Abstract
Effective planning for the medical response to a radiologic or nuclear accident is complex. Due to limited resources for medical countermeasures, the key would be to accurately triage and identify victims most likely to benefit from treatment. We had used a mouse model system to provide evidence that serum microRNAs (miRNAs) may effectively predict the impact of radiation on long-term viability of animals. Here we use non-human primates (NHPs) to demonstrate that this concept is conserved and serum miRNA signatures have the potential to serve as prediction biomarkers for radiation-induced fatality in a human population. We identified a signature of seven miRNAs that are altered by irradiation in both mice and NHPs. Genomic analysis of these conserved miRNAs revealed that there is a combination of seven transcription factors that are predicted to regulate these miRNAs in human, mice, and NHPs. Moreover, a combination of three miRNAs (miR-133b, miR-215, and miR-375) can identify, with nearly complete accuracy, NHPs exposed to radiation versus unexposed NHPs. Consistent with historical data, female macaques appeared to be more sensitive to radiation, but the difference was not significant. Sex-based stratification allowed us to identify an interaction between gender and miR-16-2 expression, which affected the outcome of radiation exposure. Moreover, we developed a classifier based on two miRNAs (miR-30a and miR-126) that can reproducibly predict radiation-induced mortality. Altogether, we have obtained a 5-miRNA composite signature that can identify irradiated macaques and predict their probability of survival.
Introduction
The threat of ‘dirty bombs’ and the possibility of the use of improvised nuclear devices are growing with the changing global socio-political scenario and politically turbulent situations in nuclear warhead capable states. Hence, radiation exposure during terrorist events, industrial accidents, or natural disasters (such as the nuclear disaster after the tsunami in Japan) is a current and continuing threat for the future. The most important characteristic of such radiation exposure is the heterogeneity of the dose delivered to different parts of the body, and the prominence of symptoms depends on the magnitude of damage to the organs resulting in complex patho-physiological features (1–3). Effective planning for the medical response to a radiologic or nuclear incident is challenging and requires an in-depth understanding of how medical triage and treatment will occur. Current use of diagnostic screening to estimate the dose of accidental radiation exposure is mainly based on three factors: time to onset of radiation sickness, kinetics of lymphocyte depletion, and analysis of chromosomal abnormalities (2, 4). These assays are time-consuming and are often not quantitative enough to draw definitive conclusions. Because acute radiation syndrome (ARS) is a multi-organ syndrome (2), it is unlikely that a single biodosimetry assay can be used for rapid detection of radiation exposure and monitoring of the clinical symptoms and organ dysfunction in ARS. Therefore, complementary assays, such as the lymphocyte depletion kinetic assay can be combined with cytogenetic assays [for example, dicentric chromosome assay and micronucleus assay] for dosimetry (2, 5–9). Although cytogenetic assays are the gold standard for radiation biodosimetry, they may have a limited dose range for detection (up to 5 Gy for dicentric chromosome assay and micronucleus assay). In a radiological mass casualty scenario, resources for biodosimetry and medical countermeasures would be limited. Therefore, the availability of rapid biodosimetry diagnostics that will allow accurate triage and identify victims most likely to benefit from treatment would be critical.
Serum microRNA (miRNAs) fall under the emerging “omic” biodosimetry assays and can be measured by simple technology that may effectively determine whether an individual was exposed to radiation (and if so, whether the dose was sublethal or lethal) and predict long-term survival of exposed individuals (10). With current progress in miniaturization of quantitative PCR or multiplexed probe-based assays, a miRNA-based assay has the potential to become a point-of-care technology that can be performed outside the laboratory. It can allow shorter lag time post-exposure before the first sample can be taken compared to lymphocyte depletion kinetics and DNA damage assay using γ-H2AX, and it can provide results within 12 to 24 h after exposure. We successfully profiled and identified a panel of serum miRNAs in mice that were differentially expressed in response to total body irradiation (TBI) and correlated well with injury at sub-lethal and lethal doses (11). A key issue we addressed is the correlation of serum miRNAs with the impact of radiation, specifically with hematopoietic injury and animal viability. Working with a narrow dose range, we identified serum miRNAs that distinguish between sub-lethal (6.5 Gy) and lethal (8 Gy) exposure of C57BL/6J mice (11). This is of paramount importance because during a radiologic emergency, doses sustained will almost never be in specific increments, and distinction between lethal and sub-lethal doses is the key challenge. Moreover, this miRNA signature also predicted the impact of radiation on animal survival after pre-treatment with the radioprotective agent amifostine or mitigation using bone marrow-derived stromal cell transplantation. However, the key question that emerged from this study is whether this concept is evolutionarily conserved, that is, whether serum miRNA signatures can predict radiation-induced mortality in healthy primates and ultimately humans.
To address this question, we conducted a systematic analysis of serum miRNAs in healthy NHPs exposed to TBI alone or in combination with a radioprotective agent, gamma-tocotrienol (GT3) (12). Validation of the initial analysis was conducted in an independent cohort of NHPs. We discovered a signature of seven serum miRNAs that are altered with TBI in both mice and NHPs. Due to the lack of experimental data from healthy humans, we adopted an in silico approach to compare the loci coding for these miRNAs in mice, NHPs, and humans. The overall sequence homology in the regulatory elements upstream of these miRNAs was moderate across the three species. However, there was a combination of seven transcription factors that are predicted to regulate these miRNAs in humans, mice, and NHPs. A combination of three miRNAs (miR-133b, miR-215, and miR-375) was able to distinguish NHPs exposed to radiation versus unexposed NHPs with complete accuracy, even in the presence of a radioprotector, GT3. Three miRNAs (miR-30a, miR-126, and miR-375) correlated with the radioprotective properties of GT3 after TBI. Next, we observed a trend (difference was not statistically significant) in radiosensitivity of the animals based on sex, with male NHPs more likely to survive the same dose of radiation relative to their female counterparts. Accounting for this potential sex bias, we showed that miR-16-2 has a sex-specific association with radiation-induced mortality, that is, rhesus macaques that died showed on average 2-fold lower expression of miR-16-2 than survivors of the same sex. This hinted at the necessity of incorporating sex into predictive models for survival after high dose irradiation. Using multivariate logistic regression with sex as a confounding variable, we created a statistical model using miR-30a and miR-126, which allowed efficient prediction of radiation-induced mortality. Combining these results with our irradiation detection model, we have obtained a 5-miRNA signature that allows us to identify irradiated macaques and predict their probability of survival.
Results
Serum MiRNA Profiling and Identification of Radiation-responsive MiRNAs in Macaques
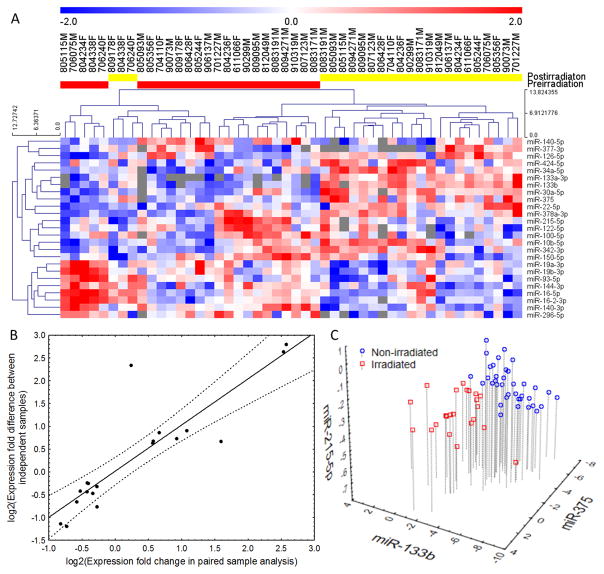
To analyze serum miRNAs altered by radiation in macaques during initial analysis, blood was drawn from 48 macaques a week before TBI and 24 h after radiation exposure. Pre-irradiation and post-irradiation samples were paired for analysis of radiation-responsive miRNAs. Furthermore, to correlate the serum miRNAs with radiation impact, the radioprotective agent, GT3, was administered 24 h before irradiation in 24 macaques (13). Both GT3- and vehicle-treated groups were divided into 3 dose levels: 5.8 Gy (LD30/60), 6.5 Gy (LD50/60), and 7.2 Gy (LD70/60). The LD represents expected lethality without any supportive care (antibiotics, blood products, and i.v. fluids) to allow for a better demonstration of the radioprotective effects of GT3 and to closely simulate a mass casualty scenario where these products are likely to be limited. A total of 651 miRNAs were detectable in at least one sample. The average number of miRNAs detected per sample was 215+/−44.8, and after filtering for missing data, 174 miRNAs were eligible for analysis. The flowchart of the datasets, comparisons, and validation steps (fig. S1) and raw data from the profiling experiment are presented as supplementary material (table S1). Paired pre-irradiation and post-irradiation samples were compared in the vehicle-treated macaques after removing the batch effect (fig. S2). This comparison identified a list of 25 miRNAs that met the significance criterion (Fig. 1A, table S2). Among these miRNAs, 12 were up-regulated after irradiation (hsa-miR-133b, hsa-miR-375, hsa-miR-377-3p, hsa-miR-34a-5p, hsa-miR-30a-5p, hsa-miR-424-5p, hsa-miR-342-3p, hsa-miR-10b-5p, hsa-miR-126-5p, hsa-miR-22-5p, hsa-miR-378a-3p, hsa-miR-133a-3p) and 13 were down-regulated (hsa-miR-215-5p, hsa-miR-122-5p, hsa-miR-150-5p, hsa-miR-144-3p, hsa-miR-140-3p, hsa-miR-16-5p, hsa-miR-19b-3p, hsa-miR-19a-3p, hsa-miR-16-2-3p, hsa-miR-93-5p, hsa-miR-296-5p, hsa-miR-140-5p, hsa-miR-100-5p). There was no significant dose dependence of the radiation-associated miRNAs, with variability in expression even within a dose (table S3). This is expected, considering that macaques are not inbred, and each individual animal may have a distinct response to the same dose of radiation. Therefore, we merged the three irradiated groups into one composite set of macaques irradiated with a lethal dose of radiation. Having identified 25 variable miRNAs in paired analysis, we validated them by comparing the post-irradiation vehicle-treated group with pre-irradiation samples allocated to receive GT3. This result yielded 19 miRNAs (table S4) that showed significant (p<0.05) association with radiation in both paired and independent comparison, convergent in the direction of expression change. The expression differences between pre-irradiation and post-irradiation samples showed nearly perfect correlation (r=0.88, p=0.02, Fig. 1B) between the paired and unpaired comparisons. Only miR-133a showed a divergent effect in the unpaired analysis, potentially because it was present in very low quantities and near the detection threshold in numerous pre-irradiation samples. Having established the set of radiation-dependent miRNAs, we used logistic regression with an Akaike information criterion-driven best subset selection to identify an efficient model of separating all non-irradiated from vehicle-treated irradiated macaques. The outcome was a model based on three miRNAs: miR-133b, miR-215, and miR-375 (Fig. 1C), which showed perfect separation of the groups with AUC (area under the ROC curve) 1.00 (95%CI 1.00–1.00) and maintained its performance in 10-fold cross validation (AUC=1.00 95%CI 1.00–1.00). Numerical parameters of the model are listed in table S5. Hosmer-Lemeshow’s test showed a very good fit of the model (p=1.00, table S6). Because some of the animals were missing data for at least one of the microRNAs evaluated during model development, 60 macaques were eligible for analysis (37 pre- and 23-post-irradiation). A reanalysis with background expression imputed for missing observations yielded convergent results, with an area under the ROC curve of 1.00 (95%CI 1.00–1.00), correctly classifying all 72 samples.
Fig. 1. MiRNA-based irradiation classifier development and performance.
(A) Hierarchical clustering plot of miRNAs detected in the sera before and after irradiation. The heatmap depicts the 25 miRNAs that were significantly differently expressed in paired analysis of pre- (red) and post-irradiation (yellow) vehicle-treated NHPs. Red color represents high expression, blue represents low expression, and the color scale depicted above the heatmap illustrates normalized expression values with units representing standard deviation scores. Numbers above each column identify individual animal IDs, with the letters M and F representing males and females. (B) Correlation between miRNA expression differences in paired (vehicle-treated post- vs pre-irradiation) and unpaired NHPs. The expression fold differences were log2 transformed, and a value of 0 on both axes corresponds to a lack of up- or downregulation. (C)Scatterplot of three miRNAs used in the final classification model for detecting irradiation: miR-133b, miR-375, and miR-215-5p. Values are presented as dCp: differences between the average amplification cycle number in the exponential product gain phase (Cp) and the Cp of an individual miRNA, with higher values representing higher expression of miRNAs.
Evolutionary Conservation of Radiation-responsive MiRNAs and Their Regulation
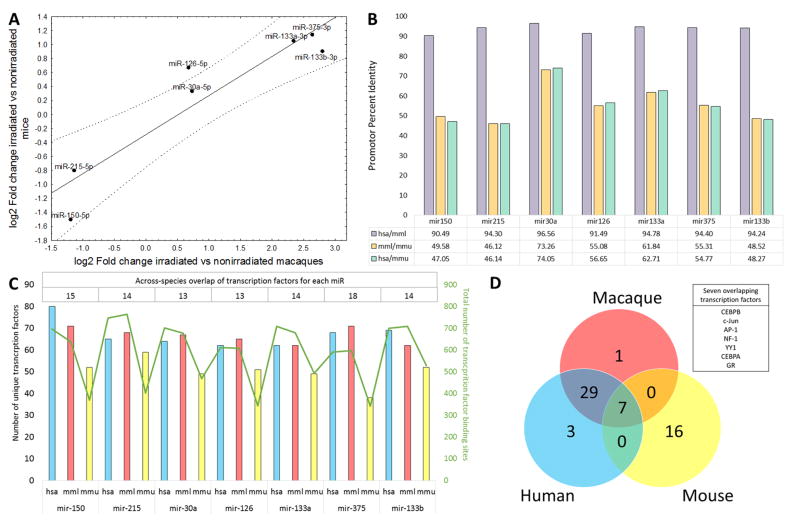
Having established that irradiated and non-irradiated NHPs may be distinguished using a serum miRNA signature, we cross-referenced the radiation-dependent miRNAs with results from our earlier work in a mouse model of irradiation (11). To do so, we reanalyzed the mouse dataset by comparing the expression of the 19 miRNAs between 10 control mice and a pool of 20 mice that received 6.5 or 8 Gy radiation exposure, which is comparable to the high, potentially lethal dose used in the macaque experiment. By correlating the fold changes in miRNA expression noted in the mouse model and 19 miRNAs significantly affected by irradiation in macaques (fig. S3), we identified seven miRNAs that showed significant (p<0.05 and FDR<0.15) associations with irradiation in both datasets and were convergent in the direction of change (Fig. 2A). The expression changes for these seven miRNAs showed strong correlation (r=0.9303; p=0.0024) in mice and macaques. This divergence is potentially due to undetectable pre-irradiation expression in the macaques. To determine whether these miRNAs were under the control of homologous transcription factors, we interrogated the promoter sequences of the miRNAs in mice, macaques, and humans for species-specific transcription factor binding sites. Positions of extracted promoter sequences are presented in table S7. An average of 55.7+/−9.0% percent identity was observed for the promoter sequences of the seven miRNAs between mice and macaques or humans (Fig. 2B). Multiple transcription factors have putative binding sites in the promoter regions of the seven miRNAs, with an average of 15 transcription factors conserved across the three species for each of the seven miRNAs (Fig. 2C and table S8). Next we focused on identifying transcription factors that were predicted to bind the promoter sequences of all seven radiation-responsive miRNAs in human, macaque, and mouse. It was very striking that despite the moderate homology in the promoter sequence across the three species, there were seven transcription factors [C/EBPalpha (CCAAT-enhancer-binding protein alpha), C/EBPbeta (CCAAT-enhancer-binding protein beta), NF-1 (Nuclear factor 1), GR (glucocorticoid receptor also known as NR3C1), YY1 (Yin Yang 1), c-Jun, and AP-1(Activator protein 1)] (Fig. 2D) that were predicted to bind the promoter sequences of all seven miRNAs in mice, macaques, and humans. A detailed list of transcription factors overlapping across the three species shown in Fig. 2D is presented in table S9. Aligned miRNA promoter sequences are presented in tables S10–16. Functionally, the miRNAs may exert their action by regulating translation. Using the MiRWalk tool, we identified validated targets for our 7 conserved microRNAs. We found that IGF1R was a target for 5 out of 7 miRNAs (mir-375, mir-133a, mir-133b, mir-215, and mir-30a). Previously, IGF1R was associated with radiation response (14, 15), along with a number of other targets listed in table S17. Nevertheless, the functional relevance of the miRNAs identified in this study has to be validated at the cellular level, because serum expression may not reflect the intracellular milieu of cells subjected to irradiation.
Fig. 2. Conservation of radiation-dependent miRNAs across the species.
(A) Correlation plot of expression differences of miRNAs significantly up- or downregulated in both the NHP and mouse datasets (reanalyzed from Acharya et al. (11)). Data are presented as log2-transformed values on both axes, with a value of 0 corresponding to a lack of change. (B) Percent identity of aligned promoter sequences between Macaca mulatta (mml), Homo sapiens (hsa), and Mus musculus (mmu) for seven selected microRNAs. (C) Number of distinct transcription factors predicted to bind to the promoter region of each microRNA in each species (left Y axis, hsa - blue bars, mml - red bars, mmu - yellow bars), total number of predicted transcription factor binding sites for each microRNA in each species (right Y axis, green line), and number of overlapping transcription factors across the species for each microRNA (top table). (D) Cross-species overlap of the transcription factors with at least one binding site present in all seven microRNA promoter sequences. Seven transcription factors (listed in the text box) had binding sites in all seven microRNAs in all analyzed species.
Correlating Expression of Conserved MiRNAs with Impact of GT3 on Radiation Injury
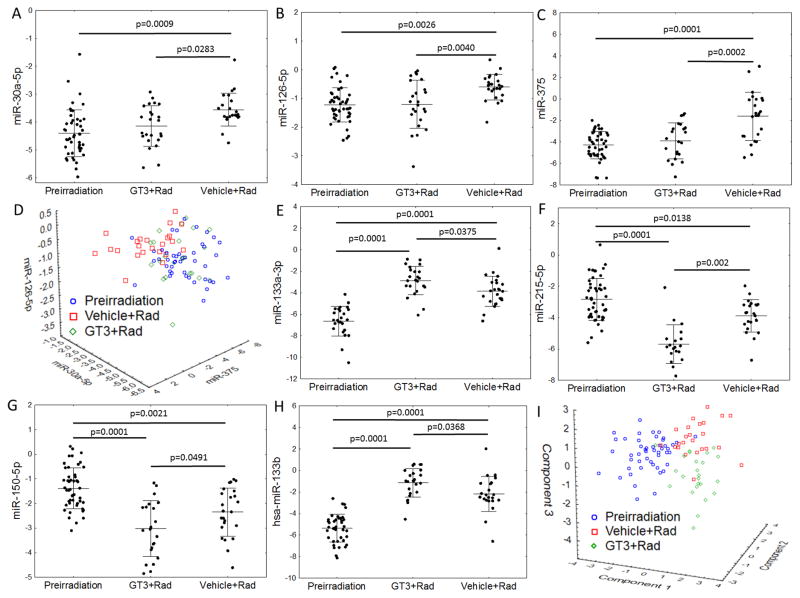
Previous publication showed that the administration of GT3 enhanced the rate of hematopoietic recovery in irradiated NHPs (13). We analyzed the correlation of the evolutionarily conserved seven miRNAs with the impact of GT3 on the radiation response of NHPs. As anticipated, the seven miRNAs were significantly (p<0.05 in all cases) altered in macaques that received vehicle control and were exposed to TBI. Serum concentrations of miR-30a, miR-126, and miR-375 correlated with the radioprotective efficacy of GT3, that is, these miRNAs in the GT3-treated irradiated cohort resembled the unirradiated animals (Fig. 3A–C). These three miRNAs could be used as indices of GT3 efficacy in protecting animals from the radiation impact because their profiles after GT3 treatment converged with those observed in the pre-irradiation macaques (Fig. 3D). However, concentrations of miR-133a, miR-215, miR-150, and miR-133b showed a greater response than observed in vehicle-treated controls (stronger upregulation for miR-133a/b and stronger down-regulation for miR-215 and miR-150 (Fig. 3E–H)). When compressed to a 3-dimensional plot using principal component analysis, GT3-treated samples stood apart from both unirradiated and untreated irradiated controls (Fig. 3I). The mechanism of GT3’s radioprotective efficacy in NHPs remains unclear, but, as published previously, GT3 boosts the recovery of the hematopoietic system after TBI (13) However, because GT3 also increases C-reactive protein (CRP) concentration after irradiation, the differential impact on radiation-associated miRNAs may be due to a complex biological function of GT3 (12). Unexpectedly, the impact of GT3 on miR-133b and miR-215 expression after irradiation outweighed that on miR-375, which allowed the model to maintain its 100% diagnostic accuracy for separating irradiated GT3-treated macaques from any of the pre-irradiation samples (fig. S4). Given the lack of differences in survival probability of GT3- or vehicle-treated macaques (log-rank p=0.6120; fig. S5), we showed that the expression of radiation-associated miRNAs is altered by GT3 but the biological role of the miRNAs or the processes regulated by GT3 and impacting their expression remain unknown.
Fig. 3. GT3 (gamma-tocotrienol) rescue’s impact on radiation-dependent miRNA expression.
Expression of the seven miRNAs that showed replicable changes in both NHPs and mice depending on the time point and treatment. Different number of data points in all figures are due to undetectable amounts of particular miRNAs at individual time points. In all cases, significant differences in a post-hoc Tukey’s test are marked with asterisks: (A) miR-30a-5p, (B) miR-126-5p, (C) miR-375. (D) A scatterplot of the expression of three miRNAs (miR-30a-5p, miR-126-5p, and miR-375) grouped by treatment. MiRNA expression in GT3-treated macaques overlapped with that in pre-irradiation samples. Quantities of four miRNAs: (E) miR-133a-3p, (F) miR-215-5p, (G) miR-150-5p, (H) miR-133b showed a more extreme radiation-induced change of expression in GT3-treated macaques in comparison with that observed in vehicle-treated samples. (I) A principal component analysis plot of the three groups. The components were extracted from the seven-miRNA subset. For the purpose of this plot, missing values of the seven miRNAs were substituted with averages.
Sex-Specific Stratification Allowing MiRNAs to Predict Radiation-Induced Mortality
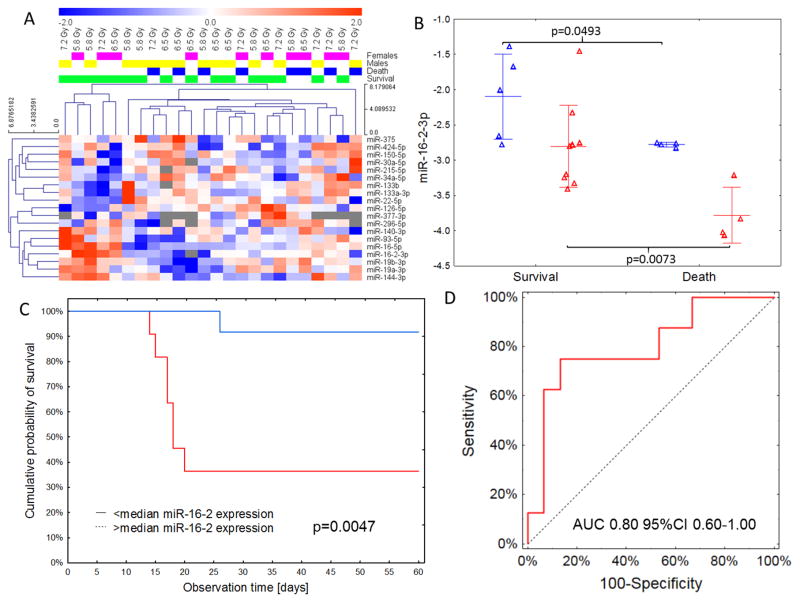
Having shown the performance of our classifier, we investigated whether miRNAs can be used to predict the radiation-induced mortality of macaques. To answer this question, we used the 19 miRNA subset of radiation-responsive miRNAs and compared post-irradiation expression in the vehicle-treated group depending on the status of death or survival. No statistically significant differences between animals that died and those that survived were noted in raw data comparisons after adjustment for multiple hypothesis testing (table S18). Epidemiological(16–18), clinical(19), and biological studies(20) have produced indications but no clear evidence of possible sex-specific differences in radiation sensitivity (endpoints: mortality and cancer). However, very few studies to date have systematically analyzed possible sex-specific differences in sensitivity to TBI (21, 22). We speculated that the lack differences of miRNA expression between surviving and non-surviving animals in our initial analysis could have been caused by a sex-specific response to TBI (table S19). Therefore, we analyzed the radiation-responsive miRNAs for correlation with mortality after stratifying for sex (Fig. 4A). To account for this effect, we adjusted the miRNA expression comparisons by sex and observed a significant association of miR-16-2 with viability (Bonferroni-adjusted p=0.02, table S18). A sex-stratified approach to outcome prediction with expression showed significant separation – macaques that did not survive TBI showed on average 2-fold lower expression of miR-16-2 than survivors of the same sex (Fig. 4B; p=0.049 in females and p=0.007 in males). This effect overlapped with lower baseline expression in males than in females, obscuring the effect if sex was not factored into the analysis. Dichotomization of the group relative to median expression of miR-16-2 in males and females showed that a strong suppression of miR-16-2 was associated with a survival probability of 35%, in comparison to 90% noted in macaques with greater than median expression of miR-16-2 (log-rank p=0.0047; Fig. 4C).
Fig. 4. Sex-specific miRNA expression and miRNA-based survival prediction.
(A) Hierarchical clustering plot of miRNAs that showed significant (p<0.05) changes of expression in both paired and unpaired comparisons depending on the outcome and sex of the macaque. No clear pattern of death/survival signature was noted, with females generally dying at a higher dose than males. (B) Sex-specific differences in miR-16-2 expression relative to outcome. Expression of miR-16-2 was lower in males (red) than females (blue), but in both sexes macaques that died showed a downregulation of serum miR-16-2. Lines represent means with standard deviations. (C) Kaplan-Meier curves showing survival relative to miR-16-2 expression. Higher (>median) expression of miR-16-2 was associated with a Hazard Ratio of 0.08 (95%CI 0.01–0.69) of dying during the 60-day post-irradiation period. (D) ROC curve for the miR-30a/miR-126/sex classification model predicting the outcome of irradiation (AUC = 0.80 95%CI 0.60–1.00).
A Serum MiRNA Signature to Predict Radiation-induced Mortality in Primates
Taking into account information on the potential impact of sex on post-irradiation microRNA expression and survival, we constructed a logistic regression model allowing us to predict the outcome of the 24 vehicle-treated irradiated macaques. The classifier was based on expression values of two microRNAs: miR-30a and miR-126 (univariate Kaplan-Meier curves for high and low expression of miR-30a and miR-126 are presented in fig. S6A and S6B) adjusted for the specimen’s sex. The area under the ROC curve of the model was 0.80 (95%CI 0.60–1.00) and 0.74 (95%CI 0.51–0.97) in 6-fold cross-validation; Fig. 4D). The model’s sensitivity and specificity equaled 75.0% and 86.7%, respectively, with a very good model fit throughout the predicted probability range (Hosmer-Lemeshow test p=0.64). After building this model, we had a total of five miRNAs needed to identify irradiated macaques and predict the outcome: miR-133b, miR-215, miR-375, miR-126, and miR-30a. However, to make the models applicable, relative miRNA expression needed to be quantified against a small set of stable normalizer miRNAs rather than the average of the expression array. To identify the best normalizers, we used the Normfinder (23) algorithm and picked two of the miRNAs with the highest stability: miR-142-5p and miR-320a (table S20). We then reanalyzed normalized data for miR-142/miR-320a and reestimated classifier parameters (table S5). Performance of the irradiation-detection classifier and survival classifier remained very good, with AUC 0.99 (95%CI 0.98–1.00) for irradiation and 0.77 (95%CI 0.56–1.00) for survival prediction. The association of miR-16-2 persisted for both death (p=0.0116) and sex (p=0.0003) and was convergent with the direction of changes observed in the array average-normalized dataset (figure S7).
Validation in an independent cohort of macaques
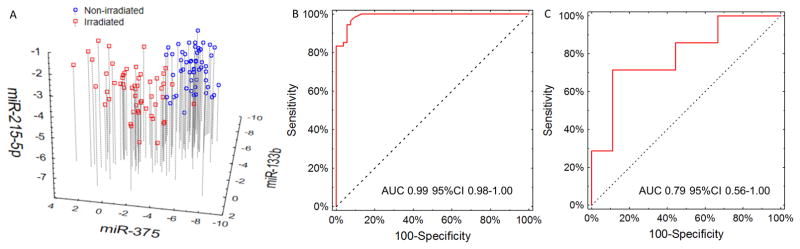
We measured the expression of evolutionarily conserved miRNAs associated with radiation exposure and survival using qPCR in 54 male macaques recruited during a separate, time-unrelated study before and 24 h after irradiation with a dose of 7.2 Gy. Raw Cp values were normalized against the average of miR-142 and miR-320a. Paired comparisons of pre- and post-irradiation expression showed that all but miR-215 (p=0.08) differed significantly (p<0.05), with the direction of change being identical to that in the primary cohort (table S21). The magnitude of differences between pre- and post-irradiation expression of microRNAs in the validation group correlated strongly (r=0.89; p=0.0069) with those observed in the primary one (fig. S8). Hierarchical clustering showed strong grouping of pre- and post-irradiation samples (fig. S9). Deployment of the irradiation signature based on miR-133b, miR-375, and miR-215 on the validation dataset yielded excellent performance (Fig. 5A), with an area under the ROC curve of 0.99 (0.98–1.00, Fig. 5B). Sensitivity of the classifier reached 95% (3/54 false negatives) with a specificity of 95% (3/54 false positives). In the validation cohort, we had survival data for only 16 macaques. In fig. S10A–C, we present individual survival curves for miR-30a, miR-126, and miR-16-2 in the validation group. Testing of survival signature yielded an area under the ROC curve of 0.79 (95%CI 0.56–1.00) for the model based on expression of miR-30a, miR-126, and gender (Figure 5C). This suggests that by using only 5 miRNAs (miR-133b, miR-215, miR-375, miR-30a, and miR-126) and two normalizers (miR-142 and miR-320a), we can successfully detect exposure to irradiation with a lethal dose during the preceding 24 h and estimate the probability of radiation-induced mortality. The triage protocol using the 5-miRNA signature is presented in fig. S11.
Fig. 5. Performance of the 5-miR irradiation/outcome classifier.
Expression of miR-133b, miR-215, miR-375, miR-30a, and miR-126 was renormalized against the average of two selected miRNAs (miR-142 and miR-320a). (A) Scatterplot of miR-133b/miR-215/miR-375 classifier for irradiation after renormalization to miR-142 and miR-320d. (B) ROC curve showing the performance of the classifier in the validation group. The area under the ROC curve equaled 0.99 95%CI 0.98–1.00. (C) ROC curve depicting the performance of the classification model for predicting death due to irradiation in the validation group. Area under the ROC curve equaled 0.79 95%CI 0.56–1.00.
Discussion
In recent years, circulating miRNAs have emerged as promising biomarkers for different pathological conditions, including radiation exposure. The inherent stability of serum miRNAs under harsh conditions and reproducible quantification in individuals of the same species make miRNAs attractive candidates for use as non-invasive biomarkers. We discovered miRNA signatures that effectively distinguished between control, sub-lethal (low dose), sub-lethal (high dose), and lethal TBI cohorts in mouse models within 24 h after radiation and correlated these signatures with the radiation-induced loss of hematopoietic stem cells (HSC). At 24 h after exposure, this serum miRNA signature distinguished fatal versus non-fatal hematopoietic injury in mice (11). The most important observation from our study was that serum miRNAs correlate with the impact of radiation in mice and can predict radiation-induced mortality. Here we take the next step as we analyze serum miRNAs in NHPs, which is the closest we can get experimentally to healthy humans. Unlike the mice, the NHPs are not inbred, and the dose response is not as clear cut, because each individual NHP has a distinct response to radiation. All doses used in our study are considered lethal because one or more NHPs are dying. However, the key is to predict which individuals are more likely to survive the exposure to radiation. We have identified radiation-responsive serum miRNAs and observed that a subset of these miRNAs is evolutionarily conserved. We found that miR-150, which was the most consistent marker for radiation exposure in mice, was surpassed by a combination of three miRNAs (miR-133b/miR-215/miR-375) that distinguished irradiated and un-irradiated macaques with complete accuracy. The serum concentrations of miR-133b/miR-215/miR-375 continued to accurately classify irradiated versus un-irradiated animals even after treatment with GT3, a promising radioprotector. A combination of these miRNAs may potentially emerge as a biomarker to precisely identify individuals exposed to radiation during or after an accidental radiation exposure incident.
To explore the relevance of our results in humans, we used an in silico approach and analyzed the genetic elements regulating these conserved miRNAs. The promoter regions had ~50% homology, but there were ~15 transcription factors for each of the seven miRNAs that were conserved in all three species. The most striking observation was that there are 7 transcription factors that are predicted to regulate all seven conserved radiation-responsive miRNAs in mice, macaques, and humans. GT3 has a radioprotective efficacy in NHPs. The small number of animals in each group and overall low mortality in the experiments with GT3 may be the reason why GT3 did not demonstrate a significant survival benefit in the macaques exposed to radiation. Lower mortality in controls results in a small difference compared to the drug-treated group, and such experiments need a large sample size to observe statistically significant differences between groups. However, three (miR-30a, miR-126, and miR-375) of the seven conserved radiation-responsive serum miRNAs correlated with GT3, that is, the expression of miR-30a, miR-126, and miR-375 was unchanged by irradiation only when GT3 had been administered before exposure. Therefore, it is feasible that these miRNAs may serve as biomarkers for GT3 efficacy, particularly because two of them, miR-30a and miR-126, were reproducibly associated with the outcome of irradiation as well as with GT3 response.
Early studies in mice have indicated females were, in general, 1.5 times more sensitive to X-ray irradiation than males of the same strain, suggesting a constant effect of sex (21). However for humans, due to the limited data availability with regard to both clinical studies and biological experiments, as well as sometimes conflicting findings of epidemiological studies, it has not been possible to make unequivocal statements about sex-specific differences in radiation sensitivity. There have been no systematic studies in NHPs that demonstrate the correlation of sex with radiation-induced mortality. Our results are consistent with the mouse studies showing that TBI-induced mortality is more pronounced in female macaques. However, by taking sex into account, we discovered that the decrease of a single miRNA, miR-16-2, shows a sex-specific pattern of expression differing between macaques that experience fatal versus non-fatal radiation injury. Furthermore, by combining the radiation biomarker miRNAs (miR-133b, miR-215, and miR-375), the survival indicators (miR-30a and miR-126), and normalizer miRNAs (miR-142 and miR-320a) into a compound classifier, we can successfully detect exposure to radiation and accurately estimate the probability of death within 24 h after exposure (table S5). Considering that these results were generated using a large number of NHPs, we anticipate that the results would extrapolate to a human radiation exposure scenario.
A limitation of our study is the restricted ability to experimentally validate results in NHPs. For example, it is striking that the same seven transcription factors are predicted to regulate the radiation-responsive serum miRNAs in humans, macaques, and mice. However, these results need to be experimentally validated, and our ability to do so has been limited for the following reasons. First, we don’t know the source (cell type) of the serum miRNAs, and second, only a limited range of experiments are feasible with macaques. For this study, we used existing serum samples from NHPs collected before and after irradiation. Although the number of samples was large enough to obtain the results reported here, these should be further validated with a larger number of independent NHP samples to account for potential sources of variation in the real world – tissue handling, varying collection times, and technical factors.
Materials and Methods
Study Design
The aim of the study was to identify miRNA signatures for efficient detection of high-dose irradiation and radiation-induced mortality in primates. The study used 48 animal samples from a study with GT3 described in a previous publication [13]. In short, the animals were subjected to randomization and given GT3 or vehicle before irradiation with three dose levels. No blinding was introduced at that stage. Serum samples were collected before and after irradiation, and after RNA isolation, miRNA expression was quantified by qPCR. After refining the signatures for irradiation detection and survival prediction, a replication cohort of additional 54 male macaques from a separate, blinded study was used to test the performance of the developed models.
Animals
Healthy naïve rhesus macaques (Macaca mulatta, Chinese sub strain, N=48) for initial analysis were obtained from Primate Products, Inc. and quarantined for 6 – 7 weeks before the start of the experiment. Clinically healthy male and female rhesus macaques 3 – 5 years of age, weighing 3.6 to 8.4 kg, were used in this study. All NHPs were individually housed in stainless steel cages in environmentally controlled rooms maintained at 22 °C ± 2 ºC, 30 – 70% relative humidity, 10 – 15 air change cycles per h, and 12 h light:12 h dark cycle. Animals were fed primate diet (Harlan Teklad T.2050 diet) twice daily and they received drinking water ad libitum. All the animals received enrichment food (fresh fruits and vegetables, prima treats, peanuts, marshmallows etc.) once a day Monday – Friday. They received mirrors, toys, and challenge balls for enrichment. TVs were used for sensory enrichment for 4 – 5 h at least 3 times a week. All the animals were able to see, hear, and/or touch their conspecifics through the cages. All animals were serologically negative for Macacine herpesvirus 1 (Herpes B virus), Simian retrovirus (SRV), Simian T-cell leukemia virus (STLV), and Simian immunodeficiency virus (SIV). They were vaccinated for measles and confirmed to have positive antibody titers, and they tested negative for tuberculin. Animals were stratified by sex and by body weight increases during the quarantine period and then assigned to different treatment groups. This animal study was conducted in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-International.
Validation group
The independent validation cohort consisted of additional 54 male rhesus macaques (Macaca mulatta, Chinese sub strain; 3.5 – 5.5 years of age, weighing 4.0 – 8.0 kg) procured from National Institute of Health Animal Facility. All other details for this cohort were the same as described above for the initial study analysis. Blood samples were collected for serum one week before and 24 h after irradiation as in the primary cohort. Out of 54 irradiated animals in the validation cohort, 16 were used for the survival study, making them eligible for validation of the survival signature.
Drug Preparation and Administration
GT3 (50 or 100 mg/ml) in 5% Tween-80 in saline was purchased from Yasoo Health Inc. The quantity of GT3 for each NHP was based on individual NHP weight. Drug and vehicle were administered at the dorsal scapular area (between the shoulder blades). The area surrounding the injection site was shaved at least 48 h before the administration of drug, so the site could be easily observed for any adverse skin reactions such as rash/eruption, inflammation, irritation, or abscess formation after GT3 or vehicle administration. Immediately before drug injection, the site was wiped with 70% isopropyl rubbing alcohol and allowed to air dry; the drug was administered sc using a 3 ml disposable luer-lock syringe with a 25-gauge 5/8 inch needle.
Radiation Exposure
A maximum of 4 NHPs in the primary cohort and 14 animals in the validation cohort were irradiated per day. Food was withheld from each animal for about 12–18 h before exposure to minimize the occurrence of radiation-induced vomiting. Approximately 30 – 45 min before irradiation, 10 – 15 mg/kg of ketamine hydrochloride was intramuscularly administered to NHPs for sedation, then placed in custom-made Plexiglas irradiation boxes and secured in a seated position. Two NHPs at a time were placed on the irradiation platform facing away from each other and exposed with a midline dose of 5.8, 6.5, or 7.2 Gy at a dose rate of 0.6 Gy/min. To deliver the precise dose, NHPs’ abdominal widths were measured with digital calipers. Animals were observed throughout the irradiation procedure via in-room cameras. After irradiation, animals were returned to the transport cart and to their cages in the housing area and monitored for recovery from the procedure. Dose rate measurements were based primarily on the alanine/EPR (electron paramagnetic resonance) system, currently accepted as one of the most accurate methods for relatively high radiation doses and used for comparisons between national metrology institutions(24). The calibration curves (EMXmicro spectrometer, Bruker Corp.) used in dose measurements at Armed Forces Radiobiology Research Institute (AFRRI) are based on standard alanine calibration sets purchased from the US National Institute of Standards and Technology (NIST). The alanine dosimeters obtained from NIST had been calibrated in terms of absorbed dose to water using the US National Standard Radiation Sources. At AFRRI, identical alanine dosimeters were placed midline within NHP phantoms (Plexiglas cylinders 6.9, 10, 12.5 cm in diameter and 34.5 cm length) and irradiated to approximately 100 Gy. Measurement of their EPR signals using the calibration curve constructed with alanine dosimeters from NIST provided dose rates to water in the core bodies of NHP. A small correction (0.9906) was subsequently applied to adjust for the difference in mass energy absorption coefficients between water and soft tissue.
To assess the radioprotective efficacy of GT3 as a radiation countermeasure, 48 NHPs from the initial analysis cohort were divided into 3 groups of 16 animals, and each group was exposed to the following lethal doses of radiation: 5.8 Gy (LD30/60), 6.5 Gy (LD50/60), and 7.2 Gy (LD70/60) (0.6 Gy/min). GT3 was administered to the NHPs subcutaneously, 24 h before radiation exposure.
Those NHPs exposed to 5.8 Gy were further divided into two groups of 8 NHPs each, with one group receiving 37.5 mg/kg GT3 (N=4) or vehicle (N=4) and the other receiving 75 mg/kg (N=4) or vehicle (N=4). All animals exposed to 6.5 and 7.2 Gy received 37.5 mg/kg GT3 or vehicle. In all experimental groups, 50% of animals were injected with GT3, and the remaining 50% were controls (vehicle-treated).
All 54 animals in the validation cohort did not receive any test drug before blood collection, which took place at either −7 d or + 24 h. Similarly, the 16 animals in the validation cohort used for the survival study did not receive any test drug during the course of the study.
Blood Sample Collection from NHPs
Blood was collected from chair-restrained NHPs via the saphenous vein located superficially on the caudal aspect of the lower leg, distal to the knee. At 48 h before the first blood collection, the hairs surrounding the collection site were shaved using an electric cordless shaver. For venipuncture, a tourniquet was applied to the hind leg below the knee, and the site was cleaned using a 70% isopropyl alcohol wipe and dried with sterile gauze. The desired volume of blood was collected with a 3 ml disposable luer-lock syringe with a 25-gauge needle. For serum collection, the blood sample was transferred to Capiject serum separator tubes (3T-MG; Terumo Medical Corp.) and allowed to clot for 30 min, then centrifuged at 400 x g for 10 min. The serum samples were stored at −70 ºC until use.
Sample Selection and Study Protocol
Samples evaluated within this study for initial analysis and validation originated from different studies conducted at different times (at least two years apart). Blood had been drawn at multiple time points in the original protocol, but for the purpose of identifying early irradiation biomarkers and predictors of survival, we profiled microRNA expression in the −7 days sample and the one collected 24 h after irradiation.
MicroRNA Profiling
MiRNA profiling was performed as described previously (11, 25) using quantitative PCR panels from Exiqon with LNA-modified primers. PCR panels are accurate and convergent with miRNA sequencing data, provided that the number of copies of a miRNA exceeds 10 per million (26). Because no macaque-specific miRNA panels were available at the time of the study, human panels I and II were used instead, and qPCR signals were decoded using Homo sapiens miRNA annotations. Profiling was performed at Exiqon and all the reagents for the RNA isolation were obtained from Exiqon. After blood collection from the macaques, the samples were centrifuged at 1000 x g for 15 min to separate the serum. Before profiling, serum samples were thawed on ice and centrifuged at 3000 x g for 5 min in a 4 °C microcentrifuge. An aliquot of 200 μl (75 μl serum+ 125 μl water) per sample was transferred to a new microcentrifuge tube, and 60 μl of Lysis Solution BF containing 1 μg carrier RNA per 60 μl Lysis Solution BF and RNA spike-in template mixture was added to the sample. The total amount of RNA used in the analysis was 65 ng. The tube was vortexed and incubated for 3 min at room temperature, followed by addition of 20 μl Protein Precipitation Solution BF. The tube was vortexed, incubated for 1 min at room temperature, and centrifuged at 11,000 x g for 3 min. The clear supernatant was transferred to a new collection tube, and 270 μl isopropanol was added. The solution was vortexed and transfer to a binding column. The column was incubated for 2 min at room temperature and emptied using a vacuum-manifold. A 100 μl wash solution 1 BF was added to the columns. The liquid was removed using a vacuum-manifold, 700 μl wash solution 2 BF was added, and the column was spun at 11.000 x g to dry the columns entirely. The dry columns were transferred to a new collection tube, and 50 μl RNase-free H2O was added directly to the membrane of the spin column. The column was incubated for 1 min at room temperature before centrifugation at 11,000 x g. The RNA was stored in a −80 °C freezer. 19 μl RNA was reverse transcribed in 95 μl reactions using the miRCURY LNA Universal RT microRNA PCR, Polyadenylation and cDNA synthesis kit (Exiqon). cDNA was diluted 50 x and assayed in 10 μl PCR reactions according to the protocol for miRCURY LNA Universal RT microRNA PCR; each microRNA was assayed once by qPCR on the microRNA Ready-to-Use PCR, Panel I+II using ExiLENT SYBR Green master mix. Negative controls excluding template from the reverse transcription reaction were analyzed the same way as the samples. The amplification was performed in a LightCycler 480 Real-Time PCR System (Roche) in 384-well plates. The amplification curves were analyzed using the Roche LC software, both for determination of Cp (by the 2nd derivative method) and for melting curve analysis.
Ethics Statement
All procedures involving animals were approved by Armed Forces Radiobiology Research Institute (AFRRI) Institutional Animal Care and Use Committee (IACUC) and second tier Department of Defense Animal Care and Use Review Office (ACURO). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (27).
Statistical Analysis
Preprocessing of Profiling Data
The dataset was filtered for miRNAs that were present in 50% of both pre-irradiation and postirradiation samples, with an additional restriction that the miRNA should be detectable in at least 2 samples from every drug/dose/time point block. Missing data were imputed with 37 cycles as a threshold for miRNA detection. After filtering the dataset for miRNA detection frequency, we evaluated it for potential batch effects resulting from the use of different generation Exiqon arrays and removed them using ComBat – an empirical Bayes method of identifying and removing batch effect (28) (fig. S2).
Statistical Methods
Principal component analysis was used for batch effect evaluation. Hierarchical clustering with Euclidean distance was used for visualization of the dataset. Comparisons of miRNA in paired pre-irradiation and post-irradiation samples were performed using a paired t-test. Statistical significance for this step was stated if a given miRNA differed with a p value <0.05 and showed a false discovery rate (calculated using the Benjamini-Hochberg method) <0.15. This fairly relaxed significance criterion was established to account for pairs dropping out of the analysis if a single pre- or post-irradiation sample yielded no miRNA detection. The validation with independent samples was done using a standard t-test under the same threshold for significance as in the paired analysis (p<0.05; FDR<0.15). The same threshold was used for identifying miRNAs associated with survival. Sex-adjusted comparisons were performed using an analysis of covariance model. Logistic regression models for irradiation detection and outcome prediction were created using an Akaike information criterion-driven best subset variable selection. Goodness of fit was evaluated using the Hosmer-Lemeshow test (29). Diagnostic accuracy of the model was evaluated by calculating the area under the Receiver Operating Characteristics (ROC) curve (AUC). The regression models for radiation effect identification and outcome prediction were created using 10-fold cross validation and 6-fold cross validation, depending on the number of samples in the two analyses [n=72 for the irradiation model (all 48 pre-irradiation and 24 vehicle-treated post-irradiation) and n=24 for the survival model (only vehicle-treated post-irradiation), respectively]. Given that we needed to use all samples for the analysis, adjusting for batch effect and filtering the variables before model development, a full cross-validation as defined in Simon R.M. et al. (30) was not performed. Multiple group comparisons were performed with one way analysis of variance, followed by Tukey’s HSD (honest significant difference) post hoc test for between-group comparisons. Kaplan-Meier curves were used to present the outcome, with log-rank test used to confirm statistical significance for differences in cumulative probabilities of survival between the groups with different miRNA expression. Reanalysis of the Acharya et al (11) dataset was done using a standard t-test. Correlations were evaluated using Pearson’s correlation coefficient. Log-rank test was used for univariate survival analysis. 95% Confidence Intervals were computed for AUCs, and all tests were two sided. Threshold for significance for univariate comparisons was established at p<0.05. All analyses were done using Statistica 12.5 (Statsoft) and MultiExperiment Viewer (Dana-Farber Cancer Institute).
Bioinformatic Analyses
Promoter sequences of miRNAs that showed significant changes in the same direction in both macaque and mice datasets were extracted from GenBank using UCSC Genome Browser (31) for Macaca mulatta (Nov.2015 (BCM Mmul_8.0.1/rheMac8)), Mus musculus (Dec. 2011 (GRCm38/mm10)), and Homo sapiens (Dec. 2013 (GRCh38/hg38)) (table S7). For intragenic miRNAs, the analysis of promoter transcription factor binding sites was performed for both the miRNA itself and an upstream region of the gene in which the miRNA was located. In each case, the sequences of 1500 base pairs upstream to 5′ end of gene sequence were extracted and imported into PROMO (32), a tool that uses the TRANSFAC database (33) to identify specific transcription factor binding sites in a given sequence and species. For Macaca mulatta, we used primate transcription factors and binding sites because Macaca mulatta data were not available. For Mus musculus, we used mouse transcription factors and binding sites, and for Homo sapiens only human ones. The output (transcription factors binding to the interrogated genomic sequences from the three species) was cross-referenced to identify common regulatory elements present on a miRNA basis and universal for all miRNAs with similar patterns of change after radiation. To check the conservation of promoter sequences across the three analyzed species, we used Clustal Omega (34), an online multiple sequence alignment program which is able to compare three or more sequences by usage of seeded guide trees and Hidden Markov Model profile-profile techniques. Alignment of the three sequences was showed in Clustal without numbers format (tables S10–16). Percent identity of each pair of promoter sequences between the species was calculated.
Supplementary Material
Table S1. Raw expression data from miRNA qPCR profiling (separate xls file).
Table S10. Promoter sequences of miR-30a (separate txt file).
Table S11. Promoter sequences of miR-126 (separate txt file).
Table S12. Promoter sequences of miR-375 (separate txt file).
Table S13. Promoter sequences of miR-150 (separate txt file).
Table S14. Promoter sequences of mir-133b (separate txt file).
Table S15. Promoter sequences of miR-133a (separate txt file).
Table S16. Promoter sequences of miR-215 (separate txt file).
Table S2. Paired sample miRNA expression comparisons (separate xls file).
Table S8. Transcription factor binding sites in miRNA promoters of human, macaque, and mouse genomes (separate xls file).
Table S17. Validated gene targets of radiation-associated evolutionarily-conserved miRNAs (separate xls file).
Table S20. Selection of best normalizer miRNAs (separate xls file).
Table S3. miRNA expression in macaques irradiated with different doses.
Table S4. Independent sample miRNA expression comparisons.
Table S5. Numerical parameters of logistic regression models.
Table S6. Goodness of fit evaluation of the irradiation detection model.
Table S7. Promoter sequences of miRNAs with similar patterns of radiation-induced expression changes.
Table S9. Overlap of transcription factor binding sites across the three species.
Table S18. Comparisons of miRNA expression depending on outcome.
Table S19. Differences of outcome frequencies depending on sex.
Table S21. Differences of radiation-induced miRNA expression in the validation group.
Fig. S1. Flowchart of miRNA expression comparisons.
Fig. S2. Batch effect removal.
Fig. S3. Overlap of radiation-induced changes of miRNA expression in the mouse and macaque experiments.
Fig. S4. Performance of the 2-miRNA irradiation classifier on vehicle and GT3-treated samples.
Fig. S5. Survival of vehicle and GT3-treated macaques.
Fig. S6. Survival curves for miRNAs retained in the survival prediction model.
Fig. S7. Differences of miR-16-2 in data normalized to miR-142/miR-320a average.
Fig. S8. Correlation of miRNA expression between the primary and validation groups.
Fig. S9. Hierarchical clustering of miRNA expression in the validation group.
Fig. S10. Survival curves for miR-30a, miR-126, and miR-16-2 in the validation group.
Fig. S11. The 5-miRNA diagnostic protocol for irradiation detection and outcome prediction.
Acknowledgments
The views expressed do not necessarily represent the opinions or policies of the Armed Forces Radiobiology Research Institute, F. Edward Hébert School of Medicine, the Uniformed Services University of the Health Sciences, the Department of Defense, or the United States Government.
Funding: DC is supported by R01 AI101897-01 (NIAID) and R01CA142698-07 (NCI), Leukemia and Lymphoma Society Scholar Grant, Claudia Adams Barr Program for Innovative Cancer Research, Robert and Deborah First Family Fund and Tina Brozman Foundation. VKS received grant support from Defense Threat Reduction Agency (CBM.RAD.01.10.AR.005) and Congressionally Directed Medical Research Program (W81XWH-15-C-0117, JW140032), Department of Defense, USA.
Footnotes
Author contributions: WF conducted all the statistical analysis with help from BM and KM. PK independently validated the analysis. The NHP irradiation and subsequent blood analysis was done under the supervision of VKS; the serum miRNA processing and analysis was conducted in DC’s laboratory with help from GC. The manuscript was written by WF, VKS, and DC with input from PK and CG.
Competing interests: The authors have declared that they do not have any competing interests.
References and Notes
- 1.Chin FK. Scenario of a dirty bomb in an urban environment and acute management of radiation poisoning and injuries. Singapore Med J. 2007;48:950–957. [PubMed] [Google Scholar]
- 2.Waselenko JK, et al. Medical management of the acute radiation syndrome: recommendations of the Strategic National Stockpile Radiation Working Group. Ann Intern Med. 2004;140:1037–1051. doi: 10.7326/0003-4819-140-12-200406150-00015. [DOI] [PubMed] [Google Scholar]
- 3.Coleman CN, Stone HB, Moulder JE, Pellmar TC. Medicine. Modulation of radiation injury. Science. 2004;304:693–694. doi: 10.1126/science.1095956. [DOI] [PubMed] [Google Scholar]
- 4.Dainiak N. Radiation response: Changing concepts and emerging paradigms. Exp Hematol. 2003;31:435–436. doi: 10.1016/s0301-472x(03)00077-8. [DOI] [PubMed] [Google Scholar]
- 5.Dainiak N, Waselenko JK, Armitage JO, MacVittie TJ, Farese AM. The hematologist and radiation casualties. Hematology Am Soc Hematol Educ Program. 2003:473–496. doi: 10.1182/asheducation-2003.1.473. [DOI] [PubMed] [Google Scholar]
- 6.Parker DD, Parker JC. Estimating radiation dose from time to emesis and lymphocyte depletion. Health Phys. 2007;93:701–704. doi: 10.1097/01.HP.0000275289.45882.29. [DOI] [PubMed] [Google Scholar]
- 7.Goans RE, Holloway EC, Berger ME, Ricks RC. Early dose assessment following severe radiation accidents. Health Phys. 1997;72:513–518. doi: 10.1097/00004032-199704000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan JM, et al. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 2013;105:540–554. doi: 10.1097/HP.0b013e31829cf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinto MM, Santos NF, Amaral A. Current status of biodosimetry based on standard cytogenetic methods. Radiat Environ Biophys. 2010;49:567–581. doi: 10.1007/s00411-010-0311-3. [DOI] [PubMed] [Google Scholar]
- 10.Singh VK, Newman VL, Romaine PL, Hauer-Jensen M, Pollard HB. Use of biomarkers for assessing radiation injury and efficacy of countermeasures. Expert Rev Mol Diagn. 2016;16:65–81. doi: 10.1586/14737159.2016.1121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acharya SS, et al. Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci Transl Med. 2015;7:287ra269. doi: 10.1126/scitranslmed.aaa6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh VK, Hauer-Jensen M. gamma-Tocotrienol as a Promising Countermeasure for Acute Radiation Syndrome: Current Status. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17050663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh VK, et al. Radioprotective Efficacy of Gamma-Tocotrienol in Nonhuman Primates. Radiat Res. 2016;185:285–298. doi: 10.1667/RR14127.1. [DOI] [PubMed] [Google Scholar]
- 14.Raju U, et al. Inhibition of EGFR or IGF-1R signaling enhances radiation response in head and neck cancer models but concurrent inhibition has no added benefit. Cancer Med. 2015;4:65–74. doi: 10.1002/cam4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chitnis MM, et al. IGF-1R inhibition enhances radiosensitivity and delays double-strand break repair by both non-homologous end-joining and homologous recombination. Oncogene. 2014;33:5262–5273. doi: 10.1038/onc.2013.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charles MW. Studies of mortality of atomic bomb survivors. Report 13: Solid cancer and noncancer disease mortality: 1950–1997. J Radiol Prot. 2003;23:457–459. doi: 10.1088/0952-4746/23/4/m02. [DOI] [PubMed] [Google Scholar]
- 17.Preston DL, Shimizu Y, Pierce DA, Suyama A, Mabuchi K. Studies of mortality of atomic bomb survivors. Report 13: Solid cancer and noncancer disease mortality: 1950–1997. Radiat Res. 2003;160:381–407. doi: 10.1667/rr3049. [DOI] [PubMed] [Google Scholar]
- 18.Preston DL, Shimizu Y, Pierce DA, Suyama A, Mabuchi K. Studies of mortality of atomic bomb survivors. Report 13: solid cancer and noncancer disease mortality: 1950–1997. 2003. Radiat Res. 2012;178:AV146–172. doi: 10.1667/rrav12.1. [DOI] [PubMed] [Google Scholar]
- 19.Borgmann K, Dikomey E, Petersen C, Feyer P, Hoeller U. Sex-specific aspects of tumor therapy. Radiat Environ Biophys. 2009;48:115–124. doi: 10.1007/s00411-009-0216-1. [DOI] [PubMed] [Google Scholar]
- 20.Kovalchuk O, Ponton A, Filkowski J, Kovalchuk I. Dissimilar genome response to acute and chronic low-dose radiation in male and female mice. Mutat Res. 2004;550:59–72. doi: 10.1016/j.mrfmmm.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Holland JM, Mitchell TJ. The relationship of strain, sex, and body weight to survival following sublethal whole-body x-irradiation. Radiat Res. 1976;66:363–372. [PubMed] [Google Scholar]
- 22.Carnes BA, Gavrilova N, Grahn D. Pathology effects at radiation doses below those causing increased mortality. Radiat Res. 2002;158:187–194. doi: 10.1667/0033-7587(2002)158[0187:peardb]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 23.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 24.Anton M. Uncertainties in alanine/ESR dosimetry at the Physikalisch-Technische Bundesanstalt. Phys Med Biol. 2006;51:5419–5440. doi: 10.1088/0031-9155/51/21/003. [DOI] [PubMed] [Google Scholar]
- 25.Fendler W, et al. Differential regulation of serum microRNA expression by HNF1beta and HNF1alpha transcription factors. Diabetologia. 2016;59:1463–1473. doi: 10.1007/s00125-016-3945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mestdagh P, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11:809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 27.Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. 8. The National Academies Press; 2011. [PubMed] [Google Scholar]
- 28.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 29.Hosmer DWL. Stanley Applied Logistic Regression. New York: Wiley; 2013. [Google Scholar]
- 30.Simon RM, Subramanian J, Li MC, Menezes S. Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Brief Bioinform. 2011;12:203–214. doi: 10.1093/bib/bbr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Messeguer X, et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 33.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Raw expression data from miRNA qPCR profiling (separate xls file).
Table S10. Promoter sequences of miR-30a (separate txt file).
Table S11. Promoter sequences of miR-126 (separate txt file).
Table S12. Promoter sequences of miR-375 (separate txt file).
Table S13. Promoter sequences of miR-150 (separate txt file).
Table S14. Promoter sequences of mir-133b (separate txt file).
Table S15. Promoter sequences of miR-133a (separate txt file).
Table S16. Promoter sequences of miR-215 (separate txt file).
Table S2. Paired sample miRNA expression comparisons (separate xls file).
Table S8. Transcription factor binding sites in miRNA promoters of human, macaque, and mouse genomes (separate xls file).
Table S17. Validated gene targets of radiation-associated evolutionarily-conserved miRNAs (separate xls file).
Table S20. Selection of best normalizer miRNAs (separate xls file).
Table S3. miRNA expression in macaques irradiated with different doses.
Table S4. Independent sample miRNA expression comparisons.
Table S5. Numerical parameters of logistic regression models.
Table S6. Goodness of fit evaluation of the irradiation detection model.
Table S7. Promoter sequences of miRNAs with similar patterns of radiation-induced expression changes.
Table S9. Overlap of transcription factor binding sites across the three species.
Table S18. Comparisons of miRNA expression depending on outcome.
Table S19. Differences of outcome frequencies depending on sex.
Table S21. Differences of radiation-induced miRNA expression in the validation group.
Fig. S1. Flowchart of miRNA expression comparisons.
Fig. S2. Batch effect removal.
Fig. S3. Overlap of radiation-induced changes of miRNA expression in the mouse and macaque experiments.
Fig. S4. Performance of the 2-miRNA irradiation classifier on vehicle and GT3-treated samples.
Fig. S5. Survival of vehicle and GT3-treated macaques.
Fig. S6. Survival curves for miRNAs retained in the survival prediction model.
Fig. S7. Differences of miR-16-2 in data normalized to miR-142/miR-320a average.
Fig. S8. Correlation of miRNA expression between the primary and validation groups.
Fig. S9. Hierarchical clustering of miRNA expression in the validation group.
Fig. S10. Survival curves for miR-30a, miR-126, and miR-16-2 in the validation group.
Fig. S11. The 5-miRNA diagnostic protocol for irradiation detection and outcome prediction.