Abstract
Background
A range of resistance loci against different races of Xanthomonas oryzae pv. oryzae (Xoo), the pathogen causing bacterial blight (BB) disease of rice, have been discovered and characterized. Several have been deployed in modern varieties, however, due to rapid evolution of Xoo, a number have already become ineffective. The continuous “arms race” between Xoo and rice makes it imperative to discover new resistance loci to enable durable deployment of multiple resistance genes in modern breeding lines. Rice diversity panels can be exploited as reservoirs of useful genetic variation for bacterial blight (BB) resistance. This study was conducted to identify loci associated to BB resistance, new genetic donors and useful molecular markers for marker-assisted breeding.
Results
A genome-wide association study (GWAS) of BB resistance using a diverse panel of 285 rice accessions was performed to identify loci that are associated with resistance to nine Xoo strains from the Philippines, representative of eight global races. Single nucleotide polymorphisms (SNPs) associated with differential resistance were identified in the diverse panel and a subset of 198 indica accessions. Strong associations were found for novel SNPs linked with known bacterial blight resistance Xa genes, from which high utility markers for tracking and selection of resistance genes in breeding programs were designed. Furthermore, significant associations of SNPs in chromosomes 6, 9, 11, and 12 did not overlap with known resistance loci and hence might prove to be novel sources of resistance. Detailed analysis revealed haplotypes that correlated with resistance and analysis of putative resistance alleles identified resistant genotypes as potential donors of new resistance genes.
Conclusions
The results of the GWAS validated known genes underlying resistance and identified novel loci that provide useful targets for further investigation. SNP markers and genetic donors identified in this study will help plant breeders in improving and diversifying resistance to BB.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-017-0147-4) contains supplementary material, which is available to authorized users.
Keywords: Genome-wide association, Oryza sativa, Xanthomonas oryzae, bacterial blight (BB), disease resistance, genotyping-by-sequencing (GBS), SNPs
Background
Bacterial blight (BB) disease, caused by Xanthomonas oryzae pv. oryzae (Xoo), is one of the most widespread and economically important diseases of rice. In Asia, continuous rice cultivation has resulted in highly virulent Xoo races that cause yield reductions of up to 50% in certain environments (Mew et al. 1993). Diversity of Xoo was characterized across Asia and revealed five distinct clusters composed of seven pathotypes (Adhikari et al. 1995). Pathotype 1 was widespread in Malaysia, Philippines and Korea. X oo populations in Indonesia mainly belonged to pathotype 3, while pathotypes 4 and 6 were common in Nepal, India, and the Philippines (Adhikari et al. 1995). A more recent comparative genomic study on the 10 major Xoo races from the Phillipines clustered them in three major lineages, PX-A (Races 1, 3c, 4, 9b and10), PX-B (Races 2, 5, 7 and 8) and PX-C (Race 6, strain PXO99) (Quibod et al. 2016). Though these were distinct from African lineages, they were suggested to be conserved across Asia. Historical data furthermore revealed two major shifts in the distributions of Xoo races in the Philippines. While race 1 was predominant in early 1970s, it was replaced by race 2 in the 1980s-1990s. Since 1992 race 9b has expanded and become the predominant race in the 2010s (Quibod et al. 2016).
As distributions are changing and new virulent Xoo races are emerging, host plant resistance remains a key component of BB disease management (Adhikari et al. 1998). Considerable efforts have been mounted towards breeding for resistance against Xoo. Genetic mapping studies for BB resistance have led to the identification of 40 Xa resistance loci (Nino-Liu et al. 2006; Verdier et al. 2011; Bhasin et al. 2012; Zhang et al. 2015; Kim et al. 2015), most of which were directly derived from Oryza sativa accessions. Nine genes have been cloned, namely Xa1, Xa3/Xa26, xa5, Xa10, xa13, Xa21, Xa23, xa25, and Xa27 (Nino-Liu et al. 2006; Liu et al. 2011; Tian et al. 2014; Wang et al. 2014b).
Five BB resistance loci (xa5, Xa7, xa13, xa24 and xa34) originated from aus genotypes (Sidhu et al. 1978; Ogawa et al. 1987; Khush and Angeles 1999; Chen et al. 2011). Among these loci, the recessive xa5, encoding the gamma subunit of transcription factor IIA, is the only gene to be linked with resistance to Race 9 and furthermore confers broad-spectrum resistance to Philippine races 1, 2, 3, 5, 7, 8, and 10 (Iyer and McCouch 2004; Nino-Liu et al. 2006). Xa7 is a dominantly inherited TAL effector R gene that recognizes TAL effector proteins (Römer et al. 2009) and confers to race 1 (PXO61), race 2 (PXO86), and race 3 (PXO79) (Sidhu et al. 1978). Resistance to race 6 is recessively conferred by xa13, a nodulin MtN3 gene (Ogawa et al. 1987; Chu et al. 2006a, b) and xa24 (Khush and Angeles 1999).
Ten BB resistance loci (Xa2, Xa4, Xa11, Xa14, Xa16, Xa18, xa25, Xa26, xa28 and Xa39) originated from indica donors (Verdier et al. 2011; Zhang et al. 2015). Xa4 is a dominant resistance locus originally identified from TKM6 (Ogawa et al. 1986). It has been widely used in rice breeding programs (Mew 1987) that led to the emergence of new virulent race 2 and race 3 strains (Mew et al. 1992). Despite its breakdown, Xa4 acts as a recessive QTL, exhibiting epistatic or additive effects when combined with other resistance genes (Li et al. 2001). Xa14 is a dominant resistance locus against race 5 which was found in Taichung Native 1 (TN1) by Taura et al. (1987). The recessive xa25 from Minghui 63 (Chen et al. 2002), encodes a nodulin MtN3 family protein essential for reproductive development and rice-Xoo interaction (Yuan and Wang 2013), and confers a high level of resistance to race 9a (PXO339) (Liu et al. 2011).
Nine dominant Xa loci were derived from wild relatives of rice (Xa21, Xa23, Xa27, Xa29, Xa30, Xa32, Xa35, Xa38 and Xa40) (Song et al. 1995; Zhang et al. 2001; Gu et al. 2004; Tan et al. 2004; Cheema et al. 2008; Zheng et al. 2009; Guo et al. 2010; Bhasin et al. 2012; Kim et al. 2015). Notably Xa21 encoding a receptor kinase-like protein involved in the recognition of pathogen effectors and the activation of defense response (Song et al. 1995) was the first cloned R gene for BB resistance. Originating from Oryza longistaminata, it has been widely introduced to popular varieties (Verdier et al. 2011), conferring broad-spectrum resistance to Philippine Xoo races (Ikeda et al. 1990; Khush, et al. 1990).
The continuous mono-cropping and deployment of rice cultivars with a narrow genetic base has provided high selection pressure for emerging virulent strains. Cultivars with single major genes for resistance are prone to resistance breakdown due to pathogen variation and evolution. Thus, pyramiding of resistance genes into individual breeding lines is a strategy to provide more durable resistance (Gnanamanickam et al. 1999; McDowell and Woffenden 2003). Though pyramiding of known loci is a promising approach for disease management, novel sources of resistance are required to keep the upper hand in the continuous plant-pathogen “arms race”. Thus, the search for new genes is crucial to reinforce host resistance breeding. Rice has a wide range of genetic diversity and global germplasm collections, serving a rich source of novel resistance loci. Useful genes for biotic stress resistance were classically identified through biparental mapping populations using landraces and wild ancestors as donors (Ikeda et al. 1990).
More recently, the advent of high-density molecular marker platforms in conjunction with carefully assorted diversity panels has enabled the exploration of agronomic traits through GWAS and allele mining. GWAS facilitates the discovery of novel alleles and allele combinations that are useful in crop improvement (Zhu et al. 2008; Tung et al. 2010). Association studies on sheath blight (Jia et al. 2012) and blast (Wang et al. 2014a, b) disease resistance in rice to identify resistance loci and novel functional candidate genes exemplifies the power of GWAS. With advances in genomics tools, this study aims to identify resistance loci associated with BB resistance and useful SNP markers to track these loci, and determine new genetics donors through GWAS.
Results
Genetic Diversity And Population Structure
Based on genetic diversity, 285 accessions were selected as initial core panel from an original collection of 380 genotypes (Additional file 1: Table S1). The panel comprised 208 breeding lines and varieties, and 77 landraces. Phylogenetic analysis featured five distinct clusters, namely three clusters of indica, one cluster for aus, and a cluster containing japonica and aromatic rice varieties (Additional file 2: Figure S1a). Mean genetic distance within the three indica clusters was 0.16 while the aus and japonica/aromatic clusters displayed 0.24 and 0.09 mean genetic distance, respectively. Population differentiation (Fst) of indica genotypes from aus genotypes is 0.13 while 0.20 from japonica/aromatic. Overall, differentiation based on global unbiased Fst of 0.10 across the five clusters was determined. Principal component analysis separated the 285 diverse panel into five subgroups. The indica subset (198 genotypes) was differentiated by PC = 3 (Additional file 2: Figure S1d).
BB Resistance Screening
Nine Philippine strains (PXO) representing eight different races of Xanthomonas oryzae pv. oryzae (Table 1) were used to screen the 285 genotypes and a wide range of disease resistance was observed (Fig. 1; Additional file 1: Table S1). A set of 17 near isogenic lines (NILs) in IR24 background, having single or two to four pyramided Xa genes (Huang et al. 1997) were included in the panel to serve as controls of known disease reactions. Results of the BB resistance screening mostly conformed to the expected reactions of the NILs. Out of the 285 genotypes, IRBB59, IRBB60, IRBB66, CBB23, IR09F154, IR08N134, and IR72890-81-3-2-2 exhibited resistance to all nine Xoo races. Several genotypes exhibited resistance against the most virulent Xoo race, Race 6 (PXO99), namely IRBB59, IR09F154, IRBB60, IRBB23, CBB23, IRBB66, IR08N134, IR 72890-81-3-2-2, IR08N104, IR09N495, IR06A147, and PSB Rc88 (Additional file 1: Table S1). Some of these resistant genotypes are known to possess xa13 and Xa21 or combination of three to four Xa genes, conferring resistance to PXO99 (Huang et al. 1997). CBB23 and IRBB23 possess the Xa23 gene that confers broad resistance at all developmental stages of rice and high resistance to PXO99 (Table 1) (Zhang et al. 2001).
Table 1.
Nine strains of Xanthomonas oryzae pv. oryzae used in this study
| Race | Straina | Genes Conferring Resistanceb |
|---|---|---|
| 1 | PXO61 | Xa4,xa5,Xa7,Xa21,Xa22,Xa23,Xa26,Xa32 |
| 2 | PXO86 | xa5,Xa7,Xa10,Xa21,Xa23,Xa27,xa28 |
| 3b | PXO79 | xa5,Xa7,Xa21,Xa23 |
| 4 | PXO71 | Xa4,Xa21,Xa23,xa24,Xa32 |
| 5 | PXO112 | Xa4,xa5,Xa7,xa8,Xa10,Xa14,Xa21,Xa23,Xa27,xa28,Xa31,Xa32,Xa35 |
| 6 | PXO99 | xa13,Xa21,Xa23,xa24,Xa30,Xa32 |
| 9a | PXO339 | xa5,Xa21,Xa23,xa25,Xa32,Xa35 |
| 9b | PXO349 | xa5,Xa21,Xa23,Xa25 |
| 10 | PXO341 | Xa4,xa5,Xa7,Xa21,Xa23 |
Fig. 1.
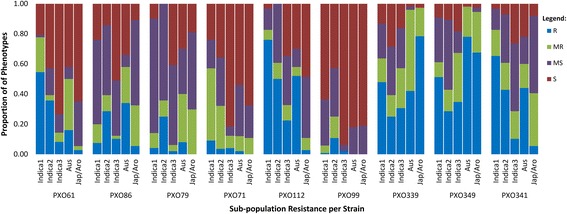
Sub-population resistance of the 285 diverse germplasm to nine representative strains of Xanthomonas oryzae pv. oryzae
Rice varieties belonging to the aus group also displayed high levels of resistance against Xoo. Aus 299, Dular, Aus Bak Tulsi, Ase Pulu Jawa, Aus 298, Makalioka 34, Kalimekri 77-5, DV 86, Chengri, Aus 307, Khaiyan, and DV85 are aus genotypes that were found to consistently exhibit moderate to high resistance across eight Xoo races. Notably most of the aus genotypes (96%) in this study exhibited resistance to both strains of race 9 (PXO339 and PXO349) (Fig. 1; Additional file 1: Table S1).
Genome-wide Association Analysis
The 285 genotypes in the diverse panel exhibited a distinct population structure (Additional file 2: Figure S1) which can cause false positives if not corrected (Zhu et al. 2008). Initial results using unified Mixed Model that incorporate principal component Analysis (PCA) showed ambiguous peaks in the Manhattan plots that could have been an effect of residual population structure not fully corrected by Q + K (data not shown). Thus, 37 japonica/aromatic genotypes were excluded. A total of 248 samples composed of indica and aus genotypes (henceforth “indica/aus panel”) were retained for subsequent genome wide analysis that was performed using 40,840 SNPs and phenotyping information for nine Xoo strains. Furthermore, separate analyses for 198 indica genotypes (henceforth “indica subset”) based on 40,396 SNPs were conducted. Manhattan plots for the two separate analyses showed similar trends between the indica/aus panel and the indica subset except for PXO86, PXO99 and PXO341 (Additional file 3: Figure S2 and Additional file 4: Figure S3). Peaks exclusively detected in the indica/aus panel were noticeable in the centromere region of chromosome 10 for PXO86 (Additional file 3: Figure S2b and Additional file 4: Figure S3b), and short arm of chromosome 5 and long arm of chromosome 12 for PXO341 (Additional file 3: Figure S2i and Additional file 4: Figure S3i). However, more distinct peaks for PXO99 were observed in the analysis of the indica subset and the detection of the xa13 region in chromosome 8 (Additional file 3: Figure S2f and Additional file 4: Figure S3f).
Significant Hits Overlapping Known Resistance Genes
GWAS identified SNPs that were consistently associated with resistance against several strains. Known genes and loci (Xa) conferring resistance to specific Xoo races (Table 1) were overlaid with the Manhattan plots (Additional file 3: Figure S2 and Additional file 4: Figure S3). Among the known Xa loci, the regions of Xa4, xa5, Xa7, xa13, Xa14, Xa21, and xa25 (Wang et al. 2001; Blair et al. 2003; Chen et al. 2008; Chu et al. 2006a, b; Bao et al. 2010; Song et al. 1995; Chen et al. 2002) were identified as overlapping with significant SNPs in this study (Additional file 3: Figure S2, Additional file 4: Figure S3 and Additional file 5: Table S2).
Significant SNP loci exclusively associated with resistance to PXO112 in the indica panel (Additional file 5: Table S2) clustered in the flanking region of Xa14 (Bao et al. 2010) that ranges from 30,063,575 bp-32,807,203 bp in the long arm of chromosome 4 (Additional file 4: Figure S3e). On the other hand, SNP loci located in the short arm of chromosome 5 namely, S5_440644 and S5_453169 were highly associated with resistance to all seven strains of Xoo (Additional file 3: Figure S2, Additional file 4: Figure S3 and Additional file 5: Table S2). S5_453169 was identified by MLMM (p-value of 5.95−17) to be a cofactor for resistance against PXO61 and PXO349. S5_440644 overlaps with a transcription initiation factor IIA gamma chain (LOC_Os05g01710), a similar gene that encodes the recessive xa5 gene (Fig. 2).
Fig. 2.

Highly associated SNPs with resistance to Xoo Race 3b (PXO79) and overlapping the xa5 gene. a Manhattan plot based on Efficient Mixed-Model Association eXpedited Model (EMMAX) where x-axis shows the SNPs along each chromosome and y-axis is the − log10 (P-value) for the association. Significant SNPs are those beyond the red line having P-value < 1 × 10−5. b LD (r2) and haplotype blocks based Expectation Maximization (EM) algorithm. c Significant SNP S5_440644 overlapping the region of LOC_Os05g01710, a transcription initiation factor IIA gamma chain
On the long arm of chromosome 6 a cluster of SNPs was associated with resistance against PXO79 (Additional file 5: Table S2). Among them S6_28100960 accounted for 11% of the phenotypic variation and was approximately 68 kb away from the ID7 marker for Xa7 (Zhang et al. 2009a). Furthermore, S6_28100960 was a cofactor for resistance to PXO79 (race 3b) in MLMM, confirming previous reports that Xa7 confers differential resistance against race 1, 2, 3, and 5 during flowering and later growth stages (Lee et al. 2003). A congruent peak for PXO61 and PXO86 was just below the significance threshold (Additional file 3: Figure S2 and Additional file 4: Figure S3).
For PXO99 (race 6), pronounced differences in the Manhattan plots were found between association analyses for the indica/aus panel and the indica subset. Significant SNPs clustering the region of the recessive resistance gene xa13 were detected exclusively in the indica subset (Additional file 4: Figure S3f and Additional file 5: Table S2). Two distinct peaks were observed flanking the actual physical position of xa13 (26,725,898-26,728,795 bp) in the long arm of chromosome 8 (Chu et al. 2006a, b). Both peaks (25,954,303 -26,044,528 bp and 27,223,555-27,663,433 bp) were both approximately 500 kb away from xa13 (DQ421394). Despite xa13 having originated from an aus variety BJ1 (Ogawa et al. 1987), association analysis of PXO99 resistance in the indica/aus panel (50 of which are aus) revealed no significant associations at the long arm of chromosome 8 (Additional file 3: Figure S2f). Instead, distinct peaks in the short arm of chromosome 5 (xa5) and in the long arm of chromosome 11 (Xa21) (Additional file 3: Figure S2f) were associated with resistance to PXO99. It was pointed out by Li et al. (2001) that xa13 strongly interacts with other Xa genes such as xa5, Xa4 and Xa21. Besides, other genes masking xa13, allele frequency and effects of population structure not accounted for by the Q and K matrices may have affected associations in the indica/aus panel.
Among the twelve chromosomes of rice, chromosome 11 is well known as a hotspot of BB resistance genes (Khush and Brar 2001; Khush and Virk 2005; Ghazi et al. 2009). In the indica/aus panel S11_21190115 and S11_21246561, proximal to Xa21 (21,273,533 -21,277,443 bp) (Ronald et al. 1992; Song et al. 1995), were highly associated with resistance to PXO61, PXO86, PXO79, PXO112, PXO99, and PXO349 (Additional file 5: Table S2). Furthermore, S11_21190115 SNP was identified as a cofactor for PXO99 resistance in the indica subset. Six significant SNPs (S11_27007623 to S11_27831021) were flanking the region of the RM224 marker (27,673,250-27,673,372 bp) linked to the Xa4 region (Wang et al. 2001; Sun et al. 2003) (Fig. 3 and Additional file 5: Table S2). Among them, S11_27603799 accounted for 29% of phenotypic variance and was a cofactor for resistance to PXO61, PXO71, PXO112 and PXO341 based on MLMM (p-value of 4.66−25). The three most downstream SNPs (S11_27672709, S11_27677853 and S11_27831021) overlapped with the recently fine-mapped Xa4 region containing NBS-LRR and WRKY gene family members (Hur et al. 2016). The resistance reactions associated with these SNPs including 2 additional upstream SNPs (S11_27007623 and S11_27007628) agreed with earlier reports for Xa4 (Nino-Liu et al. 2006). Association analyses of resistance to PXO339 (race 9a) and PXO349 (race 9b) showed similar patterns in the Manhattan plots (Additional file 3: Figure S2 and Additional file 4: Figure S3). A wide peak covering 13 Mb to 17 Mb in the centromeric region of chromosome 12 was observed in the association with resistance to PXO339 and PXO349 (Additional file 3: Figure S2 and Additional file 4: Figure S3). Two significant SNPs in this region, S12_17305618 and S12_17305619, accounted for 15% of phenotypic variance (Additional file 5: Table S2) and overlapped with xa25 (LOC_Os12g29220), a gene mapped in the centromeric region of chromosome 12, which confers resistance against PXO339 (Chen et al. 2002).
Fig. 3.

Highly associated SNPs with resistance to Xoo Race 4 (PXO71) and flanking Xa4 gene. a Manhattan plot based on Efficient Mixed-Model Association eXpedited Model (EMMAX) where x-axis shows the SNPs along each chromosome and y-axis is the − log10 (P-value) for the association. Significant SNPs are those beyond the red line having P-value < 1 × 10−5. b LD matrix (r2) and haplotype blocks based on Expectation Maximization (EM) algorithm. c Significant SNPs (S11_27603791 and S11_27603799) overlapping the exon region of LOC_Os11g45620 (rust-resistance protein Lr21), flanking the Xa4 locus
Significant Hits Overlapping Putative Novel Resistance Loci
At a 0.05 Bonferroni-corrected cut-off, and taking local LD into account, 8 peaks, consisting of a total of 33 SNPs on chromosomes 6, 9, 11 and 12 (Additional file 6: Table S3 and Additional file 7: Figure S4), were discovered that did not overlap with previously characterized Xa loci (Additional file 3: Figure S2 and Additional file 4: Figure S3). In most cases the minor allele could be associated with resistance (Additional file 6: Table S3), though in some cases the R allele was ambiguous due to high heterozygosity of the SNP.
Notably four out of the 8 peaks, qPXO79_6-1 to -4, were specific for resistance against race 3B (PXO79) and clustered between 21.5 Mb and 23.1 Mb on the long arm of chromosome 6. Interestingly qPXO79_6-1 was specific for the indica panel, while qPXO79_6-2 to -4 were specific for the indica/aus. For all 18 SNPs it was the aus-enriched allele that was associated with resistance. While the R alleles for qPXO79_6-4 were minor in the indica/aus panel with frequencies of 10–21%, they were highly enriched in the aus sub-pool of the indica/aus panel with frequencies of 34–82%. Congruently the corresponding R alleles were only found at 5–10% globally in the 3000 Rice Genomes Project (3k genomes), while they were enriched to 37–97% in the aus subpool of the 3k genomes.
qPXO79_6-1 spanned 4 SNPs across 148 kb at 21.4 Mb and included a Putative Serine Carboxypeptidase (LOC_Os06g36570) and an ABC transporter (LOC_Os06g36650) as high priority candidate genes. Serine carboxypeptidases have been implicated in chemical defense against microbes peptide transporter PTR2 (LOC_Os06g38294) as likely candidate gene. PTRs have been shown to be involved in disease resistance (Karim et al. 2007) and chemical defense (Nour-Eldin et al. 2012). Across the 3k genomes, LOC_Os06g38294 featured 11 non-synonymous (NS) SNPs. qPXO79_6-4 spanning 15 kb at 23 Mb (Mugford et al. 2009) and allele mining across the 3 k genomes for LOC_Os06g36570 revealed 21 non-synonymous NS SNPs. Likewise, ABC transporters (Krattinger et al. 2009) have been shown to contribute to disease resistance and LOC_Os06g36650 contained 21 NS SNPs. The 2 SNPs of qPXO79_6-2 spanning 69 kb at 21.8 Mb contained a metal cation transporter (LOC_Os06g37010). The 9 SNPs of qPXO79_6-3 spanning 18 kb at 22.6 Mb contained a peptide transporter PTR2 (LOC_Os06g38294) as likely candidate gene. PTRs have been shown to be involved in disease resistance (Karim et al. 2007) and chemical defense (Nour-Eldin et al. 2012). Across the 3 k genomes LOC_Os06g38294 featured 11 NS SNPs. qPXO79_6-4 spanning 15 kb at 23.1 Mb consisted of 3 SNPs and featured a receptor-like protein kinase (LOC_Os06g38990) as likely candidate gene.
On the long arm of chromosome 9‚ three SNPs within 40 bp (qPXO339/349_9-1) associated with resistance to race 9A and B (PXO339 and PXO349) in both the indica/aus and the indica panel. They were found at MAFs of 19% and clustered in the first exon of LOC_Os09g38510, which annotated as a purine permease. Purine permeases, implicated in cytokine transporters (Qi and Xiong 2013), have not been described in the context of disease resistance. Linkage of these SNPs with neighboring polymorphisms was weak and high heterozygosity made it difficult to assign the R allele.
Similarly, on the long arm of chromosome 11, qPXO339/349_11-1, a very narrow peak of of five SNPs within 60 bp and overlapping with the second exon of a palmitoyltransferase TIP1 (LOC_Os11g34860) associated with race 9A and B (PXO339 and PXO349) resistance in both panels. Palmitoyltransferases have been reported to be involved in pathogen-induced programmed cell death (Saucedo-Garcia et al. 2011). Although linkage with other SNPs in the region was low and heterozygosity high, S11_20426472 was highly significant (p-value 3.42−17) and determined in MLMM to affect multiple loci underlying resistance.
At 6.4 Mb, a 24 kb peak of four SNPs (qPXO86_11-1), was found to be associated with resistance to race 2 (PXO86). qPXO86_11-1 was limited to the indica/aus set, suggesting aus lines to be donors. While the R alleles were minor in the diverse set with frequencies of 6–15% they were enriched to 46–71% in the aus subpool. In the 3 k genomes, the MAF was only 4–6% globally, but enriched to 21–47% in the aus. The region contained one NBS-LRR disease resistance (LOC_Os11g11550) and one NB-ARC domain protein (LOC_Os11g11580). NBS-LRR and NB-ARC-domain proteins are classical resistance proteins involved in pathogen detection and host defense (DeYoung and Innes 2006) in gene-for-gene interactions.
qPXO79/112/341_12-1 on the long arm of chromosome 12 was associated with resistance to race 3B, 5 and 10. It comprised of 2 tightly linked SNPs separated by 48 kb and was specific for the indica/aus set. The rare R alleles were found at only 6% in the indica/aus set, but enriched to 25% and 26% in the aus subpool. Across the 3 k genomes MAF was 2–3% only but within the aus was 26% and 30%. qPXO79/112/341_12-1 contained three cysteine-rich receptor-like protein kinases, a gene family previously implicated in disease resistance through activation of hypersensitive response upon pathogen recognition (Chen et al. 2004).
Linkage Disequilibrium, Allele And Haplotype Analysis
Pairwise LD in the regions of Xa genes showed a relatively large LD in the indica subset (Additional file 8: Figure S5). LD decayed (r2 < 0.6) at ~400 kb in the distal end of chromosome 11 (Xa4 region). A larger LD was observed in the region of xa13 in chromosome 8 where LD started to decay around 1000 kb. A more extensive LD at ~1500 kb was observed in the putative xa25 region, found in the centromere of chromosome 12. However, an exception was observed in the xa5 region which started to decay at around 100 kb. While local LD across Xa regions were extensive, average chromosomal LD is weaker and decays within 100–200 kb (Additional file 9: Figure S6).
To identify SNP markers useful for tracking resistance genes, significant SNPs that are flanking or closely linked to a specific Xa gene were investigated. For the seven Xa genes detected in this study, allele frequency and allele patterns of 24 SNP markers were characterized across the 198 indica panel in correlation to disease reaction to the nine Xoo strains (Additional file 10: Table S4 and Additional file 11: Table S5).
For the two SNPs flanking Xa14 (S4_31492731 and S4_31492774), some genotypes carrying the minor alleles displayed resistance to PXO112 and are also known to have other Xa genes (Additional file 10: Table S4). Thus, these two SNPs could not be used as marker to track the Xa14 locus.
The highly significant SNPs for xa5 (S5_440644 and S5_453169) were found to be in LD (r2 = 0.73), forming a haplotype block that was correlated with resistance through haplotype trend regression (Additional file 12: Table S6). All indica genotypes having the minor haplotype “AA” exhibited moderate to high resistance across seven Xoo strains excluding PXO71 and PXO99. This disease reaction pattern agreed with the expected disease reaction attributed to the xa5 gene (Nino-Liu et al. 2006). Across 5 R and 10 S lines that are also present in the 3k genomes, the R alleles of S5_440644 (A) and S5_453169 (A) fully segregate with the causal mutation of xa5 at 437499 (T to A) and the phenotype (Additional file 13: Table S7). Since none of the other NS SNP alleles in the region segregate with phenotype, it suggests that the R alleles for S5_440644 (A) and S5_453169 (A) are indeed indicative of xa5 and that the SNP at 437499 bp is indeed causal for xa5. To validate if xa5 solely accounted for resistance to PXO339 and PXO349 in aus, alleles of the two SNPs (S5_440644 and S5_453169) overlapping xa5 were further inspected (Additional file 10: Table S4). Among the 50 aus genotypes of the indica/aus panel, 20 accessions exhibited resistance to the two strains of race 9 but did not have the xa5 allele (AA allele in DV85 and DV86). Two of these genotypes, Aus299 and Aus307, exhibited resistance to race 9 and three more races in the absence of the putative xa5 allele. With this, it can be inferred that there might be some other resistance QTL in these aus genotypes conferring resistance to race 9 that, due to low allele frequencies, was not detected in the GWAS.
Of the genotypes having the minor alleles of the seven SNPs linked to Xa7, 80% exhibited moderate to high resistance against PXO86, PXO79, PXO112, and PXO341 (Additional file 10: Table S4). Indica genotypes having the “A” allele for S6_28100960 always have the minor alleles of the other six SNPs, with average LD (r2) of 0.73 among the seven SNPs (Additional file 5: Table S2 and Additional file 8: Figure S5).
Among the ten SNPs flanking the xa13 region, S8_26013849 was the most significant SNP (p-value 1.14−07). Genotypes having the homozygous allele “A” for S8_26013849 also have the homozygous minor alleles of the other eight SNPs, exhibiting resistance to PXO99, the most virulent Xoo strain in the Philippines (Additional file 10: Table S4). The xa13 locus encodes a nodulin MtN3 gene, conferring differential resistance which is induced by the recessive allele resulting from mutations in the promoter region (Chu et al. 2006).
The two significant SNPs in chromosome 11 flanking Xa21 were found useful for tracking Xa21 gene in the indica genotypes. The homozygous minor alleles of S11_21190115 (T) and S11_21246561 (G) corresponded to indica genotypes exhibiting Xa21-mediated resistance against eight Xoo strains (Additional file 10: Table S4). Genotypes having these alleles were BB NILs and breeding lines with pyramided Xa genes, explaining the minor allele frequency (MAF) in the indica subset and being rare alleles in the 3k genomes (Additional file 11: Table S5).
Contrary to the R alleles of the SNPs flanking or linked to other Xa genes, the R alleles (TAT) in the three Xa4 SNPs (S11_27603791, S11_27603799 and S11_27603835) were major alleles in the indica subset (Additional file 10: Table S4 and Additional file 11: Table S5). The Xa4 gene was broadly deployed in rice breeding programs (Mew 1987), thus explaining the high frequency of the R alleles in the indica subset, which was mainly composed of breeding lines and modern varieties. While the R allele frequencies for Xa4 SNPs were 0.63 in the indica subset, their frequencies in indica accessions of the 3k genomes were only 0.21 (Additional file 11: Table S5).
For the flanking SNPs of xa25, 95% of the genotypes having the “A” allele of S12_17305618 and “T” allele of S12_17305619 displayed resistance to both PXO339 and PXO349 (Additional file 10: Table S4). These results confirmed the earlier reports regarding the differential resistance that xa25 confer against race 9 of Xoo (Chen et al. 2002).
In many cases a trait of interest cannot be reliably tracked by a single SNP. Haplotype blocks, a pattern of variation across linked SNPs, are often more robust in establishing marker trait associations (Lorenz et al. 2010). Testing associations between phenotypes and haplotypes, instead of single SNPs is thus more appropriate for capturing interactions between SNPs and a QTL (Clark 2004), particularly when multiple alleles are present (Morris and Kaplan 2002). Genome-wide haplotype block detection across the 198 indica genotypes resulted in 3,915 haplotype blocks composed of 12,361 SNPs. Based on haplotype trend regression analysis, two significant haplotypes include SNPs that are linked to Xa4 and xa5 (Additional file 12: Table S6). As shown in Fig. 4a, the haplotype “AA” for xa5 (HB1736) was predictive of the resistant genotypes to PXO86, PXO79, and PXO99 while any genotype having “GG” or “GA” corresponds to susceptibility. For HB3693 flanking the Xa4 gene, the resistant genotypes to PXO61, PXO71, PXO112, PXO99, and PXO341 mostly exhibited the “TA” haplotype (Fig. 4b).
Fig. 4.
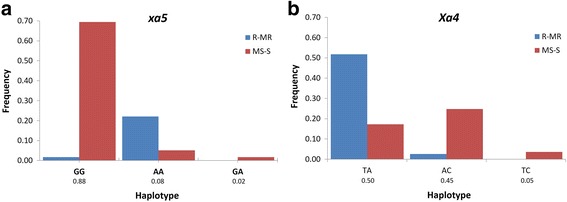
Haplotype blocks overlapping or flanking Xa genes. Values below the x-axis indicates the CHM probabilities of the haplotypes (a) Haplotype allele frequency of HB1736 overlapping the xa5 gene in terms of average disease reaction to PXO86, PXO79, and PXO99. b Haplotype allele frequency of HB3693 flanking the Xa4 gene in terms of average disease reaction to PXO61, PXO71, PXO112, PXO99, and PXO341
Among twelve highly significant haplotype blocks, two haplotype blocks (HB814 and HB3460) had SNPs that were not found to be significant in the EMMAX analysis (Additional file 12: Table S6). HB814 was found to be associated with indica genotypes’ resistance to PXO112 but SNPs in this haplotype block was not significant in the single SNP association. SNPs comprising HB814 (S2_24907121 and S2_24907123) were 1 Mb away from the most significant SNPs (25,948,468-25,948,531 bp) in chromosome 2 (Additional file 4: Figure S3e and Additional file 6: Table S3). However, looking closely at chromosome 2, the peak started to rise within the region of the HB814. Furthermore, HB3460 was composed of nine SNPs (at 2.9–3 Mb in chromosome 11). Closely inspecting Additional file 4: Figure S3d, HB3460 was the rising peak in the short arm of chromosome 11 which did not reach the threshold of P-value < 1 × 10−5. This region contained a nodulation-signaling pathway 2 protein (LOC_Os11g06180), an ATOZI1 protein (LOC_Os11g06240), a retrotransposon Ty3-gypsy subclass protein (LOC_Os11g06259) and an integral membrane protein (LOC_Os11g06310).
Discussion
The population structure of a diverse panel can affect the analysis of genotype-phenotype associations (Zhao et al. 2011). Population structure results in divergence of allele frequencies of the sub-populations. Loci which vary in allele frequencies between sub-populations can be associated with the phenotype when the means of the sub-populations diverge for a particular trait of interest (Zhang et al. 2009b). Principal component analysis of the indica/aus panel showed five components that best represent the population structure of the panel. The indica genotypes differentiated into three components.
The nine representative strains of eight Xoo races used in this study covered a wide range of virulence and prevalence and they successfully distinguished differential resistance across the indica/aus panel. Several indica genotypes having broad-spectrum resistance comprised mostly irrigated lowland lines and varieties (Additional file 1: Table S1). Other than the IRBB NILs, breeding lines IR09F154, IR08N134, IR 72890-81-3-2-2, IR05N173 and IR09N530 exhibited resistance against seven to nine Xoo strains. Most of these genotypes have xa5 and Xa4 which were introduced during the breeding process of these varieties. However, the presence alone of the xa5 gene in IR 72890-81-3-2-2 and IR05N173 would not confer resistance to PXO71 (race4) (Additional file 10: Table S4). Thus, there might be other genes underlying resistance in these genotypes.
Rice genotypes belonging to the group, such as Aus299, Dular, AusBak Tulsi, Ase Pulu Jawa, Aus298, Makalioka 34, Kalimekri 77-5, DV86, Chengri, Aus307, Khaiyan, and DV85 showed broad-spectrum resistance against Xoo. Several studies have shown that aus genotypes are valuable source of R genes for BB resistance (Sidhu et al. 1978; Khush and Angeles 1999). It is notable that most of the aus genotypes (96%) in this study exhibited resistance to both strains of race 9 (PXO339 and PXO349) (Additional file 1: Table S1). Among the Xa genes that originated from aus, xa5 is the only gene that confers resistance to race 9 (Iyer and McCouch 2004). To validate if xa5 solely accounts for resistance to PXO339 and PXO349 in aus, alleles of the two SNPs (S5_440644 and S5_453169) overlapping xa5 were closely inspected (Additional file 10: Table S4). Among the 50 aus genotypes, 20 accessions exhibited resistance to the two strains of race 9 but did not have the xa5 allele (AA allele in DV85 and DV86). Aus299 and Aus 307 exhibited resistance to race 9 and three more races in the absence of the putative xa5 allele. Thus, other resistance loci in aus besides xa5 confers resistance against strains of race 9.
Our studies confirmed the involvement of seven known Xa genes underlying differential resistance. Among the seven Xa genes detected in the GWAS, Xa4, xa5 and Xa21 are known to confer broad-spectrum resistance to Xoo (Nino-Liu et al. 2006). SNPs overlapping or linked to the regions of these genes were found to be highly associated with resistance to the expected Xoo races. Since xa5, xa13, Xa21, and xa25 are cloned (Nino-Liu et al. 2006; Liu et al. 2011), the exact physical position of these genes would ascertain that SNPs overlapping these regions could be highly predictive of these genes. Low LD between the regions of the cloned genes and the SNPs detected in GWAS would provide high resolution in tracking these genes in breeding programs.
On the contrary, Xa4, Xa7, and Xa14 loci have not yet been cloned. Despite the extensive use of Xa4 in breeding programs, successful cloning of this gene has not been published. Sun et al. (2003) have narrowed down the Xa4 locus, most significant SNPs linked to the region of the Xa4 flanking marker (RM224) overlapped with a rust-resistance protein Lr21 (LOC_Os11g45620) (Fig. 3). However, Hur et al. (2016) recently fine mapped Xa4 to a 60 kb region suggesting LOC_Os11g45930, a NBS–LRR-type disease-resistance protein or LOC_Os11g45930, a hypothetical protein as candidates, which lie between SNPs S11_27677853 and S11_27831021, the most downstream of a cluster of 8 significant SNPs overlapping with Xa4. Seven significant SNPs were flanking the region of Xa7 described by Zhang et al. (2009a). Several candidate genes were identified in the two fine mapping studies of Xa7 (Chen et al. 2008; Zhang et al. 2009a). Among these candidate genes, BTB/POZ and Nramp6, known to be involved in plant resistance (Chen et al. 2008), were within the 84 kb region comprising the SNPs detected in GWAS.
Bao et al. (2010) have mapped the Xa14 locus between two SSRs having a 300 kb interval. SNPs flanking this mapped region were linked to ATBAG1 (LOC_Os04g52890), OsFBW1-F-box family protein/WD-40 repeat family protein (LOC_Os04g52870) and an ABC transporter (LOC_Os04g52900). These genes were also predicted genes for a blast resistance gene Pikahei-1(t) mapped in chromosome 4 (Xu et al. 2008). F-Box and ATBAG proteins were reported to be involved in defense-programmed cell death (Kang et al. 2006; van den Burg et al. 2008) while ABC transporters constitutively induce defense pathways in plants (Chauhan et al. 2015).
Aside from the seven Xa genes detected in the GWAS, 33 SNPs clustering on four chromosomes showed strong associations with BB resistance. Among the 8 novel resistance loci identified five appeared to be derived from the Aus subpool, underlining the importance of this group as a reservoir for BB resistance donors. qPXO79_6-2 to -4 cluster on the long arm of chromosome 6 over a range of 1.3 Mb and conferred resistance to race 3B. The discovered markers display a high level of specificity for the qPXO79_6 loci and could directly be deployed as foreground markers in marker assisted backcrossing (MABC) schemes. At the caveat of introducing linkage drag it would be most feasible to introgress the whole region of 1.3 Mb from a suitable donor within the Aus pool used in this study. Likewise the rare Aus specific alleles of qPXO86_11-1 and qPXO79/112/341_12-1 could also readily be converted into markers for MABC.
Two loci, qPXO339/349_9-1 and qPXO339/349_11-1, were discovered to confer resistance to race 9A and 9B. New sources of resistance to race 9 of Xoo would be valuable. Despite not causing epidemics on rice fields planted with rice varieties having Xa7, race 9, particularly race 9b, have persisted at high frequency since the 1990s until present (Ponciano et al. 2004, Quibod et al., 2016). Significant SNPs for both qPXO339/349_9-1 and qPXO339/349_11-1, however, cluster within less than 100 bp, which is too low for reported rice LD. Furthermore, heterozygosity among these SNPs in our panels were very high. Though the corresponding SNP can be found in the 3k genomes, they are not part of the Filtered (4.8 M) or Base (18 M) sets and display similarly high heterozygosity globally, which might indicate wrong mapping due to larger structural variation in the region. Taken together this suggested that both loci should be regarded with reservations as to their accuracy of prediction and utility in resistance breeding.
Nearly half of the identified candidate genes within the 8 novel GWAS peaks code for families that are classically associated with disease resistance such NBS-LRR proteins, NB-ARC domain proteins and receptor-like protein kinases. They are typical R proteins that interact with Avr proteins in a gene-by-gene-interaction. In conjunction with the observed high non-synonymous natural variation within the global 3k genomes pool, it makes them high priority candidates worthy of further investigation.
The other half roughly fall into the category of chemical defense through antimicrobial metabolite biosynthesis or transport. ABC transporters have been demonstrated to transport a plethora of secondary metabolites, many of which are toxic to microbes and pests (Kretzschmar et al. 2011). Despite their annotation as peptide transporters PTRs have been shown to transport antimicrobial compounds such as glucosinolates (Nour-Eldin et al. 2012) and plant hormones (Kanno et al. 2012). Likewise, purine permeases transport hormone like secondary metabolites. Serine carboxypeptidases have been shown to act as acyltransferases in terpenoid (Mugford et al., 2009) and phenolic compound (Bontpart et al. 2015) synthesis, many of which act as toxins or deterrents against pests and diseases. Aminotransferases are similarly involved in the biosynthesis of antimicrobial compounds (Ding et al. 2016). Though less specific and often associated with partial resistance only, disease resistance through antimicrobial compounds has been observed to be durable, since resistance breakdown through deleterious Avr mutations is not possible. Rather the pathogen has to evolve to catabolize the deterring or toxic compounds. Since candidate gene prioritization was based on the Nipponbare reference and annotation in the respective regions, it can not be ruled out that additional gene models specific to indica and/or aus were overlooked. Validation of candidate genes will thus depend on comparative genomics among a set of relevant indica and aus references and transgenic approaches that knock out the R-alleles in the donor background or transfer the R-alleles into recipient backgrounds.
Knowledge about the extent of LD decay is crucial for association analysis and determines its resolution (Remington et al. 2001). LD pattern was quite consistent across chromosomes and decays within 100–200 kb. (Additional file 9: Figure S6). Indica genotypes, comprising 69% of the indica/aus panel, exhibited a wide range of resistance across the nine Xoo strains. Thus, it was of interest to assess the LD, allele frequencies, and haplotypes across genomic regions that are “hotspots” of resistance-associated loci. Lower LD makes mapping resolution higher, allowing isolation of functional variation (Burke et al. 2007). Contrary to the observed average chromosomal LD, regions flanking Xa genes have more extensive LD. Among the surrounding regions of Xa genes, only the xa5 region has the least extent of LD of approximately 100 kb which is similar to what Garris et al. (2003) has estimated for the xa5 region (Additional file 8: Figure S5b). For the distal end of chromosome 11 (26.5–28.9 Mb), the LD extended up to 400 kb. In this region, Xa genes such as Xa3/Xa26 (Yang et al. 2003; Xiang et al. 2006), Xa4, and Xa22(t) were tightly linked (Wang et al. 2001; Wang et al. 2003). This part of chromosome 11 is known to be a hotspot of resistance genes (The Rice Chromosomes 11 and 12 Sequencing Consortia 2005), and may have been selected for continuously for breeding, explaining a relatively strong LD. Furthermore, the xa25 region had the most extensive LD, reaching up to ~1500 kb (r2 = 0.6). The strong LD in the xa25 region may be explained by the centromeric location, in which recombination is low (Remington et al. 2001). The relatively stronger LD observed in these regions could be attributed to the relatedness of breeding lines and varieties which composed about 72% of the diverse panel used.
Allele frequency and allele patterns of the 27 SNPs for the seven Xa genes detected were characterized to identify markers useful for marker-aided selection. Alleles of the SNPs for the seven Xa genes corresponding to expected resistant phenotype have minor allele frequencies, except for SNPs in the Xa4 region (Additional file 10: Table S4). Homozygous minor alleles predictive of Xa7-, xa13-, and Xa21-mediated resistance behaved like haplotypes in the indica subset even though these SNPs were not detected as haplotype blocks. Table 2 summarizes the SNP markers and alleles predictive of the resistant phenotype mediated by respective Xa genes. Among the seven Xa genes detected, putative resistant alleles for Xa4 are the most predominant alleles in indica breeding lines and varieties, followed by xa25 and xa5. Genes such as Xa4 and xa5 are known to confer broad-spectrum resistance which explains their extensive use in breeding programs. However, it is surprising to note that resistant allele of xa25 was found to be predominant in breeding lines and varieties. Minghui 63, the donor of xa25, is an indica rice restorer line (Chen et al., 2002). It can be inferred that breeders have selected for the xa25 locus unconsciously. Furthermore, the comparison of the R allele frequencies for the 24 SNPs for the seven Xa genes in the 1,788 indica genotypes of the 3k genomes (Alexandrov et al. 2015) augments the potential of these SNPs to detect resistance gene alleles (Additional file 11: Table S5).
Table 2.
Most promising SNPs for marker development
| Resistance Gene | SNP | Chromosome | Position | Resistant Allelea | Breeding Line and Variety having R Allele (%)b |
|---|---|---|---|---|---|
| xa5 | S5_440644 | 5 | 440644 | A | 10 |
| xa5 | S5_453169 | 5 | 453169 | A | 10 |
| Xa7 | S6_28100960 | 6 | 28100960 | A | 1 |
| Xa7 | S6_28158893 | 6 | 28158893 | G | 2 |
| Xa7 | S6_28160615 | 6 | 28160615 | G | 2 |
| Xa7 | S6_28171883 | 6 | 28171883 | C | 2 |
| Xa7 | S6_28173425 | 6 | 28173425 | A | 2 |
| xa13 | S8_26013849 | 8 | 26013849 | A | 2 |
| xa13 | S8_26044514 | 8 | 26044514 | A | 2 |
| xa13 | S8_26044528 | 8 | 26044528 | C | 2 |
| xa13 | S8_27319974 | 8 | 27319974 | C | 2 |
| xa13 | S8_27520607 | 8 | 27520607 | G | 2 |
| xa13 | S8_27536844 | 8 | 27536844 | C | 2 |
| xa13 | S8_27641374 | 8 | 27641374 | A | 2 |
| xa13 | S8_27663433 | 8 | 27663433 | C | 2 |
| Xa21 | S11_21190115 | 11 | 21190115 | T | 4 |
| Xa21 | S11_21246561 | 11 | 21246561 | G | 3 |
| Xa4 | S11_27603791 | 11 | 27603791 | T | 60 |
| Xa4 | S11_27603799 | 11 | 27603799 | A | 59 |
| xa25 | S12_17305618 | 12 | 17305618 | A | 15 |
| xa25 | S12_17305619 | 12 | 17305619 | T | 72 |
a Allele corresponding to resistant phenotype based on 198 indica genotypes
b Percentage of indica breeding lines and varieties having the putative resistant allele
To determine polymorphisms linked to resistance QTLs that were not captured by single SNP associations, haplotype blocks in the indica genotypes were also assessed. Of the 3,915 haplotype blocks detected, 12 haplotype blocks were highly correlated with differential resistance, two of which correspond to the xa5 and Xa4 genes. HB1736 (xa5) and HB3693 (Xa4) demonstrated usefulness to predict resistant phenotypes (Fig. 4). For the rest of the haplotype blocks, involvement of the genes to which these haplotypes overlap should be resolved. Thorough analysis of genotypes having haplotypes correlated with resistance is needed to fully realize the value of these genetic variations in BB resistance breeding.
Conclusion
The results of this GWAS have pinpointed resistance loci conferring differential resistance to the nine representative strains of Xoo in the indica/aus panel and the indica subset. The use of SNPs from genotyping-by-sequencing is a powerful tool to elucidate disease resistance in rice. Differences in the resistance loci detected using two germplasm sets demonstrate the differentiation of loci underlying resistance in terms of the distinct population structure in rice. These results present a wealth of SNPs highly associated with differential Xoo resistance that would enable the development of SNP markers for marker-aided selection and tracking of known Xa genes. Efficient tracking of Xa genes in the breeding pipeline provides breeders insight on which varieties to deploy to specific areas having varying Xoo race population. Considering the predominant pathogen race ensures wiser strategies to maintain durability of Xa genes deployed in the field. To fully understand the value of these loci in breeding for BB resistance the potentially novel loci identified in this study should be further validated through expression profiling, and development of biparental populations. Moreover, genotypes having no resistance alleles for specific Xa QTLs but exhibit high degree of resistance serve as new sources of resistance loci to plant breeders to diversify the genetic base of core breeding sets. The use of aus genotypes for a more thorough analysis is recommended to identify potentially novel loci that were not detected in this study.
Methods
Selection Of Diverse Panel
An initial diverse panel of 380 genotypes composed of breeding lines, varieties, and landraces was assembled from accessions obtained from the IRRI T.T. Chang Genetic Resources Center and the IRRI Plant Breeding division based on key morpho-agronomic traits and overall field performance. Using 384 SNPs from Illumina BeadXpress system, the 380 genotypes were reduced to 285 genotypes after genetic diversity and population structure analyses using the software packages Powermarker (Liu and Muse 2005), and Structure 2.2 (Pritchard et al. 2000), respectively.
Leaf Sampling And Genotyping
For reducing the size of the initial diverse panel, leaf samples of the 380 diverse germplasm were collected for SNP genotyping. DNA samples were extracted using the modified CTAB method (Murray and Thompson 1980) and were standardized to 50 ng/μl for Illumina 384-plex BeadXpress SNP genotyping. The 380 genotypes were genotyped using the 384-plex indica x indica SNP set (VC0013033-OPA) on Illumina BeadXpress system following the Golden Gate Genotyping protocol (Illumina, CA, USA) and allele calling based on the procedure described by Thomson et al. (2012) and Wright et al. (2010).
For the selected panel of 285 genotypes, two to three healthy young leaves were collected from representative tagged plants after the BB resistance screening. The collected leaf samples were lyophilized using the Martin Christ Freeze Dryer at −50° and 0.42 mbar for 48 h, and were ground using the Qiagen Tissue Lyser II. Genomic DNA was extracted using the Qiagen DNeasy Plant Mini Kit. The DNA samples were quantified using the NanoDrop Reader ND8000 and then standardized to 100 ng/μl as required for the preparation of genomic library for GBS.
Genotyping-by-Sequencing (GBS)
The preparation of the 96-plex genomic libraries for GBS was done in the Institute of Genomic Diversity, Cornell University (Ithaca, NY, USA) following the protocol developed by (Elshire et al. 2011). The purified 96-plex GBS libraries were loaded into the Experion automated electrophoresis station to evaluate fragment sizes. Libraries having no or minimal adapter dimers (fragments of approximately 128 bp in length) and having majority of other DNA fragments between 170 and 350 bp were considered for sequencing. Then, single-end sequencing (86 bp reads) of 96-plex library per flow cell channel were performed on a HiSeq 2000 (Illumina, Inc.) at Cornell University (Ithaca, NY, USA).
Fastq files were processed using TASSEL-GBS analysis pipeline (Version 3.0.147), an extension to the Java program TASSEL (Bradbury et al. 2007). Among the 843,898,432 reads generated from the three lanes of HiSeq, 12,937,086 tags were generated from the TASSEL-GBS Discovery pipeline described by Glaubitz et al. (2014). Tags were aligned to the Nipponbare reference MSU version7 (Kawahara et al. 2013). SNPs were called from aligned tags of at least 10% of the taxa, generating 640,783 SNPs in the hapmap unfiltered file. Post SNP call filtering was also done by retaining SNPs with MAF >0.01 and 80% calling, having 245,669 in the hapmap filtered file.
BB Resistance Screening
The experiment was carried out in a contained screenhouse facility to prevent the spread of inoculum and was done using RCBD split-plot with nested subplot design. The nine Xoo strains were the main plots and in each main plot, a sub-plot of 285 genotypes was nested into early maturing and medium maturing genotypes. The BB resistance screening was performed 45 days after sowing. A total of nine strains representing eight different races of Xanthomonas oryzae pv. oryzae were used, namely: PX061 (Race1), PXO86 (Race2), PXO79 (Race3B), PXO71 (Race4), PXO112 (Race5), PXO99 (Race6), PXO339 (Race9A), PXO349 (Race9B) and PXO341 (Race10). Inoculation of the rice plants was done through cutting of 1–2 cm of the leaf tip with a pair of scissors dipped in bacterial suspension (Kauffman et al. 1973). The screening was replicated three times over time. For each replicate, five leaves of two plants per entry were inoculated with Xoo strains. Lesion lengths were measured 14 days after inoculation. Genotypes having lesion lengths ranging 1–5 cm were rated as resistant (R), 6–10 cm were rated as medium-resistant (MR), 10–15 cm were rated as medium-susceptible (MS), and those having greater than 15 cm were rated as susceptible (S).
The lesion length data generated from the BB resistance screening was averaged across sampling units per entry of each replication. Kolmogorov-Smirnov test for normality of data, analysis of variance of the rice genotypes for the nine strains of Xoo using the mixed model analysis of SAS (PROC MIXED), and least square means were generated using SAS version 9.3. LS means generated across three replications were used for association analysis.
Genetic Diversity, Population Structure, Marker Filtering And Association Analysis
The genetic distance of the 285 genotypes was computed based on Tamura-Nei model implemented in MEGA 6 (Tamura et al. 2013) using 1,193 SNPs (MAF >0.05 and 100% calling) from GBS. In addition, population differentiation Fst values were generated from PowerMarker version 3.25 (Liu and Muse 2005) while differentiation based on global unbiased Fst (Weir and Cockerham 1984) was calculated using the Hierfstat package implemented in R (Goudet 2005). Principal component analysis was performed using SNP & Variation Suite v8.4 software (Golden Helix, Inc., Bozeman, MT, www.goldenhelix.com).
TASSEL version 3.150 software (Bradbury et al. 2007) was used for manipulating and filtering SNPs for genome-wide association analysis. Initial dataset was filtered based on MAF >0.05 and 95% call rate. A unified mixed-model approach was deployed to account for population structure and familial relatedness (Yu et al. 2006; Price et al. 2006).
Compressed mixed linear model (MLM) was used to analyze association, considering population structure (Q) and relatedness or kinship (K) to reduce spurious associations (Yu et al. 2006). SNP loci with P-values < 1 × 10−5 were considered significant. Preliminary association results showed ambiguity that residual population structure has not fully corrected by Q + K. Thus, the 37 japonica/aromatic genotypes were removed from further genome-wide association analysis, retaining 248 indica and aus genotypes.
Marker-trait association of the 248 genotypes was performed using 40,840 SNPs (90% calling with MAF ≥ 0.05) using SNP & Variation Suite v8.4 software (Golden Helix, Inc., Bozeman, MT, http://www.goldenhelix.com/). Mixed linear models such as Efficient Mixed-Model Association eXpedited Model (EMMAX) (Kang et al. 2008; Vilhjalmsson 2012) and Multi-Locus Mixed Model (MLMM) (Segura et al. 2012; Vilhjalmsson 2012) were used in the analysis Principal component analysis was performed using 10 components, with PC = 5 as the optimal component for the Q matrix. Kinship matrix (K) was generated using Genomic Best Linear Unbiased Predictor (GBLUP) (Van Raden 2008). Q + K matrices were incorporated in the MLM association analysis. Single-locus mixed model (EMMAX) as described by Kang et al. (2010) was done to determine loci associated to resistance against the nine Xoo strains assuming all loci have a small effect on the trait. SNPs having P-values < 1 × 10−5 and FDR corrected values of < 0.05 were considered significant in case they overlapped with known resistance genes (Additional file 5: Table S2). For potentially new loci (Additional file 6: Table S3) a more stringent Bonferroni correction of 0.05 was applied to avoid false positive detection. Furthermore, to determine which loci are cofactors and accounts for large effect on the Xoo resistance, MLMM was performed. The MLMM algorithm assumes that multiple loci are associated with resistance, and is a stepwise EMMAX, which re-computes genetic and error variance each step (Segura et al. 2012). Of the 10 steps implemented in MLMM, steps identified to be optimal based on Bonferroni correction was taken account, and p-values of loci identified to be cofactors were noted. Since population structure may greatly affect functional variation underlying resistance to BB, separate association analysis of 198 indica genotypes using 40,396 SNPs and 3 principal components were carried out.
Bioinformatics Analysis
The physical positions of BB resistance genes were based on the published position of cloned genes. While for those genes that were not cloned, information of the published flanking markers was used for the ePCR tool of NCBI (Rotmistrovsky et al. 2004) to ascertain the putative position of the genes. To identify genes that overlap and that are closely linked to the significant SNPs, ± 50 bp and ± 150 kb region of the SNP were conservatively considered, respectively. The list of gene annotations was derived from the IRRI Galaxy tool (http://galaxy.irri.org/) linked to the MSU website (http://rice.plantbiology.msu.edu/). In addition, the Rice SNP-Seek Database (Alexandrov et al. 2015) was used to investigate the allele frequencies of significant SNPs in the 1,788 indica subset of the 3k genomes and extract non-synonymous SNPs for specific target regions.
Linkage Disequilibrium, Allele And Haplotype Analysis
Detailed analysis of SNP alleles, linkage disequilibrium (LD) and haplotype blocks for genomic regions associated with differential resistance to nine Xoo strains was done using 198 indica genotypes. Alleles of significant SNPs flanking/overlapping the seven known Xa genes were investigated to determine which allele is predictive of the resistant phenotype to Xoo.
Local LD and haplotype analysis were performed using Expectation Maximization (EM) algorithm (Dempster et al. 1977; Excoffier and Slatkin 1995) and the Composite Haplotype Method (CHM) (Weir 1996) on SVS 8.4. Average pair-wise LD (r2) was taken account and LD decay (r2 < 0.6) was characterized in regions of highly significant SNPs. Furthermore, haplotype blocks were detected on 160 kb window with maximum 30 markers per window, having 50 maximum iterations, and 90% confidence. Haplotype blocks that are correlated to differential resistance were identified using the full model linear haplotype trend regression with Bonferroni adjustment.
Additional files
Information on the 285 diverse genotypes used in this study. (XLSX 34 kb)
Phylogenetic and population structure of the 285 diverse panel. a) Radial tree of 285 diverse germplasm based on Tamura-Nei model (MEGA 6). Principal component analysis of b) 285 diverse germplasm c) 248 indica and aus genotypes, and d) 198 indica genotypes (Golden Helix SVS). (PPTX 318 kb)
Genome-wide association analysis of bacterial blight resistance to nine Xoo strains in 248 genotypes based on Efficient Mixed-Model Association eXpedited Model (EMMAX). Manhattan plots for nine Xoo strains (a) PXO61, (b) PXO86, (c) PXO79, (d) PXO71, (e) PXO112, (f) PXO99, (g) PXO339, (h) PXO349, and (i) PXO341. X-axis shows the SNPs along each chromosome; y axis is the − log10 (P-value) for the association. Significant SNPs are those beyond the red line having P-value < 1 × 10 −5. Quantile-quantile plots for nine Xoo strains (j) PXO61, (k) PXO86, (l) PXO79, (m) PXO71, (n) PXO112, (o) PXO99, (p) PXO339, (q) PXO349, and (r) PXO341. (PPTX 720 kb)
Genome-wide association analysis of bacterial blight resistance to nine Xoo strains in 198 indica genotypes based on Efficient Mixed-Model Association eXpedited Model (EMMAX). Manhattan plots for nine Xoo strains (a) PXO61, (b) PXO86, (c) PXO79, (d) PXO71, (e) PXO112, (f) PXO99, (g) PXO339, (h) PXO349, and (i) PXO341. X-axis shows the SNPs along each chromosome; y axis is the − log10 (P-value) for the association. Significant SNPs are those beyond the red line having P-value < 1 × 10 −5. Quantile-quantile plots for nine Xoo strains (j) PXO61, (k) PXO86, (l) PXO79, (m) PXO71, (n) PXO112, (o) PXO99, (p) PXO339, (q) PXO349, and (r) PXO341. (PPTX 521 kb)
Significant GWAS SNP hits that are flanking/overlapping with known resistance genes for bacterial blight. (XLSX 92 kb)
Significant GWAS SNP hits that are potentially novel loci for bacterial blight resistance. (XLSX 80 kb)
Local pair-wise linkage disequilibrium (LD) in GWAS hits overlapping putative novel loci. LD (r2) measure was based on Composite Haplotype Method (CHM). Highly significant SNPs corresponding to Additional file 6: Table S3 are highlighted by black arrows. All SNPs in the region including 8 SNPs upstream of the first significant SNP and 8 SNPs downstream of the last significant SNP are shown.in putatively novel peaks are encircled in red. a) SNPs found in chr6: 21.26–21.96 Mb. b) SNPs found in chr6 22.49–23.23 Mb. c.) SNPs found in chr9: 22.16–22.23 Mb. d) SNPs found in chr11: 6.33 M–6.45 Mb. e) SNPs found in chr11: 20.18–20.50 Mb. f) SNPs found in chr12: 25.69–25.76 Mb. (PPTX 2022 kb)
Linkage Disequilibrium (LD) Decay in GWAS hits overlapping/flanking Xa regions. LD (r2) measure was based on Composite Haplotype Method (CHM). a.) SNPs flanking Xa14 (chr 4: 31–32.9 Mb). b.) SNPs flanking xa5 (chr 5: 200–900 kb). c.) SNPs flanking Xa7 (chr 6: 26–28.6 Mb). d.) SNPs flanking xa13 (chr 8: 25–27.9 Mb). e.) SNPs flanking Xa21 (chr 11: 20.5–22 Mb). f.) SNPs flanking Xa4 (chr 11: 26.5–28.9 Mb). g.) SNPs flanking xa25 (chr 12: 13–17.6 Mb). (PPTX 1768 kb)
Linkage disequilibrium (LD) of adjacent SNPs per chromosome. LD (r2) measure of adjacent pairs was based on Composite Haplotype Method (CHM). (PPTX 324 kb)
Genotype matrix of 248 genotypes across GWAS SNPs flanking/linked to Xa genes. (XLSX 152 kb)
Comparison of allele frequency of SNPs overlapping/flanking Xa genes between GWAS 198 indica genotypes and 1788 indica subset in 3 k genomes. (XLSX 46 kb)
Most significant haplotypes associated with resistance to nine Xoo strains. (XLSX 12 kb)
Comparison of xa5 GWAS SNPs with SNPs derived from 3k Genomes. (XLSX 70 kb)
Acknowledgements
We thank Crisostomo Dizon, Socorro Carandang, Grace Cariño, Maria Ymber Reveche and Yoojin Lee for the support in the completion of the phenotyping and genotyping. We would also like to extend our gratitude to the staff of the Plant Pathology Laboratory for the technical assistance, and to Ramil Mauleon, Kenneth McNally, and Millicent Sanciangco for the discussions and advice regarding the analysis.
Funding
This research was supported by The Global Rice Science Partnership (GRiSP).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
The raw data is publically available with IRRI: https://s3-ap-southeast-1.amazonaws.com/oryzasnp-atcg-irri-org/pub-data/248Entries_40840SNPs_inorder_21May2015_v2.zip.
Authors’ contributions
CJDE designed the experiment, analyzed the data and wrote the manuscript. ET grew plants, performed BB phenotyping, and revised the manuscript. VMJ, and JD performed bioinformatics support and developed Galaxy tools. DNL grew plants and performed BB phenotyping. EGNM performed the LD analysis. CVC selected the diagnostic Xoo strains for the phenotyping, provided information on resistance genes, and revised the manuscript. EA, MGD, and MM revised the initial manuscript draft. MSD, MT and TK supervised the conduct of the experiment, data analysis and revised the manuscript. All authors contributed to the completion of draft, read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- Avr
Avirulence
- BB
Bacterial blight
- CHM
Composite haplotype method
- EM
Expectation maximization
- EMMAX
Efficient mixed-model association expedited model
- GBLUP
Genomic best linear unbiased predictor
- GBS
Genotyping-by-sequencing
- GWAS
Genome-wide association study
- HPRR
Host pattern recognition receptors
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- MLM
Mixed linear model
- MLMM
Multi-locus mixed model
- NIL
Near isogenic line
- PAMP
Pathogen-associated molecular patterns
- PCA
Principal components analysis
- QTL
Quantitative trait loci
- SNP
Single nucleotide polymorphisms
- Xoo
Xanthomonas oryzae pv. Oryzae
References
- Adhikari TB, Cruz C, Zhang Q, Nelson RJ, Skinner DZ, Mew TW, Leach JE. Genetic diversity of Xanthomonas oryzae pv. oryzae in Asia. Appl Environ Microb. 1995;61(3):966–971. doi: 10.1128/aem.61.3.966-971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari TB, Basnyat RC, Mew TW. Virulence of Xanthomonas oryzae pv. oryzae on rice lines containing single resistance genes and gene combinations. Plant Dis. 1998;83(1):46–50. doi: 10.1094/PDIS.1999.83.1.46. [DOI] [PubMed] [Google Scholar]
- Alexandrov N, Tai S, Wang W, Mansueto L, Palis K, Fuentes RR, Ulat VJ, Chebotarov D, Zhang G, Li Z, Mauleon R, Hamilton RS, McNally KL. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43(D1):D1023–D1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao SY, Tan MP, Lin XH. Genetic mapping of a bacterial blight resistance gene Xa14 in rice. Acta Agronomica Sinica. 2010;36(3):422–427. doi: 10.3724/SP.J.1006.2010.00422. [DOI] [Google Scholar]
- Bhasin H, Bhatia D, Raghuvanshi S, Lore JS, Sahi GK, Kaur B, Vikal Y, Singh K. New PCR-based sequence-tagged site marker for bacterial blight resistance gene Xa38 of rice. Mol Breeding. 2012;30:607–611. doi: 10.1007/s11032-011-9646-y. [DOI] [Google Scholar]
- Blair MW, Garris AJ, Iyer AS, Chapman B, Kresovich S, McCouch SR. High resolution genetic mapping and candidate gene identification at the xa5 locus for bacterial blight resistance in rice (Oryza sativa L.) Theor Appl Genet. 2003;107(1):62–73. doi: 10.1007/s00122-003-1231-2. [DOI] [PubMed] [Google Scholar]
- Bontpart T, Cheynier V, Ageorges A, Terrier N. Research Review: BADH or SCPL acyltransferase? What dilemma for acylatoinn in the world of plant phenolic compounds. New Phytol. 2015;208:696–707. doi: 10.1111/nph.13498. [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens Terry M, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- DeYoung BJ, Roger WI (2006) Plant NBS-LRR proteins in pathogen sensing and host defense. Nature Immunology 7(12):1243–1249. [DOI] [PMC free article] [PubMed]
- Burke JM, Burger JC, Chapman MA. Crop evolution : from genetics to genomics. Curr Opin Genet Dev. 2007;17:525–532. doi: 10.1016/j.gde.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Chauhan H, Boni R, Bucher R, Kuhn B, Buchmann G, Sucher J, Selter LL, Hansel G, Kumlehn J, Bigler L, Glauser G, Wicker T, Krattinger SG, Keller B. The wheat resistance gene Lr34 results in the constitutive induction of multiple defense pathways in transgenic barley. Plant J. 2015;84(1):202–215. doi: 10.1111/tpj.13001. [DOI] [PubMed] [Google Scholar]
- Cheema KK, Grewal NK, Vikal Y, Sharma R, Lore JS, Das A, Bhatia D, Mahalan R, Gupta V, Bharaj TS, Singh K. A novel bacterial blight resistance gene from Oryza nivara mapped to 38 kb region on chromosome 4 L and transferred to Oryza sativa L. Genet Res. 2008;90(5):397–407. doi: 10.1017/S0016672308009786. [DOI] [PubMed] [Google Scholar]
- Chen H, Wang S, Zhang Q. New gene for bacterial blight resistance in rice located on chromosome 12 identified from Minghui 63, an elite restorer line. Phytopathology. 2002;92(7):750–754. doi: 10.1094/PHYTO.2002.92.7.750. [DOI] [PubMed] [Google Scholar]
- Chen K, Fan B, Du L, Chen Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Molecular Biology. 2004;56(2):271–283. doi: 10.1007/s11103-004-3381-2. [DOI] [PubMed] [Google Scholar]
- Chen S, Huang Z, Zeng L, Yang J, Liu Q, Zhu X. High-resolution mapping and gene prediction of Xanthomonas Oryzae pv. Oryzae resistance gene Xa7. Mol Breeding. 2008;22(3):433–441. doi: 10.1007/s11032-008-9187-1. [DOI] [Google Scholar]
- Chen S, Liu X, Zeng L, Ouyang D, Yang J, Zhu X. Genetic analysis and molecular mapping of a novel recessive gene xa34(t) for resistance against Xanthomonas oryzae pv. oryzae. Theor Appl Genet. 2011;122(7):1331–1338. doi: 10.1007/s00122-011-1534-7. [DOI] [PubMed] [Google Scholar]
- Chu Z, Fu B, Yang H, Xu C, Li Z, Sanchez A, Park YJ, Bennetzen JL, Zhang Q, Wang S. Targeting xa13, a recessive gene for bacterial blight resistance in rice. Theor Appl Genet. 2006;112(3):455–461. doi: 10.1007/s00122-005-0145-6. [DOI] [PubMed] [Google Scholar]
- Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, Zhang Q, Wang S. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Gene Dev. 2006;20:1250–1255. doi: 10.1101/gad.1416306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG. The role of haplotypes in candidate gene studies. Genet Epidemiol. 2004;27(4):321–333. doi: 10.1002/gepi.20025. [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin D. Maximum likelihood from incomplete data via the EM algorithm. J Royal Stat Soc B. 1977;39:1–38. [Google Scholar]
- Ding P, Rekhter D, Ding Y, Feussner K, Busta L, Haroth S, Xu S, Li X, Jetter R, Feussner I, Zhang Y. Characterization of a pipecolic acid biosynthesis pathway required for systemic acquired resistance. Plant Cell. 2016;28(10):2603–2615. doi: 10.1105/tpc.16.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS One. 2011;6(5):e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M. Maximum-Likelihood-Estimation of Molecular Haplotype Frequencies in a Diploid Population. Mol Bio Evol. 1995;12(5):921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Garris AJ, McCouch SR, Kresovich S. Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.) Genetics. 2003;165(2):759–769. doi: 10.1093/genetics/165.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi IA, Srivastava PS, Dalal V, Gaikwad K, Singh AK, Sharma TR, Singh NK, Mohapatra T. Physical mapping, expression analysis and polymorphism survey of resistance gene analogues on chromosome 11 of rice. J Bioscience. 2009;34(2):251–261. doi: 10.1007/s12038-009-0029-z. [DOI] [PubMed] [Google Scholar]
- Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS One. 2014;9(2):e90346. doi: 10.1371/journal.pone.0090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanamanickam SS, Brindha V, Priyadarisini NNN, Vasudevan P, Kavitha S. An overview of bacterial blight disease of rice and strategies for its management. Curr Sci. 1999;77(11):1435–1443. [Google Scholar]
- Goudet J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Molecular Ecology Notes. 2005;5:184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- Gu K, Tian D, Yang F, Wu L, Sreekala C, Wang D, Wang GL, Yin Z. High-resolution genetic mapping of Xa27 (t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor Appl Genet. 2004;108:800–807. doi: 10.1007/s00122-003-1491-x. [DOI] [PubMed] [Google Scholar]
- Guo S, Zhang D, Lin X. Identification and mapping of a novel bacterial blight resistance gene xa35(t) originated from oryza minuta. Scientia Agricultura Sinica. 2010;43(13):2611–2618. [Google Scholar]
- Huang N, Angeles ER, Domingo J, Magpantay G, Singh S, Zhang G, Kumaravadivel N, Bennett J, Khush GS. Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor Appl Genet. 1997;95:313–320. doi: 10.1007/s001220050565. [DOI] [Google Scholar]
- Hur Y, Cho J, Park H, Noh T, Park D, Lee JY, Sohn Y, Shin D, Song YC, Kwaon Y, Lee J. Pyramiding of two rice bacterial blight resistance genes, Xa3 and Xa4, and a closely linked cold-tolerance QTL on chromosome 11. Theor Appl Genet. 2016;129:1861–1871. doi: 10.1007/s00122-016-2744-9. [DOI] [PubMed] [Google Scholar]
- Ikeda R, Khush GS, Tabien RE. A new resistance gene to bacterial blight derived from O. longistaminata. Jpn J Breed. 1990;40:280–281. [Google Scholar]
- Iyer AS, Mccouch SR. The Rice Bacterial Blight Resistance Gene xa5 Encodes a Novel Form of Disease Resistance. Mol Plant Microbe In. 2004;17(12):1348–1354. doi: 10.1094/MPMI.2004.17.12.1348. [DOI] [PubMed] [Google Scholar]
- Jia L, Yan W, Zhu C, Agrama H, Jackson A, Yeater K, Li X, Huang B, Hu B, McClung A, Wu D. Allelic analysis of sheath blight resistance with association mapping in rice. PLoS One. 2012;7(3):e32703. doi: 10.1371/journal.pone.0032703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CH, Jung WY, Kang YH, Kim JY, Kim DG, Jeong JC, Baek DW, Jin JB, Lee YK, Kim MO, Chung WS, Mengiste T, Koiwa H, Kwak SS, Bahk JD, Lee SY, Nam JS, Yun DJ, Cho MJ. AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 2006;13(1):84–95. doi: 10.1038/sj.cdd.4401712. [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly M, Eskin E. Efficient Control of Population Structure in Model Organism Association Mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Prod Natl Acad Sci. 2012;24:9653–9658. doi: 10.1073/pnas.1203567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S, Holmström K-O, Mandal A, Dahl P, Hofmann S, Brader G, Palva E, Pirhonen M. AtPTR3, a wound-induced peptide transporter needed for defence against virulent bacterial pathogens in in Arabidopsis. Planta. 2007;225:1431–1445. doi: 10.1007/s00425-006-0451-5. [DOI] [PubMed] [Google Scholar]
- Kauffman HE, Reddy PK, Hiesh SPY, Merca SD. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz D, Tanaka T, Wu J, Zhou S, Childs KL, Davidson MR, Lin H, Quesda-Ocampo L, Viallancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6(4). doi:10.1186/1939-8433-6-4 [DOI] [PMC free article] [PubMed]
- Khush GS, Angeles ER. A new gene for resistance to race 6 of bacterial blight in rice, Oryza sativa L. Rice Genet Newslet. 1999;16:92–93. [Google Scholar]
- Khush GS, Brar DS. Rice genetics from Mendel to functional genomics. Rice Genetics. 2001;4:3–25. [Google Scholar]
- Khush GS, Virk PS. IR varieties and their impact. Los Baños: International Rice Research Institute; 2005. [Google Scholar]
- Khush GS, Bacalangco E, Ogawa T. A New Gene for Resistance to Bacterial Blight from O. longistaminata. Rice Genet Newslet. 1990;7:121–122. [Google Scholar]
- Kim SM, Suh JP, Qin Y, Noh TH, Reinke RF, Jena KK. Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa L.) Theor Appl Genet. 2015;128(10):1933–1943. doi: 10.1007/s00122-015-2557-2. [DOI] [PubMed] [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323(5919):1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- Kretzschmar T, Burla B, Lee Y, Martinoia E, Nagy R. Functions of ABC transporters in plants. Essays Biochem. 2011;50:145–160. doi: 10.1042/bse0500145. [DOI] [PubMed] [Google Scholar]
- Lee KS, Rasabandith S, Angeles ER, Khush GS. Inheritance of resistance to bacterial blight in 21 cultivars of rice. Phytopathology. 2003;93(2):147–152. doi: 10.1094/PHYTO.2003.93.2.147. [DOI] [PubMed] [Google Scholar]
- Li ZK, Sanchez A, Angeles E, Singh S, Domingo J, Huang N, Khush GS. Are the dominant and recessive plant disease resistance genes similar? A case study of rice R genes and Xanthomonas oryzae pv. oryzae races. Genetics. 2001;159(2):757–765. doi: 10.1093/genetics/159.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21(9):2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Liu Q, Yuan M, Zhou Y, LiX XJ, Wang S. A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 2011;34(11):1958–1969. doi: 10.1111/j.1365-3040.2011.02391.x. [DOI] [PubMed] [Google Scholar]
- Lorenz AJ, Hamblin MT, Jannink JL. Performance of single nucleotide polymorphisms versus haplotypes for genome-wide association analysis in barley. PLoS One. 2010;5(11):e14079. doi: 10.1371/journal.pone.0014079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell JM, Woffenden BJ. Plant disease resistance genes: recent insights and potential applications. Trends Biotechnol. 2003;21(4):178–183. doi: 10.1016/S0167-7799(03)00053-2. [DOI] [PubMed] [Google Scholar]
- Mew TW. Current Status and Future Prospects of Research on Bacterial Blight of Rice. Annu Rev Phytopathol. 1987;25:359–382. doi: 10.1146/annurev.py.25.090187.002043. [DOI] [Google Scholar]
- Mew TW, Vera Cruz CM, Medalla ES. Changes in race frequency of Xanthomonas oryzae pv. oryzae in response to rice cultivars planted in the Philippines. Plant Dis. 1992;76:1029–1032. doi: 10.1094/PD-76-1029. [DOI] [Google Scholar]
- Mew TW, Alvarez AM, Leach JE, Swings J. Focus on Bacterial Blight of Rice. Plant Dis. 1993;77:5–12. doi: 10.1094/PD-77-0005. [DOI] [Google Scholar]
- Morris R, Kaplan N. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet Epidemiol. 2002;23(3):221–233. doi: 10.1002/gepi.10200. [DOI] [PubMed] [Google Scholar]
- Mugford ST, Qi X, Bakht S, Hill L, Wegel E, Hughes RK, Papadopoulou K, Melton R, Philo M, Sainsbury F, Lomonossoff GP, Roy AD, Goss RJM, Osbourn A. A Serine Carboxypeptidase-Like Acyltransferase Is Required for Synthesis of Antimicrobial Compounds and Disease Resistance in Oats. The Plant Cell. 2009;21(8):2473–2484. doi: 10.1105/tpc.109.065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucl Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nino-Liu DO, Ronald PC, Bogdanove AJ. Xanthomonas oryzae pathovars : model pathogens of a model crop. Mol Plant Pathol. 2006;7(5):303–324. doi: 10.1111/j.1364-3703.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin H, Andersen T, Burow M, Madsen S, Jørgensen OC, Dreyer I, Hedrich R, Geiger D, Halkier B. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature. 2012;488:532–534. doi: 10.1038/nature11285. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Busto GA, Yamamoto T, Khush GS, Mew TW. Grouping of rice varieties based on reaction to four Philippine races of Xanthomonas campestris pv. oryzae. Rice Genet Newslett. 1986;3:84–86. [Google Scholar]
- Ogawa T, Lin L, Tabien RE, Khush GS. A new recessive gene for resistance to bacterial blight of rice. Rice Genet Newsl. 1987;4:98–100. [Google Scholar]
- Ponciano G, Webb K, Bai J, Vera Cruz C, Leach JE. Molecular characterization of the avrXa7 locus from Xanthomonas oryzae pv. oryzae field isolates. Physiol Mol Plant P. 2004;64(3):145–153. doi: 10.1016/j.pmpp.2004.08.001. [DOI] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens S, Donnelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Xiong L. Characterization of a purine permease family gene OsPUP7 involved in growth and development control in rice. J Integr Plant Biol. 2013;55(11):111–935. doi: 10.1111/jipb.12101. [DOI] [PubMed] [Google Scholar]
- Quibod IL, Perez-Quintero A, Booher NJ, Dossa GS, Grande G, Szurek B, Vera Cruz C, Bogdanove AJ, Oliva R (2016) Effector Diversification Contributes to Xanthomonas oryzae pv. oryzae Phenotypic Adaptation in a Semi-Isolated Environment. Scientific Reports 6:34137. [DOI] [PMC free article] [PubMed]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, Kresovich S, Goodman MM, Buckler ES. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proceedings of the National Academy of Sciences. 2001;98(20):11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer P, Recht S, Lahaye T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc Natl Acad Sci U S A. 2009;106(48):20526–20531. doi: 10.1073/pnas.0908812106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald PC, Albano B, Tabien R, Abenes L, Wu KS, McCouch S, Tanksley SD. Genetic and physical analysis of the rice bacterial blight disease resistance locus, Xa21. Mol Gen Genet. 1992;236(1):113–120. doi: 10.1007/BF00279649. [DOI] [PubMed] [Google Scholar]
- Rotmistrovsky K, Jang W, Schuler GD. A web server for performing electronic PCR. Nucleic Acids Res. 2004;32:108–112. doi: 10.1093/nar/gkh450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-Garcia M, Guevara-Garcia A, Gonzalez-Solis A, Cruz-Garcia F, Vazquez-Santana S, Markham JE, Lozano-Rosas MG, Dietrich CR, Ramos-Vega M, Cahoon EB, Gavilanes-Ruiz M. MPK6, sphinganine and the LCB2a gene from serine palmitoyltransferase are required in the signaling pathway that mediates cell death induced by long chain bases in Arabidopsis. New Phytol. 2011;191(4):943–957. doi: 10.1111/j.1469-8137.2011.03727.x. [DOI] [PubMed] [Google Scholar]
- Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, Nordborg M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet. 2012;44:825–830. doi: 10.1038/ng.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu JS, Khush GS, Mew TW. Genetic analysis of bacterial blight resistance in seventy-four cultivars of rice, Oryza sativa L. Theor Appl Genet. 1978;53(3):105–111. doi: 10.1007/BF00272687. [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen L, Zhai W, Kim HS, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270(5243):1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Sun X, Yang Z, Wang S, Zhang Q. Identification of a 47-kb DNA fragment containing Xa4, a locus for bacterial blight resistance in rice. Theor Appl Genet. 2003;106(4):683–687. doi: 10.1007/s00122-002-1117-8. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GX, Ren X, Weng QM, Shi ZY, Zhu LL, He GC. Mapping of a new resistance gene to bacterial blight in rice line introgressed from Oryza officinalis. Acta Genet Sin. 2004;31:724–729. [PubMed] [Google Scholar]
- Taura S, Ogawa T, Tabien RE, Khush GS, Yoshimura A, Omura T. The specific reaction of Taichung Native 1 to Philippine race of bacterial blight and inheritance of resistance resistance to race 5 (PX0112) Rice Genet Newslett. 1987;4:101–102. [Google Scholar]
- The Rice Chromosomes 11 and 12 Sequencing Consortia The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol. 2005;3(20):1–18. doi: 10.1186/1741-7007-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson MJ, Zhao K, Wright M, McNally KL, Rey J, Tung C-W, Reynolds A, Scheffler B, Eizanga G, McClung A, Kim H, Ismail A, de Ocampo M, Mojica C, Reveche M, Dilla-Ermita C, Mauleon R, Leung H, Bustamante C, McCouch SR. High-throughput single nucleotide polymorphism genotyping for breeding applications in rice using the BeadXpress platform. Mol Breeding. 2012;29(4):875–886. doi: 10.1007/s11032-011-9663-x. [DOI] [Google Scholar]
- Tian D, Wang J, Zeng X, Gu K, Qiu C, Yang X, Zhou Z, Goh M, Lou Y, Murata-Hori M, White FF, Yin Z. The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell. 2014;26(1):497–515. doi: 10.1105/tpc.113.119255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C, Zhao K, Wright MH, Ali ML, Jung J, Kimball J, Scheffler B, Tyagi W, Thomson M, McNally K, Leung H, Kim H, Ahn S-A, Reynolds A, Scheffler B, Eizanga G, McClung A, Bustamante C, McCouch SR. Development of a Research Platform for Dissecting Phenotype – Genotype Associations in Rice (Oryza spp.) Rice. 2010;3:205–217. doi: 10.1007/s12284-010-9056-5. [DOI] [Google Scholar]
- van den Burg HA, Tsitsigiannis DI, Rowland O, Lo J, Rallapalli G, Maclean D, Takken FL, Jones JD. The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell. 2008;20(3):697–719. doi: 10.1105/tpc.107.056978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raden P. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91(11):4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Verdier V, Cruz CV, Leach JE. Controlling rice bacterial blight in Africa: needs and prospects. J Biotechnol. 2011;159(4):320–328. doi: 10.1016/j.jbiotec.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Vilhjalmsson B (2012) ‘mixmogam’ https://github.com/bvilhjal/mixmogam. Commit a40f3e2c95. Accessed 15 April 2016.
- Wang W, Li X, Wing RA, Zhu L, Luo M, Jiang G, Chen X, Li X, Wing RA, Zhu L. Chromosome landing at the bacterial blight resistance gene Xa4 locus using a deep coverage rice BAC library. Mol Genet Genomics. 2001;265(1):118–125. doi: 10.1007/s004380000382. [DOI] [PubMed] [Google Scholar]
- Wang C, Tan M, Xu X, Wen G, Zhang D, Lin X. Localizing the bacterial blight resistance gene, Xa22(t), to a 100-kilobase bacterial artificial chromosome. Phytopatholy. 2003;93(10):1258–1562. doi: 10.1094/PHYTO.2003.93.10.1258. [DOI] [PubMed] [Google Scholar]
- Wang C, Yang Y, Yuan X, Xu Q, Feng Y, Yu H, Wang Y, Wei X. Genome-wide association study of blast resistance in indica rice. Plant Biol. 2014;14(1):311. doi: 10.1186/s12870-014-0311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhang X, Fan Y, Gao Y, Zhu Q, Zheng C, Qin T, Li Y, Che J, Zhang M, Yang B, Liu Y, Zhao K. Xa23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol Plant. 2014 doi: 10.1093/mp/ssu132. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sunderland: Sinauer Associates; 1996. p. 445. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- Wright MH, Tung C-W, Zhao K, Reynolds A, McCouch SR, Bustamante CD. ALCHEMY: a reliable method for automated SNP genotype calling for small batch sizes and highly homozygous populations. Bioinformatics. 2010;26(23):2952–2960. doi: 10.1093/bioinformatics/btq533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Cao Y, Xu C, Li X, Wang S. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. TAG Theor Appl Genet. 2006;113(7):1347–1355. doi: 10.1007/s00122-006-0388-x. [DOI] [PubMed] [Google Scholar]
- Xu X, Chen H, Fujimura T, Kawasaki S. Fine mapping of a strong QTL of field resistance against rice blast, Pikahei-1(t), from upland rice Kahei, utilizing a novel resistance evaluation system in the greenhouse. Theor Appl Genet. 2008;117(6):997–1008. doi: 10.1007/s00122-008-0839-7. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sun X, Wang S, Zhang Q. Genetic and physical mapping of a new gene for bacterial blight resistance in rice. Theor Appl Genet. 2003;106(8):1467–1472. doi: 10.1007/s00122-003-1205-4. [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley J, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38(2):203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Yuan M, Wang S. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant. 2013;6(3):665–674. doi: 10.1093/mp/sst035. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang CL, Zhao KJ, Zhao YL, Vera Cruz CM, Zhu XD, Li DY, Jiang QX. The effectiveness of advanced rice lines with new resistance gene Xa23 to rice bacterial blight. Rice Genet Newslett. 2001;18:71–72. [Google Scholar]
- Zhang Y, Wang J, Pan J, Gu Z, Chen X, Jin Y, Liu F, Zhang H, Ma B. Identification and molecular mapping of the rice bacterial blight resistance gene allelic to Xa7 from an elite restorer line Zhenhui 084. Eur J Plant Pathol. 2009;125(2):235–244. doi: 10.1007/s10658-009-9478-3. [DOI] [Google Scholar]
- Zhang Z, Buckler ES, Casstevens TM, Bradbury PJ. Software engineering the mixed model for genome-wide association studies on large samples. Brief Bioinform. 2009;10(6):664–675. doi: 10.1093/bib/bbp050. [DOI] [PubMed] [Google Scholar]
- Zhang F, Zhuo DL, Zhang F, Huang LY, Wang WS, Xu JL, Vera Cruz C, Li Z-K, Zhou YL. Xa39, a novel dominant gene conferring broad-spectrum resistance to Xanthomonas oryzae pv. oryzae in rice. Plant Pathol. 2015;64(3):568–575. doi: 10.1111/ppa.12283. [DOI] [Google Scholar]
- Zhao K, Tung C, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, McClung AM, Bustamante CD, Mccouch SR. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C-K, Wang C-L, Yu Y-J, Liang Y-T, Zhao K-J. Identification and Molecular Mapping of Xa32(t), a Novel Resistance Gene for Bacterial Blight (Xanthomonas oryzae pv. oryzae) in Rice. Acta Agronomica Sinica. 2009;35(7):1173–1180. [Google Scholar]
- Zhu C, Gore M, Buckler ES, Yu J. Status and Prospects of Association Mapping in Plants. The Plant Genome. 2008;1(1):5–20. doi: 10.3835/plantgenome2008.02.0089. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information on the 285 diverse genotypes used in this study. (XLSX 34 kb)
Phylogenetic and population structure of the 285 diverse panel. a) Radial tree of 285 diverse germplasm based on Tamura-Nei model (MEGA 6). Principal component analysis of b) 285 diverse germplasm c) 248 indica and aus genotypes, and d) 198 indica genotypes (Golden Helix SVS). (PPTX 318 kb)
Genome-wide association analysis of bacterial blight resistance to nine Xoo strains in 248 genotypes based on Efficient Mixed-Model Association eXpedited Model (EMMAX). Manhattan plots for nine Xoo strains (a) PXO61, (b) PXO86, (c) PXO79, (d) PXO71, (e) PXO112, (f) PXO99, (g) PXO339, (h) PXO349, and (i) PXO341. X-axis shows the SNPs along each chromosome; y axis is the − log10 (P-value) for the association. Significant SNPs are those beyond the red line having P-value < 1 × 10 −5. Quantile-quantile plots for nine Xoo strains (j) PXO61, (k) PXO86, (l) PXO79, (m) PXO71, (n) PXO112, (o) PXO99, (p) PXO339, (q) PXO349, and (r) PXO341. (PPTX 720 kb)
Genome-wide association analysis of bacterial blight resistance to nine Xoo strains in 198 indica genotypes based on Efficient Mixed-Model Association eXpedited Model (EMMAX). Manhattan plots for nine Xoo strains (a) PXO61, (b) PXO86, (c) PXO79, (d) PXO71, (e) PXO112, (f) PXO99, (g) PXO339, (h) PXO349, and (i) PXO341. X-axis shows the SNPs along each chromosome; y axis is the − log10 (P-value) for the association. Significant SNPs are those beyond the red line having P-value < 1 × 10 −5. Quantile-quantile plots for nine Xoo strains (j) PXO61, (k) PXO86, (l) PXO79, (m) PXO71, (n) PXO112, (o) PXO99, (p) PXO339, (q) PXO349, and (r) PXO341. (PPTX 521 kb)
Significant GWAS SNP hits that are flanking/overlapping with known resistance genes for bacterial blight. (XLSX 92 kb)
Significant GWAS SNP hits that are potentially novel loci for bacterial blight resistance. (XLSX 80 kb)
Local pair-wise linkage disequilibrium (LD) in GWAS hits overlapping putative novel loci. LD (r2) measure was based on Composite Haplotype Method (CHM). Highly significant SNPs corresponding to Additional file 6: Table S3 are highlighted by black arrows. All SNPs in the region including 8 SNPs upstream of the first significant SNP and 8 SNPs downstream of the last significant SNP are shown.in putatively novel peaks are encircled in red. a) SNPs found in chr6: 21.26–21.96 Mb. b) SNPs found in chr6 22.49–23.23 Mb. c.) SNPs found in chr9: 22.16–22.23 Mb. d) SNPs found in chr11: 6.33 M–6.45 Mb. e) SNPs found in chr11: 20.18–20.50 Mb. f) SNPs found in chr12: 25.69–25.76 Mb. (PPTX 2022 kb)
Linkage Disequilibrium (LD) Decay in GWAS hits overlapping/flanking Xa regions. LD (r2) measure was based on Composite Haplotype Method (CHM). a.) SNPs flanking Xa14 (chr 4: 31–32.9 Mb). b.) SNPs flanking xa5 (chr 5: 200–900 kb). c.) SNPs flanking Xa7 (chr 6: 26–28.6 Mb). d.) SNPs flanking xa13 (chr 8: 25–27.9 Mb). e.) SNPs flanking Xa21 (chr 11: 20.5–22 Mb). f.) SNPs flanking Xa4 (chr 11: 26.5–28.9 Mb). g.) SNPs flanking xa25 (chr 12: 13–17.6 Mb). (PPTX 1768 kb)
Linkage disequilibrium (LD) of adjacent SNPs per chromosome. LD (r2) measure of adjacent pairs was based on Composite Haplotype Method (CHM). (PPTX 324 kb)
Genotype matrix of 248 genotypes across GWAS SNPs flanking/linked to Xa genes. (XLSX 152 kb)
Comparison of allele frequency of SNPs overlapping/flanking Xa genes between GWAS 198 indica genotypes and 1788 indica subset in 3 k genomes. (XLSX 46 kb)
Most significant haplotypes associated with resistance to nine Xoo strains. (XLSX 12 kb)
Comparison of xa5 GWAS SNPs with SNPs derived from 3k Genomes. (XLSX 70 kb)
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.
The raw data is publically available with IRRI: https://s3-ap-southeast-1.amazonaws.com/oryzasnp-atcg-irri-org/pub-data/248Entries_40840SNPs_inorder_21May2015_v2.zip.


