Abstract
Stabilizing the folded state of metastable and/or aggregation-prone proteins through exogenous ligand binding is an appealing strategy to decrease disease pathologies brought on by protein folding defects or deleterious kinetic transitions. Current methods of examining ligand binding to these marginally stable native states are limited, because protein aggregation typically interferes with analysis. Here, we describe a rapid method for assessing the kinetic stability of folded proteins and monitoring the effects of ligand stabilization for both intrinsically stable proteins (monomers, oligomers, multi-domain) and metastable proteins (e.g. low Tm) that uses a new GroEL chaperonin-based biolayer interferometry (BLI) denaturant-pulse platform. A kinetically controlled denaturation isotherm is generated by exposing a target protein immobilized on a BLI biosensor to increasing denaturant concentrations (urea or GnHCl) in a pulsatile manner to induce partial or complete unfolding of the attached protein population. Following the rapid removal of the denaturant, the extent of hydrophobic unfolded/partially folded species that remain is detected by increased GroEL binding. Since this kinetic denaturant pulse is brief, the amplitude of the GroEL binding to the immobilized protein depends on the duration of exposure to denaturant, the concentration of denaturant, wash times, and the underlying protein unfolding/refolding kinetics; fixing all other parameters and plotting GroEL binding amplitude versus denaturant pulse concentration results in a kinetically controlled denaturation isotherm. When folding osmolytes or stabilizing ligands are added to the immobilized target proteins before and during the denaturant pulse, the diminished population of unfolded/partially folded protein is manifested by a decreased GroEL binding and/or a marked shift in these kinetically controlled denaturation profiles to higher denaturant concentrations. This particular platform approach can be used to identify small molecules/solution conditions that can stabilize or destabilize thermally stable proteins, multi-domain proteins, oligomeric proteins, and most importantly, aggregation prone metastable proteins.
Rapidly assessing protein stability and determining potential ligands and stabilizers that bind to metastable proteins is a challenging area in protein science. To identify potential small molecule stabilizers or stabilizing solutions, one must document the effects these compounds or solutions have on protein structure, dynamics, and more commonly thermal or chemical stability. There are numerous general approaches available to assess protein stability. Solution phase protein stability measurements usually rely on monitoring thermally or chemically induced mid-point transition shifts of the protein in the presence or absence of stabilizing (or destabilizing) ligands or solutions. Methods such as differential scanning calorimetry,1 changes in light scattering2, global spectroscopic signatures (CD spectroscopy) or other direct physical measurements are commonly used to identify stabilizers of protein structure. Current high-throughput strategies that typically identify protein stabilization conditions rely on kinetically perturbing these states in solution using thermal or chemical denaturation. Potential stabilizing ligands/solutions are tested by determining how they delay the measured perturbation. Specifically, these methods often rely on detecting shifts in solution isotherms due to ligand or solution stabilization.
A majority of the most popular protein stability measurements in solution use spectroscopic measurements which depend on a combination of thermal perturbation coupled with fluorescence or optical methods (e.g. thermofluor, aggregation assays). Currently, the most frequently used high throughput screening approach to identify potential protein stabilizers is differential scanning fluorimetry (DSF). This protein stability method entails a gradual heating of the protein solution and monitoring the amount of fluorescent dye that binds to the exposed hydrophobic regions as the protein undergoes a partial unfolding transition to a molten globular state.3 If a stabilizing ligand is present, the shift toward the denatured state is delayed, resulting in a shifted protein unfolding profile. The stability of a test protein in solution is usually measured in a gradual manner, using a thermal scan rate of 1°C/min or incubating the protein with various denaturants until equilibration is reached. This method can be conveniently executed with high density microplates using thermal cyclers. Direct binding evidence between well-behaved target proteins and small molecules is easily discerned through protein melt kinetic transition (Tm) shifts.4 Compounds promoting protein stability have been extensively characterized or confirmed using DSF as screens for a number of drug targets5–8. This solution based method examines protein kinetic Tm when incubated with small molecule ligands. Unfortunately, the hydrophobic binding dyes used in these types of assays, such as SPYRO Orange, contribute to the protein unfolding reactions, subsequently shifting folded protein population to denaturation or aggregation. Therefore, caution is needed to minimize the pre-incubation time between the dye and target protein9, 10. Also, many test compounds have intrinsic absorbance interferences with spectroscopic measurements. In addition, ligand rank order and binding affinities derived from Tm shift cannot be easily translated to those at physiological temperatures due to the lack of knowledge of other thermodynamic parameters, such as enthalpy and heat capacity changes.
In contrast to thermal denaturation, equilibrium chemical denaturation is an alternative to evaluate protein stability and binding constants at low temperatures (lower than Tm or physiological temperature)11, 12. Equilibrium chemical denaturation performed on automated platforms (such as AVIA 2304) relies on changes in intrinsic or extrinsic fluorescence of target proteins during unfolding process as a measurable parameter.13 However, as observed with the thermal denaturation protocols, the equilibrium chemical denaturation processes in solution can also result in aggregate formation, particularly as proteins approach intermediate denaturant conditions where aggregation prone partially folded species become highly populated.
There are some other protein specific caveats that prohibit wide spread use of these methods for a broader range of proteins. The gradual thermal denaturation approach works best for well-behaved proteins which produce smooth reproducible kinetically controlled denaturation profiles. Unfortunately, oligomeric proteins or multi-domain proteins sometimes fail to produce recognizable denaturation profiles, often reporting multi-transitions when subunits or domains fail to melt cooperatively due to aggregation, making data interpretation difficult. Thermal based assays also fail for thermally stable oligomers such as transthyretin (TTR) because no signal is generated during thermal ramps under physiological solution conditions. On the other end of the stability spectrum, the solution based differential scanning fluorometry or chemical denaturation assays are also problematic in situations when the protein is marginally stable because aggregation is even more problematic at physiological temperatures (i.e. low equilibrium Tm). This latter class of proteins includes many missense mutants that are responsible for misfolding diseases.9 In addition, the method often requires a large quantity of protein in order to obtain a visible melt transition for large screening sets. It is also very challenging to use thermal denaturation approaches to monitor the stability of membrane proteins because the presence of the lipid/solubilization medium itself leads to extensive hydrophobic interactions generating uninterpretable signals.
For the aggregation based approaches, potential test stabilizers may also inhibit the aggregation reactions directly rather than physically stabilizing the protein of interest.14 For example, generation of smaller aggregates reduce the light scattering signal yielding false positive results. Furthermore, protein aggregation based assays are notorious for their slow kinetic readouts, particularly during oligomer dissociation processes.15, 16 Aggregation assays based on the binding ability of dyes such as thioflavin T also suffer from lack of complete specificity in dye binding, are subject to false negatives and include instances where the compounds stabilize folding intermediates (misfolded) without stabilizing the native protein. Problems can also arise from the physical properties of the protein stabilizer (e.g. time dependent self-aggregation, optical interference) where spectroscopic interferences may compromise specific enzyme assays.
To assess stability of slow dissociating oligomers or metastable proteins, there is a need for a general rapid perturbation system that avoids aggregation while permitting assessment of ligand-induced stabilization of these types of proteins. Here we present a simple protein kinetic stability evaluation system using a GroEL chaperonin based, label-free BLI approach that avoids general aggregation, allowing detection of partially folded transient populations immobilized on biosensor surfaces. This detection system builds upon the inherent process where the nucleotide-free GroEL chaperonin can promiscuously bind and detect partially folded proteins that have exposed hydrophobic patches. Normally, this chaperonin can allow proteins to acquire their functionally folded conformations by binding these transient forms, unfolding and releasing them to fold either inside a hydrophobic cavity or in the case of large protein, releasing them back into solution. Both the encapsulated folding (“cis”) and release (“trans”) folding processes involve elaborate allosteric ATP dependent cycling mechanisms17. The protein stability assessment system described here uses the high-affinity nucleotide-free form of the GroEL chaperonin state that has the ability to tightly bind partially folded proteins (molten globules) or unfolded states as hydrophobic regions become exposed to solvent. The substrate binding interactions with the high affinity GroEL chaperonin often has binding affinities that approach nM to sub nM Kds18. Most importantly, dynamic unfolding/refolding equilibria can populate protein states that can partition onto the nucleotide free chaperonin but this partitioning reaction is suppressed when either small molecule stabilizers (e.g. natural ligands) or stabilizing conditions (i.e. folding osmolytes/non-oxidizing conditions) shift the natural dynamic equilibria toward folded states19–22. In most cases, perturbing the initial unfolding↔ refolding equilibria with minimal temperature elevation or low denaturant conditions is enough to generate higher partially unfolded populations that kinetically partition onto the high affinity chaperonin9, 21. For the system illustrated herein, transitions toward partially folded conformers capable of binding GroEL are dictated by the intrinsic stability and the solution conditions which stabilize or destabilize the immobilized target protein. This work describes the development of a broad based chaperonin-BLI detection system capable of assessing the kinetic stability of almost any folded protein species.
MATERIALS and METHODS
Ultra pure (MB grade) guanidine hydrochloride (GnHCl) was purchased from USB corporation (Cleveland, OH). Urea (ACS grade), along with Tween-20, was obtained from Fisher Scientific (Pittsburgh, PA). Resveratrol was obtained from Tocris (Bristol, UK). Glutathione (GSH), ethacrynic acid (EA), and diethyldithiocarbamate (DDC) were purchased from Sigma-Aldrich (St. Louis, MO). GT1b (bovine brain) was purchased from Enzo LifeSciences as a lyophilized powder and resuspended in chloroform at 20 mg/ml (9.1mM). Working dilutions were made by two 1/10 serial dilutions in Millipore water. Amine reaction second-generation (AR2G) biosensors and Ni-NTA biosensors were procured from forteBIO (Menlo Park, CA).
The 53BP2 (490 – 498), p53 - Binding Loop (CDB3) p53 binding peptide (NH2-REDEDEIEW-COOH -MW 1220.2 Da) was purchased from Peptide 2.0 (Chantilly, VA) and stored at −20°C prior to use.
A monoclonal IgG was obtained as a gift from forteBIO (Menlo Park, CA) and His-tagged MalZ was purified according to a previous procedure23. The von Willebrand factor (vWF) A1-A2-A3 protein with a His tag on the A3 domain was expressed and purified following the methods outlined in Tischer et al24. Schistosoma japonicum glutathione S transferase (SjGST) was obtained from Genscript (Piscataway, NJ) while His-tagged GST was obtained from EMD Millipore (Temecula, CA). His-tagged recombinant human transthyretin and superoxide dismutase were obtained from Sino Biological Inc. (Beijing, China). A His-SUMO-tagged nucleotide binding domain of cystic fibrosis transmembrane regulator (CFTR NBD1) was obtained as a gift from the Cystic Fibrosis Foundation. Maltodextrin glucosidase (MalZ) plasmid PCS19 containing the malZ gene was a generous gift from Professor Winfried Boos (University of Konstanz, Germany).
Chaperonin purification and storage
Highly purified GroEL was obtained using the purification scheme outlined in a previous work25. Since GroEL does not contain tryptophan, purity was determined by following the diminishing contributions from tryptophan (proteins or peptides) as assessed by contributions from contaminant tryptophan fluorescence (excitation at 297 nm) or by noting the indole contributions using second derivative UV analysis26. It is crucial to obtain highly pure GroEL since small amounts of contaminating proteins and peptides diminish the effectiveness of protein capture and binding. In addition, highly purified GroEL has a tendency to slowly dissociate into heptamers and monomers, where monomers can bind to remaining oligomeric GroEL, further compromising the GroEL preparation. To avoid dissociation of the GroEL tetradecamer, the purified GroEL lots are stored in 50% glycerol. This storage solution is removed prior to immediate GroEL use and replaced with a GroEL buffer (preferred in protein refolding assays) containing 50 mM KCl, 10 mM MgCl2, 50 mM Tris HCl pH 7.5 at 25°C. For elevated temperatures (37°C) the Tris HCl buffer can be replaced with 50 mM HEPES but the pH decline is only to 7.2 for the Tris buffer.
Maltodextrin glucosidase purification
MalZ was produced and purified according to the protocol outlined by Paul et al.23 The PCS19 plasmid contained the malZ gene under the control of an isopropyl β-D-1-thiogalactopyranoside (IPTG) inducible T5 promoter and ampicillin resistance selection marker. Escherichia coli strain BL-21 was used for protein overexpression. The purification of the N-terminal His-tagged MalZ was carried out by nickel affinity chromatography. A 20 mM sodium phosphate buffer containing 500 mM NaCl was used for lysis and Ni column equilibration. The bound MalZ protein was eluted with a linear imidazole gradient with MalZ eluting near 200 mM. Appropriate fractions of MalZ were evaluated using SDS-PAGE gels stained with coomassie blue. Fractions corresponding to pure single band fractions were pooled27.
TeNT subcloning, expression and purification
DNA encoding tetanus neurotoxin (TeNT) was cloned into a modified pET28a expression vector (Merck KGaA, Darmstadt, Germany) such that the resulting fusion protein contained an N-terminal His6-tag and a C-terminal Strep-tag. To generate a catalytically inactive form of TeNT for use in downstream experiments, two point mutations within the light chain (R372A and Y375F) were generated using the QuikChange II Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA). Recombinant TeNT was expressed in E. coli BL-21 AI cells and purified by sequential chromatography on Ni-NTA agarose, Sephacryl S-200, and Strep-Tactin sepharose. Peak fractions from the Strep-Tactin sepharose column were concentrated to ~1.5 mg/ml using an Amicon filtration device (YM-100 type filter) and dialyzed into TeNT buffer (30 mM HEPES-NaOH, 500 mM NaCl, pH 7.6), and stored at −80 °C until use. A typical preparation yielded 1–3 mg of purified toxin/liter of batch culture.
p53 subcloning, expression, and purification
A plasmid with the gene for TP53 aka p53 (AY891243) was obtained from the Harvard PlasmID Database (HsCD00001269). The DNA Binding Domain (DBD) of p53 (residues 94–312) was subcloned into pTBMalE provided by Dr. Fei P Gao. The resulting construct had an N terminus His tag followed by maltose binding protein (MBP), a TEV recognition sequence and p53 DBD. The plasmid was transformed into BL21(DE3) competent E. coli cells obtained from New England BioLabs (Ipswich, MA). For expression, a 10 ml preculture was grown overnight in LB Broth with 100 μg/ml ampicillin from Fisher Scientific (Fair Lawn, NJ). One liter of LB with ampicillin was inoculated with the preculture and induced with 0.24 g of IPTG from EMD Millipore (Billerica, MA) when OD600 reached 0.6. Cells were grown overnight at 21°C, then harvested by centrifugation at 5,000 rpm and resuspended in 50 mM Tris (Fisher Scientific, Fair Lawn, NJ), pH 7.2, 1 mM DTT (Applichem GmbH, Darmstadt, Germany), and 50mM imidazole (Acros, NJ). Protein was purified by affinity chromatography with Ni-NTA Agarose (Thermo Scientific, Rockford, IL) and was concentrated with an Amicon Centrifugal filter unit from Merck Millipore (Co. Cork, Ireland). Protein was buffer exchanged into 50 mM Tris pH 7.2 with10 mM DTT using a PD10 Desalting Column (GE Healthcare, Buckinghamshire, UK) and concentrated again to ~1–5mg/ml, followed by subsequent storage at −20°C in 50% glycerol (Fisher Scientific, Fair Lawn, NJ).
Biolayer interferometry
In most instances, preliminary biolayer interferometry experiments were performed on a single channel BLItz® instrument (forteBIO, Menlo Park, CA) to establish conditions (loading, GroEL concentrations, buffers, etc.) and were later carried out on an Octet® RED96 instrument (forteBIO, Menlo Park, CA) unless otherwise stated. Biolayer interferometry is an all-optical technique measuring biomolecular interactions. The biosensor tip is a glass fiber optic system that measures the changes in the reflected white light interference patterns between a reference layer and a test biolayer. Any shift in the interference pattern of the reflected white light obtained at the detector indicates changes in the number of bound molecules. This change can be monitored in real time, resulting in a phase shift reflecting both the kinetics of the binding process and an amplitude reflecting the binding affinity28. The temperature of measurement was set at 25°C unless otherwise specified. The loading of the protein substrate was optimized to avoid crowding on the biosensor surface and was within a linear range of BLI signal to protein concentrations (Figure S1). Shaking platforms were set at 2200 rpm and 1000 rpm, for single channel BLItz and automated Octet system, respectively. At excess GroEL concentrations, the binding signals stabilize at these two shaker settings, diminishing mass transport effects. Sample volumes in all steps were provided in 250 μl aliquots in 500 μl black microcentrifuge tubes for the BLItz single channel unit and 200 μl aliquots for the 96-well formats with the automated Octet system. Ni-NTA tips were regenerated following a previously established protocol.29 All the protein targets described in this work, except IgG, were immobilized onto Ni-NTA tips via the strong and highly specific interaction between His tag and nickel ions. In contrast, immobilization of the IgG molecules was accomplished using a standard 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)/N-hydroxysuccinimide (NHS) (ThermoFisher Scientific, Waltham, MA) protocol30, 31 and amine reactive second generation (AR2G) biosensors. The IgG biosensors were manufactured and supplied by forteBio.
Optimizing protein loading onto BLI biosensors
Protein loading (e.g. GST as an example - Supplemental Figure S1) onto the biosensor surface was within the lower end of the linear response range (signal vs. protein concentration). Using lower protein concentrations diminishes tip surface overcrowding and intra-surface protein interactions. Overcrowding could result in self-association between loaded protein molecules, leading to a diminished GroEL binding signal. Since the loading of the protein and the concomitant GroEL binding was kept within a linear response range, one could correct for slight GroEL binding variations per protein load by performing a load correction based on the valid assumption that the GroEL binding signal was directly proportional to the amount of protein loaded/partially unfolded (Supplemental Figures S1 and S2). At high GroEL concentrations, the sensorgram traces would rise rapidly and approach a saturated signal amplitude due to favorable collisional rates and optimally attached (within linear loading range) target proteins.
BLI denaturant pulse method to generate kinetically controlled denaturation isotherms
The kinetic denaturant pulse method (illustrated in Figure 1) was performed using the following general steps: each protein target was individually loaded onto specific affinity biosensor tips followed by a buffer wash step (with or without a ligand) to establish a buffer baseline; biosensor tips were subsequently exposed to a series of increasing denaturant solutions (with or without a ligand) for a defined incubation (pulse) time (3,5, or 10 min) followed by a dip of the probe into a brief wash solution (usually original loading buffer) to remove any adhering denaturant for anywhere between ~ 5 sec to 20 sec. Following the brief wash, the biosensor tips were immersed into solutions containing GroEL where chaperonin binding to denatured protein populations that were present on the tip resulted in a time-dependent kinetic rise in the BLI sensorgram. For experiments performed on the automated Octet (as was the case for GST, TTR, SOD, and vWF), Ni-NTA tips were regenerated at the end of each denaturant pulse and return cycle following an established regeneration procedure27. The TeNT protein was not regenerated and fresh NiNTA tips were used for each replicate of constructing the TeNT kinetically controlled denaturation isotherm. For the regeneration procedure, Ni-NTA tips with protein-GroEL complex were first cycled through 10 mM glycine at pH 1.7 followed by neutralization buffer (1× kinetics buffer, forteBIO) for 3 times, with each pulse lasting 5 s. The NTA regenerated tips were then recharged with Ni by immersing the tips into 10 mM NiCl2 for 1 min. If regeneration protocols cannot be implemented, then new tips should be used, provided that loading amplitudes continue to remain constant (e.g. no compromise in protein integrity). Regenerated surface was re-used to bind His-tagged protein for the next round of experiments. The reloading was highly reproducible for proteins that remained stable throughout the regeneration procedures, varying by only a ~3–5 % standard deviation as assessed by noting the variation of the final loading signal amplitudes (see Figure S2, S4). As a necessary control, any evidence of stabilizer dissociation sensorgram traces (non-specific or specific declines if observed) in the absence of GroEL were always subtracted from GroEL association phase to create the final corrected sensorgrams (corrected kinetics and amplitude).
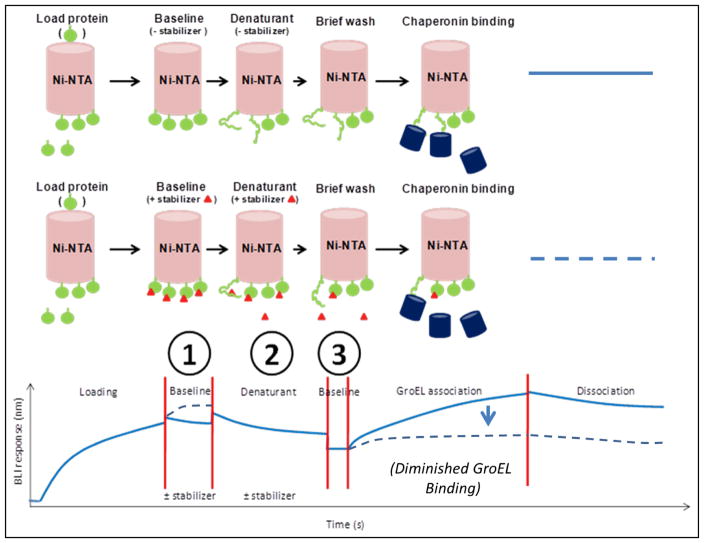
Figure 1.
Schematic illustration of a generic denaturation pulse platform for identification of kinetic protein stabilizers. Key steps include target protein loading onto biosensors followed by dipping into a buffer with or without a potential stabilizer (baseline), dipping into a chaotropic solution with or without a potential stabilizer, a wash step to remove denaturant (and compound if any) and subsequent GroEL association with protein species that expose hydrophobic residues. The following steps highlight the processes that can be varied, including 1) pretreatment time with a potential stabilizer to ensure adequate binding, 2) denaturation time to increase unfolded population and 3) wash time to remove denaturant and compound. In the presence of a stabilizer (red triangle) prior to and during the denaturant pulse phase, the GroEL binding amplitude and kinetics is diminished (dotted line). As a necessary control, profiles with any evidence of stabilizer dissociation in the absence of GroEL need to be subtracted from GroEL association phase.
Generating MalZ kinetically controlled denaturation profiles in presence of urea and osmolyte stabilizers
His-tagged MalZ at a concentration of 80 –100 μM was prepared with reduced imidazole concentration by buffer exchange and concentration on an Amicon 30 kDa centrifugal concentrator. The buffer used for BLI experiments consisted of 20 mM sodium phosphate, 20 mM MgCl2, 10 mM KCl at pH 7.4. All MalZ experiments were performed using the single channel BLItz system, and under these conditions, MalZ did not show dissociation from Ni-NTA biosensors. To construct a kinetically controlled denaturation profile of the MalZ protein, His-tagged natively folded MalZ constructs at a final concentration of 1 μM were loaded onto Ni-NTA tips over a 5-min time window. MalZ bound to these Ni-NTA tips were subjected to separate but increasing urea concentrations in a 5-min pulse and after a 10-s wash a GroEL refolding buffer (50 mM Tris, 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, pH 7.5). GroEL binding to the increasingly unfolded yet still attached MalZ population was measured. Solid urea was added to refolding buffer to achieve final concentrations of 0, 1, 2, 3, 4, 5, 6, 7 and 8 M. The same solution procedures were used with the denaturation series where urea solutions also contained stabilizing osmolytes 1M sucrose or 4 M glycerol. The resulting GroEL binding amplitudes were plotted against the respective urea concentrations to generate denaturation isothermal transitions and were compared with the kinetic isotherms generated with the urea denaturant alone. His-tagged GST attached to the NiNTA biosensor was tested for functional integrity through a titration with the known inhibitor, tannic acid, using the sensitive OctetRed 96 BLI instrument. This instrument is used to evaluate binding of small molecules >150Da to immobilized proteins. The results from this assessment indicate that the estimated binding affinity of the immobilized GST for tannic acid is ~ 17 ± 2.3 nM, well within the same order of magnitude as the in solution binding constant range (49 nM) (See results in Figure S1C).
Effect of denaturation time on His-tagged GST
As a model protein, His-tagged GST was used to explore the effect of denaturation times on its kinetic transition. The loading of GST showed a linear nm shift throughout a GST concentration range of 0.05 to 1 mg/ml. At a response signal of 1 nm loading (~ 0.25 mg/ml GST), the GroEL concentration was varied to determine the binding saturation. The GroEL concentration that was chosen for the GST kinetic denaturant pulse experiments was 0.5 μM tetradecamer. The denaturant pulse times were varied from 3, 5 and 10 minutes for GST alone and GST that was covalently modified with EA/GSH adduct. Loading of GST was relatively constant throughout the repeated runs (Figure S2) and the GroEL binding amplitudes were normalized. The raw data traces and load corrected traces revealed the same trend in GroEL binding responses (Figure S2).
Ligand induced stability assessment of oligomeric proteins (TTR and SOD) dimers
The thermal stability of TTR and SOD dimers is often difficult to assess due to the inherent stability of these oligomers. A known TTR stabilizer, resveratrol, and a known SOD1 inhibitor, DDC (chelates Cu metal ligand), both applied at 200 μM, were respectively added into both the baseline step before and during the denaturant pulse step. Following a 5s buffer wash to remove denaturant and the compound ligand solution, GroEL binding to the immobilized target oligomers was monitored after the protein loaded biosensors were stabilized or destabilized due to ligand binding or metal ligand removal respectively.
Evaluation of metastable proteins -CFTR-NBD1 and p53 kinetic stability assessments
A) CFTR-NBD1
Wild type nucleotide binding domain (NBD1) fragment of the cystic fibrosis transmembrane regulator engineered with an N-terminal His SUMO tag was loaded onto Ni- NTA tips at a concentration of 0.01 mg/ml (optimized loading amplitudes to avoid intra-surface aggregation). Since CFTR-NBD1 is metastable in the absence of a stabilizing ligand, ATP was added to maintain CFTR-NBD1 stability during the loading phase and loading amplitudes remained essentially constant throughout the experimentation on both BLItz and the Octet system. Specifically, for the Octet system, a 0.01 mg/ml CFTR-NBD1 solution (diluted in 50 mM Tris-HCl, 150 mM NaCl, 3 mM MgCl2, pH 8.0) was prepared and dispensed into one column in a 96-well solid bottom plate at a volume of 200 μl per well. Octet chamber temperature was maintained at 25°C and the plate containing CFTR-NBD1 protein solutions was kept in the chamber for the entire duration of the experiment. Ni-NTA tips were hydrated in the aforementioned CFTR-NBD1 buffer in order to establish a baseline before protein loading. Loading magnitude was observed for a 5-min period of time before dipping the tips into CFTR-NBD1 buffer again to re-establish baseline. The same set of Ni-NTA tips were regenerated and re-used to load CFTR-NBD1 repeatedly within a 40-min time window. Separately, the same experiment was performed using CFTR-NBD1 solutions that contained 2 mM ATP, a natural stabilizing ligand for the protein. Weakly binding ATP that was bound to CFTR-NBD1 was essentially washed away during the buffer wash step and the denaturant pulse step. Comparisons of CFTR-NBD1 kinetic stabilities in the presence and absence of GTP in the postload step and denaturant pulse step were performed using the single channel BLItz system. GTP, a known nucleotide stabilizer, does not bind to GroEL18 nor does this nucleotide facilitate release and folding of GroEL bound protein substrates. Both the initial GroEL binding amplitude differences and a limited kinetic isothermal denaturant curve were obtained for CFTR-NBD1 in absence and presence GTP.
B) Testing Osmolyte and CBD3 peptide stabilization of p53 core domain
The His-tagged maltose binding protein (MBP) - p53 core domain protein was loaded at concentrations of 0.1 μM, which was well within the linear concentration range for this protein (results in a 1 nm amplitude deflection during loading). The GroEL concentration (tetradecamer) used for binding immobilized p53 was 0.15 μM (yields ~ 1 nm deflection when binding completely denatured p53, which was achieved upon pulse exposure to 6 M GnHCl). The osmolyte stabilization assessment was performed on the automated Octet BLI system using the denaturant pulse protocols (25°C). The potential stabilization effect due to CDB3 peptide binding to the p53 core was assessed by monitoring the potential decline of GroEL binding to native and pulse denatured p53. The effects of CDB3 peptide binding/stabilization of p53 on the GroEL interactions after the denaturant pulse step were performed on the single channel BLItz system.
Developing kinetic stability isotherm for multidomain proteins
A) IgG kinetically controlled denaturation and stabilization
To generate kinetically controlled denaturation profiles of a known multidomain protein, immobilized IgG (randomly attached via reactive amine coupling- forteBIO manufactured Ab biosensor) was briefly exposed to increasing denaturant concentrations (5 min) using either urea or GnHCl at pH 7.5 in GroEL refolding buffer. After the denaturation pulse, tips were washed in refolding buffer for 1 min followed by exposure to a 1 μM (tetradecamer) GroEL solution using the same buffer. The amount of bound GroEL represents the amount of destabilization of IgG, and the amplitudes were measured at each denaturant concentration. GroEL binding amplitudes were plotted as a function of denaturant concentration to generate a kinetically controlled denaturation isotherm. In a separate set of experiments, immobilized IgG was exposed to mixtures of 4 M urea and 2–4 M of a number of folding osmolyte/protein stabilizer solutions to mimic an excipient screen. Following the urea denaturant pulse ± stabilizers and a 1 min wash, the GroEL binding sensorgrams were recorded.
B) von Willebrand Factor (triple A domain) - constructing kinetically controlled denaturation isotherms using varying denaturation times
To determine if the kinetic denaturant pulse method could be used to detect the stability of the von Willebrand Factor (vWF), a multidomain protein containing 3 individual integrin like domains (triple A domains) connected by flexible linkers, vWF biosensors were constructed to generate and compare a kinetically controlled denaturation isotherm with the multi-phase denaturation transitions that are observed during the solution equilibrium denaturation of vWF.32 After determining the linear range for loading His-tagged wild type vWF (attached through the A3 domain), 0.6 μM of wild type vWF triple A domain was loaded onto Ni-NTA biosensor tips over a 5 minute time period. After a brief wash 30s wash, biosensor tips were exposed to a urea denaturant pulse for 1, 5, or 10 minutes. After a second wash 10s to remove residual denaturant, tips were dipped into refolding buffer containing a 0.5 μM GroEL final concentration. The GroEL binding amplitude, recorded after each pulse time was plotted as a function of denaturant pulse concentration, and the kinetically controlled denaturation isotherms for each denaturant pulse time were constructed.
C) Generating TeNT kinetically controlled denaturation profile with and without a natural ligand
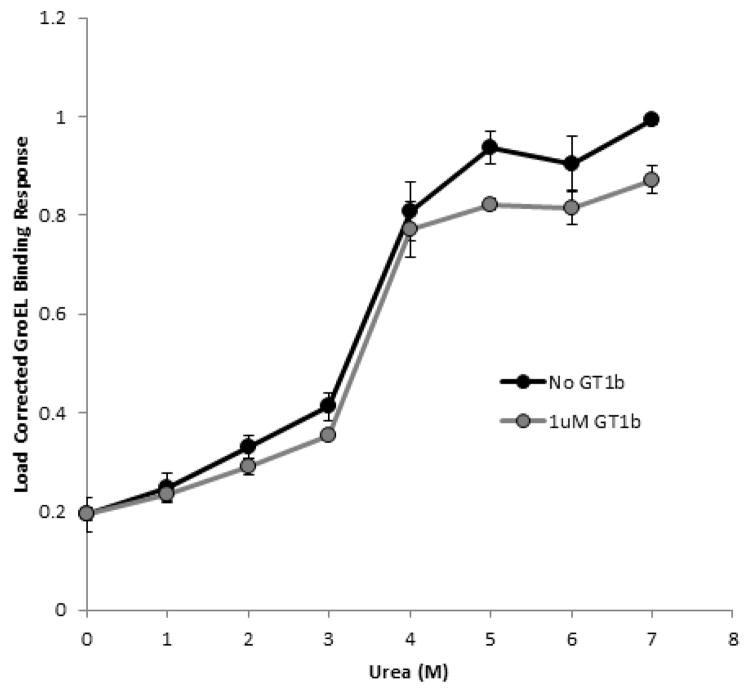
TeNT was supplied with an N-terminal His-tag at 1.5mg/ml in TeNT buffer (30 mM HEPES, 500 mM NaCl, pH 7.6). This buffer was used during the biolayer interferometry experiments for baselines, protein loading, and urea denaturation. TeNT denaturant pulse isotherms were generated on the Octet Red96 system using non-binding black well plates, containing 230 μl/well. After finding the linear response range for loading TeNT protein (linear from 2.5 μg/ml up to 0.75 mg/ml), a concentration of 7.5 μg/ml (50 nM) (forteBIO recommends lower range) was chosen to generate the kinetically controlled denaturation isotherm. Ni-NTA biosensors were used to immobilize TeNT for the assay. Once bound to the biosensors, the protein was denatured for 300 seconds in urea at a concentration of 0, 1, 2, 3, 4, 5, 6, and 7 M in refolding buffer. After denaturation, the tip was briefly washed for 10 seconds in TeNT buffer. GroEL, at 0.5 μM in refolding buffer, was allowed to bind to the denatured TeNT for 300 seconds. Following GroEL association, GroEL/TeNT complex was allowed to dissociate for 300 seconds in refolding buffer. GroEL signal was load corrected by dividing the amplitude of GroEL binding by the amplitude of TeNT loading. To determine the association of GroEL to partially unfolded TeNT, the load corrected GroEL signal was plotted against urea concentration. To examine GT1b effect on TeNT stability, the above protocol was slightly modified: 1) an additional step consisting of 1 μM GT1b in TeNT buffer was included before the urea denaturation pulse step to allow for ligand association, 2) GT1b was present in the urea solutions at 1 μM, and 3) the 10-second wash after the denaturant pulse was split into two 5-second washes, the first with 1 μM GT1b and the second with just TeNT buffer.
RESULTS
Using the GroEL chaperonin to detect partially folded states immobilized on surfaces
The use of the high-affinity nucleotide-free GroEL chaperonin system as a detector of partially folded or misfolded proteins on immobilized surfaces stems from the frequent observation that GroEL chaperonins (naturally occurring or overexpressed) bind to misfolded proteins. Interestingly, the GroEL chaperonin is a common contaminant protein binding to the affinity tagged protein isolated on affinity column matrices33 due to the fact that GroEL binds to affinity tagged proteins that remain partially folded. To avoid in vivo aggregation, methods were developed to construct chimeric proteins where difficult folding domains could be stabilized by attaching an engineered easy-to-fold chimeric affinity tag (e.g. SUMO, glutathione S-transferase, maltose binding protein, etc.). Even when the affinity tag readily folds and binds to the biospecific tag, the protein of interest may still fail to completely fold and bury its hydrophobic surface, allowing the high affinity nucleotide-free chaperonin to readily bind to the protein while immobilized on the biospecific affinity surface3. Previous work indicates that the rates of folding of immobilized protein can be facilitated by adding specific folding osmolytes, increasing temperature or including small molecule stabilizers to enhance proper folding and diminish GroEL protein binding34.
Eisenstein, Yoshita, and others pioneered the use of the nucleotide free GroEL chaperonin as a detection system for folding status of attached protein substrates with surface plasmon resonance (SPR). They monitored chaperonin binding to partially folded protein substrates attached on immobilized surfaces to eventually follow the kinetics of the ATP/GroES induced release of the GroEL chaperonin from the surface 35, 36. If the dynamic folding equilibria of a protein results in substantial populations of metastable folded states, this particular immobilization approach can also be used to examine possible ligand or solution induced stabilization shift away from the metastable population14.
In practice, the chaperonin detection of dynamic partially-folded states that are immobilized using label-free surface approaches can also applicable using an SPR based approach. However, SPR is a microfluidic flow-based system with most common throughputs of around 20 complete reactions (titration analysis) per day. In contrast to the SPR system, BLI does not involve microfluidics and the sensor surfaces are inexpensive compared with SPR. Employing dip and read solution changes rather than relying on microfluidic approaches greatly simplifies the experimental setup particularly in the precision of changing denaturant pulse times, changing wash solutions and obtaining reproducible signals following multiple regeneration procedures. The Octet Red384 systems BLI throughput can accommodate 16 protein immobilized biosensors simultaneously in 384-well plates while monitoring interactions in real time and extracting kinetic parameters, such as kon, koff and KD. Newer BLI instruments such as the Octet HDX series can sample up to 32, 48 or 96 individual wells with one reading (i.e. 32, 48 or 96 biosensor channels). In many instances, the BLI and SPR results are nearly identical. However, there have been numerous instances where denatured proteins interact with the activated carboxymethyldextran SPR surfaces particularly during pH dependent denaturation.37 In addition, the integrity and function of the microfluidic flow system channels of the SPR instrumentation are compromised by protein aggregation. Unlike the BLI system, this particular flaw in the SPR system interferes with the evaluation of the protein loading reproducibility. Also, unlike the SPR system, BLI biosensors are useful for probing the contents of whole cell extracts. Furthermore, it is much easier to recover evaluate and actually visualize macromolecular complexes that form on the BLI surfaces if one can reverse the attachment chemistries. For example large complexes as well as GroEL-protein complexes can be readily observed by electron microscopy by reversing the attachment on the biosensor surface31, 38.
In the examples that follow, dynamic populations of folding intermediates are generated by immersing target proteins attached to BLI biosensors into a denaturant solution for a defined period of time (defined as a denaturant pulse). In all of these instances, the amount of GroEL binding to the partially folded immobilized substrate is predicted to directly correlate with the extent and number of partially folded/unfolded protein molecules that remain on the BLI biosensor once the denaturant is removed and the biosensor tip is immersed in a GroEL solution (GroEL binding step) (Figure 1).
Extending stability and folding analysis for a His-tagged recombinant protein MalZ
The first example of examining GroEL binding to a large immobilized protein uses the E. coli protein MalZ, which has been shown to be efficiently folded by the GroE system. MalZ is a 69 kDa monomeric enzyme functioning to degrade maltodextrins to shorter oligosaccharides39, and has also been used to examine GroEL folding of larger proteins that are unable to be sequestered inside a GroEL-GroES cavity. Chaudhuri and colleagues demonstrated that MalZ appears to fold from GroEL by a trans mechanism with the assistance of GroEL/GroES23. To generate a kinetically controlled denaturation profile, His-tagged MalZ was immobilized onto a series of Ni-NTA biosensor tips, and each one was briefly immersed in varying denaturant concentrations (Figure 2). The urea denaturant was subsequently washed away with a 10-sec wash step, and the population of unfolded/partially folded species on the tip at the time of measurement was assessed by monitoring the chaperonin binding amplitude recorded at 5 min. The amplitude of chaperonin binding is directly correlated with amount of folded versus unfolded/partially folded protein populations that are present on the biosensor surface at the time of measurement, resulting in an increasing amplitude change as denaturant concentration increases.. Any kinetic delay in unfolding due to the presence of any stabilizing conditions (ligands or inclusion of folding stabilizers) is predicted to slow the kinetics of unfolding of the immobilized protein populations. The observed decrease in the chaperonin binding amplitude with increasing denaturant concentration agrees with this prediction (Figure 2). The presence of folding osmolytes, such as 4 M glycerol or 1 M sucrose within each denaturant pulse solution resulted in a diminished chaperonin binding at the time of measure, indicating that a lower percentage of the attached protein had unfolded during the duration of the pulse. As the denaturant concentrations increased further, the GroEL binding amplitudes in the presence of the osmolytes did approach the same maximum GroEL binding amplitude as was observed for MalZ biosensors that were not stabilized by osmolytes (Figure 2). It was important to note that each tip was removed from the osmolyte/denaturant pulse solution and was washed briefly (10 s) to remove the denaturant/osmolyte mixture before immersing into buffer containing the chaperonin. This procedure insures that bulk osmolyte removal is complete and will not interfere with the chaperonin binding to the partially folded substrate. Separate control experiments indicated the immobilized MalZ that was completely denatured at high urea concentrations at 25°C refolds very slowly (t1/2 at 25°C ~ 10 min) relative to the wash times that were used to remove adhering denaturant or ligand solution (data presented in publication currently in minor revision, Pastor et al., 2016). It was also shown that the refolded MalZ immobilized on and released from Ni-NTA surfaces was functionally active and yields were substantially improved compared with solution refolding. These control measurements indicated that no substantial refolding is detected (i.e. chaperonin binding amplitudes remain high) if two separate MalZ attached tips were compared before and after a 10-second wash period.
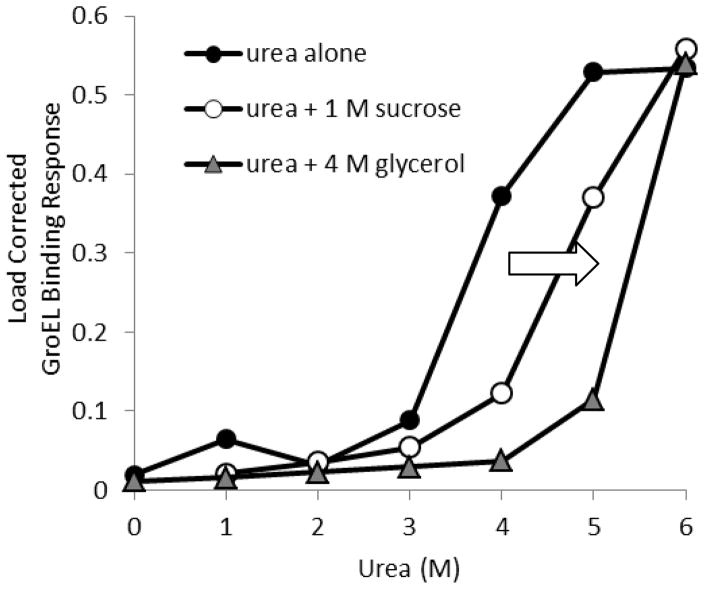
Figure 2.
Kinetic stabilization of MalZ in presence of different osmolytes. The MalZ protein was loaded onto Ni-NTA biosensor tips at 1 μM, and loading was consistent throughout the titration experiment. Each MalZ tip was exposed to various concentrations of urea, and GroEL binding amplitude was measured after 5 min. A separate set of MalZ attached tips were exposed to both urea and sucrose or glycerol. This figure denotes the GroEL binding response as a function of urea concentration with or without osmolytes. Diminished GroEL binding amplitudes were observed at each urea titration point upon the addition of the stabilizing osmolytes (1 M sucrose or 4 M glycerol), indicating a delay in the kinetically controlled denaturation isotherms, which results in an apparent stabilization of MalZ. A clear rightward shift in the GroEL binding amplitude response (denoted by arrow).
Potential for small molecule stabilizer development and lead compound validation
To illustrate the general utility of this platform in identifying or validating small molecule stabilizers and eventually accelerate pharmacological chaperone drug discovery for aggregation prone protein folding diseases, we tested a set of model or disease relevant protein targets which had different well-established inhibitors or stabilizers. As stated in the introduction, proteins involved in folding diseases are difficult to study because they are aggregation prone under physiological conditions. To facilitate the search for stabilizers while preventing aggregation, the proteins presented in the following sections were immobilized on BLI biosensors to evaluate their propensity to bind the chaperonin GroEL as they were denaturant transitioned to partially folded or fully unfolded populations.
A) Glutathione S-Transferase (GST)
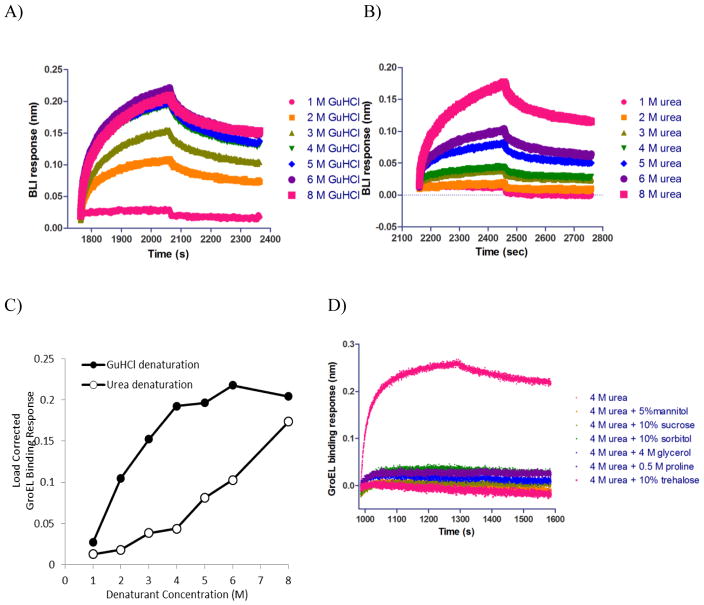
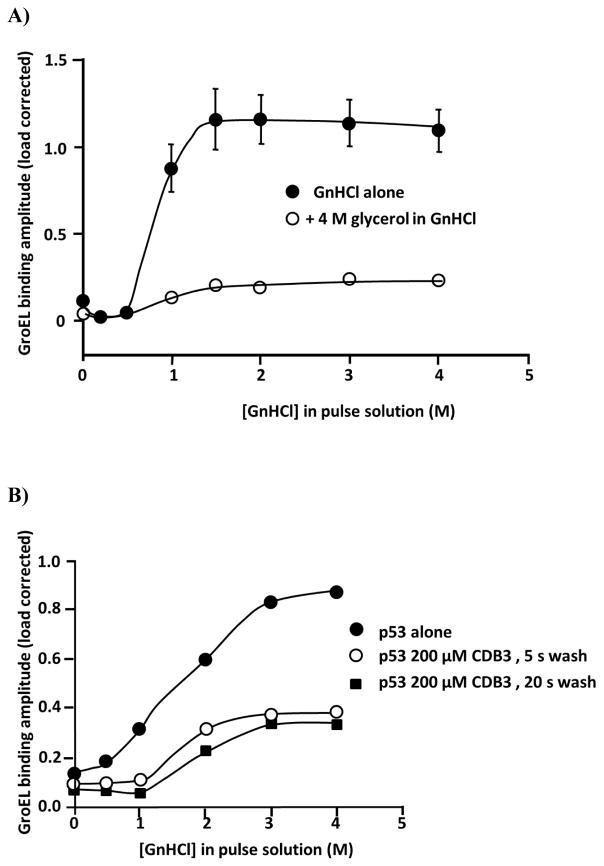
Glutathione S-transferase (GST) was initially chosen to demonstrate the presence of general protein stabilizers through the detection of shifts in the kinetically controlled denaturation isotherms. GST is one of the most commonly applied purification tags as well as an enzyme involved in phase II biotransformation of xenobiotics in different organisms.4 GSTs have been extensively associated with cancer, and their inhibitors exhibit potentiation in cancer therapy. GST protein structures are typically represented by either a homo or heterodimer with an active site in each two-domain containing subunit (N and C-terminal domains). A highly conserved GSH binding site (G site) is located at the N-terminal domain while a larger hydrophobic substrate binding site (H site) resides in the C-terminal domain. At least two main classes of GST inhibitors exist, including glutathione (GSH, GST native ligand) mimetics (i.e. GSH competitive inhibitors) or compounds forming adducts with GSH (non-competitive with respect to both G and H sites). The Compound EA is a covalent inhibitor of GST with a reported IC50 in the submicromolar range against SjGST.40 Initially, a non-tag containing version of GST was immobilized onto amine reactive tips through EDC/NHS chemistry. In spite of sufficient concentration load optimization, this particular non-specific attachment scheme failed to generate highly reproducible kinetically controlled denaturation curves (data not shown). This may not be surprising as prior reports have shown that specific and homogenous orientation of immobilized protein molecules relative to the sensor surface could be crucial in maintaining the integrity of protein function and activity, particularly for small proteins.41 When a His-tagged version of GST was immobilized on Ni-NTA biosensors using an Octet system, the controlled loading and multiple regeneration protocol resulted in highly reproducible kinetically controlled denaturation isotherms. In addition, this particular His-tagged attachment scheme appears to provide a homogeneous orientated binding surface for GroEL. To ensure GST was optimally oriented on the tip and functional, the His-tagged GST protein was titrated with the glutathione inhibitor, tannic acid, using the OctetRed 96 BLI system. The Kd values that were obtained agreed with previously published values (see Figure S1C and reference in Legend). This same His-tagged version of GST was loaded onto Ni-NTA tips with both protein loading and GroEL concentration optimized (this early optimization work was done on BLItz, Figure S1A and B). Since this is a kinetically controlled denaturation process, the extent of GroEL binding is expected to vary with the duration of the denaturant pulse. A comparison of the kinetically controlled denaturation curves at varying denaturant pulse times through the denaturant concentration range (GnHCl, 0–4 M) shows an increase in the GroEL binding magnitude in the ligand–free GST sample compared with the GST sample that is modified with the covalent inhibitor/stabilizer EA.
Curiously, the initial 3 min denaturation pulse failed yield any separation between EA covalently modified GST and unliganded GST. To determine if any differences in GST stability could be observed, the time of the denaturant pulse was progressively increased. The His-GST kinetically controlled denaturation isotherms were examined as a function of denaturant pulse exposure times of 3, 5, or 10 min (Figure 3A). As the denaturant time increases, the kinetic denaturant curves for the ligand-free GST showed a distinct left shift toward earlier destabilization. Understandably, more immobilized protein should unfold as the time period of the denaturation pulse is extended. EA forms a covalent conjugate with GSH (either enzymatically or non-enzymatically), and the resulting adduct is not only a more potent inhibitor of GST, but also covalently modifies the enzyme’s active site inducing a larger thermal stabilization42. For these experiments, EA was present at 200 μM with 2 mM GSH in both the baseline step preceding the denaturant step and during the denaturant pulse step. Unlike the wild type ligand free GST, the GroEL binding amplitudes for the EA-GST biosensors showed very little change in the kinetically controlled denaturation isotherms as the denaturant pulse time increased from 3 to 10 min (compare 3 min EA-GSH GST profile in Figure 3B with 10 min EA-GSH GST profile in Figure 3C). Both showed significant delays in GroEL binding particularly at the low denaturant concentration (e.g. compare amplitudes in the 1 – 1.5 M GnHCl range)
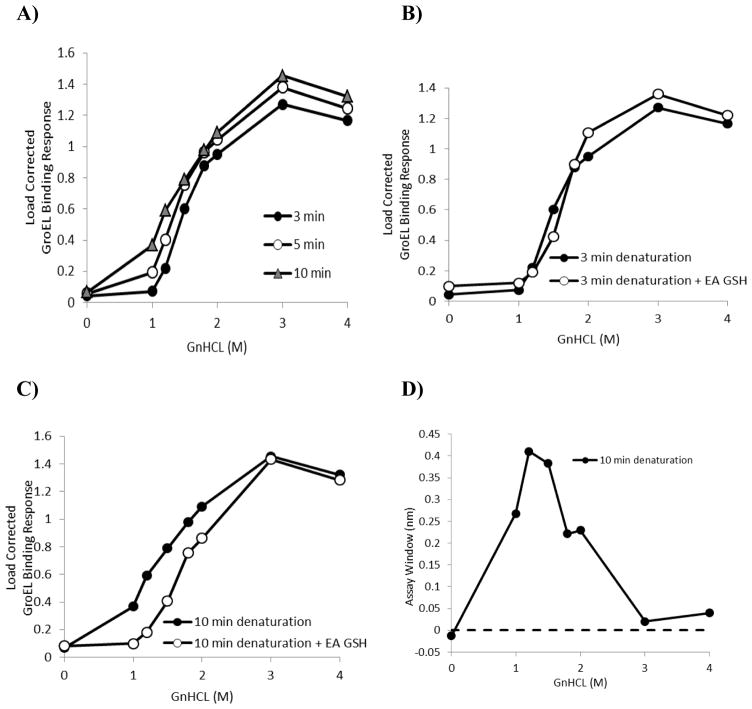
Figure 3.
Kinetically controlled denaturation profiles exhibiting the effect of denaturation times on the GroEL binding amplitudes. A) Kinetically controlled denaturation of His-GST under various denaturation times (3, 5 and 10 mins). B) Kinetically controlled denaturation of His-GST in absence (closed circles) and presence (open circles) of 200 μM EA and 2 mM GSH with 3 min denaturation time. C) Kinetically controlled denaturation of His-GST in absence (closed circles) and presence (open circles) of 200 μM ethacrynic acid and 2 mM GSH with 10 min denaturation time. D) Differences in GroEL binding amplitudes derived from stabilizer bound and stabilizer free kinetically controlled denaturation profiles the 10 minute denaturation times to indicate that stabilizer induced differences were optimal at the lower denaturant concentration range.
The ligand-induced shift in the kinetic denaturant titration curve obtained at different denaturant exposure times shows the covalent EA adduct led to a decrease in the GroEL binding suggesting that the EA modification leads to an observable kinetic stabilization of GST. Specifically, when 200 μM EA along with 2 mM GSH were preincubated with immobilized GST prior to the GnHCl denaturation, there was little difference (no reduction) in GroEL binding throughout the GnHCl titration range after the 3 min denaturation pulse (Figure 3B) but that this difference between wild type and EA modified GSH was more apparent after the 10 min (see kinetically controlled denaturation pulse (Figure 3C). Furthermore, the differences between the load corrected GroEL binding responses were plotted as a function of denaturant concentration (Figure 3D). The difference seemed to reach its maximum near 1.5 to 2 M GnHCl. Curiously, the apparent kinetically controlled transition slope appears to be sharper for the EA-GSH modified protein compared with the wild type GST after a 10 min denaturant pulse.
B) Transthyretin (TTR)
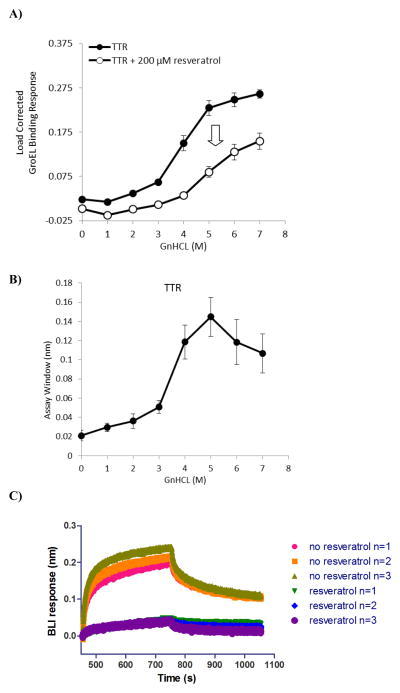
To demonstrate the potential of using this platform to screen for pharmacological chaperones of oligomers, the kinetically controlled denaturation profiles of important disease relevant oligomeric proteins were subsequently evaluated using this BLI denaturant pulse procedure. One such target considered is TTR, which is prone to fibrillogenesis for a number of missense mutants as the oligomer dissociates into monomers, or when the native oligomer is destabilized at low pH conditions and high salt concentrations in vitro.43 Wild type TTR and over 80 mutants have all been found to be capable of amyloid formation and are linked to a group of amyloid diseases, including senile systemic amyloidosis (SSA), familial amyloid polyneuropathy (FAP) and familiar amyloid cardiomyopathy (FAC).43 TTR, a 55 kDa homotetramer, carries thyroid hormone thyroxine (T4) and retinol binding protein in blood and cerebrospinal fluid. TTR possesses two distinct dimer-dimer interfaces, a robust interface stabilized by hydrogen bonding and a weaker dimer-dimer interface where its two T4 binding sites are located.16, 44 Currently, kinetic stabilization of TTR native state conformation by ligand binding to the T4 binding site(s) represents an effective strategy to prevent TTR dissociation of either tetramers or dimers,44 with the goal to prevent amyloidogenesis. In the past, several different compounds, identified by either through rational design45, 46 (including substructure combination strategy)47 or high throughput screening,48 have been reported to stabilize TTR native state, 15, 49–52 with their stabilization effect correlating to decreasing subunit dissociation constants. In this study, a dimeric form of the protein was captured onto Ni-NTA tips through the engineered N-terminal His affinity tag. After loading densities were optimized (following general protocol used for His-tagged GST Figure S1), kinetically controlled denaturation isotherms were generated for the protein in the absence and presence of the stabilizer resveratrol (Figure 4A). Resveratrol is a natural polyphenol and an antioxidant found in red wine and several plants. The trans isomer of resveratrol preferentially binds to the T4 binding site, reducing TTR fibril formation45. The BLI denaturation pulse experiments showed that there was a decrease in the GroEL binding amplitudes in the kinetically controlled transition of the wild type TTR when 200 μM resveratrol was included both prior to and during the denaturant pulse (Figure 4A). This observation is in line with the prediction that resveratrol binding should stabilize or kinetically slow the unfolding of TTR against GnHCl unfolding. In results similar to that demonstrated with GST, the assay window that was generated from differences in raw data load corrected GroEL binding response (Figure 4B) shows that the largest difference of the kinetic response in the presence of the stabilizer resveratrol was achieved at the 4–5 M GnHCl pulse range. The raw sensorgram data shows a dramatic decline in GroEL binding (Figure 4C) when the biosensor immobilized TTR is denaturant pulsed at 4 M GnHCl for 5 minutes.
Figure 4.
Detection of resveratrol stabilization effect on TTR: Kinetically controlled denaturation of His-tagged TTR in the absence (closed circles) and presence (open circles) of a known TTR stabilizer, resveratrol. Experiments were performed on the Octet, with each data point comprising 3 separate measurements and error bars representing standard deviation. A reduced GroEL binding signal was observed at each GnHCl titration point when TTR was pretreated with resveratrol, indicating the compound’s stabilizing effect on the protein delays the unfolding of TTR. B) The differences between the wild type and resveratrol stabilized kinetic isotherms were generated to determine an optimal assay window. The optimal GnHCl concentration to readily observe stabilizer effects was in the 4–5 M GnHCl pulse range. C) Aligned (aligned to zero) sensorgram traces for three sample automated runs showing association (GroEL binding to TTR that had been pretreated with or without resveratrol) and dissociation steps after a 4 M GnHCl pulse.
C) Superoxide dismutase 1 (SOD1)
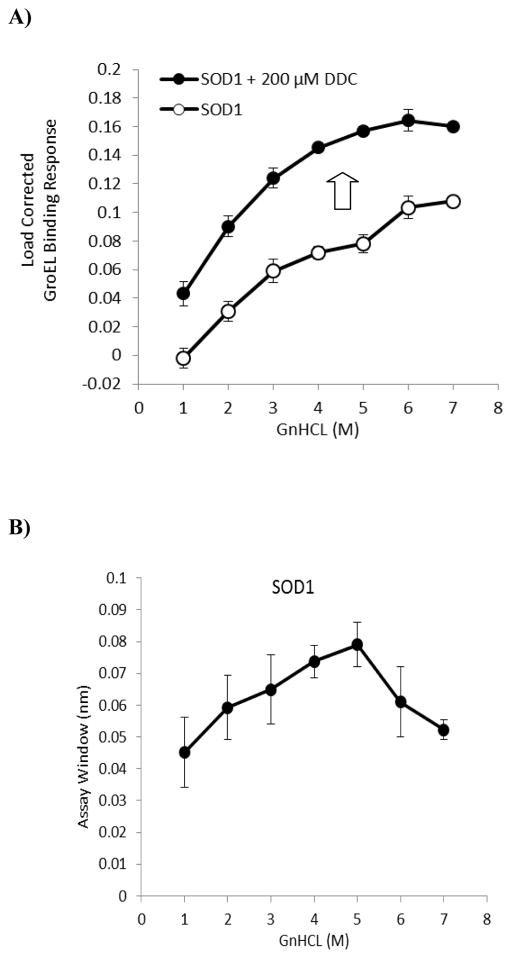
Another oligomeric disease relevant target tested is dimeric human copper-zinc SOD1. Biochemically, superoxide dismutase catalyzes the conversion of superoxide into oxygen and hydrogen peroxide53, thus serving as an antioxidant defense for the cellular components against reactive oxygen species. Structurally, eukaryotic SOD1 is a homodimer with high thermal stability (melting temperature reported to be above 80°C, when fully metallated)54. Each protein subunit contains two metal binding sites, and each subunit is further stabilized by the presence of an intramolecular disulfide bond54. Superoxide dismutase can be stabilized by binding different metal ions, including copper/zinc, manganese or nickel. In humans, there are three types of superoxide dismutases, with SOD1 and SOD3 both containing copper and zinc while SOD2 has manganese in its reactive center. The linkage between over 100 mutations in human SOD1 (hSOD1) and familiar amyotrophic lateral sclerosis (FALS; ALS is also commonly known as Lou Gehrig’s disease)55 was first reported ~20 years ago,56 with A4V and G93A being two most common mutations. Evidence provided by crystal structures of mutant SOD1 suggests a structurally defective form of SOD dimer with limited catalytic activity.56 The ability of SOD1 mutants to form higher order structures, perhaps due to dimer dissociation, that in turn could eventually lead to motor neuron death makes hSOD1 an intensely interesting target to study for our better understanding of the mechanism and pathology of FALS. Specifically, the steps involved in the formation of deleterious SOD1 aggregates may include demetallation, reduction in disulfide bond formation,57 both side reactions that destabilize the SOD dimer.
As the metal cofactors contributed significantly to the kinetic stability of SOD1 (likely through protein restructuring as structural differences exist between the apoprotein and holoprotein),6, 58, 59 we constructed and compared the kinetically controlled denaturation profiles of a His-tagged dimeric SOD1 in absence and presence of a known SOD1 inhibitor, diethyldithiocarbamate (DDC), that chelates metal away from SOD1.60 The addition of 200 μM DDC significantly destabilized SOD1 as seen from a leftward shift and increased GroEL binding amplitudes compared to the native protein (Figure 5A). Predictably, the raw chaperonin binding amplitude for the demetalated form was significantly higher than the untreated SOD1 protein showing that one could observe destabilization due to a loss in ligand/cofactor. The difference between the chaperonin binding with the metal free vs metal bound SOD shows a fairly even difference between the apo-SOD and the Cu2+ bound form throughout the range of GnHCl pulse concentrations used (Figure 5B). Since these denaturant pulse profiles were kinetically controlled, our observations agreed with previous kinetic data that showed an acceleration in unfolding rates for demetalated SOD compared to metalated protein in an acid induced denaturation experiment.6 Cupric ion replenishment to wild type SOD1 pretreated with DDC showed restored stability, similar to wild type SOD1 (Supplemental Figure 3).
Figure 5.
Assessment of the stability of SOD1 and effect of a known metal chelator DDC on SOD1 stability. A) Kinetically controlled denaturation profiles of SOD1 in absence and presence of 200 μM DDC. Experiments were performed on the Octet with 3 replicates for each data point. Error bars represent standard deviation. A higher GroEL binding response was obtained at each GnHCl denaturant concentration when SOD1 was exposed to 200 μM DDC, a known inhibitor of SOD1 to destabilize the protein via metal chelation mechanism (denoted by arrow). B) The differences between the wild type SOD1 and the destabilized Cu2+ chelated apo SOD1 kinetic isotherms were generated to determine an optimal assay window. In this instance, the differences showed a slight maximum in the 4–5 M GnHCl pulse range.
Evaluating the kinetic stability of marginally stable proteins
A) Cystic Fibrosis Transmembrane regulator Nucleotide binding Domain (CFTR-NBD1)
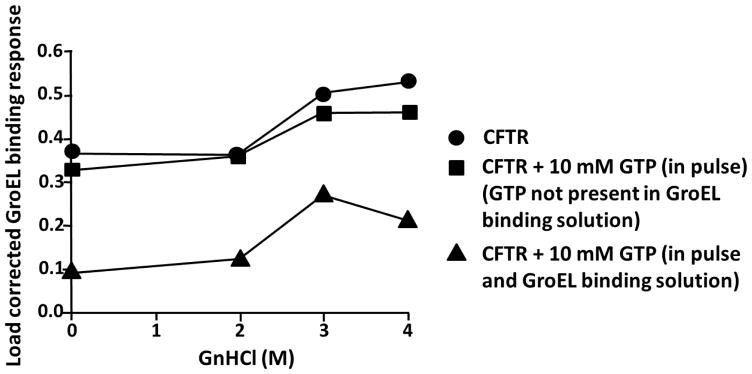
Misfolding and subsequent mistrafficking of CFTR is the source of another prominent folding disease. Lack of functional CFTR due to over 1500 mutations identified to date is the cause of cystic fibrosis, a lethal genetic disease, most commonly found in Caucasians. CFTR contains two nucleotide binding domains (NBDs) and two transmembrane domains (TMD), and one common CFTR deletion mutation ΔF508 occurs on NBD1, adversely affecting post translational folding and transport of the protein to the cell surface61. In solution, the CFTR-NBD162 fragment is stabilized by nucleotide binding by ATP or GTP63. NBD1 has been considered as a surrogate target for the search of small molecule stabilizers for CFTR since the most prominent single site mutation causing cystic fibrosis is located within this domain (Δ F508). This fragment has a low Tm and readily unfolds and aggregates at room temperature which explains why designing assays to search or validate stabilizers of this fragment has been problematic. In contrast to the stable automated loading of protein substrates GST, TTR, and SOD1 (see methods, Figures S2), CFTR-NBD1 protein showed a lack of stable loading with time, most likely due to its temperature liable nature at room temperature. To stabilize this thermally liable protein fragment prior to immobilization on BLI biosensors, the intrinsic ligand ATP (applied at 1 mM, a concentration ~20 times higher than its Kd64) was added to the loading solution. As predicted, (Figure S4A), CFTR-NBD1 exhibited more consistent load amplitudes over time when ATP was present. Once the CFTR biosensor was dipped into buffer without ATP, the stabilizing effect of ATP on NBD1 was removed since ATP binding was relatively weak and ATP dissociation readily occurred in the buffer wash pulse steps. Since the nucleotide free form was aggregation prone, the removal of ATP from the His-SUMO- tagged CFTR-NBD1 fragment was predicted to be able to bind GroEL at room temperature. As shown in Figure 6, GroEL exhibited significant binding to the immobilized CFTR-NBD1 protein fragment on the BLI biosensor at room temperature alone, without the need for a denaturant pulse. Since the nucleotide binding to CFTR has an affinity in the μM range, there was little effect on the binding magnitude when CFTR-NBD1 was pre-incubated with GTP because this ligand was washed away from CFTR during the 1 min wash step prior to the GroEL binding step. The stability of this fragment is particularly liable since ligand induced stability of the CFTR fragment is also lost once GTP rapidly dissociates from the NBD1 fragment65. Since previous experiments have shown that GTP does not interfere with GroEL capture and release66, adding GTP to the GroEL buffer should show a decrease in GroEL binding to the CFTR biosensor. As predicted, a clear reduction in GroEL binding magnitude was observed (Figure 6). This set of data agreed well with our prior observation where a chaperonin bead based setup was utilized, and GTP presence prevented CFTR-NBD1 partitioning to GroEL that was immobilized on sepharose beads66. The ligand response also supports the contention that the Ni-NTA immobilized Sumo-CFTR protein is functional in nucleotide binding.
Figure 6.
Kinetically controlled denaturation of His-SUMO-CFTR with and without pretreatment with its native ligand GTP. The trace highlighted by closed circles represents a kinetically controlled denaturation isotherm of CFTR. In the line containing the closed squares, immobilized CFTR was exposed to 10 mM GTP followed by various concentrations of GnHCl with 10 mM GTP. Both denaturant and GTP were subsequently washed off before dipping into a GroEL solution. In contrast, in the bottom trace highlighted by the filled triangles, all other steps were performed as before the same except the GroEL solution also contained the 10 mM GTP.
Thus it appears that a variation to the denaturant pulse method might be more applicable for protein fragments, particularly if the protein is inherently metastable (i.e. rapidly fluctuates between folded and partially folded forms). In this instance, one can simultaneously test ligands provided that they do not interfere with GroEL capture and release9, 66. One can simply determine if GroEL binding is diminished when the immobilized protein is incubated with GroEL and the test ligand in question. In this instance, one needs to run a secondary assay with another protein system to rule out ligand interference with GroEL binding. There are two options that one could follow. As one example, one could use any other immobilized protein that completely unfolds during a high denaturant pulse as a comparative test run (± test ligand). In another example (used in this paper), the GroEL dependent refolding of the unfolded green fluorescent protein was examined in the absence or presence of the test ligand stabilizer GTP. No differences in folding yields were observed nor does GTP interfere with the intrinsic fluorescence of GFP14.
B) DNA binding core of tumor suppressor p53
Another protein domain fragment that exhibits a low Tm (i.e. significant denaturation at body temperature) and where missense mutations result in disease is found in the DNA binding core domain of the general tumor suppressor protein p5367. As noted in the method development section, the first step in constructing valid and reproducible kinetically controlled denaturation isotherms involves demonstrating that loading of the protein onto BLI Ni-NTA biosensor surfaces remains constant. Unlike the case with the metastable CFTR fragment, there are no easily dissociable ligands that can prevent aggregation of p53 long enough to ensure constant loading onto the biosensor tip. However, one possible method to limit p53 aggregation in order to improve reproducibility of p53 loading onto the biosensor is to stabilize p53 DNA binding domain with an osmolyte stabilizer solution in the loading well. Of the osmolytes tested, glycerol seemed to yield the most uniform loading amplitudes (Figure S3B). Once constant loading of this normally metastable protein was achieved (in loading buffer containing 4 M glycerol), the multiple kinetically controlled denaturation profiles generated by the denaturant pulse showed that this protein begins to kinetically denature at lower denaturant concentrations. When glycerol was added into the denaturant pulse solution (and removed during wash phase), the overlaid GroEL binding amplitudes were dramatically diminished, verifying that glycerol was a good kinetic stabilizer of the p53 DNA binding core domain (Figure 8A).
Figure 8.
Generation of IgG kinetic isothermal denaturation curves and assessing osmolyte stabilization effect: A) Aligned sensorgram traces (raw data) showing GroEL association and dissociation steps after GnHCl pulse. GroEL binding showed a GnHCl concentration dependent response. The higher GnHCl concentration that was used in a previous denaturation step, the larger the GroEL binding response was. GroEL binding signal appeared to approach saturation with 5 M GnHCl denaturation (GnHCl was titrated from 1 to 8 M). B) Aligned sensorgram traces (raw data) showing GroEL association and dissocation steps after urea pulse. GroEL binding also exhibited a urea concentration dependent response. Higher urea concentration led to higher GroEL binding signal. GroEL binding signal kept increasing within the urea concentration range that was titrated (1–8 M). C) Comparative GroEL binding responses as a function of GuHCl (black filled circles) or urea denaturant concentration (open circles). D) Aligned sensorgram traces (raw data) showing kinetic stabilization of IgG by different osmolytes/excipients.
The tumor suppressor p53 DNA binding core domain has been the focus of a large number of stability studies because many cancer outcomes are linked to missense mutants of this protein that misfold67. Thus, the search for small molecules and peptides that stabilize this protein through specific binding mechanisms is an attractive approach to reverse misfolding and restore normal p53 function68. One such molecule that binds and stabilizes the core domain (through the DNA binding surface) is a peptide that has been rationally designed from the p53 core domain - 53BP2 complex interface resolved from the x-ray crystallographic structures by Fresht and colleagues69. They found that the binding of the CDB3 peptide stabilizes the p53 core domain, induces refolding of equilibrium denatured p53 and restores sequence-specific DNA binding to I95T, a highly destabilized p53 mutant. In addition, CDB3 peptide binding to wild type and a mutant p53 resulted in thermal stabilization as assessed by differential scanning calorimetry measurements.
To examine the effect of this stabilization on the denaturant pulse p53 kinetic unfolding transition, the CDB3 peptide was added at a concentration of 200 μM prior to and during the denaturant pulse phase. Although the binding affinity diminishes in the presence of increasing denaturant,69 it is predicted that the binding of the peptide to p53 on the BLI biosensor should slow the kinetics of unfolding of p53 and significantly diminish the GroEL binding, consistent with the stabilization of p53 by this peptide. Here, the peptide binding experiments were done on BLItz, a system that is insensitive to small molecule binding interactions (> 10 kDa sensitivity). The low GroEL binding response that is observed when the stabilizing peptide is present in the denaturant pulse step remains the same regardless of the time of the wash pulse (no peptide) (Figure 8B). These results illustrate that the CDB3 peptide binding appears to stabilize the p53 core domain and diminish general chaperonin binding. As with the weak binding of GTP nucleotide to the CFTR NBD1 binding domain, in these instances there is not a dramatic rightward shift toward higher denaturant concentrations with weaker binding ligands. Again, these experiments illustrate the utility for using this method to identify stabilizers that do not remain tightly bound to the protein after the denaturant pulse is completed (in this instance 5 min).
Assessing the kinetic stability of multiple domain proteins
A) Generating a kinetic controlled denaturation isotherm using an immobilized IgG as a model probe
It has been repeatedly demonstrated that partially folded proteins as large as antibodies easily bind to GroEL70, 71. It is known that IgG shows more complex unfolding by equilibrium monitored methods72. To determine a kinetic isothermal denaturation curve for the multi-domain IgG molecule, a set of hydrated amine coupling human IgG coupled fully functional biosensor tips (supplied by forteBIO) were simultaneously exposed to increasing concentrations of either urea or GnHCl during a 5-min denaturant pulse. The tips were then removed from these denaturant conditions and dipped into a buffer solution (1 min) to remove residual denaturant. The washed tips were then dipped into a GroEL solution (1 μM tetradecamer), and binding of GroEL to partially folded IgG population produced a signal correlating with the amount of partially folded protein present on the tip surface at the time of assay. The kinetically controlled denaturation isotherms generated for IgG show a progressive rise in the GroEL binding signal with increasing denaturant concentration. Predictably, the individual GroEL binding signals following the urea induced unfolding lagged the stronger GnHCl kinetic denaturant pulse profile (Figure 8A for GnHCl and Figure 8B for urea) but both approached nearly the same maximum GroEL binding amplitude signal at high denaturant pulse concentrations (Figure 8C).
To demonstrate that one could rapidly identify potential solution stabilizers which are predicted to slow IgG unfolding, a series of known antibody excipient stabilizers such as polyols (mannitol, sucrose, glycerol and trehalose) or amino acid osmolytes (proline) were then mixed with the 4 M urea solutions (final urea concentration) using the multi-well Octet system, and the denaturant pulse was again implemented. In all instances, GroEL binding to the tip following the brief buffer wash (removes osmolyte and denaturant) was significantly reduced (Figure 8D) within the time frame of the 5 min denaturant pulse, indicating that, as predicted, the excipient/stabilizer conditions slowed the unfolding reaction of the attached IgG. Curiously, these experiments showed that the immobilized antibody did not completely refold to its original native conformation even after the 1 min wash, reflected by the observation a small amount of GroEL binding was observed. Since the stabilizing excipients were also washed and diluted away with the buffer exchange and the GroEL incubation steps (wash and binding volumes 200 μl see scheme in Figure 1, Steps 2 and 3), osmolyte interference with GroEL binding would not be possible.
B) vWF kinetically controlled denaturation profile comparison to equilibrium denaturation
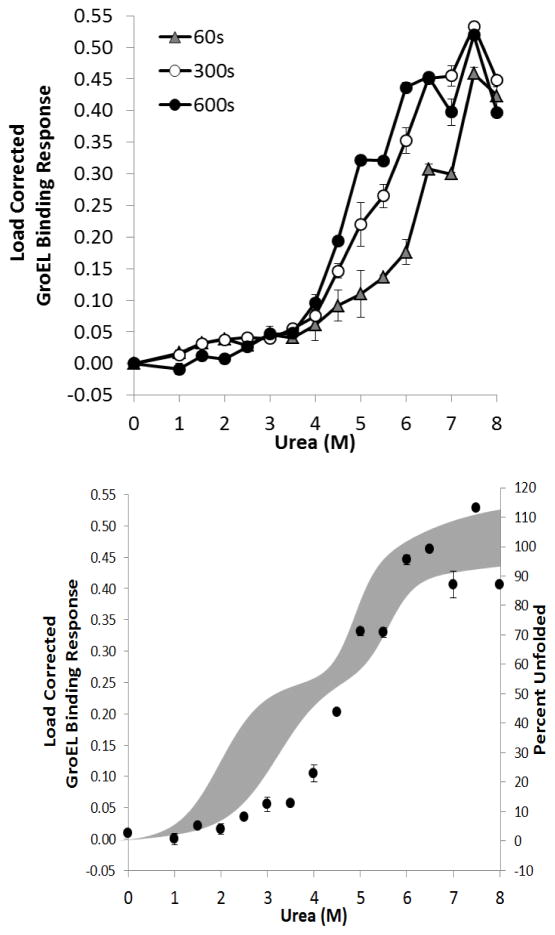
von Willebrand disease is a bleeding disorder affecting roughly 1% of the world population. This hereditary disease is caused by a deficiency or mutation in the von Willebrand Factor (vWF), a plasma glycoprotein multimer which initiates platelet adhesion at sites of vascular injury. Under high shear stress, vWF unravels and binds to platelets and collagen to create a plug to stop bleeding. The recombinant multimeric VWF has three A domains connected by flexible linkers. The A3 domain binds to collagen exposed during vascular injury, the A2 domain contains a proteolytic site which helps to regulate clot size, and the A1 domain binds to platelets via the GP1Bα receptor. We assessed the kinetic stability of the vWF WT triple domain using GroEL-BLI denaturation pulse procedures. In this instance, the A3 domain contains a His-tag that can be attached to the Ni-NTA biosensor surface and mimics the attachment that naturally occurs with this triple domain protein. This same attachment scheme was used to attach the His-tagged A3 domain onto Cu2+ coated surfaces for rheodynamic analysis for platelet adherence and pause time measurements24. The GroEL binding signal, plotted as a function of the increasing denaturant pulse concentration, increased with increasing denaturant concentration and resulted in a clear kinetic stability isotherm (Figure 9A). In comparing the kinetic denaturant pulse with the equilibrium denaturation profiles, it appeared that kinetic signals related to A1 and A2 domains did not show stable kinetic unfolding detected by GroEL after a 10 min denaturant pulse followed by the brief (10 sec) wash. Curiously, at high denaturant concentrations (~5 M urea), the equilibrium denaturation profiles showed unfolding transitions first followed by what appears to be domain A3 unfolding (Figure 9B). As the denaturation pulse times increase, a progressive increase in GroEL binding was observed and at longer denaturant pulse incubation times (10 min), the kinetically controlled denaturation isotherm approached the previously determined circular dichroism (CD) derived equilibrium isotherm equilibrium overlay at the higher denaturant concentrations32.
Figure 9.
A) Kinetic stability isotherm of von Willebrand factor triple A domain generated with BLI denaturant pulse assay:time dependent left shift of load corrected GroEL binding response as a function of denaturant concentration. B) Comparison of the kinetically controlled denaturation profile generated under10 minute kinetic denaturant pulse (black circles) to equilibrium denaturation profile with 95% confidence interval (grey shaded region).
C) Tetanus neurotoxin (TeNT) kinetic denaturant pulse
TeNT is a bacterial protein toxin produced by Clostridium tetani which induces spastic paralysis in mammals following receptor mediated endocytosis and subsequent escape from endosomes. To accomplish this end, the middle domain of the toxin undergoes acid-induced refolding and membrane insertion to translocate the enzymatically active portion of the toxin into the cell cytosol. The intrinsic response of the toxin to acidic conditions at the membrane interface is a crucial pre-transition that eventually allows for membrane insertion and transport of the enzymatic portion of the toxin into the cell. Predictably, this protein aggregates in solution when it is subjected to denaturing conditions, and it has been difficult to produce reproducible cooperative transitions to evaluate the thermodynamic parameters of this protein during global unfolding. In addition, this protein is somewhat metastable even at low temperature because it begins to aggregate at low temperatures (37°C) further complicating one’s ability to study the mechanisms of membrane insertion. Thus it is of interest to determine if this aggregation prone protein can generate kinetically controlled denaturation transitions while immobilized on a BLI biosensor.
To evaluate the kinetic stability of the soluble TeNT, the protein was immobilized onto Ni-NTA BLI biosensors through the N-terminal His-tag. In addition to the standard denaturant pulse, the kinetically controlled denaturation assay was run in the presence of GT1b to assess if this natural TeNT ligand would stabilize the toxin. GT1b is a ganglioside, a glycolipid enriched in neuronal membranes, which binds two distinct sites on the receptor binding domain of TeNT which recognizes either the conserved carbohydrate backbone of gangliosides or the terminally linked sialic acid residues73. The concentration used for the experiments was above the Kds (nanomolar range) for the receptor binding sites but below the critical micelle concentration (CMC at ~30 μM) for the ganglioside74. Previous observations with immobilized anthrax toxin protective antigen pre-pore indicate that the large conformational transitions to the membrane insertion competent pore generated by physiological temperatures (37°C) coupled with 1 M urea incubations70 could be easily observed using BLI technologies31. Therefore, it is conceivable that the soluble TeNT toxin may undergo relevant pre-insertion transitions from soluble to membrane inserted species under low denaturant conditions. Understandably, this kind of transition is predicted to expose hydrophobic patches that can bind the promiscuous chaperonin, GroEL.
In Figure 10, the kinetic denaturant pulse isotherms with and without GT1b overlapped and showed a large denaturation event (increased GroEL binding) occurring at the 3 and 4 M urea pulse concentrations. No recognizable transitions occurred during the low denaturant pulse conditions although there was a small noticeable rise in progressive chaperonin binding from 0 to 3 M urea. Additionally and perhaps importantly, there was a clear binding of GroEL to the immobilized TeNT under the zero denaturant condition indicating that this protein is inherently metastable. The transition that was observed was evidenced by the signal increasing from roughly 30% to 70% of maximal signal. The kinetic isotherms of the attached toxin show clear reproducible transitions in the absence and presence of GT1b. Although a rightward shift was expected if a strong stabilizer was present, this was not readily observed at the concentration of GT1b tested. Instead, the addition of the presumed stabilizer led to a consistently reproducible (3 runs overlaid) yet small decrease in the binding amplitude. This decline in GroEL binding amplitude was not as dramatic as in the case with p53 or CFTR NBD1. This indicated the presence of GT1b at this concentration did not confer a substantial delay or decrease in the GroEL binding amplitude during the denaturant pulse of TeNT immobilized through the His-tagged catalytic domain.
Figure 10.
Clostridia tetani neurotoxin (TeNT) denaturation profile. His-tagged TeNT (through light chain catalytic domain) (at 0.2 μM concentration for 1 nm loading deflection) was attached onto Ni-NTA tips, and a denaturant pulse procedure was performed. The kinetic isotherms were generated in the absence (black circles) and presence (gray circles) of 1 μM GT1b. Control sensorgrams (no GroEL) exhibiting no contributions from GT1b dissociation were observed over the time course of the GroEL binding phase. A small amount of GroEL binding to the immobilized TeNT was detected under non-denaturing conditions (0 M urea).
DISCUSSION
This work describes the successful development of a broad-based method to assess protein kinetic stability of inherently stable, metastable, and aggregation prone proteins using a chemical denaturation pulse method coupled with a biosensor surface-based chaperonin detection system. The automated platform facilitates the acquisition of highly-reproducible chaperonin-dependent detection of partially folded protein populations within physiological temperature ranges. This platform can rapidly evaluate protein stability of minute protein samples immobilized on biosensor surfaces. Detection of partially destabilized proteins is accomplished with a high-affinity nucleotide-free chaperonin that promiscuously binds to any protein that exposes hydrophobic patches. Most importantly, this platform technology can identify stabilizers of the native fold that diminishes the formation of hydrophobic partially unfolded populations prior to aggregation. The assay principle is simple as it is based on and is an extension of previous solution based methods where a reversal of misfolded/partially folded states by global ligand stabilizers have been demonstrated to decrease chaperonin binding19, 21. The method outlined herein accelerates the detection process because chaperonin binding to the target protein immobilized on the BLI biosensor predictably decreases in the presence of protein stabilizers. This is a particularly useful approach for identifying and validating correctors/stabilizers of difficult, aggregation prone misfolding disease proteins.
There are currently many different protein stability assays that have been developed to identify ligand enhanced stability. The most commonly used method involves thermally perturbing a protein in solution and monitoring the progressive exposure of hydrophobic patches with fluorescence dye binding using differential fluorescence scanning methods. This approach is also a kinetic stability measurement. Though popular, there are a number of protein classes that have proven difficult to be evaluated by this method. These include oligomeric proteins, thermally stable proteins, aggregation-prone proteins and most importantly, metastable proteins that exist in both folded and partially folded forms9. This latter class includes many missense proteins where misfolding results in the manifestation of human disease.
Previous work from this laboratory utilized a reverse approach where the chaperonin is directly coupled onto the biosensor to detect and capture transient preaggregate species present within concentrated therapeutic protein solutions38. These transient preaggregates exist in low concentrations prior to the formation of larger dimeric or other subsequently formed aggregate populations and are thought to represent the “seeds of destruction” that degrade therapeutic protein formulations. Currently, protein formulation integrity is frequently evaluated by particle size dependent methods where small or large aggregate populations are detected after they form. One drawback of using the approach where GroEL is immobilized on BLI biosensors occurs when significant protein aggregation rapidly forms or is present before the chaperonin biosensor is dipped into the protein solution. In particular, extensive aggregation prior to probing the solution with the biosensor diminishes the biosensor signal and correlates with decreases in chaperonin biosensor capture of large aggregates. The simple fix to avoiding large-scale solution-based aggregation interferences is to immobilize the target protein to the BLI biosensor surfaces and evaluate the presence of partially folded populations with chaperonin binding from solution with or without the denaturant pulse procedure.
The BLI chaperonin denaturant pulse system uses rationally designed affinity tags that immobilize and orient the target protein or target protein domain away from the biosensor surface to augment chaperonin binding while avoiding protein aggregation. Properly loaded proteins that are homogenous and specifically oriented present adequate populations of immobilized proteins on the BLI biosensor tip surface that minimize or abolish intra-surface protein aggregation prior to GroEL binding. In this work, the robust, broad based applicability of the BLI chaperonin-detection platform was tested with a host of monomeric, dimeric, multi-domain and metastable proteins rather than focusing on just one class of target protein.
The most common linkage for immobilization of the target proteins used in this approach is through engineered His-tags that can be immobilized onto Ni-NTA biosensor tips. The denaturant pulse approach can be used to perturb the folded protein without diminishing Ni chelate binding affinities. A simple PubMed search indicates that this is a common methodology for capturing and sometimes refolding proteins on Ni-NTA surfaces from inclusion bodies (119 hits since 1996). For the BLI approach, this attachment is also advantageous because one can regenerate the biosensor tip Ni surface and automate the regeneration process to obtain precise replicates with highly reproducible signal responses. The denaturant pulse chaperonin BLI assay is routinely accomplished with small sample sizes (often μmolar to sub μmolar quantities) provided that the amount of loaded target protein onto the 600 micron tip remains constant. Importantly, very little non-specific binding of our highly purified chaperonin preparations is observed with the Ni-NTA biosensors. To avoid non-specific binding, optimal chaperonin purity and tetradecameric stability are absolutely necessary for this assay. For instance, we observed that impure or structurally defective chaperonin samples containing protein impurities (bound peptides) or monomeric GroEL subunits showed significant non-specific background binding for a wide variety of BLI biosensor surfaces. Optimized sample loading also decreases the relatively small non-specific binding of GroEL (~ 0.1 nm) to undetectable signal levels. When the target protein loading is maintained at the lower portion of the linear response of the biosensor signal (Supplemental Figure 1), considering the size of the GroEL binding site (~ 45 Å), the assumption that GroEL will bind a single immobilized polypeptide for partially folded protein domains is valid. The GroEL signal can be corrected for slight differences in loading of the target protein (load corrected GroEL binding response).
To obtain reproducible data using the automated platforms, the timing of the loading, denaturant pulse, wash times to replace buffers and immersion into the GroEL binding buffers are precisely controlled. For example, the nm amplitude shifts that result from protein loading and GroEL binding reaction are reliably reproduced with timed data collection. Within the linear response region, the GroEL binding amplitudes are directly proportional to protein loading, and are further corrected by protein loading (i.e. GroEL nm amplitude/protein load nm amplitude). Additionally, binding the high affinity GroEL chaperonin to partially folded populations arrests refolding provided that the Kd of GroEL is in the nanomolar range or below (reviewed in Fenton and Horwich, 1997).18 As the denaturant concentration in the denaturant pulse reaction increases, the highest GroEL binding signals occur when the entire population of the attached target protein is completely denatured. Since this is a pulsatile denaturant exposure rather than an equilibrium process, this method can only be used to assess the kinetic stability of proteins.
The success of this approach depends on a number of crucial parameters that result in reproducible kinetically controlled denaturation isotherms. One element that must be maintained, particularly as the biosensor surfaces are regenerated multiple times, is that the loading of the target protein (reported as nm shift amplitudes) remains constant. The channel to channel reproducibility data presented were very good (signal variation of a ~ 3% standard deviation (S.D.) over 9 separate regeneration cycles - see Figure S2) for stable proteins. Instances of poor reproducibility were only observed if the target protein sample degraded (e.g. aggregated) over time in the sample loading wells. There are multiple solutions to this problem. In the most extreme cases (protein is only stable for a short period of time and is unable to be stabilized), fresh biosensor tips along with freshly prepared protein samples were used, forgoing regeneration. Alternatively, the most common successful method to stabilize the target protein in the loading wells is to include polyol osmolytes (glycerol or sucrose) to avoid or slow sample aggregation/misfolding (see Figure S4 panel B for p53 loading). It is noteworthy that the use of folding osmolyte solutions often prevents misfolding and aggregation during long term storage and has often been the additive of choice to the prevent misfolding of metastable proteins (both soluble and membrane bound) involved in various folding diseases75–79. The ability to rapidly detect the rescue of incorrect folding and stabilization of metastable proteins by folding osmolytes with the chaperonin-BLI denaturant pulse system is a useful prerequisite in identifying and evaluating mutants for disease therapeutic development. The demonstration that osmolytes can rescue misfolding is an important primary assay where positive reversal of misfolding of the target protein makes it a candidate for reversing misfolding through small molecule stabilizer approaches. Hence, the ability to evaluate metastable proteins and even protein fragments (e.g. mimicking metastable folds that appear during protein translation such as CFTR-NBD1 fragment) with this method while avoiding aggregation significantly broadens the applicability of this method.
There are some limits to the denaturant pulse BLI chaperonin biosensor method. For instance, the evaluation of the entire global stability of multi-domain proteins is diminished because the orientation of particular domains at the biosensor surface restricts the interaction of GroEL with the unfolding protein. For example, in this work, the p53 DNA binding domain was immobilized to the BLI biosensor surface through a His-tagged MBP. While this orientation results in preferred interactions of the exposed p53 domain with the chaperonin, the stability of the attached MBP domain is refractory to GroEL binding (i.e. no effect on p53 stability when the MBP is stabilized by maltose binding). The immobilization of proteins with three domains such as large antibodies and the tetani neurotoxin only enables us to evaluate those domains that are orientated in a way that are accessible to GroEL binding. In both examples, the GroEL binding denaturant pulse approach yielded single kinetic transitions. Interestingly and unexpectedly however, the multi-domain protein vWF A1-A2-A3 that is attached to the Ni-NTA surface through a His tag on the A3 domain shows a kinetic transition profile that is more complex, complete with a transition that correlates with the unfolding of the A3 domain (Figure 9). It is possible that flexible linkers between all three of these integrin-like domains may allow for interactions of domain A1 with domain A3 even while the protein is immobilized. Transitions that associate with the A1 and A2 domain unfolding do not dominate the initial kinetic isotherms even during longer denaturant pulse times. Compared with the equilibrium denaturation isotherm, the small amount of chaperonin binding at low denaturant concentrations may result from either slow unfolding kinetics (perhaps due to domain-domain interactions within the A1-A2-A3 linkages) or fast refolding of the extended A1 domain. This can be determined by extending the denaturant pulse times beyond those used in the present experiments (Figure 9). If the domains that interact with GroEL rapidly refold during the short wash steps (10–30s), one would not observe any progressive increases in GroEL binding signal as a function of time of denaturation under lower denaturant conditions.
Provided that the immobilized target protein domains are accessible to the GroEL binding site, this type of analysis (i.e. varying denaturation times) is useful for evaluating potential kinetic stability changes for mutant proteins. Missense or single amino acid changes that globally destabilize the target protein are predicted to result in higher initial GroEL binding following exposure to lower denaturant pulse concentrations. In fact, the two metastable proteins, CFTR NBD1 fragment and p53 DNA binding domain, show reproducible GroEL binding in the absence of a denaturant pulse at 25°C (Figures 6 & 7 at zero denaturant). As expected, GroEL binding under zero denaturant conditions increases if the BLI probe solution temperatures are elevated to 37°C (data not shown).
Figure 7.
Kinetic stabilization effect on p53 by osmolyte or CDB3 on the kinetically controlled denaturation A) Wild type p53 was attached to BLI biosensors and subjected to a 5 min denaturation pulse. When 4 M glycerol was included in the denaturant pulse phase, the GroEL binding signal to the p53 loaded tip was significantly reduced. Glycerol was washed away prior to performing the GroEL binding phase. The data was load corrected to account for slight variations in p53 loading (> 4% standard deviation). B) CDB3 peptide at a final concentration of 200 μM was added both at an initial wash step after p53 was loaded onto the BLI biosensor and in the following denaturant pulse step. After the biosensor was removed from the denaturant solution and dipped into the loading buffer (to wash away denaturant and excess CDB3 peptide) for a 5 or 20-sec wash step, the biosensor was then immersed into the GroEL buffer containing 0.15 μM GroEL.
The kinetic transition of the aggregation prone TeNT is a perfect example where this method is useful in evaluating toxin stabilization conditions, potentially leading to the development and validation of small molecules that prevent toxin dependent transitions (novel anti-toxins) prior to cell insertion and toxin translocation. Curiously, with the TeNT toxin, a small amount of specific chaperonin binding to the attached toxin is observed prior to implementation of the denaturant pulse assay in the absence and presence of low concentrations of GT1b (Figure 10). Since this toxin slowly aggregates at room temperature without activation, the chaperonin apparently binds to this protein even before any extensive denaturation occurs (Figure 10). Previous differential scanning calorimetry studies indicate that there are two distinct transitions that can be observed during thermal denaturation but these transitions were not reversible80.
In solution, the TeNT toxin will slowly partition onto the chaperonin under physiological temperature conditions and distinct chaperonin (800 kDa)-TeNT toxin (150 kDa) complexes can be readily observed using electron microscopy (image data will be presented in a later publication). Loading biosensor tips at lower temperatures (< 20°C) diminishes the slow aggregation reaction and results in constant reproducible loading amplitudes within the time frame of the multiple run experiments. Likewise, polyol osmolytes such as glycerol or sucrose also prevent aggregation of TeNT or other toxins such as the anthrax protective antigen even under low pH environments that drive unfolding/refolding transitions of these toxins to membrane insertable forms. The TeNT toxin is immobilized onto Ni-NTA tips through the N terminal light chain domain (active protease) that orientates the translocation domain (heavy chain) and the GT1b receptor binding domain toward interacting with GroEL in solution. At this time, it is impossible to determine which of these latter two domains GroEL binds. Since the interaction of partially folded or hydrophobic proteins with GroEL is promiscuous, it is possible that both of these domains may be interacting with GroEL.
The versatility of this platform technology will help researchers determine how ligand interactions influence the kinetic stability of target proteins while avoiding deleterious aggregation side reactions. This approach is of considerable interest to the protein stability field. For example, the presence of tight binding ligands will both delay and even diminish the chaperonin binding after the denaturant pulse phase (Figure 1). Including high concentrations of weak binding ligands in the denaturant pulse phase appears to slow the general protein unfolding reaction as evidenced by the diminished chaperonin binding or shifted kinetic isotherms. This effect is readily apparent when folding osmolytes are included in the denaturant pulse steps (Figure 2 & 8). If the chaperonin binds to the partially folded mutant protein in the absence of a denaturant pulse (e.g. CFTR-NBD1 with low Tm), one can directly incubate protein with stabilizer candidates (before and during GroEL binding step) and evaluate the chaperonin binding directly provided that the added ligand does not interfere with chaperonin binding interaction (Figure 6).
Future developments for this methodology will expand ligand discovery platforms. In particular, the sensitivity of this automated BLI systems will allow one to perform ligand titrations prior to the stabilization assessment by the denaturant pulse. This approach can dramatically accelerate the throughput of protein-ligand binding/stabilization evaluations. With the current available Octet system, 16 biosensors can operate simultaneously to process 384 samples with kinetic characterization in a few hours. A commonly evaluated factor in high throughput screen is Z′ factor, a dimensionless parameter to reflect assay window and data variation81. In our preliminary study using TTR as an example, based on the assay window established at 4 M GnHCl denaturation (Figure 4B), a satisfactory Z′ of above 0.5 was estimated for a sample size of 3. In another example using SOD1, a Z′ of 0.73 was achieved using the assay window obtained at 4 M GnHCl (Figure 5B). Since one can adjust the loading of target protein and the denaturant pulse times, this signal separation between fully stabilized protein (osmolyte stabilizer baseline e.g. Figure 7A) and the maximum chaperonin binding signal can be further optimized. Thus, it is further expected that this approach is amenable with robotic integration and high throughput screening. Using a newer state of the art robotic 96 pin biosensor Octet HDX 384 systems with the described a two minute chaperonin binding window (enough time of single point measurement in assay window with an optimal Z factor) can allow one to process between 1,500 to 3,000 samples over an 8 hour time period. If larger protein amounts are available along with larger ligand libraries, it may be useful to implement aggregation based prescreens to pare down in silico candidates prior to performing actual titration and stabilization assessments. It is important to note that small molecule prevention of large scale aggregation may not lead to stabilization of the native fold82, 83. Once a select set of test compounds are identified that prevent large scale aggregation, the hit compounds can be evaluated to determine direct binding and stabilization of the target protein with using single platform denaturant-pulse chaperonin BLI biosensors. This type of analysis could sort these compounds into different categories based on their binding affinity and kinetics, such as strong binders and weak binders (slow kon and koff vs. fast kon and koff). Subsequently, the denaturant pulse conditions (time of denaturant pulse or including the candidate compound in GroEL solution) and wash times could be varied to classify strong and weak kinetic stabilizers. Weaker affinity ligands could be further developed through rational chemical modification approaches to develop stronger ligand interactions, particularly if the protein binding site is known. Performing small molecule titrations followed by protein stabilization assessments on the same biosensor surface will significantly accelerate small molecule drug discovery pipelines.
Finally, as screening procedures have been expanded to examine large chemical libraries, there are numerous instances where ligand binding results in a destabilization of the target protein rather than stabilization84. This was easily shown to be the case in our current work when the superoxide dismutase dimer was destabilized after Cu2+ was removed from the protein (Figure 5A). Using the target protein titration and chaperonin detected stabilization approaches with this single platform can help determine if ligand binding events lead to protein stabilization or destabilization under physiological conditions. The latter phenomenon will be valuable for designing specific drugs that target metastable oncogenic proteins to destabilize target proteins, which may in turn accelerate clearance rates in vivo.
It is of interest to note that this method can be used to examine instances where kinetic stability of a protein becomes substantial. For example, the kinetic stability of small protein samples can be examined in the presence of increasingly stronger denaturants (urea to GnHCl to GnSCN) or tight binding ligands (e.g. increased protein stability due to binding of a reduced heme in some cytochrome proteins85. For extremely stable proteins, one has the unique ability of transferring the biosensor with the attached protein from the BLI instrumentation to incubate the biosensor tips in solutions for long term denaturation and reevaluating the chaperonin binding detection at later time intervals. This latter approach is relevant provided that the protein linkage to the biosensor remains stable.
In this work, we have primarily focused on examining the GroEL binding amplitudes at a specific time point to generate the kinetically controlled denaturation isotherms. Due to the complexity of unfolding at various denaturant concentrations, coupled with the heterogeneity in protein folding intermediates that are observed to bind to GroEL86, the kinetics of GroEL binding and dissociation to the species generated at lower GnHCl concentrations (smaller amplitudes) may be different from the kinetics of GroEL binding to the species that was more substantially denatured (larger endpoint amplitudes). This may indicate that the species that are being recognized by GroEL after the denaturant is washed away may be intrinsically different for low vs high denaturant pulse exposures. Thus, there is the real possibility that the initial species generated after low denaturant concentration pulses may certainly be exposing slightly different (i.e. smaller) hydrophobic surfaces compared with those that are generated after higher denaturant pulses. Indeed, preliminary evaluation of the binding and dissociation kinetics of GroEL binding (GroEL binding is not rate limiting) for the multi-domain protein vWF show that the kinetic traces have to be fit to different kinetic models when the protein is exposed to low vs high denaturant concentrations. This latter situation may be more common for multidomain proteins that exhibit non-two state global unfolding behavior. It is clear however; that this method should be able to uncover kinetic stability differences (in the form of different kinetically controlled denaturation isotherms) of mutant proteins compared their wild type versions, particularly those that are result in misfolding diseases.
In summary, the chaperonin based BLI denaturant pulse procedure is a highly versatile platform that can readily assess and validate ligand stabilization or destabilization for stable monomers, oligomers and aggregation-prone metastable proteins. Ultimately this approach has the capability of identifying compounds that can reverse or even enhance partial folding or misfolding under near physiological conditions. It is useful to evaluate the efficacy of small molecule stabilizers under physiological conditions, especially for missense protein folding mutants because one may be able to avoid excessive interactions with or partitioning onto chaperone proteins. Alternatively, increased chaperone interactions with mutants that bind specific small molecule destabilizers may result in rapid clearance. In both instances, discovering and validating specific small molecule interactors with target proteins will enhance our ability to modulate the lifetime of these folded mutants in vivo, which in turn, may have profound effects on folding disease outcomes.
Supplementary Material
Acknowledgments
This work was supported by grant funds from NIH R01AI090085 (MTF), a Proof of Concept Grant Kansas University Innovation Center grant (MTF), KBOR State of Kansas EPSCoR funds (MTF and JK), the J.R. and Inez Jay fund (JK), the Institutional Development Award (IDeA) grant NIH P20GM103418 from the National Institute of General Medical Sciences (JK) and NIH Grant HL109109 from National Heart Lung and Blood Institute (MA).
ABBREVIATIONS
- BLI
Biolayer interferometry
- CDB3
Binding Loop p53 binding peptide (NH2-REDEDEIEW-COOH)
- CFTR-NBD1
Cystic Fibrosis transmembrane regulator- nucleotide binding domain 1
- EA
Ethacrynic acid
- MalZ
Maltodextrin glucosidase
- GnHCl
Guanidine hydrochloride
- GST
Glutathione-S-transferase
- GSH
Glutathione
- GT1b
Trisialoganglioside-GT1b
- SOD1
Superoxide dismutase
- TeNT
Tetanus neurotoxin
- TTR
Transthyretin
- vWF
von Willebrand Factor
References
- 1.Johnson CM. Differential scanning calorimetry as a tool for protein folding and stability. Archives of biochemistry and biophysics. 2013;531:100–109. doi: 10.1016/j.abb.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Kopec J, Schneider G. Comparison of fluorescence and light scattering based methods to assess formation and stability of protein–protein complexes. Journal of structural biology. 2011;175:216–223. doi: 10.1016/j.jsb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature protocols. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 4.Lea WA, Simeonov A. Differential scanning fluorometry signatures as indicators of enzyme inhibitor mode of action: case study of glutathione S-transferase. PloS one. 2012;7:e36219. doi: 10.1371/journal.pone.0036219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ, Wasney GA, Yeung R, Arrowsmith C, Ball LJ, Berglund H. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proceedings of the National Academy of Sciences. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch SM, Boswell SA, Colon W. Kinetic stability of Cu/Zn superoxide dismutase is dependent on its metal ligands: implications for ALS. Biochemistry. 2004;43:16525–16531. doi: 10.1021/bi048831v. [DOI] [PubMed] [Google Scholar]
- 7.Matulis D, Kranz JK, Salemme FR, Todd MJ. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry. 2005;44:5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- 8.Auld DS, Lovell S, Thorne N, Lea WA, Maloney DJ, Shen M, Rai G, Battaile KP, Thomas CJ, Simeonov A. Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124. Proceedings of the National Academy of Sciences. 2010;107:4878–4883. doi: 10.1073/pnas.0909141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Correia AR, Naik S, Fisher MT, Gomes CM. Probing the kinetic stabilities of Friedreich’s ataxia clinical variants using a solid phase GroEL chaperonin capture platform. Biomolecules. 2014;4:956–979. doi: 10.3390/biom4040956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelhard M, Evans PA. Kinetics of interaction of partially folded proteins with a hydrophobic dye: evidence that molten globule character is maximal in early folding intermediates. Protein Science. 1995;4:1553–1562. doi: 10.1002/pro.5560040813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freire E, Schön A, Hutchins BM, Brown RK. Chemical denaturation as a tool in the formulation optimization of biologics. Drug discovery today. 2013;18:1007–1013. doi: 10.1016/j.drudis.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schön A, Brown RK, Hutchins BM, Freire E. Ligand binding analysis and screening by chemical denaturation shift. Analytical biochemistry. 2013;443:52–57. doi: 10.1016/j.ab.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown R, Freire E, Hutchins B. Optimizing Biologics Stability Testing 2013 [Google Scholar]
- 14.Naik S, Zhang N, Gao P, Fisher MT. On the design of broad based screening assays to identify potential pharmacological chaperones of protein misfolding diseases. Current topics in medicinal chemistry. 2012;12:2504–2522. doi: 10.2174/1568026611212220006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulawa CE, Connelly S, Devit M, Wang L, Weigel C, Fleming JA, Packman J, Powers ET, Wiseman RL, Foss TR, Wilson IA, Kelly JW, Labaudiniere R. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson SM, Wiseman RL, Sekijima Y, Green NS, Adamski-Werner SL, Kelly JW. Native state kinetic stabilization as a strategy to ameliorate protein misfolding diseases: a focus on the transthyretin amyloidoses. Accounts of chemical research. 2005;38:911–921. doi: 10.1021/ar020073i. [DOI] [PubMed] [Google Scholar]
- 17.Saibil HR, Fenton WA, Clare DK, Horwich AL. Structure and allostery of the chaperonin GroEL. Journal of molecular biology. 2013;425:1476–1487. doi: 10.1016/j.jmb.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 18.Fenton WA, Horwich AL. GroEL-mediated protein folding. Protein Science. 1997;6:743–760. doi: 10.1002/pro.5560060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viitanen PV, Donaldson GK, Lorimer GH, Lubben TH, Gatenby AA. Complex interactions between the chaperonin 60 molecular chaperone and dihydrofolate reductase. Biochemistry. 1991;30:9716–9723. doi: 10.1021/bi00104a021. [DOI] [PubMed] [Google Scholar]
- 20.Smith KE, Fisher MT. Interactions between the GroE chaperonins and rhodanese. Multiple intermediates and release and rebinding. The Journal of biological chemistry. 1995;270:21517–21523. doi: 10.1074/jbc.270.37.21517. [DOI] [PubMed] [Google Scholar]
- 21.Smith KE, Voziyan PA, Fisher MT. Partitioning of rhodanese onto GroEL. Chaperonin binds a reversibly oxidized form derived from the native protein. The Journal of biological chemistry. 1998;273:28677–28681. doi: 10.1074/jbc.273.44.28677. [DOI] [PubMed] [Google Scholar]
- 22.Tieman BC, Johnston MF, Fisher MT. A comparison of the GroE chaperonin requirements for sequentially and structurally homologous malate dehydrogenases: the importance of folding kinetics and solution environment. The Journal of biological chemistry. 2001;276:44541–44550. doi: 10.1074/jbc.M106693200. [DOI] [PubMed] [Google Scholar]
- 23.Paul S, Singh C, Mishra S, Chaudhuri TK. The 69 kDa Escherichia coli maltodextrin glucosidase does not get encapsulated underneath GroES and folds through trans mechanism during GroEL/GroES-assisted folding. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21:2874–2885. doi: 10.1096/fj.06-7958com. [DOI] [PubMed] [Google Scholar]
- 24.Tischer A, Madde P, Moon-Tasson L, Auton M. Misfolding of vWF to pathologically disordered conformations impacts the severity of von Willebrand disease. Biophysical journal. 2014;107:1185–1195. doi: 10.1016/j.bpj.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voziyan PA, Fisher MT. Chaperonin-assisted folding of glutamine synthetase under nonpermissive conditions: off-pathway aggregation propensity does not determine the co-chaperonin requirement. Protein science: a publication of the Protein Society. 2000;9:2405–2412. doi: 10.1110/ps.9.12.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher MT. Promotion of the in vitro renaturation of dodecameric glutamine synthetase from Escherichia coli in the presence of GroEL (chaperonin-60) and ATP. Biochemistry. 1992;31:3955–3963. doi: 10.1021/bi00131a010. [DOI] [PubMed] [Google Scholar]
- 27.Paul S, Punam S, Chaudhuri TK. Chaperone-assisted refolding of Escherichia coli maltodextrin glucosidase. FEBS journal. 2007;274:6000–6010. doi: 10.1111/j.1742-4658.2007.06122.x. [DOI] [PubMed] [Google Scholar]
- 28.Abdiche Y, Malashock D, Pinkerton A, Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal Biochem. 2008;377:209–217. doi: 10.1016/j.ab.2008.03.035. [DOI] [PubMed] [Google Scholar]
- 29.31, f. T. N. Ni-NTA Biosensor Kinetic Assays.
- 30.26, f. T. N. Dip and Read Amine Reactive Second-Generation (AR2G) Biosensors.
- 31.Naik S, Brock S, Akkaladevi N, Tally J, McGinn-Straub W, Zhang N, Gao P, Gogol EP, Pentelute BL, Collier RJ, Fisher MT. Monitoring the kinetics of the pH-driven transition of the anthrax toxin prepore to the pore by biolayer interferometry and surface plasmon resonance. Biochemistry. 2013;52:6335–6347. doi: 10.1021/bi400705n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auton M, Cruz MA, Moake J. Conformational stability and domain unfolding of the Von Willebrand factor A domains. Journal of molecular biology. 2007;366:986–1000. doi: 10.1016/j.jmb.2006.10.067. [DOI] [PubMed] [Google Scholar]
- 33.Apetri AC, Horwich AL. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17351–17355. doi: 10.1073/pnas.0809794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katayama H, McGill M, Kearns A, Brzozowski M, Degner N, Harnett B, Kornilayev B, Matkovic-Calogovic D, Holyoak T, Calvet JP, Gogol EP, Seed J, Fisher MT. Strategies for folding of affinity tagged proteins using GroEL and osmolytes. Journal of structural and functional genomics. 2009;10:57–66. doi: 10.1007/s10969-008-9053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Z, Eisenstein E. Nucleotide binding-promoted conformational changes release a nonnative polypeptide from the Escherichia coli chaperonin GroEL. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:1977–1981. doi: 10.1073/pnas.93.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murai N, Taguchi H, Yoshida M. Kinetic analysis of interactions between GroEL and reduced alpha-lactalbumin. Effect of GroES and nucleotides. The Journal of biological chemistry. 1995;270:19957–19963. doi: 10.1074/jbc.270.34.19957. [DOI] [PubMed] [Google Scholar]
- 37.Paynter S, Russell DA. Surface plasmon resonance measurement of pH-induced responses of immobilized biomolecules: conformational change or electrostatic interaction effects? Analytical biochemistry. 2002;309:85–95. doi: 10.1016/s0003-2697(02)00255-5. [DOI] [PubMed] [Google Scholar]
- 38.Naik S, Kumru OS, Cullom M, Telikepalli SN, Lindboe E, Roop TL, Joshi SB, Amin D, Gao P, Middaugh CR. Probing structurally altered and aggregated states of therapeutically relevant proteins using GroEL coupled to bio-layer interferometry. Protein Science. 2014;23:1461–1478. doi: 10.1002/pro.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tapio S, Yeh F, Shuman HA, Boos W. The malZ gene of Escherichia coli, a member of the maltose regulon, encodes a maltodextrin glucosidase. The Journal of biological chemistry. 1991;266:19450–19458. [PubMed] [Google Scholar]
- 40.Yasgar A, Shultz J, Zhou W, Wang H, Huang F, Murphy N, Abel EL, DiGiovanni J, Inglese J, Simeonov A. A high-throughput 1,536-well luminescence assay for glutathione S-transferase activity. Assay and drug development technologies. 2010;8:200–211. doi: 10.1089/adt.2009.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holtz B, Wang Y, Zhu XY, Guo A. Denaturing and refolding of protein molecules on surfaces. Proteomics. 2007;7:1771–1774. doi: 10.1002/pmic.200700053. [DOI] [PubMed] [Google Scholar]
- 42.Johansson A-S, Stenberg G, Widersten M, Mannervik B. Structure-activity relationships and thermal stability of human glutathione transferase P1-1 governed by the H-site residue 105. Journal of molecular biology. 1998;278:687–698. doi: 10.1006/jmbi.1998.1708. [DOI] [PubMed] [Google Scholar]
- 43.Hornberg A, Hultdin UW, Olofsson A, Sauer-Eriksson AE. The effect of iodide and chloride on transthyretin structure and stability. Biochemistry. 2005;44:9290–9299. doi: 10.1021/bi050249z. [DOI] [PubMed] [Google Scholar]
- 44.Foss TR, Wiseman RL, Kelly JW. The pathway by which the tetrameric protein transthyretin dissociates. Biochemistry. 2005;44:15525–15533. doi: 10.1021/bi051608t. [DOI] [PubMed] [Google Scholar]
- 45.Klabunde T, Petrassi HM, Oza VB, Raman P, Kelly JW, Sacchettini JC. Rational design of potent human transthyretin amyloid disease inhibitors. Nature structural biology. 2000;7:312–321. doi: 10.1038/74082. [DOI] [PubMed] [Google Scholar]
- 46.Green NS, Palaninathan SK, Sacchettini JC, Kelly JW. Synthesis and characterization of potent bivalent amyloidosis inhibitors that bind prior to transthyretin tetramerization. Journal of the American Chemical Society. 2003;125:13404–13414. doi: 10.1021/ja030294z. [DOI] [PubMed] [Google Scholar]
- 47.Choi SRN, Connelly S, Johnson SM, Wilson IA, Kelly JW. A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity. Journal of the American Chemical Society. 2009;132:1359–1370. doi: 10.1021/ja908562q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alhamadsheh MM, Connelly S, Cho A, Reixach N, Powers ET, Pan DW, Wilson IA, Kelly JW, Graef IA. Potent kinetic stabilizers that prevent transthyretin-mediated cardiomyocyte proteotoxicity. Science translational medicine. 2011;3:97ra81. doi: 10.1126/scitranslmed.3002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razavi H, Palaninathan SK, Powers ET, Wiseman RL, Purkey HE, Mohamedmohaideen NN, Deechongkit S, Chiang KP, Dendle MT, Sacchettini JC, Kelly JW. Benzoxazoles as transthyretin amyloid fibril inhibitors: synthesis, evaluation, and mechanism of action. Angewandte Chemie. 2003;42:2758–2761. doi: 10.1002/anie.200351179. [DOI] [PubMed] [Google Scholar]
- 50.Hammarstrom P, Wiseman RL, Powers ET, Kelly JW. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003;299:713–716. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 51.Miller SR, Sekijima Y, Kelly JW. Native state stabilization by NSAIDs inhibits transthyretin amyloidogenesis from the most common familial disease variants. Laboratory investigation; a journal of technical methods and pathology. 2004;84:545–552. doi: 10.1038/labinvest.3700059. [DOI] [PubMed] [Google Scholar]
- 52.Purkey HE, Dorrell MI, Kelly JW. Evaluating the binding selectivity of transthyretin amyloid fibril inhibitors in blood plasma. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5566–5571. doi: 10.1073/pnas.091431798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) The Journal of biological chemistry. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 54.Rakhit R, Chakrabartty A. Structure, folding, and misfolding of Cu,Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochimica et biophysica acta. 2006;1762:1025–1037. doi: 10.1016/j.bbadis.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Ray SS, Nowak RJ, Brown RH, Jr, Lansbury PT., Jr Small-molecule-mediated stabilization of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants against unfolding and aggregation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3639–3644. doi: 10.1073/pnas.0408277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng HX, Hentati A, Tainer JA, Iqbal Z, Cayabyab A, Hung WY, Getzoff ED, Hu P, Herzfeldt B, Roos RP, et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 57.Arnesano F, Banci L, Bertini I, Martinelli M, Furukawa Y, O’Halloran TV. The unusually stable quaternary structure of human Cu,Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. The Journal of biological chemistry. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 58.Potter SZ, Zhu H, Shaw BF, Rodriguez JA, Doucette PA, Sohn SH, Durazo A, Faull KF, Gralla EB, Nersissian AM, Valentine JS. Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. Journal of the American Chemical Society. 2007;129:4575–4583. doi: 10.1021/ja066690+. [DOI] [PubMed] [Google Scholar]
- 59.Mei G, Rosato N, Silva N, Jr, Rusch R, Gratton E, Savini I, Finazzi-Agro A. Denaturation of human Cu/Zn superoxide dismutase by guanidine hydrochloride: a dynamic fluorescence study. Biochemistry. 1992;31:7224–7230. doi: 10.1021/bi00147a003. [DOI] [PubMed] [Google Scholar]
- 60.Heikkila RE, Cabbat FS, Cohen G. In vivo inhibition of superoxide dismutase in mice by diethyldithiocarbamate. The Journal of biological chemistry. 1976;251:2182–2185. [PubMed] [Google Scholar]
- 61.Patrick AE, Thomas PJ. Development of CFTR Structure. Frontiers in pharmacology. 2012;3:162. doi: 10.3389/fphar.2012.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, Hunt JF, Kearins MC, Lorimer D, Maloney PC, Post KW, Rajashankar KR, Rutter ME, Sauder JM, Shriver S, Thibodeau PH, Thomas PJ, Zhang M, Zhao X, Emtage S. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. The EMBO journal. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Travis SM, Carson MR, Ries DR, Welsh MJ. Interaction of nucleotides with membrane-associated cystic fibrosis transmembrane conductance regulator. The Journal of biological chemistry. 1993;268:15336–15339. [PubMed] [Google Scholar]
- 64.Qu BH, Strickland EH, Thomas PJ. Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. The Journal of biological chemistry. 1997;272:15739–15744. doi: 10.1074/jbc.272.25.15739. [DOI] [PubMed] [Google Scholar]
- 65.Yike I, Ye J, Zhang Y, Manavalan P, Gerken TA, Dearborn DG. A recombinant peptide model of the first nucleotide-binding fold of the cystic fibrosis transmembrane conductance regulator: comparison of wild-type and delta F508 mutant forms. Protein science: a publication of the Protein Society. 1996;5:89–97. doi: 10.1002/pro.5560050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naik S, Haque I, Degner N, Kornilayev B, Bomhoff G, Hodges J, Khorassani AA, Katayama H, Morris J, Kelly J, Seed J, Fisher MT. Identifying protein stabilizing ligands using GroEL. Biopolymers. 2010;93:237–251. doi: 10.1002/bip.21319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bullock AN, Henckel J, DeDecker BS, Johnson CM, Nikolova PV, Proctor MR, Lane DP, Fersht AR. Thermodynamic stability of wild-type and mutant p53 core domain. Proceedings of the National Academy of Sciences. 1997;94:14338–14342. doi: 10.1073/pnas.94.26.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parrales A, Iwakuma T. Targeting oncogenic mutant p53 for cancer therapy. Frontiers in oncology. 2015;5 doi: 10.3389/fonc.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Friedler A, Hansson LO, Veprintsev DB, Freund SM, Rippin TM, Nikolova PV, Proctor MR, Rüdiger S, Fersht AR. A peptide that binds and stabilizes p53 core domain: chaperone strategy for rescue of oncogenic mutants. Proceedings of the National Academy of Sciences. 2002;99:937–942. doi: 10.1073/pnas.241629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katayama H, Janowiak BE, Brzozowski M, Juryck J, Falke S, Gogol EP, Collier RJ, Fisher MT. GroEL as a molecular scaffold for structural analysis of the anthrax toxin pore. Nature structural & molecular biology. 2008;15:754–760. doi: 10.1038/nsmb.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buchner J, Brinkmann U, Pastan I. Renaturation of a single-chain immunotoxin facilitated by chaperones and protein disulfide isomerase. Bio/technology. 1992;10:682–685. doi: 10.1038/nbt0692-682. [DOI] [PubMed] [Google Scholar]
- 72.Lazar KL, Patapoff TW, Sharma VK. MAbs. Taylor & Francis; 2010. Cold denaturation of monoclonal antibodies; pp. 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen C, Baldwin MR, Barbieri JT. Molecular Basis for Tetanus Toxin Coreceptor Interactions†. Biochemistry. 2008;47:7179–7186. doi: 10.1021/bi800640y. [DOI] [PubMed] [Google Scholar]
- 74.Burns JR, Baldwin MR. Tetanus neurotoxin utilizes two sequential membrane interactions for channel formation. Journal of Biological Chemistry. 2014;289:22450–22458. doi: 10.1074/jbc.M114.559302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown CR, Hong-Brown LQ, Biwersi J, Verkman A, Welch WJ. Chemical chaperones correct the mutant phenotype of the ΔF508 cystic fibrosis transmembrane conductance regulator protein. Cell stress & chaperones. 1996;1:117. doi: 10.1379/1466-1268(1996)001<0117:ccctmp>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morello J-P, Bouvier M, Petäjä-Repo UE, Bichet DG. Pharmacological chaperones: a new twist on receptor folding. Trends in pharmacological sciences. 2000;21:466–469. doi: 10.1016/s0165-6147(00)01575-3. [DOI] [PubMed] [Google Scholar]
- 77.Papp E, Csermely P. Molecular Chaperones in Health and Disease. Springer; 2006. Chemical chaperones: mechanisms of action and potential use; pp. 405–416. [DOI] [PubMed] [Google Scholar]
- 78.Leandro P, Gomes CM. Protein misfolding in conformational disorders: rescue of folding defects and chemical chaperoning. Mini reviews in medicinal chemistry. 2008;8:901–911. doi: 10.2174/138955708785132783. [DOI] [PubMed] [Google Scholar]
- 79.Bolen DW, Rose GD. Structure and energetics of the hydrogen-bonded backbone in protein folding. Annu Rev Biochem. 2008;77:339–362. doi: 10.1146/annurev.biochem.77.061306.131357. [DOI] [PubMed] [Google Scholar]
- 80.Krell T, Greco F, Nicolaï MC, Dubayle J, Renauld-Mongénie G, Poisson N, Bernard I. The use of microcalorimetry to characterize tetanus neurotoxin, pertussis toxin and filamentous haemagglutinin. Biotechnology and applied biochemistry. 2003;38:241–251. doi: 10.1042/BA20030089. [DOI] [PubMed] [Google Scholar]
- 81.Zhang J-H, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. Journal of biomolecular screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 82.Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. Journal of Biological Chemistry. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 83.Sabbaghian M, Ebrahim-Habibi A, Hosseinkhani S, Ghasemi A, Nemat-Gorgani M. Prevention of thermal aggregation of an allosteric protein by small molecules: Some mechanistic insights. International journal of biological macromolecules. 2011;49:806–813. doi: 10.1016/j.ijbiomac.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 84.Cimmperman P, Baranauskienė L, Jachimovičiūtė S, Jachno J, Torresan J, Michailovienė V, Matulienė J, Sereikaitė J, Bumelis V, Matulis D. A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophysical journal. 2008;95:3222–3231. doi: 10.1529/biophysj.108.134973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fisher MT. Differences in thermal stability between reduced and oxidized cytochrome b-562 from Escherichia coli. Biochemistry. 1991;30:10012–10018. doi: 10.1021/bi00105a028. [DOI] [PubMed] [Google Scholar]
- 86.Elad N, Farr GW, Clare DK, Orlova EV, Horwich AL, Saibil HR. Topologies of a substrate protein bound to the chaperonin GroEL. Molecular cell. 2007;26:415–426. doi: 10.1016/j.molcel.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.