Abstract
Our understanding of the cellular mechanisms by which animals regulate their response to starvation is limited, despite the strong relevance of the problem to major human health issues. The L1 diapause of Caenorhabditis elegans, where first-stage larvae arrest in response to a food-less environment, is an excellent system to study this mechanism. We found, through genetic manipulation and lipid analysis, that biosynthesis of ceramide, particularly those with longer fatty acid side chains, critically impacts animal survival during L1 diapause. Genetic interaction analysis suggests that ceramide may act in both insulin-IGF-1 signaling (IIS)-dependent and IIS-independent pathways to affect starvation survival. Genetic and expression analyses indicate that ceramide is required for maintaining the proper expression of previously characterized starvation-responsive genes, genes that are regulated by the IIS pathway and tumor suppressor Rb, and genes responsive to pathogen. These findings provide an important insight into the roles of sphingolipid metabolism, not only in starvation response, but also in aging and food-response-related human health problems.
Keywords: serine palmitoyltransferase, Rb, pathogen, hyl-1, daf-16
DURING evolution, organisms have developed complex mechanisms to adapt to food-deprived environments. Individual cells respond to starvation by modulating intracellular signaling to maintain basal cellular activities and survive long periods of starvation (Caro-Maldonado and Munoz-Pinedo 2011; Hardie 2011; Jonker et al. 2012). The study of an animal’s response to starvation-induced stress is highly relevant to human health and medicine. For example, the study of signaling pathways and downstream events involved in the starvation response has had a major impact on the research of aging (Finch and Ruvkun 2001; Kenyon 2010) and obesity (Hoehn et al. 2009; Wells and Siervo 2011). Understanding the regulation of the starvation response is also closely related to our treatment of cancers (Levine and Puzio-Kuter 2010; Lee et al. 2012).
The nematode Caenorhabditis elegans presents a powerful model system for genetic and genomic analysis of the starvation response in animals. C. elegans responds to food deprivation by altering reproductive developmental growth at various larval stages (Fielenbach and Antebi 2008; Baugh 2013). When first larval stage (L1) animals encounter a food-free environment, they arrest development and reproductive growth (L1 diapause), and survive in this state for over 3 weeks. When nutrients are reintroduced, animals are capable of exiting the diapause and resuming larval development. Gene expression changes defined as the “starvation-induced transcriptome,” and “refeeding induced transcriptome,” have been systematically characterized (Baugh et al. 2009).
The roles of the insulin-IGF-1 signaling (IIS) and AMPK pathways in starvation are conserved among organisms ranging from yeast, worms, and flies, to mice (Baugh and Sternberg 2006; Narbonne and Roy 2009; Hardie 2011). A transient receptor potential vanilloid (TRPV) channel and microRNAs were shown to modulate L1 diapause by regulating the IIS-dependent or -independent pathways (Lee and Ashrafi 2008; Zhang et al. 2011b). The interneurons AIY and AIB provide systemic control of the starvation response during L1 arrest partly through amino acid sensation (Kang and Avery 2009). We previously reported that tumor suppressor Rb critically impacts survival during L1 diapause by promoting the starvation-induced transcriptome, and repressing the refeeding induced transcriptome (Cui et al. 2013), which affect the activity of multiple regulatory pathways. Identifying and analyzing new factors involved in the process continues to be important for us to understand how different signals and regulatory pathways coordinately promote long-term survival of animals in response to starvation. In particular, the roles of lipid metabolites in the process have not been well characterized.
Ceramide has been studied for its role in apoptosis and the response to certain stresses such as anoxia, cytokines, toxins, and chemotherapeutic agents (Deng et al. 2008; Hannun and Obeid 2008; Menuz et al. 2009; Nikolova-Karakashian and Rozenova 2010; Mosbech et al. 2013; Cutler et al. 2014; Liu et al. 2014). Most of these studies were carried out in nutrient-rich conditions, while the roles of ceramide under fasting conditions in whole organisms remain to be investigated. In this study, we investigate the roles of ceramide biosynthesis in C. elegans survival during starvation-induced L1 diapause.
Materials and Methods
Strains
Strains were cultured and maintained at 20° unless specified otherwise. Strains lin-35(n745), lagr-1(gk327), hyl-1(ok976), hyl-2(ok976), sptl-2(ok2753), and sptl-3(ok1927) were outcrossed five times to wild-type Bristol N2. The following strains were used in this study: daf-16(mu86), age-1(hx546), asm-2(tm3746), asm-3(tm2384), sms-1(tm2660), sms-2(tm2757), sms-3(tm4022), cgt-1(tm1027), cgt-3(tm504), sphk-1(ok1097), asah-2(ok564), cerk-1(ok1252), unc-31(e928), zIs356 [daf-16p::DAF-16a/b::GFP; rol-6], and adIs2122 carrying lgg-1:GFP.
Plasmid construction and transgenic animals
For tissue-specific expression, promoters of rgef-1 (pan-neurons), ges-1 (intestine), myo-2 (pharynx muscle), and myo-3 (body wall muscle) were amplified from N2 genomic DNA, and cloned into pPD95.77. The hyl-1 genomic DNA fragment from the translational start codon to the stop codon was amplified from N2 genomic DNA. It was then placed behind the specific promoter and followed by the unc-54 3′-UTR. Each DNA construct (50 ng/μl) was coinjected with the sur-5::dsRed (25 ng/μl) into lagr-1(gk327); hyl-1(ok976) animals to create three or more extrachromosomal lines. The myo-2P:hyl-1 and myo-3P:hyl-1 constructs were combined to create muscle-specific expression lines.
L1 starvation survival assay and statistical analysis
The L1 starvation assay was done following the protocol described previously (Lee and Ashrafi 2008; Cui et al. 2013; Zhang et al. 2011b). Survival curves were drawn based on three or more independent experiments. To perform the basic starvation survival analysis, we simulated the survival rate of each genotype to 100 arbitrary “individual worms.” The mean survival rate of individual replicates was calculated through the OASIS software available at http://sbi.postech.ac.kr/oasis (Yang et al. 2011). The average of the mean survival rate of all individual replicates for each strain was calculated. The mean survival rates of individual replicates were used to assess the difference between different strains or conditions. The statistical analyses (P value) to assess the difference between the mean survival rates were conducted using Student’s t-test
RNAi by feeding
A zip-2 feeding RNAi strain was obtained from the C. elegans ORF-RNAi library (Rual et al. 2004). Control RNAi was the L4440 RNAi feeding vector (Addgene; A. Fire, Stanford University School of Medicine, Stanford, CA) without any C. elegans DNA insert. Synchronized L1s were fed on zip-2(RNAi) or control RNAi plates. The resulting gravid adults were bleached, and the L1 starvation survival of their progeny was measured.
Caenorhabditis elegans total lipid extraction
Synchronized L1 larvae (30 hr after bleach treatment) of wild type Bristol N2, sptl-2(lf), and lagr-1(lf); hyl-1(lf), were obtained using the same procedure as in L1 starvation survival assay. Around 900,000 L1s for each strain were collected as one experimental sample. Worm pellets were subjected to three cycles of freezing in liquid nitrogen and thawing followed by sonication. Lipid extraction was carried out in the presence of prespiked internal standards (Avanti Polar Lipids, Catalog#: LM6002) with 2:1 methanol and chloroform at 48° for 24 hr, followed by 15 min sonication at 37°, as described previously (Zhang et al. 2011a). The resulting samples were back-extracted with chloroform. After centrifugation, the lower organic phase was collected, washed once with artificial upper phase (chloroform/methanol/H2O; 3:48:47), and dried under nitrogen gas.
Ceramide profiling by ESI-mass spectrometry
Lipid extracts were dissolved in isopropanol:hexane:100 mM ammonium acetate (58:40:2) with 1% formic acid added, and subjected to quantitative lipid analysis using a 4000 Q-Trap mass spectrometer (AB Sciex). Samples were infused at a flow rate of 8 μl/min using a syringe pump (Harvard Apparatus). Ceramides were detected using precursor ion scans in the positive mode for the 250.3 μ fragment of D17:1 sphingoid bases. For mutants and wild type control animals, the extracts from two different batches of culture were analyzed. The relative amounts of total ceramide species were calculated relative to the number of worms.
RNA isolation
Synchronized L1 worms were obtained as above. Animal samples were collected 30 hr after bleaching. Total RNA was isolated using Trizol (Invitrogen) as per the manufacturer’s protocol, and then treated with Turbo DNase (Ambion), followed by RNA cleanup using an RNeasy Mini Kit (Qiagen).
Affymetrix microarray analysis
Analyses were performed using Affymetrix Genechip Arrays for C. elegans as per the manufacturer’s protocol at the Genomics and Microarray Core in the University of Colorado Denver. Biological replicates were analyzed in triplicate for wild type (Bristol N2) and lagr-1(gk327); hyl-1(ok976) (CerS(rf)). Microarray analyses were performed with GeneSifter web-based software (VizX Labs), using the GC robust multi-array average (RMA) algorithm, and analyzed by applying a statistical t-test: P < 0.01 for lagr-1(gk327); hyl-1(ok976) analysis with a threshold of twofold ratio of dysregulation. Gene lists were curated by cross-referencing with WormBase (http://www.wormbase.org, release WS230). When annotation indicated that a single probe correlated with multiple genes, all such genes were excluded from our final lists. Curated gene lists are included in Supplemental Material, Table S2A. The microarray gene expression data are available at http://www.ncbi.nlm.nih.gov/geo/ (accession number GSE84894).
Tissue enrichment analysis
Tissue enrichment analysis was carried out with the Tissue Expression Predictions for C. elegans program, version 1.0. (http://worm-tissue.princeton.edu/search/multi) (Chikina et al. 2009).
Gene ontology analysis
We performed gene ontology (GO) analysis by using the online tool (www.geneontology.org). Of the 272 ceramide-regulated genes, 213 genes had annotations to Biological Process GO terms. GO categories were retained if their Bonferroni-corrected P values were <5%. P-value was calculated by the binomial statistics.
Overlap analysis among different sets of genes
Four available datasets were used for overlap analysis with the list of ceramide-regulated genes identified in our microarray data analysis. (1) Two classes of daf-16-responsive genes were reported in Tepper et al. (2013). (2) lin-35/Rb regulated genes during L1 starvation were reported in Cui et al. (2013). (3) The FedUP and StarvUP gene lists obtained from www.wormbase.org were originally submitted by R. Baugh based on the published paper (Baugh et al. 2009) (http://www.wormbase.org/species/c_elegans/expression_cluster/WBPaper00032948:FedUp#0154-10; http://www.wormbase.org/species/c_elegans/expression_cluster/WBPaper00032948: StarveUp1#0154–10). (4) The Pseudomonas aeruginosa PA14 induced genes represent the pathogen-response genes (Troemel et al. 2006).
The hypergeometric probability test for statistical significance of the overlap between two sets of genes was calculated by using software provided by J. Lund accessible at http://nemates.org/MA/progs/overlap_stats.html. The total number of genes represents the overlap genes from the original data sets that the two sets of genes were generated from. The test gave rise to a representation factor (rf) and the probability (p) of finding an overlap of x genes. The representation factor is the number of overlapping genes divided by the expected number of overlapping genes drawn from two independent groups. A representation factor >1 indicates more overlap than expected of two independent groups, and a representation factor <1 indicates less overlap than expected. Expected overlap was determined by multiplying the number of genes dysregulated in data set 1 by the number dysregulated in data set 2, and then dividing by the total number of genes that were detectable (for microarray datasets) and present in both datasets.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Strains are available upon request.
Results
Reducing ceramide synthesis severely impairs animals’ ability to survive L1 starvation
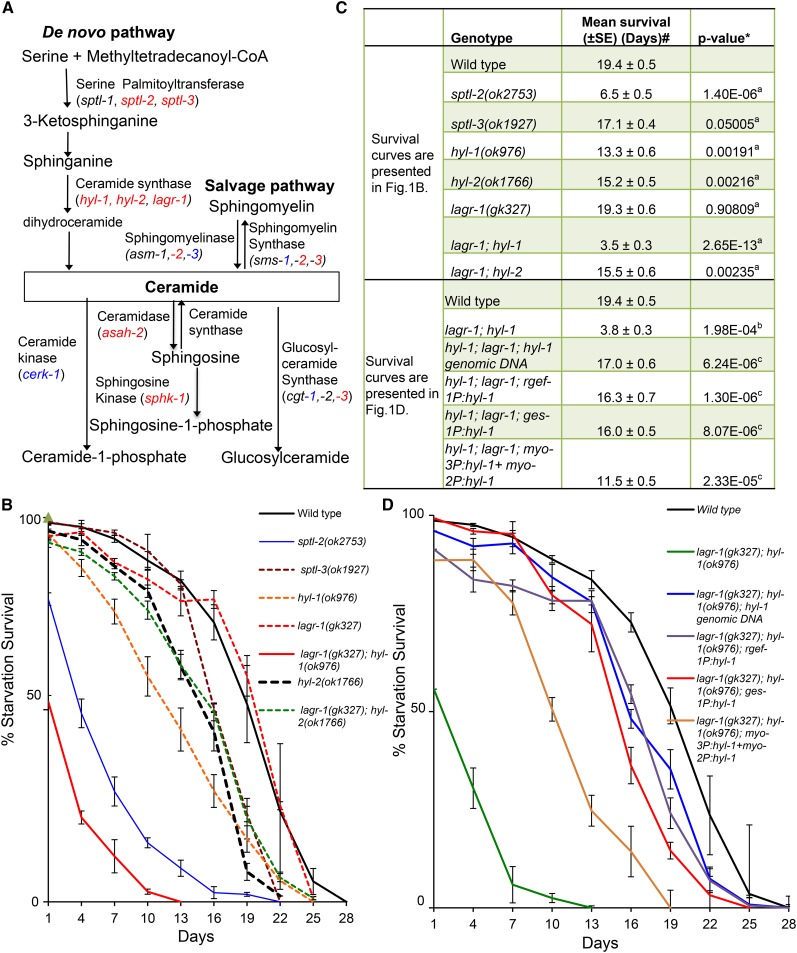
To understand the role of ceramide in regulating animal starvation survival, we screened all available, viable, C. elegans mutants in the ceramide metabolism pathways for altered L1 starvation survival rates (Cui et al. 2013) (Figure 1A and Table S1), and found many such mutants were significantly short-lived during L1 starvation (Figure 1, A–C, Figure S1, A–C, and Table S1). Specifically, animals with mutations in the de novo ceramide biosynthesis pathway were sensitive to starvation-induced stress (Figure 1, B–C and Table S1). sptl-2 and sptl-3 encode two of the three serine-palmitoyltransferase (SPT) enzymes, and hyl-1 and hyl-2 encode two of the three ceramide synthase (CerS) enzymes (Deng et al. 2008) in C. elegans. Deletion mutants of each of these four genes resulted in significantly reduced L1 starvation survival rates (Figure 1, B and C). Although a deletion mutant of the third ceramide synthase gene, lagr-1, displayed no observable defect in L1 starvation survival, it enhanced the defect of the hyl-1 mutant in a lagr-1; hyl-1 double-deletion mutant (Figure 1, B and C). Because all deletions mentioned above truncate a key functional domain, they are most likely loss-of-function [referred to as (lf) hereafter] mutants.
Figure 1.
Ceramide functions to promote L1 starvation survival. (A) Simplified diagram to illustrate the C. elegans orthologs of genes involved in ceramide biosynthesis and metabolism. Deletion mutations of genes in red, but not genes in blue, displayed a significant defect in L1 starvation survival (see Table S1 for raw data and statistical analysis). Genes that are not highlighted with red or blue color are not included in L1 starvation study due to unavailable mutants, or the mutation of that gene resulting larval lethality. (B) Survival rates of ceramide synthesis reduction mutants [lagr-1(gk327); hyl-1(ok976) is referred as CerS(rf) in all figures]. Percentage survival is defined as the percentage of worms surviving to the third larval stage and beyond on NGM plates with OP50 bacteria after incubation in S-basal buffer in the absence of food for the indicated time. Data of each strain represent the mean of three or more independent biological replicates. Errors bars are the SEs at each time point indicated. (C) The mean survival rate of individual replicates was calculated through OASIS software available at http://sbi.postech.ac.kr/oasis (Yang et al. 2011). #The average of the mean survival rate of all individual replicates for each strain is presented here. *The statistical analyses (P value) to assess the difference between the mean survival rates were conducted using Student’s t-test. a,b,cP values indicate the significance of the difference from wild type in (B) (aP), wild type in (D) (bP) and lagr-1(gk327); hyl-1(ok976) mutant animals in (D) (cP), respectively. (D) Starvation survival rate of the CerS(rf) L1 mutant animals carrying extrachromosomal arrays expressing the wild-type hyl-1 gene driven by its own promoter, and three other tissue specific promoters [intestine (ges-1), pan-neurons (rgef-1), and muscle (myo-2/myo-3)]. Transgenic animals were scored on the basis of the expression of the sur-5P:dsRed coinjection marker with a Leica fluorescence microscope. Percentage survival is defined as the percentage of animals surviving to the third larval stage and beyond on food after L1 worms were starved in S-basal buffer for the indicated time. The average from multiple independent transgenic lines for each genotype is reported with the SEM for each time point (±SEM). The starvation survival data for wild-type animals are the same as that presented in (B). Raw data and the statistical analysis data for individual starvation survival experiments for (B) and (D) are presented in Table S1.
Ceramides are essential to animal development. Depletion of ceramides by completely eliminating the key enzymes in the ceramide synthesis pathway results in larval lethality. Examples such as sptl-1(RNAi), hyl-1(lf); hyl-2(lf) double mutants and cgt-1(RNAi); cgt-3(RNAi) double RNAi have been shown to cause strong larval lethal phenotypes (Menuz et al. 2009; Seamen et al. 2009; Nomura et al. 2011). The above tested mutants represent reduction, but not elimination, of the function at the corresponding enzymatic steps. Therefore, we refer to the lagr-1(lf): hyl-1(lf) double mutants as CerS(rf). The sptl-2(lf) and CerS(rf) mutants showed a significantly reduced survival rate at day 1 (Figure 1B), which was not due to embryonic lethality or early larval lethality.
Tissue-specific expression suggests a nonautonomous role of the ceramide synthase
HYL-1 is expressed in the pharynx, intestine, and nervous system, while LAGR-1 is expressed in larval and adult pharyngeal muscle (Hunt-Newbury et al. 2007). The full-length hyl-1 genomic DNA restored L1 starvation survival of the CerS(rf) mutants (Figure 1, C and D). We next examined whether ceramide synthases act mainly in a particular tissue for the L1 starvation survival function. Specifically, we asked whether expressing HYL-1 in one of the three major tissues (intestine, neurons, or muscle), using transgenes driven by tissue-specific promoters would be sufficient to restore the L1 starvation survival in the CerS(rf) animals. The expression of HYL-1 in either the intestine (ges-1 promoter) or neurons (rgef-1 promoter) could effectively rescue the defect of CerS(rf) animals (Figure 1, C and D). Expression of HYL-1 in the pharynx (myo-2 promoter) and body wall muscle (myo-3 promoter) partially restored the L1 starvation survival of CerS(rf) animals (Figure 1, C and D and Table S1). The partial rescue from muscle-specific expression of HYL-1 may be due to lower expression of HYL-1 or may be due to the inefficiency of ceramide export from muscle cells. These results at least suggest that ceramide synthesis in multiple tissues can protect C. elegans from starvation stress.
Reduction of very long fatty acyl chain ceramide levels correlates with reduced L1 starvation survival
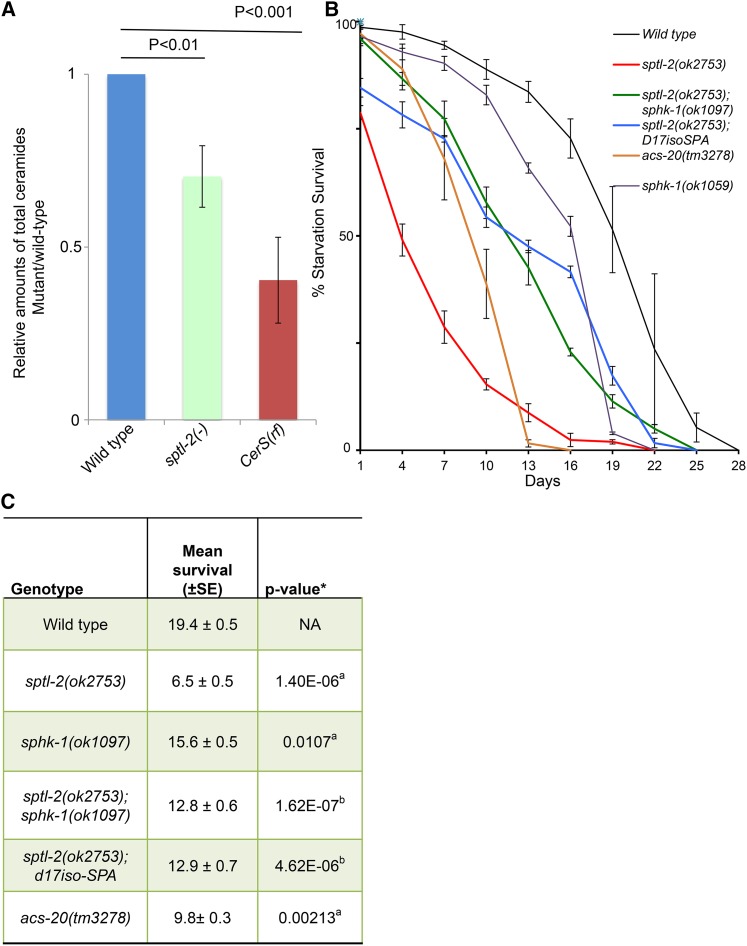
Ceramides are produced from sphinganine and fatty acyl-CoAs by the actions of multiple CerS, each of which has a preference for a specific fatty acyl-CoA (Grosch et al. 2012). The hyl-1(lf) and hyl-2(lf) mutants displayed differential starvation survival phenotypes, with the hyl-1 mutant being more sensitive to starvation stress. [The mean survival rate was 13.3 ± 0.6 for hyl-1(lf) and 15.2 ± 0.5 for hyl-2(lf). P value for the difference of these two mutants by the Student’s t-test is 0.034 (Figure 1B)]. We therefore quantified the major ceramide species in starved L1 worms of wild type, sptl-2(lf), and lagr-1(lf); hyl-1(lf) [CerS(rf)] mutants by electrospray ionization mass spectrometry (ESI-MS). The total ceramide levels of either sptl-2(lf) or CerS(rf) animals were significantly lower than those of wild-type animals (Figure 2A). Furthermore, it has been reported that hyl-1(lf) worms expressed significantly lower C25 and C26 ceramides, but more C21 and C22 ceramides compared to wild type (Menuz et al. 2009). These data suggest that levels of total ceramides, especially those with very long fatty acyl chains, promote survival of animals under starved conditions. Consistent with this hypothesis, disrupting the acs-20 gene, which encodes an acyl-coA synthetase that has been shown to incorporate exogenous very long chain (C26:0) fatty acids into sphingolipids (Kage-Nakadai et al. 2010), significantly shortened starvation survival (Figure 2, B and C).
Figure 2.
L1 starvation survival is reduced when ceramide levels are decreased, but it can be partially rescued with sphingoid base supplementation. (A) Ceramide levels are decreased in both sptl-2(lf) and lagr-1(lf); hyl-1(lf) double mutants [referred to CerS(rf)]. Total ceramide levels of mutants relative to that of wild type are shown in columns. Errors bars represent SE at each time point indicated. P values were calculated by Student’s t-test. (B) Survival rates of wild type, sptl-2(lf) ± dietary supplementation, sptl-2(lf); sphk-1(ok1097), and two acs-20 mutants. The sptl-2(lf) defect was significantly suppressed by both dietary supplementation of 250 nM iso-branched d17iso-sphinganine (d17iso-SPA) and sphk-1(ok1097) mutation. Percentage survival is defined as the percentage of animals surviving to the third larval stage and beyond on food after L1 worms were starved in S-basal buffer for the indicated time. The starvation survival data for wild type and sptl-2(lf) are the same as that presented in Figure 1B. (C) The mean survival rate of individual replicates was calculated through OASIS software. The average of the mean survival rate of all-individual replicates for each strain is presented here. *The statistical analyses (P value) to assess the difference between the mean survival rates were conducted using Student’s t-test. a,bP values indicate the significance of the difference from wild type (aP) and sptl-2(ok2753) mutant animals (bP), respectively. Raw data and detailed statistical analysis data for individual starvation survival experiments for figure 1B are presented in Table S1.
Dietary supplementation with sphingoid bases significantly rescues the reduced L1 starvation survival of sptl-2(lf)
Short fatty acyl chain ceramides (C6 and C8) are soluble in DMSO, but are toxic to animals in high concentrations, and may not be physiologically relevant. The long-acyl chain ceramides (C16–25) are insoluble in either DMSO or aqueous solution, rendering dietary supplement analysis difficult. Indeed, we failed to rescue the L1 starvation survival of either sptl-2(lf) or CerS(rf) mutants with dietary supplementation of ceramides containing various lengths of fatty acyl chains. In contrast, the sphingoid bases (sphinganine), which are ceramide precursors and downstream products of the serine palmitoyltransferase, have better solubility in DMSO and aqueous solution. In C. elegans, the majority of sphingoid bases are derived from monomethyl branched-chain fatty acids (C15ISO and C17ISO) (Zhu et al. 2013). We then examined dietary supplementation with a custom synthesized d17iso-SPA (sphinganine), and found it was able to partially rescue the L1 starvation survival of sptl-2(lf) (Figure 2, B and C). This result may also suggest that intestinal ceramides play prominent roles in promoting starvation survival. Furthermore, sphk-1(ok1097), a null allele of the sphingosine kinase that causes animals to lose the ability to convert sphingosine to sphingosine-1-phosphate, significantly rescued the L1 starvation survival of sptl-2(lf), even though the sphk-1(ok1097) mutation alone causes only a modest reduction in L1 starvation survival (Figure 2, B and C). These data indicate that sphingosines, the ceramide precursors, are critical for L1 starvation survival, whereas sphingosine-1-phosphate may also have a modest role in the process.
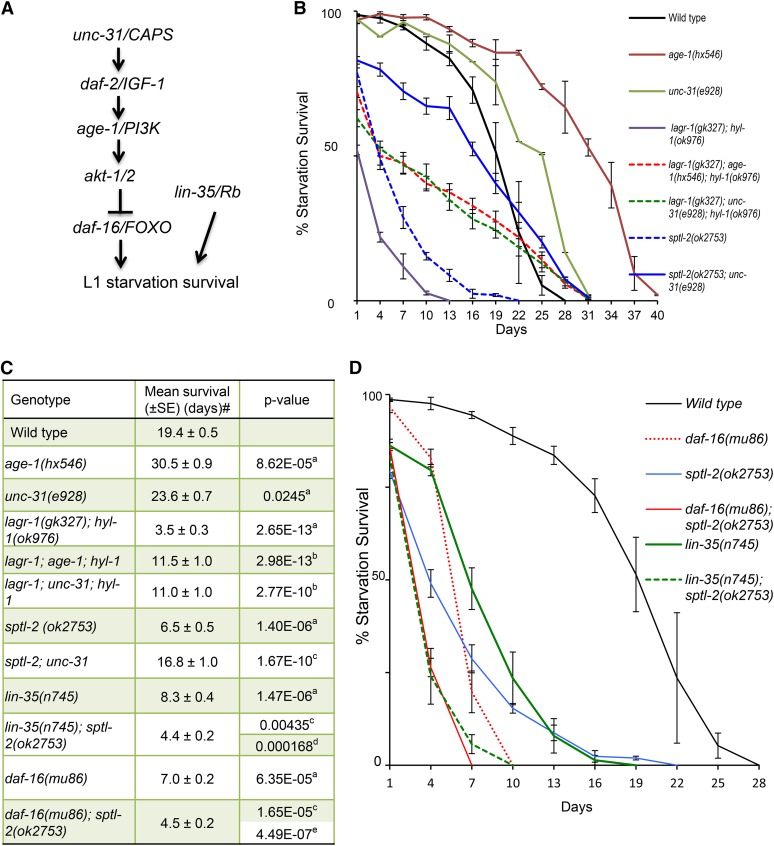
Change in IIS pathway significantly affects starvation survival of CerS(rf) animals
Previous studies have indicated that the IIS pathway critically regulates L1 starvation survival (Baugh and Sternberg 2006; Lee and Ashrafi 2008; Zhang et al. 2011b). A loss-of-function (lf) mutation of unc-31/CAPS and a reduction-of-function (rf) allele of age-1/PI3K were shown to extend L1 starvation survival (Lee and Ashrafi 2008; Zhang et al. 2011b) (Figure 3, A and B), while lf mutations in daf-18/Pten and daf-16/FOXO, both negative regulators of the pathway, shortened L1 starvation survival and lifespan (Baugh and Sternberg 2006; Kenyon 2010; Cui et al. 2013) (Figure 3A). We thus analyzed the genetic interactions of CerS with unc-31, age-1, and daf-16. We found that the starvation survival defects associated with both CerS(rf) mutants were partially but significantly suppressed by both unc-31(lf) and age-1(rf) mutations (Figure 3, B and C). The survival rates of CerS(rf); unc-31(lf) and CerS(rf); age-1(rf) mutants were more than threefold higher than that of CerS(rf), but more than onefold lower than that of unc-31(lf) and age-1(rf). These partial suppression data suggest that ceramide may potentially function both upstream of the IIS pathway and through IIS-independent mechanisms. To further examine their functional relationship, we combined the sptl-2(lf) allele with a daf-16(lf) allele, and found that the mean and maximum L1 starvation survival rates of this strain were significantly lower than that of either single mutant (Figure 3, C and D). A similar functional relationship is also observed in sptl-2(lf); lin-35/Rb(lf) double mutants (Figure 2D and Figure 3C). Therefore, neither the IIS pathway nor Rb is the sole major target of ceramide for L1 starvation survival.
Figure 3.
Ceramide may regulate L1 starvation survival by affecting both the Rb and IIS pathway-dependent and -independent functions. (A) A simplified diagram of the Rb and IIS pathway and their relationship with L1 starvation survival. (B) Survival curves showing that the L1 starvation survival defect of the CerS(rf) mutant and the sptl-2(lf) mutant was partially rescued by unc-31(lf) and age-1(rf). Percentage survival is defined as the percentage of animals surviving to the third larval stage and beyond on food after L1 worms were starved in S-basal buffer for the indicated time. Errors bars represent SE at each time point indicated. The starvation survival data for wild type, CerS(rf), and sptl-2(lf) are the same as that presented in Figure 1B. (C) Table of mean survival and P-values for survival curves presented in (B) and (D). # The average of the mean survival rate of all individual replicates for each strain is presented here. The combination of sptl-2(lf) with daf-16(lf) or lin-35/Rb(lf) resulted in more severe defects than each single mutant. a,b,c,d,eP values indicate the significance of the difference from wild type (aP), CerS(rf) (bP), sptl-2(ok2753) (cP), lin-35(n745) (dP), and daf-16(mu86) (eP) respectively. (D) Survival curves showing the defect of sptl-2(lf) was enhanced by either daf-16(lf) or lin-35/Rb(lf). % Survival is defined as the percentage of animals surviving to the third larval stage and beyond on food after L1 worms were starved in S-basal buffer for the indicated time. Errors bars represent SE at each time point indicated. The starvation survival data for wild type and sptl-2(lf) are the same as that presented in Figure 1B. Raw data and statistical analyses for individual starvation survival experiments for Figure 3 are presented in Table S1.
Transcriptional profiles of the CerS(rf) mutant during L1 starvation
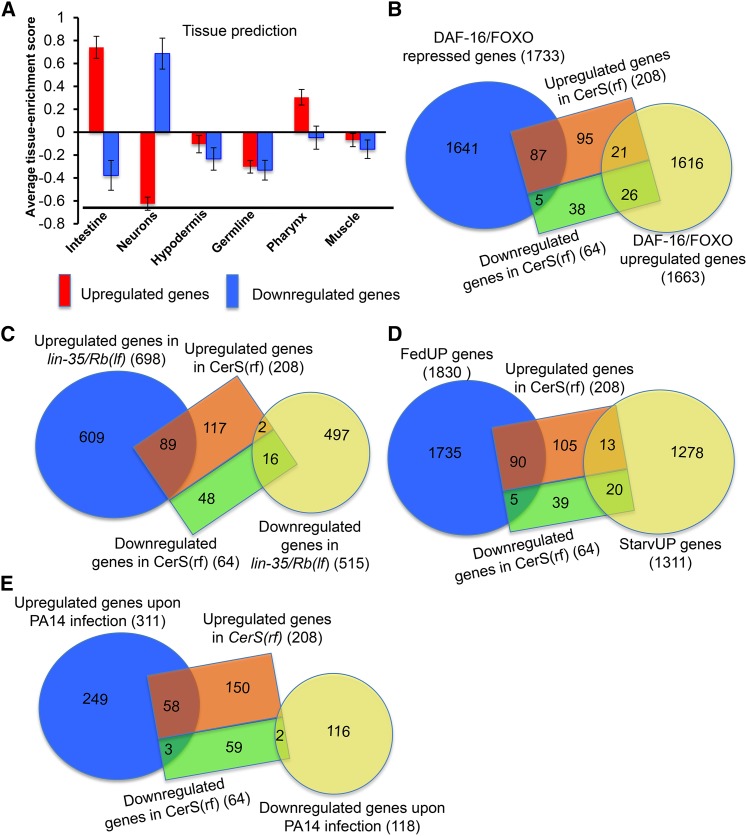
To learn more about functions downstream of ceramide in regulating L1 diapause, we compared the global gene-expression profiles of the CerS(rf) mutant and wild type animals using high-density oligonucleotide microarrays. Our microarray data analysis revealed that at a P < 0.01 and twofold cut-off, 272 genes were dysregulated in the CerS(rf) mutant (Table S2A). Of the 272 dysregulated genes, 77% (208/272) were upregulated, and 33% (64/272) were downregulated in the CerS(rf) mutant during L1 diapause. Tissue enrichment predications suggested that upregulated genes are enriched in the intestine, and downregulated genes are enriched in neurons (Figure 4A). GO analysis revealed that “immune response,” “response to defense,” and “response to stress” descriptors were highly enriched among upregulated genes (Table S3).
Figure 4.
Ceramide synthases regulate the expression of genes regulated by the IIS-pathway, Rb, feeding, starvation condition, and genes induced by pathogen. (A) Tissue enrichment analysis. Tissue enrichment scores were calculated by the online program Tissue Expression Prediction for C. elegans (Chikina et al. 2009). (B) Diagram showing the overlap between dysregulated genes in CerS(rf) and DAF-16/FOXO responsive genes (Tepper et al. 2013). The lists of overlapping genes are included in Table S2, B and C. The statistical significance of the overlap is presented in Table S4. (C) Diagram showing the overlap between dysregulated genes in CerS(rf) and lin-35/Rb (Cui et al. 2013). The list of overlapping genes is included in Table S2D. The statistical significance of the overlap is presented in Table S4. (D) Diagram showing the overlap between dysregulated genes in CerS(rf) and FedUP genes or StarvUP genes (Baugh et al. 2009). The lists of overlapping genes are included in Table S2, F and G. The statistical significance the overlap is presented in Table S4. (E) Diagram showing the overlap between dysregulated genes in CerS(rf) and previously identified pathogen (P. aeruginosa, PA14)-inducible genes (Troemel et al. 2006). The list of overlapping genes is included in Table S2H. The statistical significance of the overlap is presented in Table S4.
Ceramide affects the expression of daf-16/FOXO- and Rb-regulated genes
We further analyzed the functional relationship between ceramide biosynthesis and the IIS pathway by comparing our microarray data with other relevant transcriptional profiles. The comparison between CerS(rf) affected genes and daf-16/FOXO responsive genes (Murphy et al. 2003; Tepper et al. 2013) identified 139 overlapping genes, which is 2.5-fold higher than expected by chance (P < 1.66 e−30) (Figure 4B, Table S2, B and C, and Table S4). Among these 139 genes, 92 are repressed by daf-16/FOXO, and 47 are upregulated by daf-16/FOXO. Of the 92 overlapping genes that are repressed by daf-16/FOXO, 87 were upregulated in the CerS(rf) mutant, which is 4.1-fold higher than expected by chance (P < 7.39 e−33) (Table S2C and Table S4). These results indicated that there are a significant number of genes affected by both ceramide and the IIS-DAF-16 pathways, suggesting that ceramide acts in part through IIS to affect DAF-16 targets, which is consistent with the suggestion from above genetic interaction data that ceramide may partially function upstream of the IIS pathway to regulate L1 starvation survival.
Rb has been shown to play an important role in regulating the starvation-responsive transcriptome (Cui et al. 2013). We thus compared gene expression between CerS(rf) and Rb(lf) mutations. A total of 107 genes significantly changed their expression in both ceramide synthase and lin-35/Rb mutants, which is 5.5-fold higher than expected by random chance (P < 6.77 e−52), and equal to 39% of the total dysregulated genes in CerS(rf) (Figure 4C, Table S2D and Table S4). Of the overlapping genes, 98% (104/107) were changed in the same directions in both mutants during L1 starvation, with 85% of these genes (89/105) upregulated in both mutants, suggesting that ceramide and Rb normally repress the expression of these genes (Table S2D). Further comparisons revealed that 59% (63/107) of the overlapping genes between CerS(rf) and Rb(lf) mutants were daf-16/FOXO-responsive genes (Table S2E). Of these, 73% (46/63) were upregulated in daf-16/FOXO(lf), CerS(rf), and Rb(lf) mutants (Table S2E), suggesting that ceramide, lin-35/Rb, and daf-16/FOXO repress common targets in response to starvation stress during L1 diapause.
Ceramide is important for maintaining starvation-induced gene expression dynamics
To gain further insight into how ceramide impacts starvation-response related gene expression, we further compared the transcriptome of CerS(rf) mutant animals during L1 diapause with previously described FedUP and StarvUP genes (Baugh et al. 2009). FedUP genes are expressed at higher levels when animals hatch in the presence of food, and were proposed to promote reproductive growth. In contrast, StarvUP genes are expressed at higher levels when animals hatch in the absence of food (starvation), and were proposed to support an animal’s survival during starvation. Of the total 272 genes dysregulated by CerS(rf), we found 95 genes (35%) are FedUP genes (Figure 4D, Table S2F, and Table S4), and 90 of these 95 genes were upregulated in the CerS(rf) mutant, indicating that ceramide functions to repress these genes during L1 diapause in wild type. Furthermore, 33 of the genes (12% of the total 272 genes) dysregulated by CerS(rf) were StarvUP genes (Figure 4D, Table S2G, and Table S4). Of these 33 genes, 20 were downregulated in the CerS(rf) mutant, indicating that ceramide promotes the expression of these genes during L1 diapause in wild type. Therefore, like Rb(lf), CerS(rf) alters the “starvation transcriptome” toward a “feeding transcriptome” (Cui et al. 2013).
Many pathogen-inducible genes are repressed by ceramide during L1 diapause
GO analysis of the 272 dysregulated genes in the CerS(rf) mutant showed that the “immune response,” in particular the “innate immune response,” is the most enriched GO category based on P-value (Table S3). Therefore, we compared our CerS(rf) microarray datasets with pathogen-responsive gene datasets. Specifically, when the dataset of CerS(rf) was compared against the 4-hr exposure to the P. aeruginosa strain PA14 (Troemel et al. 2006), we identified 63 dysregulated genes in CerS(rf) that were PA14_4 hr-responsive genes, which is dramatically greater than expected by chance. Of these 63 genes, 59 were upregulated in both the CerS(rf) mutant and PA14-induced gene list (Figure 4E, Table S2H, and Table S4). These results suggest that ceramide repressed the expression of these pathogen-inducible genes for the benefit of starvation survival. A role of repressing a large number of pathogen- and toxin-inducible genes during L1 diapause was also identified for Rb in a previous report (Cui et al. 2013). As described earlier, nearly half of the CerS(rf) affected genes were found to overlap with Rb(lf) affected genes (Figure 4A and Table S2D). These shared activities support the idea that repressing pathogen-inducible genes is vital for protecting starvation survival. Many pathogen-inducible genes encode proteins with antimicrobial or detoxification roles (Hoeven et al. 2012). While they function to protect animals against various environmental threats, they are likely to be harmful to the starvation-induced response for long-term survival. However, our data do not exclude the possibility that these genes are simply unresponsive to starvation stress; they are not necessarily repressed to enhance starvation survival.
Among the 272 genes dysregulated in CerS(rf) mutants, 12 are listed in a compendium of 934 predicted C. elegans transcription factors (referred to as wTF2.0) (Reece-Hoyes et al. 2005) (Table S5). Of the nine upregulated transcription factors, zip-2, encoding a bZIP transcription factor, was upregulated by threefold in CerS(rf) mutants at P < 0.0025. A previous study indicated the role of zip-2 in regulating an early response to P. aeruginosa infection in C. elegans, and that the induction of 25 P. aeruginosa infection response genes is ZIP-2 dependent (Estes et al. 2010). We found that 18 out of these 25 genes were highly induced in starved L1 animals of the CerS(rf) mutant (Table S6), suggesting that the increase in zip-2 expression may partially mediate the impact of CerS(rf) on the expression of pathogen response genes, which implicates a negative role of ZIP-2 in the impact of ceramide on L1 starvation survival. This negative role is also consistent with our further genetic analysis of the zip-2 gene (Figure S2). However, the genetic data also suggest that zip-2 may promote other cellular processes needed for starvation survival (Figure S2), which is consistent with the impact of zip-2(lf) on the expression of a large number of genes (Estes et al. 2010).
Discussion
In this study, we described a strong impact of ceramide deficiency on animal survival during L1 diapause. Ceramide is an important secondary signaling molecule generated in response to multiple extracellular stimuli, such as DNA damage, cytokines, and growth factors, to regulate multiple cellular events like apoptosis, cell senescence, the cell cycle, and differentiation (Hannun and Obeid 2008). While studies using cell culture systems have contributed the most to our current understanding of the roles of ceramide in these cellular processes, the analysis of ceramide function in stress responses using animal models has been limited. Our findings suggest that ceramide may function as a secondary messenger of the food-deprivation signal upstream of multiple stress response pathways.
We showed that three ceramide synthases have differential roles in the L1 starvation response. hyl-1 has the most prominent role in starvation survival, whereas lagr-1 has the weakest role, based on single and double mutant analyses (Figure 1). It has been shown that hyl-1 is mainly responsible for synthesizing ceramides containing very-long-fatty-acyl chains (C25/C26) (Menuz et al. 2009). There are six mammalian CerS enzymes (CerS1–6) that vary in their spatiotemporal expression patterns and their abilities to produce ceramides with different chain lengths (reviewed by Grosch et al. 2012; Mullen et al. 2012). Equilibrium between very-long and long-chain ceramides is also thought to be important for normal cellular physiology (Grosch et al. 2012). Further study is needed to identify the chain-length dependence of ceramide target proteins in order to explain the molecular basis for how different ceramide species play differential roles in animal starvation survival.
Our analysis of ceramide-regulated genes during L1 diapause, and previous study on the function of Rb in the process, indicated that gene expression dynamics induced by starvation share common features, as well as possess distinct features from that induced by other environmental stresses, such as pathogen-induced responses. The IIS pathway has been shown to play prominent roles in animal responses to many different stresses (Baugh and Sternberg 2006; Evans et al. 2008). However, unlike in other stressed situations when animals need to counter toxic threats, food or nutrient deprivation presents a distinct physiological challenge to cells and tissues, and thus demands unique changes in gene regulation. Our previous studies of miRNA functions also indicate sharply different roles of major miRNA functions in the intestine for starvation survival compared to that during pathogen responses (Zhang et al. 2011b; Kudlow et al. 2012).
A previous study showed the impact of ceramide biosynthesis on mitochondrial functions in stress response under well-fed conditions (Liu et al. 2014). We also previously reported that Rb regulates the L1 starvation response partly by promoting the expression of many mitochondrial respiratory chain (MRC) proteins (Cui et al. 2013). However, we found that the expression of most of these MRC proteins are not significantly affected by Cer(rf) mutations (Table S2A), suggesting that, under starved conditions, the impact of ceramide on mitochondrial function may be limited.
The benefit of ceramide on starvation survival would seem to be consistent with a potential positive role of ceramide molecules in lifespan extension. Many known lifespan regulators, such as daf-16 and mir-71, play positive roles in both lifespan extension and L1 starvation survival (Baugh and Sternberg 2006; Kenyon 2010; Pincus et al. 2011; Zhang et al. 2011b; Boulias and Horvitz 2012). However, a previous publication showed that C. elegans strains with mutations in both hyl-1 and lagr-1 genes, like our CerS(rf) mutants, displayed autophagy-dependent lifespan extension under well-fed conditions (Mosbech et al. 2013). It has also been shown that, while the hyl-2(lf) mutant was sensitive to anoxia, the hyl-1(lf) mutant was resistant to anoxia (Menuz et al. 2009). These findings suggest that the poor L1 survival rate of CerS(rf) animals is not due to a general sickness, or a nonspecific sensitivity to all stresses. In addition, these observations also raise an interesting question regarding whether, under well-fed conditions, ceramide has roles that are distinct from roles observed under starved conditions. It has also been previously reported that ceramide inhibits insulin sensitivity in mammals under nutrient-rich conditions, which potentially renders individuals at risk for diabetes and cardiovascular disease (Chavez and Summers 2012). More investigation is clearly needed to learn about the mechanisms underlying the benefits and detriments of ceramide under both starved and well fed conditions.
The appropriate level of autophagy is required for optimal starvation survival of C. elegans (Kang and Avery 2009). To investigate whether an abnormal level of autophagy contributes to the poor starvation response of mutants in the ceramide synthesis pathway, we examined the level of autophagic activation in sptl-2(lf) animals. There was no significant difference in the autophagic response between sptl-2(lf) and wild-type animals during L1 starvation (Figure S3), suggesting that sptl-2(lf) affects L1 starvation survival mainly through autophagy-independent mechanisms.
Ceramide has been previously suggested to function as a tumor-suppressor lipid (Morad and Cabot 2013). Our study reveals that ceramide promotes animal starvation survival. Previous studies also showed that tumor suppressor genes, daf-16/Foxo, daf-18/Pten, and lin-35/Rb are required for promoting C. elegans starvation survival (Baugh and Sternberg 2006; Cui et al. 2013). Starvation or fasting has recently been investigated in cancer therapy (Naveed et al. 2014; Cangemi et al. 2016), as tumor cells are thought to be more susceptible to starvation due to the Warburg effect (Iansante et al. 2015). Results from this study and previous work using C. elegans may suggest that cancer cells, that commonly harbor mutations in tumor suppressor genes, are highly sensitive to starvation stress.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.194282/-/DC1.
Acknowledgments
We thank S. Mitani and the Caenorhabditis Genetics Center (CGC) [supported by National Institutes of Health (NIH) P40 OD010440] for strains and materials; M. Kniazeva, H. Zhu, A. Sewell, and S. Fechtner for assistance; A. Sewell for editing; and W. Wood and Han laboratory members for discussions. This study was supported by the Howard Hughes Medical Institute and NIH (RO1GM37869).
Footnotes
Communicating editor: M. V. Sundaram
Literature Cited
- Baugh L. R., 2013. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194: 539–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Sternberg P. W., 2006. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 16(8): 780–785. [DOI] [PubMed] [Google Scholar]
- Baugh L. R., Demodena J., Sternberg P. W., 2009. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324(5923): 92–94. [DOI] [PubMed] [Google Scholar]
- Boulias K., Horvitz H. R., 2012. The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 15(4): 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangemi A., Fanale D., Rinaldi G., Bazan V., Galvano A., et al. , 2016. Dietary restriction: could it be considered as speed bump on tumor progression road? Tumour Biol. 37(6): 7109–7118. [DOI] [PubMed] [Google Scholar]
- Caro-Maldonado A., Munoz-Pinedo C., 2011. Dying for something to eat: how cells respond to starvation. Open Cell Signal. J. 3: 42–51. [Google Scholar]
- Chavez J. A., Summers S. A., 2012. A ceramide-centric view of insulin resistance. Cell Metab. 15(5): 585–594. [DOI] [PubMed] [Google Scholar]
- Chikina M. D., Huttenhower C., Murphy C. T., Troyanskaya O. G., 2009. Global prediction of tissue-specific gene expression and context-dependent gene networks in Caenorhabditis elegans. PLOS Comput. Biol. 5(6): e1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Cohen M. L., Teng C., Han M., 2013. The tumor suppressor Rb critically regulates starvation-induced stress response in C. elegans. Curr. Biol. 23(11): 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler R. G., Thompson K. W., Camandola S., Mack K. T., Mattson M. P., 2014. Sphingolipid metabolism regulates development and lifespan in Caenorhabditis elegans. Mech. Ageing Dev. 143–144: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Yin X., Allan R., Lu D. D., Maurer C. W., et al. , 2008. Ceramide biogenesis is required for radiation-induced apoptosis in the germ line of C. elegans. Science 322(5898): 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes K. A., Dunbar T. L., Powell J. R., Ausubel F. M., Troemel E. R., 2010. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 107(5): 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E. A., Chen W. C., Tan M. W., 2008. The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging Cell 7(6): 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N., Antebi A., 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22(16): 2149–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch C. E., Ruvkun G., 2001. The genetics of aging. Annu. Rev. Genomics Hum. Genet. 2: 435–462. [DOI] [PubMed] [Google Scholar]
- Grosch S., Schiffmann S., Geisslinger G., 2012. Chain length-specific properties of ceramides. Prog. Lipid Res. 51(1): 50–62. [DOI] [PubMed] [Google Scholar]
- Hannun Y. A., Obeid L. M., 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9(2): 139–150. [DOI] [PubMed] [Google Scholar]
- Hardie D. G., 2011. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 25(18): 1895–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn K. L., Salmon A. B., Hohnen-Behrens C., Turner N., Hoy A. J., et al. , 2009. Insulin resistance is a cellular antioxidant defense mechanism. Proc. Natl. Acad. Sci. USA 106(42): 17787–17792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeven R., McCallum K. C., Cruz M. R., Garsin D. A., 2012. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog. 7(12): e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., et al. , 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 5(9): e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iansante V., Choy P. M., Fung S. W., Liu Y., Chai J.-G., et al. , 2015. PARP14 promotes the Warburg effect in hepatocellular carcinoma by inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat. Commun. 6: 7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker J. W., Suh J. M., Atkins A. R., Ahmadian M., Li P., et al. , 2012. A PPARγ–FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature 485(7398): 391–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kage-Nakadai E., Kobuna H., Kimura M., Gengyo-Ando K., Inoue T., et al. , 2010. Two very long chain fatty acid acyl-CoA synthetase genes, acs-20 and acs-22, have roles in the cuticle surface barrier in Caenorhabditis elegans. PLoS One 5(1): e8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C., Avery L., 2009. Systemic regulation of starvation response in Caenorhabditis elegans. Genes Dev. 23(1): 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. J., 2010. The genetics of ageing. Nature 464(7288): 504–512. [DOI] [PubMed] [Google Scholar]
- Kudlow B. A., Zhang L., Han M., 2012. Systematic analysis of tissue-restricted miRISCs reveals a broad role for microRNAs in suppressing basal activity of the C. elegans pathogen response. Mol. Cell 46(4): 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., Ashrafi K., 2008. A TRPV channel modulates C. elegans neurosecretion, larval starvation survival, and adult lifespan. PLoS Genet. 4(10): e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Raffaghello L., Longo V. D., 2012. Starvation, detoxification, and multidrug resistance in cancer therapy. Drug Resist. Updat. 15(1–2): 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A. J., Puzio-Kuter A. M., 2010. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330(6009): 1340–1344. [DOI] [PubMed] [Google Scholar]
- Liu Y., Samuel B. S., Breen P. C., Ruvkun G., 2014. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508(7496): 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz V., Howell K. S., Gentina S., Epstein S., Riezman I., et al. , 2009. Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science 324(5925): 381–384. [DOI] [PubMed] [Google Scholar]
- Morad S. A. F., Cabot M. C., 2013. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 13(1): 51–65. [DOI] [PubMed] [Google Scholar]
- Mosbech M. B., Kruse R., Harvald E. B., Olsen A. S., Gallego S. F., et al. , 2013. Functional loss of two ceramide synthases elicits autophagy-dependent lifespan extension in C. elegans. PLoS One 8(7): e70087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen T. D., Hannun Y. A., Obeid L. M., 2012. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem. J. 441(3): 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S., et al. , 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424(6946): 277–283. [DOI] [PubMed] [Google Scholar]
- Narbonne P., Roy R., 2009. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 457(7226): 210–214. [DOI] [PubMed] [Google Scholar]
- Naveed S., Aslam M., Ahmad A., 2014. Starvation based differential chemotherapy: a novel approach for cancer treatment. Oman Med. J. 29(6): 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova-Karakashian M. N., Rozenova K. A., 2010. Ceramide in stress response. Adv. Exp. Med. Biol. 688: 86–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K. H., Murata D., Hayashi Y., Dejima K., Mizuguchi S., et al. , 2011. Ceramide glucosyltransferase of the nematode Caenorhabditis elegans is involved in oocyte formation and in early embryonic cell division. Glycobiology 21(6): 834–848. [DOI] [PubMed] [Google Scholar]
- Pincus Z., Smith-Vikos T., Slack F. J., 2011. MicroRNA predictors of longevity in Caenorhabditis elegans. PLoS Genet. 7(9): e1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes J. S., Deplancke B., Shingles J., Grove C. A., Hope I. A., et al. , 2005. A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 6(13): R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J.-F., Ceron J., Koreth J., Hao T., Nicot A.-S., et al. , 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14(10B): 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamen E., Blanchette J. M., Han M., 2009. P-type ATPase TAT-2 negatively regulates monomethyl branched-chain fatty acid mediated function in post-embryonic growth and development in C. elegans. PLoS Genet. 5(8): e1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper R. G., Ashraf J., Kaletsky R., Kleemann G., Murphy C. T., et al. , 2013. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154(3): 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E. R., Chu S. W., Reinke V., Lee S. S., Ausubel F. M., et al. , 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2(11): e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. C., Siervo M., 2011. Obesity and energy balance: is the tail wagging the dog? Eur. J. Clin. Nutr. 65(11): 1173–1189. [DOI] [PubMed] [Google Scholar]
- Yang J. S., Nam H. J., Seo M., Han S. K., Choi Y., et al. , 2011. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6(8): e23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Abraham N., Khan L. A., Hall D. H., Fleming J. T., et al. , 2011a Apicobasal domain identities of expanding tubular membranes depend on glycosphingolipid biosynthesis. Nat. Cell Biol. 13(10): 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zabinsky R., Teng Y., Cui M., Han M., 2011b microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc. Natl. Acad. Sci. USA 108(44): 17997–18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Shen H., Sewell A. K., Kniazeva M., Han M., 2013. A novel sphingolipid-TORC1 pathway critically promotes postembryonic development in Caenorhabditis elegans. eLife 2: e00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Strains are available upon request.