Abstract
Aims: Sirtuins connect energy generation and metabolic stress to the cellular acetylome. Currently, only the mitochondrial sirtuins (SIRT3–5) and SIRT1 have been shown to direct mitochondrial function; however, Aims: NAD-dependent protein deacetylase sirtuin-2 (SIRT2), the primary cytoplasmic sirtuin, is not yet reported to associate with mitochondria.
Results: This study revealed a novel physiological function of SIRT2: the regulation of mitochondrial function. First, the acetylation of several metabolic mitochondrial proteins was found to be altered in Sirt2-deficient mice, which was, subsequently, validated by immunoprecipitation experiments in which the acetylated mitochondrial proteins directly interacted with SIRT2. Moreover, immuno-gold electron microscopic images of mouse brains showed that SIRT2 associates with the inner mitochondrial membrane in central nervous system cells. The loss of Sirt2 increased oxidative stress, decreased adenosine triphosphate levels, and altered mitochondrial morphology at the cellular and tissue (i.e., brain) level. Furthermore, the autophagic/mitophagic processes were dysregulated in Sirt2-deficient neurons and mouse embryonic fibroblasts.
Innovation: For the first time it is shown that SIRT2 directs mitochondrial metabolism.
Conclusion: Together, these findings support that SIRT2 functions as a mitochondrial sirtuin, as well as a regulator of autophagy/mitophagy to maintain mitochondrial biology, thus facilitating cell survival. Antioxid. Redox Signal. 26, 849–863.
Keywords: : sirtuins, mitochondria, SIRT2, mitophagy, autophagy, metabolism, ROS
Introduction
Sirtuins have received significant attention since the discovery that the yeast sirtuin (silent information regulator 2 or Sir2) gene extends yeast lifespan (28). Sirtuins are NAD+-dependent protein deacetylases and have been implicated in the regulation of metabolism, stress responses, and aging (22). Activation of sirtuins is believed to be beneficial not only for diseases related to metabolism, such as cancer, type II diabetes, and obesity, but also for neurodegenerative diseases, such as Alzheimer's disease and Parkinson's disease (6, 16, 21). This is, in part, because sirtuins regulate the activity of mitochondria, the powerhouses of cells, by deacetylating mitochondria-related proteins.
Seven sirtuins (SIRT1–7) are present in mammals. They target distinct protein substrates and are located in distinct subcellular compartments: SIRT1, 6, and 7 are found primarily in the nucleus; SIRT3, 4, and 5 are called “mitochondrial sirtuins” given their mitochondrial location. NAD-dependent protein deacetylase sirtuin-2 (SIRT2) not only is primarily cytosolic but also may enter the nucleus during the G2/M phase (26). SIRT2 participates in the modulation of multiple and diverse biological processes, such as cell cycle control (30), genomic integrity (10), microtubule dynamics (33, 36), cell differentiation (3), metabolic networks (27), and autophagy (46), by deacetylating lysines on histones, α-tubulin, and many other proteins. The functional consequences of deacetylation vary with the relative position of the lysine residue within the target protein, and they may either increase or decrease activity, affinity, stability, or complex formation (12).
Innovation.
For the first time it is shown that SIRT2 direct mitochondrial metabolism demonstrated by mitochondrial candidate proteins directly interact with SIRT2 (Fig 1c,d); immuno-gold electron microscopic images support that SIRT2 associates with inner mitochondrial membrane (Fig 2f) and the loss of Sirt2 increased oxidative stress (Fig 4). These findings integrate several major concepts relevant to the fields of mitochondrial physiology and biochemistry, longevity, and molecular and cellular energy homeostasis.
Post-translational modifications such as acetylation are known to regulate protein function (5), and in the mitochondria, more than 20% of proteins are acetylated in proteomic surveys (31). In addition, dietary interventions, such as caloric restriction or time-restricted fasting, activate sirtuins and deacetylate multiple mitochondrial proteins, indicating that acetylation is linked to mitochondrial dynamics (i.e., metabolism, biogenesis, stress response, mitophagy, and apoptosis). Together, these observations suggest that protein acetylation is a critical post-translational modification, and as such, mitochondrial protein acetylation may serve as a potential therapeutic target for the treatment of mitochondria-related disorders. Sirtuins appear to be a family of fidelity proteins that respond to energy conditions, such as fasting, by reprogramming cells to improve their energy efficiency. As such, it is proposed that sirtuins sense organismal nutrient status and reprogram cellular and mitochondrial metabolism to match. If this energy coordination is disrupted, aberrant mitochondrial reprogramming ensues, creating metabolic stress as well as cell damage that permits mammary carcinogenesis (8, 9, 20, 29). This metabolic reprogramming by sirtuins activates reparative cellular processes that respond to metabolic stress within the cells, possibly preventing, or more likely delaying, aging-related conditions, such as neurodegeneration, insulin resistance, cardiovascular disease, and carcinogenesis.
Several sirtuins, including the nuclear SIRT1, regulate the activity of mitochondria, either directly or indirectly, by modifying mitochondria-related proteins (1). However, whether SIRT2 can regulate mitochondrial function remains unknown (24). Interestingly, mice lacking Sirt2 do not show any changes in longevity or body weight, as compared with wild-type mice (45). Current studies of Sirt2−/− mice and mouse embryonic fibroblasts (MEFs) showed that multiple protein targets in the mitochondria exhibited acetylation changes compared with Sirt2+/+ mice and MEFs. To further investigate the possible role of SIRT2 in mitochondria, we assessed SIRT2 localization and the morphological/functional changes in the mitochondria of Sirt2-deficient mice and MEFs. We found that SIRT2 can localize to mitochondria and directly interact with mitochondrial proteins. In addition, the loss of Sirt2 increased markers of oxidative stress, reduced adenosine triphosphate (ATP) production, and impaired autophagic/mitophagic processes in both the mouse brain and MEFs.
Results
The loss of Sirt2 alters the acetylation level of mitochondrial proteins
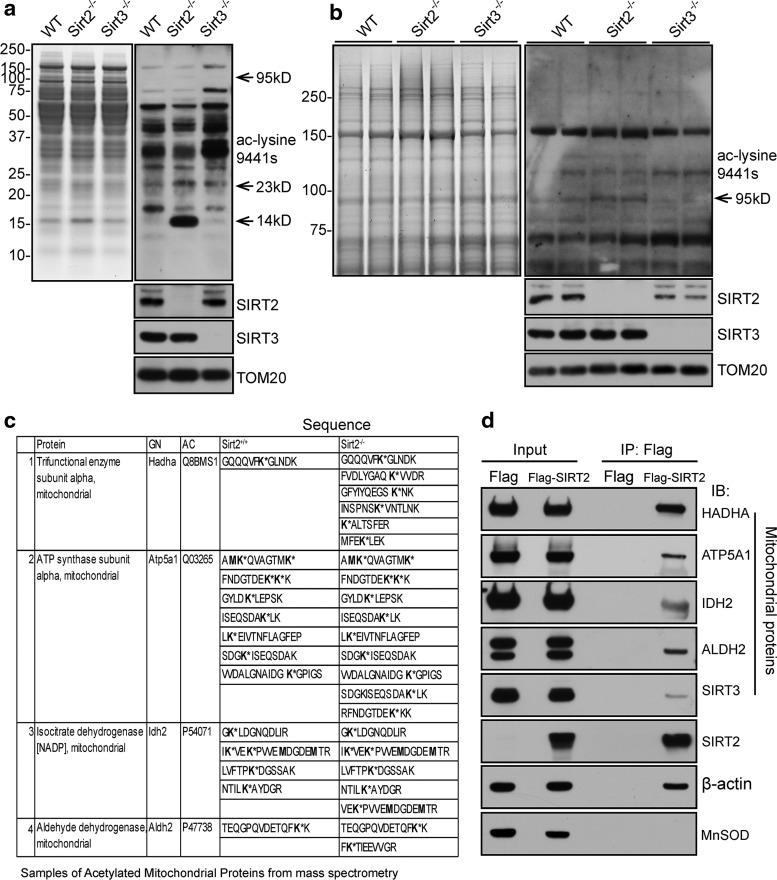
To identify possible SIRT2 targets, lysates of striatum from adult wild-type (Sirt2+/+) and Sirt2 knockout (Sirt2−/−) mice were subjected to proteolytic digestion with trypsin and, subsequently, immunoprecipitation (IP)ed with an anti-acetyl-lysine antiserum. The acetyl-lysine-containing peptide samples were analyzed by mass spectrometry. Interestingly, a significant number of mitochondrial proteins (20 mitochondrial proteins) were found among the proteins for which acetylation was changed (total 49 proteins) in Sirt2 knockout samples relative to controls (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/ars). Similar results were observed in Sirt2−/− and Sirt2+/+ MEFs based on unpublished data in the lab. The acetylation levels of some specific bands, such as ∼14, ∼23, and ∼95 kDa, were significantly increased in the absence of SIRT2 (Fig. 1a, b). To identify the specific target proteins of SIRT2, mitochondrial proteins with unique/extra acetylated lysines in Sirt2−/− samples, as compared with Sirt2+/+, were summarized in Figure 1c, and many were mitochondrial proteins involved in energy metabolism. Finally, anti-Flag IP experiments done on 293T cells confirmed a direct interaction between SIRT2 and these proteins (Fig. 1d) as well as endogenous IP experiments from cortex tissues (Supplementary Fig. S1), implying that they may be legitimate SIRT2 deacetylation targets.
FIG. 1.
The loss of Sirt2 alters the acetylation level of mitochondrial proteins. (a, b) Mitochondrial extracts from the striatum of isogenic, matched 6 month-old wild-type and Sirt2−/− mice, as well as from mice lacking Sirt3 as a control, were separated by SDS-PAGE and either stained with Coomassie blue (left panel) or immunoblotted with pan anti-acetyl (Cell Signaling, Inc.), SIRT2, and SIRT3 antibodies. TOM20 was used as the loading control. To better visualize the band at 95 kDa, the samples from (a) and one more set sample were loaded on the gel (b) with a longer running time and longer exposure. (c) Candidate mitochondrial proteins exhibit higher acetylation in Sirt2−/− as compared with the wild-type samples. Acetylated peptides found in Sirt2 wild-type and knockout MEF extracts after mass spectrometry. (d) Lysates from 293T cells infected with Flag-only or Flag-SIRT2 were harvested, IPed with an anti-Flag antibody, and, subsequently, immunoblotted with antibodies against HADHA, ATP5A1, IDH2, ALDH2, SIRT3, SIRT2, or MnSOD. β-Actin was used as the loading control. All experiments were done in triplicate. Representative images are shown. ALDH2, aldehyde dehydrogenase; ATP5A1, ATP synthase subunit alpha; HADHA, trifunctional enzyme subunit alpha; IDH2, isocitrate dehydrogenase 2 (NADP+); IP, immunoprecipitation; MEF, mouse embryonic fibroblast; MnSOD, manganese superoxide dismutase; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; SIRT2, NAD-dependent protein deacetylase sirtuin-2; TOM20, mitochondrial import receptor subunit TOM20.
SIRT2 is localized to mitochondria
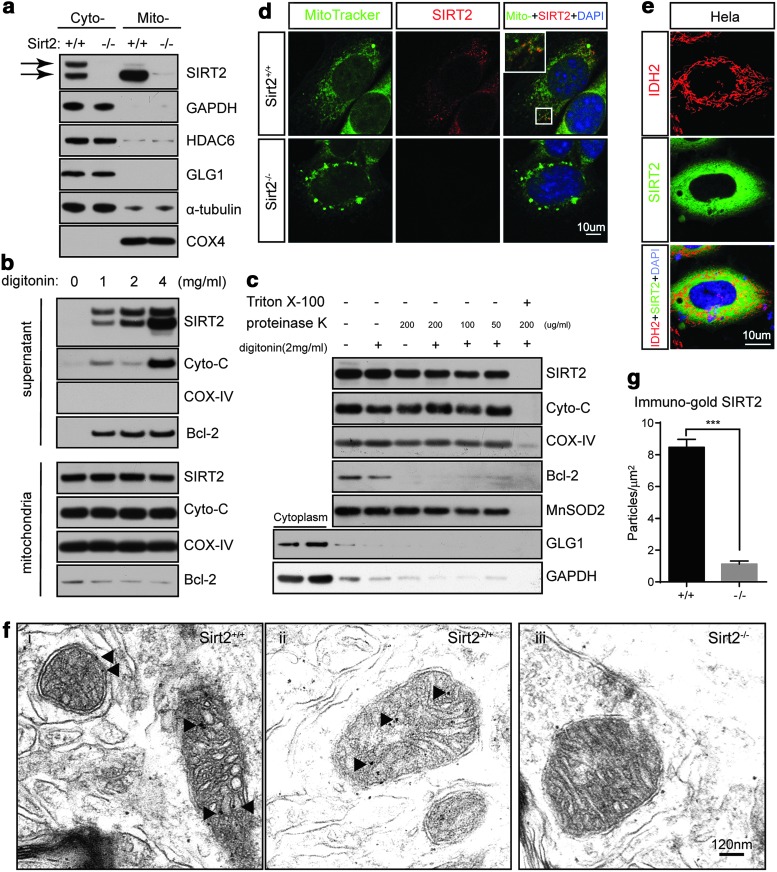
Since tissue and cells lacking Sirt2 exhibit altered mitochondrial protein acetylation, we examined whether SIRT2 localizes to mitochondria by isolating mitochondrial lysate from brains of wild-type and Sirt2−/− mice. SIRT2 can be easily detected in mitochondrial lysates from brain tissues where COX-IV, GAPDH, and GLG1 immunoblotting are markers for mitochondrial, cytoplasmic, and Golgi protein, respectively (Fig. 2a). To further examine the sub-mitochondrial localization of SIRT2, purified mitochondria were treated with digitonin. Bcl-2 is highly sensitive to digitonin treatment due to its location on the surface of mitochondria, whereas the intermembrane protein COX-IV is insensitive to (i.e., protected from) digitonin by its location (Fig. 2b). With the increasing concentration of digitonin, more SIRT2 signal can be detected in the supernatants, suggesting that some SIRT2 are localized at the surface of the mitochondria (Fig. 2b top). Consistently, a strong SIRT2 signal can be detected in the pellets (Fig. 2b bottom). To further investigate the localization of SIRT2 in the mitochondria, we treated the mitochondria with digitonin and in the next protease protection assay. In comparison, SIRT2 was moderately sensitive to proteinase K (Fig. 2c). All proteins were degraded when Triton X-100 was added, which resulted in complete disruption of the mitochondrial membrane (Fig. 2c lane 7). This indicates that the difference in sensitivity to proteinase K is due to differential localization and not inherent proteolytic susceptibility, suggesting that SIRT2 is associated with the outer mitochondrial membrane and the inner membrane or matrix.
FIG. 2.
Mitochondrial localization of SIRT2. (a) Cytoplasmic and mitochondrial fractions were isolated from the CNS cortex Sirt2+/+ and Sirt2−/− mice at 24 months of age. Samples were subsequently separated and immunoblotted with anti-SIRT2, COX-4, GLG1, and HDAC6 antibodies. GAPDH and α-tubulin were used as the loading control. (b) Mitochondria fractions were treated with digitonin in Hypotonic buffer at 1, 2, and 4 mg/ml 10 min on ice. The supernatants [(b), top] and mito-pellets [(b), bottom] were collected and Western blot was performed with SIRT2, cytochrome C, Bcl-2, and COX-IV. (c) Mitochondria were treated with digitonin (2 mg/ml) and followed by the proteinase K assay. Mito-pellets were collected and immunoblotted with anti-SIRT2, cytochrome C (intermembrane space), COX-IV (matrix), Bcl-2 (outer mitochondrial membrane), MnSOD (matrix), GLG1 (Golgi), and GAPDH. (d) Wild-type and Sirt2−/− MEFs were stained with MitoTracker as well as SIRT2 and DAPI, and representative IFC images are shown. Scale bar: 10 μm. (e) HeLa cells were stained with anti-IDH2 (to visualize mitochondria) and SIRT2 antibodies as well as DAPI, and representative IFC images are shown. Scale bar: 10 μm. (f) Sections from the neocortex of wild-type and Sirt2−/− mice stained with rabbit anti-SIRT2 (Proteintech) antibody and donkey anti-rabbit immunoglobulin G conjugated onto 10-nm gold particles followed by immunoelectron microscopy. Arrowheads point to the inner membrane. Scale bars: 120 nm. All experiments were done in triplicate. Representative images are shown. (g) Immunogold particles were counted from at least six electron microscopy images for each genotype. Error bars represent one standard deviation from the mean. ***p < 0.001. CNS, central nervous system; DAPI, 4′,6-diamidino-2-phenylindole; IFC, immunofluorescent.
SIRT2 mitochondrial localization was also shown by co-staining SIRT2 with MitoTracker in Sirt2+/+ MEFs (Fig. 2d). The SIRT2 distribution pattern in the HeLa cells overexpressing SIRT2 shows diffuse cytosolic distribution (Fig. 2e), whereas MEFs show a somewhat punctate pattern that appears to localize with MitoTracker to different degrees. These results suggested that SIRT2 may be present in the mitochondria, at least in some part, and based on these results more rigorous experiments were done. In addition, HeLa cells transfected with SIRT2 and co-stained with SIRT2 and isocitrate dehydrogenase 2 (NADP+) (IDH2) showed that although SIRT2 is mainly cytoplasmic, it partially co-localizes with the mitochondria (Fig. 2e). Finally, post-embedding electron microscopy with a rabbit anti-SIRT2 gold-particle-conjugated antibody appears to show that SIRT2 is present in the mitochondria and appears to suggest that it is localized to the inner mitochondrial membranes in the brains of control mice (Fig. 2f, g), but not in mice lacking Sirt2.
Morphological changes in mitochondria are associated with the loss of Sirt2
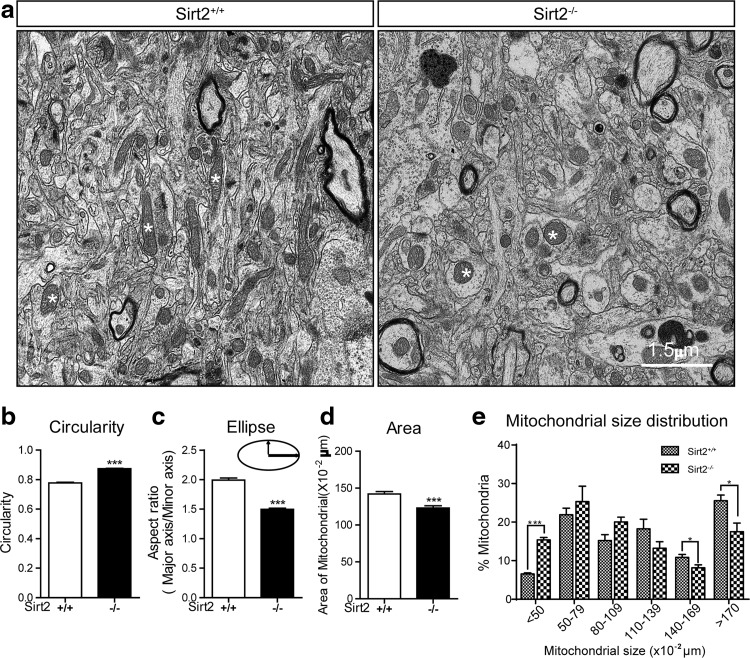
Next, we compared electron microscopic images of cortex tissues from wild-type and Sirt2−/− mice. There were significant differences in mitochondrial size and morphology. Specifically, the mitochondria were rounder (Fig. 3a, left vs. right panels) and appeared less oblong (Supplementary Fig. S2, left vs. right panels). Subsequent analyses showed that the circularity (Fig. 3b) and the aspect ratio (Fig. 3c) of mitochondria in the Sirt2−/− cortex were significantly different than in an Sirt2+/+ cortex. In addition, the mean size of the mitochondria was reduced by deletion of Sirt2 (Fig. 3d). Finally, we found an increase in the number of mitochondria with area <50 × 10−2 μm2 and a decrease in the number of mitochondria whose area was 140 × 10−2 μm2 or larger in the Sirt2−/− mice, as compared with the wild-type mice (Fig. 3e).
FIG. 3.
Loss of Sirt2 makes mitochondria rounder and smaller relative to controls. (a) Electron micrograph of mitochondria in neocortex of isogenic, 24 month-old wild-type and Sirt2−/− mice. Asterisks point to representative mitochondria. Scale bar: 1.5 μm. (b–d) Bar graph depicts the quantification of mitochondrial circularity, aspect ratio (ellipticity), and total area (n = 3 animals per genotype; n > 250 mitochondria per animal). (e) Histogram of mitochondrial size distribution in μm: (i) <50 × 10−2, (ii) 50–79 × 10−2, (iii) 60–109 × 10−2, (iv) 110–139 × 10−2, (v) 140–169 × 10−2, and (vi) >170 × 10−2. Data presented are the mean ± SEM (n > 800 for each genotype). *p < 0.05, ***p < 0.001. All experiments were done in triplicate. Representative images are shown. Error bars represent SEM. SEM, standard error of the mean.
These results were extended to an in vitro model system of wild-type and Sirt2−/− MEFs that were, subsequently, co-stained with MitoTracker (Supplementary Fig. S3A) or transiently transfected with IDH2 (Supplementary Fig. S3B) and analyzed by laser confocal microscopy. Mitochondrial distribution was strikingly different in Sirt2−/− MEFs, which exhibited sparse and swollen mitochondria (Supplementary Fig. S3B). The decrease in mitochondrial mass in MEFs lacking Sirt2 was confirmed by immunoblotting with an anti-COX-IV antibody (Supplementary Fig. S3C), a mitochondrial marker protein that was also quantified (Supplementary Fig. S3D). Finally, PGC1α protein levels were identical in wild-type and Sirt2−/− MEFs (Supplementary Fig. S3E) and brain tissue (Supplementary Fig. S3F, G).
However, PGC1α transcription activity may be regulated through SIRT2 deacetylase activities. The downstream genes Mfn1, Mfn2, and Opa1 (mRNA) are found to be downregulated in Sirt2−/− mice (Supplementary Fig. S4A–C). We further found that the level of acetylated PGC1α was significantly increased in Sirt2−/− mice (Supplementary Fig. S4F, G). Compared with Sirt2+/+ MEFs, PGC1α distribution in MEFs was strikingly different in Sirt2−/− MEFs, which exhibited big punctates in the perinuclear space, whereas PGC1α was evenly distributed in the Sirt2+/+ MEFs (Supplementary Fig. S4H). Further analysis showed that there were fewer particles in the Sirt2−/− nucleus than the wild type. These results suggest that acetylation, due to loss of SIRT2, may prevent PGC1α from entering the nucleus and activating transcription, which contributes to the difference in mitochondrial size and morphology.
Loss of Sirt2 increases the oxidative stress and decreases ATP production
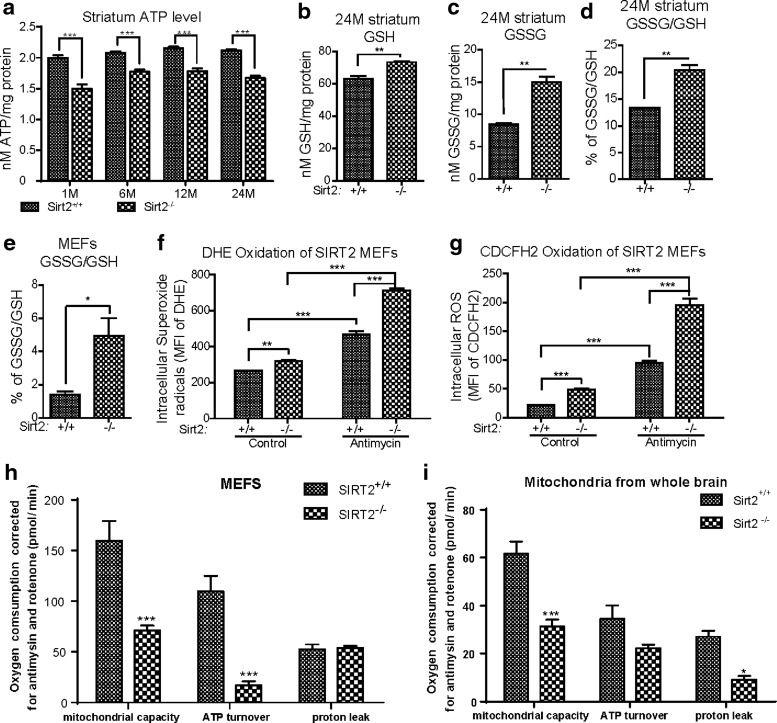
A critical role of the mitochondria is to generate ATP from ADP and organophosphate via oxidative phosphorylation, which generates reactive oxygen species (ROS) as a byproduct of cellular respiration. To investigate whether SIRT2 plays a role in mitochondrial physiology, we measured ATP levels as well as mitochondrial ROS. The striata of mice lacking Sirt2 exhibited decreased ATP levels at 1, 6, 12, and 24 months of age (Fig. 4a). In addition, total glutathione (GSH; Fig. 4b) and oxidized glutathione (GSSG; Fig. 4d) were increased in tissues lacking Sirt2. In addition, the GSSG-to-GSH ratio was significantly increased at 24 months (Fig. 4d) as well as at 12 months (Supplementary Fig. S5), suggesting an increased intracellular oxidative potential. Similar results were obtained from Sirt2−/− MEFs (Fig. 4e). The steady-state levels of intracellular superoxide (Fig. 4f) as well as ROS (Fig. 4g) were also elevated in Sirt2−/− MEFs. In addition, when the same cells were treated with antimycin A (a complex III inhibitor), both dihydroethidium (DHE) and 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate (CDCFH2) oxidation were significantly higher in MEFs lacking Sirt2 (Fig. 4f, g). Mitochondrial ATP turnover and respiration capacity was decreased in Sirt2−/− MEFs (Fig. 4h) as well as in whole brain tissues from mice lacking Sirt2 (Fig. 4i). Overall, these results suggest that a deficiency of Sirt2 impairs the function of mitochondria (such as ATP production) and induces increased steady-state and stress-induced levels of intracellular superoxides and hydroperoxides.
FIG. 4.
Loss of Sirt2 both in vitro and in vivo significantly increases oxidative stress as well as decreases cellular ATP. (a) The CNS striata from wild-type (Sirt2+/+) and Sirt2−/− mice at 1, 6, 12, and 24 months of age were harvested and used to determine ATP levels as previously shown. (b–d) The CNS striatum samples from Sirt2+/+ and Sirt2−/− mice at 24 months of age were also used to determine (b) GSH levels, (c) GSSG levels, and (d) the GSH/GSSG ratio. (e) Sirt2+/+ and Sirt2−/− MEFs were isolated, and GSH and GSSG levels were measured and, subsequently, used to determine the GSH/GSSG ratio. (f) Sirt2+/+ and Sirt2−/− MEFs without or with exposure to 5 μM of antimycin for 3 h were isolated, and superoxide levels were monitored by DHE oxidation as compared with control, untreated cells (Cont). For all DHE oxidation experiments, the results were the normalized MFI for three independent replicates. (g) Sirt2+/+ and Sirt2−/− MEFs without or with exposure to 5 μM of antimycin for 3 h were isolated, and CDCFH2 oxidation was determined as previously shown. All experiments were done in triplicate. Error bars represent one standard deviation from the mean. *p < 0.05, **p < 0.01, and ***p < 0.001. (h, i) Sirt2 deletion decreases mitochondrial ATP turnover, mitochondrial respiration capacity, and proton leak in Sirt2−/− MEFs cells and isolated mitochondria. Fifty thousand cells or equal concentrations of isolated mitochondrial pellets from whole brains were plated on a 24-well XF24 cell culture microplate overnight. Oligomycin, CCCP, and antimycin/rotenone mixture were sequentially added to measure OCR in the XF24 analyzer from Seahorse Bioscience (n = 5). The basal respiration rate was determined by the difference between the starting OCR and the OCR after adding antimycin/rotenone mixture. The ATP turnover rate was determined by the difference between the starting OCR and the OCR after adding oligomycin. The proton leak rate was determined by the difference between the OCR after adding oligomycin and the OCR after adding antimycin/rotenone mixture. Error bars represent one standard deviation from the mean. ***p < 0.001. ATP, adenosine triphosphate; CCCP, carbonyl cyanide m-chlorophenyl hydrazone; CDCFH2, 5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate; DHE, dihydroethidium; GSH, glutathione; GSSG, oxidized glutathione; MFI, mean fluorescence intensity; OCR, oxygen consumption rate.
Defective mitophagy in cells genetically lacking Sirt2 neurons
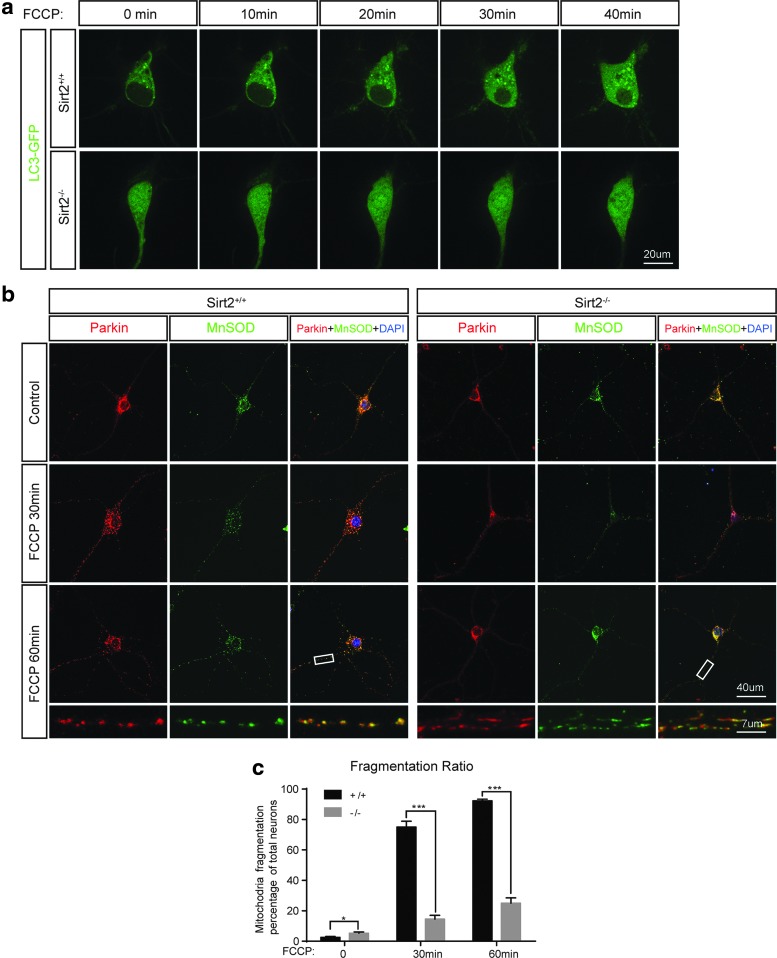
As previously described, the loss of Sirt2 resulted in a series of mitochondrial morphological and functional deficits. Thus, it seemed reasonable to examine whether those mitochondria are also properly degraded. Mitophagy represents a selective type of autophagy that removes or degrades damaged mitochondria (13). To this end, we tested how the mitophagy process works in cells lacking Sirt2. Specifically, Sirt2+/+ and Sirt2−/− murine hippocampal neurons were transfected with green fluorescent protein (GFP)-microtubule-associated protein 1 light chain 3 beta (LC3B) and, subsequently, treated with carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), a protonophore (H+ ionophore) and oxidative phosphorylation uncoupler. Interestingly, FCCP induced the formation of GFP-LC3B puncta in wild-type neurons (Fig. 5a upper panels, Supplementary Video S1), but not in the Sirt2−/− neurons (Fig. 5a lower panels, Fig. 5a upper panels, Supplementary Video S2). In addition, wild-type and Sirt2−/− cells were co-stained for Parkin, a cytosolic E3-ubiquitin ligase, and manganese superoxide dismutase (MnSOD). FCCP treatment induced a minor recruitment of Parkin to mitochondria, which is a necessary step in mitophagy, as compared with wild-type cells (Fig. 5b right panels). Meanwhile, FCCP treatment led to short and swollen mitochondria in the control but fewer in Sirt2−/− cells (Fig. 5b right panels). Five independent wild-type and Sirt2−/− primary hippocampus neuron cultures per condition were analyzed and quantified (Fig. 5c). As a control, wild-type and Sirt2−/− primary hippocampal neuron cultures were transfected with GFP and exposed to FCCP for 30 min. These experiments demonstrate no change in the cytoplasmic compartment, showing that the changes in staining are due to mitochondrial fragmentation (Supplementary Fig. S6). These results suggest that damaged mitochondria remain unmarked for turnover via mitophagy in cells lacking Sirt2.
FIG. 5.
Loss of Sirt2 impairs LC3B and Parkin recruitment in primary neurons. (a) Primary hippocampal neurons from Sirt2+/+ and Sirt2−/− P0 pups were transiently transfected with GFP–LC3B and treated with 10 nM FCCP for up to 1 h, followed by live imaging both before and after treatment via immunofluorescence microscopy. (b) Primary hippocampal neurons from Sirt2+/+ and Sirt2−/− P0 pups were treated with 10 nM FCCP for 30 min and 60 min, and they were subjected to immunofluorescence staining and microscopy. Mitochondria were visualized by MnSOD (green). Scale bar: 40 μm (top three panels) or 7 μm (bottom panel). All experiments were done in triplicate. Representative images are shown. (c) Percentage of primary hippocampus neurons with mitochondrial fragmentation after FCCP for 30 min or 60 min treatment. Five independent Sirt2+/+ and Sirt2−/− cultures per condition were analyzed. *p < 0.05, ***p < 0.001. FCCP, carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone; GFP, green fluorescent protein; LC3B, microtubule-associated protein 1 light chain 3 beta; Parkin, E3 ubiquitin-protein ligase parkin.
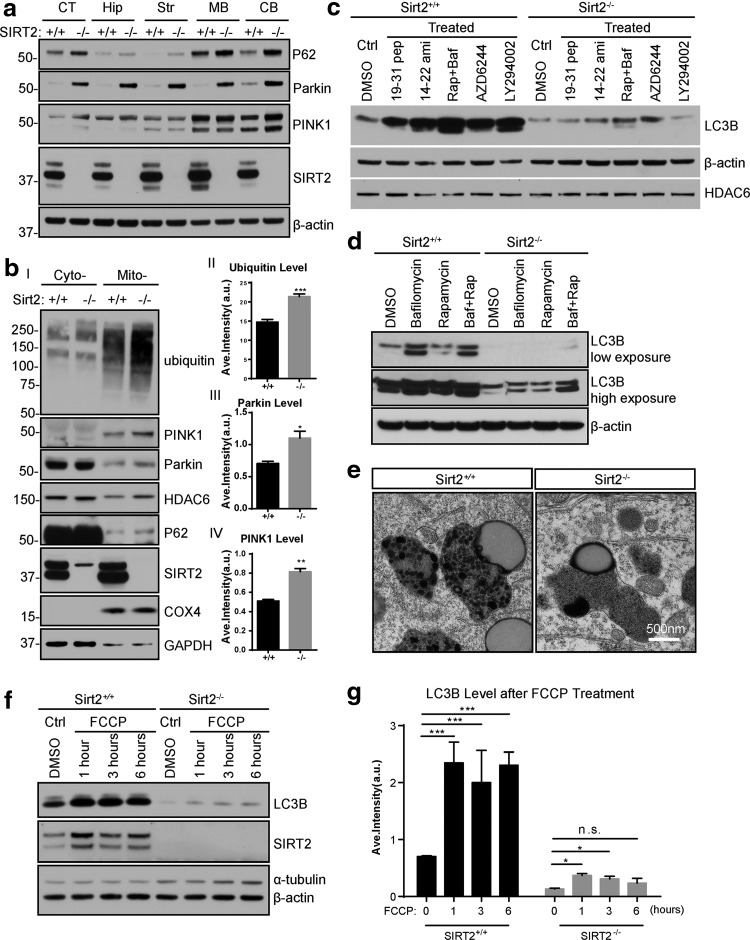
The serine/threonine-protein kinase PINK1 (PINK1)/Parkin pathway is an important sensor of mitochondrial damage and/or dysfunction and a mediator of mitochondrial quality control processes (44). In the brains lacking Sirt2, an increase of total (Fig. 6a) and mitochondrial (Fig. 6b) Parkin, and to a lesser extent, PINK1, was observed, as well as an increase in the steady-state level of p62. The whole brain mitochondria from three mice per genotype were used to quantify the immunostaining data for ubiquitin, Parkin, and PINK protein levels (see Fig. 6b, directly to the right of the immunoblots). The observed increase in p62 and PINK1/Parkin levels indicates that mitophagy/autophagy-mediated destabilization of p62, PINK1/Parkin is impaired and possibly the clearance of these proteins is blocked during the mitophagy process.
FIG. 6.
Loss of Sirt2 impairs mitophagy in mice and MEFs. (a) Brains of wild-type and Sirt2-/- mice were harvested (CT, cortex; Hip, hippocampus; Str, striatum; MB, mid-brain; and CB, cerebellum) and immunoblotted with anti-P62, Parkin, PINK1, and SIRT2 antibodies. β-actin was used as the loading control. (b) Wild-type and Sirt2−/− cytoplasmic and mitochondrial brain fractions were harvested and, subsequently, immunoblotted with anti-ubiquitin, PINK1, Parkin, HDAC6, P62, SIRT2, and COX-IV antibodies. GAPDH was used as the loading control. Bar graph quantifies the relative expression levels of ubiquitin (II), Parkin (III), and PINK1 (IV). N = 3 per genotype. *p < 0.05, **p < 0.01, and ***p < 0.001. (c) Wild-type and Sirt2−/− MEFs were treated with agents that inhibit: (i) protein kinase C (19–31 pep-Protein kinase C Fragment 19–31 Amid); (ii) protein kinase A (14–22 ami-Protein Kinase A inhibitor fragment 14–22); (iii) mTOR (Rap-Rapamycin); (iv) vacuolar H+ ATPase (Baf-Bafilomycin A1); (v) MAPK/ERK kinase (AZD6244-Selumetinib); and (vi) phosphoinositide 3-kinase (LY294002). Samples were harvested after 4 h, and extracts were immunoblotted with anti-LC3B. β-Actin and HDAC6 were used as the loading control. (d) Sirt2+/+ and Sirt2−/− MEFs were treated with Bafilomycin A1, Rapamycin, or both (Baf+Rap) and harvested, and after 4 h, extracts were immunoblotted with anti-LC3B. β-Actin was used as the loading control. (e) Neocortex sections from brains of wild-type and Sirt2−/− 24 month-old isogenic, matched mice. An electron micrograph of residual bodies (lipofuscin granules) is shown. Scale bars: 500 nm. All experiments were done in triplicate. Representative images are shown. (f) Wild-type and Sirt2−/− MEFs were treated with FCCP for 1, 3, and 6 h, harvested, and immunoblotted with anti-LC3B, SIRT2 antibodies. α-Tubulin was used as the loading control. (g) Bar graphs quantify the relative expression levels of LC3B. N = 3. *p < 0.05, ***p < 0.001. n.s., non-significant; PINK1, serine/threonine-protein kinase PINK1.
To identify the response of cells to external stimuli, we measured and compared LC3B levels as well as the effect of specific inhibitors of various intracellular signaling pathways. Wild-type MEFs displayed an increase in LC3B levels in response to inhibitors of protein kinase C (protein kinase C fragment 19–31 amide), protein kinase A (protein kinase A inhibitor fragment 14–22), MAPK/ERK kinase (AZD6244), phosphoinositide 3-kinase (2-(4-morpholinyl)-8-phenyl-1(4H)-benzopyran-4-one hydrochloride [LY294002]), mammalian target of rapamycin (rapamycin), and vacuolar H+ ATPase (bafilomycin A1; Fig. 6c). In contrast, a minor increase of LC3B was observed in MEFs lacking Sirt2 (Fig. 6c), providing further demonstration of the impaired autophagosome formation due to lack of Sirt2.
To define the flux through the macroautophagic pathway, MEFs were treated with 100 nM bafilomycin A1, 1 mM rapamycin, or both for 6 h (Fig. 6d). An increase in the ratio of LC3B-II to LC3B-I reflects the accumulation of autophagosome in cells. In wild-type MEFs, bafilomycin increased both species of LC3B, as expected, as autophagy is blocked after autophagosome formation and before the autophagosome can fuse with the lysosome. In addition, rapamycin treatment increased levels of LC3B-II, indicating increased autophagy (Fig. 6d). In contrast, these agents barely increased the LC3B-II species in Sirt2−/− MEFs, suggesting that autophagy is impaired in that context. In addition, residual bodies (lipofuscin granules), vesicles containing indigestible materials from lysosomal digestion, were abnormal in Sirt2−/− cortex motor neurons (Fig. 6e) whereas wild-type controls displayed numerous round particles that were distributed in residual bodies. However, no obvious particles were found in Sirt2−/− neurons (Fig. 6e). Finally, LC3B was slightly increased in Sirt2−/− MEFs after being treated with FCCP (Fig. 6f) and quantified (Fig. 6g). Taken together, these data strongly suggest that loss of Sirt2 impairs autophagy/mitophagy processes.
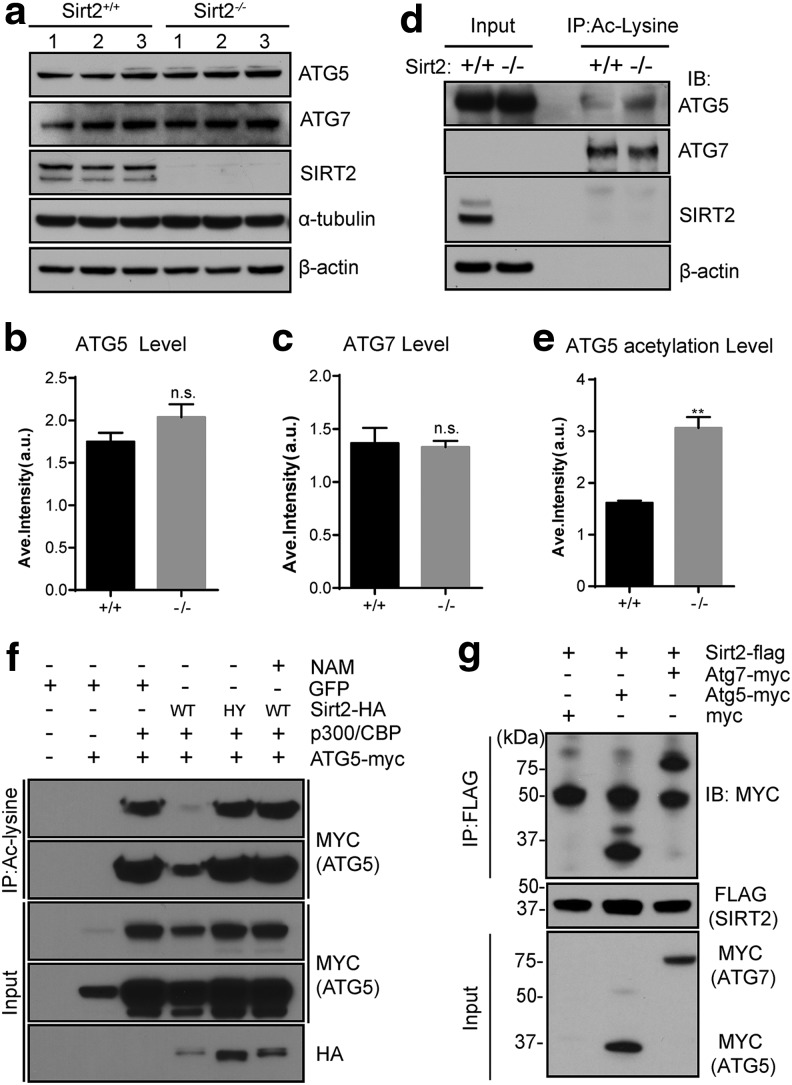
Mitochondrial mitophagy factor, autophagy protein 5 is an SIRT2 downstream deacetylation target
Autophagy protein 5 (ATG5) is an E3 ubiquitin ligase that is involved in autophagosome elongation, leading to autophagy. ATG5 is activated by autophagy protein 7 (ATG7) and forms a complex with ATG12/ATG16L1 that induces LC3B-I conjugation to phosphatidylethanolamine to form LC3B-II. Thus, it seemed reasonable to determine whether the dysregulation of ATG5 and/or ATG7 plays a role in the later “maturation” step of autophagosome degradation in cells lacking Sirt2. Although no change in ATG5 and ATG7 protein expression was observed in MEFs lacking Sirt2 (Fig. 7a) and quantified (Fig. 7b, c), a difference in ATG5 acetylation was observed in lysates from the cortex from Sirt2−/− mice (Fig. 7d) and quantified (Fig. 7e). A tissue culture co-transfection deacetylation assay showed that wild-type SIRT2 (HA-Sirt2), but not HA-SIRT2-HY, a deacetylase null mutant, deacetylated Myc-ATG5 (Fig. 7f, lane 4 vs. lane 5) and that the deacetylation by HA-SIRT2 was inhibited by nicotinamide (lane 6). These results suggest that ATG5 is a legitimate SIRT2 deacetylation target. Finally, Flag-SIRT2 and Myc-ATG5 were co-transfected into 293T cells, and lysates IPed with an anti-Flag antibody and immunoblotted with an anti-Myc antibody showed that SIRT2 interacts with ATG5 (Fig. 7g).
FIG. 7.
ATG5 can interact with and is deacetylated by SIRT2. (a) Sirt2+/+ and Sirt2−/− MEF protein extracts were separated by SDS-PAGE and immunoblotted with anti-ATG5, ATG7, and SIRT2 antibodies. β-Actin and α-tubulin were used as the loading control. (b) Bar graphs quantify the relative expression levels of ATG5. N = 3. (c) Bar graphs quantify the relative expression levels of ATG7. N = 3. (d) Whole brain lysates from Sirt2+/+ and Sirt2-/- mice were harvested, IPed with an anti-acetyl lysine antibody, and, subsequently, immunoblotted with anti-ATG5, ATG7, and SIRT2 antibodies. β-Actin was used as the loading control. (e) Bar graphs quantify the relative ATG5 acetylation levels. N = 3. **p < 0.01. (f) SIRT2 deacetylates ATG5 in vitro. HEK 293T cells were transfected with wild-type HA-SIRT2 or the deacetylation null mutant (HA-SIRT2-HY) gene, p300/CBP, and/or ATG5-myc, and 48 h later, protein extracts were harvested. Lysates were, subsequently, IPed with a pan-anti-acetyl-lysine antibody and immunoblotted with anti-MYC and HA antibodies. (g) SIRT2 physically interacts with ATG5 and ATG7. HEK 293T cells were transiently transfected to express Flag-tagged SIRT2 and Myc-tagged ATG5 or ATG7 or Myc-only plasmid, and cell extracts were IPed with an anti-Flag antibody and immunoblotted with an anti-Myc antibody. All experiments were done in triplicate. Representative images are shown. ATG5, autophagy protein 5; ATG7, autophagy protein 5.
Discussion
In this study, we propose that SIRT2 not only is a cytosolic protein but also localizes to the mitochondria. In addition, it appears that SIRT2 has unique mitochondrial targets and directs autophagy/mitophagy-related components. We have also shown that central nervous system (CNS) tissues have altered mitochondrial morphology and dysregulated mitochondrial redox status. Further investigation is required to reveal how SIRT2 directs the mitochondrial acetylome.
Sirtuins, and specifically the yeast silent mating-type information regulation 2 (Sir2) gene, were initially discovered in yeast and Caenorhabditis elegans and these genes direct organismal longevity, partly by suppressing toxic rDNA formation (15). Enforced Sir2 expression in C. elegans shifts the survival curve inflection point to the right because of the prolonged lifespan (28, 42). Although similar results have not been observed in murine models investigating mammalian sirtuins (2, 7), it has been shown that mice lacking a sirtuin gene exhibit illnesses that are similar to those observed in older humans (4, 17, 18, 34). Although the underlying mechanism for this effect in mammalian cells remains to be fully determined, it seems likely that one function of these proteins is to reprogram the cell to adapt to different nutrient statuses (32, 40). For example, sirtuins appear to sense energy availability and need and can direct specific adaptive cellular processes, such as energy production (i.e., ATP), to match energy consumption (20, 37, 47) as a means of maintaining energy homeostasis (11, 41). Thus, it is proposed that the evolutionarily conserved solution to survival on decreased nutrient availability is to improve or increase the efficiency of energy generation and usage, which is a reparative cellular reprogramming process that also favors increased longevity (14, 19, 21).
It has previously been shown that non-mitochondrial sirtuins, specifically SIRT1, can direct mitochondrial metabolism (25). In this study, we show that the cytoplasmic sirtuin SIRT2 also directs mitochondrial metabolism and relocates to the mitochondria in CNS cells. This localization of SIRT2 may allow it to deacetylate its mitochondrial targets, thereby regulating the activity of mitochondria, such as energy production, ROS, and mitophagy initiation.
To further investigate the functional influence of SIRT2 on mitochondria, Sirt2 knockout mice/MEFs were utilized in this study. Sirt2 deficiency reduced ATP production and increased oxidative stress, showing that SIRT2 is a positive regulator of mitochondria. The decreased efficiency of the Sirt2−/− mitochondria was accompanied by morphological changes in mice and MEFs lacking Sirt2. Morphological changes in mitochondria, such as fragmentation, have been previously described as a signal of impairment. Mitochondrial fragmentation results in a smaller average size (30) and involves a transition from a spaghetti-like network to distinct, round, and fragmented mitochondria (10) or a condensed conformation (27), which is an early event of apoptosis. The mitochondria in the Sirt2 knockout mice of our study were smaller and rounder, and in the Sirt2 knockout MEFs they were condensed and swollen. These morphological changes are not as severe as those characterizing fragmentation, suggesting that the mitochondria in Sirt2−/− cells are impaired but have not reached the physiological threshold that is necessary to initiate apoptosis.
The selective degradation of impaired mitochondria (mitophagy) prevents them from releasing oxidants and triggering apoptosis (43). Mutations in Parkin or PINK1 are the leading cause of recessive parkinsonism (23). A mechanistic study showed that Parkin and PINK1 were involved in the degradation of depolarized mitochondria via autophagy (mitophagy) (35) to protect cells from stress-induced mitochondrial dysfunction. The increased Parkin and Pink1 in Sirt2 knockout mice suggests that this mitochondrial quality control system had been activated, supporting the idea that the mitochondria in Sirt2−/− mice are impaired.
Whether the mitochondrial dysfunction in Sirt2 knockout cells is caused directly by the absence of SIRT2 in mitochondria, indirectly by the defective autophagic process, or a mix of both remains to be defined in detail. However, this study supports a novel role for SIRT2 in maintaining mitochondrial biology.
Materials and Methods
Animals
Sirt2 null mutant mice were generated in our laboratory (30). All of the mice were housed in a facility with a 12-h light/dark cycle and fed a regular diet ad libitum. All mouse work follows the guidelines approved by the Institutional Animal Care and Use Committees of the National Institute of Child Health and Human Development, NIH.
Genotyping
Genomic DNA was prepared from tail biopsies using Direct PCR Lysis Reagent (Viagen Biotech) and subjected to polymerase chain reaction (PCR) amplification using specific sets of PCR primers for genotyping. Three primers were employed as follows: primer A, 5′-GAC TGG AAG TGA TCA AAG CTC-3′; primer B, 5′-CAG GGT CTC ACG AGT CTC ATG-3′; and primer C, 5′-TCA AAT CTG GCC AGA ACT TAT G-3′. Primers A, B, and C are located within introns 4, 7, and 8, respectively. Primers B and C amplify the wild-type allele (200 bp) and floxed allele (255 bp). The combination of primers A and C amplifies the deleted allele (350 bp).
Cell culture (cell lines, Sirt2 null MEFs, primary neurons)
HEK-293T, HeLa, wild type, and Sirt2−/− MEF lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml) in a 37°C incubator with 5% CO2 and 6% oxygen, except when otherwise noted. Mouse primary hippocampal neuron cultures were prepared from hippocampus of newborn (postnatal day 0) pups (38). In brief, neurons were dissociated by papain buffer (Worthington Biochemicals) and were then placed on poly-d-lysine slides (BD) or plates in Basal Eagle Medium containing B27, N2, 1 M l-glutamine, and penicillin/streptomycin (all from Invitrogen). Arabinosylcytosine (Sigma-Aldrich) was used to inhibit glial cell growth.
Transfection
Primary hippocampal neurons were transfected at P4 days in vitro using the calcium phosphate method. Neuronal cultures were incubated with the DNA-calcium phosphate precipitate for ∼1.5 h. The precipitate was then dissolved by the incubation of the cells in a medium that had been pre-equilibrated in a 10% CO2 incubator. The cells were, in turn, transferred to their original conditioned medium and fixed or subjected to live imaging at about 48 h after transfection. All cell line and MEF transfections were performed using FuGENE HD (Roche). Flag-tagged IDH2, -HA- or Flag-tagged SIRT2, Myc-tagged ATG5, Myc-tagged ATG7, and GFP-tagged LC3B expression constructs were purchased from Addgene Technologies. Flag-tagged IDH2 was used to visualize mitochondria. GFP-tagged LC3B was used to visualize autophagosome formation in live imaging.
Tissue fractionation and Western blot
Liver, cortex, hippocampus, striatum, midbrain, and cerebrum tissues were homogenized with 10 volumes of radioimmunoprecipitation assay buffer (RIPA) buffer plus protease and phosphatase inhibitor cocktails, 1 μM trichostatin A (TSA), and 10 mM nicotinamide, and they were centrifuged at 10,000 g for 10 min. Protein concentrations in supernatant were measured by bicinchoninic acid assay (BCA; Thermo Fisher Scientific). Proteins were separated by 4–12% NuPage Bis-Tris polyacrylamide gel electrophoresis (Invitrogen) using MES running buffer (Invitrogen). After transfer to nitrocellulose membranes, the membranes were immunoblotted at 4°C with the appropriate dilutions of one of the following primary antibodies: acetyl-lysine (Cell Signaling, 9441s), SIRT2 (Proteintech), SIRT3 (Cell Signaling), mitochondrial import receptor subunit TOM20 (Proteintech), COX-IV (Cell Signaling), trifunctional enzyme subunit alpha (Santa Cruz Biotechnology, Inc), IDH2 (Proteintech), ATP synthase subunit alpha (Invitrogen), aldehyde dehydrogenase (Proteintech), MnSOD (Millipore), BCL-2 (Santa Cruz), β-actin (Cell Signaling), β-tubulin (Sigma), GAPDH (Millipore), ubiquitin (DAKO), LC3B (Cell Signaling), P62 (Santa Cruz), Parkin (Santa Cruz), PINK1 (Novus), ATG5 (Cell Signaling), ATG7 (Cell Signaling), HDAC6 (Cell Signaling), Flag (Sigma), Myc (Millipore), or HA (Sigma). Signals were visualized by enhanced chemiluminescence development and quantified with ImageJ.
Oxygen consumption rate and extracellular acidification rate analysis
For oxygen consumption rate (OCR) analysis using mouse embryonic fibroblasts (MEFs) cells, 50,000 Sirt2 wild-type and Sirt2−/− MEFs were seeded on an XF-24 microplate (Seahorse Biosciences) and incubated with DMEM containing 10% FBS overnight. Then, 2.5 μM oligomycin A (ATP synthase inhibitor; Sigma), 10 μM carbonyl cyanide m-chlorophenyl hydrazone (mitochondrial uncoupler; Sigma), and 2 μM antimycin/rotenone (complex I/III inhibitors; Sigma) were sequentially added into different ports of the same Seahorse cartridge. Each analysis was replicated five times, and the results were corrected for antimycin and rotenone. For OCR analysis using brain mitochondria, equal concentrations of isolated mitochondrial pellets were attached to an XF24 plate that was pre-treated with CellTak (Corning) following the manufacturer's protocol before OCR analysis.
Mass spectrometry sample preparation and analysis
Striatum of 8 month-old Sirt2+/+ and Sirt2−/− mice were homogenized with IP buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM ethylenediaminetetraaceticacid [EDTA], 0.1% NP-40, 5% glycerol) with protease and phosphatase inhibitor cocktails, 1 μM TSA, and 10 mM nicotinamide. The samples were digested by 12.5 ng/μl proteomics-grade trypsin (Sigma; T6567) at a ratio of 1:40 enzyme to protein. Digested peptides were re-IPed with acetyl-lysine-conjugated beads (Immunchem; ICP0388) and eluted. The resulting peptides were then desalted by solid-phase extraction (Sep-pak C18 cartridges; Waters Corporation). The samples were first acidified with 0.1% trifluoroacetic acid (TFA; Fluka; 40967) and loaded via a syringe onto the Sep-pak SPE material. After sample loading, the cartridges were washed with 0.1% TFA, and they were eluted stepwise with 10–80% acetonitrile/0.1% TFA. Liquid chromatography-mass spectrometry analysis of the peptides was performed using an LTQ-Orbitrap mass spectrometer (Thermo Scientific) that was equipped with a nanospray source and an Eksigent NanoLC 1D Plus and AS1 Autosampler.
Separation of mitochondrial and cytosolic fractions
Mouse tissues (cortex and liver) were rapidly collected, washed, and minced in ice-cold phosphate-buffered saline (PBS) with inhibitor cocktails, 1 μM TSA, and 10 mM nicotinamide. The tissues were then homogenized in hypotonic buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES] pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 250 mM sucrose) with a glass-Teflon motorized homogenizer. Homogenates were centrifuged for 10 min at 700 g at 4°C; then, the supernatants were centrifuged at 6800 g for 10 min. The resulting pellet was used as the mitochondrial fraction, and the supernatant was used as the cytosolic fraction (39). The mitochondrial fraction was washed with hypotonic buffer three times before resuspension in RIPA buffer. For Western blots, the antibodies used were as follows: SIRT2 (Sigma), COX-IV (Cell Signaling), GLG1 (Sigma), and GAPDH (Millipore).
Proteinase K +/− digitonin treatment of mitochondrial fraction
The mitochondrial fraction was resuspended in buffer (10 mM HEPES pH 7.4, 10 mM KCl, 1.5 mM MgCl2, and 1 mM EDTA) at 4°C, followed by centrifugation at 10,000 g for 6 min. The enriched mitochondrial pellet was then resuspended in hypotonic buffer that was treated with digitonin at 1, 2, and 4 mg/ml for 10 min on ice, and this was followed by centrifugation at 12,000 g for 10 min on ice. The supernatants and mito-pellets were collected and run on a Western blot, respectively. SIRT2, Cytochrome C, Bcl-2, and COX-IV antibodies were used. To further investigate the localization of SIRT2 in the mitochondria, mitochondria fractions were treated with digitonin at 2 mg/ml in the next protease protection assay. To remove digitonin, the mito-pellets were washed three additional times with ice-cold HB. Subsequently, the mito-pellets were used for the protease protection assay. Before the proteinase K protection assay, an aliquot of mitochondria was incubated with 1% (w/v) Triton X-100 in HB on ice for 3 min. Two hundred micrograms of mitochondria that were treated or untreated with Triton X-100 was incubated for 15 min at 37°C with 0, 50, 100, or 200 μg/ml (final concentration) proteinase K and dissolved in HB (final volume of 100 μl). The digest was then precipitated with trichloroacetic acid and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
IP and immunoblotting
Cells were harvested and lysed for 30 min on ice in IP buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 5% glycerol) with protease and phosphatase inhibitor cocktails, 1 μM TSA, and 10 mM nicotinamide. Samples were quantified with BCA assays. For IP, cell lysates were incubated with protein-specific antibodies or normal rabbit immunoglobulin G (IgG; Santa Cruz Biotechnology) overnight at 4°C, followed by incubation with protein A/G agarose beads (Millipore) for 3 h at 4°C and with IP lysis buffer that was supplemented with protease inhibitors. For Western blots, the antibodies used were as follows: Flag (Sigma), HA (Sigma), actin (Sigma), tubulin (Sigma), SIRT2 (Sigma), acetyl-lysine (Cell signaling), and GAPDH (Millipore).
Immunofluorescence sample preparation and image acquisition
Cells seeded on glass coverslips were fixed in 4% paraformaldehyde and then blocked with 1% bovine serum albumin (BSA) and 10% normal goat serum in PBS. Cells were incubated with anti-IDH2 (Cell Signaling) or anti-Flag and anti-SIRT2 (Cell Signaling) antibody in PBS followed by incubation with goat-rabbit IgG conjugated with Alexa Fluor 488 and 546 (Invitrogen) in PBS with 5% goat serum. Cells were washed in PBS, mounted, and imaged with a fluorescence microscope. Fluorescence images were captured using a laser scanning confocal microscope (Nikon A1R). The paired images in all the figures were collected at the same gain and offset settings. Post-collection processing was applied uniformly to all paired images. The images were either presented as a single optic layer after acquisition in z-series stack scans from individual fields or displayed as maximum intensity projections to represent confocal stacks.
Live-cell imaging
Primary hippocampal neurons transfected with GFP-tagged LC3B were grown in Basal Eagle Medium containing B27, N2, and 1 M l-glutamine (Sigma) on glass-bottomed dishes. Two days after transfection, the glass-bottomed dish was mounted onto the microscope stage (Andor Spinning Disk Confocal), which was equipped with an OKO labs stage CO2 incubator that maintained the dish at 37°C with 5% CO2. Images were acquired using an inverted Nikon Ti Perfect microscope.
Mitochondrial immunoelectron microscopy
Procedures for the post-embedding immunoelectron microscopy were performed at Northwestern University's Center for Advanced Microscopy. Briefly, mice were perfused through the heart with 4% formaldehyde in phosphate buffer. Brain tissues were removed, submerged in 30% sucrose for 48 h, and sectioned at 100 μm thickness using a Leica Vibratome. After permeabilizing with 0.1% saponin and 5% normal goat serum in PBS and blocking of the nonspecific binding sites with 1% BSA and 10% normal goat serum in PBS, SIRT2 was detected with primary antibody (ProteinTech) and goat-anti-rabbit IgG-gold (10 nm; EMS), and it was, subsequently, enhanced by R-Gent Silver Enhancement kit (EMS) according to the manufacturer's protocols. Tissue samples were cut into small blocks and immersed overnight at 4°C in a fixative containing 4% paraformaldehyde and 1% glutaraldehyde in phosphate buffer. The sections were embedded in LR white according to standard protocols. Ultrathin sections (70 nm) were cut on Formvar-coated nickel grids. After contrasting with uranyl acetate and lead citrate, the sections were inspected using an FEI Tecnai Spirit G2 electron microscope.
For standard transmission electron microscopy, mice were perfused through the heart with 4% formaldehyde and 1% glutaraldehyde in phosphate buffer. After preparation of 100-μm-thick sections, the sections were cut into small blocks and embedded, and ultrathin sections were prepared according to standard protocols. The sections were inspected and imaged as described earlier. For the quantitative assessment of mitochondrial size, circularity, and ellipse, images were exported to Image J (NIH) for imaging analyses. Areas of interest were first selected by freehand selection tools and then subjected to measurement by area, perimeter, and ellipse. The mean area, circularity, and ellipse of 800+ mitochondria were analyzed in each genotype.
ATP measurement
The ATP concentrations present in striata of 1-, 6-, 12-, and 24 month-old Sirt2−/− and Sirt2+/+ mice and MEFs were determined using the ATP determination kit from Molecular Probes. Samples were boiled for 5 min before analysis. Luminescence was read on a Synergy HT microplate reader (BioTek), and values were calculated based on an ATP standard curve.
Thiol analysis
Striata of 24 month-old Sirt2+/+ and Sirt2−/− mice or corresponding MEFs were lysed in 1.34 mM diethylenetriaminepenta-acetic acid (DETAPAC; Sigma) that was dissolved in sodium phosphate buffer (containing 143 mM sodium phosphate [Sigma] and 6.3 mM EDTA [Sigma]). Then, an equal amount of 5% 5-sulfosalicylic acid (SSA; Sigma) was added to the sample lysate. Fifty microliter of the sample lysate was mixed with 700 μl 0.298 mM NADPH (Sigma) dissolved in sodium phosphate buffer, 100 μl 6 mM 5,5′-dithio-bis-2-nitrobenzoic acid (Sigma) dissolved in sodium phosphate buffer, 50 μl 0.023 U/μl glutathione reductase (Sigma) dissolved in water, and 100 μl water. The production of GSH was monitored by reading the absorbance at 412 nm every 15 s for 2.5 min using a DU 800 spectrophotometer (Beckman), and the rate was compared with a standard curve of GSH. For GSSG, 2 μl of 2-vinylpyridine (Sigma) dissolved in 50% ethanol was added to the sample lysed with 1.34 mM DETAPAC for 1 h before an equal amount of 5% SSA was added, and the GSSG concentration was determined using the procedures used for GSH measurement.
Measurement of intracellular hydroperoxides and superoxide
Steady-state levels of hydroperoxides were estimated using the oxidation sensitive fluorescent dye CDCFH2 purchased from Molecular Probes. Steady-state levels of superoxide were estimated using the fluorescent dye DHE, which was also from Molecular Probes. MEFs were trypsinized, washed once with PBS, and labeled with CDCFH2 (10 μg/ml, in 0.1% dimethyl sulfoxide [DMSO], 15 min) or DHE (10 μM, in 0.1% DMSO, 40 min) at 37°C. After labeling, cells were kept on ice. Samples were analyzed using an FACScan flow cytometer (Becton Dickinson Immuno-cytometry System, Inc.) (excitation at 488 nm, emission at 530 nm). The mean fluorescence intensity (MFI) of 10,000 cells was analyzed in each sample and corrected for auto-fluorescence from unlabeled cells. The MFI data were normalized to control levels.
Statistical analysis
Unpaired Student's t-tests and one-way analysis of variance with post hoc analyses were performed via GraphPad Prism 5 (Graphpad Software, Inc.).
Supplementary Material
Abbreviations Used
- ALDH2
aldehyde dehydrogenase
- ATG5
autophagy protein 5
- ATG7
autophagy protein 7
- ATP
adenosine triphosphate
- ATP5A1
ATP synthase subunit alpha
- BCA
bicinchoninic acid assay
- BSA
bovine serum albumin
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
- CDCFH2
5-(and-6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate
- CNS
central nervous system
- DAPI
4′,6-diamidino-2-phenylindole
- DETAPAC
diethylenetriaminepenta-acetic acid
- DHE
dihydroethidium
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethyl sulfoxide
- EDTA
ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- GSH
glutathione
- GSSG
oxidized glutathione
- HADHA
trifunctional enzyme subunit alpha
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IDH2
isocitrate dehydrogenase 2 (NADP+)
- IFC
immunofluorescent
- IgG
immunoglobulin G
- IP
immunoprecipitation
- LC3
microtubule-associated protein 1 light chain 3 beta
- MEF
mouse embryonic fibroblast
- MFI
mean fluorescence intensity
- MnSOD
manganese superoxide dismutase
- OCR
oxygen consumption rate
- Parkin
E3 ubiquitin-protein ligase parkin
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PINK1
serine/threonine-protein kinase PINK1
- RIPA
radioimmunoprecipitation assay buffer
- ROS
reactive oxygen species
- SEM
standard error of the mean
- Sir2
silent information regulator 2
- SIRT2
NAD-dependent protein deacetylase sirtuin-2
- SSA
5-sulfosalicylic acid
- TFA
trifluoroacetic acid
- TSA
trichostatin A
Acknowledgments
D.G. is supported by NCI-1R01CA152601-01, 1R01CA152799-01A1, 1R01CA168292-01A1, 1R01CA16383801A1, and BC093803. Melissa Stauffer, PhD, of Scientific Editing Solutions, provided editorial assistance. Imaging work was performed at the Northwestern University Center for Advanced Microscopy that was generously supported by NCI CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center.
Author Disclosure Statement
No competing financial interest exist.
References
- 1.Aquilano K, Vigilanza P, Baldelli S, Pagliei B, Rotilio G, and Ciriolo MR. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem 285: 21590–21599, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban RS, Nemoto S, and Finkel T. Mitochondria, oxidants, and aging. Cell 120: 483–495, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Beirowski B, Gustin J, Armour SM, Yamamoto H, Viader A, North BJ, Michan S, Baloh RH, Golden JP, Schmidt RE, Sinclair DA, Auwerx J, and Milbrandt J. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proc Natl Acad Sci U S A 108: E952–E961, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalkiadaki A. and Guarente L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer 15: 608–624, 2015 [DOI] [PubMed] [Google Scholar]
- 5.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, and Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840, 2009 [DOI] [PubMed] [Google Scholar]
- 6.Donmez G. and Guarente L. Aging and disease: connections to sirtuins. Aging Cell 9: 285–290, 2010 [DOI] [PubMed] [Google Scholar]
- 7.Finkel T, Deng CX, and Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature 460: 587–591, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, and Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell 19: 416–428, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finley LW. and Haigis MC. Metabolic regulation by SIRT3: implications for tumorigenesis. Trends Mol Med 18: 516–523, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, and Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell 1: 515–525, 2001 [DOI] [PubMed] [Google Scholar]
- 11.Gius D. and Spitz DR. Redox signaling in cancer biology. Antioxid Redox Signal 8: 1249–1252, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Glozak MA, Sengupta N, Zhang X, and Seto E. Acetylation and deacetylation of non-histone proteins. Gene 363: 15–23, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Gomes LC. and Scorrano L. Mitochondrial morphology in mitophagy and macroautophagy. Biochim Biophys Acta 1833: 205–212, 2013 [DOI] [PubMed] [Google Scholar]
- 14.Guarente L. Calorie restriction and SIR2 genes—towards a mechanism. Mech Ageing Dev 126: 923–928, 2005 [DOI] [PubMed] [Google Scholar]
- 15.Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol 72: 483–488, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Guarente L. Mitochondria—a nexus for aging, calorie restriction, and sirtuins? Cell 132: 171–176, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarente L. Aging research-where do we stand and where are we going? Cell 159: 15–19, 2014 [DOI] [PubMed] [Google Scholar]
- 18.Guarente L. The many faces of sirtuins: sirtuins and the Warburg effect. Nat Med 20: 24–25, 2014 [DOI] [PubMed] [Google Scholar]
- 19.Guarente L. and Picard F. Calorie restriction—the SIR2 connection. Cell 120: 473–482, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Haigis MC, Deng CX, Finley LW, Kim HS, and Gius D. SIRT3 is a mitochondrial tumor suppressor: a scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res 72: 2468–2472, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haigis MC. and Guarente LP. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev 20: 2913–2921, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Haigis MC. and Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol 5: 253–295, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy J. Genetic analysis of pathways to Parkinson disease. Neuron 68: 201–206, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He W, Newman JC, Wang MZ, Ho L, and Verdin E. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab 23: 467–476, 2012 [DOI] [PubMed] [Google Scholar]
- 25.Hirschey MD, Shimazu T, Capra JA, Pollard KS, and Verdin E. SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2. Aging (Albany NY) 3: 635–642, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houtkooper RH, Pirinen E, and Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol 13: 225–238, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, and Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell 43: 33–44, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaeberlein M, McVey M, and Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13: 2570–2580, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, and Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17: 41–52, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, Ji J, Wang XW, Park SH, Cha YI, Gius D, and Deng CX. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20: 487–499, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, and Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23: 607–618, 2006 [DOI] [PubMed] [Google Scholar]
- 32.Lantier L, Williams AS, Williams IM, Yang KK, Bracy DP, Goelzer M, James FD, Gius D, and Wasserman DH. SIRT3 is crucial for maintaining skeletal muscle insulin action and protects against severe insulin resistance in high-fat-fed mice. Diabetes 64: 3081–3092, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maxwell MM, Tomkinson EM, Nobles J, Wizeman JW, Amore AM, Quinti L, Chopra V, Hersch SM, and Kazantsev AG. The sirtuin 2 microtubule deacetylase is an abundant neuronal protein that accumulates in the aging CNS. Hum Mol Genet 20: 3986–3996, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagawa T. and Guarente L. SnapShot: sirtuins, NAD, and aging. Cell Metab 20: 192–192e1, 2014 [DOI] [PubMed] [Google Scholar]
- 35.Narendra D, Walker JE, and Youle R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb Perspect Biol 4: pii: 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.North BJ, Marshall BL, Borra MT, Denu JM, and Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11: 437–444, 2003 [DOI] [PubMed] [Google Scholar]
- 37.Ozden O, Park SH, Wagner BA, Yong Song H, Zhu Y, Vassilopoulos A, Jung B, Buettner GR, and Gius D. SIRT3 deacetylates and increases pyruvate dehydrogenase activity in cancer cells. Free Radic Biol Med 76: 163–172, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parisiadou L, Yu J, Sgobio C, Xie C, Liu G, Sun L, Gu XL, Lin X, Crowley NA, Lovinger DM, and Cai H. LRRK2 regulates synaptogenesis and dopamine receptor activation through modulation of PKA activity. Nat Neurosci 17: 367–376, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, Lombard DB, and Zhao Y. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell 50: 919–930, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pillai VB, Samant S, Sundaresan NR, Raghuraman H, Kim G, Bonner MY, Arbiser JL, Walker DI, Jones DP, Gius D, and Gupta MP. Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nat Commun 6: 6656, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoyanovsky DA, Tyurina YY, Tyurin VA, Anand D, Mandavia DN, Gius D, Ivanova J, Pitt B, Billiar TR, and Kagan VE. Thioredoxin and lipoic acid catalyze the denitrosation of low molecular weight and protein S-nitrosothiols. J Am Chem Soc 127: 15815–15823, 2005 [DOI] [PubMed] [Google Scholar]
- 42.Tanny JC, Dowd GJ, Huang J, Hilz H, and Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99: 735–745, 1999 [DOI] [PubMed] [Google Scholar]
- 43.Twig G. and Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal 14: 1939–1951, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitworth AJ. and Pallanck LJ. The PINK1/Parkin pathway: a mitochondrial quality control system? J Bioenerg Biomembr 41: 499–503, 2009 [DOI] [PubMed] [Google Scholar]
- 45.Yu J. and Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann N Y Acad Sci 1173 Suppl 1: E10–E19, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, and Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol 12: 665–675, 2010 [DOI] [PubMed] [Google Scholar]
- 47.Zhu Y, Yan Y, Principe DR, Zou X, Vassilopoulos A, and Gius D. SIRT3 and SIRT4 are mitochondrial tumor suppressor proteins that connect mitochondrial metabolism and carcinogenesis. Cancer Metab 2: 15, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.