Abstract
The association of historical plague pandemics with Yersinia pestis remains controversial, partly because the evolutionary history of this largely monomorphic bacterium was unknown. The microevolution of Y. pestis was therefore investigated by three different multilocus molecular methods, targeting genomewide synonymous SNPs, variation in number of tandem repeats, and insertion of IS100 insertion elements. Eight populations were recognized by the three methods, and we propose an evolutionary tree for these populations, rooted on Yersinia pseudotuberculosis. The tree invokes microevolution over millennia, during which enzootic pestoides isolates evolved. This initial phase was followed by a binary split 6,500 years ago, which led to populations that are more frequently associated with human disease. These populations do not correspond directly to classical biovars that are based on phenotypic properties. Thus, we recommend that henceforth groupings should be based on molecular signatures. The age of Y. pestis inferred here is compatible with the dates of historical pandemic plague. However, it is premature to infer an association between any modern molecular grouping and a particular pandemic wave that occurred before the 20th century.
Keywords: insertion element, SNP, variable number tandem repeats, pandemic, molecular clock
Plague decimated the human population of Europe and North Africa during two pandemic waves called Justinian's plague (541-767 anno Domini) and the Black Death (1346-19th century). Clinical symptoms during those pandemics resemble those associated with modern plague, whose etiological agent is the Gram-negative bacillus, Yersinia pestis (1). Modern plague achieved global importance after 1894, when Y. pestis was disseminated by marine shipping from Hong Kong during a third pandemic wave.
Y. pestis is often subdivided into three classical biovars. The bacteria from the third pandemic are unable to ferment glycerol and are grouped in biovar Orientalis. Some isolates from Central Asia cannot reduce nitrate and are designated biovar Medievalis, whereas still others from East Asia and Africa, called biovar Antiqua, can ferment glycerol and reduce nitrate (2). Based on a correlation between the current geographical sources of the biovars and the inferred sources of historical plague, Devignat (2) suggested that Antiqua caused Justinian's plague and Medievalis caused the Black Death. Each of the biovars seems to be distinct according to the genomic patterns of IS100 insertion elements, supernumerary DNA islands, or multilocus variable number of tandem repeat analysis (MLVA) (3-8). However, direct evidence uniquely associating any of the biovars with historical plague is lacking. Furthermore, Devignat's correlations between geography and history are based exclusively on the three classical biovars and do not take into account isolates of “atypical” Y. pestis that do not fit into the classical biovars.
One such group of atypical Y. pestis, called pestoides, causes disease in a variety of rodents in Central Asia (9) and, unlike the three classical biovars, can ferment rhamnose and melibiose. Some enzootic Y. pestis isolates from a wide variety of rodents in China also do not readily fit into the classical biovars (7), resulting in a new biovar designation, Microtus, for Y. pestis that do not cause disease in larger mammals and cannot reduce nitrate or ferment arabinose (10). Even the belief that historical plague was caused by Y. pestis has been challenged repeatedly because of a different epidemiology from that of modern plague in India (11-15). A causal association between Y. pestis and historical plague is suggested by the PCR amplification of ancient Y. pestis DNA fragments from skeletons dating between the 13th century and 1722 (16, 17), but independent confirmation of these results has not been possible (18).
An understanding of the evolutionary history and population structure of Y. pestis might help resolve whether historical plague could have been caused by Y. pestis. However, Y. pestis, like other young pathogens (19-22), has evolved too recently to allow the accumulation of extensive sequence diversity. Indeed, no sequence polymorphisms were detected in six gene fragments from 36 isolates from the three classical biovars, indicating that Y. pestis evolved from Yersinia pseudotuberculosis within the last 1,500-20,000 years (3). Deducing the evolutionary history of a species with so little sequence diversity is difficult, especially when markers with high mutation rates are used that may yield inaccurate branch orders caused by homoplasies and irregular molecular clock rates. Such inaccurate branch orders are method-specific and can be recognized by comparing the results from independent methods with different clock rates. We therefore investigated the evolutionary history of Y. pestis by three independent high-resolution methods that have been applied to monomorphic species: synonymous SNPs (sSNPs) defined by genome scanning (22, 23), MLVA (24, 25), and screening for the presence of IS100 at defined locations (4).
Methods
Bacterial Strains. We examined 156 Y. pestis strains isolated from humans, fleas, and small rodents on various continents between 1946 and 1998 (Table 1, which is published as supporting information on the PNAS web site). They included isolates that had been assigned to pestoides (9 isolates) or the biovars Orientalis (94 isolates), Medievalis (27 isolates), Antiqua (25 isolates), or Microtus (1 isolate) by standard tests. Y. pseudotuberculosis isolates of sero-types I (8 isolates), II (2 isolates), III (1 isolate), IV (2 isolates) and V (1 isolate) also were examined.
napA. The entire napA gene was PCR-amplified from Y. pseudotuberculosis strain IP32953 (primers: AGTGCCAAGCTTTCAGGCCACTACCCGTTCCAG and CATCACGGATCCATGAAACTCAGTCGCCGGGACGG), digested with BamHI plus HindIII, ligated into the corresponding multicloning site at 146/207 of expression vector pQE30 (Qiagen, Valencia, CA), and cloned into Escherichia coli SCS1. One resulting recombinant plasmid (pBE696), which contains the expected insert according to sequencing, was used for complementation of the inability to reduce nitrate.
To screen for the napA613 mutation, a 430-bp product was PCR-amplified (primers: GTCAGCACGTAATCTGGATG and GATGGGTTGGCCGTAAGCCA) (annealing temperature: 54°), followed by sequencing of the internal 155-bp product (napA positions 562-716) at 58° from both strands (primers: TTGTATGGCGTCCTCGGTTG and TTCGTAAGTGGAGAGGACGG). The napA613 mutation results in a unique MboII site that also can be used for rapid screening.
MLVA. A total of 43 loci were screened for size variation of fluorescently labeled PCR amplicons, as described (26). Fragments of common sizes were inferred to represent homologous alleles, and the inability to amplify a PCR product was scored as missing data.
IS100 Typing. A total of 31 genomic locations that contain IS100 were identified by BLAST searches of the genome of strain CO92 (27) (molecular group 1.ORI). Eight additional locations where IS100 is integrated into the chromosome of strains IP554 (1.ANT) and IP564 (2.MED) but not that of CO92 were identified by inverse PCR as follows. Chromosomal DNA was ligated after digestion with eight endonucleases lacking target sequences in IS100 (BamHI, ClaI, HindIII, StyI, BfaI, DraI, KpnI, or NcoI). Fragments flanking IS100 were PCR-amplified by using oligonucleotide primers within IS100 (CTACTCATTCCCTGCTTGTGCA and TAGCAGAAGCTAATCCTGAG) and cloned into vector pCR2.1 (Invitrogen) in E. coli INVαF′. PCR amplification using M13 reverse and T7 promoter universal primers identified 125 inserts of unique sizes among 1,375 transformants, whose sequences then were compared to the genome of CO92.
Oligonucleotide primers that flank each of the 39 insertion sites by ≈100 bp were used for PCRs. Sizing of the PCR amplicons by agarose gel electrophoresis indicated whether an IS100 insertion was present (≈2,200 bp) or absent (≈200 bp), and the inability to amplify a PCR product was scored as missing data. Data on 11 locations are presented here (Fig. 5 and Table 2, which are published as supporting information on the PNAS web site); the other locations were excluded because they yielded similar results to the 11 locations or were characterized by high frequencies of missing data or homoplasies. Note that the inability to amplify Y45 in Y. pseudotuberculosis reflects the absence of an IS1541 insertion that contains the target site for that particular IS100 insertion.
Genomic Analyses. Reciprocal-best fasta hits with >40% predicted amino acid identity over >80% of the protein length were used to identify 3,283 potential orthologous coding sequences (CDSs) from pairwise comparisons of the genomes of 91001 (0.PE4) (28), CO92 (1.ORI) (27), and KIM (2.MED) (29). These CDSs were then screened for sSNPs. We excluded sSNPs in 30 CDSs that were within regions of low sequence complexity, within CDSs with multiple paralogs, or where the CDS was lacking in Y. pseudotuberculosis IP32953 (GenBank accession no. NC_006155) according to pairwise blast analyses. Three more putative sSNPs in the CO92 genome and one within the KIM genome were excluded because they reflected sequencing errors, leaving 76 sSNPs in 3,250 orthologous CDS (Tables 3-5, which are published as supporting information on the PNAS web site). Four additional sSNPs and 11 nonsynonymous changes were identified during our screening procedures (Tables 6 and 7, which are published as supporting information on the PNAS web site).
sSNP Screening. PCR products spanning sSNPs were amplified over 25 cycles in 25-μl volumes, containing 5 ng of DNA from each of 1-4 test strains plus a reference strain (CO92, IP520, or 91001), polymerase (1.25 units, Optimase, Transgenomic, Omaha, NE), as well as specific primers (Table 8, which is published as supporting information on the PNAS web site). PCR products were analyzed by using denaturing HPLC with a DNA-SepR Cartridge, (WaveR Nucleic Acid Fragment Analysis System, Transgenomic) at the temperatures indicated in Table 8.
Phylogenetic Methods. Data were stored as numerical character sets in bionumerics 4.0 (Applied Maths, Sint-Martens-Latem, Belgium), which was also used to calculate Hamming distance matrices of the number of shared alleles between isolates. paup*4.0 (30) was used for parsimony analysis, and mega 2.0 (31) was used for neighbor joining.
Results
Pestoides and Microtus Belong to Y. pestis. Because of their ability to ferment melibiose and rhamnose, it was unclear whether pestoides were more closely related to Y. pseudotuberculosis or Y. pestis (32). We therefore sequenced six housekeeping gene fragments from nine pestoides isolates. These fragments are identical among the classical Y. pestis biovars but variable in Y. pseudotuberculosis (3). The pestoides sequences were identical to those from Y. pestis. Similarly, in silico analyses of the genome (28) of biovar Microtus strain 91001 also yielded sequences identical to those from Y. pestis, except for a homopolymeric stretch of seven adenines in manB, which contains only six adenines in other pestis isolates. Thus, despite phenotypic differences, pestoides and Microtus belong to Y. pestis.
Genomic Branch Order and Age. Pairwise comparisons of the three genomic sequences from Y. pestis that are currently available (27-29) revealed 76 conservative sSNPs within 3,250 orthologous CDSs. For each sSNP, the ancestral nucleotide was deduced on the basis that it was identical with the Y. pseudotuberculosis genome. The alternative nucleotides present at those positions in other genomes represent mutations that have arisen by microevolution since descent from Y. pseudotuberculosis. According to this criterion, most of the sSNPs arose along the branches leading to 91001 (Microtus, 27 sSNPs), CO92 (Orientalis, 20 sSNPs), or KIM (Medievalis, 15 sSNPs). However, 14 sSNPs were informative about branch order: all 14 grouped Y. pseudotuberculosis with 91001 and the same mutated nucleotides were found in KIM and CO92 (Fig. 1). These results demonstrate that Y. pestis initially evolved from Y. pseudotuberculosis along one branch, called branch 0, from which 91001 split off, before splitting into branch 1 (CO92) and branch 2 (KIM).
Fig. 1.
Age of Y. pestis. sSNPs were identified by pairwise genome comparisons between 91001 (0.PE4), CO92 (1.ORI), and KIM (2.MED). For each sSNP, one of the alternative nucleotides is present at the corresponding position within the genome of Y. pseudotuberculosis strain IP32953. sSNPs on branch 0 (Table 4) were identical in IP32953 and 91001 and also identical in KIM and CO92, but differed between these pairs. Other sSNPs were unique to the branches, as indicated. To calculate ages, the number of sSNPs was divided by the 777,520 potential sSNPs within the 3,250 homologous gene pairs, and that distance was then divided by the molecular clock rate of 3.4 × 10-9 per year.
We previously calculated (3) the age of Y. pestis as 1,500-20,000 years on the basis of a lack of sequence diversity in the six gene fragments described above. Those age calculations were based on two estimates of mutation clock rates, a short-term rate derived from laboratory experiments with E. coli (33) and a long-term rate based on the divergence time between E. coli and Salmonella enterica Typhimurium (34). Unfortunately, neither clock rate estimate was applicable to the genomic analyses. The short-term rate is inappropriate because it measures all mutations, most of which are rapidly lost because of drift, whereas the sSNPs described here represent fixed nucleotides that were uniform within populations (see below). The long-term rate is appropriate but incorrect, because it ignored the fact that the time since separation of two organisms is only half of the elapsed time during which mutations have accumulated. The correct synonymous mutation rate between E. coli and Typhimurium is the synonymous distance between them (0.94) (35) divided by twice the time since these organisms separated (140 million years) (36), or 3.4 × 10-9 per year. The frequency of sSNPs per potential sSNP divided by that rate then yields the age estimates for Y. pestis that are shown in Fig. 1. We estimate that 13,000 years of evolutionary history separate CO92 and KIM and that the time since 91001 separated from branch 0 is longer (10,000 years) than since CO92 or KIM diverged from their common ancestor (average of 6,500 years).
Molecular Groupings. sSNPs could be useful for epidemiological or forensic purposes as molecular markers for specific populations within Y. pestis. Therefore, 40 sSNPs in 38 gene fragments (total length of 11.2 kb) that marked branches 0, 1, or 2 (Tables 3 and 4) were screened among 105 diverse isolates of Y. pestis by dHPLC (Fig. 6, which is published as supporting information on the PNAS web site). Four additional sSNPs were identified by these procedures (Table 6), for a total of 44. The nucleotides at these 44 positions are identical among Orientalis isolates, except that sSNP s34 is specific to CO92 and s36 is specific for a different Orientalis isolate. However, although most (Medievalis) isolates that cannot reduce nitrate were indistinguishable from KIM (Fig. 6), others were very different.
These and other discrepancies (see below) between classical biovar designations and molecular groupings stimulated us to devise a nomenclature that is based on molecular relatedness but includes mnemonic biovar designations to facilitate the transition. The group of bacteria related to Orientalis is referred to as 1.ORI to reflect the association of the Orientalis phenotype with branch 1 and classical Medievalis isolates are referred to as 2.MED (Figs. 2 and 3). Antiqua isolates split into distinct groups on each of branches 1 and 2, designated 1.ANT and 2.ANT, which were isolated in Africa and East Asia, respectively. Branch 0 includes almost all pestoides isolates (groups 0.PE1, 0.PE2, and 0.PE3) as well as the Microtus isolate, 91001 (0.PE4).
Fig. 2.
Evolutionary branch order within Y. pestis. (a-d) Simplified branch order of the major groups as indicated by sSNPs (a), MLVA (b), and IS100 insertions (c and d), based on data in Figs. 3, 6, and 7. The primary inconsistencies between a and b-d are indicated in orange and purple. The differences in branch order between c and d reflect different interpretation of insertion events (green text). Nodes along branches are indicated by circles, the sizes of which indicate the number of isolates. (e) Consensus evolutionary order of IS100 insertions (Yxx) and synonymous mutations (sxx). The diagram also indicates the inferred order of phenotypic changes (Rha-, Mel-, and Nit-) and nutritional mutations (glpD, napA316), except for the Nit- isolates in 2.ANT, which are not indicated. Sources of isolates according to grouping: 0.PE1, former Soviet Union (4 isolates); 0.PE2, former Soviet Union (3 isolates); 0.PE3, Africa (1 isolate); 0.PE4, China (1 isolate); 1.ANT, Africa (21 isolates); 1.ORI, global (95 isolates); 2.ANT, East Asia (5 isolates); and 2.MED, Kurdistan (26 isolates).
Fig. 3.
Relationships among 104 isolates according to MLVA. A neighbor-joining dendrogram was constructed from Hamming distances based on 43 variable number of tandem repeat loci. Individual isolates are shown except within 1.ORI (58 isolates) and pseudoTB (Y. pseudotuberculosis; 9 isolates), which were collapsed. Numbers within the dendrogram indicate high (>50%) bootstrap values associated with individual nodes. Group assignments according to sSNPs and the ability to reduce nitrate and ferment particular sugars (glycerol, rhamnose, and melibiose) are indicated at the right. For groups with mixed phenotypes, the predominant phenotype is indicated first. Exceptional strains were: 1.ORI Gly+, strain Nich51; 2.MED Gly- Mel+, pestoides J; and 2.ANT.a Nit-, Harbin 35, Nicholisk 41.
A strong discovery bias affects the particular sSNPs that were used for screening because they were defined by a comparison between only three genomes (0.PE4, 1.ORI, and 2.MED). As a result, the current set of sSNPs can indicate the branch order and time of separation for molecular groups from which genome sequences are not (yet) available (0.PE1-0.PE3, 1.ANT, and 2.ANT), but is not particularly informative about their genetic diversity and age (37). Therefore, we screened Y. pestis by an independent approach, MLVA, which should yield neutral estimates of the pairwise genetic distances between all isolates. MLVA of 43 variable number of tandem repeats detected 102 unique patterns among 104 isolates of Y. pestis and Y. pseudotuberculosis. After phylogenetic clustering, the patterns clustered together in molecular groups that were consistent with those found by sSNP analysis (Fig. 3), except that all branch lengths were relatively long. The branch order of a neighbor-joining dendrogram indicated that 2.MED and 2.ANT represent sister clades, as do 0.PE1, 0.PE2, and 0.PE3, consistent with the sSNP data (Fig. 3). However, unlike the three branch structure described above, 1.ANT was more distinct from 1.ORI than are 2.MED/2.ANT, and 0.PE4 did not cluster together with 0.PE1-0.PE3 (Figs. 2b and 3). Similar results were obtained when the MLVA data were analyzed with other clustering algorithms (data not shown).
To resolve differences between discrepant branch orders, we applied still a third molecular grouping method, namely the presence or absence of the IS100 insertion element at 11 distinct genomic locations (Fig. 5 and Fig. 7, which is published as supporting information on the PNAS web site). Except for 0.PE1, 0.PE2, and 0.PE4, which were not distinguished by this method, the same molecular groups were found within 131 isolates as with the other two methods. The IS100 results confirmed the split between branches 1 and 2 (Fig. 2) and revealed minor subdivisions within 1.ANT (1.ANT.a and 1.ANT.b) and 2.ANT (2.ANT.a and 2.ANT.b) that were consistent with the results from MLVA. However, branch 0 was lacking in the most parsimonious interpretation (Fig. 2d) and first reappeared in a less parsimonious interpretation involving one more step (Fig. 2c). According to the latter interpretation, an insertion of IS100 at Y23 predated the separation of all Y. pestis molecular groups but was subsequently lost by excision during the evolution of branch 2. We conclude that the molecular groupings represent major populations and that the patterns of descent within Y. pestis correspond to a three branch structure. Characteristic sSNPs and changes in IS100 patterns are summarized in a consensus tree containing eight populations and six subpopulations that is shown in Fig. 2e.
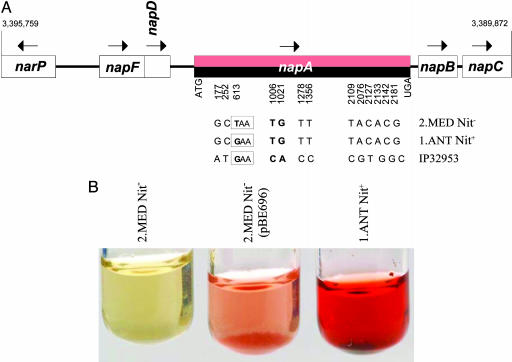
A Signature Mutation in napA. According to the data presented here and by others (8, 10), the inability to reduce nitrate is common to distantly related organisms in 2.MED, 0.PE1, 0.PE4, and 2.ANT (3/5 isolates). We found that the sequence of the entire nap operon is identical between strains IP564 (2.MED), IP554 (1.ANT), and CO92 (1.ORI), except for a premature stop codon in IP564 (Fig. 4A) within the napA gene, which encodes a periplasmic nitrate reductase. This stop codon, which we designated napA613, prevents IP564 from reducing nitrate because nitrate reduction was restored by complementation with an intact napA gene from Y. pseudotuberculosis strain IP32953 (Fig. 4B).
Fig. 4.
The napA613 mutation results in the inability to reduce nitrate. (A) Organization of the nap operon in Y. pestis. The only sequence differences between a 2.MED Nit- strain (IP564) and a 1.ANT Nit+ strain (IP554) within 5.9 kb spanning the nap operon was napA613, a stop codon. The predicted NapA protein from Y. pseudotuberculosis IP32953 differs by two other amino acids encoded by the nucleotides in bold type. (B) Complementation of nitrate reduction. Transformation of plasmid pBE696, containing the napA gene from IP32953, into 2.MED strains IP519 or IP616 (data not shown) restores their ability to reduce nitrate, as indicated by the red color of the growth medium.
The napA613 mutation is a diagnostic marker for 2.MED, and an inability to reduce nitrate by some isolates from other groups has a different genetic basis. For example, 2.ANT.b strain IP546 (Nepal) was originally classified as Medievalis because it is impaired in nitrate reduction. However, IP546 possesses a WT napA sequence and, upon reexamination, we found that IP546 does reduce nitrate weakly on extended cultivation (Fig. 3). In contrast, modern stocks of 1.ANT strain IP566 do not reduce nitrate because of a deletion, acquired in the laboratory, which encompasses the napA gene. IP566 did reduce nitrate originally, as expected for 1.ANT strains, and older DNA preparations yielded a weak napA PCR product. Finally, one 2.MED isolate, pestoides J, has been designated pestoides because it ferments melibiose (but not glycerol). In this study, we found napA613 in 24 2.MED isolates (Table 1), including pestoides J, but not in 98 other strains, including seven from 0.PE1, 0.PE4, or 2.ANT that do not reduce nitrate. Similar results have recently been published by other investigators (8, 10).
Discussion
Populations Versus Biovars. We propose that Y. pestis should be subdivided into populations based on molecular groupings, eight of which are defined here, rather than biovar. The same eight molecular groupings were detected among 156 isolates by three independent methods, except that 0.PE1, 0.PE2, and 0.PE4 were not distinguished by IS100 typing. Assignments to these groupings were unambiguous and consistent for the 60 isolates that were tested by all three methods (Table 1), with only minor exceptions (Supporting Text, which is published as supporting information on the PNAS web site). We infer that these molecular grouping represent distinct bacterial populations. Independent support for the existence of these populations also can be deduced from other molecular analyses, which have examined subsets of the diversity examined here (3-8).
The populations are only partially compatible with the classical phenotypic categories designated as biovars. An inability to reduce nitrate, the hallmark of biovar Medievalis, is found among isolates from groups 2.MED, 2.ANT, 0.PE1, and 0.PE4, probably because of multiple, independent molecular events. Similarly, biovar Antiqua includes unrelated organisms from 1.ANT and 2.ANT that can ferment glycerol and reduce nitrate. Finally, the designation pestoides for organisms that can ferment melibiose and rhamnose combines a variety of diverse organisms from 0.PE1, 0.PE2, and 0.PE3. Thus, biovars are not necessarily monophyletic and should not be used for evolutionary or taxonomic purposes.
Molecular groupings also are not necessarily a reliable indicator of phenotype. One 2.MED isolate (pestoides J) was unable to ferment glycerol and, concordant with other results (4), 1.ORI includes one isolate (Nich51) that can ferment glycerol. Similarly, some 2.ANT isolates can reduce nitrate, whereas others cannot. The multilocus molecular markers that are defined here provide the basis for a common language for classifying the diversity and relatedness among isolates from distinct geographical areas, such as enzootic isolates from the former Soviet Union and China. These isolates manifest extensive phenotypic diversity but their genetic relationships remain unresolved (7, 9). For example, molecular tests could be used to determine whether Central Asian isolates that were previously designated as altaica and hissarica (9) belong to the same population (0.PE4) as Microtus strain 91001 from China (10), with which they share phenotypic properties. Many Central and East Asian isolates probably will fall into the populations described here, whereas others may quite possibly define new groupings.
Detecting Phylogenetic Structure in a Highly Monomorphic Species. Each of the three screening methods used here has distinct advantages and disadvantages for deducing the phylogenetic structure of Y. pestis. MLVA was the most discriminatory but the boundaries of population groupings were somewhat ambiguous. Furthermore, the high mutation rate of variable number of tandem repeat loci resulted in very long branch lengths, with corresponding problems for tree reconstruction. As a result, MLVA did not correctly detect the binary split between branches 1 and 2. We hoped that IS100 analyses would combine adequate discrimination with reliable classification. However, the most parsimonious tree was partially wrong because of hotspots for genomic rearrangements and excision events at the Y23 and Y36 loci (data not shown), and the IS100 analysis also suffered from a higher proportion of missing data (0.04 versus 0.02 for either sSNPs or MLVA). Although it is conceivable that screening additional genomic locations would have resulted in more reliable conclusions, our unpublished data do not support this possibility. Four additional locations that we analyzed in detail were difficult to interpret because of high homoplasy levels and still other locations could not be reliably amplified from numerous isolates (data not shown). Thus, IS100 analyses are probably not ideal for classification and phylogeny of Y. pestis.
Of the three methods, sSNP analyses are the easiest to interpret from an evolutionary viewpoint. No homoplasies were detected, and most branches were supported by multiple, independent sSNPs. However, Y. pestis is so monomorphic that three complete genome sequences of 4.5 MB differed by only 76 conservative sSNPs, most of which were specific for the 1.ORI, 2.MED, and 0.PE4 populations represented by the three genomes. A definitive sSNP-based classification will probably only be possible after at least one genome has been sequenced from each of the other five populations. For the moment, the sSNP-based resolution within branch 0, 1.ANT, and 2.ANT is scanty, and the best current estimates of genetic diversity within these populations are given by the MLVA and IS100 data. As a result, the evolutionary branch order along branch 0 should be considered as a working hypothesis for subsequent investigations.
With time, as additional genomes are sequenced, sSNP analysis may become the method of choice for determining the evolutionary branch structure and molecular groupings within highly uniform species. Genotyping of bacteria might then be efficiently performed by a hierarchical approach (38) in which molecular markers for the branch structure are used to group bacteria into populations before using more variable methods with higher resolution, such as high-throughput SNP typing, whole gene microarrays (6, 7), or MLVA, for subdivision into genotypes. Although multiple nonsynonymous polymorphisms were found here (Table 7), the frequency of nonsynonymous SNPs was only slightly higher than the frequency of SNPs within our pairwise genome comparisons. Similarly, only 14-16 genotypes were detected by whole gene microarrays (6, 7). In contrast, MLVA might be particularly suitable for genotyping within a hierarchical approach because it distinguished 102 patterns among 104 isolates and correlated strongly with geographical source within 1.ORI (Fig. 8, which is published as supporting information on the PNAS web site).
History of Pandemics. We previously suggested that Y. pestis may have evolved in Africa shortly before Justinian's plague of 541 anno Domini (3). Instead, >10,000 years have elapsed since 0.PE4 split from branch 0 (Fig. 1) and Y. pestis probably spread globally long before Justinian's plague, as indicated by the isolation of representatives of branch 0 from the former Soviet Union (0.PE1 and 0.PE2), China (0.PE4), and Africa (0.PE3). Furthermore, it is possible that Y. pestis arose in Asia, where all three branches (0.PE1, 0.PE2, 0.PE4, 1.ORI, and 2.ANT) are found, rather than Africa, from which branch 2 has not been isolated. High diversity is often a good indicator of the geographical source of microbes.
Devignat (2) suggested on the basis of geographical sources and epidemiological observations that each of the three biovars was responsible for an independent pandemic wave. The age estimates presented here confirm that Y. pestis is old enough to have caused historical pandemics of plague. And the epidemiological data supporting an association of pandemic plague since the mid-1890s with Orientalis clearly implicate 1.ORI as the cause of the third pandemic. However, a putative association of older pandemics with unique biovars is not interpretable, especially because biovars Medievalis and Antiqua are polyphyletic, and because Y. pestis contains eight populations, many more than are needed to account for three pandemic waves.
One could attempt to refine Devignat's hypothesis by associating Justinian's plague and the Black Death with specific populations, such as 1.ANT and 2.MED, respectively. The following considerations argue against such a refinement. The frequent current isolation of 1.ANT from Africa does not necessarily indicate that it existed there 1,500 years ago. Even if 1.ANT did exist in Africa at the time, other Y. pestis groupings may have caused Justinian's plague, particularly because 0.PE3 strain Angola was also isolated from Africa. The Black Death did begin in Central Asia, and 2.MED isolates have been collected in “Kurdistan” (Table 1) (corresponding to areas in Iran, Iraq, and Turkey) and China (10). However, Central Asia also includes parts of the former Soviet Union where 0.PE1 and 0.PE2 were isolated. Also, 2.MED is possibly too young to have caused the Black Death, because it is as uniform as 1.ORI, whose lack of diversity probably reflects clonal expansion over only 100 years. Thus, Devignat's hypothesis is no longer convincing, and we can only hope for direct data from ancient DNA (16, 17). The molecular signatures described here might facilitate such studies and were indeed originally designed for that purpose.
The history of plague and the population structure of Y. pestis are difficult to elucidate, because most cases of human disease occurred before the introduction of microbiology, modern disease is most frequent in areas that are remote from centers of molecular biology, and the causative organism is so unusually monomorphic. The results presented here provide a foundation for historical analyses, as well as a precise terminology based on molecular signatures that can be used for future epidemiological investigations. We also have readdressed the association between historical disease and modern isolates while providing technology that can hopefully supply a solid basis for future investigations of that association.
Note Added in Proof: Independent amplification of Y. pestis-specific DNA from Justinian's plague has now been reported (39). Y. pestis-specific DNA from Justinian's plague and the Black Death has been shown to most closely resemble biovar Orientalis (40).
Supplementary Material
Acknowledgments
We gratefully acknowledge technical support by Ying Liu, Matt Van Ert, and Tatum Simonson, extensive discussions with Edward C. Holmes (Oxford University, Oxford), the generous gift of DNA strain 91001 by Ruifu Yang (Institute of Microbiology and Epidemiology, Beijing), and financial support from the Deutsche Forschungsgemeinschaft (Grant Ac 36/9-3) and the National Institutes of Health-National Institute of General Medical Sciences (Grant R01-GM060795).
Author contributions: M.A., G.M., J.P., E.C., and P.K. designed research; G.M., P.Z., I.D., B.K., A.J.V., C.J.A., W.R.E., V.C.-F., L.E.L., and E.C. performed research; P.W., E.C., and P.K. contributed new reagents/analytic tools.; M.A., G.M., T.W., D.M.W., N.R.T., E.C., and P.K. analyzed data; and M.A., D.M.W., and P.K. wrote the paper.
Abbreviations: MLVA, multilocus variable number of tandem repeat analysis; sSNP, synonymous SNP; CDS, coding sequence.
References
- 1.Yersin, A. (1894) Ann. Inst. Pasteur 2, 428-430. [Google Scholar]
- 2.Devignat, R. (1951) Bull. W. H. O. 4, 247-263. [PMC free article] [PubMed] [Google Scholar]
- 3.Achtman, M., Zurth, K., Morelli, G., Torrea, G., Guiyoule, A. & Carniel, E. (1999) Proc. Natl. Acad. Sci. USA 96, 14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motin, V. L., Georgescu, A. M., Elliott, J. M., Hu, P., Worsham, P. L., Ott, L. L., Slezak, T. R., Sokhansanj, B. A., Regala, W. M., Brubaker, R. R., et al. (2002) J. Bacteriol. 184, 1019-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radnedge, L., Agron, P. G., Worsham, P. L. & Andersen, G. L. (2002) Microbiology 148, 1687-1698. [DOI] [PubMed] [Google Scholar]
- 6.Hinchliffe, S. J., Isherwood, K. E., Stabler, R. A., Prentice, M. B., Rakin, A., Nichols, R. A., Oyston, P. C., Hinds, J., Titball, R. W. & Wren, B. W. (2003) Genome Res. 13, 2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou, D., Han, Y., Song, Y., Tong, Z., Wang, J., Guo, Z., Pei, D., Pang, X., Zhai, J., Li, M., et al. (2004) J. Bacteriol. 186, 5138-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pourcel, C., Andre-Mazeaud, F., Neubauer, H., Ramisse, F. & Vergnaud, G. (2004) BMC Microbiol. 4, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anisimov, A. P., Lindler, L. E. & Pier, G. B. (2004) Clin. Microbiol. Rev. 17, 434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou, D., Tong, Z., Song, Y., Han, Y., Pei, D., Pang, X., Zhai, J., Li, M., Cui, B., Qi, Z., et al. (2004) J. Bacteriol. 186, 5147-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch, A. (1881) in Handbuch der Historisch-Geographischen Pathologie (Verlag von Ferdinand Enke, Stuttgart), Vol. I, pp. 349-384. [Google Scholar]
- 12.Karlsson, G. (1996) J. Med. Hist. 22, 263-284. [Google Scholar]
- 13.Cohn, S. K., Jr. (2002) The Black Death Transformed: Disease and Culture in Early Renaissance Europe (Arnold, London).
- 14.Wood, J. W., Ferrell, R. J. & Dewitte-Avina, S. N. (2003) Hum. Biol. 75, 427-448. [DOI] [PubMed] [Google Scholar]
- 15.Twigg, G. (2003) Local Popul. Stud. 71, 40-52. [PubMed] [Google Scholar]
- 16.Drancourt, M., Aboudharam, G., Signoli, M., Dutour, O. & Raoult, D. (1998) Proc. Natl. Acad. Sci. USA 95, 12637-12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raoult, D., Aboudharam, G., Crubezy, E., Larrouy, G., Ludes, B. & Drancourt, M. (2000) Proc. Natl. Acad. Sci. USA 97, 12800-12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert, M. T., Cuccui, J., White, W., Lynnerup, N., Titball, R. W., Cooper, A. & Prentice, M. B. (2004) Microbiology 150, 341-354. [DOI] [PubMed] [Google Scholar]
- 19.Sreevatsan, S., Pan, X., Stockbauer, K., Connell, N. D., Kreiswirth, B. N., Whittam, T. S. & Musser, J. M. (1997) Proc. Natl. Acad. Sci. USA 94, 9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keim, P., Klevytska, A. M., Price, L. B., Schupp, J. M., Zinser, G., Smith, K. L., Hugh-Jones, M. E., Okinaka, R., Hill, K. K. & Jackson, P. J. (1999) J. Appl. Microbiol. 87, 215-217. [DOI] [PubMed] [Google Scholar]
- 21.Kidgell, C., Reichard, U., Wain, J., Linz, B., Torpdahl, M., Dougan, G. & Achtman, M. (2002) Infect. Genet. Evol. 2, 39-45. [DOI] [PubMed] [Google Scholar]
- 22.Joy, D. A., Feng, X., Mu, J., Furuya, T., Chotivanich, K., Krettli, A. U., Ho, M., Wang, A., White, N. J., Suh, E., et al. (2003) Science 300, 318-321. [DOI] [PubMed] [Google Scholar]
- 23.Gutacker, M. M., Smoot, J. C., Migliaccio, C. A., Ricklefs, S. M., Hua, S., Cousins, D. V., Graviss, E. A., Shashkina, E., Kreiswirth, B. N. & Musser, J. M. (2002) Genetics 162, 1533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keim, P., Price, L. B., Klevytska, A. M., Smith, K. L., Schupp, J. M., Okinaka, R., Jackson, P. J. & Hugh-Jones, M. E. (2000) J. Bacteriol. 182, 2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klevytska, A. M., Price, L. B., Schupp, J. M., Worsham, P. L., Wong, J. & Keim, P. (2001) J. Clin. Microbiol. 39, 3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girard, J. M., Wagner, D. M., Vogler, A. J., Keys, C., Allender, C. J., Drickamer, L. C. & Keim, P. (2004) Proc. Natl. Acad. Sci. USA 101, 8408-8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T., Prentice, M. B., Sebaihia, M., James, K. D., Churcher, C., Mungall, K. L., et al. (2001) Nature 413, 523-527. [DOI] [PubMed] [Google Scholar]
- 28.Song, Y., Tong, Z., Wang, J., Wang, L., Guo, Z., Han, Y., Zhang, J., Pei, D., Zhou, D., Qin, H., et al. (2004) DNA Res. 11, 179-197. [DOI] [PubMed] [Google Scholar]
- 29.Deng, W., Burland, V., Plunkett, G., III, Boutin, A., Mayhew, G. F., Liss, P., Perna, N. T., Rose, D. J., Mau, B., Zhou, S., et al. (2002) J. Bacteriol. 184, 4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swofford, D. L. (1998) paup*: Phylogenetic Analysis Using Parsimony and Other Methods (Sinauer, Sunderland, MA), version 4.0 beta.
- 31.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 32.Englesberg, E. (1957) J. Bacteriol. 73, 641-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guttman, D. S. & Dykhuizen, D. E. (1994) Science 266, 1380-1383. [DOI] [PubMed] [Google Scholar]
- 34.Whittam, T. S. (1996) in Escherichia coli and Salmonella, eds. Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), Vol. 2, pp. 2708-2720. [Google Scholar]
- 35.Sharp, P. M. (1991) J. Mol. Evol. 33, 23-33. [DOI] [PubMed] [Google Scholar]
- 36.Ochman, H. & Wilson, A. C. (1987) J. Mol. Evol. 26, 74-86. [DOI] [PubMed] [Google Scholar]
- 37.Pearson, T., Busch, J. D., Ravel, J., Read, T. D., Roton, S. D., U'Ren, J. M., Simonson, T. S., Kachur, S. M., Leadem, R. R., Cardon, M. L., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 13536-13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keim, P., Van Ert, M. N., Pearson, T., Vogler, A. J., Huynh, L. Y. & Wagner, D. M. (2004) Infect. Genet. Evol. 4, 205-213. [DOI] [PubMed] [Google Scholar]
- 39.Wiechmann, I. & Grupe, G. (2004) Am. J. Phys. Anthropol. 126, 48-55. [DOI] [PubMed] [Google Scholar]
- 40.Drancourt, M., Roux, V., Dang, L. V., Tran-Hung, L., Castex, D., Chenal-Francisque, V., Ogata, H., Fournier, P.-E., Crubézy, E. & Raoult, D. (2004) Emerging Inf. Dis. 10, 1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.