Abstract
A master sex-determining gene, the Y chromosome-linked anti-Müllerian hormone (amhy) gene, has been described in two New World atheriniform species but little is known on the distribution, evolution, and function(s) of this gene in other Atheriniformes. Interestingly, amhy has been found to coexist with temperature-dependent sex determination (TSD), providing a unique opportunity to explore the interplay between genotypic and environmental sex determination. In this study, the search for an amhy homolog was extended to an Old World atheriniform, the cobaltcap silverside Hypoatherina tsurugae (Atherinidae). The full sequences, including the coding and noncoding regions, of the autosomal amh (amha) and a putative amhy were obtained. The deduced Amha and Amhy proteins comprised 511 and 340 amino acids (aa), respectively. PCR analysis with genomic DNA from wild adults and from laboratory-reared juveniles revealed a high, but not complete association of ∼95% between amhy and maleness. The spatiotemporal expression of amhy and amha during gonadal sex differentiation was analyzed by qRT-PCR and in situ hybridization (ISH). amhy transcription (in amhy-positive larvae) started before and peaked during histological differentiation of the gonads whereas amha was negligible during the same period in both genotypes. These results demonstrate that the amhy, although with some structural differences in relation to the amhy of some New World atheriniforms, is strongly associated with maleness and probably important for testicular development in this Old World atheriniform. Thus, amhy is a candidate sex determination gene in cobaltcap silverside and it will be key to scrutinize the mechanism of sex determination in this species.
Keywords: Atheriniformes, amhy, fish, Hypoatherina tsurugae, sex determination
In recent years, a growing number of genes have been identified as major triggers of sex determination in teleosts (Hattori et al. 2013; Kikuchi and Hamaguchi 2013; Takehana et al. 2014; and other references below). It is now evident that sex-determining genes in fishes are not restricted to transcription factors as there are reports also implicating members of the TGF-β superfamily and even an immune-related gene with this function. Interestingly, the degree of conservation of these genes apparently varies greatly with the taxonomic group. For instance, while most salmonids share a common sex-determining gene (e.g., sdY, Yano et al. 2012, 2013), in species of the genus Oryzias there are a variety of genes with this function (e.g., dmy/dmrt1bY, Matsuda et al. 2002; gsdfY, Myosho et al. 2012; sox3Y, Takehana et al. 2014).
In silversides, a homolog of the Y chromosome-linked duplication of the amhy gene, first discovered in Patagonian pejerrey (Odontesthes hatcheri; Atherinopsidae; Hattori et al. 2012), was found to be functional also in the congeneric species O. bonariensis (Yamamoto et al. 2014). Nevertheless, gonadal fate in these two species is affected also by the temperature experienced during a critical period of sex determination early in life (TSD; Strüssmann et al. 1997), sometimes overriding the genetic predisposition of an individual and giving rise to phenotypic–genotypic mismatches in relation to amhy even at environmentally relevant temperatures. These facts demonstrate the coexistence of two opposing sex determination systems in this group of fishes and suggest that their genetic trigger of sex determination may be functional only within a given thermal range (Yamamoto et al. 2014). TSD has also been reported in several other atherinopsids (genera Menidia and Chirostoma; Strüssmann et al. 2010; Corona-Herrera et al. 2016) and is conceivably widespread in this taxon (Strüssmann and Patiño 1999). However, in contrast to the abundance of information on atherinopsids, there is little or no knowledge on the sex determination system and evolution of the amhy gene in other atheriniform families.
The phylogenetic relationships within Atheriniformes are still controversial and different authors describe it as containing between six and nine families (Sparks and Smith 2004; Nelson 2006; Froese et al. 2012). Until recently, atherinopsids (known as “New World silversides”) were included in the family Atherinidae, which is now reserved for the “Old World silversides.” These two families comprise numerous species that inhabit coastal marine, estuarine, and freshwater environments. They are generally small-sized and form large schools that represent an important forage for upper trophic predators worldwide (Bloom et al. 2012). This fundamental role in coastal ecosystems and the possibility that many of them display TSD, which renders species particularly vulnerable to global warming and changing climatic patterns (Janzen 1994; Miller et al. 2004; Hulin et al. 2009), make it urgent that we obtain knowledge of their sex-determining systems and monitor possible environmental effects on population sex ratios and stability.
In this study, we probed the presence of the amhy gene and its role in testis determination in a population of the cobaltcap silverside Hypoatherina tsurugae, a marine atherinid from the Northwest Pacific Ocean. This study was conceived with two basic aims. The first was to expand our understanding on the distribution of amhy among atheriniforms. The second was to provide the scientific basis for using amhy as a marker of genetic sex tendency in studies on the molecular mechanism of sex determination of this species.
Materials and Methods
Collection of wild specimens
Sexually mature adult cobaltcap silversides were collected by hand net on July 2014 in Tokyo Bay (Chiba, Japan). The gonadal sex of 81 individuals was assessed by careful stripping of gametes and eight fish of each sex were randomly selected for cloning of amh genes (see details below). The remaining fish (48 females and 17 males) were stocked in a 500 L circular tank at the Tateyama Station, Field Science Center of Tokyo University of Marine Science and Technology (Chiba, Japan) and used as broodstock fish to obtain gametes and offspring for further experiments (see below).
Cloning of autosomal amh (amha) and Y chromosome-linked amh (amhy)
Genomic DNA was extracted from the caudal fin tissue of one sexually mature male following the protocol described by Aljanabi and Martinez (1997) and subjected to PCR amplification using degenerate primers designed based on O. hatcheri amha. To determine the complete open reading frame for cobaltcap silverside amha, total RNA was isolated from testis using TRIzol (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions and 1 µg of total RNA per sample was reverse transcribed using SuperScript III (Thermo Fisher Scientific) with Oligo-(dT) primers (Merk Millipore, Darmstadt, Germany) in 20 μl reactions. RT-PCR, genome walking, and 5′- and 3′-RACE PCRs using a Smart RACE cDNA amplification kit (Takara Bio, Shiga, Japan) were then performed according to the manufacturer’s protocol. The PCR conditions and specific primers used in each reaction are listed in Supplemental Material, Table S1 and Table S2 in File S1.
Based on the amha full sequence, several primer sets flanking intronic sequences were designed in coding regions and used to amplify a Y chromosome-linked amhy in this species. This strategy was based on the differences between amhy and amha genes in O. hatcheri and O. bonariensis, whereby an insertion of ∼500 bases specific to amhy is found in the third intron. Genomic DNA was isolated from the caudal fin of 16 adult fish, eight females and eight males, following the protocol described above and subjected to PCR amplification. One set of the primers designed in the first and fifth exons (Table S1 in File S1; Amh 613 F and Amh 35 R) amplified two fragments. The larger fragment was present in both sexes whereas the smaller one, a putative amhy, was present only in males. The smaller fragment was purified, cloned, and sequenced as described above. To obtain the full genomic and cDNA sequences of the putative amhy, genome walking and RACE PCR were conducted according to the same protocols as for amha cloning.
The specific amplicons from each PCR reaction were purified, cloned, and sequenced in an ABI PRISM 3100 capillary sequencer (Thermo Fisher Scientific) using the BigDye Terminator method. Sequences were then analyzed by the GENETYX version 11.0 (GENETYX, Tokyo, Japan) software.
Phylogenetic analysis of amh sequences
The predicted aa sequences of H. tsurugae Amha (exons I to VII) and Amhy (exons I, IV, VI, and VII) were compared with Amh sequences of other species available in GenBank using the software GENETYX version 11.0. Multiple alignments were performed using Clustal W in MEGA software version 5.2 (Tamura et al. 2011). The sequences for O. bonariensis Amha and Amhy, O. hatcheri Amha and Amhy, Dicentrarchus labrax Amh, Oreochromis niloticus Amh, and Danio rerio Amh were used in the comparison, and Xenopus laevis Amh was used as the outgroup. Phylogenetic trees were generated by Neighbor-Joining (Saitou and Nei 1987), Maximum Parsimony, and Maximum Likelihood methods with 10,000 bootstrap replicates each to determine the confidence of tree topology. Analyses were performed by the MEGA software (v. 5.2).
Sex-association analysis by amhy amplification in wild specimens
All wild-caught fish were screened for the presence of amhy by PCR analysis using the same primers (Table S1 in File S1; Amh 613 F and Amh 35 R) and conditions described previously. Animals carrying the amhy gene (amhy positives) were represented by amhy+ and those without it by amhy−.
Testing of Mendelian inheritance and determination of parental genotype
To test the Mendelian inheritance of amhy and determine the exact parental genotype, we performed artificial insemination with gametes from four amhy− females and four amhy+ males in single-pair crosses. Fertilized eggs from each of the crosses were incubated separately until analysis. Randomly-chosen eyed-egg stage embryos (n = 38–45) from each cross were analyzed by amhy amplification following procedures described above.
Rearing of larvae for gene expression analysis and gonadal histology
Fertilized eggs were obtained by natural spawning using wild-caught adult individuals (48 females and 17 males) that were reared overnight in the laboratory. Fertilized eggs were collected the next morning from the bottom of the tank and incubated in flowing sea water until hatching. We could not ascertain how many females and males actually contributed fertilized eggs. Approximately 500 hatchlings (10–13 d postfertilization) were stocked in two 30 L tanks kept at 22°, the average temperature during the spawning season of H. tsurugae in Tateyama Bay, and reared for up to 12 wk. The tanks were supplied with filtered natural sea water at a rate of 100 ml/min. Larvae were fed rotifers Branchionus rotundiformis and Artemia nauplii from the first day to satiation twice daily and gradually weaned onto powdered marine fish food (AQUEON, Franklin, WI) from the fifth week of the experiment.
Fish were sampled biweekly from 0 to 10 wk after hatching (wah) for gene expression analyses and gonadal histology. The remaining larvae were sampled at the end of the rearing experiment to determine the sex ratio. The trunks of the fish were stored in RNA later (Thermo Fisher Scientific) (n = 8) or in Bouin’s solution (n = 8) for gene expression analyses and gonadal histology, respectively, at each time point. Samples in RNA later were stored at −80° until use. Bouin-fixed samples were rinsed three times with phosphate-buffered saline, transferred into 70% ethanol, and stored at 4° until use. All larvae were fin-clipped for amhy genotyping as described above.
Histological analysis of gonadal sex differentiation and sex ratio
Trunk samples were dehydrated through an ascending ethanol series (70, 90, 99, and 100%), cleared in xylene, embedded in Paraplast Plus (McCormick Scientific, St. Louis, MO), sectioned serially with a thickness of 5 µm, and stained with hematoxylin and eosin. Stages of gonadal sex differentiation were determined by light microscopy using histological criteria for another atheriniform, the pejerrey O. bonariensis (Ito et al. 2005; Strüssmann and Ito 2005).
Expression analyses by qRT-PCR and ISH
Total RNA extraction and cDNA synthesis were performed following previous studies (Yamamoto et al. 2014). The expression level of mRNA transcripts was analyzed by qRT-PCR using specific primers designed for amha and amhy loci. The β-actin gene was taken as an endogenous control because of its stability during the sex determination/differentiation period (Figure S1 and File S2). All primer sets and their respective conditions are listed in Table S1 and Table S2 in File S1.
The ISH analysis used trunks of amhy+ larvae collected before (4 wah) and after (8 wah) the onset of histological differentiation of the gonads. Ovaries from adult amhy− specimens were used to confirm the binding specificity of the amha-specific probe (see the Results section). Samples were fixed and processed as per the protocol mentioned above. We were not able to develop an amhy-specific probe so hybridizations were conducted using a 775 bp amhy probe [nucleotides (nt) +207 to +982; exons VI to VII; and 93.5% identity with the respective sequence for amha] that recognized both loci and a 523 bp amha-specific probe designed in the amha-specific region (nt −22 to +501; exons I to III; and 17.2% of identity with amhy). ISH was performed as described previously (Yamamoto et al. 2011). Briefly, sense and antisense RNA probes were transcribed in vitro using digoxigenin-labeled UTP (Roche Diagnostics, Basel, Switzerland) and T7 RNA polymerase (Roche Diagnostics). Sections for ISH were permeabilized with 5 mg/ml proteinase K at 37° for 10 min. The sections were subsequently acetylated and incubated with a 0.0125–0.2 mg/ml RNA probe. After hybridization at 65° for 16 hr, the sections were washed and unbound probes were digested using 20 mg/ml RNase A to reduce background signals. The sections were then placed in blocking solution (Roche Diagnostics) at room temperature for 1 hr and incubated with the Fab fragment of an anti-DIG-alkaline phosphatase-conjugated antibody (Roche Diagnostics) diluted 1:2000 with blocking solution at 25° for 1 hr. The sections were washed and specific signals were detected by NBT/BCIP (Roche Diagnostics) according to the recommendations of the manufacturer. The signal detections by NBT/BCIP were stopped when the specific signals of antisense probes appeared.
Statistical analysis
The significance of the association between genotype (amhy+ or amhy−) and sex phenotype, and of deviations from a 1:1 genotypic sex ratio, were determined by the Yates’ continuity corrected Chi-square test, whereas that of the differences in gene expression between groups was analyzed by ANOVA followed by the Tukey test using GraphPad Prism (v.6.0; GraphPad Software, San Diego, CA). Differences in gene expression were considered as statistically significant at P < 0.05.
Data availability
DNA sequences: GenBank accessions; H. tsurugae Amha (KU664386) and Amhy (KU664387), O. bonariensis Amha (AHG98063.1) and Amhy (AAV31752.2), O. hatcheri Amha (AEE60845.1) and Amhy (ABF47515.2), D. labrax Amh (CAJ78431.1), O. niloticus Amh (ABS58513.1), D. rerio Amh (NP001007780.1), and X. laevis Amh (BAO04196.1).
Results
Isolation of amh paralogues in H. tsurugae
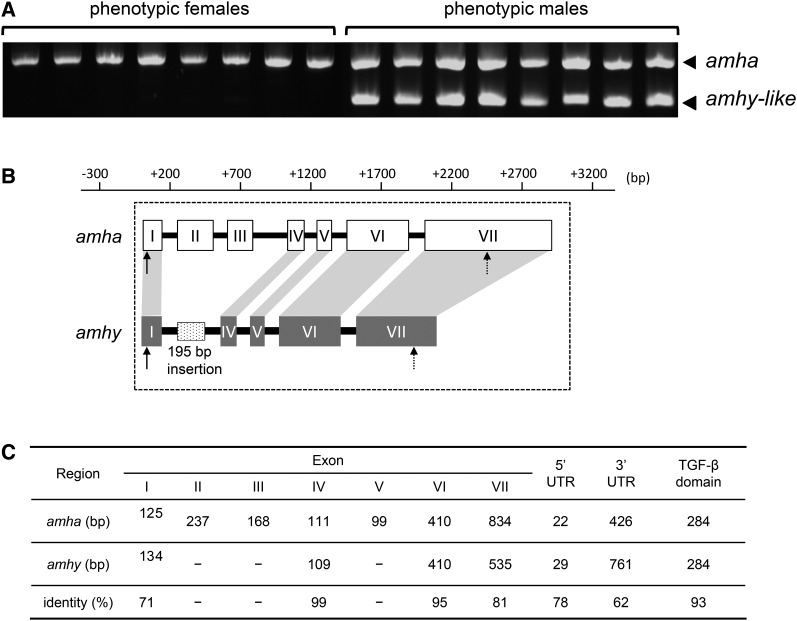
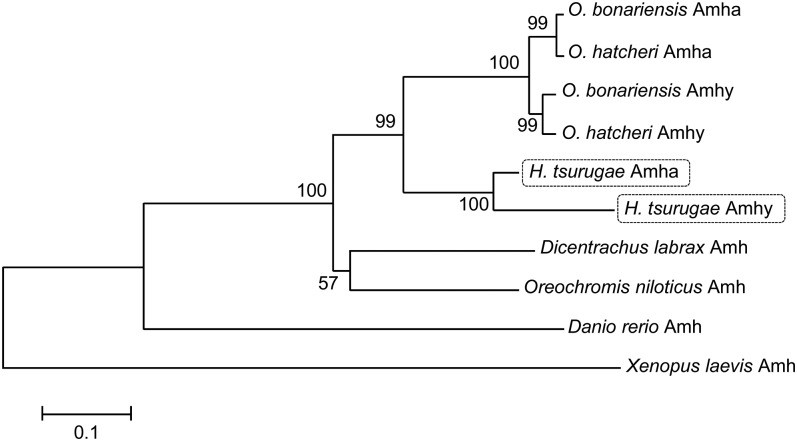
Two amh genes were cloned and isolated in H. tsurugae. One was detected in all individuals regardless of sex (Figure 1A) and for this reason was named Hts-amha, for H. tsurugae amh on autosomes. The cDNA sequence has 2015 nt and seven exons (Figure 1B). The other was detected only in phenotypic males (Figure 1A) and was named Hts-amhy for its high association with the Y chromosome as in O. hatcheri (Hattori et al. 2012) and O. bonariensis (Yamamoto et al. 2014). The full-length Hts-amhy cDNA sequence comprises 1838 nt and only four exons (Figure 1B). The homologs of amha exons II and III were absent in amhy. In contrast, an insertion of 195 bp was detected between exons I and IV when compared to the amha gene structure. The homolog of amha exon V was detected in genomic DNA sequence but not in cDNA sequence. The lowest and highest nt identity values were found for exons I and IV, respectively (Figure 1C). The deduced aa sequences of Amha (511 aa) and Amhy (340 aa) shared 91% of identity. Both the amha and amhy genes contained the TGF-β domain with seven canonical cysteine residues, which form disulfide bonds necessary for dimer formation. Phylogenetic analyses of Amha and Amhy aa sequences of H. tsurugae and other species available in the NCBI database using X. laevis as an outgroup revealed that H. tsurugae amhy and amha form a different clade from that of Odontesthes species amhy and amha (Figure 2, Figure S2 and File S2).
Figure 1.
Isolation, cloning and characterization of amha and amhy in H. tsurugae. (A) Polymerase chain reaction-amplified amha in both male and female wild specimens (upper band) and amhy amplified only in males (lower band). (B) Comparison of full-length gene structure of amha and amhy in H. tsurugae. Compared to amha, the amhy gene of H. tsurugae is shorter, lacks exons II and III, and contains a specific insertion of 195 bp at the position of exons II and IV. Exon V is present in the genomic sequence but it is not transcribed. (C) Identity values of nucleotide sequence between amha and amhy exons, untranslated regions (UTRs), and the transforming growth factor-β (TGF-β) domain.
Figure 2.
Phylogenetic analysis (neighbor-joining tree) of the amino acid sequences of H. tsurugae Amha and Amhy in relation to other species. Numbers indicate bootstrap values based on 10,000 replicates.
Association of amhy genotype and phenotypic sex in wild and laboratory-reared fish
Adult specimens of H. tsurugae collected from Tokyo Bay showed high concordance between phenotypic sex and the presence/absence of amhy. For instance, 96% of the fish bearing testes (males) and 91.1% of the fish bearing ovaries (females) were amhy+ and amhy−, respectively (Table 1; wild-caught fish; combined association of 92.6%; P < 0.0001). Laboratory-reared fish kept at 22° during the period of gonadal sex differentiation also showed a high association between phenotypic and genotypic sex (Table 1; laboratory-reared fish; P < 0.0001). In the progeny test of four single-pair crosses, the ratios of amhy− and amhy+ in the progeny did not deviate significantly from 1:1 in any of the crosses (Table 2), supporting the Mendelian inheritance of the amhy gene and indicating that all males used for single-pair crosses were heterozygous (amhy+/−) for the amhy gene.
Table 1. Relationship between genotype (presence/absence of amhy) and the phenotypic sex in wild-caught and laboratory-reared (rearing at 22° during the period of sex determination/differentiation) H. tsurugae.
| Phenotypic Sex | Genotype | Total | |
|---|---|---|---|
| amhy+ (%) | amhy− (%) | ||
| Wild-caught (P < 0.0001)a | |||
| Testis | 24 (96.0) | 1 (4.0) | 25 (31.9%) |
| Ovary | 5 (8.9) | 51 (91.1) | 56 (69.1%) |
| Total | 29 (35.8) | 52 (64.2) | 81 |
| Laboratory-reared (P < 0.0001)a | |||
| Testis | 26 (96.0) | 1 (4.0) | 27 (57.4%) |
| Ovary | 4 (20.0) | 16 (80.0) | 20 (42.6%) |
| Total | 30 (63.8) | 17 (36.2) | 47 |
Significant association between genotype and sex phenotype (Yates’ continuity corrected Chi-square test).
Table 2. Frequency of amhy+ and amhy− genotypes in progenies from four single-pair crosses of amhy− females and amhy+ males.
| Crossa | Genotype | Total | |
|---|---|---|---|
| amhy+ (%) | amhy− (%) | ||
| A | 21 (46.7) | 24 (53.3) | 45 |
| B | 16 (35.6) | 29 (64.4) | 45 |
| C | 21 (51.2) | 20 (49.8) | 41 |
| D | 20 (52.6) | 18 (47.4) | 38 |
| Total | 78 (46.2) | 91 (53.8) | 169 |
The sex ratios of all progenies do not deviate significantly from 1:1.
Expression analyses of amhy and amha during gonadal sex differentiation
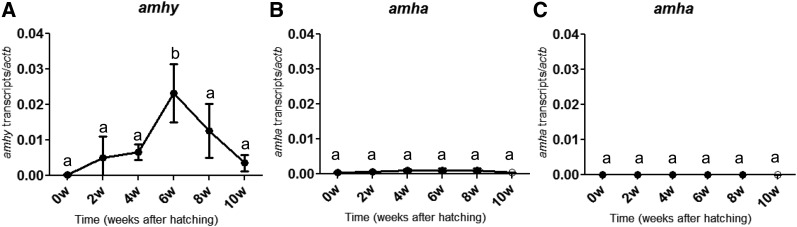
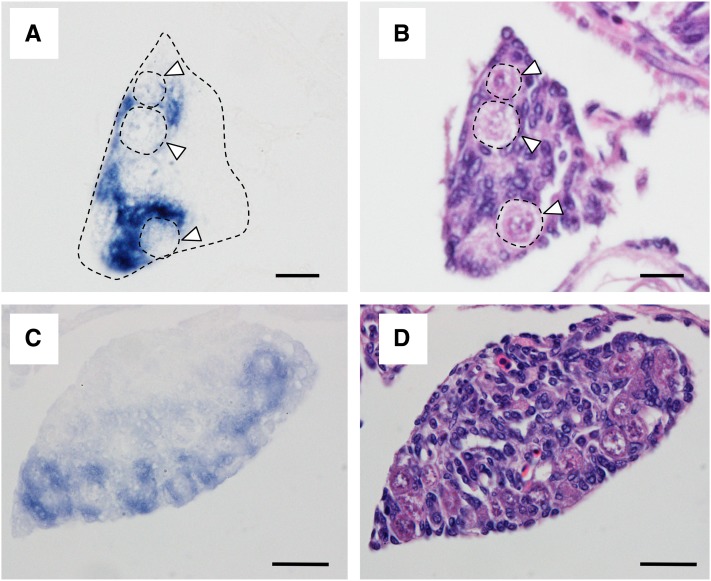
The qRT-PCR analysis of amhy+ individuals revealed the presence of amhy transcripts between 2 and 10 wah with a significant peak at 6 wah (Figure 3A). In contrast, the levels of amha expression were extremely low in both genotypes (Figure 3, B and C). ISH signals with the amhy probe (that potentially also detects amha) were detected in undifferentiated gonads (4 wah; Figure 4A, Figure S3A and File S2) and differentiating testes (8 wah; Figure 4C) of amhy+ larvae. Signals were found in presumptive Sertoli cells surrounding germ cells at the ventral side of the gonads (Figure 4C). Since ISH signals with the amha-specific probe were almost undetectable in undifferentiated gonads (Figure S3B), it can be surmised by exclusion that the signals obtained at this stage with the amhy probe represented mostly amhy transcripts. This hypothesis is also supported by the qRT-PCR results described above. In addition, the binding specificity of the amha probe was confirmed in ovaries from adult amhy− specimens (Figure S3D).
Figure 3.
Expression profiles of amhy (A) and amha (B) in amhy+ genotype and amha (C) in amhy− genotype during gonadal sex differentiation. Values represent the mean ± SEM of 3–6 fish per time point (w, weeks). Symbols with the same letter indicate groups that are not significantly different between time points.
Figure 4.
Localization of mRNAs using an amhy riboprobe by in situ hybridization in undifferentiated [4 wah (weeks after hatching), (A)] and differentiated [8 wah, (C)] gonads. Adjacent sections were stained with hematoxylin and eosin [4 wah, (B) and 8 wah, (D)]. Arrowheads indicate germ cells. Bars represent 10 μm (A and B) and 20 μm (C and D).
Discussion
In this study, we investigated the occurrence of two amh paralogs and their possible roles in sex determination of the atheriniform H. tsurugae. One locus was termed amha for its occurrence in individuals of both sexes, whereas the other was found predominantly in males (see Discussion below) and for this reason was denominated as amhy. Although the aa sequences of both loci shared 91% identity, a comparative structural analysis revealed the absence of exons II, III, and V in the cDNA sequence of amhy, resulting in a truncated gene. Interestingly, exon V is found in the genomic DNA sequence but it is not transcribed together with other exons. The structure of the C-terminus including the TGF-β domain with seven cysteine knots, which form the disulfide bonds required for protein homodimerization (Vitt et al. 2001), was conserved in amhy locus and shared 93% aa identity with the same domain of amha. On the other hand, the N-terminus is probably not structurally complete because only exons I and IV are present. In general, biological activity in amh is mainly related to the C-terminus (Massagué 1990; Pfennig et al. 2015), but there are also cases such as human AMH in which the N-terminus actually enhances the activity of the C-terminus (Wilson et al. 1993). In the case of H. tsurugae, the integrity of the TGF-β domain suggests that amhy might be able to bind to the AmhrII (Amh receptor type II) and thus activate the downstream pathway of testis differentiation.
Sex-association analysis using wild adults and captive-reared juveniles (22°) showed a high but not complete association between the presence and absence of amhy with maleness and femaleness, respectively. The reason for the phenotype–genotype mismatches is still not clear. It could be that amhy is only distantly linked to the sex-determining locus and that these fish are simply recombinants. However, a more plausible explanation is that sex determination in H. tsurugae is affected by water temperature as in many other atheriniforms (Strüssmann and Patiño 1999; Corona-Herrera et al. 2016). In fact, ongoing experiments have provided preliminary evidence of thermal modulation of sex determination in this species (K. Miyoshi, C.A. Strüssmann, and Y., Yamamoto, unpublished data), which in other atheriniforms has been shown to cause phenotypic–genotypic mismatches (Yamamoto et al. 2014).
The analysis of mRNA expression during larval development showed that amhy transcripts were restricted to amhy+ individuals. The expression of amhy was detected from before the appearance of the first signs of histological sex differentiation in presumptive Sertoli cells surrounding germ cells in the undifferentiated gonad and was maintained during testis differentiation. In contrast, amha showed low, basal expression levels in both genotypes during the same period. This is similar to the pattern described for O. hatcheri (Hattori et al. 2012) and different to that of O. bonariensis, where amha is coexpressed with amhy during the critical period of sex determination (Yamamoto et al. 2014). It has been reported that sex determination in O. bonariensis shows higher temperature sensitivity than that of O. hatcheri (Strüssmann et al. 1997). The high thermosensitivity at both high and low temperatures in the former species could be related to the profile of amha, which increases before the appearance of sex-specific histological differences not only in XY genotypes but also during masculinization of XX individuals (Yamamoto et al. 2014). If this is true, one might expect only moderate effects of temperature on sex ratios in H. tsurugae as is the case of O. hatcheri. Studies on the effects of temperature on the sex ratios and expression profiles of amha and amhy of H. tsurugae are currently underway.
Although these results suggest that amhy is a candidate sex determination gene in H. tsurugae as in other atheriniforms, it is not yet a foregone conclusion that this gene is an ortholog of the amhys of O. hatcheri and O. bonariensis. On the contrary, so far the results of phylogenetic analysis place it in a separate clade with other amhys, suggesting that it may have evolved independently. Sex-determining genes are known to show recurrent and independent evolution in teleosts (e.g., as exemplified in medaka species; Matsuda et al. 2002; Myosho et al. 2012; Takehana et al. 2014). In fact, recent reports implicating amh/AMH as a candidate sex-determining gene in unrelated taxa such as tilapia O. niloticus (Eshel et al. 2014; Li et al. 2015) and the marsupial mammal Ornithorhynchus anatinus (Cortez et al. 2014) lend support to the notion that this gene has a high probability of being recruited as a key genetic player of sex determination. Hence, it is possible that a coincidental de novo appearance of amhy has occurred in H. tsurugae. Nevertheless, the hypothesis of conservation cannot be fully ruled out at this point. For example, given the particularly fast evolutionary rates of sex-determining genes in relation to their autosomal paralogues (Mawaribuchi et al. 2012) and the relatively large genetic distance of Atherinopsidae and Atherinidae families (Bloom et al. 2012), it could be that the amhys in H. tsurugae and Odontesthes have accumulated enough structural changes as to make them lose the characteristics they once had in common. The absence of some exons in the truncated amhy of H. tsurugae could be such a case. This issue will only be clarified by analyzing relevant sequences for other atheriniform families besides Atherinopsidae and Atherinidae.
In conclusion, this study demonstrated that amhy, although with some structural differences in relation to the amhy of some New World atheriniforms (Hattori et al. 2012; Yamamoto et al. 2014), is also present in an Old World atheriniform. The high expression of amhy early in larval development and the high association with maleness in captive-reared and wild animals make H. tsurugae amhy a sex-determining gene candidate. The finding of a putative Y chromosome-specific marker will be extremely useful for monitoring the effects of environmental factors and anthropogenic influences on sex determination in this species. For example, ongoing studies suggest the occurrence of TSD in H. tsurugae (K. Miyoshi, C.A. Strüssmann, and Y. Yamamoto, unpublished data), which might place this species at higher risk of sex reversal and skewed sex ratios within a scenario of global warming and/or climate change. Moreover, species presenting both temperature-dependent and genotypic sex determination mechanisms such as H. tsurugae may be suitable as early-warning bioindicators of the effects of global warming on fish reproduction.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.042697/-/DC1.
Acknowledgments
We thank Y. Zhang and M. Sarida (Tokyo University of Marine Science and Technology) for their assistance with this work. We also thank M. Yokota (Tokyo University of Marine Science and Technology) for his kind assistance with phylogenetic analyses. This work was supported by the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (KAKENHI) to Y.Y. (15K18728) and C.A.S. (26241018) and FAPESP to R.S.H. (2009/15877-0).
Footnotes
Communicating editor: R. Houston
Literature Cited
- Aljanabi S. M., Martinez I., 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25: 4692–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom D. D., Unmack P. J., Gosztonyi A. E., Piller K. R., Lovejoy N. R., 2012. It’s a family matter: molecular phylogenetics of Atheriniformes and the polyphyly of the surf silversides (family: Notocheiridae). Mol. Phylogenet. Evol. 62: 1025–1030. [DOI] [PubMed] [Google Scholar]
- Corona-Herrera G. A., Tello-Ballinas J. A., Hattori R. S., Martínez-Palacios C. A., Strüssmann C. A., et al. , 2016. Gonadal differentiation and temperature effects on sex determination in the freshwater pike silverside Chirostoma estor Jordan 1880. Environ. Biol. Fishes 99: 463–471. [Google Scholar]
- Cortez D., Marin R., Toledo-Flores D., Froidevaux L., Liechti A., et al. , 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508: 488–493. [DOI] [PubMed] [Google Scholar]
- Eshel O., Shirak A., Dor L., Band M., Zak T., et al. , 2014. Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genomics 15: 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froese R., Zeller D., Kleisner K., Pauly D., 2012. What catch data can tell us about the status of global fisheries. Mar. Biol. 159: 1283–1292. [Google Scholar]
- Hattori R. S., Murai Y., Oura M., Masuda S., Majhi S. K., et al. , 2012. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. USA 109: 2955–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori R. S., Strüssmann C. A., Fernandino J. I., Somoza G. M., 2013. Genotypic sex determination in teleosts: insights from the testis-determining amhy gene. Gen. Comp. Endocrinol. 192: 55–59. [DOI] [PubMed] [Google Scholar]
- Hulin V., Delmas V., Girondot M., Godfrey M. H., Guillon J. M., 2009. Temperature-dependent sex determination and global change: are some species at greater risk? Oecologia 160: 493–506. [DOI] [PubMed] [Google Scholar]
- Ito L. S., Yamashita M., Takashima F., Strüssmann C. A., 2005. Dynamics and histological characteristics of gonadal sex differentiation in Pejerrey (Odontesthes bonariensis) at feminizing and masculinizing temperatures. J. Exp. Zool. 303A: 504–514. [DOI] [PubMed] [Google Scholar]
- Janzen F. J., 1994. Climate change and temperature-dependent sex determination in reptiles. Proc. Natl. Acad. Sci. USA 91: 7487–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Hamaguchi S., 2013. Novel sex-determining genes in fish and sex chromosome evolution. Dev. Dyn. 242: 339–353. [DOI] [PubMed] [Google Scholar]
- Li M., Sun Y., Zhao J., Shi H., Zeng S., et al. , 2015. A tandem duplicate of Anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile tilapia, Oreochromis niloticus. PLoS Genet. 11: e1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J., 1990. The transforming growth factor-β family. Annu. Rev. Cell Biol. 6: 597–641. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C., et al. , 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. [DOI] [PubMed] [Google Scholar]
- Mawaribuchi S., Yoshimoto S., Ohashi S., Takamatsu N., Ito M., 2012. Molecular evolution of vertebrate sex-determining genes. Chromosome Res. 20: 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D., Summers J., Silber S., 2004. Environmental vs. genetic sex determination: a possible factor in dinosaur extinction? Fertil. Steril. 81: 954–964. [DOI] [PubMed] [Google Scholar]
- Myosho T., Otake H., Masuyama H., Matsuda M., Kuroki Y., et al. , 2012. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191: 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. S., 2006. Fishes of the World. John Wiley & Sons, Hoboken NJ. [Google Scholar]
- Pfennig F., Standke A., Gutzeit H. O., 2015. The role of Amh signaling in teleost fish – multiple functions not restricted to the gonads. Gen. Comp. Endocrinol. 223: 87–107. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M., 1987. The neighbor–joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sparks J. S., Smith W. L., 2004. Phylogeny and biogeography of the Malagasy and Australasian rainbowfishes (Teleostei: Melanotaenioidei): Gondwanan vicariance and evolution in freshwater. Mol. Phylogenet. Evol. 33: 719–734. [DOI] [PubMed] [Google Scholar]
- Strüssmann C. A., Ito L. S., 2005. Where does gonadal sex differentiation begin? Gradient of histological sex differentiation in the gonads of Pejerrey, Odontesthes bonariensis (Pisces, Atherinidae). J. Morphol. 265: 190–196. [DOI] [PubMed] [Google Scholar]
- Strüssmann C. A., Patiño R., 1999. Sex determination, environmental, pp. 402–409 in Encyclopedia of Reproduction, edited by Knobil E., Neill J. D. Academic Press, New York. [Google Scholar]
- Strüssmann C. A., Saito T., Usui M., Yamada H., Takashima F., 1997. Thermal thresholds and critical period of thermolabile sex determination in two atherinid fishes Odontesthes bonariensis and Patagonina hatcheri. J. Exp. Zool. 278: 167–177. [Google Scholar]
- Strüssmann C. A., Conover D. O., Somoza G. M., Miranda L. A., 2010. Implications of climate change for the reproductive capacity and survival of New World silversides (family Atherinopsidae). J. Fish Biol. 77: 1818–1834. [DOI] [PubMed] [Google Scholar]
- Takehana Y., Matsuda M., Myosho T., Suster M. L., Kawakami K., et al. , 2014. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 5: 4157. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitt U. A., Hsu S. Y., Hsueh A. J., 2001. Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Mol. Endocrinol. 15(5): 681–694. [DOI] [PubMed] [Google Scholar]
- Wilson C A., di Clemente N., Ehrenfels C., Pepinsky R. B., Josso N., et al. , 1993. Müllerian inhibiting substance requires its N-terminal domain for maintenance of biological activity, a novel finding within the transforming growth factor-β superfamily. Mol. Endocrinol. 7: 247–257. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Luckenbach J. A., Middleton M. A., Swanson P., 2011. The spatiotemporal expression of multiple coho salmon ovarian connexin genes and their hormonal regulation in vitro during oogenesis. Reprod. Biol. Endocrinol. 9: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Zhang Y., Sarida M., Hattori R. S., Strüssmann C. A., 2014. Coexistence of genotypic and temperature dependent sex determination in pejerrey Odontesthes bonariensis. PLoS One 9: e102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A., Guyomard R., Nicol B., Jouanno E., Quillet E., et al. , 2012. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22: 1423–1428. [DOI] [PubMed] [Google Scholar]
- Yano A., Nicol B., Jouanno E., Quillet E., Fostier A., et al. , 2013. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol. Appl. 6: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: GenBank accessions; H. tsurugae Amha (KU664386) and Amhy (KU664387), O. bonariensis Amha (AHG98063.1) and Amhy (AAV31752.2), O. hatcheri Amha (AEE60845.1) and Amhy (ABF47515.2), D. labrax Amh (CAJ78431.1), O. niloticus Amh (ABS58513.1), D. rerio Amh (NP001007780.1), and X. laevis Amh (BAO04196.1).