Abstract
Bacterial type IV secretion (T4S) systems mediate the transfer of macromolecular substrates into various target cells, e.g., the conjugative transfer of DNA into bacteria or the transfer of virulence proteins into eukaryotic host cells. The T4S apparatus VirB of the vascular tumor-inducing pathogen Bartonella henselae causes subversion of human endothelial cell (HEC) function. Here we report the identification of multiple protein substrates of VirB, which, upon translocation into HEC, mediate all known VirB-dependent cellular changes. These Bartonella-translocated effector proteins (Beps) A-G are encoded together with the VirB system and the T4S coupling protein VirD4 on a Bartonella-specific pathogenicity island. The Beps display a modular architecture, suggesting an evolution by extensive domain duplication and reshuffling. The C terminus of each Bep harbors at least one copy of the Bep-intracellular delivery domain and a short positively charged tail sequence. This biparte C terminus constitutes a transfer signal that is sufficient to mediate VirB/VirD4-dependent intracellular delivery of reporter protein fusions. The Bep-intracellular delivery domain is also present in conjugative relaxases of bacterial conjugation systems. We exemplarily show that the C terminus of such a conjugative relaxase mediates protein transfer through the Bartonella henselae VirB/VirD4 system into HEC. Conjugative relaxases may thus represent the evolutionary origin of the here defined T4S signal for protein transfer into human cells.
Keywords: conjugative relaxase, effector protein, endothelial cell, protein translocation, antiapoptosis
Bacterial type IV secretion (T4S) systems are versatile transporters ancestrally related to bacterial conjugation machines. Present-day functions of T4S systems include (i) DNA transfer into bacterial or plant cells by cell-to-cell contact, (ii) protein delivery into mammalian or plant cells by cell-to-cell contact, (iii) DNA release to or uptake from the extracellular milieu, and (iv) release of multisubunit protein toxins to the extracellular milieu (1, 2). The prototypic T4S system for interkingdom substrate transfer is the VirB apparatus (encoded by virB1-virB11) and associated T4S coupling protein VirD4 of the phytopathogen Agrobacterium tumefaciens (At). This VirB/VirD4 T4S system mediates transfer of all components of the so called T-DNA complex, which is composed of protein substrates (VirD2 and VirE2) and single-stranded DNA (T-DNA), into plant cells (3). Intracellular delivery of solely protein substrates subverting host cell function (effector proteins) is considered to represent the primary function of T4S systems in human pathogenic bacteria (2). Examples include the Cag system of the gastric pathogen Helicobacter pylori (Hp), which translocates the CagA effector protein into gastric epithelial cells (4), and the Dot/Icm system of the Legionnaires disease agent Legionella pneumophila (Lp), which translocates multiple effector proteins into infected macrophages (5, 6). Although reporter protein fusions with subdomains of T4S substrates of At VirB/VirD4 or Lp Dot/Icm have indicated the requirement of C-terminal sequences for interkingdom protein transfer (5, 7, 8), no conserved T4S signal has been defined yet (1, 2).
Bartonella henselae (Bh) is a zoonotic pathogen causing a broad range of clinical manifestations in humans, including cat-scratch disease, bacillary angiomatosis-peliosis, bacteremia with fever, and neuroretinitis. Bacillary angiomatosis-peliosis is characterized by the formation of vasoproliferative tumors, which result from bacterial colonization and activation of human endothelial cell (HEC) (9). VirB, a T4S system closely related to conjugative DNA-transfer systems of α-proteobacterial plasmids (10), is a major virulence determinant of Bh for subversion of HEC function. VirB-dependent changes of HEC include (i) massive cytoskeletal rearrangements resulting in cell-surface aggregation and uptake of large bacterial aggregates by a defined structure termed the invasome; (ii) induction of a proinflammatory phenotype by activation of NF-κB, resulting in surface expression of the cell adhesion molecules ICAM-1 and E-selectin and secretion of the proinflammatory cytokine IL-8; (iii) increased cell survival by inhibition of early and late events of apoptosis (caspase activation and DNA fragmentation, respectively); and (iv) cytostatic or even cytotoxic effects at high infection doses, which interfers with a potent VirB-independent mitogenic activity of Bh (11).
Here, we report the identification of the genes encoding the T4S coupling protein VirD4 and seven putative effector proteins [Bartonella-translocated effector proteins (Beps) A-G]. We provide evidence that VirD4 and at least one of the effector proteins mediates all VirB-dependent phenotypes in HEC. Furthermore, we exemplarily show BepD to be translocated into HEC in a VirB/VirD4-dependent manner. Based on sequence homology between all seven Beps, we functionally define the signal for VirB/VirD4-dependent protein transfer and propose its evolutionary origin from conjugative relaxases of bacterial conjugation systems.
Materials and Methods
Bacterial Strains, Cell Lines, and Growth Conditions. Bh and Escherichia coli strains were grown as described in ref. 11, and At C58 was grown on plates containing Luria-Bertani medium plus agar at 28°C overnight. Table 1, which is published as supporting information on the PNAS web site, lists all the strains used in this study. Human umbilical vein endothelial cells (HUVEC) were isolated and cultured as described in ref. 12. The endothelial cell line Ea.hy926 resulting from a fusion of HUVEC and the lung carcinoma cell line A549 were cultured as reported in ref. 13.
DNA Sequencing and Plasmid Construction. Sequencing of the bep region of Bh ATCC 49882T was performed from a cosmid library by using a primer walking strategy, starting with primers used for the sequencing of virD4 of Bartonella tribocorum (10). Details are described in Supporting Materials and Methods and Table 2, which are published as supporting information on the PNAS web site. The resulting sequence has been deposited in GenBank under accession no. AJ556988. The nuclear localization signal (NLS)-Cre-Bep fusion protein-expressing vectors (pRS49-pRS124), the Cre-sensor vector (pRS56), and the BepD expression vector (pPG104) were constructed by multiple cloning steps. Sequences of oligonucleotides (Table 2), sources of gene cassettes, and further details of cloning steps are given in Supporting Materials and Methods. Briefly, for the expression of NLS-Cre-Bep fusion proteins in the bacteria, we first constructed pRS40, which contains the coding sequence for an NLS-Cre fusion protein under the control of the taclac promoter. Sequences of interest of the bep genes were amplified from genomic DNA and cloned into the region encoding the C terminus of the NLS-Cre gene in pRS40, providing vectors for inducible expression of NLS-Cre-Bep fusion proteins (pRS49-pRS124). pRS56 was constructed for generation of cell line Ea.hy926/pRS56-c#B1, and it contains the successive arrangement of a loxH site, a neomycin phosphotransferase (neo) gene followed by a terminator, a loxP site, and an egfp gene encoding GFP. To express FLAG-tagged BepD, we first constructed a vector containing the coding sequence for the FLAG tag following the starting methionine (MDYKDDDDK) under the control of the taclac promoter (pPG100). bepD was amplified from genomic DNA and cloned downstream of the FLAG tag in pPG100, which yielded pPG104.
Construction of In-Frame Deletions and Complementation of the Deletion Mutants. In-frame deletion mutants of Bh RSE247 were generated by a two-step gene replacement procedure as described in refs. 10 and 11. The ΔvirD4 mutant contains an in-frame deletion of 1.63 kb in virD4. The ΔbepB-G strain carries a 14.33-kb chromosomal deletion resulting in a 51-bp cryptic ORF composed of a 5′ sequence of bepB and a 3′ sequence of bepG. To construct the ΔbepA-G strain, a 1.49 kb in-frame deletion in bepA was introduced into the ΔbepB-G strain, which resulted in a remaining 144-bp cryptic ORF composed of 5′ and 3′ sequences of bepA. Further details are provided in Supporting Materials and Methods.
Caspase Activity, IL-8 Secretion, and Proliferation. The infection of HEC and the determination of caspase-3 and caspase-7 activity [multiplicity of infection (moi) = 100], secretion of IL-8 (moi = 300), and cell proliferation (moi = 30) were carried out as described in ref. 11.
Immunocytochemical Stainings and Immunoprecipitation. HEC were infected with Bh strains, stained for F-actin, total bacteria, and extracellular bacteria or anti-FLAG M2. To assess the tyrosine phosphorylation of BepD upon translocation by the T4S system, Ea.hy926 cells were infected with Bh strains expressing FLAGtagged BepD. Cells were subsequently lysed, and the FLAG-tagged BepD was immunoprecipitated with anti-FLAG agarose and probed with antiphosphotyrosine antibody in a Western blot. Experimental details are described in Supporting Materials and Methods.
Cre Recombinase Reporter Assay for Translocation (CRAFT). CRAFT (7, 8) was used to monitor the translocation of NLS-Cre-Bep fusion proteins from Bh into Ea.hy926 cells stably transfected with pRS56 (clone Ea.hy926/pRS56-c#B1). After transport to the nucleus, the fusion protein recombines two lox sites in pRS56, thereby excising neo and the terminator, which resulted in expression of eGFP. Briefly, Ea.hy926/pRS56-c#B1 were infected with Bh strains harboring plasmids containing NLS-Cre-Bep fusions, trypsinized after 120 h, and analyzed by flow cytometry. To monitor the stability of NLS-Cre-Bep fusions in Bh, steady-state protein levels in total lysates of bacteria grown on isopropyl β-d-thiogalactoside-containing medium were determined by immunoblotting with anti-Cre antibodies. Experimental details are described in Supporting Materials and Methods.
Bioinformatic Analysis. The putative C-terminal transfer domains of Bh BepA-G were aligned by clustalw. The alignment was further edited manually, and a hidden Markov model was built thereof. By using this model, we queried the UniProt database (14) as described in Supporting Materials and Methods. Sequences of interest were aligned, and a neighbor-joining tree was generated as described in Supporting Materials and Methods.
Results
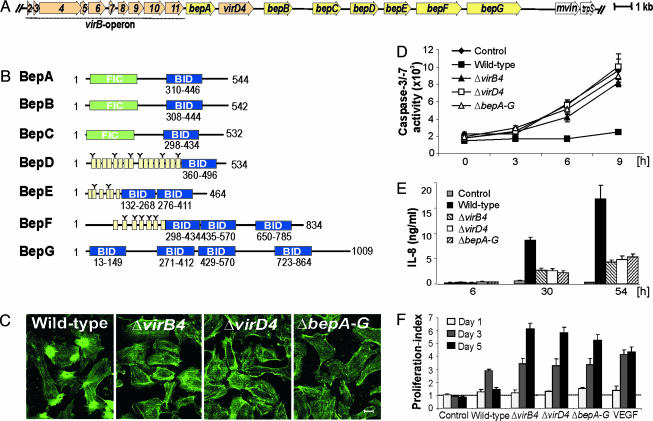
Bh Carries a Pathogenicity Island (PAI) Encoding the VirB/VirD4 T4S System and Seven Putative Protein Substrates. Assuming functional clustering of the operon encoding the previously described VirB apparatus (virB2-virB11) (15) with genes encoding further T4S-related functions, we sequenced 23,294 base pairs that were downstream of virB11 (Fig. 1A) (GenBank accession no. AJ556988). Among the 10 genes encoded by this region, only the distal mviN and trpS are present in the chromosome of related α-proteobacteria, suggesting that these genes belong to the ancestral core genome (16). A cryptic prophage integrase gene upstream of mviN indicates that the flanking region may have been acquired by horizontal gene transfer (16). Based on criteria defined by Hacker et al. (17), the virB operon and the eight downstream-located genes may constitute a PAI. The second gene downstream of virB11 encodes the T4S coupling protein VirD4. The remaining seven genes of the PAI code for putative VirB/VirD4-translocated effector proteins, which we termed BepA-G. Sequence analysis revealed a modular domain structure for BepA-G (Fig. 1B). BepA-C are homologues carrying an N-terminal filamentation induced by cAMP (Fic) domain, which is implicated in bacterial cell division (18) and is conserved in many bacterial species (Fig. 1B and Fig. 5, which is published as supporting information on the PNAS web site). The N-terminal regions of BepD-F contain repeated tyrosine-containing peptide sequences that resemble tyrosine-phosphorylation motifs (e.g., EPLYA, Fig. 1B and Fig. 6, which is published as supporting information on the PNAS web site). Strikingly, all Beps share at least one copy of a domain of ≈140 aa in their C-terminal region (Fig. 1B and Fig. 7, which is published as supporting information on the PNAS web site). This domain was suspected to be involved in Bep translocation and was thus designated the Bep intracellular delivery (BID) domain. In addition to the BID domain, the C termini of BepA-G contain short unconserved tail sequences rich in positively charged residues, each carrying a net positive charge (Table 3, which is published as supporting information on the PNAS web site).
Fig. 1.
The Beps mediate VirB/VirD4-dependent invasion, antiapoptotic protection, proinflammatory activation, and control of proliferation of HEC. (A) Structure of the virB/virD4/bep locus encoding the VirB components (VirB2-VirB11), the T4S coupling protein (VirD4), and seven putative effector proteins (BepA-G). (B) Domain structure of BepA-G. Yellow boxes represent tyrosine-containing sequence repeats resembling tyrosine-phosphorylation motifs (indicated by Y). (C) VirB4/VirD4/Bep proteins are required for mediating characteristic actin rearrangements, which result in uptake of Bh aggregates by means of invasomes. HUVEC infected with the indicated Bh strains were stained for F-actin. (Scale bar, 10 μm.) (D) VirB4/VirD4/Bep proteins are required for antiapoptosis. Caspase-3/7 activity of HUVEC was measured after infection with the indicated Bh strains for 24 h, followed by induction of apoptosis by actinomycin D for the indicated times. (E) VirB/VirD4/Bep proteins are required for NF-κB-dependent proinflammatory activation. HUVEC were infected for the indicated time with the indicated Bh strains, followed by quantification of IL-8 in the culture medium. (F) VirB4/VirD4/Bep proteins are required for controlling Bh-stimulated HUVEC proliferation. HUVEC infected with the indicated Bh strains were counted at the indicated time points, and proliferation indices were calculated. (D-F) Triplicate samples ± standard deviation.
All Known VirB-Dependent Cellular Phenotypes of HEC Require VirD4 and at Least One of the Putative Effector Proteins BepA-G. To test whether VirD4 and BepA-G contribute to VirB-mediated virulence, we generated nonpolar in-frame deletion mutants (ΔvirD4 and ΔbepA-G, the latter mutant being constructed by sequential deletion of bepB-G and bepA) and compared them with the isogenic ΔvirB4 mutant and wild-type strain with respect to known VirB-dependent phenotypes of Bh-infected HEC (11). Opposed to wild type, all three deletion mutants were deficient for triggering (i) the formation of the characteristic F-actin rearrangements associated with invasome-mediated invasion (Fig. 1C and Table 4, which is published as supporting information on the PNAS web site), (ii) the inhibition of apoptotic cell death triggered by actinomycin D as measured by caspase-3/7 activity (Fig. 1D), (iii) the activation of an NF-κB-dependent proinflammatory response determined by quantification of secreted IL-8 in the culture medium (Fig. 1E), and (iv) cytostatic/cytotoxic effects interfering with the VirB-independent mitogenic activity of Bh as measured by cell counting (Fig. 1F). We conclude that all known VirB-mediated phenotypes of HEC require the T4S coupling protein VirD4 and at least one of the putative effector proteins BepA-G. Moreover, these data suggest that most likely all genes encoded by the virB/virD4/bep PAI of Bh have functions related to T4S.
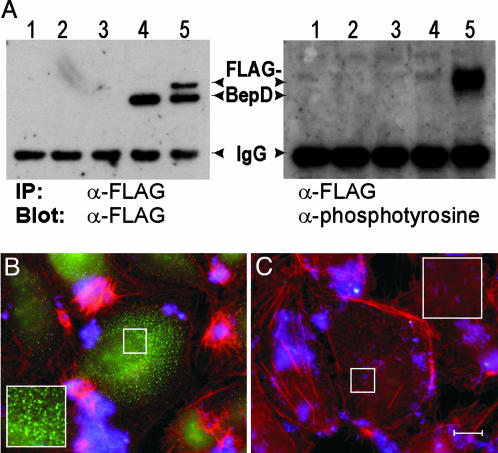
BepD Is Translocated into HEC in a VirB-Dependent Manner. Next, we tested whether BepD, one of the three putative substrates for host cell tyrosine kinases among the Beps (Fig. 1B), becomes tyrosine-phosphorylated during infection of HEC. Phosphorylation by host cell tyrosine kinases was previously used to demonstrate translocation of bacterial proteins into human cells (19). We show that FLAG-epitope-tagged BepD becomes tyrosine-phosphorylated during HEC infection when expressed in wild type but not in the ΔvirB4 mutant (Fig. 2A Right). Tyrosine phosphorylation coincided with a prominent shift in electrophoretic mobility (Fig. 2 A Left), suggesting additional protein modification. Immunocytochemistr y revealed a VirB4-dependent punctuate staining pattern of FLAG-BepD in the host cell cytoplasm (Fig. 2 B and C). Together, these data demonstrate VirB-dependent translocation of BepD into HEC.
Fig. 2.
BepD becomes tyrosine-phosphorylated after VirB4-dependent translocation into HEC. (A) VirB4-dependent translocation of BepD into HEC results in tyrosine phosphorylation and a coincident reduction in electrophoretic mobility. Total protein extracts of Ea.hy926 cells uninfected (lane 1) or infected with ΔvirB4 (lane 2), wild type (lane 3), ΔvirB4/pPG104 (lane 5), or wild type/pPG104 were prepared. FLAG-BepD encoded by pPG104 was immunoprecipitated with anti-FLAG antibodies, separated by SDS/PAGE, and immunoblotted with anti-FLAG (Left) or anti-phosphotyrosine antibodies (Right). (B and C) Immunocytochemical detection of FLAG-BepD after VirB/VirD4-mediated translocation into HEC. Ea.hy926 cells were infected with wild-type (B) or the ΔvirB4 mutant (C), each harboring pPG104. Specimens were immunocytochemically stained for the FLAG epitope (green), F-actin (red), and bacteria (blue). (Scale bar, 10 μm.)
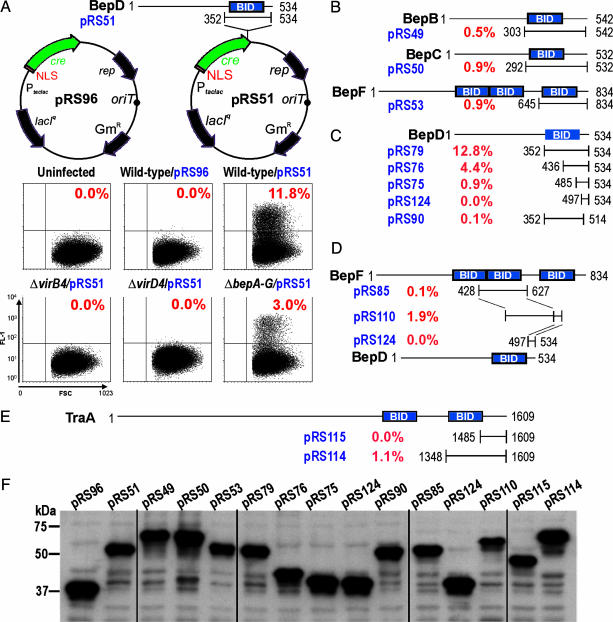
Delineation of the Bipartite T4S Signal of the Beps. To delimit the BepD translocation signal and to demonstrate translocation of other Beps, we adapted CRAFT, a reporter assay originally designed to detect translocation of bacterial effector proteins into plant cells (8) (Fig. 3A and Fig. 8, which is published as supporting information on the PNAS web site). After infection of the Cre-tester cell line Eahy.926/pRS56-c#B1 with Bh strains expressing NLS-Cre-recombinase fusion proteins, the percentage of GFP-positive cells (gpc) as determined by FACS analysis was used as a relative measure for the efficiency of protein transfer from Bh into HEC. Expression of an NLS-Cre-recombinase fusion protein in wild-type Bh resulted in 0.0% gpc and was thus negative in this assay (Fig. 3A, pRS96). In contrast, NLS-Cre fused to the C-terminal 183 aa of BepD (BID domain plus a short positively charged tail sequence, pRS51) was efficiently translocated from wild-type (11.8% gpc) and ΔbepA-G (3.0% gpc), whereas no translocation occurred from ΔvirB4 or ΔvirD4 strains (0.0% gpc) (Fig. 3 A and F). Hence, this heterologous fusion protein was translocated in a VirB4/VirD4-dependent, and essentially Bep-independent, manner. Similar as for BepD, NLS-Cre fusions to the BID domain-containing C terminus of BepB, BepC, and BepF were translocated into HEC, albeit at lower frequency (Fig. 3 B and F). Taken together, we provide evidence for a functional T4S signal in the C terminus of four Bep proteins (BepB, BepC, BepD, and BepF).
Fig. 3.
The C-terminal translocation signal of Beps mediates VirB/VirD4-dependent protein transfer into HEC. Protein transfer was determined by CRAFT. The Cre-tester cell line Ea.hy926/pRS56-c#B1 was infected with the indicated Bh strains expressing different NLS-Cre fusion proteins (plasmid names are indicated in blue in A-E or black in F). The region of a given Bep fused to the C terminus of NLS-Cre is specified by the respective first and last amino acids (except for pRS96, which expresses only NLS-Cre). Percentages of GFP-positive cells as determined by FACS analysis are indicated in red. (A) NLS-Cre fused to the C-terminal 183 aa of BepD translocates efficiently into HEC in a VirB/VirD4-dependent manner. Dot blots of forward scatter (FSC) and GFP fluorescence (FL-1) are shown for the indicated Bh strains. (B) Relative translocation efficiency mediated by the BID domain of BepB, BepC, and BepF. (C) The signal for VirB/VirD4-dependent translocation into HEC is bipartite, composed of the BID domain and an adjacent unconserved C-terminal tail. (D) Creation of an efficient bipartite translocation signal by fusing a BID domain of BepF and the C-terminal tail of BepD. (E) The C terminus of the relaxase TraA of At plasmid pATC58 contains a BID domain and mediates efficient protein transfer from Bh into HEC. (F) Steady-state NLS-Cre fusion protein levels in Bh grown on isopropyl β-d-thiogalactoside-containing medium.
To further delimit the T4S signal contained in the 183-aa C-terminal fragment of BepD, we performed a deletion analysis (Fig. 3 C and F). C-terminal deletion of 20 aa of the short positively charged C-terminal tail sequence almost completely abolished translocation (0.1% gpc). Stepwise deletion of the BID domain from the N terminus resulted in a gradual reduction of translocation efficiency. Together, these data suggest a bipartite translocation signal at the C terminus, composed of a BID domain and a short positively charged tail sequence. As illustrated in Fig. 3D, this notion was supported by the success in creating an efficient translocation signal (1.9% gpc) via fusion of a translocation-inefficient BID domain of BepF (0.1% gpc) with the translocation-deficient positively charged tail of BepD (0.0% gpc). Notably, all NLS-Cre-Bep fusion proteins analyzed by CRAFT displayed comparable steady-state protein levels in bacteria (Fig. 3F), indicating that the low translocation efficiency observed for several fusion proteins does not result from protein instability but rather reflects the absence of an appropriate T4S signal.
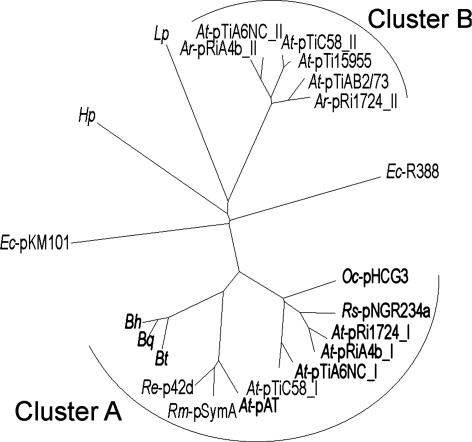
Identification of BID Domains in Conjugative Relaxases and Demonstration of Their Function As T4S Signal for the Bh VirB/VirD4 System. To search for other proteins containing a BID domain, we queried the UniProt database with a hidden Markov model (20) generated from an alignment of all BID domains of BepA-G (Fig. 7 and Table 5, which is published as supporting information on the PNAS web site). Among the 40 top hits are 27 hits within putative T4S substrates. These hits include BepA-G of Bh and their homologues in Bartonella quintana, annotated as hypothetical proteins in the recently published genome sequences (16), as well as Fic-1, which is a BepA homologue in Bartonella tribocorum (10). The other hits in putative T4S substrates are all in relaxases of conjugative plasmids found in various α-proteobacteria. The plasmid-borne conjugation systems associated with these conjugative relaxases are closely related to each other as well as to the Bh VirB/VirD4 system (10), as indicated by clustering in one clade of a phylogenetic tree for VirD4/TraG-like T4S coupling proteins (Fig. 4, cluster A). Interestingly, no BID domain was found in protein substrates of agrobacterial T-DNA transfer systems (VirB/VirD4), which cluster in a separate clade of the VirD4/TraG phylogram (Fig. 4, cluster B), or in the T4S substrates of Lp or Hp. For the AvhB/TraG conjugation system of At plasmid pAtC58 (21), we show that the C terminus of its relaxase harbors a BID domain and a positively charged tail sequence, which efficiently directs VirB/VirD4-dependent protein transfer from Bh into HEC (Fig. 3E, pRS114), whereas the positively charged tail alone did not result in detectable transfer activity (Fig. 3E, pRS115). These data suggest that the Bh VirB/VirD4/Bep protein transfer system evolved rather recently from one of the wide-spread conjugative plasmid-transfer systems in α-proteobacteria and that the bipartite transfer signals in the substrates of these T4S systems are functionally interchangeable.
Fig. 4.
The coupling proteins (VirD4/TraG) of T4S systems containing a BID domain in their protein substrate(s) form a distinct phylogenetic cluster. This cluster is formed by Bartonella VirB/VirD4 systems and α-proteobacterial conjugative DNA transfer systems (cluster A) and does not contain agrobacterial T-DNA transfer systems (cluster B). VirD4/TraG protein sequences were extracted from the Uniprot database and then aligned and diagrammed as an unrooted neighbor-joining radial tree. T4S systems containing a BID domain in one of their substrate(s) are marked in bold (compare with Table 5). The following sequences (with corresponding accession numbers) are included. At: plasmid pAT (Q8UKJ4), pTiC58 (Q44346 and P18594), pTiA6NC (Q44360 and P09817), pRi1724 (Q9F5E3 and Q9F585), pTi15955 (Q8VLK3), and pTiAB2/73 (Q8VT85); Agrobacterium rhizogenes: pRiA4b (Q93UY7 and P13464); Bh (Q6G2A8); Bartonella quintana (Q6FYV9); Bartonella tribocorum (Q8GJ55); Escherichia coli: pKM101 (Q46706) and R388 (Q04230); Hp (Q75XB9); Lp (Q9RLR2); Oligotropha carboxydovorans (Q6LB53); Rhizobium etli: p42d (Q8KL68); Rhizobium meliloti: pSymA (Q92ZI3); and Rhizobium spp.: pNGR234a (P55421).
Discussion
In this study, we characterized a PAI encoding presumably all proteins related to the function of a pathogenesis-related T4S system in Bh. In addition to the previously described T4S apparatus VirB (VirB2-VirB11) (11, 15), this PAI encodes the T4S coupling protein, VirD4, and seven T4S substrates termed BepA-G. Deletion of either virD4 or the complete set of bep genes (bepA-G) resulted in a similar phenotype as that described for deletion of virB4 (11); i.e., these mutants are deficient for subverting multiple HEC functions related to the cytoskeleton and to inflammation, apoptosis, and proliferation. The essential role of VirD4 for mediating VirB-dependent host cellular changes is consistent with the proposed function as T4S coupling protein, representing the interface between the T4S apparatus and the translocated substrates (1). The loss of all known VirB/VirD4-dependent HEC changes in the ΔbepA-G mutant indicates that BepA-G may comprise the complete set of VirB/VirD4-translocated effector proteins. Preliminary data from our laboratory suggest that the specific contribution of individual Beps to the complex VirB/VirD4-dependent phenotypic changes of HEC can be assessed by their expression, either alone or in combination, in the effector-free ΔbepA-G mutant background (M.C.S., P.G., and C.D., unpublished data).
The recently published comparative genome analysis of Bh and Bartonella quintana revealed that the virB/virD4/bep PAI characterized herein is present in both Bartonella genomes but not in any other published genome sequence. However, in contrast to the highly conserved virB/virD4 loci, the bep loci display a high degree of plasticity, including signatures of gene duplication and degradation (data not shown) as well as intragenic domain duplication and intragenic or intergenic domain reshuffling (Fig. 1B). As a result, the domain structure of the Beps is highly modular. The N termini of BepA-C are composed of a domain (Fic) conserved in many bacterial species that is considered to be involved in cell division (18). The N termini of BepD-F contain short repeated peptide sequences containing conserved putative tyrosine phosphorylation motifs (i.e., EPLYA) similar to the EPIYA motif of the CagA effector protein of Hp known to be phosphorlyated by human Src-family kinases (4, 22). Consistently, we show BepD to become tyrosine-phosphorylated upon T4S-dependent transfer into HEC. Taken together, the N termini of the Beps are highly divergent and may primarily serve effector functions within HEC. In the C-terminal region of all Beps, we could define at least one copy of a 142-aa domain called BID. An unconserved, positively charged tail sequence at the C terminus and the proximal BID domain was shown here to represent a bipartite T4S signal that mediates VirB/VirD4-dependent protein transfer into HEC. This finding is in agreement with a requirement of C-terminal sequences for interkingdom transfer of T4S substrates of At and Lp (5, 7, 8).
A hidden Markov model of the BID domain alignment from Bh allowed us to search for other proteins containing a similar domain. A large proportion of the top hits were indeed within putative T4S substrates, including all Bep homologues of bartonellae as well as the conjugative relaxases of plasmid-borne bacterial conjugation systems present in various α-proteobacteria. Conjugative relaxases direct the transfer of plasmid DNA by first cleaving and covalently attaching to one DNA strand, followed by transport of the resulting protein-DNA conjugate by the plasmid-encoded T4S system (23). In this process, the specific interaction between the relaxase and the T4S coupling protein is thought to initiate the transport through the membrane-spanning T4S channel (24). The BID domain has likely evolved in the relaxases of α-proteobacterial conjugation systems before horizontal transfer occurred into a progenitor of Bartonella. A phylogenetic analysis of the T4S coupling proteins (VirD4/TraG) of representative T4S systems indeed revealed that the coupling proteins of T4S systems with a BID domain in their substrate(s) form a distinct cluster. This finding suggests coevolution of the coupling protein and the T4S signal, which is consistent with the finding that coupling proteins and T4S substrates physically interact (24-26). The absence of a BID domain in the substrates of other T4S systems (e.g., of the agrobacterial VirB/VirD4 system, the Hp Cag system, and the Lp Dot/Icm system) suggests that a different signal mediates protein transfer by these T4S systems.
We show that the BID domain and short positively charged C-terminal tail of the conjugative relaxase (TraA) of the At pAtC58 conjugation system AvhB/TraG is functional for mediating VirB/VirD4-dependent protein transfer from Bh into HEC. The T4S signals of these related T4S systems involved either in interbacterial DNA transfer or interkingdom protein transfer are thus interchangeable. This finding makes it tempting to speculate that conjugative relaxases are also transported by the Bh VirB/VirD4 system into HEC when they are covalently attached to their single-stranded DNA substrate, similar to the interkingdom DNA transfer by the At VirB/VirD4 system into plant cells. T4S-mediated DNA transfer from virulence-attenuated Bh in human cells could have important applications for gene therapy and vaccination and should thus be an interesting subject for future investigations.
Supplementary Material
Acknowledgments
We thank P. J. J. Hooykaas for helpful suggestions and H. L. Saenz, C. Thompson, and G. Cornelis for critical reading of the manuscript. This work was supported by Swiss National Science Foundation Grant 3100-061777 (to C.D.).
Author contributions: C.D. designed research; R.S., P.G., T.A.R., M.C.S., G.S., and I.C. performed research; P.G. and C.D. analyzed data; and C.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: T4S, type IV secretion; Bep, Bartonella-translocated effector protein; Hp, Helicobacter pylori; Lp, Legionella pneumophila; At, Agrobacterium tumefaciens; Bh, Bartonella henselae; HEC, human endothelial cell; HUVEC, human umbilical vein endothelial cell; NLS, nuclear localization signal; CRAFT, Cre recombinase reporter assay for translocation; PAI, pathogenicity island; gpc, GFP-positive cells; BID, Bep intracellular delivery.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ556988).
References
- 1.Cascales, E. & Christie, P. J. (2003) Nat. Rev. Microbiol. 1, 137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding, Z., Atmakuri, K. & Christie, P. J. (2003) Trends Microbiol. 11, 527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christie, P. J. (2001) Mol. Microbiol. 40, 294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odenbreit, S., Puls, J., Sedlmaier, B., Gerland, E., Fischer, W. & Haas, R. (2000) Science 287, 1497-1500. [DOI] [PubMed] [Google Scholar]
- 5.Luo, Z. Q. & Isberg, R. R. (2004) Proc. Natl. Acad. Sci. USA 101, 841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagai, H., Kagan, J. C., Zhu, X., Kahn, R. A. & Roy, C. R. (2002) Science 295, 679-682. [DOI] [PubMed] [Google Scholar]
- 7.Vergunst, A. C., van Lier, M. C., den Dulk-Ras, A. & Hooykaas, P. J. (2003) Plant Physiol. 133, 978-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vergunst, A. C., Schrammeijer, B., den Dulk-Ras, A., de Vlaam, C. M., Regensburg-Tuink, T. J. & Hooykaas, P. J. (2000) Science 290, 979-982. [DOI] [PubMed] [Google Scholar]
- 9.Dehio, C. (2003) Curr. Opin. Microbiol. 6, 61-65. [DOI] [PubMed] [Google Scholar]
- 10.Schulein, R. & Dehio, C. (2002) Mol. Microbiol. 46, 1053-1067. [DOI] [PubMed] [Google Scholar]
- 11.Schmid, M. C., Schulein, R., Dehio, M., Denecker, G., Carena, I. & Dehio, C. (2004) Mol. Microbiol. 52, 81-92. [DOI] [PubMed] [Google Scholar]
- 12.Dehio, C., Meyer, M., Berger, J., Schwarz, H. & Lanz, C. (1997) J. Cell Sci. 110, 2141-2154. [DOI] [PubMed] [Google Scholar]
- 13.Kempf, V. A., Schaller, M., Behrendt, S., Volkmann, B., Aepfelbacher, M., Cakman, I. & Autenrieth, I. B. (2000) Cell. Microbiol. 2, 431-441. [DOI] [PubMed] [Google Scholar]
- 14.Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., et al. (2004) Nucleic Acids Res. 32, D115-D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padmalayam, I., Karem, K., Baumstark, B. & Massung, R. (2000) DNA Cell. Biol. 19, 377-382. [DOI] [PubMed] [Google Scholar]
- 16.Alsmark, C. M., Frank, A. C., Karlberg, E. O., Legault, B. A., Ardell, D. H., Canback, B., Eriksson, A. S., Naslund, A. K., Handley, S. A., Huvet, M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 9716-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hacker, J., Hentschel, U. & Dobrindt, U. (2003) Science 301, 790-793. [DOI] [PubMed] [Google Scholar]
- 18.Kawamukai, M., Matsuda, H., Fujii, W., Nishida, T., Izumoto, Y., Himeno, M., Utsumi, R. & Komano, T. (1988) J. Bacteriol. 170, 3864-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stein, M., Rappuoli, R. & Covacci, A. (2000) Proc. Natl. Acad. Sci. USA 97, 1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonnhammer, E. L., Eddy, S. R., Birney, E., Bateman, A. & Durbin, R. (1998) Nucleic Acids Res. 26, 320-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen, L., Chen, Y., Wood, D. W. & Nester, E. W. (2002) J. Bacteriol. 184, 4838-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selbach, M., Moese, S., Hauck, C. R., Meyer, T. F. & Backert, S. (2002) J. Biol. Chem. 277, 6775-6778. [DOI] [PubMed] [Google Scholar]
- 23.Llosa, M., Gomis-Ruth, F. X., Coll, M. & de la Cruz, F. (2002) Mol. Microbiol. 45, 1-8. [DOI] [PubMed] [Google Scholar]
- 24.Schroder, G., Krause, S., Zechner, E. L., Traxler, B., Yeo, H. J., Lurz, R., Waksman, G. & Lanka, E. (2002) J. Bacteriol. 184, 2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szpirer, C. Y., Faelen, M. & Couturier, M. (2000) Mol. Microbiol. 37, 1283-1292. [DOI] [PubMed] [Google Scholar]
- 26.Llosa, M., Zunzunegui, S. & de la Cruz, F. (2003) Proc. Natl. Acad. Sci. USA 100, 10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.