Abstract
Anti-angiogenic therapies have failed to confer survival benefits in patients with metastatic breast cancer (mBC). However, to date there has not been an inquiry into roles for acquired versus innate drug resistance in this setting. In this study, we report roles for these distinct phenotypes in determining therapeutic response in a murine model of mBC resistance to the anti-angiogenic tyrosine kinase inhibitor sunitinib. Using tumor measurement and vascular patterning approaches, we differentiated tumors displaying innate versus acquired resistance. Bioluminescent imaging of tumor metastases to the liver, lungs and spleen revealed that sunitinib administration enhances metastasis, but only in tumors displaying innate resistance to therapy. Transcriptomic analysis of tumors displaying acquired versus innate resistance allowed the identification of specific biomarkers, many of which have a role in angiogenesis. In particular, aquaporin-1 upregulation occurred in acquired resistance, mTOR in innate resistance, and pleiotrophin in both settings, suggesting their utility as candidate diagnostics to predict drug response or to design tactics to circumvent resistance. Our results unravel specific features of antiangiogenic resistance, with potential therapeutic implications.
Introduction
The use of antiangiogenic drugs to treat metastatic breast cancer has had limited success in the clinic. The monoclonal anti-VEGF antibody bevacizumab, when combined with various chemotherapy regimens has delivered modest but significant progression free survival (PFS) improvements in this setting, the IMELDA phase III clinical trial reporting a 7.6 and 15.3 month improvement in PFS and overall survival (OS) respectively (1), with other trials showing more modest improvements (2–4). Antiangiogenic, small molecule oral receptor tyrosine kinase inhibitors (RTKIs), however, have failed completely in this setting (5–7) with some reports linking their use with an adverse impact on patient survival (6). This may be in part due to an increased frequency of adverse events, leading to more temporary discontinuations of therapy. However, evidence is mounting to suggest that antiangiogenic RTKIs could also promote metastasis following treatment, contributing to the failure of the drugs to improve PFS or OS (8,9).
It has been suggested that anti-angiogenic RTKIs have failed as a treatment for breast cancer due to a limited dependence on angiogenesis for tumour growth and possible angiogenic growth factor redundancy in breast cancer, facilitating the rapid acquisition of resistance (10). This resistance takes two forms. Some tumours show no objective response in terms of tumour growth and are termed innately resistant, whilst others progress after a short period of stasis or shrinkage. These tumours are termed as having acquired resistance (reviewed in (11)). In order to investigate and compare the differences between these two forms of resistance and the impact this has on metastasis, an in vivo model of metastatic breast cancer resistance to the anti-angiogenic drug sunitinib, was developed.
Sunitinib malate is a multi-target oral tyrosine kinase receptor (RTK) inhibitor (12), targeting both pro-angiogenic platelet derived growth factor receptors α and β (PDGFRα/β) and vascular endothelial growth factor receptors (VEGFRs) on endothelial cells as well as some pro-tumourogenic growth factor receptors including the RET proto-oncogene. Despite initially promising results in phase II clinical trials (13,14) sunitinib has thus far failed to demonstrate utility in the metastatic and triple negative setting in phase III trials (15–17). Efforts continue however, in clinical trials, to investigate its utility in breast cancer, in combination with Crizotinib, Doxorubicin, or Cyclophophamide (Information retrieved from: http://clinicaltrials.gov and http://www.cancer.gov [accessed: 02.11.2016]).
In this study, tumour growth patterns resembling clinically characterised acquired and innate resistance were observed, after daily sunitinib treatment. Further analysis revealed that tumours, displaying acquired or innate resistance, has distinct patterns of vascular and metastasis formation and that sunitinib treatment does enhance metastasis, but only in the innately resistant setting. Comparative transcriptomic analysis of these tumours and their vasculature revealed that resistance was gained primarily via the utilisation of non-inhibited angiogenic pathways and this process was mapped over time. A number of possible markers of acquired and innate resistance were also identified.
Materials and Methods
Generation of a stable 4T1-Luc cell line
Phoenix-Ampho cells (see: https://web.stanford.edu/group/nolan/_OldWebsite/retroviral_systems/phx.html) were transfected with MSCV-LUC plasmid DNA (18), using lipofectamine 2000 (Invitrogen) and cultured to generate MSCV-LUC plasmid containing retrovirus. The MSCV-LUC retroviral media was used to transduce 4T1 cells (ATCC), which were positively selected using puromycin. The 4T1 cell line was obtained from the American Type Culture Collection (ATCC), resuscitated from early passage liquid nitrogen stocks, treated as described in (18) and used in this experiment less than 2 months after the re-initiation of culture. Cells were tested negative for mycoplasma contamination. ATCC uses morphology, karyotyping, and PCR based approaches to confirm the identity of human cell lines.
Animal experiments
Mice were handled and treated in accordance with British home office requirements (Licence number, PPL. 40/3339). 2.5x105 4T1-Luc cells suspended in Optimem (Gibco) and in a volume of 100 μL, were injected into the third mammary fat pad of anaesthetised 8-week-old female Balb/C mice. After a 1-week tumour establishment period, mice received either 40 mg sunitinib (Selleck Chemicals) per kg body weight in PBS and 3.72% DMSO or the drug vehicle only. 2-3 times a week, tumour bearing mice were IP injected with 150 mg/Kg D-luciferin (PerkinElmer), left for 5 minutes, then imaged over a period of 30 minutes and the peak average radiance of bioluminescence reading for each region of interest calculated. At the experimental endpoint the same procedure was used, but the mice were culled after a 10 minute incubation. Selected organs were dissected and imaged using the IVIS imaging system (PerkinElmer). Luminescent quantification was performed on the two opposite sides of each organ and the results averaged. In order to avoid bioluminescence crossover between experiments, In vivo and ex vivo imaging were not performed on the same day, therefore final day tumour measurements were not taken by in vivo imaging.
Immunofluorescent staining
Immunofluorescent staining of frozen mouse tissue was performed using 2 μg/ml rabbit polyclonal antisera to mouse AQP1 (Boster Immunoleader) and 75 ng/ml rat monoclonal antisera to mouse PECAM-1 as primary antibodies. Antibody binding was then detected using 4 μg/ml goat polyclonal antisera to rat IgGs conjugated to alexafluor 546 (A11081, Invitrogen) and 4 μg/ml donkey polyclonal antisera to rabbit IgGs conjugated to alexafluor 488 (A21206, Invitrogen) and mounted in ProLong Gold mounting media containing DAPI (Invitrogen). Quantification of fluorescence was conducted using the ImageJ software package (19).
Quantitative real-time PCR
RNA isolation was performed using the miRNeasy mini kit (Qiagen) and complementary DNA generated using the High Capacity cDNA Reverse Transcription kit (Invitrogen). Quantitative real time PCR was performed using the Exiqon universal probe system (Roche) as previously described (20). Primer sequences are provided in Supplementary Table 1. The Delta-Delta Ct method was used to compare the expression levels between samples and β-actin was used to standardise expression unless otherwise stated.
Endothelial isolation using PECAM-1 conjugated magnetic beads
Tumour tissue was minced and digested in DMEM containing 2 mg ml-1 collagenase type V (Sigma), 7.4 mg ml-1 actinomycin D (Sigma) and 30 kU ml-1 DNase I (Qiagen), shaking at 37°C for 1 hour. Endothelial cells were isolated from the digested single cell suspension by positive magnetic selection using 1x107/g sheep anti-rat IgG coated M-280 Dynabeads® (Invitrogen) conjugated to 2.5 μg rat anti-mouse PECAM-1 antibody (MEC13.3 clone, BD Falcon).
Microarray and analysis
RNA was isolated from the magnetic bead isolated endothelium, reverse transcribed to cDNA, transcribed, amplified and labelled with Cy3 (Low input quick amp labelling kit, Agilent). Labelled cRNA samples were then hybridized to SurePrint G3 Mouse Gene Expression v2 8x60K microarray chips (Agilent). The R programming language (Lucent Technologies), marray (21) and the Limma (Bioconductor) plug-in were used to subtract background, quantile normalize probe signal intensities and perform differential gene expression analyses on the microarray data. Raw and processed data from this analysis are deposited in the Gene Expression Omnibus (GEO) repository (accession number: GSE84048).
Results
In order to investigate tumour responses to sunitinib, an orthotopic model of metastatic breast cancer was set up using 4T1 cells engineered to express luciferase (4T1-luc). This enabled tumour growth and metastasis to be tracked by bioluminescence. Briefly, 4T1-luc cells were injected into the 3rd mammary fat pad of female Balb/c mice. The tumours were grown for 7 days prior to the start of treatment with 40 mg/kg sunitinib, or vehicle only control. Once tumours reached 1300 mm3 in size, mice were culled and tumours collected.
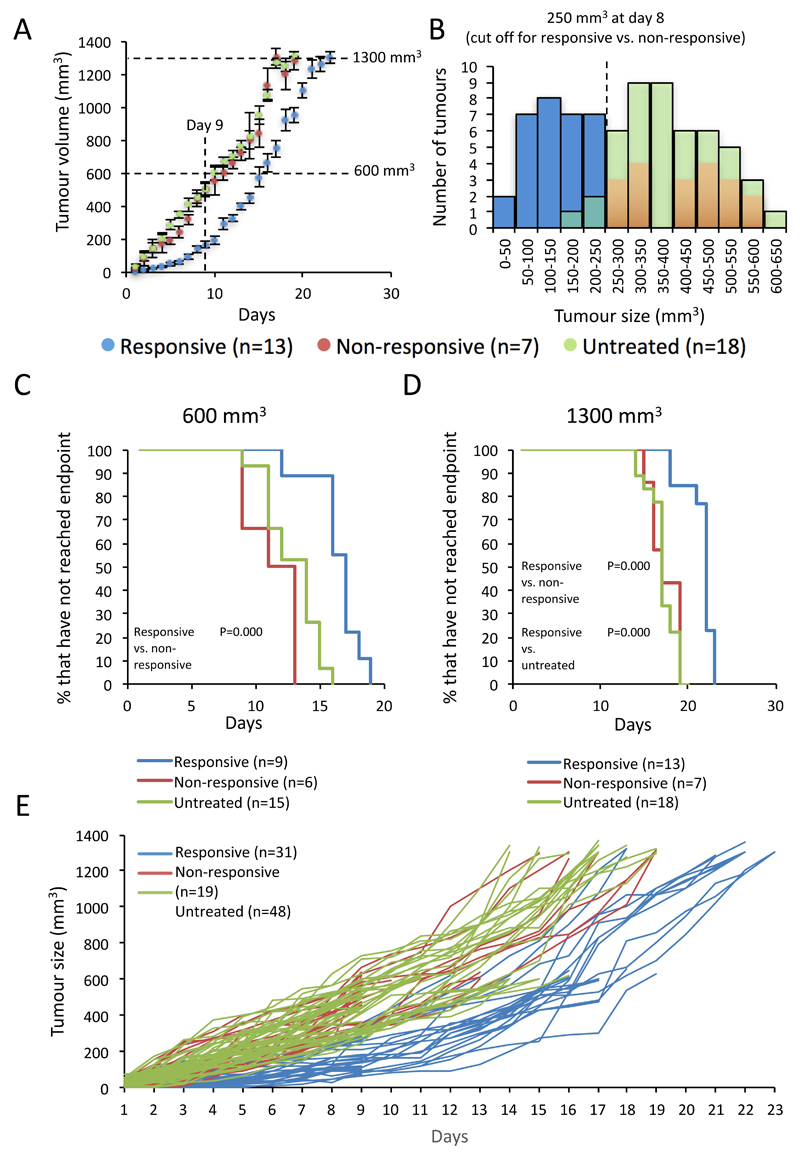
Daily caliper measurements were performed to track the development of these tumours. This analysis revealed that whilst untreated tumours grew at a fairly linear rate throughout the experiment, sunitinib treated tumours appeared to fall into two groups. Some showed no response to sunitinib treatment and shared a similar growth profile with the untreated tumours. These were classified as non-responsive. Others had retarded growth for the first 9 days, after which the tumours grew at a similar rate to the other two groups (Figure 1A). In order to determine whether these two treated groups could be considered distinct, the distribution of tumour sizes at day 8, the point of greatest disparity between responsive and non-responsive tumour growth, was investigated for this and subsequent experiments (Figure 1B). This analysis showed that whilst the untreated tumours appeared to follow a close to normal distribution, the treated group had two populations, one considerably smaller than the majority of untreated tumours (responsive) and the other with similar tumour size and distribution to the untreated group (non-responsive). The best point of distinction between the two groups was determined to be whether they were larger or smaller than 250 mm3 at day 8, therefore this cut-off point was selected. Of note, the term “responsive”, used here and later in the manuscript, refers to the cohort from which the tumour is derived, based on its growth pattern, not its current resistance status. Therefore “responsive” tumours collected at 1300 mm3 display sunitinib resistant behaviour, but only after an initial sensitive phase.
Figure 1. Sunitinib treated 4T1 tumours display both acquired and innate resistance.
A, Tumour growth curves from the initial sunitinib drug trials, with endpoint set at 1300 mm3 (mean ± SEM ). Measurements begin one week after tumour inoculation and on the day sunitinib treatment began. Subsequent experimental endpoints were set based on these growth curves and their intersections with this data are shown. B, histogram plot showing the distribution of tumour sizes at day 8 of treatment. Sunitinib treated tumours exceeding 250 mm3 in size were identified as falling into the non-responsive cohort. Sunitinib treatment significantly retards growth of responsive tumours. C & D, Kaplan-Meier comparative analysis of time to endpoint of tumours grown to 600 mm3 and 1300 mm3. Log ranks statistical analysis of significant results is shown. E, spider plot of the growth curves of all tumours used in the experiment. Throughout this figure n-numbers are as shown and tumour cohorts are coloured as follows (Responsive – blue, Non-responsive – red, Untreated – green).
In summary the experiment generated 3 cohorts of tumours, (i) naïve/ untreated, (ii) those that showed no response to sunitinib, modelling innate resistance and (iii), those that after an initial sensitive period became resistant to sunitinib growth inhibition, mirroring the response pattern of acquired or adaptive resistance, encountered in the clinic.
In order to investigate this further, tumours were harvested at key time and size points for analysis. The tumours of a cohort of 15 treated and 15 untreated mice were collected at day 9 (the end of the treatment sensitive period in the responsive group) and at 600 mm3 (a size point at which all initially responsive tumours are growing at the same rate as their untreated counterparts) (Figure 1D). Of note, in each experiment there was a roughly 60/40% split between responsive and non-responsive tumours and by log-ranks statistical analysis it was determined that sunitinib treatment had a significant effect on the time it took responsive tumours to grow to both 600 mm3 and 1300mm3, versus non-responsive or untreated tumours, confirming that sunitinib had a significant, if transient, effect on tumour growth, restricted to the responsive group (Figures 1C and D). In order to reduce the impact of transient environmental or mouse batch effects on results, the data from each time-point was collated from three separate experiments, with results consistent between experiments (Figures 1E).
Differential sunitinib mediated effects on tumour vascular patterning
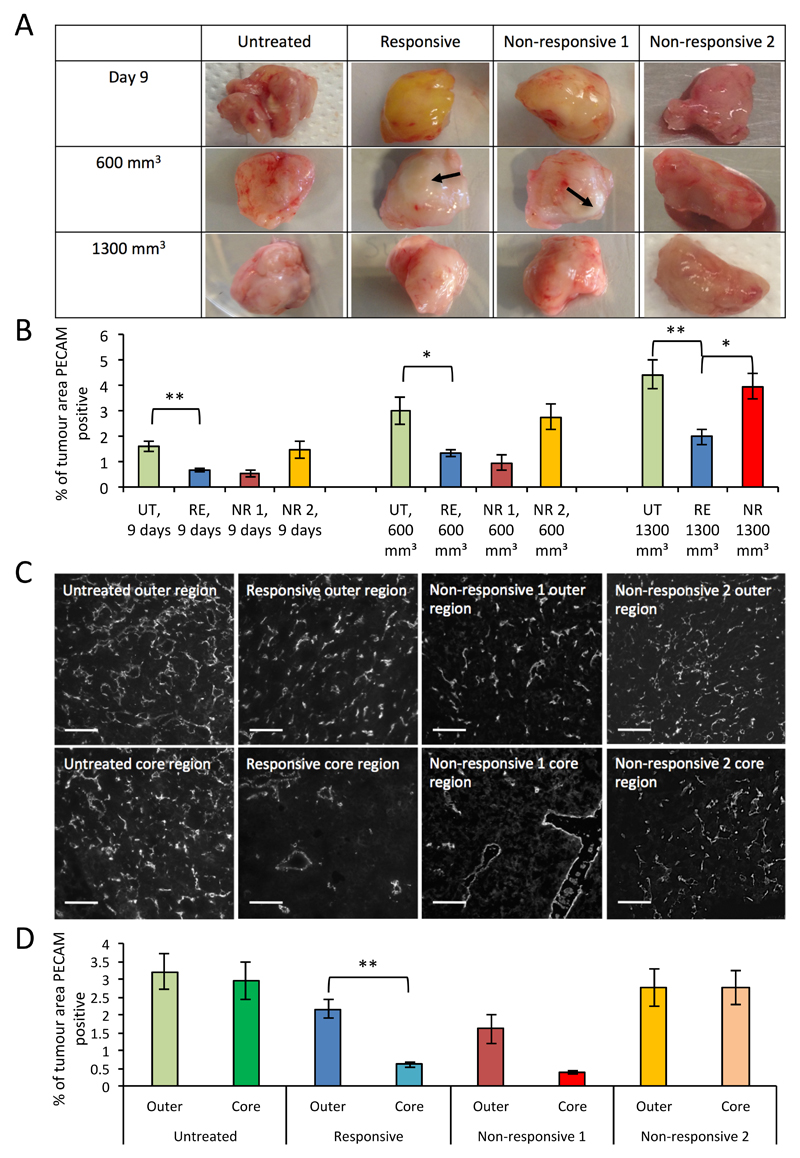
Tumours collected at either day 9, 600 mm3 or 1300 mm3 in size were assessed both macro- and microscopically for effects of sunitinib treatment on vascular patterning (Figure 2). This assessment revealed distinct patterns of vascularisation in each cohort. Untreated tumours appeared macroscopically well vascularised throughout the experiment (Figure 2A) and showed modest increases in vascularity (% of microscopic tumour area PECAM-1 positive) (Figure 2B). Responsive sunitinib treated tumours were macroscopically avascular at day 9, by 600mm3 they had an apparent avascular core, possibly derived from growth up to day 9, around which was a crest of greater vascularity, which by 1300 mm3 had encompassed the entire outside of the tumour. Of note, the yellow hue clearly observable on all responsive day 9 tumours, presumably from the accumulation of the yellow drug sunitinib in the tumour, had disappeared by 600 mm3 in all but one tumour. This suggests that the drug had been removed or could not gain entry to the core of the tumour at this time point, possibly contributing to drug resistance (Figure 2A). Intriguingly the non-responsive cohort appeared to split into two groups, one macroscopically resembling the responsive group (day 9 n=3, 600 mm3 n=2) and another macroscopically resembling the untreated group (day 9 n=3, 600 mm3 n=4). This hinted at a possible further subdivision of cohorts, in that some non-responsive tumours did not respond to sunitinib at all and appeared to be untreated, whilst others showed signs of sunitinib induced vascular inhibition but continued to grow regardless, possibly due to support from their surrounding environment (Figure 2A). Microscopic assessment of vascular density between the two non-responsive populations corroborated these observations (Figure 2B). Assessment of the comparative level of vascularity found in the outer and core regions of 600 mm3 tumours supported observations made on the macroscopic level, in that untreated tumours had a consistent level of vascularity, responsive tumours had a clear reduction of vascularity in the core of the tumour and non-responsive tumours resembled one or the other of these groups (Figure 2C and D).
Figure 2. Sunitinib treatment reduces vascularity and impacts vascular patterning of 4T1 tumours.
A, representative images of tumours from each cohort and experimental endpoint, showing macroscopic vascular patterning, not to scale. Avascular regions marked with arrows. B, bar chart of vascular density of tumours from each cohort and experimental endpoint, determined by the average percentage of PECAM immunofluorescent staining across 10 fields of view (mean ± SEM, Mann-Whitney, ** p<0.01, * p<0.05, n-numbers: Day 9, responsive (RE)=9, non-responsive 1 (NR 1)=3, non-responsive 2 (NR 2)=3, untreated (UT)=15; 600mm3, RE=9, NR 1=2, NR 2=4, UT=15; 1300mm3, RE=13, NR=7, UT=18.). C, representative images of immunofluorescent PECAM-1 staining in the core and outer regions of tumours from each cohort of tumours, taken at 600 mm3. D, bar chart of vascular density in the core and outer regions of tumours from each cohort, taken at 600 mm3, determined by the average percentage of PECAM immunofluorescent staining across 5 fields of view (mean ± SEM, statistics and n-numbers as in B).
Sunitinib enhances metastasis in innately resistant tumours
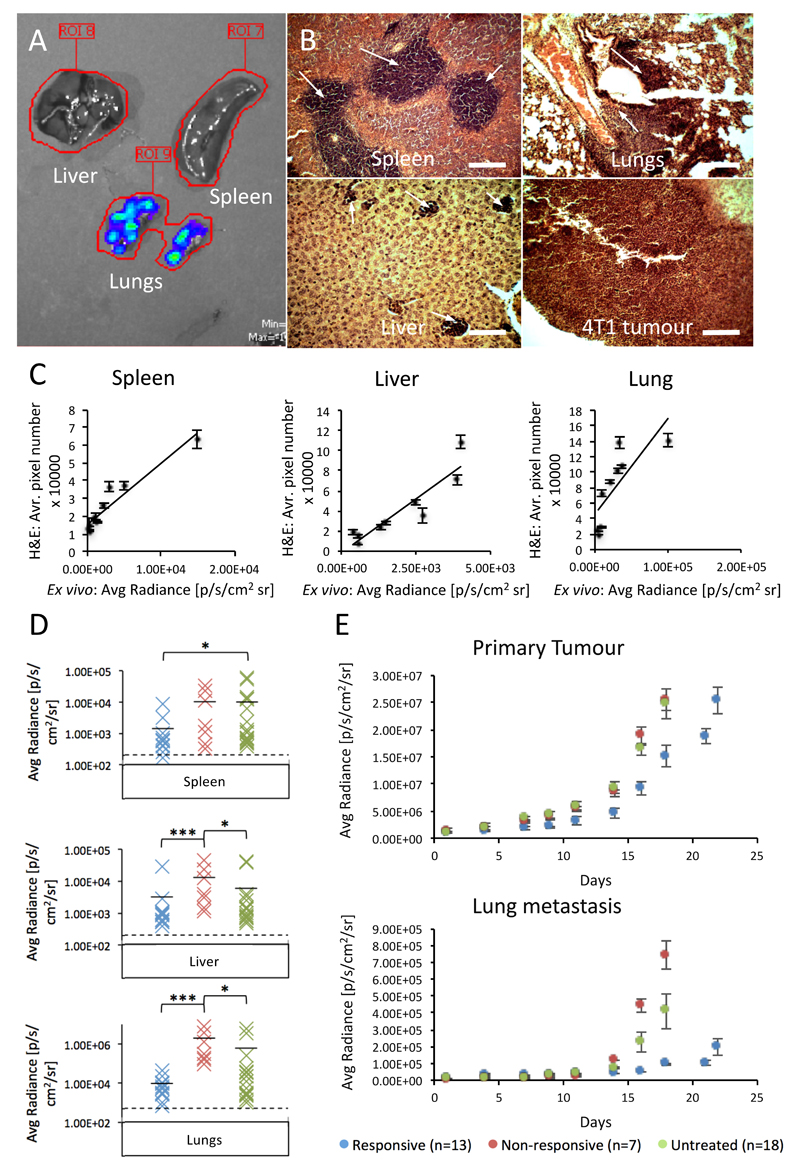
Anti-angiogenic drugs have been reported to enhance tumour metastasis in certain circumstances (11). The utility of ex vivo bioluminescent imaging of resected organs for signs of metastasis was investigated as a reliable method for quantifying metastasis. Key sites of 4T1 metastasis (the liver, lungs and spleen) were probed by both ex vivo bioluminescent imaging and H&E staining (Figure 3A and B). The apparent metastatic burden, from 10 mice where tumours had been harvested at 1300 mm3, was quantified by each method and correlated. There was a good level of correlation of metastatic burden between the two approaches on an individual organ level (Figure 3C).
Figure 3. Sunitinib treatment enhances 4T1 tumour metastasis, but only in the innately resistant setting.
A, representative image of liver, spleen and lung whole organs undergoing bioluminescent imaging by the IVIS (overlay of blue-green-red colouring represents bioluminescence of increasing intensity. B, representative images of H&E staining of 4T1 tumour and metastasis in spleen, lungs and liver (metastasis marked by white arrows). Scale bar = 100 μm. C, correlation between metastatic burden, as determined by measurement of average metastatic area across 10 fields of view, in organs stained by H&E and by whole organ bioluminescent pixel density, in spleen, liver and lung tissues (mean ± SEM, n=10). D, distribution plots of bioluminescence from each organ and cohort at the 1300 mm3 endpoint. Cohorts are coloured as follows (Responsive – blue, Non-responsive – red, Untreated – green). The level of background auto-fluorescence measured by imaging of organs from mice with no tumour is also displayed (dashed line). Statistical analysis: Mann-Whitney, *** p<0.001, * p<0.05. E, longitudinal bioluminescent quantification of primary and secondary lung tumour development, generated by bioluminescent imaging of the whole mouse with regions of interest drawn over the tumour and lung areas (mean ± SEM, n-numbers as displayed).
This analysis revealed that the liver and lungs of mice with non-responsive primary tumours developed a significantly greater metastatic burden compared to those of both responsive and untreated cohorts at 1300 mm3, whereas in the spleen the untreated cohort developed the greatest metastatic burden (Figure 3D).
It is unclear from this analysis whether metastasis in the responsive group is curtailed by retarded seeding at the sites of metastasis, or by sunitinib induced inhibition of growth once seeded. A key unknown is the timing of metastasis seeding. In order to investigate this, longitudinal measurements of primary and secondary lung tumour development were taken by bioluminescent imaging of mice, where tumours were allowed to develop to 1300 mm3. Analysis of the primary tumour revealed a similar growth pattern between cohorts to the caliper measurements (Figure 3E). Assessment of bioluminescence emanating from the lung area revealed that metastases reach a measurable level markedly earlier in the non-responsive and untreated cohorts than the responsive cohort (day 14 vs. 18) and that metastasis develops rapidly in these two cohorts, whereas in the responsive cohort metastasis growth is slow even once detectable. This suggests that both metastasis seeding and growth is retarded in the responsive group.
Sunitinib resistance inheritance
These metastasis findings beg the question as to whether sunitinib resistance in this model is induced by environmental factors (as appears to be the case in the responsive group, where tumours revert to a sensitive state once metastasised) or by the development or existence of intrinsic drug resistance within the tumour cells themselves (as would be suggested by the metastatic pattern of the non-responsive tumours). In order to investigate whether tumour cells removed from their immediate environment maintain resistance, tumours from the responsive and non-responsive cohorts were removed at 1300 mm3, minced and tumour cells cultured for two weeks, prior to re-implantation in the mammary fat pad subjection to the same experimental conditions as before. The resultant tumours derived from the responsive tumour donor cells displayed an only slightly greater propensity to innate resistance than tumours derived from naïve 4T1 cells (1/5 vs. 0/5), whereas those derived from an innately resistant tumour displayed a markedly greater propensity (3/5) (Supplementary figure 1). These finding could suggest that some level of resistance, intrinsic to the tumour is retained even after re-implantation in innately resistant tumours, in agreement with the metastasis data.
Murine tumour endothelial isolation and microarray analysis
In order to investigate the impact sunitinib therapy had on the tumour vessels, endothelium was isolated from the untreated, responsive and non-responsive cohorts, using anti-PECAM magnetic bead isolation (Supplementary figure 2A). PECAM-1 is also a marker of a small subset of leukocytes, therefore RTqPCR was performed on the isolates to confirm specific endothelial enrichment. Relative expression of markers of leukocytes (CD11b), macrophages (CD68), epithelial cells (EPCAM), smooth muscle cells (PDGFRA) and endothelium (PECAM), was assessed between matched endothelial isolates and endothelial depleted fractions, by RTqPCR. This analysis confirmed that PECAM expression alone was enriched 25-30 fold in the endothelial isolates, suggesting very good endothelial enrichment (Supplementary figure 2B).
Two tumour bulk samples each, from the responsive and untreated cohorts harvested at day 9 and 600 mm3, along with four representative endothelial isolate samples and bulk samples from each of the responsive, non-responsive and untreated cohorts taken at 1300 mm3, were selected for microarray transcriptomic analysis (individual growth curves shown in Supplementary figure 3).
The impact of sunitinib treatment on tumour and endothelial gene expression
Microarray analysis of the selected samples facilitated the transcriptional characterisation of the tumour and associated endothelium, from naïve (untreated), innately resistant (non-responsive) and adaptively resistant (responsive) tumours, at key stages of resistance acquisition (day 9, 600 mm3 and 1300 mm3). This was done by the investigation of the comparative expression profile between these cohorts, of genes known to be associated with sunitinib response (Supplementary tables 2 and 3), metastasis (Supplementary tables 4 and 5), and endothelial migration (Supplementary table 6 and Table 1).
Table 1.
Expression change of genes that enhance endothelial migration in EC isolates from tumours harvested at 1300 mm3. Log2 fold change in gene expression shown.
| Gene ID | Gene symbol | GeneBank accession no. | Effect on endothelial migration | Non-responsive vs. Untreated EC | Responsive vs. Untreated EC | Non-responsive vs. Responsive EC |
|---|---|---|---|---|---|---|
| Leptin | LEP | NM_008493 | Increased | -2.24 | -0.11 | -2.08 |
| SH2 domain protein 2A | SH2D2A | NM_021309 | Increased | -2.09 | 0.02 | -1.96 |
| Interleukin 17A | IL17A | NM_010552 | Increased | -1.91 | -0.54 | -1.29 |
| Colony stimulating factor 2 | CSF2 | NM_009969 | Increased | -1.53 | -0.90 | -0.56 |
| Prostaglandin-endoperoxide synthase 2 | PTGS2 | NM_011198 | Increased | -1.42 | -0.46 | -0.97 |
| Chemokine (C-X-C motif) ligand 1 | CXCL1 | NM_008176 | Increased | -1.31 | -0.39 | -0.91 |
| Sphingosine-1-phosphate receptor 3 | S1PR3 | NM_010101 | Increased | -1.31 | 0.10 | -1.41 |
| Chemokine (C-X-C motif) ligand 12 | CXCL12 | NM_001012477 | Increased | -1.28 | 0.86 | -2.14 |
| Hyaluronan synthase 3 | HAS3 | NM_008217 | Increased | -1.16 | 0.04 | -1.16 |
| GATA binding protein 1 | GATA1 | NM_008089 | Increased | -1.14 | -0.78 | -0.31 |
| Elastin | ELN | NM_007925 | Increased | -1.07 | 0.02 | -1.07 |
| Insulin-like growth factor binding protein 3 | IGFBP3 | NM_008343 | Increased | -1.03 | 0.57 | -1.60 |
| Thrombomodulin | THBD | NM_009378 | Increased | -0.79 | 0.68 | -1.49 |
| Protein tyrosine phosphatase 4a3 | PTP4A3 | NM_008975 | Increased | -0.78 | 0.22 | -1.00 |
| Pleckstrin homology domain containing, G5 | PLEKHG5 | NM_001004156 | Increased | -0.76 | 0.51 | -1.27 |
| Bone morphogenetic protein 6 | BMP6 | NM_007556 | Increased | -0.70 | 0.49 | -1.12 |
| Phosphodiesterase 2A, cGMP-stimulated | PDE2A | NM_001143848 | Increased | -0.64 | 0.52 | -1.15 |
| Placental growth factor | PGF | NM_008827 | Increased | -0.61 | 0.39 | -1.01 |
| Melanoma cell adhesion molecule | MCAM | NM_023061 | Increased | -0.37 | 0.69 | -1.08 |
| Endothelial cell-specific adhesion molecule | ESAM | NM_027102 | Increased | -0.35 | 1.07 | -1.45 |
| Endothelial-specific receptor tyrosine kinase | TEK | NM_013690 | Increased | -0.26 | 0.81 | -1.14 |
| Nitric oxide synthase 3, endothelial cell | NOS3 | NM_008713 | Increased | -0.22 | 0.78 | -1.11 |
| Phospholipase C, gamma 1 | PLCG1 | AK169695 | Increased | 0.06 | 1.05 | -1.05 |
| Endothelin 1 | EDN1 | NM_010104 | Increased | 0.07 | 1.02 | -1.00 |
| Gastrin releasing peptide | GRP | NM_175012 | Increased | 0.49 | -0.73 | 1.05 |
| Mechanistic target of rapamycin | MTOR | NM_020009 | Increased | 1.03 | 0.18 | -0.01 |
| Arachidonate 12-lipoxygenase | ALOX12 | NM_007440 | Increased | 1.06 | 0.63 | 0.47 |
| Nuclear factor of activated T cells C3 | NFATC3 | NM_010901 | Increased | 1.12 | -0.14 | 1.40 |
| Integrin alpha 4 | ITGA4 | NM_010576 | Increased | 1.12 | -0.07 | 1.19 |
| 5-hydroxytryptamine (serotonin) receptor 7 | HTR7 | FM178516 | Increased | 1.15 | 0.03 | 1.10 |
| Tenascin N | TNN | NM_177839 | Increased | 1.17 | 0.32 | 0.88 |
| Activating transcription factor 2 | ATF2 | NM_001025093 | Increased | 1.18 | -0.01 | 1.21 |
| Heparanase | HPSE | NM_152803 | Increased | 1.19 | 0.43 | 0.74 |
| Pleiotrophin | PTN | NM_008973 | Increased | 1.19 | 1.54 | -0.45 |
| Collagen and calcium binding EGF domains 1 | CCBE1 | NM_178793 | Increased | 1.22 | -0.09 | 1.33 |
| Inhibitor of DNA binding 1 | ID1 | NM_010495 | Increased | 1.28 | 0.67 | 0.58 |
| Fibroblast growth factor 2 | FGF2 | AY027558 | Increased | 1.35 | -0.02 | 1.33 |
| Teratocarcinoma-derived growth factor 1 | TDGF1 | NM_011562 | Increased | 1.89 | -0.24 | 2.14 |
Comparative analysis of genes associated with sunitinib response
A list of genes associated with sunitinib response was compiled from the Ingenuity online database and their expression analysed between arrays. This analysis revealed that at day 9, tumours from the “responsive” cohort displayed an expression profile in line with that predicted of response to sunitinib therapy, when compared to the untreated cohort. All seven genes with expression >2 fold changed, were altered in line with predicted sunitinib response (Supplementary table 2). This response profile was lost however, at later time points, in line with the loss of sunitinib induced growth retardation in the responsive cohort, discussed previously.
This analysis when applied to endothelial isolates from the responsive, non-responsive and untreated cohorts, at 1300 mm3 agreed with the bulk tumour data, as regards the responsive (but now having acquired resistance) cohort, showing no obvious sensitivity to sunitinib treatment, in terms of gene expression (Supplementary table 3). Conversely, gene expression of sunitinib target genes was reduced in the non-responsive cohort, relative to the responsive cohort, suggesting that despite displaying innate resistance from the outset of the experiment, in terms of growth profile, on the transcriptomic level the endothelium of this cohort were more sensitive to sunitinib treatment than the responsive cohort (Supplementary table 3).
Comparative analysis of genes associated with metastasis
Metastasis signalling in the responsive tumours appeared to be quite strongly inhibited at day 9, when compared to the untreated cohort at the same time period (Supplementary table 4). Fifteen known or predicted pro-metastatic genes were >2 fold down-regulated by sunitinib treatment at this stage. Five pro-metastatic genes were >2 fold up-regulated at this stage in the treated tumours, however, the pattern of signalling was primarily inhibitory to metastasis. This expression pattern was progressively reversed at subsequent harvesting points, with a third of >2 fold altered pro-metastatic genes up-regulated at 600 mm3 and three quarters at 1300 mm3. In the non-responsive cohort too the majority of pro-metastatic genes >2 fold altered were up-regulated at 1300 mm3, when compared to the untreated cohort (Supplementary table 5). A number of pro-metastatic genes are also downregulated in this setting, suggesting the inhibition of certain metastatic pathways, possibly induced by continued sunitinib sensitivity, in agreement with supplementary table 3. Intriguingly, when comparing the two treated cohorts, it was the responsive group that displayed the slightly more pro-metastatic profile, with five genes stimulatory to metastasis, up-regulated in the responsive group, over the non-responsive cohort. This data overall agrees with the observed metastatic profile of the tumours discussed previously, with sunitinib initially inhibiting metastasis up to day 9, and even 600 mm3, but by 1300 mm3 metastasis and the signalling for it, was enhanced by sunitinib treatment.
Comparative analysis of genes associated with endothelial migration
In order to explore the effect sunitinib treatment had on pro-angiogenic gene expression over the course of the experiment, a list of genes known to enhance endothelial migration (a key component of angiogenesis) was compiled and their expression investigated. At day 9 pro-angiogenic gene expression was primarily inhibited in the responsive group versus the untreated group, with the majority of >2 fold altered genes being down-regulated at this stage, presumably in response to sunitinib angiogenesis inhibition (Supplementary table 6). This pattern of inhibition was lost at later time points, with even a few pro-angiogenic genes showing enhanced expression in the treated group. This data is in agreement with the observed vascularisation patterns reported in Figure 2. At 1300 mm3 genes stimulatory to endothelial migration were both up and down-regulated in the endothelium of non-responsive tumours, versus the untreated cohort (Table 1).This suggests that despite the observed similarity in tumour growth and vascularisation between the groups, the methods of vascularisation could be quite distinct. Therefore, despite sensitivity to sunitinib signalling inhibition potentially being maintained, as reported in Supplementary table 3, the use of alternative angiogenic pathways permits the continued growth and vascularisation of this cohort. The responsive group on the other-hand displayed an endothelial expression pattern primarily stimulatory to endothelial migration with the expression of pro-angiogenic molecules, such as endothelial cell specific adhesion molecule (ESAM), endothelin 1 (EDN1) and pleiotrophin (PTN) enhanced versus the untreated group. Endothelial migration signalling appears to be enhanced in the responsive group at the 1300 mm3, beyond that found in the other cohorts, but not via the same alternative pathways utilised by the non-responsive group.
The selection and validation of acquired resistance markers
In order to investigate and validate this observed up-regulation of sunitinib targeted pathways and angiogenesis in general, in the vessels of the initially responsive cohort, a matrix comparing responsive tumour endothelium to untreated tumour endothelium was set up. This analysis identified a strikingly large number of angiogenic genes >2 fold up-regulated in the responsive group, including aquaporin 1 (AQP1) and angiopoietin 2 (ANGPT2) among others (Table 2). Interestingly the RET proto-oncogene and VEGF receptor 2, targets of sunitinib inhibition were also both up-regulated in the responsive group, possibly suggesting a mechanism of resistance by the up-regulation of the target genes (Table 2).
Table 2.
Genes significantly enriched in the microarray analysis of endothelial isolates from responsive vs. untreated tumours harvested at 1300 mm3. RTqPCR selection of genes of interest for further analysis is also shown. Gene expression was normalised to β-actin and PECAM (n=4). Selected genes exhibited >3 fold enrichment in responsive tumours normalised to both β-actin and PECAM and had at least 5% of the expression of β-actin. Rejected genes are highlighted in black.
| Microarray | RTqPCR | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene ID | Gene Symbol | GeneBank accession no. | Fold change | P-value | β-actin vs. target fold expression change | PECAM vs. target fold expression change | Expression level relative to β-actin (%) | Selected genes |
| Prolactin receptor | PRLR | NM_011169 | 4.32 | 0.01 | 9.81 | 10.2 | 5.23 | PRLR |
| Pleiotrophin | PTN | NM_008973 | 2.91 | 0.01 | 2.25 | 2.72 | 0.01 | |
| Aquaporin 1 | AQP1 | NM_007472 | 2.76 | 0.00 | 4.93 | 4.19 | 240.42 | AQP1 |
| Ret proto-oncogene | RET | NM_001080780 | 2.4 | 0.00 | 9.02 | 7.85 | 0.27 | |
| Angiopoietin 2 | ANGPT2 | NM_007426 | 2.34 | 0.00 | 5.75 | 3.57 | 41.39 | ANGPT2 |
| Duffy blood group, chemokine receptor | DARC | NM_010045 | 2.3 | 0.01 | 6.79 | 5.66 | 30.83 | DARC |
| Leptin receptor | LEPR | NM_001122899 | 2.3 | 0.00 | 5.44 | 3.75 | 2.26 | |
| Endothelial cell surface expressed chemotaxis regulator | ECSCR | NM_001033141 | 2.25 | 0.00 | 1.86 | 1.7 | 0.89 | |
| Tetraspanin 7 | TSPAN7 | NM_019634 | 2.25 | 0.00 | 0.5 | 0.64 | 0.03 | |
| Stanniocalcin 2 | STC2 | NM_011491 | 2.12 | 0.00 | 27.22 | 18.02 | 0.04 | |
| Endothelial cell-specific adhesion molecule | ESAM | NM_027102 | 2.1 | 0.00 | 2.49 | 1.74 | 10.33 | |
| Multimerin 2 | MMRN2 | NM_153127 | 2.06 | 0.00 | 3.23 | 2.59 | 11.58 | MMRN2 |
| Vascular endothelial growth factor receptor 2 | VEGFR2 | NM_010612 | 2.06 | 0.00 | 4.69 | 3.49 | 11.65 | VEGFR2 |
| Endothelin 1 | EDN1 | NM_010104 | 2.03 | 0.00 | 3.04 | 2.41 | 1.98 | |
In order to validate the differential expression of the 14 candidate genes arising from the microarray analysis (Table 2), RTqPCR analysis comparing the expression level of each of these genes between responsive EC and untreated EC, used for the microarray analysis, was performed (Table 2). Genes warranting further investigation were selected based on fold expression change, normalised to β-actin (per cell level) and PECAM-1 (per endothelial cell level), as well as on gene expression level relative to β-actin. Genes identified by RTqPCR analysis, with a fold enrichment of >3 times and expressed at >5% of the expression level of β-actin, were taken forward (Table 2). This left ANGPT2, AQP1, DARC, MMRN2, PRLR and VEGFR2, as the key genes of interest for further investigation.
Expression changes of candidate markers of acquired resistance over time
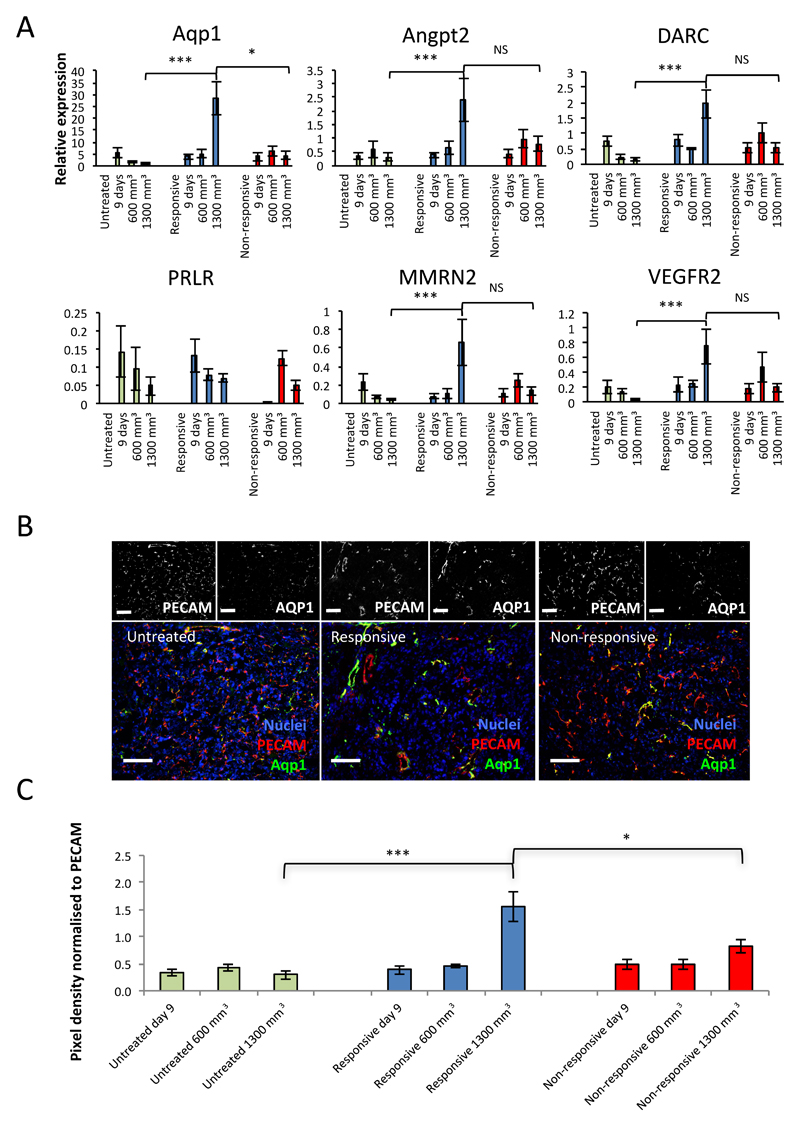
In order to investigate the expression of the candidate genes at key stages of tumour development, in the full set of isolates of different cohorts, RTqPCR was performed on endothelial isolates from day 9, 600 mm3 and 1300 mm3, in the responsive, non-responsive and untreated cohorts (Figure 4A). This analysis revealed a significant shift in the expression of all the candidate genes, except for PRLR, between the responsive and untreated tumour endothelium at 1300 mm3. Aquaporin-1 alone had an additional significant shift in expression between the responsive and non-responsive cohorts at 1300 mm3, marking it out as a key distinguishing gene, enriched in the responsive cohort alone, at this time point.
Figure 4. Aquaporin is significantly enriched in the vessels of responsive tumours over those of untreated and non-responsive tumours.
A, RTqPCR for the relative expression of the six genes of interest in endothelial isolates from untreated, responsive and non-responsive tumours harvested at 9 days, 600 mm3 and 1300 mm3 (mean expression ±SEM, *** p<0.001, * p<0.05, NS – Not Significant, Mann-Whitney). B, representative images of AQP1 staining in untreated, responsive and non-responsive tumours by immunofluorescence (IF). Black and white split channel and colour merged channel images of tumours triple stained by IF for DAPI (nuclei, blue), PECAM-1 (vessels, red) and AQP1 (green). C, quantitation of pixel density of staining by IF for AQP1 standardised to PECAM-1 staining (mean ± SEM, *** p<0.001, * p<0.05, Mann-Whitney, n-numbers: Day 9, responsive (R)=5, non-responsive (NR)=5, untreated (UT)=10; 600mm3, R=4, NR=6, UT=10; 1300mm3, R=12, NR=7, UT=17, 10 fields of view each).
In order to further validate aquaporin 1 as a specific marker of acquired resistance on the protein level, immunohistochemistry and immunofluorescence was conducted, comparing marker expression in sections cut from responsive, non-responsive and untreated cohort tumours, harvested at day 9, 600 mm3 and 1300 mm3. The analysis compared the optical density of marker fluorescence (green channel), normalised to the PECAM-1 fluorescence (red channel), between the groups of tumour samples. In this way it was confirmed that AQP1 was enriched in the vessels of responsive tumours specifically at the 1300 mm3 stage (Figure 4B and C).
Discussion
4T1 tumour cells engineered to express luciferase were used to set up a model of breast cancer tumour growth and metastasis. The effect daily sunitinib treatment had on these tumours was investigated. This study identified 4T1 tumours responding to treatment in three distinct manners. One group were initially sensitive to therapy, with retarded tumour growth for the first 9 days of treatment, after which tumour growth followed the same rate as the untreated cohort. These tumours displayed marked devascularisation during the period of inhibition, and evidence of revascularisation once insensitive. This cohort additionally displayed significantly reduced metastasis over the period of the experiment. Transcriptomic analysis of this cohort showed the initial down-regulation of various signalling molecules targeted by sunitinib inhibition, angiogenic and metastatic signalling molecule expression was also reduced relative to the untreated cohort, but only up to day 9. After this point, at the 600 and 1300 mm3 size-points, the downregulation of these pathways was lost or even reversed. This profile was suggestive of an acquired loss of sensitivity to sunitinib therapy after day 9, in both tumour growth rate and signalling
A second cohort, showed insensitivity to the sunitinib treatment, with no growth retardation relative to the untreated cohort. This cohort displayed significantly enhanced metastatic growth in the liver and lungs. Transcriptomic analysis of this group however, showed the down-regulation of certain signalling molecules in line with sunitinib induced inhibition. This suggests that the tumours were still sensitive to sunitinib therapy at the 1300 mm3 size point and that there was an innate redundancy for sunitinib targeted pathways in the growth and vascularisation of these tumours. This cohort of tumours appeared to utilise a separate, uninhibited mechanism for continued tumour growth and metastasis and therefore had an innate resistance to sunitinib therapy which was shown to have some level of heritability after being re-derived in culture, although greater n-numbers are required to definitively show this.
The transcriptional profile of acquired and innate resistance
The existence of a cohort of tumours displaying no response to sunitinib is well established in the clinic. A recent phase III clinical trial investigating sunitinib in combination with docetaxel as a treatment for advanced breast cancer, reported a response rate of only 55% (17), while Barrios et al., 2010 (22) reported an objective response rate of 11% with sunitinib used as a monotherapy in HER2-negative advanced breast cancer. The identification of markers that will allow the prediction of which tumours are likely to respond to sunitinib, has the potential to considerably improve the use and effectiveness of this drug.
Innate resistance
The angiogenic profile of the non-responsive tumours appeared quite distinct from the untreated and responsive cohorts. The expression of nine pro-angiogenic genes were at least 2 fold reduced in the non-responsive cohort, including leptin (LEP), the reduction of which, significantly correlates with response to sunitinib in RCC and prostate cancer (23,24), chemokine ligand-1 (CXCL1), the release of which is induced by VEGF signalling (25) and S1P receptor 3, an endothelial mitogen receptor that operates synergistically with PDGFR-β and is known to be down-regulated by sunitinib treatment in breast cancer (26). This profile is suggestive of active sunitinib inhibition.
The expression of twelve other proangiogenic genes, were enhanced in the non-responsive tumours, including pleiotrophin (PTN), an angiogenic cytokine, highly expressed in 60% of breast cancers (27). Additionally the expression of PTN has been shown to be specifically enhanced in response to VEGF signalling blockade, in three separate pre-clinical tumour models (27), suggesting that it may form part of an adaptive response to VEGF targeted therapies. Intriguingly PTN is additionally up-regulated in the vessels of initially responsive tumours at 1300 mm3. This suggests that PTN may be playing an important role in mediating the evasion of sunitinib angiogenic blockade in both cohorts, either as an acquired resistance mechanism in the responsive group, or an innate one in the non-responsive group. The role of PTN in breast cancer resistance to sunitinib warrants further investigation.
Mechanistic target of rapamycin (mTOR), a pro-angiogenic protein kinase, whose phosphorylation is known to be inhibited by sunitinib (28), was also up-regulated in the non-responsive cohort. MTOR is known to enhance pro-angiogenic hypoxia inducible factor (HIF) signalling (29) and therefore may also be playing a role in mediating sunitinib resistance. Likewise, FGF2, an angiogenic factor extensively associated with antiangiogenic resistance (reviewed in (11)), was upregulated in the non-responsive cohort specifically. FGF2 has been shown to directly stimulate endothelial proliferation and capillary tube formation in the presence of sunitinib (30), suggesting that FGF could be playing a role in mediating the treatment resistant vascularisation and tumour growth observed in the non-responsive cohort.
Acquired resistance
Whereas innately resistant tumours appeared to display a distinct angiogenic expression profile, leading to insensitivity to sunitinib treatment, tumours that displayed acquired resistance in this investigation, instead showed initial responsiveness, characterised by the inhibition of key sunitinib targeted pathways, followed by a gradual reversal of this inhibition. This resulted not in a transition to the alternate angiogenic profile of the non-responsive cohort, but rather a loss of sensitivity in the targeted pathways to sunitinib blockade, leading to constitutive activation. This cohort developed an expression profile progressively more similar to the untreated cohort. This finding is not without precedent, Sakai et al., 2013 (31) generated a sunitinib resistant RCC cell line, through prolonged treatment with sunitinib. They found that the cells acquired resistance via the constitutive activation of target signal transduction pathways. It is possible then that this phenomenon was due to mutations in the target pathways, leaving them insensitive to sunitinib inhibition. This offers a survival benefit to the cells effected and is propagated throughout the tumour. This mechanism of acquired resistance has been observed in gastrointestinal tumours treated with sunitinib, but only after approximately a year of response (32). It seems unlikely then that 9 days would be sufficient time for tumours to acquire such mutations. Additionally, when these tumours were re-implanted after two weeks in culture they formed treatment sensitive tumours suggesting this form of resistance is unstable and reversible, in agreement with (33–35). It is therefore unlikely to be induced by genetic mutations.
The loss of sunitinib signalling sensitivity was coupled in the tumour with the up-regulation of pro-angiogenic genes. RTqPCR, IHC and IF validation of this profile, identified AQP1, ANGPT2, DARC, MMRN2 and VEGFR2, to be significantly up-regulated at 1300 mm3, in the responsive cohort alone and enhanced AQP1 expression to be a distinct marker of acquired resistance in this experiment.
AQP1 is a widely expressed cell surface water channel, important for water transfer in the kidney (36), but with a key function in many cells, including endothelium, of allowing the rapid transit of water across the plasma membrane, facilitating the increase in cellular volume critical for cell proliferation (37). Increased AQP1 expression has been reported in a range of solid tumours (38), as well as being shown to correlate with microvessel density in ovarian cancer (39). Hypoxia and increased hypoxia inducible factor 1 (HIF-1) activity has been implicated as a key regulator of AQP1 expression in endothelial cells (40). The responsive tumour specific expression of AQP1 as well as a number of other hypoxia responsive genes, including ANGPT2 (41) and VEGFR2 (42), may suggest a heightened level of hypoxia, in this cohort, caused by sunitinib induced vascular inhibition. Hypoxia therefore, may have played a role in driving the pro-angiogenic profile of expression observed in the responsive cohort. The role of hypoxia and AQP1 expression in driving the acquisition of sunitinib resistance warrants further investigation.
The effect of sunitinib therapy on metastasis is dependent on initial tumour response
The presence of tumour metastases are one of the major risk factors for death in all cancers and anti-angiogenic therapies, including sunitinib, have been implicated in increasing the risk of this eventuality. Both Yin et al., 2014 (8) and Welti et al., 2012 (9), report enhanced lung metastasis and tumour infiltration in models of breast cancer. Admittedly the tumour cells in Welti et al., 2012 (9) were injected intravenously, therefore were already circulating and did not have to escape the tumour. The observations detailed in this paper further our understanding.
Metastasis to the liver and lungs was significantly enhanced by sunitinib treatment at the 1300 mm3 stage, but only in the innately resistant cohort. Unlike the responsive cohort, where metastasis was either unchanged or reduced relative to the untreated cohort, the majority of the non-responsive cohort displayed no devascularisation. This suggests that where tumour cell escape is not curtailed by reduced vascularity, tumour cell seeding to distant organs is enhanced by sunitinib. Some potential mechanisms by which might occur have been reported including, the up-regulation of angiogenesis and metastasis associated cytokines and growth factors in response to treatment (43), the mobilisation of bone-marrow derived cells generating a pre-metastatic niche (44) and finally high dose treatment with sunitinib has been reported to lead to pericyte depletion, likely through PDGF signalling inhibition, in lung vasculature, which is correlated with enhanced breast tumour seeding (45). This latter observation is supported by pericyte depletion studies in which they found that loss of pericytes in advanced tumours, inhibited growth but enhanced lung metastasis (46).
Upon seeding metastasis growth from innately resistant tumours also appears to be resistant to sunitinib inhibition to a far greater level than those from acquired resistance tumours, as shown by IVIS longitudinal measurements of lung metastasis. This partially agrees with Guerin et al., 2013 (47), who showed sunitinib to be ineffective when treating established visceral metastases, formed after removal of the primary tumour, even when the primary tumours were treatment sensitive.
Intriguingly despite reduced physical metastasis from acquired resistant tumours, metastatic gene expression was enhanced beyond the other two cohorts. It Is possible that in this cohort, which shows by some distance the greatest treatment induced restriction on vascular development, leading to retarded metastasis in the short term, adaption to a metastatic profile of expression in strongly selected in the interest of survival and to achieve vascular sufficiency as described in (11).
This study describes an approach to investigate and compare different forms of sunitinib resistance, by assigning treated tumours to groups based on the timing of resistance acquisition. This allowed an investigation of the contrasting molecular mechanisms governing innate and acquired resistance to sunitinib and the effect this has on metastasis development. The broad pattern of the data was that where primary tumour growth was retarded and vascular production inhibited, metastasis was slowed by sunitinib therapy. On the other hand, when the primary tumour was non-responsive to treatment the presence of sunitinib enhanced the tumour metastasis. This data further highlights the need for reliable markers for the prediction of sunitinib response, as inappropriate treatment could potentially not only waste resources, but also enhance metastasis.
Supplementary Material
Financial Support
All authors received funds from the University of Birmingham CRUK Cancer Center.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Gligorov J, Doval D, Bines J, Alba E, Cortes P, Pierga J-Y, et al. Lancet Oncol. Vol. 15. Elsevier; 2014. Maintenance capecitabine and bevacizumab versus bevacizumab alone after initial first-line bevacizumab and docetaxel for patients with HER2-negative metastatic breast cancer (IMELDA): a randomised, open-label, phase 3 trial; pp. 1351–60. [DOI] [PubMed] [Google Scholar]
- 2.Patel NS, Li J-L, Generali D, Poulsom R, Cranston DW, Harris AL. Up-regulation of delta-like 4 ligand in human tumor vasculature and the role of basal expression in endothelial cell function. Cancer Res. 2005;65:8690–7. doi: 10.1158/0008-5472.CAN-05-1208. [DOI] [PubMed] [Google Scholar]
- 3.Ebos JML, Kerbel RS. Antiangiogenic therapy: impact on invasion, disease progression, and metastasis. Nat Rev Clin Oncol. 2011;8:210–21. doi: 10.1038/nrclinonc.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minckwitz von G, Minckwitz von G, Puglisi F, Puglisi F, Cortes J, Cortes J, et al. Lancet Oncol. Vol. 15. Elsevier; 2014. Bevacizumab plus chemotherapy versus chemotherapy alone as second-line treatment for patients with HER2-negative locally recurrent or metastatic breast cancer after first-line treatment with bevacizumab plus chemotherapy (TANIA): an open-label, randomised phase 3 trial; pp. 1269–78. [DOI] [PubMed] [Google Scholar]
- 5.Crown JP, Dieras V, Staroslawska E, Yardley DA, Bachelot T, Davidson N, et al. Phase III Trial of Sunitinib in Combination With Capecitabine Versus Capecitabine Monotherapy for the Treatment of Patients With Pretreated Metastatic Breast Cancer. JCO American Society of Clinical Oncology. 2013;31:2870–8. doi: 10.1200/JCO.2012.43.3391. [DOI] [PubMed] [Google Scholar]
- 6.Barrios C, Liu M, Lee S, Vanlemmens L, Ferrero J, Tabei T, et al. Phase III Randomized Trial of Sunitinib (SU) vs. Capecitabine (C) in Patients (Pts) with Previously Treated HER2-Negative Advanced Breast Cancer (ABC) Cancer Res. American Association for Cancer Research. 2009;69:46–6. doi: 10.1007/s10549-010-0788-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfizer. Pfizer Discontinues One Sunitinib Phase 3 Trial in Advanced Breast Cancer; Other Advanced Breast Cancer Trials Continue [Internet] Pfizer. Springer International Publishing; 2009. [cited 2015 Oct 23]. Available from: http://link.springer.com/10.2165/00128415-201013000-00008. [Google Scholar]
- 8.Yin T, He S, Ye T, Shen G, Wan Y, Wang Y. Translational Oncology. Vol. 7. Elsevier; 2014. Antiangiogenic therapy using sunitinib combined with rapamycin retards tumor growth but promotes metastasis; pp. 221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welti JC, Powles T, Foo S, Gourlaouen M, Preece N, Foster J, et al. Contrasting effects of sunitinib within in vivo models of metastasis. Angiogenesis. 2012;15:623–41. doi: 10.1007/s10456-012-9291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerbel RS. Breast. 20 Suppl 3. Elsevier; 2011. Reappraising antiangiogenic therapy for breast cancer; pp. S56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roskoski R. Sunitinib: a VEGF and PDGF receptor protein kinase and angiogenesis inhibitor. Biochemical and Biophysical Research Communications. 2007;356:323–8. doi: 10.1016/j.bbrc.2007.02.156. [DOI] [PubMed] [Google Scholar]
- 13.Miller KD, Burstein HJ, Elias AD, Rugo HS, Cobleigh MA, Pegram MD, et al. Phase II study of SU11248, a multitargeted receptor tyrosine kinase inhibitor (TKI), in patients (pts) with previously treated metastatic breast cancer (MBC) ASCO Meeting Abstracts. 2005;23:563. [Google Scholar]
- 14.Burstein HJ, Elias AD, Rugo HS, Cobleigh MA, Wolff AC, Eisenberg PD, et al. Phase II study of sunitinib malate, an oral multitargeted tyrosine kinase inhibitor, in patients with metastatic breast cancer previously treated with an anthracycline and a taxane. JCO. 2008;26:1810–6. doi: 10.1200/JCO.2007.14.5375. [DOI] [PubMed] [Google Scholar]
- 15.Curigliano G, Pivot X, Cortés J, Elias A, Cesari R, Khosravan R, et al. Randomized phase II study of sunitinib versus standard of care for patients with previously treated advanced triple-negative breast cancer. The Breast. 2013;22:650–6. doi: 10.1016/j.breast.2013.07.037. [DOI] [PubMed] [Google Scholar]
- 16.Crown J, Dieras V, Staroslawska E, Yardley DA, Davidson N, Bachelot TD, et al. Phase III trial of sunitinib (SU) in combination with capecitabine (C) versus C in previously treated advanced breast cancer (ABC) ASCO Meeting Abstracts. 2010;28:LBA1011. [Google Scholar]
- 17.Bergh J, Bondarenko IM, Lichinitser MR, Liljegren A, Greil R, Voytko NL, et al. First-line treatment of advanced breast cancer with sunitinib in combination with docetaxel versus docetaxel alone: results of a prospective, randomized phase III study. JCO. 2012;30:921–9. doi: 10.1200/JCO.2011.35.7376. [DOI] [PubMed] [Google Scholar]
- 18.Fang L, Lee VC, Cha E, Zhang H, Hwang ST. CCR7 regulates B16 murine melanoma cell tumorigenesis in skin. Journal of Leukocyte Biology. 2008;84:965–72. doi: 10.1189/jlb.1107776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider CA, Rasband WS, Eliceiri KW. Nat Meth. Vol. 9. Nature Publishing Group; 2012. NIH Image to ImageJ: 25 years of image analysis; pp. 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong L-J, Heath VL, Sanderson S, Kaur S, Beesley JFJ, Herbert JMJ, et al. ECSM2, an endothelial specific filamin a binding protein that mediates chemotaxis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2008;28:1640–6. doi: 10.1161/ATVBAHA.108.162511. [DOI] [PubMed] [Google Scholar]
- 21.Yang YH, Paquet A, Dudoit S. marray: Exploratory analysis for two-color spotted microarray data [Internet] 2009. [cited 2015 Sep 11]. R package version. Available from: http://www.bioconductor.org/packages/release/bioc/html/marray.html. [Google Scholar]
- 22.Barrios CH, Liu M-C, Lee SC, Vanlemmens L, Ferrero J-M, Tabei T, et al. Phase III randomized trial of sunitinib versus capecitabine in patients with previously treated HER2-negative advanced breast cancer. Breast Cancer Res Treat. 2010;121:121–31. doi: 10.1007/s10549-010-0788-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebos JML, Mastri M, Hudson JM, Lee CR, Tracz A, Attwood K, et al. Effect of the timing of sunitinib administration on the predictive value of biomarkers in renal cell cancer (mRCC) ASCO Meeting Abstracts. 2015;33:11096. [Google Scholar]
- 24.Dror Michaelson M, Regan MM, Oh WK, Kaufman DS, Olivier K, Michaelson SZ, et al. Phase II study of sunitinib in men with advanced prostate cancer. Annals of Oncology. 2009;20:913–20. doi: 10.1093/annonc/mdp111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo H-M, Shieh J-M, Chen C-L, Tsou C-J, Wu W-B. Vascular Endothelial Growth Factor Induces CXCL1 Chemokine Release via JNK and PI-3K-Dependent Pathways in Human Lung Carcinoma Epithelial Cells. International Journal of Molecular Sciences 2013, Vol 14, Pages 10090–10106. Multidisciplinary Digital Publishing Institute. 2013;14:10090–106. doi: 10.3390/ijms140510090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mousseau Y, Mollard S, Faucher-Durand K, Richard L, Nizou A, Cook-Moreau J, et al. Fingolimod potentiates the effects of sunitinib malate in a rat breast cancer model. Breast Cancer Res Treat. 2011;134:31–40. doi: 10.1007/s10549-011-1903-6. [DOI] [PubMed] [Google Scholar]
- 27.Lynn KD, Roland CL, Brekken RA. VEGF and pleiotrophin modulate the immune profile of breast cancer. Cancers (Basel) 2010;2:970–88. doi: 10.3390/cancers2020970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito Y, Tanaka Y, Aita Y, Ishii KA, Ikeda T, Isobe K, et al. Sunitinib induces apoptosis in pheochromocytoma tumor cells by inhibiting VEGFR2/Akt/mTOR/S6K1 pathways through modulation of Bcl-2 and BAD. AJP: Endocrinology and Metabolism. 2012;302:E615–25. doi: 10.1152/ajpendo.00035.2011. [DOI] [PubMed] [Google Scholar]
- 29.Radulovic S, Bjelogrlic SK. Sunitinib, sorafenib and mTOR inhibitors in renal cancer. J BUON. 2007;12 Suppl 1:S151–62. [PubMed] [Google Scholar]
- 30.Welti JC, Gourlaouen M, Powles T, Kudahetti SC, Wilson P, Berney DM, et al. Fibroblast growth factor 2 regulates endothelial cell sensitivity to sunitinib. Oncogene. 2011;30:1183–93. doi: 10.1038/onc.2010.503. [DOI] [PubMed] [Google Scholar]
- 31.Sakai I, Miyake H, Fujisawa M. Acquired resistance to sunitinib in human renal cell carcinoma cells is mediated by constitutive activation of signal transduction pathways associated with tumour cell proliferation. BJU Int. 2013;112:E211–20. doi: 10.1111/j.1464-410X.2012.11655.x. [DOI] [PubMed] [Google Scholar]
- 32.Guo T, Hajdu M, Agaram NP, Shinoda H, Veach D, Clarkson BD, et al. Mechanisms of sunitinib resistance in gastrointestinal stromal tumors harboring KITAY502-3ins mutation: an in vitro mutagenesis screen for drug resistance. Clin Cancer Res. 2009;15:6862–70. doi: 10.1158/1078-0432.CCR-09-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang TC, Man S, Xu P, Francia G, Hashimoto K, Emmenegger U, et al. Development of a resistance-like phenotype to sorafenib by human hepatocellular carcinoma cells is reversible and can be delayed by metronomic UFT chemotherapy. Neoplasia. 2010;12:928–40. doi: 10.1593/neo.10804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Bhasin M, Schor-Bardach R, Wang X, Collins MP, Panka D, et al. Resistance of renal cell carcinoma to sorafenib is mediated by potentially reversible gene expression. In: Chammas R, editor. PLoS ONE. Vol. 6. Public Library of Science; 2011. p. e19144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammers HJ, Verheul HM, Salumbides B, Sharma R, Rudek M, Jaspers J, et al. Reversible epithelial to mesenchymal transition and acquired resistance to sunitinib in patients with renal cell carcinoma: evidence from a xenograft study. Mol Cancer Ther. 2010;9:1525–35. doi: 10.1158/1535-7163.MCT-09-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maunsbach AB, Marples D, Chin E, Ning G, Bondy C, Agre P, et al. Aquaporin-1 water channel expression in human kidney. J Am Soc Nephrol. 1997;8:1–14. doi: 10.1681/ASN.V811. [DOI] [PubMed] [Google Scholar]
- 37.Galán-Cobo A, Ramírez-Lorca R, Toledo-Aral JJ, Echevarría M. Aquaporin-1 Plays Important Role in Proliferation by Affecting Cell Cycle Progression. J Cell Physiol. 2015 doi: 10.1002/jcp.25078. [DOI] [PubMed] [Google Scholar]
- 38.Mobasheri A, Airley R, Hewitt SM, Marples D. Heterogeneous expression of the aquaporin 1 (AQP1) water channel in tumors of the prostate, breast, ovary, colon and lung: a study using high density multiple human tumor tissue microarrays. Int J Oncol. 2005;26:1149–58. [PubMed] [Google Scholar]
- 39.Yang JH, Shi YF, Chen XD, Qi WJ. The influence of aquaporin-1 and microvessel density on ovarian carcinogenesis and ascites formation. Int J Gynecol Cancer. 2006;16 Suppl 1:400–5. doi: 10.1111/j.1525-1438.2006.00476.x. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka A, Sakurai K, Kaneko K, Ogino J, Yagui K, Ishikawa K, et al. The role of the hypoxia-inducible factor 1 binding site in the induction of aquaporin-1 mRNA expression by hypoxia. DNA Cell Biol. 2011;30:539–44. doi: 10.1089/dna.2009.1014. [DOI] [PubMed] [Google Scholar]
- 41.Tsuzuki T, Okada H, Cho H, Shimoi K, Miyashiro H, Yasuda K, et al. Divergent regulation of angiopoietin-1, angiopoietin-2, and vascular endothelial growth factor by hypoxia and female sex steroids in human endometrial stromal cells. Eur J Obstet Gynecol Reprod Biol. 2013;168:95–101. doi: 10.1016/j.ejogrb.2012.12.040. [DOI] [PubMed] [Google Scholar]
- 42.Collet G, Lamerant-Fayel N, Tertil M, Hafny-Rahbi El B, Stepniewski J, Guichard A, et al. Hypoxia-regulated overexpression of soluble VEGFR2 controls angiogenesis and inhibits tumor growth. Mol Cancer Ther. 2014;13:165–78. doi: 10.1158/1535-7163.MCT-13-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebos JML, Lee CR, Christensen JG, Mutsaers AJ, Kerbel RS. Multiple circulating proangiogenic factors induced by sunitinib malate are tumor-independent and correlate with antitumor efficacy. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17069–74. doi: 10.1073/pnas.0708148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okazaki T, Ebihara S, Asada M, Kanda A, Sasaki H, Yamaya M. Granulocyte colony-stimulating factor promotes tumor angiogenesis via increasing circulating endothelial progenitor cells and Gr1+CD11b+ cells in cancer animal models. Int Immunol. 2006;18:1–9. doi: 10.1093/intimm/dxh334. [DOI] [PubMed] [Google Scholar]
- 45.Welti JC, Powles T, Foo S, Gourlaouen M, Preece N, Foster J, et al. Contrasting effects of sunitinib within in vivo models of metastasis. Angiogenesis. 2012;15:623–41. doi: 10.1007/s10456-012-9291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keskin D, Kim J, Cooke VG, Wu C-C, Sugimoto H, Gu C, et al. Targeting vascular pericytes in hypoxic tumors increases lung metastasis via angiopoietin-2. Cell Rep. 2015;10:1066–81. doi: 10.1016/j.celrep.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guerin E, Man S, Xu P, Kerbel RS. A model of postsurgical advanced metastatic breast cancer more accurately replicates the clinical efficacy of antiangiogenic drugs. Cancer Res. 2013;73:2743–8. doi: 10.1158/0008-5472.CAN-12-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.