Abstract
Age-based inheritance of centrosomes in eukaryotic cells is associated with faithful chromosome distribution in asymmetric cell divisions. During Saccharomyces cerevisiae ascospore formation, such an inheritance mechanism targets the yeast centrosome equivalents, the spindle pole bodies (SPBs) at meiosis II onset. Decreased nutrient availability causes initiation of spore formation at only the younger SPBs and their associated genomes. This mechanism ensures encapsulation of nonsister genomes, which preserves genetic diversity and provides a fitness advantage at the population level. Here, by usage of an enhanced system for sporulation-induced protein depletion, we demonstrate that the core mitotic exit network (MEN) is involved in age-based SPB selection. Moreover, efficient genome inheritance requires Dbf2/20-Mob1 during a late step in spore maturation. We provide evidence that the meiotic functions of the MEN are more complex than previously thought. In contrast to mitosis, completion of the meiotic divisions does not strictly rely on the MEN whereas its activity is required at different time points during spore development. This is reminiscent of vegetative MEN functions in spindle polarity establishment, mitotic exit, and cytokinesis. In summary, our investigation contributes to the understanding of age-based SPB inheritance during sporulation of S. cerevisiae and provides general insights on network plasticity in the context of a specialized developmental program. Moreover, the improved system for a developmental-specific tool to induce protein depletion will be useful in other biological contexts.
Keywords: conditional degron, differential centrosome inheritance, meiotic plaque formation, meiosis, spindle polarity
DIFFERENTIAL inheritance of centrosomes or corresponding structures can be observed in many organisms ranging from simple, unicellular fungi to mammals (Pereira et al. 2001; Yamashita et al. 2007; Wang et al. 2009; Conduit and Raff 2010; Januschke et al. 2011; Izumi and Kaneko 2012; Salzmann et al. 2014). The underlying spindle polarity is based on distinct functional qualities of the spindle poles and is important for high fidelity of genome inheritance during asymmetric cell divisions (Miller and Rose 1998; Beach et al. 2000; Piel et al. 2000; Liakopoulos et al. 2003; Rebollo et al. 2007; Rusan and Peifer 2007; Wang et al. 2009; Januschke et al. 2013; Lerit and Rusan 2013). One of the best-studied model organisms for spindle polarity is the yeast Saccharomyces cerevisiae. Cells of S. cerevisiae divide asymmetrically by budding. Alignment of the mitotic spindle with the mother–daughter axis requires coordinated interactions of astral microtubules (aMT) with polarized actin cables at the bud neck and the bud cortex (Shaw et al. 1997; Beach et al. 2000; Liakopoulos et al. 2003; Sheeman et al. 2003). The intrinsic result of spindle polarity in S. cerevisiae is an age-based inheritance mechanism of the spindle pole bodies (SPBs), the sole microtubule organizing centers equivalent to centrosomes of higher eukaryotes. The predominantly conservative SPB duplication between G1 and S phase results in an older SPB and a younger SPB, which consists mostly of newly synthesized proteins (Adams and Kilmartin 1999; Menendez-Benito et al. 2013). This younger SPB is retained in the mother cell whereas the old SPB migrates into the bud (Pereira et al. 2001).
During gametogenesis of S. cerevisiae, which is called sporulation, the situation is even more complex due to the higher number of genomes that must be faithfully distributed. In this developmental program, spore formation is coupled to meiotic divisions resulting in the formation of four haploid genomes encapsulated by spore walls and contained within the remnants of the former mother cell, then called an ascus (Esposito and Klapholz 1981). During meiosis, SPBs are duplicated twice, which results in four SPBs of three different generations: one old, one of intermediate age, and two young SPBs. The meiotic divisions show no obvious asymmetry; yet, establishment of meiotic spindle polarity has been demonstrated by an age-based hierarchy of SPB inheritance during spore formation (Taxis et al. 2005). Sporulation is initiated in cells deprived of a fermentable carbon source as well as a nitrogen source in the presence of a nonfermentable carbon source such as acetate (Freese et al. 1982). Timing and progression of meiosis and spore formation is controlled by a transcriptional cascade; early, middle, midlate, and late gene expression events can be distinguished (Kawaguchi et al. 1992; Chu et al. 1998). Initiation of spore formation takes place around the onset of meiosis II at the cytoplasmic faces of the SPBs by substitution of the γ-tubulin complex and its receptor Spc72 with meiotic plaques (MPs) (Davidow et al. 1980; Knop and Schiebel 1998; Knop and Strasser 2000). The MPs are composed of the essential components Mpc54, Mpc70/Spo21, and Spo74, as well as the auxiliary, stabilizing factor Ady4 (Knop and Strasser 2000; Nickas et al. 2003; Mathieson et al. 2010).
The MPs serve as nucleation platforms and anchors for de novo formation of the prospore membranes (PSM), which derive from fusion of secretory vesicles and grow around the nuclear lobes and parts of the cytoplasm (Neiman 1998; Nakanishi et al. 2006; Mathieson et al. 2010). A protein coat consisting of Ssp1, Ady3, Irc10, and Don1 covers the leading edge of the growing PSMs (Knop and Strasser 2000; Nickas and Neiman 2002; Lam et al. 2014). The protein Ssp1 is essential for PSM formation; it is required for localization of the other proteins to the leading edge and to maintain the opening of the PSMs until the end of meiosis II (Moreno-Borchart et al. 2001; Lam et al. 2014). The cytokinetic event of PSM closure occurs after spindle breakdown and depends on the removal of Ssp1 from the leading edge (Maier et al. 2007; Diamond et al. 2009; Paulissen et al. 2016). Finally, the four spore wall layers (mannan, β-glucan, chitosan, and dityrosine) are synthesized consecutively within the lumen of the PSMs, resulting in the protection of spores against harsh environmental conditions, whereas the remnants of the mother cell mature to form the ascus (Coluccio et al. 2004, 2008; Eastwood et al. 2012).
Spindle polarity plays an essential role in spore number control. This means that sporulating S. cerevisiae cells regulate the number of MPs and, thus, spores in response to the available nutrients. Reduction of, e.g., acetate leads to a decrease in MP protein levels, which results in modification of selected SPBs with an MP and the formation of less than four spores per cell (Davidow et al. 1980; Nickas et al. 2004; Taxis et al. 2005; Gordon et al. 2006). In this case, SPB inheritance is not random but linked to the age of the SPB; the two young SPBs are preferred over the older ones and the oldest SPB has the least chance to be incorporated into a spore. This mechanism maximizes intra-ascus mating by the inheritance of nonsister genomes originating from the two different meiosis II spindles. Thus, beneficial heterozygosities are preserved, which provide fitness advantages at the population level (Taxis et al. 2005; Sellis et al. 2016).
How meiotic cells discriminate between the different SPBs and generate a signal for MP formation is still an open question. Once the process is initiated, MP components self-organize into the mature MP, which is thought to be a crystal-like structure reminiscent of the central plaque of the SPB (Bullitt et al. 1997; Taxis et al. 2005). The current model is that the MP grows rapidly due to a positive feedback mechanism until saturation is reached. MP formation happens in a consecutive fashion with delayed maturation at older SPBs (Taxis et al. 2005). Moreover, age-based selection of SPBs relies on the outer plaque proteins Nud1 as well as Spc72; potentially, these proteins link the presence of aMTs to differences between the SPBs (Gordon et al. 2006).
During vegetative growth of S. cerevisiae, several factors and pathways are involved in the establishment and maintenance of cell and spindle polarity. Among them is the mitotic exit network (MEN), an equivalent to the metazoan hippo tumor suppressor pathway (Hergovich and Hemmings 2012). In mitosis, the essential function of the MEN takes place in late anaphase by integration of temporal cues of mitotic progression with spatial signals of spindle positioning to control the release of Cdc14 to the cytoplasm (Shou et al. 1999; Visintin et al. 1999; Bardin et al. 2000; Adames et al. 2001; Hu et al. 2001). The main role of the phosphatase Cdc14 is to counteract Cdk1 activity, thereby allowing the cell to exit mitosis and to reenter G1 phase (Jaspersen et al. 1998; Visintin et al. 1998; Mohl et al. 2009). However, the MEN also fulfils functions before and after mitotic exit: during metaphase, it is required for the establishment of spindle polarity by targeting Kar9 localization to aMTs nucleated at the old SPB, and after exit from mitosis the network acts directly on several proteins at the bud neck to promote cytokinesis (Meitinger et al. 2010, 2013; Hotz et al. 2012a,b).
The core MEN consists of the small GTPase Tem1, the PAK (p21-activated kinase)-like kinase Cdc15, the downstream NDR (nuclear Dbf2-related) kinases Dbf2 and Dbf20, Mob1, and the SPB outer plaque protein Nud1. Mob1 forms complexes with the paralogs Dbf2 or Dbf20 and acts as a coactivator; Nud1 serves as signaling scaffold (Gruneberg et al. 2000; Lee et al. 2001; Mah et al. 2001; Visintin and Amon 2001; Rock et al. 2013). Until late anaphase, Tem1 is kept in its inactive GDP-bound form by the bipartite GAP (GTPase activating protein) Bfa1-Bub2 (Geymonat et al. 2002; Fraschini et al. 2006; Caydasi et al. 2012). To activate the MEN, the polo-like kinase Cdc5 inhibits the GAP activity of Bfa1-Bub2 by phosphorylation, a function that is antagonized by the spindle position checkpoint kinase Kin4 (Hu et al. 2001; Hu and Elledge 2002; Geymonat et al. 2003; Park et al. 2004; Pereira and Schiebel 2005; Maekawa et al. 2007; Kim et al. 2012). As the daughter cell-directed SPB passes the bud neck, Kin4 is inhibited by Lte1, which localizes specifically to the bud (Geymonat et al. 2009; Bertazzi et al. 2011; Falk et al. 2011). This triggers activation of Tem1 and Cdc5 at the daughter-localized SPB and leads to SPB recruitment of Cdc15, which activates the Dbf2-Mob1 kinase complex resulting in sustained release of Cdc14 from the nucleolus to the cytoplasm (Asakawa et al. 2001; Visintin and Amon 2001; Mohl et al. 2009; Rock and Amon 2011; Valerio-Santiago and Monje-Casas 2011; Falk et al. 2016; Gryaznova et al. 2016).
During sporulation, MEN activity has been detected mostly during the second meiotic division (Attner and Amon 2012). Phenotypic analyses on Cdc15 mutants showed participation in PSM formation, exit from meiosis II, and cytokinesis (Kamieniecki et al. 2005; Pablo-Hernando et al. 2007; Diamond et al. 2009; Attner and Amon 2012). Furthermore, several mechanistic differences between mitotic and meiotic cell divisions were reported: First, MEN activity in meiosis neither requires the scaffold protein Nud1 nor localization of its components to an SPB (Attner and Amon 2012). Second, the GTPase Tem1 and its GAP complex Bfa1-Bub2 are dispensable for activation of the kinases Cdc15 and Dbf2/20-Mob1, as well as spore formation (Gordon et al. 2006; Attner and Amon 2012). Third, Dbf20 is the predominantly active NDR kinase and needs Cdc15 activity to associate with the Mob1 coactivator (Attner and Amon 2012).
Here, we report multiple meiotic roles of the MEN in the regulation of SPB inheritance, meiotic plaque numbers, and cytokinesis during sporulation of S. cerevisiae. At the transition from meiosis I to meiosis II, Cdc15 exhibits an inhibitory function on MP formation, while Cdc15 and the terminal Dbf2-Mob1 and Dbf20-Mob1 kinase complexes are involved in the establishment of meiotic spindle polarity. After meiosis II, Cdc15 functions independently in PSM closure, whereas Dbf2-Mob1 and Dbf20-Mob1 are necessary for efficient spore maturation.
Materials and Methods
Yeast strains and plasmids
All yeast strains used in this study are derivatives of SK1 strains YKS32 (Knop and Strasser 2000) and LH177 (Huang et al. 2005) and their relevant genotypes are described in Supplemental Material, Table S2 in File S1. The control strains used during the experiments are mentioned by name in the figure legends. The strains have the same genotype as the mutant strains [e.g., auxotrophy markers and tobacco etch virus (TEV) protease expression], but are lacking the modification of the target gene with a sid-tag (sporulation-induced depletion). PCR-based manipulation of target genes at the chromosomal locus was performed as previously described (Janke et al. 2004; Taxis and Knop 2012). Yeast transformation was done by the lithium acetate method (Schiestl and Gietz 1989). Correct integration of the PCR products was tested by PCR on chromosomal DNA and, if applicable, by immunodetection of the fusion proteins. Genetic manipulations were introduced into haploid strains; diploids were obtained by mating of respective haploids. Alternatively, diploids were obtained by tetrad dissection of heterozygous strains and subsequent mating of haploids. For creation of YCR329, the PIME2-pTEV+-TDIT1::kanMX4 cassette of pCR107 was amplified with primers containing homologous sequences for HIS3 or TRP1. For creation of YCR370, the kanMX4 resistance markers of YAB12 and a MATα haploid of YCT716 were substituted by the PCR-amplified hphNT1 cassette of pFA6a-hphNT1.
Standard protocols were used for yeast cultivation (Sherman 2002). In general, strains with stable integrations were grown on YPD, selection for antibiotics resistances was performed on YPD (1% yeast extract, 2% peptone, and 2% glucose) supplemented with 100 µg/liter ClonNAT, 200 µg/liter G418 or 300 µg/liter HygromycinB, selection for auxotrophy markers was done on synthetic complete (SC) medium with 2% glucose lacking uracil, leucine, histidine, tryptophan, or lysine. Growth assays were performed with 1:5 serial dilutions using an initial OD600 of 0.008. For fluorescence microscopy of vegetatively growing cells, low-fluorescence medium with 2% glucose was used (Usherenko et al. 2014).
Plasmids were constructed by standard methods (Ausubel et al. 2001) and are listed with their respective genotypes in Table S3 in File S1. Yeast codon-optimized yomNeonGreen (Shaner et al. 2013) was synthesized by GeneArt (Life Technologies, Carlsbad, CA). To create pCR93, pmTurquoise2-C1 (Goedhart et al. 2012) was used as template to create three cassettes with overlapping linker sequences by PCR; these fragments were then joined and cloned into the TADH1-containing backbone of pYM12 by Gibson assembly (Gibson et al. 2009). For creation of pCR87 and pCR124 from pKT178-yomTagRFP-T and pKT146-yomTagRFP-T (Lee et al. 2013), the open reading frames of the genes were changed by site-directed mutagenesis within a linker sequence to make the plasmids compatible with S3-primers (Janke et al. 2004). pCR118 was created by ligation of a PCR product containing the first 2250 bp of CDC15 into pDS89, subsequent site-directed mutagenesis of the psd coding region to yield the psdE139N variant (Usherenko et al. 2014), and finally replacement of the ADH1 promoter by the one of SPO74 from pCR6.
Sporulation experiments
Synchronous sporulation in liquid culture was performed as described before (Taxis et al. 2005). For the plasmid-based experiments of sporulation-specific expression of CDC15ΔC-psdE139N, the glycerol step was skipped and SC instead of YP medium was used; liquid precultures were grown in ventilated, clear cell culture flasks and illuminated by blue light LEDs (465 nm) with a photon flux of 30 µmol m−2 sec−1; cells were incubated in the dark during sporulation. For live-cell microscopy, cells were attached to poly-L-lysine coated glass-bottom dishes (MatTek Corporation, Ashland, MA) 4 hr after induction of sporulation, washed twice with 0.01% potassium acetate (KOAc), and covered with 0.1% KOAc or water. Total sporulation was checked after 2 days.
To assess spore numbers, sporulation on solid medium with 1% KOAc and DNA staining with Hoechst 33342 were performed as previously described (Jungbluth et al. 2012). Experiments on sister dyad formation were done in the same way skipping cell fixation and DNA staining.
Microscopy
Microscopy was performed with an Axiovert 200M inverse microscope (Zeiss [Carl Zeiss], Thornwood, CA) equipped with a 1394 ORCA ERA CCD camera (Hamamatsu Photonics, Hamamatsu City, Japan), filter sets for DAPI, cyanGFP, enhanced GFP (EGFP), yellow fluorescent protein (YFP), and rhodamine (Chroma Technology, Bellows Falls, VT), and a Zeiss 63 × Plan Apochromat oil lens (NA 1.4). Single-plane bright field or phase contrast images of the cells and z-stacks (0.5 or 0.3 µm z-spacing) in the appropriate fluorescence channels were recorded using the image acquisition software Volocity 5.3 (Perkin-Elmer [Perkin-Elmer Cetus], Norwalk, CT). Next, 2 × 2 binning was used for time course experiments. Images were exported as 16-bit TIFFs and further processed and evaluated in ImageJ (Schneider et al. 2012). For analysis of protein localization, image stacks (0.3 µm z-spacing) of the fluorescent channels were deconvolved: first, point spread functions for each channel were computed by the ImageJ plugin PSF Generator using the Richards & Wolf model (Kirshner et al. 2013); image deconvolution was then performed on the z-stacks by the ImageJ plugin DeconvolutionLab with 25 iterations of the Richardson–Lucy algorithm (Vonesch and Unser 2008). Composites for evaluations were prepared using maximum projections of the fluorescence channels and bicubic image scaling. No image manipulations other than adjustment of histogram and background subtraction were applied.
Immunoblotting
Samples from liquid cultures were treated by alkaline lysis with subsequent trichloroacetic acid precipitation and subjected to immunoblotting as previously described (Jungbluth et al. 2010). For quantification of meiotic plaque proteins, Nud1, and Cnm67, protein samples of the time course (4–10 hr, samples taken every hour) were pooled before subjecting them to SDS-PAGE. Proteins were detected by primary antibodies specific for either hemagglutinin (HA; Sigma [Sigma Chemical], St. Louis, MO), Myc (Cell Signaling Technology, Danvers, MA), TEV protease (a kind gift of M. Ehrmann, University of Duisburg-Essen, Germany) or Tub1 (loading control; Abcam, Cambridge, UK), HRP-coupled secondary antibodies against mouse or rabbit IgG (Dianova, Hamburg, Germany; Santa Cruz Biotechnology, Santa Cruz, CA) with a Chemostar professional imaging device (INTAS, Göttingen, Germany). Representative images were prepared by inverting gray values and adjusting brightness and contrast. Quantification of the signals was done using 16-bit images and ImageJ. Protein levels were corrected for tubulin signals and normalized to the respective controls.
Quantitative fluorescence measurements
Sodium azide was immediately added to samples from liquid cultures to a final concentration of 10 mM and samples were stored on ice until the end of the experiment. Then, samples were transferred into a black, flat-bottom 96-well microplate and fluorescence intensity was recorded with a plate reader (Synergy Mx, BioTek, Winooski, VT); wavelengths were set to 485 nm excitation and 525 nm emission. A control strain (YKS32 with pRS426) was used to subtract background fluorescence.
Data evaluation and statistical tests
Data evaluation and visualization was performed in R (R Core Team 2015). Fractions of different cell species were calculated as the percent of total cells. Stacked bar plots were created using mean values. Statistical evaluations of the data shown in stacked bar plots are given per figure in Table S1. Box plots show the median and the first and third quartiles, notches span the maximum 1.5-fold of the interquartile range, and data points outside this range are shown as separate points. Numbers of independent biological replicates (n) and minimum as well as maximum counted cell numbers are stated in the figure descriptions.
Sporulation efficiency as a simplified measure of sporulation performance was calculated by the following formula:
Tests for statistical significance of differences were performed according to data type and structure: Fisher’s exact test was used for categorical data with less than two categories. Categorical data with only two categories were transformed to continuous relative values. The Shapiro–Wilk test was applied to check for normality. Accordingly, a Welch two-sample t-test or the exact Wilcoxon–Mann–Whitney test of the “coin” R-package (Hothorn et al. 2008) was used to check for statistically significant differences. To testing for differences in meiotic plaque protein levels and initial amounts of Nud1 and Cnm67, two-sided one-sample t-tests against 100% were performed.
Electron microscopy
Concentrated yeast cell suspensions were high-pressure frozen (HPF Compact 02, Wohlwend, CH) and freeze-substituted (AFS2; Leica, Wetzlar, Germany). For the 6 hr samples, a freeze-substitution medium based on acetone was used containing 0.25% osmium tetroxide, 0.2% uranyl acetate, and 5% ddH2O; medium for the 9 hr samples contained additionally 0.1% KMnO4. All samples were freeze-substituted according to the following protocol: −90° for 20 hr, from −90 to −60° for 1 hr, −60° for 8 hr, −60 to −30° for 1 hr, −30° for 8 hr, and −30 to 0° for 1 hr. At 0°, samples were washed three times with acetone before a 1:1 mixture of Epon 812 substitute resin (Fluka Chemical, Buchs, Switzerland) and acetone was applied at room temperature for 2 hr. The 1:1 mixture was substituted with pure resin to impregnate the samples overnight. After another substitution with fresh Epon, samples were polymerized at 60° for 2 days. The samples containing polymerized Epon blocks were then trimmed with razor blades and cut to 50 nm ultrathin sections using an ultramicrotome (UC7; Leica) and a diamond knife (Diatome, Biel, Switzerland). Sections were applied onto 100 mesh copper grids coated with pioloform. For additional contrast, mounted sections were poststained with 2% uranyl acetate for 20–30 min and subsequently with lead citrate for another 1–2 min. The sections were finally analyzed and imaged using a JEM-2100 transmission electron microscope (JEOL, Tokyo, Japan) equipped with a 2k × 2k F214 fast-scan CCD camera (TVIPS, Gauting, Germany).
Data availability
Strains and reagents are available on request. All data that we used for our investigations are included in the manuscript or in the Supplemental Material.
Results
Enhancement of a system for sporulation-specific protein depletion
Most of the proteins of the MEN fulfill essential roles in vegetative growth of S. cerevisiae. Therefore, a reliable and generic method to create loss-of-function mutants of essential proteins during meiosis was necessary. We decided to use a system relying on N-terminal modification of the target proteins with a genetically-encoded protein tag, TDegF, which contains an inactive degradation-inducing sequence (degron). By use of the meiosis-specific IME2 promoter, a chromosomally-encoded TEV protease with enhanced processivity (pTEV+) is produced exclusively during sporulation and activates the degron by cleavage of the tag. This leads to depletion of the target proteins during sporulation (Taxis et al. 2009; Jungbluth et al. 2010, 2012). However, we changed the existing method in two points, as we were not able to achieve sufficient meiosis-specific downregulation of Cdc15 with this method (data not shown). We replaced the constitutive promoters (PADH1 or PCYC1) that control synthesis of the degron-tagged proteins with a promoter that is active only during vegetative growth (PMCD1; Klein et al. 1999; Clyne et al. 2003). Additionally, we exchanged the CYC1 terminator of the TEV protease construct (pTEV+) by the DIT1 terminator. This terminator conferred about fourfold higher expression levels of a tester construct than the CYC1 terminator in logarithmically growing cells and was shown to enhance expression under diverse starvation conditions (Yamanishi et al. 2013; Ito et al. 2013). We envisioned that these changes should lead to robust downregulation of target proteins specifically during meiosis; the chromosomal tagging construct PMCD1-GFP-TDegF-3HA was termed sid-tag (Figure 1A). To check the effect of the terminator exchange in combination with the meiosis-specific IME2 promoter, we compared meiotic expression of a PIME2-GFP-pTEV+ construct with terminator sequences of either CYC1 or DIT1. Indeed, the expression of the PIME2-GFP-pTEV+-TDIT1 construct was two- to threefold higher than that of the PIME2-GFP-pTEV+-TCYC1 construct (Figure 1B). Next, a PIME2-pTEV+-TDIT1 construct was integrated at two chromosomal loci (HIS3 and TRP1) to ensure efficient meiotic production of the pTEV+ protease.
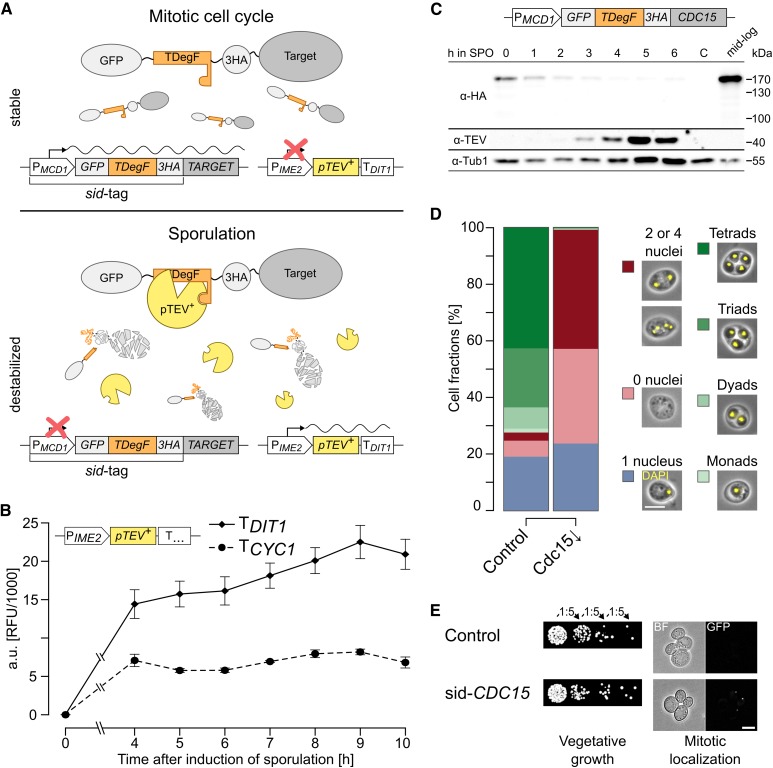
Figure 1.
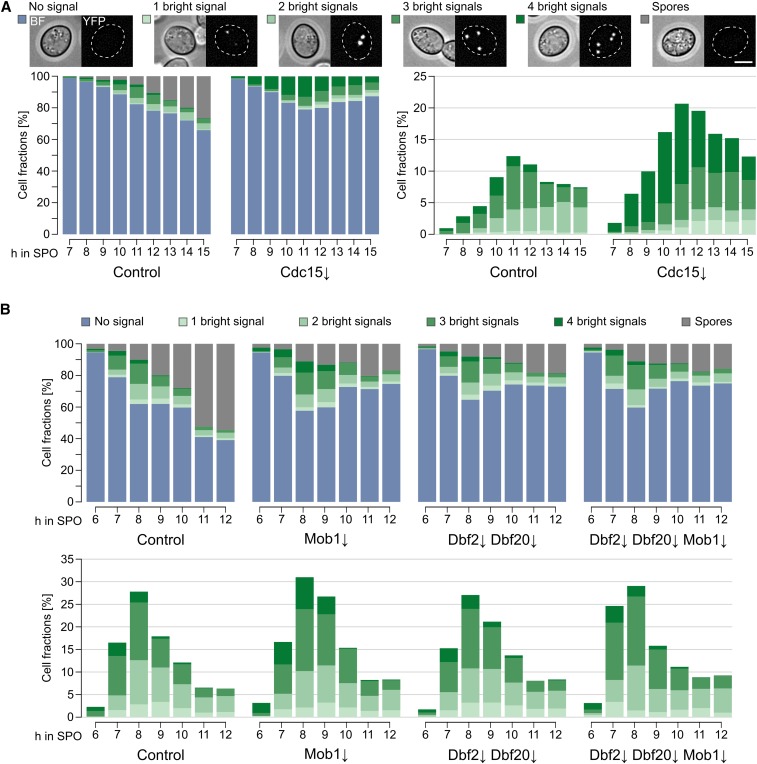
Enhanced sporulation-induced protein depletion (A) Principle of sporulation-induced protein depletion (sid). The sid-tag is fused to the 5′-end of the target gene and substitutes the original promoter with the mitosis-specific MCD1 promoter and the GFP-TDegF [tobacco etch virus (TEV) protease-dependent degron]-3HA-tag. At one or more chromosomal loci, a functional cassette of the meiosis-specific IME2 promoter, pTEV+ gene, and DIT1 terminator is integrated. During vegetative growth, the target gene is expressed and stable. Upon switch to sporulation conditions, the MCD1 promoter is downregulated and pTEV+ is produced after initiation of meiosis. TEV cleavage of TDegF leads to activation of an N-degron and degradation of the target protein by the ubiquitin–proteasome system. (B) Terminator dependency of GFP-pTEV+ production during sporulation. Diploid yeast cells with high-copy plasmids bearing either PIME2-GFP-pTEV+-TCYC1 or PIME2-GFP-pTEV+-TDIT1 were subjected to sporulation conditions. Expression of the constructs was followed by fluorimeter measurements, an empty vector control allowed background subtraction (TCYC1: n = 2, TDIT1: n = 9; mean ± SEM). (C) Sporulation-induced depletion of Cdc15. Diploid sid-CDC15 cells with four integrated copies of PIME2-pTEV+-TDIT1 (YCR332) were subjected to sporulation conditions. Samples for immunoblotting were taken at the indicated time points as well as during midlog growth phase, a strain without sid-tag (taken at 0 hr in sporulation medium) served as negative control [(C) YCR329]. Anti-HA, anti-TEV, and anti-Tub1 (loading control) antibodies were used for detection. (D) Sporulation of the Cdc15 depletion mutant. The Cdc15 depletion strain (Cdc15↓; YCR332) was sporulated together with a control strain (YCR329) on solid sporulation medium with 1% acetate. Cells were classified according to their morphology and number of nuclei. Bar plot shows average results of at least six biological replicates, between 163 and 516 cells were counted per replicate. Please see Table S1 for statistical evaluations. Bar, 5 µm. (E) Characterization of the sid-CDC15 strain during vegetative growth. sid-CDC15 cells (YCR332) were subjected to a serial dilution growth assay (left) as well as fluorescence microscopy for mitotic localization. Bar, 5 µm. YCR329 was used as control.
The essential MEN kinase Cdc15 was used as target to test the efficacy of the modified depletion system during sporulation and check for negative side effects on vegetative growth. Remarkably, Cdc15 was efficiently depleted shortly after induction of meiosis (Figure 1C). Compared to a logarithmically growing culture, protein levels were already reduced in the presporulation culture (0 hr). Similar observations have been made before with the MCD1 promoter; it may be that the number of M phase cells is reduced in cultures growing in medium containing the poor carbon source acetate (Klein et al. 1999). As expected from earlier studies (Kamieniecki et al. 2005; Pablo-Hernando et al. 2007), Cdc15 depletion resulted in a massive sporulation defect; cells underwent the meiotic divisions but did not form spore-containing asci (Figure 1D). Instead, cells arrested with either two, four, or no distinct nuclei. This observation was in agreement with earlier studies, which suggested a role for Cdc15 in exit of meiosis II, PSM growth, and closure, as well as spore wall maturation (Kamieniecki et al. 2005; Pablo-Hernando et al. 2007; Diamond et al. 2009; Attner and Amon 2012). The high number of sid-CDC15 cells without detectable nuclei prompted us to use a genetically-encoded, fluorescently-labeled variant of histone H2B (Htb2-RFP) as an in vivo marker in the CDC15 mutant. Also, in this case, a high fraction of cells without detectable nuclei were observed after sporulation of sid-CDC15 cultures (Figure S1 in File S2). This shows that the loss of signal was not only evoked by incomplete staining, but that a considerable fraction of cells seemed to fragment their nuclei during sporulation. At the same time, chromosomal fusion of the sid-tag to CDC15 did not impair vegetative growth or localization of sid-Cdc15 to the SPB in mitotic anaphase (Figure 1E). These results imply that the modifications of the meiosis-specific depletion system increased the usability of the method. However, we did not directly test dependence of sid-Cdc15-depletion on TEV protease activity and cannot exclude that some destabilization of sid-tagged proteins during sporulation may occur independently of TEV protease activity. Indeed, it has been observed previously that TEV protease-independent reduction of target protein abundance is possible; yet efficient target protein depletion usually required the presence of the TEV protease (Taxis et al. 2009; Jungbluth et al. 2010).
Dbf2-Mob1 and Dbf20-Mob1 are required for efficient spore formation
To gather information about a putative function of the Dbf2-Mob1 and Dbf20-Mob1 complexes during sporulation, we first sought to examine the meiotic localization of Dbf2 and Dbf20 fused to GFP. In line with previous results on Cdc15 and Mob1 localization and the lack of Nud1 requirement for meiotic MEN activity (Attner and Amon 2012), we did not observe specific localization of Dbf2 and Dbf20 during sporulation (Figure S2 in File S2).
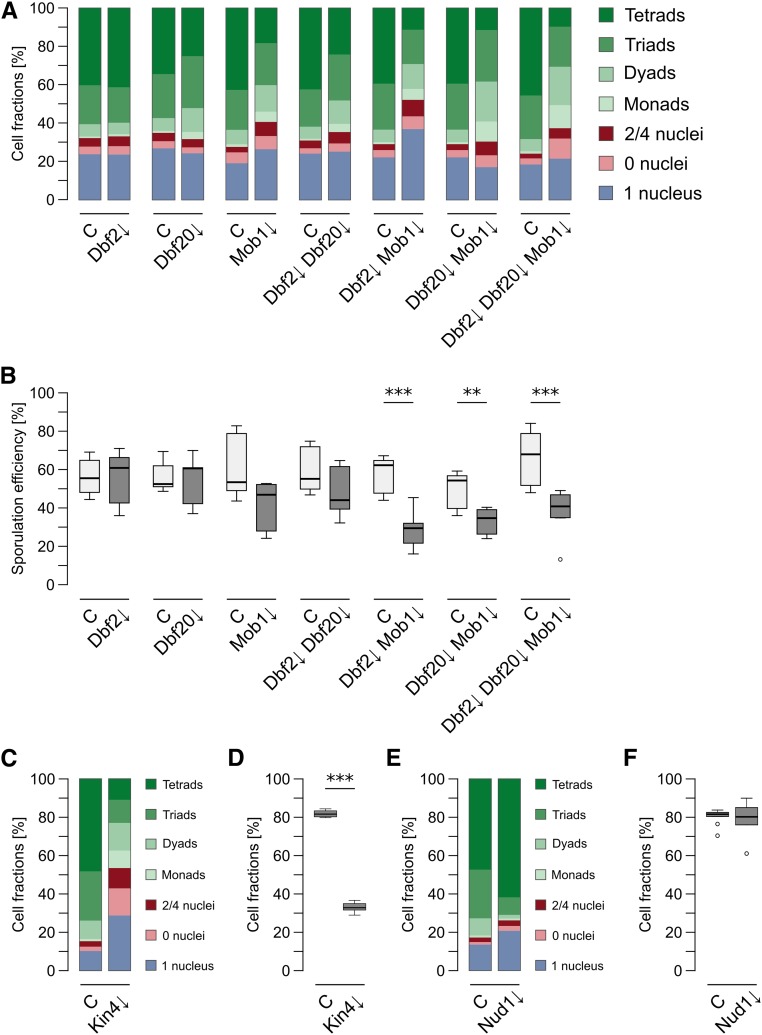
We then investigated the role of the Dbf2-Mob1 and Dbf20-Mob1 complexes during sporulation by creation of depletion mutants for the single proteins (Dbf2↓, Dbf20↓, and Mob1↓) or different combinations (Dbf2↓ Dbf20↓, Dbf2↓ Mob1↓, Dbf20↓ Mob1↓, and Dbf2↓ Dbf20↓ Mob1↓). The efficiency of protein depletion and pTEV+ expression was checked by western blot analysis during the first 6 hr of sporulation for the single mutants and the triple mutant. All targets were undetectable 4 hr after induction of sporulation (Figure S3A in File S2). During vegetative growth, no negative effect was observed in any of the strains (Figure S3, B and C in File S2). Subsequently, we used these strains to assess spore formation. In short, a clear reduction of spore numbers compared to the control strain was observable for the Mob1↓ mutant, all double mutants, and the triple mutant (Figure 2A). Depletion of Dbf2 alone had no effect on sporulation and depletion of Dbf20 caused only a mild phenotype. Generally, the mutants displayed a decrease in the number of tetrads and an increase of dyads and monads. The strongest effects were observed for the Dbf2↓ Mob1↓ and Dbf20↓ Mob1↓ double mutants as well as the Dbf2↓ Dbf20↓ Mob1↓ triple mutant, which was also depicted by their sporulation efficiencies (Figure 2B). Notably, even the Dbf2↓ Dbf20↓ Mob1↓ triple mutant did not block spore formation as completely as Cdc15 depletion did. While most mutants showed an impact on spore numbers, they still reacted to lowered acetate concentrations by further decreasing spore numbers (data not shown).
Figure 2.
Dbf2-Mob1 and Dbf20-Mob1 are required for efficient sporulation. (A) Impaired spore formation in strains with deficiencies in mitotic exit network (MEN) kinase complexes. Depletion mutants for the indicated proteins (Dbf2↓: YCR359; Dbf20↓: YCR344; Mob1↓: YCR333; Dbf2↓ Dbf20↓: YCR363; Dbf2↓ Mob1↓: YCR447; Dbf20↓ Mob1↓: YCR450; and Dbf2↓ Dbf20↓ Mob1↓: YCR446) were sporulated together with a control strain [(C); harboring PIME2-pTEV+-TDIT1; YCR329] as described for Figure 1D. Cells were classified according to morphology and number of nuclei. Stacked bars represent the means of at least six independent biological replicates, between 137 and 577 cells per replicate were counted. Statistical analysis of the values for the different species and strains are shown in Table S1. (B) MEN kinase mutants show reduced sporulation efficiency. Sporulation efficiencies were calculated for the experiments described in (A). Statistical significance of the differences was checked by Wilcoxon–Mann–Whitney tests against controls (n ≥ 6; ** P ≤ 0.01, *** P ≤ 0.001 in Wilcoxon–Mann–Whitney tests). (C) Impaired spore formation in a Kin4↓ strain. Sporulation of a Kin4↓ strain (YAA215) was compared to a control strain (YAA146) as described for (A) (n ≥ 6; 94–423 cells per replicate). (D) Sporulation efficiencies of the Kin4↓ strain. Sporulation efficiency was calculated for the experiment shown in (C). Statistical significance of differences to the control strain was assessed by Wilcoxon–Mann–Whitney tests (n ≥ 6; *** P ≤ 0.001). (E) Sporulation profile of the Nud1↓ strain. A Nud1↓ (YAA216) and control [(C); YAA146] strain were sporulated as described for (A) (n ≥ 6; 130–780 cells per replicate). (F) Sporulation efficiency of the Nud1↓ strain. Sporulation efficiency was calculated for the experiment shown in (E). No statistical significant differences to the control were found in Wilcoxon–Mann–Whitney tests (n ≥ 6; P ≥ 0.05). Please see Table S1 for all statistical evaluations.
It has been shown that, during sporulation, Tem1 and its GAP complex Bfa1-Bub2 are dispensable for MEN activation (Kamieniecki et al. 2005; Gordon et al. 2006), whereas no direct analysis of the MEN inhibitor Kin4 has been undertaken during sporulation (D’Aquino et al. 2005; Pereira and Schiebel 2005; Maekawa et al. 2007; Gryaznova et al. 2016). Thus, we tested this kinase for a function in sporulation since it has been shown to directly phosphorylate Cdc15 in vitro (Ptacek et al. 2005). Fluorescently-labeled Kin4 displayed no specific localization during sporulation whereas the localization during vegetative growth was found to be as reported in the literature (Figure S4, A and B in File S2; D’Aquino et al. 2005; Pereira and Schiebel 2005). Next, we generated a sid-KIN4 mutant strain that allowed efficient depletion of the protein during sporulation; the tag had no impact on the mitotic function of Kin4 (Figure S5, A–C in File S2). Remarkably, Kin4 depletion caused severe reduction of spore numbers and sporulation efficiency (Figure 2, C and D). This shows that this mitotic MEN regulator is necessary for efficient spore formation.
To analyze putative functions of the MEN in SPB selection, we created a sid-NUD1 strain. Previously, it has been observed that a temperature-sensitive allele of this SPB outer plaque protein (nud1-2) induces randomization of SPB inheritance (Gordon et al. 2006). However, pronounced defects in SPB selection and genome inheritance were already found at permissive temperatures. Therefore, we reinvestigated Nud1 function by using the sporulation-induced depletion mutant. The protein was fully functional during vegetative growth and quickly depleted upon induction of sporulation (Figure S6, A–C in File S2). Surprisingly, in contrast to the nud1-2 allele, depletion of Nud1 induced higher spore numbers per ascus accompanied by a modest increase in unsporulated cells, whereas the sporulation efficiency was not significantly increased (Figure 2, E and F). We considered that SPB-associated Nud1 might be inaccessible for degradation resulting in depletion of only cytoplasmic Nud1, whereas a small fraction of SPB-associated Nud1 might be sufficient for its function. Hence, we applied the depletion system on the SPB protein Cnm67, which links Nud1 to the central plaque (Schaerer et al. 2001). No side effects were found during vegetative growth and depletion kinetics of Cnm67 were comparable to those of Nud1 (Figure S6, A–C in File S2). Depletion of Cnm67 completely blocked spore formation (Figure S6D in File S2). These results make it less likely that residual Nud1 fractions at the SPBs are causing the observed phenotype in Nud1↓ cells.
The MEN influences meiotic genome inheritance
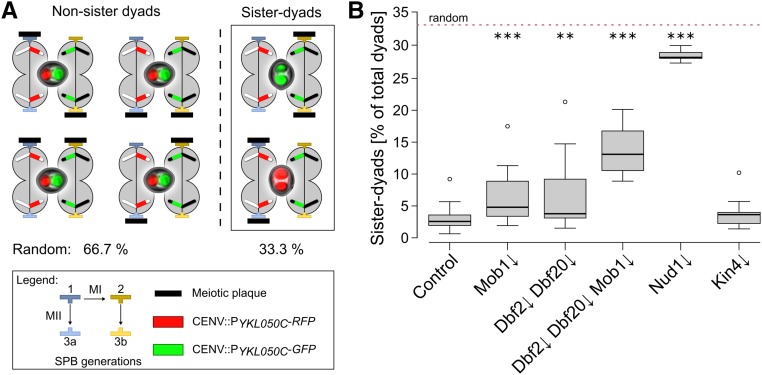
Next, we investigated genome inheritance during yeast meiosis in the Mob1↓, Dbf2↓ Dbf20↓, Dbf2↓ Dbf20↓ Mob1↓, and Kin4↓, as well as the Nud1↓, mutants. In dyads, the preference of the young SPBs for modification with an MP leads to nearly exclusive packaging of genomes from different meiosis II spindles. While without regulation, the theoretical random fraction of nonsister dyads would be 66.7%, this preference results in the formation of ∼95% nonsister dyads, containing homologous and not sister chromosomes in the two spores (Davidow et al. 1980; Okamoto and Iino 1981; Nickas et al. 2004; Taxis et al. 2005; Gordon et al. 2006). To investigate whether genome inheritance was disturbed in the different mutants, we used a yeast strain that allows assessment of sister-chromosome segregation (Gordon et al. 2006). This diploid strain harbors a centromere-linked gene encoding for RFP on one copy of chromosome V and one for GFP on the other. Both genes are expressed only after PSM closure. The two different fluorescent proteins permit discrimination of nonsister and sister dyads, depending on the fluorescent label distribution within the spores of a dyad (Figure 3A). Due to the complete lack of spores in the Cdc15↓ mutant, the analysis could not be performed with this strain. In the control strain, we observed a fraction of sister dyads below 5%, comparable with the results of an earlier study (Figure 3B; Gordon et al. 2006). The Nud1↓ strain displayed a nearly random distribution of sister and nonsister dyads as reported for the nud1-2 allele (Gordon et al. 2006). Both, the Mob1↓ and the Dbf2↓ Dbf20↓ strains produced significantly more sister dyads than the control and this fraction was further increased in the Dbf2↓ Dbf20↓ Mob1↓ mutant; however, the observed defects in nonsister dyad formation were much weaker than in the Nud1↓ mutant. In contrast to the core MEN components, depletion of Kin4 had no impact on the formation of nonsister dyads. Additionally, the assay allowed detection of chromosome segregation defects indicated by spores with either both or no fluorophores. Yet, the Mob1↓, Dbf2↓ Dbf20↓, Dbf2↓ Dbf20↓ Mob1↓, and Kin4↓ strains displayed no chromosome segregation defects in contrast to the Nud1↓ mutant (Figure S7, A–C in File S2). These results show that inactivation of the terminal MEN kinase modules partially interferes with faithful genome inheritance but not chromosome segregation. In contrast, depletion of Kin4—despite its massive spore number effects—does not affect genome inheritance. Overall, our results on the Nud1↓ mutant point to a negative role of this protein on spore numbers in general, opposed by a requirement of Nud1 for efficient establishment of meiotic spindle polarity and chromosome segregation.
Figure 3.
The MEN is required for efficient genome inheritance during meiosis. (A) Principle of a genome inheritance assay. A strain background heterozygous for the spore-autonomous markers CENV∷PYKL050C-RFP and CENV∷PYKL050C-GFP allows discrimination between sister and nonsister chromosome inheritance by fluorescence microscopy. The scheme depicts the different possibilities of SPB modification with two meiotic plaques and images of the resulting dyads together with the theoretical probabilities for sister and nonsister dyad distribution in case of random SPB selection. (B) Faithful genome inheritance depends on the MEN. Indicated mutants were created in the strain background shown in (A). Strains (control: YCR481, Mob1↓: YCR482, Dbf2↓ Dbf20↓: YCR483, Mob1↓ Dbf2↓ Dbf20↓: YCR525, Nud1↓: YCR699, and Kin4↓: YCR686) were sporulated on solid sporulation medium with 0.1% acetate and different dyad types were evaluated. Statistical significance of differences was tested by Wilcoxon–Mann–Whitney tests (n ≥ 8; ** P ≤ 0.01, *** P ≤ 0.001; 88–384 cells per replicate). MEN, mitotic exit network; RFP, red fluorescent protein; SPB, spindle pole body.
The MEN affects meiotic SPB selection
The observed effect on genome inheritance in the core MEN mutants could be due to a defect in age-based inheritance of SPBs caused by an altered pattern of MP formation. To test this possibility, we used a strain with the moderately slow-maturing tagRFP-T (maturation half-time: ∼100 min; Shaner et al. 2008) as a fluorescent timer fused to the integral SPB central plaque component Spc42 together with Mpc54-YFP as the MP marker. This allowed the correlation of SPB age with MP formation. Due to the maturation kinetics of tagRFP-T, three modification patterns were distinguished in cells with two mature MPs: modification of the two third-generation SPBs, modification of a first- or second-generation and one third-generation SPB, as well as modification of the first- and second-generation SPBs.
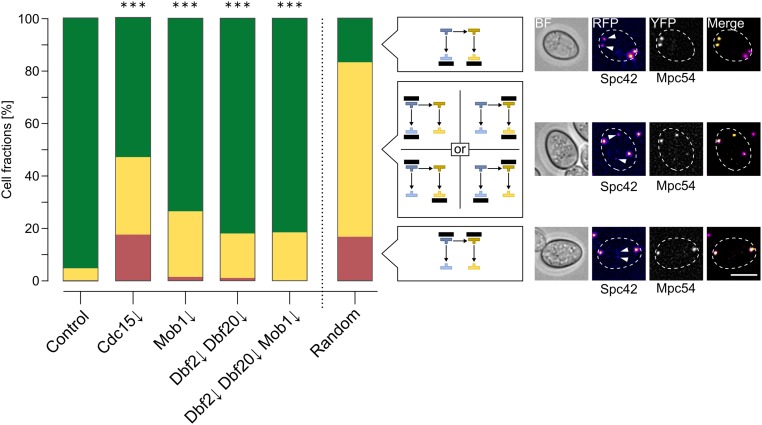
We performed this assay with the Cdc15↓, Mob1↓, Dbf2↓ Dbf20↓, and Dbf2↓ Dbf20↓ Mob1↓ mutants. For a completely random selection, the expected fractions would be 16.7% for modification of only the third-generation SPBs, 16.7% for selected first- and second-generation SPBs, while selection of one first- or second-generation SPB together with one third-generation SPB would occur in 66.7% of the cells. However, selection of SPBs for modification with MPs has been shown to be highly regulated; the two third-generation SPBs are by far preferred over the older ones. In unperturbed cells with two MPs, ∼95% of the cells modify the two youngest SPBs with MPs (Taxis et al. 2005; Gordon et al. 2006). We found similar values for the control strain in our experiments (Figure 4). Strikingly, all four tested mutants exhibited significantly lower percentages of cells with MPs at the youngest SPBs. In the Mob1↓ mutant, 25% of the cells showed modification of one older and one younger SPB; in the Dbf2↓ Dbf20↓ double mutant 17%, and in the Dbf2↓ Dbf20↓ Mob1↓ triple mutant 18% of cells displayed this pattern. This phenotype was even more severe in the Cdc15↓ mutant (30%) accompanied by MP formation at the oldest SPBs in 17% of the cells, a situation rarely found in the other strains. In summary, the results demonstrate a function of the MEN in SPB selection during sporulation, with a more pronounced role of Cdc15 than Dbf2, Dbf20, and Mob1 in this process.
Figure 4.
MEN kinases are involved in age-based SPB selection during meiosis. Live-cell microscopy images of cells (control: YCR555, Cdc15↓: YCR690, Mob1↓: YCR556, Dbf2↓ Dbf20↓: YCR557, and Dbf2↓ Dbf20↓ Mob1↓: YCR558) with Spc42-tagRFP-T (marker for SPB age) and MPC54-YFP (MP marker). Sporulation was induced with 1% potassium acetate for 4 hr; cells were then shifted for the rest of the experiment to water to increase the number of cells with < 4 meiotic plaques. Cells with two bright Mpc54-YFP signals were classified according to the SPB generations in which the MPs resided and three classes could be clearly distinguished: MPs at the youngest SPBs (green), the oldest SPBs (red), and at one younger and one older SPB (yellow). Differences to the control were all highly significant (Fisher’s exact test, P ≤ 0.001, control: 1137 cells, Cdc15↓: 132 cells, Mob1↓: 379 cells, Dbf2↓ Dbf20↓: 430 cells, and Dbf2↓ Dbf20↓ Mob1↓: 349 cells from at least four independent biological replicates). The right bar depicts distribution of cell types for a theoretical random situation. Example images show cells in BF, Spc42-RFP signals in false colors (arrow heads mark the youngest SPBs), Mpc54-YFP signals, and merges of the two fluorescence channels, cell outlines are shown as broken white lines. Bar, 5 µm. BF, brightfield channel; MEN, mitotic exit network; MP, meiotic plaque; RFP, red fluorescent protein; SPB, spindle pole body; YFP, yellow fluorescent protein.
Cdc15 influences meiotic plaque numbers
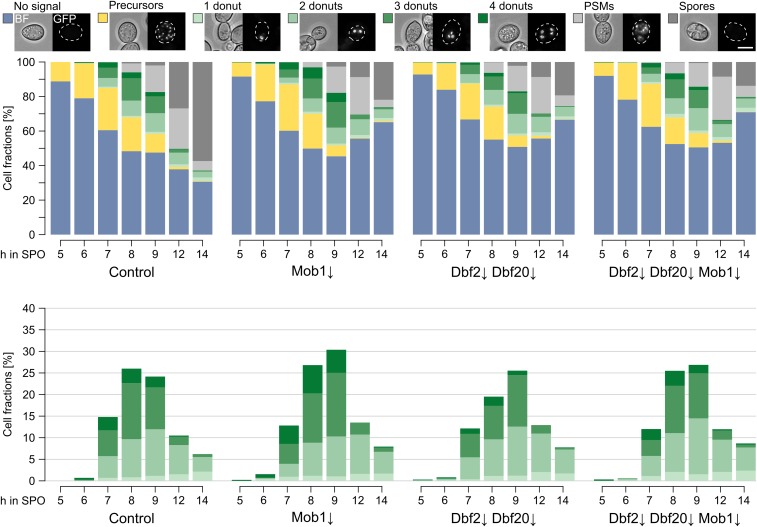
To investigate the influence of Cdc15, Dbf2, Dbf20, and Mob1 on the number of mature MPs, we performed fluorescence microscopy and followed the number of bright Mpc54-YFP signals during a sporulation time course. In accordance with a previous study that used Mpc70 as marker for MPs (Pablo-Hernando et al. 2007), we found robust localization of Mpc54 at SPBs in the Cdc15↓ strain, but no formation of refractive spores (Figure 5A). This could be explained by defects in exit from meiosis II and PSM formation and closure, which have been reported for a CDC15 mutant (Kamieniecki et al. 2005; Pablo-Hernando et al. 2007; Diamond et al. 2009). Remarkably, Cdc15 depletion led to a significant increase in the number of cells with four mature MPs under low-acetate conditions (Figure 5A and Figure S8 in File S2). In contrast, the kinetics of MP formation and number of MPs formed were comparable between cells deficient for the terminal MEN components and the control strain (Figure 5B). Most cells formed two or three MPs due to exposure of the cells to low-acetate conditions. However, the number of cells that formed refractive spores was much lower in cells depleted for Mob1; Dbf2 and Dbf20; or Dbf2, Dbf20, and Mob1. At the end of the time courses only ∼20% of the cells contained spores compared to ∼60% in control cells (Figure 5B). The time course analysis demonstrates that Cdc15, but not Dbf2, Dbf20, or Mob1, are involved in control of MP numbers. Due to the influence of Cdc15 on MP formation and the changes in age-based SPB selection of Cdc15↓ and Dbf2↓ Dbf20↓ Mob1↓ cells, we performed electron microscopy to investigate SPB morphology in sporulating cells. However, we did not observe morphological changes of SPBs and MPs in the mutants (Figure S9 in File S2).
Figure 5.
Functional separation of Cdc15 and the Dbf2-Mob1 and Dbf20-Mob1 complexes. (A) Cdc15 restricts meiotic plaque numbers and is essential for completion of sporulation. Meiotic plaque and spore formation was observed in Cdc15↓ cells (YCR357) compared to control cells (YCR356). Sporulation was induced with 1% potassium acetate for 4 hr; cells were then shifted for the rest of the experiment to water to increase the number of cells with less than four meiotic plaques. Cells were classified according to the number of punctuate bright Mpc54-YFP signals and presence of refractive spores. On the left, stacked bar plots of all cell classes are shown; on the right, only the fractions of cells with Mpc54-YFP signals (n = 5; 259–484 cells per timepoint and replicate) are shown. Examples show cells BF as well as YFP signals. Bar, 5 µm. (B) Dbf2, Dbf20, and Mob1 are involved in a late step of sporulation. Meiotic plaque formation was followed over time in the indicated mutants (control: YCR555, Mob1↓: YCR556, Dbf2↓ Dbf20↓: YCR557, and Dbf2↓ Dbf20↓ Mob1↓: YCR558). Conditions as in (A). The upper panel shows plots of all cell classes while the lower panel shows only the fractions of cells with meiotic plaques (Control, Mob1↓, and Dbf2↓ Dbf20↓: n = 4; Dbf2↓ Dbf20↓ Mob1↓: n = 2; 156–513 cells per timepoint and replicate). Bar, 5 µm. Please see Table S1 for statistical evaluations. BF, brightfield channel; SPO, sporulation medium/water; YFP, yellow fluorescent protein.
The reduction of spore numbers and defects in SPB selection in the MEN mutants could be evoked by decreased levels of MP proteins. Therefore, we measured the levels of Mpc54-9Myc, Mpc70-9Myc, and Spo74-9Myc during sporulation in the Cdc15↓, Mob1↓, Dbf2↓ Dbf20↓, and Dbf2↓ Dbf20↓ Mob1↓ mutants. This revealed no significant changes of the MP protein amounts in any of the mutant strains (Figure S10 in File S2). Moreover, we analyzed Nud1 and Cnm67 during sporulation in the same mutants (Figure S11 in File S2). Both, Cnm67 and Nud1 possess putative phosphorylation consensus sites for either Cdc15 ([S/T]X[R/K]) or Dbf2/20-Mob1 (RXXS) and have been shown to be hyperphosphorylated in a cell cycle-dependent manner (Gruneberg et al. 2000; Schaerer et al. 2001; Mah et al. 2005; Mok et al. 2010; Keck et al. 2011). Of all mutants, only the Mob1↓ strain showed differences to the control; Nud1 abundance was increased at 0 hr and during sporulation. Interestingly, a similar increase in Nud1 levels was not observed in the triple mutant. Thus, we could exclude changes in the levels of MP proteins, Nud1, or Cnm67 as the reason for impaired spore formation, dysregulation of MP numbers in Cdc15 mutants, or the defects in age-based SPB selection; however, due to the limitations of standard SDS-PAGE, we cannot rule out that, e.g., post-translational modifications of MP proteins, Nud1, or Cnm67 may alter the affinity of meiotic plaque proteins for the SPB.
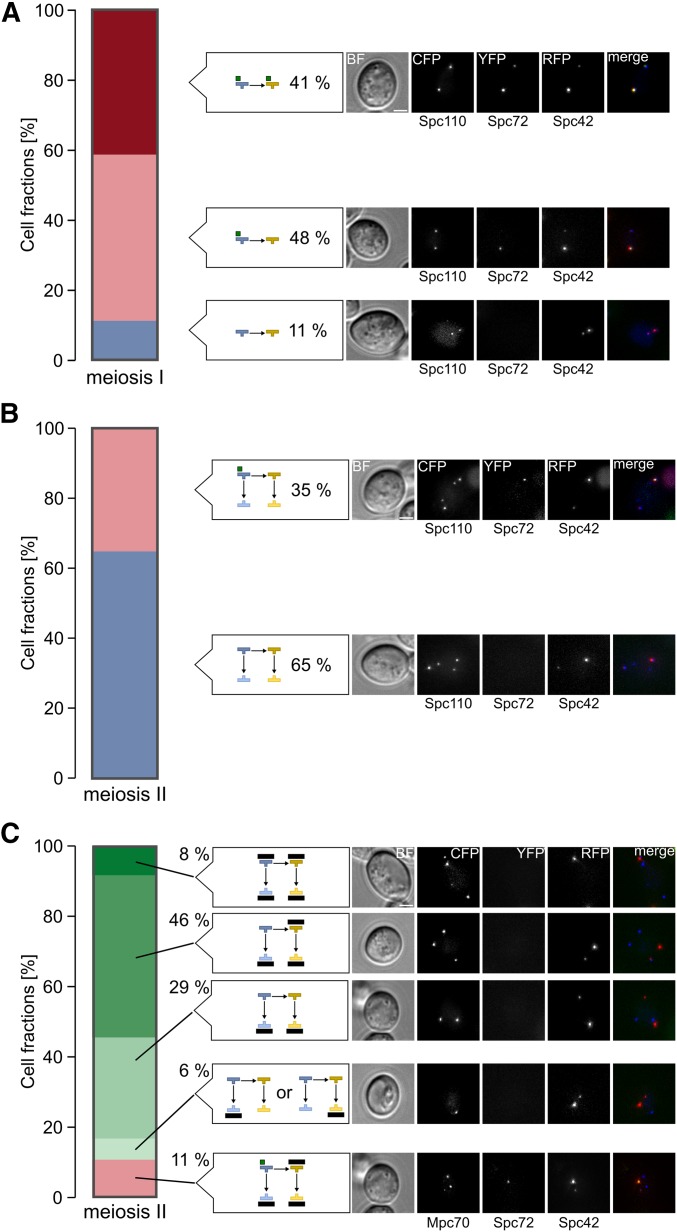
A major function of Nud1, which could link meiotic MEN function to SPB inheritance, is the nucleation of aMTs as an anchor of the γ-tubulin complex receptor Spc72 (Knop and Schiebel 1998; Gruneberg et al. 2000). As Spc72 has an influence on meiotic SPB selection (Gordon et al. 2006), we investigated Spc72 localization in cells undergoing the first and second meiotic division. During metaphase and anaphase of meiosis I, Spc72 localized to both SPBs in 41% of the cells and to the older SPB in 48% of the cells; only 11% of the cells did not show an Spc72 signal (Figure 6A). In meiosis II, most cells (65%) had no Spc72 signal, whereas in the rest of the cells Spc72 was localized at the oldest SPB (Figure 6B). Colocalization analysis with the meiotic plaque marker Mpc70 revealed that Spc72 was not present at SPBs modified with MPs. However, in 11% of the cases, Spc72 was localized to the oldest SPB whereas Mpc70 signals were detectable at the three other SPBs within the same cells (Figure 6C). These data are in agreement with a model that Spc72 might be involved in regulation of MP formation.
Figure 6.
Spc72 localization during meiosis. (A) Spc72 localizes in the majority of cells to one or two SPBs during meiosis I. Cells harboring Spc110-CFP Spc72-YFP Spc42-RFP (YCR414) were sporulated in liquid medium with 1% potassium acetate. Cells with two separated SPBs (n = 97) were evaluated with respect to localization of Spc72-YFP and SPB age. Example images for the cell types that were observed are shown on the right. Bar, 2 µm. (B) Spc72 localizes in a fraction of the cells to the oldest SPB during meiosis II. Experimental conditions as in (A). Cells with four separated SPBs (n = 108) were evaluated with respect to localization of Spc72-YFP and SPB age. Example images for the cell types that were observed are shown on the right. Bar, 2 µm. (C) Mpc70 and Spc72 localize to different SPBs in meiosis II cells. Cells harboring Mpc70-CFP Spc72-YFP Spc42-RFP (YCR459) were sporulated in liquid medium with 1% potassium acetate. Cells with four separated SPBs (n = 167) were evaluated with respect to localization of Spc72-YFP and Mpc70-CFP. Example images for cell types that were observed are shown on the right. Bar, 2 µm. BF, brightfield channel; CFP, cyan fluorescing protein; RFP, red fluorescent protein; SPB, spindle pole body; YFP, yellow fluorescent protein.
To test this hypothesis, we analyzed sporulation behavior of a sid-SPC72 strain. Sporulation-specific depletion of Spc72 was achieved at an early time point after induction of sporulation (Figure S12A in File S2) whereas growth and localization of Spc72 were not influenced by the sid-tag (Figure S12, B and C in File S2). Analysis of spore formation, sporulation efficiency, and genome inheritance did not reveal significant changes compared to the control strain (Figure S12, D–F in File S2). This is in agreement with previously published data using a different SPC72 mutant (Gordon et al. 2006). Finally, we analyzed the abundance and localization of Spc72 during sporulation in control cells and MEN depletion mutants; we observed that Spc72 levels were dropping after initiation of sporulation with a similar rate in control, Cdc15↓, and Dbf2↓ Dbf20↓ Mob1↓ cells (Figure S13A in File S2). Fluorescence microscopy experiments revealed that Spc72 localized to SPBs in all three strains regardless of whether sporulation was induced or if cells were growing vegetatively (Figure S13, B and C in File S2). During sporulation, the number of cells with Spc72 signals decreased slowly over time. These experiments make it rather unlikely that the sporulation phenotypes we observed in the MEN mutants depend on Spc72 or that Spc72 homeostasis is regulated by the MEN.
Hyperactivation of the MEN during sporulation
To obtain further insights in MEN signaling during sporulation, we used a Cdc15 gain-of-function mutant; in mitosis, expression of a truncated allele of CDC15 (CDC15ΔC, corresponding to aa 1–750) leads to hyperactivation of the MEN (Bardin et al. 2003; Rock and Amon 2011). We used this allele under control of the meiosis-specific SPO74 promoter and added a photosensitive degron module to minimize Cdc15ΔC levels during vegetative growth by blue light-dependent destabilization of the hyperactive kinase (Renicke et al. 2013). Considerable amounts of the construct accumulated specifically during meiosis and were sufficient to complement the Cdc15↓ phenotype (Figure S14, A and B in File S2). The presence of Cdc15ΔC during meiosis in addition to the endogenous wild-type protein led to a slight decrease in spore formation and sporulation efficiency (Figure S14C in File S2). However, a genome inheritance assay revealed no effect of Cdc15ΔC on nonsister dyad formation (Figure S14D in File S2). Compared to control cells, levels of the meiotic plaque protein Mpc54 were increased and the amounts of Spo74 were reduced, whereas Mpc70 was unaffected (Figure S14E in File S2). Additionally, we observed a decrease of Nud1 levels while no changes were found for Cnm67 (Figure S14F in File S2). Thus, hyperactivation of the MEN during sporulation might impinge on spore formation, most likely through limitation of Spo74, but does not cause aberrant SPB inheritance.
The terminal MEN kinases are involved in spore maturation
To directly follow PSM formation, we used a strain with the leading-edge protein Don1 fused to GFP. Again, the Cdc15↓ mutant strain formed no refractive spores, instead it accumulated cells with faint Don1-GFP signals staining whole PSMs (Figure S15 in File S2). This localization pattern has been attributed to cells that are about to close the PSM (Taxis et al. 2006; Maier et al. 2007); however, PSM numbers were not affected by Cdc15 depletion. During similar experiments with the Mob1↓, Dbf2↓ Dbf20↓, and Dbf2↓ Dbf20↓ Mob1↓ mutants under conditions favoring low spore numbers, we observed development of PSMs in all strains comparable to the control (Figure 7). Subsequently, the number of cells with Don1-GFP signal decreased, but the number of cells that formed refractive spores during the experiment was reduced in all three mutants compared to control cells. This points to a defect in spore maturation after closure of the PSM. We used EOSIN Y and Calcofluor white staining, which enables the identification of putative defects in assembly of the two outer spore wall layers, chitosan and dityrosine (Lin et al. 2013); however, asci of the Dbf2↓ Dbf20↓ Mob1↓ mutant strain showed the same staining pattern as the control strain (data not shown).
Figure 7.
Mob1↓, Dbf2↓, and Dbf20↓ mutants show a defect at a step that is postmeiotic and prior to spore wall formation. PSM formation in Mob1↓ (YCR537), Dbf2↓ Dbf20↓ (YCR538), and Dbf2↓ Dbf20↓ Mob1↓ (YCR539) mutants and a control strain (YCR536) during a sporulation time course under conditions favoring low spore numbers. Don1-GFP was used as marker for the leading edge of PSMs. By correlation of fluorescent and BF images cells prior to meiosis I (no distinct Don1-GFP signals; blue), prior to meiosis II (Don1-GFP at precursor vesicles; yellow), and in meiosis II with one to four donuts (Don1-GFP signals at the leading edges of PSMs; light to dark green), cells at/after closure of the PSMs (Don1-GFP signals at whole PSMs; light gray) and cells with refractive spores (dark gray) could be distinguished. Left panel shows fractions of all cell classes, right panel only cells with donuts (n = 4; 63–1017 cells per timepoint and replicate). Bar, 5 µm. Please see Table S1 for statistical evaluations. BF, brightfield channel; PSM, prospore membrane; SPO, sporulation medium/water.
To further investigate the late sporulation defects, we performed electron microscope analysis of Cdc15↓ and Dbf2↓ Dbf20↓ Mob1↓ strains. No difference to control cells was apparent concerning PSM formation and spore maturation for Dbf2↓ Dbf20↓ Mob1↓ cells (Figure S16, A, B, E, and F in File S2). However, unusual structures prior to PSM closure were observed in a minority (< 10%) of Cdc15↓ cells at that stage (Figure S16, C and D in File S2). The changes are reminiscent of the phenotypes of enlarged PSM lumen and premature spore wall formation described for the vps13Δ single and are1Δ are2Δ dga1Δ lro1Δ quadruple mutants (Park and Neiman 2012; Hsu et al. 2017).
Discussion
During sporulation of diploid S. cerevisiae cells, spindle polarity results in preferential inheritance of newly formed SPBs if less than four spores are formed (Gordon et al. 2006). The decision of how many and which SPBs and their associated genomes will be incorporated into spores takes place at the onset of meiosis II by formation of MPs at selected SPBs (Knop and Strasser 2000; Taxis et al. 2005). Our results demonstrate that the MEN is involved in this regulatory step: Cdc15, as well as Dbf2/20-Mob1, activity is necessary for age-based selection of SPBs by modification with MPs. Moreover, Cdc15 is involved in control of MP numbers and is essential for spore formation, whereas inactivation of Dbf2/20-Mob1 results in partial defects at a late step in spore formation.
Parallels between mitotic and meiotic functions of the MEN
In mitosis, the MEN is required for the establishment of spindle polarity (Hotz et al. 2012a,b), exit from mitosis, and cytokinesis, and its activation mechanism is tightly linked to the SPB by its scaffold protein Nud1 [reviewed in Juanes and Piatti (2016)]. During the developmental program of sporulation, a rewiring of the MEN takes place. Activity of the MEN kinases is independent of SPB localization, Dbf20 is more abundant and exhibits higher activity than Dbf2, and its association with Mob1 is Cdc15-dependent (Attner and Amon 2012). Similar to mitosis, the MEN is involved in anaphase II release of Cdc14, in exit of meiosis II, and cytokinesis (Kamieniecki et al. 2005; Pablo-Hernando et al. 2007; Diamond et al. 2009; Attner and Amon 2012).
This study provides evidence that Dbf2/20-Mob1 are involved in spore formation. In contrast to Cdc15 inactivation, depletion of the terminal MEN kinases did not result in a complete failure of sporulation, but in random spore abortion. This implies that Cdc15 does not rely exclusively on Dbf2/20-Mob1 during sporulation. Furthermore, the sporulation assays suggest partially redundant functions of Dbf2 and Dbf20. Surprisingly, Mob1↓ or Dbf2↓ Dbf20↓ mutants had a much weaker sporulation defect than the triple mutant. Thus, the coactivator Mob1 and/or the kinases Dbf2/20 might form sporulation-specific complexes with yet unknown binding partners. In mammals and flies, NDR kinases of different pathways have been found to share one Mob coactivator while some of them interact with a second Mob protein (Bichsel et al. 2004; He et al. 2005; Hergovich et al. 2005, 2006; Kohler et al. 2010; Nishio et al. 2012; Couzens et al. 2013; Gomez et al. 2015). During sporulation of S. cerevisiae, candidates could be components of the second hippo-like signaling pathway in S. cerevisiae, the RAM (regulation of Ace2p activity and cellular morphogenesis) pathway. Mob1 has been shown to form heterodimers with its RAM homolog Mob2 in vitro and in vivo (Mrkobrada et al. 2006); Mob2 also displayed genetic interactions with Dbf2 (Costanzo et al. 2016). Interestingly, physical interaction of Mob1 with the RAM NDR kinase Cbk1 has been observed in a yeast two-hybrid assay (Ito et al. 2001).
Moreover, Don1 localization experiments might suggest two roles of the MEN after exit from meiosis II. Accumulation of faint Don1-GFP on whole PSMs in Cdc15↓ cells is in line with previously published defects in PSM closure, most likely due to a failure in removal of Ssp1 from the leading edge of the PSM (Maier et al. 2007; Diamond et al. 2009; Paulissen et al. 2016). In the mutants depleted for Dbf2, Dbf20, and Mob1, Don1-GFP signals disappeared from the PSMs with control-like kinetics. Yet, these mutants formed lower fractions of cells with mature spores. This might indicate a defect at a later time point compared to the Cdc15 mutant (Coluccio et al. 2004).
Regulation of the MEN during sporulation
An interesting question is the intrinsic activation mechanism of the MEN. Lack of the GTPase Tem1 as well as its GAP Bfa1-Bub2, essential for signal integration and MEN function in mitosis, have no obvious effect on sporulation (Gordon et al. 2006; Attner and Amon 2012). The main mitotic Tem1 function is localization of Cdc15 to the SPB; consequently, Nud1-dependent SPB association is dispensable for meiotic MEN activity (Visintin and Amon 2001; Rock and Amon 2011; Attner and Amon 2012). Recently, a mechanism for activation of one core mammalian Hippo pathway consisting of Mst2, Lats1, and hMob1 has been proposed in which hMob1 acts as scaffold for the kinases (Ni et al. 2015). There, the Cdc15 ortholog Mst2 autophosphorylates multiple residues within its linker region to allow hMob1 docking. This leads to a conformational change allowing recruitment of the NDR kinase Lats1 and formation of a ternary complex, which then permits direct activation of Lats1 by Mst2. The high conservation of Hippo signaling provokes the question of whether a similar activation mechanism is in place in budding yeast meiosis. Like Mst2, Cdc15 is capable of autophosphorylation and is required for Dbf2/20-Mob1 interaction (Jaspersen et al. 1998; Asakawa et al. 2001; Ptacek et al. 2005; Attner and Amon 2012). Moreover, the Nud1-independent activation of Dbf2/20-Mob1 (Attner and Amon 2012) suggests a comparable mechanism for the MEN in meiosis.
Known upstream regulators of the MEN during mitosis are the kinases Cdc5 and Kin4 (D’Aquino et al. 2005; Pereira and Schiebel 2005; Maekawa et al. 2007; Falk et al. 2016; Gryaznova et al. 2016). We observed SPB and nuclear localization of the Polo-like kinase Cdc5 during the meiotic divisions until prospores became visible (data not shown). A comparable localization pattern was correlated with Cdc5 functions during meiotic SPB duplication (Attner et al. 2013). Analysis of Cdc5, Spc42, and Don1 localization did not reveal a connection between SPB localization of Cdc5 with the number of PSMs (data not shown). Thus, it is unlikely that Cdc5 localization is directly linked to MEN-dependent SPB selection. The data on depletion of the mitotic Cdc5 antagonist Kin4 indicate that this kinase is required for efficient sporulation but not for SPB selection and genome inheritance. Differences between Kin4 and MEN mutants in the latter assays and the dispensability of Bfa1-Bub2 for spore formation (Gordon et al. 2006) suggest that Kin4 acts in a different pathway.
SPB-associated functions of the MEN during sporulation
In mitosis, establishment of spindle polarity depends on the regulation of Kar9 localization toward the old SPB by the MEN (Leisner et al. 2008; Hotz et al. 2012a,b). Our results indicate that meiotic spindle polarity is also regulated by the MEN. However, it is unlikely that Kar9 is involved in this process since it is not required for faithful SPB selection (Gordon et al. 2006). Although Cdc15 and Dbf2/20-Mob1 are not found at the SPBs (Figure S2 in File S2; Attner and Amon 2012), they contribute to the selection of the younger SPBs for MP formation. This function correlates with a subtle increase in activity of Dbf2/20-Mob1 at the transition from meiosis I to meiosis II reported by a previous study (Attner and Amon 2012). Thus, similar to mitosis, low activity might be sufficient for MEN function in SPB selection, whereas high activity seems to be required for cytokinesis after meiosis II (Visintin and Amon 2001; Bardin et al. 2003; Hotz et al. 2012a,b).
Age-based SPB selection was affected in all MEN mutants that we tested; however, Cdc15 depletion resulted in the strongest phenotype. This could imply a functional separation between Cdc15 and Dbf2/20-Mob1, similarly to the distinct phenotypes in late sporulation. We cannot fully rule out that potential differences in depletion efficiencies had an impact on our experiments. The effects on SPB selection of Dbf2/20-Mob1 mutants were lower than that of the Nud1-2 mutant, whereas Cdc15 depletion caused similar randomization (Figure 4; Gordon et al. 2006). This effect could be connected to the increased MP formation found in this mutant that might result in an additive effect concerning age-based SPB selection.
The kinase Cdc15 is involved in the regulation of MP numbers, most likely independently from Dbf2/Dbf20-Mob1. In the Cdc15↓ strain, MP protein levels were unchanged, yet significantly more cells with four MPs were observed. Strikingly, this increase was prominent in cells relatively early and the ratio of cells with four MPs vs. cells with one/two/three MPs decreased during the time course (Figure 5A and Figure S8 in File S2), which argues against an accumulation of MPs due to a meiotic block or defects in MP disassembly. We cannot formally exclude the possibility that a very small amount of Dbf2/20-Mob1 remains in our depletion mutant and is active, for example, in the regulation of MP formation at the onset of meiosis II. However, the age-based SPB selection phenotype we observed in this mutant makes it rather unlikely that small amounts of remaining Dbf2/20-Mob1 affect only SPB selection and not the regulation of MP numbers, which both take place during onset of meiosis II. The kinetics of MP and PSM formation in the mutants are not different from those of control cells, which argues for an early regulatory defect (SPB selection) rather than problems at later stages. The most straightforward model based on our experimental data concerning MEN function during MP formation would be a functional separation within the MEN: Cdc15 is involved in regulation of MP numbers and SPB selection, whereas Dbf2/20-Mob1 is only required for the latter process.
The phenotype observed in Cdc15↓ cells is reminiscent of an ADY1 deletion mutant. There, MP levels were not affected, but MP numbers were strongly reduced (Deng and Saunders 2001; Jungbluth et al. 2012). Recently, the Ady1-interacting kinase Hrr25 was shown to be involved in MP formation (Argüello-Miranda et al. 2017). Although partially opposing in their effects, all mutants seem to impact on the ability of MP components to form a mature MP.
Surprisingly, we found a positive effect of Nud1 depletion on spore numbers, conversely to that of the nud1-2 allele (Gordon et al. 2006), pointing to an inhibitory function of Nud1 on MP formation. We cannot fully explain the phenotypic differences of the two mutants. Similar to meiosis-specific Nud1 depletion, it can be assumed that Nud1-2 dissociates from the SPB at restrictive temperature as shown in vegetative cells (Gruneberg et al. 2000). However, the timing of Nud1 inactivation as well as experimental conditions were different, which could account for the dissimilar sporulation results. Remarkably, effects on genome inheritance were comparable for both mutants (Figure 3B; Gordon et al. 2006). Although different groups have published evidence that Nud1 localizes to the SPB during meiosis II, the number of SPBs with Nud1 signals varied in these reports (Knop and Strasser 2000; Nickas et al. 2004; Attner and Amon 2012). Thus, an inhibitory function of Nud1 on MP formation cannot be excluded by the available experimental data.
A negative effect of Nud1 on spore formation might also explain the unexpected phenotypic enhancement observed in most experiments for the Mob1↓ mutant compared to the Dbf2↓ Dbf20↓ double mutant, as Nud1 levels were increased in cells depleted only for Mob1. Functional connection of Mob1 to Mps1 that is involved in SPB homeostasis might provide an explanation for these findings (Winey et al. 1991; Luca and Winey 1998; Elserafy et al. 2014; Burns et al. 2015).
Is meiotic SPB selection regulated by a dynamic process?
Two opposing processes could occur at the transition from anaphase I to metaphase II: removal of aMTs and formation of MPs. Differences in aMT nucleation between the old and the new SPB have been observed in mitosis and impact on mitotic spindle polarity (Juanes et al. 2013). Involvement of Spc72 in age-based SPB selection and localization of Spc72 to SPBs during meiosis I has been reported before (Gordon et al. 2006). Here, we expanded the Spc72 localization data with respect to SPB age and MP formation. The model that, in a short time window during meiosis, one or two older SPBs possess aMTs (or Spc72) while the new SPBs do not is in agreement with our data (Figure 6, A and B). A necessity to remove aMTs (or Spc72) from an older SPB before MP formation is permitted would explain the slight delay in MP maturation at older SPBs (Taxis et al. 2005). Indeed, we did not observe the MP marker Mpc70 together with the aMT marker Spc72 at the same SPB (Figure 6C). Limiting amounts of MP components and a positive feedback loop in MP maturation might then result in the observed SPB selection patterns. Nud1 or Cdc15 inactivation impinge on this process to a similar extent (Figure 4; Gordon et al. 2006) and Nud1 functions in aMT nucleation (Knop and Schiebel 1998; Gruneberg et al. 2000). However, as neither Cdc15 nor Dbf2/20-Mob1 affect Spc72 removal during meiosis, a MEN-dependent mechanism that regulates timely removal of Spc72 from the SPB is unlikely. Thus, the MEN pathway must act through a so far unidentified target to influence meiotic plaque formation.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.194522/-/DC1.
Acknowledgments
We thank H.-U. Mösch and J. Freitag for helpful discussions, M. Knop, T. W. J. Gadella, K. Thorn, and M. Ehrmann for reagents, as well as U. Maier for discussions and his generous sharing of equipment. D. Störmer and Marion Debus provided excellent technical assistance. This work was supported by the DFG grants GK1216 “Intra- and Intercellular Transport and Communication” and TA320/3-1.
Footnotes
Communicating editor: O. Cohen-Fix
Literature Cited
- Adames N. R., Oberle J. R., Cooper J. A., 2001. The surveillance mechanism of the spindle position checkpoint in yeast. J. Cell Biol. 153: 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams I. R., Kilmartin J. V., 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145: 809–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüello-Miranda O., Zagoriy I., Mengoli V., Rojas J., Jonak K., et al. , 2017. Casein kinase 1 coordinates cohesin cleavage, gametogenesis, and exit from M Phase in Meiosis II. Dev. Cell 40: 37–52. [DOI] [PubMed] [Google Scholar]
- Asakawa K., Yoshida S., Otake F., Toh-e A., 2001. A novel functional domain of Cdc15 kinase is required for its interaction with Tem1 GTPase in Saccharomyces cerevisiae. Genetics 157: 1437–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attner M. A., Amon A., 2012. Control of the mitotic exit network during meiosis. Mol. Biol. Cell 23: 3122–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attner M. A., Miller M. P., Ee L., Elkin S. K., Amon A., 2013. Polo kinase Cdc5 is a central regulator of meiosis I. Proc. Natl. Acad. Sci. USA 110: 14278–14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., et al. , (Editors), 2001 Current Protocols in Molecular Biology. John Wiley & Sons, Inc., Hoboken, New Jersey. [Google Scholar]
- Bardin A. J., Visintin R., Amon A., 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102: 21–31. [DOI] [PubMed] [Google Scholar]
- Bardin A. J., Boselli M. G., Amon A., 2003. Mitotic exit regulation through distinct domains within the protein kinase Cdc15. Mol. Cell. Biol. 23: 5018–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D. L., Thibodeaux J., Maddox P., Yeh E., Bloom K., 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 10: 1497–1506. [DOI] [PubMed] [Google Scholar]
- Bertazzi D. T., Kurtulmus B., Pereira G., 2011. The cortical protein Lte1 promotes mitotic exit by inhibiting the spindle position checkpoint kinase Kin4. J. Cell Biol. 193: 1033–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichsel S. J., Tamaskovic R., Stegert M. R., Hemmings B. A., 2004. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J. Biol. Chem. 279: 35228–35235. [DOI] [PubMed] [Google Scholar]
- Bullitt E., Rout M. P., Kilmartin J. V., Akey C. W., 1997. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell 89: 1077–1086. [DOI] [PubMed] [Google Scholar]
- Burns S., Avena J. S., Unruh J. R., Yu Z., Smith S. E., et al. , 2015. Structured illumination with particle averaging reveals novel roles for yeast centrosome components during duplication. Elife 4: e08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caydasi A. K., Lohel M., Grünert G., Dittrich P., Pereira G., et al. , 2012. A dynamical model of the spindle position checkpoint. Mol. Syst. Biol. 8: 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., et al. , 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- Clyne R. K., Katis V. L., Jessop L., Benjamin K. R., Herskowitz I., et al. , 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5: 480–485. [DOI] [PubMed] [Google Scholar]
- Coluccio A., Bogengruber E., Conrad M. N., Dresser M. E., Briza P., et al. , 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 3: 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio A. E., Rodriguez R. K., Kernan M. J., Neiman A. M., 2008. The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS One 3: e2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P. T., Raff J. W., 2010. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20: 2187–2192. [DOI] [PubMed] [Google Scholar]
- Costanzo, M., B. VanderSluis, E. N. Koch, A. Baryshnikova, C. Pons et al., 2016 A global genetic interaction network maps a wiring diagram of cellular function. Science 353: aaf1420. [DOI] [PMC free article] [PubMed]
- Couzens A. L., Knight J. D. R., Kean M. J., Teo G., Weiss A., et al. , 2013. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal. 6: rs15. [DOI] [PubMed] [Google Scholar]
- D’Aquino K. E., Monje-Casas F., Paulson J., Reiser V., Charles G. M., et al. , 2005. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol. Cell 19: 223–234. [DOI] [PubMed] [Google Scholar]
- Davidow L. S., Goetsch L., Byers B., 1980. Preferential occurrence of nonsister spores in two-spored asci of SACCHAROMYCES CEREVISIAE: evidence for regulation of spore-wall formation by the spindle pole body. Genetics 94: 581–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Saunders W. S., 2001. ADY1, a novel gene required for prospore membrane formation at selected spindle poles in Saccharomyces cerevisiae. Mol. Biol. Cell 12: 2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond A. E., Park J.-S., Inoue I., Tachikawa H., Neiman A. M., 2009. The anaphase promoting complex targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol. Biol. Cell 20: 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood M. D., Cheung S. W. T., Lee K. Y., Moffat J., Meneghini M. D., 2012. Developmentally programmed nuclear destruction during yeast gametogenesis. Dev. Cell 23: 35–44. [DOI] [PubMed] [Google Scholar]
- Elserafy M., Šarić M., Neuner A., Lin T., Zhang W., et al. , 2014. Molecular mechanisms that restrict yeast centrosome duplication to one event per cell cycle. Curr. Biol. 24: 1456–1466. [DOI] [PubMed] [Google Scholar]
- Esposito R. E., Klapholz S., 1981. Meiosis and ascospore development. Cold Spring Harb. Monogr. Arch. 11: 211–287. [Google Scholar]
- Falk J. E., Chan L. Y., Amon A., 2011. Lte1 promotes mitotic exit by controlling the localization of the spindle position checkpoint kinase Kin4. Proc. Natl. Acad. Sci. USA 108: 12584–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J. E., Tsuchiya D., Verdaasdonk J., Lacefield S., Bloom K., et al. , 2016. Spatial signals link exit from mitosis to spindle position. Elife 5: e14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., D’Ambrosio C., Venturetti M., Lucchini G., Piatti S., 2006. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J. Cell Biol. 172: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese E. B., Chu M. I., Freese E., 1982. Initiation of yeast sporulation of partial carbon, nitrogen, or phosphate deprivation. J. Bacteriol. 149: 840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Smith S. J. M., Wheatley E., Rittinger K., et al. , 2002. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 277: 28439–28445. [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Walker P. A., Johnston L. H., Sedgwick S. G., 2003. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J. Biol. Chem. 278: 14591–14594. [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., de Bettignies G., Sedgwick S. G., 2009. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J. Cell Biol. 187: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Goedhart J., von Stetten D., Noirclerc-Savoye M., Lelimousin M., Joosen L., et al. , 2012. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 3: 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez V., Gundogdu R., Gomez M., Hoa L., Panchal N., et al. , 2015. Regulation of DNA damage responses and cell cycle progression by hMOB2. Cell. Signal. 27: 326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon O., Taxis C., Keller P. J., Benjak A., Stelzer E. H. K., et al. , 2006. Nud1p, the yeast homolog of Centriolin, regulates spindle pole body inheritance in meiosis. EMBO J. 25: 3856–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Campbell K., Simpson C., Grindlay J., Schiebel E., 2000. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 19: 6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryaznova Y., Koca Caydasi A., Malengo G., Sourjik V., Pereira G., 2016. A FRET-based study reveals site-specific regulation of spindle position checkpoint proteins at yeast centrosomes. Elife 5: e14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Emoto K., Fang X., Ren N., Tian X., et al. , 2005. Drosophila Mob family proteins interact with the related tricornered (Trc) and warts (Wts) kinases. Mol. Biol. Cell 16: 4139–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Hemmings B. A., 2012. Hippo signalling in the G2/M cell cycle phase: lessons learned from the yeast MEN and SIN pathways. Semin. Cell Dev. Biol. 23: 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Bichsel S. J., Hemmings B. A., 2005. Human NDR kinases are rapidly activated by MOB proteins through recruitment to the plasma membrane and phosphorylation. Mol. Cell. Biol. 25: 8259–8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Schmitz D., Hemmings B. A., 2006. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem. Biophys. Res. Commun. 345: 50–58. [DOI] [PubMed] [Google Scholar]
- Hothorn T., Hornik K., van de Wiel M. A., Zeileis A., 2008. Implementing a class of permutation tests: the coin package. J. Stat. Softw. 28: 1–23.27774042 [Google Scholar]
- Hotz M., Leisner C., Chen D., Manatschal C., Wegleiter T., et al. , 2012a Spindle pole bodies exploit the mitotic exit network in metaphase to drive their age-dependent segregation. Cell 148: 958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz M., Lengefeld J., Barral Y., 2012b The MEN mediates the effects of the spindle assembly checkpoint on Kar9-dependent spindle pole body inheritance in budding yeast. Cell Cycle 11: 3109–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T., Chen R., Cheng Y., Wang C., 2017. Lipid droplets are central organelles for meiosis II progression during yeast sporulation. Mol. Biol. Cell 28: 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Elledge S. J., 2002. Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints. Cell Cycle 1: 351–355. [PubMed] [Google Scholar]
- Hu F., Wang Y., Liu D., Li Y., Qin J., et al. , 2001. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107: 655–665. [DOI] [PubMed] [Google Scholar]
- Huang L. S., Doherty H. K., Herskowitz I., 2005. The Smk1p MAP kinase negatively regulates Gsc2p, a 1,3-beta-glucan synthase, during spore wall morphogenesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102: 12431–12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., et al. , 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Yamanishi M., Ikeuchi A., Imamura C., Tokuhiro K., et al. , 2013. Characterization of five terminator regions that increase the protein yield of a transgene in Saccharomyces cerevisiae. J. Biotechnol. 168: 486–492. [DOI] [PubMed] [Google Scholar]
- Izumi H., Kaneko Y., 2012. Evidence of asymmetric cell division and centrosome inheritance in human neuroblastoma cells. Proc. Natl. Acad. Sci. USA 109: 18048–18053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., et al. , 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962. [DOI] [PubMed] [Google Scholar]
- Januschke J., Llamazares S., Reina J., Gonzalez C., 2011. Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Reina J., Llamazares S., Bertran T., Rossi F., et al. , 2013. Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nat. Cell Biol. 15: 241–248. [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Charles J. F., Tinker-Kulberg R. L., Morgan D. O., 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 9: 2803–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juanes M. A., Piatti S., 2016. The final cut: cell polarity meets cytokinesis at the bud neck in S. cerevisiae. Cell. Mol. Life Sci. 73: 3115–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juanes M. A., Twyman H., Tunnacliffe E., Guo Z., ten Hoopen R., et al. , 2013. Spindle pole body history intrinsically links pole identity with asymmetric fate in budding yeast. Curr. Biol. 23: 1310–1319. [DOI] [PubMed] [Google Scholar]
- Jungbluth M., Renicke C., Taxis C., 2010. Targeted protein depletion in Saccharomyces cerevisiae by activation of a bidirectional degron. BMC Syst. Biol. 4: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungbluth M., Mösch H.-U., Taxis C., 2012. Acetate regulation of spore formation is under the control of the Ras/cyclic AMP/protein kinase A pathway and carbon dioxide in Saccharomyces cerevisiae. Eukaryot. Cell 11: 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamieniecki R. J., Liu L., Dawson D. S., 2005. FEAR but not MEN genes are required for exit from meiosis I. Cell Cycle 4: 1093–1098. [PubMed] [Google Scholar]
- Kawaguchi H., Yoshida M., Yamashita I., 1992. Nutritional regulation of meiosis-specific gene expression in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 56: 289–297. [Google Scholar]
- Keck J. M., Jones M. H., Wong C. C. L., Binkley J., Chen D., et al. , 2011. A cell cycle phosphoproteome of the yeast centrosome. Science 332: 1557–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Luo G., Bahk Y. Y., Song K., 2012. Cdc5-dependent asymmetric localization of bfa1 fine-tunes timely mitotic exit. PLoS Genet. 8: e1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshner H., Aguet F., Sage D., Unser M., 2013. 3-D PSF fitting for fluorescence microscopy: implementation and localization application. J. Microsc. 249: 13–25. [DOI] [PubMed] [Google Scholar]
- Klein F., Mahr P., Galova M., Buonomo S. B., Michaelis C., et al. , 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103. [DOI] [PubMed] [Google Scholar]
- Knop M., Schiebel E., 1998. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 17: 3952–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Strasser K., 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19: 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R. S., Schmitz D., Cornils H., Hemmings B. A., Hergovich A., 2010. Differential NDR/LATS interactions with the human MOB family reveal a negative role for human MOB2 in the regulation of human NDR kinases. Mol. Cell. Biol. 30: 4507–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C., Santore E., Lavoie E., Needleman L., Fiacco N., et al. , 2014. A visual screen of protein localization during sporulation identifies new components of prospore membrane-associated complexes in budding yeast. Eukaryot. Cell 13: 383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lim W. A., Thorn K. S., 2013. Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PLoS One 8: e67902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. E., Frenz L. M., Wells N. J., Johnson A. L., Johnston L. H., 2001. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 11: 784–788. [DOI] [PubMed] [Google Scholar]
- Leisner C., Kammerer D., Denoth A., Britschi M., Barral Y., et al. , 2008. Regulation of mitotic spindle asymmetry by SUMO and the spindle-assembly checkpoint in yeast. Curr. Biol. 18: 1249–1255. [DOI] [PubMed] [Google Scholar]
- Lerit D. A., Rusan N. M., 2013. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D., Kusch J., Grava S., Vogel J., Barral Y., 2003. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112: 561–574. [DOI] [PubMed] [Google Scholar]
- Lin C. P.-C., Kim C., Smith S. O., Neiman A. M., 2013. A highly redundant gene network controls assembly of the outer spore wall in S. cerevisiae. PLoS Genet. 9: e1003700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F. C., Winey M., 1998. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell 9: 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Priest C., Lechner J., Pereira G., Schiebel E., 2007. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J. Cell Biol. 179: 423–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah A. S., Jang J., Deshaies R. J., 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 98: 7325–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah A. S., Elia A. E. H., Devgan G., Ptacek J., Schutkowski M., et al. , 2005. Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening. BMC Biochem. 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier P., Rathfelder N., Finkbeiner M. G., Taxis C., Mazza M., et al. , 2007. Cytokinesis in yeast meiosis depends on the regulated removal of Ssp1p from the prospore membrane. EMBO J. 26: 1843–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson E. M., Schwartz C., Neiman A. M., 2010. Membrane assembly modulates the stability of the meiotic spindle-pole body. J. Cell Sci. 123: 2481–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F., Petrova B., Lombardi I. M., Bertazzi D. T., Hub B., et al. , 2010. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J. Cell Sci. 123: 1851–1861. [DOI] [PubMed] [Google Scholar]
- Meitinger F., Palani S., Hub B., Pereira G., 2013. Dual function of the NDR-kinase Dbf2 in the regulation of the F-BAR protein Hof1 during cytokinesis. Mol. Biol. Cell 24: 1290–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez-Benito V., van Deventer S. J., Jimenez-Garcia V., Roy-Luzarraga M., van Leeuwen F., et al. , 2013. Spatiotemporal analysis of organelle and macromolecular complex inheritance. Proc. Natl. Acad. Sci. USA 110: 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K., Rose M. D., 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140: 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl D. A., Huddleston M. J., Collingwood T. S., Annan R. S., Deshaies R. J., 2009. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J. Cell Biol. 184: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok J., Kim P. M., Lam H. Y. K., Piccirillo S., Zhou X., et al. , 2010. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3: ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Borchart A. C., Strasser K., Finkbeiner M. G., Shevchenko A., Shevchenko A., et al. , 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20: 6946–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrkobrada S., Boucher L., Ceccarelli D. F. J., Tyers M., Sicheri F., 2006. Structural and functional analysis of Saccharomyces cerevisiae Mob1. J. Mol. Biol. 362: 430–440. [DOI] [PubMed] [Google Scholar]
- Nakanishi H., Morishita M., Schwartz C. L., Coluccio A., Engebrecht J., et al. , 2006. Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J. Cell Sci. 119: 1406–1415. [DOI] [PubMed] [Google Scholar]
- Neiman A. M., 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Zheng Y., Hara M., Pan D., Luo X., 2015. Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling. Genes Dev. 29: 1416–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas M. E., Neiman A. M., 2002. Ady3p links spindle pole body function to spore wall synthesis in Saccharomyces cerevisiae. Genetics 160: 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas M. E., Schwartz C., Neiman A. M., 2003. Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot. Cell 2: 431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas M. E., Diamond A. E., Yang M.-J., Neiman A. M., 2004. Regulation of spindle pole function by an intermediary metabolite. Mol. Biol. Cell 15: 2606–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M., Hamada K., Kawahara K., Sasaki M., Noguchi F., et al. , 2012. Cancer susceptibility and embryonic lethality in Mob1a/1b double-mutant mice. J. Clin. Invest. 122: 4505–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S., Iino T., 1981. Selective abortion of two nonsister nuclei in a developing ascus of the hfd-1 mutant in Saccharomyces cerevisiae. Genetics 99: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pablo-Hernando M. E., Arnaiz-Pita Y., Nakanishi H., Dawson D., del Rey F., et al. , 2007. Cdc15 is required for spore morphogenesis independently of Cdc14 in Saccharomyces cerevisiae. Genetics 177: 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-S., Neiman A. M., 2012. VPS13 regulates membrane morphogenesis during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 125: 3004–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-E., Park C. J., Sakchaisri K., Karpova T., Asano S., et al. , 2004. Novel functional dissection of the localization-specific roles of budding yeast polo kinase Cdc5p. Mol. Cell. Biol. 24: 9873–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulissen S. M., Slubowski C. J., Roesner J. M., Huang L. S., 2016. Timely closure of the prospore membrane requires SPS1 and SPO77 in Saccharomyces cerevisiae. Genetics 203: 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E., 2005. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol. Cell 19: 209–221. [DOI] [PubMed] [Google Scholar]
- Pereira G., Tanaka T. U., Nasmyth K., Schiebel E., 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20: 6359–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C. L., Bornens M., 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., et al. , 2005. Global analysis of protein phosphorylation in yeast. Nature 438: 679–684. [DOI] [PubMed] [Google Scholar]
- R Core Team , 2015. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rebollo E., Sampaio P., Januschke J., Llamazares S., Varmark H., et al. , 2007. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell 12: 467–474. [DOI] [PubMed] [Google Scholar]
- Renicke C., Schuster D., Usherenko S., Essen L.-O., Taxis C., 2013. A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem. Biol. 20: 619–626. [DOI] [PubMed] [Google Scholar]
- Rock J. M., Amon A., 2011. Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev. 25: 1943–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J. M., Lim D., Stach L., Ogrodowicz R. W., Keck J. M., et al. , 2013. Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes. Science 340: 871–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N. M., Peifer M., 2007. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 177: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzmann V., Chen C., Chiang C.-Y. A., Tiyaboonchai A., Mayer M., et al. , 2014. Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Mol. Biol. Cell 25: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerer F., Morgan G., Winey M., Philippsen P., 2001. Cnm67p is a spacer protein of the Saccharomyces cerevisiae spindle pole body outer plaque. Mol. Biol. Cell 12: 2519–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D., 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16: 339–346. [DOI] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellis D., Kvitek D. J., Dunn B., Sherlock G., Petrov D. A., 2016. Heterozygote advantage is a common outcome of adaptation in Saccharomyces cerevisiae. Genetics 203: 1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Lin M. Z., McKeown M. R., Steinbach P. A., Hazelwood K. L., et al. , 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5: 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Lambert G. G., Chammas A., Ni Y., Cranfill P. J., et al. , 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10: 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S. L., Yeh E., Maddox P., Salmon E. D., Bloom K., 1997. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 139: 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., et al. , 2003. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13: 364–372. [DOI] [PubMed] [Google Scholar]
- Sherman F., 2002. Getting started with yeast. Methods Enzymol. 41: 3–41. [DOI] [PubMed] [Google Scholar]
- Shou W., Seol J. H., Shevchenko A., Baskerville C., Moazed D., et al. , 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233–244. [DOI] [PubMed] [Google Scholar]
- Taxis C., Knop M., 2012. TIPI: TEV protease-mediated induction of protein instability, edited by J. R. Dohmen and M. Scheffner. Methods Mol. Biol. 832: 611–26. [Google Scholar]
- Taxis C., Maeder C., Reber S., Rathfelder N., Miura K., et al. , 2006. Dynamic organization of the actin cytoskeleton during meiosis and spore formation in budding yeast. Traffic 7: 1628–1642. [DOI] [PubMed] [Google Scholar]
- Taxis C., Stier G., Spadaccini R., Knop M., 2009. Efficient protein depletion by genetically controlled deprotection of a dormant N-degron. Mol. Syst. Biol. 5: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usherenko S., Stibbe H., Muscò M., Essen L.-O., Kostina E. A., et al. , 2014. Photo-sensitive degron variants for tuning protein stability by light. BMC Syst. Biol. 8: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio-Santiago M., Monje-Casas F., 2011. Tem1 localization to the spindle pole bodies is essential for mitotic exit and impairs spindle checkpoint function. J. Cell Biol. 192: 599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Amon A., 2001. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell 12: 2961–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E. S., Prinz S., Tyers M., et al. , 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 2: 709–718. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang E. S., Amon A., 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398: 818–823. [DOI] [PubMed] [Google Scholar]
- Vonesch C., Unser M., 2008. A fast thresholded landweber algorithm for wavelet-regularized multidimensional deconvolution. IEEE Trans. Image Process. 17: 539–549. [DOI] [PubMed] [Google Scholar]
- Wang X., Tsai J.-W., Imai J. H., Lian W.-N., Vallee R. B., et al. , 2009. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461: 947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Goetsch L., Baum P., Byers B., 1991. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 114: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi M., Ito Y., Kintaka R., Imamura C., Katahira S., et al. , 2013. A genome-wide activity assessment of terminator regions in Saccharomyces cerevisiae provides a ″terminatome″ toolbox. ACS Synth. Biol. 2: 337–347. [DOI] [PubMed] [Google Scholar]
- Yamashita Y. M., Mahowald A. P., Perlin J. R., Fuller M. T., 2007. Asymmetric inheritance of mother vs. daughter centrosome in stem cell division. Science 315: 518–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and reagents are available on request. All data that we used for our investigations are included in the manuscript or in the Supplemental Material.