Abstract
Fungi play a major role in the global carbon cycle because of their ability to utilize plant biomass (polysaccharides, proteins, and lignin) as carbon source. Due to the complexity and heterogenic composition of plant biomass, fungi need to produce a broad range of degrading enzymes, matching the composition of (part of) the prevalent substrate. This process is dependent on a network of regulators that not only control the extracellular enzymes that degrade the biomass, but also the metabolic pathways needed to metabolize the resulting monomers. This review will summarize the current knowledge on regulation of plant biomass utilization in fungi and compare the differences between fungal species, focusing in particular on the presence or absence of the regulators involved in this process.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-017-0841-x) contains supplementary material, which is available to authorized users.
Keywords: Plant biomass degradation, Transcription regulation, CAZy genes, Fungi, Bioeconomy
Background
Plant biomass is the most abundant carbon source on Earth and therefore is of major importance for ecology and the global carbon cycle. Fungi are highly efficient degraders of plant biomass. Due to the complexity and heterogeneity of plant biomass (see “Plant biomass composition and its degradation”) (Table 1), fungi have developed a complex and efficient degrading machinery, consisting of a large set of hydrolytic and oxidative enzymes. These enzymes are not only involved in saprobic degradation of plant biomass, but also in other types of plant–fungal interaction, such as pathogenicity and symbiosis (mycorrhizae) or parasitism.
Table 1.
Composition of plant biomass polymers.
Based on Kowalczyk et al. [37]
| Polymer type | Polymer | Monomers |
|---|---|---|
| Cellulose | d-Glucose | |
| Hemicellulose | Xylan | d-Xylose |
| Glucuronoxylan | d-Glucuronic acid, d-xylose | |
| Arabinoglucuronoxylan | d-Xylose, l-arabinose | |
| Arabinoxylan | d-Xylose, l-arabinose | |
| Galacto(gluco)mannan | d-Glucose, d-mannose, d-galactose | |
| Mannan/galactomannan | d-Mannose, d-galactose | |
| Xyloglucan | d-Glucose, d-xylose, d-fructose, d-galactose | |
| β(1,3)/(1,4)-glucan | d-Glucose | |
| Pectin | Homogalacturonan | d-Galacturonic acid |
| Xylogalacturonan | d-Galacturonic acid, d-xylose | |
| Rhamnogalacturonan I | d-Galacturonic acid, l-rhamnose, d-galactose, l-arabinose, ferulic acid, d-glucuronic acid | |
| Rhamnogalacturonan II | d-Galacturonic acid, l-rhamnose, d-galactose, l-arabinose, l-fucose, d-glucose, d-manno-octulosonic acid (KDO), d-lyxo-heptulosaric acid (DhA), d-xylose, d-apiose, l-acetic acid | |
| Inulin | d-Fructose, d-glucose | |
| Starch | Amylose | d-Glucose |
| Amylopectin | d-Glucose | |
| Various gums | d-Galacturonic acid, l-rhamnose, d-galactose, l-arabinose, d-xylose, l-fucose (depending on the specific gum type) | |
| Lignin | Monolignols: ρ-coumaryl alcohol, coniferyl alcohol, sinapyl alcohol |
However, plant biomass is also the source of many food and industrial products that are used in our society. This has resulted in a broad interest from researchers and industry in the enzymatic degradation of plant biomass, starting with the first application of these degrading enzymes in the beginning of the last century (e.g., hydrolysis of starch and maltose [1]). Since then many applications have been developed in several industrial fields, such as food and feed, pulp and paper, detergents, textile, crop protection, and more recently biofuels and biochemicals [2, 3].
Currently, only a small number of Ascomycetes (including Aspergillus spp. and Trichoderma reesei) have been developed for industrial applications [4], although a larger group is being used as a source of novel enzymes. The major companies producing these fungal enzymes cocktails, such as Novozymes (http://www.novozymes.com), DSM (http://www.dsm.com), Roal Oy (http://www.roal.fi), AB Enzymes (http://www.abenzymes.com), and DuPont (http://www.dupont.com), have selected these fungi, due to their good fermentation properties and high level of protein production. However, enzymatic hydrolysis (i.e., saccharification) is still one of the major bottlenecks in the biorefinery process, due to the recalcitrance of plant cell wall and the insufficient efficiency of current enzymatic cocktails to convert all the biomass into the desired products. Strategic improvements are required, and recent developments in fungal research (genomics, transcriptomics and proteomics) have provided a much deeper insight into the mechanisms/strategies of fungi related to plant biomass utilization.
In general, regulation of fungal gene expression related to plant biomass utilization occurs by activation of gene expression via specific inducers (see “Induction of plant biomass utilization”) in balance with repression of gene expression via carbon catabolite repression (CCR) (see “Carbon catabolite repression: CreA/1”). The sensing of inducers starts a signaling pathway resulting in the activation of transcriptional regulators, which is followed by the production of plant biomass-degrading enzymes as well as the metabolic pathways to use the accumulating sugar monomers [5].
Several transcription factors (TFs) involved in plant biomass have been characterized from the fungal kingdom, mainly belonging to zinc binuclear cluster family [6] (see “Transcription factors directly involved in plant biomass degradation”) (Table 2). However, only a few regulators are conserved across the fungal kingdom, such as the carbon catabolite repressor CreA, while the majority of them are restricted to subgroups of fungal species [6] (Additional file 1). The presence of a regulator in a larger group of fungi also does not guarantee that its function is fully conserved: a phenomenon known as “transcriptional rewiring” [7]. Differences in target genes and induction have been observed, such as for the well-studied case of XlnR (see “The (hemi-)cellulolytic regulator XlnR/XYR1/XLR1”) [8] or Gal4 in yeasts [9]. Evolution of fungal TFs appears to be ruled mainly by transcriptional rewiring and divergence as consequence of duplication or horizontal gene transfer (also from bacteria and viruses), while parallel or convergent evolution appears to be relatively rare [10–12]. In addition, TFs need also to have access to chromatin (which is controlled by other factors, such as methylation, acetylation, and histone modification) [13–15]. This adds a second level of complexity in gene regulation ruled by chromatin access and epigenetics.
Table 2.
Comparison of transcription factors involved in plant biomass utilization
| TF | Class | Binding site | Function | Sections | Fungi | References | |
|---|---|---|---|---|---|---|---|
| Main | Secondary | ||||||
| XlnR/XLR1/XYR1 | Zn2Cys6 | GGCTRRR or GGC(A/T)3 | (Hemi)-cellulose utilization | Unknown | “The (hemi-)cellulolytic regulator XlnR/XYR1/XLR1” | Aspergillus spp., Trichoderma spp., Fusarium spp., N. crassa, M. oryzae, P. canescens, T. cellulolyticus | [8, 84*] |
| AraR | Zn2Cys6 | Unknown | l-Arabinose utilization | Unknown | “l-Arabinose-responsive regulators (AraR and ARA1)” | Aspergillus spp. | [55, 112*] |
| ARA1 | Zn2Cys6 | Unknown | l-Arabinose utilization | Unknown | “l-Arabinose-responsive regulators (AraR and ARA1)” | Magnaporthe oryzae | [102*] |
| CLR-1/ClrA | Zn2Cys6 | CGGN5CGGNCCG | Cellulose utilization | Unknown | “Neurospora crassa cellulose regulators CLR-1 and CLR-2 and their homologs” | N. crassa, Aspergillus spp. | [98*, 114, 119] |
| CLR-2/ClrB/ManR | Zn2Cys6 | CGGN11CGG or YAGAAT | Cellulose utilization | Unknown | “Neurospora crassa cellulose regulators CLR-1 and CLR-2 and their homologs” | N. crassa, Aspergillus spp., T. cellulolyticus, P. oxalicum | [98*, 114, 119] |
| ACE2 | Zn2Cys6 | GGCTAATAA or GGC(T/A)4 or XAE | Cellulose utilization | Unknown | “Specific activators of cellulase gene expression in T. reesei: ACE2 and ACE3” | Trichoderma reesei | [126]* |
| ACE3 | Zn2Cys6 | Unknown | Cellulose utilization | Unknown | “Specific activators of cellulase gene expression in T. reesei: ACE2 and ACE3” | Trichoderma reesei | [121]* |
| AmyR | Zn2Cys6 | CGGN8(C/A)GG or CGGAAATTTAA | Starch utilization | Unknown | “Amylolytic regulation: AmyR and MalR” | Aspergillus spp. | [129–131] |
| MalR | Zn2Cys6 | Unknown | Maltose utilization | Unknown | “Amylolytic regulation: AmyR and MalR” | Aspergillus spp. | [128, 137*] |
| BglR/COL-26 | Zn2Cys6 | Unknown | Sugar sensing | BGL repressor | “Glucose-sensing regulator: BglR and COL-26” | Trichoderma reesei, Neurospora crassa | [99, 118, 139*] |
| ClbR | Zn2Cys6 | CGG OR CCG | Cellobiose utilization | Unknown | “ClbR: cellobiose response regulator” | Aspergillius aculeatus | [17, 140*] |
| RhaR | Zn2Cys6 | Unknown | l-Rhamnose utilization | Unknown | “Pectinolytic regulation: RhaR, GaaR, AraR and GaaX” | Aspergillus spp. | [42, 144*] |
| GaaR | Zn2Cys6 | TCCNCCAAT | Galacturonic acid utilization | Unknown | “Pectinolytic regulation: RhaR, GaaR, AraR and GaaX” | Botrytis cinerea, Aspergillus niger | [146, 147*] |
| InuR | Zn2Cys6 | CGGN8CGG | Inulin utilization | Unknown | “Inulinolytic regulation: InuR” | Aspergillus niger | [37, 148*] |
| GalX | Zn2Cys6 | Unknown | d-Galactose utilization | Unknown | “d-Galactose-responsive regulators: GalR and GalX” | Aspergillus spp. | [152*] |
| GalR | Zn2Cys6 | Unknown | d-Galactose utilization | Unknown | “d-Galactose-responsive regulators: GalR and GalX” | Aspergillus nidulans | [113, 149*] |
| GaaX | Unknown | Unknown | Galacturonic acid repressor | Unknown | “Pectinolytic regulation: RhaR, GaaR, AraR and GaaX” | Aspergillus niger | [146] |
| CreA/CRE1 | Cys2His2 | SYGGRG | Carbon catabolite repression | Unknown | “Carbon catabolite repression: CreA/1” | Aspergillus spp., Trichoderma spp., N. crassa, etc. | [156, 157*] |
| ACE1 | Cys2His2 | Unclear | Cellulase repression | Unknown | “Activator of cellulase expression 1 in T. reesei and hemicellulase regulator 1 in N. crassa” | Trichoderma reesei | [125, 198] |
| HCR-1 | Cys2His2 | Unknown | Hemicellulase repressor | Unknown | “Activator of cellulase expression 1 in T. reesei and hemicellulase regulator 1 in N. crassa” | Neurospora crassa | [199] |
| McmA | MADS-box | CC(A/T)6GG | Cellulase regulation | Unknown | “McmA” | Aspergillus nidulans | [124*] |
| XPP1 | E-box–HLH | WCTAGW + AGAA | 1°–2° metabolism switch | Xylanases repressor | “Xylanase promoter-binding protein (XPP1)” | Trichoderma reesei | [206, 207] |
| VIB1 | p53-like | Unknown | C-derepression | CLR-2 induction, cellulases induction | “The (hemi-)cellulolytic regulator XlnR/XYR1/XLR1” | Neurospora crassa | [99] |
| HAP complex | CBF | CCAAT | Chromatin remodeling, respiratory metabolism | CAZy regulation | “Chromatin access” | Aspergillus spp., N. crassa, Trichoderma spp., etc. | [213–216] |
| WC-1/BLR1 | GATA | WGATAR | Blue light/UV-A response, circadian rhythms | 1° metabolism response, (hemi)-cellulose utilization | “Light effect” | Neurospora crassa, Trichoderma spp. | [221, 235, 241] |
| WC-2/BLR2 | GATA | WGATAR | Blue light/UV-A response, circadian rhythms | 1° metabolism response, (hemi)-cellulose utilization | “Light effect” | Neurospora crassa, Trichoderma spp. | [221, 234, 235] |
| VeA/VEL1 | Velvet | Unknown | Light response | 2° metabolism response, (a)sexual development | “Light effect” | Aspergillus spp., Trichoderma spp., N. crassa, Fusarium spp., etc. | [204, 205, 224] |
| VelB | Velvet | Unknown | Light response | 2° metabolism | “Light effect” | Aspergillus spp. | [212, 224] |
| AreA/NIR2 | Cys2Cys2 | HGATAR | N-assimilation/sensing | Chromatin remodeling | “TFs involved in nitrogen and pH regulation” | Neurospora crassa, Aspergillus spp. | [261, 262, 267] |
| AreB | GATA | Unknown | Nitrogen metabolite repression | Morphology and asexual development | “TFs involved in nitrogen and pH regulation” | Colletotrichum gloeosporioides, Aspergillus spp. | [258, 267, 268] |
| NmrA/1 | Rossmann fold | Unknown | Nitrogen metabolite repression | Unknown | “TFs involved in nitrogen and pH regulation” | Aspergillus spp., Neurospora crassa | [260, 266, 267] |
| NirA/NIT-4 | Zn2Cys6 | Unknown | Nitrate pathway | Unknown | “TFs involved in nitrogen and pH regulation” | Aspergillus spp., Neurospora crassa | [263, 264] |
| PacC/1 | Cys2His2 | GCCARG | Alkaline pH response | Unknown | “TFs involved in nitrogen and pH regulation” | Aspergillus spp., T. reesei, N. crassa | [271, 276, 283] |
| PacX | Zn2Cys6 | Unknown | PacC repressor | Unknown | “TFs involved in nitrogen and pH regulation” | Aspergillus nidulans | [282] |
References reported here are the most relevant (for more see, specific sections). References with* are the first characterization in fungi and selected as protein reference for orthologous clustering searching
Regulation of gene expression directly affects the composition of the resulting enzyme mixtures and is therefore highly relevant for applications [16], resulting in detailed studies in several species, such as Aspergillus spp. [17–19], T. reesei [20–23], Neurospora crassa [24], and Penicillium oxalicum [25, 26]. This review will discuss the knowledge of the main ascomycetous transcriptional regulators directly involved in plant biomass utilization, focusing in particular on the regulatory differences among Ascomycetes. In addition, we will summarize the knowledge about regulatory factors indirectly involved in plant biomass utilization.
Plant biomass composition and its degradation
Plant biomass consists mainly of polysaccharides, but also contains proteins and the aromatic polymer lignin. Its precise composition is highly complex and varies depending on plant species and tissue, season, and geographic location. Plant polysaccharides (Table 1) can be divided into plant cell wall polysaccharides (e.g., cellulose, hemicelluloses, pectins) and storage polysaccharides (e.g., starch, inulin, gums) [5]. They consist of many monomeric components that are attached to each other through a variety of linkages. Plant cell wall polysaccharides are not only linked to each other, but also to the aromatic polymer lignin, providing the main strength and structure for the plant cell wall, and serving as a defense against microbial attack.
Cellulose is the most abundant plant cell wall polysaccharide, consisting of a linear chain of β-1,4-linked d-glucose residues, organized in bundles called microfibrils [27]. Hemicelluloses are more diverse in nature, and they are classified in three main types, depending on the backbone: xylan (β-1,4-linked d-xylose), xyloglucan (β-1,4-linked d-glucose), and mannan (β-1,4-linked d-mannose). These backbones are interrupted, branched, or decorated with several different monomers or chains, resulting in different variants of these polymers [28, 29]. Pectin is the third polysaccharide in plant cell wall, but its amount depends on the plant species and tissue. This polysaccharide consists of four substructures with galacturonic acid as the main monomeric component: homogalacturonan (HGA), xylogalacturonan (XGA), and the more complex rhamnogalacturonan I (RG-I) and II (RG-II) [2].
Starch is one of the main storage polysaccharides, consisting of an α-1,4-linked polymer (amylose) of d-glucose residues which can be branched at α-1,6-linked points (amylopectin) [30, 31]. Another major storage polysaccharide is inulin that consists of a branched β-2,1-linked chain of d-fructose with a terminal d-glucose residue [32, 33]. Gums are another varied group of storage polysaccharides, containing many structures. Some of them (e.g., locust bean gum, guar gum) are similar to cell wall galactomannan [2, 34].
Lignin is the most complex and recalcitrant heteropolymer, consisting of aromatic alcohols known as monolignols, built from three phenylpropanoid precursors: ρ-coumaryl alcohol, coniferyl alcohol, and sinapyl alcohol. The exact structure of lignin varies depending on plant species or tissue [2, 35].
In order to release the monomers present in those complex structures, the simultaneous action of several plant biomass-degrading enzymes are required.
Those enzymes have been classified into families in the Carbohydrate-Active enZymes database (CAZy) [36] (http://www.cazy.org), based on their amino acid sequences. CAZy contains six main groups: glycoside hydrolases (GH), glycosyltransferases (GT), polysaccharide lyases (PL), carbohydrate esterases (CE), auxiliary activities (AA). and carbohydrate-binding modules (CBMs). In each group, there are several families, some containing only a single known enzyme activity (e.g., GH67), while others contain several activities (e.g., GH43) and are therefore not an immediate indication of enzyme function [2].
Induction of plant biomass utilization
In order to utilize plant biomass, fungi need to recognize its components and subsequently induce the production of plant biomass-degrading enzymes, as well as the metabolic pathways needed to convert these sugars into energy and/or bioproducts. These signal compounds are commonly defined as inducer, but polysaccharides are too large to enter fungal cells and therefore probably cannot act directly as inducers. Fungi likely recognize the presence of complex polymers through low molecular weight compounds derived from them, such as monosaccharides or disaccharides. As these simple compounds are rare in the environment, their presence is an indication for the fungus that specific polysaccharides might be present. When one of these simple compounds is sensed by the fungus, a signaling pathway results in the activation of one or more transcriptional activators (see “Transcription factors directly involved in plant biomass degradation”, “Other factors affecting plant biomass utilization”), which (in most of the cases) enters the nucleus and triggers the expression of its target genes, encoding hydrolytic/oxidative enzymes (CAZymes), as well as the metabolic pathways needed to utilize the available C-source [37].
The main theory is that inducers are produced by hydrolysis of polysaccharides by small amounts of constitutively produced enzymes. These degradation products might then be further modified by transglycosylation [38–40]. According to an integrating hypothesis, proposed more than one decade ago, some genes encoding plant biomass-degrading enzymes are induced by carbon starvation, acting as scouting enzymes playing a foraging role under these conditions. They release the inducing molecules that trigger the main hydrolytic response [41]. These phenomena have been observed in Aspergillus niger in response to pectin [42], starch [43], and wheat straw [44].
Different inducers for (hemi-)cellulases have been described in fungi: d-xylose in A. niger [45], cellobiose in Aspergillus oryzae [46], gentiobiose in Penicillium purpurogenum [47], and sophorose in Aspergillus terreus and T. reesei [48, 49]. Amore et al. [50] have summarized all known inducers and related induced genes in the three model fungi A. niger, T. reesei, and N. crassa. Some of the more interesting cases are discussed below. However, in the majority of the cases, the exact structure of the inducer is unknown, and “inducer” is often used to refer to the mono- or disaccharide which triggers the initial transcription of the target genes.
Commonly, it is believed that in Aspergillus spp., the (hemi)-cellulolytic systems are strictly coregulated via the monomeric inducer d-xylose [51–53] and/or cellulose-derived disaccharides [46, 53], while the respective systems in T. reesei and N. crassa are more distinctively regulated. It has been shown that l-arabitol is the inducer of the arabinanolytic and xylanolytic system in Aspergilli [54–56], while in T. reesei, only l-arabinose and not l-arabitol can trigger these effects [57]. It has been demonstrated in A. niger [58, 59] and T. reesei [57, 60] that d-xylose induction is concentration-dependent: at very low concentrations, it acts as an inducer for xylanases, while at higher concentrations as a repressor through CreA/Cre1.
In T. reesei, pure saccharides such as d-xylose, xylobiose, sophorose, lactose, and galactose can act as inducers for (hemi)-cellulase production, but each one can activate only part of these systems [50, 61, 62]. Therefore, induction of (hemi-)cellulases in T. reesei appears different and more diverse than in other fungi [57], such as N. crassa or A. niger, and a general model for the substrates recognition and the induction of (hemi)-cellulolytic systems in T. reesei has been suggested [63]. While every fungus appears to have a best (natural) inducer for (hemi-)cellulase production, such as d-xylose in Aspergillus, sophorose in T. reesei [64] or cellobiose in N. crassa [65], other compounds can also have an inducing effect. Some examples of this are d-galactose in N. crassa and cellobiose and lactose in T. reesei [66, 67].
At the industrial level, lactose is a promising inducer for (hemi)-cellulase production in T. reesei [67], because it is the only soluble and also cheap inducer of carbon source ever shown, at least in this fungus.
Although the exact mechanism of this induction is not known yet, xyr1 is involved in this process [68, 69]. In contrast, the induction of pectinolytic genes is more complex: some of them are constitutively expressed, while others are specifically induced, as studied in Aspergilli [42, 70, 71]. These constitutively expressed genes probably encode the scouting enzymes that are needed to liberate some monomeric sugars from pectin, such as galacturonic acid, rhamnose, arabinose, galactose, or xylose, which induce the other pectinolytic genes through specific regulatory systems [70, 71].
The degradation of crude plant biomass is more complex than that of simple polysaccharides, requiring a network of TFs which respond to different inducers. These regulators need to act together in a coordinated manner to express the right enzymes over time, depending on the substrate and the fungal species. In the literature, only few studies have covered these aspects, but recently, expression profiles of a “large” time course of CAZy and pathway genes have been reported in Aspergilli [72–74], Trichoderma reesei [75], Neurospora crassa [39], and Myceliophthora thermophila [76] or mixed cultures [77].
As de Souza et al. [78] have shown, A. niger during growth on steam-exploded sugarcane bagasse (SEB) prefers to use first d-glucose, followed by d-xylose, and finally l-arabinose, affecting enzyme production over time. In this system, the transcription factor XlnR (xylanolytic regulator, see “The (hemi-)cellulolytic regulator XlnR/XYR1/XLR1”) has been confirmed as the main regulator for the expression of genes encoding complex substrates-degrading enzymes, overruling effects of AraR (arabinanolytic regulator; see “l-Arabinose-responsive regulators (AraR and ARA1)”). Surprisingly and in contrast to previous reports, XlnR and AraR have an overlapping role in promoting the expression of some degrading enzyme-encoding genes depending on the substrate (SEB or monosaccharides mix), such as bglA/4 (β-glucosidases), agdA/B (α-glucosidases), eglA (endoglucanase), lacA (α-galactosidase), aglB (β-galactosidase), axhA (arabinoxylan arabinofuranohydrolase). and xlnD (β-endoxylanase) [78]. Interestingly, Alazi et al. [79] showed that some pectinolytic genes are more expressed at late time points, suggesting that they are induced by starvation or derepressed conditions. In addition, Daly et al. [74] showed that the type of pretreatment of the plant biomass (wheat straw in this case) will also affect the expression of CAZy genes over time, reflecting the accessibility/availability of saccharides/polymers due to the different pretreatments.
All these data suggest that the presence of a fine-tuned and complex crosstalk between inducers and regulatory systems involved in plant biomass utilization in filamentous fungi still remains to be fully understood.
Transcription factors directly involved in plant biomass degradation
Many fungal transcription factors have been described to be directly involved in the regulation of plant biomass utilization (Additional file 1). Their number has increased rapidly in the last years, due to deeper studies and novel tools (“Omics” Era) (i.e., transcriptomics and bioinformatics), providing new insights in this field.
So far, the majority of these TFs belong to the zinc cluster family. This TF family is characterized by the presence of zinc finger(s) in their binding domains, and it is classified in “fold groups” based on the overall shape of the protein backbone in the folded domain, which depends and on the amounts of cysteine and histidine residues. The majority of positive regulators appear to belong to the Zn2Cys6 class, while the repressors to the Cys2His2 class.
Interestingly, several TFs are involved in cellulose degradation (CLR-1/2, XlnR, ACE2/3, ClbR, and McmA), suggesting overlapping function and/or fine tune regulation, depending on the species. Table 2 wants to give an overview to the known TFs and their role in carbon utilization although differences have been observed between fungal species. Figure 1 represents the network of these regulators, based on (and combining) the current knowledge of model systems, such as Aspergilli, Neurospora crassa and Trichoderma reesei. This schema is a general overview and needs to be adapted to every fungal species regarding the presence/absence, function(s). and possible interaction of each regulator.
Fig. 1.
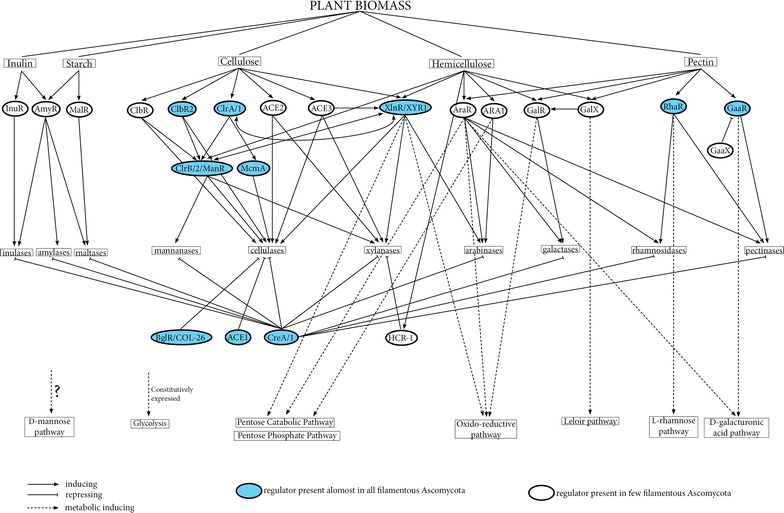
Overview of the regulators-network involved in plant biomass degradation. It is based on (and combining) the current knowledge of the model system Aspergillus, Neurospora crassa and Trichoderma reesi
(This schema needs to be adapted to every fungal species, regarding the presence/absence, function(s). and interaction of/with each regulators)
Phylogenomic analysis was performed to study TF distribution along fungal kingdom following the pipeline of Todd et al. [6]. In summary, we used the same genome set to calculate genome scale protein ortholog clusters using OrthoMCL. The all-vs-all BlastP search required by OrthoMCL was carried out in a grid of 500 computers by parallel fashion. Clusters were detected according to [80], using inflction factor 1, E value cutoff 1 E−3, percentage match cutoff 60% as for identification of distant homologs [81]. Orthologs clusters based on already known TF (Table 2) were extracted and grouped according to their TF domains, such as Zn2Cys6, Cys2His2, MADS box, and basic helix-loop-helix (bHLH). Manual curation of the groups was performed by aligning the amino acid sequences with a suitable outgroup using MAFFT [82], and false positive were removed. Maximum likelihood trees (Additional files 2–5) were generated using MEGA5 [83] with 100 bootstraps and manually curated by refining the alignments (Additional file 1).
This approach allowed us to identify a new putative TF, named ClbR3 (paralog of ClbR/2) (see “ClbR: cellobiose response regulator”) (Fig. 2), which is present in just a few species (Additional file 2).
Fig. 2.

Maximum likelihood phylogenetic tree of ClbR paralogs
Zn2Cys6 transcription factors
The (hemi-)cellulolytic regulator XlnR/XYR1/XLR1
Aspergillus niger xlnR was the first identified transcriptional factor [84] involved in the regulation of (hemi)-cellulose utilization and appears to be the main regulator for this process. Orthologs are present in nearly all filamentous Ascomycetes, confirming its key role in plant biomass utilization [6, 85]. This regulator has been well characterized in several fungi, such as Aspergillus spp. (xlnR) [46, 84, 86], T. reesei and Trichoderma atroviride (xyr1) [63, 87], N. crassa (xlr-1) [88], F. graminearum (xyr1) [89], Fusarium oxysporum (xlnR) [90], Magnaporthe oryzae (xlr1) [91], Penicillium canescens (xlnR) [92], and Talaromyces cellulolyticus (xlnR) [93], but its precise role differs depending on the species [8]. These differences include their set of target genes and binding efficiency, while the binding motif appears to be almost the same in all fungi, with the general consensus sequence GGCTRR (Table 2). In all fungi, this regulator controls d-xylose catabolism and the xylanolytic system [51, 63, 88–90], with the exception of M. oryzae [91] and P. canescens [92]. In M. oryzae, xlr1 controls mainly d-xylose catabolism, while the xylanolytic system appears to be controlled not only by this regulator, but also by another unknown regulatory network [8]. The opposite appears to be the case in P. canascens, where XlnR is involved only in xylan degradation and not in d-xylose catabolism [92].
In Aspergilli [52, 86, 94, 95], except Aspergillus aculeatus [96], and T. reesei [63, 97] this regulator also controls the cellulolytic system, while this appears not to be the case in other species. So far, T. reesei is the only fungus with a clear XYR1 dependent regulation of the cellulolytic system [8]. In N. crassa, xlr-1 can only modulate the expression levels of a few cellulases, suggesting that other TFs are more important [88] such as clr-1 and clr-2 [98] (see “Neurospora crassa cellulose regulators CLR-1 and CLR-2 and their homologs”). Interestingly in this fungus, XLR-1 induces also the TF VIB1, which represses glucose sensing and CCR (in a CRE-1-independent manner), under starvation inducing cellulase production by CLR-2 activation [99].
Moreover, T. reesei appears to be the only fungus that extended the role of xyr1 also to regulating some genes of the arabinanolytic system. While abf2 and bxl1 are under its control, this is not the case for abf1 and abf3. It is likely that another arabinanolytic transcription factor exists, that controls abf1 and abf3 [100, 101]. Recently, a new arabinose-responsive regulator was described in M. oryzae, that has orthologs only in Sordariomycetes and Leotiomycetes [102] (see “l-Arabinose-responsive regulators (AraR and ARA1)”).
Interestingly, regulation of the d-xylose reductase-encoding gene by XYR1 in T. reesei implicates control of XYR1 on d-galactose, lactose, l-arabinose, and d-xylose catabolism, due to the broad specificity and metabolic role of this enzyme [28, 63, 69, 100, 103].
Expression of most xlnR/xyr1/xlr1 orthologs is not specifically induced [57, 58, 104, 105], but rather controlled by CCR (see “The (hemi-)cellulolytic regulator XlnR/XYR1/XLR1”). The only reported exception is T. reesei, where xyr1 expression is clearly induced by lactose and cellulose [68, 69], but not during growth on xylan [106]. The mechanism by which xyr1 responds to these inducers is still not fully understood, but appears to be linked to the nuclear import of XYR1 by the ß-importin KAP8 [107].
In T. reesei, the binding of XYR1 to his target genes appears to be more complex than in Aspergillus spp. [94]. XYR1 binds not only to the GGCTAA motif, arranged as double and inverted sites (separated by 10–12 bp) but also with a single GGC(A/T)3 motif [108, 109]. Interestingly, this last binding motif is present as single site in upstream region in all the XYR1-regulated genes [109]. While XYR1 appears to dimerize before binding to the DNA, dimerization is not essential for function, and it appears that posttranslational modification of XYR1 is the major mechanism governing (hemi)cellulase genes expression [106]. In addition to this, it has been shown that a strongly elevated basal transcription level of xyr1 [69] and/or mutation in its regulatory domain [23] are responsible for the phenotype of T. reesei hyper-producer strains. In T. reesei, the shift between cytoplasm and nucleus is the key mechanism to switch the expression of target genes on/off. Under induction conditions, de novo biosynthesis and rapid nuclear import of XYR1 occur, while the termination of induction results in its rapid nuclear degradation [110]. This is similar to A. niger, where XlnR is inactive in the cytoplasm and nuclear import occurs under d-xylose induction [104].
Although XlnR orthologs are present in almost all filamentous Ascomycota, the set of genes controlled by this regulator is highly species specific [8] and appears to be linked to the lifestyle of the species. During evolution, it appears that this regulator had gradually lost functions, which led to adaptation to a more specialized biotype, as was observed in the pathogenic fungi (Fusarium spp. and M. oryzae) with the loss of cellulolytic function or in Trichoderma spp. [87] with the loss of mycoparasitism function from the “ancient” specie T. atroviride and the “most recent” T. reesei [87, 111]. Considering the composition of plant biomass (cellulose and hemicellulose are often present together) we can hypnotized that pathogenic fungi split the cellulolytic and xylanolytic function, most likely, to better control the host infection. In that sense, the saprobic Talaromyces cellulolyticus is a surprising exception, without an apparent biological explanation that deserves deeper investigation. Despite their taxonomic distance and different lifestyles, the functions of XlnR in Fusarium genus and T. cellulolyticus appear similar in that it regulates xylanase but not cellulase production [93].
l-Arabinose-responsive regulators (AraR and ARA1)
In Aspergillus spp., the arabinanolytic system is under control of AraR (homolog of XlnR), present only in the family Trichocomaceae. It appears to have originated from a gene duplication event from XlnR after this family split from the other filamentous Ascomycetes [112].
It has been reported that XlnR and AraR control distinct sets of genes in response to the presence of d-xylose and l-arabinose, respectively, but also interact with each other [112, 113]. Both regulators are involved in the regulation of genes encoding (hemi)-cellulases, as well as enzymes from the pentose catabolic pathway (PCP). and pentose phosphate pathway (PPP) [55]. In the absence of one regulator, the other can partially compensate for this loss [112]. The regulation of the PCP by AraR has been shown to differ inside the Aspergillus genus, both in expression and growth profiles, suggesting evolutionary changes in Aspergilli regarding pentose utilization [56]. Recently a new l-arabinose-responsive regulator, ARA1, has been described in M. oryzae. Despite having no significant sequence similarity to AraR, ARA1 appears to be the functional analog of AraR, controlling the arabinanolytic system as well as l-arabinose catabolism. A preliminary phylogenetic analysis shows that it is present only in the Sordariomycetes and Leotiomycetes (Additional file 2) and confirms that there is no significant sequence similarity to AraR [102]. This suggests that AraR and ARA1 are an example of parallel evolution in these fungal clades, which appears to be a relatively uncommon phenomenon for regulators in Ascomycetes.
Neurospora crassa cellulose regulators CLR-1 and CLR-2 and their homologs
Two transcription factors, CLR-1 and CLR-2, have been identified in N. crassa that are essential regulators of cellulolytic, but not hemicellulolytic gene expression [98]. Deletion of clr-1 or clr-2 is essential for growth and the complete loss of cellulase activity on cellulose, but not on xylan [98].
Homologs of these regulators are present in almost all genomes of filamentous Ascomycetes (Additional file 1). Despite differences in function have been reported, this prevalence suggests partially conserved regulatory mechanisms involved in cellulose utilization [98, 114]. These regulators have been fully characterized in N. crassa (CLR-1/2) and partially in A. nidulans (ClrA/B) [98, 115], A. niger [114], and A. oryzae (ManR) [116, 117], but only poorly in Talaromyces cellulolyticus (tCLB2) [118] and Penicillium oxalicum (CLR2) [25], showing both similarities and differences.
ClrA/1 appears to have a conserved role in cellulose-sensing, at least in N. crassa and A. nidulans [115]. The presence of cellulose or its products (i.e., cellobiose), results in the activation of ClrA/1, which induces expression of genes necessary for efficient import and utilization of cellobiose, including cellodextrin transporters and β-glucosidases, as well as clrB/2. Then ClrB/2 directly induces expression of cellulases in both fungi, while only a few selected hemicellulases are induced in N. crassa [98, 119].
The roles of these two TFs in other fungi seem to differ from what has been shown in N. crassa. Particularly in Aspergillus spp. ClrB (ManR) appears to have different functions compared to N. crassa CLR-2, and ClrA is not required for the induction of cellulases or other GH-encoding genes [114]. T. reesei lacks a clear homolog of CLR-1, indicating a different strategy for cellulase regulation/sensing in these species [120].
In A. oryzae, the CLR-2 ortholog was characterized as a regulator of mannan-degrading enzymes, and consequently named ManR [116]. Only later on, it has been shown to also control the cellulolytic system in this fungus [117]. Surprisingly, this protein is no longer present in the latest version of the A. oryzae genome and therefore not included in Additional files 1 and 2 in this study.
CLR-2/B is absolutely necessary for cellulolytic activity in N. crassa, A. nidulans [115], A. niger [114], A. oryzae [117], and P. oxalicum [25], but not in T. reesei [121] and T. cellulolyticus [118]. In fact in T. cellulolyticus, cellulase and xylanase production appears to be under control of the Zn2Cys6 TF TctA (Fusarium ctf1B homolog, involved in cutinase induction) [118], confirming a different regulation of (hemi)-cellulase in this species. Interestingly, in N. crassa CLR-1/2 regulates the expression of other main TF-encoding genes, necessary for the lignocellulose utilization, such as xlr-1, vib-1, col-26 (bglR homolog; see “Amylolytic regulation: AmyR and MalR”), sah-2 and hac-1 involved in (hemi)cellulase production and cpc-1 and the homolog of tamA involved in amino acid/nitrogen metabolism [119]. The opposite is the case in the Aspergilli, where ClrA and ClrB appear to be regulated by the main (hemi)-cellulolytic regulator XlnR [114].
The binding sites of CLR-1 and CLR-2 (Table 2) have been recently identified in N. crassa, as well as their mechanism. They act as a homocomplex and not as a heterocomplex as previously supposed [119]. Transcriptional analysis of clr-2/B mutants showed that only a relatively small group of cellulolytic genes have a strictly conserved dependence on CLR-2/B. This conservation between Aspergillus and Neurospora, which are only distantly related, suggests that this cellulase set is of major importance for cellulose utilization strategies in ascomycete filamentous fungi, but with a different regulatory mechanism [98, 115].
Recently Coradetti et al. [115] showed that it is possible to produce and secrete cellulases under noninducing conditions by misexpression of CLR-2 in N. crassa, but not of ClrB in A. nidulans. This could be explained by the fact that N. crassa CLR-2 is fully competent to drive transcription of its targets under noninducing conditions, while Aspergillus ClrB requires other factors, such as XlnR [51, 122], ClbR [17], or McmA [123, 124]. It suggests diversity in the signaling pathways that activate cellulases through XlnR and ClrB in filamentous fungi [98].
Specific activators of cellulase gene expression in T. reesei: ACE2 and ACE3
In T. reesei, the cellulase machinery is tightly regulated, involving other TFs in addition to XYR1: the repressor ACE1 [125] (see “Activator of cellulase expression 1 in T. reesei and hemicellulase regulator 1 in N. crassa”) and the two activators ACE2 [126] and ACE3 [121]. The latter two are classical Zn2Cys6 transcription factors, while ACE1 is a Cys2His2-type regulator (see “Activator of cellulase expression 1 in T. reesei and hemicellulase regulator 1 in N. crassa”).
ACE2 is acting as activator of cellulases and hemicellulases (mainly xylanases) in T. reesei. According to literature this regulator is considered as unique of the Trichoderma genus, but our phylogenetic analysis (Additional file 1) shows the presence of homologs also in some other Sordariomycetes. This suggests that Trichoderma spp., as well as few other Sordariomycetes, evolved a different regulatory system for (hemi)-cellulase production, compared to other species [3, 62].
Interestingly, ACE2 binds to the same promoter motif GGC(T/A)4 as XYR1 [109], but a second binding site GGGTAAATTGG was found in the xylanase-activating element sequence (XAE) in the xyn2 promoter. It has been proposed that phosphorylation and dimerization are prerequisites for the binding of ACE2 to its target promoters [3].
When cellulose is the only carbon source, deletion of ace2 in T. reesei resulted in the reduction of expression of all main cellulase-encoding genes (cbh1, cbh2, egl1, and egl2) and the xylanase-encoding gene xyn2, but not xyn1. This indicates that ACE2 acts as an activator of these genes, even though other factors also play a role in their induction. Sophorose induction is not affected by deletion of ace2, which suggests that the sophorose and cellulose inductions are at least partially mediated by different mechanisms [126]. Würleitner et al. [127] showed that in addition to ACE2, the HAP complex (HAP2/3/5) regulates xyn2 expression in xylobiose-induced cultures, acting through the XAE sequence in the gene’s promoter.
Expression of ace2 appears to be necessary for the formation of high levels of cellulase. ace2 is induced by lactose and appears to not be affected by CCR, but similarly to XYR1, requires CRE1 for full induction during growth on lactose [69].
Recently, a novel regulator of lignocellulose degradation has been described in T. reesei: the activator of cellulase expression 3 (ACE3) [121]. Like ACE2, it is a typical Zn2Cys6 transcription factor and acts as positive regulator of cellulases and (partially) xylanases, both in direct and indirect manners, by regulating xyr1 transcription. This suggests that ace3 overexpression can both directly and indirectly through xyr1 improve cellulolytic and xylanolytic gene expressions. For these reasons, it is considered a master regulator of cellulolytic and a modulator of xylanolytic genes [121].
Interestingly, our phylogenetic analysis (Additional file 2) shows that ACE3 is spread among the whole fungal kingdom (including Basidiomycota). Unfortunately it has not been characterized yet in other species to demonstrate its role in fungal plant biomass utilization.
Amylolytic regulation: AmyR and MalR
Two positive transcription factors have been reported to be involved in starch and maltose utilization in Aspergilli, AmyR and MalR, regulating amylase genes and maltose-utilizing (MAL) cluster genes, respectively [128]. AmyR is one of the first TF identified to be involved in plant biomass degradation in 1999 in A. oryzae [129], while MalR has been characterized more recently [128]. AmyR is well characterized in several Aspergillus species, such as A. nidulans, A. oryzae and A. niger [129–131]. Its orthologs were found in several Ascomycetes (Additional file 1), but its function has not been analyzed in these fungi.
Deletion of this regulator results in reduced or impaired growth on starch and/or maltose, due to insufficient production of the enzymes to convert these saccharides to d-glucose. A. nidulans AmyR shows 72 and 75% structural homology with its orthologs from A. oryzae and A. niger, respectively [132], and their DNA-binding domains are 100% identical to each other, which suggest that they recognize the identical DNA sequence CGGN8(C/A)GG with the A or C depending on the species. Most likely AmyR binds as a dimer with strong affinity to a motif with the two CGG triplets [133]. In addition, it binds also the CGGAAATTTAA sequence in amylase promoters in A. oryzae [134]. Two AmyR molecules are necessary to bind this sequence by recognizing the CGG triplet at the 5′-end and the AGG triplet just downstream of the sequence [134]. AmyR requires translocation to the nucleus to be activ). and the MH4 domain in the C-terminal region is essential to its cytoplasmic localization [135]. In fact, its deletion resulted in a constitutive nuclear localization of AmyR (and consequently it is constitutive active) in A. nidulans and A. oryzae [128]. However,, there are differences in the amylolytic regulation in these species. A. niger AmyR appears to be involved in the utilization of a broader range of oligo- and polysaccharides compared to other Aspergilli [131]. Deletion of amyR in A. nidulans results in no growth on maltose or starch, while in A. oryzae it only reduced growth on starch. The genes under control of AmyR appear to differ in these two strains [86, 131, 136], maybe due to the presence of the additional MAL cluster in A. oryzae [137]. This consists of a second maltose-responsive regulator (malR) [128, 137], a maltose permease (malP), and a maltase (malT) [128, 137]. This MAL cluster has been found in other, but not all Aspergilli, such as A. fumigatus, A. flavus, A. clavatus, and A. fischeri [138].
Deletion of malR in A. oryzae resulted not only in a growth defect on maltose and reduced malP and malT expression [137], but also in poor growth on starch and a reduced α-amylase activity on maltose but not on isomaltose [128]. Expression of amyR is induced by starch, maltos). and isomaltose (strongest), while malR in induced only by maltose and earlier in time than AmyR.
In contrast to AmyR, MalR is constitutively localized in the nucleus, probably due to the absence of the MH4 domain in its sequence [128]. All of this suggests that MalR is essential for the utilization of maltose and subsequent production of isomaltose as an inducer for AmyR activation in this fungus [128]. However, the mechanism of MalR activation is fully unknown and needs to be investigated deeper.
Glucose-sensing regulators: BglR and COL-26
BglR was initially identified in T. reesei as a regulator only of β-glucosidases genes with the exception of bgl1, which is seemingly under control of xyr1 [139]. It is phylogenetically related to AmyR (Additional file 1).
Deletion of bglR increased cellulase production during cellobiose growth, possibly due the inability to produce a glucose signal for CCR. In N. crassa, the homolog of BglR, COL-26, regulates glucose sensing and metabolism, separately from CRE1-mediated CCR [99]. BglR is well conserved among filamentous Ascomycota, mainly in plant pathogen species which require cellulase production for a successful host infection, such as Fusarium spp., Magnaporthe spp., Verticillium spp., Botrytis spp., Alternaria spp., Septoria spp., etc. This suggests that BglR could be a main TF for those fungi which require a functional and accurate cellulose/glucose sensing for survival and/or virulence [139].
The bglR homolog in T. cellulolyticus (tbgA) does not control the cellulolytic system, but only affects part of the xylanolytic system. However, this needs to be confirmed by further investigation [118].
Much remains to be learned about BglR/COL-26 function, but it is likely to play a key role in cellulase overproduction in cellobiose cultures [139], mainly due to the involvement in glucose sensing [99].
ClbR: cellobiose response regulator
Recently, the TF ClbR has been described in A. aculeatus, which is involved in the early phase of cellobiose and cellulose induction through both XlnR-independent and XlnR-dependent signaling pathways [140]. It regulates cbhI, cmc2, xynIa (XlnR independent), cmc1, and xynIb (XlnR-dependent). However, all these genes are still induced in a ΔclbR strain, suggesting that others regulators, such as ClrA/B, are also involved [140] (see “Neurospora crassa cellulose regulators CLR-1 and CLR-2 and their homologs”). In fact, cellobiose-induction and XlnR-independent expression are under the control of both ManR (Clr2/B homolog) and ClbR [120].
Overexpression of ClbR in A. aculeatus resulted in an increase of a subset of xylanolytic and cellulolytic activities. In contrast, the cellobiohydrolase Cel7b was decreased, showing that the effects of ClbR overexpression are strictly depending on the type of enzyme. This suggests that ClbR is involved in diverse signaling pathways to regulate the expression of cellulose-degrading enzymes in A. aculeatus [17].
ClbR orthologs are only present in the Eurotiales, including Aspergillus, Penicillium, and Talaromyces species, while ClbR2 (ClbR paralog, 42% amino acid identity) orthologs are also found in other fungal orders and classes, including Sordariomycetes. Preliminary results in A. aculeatus, showed that ClbR2 regulates only cbhI, cmc2, and manR, and appears to compete with ClbR for the same DNA-binding region in the promoter of its target genes. ClbR2 is currently under investigation, in order to clarify its function, particularly in relation to ClbR [141].
Interestingly, some Eurotiales and Hypocreales (particularly Trichoderma spp.) genomes contain a third paralog with unknown function, named ClbR3 (Additional file 2; Fig. 2). The prevalence of these three TFs suggests that ClbR2 is the common ancestral TF from which ClbR and ClbR3 have originated, through (probably) duplication events after the divergence of those orders.
Pectinolytic regulation: RhaR, GaaR, AraR, and GaaX
The heterogeneity and complexity of the pectin structure suggests that regulation of pectinolytic gene systems could be much more complex than of the other polysaccharides, which is confirmed by the complex of regulators identified so far.
Wubben et al. [142] postulated that Botrytis cinerea endopolygalacturonases genes are regulated by four systems: basal expression, induction by pectin monomers, glucose repression and modulation by pH.
A similar model has been proposed in Aspergilli, where several TFs are involved and the expression profile of pectinolytic genes changes over time [70, 71, 143].
In Aspergillus spp., the system is controlled at least by AraR, responding to l-arabinose (see “The (hemi-)cellulolytic regulator XlnR/XYR1/XLR1”), RhaR, responding to l-rhamnose [37, 42, 144], and GalR/X, responding to d-galactose [113] (section “Inulinolytic regulation: InuR”) and the unknown ferulic acid regulator [143], while the d-galacturonic acid (GA) utilization is controlled by the novel activator GaaR [79] and the repressor GaaX [145].
Aspergillus niger RhaR controls the expression of the genes involved in l-rhamnose catabolism and only few pectinolytic genes, mainly related to the degradation of rhamnogalacturonan I [86]. The RhaR regulator is well distributed among Ascomycetes (phylogenetic see Additional file 1) [6], but it is not characterized yet outsides the Aspergillus genus. A novel d-galacturonic acid regulator has been recently characterized in B. cinerea (BcGaaR) [146] and A. niger (GaaR) [79]. It is essential for growth on d-galacturonic acid (GA), polygalacturonic acid (PGA). and (partially) pectins, regulating most of the pectinolytic genes, GA-transporters, as well as d-galacturonic acid catabolism. The residual growth reported in A. niger ΔgaaR during growth on pectin, can be easily explained by the presence of alternative regulation mechanisms independent of GaaR and/or by growth on other sugars from pectin, such as l-arabinose, d-galactose, d-xylose or l-rhamnose, which are regulate by different TFs (AraR, GalR/X, XlnR, RhaR etc.) [79]. This TF binds to the GARE motif (TCCNCCAAT) of the target genes in both species, and it is imported into the nucleus during induction, similarly to XlnR/XYR1 in A. niger and T. reesei (see “The (hemi-)cellulolytic regulator XlnR/XYR1/XLR1”). In addition to this, in A. niger the repressor GaaX appears to inactivate GaaR in the absence or at low levels of intracellular GA, ensuring a rapid response to the presence of GA as it does not require de novo synthesis of GaaR [145].
Homologs of GaaR are present in most filamentous Ascomycetes, including industrially relevant genera, such as Aspergillus spp. and Trichoderma spp. (Additional file 2). GaaX is located next to gaaR, and this organization appears to be conserved in most of Ascomycota species analyzed [145], suggesting that also the regulation of GA utilization is conserved across filamentous Ascomycota.
Inulinolytic regulation: InuR
InuR [147] appears to be a homolog of AmyR, and they likely have originated from a common ancestor [37, 43]. Additional support for this is that the InuR putative DNA-binding site (CGGN8CGG) seems to be identical to the DNA-binding site of AmyR [43]. Only when both inuR and amyR are deleted growth is abolished on inulin in A. niger, suggesting coregulation by these regulators of inulin conversion [147]. InuR regulates inulinolytic genes (inuA and inuE) and sucrose metabolism (sucA and sucB), as well as the (putative) transporters related to these sugars. Our phylogenetic analysis (Additional file 1) shows that it is mainly present in Aspergilli and related species. F. oxysporum appears to have four homologs of InuR, consisting of two pairs of identical TFs, suggesting different functions in this fungus.
d-Galactose-responsive regulators: GalR and GalX
So far, d-galactose-responsive regulators have been described only in aspergilli (GalX and GalR) and yeasts, e.g., Saccharomyces cerevisiae (Gal4, Rtg1 and Rtg3) [9]. Those TFs do not appear to have a common ancestor (Additional file 1) as they share low sequence homology [148]. GalX and GalR share only 12% amino acid identity, while GAL4 shares 11% aa identity with GalR and 20% with GalX. Moreover, high variation within their DNA binding domains has been reported [148]. In agreement with this, differences in d-galactose utilization have been observed among the Aspergilli and even more between aspergilli and S. cerevisiae [148–150].
GalX is present in the whole genus, regulating the oxido-reductive d-galactose catabolic pathway [151], while GalR is only present in section Nidulantes (e.g., A. nidulans, A. versicolor, A. sydowii [113], controlling the Leloir pathway. So far no regulators have been reported to control this pathway in other Aspergilli. Interestingly, GalR appears to be under control of GalX in A. nidulans, and it has been shown that in this fungus d-galactose catabolism is not only regulated by this two TFs, but also includes involvement of XlnR and AraR [113]. Considering that d-galactose, l-arabinose and d-xylose are often present together in nature, this coregulation is not surprising.
Although both GalX and GalR have so far been mainly shown to be involved in d-galactose catabolism, there are some indications for an involvement in d-galactose release from polysaccharides [113]. Particularly GalR appears to trigger the expression of one α-galactosidase (AGL). However, for full control of this gene, cooperation with AraR is required [151].
Phylogenetic analysis (Additional file 1) showed that GalX and GalR cluster separately, disclaiming the possibility of gene duplication and no orthologs are present outside this genus. In other species, the presence of specific d-galactose-responsive regulator(s) is not clear yet and currently under investigation, particularly in T. reesei [61, 152]. In this fungus, a lactose/d-galactose response has been reported that is mediated by XYR1 and other(s) unidentified TF(s), possibly including CLR2 [67, 68]. This suggests that most likely a network of TFs is involved instead a single specific TF in this fungus [68].
Differences in growth on d-galactose and its regulation have been reported among Aspergilli, suggesting a different evolution related to d-galactose utilization in this genus. For example, A. niger cannot grow on d-galactose from spores, due to the absence of d-galactose uptake during germination [153], while A. nidulans, A. sydowii and A. versicolor can use this sugar as a sole carbon source (http://www.fung-growth.org). This suggests that GalR (present in these species, but not in A. niger), may be required for expression of genes encoding d-galactose transporters during germination [113].
Cys2His2 transcription factors
Carbon catabolite repression: CreA/1
Carbon catabolite repression (CCR) is a universal regulatory system which prevents wasting energy on the production of extracellular enzymes, as well as metabolic routes that are not needed. In relation to plant biomass conversion, this means that when there is sufficient monosaccharide already present, there is no need to produce enzymes to release more monomers from the polysaccharides, and therefore CreA/CRE1 represses the expression of the genes encoding these enzymes [62, 154–156]. Sugar sensing (mainly coordinated via cAMP-dependent PKA pathway and phosphorylation of glucose) induces CCR which repress the utilization of alternative carbon sources [157]. In fungi, CCR occurs mainly through the well conserved Mig1/CreA/CRE1/Cre1 Cys2–His2 double zinc finger TF. So far this TF is the only one which is conserved throughout the fungal kingdom (Additional file 3), suggesting a conserved mechanism for CCR in fungi. It is well studied in S. cerevisiae and filamentous Ascomycota, particularly in Aspergillus spp., T. reesei and N. crassa, but not yet in Basidiomycota.
So far all binding sites of CreA/1 described are two closely spaced SYGGRG, and repression occurs only through this double-binding sites. However, data from in vitro studies suggested that only one site is required, as has been shown in cbh2 promoter in T. reesei [158].
In general, deletion of creA/1 leads to derepression of transcription of (hemi)-cellulolytic genes and other genes involved in polymeric carbon sources, under both repressing and inducing conditions [69, 155, 159, 160].
In aspergilli and N. crassa, CreA/1 directly represses mainly expression of the genes encoding enzymes involved in plant cell wall degradation, such as (hemi)-cellulases [159, 161] or the TF(s) involved in this process [160, 162], while this has not been observed in T. reesei. In this fungus, CRE1 appears to mainly “switch on/off” the transport of inducers/repressor depending on the growth conditions [45, 163, 164] and consequently the TFs which should trigger the expression of the (hemi)-cellulolytic systems.
Although all main TFs, including xlnR/xyr1/xlr1, appear to be under control of CCR and have CreA/CRE1 and/or ACEI (see “Activator of cellulase expression 1 in T. reesei and hemicellulase regulator 1 in N. crassa”) binding sites in their promoter region, evidence of direct regulation only exists for a few cases. A direct effect has been reported for xlnR/xyr1 in A. nidulans [162], A. niger [59], T. reesei [106, 155, 163] and T. koningii (208), with ace2 and ace1 in T. reesei [69], and with amyR in A. nidulans [136, 165] and N. crassa [159].
However, during repressing condition in T. reesei [106] and T. koningii [166], the main target of CRE1 is XYR1, while in N. crassa XLR-1 appears to be regulated mainly by other CCR mechanisms [88]. Moreover, in T. reesei CRE1 acts together with other regulatory proteins in a coordinated manner to promote CCR and optimize C-source utilization. This ensures a good adaptation to different growth condition [163].
Trichoderma reesei appears to be the only fungus in which full induction of xyr1 and ace2 requires the positive action of CRE1, at least during growth on lactose [69]. In T. reesei RUT-C30, partial truncation of CRE1 (CRE1-96, which lacks one zinc finger) leads to an increase of cellulase production, while a full deletion has a less pronounced effect [167]. CRE1-96 controls a more open chromatin at three levels: direct action on the promoters of its target genes, and in an indirect way by increasing the transcription of a chromatin-remodeling protein HTF1. The third level is the loss of the autoregulatory function of CRE1, leading to high transcript levels of cre1-96 [167]. RUT-C30 is an industrial cellulase hyper-producer strains obtain by several rounds of random mutagenesis [21], and is still under investigation to clarify what exactly enhances cellulase production.
Beside transcriptional regulation, CCR appears to act also through chromatin remodeling. In A. nidulans and T. reesei CreA/1 directly affects chromatin structure (packaging, nucleosome position, acetylation etc.) during repression condition. Particularly, T. reesei CRE1 is involved in organizing the local chromatin structure (packaging) or nucleosome positioning in xyr1 promoter and cellulases cbh1 and cbh2 during repressing conditions, and its loss leads to a less dense chromatin structure during CCR [168–170]. Similarly to this, in A. nidulans CreA is involved in chromatin remodeling, through histone deacetylation [171].
A CreA/1 deletion also results in impaired colony morphology in almost all analyzed fungi [159, 172–174], such as smaller and more compressed colonies, with fewer aerial hyphae and spores. In addition, no deletion strains have been obtained in F. oxysporum, P. chrysogenum and M. oryzae, suggesting that this mutation is lethal in these species [175]. The only exceptions to this impaired morphology in deletion strains are Alternaria citri and Alternaria brassicola, suggesting that this genus regulates expression of cell wall-degrading enzymes in a novel manner [176, 177].
CreA/1 is also involved in many other processes, such as penicillin production in P. chrysogenum [178], mycoparasitism in T. harzianum [179], nitrogen and amino acid transport/metabolism in A. nidulans and T. reesei [160], and environmental pH control in pathogenic fungi [175]. All these suggest that CreA/1 have broader function then initially known.
The mechanism of CCR is more complex in filamentous fungi compared with S. cerevisiae, reflecting the differences in lifestyle and the ability of filamentous fungi to utilize a broad range of C-sources, such as pentoses.
In contrast to S. cerevisiae, CCR in filamentous fungi is not only regulated by glucose, but also by high concentrations of other monosaccharides and by nuclear localization [156]. In Sclerotinia sclerotiorum [180] and T. reesei [110], nuclear localization of Cre1/A is glucose dependent, while in A. nidulans, this is not the case [181]. Also the transcription level of cre1/A differs between species. In T. reesei and A. nidulans, cre1/A are negatively autoregulated and transcribed at lower level in repressing conditions [110, 182, 183], while in Acremonium chrysogenum, cre1 transcription is dependent on glucose concentration [184]. In B. cinerea and Gibberella fujikuroi, creA expression is C-source independent [185].
Moreover, in filamentous fungi, carbon catabolite derepression occurs only in the presence of inducers and during carbon limitation or metabolic stress [156], while it is not so in the case in S. cerevisiae. In addition to d-glucose, high concentrations of other simple C-sources trigger CCR through nuclear localization of CreA/1 in filamentous fungi, such as cellobiose and xylose in A. nidulans, or ethanol in F. oxysporum [156].
In both S. cerevisiae and filamentous fungi, phosphorylation of CreA/1, mediated by SnfA/1, is a key for its re-localization during derepression conditions. In A. nidulans SnfA is essential for removal of CreA from the nucleus [186], but not in S. sclerotiorum, although it is still necessary for Cre1 derepression [187]. In contrast to other lignocellulolytic fungi, in T. reesei, CRE1 is positively regulated by phosphorylation under repressing conditions [188]. Moreover, SNF1 (SnfA orthologs) is not able to phosphorylate CRE1 in vivo in T. reesei, while it is able to phosphorylate Mig1 when heterologously expressed in yeast. These differences, suggests that CCR has evolved differently in T. reesei compared to other fungal species [156].
In A. nidulans, A. oryzae, and T. reesei, CreA/1 forms an essential complex for CCR with the deubiquitinating enzyme CreB/2 [189–191], the WD40 motif-containing protein CreC [192] and HECT-type ubiquitin ligase-interacting protein CreD [181, 193]. These additional proteins influence CreA/1 stability and proteosomal degradation, as well providing a link between ubiquitination and phosphorylation in protein regulation and stability. In A. nidulans [194] and F. oxysporum [195], F-box proteins (particularly the unique fungal protein FbxA) also are involved in hemi-cellulase production during CCR. In conclusion, carbon catabolite repression in fungi is not only regulated by CreA/1, but F-box, and additional-Cre proteins introduce a new level of complexity to this system [156].
Activator of cellulase expression 1 in T. reesei and hemicellulase regulator 1 in N. crassa
In contrast to its initially assigned name, in T. reesei and T. koningii, ACE1 acts as a repressor for both cellulase and xylanase production [106, 125, 196, 197]. In contrast, in Talaromyces cellulolyticus, its homologous gene tacA appears to act as an inducer of cellulases and xylanases, as well as cutinases. However, the TacA protein shows a low similarity to ACE1 except for the zinc finger domain (this is confirmed also from BlastP analysis, data not shown), suggesting that it is a novel transcriptional regulator protein [118]. Homologs of ACE1 have been found in almost all filamentous Ascomycota, but very few have been characterized.
The ACE1 protein contains three Cys2His2-type zinc fingers and was shown to bind in vitro to eight sites in the cbh1 promoter, all of which contain the core AGGCA sequence [197]. This core sequence is found in nearly all cellulase promoters in Trichoderma spp., but their functions are not completely clear. In the xyn1 promoter, ACE1 binds to two GGCTAA motifs, competing with the positive regulator XYR1 [108].
In sophorose- and cellulose-induced cultures of T. reesei, the ace1 deletion resulted in an increase in the expression of genes encoding all the main cellulases (such as cbh1, cbh2, egl1, and egl2) and hemicellulases (such as xyn1 and xyn2) [197]. Despite its repressor role, ace1 transcription is induced by lactose, and repressed by CRE1-mediated CCR [69].
All ace1 deletion strains show strong impaired growth on d-sorbitol, suggesting additional targets and a more general regulatory role than expression of cellulase- and hemicellulose-encoding genes [62, 197]. In A. nidulans, a gene (stzA) encoding a highly similar protein to ACE1 is involved in abiotic stress response, such as sensitivity to salt and DNA damaging agents [198]. According to our phylogeny, this appears to be a clear example or divergent evolution of a TF (Additional file 3).
Recently in N. crassa a novel Cys2His2-type zinc fingers TF involved in hemicellulase regulation, HCR-1, has been characterized [199]. This regulator acts as a hemicellulase repressor (affecting mainly xylanases) during growth on l-arabinose or xylan. It appears to be conserved across several cellulolytic fungi but not characterized yet in other species, suggesting a conserved role in lignocellulose degradation. Unfortunately, very little is currently known about its function(s), and deeper investigations are required, particularly regarding its mechanism and role in the regulatory network [199].
Transcription factors without zinc finger: MADS-box
McmA
MADS-box proteins are a well conserved family of transcription factors in eukaryotic organisms, controlling a broad range a cellular function through interaction with their cofactors, such as primary metabolism, cell cycle, and cell identity. Based on the amino acid sequence of the conserved MADS-box domain, these protein are classified into two types: SRF-like (Serum Response Factor) (type I) and MEF2 (Myocyte Enhancer Factor2) (type II). Only few MADS-box proteins have been reported in fungi, mainly in the SRF-like subfamily [200].
The MADS-box motif, of SRF proteins, generally binds to the consensus sequence CC(A/T)6GG (the CArG box) on the target genes [201, 202]. The MAD-domain is also responsible for the nuclear localization, DNA-binding specificity, accessory factor binding and dimerization of the protein [200, 203].
In A. nidulans, the MADS-box protein McmA (similar to S. cerevisiae Mcm1) has been shown to positively control cellulase expression, probably through interaction with ClrB/ManR [120, 124]. McmA and ClrB both bind to the cellulose-responsive element (CeRE) of the endoglucanase eglA promoter in a cooperative manner [124], confirming the common claim [201, 204, 205] that MADS-box proteins have a strong ability to interact with others proteins, integrating different biological processes. In contrast, in T. cellulolyticus, the mcmA homologous tmcA appears to have only small effects on cellulase production [118].
Other factors affecting plant biomass utilization
Additional factors affect plant biomass utilization by fungi, such as temperature, pH, light, nitrogen sources, and the access to heterochromatin. These phenomena are poorly studied in relation to plant biomass utilization, but are crucial to fully understand the regulation of this process. In addition, their signal pathways are not clarified yet, and most likely, they are involved in the transduction of many different processes. Here we present a short overview of these factors.
Xylanase promoter-binding protein (XPP1)
Recently a novel transcription factor has been characterized in T. reesei, named xylanase promoter-binding protein (XPP1) [206]. It acts as a repressor for xylanase-encoding genes (xyn1, xyn2, and bxl2) during growth on d-glucose or high d-xylose concentration at later cultivation stages, while it has no effect on cellulase or d-xylose catabolism [206]. In addition, recently it has been shown that XPP1 is a switch between primary and secondary fungal metabolism (mainly repressing secondary metabolism), suggesting that its role in regulating xylanases may just be a secondary effect [207].
XPP1 is a basic helix-loop-helix TF (bHLH) with E-box domain, which typically would bind to a hexameric palindrome 5′-CANNTG-3′ [208]. Experimental observations suggest that most likely the actual binding is composed of the hexameric palindrome 5′-WCTAGW-3′ together with an inverted AGAA-repeat [206]. Phylogenetic analysis shows that this regulator is only present in Sordariomycetes (Additional file 5).
Chromatin access
Access to heterochromatin is involved in the control of gene expression and therefore also in plant biomass utilization. It is mainly organized through the CCAAT box (Hap complex), methylation (mainly through LaeA/1 [209–212]). and acetylation levels. The CCAAT box complex is believed to be necessary for the generation of an open chromatin structure, which enables full transcriptional activation of certain promoters [158, 213].
CCAAT sequences are present in the 5′ regions of approximately 30% of all eukaryotic genes. The Hap complex is the first CCAAT-binding complex described in Saccharomyces cerevisiae, and it consists of the Hap2, Hap3, Hap4, and Hap5 proteins. Homologs have been identified in several organisms, such as HAP2-3-5 in T. reesei, Hap5 in N. crassa and AnCF from A. nidulans [214].
CCAAT sequences have been found not only in the promoters of respiratory genes [154], but also in promoters of many cellulase- and hemicellulase-encoding genes as well as in the promoters of the ligninolytic genes of several fungi. This sequence in promoter regions appears to be essential for gene expression, as was reported for cbh2 in T. reesei [215]. This reduction in gene expression occurred either at the basal level or in response to specific induction signals, indicating that the CCAAT motif cooperates with other specific elements to affect transcription [216].
An opposite result was obtained when the CCAAT sequence in the XAE in the T. reesei xyn2 promoter was mutated, resulting in a slight increase in xyn2 transcription in glycerol and xylan grown cultures [127]. Moreover in T. reesei the HAP complex, CRE1 and an unknown GTAATA-binding protein affect nucleosome positioning, influencing the accessibility to the TATA box for transcription initiation of cbh2 [158]. More recently and confirming this, Cre1/A has been shown to directly affect the chromatin structure (packaging, nucleosome position, acetylation etc.) during repression condition in A. nidulans and T. reesei (see “Carbon catabolite repression: CreA/1”) [168, 171].
Acetylation levels influence the access to chromatin and consequently the expression of genes. In T. reesei the histone acetyltransferase GCN5 has been deleted, which is involved in chromatin modification by catalyzing the acetylation of specific lysine residues within the N-terminal tails of the core histones. This deletion severely affects acetylation levels resulting in impaired growth, morphogenesis, and expression of cellulase-encoding genes [217].
Light effect
Filamentous fungi can rapidly react to light [218–221], affecting morphology, development and primary and secondary metabolism [222–226], as well as sexual and asexual reproduction [227, 228].
Consequently light influences the lignocellulose degradation in many fungi, such as T. reesei [229–232], T. atroviride [233, 234], N. crassa [119, 235], and Aspergilli [236–238], regulating at the (post)-transcriptional level sugar uptake, carbon catabolism and production of hydrolytic enzymes [223, 239]. For example, in T. atroviride, it has been shown that light strongly influences growth on diverse C-sources [240].
Two light-responsive complexes have been described so far in fungi. The White collar complex (WCC) [239, 241] together with VIVID (VVD) [218] mainly responds to blue light/UV-A through photoreceptors WC1 and WC2 (N. crassa) (BLR1, BLR2 in T. reesei) [219, 235]. These photoreceptors regulate metabolic pathways in response to light [242] in T. reesei [219] and N. crassa [235]. In N. crassa, it has been shown that WWC controls several TF depending on circadian rhythms (Dawn- and dusk-phases), including the glucose-dependent repressor CSP1, important for (hemi)-cellulase production [243]. The other system is the heterotrimeric VELVET complex [211, 224], consisting of VeA—the velvet-like protein VelB—and LaeA, which controls secondary metabolism as well as sexual and asexual reproduction in several species [212, 224, 244–246]. Interestingly, T. reesei and T. atroviride LAE1 (LaeA ortholog) and VEL1 (VeA ortholog) are essential for the expression of (hemi)-cellulase-encoding genes [211, 233, 247], while this was not observed in other species, such as Aspergilli [248, 249].
These light-responsive systems are in general highly conserved in Ascomycetes, but some species do not have VVD orthologs, such as Aspergillus spp. [236], indicating that they are also differentially organized [219].
These data confirm the crosstalk between nutrition, circadian clock and light response in filamentous fungi [119, 220, 231, 240].
TFs involved in nitrogen and pH regulation
Plant biomass utilization is also affected by nitrogen sources, environmental pH and Reactive Oxygen Species (ROS).
Similarly to CCR, Nitrogen Metabolite Repression (NMR) enables preferential utilization of easily assimilated N-sources (ammonium or glutamine) instead of energetically less-favored ones (i.e., nitrate), preventing wasting energy [250, 251]. This mechanism will also negatively affect hydrolytic enzyme production during nitrogen starvation, as well as carbon and secondary metabolism, in several fungi, such as A. nidulans [252, 253], Trichoderma spp. [234, 254], and N. crassa [50, 255–257] or Fusarium spp. [250]. This effect is well studied in A. nidulans and N. crassa, and occurs through 3 key TFs: the activator AreA/NIT-2 (A. nidulans and N. crassa, respectively) [253, 257], and two repressors AreB [258] and NmrA/1 [259, 260].
AreA belongs to GATA family with Cys2Cys2-binding-domain [261], and it is considered the main general nitrogen status-sensing regulator: under N-limiting condition or starvation activates a broad range of genes involved in the utilization of alternative N-sources, such as catabolic genes and permeases [262]. AreA activates directly the transcription of target genes, stimulates the pathway-specific TF(s) necessary to metabolize only the “secondary” N-source present (i.e., NirA [263] in the presence of nitrate) [264], and remodels the chromatin [265] and increases histone acetylation [252]. The activity of AreA is modulated at the post-transcriptional/translational level by its corepressor NmrA through direct binding during N-sufficient conditions [259, 266].
Another GATA repressor is AreB, coding for three distinct proteins [258]. It modulates AreA activity, repressing Area-dependent nitrogen catabolic genes under C-limiting conditions, probably through DNA-binding competition [267]. In addition, it acts also pleiotropically, regarding growth, conidial germination and asexual development [268].
Macios et al. [267] showed that these 3 main TFs negatively regulate arginine catabolism (used as N and C-source) in A. nidulans. In this system, NmrA appears to modulate AREA and AREB activities in response to the carbon status of the cell [267], confirming the crosstalk between Carbon and Nitrogen regulation proposed by Lockington et al. [269].
This crosstalk can explain the negative effects of N-starvation on hydrolytic enzyme (CAZy) production observed and described above. Those regulators mediate chromatin remodeling, in particular during nitrogen starvation, affecting the CAZyme production in A. nidulans [250, 252]. Particularly AreA appears to have a major role in this and cellulase-encoding genes in A. nidulans contain potential binding sites for the global carbon and nitrogen regulatory TFs, such as CreA, XlnR and AreA, in their promoter [269]. This supports the existence of a link between regulation of carbon and nitrogen metabolism in fungi [269].
Fungi also need to adapt to ambient pH changes [270], mediated through an elaborate signal transduction network to allow the proper physiological response, such as production of specific hydrolytic enzymes, growth, cellular transport, development, and pathogenicity/virulence [271–277].
The pH regulation has been well studied in A. nidulans [271, 278] and Saccharomyces cerevisiae and reviewed previously [279]. In summary, the fungal response to pH occurs through the Pal signal-sensing pathway and the key TF PacC (Cys2His2 type), both highly conserved in fungi [271]. The active form of PacC (PacC27) induces the transcription of alkaline-expressed genes (including pacC itself) and represses transcription of acid-expressed genes [280, 281], including lignocellulolytic enzymes. Recently, another factor involved in pH response has been described: the transcription factor PacX [282]. This is a repressor of PacC, but its exact mechanism and function are not clear yet. In addition, PacC not only regulates (hemi)-cellulase production directly by binding the promoter regions of the corresponding genes, but mainly indirectly via modulation of the activity of the transcriptional regulators involved in their production, such as XlnR [283] and ClrB [284] or/and uncharacterized TF(s) as shown in T. reesei [285]. Particularly in T. reesei PAC1 (homolog of PacC) increases cellulase production by induction of xyr1 and ace2 expression [276]. This indicates that the pH-regulation, besides its conservation across fungi, may differ depending on the species.
Conclusion
Due to the complexity of plant biomass, fungi have evolved different strategies to utilize all or some of its components as carbon source. They need a broad set of degrading enzymes as well as diversified metabolic pathways, which are under control of several regulators. An additional level of complexity is the crosstalk between regulatory systems. Moreover, regulatory systems are poorly conserved in fungi. This is likely caused by the need of different fungal species to adapt to their specific biotope such as forest, crops, herbivore dung, and gut etc. On top of that, the lifestyle of every fungus (such as saprobe, pathogen, symbiont, etc.) adds a second level of adaptation, affecting gene regulation. Example of this complex adaptation is the transcriptional rewiring, where a single TF has different functions depending on the species.
With the availability of an ever-increasing number of fungal genomes, transcriptomes, and proteomes, the differences between regulatory systems in fungi related to in plant biomass utilization have become more evident. This study demonstrated both similarities and differences between fungal species, as well as the much higher level of complexity of and interaction between the regulatory systems than was previously assumed. With the addition of new regulators, our understanding of plant biomass strategies of fungi increases, enabling a more fine-tuned manipulation of enzyme production in industrial settings. This will not only benefit the development of better enzyme cocktails for the production of biofuel and biochemical, but also many other industrial sectors, such as pulp and paper, food and feed, and textiles.
Additional files
Additional file 1. Prevalence of transcription factors in Fungi.
Additional file 2. Maximum likelihood tree of Zn2Cys6 transcription factors (TFs). In bold are the species for which the TF has been characterized, while in bold and larger font are the species in which the TF was first discovered.
Additional file 3. Maximum likelihood tree of Cys2Hys2 transcription factors (TFs). In bold are the species for which the TF has been characterized, while in bold and larger font are the species in which the TF was first discovered.
Additional file 4. Maximum likelihood tree of MADS BOX (McmA) transcription factors (TFs). In bold are the species for which the TF has been characterized, while in bold and larger font are the species in which the TF was first discovered.
Additional file 5. Maximum likelihood tree of bHLH (XPP1) transcription factors (TFs). In are the species for which the TF has been characterized, while in bold and larger font are the species in which the TF was first discovered.
Authors’ contributions
RPDV coordinated the manuscript. TB wrote the manuscript and performed phylogenetic analysis. MVAP and MZ performed bioinformatics analysis. RDPV and BS reviewed the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Funding
The large-scale genome mining was carried out on the Dutch national e-infrastructure with the support of SURF Foundation (e-infra130078). TB and MVAP were supported by a grant of the Dutch Technology Foundation STW, the Applied Science division of NWO, and the Technology Program of the Ministry of Economic Affairs 016.130.609 to RPdV. MZ was supported by a grant from the Netherlands Organisation for Scientific Research (NWO) and the Netherlands Genomics Initiative 93511035 to RPdV.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- TF
transcription factor
- CAZy
Carbohydrate-Active enZymes database
- HGA
homogalacturonan
- XGA
xylogalacturonan
- RG
rhamnogalacturonan
- GH
glycoside hydrolases
- GT
glycosyltransferases
- PL
polysaccharide lyases
- CE
carbohydrate esterases
- AA
auxiliary activities
- CBM
carbohydrate-binding modules
- bHLH
basic helix-loop-helix
- HECT
homologous to the E6-AP carboxyl terminus
- WD40
beta-transducin repeat
- CCR
carbon catabolite repression
- XAE
xylanase-activating element
- ROS
reactive oxygen species
- SRF
serum response factor
- MEF2
myocyte enhancer factor2
- CeRe
cellulose-responsive element
- WCC
white collar complex
- VVD
VIVID complex
- cAMP
cyclic AMP
- PKA
protein kinase A
- PCP
pentose catabolic pathway
- PPP
pentose phosphate pathway
- ABF
arabinofuranosidase
- BXL
β-xylosidase
- XYN
xylanase
- CBH
cellobiohydrolase
- EGL
endoglucanase
- BGL
β-glucosidase
- CMC
carboxymethyl cellulase
- AGL
α-galactosidase
- INU
inulase
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-017-0841-x) contains supplementary material, which is available to authorized users.
Contributor Information
Tiziano Benocci, Email: t.benocci@westerdijkinstitute.nl.
Maria Victoria Aguilar-Pontes, Email: v.aguilar@westerdijkinstitute.nl.
Miaomiao Zhou, Email: m.zhou@westerdijkinstitute.nl.
Bernhard Seiboth, Email: bernhard.seiboth@tuwien.ac.at.
Ronald P. de Vries, Phone: +31 302122600, Email: r.devries@westerdijkinstitute.nl
References
- 1.Okada S. On the optimal conditions for the proteoclastic action of Taka-diastase. Biochem J. 1916;10:130–136. doi: 10.1042/bj0100130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makela MR, Donofrio N, de Vries RP. Plant biomass degradation by fungi. Fungal Genet Biol. 2014;72:2–9. doi: 10.1016/j.fgb.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Stricker AR, Mach RL, de Graaff LH. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei) Appl Microbiol Biotechnol. 2008;78:211–220. doi: 10.1007/s00253-007-1322-0. [DOI] [PubMed] [Google Scholar]
- 4.Pariza MW, Johnson EA. Evaluating the safety of microbial enzyme preparations used in food processing: update for a new century. Regul Toxicol Pharmacol. 2001;33:173–186. doi: 10.1006/rtph.2001.1466. [DOI] [PubMed] [Google Scholar]
- 5.de Vries RP, Visser J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microb Mol Biol Rev. 2001;65:497–522. doi: 10.1128/MMBR.65.4.497-522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Todd RB, Zhou M, Ohm RA, Leeggangers HA, Visser L, de Vries RP. Prevalence of transcription factors in ascomycete and basidiomycete fungi. BMC Genom. 2014;15:214. doi: 10.1186/1471-2164-15-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rokas A, Hittinger CT. Transcriptional rewiring: the proof is in the eating. Curr Biol. 2007;17:R626–R628. doi: 10.1016/j.cub.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 8.Klaubauf S, Narang HM, Post H, Zhou M, Brunner K, Mach-Aigner AR, Mach RL, Heck AJ, Altelaar AF, de Vries RP. Similar is not the same: differences in the function of the (hemi-)cellulolytic regulator XlnR (Xlr1/Xyr1) in filamentous fungi. Fungal Genet Biol. 2014;72:73–81. doi: 10.1016/j.fgb.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Dalal CK, Zuleta IA, Mitchell KF, Andes DR, El-Samad H, Johnson AD. Transcriptional rewiring over evolutionary timescales changes quantitative and qualitative properties of gene expression. Elife. 2016;5:e18981. doi: 10.7554/eLife.18981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shelest E. Transcription factors in fungi. FEMS Microbiol Lett. 2008;286:145–151. doi: 10.1111/j.1574-6968.2008.01293.x. [DOI] [PubMed] [Google Scholar]
- 11.Munoz A, Santos Munoz D, Zimin A, Yorke JA. Evolution of transcriptional networks in yeast: alternative teams of transcriptional factors for different species. BMC Genom. 2016;17:826. doi: 10.1186/s12864-016-3102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Mendoza A, Sebe-Pedros A, Sestak MS, Matejcic M, Torruella G, Domazet-Loso T, Ruiz-Trillo I. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc Natl Acad Sci USA. 2013;110:E4858–E4866. doi: 10.1073/pnas.1311818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brosch G, Loidl P, Graessle S. Histone modifications and chromatin dynamics: a focus on filamentous fungi. FEMS Microbiol Rev. 2008;32:409–439. doi: 10.1111/j.1574-6976.2007.00100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Jin G, Zhou X. Modeling the relationship of epigenetic modifications to transcription factor binding. Nucleic Acids Res. 2015;43:3873–3885. doi: 10.1093/nar/gkv255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soyer JL, El Ghalid M, Glaser N, Ollivier B, Linglin J, Grandaubert J, Balesdent MH, Connolly LR, Freitag M, Rouxel T, Fudal I. Epigenetic control of effector gene expression in the plant pathogenic fungus Leptosphaeria maculans. PLoS Genet. 2014;10:e1004227. doi: 10.1371/journal.pgen.1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aghcheh RK, Kubicek CP. Epigenetics as an emerging tool for improvement of fungal strains used in biotechnology. Appl Microbiol Biotechnol. 2015;99:6167–6181. doi: 10.1007/s00253-015-6763-2. [DOI] [PubMed] [Google Scholar]
- 17.Kunitake E, Kawamura A, Tani S, Takenaka S, Ogasawara W, Sumitani J, Kawaguchi T. Effects of clbR overexpression on enzyme production in Aspergillus aculeatus vary depending on the cellulosic biomass-degrading enzyme species. Biosci Biotechnol Biochem. 2015;79:488–495. doi: 10.1080/09168451.2014.982501. [DOI] [PubMed] [Google Scholar]
- 18.MacCabe AP, Orejas M, Tamayo EN, Villanueva A, Ramon D. Improving extracellular production of food-use enzymes from Aspergillus nidulans. J Biotechnol. 2002;96:43–54. doi: 10.1016/S0168-1656(02)00036-6. [DOI] [PubMed] [Google Scholar]
- 19.Valkonen M, Ward M, Wang H, Penttila M, Saloheimo M. Improvement of foreign-protein production in Aspergillus niger var. awamori by constitutive induction of the unfolded-protein response. Appl Environ Microbiol. 2003;69:6979–6986. doi: 10.1128/AEM.69.12.6979-6986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubicek CP, Mikus M, Schuster A, Schmoll M, Seiboth B. Metabolic engineering strategies for the improvement of cellulase production by Hypocrea jecorina. Biotechnol Biofuels. 2009;2:19. doi: 10.1186/1754-6834-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson R, Nevalainen H. Trichoderma reesei RUT-C30—thirty years of strain improvement. Microbiology. 2012;158:58–68. doi: 10.1099/mic.0.054031-0. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y, Duarte AV, van den Brink J, Wiebenga A, Zou G, Wang C, de Vries RP, Zhou Z, Benoit I. Enhancing saccharification of wheat straw by mixing enzymes from genetically-modified Trichoderma reesei and Aspergillus niger. Biotechnol Lett. 2016;38:65–70. doi: 10.1007/s10529-015-1951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derntl C, Gudynaite-Savitch L, Calixte S, White T, Mach RL, Mach-Aigner AR. Mutation of the Xylanase regulator 1 causes a glucose blind hydrolase expressing phenotype in industrially used Trichoderma strains. Biotechnol Biofuels. 2013;6:62. doi: 10.1186/1754-6834-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildebrand A, Szewczyk E, Lin H, Kasuga T, Fan Z. Engineering Neurospora crassa for improved cellobiose and cellobionate production. Appl Environ Microbiol. 2015;81:597–603. doi: 10.1128/AEM.02885-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao G, Li Z, Gao L, Wu R, Kan Q, Liu G, Qu Y. Redesigning the regulatory pathway to enhance cellulase production in Penicillium oxalicum. Biotechnol Biofuels. 2015;8:71. doi: 10.1186/s13068-015-0253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, Zou G, Zhang L, de Vries RP, Yan X, Zhang J, Liu R, Wang C, Qu Y, Zhou Z. The distinctive regulatory roles of PrtT in the cell metabolism of Penicillium oxalicum. Fungal Genet Biol. 2014;63:42–54. doi: 10.1016/j.fgb.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Kolpak FJ, Blackwell J. Determination of the structure of cellulose II. Macromolecules. 1976;9:273–278. doi: 10.1021/ma60050a019. [DOI] [PubMed] [Google Scholar]
- 28.Khosravi C, Benocci T, Battaglia E, Benoit I, de Vries RP. Sugar catabolism in Aspergillus and other fungi related to the utilization of plant biomass. Adv Appl Microbiol. 2015;90:1–28. doi: 10.1016/bs.aambs.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 29.van den Brink J, de Vries RP. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. 2011;91:1477–1492. doi: 10.1007/s00253-011-3473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoover R, Sosulski FW. Composition, structure, functionality, and chemical modification of legume starches: a review. Can J Physiol Pharmacol. 1991;69:79–92. doi: 10.1139/y91-012. [DOI] [PubMed] [Google Scholar]
- 31.Mischnick P, Momcilovic D. Chemical structure analysis of starch and cellulose derivatives. Adv Carbohydr Chem Biochem. 2010;64:117–210. doi: 10.1016/S0065-2318(10)64004-8. [DOI] [PubMed] [Google Scholar]
- 32.Ritsema T, Smeekens S. Fructans: beneficial for plants and humans. Curr Opin Plant Biol. 2003;6:223–230. doi: 10.1016/S1369-5266(03)00034-7. [DOI] [PubMed] [Google Scholar]
- 33.Roberfroid MB. Introducing inulin-type fructans. Br J Nutr. 2005;93(Suppl 1):S13–S25. doi: 10.1079/BJN20041350. [DOI] [PubMed] [Google Scholar]
- 34.Verbeken D, Dierckx S, Dewettinck K. Exudate gums: occurrence, production, and applications. Appl Microbiol Biotechnol. 2003;63:10–21. doi: 10.1007/s00253-003-1354-z. [DOI] [PubMed] [Google Scholar]
- 35.Makela MR, Marinovic M, Nousiainen P, Liwanag AJ, Benoit I, Sipila J, Hatakka A, de Vries RP, Hilden KS. Aromatic metabolism of filamentous fungi in relation to the presence of aromatic compounds in plant biomass. Adv Appl Microbiol. 2015;91:63–137. doi: 10.1016/bs.aambs.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Carbohydrate-Active Enzymes server [http://afmb.cnrs-mrs.fr/CAZY/].
- 37.Culleton H, McKie V, de Vries RP. Physiological and molecular aspects of degradation of plant polysaccharides by fungi: what have we learned from Aspergillus? Biotechnol J. 2013;8:884–894. doi: 10.1002/biot.201200382. [DOI] [PubMed] [Google Scholar]
- 38.Kubicek CP, Messner R, Gruber F, Mach RL, Kubicek-Pranz EM. The Trichoderma cellulase regulatory puzzle: from the interior life of a secretory fungus. Enzyme Microb Technol. 1993;15:90–99. doi: 10.1016/0141-0229(93)90030-6. [DOI] [PubMed] [Google Scholar]
- 39.Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JH, Glass NL. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci USA. 2009;106:22157–22162. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carle-Urioste JC, Escobar-Vera J, El-Gogary S, Henrique-Silva F, Torigoi E, Crivellaro O, Herrera-Estrella A, El-Dorry H. Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J Biol Chem. 1997;272:10169–10174. doi: 10.1074/jbc.272.15.10169. [DOI] [PubMed] [Google Scholar]
- 41.Foreman PK, Brown D, Dankmeyer L, Dean R, Diener S, Dunn-Coleman NS, Goedegebuur F, Houfek TD, England GJ, Kelley AS, et al. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J Biol Chem. 2003;278:31988–31997. doi: 10.1074/jbc.M304750200. [DOI] [PubMed] [Google Scholar]
- 42.de Vries RP, Jansen J, Aguilar G, Parenicová L, Benen JAE, Joosten V, Wulfert F, Visser J. Expression profiling of pectinolytic genes from Aspergillus niger. FEBS Lett. 2002;530:41–47. doi: 10.1016/S0014-5793(02)03391-4. [DOI] [PubMed] [Google Scholar]
- 43.Yuan XL, van der Kaaij RM, van den Hondel CA, Punt PJ, van der Maarel MJ, Dijkhuizen L, Ram AF. Aspergillus niger genome-wide analysis reveals a large number of novel alpha-glucan acting enzymes with unexpected expression profiles. Mol Genet Genom. 2008;279:545–561. doi: 10.1007/s00438-008-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delmas S, Pullan ST, Gaddipati S, Kokolski M, Malla S, Blythe MJ, Ibbett R, Campbell M, Liddell S, Aboobaker A, et al. Uncovering the genome-wide transcriptional responses of the filamentous fungus Aspergillus niger to lignocellulose using RNA sequencing. PLoS Genet. 2012;8:e1002875. doi: 10.1371/journal.pgen.1002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Souza WR, de Gouvea PF, Savoldi M, Malavazi I, de Souza Bernardes LA, Goldman MH, de Vries RP, de Castro Oliveira JV, Goldman GH. Transcriptome analysis of Aspergillus niger grown on sugarcane bagasse. Biotechnol Biofuels. 2011;4:40. doi: 10.1186/1754-6834-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marui J, Kitamoto N, Kato M, Kobayashi T, Tsukagoshi N. Transcriptional activator, AoXlnR, mediates cellulose-inductive expression of the xylanolytic and cellulolytic genes in Aspergillus oryzae. FEBS Lett. 2002;528:279–282. doi: 10.1016/S0014-5793(02)03328-8. [DOI] [PubMed] [Google Scholar]
- 47.Kurasawa T, Yachi M, Suto M, Kamagata Y, Takao S, Tomita F. Induction of cellulase by gentiobiose and its sulfur-containing analog in Penicillium purpurogenum. Appl Environ Microbiol. 1992;58:106–110. doi: 10.1128/aem.58.1.106-110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hrmová M, Petráková E, Biely P. Induction of cellulose- and xylan-degrading enzyme systems in Aspergillus terreus by homo- and heterodisaccharides composed of glucose and xylose. J Gen Microbiol. 1991;137:541–547. doi: 10.1099/00221287-137-3-541. [DOI] [PubMed] [Google Scholar]
- 49.Mandels M, Parrish FW, Reese ET. Sophorose as an inducer of cellulase in Trichoderma viride. J Bacteriol. 1962;83:400–408. doi: 10.1128/jb.83.2.400-408.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amore A, Giacobbe S, Faraco V. Regulation of cellulase and hemicellulase gene expression in fungi. Curr Genom. 2013;14:230–249. doi: 10.2174/1389202911314040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Peij N, Gielkens MMC, de Vries RP, Visser J, de Graaff LH. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol. 1998;64:3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasper AA, Visser J, de Graaff LH. The Aspergillus niger transcriptional activator XlnR, which is involved in the degradation of the polysaccharides xylan and cellulose, also regulates d-xylose reductase gene expression. Mol Microbiol. 2000;36:193–200. doi: 10.1046/j.1365-2958.2000.01843.x. [DOI] [PubMed] [Google Scholar]
- 53.Gielkens MM, Dekkers E, Visser J, de Graaff LH. Two cellobiohydrolase-encoding genes from Aspergillus niger require d-xylose and the xylanolytic transcriptional activator XlnR for their expression. Appl Environ Microbiol. 1999;65:4340–4345. doi: 10.1128/aem.65.10.4340-4345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikolaev I, Farmer Hansen S, Madrid S, de Vries RP. Disruption of the l-arabitol dehydrogenase encoding gene in Aspergillus tubingensis results in increased xylanase production. Biotechnol J. 2013;8:905–911. doi: 10.1002/biot.201200256. [DOI] [PubMed] [Google Scholar]
- 55.Battaglia E, Zhou M, de Vries RP. The transcriptional activators AraR and XlnR from Aspergillus niger regulate expression of pentose catabolic and pentose phosphate pathway genes. Res Microbiol. 2014;165:531–540. doi: 10.1016/j.resmic.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 56.Battaglia E, Hansen SF, Leendertse A, Madrid S, Mulder H, Nikolaev I, de Vries RP. Regulation of pentose utilisation by AraR, but not XlnR, differs in Aspergillus nidulans and Aspergillus niger. Appl Microbiol Biotechnol. 2011;91:387–397. doi: 10.1007/s00253-011-3242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herold S, Bischof R, Metz B, Seiboth B, Kubicek CP. Xylanase gene transcription in Trichoderma reesei is triggered by different inducers representing different hemicellulosic pentose polymers. Eukaryot Cell. 2013;12:390–398. doi: 10.1128/EC.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Vries RP, Visser J, de Graaff LH. CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res Microbiol. 1999;150:281–285. doi: 10.1016/S0923-2508(99)80053-9. [DOI] [PubMed] [Google Scholar]
- 59.Mach-Aigner AR, Omony J, Jovanovic B, van Boxtel AJ, de Graaff LH. d-Xylose concentration-dependent hydrolase expression profiles and the function of CreA and XlnR in Aspergillus niger. Appl Environ Microbiol. 2012;78:3145–3155. doi: 10.1128/AEM.07772-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mach-Aigner AR, Pucher ME, Mach RL. d-Xylose as a repressor or inducer of xylanase expression in Hypocrea jecorina (Trichoderma reesei) Appl Environ Microbiol. 2010;76:1770–1776. doi: 10.1128/AEM.02746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karaffa L, Fekete E, Gamauf C, Szentirmai A, Kubicek CP, Seiboth B. d-Galactose induces cellulase gene expression in Hypocrea jecorina at low growth rates. Microbiology. 2006;152:1507–1514. doi: 10.1099/mic.0.28719-0. [DOI] [PubMed] [Google Scholar]
- 62.Aro N, Pakula T, Penttilä M. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol Rev. 2005;29:719–739. doi: 10.1016/j.femsre.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 63.Stricker AR, Grosstessner-Hain K, Wurleitner E, Mach RL. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot Cell. 2006;5:2128–2137. doi: 10.1128/EC.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeilinger S, Mach R, Schindler M, Herzog P, Kubicek CP. Different inducibility of expression of the two xylanase genes xyn1 and xyn2 in Trichoderma reesei. J Biol Chem. 1996;271:25624–25629. doi: 10.1074/jbc.271.41.25624. [DOI] [PubMed] [Google Scholar]
- 65.Znameroski EA, Coradetti ST, Roche CM, Tsai JC, Iavarone AT, Cate JH, Glass NL. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proc Natl Acad Sci USA. 2012;109:6012–6017. doi: 10.1073/pnas.1118440109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dos Santos Castro L, Pedersoli WR, Antonieto AC, Steindorff AS, Silva-Rocha R, Martinez-Rossi NM, Rossi A, Brown NA, Goldman GH, Faca VM, et al. Comparative metabolism of cellulose, sophorose and glucose in Trichoderma reesei using high-throughput genomic and proteomic analyses. Biotechnol Biofuels. 2014;7:41. doi: 10.1186/1754-6834-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ivanova C, Baath JA, Seiboth B, Kubicek CP. Systems analysis of lactose metabolism in Trichoderma reesei identifies a lactose permease that is essential for cellulase induction. PLoS ONE. 2013;8:e62631. doi: 10.1371/journal.pone.0062631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bischof R, Fourtis L, Limbeck A, Gamauf C, Seiboth B, Kubicek CP. Comparative analysis of the Trichoderma reesei transcriptome during growth on the cellulase inducing substrates wheat straw and lactose. Biotechnol Biofuels. 2013;6:127. doi: 10.1186/1754-6834-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Portnoy T, Margeot A, Seidl-Seiboth V, Le Crom S, Ben Chaabane F, Linke R, Seiboth B, Kubicek CP. Differential regulation of the cellulase transcription factors XYR1, ACE2, and ACE1 in Trichoderma reesei strains producing high and low levels of cellulase. Eukaryot Cell. 2011;10:262–271. doi: 10.1128/EC.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vries RP, Pařenicová L. Regulation of pectinolytic gene expression in Aspergillus. In: Voragen F, Schols H, Visser R, editors. Advances in pectin and pectinase research. Dordrecht: Springer Netherlands; 2003. pp. 169–181. [Google Scholar]
- 71.Martinez-Trujillo A, Aranda JS, Gomez-Sanchez C, Trejo-Aguilar B, Aguilar-Osorio G. Constitutive and inducible pectinolytic enzymes from Aspergillus flavipes FP-500 and their modulation by pH and carbon source. Braz J Microbiol. 2009;40:40–47. doi: 10.1590/S1517-83822009000100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saykhedkar S, Ray A, Ayoubi-Canaan P, Hartson SD, Prade R, Mort AJ. A time course analysis of the extracellular proteome of Aspergillus nidulans growing on sorghum stover. Biotechnol Biofuels. 2012;5:52. doi: 10.1186/1754-6834-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pullan ST, Daly P, Delmas S, Ibbett R, Kokolski M, Neiteler A, van Munster JM, Wilson R, Blythe MJ, Gaddipati S, et al. RNA-sequencing reveals the complexities of the transcriptional response to lignocellulosic biofuel substrates in Aspergillus niger. Fungal Biol Biotechnol. 2014;1:1–14. doi: 10.1186/s40694-014-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Daly P, van Munster JM, Blythe MJ, Ibbett R, Kokolski M, Gaddipati S, Lindquist E, Singan VR, Barry KW, Lipzen A, et al. Expression of Aspergillus niger CAZymes is determined by compositional changes in wheat straw generated by hydrothermal or ionic liquid pretreatments. Biotechnol Biofuels. 2017;10:35. doi: 10.1186/s13068-017-0700-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hakkinen M, Arvas M, Oja M, Aro N, Penttila M, Saloheimo M, Pakula TM. Re-annotation of the CAZy genes of Trichoderma reesei and transcription in the presence of lignocellulosic substrates. Microb Cell Fact. 2012;11:134. doi: 10.1186/1475-2859-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolbusz MA, Di Falco M, Ishmael N, Marqueteau S, Moisan MC, Baptista Cda S, Powlowski J, Tsang A. Transcriptome and exoproteome analysis of utilization of plant-derived biomass by Myceliophthora thermophila. Fungal Genet Biol. 2014;72:10–20. doi: 10.1016/j.fgb.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 77.Daly P, van Munster JM, Kokolski M, Sang F, Blythe MJ, Malla S, de Castro Oliveira VJ, Goldman GH, Archer DB. Transcriptomic responses of mixed cultures of ascomycete fungi to lignocellulose using dual RNA-seq reveal inter-species antagonism and limited beneficial effects on CAZyme expression. Fungal Genet Biol. 2016;102:4–21. doi: 10.1016/j.fgb.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de Souza WR, Maitan-Alfenas GP, de Gouvea PF, Brown NA, Savoldi M, Battaglia E, Goldman MH, de Vries RP, Goldman GH. The influence of Aspergillus niger transcription factors AraR and XlnR in the gene expression during growth in d-xylose, l-arabinose and steam-exploded sugarcane bagasse. Fungal Genet Biol. 2013;60:29–45. doi: 10.1016/j.fgb.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 79.Alazi E, Niu J, Kowalczyk JE, Peng M, Aguilar Pontes MV, van Kan JA, Visser J, de Vries RP, Ram AF. The transcriptional activator GaaR of Aspergillus niger is required for release and utilization of d-galacturonic acid from pectin. FEBS Lett. 2016;590:1804–1815. doi: 10.1002/1873-3468.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boekhorst J, Snel B. Identification of homologs in insignificant blast hits by exploiting extrinsic gene properties. BMC Bioinform. 2007;8:356. doi: 10.1186/1471-2105-8-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Peij NN, Visser J, de Graaff LH. Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Mol Microbiol. 1998;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- 85.Battaglia E. Regulation and diversity of plant polysaccharide utilisation in fungi. Dissertation. University Utrecht, Ph.D. thesis Biologie; 2011.
- 86.Kowalczyk JE, Benoit I, de Vries RP. Regulation of plant biomass utilization in Aspergillus. Adv Appl Microbiol. 2014;88:31–56. doi: 10.1016/B978-0-12-800260-5.00002-4. [DOI] [PubMed] [Google Scholar]
- 87.Reithner B, Mach-Aigner AR, Herrera-Estrella A, Mach RL. The transcriptional regulator Xyr1 of Trichoderma atroviride supports the induction of systemic resistance in plants. Appl Environ Microbiol. 2014;80:5274–81. [DOI] [PMC free article] [PubMed]
- 88.Sun J, Tian C, Diamond S, Glass NL. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot Cell. 2012;11:482–493. doi: 10.1128/EC.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brunner K, Lichtenauer AM, Kratochwill K, Delic M, Mach RL. Xyr1 regulates xylanase but not cellulase formation in the head blight fungus Fusarium graminearum. Curr Genet. 2007;52:213–220. doi: 10.1007/s00294-007-0154-x. [DOI] [PubMed] [Google Scholar]
- 90.Calero-Nieto F, Di Pietro A, Roncero MI, Hera C. Role of the transcriptional activator XlnR of Fusarium oxysporum in regulation of xylanase genes and virulence. Mol Plant Microbe Interact. 2007;20:977–985. doi: 10.1094/MPMI-20-8-0977. [DOI] [PubMed] [Google Scholar]
- 91.Battaglia E, Klaubauf S, Vallet J, Ribot C, Lebrun M-H, de Vries RP. Xlr1 is involved in the transcriptional control of the pentose catabolic pathway, but not hemi-cellulolytic enzymes in Magnaporthe oryzae. Fungal Genet Biol. 2013;57:76–84. doi: 10.1016/j.fgb.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 92.Serebrianyi VA, Sinitsyna OA, Fedorova EA, Okunev ON, Bekkarevich AO, Sokolova LM, Vinetskii IuP, Sinitsyn AP. Synthesis of xylanolytic enzymes and other carbohydrases by the fungus Penicillium canescens: transformants with an altered copy number of the gene xlnR and an encoding transcriptional activator. Prikl Biokhim Mikrobiol. 2006;42:665–673. [PubMed] [Google Scholar]
- 93.Fujii T, Inoue H, Ishikawa K. Characterization of the xylanase regulator protein gene, xlnR, in Talaromyces cellulolyticus (formerly known as Acremonium cellulolyticus) Biosci Biotechnol Biochem. 2014;78:1564–1567. doi: 10.1080/09168451.2014.923298. [DOI] [PubMed] [Google Scholar]
- 94.Noguchi Y, Sano M, Kanamaru K, Ko T, Takeuchi M, Kato M, Kobayashi T. Genes regulated by AoXlnR, the xylanolytic and cellulolytic transcriptional regulator, in Aspergillus oryzae. Appl Microbiol Biotechnol. 2009;85:141–154. doi: 10.1007/s00253-009-2236-9. [DOI] [PubMed] [Google Scholar]
- 95.Andersen MR, Vongsangnak W, Panagiotou G, Salazar MP, Lehmann L, Nielsen J. A trispecies Aspergillus microarray: comparative transcriptomics of three Aspergillus species. Proc Natl Acad Sci USA. 2008;105:4387–4392. doi: 10.1073/pnas.0709964105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tani S, Kanamasa S, Sumitani J, Arai M, Kawaguchi T. XlnR-independent signaling pathway regulates both cellulase and xylanase genes in response to cellobiose in Aspergillus aculeatus. Curr Genet. 2012;58:93–104. doi: 10.1007/s00294-012-0367-5. [DOI] [PubMed] [Google Scholar]
- 97.Dos Santos Castro L, de Paula RG, Antonieto AC, Persinoti GF, Silva-Rocha R, Silva RN. Understanding the role of the master regulator XYR1 in Trichoderma reesei by global transcriptional analysis. Front Microbiol. 2016;7:175. doi: 10.3389/fmicb.2016.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, Glass NL. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci USA. 2012;109:7397–7402. doi: 10.1073/pnas.1200785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xiong Y, Sun J, Glass NL. VIB1, a link between glucose signaling and carbon catabolite repression, is essential for plant cell wall degradation by Neurospora crassa. PLoS Genet. 2014;10:e1004500. doi: 10.1371/journal.pgen.1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Akel E, Metz B, Seiboth B, Kubicek CP. Molecular regulation of arabinan and l-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei) Eukaryot Cell. 2009;8:1837–1844. doi: 10.1128/EC.00162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seiboth B, Metz B. Fungal arabinan and l-arabinose metabolism. Appl Microbiol Biotechnol. 2011;89:1665–1673. doi: 10.1007/s00253-010-3071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Klaubauf S, Zhou M, Lebrun MH, de Vries RP, Battaglia E. A novel l-arabinose-responsive regulator discovered in the rice-blast fungus Pyricularia oryzae (Magnaporthe oryzae) FEBS Lett. 2016;590:550–554. doi: 10.1002/1873-3468.12070. [DOI] [PubMed] [Google Scholar]
- 103.Seiboth B, Gamauf C, Pail M, Hartl L, Kubicek CP. The d-xylose reductase of Hypocrea jecorina is the major aldose reductase in pentose and d-galactose catabolism and necessary for beta-galactosidase and cellulase induction by lactose. Mol Microbiol. 2007;66:890–900. doi: 10.1111/j.1365-2958.2007.05953.x. [DOI] [PubMed] [Google Scholar]
- 104.Hasper AA, Trindade LM, van der Veen D, van Ooyen AJ, de Graaff LH. Functional analysis of the transcriptional activator XlnR from Aspergillus niger. Microbiol. 2004;150:1367–1375. doi: 10.1099/mic.0.26557-0. [DOI] [PubMed] [Google Scholar]
- 105.Noguchi Y, Tanaka H, Kanamaru K, Kato M, Kobayashi T. Xylose triggers reversible phosphorylation of XlnR, the fungal transcriptional activator of xylanolytic and cellulolytic genes in Aspergillus oryzae. Biosci Biotechnol Biochem. 2011;75:953–959. doi: 10.1271/bbb.100923. [DOI] [PubMed] [Google Scholar]
- 106.Mach-Aigner AR, Pucher ME, Steiger MG, Bauer GE, Preis SJ, Mach RL. Transcriptional regulation of xyr1, encoding the main regulator of the xylanolytic and cellulolytic enzyme system in Hypocrea jecorina. Appl Environ Microbiol. 2008;74:6554–6562. doi: 10.1128/AEM.01143-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ghassemi S, Lichius A, Bidard F, Lemoine S, Rossignol MN, Herold S, Seidl-Seiboth V, Seiboth B, Espeso EA, Margeot A, Kubicek CP. The ss-importin KAP8 (Pse1/Kap121) is required for nuclear import of the cellulase transcriptional regulator XYR1, asexual sporulation and stress resistance in Trichoderma reesei. Mol Microbiol. 2015;96:405–418. doi: 10.1111/mmi.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rauscher R, Wurleitner E, Wacenovsky C, Aro N, Stricker AR, Zeilinger S, Kubicek CP, Penttila M, Mach RL. Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina. Eukaryot Cell. 2006;5:447–456. doi: 10.1128/EC.5.3.447-456.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Furukawa T, Shida Y, Kitagami N, Mori K, Kato M, Kobayashi T, Okada H, Ogasawara W, Morikawa Y. Identification of specific binding sites for XYR1, a transcriptional activator of cellulolytic and xylanolytic genes in Trichoderma reesei. Fungal Genet Biol. 2009;46:564–574. doi: 10.1016/j.fgb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 110.Lichius A, Seidl-Seiboth V, Seiboth B, Kubicek CP. Nucleo-cytoplasmic shuttling dynamics of the transcriptional regulators XYR1 and CRE1 under conditions of cellulase and xylanase gene expression in Trichoderma reesei. Mol Microbiol. 2014;94(5):1162–1178. doi: 10.1111/mmi.12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, Casas-Flores S, Horwitz BA, Mukherjee PK, et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011;12:R40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Battaglia E, Visser L, Nijssen A, van Veluw GJ, Wosten HA, de Vries RP. Analysis of regulation of pentose utilisation in Aspergillus niger reveals evolutionary adaptations in Eurotiales. Stud Mycol. 2011;69:31–38. doi: 10.3114/sim.2011.69.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kowalczyk JE, Gruben BS, Battaglia E, Wiebenga A, Majoor E, de Vries RP. Genetic Interaction of Aspergillus nidulans galR, xlnR and araR in regulating d-galactose and l-arabinose release and catabolism gene expression. PLoS ONE. 2015;10:e0143200. doi: 10.1371/journal.pone.0143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Raulo R, Kokolski M, Archer DB. The roles of the zinc finger transcription factors XlnR, ClrA and ClrB in the breakdown of lignocellulose by Aspergillus niger. AMB Express. 2016;6:5. doi: 10.1186/s13568-016-0177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Coradetti ST, Xiong Y, Glass NL. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiologyopen. 2013;2:595–609. doi: 10.1002/mbo3.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ogawa M, Kobayashi T, Koyama Y. ManR, a novel Zn(II)(2)Cys(6) transcriptional activator, controls the beta-mannan utilization system in Aspergillus oryzae. Fungal Genet Biol. 2012;49(12):987–995. doi: 10.1016/j.fgb.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 117.Ogawa M, Kobayashi T, Koyama Y. ManR, a transcriptional regulator of the beta-mannan utilization system, controls the cellulose utilization system in Aspergillus oryzae. Biosci Biotechnol Biochem. 2013;77:426–429. doi: 10.1271/bbb.120795. [DOI] [PubMed] [Google Scholar]
- 118.Fujii T, Inoue H, Ishikawa K. Decreased cellulase and xylanase production in the fungus Talaromyces cellulolyticus by disruption of tacA and tctA genes, encoding putative zinc finger transcriptional factors. Appl Biochem Biotechnol. 2015;175(6):3218–3229. doi: 10.1007/s12010-015-1497-2. [DOI] [PubMed] [Google Scholar]
- 119.Craig JP, Coradetti ST, Starr TL, Glass NL. Direct target network of the Neurospora crassa plant cell wall deconstruction regulators CLR-1, CLR-2, and XLR-1. MBio. 2015;6:e01452–e01515. doi: 10.1128/mBio.01452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tani S, Kawaguchi T, Kobayashi T. Complex regulation of hydrolytic enzyme genes for cellulosic biomass degradation in filamentous fungi. Appl Microbiol Biotechnol. 2014;98:4829–4837. doi: 10.1007/s00253-014-5707-6. [DOI] [PubMed] [Google Scholar]
- 121.Hakkinen M, Valkonen MJ, Westerholm-Parvinen A, Aro N, Arvas M, Vitikainen M, Penttila M, Saloheimo M, Pakula TM. Screening of candidate regulators for cellulase and hemicellulase production in Trichoderma reesei and identification of a factor essential for cellulase production. Biotechnol Biofuels. 2014;7:14. doi: 10.1186/1754-6834-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hasper AA, Dekkers E, van Mil M, van de Vondervoort PJ, de Graaff LH. EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl Environ Microbiol. 2002;68:1556–1560. doi: 10.1128/AEM.68.4.1556-1560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li N, Kunitake E, Endo Y, Aoyama M, Kanamaru K, Kimura M, Kato M, Kobayashi T. Involvement of an SRF-MADS protein McmA in regulation of extracellular enzyme production and asexual/sexual development in Aspergillus nidulans. Biosci Biotechnol Biochem. 2016;80:1820–1828. doi: 10.1080/09168451.2016.1146074. [DOI] [PubMed] [Google Scholar]
- 124.Yamakawa Y, Endo Y, Li N, Yoshizawa M, Aoyama M, Watanabe A, Kanamaru K, Kato M, Kobayashi T. Regulation of cellulolytic genes by McmA, the SRF-MADS box protein in Aspergillus nidulans. Biochem Biophys Res Commun. 2013;431:777–782. doi: 10.1016/j.bbrc.2013.01.031. [DOI] [PubMed] [Google Scholar]
- 125.Saloheimo A, Aro N, Ilmen M, Penttila M. Isolation of the ace1 gene encoding a Cys(2)–His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J Biol Chem. 2000;275:5817–5825. doi: 10.1074/jbc.275.8.5817. [DOI] [PubMed] [Google Scholar]
- 126.Aro N, Saloheimo M, Ilmén M, Penttilä M. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J Biol Chem. 2001;276:24309–24314. doi: 10.1074/jbc.M003624200. [DOI] [PubMed] [Google Scholar]
- 127.Wurleitner E, Pera L, Wacenovsky C, Cziferszky A, Zeilinger S, Kubicek CP, Mach RL. Transcriptional regulation of xyn2 in Hypocrea jecorina. Eukaryot Cell. 2003;2:150–158. doi: 10.1128/EC.2.1.150-158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Suzuki K, Tanaka M, Konno Y, Ichikawa T, Ichinose S, Hasegawa-Shiro S, Shintani T, Gomi K. Distinct mechanism of activation of two transcription factors, AmyR and MalR, involved in amylolytic enzyme production in Aspergillus oryzae. Appl Microbiol Biotechnol. 2015;99:1805–1815. doi: 10.1007/s00253-014-6264-8. [DOI] [PubMed] [Google Scholar]
- 129.Petersen KL, Lehmbeck J, Christensen T. A new transcriptional activator for amylase genes in Aspergillus. Mol Gen Genet. 1999;262:668–676. doi: 10.1007/s004380051129. [DOI] [PubMed] [Google Scholar]
- 130.Gomi K, Akeno T, Minetoki T, Ozeki K, Kumagai C, Okazaki N, Iimura Y. Molecular cloning and characterization of a transcriptional activator gene, amyR, involved in the amylolytic gene expression in Aspergillus oryzae. Biosci Biotechnol Biochem. 2000;64:816–827. doi: 10.1271/bbb.64.816. [DOI] [PubMed] [Google Scholar]
- 131.Vankuyk PA, Benen JA, Wosten HA, Visser J, de Vries RP. A broader role for AmyR in Aspergillus niger: regulation of the utilisation of d-glucose or d-galactose containing oligo- and polysaccharides. Appl Microbiol Biotechnol. 2012;93:285–293. doi: 10.1007/s00253-011-3550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tani S, Itoh T, Kato M, Kobayashi T, Tsukagoshi N. In vivo and in vitro analyses of the AmyR binding site of the Aspergillus nidulans agdA promoter; requirement of the CGG direct repeat for induction and high affinity binding of AmyR. Biosci Biotechnol Biochem. 2001;65:1568–1574. doi: 10.1271/bbb.65.1568. [DOI] [PubMed] [Google Scholar]
- 133.Wang P, Kojima T, Kobayashi T, Nakano H. Comprehensive analysis of the DNA-binding specificity of an Aspergillus nidulans transcription factor, AmyR, using a bead display system. Biosci Biotechnol Biochem. 2012;76:1128–1134. doi: 10.1271/bbb.110949. [DOI] [PubMed] [Google Scholar]
- 134.Ito T, Tani S, Itoh T, Tsukagoshi N, Kato M, Kobayashi T. Mode of AmyR binding to the CGGN8AGG sequence in the Aspergillus oryzae taaG2 promoter. Biosci Biotechnol Biochem. 2004;68:1906–1911. doi: 10.1271/bbb.68.1906. [DOI] [PubMed] [Google Scholar]
- 135.Makita T, Katsuyama Y, Tani S, Suzuki H, Kato N, Todd RB, Hynes MJ, Tsukagoshi N, Kato M, Kobayashi T. Inducer-dependent nuclear localization of a Zn(II)(2)Cys(6) transcriptional activator, AmyR, in Aspergillus nidulans. Biosci Biotechnol Biochem. 2009;73:391–399. doi: 10.1271/bbb.80654. [DOI] [PubMed] [Google Scholar]
- 136.Murakoshi Y, Makita T, Kato M, Kobayashi T. Comparison and characterization of alpha-amylase inducers in Aspergillus nidulans based on nuclear localization of AmyR. Appl Microbiol Biotechnol. 2012;94:1629–1635. doi: 10.1007/s00253-012-3874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hasegawa S, Takizawa M, Suyama H, Shintani T, Gomi K. Characterization and expression analysis of a maltose-utilizing (MAL) cluster in Aspergillus oryzae. Fungal Genet Biol. 2010;47:1–9. doi: 10.1016/j.fgb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 138.Vongsangnak W, Salazar M, Hansen K, Nielsen J. Genome-wide analysis of maltose utilization and regulation in aspergilli. Microbiology. 2009;155:3893–3902. doi: 10.1099/mic.0.031104-0. [DOI] [PubMed] [Google Scholar]
- 139.Nitta M, Furukawa T, Shida Y, Mori K, Kuhara S, Morikawa Y, Ogasawara W. A new Zn(II)(2)Cys(6)-type transcription factor BglR regulates beta-glucosidase expression in Trichoderma reesei. Fungal Genet Biol. 2012;49:388–397. doi: 10.1016/j.fgb.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 140.Kunitake E, Tani S, Sumitani J, Kawaguchi T. A novel transcriptional regulator, ClbR, controls the cellobiose- and cellulose-responsive induction of cellulase and xylanase genes regulated by two distinct signaling pathways in Aspergillus aculeatus. Appl Microbiol Biotechnol. 2013;97:2017–2028. doi: 10.1007/s00253-012-4305-8. [DOI] [PubMed] [Google Scholar]
- 141.Tani SEK, Kawamura A, Sumitani J, Kawaguchi T. Functional analysis of ClbR and ClbR2 controlling the cellulosic biomass degrading enzyme gene expression in Aspergillus aculeatus. In: The eleventh international Aspergillus meeting, Asperfest 11, vol. 75. Spain: Sevilla; 2014.
- 142.Wubben JP, ten Have A, van Kan JAL, Visser J. Regulation of endopolygalacturonase gene expression in Boytritis cinerea by galacturonic acid, ambient pH and carbon catabolite repression. Curr Genet. 2000;37:152–157. doi: 10.1007/s002940050022. [DOI] [PubMed] [Google Scholar]
- 143.Schols HA, Visser RGF, Voragen AGJ. Pectins and Pectinases. Wageningen: Wageningen Academic Publishers; 2009. [Google Scholar]
- 144.Gruben BS, Zhou M, Wiebenga A, Ballering J, Overkamp KM, Punt PJ, de Vries RP. Aspergillus niger RhaR, a regulator involved in l-rhamnose release and catabolism. Appl Microbiol Biotechnol. 2014;98:5531–5540. doi: 10.1007/s00253-014-5607-9. [DOI] [PubMed] [Google Scholar]
- 145.Niu J, Alazi E, Reid ID, Arentshorst M, Punt PJ, Visser J, Tsang A, Ram AF. An evolutionarily conserved transcriptional activator-repressor module controls expression of genes for d-galacturonic acid utilization in Aspergillus niger. Genetics. 2017;205:169–183. doi: 10.1534/genetics.116.194050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang L, Lubbers RJ, Simon A, Stassen JH, Vargas Ribera PR, Viaud M, van Kan JA. A novel Zn2 Cys6 transcription factor BcGaaR regulates d-galacturonic acid utilization in Botrytis cinerea. Mol Microbiol. 2016;100:247–262. doi: 10.1111/mmi.13314. [DOI] [PubMed] [Google Scholar]
- 147.Yuan XL, Roubos JA, van den Hondel CA, Ram AF. Identification of InuR, a new Zn(II)2Cys6 transcriptional activator involved in the regulation of inulinolytic genes in Aspergillus niger. Mol Genet Genom. 2008;279:11–26. doi: 10.1007/s00438-007-0290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Christensen U, Gruben BS, Madrid S, Mulder H, Nikolaev I, de Vries RP. Unique regulatory mechanism for d-galactose utilization in Aspergillus nidulans. Appl Environ Microbiol. 2011;77:7084–7087. doi: 10.1128/AEM.05290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li Y, Chen G, Liu W. Multiple metabolic signals influence GAL gene activation by modulating the interaction of Gal80p with the transcriptional activator Gal4p. Mol Microbiol. 2010;78:414–428. doi: 10.1111/j.1365-2958.2010.07343.x. [DOI] [PubMed] [Google Scholar]
- 150.Weinhandl K, Winkler M, Glieder A, Camattari A. Carbon source dependent promoters in yeasts. Microb Cell Fact. 2014;13:5. doi: 10.1186/1475-2859-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gruben BS, Zhou M, de Vries RP. GalX regulates the d-galactose oxido-reductive pathway in Aspergillus niger. FEBS Lett. 2012;586:3980–3985. doi: 10.1016/j.febslet.2012.09.029. [DOI] [PubMed] [Google Scholar]
- 152.Stricker AR, Steiger MG, Mach RL. Xyr1 receives the lactose induction signal and regulates lactose metabolism in Hypocrea jecorina. FEBS Lett. 2007;581:3915–3920. doi: 10.1016/j.febslet.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 153.Fekete E, de Vries RP, Seiboth B, vanKuyk PA, Sandor E, Fekete E, Metz B, Kubicek CP, Karaffa L. d-Galactose uptake is nonfunctional in the conidiospores of Aspergillus niger. FEMS Microbiol Lett. 2012;329:198–203. doi: 10.1111/j.1574-6968.2012.02524.x. [DOI] [PubMed] [Google Scholar]
- 154.Ronne H. Glucose repression in fungi. Trends Genet. 1995;11:12–17. doi: 10.1016/S0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 155.Portnoy T, Margeot A, Linke R, Atanasova L, Fekete E, Sandor E, Hartl L, Karaffa L, Druzhinina IS, Seiboth B, et al. The CRE1 carbon catabolite repressor of the fungus Trichoderma reesei: a master regulator of carbon assimilation. BMC Genom. 2011;12:269. doi: 10.1186/1471-2164-12-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brown NA, Ries LN, Goldman GH. How nutritional status signalling coordinates metabolism and lignocellulolytic enzyme secretion. Fungal Genet Biol. 2014;72:48–63. doi: 10.1016/j.fgb.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 157.Ruijter GJG, Visser J. Carbon repression in Aspergilli. FEMS Microbiol Lett. 1997;151:103–114. doi: 10.1111/j.1574-6968.1997.tb12557.x. [DOI] [PubMed] [Google Scholar]
- 158.Zeilinger S, Schmoll M, Pail M, Mach RL, Kubicek CP. Nucleosome transactions on the Hypocrea jecorina (Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Mol Genet Genom. 2003;270:46–55. doi: 10.1007/s00438-003-0895-2. [DOI] [PubMed] [Google Scholar]
- 159.Sun J, Glass NL. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS ONE. 2011;6:e25654. doi: 10.1371/journal.pone.0025654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ries LN, Beattie SR, Espeso EA, Cramer RA, Goldman GH. Diverse regulation of the CreA carbon catabolite repressor in Aspergillus nidulans. Genetics. 2016;203:335–352. doi: 10.1534/genetics.116.187872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.de Vries RP, van de Vondervoort PJ, Hendriks L, van de Belt M, Visser J. Regulation of the alpha-glucuronidase-encoding gene (aguA) from Aspergillus niger. Mol Genet Genom. 2002;268:96–102. doi: 10.1007/s00438-002-0729-7. [DOI] [PubMed] [Google Scholar]
- 162.Tamayo EN, Villanueva A, Hasper AA, de Graaff LH, Ramon D, Orejas M. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet Biol. 2008;45:984–993. doi: 10.1016/j.fgb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 163.Antonieto AC, Dos Santos Castro L, Silva-Rocha R, Persinoti GF, Silva RN. Defining the genome-wide role of CRE1 during carbon catabolite repression in Trichoderma reesei using RNA-Seq analysis. Fungal Genet Biol. 2014;73:93–103. doi: 10.1016/j.fgb.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 164.Ries L, Pullan ST, Delmas S, Malla S, Blythe MJ, Archer DB. Genome-wide transcriptional response of Trichoderma reesei to lignocellulose using RNA sequencing and comparison with. BMC Genom. 2013;14:541. doi: 10.1186/1471-2164-14-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tani S, Katsuyama Y, Hayashi T, Suzuki H, Kato M, Gomi K, Kobayashi T, Tsukagoshi N. Characterisation of the amyR gene encoding a transcriptional activator for the amylase genes in Aspergillus nidulans. Curr Genet. 2001;39:10–15. doi: 10.1007/s002940000175. [DOI] [PubMed] [Google Scholar]
- 166.Wang S, Liu G, Yu J, Tian S, Huang B, Xing M. RNA interference with carbon catabolite repression in Trichoderma koningii for enhancing cellulase production. Enzyme Microb Technol. 2013;53:104–109. doi: 10.1016/j.enzmictec.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 167.Mello-de-Sousa TM, Gorsche R, Rassinger A, Pocas-Fonseca MJ, Mach RL, Mach-Aigner AR. A truncated form of the carbon catabolite repressor 1 increases cellulase production in Trichoderma reesei. Biotechnol Biofuels. 2014;7:129. doi: 10.1186/s13068-014-0129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mello-de-Sousa TM, Rassinger A, Derntl C, Pocas-Fonseca MJ, Mach RL, Mach-Aigner AR. The relation between promoter chromatin status, Xyr1 and cellulase ex-pression in Trichoderma reesei. Curr Genom. 2016;17:145–152. doi: 10.2174/1389202917666151116211812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mello-de-Sousa TM, Rassinger A, Pucher ME, dos Santos Castro L, Persinoti GF, Silva-Rocha R, Pocas-Fonseca MJ, Mach RL, Nascimento Silva R, Mach-Aigner AR. The impact of chromatin remodelling on cellulase expression in Trichoderma reesei. BMC Genom. 2015;16:588. doi: 10.1186/s12864-015-1807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ries L, Belshaw NJ, Ilmen M, Penttila ME, Alapuranen M, Archer DB. The role of CRE1 in nucleosome positioning within the cbh1 promoter and coding regions of Trichoderma reesei. Appl Microbiol Biotechnol. 2014;98:749–762. doi: 10.1007/s00253-013-5354-3. [DOI] [PubMed] [Google Scholar]
- 171.Garcia I, Gonzalez R, Gomez D, Scazzocchio C. Chromatin rearrangements in the prnD–prnB bidirectional promoter: dependence on transcription factors. Eukaryot Cell. 2004;3:144–156. doi: 10.1128/EC.3.1.144-156.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shroff RA, O’Connor SM, Hynes MJ, Lockington RA, Kelly JM. Null alleles of creA, the regulator of carbon catabolite repression in Aspergillus nidulans. Fungal Genet Biol. 1997;22:28–38. doi: 10.1006/fgbi.1997.0989. [DOI] [PubMed] [Google Scholar]
- 173.Nakari-Setala T, Paloheimo M, Kallio J, Vehmaanpera J, Penttila M, Saloheimo M. Genetic modification of carbon catabolite repression in Trichoderma reesei for improved protein production. Appl Environ Microbiol. 2009;75:4853–4860. doi: 10.1128/AEM.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fujii T, Inoue H, Ishikawa K. Enhancing cellulase and hemicellulase production by genetic modification of the carbon catabolite repressor gene, creA, in Acremonium cellulolyticus. AMB Express. 2013;3:73. doi: 10.1186/2191-0855-3-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bi F, Barad S, Ment D, Luria N, Dubey A, Casado V, Glam N, Minguez JD, Espeso E, Fluhr R, Prusky D. Carbon regulation of environmental pH by secreted small molecules that modulate pathogenicity in phytopathogenic fungi. Mol Plant Pathol. 2015;17:1178–1195. doi: 10.1111/mpp.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Katoh H, Ohtani K, Yamamoto H, Akimitsu K. Overexpression of a gene encoding a catabolite repression element in Alternaria citri causes severe symptoms of black rot in citrus fruit. Phytopathology. 2007;97:557–563. doi: 10.1094/PHYTO-97-5-0557. [DOI] [PubMed] [Google Scholar]
- 177.Cho Y, Kim KH, La Rota M, Scott D, Santopietro G, Callihan M, Mitchell TK, Lawrence CB. Identification of novel virulence factors associated with signal transduction pathways in Alternaria brassicicola. Mol Microbiol. 2009;72:1316–1333. doi: 10.1111/j.1365-2958.2009.06689.x. [DOI] [PubMed] [Google Scholar]
- 178.Cepeda-Garcia C, Dominguez-Santos R, Garcia-Rico RO, Garcia-Estrada C, Cajiao A, Fierro F, Martin JF. Direct involvement of the CreA transcription factor in penicillin biosynthesis and expression of the pcbAB gene in Penicillium chrysogenum. Appl Microbiol Biotechnol. 2014;98:7113–7124. doi: 10.1007/s00253-014-5760-1. [DOI] [PubMed] [Google Scholar]
- 179.Lorito M, Mach RL, Sposato P, Strauss J, Peterbauer CK, Kubicek CP. Mycoparasitic interaction relieves binding of the Cre1 carbon catabolite repressor protein to promoter sequences of the ech42 (endochitinase-encoding) gene in Trichoderma harzianum. Proc Natl Acad Sci USA. 1996;93:14868–14872. doi: 10.1073/pnas.93.25.14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Vautard-Mey G, Cotton P, Fevre M. Expression and compartmentation of the glucose repressor CRE1 from the phytopathogenic fungus Sclerotinia sclerotiorum. Eur J Biochem. 1999;266:252–259. doi: 10.1046/j.1432-1327.1999.00857.x. [DOI] [PubMed] [Google Scholar]
- 181.Roy P, Lockington RA, Kelly JM. CreA-mediated repression in Aspergillus nidulans does not require transcriptional auto-regulation, regulated intracellular localisation or degradation of CreA. Fungal Genet Biol. 2008;45:657–670. doi: 10.1016/j.fgb.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 182.Strauss J, Horvath HK, Abdallah BM, Kindermann J, Mach RL, Kubicek CP. The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post-transcriptional level. Mol Microbiol. 1999;32:169–178. doi: 10.1046/j.1365-2958.1999.01341.x. [DOI] [PubMed] [Google Scholar]
- 183.Ilmen M, Thrane C, Penttila M. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol Gen Genet. 1996;251:451–460. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- 184.Jekosch K, Kuck U. Loss of glucose repression in an Acremonium chrysogenum beta-lactam producer strain and its restoration by multiple copies of the cre1 gene. Appl Microbiol Biotechnol. 2000;54:556–563. doi: 10.1007/s002530000422. [DOI] [PubMed] [Google Scholar]
- 185.Tudzynski B, Liu S, Kelly JM. Carbon catabolite repression in plant pathogenic fungi: isolation and characterization of the Gibberella fujikuroi and Botrytis cinerea creA genes. FEMS Microbiol Lett. 2000;184:9–15. doi: 10.1111/j.1574-6968.2000.tb08982.x. [DOI] [PubMed] [Google Scholar]
- 186.Brown NA, de Gouvea PF, Krohn NG, Savoldi M, Goldman GH. Functional characterisation of the non-essential protein kinases and phosphatases regulating Aspergillus nidulans hydrolytic enzyme production. Biotechnol Biofuels. 2013;6:91. doi: 10.1186/1754-6834-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Vautard-Mey G, Fevre M. Mutation of a putative AMPK phosphorylation site abolishes the repressor activity but not the nuclear targeting of the fungal glucose regulator CRE1. Curr Genet. 2000;37:328–332. doi: 10.1007/s002940050535. [DOI] [PubMed] [Google Scholar]
- 188.Cziferszky A, Mach RL, Kubicek CP. Phosphorylation positively regulates DNA binding of the carbon catabolite repressor Cre1 of Hypocrea jecorina (Trichoderma reesei) J Biol Chem. 2002;277:14688–14694. doi: 10.1074/jbc.M200744200. [DOI] [PubMed] [Google Scholar]
- 189.Lockington RA, Kelly JM. Carbon catabolite repression in Aspergillus nidulans involves deubiquitination. Mol Microbiol. 2001;40:1311–1321. doi: 10.1046/j.1365-2958.2001.02474.x. [DOI] [PubMed] [Google Scholar]
- 190.Hunter AJ, Morris TA, Jin B, Saint CP, Kelly JM. Deletion of creB in Aspergillus oryzae increases secreted hydrolytic enzyme activity. Appl Environ Microbiol. 2013;79:5480–5487. doi: 10.1128/AEM.01406-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Denton JA, Kelly JM. Disruption of Trichoderma reesei cre2, encoding an ubiquitin C-terminal hydrolase, results in increased cellulase activity. BMC Biotechnol. 2011;11:103. doi: 10.1186/1472-6750-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lockington RA, Kelly JM. The WD40-repeat protein CreC interacts with and stabilizes the deubiquitinating enzyme CreB in vivo in Aspergillus nidulans. Mol Microbiol. 2002;43:1173–1182. doi: 10.1046/j.1365-2958.2002.02811.x. [DOI] [PubMed] [Google Scholar]
- 193.Boase NA, Kelly JM. A role for creD, a carbon catabolite repression gene from Aspergillus nidulans, in ubiquitination. Mol Microbiol. 2004;53:929–940. doi: 10.1111/j.1365-2958.2004.04172.x. [DOI] [PubMed] [Google Scholar]
- 194.Colabardini AC, Humanes AC, Gouvea PF, Savoldi M, Goldman MH, Kress MR, Bayram O, Oliveira JV, Gomes MD, Braus GH, Goldman GH. Molecular characterization of the Aspergillus nidulans fbxA encoding an F-box protein involved in xylanase induction. Fungal Genet Biol. 2012;49:130–140. doi: 10.1016/j.fgb.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 195.Jonkers W, Rep M. Mutation of CRE1 in Fusarium oxysporum reverts the pathogenicity defects of the FRP1 deletion mutant. Mol Microbiol. 2009;74:1100–1113. doi: 10.1111/j.1365-2958.2009.06922.x. [DOI] [PubMed] [Google Scholar]
- 196.Wang SW, Xing M, Liu G, Yu SW, Wang J, Tian SL. Improving cellulase production in Trichoderma koningii through RNA interference on ace1 gene expression. J Microbiol Biotechnol. 2012;22:1133–1140. doi: 10.4014/jmb.1112.12037. [DOI] [PubMed] [Google Scholar]
- 197.Aro N, Saloheimo M, Ilmén M, Penttilä M. ACEI, of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Environ Microbiol. 2003;69:56–65. doi: 10.1128/AEM.69.1.56-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chilton IJ, Delaney CE, Barham-Morris J, Fincham DA, Hooley P, Whitehead MP. The Aspergillus nidulans stress response transcription factor StzA is ascomycete-specific and shows species-specific polymorphisms in the C-terminal region. Mycol Res. 2008;112:1435–1446. doi: 10.1016/j.mycres.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 199.Li J, Lin L, Li H, Tian C, Ma Y. Transcriptional comparison of the filamentous fungus Neurospora crassa growing on three major monosaccharides d-glucose, d-xylose and l-arabinose. Biotechnol Biofuels. 2014;7:31. doi: 10.1186/1754-6834-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Messenguy F, Dubois E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene. 2003;316:1–21. doi: 10.1016/S0378-1119(03)00747-9. [DOI] [PubMed] [Google Scholar]
- 201.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 202.Wu W, Huang X, Cheng J, Li Z, de Folter S, Huang Z, Jiang X, Pang H, Tao S. Conservation and evolution in and among SRF- and MEF2-type MADS domains and their binding sites. Mol Biol Evol. 2011;28:501–511. doi: 10.1093/molbev/msq214. [DOI] [PubMed] [Google Scholar]
- 203.De Bodt S, Raes J, Van de Peer Y, Theissen G. And then there were many: MADS goes genomic. Trends Plant Sci. 2003;8:475–483. doi: 10.1016/j.tplants.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 204.Marsch-Martinez N, Wu W, de Folter S. The MADS symphonies of transcriptional regulation. Front Plant Sci. 2011;2:26. doi: 10.3389/fpls.2011.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Veron AS, Kaufmann K, Bornberg-Bauer E. Evidence of interaction network evolution by whole-genome duplications: a case study in MADS-box proteins. Mol Biol Evol. 2007;24:670–678. doi: 10.1093/molbev/msl197. [DOI] [PubMed] [Google Scholar]
- 206.Derntl C, Rassinger A, Srebotnik E, Mach RL, Mach-Aigner AR. Xpp1 regulates the expression of xylanases, but not of cellulases in Trichoderma reesei. Biotechnol Biofuels. 2015;8:112. doi: 10.1186/s13068-015-0298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Derntl C, Kluger B, Bueschl C, Schuhmacher R, Mach RL, Mach-Aigner AR. Transcription factor Xpp1 is a switch between primary and secondary fungal metabolism. Proc Natl Acad Sci USA. 2017;114:E560–E569. doi: 10.1073/pnas.1609348114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chaudhary J, Skinner MK. Basic helix-loop-helix proteins can act at the E-box within the serum response element of the c-fos promoter to influence hormone-induced promoter activation in Sertoli cells. Mol Endocrinol. 1999;13:774–786. doi: 10.1210/mend.13.5.0271. [DOI] [PubMed] [Google Scholar]
- 209.Fekete E, Karaffa L, Karimi Aghcheh R, Nemeth Z, Fekete E, Orosz A, Paholcsek M, Stagel A, Kubicek CP. The transcriptome of lae1 mutants of Trichoderma reesei cultivated at constant growth rates reveals new targets of LAE1 function. BMC Genom. 2014;15:447. doi: 10.1186/1471-2164-15-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Karimi-Aghcheh R, Bok JW, Phatale PA, Smith KM, Baker SE, Lichius A, Omann M, Zeilinger S, Seiboth B, Rhee C, et al. Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 (Bethesda). 2013;3:369–378. doi: 10.1534/g3.112.005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Karimi Aghcheh R, Nemeth Z, Atanasova L, Fekete E, Paholcsek M, Sandor E, Aquino B, Druzhinina IS, Karaffa L, Kubicek CP. The VELVET A orthologue VEL1 of Trichoderma reesei regulates fungal development and is essential for cellulase gene expression. PLoS ONE. 2014;9:e112799. doi: 10.1371/journal.pone.0112799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, Braus-Stromeyer S, Kwon NJ, Keller NP, Yu JH, Braus GH. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 213.Narendja FM, Davis MA, Hynes MJ. AnCF, the CCAAT binding complex of Aspergillus nidulans, is essential for the formation of a DNase I-hypersensitive site in the %’ region of the amdS gene. Mol Cell Biol. 1999;19:6523–6531. doi: 10.1128/MCB.19.10.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zeilinger S, Mach R, Kubicek CP. Two adjacent protein binding motifs in the cbh2 (Cellobiohydrolase-II encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J Biol Chem. 1998;273:34463–34471. doi: 10.1074/jbc.273.51.34463. [DOI] [PubMed] [Google Scholar]
- 216.Steidl S, Papagiannopoulos P, Litzka O, Andrianopoulos A, Davis MA, Brakhage AA, Hynes MJ. AnCF, the CCAAT binding complex of Aspergillus nidulans, contains products of the hapB, hapC, and hapE genes and is required for activation by the pathway-specific regulatory gene amdR. Mol Cell Biol. 1999;19:99–106. doi: 10.1128/MCB.19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Xin Q, Gong Y, Lv X, Chen G, Liu W. Trichoderma reesei histone acetyltransferase Gcn5 regulates fungal growth, conidiation, and cellulase gene expression. Curr Microbiol. 2013;67:580–589. doi: 10.1007/s00284-013-0396-4. [DOI] [PubMed] [Google Scholar]
- 218.Schafmeier T, Diernfellner AC. Light input and processing in the circadian clock of Neurospora. FEBS Lett. 2011;585:1467–1473. doi: 10.1016/j.febslet.2011.03.050. [DOI] [PubMed] [Google Scholar]
- 219.Tisch D, Schmoll M. Targets of light signalling in Trichoderma reesei. BMC Genom. 2013;14:657. doi: 10.1186/1471-2164-14-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Schmoll M. Assessing the relevance of light for fungi: implications and insights into the network of signal transmission. Adv Appl Microbiol. 2011;76:27–78. doi: 10.1016/B978-0-12-387048-3.00002-7. [DOI] [PubMed] [Google Scholar]
- 221.Dasgupta A, Fuller KK, Dunlap JC, Loros JJ. Seeing the world differently: variability in the photosensory mechanisms of two model fungi. Environ Microbiol. 2016;18:5–20. doi: 10.1111/1462-2920.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sancar G, Sancar C, Brugger B, Ha N, Sachsenheimer T, Gin E, Wdowik S, Lohmann I, Wieland F, Hofer T, et al. A global circadian repressor controls antiphasic expression of metabolic genes in Neurospora. Mol Cell. 2011;44:687–697. doi: 10.1016/j.molcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 223.Tisch D, Schmoll M. Light regulation of metabolic pathways in fungi. Appl Microbiol Biotechnol. 2010;85:1259–1277. doi: 10.1007/s00253-009-2320-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bayram O, Braus GH. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev. 2012;36:1–24. doi: 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- 225.Kim H, Son H, Lee YW. Effects of light on secondary metabolism and fungal development of Fusarium graminearum. J Appl Microbiol. 2014;116:380–389. doi: 10.1111/jam.12381. [DOI] [PubMed] [Google Scholar]
- 226.Hurley JM, Loros JJ, Dunlap JC. The circadian system as an organizer of metabolism. Fungal Genet Biol. 2016;90:39–43. doi: 10.1016/j.fgb.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Bazafkan H, Dattenbock C, Bohmdorfer S, Tisch D, Stappler E, Schmoll M. Mating type dependent partner sensing as mediated by VEL1 in Trichoderma reesei. Mol Microbiol. 2015;96:1103–1118. doi: 10.1111/mmi.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chen CL, Kuo HC, Tung SY, Hsu PW, Wang CL, Seibel C, Schmoll M, Chen RS, Wang TF. Blue light acts as a double-edged sword in regulating sexual development of Hypocrea jecorina (Trichoderma reesei) PLoS ONE. 2012;7:e44969. doi: 10.1371/journal.pone.0044969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Castellanos F, Schmoll M, Martinez P, Tisch D, Kubicek CP, Herrera-Estrella A, Esquivel-Naranjo EU. Crucial factors of the light perception machinery and their impact on growth and cellulase gene transcription in Trichoderma reesei. Fungal Genet Biol. 2010;47:468–476. doi: 10.1016/j.fgb.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 230.Gyalai-Korpos M, Nagy G, Mareczky Z, Schuster A, Reczey K, Schmoll M. Relevance of the light signaling machinery for cellulase expression in Trichoderma reesei (Hypocrea jecorina) BMC Res Notes. 2010;3:330. doi: 10.1186/1756-0500-3-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tisch D, Schuster A, Schmoll M. Crossroads between light response and nutrient signalling: ENV1 and PhLP1 act as mutual regulatory pair in Trichoderma reesei. BMC Genom. 2014;15:425. doi: 10.1186/1471-2164-15-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Schmoll M, Franchi L, Kubicek CP. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (Anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot Cell. 2005;4:1998–2007. doi: 10.1128/EC.4.12.1998-2007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Karimi Aghcheh R, Druzhinina IS, Kubicek CP. The putative protein methyltransferase LAE1 of Trichoderma atroviride is a key regulator of asexual development and mycoparasitism. PLoS ONE. 2013;8:e67144. doi: 10.1371/journal.pone.0067144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Bazafkan H, Tisch D, Schmoll M. Chapter 20—regulation of glycoside hydrolase expression in Trichoderma. In: Gupta VK, Herrera-Estrella MS, Druzhinina RSU, Tuohy MG, editors. Biotechnology and biology of trichoderma. Amsterdam: Elsevier; 2014. pp. 291–308. [Google Scholar]
- 235.Schmoll M, Tian C, Sun J, Tisch D, Glass NL. Unravelling the molecular basis for light modulated cellulase gene expression—the role of photoreceptors in Neurospora crassa. BMC Genom. 2012;13:127. doi: 10.1186/1471-2164-13-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Purschwitz J, Muller S, Fischer R. Mapping the interaction sites of Aspergillus nidulans phytochrome FphA with the global regulator VeA and the White Collar protein LreB. Mol Genet Genom. 2009;281:35–42. doi: 10.1007/s00438-008-0390-x. [DOI] [PubMed] [Google Scholar]
- 237.Purschwitz J, Muller S, Kastner C, Schoser M, Haas H, Espeso EA, Atoui A, Calvo AM, Fischer R. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr Biol. 2008;18:255–259. doi: 10.1016/j.cub.2008.01.061. [DOI] [PubMed] [Google Scholar]
- 238.Idnurm A, Heitman J. Ferrochelatase is a conserved downstream target of the blue light-sensing White collar complex in fungi. Microbiology. 2010;156:2393–2407. doi: 10.1099/mic.0.039222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Proietto M, Bianchi MM, Ballario P, Brenna A. Epigenetic and posttranslational modifications in light signal transduction and the circadian clock in Neurospora crassa. Int J Mol Sci. 2015;16:15347–15383. doi: 10.3390/ijms160715347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Friedl MA, Kubicek CP, Druzhinina IS. Carbon source dependence and photostimulation of conidiation in Hypocrea atroviridis. Appl Environ Microbiol. 2008;74:245–250. doi: 10.1128/AEM.02068-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Hurley J, Loros JJ, Dunlap JC. Dissecting the mechanisms of the clock in Neurospora. Methods Enzymol. 2015;551:29–52. doi: 10.1016/bs.mie.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Schmoll M, Dattenbock C, Carreras-Villasenor N, Mendoza-Mendoza A, Tisch D, Aleman MI, Baker SE, Brown C, Cervantes-Badillo MG, Cetz-Chel J, et al. The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiol Mol Biol Rev. 2016;80:205–327. doi: 10.1128/MMBR.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sancar C, Sancar G, Ha N, Cesbron F, Brunner M. Dawn- and dusk-phased circadian transcription rhythms coordinate anabolic and catabolic functions in Neurospora. BMC Biol. 2015;13:17. doi: 10.1186/s12915-015-0126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Calvo AM. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet Biol. 2008;45:1053–1061. doi: 10.1016/j.fgb.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 245.Lopez-Berges MS, Hera C, Sulyok M, Schafer K, Capilla J, Guarro J, Di Pietro A. The velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Mol Microbiol. 2013;87:49–65. doi: 10.1111/mmi.12082. [DOI] [PubMed] [Google Scholar]
- 246.Schumacher J, Simon A, Cohrs KC, Traeger S, Porquier A, Dalmais B, Viaud M, Tudzynski B. The VELVET complex in the gray mold fungus Botrytis cinerea: impact of BcLAE1 on differentiation, secondary metabolism, and virulence. Mol Plant Microbe Interact. 2015;28:659–674. doi: 10.1094/MPMI-12-14-0411-R. [DOI] [PubMed] [Google Scholar]
- 247.Seiboth B, Karimi RA, Phatale PA, Linke R, Hartl L, Sauer DG, Smith KM, Baker SE, Freitag M, Kubicek CP. The putative protein methyltransferase LAE1 controls cellulase gene expression in Trichoderma reesei. Mol Microbiol. 2012;84:1150–1164. doi: 10.1111/j.1365-2958.2012.08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Linde T, Zoglowek M, Lubeck M, Frisvad JC, Lubeck PS. The global regulator LaeA controls production of citric acid and endoglucanases in Aspergillus carbonarius. J Ind Microbiol Biotechnol. 2016;43:1139–1147. doi: 10.1007/s10295-016-1781-3. [DOI] [PubMed] [Google Scholar]
- 249.Niu J, Arentshorst M, Nair PD, Dai Z, Baker SE, Frisvad JC, Nielsen KF, Punt PJ, Ram AF. Identification of a classical mutant in the industrial host Aspergillus niger by systems genetics: LaeA is required for citric acid production and regulates the formation of some secondary metabolites. G3 (Bethesda) 2016;6:193–204. doi: 10.1534/g3.115.024067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tudzynski B. Nitrogen regulation of fungal secondary metabolism in fungi. Front Microbiol. 2014;5:656. doi: 10.3389/fmicb.2014.00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Marzluf GA. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Berger H, Basheer A, Bock S, Reyes-Dominguez Y, Dalik T, Altmann F, Strauss J. Dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol Microbiol. 2008;69:1385–1398. doi: 10.1111/j.1365-2958.2008.06359.x. [DOI] [PubMed] [Google Scholar]
- 253.Arst HN, Jr, Cove DJ. Nitrogen metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1973;126:111–141. doi: 10.1007/BF00330988. [DOI] [PubMed] [Google Scholar]
- 254.Gautam SP, Bundela PS, Pandey AK, Khan J, Awasthi MK, Sarsaiya S. Optimization for the production of cellulase enzyme from municipal solid waste residue by two novel cellulolytic fungi. Biotechnol Res Int. 2011;2011:810425. doi: 10.4061/2011/810425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Goncalves RD, Cupertino FB, Freitas FZ, Luchessi AD, Bertolini MC. A genome-wide screen for Neurospora crassa transcription factors regulating glycogen metabolism. Mol Cell Proteom. 2011;10(M111):007963. doi: 10.1074/mcp.M111.007963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Fu YH, Marzluf GA. nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol. 1990;10:1056–1065. doi: 10.1128/MCB.10.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Fu YH, Marzluf GA. Characterization of nit-2, the major nitrogen regulatory gene of Neurospora crassa. Mol Cell Biol. 1987;7:1691–1696. doi: 10.1128/MCB.7.5.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Conlon H, Zadra I, Haas H, Arst HN, Jr, Jones MG, Caddick MX. The Aspergillus nidulans GATA transcription factor gene areB encodes at least three proteins and features three classes of mutation. Mol Microbiol. 2001;40:361–375. doi: 10.1046/j.1365-2958.2001.02399.x. [DOI] [PubMed] [Google Scholar]
- 259.Andrianopoulos A, Kourambas S, Sharp JA, Davis MA, Hynes MJ. Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J Bacteriol. 1998;180:1973–1977. doi: 10.1128/jb.180.7.1973-1977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tomsett AB, Dunn-Coleman NS, Garrett RH. The regulation of nitrate assimilation in Neurospora crassa: the isolation and genetic analysis of nmr-1 mutants. Mol Gen Genet. 1981;182:229–233. doi: 10.1007/BF00269662. [DOI] [PubMed] [Google Scholar]
- 261.Kudla B, Caddick MX, Langdon T, Martinez-Rossi NM, Bennett CF, Sibley S, Davies RW, Arst HN., Jr The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990;9:1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Caddick MX. Nitrogen metabolite repression. Prog Ind Microbiol. 1994;29:323–353. [PubMed] [Google Scholar]
- 263.Burger G, Tilburn J, Scazzocchio C. Molecular cloning and functional characterization of the pathway-specific regulatory gene nirA, which controls nitrate assimilation in Aspergillus nidulans. Mol Cell Biol. 1991;11:795–802. doi: 10.1128/MCB.11.2.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Berger H, Pachlinger R, Morozov I, Goller S, Narendja F, Caddick M, Strauss J. The GATA factor AreA regulates localization and in vivo binding site occupancy of the nitrate activator NirA. Mol Microbiol. 2006;59:433–446. doi: 10.1111/j.1365-2958.2005.04957.x. [DOI] [PubMed] [Google Scholar]
- 265.Muro-Pastor MI, Gonzalez R, Strauss J, Narendja F, Scazzocchio C. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J. 1999;18:1584–1597. doi: 10.1093/emboj/18.6.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lamb HK, Ren J, Park A, Johnson C, Leslie K, Cocklin S, Thompson P, Mee C, Cooper A, Stammers DK, Hawkins AR. Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis. Protein Sci. 2004;13:3127–3138. doi: 10.1110/ps.04958904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Macios M, Caddick MX, Weglenski P, Scazzocchio C, Dzikowska A. The GATA factors AREA and AREB together with the co-repressor NMRA, negatively regulate arginine catabolism in Aspergillus nidulans in response to nitrogen and carbon source. Fungal Genet Biol. 2012;49:189–198. doi: 10.1016/j.fgb.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 268.Wong KH, Hynes MJ, Todd RB, Davis MA. Deletion and overexpression of the Aspergillus nidulans GATA factor AreB reveals unexpected pleiotropy. Microbiology. 2009;155:3868–3880. doi: 10.1099/mic.0.031252-0. [DOI] [PubMed] [Google Scholar]
- 269.Lockington RA, Rodbourn L, Barnett S, Carter CJ, Kelly JM. Regulation by carbon and nitrogen sources of a family of cellulases in Aspergillus nidulans. Fungal Genet Biol. 2002;37:90–196. doi: 10.1016/S1087-1845(02)00504-2. [DOI] [PubMed] [Google Scholar]
- 270.Selvig K, Alspaugh JA. pH response pathways in fungi: adapting to host-derived and environmental signals. Mycobiology. 2011;39:249–256. doi: 10.5941/MYCO.2011.39.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Rossi A, Cruz AH, Santos RS, Silva PM, Silva EM, Mendes NS, Martinez-Rossi NM. Ambient pH sensing in filamentous fungi: pitfalls in elucidating regulatory hierarchical signaling networks. IUBMB Life. 2013;65:930–935. doi: 10.1002/iub.1217. [DOI] [PubMed] [Google Scholar]
- 272.Landraud P, Chuzeville S, Billon-Grande G, Poussereau N, Bruel C. Adaptation to pH and role of PacC in the rice blast fungus Magnaporthe oryzae. PLoS ONE. 2013;8:e69236. doi: 10.1371/journal.pone.0069236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ment D, Alkan N, Luria N, Bi FC, Reuveni E, Fluhr R, Prusky D. A role of AREB in the regulation of PACC-dependent acid-expressed-genes and pathogenicity of Colletotrichum gloeosporioides. Mol Plant Microbe Interact. 2015;28:154–166. doi: 10.1094/MPMI-09-14-0252-R. [DOI] [PubMed] [Google Scholar]
- 274.Bertuzzi M, Schrettl M, Alcazar-Fuoli L, Cairns TC, Munoz A, Walker LA, Herbst S, Safari M, Cheverton AM, Chen D, et al. The pH-responsive PacC transcription factor of Aspergillus fumigatus governs epithelial entry and tissue invasion during pulmonary aspergillosis. PLoS Pathog. 2014;10:e1004413. doi: 10.1371/journal.ppat.1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Zhang T, Sun X, Xu Q, Candelas LG, Li H. The pH signaling transcription factor PacC is required for full virulence in Penicillium digitatum. Appl Microbiol Biotechnol. 2013;97:9087–9098. doi: 10.1007/s00253-013-5129-x. [DOI] [PubMed] [Google Scholar]
- 276.He R, Ma L, Li C, Jia W, Li D, Zhang D, Chen S. Trpac1, a pH response transcription regulator, is involved in cellulase gene expression in Trichoderma reesei. Enzyme Microb Technol. 2014;67:17–26. doi: 10.1016/j.enzmictec.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 277.Campos Antonieto AC, Ramos Pedersoli W, Dos Santos Castro L, da Silva Santos R, Cruz AH, Nogueira KM, Silva-Rocha R, Rossi A, Silva RN. Deletion of pH regulator pac-3 affects cellulase and xylanase activity during sugarcane bagasse degradation by Neurospora crassa. PLoS ONE. 2017;12:e0169796. doi: 10.1371/journal.pone.0169796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Dorn G. Phosphatase mutants in Aspergillus nidulans. Science. 1965;150:1183–1184. doi: 10.1126/science.150.3700.1183. [DOI] [PubMed] [Google Scholar]
- 279.Penalva MA, Tilburn J, Bignell E, Arst HN., Jr Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 2008;16:291–300. doi: 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 280.Tilburn J, Sarkar S, Widdick DA, Espeso EA, Orejas M, Mungroo J, Penalva MA, Arst HN., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Negrete-Urtasun S, Reiter W, Diez E, Denison SH, Tilburn J, Espeso EA, Penalva MA, Arst HN., Jr Ambient pH signal transduction in Aspergillus: completion of gene characterization. Mol Microbiol. 1999;33:994–1003. doi: 10.1046/j.1365-2958.1999.01540.x. [DOI] [PubMed] [Google Scholar]
- 282.Bussink HJ, Bignell EM, Munera-Huertas T, Lucena-Agell D, Scazzocchio C, Espeso EA, Bertuzzi M, Rudnicka J, Negrete-Urtasun S, Penas-Parilla MM, et al. Refining the pH response in Aspergillus nidulans: a modulatory triad involving PacX, a novel zinc binuclear cluster protein. Mol Microbiol. 2015;98:1051–1072. doi: 10.1111/mmi.13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.de Graaff LH, van den Broeck HC, van Ooijen AJ, Visser J. Regulation of the xylanase-encoding xlnA gene of Aspergillus tubigensis. Mol Microbiol. 1994;12:479–490. doi: 10.1111/j.1365-2958.1994.tb01036.x. [DOI] [PubMed] [Google Scholar]
- 284.Kunitake E, Hagiwara D, Miyamoto K, Kanamaru K, Kimura M, Kobayashi T. Regulation of genes encoding cellulolytic enzymes by Pal-PacC signaling in Aspergillus nidulans. Appl Microbiol Biotechnol. 2016;100:3621–3635. doi: 10.1007/s00253-016-7409-8. [DOI] [PubMed] [Google Scholar]
- 285.Hakkinen M, Sivasiddarthan D, Aro N, Saloheimo M, Pakula TM. The effects of extracellular pH and of the transcriptional regulator PACI on the transcriptome of Trichoderma reesei. Microb Cell Fact. 2015;14:63. doi: 10.1186/s12934-015-0247-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Prevalence of transcription factors in Fungi.
Additional file 2. Maximum likelihood tree of Zn2Cys6 transcription factors (TFs). In bold are the species for which the TF has been characterized, while in bold and larger font are the species in which the TF was first discovered.
Additional file 3. Maximum likelihood tree of Cys2Hys2 transcription factors (TFs). In bold are the species for which the TF has been characterized, while in bold and larger font are the species in which the TF was first discovered.
Additional file 4. Maximum likelihood tree of MADS BOX (McmA) transcription factors (TFs). In bold are the species for which the TF has been characterized, while in bold and larger font are the species in which the TF was first discovered.
Additional file 5. Maximum likelihood tree of bHLH (XPP1) transcription factors (TFs). In are the species for which the TF has been characterized, while in bold and larger font are the species in which the TF was first discovered.


