Abstract
As the “human microbiome era” continues, there is an increasing awareness of our resident microbiota and its indispensable role in our increased fitness as holobionts. However, the host-microbe relationship is not so clearly defined for some human symbionts. Here we discuss examples of “accidental pathogens”, meaning previously non-pathogenic and/or environmental microbes thought to have inadvertently experienced an evolutionary shift towards pathogenicity. For instance, symbionts such as Helicobacter pylori and JC Polyomavirus have been shown to accompany humans since prehistoric times and are still abundant in extant populations as part of the microbiome. And yet, the relationship between a subgroup of these microbes and their human hosts seems to have changed with time, and they have recently gained notoriety as gastrointestinal and neuropathogens, respectively. On the other hand, environmental microbes such as Legionella spp. have recently experienced a shift in host range and are now a major problem in industrialized countries as a result of artificial ecosystems. Other variables involved in this accidental phenomenon could be the apparent change or reduction in the diversity of human-associated microbiota because of modern medicine and lifestyles. All of this could result in an increased prevalence of “accidental pathogens” in the form of emerging pathogens.
Keywords: paleomicrobiology, pathogenicity, symbiosis
Potential pathogens or ancient symbionts?
In a world dominated by microbes, our ancestors evolved alongside an outstandingly diverse ancient microbiota (see Figure 1 and Figure Box 1). Symbiotic relationships between animals, microbes and viruses have been observed as far back in animal evolution history as when Hydras made their appearance; and these interactions currently span across all types of life forms (1–3). It's clear that these ancient relationships have shaped the evolution of both the host and symbionts, although the selective pressures governing these processes remain unclear (4–6).
Fig. 1. Endosymbiosis: Homage to Lynn Margulis. Artist: Shoshanah Dubiner.
This painting illustrates a portion of the incredible microbial complexity that existed on this planet when animals evolved, and thus the microbial soup in which all organisms developed. (Image courtesy of the artist. Image credits: Endosymbiosis: Homage to Lynn Margulis, Shoshanah Dubiner, 2012. www.cybermuse.com.)
As holobionts (see Table 1), our associated microbiota allows us to broaden our intrinsic capabilities and increase our survival fitness (7). From our postnatal development (8, 9) to post-mortem decomposition (10, 11) the human body is thoroughly influenced by, and most times highly dependent on, its associated microbiome and virome (see Table 1). The beneficial effects a stable microbiota (see Table 1) has on host fitness has been extensively reviewed, as these microbes often protect from disease (12–14), aid in food digestion (15–17), increment nutrient absorption and vitamin production (18–21), propel the maturation of host immune systems (22–25) and organ development (26), among many other attributes. However, an individual's microbiota is dynamic, always adjusting in response to changes in the needs and lifestyle of the host environment through time (27–29). In this chapter we will discuss examples of environmental and nonpathogenic human-associated microbes believed to have “accidentally” undergone a shift in their life cycles towards pathogenicity as a response to changes induced by or within their human hosts.
Table 1. Definition of terms used from microbiology and virology.
| Term | Definition | References |
|---|---|---|
| Microbiota | Microorganisms associated to a habitat or host. | (30, 31) |
| Holobiont | The total collection of life present in an organism, including the host and its associated microorganisms, viruses and phages. | (7) |
| Microbiome | The collection of microbial genomes present in a habitat or host | (32, 33) |
| Virome | The collection of virus and phage genomes in a habitat or host | (34) |
a) Helicobacter pylori
Microbial communities within holobionts are not stagnant, and population shifts within these communities are common (35, 36). However, a portion of these changes in the microbiota (abrupt or not) could result in detrimental effects to the host when mutualisms go awry (37). Helicobacter pylori is suspected to be one such case. H. pylori normally colonizes humans during infancy, a colonization that persists throughout the individual's life (38). Parent-child transmission is the most common manner (39), although people inhabiting the same area may acquire the microbe horizontally as well (40). Most H. pylori infections are asymptomatic (41–43). Interestingly however, under circumstances that still remain unclear H. pylori infections may predispose the host to gastrointestinal problems including duodenal ulcers and increased acid production (44, 45). Moreover, in certain populations H. pylori has been linked to gastric carcinoma (41, 46); it is therefore currently characterized as a group I carcinogen by the International Agency for Research on Cancer (47). Molecular methods of detecting and differentiating Helicobacter pylori strains are usually based on PCR amplification of DNA encoding for 16srRNA, H.pylori-specific surface proteins, pathogenicity (e.g., ureases ureA and ureC) and microsatellites in the genome (48, 49).
H. pylori has been intrinsically associated to the human gut since our ancestors first migrated from Africa approximately 60,000 years ago (50). It is even theorized that this bacterium has accompanied our species since well before that time. A strong phylogeographic association has always existed between H. pylori strains during their co-evolution alongside ancient and modern human populations (51, 52). In fact, modern Helicobacter pylori strains are currently grouped into various geographical populations as a function of global human demographics and migrations (52). This strong phylogenetic association is mainly due to the bacterium's high genetic diversity as well as its mode of transmission (53). The high genomic diversity observed among H. pylori strains is associated to the bacterium's elevated mutation and recombination rates compared to other bacteria (54–57).
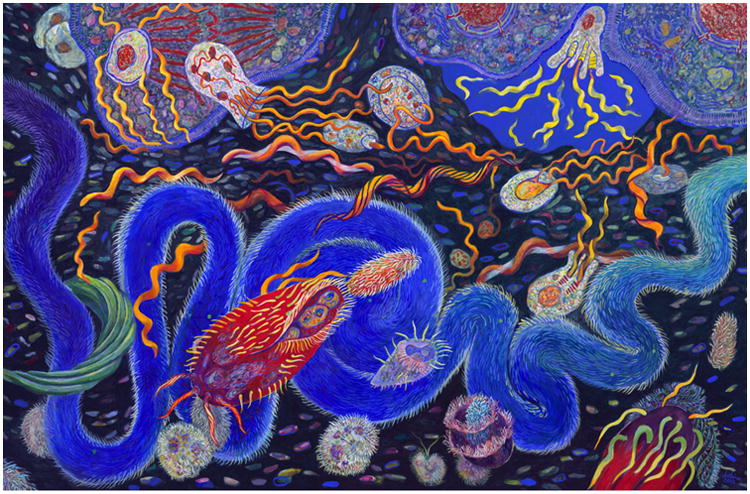
As a result of these characteristics, Helicobacter pylori has long been a prime candidate to determine past migrations (58–63), possible admixture (64) and genetic ancestry (65, 66) of current and ancient cultures. For instance, genetic admixture and past migrations are evident with the widespread presence of European H. pylori strains in North and South America, which are possibly remnants of massive colonial expansions, while in comparison rural Amerindian communities commonly harbor strains of Asian origins, products of pre-Columbian migrations to the New World (67–69). Moreover, H. pylori strains associated to African subpopulations (hspWAfrica and hpNEAfrica) are commonly found in the United States (70), and were most likely introduced during the slave trade era. The genetic ancestry of individuals can also be assessed via H. pylori genotyping. In these ways, the identification of H. pylori genotypes could help microbial forensics determine the cultural origins of a human specimen under study. For example, although modern Europeans are mostly colonized by recombinant hpEurope strains, a 5300-year-old Southern European mummy (named Ötzi the “Iceman”) was found to exclusively harbor an ancestral H. pylori strain of Asian origin named hpAsia2 (Figure 2) (71). Lack of similarity to European strains indicates that ancestral strains such as the hpEurope group settled in the population a mere several thousand years ago. Lack of genome hybridization in this ancient strain also suggested ancestral African strains (such as hpAfrica) had not been largely introduced into these European populations at that time.
Figure 2. Helicobacter pylori in Ötzi the 5300 year-old Iceman.
Reads specific to Helicobacter pylori were detected via metagenomic analysis of DNA extracted from different regions of the gastrointestinal tract of Ötzi, the Tyrolean Iceman. In this figure, the area where the muscle control sample was obtained is highlighted as a diamond (picture on the left), whereas the gastrointestinal sampling sites are marked in the radiographic image using the following legend: asterisk, stomach content; circle, small intestine; square, upper large intestine; triangle, lower large intestine. The number of Helicobacter-specific reads per million metagenomic reads is indicated by the colored gradient bar on the right. (Credits: Maixner et al, Science, 2016. Fig.1 (71)).
If Helicobacter pylori have accompanied us throughout human history, has this relationship changed through time? It is unknown whether H. pylori infections in our ancestors were problematic. However, the asymptomatic presence of ancestral Asian strains in the guts of rural Amerindian communities in the Amazon and Peru (62, 65) suggests a previous, mutually-tolerated, or even possibly beneficial, relationship with Asian cultures that migrated to the area in prehistoric times (72–74). Nevertheless, this poses the question why the current shift to a more pathogenic dynamic with its host? As rural populations have been associated to higher microbial diversity (75–77), one possibility could be linked to the evident loss of the microbial diversity associated to our human ancestors as a result of modernization, cosmopolitan diets and exposure to antimicrobials (Fig.2) (78–82). This is supported by studies showing that hyper acidity and/or medical treatments that reduce the diversity in the gut microbiota allow H. pylori to germinate and proliferate without competition, causing an array of chronic diseases such as gastric inflammation and peptic ulcer disease (44, 45, 83, 84). In addition, a decreased diversity in the individual human microbiome has been associated to higher probabilities of developing an array of diseases, including obesity and diabetes (17, 85, 86). However, only some Helicobacter strains carry virulence genes, commonly associated to the acquisition of pathogenicity islands, including disease-linked genes, such as CagA and VacA which encode for cytotoxins (55, 87–89).
c) JC virus
Human Polyomavirus JC virus is also believed to be an ancestral symbiont that has co-evolved with humans throughout history. Similar to Helicobacter pylori, primary JC Polyomavirus (JCV) infections normally occur during childhood and are persistent throughout the host's life (90). In humans and other animals, possible JCV reservoirs include the kidneys, bone marrow, lymphoid organs and brain tissue (91–96). JC viruses are secreted for long periods of time in the urine and feces of infected individuals (97–100). Because of this, JCV transmission is believed to occur via the fecal-oral route and exposure to sewage-contaminated waters (101, 102). Parent-child or transmission between family members is also common (103, 104). For these reasons, Microbial Source Tracking (MST) methods previously proposed JCV as an indicator of human fecal contamination in water reservoirs (105, 106). However, the possibility of other routes of transmission still remains unclear, including trans-placental transmission during pregnancy (107). JCV strains do not seem to be commonly transmissible between populations with established infections (108), therefore individuals co-habiting the same region usually have latent infections of the same JCV genotypes, whereas foreigners currently living in the area usually harbor a different strain from the established population.
Currently, JCV genotypes infect over 80% of the human population worldwide (109). JCV is identified and genotyped by PCR amplification of its VP1 major capsid protein and T antigen genes (106, 110). The distribution of the distinct JCV genotypes across the globe usually correlates with the presence of particular human populations in each geographical region. In fact, the dominance of certain genotypes in distinct ethnic groups suggests an ancient association with these populations (111). Although its limitations as a human marker have been argued (112), its strong host co-demography has also resulted in JCV to be considered a convenient historical and forensic biomarker to elucidate migrations, genetic heritage and admixture in modern and ancient human populations (Figure 4) (113–115). For instance, the complex genetic history of African American communities in the United States is reflected with the presence of JCV genotypes associated to European (Types 1 and 4) and African regions (Type 3) in these individuals (116). Similarly, JCV genotypes present in native Japanese communities (CY and MY) are shared among subsequent Japanese generations, even those that migrated to other continents (104). Interestingly, the Japanese MY genotype has been detected among several South American Amerindian cultures (117) and was presumably introduced to the region by pre-Columbian ethnic groups from the Pacific. In addition, preliminary forensic studies in modern Puerto Ricans revealed the presence of seven JCV genotypes, originating from Spain, Africa and Asia (118). In this case, detected JCV genotypes also mirrored the known migration and genetic history of the population, as the Asian strain detected in these individuals was probably obtained from indigenous cultures migrating to the Caribbean in ancient times (119–122). Finally, JCV strains Type 2 and 7 (of Asian origin) were detected in rural communities located in the highlands of Papa New Guinea, whereas populations in Guam only had one Asian strain (123). This again associates to the settlement of different areas of the Pacific by people coming from various Asian continents.
Figure 4. Forensic studies with JCV DNA suggest the expansion of Homo sapiens from prehistoric Africa occurred as a two-migrations model.
As Pavesi shows in his model, two out-of-Africa migrations were suggested by currently characterized JCV subtypes. The first migration, represented with a solid line, is compatible with that previously suggested by human genes. The second migration, traced with a dashed line, indicates an additional route of expansion suggested by JCV, but that is undetectable using only human genes. Credits: Pavesi, J Virol, 2005 (135).
Historically, JCV has received much attention in clinical settings due to its isolation from the brain of a patient with progressive multifocal leukoencephalopathy (124) and subsequent associations with severe brain tumors in immunosuppressed patients (125–130). However, in the immune-competent JCV infections are often asymptomatic (131–133). In addition, whole-genome phylogenetic analyses of JCV genotypes suggested JCV type 6 is an ancestral variant that colonized humans before their exodus from Africa, and has since diverged into two distinct evolutionary lineages, each with multiple subpopulations (134). This suggests that the relationship resulting of the co-evolution between our species and JCV was initially not meant to be detrimental to the host, but is being pushed towards a pathogenic cycle as a result of other unknown influences.
Environmental microbes turned human nightmares
a) Mycobacterium tuberculosis
Tuberculosis (TB) is a disease caused by a member of the Mycobacterium genus, an obligate pathogen that can infect any organ of the body but is usually associated to the highly oxygenated tissue (e.g., lungs). TB, particularly pulmonary tuberculosis, has been a global threat to both humans and animals for ages. It is currently present in a third of the world's population (136), in many cases as latent infections with multi-drug resistance (137). Currently, cases are commonly linked to diabetes, tobacco use, AIDS and/or deteriorated social conditions (138). Today, most TB cases are caused by Mycobacterium tuberculosis (Mtb), one of many Mycobacterium species forming the Mycobacterium tuberculosis complex of organisms (Mtb C), which also includes Mycobacterium africanum, Mycobacterium bovis, Mycobacterium microti, and Mycobacterium canetti. (139). Mycobacterium bovis is also a causing agent of TB in humans and other organisms (Division of Tuberculosis Elimination 2014; Cosivi et al. 1999).
i. Past and current ‘tuberculli’
MTb is usually identified by Restriction Fragment Length Polymorphism (RFLP) of its IS6110 insertion sequence (142). However, because of their essentially clonal genomes, Mtb strains are usually differentiated among each other via CRISPR spoligotyping (143) and profiles of mycobacterial interspersed repetitive units present in variable number tandem repeats (MIRU-VNTR) (144). Whole-genome sequencing is also used to identify Mtb strains. These techniques have proven essential to microbial forensics when tracking outbreak origins. For example, from May 2006 and December 2008, a tuberculosis outbreak was observed in a medium-size community in British Columbia, Canada; it affected 41 individuals with an incidence rate of 6.4 cases per 100,000 population (145). None were classified as foreign-born or human immunodeficiency virus-infected individuals. In this case, whole genome sequencing of 36 M. tuberculosis isolates found the strains were essentially identical, indicating a clonal outbreak (146). However, in-depth phylogenetic analyses showed the presence of two major genetic lineages, suggesting it was actually two clonal outbreaks that occurred simultaneously. Both lineages had been sporadically detected in the area since at least 2005. Epidemiological, social-network and molecular forensic analyses determined the Index patient was an adult with an asymptomatic pulmonary Mtb infection that had been untreated for 8 months.
In another study, genotyping of Mtb strains involved in a 4-fold increase in tuberculosis cases in Indiana from 1996-1999 determined the DNA fingerprints were highly similar between the strains detected throughout the outbreak (147). Findings suggested inadequate handling of the latent risks driving the transmission of the pathogen was responsible for the spread throughout the community. Thus, spread was mainly due to lack of identification of infected individuals and persons at highest risk of subsequent infection. Another example of contact tracing in tuberculosis outbreaks was conducted in a high school in Alabama, where a 13 year-old was found to be the source patient that infected 30 percent of students and staff attending the same school (148). Transmission via exposure of the same classroom and school as source patient resulted in around 200 new cases of tuberculin reactivity and 6 cases of active tuberculosis.
Currently, phylogenetic studies have classified 6 main lineages grouping strains from (i) Europe and Americas, (ii) East Asia, (iii) India and East Africa, (iv) West Africa, (v) the Indian Ocean and (vi) the Philippines as defined major clusters (149). The dispersal of each main lineage seems to correspond to the migration patterns of human populations from that geographic area (150). This may explain why pulmonary and spinal TB's notoriety were so well documented across many ancient civilizations. In ancient Greece it was known as ‘phthisis’ (151, 152); it was also recorded in ancient India, Asia and Turkey (153, 154). Tuberculosis-associated “hunchbacks” were also represented throughout Egyptian art (155). Although thousands of years have passed since the first recorded accounts of the disease, ancestral and current strains remain very similar. The oldest human-associated Mtb genetically confirmed and characterized to date is from a Neolithic population in the Mediterranean (156). This 9,000 year-old Mtb showed high genome similarity to extant strains. In fact, forensic studies have identified an ancient composite genome present in modern variants (157). This composite genome is potentially the product of a bottleneck event or high rates of intraspecies horizontal transfer in MtbC early history, and it is remarkably still detectable even despite recent genome restructuring. Moreover, ancient and extant Mtb strains share common deletion events of TbD1 in their genome, again suggesting an ancient common ancestor (158).
ii. Origins of the ‘White Plague’
Previous hypotheses suggested that TB epidemics began in 18th century Europe and spread to Africa and the Americas (159, 160). However, this speculation was mainly due to forensic studies not showing confirmed tuberculosis cases in ancient Africa and the New World, especially compared to the high number of Mtb-infected specimens in pre-Columbian Europe. Nevertheless, studies detected MtbC in pre-Columbian individuals from Peru (161). Although some consider the sample dates controversial (162, 163), these findings advocate the presence of the tuberculosis pathogen prior to European contact in South America.
Current data indicates MtbC originated in East Africa, where an ancestral precursor of MTB infected hominids around 3 million years ago (164). This hypothesis is known as “Out-Of-And-Back-To-Africa” for human-associated MtbC, in which this precursor strain would have accompanied and evolved alongside mankind as we migrated from Africa. As of yet, this has not been detected in African hominid skeletons from that time period (although several Egyptian mummies have been confirmed as MtbC-positive (155, 165, 166)). However, the high genomic similarity between ancient and extant Mtb strains clearly supports the hypothesis of a long-term co-evolution with its human hosts. In addition, the geographic location of modern Mtb strains also suggest human MtbC originated in Africa, as this is the only place where all six human MtbC major lineages have been detected (149).
One question remains however: was MtbC originally a human pathogen? Some have argued that MtbC was zoonotic long before its contact with humans; acquiring its infectivity to humans posteriorly (159, 168). Interestingly, forensic studies showed the MtbC strain detected in pre-Columbian Peruvians had higher similarity to strains infecting seals than human-associated modern strains. On the other hand, recent data suggests that MtbC could have initially been a free-living microbe that slowly acquired strategies allowing it to persist in humans and animals (169). This idea is supported by the vast amount of virulence factors detected in pristine environments (Søborg et al. 2013; Gonzalez et al in preparation) as well as the diversity of Mycobacterium species commonly observed in the environment, especially in water and soil (for a review on this subject please see: Hruska & Kaevska 2012). Furthermore, M. bovis and M. tuberculosis are capable of long-term survival in soil and also maintain their virulence throughout soil exposure (172). But if human MtbC was initially an environmental microbe, how did it end up infecting humans? One hypothesis for transmission is known as the Pleistocene campfire theory, and proposes that MtbC was acquired by ancient humans as a result of social interactions around wood fires (173). This possibility has a certain appeal, as the inhalation of carcinogens present in wood smoke lowers our immunological defenses (174, 175) and could have thus paved way for those first MtbC infections in humans. Notably, despite our best intentions at deciphering the history of TB in humans one thing is clear: further studies on ancient and extant strains are needed to effectively elucidate the evolution and epidemiology of this ancient human pathogen.
b) Toxoplasma gondii
Toxoplasma gondii is the protozoan responsible for latent toxoplasmosis, which infects over 20% of the population in the United States of America. Although it can infect many warm-blooded vertebrates as intermediate hosts, only infecting cells lining the gastrointestinal tract of a felid allows the pathogen to mature and reproduce (176). Humans are usually infected by T. gondii via exposure to feces-contaminated soil, congenital transmission (177) or consumption of uncooked contaminated meat (178). Latent toxoplasmosis is usually considered asymptomatic in the immunocompetent. However, T. gondii infections notoriously alter host brain and behavior to its own advantage (179). For example, in rats T. gondii causes a lack of fear towards cats (180, 181) and even causes rodents to be sexually aroused by their feline predators (182). This is presumably to increase the chances of the intermediate host to be eaten by the parasite's final host, thus allowing the brain T. gondii to complete its life cycle. In other animals, such as goats, toxoplasmosis has been linked to abortions and perinatal death (183, 184). In humans, latent toxoplasmosis has been linked to an apparent increase in host neuroticism (185) and suicide attempts (186), among other behavioral manipulations (187).
Toxoplasmosis is usually serologically diagnosed using IgG, IgM, IgA, IgE antibody titers specific to T. gondii (188). Agglutination tests are also common. Molecular characterization of T. gondii is done by PCR of its B1 gene, and strain genotyping is conducted based on microsatellite markers such as TUB2, W35, TgMA, B18 and B17, among others (189). Initial studies on T. gondii populations suggested they were clonal from one ancestor. On the other hand, in-depth analyses discovered genetic diversity between strains (190, 191) and phylogenetic analyses suggest this pathogen's history includes both clonal and sexual strain propagation (191). Although this pathogen is highly prevalent in many communities, documented, in-depth investigations of toxoplasmosis outbreaks are not common. There are some exceptions however. For instance, source tracing of a toxoplasmosis outbreak in British Columbia, Canada determined transmission of the disease occurred via a contaminated municipal drinking water tank (192). A village in Patam, Burma, suffered a toxoplasmosis outbreak in late 2003, which resulted in the deaths of an adult, a neonate and a fetus. Although no epidemiological sources were identified, studies determined that clonal isolates from one atypical T. gondii strain were responsible for the outbreak (193). Similarly, an outbreak that resulted in 155 reported cases of toxoplasma in Brazil was also studied (194). In this case, unfiltered, municipally treated water was determined as the epidemiological source of infection. All strains implicated in the outbreak expressed SAG-2 parasite-specific antigens and were classified as the same genotype (type I). However, there was insufficient genomic information to determine if this was a clonal outbreak. Outbreaks have also been observed in Hawaiian monk seals after exposure to marine water contaminated with feline feces (195).
c) Legionella pneumophila
Legionella pneumophila is a facultative intracellular parasite infecting protozoa in freshwater and soil (196, 197) (for a review see, Bitar et al. 2004). In recent years however, L. pneumophila has been prevalent in man-made water systems, including those found in cars (199), water distribution systems and buildings (Center for Disease Control and Prevention, 2016). In these reservoirs, L. pneumophila infects amoeba that upon inhalation act as a “Trojan horse” in humans (200, 201). Due to constant exposure to these water systems in infected infrastructures, cases of Legionellosis are becoming common in human populations (202). For example, in 1994 fifty cases of Legionnaires' disease were reported among passengers of a cruise ship; cases were posteriorly linked to a contaminated whirlpool spa on the ship (203). In 2005, an outbreak was observed in an extended care facility in Ontario, Canada, resulting in 23 deaths (204). The source of the outbreak was identified as a contaminated air conditioning cooling tower. Genetic assays determined high similarity between clinical strains associated to the outbreak and environmental strains from the outbreak site.
In a way, the adaptations L. pneumophila obtained during its co-evolution with its protozoan hosts accidentally played a key role in its recent host switch towards humans (205). This is because once it is inside the human body, L. pneumophila is now capable of infecting its host's macrophages (206, 207). This is mainly due to similarities between the microbe's environmental amoeba host and human macrophages (208, 209). In addition, the same genes are expressed by L. pneumophila during the infection of both host cells (210). Also, L. pneumophila protects itself by hijacking and replicating in membrane-bound compartments, vesicles or “vacuoles” within both host cells (for a review, see Isberg et al. 2009). Given the known evidence of its epidemiology, it is highly likely that this bacterium is actually an “accidental human pathogen” (212). Thus, Legionella pneumophila is an excellent example of how our industrialized environment is capable of providing the stage for a previously environmental, non-pathogenic microbe to shift towards humans as hosts.
Concluding remarks
We are slowly beginning to understand the complexity driving host-microbe interactions and the shifts in the microbiome and virome associated to changes in the host environment. Studies in extant individuals indicate that several current diseases are triggered by the use of antimicrobials, eliminating microbial competition and allowing for potentially pathogenic microbes to increase in numbers within the host. This is especially true for microbes possessing virulence factors that depend on quorum sensing molecules to be expressed (214, 215). As holobionts, we seem to have eliminated a portion of our associated microbes across evolutionary time. Indeed, it is possible that the “opportunistic pathogens” discussed in this section could have been a part of a previously beneficial symbioses with humans that went awry, thus rendering these microbes “accidental pathogens”. On the other hand, some of the beneficial symbiotic relationships between ancient humans and microbes still persist as such to this day. Many of these microbes have co-diversified alongside their human host populations since then. It remains unclear, however, which selective pressures have shaped these relationships. Such “micro-evolution” stories now present high phylogeographical correlations to specific human populations.
Many aspects surrounding host-microbe interactions remain unclear; but many unknowns are being explained using molecular forensics for the first time. One thing is clear however; the mutualism-parasitism continuum that governs a microbe-host relationship through time is now seen as something complex and dynamic, where a microbe can change its effect and relationship with the host in response to environmental stimuli. This in turn results in a microbial version of the Red Queen effect (216), a constant competition between the members of this symbiosis, where the host tries to control symbiont populations and the symbiont evolves ways of circumventing these measures (217). Over time, we seem to have lost many of these ancient symbioses with our microbial partners. This could be due to migrations, shifts in diets or lifestyle changes, resulting in a less diverse gut microbiome in modern humans. Symbionts can indirectly protect the host from pathogen infection and colonization through constant activation of the immune system and alteration of cell morphology. Cross-protection has also been frequently observed as a result of particular virus infections. For instance, herpes viruses, which persist as latent infections ubiquitous in modern humans, seem to give the host immunity towards the pathogens Listeria monocytogenes and Yersinia pestis (218). It is apparent that the disappearance of these ancestral human microbiota may have helped shift the ‘mutualism-parasitism continuum’, inducing in the human host a higher predisposal to several of today's widespread diseases. Our ability to carry out molecular microbial forensic studies is elucidating many of the previously undetermined interactions between humans and emerging accidental pathogens.
Figure 3. A lower fecal microbiome diversity associated to individuals from industrialized cultures.
Dominguez-Bello and her team compared the bacterial diversity in feces from cultures with hunter-gatherer lifestyles compared to progressively more industrialized cultures. A) Phylogenetic diversity in feces from Yanomami and Guahibo Amerindians, Malawians and U.S. individuals. A higher bacterial diversity was detected in feces from the Yanomami, an isolated, rural indigenous culture inhabiting the Amazon. In comparison, a slightly decreased fecal diversity was found in Guahibo Amerindians. However, a major decrease was detected in the diversity of the fecal microbiota in U.S. subjects. A pronounced decrease was also detected in the functional profiles of fecal microbiomes from U.S. subjects compared to cultures with more traditional lifestyles (figure not shown). (Credits: Clemente, et al. Sci Adv. 2015. Fig.1A.).
B) Key differential bacterial groups between fecal microbiomes from Yanomami and Guahibo Amerindians, Malawians and U.S. subjects. (Credits: Clemente, et al. Sci Adv. 2015. Fig.1C).
C) Functional diversity in feces from Yanomami and Guahibo Amerindians, Malawians and U.S. individuals. As expected, a higher overall functional diver(Credits: Clemente, et al. Sci Adv. 2015. Fig.2A.).
D) Comparison of major metabolic pathways detected in fecal microbiomes from Yanomami and Guahibo Amerindians, Malawians and U.S. subjects. (Credits: Clemente, et al. Sci Adv. 2015. Fig.2C).
Figure 5. Origin of human-associated Mycobacterium tuberculosis.
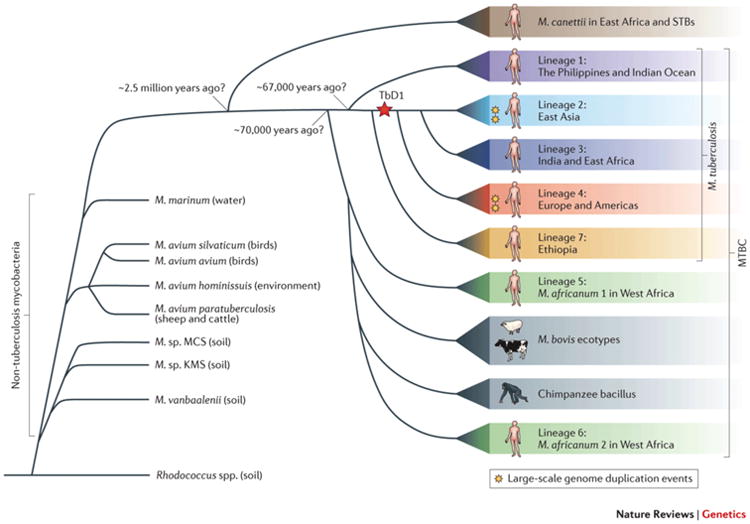
Although its exact ancestral history remains unresolved, recent studies clearly suggest Mycobacterium tuberculosis was associated to humans previous to their expansion from prehistoric Africa. However, M. tuberculosis is believed to have been an environmental microbe long before its association to ancient humans. This figure was taken from Galagan, Nature Reviews Genetics, 2014, and depicts a summary of the conclusions implied by current phylogenetic literature on the evolution of M. tuberculosis and other members of the Mycobacterium tuberculosis complex (167).
Figure 6. Legionella pneumophila infections in environmental amoebae and human macrophages.
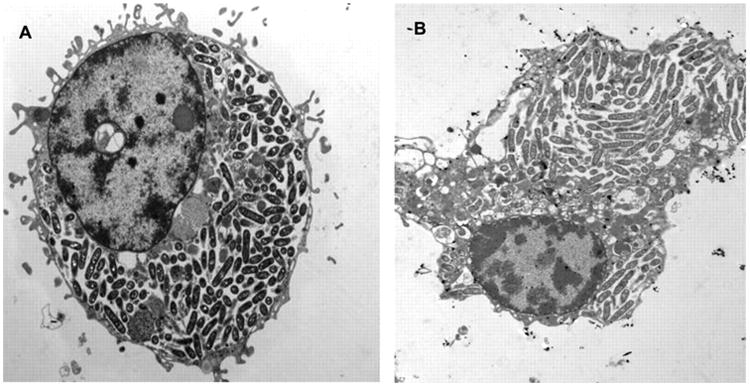
Electron micrographs of U937 macrophages (A) and A. polyphaga (B) infected by L. pneumophila (strain AA100) at 24 h. Image taken from Molmeret et al, AEM, 2005 (213).
References
- 1.Fraune S, Bosch TCG. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci U S A. 2007;104(32):13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grasis JA, et al. Species-specific viromes in the ancestral holobiont hydra. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0109952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Werren JH, O'Neill SL. Influential Passengers: Inherited Microorganisms and Arthropod Reproduction. Oxford University Press; 1997. The evolution of heritable symbionts; pp. 3–41. [Google Scholar]
- 4.Franzenburg S, et al. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc Natl Acad Sci. 2012;109(47):19374–19379. doi: 10.1073/pnas.1213110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch TCG. Cnidarian-Microbe Interactions and the Origin of Innate Immunity in Metazoans. Annu Meview Microbiol. 2013;67(June):499–518. doi: 10.1146/annurev-micro-092412-155626. [DOI] [PubMed] [Google Scholar]
- 6.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci U S A. 2007;104(49):19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol Rev. 2008;32(5):723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 8.Marcobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota: Human milk oligosaccharide consumption. Clin Microbiol Infect. 2012;18:12–15. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Leoz MLA, et al. Human Milk Glycomics and Gut Microbial Genomics in Infant Feces Shows Correlation between Human Milk Oligosaccharides and Gut Microbiota: A Proof-of-Concept Study. J Proteome Res. 2014 doi: 10.1021/pr500759e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finley SJ, Benbow ME, Javan GT. Microbial communities associated with human decomposition and their potential use as postmortem clocks. Int J Legal Med. 2015;129(3):623–632. doi: 10.1007/s00414-014-1059-0. [DOI] [PubMed] [Google Scholar]
- 11.Javan GT, Finley SJ, Abidin Z, Mulle JG. The Thanatomicrobiome: A Missing Piece of the Microbial Puzzle of Death. Front Microbiol. 2016;7(February):225. doi: 10.3389/fmicb.2016.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenthal M, Goldberg D, Aiello A, Larson E, Foxman B. Skin microbiota: Microbial community structure and its potential association with health and disease. Infect Genet Evol. 2012;2011(5):839–848. doi: 10.1016/j.meegid.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schommer NN, Gallo RL. Structure and function of the human skin microbiome. Trends Microbiol. 2013;21(12):660–668. doi: 10.1016/j.tim.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stecher B, Hardt WD. The role of microbiota in infectious disease. Trends Microbiol. 2008;16(3):107–114. doi: 10.1016/j.tim.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, et al. A Genomic View of the Human-Bacteroides thetaiotaomicron Symbiosis. Science (80-) 2003;299(5615):2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 16.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11(7):497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 17.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 18.Jumpertz R, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krajmalnik-Brown R, Zehra-Esra I, Dae-Wook K, DiBaise JK. Effects of Gut Microbes on Nutrient Absorption and Energy Regulation. Nutr Clin Pr. 2013;27(2):201–214. doi: 10.1177/0884533611436116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semova I, et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12(3):277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeBlanc JG, et al. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr Opin Biotechnol. 2013;24(2):160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazmanian SK, Cui HL, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69(5):1046–1051. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 25.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(May):313–324. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99(24):15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minot S, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21(10):1616–25. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12(5):R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gajer P, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4(132):132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson CM, Beard BL, Roden EE. The Iron Isotope Fingerprints of Redox and Biogeochemical Cycling in Modern and Ancient Earth. Annu Rev Earth Planet Sci. 2008;36(1):457–493. [Google Scholar]
- 31.Beraldi-Campesi H. Early life on land and the first terrestrial ecosystems. Ecol Process. 2013;2(1):1. [Google Scholar]
- 32.Ursell LK, Metcalf J, Wegener Parfrey L, Knight R. Defining the Human Microbiome. Nutr Rev. 2012;70(Suppl 1):S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turnbaugh PJ, et al. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haynes M, Rohwer F. The Human Virome. In: Nelson KE, editor. Metagenomics of the Human Body. Springer; New York, New York, NY: 2011. pp. 63–77. [Google Scholar]
- 35.Flores GE, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15(12):531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shade A, Caporaso JG, Handelsman J, Knight R, Fierer N. A meta-analysis of changes in bacterial and archaeal communities with time. ISME J. 2013;7(8):1493–506. doi: 10.1038/ismej.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cani PD, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58(8):1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blaser MJ, Atherton JC. Helicobacter pylori persistence: Biology and disease. J Clin Invest. 2004;113(3):321–333. doi: 10.1172/JCI20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman KJ, Correa P. The Transmission of Helicobacter pylori. A Critical Review of the Evidence. Int J Epidemiol. 1995;24(5):875–887. doi: 10.1093/ije/24.5.875. [DOI] [PubMed] [Google Scholar]
- 40.Delport W, Cunningham M, Olivier B, Preisig O, Van Der Merwe SW. A population genetics pedigree perspective on the transmission of Helicobacter pylori. Genetics. 2006;174(4):2107–2118. doi: 10.1534/genetics.106.057703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dooley CP, et al. Prevalence of Helicobacter pylori Infection and Histologic Gastritis in Asymptomatic Persons. N Engl J Med. 1989;321(23):1562–1566. doi: 10.1056/NEJM198912073212302. [DOI] [PubMed] [Google Scholar]
- 42.Holcombe C, Omotara BA, Eldridge DJ, Jones DM. H. pylori, the Most Common Bacterial Infection in Africa: A Random Serological Study. Am J Gastroenterol. 1992;87(1):28–30. [PubMed] [Google Scholar]
- 43.Ahmad MM, et al. Prevalence of Helicobacter Pylori in Asymptomatic Population a Pilot Serological Study in Bangladesh. J Epidemiol. 1997;7(4):251–254. doi: 10.2188/jea.7.251. [DOI] [PubMed] [Google Scholar]
- 44.Moss SF, Calam J, Legon S, Bishop AE, Polak JM. Effect of Helicobacter pylori on gastric somatostatin in duodenal ulcer disease. Lancet. 1992;340(8825):930–932. doi: 10.1016/0140-6736(92)92816-x. [DOI] [PubMed] [Google Scholar]
- 45.Sipponen P, Hyvarinen H. Role of Helicobacter pylori in the Pathogenesis of Gastritis, Peptic Ulcer and Gastric Cancer. Scand J Gastroenterol. 1993;28:407–425. doi: 10.3109/00365529309098333. [DOI] [PubMed] [Google Scholar]
- 46.Miwa H, Go MF, Sato N. H. pylori and gastric cancer: the Asian enigma. Am J Gastroenterol. 2002;97(5):1106–1112. doi: 10.1111/j.1572-0241.2002.05663.x. [DOI] [PubMed] [Google Scholar]
- 47.Yamada T, et al. Helicobacter pylori in peptic ulcer disease. Jama. 1994;272(1):65–69. [PubMed] [Google Scholar]
- 48.Lu J, et al. Comparison of Five PCR Methods for Detection of Helicobacter pylori DNA in Gastric Tissues. J Clin Microbiol. 1999;37(3):772–774. doi: 10.1128/jcm.37.3.772-774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Israel DA, et al. Helicobacter pylori strain-specific differences in genetic content, identified by microarray, influence host inflammatory responses. J Clin Invest. 2001;107(5):611–620. doi: 10.1172/JCI11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linz B, et al. An African origin for the intimate association between humans and Helicobacter pylori_. Nature. 2007;445:915–918. doi: 10.1038/nature05562. 1476–4687 (Electronic) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montano V, et al. Worldwide Population Structure, Long Term Demography, and Local Adaptation of Helicobacter pylori. Genetics. 2015;200(July):947–963. doi: 10.1534/genetics.115.176404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janssen PJ, Audit B, Ouzounis Ca. Strain-specific genes of Helicobacter pylori: distribution, function and dynamics. Nucleic Acids Res. 2001;29(21):4395–4404. doi: 10.1093/nar/29.21.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki R, Shiota S, Yamaoka Y. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. NIH Public Access. 2013;12(2):203–213. doi: 10.1016/j.meegid.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gressmann H, et al. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 2005;1(4):0419–0428. doi: 10.1371/journal.pgen.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martínez-Carrillo DN, et al. Helicobacter pylori vacA and cagA genotype diversity and interferon gamma expression in patients with chronic gastritis and patients with gastric cancer. Rev Gastroenterol México (English Ed. 2014;79(4):220–228. doi: 10.1016/j.rgmx.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Kraft C, Suerbaum S. Mutation and recombination in Helicobacter pylori: Mechanisms and role in generating strain diversity. Int J Med Microbiol. 2005;295(5):299–305. doi: 10.1016/j.ijmm.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Kersulyte D, Chalkauskas H, Berg DE. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31(1):31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- 58.Falush D, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–5. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 59.Breurec S, et al. Evolutionary history of helicobacter pylori sequences reflect past human migrations in southeast Asia. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0022058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaoka Y. Helicobacter pylori typing as a tool for tracking human migration. Clin Microbiol Infect. 2009;15(9):829–834. doi: 10.1111/j.1469-0691.2009.02967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moodley Y, Linz B. Helicobacter pylori sequences reflect past human migrations. Genome Dyn. 2009;6(January 2016):62–74. doi: 10.1159/000235763. [DOI] [PubMed] [Google Scholar]
- 62.Kersulyte D, et al. Helicobacter pylorifrom Peruvian Amerindians: Traces of human migrations in strains from remote Amazon, and genome sequence of an Amerind strain. PLoS One. 2010;5(11) doi: 10.1371/journal.pone.0015076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dominguez-Bello MG, Blaser MJ. The Human Microbiota as a Marker for Migrations of Individuals and Populations. Annu Rev Anthropol Vol 40. 2011;40:451–474. [Google Scholar]
- 64.Devi SM, et al. Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island. BMC Genomics. 2006;7:191. doi: 10.1186/1471-2164-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghose C, et al. East Asian genotypes of Helicobacter pylori strains in Amerindians provide evidence for its ancient human carriage. Proc Natl Acad Sci U S A. 2002;99(23):15107–11. doi: 10.1073/pnas.242574599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wirth T, et al. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proc Natl Acad Sci U S A. 2004;101(14):4746–4751. doi: 10.1073/pnas.0306629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamaoka Y, et al. Helicobacter pylori in North and South America before Columbus. FEBS Lett. 2002;517(1–3):180–184. doi: 10.1016/s0014-5793(02)02617-0. [DOI] [PubMed] [Google Scholar]
- 68.Molina-Castro SE, Herrera D, Malespin-Bendana W, Ramirez V, Une C. The geographic origin of Helicobacter pylori isolated from Costa Rican patients. Gut Microbes. 2014;5(4):517–521. doi: 10.4161/gmic.32148. [DOI] [PubMed] [Google Scholar]
- 69.Ghose C, Perez-Perez GI, Van Doorn LJ, Domínguez-Bello MG, Blaser MJ. High frequency of gastric colonization with multiple Helicobacter pylori strains in Venezuelan subjects. J Clin Microbiol. 2005;43(6):2635–2641. doi: 10.1128/JCM.43.6.2635-2641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Breurec S, et al. Impact of human migrations on diversity of helicobacter pylori in Cambodia and New Caledonia. Helicobacter. 2013;18(4):249–261. doi: 10.1111/hel.12037. [DOI] [PubMed] [Google Scholar]
- 71.Maixner F, et al. The 5300-year-old Helicobacter pylori genome of the Iceman. Science (80-) 2016;351(6269):162–165. doi: 10.1126/science.aad2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoffecker JF, Powers WR, Goebel T. The colonization of beringia and the peopling of the new world. Science (80-) 1993;259(5091):46–53. doi: 10.1126/science.259.5091.46. [DOI] [PubMed] [Google Scholar]
- 73.Achilli A, et al. Reconciling migration models to the Americas with the variation of North American native mitogenomes. Proc Natl Acad Sci U S A. 2013;110(35):14308–13. doi: 10.1073/pnas.1306290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dillehay TD. Probing deeper into first American studies. Proc Natl Acad Sci U S A. 2009;106(4):971–8. doi: 10.1073/pnas.0808424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martínez I, et al. The Gut Microbiota of Rural Papua New Guineans: Composition, Diversity Patterns, and Ecological Processes. Cell Rep. 2015;11(4):527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 76.Blaser MJ, et al. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J. 2012;7(1):85–95. doi: 10.1038/ismej.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clemente JC, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1(3):e1500183–e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mane SP, et al. Host-interactive genes in Amerindian Helicobacter pylori diverge from their old world homologs and mediate inflammatory responses. J Bacteriol. 2010;192(12):3078–3092. doi: 10.1128/JB.00063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reinhard KJ, et al. Understanding the Pathoecological Relationship between Ancient Diet and Modern Diabetes through Coprolite Analysis. Curr Anthropol. 2012;53:506–512. [Google Scholar]
- 80.Walter J, Ley R. The Human Gut Microbiome: Ecology and Recent Evolutionary Changes. Annu Rev Microbiol. 2011;65(1):411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 81.Contreras M, et al. Human gut microbiome viewed across age and geography. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoffmann C, et al. Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Graham DY. Helicobacter pylori: Its epidemiology and its role in duodenal ulcer disease. J Gastroenterol Hepatol. 1991;6:105–113. doi: 10.1111/j.1440-1746.1991.tb01448.x. [DOI] [PubMed] [Google Scholar]
- 84.Valle J, Kekki M, Sipponen P, Ihamaki TSM. Long-Term Course and Consequences of Helicobacter pylori gastritis. Scand J Gastroenterol. 1996;31(6):546–550. doi: 10.3109/00365529609009126. [DOI] [PubMed] [Google Scholar]
- 85.Tilg H, Kaser A. Review series Gut microbiome, obesity, and metabolic dysfunction. J Clin Investig. 2011;121(6):2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsai F, Coyle WJ. The Microbiome and Obesity : Is Obesity Linked to Our Gut Flora? Curr Gastroenterol Rep. 2009 doi: 10.1007/s11894-009-0045-z. [DOI] [PubMed] [Google Scholar]
- 87.Suzuki R, Shiota S, Yamaoka Y. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infect Genet Evol. 2012;12(2):203–213. doi: 10.1016/j.meegid.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nilsson C, et al. Correlation between cag Pathogenicity Island Composition and Helicobacter pylori- Associated Gastroduodenal Disease Correlation between cag Pathogenicity Island Composition and Helicobacter pylori- Associated Gastroduodenal Disease. Infect Immun. 2003;71(11):6573–6581. doi: 10.1128/IAI.71.11.6573-6581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Censini S, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Genetics. 1996;93(December):14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ling PD, et al. The Dynamics of Herpesvirus and Polyomavirus Reactivation and Shedding in Healthy Adults : A 14-Month Longitudinal Study. J Infect Dis. 2003;187(15 May):1571–1580. doi: 10.1086/374739. [DOI] [PubMed] [Google Scholar]
- 91.Kitamura T, et al. Persistent JC Virus (JCV) Infection Is Demonstrated by Continuous Shedding of the Same JCV Strains. J Clin Microbiol. 1997;35(5):1255–1257. doi: 10.1128/jcm.35.5.1255-1257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Caldarelli-Stefano R, et al. Detection and typing of JC virus in autopsy brains and extraneural organs of AIDS patients and nonimmunocompromised individuals. J Neurovirol. 1999;5(2):125–33. doi: 10.3109/13550289909021994. [DOI] [PubMed] [Google Scholar]
- 93.Loeber G, Dörries K. DNA rearrangements in organ-specific variants of polyomavirus JC strain GS. J Virol. 1988;62(5):1730–1735. doi: 10.1128/jvi.62.5.1730-1735.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Monaco MC, Jensen PN, Hou J, Durham LC, Major EO. Detection of JC virus DNA in human tonsil tissue: evidence for site of initial viral infection. J Virol. 1998;72(12):9918–23. doi: 10.1128/jvi.72.12.9918-9923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rieckmann P, Michel U, Kehrl JH. Regulation of JC virus expression in B lymphocytes. J Virol. 1994;68(1):217–22. doi: 10.1128/jvi.68.1.217-222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arthur R, a YR, Dagostin S, Shah KV. Detection of BK Virus and JC Virus in Urine and Brain Tissue by the Polymerase Chain Reaction. J Clin Microbiol. 1989;27(6):1174–1179. doi: 10.1128/jcm.27.6.1174-1179.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hundesa A, Motes De CM, Bofill-mas S, Albinana-gimenez N, Girones R. Identification of Human and Animal Adenoviruses and Polyomaviruses for Determination of Sources of Fecal Contamination in the Environment. 2006;72(12):7886–7893. doi: 10.1128/AEM.01090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rudick Ra, et al. Assessment of JC virus DNA in blood and urine from natalizumab-treated patients. Ann Neurol. 2010;68(3):304–10. doi: 10.1002/ana.22107. [DOI] [PubMed] [Google Scholar]
- 99.Kokkinos Pa, Ziros PG, Mpalasopoulou A, Galanis A, Vantarakis A. Molecular detection of multiple viral targets in untreated urban sewage from Greece. Virol J. 2011;8:195. doi: 10.1186/1743-422X-8-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mcquaig SM, et al. Quantification of Human Polyomaviruses JC Virus and BK Virus by TaqMan Quantitative PCR and Comparison to Other Water Quality Indicators in Water and Fecal Samples Quantification of Human Polyomaviruses JC Virus and BK Virus by TaqMan Quantitative PCR and. Appl Environ Microbiol. 2009;75(11):3379. doi: 10.1128/AEM.02302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bofill-Mas S, Girones R. Role of the environment in the transmission of JC virus. J Neurovirol. 2003;9(1):54–58. doi: 10.1080/13550280390195306. [DOI] [PubMed] [Google Scholar]
- 102.Berger JR, et al. JC virus detection in bodily fluids: clues to transmission. Clin Infect Dis. 2006;43(1):e9–e12. doi: 10.1086/504947. [DOI] [PubMed] [Google Scholar]
- 103.Kunitake T, et al. Parent-to-child transmission is relatively common in the spread of the human polyomavirus JC virus. J Clin Microbiol. 1995;33(6):1448–1451. doi: 10.1128/jcm.33.6.1448-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suzuki M, et al. Asian genotypes of JC virus in Japanese-Americans suggest familial transmission. J Virol. 2002;76(19):10074–10078. doi: 10.1128/JVI.76.19.10074-10078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Albinana-Gimenez N, Girones R, Miagostovich MP, Calgua B, Huguet JM. Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants. Water Res. 2009;43:2011–2019. doi: 10.1016/j.watres.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 106.Hundesa A, Motes CM De, Bofill-mas S, Albinana-gimenez N, Girones R. Identification of Human and Animal Adenoviruses and Polyomaviruses for Determination of Sources of Fecal Contamination in the Environment. Appl Environ Microbiol. 2006;72(12):7886–7893. doi: 10.1128/AEM.01090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boldorini R, et al. Latent human polyomavirus infection in pregnancy: investigation of possible transplacental transmission. Pathology. 2008;40(1):72–7. doi: 10.1080/00313020701716458. [DOI] [PubMed] [Google Scholar]
- 108.Kato A, et al. Lack of evidence for the transmission of JC polyomavirus between human populations. Arch Virol. 1997;142(5):875–882. doi: 10.1007/s007050050125. [DOI] [PubMed] [Google Scholar]
- 109.Knowles WA. Discovery and Epidemiology of the Human Polyomaviruses BK Virus (BKV) and JC Virus (JCV) In: Ahsan N, editor. Polyomaviruses and Human Diseases. Springer-Verlag; New York: 2005. pp. 1–25. [DOI] [PubMed] [Google Scholar]
- 110.Agostini RT, Ryschkewitsch CF, Stoner GL. Genotype Profile of Human Polyomavirus JC Excreted in Urine of Immunocompetent Individuals. J Clin Microbiol. 1996;34(1):159–164. doi: 10.1128/jcm.34.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kersulyte D, et al. Differences in Genotypes of Helicobacter pylori from Different Human Populations. J Bacteriol. 2000;182(11):3210–3218. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Holmes EC, Holmes EC. Evolutionary history and phylogeography of human viruses. Annu Rev Microbiol. 2008;62:307–28. doi: 10.1146/annurev.micro.62.081307.162912. [DOI] [PubMed] [Google Scholar]
- 113.Kitchen A, Miyamoto MM, Mulligan CJ. Utility of DNA viruses for studying human host history: Case study of JC virus. Mol Phylogenet Evol. 2008;46(2):673–682. doi: 10.1016/j.ympev.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 114.Wooding S. Do human and JC virus genes show evidence of host-parasite codemography? Infect Genet Evol. 2001;1(1):3–12. doi: 10.1016/s1567-1348(01)00002-8. [DOI] [PubMed] [Google Scholar]
- 115.Sugimoto C, et al. Typing of urinary JC virus DNA offers a novel means of tracing human migrations. Proc Natl Acad Sci U S A. 1997;94(17):9191–9196. doi: 10.1073/pnas.94.17.9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chima SC, et al. Polyomavirus JC Genotypes in an Urban United States Population Reflect the History of African Origin and Genetic Admixture in Modern African Americans. Hum Biol. 2014;72(5):837–850. [PubMed] [Google Scholar]
- 117.Zheng HY, et al. Phylogenetic relationships among JC virus strains in Japanese/Koreans and Native Americans speaking Amerind or Na-Dene. J Mol Evol. 2003;56(1):18–27. doi: 10.1007/s00239-002-2376-3. [DOI] [PubMed] [Google Scholar]
- 118.Cobo MF, et al. Reconstructing Population History Using JC Virus : Amerinds, Spanish, and Africans in the Ancestry of Modern Puerto Ricans. Hum Biol. 2001;73(3):385–402. doi: 10.1353/hub.2001.0032. [DOI] [PubMed] [Google Scholar]
- 119.Narganes-Sorde Y, Chanlatte-Baik L, Garcia-Padilla A. La Cultura Saladoide en Puerto Rico. Museo de Historia, Antropologia y Arte: Universidad de Puerto Rico, Recinto de Rio Piedras, Universidad de Puerto Rico; 2002. [Google Scholar]
- 120.Giovas CM, Fitzpatrick SM. Prehistoric migration in the Caribbean: past perspectives, new models and the ideal free distribution of West Indian colonization. World Archaeol. 2014;46(4):569–589. [Google Scholar]
- 121.Chanlatte-Baik L, Narganes-Storde YM. Archaeological Evaluation of Sorce La Hueca, Vieques. Vieques, Puerto Rico: 2002. [Google Scholar]
- 122.Pagán-Jiménez JR. El mundo vivido por los antiguos pobladores indígenas Huecoide en Las Antillas nororientales (circa 300 aC-500 dC) 2009:1–136. [Google Scholar]
- 123.Ryschkewitsch CF, et al. Human polyomavirus JC variants in Papua New Guinea and Guam reflect ancient population settlement and viral evolution. Microbes Infect. 2000;2(9):987–996. doi: 10.1016/s1286-4579(00)01252-1. [DOI] [PubMed] [Google Scholar]
- 124.Padgett B, Zurhein G, Walker D, Eckroade R, Dessel B. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;297(7712):1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 125.Chen NN, et al. Cooperative action of cellular proteins YB-1 and Pura with the tumor antigen of the human JC polyomavirus determines their interaction with the viral lytic control element. Proc Natl Acad Sci. 1995;92(February):1087–1091. doi: 10.1073/pnas.92.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Imperiale MJ. The Human Polyomaviruses, BKV and JCV: Molecular Pathogenesis of Acute Disease and Potential Role in Cancer. Virology. 2000;267(1):1–7. doi: 10.1006/viro.1999.0092. [DOI] [PubMed] [Google Scholar]
- 127.Drachenberg CB, Hirsch HH, Ramos E, Papadimitriou JC. Polyomavirus disease in renal transplantation: Review of pathological findings and diagnostic methods. Hum Pathol. 2005;36(12):1245–1255. doi: 10.1016/j.humpath.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 128.Del Valle L, et al. Detection of JC Polyomavirus DNA sequences and cellular localization of T-antigen and agnoprotein in oligodendrogliomas. Clin Cancer Res. 2002;8(11):3332–3340. [PubMed] [Google Scholar]
- 129.Hogan TF, Padgett BL, Walker DL, Borden EC, Frias Z. Survey of human polyomavirus (JCV, BKV) infections in 139 patients with lung cancer, breast cancer, melanoma, or lymphoma. Prog Clin Biol Res. 1983;105:311–324. [PubMed] [Google Scholar]
- 130.Padgett BL, Rogers CM, Walker DL. JC virus, a human polyomavirus associated with progressive multifocal leukoencephalopathy: additional biological characteristics and antigenic relationships. Infect Immun. 1977;15(2):656–662. doi: 10.1128/iai.15.2.656-662.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yogo Y, et al. Isolation of a Possible Archetypal JC Virus DNA Sequence from Nonimmunocompromised Individuals. J Virol. 1990;64(6):3139. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Karalić D, Lazarević I, Ćupić M, Jovanović T. The prevalence of human polyomaviruses in urine samples of immunocompetent individuals in the Serbian population. Arch Biol Sci. 2012;64(4):1383–1388. [Google Scholar]
- 133.Jeong BH, Lee KH, Choi EK, Kim K, Kim YS. Genotyping of the JC Virus in Urine Samples of Healthy Korean Individuals. J Med Virol. 2004;72(2):281–289. doi: 10.1002/jmv.10568. [DOI] [PubMed] [Google Scholar]
- 134.Pavesi A. African origin of polyomavirus JC and implications for prehistoric human migrations. J Mol Evol. 2003;56(5):564–572. doi: 10.1007/s00239-002-2425-y. [DOI] [PubMed] [Google Scholar]
- 135.Pavesi A. Utility of JC polyomavirus in tracing the pattern of human migrations dating to prehistoric times. J Gen Virol. 2005;86(5):1315–1326. doi: 10.1099/vir.0.80650-0. [DOI] [PubMed] [Google Scholar]
- 136.Parpia A, Ndeffo Mbah M, Wenzel N, Galvani A. Effects of response to the 2014–2015 Ebola outbreak on deaths from malaria, HIV, and tuberculosis, West Africa. Emerg Infect Dis. 2016;22(3) doi: 10.3201/eid2203.150977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cohn DL, Bustreo F, Raviglione MC. Drug-Resistant Tuberculosis : Review of the Worldwide Situation and the WHO / IUATLD Global Surveillance Project. Clin Infect Dis. 1997;24:121–130. doi: 10.1093/clinids/24.supplement_1.s121. [DOI] [PubMed] [Google Scholar]
- 138.Jaramillo E, Williams BG, Dye C, Raviglione M. Drivers of tuberculosis epidemics : The role of risk factors and social determinants. Soc Sci Med. 2009;68(12):2240–6. doi: 10.1016/j.socscimed.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 139.Glickman MS, Jacobs WR., Jr Microbial Pathogenesis of <em>Mycobacterium tuberculosis:</em> Dawn of a Discipline. Cell. 2016;104(4):477–485. doi: 10.1016/s0092-8674(01)00236-7. [DOI] [PubMed] [Google Scholar]
- 140.Cosivi O, Grange JM, Daborn CJ, Raviglione MC, Fujikura T. Zoonotic Tuberculosis due to Mycobacterium bovis in Developing Countries. Emerg Infect Dis. 1999;4(1):59–70. doi: 10.3201/eid0401.980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Center for Disease Control; [Accessed October 10 2016]. 2014. Division of Tuberculosis Elimination. https://www.cdc.gov/tb/statistics/default.htm. [Google Scholar]
- 142.van Embden JD, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting : recommendations for a standardized Strain Identification of Mycobacterium tuberculosis by DNA Fingerprinting : Recommendations for a Standardized Methodology. J Clin Microbiol. 1993;31(2):406–9. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Streicher EM, et al. Spoligotype Signatures in the Mycobacterium tuberculosis Complex. J Clin Microbiol. 2007;45(1):237–240. doi: 10.1128/JCM.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Homolka S, Niemann S, Gagneux S. Genotyping of Genetically Monomorphic Bacteria : DNA Sequencing in Mycobacterium tuberculosis Highlights the Limitations of Current Methodologies. PLoS One. 2009;4(11):e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Minister of Public Works Government Services C. Tuberculosis in Canada 2007 (catalog no HP37-5/2007E-PDF) Ottawa, Canada: 2009. [Google Scholar]
- 146.Gardy J, et al. Whole-Genome Sequencing and Social- Network Analysis of a Tuberculosis Outbreak. N Engl J Med. 2011;364(8) doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 147.Fitzpatrick LK, et al. A Preventable Outbreak of Tuberculosis Investigated through an Intricate Social Network. Clin Infect Dis. 2001 Oct;30333:1801–1806. doi: 10.1086/323671. [DOI] [PubMed] [Google Scholar]
- 148.Sacks JJ, Brenner ER, Breeden DEEC, Anders HM, Parker RL. Epidemiology of a Tuberculosis Outbreak in a South Carolina Junior High School. AJPH. 1985;75(4):361–365. doi: 10.2105/ajph.75.4.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hershberg R, et al. High Functional Diversity in Mycobacterium tuberculosis Driven by Genetic Drift and Human Demography. PLoS Biol. 2008;6(12) doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007 May;7:328–37. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 151.Frith J, Villemin A. History of Tuberculosis. Part 1 - Phthisis, consumption and the White Plague. J Mil Veterans' Heal. 2014;22(2):29–35. [Google Scholar]
- 152.Daniel TM. The history of tuberculosis. Respir Med. 2006;100:1862–1870. doi: 10.1016/j.rmed.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 153.Spigelman M, Matheson C, Lev G, Greenblatt C, Donoghue HD. Confirmation of the presence ofMycobacterium tuberculosis complex-specific DNA in three archaeological specimens. Int J Osteoarchaeol. 2002;12(6):393–401. [Google Scholar]
- 154.Kerley ER. Diseases in Antiquity: A Survey of the Diseases, Injuries, and Surgery of Early Populations. Science (80-) 1968;161(3844):875–876. [Google Scholar]
- 155.Morse D, Brothwell DR, Ucko PJ. Tuberculosis in Ancient Egypt. Am Rev Respir Dis. 1964;90(4):524–541. doi: 10.1164/arrd.1964.90.4.524. [DOI] [PubMed] [Google Scholar]
- 156.Hershkovitz I, et al. Detection and Molecular Characterization of 9000-Year- Old Mycobacterium tuberculosis from a Neolithic Settlement in the Eastern Mediterranean. PLoS One. 2008;3(10):1–6. doi: 10.1371/journal.pone.0003426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Marmiesse M, et al. Ancient Origin and Gene Mosaicism of the Progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1(1):e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Brosch R, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci. 2002;99(6):3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stead WW. The origin and erratic global spread of tuberculosis. How the past explains the present and is the key to the future. Clin Chest Med. 1997;18(1):65–77. doi: 10.1016/s0272-5231(05)70356-7. [DOI] [PubMed] [Google Scholar]
- 160.Stead WW, et al. When did Mycobacterium tuberculosis infection first occur in the New World? An important question with public health implications. Am J Respir Crit Care Med. 1995;151(4):1267–1268. doi: 10.1164/ajrccm/151.4.1267. [DOI] [PubMed] [Google Scholar]
- 161.Salo WL, Aufderheide aC, Buikstra J, Holcomb Ta. Identification of Mycobacterium tuberculosis DNA in a pre-Columbian Peruvian mummy. Proc Natl Acad Sci U S A. 1994;91(6):2091–4. doi: 10.1073/pnas.91.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Smith NH. A Re-Evaluation of prototuberculosis. PLoS Pathog. 2006;2(9):e98. doi: 10.1371/journal.ppat.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Smith NH, Hewinson RG, Kremer K, Brosch R, Gordon SV. Myths and misconceptions: the origin and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol. 2009;7(7):537–544. doi: 10.1038/nrmicro2165. [DOI] [PubMed] [Google Scholar]
- 164.Brites D, Gagneux S. Co-evolution of Mycobacterium tuberculosis and Homo sapiens. Immunol Rev. 2015;264(1):6–24. doi: 10.1111/imr.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zink AR, et al. Characterization of Mycobacterium tuberculosis Complex DNAs from Egyptian Mummies by Spoligotyping. J Clin Microbiol. 2003;41(1):359. doi: 10.1128/JCM.41.1.359-367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cave AJE, Demonstrator A. The evidence for the incidence of tuberculosis in ancient Egypt. Br J Tuberc. 1939;33(3):142–152. [Google Scholar]
- 167.Galagan JE. Genomic insights into tuberculosis. Nat Rev Genet. 2014;15(5):307–320. doi: 10.1038/nrg3664. [DOI] [PubMed] [Google Scholar]
- 168.Bos KI, et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature. 2014;514(7523):494–497. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chisholm RH, Tanaka MM, Chisholm RH. The emergence of latent infection in the early evolution of Mycobacterium tuberculosis. Proc R Soc B. 2016;283(20160499) doi: 10.1098/rspb.2016.0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Søborg DA, Hendriksen B, Kilian M. Widespread Occurrence of Bacterial Human Virulence Determinants in Soil and Freshwater Environments. Appl Environ Microbiol. 2013;79(18):5488–5497. doi: 10.1128/AEM.01633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hruska K, Kaevska M. Mycobacteria in water, soil, plants and air : a review. Vet Med (Praha) 2012;57(12):623–679. [Google Scholar]
- 172.Ghodbane R, Medie FM, Lepidi H, Nappez C, Drancourt M. Long-term survival of tuberculosis complex mycobacteria in soil. Microbiology. 2016;160(2014):496–501. doi: 10.1099/mic.0.073379-0. [DOI] [PubMed] [Google Scholar]
- 173.Chisholm RH, Trauer JM, Curnoe D, Tanaka MM. Controlled fire use in early humans might have triggered the evolutionary emergence of tuberculosis. PNAS. 2016;113(32):9051–9056. doi: 10.1073/pnas.1603224113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Brown DR, Alderman N. North Haven: 2010. The dangers to health from outdoor wood furnaces. Available at: www.ehhi.org. [Google Scholar]
- 175.Naeher LP, et al. Woodsmoke Health Effects: A Review. Inhal Toxicol. 2007;19(1):67–106. doi: 10.1080/08958370600985875. [DOI] [PubMed] [Google Scholar]
- 176.Global Health - Division of Parasitic Diseases. Center for Disease Control. [Accessed October 10 2016];2015 http://www.cdc.gov/parasites/toxoplasmosis/biology.html.
- 177.Tenter AM, Heckeroth AR, Weiss LM. Toxoplasma gondii: from animals to humans. Int J Parasitol. 2000;30(12–13):1217–1258. doi: 10.1016/s0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pereira KS, Franco RMB, Leal DAG. Transmission of toxoplasmosis (Toxoplasma gondii) by foods. Adv Food Nutr Res. 2010;60:1–19. doi: 10.1016/S1043-4526(10)60001-0. [DOI] [PubMed] [Google Scholar]
- 179.Flegr J. Effects of Toxoplasma on Human Behavior. Schizophr Bull. 2007;33(3):757–760. doi: 10.1093/schbul/sbl074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Vyas A, Kim S, Giacomini N, Boothroyd JC, Sapolsky RM. Behavioral changes induced by Toxoplasma infection of rodents are highly specific to aversion of cat odors. PNAS. 2007;104(15):6442–47. doi: 10.1073/pnas.0608310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ingram WM, Goodrich LM, Robey EA, Eisen MB. Mice Infected with Low-Virulence Strains of Toxoplasma gondii Lose Their Innate Aversion to Cat Urine, Even after Extensive Parasite Clearance. PLoS One. 2013;8(9):1–6. doi: 10.1371/journal.pone.0075246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.House PK, Vyas A, Sapolsky R. Predator Cat Odors Activate Sexual Arousal Pathways in Brains of Toxoplasma gondii Infected Rats. PLoS One. 2011;6(8):8–11. doi: 10.1371/journal.pone.0023277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nurse G, Lenghaus C. An outbreak of Toxoplasma gondii abortion, mummification and perinatal death in goats. Aust Vet J. 1986;63(1):1983–1985. doi: 10.1111/j.1751-0813.1986.tb02869.x. [DOI] [PubMed] [Google Scholar]
- 184.Munday BL, Mason RW. Toxoplasmosis as a cause of perinatal death in goats. Aust Vet J. 1979;55(10):485–487. doi: 10.1111/j.1751-0813.1979.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 185.Webster JP, Kaushik M, Bristow GC, McConkey GA. Toxoplasma gondii infection, from predation to schizophrenia: can animal behaviour help us understand human behaviour? J Exp Biol. 2013;216(1):99–112. doi: 10.1242/jeb.074716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Arling TA, et al. Toxoplasma gondii antibody titers and history of suicide attempts in patients with recurrent mood disorders. J Nerv Ment Dis. 2009;197(12):905–908. doi: 10.1097/NMD.0b013e3181c29a23. [DOI] [PubMed] [Google Scholar]
- 187.Lafferty KD. Can the common brain parasite, Toxoplasma gondii, influence human culture ? Proc R Soc B. 2006;273(August):2749–2755. doi: 10.1098/rspb.2006.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Switaj K, Master A, Skrzypczak M, Zaborowski P. Recent trends in molecular diagnostics for Toxoplasma gondii infections. Clin Microbiol Infect. 2005;11(3):170–176. doi: 10.1111/j.1469-0691.2004.01073.x. [DOI] [PubMed] [Google Scholar]
- 189.Liu Q, Wang ZD, Huang SY, Zhu XQ. Diagnosis of toxoplasmosis and typing of Toxoplasma gondii. Parasit Vectors. 2015;8:292. doi: 10.1186/s13071-015-0902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Blackston CR, et al. High-Resolution Typing of Toxoplasma gondii Using Microsatellite Loci. J Parasitol. 2001;87(6):1472–1475. doi: 10.1645/0022-3395(2001)087[1472:HRTOTG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 191.Demar M, Carme B, Darde ML, Ajzenberg D, Ban AL. Genetic diversity, clonality and sexuality in Toxoplasma gondii. Int J Parasitol. 2004;34:1185–1196. doi: 10.1016/j.ijpara.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 192.Bowie WR, et al. Outbreak of toxoplasmosis associated with municipal drinking water. Lancet. 1997;350(Jul 19):173–177. doi: 10.1016/s0140-6736(96)11105-3. [DOI] [PubMed] [Google Scholar]
- 193.Demar M, et al. Fatal Outbreak of Human Toxoplasmosis along the Maroni River : Epidemiological, Clinical, and Parasitological Aspects. Clin Infect Dis. 2007;45(October):e88–e95. doi: 10.1086/521246. [DOI] [PubMed] [Google Scholar]
- 194.Moura L de, et al. Waterborne Toxoplasmosis Brazil, from Field to Gene. Emerg Infect Dis. 2006;12(2):2–5. doi: 10.3201/eid1202.041115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Honnold SP, Braun R, Scott DP, Sreekumar C, Dubey JP. Toxoplasmosis in a Hawaiian monk seal (Monachus schauinslandi) J Parasitol. 2005;91(3):695–697. doi: 10.1645/GE-469R. [DOI] [PubMed] [Google Scholar]
- 196.Fliermans CB, et al. Ecological Distribution of Legionella pneumophila. Appl Environ Microbiol. 1981;41(1):9–16. doi: 10.1128/aem.41.1.9-16.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33(12):1179–83. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bitar DM, Molmeret M, Abu Kwaik Y. Molecular and cell biology of Legionella pneumophila. Int J Med Microbiol. 2004;293(7–8):519–527. doi: 10.1078/1438-4221-00286. [DOI] [PubMed] [Google Scholar]
- 199.Jurcev-Savicevic A, et al. Bus water storage tank as a reservoir of Legionella pneumophila. Coll Antropol. 2014;38(3):1033–1037. [PubMed] [Google Scholar]
- 200.Zbikowska E, Kletkiewicz H, Walczak M, Burkowska A. Coexistence of Legionella pneumophila bacteria and free-living amoebae in lakes serving as a cooling system of a power plant. Water Air Soil Pollut. 2014;225(8) doi: 10.1007/s11270-014-2066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Carvalho FRS, Foronda AS, Pellizari VH. Detection of Legionella pneumophila in water and biofilm samples by culture and molecular methods from man-made systems in Sao Paulo, Brazil. Brazilian J Microbiol. 2007;38(4):743–751. [Google Scholar]
- 202.Yu VL, et al. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J Infect Dis. 2002;186(1):127–128. doi: 10.1086/341087. [DOI] [PubMed] [Google Scholar]
- 203.Jernigan DB, et al. Outbreak of Legionnaires' disease among cruise exposed to a contaminated whirlpool spa ship passengers. Lancet. 1996;347(February 24):60–64. doi: 10.1016/s0140-6736(96)91137-x. [DOI] [PubMed] [Google Scholar]
- 204.Gilmour MW, et al. Molecular typing of a Legionella pneumophila outbreak in Ontario, Canada. J Med Microbiol. 2007;56:336–341. doi: 10.1099/jmm.0.46738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Salah IB, Ghigo E, Drancourt M. Free-living amoebae, a training field for macrophage resistance of mycobacteria. Clin Microbiol Infect. 2009;15(10):894–905. doi: 10.1111/j.1469-0691.2009.03011.x. [DOI] [PubMed] [Google Scholar]
- 206.Horwitz Silverstein. Legionnaires' Disease Bacterium (Legionella pneumophila) Multiplies Intracellularly in Human Monocytes. J Clin Invest. 1980;66(3):441–450. doi: 10.1172/JCI109874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Horwitz MA. The Legionnaires' Disease Bacterium (Legionella Pneumophila) Inhibits Phagosome-Lysosome Fusion in Human Monocytes. J Exp Med. 1983;158(December):2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Patrizia M, et al. Protozoa and human macrophages infection by Legionella pneumophila environmental strains belonging to different serogroups. Arch Microbiol. 2013;195(2):89–96. doi: 10.1007/s00203-012-0851-9. [DOI] [PubMed] [Google Scholar]
- 209.Gao LY, Harb OS, Kwaik YA. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect Immun. 1997;65(11):4738–4746. doi: 10.1128/iai.65.11.4738-4746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Segal G, Shuman HA. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999;67(5):2117–2124. doi: 10.1128/iai.67.5.2117-2124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Isberg, O'Connor, Heidtman The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7(1):13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Steinert M, Hentschel U, Hacker J. Legionella pneumophila: An aquatic microbe goes astray. FEMS Microbiol Rev. 2002;26(2):149–162. doi: 10.1111/j.1574-6976.2002.tb00607.x. [DOI] [PubMed] [Google Scholar]
- 213.Molmeret M, Horn M, Wagner M, Santic M. Amoebae as Training Grounds for Intracellular Bacterial Pathogens MINIREVIEW. Appl envriomental Microbiol. 2005;71(1):20–28. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Miller MB, Bassler BL. Quorum Sensing in Bacteria. Annu Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 215.Deep A, Chaudhary U, Gupta V. Quorum sensing and Bacterial Pathogenicity: From Molecules to Disease. J Lab Physicians. 2011;3(1):4–11. doi: 10.4103/0974-2727.78553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.VanValen L. A New Evolutionary Law. Evol Theory. 1973;1:1–30. [Google Scholar]
- 217.Herre EA, Knowlton N, Mueller UG, Rehner SA. The evolution of mutualisms: Exploring the paths between conflict and cooperation. Trends Ecol Evol. 1999;14(2):49–53. doi: 10.1016/s0169-5347(98)01529-8. [DOI] [PubMed] [Google Scholar]
- 218.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447(7142):326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]