Abstract
Responses of photosynthetic organisms to sulfur starvation include (i) increasing the capacity of the cell for transporting and/or assimilating exogenous sulfate, (ii) restructuring cellular features to conserve sulfur resources, and (iii) modulating metabolic processes and rates of cell growth and division. We used microarray analyses to obtain a genome-level view of changes in mRNA abundances in the green alga Chlamydomonas reinhardtii during sulfur starvation. The work confirms and extends upon previous findings showing that sulfur deprivation elicits changes in levels of transcripts for proteins that help scavenge sulfate and economize on the use of sulfur resources. Changes in levels of transcripts encoding members of the light-harvesting polypeptide family, such as LhcSR2, suggest restructuring of the photosynthetic apparatus during sulfur deprivation. There are also significant changes in levels of transcripts encoding enzymes involved in metabolic processes (e.g., carbon metabolism), intracellular proteolysis, and the amelioration of oxidative damage; a marked and sustained increase in mRNAs for a putative vanadium chloroperoxidase and a peroxiredoxin may help prolong survival of C. reinhardtii during sulfur deprivation. Furthermore, many of the sulfur stress-regulated transcripts (encoding polypeptides associated with sulfate uptake and assimilation, oxidative stress, and photosynthetic function) are not properly regulated in the sac1 mutant of C. reinhardtii, a strain that dies much more rapidly than parental cells during sulfur deprivation. Interestingly, sulfur stress elicits dramatic changes in levels of transcripts encoding putative chloroplast-localized chaperones in the sac1 mutant but not in the parental strain. These results suggest various strategies used by photosynthetic organisms during acclimation to nutrient-limited growth.
Sulfur (S) is an essential element present in proteins, lipids, and various metabolites. It is critical for the association of metal ions to proteins (electron carriers and redox controllers) and is a component of metabolites that function in photoprotection (14, 29) and signal transduction (such as in symbiosis) (45). Because most organisms have limited S storage, their growth and development is dependent upon a continuous supply of this nutrient from the environment. The majority of accessible S in soil solutions is in the form of the SO42− anion. However, in some cases the majority of soil SO42− may not be readily available to the biota, since the SO42− anion is often complexed with cations as insoluble salts that are tightly adsorbed onto the surface of soil particles or exists as a soluble anion that rapidly leaches through the soil matrix. Furthermore, a large proportion of soil SO42− may be covalently bonded to organic molecules in the form of sulfate esters and sulfonates.
The acquisition of SO42− by plants and microorganisms is facilitated by specific transport systems. Following uptake, the anion is either used for the direct sulfation of compounds or is reduced and converted to cysteine and methionine, which is incorporated into both proteins and specific cellular metabolites (1, 25, 41, 49). Cysteine is also an important building block for the synthesis of both glutathione and phytochelatins. The former serves as a source of reductant for many physiological processes and as an antioxidant that enables cells to withstand oxidative stress (34, 35), while the latter helps combat heavy metal toxicity (16, 63).
S can be limiting in the environment and strongly influence ecosystem composition. It may also limit plant productivity in certain agricultural settings (27, 28, 57), which can result in reduced quality and yields of seeds, and upon severe S limitation, the growth of the plant may be stunted. We have used the unicellular alga Chlamydomonas reinhardtii to identify and elucidate S limitation responses. C. reinhardtii synthesizes a prominent, extracellular arylsulfatase (ARS) in response to S limitation (26, 44). An ARS associated with the proteinaceous cell wall of the alga has been purified to homogeneity and characterized (12). This ARS polypeptide has at least three O-linked oligosaccharides, is very stable in the extracellular space (stability may be conferred on the polypeptide by oligosaccharide decorations), and is synthesized as a preprotein with a signal sequence that is cleaved as it is exported from the cell (11). The extracellular location of ARS allows it to hydrolyze soluble SO42− esters in the medium, releasing free SO42− for assimilation by the cells. Two ARS genes arranged in tandem, but in the opposite orientation on the C. reinhardtii genome, have been described (11, 40), but the gene family may contain a number of additional members.
There are several transcripts, in addition to those encoding ARS, that are synthesized by C. reinhardtii in response to S deprivation. Some of these transcripts encode enzymes involved in the uptake and assimilation of SO42− (62). Levels of mRNAs for enzymes of the SO42− assimilation pathway in plants also increase during S starvation (15, 42, 47, 52). S starvation of C. reinhardtii causes a dramatic increase in the Vmax and a decrease in the Km for SO42− transport (61). Eukaryotic organisms have multiple systems for SO42− uptake, and generally high-affinity SO42− uptake increases when they are deprived of S (3, 4, 17, 24, 61). Recently, a C. reinhardtii gene encoding a putative SO42− transporter was isolated and appears to be activated during S-limited growth (J. Davies, personal communication), but genes encoding potential SO42− transporters (designated SULTR1, SULTR2, and SULTR3) have also been identified in cDNA and genomic databases (http://genome.jgi-psf.org/chlre2/chlre2.home.html). The subcellular locations of these putative protein products have not been determined. Furthermore, in C. reinhardtii there are prominent extracellular polypeptides of apparent molecular masses of 76 kDa (designated extracellular polypeptide 76, or ECP76) and 84 kDa (ECP84) that have been shown to be synthesized in response to S deprivation (51). The deduced ECP76 and ECP84 polypeptide sequences have significant similarity to those of various cell wall proteins but contain either no or few S-containing amino acids (between mature ECP76 and ECP84 there is only a single S-containing amino acid). These results suggest that a highly regulated process tailors the protein-rich cell wall of C. reinhardtii for S deprivation.
There are a number of factors that control the assimilation of SO42− and the regulation of genes involved in that assimilation. O-acetylserine, the carbon backbone used for the synthesis of cysteine, functions as a positive effector for the transcription of genes encoding enzymes that participate in the uptake and assimilation of SO42− (22) in both bacteria and vascular plants (20, 50). Increases in O-acetylserine levels appear to occur when Arabidopsis thaliana plants are starved for S or provided with excess N (19). In C. reinhardtii, specific regulatory elements, including SAC1 and SAC3, control S deprivation responses (8-10). A number of physiological responses of C. reinhardtii to S limitation require the SAC1 protein (8). The sac1 mutant of C. reinhardtii exhibits abnormal SO42− uptake and is unable to synthesize extracellular ARS, as well as other extracellular proteins, in response to S deprivation. Furthermore, there was little or no induction of ECP76, ECP84, and ATS1 (encoding ATP sulfurylase, the enzyme that activates free SO42−, allowing reduction to occur) genes in a sac1 null mutant background (51, 62). The responses of the sac1 mutant to P and N limitation appear normal. The inability of the sac1 strain to properly respond to S limitation is also reflected in a rapid decline in the viability of mutant cells following exposure to S deprivation (8); the decreased viability is sensitive to environmental light levels and the activity of photosynthetic electron transport. Since the sac1 mutant cannot properly control photosynthetic activity during S deprivation, it may accumulate reactive oxygen species, which could cause extensive cellular damage, and the electron transport chain would become hyperreduced (e.g., a highly reduced plastoquinone pool), which could have adverse affects on the control of metabolic processes.
Interestingly, the SAC1 gene product is predicted to be a polypeptide with similarity to anion transporters (reference 9 and our unpublished work). The deduced SAC1 polypeptide sequence and phenotype of the sac1 mutant have some similarities with the Snf3 polypeptide of yeast and the phenotype of the snf3 mutant, respectively. Snf3 is a yeast transporter-like regulatory protein that governs expression of genes involved in hexose utilization (37, 38). The putative regulatory functions of SAC1 and Snf3 raise the possibility that polypeptides that originally functioned in binding and transporting various substrates into cells may have evolved into regulatory elements.
In this study we used microarray analyses to obtain a genome-based picture of C. reinhardtii responses to S limitation and to determine how the sac1 lesion alters S deprivation-triggered changes in gene expression. The results presented are discussed in the context of physiological changes that may be elicited by S limitation and the regulatory circuits that may operate during S starvation.
MATERIALS AND METHODS
Cell culture.
The C. reinhardtii arginine auxotroph CC425 and the sac1 mutant (carrying ars5-5) (8) were cultured in S-replete, Tris-acetate-phosphate (TAP) medium to mid-logarithmic phase on a rotating platform (120 rpm) and continuously illuminated (80 μmol of photon m−2 s−1) at 25°C. To assay global changes in transcript levels in response to S deprivation, the cells were harvested at room temperature (5,000 × g, 5 min), washed twice with TAP medium without S, and resuspended in TAP medium without S to 5 × 106 to 10 × 106 cells per ml. The cultures were maintained on a rotating platform, illuminated with 80 μmol of photon m−2 s−1 for 2, 4, 8, 12, and 24 h, and then rapidly cooled in liquid N2 and harvested by centrifugation (5,000 × g, 5 min, 4°C).
RNA preparation.
Cell pellets were resuspended in 3 ml of extraction buffer (50 mM Tris, 300 mM NaCl, 0.5 mM EDTA, 2% sodium dodecyl sulfate [SDS], pH 8.0, that had been treated with 0.1% diethyl pyrocarbonate), followed by the addition of 5 μl of proteinase K (40 μg/ml). The suspension was gently agitated for 30 min at room temperature and then extracted with an equal volume of phenol-chloroform (1:1) until the aqueous phase became clear. Nucleic acid in the aqueous phase was precipitated with 2 volumes of ethanol, dried, and dissolved in diethyl pyrocarbonate-treated double-distilled H2O (ddH2O). RNA was purified by adding an equal volume of 4 M LiCl to the nucleic acid solution, incubating the solution on ice for 8 h, and collecting the precipitated RNA by centrifugation at 10,000 × g for 15 min. Two RNA preparations for each time point were generated from independent experiments. RNA samples were further purified with the RNeasy MiniKit (QIAGEN, Valencia, Calif.).
Fluorescent labeling of probe.
Total RNA served as the template for labeling cDNA by direct incorporation of Cy3-dUTP or Cy5-dUTP (Amersham Pharmacia Biotech, Piscataway, N.J.) by using reverse transcription. RNA (10 μg total) was denatured at 70°C for 10 min in the presence of 1 μg of Oligo(dT)12-18 (Invitrogen, Carlsbad, Calif.) in a reaction volume of 23 μl. The reaction was cooled on ice prior to the addition of 8 μl of 5× Superscript II reverse transcriptase buffer (Invitrogen), 1 μl of 0.1 M dithiothreitol, 4 μl of deoxynucleoside triphosphates (10 mM dTTP, 25 mM dATP, 25 mM dGTP, and 25 mM dCTP), 2 μl of Superscript II reverse transcriptase (200 U/μl), and 2 μl of 1 mM Cy3-dUTP or Cy5-dUTP. The reaction was incubated at 42°C in a thermocycler (MJ Research, Waltham, Mass.) for 2 h and then stopped by the addition of 2 μl of 500 mM EDTA and 2 μl of 500 mM NaOH. The alkalinized solution was incubated at 70°C for 10 min to degrade RNA prior to neutralization by the addition of 2 μl of 500 mM HCl. The Cy5-dUTP-labeled probe from each time point following the initiation of starvation was combined with the Cy3-dUTP-labeled control RNA (zero time; unstarved cells), purified with a QiaQuick PCR purification kit (QIAGEN), and recovered in 17 μl of ddH2O. Incorporation of the fluorescent dyes into cDNA was visualized with a Typhoon 8600 (following electrophoresis of the cDNA on a 0.8% agarose gel). It should be emphasized that the reference RNA was isolated from our parental strain (CC425) prior to S deprivation (0 h). Samples prepared from S-deprived (2, 4, 8, 12, and 24 h) CC425 and the unstarved (0 h) and starved (2, 4, 8, 12, and 24 h) sac1 mutant were compared to this sample. The dye labeling was reversed in a separate experiment, and the experiments were duplicated with independently isolated RNA samples.
Microarray preparation.
C. reinhardtii cDNA clones from the Core (TAP light, TAP dark, HS+CO2, and HS) and Stress I Libraries (NO3− to NH4+ [30 min, 1 h, 4 h], NH4+ to NO3− [30 min, 1 h, 4 h], TAP without N [30 min, 1 h, 4 h], TAP without S [30 min, 1 h, 4 h], and TAP without P [4 h, 12 h, 24 h]) were sequenced (48), and the sequences generated from the 3′ ends were assembled into 2,761 unique contigs that were used to construct a microarray. Further details of the “2.7-k array ” are given at the website http://nostoc.stanford.edu/jeff/lab/chlamyarray/index.html and in both Table 1 and Table S1 in the supplemental material. The 3′ sequences were amplified with a universal primer (forward primers: 631u24, 5′-CGACTCACTATAGGGCGAATTGGG-3′, 1 bp longer than the T7 primer; or M13-21, 5′-TGTAAAACGACGGCCAGT-3′) and a specific reverse primer designed to anneal to the first strand of each clone in a region 200 to 500 bp away from 3′ end of the transcript. PCRs were performed in a 96-well format according to the protocol 94°C for 2.5 min, 94°C for 30 s, 62°C or 50°C for 30 s, and 72°C for 1 min for 40 cycles. The quality and specificity of the amplification products were determined by electrophoresis on a 1% agarose gel followed by visualization of nucleic acid by ethidium bromide staining; 96.6% of the clones gave a single product of the expected size. The PCR products were cleaned by ethanol precipitation and dissolved in 40 μl of 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 5 μl was transferred from the 96-well microtiter plates to printing plates, which were in the 384-well microtiter format (MJ Research). Printing plates were wrapped with moistened paper towels, sealed with Saran Wrap, and stored at −20°C until they were used. PCR products were arrayed from the 384-well microtiter plates onto CMT-gamma amino propyl silane slides coated with γ amino propyl silane (Corning Microarray Technology, Corning, N.Y.) by using an Omnigrid microarrayer (GeneMachines, San Carlos, Calif.) with ChipMaker 2 pins (TeleChem International, Sunnyvale, Calif.). Array spots were ∼150 μm in diameter, and the center-to-center distance was 212.5 μm. Four complete cDNA sets were printed onto each slide, with each set consisting of eight subsets of 349 spots each (18 rows by 20 columns). Printed slides were baked at 80°C for 2 h and cross-linked by UV irradiation in the Stratalinker 1800 (Stratagene, La Jolla, Calif.) at a total power of 300 mJ. The arrays were blocked by incubating them for 15 min in a succinic anhydride-NaBO4− solution. This solution was freshly prepared by dissolving 5 g of succinic anhydride (Sigma, St. Louis, Mo.) in 315 ml of N-methyl-pyrrolidinone and adding 35 ml of 0.2 M NaBO4− (boric acid dissolved in ddH2O and made pH 8.0 with NaOH). The arrays were then soaked for 2 min with gentle agitation in 95°C ddH2O, immersed in 95% ethanol (high-performance liquid chromatography grade) for 1 min, and dried by centrifugation for 5 min in a SpeedVac Plus model SC210A (Savant, Holbrook, N.Y.). The arrays were stored in a desiccator at room temperature until they were used.
TABLE 1.
Transcripts showing a ≥3-fold change in abundance during sulfur deprivationa
| Clone (Fig. 3 panel)b | ACEc | Name | Gene annotation | Direction of change in transcript level
|
Transcript level (fold increase) for
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC425
|
sac1 mutant
|
|||||||||||||||||||||
| CC425d | sac1d | 2 h | 4 h | 8 h | 12 h | 24 h | 0 h | 2 h | 4 h | 8 h | 12 h | 24 h | ||||||||||
| Sulfur metabolism | ||||||||||||||||||||||
| ′963038E01 | 1936 | ARSI | Arylsulfatase | +++++ | ...... | 8.5 | 17.52 | 42.81 | 41.59 | 3.39 | 1.32 | 1.1 | 0.8 | 0.77 | 0.99 | NA | ||||||
| ′894069D01 | 9009 | SIRI | Sulfite reductase | +++++ | −+.++. | 1.48 | 1.63 | 2.05 | 2.62 | 3.16 | 0.39 | 3.30 | 1.42 | 1.24 | 1.47 | 1.20 | ||||||
| ′963038A06 | 4494 | Sulfite reductase | +++++ | .+++++ | 2.69 | 2.48 | 3.18 | 3.59 | 2.39 | 0.75 | 2.37 | 1.85 | 1.76 | 1.91 | 1.89 | |||||||
| ′963042H10 | 8444 | SIR3 | Sulfite reductase, bacterial type | ..... | .+.−.. | 0.94 | 1.03 | 1.11 | 0.96 | 0.99 | 0.93 | 2.16 | NAe | 0.49 | 1.72 | NA | ||||||
| ′894033F10 | [7588] | ATS1 | ATP sulfurylase | +++++ | .++++. | 4.92 | 5.29 | 8.62 | 10.58 | 7.69 | 1.05 | 2.65 | 2.43 | 2.04 | 1.7 | 0.9 | ||||||
| ′963046C09 | 1668 | ATS2 | ATP sulfurylase | +++++ | ...... | 3.63 | 4.69 | 5.91 | 8.64 | 7.74 | 0.9 | 1.61 | 1.23 | 0.85 | 1.35 | 0.42 | ||||||
| ′894044E12 | 9443 | APR | 5′-Adenylylsulfate reductase | +++++ | ...... | 2.35 | 1.57 | 1.31 | 1.90 | 1.34 | 1.01 | 0.90 | 0.85 | 0.99 | 1.21 | 0.97 | ||||||
| ′894077C01 | 1669 | AKN2 | APS kinase | ...+. | ...... | 0.77 | 1.17 | 0.70 | 1.42 | 1.07 | 0.57 | NA | NA | NA | NA | NA | ||||||
| ′963045D11 | 3138 | SAT1 | Serine O-acetyltransferase | +++++ | −.+.++ | 6.69 | 6.25 | 10.59 | 14.77 | 9.06 | 0.88 | 1.77 | 1.44 | 1.24 | 1.44 | 2.18 | ||||||
| ′963024B05 | 8367 | OASTL4 | O-Acetylserine(thiol)lyase | +++++ | −++++. | 6.01 | 7.00 | 10.03 | 9.16 | 8.3 | 0.77 | 2.34 | 2.29 | 2.59 | 2.23 | 1.17 | ||||||
| ′963076G10 | 6009 | CDO1 | Cysteine dioxygenase | +++++ | −++++. | 5.94 | 5.68 | 7.75 | 8.9 | 7.54 | 0.71 | 2.29 | 1.92 | 2.01 | 2.01 | 0.71 | ||||||
| ′963035D06 | 8168 | SQDI | UDP-SQi synthase | +++++ | .+++++ | 5.63 | 6.64 | 7.73 | 9.5 | 9.57 | 0.97 | 4.43 | 2.4 | 2.45 | 2.36 | 4.2 | ||||||
| ′894101C06 | Nonef | SQD2a | UDP-SQj; diacylglycerol SQj transferase | +++++ | −++++. | 5.16 | 6.13 | 8.44 | 8.31 | 6.89 | 0.83 | 1.69 | 2.35 | 1.9 | 1.75 | 1.12 | ||||||
| ′963017H04 | [5161] | ECP76 | Extracellular polypeptide 76 | +++++ | −...++ | 4.83 | 9.49 | 34.07 | 22.14 | 21.45 | 0.85 | 1.12 | 0.78 | 1.28 | 1.41 | 1.28 | ||||||
| ′894020C12 | 5894 | SBDP | Selenium-binding protein | +++++ | ++++++ | 7.89 | 5.52 | 9.66 | 8.23 | 5.19 | 1.29 | 1.7 | 1.53 | 1.39 | 1.52 | 1.6 | ||||||
| ′963027A09 | 5894 | SBDP | Selenium-binding protein | +++++ | −+.++. | 12.72 | 14.16 | 16.13 | 20.4 | 15.71 | 0.74 | 1.75 | 1.53 | 1.57 | 1.86 | 0.89 | ||||||
| ′963046G02 | 7825 | SACI-like proteins (2 overlapping genes) | +++++ | −+++++ | 27.52 | 18.06 | 40.38 | 45.9 | 26.56 | 0.77 | 3.66 | 2.13 | 1.86 | 2.41 | 1.63 | |||||||
| ′894081F11 | [7825] | SAC1-like protein | +++++ | −++.+. | 60.83 | 48.89 | 37.09 | 84.46 | 53.56 | 0.64 | 1.62 | 2.12 | 2.34 | 2.95 | 1.23 | |||||||
| Photosynthesis | ||||||||||||||||||||||
| ′963024B11 (C) | 5736 | LHCA1 | Light-harvesting complex, PS I | .++−− | −−−−−− | 1.21 | 1.38 | 1.2 | 0.7 | 0.39 | 0.38 | 0.41 | 0.25 | 0.25 | 0.26 | 0.06 | ||||||
| ′963047H05 (C) | 2886 | LHCA2 | Light-harvesting complex, PS I | ...−− | −−−−−− | 1.27 | 1.16 | 1.1 | 0.58 | 0.3 | 0.39 | 0.2 | 0.16 | 0.15 | 0.09 | 0.03 | ||||||
| ′894033H06 (C) | 4209 | LHCA3 | Light-harvesting complex, PS I | ...−− | −−−−−− | 1.04 | 1.13 | 1.09 | 0.55 | 0.22 | 0.37 | 0.31 | 0.22 | 0.17 | 0.12 | 0.01 | ||||||
| ′963042A01 (C) | 9189 | LHCA5 | Light-harvesting complex, PS I | +++.− | −−−−−− | 1.55 | 1.67 | 1.28 | 0.85 | 0.39 | 0.34 | 0.38 | 0.28 | 0.26 | 0.16 | 0.02 | ||||||
| ′894041D11 (C) | 7340 | LHCA5 | Light-harvesting complex, PS I | ...−− | −−−−−− | 1.22 | 1.67 | 1.19 | 0.69 | 0.45 | 0.66 | 0.37 | 0.29 | 0.29 | 0.32 | 0.22 | ||||||
| ′894087C09 (C) | 4579 | LHCA8 | Light-harvesting complex, PS I | +++.− | −−−−−. | 1.46 | 1.42 | 1.36 | 0.84 | 0.36 | 0.86 | 0.56 | 0.33 | 0.37 | 0.42 | 0.41 | ||||||
| ′894044B07 (C) | 5739 | LHCA6 | Light-harvesting complex, PS I | +++−− | −−−−−− | 1.2 | 1.15 | 1.21 | 0.68 | 0.41 | 0.69 | 0.57 | 0.39 | 0.32 | 0.26 | 0.1 | ||||||
| ′894078C01 (C) | ′[6907] | LHCA9 | Light-harvesting complex, PS I | +++−− | .−−−−− | 1.28 | 1.27 | 1.22 | 0.79 | 0.26 | 0.78 | 0.34 | 0.39 | 0.33 | 0.18 | 0.01 | ||||||
| ′894076B06 (C) | 7340 | LHCA9 | Light-harvesting complex, PS I | +..−− | −−−−−. | 1.2 | 1.13 | 1.18 | 0.67 | 0.37 | 0.47 | 0.19 | 0.13 | 0.11 | 0.1 | NA | ||||||
| ′963069C06 (C) | ′[8250] | LHCBM1 | Light-harvesting complex, PS II | +++.− | −−−−−− | 1.26 | 1.46 | 1.33 | 1.11 | 0.41 | 0.54 | 0.19 | 0.23 | 0.17 | 0.07 | 0.00 | ||||||
| ′894080G01 (C) | 1951 | LHCBM3 | Light-harvesting complex, PS II | .+−.− | −−−−−− | NA | 1.18 | 0.79 | 0.84 | 0.52 | 0.57 | 0.25 | 0.19 | 0.14 | 0.09 | 0.01 | ||||||
| ′894062E07 (C) | 7231 | LHCB4 | Light-harvesting complex, PS II | +++.− | −−−−−− | 1.38 | 1.34 | 1.49 | 0.95 | 0.73 | 0.82 | 0.52 | 0.36 | 0.47 | 0.39 | 0.07 | ||||||
| ′894052A01 (C) | 1436 | LHCB5 | CP26, minor light-harvesting complex, PS II | +++−− | −−−−−− | 1.21 | 1.3 | 1.36 | 0.69 | 0.63 | 0.55 | 0.28 | 0.22 | 0.22 | 0.18 | 0.07 | ||||||
| ′894097E05 (E) | 5770 | LhcSR2 | Light-harvesting family polypeptide | +++++ | .+.... | 10.22 | 13.94 | 19.99 | 20.54 | 22.38 | 0.82 | 1.4 | 0.67 | NA | 1.62 | NA | ||||||
| ′894005B12 | ′[8181] | LIL | Light-harvesting family polypeptide | +..−. | .....− | 1.55 | 0.97 | 0.85 | 0.58 | 0.94 | 1.00 | 0.7 | 0.74 | 0.87 | 0.72 | 0.38 | ||||||
| ′963047E03 (A) | ′[2510] | PSAD | PS I subunit IV | ....− | −−−−−− | 1.16 | 1.13 | 1.18 | 1.01 | 0.65 | 0.84 | 0.45 | 0.43 | 0.44 | 0.29 | 0.03 | ||||||
| ′894083B07 (A) | ′[3606] | PSAE | PS I subunit V | ...−− | −−−−−− | 1.03 | 0.9 | 1.00 | 0.69 | 0.24 | 0.75 | 0.36 | 0.36 | 0.44 | 0.28 | 0.03 | ||||||
| ′894041H01 (A) | 1029 | PSAF | PS I subunit III | .++.− | −−−−−− | 1.07 | 1.21 | 1.31 | 0.91 | 0.42 | 0.69 | 0.57 | 0.49 | 0.44 | 0.37 | 0.08 | ||||||
| ′894065A07 (A) | ′[4807] | PSAG | PS I subunit G | .−−−− | −−−−−− | 1.05 | 0.88 | 0.96 | 0.57 | 0.24 | 0.63 | 0.24 | 0.23 | 0.17 | 0.1 | 0.03 | ||||||
| ′894100A05 (A) | ′[4807] | PSAG | PS I subunit G | ...−− | −−−−−− | 1.03 | 1.15 | 1.01 | 0.63 | 0.27 | 0.63 | 0.27 | 0.24 | 0.17 | 0.11 | 0.05 | ||||||
| ′894014A05 (A) | 6118 | PSAH. | PS I subunit H | +++.− | .−−−−− | 1.17 | 1.26 | 1.21 | 0.86 | 0.36 | 0.74 | 0.49 | 0.63 | 0.6 | 0.4 | 0.16 | ||||||
| ′894086C09 (A) | 4991 | PSAK | PS I subunit K | ...−− | .−−.−− | 0.99 | 0.7 | 0.98 | 0.57 | 0.41 | 1.03 | 0.51 | 0.54 | 0.51 | 0.42 | 0.39 | ||||||
| ′894004A09 (A) | ′[7107] | PSAL | PS I subunit L | ...−− | −−−−−− | 1.09 | 1.1 | 1.13 | 0.8 | 0.4 | 0.55 | 0.39 | 0.39 | 0.33 | 0.32 | 0.16 | ||||||
| ′894019E07 (A) | 683 | PSAO | PS I subunit O | ...−−. | .−−−−−− | 1.19 | 1.13 | 1.19 | 0.65 | 0.27 | 0.81 | 0.31 | 0.3 | 0.24 | 0.16 | 0.03 | ||||||
| ′894068A11 (B) | 3356 | PSBO | OEE1, oxygen evolution enhancer 1 | .++.− | −−−−−− | 1.08 | 1.22 | 1.18 | 0.9 | 0.58 | 0.69 | 0.36 | 0.35 | 0.42 | 0.32 | 0.09 | ||||||
| ′894006E05 (B) | 7935 | PSBP1 | OEE2, oxygen evolution enhancer 2 | ...−− | −−−−−− | 1.02 | 1.00 | 0.94 | 0.64 | 0.66 | 0.78 | 0.27 | 0.26 | 0.32 | 0.23 | 0.07 | ||||||
| ′963041E04 (B) | [5978] | PSBQ | OEE3, oxygen evolution enhancer 3 | ..+−. | −.−−−− | 0.93 | 1.09 | 1.22 | 0.86 | 0.93 | 0.75 | 0.15 | 0.4 | 0.48 | 0.31 | 0.07 | ||||||
| ′894035D06 | 2796 | PSBP3 | Similar to OEE2 | −.−.− | .+.... | 0.73 | 0.84 | 0.81 | 0.99 | 0.82 | 1.04 | 1.14 | 1.00 | 1.04 | 1.08 | 1.02 | ||||||
| ′894084F07 | ′[3599] | PSBR | 10-kDa PS II polypeptide | ...+. | ...... | 1.24 | 1.19 | 1.31 | 1.67 | 1.37 | 1.13 | 1.31 | 1.26 | 1.21 | 1.25 | 1.25 | ||||||
| ′894069B11 | 6084 | PSBW | PS II reaction center W protein | ..+.− | .−−−−− | 0.97 | 1.21 | 1.27 | 1.01 | 0.68 | 1.06 | 0.53 | 0.52 | 0.61 | 0.46 | 0.14 | ||||||
| ′894072A03 | ′[2051] | PSB28 | Ycf79, also called PsbW-like | −−−−− | ..−−−− | 0.78 | 0.76 | 0.56 | 0.64 | 0.77 | 1.02 | 0.90 | 0.84 | 0.78 | 0.81 | 0.79 | ||||||
| ′894100F04 (D) | ′[5600] | PETC | Cytochrome b6fRieske subunit | +..−− | ...−−− | 1.22 | 1.01 | 0.85 | 0.49 | 0.47 | 0.83 | 0.65 | 0.54 | 0.47 | 0.43 | 0.33 | ||||||
| ′963053C08 (D) | 9358 | PETM | Cytochrome b6f M subunit | ...−− | ...−−− | 1.27 | 1.45 | 1.25 | 0.6 | 0.42 | 1.49 | 0.57 | 0.68 | 0.51 | 0.43 | 0.15 | ||||||
| ′894089E08 (D) | 5858 | PETN | Cytochrome b6f N subunit | ...−− | ..−−−− | 1.03 | 1.10 | 0.84 | 0.48 | 0.37 | 1.11 | 0.83 | 0.63 | 0.57 | 0.46 | 0.36 | ||||||
| ′963092G08 (D) | 5888 | PETO | Cytochrome b6f subunit V | −−−−− | −−−−−. | 0.86 | 0.78 | 0.55 | 0.34 | 0.5 | 0.74 | 0.38 | 0.23 | 0.23 | 0.1 | NA | ||||||
| ′894002C07 (D) | 5888 | PETO | Cytochrome b6f subunit V | .−−−−− | −−−−− | 0.94 | 0.75 | 0.64 | 0.38 | 0.46 | 0.61 | 0.36 | 0.23 | 0.24 | 0.12 | 0.02 | ||||||
| ′894069E01 (D) | 5927 | PETE | Plastocyanin (PCY1) | −.−−− | .−−−−− | 0.8 | 0.81 | 0.64 | 0.58 | 0.18 | 0.76 | 0.38 | 0.27 | 0.16 | 0.1 | 0.02 | ||||||
| ′894017C09 (D) | 1713 | PETF1 | Ferredoxin | −−−−− | +−−−−− | 0.77 | 0.62 | 0.39 | 0.23 | 0.2 | 1.34 | 0.28 | 0.33 | 0.26 | 0.18 | 0.04 | ||||||
| ′963046B11 (D) | 4881 | PETF5 | Ferredoxin | −−−−−. | −−−−−− | 0.04 | 0.05 | 0.04 | 0.04 | 0.05 | 0.13 | 0.09 | 0.23 | 0.18 | 0.08 | 0.06 | ||||||
| ′963025F07 (D) | 6292 | FNR | Ferredoxin-NADP reductase | ..−−− | −−−−−− | 1.08 | 0.87 | 0.63 | 0.26 | 0.21 | 0.6 | 0.32 | 0.17 | 0.2 | 0.12 | 0.15 | ||||||
| ′963048F01 | 7947 | FTRV | Ferredoxin-thioredoxin reductase variable chain | −−−−− | .−−−−− | 0.83 | 0.67 | 0.67 | 0.62 | 0.54 | 1.04 | 0.21 | 0.54 | 0.6 | 0.52 | 0.25 | ||||||
| ′894006G05 | 8211 | ATPC | ATP synthase gamma chain, short transcript | ++.−− | .−−−−− | 1.18 | 1.27 | 1.05 | 0.67 | 0.59 | 0.87 | 0.85 | 0.78 | 0.71 | 0.74 | 0.64 | ||||||
| ′894041H06 | 6174 | ATPC | ATP synthase gamma chain, longer transcript | +..−− | .−...− | 1.23 | 1.05 | 0.97 | 0.36 | 0.41 | 0.71 | 0.54 | NA | NA | NA | 0.38 | ||||||
| ′894073D05 | 8211 | ATPC | ATP synthase gamma chain, longer transcript | +.−−− | −−.−−− | 1.21 | 1.02 | 0.91 | 0.35 | 0.49 | 0.88 | 0.68 | 0.78 | 0.64 | 0.60 | 0.50 | ||||||
| ′963039D06 | 6174 | ATPC | ATP synthase gamma chain, longest transcript | +..−− | ....−. | 1.52 | 1.11 | 1.13 | 0.69 | 0.60 | 0.89 | 1.69 | 0.53 | 0.44 | 0.44 | NA | ||||||
| ′894021B01 | ′[6167] | ATPD | ATP synthase delta chain | +++−− | +.+.−. | 1.39 | 1.55 | 1.40 | 0.49 | 0.66 | 1.39 | 0.96 | 1.26 | 1.01 | 0.81 | 0.90 | ||||||
| ′963089H03 | 302 | ATPG | ATP synthase CFO subunit II | +++−− | +−..−− | 1.74 | 1.63 | 1.35 | 0.59 | 0.45 | 1.49 | 0.90 | 1.11 | 0.84 | 0.59 | 0.42 | ||||||
| Carbon metabolism | ||||||||||||||||||||||
| ′894022H12 | 2613 | PRK | Phosphoribulokinase | ..−−− | .−−−−− | 1.11 | 1.00 | 0.76 | 0.37 | 0.37 | 1.04 | 0.43 | 0.35 | 0.44 | 0.34 | 0.05 | ||||||
| ′963079E12 | ′[8511] | RBCS1/2 | Ribulose bisphosphate carboxylase small chain | .++.− | +−−−−− | NA | 1.25 | 1.28 | 1.03 | 0.63 | 1.99 | 0.76 | 0.76 | 0.75 | 0.45 | 0.19 | ||||||
| ′894098E09 | 1399 | TPI | Chloroplast triosephosphate isomerase | −−−−− | −−−.−− | 0.57 | 0.55 | 0.51 | 0.69 | 0.61 | 0.57 | 0.6 | 0.47 | 0.27 | 0.47 | 0.41 | ||||||
| ′963014G05 | 6790 | SERA, putative d-3-phosphoglycerate dehydrogenase | +++++ | −+.++. | 2.06 | 2.77 | 4.03 | 5.15 | 3.3 | 0.65 | 1.71 | 1.09 | 1.47 | 1.84 | 0.61 | |||||||
| ′963028B11 | 6790 | SERA, putative d-3-phosphoglycerate dehydrogenase | +++++ | .+.++− | 2.06 | 2.66 | 4.46 | 4.38 | 3.1 | 0.35 | 1.93 | 0.96 | 1.5 | 1.88 | 0.47 | |||||||
| ′894006B02 | 4808 | ALD3, fructose-1,6-bisphosphate aldolase | ++++. | ++++.. | 2.05 | 2.69 | 3.6 | 3.08 | 1.22 | 1.35 | 1.8 | 2.14 | 2.07 | 1.25 | 0.9 | |||||||
| ′894040F03 | 9404 | PGK | Phosphoglycerate kinase | −−−−− | .−−−−− | 0.82 | 0.81 | 0.73 | 0.59 | 0.6 | 0.91 | 0.32 | 0.27 | 0.27 | 0.18 | 0.05 | ||||||
| ′894082E07 | 6137 | Putative transketolase | ..−−− | −−−−−− | 0.96 | 0.91 | 0.61 | 0.47 | 0.53 | 0.67 | 0.45 | 0.37 | 0.41 | 0.45 | 0.22 | |||||||
| ′894022G03 | 5947 | PCK | Phosphoenolpyruvate carboxykinase | ++.−− | ....−− | 1.31 | 1.12 | 1.07 | 0.5 | 0.35 | 1.03 | 0.81 | 0.91 | 0.74 | 0.47 | 0.16 | ||||||
| ′963017B07 | ′[6111] | GND | 6-Phosphogluconate dehydrogenase | +++++ | .+++++ | 2.25 | 3.07 | 4.47 | 6.76 | 6.55 | 1.41 | 4.95 | 3.93 | 4.85 | 5.63 | 5.33 | ||||||
| ′894037H05 | 4431 | TAL1 | Transaldolase | +++.− | −..−−. | 2.98 | 3.16 | 2.38 | 1.14 | 0.59 | 0.81 | 0.86 | 0.92 | 0.5 | 0.28 | NA | ||||||
| ′894029C08 | 6695 | Similar to PGLS (6-phosphogluconolactonase) | +++++ | −..−.− | 2.87 | 3.23 | 3.78 | 4.44 | 2.61 | 0.58 | 0.85 | 0.76 | 0.86 | 0.96 | 0.67 | |||||||
| ′894080B03 | 1974 | GAP1 | Cytosolic glyceraldehyde 3-P dehydrogenase | −−+++ | +−+++− | 0.42 | 0.69 | 1.79 | 2.44 | 1.84 | 1.29 | 0.51 | 1.69 | 2.31 | 2.13 | 0.37 | ||||||
| ′963029F05 | 6610 | SHMT1 | Serine hydroxymethyltransferase | +++++ | .+++++ | 1.39 | 1.4 | 1.33 | 1.32 | 1.5 | 0.8 | 1.71 | 2.29 | 1.98 | 1.88 | 2.49 | ||||||
| ′963036C03 | 6493 | GLX1, putative glyoxal oxidase | −−−−− | −−−−−. | 0.03 | 0.09 | 0.04 | 0.04 | 0.05 | 0.11 | 0.04 | 0.03 | 0.02 | 0.02 | NA | |||||||
| ′963070H06 | ′[9345] | SDC1, serine decarboxylase | +.−.. | −−−−−− | 1.24 | 0.95 | 0.78 | 0.81 | 0.97 | 0.44 | 0.4 | 0.38 | 0.34 | 0.37 | 0.31 | |||||||
| ′963045H04 | 5544 | ACS3, putative acetyl-coenzyme A (CoA) synthetase | +++−− | ++++.− | 3.15 | 2.58 | 1.47 | 0.78 | 0.3 | 1.68 | 1.55 | 1.43 | 1.14 | 0.68 | 0.08 | |||||||
| ′894038D07 | 7098 | PFL, pyruvate formate lyase | −−−−− | −−−−−− | 0.21 | 0.21 | 0.31 | 0.34 | 0.32 | 0.25 | 0.06 | 0.3 | 0.42 | 0.31 | 0.06 | |||||||
| ′963048D10 | 1813 | STA2 | Granule-bound starch synthase I | +++++ | ....+− | 3.25 | 4.04 | 6.16 | 4.96 | 5.07 | 0.85 | 1.41 | 1.5 | 1.4 | 1.58 | 0.19 | ||||||
| ′894008B02 | 7333 | GLPV | Glycogen/starch phosphorylase | .++++ | ..++++ | 0.99 | 1.25 | 2.34 | 3.11 | 4.17 | 0.84 | 1.07 | 1.82 | 2.07 | 2.08 | 2.08 | ||||||
| Respiration alternative electron transfer pathways, ATPases, transporters
|
||||||||||||||||||||||
| ′894024D07 | 8491 | COX2a | Cytochrome oxidase subunit IIa, N terminus | −.++− | −−..−− | 0.77 | 1.03 | 1.41 | 1.38 | 0.93 | 0.77 | 0.4 | 0.81 | 0.92 | 0.73 | 0.38 | ||||||
| ′894102D01 | ′[7853] | COX2a | Cytochrome oxidase subunit IIb, C terminus | −.+++ | +.+++− | 0.76 | 1.02 | 1.10 | 1.51 | 1.18 | 1.17 | 1.14 | 1.37 | 1.40 | 1.19 | 0.88 | ||||||
| ′894076G10 | 6296 | COX90 | Cytochrome oxidase, Chlamydomonas-specific subunit | −..+− | −−−−−− | 0.72 | 1.03 | 1.09 | 1.28 | 0.76 | 0.93 | 0.43 | 0.72 | 0.67 | 0.66 | 0.27 | ||||||
| ′963044H03 | 1715 | ATP6 | Subunit 6 mitochondrial ATP synthase | −..−− | −−−−−− | 0.86 | 0.94 | 0.99 | 0.82 | 0.53 | 0.51 | 0.48 | 0.64 | 0.68 | 0.68 | 0.37 | ||||||
| ′894008C03 | 1860 | AOX4 | Chloroplast plastoquinol-oxygen oxidoreductase | +++++ | ++++++ | 2.1 | 2.24 | 3.28 | 3.33 | 9.52 | 1.58 | 5.68 | 6.39 | 7.52 | 7.35 | 4.16 | ||||||
| ′894076B10 | 4448 | HYD1 | Chloroplast Fe-hydrogenase | −−−−− | −−−−−− | 0.27 | 0.25 | 0.19 | 0.23 | 0.22 | 0.56 | 0.16 | 0.17 | 0.24 | 0.14 | 0.22 | ||||||
| ′894044F09 | 8476 | AOX1 | Mitochondrial alternative oxidase | −−−.. | .−−−−− | 0.57 | 0.59 | 0.77 | 0.89 | 1.14 | 0.76 | 0.25 | 0.5 | 0.66 | 0.58 | 0.2 | ||||||
| ′894072B06 | ′[4402] | Mitochondrial fatty acid carrier/uncoupling protein | ++.−− | ++++.. | 2.83 | 3.48 | 1.41 | 0.67 | 0.29 | 5.35 | 10.56 | 5.29 | 1.89 | 0.67 | NA | |||||||
| ′894059B10 | ′[3328] | MPC1, mitochondrial phosphate transporter | +++++ | .++++. | 1.66 | 1.9 | 2.52 | 2.57 | 1.42 | 1.12 | 2.61 | 3.05 | 1.8 | 1.47 | 1.15 | |||||||
| ′894026D10 | 4026 | Plastid ATP/ADP transporter | −−..− | −−.−−− | 0.73 | 0.79 | 1.05 | 0.98 | 0.81 | 0.35 | 0.38 | 0.53 | 0.49 | 0.6 | 0.33 | |||||||
| ′894092F04 | 2863 | Plastid ATP/ADP transporter | ..... | −....− | 0.73 | 0.85 | 1.04 | 1.08 | 1.15 | 0.42 | 0.8 | 0.56 | 0.54 | 0.7 | 0.42 | |||||||
| ′894063B07 | 3927 | Plasma membrane proton ATPase | −−−−− | −.−−.− | 0.32 | 0.3 | 0.38 | 0.48 | 0.53 | 0.79 | 0.41 | 0.55 | 0.68 | 0.73 | 0.7 | |||||||
| ′894040E11 | 592 | Vacuolar ATP synthase, subunit A | ..... | .....+ | 1.2 | 1.77 | 1.08 | 1.12 | 1.19 | 0.76 | 1.35 | 1.14 | 1.25 | 1.5 | 1.78 | |||||||
| ′894018B07 | 4771 | ATPvE | Vacuolar ATPase chain E | .+.++ | .+++++ | 1.05 | 1.15 | 1.05 | 1.18 | 1.44 | 1.03 | 1.72 | 1.42 | 1.24 | 1.31 | 1.44 | ||||||
| ′894022G07 | ′[4453] | ATPvL1 | Vacuolar ATPase proteolipid subunit | ..+++ | .+++++ | 1.26 | 1.40 | 1.42 | 1.70 | 2.01 | 1.15 | 1.38 | 1.55 | 1.51 | 1.68 | 1.60 | ||||||
| ′963017F05 | ′[4552] | Proton-translocating, vacuolar pyrophosphatase | −−..− | −−−−−− | 0.84 | 0.73 | 0.99 | 0.91 | 0.45 | 0.61 | 0.52 | 0.43 | 0.56 | 0.65 | 0.34 | |||||||
| ′894054D08 | ′[7558] | Putative transporter/GTP binding | ++.++ | ...... | 1.28 | 1.37 | 1.15 | 1.46 | 1.32 | 1.18 | 1.31 | 1.28 | 1.12 | 1.23 | 1.24 | |||||||
| ′894071E08 | ′[4402] | Similar to mitochondrial substrate carrier | +++−− | +++... | 3.63 | 2.92 | 1.66 | 0.43 | 0.17 | 3.74 | 2.36 | 4.48 | 1.4 | 0.46 | NA | |||||||
| ′894099G11 | ′3494 | PTB4, phosphate transporter | ..... | −−−−−− | 1.07 | 1.04 | 1.04 | 1.07 | 0.96 | 0.74 | 0.55 | 0.47 | 0.43 | 0.62 | 0.35 | |||||||
| Oxidative stress, chaperones, proteolysis
|
||||||||||||||||||||||
| ′894081G12 | 3424 | FSD1 | Iron chloroplast superoxide dismutase | +++++ | −+++++ | 1.58 | 1.6 | 1.78 | 1.98 | 2.83 | 0.87 | 1.4 | 1.89 | 1.92 | 2.19 | 3.11 | ||||||
| ′963046E04 | 5826 | GPX1 | Glutathionine peroxidase | +++++ | .+++++ | 3.14 | 3.32 | 3.17 | 2.6 | 2.7 | 1.2 | 2.53 | 3.72 | 4.15 | 3.14 | 2.2 | ||||||
| ′894082E09 | 6986 | Thioredoxin peroxidase/hydroperoxide reductase | +.+++ | +++.+. | 1.95 | 1.49 | 2.43 | 2.07 | 1.96 | 1.18 | 1.92 | 2.14 | 1.78 | 1.95 | 1.16 | |||||||
| ′963032D03 | 5314 | Putative vanadium chloroperoxidase | +++++ | .+.... | 15.15 | 12.76 | 46.99 | 40.92 | 5.64 | 0.92 | 1.56 | 0.57 | 0.83 | 1.16 | 0.58 | |||||||
| ′963025C05 | 4324 | Putative peroxiredoxin Q | +++++ | ...−−+ | 2.33 | 3.71 | 4.75 | 2.24 | 4.35 | 0.96 | NA | 0.97 | 0.23 | 0.25 | 3.9 | |||||||
| ′894062A07 | 8645 | PDX1, pyridoxin biosynthesis protein | ++++. | .+++++ | 1.71 | 1.71 | 1.7 | 1.63 | 1.00 | 0.86 | 2.64 | 2.35 | 1.9 | 1.98 | 2.49 | |||||||
| ′894024F09 | 5909 | HSP90A | Heat shock protein 90, cytosolic | −−−−− | −−−−−− | 0.2 | 0.32 | 0.36 | 0.46 | 0.73 | 0.45 | 0.21 | 0.52 | 0.59 | 0.57 | 0.58 | ||||||
| ′963036F10 | 4194 | HSP70C | Mitochondrial HSP70 chaperone | ..−−+ | .....+ | 0.88 | 0.90 | 0.69 | 0.79 | 0.91 | 1.06 | 1.16 | 1.13 | 1.07 | 1.12 | 1.20 | ||||||
| ′894078G05 | 4517 | HSP70B | Chloroplast HSP70 chaperone | +++++ | −.++++ | 1.23 | 1.62 | 1.34 | 1.49 | 1.38 | 0.84 | 0.96 | 1.53 | 1.64 | 1.49 | 1.77 | ||||||
| ′963026B05 | 7505 | CGE1 | Cochaperone of chloroplast HSP70 | ....+ | .....+ | 1.35 | 0.79 | 0.79 | 0.91 | 1.81 | 1.02 | 0.87 | 1.67 | 1.13 | 1.27 | 2.97 | ||||||
| ′963041D04 | 1067 | VIPP1 | Membrane-associated 30-kDa chloroplast protein | +++++ | .+++++ | 1.96 | 2.06 | 2.13 | 1.87 | 2.80 | 1.10 | 4.46 | 5.10 | 3.94 | 8.75 | 3.37 | ||||||
| ′894038F08 | 8733 | HCF136 | PS II stability/assembly factor | +++++ | −++.+. | 4.46 | 3.89 | 4.11 | 4.76 | 3.68 | 0.76 | 5.58 | 1.7 | 3.3 | 2.78 | 1.4 | ||||||
| ′894044H01 | 2747 | ALB3.1 | Required for LHC integration | ++.−− | ...... | 1.81 | 1.44 | 0.86 | 0.82 | 0.81 | 0.84 | 1.33 | 0.74 | 0.81 | 0.82 | 0.62 | ||||||
| ′963035G03 | ′[4315] | CPN60A | Alpha subunit Rubisco-binding protein, GroEL | −−−−− | −−−−.. | 0.29 | 0.26 | 0.17 | 0.27 | 0.42 | 0.7 | 0.31 | 0.53 | 0.68 | 0.7 | 0.72 | ||||||
| ′894062E01 | 5669 | CPN60B | Beta subunit Rubisco-binding protein, GroEL | −−−−− | ...... | 0.35 | 0.23 | 0.22 | 0.3 | 0.44 | 0.64 | 0.21 | 0.73 | 0.76 | 0.9 | 1.15 | ||||||
| ′894039G09 | 1845 | GROES-like protein | ..−−− | ...... | 0.9 | 0.96 | 0.63 | 0.86 | 0.85 | 1.19 | 1.39 | 1.36 | 1.32 | 1.42 | 1.46 | |||||||
| ′894076H02 | ′[4562] | Rubisco activase | +++.+ | +−−+.− | 1.90 | 1.77 | 1.42 | 1.07 | 1.39 | 1.77 | 0.49 | 0.68 | 1.36 | 1.06 | 0.44 | |||||||
| ′963050F02 | 3862 | HSP22C | Chloroplast 22-kDa heat shock protein | .−... | .+...+ | 0.77 | 0.73 | 1.13 | 0.88 | 1.17 | 1.10 | 2.21 | 1.25 | 1.52 | 2.09 | 2.96 | ||||||
| ′963068F01 | 5588 | HSP22E | Chloroplast 22-kDa heat shock protein | ++++. | −+++++ | 1.74 | 2.68 | 2.39 | 2.14 | 1.1 | 0.37 | 1.33 | 3.55 | 4.06 | 5.39 | 18.74 | ||||||
| ′894096A07 | ′[7754] | HSP22F | Chloroplast 22-kDa heat shock protein | ++++. | −+++++ | 1.61 | 3.23 | 1.83 | 2.42 | 0.89 | 0.49 | 1.8 | 5.11 | 5.69 | 8.22 | 42.22 | ||||||
| ′894008D03 | 2231 | Cytosolic peptidyl-prolyl cis-trans isomerase | −−−−− | .−.−−− | 0.33 | 0.39 | 0.43 | 0.42 | 0.34 | 1.03 | 0.53 | 0.68 | 0.67 | 0.7 | 0.37 | |||||||
| ′894086F07 | 344 | Chloroplast peptidyl-prolyl cis-trans isomerase | −−−−− | ..−−−− | 0.31 | 0.43 | 0.34 | 0.41 | 0.32 | 1.08 | 0.72 | 0.63 | 0.72 | 0.8 | 0.46 | |||||||
| ′963042H09 | 344 | Chloroplast peptidyl-prolyl cis-trans isomerase | −−−−− | .−−−−− | 0.42 | 0.51 | 0.37 | 0.44 | 0.30 | 1.04 | 0.54 | 0.57 | 0.77 | 0.71 | 0.50 | |||||||
| ′894005A06 | ′[2563] | Chloroplast peptidyl-prolyl cis-trans isomerase | −−−−− | −−−−−− | 0.59 | 0.46 | 0.36 | 0.36 | 0.26 | 0.72 | 0.27 | 0.33 | 0.31 | 0.33 | 0.18 | |||||||
| ′894037G11 | 8546 | AAA-type ATPase, possibly mitochondrial | +++++ | .+++++ | 3.62 | 3.05 | 3.03 | 3.79 | 1.91 | 1.11 | 2.26 | 2.27 | 2.19 | 2.53 | 1.79 | |||||||
| ′894006G02 | 5487 | PAF1 (proteasome alpha-6 subunit) | ..... | .....+ | 0.91 | 1.03 | 0.87 | 0.96 | 0.77 | 0.75 | 2.03 | 1.38 | 1.48 | 2.54 | 3.00 | |||||||
| ′894014B04 | 5527 | UBC2, ubiquitin-conjugating enzyme E2 | ..... | ..++++ | 0.91 | 1.21 | 0.88 | 1.03 | 0.98 | 0.68 | 1.34 | 2.04 | 1.63 | 1.7 | 2.22 | |||||||
| ′894080E10 | 4988 | UBC2, ubiquitin-conjugating enzyme E2 | −−−+. | ..++++ | 0.66 | 0.88 | 0.90 | 1.21 | 0.92 | 1.07 | 1.07 | 1.07 | 1.96 | 1.85 | 2.48 | |||||||
| ′894008E11 | 7585 | SKP1 E3 ubiquitin ligase; similar to Skp1 | −−... | .+++++ | 0.78 | 0.89 | 0.83 | 0.92 | 0.9 | 1.1 | 2.23 | 2.37 | 2.1 | 1.96 | 2.83 | |||||||
| ′963033H10 | 5803 | DEGPd | Chloroplast serine endoprotease | ++++. | .+++++ | 1.77 | 1.99 | 1.76 | 1.64 | 1.05 | 0.89 | 2.29 | 3.14 | 2.07 | 1.68 | 1.72 | ||||||
| ′963014H04 | 8642 | Cysteine protease | +++++ | ...+++ | 2.21 | 2.26 | 2.92 | 3.03 | 3.12 | 0.97 | 1.33 | 1.4 | 1.72 | 1.88 | 2.07 | |||||||
| ′963078B03 | 3753 | Cathepsin Z precursor | −−−−. | ++++++ | 0.83 | 0.63 | 0.54 | 0.84 | 0.97 | 1.47 | 2.14 | 2.08 | 2.38 | 2.92 | 5.47 | |||||||
| ′894024A08 | 6171 | Aspartic proteinase, delta subunit | +++++ | .+++++ | 3.47 | 4.57 | 5.87 | 4.97 | 4.26 | 0.66 | 1.62 | 2.18 | 2.31 | 2.43 | 2.46 | |||||||
| ′963065A08 | 7713 | Aspartyl aminopeptidase | +++++ | −+.... | 7.07 | 10.3 | 13.34 | 15.81 | 9.27 | 0.71 | 1.82 | 1.18 | 1.28 | 0.88 | 0.67 | |||||||
| ′894082D09 | 4868 | LCI5 | +++++ | .−−... | 4.52 | 3.38 | 3.31 | 1.73 | 1.57 | 1.28 | 0.58 | 0.57 | 0.98 | 1.03 | 0.79 | |||||||
| Other metabolic and/or biosynthetic processes
|
||||||||||||||||||||||
| ′894007D09 | None | CYB5-1 | .+−−− | .+.... | 1.06 | 1.27 | 0.8 | 0.81 | 0.69 | 1.36 | 1.96 | 1.48 | 1.11 | 1.39 | 1.2 | |||||||
| ′894042G02 | ′[1451] | ASSD, aspartate-semialdehyde dehydrogenase | ++−−− | .+++++ | 1.28 | 1.18 | 0.68 | 0.52 | 0.31 | 1.12 | 1.77 | 2.32 | 1.57 | 1.38 | 1.75 | |||||||
| ′894082E03 | 6330 | CPX1 | −−−−− | −−−−−− | 0.39 | 0.27 | 0.26 | 0.22 | 0.43 | 0.54 | 0.36 | 0.31 | 0.39 | 0.36 | 0.3 | |||||||
| ′894102G04 | ′[3236] | Putative Mg chelatase subunit | +++++ | .+...+ | 2.18 | 1.83 | 1.36 | 1.36 | 1.56 | 1.06 | 1.73 | 1.32 | 1.42 | 1.53 | 1.72 | |||||||
| ′894024D10 | 150 | CHLH1 | +++++ | −+.... | 3.33 | 3.00 | 3.32 | 1.85 | 1.62 | 0.86 | 1.51 | 1.26 | 1.1 | 1.18 | 0.59 | |||||||
| ′894093F09 | 2392 | CHL27A | CTH1, Mg-ProtoIX monomethyl ester cyclase | +++++ | +++++. | 17.96 | 17.44 | 17.28 | 11.54 | 4.97 | 3.07 | 8.6 | 4.75 | 8.52 | 8.51 | 2.77 | ||||||
| ′894066E11 | 2088 | CHL27B | CRD1, Mg-ProtoIX monomethyl ester cyclase | −−−−− | −−−−−− | 0.09 | 0.12 | 0.1 | 0.08 | 0.08 | 0.2 | 0.11 | 0.18 | 0.13 | 0.12 | 0.02 | ||||||
| ′894038E11 | 2279 | CUTA1 | Copper-binding protein, chloroplast | −−−−− | .+++++ | 0.66 | 0.7 | 0.62 | 0.6 | 0.5 | 0.83 | 3.8 | 3.08 | 3.03 | 3.43 | 2.41 | ||||||
| ′894086H06 | 5963 | GLN1, glutamine synthetase | +.+++ | ...... | 1.57 | 1.38 | 2.02 | 1.54 | 1.8 | 0.68 | 1.2 | 1.49 | 1.75 | 1.92 | 1.77 | |||||||
| ′894006D02 | 6255 | GLN2, chloroplast glutamine synthetase | ..++. | ...... | 1.01 | 1.23 | 1.54 | 1.53 | 1.24 | 1.16 | 0.8 | 1.33 | 1.7 | 1.37 | 1.47 | |||||||
| ′894044E11 | 7939 | ASNS, asparagine synthase | ++.+− | −...++ | 1.39 | 1.71 | 1.12 | 1.6 | 0.6 | 0.79 | 1.73 | 1.54 | 1.37 | 1.49 | 2.21 | |||||||
| ′963047G05 | 9088 | NADB, l-aspartate oxidase | ++++. | −+++++ | 1.66 | 1.39 | 1.32 | 1.33 | 1.18 | 0.65 | 7.15 | 3.17 | 2.31 | 3.1 | 1.83 | |||||||
| ′894039C12 | 651 | HDH, histidinol dehydrogenase | ...++ | ...... | 1.28 | 1.1 | 0.98 | 1.46 | 1.53 | 1.25 | 1.15 | 1.16 | 1.14 | 1.28 | 1.3 | |||||||
| ′894014G02 | 976 | Putative lumenal 17.4-kDa protein | −−..− | −−−−−− | 0.79 | 0.81 | 0.86 | 0.88 | 0.8 | 0.88 | 0.4 | 0.32 | 0.52 | 0.58 | 0.33 | |||||||
| ′963046C03 | 5100 | Putative lumenal polypeptide | +++.− | −−−−−− | 1.33 | 1.36 | 1.45 | 1.09 | 0.83 | 0.82 | 0.53 | 0.53 | 0.59 | 0.38 | 0.1 | |||||||
| ′894083E11 | ′[8722] | ACS3, putative acetyl-CoA synthetase | +++++ | ++++++ | 4.19 | 7.46 | 16.34 | 26.09 | 29.73 | 3.13 | 7.25 | 15.04 | 11.00 | 13.88 | 8.88 | |||||||
| ′894086H03 | ′[7647] | SMT1, similar to sterol-C24-methyltransferase | −−−−− | −−−−−− | 0.58 | 0.34 | 0.26 | 0.27 | 0.48 | 0.55 | 0.55 | 0.5 | 0.37 | 0.36 | 0.18 | |||||||
| ′894006B03 | ′[9318] | DES6, omega-6 desaturase | ++++. | −−−−−− | 2.11 | 2.51 | 3.47 | 2.85 | 0.86 | 0.67 | 0.32 | 0.61 | 0.66 | 0.47 | 0.26 | |||||||
| ′894055F12 | ′[9376] | LOX1-like lipoxygenase | −−−−− | −−−−−− | 0.14 | 0.19 | 0.19 | 0.2 | 0.21 | 0.42 | 0.08 | 0.07 | 0.2 | 0.12 | 0.08 | |||||||
| ′894081C10 | ′[174] | NNT, NAD transhydrogenase | −−−−− | −−−−−− | 0.42 | 0.48 | 0.8 | 0.81 | 0.48 | 0.26 | 0.06 | 0.24 | 0.29 | 0.14 | 0.03 | |||||||
| ′963047D02 | 4325 | SAS1, S-adenosylmethionine synthetase | −−−−− | −.−−−− | 0.84 | 0.53 | 0.33 | 0.28 | 0.3 | 0.51 | 0.64 | 0.41 | 0.59 | 0.64 | 0.42 | |||||||
| ′894062A09 | 6078 | THIH, thiazole biosynthesis protein | −−−−− | −−−−−− | 0.08 | 0.08 | 0.13 | 0.2 | 0.18 | 0.25 | 0.05 | 0.16 | 0.24 | 0.13 | 0.06 | |||||||
| ′963041C09 | 2889 | THI4, putative thiamine biosynthesis protein | −−−−− | −−−−−− | 0.42 | 0.44 | 0.39 | 0.29 | 0.15 | 0.31 | 0.05 | 0.12 | 0.18 | 0.12 | 0.01 | |||||||
| ′894057E11 | ′[6582] | HCP, hydroxylamine reductase | −−−−− | −−−−−− | 0.18 | 0.23 | 0.19 | 0.27 | 0.37 | 0.64 | 0.29 | 0.39 | 0.44 | 0.54 | 0.43 | |||||||
| ′894018F09 | 6945 | Putative HCP-like hydroxylamine reductase | −−−−− | −−−−−− | 0.15 | 0.17 | 0.29 | 0.39 | 0.42 | 0.38 | 0.05 | 0.08 | 0.17 | 0.18 | 0.14 | |||||||
| ′963036C07 | 959 | DS2, 3-deoxy-d-arabinoheptulosonate 7-phosphate synthase | +++++ | .+..++ | 2.26 | 3.01 | 5.33 | 3.4 | 1.57 | 1.21 | 1.77 | 1.98 | 1.86 | 1.59 | 1.86 | |||||||
| ′963035C07 | 955 | Putative carbamoyl phosphate synthetase | .−... | −..... | 0.92 | 0.79 | 1.02 | 1.01 | 0.93 | 0.76 | 0.89 | 1.43 | 1.7 | 1.59 | 0.96 | |||||||
| ′894097B09 | 2841 | IGPD (imidazoleglycerol-phosphate dehydratase) | .+... | −.+..+ | 1.04 | 1.17 | 0.91 | 1.15 | 1.03 | 0.7 | 1.76 | 1.31 | 1.54 | 1.96 | 1.84 | |||||||
| ′894103C12 | ′[2773] | SAH1, S-adenosyl homocysteine hydrolase | −−−−− | −−−−−− | 0.46 | 0.3 | 0.28 | 0.12 | 0.06 | 0.25 | 0.69 | 0.39 | 0.64 | 0.69 | 0.27 | |||||||
| ′894056F03 | ′[6078] | THIH-like protein, thiazole biolsynthesis | −−−−− | ...... | 0.17 | 0.25 | 0.16 | 0.42 | 0.38 | 1.03 | 1.03 | 0.94 | 0.79 | 0.83 | 1.07 | |||||||
| ′894096H02 | 1312 | HISC2, putative histidinol-phosphate transaminase | −.−.−. | −+++++ | 0.82 | 0.96 | 0.89 | 1.22 | 0.88 | 0.73 | 2.26 | 2.25 | 2.28 | 2.53 | 2.88 | |||||||
| ′963030A07 | 2652 | FER1 | FER1, ferritin | −−−−− | −−−−−− | 0.77 | 0.61 | 0.68 | 0.58 | 0.29 | 0.56 | 0.54 | 0.42 | 0.4 | 0.43 | 0.17 | ||||||
| ′963042H06 | 6157 | ACH, aconitate hydratase | ++.−− | ...−−− | 1.65 | 1.2 | 0.9 | 0.74 | 0.63 | 0.94 | 0.94 | 0.93 | 0.63 | 0.63 | 0.27 | |||||||
| ′963044D08 | 2010 | APOC, apospory-associated protein | ...−− | .−−−−− | 1.01 | 1.07 | 1.13 | 0.73 | 0.54 | 0.8 | 0.57 | 0.56 | 0.34 | 0.33 | 0.25 | |||||||
| ′894037G08 | 8636 | ALS, acetolactate synthase | +++++ | ...... | 1.39 | 1.29 | 1.47 | 1.55 | 1.33 | 1.14 | 1.23 | 1.23 | 1.25 | 1.35 | 1.38 | |||||||
| ′894064A10 | 468 | ILV3, dihydroxy-acid dehydratase | −−−.− | ..++++ | 0.49 | 0.55 | 0.43 | 0.8 | 0.57 | 0.83 | 1.41 | 1.98 | 2.00 | 2.07 | 3.88 | |||||||
| ′894080E01 | 773 | Putative acetyl-CoA carboxylase subunit | .−−−+ | −.−−−− | 0.96 | 0.78 | 0.75 | 0.69 | 1.33 | 0.83 | 0.49 | 0.33 | 0.5 | 0.54 | 0.38 | |||||||
| ′894062A07 | 8645 | PDX1, pyridoxine biosynthesis protein | ++++. | .+++++ | 1.71 | 1.71 | 1.7 | 1.63 | 1.00 | 0.86 | 2.64 | 2.35 | 1.9 | 1.98 | 2.49 | |||||||
| ′894102B02 | ′[2302] | Putative Igr3p cargo protein | −−−−− | +....− | 0.28 | 0.29 | 0.32 | 0.33 | 0.32 | 3.95 | NA | NA | NA | NA | 0.72 | |||||||
| ′894019G08 | 4767 | Adenylosuccinate synthetase (AMP biosynthesis) | −−−−− | −−.−.. | 0.3 | 0.33 | 0.32 | 0.45 | 0.4 | 0.76 | 0.62 | 0.84 | 0.87 | 0.92 | 1.01 | |||||||
| ′894068F05 | 415 | dTDP-glucose 4-6-dehydratase | −−−−− | −−.−−− | 0.5 | 0.46 | 0.6 | 0.6 | 0.43 | 0.9 | 0.39 | 0.5 | 0.62 | 0.66 | 0.28 | |||||||
| Signal transduction, transcription, translation
|
||||||||||||||||||||||
| ′894096C06 | 467 | Sensory opsin A | −−−−− | −−−−−− | 0.63 | 0.56 | 0.74 | 0.79 | 0.67 | 0.59 | 0.38 | 0.59 | 0.69 | 0.63 | 0.07 | |||||||
| ′963038D05 | 7033 | RABF1, RabH/Rab5 type of small GTPase | +++++ | .+++++ | 1.49 | 1.73 | 1.87 | 2.2 | 2.07 | 0.99 | 2.2 | 2.23 | 1.9 | 1.74 | 1.64 | |||||||
| ′963048H07 | ′[6103] | GBLP, guanine nucleotide binding protein | ....+ | ...++. | 0.93 | 1.07 | 1.09 | 1.01 | 1.44 | 1.42 | 1.3 | 1.53 | 1.63 | 1.47 | 1.46 | |||||||
| ′894006F01 | ′[9232] | SKS1-like protein kinase | −−−−− | .−−−−− | 0.54 | 0.53 | 0.58 | 0.43 | 0.37 | 0.77 | 0.2 | 0.4 | 0.42 | 0.36 | 0.18 | |||||||
| ′963045F12 | ′[2130] | PP1, serine/threonine protein phosphatase | −−−−− | −−−−−− | 0.45 | 0.42 | 0.44 | 0.54 | 0.55 | 0.53 | 0.31 | 0.4 | 0.45 | 0.45 | 0.31 | |||||||
| ′894006C11 | 7297 | XPO1, exportin- | −.−.. | −−−−.− | 0.72 | 0.9 | 0.83 | 0.81 | 0.84 | 0.45 | 0.49 | 0.12 | 0.36 | 0.25 | 0.36 | |||||||
| ′894006C08 | 7660 | GBP1p, G-strand-binding protein | −−−−− | −−−−−− | 0.24 | 0.17 | 0.13 | 0.2 | 0.15 | 0.59 | 0.22 | 0.25 | 0.26 | 0.29 | 0.3 | |||||||
| ′963046C02 | 8379 | Putative nucleosome assembly protein 1-like protein | +++++ | ....++ | 2.24 | 2.47 | 2.86 | 3.96 | 4.34 | 0.98 | 1.08 | 1.19 | 1.26 | 1.32 | 1.37 | |||||||
| ′894027A08 | None | ATP dependent helicase | −−−−− | .−.... | 0.28 | 0.19 | 0.27 | 0.41 | 0.37 | 0.74 | 0.63 | 0.73 | 0.73 | 0.75 | 0.84 | |||||||
| ′963047C11 | 5558 | Putative transcription factor IIB | −−−−− | +...+. | 0.22 | 0.28 | 0.3 | 0.37 | 0.3 | 2.69 | 1.22 | NA | 1.15 | 3.12 | NA | |||||||
| ′963087F05 | 7852 | Probably DNA-binding protein | −−−−− | −−.... | 0.4 | 0.31 | 0.26 | 0.32 | 0.35 | 0.75 | 0.5 | 0.83 | 1.05 | 1.01 | 1.14 | |||||||
| ′894081G03 | 8592 | Potential RNA-binding protein | ++.−− | .−.−−− | 1.56 | 1.29 | 1.08 | 0.8 | 0.68 | 0.99 | 0.73 | 0.76 | 0.55 | 0.53 | 0.26 | |||||||
| ′894010G03 | 8151 | Similar to Arabidopsis SC35-like splicing factor SCL35 | −−−−− | −−−−−− | 0.39 | 0.47 | 0.55 | 0.54 | 0.39 | 0.48 | 0.27 | 0.3 | 0.32 | 0.41 | 0.31 | |||||||
| ′963079B08 | 6042 | EFIA1, putative eukaryotic translation initiation factor | ..−−− | ...+++ | 0.94 | 0.88 | 0.55 | 0.61 | 0.63 | 0.92 | 1.25 | 1.43 | 1.65 | 1.53 | 1.72 | |||||||
| ′894081B08 | 7498 | EFIA2, putative elongation factor 1 a 2 | +.+++ | ++++++ | 1.24 | 1.23 | 1.6 | 1.44 | 1.13 | 1.86 | 2.32 | 2.67 | 2.4 | 2.45 | 2.38 | |||||||
| ′963077H03 | 1273 | Protein translation factor, similar to SUI1 | ..... | −+.+++ | 1.18 | 1.25 | 1.12 | 0.97 | 0.9 | 1.19 | 1.72 | 1.44 | 1.93 | 2.29 | 5.69 | |||||||
| ′894080F07 | 5613 | NOP56-like, nucleolar rRNA processing protein | −−−−− | ...... | 0.39 | 0.49 | 0.28 | 0.48 | 0.4 | 1.23 | 0.74 | 1.36 | 1.06 | 1.27 | 1.24 | |||||||
| ′963030C08 | 8384 | Fibrillarin-related nucleolar RNA-binding protein | −−−−− | .−...+ | 0.22 | 0.25 | 0.21 | 0.52 | 0.34 | 0.89 | 0.27 | 0.75 | 0.84 | 1.07 | 1.33 | |||||||
| ′894069A01 | 6666 | HMGB1, high-mobility-group DNA-binding protein | −−−−− | −−−−−− | 0.48 | 0.57 | 0.67 | 0.64 | 0.25 | 0.45 | 0.44 | 0.56 | 0.52 | 0.47 | 0.37 | |||||||
| ′963047E09 | 115 | NHP2, high-mobility-group nucleolar protein | −−−−− | .−...+ | 0.38 | 0.39 | 0.36 | 0.57 | 0.46 | 1.14 | 0.41 | 1.23 | 1.09 | 1.25 | 1.61 | |||||||
| Cytoplasmic and chloroplast ribosomal proteinsg
|
||||||||||||||||||||||
| ′894081C01 | 6021 | RPS3a-like | .++++ | ++++++ | 1.02 | 1.21 | 1.42 | 1.63 | 1.7 | 2.69 | 1.79 | 2.66 | 2.49 | 2.63 | 4.00 | |||||||
| ′963045C05 | 448 | RPS5 | .+.++ | +−.... | 0.98 | 1.15 | 1.02 | 1.31 | 1.31 | 1.28 | 0.51 | 0.84 | 0.77 | 0.72 | 0.73 | |||||||
| ′894070B04 | 2212 | RPL22 | ....+ | ++++.+ | 0.97 | 1.02 | 0.91 | 1.14 | 1.37 | 1.96 | 1.54 | 2.17 | 1.74 | 1.42 | 1.94 | |||||||
| ′963038E05 | 2202 | RPS20 | ..... | +..... | 1.11 | 0.96 | 1.06 | 0.97 | 1.29 | 2.03 | 0.97 | 1.23 | 1.42 | 1.22 | 1.19 | |||||||
| ′894011F03 | ′[8867] | RPS16 | −.−++ | .++.++ | 0.72 | 1.06 | 0.4 | 1.98 | 2.77 | 1.26 | 3.91 | 4.88 | 1.26 | 2.81 | 2.85 | |||||||
| ′894006G10 | ′[9571] | RPS17 | ...++ | +....+ | 1.00 | 1.09 | 1.1 | 1.24 | 1.35 | 2.42 | 0.96 | 1.57 | 1.81 | 1.66 | 2.24 | |||||||
| ′894064E10 | 5820 | RPS19 | ....+ | +.++++ | 0.87 | 1.07 | 1.01 | 1.21 | 1.54 | 2.08 | 1.62 | 1.97 | 1.86 | 1.83 | 1.79 | |||||||
| ′894037B07 | 8122 | RPS25 | ..... | +..... | 1.17 | 0.95 | 1.19 | 1.05 | 1.13 | 2.00 | 0.7 | 1.25 | 1.23 | 1.09 | 1.00 | |||||||
| ′894014F12 | 4106 | RPS30 | .++++ | ++++++ | 1.01 | 1.27 | 1.22 | 1.65 | 1.7 | 4.07 | 1.68 | 2.4 | 2.48 | 2.04 | 2.22 | |||||||
| ′894017F08 | 5393 | RPS27 | ....+ | +.++++ | 0.92 | 1.33 | 1.28 | 1.22 | 1.25 | 1.89 | 1.31 | 1.76 | 2.1 | 1.97 | 2.36 | |||||||
| ′963036H09 | 6746 | RPL6 | ..+.. | +..++. | 1.24 | 1.09 | 1.36 | 1.04 | 1.07 | 1.46 | 1.68 | 1.8 | 2.23 | 2.1 | 1.93 | |||||||
| ′894027B04 | 2452 | RPL9 | ...+. | +..+.. | 1.00 | 1.02 | 1.02 | 1.24 | 1.07 | 2.72 | 1.16 | 1.49 | 1.69 | 1.49 | 1.6 | |||||||
| ′963038C05 | ′[3133] | RPL10 | ....+ | ..++++ | 0.92 | 0.94 | 0.99 | 0.93 | 1.15 | 1.65 | 1.28 | 1.81 | 1.75 | 1.58 | 2.19 | |||||||
| ′894065B09 | 9108 | RPL11 | ....+ | +..++. | 1.11 | 0.79 | 1.14 | 1.13 | 1.29 | 2.56 | 0.93 | 1.25 | 1.73 | 1.59 | 1.57 | |||||||
| ′894064D04 | 3624 | RPL12 | ...++ | +..+++ | 0.95 | 1.03 | 0.97 | 1.23 | 1.32 | 2.35 | 1.16 | 1.57 | 1.67 | 1.54 | 1.99 | |||||||
| ′894019C08 | 7238 | RPL15 | ..+.+ | +...+. | 1.2 | 1.2 | 1.45 | 1.29 | 1.46 | 3.48 | 1.26 | 1.76 | 1.96 | 1.85 | 1.5 | |||||||
| ′963063C08 | 3 | RPL17 | ..... | +..... | 1.07 | 0.86 | 0.99 | 0.96 | 1.17 | 2.05 | 0.7 | 0.93 | 1.29 | 1.11 | 1.22 | |||||||
| ′894039B07 | 2305 | RPL23 | ....+ | +..... | 1.07 | 0.94 | 1.11 | 1.16 | 1.25 | 2.09 | 0.66 | 1.07 | 1.63 | 1.22 | 1.1 | |||||||
| ′963048E03 | 2072 | RPL24 | ....+ | +..... | 1.11 | 0.86 | 0.98 | 1.13 | 1.33 | 2.27 | 0.84 | 1.39 | 1.63 | 1.3 | 1.31 | |||||||
| ′963017B08 | ′[8598] | RPL27 | ...++ | +..... | 1.06 | 0.96 | 1.00 | 1.12 | 1.11 | 1.85 | 0.9 | 1.27 | 1.28 | 1.2 | 1.21 | |||||||
| ′894006A06 | 111 | RPL30 | ....+ | +..... | 1.1 | 0.98 | 1.06 | 1.12 | 1.67 | 3.35 | 0.97 | 1.39 | 1.9 | 1.52 | 1.5 | |||||||
| ′894066H09 | 6057 | RPL35 | −−−−− | .−.... | 0.48 | 0.41 | 0.31 | 0.3 | 0.55 | 0.9 | 0.49 | 0.7 | 0.77 | 0.8 | 1.22 | |||||||
| ′894026G08 | 2805 | RPSCL1 | −−−−− | −−−−−− | 0.25 | 0.2 | 0.21 | 0.19 | 0.21 | 0.32 | 0.1 | 0.11 | 0.13 | 0.12 | 0.05 | |||||||
| ′894044G05 | 5258 | RPSCL3 | −−−−− | .−−−.+ | 0.38 | 0.41 | 0.19 | 0.29 | 0.52 | 0.95 | 0.76 | 0.74 | 0.83 | 0.95 | 1.27 | |||||||
| ′894037C03 | 9418 | RPSCL13 | −−−−− | ...... | 0.46 | 0.33 | 0.32 | 0.34 | 0.62 | 1.02 | 0.61 | 0.71 | 0.87 | 1.01 | 1.4 | |||||||
| ′894002G05 | 825 | RPSCL20 | −−−−− | .−−−.. | 0.37 | 0.27 | 0.19 | 0.28 | 0.49 | 1.14 | 0.38 | 0.51 | 0.77 | 0.78 | 0.88 | |||||||
| ′963039A05 | 8056 | RPLCL1 | −−−−− | .−.−.+ | 0.35 | 0.29 | 0.2 | 0.27 | 0.59 | 0.96 | 0.59 | 0.82 | 0.82 | 0.99 | 1.94 | |||||||
| ′894037B11 | 2083 | RPLCL6 | −−−−− | .−.... | 0.47 | 0.38 | 0.25 | 0.27 | 0.54 | 1.3 | 0.47 | 0.64 | 0.85 | 0.99 | 1.26 | |||||||
| ′894089F04 | 4407 | RPLCL9 | −−−−− | +...−+ | 0.48 | 0.48 | 0.26 | 0.3 | 0.54 | 1.25 | 1.01 | 0.88 | 1.00 | 0.88 | 1.2 | |||||||
| ′963032C10 | 3366 | RPLCL12 | −−−−− | −−−−.. | 0.34 | 0.23 | 0.14 | 0.18 | 0.4 | 0.93 | 0.49 | 0.54 | 0.69 | 0.86 | 1.15 | |||||||
| ′894055G04 | ′[2967] | RPLCL18 | −−−−− | +−−−.+ | 0.4 | 0.36 | 0.2 | 0.24 | 0.5 | 1.3 | 0.53 | 0.74 | 0.8 | 0.93 | 1.48 | |||||||
| ′894050B05 | ′[6564] | RPLCL28 | −−−−− | +−−..+ | 0.34 | 0.31 | 0.2 | 0.3 | 0.68 | 1.14 | 0.57 | 0.74 | 1.02 | 1.06 | 2.01 | |||||||
| ′894073D04 | ′9202 | RPLCL31 | −−−−− | +....+ | 0.46 | 0.45 | 0.24 | 0.29 | 0.54 | 1.14 | 0.74 | 0.82 | 0.94 | 1.02 | 1.2 | |||||||
| Putative structural, surface, and matrix proteins
|
||||||||||||||||||||||
| ′894017B12 | 7239 | VFL2, centrin (caltractin) | −−−−− | +−.−−− | 0.65 | 0.54 | 0.66 | 0.61 | 0.74 | 1.51 | 0.42 | 0.6 | 0.59 | 0.54 | 0.34 | |||||||
| ′894032G12 | 7345 | TUA2, a tubulin 2 | −−−−− | ..−−.− | 0.38 | 0.34 | 0.28 | 0.36 | 0.33 | 1.29 | 0.48 | 0.5 | 0.58 | 0.68 | 0.57 | |||||||
| ′894006H07 | 82 | CHLRE_650068, microtubule-associated protein | +++++ | ++++++ | 2.73 | 3.08 | 2.93 | 3.08 | 3.3 | 1.31 | 5.63 | 5.27 | 5.43 | 5.59 | 8.45 | |||||||
| ′963046E08 | 8405 | TUA1, alpha tubulin 1 | −−−−− | −−−−−. | 0.46 | 0.32 | 0.3 | 0.29 | 0.26 | 0.77 | 0.06 | 0.08 | 0.12 | 0.07 | NA | |||||||
| ′894056C11 | ′[7442] | TUB1, beta tubulin 1 | −−−−− | +−−−−− | 0.27 | 0.19 | 0.21 | 0.27 | 0.16 | 1.18 | 0.1 | 0.22 | 0.23 | 0.24 | 0.02 | |||||||
| ′894093H02 | ′[3445] | TUB2, beta tubulin G1 | −−−−− | +..−−. | 0.54 | 0.36 | 0.29 | 0.23 | 0.12 | 4.16 | 0.81 | 0.56 | 0.35 | 0.16 | NA | |||||||
| ′894056E01 | ′[1531] | FAP15, flagellar proteome | −−−−− | −−−−−. | 0.24 | 0.24 | 0.37 | 0.39 | 0.39 | 0.49 | 0.03 | 0.07 | 0.2 | 0.06 | NA | |||||||
| ′894083H05 | ′[285] | SYP71, syntaxin | ++.++ | .++++. | 1.37 | 1.27 | 1.3 | 1.65 | 1.82 | 1.19 | 1.9 | 2.57 | 2.42 | 2.44 | 1.32 | |||||||
| ′963026C08 | 568 | FLA14, dynein light chain | −−−−− | +−..−− | 0.36 | 0.33 | 0.35 | 0.36 | 0.31 | 1.85 | 0.67 | 0.74 | 0.86 | 0.59 | 0.32 | |||||||
| ′963077B03 | 2780 | FMG1b, flagellar membrane glycoprotein 1b | −−−−− | −−−−−− | 0.19 | 0.17 | 0.17 | 0.18 | 0.12 | 0.16 | 0.07 | 0.02 | 0.02 | 0.04 | 0.01 | |||||||
| ′963046G04 | 835 | Putative cell adhesion protein | +++++ | ...... | 3.34 | 2.1 | 3.96 | 4.01 | 5.51 | 1.13 | 1.25 | 1.27 | 1.18 | 1.28 | 1.36 | |||||||
| ′963024H07 | 8951 | KATA, kinesin-like motor protein | +.++. | ...... | 1.95 | 1.14 | 3.24 | 2.11 | 1.41 | 0.84 | 0.68 | 0.88 | 1.14 | 1.05 | 0.95 | |||||||
| ′963046H09 | 7877 | Putative sulfated surface glycoprotein | .++.− | −−−−−− | 1.19 | 1.32 | 1.12 | 0.89 | 0.83 | 0.34 | 0.34 | 0.32 | 0.36 | 0.44 | 0.09 | |||||||
| ′894078A07 | 1239 | Similar to pherphorin of Volvox | ....− | −−−−−− | 0.91 | 1.04 | 1.23 | 0.86 | 0.2 | 0.37 | 0.17 | 0.19 | 0.24 | 0.3 | 0.17 | |||||||
| ′963029B12 | 1971 | GAS31, hydroxyproline-rich glycoprotein | −−−−− | −−.−−. | 0.4 | 0.36 | 0.37 | 0.31 | 0.2 | 0.62 | 0.35 | NA | 0.26 | 0.25 | NA | |||||||
| ′894068G03 | ′[6102] | Putative prolyl 4-hydroxylase, a subunit | −−−−− | +−.−−− | 0.15 | 0.18 | 0.19 | 0.23 | 0.31 | 1.27 | 0.37 | 0.99 | 0.78 | 0.31 | 0.14 | |||||||
| ′894068C01 | 4654 | Putative prolyl 4-hydroxylase, a subunit | −−−−− | −−−−−− | 0.04 | 0.05 | 0.04 | 0.05 | 0.08 | 0.21 | 0.04 | 0.08 | 0.11 | 0.05 | 0.01 | |||||||
| ′894042G04 | 7452 | Putative prolyl 4-hydroxylase, a subunit | −−−−− | ...−.. | 0.07 | 0.19 | 0.07 | 0.1 | 0.1 | 0.15 | 1.02 | 0.56 | 0.84 | 0.79 | 0.88 | |||||||
| ′894101C02 | [428] | Putative prolyl 4-hydroxylase, a subunit | −−−−− | −−.−−− | 0.17 | 0.24 | 0.19 | 0.16 | 0.19 | 0.48 | 0.13 | 0.28 | 0.25 | 0.2 | 0.05 | |||||||
| Unknownh | ||||||||||||||||||||||
| ′963016B04 | 1549 | Unknown | +++++ | ..++++ | 8.95 | 10.94 | 18.57 | 22.72 | 12.48 | 1.04 | NA | 4.17 | 3.29 | 3.72 | 1.4 | |||||||
| ′963033D03 | None | Unknown | +++++ | −+++++ | 13.46 | 15.01 | 22.74 | 23.51 | 22.3 | 0.39 | 4.5 | 5.34 | 4.58 | 3.85 | 2.16 | |||||||
| ′963032A08 | 6781 | Unknown | +++++ | .+++++ | 35.69 | 30.83 | 59.23 | 50.06 | 34.64 | 0.68 | 22.69 | 16.05 | 17.99 | 21.9 | 19.76 | |||||||
| ′963047F05 | 2729 | Unknown | +++++ | .++++. | 2.07 | 4.42 | 16.00 | 18.91 | 15.01 | 1.05 | 1.74 | 5.6 | 4.76 | 3.56 | 1.21 | |||||||
| ′963096E10 | 1549 | Unknown | +++++ | .+++++ | 16.98 | 21.88 | 32.67 | 35.42 | 23.93 | 0.95 | 3.61 | 3.89 | 3.32 | 4.62 | 1.47 | |||||||
| ′894063D10 | 3295 | Unknown, starch-binding domain of glycoside hydrolase | +++++ | .+.+++ | 5.18 | 4.33 | 16.54 | 14.59 | 13.15 | 0.92 | 1.54 | 1.16 | 1.53 | 1.71 | 1.63 | |||||||
| ′963027G01 | 6569 | Unknown | +++++ | ..−−.+ | 1.56 | 4.35 | 16.00 | 23.01 | 18.73 | 1.01 | 1.4 | 0.35 | 0.31 | 1.36 | 5.06 | |||||||
| ′963063B08 | 5533 | Unknown | +++++ | −+++++ | 10.05 | 17.49 | 23.73 | 21.99 | 18.27 | 0.41 | 2.96 | 4.27 | 3.85 | 3.25 | 2.39 | |||||||
| ′963068A11 | None | Unknown | +++++ | .+++++ | 6.97 | 8.97 | 17.49 | 18.08 | 12.56 | 1.24 | 4.32 | 2.15 | 1.7 | 2.8 | 6.36 | |||||||
| ′963032A01 | ′9229 | Unknown proline-rich protein | +++++ | ++++++ | 5.4 | 6.85 | 18.01 | 23.03 | 24.96 | 1.26 | 4.61 | 7.67 | 6.57 | 10.67 | 8.17 | |||||||
The ≥3-fold change is for at least two points in either CC425 and/or the sac1 mutant. Some genes that did not satisfy these criteria but were discussed in the text are also included. This table also has a finer partitioning of the genes into categories than Table S1 in the supplemental material, but the genes can be tracked by the clone ID numbers.
The identifier of the array element, based on the cDNA clone used as a template for PCR amplification. Clone numbers followed by a letter in parentheses belong to a category used to generate Fig. 3A through E, as indicated.
The identification number in the 20021010 assembly of ESTs for a predicted unique gene (ACE) in which this clone is a member. The bracketed numbers are for clones not used in the ACE assembly but that match a specific ACE as determined by BLAST analysis.
Difference in transcript abundance (+ or −; a dot indicates no statistically significant change) relative to RNA from CC425 at time zero. The five symbols represent 2, 4, 8, 12, and 24 h after initiation of sulfur starvation of CC425; the data for the sac1 mutant include a comparison of the sac1 zero time point (initial symbol) with the wild-type zero time point.
NA, not enough high-quality data for statistical tests.
The read did not match any of the ACEs and thus was not included in the assembly of reads.
RP, ribosomal protein; L, large subunit; S, small subunit; CL, chloroplast; the number represents the molecular mass of the subunit.
A selection of 10 of the 187 “unknowns” that satisfied the ≥3-fold selection criterion. The abundance of the transcripts from these genes within the unknown category exhibited the greatest increase following S deprivation.
SQ, sulfoquinovose.
SQ, sulfoquinovosyl.
Hybridization, washing, and scanning.
Arrays were prehybridized in 3× SSC-0.1% SDS-10-mg/ml bovine serum albumin for 20 min at 50°C, followed by immersion of the slides into ddH2O and then isopropanol for 2 min each. Slides were dried by centrifugation for 5 min in a SpeedVac. The hybridization solution was prepared by adding 15 μl of 2× hybridization buffer [6× SSC, 0.2% SDS, 0.4-μg/μl poly(dA), 0.4-μg/μl tRNA] to 15 μl of the labeled probes, followed by passage of the probe through a QiaQuick PCR purification column. The probe solution was incubated at 95°C for 3 min, cooled to room temperature, applied to a prehybridized microarray, covered with a LifterSlip (Erie Scientific Company, Portsmouth, N.H.), placed in a hybridization chamber, and hybridized at 50°C for 16 h. Following hybridization, the slides were washed for 5 min each in successive solutions A (2× SSC, 0.1% SDS), B (1× SSC), and C (0.05× SSC) at room temperature and dried by centrifugation in a SpeedVac Plus model SC210A (Savant) for 5 min. The arrays were scanned at 532 and 635 nm in an Axon GenePix 4000A scanner (Foster City, Calif.) at 10-μm resolution. Photomultiplier tube voltages were adjusted to minimize background and saturation of the hybridization signals. Images of the fluorescence at 532 nm for Cy3 and 635 nm for Cy5 were recorded and analyzed for eight complete sets of all cDNAs (four sets on each of two slides). RNA samples used to synthesize the probes that were hybridized to the slides were from independent experiments.
Data selection and analysis.
Microarray images representing spot intensities from scanned slides were imported into the GenePix Pro 3.0 program (Axon Instruments) and quantified. Spot positions were defined according to a predefined microarray layout that was subsequently adjusted by eye to help optimize spot recognition. Spot signals that were distorted by dust or locally high backgrounds were not included in the analyses. The data were imported into the Stanford Microarray Database (13, 46) and normalized by using that database's standard computed algorithm (for details, see http://genome-www.stanford.edu/microarray/). The experiment identification numbers within the database are 31781 to 31790, 37207, 37266 to 37269, 37271 to 37276, and 37356.
Only those spots with an intensity/background ratio of >2 in either channel, and with a minimum normalized net intensity for the median (channel 1 net median intensity or normalized channel 2 net median intensities) of >350 in at least one of the two channels, were included in the analysis. Log2 of the 635 nm/532 nm normalized ratio for the median was retrieved. Genes for which the transcripts appeared to increase or decrease by ≥3-fold (averaged ratio per array over the duplicate samples) during S deprivation, for ≥4 of the 8 array sets analyzed, and for at least two of the time points following the imposition of S deprivation were chosen for further analysis. Gene annotations were retrieved from the Unigene Set stored in the Chlamydomonas EST database (http://www.biology.duke.edu/chlamy_genome/unigene.html) and further confirmed manually by BLAST analyses by using the Chlamydomonas Genome Sequence Database (http://genome.jgi-psf.org/chlre2/chlre2.home.html), the predicted gene models within the database, and NCBI protein databases. All alignments were visually inspected.
At each time point, for each gene, a one-way t test was conducted to assess the significance of the relative transcript abundance levels. Each t test considered the unaveraged (but normalized) log2 data for each gene. The following t-score threshold levels were set: for α = 0.05, df(1), t = 12.6; and for α = 0.10, df(7), t = 2.8. The 2.8 t value represents the least stringent significance measure, where each sample is considered to be independent of all others. The 12.6 t value represents the most stringent significance measure, where the test is considered to have only two independent samples. The + and − patterns displayed in Table 1 depict the least stringent threshold (t = 2.8), although both are reported in the larger Table S1 in the supplemental material.
A significance analysis of microarrays (SAM) (39, 55) was also used to correct for false discovery rates. All eight replicates (two biological replicates by four on-array replicates) at each time point were analyzed separately, and the median false discovery rate was set as close to 1 as possible in order to compare results between time points. The SAM results, which are reported in Tables S3 and S4 in the supplemental material, were largely congruent with the results reported in Table 1, although SAM selected fewer genes.
RESULTS
Modulation of transcript abundance in response to S deprivation.
Among the 2,761 cDNA fragments included on the microarray, 2,565 were amplified as 3′ fragments of cDNAs and had the expected sizes, while the characteristics of the remaining 196 PCR products were not acceptable (no band, multiple bands, or smearing along the lane) and were excluded from analysis. The number of genes represented on the array is probably about one-sixth of those present on the C. reinhardtii genome and may be biased toward highly expressed genes, as the microarray was designed from expressed sequence tags (ESTs) selected at an early stage of the EST sequencing project (48). Since the cDNA library from S-starved cells was among the first to be sequenced, there is also presumably some bias toward genes whose transcripts increase during S starvation. The RNA used for synthesizing labeled cDNA to probe the array was isolated from cells at five different times (2, 4, 8, 12, and 24 h) following the imposition of S deprivation. We analyzed both the parental strain, CC425 (Chlamydomonas Genetics Center), and the sac1 mutant (carrying ars5-5; equivalent to CC3794; this strain was generated by mutagenesis of CC425). The reference for all time points for the parental cells and the sac1 mutant (including the sac1 zero time) was RNA from the parental strain isolated at time zero (after pelleting the cells by centrifugation and just as SO42− was eliminated from the medium).
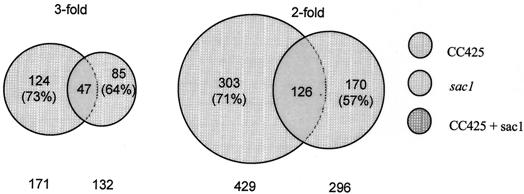
There were 171 cDNAs for CC425 and 132 for the sac1 mutant for which transcripts exhibited a ≥3-fold alteration (ratios were either ≥3 or ≤0.33) for at least 4 of the 8 array sets analyzed and two of the five time points sampled following the imposition of S deprivation (Fig. 1, left panel). Forty-seven of the selected clones were common to CC425 and the sac1 mutant. If the cutoff was set to select genes for which transcript abundance changed ≥2-fold (Fig. 1, right panel), more genes were selected, but the proportion of regulated genes for both strains remained similar. All of the CC425 and sac1 mutant common transcripts were regulated in a similar direction (e.g., those that increased in CC425 also increased in the sac1 mutant), although the kinetics and the extent of the changes could be somewhat different.
FIG. 1.
Interloping diagrams showing the number of genes from CC425 (left circles) and the sac1 mutant (right circles) that respond to S-deprivation conditions. Left panel, transcript levels altered ≥3-fold; right panel, transcript levels altered ≥2-fold.
These results demonstrate that S deprivation leads to extensive changes in transcript abundance (transcripts from >20% of the CC425 genes represented on the array exhibit a change of twofold or more), reflecting a change in the physiological state of the cell. Furthermore, while the sac1 mutant is defective for the control of a number of genes, there are still 30% of the sulfur-responsive genes in CC425 (126 out of 429) that appear to be properly regulated in this strain. We performed further analyses of those cDNAs for which the transcripts changed ≥3-fold; the levels of all of these transcripts at the different times following S deprivation are shown in Table 1. This table also contains the data for a number of other genes, providing information relevant to the discussion of S deprivation responses. The data presented are an average of eight replicates in two separate experiments. The direction of change is indicated in the table for each time point (by + or −) when judged significant based on a t test. The ratio values for all of the genes represented on the array is available in the supplemental material (Table S1).
Genes involved in sulfur assimilation.
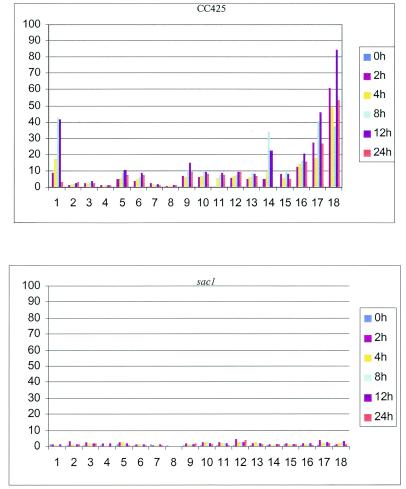
Most transcripts for genes encoding polypeptides involved in the acquisition or assimilation of SO42− increased following the imposition of S deprivation conditions. Relative transcript abundance for all of the genes in this category is presented in Table 1 (under the heading Sulfur metabolism); the data are also visually depicted in a bar graph presented as Fig. 2. Genes from this group that have been previously studied encode ARS1, ATP sulfurylase (ATS1), and ECP76; transcripts from these genes were previously shown to increase in response to S deprivation (7, 51, 62). Elevated levels of these transcripts were observed by 2 h following the imposition of S deprivation. Temporal changes in the levels of these transcripts during S deprivation, as determined from analyses of the microarrays, were consistent with the evaluations of transcript levels by RNA blot hybridizations (7, 51, 62). This category also contains genes for all steps of the sulfate assimilation pathway, including two distinct sulfite reductases (SIR1 and a distinct unannotated SIR gene; note that the transcript from the SIR3 gene, which encodes another type of sulfite reductase, did not increase), the two ATP sulfurylases, and adenylyl sulfate reductase. Surprisingly, the single adenosine-S-phosphosulfate kinase gene present in the genome was not induced, suggesting that 3′-phosphoadenosine-5′-phosphosulfate synthesis does not become limiting under sulfur stress conditions. Transcripts for UDP-sulfoquinivoso synthase (SQD1) and for a sulfolipid synthase (SQD2a), which probably utilizes sulfite for the synthesis of sulfolipids in the thylakoid membranes, increase during S starvation. In addition, the transcripts for serine acetyltransferase (SAT) and O-acetylserine(thiol)lyase (OASTL), which are involved in the biosynthesis of cysteine, are upregulated, as is the transcript for cysteine dioxygenase (963076G10), which converts cysteine to cysteine sulfinic acid (58) and may be involved in reclaiming S from amino acids. Furthermore, there is a rise in the levels of transcripts encoding a selenobinding protein (SBDP), which may be critical for sequestering selenate (by binding it and depositing it in the vacuole and/or facilitating its excretions), a potentially toxic SO42− analog. Selenate would be more likely to accumulate in S-starved cells since the transport machinery of such cells would have a high affinity for SO42− (and potentially selenate) and an elevated capacity for its uptake. It is also possible that the SBDP is involved in acclimation of cells to selenate deprivation (most selenate in the culture medium would originate as contamination from the sulfate stock solution), allowing for efficient acquisition when the levels of selenate are low in the environment. Recent work strongly suggests the presence of a number of selenoproteins in C. reinhardtii (36). The transcripts corresponding to the two array elements for SBDP (clone identification numbers 894020C12 and 963027A09) behave similarly and appear to represent the same gene. Finally, two genes coding for SAC1-like polypeptides are also induced (SAC1 itself is not on the array).
FIG. 2.
Bar graph showing changes in transcript levels for genes involved in sulfur metabolism in CC425 (top graph) and the sac1 mutant (bottom graph) during S deprivation. The responses, shown as the change (n-fold) relative to RNA from CC425 at time zero (which is set at 0), at the different times following S deprivation are given in different colors, as indicated on the graph, and the different genes represented encode the following proteins: 1, arylsulfatase (ARS1); 2, sulfite reductase (SIR1); 3, sulfite reductase (no gene or protein designation given); 4, sulfite reductase (SIR3); 5, ATP sulfurylase (ATS1); 6, ATP sulfurylase (ATS2); 7, 5′-adenylyl sulfate reductase (APR); 8, adenosine 5′-phosphosulfate kinase (AKN2); 9, serine O-acetyltransferase (SAT1); 10, O-acetyserine(thio)lyase (OASTL4); 11, cysteine dioxygenase (CDO1); 12, UDP-sulfoquinovose synthase (SQD1); 13, UDP-sulfoquinovose:diacylglycerol sulfoquinovosyltransferase (SQD2a); 14, extracellular polypeptide 76 (ECP76); 15, selenium-binding protein (SBDP); 16, selenium-binding protein (SBDP) (15 and 16 represent the same gene); 17, SAC1-like protein; and 18, SAC1-like protein.
These results demonstrate that transcripts from most genes involved in the acquisition, assimilation, and utilization of SO42− (almost all of those known to be on the array; note that none of the sulfate transporter genes are on this array) increase during S deprivation and that SO42− is likely scavenged from external and internal stores. Furthermore, as observed in the lower panel of Fig. 2, there was very little increase in the levels of many of these transcripts in the sac1 mutant, suggesting that these genes may be under the control of SAC1 (either directly or indirectly).
Photosynthesis genes.
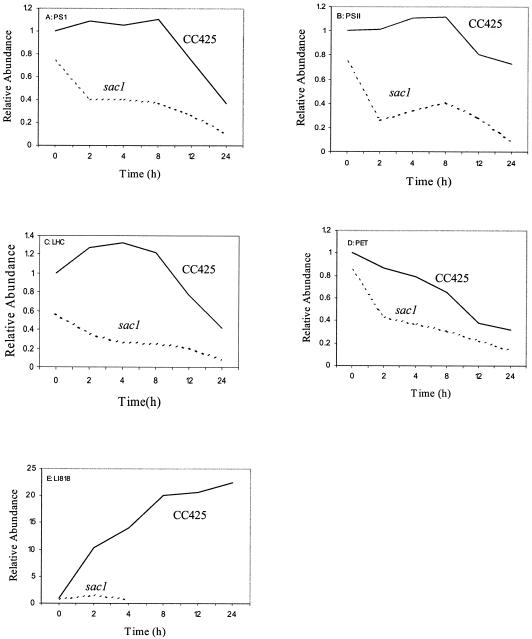
Many transcripts encoding proteins involved in photosynthesis decline by 12 to 24 h of S deprivation in both wild-type cells and the sac1 mutant. This decline is often more severe in the sac1 mutant strain, although the levels of these transcripts are somewhat lower in the mutant strain even under nutrient-replete conditions. The graphs presented in Fig. 3 represent an average of the change in transcript abundances of genes encoding subunits of the different complexes of the photosynthetic apparatus; the values for the transcript levels used in these analyses are presented in Table 1 (under Photosynthesis) and Table S1 in the supplemental material. On average, the levels of transcripts for genes encoding subunits of photosystem I (PS I) (Fig. 3A; Table 1), PS II (Fig. 3B; Table 1), cytochrome b6f (Fig. 3C; Table 1), light-harvesting complex (LHC) polypeptides (Fig. 3D; Table 1), and chloroplast ATP synthase (Table 1) either increase slightly or stay approximately the same as the level observed for nutrient-replete CC425 cells during the first 8 h of S deprivation. However, after 12 and 24 h, the transcript levels usually decline to 25 to 50% of the prestress level (Fig. 3). The only exceptions on the whole microarray were the PSBR and PSBP3 genes (the changes in transcript levels for these genes did not make the threefold cutoff but were added to Table 1), coding for PS II polypeptides that remained nearly constant in both strains over the time course. For PS II and cytochrome b6f, some of the genes (PSB28 and PETO) declined before the others, suggesting that they may play a role in the inactivation or disassembly of these complexes under stress conditions.
FIG. 3.
Influence of S deprivation on the abundance of transcripts encoding proteins that function in photosynthesis in both CC425 and the sac1 mutant strains. The average values represented on the graphs are for each time point for all of the transcripts listed in the different categories, given below. All of the values are relative to the time zero value of RNA from CC425, which is set at 1. Transcripts encode the following proteins. (A) PS I: PSAD (963047E03), PSAE (894083B07), PSAF (894041H01), PSAG (894065A07 and 894100A05), PSAH (894014A05), PSAK (894086C09), PSAL (894004A09), and PSAO (894019E07). (B) PS II (oxygen-evolving complex): PSBO (894068A11), PSBP (894006E05), and PSBQ (963041E04). (C) LHC: LHCA1 (963024B11), LHCA2 (963047H05), LHCA3 (894033H06), LHCA5 (963042A01 and 894041D11), LHCA6 (894044B07), LHCA7 (894076B06), LHCA8 (894087C09), LHCA9 (894078C01), LHCBM1 (963069C06), LHCBM3 (894080G01), LHCB (894052A01), and LHCB4 (894062E07). (D) Photosynthetic electron transport: PETC (894100F04), PETE (894069E01), PETN (894089E08), PETF1 (894017C09), PETF5 (963046B11), PETM (963053C08), PETO (963092G08 and 894002C07), and FNR (963025F07). (E) LI818r-1 (894097E05).
Interestingly, nearly all of the transcripts encoding components of the photosynthetic complexes exhibited an earlier and more pronounced decline in the sac1 mutant than in the parental strain, and this decline could be highly significant even after 2 h of S starvation. These results suggest that SAC1 may influence the stoichiometry of complexes involved in the photosynthetic electron transport chain, possibly in an indirect manner. The overall rapid downregulation of photosynthesis genes during S deprivation of the sac1 mutant may reflect the extreme stress that this strain experiences as it is unable to acclimate to S deprivation.
In contrast to the results described above, the level of transcripts for LhcSR2 (LI818r-1), an unusual member of the LHC gene family with unknown functions, increased dramatically in CC425 but not in the sac1 mutant during S deprivation (Fig. 3E; Table 1). Some of the LhcSR2 transcript data for the sac1 mutant is not shown in the figure because the normalized net intensity for the median is below 350 pixels for both the red and green channels, the criterion used to filter dim spots from further analysis. One aspect of the SAC1-dependent responses may be to restructure the light-harvesting apparatus to accommodate a reduction in electron flow caused by S deprivation; LhcSR2 may be involved in this process.
Reorganization of C metabolism.
A decline in photosynthetic electron transfer is an early event in the suite of responses exhibited by C. reinhardtii during S starvation, occurring even before the major complexes cease to be synthesized. This result is exemplified by early declines in the levels of mRNAs encoding plastocyanin, two ferredoxins, and a ferredoxin-thioredoxin reductase subunit. In addition, transcripts for polypeptides associated with the Calvin-Benson cycle also decline during S deprivation (PRK, RBCS1/2, TPI, and PGK), while the level of serine hydroxylmethyltransferase mRNA becomes elevated, suggesting a stimulation of photorespiration, which can participate in photoprotection (59).
There is an increase in the level of transcripts encoding polypeptides of the oxidative pentose phosphate cycle (GND, TAL1, and 6-phosphogluconolactonase) and cytosolic glyceraldehyde 3-P dehydrogenase after a transient decline. In addition, two types of complementary changes are observed that tend to reduce the redox pressure on the photosynthetic electron transfer chain, a predictable effect of cessation of growth in an environment in which the cells are absorbing considerable excitation energy. First, transcripts for polypeptides that function in starch synthesis accumulate; these include starch synthase and starch phosphorylase. Second, the AOX4 (or PTOX1) transcript, which encodes a chloroplast alternative oxidase, rapidly increases. This enzyme diverts electrons away from the photosynthetic electron transfer chain when carbon fixation is limited (5, 6). Together, these responses may serve as electron valves that help control redox poising in the thylakoid membrane and allow for the maintenance of an appropriate cellular redox environment. Not all alternative electron pathways are stimulated during S deprivation, as demonstrated by the findings that levels of transcripts encoding the mitochondrial AOX1 and the chloroplast Fe-hydrogenase declined.
Stress genes.
Several transcripts involved in limiting the potential damage caused by oxidative stress increase in both CC425 and the sac1 mutant following exposure of the cells to S deprivation. Within this group are transcripts encoding glutathione peroxidase (GPX1; 963046E06), the chloroplast iron superoxide dismutase (FSD1; 894081G12), a putative thioredoxin/periredoxin (894082E09), and serine hydroxymethyltransferase (SHMT1; 963029F05) (Table 1). GPX helps limit oxidative damage by catalyzing the reduction of H2O2, lipid peroxides, and organic hydroperoxides, while the chloroplast-localized FSD1 catalyzes the dismutation of superoxides which can form when the photosynthetic apparatus is absorbing excess excitation energy.
Metabolic stress usually triggers activation of the chaperone systems, which function in protein repair and remodeling. However, each chaperone system will react differently, depending on its functions, substrates, and subcellular localization. Compared to the relatively stable transcript levels associated with mitochondrial HSP70C, the modest increase in mRNA associated with the chloroplast HSP70B isoform may be significant with respect to the response of the organelle to stress conditions. The observed increase in transcript accumulation for VIPP1 (vesicle-inducing protein in plastids), a coiled-coiled protein involved in the biogenesis of thylakoid membranes (23), may be indicative of increased turnover of thylakoid lipids and proteins during S deprivation. The transcript for HCF136, a chaperone of unknown function present in the thylakoid lumen and necessary for PS II biogenesis (30), significantly increases, as do transcripts encoding two of the three chloroplast 22-kDa heat shock proteins. These HSP22 polypeptides are similar in sequence and are encoded by genes oriented head-to-head on the genome. In contrast, the transcript encoding the CPN60 subunits of the chloroplast chaperonin exhibits a marked decline, suggesting a reduction in chloroplast translation and the synthesis of organellar complexes. The levels of transcripts for several peptidyl-prolyl cis-trans isomerases of the cyclophilin family, involved in protein folding and other functions, are reduced. Note also the dramatic decrease in the level of mRNA for the cytosolic HSP90A. The transcripts encoding several proteases of the aspartic, cysteine, and serine (a chloroplast DEGP) types increase in both CC425 and the sac1 mutant during S deprivation (Table 1 and Table S1 in the supplemental material). These findings could reflect initiation of specific protease-dependent regulatory processes and/or the need to redistribute the amino acid resources of the cell. Furthermore, the finding that the transcript levels for a number of these genes appear to be properly regulated in the sac1 mutant, at least over a short term following the initiation of S deprivation, again suggests that some regulatory aspects of the acclimation response still operate in the sac1 mutant.
There are also several transcripts encoding stress-related polypeptides that attain high levels in CC425 but not the sac1 mutant. One of these transcripts encodes a putative vanadium chloroperoxidase (963032D03), an enzyme that reduces alkyl peroxides, potentially repairing damaged molecules in the cell (56). Other transcripts that exhibit a SAC1-dependent increase encode an aspartyl aminopeptidase (963065A08), LCI5 (894082D09), which is also elevated during CO2 limitation, and the nucleosome assembly protein I-like protein (963046C02). The levels of these transcripts may be directly controlled by SAC1, like those of most of the sulfur assimilation genes.
There are also stress-associated transcripts that increase in the sac1 mutant but not in CC425. An increase in some of these might reflect the fact that the mutant strain cannot acclimate to S deprivation, which could result in an extreme stress response. Interestingly, some of the transcripts in this category encode components of the ubiquitin and proteasome protein degradation machinery. There is an increase in levels of transcripts encoding two E2 ubiquitin-conjugating enzymes, a SKP1-like E3 ubiquitin ligase and the proteosome α-6 subunit (PAF1) (Table 1 and Table S1 in the supplemental material). In the case of the chloroplast 22-kDa heat shock proteins, the increase in their mRNA levels is much stronger in the sac1 mutant than in CC425; the increase is as high as 19- and 42-fold after 24 h of S deprivation in the mutant strain. Previous studies have suggested that the absorption of excitation energy by the photosynthetic apparatus is lethal to the sac1 mutant strain during S deprivation, and death of the cells is observed between 1 and 2 days after they are transferred to medium devoid of S. In contrast, wild-type cells survive under these conditions for prolonged periods of time (8). The sharp increases in HSP22E and HSP22F transcript abundance occur when the cells are beginning to lose viability.
Other significant changes.
Examination of Table 1 shows that there are significant declines in the levels of many transcripts encoding chloroplast ribosomal polypeptides and that this decline is often more pronounced in CC425 than in the sac1 mutant. Another intriguing case relates to the CHL27A and CHL27B polypeptides (also known as CTH1 and CRD1 [31, 33]), two isoforms of the oxidative Mg-ProtoIX monomethyl ester cyclase involved in chlorophyll synthesis (53). While transcripts for the latter severely decline in both strains, transcripts for the former markedly increase, even after 2 h of starvation. This increase is reduced to some extent in the sac1 mutant, suggesting that SAC1 participates in regulation of the abundance of this mRNA. The expression patterns of these genes appear to be orthogonal, with CHL27B being expressed in Cu-limited, hypoxic conditions and CHL27A being expressed in Cu-replete, oxygenic conditions (33). Although the CHL27B and CHL27A polypeptides are very similar and appear to have similar functions, they are not able to completely substitute for each other with respect to the formation of pigment-protein complexes in the photosynthetic apparatus (32). CHL27B and CHL27A may have distinct functions in the delivery of chlorophyll to different complexes of the photosynthetic apparatus and help tailor pigment binding and photosynthetic function to different environmental conditions.
Another intriguing link with Cu metabolism is provided by the observation that the transcript for CUTA1, a chloroplast copper-binding protein, is reduced in CC425 but significantly elevated in the sac1 mutant. Also, while levels of transcripts for many regulatory factors decrease during S deprivation, some, including those encoding an AAA-type ATPase of unknown function (894037G11), an elongation factor (894081B08), and a RAB-type GTPase (963038D05), appear to increase. It should be emphasized that the function of these potential gene products need to be more clearly established before they can be ascribed a role in the acclimation process.
Finally, it is interesting that numerous transcripts for genes encoding polypeptides of unknown function increase dramatically during S deprivation. The full list of those genes whose transcripts satisfied the ≥3-fold filter is given in Table S2 in the supplemental material. A short list of 10 of the unknowns that exhibit very high transcript levels are given in Table 1 and include 963016B04 (23-fold at 12 h), 963033D03 (24-fold at 12 h), 963032A08 (50-fold at 12 h), 963047F05 (18-fold at 12 h), 963096E10 (35-fold at 12 h), 894063D10 (15-fold at 12 h), 963027G01 (23-fold at 12 h), 963063B08 (22-fold at 12 h), 963068A11 (18-fold at 12 h), and 963032A01 (23-fold at 12 h). There are many other transcripts that exhibit statistically significant increases in abundance during S deprivation, and some of these appear to be under the control of SAC1 (e.g., 894063D10). This result emphasizes how little we know about acclimation processes and also presents us with the challenge of identifying these unknown genes and defining their function in the acclimation process.
DISCUSSION
Examination of gene expression using high-density DNA microarrays is a potent means of examining how organisms modulate gene activity in response to environmental conditions. We have used this technology to explore the ways in which C. reinhardtii acclimates to S deprivation. While there are approximately 2,565 genes represented on the arrays used for these experiments (slightly less because of some duplications on the array), the array elements were derived from cDNAs defined in recombinant libraries from cells grown both under nutrient-replete conditions (Core Library) and under S, phosphorus, and nitrogen starvation conditions (Stress I Library) (48). Therefore, the microarray was likely to contain a number of stress-responsive genes. This probability was substantiated by the findings that both the ARS and ECP76 genes were represented on the array (present only in the Stress 1 and not the Core Library); these genes only become active when C. reinhardtii cells are starved for S (8, 11, 51). The presence of numerous stress-related array elements was also substantiated by the results of the experiments in which 171 of the represented genes exhibited a ≥3-fold change in gene expression in CC425 after the cells were placed into medium devoid of S; the levels of transcripts encoded by many of these genes increased.
The patterns of gene expression in C. reinhardtii cells following S deprivation reveal both the general and specific responses that enable a cell to survive extended periods of time under specific deprivation conditions. Furthermore, the use of mutant strains with aberrant acclimation responses that die much more rapidly than wild-type cells following the imposition of the stress are beginning to reveal extreme responses of cells when their normal acclimation responses become compromised. This is the case for the sac1 mutant, which exhibits rapid death (relative to wild-type cells) and an inability to accumulate the ARS transcript and transcripts encoding many other polypeptides during S deprivation (8).
There are a number of different generalizations that can be made from the data presented in this study. First, the levels of transcripts for many genes change substantially during S deprivation. A number of the transcripts that are extremely sensitive to S deprivation encode proteins involved in the conservation, acquisition, and assimilation of SO42−, including ARS, ECP76, ATS, SAT, OASTL, polypeptides involved in sulfolipid biosynthesis, a selenium-binding protein, and SAC1-like polypeptides (which may be involved in regulation or in the transport of SO42− into the cell). Transcripts encoding a number of these polypeptides increase to well over 10-fold the level observed in nutrient-sufficient cells. However, a number of these transcripts are not detected in nutrient-replete cells (e.g., this is the case for the ARS and ECP76 transcripts), which causes imprecision in the quantification, making the ratio values a semiquantitative estimate. Based on our experience, the increase measured in the microarray analyses is often less than that determined by using real time reverse transcription-PCR (54). Furthermore, while in many instances changes in transcript levels reflect changes in corresponding protein and activity levels, as has been shown for ARS and ECP76 (8, 51), this correlation may not always hold. For example, even though transcripts encoding the sulfolipid biosynthesis enzymes SQD1 and SQD2a accumulate during sulfur deprivation (Table 1), the sulfolipid content of C. reinhardtii membranes appears to decline (K. Sugimoto and M. Tsuzuki [Tokyo University of Pharmacy and Life Science], personal communication).
Many of the genes, including ARS1, ATS2, APR, SQD2a, and ECP76, that are sensitive to S-limited growth conditions appear to be under SAC1 control, since there is no significant increase in their transcript levels in the sac1 mutant at any time point analyzed, even the early 2-h and 4-h time points. In other cases (SAT1, OASTL4, ATS1, SIR1, CDO1, SQD1, SBDP, and genes encoding SAC1-like proteins) an increase in transcript levels is observed in the sac1 mutant, but the increase is generally much more modest and less sustained than that in wild-type cells. These results suggest that SAC1 may play a direct role in the control of a number of genes. While other regulatory mechanisms may result in some changes in transcript levels during the early phase of starvation, sustained expression requires a functional SAC1 gene and may reflect the fact that SAC1 is required to maintain cell vitality. There are a number of other transcripts whose levels are potentially controlled by SAC1, some of which encode proteins that might function in the chloroplast, including a putative vanadium chloroperoxidase (although the similarity to other vanadium chloroperoxidases is not that high), aspartyl amino peptidase, and a specific light-harvesting polypeptide (LhcSR2; see below).
While the levels of transcripts from a number of genes dramatically change in CC425 and are no longer properly regulated in the sac1 mutant, it is clear that other genes are properly controlled in both the parental and the mutant strains. For example, transcripts for genes encoding the chloroplast quinol oxidase, the glutathione peroxidase, and the putative periredoxin Q increase to a similar extent in CC425 and the sac1 mutant strain during the 24 h of S deprivation. Hence, regulatory elements other than SAC1 are involved in modulating S deprivation-elicited changes in gene expression in C. reinhardtii. This result is not surprising, since there are mutations in at least four other loci that result in a sac1 mutant-like phenotype (J. Davies, S. Pollock, W. Pootakham, and A. R. Grossman, unpublished data). Isolating the altered genes and examining how lesions in these genes modulate global transcript levels during S deprivation will help define the nature of the gene products and their potential relationships with each other.
There are also many polypeptides for which the transcript levels dramatically decline in CC425 during S deprivation. Perhaps most striking is the finding that a number of transcripts encoding proteins involved in photosynthesis and chloroplast ribosomal polypeptides decline markedly in CC425 during S deprivation. In the case of the chloroplast ribosomal protein transcripts, the decrease is generally not nearly as great in the sac1 mutant. These results suggest that it might be critical to control the activity of the chloroplast ribosomes during S deprivation and that such control may be conferred to ribosomes by changing the complement of ribosomal proteins. Changes in ribosomal proteins have also been associated with nitrogen starvation (21). Similarly, many transcripts for proteins involved in photosynthesis decline in both CC425 and the sac1 mutant, but the decline is generally more precipitous in sac1 mutant cells. Such transcripts include those encoding polypeptides of PS I, PS II, the photosynthetic electron transport chain, and the LHC. While the reason for the earlier decline in the sac1 mutant is not clear, it may reflect the inability of the cells to scavenge S from the environment and tune cellular metabolism to decreased nutrient availability, which could elicit a highly stressed condition and nonspecific degradation of many of the less stable transcripts.
A highly stressed state of the S-depleted sac1 mutant is also suggested by elevated levels of specific transcripts encoding proteins associated with stress conditions. For example, the sac1 mutant exhibits a dramatic increase in transcripts encoding two putative chloroplast 22-kDa heat shock proteins (18); the increase in transcripts encoding these polypeptides in CC425 is much less pronounced. This type of chaperone is involved in preventing protein denaturation and aggregation in plastids. Furthermore, sac1 mutant cells exhibit an increase in transcripts for proteins involved in proteolysis relative to CC425 cells, although the increases are not that marked. Some of these transcripts encode proteins of the ubiquitin and proteosome systems, including two E2 ubiquitin-conjugating enzymes, an E3 ubiquitin ligase, and a proteasome α-6 subunit (Table 1). There is also a significant increase in a cathepsin Z protease.
While, as mentioned previously, transcripts for many PS and LHC polypeptides decrease to some extent in CC425 and more dramatically in the sac1 mutant during S deprivation, there is one exception to this trend. The transcript encoding the LhcSR2 polypeptide increases in CC425 nearly 20-fold relative to the level observed in unstarved cells. This increase is not observed in the sac1 mutant (indeed, the level of the transcript is so low in the sac1 mutant at some of the time points following S deprivation that we could not reliably quantify it). The transcript for this member of the LHC gene family has been previously shown to be transiently expressed following the transfer of cells from darkness to light, which would cause high light stress (2, 43). Hence, the organism is retailoring the photosynthetic apparatus to accommodate S deprivation conditions. The inability of the sac1 mutant to scavenge S and restructure its metabolic machinery (e.g., the photosynthetic apparatus) during S deprivation may cause severe functional and structural damage, especially within chloroplasts (which may be reflected in the increase in the HSP22-related polypeptide). This conjecture is supported by the facts that nutrient stress causes dramatic changes in the activity of the electron transport chain (60) and that the rapid death of the sac1 mutant during S deprivation can be ameliorated by placing the strain in the dark or blocking photosynthetic electron flow with specific inhibitors (8).
In sum, experiments presented in this report augment our understanding of how photosynthetic organisms respond to S deprivation and provide clues concerning processes that are important for acclimation. This work also highlights the fact that there are many genes of unknown function whose transcripts markedly increase in response to S deprivation. Furthermore, the study has revealed processes triggered when cells are unable to acclimate to the limitation conditions and as cell viability declines. It is now important to try to understand some of the other changes in transcript levels that have been observed (for which there are no obvious explanations) and whether all of these changes are observed at the level of transcript abundance that reflects changes in protein accumulation and activity.
Supplementary Material
Acknowledgments
We acknowledge NSF MCB 9975765 and MCB 0235878 for supporting the work presented in this report.
We thank people at the Stanford Center for Genome Sequence and Technology and the Arabidopsis Functional Genomics Consortium under the direction of Shauna Somerville for providing both strong technical support and helpful discussions; we are especially indebted to Bi-huei Hou, Katrina Ramonell, Lorne Rose, and Sue Thayer. Finally, we are grateful to Sufang Zhang for help with analyzing the data, Senior Scientific Programmer Jeremy Gollub for help in using the Stanford Microarray Database, and members of the team of investigators affiliated with the C. reinhardtii genome project, including David Stern, Pete Lefebvre, and Carolyn Silflow, for helping to make our efforts fruitful.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
This is Carnegie Institution Publication no. 1626.
REFERENCES
- 1.Bick, J. A., and T. Leustek. 1998. Plant sulfur metabolism: the reduction of sulfate to sulfite. Curr. Opin. Plant Biol. 1:240-244. [DOI] [PubMed] [Google Scholar]
- 2.Burssens, S., J. de Almeida Engler, T. Beeckman, C. Richard, O. Shaul, P. Ferreira, M. Van Montagu, and D. Inze. 2000. Developmental expression of the Arabidopsis thaliana CycA2;1 gene. Planta 211:623-631. [DOI] [PubMed] [Google Scholar]
- 3.Cherest, H., J.-C. Davidian, D. Thomas, V. Benes, W. Ansorge, and Y. Surdin-Kerjan. 1997. Molecular characterization of two high affinity sulfate transporters in Saccharomyces cerevisiae. Genetics 145:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarkson, D., F. Smith, and P. Van den Berg. 1983. Regulation of sulfate transport in a tropical legume, Macroptillium atropurpurum cv. Sirato. J. Exp. Bot. 34:1463-1483. [Google Scholar]
- 5.Cournac, L., E. M. Josse, T. Joet, D. Rumeau, K. Redding, M. Kuntz, and G. Peltier. 2000. Flexibility in photosynthetic electron transport: a newly identified chloroplast oxidase involved in chlororespiration. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1447-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cournac, L., K. Redding, J. Ravenel, D. Rumeau, E. M. Josse, M. Kuntz, and G. Peltier. 2000. Electron flow between photosystem II and oxygen in chloroplasts of photosystem I-deficient algae is mediated by a quinol oxidase involved in chlororespiration. J. Biol. Chem. 275:17256-17262. [DOI] [PubMed] [Google Scholar]
- 7.Davies, J., and A. R. Grossman. 1994. Sequences controlling transcription of the Chlamydomonas reinhardtii β2-tubulin gene after deflagellation and during the cell cycle. Cell Mol. Biol. 14:5165-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, J., F. Yildiz, and A. R. Grossman. 1996. Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 15:2150-2159. [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, J. D., and A. R. Grossman. 1998. Responses to deficiencies in macronutrients, p. 613-635. In J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), The molecular biology of Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 10.Davies, J. P., F. H. Yildiz, and A. R. Grossman. 1999. Sac3, an Snf1-like serine/threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell 11:1179-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Hostos, E. L., J. Schilling, and A. R. Grossman. 1989. Structure and expression of the gene encoding the periplasmic arylsulfatase of Chlamydomonas reinhardtii. Mol. Gen. Genet. 218:229-239. [DOI] [PubMed] [Google Scholar]
- 12.de Hostos, E. L., R. K. Togasaki, and A. R. Grossman. 1988. Purification and biosynthesis of a derepressible periplasmic arylsulfatase from Chlamydomonas reinhardtii. J. Cell Biol. 106:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gollub, J., C. A. Ball, G. Binkley, J. Demeter, D. B. Finkelstein, J. M. Hebert, T. Hernandez-Boussard, H. Jin, M. Kaloper, J. C. Matese, M. Schroeder, P. O. Brown, D. Botstein, and G. Sherlock. 2003. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 31:94-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, A. S., R. G. Alscher, and D. F. McCune. 1990. Ozone exposure, glutathione levels and photosynthesis in hybrid poplar, p. 195-197. In H. Rennenberg, C. Brunold, L. J. Dekoli, and I. Stulen (ed.), Sulfur nutrition and sulfur assimilation in higher plants. SPB Academic Publishers, The Hague, The Netherlands.
- 15.Gutierrez-Marcos, J., M. Roberts, E. Campbell, and J. Wray. 1996. Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc. Natl. Acad. Sci. USA 93:13377-13382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall, J. L. 2002. Cellular mechanisms for heavy metal detoxification and tolerance. J. Exp. Bot. 53:1-11. [PubMed] [Google Scholar]
- 17.Hawkesford, M. J., J.-C. Davidian, and C. Grignon. 1993. Sulphate/proton cotransport in plasma-membrane vesicles isolated from roots of Brassica napus L.: increased transport in membranes isolated from sulphur-starved plants. Planta 190:297-304. [Google Scholar]
- 18.Ish-Shalom, D., K. Kloppstech, and I. Ohad. 1990. Light regulation of the 22 kd heat shock gene transcription and its translation product accumulation in Chlamydomonas reinhardtii. EMBO J. 9:2657-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, H., M. Y. Hirai, H. Hayashi, M. Chino, S. Naito, and T. Fujiwara. 1999. Role of O-acetyl-l-serine in the coordinated regulation of the expression of a soybean seed storage-protein gene by sulfur and nitrogen nutrition. Planta 209:282-289. [DOI] [PubMed] [Google Scholar]
- 20.Koprivova, A., M. Suter, R. O. den Camp, C. Brunhold, and S. Kopriva. 2000. Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol. 122:737-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraig, E., J. E. Haber, and M. Rosbash. 1982. Sporulation and rna2 lower ribosomal protein mRNA levels by different mechanisms in Saccharomyces cerevisiae. Mol. Cell. Biol. 2:1199-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kredich, N. M. 1992. The molecular basis for positive regulation of cys promoters in Salmonella typhimurium and Escherichia coli. Mol. Microbiol. 6:2747-2753. [DOI] [PubMed] [Google Scholar]
- 23.Kroll, D., K. Meierhoff, N. Bechtold, M. Kinoshita, S. Westphal, U. C. Vothknecht, J. Soll, and P. Westhoff. 2001. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. USA 98:4238-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lass, B., and C. I. Ullrich-Eberius. 1984. Evidence for proton/sulfate cotransport and its kinetics in Lemna gibba G1. Planta 161:53-60. [DOI] [PubMed] [Google Scholar]
- 25.Leustek, T., and K. Saito. 1999. Sulfate transport and assimilation in plants. Plant Physiol. 120:637-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lien, T., and Ø. Schreiner. 1975. Purification of a derepressible arylsulfatase from Chlamydomonas reinhardtii. Properties of the enzyme in intact cells and in purified state. Biochim. Biophys. Acta 384:168-179. [DOI] [PubMed] [Google Scholar]
- 27.Mahler, R. J., and R. L. Maples. 1987. Effect of sulfur additions on soil and the nutrition of wheat. Commun. Soil Sci. Plant Anal. 18:653-673. [Google Scholar]
- 28.Mahler, R. J., and R. L. Maples. 1986. Responses of wheat to sulfur fertilization. Commun. Soil Sci. Plant Anal. 17:975-988. [Google Scholar]
- 29.Meister, A., and M. E. Anderson. 1983. Glutathione. Annu. Rev. Biochem. 52:711-760. [DOI] [PubMed] [Google Scholar]
- 30.Meurer, J., H. Plucken, K. V. Kowallik, and P. Westhoff. 1998. A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 15:5286-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moseley, J., J. Quinn, M. Eriksson, and S. Merchant. 2000. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 219:2139-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moseley, J. L., T. Allinger, S. Herzog, P. Hoerth, E. Wehinger, S. Merchant, and M. Hippler. 2002. Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21:6709-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moseley, J. L., M. D. Page, N. P. Alder, M. Eriksson, J. Quinn, F. Soto, S. M. Theg, M. Hippler, and S. Merchant. 2002. Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14:673-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niyogi, K. K. 1999. Photoprotection revisited: genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:333-359. [DOI] [PubMed] [Google Scholar]
- 35.Noctor, G., A.-C. Arisi, L. Jouanin, K. J. Kunert, H. Rennenberg, and C. Foyer. 1998. Glutathione: biosynthesis, metabolism and relationship to stress tolerance explored in transgenic plants. J. Exp. Bot. 49:623-647. [Google Scholar]
- 36.Novoselov, S. V., M. Rao, N. V. Onoshko, H. Zhi, G. V. Kryukov, Y. Xiang, D. P. Weeks, D. L. Hatfield, and V. N. Gladyshev. 2004. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J. 21:3681-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ozcan, S., J. Dover, and M. Johnston. 1998. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17:2566-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozcan, S., J. Dover, A. G. Rosenwald, S. Wolf, and M. Johnston. 1996. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 93:12428-12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan, W. 2002. A comparative review of statistical methods for discovering differentially expressed genes in replicated microarray experiments. Bioinformatics 18:546-554. [DOI] [PubMed] [Google Scholar]
- 40.Ravina, C. G., C.-I. Chang, G. P. Tsakraklides, J. P. McDermott, J. M. Vega, T. Leustek, C. Gotor, and J. P. Davies. 2002. The sac mutants of Chlamydomonas reinhardtii reveal transcriptional and posttranscriptional control of cysteine biosynthesis. Plant Physiol. 130:2076-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito, K. 2000. Regulation of sulfate transport and synthesis of sulfur-containing amino acids. Curr. Opin. Plant Biol. 3:188-195. [PubMed] [Google Scholar]
- 42.Saito, K., K. Inoue, R. Fukushima, and M. Noji. 1997. Genomic structure and expression analyses of serine acetyltransferase gene in Citrullus vulgaris (watermelon). Gene 189:57-63. [DOI] [PubMed] [Google Scholar]
- 43.Savard, F., C. Richard, and M. Guertin. 1996. The Chlamydomonas reinhardtii LI818 gene represents a distant relative of the cabI/II genes that is regulated during the cell cycle and in response to illumination. Plant Mol. Biol. 32:461-473. [DOI] [PubMed] [Google Scholar]
- 44.Schreiner, Ø., T. Lien, and G. Knutsen. 1975. The capacity for arylsulfatase synthesis in synchronous and synchronized cultures of Chlamydomonas reinhardtii. Biochim. Biophys. Acta 384:180-193. [DOI] [PubMed] [Google Scholar]
- 45.Schultze, M., B. Quiclet-Sire, E. Kondorosi, H. Virelizer, J. N. Glushka, G. Endre, S. D. Gero, and A. Kondorosi. 1992. Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proc. Natl. Acad. Sci. USA 89:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibagaki, N., A. Rose, J. P. McDermott, T. Fujiwara, H. Hayashi, T. Yoneyama, and J. P. Davies. 2002. Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 29:475-486. [DOI] [PubMed] [Google Scholar]
- 48.Shrager, J., C. Hauser, C. W. Chang, E. H. Harris, J. Davies, J. McDermott, R. Tamse, Z. Zhang, and A. R. Grossman. 2003. Chlamydomonas reinhardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol. 131:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, F., A. L. Rae, and M. J. Hawkesford. 2000. Molecular mechanisms of phosphate and sulphate transport in plants. Biochem. Biophys. Res. Commun. 1465:236-245. [DOI] [PubMed] [Google Scholar]
- 50.Smith, F. W., M. J. Hawkesford, P. M. Ealing, D. T. Clarkson, P. J. Vanden Berg, A. R. Belcher, and A. G. Warrilow. 1997. Regulation of expression of a cDNA from barley roots encoding a high affinity sulphate transporter. Plant J. 12:875-884. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi, H., C. E. Braby, and A. R. Grossman. 2001. Sulfur economy and cell wall biosynthesis during sulfur limitation of Chlamydomonas reinhardtii. Plant Physiol. 127:665-673. [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi, H., M. Yamazaki, N. Sasakura, A. Watanabe, T. Leustek, J. de Almeida Engler, G. Engler, M. Van Montagu, and K. Saito. 1997. Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate starved roots plays a central role in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94:11102-11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tottey, S., M. A. Block, M. Allen, T. Westergren, C. Albrieux, H. V. Scheller, S. Merchant, and P. E. Jensen. 2003. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 100:16119-16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tu, C.-J., J. Shrager, R. L. Burnap, B. L. Postier, and A. R. Grossman. 2004. Consequences of a deletion in dspA on transcript accumulation in Synechocystis sp. strain PCC6803. J. Bacteriol. 186:3889-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vollenbroek, E. G., L. H. Simons, J. W. van Schijndel, P. Barnett, M. Balzar, H. Dekker, C. van der Linden, and R. Wever. 1995. Vanadium chloroperoxidases occur widely in nature. Biochem. Soc. Trans. 23:267-271. [DOI] [PubMed] [Google Scholar]
- 57.Warman, P. R., and H. G. Sampson. 1994. Effect of sulfur additions on the yield and elemental composition of canola and spring wheat. J. Plant Nutr. 17:1817-1825. [Google Scholar]
- 58.Wilkinson, L. J., and R. H. Waring. 2002. Cysteine dioxygenase: modulation of expression in human cell lines by cytokines and control of sulphate production. Toxicol. In Vitro 16:481-483. [DOI] [PubMed] [Google Scholar]
- 59.Wingler, A., P. J. Lea, W. P. Quick, and R. C. Leegood. 2000. Photorespiration: metabolic pathways and their role in stress protection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:1517-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wykoff, D., J. Davies, and A. Grossman. 1998. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117:129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yildiz, F., J. P. Davies, and A. R. Grossman. 1994. Characterization of sulfate transport in Chlamydomonas reinhardtii during sulfur-limited and sulfur-sufficient growth. Plant Physiol. 104:981-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yildiz, F. H., J. P. Davies, and A. R. Grossman. 1996. Sulfur availability and the SAC1 gene control adenosine triphosphate sulfurylase gene expression in Chlamydomonas reinhardtii. Plant Physiol. 112:669-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zenk, M. H. 1996. Heavy metal detoxification in higher plants—a review. Gene 179:21-30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.