Abstract
The discovery of neuropeptides has resulted in an increased understanding of novel regulatory mechanisms of certain physiological phenomena. Here we identify a novel neuropeptide of 36 amino-acid residues in rat brain as an endogenous ligand for the orphan G protein-coupled receptor FM-4/TGR-1, which was identified to date as the neuromedin U (NMU) receptor, and designate this peptide ‘neuromedin S (NMS)' because it is specifically expressed in the suprachiasmatic nuclei (SCN) of the hypothalamus. NMS shares a C-terminal core structure with NMU. The NMS precursor contains another novel peptide. NMS mRNA is highly expressed in the central nervous system, spleen and testis. In rat brain, NMS expression is restricted to the core of the SCN and has a diurnal peak under light/dark cycling, but remains stable under constant darkness. Intracerebroventricular administration of NMS in rats activates SCN neurons and induces nonphotic type phase shifts in the circadian rhythm of locomotor activity. These findings suggest that NMS in the SCN is implicated in the regulation of circadian rhythms through autocrine and/or paracrine actions.
Keywords: circadian rhythm, endogenous ligand, neuropeptide, orphan G protein-coupled receptor, suprachiasmatic nucleus
Introduction
G protein-coupled receptors (GPCRs) constitute a large protein superfamily, and share the seven-transmembrane motif as common structure. The progress of human genome sequencing has revealed the existence of several hundred orphan GPCRs, for which ligands have not yet been identified (Vassilatis et al, 2003). GPCRs play crucial roles in cell-to-cell communication involved in a variety of physiological phenomena, and are the most common target of pharmaceutical drugs. Therefore, identification of endogenous ligands for orphan GPCRs will lead to clarification of novel regulatory mechanisms of physiological phenomena and to novel drug targets. Recently, many bioactive molecules have been discovered or identified as endogenous ligands of orphan GPCRs using a reverse-pharmacological technique (Civelli et al, 2001).
Neuromedin U (NMU), initially isolated from porcine spinal cord by our group, is a brain-gut neuropeptide that has a potent activity to cause uterine smooth muscle contraction (Minamino et al, 1985). The physiological roles of NMU in the central nervous system (CNS) have been poorly understood, because the NMU receptor in the CNS has not been identified. In contrast, the peripheral activities of NMU have been well characterized; these include contraction of smooth muscle (Minamino et al, 1985), elevation of arterial blood pressure (Minamino et al, 1985), alternation of intestinal ion transport (Brown and Quito, 1988) and control of local blood flow (Sumi et al, 1987; Gardiner et al, 1990). Recently, NMU was identified as an endogenous ligand for two orphan GPCRs, FM-3/GPR66 and FM-4/TGR-1, which were then designated the NMU receptors type-1 (NMU1R) and type-2 (NMU2R), respectively (Fujii et al, 2000; Hedrick et al, 2000; Hosoya et al, 2000; Howard et al, 2000; Kojima et al, 2000; Raddatz et al, 2000). The FM-3/GPR66 mRNA exhibits wide distribution in peripheral tissues. In contrast, the FM-4/TGR-1 mRNA is mainly expressed in the CNS, especially in the hypothalamic paraventricular nucleus (PVN). These reports have provided novel insights into the function of NMU in the CNS. The central NMU suppresses feeding (Howard et al, 2000; Kojima et al, 2000), regulates energy homeostasis (Nakazato et al, 2000; Hanada et al, 2003), affects the cardiovascular system (Chu et al, 2002) and induces the release of stress-mediating molecules (Hanada et al, 2001; Ozaki et al, 2002; Wren et al, 2002). Our detailed analysis by RT–PCR recently demonstrated that the FM-4/TGR-1 mRNA is expressed not only in the PVN but also in the suprachiasmatic nucleus (SCN) of the hypothalamus (Nakahara et al, 2004). However, the functional role of FM-4/TGR-1 in the SCN remains unclear.
The SCN is the site of the master circadian pacemaker in mammals, which governs the circadian rhythm of behavioral and physiological processes. The pacemaker generates the near-24-h period of the circadian rhythm by an autoregulatory transcription/translation feedback loop composed of clock-gene families of transcription factors, and is entrained to the 24-h daily cycle by periodic environmental cues, such as light/dark cycle and temperature, which are typical examples of photic and nonphotic signals, respectively (Lowrey and Takahashi, 2000; Reppert and Weaver, 2001, 2002). The SCN is functionally and anatomically divided into ‘core' and ‘shell' regions, which correspond to the ventrolateral and dorsomedial portions of the SCN, respectively (Moore et al, 2002). The SCN core and shell play roles in circadian entrainment and spontaneous generation of strong rhythm, respectively (Hamada et al, 2001). Several neuropeptides participate in the regulation of the circadian pacemaker within the SCN (Reppert and Weaver, 2001). For example, vasoactive intestinal polypeptide (VIP), which is expressed in the SCN core, plays roles in both photic entrainment and maintenance of the circadian rhythm within the SCN (Piggins and Cutler, 2003). Therefore, the expression of FM-4/TGR-1 within the SCN strongly suggests that the endogenous ligand for its receptor is involved in the circadian oscillator system.
Based on the sequences obtained by a homology search of genomic and expressed sequence tag (EST) databases with known GPCR, our group independently identified FM-4/TGR-1 as an orphan GPCR, and have searched for its endogenous ligand. Here we report the identification of a novel neuropeptide, neuromedin S (NMS), which was purified from rat brain extracts as an endogenous ligand for FM-4/TGR-1 using a reverse-pharmacological technique. The NMS mRNA is specifically and rhythmically expressed in the SCN core in rat brain with a diurnal peak. Intracerebroventricular (ICV) administration of NMS in rats activates SCN neurons and causes nonphotic type phase shifts in the circadian rhythm of locomotor activity. Our data suggest that NMS is a candidate for a nonphotic entrainment factor of circadian rhythm and that NMS acts through its receptor in an autocrine and/or paracrine manner within the SCN.
Results
Purification of NMS
To identify endogenous ligands for orphan GPCRs, we have purified natural peptides from tissue extracts using a reverse-pharmacological technique (Kojima et al, 1999, 2000). While searching for endogenous ligands of the orphan GPCR FM-4/TGR-1 (identified to date as NMU2R), gel filtration of rat brain extracts revealed two agonist activities capable of increasing intracellular calcium ion concentration ([Ca2+]i) in CHO cells expressing FM-4/TGR-1 (Figure 1A, P1 and P2). P2 activity was eluted in fractions with a relative molecular mass (Mr) of 2500, corresponding to the Mr of rat NMU (2642.9), and NMU was purified from these fractions (data not shown). In contrast, P1 activity was found at a larger Mr (∼4000) by gel filtration chromatography and was more hydrophobic by reverse-phase HPLC (RP-HPLC) compared to P2 activity, indicating that the rat brain contains another endogenous ligand for FM-4/TGR-1. Therefore, P1 activity was purified to homogeneity by successive HPLC steps (Figure 1B, upper panel). The final yield of the purified peptide was approximately 2.5 pmol from 420 rat brains (510 g).
Figure 1.
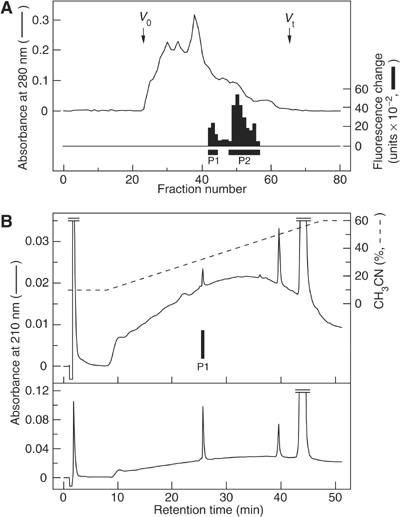
Purification of NMS. (A) Gel filtration on a Sephadex G-50 (fine) column of the SP-III fraction from 510 g of rat brain. Black bars indicate fluorescence change due to [Ca2+]i increase in CHO/FM-4. Active fractions containing NMS (P1) and NMU (P2) are indicated. V0, void volume; Vt, total volume. (B) Chromatographic comparison using RP-HPLC of natural NMS (upper panel) and synthetic NMS (lower panel). The black bar indicates [Ca2+]i -increasing activity (upper panel). Each peptide was applied to a Chemcosorb 3ODS-H column. The flow rate was 0.2 ml/min. Solvent system, a linear gradient elution from (A) to (B) for 40 min. H2O:CH3CN:10% TFA for (A) was 90:10:1, for (B) 40:60:1 (v/v).
Structure determination of NMS
The partial N-terminal amino-acid sequence of the purified peptide was determined by a protein sequencer to be LPRLLHTDSRMATIDFPKK. To elucidate the complete amino-acid sequence of this peptide, human, rat and mouse cDNAs encoding the purified peptide were isolated from the brain of each species by PCR. The rat cDNA encoded a 152-residue protein containing features characteristic of an N-terminal signal peptide (von Heijne, 1986) (Figure 2A). This preproprotein contained four potential processing sites to be cleaved by subtilisin-like proprotein convertases (Rouille et al, 1995). The N-terminal sequence of the purified peptide directly followed the third processing site, indicating that the purified peptide is generated by cleavage at the third and fourth sites. The fourth processing site contained Gly 145, which presumably serves as an amide donor for C-terminal amidation (Rouille et al, 1995). All characteristic structures described above were conserved in the preproproteins of human and mouse. We therefore deduced the primary structure of the newly purified peptide to be LPRLLHTDSRMATIDFPKKDPTTSLGRPFFLFRPR N-NH2, and designated this 36-residue peptide NMS. Mass spectrometric analysis revealed that the observed monoisotopic m/z value of the purified peptide (4240.2) was very close to the theoretically predicted value for rat NMS (4240.3). Moreover, the synthetic NMS had an identical retention time to the natural peptide on RP-HPLC (Figure 1B), suggesting that the deduced primary structure is identical to that of the natural peptide. The N-terminal portion of NMS has no sequence homology to previously known peptides or proteins. On the other hand, the C-terminal amidated seven-residue sequence of NMS is identical to that of NMU (Minamino et al, 1985) (Figure 2B); this sequence has been found to be essential for NMU receptor binding (Minamino et al, 1985). NMS is not a splice variant of NMU, because the human NMS and NMU genes were mapped to chromosomes 2q11.2 and 4q12, respectively, by a blast search of the genome database. The structural organization and putative promoter region of the NMS gene were also analyzed computationally (see Supplementary data and Supplementary Figure S1). The four proteolytic processing sites described above in the NMS proprotein are conserved in the NMU proprotein (Lo et al, 1992) (Figure 2C), and the amino-acid sequences between the first and second processing sites show high homology to each other, indicating the existence of other novel 34- and 33-residue neuropeptides produced from the NMS and NMU proproteins, respectively, by processing at the first and second sites.
Figure 2.
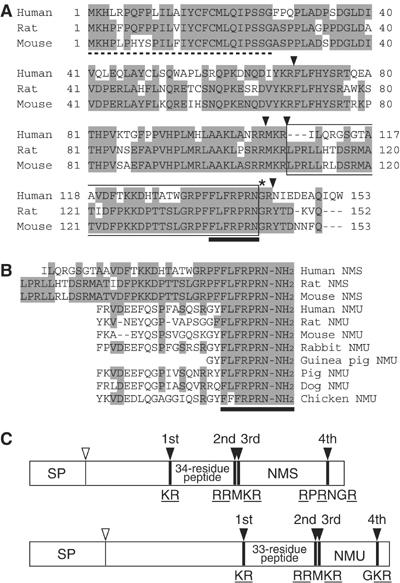
Structure of NMS. (A) Amino-acid sequences of human, rat and mouse prepro-NMS. Identical residues are shaded. The dotted line denotes the predicted signal peptide. The arrowheads indicate proteolytic processing sites. The asterisk shows a glycine residue, which serves as an amide donor for C-terminal amidation. NMS sequences are boxed. Sequences conserved between NMS and NMU are indicated by a solid underline. The sequence data for the human, rat and mouse NMS cDNAs have been submitted to the DDBJ/EMBL/GenBank databases under accession nos. AB164464, AB164465 and AB164466, respectively. (B) Sequence comparison of NMS and NMU. Human, rat and mouse NMS and NMU sequences are aligned. Residues identical between peptides are shaded. Conserved core sequences are indicated by a solid underline. (C) Schematic representation of the preproproteins of rat NMS and NMU. The preproproteins of NMS and NMU are represented by boxes divided into protein domain, proportional to their length. The open and filled arrowheads indicate cleavage sites by signal peptidase and proprotein convertase, respectively. The sequences of the proteolytic processing sites are shown. The basic amino-acid residues recognized by proprotein convertase are underlined. SP, signal peptide.
Pharmacological characterization of NMS
Because NMS and NMU share a core structure that is required for binding to receptors, the interaction of NMS with FM-3/GPR66 and FM-4/TGR-1 was examined using synthetic peptides. Human NMS induced dose-dependent, robust increases in [Ca2+]i in CHO cells expressing either FM-3/GPR66 or FM-4/TGR-1 (CHO/FM-3 or CHO/FM-4, respectively), with half-maximal response concentrations (EC50) of 6.5 × 10−11 and 9.1 × 10−11 M, respectively (Figure 3A and B). Rat NMS and NMU, and human NMU also showed comparable potency and efficacy for these receptors. Neither NMS nor NMU induced a response in CHO cells transfected with vector alone (data not shown). Competitive radioligand binding analysis showed high-affinity binding of NMS to receptors. Binding of [125I-Tyr0]-human NMS to FM-3/GPR66 was qinhibited in a dose-dependent manner by human NMS and NMU, with similar inhibition constants (IC50) of 1.2 × 10−9 M (Figure 3C). On the other hand, human NMS had a higher binding affinity to FM-4/TGR-1 than did NMU (IC50=1.0 × 10−10 and 6.8 × 10−10 M, respectively) (Figure 3D). These data indicate that NMS, as well as NMU, is a cognate ligand for both FM-3/GPR66 and FM-4/TGR-1.
Figure 3.
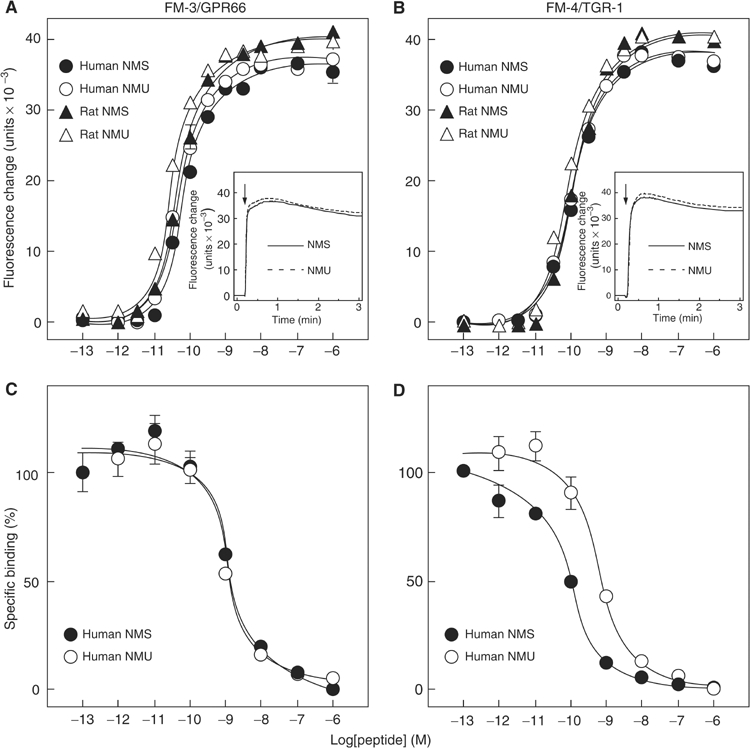
Pharmacological characterization of synthetic NMS using human FM-3/GPR66 and FM-4/TGR-1 stably expressed in CHO cells. (A, B) Dose-response relationships of [Ca2+]i change for human NMS (filled circle), human NMU (open circle), rat NMS (filled triangle) and rat NMU (open triangle) in CHO/FM-3 (A) and CHO/FM-4 (B) cells. Data points are means±s.e.m. of triplicates for each experiment. Insets show the time course of [Ca2+]i changes induced by human NMS (solid line) and human NMU (dotted line). Each peptide (10−8 M) was added at the time indicated by the arrow. (C, D) Competitive radioligand binding analysis. [125I-Tyr0]-human NMS binding to FM-3/GPR66 (C) and FM-4/TGR-1 (D) was displaced by increasing concentrations of human NMS (filled circle) and human NMU (open circle). Data were determined in triplicate.
In addition, the activities of NMS and NMU were compared in chick rectum contraction and rat systemic blood pressure assays. NMS induced contraction of chick rectum and elevation of rat systemic blood pressure with activities similar to NMU (see Supplementary Figure S2).
Expression analysis of NMS
We investigated the tissue distribution of the NMS mRNA in various rat tissues using quantitative RT–PCR. The NMS mRNA was expressed mainly in the CNS, spleen and testis (Figure 4A). The highest level of NMS gene expression was found in the hypothalamus. Interestingly, the NMS mRNA was expressed predominantly in the SCN, with only very slight expression in other brain regions (Figure 4B). In situ hybridization histochemistry also showed that the NMS mRNA expression was restricted to the SCN in the rat brain (Figure 4C). No hybridization signal was observed in other brain regions. In the rat, the SCN is divided into core, where the neuropeptide VIP is expressed, and shell, where the neuropeptide arginine vasopressin (AVP) is expressed (Moore et al, 2002). The NMS mRNA was expressed in the SCN core (Figure 4D–F), in a similar manner to the VIP mRNA. This unique expression pattern of NMS in the brain and peripheral tissues led us to speculate that NMS may play important roles in tissue-specific physiological functions, such as circadian rhythm regulation in the SCN, immune response in the spleen and spermatogenesis in the testis. We thus examined the possible role of NMS in the circadian oscillation systems of the SCN.
Figure 4.
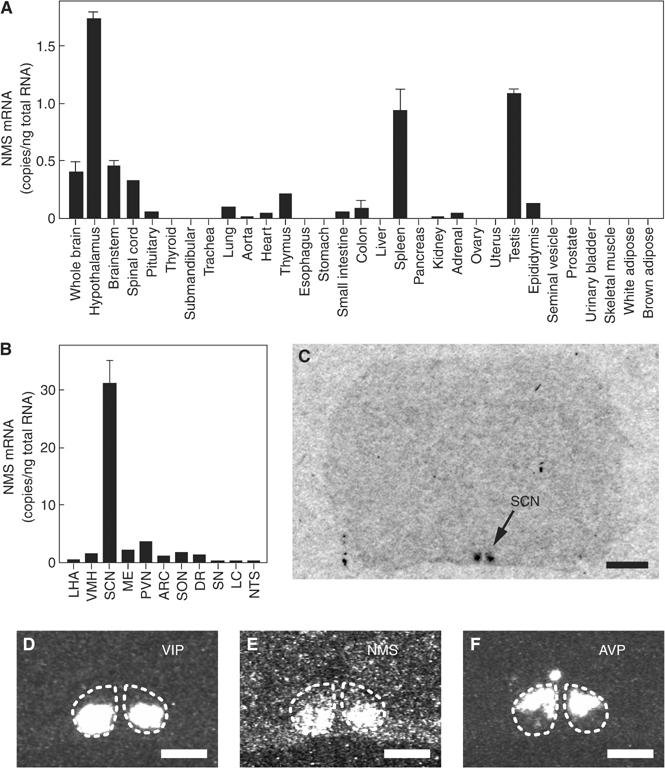
Expression studies of rat NMS. (A, B) Quantitative RT–PCR analysis of the NMS mRNA in a rat multiple-tissue cDNA panel. Each column represents the mean±s.e.m. of triplicate experiments. LHA, lateral hypothalamic area; VMH, ventromedial hypothalamus; SCN, suprachiasmatic nucleus; ME, median eminence; PVN, paraventricular nucleus; ARC, arcuate nucleus; SON, supraoptic nucleus; DR, dorsal raphe; SN, substantia nigra; LC, locus coeruleus; NTS, nucleus of the solitary tract. (C) Autoradiogram of the NMS mRNA expression in a coronal section of the rat brain. Scale bar, 2 mm. (D–F) Distribution of VIP (D), NMS (E) and AVP (F) mRNA in the rat SCN. Serial sections were used. The SCN is indicated with a dotted line. Scale bar, 500 μm.
Expression pattern of NMS within the SCN
We examined the time-dependent profile of the NMS mRNA expression in the rat SCN. Under 12-h light/dark cycles, a pronounced rhythmic expression of the NMS mRNA was observed (ANOVA, F(3,10)=11.287, P<0.005), with the largest decrease occurring during the dark period, followed by a gradual increase until the late light period at ZT11 (Zeitgeber time, ZT; ZT0 is lights on and ZT12 is lights off) (Figure 5A). Under conditions of constant darkness, no NMS mRNA oscillation was observed (ANOVA, F(3,11)=1.170, P>0.3) (Figure 5B). These data indicate that the expression of NMS is not under the control of clock-gene families that generate circadian rhythm by an autoregulatory transcription/translation feedback loop (Lowrey and Takahashi, 2000; Reppert and Weaver, 2001, 2002). We next examined the NMS mRNA level within the SCN in response to a light pulse under conditions of constant darkness. The expression of the NMS mRNA was significantly decreased 1 h after the light exposure at CT6 (circadian time, CT; CT0–12 is subjective day and CT12–24 is night) (Figure 5C). We also assessed the effect of a dark pulse on NMS expression within the SCN of rats maintained under light/dark cycling. When rats were exposed to a dark pulse at ZT6, the NMS mRNA level tended to be suppressed (Figure 5D).
Figure 5.
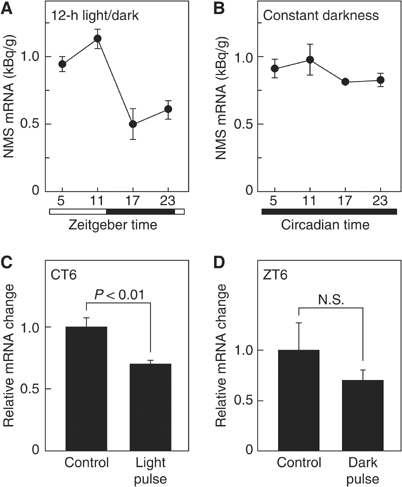
Expression pattern of rat NMS within the SCN. (A, B) Temporal expression profiles of the NMS mRNA. Animals were maintained under 12-h light/dark cycles (A) or constant darkness for 2 days (B). The amount of NMS mRNA was quantified by in situ hybridization analysis. The experiments were performed side by side. Data represent the means±s.e.m. of 3–4 animals. Open and filled horizontal bars indicate light and dark periods, respectively. (C, D) Responses of NMS expression to light exposure under conditions of constant darkness and to a dark pulse during the light period of a 12-h light/dark cycle. Animals maintained in constant darkness for 2 days were exposed to a light pulse for 30 min at CT6 (C), or animals maintained under 12-h light/dark cycles were exposed to a dark pulse for 30 min at ZT6 (D). Brain samples were collected 1 h after exposure to the light or dark pulse. Data are presented as the means±s.e.m. of 3–4 animals, and are shown as relative changes in NMS mRNA levels.
Intracerebroventricularly administered NMS induces phase shifts in the circadian rhythm of locomotor activity
In the SCN, two types of receptors for NMS were expressed, although the FM-4/TGR-1 mRNA was more highly expressed than the FM-3/GPR66 mRNA (23.78±2.61 and 0.27±0.006 copies/ng of total RNA, respectively). We therefore examined the effect of ICV administration of NMS on the circadian rhythm of locomotor activity in rats maintained under constant darkness. When injected at CT3–9, 1 nmol of rat NMS phase-advanced the rhythm of locomotor activity (Figure 6A and E), whereas administration at CT22–24 induced a phase delay (Figure 6D and E). No phase-shifting effect was observed during either early or middle subjective night (Figure 6B, C and E). The phase-response curve (PRC) for NMS fluctuated significantly (ANOVA, F(14,28)=2.549, P<0.05). These effects were dependent on both phase and dose (Figure 6E and F). Saline administration did not elicit a phase shift under the same conditions (data not shown). Circadian period length was not influenced by ICV administration of NMS at any time. In addition, after ICV administration of NMS, the expression of c-Fos protein, a marker of neuronal activation, was increased within the SCN compared with saline-administered control rats (Figure 6G and H). The c-Fos protein was preferentially expressed on the core of the SCN. To further test the effects of NMS on SCN function, NMS was microinjected directly into the SCN. Intranuclearly administered NMS also induced phase advance and delay in the circadian rhythm of locomotor activity at CT6 and CT23, respectively (see Supplementary Figure S3). Saline administration had no effect. Taken together, these data suggest that endogenous NMS acts through its receptors to have a strong effect on SCN function.
Figure 6.
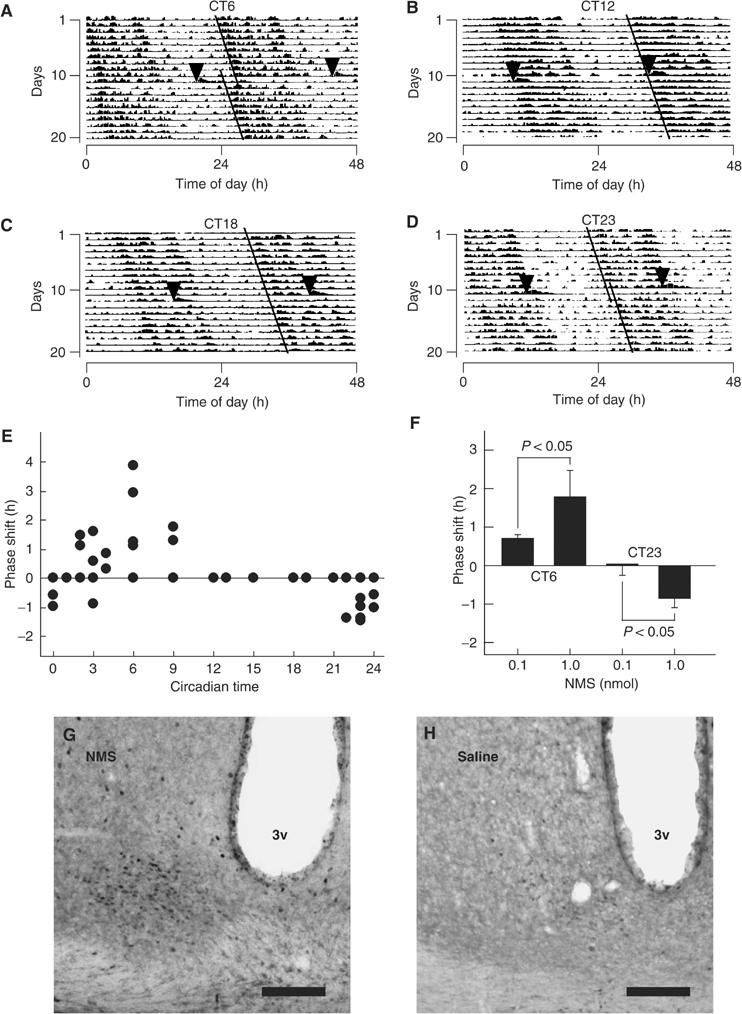
In vivo experiments with rat NMS. (A–D) Phase shifts of the circadian rhythm of locomotor activity induced by ICV administration of NMS. Representative doubled-plotted actograms of locomotor activity are shown. Rats were ICV administered with rat NMS (1 nmol) at CT6 (A), CT12 (B), CT18 (C) and CT23 (D). Each arrowhead indicates the time of NMS administration. Each line shows the regression lines drawn based on the daily onset of locomotor activity before and after administration. (E) Phase-response plot for ICV administration of 1 nmol NMS. The plus and minus values indicate phase advance and delay, respectively. n=3–5. The data points at CT12, 13, 15, 18 and 19 overlap. (F) Dependence of circadian rhythm phase shift on NMS administration. ICV administration of NMS at CT6 and CT23 induced significant phase advance and delay, respectively, in a dose-dependent manner. n=4–5 per group. (G, H) Induction of c-Fos protein expression within the SCN following ICV administration of NMS. Rats were administered at CT6 with 1 nmol NMS (G) or saline (H). 3v, third ventricle. Scale bar, 150 μm.
Discussion
In this study, we have purified a novel neuropeptide, NMS, as an endogenous ligand for the orphan GPCR FM-4/TGR-1 (designated to date as NMU2R). Pharmacological characterization using recombinant receptors showed that NMS is a cognate ligand for both FM-3/GPR66 and FM-4/TGR-1, indicating that NMS shares these receptors with NMU, which is a brain-gut neuropeptide originally isolated from porcine spinal cord by our group (Minamino et al, 1985). NMS shares a C-terminal core structure with NMU. The nucleotide sequences of the NMS and NMU cDNAs show low homology (53%) to each other. Genome database searches showed that, similar to the NMU gene (GenBank accession numbers AF222755–AF222765), the NMS gene is composed of 10 exons and the exon–intron boundaries in the NMS proprotein are comparably conserved with those in the NMU proprotein. Moreover, the domain structures of both proproteins are also similar, because the proteolytic processing sites are conserved. The structural similarities of NMS and NMU on the gene and proprotein levels were observed among mouse, rat and human, indicating that the divergence of the two genes encoding these peptides preceded the rodent/primate split during the process of evolution. In addition, the NMS proprotein contains another novel 34-residue peptide, similar to the novel 33-residue peptide that Lo et al (1992) speculated to exist in the NMU proprotein. Indeed, we purified such an endogenous 33-residue peptide derived from the NMU proprotein from rat brain and small intestine extracts. Both synthetic 33- and 34-residue peptides derived from the NMU and NMS proproteins show potent prolactin-releasing activity upon ICV administration in rats (Mori et al, in preparation).
Pharmacological characterization using CHO cells expressing receptors indicated that NMS and NMU share quite similar potency and efficacy for both recombinant FM-3/GPR66 and FM-4/TGR-1; these results were confirmed with another expression system using HEK293 cells (data not shown). However, it is interesting that ICV-administered NMS induced more potent phase shifting of circadian rhythm and suppression of feeding than ICV-administered NMU (Nakahara et al, 2004; Ida et al, in preparation), whereas both peptides showed similar effects on the contractile activities of chick rectum, and on elevation of systemic blood pressure. These results indicate that NMS showed more potent activity than NMU in the CNS but not in peripheral tissues. The pharmacological effects of NMS and NMU were different in the CNS, whereas the two peptides showed the same activity in peripheral tissues and in in vitro experiments. The molecular mechanisms of this difference remain unclear. The functional activity of receptors in the CNS may be modulated by unknown modifying molecules, such as receptor-activity-modifying proteins (McLatchie et al, 1998).
Because NMU is an anorexigenic neuropeptide involved in the central regulation of feeding behavior (Howard et al, 2000; Kojima et al, 2000; Hanada et al, 2004), NMS may also play a role in feeding regulation. However, unlike NMU, the NMS mRNA is expressed at only a low level in the PVN and arcuate nuclei, which are closely implicated in the regulation of this process (Schwartz et al, 2000). Further investigation of the physiological significance of NMS in the regulation of feeding behavior is needed, because the NMS gene was mapped to chromosome 2q11.2 in human, and this locus is consistent with one potential location of quantitative trait loci implicated in obesity (Barsh et al, 2000).
It has been well established that the circadian rhythm is generated by an endogenous pacemaker located in the SCN, and that precise, rhythmic oscillation of this pacemaker is coordinated by various neurochemical substances (Lowrey and Takahashi, 2000; Reppert and Weaver, 2001, 2002). The data obtained in this study strongly suggest that NMS functions as a regulator of the circadian pacemaker in an autocrine and/or paracrine manner within the SCN. The circadian rhythm can be phase shifted by photic and nonphotic stimuli, both of which contribute to important mechanisms in the entrainment of the circadian pacemaker (Mrosovsky, 1996; Lowrey and Takahashi, 2000). The SCN core receives photic and nonphotic environmental signals from the retina, the intergeniculate leaflet (IGL) of the lateral geniculate nuclei (LGN) and the median raphe of the brainstem, and consequently plays a crucial role in circadian entrainment (Reppert and Weaver, 2001). Several neuropeptides are implicated in the mechanism of circadian entrainment. For example, pituitary adenylate cyclase-activating polypeptide conveys photic signal via the retinohypothalamic tract (Hannibal, 2002). Neuropeptide Y in the geniculohypothalamic tract plays a role in mediating nonphotic signal from the IGL (Harrington, 1997). Neuropeptides intrinsic to the SCN also regulate the phase of the circadian rhythm. The VIP–VPAC2 receptor system is important for photic entrainment and maintenance of the circadian rhythm (Piggins and Cutler, 2003). VPAC2 receptor-deficient mice are incapable of sustaining normal circadian rhythm of rest/activity behavior (Harmar et al, 2002). Loss of VIP/peptide histidine isoleucine in mice is sufficient to produce severe disruptions in the generation of circadian oscillations and the synchronization of these rhythms to the environment (Colwell et al, 2003). VIP is expressed in the SCN core and is released into the SCN shell to entrain the pacemaker. Similar to light pulses, application of VIP into the SCN results in a phase shift of the circadian rhythm during subjective night in vivo and in vitro (Piggins and Cutler, 2003). On the other hand, a neurotransmitter γ-aminobutylic acid, located in the SCN, LGN and IGL, is involved in nonphotic entrainment of the circadian rhythm (Turek and Van Reeth, 1988; Harrington, 1997). However, no SCN-intrinsic neuropeptides involved in this process have yet been identified. The expression of NMS in the SCN core suggests its involvement in circadian entrainment. In contrast to VIP, NMS induced phase shifts of the circadian rhythm of locomotor activity when tested during subjective day, and the PRC for NMS is very similar to that for nonphotic stimuli (Mrosovsky, 1996). These data suggest that NMS is a candidate for a nonphotic entrainment factor intrinsic to the SCN. Moreover, NMS, as well as VIP, may be involved in maintenance of the circadian rhythm.
The expression of NMS mRNA fluctuated within the SCN under light/dark cycling, but not under conditions of constant darkness. The expression was high during daytime and low at night, indicating that NMS expression is influenced by the light conditions. However, the NMS mRNA oscillation under light/dark cycling is not simply due to an enhancement of NMS gene expression by light, because the peak level of NMS mRNA during the light period is approximately the same as the levels throughout the day under conditions of constant darkness. Moreover, the NMS mRNA levels under constant darkness are not increased by light exposure at CT6, whereas the expression level is high during ZT5–11. Therefore, NMS expression is likely suppressed during the dark period of the light/dark cycle; this is consistent with the fact that a dark pulse at ZT6 tended to suppress the expression of NMS mRNA. It is well known that a cAMP responsive element (CRE) in the promoter regions of light responsive genes plays a crucial role in induction of gene expression by light in the SCN (Reppert and Weaver, 2001). The phosphorylation of CRE binding protein (CREB) at Ser 133 is required to activate the transcription of genes containing CRE (Gonzalez and Montminy, 1989). The light exposure induces CREB phosphorylation in the SCN during subjective night (CT19), but not during subjective day (CT6), suggesting that the components of the circadian pacemaker that gate light-sensitive molecular responsiveness in the SCN may act upstream of phosphorylation of CREB (Ginty et al, 1993). Computational analysis did not identify a CRE within the putative promoter region of the NMS gene (see Supplementary data). Therefore, it is natural that photic induction of the NMS expression at CT6 is not observed in the SCN. These data suggest that the expression of the NMS mRNA within the SCN is regulated indirectly by light under light/dark cycling. In addition, no CACGTG E-box element is found in the promoter region of the NMS gene (see Supplementary data). This element is closely involved in intrinsic circadian rhythmicity of gene expression (Reppert and Weaver, 2001). Therefore, the absence of a CACGTG E-box is consistent with the observation that no NMS mRNA oscillation is found under conditions of constant darkness, and supports the suggestion that the expression of NMS is not under the control of clock-gene families. As described above, NMS expression within the SCN shows a unique profile in response to light conditions. In particular, the NMS mRNA levels are suppressed by a light pulse at CT6. At present, it is not clear how the expression of the NMS mRNA is transcriptionally regulated in the SCN. The elucidation of this mechanism may provide novel insights into the regulatory mechanism of circadian oscillator systems.
Recently, our group reported that NMU is involved in the circadian oscillator system (Nakahara et al, 2004). Because NMS induces a circadian phase shift through same receptor with NMU within the SCN, it is no wonder that ICV administration of NMU also induces phase shifts. However, NMU is expressed in many discrete nuclei of the hypothalamus and the caudal brainstem (Howard et al, 2000), whereas NMS expression is restricted to the SCN. Furthermore, the magnitude of the phase shift induced by NMU is smaller than that caused by NMS. These facts suggest that NMS is a more specific and effective regulator of the circadian oscillator system than NMU.
The shapes of the NMS and NMU PRCs were subtly different. The magnitude of phase advance induced by NMS was larger than the magnitude of phase delay. In contrast, when NMU was ICV injected, phase delay was stronger than phase advance (Nakahara et al, 2004). Moreover, the maximum phase delays were caused by NMS and NMU when injected at CT23 and CT0, respectively, while the maximum phase advances were elicited by these peptides when injected at the same time (CT6). Two types of receptors for NMS and NMU are expressed in the SCN. The FM-4/TGR-1 mRNA has a nocturnal peak in its expression rhythm, whereas the expression of the FM-3/GPR66 mRNA peaks during daytime (Nakahara et al, 2004). Moreover, the receptor activities to the respective peptides may be modulated in the CNS as described above. Therefore, the difference in the PRC shapes for NMS and NMU may be due to the timing of receptor expression and different affinities of the receptors to the respective peptides in the CNS. On the other hand, the expression levels of NMS and NMU in the SCN are high during daytime and low at night. These rhythms are similar to the rhythm of FM-3/GPR66, and opposite to the rhythm of FM-4/TGR-1. Therefore, it is important to determine the endogenous peptide/receptor pairs and their efficiencies to elucidate the functional roles of NMS and NMU in the circadian oscillator system of the SCN. However, this is extremely difficult, because there are currently very few tools, such as specific antagonists, for analysis. This problem will be solved by engineering mice deficient for NMS, NMU or their receptors.
Light- and photic-related stimuli strongly elicit the expression of c-Fos protein in the SCN with a photic type phase shift (Castel et al, 1997). In contrast, c-Fos protein expression is not associated with phase shifts induced by nonphotic stimuli and related-substances, such as triazolam (Zhang et al, 1993). However, in our previous study (Nakahara et al, 2004) and in this study, ICV-administered NMS and NMU induced c-Fos expression in the SCN core, even though these peptides cause a nonphotic type phase shift. These data suggest that NMS and/or NMU may also be involved in SCN functions other than nonphotic entrainment of the circadian rhythm. On the other hand, there is a report that nonphotic manipulations induce the expression of c-Fos protein in rat SCN (Edelstein and Amir, 1995). Therefore, further analysis is necessary to elucidate the significance of c-Fos expression in the SCN induced by ICV-administered NMS and NMU in the SCN.
Our group has previously demonstrated the specific expression of NMU in the SCN core (Nakahara et al, 2004). In contrast, Graham et al (2003) reported that the NMU mRNA is expressed in the shell of the SCN by in situ hybridization histochemistry. This disagrees with our results. We analyzed the distribution of NMU by immunohistochemistry using an antibody against rat NMU. Because this antibody was raised against the C-terminal amidated seven-residue structure of rat NMU, which is completely conserved in rat NMS (Honzawa et al, 1990), it is able to equally recognize both NMU and NMS, suggesting that the immunoreactivity we observed in the SCN core probably reflects the distribution of NMS. Therefore, we accept the specific expression of NMU in the SCN shell, although further analysis of the distribution of these peptides within the SCN is needed.
NMS and NMU probably play distinct roles in the circadian oscillator system. NMS may be involved in the regulation of circadian rhythm as described above, because the expression of the NMS mRNA in the core region of the SCN is not regulated by clock-gene families, indicating that NMS functions upstream of the endogenous circadian pacemaker. On the other hand, the expression of the NMU mRNA shows a circadian rhythm in the SCN shell of rats maintained under constant darkness (Graham et al, 2003; Nakahara et al, 2004), indicating that NMU, unlike NMS, is controlled by the endogenous pacemaker. Therefore, NMU may act either as a part of a central clock mechanism or as an output signal from the SCN, such as transforming growth factor-α (Kramer et al, 2001) and prokineticin 2 (Cheng et al, 2002). This speculation is supported by the phenotype of some NMU-deficient mice that have shown arrhythmicity in feeding behavior and locomotor activity (Hanada et al, 2004; Kojima et al, in preparation).
In conclusion, we have identified a novel 36-residue neuropeptide NMS, which was purified from rat brain extracts. The restricted expression of NMS in the SCN core and the ability of NMS to shift the phase of the circadian rhythm demonstrate that this newly identified peptide is important for the regulation of circadian rhythm. NMS is a candidate for a nonphotic entrainment factor of circadian rhythm. Our identification and characterization of NMS provide a foundation on which further research can build to provide novel insights into the regulatory mechanism of circadian rhythm.
Materials and methods
Establishment of cells expressing orphan GPCRs and assay system
Full-length cDNAs of human FM-3/GPR66 (GenBank accession number BC036543; residues 209–1420) and FM-4/TGR-1 (AF242874; residues 1–1240) were cloned into pcDNA3.1 vectors (Invitrogen) and transfected into CHO cells. The stably expressing cell lines (CHO/FM-3-14 and CHO/FM-4-16) that showed the highest change in [Ca2+]i induced by human NMU were used in this study. The calcium-mobilization assay was carried out using the FLIPR system (Molecular Device) as described previously (Kojima et al, 1999). The test samples were dissolved in assay buffer containing 1% bovine serum albumin (BSA).
Purification of NMS
The peptide fraction (SP-III) was extracted from 420 rat brains (510 g) as described previously (Kojima et al, 1999), and then fractionated on a Sephadex G-50 gel filtration column (2.9 × 145 cm; Amersham Biosciences). A portion of each fraction equivalent to 1 g of tissue was subjected to assay using CHO/FM-4 cells. The active fraction was separated by carboxymethyl ion exchange HPLC on a TSK-gel CM-2SW column (4.6 × 250 mm; Tosoh) at pH 6.4. The activity was further purified by fractionation on the same column at pH 4.8. Finally, a single active peak was purified by successive RP-HPLCs on a Symmetry 300 C18 column (3.9 × 150 mm; Waters), a diphenyl column (2.1 × 150 mm, 219TP5215; Vydac) and a Chemcosorb 3ODS-H column (2.1 × 75 mm; Chemco). The primary structure was determined using a protein sequencer (494cLC; Applied Biosystems). The correct mass of the purified peptide was verified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using a Voyager-DE PRO (Applied Biosystems).
Cloning of mouse, rat and human prepro-NMS cDNA
A tblastn search of the GenBank database was performed using the partial primary sequence of the purified peptide, and one mouse EST sequence (GenBank accession number BQ175462) was obtained. Based on this sequence, a full-length cDNA encoding mouse prepro-NMS was cloned from mouse hypothalamic poly(A)+ RNA by RNA ligase-mediated rapid amplification of cDNA ends (RACE) using a GeneRacer kit (Invitrogen). The entire coding region of the cDNA was also amplified by nested PCR. The GenBank database was searched again using the mouse cDNA sequence, and the genomic sequences for the human and rat (AC068538 and AC096486, respectively) homologs were obtained. The entire coding regions of the human and rat cDNAs were amplified by nested PCR using human and rat brain Marathon Ready cDNAs (Clontech). All PCR reactions were conducted using the high-fidelity Pyrobest DNA polymerase (TaKaRa). The cDNA sequences were determined from six independent clones and were all completely identical to one another. The primer sequences used for cDNA cloning are listed in Tables I and II.
Table 1.
Primer sequences used in the cloning of mouse NMS cDNA by RACE
| Name | Sequence |
|---|---|
| 5′-RACE first | 5′-GGAAAAATGGCCTCCCCAGG-3′ |
| 5′-RACE nested | 5′-CCCAGGCTGGTAGTAGGATC-3′ |
| 3′-RACE first | 5′-GGGAAATGCTCATCACCTCTCTGAACAG-3′ |
| 3′-RACE nested | 5′-TCTGTGGTCTGCAAAGAGAATCCAGAGC-3′ |
Table 2.
Primer sequences used in the cloning of mouse, rat and human NMS cDNAs by nested PCR
| Species | PCR | Orientation | Sequence |
|---|---|---|---|
| Mouse | First | (+) | 5′-TGCTCATCACCTCTCTGAACAG-3′ |
| (−) | 5′-GCATGGGACAACAGAAATATGGC-3′ | ||
| Nested | (+) | 5′-CTGTGGTCTGCAAAGAGAATCC-3′ | |
| (−) | 5′-CTCCAAAGGCGCACACCGTCTG-3′ | ||
| Rat | First | (+) | 5′-CTCTGCCTCTGGACCCTCG-3′ |
| (−) | 5′-GCGTGGGACAGCAGAAATATGGT-3′ | ||
| Nested | (+) | 5′-CTCATCTGTGGTCTGCAAAGAG-3′ | |
| (−) | 5′-CTCCAAAGATGCACACTGTCTT-3′ | ||
| Human | First | (+) | 5′-TGTCCTAGCAGCACCTGTCTGTG-3′ |
| (−) | 5′-GAAGGCCTCATCAGATCCAG-3′ | ||
| Nested | (+) | 5′-TGGCCAGCAAGGAGAAACCAGAC-3′ | |
| |
|
(−) |
5′-CAGATCCAGCTTTCTTTCACC-3′ |
| (+), sense; (−), antisense. |
Receptor binding assay
Binding analysis was performed as described previously with slight modification (Gottschall et al, 1990). Plasma membranes isolated from CHO/FM-3 and CHO/FM-4 cells were incubated with 50 pM [125I-Tyr0]-human NMS and increasing concentrations of competitive peptide at room temperature for 2 h in binding buffer containing 0.05% CHAPS and 1% BSA. Bound and free ligands were separated by filtration, and the radioactivity was measured using a TopCount liquid scintillation counter (Packard).
Quantitative RT–PCR
RNA was isolated from 8-week-old Wistar rats, and various brain regions were dissected as reported previously (Murakami and Takahashi, 1983). Quantitative RT–PCR was conducted with a LightCycler system (Roche) using a LightCycler-FastStart DNA Master SYBR Green I kit (Roche). The primer set used for rat NMS was 5′-CTCATCTGTGGTCTGCAAAGAG-3′ and 5′-GCATACAGAAGCAGTAGATGAC-3′. Known amounts of rat NMS cDNA were used to obtain a standard curve. Rat GAPDH mRNA was also measured as an internal control.
In situ hybridization histochemistry
In situ hybridization histochemistry was carried out on 12 μm coronal sections of whole rat brains as described previously (Ozaki et al, 2002). The following 35S-labeled antisense oligonucleotides were used: 5′-TGAGGAGGGGATCTGTAGCATACAGAAGCA-3′ for NMS; 5′-GTGTCGTTTGACCGGCACGGGGTCTTCCG-3′ for VIP; and 5′-CAGCTCCCGGGCTGGCCCGTCCAGCT-3′ for AVP. Hybridized sections were apposed to autoradiography film. The autoradiographic images were quantified by an MCID imaging analyzer (Imaging Research) using a 14C standard. Data were normalized with respect to the differences between signal intensities in equivalent areas of the SCN. The time-dependent profiles of expression were analyzed using one-way ANOVA followed by Scheffe's multiple comparisons test. The effects of light and dark pulses on expression levels were evaluated using one-way ANOVA with the post hoc Fisher's test.
Recording of rhythm of locomotor activity
Male Wistar rats (300–350 g) were used in this study. ICV administration was performed as described previously (Ida et al, 1999). Locomotor activity was measured as described previously (Nakahara et al, 2004). The phase shift in the rhythm of locomotor activity was calculated based on the distance between the two regression lines representing the daily onset of locomotor activity for at least 10 days before and after ICV administration of NMS or saline. The onset of locomotor activity was set as CT12. Phase-response data were evaluated using one-way ANOVA followed by Scheffe's multiple comparisons test. Dose-dependent effects were evaluated using one-way ANOVA with the post hoc Fisher's test.
Immunostaining for c-Fos protein
Rats were maintained under constant darkness for 2 weeks, and ICV administration of 1 nmol NMS or saline was performed at CT6. At 90 min after administration, rats were perfused with 2% paraformaldehyde. Immunostaining for c-Fos protein was performed as described previously (Date et al, 1999).
Supplementary Material
Supplementary Data
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Table SI
Supplementary Table SII
Supplementary Table SIII
Acknowledgments
We thank JT Pearson for editing the manuscript, and H Kuwahara, Y Egi, H Okumura and M Miyazaki for technical assistance. This work was supported in part by the Program for Promotion of Fundamental Studies in Health Sciences of Pharmaceuticals and Medical Devices Agency (to KK), grant-in-aids from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to KK, MM and MK), and the Program for the Promotion of Basic Research Activities for Innovative Bioscience (to NM and MK).
References
- Barsh GS, Farooqi IS, O'Rahilly S (2000) Genetics of body-weight regulation. Nature 404: 644–651 [DOI] [PubMed] [Google Scholar]
- Brown DR, Quito FL (1988) Neuromedin U octapeptide alters ion transport in porcine jejunum. Eur J Pharmacol 155: 159–162 [DOI] [PubMed] [Google Scholar]
- Castel M, Belenky M, Cohen S, Wagner S, Schwartz WJ (1997) Light-induced c-Fos expression in the mouse suprachiasmatic nucleus: immunoelectron microscopy reveals co-localization in multiple cell types. Eur J Neurosci 9: 1950–1960 [DOI] [PubMed] [Google Scholar]
- Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou QY (2002) Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 417: 405–410 [DOI] [PubMed] [Google Scholar]
- Chu C, Jin Q, Kunitake T, Kato K, Nabekura T, Nakazato M, Kangawa K, Kannan H (2002) Cardiovascular actions of central neuromedin U in conscious rats. Regul Peptides 105: 29–34 [DOI] [PubMed] [Google Scholar]
- Civelli O, Nothacker HP, Saito Y, Wang Z, Lin SH, Reinscheid RK (2001) Novel neurotransmitters as natural ligands of orphan G-protein-coupled receptors. Trends Neurosci 24: 230–237 [DOI] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA (2003) Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol 285: R939–R949 [DOI] [PubMed] [Google Scholar]
- Date Y, Ueta Y, Yamashita H, Yamaguchi H, Matsukura S, Kangawa K, Sakurai T, Yanagisawa M, Nakazato M (1999) Orexins, orexigenic hypothalamic peptides, interact with autonomic, neuroendocrine and neuroregulatory systems. Proc Natl Acad Sci USA 96: 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein K, Amir S (1995) Non-photic manipulations induce expression of Fos protein in the suprachiasmatic nucleus and intergeniculate leaflet in the rat. Brain Res 690: 254–258 [DOI] [PubMed] [Google Scholar]
- Fujii R, Hosoya M, Fukusumi S, Kawamata Y, Habata Y, Hinuma S, Onda H, Nishimura O, Fujino M (2000) Identification of neuromedin U as the cognate ligand of the orphan G protein-coupled receptor FM-3. J Biol Chem 275: 21068–21074 [DOI] [PubMed] [Google Scholar]
- Gardiner SM, Compton AM, Bennett T, Domin J, Bloom SR (1990) Regional hemodynamic effects of neuromedin U in conscious rats. Am J Physiol 258: R32–R38 [DOI] [PubMed] [Google Scholar]
- Ginty DD, Kornhauser JM, Thompson MA, Bading H, Mayo KE, Takahashi JS, Greenberg ME (1993) Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science 260: 238–241 [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR (1989) Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59: 675–680 [DOI] [PubMed] [Google Scholar]
- Gottschall PE, Tatsuno I, Miyata A, Arimura A (1990) Characterization and distribution of binding sites for the hypothalamic peptide, pituitary adenylate cyclase-activating polypeptide. Endocrinology 127: 272–277 [DOI] [PubMed] [Google Scholar]
- Graham ES, Turnbull Y, Fotheringham P, Nilaweera K, Mercer JG, Morgan PJ, Barrett P (2003) Neuromedin U and neuromedin U receptor-2 expression in the mouse and rat hypothalamus: effects of nutritional status. J Neurochem 87: 1165–1173 [DOI] [PubMed] [Google Scholar]
- Hamada T, LeSauter J, Venuti JM, Silver R (2001) Expression of period genes: rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J Neurosci 21: 7742–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada R, Date Y, Shimbara T, Sakihara S, Murakami N, Hayashi Y, Kanai Y, Suda T, Kangawa K, Nakazato M (2003) Central action of neuromedin U via corticotropin-releasing hormone. Biochem Biophys Res Commun 311: 954–958 [DOI] [PubMed] [Google Scholar]
- Hanada R, Nakazato M, Murakami N, Sakihara S, Yoshimatsu H, Toshinai K, Hanada T, Suda T, Kangawa K, Matsukura S (2001) A role for neuromedin U in stress response. Biochem Biophys Res Commun 289: 225–228 [DOI] [PubMed] [Google Scholar]
- Hanada R, Teranishi H, Pearson JT, Kurokawa M, Hosoda H, Fukushima N, Fukue Y, Serino R, Fujihara H, Ueta Y, Ikawa M, Okabe M, Murakami N, Shirai M, Yoshimatsu H, Kangawa K, Kojima M (2004) Neuromedin U has a novel anorexigenic effect independent of the leptin signaling pathway. Nat Med 10: 1067–1073 [DOI] [PubMed] [Google Scholar]
- Hannibal J (2002) Neurotransmitters of the retino-hypothalamic tract. Cell Tissue Res 309: 73–88 [DOI] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH (2002) The VPAC2 receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109: 497–508 [DOI] [PubMed] [Google Scholar]
- Harrington ME (1997) The ventral lateral geniculate nucleus and the intergeniculate leaflet: interrelated structures in the visual and circadian systems. Neurosci Biobehav Rev 21: 705–727 [DOI] [PubMed] [Google Scholar]
- Hedrick JA, Morse K, Shan L, Qiao X, Pang L, Wang S, Laz T, Gustafson EL, Bayne M, Monsma FJ Jr (2000) Identification of a human gastrointestinal tract and immune system receptor for the peptide neuromedin U. Mol Pharmacol 58: 870–875 [DOI] [PubMed] [Google Scholar]
- Honzawa M, Sudoh T, Minamino N, Kangawa K, Matsuo H (1990) Neuromedin U-like immunoreactivity in rat intestine: regional distribution and immunohistochemical study. Neuropeptides 15: 1–9 [DOI] [PubMed] [Google Scholar]
- Hosoya M, Moriya T, Kawamata Y, Ohkubo S, Fujii R, Matsui H, Shintani Y, Fukusumi S, Habata Y, Hinuma S, Onda H, Nishimura O, Fujino M (2000) Identification and functional characterization of a novel subtype of neuromedin U receptor. J Biol Chem 275: 29528–29532 [DOI] [PubMed] [Google Scholar]
- Howard AD, Wang R, Pong SS, Mellin TN, Strack A, Guan XM, Zeng Z, Williams DL Jr, Feighner SD, Nunes CN, Murphy B, Stair JN, Yu H, Jiang Q, Clements MK, Tan CP, Mckee KK, Hreniuk DL, McDonald TP, Lynch KR, Evans JF, Austin CP, Caskey CT, Van der Ploeg LHT, Liu Q (2000) Identification of receptors for neuromedin U and its role in feeding. Nature 406: 70–74 [DOI] [PubMed] [Google Scholar]
- Ida T, Nakahara K, Katayama T, Murakami N, Nakazato M (1999) Effect of lateral cerebroventricular injection of the appetite-stimulating neuropeptide, orexin and neuropeptide Y, on the various behavioural activities of rats. Brain Res 821: 526–529 [DOI] [PubMed] [Google Scholar]
- Kojima M, Haruno R, Nakazato M, Date Y, Murakami N, Hanada R, Matsuo H, Kangawa K (2000) Purification and identification of neuromedin U as an endogenous ligand for an orphan receptor GPR66 (FM3). Biochem Biophys Res Commun 276: 435–438 [DOI] [PubMed] [Google Scholar]
- Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K (1999) Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 402: 656–660 [DOI] [PubMed] [Google Scholar]
- Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, Weitz CJ (2001) Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science 294: 2511–2515 [DOI] [PubMed] [Google Scholar]
- Lo G, Legon S, Austin C, Wallis S, Wang Z, Bloom SR (1992) Characterization of complementary DNA encoding the rat neuromedin U precursor. Mol Endocrinol 6: 1538–1544 [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS (2000) Genetics of the mammalian circadian system: photic entrainment, circadian pacemaker mechanisms, and posttranslational regulation. Annu Rev Genet 34: 533–562 [DOI] [PubMed] [Google Scholar]
- McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, Solari R, Lee MG, Foord SM (1998) RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature 393: 333–339 [DOI] [PubMed] [Google Scholar]
- Minamino N, Kangawa K, Matsuo H (1985) Neuromedin U-8 and U-25: novel uterus stimulating and hypertensive peptides identified in porcine spinal cord. Biochem Biophys Res Commun 130: 1078–1085 [DOI] [PubMed] [Google Scholar]
- Moore RY, Speh JC, Leak RK (2002) Suprachiasmatic nucleus organization. Cell Tissue Res 309: 89–98 [DOI] [PubMed] [Google Scholar]
- Mrosovsky N (1996) Locomotor activity and non-photic influences on circadian clocks. Biol Rev 71: 343–372 [DOI] [PubMed] [Google Scholar]
- Murakami N, Takahashi K (1983) Circadian rhythm of adenosine-3′, 5′-monophosphate content in suprachiasmatic nucleus (SCN) and ventromedial hypothalamus (VMH) in the rat. Brain Res 276: 297–304 [DOI] [PubMed] [Google Scholar]
- Nakahara K, Hanada R, Murakami N, Teranishi H, Ohgusu H, Fukushima N, Moriyama M, Ida T, Kangawa K, Kojima M (2004) The gut-brain peptide neuromedin U is involved in the mammalian circadian oscillator system. Biochem Biophys Res Commun 318: 156–161 [DOI] [PubMed] [Google Scholar]
- Nakazato M, Hanada R, Murakami N, Date Y, Mondal MS, Kojima M, Yoshimatsu H, Kangawa K, Matsukura S (2000) Central effects of neuromedin U in the regulation of energy homeostasis. Biochem Biophys Res Commun 277: 191–194 [DOI] [PubMed] [Google Scholar]
- Ozaki Y, Onaka T, Nakazato M, Saito J, Kanemoto K, Matsumoto T, Ueta Y (2002) Centrally administered neuromedin U activates neurosecretion and induction of c-fos messenger ribonucleic acid in the paraventricular and supraoptic nuclei of rat. Endocrinology 143: 4320–4329 [DOI] [PubMed] [Google Scholar]
- Piggins HD, Cutler DJ (2003) The roles of vasoactive intestinal polypeptide in the mammalian circadian clock. J Endocrinol 177: 7–15 [DOI] [PubMed] [Google Scholar]
- Raddatz R, Wilson AE, Artymyshyn R, Bonini JA, Borowsky B, Boteju LW, Zhou S, Kouranova EV, Nagorny R, Guevarra MS, Dai M, Lerman GS, Vaysse PJ, Branchek TA, Gerald C, Forray C, Adham N (2000) Identification and characterization of two neuromedin U receptors differentially expressed in peripheral tissues and the central nervous system. J Biol Chem 275: 32452–32459 [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR (2001) Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol 63: 647–676 [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418: 935–941 [DOI] [PubMed] [Google Scholar]
- Rouille Y, Duguay SJ, Lund K, Furuta M, Gong Q, Lipkind G, Oliva AA Jr, Chan SJ, Steiner DF (1995) Proteolytic processing mechanisms in the biosynthesis of neuroendocrine peptides: the subtilisin-like proprotein convertases. Front Neuroendocrinol 16: 322–361 [DOI] [PubMed] [Google Scholar]
- Schwartz MW, Woods SC, Porte D Jr, Seeley RJ, Baskin DG (2000) Central nervous system control of food intake. Nature 404: 661–671 [DOI] [PubMed] [Google Scholar]
- Sumi S, Inoue K, Kogire M, Doi R, Takaori K, Suzuki T, Yajima H, Tobe T (1987) Effect of synthetic neuromedin U-8 and U-25, novel peptides identified in porcine spinal cord, on splanchnic circulation in dogs. Life Sci 41: 1585–1590 [DOI] [PubMed] [Google Scholar]
- Turek FW, Van Reeth O (1988) Altering the mammalian circadian clock with the short-acting benzodiazepine, triazolam. Trends Neurosci 11: 535–541 [DOI] [PubMed] [Google Scholar]
- Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, Bergmann JE, Gaitanaris GA (2003) The G protein-coupled receptor repertories of human and mouse. Proc Natl Acad Sci USA 100: 4903–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G (1986) A new method for predicting signal sequence cleavage site. Nucleic Acids Res 14: 4683–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren AM, Small CJ, Abbott CR, Jethwa PH, Kennedy AR, Murphy KG, Stanley SA, Zollner AN, Ghatei MA, Bloom SR (2002) Hypothalamic actions of neuromedin U. Endocrinology 143: 4227–4234 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Van Reeth O, Zee PC, Takahashi JS, Turek FW (1993) Fos protein expression in the circadian clock is not associated with phase shifts induced by a nonphotic stimulus, triazolam. Neurosci Lett 164: 203–208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Table SI
Supplementary Table SII
Supplementary Table SIII


