Abstract
Steroid receptor coactivator-3 (SRC-3/AIB1) is a coactivator for nuclear receptors and other transcription factors and an oncogene that contributes to growth regulation and development of mammary and other tumor types. Because of its biological functions, it is important to identify genes regulated by SRC-3. However, because coactivators do not bind DNA directly, extensive work is required to determine whether genes identified by RNA profiling approaches are direct or indirect targets. Here, we report the use of chromatin immunoprecipitation (ChIP)-based assays that involve genomic mapping and computational analyses of immunoprecipitated DNA to identify SRC-3-binding target genes in estradiol (E2)-treated MCF-7 breast cancer cells. We identified 18 SRC-3 genomic binding sites and demonstrated estrogen receptor-α (ERα) binding to all of them. Both E2-dependent and -independent SRC-3/ERα-binding sites were identified. RNA polymerase II ChIP assays were used to determine the correlation between SRC-3 and ERα binding and recruitment of the transcriptional machinery. These assays, in conjunction with analyses of RNA obtained from E2-treated cells, lead to the identification of SRC-3/ERα-associated genes. The ability of SRC family coactivators to regulate the expression of one of these genes, PARD6B/Par6, was confirmed by using cells individually depleted of SRC-1, SRC-2, or SRC-3 by small interfering RNA. The method described herein can be used to identify genes regulated by non-DNA-binding factors, such as other coactivators or corepressors, as well as DNA-binding transcription factors, and provides information on their binding location that can accelerate further gene characterization.
Keywords: chromatin immunoprecipitation assay, estrogen receptor, transcription
Nuclear receptors, including estrogen receptor-α (ERα), are transcription factors that control processes important for development, homeostasis, and reproduction (1). The ERs bind to specific DNA sequences that serve as enhancers, and recruit a range of coactivators important for stimulation of gene expression (2, 3). Many ERα coactivators have been identified, and numerous publications substantiate the ability of the steroid receptor coactivator (SRC) family to participate in the regulation of ERα-dependent gene expression (4).
There are three members of the SRC (p160) family of coactivators, SRC-1/NCoA-1, SRC-2/TIF2/GRIP1/NCoA-2, and SRC-3/ACTR/pCIP/RAC3/TRAM-1/AIB1. They share significant structural homology and function in a similar fashion to stimulate the transcription of nuclear receptor target genes in transient transfection experiments (4, 5). However, small interfering RNA (siRNA) depletion experiments suggest that different nuclear receptors have preferences for specific members of the SRC family (6), and the nature of target gene promoters influences the relative ability of p160s to stimulate ERα-dependent gene expression (7, 8). These findings are consistent with the distinct phenotypes of SRC coactivator knockout mice (reviewed in ref. 5). For example, SRC-3-, but not SRC-1- or SRC-2-, null mice exhibit growth retardation (9, 10). In addition to controlling growth during development, increased SRC-3/AIB1 expression has been implicated in human breast cancer (11, 12). Moreover, SRC-3 overexpression in mice leads to increased tumorigenesis (13), whereas ablation significantly impairs mammary tumorigenesis (14), indicating that SRC-3 is an oncogene.
Because the SRC-3/AIB1 and estrogen pathways that contribute to breast cancer may not be identical (14), an understanding of how each contributes to tumorigenesis is important. To this end, RNA profiling and bioinformatic approaches have been used to investigate the molecular pathways by which ER ligands affect breast cancer cells (15–20), and it is clear that ER target genes are subject to diverse modes of regulation. Although this is partially due to different modes of ER interaction with DNA (21), it is expected that coactivators contribute significantly to this diversity (7, 8).
Our understanding of coactivator target genes is limited, and has been obtained primarily through analysis of gene expression after overexpression or depletion of coactivator protein. For example, cells/tissues obtained from SRC-3 null mice showed reduced expression of NF-κB target genes (22, 23) and of genes associated with the insulin-like growth factor-1/Akt/mTOR pathways (13, 14). A recently published microarray study also described alterations in mRNA expression levels associated with siRNA-mediated depletion of AIB1/SRC-3 in unstimulated MCF-7 cells (24). However, as for all microarray analyses, it is not known whether the differentially regulated genes are direct or indirect transcriptional targets of SRC-3. Moreover, because coactivators do not bind DNA directly, computational analyses cannot be used to search for coactivator binding sites in the vicinity of genes encoding differentially regulated mRNAs. Here we present an approach to identify primary coregulator target genes based on ChIP-based assays that involve sequence and bioinformatic analyses of large numbers of isolated DNA fragments, and report the identification of SRC-3 target genes in MCF-7 breast cancer cells.
Materials and Methods
Cell Culture. MCF-7 cells were maintained in DMEM supplemented with 10% FBS, nonessential amino acids, and antibiotic/antimycotic. For stimulation of cells with either 17β-estradiol (E2; Sigma) or ICI 182,780 (ICI; Tocris Cookson, Bristol, U.K.), cells were plated before stimulation in phenol-red free DMEM containing stripped FBS for 48–72 h.
Chromatin Immunoprecipitation (ChIP)-Based Assays. Genpathway's FactorPath and TranscriptionPath methods (see Fig. 5, which is published as supporting information on the PNAS web site) started with ChIP, which was performed as described (25). Protein G-agarose (for goat polyclonal anti-SRC-3) and protein A-agarose (for rabbit polyclonal anti-ERα and anti-pol II) were from Invitrogen. Antibodies were from Santa Cruz Biotechnology.
Cloning, Sequencing, and Analysis of Tags. Genpathway's Discovery approach was used to analyze immunoprecipitated DNA fragments. DNA was amplified (26), and purified PCR products were subjected to a random-priming/extension and gel-purification protocol (27) to generate a library of 40- to 50-bp “tags,” which were concatemerized and cloned as 10- to 20-mers into the pAMP10 vector (Invitrogen) for sequencing. Individual tag sequences were aligned to the human genome by using megablast version 2.2.8. Low-scoring alignments were eliminated, as were tags that aligned to multiple locations. Tags located within 1,200 bp of each other were grouped into clusters, and 1 kb of genomic sequence flanking both sides of the clustered alignments were searched for putative estrogen response elements (EREs) with both MatInspector (www.genomatix.de) and dragon ere finder version 2.0 (http://sdmc.lit.org.sg/ERE-V2/index) using default parameters. To confirm candidate SRC-3-binding sites by Genpathway's FactorPath Query method, PCR primers targeting a region within 200 bp of each selected alignment or cluster (Table 2, which is published as supporting information on the PNAS web site) were used to measure the amount of this sequence in immunoprecipitated samples by quantitative real-time PCR (Q-PCR) using SYBR Green-based detection (Bio-Rad). Experimental Q-PCR values were normalized against values obtained for 25 ng of input DNA with the same primer set. The same method was used to determine ERα binding. The density of RNA polymerase II (pol II) binding was determined by an analogous method (Genpathway's TranscriptionPath Query) using an antibody against the largest subunit of this complex, except that for sites located upstream from known genes (CAP2, PARD6B, FLJ20294, IER3, CBWD2), additional primers were designed to target a region inside the gene (Table 3, which is published as supporting information on the PNAS web site).
Inhibition of Coactivator Expression with siRNA. A total of 3 × 105 MCF-7 cells per well of a six-well multiplate were transfected with 10 pmol of siRNA (Ambion) by using Oligofectamine transfection reagent (Invitrogen). The siRNA sequences for luciferase, SRC-1, SRC-2, and SRC-3 have been described and validated (6, 28). Ligand treatments were added 48 h thereafter.
Western Blot Analysis. Cells were lysed in buffer (50 mM Tris, pH 7.5/150 mM NaCl/5 mM EDTA/0.5% Nonidet P-40 and a protease inhibitor tablet; Roche Applied Sciences), and 5 μg (for SRC-3) or 35 μg (for SRC-1 and SRC-2) of the resulting supernatants were resolved by SDS/PAGE and subjected to Western blot analysis using primary antibodies against SRC-1, SRC-2, SRC-3, or actin (Chemicon), followed by the appropriate horseradish peroxidase-conjugated secondary antibodies and ECL Plus detection reagent (Amersham Pharmacia).
Reverse-Transcription Q-PCR. Total RNA was extracted with Trizol (Invitrogen), and reverse transcribed with SuperScript RNaseH-(Invitrogen). Q-PCR was used to quantitate cDNAs by using an ABI Prism 7700 detection system (Applied Biosystems) with SYBR Green as the fluorescent dye; primer sequences are available in Table 4, which is published as supporting information on the PNAS web site. Gene expression values were calculated based on the comparative ΔΔCT method (Applied Biosystems User Bulletin no. 2) and normalized to values obtained for 18S ribosomal RNA.
Results
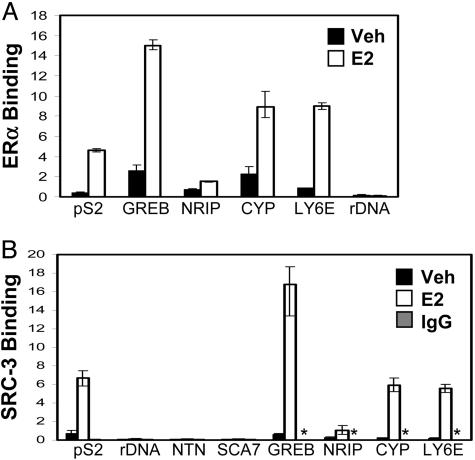
Because the SRC-3 oncogene is an important ERα coactivator, and estrogen exposure increases breast cancer risk (29), we sought to identify SRC-3 target genes in breast cancer cells treated with E2. To validate our approach, the ability of ERα and SRC-3 ChIP-based assays to detect a known SRC-3 target gene (pS2/trefoil factor 1; refs. 30 and 31) as well as several known E2/ER target genes [GREB1 (32), NRIP1 (16–19), CYP1B1 (15, 33), and LY6E (18, 20)] was determined. Q-PCR using primers flanking known EREs (Table 2) demonstrated concordant patterns of E2-stimulated interaction of ERα (Fig. 1A) and SRC-3 (Fig. 1B) for all genes. With the SRC-3 antibody, enrichment of pS2 ERE sequences over 18S ribosomal DNA (rDNA) was >50-fold. The lack of ERα and SRC-3 interaction with rDNA and with sites within the netrin G1 (NTNG1) gene and downstream of the ataxin 7 (SCA7) gene served as negative controls. Signals obtained in reactions with a control IgG antibody were insignificant (Fig. 1B).
Fig. 1.
Demonstration of ERα (A) and SRC-3 (B) binding to known ERE-containing promoters. MCF-7 cells were treated with vehicle (Veh; 0.01% ethanol) or 10 nM E2 for 1 h and processed for ChIP assays. Control Ig (IgG) immunoprecipitations were performed from a 1:1 mixture of vehicle- and E2-treated cell chromatin. Asterisks indicate samples that were not assayed. Results shown are from a single experiment, and error bars indicate maximal and minimal values of triplicate Q-PCRs. Assays were repeated with different cell cultures, and results were reproducible.
A discovery approach for the identification of SRC-3 binding sites in the genome was initiated, and of the 7,017 tags representing SRC-3 immunoprecipitated DNA that were sequenced, 4,853 produced acceptable genomic alignments. These tags generated ≈70 clusters consisting of two or more distinct alignments within 1,200 bp (class A clusters). Because ChIP fragments were <600 bp, 1,200 bp was set as the maximal distance between two tags representing the same binding site. In addition, there were ≈600 single alignments that mapped between -10 and +5 kb relative to transcription start sites.
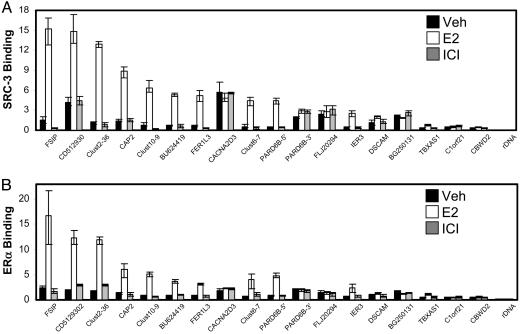
The binding of SRC-3 to the identified genomic locations in cells treated with vehicle, E2, or ICI was confirmed by Q-PCR using PCR primers encompassing genomic segments within ≈200 nt of the putative SRC-3-binding sites. Of the ≈70 clusters, 30 were selected for further testing (preference was given to clusters with more than two tags, located in putative promoter regions, and/or containing a predicted ERE nearby), and 15 of those (50%) were confirmed as strong SRC-3-binding sites (Q-PCR signal >5-fold above the background signal obtained for the rDNA, NTNG1, and SCA7 sites). From the group of 600 single-hit alignments and clusters consisting of multiple identical or almost identical tags (class B clusters), an additional 35 sites were selected for further testing as above, and three (8.5%) were confirmed as strong SRC-3-binding sites. Therefore, a total of 18 strong, previously undescribed SRC-3 binding sites were identified (Fig. 2A and Table 1). An additional 11 sites were identified as “weak” SRC-3-binding sites because the Q-PCR signal was only 2- to 5-fold above the background signal obtained for the negative control sites (Table 5, which is published as supporting information on the PNAS web site). The genomic locations of the SRC-3 clusters are provided in Table 6, which is published as supporting information on the PNAS web site.
Fig. 2.
Confirmation of SRC-3 and ERα binding to the 18 sites identified by the Discovery approach. Chromatin from MCF-7 cells treated with vehicle, 10 nM E2, or 1 μM ICI for 2 h was immunoprecipitated with anti-SRC-3 (A) or anti-ERα (B) antibodies, and the enrichment of candidate SRC-3-binding sites was determined by Q-PCR. Values for the 18 sites were >5-fold higher than the value for SRC-3 binding to rDNA (Right). Results shown are from single experiments; error bars indicate maximal and minimal values of triplicate Q-PCRs. All binding assays were repeated with cell cultures treated with ligands for 1 h, and similar results were obtained.
Table 1. List of identified strong SRC-3 binding sites.
| Cluster Information
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Binding site ID | Nearest gene, mRNA, EST, or genomic location | GenBank accession no. | Distance to gene start, bp | No. of tags | No. of Distinct tags | Length, nt | Class* | ERE† |
| FSIP1 | Fibrous sheath interacting protein 1 | NM152597 | 114,099 | 2 | 2 | 697 | A | ERE |
| CD512930 | IMAGE:30393829 | 1,629 | 2 | 2 | 67 | A | ERE | |
| Clust2–36 | chr2: 240729443‡ | NA | 2 | 2 | 69 | A | ERE | |
| CAP2 | Adenylyl cyclase-associated protein 2 | NM006366 | -3,888 | 3 | 1 | 43 | B | ERE |
| Clust10–9 | chr10: 43989225 (intron of EST BI019229)‡ | NA | 3 | 3 | 179 | A | ERE | |
| BU624419 | UI-H-FG1-bgk-i-13–0-UI | -5,893 | 3 | 2 | 83 | A | ERE | |
| FER1L3 | Myoferlin isoform a; dysferlin | NM013451 | 47,362 | 2 | 2 | 535 | A | ERE |
| CACNA2D3 | Ca channel, voltage-dependent, α2/Δ3 subunit | NM018398 | 230,660 | 2 | 2 | 109 | A | |
| Clust6–7 | chr6: 53437853‡ | NA | 2 | 2 | 74 | A | 2 ERE | |
| PARD6B 5′ | Par-6 partitioning defective 6 homolog-β | XM030559 | -4,078 | 4 | 2 | 77 | A | ERE |
| PARD6B 3′ | par-6 partitioning defective 6 homolog-β | XM030559 | +7,975 from 3′ end | 4 | 3 | 437 | A | (ERE) |
| FLJ20294 | Hypothetical protein FLJ20294 | NM017749 | -2,721 | 2 | 2 | 71 | A | (ERE) |
| IER3 | Immediate early response 3 | NM003897 | -7,187 | 1 | 1 | 43 | NA | ERE |
| DSCAM, AF401032 | Down syndrome cell adhesion molecule | NM001389 | 467,139, -3,087 | 6 | 1 | 47 | B | |
| BG250131 | IMAGE:4470585 | -50 to -1,144 | 4 | 3 | 1,091 | A | ||
| TBXAS1 | Thromboxane A synthase 1 (LOC389562) | BC014117 | -6,868 | 2 | 2 | 335 | A | ERE |
| C1orf21 | Chromosome 1 open reading frame 21 | NM030806 | 86,467 | 4 | 2 | 67 | A | |
| CBWD2 | COBW domain-containing protein 2 | NM172003 | -741, -1,096 | 3 | 2 | 399 | A | ERE |
NA, not applicable.
Class A clusters consist of two or more distinct tags with different or only partially overlapping sequences; class B clusters consist of three or more identical or almost identical tags. Class B clusters could arise during amplification steps in the cloning protocol and may represent single alignments
ERE, ERE within 500 bp of nearest tag; (ERE), ERE within 500–1,000 bp of nearest tag
Midpoint of cluster, May 2004 human reference sequence NCBI Build 35 (hg17)
A similar query-based approach using an ERα antibody confirmed all 18 of the identified genomic DNA fragments as ER-binding sites (Fig. 2B). The relative binding patterns for SRC-3 and ERα were very similar. At 11 of these locations, E2, but not the pure ICI antagonist, promoted both ERα and SRC-3 binding, and all of these sites had a predicted ERE within 500 bp (Tables 1 and 6). Interestingly, five of the ligand-insensitive sites lacked a predicted ERE within 500 bp (CACNA2D3, PARD6B-3′, FLJ20294, BG250131, C1orf21). Only the ERE-less DSCAM site and the ERE-containing CBWD2 site could not clearly be classified into either of these categories: DSCAM showed only minimal E2 induction, and CBWD2 showed no significant E2 induction. It is noteworthy that five sites are located within genes (FSIP1, FER1L3, CACNA2D3, DSCAM, and C1orf21), one is downstream (PARD6B-3′), and three are not in proximity of any known gene or mRNA (Clust2–36, Clust10–9, and Clust6–7). Of the 18 sites/genes, only IER3 is known to be regulated by ER ligands (18, 34).
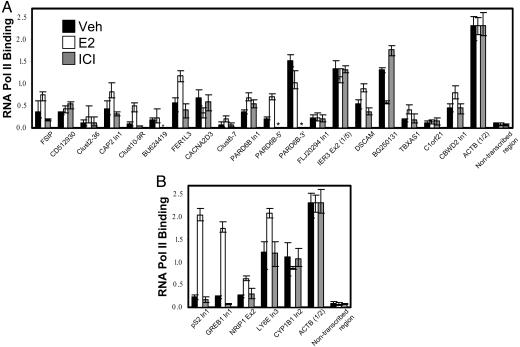
A ChIP-based approach using an antibody against pol II was used to determine the level of transcription at the identified SRC-3-binding sites or nearby genes. The signal obtained for the constitutively expressed β-actin (ACTB) gene, which served as a positive control, was very robust (Fig. 3A). Estradiol-enhanced pol II association was detected within the FSIP1, CAP2, FER1L3, PARD6B (In1), DSCAM, and CBWD2 genes, suggesting that E2-induced SRC-3 or ERα binding positively regulates their transcription. Estradiol-inducible pol II binding also was detected at genomic sites where no RNA had previously been annotated (Clust2–36, Clust6–7, Clust10–9, PARD6B-5′, TBXAS1). The ICI antagonist did not promote pol II binding at any location. Notably, the SRC-3/ERα-binding sites for three of the E2-stimulated genes (FER1L3, FSIP1, DSCAM) are located 47–467 kb downstream from their promoters (Table 1). In addition to several hormone-independent sites (e.g., CD512930, BU624419, and C1orf21), negative regulation of pol II binding to the BG250131, CACNA2D3, and PARD6B-3′ sites after E2 treatment was observed. It may be significant that SRC-3 and ERα binding to these sites, which lack an ERE within 500 bp, is ligand-independent. The E2-stimulated association of pol II with known E2/ER-target genes (pS2, GREB1, NRIP1, and LY6E) served as positive controls (Fig. 3B). Surprisingly, no increased association of pol II with the CYP1B1 gene was observed even though its mRNA was reported to be induced by E2 (33). Although this apparent discrepancy could be due to experimental conditions, it also is possible that E2-induced CYP1B1 mRNA expression is achieved posttranscriptionally.
Fig. 3.
Measurement of pol II binding at or near previously unknown (A) and known (B) SRC-3/ERα binding sites. Chromatin from vehicle-, 10 nM E2-, or 1 μM ICI-treated MCF-7 cells (2 h) was immunoprecipitated with anti-pol II antibodies, and binding of pol II to the candidate SRC-3-binding sites or nearby gene sequences was determined by Q-PCR. Sequences amplified by Q-PCR were the same as for Fig. 2, except where specified after the name (e.g., by “In1” for intron 1 or “Ex2” for exon 2). Signals were normalized to those obtained for the β-actin housekeeping gene (ACTB; assayed in intron 3). Values for IER3 Ex2 and ACTB were divided by 5 and 2, respectively (indicated as “1/5” and “1/2”), to reduce their values into the range of the other Q-PCR signals. Asterisks indicate ICI-treated samples that were not assayed. Error bars indicate maximal and minimal values of triplicate Q-PCRs. All assays were repeated with independently treated cell cultures, and similar results were obtained.
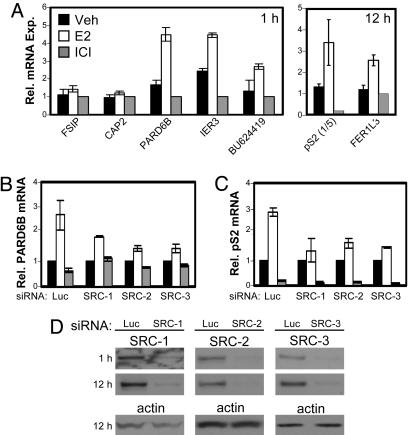
Expression of mRNAs for several of the recently discovered SRC-3-associated genes, selected on the basis of E2-sensitive SRC-3, ERα, and pol II binding, and for the known ER target genes, pS2 and IER3, was evaluated after 1 and 12 h of ligand treatment. Values obtained for the time point with the greatest induction are shown in Fig. 4A. Estradiol-induced mRNA expression of IER3 and pS2, as well as of FER1L3, PARD6B, and BU624419, confirmed that the latter are E2/ER regulated target genes. In contrast, there was no significant induction of FSIP1 or CAP2 mRNAs. Finally, a siRNA approach was used to verify that one of the above genes, PARD6B, was regulated by SRC-3, and to investigate whether SRC-1 or SRC-2 might also control this gene's expression. Depletion of each of the p160 coactivators partially inhibited the ability of E2 to increase PARD6B (Fig. 4B) and pS2 (Fig. 4C) mRNA levels, indicating that all three SRCs contribute to E2-stimulated gene transcription. These results are consistent with previous reports of the effect of depleting p160 coactivator expression on pS2 mRNA levels and the known interactions of each of the SRC family coactivators with the pS2 promoter (31, 35, 36).
Fig. 4.
Relative mRNA expression of selected SRC-3 target genes. (A) MCF-7 cells were treated with vehicle, E2, or ICI for 1 (Left)or12(Right) h, and mRNA levels were measured by reverse-transcription Q-PCR. Values were normalized to 18S RNA levels and plotted relative to the signal obtained for ICI-treated samples. Expression of PARD6B (B) and pS2 (C) was assessed in cells treated with siRNA directed against luciferase (Luc), SRC-1, SRC-2, or SRC-3. Values were normalized to the corresponding 18S rRNA values and expressed as fold change relative to vehicle-treated samples. Data are the average of two independent experiments with error bars representing the range of the two values. (D) Demonstration of effective siRNA inhibition of SRC-1 (Left), SRC-2 (Center), and SRC-3 (Right) expression measured by Western blot. Protein samples were prepared from cells treated with 10 nM E2 for 1 or 12 h. (Lower) Actin expression used as a loading control.
Discussion
Here, we present the identification of direct target genes of the SRC-3 coactivator through a recently developed approach that employs sequencing and mapping of genomic DNA fragments obtained by SRC-3 ChIP assays. Although microarrays have been widely used to identify mRNAs that are influenced by a perturbation of interest (e.g., hormonal stimulation, knockout of a specific gene), there are limitations to this approach. First, detection depends on mRNA expression levels that must be sufficient for detection. Moreover, mRNA levels can be influenced by factors other than those controlling their synthesis (i.e., heterogeneous nuclear RNA processing or altered mRNA stability). Secondly, microarray approaches do not distinguish between direct or indirect target genes, and additional experimentation involving protein synthesis inhibitors or “ChIP scanning” (37) is required to make this determination. Third, microarray approaches are inherently biased because only the expression of preselected targets can be determined. Finally, once target genes have been identified, extensive effort is required to analyze potential promoter sequences for binding sites for the trans-acting factors suspected to be involved in regulation. Typically, only 2–3 kb of upstream sequence is examined, so that regulatory elements located further upstream, within, or downstream of the target gene are likely to be missed. Moreover, this analysis is not possible for regulatory molecules such as coactivators that do not bind directly to DNA.
In contrast, our approach of ChIP in combination with sequencing and mapping of isolated genomic DNAs provides a method for detecting direct target genes that does not depend on mRNA expression. Moreover, the isolated DNA tags are mapped against the entire human genome instead of against preselected sequences. This approach contrasts with a recent report on the use of ChIP in combination with promoter microarrays to identify target genes of the C/EBPβ transcription factor (38). In addition, unbiased location of the coactivator interaction sites can be determined within fairly small regions. Indeed, our data suggest that SRC-3-interaction sites can be located large distances upstream from, within, or even downstream of their associated genes. Finally, in comparison to a microarray approach that requires altering SRC-3 expression, this methodology allows the identification of SRC-3-target genes in cells without perturbing endogenous SRC-3 expression. This ability reduces the likelihood of identifying false positives related to coactivator overexpression or of missing target genes because of the compensatory function of other members of the SRC family under depletion conditions.
Analyses of gene transcription patterns suggest that E2-stimulated interaction of SRC-3 and ERα with genomic fragments is associated with diverse effects on pol II recruitment (i.e., increased, decreased, and no change in binding). It should be noted that our binding analyses detected both nonphosphorylated and phosphorylated forms of pol II and were performed after 2 h of E2 or ICI treatment, and different time points could yield different results. In addition, it is still important to verify changes in mRNA expression, especially for several of the recently discovered sites where it is not clear which (if any) mRNA is regulated.
Based on the association of SRC-3, ERα, and pol II with DNA fragments in proximity to known genes, we verified that FER1L3, a gene predicted to encode a transmembrane protein (39), PARD6B (see below), and an unnamed gene represented by the EST BU624419, are E2-sensitive target genes. We also confirmed the regulation of IER3 (also known as IEX3), a gene involved in regulating apoptosis and cell cycle progression (40–42), by E2/ER. It is interesting to note that the association of ERα and SRC-3 with the CACNA2D3 gene and the locus associated with the BG250131 mRNA showed decreased pol II binding, suggesting that ERα and its SRC-3 coactivator may be involved in negative regulation of transcription. The E2-stimulated association of ERα, SRC-3, and pol II with Clust2–36, Clust10–9, and Clust6–7 indicates that responsive genes residing near or within these clusters remain to be identified. Likewise, sites located more than 10 kb inside of genes may in fact regulate hitherto unknown genes nested within. For example, the DSCAM site, which is located 467 kb from the DSCAM 5′ end, is also 3 kb upstream of an unnamed mRNA, AF401032, inside DSCAM. It should also be noted that 157 of the ≈600 genes identified by single-tag alignments mapping -10 to +5 kb relative to gene transcription start sites were also present on the list generated by a computer search for genes with an ERE located within the same 15-kb region (20). It is highly likely that additional sequencing of SRC-3-immunoprecipitated DNA tags would increase the number of clusters and, therefore, the number of target genes available for future study.
Based on the ability of E2 to stimulate binding of SRC-3, ERα, and pol II with PARD6B/Par6 genomic DNA, we assessed the regulation of PARD6B mRNA in SRC-depleted MCF-7 cells. PARD6B is the human homolog of a cell polarity determinant in Caenorhabditis elegans (PAR-6), originally identified in a study of asymmetric cell division that revealed a series of partitioning-defective genes important for establishing anterior/posterior polarity (43). More recently, PARD6B has been shown to bind to the GTPases Rac and Cdc42, and atypical protein kinase C. Overexpression of this protein negatively regulates the formation of tight junctions at epithelial cell–cell contacts (44–46), inhibits insulin signaling (47), and has been implicated in cell transformation (48). Evaluation of the relationship of PARD6B to the oncogenic potential of SRC-3 awaits further investigation.
In conclusion, the use of a ChIP-based approach in combination with mapping the genomic locations of immunoprecipitated DNA provides a method for direct identification of target genes of coregulator factors that do not directly bind to DNA. This method should be broadly applicable to the identification of genes regulated by other coactivators and corepressors, thus providing detailed information on the location of regulatory elements and enhancing subsequent gene expression analyses.
Supplementary Material
Acknowledgments
We thank Dr. Mary Warren (Genpathway) for helpful guidance and discussions and Dr. Benjamin Monderer (Genpathway) for developing the sequence analysis software. This work was supported by National Institutes of Health Grants DK53002 (to C.L.S.), HD07857 and HD08818 (to B.W.O.), and R43CA094564 (to P.L.) and a Welch Foundation grant (to B.W.O.). E.M.S. was supported by Susan G. Komen Breast Cancer Foundation Award PDF0402260.
Abbreviations: ERα, estrogen receptor-α; SRC, steroid receptor coactivator; siRNA, small interfering RNA; E2, 17β-estradiol; ChIP, chromatin immunoprecipitation; ERE, estrogen response element; Q-PCR, quantitative real-time PCR; pol II, RNA polymerase II; ICI, ICI 182,780.
References
- 1.Nilsson, S., Mäkelä, S., Treuter, E., Tujague, M., Thomsen, J., Andersson, G., Enmark, E., Pettersson, K., Warner, M. & Gustafsson, J.-Å. (2001) Physiol. Rev. 81, 1535-1565. [DOI] [PubMed] [Google Scholar]
- 2.Glass, C. K. & Rosenfeld, M. G. (2000) Genes Dev. 14, 121-141. [PubMed] [Google Scholar]
- 3.McKenna, N. J. & O'Malley, B. W. (2002) Cell 108, 465-474. [DOI] [PubMed] [Google Scholar]
- 4.McKenna, N. J., Lanz, R. B. & O'Malley, B. W. (1999) Endocr. Rev. 20, 321-344. [DOI] [PubMed] [Google Scholar]
- 5.Xu, J. & Li, Q. (2003) Mol. Endocrinol. 17, 1681-1692. [DOI] [PubMed] [Google Scholar]
- 6.Li, X., Wong, J., Tsai, S. Y., Tsai, M.-J. & O'Malley, B. W. (2003) Mol. Cell. Biol. 23, 3763-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barkhem, T., Haldosén, L.-A., Gustafsson, J.-Å. & Nilsson, S. (2002) Mol. Endocrinol. 16, 2571-2581. [DOI] [PubMed] [Google Scholar]
- 8.Klinge, C. M., Jernigan, S. C., Mattingly, K. A., Risinger, K. E. & Zhang, J. (2004) J. Mol. Endocrinol. 33, 387-410. [DOI] [PubMed] [Google Scholar]
- 9.Wang, Z., Rose, D. W., Hermanson, O., Liu, F., Herman, T., Wu, W., Szeto, D., Gleiberman, A., Krones, A., Pratt, K., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 13549-13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu, J., Liao, L., Ning, G., Yoshida-Komiya, H., Deng, C. & O'Malley, B. W. (2000) Proc. Natl. Acad. Sci. USA 97, 6379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anzick, S. L., Kononen, J., Walker, R. L., Azorsa, D. O., Tanner, M. M., Guan, X. Y., Sauter, G., Kallioniemi, O. P., Trent, J. M. & Meltzer, P. S. (1997) Science 277, 965-968. [DOI] [PubMed] [Google Scholar]
- 12.Shou, J., Massarweh, S., Osborne, C. K., Wakeling, A. E., Ali, S., Weiss, H. & Schiff, R. (2004) J. Natl. Cancer Inst. 96, 926-935. [DOI] [PubMed] [Google Scholar]
- 13.Torres-Arzayus, M. I., de Mora, J. F., Yuan, J., Vazquez, F., Bronson, R., Rue, M., Sellers, W. R. & Brown, M. (2004) Cancer Cell 6, 263-274. [DOI] [PubMed] [Google Scholar]
- 14.Kuang, S.-Q., Liao, L., Zhang, H., Lee, A. V., O'Malley, B. W. & Xu, J. (2004) Cancer Res. 64, 1875-1885. [DOI] [PubMed] [Google Scholar]
- 15.Frasor, J., Stossi, F., Danes, J. M., Komm, B., Lyttle, C. R. & Katzenellenbogen, B. S. (2004) Cancer Res. 64, 1522-1533. [DOI] [PubMed] [Google Scholar]
- 16.Frasor, J., Danes, J. M., Komm, B., Chang, K. C. N., Lyttle, C. R. & Katzenellenbogen, B. S. (2003) Endocrinology 144, 4562-4574. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, A., Yoshida, N., Omoto, Y., Oguchi, S., Yamori, T., Kiyama, R. & Hayashi, S. (2002) J. Mol. Endocrinol. 29, 175-192. [DOI] [PubMed] [Google Scholar]
- 18.Soulez, M. & Parker, M. G. (2001) J. Mol. Endocrinol. 27, 259-274. [DOI] [PubMed] [Google Scholar]
- 19.Lin, C.-Y., Ström, A., Vega, V. B., Kong, S. L., Yergey, A. L., Thomsen, J. S., Chan, W. C., Doray, B., Bangarusamy, D. K., Ramasamy, A., et al. (2004) Genome Biol. 5, R66.1-R66.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourdeau, V., Deschênes, J., Métivier, R., Nagai, Y., Nguyen, D., Bretschneider, N., Gannon, F., White, J. H. & Mader, S. (2004) Mol. Endocrinol. 18, 1411-1427. [DOI] [PubMed] [Google Scholar]
- 21.O'Lone, R., Frith, M. C., Karlsson, E. K. & Hansen, U. (2004) Mol. Endocrinol. 18, 1859-1875. [DOI] [PubMed] [Google Scholar]
- 22.Wu, R.-C., Qin, J., Hashimoto, Y., Wong, J., Xu, J., Tsai, S. Y., Tsai, M.-J. & O'Malley, B. W. (2002) Mol. Cell. Biol. 22, 3549-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu, R. C., Qin, J., Yi, P., Wong, J., Tsai, S. Y., Tsai, M. J. & O'Malley, B. W. (2004) Mol. Cell 15, 937-949. [DOI] [PubMed] [Google Scholar]
- 24.Oh, A., List, H.-J., Reiter, R., Mani, A., Zhang, Y., Gehan, E., Wellstein, A. & Riegel, A. T. (2004) Cancer Res. 64, 8299-8308. [DOI] [PubMed] [Google Scholar]
- 25.Soutoglou, E. & Talianidis, I. (2002) Science 295, 1901-1904. [DOI] [PubMed] [Google Scholar]
- 26.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290, 2306-2309. [DOI] [PubMed] [Google Scholar]
- 27.Singer, B. S., Shtatland, T., Brown, D. & Gold, L. (1997) Nucleic Acids Res. 25, 781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shang, Y. & Brown, M. (2002) Science 295, 2465-2468. [DOI] [PubMed] [Google Scholar]
- 29.The Endogenous Hormones and Breast Cancer Collaborative Group (2002) J. Natl. Cancer Inst. 94, 606-616. [DOI] [PubMed] [Google Scholar]
- 30.Shang, Y., Hu, X., DiRenzo, J., Lazar, M. A. & Brown, M. (2000) Cell 103, 843-852. [DOI] [PubMed] [Google Scholar]
- 31.Shao, W., Keeton, E. K., McDonnell, D. P. & Brown, M. (2004) Proc. Natl. Acad. Sci. USA 101, 11599-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh, M. G., Thompson, D. A. & Weigel, R. J. (2000) Cancer Res. 60, 6367-6375. [PubMed] [Google Scholar]
- 33.Tsuchiya, Y., Nakajima, M., Kyo, S., Kanaya, T., Inoue, M. & Yokoi, T. (2004) Cancer Res. 64, 3119-3125. [DOI] [PubMed] [Google Scholar]
- 34.Semlali, A., Oliva, J., Badia, E., Pons, M. & Duchesne, M.-J. (2004) J. Steroid Biochem. Mol. Biol. 88, 247-259. [DOI] [PubMed] [Google Scholar]
- 35.Cavarretta, I. T. R., Mukopadhyay, R., Lonard, D. M., Cowsert, L. M., Bennett, C. F., O'Malley, B. W. & Smith, C. L. (2002) Mol. Endocrinol. 16, 253-270. [DOI] [PubMed] [Google Scholar]
- 36.Burakov, D., Crofts, L. A., Chang, C.-P. B. & Freedman, L. P. (2002) J. Biol. Chem. 277, 14359-14362. [DOI] [PubMed] [Google Scholar]
- 37.Wang, J.-C., Kakefuda, D., Nonaka, D. F., Khodabakhsh, D. B., Haqq, C. & Yamamoto, K. R. (2004) Proc. Natl. Acad. Sci. USA 101, 15603-15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedman, J. R., Larris, B., Le, P. P., Harshani Peiris, T., Arsenlis, A., Schug, J., Tobias, J. W., Kaestner, K. H. & Greenbaum, L. E. (2004) Proc. Natl. Acad. Sci. USA 101, 12986-12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Britton, S., Freeman, T., Vafiadaki, E., Keers, S., Harrison, R., Bushby, K. & Bashir, R. (2000) Genomics 68, 313-321. [DOI] [PubMed] [Google Scholar]
- 40.Arlt, A., Grobe, O., Sieke, A., Kruse, M. L., Folsch, U. R., Schmidt, W. E. & Schafer, H. (2001) Oncogene 20, 69-76. [DOI] [PubMed] [Google Scholar]
- 41.Grobe, O., Arlt, A., Ungefroren, H., Krupp, G., Folsch, U. R., Schmidt, W. E. & Schafer, H. (2001) FEBS Lett. 494, 196-200. [DOI] [PubMed] [Google Scholar]
- 42.Schilling, D., Pittelkow, M. R. & Kumar, R. (2001) Oncogene 20, 7992-7997. [DOI] [PubMed] [Google Scholar]
- 43.Nelson, W. J. & Grindstaff, K. K. (1997) Curr. Biol. 7, R562-R564. [DOI] [PubMed] [Google Scholar]
- 44.Joberty, G., Petersen, C., Gao, L. & Macara, I. G. (2000) Nat. Cell Biol. 2, 531-539. [DOI] [PubMed] [Google Scholar]
- 45.Noda, Y., Takeya, R., Ohno, S., Naito, S., Ito, T. & Sumimoto, H. (2001) Genes Cells 6, 107-119. [DOI] [PubMed] [Google Scholar]
- 46.Gao, L., Joberty, G. & Macara, I. G. (2002) Curr. Biol. 12, 221-225. [DOI] [PubMed] [Google Scholar]
- 47.Weyrich, P., Kapp, K., Niederfellner, G., Melzer, M., Lehmann, R., Häring, H.-U. & Lammers, R. (2004) Mol. Endocrinol. 18, 1287-1300. [DOI] [PubMed] [Google Scholar]
- 48.Qiu, R. G., Abo, A. & Steven, M. G. (2000) Curr. Biol. 10, 697-707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.