Abstract
The double-stranded RNA (dsRNA)-dependent protein kinase (PKR) is induced as part of the IFN response in mammals and acts to shut down protein synthesis by the phosphorylation of eukaryotic initiation factor 2α (eIF2α). In fish, a PKR-like kinase activity has been detected, but the enzyme responsible has eluded characterization. Here, we describe a PKR-like kinase from zebrafish. Phylogenetic analysis shows that the C-terminal kinase domain is more closely related to the kinase domain of PKR than to any of the other three known eIF2α kinases. Surprisingly, instead of the two dsRNA binding domains found at the N terminus of PKR, there are two Zα domains. Zα domains specifically bind dsDNA and RNA in the left-handed Z conformation, often with high affinity. They have been found previously in two other IFN-inducible proteins, the dsRNA editing enzyme, ADAR1, and Z-DNA binding protein 1 (ZBP1), as well as in the poxvirus virulence factor, E3L. This previously undescribed kinase, designated PKZ (protein kinase containing Z-DNA binding domains), is transcribed constitutively at low levels and is highly induced after injection of poly(inosinic)–poly(cytidylic) acid, which simulates viral infection. Binding of Z-DNA by the Zα domain of PKZ was demonstrated by circular dichroism. PKZ inhibits translation in transfected cells; site-directed mutagenesis indicates that this inhibition depends on its catalytic activity. Identification of a gene combining Zα domains with a PKR-like kinase domain strengthens the hypothesis that the ability to bind left-handed nucleic acid plays a role in the host response to viruses.
Keywords: E3L, interferon response, viral infection, Z-RNA, Zα domain
Double-stranded RNA (dsRNA)-dependent protein kinase (PKR) mediates the antiviral and antiproliferative activity of interferons. PKR can be directly activated by dsRNA, which is produced in virally infected cells, and by immunostimuli such as cytokines or lipopolysaccharide (reviewed in refs. 1 and 2). When dsRNA binds to the two N-terminal dsRNA-binding domains of PKR, it is believed to induce a conformational change in the protein, which uncovers the catalytic site and allows dimerization of PKR. These processes lead to PKR autophosphorylation and activation (3).
The best-characterized function of PKR is the phosphorylation of the α subunit of eukaryotic translation initiation factor 2 (eIF2α), which inhibits protein synthesis (4). In addition, PKR induces proinflammatory genes by activating the NF-κB pathway and acts as an RNA sensor, thereby regulating expression of specific genes by the regulation of mRNA splicing and translation (1, 5). Phosphorylation of eIF2α has been described in rainbow trout cells after infection with infectious pancreatic necrosis virus or incubation with poly(inosinic)–poly(cytidylic) acid [poly(I:C)], which mimics viral dsRNA (6). The protein responsible for this PKR-like activity in fish has not been identified.
In addition to PKR, other kinases have been described that phosphorylate eIF2α at serine 51: heme-regulated eIF2α kinase (HRI), general control of nitrogen metabolism kinase 2 (GCN2), and endoplasmic reticulum eIF2α kinase (PEK). These kinases share conserved kinase domains but have different regulatory domains (4).
During viral infection, phosphorylation of eIF2α by PKR prevents the synthesis of viral proteins and halts further viral replication. To circumvent this response, viruses have evolved proteins that inhibit PKR function. One of these is the vaccinia virus IFN resistance gene E3L, which competitively inhibits PKR activity (7). Orthologues are found in most chordopoxviridae. E3L consists of an N-terminal Z-DNA binding (Zα) domain and a C-terminal dsRNA binding (RBM) domain. Only the C-terminal half is required to overcome the IFN response in tissue culture cells. However, the ability of the Zα domain to bind left-handed Z-DNA is essential for pathogenicity in mice (8, 9). The RBM of E3L acts competitively to inhibit the activity of PKR and other proteins that contain RBMs (7, 10, 11). E3L also may act competitively against a group of host proteins that bind to the Z-conformation (12). It is therefore possible that the ability to bind right-handed dsRNA and the ability to bind Z-DNA or Z-RNA (13) are used in similar contexts in host–viral interactions.
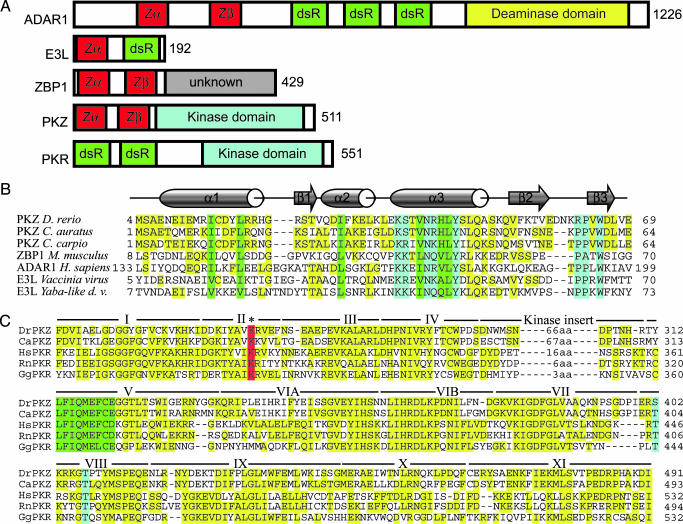
Left-handed Z-DNA is a higher-energy form of the double helix, which can be stabilized by negative supercoiling or the binding of a conformation-specific protein. Z-DNA formation is associated with transcriptional activity, which results in a local increase in negative supercoiling (14). Z-DNA forming sequences can either enhance or repress promoter activity, probably by modulating the local architecture of transcriptional control regions (15, 16). The family of Zα proteins (Fig. 1) recognizes the characteristic features of Z-DNA. Three members of this family have been identified previously: the RNA editing enzyme ADAR1 (17) (found in vertebrates), Z-DNA binding protein 1 (ZBP1/DLM-1) (thus far identified in mammals; ref. 18), and the poxvirus virulence factor, E3L (19) (Fig. 1 A). In eukaryotic proteins, Z-DNA binding domains are arranged in pairs, separated by a linker of variable size (12, 20) (see Fig. 7, which is published as supporting information on the PNAS web site). The N-terminal Zα domain of the pair is more highly conserved between ADAR1 and ZBP1 and with the single Zα of E3L, whereas the second, called Zβ, is more variable. Not all Zβ domains tested bind to Z-DNA when isolated; however, the presence of Zβ modulates Zα binding in the context of the larger domain in hADAR1 (20). Cocrystallization of Zα domains from hADAR1 (21), mZBP1 (18), and yabE3L (19) bound to Z-DNA has revealed that Z-DNA is recognized in a conformation-specific manner. No contacts are made that support a sequence-specific interaction between Zα and DNA. Although the overall sequence identity between the Z-DNA binding domains is only ≈25%, critical amino acids that contact Z-DNA or make up the hydrophobic core are highly conserved between the different proteins. dsRNA also can adopt the left-handed Z conformation. In contrast to the lower-energy, right-handed forms of RNA and DNA, which are different from each other, Z-RNA is virtually indistinguishable from Z-DNA. The only major change is the presence of the 2′-hydroxyl group, which is not exposed. It has been shown that Zα can bind Z-RNA specifically and that binding of Zα is sufficient to stabilize r(CG)6 in the Z conformation (13). Although we speak of the Z-DNA binding activity of Zα domains, in no case has the natural substrate been determined to be either RNA or DNA.
Fig. 1.
Proteins containing Zα domain. (A) Domain organization is shown schematically. Numbers to the right indicate the number of residues in human ADAR1, vaccinia virus E3L, human ZBP-1, zebrafish PKZ, and human PKR. Domains containing a Zα motif (Zα and Zβ) are shown in red. dsRNA binding domains are in green. The deaminase domain of ADAR1 (yellow) and the kinase domains of PKR and PKZ (light blue) are labeled. The function of the C-terminal region of ZBP-1 (gray) is unknown. (B) Comparison of Zα domains from PKZ of three fish species with the Zα domains of mouse ZBP-1, human ADAR1, vaccinia virus E3L, and yaba-like disease virus E3L. The crystal structures of the three domains, hZαADAR1, mZαZBP-1, and yabZαE3L, have been determined complexed with Z-DNA. Residues that make contact with Z-DNA, or the analogous residues in PKZ, are boxed in light blue. Residues that form the hydrophobic core are boxed in green. Residues that are neither DNA-contacting nor structural, but that match the consensus sequence, are highlighted in yellow. Sequences are bracketed by residue numbers relative to the translational start. Full genus, species, and accession numbers are listed in Table 1. For a more extensive comparison, see Fig. 7. (C) Alignment of the kinase domains from fish PKZ and selected mammalian PKRs. The kinase subdomains as defined by Hanks and Hunter (40) are marked with roman numerals. The position of the kinase inserts, specific for eIF2α kinases, and their sizes, are indicated between subdomains IV and V. A conserved lysine, which is essential for kinase activity, is boxed in red. The residues that are autophosphorylated in PKR and the analogous residues in PKZ are boxed in light blue. The highly conserved hydrophobic region is boxed in green. Other residues that match consensus are highlighted in yellow.
Here, we describe the identification and characterization of a gene from zebrafish closely related to mammalian PKR. The predicted gene product contains Z-DNA binding domains in the N terminus in place of dsRNA binding domains. A similar gene recently was described from goldfish (22). These findings support a role for Z-DNA binding in the fish host antiviral response and provide a previously undescribed way to investigate the role of the Z conformation in the cell.
Materials and Methods
For details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Identification, Cloning, and Expression of PKZ. Zebrafish [Danio rerio (dr)] were injected i.p. with 10 μg/g of bodyweight poly(I:C) (Sigma) in sterile PBS. After 12 h, animals were killed and tissues were prepared. Total RNA was prepared by the TRIzol method (Invitrogen). RNA was reverse-transcribed and cloned into pCR2.1 (Invitrogen). Expression patterns were analyzed by RT-PCR. PKZ inserts were cloned into the eukaryotic expression vector pcDNA3.1/Myc-His(–)A (Invitrogen). For expression of Z-DNA binding domains in Escherichia coli, PCR products were cloned into pET-28a (Novagen).
Circular Dichroism (CD) Assay of Z-DNA Binding. drZαPKZ was purified by using methods described in ref. 20. The CD assay in the presence of cobalt hexamine was performed as described in ref. 23. Cobalt hexamine was titrated with DNA until an equilibrium intermediate between B and Z was reached. Protein was added, and the effect on the equilibrium was observed.
Transient Transfections, Luciferase Activity, and Western Blot. For the luciferase assay, HEK293T and CHO cells (both obtained from American Type Culture Collection) were cotransfected by using poly(ethylenimine) (Qbiogene, Irvine, CA) with 2.5 μg of luciferase plasmid pGL3 promoter (Promega) and 0.5 μg of the respective pcDNA3.1 expression vector. For each plasmid, triplicate transfections were performed. After 16 h, cells were harvested, and luciferase activity was determined by using the luciferase detection kit (Promega).
For Western blot analysis, HEK293T cells were transfected with 3 μg of the indicated expression vector. Cells were harvested after 46 h, and total lysate was analyzed by SDS/PAGE followed by bidirectional blotting. Paired membranes were silver-stained or probed with anti-His Antibody (Dianova, Hamburg, Germany).
Phylogenetic Analysis. Phylogenetic analysis was applied to 42 sequences of eIF2α-kinases containing the kinase subdomains I–XI but without kinase inserts, including the zebrafish PKZ (see Table 1, which is published as supporting information on the PNAS web site). Both nucleic acid and protein sequences were used in the analysis. Phylogenetic analysis was performed on the computational cluster of the College of Biology and Agriculture at Brigham Young University on both data sets by using maximum parsimony and Bayesian Markov chain Monte Carlo approaches (http://babeast.byu.edu) (see Table 2, which is published as supporting information on the PNAS web site).
Results
To identify previously undescribed Z-DNA binding proteins, we searched expressed sequence tag (EST) databases at the National Center for Biotechnology Information using protein query vs. translated database (tblastn, www.ncbi.nlm.nih.gov), with ratZαZBP1 as a probe. We identified two EST clones derived from a single zebrafish gene, in which the amino acids critical for Z-DNA binding were conserved in the deduced sequence. When the complete ESTs were sequenced, we identified a 1.6-kb ORF predicted to encode a 58-kDa protein (diagrammed in Fig. 1; see also Fig. 8, which is published as supporting information on the PNAS web site). Two Zα-like domains (Zα and Zβ) are present in the N terminus; a kinase domain was identified in the C terminus. Because of this combination, the gene was named protein kinase containing Z-DNA binding domains (PKZ). Databank searches identified a sequence (GenBank accession no. AAP49830) from goldfish (Carassius auratus) with 69% sequence identity over the complete ORF, which was cloned as one of several genes induced from cells incubated with inactivated grass carp hemorrhage virus by subtractive hybridization (22). In addition, we identified a partially sequenced, highly similar EST from the common carp (Cyprinus carpio; accession no. AU301066). These are likely to be the PKZ orthologues from these species.
The Zα and Zβ domains of drPKZ were aligned to the Z-DNA binding domains of ZBP1 and ADAR1 from different species, E3L from poxviruses, and PKZ from goldfish and common carp, the latter represented by a partially sequenced EST containing the complete Zα and Zβ domains (Figs. 1B and 7). The Zα domains are only moderately conserved in overall sequence but retain important features. At five positions (K37, N41, L44, Y45, and W65, numbered with respect to PKZ), the amino acids are completely conserved in Zα (Fig. 1B). At four other positions (I14, L18, I29, and V40), hydrophobic amino acids are consistently present. These conserved residues are either involved in Z-DNA recognition (light blue) or form the hydrophobic protein core (green) as seen in the cocrystal structures of mZαZBP1:Z-DNA, hZαADAR1:Z-DNA, and yabZαE3L:Z-DNA complexes. These residues are less well conserved but are still recognizable in Zβ. The linker between Zα and Zβ is conserved neither in sequence nor size, ranging from 16 to 91 aa.
The alignment of the kinase domain of PKZ with PKR is shown in Fig. 1C along with the subdomain structure. The kinase insert domain of drPKZ is 78 aa long, with a calculated pI of 3.3. A conserved lysine (marked in red), which is important for catalytic activity in PKR, is found at position 199. Sites that are autophosphorylated in human PKR (Thr-446 and Thr-451) and are important for kinase activity (3) are marked (light blue). At position 402 in PKZ (corresponding to Thr-446 in human PKR) a serine is found, a conservative change that allows for autophosphorylation, whereas Thr-407 (corresponding to Thr-451 in human PKR) is conserved.
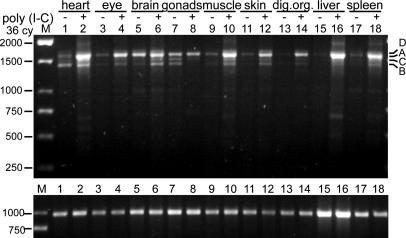
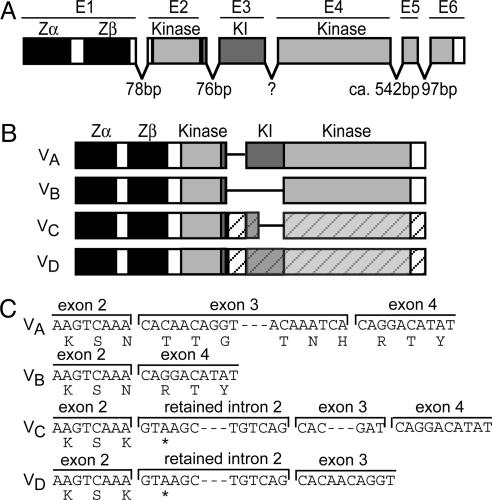
PKZ is constitutively expressed at low to moderate levels in a variety of tissues (Fig. 2; see also Fig. 9, which is published as supporting information on the PNAS web site). Several variants, probably generated by alternative splicing, are observed, with some tissue specificity of expression. Variants A–D (named in order of decreasing abundance) were cloned and sequenced (Fig. 3 B and C; see also Fig. 10, which is published as supporting information on the PNAS web site). PKZ is made up of six exons (Fig. 3). Variant A codes for the entire protein (Fig. 3 B and C). The second most abundant variant, B, lacks the kinase insert domain. Variants C and D retain an intron providing a premature stop codon and are predicted to encode truncated proteins.
Fig. 2.
Expression pattern of PKZ 12 h after induction by poly(I:C). RT-PCR was performed on the indicated tissues, by using primers covering the complete ORF. The constitutively expressed receptor for activated C kinase 1 (RACK1) is shown as a loading control at the bottom. Expression patterns in tissues from untreated animals are in Fig. 9.
Fig. 3.
Identification of splice variants and genomic organization of PKZ. (A) The exon/intron structure is shown, superimposed on the known domains. Zα and Zβ are encoded by a single exon (E1). The largest part of kinase insert (KI) is encoded by exon 3. Intron sizes were determined by PCR and partial sequencing. The size of intron 3 could not be determined, and genomic organization should be seen as provisional. (B) The structures of splice variants A–D are depicted. In variants C and D, retained intron 2 provides a premature stop codon, which terminates the reading frame as indicated by the hatched bars. (C) The nucleotide and deduced amino acid sequences of variants A–D in the region of intron 2 are shown. The stop codon (TAA) at the beginning of retained intron 2 is marked by an asterisk in the amino acid sequence. The complete sequences of the splice variants, as well as several alleles, are shown in Fig. 10.
PKR is induced by viral infection, poly(I:C) treatment, or interferons (1, 2). To determine whether drPKZ shares this behavior, viral infection was simulated by intraperiteonal injection of poly(I:C). After 12 h, tissues were prepared from PBS only (control) and poly(I:C)-injected animals. Semiquantitative RT-PCR demonstrated that drPKZ is highly induced in heart, liver, spleen, skeletal muscle, eye, skin, and digestive organs (Fig. 2). Variant A, containing the complete ORF, is most strongly up-regulated.
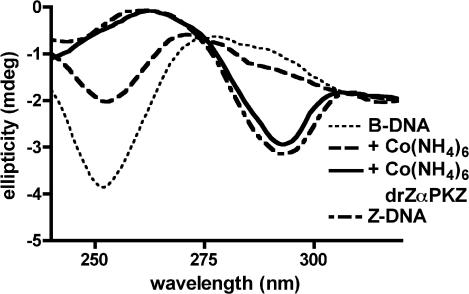
The Z-DNA binding activity of the Zα domain from drPKZ was demonstrated by observing the change in the B-Z equilibrium of poly(dG-dC) in the presence of protein. The B conformation of poly(dG-dC) can be distinguished from the Z form by CD (Fig. 4) (24). drZαPKZ was tested for the ability to convert poly(dG-dC) into the Z form in the presence of low amounts of cobalt hexamine. This assay has been used previously to verify the in vitro Z-DNA binding activity of vacinnia virus ZαE3L (23). Fig. 4 shows that the addition of drZαPKZ shifts the equilibrium to the Z conformation.
Fig. 4.
Binding of drZαPKZ to Z-DNA as shown by CD. The spectra of B-DNA (dotted line) and Z-DNA stabilized with 4 M NaCl (dashed line) are shown as controls. Cobalt hexamine was added to stabilize some of the DNA in the Z-form (dashed-dotted line). Protein was added to a final ratio of 1:2.5 (protein/base pair). The protein-bound DNA assumes the Z conformation (solid line). The kinetics of this reaction are shown in Fig. 11, which is published as supporting information on the PNAS web site.
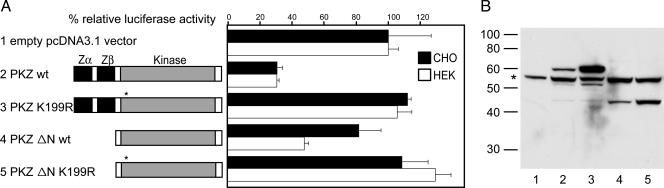
The ability of PKR to inhibit the expression of a cotransfected reporter gene in cell culture is an established method to assess PKR function (25, 26). HEK293T and CHO cells were cotransfected with the luciferase reporter plasmid pGL3promoter and with several drPKZ expression vectors containing a C-terminal myc-His tag for detection of expression. Cells were harvested at an early time point (16 h) after transfection, to avoid secondary effects perturbing this system; luciferase activity and total protein were determined. Protein content varied <10% between different experiments (data not shown). Wild-type (WT) drPKZ strongly down-regulated luciferase activity in both cell lines (Fig. 5A). A point mutation K296R (marked in red in Fig. 1C) in human PKR retained the ability to bind eIF2α (25) but abolished enzymatic activity (27). The analogous drPKZ mutant K199R was not active as shown by its failure to inhibit luciferase expression: expression was comparable with that seen with the empty vector. The enzyme missing the Zab domain (PKZ ΔN) was less effective than WT at inhibiting luciferase activity. PKZ ΔN was consistently less active in CHO than in HEK293T cells. The K199R mutation completely abolished activity of PKZ ΔN as well as the WT. To exclude the possibility that the observed effects were due to weaker expression of mutants, we looked at protein concentration by Western blot analysis using an anti-His antibody (Fig. 5B). A nonspecific band was observed in all lanes (asterisk). In WT drPKZ-transfected cells, a specific 60-kDa band was detected (lane 2), which fits well with the predicted molecular weight of 61 kDa. The kinase inactive mutants were expressed at a much higher level than WT (compare lanes 2 and 3, and 4 and 5). The N-terminal truncated proteins were detected as 44-kDa bands (lanes 4 and 5). In lanes 2 and 3, by using full-length constructs, smaller specific bands were detected at 53 and 44 kDa at low levels. These bands may represent fragments proteolytically cleaved after the Zβ and Zα domains, respectively.
Fig. 5.
Inhibition of protein synthesis by PKZ. (A) HEK293T and CHO cells were cotransfected with 2.5 μg of the luciferase plasmid pGL3promoter and 0.5 μg of the expression vector pcDNA3.1 containing the indicated inserts. Luciferase activity was normalized for protein content. A control experiment by using the empty pcDNA3.1 vector was used as 100%. An asterisk denotes the position in the constructs of the K199R mutation that abolishes kinase activity. Each bar is the average of three independent experiments. (B) Western blot analysis of HEK293T cells transfected with the expression constructs shown in A. His-tagged proteins were detected by an anti-His-tag antibody. The asterisk marks a nonspecific band detected in all lanes. Silver staining demonstrated comparable loading of proteins (data not shown).
Several families of eIF2α kinases are found in eukaryotes. Phylogenetic analysis of most members of these families found in the databases, as well as the fish PKZ sequences, resulted in two trees, a Bayesian maximum a posteriori tree derived from the nucleotide data (Fig. 6) and a strict consensus maximum parsimony tree of the protein data (data not shown).
Fig. 6.
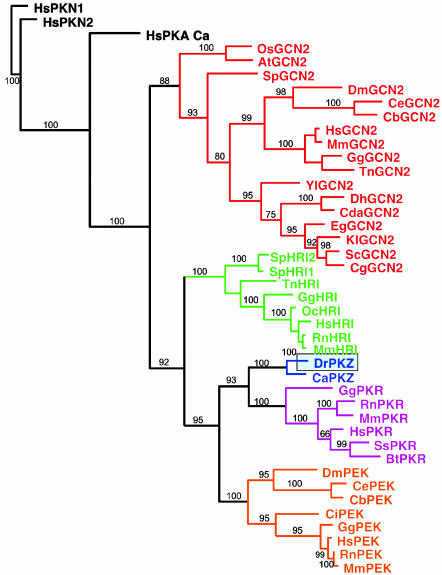
Evolutionary relationship between PKZ and other protein kinases, shown as a maximum a posteriori phylogram of Bayesian analysis. Posterior probabilities were converted to percentages and are shown above the branches.
Both trees recovered four basic clades, where the GCN2 kinases are sistergroup to the clade formed by HRI, PKR/PKZ, and PEK kinases. The zebrafish kinase (DrPKZ) is nested with the goldfish kinase (CaPKZ), a relationship receiving high support values (100% bootstrap and 100% posterior probability) in all analyses. Both of the PKZ kinases form a monophyletic sisterclade to the PKR kinases, indicating a close relationship.
Comparison of the sequence of drPKZ, as well as the goldfish (C. auratus) orthologue, with other eIF2a kinases clearly shows that PKZ belongs to the PKR family (Fig. 6). Database searches also have allowed us to identify with some confidence the zebrafish members of other families represented as ESTs as follows with their respective GenBank accession nos.: PEK (BM777743) and HRI (AW115606, CK692467, and BQ262919).
Discussion
eIF2α is highly conserved between human and zebrafish, displaying 94% sequence identity on the protein level (6). More than 100 aa are identical surrounding the conserved Ser-51 that is phosphorylated by eIF2α kinases. Treatment of the rainbow trout cell line RTG-2 with poly(I:C) or infectious pancreatic necrosis virus leads to phosphorylation of eIF2α by a kinase not further characterized (6). This activity is the same as that of PKR in mammalian cells.
The results presented here indicate that PKZ is closely related to mammalian PKR and is likely to fulfill the same function. PKZ has the overall structure of an eIF2α kinase, including the kinase insert domain. The motif LFIQMEF, which is essential for kinase activity (25), is conserved between PKZ and PKR. PKZ performs well in an assay of inhibition of protein synthesis, a standard assay for PKR activity. Furthermore, PKZ is active in mammalian cells in a manner similar to PKR. A mutation of the lysine conserved in all kinases results in loss of PKZ activity. Moreover, like PKR, which is induced by immunostimuli, PKZ is strongly induced by the treatment of goldfish cells (28) with viruses and by poly(I:C) in zebrafish. Construction of an evolutionary tree shows PKZ grouped with mammalian PKR. At present, we conclude that PKZ fulfills the function of PKR in zebrafish and that the Z-DNA binding domains may substitute for the dsRNA binding domains found in mammalian PKR.
A unique feature of eIF2α kinases is the kinase insert domain, which links subdomains IV and V. A possible clue to the function of the insert domain comes from its high acidity: this domain may react with a target protein containing basic domains. The insert domain is essential for phosphorylation of eIF2α (29); however, it varies greatly in length even between orthologues. The length of the insert ranges from 15–30 aa in PKR to >200 in PEK (Table 1). The zebrafish PKZ insert is 78 aa, intermediate among eIF2α kinases. Splice variant B, the second most abundant PKZ variant, lacks most of the kinase insert, leaving only 15 aa, the same length as found in porcine and bovine PKR. The kinase insert domain is important for eIF2α phosphorylation (29). Therefore, we speculate that different PKZ isoforms may phosphorylate different targets.
The functions of the Z-DNA binding domains of PKZ are yet to be explored. Many viruses contain potential Z-DNA- or Z-RNA-forming sequences, some of which have been shown to be essential for enhancing activity, for example sequences in the simian virus 40 enhancer (30). The Zα domains may target PKZ and thus influence gene expression, just as PKR regulates mRNA splicing of tumor necrosis factor-α (31) and translation of IFN-γ (32).
Z binding also may be involved in the regulation of PKZ activity. In PKR, the two dsRNA binding domains are important in the regulation of kinase activity. It has been proposed that, in the absence of dsRNA, these domains fold back on the kinase domain and suppress kinase activity. A conformational change is induced upon binding of dsRNA, which leads to PKR activation by autophosphorylation and dimerization (3). A similar model could be applied to PKZ.
Another interesting observation is the identification of variants C and D, each of which retains an intron containing a premature stop codon. The truncated PKZ isoform is predicted to be devoid of kinase activity because important kinase subdomains are missing; this isoform could play a dominant negative role. Intriguingly, a similar situation is seen in human PKR. A splice variant (PKRΔE7) exists in which exon 7 is spliced out, the reading frame is shifted, and a premature stop codon results (33). The truncated protein, containing only the dsRNA binding domains, inhibits both eIF2α and PKR autophosphorylation and relieves translational inhibition by WT PKR (33). Splice variants that are predicted to encode for truncated proteins containing only the N terminus also have been identified for ZBP1 (12).
Expression of WT PKR in mammalian cells has been shown to decrease reporter protein activity, probably due to the inhibition of translation (25, 33, 34). Like PKZ, enzymatically inactive PKR is more highly expressed than WT (25, 29). In contrast to PKZ, K296R mutants in PKR stimulate reporter protein expression 2- to 6-fold, perhaps by interacting with endogenous PKR leading to dominant negative inhibition or by sequestration of dsRNA preventing PKR activation (26, 29, 35, 41). This finding is in agreement with the observation that the dominant negative form, PKRΔE7, induces reporter enzyme activity in HeLa cells but not in PKR-deficient cells (33). PKZ lacks dsRNA binding domains and thus may not compete for dsRNA and inhibit endogenous PKR.
Poxviruses have a protein, called E3L in vaccinia virus, which is required to overcome the host IFN response. This protein contains an N-terminal Zα domain and a C-terminal dsRNA binding domain. The N terminus is required for pathogenicity of vaccinia virus in mice (8). Domain swap experiments, in which viral pathogenicity was restored by the replacement of E3LZα with ZBP1Zα or ADAR1Zα, as well as mutational analyses demonstrated that the ability to bind nucleic acid in the Z conformation is essential for E3L activity (9). This finding suggests that Z binding activity plays a role in viral infection. Further evidence for a connection between vertebrate Z-DNA binding proteins and infection is indicated by their inducibility by immunostimulation. ZBP1 is strongly induced by interferons and LPS (ref. 36 and N.D., unpublished data) and its expression is tightly associated with inhibition of hepatitis B virus replication (37). Human ADAR1 has differently regulated isoforms: a constitutively expressed isoform lacking the Zα domain and a larger, IFN-induced form, which contains Zα (38). These observations are extended by the results presented here that PKZ, the third vertebrate Z-DNA binding protein known to date, is strongly induced by poly(I:C) in many organs and by the recent finding that its goldfish orthologue is induced by grass carp hemorrhage virus in cultured cells (28).
Because GCN2 is found in a wide variety of organisms, including fungi, metazoa, and plants, it has been suggested to be the founding member of the eIF2α kinase family (39). According to our phylogenetic analysis, GCN2 and the other eIF2α kinases are sistergroups, probably derived from a common ancestor. HRI appears to be ancestral to PEK and PKR/PKZ, the latter of which are sistergroups. Both of the PKZ genes form a monophyletic sisterclade to the PKR genes, indicating a close relationship. It is interesting to note that the species studied to date contain either PKR or PKZ but not both. The presence of nucleic acid binding domains seems to be important for the function of these related kinases.
Nevertheless, we are left with a fundamental unanswered question: why do fish PKR-like activities employ left-handed Z-DNA (or Z-RNA) binding domains, whereas the mammalian enzymes use dsRNA-binding domains? It might be a reflection of a different suite of viruses challenging fish as compared with those challenging mammals. Further work will be needed to address these ideas.
Supplementary Material
Acknowledgments
We thank Thomas Schwartz for helpful discussions and careful reading of the manuscript and Andrea Brunngartner and Daniel Saupe for technical assistance. This work has been supported by grants from the National Institutes of Health (to A.R.).
Abbreviations: dr, Danio rerio; eIF2α, eukaryotic translation initiation factor 2; poly(I:C), poly(inosinic)–poly(cytidylic) acid; PKR, dsRNA-dependent protein kinase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AJ852018 (PKZa Va), AJ852019 (PKZb Va), AJ634742 (PKZc Va), AJ852020 (PKZd Va), AJ852021 (PKZe Va), AJ852022 (PKZf Va), AJ852023 (PKZg Va), AJ852024 (PKZh Vb), AJ852025 (PKZi Vd), AJ852026 (PKZc Vd), and AJ852027 (PKZh Vc)].
References
- 1.Williams, B. R. (1999) Oncogene 18, 6112–6120. [DOI] [PubMed] [Google Scholar]
- 2.Samuel, C. E. (2001) Clin. Microbiol. Rev. 14, 778–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, F., Romano, P. R., Nagamura-Inoue, T., Tian, B., Dever, T. E., Mathews, M. B., Ozato, K. & Hinnebusch, A. G. (2001) J. Biol. Chem. 276, 24946–24958. [DOI] [PubMed] [Google Scholar]
- 4.Dever, T. E. (1999) Trends Biochem. Sci. 24, 398–403. [DOI] [PubMed] [Google Scholar]
- 5.Kaempfer, R. (2003) EMBO Rep. 4, 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garner, J. N., Joshi, B. & Jagus, R. (2003) Dev. Comp. Immunol. 27, 217–231. [DOI] [PubMed] [Google Scholar]
- 7.Chang, H. W. & Jacobs, B. L. (1993) Virology 194, 537–547. [DOI] [PubMed] [Google Scholar]
- 8.Brandt, T. A. & Jacobs, B. L. (2001) J. Virol. 75, 850–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, Y. G., Muralinath, M., Brandt, T., Pearcy, M., Hauns, K., Lowenhaupt, K., Jacobs, B. L. & Rich, A. (2003) Proc. Natl. Acad. Sci. USA 100, 6974–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, H. W., Watson, J. C. & Jacobs, B. L. (1992) Proc. Natl. Acad. Sci. USA 89, 4825–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia, M. A., Guerra, S., Gil, J., Jimenez, V. & Esteban, M. (2002) Oncogene 21, 8379–8387. [DOI] [PubMed] [Google Scholar]
- 12.Rothenburg, S., Schwartz, T., Koch-Nolte, F. & Haag, F. (2002) Nucleic Acids Res. 30, 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown, B. A., II, Lowenhaupt, K., Wilbert, C. M., Hanlon, E. B. & Rich, A. (2000) Proc. Natl. Acad. Sci. USA 97, 13532–13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wölfl, S., Wittig, B., Dorbic, T. & Rich, A. (1997) Biochim. Biophys. Acta 1352, 213–221. [DOI] [PubMed] [Google Scholar]
- 15.Liu, R., Liu, H., Chen, X., Kirby, M., Brown, P. O. & Zhao, K. (2001) Cell 106, 309–318. [DOI] [PubMed] [Google Scholar]
- 16.Rothenburg, S., Koch-Nolte, F., Rich, A. & Haag, F. (2001) Proc. Natl. Acad. Sci. USA 98, 8985–8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbert, A., Alfken, J., Kim, Y. G., Mian, I. S., Nishikura, K. & Rich, A. (1997) Proc. Natl. Acad. Sci. USA 94, 8421–8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz, T., Behlke, J., Lowenhaupt, K., Heinemann, U. & Rich, A. (2001) Nat. Struct. Biol. 8, 761–765. [DOI] [PubMed] [Google Scholar]
- 19.Ha, S. C., Lokanath, N. K., Van Quyen, D., Wu, C. A., Lowenhaupt, K., Rich, A., Kim, Y. G. & Kim, K. K. (2004) Proc. Natl. Acad. Sci. USA 101, 14367–14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz, T., Lowenhaupt, K., Kim, Y. G., Li, L., Brown, B. A., II, Herbert, A. & Rich, A. (1999) J. Biol. Chem. 274, 2899–2906. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz, T., Rould, M. A., Lowenhaupt, K., Herbert, A. & Rich, A. (1999) Science 284, 1841–1845. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, Y. B., Zhang, Q., Xu, D., Hui, C. & Gui, J. F. (2003) Chin. Sci. Bull. 48, 581–588. [Google Scholar]
- 23.Kim, Y. G., Lowenhaupt, K., Oh, D. B., Kim, K. K. & Rich, A. (2004) Proc. Natl. Acad. Sci. USA 101, 1514–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pohl, F. M. & Jovin, T. M. (1972) J. Mol. Biol. 67, 375–396. [DOI] [PubMed] [Google Scholar]
- 25.Cai, R. & Williams, B. R. (1998) J. Biol. Chem. 273, 11274–11280. [DOI] [PubMed] [Google Scholar]
- 26.Kaufman, R. J., Davies, M. V., Pathak, V. K. & Hershey, J. W. (1989) Mol. Cell. Biol. 9, 946–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katze, M. G., Wambach, M., Wong, M. L., Garfinkel, M., Meurs, E., Chong, K., Williams, B. R., Hovanessian, A. G. & Barber, G. N. (1991) Mol. Cell. Biol. 11, 5497–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, C. Y., Zhang, Y. B., Huang, G. P., Zhang, Q. Y. & Gui, J. F. (2004) Fish Shellfish Immunol. 17, 353–366. [DOI] [PubMed] [Google Scholar]
- 29.Craig, A. W., Cosentino, G. P., Donze, O. & Sonenberg, N. (1996) J. Biol. Chem. 271, 24526–24533. [DOI] [PubMed] [Google Scholar]
- 30.Nordheim, A. & Rich, A. (1983) Nature 303, 674–679. [DOI] [PubMed] [Google Scholar]
- 31.Osman, F., Jarrous, N., Ben-Asouli, Y. & Kaempfer, R. (1999) Genes Dev. 13, 3280–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben-Asouli, Y., Banai, Y., Pel-Or, Y., Shir, A. & Kaempfer, R. (2002) Cell 108, 221–232. [DOI] [PubMed] [Google Scholar]
- 33.Li, S. & Koromilas, A. E. (2001) J. Biol. Chem. 276, 13881–13890. [DOI] [PubMed] [Google Scholar]
- 34.Wu, S. & Kaufman, R. J. (1996) J. Biol. Chem. 271, 1756–1763. [DOI] [PubMed] [Google Scholar]
- 35.Terenzi, F., deVeer, M. J., Ying, H., Restifo, N. P., Williams, B. R. & Silverman, R. H. (1999) Nucleic Acids Res. 27, 4369–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu, Y., Comella, N., Tognazzi, K., Brown, L. F., Dvorak, H. F. & Kocher, O. (1999) Gene 240, 157–163. [DOI] [PubMed] [Google Scholar]
- 37.Wieland, S. F., Vega, R. G., Muller, R., Evans, C. F., Hilbush, B., Guidotti, L. G., Sutcliffe, J. G., Schultz, P. G. & Chisari, F. V. (2003) J. Virol. 77, 1227–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.George, C. X. & Samuel, C. E. (1999) Proc. Natl. Acad. Sci. USA 96, 4621–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berlanga, J. J., Santoyo, J. & De Haro, C. (1999) Eur. J. Biochem. 265, 754–762. [DOI] [PubMed] [Google Scholar]
- 40.Hanks, S. K. & Hunter, T. (1995) FASEB J. 9, 576–596. [PubMed] [Google Scholar]
- 41.Benkirane, M., Neuveut, C., Chun, R. F., Smith, S. M., Samuel, C. E., Gatignol, A. & Jeang, K. T. (1997) EMBO J. 16, 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.