Summary
The pro‐inflammatory cytokine interferon‐γ (IFN‐γ) is critical for activating innate and adaptive immunity against tumours and intracellular pathogens. Interferon‐γ is secreted at the fetal–maternal interface in pregnant women and mice. The outer layer of the placenta in contact with maternal blood is composed of semi‐allogeneic trophoblast cells, which constitute the fetal component of the fetal–maternal interface. The simultaneous presence of pro‐inflammatory IFN‐γ and trophoblast cells at the fetal–maternal interface appears to represent an immunological paradox, for trophoblastic responses to IFN‐γ could potentially lead to activation of maternal immunity and subsequent attack of the placenta. However, our previous studies demonstrate that IFN‐γ responsive gene (IRG) expression is negatively regulated in human and mouse trophoblast cells. In human cytotrophoblast and trophoblast‐derived choriocarcinoma cells, janus kinase signalling is blocked by protein tyrosine phosphatases (PTPs), whereas in mouse trophoblast, histone deacetylases (HDACs) inhibit IRG expression. Here, we used genome‐wide transcriptional profiling to investigate the collective roles of PTPs and HDACs on regulation of IRG expression in human choriocarcinoma cells. Logic‐rules were optimized to derive regulatory modes governing gene expression patterns observed upon different combinations of treatment with PTP and HDAC inhibitors. The results demonstrate that IRGs can be divided into several categories in human choriocarcinoma cells, each of which is subject to distinct mechanisms of repression. Hence, the regulatory modes identified in this study suggest that human trophoblast and choriocarcinoma cells may evade the potentially deleterious consequences of exposure to IFN‐γ by using several overlapping mechanisms to block IRG expression.
Keywords: choriocarcinoma, interferon‐γ signalling, logics, pervanadate, valproic acid
Abbreviations
- P
Pervanadate
- V
Valproic acid
Introduction
The pro‐inflammatory cytokine interferon‐γ (IFN‐γ) plays important roles in diverse cellular processes that include the activation of innate and adaptive immune responses against pathogens and tumours, inhibition of cell proliferation, and induction of apoptosis.1 The functions of IFN‐γ are facilitated through the up‐regulation of over 300 genes.1, 2 Activation of IFN‐γ responsive gene (IRG) expression is mediated through the Janus kinase and signal transducer and activator of transcription 1 (JAK–STAT1) pathway.1, 2 Interaction of IFN‐γ with its cell surface receptor leads to activation of the receptor‐associated kinases JAK1 and JAK2, which phosphorylate monomers of the transcription factor STAT1 that are present in the cytoplasm. Phosphorylated STAT1 (pSTAT1) homodimerizes, translocates to the nucleus, and activates transcription of IRGs that contain γ‐activating sequences in their promoters.1, 2 Expression of IRG is also subject to negative control by multiple distinct molecules that include protein tyrosine phosphatases (PTPs), suppressors of cytokine signalling‐1 (SOCS‐1) and protein inhibitors of activated STAT (PIAS).3 The PTPs repress IFN‐γ signalling both by antagonizing the activities of the JAKs, and dephosphorylating pSTAT1 present in the nucleus. SOCS‐1 also inhibits JAK activity, whereas PIAS family members block pSTAT1‐mediated transcriptional activation.3 These repressor molecules are critical for preventing excessive IRG expression that can lead to deleterious inflammatory reactions at distal sites.
Interestingly, IFN‐γ is secreted at the fetal–maternal interface in pregnant mice and women. In pregnant mice, IFN‐γ is essential for remodelling the spiral arteries within the pregnant uterus to facilitate increased blood flow to the fetus and for maintaining the decidua.4 However, increased levels of IFN‐γ can lead to pregnancy loss in certain strains of mice.5, 6, 7 Furthermore, alterations in IFN‐γ expression have been observed in pre‐eclampsia and recurrent miscarriage in pregnant women.8, 9, 10 Hence, precise modulation of IFN‐γ expression may be critical for successful pregnancy.
The outer layer of the human placenta in direct contact with maternal blood and tissues is composed of semi‐allogeneic trophoblast cells, which perform numerous functions that are critical for successful pregnancy, such as gas, nutrient and waste exchange, hormone production, and formation of a protective barrier against infection and attack by the maternal immune system.11 There are several subpopulations of human trophoblast, including the syncytiotrophoblast, a continuous, multinucleate layer that is in contact with maternal blood within the intravillous space, and the extravillous trophoblast, which invade into the uterine endothelium and play a role in remodelling the uterine spiral arteries.11 The syncytiotrophoblast and extravillous trophoblast are both derived from villous cytotrophoblast cells.
The simultaneous presence of semi‐allogeneic trophoblast cells and pro‐inflammatory IFN‐γ at the fetal–maternal interface appears to represent an immunological conundrum, for trophoblastic responses to IFN‐γ could potentially lead to activation of the maternal immune system, and subsequent attack of the placenta. We previously demonstrated that human trophoblast‐derived choriocarcinoma cells and term villous cytotrophoblasts are hyporesponsive to IFN‐γ due to compromised activation of the JAK–STAT pathway, suggesting that trophoblast cells have evolved mechanisms for circumventing potentially deleterious consequences of exposure to IFN‐γ.12 Co‐treatment of choriocarcinoma cells with IFN‐γ and the PTP inhibitor pervanadate (P) resulted in enhanced STAT1 phosphorylation and significantly increased expression of IRGs such as IRF1 and guanylate‐binding proteins (GBPs) relative to IFN‐γ treatment alone.12 However, expression of the IRG LMP7 was not substantially changed by IFN‐γ and P co‐treatment of choriocarcinoma cells, suggesting that some IRGs may be subject to additional levels of control in these cells. A subsequent study revealed that the JAK–STAT1 pathway is activated by IFN‐γ in mouse trophoblast cells, but transcriptional activation of the IRGs is blocked by histone deacetylases (HDACs).13
In the current study, we investigated the collective roles of PTPs and HDACs on regulation of IRG expression in human choriocarcinoma cells by genome‐wide transcriptional profiling. Logic‐rules were optimized to derive rules governing gene expression patterns observed upon different combinations of treatment with PTP and HDAC inhibitors. The data reveal that IRGs can be divided into distinct subsets that are differentially modulated by co‐treatment of Jar cells with IFN‐γ and PTP versus HDAC inhibitors, respectively. Furthermore, promoter analysis of the genes governed by the rules identifies transcription factor binding sites associated with the different gene subsets. Hence, the regulatory modes identified in this study provide insights into the complex regulation of inflammatory pathways at the fetal–maternal interface, as well as mechanisms that choriocarcinoma cells may use to promote their survival.
Methods
Cell culture and reagents
Jar choriocarcinoma and HeLa cervical carcinoma cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured as previously described.12 Human IFN‐γ (I) was purchased from PBL Biomedical Laboratories (Piscataway, NJ) and used at a concentration of 200 U/ml. Sodium orthovanadate (S6508 450243), hydrogen peroxide (31642) and bovine liver catalase (C1345) were purchased from Sigma‐Aldrich (St Louis, MO). Pervanadate (P) was generated as previously described and used at a concentration of 100 μm.12 Valproic acid (V) was purchased from Calbiochem (San Diego, CA), reconstituted in water and used at 5 mm. Briefly, 1·5 million Jar cells and 2 million HeLa cells were plated on 60 mm2 dishes, and 24 hr later, the cells were treated with IFN‐γ or the drug combinations. After 16 hr, the treatment cells were harvested in Trizol. The above procedure was repeated four times.
RNA isolation and RNA seqencing
RNA was isolated from four sets of experiments using TRIzol (Invitrogen Life Technologies, Carlsbad, CA) as specified by the manufacturer. RNA concentrations were determined with the NanopDrop 1000 spectrophotometer (NanoDrop, Wilmington, DE) and RNA quality was assessed with the Agilent Bioanalyzer (Agilent, Santa Clara, CA). The TruSeq RNA Sample Preparation Kit V2 (Illumina, San Diego, CA) was used for next‐generation sequencing library construction per the manufacturer's protocols. Briefly, mRNA was purified from 100 ng total RNA with oligo‐dT magnetic beads and fragmented. First‐strand cDNA synthesis was performed with random hexamer priming followed by second‐strand cDNA synthesis. End repair and 3’ adenylation were then performed on the double‐stranded cDNA. Illumina adaptors were ligated to both ends of the cDNA, which was then purified by gel electrophoresis and amplified with PCR primers specific to the adaptor sequences to generate amplicons of approximately 200–500 bp in size. The amplified libraries were hybridized to the Illumina single end flow cell and amplified using the cBot (Illumina, San Diego, CA) at a concentration of 8 pm per lane. Single end reads of 100 nt were generated for each sample and aligned to the organism‐specific reference genome. The raw and cleaned sequence data are available in NCBI's Sequence Read Archive (SRA) (id: SRP095402).
RNA‐sequencing data analysis and inference of logic rules
RNA‐Seq fastq14 data were mapped to a reference genome obtained from ensembl 15 by tophat (v2.1.0)16 using the high‐throughput short read aligner bowtie (v1.01).17 The htseq‐count script from htseq (v0.6.1) was then used to generate raw count data from the accepted hits output of tophat upon its conversion into a .sam file using samtools (v0.1.19).18 The Bioconductor package biomart (v2.27.2)19 was used for conversion from ensemble gene IDs to official gene symbols. The Bioconductor package DEseq2 (v1.10.1) was used to perform differential sequence analysis.
The differential sequence analysis of raw count data from four experiments for each treatment was performed relative to the untreated controls. Genes were considered differentially expressed if q‐value (P‐value corrected for multiple testing) < 0·05 and a log2 |fold change| ≥ 1. Volcano plots and heat maps describing the data were generated using ggplots2 (v2.0.0).20
Inference of logic rules
To investigate the impact of different combinations of treatments and interactions between them, we decided to use algebraic logic where relationship between inputs (different treatments) and output (gene expression) can be defined by logic rules. Typically, differential expression of the genes is represented by −1 (down‐regulated), 0 (not differentially expressed) or 1 (up‐regulated) in response to the treatment. In this study, gene expression was studied in the case of five treatments namely I, V, P, IV and IP. If we assume that the effect of joint treatments on the gene expression is independent of single treatment there would be 35 = 243 possible gene‐expression patterns. These 243 bit strings were enumerated using the balanced ternary system and were mapped to the observed differential expression pattern. The unique patterns were translated into signed logic rules to improve interpretation of the truth‐tables.
Transcription factor target identification
The table mapping genes to transcription factor binding sites (TFBS) was created in R using the databases ‘JASPAR_CORE’ and ‘JASPAR_2014’ from JASPAR.21 JASPAR databases were accessed using the Biocondcutor package motifDb.22 The UCSC hg19, mm10 and rn6 genomes were obtained using the bioconductor package bsgenome.hsapiens.ucsc.hg19,23 bsgenome.mmusculus.ucsc.mm10 23 and bsgenome.rnorvegicus.ucsc.rn6,24 respectively. The Bioconductor package genomicfeatures 25 was used to extract official gene symbols and the sequence 2 kb upstream and downstream from the promoter. The function matchPWM found in the Bioconductor package biostrings 26 was used to perform the match algorithm on a given PWM (position weight matrix) from JASPAR and a gene's 2 kb upstream or downstream promotor sequence for each TFBS and each gene. A cut‐off value of 85% was used for matchPWM function. The genes regulated by the same TFBSs across three species were used for the enrichment analysis. The enrichment analysis was performed using Hypergeometric test and P‐value corrected using the Benjamini & Hochberg method.27
Results
Global characterization of suppression of IFN‐γ responsive genes in Jar cells
Previous work demonstrated that Jar choriocarcinoma cells are hypo‐responsive to IFN‐γ due to impaired activation of the JAK–STAT1 pathway.12 To completely characterize IRGs, genome‐wide transcriptional profiling using RNA‐sequencing was performed following stimulation of Jar and HeLa cells for 16 hr with 200 U/ml IFN‐γ in four separate experiments. HeLa cells were used as positive control because of their ability to induce a classical IFN‐γ‐induced response as documented previously.12, 13, 27 In HeLa cells, there were 364 and 35 up‐regulated and down‐regulated IRGs, respectively. As expected from previous observations, IFN‐γ treatment of Jar cells modulated very few genes. Specifically, 45 IRGs were up‐regulated in Jar cells, and none were down‐regulated (Fig. 1a). Of the 45 genes, only two, ADAMTS6 and FST, were differentially expressed/up‐regulated in Jar cells but not in HeLa cells, suggesting cell‐specific expression patterns. Indeed, FST produces Follistatin, a single‐chain gonadal protein that specifically inhibits follicle‐stimulating hormone release.28 Note that the 43 genes up‐regulated in both Jar and HeLa cells exhibited significantly higher fold changes in HeLa cells compared with Jar cells, suggesting that expression of all IRGs is suppressed in Jar cells (Fig. 1b).
Figure 1.
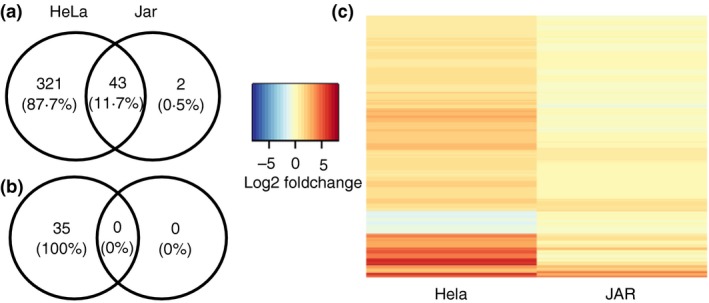
Jar cells suppress interferon‐γ (IFN‐γ) responsive genes compared with HeLa cells: The number of (a) up‐regulated and (b) down‐regulated genes upon IFN‐γ stimulation of HeLa and Jar cells are represented by a Venn diagram. (c) Heatmap representing mean of Log2 (Fold change) of differentially expressed genes in IFN‐γ stimulated HeLa and Jar cells relative to the unstimulated cells. Colourmap ranging from blue to red represents low to high mean of the fold‐change from four independent experiments. Differential expression compared four IFN‐γ‐stimulated samples with four unstimulated samples and was defined as a mean of the absolute fold change of at least two relative to the unstimulated cells and a significant (q < 0·05) change in expression by DESeq2.
Alleviation of suppression of IFN‐γ responsive genes upon inhibition of protein tyrosine phosphatases and histone deacetylases
To comprehensively evaluate the relative roles(s) of PTPs and HDACs on IRG expression in choriocarcinoma cells, genome‐wide transcriptional profiling using RNA‐sequencing was performed after culturing Jar cells for 16 hr in the presence and absence of 200 U/ml IFN‐γ, in conjunction with the PTP inhibitor Pervanadate (P) and the histone deacetylase inhibitor Valproic acid (V). Differential expression (DE) analysis identified 5635 genes from four comparisons between different combinations of treatments compared with the untreated control (Fig. 2).
Figure 2.
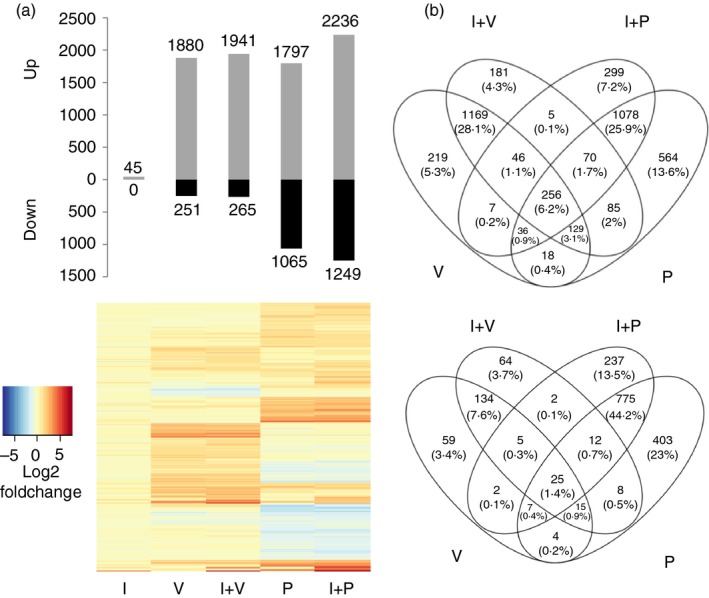
The transcription response of Jar cells upon inhibition of protein tyrosine phosphatases (PTPs) and histone deacetylases (HDACs): (a) Differentially expressed genes (upper panel: absolute fold change, ≥ 2; false discovery rate < 0·05) and their fold changes (lower panel) were measured for each treatment by comparing their levels of expression (x‐axis) with unstimulated cells (horizontal axis). Colour map ranging from blue to red depicts low to high log2 (fold change). (b) Venn diagram showing overlap between the up‐regulated (upper panel) and down‐regulated (bottom panel) genes upon treatments with V (valproic acid, which is an HDAC inhibitor), I (human interferon‐γ) +V, P (pervanadate, which inhibits PTPs) and I+P.
There was a strong overlap between genes induced by the inhibitors (V or P) and by inhibitors + IFN‐γ co‐treatments. Interestingly, there was minimal overlap between genes altered by IFN‐γ + V co‐treatment versus IFN‐γ + P co‐treatment (1·7% up‐regulated and 0·7% down‐regulated), suggesting that distinct mechanisms are involved in the control of IRGs in Jar cells. Overlap across all four conditions was 6·2% and 1·4% in up‐regulated and down‐regulated genes, respectively. V treatment led to more up‐regulation (88% out of the total number of differentially expressed genes by V and V+I) of the genes than down‐regulation, compared with P treatment (63%) (Fig. 2b).
Among 399 DE IRGs in HeLa, 94 and 85 genes were differentially expressed by treatment with V or P alone, respectively. Stimulation with IFN‐γ along with the V or P increased these numbers to 173 (43%) and 229 (57%), respectively. However, inhibitor treatment did not lead to modulation of all 399 genes, indicating that the suppression observed in Jar cells was not completely alleviated by these inhibitors. Moreover, nine IRGs that were up‐regulated in HeLa were down‐regulated in Jar (e.g. PEG10, TNFRSF10D) and 11 IRGs that were down‐regulated in HeLa were up‐regulated in Jar (e.g. FOSL1, ASAP3).
Regulatory modes induced by IFN‐γ stimulation in Jar cells
To investigate the regulatory modes in Jar cells logic rules were written to describe the up‐/down‐regulation patterns of 5635 DE genes (refer to Materials and methods section). The responses of HeLa IRGs across five treatments in Jar cells were defined by 30 logic rules (Fig. 3). Two rules, f 5 and f 7 each governing only one gene, suggest inhibition of IFN‐γ‐mediated stimulation by P and V, respectively. Particularly, IFN‐γ‐mediated activation of Toll‐like receptor 3 and interferon regulatory factor 9 (IRF9) was inhibited by P and V, respectively, in Jar cells. Suppression of IRF9 expression following treatment with V is in line with the known inhibitory role of STAT1 acetylation.29
Figure 3.
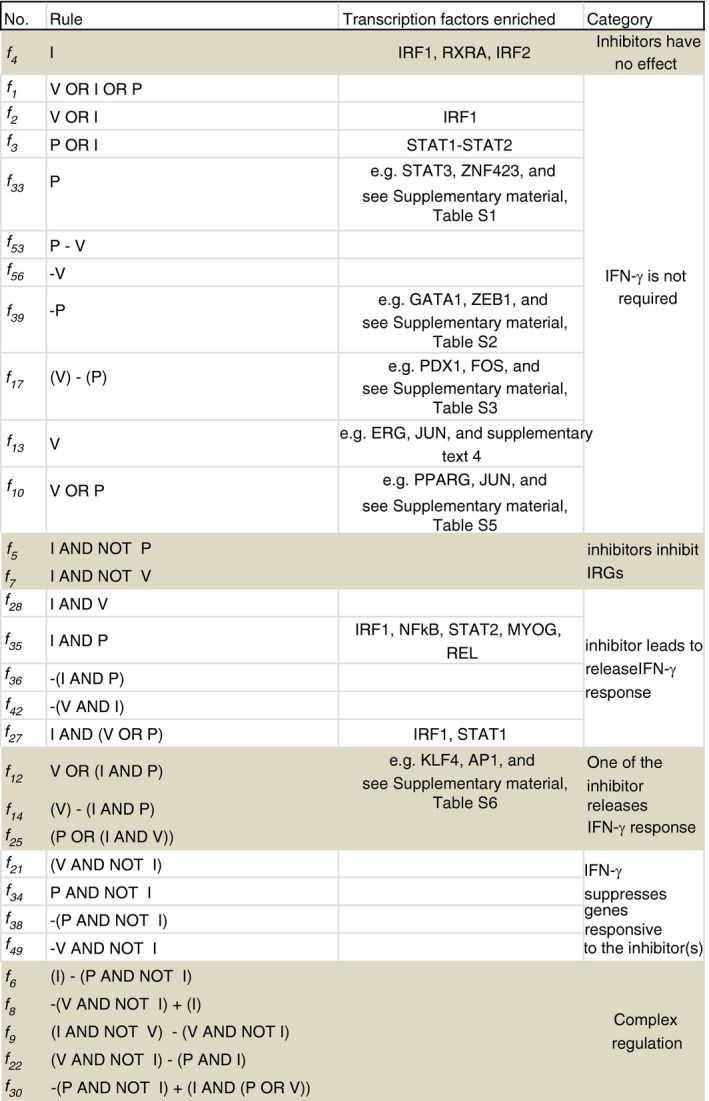
Regulatory modes induced by interferon‐γ (IFN‐γ) upon treatments with the inhibitors: Rules (second column) describing differential expression patterns of 399 HeLa IFN‐γ responsive gene (IRGs) upon stimulation of Jar cells with human interferon‐γ (I), valproic acid (V), pervanadate (P), I+V and I+P, and binding sites of the transcription factors (third column) enriched in the gene clusters. [Colour figure can be viewed at wileyonlinelibrary.com]
Interestingly, 10 rules indicate that inhibitors alone in the absence of IFN‐γ can modulate the expression of IRGs in Jar cells. These responses are regulated by several transcription factors such as IRF1, PPARG (Peroxisome proliferator‐activated receptor gamma). Five rules represent dependence on IFN‐γ, even in the presence of the inhibitors, suggesting that V and P can indeed alleviate the suppression of IRGs as suggested previously.30 However, the absence of significant enrichment of transcription factor binding sites in most of these gene clusters suggests that the regulatory modes are not driven by known transcription factors. It is possible that multiple transcription factors might be involved in regulation of these genes. The exceptions were IRF1 or STAT1 regulated f 27 and STAT1‐STAT2 regulated f 35. The f 35 rule represents genes requiring stimulation by IFN‐γ in addition to treatment by P, suggesting a critical role of IFN‐γ. Indeed, several well‐known IRGs such as STAT2, CXCL family genes, and TLR genes are regulated by rule f 35. Interestingly, the negative regulator SOCS1, but not SOCS3 or SOCS6, is also governed by f 35, suggesting its specific activation by IFN‐γ.
Three rules identify IFN‐γ‐dependent and independent modes of regulation induced by the two inhibitors. For example, f 14 defines regulation of 48 genes, e.g. cAMP‐dependent protein kinase inhibitor and zinc finger protein 540, which are up‐regulated by V alone and are down‐regulated by a joint treatment with IFN‐γ and P. Finally, four rules suggest that IFN‐γ suppresses the effect of one inhibitor but stimulates the modulation by the other inhibitor. For example, rule f 31 governs genes that are down‐regulated in the presence of IFN‐γ and P, but are up‐regulated by IFN‐γ and V. However, these clusters of mixed regulation were too small for further promoter analysis.
Systems level regulation by IFN‐γ, P and V in Jar cells
The patterns of all the DE genes identified in Jar cells upon comparison of the gene expression observed by combinations of the treatments with the untreated control could be defined by 59 rules (Fig. 4a). However, only four rules were necessary to describe 50% of the 5635 DE genes. These rules are: f 33 = P (1051 genes), f 13 = V (974 genes), f 39 = −P (659 genes) and f 35 = I AND P (556 genes). Three out of the four rules were independent of IFN‐γ. Exclusive treatment with inhibitors led to differential expression of 2684 genes, suggesting that an increase in phosphatase and deacetylase activities up‐regulate these genes. This is not surprising since the inhibitors used in this study block the activities of all PTPs and nuclear HDACs. Interestingly, 25 and 5 HeLa IRGs were governed by f 33 and f 39, respectively, suggesting that the induction of these genes is suppressed in Jar cells because of limited availability of phosphorylated signalling molecules.
Figure 4.
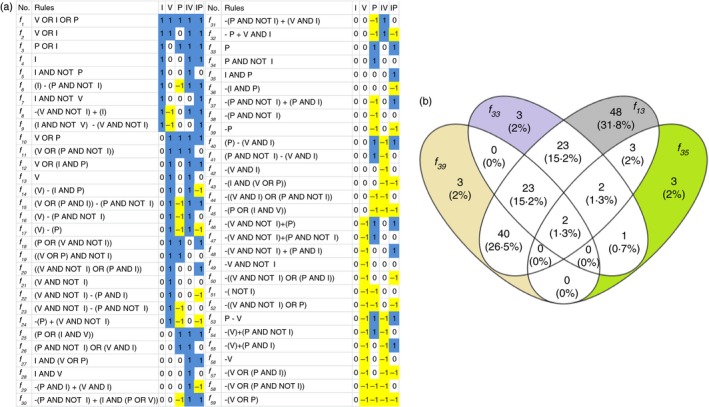
System‐wide optimization of logic rules and promoter analysis: (a) All 59 rules (second column) describing differential expression patterns (blue/1 = up‐regulation, white/0 = no significant difference and yellow/−1 = down‐regulation) of 5635 genes upon stimulation with human interferon‐γ (I), valproic acid (V), pervanadate (P), I+V and I+P (horizontal axis). (b) f 13, f 33, f 35 and f 39 governed 50% of the differentially expressed genes. The promoter analysis reveals enrichment of several transcription factor binding sites. The distinct patterns described by the f 13, f 33, f 35 and f 39 rules are expected to be governed by a subset of transcription factors unique to those gene clusters, and are depicted by grey, purple, green and yellow colours look different, not the ones described colours, respectively. [Colour figure can be viewed at wileyonlinelibrary.com]
Promoter analysis of the genes governed by the f 33 and f 39 rules was performed to identify transcription factors regulating the genes. Spil, SRF and SREBF2, were enriched in f 33 governed genes, while Foxd3, Gfilb and Ddit3 were enriched in f 39 governed genes (Fig. 4b). Acetylation of key transcription factors is known to have positive and negative roles on the IFN‐γ induced signalling pathway.29, 31 f 13, a rule governing regulation of the second largest group of genes, was exclusively induced by HDAC inhibition, and was enriched for 48 transcription factor binding sites (see Supplementary material, Table S4). Enrichment of many transcription factors suggests that HDAC inhibitors regulate genes by different mechanisms than by modulation through a limited number of specific transcription factors.
Signal processing through gene clusters
To investigate the signal processing through the 59 rules (Fig. 4a) we simplified the complex logic rules by replacing those with simpler rules, for example f 20 = ((V AND NOT I) OR (P AND I)) was simplified to f 20 = f 21 OR f 35, where f 21 = V AND NOT I and f 35 = I AND P. The gene‐regulatory network shown in Fig. 5 emerged, representing collaborations between gene modules governed by different rules. f 34, f 28, f 35 and f 21 had highest betweenness centrality, suggesting a strong influence of those gene modules in transferring the signal from I, V and P. Furthermore, the target node (nodes having no outgoing edge) f 30 = −(P AND NOT I) + (I AND (P OR V) and f 15 = (V OR (P AND I)) − (P AND NOT I) had the highest closeness centrality. The strong closeness centrality could be explained by their complex regulation leading to a smaller distance from most other nodes in the network. Furthermore, 12 critical rules were identified as master regulators that could replace all 59 rules (Fig. 6a). f 2, f 3 and f 4 were significantly enriched with targets regulated by IRF1, STAT1–STAT2 complex and IRF1, IRF2 and RXRA transcription factors, respectively. The f 2 rule suggests that the expression of IRF1 targets is regulated by HDACs. f 5, f 7, f 21, f 28 and f 34 were not enriched for any transcription factors. f 13, f 10, f 33 and f 35 were enriched for several transcription factors, which have been described in the previous sections. In general, treatment with V alone induced 49 specific transcription factors (Fig. 6b). In contrast, f 10, f 33 and f 35 induced fewer specific transcription factors (depicted in Fig. 6b).
Figure 5.
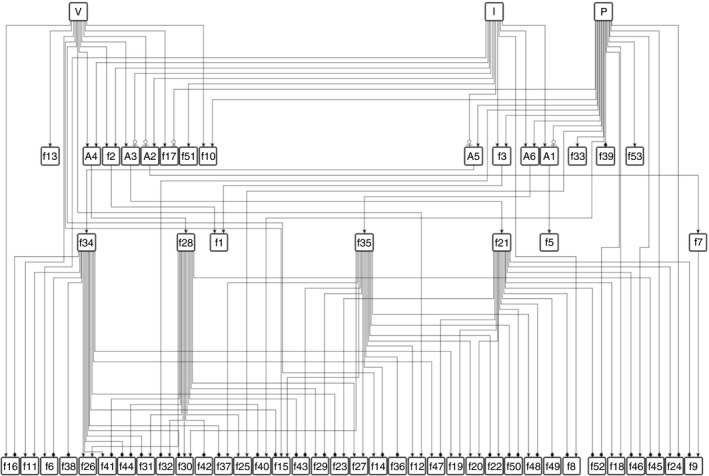
Gene‐regulatory network: interactions between gene modules is represented by a hierarchical network where node names with prefix f represent rules described in Fig. 3 and prefix A represent ‘AND’ logic. Edges with directional arrows represent  : up‐regulation,
: up‐regulation,  : inhibition and
: inhibition and  : down‐regulation.
: down‐regulation.
Figure 6.
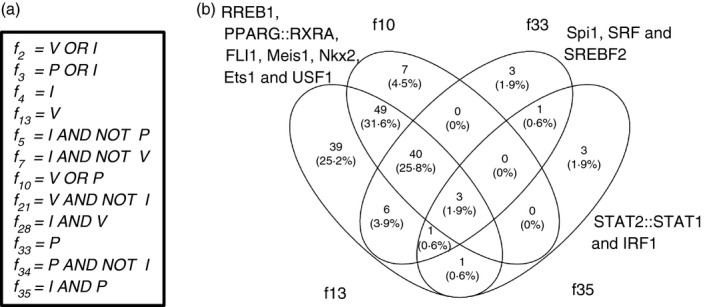
Transcription factors regulating master rules: (a) Twelve master rules were identified that could replace all the 59 rules. (b) Four of these 12 rules showed enrichment of transcription factors, indicating their critical roles in signal processing.
Discussion
Interferon‐γ‐mediated activation of the JAK–STAT1 pathway is blocked in human trophoblast‐derived choriocarcinoma cells and term villous cytotrophoblast cells, resulting in compromised phosphorylation of STAT1 and expression of IRGs.12 Pervanadate, the PTP inhibitor, enhanced IFN‐γ‐mediated STAT1 phosphorylation and significantly increased expression of the IRGs IRF1 and GBP in Jar choriocarcinoma cells, but not LMP7.12 These results suggest that additional mechanism(s) may be involved in repression of different IRGs in choriocarcinoma cells. In this study, we addressed this hypothesis by performing genome‐wide transcriptional profiling on RNAs from Jar cells treated with IFN‐γ alone, or in combination with PTP and HDAC inhibitors. The RNA seqencing data support the hypothesis that several overlapping mechanisms are involved in controlling IFN‐γ‐inducible gene expression in Jar cells.
Inference of standard logic rules provided mechanistic insights into the regulation of genes, specifically IRGs in Jar cells by P and V. f 35 = I AND P emerges as the most critical rule governing IRGs, that ranks fourth on the number of genes governed. It is also one of the rules with highest betweenness centrality, suggesting that it has critical role in regulating the downstream gene modules. The promoter analysis of the f 35 module reveals several transcription factors regulated by PTPs such as NFkB1, IRF1 and STAT2. A similar rule, f 28, described genes regulated by V, but they were fewer in number (166). However, the promoters of the affected genes were not enriched for binding sites of any specific transcription factors, suggesting that diverse molecular mechanisms might be underlying the regulation of IRGs by V.
The interactions between PTPs and HDACs in the regulation of IRGs and other genes are complex. Several regulatory modes are in consensus with previous observations by Kramer et al. showing that acetylation of STAT1 counteracts IFN‐induced STAT1 phosphorylation, nuclear translocation, DNA binding, and target gene expression.29 For example, f 32 governs a set of IRGs that is expressed upon co‐treatment with I and V, but is down‐regulated by treatment with P. Several subsets of genes are suppressed by treatment with I even in the presence of the inhibitors. Future studies will reveal if this suppression is specific to Jar cells, further supporting selective responsiveness to IFN‐γ. Regulation of gene expression could be mediated by differential binding of the co‐factors. Methylation of the respective promoters may play a role in repression of these genes because previous studies suggest extensive alterations in the patterns of DNA methylation in Jar and JEG‐3 choriocarcinoma cells.32 Alternatively, the current study reveals 19 regulatory modes of Zinc finger proteins, which are a dominant class of co‐factors that can bind to nucleic acid.33 Moreover, genes regulated by ZFP423 and/or ZNF354C were enriched in modules governed by f 10, f 12, f 13, f 16, f 17, f 33 and f 39. Interestingly, most of these modules showed up‐regulation driven by V.
Both phosphorylation and acetylation are versatile modulators of protein functions and interactions.34 Our promoter analysis reveals that V induces genes induced by wide range of transcription factors, which could be due to the influence of HDACs on chromatin conformation facilitating easier access to the transcription factors.35 These results suggest that the reduced expression of IRGs in Jar cells is, at least to some extent, regulated by the inefficient recruitment of transcription factors or co‐factors to target promoters. In contrast, all clusters responsive to P and I show enrichment of at least one of the transcription factors implicated in the activation of IRGs. As expected from the broad action of P and V in inhibition of PTPs and HDAC, respectively, both treatments stimulate several transcription factors on their own.
The current study corroborates previous observations made by PCR.12, 13 For example, IFN‐γ‐induced stimulation of genes such as GBP require inhibitors for up‐regulation in Jar cells.12, 13 The GBPs are governed by different rules, including f 27 and f 35. In addition, the CASP1, CASP8 and CASP9 genes, which are necessary for induction of apoptosis, are only modulated in Jar cells upon co‐treatment with I and P (rule f 35). In contrast, CASP7 is induced upon treatment with I even in the absence of the inhibitors (rules f 4). These results are consistent with our observation that co‐treatment of Jar cells with I and P, but not I alone, resulted in apoptosis. Additionally, the current study reveals rules of regulation for several previously unstudied genes.
Klampfer et al. previously demonstrated that HDAC inhibitors (HDACi) blocked IFN‐γ‐inducible JAK1 and STAT1 phosphorylation, and activation of IRF1 expression in human colorectal carcinoma cells, but STAT1, STAT2 and CASP7 were not affected.36 Similar effects of HDACi were observed on type I IFN‐inducible gene expression in a number of different cell types.37, 38, 39 These results directly contrasted with studies of mouse trophoblast cells, in which HDACi enhanced IFN‐γ‐inducible IRF1 expression.13 The present study confirms that IFN‐γ‐induced IRF1 mRNA expression is repressed in Jar trophoblast‐derived choriocarcinoma cells, but suppression can be alleviated by both P and the HDACi V. Collectively, these observations support the hypothesis that trophoblast cells use distinct mechanisms for regulating IFN‐γ‐responsive gene expression.
Interferon‐γ plays an integral role in immunity against many types of tumours, particularly in the immunoediting phase, during which the host immune system keeps developing tumours in check.40 Subsequent tumour progression is associated with down‐regulation of tumour responses to IFN‐γ, which ultimately enables tumours to escape host immunosurveillance.40 The current study demonstrates that choriocarcinoma cells use multiple mechanisms to block IFN‐γ‐inducible gene expression, which suggests that these tumour cells may be especially adept at evading host immune responses, particularly as they lack the expression of classical MHC molecules.41 Repression of IFN‐γ responsiveness may also be essential for semi‐allogeneic trophoblast cells to evade maternal immunity, as this phenotype was also observed in normal term, villous cytotrophoblast cells.12
Disclosures
There are no competing interests to declare.
Supporting information
Table S1. Transcription factor binding sites enriched in the gene module regulated by f 33 rule.
Table S2. Transcription factor binding sites enriched in the gene module regulated by f 39 rule.
Table S3. Transcription factor binding sites enriched in the gene module regulated by f 17 rule.
Table S4. transcription factor binding sites enriched in the gene module regulated by f 13 rule.
Table S5. Transcription factor binding sites enriched in the gene module regulated by f 10 rule.
Table S6. Transcription factor binding sites enriched in the gene module regulated by f 12 rule.
Acknowledgements
This work was supported by funds from the National Institute of Child Health and Development (R01 HD056183) and the Richard and Mae Stone Goode Foundation to S.P.M, and by an PhRMA informatics research starter award to J.T. We would also like to thank the staff of the University of Rochester Genomics Research Center (URGRC) for their expert technical assistance.
References
- 1. Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR et al Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov 2007; 6:975–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Platanias LC. Mechanisms of type‐I‐ and type‐II‐interferon‐mediated signalling. Nat Rev Immunol 2005; 5:375–86. [DOI] [PubMed] [Google Scholar]
- 3. Shuai K, Liu B. Regulation of JAK‐STAT signalling in the immune system. Nat Rev Immunol 2003; 3:900–11. [DOI] [PubMed] [Google Scholar]
- 4. Ashkar AA, Croy BA. Interferon‐γ contributes to the normalcy of murine pregnancy. Biol Reprod 1999; 61:493–502. [DOI] [PubMed] [Google Scholar]
- 5. Mattsson R, Mattsson A, Holmdahl R, Scheynius A, Van der Meide PH. In vivo treatment with interferon‐gamma during early pregnancy in mice induces strong expression of major histocompatibility complex class I and II molecules in uterus and decidua but not in extra‐embryonic tissues. Biol Reprod 1992; 46:1176–86. [DOI] [PubMed] [Google Scholar]
- 6. Clark DA, Chaouat G, Arck PC, Mittruecker HW, Levy GA. Cytokine‐dependent abortion in CBA × DBA/2 mice is mediated by the procoagulant fgl2 prothrombinase [correction of prothombinase]. J Immunol 1998; 160:545–9. [PubMed] [Google Scholar]
- 7. Clark DA, Manuel J, Lee L, Chaouat G, Gorczynski RM, Levy GA. Ecology of danger‐dependent cytokine‐boosted spontaneous abortion in the CBA × DBA/2 mouse model. I. Synergistic effect of LPS and (TNF‐α + IFN‐γ) on pregnancy loss. Am J Reprod Immunol 2004; 52:370–8. [DOI] [PubMed] [Google Scholar]
- 8. Kumar A, Begum N, Prasad S, Agarwal S, Sharma S. IL‐10, TNF‐α & IFN‐γ: potential early biomarkers for preeclampsia. Cell Immunol 2013; 283:70–4. [DOI] [PubMed] [Google Scholar]
- 9. Sargent IL, Borzychowski AM, Redman CW. Immunoregulation in normal pregnancy and pre‐eclampsia: an overview. Reprod Biomed Online 2006; 13:680–6. [DOI] [PubMed] [Google Scholar]
- 10. Moore LG, Shriver M, Bemis L, Hickler B, Wilson M, Brutsaert T et al Maternal adaptation to high‐altitude pregnancy: an experiment of nature – a review. Placenta 2004; 25(Suppl. A):S60–71. [DOI] [PubMed] [Google Scholar]
- 11. Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol 2006; 6:584–94. [DOI] [PubMed] [Google Scholar]
- 12. Choi JC, Holtz R, Petroff MG, Alfaidy N, Murphy SP. Dampening of IFN‐γ‐inducible gene expression in human choriocarcinoma cells is due to phosphatase‐mediated inhibition of the JAK/STAT‐1 pathway. J Immunol 2007; 178:1598–607. [DOI] [PubMed] [Google Scholar]
- 13. Choi JC, Holtz R, Murphy SP. Histone deacetylases inhibit IFN‐γ‐inducible gene expression in mouse trophoblast cells. J Immunol 2009; 182:6307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res 2010; 38:1767–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S et al Ensembl 2014. Nucleic Acids Res 2014; 42(Database issue):D749–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA‐Seq. Bioinformatics 2009; 25:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009; 4:1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer‐Verlag, 2009. [Google Scholar]
- 21. Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley‐Hunt R, Arenillas DJ et al JASPAR 2014: an extensively expanded and updated open‐access database of transcription factor binding profiles. Nucleic Acids Res 2014; 42(Database issue):D142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shannon P. MotifDb: An Annotated Collection of Protein‐DNA. Binding Sequence Motifs. R package version 1.10.0, 2014.
- 23. Team TBD. BSgenome.Mmusculus.UCSC.mm10: Full genome sequences for Mus musculus (UCSC version mm10). R package version 1.4.0, 2014.
- 24. Team TBD. BSgenome.Rnorvegicus.UCSC.rn6: Full genome sequences for Rattus norvegicus (UCSC version rn6). R package version 1.4.0, 2014.
- 25. Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R et al Software for computing and annotating genomic ranges. PLoS Comput Biol 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pages H, Aboyoun P, Gentleman R, DebRoy S. Biostrings: String objects representing biological sequences, and matching algorithms. R package version 2364, 2016.
- 27. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series 1995; B57:289–300. [Google Scholar]
- 28. Shimasaki S, Koga M, Esch F, Cooksey K, Mercado M, Koba A et al Primary structure of the human follistatin precursor and its genomic organization. Proc Natl Acad Sci USA 1988; 85:4218–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH et al A phosphorylation‐acetylation switch regulates STAT1 signaling. Genes Dev 2009; 23:223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lamb P, Haslam J, Kessler L, Seidel HM, Stein RB, Rosen J. Rapid activation of the interferon‐γ signal transduction pathway by inhibitors of tyrosine phosphatases. J Interferon Res 1994; 14:365–73. [DOI] [PubMed] [Google Scholar]
- 31. Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene 2007; 26:5541–52. [DOI] [PubMed] [Google Scholar]
- 32. Novakovic B, Gordon L, Wong NC, Moffett A, Manuelpillai U, Craig JM et al Wide‐ranging DNA methylation differences of primary trophoblast cell populations and derived cell lines: implications and opportunities for understanding trophoblast function. Mol Hum Reprod 2011; 17:344–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol 2001; 11:39–46. [DOI] [PubMed] [Google Scholar]
- 34. Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell 2002; 111:771–8. [DOI] [PubMed] [Google Scholar]
- 35. Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkuhler C. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res 2007; 17:195–211. [DOI] [PubMed] [Google Scholar]
- 36. Klampfer L, Huang J, Swaby LA, Augenlicht L. Requirement of histone deacetylase activity for signaling by STAT1. J Biol Chem 2004; 279:30358–68. [DOI] [PubMed] [Google Scholar]
- 37. Nusinzon I, Horvath CM. Interferon‐stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci USA 2003; 100:14742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chang HM, Paulson M, Holko M, Rice CM, Williams BR, Marie I et al Induction of interferon‐stimulated gene expression and antiviral responses require protein deacetylase activity. Proc Natl Acad Sci USA 2004; 101:9578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sakamoto S, Potla R, Larner AC. Histone deacetylase activity is required to recruit RNA polymerase II to the promoters of selected interferon‐stimulated early response genes. J Biol Chem 2004; 279:40362–7. [DOI] [PubMed] [Google Scholar]
- 40. Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol 2006; 6:836–48. [DOI] [PubMed] [Google Scholar]
- 41. Hunt JS, Orr HT. HLA and maternal–fetal recognition. FASEB J 1992; 6:2344–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Transcription factor binding sites enriched in the gene module regulated by f 33 rule.
Table S2. Transcription factor binding sites enriched in the gene module regulated by f 39 rule.
Table S3. Transcription factor binding sites enriched in the gene module regulated by f 17 rule.
Table S4. transcription factor binding sites enriched in the gene module regulated by f 13 rule.
Table S5. Transcription factor binding sites enriched in the gene module regulated by f 10 rule.
Table S6. Transcription factor binding sites enriched in the gene module regulated by f 12 rule.


