencing given a small transcriptome and smooth trends.
How complex is the “functional” transcriptome in S. pombe? In agreement with previous studies (Awan et al. 2013; Bitton et al. 2015; Stepankiw et al. 2015), our study shows that AS events are widespread in this unicellular organism. Could it be even more complex than this? Although we captured all AS events identified from prior ribosomal profiling data in meiosis, our AS events had a relatively small overlap with the AS events identified by lariat sequencing performed in log phase, diauxic shift, and heat stress (Fig. 4F). “Constitutive” AS events were detected under different conditions by different techniques and seem unlikely to be random splicing errors. AS events observed in a single condition, on the other hand, potentially include functional, condition-specific cases of AS. Additionally, we focused on isoforms with alternative internal structures, given the limitation of our cDNA library preparation strategy in capturing 5′ intact mRNAs. The diversity of AS increased substantially when we considered isoforms with precise 5′ and 3′ ends different from the annotated ones. Given the differences in AS events between the two studies, a substantially larger diversity of isoforms might be revealed in this unicellular organism when examining additional extrinsic factors like different treatments and environments. On the other hand, in plants and animals, AS events can vary across different tissues or organs. Therefore, it might be interesting to explore AS events in fission yeast with varied “developmental” status besides mitosis and meiosis, such as in young vs. aged cells or mother vs. daughter cells.
Is an isoform truly functional or just a result of splicing errors? This is a critically important question transcending the overall complexity of detected isoforms. Multiple studies have suggested that a majority of AS events are aberrant and not functional despite their prevalence (Bitton et al. 2015; Stepankiw et al. 2015). This hypothesis is supported by (1) the small number of reads supporting AS events compared to the number of reads supporting annotated splicing, (2) an increased prevalence of AS events in mutants of RNA surveillance, and (3) AS events failing to lead to meaningful translational products and ribosomes that are unlikely to be bound (Bitton et al. 2015). Our study is in line with this conclusion from the isoform perspective. Most genes have >90% reads supporting annotated isoform (Fig. 4D); the majority of AS isoforms lead to frameshifts and shorter than annotated protein sequences (Fig. 6C–E), although we describe several interesting exceptions. We additionally show that a majority of genes have correlated abundance between annotated and AS isoform during meiosis, suggesting a lack of regulated AS from the temporal perspective for these genes. Combining all these characteristics, we suspect that a large portion of the novel isoforms are not translationally active. However, the possibility of functional AS is not excluded and could be condition-specific. AS in the alp41 and qcr10 genes were specifically induced by heat or cold shock (Awan et al. 2013).
On the other hand, we find candidate novel transcripts that are likely to be functional. For example, we identified a few hundred genes that have less reads assigned to the annotated isoforms than to the AS isoforms as described here. Moreover, certain AS isoforms were dynamically regulated during meiosis, and some of them even exhibited anti-correlated expression patterns compared to corresponding annotated isoforms (Supplemental Fig. S5). Despite the lack of evidence for translation of novel AS isoforms, we suspect that AS could explain the regulation of some of these genes in meiosis. Furthermore, the combination of conservation analysis and secondary structure modeling indicates that the predicted proteins are potentially able to form stable structures. We infer the functions of some potential novel proteins through their homologs in other related organisms. Future studies are needed to confirm the existence of predicted proteins and diagnose potential physiological functions. Additionally, AS isoforms could play regulatory, noncoding roles, similar to what was observed for some of the antisense isoforms. Finally, the fact that many novel TUs encode protein isoforms not found in closely related species could represent simply a low level of “background noise” arising from aberrant splicing, but it could also represent a mechanism for the evolution of new gene function; some subset of these reading frames may represent species-specific new gene isoform birth (Carvunis et al. 2012).
Overall, these data indicate that fission yeast has a large repertoire of AS events. Although the majority may not function to produce altered proteins, some of these may form a kind of substrate for the evolution of condition-specific isoforms. The remainder could also serve as a resource for neo-functionalization or new isoform birth. Finally, this data set will be a valuable resource for studying AS as a regulatory mechanism for a given gene of interest in this widely used model organism.
Methods
General methods
Standard methods were used for growth and meiosis of S. pombe. RNA extraction and library preparation are described in Supplemental Materials. Iso-Seq pre-processing was performed on the SMRT portal website. We developed software program SpliceHunter to analyze complex splicing patterns along novel isoforms inferred from Iso-Seq reads. SpliceHunter is described in more detail below. R (R Core Team 2015) was used for downstream data analysis and plotting, for which we provide more details in Supplemental Materials.
Alternative splicing detection by SpliceHunter
Initially (step 1 in Fig. 1A), reads are mapped to a reference genome and assigned to annotated genes (Wood et al. 2012) based on matching introns, matching splice sites, or exonic overlap, in this order. If a read does not span any introns or no splice site matches any annotated splice site, the exonic overlap with annotated genes is considered. If the read alignment overlaps multiple genes, the gene with maximal overlap is chosen, provided that the overlap is significantly larger (default 1.5×) than the second largest overlap. Reads that do not overlap any annotated gene are used to infer novel TUs. Ambiguous reads that overlap multiple genes equally well (less than 1.5× difference) were excluded from further analysis.
For each gene and novel TU, all assigned reads are clustered to isoforms by their intron chains and their start and end sites (step 2 in Fig. 1A). Single-exons reads are grouped by the (potentially empty) sequence of retained annotated introns instead. While intron chains and retained introns are required to match perfectly, start and end sites of reads in the same cluster must lie within a sliding window of adjustable size. The start and end site of the isoform representing such a cluster is defined to be the most 5′ start and most 3′ end site, respectively, of any read contained in the cluster. For annotated genes, start and end sites of assigned reads that lie close (default 50 bp) to the annotated start or end site, respectively, are snapped to the annotated sites. Single-exon reads that were not assigned to an annotated gene are grouped to novel TUs simply based on their overlap.
Third (Fig. 1A, step 3), each isoform (sense) is compared to the exon-intron structure of the gene to which it was assigned to detect alternative splicing events (Supplemental Materials, Definitions 1–8). Single-exon isoforms (sense) are tested for intron retention events only. Start and end sites of isoforms that do not show any alternative splicing are annotated as truncated, novel (extends beyond the annotated site), or as annotated. Antisense transcripts are marked in the output as such. Isoforms whose introns match introns of multiple genes on the same strand are output as read-through transcripts. If the matching genes lie on different strands or different chromosomes, the isoform is reported as a fused RNA molecule.
Finally, the effect of alternative splicing events on the protein sequence is studied (step 4 in Fig. 1A). Each (truncated) isoform that has been assigned to an annotated gene is extended on both ends, consistent with the exon-intron structure of the annotated transcript. If the start or end site of the isoform falls into an intron of the annotated transcript, the isoform cannot be extended in 5′ or 3′ direction, respectively. If the (extended) isoform stretches beyond the position of the annotated start codon, the isoform is translated starting at the annotated start codon. For all isoforms whose structures imply an open reading frame, the corresponding protein sequences are output in FASTA format. For all remaining isoforms, the negative translation status is marked accordingly. For novel TUs, the longest ORF is reported.
Furthermore, for each isoform all alternative splicing events are annotated with the number of inserted or deleted amino acids and the resulting shift in reading frame. SpliceHunter aligns novel protein sequences with corresponding annotated sequences and outputs a sequence of strings with pattern Tx_y^z. T is either “D” or “I” and indicates whether the alignment gap is a deletion (“D”) or insertion (“I”), x gives the length of the gap, y the position with respect to the reference sequence, and z the frame (0,1,2) following the current gap. At the end of the string, =d+/− denotes the positive or negative distance of the active stop codon from the annotated stop codon.
When provided with samples from multiple replicates across different time points, SpliceHunter pools all samples to cluster reads to isoforms (see second step) but keeps track of the distribution of supporting FL CCS reads across time points (see 8th column in Supplemental Table S1) or (optionally) across individual samples. SpliceHunter is also able to provide certificates for each predicted isoform. Certificates list, separately for each time point, all Iso-Seq read names from the input alignment (.bam) files that are contained in the cluster of an isoform (see second step). These certificates can be used to extract from the input .bam files the Iso-Seq reads that define an isoform across different time points and to visualize its dynamics as we illustrate for two examples in Supplemental Figure S8.
Software availability
SpliceHunter is free open-source software released under the GNU GPL license and has been developed and tested on a Linux x86_64 system. SpliceHunter's source is available as Supplemental Material and at https://bitbucket.org/canzar/splicehunter.
Data access
PacBio sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE79802.
Supplementary Material
Acknowledgments
We thank Amar Klar for the S. pombe strains. We thank Haiping Hao for assistance with sequencing, and Alix Kieu and Roberto Lleras, Pacific Biosciences, for in-kind contribution of SMRT cells and suggestions for analysis. We thank Ed Miller for providing funding to the High Throughput Biology Center to acquire the PacBio sequencer, and Steven Salzberg and Kai Kammers for helpful discussions.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.208041.116.
References
- Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, Williams BA, van Bakel H, Schadt EE, Reijo-Pera RA, Underwood JG, et al. 2013. Characterization of the human ESC transcriptome by hybrid sequencing. Proc Natl Acad Sci 110: E4821–E4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan AR, Manfredo A, Pleiss JA. 2013. Lariat sequencing in a unicellular yeast identifies regulated alternative splicing of exons that are evolutionarily conserved with humans. Proc Natl Acad Sci 110: 12762–12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitton DA, Atkinson SR, Rallis C, Smith GC, Ellis DA, Chen YY, Malecki M, Codlin S, Lemay JF, Cotobal C, et al. 2015. Widespread exon skipping triggers degradation by nuclear RNA surveillance in fission yeast. Genome Res 25: 884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. 2013. Dynamic integration of splicing within gene regulatory pathways. Cell 152: 1252–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis A, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B. 2012. Proto-genes and de novo gene birth. Nature 487: 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF. 2007. Non-coding RNAs: lost in translation? Gene 386: 1–10. [DOI] [PubMed] [Google Scholar]
- Dong L, Liu H, Zhang J, Yang S, Kong G, Chu JS, Chen N, Wang D. 2015. Single-molecule real-time transcript sequencing facilitates common wheat genome annotation and grain transcriptome research. BMC Genomics 16: 1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CD, Mata J. 2014. The translational landscape of fission-yeast meiosis and sporulation. Nat Struct Mol Biol 21: 641–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, et al. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323: 133–138. [DOI] [PubMed] [Google Scholar]
- Eser P, Wachutka L, Maier KC, Demel C, Boroni M, Iyer S, Cramer P, Gagneur J. 2016. Determinants of RNA metabolism in the Schizosaccharomyces pombe genome. Mol Syst Biol 12: 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish W, States DJ. 1993. Identification of protein coding regions by database similarity search. Nat Genet 3: 266–272. [DOI] [PubMed] [Google Scholar]
- Gordon SP, Tseng E, Salamov A, Zhang J, Meng X, Zhao Z, Kang D, Underwood J, Grigoriev IV, Figueroa M. 2015. Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS One 10: e0132628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A, Schattner P, Polacek N. 2005. Non-coding RNAs: hope or hype? Trends Genet 21: 289–297. [DOI] [PubMed] [Google Scholar]
- Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. 2012. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7: 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G. 2010. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11: 345–355. [DOI] [PubMed] [Google Scholar]
- Laufman O, Ben Yosef R, Adir N, Manor H. 2005. Cloning and characterization of the Schizosaccharomyces pombe homologs of the human protein Translin and the Translin-associated protein TRAX. Nucleic Acids Res 33: 4128–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 28: 215–220. [DOI] [PubMed] [Google Scholar]
- Lee Y, Rio DC. 2015. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem 84: 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JA, Johnson NV, Gross SM, Schnable J, Meng X, Wang M, Coleman-Derr D, Lindquist E, Wei C, Kaeppler S. 2014. A near complete snapshot of the Zea mays seedling transcriptome revealed from ultra-deep sequencing. Sci Rep 4: 4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Castellanos C, Blanco M, Rozalén AE, Pérez-Hidalgo L, García AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P. 2005. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol 15: 2056–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bähler J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147. [DOI] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CW. 2008. Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem Sci 33: 385–393. [DOI] [PubMed] [Google Scholar]
- Minoche AE, Dohm JC, Schneider J, Holtgräwe D, Viehöver P, Montfort M, Sörensen TR, Weisshaar B, Himmelbauer H. 2015. Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol 16: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kayne PS, Moerschell RP, Tsunasawa S, Gribskov M, Colavito-Shepanski M, Grunstein M, Sherman F, Sternglanz R. 1989. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J 8: 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Tu K, Wang Z, Song S, Wu H, Xie B, Scott KC, Grewal SI, Gao Y, Zhu J. 2010. The prevalence and regulation of antisense transcripts in Schizosaccharomyces pombe. PLoS One 5: e15271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Rhoads A, Au KF. 2015. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13: 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D, Tilgner H, Grubert F, Snyder M. 2013. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol 31: 1009–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepankiw N, Raghavan M, Fogarty EA, Grimson A, Pleiss JA. 2015. Widespread alternative and aberrant splicing revealed by lariat sequencing. Nucleic Acids Res 43: 8488–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Underwood JG, Tseng E, Holloway AK. 2014. Long-read sequencing of chicken transcripts and identification of new transcript isoforms. PLoS One 9: e94650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Grubert F, Sharon D, Snyder MP. 2014. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc Natl Acad Sci 111: 9869–9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Jahanbani F, Blauwkamp T, Moshrefi A, Jaeger E, Chen F, Harel I, Bustamante CD, Rasmussen M, Snyder MP. 2015. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat Biotechnol 33: 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB. 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J. 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243. [DOI] [PubMed] [Google Scholar]
- Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, et al. 2012. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 40: D695–D699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.