Abstract
Incipient Alzheimer’s Disease (AD) is characterized by a slow onset of clinical symptoms, with pathological brain changes starting several years earlier. Consequently, it is necessary to first understand and differentiate age-related changes in brain regions in the absence of disease, and then to support early and accurate AD diagnosis. However, there is poor understanding of the initial stage of AD; seemingly healthy elderly brains lose matter in regions related to AD, but similar changes can also be found in non-demented subjects having mild cognitive impairment (MCI). By using a Linear Mixed Effects approach, we modelled the change of 166 Magnetic Resonance Imaging (MRI)-based biomarkers available at a 5-year follow up on healthy elderly control (HC, n = 46) subjects. We hypothesized that, by identifying their significant variant (vr) and quasi-variant (qvr) brain regions over time, it would be possible to obtain an age-based null model, which would characterize their normal atrophy and growth patterns as well as the correlation between these two regions. By using the null model on those subjects who had been clinically diagnosed as HC (n = 161), MCI (n = 209) and AD (n = 331), normal age-related changes were estimated and deviation scores (residuals) from the observed MRI-based biomarkers were computed. Subject classification, as well as the early prediction of conversion to MCI and AD, were addressed through residual-based Support Vector Machines (SVM) modelling. We found reductions in most cortical volumes and thicknesses (with evident gender differences) as well as in sub-cortical regions, including greater atrophy in the hippocampus. The average accuracies (ACC) recorded for men and women were: AD-HC: 94.11%, MCI-HC: 83.77% and MCI converted to AD (cAD)-MCI non-converter (sMCI): 76.72%. Likewise, as compared to standard clinical diagnosis methods, SVM classifiers predicted the conversion of cAD to be 1.9 years earlier for females (ACC:72.5%) and 1.4 years earlier for males (ACC:69.0%).
Introduction
AD is a disease with both brain pathological processes and clinical decline occurring gradually, with dementia representing the last stage of many years of accumulation of these pathological changes. Changes begin several years before the onset of clinical symptoms and there is strong supporting evidence that it is related to the early abnormal production of β−amyloid (Aβ) peptide in the Cerebrospinal fluid (CSF). β− amyloidosis and the increase of CSF- τ protein precede neuronal dysfunction and neurodegeneration; and all of these precede cognitive changes and correlate with clinical symptom severity [1, 2]. Neurodegeneration begins with a typical pattern of early neurofibrillary tangles in medial temporal lobe structures, mainly entorhinal cortex and hippocampus, and subsequently extending throughout most of the temporal lobe and the posterior cingulate. Finally, this pattern involves extensive cortical regions, especially parietal, prefrontal and orbitofrontal; and in an advance stage, change in several brain structures correlate closely with changes in cognitive [3, 4]. In addition to the foregoing, it is known that the apolipoprotein E (APOE) ϵ4 allele is the most prevalent genetic risk factor for AD [5]. Carriers of ϵ4 attain AD at an earlier age than non-carriers [1, 5]; this may be due to ϵ4 being involved in early Aβ plaque concentration and neuro-degeneration. However, in spite of these findings, it is important to be clear that ϵ4 carriers merely inherit an increased risk of developing the disease, but not all people with AD are ϵ4 carriers, and not all ϵ4 carriers will develop the disease.
Indeed, to understand the specific effects of AD on brain structures, it is important to study and differentiate their age-related changes in the absence of disease. Ageing causes changes at all levels, from molecular to morphological, in the brain which include shrinking. Studies have demonstrated that seemingly healthy subjects lose brain matter over time and brain age-related changes and function are not uniform across the whole brain or over subjects. The volume of the brain decreases with age at a rate of approximately 0.2–0.5% per year [6] and this rate might be even greater over the age of 70 [7]. Regionally, ageing-related atrophy has been observed across many of the cortical regions [6, 8, 9] with a prominent decline in the prefrontal cortex and the slight decline of the temporal cortex and parahippocampal cortex [8]. Decline has been also found in several subcortical structures including the caudate nucleus, amygdala, cerebellum and hippocampus [6, 9], the latter being the most studied structure, with annual atrophy rate of about 2.0% [6]. Studies on cross-sectional and longitudinal MRI have found significant correlations between gender and cortical and subcortical regions, but there are inconsistencies between the results [8, 9]. Finally, another factor to consider with regard to the effects of ageing on the brain and cognition is the influence of socio-demographic characteristics. Both higher levels of education or occupational attainment may act as a protective factor. For example, studies have found that higher levels of education may increase the brain reserve, leading to larger brain structures, which can help to counter the effects of brain atrophy [9, 10].
However, many of these age-related changes are shared by neurodegenerative diseases. Some studies have found that cognitively normal elderly (HC) have Aβ deposition in the brain with similar levels of the substances observed in subjects with dementia due to AD [11]. Similarly, despite an increase in τ protein having been seen in AD as compared to HC, τ deposition is found in other neurodegenerative diseases referred to as tauopathies [12]. At the macroscopic level, ageing has an effect on brain morphology with atrophy caused by dendritic pruning and loss of synapses and neurons. Part of this atrophy occurs in areas vulnerable to AD, while other changes have been observed in areas less characteristic of early-stage AD [6, 8, 9]. Because of the shared biochemical and morphological characteristics, it must be pointed out that it is a complex task to discriminate some of these age-related changes in healthy elderly subjects from subjects affected with early stage of AD.
In clinical practice, neuropsychological tests such as, the Clinical Dementia Rating (CDR) [13], the Mini-Mental Examination Score (MMSE) [14] and the Alzheimer’s Disease Assessment Scale cognitive subscale are used to monitor AD progression or treatment efficacy. However, although these tests unquestionably reflect an important aspect of disease progression (i.e. functional impairment), they also have several limitations such as relatively low specificity [15] and reliability [16]. During the past few years, there has been a considerable effort to identify additional biomarkers as early indicators of pathological changes to improve the accuracy of the clinical diagnosis of possible/probable AD and the prediction of disease progression from Mild Cognitive Impairment (MCI) which is a symptomatic predementia phase of AD [17]. In spite of CSF-Aβ and CSF-τ have been suggested as the most informative AD biomarkers [18]; several studies also suggested their combination with other features types, such as Magnetic Resonance Imaging (MRI)-based biomarkers, to increase early prediction accuracy [19]. Currently, neuroimaging and specifically structural MRI biomarkers support earlier and more precise diagnosis and measurement of progression. MRI Biomarkers of atrophy of medial temporal regions have been validated for early diagnosis of subjects at the MCI stage. Furthermore, MRI images used to measure rates of whole-brain and hippocampal region are considered powerful markers of progression of neurodegeneration being used as outcomes in clinical trials [4].
Alternatively, studies based on high dimensional methods have approached the HC/MCI/AD early prediction problem of automatically classifying brain regions and subjects from structural MRI-based biomarkers. These biomarkers allow the careful examination of morphological changes and their correlation with the progression of cognitive impairment. Multivariate techniques such as Orthogonal Partial Least Squares (OPLS), Support Vector Machine (SVM) and Relevance Vector Machine (RVM), among others, have enabled the correlation and variance of these biomarkers to be evaluated in a more easily interpreted way and with greater statistical power as compared to univariate approaches. Likewise, MRI biomarkers have been used in combination with other types of biomarkers, such as Positron Emission Tomography (PET)-based features, CSF-based biomarkers, Genetic-based biomarkers and socio-demographic and neuropsychological features to improve early diagnosis performance. Recent studies have proposed an age estimation framework by training RVM for regression (RVR) from MRI-based biomarkers of healthy subjects, which was used to estimate the age of MCI and AD subjects and to recognize faster brain atrophy [20, 21] and to predict the conversion from MCI to AD [22]. Other studies have proposed methods to calculate an MRI-based AD severity index from cross-sectional [23] and longitudinal studies [24], where multivatiate models based on OPLSR were built from HC and AD subject data. Models were applied to MCI subjects for early prediction of conversion to AD through subject classification as either healthy control-like or as AD-like. Besides the above-mentioned methods, studies have also applied SVM algorithms to improve prediction quality by selecting the most significant brain structures or other feature types in HC, MCI and AD subjects [3, 19, 25–29]; and to discriminate between these subject groups [3, 19, 25–28, 30, 31]. However, in spite of applied multivariate techniques and the features used, all these studies are focused mainly on three classification experiments: (1) HC vs AD, (2) HC vs MCI and (3) MCI vs AD; where the last one is mainly directed at discriminating between the stable MCI (sMCI, MCI who had not converted to AD) vs converted to AD (cAD, subjects who had converted to AD). For HC vs AD, the methods mentioned achieved high accuracy values (up to 94.9% sensitivity and 96.33% specificity); and from 78% to 85% on MCI vs AD. However, for the detection of prodromal AD (sMCI vs cAD), the sensitivity is substantially lower (below 60%) for most methods.
In this study, we aimed to characterize the age-related changes in brain structures and to identify the variant (vr) and quasi-variant (qvr) MRI biomarkers to build ageing-based null models. Null models were built from identified HC subjects (n = 46) with a normal CSF-profile. Most importantly, we addressed the HC/MCI/AD subject classification and the early prediction of conversion to AD, by using these null models to estimate the age-related values of vr and qvr MRI biomarkers for longitudinal data of HC (n = 161), MCI (n = 209) and AD (n = 331) subjects. Residuals were then calculated as deviation scores of observed MRI-based biomarkers from estimated normal MRI-based biomarkers. Support vector machines (SVM) were used to build residual-based classifiers for three experiments: MCI vs HC, AD vs MCI and AD vs HC. The advancement for early disease prediction was calculated as the number of years that the proposed method leads in predicting the last known subject diagnostic. Data used in this study was obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) study (adni.loni.usc.edu).
Data
ADNI cohorts
The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and AD. For up-to-date information, see www.adni-info.org.
The complete ADNI project enrolled 819 adult subjects, aged 55—90 years, and recruited from over 50 sites across the United States and Canada. These subjects met entry criteria for a clinical diagnostic at baseline (see S1 Appendix) of late MCI (LMCI, n = 398), early probable AD (n = 193) and control normal (CN, n = 229) [32]. ADNI has been used by many number of publications focused on the characterization of age-related brain changes [6, 33] and the early prediction of conversion to AD [19, 23, 31, 34, 35]; a recent review has been published by Weiner and colleagues in Alzheimer & Dementia journal [36]. Details about the procedures for selection of participants and the full study protocol have been presented in [16, 37]. Also, details of the acquisition of structural MRIs of the participants can be found in ADNI project site (adni.loni.usc.edu).
CSF biomarkers
The ADNI CSF-based biomarkers were measured for Aβ 1 to 42 peptide (CSF-Aβ), total tau (CSF-τ) and τ phosphorylated at the theorine 181 P-τ181P concentrations using the xMAP platform (Luminex Corp, Austin, Texas) and INNO-BIA AlzBio3 research-use-only reagents. Biomarkers analysis applied by ADNI are described in detail at http://adni.loni.usc.edu/methods/biomarker-analysis/. CSF biomarkers set was conformed by CSF-Aβ and CSF-τ, in pg/mL.
MRI-based Biomarkers
MRI-based biomarkers used in this study correspond to measured of neuro-degeneration available in ADNI at 5 years follow-up. These biomarkers were obtained with the FreeSurfer image analysis suite (version 5.1.0) developed by Martinos Center for Biomedical Imaging and freely available at the “Freesurfer wiki site” (http://surfer.nmr.mgh.harvard.edu/fswiki). FreeSurfer has been used in several studies dedicated to automatically obtaining MRI-based measures from whole brain or specific regions to sucessfully predict the early conversion from MCI to AD subjects [19, 23, 24, 31, 34, 38, 39]. Technical details of the FreeSurfer processing pipeline have been described in prior publications [40, 41]. Biomarkers with missing values for most samples were discarded. Likewise, we only included the ones correctly processed and available for at least two time points. In this sense, we included an unbalanced longitudinal data of 166 longitudinal MRI-based biomarkers measured at multiple time points: baseline, 6, 12, 18, 24, 36, 48 and 60 months. These biomarkers correspond to cortical volume (CV), cortical thickness average (TA) and the volume estimates of a wide range of sub-cortical structures (SV).
Subjects and inclusion criteria
Participants were selected from original ADNI study if they met the following criteria (at the time of the study, April, 2015): (1) Had all selected longitudinal MRI images correctly processed (2) Had completed demographic and neuropsychological data and were clinically diagnosed at each visitation. In total, longitudinal data of 747 subjects (215 CN, 366 LMCI and 166 AD) were studied. Demographic details of the studied cohort are given in Table in S1 Table. Summaries are grouped by gender and correspond to baseline stage. It should be noted that just 164 women and 236 men have available measures of CSF-based biomarkers.
Methods
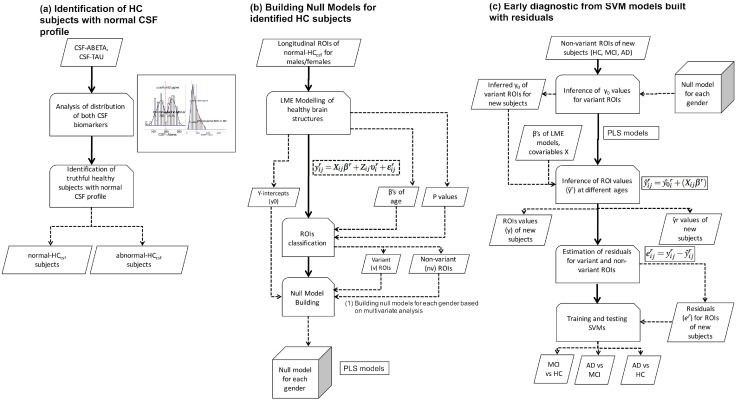
This study was divided into three main stages: (1) Subject classification, (2) Building the ageing-based null models, and (3) Residuals-based early prediction of conversion to MCI/AD and HC/MCI/AD classification with SVM classifiers. Fig 1 illustrates a schematic diagram of the proposed framework.
Fig 1. Proposed framework.
(A) HC subjects with normal CSF profile are identified from cutoff values calculated from CSF biomarkers distributions. (B) Longitudinal ROIs of these subjects are modelled using LME approach, variant (vr) and quasi-variant (qvr) ROIs and Y-intercepts (y0) ROIs values are identified and then null models for both genders are built from these values by applying multivariate modelling. (C) qvr ROIs values of new HC, MCI and AD subjects are used within null models to infer the y0 values of vr ROIs. Estimated ROIs values () at different ages are estimated by linear regression by using y0 and β coefficients of age and education. Residuals are calculated as the difference ; and finally, SVM classifiers are trained for subject classification and addressing the early diagnosis problem: HC vs MCI, MCI vs. AD and HC vs AD. The full workflow of last two stages is applied separately for each gender.
Classification of Subjects
Since the diagnosis of MCI and AD is progressive, in addition to ADNI clinical assessment established at every visitation (dxage), we aim to control the last ADNI clinical diagnosis of each subject at time of this study, by building a time-invariant diagnosis variable (dxlast). This variable labels subjects according the following classes: (1) stable HC (sHC), (2) stable MCI (sMCI), (3) converted to MCI (cMCI), (4)stable AD (sAD) and (5) converted to AD (cAD). Because previous studies have found ADNI participants with abnormal concentrations of CSF-based biomarkers even in healthy elderly subjects [18, 42], we decided to study the subjects with available CSF data (see Table in S1 Table) and to discriminate the ones with normal CSF-profile from the ones with abnormal concentrations. For this propose, we investigated the cut-off values previously established by [18] (CSF-Aβ: 192pg/ml, CSF-τ: 93pg/ml) [18] from their study with ADNI-independent autopsy-based samples, on our samples at long-term follow-up times (84 months). As many of these subjects have more than one measurement available for both CSF-based biomarkers, the normal or abnormal profile was evaluated for all observations. In this way, we expected to classify these subjects through the other time-invariant feature (dxcsf), which combines their dxlast state and their CSF profile. Table 1 gives a brief description of all diagnosis variables used in this study. Once the subjects were characterized according to dxlast and dxcsf, two cohorts were established: (1) The null model cohort, integrated by the stable HC subjects with normal CSF profile (normal-HCcsf); and (2) The early prediction cohort, integrated by the remaining subjects who did not meet the previous condition.
Table 1. Classification results with current diagnostic.
| Source | Diagnostic variable | Class | Class description |
|---|---|---|---|
| ADNI | dxbl | CN | Control normal at baseline |
| LMCI | Late MCI at baseline | ||
| AD | Early probable AD at baseline | ||
| dxage | NL | Subjects diagnosed as stable normal at current visitation | |
| NL to MCI | Subjects diagnosed as MCI at current visit who previously were NL | ||
| NL to Dementia | Subjects diagnosed as dementia due to AD at current visit who previously were NL | ||
| MCI | Subjects diagnosed as stable MCI at current visit who previously were also MCI | ||
| MCI to Dementia | Subjects diagnosed as dementia due to AD at current visit who previously were MCI | ||
| Dementia | Subjects diagnosed as stable dementia due to AD at current visit who previously were also MCI | ||
| Our study | dxlast | sHC | Subjects labelled as HC who remained like HC in all follow-up visits (who did not become MCI or AD) |
| sMCI | the MCI subject who did not become AD | ||
| cMCI | Subjects initially labelled as HC who subsequently have converted to MCI | ||
| sAD | Subjects who remained like probable or possible AD all the follow-up visits | ||
| cAD | Subjects labelled as HC or MCI who subsequently have converted to probable or possible AD | ||
| dxcsf | normal-HCcsf | sHC subjects with normal CSF profile | |
| abnormal-HCcsf | sHC subjects with abnormal CSF profile | ||
| normal-MCIcsf | sMCI and cMCI subjects with normal CSF profile | ||
| abnormal-MCIcsf | sMCI and cMCI subjects with abnormal CSF profile | ||
| normal-ADcsf | sAD and cAD subjects with normal CSF profile | ||
| abnormal-ADcsf | sAD and cAD subjects with abnormal CSF profile |
Building the ageing-based null models
In this stage, we analyzed data from the null model cohort. Every MRI-based biomarker of the normal-HCcsf group was standardized to have zero mean and unit variance. Here, we also refer to an MRI-based biomarker as a Region of Interest (ROI) when we refer to the modelling process.
Modelling temporal change in ROIs
To visualize the between-subject and within-group variabilities on normal-HCcsf, ROIs were represented by quartiles. The change in cortical and subcortical brain regions over 5 years was calculated by applying the LME approach for every ROI. The subject age (age) at each observation and the years of education (educ) were included as covariates in all models. Because we hypothesize that there are important individual-level effects and believe that subjects have similar rates of change over time, we fitted random intercepts LME models. In this type of model, the measured value of ROI r defined as yij is assumed to have a set of parameters β, fixed across subjects. In addition, for each individual i, a set of random parameters υi is assigned that models the deviation from the fixed effect β. For i = 1, …, n, each model reads as follows:
| (1) |
where, is the standardized value of ROI r measured for the ith subject in the jth observation; i = 1, …, n subjects, j = 1, …, ni available observations for subject i and r = 1, …, nr, nr = 166 ROIs. Xij is a ni x p design matrix, where p is the number of covariates (age, educ and the constant term of 1’s) on the jth observation of ith subject. βr is the p x 1 vector of unknown fixed effects or regressor’s coefficients, which are: (coefficient for constant term or Intercept), (coefficient for age) and (coefficient for educ). Zij is a known design matrix of size ni x q, where q is the number of random effects for the jth observation of subject i. is the q × 1 vector of unknown random effects coefficients ∼Nq(0, ψ) for subject i measured for ROI r. is the ni x1 residual vector of errors ∼Nni(0, σ2 λi) for the jth observation in subject i measured for ROI k. ψ is the q × q covariance matrix for the random effects. 0, σ2 λi is the ni x ni covariance matrix for the errors in subject i.
LME modelling was performed using the lme4 package available for R [43]. See S1 Appendix for further detail of the specific LME approach.
Identification of variant and quasi-variant ROIs
The predictor estimates of LME models were interpreted the same way as the coefficients from a traditional regression. For instance, a one year increase in the regressor age corresponded to the effect of age (βa) increase or decrease in the outcome. Taking into account the βa’s, we classified as variant (vr) ROIs the ones that had both an annual change greater than 1% on ROI standard deviation and a significant change at p-values ≤ 0.05. The ROIs with annual change less than or equal to 1% were considered as quasi-variant (qvr), where all of these did not have a significant change.
Inference of variant ROIs y-intercept values from quasi-variant ROIs
The Y-intercept value (y0) from each ROI, which represents the subject-specific ROIs measure at basal stage (age = 0), was directly obtained from all LME models. We assumed that for healthy elderly people, the qvr ROIs values remains basically the same along time, even at basal stage, which is not true in the vr. We also assumed there are a strong correlation between the y0 set of both ROI types. Therefore, we built the HC null model based on the Partial Least Squares Regression (PLSR) algorithm [44] to infer the set of y0’s for vr ROIs in function of the set of y0’s for qvr ROIs. PLSR is a linear algorithm particularly useful to analyze data with strongly collinear (correlated), noisy, and numerous predictors variables, and also simultaneously model several response variables [45]. There are several algorithms proposed to implement the PLSR model. In this study, we use the kernel algorithm [46] available in the pls package available for R [47]. To determine the optimal number of components to take into account, it was used leave-one-out (LOO) cross-validation method available in this package. LOO calculates potential models excluding one observation at a time. See S1 Appendix for a short PLSR description.
Early disease prediction based on residuals
In this stage, we addressed the subject classification and early disease prediction problems using data from the early prediction cohort (HC, MCI and AD), unused in the previous stage. ROIs of these subjects were standardized according to the mean and standard deviation of the null model cohort.
Inference of age-associated ROIs values for new subjects
For each subject, the y0’s of vr ROIs were inferred from the y0’s of qvr ROIs using the same PLSR model described in previous section. Once the y0 set for both ROIs types was completed, we used these values in combination with the vector of coefficients β, see Eq (1) and the observed covariates (age and educ), to infer the expected values of each ROI () according to Eq (2). Here,the inferred value represents the value that should measure the ROI at age ≠ 0 whether the subject is healthy or not.
| (2) |
where represents the inferred or predicted value of the rth ROI variant on the jth observation for the ith subject. Xij is the design matrix with observed covariates at age ≠ 0 without the constant term. βr is the vector of calculated fixed effects of ROI r obtained from its LME model, according Eq (1).
Computing the residuals of variant ROIs
In order to get a meaningful deviation value of ROIs from an inferred healthy subject-specific trend, the difference between the estimated () and the true () ROIs values, here called residual (e), was computed. The residuals for each ROI were calculated as following:
| (3) |
where and are the observed and inferred values, respectively, for each ROI r measured on the ith subject in the jth observation. The e’s for all ROIs were stored in a matrix E.
In S1 Appendix we describe a hypothetical example of how we have used the LME and PLSR approaches to infer the ROI values at basal stage and over time; and finally, the residuals.
Diagnosis prediction using Support Vector Machines
In this stage, we used the matrix of residuals E to address two problems. The first one was focused on the subject classification, where the vector of class labels, used as the outcome, was the diagnostics at the time of the visit, dxage (Table 1). The second problem was the early disease prediction, here, we trained a classifier that predicts the future diagnosis of the subjects given their current clinical tests, i.e. what is the expected diagnostic of the subject some years after the current visit. The vector of class labels used to train this classifier was dxlast (Table 1). In this case, as the feature set used was obtained in previous visits of the subject, the classifier learns to predict the future outcome of a subject, given his present state.
For both problems, we performed three experiments focused on the binary classification problems: (1) HC vs AD, (2) HC vs MCI and (3) MCI vs AD. The MCI vs AD experiment in the early prediction problem was focused on addressing the problem of prodromal stage of AD, by comparing the sMCI (stable MCI over all visitations) with the subject initially diagnosed as MCI who became to AD over time (cAD). Likewise, for each problem, two configurations of the feature set have been tested. Let Ev be the subset of matrix E where only the columns of either the vr ROIs or the qvr ROIs whose residuals are different from zero were included. The first feature set used in each experiment, F1, included the residuals from matrix Ev plus the age (age of the subject at the time of the visit). The second feature set used, F2, includes the information in F1 together with the results from the CDR global score (CDRGLOBAL) and MMSE tests. The goal of testing two training sets was to assess the influence of the neuropsychological tests in the quality of diagnostic as opposed to the use of ROI residuals with age only.
The classification method used to carry out each experiment is a Support Vector Machine (SVM), configured with a Gaussian radial kernel and a misclassification cost parameter C = 1. Each experiment has been evaluated using a ten-fold cross validation with a specific constraint: as ADNI is a longitudinal database, there may be multiple samples per subject (multiple visits). The subjects were assigned to either the training or the test subsets, in order to perform a fair evaluation. On each cross validation run, 60% of the subjects weren assigned to the training subset and the remaining 40% to the test subset.
Because, it is known that the rate of atrophy increases with age, for studying the development of our method for early disease prediction, SVM classifiers were built on age groups. The ADNI data used to evaluate the classification for males and females are summarized in tables in S2 and S3 Tables, respectively. These tables provide demographic information and the number of measurements and diagnoses by each age group.
Since previous studies have suggested that there are gender differences in brain atrophy with aging [8, 9, 48–50], the full workflow in building null models and early diagnostic from SVM models was applied separately for each gender.
Results
Healthy elderly subjects with normal CSF profile
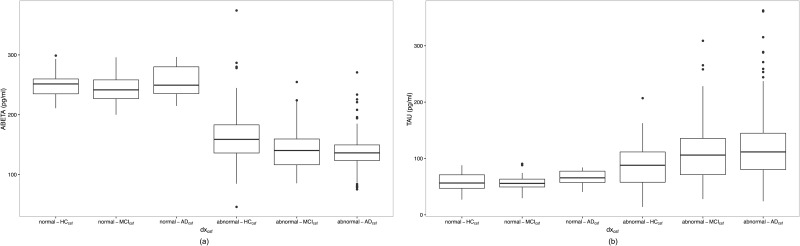
Analysis of CSF-Aβ and CSF-τ biomarkers showed us that their distributions were not normal for all diagnosis groups (see figure in S2 Fig). A bimodal distribution was observed in CSF-Aβ levels within each diagnosis group. CSF-τ values in CN group showed a unimodal normal distribution. We confirmed cut-off values determined by [18], classifying as normal the CSF profile of subjects who fulfilled both conditions: CSF-Aβ > = 192pg/ml and CSF-τ ≤ 93pg/ml. From CSF profiles, we identified 46 normal-HCcsf, 33 normal-MCIcsf, 11 normal-ADcsf, 63 abnormal-HCcsf, 75 abnormal-MCIcsf and 172 non-ADcsf subjects. Figure in S3 Fig shows CSF-τ concentration versus CSF-Aβ concentration for these six groups, where the blue dots represent the HC subjects with normal CSF profile used to build the null models for men and women. Furthermore, in Fig 2 is shown the last available measurements of CSF biomarkers concentrations for the studied ADNI subjects grouped according dxcsf. An increase in τ values was observed when comparing normal-HCcsf and subjects with abnormal CSF-profile, and when comparing abnormal-HCcsf with MCI and AD subjects with abnormal profile. With respect to Aβ levels, we observed a reduction of levels when comparing normal-HCcsf with abnormal-HCcsf and the remaining groups with abnormal profile.
Fig 2. Boxplot of CSF biomarkers concentrations for dxcsf diagnostic groups.
(a) CSF-Aβ1 − 42; and (b) CSF-τ.
In total, 226 samples were available for the 46 normal-HCcsf subjects (males: 22, females: 24). Table 2 provides their demographic information at baseline. Age, education and MMSE scores were similar across gender, but CSF concentrations and CDRGLOBAL were different between men and women. Furthermore, only three subjects (2 males and 1 female) were carriers of APOE-ϵ4 at allele 1. More detail of demographic and cognitive measures are given in S1 Appendix.
Table 2. Baseline statistical descriptors of HC subjects selected for null models building.
| Female | Male | |
|---|---|---|
| N = 24 | N = 22 | |
| age | 71.2 75.1 77.7 (74.7± 5.3) | 70.4 71.8 74.0 (72.7± 6.0) |
| education | 65.0 70.0 80.0 (0.73± 0.09) | 70.0 82.5 90.0 (0.81±0.15) |
| APOE-ϵ4 | ||
| 0 | 95.8% | 90.9% |
| 1 | 4.2% | 9.1% |
| MMSE | 29.0 29.0 30.0 (29.25± 0.67) | 28.0 29.0 30.0 (28.54± 1.50) |
| CDRGLOBAL | ||
| 0 | 100% | 82% |
| 0.5 | 0% | 18% |
| CSF-Aβ | 234.9 248.5 256.0 (247.23± 20.14) | 235.0 257.9 268.2 (253.22± 21.69) |
| CSF-τ | 47.9 55.260.9 (55.41± 15.10) | 47.0 59.9 73.8 (59.84± 16.83) |
Values of continuous variables are represented by the lower, the median and the upper quartiles; and the mean± standard deviation in parentheses.
Ageing-based variant (vr) and quasi-variant (qvr) ROIs
A study of within-subject and between-subject variabilities, using the box plots representation, showed us that there are significant between-subject variability and strong gender effect for several ROIs; for example, the left hippocampal volume (see Figure in S4 Fig). However, for other ROIs their change was not very evident. From LME modelling, we identified 97 vr ROIs for males and 109 vr ROIs for females. Regarding quasi-variant ROIs, we found 69 qvr for males and 57 qvr for females. The figure in S5 Fig shows examples of fitted LME models for both types of ROIs within each gender. In a similar way to these examples, the βa’s of the remaining vr ROIs were observed as being close to zero, but for qvr ROIs, their slopes were not. The figure in S6 Fig shows the summary of ageing-associated differences of biomarkers in men and women. The results suggest important ageing-related reductions in neocortical and subcortical regions and ventricular expansion, with several gender-specific significances. Reductions are observed in the most of cortical volumes and cortical thickness, where men and women showed a similar degree of global thinning. Some of these regions showed prominent atrophy while others showed a more conservative change; for example, the entorhinal volume was observed to be significantly reduced in females but not in males. In relation to the subcortical regions, we observed several gender-specific differences, some of these are: significant volumetric reductions in both hemispheres of hippocampal volume for both males and females; ventricular expansion in both genders, but just the change of fourth ventricle volume was not significant; the volume of the optic chiasm region was significantly increased in females, but it was not significant in males; a reduction in amygdala volume was significant in both hemispheres of the male brain but it was not significant in females; and, in general, the volume of bilateral corpus callosum regions are observed as thinned, but without significant differences between the genders.
Subject classification based on ROI residuals
The table in S4 Fig shows the average prediction accuracy (ACC), sensitivity (SEN) and specificity (SPE) of the test for both the F1 and F2 training set configurations. Although the performance of models with the F2 set was the best for all cases, the experiments performed on the F1 set had also good results. Table 3 compares the prediction accuracy of the proposed method (by averaging the accuracy for both genders) with existing methods, which have been evaluated on the ADNI dataset and other data sources. Note that we only showed the most significant results of those studies, mainly focusing on MRI and its combination with other features.
Table 3. Comparison of methods performances focused on subject classification (%).
| Method | AD vs HC | MCI vs HC | cAD vs sMCI | Data used | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC | SEN | SPE | ACC | SEN | SPE | ACC | SEN | SPE | ||
| Kloeppel et al. [30] | 95.0 | 95.0 | 95.0 | - | - | - | - | - | - | MRI |
| Vemuri et al. [3] | - | 86.0 | 86.0 | - | - | - | - | - | - | MRI |
| - | 88.0 | 90.0 | - | - | - | - | - | - | MRI,age,gender | |
| - | 86.0 | 92.0 | - | - | - | - | - | - | MRI,age,gender,APOE | |
| Cui et al. [19] | - | - | - | - | - | - | 67.13 | 96.43 | 48.28 | NM,CSF,MRI |
| - | - | - | - | - | - | 62.24 | 92.86 | 42.53 | NM,MRI | |
| - | - | - | - | - | - | 62.24 | 57.14 | 65.52 | MRI | |
| Cuingnet et al. [31] | - | 81.0 | 95.0 | - | - | - | - | 57.0 | 78.0 | MRI |
| Zhang et al. [25]* | 93.3 | - | - | 83.2 | - | - | 73.9 | 68.6 | 73.6 | MRI,PET,CSF |
| Suk et al. [28] | 95.9 | - | - | 85.0 | - | - | 75.8 | - | - | MRI,PET |
| Jie et al. [26] | 95.03 | 94.90 | 95.00 | 79.27 | 85.86 | 66.64 | 68.94 | 64.65 | 71.79 | MRI,FDG-PET |
| Gaser et al. [22] | - | - | - | - | - | - | 75.00 | 71.00 | 84.00 | MRI-based age |
| Spulber et al. [23] | 88.4 | 86.1 | 90.4 | - | - | - | 67.7 | 69.6 | 66.8 | MRI-based index |
| Aguilar et al. [24] | - | 92.0 | 75.0 | - | - | - | - | 92.0 | 47.0 | MRI-based index |
| Liu et al. [27] | 94.37 | 94.71 | 94.04 | 78.8 | 84.85 | 67.06 | 67.83 | 64.88 | 70.0 | MRI,PET |
| Suk et al. [29] | 92.38 | 91.54 | 94.56 | 84.24 | 99.58 | 53.79 | 72.42 | 36.70 | 90.98 | MRI |
| 93.35 | 94.65 | 95.22 | 85.67 | 95.37 | 65.87 | 75.92 | 48.04 | 95.23 | MRI,PET | |
| *Proposed method (F1) | 89.22 | 90.73 | 86.85 | 77.08 | 95.53 | 26.98 | 70.30 | 64.26 | 75.14 | MRI-based residuals, age |
| *Proposed method (F2) | 94.11 | 96.54 | 90.28 | 83.77 | 93.55 | 57.55 | 76.72 | 70.77 | 81.62 | MRI-based residuals, age, MMSE,CDRGLOBAL |
* Results of this method correspond to the average of performance recorded for men and women. MRI, Magnetic Resonance Imaging-based features; CSF; Cerebral Spinal Fluid-based biomarkers; NM: Neuro-psychological measures; PET, Positron Emission Tomography-based features; FDG-PET, [18F]fluorodeoxyglucose uptake measured in PET; MRI-based age, individual estimated age computed from MRI images; MRI-based index, individual severity index computed from MRI images; MMSE, Mini-Mental Clinical Dementia Rating; CDRGLOBAL, CDR Global Score.
Early prediction of subject diagnostics based on ROI residuals
The table in S5 Table shows the performance of models built for early prediction of disease progression. As in previous results, although models built with the F2 configuration have obtained slightly better accuracies than the ones built on the F1 configuration, both performances are very satisfactory. Table 4 shows the average number of years this method leads in predicting the subject diagnosis. This advancement in the prediction was only possible in subjects whose dxage (diagnostic at each visitation) was different from their dxlast diagnosis (last known diagnostic). Therefore, only these subjects were taken into account and some age groups lack enough data to be shown in this table. For females, the prediction of conversion from MCI to AD was up to 1.9 years earlier (60–64 age group); and for males, up to 1.73 years earlier (80–84 age group). However, the greatest lead was obtained in the early prediction from HC to MCI in females, this being 2.89 years earlier.
Table 4. Last known diagnostic prediction advancement.
| Advancement for early prediction in years | |||||||
|---|---|---|---|---|---|---|---|
| Experiment | 60–64 yrs. | 65–69 yrs. | 70–74 yrs. | 75–79 yrs. | 80–84 yrs. | 85–89 yrs. | |
| Females | AD vs HC | 1.772 (85.73) | 1.596(86.05) | 1.335(86.79) | 1.745(88.93) | 1.523(90.21) | 1.245(89.67) |
| MCI vs HC | N/A | N/A | 2.888(74.42) | 2.071(76.08) | 2.381(78.40) | N/A | |
| AD vs MCI | 1.982(69.23) | 1.539(70.04) | 1.307(69.55) | 1.751(71.37) | 1.509(72.41) | 1.433(71.28) | |
| Males | AD vs HC | 1.232(84.27) | 1.801(85.30) | 1.667(86.91) | 1.327(87.75) | 1.775(89.44) | 1.006(89.52) |
| MCI vs HC | 1.257(72.47) | N/A | 1.739(77.29) | 2.529(78.32) | 2.686(78.59) | 1.439(79.81) | |
| AD vs MCI | 1.448(68.95) | 1.697(69.44) | 1.679(70.22) | 1.372(71.18) | 1.733(73.46) | 1.011(73.06) | |
N/A means there are not enough samples in the age interval for binary classification. (ACC) represents the prediction accuracy of F2 method in %
Along with the time advancement in the prediction, Table 4 also shows the average prediction accuracy for that differential diagnostic stratified by gender and age group. Note that differences between accuracies of Tables 3 and 4 are due to the last one included a few samples, namely those that fit the conditions of five-year age group stratification and whose last known diagnostic differs from the diagnostic at the time of the test in the given age group.
Discussion and Conclusions
This study has presented a framework for building ageing-associated null models from longitudinal MRI-based biomarkers, as well as subject classification and early diagnosis from residuals calculated through these null models.
Analysis of CSF biomarkers distributions allowed us to confirm the cut-offs values established by [18] and to establish criteria for CSF profiling. We found subjects clinically diagnosed as CN at baseline with abnormal CSF profile, which is consistent with previous studies where the presence of possible AD pathology has been found in ADNI control subjects [18, 51, 52]. These findings may be because ADNI subject diagnosis is made independently of the CSF biomarkers values (see S1 Appendix).
We modelled the longitudinal change of an extensive set of MRI-based biomarkers obtained from cortical and subcortical regions. Studies have shown that early diagnosis methods using the whole brain or the whole cortex reached higher specificity (over 90%) than those based on the specific regions like the hippocampus (from 63% to 84%) [31, 53, 54]. LME modelling allowed to classify biomarkers as variant or quasi-variant ROIs, and to build null models for ageing-related changes in men and women. As in previous studies [6, 8, 9], our results (see Figure in S6 Fig) show that part of these changes occur in brain areas related with AD. We found reductions for most thickness and volume biomarkers of cortical regions [8] in both genders. Similar changes were also found in subcortical regions including the greater atrophy in the hippocampus and regions of the corpus callosum.
Null models were carried out by making assumptions of correlation between the y-intercepts values (at basal stage) of variant and quasi-variant ROIs, where the first ones were explained in function of last ones using PLSR. By using these null models and LME β coefficients, we calculated residuals, which were established as differences between the observed ROIs and age-related inferred ROIs. These residuals were computed for new cohort of HC, MCI and subjects; and used for training and testing SVM models to address subject classification and early disease prediction.
As of the date of this study, we were unable to find studies where ageing-related null models and residuals-based classifiers were applied to early diagnosis. The performance obtained in all experiments suggests that the proposed method of obtaining the ROI residuals and their use to train SVM predictors is useful to support the early diagnosis problem, the fundamental challenge in AD research. LME modelling of MRI-based biomarkers was only applied by fitting age and years of education, but future work could assess the impact of different available feature types such as other functional neuroimaging biomarkers, genetic factors, biological markers and other clinical and neuropsychological assessment.
The main contributions of the residual-based classifier presented in this paper are: (a) From a longitudinal study of 5 years follow-up, the ability to predict the future diagnostic of the subjects up to 2.88 years earlier than the standard clinical procedure. (b) Use of relatively common clinical tests such as MRI and neuropsychological tests, as opposed to methods that rely on more expensive or invasive tests such as PET-based, CSF-based and Genotype-based biomarkers. (c) HC vs AD: highest sensitivity among the state of the art methods; a classification accuracy of 94%, higher than all MRI-only methods except for [30]. (d) sMCI vs cAD: highest classification accuracy among the state of the art methods. (e) In most experiments, the sensitivity (the ability of a predictor to correctly classify a subject as ‘diseased’) was higher than the specificity (the ability of predictor to correctly classify a subject as ‘disease-free’). This may be due to the fact that ADNI clinical diagnosis is based on neuro-psychological tests, but neurodegeneration occurs many years before the onset of clinical symptoms. Possibly, residual-based SVM predictors may determine that subjects are into early stages of disease (MCI and prodromal AD) but this finding is not consistent with clinical diagnosis because the subject does not yet present clinical symptoms. Abnormalities of CSF profile observed on several subjects diagnosed as HC and MCI (see Figure in S3 Fig) may support this hypothesis.
The use of MMSE and CDRGLOBAL tests (F2 method) yields significantly better results than using the residuals alone (F1 method). These are two of the most common neuropsychological tests routinely applied to patients in the primary clinical practice. However this should not be seen as a mere contribution of MMSE and CDRGLOBAL, as these tests on their own have several limitations such as relatively low specificity and reliability. However, they complement and enhance the present method here without adding a significant cost or invasive clinical tests.
Despite promising results, there are several limitations to our study. Firstly, data used here correspond to research participants, who meet the inclusion and exclusion criteria established by ADNI, and thus are not from general population. Secondly, the available observations of CSF biomarkers do not correspond in number or time points with the available MRI-based observations. In most cases we had just CSF values at the baseline stage, so it is impossible to track the reliability of CSF profile at the final stages. Due to this, it is important to emphasize that subject classifications based on CSF profile are applicable to the ADNI study subjects, but not necessarily to individuals in other settings. Thirdly, like CSF biomarkers, we had substantial missing data for MRI-based biomarkers measured for 46 HC subjects during the 60-month follow-up period. Although a LME approach can handle missing time points within individuals, this issue may limit our ability to make inferences about the age-related changes in these biomarkers. Finally, the proposed methodology to obtain a null model of healthy elderly people is still at the early stages of the development and evaluation process, it is necessary to test the null model in populations different from those used in model development.
Supporting Information
(PDF)
HC and AD are hypothetical subjects. P1, P2 and P3 are observations of each ROI y at three different ages (a1, a2 and a3). Black lines, healthy population regression line calculated from LME. , vertical y-intercept value of healthy population. Blue and red lines, individual regression lines estimated by assuming both as healthful. Points , and , inferred ’s for the three ages. and , the subject-specific y-intercepts estimated for HC and AD subjects, respectively. and of vr ROI are inferred from the and of qvr ROI through PLSR model. βa, slope or rate change of standard deviation of ROI per unit of age. ϵHC1, ϵHC2, ϵHC3, ϵAD1, ϵAD2 and ϵAD3, the residuals of each observation with respect to the estimated individual regression lines.
(PDF)
(a) CSF-Aβ. (b) CSF-τ. △: 8 of 113 CN subjects were converted to MCI and 1 CN subject was converted to AD. ○: 88 of 94 LMCI were converted to AD and 5 were re-converted to HC at follow-up visits. Dotted vertical lines within each diagnosis are the determined cutoff concentrations. Figure shows us that several subjects with abnormal CSF profile classified as CN by ADNI at baseline clinical assessment were converted to MCI or AD (triangle dots) along time. Likewise, some subjects classified like ‘LMCI’ by ADNI were converted to AD later (open circles).
(PDF)
Dots represent the last CSF biomarker measured for subjects available at April, 2015. Vertical and horizontal dashed lines split normal CSF-profile from abnormal profile. Null models for characterization of healthy brain structures were built from samples labelled with blue dots.
(PDF)
High between-subject variability is evident, e.g., by comparing subject 099_S_0533 with subject 133_S_0488. Likewise, there is a strong indication of gender effect over hippocampal volume, female volumes are less than the male ones. Note that we standarized every MRI-based biomarker to have zero mean and unit variance.
(PDF)
(a) Left hippocampal volume classified as variant (vr) ROI; where the slope of trajectories is not close to zero. (b) Left caudate volume classified as quasi-variant (qvr) ROI; where the slope of trajectories is close to zero. Note that for both regions, the y-intercept values vary between subjects, but the slope value of each ROI is the same for all subjects.
(PDF)
(a) Males. (b) Females. Biomarkers are coloured according their change type: blue for vr regions and red for the qvr ones. The size of biomarkers with significant P-value ≤ 0.05 are bigger than the not significant. Dotted vertical lines separate the increased biomarkers (positive beta values) from the reduced ones (negative beta values). ROIs are represented according to their location into brain hemisphere (lh:left, rh: right or bilateral). SV: Subcortical Volume, CV: Cortical volume, SA: Surface Area. lh: left hemisphere. rh: right hemisphere.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). Data used in preparation of this article were obtained from the ADNI (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Data Availability
All MRI-based biomarkers, clinical and demographic information were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu) and are public and freely accessible.
Funding Statement
This work was supported by TEC2014-60337-R and PSI2013-47724-P grants from the Ministerio de Economía y Competitividad (MINECO). CIBER of Bioengineering, Biomaterials and Nanomedicine is an initiative of ISCIII. Authors are part of the 2014SGR-1063 consolidated research group of the Generalitat de Catalunya, Spain. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jack CR, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010. January;9(1):119–128. Available from: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dubois B, Hampel H, Feldman HH, Scheltens P, Aisen P, Andrieu S, et al. Preclinical Alzheimer’s disease: Definition, natural history, and diagnostic criteria; 2016. 10.1016/j.jalz.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vemuri P, Gunter JL, Senjem ML, Whitwell JL, Kantarci K, Knopman DS, et al. Alzheimer’s disease diagnosis in individual subjects using structural MR images: validation studies. Neuroimage. 2008. February;39(3):1186–1197. Available from: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frisoni GB, Fox NC, Jack CR, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nature reviews Neurology. 2010;6(2):67–77. Available from: http://www.nature.com.ezproxy.usc.edu.au:2048/nrneurol/journal/v6/n2/full/nrneurol.2009.215.html. 10.1038/nrneurol.2009.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer’s & dementia: the journal of the Alzheimer’s Association. 2010. May;6(3):265–73. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2868595{&}tool=pmcentrez{&}rendertype=abstract. 10.1016/j.jalz.2010.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, et al. One-year brain atrophy evident in healthy aging. J Neurosci. 2009. December;29(48):15223–15231. Available from: 10.1523/JNEUROSCI.3252-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peters R. Ageing and the brain. Postgraduate medical journal. 2006. February;82(964):84–8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2596698{&}tool=pmcentrez{&}rendertype=abstract. 10.1136/pgmj.2005.036665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salat D, Buckner RL, Snyder AZ, Greve DN, Desikan RS, Busa E, et al. Thinning of the cerebral cortex in aging. Cerebral Cortex. 2004;14:721–730. 10.1093/cercor/bhh032 [DOI] [PubMed] [Google Scholar]
- 9. Jiang J, Sachdev P, Lipnicki DM, Zhang H, Liu T, Zhu W, et al. A longitudinal study of brain atrophy over two years in community-dwelling older individuals. Neuroimage. 2014. February;86:203–211. Available from: 10.1016/j.neuroimage.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 10. Liu Y, Julkunen V, Paajanen T, Westman E, Wahlund LO, Aitken A, et al. Education increases reserve against Alzheimer’s disease-evidence from structural MRI analysis. Neuroradiology. 2012;54(9):929–938. 10.1007/s00234-012-1005-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morris GP, Clark IA, Vissel B. Inconsistencies and controversies surrounding the amyloid hypothesis of Alzheimer’s disease. Acta Neuropathol Commun. 2014;2:135 Available from: 10.1186/s40478-014-0135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolfe MS. The role of tau in neurodegenerative diseases and its potential as a therapeutic target. Scientifica. 2012;2012:796024 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3820460{&}tool=pmcentrez{&}rendertype=abstract. 10.6064/2012/796024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morris J. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. 10.1212/WNL.43.11.2412-a [DOI] [PubMed] [Google Scholar]
- 14. Folstein M, Folstein S, McHugh P. Mini-mental state. A practical method for grading the cognitive state of patients for the clinician. Journal of Psychiatric Research. 1975;12(3):189–198. 10.1016/0022-3956(75)90026-6 [DOI] [PubMed] [Google Scholar]
- 15. Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, et al. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimers Dement. 2005. July;1(1):55–66. Available from: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, et al. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005. November;15(4):869–77, xi–xii. Available from: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011. May;7(3):270–279. Available from: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009. April;65(4):403–413. 10.1002/ana.21610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cui Y, Liu B, Luo S, Zhen X, Fan M, Liu T, et al. Identification of conversion from mild cognitive impairment to Alzheimer’s disease using multivariate predictors. PLoS One. 2011;6(7):e21896 10.1371/journal.pone.0021896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Franke K, Ziegler G, Klöppel S, Gaser C. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage. 2010. April;50(3):883–92. Available from: http://www.sciencedirect.com/science/article/pii/S1053811910000108. [DOI] [PubMed] [Google Scholar]
- 21. Franke K, Gaser C. Longitudinal changes in individual BrainAGE in healthy aging, mild cognitive impairment, and Alzheimer’s disease. Journal of Gerontopsychology and Geriatric Psychiatry. 2012;25(4):235–45. [Google Scholar]
- 22. Gaser C, Franke K, Klöppel S, Koutsouleris N, Sauer H. BrainAGE in Mild Cognitive Impaired Patients: Predicting the Conversion to Alzheimer’s Disease. PLoS ONE. 2013;8(6). 10.1371/journal.pone.0067346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spulber G, Simmons A, Muehlboeck JS, Mecocci P, Vellas B, Tsolaki M, et al. An MRI-based index to measure the severity of Alzheimer’s disease-like structural pattern in subjects with mild cognitive impairment. Journal of internal medicine. 2013. April;273(4):396–409. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3605230{&}tool=pmcentrez{&}rendertype=abstract. 10.1111/joim.12028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aguilar C, Muehlboeck JS, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, et al. Application of a MRI based index to longitudinal atrophy change in Alzheimer disease, mild cognitive impairment and healthy older individuals in the AddNeuroMed cohort. Frontiers in aging neuroscience. 2014. January;6:145 10.3389/fnagi.2014.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang D, Shen D, Alzheimer’s Disease Neuroimaging Initiative. Multi-Modal Multi-Task Learning for Joint Prediction of Multiple Regression and Classification Variables in Alzheimer’s Disease. Neuroimage. 2012;59(2):895–907. 10.1016/j.neuroimage.2011.09.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jie B, Zhang D, Cheng B, Shen D. Manifold regularized multi-task feature selection for multi-modality classification in Alzheimer’s disease In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013. Springer; 2013. p. 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu F, Wee CY, Chen H, Shen D. Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer’s Disease and mild cognitive impairment identification. NeuroImage. 2014;84:466–475. 10.1016/j.neuroimage.2013.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suk HI, Shen D. Deep Learning-Based Feature Representation for AD/MCI Classification. Med Image Comput Comput Assist Interv. 2013;16(2):583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suk HI, Lee SW, Shen D, Initiative ADN, et al. Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. NeuroImage. 2014;101:569–582. 10.1016/j.neuroimage.2014.06.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, et al. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008. March;131(Pt 3):681–689. Available from: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert MO, et al. Automatic classification of patients with Alzheimer’s disease from structural MRI: A comparison of ten methods using the ADNI database. Multivariate Decoding and Brain Reading. 2011;56(2):766–781. Available from: http://www.sciencedirect.com/science/article/pii/S1053811910008578. [DOI] [PubMed] [Google Scholar]
- 32. Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI). Neurology. 2010;74(3):201–209. Available from: http://www.neurology.org/content/74/3/201.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fjell AM, McEvoy L, Holland D, Dale AM, Walhovd KB, ADNI. Brain changes in older adults at very low risk for Alzheimer’s disease. J Neurosci. 2013. May;33(19):8237–8242. Available from: 10.1523/JNEUROSCI.5506-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Desikan R, Cabral H, Hess C, Dillon W, Glastonbury C, Weiner M, et al. Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer’s disease. Brain. 2009;132(8):2048–2057. 10.1093/brain/awp123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Desikan RS, Cabral HJ, Settecase F, Hess CP, Dillon WP, Glastonbury CM, et al. Automated MRI measures predict progression to Alzheimer’s disease. Neurobiol Aging. 2010. August;31(8):1364–1374. 10.1016/j.neurobiolaging.2010.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimer’s and Dementia. 2012;8(1, Supplement):S1–S68. 10.1016/j.jalz.2011.09.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jack C, Bernstein M, Fox N. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27(4):685–691. 10.1002/jmri.21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, et al. The cortical signature of Alzheimer’s disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex. 2009. March;19(3):497–510. Available from: 10.1093/cercor/bhn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cho Y, Seong JK, Jeong Y, Shin SY, Initiative ADN. Individual subject classification for Alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. Neuroimage. 2012. February;59(3):2217–2230. Available from: 10.1016/j.neuroimage.2011.09.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Desikan R, Sigonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–980. Available from: http://www.sciencedirect.com/science/article/pii/S1053811906000437. [DOI] [PubMed] [Google Scholar]
- 41. Fischl B. FreeSurfer. Neuroimage. 2012. January;Available from: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tosun D, Schuff N, Truran-Sacrey D, Shaw LM, Trojanowski JQ, Aisen P, et al. Relations between brain tissue loss, CSF biomarkers, and the ApoE genetic profile: a longitudinal MRI study. Neurobiol Aging. 2010. August;31(8):1340–1354. 10.1016/j.neurobiolaging.2010.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software. 2015;67(1):1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- 44. Höskuldsson A. PLS regression methods. Journal of Chemometrics. 1988;2(3):211–228. Available from: 10.1002/cem.1180020306. [DOI] [Google Scholar]
- 45. Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems. 2001;58(2):109–130. {PLS} Methods. Available from: http://www.sciencedirect.com/science/article/pii/S0169743901001551. [Google Scholar]
- 46. Dayal BS, MacGregor JF. Improved PLS algorithms. Journal of Chemometrics. 1997;11(1):73–85. Available from: http://onlinelibrary.wiley.com/doi/10.1002/(SICI)1099-128X(199701)11:1%3C73::AID-CEM435%3E3.0.CO;2-%23/abstract. [Google Scholar]
- 47. Mevik BH, Wehrens R, Liland KH. pls: Partial Least Squares and Principal Component regression; 2013. R package version 2.4-3 Available from: http://CRAN.R-project.org/package=pls. [Google Scholar]
- 48. Coffey CE, Lucke JF, Saxton JA, Ratcliff G, Unitas LJ, Billig B, et al. Sex differences in brain aging: a quantitative magnetic resonance imaging study. Arch Neurol. 1998;55(2):169–79. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9482358. 10.1001/archneur.55.2.169 [DOI] [PubMed] [Google Scholar]
- 49. Xu J, Kobayashi S, Yamaguchi S, Iijima K, Okada K, Yamashita K. Gender effects on age-related changes in brain structure. AJNR Am J Neuroradiol. 2000;21(1):112–118. [PMC free article] [PubMed] [Google Scholar]
- 50. Király A, Szabó N, Tóth E, Csete G, Faragó P, Kocsis K, et al. Male brain ages faster: the age and gender dependence of subcortical volumes. Brain Imaging and Behavior. 2015;p. 1–10. Available from: 10.1007/s11682-015-9468-3. [DOI] [PubMed] [Google Scholar]
- 51. Toledo JB, Xie SX, Trojanowski JQ, Shaw LM. Longitudinal change in CSF Tau and ABeta biomarkers for up to 48 months in ADNI. Acta Neuropathologica. 2013;126(5):659–670. 10.1007/s00401-013-1151-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. De Meyer G, Shapiro F, Vanderstichele H, Vanmechelen E, Engelborghs S, De Deyn PP, et al. Diagnosis-independent Alzheimer disease biomarker signature in cognitively normal elderly people. Arch Neurol. 2010. August;67(8):949–956. 10.1001/archneurol.2010.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chupin M, Gérardin E, Cuingnet R, Boutet C, Lemieux L, Lehéricy S, et al. Fully automatic hippocampus segmentation and classification in Alzheimer’s disease and mild cognitive impairment applied on data from ADNI. Hippocampus. 2009. June;19(6):579–587. Available from: 10.1002/hipo.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Das SR, Avants BB, Pluta J, Wang H, Suh JW, Weiner MW, et al. Measuring longitudinal change in the hippocampal formation from in vivo high-resolution T2-weighted MRI. Neuroimage. 2012. April;60(2):1266–1279. Available from: 10.1016/j.neuroimage.2012.01.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
HC and AD are hypothetical subjects. P1, P2 and P3 are observations of each ROI y at three different ages (a1, a2 and a3). Black lines, healthy population regression line calculated from LME. , vertical y-intercept value of healthy population. Blue and red lines, individual regression lines estimated by assuming both as healthful. Points , and , inferred ’s for the three ages. and , the subject-specific y-intercepts estimated for HC and AD subjects, respectively. and of vr ROI are inferred from the and of qvr ROI through PLSR model. βa, slope or rate change of standard deviation of ROI per unit of age. ϵHC1, ϵHC2, ϵHC3, ϵAD1, ϵAD2 and ϵAD3, the residuals of each observation with respect to the estimated individual regression lines.
(PDF)
(a) CSF-Aβ. (b) CSF-τ. △: 8 of 113 CN subjects were converted to MCI and 1 CN subject was converted to AD. ○: 88 of 94 LMCI were converted to AD and 5 were re-converted to HC at follow-up visits. Dotted vertical lines within each diagnosis are the determined cutoff concentrations. Figure shows us that several subjects with abnormal CSF profile classified as CN by ADNI at baseline clinical assessment were converted to MCI or AD (triangle dots) along time. Likewise, some subjects classified like ‘LMCI’ by ADNI were converted to AD later (open circles).
(PDF)
Dots represent the last CSF biomarker measured for subjects available at April, 2015. Vertical and horizontal dashed lines split normal CSF-profile from abnormal profile. Null models for characterization of healthy brain structures were built from samples labelled with blue dots.
(PDF)
High between-subject variability is evident, e.g., by comparing subject 099_S_0533 with subject 133_S_0488. Likewise, there is a strong indication of gender effect over hippocampal volume, female volumes are less than the male ones. Note that we standarized every MRI-based biomarker to have zero mean and unit variance.
(PDF)
(a) Left hippocampal volume classified as variant (vr) ROI; where the slope of trajectories is not close to zero. (b) Left caudate volume classified as quasi-variant (qvr) ROI; where the slope of trajectories is close to zero. Note that for both regions, the y-intercept values vary between subjects, but the slope value of each ROI is the same for all subjects.
(PDF)
(a) Males. (b) Females. Biomarkers are coloured according their change type: blue for vr regions and red for the qvr ones. The size of biomarkers with significant P-value ≤ 0.05 are bigger than the not significant. Dotted vertical lines separate the increased biomarkers (positive beta values) from the reduced ones (negative beta values). ROIs are represented according to their location into brain hemisphere (lh:left, rh: right or bilateral). SV: Subcortical Volume, CV: Cortical volume, SA: Surface Area. lh: left hemisphere. rh: right hemisphere.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All MRI-based biomarkers, clinical and demographic information were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu) and are public and freely accessible.