Abstract
Androgen receptor splice variants (AR-Vs) are implicated in resistance of prostate cancer to androgen-directed therapies. When expressed alone in cells, some AR-Vs (e.g., AR-V7) localize primarily to the nucleus, whereas others (e.g., AR-V1, AR-V4, and AR-V6) localize mainly to the cytoplasm. Significantly, the latter are often co-expressed with the nucleus-predominant AR-Vs and the full-length AR (AR-FL). An important question to be addressed is whether the cytoplasmic-localized AR-Vs play a role in castration-resistant prostate cancer (CRPC) through interaction with the nucleus-predominant AR-Vs and AR-FL. Here, it is demonstrated that AR-V1, -V4, and -V6 can dimerize with both AR-V7 and AR-FL. Consequently, AR-V7 and androgen-bound AR-FL induced nuclear localization of AR-V1, -V4, and -V6, and these variants, in turn, mitigated the ability of the anti-androgen enzalutamide to inhibit androgen-induced AR-FL nuclear localization. Interestingly, the impact of nuclear localization of AR-V4 and -V6 on AR transactivation differs from that of AR-V1. Nuclear localization leads to an increased ability of AR-V4 and -V6 to transactivate both canonical AR targets and AR-V-specific targets and to confer castration-resistant cell growth. However, while AR-V1, which lacks inherent transcriptional activity, appears to activate AR-FL in an androgen-independent manner, it significantly antagonizes AR-V7 transactivation. Together, these data demonstrate that the complex interactions among different AR-Vs and AR-FL play a significant role in castration resistant disease.
Implications
This study suggests important consequences for clinical castration resistance due to simultaneous expression of AR-FL and AR-Vs in patient tumors and suggests that dissecting these interactions should help develop effective strategies to disrupt AR-V signaling.
Keywords: androgen receptor, splice variant, dimerization, prostate cancer, castration resistance
INTRODUCTION
Prostate cancer is the second leading cause of cancer mortality in men in the United States (1). Androgen deprivation therapy (ADT), which disrupts androgen receptor (AR) signaling through androgen depletion or an anti-androgen, is the first-line treatment for advanced prostate cancer (2–4). ADT initially results in a favorable clinical response. However, prostate cancer invariably progresses to incurable castration-resistant prostate cancer (CRPC) (2–4). Resurgent AR activity is increasingly recognized as a pivotal driver for castration-resistant progression (2–4). This led to the development of two next-generation agents of androgen-directed therapy, the androgen biosynthesis inhibitor abiraterone and the potent AR antagonist enzalutamide (5,6). These agents prolong the survival of patients with metastatic CRPC, but both de novo and acquired resistance to the two drugs are common (7–10). One potential mechanism of AR reactivation after androgen-directed therapies, including abiraterone and enzalutamide, is the synthesis of C-terminally truncated AR variants (AR-Vs), through alternative RNA splicing (2,4).
The full-length AR (AR-FL) is composed of an N-terminal domain, a central DNA-binding domain, a hinge region, and a C-terminal ligand-binding domain (11). Due to insertions of cryptic exons downstream of the DNA-binding domain, AR-V transcripts lack the reading frame for the ligand-binding domain (Figure S1) (2,4). Nonetheless, because the majority of the AR-Vs retain the DNA-binding domain and the N-terminal domain, which contains the most critical transactivation domain of the receptor (AF-1), many AR-Vs display constitutive activity (12–18). AR-V7 (aka AR3) and ARv567es (aka AR-V12) are two major AR-Vs expressed in clinical specimens (14,15,17,19–26). They activate target-gene expression in a ligand-independent manner and promote castration-resistant growth of prostate cancer cells in vitro and in vivo (12,14–18). Strikingly, patients with high levels of AR-V7 mRNA or nuclear AR-V7 protein or detectable expression of ARv567es mRNA in prostate tumors have a shorter survival than other CRPC patients (20,23,25). Moreover, the expression of AR-V7 in circulating tumor cells of patients with metastatic CRPC is associated with resistance to both abiraterone and enzalutamide (19). AR-V7 and ARv567es have also been shown to be insensitive to taxane inhibition of nuclear translocation and confer resistance of CRPC to taxanes in preclinical models (27–29).
Nuclear import of AR is essential for its genomic function. AR-Vs have variable capability in nuclear localization. For example, when expressed alone, AR-V7 and ARv567es localize predominantly to the nucleus (12,15–17,30), AR-V1 (aka AR4), AR-V4 (aka AR 1/2/3/2b, AR5), AR-V6, and AR-V9, in general, localize mainly to the cytoplasm (12,16,18), and AR8 localizes primarily to the plasma membrane (31). We previously reported that dimerization is essential for the nucleus-predominant AR-Vs, AR-V7 and ARv567es, to function and that the dimerization between these AR-Vs and AR-FL induces AR-FL nuclear localization and activates AR-FL in an androgen-independent manner (30,32). Compared to AR-V7 and ARv567es, the non-nucleus-predominant AR-Vs are much less characterized in terms of their functions and clinical relevance. A recent single-cell RNA-seq analysis of 73 circulating tumor cells from 11 CRPC patients showed that 78% and 43% of these cells express AR-FL and at least one AR-V, respectively (22). Importantly, all the AR-V-expressing cells were also found to express AR-FL (22). In addition, the non-nucleus-predominant AR-Vs were found to be always co-expressed with one of the nucleus-predominant AR-Vs (22). Moreover, the frequencies of expression of the lesser-known AR-Vs in clinical prostate cancer specimens, although lower than that of AR-V7, are quite high (24). For example, at least 7 of these variants have an incidence of >8% (16.6% ± 3.2%) in the hormone-naïve prostate cancer samples in the TCGA dataset, and at least 12 have an incidence of >25% (55.8% ± 4.8%) in the metastatic CRPC samples in the SU2C cohort (24). Given their high frequencies of expression, the simultaneous expression of the different types of AR-Vs and AR-FL, as well as AR-V7 being the most abundantly-expressed AR-V in patient tumors (24,26), we set out to investigate the influences of AR-V7 and AR-FL on the subcellular localization and transcriptional activities of three non-nucleus-predominant AR-Vs, AR-V4, -V6, and -V1, and the impact of these interactions on castration resistance.
MATERIALS AND METHODS
Cell Lines and Reagents
LNCaP, PC-3, DU145, 22Rv1, and HEK-293T cells were obtained from the American Type Culture Collection, and cultured as described (33). All the cell lines were authenticated on April 1, 2015, by the method of short tandem repeat profiling at the Genetica DNA Laboratories. Enzalutamide was purchased from Selleck Chemicals (Houston, TX). The following antibodies were used in Western blot analyses: anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Millipore), anti-AR (N-20, Santa Cruz), anti-AR-V7 (Precision Antibody), and anti-FLAG (M2, Sigma).
Plasmid Construction
The coding regions of AR-V1, -V4, and -V6 were PCR amplified from 22Rv1 cell cDNA and cloned into the pGEM-T EASY TA-cloning vector separately (Promega). Their expression constructs were generated by subcloning the respective coding region from the TA- plasmids into the pLVX-puro vector (Clontech). The FLAG-tagged AR-V constructs were generated by adding 3 tandem FLAG epitopes (DYKDHDG-DYKDHDI-DYKDDDDK) in front of the AR-V genes, and the NLS-AR-V6 construct was generated by adding the nuclear localization signal sequence (PKKKRKV) before the AR-V6 gene. BRET-fusion constructs of AR-V1, -V4, and -V6 were generated by subcloning the AR-V1, -V4, and -V6 cDNA from the respective TA-plasmids into the BamHI and XbaI sites of the pcDNA3.1-RLuc8.6 and TurboFP635 vectors (34). All plasmids were sequence verified. The sequences of the primers used for PCR cloning are listed in the Supplementary Table.
Reporter Gene Assay
LNCaP and PC-3 cells were transfected by using the Lipofectamine 3000 (Invitrogen) and TransIT-2020 (Mirus), respectively, and TurboFect (Thermo) was used for transfection of DU145 and HEK-293T cells. Reporter gene assay was performed as previously described (35) with either an androgen-responsive element-luciferase plasmid (ARE-luc) containing three ARE regions ligated in tandem to the luciferase reporter or a luciferase construct driven by three repeats of an AR-V-specific promoter element of the ubiquitin-conjugating enzyme E2C (UBE2C) gene (UBE2C-luc) (32). To ensure an even transfection efficiency, we conducted the transfection in bulk, and then split the transfected cells for luciferase assay.
Immunofluorescence (IF) Staining and Confocal Fluorescence Microscopy
Cells were transfected with indicated plasmids on Poly-D-Lysine-coated chambered coverglass (Thermo) and cultured in phenol red-free medium supplemented with 10% charcoal-stripped fetal bovine serum. For IF staining, at 48 hr after transfection, cells were fixed with 4% paraformaldehyde, and incubated with a pan-AR antibody (N-20, Santa Cruz; 1:500) or a FLAG antibody (M2, Sigma; 1:1000) overnight at 4°C and subsequently with Alexa Fluor 488- or 594-conjugated secondary antibody (Invitrogen; 1:1000) for 1 hr at room temperature in the dark. Nuclei were then stained with 4′,6-diamidino-2-phenylindole (DAPI). A Nikon ECLIPSE Ti system with a 40X oil-immersion objective was used for confocal imaging. An average of 6 fields with ~10 cells per field was captured for each group, and image analysis was carried out with the use of the NIH Image J Software. The intensities of the nucleus and the whole cell fluorescence signals were quantitated for calculation of the percentage of nuclear localization.
Quantitative Reverse Transcription-PCR (qRT-PCR) and Cell Growth Assay
The qRT-PCR procedure was described previously (36), and the qPCR primer-probe sets were from IDT. The Sulforhodamine (SRB) assay was used to determine cell growth (37). To ensure an even transfection efficiency, we conducted the transfection in bulk and then split the transfected cells for qRT-PCR and SRB assays.
Bioluminescence resonance energy transfer (BRET) Assay
The BRET assay was performed as we previously described (32) in cells that were either transfected with an RLuc BRET fusion plasmid or co-transfected with an RLuc and a TFP BRET fusion plasmid. The BRET ratio was calculated as follows: BRET ratio = (emission at 635 nm)/(emission at 528 nm) – (emission at 635 nm RLuc only)/(emission at 528 nm RLuc only).
Statistical Analysis
The Student’s two-tailed t test was used to determine the mean differences between two groups. P < 0.05 is considered significant. Data are presented as mean ± SEM.
RESULTS
AR-V7 induces nuclear localization of AR-V4 and -V6
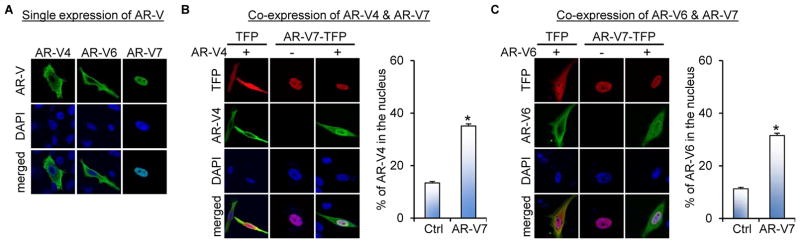
We first studied the effect of AR-V7 on AR-V4 and -V6 subcellular localization. To this end, we expressed FLAG-tagged AR-V4 or -V6 with or without AR-V7-turbo-red-fluorescent-protein (AR-V7-TFP) in the AR-null PC-3 cells. Consistent with a previous report (16), AR-V4 and -V6 were found to localize mainly in the cytoplasm (Figure 1A) with detectable signal in the nucleus (Figures 1B and 1C), whereas AR-V7 localized predominantly to the nucleus. Co-expression of AR-V7 with AR-V4 or -V6 led to increased nuclear AR-V4 and -V6 accumulation without affecting AR-V7 localization (Figures 1B and 1C). This observation suggests that AR-V7 facilitates nuclear localization of AR-V4 and -V6.
Figure 1. AR-V7 facilitates AR-V4 and AR-V6 nuclear localization.
Confocal fluorescence microscopy of AR-V4 and AR-V6 subcellular localization when expressed alone (A) or when co-expressed with AR-V7 in PC-3 cells (B & C). Right panels in B & C, quantitation of % of nuclear AR-V4 or -V6 expression. The FLAG-tagged AR-V4 or -V6 expression construct was transfected with or without the AR-V7-TFP plasmid into PC-3 cells under androgen-deprived condition, and immunofluorescence (IF) staining with an anti-FLAG antibody was conducted at 48 hr after transfection. DAPI was used for nuclear staining. *, P < 0.05 from the control group.
Androgen-bound AR-FL induces nuclear localization of AR-V4 and -V6
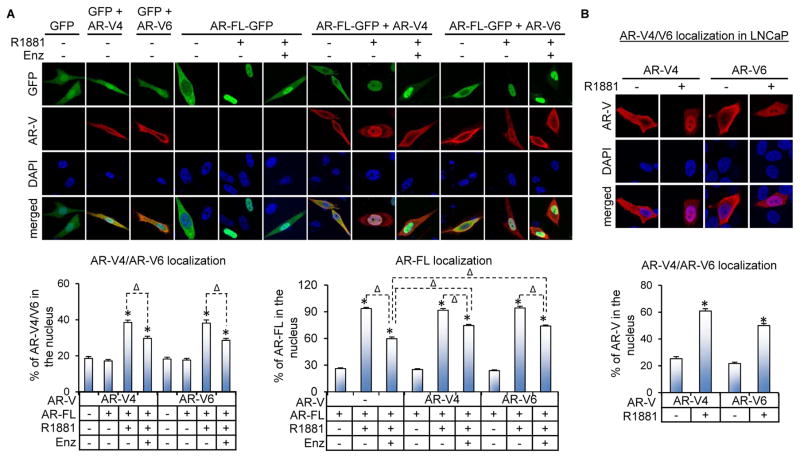
We next investigated the impact of AR-FL on the subcellular localization of AR-V4 and -V6 by expressing FLAG-tagged AR-V4 or -V6 with AR-FL-green-fluorescent-protein (AR-FL-GFP) in PC-3 cells or alone in the AR-FL-expressing LNCaP cells. Co-expression with AR-FL did not affect the subcellular localization of either AR-FL or AR-V4/V6 in androgen-deprived condition (Figure 2A). The addition of androgen in AR-FL-expressing cells, however, not only resulted in increased nuclear localization of AR-FL but also of AR-V4 and -V6 (Figure 2), and this effect was inhibited by the anti-androgen enzalutamide (Figure 2A). Importantly, the androgen-dependent nuclear localization of AR-V4 and -V6 was not observed in the absence of AR-FL (data not shown), suggesting that the effect of androgen on AR-V4 and -V6 localization was mediated through AR-FL. Moreover, while AR-V4 and -V6 could not retain AR-FL in the cytoplasm when cells were treated with androgen, they rendered enzalutamide less effective in inhibiting androgen-induced AR-FL nuclear translocation (Figure 2A). Taken together, the data indicate that androgen-bound AR-FL can induce nuclear localization of AR-V4 and -V6 and that AR-V4 and -V6, in turn, can mitigate the ability of anti-androgens to inhibit androgen-induced AR-FL nuclear localization.
Figure 2. Androgen-bound AR-FL induces nuclear translocation of AR-V4 and AR-V6.
A. Confocal fluorescence microscopy of AR-V4 and AR-V6 subcellular localization when co-expressed with AR-FL in PC-3 cells. The FLAG-tagged AR-V4 or -V6 expression construct was transfected with or without the AR-FL-GFP plasmid into PC-3 cells under androgen-deprived condition. At 40 hr after transfection, the cells were treated with or without 1 nM R1881 in the presence or absence of 10 μM enzalutamide (Enz) for 6 hr. B. Subcellular localization of AR-V4 and AR-V6 in LNCaP cells. The FLAG-tagged AR-V4 or -V6 expression construct was transfected into LNCaP cells under androgen-deprived condition. At 40 hr after transfection, the cells were treated with or without 1 nM R1881 for 6 hr. IF staining with an anti-FLAG antibody was conducted for AR-V detection. Bottom panels, quantitation of % of nuclear AR-FL, AR-V4, or AR-V6 expression. *, P < 0.05 from the no-R1881 control. Δ, P < 0.05.
AR-V4 and -V6 dimerize with AR-V7 and AR-FL
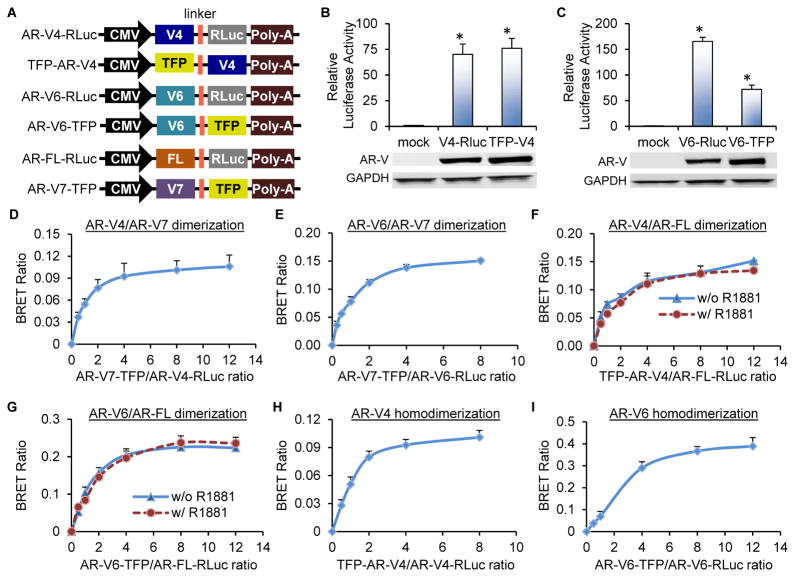
We next used the bioluminescence resonance energy transfer (BRET) assay to determine whether the above observations were due to direct dimerization between AR-V4 or -V6 and AR-V7 or AR-FL. BRET is based on energy transfer from an energy donor to an energy acceptor when the donor and acceptor are brought into close proximity by their fused proteins. In the BRET6 system that we adopted in our study, the energy donor is the RLuc8.6 Renilla luciferase (RLuc) protein, and the energy acceptor is TFP (34). Since BRET depends on the relative orientation of the fusion proteins, we generated all possible combinations of N- and C-terminal fusions through cloning the AR-V4, -V6, -V7, or -FL cDNA either in front of or after RLuc or TFP. Different pairs of the fusion protein constructs were transfected into the AR-null HEK-293T cells (to avoid confounding effect of endogenous AR), and the fusion constructs showing the highest BRET signals (Figure 3A) were chosen for further analysis. The expression and trans-activating abilities of the fusion proteins were validated by Western blotting and reporter gene analysis, respectively (Figures 3B & 3C and Reference (32)), indicating that the AR-Vs and AR-FL are functional in the BRET fusion protein context.
Figure 3. AR-V4 and AR-6 dimerize with AR-V7 and AR-FL and also homodimerize.
A. Schematic diagram of the constructs used in the BRET assay. RLuc, RLuc8.6 luciferase; TFP, TurboFP635 fluorescent protein. B & C. Luciferase assay showing AR trans-activating activity in HEK-293T cells co-transfected with the indicated BRET construct and the ARE-luc plasmid. Lower panels, Western blotting confirmation of AR-V expression. D–I. BRET saturation curves showing AR-V4/AR-V7, AR-V6/AR-V7, AR-V4/AR-FL, and AR-V6/AR-FL heterodimerization as well as AR-V4 and AR-V6 homodimerization. Indicated BRET fusion constructs were co-transfected into HEK-293T cells at different ratios. Cells were cultured under androgen-deprived condition unless specified. R1881, 1 nM. *, P < 0.05 from mock control.
The BRET saturation curves for different combinations of the BRET fusion proteins in HEK-293T cells are shown in Figures 3D–3I. The BRET ratios increased hyperbolically and rapidly saturated with the increase in the ratio of the energy acceptor to the energy donor, indicating specific protein-protein interactions (38). Thus, both AR-V4 and -V6 can dimerize with AR-V7 and AR-FL. Interestingly, the dimerization between AR-V4 or -V6 and AR-FL is independent of androgen (Figures 3F and 3G). In addition to heterodimerization with AR-V7 and AR-FL, AR-V4 and -V6 can also homodimerize (Figures 3H and 3I).
AR-V4 and -V6 nuclear localization promotes castration resistance
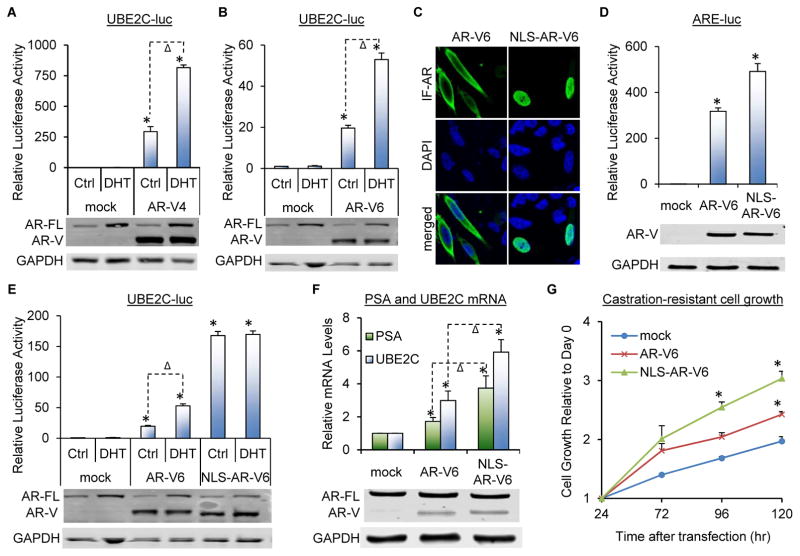
To study the functional significance of AR-V4 and -V6 nuclear localization, we first analyzed the effect of androgen on the ability of AR-V4 and -V6 to transactivate the ubiquitin-conjugating enzyme E2C (UBE2C) promoter that we previously demonstrated to be specific for AR-V activation and not subjected to AR-FL modulation (32). As shown in Figures 4A, 4B, and S2, both AR-V4 and -V6 displayed constitutive trans-activating activity when expressed in LNCaP or C4-2 cells, and this activity was induced by androgen. On the other hand, androgen was not able to alter the trans-activating activity of AR-V6 when AR-FL was not present (Figure S3), indicating that the increased AR-V transactivation was induced by androgen-bound AR-FL. We then added a nuclear localization signal (39) (NLS) to the 5′-end of AR-V6 (NLS-AR-V6), and found that this nucleus-localized AR-V6 (Figures 4C) had improved ability to transactivate both canonical and AR-V-specific targets (Figures 4D–4F and S2B) and to promote castration-resistant growth of prostate cancer cells (Figure 4G). In addition, consistent with the ability of NLS-AR-V6 to localize primarily to the nucleus constitutively, androgen was not able to further induce NLS-AR-V6 transactivation (Figures 4E and S2B). Taken together, the data indicate that AR-V4 and AR-V6 nuclear localization can induce AR transactivation and promote castration-resistant cell growth.
Figure 4. AR-V4/V6 nuclear translocation leads to induced AR transactivation and castration-resistant cell growth in LNCaP cells.
A&B. Luciferase assay showing androgen inducing AR-V transactivation in LNCaP cells co-transfected with AR-V4 (A) or AR-V6 (B) and the UBE2C-luc construct. C. Confocal fluorescence microscopy of IF-stained AR-V6 or NLS-AR-V6 expressed in PC-3 cells. D&E. Luciferase assay showing the addition of NLS (nuclear localization signal) increasing AR-V transactivation. AR-V6 or NLS-AR-V6 was co-transfected with either the ARE-luc plasmid in PC-3 cells (D) or the UBE2C-luc construct in LNCaP cells. F&G. qRT-PCR and SRB assays showing the addition of NLS increasing the ability of AR-V6 to induce target gene expression (F) and to promote castration-resistant growth of LNCaP cells (G). Western blotting confirmed AR-V expression. Cells were cultured under androgen-deprived condition unless specified. DHT, 1 nM for 24 hr. *, P < 0.05 from mock control. Δ, P < 0.05.
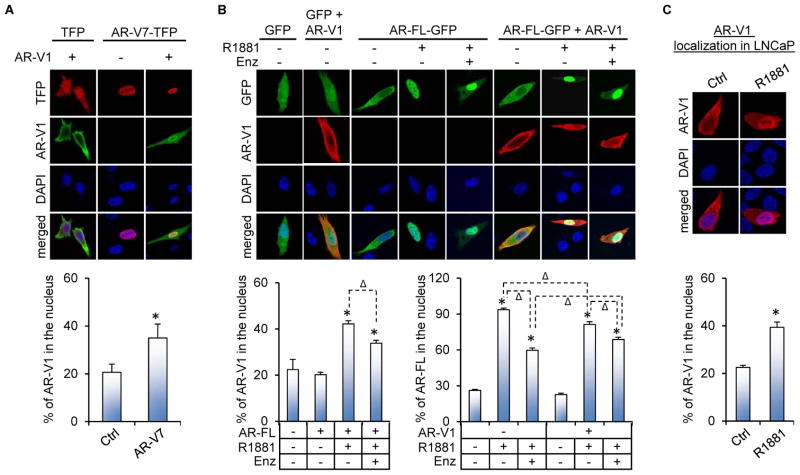
AR-V7 and androgen-bound AR-FL also induce AR-V1 nuclear localization
We next characterized the interactions between AR-V7 or AR-FL and AR-V1. We specifically chose AR-V1 to study because it localizes to both the nucleus and the cytoplasm, (Figure S4 and References (12,16,18)), has been shown to abrogate the ability of AR-V7 to confer castration-resistant cell growth (18), and has an intact D-box, which we showed previously to mediate AR-V/AR-V and AR-V/AR-FL dimerization (32). Similar to AR-V4 and -V6, AR-V1 can heterodimerize with AR-V7 and AR-FL and can also homodimerize (Figure S5). Interestingly, while AR-V1 was able to dimerize with AR-FL in the absence of androgen, androgen enhanced their dimerization (Figure S5D). This was slightly different from AR-V4 or -V6 dimerization with AR-FL, which was not modulated by androgen (Figures 3F and 3G). As a result of the dimerization, AR-V1 was piggybacked to the nucleus by AR-V7 and androgen-bound AR-FL (Figures 5A, 5B, and 5C), and the latter can be attenuated by enzalutamide (Figure 5B). Notably, while AR-V7 localization was not affected by AR-V1 (Figure 5A), AR-V1 was able to moderately inhibit androgen induction of AR-FL nuclear translocation and significantly mitigate enzalutamide activity (Figure 5B). This again slightly differs from AR-V4 and -V6, which could not retain AR-FL in the cytoplasm when cells were treated with androgen (Figure 2A).
Figure 5. AR-V7 and androgen-bound AR-FL facilitate AR-V1 nuclear localization.
A&B. Confocal fluorescence microscopy of AR-V1 subcellular localization when co-expressed with AR-V7 (A) or AR-FL (B) in PC-3 cells. C. Subcellular localization of AR-V1 in LNCaP cells. The FLAG-tagged AR-V1 expression construct was transfected with or without the AR-FL-GFP or AR-V7-TFP plasmid into PC-3 or LNCaP cells under androgen-deprived condition. At 40 hr after transfection, the cells were treated with or without 1 nM R1881 in the presence or absence of 10 μM enzalutamide (Enz) for 6 hr. IF staining with an anti-FLAG antibody was conducted for AR-V1 detection. Bottom panels, quantitation of % of nuclear AR-V1 or AR-FL expression. *, P < 0.05 from control cells. Δ, P < 0.05.
AR-V1 enhances constitutive AR transactivation but attenuates AR-V7 transactivation
AR-V1 has been shown to be inactive in the AR-null PC-3 and DU145 cells but possess high constitutive transcriptional activity in the AR-FL-expressing LNCaP cells (16,18). Concordantly, we found that, compared to AR-V4 and -V6, AR-V1 showed minimal trans-activating activity in AR-null cells (Figures 6A and 6B). However, expression of AR-V1 in LNCaP cells led to a significant activation of a canonical AR-responsive reporter and the expression of a canonical AR target, PSA (Figures 6C and 6D). Interestingly, unlike AR-V4 and -V6, AR-V1 failed to transactivate the AR-V-specific UBE2C promoter or induce UBE2C expression even when AR-FL was present (Figures 6E, 6F, 6G and S6A), indicating that AR-V1 might not have inherent transcriptional activity and that the increased canonical AR trans-activating activity was likely due to androgen-independent activation of AR-FL by AR-V1. In contrast, when co-expressed with AR-V7 in LNCaP or C4-2 cells, AR-V1 antagonized the ability of AR-V7 to transactivate the AR-V-specific UBE2C promoter (Figures 6H and S6B). Taken together, the data indicate the potential of AR-V1 to selectively activate canonical AR signaling while inhibiting AR-V-specific gene expression.
Figure 6. AR-V1 enhances constitutive canonical AR activity but attenuates AR-V7 transactivation.
A&B. Luciferase assay showing minimal ability of AR-V1 to transactivate the ARE-luc reporter in PC-3 and DU145 cells. C. Luciferase assay showing AR-V1 enhancing the constitutive canonical AR activity in LNCaP cells. Cells were co-transfected with the AR-V1 expression construct and the ARE-luc plasmid in bulk and reseeded for treatment with 1 nM DHT for 24 hr. D. qRT-PCR showing AR-V1 increasing the expression of the canonical AR target, PSA, in LNCaP cells. E&F. Luciferase assay showing inability of AR-V1 to transactivate the UBE2C-luc reporter in PC-3 and LNCaP cells. G. qRT-PCR showing inability of AR-V1 to induce the expression of UBE2C gene in LNCaP cells. H. Luciferase assay showing AR-V1 attenuating the ability of AR-V7 to transactivate the UBE2C-luc reporter in LNCaP cells. The pan-AR antibody and the AR-V7 and anti-FLAG antibodies were used to confirm AR-V expression by Western blotting in Panels A-F and Panel G, respectively. *, P < 0.05 from mock control. Δ, P < 0.05.
DISCUSSION
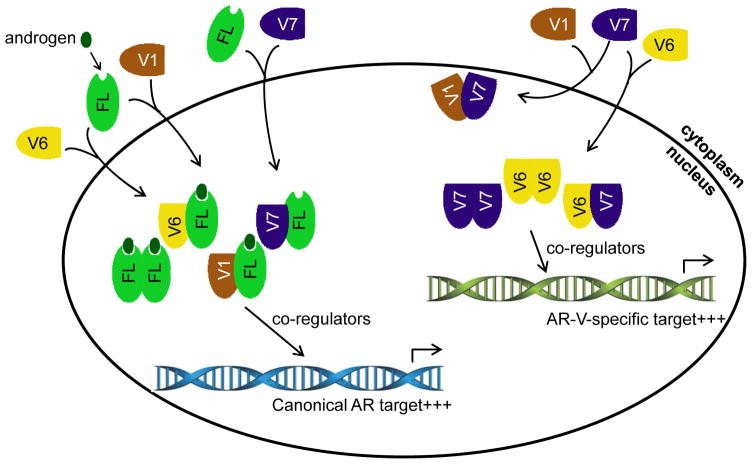
The present study represents the first to characterize the complex interactions of AR-FL as well as AR-V7 with three non-nucleus-predominant AR-Vs and the implication of these interactions on castration resistance. A schematic model of these interactions is presented in Figure 7. We showed that AR-V7 heterodimerizes with AR-V4, -V6, and -V1 and that the dimerization facilitates nuclear localization of AR-V4, -V6, and -V1. Moreover, these non-nucleus-predominant AR-Vs also dimerize with AR-FL, and the dimerization can occur in the absence of androgen. While their dimerizations do not affect the subcellular localization of either AR-FL or any of the three AR-Vs in androgen-deprived condition, AR-V4, -V6, and -V1 can be piggybacked into the nucleus by androgen-bound AR-FL. Significantly, the interactions between AR-V4, -V6, or -V1 and AR-FL mitigate the ability of the anti-androgen enzalutamide to inhibit androgen-induced AR-FL nuclear localization.
Figure 7. A schematic model of the interactions among AR-Vs and AR-FL and the impact of the interactions on AR transcriptional program.
AR-V7 and androgen-bound AR-FL induce nuclear localization of AR-V1 and -V6. AR-V7 also induces nuclear localization of AR-FL in the absence of androgen. In the nucleus, the heterodimer between AR-V and AR-FL as well as the AR-FL homodimer recruit co-regulators to transactivate canonical AR targets, and the AR-V6 and -V7 homodimers as well as AR-V6/-V7 heterodimer recruit co-regulators to activate AR-V-specific targets. AR-V1, which lacks inherent transcriptional activity, antagonizes AR-V7 transactivation. AR-V4, which is not included in this schematic model, behaves the same as AR-V6.
Although all of these three AR-Vs can be piggybacked into the nucleus by AR-V7 and androgen-bound AR-FL, the impact of nuclear localization of AR-V4 and -V6 on AR transactivation differs significantly from that of AR-V1. Nuclear localization leads to an increased ability of AR-V4 and -V6 to transactivate both canonical AR targets and AR-V-specific targets and to confer castration-resistant cell growth. In contrast, AR-V1 lacks the ability to transactivate the AR-V-specific UBE2C promoter either in the presence or absence of AR-FL but is efficient in inducing a canonical AR-responsive reporter in androgen-deprived condition when AR-FL is present. On the other hand, it antagonizes significantly the trans-activating activity of AR-V7, indicating the potential of AR-V1 to selectively activate canonical AR signaling while inhibiting AR-V-specific gene expression.
Our finding of AR-V7 facilitating AR-V4 and -V6 nuclear localization and inducing their abilities to transactivate target genes and to confer castration-resistant cell growth is reminiscent of our previous report on its ability to activate AR-FL in an androgen-independent manner (30). AR-V7 is the most abundantly expressed AR-V in clinical specimens (24,26), and its role in mediating castration resistance is strongly supported by accumulating clinical evidences (19,20,23,25). Its potential to confer resistance of CRPC to taxanes has also been demonstrated by preclinical studies (27–29). Our findings indicate that, in addition to its own transcriptional activity, AR-V7 can activate AR-FL and other AR-Vs, and the latter could be an important mechanism of its action. Our findings also suggest that, although some AR-Vs localize in large part in the cytoplasm when expressed alone and are less abundant compared to AR-V7, their clinical relevance should not be dismissed. Since these AR-Vs are co-expressed with AR-V7 in patient tumors, as a result of activation by AR-V7, these AR-Vs, collectively, may contribute significantly to clinical castration resistance and to taxane resistance possibly by providing functions and transcriptional activation in additional to the canonical AR programs in the absence of androgen. Nonetheless, this does not mean that all non-nucleus-predominant AR-Vs can be activated by AR-V7. Together with the reported ability of AR-V1 to abrogate AR-V7 induction of castration-resistant cell growth (18), our data on AR-V1 point to the existence of another scenario where AR-V7-induced nuclear translocation of the AR-Vs that lack inherent transcriptional activity could lead to inhibition of AR-V7 transactivation. In addition, another AR-V, AR8, has been shown to localize mainly to the plasma membrane even in the presence of AR-V7 (31). At least 20 AR-Vs have been identified to date. Detailed characterization of their activities in the context of other AR-Vs and AR-FL would be important for a better appreciation of their clinical relevance.
An intriguing phenomenon that we observed in this study is that, while AR-V1 may act as a dominant-negative regulator of AR-V7, it activates a canonical AR-responsive reporter in an androgen-independent manner when AR-FL is present. Since AR-FL and AR-V1 do not affect the subcellular localization of each other in androgen-deprived condition, the increased constitutive AR activity is unlikely a result of more nuclear localization of AR-V1. Considering that AR-V1 lacks inherent transcriptional activity, this increased activity is likely due to androgen-independent activation of AR-FL by AR-V1. Therefore, it is possible that AR-V1 may function as a transcriptional activator of AR-FL but as a transcriptional suppressor of AR-V7. This androgen-independent AR-FL transcriptional activator function of AR-V1, together with its ability to mitigate anti-androgen inhibition of androgen-induced AR-FL nuclear localization, indicates that AR-V1 may still play a role in mediating castration resistance. Although stable expression of AR-V1 failed to confer castration-resistant growth of LNCaP xenograft (18), further investigations, especially in different model systems, are warranted.
Like AR-V7, AR-V1, -V4, and -V6 are all truncated after exon 3, and all contain the D-box, which mediates AR-V/AR-V and AR-V/AR-FL dimerization (32) (Figure S1). They differ only in the amino acid sequence of the short C-terminal extension (Figure S1). However, they are distinctive from AR-V7 in subcellular localization and transcriptional activity, and they also differ among themselves in biological functions. AR-V4 and -V6 appear to resemble each other but are functionally distinct from AR-V1. Although all three AR-Vs can homodimerize, AR-V4 and -V6, but not AR-V1, possess inherent constitutive transcriptional activity. When interacting with AR-V7, AR-V4 and -V6 increase, but AR-V1 suppresses, AR-V transactivation. In addition, while all three AR-Vs can dimerize with AR-FL in the absence of androgen, androgen does not affect the dimerization between AR-V4/-V6 and AR-FL but enhances AR-V1 and AR-FL dimerization. This also differs significantly from AR-V7 in that androgen disrupts AR-V7 and AR-FL dimerization, i.e., only unliganded AR-FL can dimerize with AR-V7 (30). Nonetheless, all four AR-Vs can mitigate the ability of the anti-androgen enzalutamide to inhibit androgen-induced AR-FL nuclear localization. Taken together, although differing only in the C-terminal peptide sequence, these truncated AR-Vs can have distinct biological properties. A detailed functional characterization of their unique C-terminal peptide sequences and the impact on their protein structures, nucleocytoplasmic trafficking, DNA-binding capacities and specificities, as well as interactions with molecular chaperones and co-regulators would help explain these distinctions.
In summary, we demonstrated the complex interactions among different AR-Vs and AR-FL in mediating castration resistance. Since different types of AR-Vs are often simultaneously expressed and also co-expressed with AR-FL in patient tumors, our findings underscore the important role of their interactions in clinical castration resistance. The clinical relevance of certain AR-Vs may be neglected if they are viewed in isolation. Careful annotation of the expression of different AR-Vs in prostate cancer patients after initiation of androgen-directed therapies and at relapse should help clarify the role of different AR-Vs in driving the progression of the disease and help develop effective therapeutic strategies to disrupt AR-V signaling.
Supplementary Material
Acknowledgments
Funding: This work was supported by the following grants: NIH/NCI R01CA188609; NIH 1U01CA149556-01; DOD W81XWH-15-1-0439, W81XWH-12-1-0112, W81XWH-14-1-0485, and W81XWH-12-1-0275; Louisiana Cancer Research Consortium Fund; National Natural Science Foundation of China Projects 81272851 and 81430087.
We are grateful to Drs. Yun Qiu (University of Maryland) and Jun Luo (Johns Hopkins University) for the AR-V7 expression construct and the pEGFP-AR-FL plasmid, respectively, and to Dr. Sanjiv Gambhir (Stanford University) for the pcDNA3.1-RLuc8.6 and pCMV-TurboFP635 vectors.
Footnotes
Conflict of Interest: None
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Egan A, Dong Y, Zhang H, Qi Y, Balk SP, Sartor O. Castration-resistant prostate cancer: Adaptive responses in the androgen axis. Cancer Treat Rev. 2014;40:426–33. doi: 10.1016/j.ctrv.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 3.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–98. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Zhan Y, Liu X, Qi Y, Zhang G, Sartor O, et al. Splicing variants of androgen receptor in prostate cancer. Am J Clin Exp Urol. 2013;1:18–24. [PMC free article] [PubMed] [Google Scholar]
- 5.Attard G, Reid AH, Yap TA, Raynaud F, Dowsett M, Settatree S, et al. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J Clin Oncol. 2008;26:4563–71. doi: 10.1200/JCO.2007.15.9749. [DOI] [PubMed] [Google Scholar]
- 6.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–90. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. Abiraterone and Increased Survival in Metastatic Prostate Cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fizazi K, Scher HI, Molina A, Logothetis CJ, Chi KN, Jones RJ, et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2012;13:983–92. doi: 10.1016/S1470-2045(12)70379-0. [DOI] [PubMed] [Google Scholar]
- 9.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 10.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–48. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dehm SM, Tindall DJ. Androgen receptor structural and functional elements: role and regulation in prostate cancer. Mol Endocrinol. 2007;21:2855–63. doi: 10.1210/me.2007-0223. [DOI] [PubMed] [Google Scholar]
- 12.Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem. 2012;287:19736–49. doi: 10.1074/jbc.M112.352930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–77. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–13. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. The Prostate. 2011;71:1656–67. doi: 10.1002/pros.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–30. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci USA. 2010;107:16759–65. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–62. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–6. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu Y, Dai B, Ye D, Kong Y, Chang K, Jia Z, et al. Constitutively active AR-V7 plays an essential role in the development and progression of castration-resistant prostate cancer. Sci Rep. 2015;5:7654. doi: 10.1038/srep07654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welti J, Rodrigues DN, Sharp A, Sun S, Lorente D, Riisnaes R, et al. Analytical Validation and Clinical Qualification of a New Immunohistochemical Assay for Androgen Receptor Splice Variant-7 Protein Expression in Metastatic Castration-resistant Prostate Cancer. European Urology. doi: 10.1016/j.eururo.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry Christopher D, et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thadani-Mulero M, Portella L, Sun S, Sung M, Matov A, Vessella RL, et al. Androgen receptor splice variants determine taxane sensitivity in prostate cancer. Cancer Res. 2014;74:2270–82. doi: 10.1158/0008-5472.CAN-13-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang G, Liu X, Li J, Ledet E, Alvarez X, Qi Y, et al. Androgen receptor splice variants circumvent AR blockade by microtubule-targeting agents. Oncotarget. 2015;6:23358–71. doi: 10.18632/oncotarget.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin SK, Pu H, Penticuff JC, Cao Z, Horbinski C, Kyprianou N. Multinucleation and Mesenchymal-to-Epithelial Transition Alleviate Resistance to Combined Cabazitaxel and Antiandrogen Therapy in Advanced Prostate Cancer. Cancer Res. 2016;76:912–26. doi: 10.1158/0008-5472.CAN-15-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao B, Qi Y, Zhang G, Xu D, Zhan Y, Alvarez X, et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014;5:1646–56. doi: 10.18632/oncotarget.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, Guo Z, Sun F, Li W, Alfano A, Shimelis H, et al. Novel membrane-associated androgen receptor splice variant potentiates proliferative and survival responses in prostate cancer cells. J Biol Chem. 2011;286:36152–60. doi: 10.1074/jbc.M111.265124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu D, Zhan Y, Qi Y, Cao B, Bai S, Xu W, et al. Androgen Receptor Splice Variants Dimerize to Transactivate Target Genes. Cancer Res. 2015;75:3663–71. doi: 10.1158/0008-5472.CAN-15-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, Qi Y, Ge Y, Duplessis T, Rowan BG, Ip C, et al. Telomerase as an important target of androgen signaling blockade for prostate cancer treatment. Mol Cancer Ther. 2010;9:2016–25. doi: 10.1158/1535-7163.MCT-09-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dragulescu-Andrasi A, Chan CT, De A, Massoud TF, Gambhir SS. Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects. Proc Natl Acad Sci USA. 2011;108:12060–65. doi: 10.1073/pnas.1100923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhan Y, Cao B, Qi Y, Liu S, Zhang Q, Zhou W, et al. Methylselenol prodrug enhances MDV3100 efficacy for treatment of castration-resistant prostate cancer. Int J Cancer. 2013;133:2225–33. doi: 10.1002/ijc.28202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong Y, Lee SO, Zhang H, Marshall J, Gao AC, Ip C. Prostate specific antigen expression is down-regulated by selenium through disruption of androgen receptor signaling. Cancer Res. 2004;64:19–22. doi: 10.1158/0008-5472.can-03-2789. [DOI] [PubMed] [Google Scholar]
- 37.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–16. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 38.Hamdan FF, Percherancier Y, Breton B, Bouvier M. Monitoring protein-protein interactions in living cells by bioluminescence resonance energy transfer (BRET) Curr Protoc Neurosci. 2006;Chapter 5 doi: 10.1002/0471142301.ns0523s34. Unit. [DOI] [PubMed] [Google Scholar]
- 39.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.