Abstract
How do organisms respond adaptively to environmental stress? Although some gene-specific responses have been explored, others remain to be identified, and there is a very poor understanding of the system-wide integration of response, particularly in complex, multitissue animals. Here, we adopt a transcript screening approach to explore the mechanisms underpinning a major, whole-body phenotypic transition in a vertebrate animal that naturally experiences extreme environmental stress. Carp were exposed to increasing levels of cold, and responses across seven tissues were assessed by using a microarray composed of 13,440 cDNA probes. A large set of unique cDNAs (≈3,400) were affected by cold. These cDNAs included an expression signature common to all tissues of 252 up-regulated genes involved in RNA processing, translation initiation, mitochondrial metabolism, proteasomal function, and modification of higher-order structures of lipid membranes and chromosomes. Also identified were large numbers of transcripts with highly tissue-specific patterns of regulation. By unbiased profiling of gene ontologies, we have identified the distinctive functional features of each tissue's response and integrate them into a comprehensive view of the whole-body transition from one strongly adaptive phenotype to another. This approach revealed an expression signature suggestive of atrophy in cooled skeletal muscle. This environmental genomics approach by using a well studied but nongenomic species has identified a range of candidate genes endowing thermotolerance and reveals a previously unrecognized scale and complexity of responses that impacts at the level of cellular and tissue function.
Keywords: fish, microarray, stress
By disrupting homeostasis, environmental stress deleteriously affects biological function. Understanding responses to stress and identifying the underpinning mechanisms has traditionally formed an important part of cell physiology. Much attention has recently been directed at model unicellular species, particularly yeast, where a core transcriptional response to a range of different stressors has been identified (1). However, much less attention has been paid to environmental responses in animal cells, or to the differentiated tissue responses in complex higher organisms and how these tissue responses combine to form the new adaptive phenotype. Responses to environmental stress are most easily identified in species that naturally experience large and potentially debilitating fluctuations in environmental conditions, where they constitute a crucial component of both survival and fitness. Understanding the mechanisms of phenotypic response offers fundamental insights into the nature of environmental adaptation that offer new directions for the experimental manipulation of environmental tolerance.
The common carp, Cyprinus carpio L., belongs to the same Cyprinid family of fish as zebrafish, but originates in a continental climate with extremes of winter and summer. It is an economically important farmed fish, and carp species account for much of world aquaculture production. It is hardy and tolerant of a wide range of temperatures for which it exhibits an extremely plastic thermal phenotype. Responses occur just a few days or weeks after a change in temperature and include acquired tolerance of both extreme cold and heat. They also overcome direct thermal effects on function at normal temperatures leading to conserved visual, brain (2), muscle (3), and intestinal function (4). Regulation of specific candidate genes and proteins has been confirmed in liver (5) and muscle (6). In this study, we use microarray-based expression profiling to identify the transcriptional responses of common carp subjected to a progressive cooling regime.
Methods
Animals and Cold Exposure. Common carp were acclimated for 2 months at 30 ± 0.5°C. For cooling, fish were subjected to a stepped cooling regime of 1°C/h to a maximum of 7°C/day, to either 23°C, 17°C, or 10°C, over 1, 2, or 3 days, respectively, and then maintained at the colder temperature for 22 days. Control 30°C-acclimated animals were subjected to an identical handling regime. At prescribed time points, fish were sampled and RNA was isolated.
Microarray Analysis. The carp microarray was constructed from 13,349 PCR-amplified cDNA clones spotted onto poly-l-lysinecoated glass slides. The arrayed cDNA clones were randomly picked from a collection of high-quality C. carpio cDNA libraries (Table 1, which is published as supporting information on the PNAS web site). Fluorescently labeled cDNA was synthesized and compared with a reference RNA by hybridization to two arrays with reversal of the labeled fluorophores.
Data Normalization and Gene List Extraction. Array normalization and analysis used a statistical error model of fluor-reversed microarray ratios (7) (Figs. 4–6, which are published as supporting information on the PNAS web site). The list of common response genes was extracted by using a response threshold test based on an estimation of the random error contained in the response matrix. Tissue-specific responsive genes were identified by using the Significance Analysis of Microarrays method (8), which compared the expression of genes in the control animals with that of fish cooled to 17°C and 10°C through days 2–12. To estimate the percentage of genes identified by chance, 1,000 permutations of the measurements were tested, and the false discovery rate was adjusted to <1%.
cDNA Sequencing and Annotation. Arrayed clones were selected for 5′ end sequencing both randomly and on the basis that the corresponding mRNA exhibited an interesting expression profile. A total of 9,456 5′ end sequences were assembled into 6,257 groups and annotated on the basis of the results of blastx homology searches (Supporting Materials and Methods, which is published as supporting information on the PNAS web site). For clarity, expression data from redundant cDNAs that aligned in the same sequence contig are presented as averages. To assess the enrichment of a particular classes of genes in a list, the dereplicated gene list for a cluster was divided into 24 Gene Ontology (GO) database categories and the significance of their over- or underrepresentation was estimated by using Fisher's exact test (9) with a multiple testing correction (10).
Additional descriptions of methods used can be found in Supporting Materials and Methods.
Results and Discussion
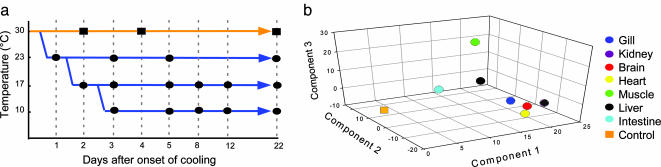
To screen for genes involved in cold acclimation, we undertook a time-course analysis of transcript expression in seven tissues (liver, brain, kidney, heart, skeletal white muscle, gill, and intestinal mucosa) of carp subjected to graded cooling regimes (cooled to 23°C, 17°C, or 10°C), and compared them with the expression profiles of fish maintained throughout at the control 30°C temperature (Fig. 1a). At each time point, five to six replicate fish were sampled and the RNA from the different tissues was isolated. Given the very large number of tissue samples arising from this design (≈630), we elected to pool the RNA from the individual animals taken at each cooling time point. However, to provide a formal estimate of the interindividual variance that exists within a pool, the RNA from the control fish was prepared individually. Changes in mRNA levels were determined by competitive hybridization to carp microarrays consisting of 13,440 cDNAs derived from a collection of carp cDNA libraries that were enriched for environmentally regulated genes.
Fig. 1.
Analysis of cold-induced gene expression. (a) Schematic diagram showing the cooling time course and sampling regime used. Warm-acclimated control fish (30°C) were sampled at three separate time points and compared with fish sampled more frequently over a 3-week time course of cooling to either 23°C, 17°C, or 10°C. (b) PCA of cold-induced tissue-expression profiles showing clear separation of the cooled and warm-acclimated samples. For PCA, the expression profile of each gene was summarized by two centroids, representing the average expression of each cDNA in the cooled fish compared with the warm-acclimated controls (arbitrarily set to 0). PCA used the entire set of cDNAs printed on the carp array and the axes represent the combinations of genes that explain most of the expression changes affected by cooling.
In total, 187 separate RNA samples were hybridized to 374 fluor-reversed microarrays. Principal component analysis (PCA) showed that 87% of the transcriptional changes associated with cooling were explained by the first three PCA components (Fig. 1b). This analysis revealed that each tissue's cooled RNA samples were clearly separable from their respective control, indicating that large and coherent transcriptional changes were induced by the shift in environmental temperature. The first component had a similar trajectory and magnitude in all tissues, suggesting the existence of a common transcriptional response. PCA also identified discrete tissue responses, with component 2 capturing a transcriptional profile shared by the liver and intestinal mucosa, and component 3 defining a response of skeletal muscle. All of these PCA components were statistically significant.
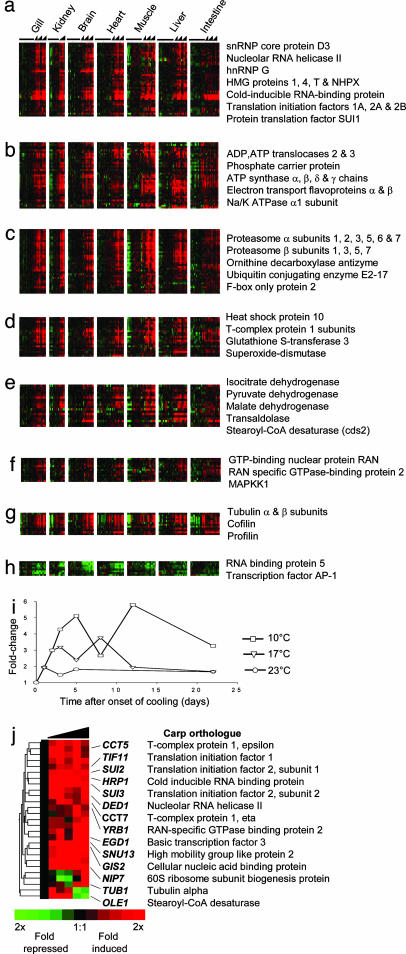
Common Response to Cold. To interpret the nature of the major component of the data that suggested a common response to cold, we identified 260 unique cDNAs that were significantly differentially expressed in all seven tissues, of which 221 had homology to previously described genes (Fig. 2a, and Fig. 7 and Table 2, which are published as supporting information on the PNAS web site). The majority (252) of transcripts increased in expression upon cooling, reflecting a basic paradigm of cold acclimation: that organisms frequently compensate for the rate-depressing effects of cold by synthesizing more enzymes to increase biochemical performance (11). In general, the cold induction of gene expression was a graded function of the thermal perturbation, and was transient. Fig. 2b illustrates this for cold-inducible RNA-binding protein (CIRBP) with peak transcript amounts on days 8 and 12 of the 17°C and 10°C cooling trajectories, respectively. The common response includes a key cold-responsive gene involved in membrane adaptation, the acyl-CoA Δ9-desaturase (5), which was previously thought to be inducible only in liver (12), but which we now discover is elevated in all tissues. To aid the overall functional interpretation of the common response, the induced genes were assigned to seven groups according to their GO annotation for biological process (13) (Fig. 2 a–g). The genes repressed in the common response did not reveal a coherent theme, in part due to the fact that so few repressed genes were identified (eight cDNAs; Fig. 2h).
Fig. 2.
Identification of a common set of cold-regulated genes. cDNAs were grouped according to the GO biological processes in which they participate. (a) Nucleic acid processing (40 genes). (b) Transport (37 genes). (c) Protein catabolism (35 genes). (d) Cell stress or molecular chaperones (21 genes). (e) Metabolism (18 genes). (f) Signaling (13 genes). (g) Cell structure (12 genes). (h) Only eight cDNAs were globally repressed by cold. Representative or notable genes found in each class are indicated. The expression of each cDNA is presented as the ratio of transcript abundance relative to its mean abundance in the control warm-acclimated samples. Each row represents a different cDNA, and each column represents the expression of the corresponding transcript in the different tissues in the control animals (left columns) or after cooling to 23°C, 17°C, and 10°C on the basis of time (right columns). For kidney, only the 17°C cooling trajectory was collected. Red indicates a relative increase in transcript abundance with cooling, and green represents a decrease. (i) mRNA levels in gill of CIRBP, a representative gene in the common response showing that increased cooling resulted in a greater induction of transcript. (j), The cold-induced expression profile of putative yeast orthologues (1) to the induced genes in the carp common response.
The largest GO category of cold-induced transcripts comprised genes involved in transcriptional regulation, RNA splicing, and translation (Fig. 2a). This group included RNA polymerase II activators (BTF3, PC4, SKP1A, and TCBE1), ribonucleoproteins involved in mRNA processing and splicing (SNRPD3, SNRPA1, HNRPG, PRPF8, SF3A2, and SFRS3), and several translation initiation factors (EIF1A, EIF2A, EIF2B, and DENR), and SUI1, a key gene that monitors the fidelity of the initiation complex (14). Cold is believed to affect translation due to increased RNA secondary structure at reduced temperatures. For example, chilled bacteria express cold shock proteins that act as RNA chaperones to reduce RNA secondary structure and rescue translation (15). Indeed, in carp we identified two genes that may alter RNA secondary structure, an RNA helicase, DDX21, and CIRBP (Fig. 2i). CIRBP showed among the largest inductions in all tissues assessed, and, given that it is also cold-responsive in mammals (16, 17), may represent a universal marker of cold exposure in higher organisms. We also detected the strong induction of a number of high-mobility group proteins (HMG1, HMGT1, HMG4, and NHPX) that modulate transcription through the alteration of chromosome conformation, suggesting that chromosomal DNA secondary structure might also be disturbed in the cold. In killifish, the expression of another high-mobility group protein, HMGB1, has also been shown to fluctuate with cycling temperature, providing further evidence that this gene family is involved in temperature responses (18). The identification of genes involved in both RNA processing and chromosomal architecture suggests an important function of cold acclimation is to regulate nucleic acid structures.
Other GO categories contained genes that operate in closely associated biochemical processes or reside in common macro-molecular complexes. For example, a large group of genes comprised those involved in ubiquitin-dependent protein catabolism and proteasomal function, and included 21 proteasome subunits and the ubiquitin-conjugating enzymes UB5A and UBCA (Fig. 2c). This finding was surprising because proteasome activation is often linked to increases in damaged proteins, a situation that is more often associated with elevated temperature rather than cold. Evidence for the cold induction of a general cell-stress response comes from a second GO class that includes genes involved in free radical protection, protein chaperoning, and apoptosis (Fig. 2d; GSTM3, MGST3, SOD2, HSP10, STCH, DAP, and TEGT). Microtubule stability is highly temperature-dependent (19), and a third GO group consists of eight different α- and β-subunits of tubulin (Fig. 2g). Also identified were four genes of the TCP-1 chaperonin complex responsible for tubulin polymerization and chaperoning nonnative proteins (Fig. 2d).
The second largest GO group included genes involved in energy charge and the mitochondrial production of ATP (Fig. 2b). This group included a number of genes that constitute complex V of the oxidative phosphorylation machinery, the α-, β-, γ-, and δ-subunits of the F1 ATP synthase complex as well as components of the nonenzymatic F0 complex (ATP5G2 and ATP5G3). Likewise, we detected the increased expression of two ADP/ATP translocases (SLC25A5 and SLC25A6) and phosphate carrier protein (SLC25A3), which cooperate to supply ADP and Pi to ATP synthase. In addition, 10 transcripts encoding components of the mitochondrial electron transport chain increased, whereas the GO functional group corresponding to metabolism included genes encoding components of the citric acid cycle (IDH2, PDHA1, and MDH1; Fig. 2e). Thus, our expression data suggest that the capacity for ATP synthesis through oxidative phosphorylation increases at reduced temperatures. This finding is consistent with observations that mitochondrial transcripts (20), as well as the number of mitochondria (21), increase in the skeletal muscle of some cooled fish. The data presented here may indicate that this response may extend beyond skeletal muscle to all tissues.
Gasch et al. (1) showed that the yeast Saccharomyces cerevisiae responds to environmental stress with a large transcriptional response. To identify shared elements in the cold responses of both carp and yeast, we identified putative yeast orthologues to the induced genes in the carp common response. Of the 252 carp genes, 107 had significant homology to yeast peptides (blastx <1e-15), and of these genes, 27 (25%) were reported to be induced by >2-fold during the 90 min that followed cooling yeast cultures from 37°C to 25°C (Fig. 2j, and Fig. 8, which is published as supporting information on the PNAS web site). Among the cold-induced yeast genes were orthologues to all three translation initiation factors induced in the carp, three of the TCP1 chaperonins, and OLE1, the orthologue for Δ9-desaturase. Notably, orthologues to the carp proteasomal subunits were absent from the yeast response to cold and instead are reported to be heat-induced (1). Conserved responses between a free-living unicellular organism such as yeast and a higher organism like carp suggest that all poikilothermic organisms may suffer some of the same fundamental cellular problems on cold exposure, and also share the same adaptive mechanisms. By contrast, divergent responses may reflect system-level and tissue-based responses that act to conserve homeostasis in the multicellular carp. Importantly, the identification of yeast homologues allows the functional significance of particular genes in cold adaptation to be directly addressed in a genetically and experimentally manipulable system. For example, the induction of CIRBP was a robust marker of cold exposure in all carp tissues, but its function is poorly understood in higher organisms. HRP1 (YOL123W), the yeast orthologue to CIRBP, is also cold-inducible and is reported to target aberrant mRNAs for decay (22). This finding leads us to speculate that CIRBP may function in the surveillance of mRNAs that are incorrectly processed due the effects of cold on the transcriptional machinery.
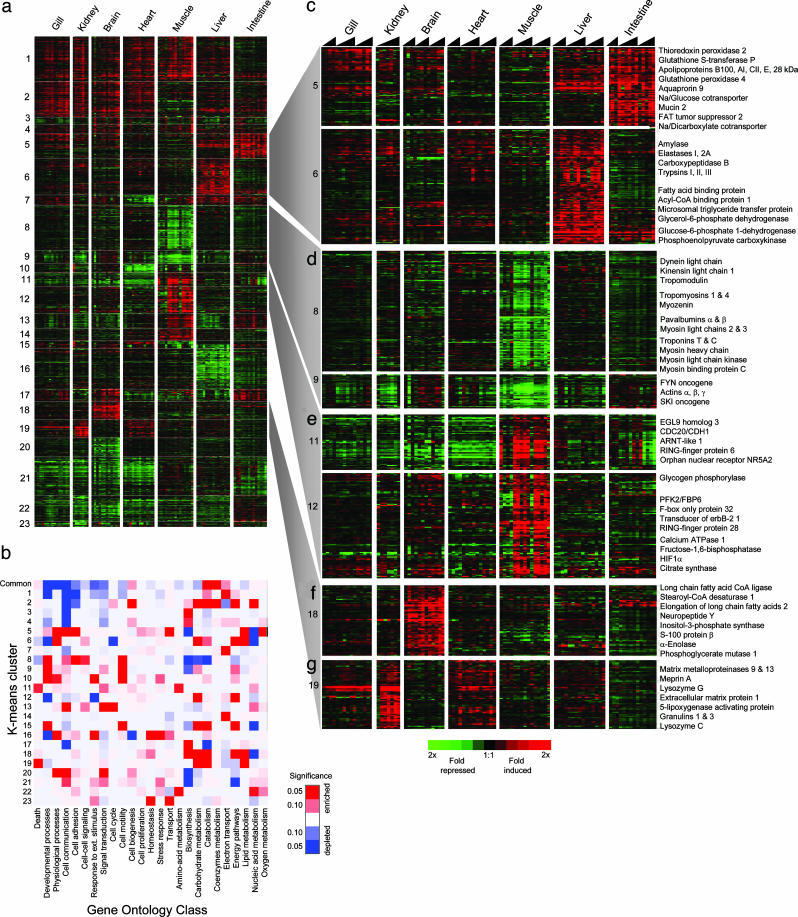
Tissue-Specific Responses to Cold. To explore tissue-specific responses, we used a published signal-to-noise statistic (8) to identify genes that exhibited significant changes in gene expression in at least one tissue of the cooled fish (Table 3, which is published as supporting information on the PNAS web site). After filtering out genes that were present in the common response, we defined an additional 3,201 cDNAs of which 1,728 had homology to previously described genes. These identified genes were grouped into 23 clusters by a K-means clustering algorithm (23) each with a characteristic pattern of tissue expression (Fig. 3a, and Fig. 9, which is published as supporting information on the PNAS web site). To interpret the functional significance of each cluster, we profiled the distribution of genes across 24 categories of GO biological process terms. A Fisher's exact test yielded a heuristic measure of the likelihood that a particular biological process was over- or underrepresented in a cluster compared with that expected by random selection from the entire list of arrayed carp cDNAs (ref. 9 and Table 4, which is published as supporting information on the PNAS web site). The limitations of this approach have been discussed (9). The resulting probability values are presented in Fig. 3b as a heat map, or GO-Matrix chart.
Fig. 3.
Variation in the expression of 1,728 cDNAs in tissues of cooled fish. The expression of each cDNA is presented as the ratio of transcript abundance in each cooled sample relative to its mean abundance in the control warm-acclimated samples. (a) Cluster diagram showing the 23 K-means clusters of cDNAs that exhibited similar expression profiles. (b) GO-Matrix chart, a pseudocolor map of the significant over- or underrepresentation (red or blue, respectively) of genes within the common response and each of the K-means clusters. Saturated colors represent P values of <0.05. Detailed views of selected clusters are as follows: liver and intestinal mucosa-induced cDNAs (c), muscle-repressed transcripts (d), muscle-induced transcripts (e), brain-induced transcripts (f), and cDNAs induced in kidney (g). Representative or notable genes found in these clusters are indicated.
Fig. 3 illustrates the complexity of cold-responsive gene-expression profiles, with the K-means clustering distinguishing tissue-specific responses from those that are shared by more than one tissue, whereas the GO-Matrix chart highlights the most distinctive biological features of each cluster. For example, cluster 5, which comprises genes up-regulated principally in the intestinal mucosa, and, to a lesser extent, liver, was enriched for genes involved in transport and oxygen-free radical metabolism. Closer examination of the cluster revealed that six different apolipoproteins (APOA1, APOA4, APOB, APOC2, APOE, and APO28kDa) and a microsomal triglyceride-transfer protein increased in both tissues, consistent with their shared participation in lipid transport, whereas the expression of solute transporters (SLC5A1, SLC13A2, and AQP9) was restricted to the intestinal mucosa (Figs. 3b and 9). The compensatory up-regulation of both active and passive pathways has long been regarded as a central feature of enterocyte adaptation to cold (24). By exploring gene lists, we have also identified evidence of increased lipid metabolism in liver and brain (clusters 5 and 19, respectively). In liver, genes involved in fatty acid metabolism (FABP1, GPD1, ME1, MTP, and ACBP) and cholesterol synthesis (ACAS, ACAT, LSS, IDI1, and EBRP) increased, whereas the expression signature in the brain suggests increased synthesis of long-chain unsaturated fatty acids (FACL1, SCD1, and ELOVL2). In liver, the adaptive significance of these changes may be to provide the lipid and cholesterol building blocks necessary for the extensive restructuring of membranes reported in cold-exposed organisms (25), or, in brain, to meet the tissue's specific demands for lipid-signaling molecules. Cluster 6 (liver, up-regulated) also included a number of cold-induced digestive enzymes (PRSS1, PRSS2, PRSS3, CPB1, ELA2A, CTRB1, AMY1, and RNASE1; Fig. 3c) consistent with the presence within the liver of pancreatic cells. By contrast, cluster 16 that largely consists of transcripts repressed in liver, and is enriched for genes involved in homeostatic physiological processes such as secreted plasma proteins (Fig. 9).
We identified expression profiles that are indicative of cold-induced structural remodelling of some tissues. For example, the transcriptional response in skeletal muscle in clusters 8 and 9 was unusual in that it was dominated by the coordinated repression of a large group of genes that comprise many structural components of the sarcomere, such as the myosin heavy and light chains, tropomyosins, and actins (Fig. 3c). This conclusion was supported by the GO-Matrix chart, which indicated that genes associated with developmental processes and cell motility were significantly enriched in these clusters. Also repressed were genes involved in muscle contraction, including, for example, calcium-binding proteins such as parvalbumins α and β, and slow-twitch troponins I and T. Of significance was the strong induction in cooled muscle of two ubiquitin ligases, FBX032 and RNF28, which target specific proteins for proteolysis at the proteasome and are reliable markers of skeletal muscle atrophy (26). In addition, expression of the oncogene SKI (Fig. 3c), which plays an important role in the proliferation of myogenic cells (27), and whose overexpression leads to hypertrophy (28), was also repressed by cold. Because skeletal muscles adapt to changes in locomotory activity and load by regulating fiber size and overall muscle mass (29), we speculate that atrophy may be the result of the depressed locomotory activity exhibited by the fish at colder temperatures (A.Y.G., unpublished work). These data suggest that a process of coordinated reductive remodelling through atrophy is induced with prolonged cold exposure in skeletal muscle. This signature was not evident in cardiac muscle, indicating that cold has distinct effects on different types of contractile tissue. The relationship between temperature and skeletal muscle atrophy may have important implications to the aquaculture of carp, both in terms of the quantity and quality of the harvested tissue.
Another prominent cluster of genes that was strongly induced in kidney (Fig. 3f) contained genes associated with the turnover of the extracellular matrix during tissue repair or remodelling. The cluster included granulins (GRN1 and GRN2) that stimulate cell invasion and tumorigenesis (30), as well as matrix metalloproteinases (MMP9, MMP13, and MEP1A) and matrix proteins (DSC1, PRG1, and SGCE). Other genes that were most significantly induced in the kidney were lysozymes C and G (Table 3). Matrix metalloproteinases were also strongly induced in gill tissue, suggesting that the process of cold acclimation of these two major ion-transporting tissues may involve structural remodelling.
The GO-Matrix chart further highlights the transcriptional regulation of both electron transport (clusters 1, 7, and 14) and energetic pathways (clusters 6, 12, 15, and 18) in the cold response. Cluster 1, which describes genes that increased in six of the seven tissues was highly enriched for electron transport genes (P = 0.00007), which, together with the common response, indicates that a central feature of cold acclimation is a compensated or enhanced capacity for oxidative phosphorylation. Another recurring theme was that the expression of genes involved in carbohydrate metabolism was altered in almost all tissues (clusters 2, 5, 12, 15, and 19). Closer examination revealed a complex pattern of response consistent with a reorganization of energy metabolism between tissues. For example, brain, and, to a lesser extent, gill and kidney, showed increased expression of most glycolytic genes, whereas skeletal muscle showed a decrease (Fig. 10, which is published as supporting information on the PNAS web site). In fast-twitch skeletal muscle, glycolysis is responsible for supplying most of the ATP needed for contraction, thus the repression of glycolytic genes in this tissue may be linked to the expression of the genes that comprise the contractile apparatus, which were found to decrease with cooling. In contrast, the transcript profile of liver suggests an activation of the pentose phosphate pathway (Fig. 10) and may supply additional NADPH for elevated lipid metabolism in the cold.
Conclusion
Our analysis demonstrates that cold exposure of a poikilotherm that naturally experiences environmental cooling involves the regulation of very large numbers of genes. Whereas the number of genes expressed by carp is unknown, our identification of 3,461 cold-regulated cDNAs, 1,949 with homology to previously described genes (221 of 260, and 1,728 of 3,201 cDNAs in the common and tissue-specific responses, respectively), suggests that cold has a pervasive effect on the transcriptome. Although additional EST sequencing may yield a revised estimate of the number of unique cDNAs present on the array, the scale of the response in carp is perhaps to be expected, given that for poikilotherms all biological processes are directly affected by changes in environmental temperature. However, some genes were particularly consistent markers of cold exposure in all tissues on the basis of their statistical significance and fold induction. These genes included well described mediators of cold adaptation such as stearoyl-CoA desaturase (5), as well as unexpected genes such as the tubulin and proteasome subunits, high-mobility group proteins, and CIRBP. Identification of these candidate genes provides focused starting points for further investigation of their role in the cold-acclimation process. In addition, we discovered a large number of strongly regulated genes with no known identity (1,473 of 3,461 cDNAs).
The complexity of the transcriptional responses of the different tissues is revealing, indicating a profound tissue specificity that matches their known differentiated roles and contribution to the cold-acclimated phenotype. Although all tissues respond with a common transcriptional response, our data also suggest quite divergent energetic and metabolic strategies among tissues, with brain modulating glycolytic activity, and liver showing a transition to lipid metabolism, whereas muscle reductively remodels its contractile apparatus. These responses, together with the other tissue responses, give rise to a complex adaptive phenotype that not only improves physiological performance in the cold but also promotes thermotolerance of extreme lethal cold.
In combining a broad, system-wide overview with insights into underlying physiological mechanisms, this data set provides a basis to explore a range of specific mechanistic hypotheses at all levels of organization, from individual biochemical pathways to the level of the whole organism. The underlying determinants of thermal plasticity are also interesting in the context of stenothermal species with restricted thermal tolerance because these species may either lack certain classes of genes or are unable to regulate their expression in response to changing temperature. Furthermore, identifying the genes and physiological processes that are subject to acclimation in eurythermal species points directly at environmentally induced lesions that may be subjected to experimental or genetic manipulation to improve cold tolerance.
Supplementary Material
Acknowledgments
We thank G. Govan and M. Hughes for expert technical assistance. This work was supported by the Natural Environment Research Council (Swindon, U.K.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PCA, principal component analysis; GO, Gene Ontology.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CA963982–CA970467 and CF660356–CF663121). The gene expression data have been deposited at the ArrayExpress database in accordance with Microarray Gene Expression Data Society recommendations (accession no. E-MAXD-1).
References
- 1.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunningham, J. R. C. & Hyde, D. (1995) J. Therm. Biol. 20, 223-229. [Google Scholar]
- 3.Watabe, S. (2002) J. Exp. Biol. 205, 2231-2236. [DOI] [PubMed] [Google Scholar]
- 4.Lee, J. A. & Cossins, A. R. (1988) Cell Tissue Res. 251, 451-456. [DOI] [PubMed] [Google Scholar]
- 5.Tiku, P. E., Gracey, A. Y., Macartney, A. I., Beynon, R. J. & Cossins, A. R. (1996) Science 271, 815-818. [DOI] [PubMed] [Google Scholar]
- 6.Hirayama, Y., Kobiyama, A., Ochiai, Y. & Watabe, S. (1998) J. Exp. Biol. 201, 2815-2820. [PubMed] [Google Scholar]
- 7.Fang, Y., Brass, A., Hoyle, D. C., Hayes, A., Bashein, A., Oliver, S. G., Waddington, D. & Rattray, M. (2003) Nucleic Acids Res. 31, e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeeberg, B. R., Feng, W., Wang, G., Wang, M. D., Fojo, A. T., Sunshine, M., Narasimhan, S., Kane, D. W., Reinhold, W. C., Lababidi, S., et al. (March 25, 2003) Genome Biol., 10.1186/gb-2003–4-4-r28. [DOI] [PMC free article] [PubMed]
- 10.Benjamini, Y. & Hochberg, Y. (1995) J. R. Stat. Soc. B 57, 289-300. [Google Scholar]
- 11.Somero, G. N. & Hochachka, P. W. (2002) Bichemical Adaptation: Mechanism and Process in Physiological Evolution (Oxford Univ. Press, Oxford).
- 12.Polley, S. D., Tiku, P. E., Trueman, R. T., Caddick, M. X., Morozov, I. Y. & Cossins, A. R. (2003) Am. J. Physiol. 284, R41-R50. [DOI] [PubMed] [Google Scholar]
- 13.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui, Y., Dinman, J. D., Kinzy, T. G. & Peltz, S. W. (1998) Mol. Cell. Biol. 18, 1506-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, W., Hou, Y. & Inouye, M. (1997) J. Biol. Chem. 272, 196-202. [DOI] [PubMed] [Google Scholar]
- 16.Nishiyama, H., Higashitsuji, H., Yokoi, H., Itoh, K., Danno, S., Matsuda, T. & Fujita, J. (1997) Gene 204, 115-120. [DOI] [PubMed] [Google Scholar]
- 17.Danno, S., Itoh, K., Matsuda, T. & Fujita, J. (2000) Am. J. Pathol. 156, 1685-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Podrabsky, J. E. & Somero, G. N. (2004) J. Exp. Biol. 207, 2237-2254. [DOI] [PubMed] [Google Scholar]
- 19.Wallin, M. & Stromberg, E. (1995) Int. Rev. Cytol. 157, 1-31. [DOI] [PubMed] [Google Scholar]
- 20.Itoi, S., Kinoshita, S., Kikuchi, K. & Watabe, S. (2003) Am. J. Physiol. 284, R153-R163. [DOI] [PubMed] [Google Scholar]
- 21.Egginton, S. & Sidell, B. D. (1989) Am. J. Physiol. 256, R1-R9. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez, C. I., Ruiz-Echevarria, M. J., Vasudevan, S., Henry, M. F. & Peltz, S. W. (2000) Mol. Cell 5, 489-499. [DOI] [PubMed] [Google Scholar]
- 23.Gasch, A. P. & Eisen, M. B. (2002) Genome Biol 3, RESEARCH0059. [DOI] [PMC free article] [PubMed]
- 24.Lee, J. A., James, P. S., Smith, M. W. & Cossins, A. R. (1991) J. Therm. Biol. 16, 7-11. [Google Scholar]
- 25.Hazel, J. R. & Williams, E. E. (1990) Prog. Lipid Res. 29, 167-227. [DOI] [PubMed] [Google Scholar]
- 26.Bodine, S. C., Latres, E., Baumhueter, S., Lai, V. K., Nunez, L., Clarke, B. A., Poueymirou, W. T., Panaro, F. J., Na, E., Dharmarajan, K., et al. (2001) Science 294, 1704-1708. [DOI] [PubMed] [Google Scholar]
- 27.Berk, M., Desai, S. Y., Heyman, H. C. & Colmenares, C. (1997) Genes Dev. 11, 2029-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costelli, P., Carbo, N., Busquets, S., Lopez-Soriano, F. J., Baccino, F. M. & Argiles, J. M. (2003) Cancer Lett. (Shannon, Irel.) 200, 153-160. [DOI] [PubMed] [Google Scholar]
- 29.Jagoe, R. T. & Goldberg, A. L. (2001) Curr. Opin. Clin. Nutr. Metab. Care 4, 183-190. [DOI] [PubMed] [Google Scholar]
- 30.He, Z. & Bateman, A. (2003) J. Mol. Med. 81, 600-612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.