ABSTRACT
Research on oral fungi has centered on Candida. However, recent internal transcribed spacer (ITS)-based studies revealed a vast number of fungal taxa as potential oral residents. We review DNA-based studies of the oral mycobiome and contrast them with cultivation-based surveys, showing that most genera encountered by cultivation have also been detected molecularly. Some taxa such as Malassezia, however, appear in high prevalence and abundance in molecular studies but have not been cultivated. Important technical and bioinformatic challenges to ITS-based oral mycobiome studies are discussed. These include optimization of sample lysis, variability in length of ITS amplicons, high intra-species ITS sequence variability, high inter-species variability in ITS copy number and challenges in nomenclature and maintenance of curated reference databases. Molecular surveys are powerful first steps to characterize the oral mycobiome but further research is needed to unravel which fungi detected by DNA are true oral residents and what role they play in oral homeostasis.
KEYWORDS: internal transcribed spacer, ITS1, microbiome, mycobiome, oral fungi, saliva
Introduction
For over a century fungi have been recognized as commensal inhabitants of the human oral cavity. A study from the first half of the twentieth century recovered yeast-like organisms from the mouth of ∼35% of subjects.1 The incidence of yeast-positive cultures was higher, ∼60%, in denture-wearers free of thrush lesions. The most consistently recovered yeast-like colonies were Monilia (now Candida) albicans. However, individuals also harbored Cryptococcus, Saccharomyces and other unidentified “Monilia.” A study by Young et al.2 yielded positive yeast cultures from the oral cavity in 48.6% of subjects (284/584). About 93% of isolates were Candida albicans; the remaining other Candida species or Cryptococcus. Candida spp. colonize the oral tissues at a very early age, with about 28% of children <2 years yielding positive cultures at multiple visits.3 These early studies established fungi as normal inhabitants of the human mouth.
Although the vast majority of research in oral mycology has focused on Candida, recent molecular studies revealed a diverse array of fungi as potential oral residents.4, 5 These results opened a new era for questions in oral mycology: Are detected fungi stable oral residents? What is the functional role of these species in the oral ecosystem? Do non-Candida fungi play a role in oral health? In this review we evaluate the classic literature and contrast it with studies that use modern high throughput sequencing to characterize the oral mycobiome. We then consider challenges to the molecular analysis of oral fungi and present an overview of pressing questions that need resolution.
Classic studies on oral mycology
Candida species and oral candidiasis
Candida albicans is the most commonly cultivated and studied fungus from the oral cavity. Candida species, especially C. albicans, have been unequivocally linked to the etiology of oral thrush (candidiasis). The mean carriage rate for C. albicans in healthy individuals has been estimated at 17.7% (range 1.9-62.3%), based on cultivation.6 Carriage rates are usually higher in hospitalized patients, subjects under cancer therapy or with other immunosuppressive conditions, and in elderly individuals.6-8 Local factors associated with increased oral carriage of Candida spp. include low salivary flow9-11, low pH2,12 and wearing a denture.11,13 The association of smoking and antibiotic intake with oral Candida colonization remains controversial with some studies showing an association,12,14,15 while others do not support a link.16,17
Species of Candida other than albicans are less frequently, but consistently, recovered from the oral cavity. These include Candida glabrata, Candida tropicalis, Candida parapsilosis, Candida dubliniensis, Candida rugosa, Candida krusei (currently Pichia kudriazevii and Issatchenkia orientalis), Candida guilliermondii (Meyerozyma guilliermondii) and Candida lusitaniae (Clavispora lusitaniae).2,18-22 Candida species other than albicans are more prevalent in immunosuppressed individuals, particularly those with a history of antifungal therapy, since species such as C. glabrata and C. krusei are naturally resistant to azoles.23,24
Despite considerable research in oral candidiasis, it is not clear why some subjects develop the condition, while others with similar risk factors remain disease-free. Oral candidiasis can present clinically as a pseudomembranous condition or as erythematous and/or hyperplastic lesions25. Most cases of oral candidiasis are associated with C. albicans, although some have been linked to C. dubliniensis, C. glabrata, C. krusei and C. tropicalis, either in isolation or more commonly as mixed infections sometimes including C. albicans.21,22 Molecular epidemiological studies show the origin of C. albicans infections is often attributable to endogenous strains.26,27 Although some resident strains of C. albicans are more pathogenic, it is not clear which specific genotypes increase risk for oral lesions.27,28 The main risk factor for oral candidiasis seems a pharmacologically-induced or naturally occurring impairment of the immune response, such as that in organ transplantation, HIV infection, cancer treatment or in specific immune defects such as autosomal dominant hyper IgE (HIES) syndrome.15,18,29 In HIES, mutations in STAT3 result in defects in T helper type 17 (Th17) cytokine production.30 Animal models suggests that the Th17/IL-17 axis is essential for immunity to oral candidiasis, exerting its effects through upregulation of proinflammatory cytokines (IL-6), neutrophil-recruiting chemokines (CXCL1 and CXCL5) and antimicrobial peptides.31 Local factors may also play roles in risk for oral candidiasis. Saliva could offer protection aiding in Candida clearance either via aggregation or by direct killing through anti-fungal proteins such as histatin 5 and calprotectin.32 Dentures probably predispose to candidiasis by disrupting the epithelial barrier and promoting biofilm formation.33
Cultivation-based studies of fungi other than Candida
Oral lesions not due to Candida are very rare and therefore few studies have focused on colonization by other genera. Cases of oral lesions associated with Cryptococcus spp., filamentous fungi (Aspergillus spp. and Zygomycetes), and dimorphic fungi (Histoplasma, Blastomyces and Coccidioides) have been reported but usually involve severe immunosuppression and disseminated infection to extra-oral sites.34-36 Histoplasma and Coccidioides are likely acquired infections. Other fungi responsible for rare oral and systemic infections are likely opportunistic with the mouth potentially serving as infection reservoir. For instance, Knighton1 and Young et al.2 detected cryptococci in the oral mucosal surfaces of healthy subjects. Aspergillus species including niger, versicolor and fumigatus have also been frequently recovered.37-39
Few broad cultivation surveys of fungi from the oral cavity exist (Fig. 2). A contributing factor was the indication that the origin of cultivated fungi other than Candida was environmental.40 More recent studies with air-exposed control plates or plating under laminar flow contradict these findings.38,41 Gomes et al.38 reported filamentous fungi in 28.3% of samples from root canals of teeth with pulp necrosis in a study including air-exposed control plates. Five Aspergillus species were identified (ustus, granulosus, niger, sydowii) and Emericella quadriluniata (sexual form of Aspergillus). Four Penicillium species (implicatum, micsynvisk, lividum and citrionigrum) were recovered. Other taxa represented were Fusarium (moniliforme and melanochorum), Aureobasidium pullulans, Exophiala jeanselmei, Eurotium amstelodame and Cladosporium sphaerospermum. In a recent, carefully-controlled, cultivation study, Monteiro da Silva et al.41 investigated oral fungi in 40 healthy subjects. Growth was evaluated at 25°C and 37°C. The most commonly isolated fungi were species of Candida (67.5%), Rhodotorula (75%), Penicillium (85%), Aspergillus (75%), Cladosporium (72.5%), Trichoderma (10%), Scedosporium (7.5%), Alternaria (5%) and Rhizopus (2.5%). More samples produced cultures at 25°C, 100% yielding molds and 92.5% yielding yeasts. At 37°C, 42.5% produced molds and 45% produced yeasts. Three factors argue against environmental contamination to explain these high recovery rates: 1) use of sterile conditions including laminal flow; 2) variability in taxa colonizing participants and 3) replication of subject-specific colonization profiles in 10 subjects at 2 additional time points, including a seasonal climatic change.
Figure 2.
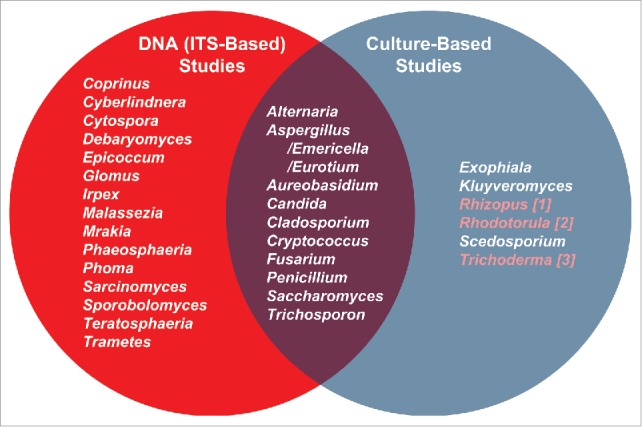
Comparison of oral mycobiome members (genus-level) as identified by molecular and culture-based approaches. Most prevalent oral genera (appearing in ∼20% of subejcts in Ghannoum et al.4 and Dupuy et al.5) DNA sequencing studies are in the left circle. Those studies38,41,42,44 appear in the right circle. The overlap in the Venn diagram illustrates the fungal genera identified by both approaches. Highlighted genera in the culture-based list are those present in our saliva data set but at less than 20% frequency.5 The molecular studies were conducted on systemically healthy individuals, while culture-based studies included both healthy and cancer patients. Samples in the molecular studies included oral rinses and unstimulated saliva, while samples for the cultivation studies included oral rinses, mucosal swabs and contents of infected root canals. Despite these differences, there is great agreement between the molecular surveys and cultivation studies with most taxa detected by cultivation also seen molecularly.
Additional studies have also recovered fungi other than Candida from a few subjects. Saccharomyces spp. and Rhodotorula rubra were isolated from oral swabs of radiotherapy patients.42 Rhodotorula mucilaginosa, Trichosporon mucoides and Cryptococcus humicolus were isolated from infected root canals and saliva.43 Also, Miranda et al.44 reported Saccharomyces cerevisiae and Kluyveromyces lactis from the tongue.
The oral mycobiome surveyed via molecular methods
Universal primers and amplicon sequencing enabled broad spectrum surveys of microbiota circumventing limitations of cultivation approaches. The most widely-used locus for mycobiota surveys is the internal transcribed spacer (ITS) between the 18S and 28S rRNA genes. The ITS includes the most rapidly-evolving ITS1, the highly conserved 5.8S and the moderately to rapidly evolving ITS2.45-47 Figure 1 shows a map of the ITS region and primer sets used in oral mycobiome studies.
Figure 1.
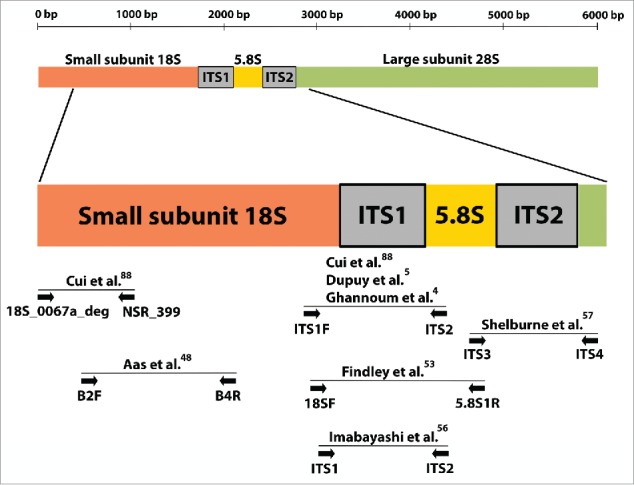
Schematic diagram of the nuclear ribosomal repeat unit in fungi. This region includes the 18S, 5.8S and 28S rRNA genes and the internal transcribed spacers (ITS) 1 and 2. Sites of universal primers used by molecular surveys of the oral mycobiome and by one landmark skin mycobiome study are indicated by arrows. As seen in the Figure, studies commonly rely on ITS1 amplification.
Aas et al.48 conducted one of the earliest amplicon-based sequencing studies of the oral mycobiome. Their primers were capable of amplifying the 18S gene from a variety of medically-important species.49 Cloned amplicons from subgingival plaque of HIV-positive subjects were Sanger sequenced. Only Candida albicans and Saccharomyces cerevisiae were found.
The first comprehensive survey of the oral mycobiome employing universal primers and high throughput sequencing analyzed oral rinse samples of 20 healthy individuals.4 Although Candida was the most frequently detected genus (75% of subjects; C. albicans identified in 40%), a diverse array of fungi were revealed as potential oral residents. Fifteen genera were present in more than 20% of subjects and designated the “core” mycobiome. A later study by the same group compared the mycobiome of 12 HIV-positive subjects to 12 controls.50 Results confirmed previously reported taxa and also showed a negative correlation between abundance of Candida and Pichia. An investigation of the in vitro interaction of Pichia and other fungi found that spent media from a Pichia farinosa strain inhibited growth of Candida, Aspergillus and Fusarium. The most commonly reported Pichia in oral mycobiome studies are Pichia guilliermondii (Candida/Meyerozyma guilliermondii) and Pichia jadinii (Cyberlindnera jadinii).4,5,51 It is not clear that Pichia farinosa is part of the oral mycobiome. Nevertheless, this study highlights the potential for fungal interactions to modulate oral mycobiome composition.
Our group recently characterized fungi in unstimulated saliva of healthy individuals using ITS1 pyrosequencing.5 Our results are in good agreement with Ghannoum et al.4 in identifying Candida, Pichia, Cladosporium/Davidiella, Alternaria/Lewia, Aspergillus/Emericella, Eurotium, Fusarium/Gibberella, Cryptococcus/Filobasidiella, and Aureobasidium. However, our study uniquely identified Malassezia as a prominent commensal in all individuals (abundance 13% to 96%) (Fig. 2). Since Malassezia species had never been reported as oral residents, we eliminated the possibility of contamination through rigorous negative controls. Malassezia are well-known skin commensals, also recoverable from respiratory tracts, and therefore oral residency is very plausible.52,53 Lack of detection in previous cultivation studies was likely due to media that do not support Malassezia growth.41 Malassezia have an incomplete fatty acid synthesis pathway requiring addition of lipids.54,55 Lack of previous molecular detection is attributable to cell lysis methods, since studies reporting Malassezia in human compartments employed very harsh protocols5,53,56,57 necessary for disrupting the thick cell walls of Malassezia. We use bead-beating with high-density 0.5 mm yttria-zyrconia beads for lysis and have noticed that even small variations in homogenization speed can greatly impact DNA yields from Malassezia (Fig. 3a). These results emphasize the need for validated protocols to improve detection of resident taxa.
Figure 3.
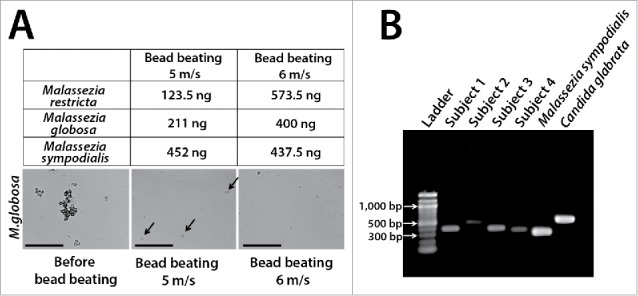
Examples of technical challenges important in the molecular evaluation of oral fungi. Table in Panel A shows yields after DNA extraction of standardized Malassezia cell suspensions (5 × 107 cells) using slightly different bead-beating speeds. DNA was extracted using yttria-zyrconia beads following a protocol developed by our group.5 Samples were subjected to bead-beating (3 × 30 sec) in an MP Biomedicals FastPrep™ instrument using a speed of 5 m/s or 6 m/s. Notice that this small variation almost doubled DNA recovery for M. restricta and M. globosa while it did not affect DNA yield for M. sympodialis. Arrows in middle phase-contrast micrograph show intact M. globosa cells after bead-beating using the 5 m/s setting. Scale bar = 50 μm. Panel B shows dramatic differences in amplicon length in the ITS1 region. Amplicons were generated from saliva samples of 4 subjects and from 2 reference strains using a published protocol.5 Such variation in amplicon length may introduce bias in abundance estimates as shorter fragments are preferentially sequenced.
A recent longitudinal microbiome characterization in a leukemia patient further corroborates our finding of Malassezia as a prominent commensal. Malassezia spp. dominated oral communities (buccal swabs) during certain time points of chemotherapeutic treatment.57 In contrast to oral samples, stool from the same individual did not show Malassezia. Other abundant fungi in the oral cavity of this patient included C. albicans, C. glabrata, Fusarium spp. and Alternaria spp. Mucur ventilosus was also detectable in oral samples prior to occurrence of invasive mucormycosis, with the oral cavity potentially serving as the infection source.
Recently, the oral mycobiome was compared between healthy subjects and those suffering oral candidiasis.56 The mycobiomes of subjects with candidiasis were dominated by C. albicans, but C. tropicalis and C. dublinensis were also detected in some subjects in moderately high abundance. Interestingly, antifungal treatment did not dramatically change the mycobiome with only C. dubliniensis decreasing while C. albicans increased proportions. The “healthy” mycobiome was not markedly different from the “candidiasis” mycobiome, both dominated by C. albicans. These results suggest that strain shifts, changes in load, altered pathogenic potential or host-related factors, rather than simple alterations in C. albicans proportions are associated with clinical symptoms.
In summary, pioneering molecular studies relying on saliva or oral rinses revealed highly diverse fungal communities in the human mouth. Detailed characterization of fungal biogeography at different oral sites is lacking. Cultivation studies suggest Candida species are more abundant in the posterior half of the tongue.1,12,58 Is there partitioning of other fungal species across oral sites? Also, are there relationships between fungi and other dental diseases? Beyond the association of Candida spp. with oral candidiasis, and suggested relationships between Candida carriage, salivary pH and caries,2,59 no further evaluation of oral health in the context of the mycobiome has been conducted.
Challenges in oral mycobiome analysis
In addition to lysis optimization, other technical and bioinformatic aspects to ITS-based surveys must be taken into account. These include differences in ITS amplicon lengths, variable rates of intra-species ITS sequence variation, variable number of ITS genomic copies among taxa and challenges in nomenclature and maintenance of comprehensively curated reference databases.
Fungal ITS amplicon lengths can differ greatly among taxa. Figure 3b shows ITS1 amplicons ranging from ∼200 to ∼600 bps in saliva and reference strains. ITS amplicon fragments from mouse and human fecal samples also vary in length (170 to 550 bp).60 Awareness of length discrepancies is especially necessary with bidirectional sequencing approaches since overlap between forward and reverse reads will differ. There is also potential bias in abundance estimates due to preferential amplification of the shorter fragments.60
The sequence variation in the fungal ITS region makes it ideal for phylogenetics. However, the degree of intra-species variability is remarkably different among fungi.45 In a comprehensive analysis, Nilsson et al.45 showed that ITS intra-species variation is above the 3% threshold in ¼ of fungal species, with ITS1 being more variable than ITS2. In practice, the combination of multiple alignment difficulties for different amplicon lengths and high intra-species sequence variation render unreliable the automated pipelines that separate sequence reads by similarity thresholds to delineate species-level operational taxonomic units (OTU)s. Therefore, most ITS-based studies employ taxonomic identification of individual sequences followed by grouping based on assigned names.
Total fungal load is a critical piece of information for clinical samples, but quantitative molecular methods used for other organisms are of limited value for fungi. While methods for fungal load using universal primers and real time PCR exist61, the interspecific (inter-species) variability in ITS copy number (few to ∼250 copies) makes overall quantitative estimates challenging. Even with species-specific primers, intraspecific copy number differences make the quantitative estimate of fungal cells or spores difficult in the absence of prior knowledge of copy numbers and relative abundance of different strains. Some species of important oral mycobiome members show discrepancies in copy number among strains. Candida albicans strains have been shown to vary ∼4-fold (56-222 copies)62, while Aspergillus fumigatus strains differ ∼2.5-fold (38-91)63.
A daunting challenge in mycobiome research is the confusing state of nomenclature. Unlike other organisms, a single fungus was permitted multiple names (reflecting asexual/sexual morphs, or diverse historical or geographical discoveries). Compounding nomenclature confusion was assignment of species to genera without phylogenetic support. These uncertainties are amplified in some oral mycobiome genera such as Candida or Cryptococcus that were depositories for varied anamorphic fungi. Unification of nomenclature is important to avoid reporting synonyms as different fungi, especially given the reliance of mycobiome studies on groupings by taxonomy. A recent goal has been to minimize nomenclature confusion through the “one name one fungus” (1N1F) initiative.64,65 International working groups are currently creating morphological, biochemical and multigene phylogenetic descriptions to inform recommendations for nomenclature (e.g. Consolidated Species Concept66). Subject to formal approvals, some recommendations have been published, which we have used to revisit nomenclature for oral cavity genera (Table 1). Our intent is not to provide a simple selection chart, but rather to alert researchers working on oral communities to the complexities of fungal nomenclature and to direct to more complete sources of information. Two major contributors to nomenclature adjustments are 1) historical assignments of different names to the same fungus and 2) new assignments due to phylogenetic circumscription. In the first instance, some synonyms arose when asexual (anamorphic) and sexual (teleomorphic) forms were assigned different names. These are resolved by reassigning members of one of the genera to the protected genus of the pair, joining other legitimate species in the phylogenetically circumscribed genus. In Table 1, Lewia joins Alternaria, Davidiella joins Cladosporium, Cochliobolus joins Curvularia. In the second instance, new phylogenetic circumscription of genera can change nomenclature. An illustration is the large, heterogeneous, genus Candida. A recent reclassification67 considered 434 Candida species that were reassigned to 15 genera and clades. Most relevant to the oral mycobiome is the delineation of the genus Candida to 31 species, including many oral members (i.e. albicans, dubliniensis, parapsilosis, tropicalis and others) that will not change names.
Table 1.
Naming Information for Genera Reported in the Human Oral Cavity. Protected/preferred names, synonyms and references are presented for an expanded list of fungal genera (this review, Dupuy et al.5). Sexual (T) and asexual (A) forms are designated. Only genus level information is presented, however it should be noted that not all species within a genus have synonyms in only one other genus and thus ultimate decisions on preferred names should be taken at the species-level. Lack of entry in the synonyms column does not imply that there are no synonyms but rather that there may be many possible synonyms. Readers are directed to the literature citations, MycoBank or Index Fungorum for extensive listings of alternate names and reassignments of species to new or different genera. Absence of entry in the basis for decision column means that information from MycoBank and/or Index Fungorum was used to infer legitimacy and currency of names.
| PROTECTED OR PREFERRED NAME | SYNONYMS | REFERENCE |
|---|---|---|
| Agaricus | ||
| Alternaria | Lewia, Allewia, Crivellia | Zhang et al.73, Rossman et al.74 |
| Aspergillus | Eurotium, Emericella | Controversial: Pitt and Taylor75; Samson et al.76 |
| Aureobasidium | ||
| Bipolaris | Cochliobolus (T) | Rossman et al.74 |
| Bullera | Bulleromyces, hannae clades | Liu et al.78 |
| Candida | Lodderomyces, several genera | Daniel et al.67 |
| Cladosporium | Davidiella (T) | Rossman et al.74 |
| Coprinus | Coprinopsis, Coprinellus | |
| Cryptococcus | Neoformans/gatti complex, Filobasidiella(T) | Liu et al.78 |
| Curvularia (A) | Pseudococclioblous | Rossman et al.74 |
| Cyberlindnera | Pichia, Candida, others | Daniel et al.67 |
| Cystofilobasidium (T) | Some Cryptococcus species | Liu et al.78 |
| Cytospora (A) | Valsa (T), Valsella, Valseutypella, Leucostoma | Rossman et al.77 |
| Debaryomyces | Daniel et al.67 | |
| Didymella | Genus Peyronellaea, Phoma | Chen et al.79 |
| Dioszegia | Dioszegia clade (includes some Cryptococcus) | Liu et al.78 |
| Epicoccum | Several Phoma | Chen et al.79 |
| Erythrobasidium | Sporobolomyces | Wang et al.80 |
| Exophiala | ||
| Filobasidium (T) | Filobasidium clade (includes some Cryptococcus) | Liu et al.78 |
| Fusarium | Gibberella | Rossman et al.81 |
| Glomus | ||
| Hanseniaspora (T) | Kloeckera (A) | Daniel et al.67 |
| Irpex | Synonyms in several genera | |
| Kluyveromyces | Kurtzman82 | |
| Lenzites | Trametes, others | |
| Leptosphaerulina | Chen et al.79 | |
| Malassezia | Wu et al.55 | |
| Mrakia | Mrakia clade (includes some Mrakiella) | Liu et al.78 |
| Naganishia | Cryptococcus albus clade | Liu et al.78 |
| Penicillium | Eupenicillium | Redhead et al.83, Zhang et al.73 |
| Phaeosphaeria | Phaeoseptoria | Rossman et al.73 |
| Phoma | Atradidymella, others | Chen et al.79 |
| Pichia | Daniel et al.67 | |
| Pisolithus | Synonyms in several genera | |
| Pyrenochaetopsis | Chen et al.79 | |
| Ramularia (T) | Mycosphaerella (A) | Quaedvlieg et al.66, Rossman et al.73 |
| Rhizocarpon | Synonyms in several genera | |
| Rhizopus | Hoffman et al.84 | |
| Rhodosporidiobolus | Rhodosporidium/Sporidiobolus Rhodotorula(A)/Rhodosporidium (T) Sporobolomyces(A)/Sporidiobolus(T) | Wang et al.80 |
| Rhodotorula | Rhodosporidium (T) in Rhodosporidium clade | Wang et al.80 |
| Saccharomyces | Kurtzman82, Daniel et al.67 | |
| Sarcinomyces | ||
| Scedosporium | Lackner and de Hoog85 | |
| Sporobolomyces (A) | Sporidiobolus (T) in Sporobolomyces clade | Wang et al.80 |
| Talaromyces (T) | Penicillium (A) | Redhead et al.83, Yilmaz et al.86 |
| Taphrina (T) | Lalaria (A) | Daniel et al.67 |
| Tausonia | Guehomyces, Trichosporon | Liu et al.78 |
| Teratosphaeria | Kirramyces, Colletogloeopsis | Quaedvlieg et al.66 Rossman et al.74 |
| Torulaspora | Kurtzman82 | |
| Trametes | Carlson et al.87 | |
| Trichoderma (A) | Hypocrea (T) | Rossman et al.87 |
| Trichosporon | ||
| Wallemia | Bargellinia |
Since mycobiome studies rely on taxonomic identification to group sequences, well-curated databases are a necessity. Publicly available repositories contain about 20% of sequences incorrectly annotated at the species level.68 Many GenBank ITS sequences are annotated as “fungi” or “uncultured fungi;” with some shown to originate from primer artifacts.5 Three commonly used databases, Findley et al.,53 UNITE69 and Fungal ITS RefSeq Targeted Loci Project (RTL) were recently evaluated using a single mouse fecal ITS library.60 Unsurprisingly, the distribution of fungi depended on the database and the authors ultimately constructed a niche-specific reference set. A recent, curated, quality-controlled database of 3400 ITS sequences covering 524 human and animal pathogens was constructed to aid in identification of causes of mycoses.70 A specific database for oral fungi would be an invaluable resource for tracking nomenclature evolution, avoiding inclusion of redundant taxa, and allowing accurate taxonomic assignments.
Ultimately, a full understanding of the roles of fungi in oral health and disease requires species level resolutions. As discussed earlier, the ITS power of discrimination for species varies widely. As a result, a number of additional gene sequences have been used in various “multigene” studies. One example uses ΕF1α (translation elongation factor), RPB2 (subunit B2 of RNA polymerase II), β-tubulin, actin, calmodulin, LSU and ITS to create more robust phylogenies and reference sequence datasets.66 One can envision that selective multigene approaches may become useful in circumstances when ITS is insufficient.
A glimpse into the future
In addition to the need to explore oral fungal biogeography is one to evaluate whether fungi detected by DNA are functional residents of the oral biocompartment, rather than a transient environmental presence. Cultivation studies with appropriate media to target growth of prevalent molecularly-detected species would help answer this question. For instance, lipid-enriched media to support growth of Malassezia would confirm their presence as true mycobiome residents. Such studies should employ rigorous laboratory conditions and negative controls to exclude environmental contaminants. Also, longitudinal studies that include seasonal changes could help separate transient environmental fungi from permanent residents.
One reason that the study of oral fungi has lagged behind that of oral bacteria is the comparatively lower biomass of fungi in the mouth. The number of cultivable yeasts ranges from 10 to 10,000 per mL of saliva, with the majority of specimens at 1,000 or less.2,41 The concentration of molds recovered from healthy subjects by cultivation of oral rinse is also low, not exceeding 10 cells per mL.41 These data sharply contrast with salivary bacterial load between 108 to 1010 16S rRNA gene copies per mL (unpublished data). Gut microbiome studies indicate that small numbers of fungi could have surprisingly strong effects on immunological responses and the composition of the bacterial microbiome.71,72 Therefore, low biomass should not deter interest in the effects of fungi in the oral ecosystem.
Finally, a combination of molecular-based clinical surveys and model-based research using in vivo and in vitro systems will be necessary to fully understand the roles of fungi in oral health. Pressing questions include: How do fungal oral commensals affect immune responses? How do oral fungi and bacteria interact? Are fungi “accessory” pathogens in oral diseases apart from their direct involvement in candidiasis? Molecular surveys are powerful first steps toward understanding the oral mycobiome, but mechanistic studies are necessary to uncover the functional roles of fungi in oral homeostasis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grant R01 DE021578 from the National Institute of Dental and Craniofacial Research (NIDCR), National Institutes of Health (NIH).
References
- [1].Knighton HT. A study of Monilia and Other Yeastlike Organisms Found in the Oral Cavity. J Dent Res 1939; 18:103-25; http://dx.doi.org/ 10.1177/00220345390180020101 [DOI] [Google Scholar]
- [2].Young G, Resca HG, Sullivan MT. The yeasts of the normal mouth and their relation to salivary acidity. J Dent Res 1951; 30:426-30; PMID:14841321; http://dx.doi.org/ 10.1177/00220345510300031901 [DOI] [PubMed] [Google Scholar]
- [3].Hannula J, Saarela M, Jousimies-Somer H, Takala A, Syrjanen R, Kononen E, Asikainen S. Age-related acquisition of oral and nasopharyngeal yeast species and stability of colonization in young children. Oral Microbiol Immunol 1999; 14:176-82; PMID:10495712; http://dx.doi.org/ 10.1034/j.1399-302X.1999.140306.x [DOI] [PubMed] [Google Scholar]
- [4].Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathogens 2010; 6:e1000713; PMID:20072605; http://dx.doi.org/ 10.1371/journal.ppat.1000713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, Dongari-Bagtzoglou A, Diaz PI, Strausbaugh LD. Redefining the Human Oral Mycobiome with Improved Practices in Amplicon-based Taxonomy: Discovery of Malassezia as a Prominent Commensal. PloS One 2014; 9:e90899; PMID:24614173; http://dx.doi.org/ 10.1371/journal.pone.0090899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Odds FC. Candida and Candidosis. Elsevier Science Health Science Division, 1988 [Google Scholar]
- [7].Cumming CG, Wight C, Blackwell CL, Wray D. Denture stomatitis in the elderly. Oral Microbiol Immunol 1990; 5:82-5; PMID:2087353; http://dx.doi.org/ 10.1111/j.1399-302X.1990.tb00232.x [DOI] [PubMed] [Google Scholar]
- [8].Wilkieson C, Samaranayake LP, MacFarlane TW, Lamey PJ, MacKenzie D. Oral candidosis in the elderly in long term hospital care. J Oral Pathol Med 1991; 20:13-6; PMID:1900531; http://dx.doi.org/ 10.1111/j.1600-0714.1991.tb00880.x [DOI] [PubMed] [Google Scholar]
- [9].Radfar L, Shea Y, Fischer SH, Sankar V, Leakan RA, Baum BJ, Pillemer SR. Fungal load and candidiasis in Sjogren's syndrome. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2003; 96:283-7; PMID:12973284; http://dx.doi.org/ 10.1016/S1079-2104(03)00224-5 [DOI] [PubMed] [Google Scholar]
- [10].Tapper-Jones L, Aldred M, Walker DM. Prevalence and intraoral distribution of Candida albicans in Sjogren's syndrome. J Clin Pathol 1980; 33:282-7; PMID:6991530; http://dx.doi.org/ 10.1136/jcp.33.3.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shimizu C, Kuriyama T, Williams DW, Karasawa T, Inoue K, Nakagawa K, Yamamoto E. Association of oral yeast carriage with specific host factors and altered mouth sensation. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2008; 105:445-51; PMID:18329579; http://dx.doi.org/ 10.1016/j.tripleo.2007.11.030 [DOI] [PubMed] [Google Scholar]
- [12].Arendorf TM, Walker DM. The prevalence and intra-oral distribution of Candida albicans in man. Archives of oral biology 1980; 25:1-10; PMID:6996654; http://dx.doi.org/ 10.1016/0003-9969(80)90147-8 [DOI] [PubMed] [Google Scholar]
- [13].Schelenz S, Abdallah S, Gray G, Stubbings H, Gow I, Baker P, Hunter PR. Epidemiology of oral yeast colonization and infection in patients with hematological malignancies, head neck and solid tumors. J Oral Pathol Med 2011; 40:83-9; PMID:20923440; http://dx.doi.org/ 10.1111/j.1600-0714.2010.00937.x [DOI] [PubMed] [Google Scholar]
- [14].McGovern JJ, Parrott RH, Emmons CW, Ross S, Burke FG, Rice EC. The effect of aureomycin and chloramphenicol on the fungal and bacterial flora of children. N Engl J Med 1953; 248:397-403; PMID:13025705; http://dx.doi.org/ 10.1056/NEJM195303052481001 [DOI] [PubMed] [Google Scholar]
- [15].Patel PK, Erlandsen JE, Kirkpatrick WR, Berg DK, Westbrook SD, Louden C, Cornell JE, Thompson GR, Vallor AC, Wickes BL, et al.. The Changing Epidemiology of Oropharyngeal Candidiasis in Patients with HIV/AIDS in the Era of Antiretroviral Therapy. AIDS Res Treat 2012; 2012:262471; PMID:22970352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Oliver DE, Shillitoe EJ. Effects of smoking on the prevalence and intraoral distribution of Candida albicans. J Oral Pathol 1984; 13:265-70; PMID:6429300; http://dx.doi.org/ 10.1111/j.1600-0714.1984.tb01424.x [DOI] [PubMed] [Google Scholar]
- [17].de Araujo Navas EA, Inocencio AC, Almeida JD, Back-Brito GN, Mota AJ, Jorge AO, Querido SM, Balducci I, Koga-Ito CY. Oral distribution of Candida species and presence of oral lesions in Brazilian leprosy patients under multidrug therapy. J Oral Pathol Med 2009; 38:764-7; PMID:19549111; http://dx.doi.org/ 10.1111/j.1600-0714.2009.00786.x [DOI] [PubMed] [Google Scholar]
- [18].Dongari-Bagtzoglou A, Dwivedi P, Ioannidou E, Shaqman M, Hull D, Burleson J. Oral Candida infection and colonization in solid organ transplant recipients. Oral Microbiol Immunol 2009; 24:249-54; PMID:19416456; http://dx.doi.org/ 10.1111/j.1399-302X.2009.00505.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cannon RD, Holmes AR, Mason AB, Monk BC. Oral Candida: clearance, colonization, or candidiasis? J Dent Res 1995; 74:1152-61; PMID:7790592; http://dx.doi.org/ 10.1177/00220345950740050301 [DOI] [PubMed] [Google Scholar]
- [20].Goncalves RH, Miranda ET, Zaia JE, Giannini MJ. Species diversity of yeast in oral colonization of insulin-treated diabetes mellitus patients. Mycopathologia 2006; 162:83-9; PMID:16897585; http://dx.doi.org/ 10.1007/s11046-006-0038-5 [DOI] [PubMed] [Google Scholar]
- [21].Redding SW, Dahiya MC, Kirkpatrick WR, Coco BJ, Patterson TF, Fothergill AW, Rinaldi MG, Thomas CR Jr. Candida glabrata is an emerging cause of oropharyngeal candidiasis in patients receiving radiation for head and neck cancer. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2004; 97:47-52; PMID:14716255; http://dx.doi.org/ 10.1016/j.tripleo.2003.09.008 [DOI] [PubMed] [Google Scholar]
- [22].Redding SW, Bailey CW, Lopez-Ribot JL, Kirkpatrick WR, Fothergill AW, Rinaldi MG, Patterson TF. Candida dubliniensis in radiation-induced oropharyngeal candidiasis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001; 91:659-62; PMID:11402278; http://dx.doi.org/ 10.1067/moe.2001.112946 [DOI] [PubMed] [Google Scholar]
- [23].Fidel PL Jr., Vazquez JA, Sobel JD. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin Microbiol Rev 1999; 12:80-96; PMID:9880475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Nagy E, Dobiasova S, Rinaldi M, Barton R, Veselov A. Global Antifungal Surveillance G. Candida krusei, a multidrug-resistant opportunistic fungal pathogen: geographic and temporal trends from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. J Clin Microbiol 2008; 46:515-21; PMID:18077633; http://dx.doi.org/ 10.1128/JCM.01915-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Williams D, Lewis M. Pathogenesis and treatment of oral candidosis. J Oral Microbiol 2011; 3: 5771; PMID:21547018; http://dx.doi.org/ 10.3402/jom.v3i0.5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].McManus BA, Coleman DC. Molecular epidemiology, phylogeny and evolution of Candida albicans. Infect Genet Evol 2014; 21:166-78; PMID:24269341; http://dx.doi.org/ 10.1016/j.meegid.2013.11.008 [DOI] [PubMed] [Google Scholar]
- [27].Lasker BA, Elie CM, Lott TJ, Espinel-Ingroff A, Gallagher L, Kuykendall RJ, Kellum ME, Pruitt WR, Warnock DW, Rimland D, et al.. Molecular epidemiology of Candida albicans strains isolated from the oropharynx of HIV-positive patients at successive clinic visits. Med Mycol 2001; 39:341-52; PMID:11556764; http://dx.doi.org/ 10.1080/714031035 [DOI] [PubMed] [Google Scholar]
- [28].Jacobsen MD, Duncan AD, Bain J, Johnson EM, Naglik JR, Shaw DJ, Gow NA, Odds FC. Mixed Candida albicans strain populations in colonized and infected mucosal tissues. FEMS Yeast Res 2008; 8:1334-8; PMID:18795958; http://dx.doi.org/ 10.1111/j.1567-1364.2008.00438.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lalla RV, Latortue MC, Hong CH, Ariyawardana A, D'Amato-Palumbo S, Fischer DJ, Martof A, Nicolatou-Galitis O, Patton LL, Elting LS, et al.. A systematic review of oral fungal infections in patients receiving cancer therapy. Support Care Cancer 2010; 18:985-92; PMID:20449755; http://dx.doi.org/ 10.1007/s00520-010-0892-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Freeman AF, Domingo DL, Holland SM. Hyper IgE (Job's) syndrome: a primary immune deficiency with oral manifestations. Oral Dis 2009; 15:2-7; PMID:19036057; http://dx.doi.org/ 10.1111/j.1601-0825.2008.01463.x [DOI] [PubMed] [Google Scholar]
- [31].Conti HR, Gaffen SL. IL-17-Mediated Immunity to the Opportunistic Fungal Pathogen Candida albicans. J Immunol 2015; 195:780-8; PMID:26188072; http://dx.doi.org/ 10.4049/jimmunol.1500909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Salvatori O, Puri S, Tati S, Edgerton M. Innate Immunity and Saliva in Candida albicans-mediated Oral Diseases. J Dent Res 2016; 95:365-71; PMID:26747422; http://dx.doi.org/ 10.1177/0022034515625222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Johnson CC, Yu A, Lee H, Fidel PL Jr., Noverr MC. Development of a contemporary animal model of Candida albicans-associated denture stomatitis using a novel intraoral denture system. Infect Immun 2012; 80:1736-43; PMID:22392931; http://dx.doi.org/ 10.1128/IAI.00019-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schwartz S, Thiel E. Images in clinical medicine. Palate destruction by Aspergillus. N Engl J Med 1997; 337:241; PMID:9227930; http://dx.doi.org/ 10.1056/NEJM199707243370405 [DOI] [PubMed] [Google Scholar]
- [35].Alcure ML, Di Hipolito Junior O, Almeida OP, Bonilha H, Lopes MA. Oral histoplasmosis in an HIV-negative patient. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2006; 101:e33-6; PMID:16448911; http://dx.doi.org/ 10.1016/j.tripleo.2005.06.028 [DOI] [PubMed] [Google Scholar]
- [36].Iatta R, Napoli C, Borghi E, Montagna MT. Rare mycoses of the oral cavity: a literature epidemiologic review. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2009; 108:647-55; PMID:19836721; http://dx.doi.org/ 10.1016/j.tripleo.2009.07.010 [DOI] [PubMed] [Google Scholar]
- [37].Fox EC, Ainsworth GC. A contribution to the mycology of the mouth. Br Med J 1958; 2:826-8; PMID:13572917; http://dx.doi.org/ 10.1136/bmj.2.5100.826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gomes C, Fidel S, Fidel R, de Moura Sarquis MI. Isolation and taxonomy of filamentous fungi in endodontic infections. J Endod 2010; 36:626-9; PMID:20307734; http://dx.doi.org/ 10.1016/j.joen.2010.01.016 [DOI] [PubMed] [Google Scholar]
- [39].Kamma JJ, Nakou M, Baehni PC. Clinical and microbiological characteristics of smokers with early onset periodontitis. J Periodontal Res 1999; 34:25-33; PMID:10086883; http://dx.doi.org/ 10.1111/j.1600-0765.1999.tb02218.x [DOI] [PubMed] [Google Scholar]
- [40].Bartels HA, Blechman H. Limitations of Mold Establishment in Oral Cavity. J Dent Res 1964; 43:136-9; PMID:14104345; http://dx.doi.org/ 10.1177/00220345640430010701 [DOI] [PubMed] [Google Scholar]
- [41].Monteiro-da-Silva F, Araujo R, Sampaio-Maia B. Interindividual variability and intraindividual stability of oral fungal microbiota over time. Med Mycol 2014; 52:498-505; PMID:24934804; http://dx.doi.org/ 10.1093/mmy/myu027 [DOI] [PubMed] [Google Scholar]
- [42].Martin MV, Al-Tikriti U, Bramley PA. Yeast flora of the mouth and skin during and after irradiation for oral and laryngeal cancer. J Med Microbiol 1981; 14:457-67; http://dx.doi.org/ 10.1099/00222615-14-4-457 [DOI] [PubMed] [Google Scholar]
- [43].Egan MW, Spratt DA, Ng YL, Lam JM, Moles DR, Gulabivala K. Prevalence of yeasts in saliva and root canals of teeth associated with apical periodontitis. Int Endod J 2002; 35:321-9; PMID:12059932; http://dx.doi.org/ 10.1046/j.1365-2591.2002.00478.x [DOI] [PubMed] [Google Scholar]
- [44].Miranda TT, Vianna CR, Rodrigues L, Monteiro AS, Rosa CA, Correa A Jr. Diversity and frequency of yeasts from the dorsum of the tongue and necrotic root canals associated with primary apical periodontitis. Int Endod J 2009; 42:839-44; PMID:19712195; http://dx.doi.org/ 10.1111/j.1365-2591.2009.01601.x [DOI] [PubMed] [Google Scholar]
- [45].Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH. Intraspecific ITS variability in the kingdom fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinform Online 2008; 4:193-201; PMID:19204817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol 1991; 66:411-53; PMID:1784710; http://dx.doi.org/ 10.1086/417338 [DOI] [PubMed] [Google Scholar]
- [47].Hershkovitz MA, Lewis LA. Deep-level diagnostic value of the rDNA-ITS region. Mol Biol Evol 1996; 13:1276-95; PMID:8896380; http://dx.doi.org/ 10.1093/oxfordjournals.molbev.a025693 [DOI] [PubMed] [Google Scholar]
- [48].Aas JA, Barbuto SM, Alpagot T, Olsen I, Dewhirst FE, Paster BJ. Subgingival plaque microbiota in HIV positive patients. J Clin Periodontol 2007; 34:189-95; PMID:17309593; http://dx.doi.org/ 10.1111/j.1600-051X.2006.01034.x [DOI] [PubMed] [Google Scholar]
- [49].Makimura K, Murayama SY, Yamaguchi H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J Med Microbiol 1994; 40:358-64; PMID:8176723; http://dx.doi.org/ 10.1099/00222615-40-5-358 [DOI] [PubMed] [Google Scholar]
- [50].Mukherjee PK, Chandra J, Retuerto M, Sikaroodi M, Brown RE, Jurevic R, Salata RA, Lederman MM, Gillevet PM, Ghannoum MA. Oral Mycobiome Analysis of HIV-Infected Patients: Identification of Pichia as an Antagonist of Opportunistic Fungi. PLoS Pathog 2014; 10:e1003996; PMID:24626467; http://dx.doi.org/ 10.1371/journal.ppat.1003996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Watkins RR, Mukherjee PK, Chandra J, Retuerto MA, Guidry C, Haller N, Paranjape C, Ghannoum MA. Admission to the Intensive Care Unit is Associated With Changes in the Oral Mycobiome. J Intensive Care Med 2016; PMID:26893317 [DOI] [PubMed] [Google Scholar]
- [52].Delhaes L, Monchy S, Frealle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, et al.. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community–implications for therapeutic management. PloS One 2012; 7:e36313; PMID:22558432; http://dx.doi.org/ 10.1371/journal.pone.0036313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Program NIHISCCS, et al.. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013; 498:367-70; PMID:23698366; http://dx.doi.org/ 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Xu J, Saunders CW, Hu P, Grant RA, Boekhout T, Kuramae EE, Kronstad JW, Deangelis YM, Reeder NL, Johnstone KR, et al.. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. PNAS 2007; 104:18730-5; PMID:18000048; http://dx.doi.org/ 10.1073/pnas.0706756104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wu G, Zhao H, Li C, Rajapakse MP, Wong WC, Xu J, Saunders CW, Reeder NL, Reilman RA, Scheynius A, et al.. Genus-Wide Comparative Genomics of Malassezia Delineates Its Phylogeny, Physiology, and Niche Adaptation on Human Skin. PLoS Genet 2015; 11:e1005614; PMID:26539826; http://dx.doi.org/ 10.1371/journal.pgen.1005614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Imabayashi Y, Moriyama M, Takeshita T, Ieda S, Hayashida JN, Tanaka A, Maehara T, Furukawa S, Ohta M, Kubota K, et al.. Molecular analysis of fungal populations in patients with oral candidiasis using next-generation sequencing. Sci Rep 2016; 6:28110; PMID:27305838; http://dx.doi.org/ 10.1038/srep28110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shelburne SA, Ajami NJ, Chibucos MC, Beird HC, Tarrand J, Galloway-Pena J, Albert N, Chemaly RF, Ghantoji SS, Marsh L, et al.. Implementation of a Pan-Genomic Approach to Investigate Holobiont-Infecting Microbe Interaction: A Case Report of a Leukemic Patient with Invasive Mucormycosis. PloS One 2015; 10:e0139851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Borromeo GL, McCullough MJ, Reade PC. Quantitation and morphotyping of Candida albicans from healthy mouths and from mouths affected by erythematous candidosis. J Med Vet Mycol 1992; 30:477-80; PMID:1287167; http://dx.doi.org/ 10.1080/02681219280000641 [DOI] [PubMed] [Google Scholar]
- [59].Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, Gonzalez M, Watson G, Krysan DJ, Bowen WH, et al.. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes the virulence of plaque-biofilms in vivo. Infect Immun 2014; 82(5):1968-81; PMID:24566629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tang J, Iliev ID, Brown J, Underhill DM, Funari VA. Mycobiome: Approaches to analysis of intestinal fungi. J Immunol Methods 2015; 421:112-21; PMID:25891793; http://dx.doi.org/ 10.1016/j.jim.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Liu CM, Kachur S, Dwan MG, Abraham AG, Aziz M, Hsueh PR, Huang YT, Busch JD, Lamit LJ, Gehring CA, et al.. FungiQuant: a broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol 2012; 12:255; PMID:23136846; http://dx.doi.org/ 10.1186/1471-2180-12-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rustchenko EP, Curran TM, Sherman F. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J Bacteriol 1993; 175:7189-99; PMID:8226665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Herrera ML, Vallor AC, Gelfond JA, Patterson TF, Wickes BL. Strain-dependent variation in 18S ribosomal DNA Copy numbers in Aspergillus fumigatus. J Clin Microbiol 2009; 47:1325-32; PMID:19261786; http://dx.doi.org/ 10.1128/JCM.02073-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Taylor JW. One Fungus = One Name: DNA and fungal nomenclature twenty years after PCR. IMA Fungus 2011; 2:113-20; PMID:22679595; http://dx.doi.org/ 10.5598/imafungus.2011.02.02.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hibbett DS, Taylor JW. Fungal systematics: is a new age of enlightenment at hand? Nat Rev Microbiol 2013; 11:129-33; PMID:23288349; http://dx.doi.org/ 10.1038/nrmicro2963 [DOI] [PubMed] [Google Scholar]
- [66].Quaedvlieg W, Binder M, Groenewald JZ, Summerell BA, Carnegie AJ, Burgess TI, Crous PW. Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 2014; 33:1-40; PMID:25737591; http://dx.doi.org/ 10.3767/003158514X681981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Daniel HM, Lachance MA, Kurtzman CP. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie van Leeuwenhoek 2014; 106:67-84; PMID:24748333; http://dx.doi.org/ 10.1007/s10482-014-0170-z [DOI] [PubMed] [Google Scholar]
- [68].Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson KH, Koljalg U. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PloS One 2006; 1:e59; PMID:17183689; http://dx.doi.org/ 10.1371/journal.pone.0000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Abarenkov K, Henrik Nilsson R, Larsson KH, Alexander IJ, Eberhardt U, Erland S, Hoiland K, Kjoller R, Larsson E, Pennanen T, et al.. The UNITE database for molecular identification of fungi–recent updates and future perspectives. New Phytol 2010; 186:281-5; PMID:20409185; http://dx.doi.org/ 10.1111/j.1469-8137.2009.03160.x [DOI] [PubMed] [Google Scholar]
- [70].Irinyi L, Serena C, Garcia-Hermoso D, Arabatzis M, Desnos-Ollivier M, Vu D, Cardinali G, Arthur I, Normand AC, Giraldo A, et al.. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database–the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med Mycol 2015; 53:313-37; PMID:25802363; http://dx.doi.org/ 10.1093/mmy/myv008 [DOI] [PubMed] [Google Scholar]
- [71].Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, et al.. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 2016; 19:865-73; PMID:27237365; http://dx.doi.org/ 10.1016/j.chom.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Mason KL, Erb Downward JR, Mason KD, Falkowski NR, Eaton KA, Kao JY, Young VB, Huffnagle GB. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun 2012; 80:3371-80; PMID:22778094; http://dx.doi.org/ 10.1128/IAI.00449-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhang N, Rossman AY, Seifert K, Bennett JW, Cai G, Cai L, Hillman B, Hyde KD, Luo J, Manamgoda D, et al.. Impacts of the International Code of Nomenclature for algae, fungi and plants (Melbourne Code) on the scientific names of plant pathogenic fungi. APSnet Feature 2013; http://dx.doi.org/10.1094/APSFeature-2013-06 [Google Scholar]
- [74].Rossman AY, Crous PW, Hyde KD, Hawksworth DL, Aptroot A, Bezerra JL, Bhat JD, Boehm E, Braun U, Boonmee S, et al.. Recommended names for pleomorphic genera in Dothideomycetes. IMA Fungus 2015; 6:507-23; PMID:26734553; http://dx.doi.org/ 10.5598/imafungus.2015.06.02.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Pitt JI, Taylor JW. Aspergillus, its sexual states and the new International Code of Nomenclature. Mycologia 2014; 106:1051-62; PMID:24871603; http://dx.doi.org/ 10.3852/14-060 [DOI] [PubMed] [Google Scholar]
- [76].Samson RA, Visagie CM, Houbraken J, Hong SB, Hubka V, Klaassen CH, Perrone G, Seifert KA, Susca A, Tanney JB, et al.. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol 2014; 78:141-73; PMID:25492982; http://dx.doi.org/ 10.1016/j.simyco.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rossman AY, Adams GC, Cannon PF, Castlebury LA, Crous PW, Gryzenhout M, Jaklitsch WM, Mejia LC, Stoykov D, Udayanga D, et al.. Recommendations of generic names in Diaporthales competing for protection or use. IMA Fungus 2015; 6:145-54; PMID:26203420; http://dx.doi.org/ 10.5598/imafungus.2015.06.01.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Liu XZ, Wang QM, Goker M, Groenewald M, Kachalkin AV, Lumbsch HT, Millanes AM, Wedin M, Yurkov AM, Boekhout T, et al.. Towards an integrated phylogenetic classification of the Tremellomycetes. Stud Mycol 2015; 81:85-147; PMID:26955199; http://dx.doi.org/ 10.1016/j.simyco.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Chen Q, Jiang JR, Zhang GZ, Cai L, Crous PW. Resolving the Phoma enigma. Stud Mycol 2015; 82:137-217; PMID:26955202; http://dx.doi.org/ 10.1016/j.simyco.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wang QM, Yurkov AM, Goker M, Lumbsch HT, Leavitt SD, Groenewald M, Theelen B, Liu XZ, Boekhout T, Bai FY. Phylogenetic classification of yeasts and related taxa within Pucciniomycotina. Stud Mycol 2015; 81:149-89; PMID:26951631; http://dx.doi.org/ 10.1016/j.simyco.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Rossman AY, Seifert KA, Samuels GJ, Minnis AM, Schroers HJ, Lombard L, Crous PW, Poldmaa K, Cannon PF, Summerbell RC, et al.. Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus 2013; 4:41-51; PMID:23898411; http://dx.doi.org/ 10.5598/imafungus.2013.04.01.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kurtzman CP. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS yeast research 2003; 4:233-45; PMID:14654427; http://dx.doi.org/ 10.1016/S1567-1356(03)00175-2 [DOI] [PubMed] [Google Scholar]
- [83].Redhead SA, Demoulin V, Hawksworth DL, Seifert KA, Turland NJ. Fungal Nomenclature at IMC10: Report of the Nomenclature Sessions. IMA Fungus 2014; 5:449-62; PMID:25734034; http://dx.doi.org/ 10.5598/imafungus.2014.05.02.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hoffmann K, Pawlowska J, Walther G, Wrzosek M, de Hoog GS, Benny GL, Kirk PM, Voigt K. The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia 2013; 30:57-76; PMID:24027347; http://dx.doi.org/ 10.3767/003158513X666259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lackner M, de Hoog GS. Parascedosporium and its relatives: phylogeny and ecological trends. IMA Fungus 2011; 2:39-48; PMID:22679587; http://dx.doi.org/ 10.5598/1imafungus.2011.02.01.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yilmaz N, Visagie CM, Houbraken J, Frisvad JC, Samson RA. Polyphasic taxonomy of the genus Talaromyces. Stud Mycol 2014; 78:175-341; PMID:25492983; http://dx.doi.org/ 10.1016/j.simyco.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Carlson A, Justo A, Hibbett DS. Species delimitation in Trametes: a comparison of ITS, RPB1, RPB2 and TEF1 gene phylogenies. Mycologia 2014; 106:735-45; PMID:24898532; http://dx.doi.org/ 10.3852/13-275 [DOI] [PubMed] [Google Scholar]
- [88].Cui L, Lucht L, Tipton L, Rogers MB, Fitch A, Kessinger C, Camp D, Kingsley L, Leo N, Greenblatt RM et al.. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am J Respir Crit Care Med 2015; 191:932-42; PMID:25603113; http://dx.doi.org/ 10.1164/rccm.201409-1583OC [DOI] [PMC free article] [PubMed] [Google Scholar]


