Abstract
Agrobacterium tumefaciens uses a type IV secretion system (T4SS) to transfer T-DNA and virulence proteins to plants. The T4SS is composed of two major structural components: the T-pilus and a membrane-associated complex that is responsible for translocating substrates across both bacterial membranes. VirB2 protein is the major component of the T-pilus. We used the C-terminal–processed portion of VirB2 protein as a bait to screen an Arabidopsis thaliana cDNA library for proteins that interact with VirB2 in yeast. We identified three related plant proteins, VirB2-interacting protein (BTI) 1 (BTI1), BTI2, and BTI3 with unknown functions, and a membrane-associated GTPase, AtRAB8. The three BTI proteins also interacted with VirB2 in vitro. Preincubation of Agrobacterium with GST-BTI1 protein decreased the transformation efficiency of Arabidopsis suspension cells by Agrobacterium. Transgenic BTI and AtRAB8 antisense and RNA interference Arabidopsis plants are less susceptible to transformation by Agrobacterium than are wild-type plants. The level of BTI1 protein is transiently increased immediately after Agrobacterium infection. In addition, overexpression of BTI1 protein in transgenic Arabidopsis results in plants that are hypersusceptible to Agrobacterium-mediated transformation. Confocal microscopic data indicate that GFP-BTI proteins preferentially localize to the periphery of root cells in transgenic Arabidopsis plants, suggesting that BTI proteins may contact the Agrobacterium T-pilus. We propose that the three BTI proteins and AtRAB8 are involved in the initial interaction of Agrobacterium with plant cells.
INTRODUCTION
Agrobacterium tumefaciens is a Gram-negative soil bacterium that can genetically transform cells of numerous dicot plant species, as well as some monocots and gymnosperms (DeCleene and DeLey, 1976). Agrobacterium can additionally transform numerous fungal species, including yeasts, ascomycetes, and basidiomycetes, as well as human cells (Bundock et al., 1995, 1999; Piers et al., 1996; de Groot et al., 1998; Gouka et al., 1999; Kunik et al., 2001). Agrobacterium elicits crown gall disease, an agronomically important disease that affects many plant species. The uncontrolled proliferation of crown gall tumors results from the transfer, integration, and expression of oncogenes encoded by the T-DNA (transferred DNA) region of the Ti-plasmid (tumor-inducing plasmid) normally resident in the bacterium. Plant transformation by Agrobacterium requires the presence of two genetic components located on the Ti-plasmid: (1) T-DNA, the genetic entity transferred into the plant genome, and (2) the virulence (vir) region composed of several loci encoding proteins mediating T-DNA processing and transfer (Zupan et al., 2000; Tzfira and Citovsky, 2002; Gelvin, 2003).
Virulence gene expression results in the production and transport of a single-stranded T-DNA (T-strand) into the host cell. Agrobacterium uses a type IV secretion system (T4SS) to deliver T-strands and several Vir proteins across the bacterial envelope; this process resembles bacterial conjugation. The type IV secretion system of Agrobacterium is encoded by 11 virB genes and virD4 that form two functional components: a filamentous pilus and a membrane-associated transporter complex (reviewed in Christie, 1997, 2001; Zupan et al., 1998; Lai and Kado, 2000; Kado, 2000; Cascales and Christie, 2003, 2004; Ding et al., 2003).
The T-pilus is a flexuous filamentous appendage that is essential for Agrobacterium virulence. Biogenesis of the T-pilus requires all 11 VirB proteins, but not VirD4 protein (Lai et al., 2000; Lai and Kado, 2002). There are at least three VirB proteins that compose the T-pilus. VirB5 and VirB7 form minor components (Schmidt-Eisenlohr et al., 1999; Sagulenko et al., 2001; Krall et al., 2002). The major component, VirB2, is translated as a 12.3-kD pro-pilin protein. A signal peptide of 47 amino acids is cleaved by a signal peptidase, generating a 7.2-kD pilin protein (Jones et al., 1996; Eisenbrandt et al., 1999; Lai et al., 2000, 2002). The 74–amino acid T-pilin is further cyclized (Eisenbrandt et al., 1999). Genetic and environmental studies showed that each virB gene is required for T-pilus biogenesis; however, only virB2 is required for pro-pilin processing and peptide cyclization (Eisenbrandt et al., 1999; Lai et al., 2000). Nonpolar virB mutants abolish T-pilin transport and T-pilus biogenesis. These experiments support a model that the VirB-specific transporter is not only used for translocating the T-complex, but also for exporting cyclic T-pilin subunits to the bacterial cell surface, perhaps as a scaffold to facilitate efficient assembly of the T-pilus (Christie, 1997; Lai and Kado, 2000).
Although the T-pilus is essential for T-DNA transfer and virulence, the molecular mechanism by which T-DNA and virulence proteins are transferred from Agrobacterium into the host cells remains unknown. An intriguing question arises as to whether the T-pilus is directly involved in the transfer of T-DNA and Vir proteins, or whether the T-pilus is only indirectly involved by mediating close cell-to-cell contact for intimate mating-pair formation. Several possible functions could be assigned to the T-pilus. First, the T-pilus could serve as a conduit for export of several components needed for virulence, including T-pilin subunits, VirE2, VirE3, VirF proteins, and single-stranded T-DNA that is piloted by the linked VirD2 protein. A recent study by Cascales and Christie (2004) demonstrated the direct contact of six of the 12 components of the VirB/D4 system of Agrobacterium, including VirB2, with the T-strand. Cascales and Christie (2004) further established the temporal order by which VirB proteins interact with the DNA as it translocates through the VirB/D4 apparatus. Second, the T-pilus could serve as a bridge to bring the bacterium and the host cell into close proximity while T-DNA is transferred into the host cell through some other transfer apparatus (Lai and Kado, 2000; Kelly and Kado, 2002). For example, Kelly and Kado (2002) described a coiled thread-like interconnection between Agrobacterium and Streptomyces lividans cells during Agrobacterium transformation. This interconnecting structure is dependent on virB genes and appears only under the same conditions as those required for T-pilus formation. A model derived from this observation is that the T-pilus may retract and subsequently draw the bacterial cell into sufficiently close contact with the host cell to permit T-DNA and Vir protein transfer to the recipient cell. Third, the T-pilus could serve as a sensor for potential mating-signal molecules from the host cell. In this regard, it would be interesting to determine whether host cell factors mediate the interaction between the donor bacterium and the recipient host during Agrobacterium-mediated transformation. Plant cells may have a receptor for the T-pilus or a pore for T-DNA transfer through the plant cell wall and plasma membrane. Escherichia coli uses a T4SS to conjugate plasmids between bacterial cells. Although proteins or lipopolysaccharides that interact with the F-pilus during conjugation of F-like plasmids have been identified (Anthony et al., 1994), no corresponding plant components for the Agrobacterium T-pilus have yet been demonstrated.
In this study, we identify plant-encoded proteins that may mediate Agrobacterium T-pilus interaction with the host cell. Using yeast two-hybrid and in vitro assays, we identified two classes of Arabidopsis proteins that interact with VirB2 protein. The first class is composed of three related proteins with previously unknown function; we term these proteins VirB2-interacting proteins (BTI). The second class is the Arabidopsis GTPase AtRAB8. Antisense (A/S), RNA interference (RNAi), and T-DNA mutagenesis experiments demonstrated that BTI and AtRAB8 proteins are involved in the early stages of Agrobacterium-mediated root transformation.
RESULTS
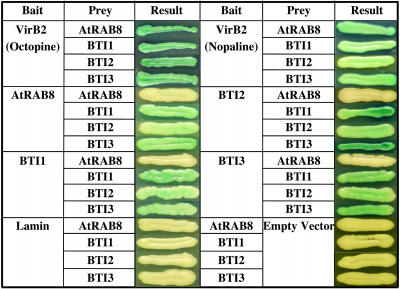
Four Arabidopsis Proteins Interact with Processed VirB2 in Yeast
We constructed a bait plasmid, pE2180, as a translational fusion of processed VirB2 (amino acids 48 to 121) fused to the LexA coding sequence of the plasmid pSST91. pE2180 alone was unable to activate transcription of the lacZ reporter gene in yeast strain YB2. We introduced into YB2 an Arabidopsis cDNA library (Ballas and Citovsky, 1997) and screened for blue colonies on selective medium containing X-Gal. We screened prey plasmids from positive colonies for interaction with the unrelated bait plasmid pSST-lamin and eliminated these interacting clones from consideration. From ∼3 × 106 primary transformants, we identified and sequenced ∼80 remaining prey plasmids. Based on DNA sequence analysis and BLASTX search results, we recurrently identified two classes of plant proteins. The first class contains the VirB2-interacting proteins composed of a three-member family of previously unknown function (BTI1 [RTNLB1]: At4g23630, BTI2 [RTNLB2]: At4g11220, BTI3 [RTNLB4]: At5g41600); the second class is the Ras-related small GTPase, AtRAB8 (At3g53610).
Interactions among BTI Proteins, AtRAB8, and Vir Proteins in Yeast
We further examined whether BTI and AtRAB8 proteins interact with VirB5, a minor component of the T-pilus, and with other virulence proteins VirB1, VirB1*, VirD2, VirE1, VirE2, and VirF. Supplemental Figure 1 online shows that the plant proteins BTI1, BTI2, BTI3, and AtRAB8 interact in yeast only with VirB2 but not with VirB1, VirB1*, VirB5, VirD2, VirE1, VirE2, or VirF. The C-terminal 47–amino acid regions of VirB2 from octopine (pTiA6)- and nopaline (pTiC58)-type Ti-plasmids are nearly identical. Consequently, the three BTI and AtRAB8 proteins interact with both of these VirB2 proteins in yeast (Figure 1).
Figure 1.
AtRAB8 and the Three BTI Proteins Interact with Processed VirB2 in Yeast.
Virulence proteins were tested for interaction with BTI proteins or AtRAB8 using a two-hybrid system as described in Methods. In addition, the three BTI proteins were tested for interaction with themselves, each other, and AtRAB8. Note that AtRAB8 interacts with the BTI proteins when used as a bait, but not when used as a prey protein. AtRAB8 does not show interaction with itself.
In addition, we tested interactions in yeast among the AtRAB8 and BTI proteins. As shown in Figure 1, the three BTI proteins interacted with each other and with themselves. AtRAB8 as a bait interacts with all three BTI proteins, but not with itself. These results suggest that the BTI and AtRAB8 proteins may form a complex in the plant cell. However, AtRAB8 as a prey protein did not interact with the BTI proteins. These findings may result from different conformations of the bait and prey fusion proteins in yeast, as previously reported for interactions between VirB7 and VirB9 (Baron et al., 1997; Ward et al., 2002).
BTI Proteins Interact with VirB2, with Each Other, and with AtRAB8 in Vitro
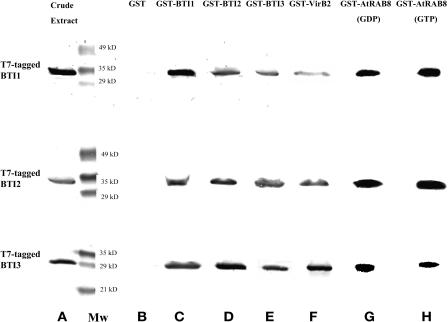
To determine whether VirB2 interacts directly with the BTI proteins, we performed glutathione-S-transferase (GST) pull-down assays using a GST-VirB2 fusion protein. We incubated lysates from E. coli expressing T7-tagged BTI proteins with a GST-VirB2 fusion protein that had been linked to glutathione-sepharose beads. After extensive washing, bound proteins were eluted and used for protein gel blot analyses with anti-T7 tag antibodies. As a negative control, GST linked to the glutathione column was similarly treated with T7-tagged proteins. Figure 2 shows the results of these experiments. All three BTI proteins interacted with the GST-VirB2 fusion protein, but not GST, in vitro. These data confirm direct interaction of VirB2 protein with the three BTI proteins. Recombinant AtRAB8 was highly insoluble when fused to a T7 tag; we therefore did not investigate its in vitro interaction with VirB2.
Figure 2.
GST-VirB2, the Three BTI Proteins, and AtRAB8 Interact with Each Other in Vitro.
Crude lysates of E. coli that contained either GST-VirB2, GST tags on the various BTI proteins, or GST-AtRAB8 were incubated with glutathione-sepharose beads, washed with binding buffer, then incubated with T7-tagged versions of BTI1, BTI2, or BTI3. After washing the beads, the bound proteins were eluted with glutathione, separated by SDS-PAGE, and protein gel blot analysis performed using anti-T7 tag antibodies. (Lane A) Crude extracts from E. coli individually expressing the T7-tagged BTI proteins analyzed using T7-tag antibodies; (Lane B) the three T7-tagged BTI proteins individually incubated with GST; (Lanes C, D, and E) the three BTI proteins individually incubated with themselves and with each other; (Lane F) GST-VirB2 protein individually incubated with the three BTI proteins; (Lane G) GST-AtRAB8 (supplemented with GDP) protein individually incubated with the three BTI proteins; (Lane H) GST-AtRAB8 (supplemented with GTP) protein individually incubated with the three BTI proteins. Mw, molecular weight markers.
To investigate further the in vitro interactions among these VirB2-interacting plant proteins, we linked GST fusions of each of the BTI proteins and AtRAB8 to glutathione-sepharose beads and incubated them with lysates from E. coli expressing T7-tagged versions of the various BTI proteins. Figure 2 shows that the three BTI proteins interacted with each other and with themselves in vitro. In addition, all three T7-tagged BTI proteins interacted with either the GTP or GDP form of GST-AtRAB8. These data indicate that each of the three BTI proteins can interact with themselves and with AtRAB8 in vitro, and further suggest the possibility that these proteins may form a complex in plants.
Mapping BTI Protein Domains Necessary for Interaction with VirB2, BTI, and AtRAB8 in Yeast
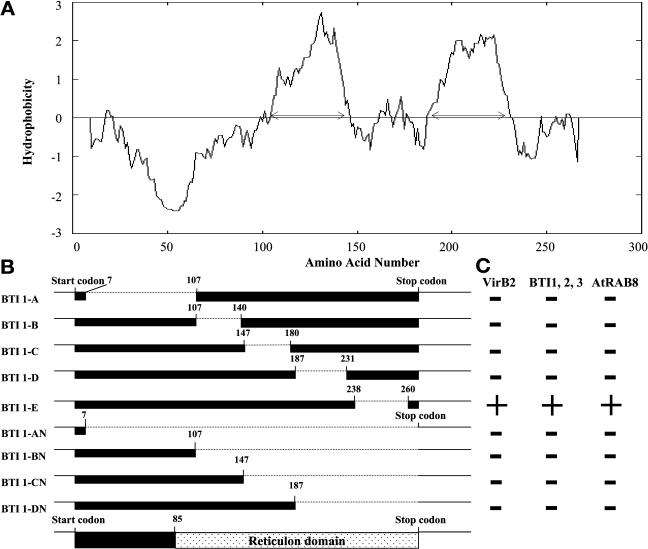
Figure 3 shows that the three BTI proteins share 56% amino acid similarity. All three proteins contain a C-terminal 150– to 201–amino acid reticulon (RTN) homology domain (RHD; Pfam PF02453; Oertle and Schwab, 2003; Oertle et al., 2003) comprising two large hydrophobic regions with an ∼66–amino acid loop in between, as determined by the BLASTP and RPSBLAST programs.
Figure 3.
Protein Sequence Alignment of the Three BTI Proteins Based on the ClustalW Program (Thompson et al., 1994).
The asterisks indicate identical amino acid residues. The three BTI proteins share 56% amino acid similarity.
Based on protein sequence analyses using the Prediction of Transmembrane Regions and Orientation program (Hofmann and Stoffel, 1993), the three BTI proteins have two or three putative transmembrane domains (Figure 4A). To determine which domain(s) of BTI1 is involved in the interaction between BTI1 and VirB2, we generated a series of BTI1 protein-deletion mutants (Figure 4B) and performed yeast two-hybrid assays using these truncated BTI1 proteins as preys and processed VirB2 protein as a bait. The BTI1-AN mutant, containing only the first seven amino acids of BTI1, served as a negative control. Of the nine truncated BTI1 proteins tested, only mutant BTI1-E interacted with VirB2, BTI1, BTI2, BTI3, and AtRAB8 (Figure 4C). Control experiments indicated that the various BTI1 mutant proteins were expressed in yeast (data not shown). These results indicate that only the C-terminal hydrophilic region is dispensable for interaction between BTI1 and the other proteins tested, either because direct interactions do not involve this C-terminal region, or because deletion of the other domains result in conformational changes in BTI1 that no longer permit interaction.
Figure 4.
Mapping BTI Protein Domains Necessary for Interaction with VirB2, BTI, and AtRAB8 in Yeast.
(A) Hydropathy profile of BTI1 protein using the Prediction of Transmembrane Regions and Orientation program (Hofmann and Stoffel, 1993). The C-terminal region of BTI1 protein contains two hydrophobic domains. The N-terminal region of BTI1 has one hydrophilic domain and is variable among the three BTI proteins. The arrows indicate the putative membrane domains of BTI1 protein.
(B) Schematic diagram of the various BTI1 protein-deletion mutants. The dotted lines indicate the deleted region of each mutant protein.
(C) Results of protein-interaction experiments in yeast. Only mutant protein BTI1-E interacts with VirB2, BTI1, BTI2, BTI3, and AtRAB8. Construction of the mutant bti1 genes and the yeast interaction assays are described in Materials and Methods.
Preincubation of Agrobacterium with GST-BTI1 Inhibits Plant Cell Transformation
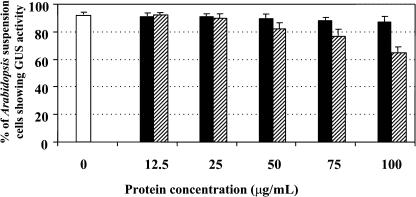
If BTI1 protein directly binds to VirB2 in the T-pilus, we should be able to inhibit plant transformation by saturating the T-pilus with BTI1 protein before cocultivation of the bacteria with plant cells. To test this hypothesis, we first induced VirB2 expression in Agrobacterium with acetosyringone, then incubated the induced cells with none or increasing concentrations of recombinant GST-BTI1 or GST. We subsequently used these bacteria to infect Arabidopsis suspension cells. Figure 5 shows that incubating Agrobacterium with increasing amounts of GST-BTI1 inhibited transformation in a concentration-dependent manner. When induced Agrobacterium cells were preincubated with 100 μg/mL (5.4 × 105 molecules/Agrobacterium cell) GST-BTI1, 68% of the Arabidopsis suspension cells were transformed compared with 93% of the suspension cells that were transformed in the absence of GST-BTI1 protein or in the presence of 100 μg/mL GST (1.1 × 106 molecules/Agrobacterium cell). We further tested whether GST-BTI1 protein affects the viability of Agrobacterium. After 24-h cocultivation, the number of viable Agrobacterium cells incubated with 100 μg/mL GST-BTI1 did not differ from that of bacteria incubated with GST or without recombinant protein (data not shown). These data indicate that the lower transformation efficiency of Arabidopsis suspension cells results from inhibition of infection by GST-BTI1, rather than a toxic effect of the recombinant protein on bacterial viability.
Figure 5.
Preincubation of Agrobacterium with GST-BTI1 Inhibits Arabidopsis Suspension Cell Transformation.
Agrobacterium At849 induced with acetosyringone was used to infect Arabidopsis suspension cells either without pretreatment (open), with bacteria pretreated with GST (solid), or GST-BTI1 (striped) before plant infection. The numbers represent the percentage of Arabidopsis suspension cells that stained blue with X-Gluc. The data are presented as the average of three experiments. Error bars = se.
Transgenic BTI1 Antisense and RNAi Plants Show Reduced Susceptibility to Agrobacterium-Mediated Root Transformation
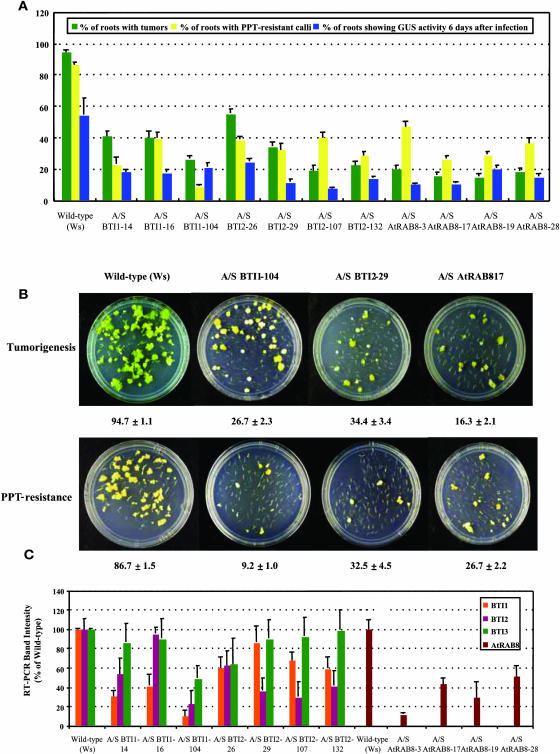
To determine if BTI1 protein plays an important role in Agrobacterium-mediated plant transformation, we generated Arabidopsis plants with a decreased level of BTI1 gene expression and subsequently tested them for susceptibility to Agrobacterium-mediated transformation. Figures 6 and 7 show the results of transformation assays for BTI1 antisense and RNAi transgenic plants, respectively. When we infected root segments with the tumorigenic strain Agrobacterium A208, the A/S and RNAi transgenic plants formed tumors at an efficiency two- to fivefold lower than that of wild-type plants (Figures 6A and 7A; see Supplemental Tables 1 and 2 online). Figures 6B and 7B show root tumorigenesis assays on sets of representative plates. We performed additional stable transformation assays to test the formation of phosphinothricin (ppt)-resistant calli on the roots of transgenic BTI1 A/S plants after infection with Agrobacterium At872 that contains a bar gene on the incoming T-DNA. These transgenic BTI1 A/S plants formed ppt-resistant calli at a two- to 10-fold lower efficiency compared with that of wild-type plants (Figure 6A; see Supplemental Table 1 online). Figure 6B shows herbicide-resistance assays on sets of representative plates.
Figure 6.
Transgenic BTI and AtRAB8 Antisense Plants Are Less Susceptible to Agrobacterium-Mediated Root Transformation Than Are Wild-Type Plants.
(A) T2 generation transgenic BTI1, BTI2, and AtRAB8 antisense plants show lower stable and transient transformation efficiencies than do wild-type plants. Green bars represent the percentage of root segments forming tumors 4 weeks after infection with the tumorigenic strain Agrobacterium A208. Yellow bars represent the percentage of root segments forming phosphinothricin-resistance calli 4 weeks after infection with Agrobacterium At872. Blue bars represent the percentage of root segments showing GUS activity 6 d after infection with Agrobacterium At849. At least 20 independent plants were tested for each transgenic line and >80 root segments were examined for each plant. Error bars = se.
(B) Representative plates of BTI1, BTI2, and AtRAB8 A/S transgenic root segments showing reduced frequency of tumor formation and ppt-resistance. The numbers below each plate indicate the average stable transformation efficiency of each line ±se.
(C) Transgenic BTI1 and BTI2 antisense plants show reduced levels of BTI transcripts. Transgenic AtRAB8 antisense (A/S) plants show reduced levels of AtRAB8 transcripts. Transcript levels of each BTI gene and AtRAB8 in A/S transgenic plants are shown as a relative percentage of that of wild-type plants. Data are shown as average values of two RT-PCR reactions from three T2 generation plants of each line. Note that antisense BTI1 and BTI2 plants show a reduced level of BTI2 and BTI1 transcripts, respectively, as well as reduced levels of transcripts targeted by the specific antisense construction. Note also that antisense BTI1 and BTI2 plants generally show a lesser reduction in BTI3 transcripts. Error bars = se.
Figure 7.
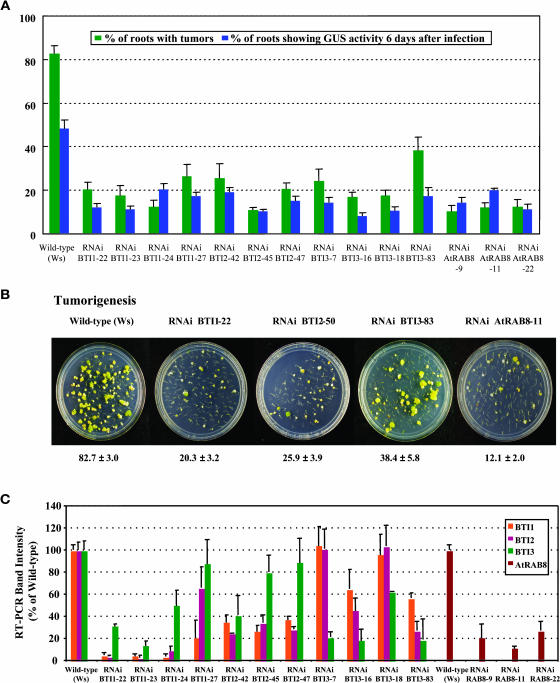
Transgenic BTI and AtRAB8 RNAi Plants Show Reduced Susceptibility to Agrobacterium-Mediated Root Transformation.
(A) T2 generation BTI1, BTI2, BTI3, and AtRAB8 RNAi transgenic plants show lower stable and transient transformation efficiencies compared with wild-type plants. Green bars represent the stable transformation efficiency. The root segments were infected with the tumorigenic strain Agrobacterium A208. Blue bars represent the percentage of root segments showing GUS activity 6 d after infection with Agrobacterium At849. At least 20 independent plants were tested for each transgenic line and >80 root segments were examined for each plant. Error bars = se.
(B) Representative plates of transgenic BTI and AtRAB8 RNAi plants showing resistance to tumor formation. The numbers below each plate indicate the average stable transformation efficiency ±se.
(C) Transgenic BTI RNAi plants show reduced levels of BTI transcripts. Transgenic AtRAB8 RNAi plants show reduced levels of AtRAB8 transcripts. Transcript levels of each BTI gene and AtRAB8 in RNAi transgenic plants are shown as a relative percentage of that of wild-type plants. Data are shown as average values of two RT-PCR reactions from three T2 generation plants of each line. Note that RNAi BTI1, BTI2, and BTI3 plants preferentially show a reduced level of transcripts targeted by the specific construction, whereas other BTI transcripts are reduced to a lesser extent. Error bars = se.
A plant resistant to Agrobacterium-mediated transformation may have transformation blocked at a step inside the nucleus, such as T-DNA integration, or at a step before nuclear import. We therefore tested the susceptibility of the BTI1 A/S and RNAi transgenic plants to transient transformation. Such transformation does not require integration of T-DNA into the plant genome (Mysore et al., 1998). When infected by Agrobacterium At849, harboring a T-DNA carrying a gusA-intron reporter gene (the intron was used to prevent expression of GUS activity in Agrobacterium; Narasimhulu et al., 1996), the A/S and RNAi transgenic plants showed a two- to threefold reduction in transient transformation frequency compared with that of wild-type plants, as indicated by staining for GUS activity using X-Gluc (Figures 6A and 7A; see Supplemental Tables 1 and 2 online). These data suggest that transformation of these transgenic plants may be blocked at an early stage, such as bacterial attachment, T-DNA and Vir protein transfer, and/or nuclear import of T-DNA and Vir proteins.
To investigate whether the transformation-resistance phenotypes correspond to decreased transcript levels of the genes targeted by antisense RNA or RNAi in these transgenic plants, we examined the transcript levels of BTI1, BTI2, and BTI3 in BTI1 A/S and RNAi transgenic plants using RT-PCR with gene-specific primers. Figures 6C and 7C show that in both BTI1 A/S and RNAi transgenic plants, BTI1 transcript levels are lower than those in wild-type plants. Because BTI1 transcripts share sequence similarity with BTI2 and BTI3 transcripts (see Supplemental Figure 2 online), BTI2 and BTI3 transcript levels are also affected by antisense and RNAi constructions targeting BTI1 transcripts.
To examine further the expression levels of BTI1 protein in the BTI1 A/S and RNAi transgenic plants, we performed protein gel blot analysis on protein extracts from root tissues of Arabidopsis plants using an antibody directed against BTI1 protein. Amino acid residues 11 to 25 of BTI1 protein were chosen to synthesize peptide F9 N11-25 (VIAPEPAVEVVERESC). This sequence was chosen from a region that is highly variable among the three BTI proteins and does not correspond to the conserved reticulon domain. Anti-BTI1 antibody does not cross-react with recombinant GST-BTI3 protein and only slightly cross-reacts with BTI2 protein (data not shown). Protein gel blot analysis demonstrated that BTI1 protein levels are greatly reduced in BTI1 A/S and RNAi transgenic plants in comparison with those found in wild-type plants (see Supplemental Figure 4 online). The reduction in BTI1 protein levels in the various transgenic lines is relatively the same as is the reduction in BTI1 transcript levels in these plants (Figures 6C and 7C; see Supplemental Figure 4 online), and correlates with the decrease in their transformation susceptibility. It should be noted, however, that in some BTI1 A/S and RNAi transgenic plants, the antisense or RNAi constructions targeted against the BTI1 mRNA additionally affected the level of BTI2 and BTI3 transcripts. Thus, it is possible that the resistance phenotype of some of these transgenic plants may result from lower expression of more than one BTI gene.
Arabidopsis Plants with T-DNA Insertions in BTI1 Show Reduced Levels of Agrobacterium-Mediated Root Transformation
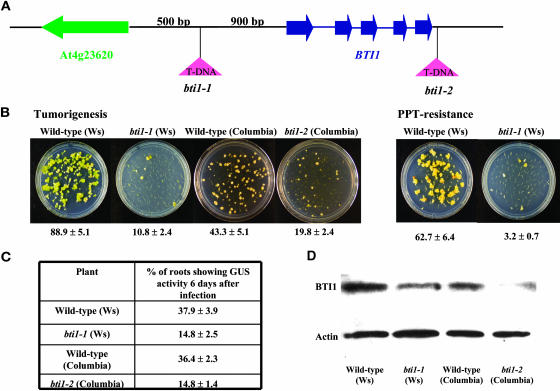
To determine the extent to which the BTI1 gene alone is involved in the Agrobacterium-mediated transformation process, we identified two T-DNA insertions in the BTI1 gene. The first insertion (bti1-1), in ecotype Ws-2, is located 900 bp upstream of the start codon of the BTI1 gene (Figure 8A). We tested the susceptibility of bti1-1 mutant plants to stable transformation. bti1-1 mutants showed lower levels of tumor formation and formed fewer ppt-resistant calli than did wild-type plants (Figure 8B). Tumors that formed on wild-type roots were mainly green teratomas, whereas tumors that formed on bti1-1 mutant plants were generally smaller yellow tumors with a few small green tumors (Figure 8B). We also performed transient transformation assays on bti1-1 mutant plants and found that bti1-1 mutants were less susceptible than were wild-type plants (Figure 8C). These data suggest that bti1-1 mutants may be blocked at an early stage of transformation. Protein gel blot analysis indicated that the level of BTI1 protein is lower in bti1-1 plants compared with that of wild-type plants (Figure 8D), and only the BTI1 transcript level, but not the BTI2 transcript level, is affected by the T-DNA insertion (data not shown). These data suggest that disruption of the BTI1 promoter region results in a lower level of BTI1 gene expression and a concomitant decrease in Agrobacterium-mediated transformation. Analysis of another T-DNA insertion in BTI1, Salk-032220 (bti1-2, ecotype Columbia), similarly showed lower BTI1 protein levels and transformation efficiency compared with that of wild-type plants (Figures 8B and 8C). The results of stable and transient transformation assays with two independent bti1 T-DNA insertion lines indicate that the rat (resistant to Agrobacterium transformation) phenotypes correlate with reduced expression of the BTI1 gene. Additionally, in A/S BTI1-16 transgenic plants, BTI1 transcript levels are relatively low, whereas BTI2 and BTI3 transcript levels are approximately the same as in wild-type plants (Figure 6C). Taken together, these data suggest that BTI1 protein is involved in the Agrobacterium transformation process.
Figure 8.
Arabidopsis Plants with T-DNA Insertions in BTI1 Show Reduced Levels of Agrobacterium-Mediated Root Transformation.
(A) Schematic representation of the region around the Arabidopsis BTI1 gene. In bti1-1 mutant plants, the T-DNA is inserted 900 bp before the start codon of the BTI1 gene. In bti1-2 mutant plants, the T-DNA is inserted 117 bp downstream of the BTI1 stop codon.
(B) bti1-1 and bti1-2 mutant plants are resistant to stable Agrobacterium-mediated root transformation. Representative plates of roots infected with Agrobacterium A208 (for tumorigenesis assays) and Agrobacterium At872 (for ppt-resistance assays) are shown. The numbers below each plate indicate average values of results from 20 individual plants ±se.
(C) bti1-1 and bti1-2 mutant plants show reduced susceptibility to transient transformation. Data are indicated as average values of results from 20 individual plants infected with Agrobacterium At849, ±se.
(D) bti1-1 and bti1-2 T-DNA–insertion mutant plants show reduced levels of BTI1 protein compared with wild-type plants. Proteins were extracted from roots and subjected to protein gel blot analysis as described in Methods. The amount of actin protein was used to show equivalent loading of each lane.
Transgenic BTI2 A/S and RNAi Plants and BTI3 RNAi Plants Are Less Susceptible to Agrobacterium-Mediated Root Transformation
BTI2 and BTI3 proteins interact with VirB2 in yeast and in vitro. In addition, the three BTI proteins interact with each other and with themselves, suggesting that BTI proteins may form a complex. Because no BTI2 and BTI3 T-DNA exon insertion mutants were available, we generated transgenic BTI2 and BTI3 A/S and RNAi plants and tested their susceptibility to Agrobacterium-mediated transformation. Figures 6A and 7A, as well as Supplemental Tables 1 and 2 online, indicate that inhibition of BTI2 and BTI3 expression also results in a rat phenotype.
Because of sequence similarity among the three BTI genes, both BTI1 and BTI3 transcript levels are affected by the BTI2 antisense and RNAi constructions in some of the BTI2 A/S and RNAi transgenic plants. In BTI2 A/S and RNAi plants, BTI1 transcript levels decreased more than that of BTI3. Because the BTI3 cDNA sequence is divergent from that of the BTI1 and BTI2 cDNA sequences, BTI1 and BTI2 transcript levels are less affected by the BTI3 RNAi construction. The resistance phenotypes of the BTI2 A/S and RNAi transgenic plants correlate with reduced expression of the BTI2 gene. Similarly, in BTI3-7 and BTI3-18 RNAi transgenic plants, the resistance phenotypes are correlated with reduced levels of BTI3 transcripts. However, in some of the BTI2 A/S, BTI2, and BTI3 RNAi transgenic plants, the resistance phenotype may result from reduced expression of two or three of the BTI genes.
Transgenic AtRAB8 A/S and RNAi Plants Are Less Susceptible to Agrobacterium-Mediated Root Transformation
Because AtRAB8 protein interacts with VirB2 in yeast, we generated transgenic AtRAB8 A/S and RNAi plants to investigate the involvement of AtRAB8 in the Agrobacterium-mediated transformation process. Figures 6 and 7, as well as Supplemental Tables 1 and 2 online, indicate that reduction in expression of AtRAB8 also results in reduced transformation efficiency.
BTI1 Protein Levels Transiently Increase after Agrobacterium Infection of Arabidopsis Suspension Cells
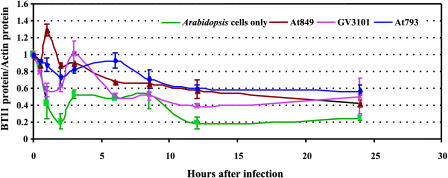
Infection of tobacco and Arabidopsis cells by transfer-competent Agrobacterium strains induces the expression of numerous plant genes (Ditt et al., 2001; Yi et al., 2002; Veena et al., 2003). To determine if Agrobacterium infection affects BTI1 protein expression, we separately infected Arabidopsis suspension cell cultures with three different Agrobacterium strains. Each strain contained the binary vector pBISN1 (Narasimhulu et al., 1996) to monitor transformation efficiency by examining GUS activity in the infected Arabidopsis cells. Agrobacterium At849 (GV3101 plus pBISN1) can transfer both T-DNA and Vir proteins to plant cells. Agrobacterium At793 contains pBISN1 but lacks a Ti plasmid; this strain cannot produce VirB2 nor transfer T-DNA or Vir proteins to plants. Agrobacterium GV3101 contains a Ti plasmid lacking a T-DNA region. Although this strain cannot transfer T-DNA, it can export VirD2, VirE2, and VirF proteins via the VirB/D4 transport apparatus. Additionally, we examined uninfected Arabidopsis suspension cells. We sampled uninfected Arabidopsis cells and cells separately cocultivated with the various Agrobacterium strains from three independent infections. The average transformation efficiency using Agrobacterium At849 was 92.4% ± 1.2%. We monitored BTI1 protein levels by protein gel blot analysis and normalized the amount of BTI1 protein in each sample to the amount of actin (see Supplemental Figure 5 online).
Figure 9 shows the expression profiles of BTI1 protein for each of the four treatments. When Arabidopsis suspension cell cultures were infected with At849, BTI1 protein levels increased by almost 1.5-fold 1 h after the start of cocultivation, after which BTI1 protein amounts gradually declined during the 24-h cocultivation period. Arabidopsis suspension cell cultures infected with Agrobacterium At793 or GV3101 did not show a similar transient increase in BTI1 protein. These results suggest that the transient increase of BTI1 protein levels after At849 infection results from T-DNA but not Vir protein transfer, and does not occur merely because of contact of the plant with Agrobacterium cells.
Figure 9.
BTI1 Protein Levels Transiently Increase after Agrobacterium Infection of Arabidopsis Suspension Cells.
Plant cells were infected with various Agrobacterium strains (or were mock-inoculated), proteins were extracted at various times, and the proteins were subjected to protein gel blot analysis using anti-BTI1 antibody. The amount of BTI1 protein in each sample was normalized to the amount of actin, as determined by protein gel blot analysis using antiactin antibody. The ratio of BTI1 protein:actin protein was normalized to 1 at 0 h. The green line indicates BTI1 protein levels of uninfected Arabidopsis suspension cell cultures. The red line indicates BTI1 protein levels of Arabidopsis suspension cells infected with Agrobacterium At849 (GV3101 containing pBISN1; this strain can transfer both T-DNA and virulence proteins). The purple line shows BTI1 protein levels of Arabidopsis suspension cells infected with Agrobacterium GV3101 (nononcogenic Agrobacterium strain containing a disarmed pTiC58 plasmid; this strain can transfer virulence proteins but not T-DNA). The blue line indicates BTI1 protein levels of Arabidopsis suspension cells infected with Agrobacterium At793 (pBISN1 in Agrobacterium A136 lacking a Ti-plasmid; this strain can transfer neither virulence proteins nor T-DNA). Data shown in the figure are average values of three independent experiments. Error bars = se.
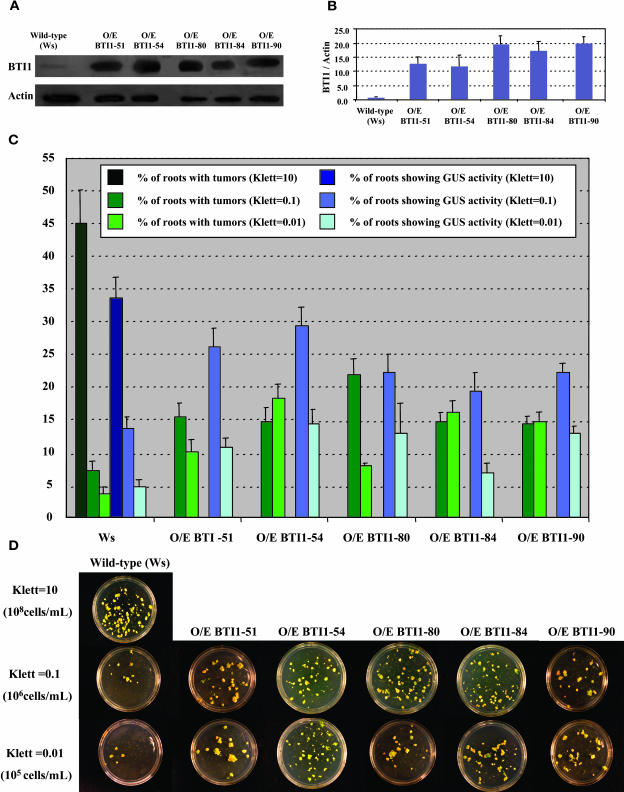
BTI1 Overexpression Increases the Susceptibility of Plants to Agrobacterium-Mediated Transformation
To determine if overexpression of BTI1 protein in plants could enhance the efficiency of Agrobacterium-mediated transformation, we generated transgenic Arabidopsis plants that overexpress BTI1 protein and performed transformation assays on them. Protein gel blot analysis indicated that many of these plants markedly overproduce BTI1 protein compared with wild-type plants (Figures 10A and 10B). Figures 10C and 10D show the results of stable and transient transformation assays. When root segments from wild-type Arabidopsis plants and transgenic BTI1 overexpressing plants were infected with various Agrobacterium strains, the BTI1 overexpressing plants showed a twofold to threefold increase in transformation efficiency.
Figure 10.
Transgenic BTI1 Overexpressing Plants Show an Increased Frequency of Agrobacterium-Mediated Root Transformation.
(A) BTI1 protein levels of transgenic BTI1 overexpressing plants were monitored by protein gel blot analysis using BTI1 antibodies.
(B) BTI1 protein levels in each sample shown in (A) were normalized to the amount of actin in the sample. The ratio of BTI1 protein:actin protein was normalized to 1 in the wild-type plant. Data are shown as average values of three protein gel blot analyses from three T2 generation plants of each line. Error bars = se.
(C) T2 Generation transgenic BTI1 overexpressing (O/E) plants show higher stable and transient transformation efficiencies than do wild-type plants. Plants were inoculated with Agrobacterium A208 (for tumorigenesis assays) or Agrobacterium At849 (for transient GUS assays) at 106 cells/mL (Klett = 0.1) or 105 cells/mL (Klett = 0.01). As a control to indicate successful transformation, roots of wild-type plants were also inoculated at 108 cells/mL (Klett = 10). At least 15 different plants were tested for each transgenic line and >80 root segments were examined for each plant. Error bars = se.
(D) Representative plates of transgenic BTI1 overexpressing (O/E) plants showing increased frequency of tumor formation at low inoculation densities (106 cells/mL [Klett = 0.1] and 105 cells/mL [Klett = 0.01]).
BTI-GFP Fusion Proteins Preferentially Localize to the Cell Periphery in Roots of Transgenic Arabidopsis Plants
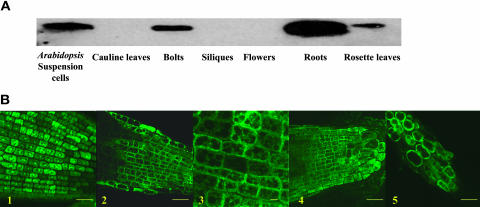
RT-PCR analysis indicated that the three BTI genes are expressed in roots, rosette leaves, and inflorescence tissues of wild-type Arabidopsis plants (data not shown). We investigated further the tissues in which BTI1 protein is expressed. Protein gel blot analyses using antibodies directed against BTI1 protein revealed that BTI1 protein accumulates in roots, rosette leaves, and the bolt regions of the inflorescence from mature Arabidopsis plants, but not in siliques, flowers, or caluine leaves. BTI1 protein is also expressed in Arabidopsis suspension cells (Figure 11A). Figure 11A shows that, as a percentage of total protein, BTI1 is expressed at relatively high levels in root tissues compared with other plant tissues.
Figure 11.
Expression Pattern of BTI Proteins in Plant Tissues and Cells.
(A) Protein gel blot analysis showing that BTI1 protein is expressed in root tissues, rosette leaves, and the bolt regions of the inflorescence of mature Arabidopsis plants, but not in siliques, flowers, or cauline leaves. BTI1 protein is also expressed in Arabidopsis suspension cells.
(B) Confocal fluorescence microscopic images of transgenic Arabidopsis root tips expressing GFP or BTI-GFP fusion proteins. (1) Single optical section of root tip cells from GFP transgenic plants. (2 and 3) Single optical sections of root tip cells from BTI1-GFP transgenic plants. (4) Single optical section of root tip cells from BTI3-GFP transgenic plants. (5) BTI1-GFP protein is not associated with the plant cell wall. BTI1-GFP transgenic plants were treated with 0.8 M mannitol for 30 to 60 min before examining by confocal microscopy. The images indicate that GFP alone localizes through the cytoplasmic region and the nucleus, whereas the BTI-GFP fusion proteins localize throughout the cytoplasm, preferentially to the cell periphery, but not the walls, of root tip cells. Bars (1, 2, 4, and 5) = 25 μm; bar (3) = 5 μm.
It is not clear whether the T-pilus penetrates the plant cell wall and plasma membrane or if the T-pilus attaches to or fuses with the plant cell surface during Agrobacterium infection. If BTI proteins interact with VirB2 protein in the T-pilus, they should localize to the plant cell surface. To address this issue, we fused BTI protein with GFP and expressed this fusion protein in transgenic Arabidopsis plants. Because BTI1 protein is expressed mainly in the root tissues and we have used root tissues to perform transformation assays, we choose roots from BTI1-GFP transgenic plants to perform further analyses. Confocal microscopic analysis indicated that GFP alone is found throughout both the cytoplasm and the nucleus (Figure 11B-1), whereas BTI1-GFP fusion protein localizes throughout the cytoplasm but preferentially to the periphery of root tip cells (Figures 11B-2 and 11B-3). Similar results were obtained with BTI2-GFP (data not shown) and BTI3-GFP transgenic plants (Figure 11B-4).
We additionally treated root tissues from BTI1-GFP transgenic plants with 0.8 M mannitol for 30 to 60 min. After such treatment, the plant cell dehydrates and the cytoplasm along with the plasma membrane separates from the cell wall. Figure 11B-5 shows that after such mild plasmolysis, BTI1-GFP is predominantly in the cytoplasmic region and does not associate with the cell wall. These results indicate that, as predicted by the presence of putative membrane spanning domains (Figure 4A), BTI1-GFP fusion protein does not localize to the plant cell wall.
DISCUSSION
Agrobacterium uses a type IV secretion system to transfer T-DNA and Vir proteins to host cells. This system is composed of two functional components: a filamentous T-pilus and a transporter complex. Although numerous recent genetic and biochemical studies have focused on the functional roles of VirB proteins in the assembly and composition of the transporter and T-pilus, little is known about the role that the T-pilus plays in T-DNA and Vir protein transfer. In this study, we initiated a search for plant proteins that interact with the T-pilus and may therefore be involved in the initial steps of T-DNA and Vir protein transfer. We used a yeast two-hybrid system to identify two classes of Arabidopsis proteins that interact with the processed portion of VirB2, the major pilin protein. The first category is composed of the three VirB2-interacting proteins. We further demonstrated that these proteins interact directly with VirB2 using GST pull-down assays. All three proteins share a C-terminal 150– to 201–amino acid reticulon homology domain (Pfam PF02453) comprising two large hydrophobic regions separated by an ∼66–amino acid loop. More than 250 reticulon-like (RTNL) genes were identified in divergent eukaryotes, fungi, plants, and animals (Oertle and Schwab, 2003; Oertle et al., 2003). No homologs of the RTN proteins have been identified in prokaryotes to date, suggesting that RTNs emerged in eukaryotes (Oertle and Schwab, 2003; Oertle et al., 2003). Based on the reticulon domain present in their C termini, Arabidopsis encodes 15 reticulon-like proteins (Oertle et al., 2003). BTI1, BTI2, and BTI3 correspond to RTNLB1, RTNLB2, and RTNLB4.
The functions of RTN1, RTN2, and RTN3 are unknown, yet all RTNs (including RTN4/Nogo) are enriched in endoplasmic reticulum membranes (van de Velde et al., 1994a, 1994b; Senden et al., 1996). Some of the RTNs also associate with cellular structures other than the endoplasmic reticulum (ER). Nogo-A localizes with Golgi markers in addition to the ER and is present in small amounts at the plasma membrane of oligodendrocytes and fibroblasts (GrandPre et al., 2000).
Because VirB2 cyclizes in Agrobacterium but not in yeast nor in E. coli, it is possible that the VirB2 fusion proteins used in our yeast two-hybrid and GST pull-down assays may not form the same conformation as does VirB2 in the Agrobacterium pilus. To investigate if the noncyclized form of VirB2 can affect transformation, we infected Arabidopsis suspension cells with Agrobacterium that were pretreated with GST-BTI1. Results from transient transformation assays revealed that GST-BTI1 inhibited transformation in a dose-dependent manner, whereas treatment with GST alone had no effect on transformation. These data suggest that GST-BTI1 may interact with the Agrobacterium T-pilus and inhibit subsequent pilin-dependent attachment to the plant.
Both transient and stable transformation efficiencies of transgenic antisense and RNAi lines targeting the various BTI genes are severely impaired, indicating that BTI proteins are involved in Agrobacterium-mediated plant transformation. Because of extensive nucleotide similarity among the BTI genes, it is difficult to gauge the relative importance of the various BTI genes for transformation. BTI1 and BTI2, especially, share a high level of nucleotide sequence identity; BTI3 is more distantly related. Antisense and RNAi plants targeting one of these genes also may have reduced expression of the other BTI genes (Figures 6 and 7). However, the antisense and RNAi data suggest that more than one BTI gene contribute to transformation competence of the plant. In the BTI3-7 RNAi and BTI3-18 RNAi transgenic plants, only the BTI3 transcript level, but not those of BTI1 and BTI2, is reduced. These plants show significantly lower stable and transient transformation efficiencies than do wild-type plants, suggesting that BTI3 is involved in the Agrobacterium-mediated root transformation process. Similar results were obtained from the BTI2-29 A/S transgenic plants, suggesting that BTI2 protein is also involved in the transformation process. Because the three BTI proteins interact with themselves and with each other in yeast and in vitro, it is possible that the three BTI proteins form a multimeric complex inside plant cells, and that this complex may be involved in the transformation process.
To test directly if BTI1 protein alone is involved in transformation, we investigated two T-DNA insertion mutants in the BTI1 gene. In the bti1-1 mutant, the T-DNA is inserted in the promoter region of the BTI1 gene. Protein gel blot analysis demonstrated that the BTI1 protein level is slightly reduced in these plants. bti1-2 has a T-DNA insertion in the 3′ untranslated region of the BTI1 gene. This mutant also showed lower BTI1 protein levels. Both bti1-1 and bti1-2 mutant plants are resistant to stable and transient transformation, indicating that BTI1 is involved in the Agrobacterium-mediated root transformation process. These data also indicate that in these mutants, BTI2 and BTI3 do not fully compensate for the functions of BTI1 protein during Agrobacterium infection. Taken together, data from our antisense, RNAi, and T-DNA insertion lines indicate that each of the three BTI proteins is involved in the transformation process.
We examined BTI1 protein levels in Arabidopsis suspension cells during Agrobacterium infection. During the first 2 h of cocultivation, BTI1 protein levels transiently increased only after infection by Agrobacterium At849, a strain that could transfer both T-DNA and virulence proteins, but not by strain GV3101 that could transfer only virulence proteins or strain At793 that could transfer neither T-DNA nor virulence proteins, indicating that this increase in BTI1 protein level may result from T-DNA transfer. This transient increase in BTI1 protein is consistent with the hypothesis that Agrobacterium may regulate the expression of host genes to assure a successful transformation process (Ditt et al., 2001; Yi et al., 2002; Veena et al., 2003). Narasimhulu et al. (1996) previously demonstrated that cocultivation of tobacco BY-2 cells for 2 h with Agrobacterium is sufficient to achieve transformation. Yusibov et al. (1994) reported that maximal T-strand accumulation in the cytoplasm of tobacco cells occurs 2 h after the initiation of cocultivation. These results suggest that T-DNA and virulence proteins can be transferred from the bacterium to plant cells within 2 h. Interestingly, the results of this study show that the level of BTI1 protein is transiently induced 30 min to 2 h after infection. This rapid response correlates well with previous studies suggesting that bacterial attachment also occurs within 1 h after the initiation of cocultivation (Matthysse et al., 1982; Neff and Binns, 1985).
BTI1 overexpression in transgenic plants increases susceptibility to Agrobacterium-mediated transformation, supporting further the correlation between BTI1 protein levels and Agrobacterium transformation efficiency. BTI1 protein levels in these transgenic plants are 10- to 20-fold higher than that of wild-type plants. These results suggest that BTI1 may be a limiting cellular factor required for Agrobacterium infection.
We used confocal fluorescence microscopy to examine the subcellular localization of BTI-GFP fusion protein in root cells of transgenic Arabidopsis plants. Our results showed that BTI-GFP protein localizes throughout the cytoplasm but preferentially to the cell periphery. Additional experiments further suggested that BTI1-GFP fusion proteins are not on the cell wall. Protein sequence analysis indicated that BTI1 protein does not have a signal peptide but contains two putative transmembrane domains. Based on previous studies of RTN proteins in other organisms and BTI protein sequence analysis, it is reasonable to speculate that BTI proteins may associate with the plasma membrane, Golgi apparatus, and/or ER. It is not clear how the Agrobacterium T-pilus contacts the plant cell surface. The results from these studies provide candidates to interact with the Agrobacterium T-pilus. It is possible, however, that other plant proteins involved in cell wall synthesis or other plant cell wall structural proteins may also participate in the initial contact of the Agrobacterium T-pilus with the plant cell surface (Zhu et al., 2003a, 2003b).
The GTPase AtRAB8 is the second type of plant protein that interacts with VirB2. Rab proteins are membrane-associated proteins that modulate tubulovesicular trafficking between compartments of the biosynthetic and endocytic pathways (Olkkonen and Stenmark, 1997; Martinez and Goud, 1998; Schimmoller et al., 1998; Moyer and Balch, 2001). Previous studies suggested that AtRAB8 is similar to RAB8 and RAB10 of mammals, to Ypt2 of the fission yeast Schizosaccharomyces pombe, and to Sec4 of the budding yeast Saccharomyces cerevisiae (Haubruck et al., 1990; Rutherford and Moore, 2002). Rab proteins are important regulators of vesicular trafficking and Sec4 is essential for post-Golgi events in yeast secretion. Rab8 regulates transport from the trans-Golgi network to the basolateral plasma membrane in epithelial cells and to the dendritic plasma membrane in cultured hippocampal neurons (Huber et al., 1993). However, little is known about the function of Rab8 in plant cells.
Transgenic AtRAB8 A/S and RNAi plants are less susceptible to transient and stable transformation, suggesting that AtRAB8 is also involved in the Agrobacterium-mediated root transformation process. AtRAB8 and BTI proteins may interact with each other in the plant cell, and this interaction may be important for transformation. Interestingly, tomato proteins homologous to AtRAB8 interact with the Pseudomonas sp avirulence factor avrPto in yeast. This interaction occurs only in the absence of the resistance protein Pto, raising the possibility that in susceptible plants, AvrPto may interfere with membrane trafficking pathways regulated by this AtRAB8 homolog (Bogdanove and Martin, 2000; Vernoud et al., 2003). Further characterization of the involvement of AtRAB8 in the Agrobacterium infection process may help to decipher the possible functions of AtRAB8 in plant cells.
Agrobacterium may use the T-pilus to mediate T-DNA and Vir protein transfer from bacteria to host cells. However, the molecular mechanism by which transport occurs is not well understood. Experiments performed in this study identified two classes of plant proteins that may participate in the initial steps of the T-DNA transfer process. We are in the process of examining whether BTI proteins and AtRAB8 are recruited to infection sites through direct interactions with Agrobacterium T-pili during Agrobacterium transformation. Further characterization of BTI and AtRAB8 proteins will provide information on how T-DNA is transferred from Agrobacterium into plant cells and how the Agrobacterium T-pilus contacts the plant cell surface.
METHODS
Plasmid Constructions
Plasmids used for the yeast two-hybrid studies are listed in Supplemental Table 3 online. Plasmids pSST91 and pGAD424 (Clontech, Palo Alto, CA) were used as vectors for the construction of the various fusions. pSST91 contains the LexA protein coding sequence under the control of the yeast ADH1 promoter. pGAD424 generates a hybrid protein that contains the sequence for the GAL4 activation domain. To construct pE2180 expressing the LexA-VirB2 bait protein, an EcoRI-PstI fragment from the C terminus of VirB2 (amino acids 48 to 121) from the Agrobacterium tumefaciens octopine-type plasmid pTiA6 was cloned into pSST91 digested with EcoRI/PstI as an in-frame fusion to the LexA coding sequence. The VirB2 open reading frame was amplified by PCR, using a high-fidelity PWO DNA polymerase (Roche, Indianapolis, IN) with the primers 5′-GGAATTCCAATCTGCGGGTGGC-3′ (forward primer) and 5′-AACTGCAGTCAACTACCGCCAGTG-3′ (reverse primer). EcoRI-PstI restriction sites were introduced at the 5′ and 3′ ends of the PCR product that maintained the reading frame of the LexA DNA binding domain. The Arabidopsis thaliana cDNA library was provided by Vitaly Citovsky of the State University of New York, Stony Brook (Ballas and Citovsky, 1997).
Yeast Two-Hybrid Screen
The yeast strain CTY10-5d was provided by Vitaly Citovsky of the State University of New York, Stony Brook. All yeast strains were cultured at 30°C in synthetic dropout (SD) medium (Ausubel et al., 2003) containing yeast nitrogen-base, glucose, and all but the selective amino acids. Yeast transformations were performed using a lithium acetate method (Golemis et al., 1994). Yeast strain YB2 was generated by transforming strain CTY10-5d with pE2180. pE2180 alone was insufficient for transcriptional activation in the two-hybrid system. The Arabidopsis cDNA library in pGAD424 was transformed into YB2 and colonies were screened for protein interaction by colony color phenotype on SD medium lacking Leu and Trp and containing the chromogenic substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Plasmids were recovered from yeast colonies containing candidate-interacting cDNAs and used for direct PCR to amplify the cDNA (Ausubel et al., 2003). The PCR was performed in a 50-μL reaction using two units of ExTaq polymerase (TaKaRa, Madison, WI) and 0.25 μM each of the forward primer 5′-TACCACTACAATGGATG-3′ and the reverse primer 5′-GTTGAAGTGAACTTGCGGGG-3′ for 36 cycles with the following program: 95°C for 1 min (one cycle); 94°C for 50 s, 58°C for 40 s, 72°C for 2.5 min (35 cycles); and 72°C for 5 min (one cycle). The cDNA insert was cloned into pCR2.1-TOPO using a TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA), sequenced, and the DNA sequence compared with sequences in the database using the BLASTX program (Altschul et al., 1990, 1997; Gish and States, 1993; Madden et al., 1996; Zhang and Madden., 1997).
GST Protein Affinity Purification and In Vitro Protein Binding Assays
The plasmids pGEX4T-1 (Amersham Biosciences, Piscataway, NJ) and pET23a (Novagen, San Diego, CA) were used to generate recombinant proteins fused in frame with the GST tag and T7 tag, respectively. The coding sequence of the BTI and AtRAB8 genes were amplified by PCR. PCR reactions were performed with PWO DNA polymerase using the following primers: (1) forward primer of BTI1, 5′-GGAATTCATGGCGGAAGAACATAAG -3′; (2) reverse primer of BTI1, 5′-GCTCTAGAATCTTTCTTCTTGTTCTT-3′; (3) forward primer of BTI2, 5′-GGAATTCATGGCGGATGAACATAAGC-3′; (4) reverse primer of BTI2, 5′-GCTCTAGAATCCTTCTTCTTGTCTTT-3′; (5) forward primer of BTI3, 5′-GGAATTCATGGTGGAAGACCACAAG-3′; (6) reverse primer of BTI3, 5′-GCTCTAGAATCCTTCTTCTTGTTCAG-3′; (7) forward primer of AtRAB8, 5′-GGAATTCATGGCGGATGAACATAAGC-3′; (8) reverse primer of AtRAB8, 5′-GCTCTAGAATCCTTCTTCTTGTCTTT-3′. The PCR amplification cycle was: 95°C for 1 min (one cycle); 94°C for 30 s, 56°C for 40 s, 72°C for 1.5 min (29 cycles); and 72°C for 5 min (one cycle). The PCR products and the coding sequence of virB2 were digested with EcoRI and XhoI, and cloned into pGEX4T-1 and/or pET23a digested with the same enzymes. Expression and purification of GST fusion proteins and affinity purification of proteins binding to GST fusion proteins were performed as described (Ausubel et al., 2003) with some minor modifications. Isopropyl β-d-thiogalactoside (IPTG)-induced E. coli BL21 (DE3) cells carrying pGEX4T-1 (GST vector control), pE2338 (GST-BTI1), pE2548 (GST-BTI2), pE2346 (GST-BTI3), pE2342 (GST-AtRAB8), or pE2348 (GST-VirB2) were collected, resuspended, and lysed by sonication in binding buffer: 20 mM Tris-HCl, pH 7.5, 75 mM KCl, 5 mM MgCl2, 50 mM NaCl, 1 mM EDTA, pH 8.0, 0.05% nonidet P-40, 10% glycerol, 1 mM DTT, and 1 mM phenylmethylsulfonylfluoride (PMSF). Binding reactions used equal volumes of crude bacterial lysates with 75 μg of glutathione-sepharose beads (Sigma Chemical, St. Louis, MO) and were incubated at 4°C for 1 to 2 h with gentle shaking. The glutathione-sepharose beads were washed three times with binding buffer and were then incubated with bacterial lysates from E. coli cells expressing the T7-tagged BTI proteins that were prepared as described above. The binding reactions were incubated again at 4°C for 1 to 2 h with gentle shaking. The beads were washed and the bound proteins were eluted with 5 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0. We loaded these protein complexes onto 12.5% SDS-polyacrylamide gels and performed protein gel blot analyses (Ausubel et al., 2003) with a 1:5000 dilution of anti-T7 tag antibodies (Novagen) to confirm the identities of these fusion proteins. For GST pull-down assays with GST-AtRAB8 and T7-tagged BTI proteins, the binding buffers were additionally supplemented with either 0.5 mM GDP or 0.5 mM GTP (Sigma Chemical). The membranes were developed with colorimetric detection methods or chemiluminescent detection methods (SuperSignal West Pico kit; Pierce, Rockford, IL) and subjected to autoradiography.
Generation of Prey Plasmids Encoding BTI1 Deletion Mutants Fused to the GAL4 Activation Domain Using Inverse PCR
The cDNA clone encoding BTI1 was cloned into the plasmid pCR2.1-TOPO using a TOPO TA Cloning Kit (Invitrogen). We used this plasmid as the template to perform inverse PCR and generated serial internal deletion mutants of BTI1. The PCR reactions were performed with PWO DNA polymerase using five different sets of PCR primers to generate nine different BTI1 deletion mutants. The PCR primer sequences are as follows: (1) F9a, 5′-TCTGGTGGTGTACTTGGTGGT-3′ (forward primer) and 5′-ATGCTTATGTTCTTCCGCCAT-3′ (reverse primer); (2) F9b, 5′-TCTAATGCCACTATGTTCATT-3′ (forward primer) and 5′-ACCACCAAGTACACCACCAGA-3′ (reverse primer); (3) F9c, 5′-TCTCTCCGTGAAATTGCATCA-3′ (forward primer) and 5′-AATGAACATAGTGGCATTAGA-3′ (reverse primer); (4) F9d, 5′-TACGACAAGTATGAAGACAAA-3′ (forward primer) and 5′-TGATGCAATTTCACGGAGAGA-3′ (reverse primer); and (5) F9e, 5′-TTCAGCAAGATCCCACTTGGG-3′ (forward primer) and 5′-TTTGTCTTCATACT TGTCGTA-3′ (reverse primer). The PCR products were generated with the following program: 95°C for 1 min (one cycle); 94°C for 50 s, 56°C for 40 s, 72°C for 2.5 min (30 cycles); and 72°C for 5 min (one cycle). The purified PCR products were digested with NheI, and the digested DNA fragments were self-ligated and transformed into competent E. coli DH10B cells (Stratagene, La Jolla, CA). The clones were sequenced, digested with the enzymes EcoRI and PstI, and the inserts cloned into the same restriction enzyme sites of pGAD424. The resulting nine different prey plasmids that encode the BTI1 deletion mutants fused with the GAL4 activation domain are listed in Supplemental Table 3 online.
Agrobacterium-Mediated Transformation of Arabidopsis Suspension Cell Cultures
Agrobacterium At849, the nontumorigenic strain GV3101 containing the binary vector pBISN1 (Narasimhulu et al., 1996), was used to transform Arabidopsis suspension cells. Agrobacterium cells were grown to a density of 2 × 109 cells/mL in AB-sucrose medium (Lichtenstein and Draper, 1986) at 30°C. Cells were harvested by centrifugation at 5000 rpm and resuspended in two volumes of induction medium (AB salts, 0.5% glucose, 2 mm sodium phosphate, 50 mm Mes, pH 5.6, 100 μm acetosyringone), and incubated at 22 to 24°C for 14 to 18 h with gentle shaking. Induced Agrobacterium cells were washed with Arabidopsis cell culture medium and incubated with or without GST or with GST-BTI1 protein for 2 h before infecting Arabidopsis suspension cell cultures. Expression and purification of GST fusion proteins and affinity purification of proteins binding to GST fusion proteins were performed as described (Ausubel et al., 2003).
Arabidopsis suspension cells (ecotype Columbia) were maintained in Murashige and Skoog medium (Gibco, Carlsbad, CA) containing 2% sucrose, 10 mg/mL thiamine-HCl, 1 mg/mL nicotinic acid, 1 mg/mL pyridoxine-HCl, 100 μg/mL myo-inositol, and 2 μg/mL 2,4-D with continuous shaking at 140 rpm at 23°C in the presence of light. Seven-day-old Arabidopsis suspension cell cultures were infected with the pretreated Agrobacterium cells (∼1000 bacterial cells per plant cell) at 22 to 24°C with gentle shaking. After 24 h cocultivation, the Arabidopsis cells were pelleted by centrifugation at 800 rpm for 5 min and washed two times with Arabidopsis suspension cell medium. The suspension cells were stained with GUS staining solution (50 mM sodium phosphate buffer, pH 7.0, 0.1% Tween 20, 3% sucrose, and 1 to 2 mM X-Gluc) overnight at 37°C to determine the efficiency of transformation. The stained cells were visualized using a light microscope and scored to determine the percentage of cells staining blue with X-Gluc.
Generation of BTI and AtRAB8 Antisense and RNAi Arabidopsis Transgenic Plants, and BTI1 Overexpression Arabidopsis Transgenic Plants
Antisense cDNA clones corresponding to BTI1, BTI2, or AtRAB8 were cloned into the KpnI and XhoI sites of the binary vector pE1775 containing a hptII (hygromycin resistance) gene as a selectable marker, generating the plasmids pE1978, pE1980, and pE1976, respectively. The cDNAs were under the transcriptional control of a superpromoter (Ni et al., 1995). The plasmids pE1978, pE1980, and pE1976 were separately transformed into the nontumorigenic strain Agrobacterium GV3101 (Koncz and Schell, 1986) to generate A/S transgenic Arabidopsis plants using a floral dip method (Clough and Bent, 1998).
To generate BTI1, BTI2, BTI3, and AtRAB8 RNAi transgenic plants, the coding sequences of the BTI1, BTI2, BTI3, and AtRAB8 genes were generated by PCR and cloned into the RNAi vector pFGC5941 (http://ag.arizona.edu/chromatin/fgc5941.html). The PCR reaction was performed in a 50-μL reaction volume using two units of PWO DNA polymerase and the following primers: (1) forward primer for BTI1, 5′-GGACTAGTGGCGCGCCATGGCGGAAGAACATAAGCATGAT-3′; (2) reverse primer for BTI1, 5′-CGGGATCCATTTAAATCTAATCTTTCTTCTTGTTCTTCAA-3′; (3) forward primer for BTI2; 5′-GGACTAGTGGCGCGCCATGGCGGATGAACATAAGCATGAA-3′; (4) reverse primer for BTI2; 5′-GAAGATCTATTTAAATCTAATCCTTCTTCTTGTCTTTCAA-3′; (5) forward primer for BTI3, 5′-GGACTAGTGGCGCGCCATGGTGGAAGACCACAAGCACGAG-3′; (6) reverse primer for BTI3, 5′-CGGGATC CATTTAAATTTAATCCTTCTTCTTGTTCAGAGC-3′; (7) forward primer for AtRAB8; 5′-GCTCTAGAGGCGCGCCATGGCTGCTCCTCCTGCTAGAGCT-3′; and (8) reverse primer for AtRAB8; 5′-GAAGATCTATTTAAATTTATGTGCCGCAACATGCTGATTT-3′ using the following program: 95°C for 1 min (one cycle); 94°C for 50 s, 56°C for 50 s, 72°C for 2.5 min (30 cycles); and 72°C for 5 min (one cycle). The PCR products were purified and digested with the restriction enzymes AscI and SwaI and cloned into the same sites of pFGC5941, resulting in the plasmid pE2086 for BTI1, pE2085 for BTI2, pE2098 for BTI3, and pE2084 for AtRAB8. The PCR product of the BTI1 gene was digested with BamHI and SpeI and cloned into the same sites of pE2086, resulting in the plasmid pE2087. The PCR product of the BTI2 gene was digested with BglII and SpeI and cloned into the BamHI-SpeI sites of pE2085, resulting in the plasmid pE2089. The PCR product of the BTI3 gene was digested with BglII and SpeI and cloned into the BamHI-SpeI sites of pE2098, resulting in the plasmid pE2088. The PCR product of the AtRAB8 gene was digested with BglII and XbaI and cloned into the BamHI-XbaI sites of pE2084, resulting in the plasmid pE2090. The plasmids pE2087, pE2089, pE2088, and pE2090 were used to generate BTI1, BTI2, BTI3, and AtRAB8 RNAi transgenic plants, respectively, using the procedures discussed above.
To generate BTI1 overexpression transgenic plants, the coding sequence of the BTI1 gene was cloned as an XbaI-KpnI fragment into the same sites of the binary vector pE1798 with a hptII (hygromycin resistance) gene as a selectable marker, resulting in the plasmid pE2186. The BTI1 gene was under the transcriptional control of a double 35S promoter of Cauliflower mosaic virus in the plasmid pE2186. pE2186 was transformed into the nontumorigenic strain Agrobacterium GV3101 (Koncz and Schell, 1986) to generate BTI1 overexpression transgenic Arabidopsis plants using a floral dip method (Clough and Bent, 1998).
Identification of bti1 T-DNA Insertion Mutant Plants from the Feldmann and Salk Collection of T-DNA Insertion Lines
A PCR-based approach similar to that described by Zhu et al. (2003b) was used to identify Arabidopsis (ecotype Ws-2) mutants containing a T-DNA insertion near BTI1. Pooled samples of DNA from 120 plants from the Feldmann T-DNA insertion library (Dellaporta et al., 1983; Feldmann and Marks, 1987) were successively assayed for insertions, followed by assay of individual plants from the pool of 20 mutants with either the right T-DNA border primer or the left T-DNA border primer paired with either the forward primer or the reverse primer of the BTI1 gene. The primer sequences are as follows: (1) left T-DNA border primer, 5′-GATGCACTCGAAATCAGCCAATTTTAGAC-3′; (2) right T-DNA border primer, 5′-TCCTTCAATCGTTGCGGTTCTGTCAGTTC-3′; (3) forward primer for BTI1, 5′-CTCGAGATGGCGGAAGAACATAAGC-3′; (4) reverse primer for BTI1, 5′-CTGCAGCTAATCTTTCTTCTTGTTC-3′. The PCR amplification cycle was: 95°C for 5 min (one cycle); 95°C for 30 s, 56°C for 1 min, 72°C for 5 min (36 cycles); and 72°C for 10 min (one cycle). Each PCR product was confirmed by DNA gel blot analysis (Ausubel et al., 2003) using a DNA fragment encoding the BTI1 protein as a probe. bti1-1 T-DNA insertion mutant plants were identified using the right T-DNA border primer and the reverse primer for the BTI1 gene. The PCR products were cloned into the plasmid pCR2.1-TOPO using a TOPO TA cloning kit, sequenced, and compared with the database using the BLASTN program (Altschul et al., 1990, 1997; Gish and States, 1993; Madden et al., 1996; Zhang and Madden., 1997).
The Arabidopsis T-DNA insertion mutant bti1-2 (ecotype Columbia CS60000) was identified using the SIGnAL T-DNA Express Arabidopsis Gene Mapping Tool (http://signal.salk.edu; Alonso et al., 2003). Seeds of bti1-2 plants were obtained from the Arabidopsis Biological Resource Center at The Ohio State University.
Agrobacterium-Mediated Stable and Transient Root Transformation Assays of BTI and AtRAB8 Antisense and RNAi Arabidopsis Transgenic Plants, and BTI1 Overexpression Arabidopsis Transgenic Plants
All Agrobacterium strains were cultured in 5 mL of yeast extract peptone (YEP) medium (Lichtenstein and Draper, 1986) supplemented with the appropriate antibiotics (rifampicin, 10 μg/mL; kanamycin, 25 μg/mL) at 30°C. An overnight bacterial culture (1 mL) was inoculated into 25 mL of YEP medium with antibiotics and grown to a density of 2 × 109 cells/mL. The bacterial cells were washed with 0.9% sodium chloride and resuspended in 0.9% sodium chloride at 2 × 108 cells/mL for stable and transient root transformation assays.
Seeds from wild-type, A/S, RNAi, overexpression transgenic plants, and T-DNA mutant plants were surface sterilized and placed on Gamborg's B5 medium (Gibco) solidified with 0.75% Bactoagar (BD Biosciences, Palo Alto, CA) containing the appropriate antibiotics (50 μg/mL kanamycin for T-DNA mutant plants, 20 μg/mL hygromycin for A/S and overexpression transgenic plants, and 10 μg/mL phosphinothricin for RNAi transgenic plants). Seedlings were transferred individually into baby food jars containing solidified B5 medium without antibiotics and grown for 3 weeks to perform transformation assays as described by Nam et al. (1997, 1998, 1999) and Zhu et al. (2003b).
For transient transformation assays, root segments were infected with Agrobacterium At849. After 2-d cocultivation, the roots were placed on callus induction medium (CIM) plates (CIM is 4.32 g/L of MS salts [Gibco], 0.5 g/L Mes, pH 5.7, 1 mL/L vitamin stock solution [0.5 mg/mL nicotinic acid, 0.5 mg/mL pyridoxine, and 0.5 mg/mL thiamine-HCl], 100 mg/L myo-inositol, 20 g/L glucose, 0.5 mg/L 2,4-D, 0.3 mg/L kinetin, 5 mg/L indole acetic acid, and 0.75% bactoagar) containing timentin (100 μg/mL equivalent to 96.8 μg/mL ticarcillin and 3.3 μg/mL clavulanic acid [GlaxoSmithKline, Research Triangle Park, NC]). CIM plates were incubated at 25°C for an additional 4 d. The roots were stained with X-Gluc staining solution for 1 d at 37°C. Roots were examined using a Nikon SMZ-10 stereoscopic microscope.
For tumorigenesis assays, root segments were transferred to solidified MS medium and infected with the tumorigenic strain Agrobacterium A208. After 2 d cocultivation at 20°C, root segments were separated and placed on solidified MS medium lacking hormones but containing timentin. The MS plates were incubated for 4 weeks at 25°C.
For transformation of root segments to phosphinothricin-resistance, root segments were infected with Agrobacterium At872, a nontumorigenic derivative of Agrobacterium GV3101 containing pCAS1. pCAS1 is a modified pGPTV-bar binary vector (Becker et al., 1992) containing a nos-bar gene as a selectable marker driven by a mannopine synthase promoter plus an octopine synthase activator. After 2 d of cocultivation, root segments were separated and transferred onto CIM plates containing timentin (100 μg/mL) and phosphinothricin (10 μg/mL). Phosphinothricin-resistant calli were scored after 4 weeks incubation at 25°C.
RNA Isolation from Arabidopsis Plants and RT-PCR Analysis
Root tissues from 3- to 4-week-old BTI and AtRAB8 antisense plants, transgenic RNAi Arabidopsis plants, and wild-type plants were ground with a liquid nitrogen-cooled pellet pestle in a 1.5-mL eppendorf tube. The ground materials were mixed with Total RNA Isolation Reagent for Liquid Samples (TRIZOL LS reagent) (Invitrogen) following the manufacturer's instructions. RNA (0.5 to 1 μg ) was used to perform RT-PCR reactions with a reverse transcription system (Promega, Madison, WI) following the manufacturer's instructions. Oligo(dT) primers were used for the reverse transcription to generate the first-strand cDNA products. A series of oligonucleotide primers was designed that in combination would amplify specifically the sense mRNA strand of BTI1, BTI2, BTI3, and AtRAB8 genes in the PCR reactions. The primer sequences were chosen based on the nucleotide sequences in the 5′-untranslated regions (UTR) and 3′-UTRs of the BTI1, BTI2, BTI3, and AtRAB8 genes. α-Tubulin transcript accumulation was used as an internal control in each RT-PCR reaction. The primer sequences are as follows: (1) forward primer for BTI1, 5′-AACTTTGGGAAGCAGCTAAAATAC-3′; (2) reverse primer for BTI1, 5′-CACGAGATTGCAAAATTCAAATAT-3′; (3) forward primer for BTI2, 5′-ATCCGAACCAACCAACGGATCAGA-3′; (4) reverse primer for BTI2, 5′-CATTGTGCTAATGCCACAAACCAC-3′; (5) forward primer for BTI3, 5′-TTCTTCTTCTCGGAGGTTTGTAGA-3′; (6) reverse primer for BTI3, 5′-AATTATCATCCATCCGTGTTCTCT-3′; (7) forward primer for AtRAB8, 5′-TCTGGTTACTGTCTTGCTTGCTTC-3′; (8) reverse primer for AtRAB8, 5′-CTGATGTCAGAGAATCCATTCCAG-3; (9) forward primer for α-tubulin, 5′-AGCCTTCCATGAGCAACTCT-3′; and (10) reverse primer for α-tubulin, 5′-CAGCACCGACCTCTTCATAA-3′. The entire amplified sample was loaded onto an agarose gel to proceed with further analyses. The DNA bands were visualized using a UVP BioImaging System (UVP, Upland, CA) and quantified using LabWorks software.
Protein Extraction from Arabidopsis Plants and Protein Gel Blot Analyses
Root tissues from 3- to 4-week-old BTI1 antisense, BTI1 RNA interference, BTI1 overexpression transgenic Arabidopsis plants, bti1 T-DNA mutants, and wild-type plants were ground with a liquid nitrogen-cooled pellet pestle in a 1.5-mL eppendorf tube. Roots, rosette leaves, flowers, sliques, caluine leaves, and flower bolts from 4- to 5-week-old Arabidopsis plants were also used to extract proteins. The ground materials were mixed with CelLytic P (Sigma Chemical) supplemented with a protease inhibitor cocktail (1:100 dilution) from Sigma. Crude plant protein extracts were isolated following the manufacturer's instructions. The final protein concentrations were determined using a BCA protein assay kit (Pierce) and spectroscopy (SPECTRA MAX PLUS 384; Molecular Devices, Sunnyvale, CA). Equal amounts of plant proteins were loaded on 12.5% SDS-polyacrylamide gels and protein gel blot analyses were performed (Ausubel et al., 2003) using a 1:1000 dilution of peptide-raised BTI1 antibody to determine the BTI1 protein level in BTI1 A/S, BTI1 RNAi, BTI1 overexpression, bti1-1, bti1-2 mutant plants, and wild-type Arabidopsis plants. In addition, we used antibodies against actin (kindly provided by Chris Staiger, Purdue University, West Lafayette, IN) at a 1:1000 dilution to perform protein gel blot analyses to normalize plant protein amounts. The membranes were developed with a chemiluminescent detection method (SuperSignal West Pico Kit; Pierce) and subjected to autoradiography. The protein bands on the x-ray films were visualized with a UVP BioImaging system and quantified using LabWorks software. The protein sequence from amino acid residues 11 to 25 of BTI1 was chosen to generate peptide F9 N11-25 (VIAPEPAVEVVERESC). This peptide was injected into the two individual rabbits to raise polyclonal antibodies (Sigma-Genosys, St. Louis, MO).
Purification of Proteins from Arabidopsis Suspension Cells Infected with Agrobacterium and Protein Gel Blot Analyses
Seven-day-old Arabidopsis suspension cell cultures were separately infected with three Agrobacterium strains (At849, At793, and GV3101 as discussed in Results). Agrobacterium cells were prepared and used to infect Arabidopsis suspension cell cultures as described above. Cocultivation of Agrobacterium cells with Arabidopsis suspension cell cultures were performed at 22 to 24°C with gentle shaking for various time periods (0, 0.5, 1,2, 3, 6, 8.5, 12, and 24 h). At each time point, plant cells were pelleted by centrifugation at 800 rpm and washed two times in Arabidopsis suspension cell medium containing timentin (100 μg/mL) followed by protein isolation. Three independent infection experiments were performed for each treatment (uninfected Arabidopsis suspension cell cultures and cultures infected with various Agrobacterium strains). Crude protein extracts were isolated and protein concentrations were determined as described above. Equal amounts of crude protein extract were loaded on 12.5% SDS-polyacrylamide gels and protein gel blot analyses were performed (Ausubel et al., 2003) with a 1:1000 dilution of peptide-raised anti-BTI1 antibodies to determine the BTI1 protein levels in infected Arabidopsis suspension cells at different time points. The membranes were developed by a chemiluminescent detection method and subjected to autoradiography. Additionally, we used antibodies against actin at a 1:1000 dilution to perform protein gel blot analyses to normalize plant protein amounts in each crude protein extract. Protein gel blot analyses results were obtained and analyzed as described above.
Generation of Arabidopsis Transgenic Plants Expressing BTI-GFP Fusion Proteins and Examination of BTI-GFP Protein Cellular Localization Using Confocal Fluorescence Microscopy
The coding sequences of the three BTI genes generated by PCR were cloned as XbaI-BglII fragments into these same sites of the T-DNA binary vector pE1457. The PCR reaction was performed in a 50-μL reaction volume using two units of high fidelity PWO DNA polymerase with a 2.5-mM dNTP mixture, 1X PWO polymerase reaction buffer (500 mM KCl, 100 mM Tris-HCl, 1% Triton X-100, and 20 mM MgCl2), and with 0.25 μM of the following primers: (1) forward primer for BTI1 gene, 5′-GCTCTAGAATGGCGGAAGAACATAAG-3′; (2) reverse primer for BTI1 gene, 5′-GAAGATCTTATCTTTCTTCTTGTTCTT-3′; (3) forward primer for BTI2 gene, 5′-GCTCTAGAATGGCGGATGAACATAAG-3′; (4) reverse primer for BTI2 gene, 5′-GAAGATCTTATCCTTCTTCTTGTCTTT-3′; (5) forward primer for BTI3 gene, 5′-GCTCTAGAATGGTGGAAGACCACAAG-3′; and (6) reverse primer for BTI3 gene, 5′-GAAGATCTTATCCTTCTTCTTGTTCAG-3′ using the following program: 95°C for 1 min (1 cycle); 94°C for 40 s, 58°C for 40 s, 72°C for 1.5 min (30 cycles); and 72°C for 5 min (1 cycle). Genes encoding the three BTI proteins without their stop codons were translationally fused to an N-terminal–GFP coding region in the plasmids pE2021 (for BTI1-GFP fusion), pE2027 (for BTI2-GFP fusion), and pE2019 (for BTI3-GFP fusion). Plasmids pE2021, pE2027, and pE2019 were subsequently electroporated into Agrobacterium GV3101 and used to generate Arabidopsis transgenic plants using a floral dip method (Clough and Bent, 1998).
Seeds from transgenic Arabidopsis plants expressing either the BTI-GFP fusion proteins or GFP were surface sterilized and placed on Gamborg's B5 medium (Gibco) with the appropriate antibiotics (kanamycin 50 μg/mL for BTI-GFP transgenic plants and phosphinothricin 10 μg/mL for GFP transgenic plants) and solidified with 0.75% Bactoagar (BD Biosciences) in vertically oriented square Petri plates. Seedlings were grown for 10 to 14 d using 16-h-light/8-h-dark conditions at 25°C before examining the subcellular localization of GFP fluorescence in root cells of Arabidopsis transgenic plants. The root tissues were examined using a Bio-Rad MRC-1024 confocal fluorescence microscope and the images were collected using the LaserSharp2000 program (Bio-Rad, Hercules, CA).
Supplementary Material
Acknowledgments
The authors thank Chien-Wei Chan for technical help in conducting some of these experiments, Chris Staiger for providing the actin antibody, Saikat Bhattacharjee for providing seeds of transgenic Arabidopsis plants expressing GFP protein, Stephen Farrand for helpful discussions, and Lan-Ying Lee and Susan Johnson for critical reading of the manuscript. This research was funded by the National Science Foundation (plant genome Grant 99-75715) to S.B.G.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Stanton B. Gelvin (gelvin@bilbo.bio.purdue.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026476.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony, K.G., Sherburne, C., Sherburne, R., and Frost, L.S. (1994). The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol. Microbiol. 13, 939–953. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (2003). Current Protocols in Molecular Biology. (New York: Wiley).
- Ballas, N., and Citovsky, V. (1997). Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl. Acad. Sci. USA 94, 10723–10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, C., Thorstenson, Y.R., and Zambryski, P.C. (1997). The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol. 179, 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20, 1195–1197. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J., and Martin, G.B. (2000). AvrPto-dependent Pto-interacting proteins and AvrPto-interacting proteins in tomato. Proc. Natl. Acad. Sci. USA 97, 8836–8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock, P., den Dulk-Ras, A., Beijersbergen, A., and Hooykaas, P.J. (1995). Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14, 3206–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundock, P., Mroczek, K., Winkler, A.A., Steensma, H.Y., and Hooykaas, P.J. (1999). T-DNA from Agrobacterium tumefaciens as an efficient tool for gene targeting in Kluyveromyces lactis. Mol. Gen. Genet. 261, 115–121. [DOI] [PubMed] [Google Scholar]
- Cascales, E., and Christie, P.J. (2003). The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales, E., and Christie, P.J. (2004). Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304, 1170–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, P.J. (1997). Agrobacterium tumefaciens T-complex transport apparatus: A paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179, 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, P.J. (2001). Type IV secretion: Intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- de Groot, M.J., Bundock, P., Hooykaas, P.J., and Beijersbergen, A.G. (1998). Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16, 839–842. [DOI] [PubMed] [Google Scholar]
- DeCleene, M., and DeLey, J. (1976). The host range of crown gall. Bot. Rev. 42, 389–466. [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version 2. Plant Mol. Biol. Rep. 1, 19–22. [Google Scholar]
- Ding, Z., Atmakuri, K., and Christie, P.J. (2003). The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 11, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditt, R.F., Nester, E.W., and Comai, L. (2001). Plant gene expression response to Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 98, 10954–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbrandt, R., Kalkum, M., Lai, E.M., Lurz, R., Kado, C.I., and Lanka, E. (1999). Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 274, 22548–22555. [DOI] [PubMed] [Google Scholar]
- Feldmann, K.A., and Marks, M.D. (1987). Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: A non-tissue culture approach. Mol. Gen. Genet. 208, 1–9. [Google Scholar]
- Gelvin, S.B. (2003). Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish, W., and States, D.J. (1993). Identification of protein coding regions by database similarity search. Nat. Genet. 3, 266–272. [DOI] [PubMed] [Google Scholar]
- Golemis, E., Gyuris, J., and Brent, R. (1994). Interaction trap/two-hybrid system to identify interacting proteins. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: John Wiley & Sons), pp. 13.14.1–13.14.17.
- Gouka, R.J., Gerk, C., Hooykaas, P.J., Bundock, P., Musters, W., Verrips, C.T., and de Groot, M.J. (1999). Transformation of Aspergillus awamori by Agrobacterium tumefaciens-mediated homologous recombination. Nat. Biotechnol. 17, 598–601. [DOI] [PubMed] [Google Scholar]
- GrandPre, T., Nakamura, F., Vartanian, T., and Strittmatter, S.M. (2000). Identification of the Nogo inhibitor of axon regeneration as a reticulon protein. Nature 403, 439–444. [DOI] [PubMed] [Google Scholar]
- Haubruck, H., Engelke, U., Mertins, P., and Gallwitz, D. (1990). Structural and functional analysis of ypt2, an essential ras-related gene in the fission yeast Schizosaccharomyces pombe encoding a Sec4 protein homologue. EMBO J. 9, 1957–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, K., and Stoffel, W. (1993). TMbase-A database of membrane spanning protein segments. Biol. Chem. Hoppe Seyler 374, 166. [Google Scholar]
- Huber, L.A., de Hoop, M.J., Dupree, P., Zerial, M., Simons, K., and Dotti, C. (1993). Protein transport to the dendritic plasma membrane of cultured neurons is regulated by rab8p. J. Cell Biol. 123, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.L., Lai, E.-M., Shirasu, K., and Kado, C.I. (1996). VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J. Bacteriol. 178, 5706–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado, C.I. (2000). The role of the T-pilus in horizontal gene transfer and tumorigenesis. Curr. Opin. Microbiol. 3, 643–648. [DOI] [PubMed] [Google Scholar]
- Kelly, B.A., and Kado, C.I. (2002). Agrobacterium-mediated T-DNA transfer and integration into the chromosome of Streptomyces lividans. Mol. Plant Pathol. 3, 125–134. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Krall, L., Wiedemann, U., Unsin, G., Weiss, S., Domke, N., and Baron, C. (2002). Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99, 11405–11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunik, T., Tzfira, T., Kapulnik, Y., Gafni, Y., Dingwall, C., and Citovsky, V. (2001). Genetic transformation of HeLa cells by Agrobacterium. Proc. Natl. Acad. Sci. USA 98, 1871–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E.M., and Kado, C.I. (2000). The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 8, 361–369. [DOI] [PubMed] [Google Scholar]
- Lai, E.M., and Kado, C.I. (2002). The Agrobacterium tumefaciens T pilus composed of cyclic T pilin is highly resilient to extreme environments. FEMS Microbiol. Lett. 210, 111–114. [DOI] [PubMed] [Google Scholar]
- Lai, E.M., Chesnokova, O., Banta, L.M., and Kado, C.I. (2000). Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 182, 3705–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E.M., Eisenbrandt, R., Kalkum, M., Lanka, E., and Kado, C.I. (2002). Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J. Bacteriol. 184, 327–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein, C., and Draper, J. (1986). Genetic engineering of plants. In DNA Cloning: A Practical Approach, Vol. 2, D.M. Glover, ed (Oxford: IRL Press), pp. 67–119.
- Madden, T.L., Tatusov, R.L., and Zhang, J. (1996). Applications of network BLAST server. Methods Enzymol. 266, 131–141. [DOI] [PubMed] [Google Scholar]
- Martinez, O., and Goud, B. (1998). Rab proteins. Biochim. Biophys. Acta. 1404, 101–112. [DOI] [PubMed] [Google Scholar]
- Matthysse, A.G., Holmes, K.V., and Gurlitz, R.H.G. (1982). Binding of Agrobacterium tumefaciens to carrot protoplasts. Physiol. Plant Pathol. 20, 27–33. [Google Scholar]
- Moyer, B.D., and Balch, W.E. (2001). Structural basis for Rab function: An overview. Methods Enzymol. 329, 3–6. [DOI] [PubMed] [Google Scholar]
- Mysore, K.S., Bassuner, B., Deng, X.-b., Darbinian, N.S., Motchoulski, A., Ream, W., and Gelvin, S.B. (1998). Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant-Microbe Interact. 11, 668–683. [DOI] [PubMed] [Google Scholar]
- Nam, J., Matthysse, A.G., and Gelvin, S.B. (1997). Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 9, 317–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, J., Mysore, K.S., and Gelvin, S.B. (1998). Agrobacterium tumefaciens transformation of the radiation hypersensitive Arabidopsis thaliana mutants uvh1 and rad5. Mol. Plant-Microbe Interact. 11, 1136–1141. [DOI] [PubMed] [Google Scholar]
- Nam, J., Mysore, K.S., Zheng, C., Knue, M.K., Matthysse, A.G., and Gelvin, S.B. (1999). Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 261, 429–438. [DOI] [PubMed] [Google Scholar]
- Narasimhulu, S.B., Deng, X.B., Sarria, R., and Gelvin, S.B. (1996). Early transcription. of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8, 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, N.T., and Binns, A.N. (1985). Agrobacterium tumefaciens interaction with suspension-cultured tomato cells. Plant Physiol. 77, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M., Cui, D., Einstein, J., Narasimhulu, S., Vergara, C.E., and Gelvin, S.B. (1995). Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 7, 661–676. [Google Scholar]
- Oertle, T., and Schwab, M.E. (2003). Nogo and its paRTNers. Trends Cell Biol. 13, 187–194. [DOI] [PubMed] [Google Scholar]
- Oertle, T., Klinger, M., Stuermer, C.A., and Schwab, M.E. (2003). A reticular rhapsody: Phylogenic evolution and nomenclature of the RTN/Nogo gene family. FASEB J. 17, 1238–1247. [DOI] [PubMed] [Google Scholar]
- Olkkonen, V.M., and Stenmark, H. (1997). Role of Rab GTPases in membrane traffic. Int. Rev. Cytol. 176, 1–85. [DOI] [PubMed] [Google Scholar]
- Piers, K.L., Heath, J.D., Liang, X., Stephens, K.M., and Nester, E.W. (1996). Agrobacterium tumefaciens-mediated transformation of yeast. Proc. Natl. Acad. Sci. USA 93, 1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, S., and Moore, I. (2002). The Arabidopsis Rab GTPase family: Another enigma variation. Curr. Opin. Plant Biol. 5, 518–528. [DOI] [PubMed] [Google Scholar]
- Sagulenko, V., Sagulenko, E., Jakubowski, S., Spudich, E., and Christie, P.J. (2001). VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183, 3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmoller, F., Simon, I., and Pfeffer, S.R. (1998). Rab GTPases, directors of vesicle docking. J. Biol. Chem. 273, 22161–22164. [DOI] [PubMed] [Google Scholar]
- Schmidt-Eisenlohr, H., Domke, N., Angerer, C., Wanner, G., Zambryski, P.C., and Baron, C. (1999). Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181, 7485–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senden, N.H., Timmer, E.D., Boers, J.E., van de Velde, H.J., Roebroek, A.J., Van de Ven, W.J., Broers, J.L., and Ramaekers, F.C. (1996). Neuroendocrine-specific protein C (NSP-C): Subcellular localization and differential expression in relation to NSP-A. Eur. J. Cell Biol. 69, 197–213. [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira, T., and Citovsky, V. (2002). Partners-in-infection: Host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol. 12, 121–129. [DOI] [PubMed] [Google Scholar]
- van de Velde, H.J., Roebroek, A.J., Senden, N.H., Ramaekers, F.C., and Van de Ven, W.J. (1994. a). NSP-encoded reticulons, neuroendocrine proteins of a novel gene family associated with membranes of the endoplasmic reticulum. J. Cell Sci. 107, 2403–2416. [DOI] [PubMed] [Google Scholar]
- van de Velde, H.J., Roebroek, A.J., van Leeuwen, F.W., and Van de Ven, W.J. (1994. b). Molecular analysis of expression in rat brain of NSP-A, a novel neuroendocrine-specific protein of the endoplasmic reticulum. Brain Res. Mol. Brain Res. 23, 81–92. [DOI] [PubMed] [Google Scholar]
- Veena, Jiang, H., Doerge, R.W., and Gelvin, S.B. (2003). Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 35, 219–236. [DOI] [PubMed] [Google Scholar]
- Vernoud, V., Horton, A.C., Yang, Z., and Nielsen, E. (2003). Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131, 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, D.V., Draper, O., Zupan, J.R., and Zambryski, P.C. (2002). Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99, 11493–11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, H., Mysore, K.S., and Gelvin, S. (2002). Expression of the Arabidopsis histone H2A–1 gene correlates with susceptibility to Agrobacterium transformation. Plant J. 32, 285–298. [DOI] [PubMed] [Google Scholar]
- Yusibov, V.M., Steck, T.R., Gupta, V., and Gelvin, S.B. (1994). Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl. Acad. Sci. USA 91, 2994–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and Madden, T.L. (1997). PowerBLAST: A new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 7, 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Nam, J., Carpita, N.C., Matthysse, A.G., and Gelvin, S.B. (2003. a). Agrobacterium-mediated root transformation is inhibited by mutation of an Arabidopsis cellulose synthase-like gene. Plant Physiol. 133, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., et al. (2003. b). Identification of Arabidopsis rat mutants. Plant Physiol. 132, 494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan, J., Muth, T.R., Draper, O., and Zambryski, P. (2000). The transfer of DNA from Agrobacterium tumefaciens into plants: A feast of fundamental insights. Plant J. 23, 11–28. [DOI] [PubMed] [Google Scholar]
- Zupan, J.R., Ward, D., and Zambryski, P. (1998). Assembly of the VirB transport complex for DNA transfer from Agrobacterium tumefaciens to plant cells. Curr. Opin. Microbiol. 1, 649–655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.