ABSTRACT
Clostridium difficile is a Gram-positive spore-forming obligate anaerobe that is a leading cause of antibiotic-associated diarrhea worldwide. In order for C. difficile to initiate infection, its aerotolerant spore form must germinate in the gut of mammalian hosts. While almost all spore-forming organisms use transmembrane germinant receptors to trigger germination, C. difficile uses the pseudoprotease CspC to sense bile salt germinants. CspC activates the related subtilisin-like protease CspB, which then proteolytically activates the cortex hydrolase SleC. Activated SleC degrades the protective spore cortex layer, a step that is essential for germination to proceed. Since CspC incorporation into spores also depends on CspA, a related pseudoprotease domain, Csp family proteins play a critical role in germination. However, how Csps are incorporated into spores remains unknown. In this study, we demonstrate that incorporation of the CspC, CspB, and CspA germination regulators into spores depends on CD0311 (renamed GerG), a previously uncharacterized hypothetical protein. The reduced levels of Csps in gerG spores correlate with reduced responsiveness to bile salt germinants and increased germination heterogeneity in single-spore germination assays. Interestingly, asparagine-rich repeat sequences in GerG’s central region facilitate spontaneous gel formation in vitro even though they are dispensable for GerG-mediated control of germination. Since GerG is found exclusively in C. difficile, our results suggest that exploiting GerG function could represent a promising avenue for developing C. difficile-specific anti-infective therapies.
IMPORTANCE
The spore-forming bacterium Clostridium difficile is a leading cause of health care-associated infections. While a subset of antibiotics can treat C. difficile infections (CDIs), the primary determinant of CDI disease susceptibility is prior antibiotic exposure, since it reduces the colonization resistance conferred by a diverse microflora. Thus, therapies that minimize perturbations to the gut microbiome should be more effective at reducing CDIs and their recurrence, the main source of disease complications. Given that spore germination is essential for C. difficile to initiate infection and that C. difficile uses a unique pathway to initiate germination, methods that inhibit distinct elements of germination could selectively prevent C. difficile disease recurrence. Here, we identify GerG as a C. difficile-specific protein that controls the incorporation of germinant signaling proteins into spores. Since gerG mutant spores exhibit germination defects and are less responsive to germinant, GerG may represent a promising target for developing therapeutics against CDI.
INTRODUCTION
The spore-forming bacterium Clostridium difficile is a leading cause of health care-associated infections and gastroenteritis-associated death worldwide (1, 2). C. difficile infections also occur in community and commercial farm settings (3–5). The Centers for Disease Control and Prevention in the United States recently designated C. difficile an urgent threat to the health care system in part because antibiotic usage is a major risk factor for C. difficile infection (6). Antibiotic treatment eliminates members of the gut microflora that normally suppress C. difficile vegetative growth and thus increases patient susceptibility to C. difficile infections (7–9). Antibiotic usage may also promote C. difficile spore germination by altering the balance of microbially generated bile salt germinants. These changes may effectively increase the infectious dose of C. difficile (8, 10–12), since spores are the primary infectious form of this obligate anaerobe (13).
During infection, C. difficile spores germinate upon sensing select bile salts in the mammalian gastrointestinal tract (10, 14); the germinating spores then transform into the toxin-secreting vegetative cells that cause disease (7). A key step during this process is the degradation of the spore cortex, a thick protective layer of modified peptidoglycan (15, 16). The cortex is critical to maintaining spore dormancy, since it keeps the spore core, which contains the cell’s genetic material, in a partially dehydrated state that minimizes cellular metabolism. Removal of this physical constraint by cortex hydrolases allows the core to fully hydrate, resume metabolism, and initiate outgrowth (15, 16).
Interestingly, the signaling process that controls cortex hydrolysis in C. difficile exhibits marked differences from the other spore-forming bacteria studied to date (17). While most spore formers use inner membrane-bound germinant receptors to trigger germination in response to amino acids and sugars (17–19), C. difficile lacks these receptors and instead uses the CspC pseudoprotease as a germinant receptor to sense bile acids (20). When C. difficile CspC senses germinant, it activates the subtilisin-like serine protease CspB to remove an inhibitory propeptide from the cortex hydrolase SleC (21). Activation of the SleC zymogen then allows the hydrolase to degrade the thick protective cortex layer (22, 23). Cortex degradation subsequently induces the release of calcium dipicolinic acid (CaDPA) from the spore core (23), which leads to full hydration of the core (23, 24).
In the model organism Bacillus subtilis, the order of cortex hydrolysis and CaDPA release is reversed: germinant binding induces CaDPA release, and CaDPA directly activates the cortex hydrolase CwlJ (16). While there is little homology in the germination signaling proteins used by B. subtilis relative to C. difficile (17–19), many C. difficile germination regulators have homologs in Clostridium perfringens (17, 18). The functions of these regulators, however, differ between the two clostridial organisms. In C. perfringens, Csp proteins are encoded as individual proteases that are functionally interchangeable, with any one of the Csps, CspA, CspB, and CspC, being able to proteolytically activate the SleC zymogen (25, 26). In contrast, C. difficile CspB and CspA are produced as a fusion protein that undergoes interdomain processing during sporulation (21, 27). Furthermore, both CspA and CspC harbor catalytic site mutations that render them inactive such that CspB is the only Csp that can proteolytically activate SleC (21). Interestingly, C. difficile CspA controls the incorporation of the CspC germinant receptor into mature spores (27), indicating that CspC and CspA have divergent functions in C. difficile relative to C. perfringens.
Since the CspA and CspC pseudoproteases are conserved only in C. difficile’s closest relatives in the Peptostreptococcaceae family (27, 28), C. difficile’s germinant signaling pathway appears to be unique relative to other organisms studied to date (17, 23). In support of this notion, we recently identified the lipoprotein GerS as another key germination regulator specific to the Peptostreptococcaceae family (22). GerS is necessary for SleC activity but is dispensable for pro-SleC cleavage. In this report, we identify a novel C. difficile-specific protein, CD0311, that is required for optimal C. difficile spore germination. Using a variety of functional assays, we determine the stage of germination impaired by loss of CD0311, herein referred to as GerG, and analyze its potential functional regions.
RESULTS
GerG is a novel regulator of C. difficile spore germination.
In previous work, we used transcriptome sequencing (RNA-Seq) analyses (29, 30) to inform the targeted mutagenesis of gerS, as it was one of the most highly induced genes during sporulation and encoded a conserved hypothetical protein (22). Using a similar rationale, we targeted gerG for mutagenesis, since it is the 10th most highly expressed, sporulation-induced gene whose function was unknown. Similar to gerS, expression of gerG is under the control of the mother cell-specific sigma factor E (σE [30, 31]) (Fig. 1A). Interestingly, GerG appears to be conserved exclusively in C. difficile, unlike previously identified germination regulators.
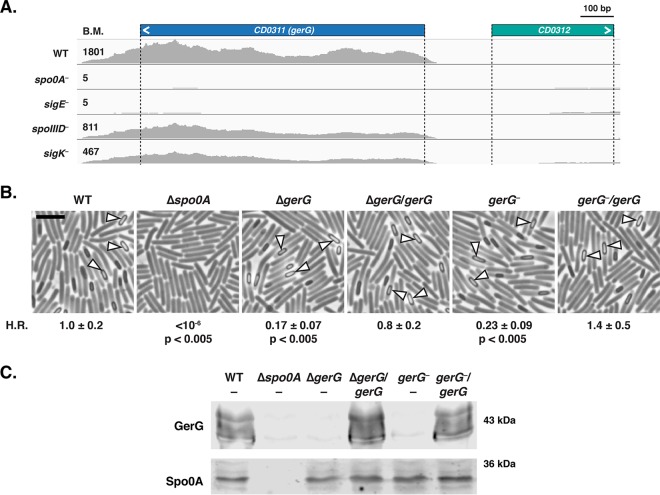
FIG 1 .
The highly expressed, σE-controlled gerG gene contributes to C. difficile heat resistance. (A) Representative image of Integrative Genomics Viewer software (75) used to visualize transcript reads of the gerG locus from RNA-Seq analyses of sporulating C. difficile cultures (30) from wild-type (WT) JIR8094, spo0A-negative, sigE-negative, spoIIID-negative, and sigK-negative strains. σE activates the transcription of spoIIID, which encodes a transcriptional regulator that activates sigK transcription (53). The angled bracket denotes the direction of transcription. The average base mean (BM) refers to the number of transcripts detected for the respective gene normalized to the length of that gene. The difference in transcript levels detected between wild-type, spoIIID-negative, and sigK-negative strains did not meet the cutoff of >4-fold change defined in reference 30. (B) Phase-contrast microscopy of sporulating cells of the wild type (630Δerm-p), the Δspo0A mutant, the ΔgerG mutant and its complement (ΔgerG/gerG), and the gerG::ermB (gerG-negative) TargeTron disruption strain and its complement (gerG-negative/gerG). The Δspo0A mutant cannot initiate sporulation (13), but phase-bright (fore)spores are visible in all other strains (white triangles). H.R. represents the heat resistance of each strain relative to the wild type as determined from three biological replicates. The means and standard deviations shown are based on three biological replicates. Statistical significance relative to the wild type was determined using one-way analysis of variance and Tukey’s test. Bar, 5 µm. (C) Western blot analysis of GerG and the Spo0A loading control (76) in sporulating cells of indicated strains.
To test whether GerG plays a role in regulating sporulation and/or germination, we constructed TargeTron mutations in gerG in the JIR8094 and 630ΔermΔpyrE strain backgrounds as well as a clean deletion of gerG in the 630ΔermΔpyrE background (see Fig. S1 in the supplemental material). The latter strain background provides the advantage of allowing mutations to be complemented in single copy on the chromosome at the pyrE locus (32), in contrast with the plasmid-based complementation (33) typically used for TargeTron mutants (34) in strains with an intact pyrE gene like JIR8094. Both the gerG deletion (Fig. 1B) and TargeTron (Fig. S2A) mutants produced phase-bright spores with no obvious defects in either the morphology or frequency of spore formation. However, when functional spore formation was measured using a heat resistance assay (29, 35), 630Δerm gerG mutants exhibited an ~4- to 5-fold defect relative to the wild type (Fig. 1B, P < 0.005), and the JIR8094 gerG TargeTron mutant exhibited an ~30-fold defect relative to the wild type (Fig. S2A, P < 0.05). Western blot analyses confirmed that gerG mutants were defective in producing GerG (Fig. 1C and S2B), while single-copy complementation of gerG restored GerG to wild-type levels (Fig. 1C). Multiple bands were detected for GerG in both strain backgrounds, suggesting that it is posttranslationally modified and/or unstable during sporulation.
Construction of gerG (CD0311) and sleC mutants. Download FIG S1, PDF file, 0.7 MB (755.4KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A TargeTron insertion in CD0311 in the JIR8094 strain background leads to a heat resistance defect. Download FIG S2, PDF file, 0.4 MB (428.3KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
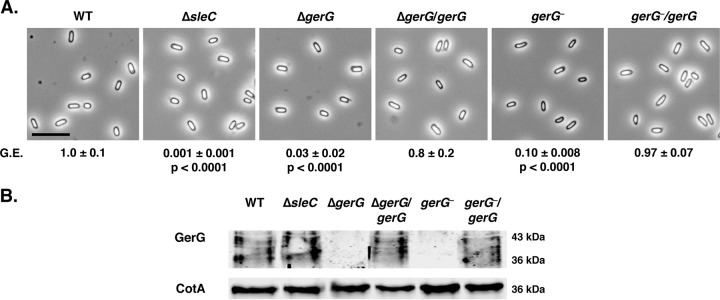
Since a recent transposon mutagenesis study indicated that insertional mutations in the gerG homolog of the epidemic strain R20291 resulted in an ~5-fold decrease in spore purification yields (36), we compared the spore purification yields of the ΔgerG mutant relative to those of its complementation strain and the wild type. No statistically significant difference in spore purification was observed for ΔgerG spores relative to wild-type 630Δerm (see Table S1 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing). In contrast, ΔgerG and gerG::ermB (gerG-negative) spores exhibited 30- and 10-fold decreases in germination efficiency relative to the wild type, respectively, when plated on brain heart infusion-supplemented (BHIS) plates containing taurocholate (TA) (Fig. 2, P < 0.0001). JIR8094 gerG::ermB spores exhibited a 500-fold defect in germination efficiency relative to the wild type (Fig. S3, P < 0.0005). JIR8094 gerG-minus may have exhibited a more severe spore germination defect than its heat resistance defect because of the high variability in sporulation levels between replicates of our heat resistance assays (Fig. S2). Regardless, these results imply that loss of GerG impairs spore germination rather than spore formation. Importantly, the germination defect of all gerG mutant spores could be complemented to wild-type levels (Fig. 2 and S4B), even though GerG was strongly overproduced when gerG was expressed from a multicopy plasmid in the JIR8094 gerG mutant (Fig. S4B). Interestingly, multiple species of GerG were detected at a lower apparent molecular weight in mature spores (Fig. 2B) than in sporulating cells (Fig. 1C), suggesting that GerG undergoes posttranslational changes during its incorporation into spores.
FIG 2 .
Optimal spore germination depends on GerG. (A) Phase-contrast microscopy of purified spores from the wild type (WT) (630Δerm-p), the ΔsleC mutant, the ΔgerG mutant and its complement, and the gerG::ermB (gerG-negative) TargeTron disruption strain and its complement. Bar, 5 µm. The germination efficiency (G.E.) of the strains relative to the wild type is shown. The means and standard deviations shown are based on three biological replicates. Statistical significance was determined relative to the wild type using one-way analysis of variance and Tukey’s test on the number of colonies formed by spores when plated on BHIS containing taurocholate. No statistically significant difference in germination efficiency was observed between the ΔgerG and gerG-negative strains. (B) Western blot analyses of GerG and a CotA loading control in purified spores of the indicated strains.
GerG is required for optimal spore germination in JIR8094. Download FIG S3, PDF file, 0.4 MB (369.8KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overexpression of cspBAC fails to rescue the germination defect of gerG-negative spores in JIR8094. Download FIG S4, PDF file, 0.5 MB (484.5KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In the germination assays shown in Fig. 2 and S3, we used a sleC deletion mutant as a negative control (Fig. S1), since sleC mutants do not hydrolyze their cortex and thus fail to germinate in at least two strain backgrounds (21, 23, 37). However, in 630Δerm, the ΔsleC mutant had only an ~3-log defect in spore germination (Fig. 2; see Table S2 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing), whereas the JIR8094 sleC::ermB TargeTron mutant (sleC-negative) exhibited an ~5-log germination defect (Fig. S3; see Table S2 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing). sleC mutant spores outgrew to produce colonies more slowly than wild-type and gerG spores when germinated on BHIS plates containing taurocholate in both strain backgrounds (data not shown). For 630ΔermΔsleC, spore outgrowth was not visible until ~16 to 20 h (>4-h delay). If spore germination was assayed earlier than this time point, 630ΔermΔsleC spores appeared to be completely defective in germination. Spore germination did not increase after prolonged incubation (>40 h) for wild-type, gerG, or sleC mutant spores in both strain backgrounds. Notably, colony formation by gerG mutant spores on BHIS-TA plates exhibited kinetics similar to those of the wild type (data not shown). These observations suggest that GerG likely does not affect germinant accessibility, in contrast with the delayed-germination phenotype observed for Bacillus gerP mutant spores, which exhibit decreased permeability to germinants (38–40).
While the ΔsleC mutant in our hands exhibited an ~3-log defect in spore germination, TargeTron disruption of sleC in 630Δerm has been reported to cause an ~4-log germination defect (37). This difference in phenotypes could be due to differences in the nature of the mutations (gene deletion versus disruption) or germination conditions. To distinguish between these possibilities, we reconstructed the sleC TargeTron mutation in the 630ΔermΔpyrE strain background (Fig. S1) and restored its pyrE locus. The resulting 630Δerm sleC::ermB (sleC-negative) mutant had an ~3-log germination defect similar to 630ΔermΔsleC, which is ~100-fold less severe than JIR8094 sleC::ermB (see Table S2 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing). The difference between our results and those of Burns et al. (37) may be due to differences in spore preparation conditions, as has been described previously (41–43). Consistent with this possibility, we observed a >10-fold variation in sleC mutant spore germination between spore preparations (see Table S3 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing).
In spite of these differences, the germination defect of all sleC mutations could be complemented in both the 630Δerm and JIR8094 strain backgrounds (see Table S2 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing), and artificial germination of 630Δerm ΔsleC and ΔgerG spores using thioglycolate and lysozyme rescued the ability of these spores to produce colonies to wild-type levels (Fig. S5). Altogether, the results suggest that there are strain-specific differences in the requirement for SleC during spore germination.
Artificial germination bypasses the need for GerG. Download FIG S5, PDF file, 0.1 MB (106.4KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GerG is required for cortex hydrolysis.
Having established that GerG is necessary for optimal spore germination, we sought to identify the stage of germination affected by loss of GerG. We first monitored changes in the optical density at 600 nm (OD600) of ΔgerG spores relative to the wild type, since the optical density of germinating spores decreases due to cortex hydrolysis and subsequent core expansion (24). When wild-type and ΔgerG/gerG spores were incubated with 1% taurocholate (19 mM) at room temperature for 45 min, their optical density decreased by ~50% (Fig. 3A); in contrast, the optical density of ΔgerG and ΔsleC spores did not change appreciably. Since changes in spore optical density in this assay depend on cortex hydrolysis (23), ΔgerG spores would appear to be defective in cortex hydrolysis.
FIG 3 .
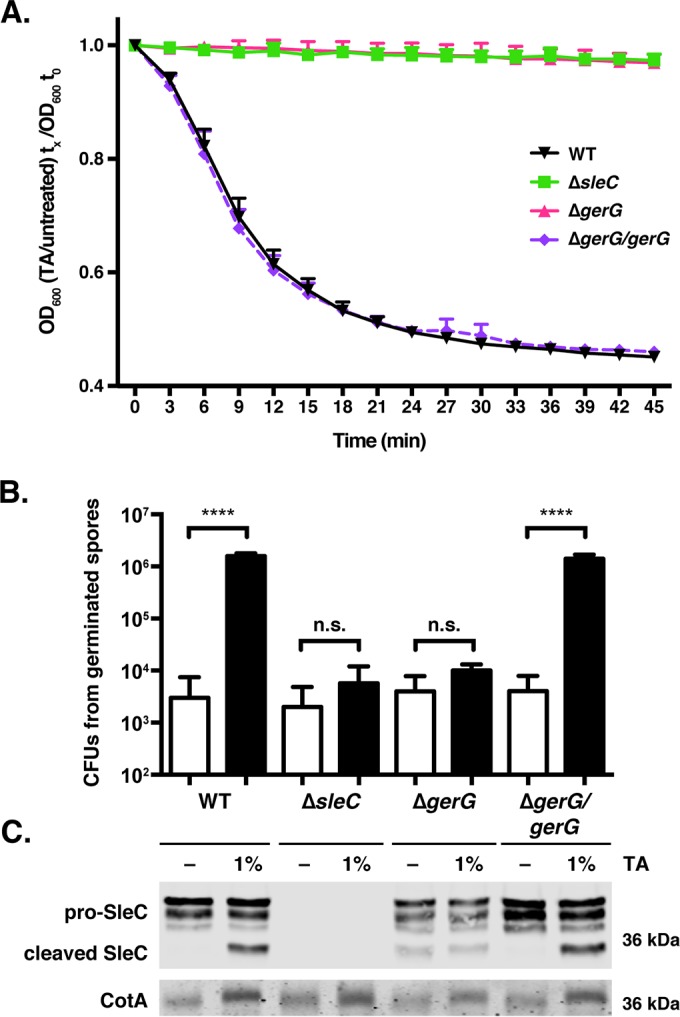
GerG is required for activation of the SleC cortex hydrolase. (A) Change in optical density at 600 nm (OD600) of the indicated spores in response to germinant. ΔsleC spores were used as a negative control, since SleC is required for cortex hydrolysis (23). OD600 measurements were taken every 3 min. Wild-type (WT) and ΔgerG/gerG spores exhibited statistically significant differences in OD600 over time relative to ΔsleC and ΔgerG spores (P < 0.0001); the latter two strains did not exhibit statistically significant differences in OD600 over time based on two-way analyses of variance using Tukey’s test. (B) CFU that arose from spores exposed to either water (white bars) or TA germinant (1%, 19 mM; black bars) for 20 min at 37°C and plated on BHIS. Statistical significance relative to the wild type was determined using a one-way analysis of variance and Tukey’s test. n.s., no statistical significance; ****, P < 0.0001. (C) Western blot analysis of SleC and CotA (loading control) in samples shown in panel B. −, exposure to water. The pro-SleC zymogen is processed in response to germinant by CspB (21).
Based on these observations, we predicted that the cortex hydrolase SleC fails to undergo proteolytic activation by CspB (21), with the caveat that gerS::ermB spores can still cleave pro-SleC despite failing to hydrolyze cortex (22). To test this hypothesis, wild-type, ΔgerG, and ΔgerG/gerG spores were exposed to germinant for 20 min at 37°C, and pro-SleC cleavage was monitored by Western blotting (Fig. 3C). Whereas wild-type and ΔgerG/gerG spores cleaved pro-SleC in response to taurocholate addition, no change in pro-SleC processing was observed in ΔgerG spores. Interestingly, low levels of processed SleC were detectable in ΔgerG spores even in the absence of germinant, indicating that GerG alters the protease susceptibility of SleC in either sporulating cells or mature spores.
To correlate SleC cleavage with spore germination and outgrowth, we measured the number of spores that had germinated after the 20-min exposure to taurocholate by plating wild-type, ΔgerG, and ΔsleC spores on medium lacking germinant. An ~1,000-fold increase in CFU was observed when wild-type and ΔgerG/gerG spores were transiently exposed to taurocholate germinant and plated on medium lacking germinant relative to untreated spores (Fig. 3B, P < 0.0001). In contrast, transient exposure of ΔgerG and ΔsleC spores to germinant did not significantly increase the number of CFU recovered on medium lacking germinant (Fig. 3B). Taken together, CspB-mediated activation of SleC cortex hydrolase activity does not occur in the absence of GerG.
GerG controls the levels of Csp germination regulators in spores.
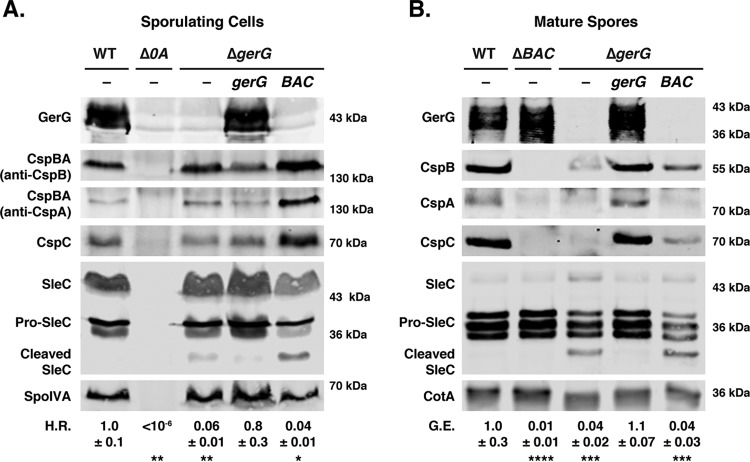
Given that the signaling pathway that leads to CspB-mediated activation of the SleC cortex hydrolase involves CspC sensing germinant (20) and CspA modulating CspC incorporation into spores (27), we tested whether GerG affects the levels of Csp proteins during sporulation. In particular, we analyzed the levels of CspBA and CspC in sporulating cells and CspB, CspA, and CspC in purified spores using immunoblotting. While CspBA and CspC levels were unchanged in sporulating ΔgerG cells (Fig. 4A), the levels of CspB, CspA, and CspC were severely reduced in ΔgerG spores relative to the wild type (Fig. 4B). Importantly, complementation of ΔgerG restored wild-type levels of Csps in spores, indicating that GerG regulates the incorporation of Csp germination regulators into spores and/or their stability in mature spores.
FIG 4 .
GerG controls Csp levels in mature spores. Western blot analyses of germination regulators in sporulating cells (A) and purified spores (B) from the wild type (WT) (630Δerm-p), the ΔgerG mutant, and the ΔgerG mutant complemented with either gerG (ΔgerG/gerG) or the cspBAC operon (ΔgerG/cspBAC). The Δspo0A mutant was used as a negative control for sporulating cells, since this strain does not sporulate (13); a ΔcspBAC mutant was used as a negative control for analyzing Csp levels in mature spores. CspBA is the primary form detected in sporulating cells, whereas CspB and CspA domains are the primary form detected in mature spores (27). SleC is detected in three major forms: full length (SleC), zymogen (pro-SleC), and proteolytically activated (cleaved SleC [21]). Antibodies against SpoIVA and CotA were used as a loading controls. H.R. represents the heat resistance of each strain relative to the wild type; G.E. refers to the germination efficiency of each strain relative to the wild type. The means and standard deviations shown are based on three biological replicates. Statistical significance relative to the wild type was determined using a one-way analysis of variance and Tukey’s test. ****, P < 0.0001; ***, P < 0.0005; **, P < 0.005; *, P < 0.05.
Based on these findings, we wondered whether overproduction of Csp family members might rescue the germination defect of ΔgerG spores. To this end, we introduced a second copy of the cspBA-cspC operon into the pyrE locus of ΔgerG (ΔgerG/BAC). The extra copy of cspBA-cspC increased CspBA and CspC levels in sporulating cells above those observed in wild type (Fig. 4). While the levels of CspB and CspC were increased in mature ΔgerG/cspBAC spores relative to ΔgerG spores (Fig. 4), they were still lower than those in the wild type. The increased Csp levels, however, did not affect sporulating cell heat resistance or spore germination efficiency in ΔgerG/cspBAC spores relative to ΔgerG spores.
Since the moderate increase in Csp levels in ΔgerG/cspBA-cspC spores may have been insufficient to restore germination to wild-type levels, we tested whether plasmid-based overexpression of cspBAC might produce enough CspBA and CspC to bypass the need for GerG to incorporate Csp family members into mature spores. We used JIR8094-based mutants in these assays because previous work from our lab showed that expression of cspBA-cspC from the multicopy pMTL83151 plasmid overproduced CspBA and CspC in sporulating cells and increased Csp levels in spores (27). Complementation of the JIR8094 gerG mutant with the pMTL83151-cspBAC plasmid markedly elevated CspBA and CspC levels in sporulating cells (Fig. S4); however, far less CspB, CspA, and CspC were detected in gerG-negative/cspBAC spores relative to cspBAC-negative/cspBAC spores (Fig. S4). Despite a marginal increase in Csp protein levels in gerG-negative/cspBAC spores relative to gerG-negative spores carrying empty vector, no change in heat resistance or germination efficiency was observed between these strains (Fig. S4). Taken together, these results highlight the role of GerG modulating the incorporation of Csp proteins into spores.
The cspBAC overexpression experiments used cspBAC deletion and TargeTron mutants in the 630Δerm and JIR8094 backgrounds, respectively, as negative controls for measuring Csp levels in spores. Similarly to our observations with sleC mutants in these two strain backgrounds, the germination phenotypes of these cspBAC mutants differed markedly. JIR8094 cspBAC-negative spores exhibited a >5-log germination defect relative to wild-type spores, while 630ΔermΔcspBAC spores displayed an ~2-log germination defect (Fig. 4 and S4). These phenotypes support the notion that 630Δerm spores germinate more readily than JIR8094 spores.
GerG mutant spores are less responsive to germinant.
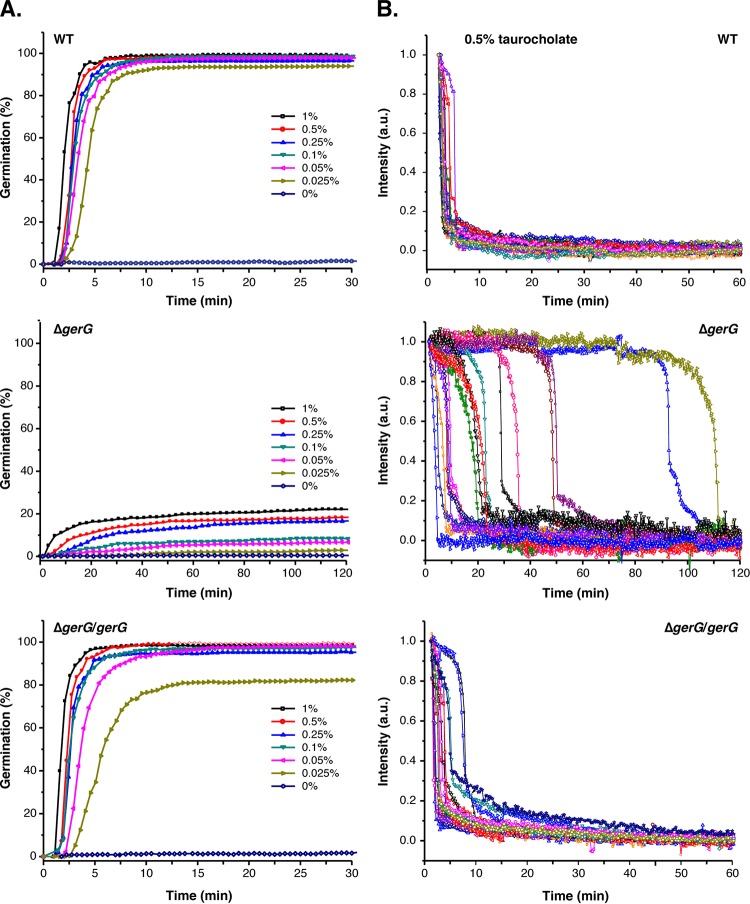
The observation that ΔgerG spores have lower levels of CspC germinant receptor prompted us to investigate whether they would be less sensitive to germinant, as the levels of transmembrane germinant receptors have been shown to control the responsiveness of Bacillus subtilis spores to germinant (44, 45). To test this hypothesis, we monitored the germination of individual ΔgerG spores in response to increasing concentrations of germinant using optical trapping (46). In this assay, the decrease in optical refractility of germinating spores is monitored on a single-spore basis over time. The decrease in germinating spore optical refractility corresponds with cortex hydrolysis and CaDPA release in C. difficile (47), similarly to analyses of bacterial spores from other species (46, 48, 49). Since spore germination dynamics can be measured on a single-spore basis, this assay allows germination heterogeneity to be assessed in contrast with bulk, population-wide measurements such as the optical density assay in Fig. 3.
When wild-type spores were incubated with the lowest concentration of germinant tested (0.025% taurocholate, 0.5 mM), 95% of the spores completed germination within 10 min at 37°C (Fig. 5; see Table S4 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing). ΔgerG/gerG complementation spores behaved similarly to the wild type, with 98% of these spores completing germination within 10 min of being exposed to 0.05% taurocholate (0.9 mM). In contrast, only 3% of ΔgerG spores completed germination after incubation with 0.025% (0.5 mM) taurocholate for 120 min. While germination was essentially maximal in wild-type and ΔgerG/gerG spores at 0.25% TA (5 mM), ΔgerG spore germination increased in a dose-dependent manner up to the maximum concentration of taurocholate tested (1%, 19 mM; Fig. 5A).
FIG 5 .
C. difficile spores are less responsive to germinant in the absence of GerG. (A) C. difficile spore germination in response to increasing concentrations of taurocholate and 15.5 mM glycine at 37°C. Germination responses were measured at the single-spore level by monitoring decreases in phase-contrast image intensity (46). A minimum of 200 spores were monitored for 30 min (wild-type [WT] [630Δerm] and ΔgerG/gerG strain) or 120 min for the ΔgerG strain. (B) Germination responses of individual C. difficile spores in the presence of 0.5% taurocholate (9 mM) and 15.5 mM glycine at 37°C. Spores were chosen at random from the subpopulation of germinating spores. The phase-contrast image intensity of a given spore was normalized to 1 based on the respective values at the first time point measured. Image intensities at the end of the experiment were set to zero. a.u., arbitrary units.
Spore germination was also more heterogeneous in ΔgerG spores than in wild-type and ΔgerG/gerG spores (Fig. 5B). When the germination dynamics of 15 individual ΔgerG spores were analyzed, germination was observed between 10 and 120 min of germinant addition. These 15 spores were chosen at random from the subset (<20%) of ΔgerG spores that germinated after exposure to 0.5% taurocholate (9 mM) for 120 min. In contrast, all the wild-type and ΔgerG/gerG spores analyzed in this manner uniformly initiated and completed germination within 10 min of germinant addition. Although ΔgerG spores exhibited greater variability in the time to initiate germination than did the wild type, ΔgerG spores completed germination with similar kinetics as the wild-type spores as evidenced by the kinetics of the sharp decrease in optical refractility. Since this decrease corresponds to cortex hydrolysis and CaDPA release (47, 50), GerG would appear to control the initiation of germination rather than the kinetics of these germination events. This finding is consistent with the observation that the ΔgerG spores form colonies at a similar rate as the wild type when germinated on BHIS plates containing taurocholate, whereas ΔsleC spores form colonies with a >4-h delay (data not shown).
To assess whether the levels of germination measured in the optical trapping assay corresponded to spore outgrowth, we determined the colony-forming capacity of wild-type, ΔgerG, and ΔgerG/gerG spores exposed to increasing concentrations of taurocholate. Spores were exposed to taurocholate for 20 min at 37°C, and the number of spores that initiated germination was enumerated by plating on BHIS plates lacking germinant. Almost all of the wild-type and ΔgerG/gerG spores initiated germination upon being exposed to 0.5% taurocholate (9 mM) for 20 min (Fig. S6), since similar numbers of CFU were produced when taurocholate-treated spores were plated on BHIS plates lacking germinant as when untreated spores were plated on BHIS plates containing taurocholate germinant (dashed line). In contrast, ΔgerG spores needed to be exposed to 10-fold-higher levels of taurocholate (>5%, 86 mM) in order to achieve maximal ΔgerG spore germination, i.e., CFU comparable to those observed when untreated ΔgerG spores were plated on BHIS containing taurocholate (dashed line). These results are largely consistent with the single-spore germination analyses, although the germination defect of ΔgerG spores relative to wild-type spores was slightly more severe in the spore outgrowth assay (15-fold decrease at 0.5% TA, P < 0.0001; Fig. S6A) than the optical trapping assay (~5-fold decrease at 0.5% TA; see Table S4 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing). Interestingly, Western blot analyses of ΔgerG spores exposed to supraphysiological concentrations of taurocholate (8, 51) did not show increased SleC cleavage even though greater levels of ΔgerG spores germinated and outgrew to form colonies on BHIS plates (Fig. S6B).
Responsiveness of gerG mutant spores to germinant. Download FIG S6, PDF file, 0.5 MB (475.6KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Asparagine-rich internal repeat regions are dispensable for GerG function.
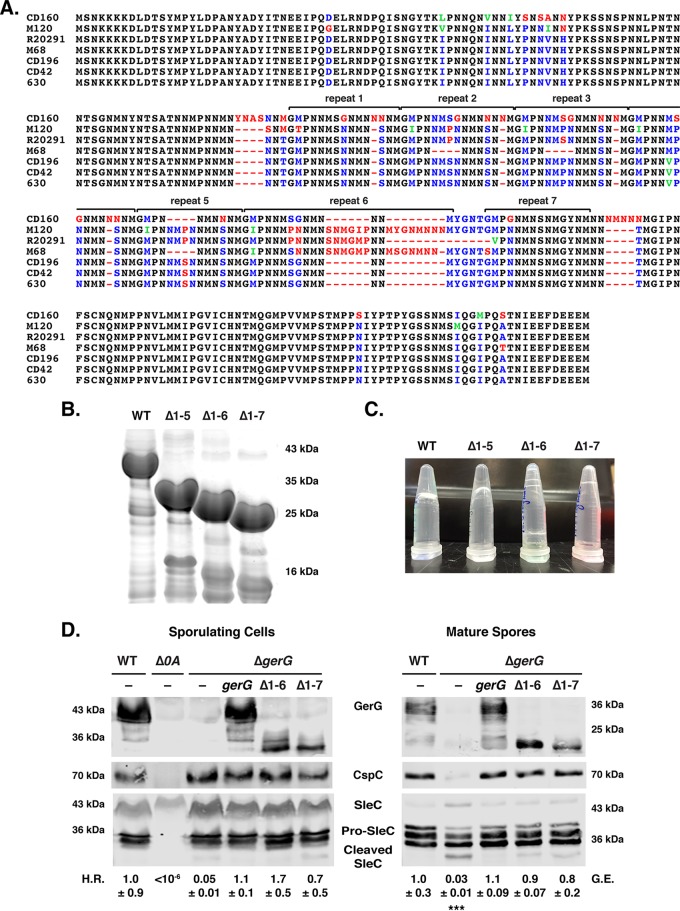
Having demonstrated that GerG modulates Csp incorporation into spores and thus appears to affect their responsiveness to germinant, we next sought to identify functional regions within this previously uncharacterized protein. GerG has a highly unusual amino acid composition: ~30% of its residues are asparagines, 15% are methionines, 11% are prolines, 8% are serines, and 6% are glycines. Most of these residues are concentrated in a central repeat region, which consists of GMPNNMSNNMNSNM repeats that vary slightly in composition and number among C. difficile strains (Fig. 6A).
FIG 6 .
The central repeat region of GerG is required for gel formation but dispensable for spore germination. (A) ClustalW alignment of GerG from selected C. difficile strains. Completely conserved residues are colored black, conserved identical residues are in blue, conserved similar residues are in green, and non-conserved residues are in red. (B) Coomassie blue staining of recombinant, affinity-purified, His-tagged GerG variants. The repeat sequences shown in panel A that were deleted are shown. WT, wild type. (C) Gel formation of GerG variants shown in panel B as determined by inverting the purified protein preparations. (D) Western blot analyses of GerG, CspC, and SleC in sporulating cells and purified spores of the wild type, the Δspo0A mutant, the ΔgerG mutant, and the ΔgerG strain complemented with either wild-type gerG or gerG encoding deletions of repeats 1 to 6 (Δ1–6) or 1 to 7 (Δ1–7). H.R. represents the heat resistance of each strain relative to the wild type; G.E. refers to the germination efficiency of each strain relative to the wild type. The means and standard deviations shown are based on three biological replicates. Statistical significance relative to the wild type was determined using a one-way analysis of variance and Tukey’s test. ***, P < 0.0005. No statistically significant difference was observed between the strains used in the heat resistance assay due to high variance in sporulation levels. No statistically significant difference in germination efficiency was observed between Δ1–6 and Δ1–7 relative to the wild type.
Interestingly, when recombinant His-tagged GerG was purified (Fig. 6B) to generate antibodies, the protein spontaneously formed a gel overnight (Fig. 6C). GerG gel formation appeared to occur in a concentration-dependent manner (see Table S5 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing), with concentrations of ~200 µM being sufficient to permit gel formation. However, since concentrating GerG-His6 did not necessarily lead to gel formation (data not shown), this phenomenon may depend on high concentrations of GerG-His6 being immobilized in close proximity on nickel affinity beads. Gel formation was observed at 4°C, 25°C, and 37°C, although it occurred more slowly at higher temperatures (data not shown).
Since asparagine-rich proteins with low-complexity regions (LCRs) are prone to aggregation (52), we tested whether deleting the asparagine-rich repeat sequences would prevent gel formation. Strain 630Δerm contains seven internal repeat regions (Fig. 6A), and deletion of the first five did not abolish gel formation (Fig. 6C; see Table S5 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing). Deletion of six or all of its repeat regions, however, prevented gel formation at >600 µM protein concentrations (Fig. 6C; see Table S5 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing).
To assess whether the repeat regions were important for GerG to regulate spore germination, we complemented ΔgerG spores with constructs lacking six (Δ1–6) or all (Δ1–7) of the repeat sequences. Remarkably, the Δ1–6 and Δ1–7 constructs fully complemented the heat resistance and germination defects of the parental ΔgerG strain (Fig. 6C). CspC levels in ΔgerG/Δ1–6 and ΔgerG/Δ1–7 strains were restored to wild-type levels by the GerG truncations, even though the truncations themselves reduced the overall levels of GerG produced (Fig. 6C) as detected by an antibody raised against recombinant GerG(Δ1–7). Taken together, our results indicate that the repeat region is dispensable for GerG function in C. difficile, even though this region mediates GerG gel formation in vitro.
DISCUSSION
The regulators controlling spore germination in C. difficile differ markedly from those identified in Bacillus and Clostridium spp. to date. In particular, the CspC germinant receptor, the CspA pseudoprotease domain, and the GerS lipoprotein appear to be unique to the Peptostreptococcaceae family. In this report, we identified GerG as a C. difficile-specific factor that is required for optimal C. difficile spore germination, further underscoring the uniqueness of the C. difficile germination pathway. We find that the levels of key players in the C. difficile germinant signaling pathway, CspA, CspB, and CspC, are dramatically reduced in gerG mutant spores (Fig. 4 and S4) and that this decrease correlates with reduced responsiveness to germinant (Fig. 5 and S6).
An obvious question that arises from our study is how GerG regulates Csp levels in mature spores. While Csp family members are produced in the mother cell (29, 31, 53), they almost certainly function in the intermembrane space between the mother cell and forespore based on the following observations. During germination, the CspB protease must access its target, SleC, which is localized in the cortex (54); thus, CspB is presumably also localized to the cortex. Additionally, CspC activates CspB in response to germinant (20) through a proposed direct binding event (27, 55), making CspC likely to also be localized to the cortex region. Finally, CspA regulates CspC incorporation into spores (27) via a putative interaction, suggesting a cortex region localization for CspA as well. In support of these predicted protein-protein interactions, it has previously been shown that subtilisin-like serine proteases can form higher-order structures (56). Taken together, we predict that the ability of Csp proteins to control C. difficile spore germination depends on their transport across the mother cell-derived outer forespore membrane. Based on these assumptions, we speculate that GerG facilitates the transport of Csp proteins into the cortex region. Consistent with this hypothesis, overproduction of the Csps fails to substantially increase their levels in mature spores (Fig. S4). Since the Csp proteins that were incorporated into these spores (gerG::ermB/cspBAC spores; Fig. S4) remained unable to function properly, we speculate that they became spore associated as the coat assembled around the forespore. Unfortunately, this hypothesis cannot be tested in the absence of fractionation methods that can specifically isolate coat proteins without also extracting proteins in the intermembrane space (e.g., cortex region) (22, 57).
While these issues remain to be resolved, our results suggest that GerG acts upstream of CspA, since loss of GerG reduces levels of all three Csp germination regulators in spores (Fig. 4), whereas loss of CspA reduces levels of CspC but not CspB (27). Another clue to GerG’s function may be the low levels of pro-SleC processing observed in dormant gerG mutant spores (Fig. 3 and 4), which suggest that GerG prevents improper associations between CspB and SleC in sporulating cells and/or mature spores. Investigating the putative binding partners of GerG should provide insight into its precise mechanism of action.
Additional insight into GerG function will likely come from delineating its functional domains. Our results indicate that the central, asparagine-rich repeat region of GerG is dispensable for spore germination under the conditions tested (Fig. 6C), even though it regulates gel formation in vitro (Fig. 6B). Interestingly, this region varies in the number of repeats (typically between 6 and 7) in expanded analyses of C. difficile GerG homologs (see Fig. S7 in the supplemental material). These observations suggest that there may be selective pressure to maintain these repeats, especially since DNA replication slippage could alter the number of repeats (see Materials and Methods). By analogy, a DNA-based mechanism for expanding regions encoding asparagine-rich sequences has been observed in the AT-rich genome of the parasite Plasmodium falciparum, and these regions have been shown to be under positive selection (58).
ClustalW alignment of expanded GerG homologs. Download FIG S7, PDF file, 0.1 MB (88.7KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The repeat region is predicted to be part of a larger region predicted to be disordered based on analyses using Predictors of Natural Disorder (PONDR [59]) (Fig. S8). Naturally disordered proteins often contain regions of low complexity, i.e., regions with little diversity in their amino acid composition, similar to GerG. Since low-complexity region (LCR) proteins can become structured upon interacting with binding partners and have been observed to have more binding partners in protein-protein interaction networks (60, 61), it is tempting to speculate that GerG may directly bind the Csp germination regulators in sporulating cells.
PONDR plot analysis of GerG. Download FIG S8, PDF file, 0.3 MB (301.6KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Interestingly, asparagine-rich proteins have also been associated with prion formation (52, 62). Bioinformatic analysis of GerG using the PLAAC (Prion-Like Amino Acid Composition [63]) application predicts that the majority of GerG constitutes a prion-forming domain (PrD, amino acids [aa] 43 to 270; Fig. S9). The capacity of GerG to self-aggregate (Fig. 6C) in a time- and concentration-dependent manner (see Table S5 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing) is consistent with a propensity to form amyloid-like aggregates (62). It will be interesting to test whether GerG has prionogenic properties and the functional relevance of such a result.
PLAAC analysis of GerG. Download FIG S9, PDF file, 0.4 MB (392.9KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Another question raised by our study is why ΔgerG spore germination is so heterogeneous (Fig. 5B). Germination heterogeneity may be related to the amount of Csps in a given ΔgerG spore, since the levels of germinant receptor have been shown in other systems to control the responsiveness of spores to germinant (44, 45). If Csp levels are the primary driver of spore germination heterogeneity, one could imagine that nongerminating gerG spores either are devoid of Csps (i.e., a bimodal distribution) or carry insufficient levels of Csps to meet the threshold level needed to initiate germination (i.e., normal distribution). Given that ΔgerG spore germination is dose dependent (Fig. 5 and S6) and that gerG mutant spores do not exhibit delayed germination, our results are more consistent with the latter possibility. We are currently developing fluorescent protein fusions to Csp germination regulators to test this hypothesis.
The reduced responsiveness of ΔgerG spores to germinant also raises the possibility that GerG could affect the ability of C. difficile to initiate disease. At the lowest concentration of germinant tested (0.025%, 0.5 mM), only 3% of ΔgerG spores germinated compared to 95% of wild-type spores after a 120-min and a 60-min exposure to germinant, respectively (see Table S3 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing). This concentration of taurocholate is equivalent to the concentrations measured in the ileum of mice prior to antibiotic treatment (8), the ileum being a location that supports C. difficile spore germination in ex vivo studies (10). It is also similar to the levels measured in the feces of patients suffering from recurrent C. difficile infection (0.55 mM [51]). Thus, the infectious dose of a gerG mutant could be diminished under conditions favorable to C. difficile infection and ultimately reduce disease severity and recurrence. Reduced responsiveness to germinant has been correlated with decreased recurrence in some studies (64–66), while others have linked it to greater disease severity, with the notion that spores do not prematurely germinate before they have reached the colon (67). Since the clinical isolates used in these studies are genetically diverse, comparing the infectious dose and virulence of wild-type and gerG mutant spores would allow this question to be resolved.
If gerG mutant spores exhibit virulence defects in animal models of infection, the uniqueness of GerG to C. difficile could be exploited for developing therapies that prevent C. difficile spore germination without disrupting the normal microbiota. Given that a healthy microbiome confers significant colonization resistance against C. difficile infection (7, 9, 12, 68), therapies that target GerG could be more effective at preventing disease recurrence. Testing this possibility would be greatly aided by further study into the functional significance and mechanism of action of this unique protein.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. difficile strains are listed in Table S6 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing and derive from two different parent strains, JIR8094 (630E) and 630ΔermΔpyrE, which are erythromycin-sensitive derivatives of the sequenced clinical isolate 630 (69). 630ΔermΔpyrE is a derivative of 630Δerm that carries a deletion in the gene pyrE, which encodes an orotate phosphoribosyltransferase required for uracil prototrophy (32). Despite their shared lineages, 630Δerm and JIR8094 exhibit marked phenotypic differences (70). C. difficile strains were typically grown on solid BHIS medium (71) supplemented with taurocholate (TA; 0.1% [wt/vol], 1.9 mM), thiamphenicol (5 to 15 µg/ml), kanamycin (50 µg/ml), cefoxitin (8 µg/ml), FeSO4 (50 µM), and/or erythromycin (10 µg/ml) as indicated. During allele-coupled exchange (32), C. difficile defined minimal medium (CDMM) was used as previously described (72) supplemented with 5-fluoroorotic acid (5-FOA) at 2 mg/ml and uracil at 5 µg/ml as needed. Cultures were grown at 37°C, under anaerobic conditions using a gas mixture containing 85% N2, 5% CO2, and 10% H2. HB101/pRK24 Escherichia coli strains were used for conjugations, and BL21(DE3) strains were used for protein production. E. coli strains (see Table S1 at https://drive.google.com/file/d/0B0M1PLMSo_vDb24yTXNKVFZQaTA/view?usp=sharing) were routinely grown at 37°C with shaking at 225 rpm in Luria-Bertani broth (LB). Medium was supplemented with chloramphenicol (20 µg/ml), ampicillin (50 µg/ml), or kanamycin (30 µg/ml) as indicated.
Details of strain construction, antibody production, Western blot analyses, and artificial germination are provided in Text S1 in the supplemental material.
Supplemental materials and methods and references. Download TEXT S1, PDF file, 0.1 MB (146.9KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sporulation.
C. difficile strains were grown from glycerol stocks on BHIS plates supplemented with TA (0.1% [wt/vol], 1.9 mM). Colonies that arose were then used to inoculate 70:30 agar plates (35, 73, 74) for ~24 h. For JIR8094 strains carrying the pMTL83151 vector, 5 µg/ml thiamphenicol was added to these plates to maintain the plasmid. Sporulating cells were harvested into phosphate-buffered saline (PBS) and washed, and sporulation was assessed by phase-contrast microscopy. The remaining sample was processed as needed.
Heat resistance assay on sporulating cells.
C. difficile strains were induced to sporulate as described above for 24 h, and functional (heat-resistant) spore formation was measured as described in reference 35. Heat resistance efficiencies were determined based on the ratio of heat-resistant cells to total cells for a given strain relative to the ratio determined for the wild type. Results are based on the average ratio obtained from a minimum of three biological replicates.
Spore purification.
After sporulation was induced on 70:30 plates for 2 to 3 days, spores were purified as previously described (22). Briefly, spores were washed ~5 times in ice-cold water, incubated overnight in water, DNase treated, purified on a HistoDenz gradient (71), and washed with water. Spore purity was evaluated using phase-contrast microscopy, and spores were stored in water at 4°C.
Germination assay.
Approximately 1 × 107 spores (0.35 OD600 units) were resuspended in 100 µl of water, and 1/10 of this mixture was serially diluted in PBS. Ten-microliter dilutions were plated onto prereduced BHIS-TA, and colonies arising from germinated spores were enumerated at ~22 h. For sleC mutant spore germination, CFU were monitored for up to 48 h. Germination efficiency represents the average number of CFU produced when spores of a given strain were plated on BHIS-TA relative to the number produced by wild-type spores. At least three biological replicates were performed using at least two independent spore preparations. The rest of the sample was processed for Western blot analyses.
SleC cleavage analysis.
SleC cleavage was induced as previously described (22) with 1% taurocholate (19 mM) for 20 min at 37°C.
Optical density analysis of spore germination.
Germination was induced as previously described (24). Briefly, spores in BHIS were exposed to 1% taurocholate (19 mM), and the OD600 was measured every 3 min for 45 min. The ratio of the OD600 measured for the TA-treated sample to that of the untreated sample for a given time point was calculated and then divided by the ratio determined at 0 min.
Germinant titration analyses.
Responsiveness of spores to low taurocholate concentrations was measured by resuspending ~4 × 107 spores (~1.4 OD600 units) in 160 µl of water. Two hundred microliters of BHIS was added to each spore suspension, and 90-µl aliquots of this suspension were added to 10 µl of either water, 1% TA, or 5% TA (to give a final concentration of 0.025% TA, 0.1% TA, or 0.5% TA, respectively). To determine responsiveness of spores to high taurocholate concentrations, ~5 × 107 spores (1.7 OD600 units) were resuspended in 280 µl of BHIS, and 50-µl aliquots of this suspension were added to 50 µl of 2%, 5%, 10%, or 20% TA (to give final concentrations of 1%, 2.5%, 5%, and 10% TA, respectively). The samples were incubated for 20 min at 37°C, and a 10 -µl aliquot was removed for 10-fold serial dilutions into PBS. Ten-microliter dilutions were plated onto BHIS to determine the number of spores that had initiated germination and on BHIS-TA plates to determine the maximum level of spore germination.
Monitoring single-spore germination by phase-contrast microscopy.
The germination of multiple individual spores was monitored by phase-contrast microscopy as described previously (46). In brief, spores were spread on the surface of a coverslip, and the coverslips were sealed and mounted to a microscope sample holder kept at a constant temperature. Preheated germinant solution was added to the spores on the coverslips, and phase-contrast images of spores were recorded at a rate of 1 frame per 15 s for 60 to 120 min by a digital charge-coupled device camera (16 bits; 1,600 by 1,200 pixels). The averaged pixel intensity of an area of 20 by 20 pixels covering each individual spore was calculated, and the image intensity of each individual spore was plotted as a function of the incubation time with a resolution of 15 s. The initial image intensity at the first time of measurement, T0, was normalized to 1, and the intensity at the end of the measurement period was normally set at 0. Invariably, the image intensity was constant for >10 min at the end of measurements. The degree of germination of spore populations was measured by simultaneously monitoring the germination of >200 individual spores by phase-contrast microscopy.
Assessment of GerG-His6 gel formation.
GerG-His6 variants were purified on Ni2+ affinity resin as previously described from 1 liter of 2YT culture (21). Briefly, culture pellets were resuspended in 25 ml lysis buffer (500 mM NaCl, 50 mM Tris-HCl, pH 7.5, 15 mM imidazole, 10% [vol/vol] glycerol) and flash frozen in liquid nitrogen. The sample was lysed by sonication, and the lysate was cleared by centrifugation prior to incubation with 1.0 ml nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (5 Prime) for 2 to 3 h. After the resin was washed with lysis buffer, the His6-tagged proteins were eluted into 325 µl of high-imidazole buffer (500 mM NaCl, 50 mM Tris, pH 7.5, 175 mM imidazole, 10% glycerol) and nutated for 5 to 10 min. Samples were pelleted at low speed, and the supernatant was removed without disturbing the pelleted resin. This procedure was repeated four times, and the concentration of protein in the supernatants was measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific); the high-imidazole buffer was used as the blank. The samples were incubated at 4°C and periodically inverted to assess gel formation. Samples were scored as forming a gel if the sample remained at the bottom of the tube when inverted and shaken. The high-imidazole buffer was stored as a stock solution at 4°C for >1 year; no gel formation was observed in the buffer during this time. Gel formation of the Ger-His6 variants was assessed in at least two independent protein purifications.
ACKNOWLEDGMENTS
We thank Joseph Sorg (Texas A&M) for generously sharing the anti-CspA antibody prior to its publication, N. Minton (University of Nottingham) for generously providing us with access to the 630ΔermΔpyrE strain and pMTL-YN1C and pMTL-YN3 plasmids for allele-coupled exchange (ACE), Marcin Dembek for directly providing these materials to us and sharing his specific protocols on ACE with us, Allan Drummond (University of Chicago) for insightful discussions on protein aggregation and gel formation, Yuzo Kevorkian for constructing the ΔcspBAC and cspBAC complementation strains, and Megan Touchette for careful reading of the manuscript.
Funding Statement
M.L.D. was supported by the Nicole J. Ferland award. Research in this paper was funded by R21AI26067 from the National Institute of Allergy and Infectious Diseases (NIAID) to Y.L., P.S., and A.S. Research in this paper benefitted from funding by award number R01GM108684 from the National Institute of General Medical Sciences (NIGMS) to A.S. A.S. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts. The content is solely the responsibility of the author(s) and does not necessarily reflect the views of the Pew Charitable Trusts, NIAID, NIGMS, or the National Institutes of Health. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Donnelly ML, Li W, Li Y, Hinkel L, Setlow P, Shen A. 2017. A Clostridium difficile-specific, gel-forming protein required for optimal spore germination. mBio 8:e02085-16. https://doi.org/10.1128/mBio.02085-16.
REFERENCES
- 1.Davies KA, Ashwin H, Longshaw CM, Burns DA, Davis GL, Wilcox MH, EUCLID Study Group. . 2016. Diversity of Clostridium difficile PCR ribotypes in Europe: results from the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID), 2012 and 2013. Euro Surveill 21:30294. doi: 10.2807/1560-7917.ES.2016.21.29.30294. [DOI] [PubMed] [Google Scholar]
- 2.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janezic S, Zidaric V, Pardon B, Indra A, Kokotovic B, Blanco JL, Seyboldt C, Diaz CR, Poxton IR, Perreten V, Drigo I, Jiraskova A, Ocepek M, Weese JS, Songer JG, Wilcox MH, Rupnik M. 2014. International Clostridium difficile animal strain collection and large diversity of animal associated strains. BMC Microbiol 14:173. doi: 10.1186/1471-2180-14-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, Harmsen WS, Zinsmeister AR. 2012. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol 107:89–95. doi: 10.1038/ajg.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moono P, Foster NF, Hampson DJ, Knight DR, Bloomfield LE, Riley TV. 2016. Clostridium difficile infection in production animals and avian species: a review. Foodborne Pathog Dis 13:647–655. doi: 10.1089/fpd.2016.2181. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/drugresistance/threat-report-2013/. [Google Scholar]
- 7.Abt MC, McKenney PT, Pamer EG. 2016. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol 14:609–620. doi: 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theriot CM, Bowman AA, Young VB. 2016. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1:e00045-15. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theriot CM, Young VB. 2015. Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu Rev Microbiol 69:445–461. doi: 10.1146/annurev-micro-091014-104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giel JL, Sorg JA, Sonenshein AL, Zhu J. 2010. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS One 5:e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koenigsknecht MJ, Theriot CM, Bergin IL, Schumacher CA, Schloss PD, Young VB. 2015. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun 83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li JZ, Young VB. 2014. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun 5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorg JA, Sonenshein AL. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol 190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Setlow P. 2003. Spore germination. Curr Opin Microbiol 6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Setlow P. 2014. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol 196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharjee D, McAllister KN, Sorg JA. 2016. Germinants and their receptors in clostridia. J Bacteriol 198:2767–2775. doi: 10.1128/JB.00405-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol 19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, Rigden DJ. 2012. Genomic determinants of sporulation in bacilli and clostridia: towards the minimal set of sporulation-specific genes. Environ Microbiol 14:2870–2890. doi: 10.1111/j.1462-2920.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francis MB, Allen CA, Shrestha R, Sorg JA. 2013. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog 9:e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams CM, Eckenroth BE, Putnam EE, Doublié S, Shen A. 2013. Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog 9:e1003165. doi: 10.1371/journal.ppat.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fimlaid KA, Jensen O, Donnelly ML, Francis MB, Sorg JA, Shen A. 2015. Identification of a novel lipoprotein regulator of Clostridium difficile spore germination. PLoS Pathog 11:e1005239. doi: 10.1371/journal.ppat.1005239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis MB, Allen CA, Sorg JA. 2015. Spore cortex hydrolysis precedes DPA release during Clostridium difficile spore germination. J Bacteriol 197:2276–2283. doi: 10.1128/JB.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donnelly ML, Fimlaid KA, Shen A. 2016. Characterization of Clostridium difficile spores lacking either SpoVAC or dipicolinic acid synthetase. J Bacteriol 198:1694–1707. doi: 10.1128/JB.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paredes-Sabja D, Setlow P, Sarker MR. 2009. The protease CspB is essential for initiation of cortex hydrolysis and dipicolinic acid (DPA) release during germination of spores of Clostridium perfringens type A food poisoning isolates. Microbiology 155:3464–3472. doi: 10.1099/mic.0.030965-0. [DOI] [PubMed] [Google Scholar]
- 26.Shimamoto S, Moriyama R, Sugimoto K, Miyata S, Makino S. 2001. Partial characterization of an enzyme fraction with protease activity which converts the spore peptidoglycan hydrolase (SleC) precursor to an active enzyme during germination of Clostridium perfringens S40 spores and analysis of a gene cluster involved in the activity. J Bacteriol 183:3742–3751. doi: 10.1128/JB.183.12.3742-3751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kevorkian Y, Shirley DJ, Shen A. 2016. Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain. Biochimie 122:243–254. doi: 10.1016/j.biochi.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yutin N, Galperin MY. 2013. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol 15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, Shen A. 2013. Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet 9:e1003660. doi: 10.1371/journal.pgen.1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pishdadian K, Fimlaid KA, Shen A. 2015. SpoIIID-mediated regulation of sigma(K) function during Clostridium difficile sporulation. Mol Microbiol 95:189–208. doi: 10.1111/mmi.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira FC, Saujet L, Tomé AR, Serrano M, Monot M, Couture-Tosi E, Martin-Verstraete I, Dupuy B, Henriques AO. 2013. The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet 9:e1003782. doi: 10.1371/journal.pgen.1003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng YK, Ehsaan M, Philip S, Collery MM, Janoir C, Collignon A, Cartman ST, Minton NP. 2013. Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS One 8:e56051. doi: 10.1371/journal.pone.0056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heap JT, Pennington OJ, Cartman ST, Minton NP. 2009. A modular system for clostridium shuttle plasmids. J Microbiol Methods 78:79–85. doi: 10.1016/j.mimet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. 2010. The ClosTron: mutagenesis in clostridium refined and streamlined. J Microbiol Methods 80:49–55. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 35.Shen A, Fimlaid KA, Pishdadian K. 2016. Inducing and quantifying Clostridium difficile spore formation. Methods Mol Biol 1476:129–142. doi: 10.1007/978-1-4939-6361-4_10. [DOI] [PubMed] [Google Scholar]
- 36.Dembek M, Barquist L, Boinett CJ, Cain AK, Mayho M, Lawley TD, Fairweather NF, Fagan RP. 2015. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio 6:e02383-14. doi: 10.1128/mBio.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burns DA, Heap JT, Minton NP. 2010. SleC is essential for germination of Clostridium difficile spores in nutrient-rich medium supplemented with the bile salt taurocholate. J Bacteriol 192:657–664. doi: 10.1128/JB.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carr KA, Janes BK, Hanna PC. 2010. Role of the gerP operon in germination and outgrowth of Bacillus anthracis spores. PLoS One 5:e9128. doi: 10.1371/journal.pone.0009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butzin XY, Troiano AJ, Coleman WH, Griffiths KK, Doona CJ, Feeherry FE, Wang G, Li YQ, Setlow P. 2012. Analysis of the effects of a gerP mutation on the germination of spores of Bacillus subtilis. J Bacteriol 194:5749–5758. doi: 10.1128/JB.01276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behravan J, Chirakkal H, Masson A, Moir A. 2000. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect access of germinants to their targets in spores. J Bacteriol 182:1987–1994. doi: 10.1128/JB.182.7.1987-1994.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen Thi Minh H, Durand A, Loison P, Perrier-Cornet JM, Gervais P. 2011. Effect of sporulation conditions on the resistance of Bacillus subtilis spores to heat and high pressure. Appl Microbiol Biotechnol 90:1409–1417. doi: 10.1007/s00253-011-3183-9. [DOI] [PubMed] [Google Scholar]
- 42.Ramirez-Peralta A, Zhang P, Li YQ, Setlow P. 2012. Effects of sporulation conditions on the germination and germination protein levels of Bacillus subtilis spores. Appl Environ Microbiol 78:2689–2697. doi: 10.1128/AEM.07908-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Voort M, Abee T. 2013. Sporulation environment of emetic toxin-producing Bacillus cereus strains determines spore size, heat resistance and germination capacity. J Appl Microbiol 114:1201–1210. doi: 10.1111/jam.12118. [DOI] [PubMed] [Google Scholar]
- 44.Cabrera-Martinez RM, Tovar-Rojo F, Vepachedu VR, Setlow P. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J Bacteriol 185:2457–2464. doi: 10.1128/JB.185.8.2457-2464.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart KA, Yi X, Ghosh S, Setlow P. 2012. Germination protein levels and rates of germination of spores of Bacillus subtilis with overexpressed or deleted genes encoding germination proteins. J Bacteriol 194:3156–3164. doi: 10.1128/JB.00405-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong L, Zhang P, Wang G, Yu J, Setlow P, Li YQ. 2011. Characterization of bacterial spore germination using phase-contrast and fluorescence microscopy, Raman spectroscopy and optical tweezers. Nat Protoc 6:625–639. doi: 10.1038/nprot.2011.307. [DOI] [PubMed] [Google Scholar]
- 47.Wang S, Shen A, Setlow P, Li YQ. 2015. Characterization of the dynamic germination of individual Clostridium difficile spores using Raman spectroscopy and differential interference contrast microscopy. J Bacteriol 197:2361–2373. doi: 10.1128/JB.00200-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang G, Zhang P, Paredes-Sabja D, Green C, Setlow P, Sarker MR, Li YQ. 2011. Analysis of the germination of individual Clostridium perfringens spores and its heterogeneity. J Appl Microbiol 111:1212–1223. doi: 10.1111/j.1365-2672.2011.05135.x. [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Zhang P, Setlow P, Li YQ. 2011. Kinetics of germination of wet-heat-treated individual spores of Bacillus species, monitored by Raman spectroscopy and differential interference contrast microscopy. Appl Environ Microbiol 77:3368–3379. doi: 10.1128/AEM.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen D, Huang SS, Li YQ. 2006. Real-time detection of kinetic germination and heterogeneity of single Bacillus spores by laser tweezers Raman spectroscopy. Anal Chem 78:6936–6941. doi: 10.1021/ac061090e. [DOI] [PubMed] [Google Scholar]
- 51.Weingarden AR, Dosa PI, DeWinter E, Steer CJ, Shaughnessy MK, Johnson JR, Khoruts A, Sadowsky MJ. 2016. Changes in colonic bile acid composition following fecal microbiota transplantation are sufficient to control Clostridium difficile germination and growth. PLoS One 11:e0147210. doi: 10.1371/journal.pone.0147210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halfmann R, Alberti S, Krishnan R, Lyle N, O’Donnell CW, King OD, Berger B, Pappu RV, Lindquist S. 2011. Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol Cell 43:72–84. doi: 10.1016/j.molcel.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, Shelyakin PV, Gelfand MS, Dupuy B, Henriques AO, Martin-Verstraete I. 2013. Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet 9:e1003756. doi: 10.1371/journal.pgen.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyata S, Kozuka S, Yasuda Y, Chen Y, Moriyama R, Tochikubo K, Makino S. 1997. Localization of germination-specific spore-lytic enzymes in Clostridium perfringens S40 spores detected by immunoelectron microscopy. FEMS Microbiol Lett 152:243–247. doi: 10.1111/j.1574-6968.1997.tb10434.x. [DOI] [PubMed] [Google Scholar]
- 55.Bhattacharjee D, Francis MB, Ding X, McAllister KN, Shrestha R, Sorg JA. 2015. Reexamining the germination phenotypes of several Clostridium difficile strains suggests another role for the CspC germinant receptor. J Bacteriol 198:777–786. doi: 10.1128/JB.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ottmann C, Rose R, Huttenlocher F, Cedzich A, Hauske P, Kaiser M, Huber R, Schaller A. 2009. Structural basis for Ca2+-independence and activation by homodimerization of tomato subtilase 3. Proc Natl Acad Sci U S A 106:17223–17228. doi: 10.1073/pnas.0907587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banawas S, Korza G, Paredes-Sabja D, Li Y, Hao B, Setlow P, Sarker MR. 2015. Location and stoichiometry of the protease CspB and the cortex-lytic enzyme SleC in Clostridium perfringens spores. Food Microbiol 50:83–87. doi: 10.1016/j.fm.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 58.Muralidharan V, Goldberg DE. 2013. Asparagine repeats in Plasmodium falciparum proteins: good for nothing? PLoS Pathog 9:e1003488. doi: 10.1371/journal.ppat.1003488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN. 2010. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta 1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradović Z. 2002. Intrinsic disorder and protein function. Biochemistry 41:6573–6582. [DOI] [PubMed] [Google Scholar]
- 61.Dyson HJ, Wright PE. 2002. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol 12:54–60. doi: 10.1016/S0959-440X(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 62.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. 2009. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lancaster AK, Nutter-Upham A, Lindquist S, King OD. 2014. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30:2501–2502. doi: 10.1093/bioinformatics/btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carlson PE Jr, Walk ST, Bourgis AE, Liu MW, Kopliku F, Lo E, Young VB, Aronoff DM, Hanna PC. 2013. The relationship between phenotype, ribotype, and clinical disease in human Clostridium difficile isolates. Anaerobe 24:109–116. doi: 10.1016/j.anaerobe.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore P, Kyne L, Martin A, Solomon K. 2013. Germination efficiency of clinical Clostridium difficile spores and correlation with ribotype, disease severity and therapy failure. J Med Microbiol 62:1405–1413. doi: 10.1099/jmm.0.056614-0. [DOI] [PubMed] [Google Scholar]
- 66.Oka K, Osaki T, Hanawa T, Kurata S, Okazaki M, Manzoku T, Takahashi M, Tanaka M, Taguchi H, Watanabe T, Inamatsu T, Kamiya S. 2012. Molecular and microbiological characterization of Clostridium difficile isolates from single, relapse, and reinfection cases. J Clin Microbiol 50:915–921. doi: 10.1128/JCM.05588-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carlson PE Jr, Kaiser AM, McColm SA, Bauer JM, Young VB, Aronoff DM, Hanna PC. 2015. Variation in germination of Clostridium difficile clinical isolates correlates to disease severity. Anaerobe 33:64–70. doi: 10.1016/j.anaerobe.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Târraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 70.Collery MM, Kuehne SA, McBride SM, Kelly ML, Monot M, Cockayne A, Dupuy B, Minton NP. 2016. What’s a SNP between friends: the influence of single nucleotide polymorphisms on virulence and phenotypes of Clostridium difficile strain 630 and derivatives. Virulence doi: 10.1080/21505594.2016.1237333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sorg JA, Dineen SS. 2009. Laboratory maintenance of Clostridium difficile. Curr Protoc Microbiol Chapter 9:Unit 9A.1. doi: 10.1002/9780471729259.mc09a01s12. [DOI] [PubMed] [Google Scholar]
- 72.Cartman ST, Minton NP. 2010. A mariner-based transposon system for in vivo random mutagenesis of Clostridium difficile. Appl Environ Microbiol 76:1103–1109. doi: 10.1128/AEM.02525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Permpoonpattana P, Tolls EH, Nadem R, Tan S, Brisson A, Cutting SM. 2011. Surface layers of Clostridium difficile endospores. J Bacteriol 193:6461–6470. doi: 10.1128/JB.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Putnam EE, Nock AM, Lawley TD, Shen A. 2013. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J Bacteriol 195:1214–1225. doi: 10.1128/JB.02181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fimlaid KA, Jensen O, Donnelly ML, Siegrist MS, Shen A. 2015. Regulation of Clostridium difficile spore formation by the SpoIIQ and SpoIIIA proteins. PLoS Genet 11:e1005562. doi: 10.1371/journal.pgen.1005562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Construction of gerG (CD0311) and sleC mutants. Download FIG S1, PDF file, 0.7 MB (755.4KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A TargeTron insertion in CD0311 in the JIR8094 strain background leads to a heat resistance defect. Download FIG S2, PDF file, 0.4 MB (428.3KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GerG is required for optimal spore germination in JIR8094. Download FIG S3, PDF file, 0.4 MB (369.8KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overexpression of cspBAC fails to rescue the germination defect of gerG-negative spores in JIR8094. Download FIG S4, PDF file, 0.5 MB (484.5KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Artificial germination bypasses the need for GerG. Download FIG S5, PDF file, 0.1 MB (106.4KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Responsiveness of gerG mutant spores to germinant. Download FIG S6, PDF file, 0.5 MB (475.6KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ClustalW alignment of expanded GerG homologs. Download FIG S7, PDF file, 0.1 MB (88.7KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PONDR plot analysis of GerG. Download FIG S8, PDF file, 0.3 MB (301.6KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PLAAC analysis of GerG. Download FIG S9, PDF file, 0.4 MB (392.9KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods and references. Download TEXT S1, PDF file, 0.1 MB (146.9KB, pdf) .
Copyright © 2017 Donnelly et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.