Abstract
Reactive oxygen species continuously assault the structure of DNA resulting in oxidation and fragmentation of the nucleobases. Both oxidative DNA damage itself and its repair mediate the progression of many prevalent human maladies. The major pathway tasked with removal of oxidative DNA damage, and hence maintaining genomic integrity, is base excision repair (BER). The aphorism that structure often dictates function has proven true, as numerous recent structural biology studies have aided in clarifying the molecular mechanisms used by key BER enzymes during the repair of damaged DNA. This review focuses on the mechanistic details of the individual BER enzymes and the association of these enzymes during the development and progression of human diseases, including cancer and neurological diseases. Expanding on these structural and biochemical studies to further clarify still elusive BER mechanisms, and focusing our efforts toward gaining an improved appreciation of how these enzymes form co-complexes to facilitate DNA repair is a crucial next step toward understanding how BER contributes to human maladies and how it can be manipulated to alter patient outcomes.
Keywords: Base excision repair, Oxidative DNA damage, DNA repair, Review
2. INTRODUCTION
2.1. Oxidative DNA damage
Reactive oxygen species (ROS) are generated as a by-product of normal mitochondrial activity, these include superoxide, hydrogen peroxide, and the hydroxyl radical. The metabolic processes that generate ROS are essential for the cell, however a tradeoff is involved, and if not properly controlled the ROS concentration can exceed the antioxidant scavenging ability of the cell. Under this cellular condition, termed oxidative stress, ROS can cause extensive damage to cellular macromolecules. The structure of DNA is especially vulnerable to damage, and it has been estimated that DNA damage occurs at a rate of 104 lesions/cell/day in humans, with oxidative DNA damage being particularly prevalent (1). Specifically, the structures of all four DNA nucleobases are susceptible to oxidative damage from ROS, with more than 100 different types of base damage being identified as products of oxidative stress (2). This base damage includes fragmented or ring-opened forms and oxidized aromatic derivatives. Several base lesions are highlighted in Figure 1 and discussed in more detail below. Generally, when the nucleobase structure is altered as the result of oxidative damage, its base-pairing properties are also altered, often leading to either transition (purine to purine or pyrimidine to pyrimidine) or transversion (purine to pyrimidine or pyrimidine to purine) mutations. Consequently, oxidative DNA damage is a major source of the mutation load that gives rise to numerous human maladies (discussed in section 6).
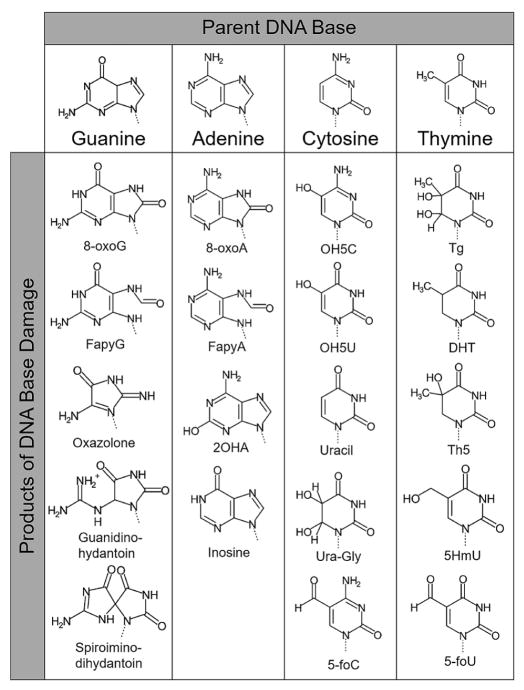
Figure 1.
A compilation of biologically significant base lesions organized by parent base. All lesions or several ROS mediated attacks, and/or other damage related to oxidative stress. Specifically, cytosine can give rise to uracil via deamination by N2O3, an oxidized nitric oxide, and a hydrolysis reaction (27).
Guanine, as the result of its low oxidation potential relative to the other nucleotides, is the most frequently oxidized nucleotide (3). A major oxidized form of guanine is 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxoG), and it arises by the introduction of an oxo group to the carbon at the C8 position and a hydrogen atom at the N7 position. Due to its stability and biological importance, 8-oxoG is one of the most extensively studied DNA lesions, and a majority of what is broadly known about how damaged DNA is handled and repaired is based on studies utilizing 8-oxoG. This lesion does not cause a significant block to DNA replication in mammalian cells, with the exception of DNA polymerase I (4). The replicative and repair DNA polymerases, including DNA polymerases δ, κ, β, λ, and γ, have been shown to efficiently insert either dATP or dCTP opposite a templating 8-oxoG in the syn- or anti- conformation, respectively. 8-oxoG(anti) can form a Watson-Crick base pair with cytosine analogously to guanine, while a rotation about its N-glycosidic bond presents the Hoogsteen edge of 8-oxoG(syn) to form a mismatched base pair with adenine (5). The stability of the syn-conformation of 8-oxoG in duplex DNA results in an increase in G:C → T:A transversion mutations during subsequent rounds of replication. The nucleotide form of guanine can also be oxidized, generating 8-oxodGTP. This provides further opportunities for 8-oxoG to arise in the genome, and once inserted by a DNA polymerase, both A:T → C:G and G:C → T:A transversions may occur during replication and repair. The 8-oxoG DNA lesion is also highly susceptible to further oxidative damage, yielding the additional mutagenic base lesions guanidinohydantoin and spiroiminodihydantoin, shown in Figure 1 (6, 7). The other major oxidative lesion of guanine, 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG), is produced by fragmentation of the purine imidazole ring (Figure 1). FapyG is also mutagenic, even slightly more so than 8-oxoG, producing predominately G:C → T:A transversions (8). FapyG is not as comprehensively characterized as 8-oxoG and we point the readers to a review focused on FapyG for more information (9).
The oxidation products of adenine are structurally similar to that of guanine, with the two major products being: 8-oxo-7,8-dihydro-2′-deoxyadenosine (8-oxoA) and 4,6-diamino-5-formamidopyrimidine (FapyA), both shown in Figure 1. The yields of 8-oxoA and FapyA are about ten-fold lower than the corresponding yields of 8-oxoG and FapyG (10). This difference in yields may be explained in part by the comparatively low oxidation potential of guanine, which can result in the transfer of an electron from guanine radicals in close proximity. The FapyA lesion is present in both normal and cancerous tissues and is the most abundant of the adenine lesions induced by radiation (11, 12). In vitro translesion synthesis experiments past FapyA have demonstrated adenine and guanine to be the most frequently incorporated bases opposite the lesion, suggesting induction of A:T → T:A and A:T → C:G transversion mutations (13). 8-oxoA, on the other hand, has been shown to induce predominately A:T → G:C transition and A:T → C:G transversion mutations (14). Both 8-oxoA and FapyA are considered less mutagenic than their guanine counterparts. Based on modeling studies, it has been proposed the less stable base stacking interactions with the oxidized adenine, as opposed to oxidized guanine, may be the cause for the reduced mutagenic potential (8, 15). A less prevalent adenine lesion, 2-hydroxydeoxyadenosine-5′-triphosphate (2OHA), Figure 1, produces mutations at a high rate through the misincorporation of 2OHA opposite cytosine, which generates G:C → A:T transitions upon replication (16).
Free radical attack of thymine occurs primarily at either the 5,6-double bond or the 5-methyl group, generating various lesions, including those shown in Figure 1 (17). The former converts the planar aromatic ring structure into a non-aromatic, non-planar structure. The most thoroughly examined oxidation product of thymine, which is generated by the ring opening attack of the 5,6-double bond, is thymine glycol (Tg). The Tg lesion is an effective block to replicative polymerases, but not to translesion DNA polymerases (η, κ, υ, β, and λ), resulting in a mutagenic signature indicative of translesion synthesis polymerases (18–21). Thymine can also be oxidized to form the 5,6-dihydrothymine (DHT), which despite being targeted for repair, does not appear to cause mutations or cytotoxicity (17). In contrast to free radical attack on the thymine 5,6-double bond, attack on the 5-methyl group generates several oxidation products with intact aromatic ring structures. These include 5-hydroxymethyluracil (5HmU), which is both mutagenic and cytotoxic (17). Mutagenicity is through T:A → C:G transitions resulting from its ability to base pair with both adenine and guanine. 5HmU base pairs with adenine utilizing Watson-Crick geometry and a stabilizing inter-residue hydrogen bond between the hydroxymethyl group and a neighboring 5′ guanine base. The mismatch with guanine, however, displays wobble base pairing stabilized by an intra-residue hydrogen bond between the hydroxymethyl and the O4 carbonyl group.
The 5,6-double bond is the sole oxidation target of cytosine, with a major oxidative product of 5-hydroxy-2′-deoxycytidine (OH5C in Figure 1), which is found in DNA both spontaneously and after exposure to ROS generating chemicals (17). Once in the DNA, OH5C is not correctly replicated past and leads to an increased frequency of C:G → T:A transition mutations compared to cytosine (17). On average, about five percent of the cytosine in human DNA is methylated, making 5-methylcytosine (5mC) a minor, but significant, component of mammalian DNA. DNA methylation is thought to play an important role in gene regulation, making the integrity of 5mC a potential player in the deregulation of genes in addition to mutagenic and cytotoxic effects. Although 5mC is a cytosine analogue, like thymine, it can be attacked by oxygen radicals at both the 5,6-double bond and the 5-methyl group. Behaving as a mixture of both cytosine and thymine in terms of oxidative damage, several oxidation products of 5mC have been described (17).
2.2. Mammalian base excision repair (BER): an overview
Considering the potential detrimental effects of ROS on DNA during oxidative stress, it is not surprising that sophisticated systems have evolved to thwart the deleterious effects of oxidative base damage. Such a system for removing 8-oxoG, termed the “GO-system”, was originally described in Escherichia coli. The GO-system involves DNA repair enzymes mutT, mutM and mutY (22). MutT attempts to cleanse oxidized nucleotides, including 8-oxodGTP and 2OHdATP, from the nucleotide pool (23, 24). If the 8-oxodGTP manages to escape this cleansing, it can be inserted into nascent DNA by a polymerase during DNA replication and repair. Therefore, additional levels of protection are necessary to protect the cell from the mutagenic effects of 8-oxodGTP and 8-oxoG that is generated within duplex DNA. The DNA glycosylase mutM removes the oxidized base when 8-oxoG is base paired to cytosine in duplex DNA, while mutY is tasked with the removal of adenine mispaired opposite 8-oxoG. The mammalian version of these enzymes (OGG1, MTH1, and MUTYH) are discussed in subsequent sections. The analogous mammalian system, for the repair of oxidatively damaged DNA bases, is primarily accomplished through the base excision repair (BER) pathway.
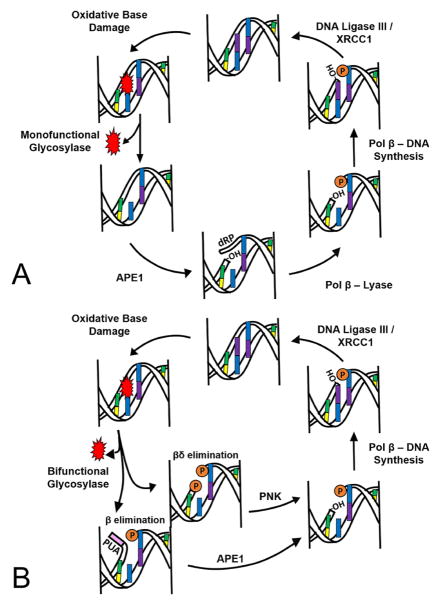
In the initiation step of BER, a damage specific DNA glycosylase identifies and removes the damaged base from the DNA. In general, DNA glycosylases can be classified as either monofunctional or bifunctional (Table 1, and further discussed in section 3.1.). Figure 2A depicts a “classical” BER cycle in which repair of the oxidized base is commenced by a monofunctional glycosylase. During this process, a multi-protein complex removes DNA lesions that would otherwise potentially block replication or increase the prevalence of disease causing mutations within the genome. In this classical case, a glycosylase removes the damaged base through cleavage of the N-glycosylic bond between the target base and deoxyribose, leaving an intact AP site (a location in DNA that has neither a purine nor a pyrimidine base). This intact AP site is further processed by human AP-endonuclease 1 (APE1), which subsequently creates a nick in the DNA 5′ of the AP site resulting in 3′-hydroxyl and 5′-deoxyribosephosphate (dRP) termini. The 5′-dRP is removed by polymerase beta’s (pol β) lyase domain, and a new nucleotide is inserted into the gap by pol β’s nucleotidyl transferase activity. With the assistance of the scaffolding protein X-ray repair cross-complimenting protein 1 (XRCC1), ligase III seals the nick and generates a repaired BER product. In comparison, a bifunctional glycosylase initiates the BER cycle by removing both the damaged base and cleaving the DNA at the phosphodiester bond 3′ of the AP site via its AP lyase activity (either β or β,δ-elimination), creating a single-strand break (Figure 2B). The 3′-α,β-unsaturated aldehyde (PUA) that results from glycosylase β-elimination can be removed by the phosphodiesterase activity of APE1. If the bifunctional glycosylase utilizes a β,δ-elimination mechanism, the resulting 3′-phosphate group will be removed by polynucleotide kinase (PNK). Both APE1 and PNK leave a hydroxyl at the 3′ terminus, allowing for subsequent gap filling by pol β and ligation by ligase III/XRCC1 to occur as described above in classical BER (Figure 2). In the particular case of clustered oxidative lesions, long-patch BER is utilized, borrowing several enzymes from DNA replication pathways including DNA pols δ and/or ε, FEN1, PCNA and DNA ligase I (25). Understanding the mechanisms involved in oxidative base damage repair by BER and how perturbations in the BER enzymes can result in human disease is the combined focus of this review.
Table 1.
A summary of prominent mammalian DNA glycosylases
| Structural Motif Superfamily |
Name | Mono/ Bifunctional |
Oxidative Stress |
Amino Acid Wedge |
Target Examples | References | |
|---|---|---|---|---|---|---|---|
| Alpha-beta fold (UDG superfamily) | Uracil-N glycosylase | UNG | Mono | Yes | Leu272 | Uracil, uracil derivatives | (45, 119, 181) |
| Single-strand-specific monofunctional uracil DNA glycosylase 1 | SMUG1 | Mono | Yes | Residues 251–260 | Uracil, uracil derivatives | (29, 46, 182) | |
| Thymine DNA glycosylase | TDG | Mono | Yes | Arg275 | Thymine mismatch, uracil, uracil derivatives | (29, 182, 183) | |
| Helix-hairpin-helix | Methyl-binding domain glycosylase 4 | MBD4 | Mono | Yes | Arg468 | Thymine mismatch, uracil, uracil derivatives | (29, 119, 184) |
| 8-OxoG DNA glycosylase 1 | OGG1 | Bi | Yes | Asn149/Asn150 | 8-oxoG, FapyA, FapyG | (33, 119, 185) | |
| MutY homolog DNA glycosylase | MUTYH | Mono | Yes | Gln48 | Adenosine opposite 8-oxoG | (186, 187) | |
| Endonuclease III-like 1 | NTHL | Bi | Yes | Gln42 | Thymine glycol, FapyG | (188, 189) | |
| 3-methyl-purine glycosylase (MPG) | 3-methyl-purine glycosylase | MPG | Mono | No | Tyr162 | 3-methyladenine, 7-methylguanine, hypoxanthine | (119, 190, 191) |
| Hairpin-2- turn-hairpin (NEIL superfamily) | Endonuclease VIII-like glycosylase 1 | NEIL1 | Bi | Yes | Arg118 | Thymine glycol, FapyG, FapyA, 8-oxoG | (47, 192–194) |
| Endonuclease VIII-like glycosylase 2 | NEIL2 | Bi | Yes | Met72 | Thymine glycol, FapyG, FapyA, 8-oxoG | (192, 194, 195) | |
| Endonuclease VIII-like glycosylase 3 | NEIL3 | Mono | Yes | No wedge | FapyA, FapyG, spiroiminodihydantoin, guanidinohydantoin | (192, 196, 197) | |
Figure 2.
A representation of the base excision repair (BER) pathway for removing oxidative damage. (A) Classical BER cycle initiated by a monofunctional glycosylase and (B) a BER cycle started by a bifunctional glycosylase. The orange “P” represents a phosphate, -OH represents a 3′-hydroxyl and a 5′-2-deoxyribose-5-phosphate is represented by dRP. The 3′-phospho-α,β-unsaturated aldehyde (PUA) is in pink.
3. IDENTIFICATION OF OXIDATIVE DNA DAMAGE DURING BER
Due to the diversity in the size and shape of DNA lesions caused by oxidative stress (Figure 1), a varied array of DNA glycosylases has evolved to maintain genomic stability and combat the various base modifications. The seemingly impossible task of identifying subtle changes in nucleotide structure, arising from DNA damage, within the billions of non-damaged nucleotides is accomplished via unique mechanisms of glycosylase-dependent damage removal. In this section, the diversity of mechanisms that DNA glycosylases utilize to detect and remove DNA damage is discussed.
3.1. Overview of mammalian DNA glycosylases
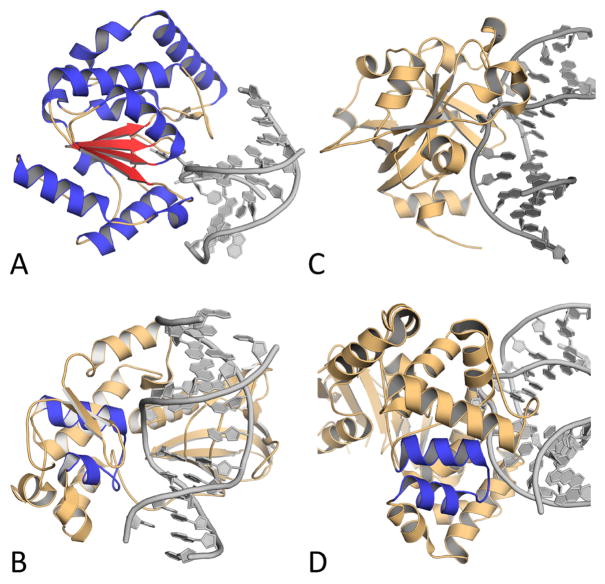
DNA glycosylases sort into four superfamilies based on their structural characteristics: uracil DNA glycosylases (UDG), helix-hairpin-helix glycosylases (HhH), 3-methyl-purine glycosylases (MPG), and endonuclease VIII-like glycosylases (NEIL). Figure 3 presents the characteristic structural fold of each superfamily. Members of the UDG superfamily all contain a characteristic α/β fold (Figure 3A) and some target the removal of uracil from the genome (26). Uracil can arise during oxidative stress when deaminated cytosine is generated by nitric oxide (27). UNG, SMUG, and TDG are members of the UDG superfamily. The HhH superfamily contains a characteristic helix transitioning to hairpin, transitioning to helix motif (Figure 3B). This family of glycosylases remove numerous lesions brought on by oxidative stress, including 8-oxoG, FapyA, FapyG, and Tg. Members of this superfamily include NTHL1, OGG1, and MUTYH (28). MPG (also known as AAG or MDG) is the sole member of the MPG fold superfamily, and sharply contrasts the other three groups. It contains none of the structural folds attributed to the other superfamilies and principally addresses alkylation rather than oxidative stress (Figure 3C). The final fold superfamily encompasses endonuclease VIII-like glycosylases (NEIL) with the structural fold of helix-two-turn-helix (Figure 3D). Members of this family include NEIL1, NEIL2, and NEIL3 (29). This group is highly relevant to oxidative stress, as it targets the common forms of oxidative base damage: 8-oxoG, FapyG, and FapyA. This redundancy in oxidative lesion targets between the HhH and NEIL superfamilies underscores the biological importance of removing oxidative DNA damage and the prevalence of these lesions in the genome.
Figure 3.
Images of representative members of each structural fold family, with the structural motifs highlighted. (A) Alpha-beta fold of UNG with alpha helices in blue and beta sheets in red. (B) Helix-two-turn-helix (H2TH) motif of NEIL1 colored in blue. (C) MPG fold containing none of the other motifs from MPG. (D) Helix-hairpin-helix (HhH) fold of OGG1 in blue. PDB accession codes are 1EMG, 5ITR, 1F6O, and 1EBM, respectively.
As shown in Figure 2, glycosylases utilize either a monofunctional or bifunctional mechanism, depending on which bonds are cleaved during the damage removal process (see Table 1). Monofunctional glycosylases do not cleave the phosphodiester backbone; only the N-glycosyl bond is cleaved during the removal of the damaged nucleobase. The mechanism, in short, can be described as a polarization of the N-glycosyl bond and an attack by an activated water molecule (29). Major members of the monofunctional mechanistic group are UNG, SMUG, MPG, MBD4, TDG, and MUTYH (30). In contrast, bifunctional glycosylases are more complex because the phosphate backbone is cleaved in addition to the N-glycosyl bond during damaged base removal (Figure 2B). The bifunctional group is further classified into β eliminators and β,δ eliminators (31). β eliminators cleave the phosphate backbone on the 3′ side of the resulting abasic site, whereas β,δ eliminators cleave the phosphate backbone on both the 5′ and 3′ sides of the abasic site, effectively forming a gap (Figure 2B). Mechanistically, for their AP lyase activity, bifunctional glycosylases use a conserved nucleophilic lysine residue to form a Schiff base, cleave the backbone, and eliminate the damaged nucleotide (29). The β eliminating glycosylases include OGG1 and NTHL1, and prominent β,δ eliminating glycosylases include NEIL1, NEIL2, and NEIL3 (31).
3.2. Detection of damaged bases by DNA glycosylases
In a sea containing billions of non-damaged nucleotides, that are in many cases only one atom different than their preferred target, the ability of glycosylases to find and excise their precise substrate has intrigued researchers for decades. Offering an idea of the scale of this daunting task, it was estimated by Friedman and Stivers that each individual glycosylase molecule must analyze 70,000 base pairs of DNA to fully cover the genome (30). The difficulty of this task is compounded by the relatively low damage rate during normal cellular conditions, as is it estimated that only one out of every 30,000 bases that are scanned is damaged (30). This challenge is primarily addressed through the ability of glycosylases to sample various conformational states depending on the stage of the reaction.
Generally, glycosylases exist in three different conformations: the “searching complex”, the “interrogation complex”, and the “excision complex”. While scanning the DNA, the primary conformation is the searching complex (30). This conformation is optimized to slide up and down the DNA helix while weakly interacting with the bases and causing a bend in the DNA reminiscent of the distorted shape created by an oxidative lesion (32). When a glycosylase encounters an oxidized base, it shifts into the interrogation complex, promoting the base to flip out of the DNA helix and into the enzyme’s substrate binding pocket. It can then distinguish whether or not the base is damaged by the specificity binding pocket and shift to a new catalytically-active conformation. The final conformation in this scheme is the excision complex, where the N-glycosyl bond is polarized and cleaved according to each individual glycosylase’s respective mechanism, followed by the eventual return to the searching complex where the genome will continue to be analyzed (30).
Single molecule imaging by atomic force microscopy (AFM) determined that TDG and OGG1 (glycosylases targeting genomic uracil and 8-oxoG, respectively) bend the DNA during the scanning procedure to enhance residence time on sites of DNA damage (32). Specifically, OGG1 caused an extreme ~70° bending of the DNA localized to the point where the protein binds the DNA. This corroborated the conclusions made from the seminal OGG1 X-ray crystal structures produced by the Verdine lab that demonstrated a similar degree of bending (33). Although glycosylase conformational changes explain how many base lesions are detected, some glycosylases do not exhibit this phenomena, necessitating additional theories for how glycosylases detect DNA damage. One such theory capitalizes on the innate interference that oxidized nucleotides have on DNA mediated charge transfer.
For DNA glycosylases that have iron-sulfur clusters, scanning the DNA through electron transfer has been proposed to further optimize DNA damage detection (34). Initial oxidation of the iron-sulfur clusters from the +2 to +3 state is sparked by electron transfer from guanine radicals that arise as precursors to the more stable guanine required for oxidative each glycosylase’s lesions during oxidative stress (Figure 1). Guanine radicals are one of the initial forms of oxidative damage, allowing this system to demonstrate an exquisite sensitivity for potential DNA damage (35). Support for the role of charge transfer in mediating glycosylase scanning was shown by measuring DNA binding affinity with highly oriented pyrolytic graphite electrochemistry, demonstrating a shift in the redox potential by 200 mV (36). This assay demonstrated that the affinity for duplex DNA increases by a factor of 1,000 when the glycosylase iron-sulfur clusters are oxidized to the +3 state (34). This implies that during times of high oxidative stress (i.e., when there would be increased DNA damage), more glycosylase molecules would be bound to the DNA to facilitate scanning. In this model, the electron from the original glycosylase is passed through DNA mediated charge transfer to a second glycosylase on the DNA. Only the pi-pi interactions of undamaged, stacked, nucleotides will allow the electron to traverse from protein to protein efficiently (37). When damage is present in the DNA, these interactions are distorted and the charge transfer fails between proteins. Thus, the rate of glycosylase disassociation is reduced and the localization to DNA damage is facilitated. This model would allow DNA mediated charge transfer to scan the DNA for damage rather than a glycosylase individually scrutinizing each base in the genome.
Experimental evidence for the charge transfer model has been observed using AFM. This assay allows for the visualization of individual molecules localized onto damaged DNA. By having both damaged and undamaged DNA in the same solution, it was observed that the glycosylases were much more attracted to the damaged strand, even though they could not repair it (34). To further test the sensitivity of DNA mediated charge transport, Barton et al. compared matched and mismatched DNA assembled on a gold electrode. They determined that a single mismatch, which distorts the DNA, greatly reduced charge transfer potential. This system of damage detection helps to effectively localize the glycosylases to regions of the genome that have undergone damage, particularly during times of high oxidative stress (38).
3.4. Excision strategies of DNA glycosylases
Glycosylases largely utilize base flipping to insert a damaged base into their active site binding pocket, where it is positioned for cleavage of the N-glycosyl bond. To perform a base flip, the DNA hydrogen bonds are broken along with the pipi interactions of bases flanking either side of the damage, swinging the base moiety 180° about the axis of the phosphate backbone (30). The flipped base or lesion then moves into a conserved binding pocket, which contains variable sequence regions that provide the specificity respective target (39). The enzyme then “wedges” an amino acid (see Table 1) into the DNA, stabilizing the resultant orphan base (39). This wedge helps to lower the energy barrier for base flipping. In TDG specifically an arginine residue (Arg275) wedges into the void left by the flipped base. Recently obtained structures show the carbonyl oxygen of Ala274 within hydrogen bonding distance of the orphan base, and Arg275 moving in to stabilize phosphates on both sides of the resulting abasic (AP) site (32, 40). Mutating the wedge amino acid in UNG caused a drastic decrease in catalytic efficiency (kcat/KM) of the cleavage reaction, demonstrating the essential nature of the wedge stabilization. See Table 1 for a description of several amino acid wedges used during glycosylase excision reactions (33, 41–47).
Although the vast majority of glycosylases exhibit similar mechanisms, alternative mechanisms do exist that are worth mentioning. For example, studies of AlkD revealed a novel mechanism for damage removal where flipping does not occur (48). AlkD is a glycosylase from Bacillus cereus that binds alkylated purines. Rather than relying on the standard nucleobase binding pocket and intercalating amino acid wedge, Trp109 and Trp187 form CH–π interactions with the nucleotide. Using this alternative mechanism, AlkD is capable of excising larger lesions because it does not need to envelop the lesion within the binding pocket. It was suggested the binding pocket may have more to do with facilitating the removal of the base rather than increasing specificity (48). Sometimes two base flips occur, as in bacterial MutY, the homolog to mammalian MUTYH (49). In this mechanism, the damaged nucleotide (often 8-oxoG) flips into the binding pocket first. After this occurs, the adenosine base pairing with the damage is flipped into a second binding pocket, where it is excised. Unlike most glycosylases, the damaged nucleotide is not removed, as this would result in loss of genetic information. Instead, it gives 8-oxoG another chance to base pair with cytosine, and then be recognized by another glycosylase (e.g. OGG1). This elegant mechanism ensures accuracy as MUTYH removes an undamaged nucleotide that would have caused a transversion mutation if it were to remain unchecked.
From conformational changes tailored to recognize the kinks in DNA formed by damaged lesions to specialized electron transfers that identify faults in the stacking of bases, recent advances have begun unraveling the enigma of lesion detection by glycosylases. It is through understanding this initial step of BER that the mammalian response to oxidative stress can be better understood.
4. FURTHER PROCESSING OF THE AP SITE
Apurinic/apyrimidinic sites (AP sites) are among the most abundant oxidative DNA damage and can occur as the result of either spontaneous hydrolysis of the N-glycosyl bond or during removal of the damaged base by a DNA glycosylase, leaving the DNA phosphate (60). backbone APE1 is a processive enzyme that intact (as described in section 3). AP sites are cytotoxic to the cell by blocking DNA replication and transcription, and are mutagenic because the bypass of AP sites by a polymerase can result in base substitutions and insertions/deletions (50, 51). Additionally, the chemical reactivity of these AP sites can result in the formation of DNA breaks as well as DNA-protein and DNA-DNA crosslinks (52). The biological significance of AP sites and their repair is highlighted by the embryonic lethality of mammalian cells lacking APE1, the major BER enzyme responsible for incising AP sites. Further reinforcing the cytotoxicity of AP sites, it is believed the antitumor agent leinamycin functions by forming AP sites (53). Moreover, expression of an inactive variant of APE1, which results in the accumulation of AP sites, has been shown to enhance the effectiveness of DNA damaging have drugs (54). Hence, the repair of AP sites by BER is a key part of maintaining genome stability.
4.1. Endo- and exo-nuclease activities of APE1
APE1 is responsible for initiating the repair of AP sites, with greater than 95% of the AP cleavage activity in HeLa cell extracts being attributed to APE1 (55). After an oxidatively damaged base is removed by a monofunctional glycosylase in the BER cycle, APE1 steps in to cleave the DNA at the phosphodiester bond 5′ of the resulting AP site, leaving behind an intermediate with a 3′-hydroxyl and 5′-dRP termini (Figure 2A). The pre-steady state kinetic description of strand incision by APE1 is that rapid catalysis is followed by slow product release (56). Rapid catalysis is vital for genomic stability given the prevalence of AP sites in the genome, while the slow catalysis step may conceal cytotoxic incised BER intermediates during DNA-damage processing. Alternatively, when the damaged base is removed by a bifunctional glycosylase it results in a nicked DNA intermediate with either a 3′-α,β-unsaturated aldehyde or a 3′-phosphate, and a 5′-phosphate. The 3′-end groups resulting from bifunctional glycosylases can be converted into a 3′-hydroxyl by APE1, resulting in an alternative intermediate with a 3′-hydroxyl and a 5′-phosphate (Figure 2B). The resulting nicked DNA BER intermediates are substrates for downstream BER processing by pol β, as described in section 4.2.
X-ray diffraction and site-directed mutagenesis experiments have shown that APE1 is composed of a rigid globular nuclease domain and a flexible N-terminal domain (57–59). The N-terminal domain is responsible for the redox activity of APE1 and is thought to mediate alternative APE1 functions, whereas the C-terminal domain is responsible for DNA binding and the AP endonuclease activity. The endonuclease domain of APE1 belongs to the phosphoesterase superfamily of enzymes that contain a common four-layered α/β sandwich structural core and bear variable loop regions and active site characteristics to provide substrate specificity binds to the DNA and slides along the strand in search of an AP site primarily through interacting with the DNA phosphate backbone (61, 62). The original crystal structures of APE1 complexed with DNA revealed a “flipped out” AP site, reminiscent of that observed in glycosylases, positioned within a baseless pocket stabilized by four loops and an α-helix, leaving an orphan base in the opposite strand (59). Specifically, APE1 is proposed to stabilize the flipped out abasic residue via a double-loop mechanism involving interactions with both the minor and the major grooves at the AP site. The other two loop domains also interact with DNA on either the 5′ or the 3′ side of the AP residue to facilitate the formation of a stable APE1:DNA complex. The role of these additional loop domain interactions and active-site residues in defining substrate specificity further characterized using site-specific APE1 variants (63, 64).
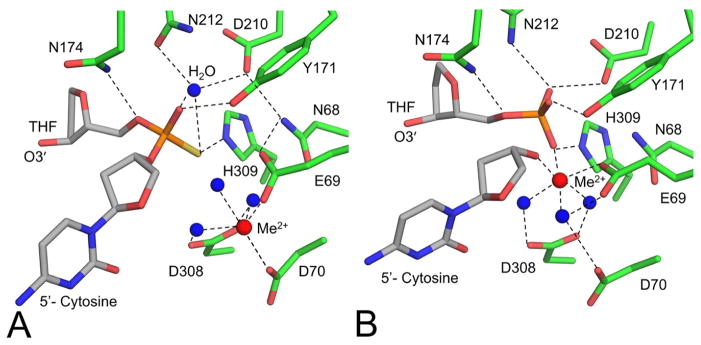
More recent high-resolution APE1 product and substrate structures have provided additional mechanistic details of the APE1 endonuclease reaction (58). Figure 4A and 4B highlight the APE1 active site residues of these structures in complex with DNA containing tetrahydrofuran (THF, a stable AP site analog). These structures further elucidated the mechanism of cleavage by APE1. Prechemistry snapshots identified a single Mg2+ is coordinated by Asp70, Asp96, and a water that is in contact with the non-bridging oxygen of the phosphate as shown in Figure 4B. Additionally, the nucleophilic water is in positon for inline attack on the phosphorus atom and is highly coordinated by Asn212 and Asp210. The structures imply a pentacovalent intermediate that is stabilized by Mg2+ and key active site contacts. It has been proposed that during catalysis the metal shifts to coordinate a phosphate non-bridging oxygen and the newly generated O3′ (58). In addition to clarifying the mechanism of APE1, the high-resolution product structure identified novel contacts that mediate product release during APE1 catalysis. Notably, product release has been kinetically demonstrated to be the rate limiting step of the reaction, and it is thought that the enzyme evolved slow product release to protect the cell from the potentially cytotoxic nicked intermediate while the DNA is passed to the next enzyme, pol β, in the BER pathway.
Figure 4.
High resolution substrate and product structures of APE1 led to a refined mechanism of strand cleavage. (A) Active site of APE1:DNA substrate complex (PDB code 5DG0). (B) Active site of APE1:DNA product complex (PDB code 5DFF). Waters are shown as blue spheres and metals are shown as red spheres. Tetrahydrofuran, a stable AP site analog, is labeled as THF.
Importantly, strand breaks with damaged 3′ DNA termini are created spontaneously by ROS and are also generated during BER reactions as intermediates, Figure 2B. Failure to resolve these damaged “dirty-ends” can lead to errors and/or blocks during DNA replication and repair, and are consequently associated with genomic instability and mutagenesis. In addition to its AP endonuclease activity described above, APE1 has also been shown to have 3′ → 5′ exonuclease activity, and it has been suggested that this exonuclease activity may be involved in cleaning up DNA dirty ends. For example, 3′-phosphoglycolate and 3′-phosphate sugar residues can be generated by either ROS or an AP lyase at a strand break. Both of these 3′-end groups can be recognized and removed by APE1 (65–67). APE1 has also been shown to remove oxidized bases and mismatches from 3′-ends of DNA (68–71). The strategy by which APE1 modifies damaged 3′-termini is likely analogous to that observed for its AP-endonuclease reactions; however, the current lack of structural evidence prevents the elucidation of the precise catalytic mechanism.
4.2. Lyase and polymerase activities of pol β
It is the job of the multifunctional enzyme pol β to further process the nicked BER intermediates generated by oxidative base removal and subsequent AP site cleavage. Pol β, at 39 kDa, is the smallest of the eukaryotic polymerases, and has both 5′-dRP lyase and nucleotidyl transferase activities. Pol β’s 8 kDa N-terminal domain contains the 5′-dRP lyase activity, while pol β’s polymerase activity resides in the 31 kDa C-terminal domain (Figure 5A). Incision of the AP site by a bifunctional glycosylase results in an intermediate with a 5′-phosphate that does not require the lyase activity of pol β (Figure 2B). In contrast, the AP site incision by APE1 generates a DNA nick bearing a 5′-dRP group that must be further tailored. The pol β lyase activity excises this dRP group and leaves a 1-nucleotide gap with a 5′-phosphate. The processed gapped DNA containing a 5′-phosphate is filled by pol β’s polymerase activity, resulting in a 3′-nick substrate that is sealed by the DNA ligase III/XRCC1 complex.
Figure 5.
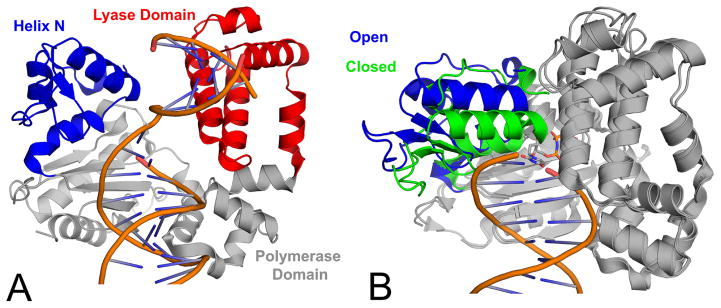
Structural overview of pol β. (A) A ribbon representation of the pol β binary complex (PDB code 3ISB) highlighting the polymerase domain in gray, lyase domain in magenta, and helix-N in blue. (B) Pol β open binary complex (PDB code 3ISB; blue helix-N) superimposed with the ternary complex (PDB code 2FMS; green helix-N). The DNA is truncated in panel B for clarity.
Pol β catalyzes the removal of the 5′-dRP from the cleaved AP site through a metal independent reaction involving lysine mediated nucleophilic attack on C1′ of the sugar to form a Schiff base intermediate (72, 73). Nucleophilic attack is followed by a β-elimination reaction and release of dRP via hydrolysis. Crystal structures of pol β bound to product and substrate DNA demonstrate that for these molecular contacts to occur the DNA is bent ~90° across the surface of the protein, positioning the DNA for optimal enzymatic activity (74). These structures also show that the 5 α-helices that constitute the lyase domain form a lysine rich pocket that engages the 5′-end of the DNA. Site-directed mutagenesis and mass spectrometry experiments both suggest that Nε of Lys72 within the helical pocket acts as a potential nucleophile in the lyase reaction (75, 76). However, a crystal structure of pol β bound to nicked DNA containing a dRP analog THF suggests that the proposed nucleophile, N of Lys72, lies further than thought from the 5′-dRP and suggests that a 120° rotation of the flexible dRP group about its 3′-phosphate occurs to reposition the lesion for catalysis (77). Importantly, if the 5′-dRP is not removed before the ligation step, the DNA ligase reaction is inhibited, which can result in abortive ligation and the formation of a 5′-AMP-dRP group. Pol β was shown to be able to remove the 5′-AMP-dRP group from substrates that mimic BER intermediates after abortive ligation using its dRP lyase activity, albeit the reaction is weaker than that for the unmodified 5′-dRP (78).
Pol β uses its DNA synthesis activity to fill the single nucleotide gap that now occupies the position where the oxidized base was, generating nicked DNA that will successively be ligated to restore the native DNA structure. Pol β binds the gapped DNA substrate in an open conformation. In this binary, gapped DNA complex, the DNA is sharply bent about 90° at the 5′-phosphodiester bond of the templating base as it enters the polymerase active site exposing the primer terminus base pair situated in the gap (79). A conformational change of the N-subdomain, and specifically helix-N, occurs upon the binding of the incoming nucleotide in which the enzyme transitions from an open to closed conformation around the nascent base pair (Figure 5B). This closed, pre-catalytic, ternary complex, primed for nucleotidyl transfer, contains both the catalytic and nucleophilic divalent cations. Of note, the role of additional metal ions has been revisited with the development of time-lapse crystallography using pol β. These non-traditional metal ions are an active area of research and their mechanistic implications are still being elucidated (80–82). Among subtle conformation changes that occur, is the repositioning of the 3′-OH of the deoxyribose at the primer terminus for in-line attack on the α-phosphate (Pα) of the incoming dNTP. Upon catalysis, pyrophosphate (PPi) is formed and then released when pol β re-opens.
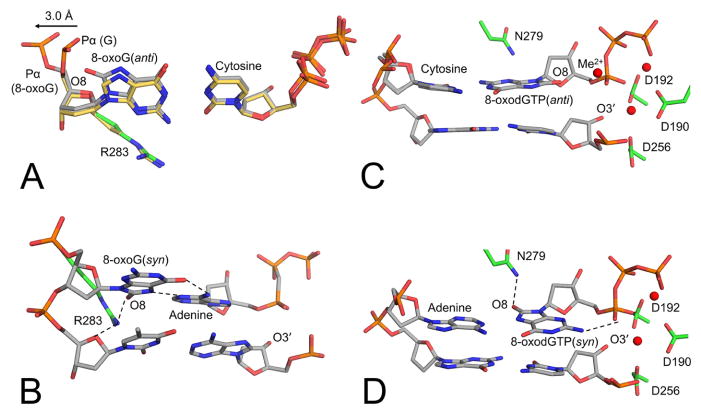
Importantly, pol β has been utilized to elucidate essential mechanistic information on how DNA polymerases handle oxidatively damaged DNA. This includes scenarios in which pol β is tasked with gap filling opposite a damaged base and when the damaged base is in the incoming nucleotide. When the damaged base, 8-oxoG, is in the templating positon of single nucleotide gapped DNA, pol β binds to form an open binary complex with the phosphate backbone, and 8-oxoG is in equilibrium between the anti- and syn- conformations (83). Nucleotide binding causes pol β to undergo closure, resulting in a stable ternary complex poised for insertion that alters the 8-oxoG conformational equilibrium. In the case of an incoming dCTP, as shown in Figure 6A, the equilibrium is shifted to the 8-oxoG(anti) conformation and is stabilized by Watson-Crick hydrogen bonding interactions and movement of the phosphodiester backbone away from the adducted oxygen (O8). In the case of an incoming dATP, the transition to 8-oxoG(syn) requires contacts with both the active site and the incoming dATP (Figure 6B). This is evidenced by the inability of the incoming dATP to stably form a Hoogsteen base-pairing interaction with 8-oxoG in the absence of the Arg283 residue, which is a key minor groove contact in the pol β active site. During subdomain closure, wild-type pol β captures 8-oxoG through hydrogen bonding interactions between Arg283 and O8 of 8-oxoG, which is only observed when 8-oxoG is in the syn-conformation. This syn-conformation is further stabilized by the incoming dATP Hoogsteen base pairing with the templating base to form the final ternary complex poised for mutagenic insertion (84). Overall, the structures reveal that the template binding pocket will permit 8-oxoG to assume either the anti- or syn- conformations and encode incorporation of an incoming cytosine or adenine, respectively. Of note, only a minor change in the phosphate backbone conformation of the templating 8-oxoG is required to relieve the steric clash of O8 with the sugar-phosphate backbone in the anti-conformation.
Figure 6.
Pol β pre-catalytic complexes demonstrating varying strategies used to accommodate the oxidized DNA lesion, 8-oxoG. (A) 8-oxoG(anti) in the templating positon opposite cytosine (PDB code 3RJI, gray carbons) overlaid with the guanine(anti):cytosine (PDB code 2FMP, yellow carbons). (B) 8-oxoG(syn):adenine with 8-oxoG in the templating position (PDB code 3RJF). (C) Incoming 8-oxodGTP(anti) opposite cytosine (PDB code 4UBC). (D) Incoming 8-oxodGTP(syn) opposite adenine (PDB code 4UAW). Metals are shown in red.
Alternatively, in the case of insertion of the oxidized nucleotide 8-oxoGTP, different structural rearrangements are needed for nucleotidyl transfer, Specifically as shown in Figure 6C and 6D (82). intermolecular stabilizing hydrogen bond between Pα with N2 occurs during the mutagenic insertion of 8-oxodGTP(syn) (Figure 6D). Alternatively, an additional divalent metal cation interacts with Pα of 8-oxodGTP(anti) during its non-mutagenic insertion opposite cytosine, effectively alleviating the clash between O8 and Pα without the Pα having to be repositioned, as observed in the templating 8-oxoG(anti) (compare Figure 6C and 6D). With either templating base, post-catalysis the hydrogen-bonding interactions between the bases are lost and the polymerase reopens, similarly to as it does with an incorrect (non-damaged, mismatch) insertion, suggesting 8-oxoGMP promotes genome instability via exposing the cytotoxic nicked DNA repair intermediate (80, 82). The biological implications for the insertion of oxidized nucleotides is an area of active research by multiple groups to target the DNA damage response in cancer cells (85–87).
4.3. Ligation and final repair of DNA
Ligation during BER occurs via a DNA ligase III/XRCC1 complex following pol β nucleotide insertion and generation of a 3′ nicked BER intermediate. If pol β were to insert an incorrectly matched or oxidatively damaged nucleotide into the gapped DNA, the resulting nicked product can potentially be passed on to DNA ligase III for nick sealing. However, the presence of the modified or mismatched base pair at the 3′-end of the nick could lead to ligation failure and formation of an abortive ligation product with a 5′-adenylate (5′-AMP) group (88–90). Furthermore, if pol β does not remove the 5′-dRP of an AP site using its lyase activity prior to the ligation step, DNA ligases can generate abortive ligation products with a 5′-adenylated dRP containing BER intermediate (78). A similar situation can occur when a ligase is confronted with DNA single-strand breaks resulting from direct oxidation, such as DNA nicks with 3′-AP sites and RNA DNA junctions that occur during ribonucleotide excision repair (65). These 5′-adenylated BER intermediates with 3′-damaged bases could potentially become mutagenic or cytotoxic, leading to abnormal DNA replication and/or double-strand breaks. Therefore, the efficient repair of the 5′-adenylated BER intermediates by DNA end- to sites processing enzymes is critical to cell viability and genomic stability during oxidative stress.
It is possible that the presence of a damaged base 3′ the DNA nick after pol β gap-filling ligation failure could lead to BER pausing. Moreover, in contrast to the insertion of a normal guanine, after 8-oxodGMP is inserted, pol β’s active site rapidly opens and the Watson-Crick base pairing interaction is lost (82, 84). This conformational change could possibly disrupt channeling of the substrate from pol β to DNA ligase in the BER pathway and consequently alter the normal coordination during repair. It is also conceivable that the BER intermediate possessing the 5′-AMP group could serve as a signaling mechanism to trigger recruitment of DNA end-processing enzymes (88). After end-processing, pol β would have another opportunity at the gap-filling is inserted, the DNA ligase would ideally be able to successfully join 5′-phosphate and 3′-OH groups to complete the repair.
Aprataxin (APTX), a member of the histidine triad (HIT) superfamily, resolves the abortive DNA ligation products by 5′-AMP removal, thereby restoring the 5′′-phosphate group at the nicked DNA terminus, and allowing another attempt at the ligation (88, 91). Alternatively, long-patch BER can be utilized to remove 5′-end-blocking lesions via FEN1’s flap excision activity (25). Other mechanisms proposed to remove blocked 5′-ends involve the lyase activity of polynucleotide kinase phosphatase (PNKP) or Ku70/80 (92, 93). Additional repair mechanisms serve to resolve the problem of the 3′-end damage that leads to abortive ligation. These enzymes include DNA glycosylases, APE1, APE2, and tyrosyl-DNA phosphodiesterase 1 (Tdp1) (65). Tdp1 is a general 3′-end-processing DNA repair enzyme that acts on mismatched 3′-ends of DNA substrates (94). As discussed in section 4.1., it has been postulated that APE1 may be involved in BER proofreading of DNA pol β. XRCC1 could also play a role in the recruitment of DNA end-processing proteins involved in reversal of impaired BER due to lack of normal coordination between pol β and DNA ligase in the last step of the BER pathway (88).
5. BER MULTI-PROTEIN CO-COMPLEXES
5.1. Scaffolding proteins
BER is a complex process that requires multiple proteins to protect the DNA damage intermediates generated during the course of repair. As would be expected with such a complex process, there are proteins that help to recruit and provide scaffolding roles. A key protein involved in the recruitment and signaling to other DNA repair proteins is poly(ADP-ribose) polymerase 1 (PARP-1). In response to oxidative stress, PARP-1 binds efficiently DNA breaks, nicks, gaps, and other lesions, which stimulates the addition of repeating ADP-ribose units, or “PAR” chains as a post-translational modification. This assembly of PAR chains by PARP-1 is achieved by transferring the ADP portion of NAD+ to an acidic residue via a unique O-glycosidic ribose-ribose bond (95). The accumulation of PAR chains, especially on PARP-1 itself (and its homolog PARP-2), promotes the recruitment of BER components such as XRCC1, APE1, pol β and DNA Ligase III (96–100). While 90% of PAR chains occur on PAR itself, the other 10% of PAR modifications are predominantly placed on histones and chromatin-associated proteins, which alter DNA super structure and increase accessibility of BER enzymes to the DNA (101–103). In addition, studies performed with purified PARP-1 and APE1 revealed that PARP-1 is able to stimulate APE1 strand incision activity (104). The multifunctional role of PARP-1 in recruiting and modifying the BER complex makes this protein a complex scaffolding-like enzyme, which has resulted in a plethora of studies aimed to elucidate its molecular functions.
The other key scaffolding protein during BER is XRCC1. XRCC1 is a non-enzymatic scaffolding protein that has been proposed to be dependent on PARP-1 recruitment in order to repair oxidative purine base damage, such as 8-oxoG (105). XRCC1 also interacts with itself and rapidly accumulates at sites of DNA damage. XRCC1 interacts with, stabilizes, and stimulates BER repair complexes through interactions with its N-terminal domain and two BRCT (BRCA1 C-terminal) domains (106–108). The N-terminal domain (NTD) binds pol β with high affinity (109). X-ray crystallography studies of XRCC1 show that the NTD of XRCC1 interacts exclusively with the thumb subdomain of pol β and that the architecture of this complex excludes direct interactions between XRCC1 and the DNA in the immediate vicinity of the gap (110). The study also revealed that an oxidized form of XRCC1-NTD exhibits enhanced affinity for pol β and suggested that the formation of a disulfide bond could regulate repair pathways involving the XRCC1/pol β complex. It has been demonstrated that for efficient ligation, XRCC1 forms a strong complex with DNA ligase III that is mediated by interactions between the C-terminal BRCT domains on both proteins (111). The structural architecture of this interaction was further explained by X-ray crystallography structures, which revealed the structural basis of BRCT-mediated dimerization (112).
5.2. Substrate channeling
Importantly, while each step of BER can be performed in isolation in vitro, biochemical and structural biology studies support a model in which the proteins of BER channel the DNA in a concerted, cooperative fashion (59, 113, 114). It is believed that BER enzymes facilitate the channeling by forming a co-complex which passes cytotoxic DNA intermediates Specifically step-by-step throught here pair path way. biochemical studies with purified human BER enzymes revealed substrate channeling from APE1 to pol β and then to DNA ligase by preloading the enzyme(s) onto DNA and initiating the reactions in the presence of a trap, designed to prevent any unbound enzyme from participation in the reaction (115, 116). After APE1 cleaved the AP site, pol β performed its dRP removal and gap-filling DNA product was channeled to the ligation step where DNA ligase catalyzed the phosphodiester bond formation between the 3′-OH and 5′-phosphate groups of the DNA nick. These studies provided the first direct evidence of substrate channeling in vitro. Additionally, a direct physical interaction between glycosylase MUTYH and APE1 has been demonstrated through co-immunoprecipitation and GST-pulldown assays, and more recently NMR chemical shift perturbation experiments were used to identify APE1 residues that contact the flexible inter-domain connector (IDC) region of MUTYH (117, 118). Looking at a subset of different glycosylases, pre-steady state fluorescence kinetic analyses indicated that the direct transfer of DNA via transient DNA glycosylase and APE1 protein-protein interactions may be universal (119). Recent fluorescence and light scattering binding studies provided direct physical evidence for the formation of a very stable complex between XRCC1 and pol β (120). Combined, these studies indicate that substrate channeling occurs in vitro under controlled conditions. It remains to be discovered if this phenomenon also occurs in vivo.
6. BER AND HUMAN DISEASE
Genetic stability is of paramount importance, not only for the avoidance of disease, but also for cell survival itself. The BER pathway, which is the cell’s primary tool for repairing oxidative DNA damage, is so vital for life that it has proven difficult to directly relate the ablation of an individual protein in the pathway to a distinct disease, as attempts to generate homozygous knockout models of many BER enzymes quickly results in the organism’s death (121). That being said, experimentalists have found ways to alter expression levels of BER enzymes, and epidemiologists have correlated aberrant BER function with a propensity towards a handful of human diseases. These studies have underscored the biological importance of BER during the oxidative stress response, as discussed here.
6.1. BER knockout models
A majority of mouse models in which both of the alleles for a single glycosylase have been knocked out produce viable offspring, and surprisingly show minimal phenotypic deviations (121). As an attempt to explain the viability of these knockouts, it has been proposed that the redundant nature of glycosylases, in terms of DNA damage targets, causes the loss of a single glycosylase to be compensated for by another. For instance, OGG1, NTHL1, NEIL1, NEIL2, and NEIL3 all target Fapy lesions (Figure 1 and Table 1). In contrast, TDG has proven to be the exception to this trend because a homozygous knockout resulted in an embryonic lethal phenotype. Increasing evidence is arising to support it is likely TDG’s epigenetic function that causes this lethality as opposed to its role during BER (122, 123). The detrimental effects were seen when glycosylase knockout models were laden with increased oxidative stress or in cases in which multiple glycosylases were knocked out. MPG (the primary glycosylase dealing with alkylated damage) knockout mice were put under conditions of high oxidative stress and exhibited an elevated susceptibility to colon cancer and a rise in DNA damage levels, possibly implicating MPG in the oxidative stress response (124). Simultaneously knocking out OGG1 and MUTYH, both of which prevent transversions caused by 8-oxoG, showed increased incidence of small intestine and lung cancer in mice. This further supports the idea that redundancy among glycosylases may explain why single knockouts are viable and underscores the notion that the biological response to oxidative stress has evolved to have redundant enzymes to handle the genomic burden that oxidative DNA damage places on the genome (125).
It has proved more difficult to create homozygous knockout models of the BER proteins downstream of glycosylases (Figure 2), which is in part explained by the lack of mechanistic redundancy for these enzymes. APE1 homozygous knockouts are embryonic lethal, for instance (121). However, a mouse model where a single allele of APE1 has been knocked out was created to determine the effects of sharply decreased APE1 expression; the study found up to a 5-fold increase in microscopic tumor formation (126). Furthermore, this mouse model presented increased liver apoptosis when exposed to 2-nitropropane, which generates oxidative stress (127). Similar to APE1, the pol beta; homozygous knockout mice have an extremely short lifespan of only a few hours (128). Instead of a knockout, a model was created with a variant of pol β (Y265C) that impaired its polymerase activity, and although a majority died quickly after birth, the survivors showed an increase in DNA damage, particularly double strand breaks. Additionally, these mutant mice were sensitive to methyl methanesulfonate (MMS), a DNA damaging agent (128). Even after expression of the mutant phenotype was ablated, the cancerous phenotype lingered, implying that the cellular transformation was due in part to mutations created by mutated pol β (129). Knocking out the scaffolding protein XRCC1 in mice is also embryonic lethal, but studies with one allele knocked out show similar phenotypes to other essential BER proteins: a largely normal phenotype with an increased sensitivity to DNA damage (130, 131). The body of work as a whole stresses the vital importance of having functional BER capacity, but also that even large reductions of functionality can be lost with little consequence during times of low oxidative stress.
6.2. BER’s role in neurological disorders
Around 20% of oxygen taken in by the body is used for cerebral activities (132). This high rate of oxygen consumption requires functional BER enzymes to repair potential lesions caused by an environment containing a high number of ROS. Alterations in BER function have been linked to Alzheimer’s disease (AD), Huntington’s disease (HD), and other neurological disorders. Patients with AD show decreased levels of OGG1 and higher levels of 8-oxoG, suggesting dysfunctional BER (133, 134). Other glycosylases have also been linked to the disease; post-mortem analyses of brain tissues from ten patients with sporadic AD showed a sharp decrease in protein levels of UNG (135). Contradictory results have been published on the role of APE1 in AD. One study, examining expression levels in the brain of ten AD patients and ten controls showed similar expression levels of active APE1 in both populations. (135). A contradictory study found increased expression levels of APE1 in the brain tissues of AD patients (136). It has been suggested that different tissue handling protocols could account for these discrepancies, and for now APE1’s involvement in the disease remains unclear (135).
Patients with AD display pol β expression levels of around 25% compared to healthy individuals within homogenized brain samples (135). In addition to expression levels, Weissman et al. analyzed overall BER pathway activity by measuring damage excision and gap filling capacities. Samples from patients with mild cognitive impairment (an AD precursor) had 0.38 times control activity, and AD was 0.12 times the control, highlighting the continued reduction of BER as AD progresses. These results are consistent with pol β knockdown models in an AD mouse that showed an increase in synaptic problems as observed in AD patients (137). An increase in PARP-1 has also been implicated in neurodegenerative diseases, along with other chronic health conditions (138–144). In cases of high oxidative stress, the increased PARP-1 response can be especially toxic in neuronal cells, causing necrosis (145). Because oxidative damage increases with age, the loss of neurons via this increase in the PARP-1 response to DNA damage has been proposed to be a source of the aging and AD phenotype (146). In both cases, PARP-1 inhibitors are a potential point of intervention to block the downstream effects of oxidative damage in the brain. These inhibitors have recently been shown the ability to modulate some effects of AD pathogenesis (147). Regulation of PARP-1’s function, via inhibitors, in DNA maintenance and nuclear signaling has made impacts in neurological disorders, and has the potential to aid in the treatments of other human diseases (148).
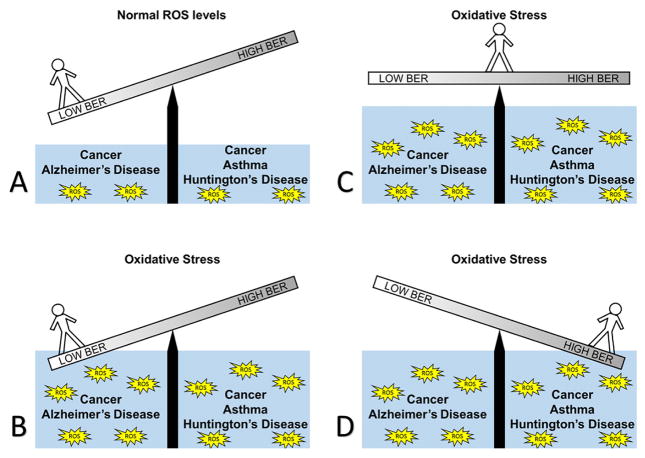
Although it appears that in AD, lower BER activity is correlated with BER expression, an entirely separate class of diseases exhibits nearly the exact opposite; namely, diseases caused by trinucleotide repeats (TNR), such as Huntington’s disease (HD). This emphasizes the importance of a balance in BER during the development of human diseases. Figure 7 illustrates the importance of the proper balance of BER activity during times of both high and low oxidative stress with an imbalance promoting adverse human health impacts. HD has been characterized by the excessive copying of trinucleotide CAG within the coding region of the HD gene, up to hundreds of thousands of times (149). The severity of the condition is dependent on the number of CAG repeats, with the longer the repeat the more debilitating the disease (149). Cell cultures isolated from HD patients were treated with hydrogen peroxide to induce oxidative stress conditions. These cells showed a marked increase in CAG expansion rate, highlighting the close relationship of this disease to oxidative stress. BER proteins were shown to promote CAG expansion in vitro using OGG1, APE1, and pol β. The in vitro results showed a direct CAG expansion in the strands containing regions with repeating CAGs compared to strands lacking the CAG repeats (149). Further exploration of repeat expansion system revealed that disrupting the coordination between pol β and FEN-1 increases repeat expansion (150). A proposed model for the mechanism of TNR expansion in HD is that two CAG repeats form a hairpin after 8-oxoG is removed from the template by BER. This hairpin escapes cleavage by FEN-1 and is stabilized by mismatch repair proteins MSH2 and MSH3. Subsequently, the nick is resealed, causing six nucleotides to be present in the DNA strand where there previously was only three. During somatic expansion, the daughter strand of DNA is replicated with the CAG repeats, and the repeat expansion cycle can continue during subsequent rounds (149). The implications for this model go beyond HD, as CAG repeats alone are responsible for many other diseases including: dentatorubral-pallidolyusian atrophy, spinal and bulbar muscular atrophy, and forms of spinocerebellar ataxia’s (151). Similar to CAG repeats, CTG repeats also cause a host of disorders, depending on where in the genome the repeat occurs. These diseases include: Fuchs endothelial corneal dystrophy, Huntington’s disease-like 2, myotonic dystrophy 1, and spinocerebellar ataxia 8 (151).
Figure 7.
An illustration of the importance of balance in BER. (A) In conditions of low oxidative stress, a person with low BER could still be disease free, as would a person with normal or high BER. (B) When oxidative stress rises, a person with low BER is at risk for disease; in this case, AD or cancer could result. (C) In conditions with high oxidative stress, a person with balanced BER can results in no disease. (D) In conditions with high oxidative stress and overactive BER, the balance shifts, and a person would have an increased risk of HD, cancer, and asthma.
Other notable diseases that underscore the intimate relationship of BER to oxidative stress include asthma and ischemia/reperfusion (I/R) events. Mouse models for asthma have shown that knocking down OGG1 decreased airway inflammation, alleviating asthmatic symptoms (152). Recent studies have further elucidated the relationship between ROS mediated inflammatory signaling and BER; namely, evidence for a role beyond damage excision for OGG1, where it forms a complex with 8-oxoG in order to activate small GTPases such as RAS and RHO (recently reviewed here (153)). During events where an organ loses blood supply, such as a heart attack or stroke, much of the tissue damage occurs during blood reperfusion in response to the flooding of oxygen to the tissue. In rats, mRNA analysis of OGG1 levels were shown to increase after an induced ischemia/reperfusion event (154). These higher expression levels of proteins involved in BER correlate with tissue and oxidative DNA base damage (155). The balanced and functional coordination of BER plays a key role in the prevention of disease within tissues that have a high oxygen exposure, such as lungs and airways, and also in tissues which require high oxygen consumption rates, such as neurological systems.
6.3. BER and Cancer
As cancer is caused by DNA mutations, it should come as no surprise that there is a link between the function of DNA repair enzymes, oxidative stress, and cancer. The maintenance of highly functional excision repair is reliant not only on the abundance of functional DNA repair proteins, but also upon the balance between these enzymes (Figure 7). Although mutations that reduce the functionality of key enzymes have been shown to predispose patients to certain cancers, epidemiological studies suggest that even single nucleotide polymorphism (SNPs) far from the active site of such enzymes can alter cancer rates. It is perhaps even more surprising to consider that the upregulation of DNA repair enzymes, such as APE1, can actually increase the rate of mutation, accentuating the fact that BER is not a risk-free procedure (156). Alternatively, in a cell’s efforts to conserve one nucleotide in a gene, single strand breaks (among other highly reactive species) are created, which pave the way for even more catastrophic double strand breaks when a balance between the repair enzymes is not maintained (see Figure 7).
Missense and nonsense mutations in MUTYH (the glycosylase that removes adenosine opposite 8-oxoG, see Table 1) can result in MUTYH-associated polyposis, which is characterized by an increased propensity towards forming polyps in the colon caused by a variety of missense and nonsense variants in the MUTYH gene (157). These polyps are often precursors to tumors, therefore the 1% of the population with MUTYH mutations are considered to be at an increased risk for developing colon cancer. Mechanistically, G:C → T:A transversion mutations are presumably brought on by elevated levels of 8-oxoG, thus highlighting the relationship between BER, oxidative stress and cancer (157). The D239Y variant of NTHL1 (Table 1) occurs in around 6.2% of the population, resulting in loss of activity, while still retaining the ability to bind the oxidatively damaged DNA. This leads to blocked replication, chromosome damage and a cancerous phenotype (158). Mutations in UNG (Table 1), a glycosylase responsible for uracil removal, can cause hyper-IgM syndrome type V (157). Symptoms of this disease include immunodeficiency, making patients highly susceptible to infections (157). Overall, epidemiological studies of human patients with mutations in glycosylase genes show higher incidents of a variety of types of cancers, despite the redundancy in the glycosylase targets.
APE1 introduces a potential degree of genomic instability by creating a single stranded break (SSB) during DNA repair (see section 4.1.). This break can be highly mutagenic if not processed properly. Therefore, not surprisingly, dysfunction in APE1 expression or activity has been associated with cancer (156). During times of higher genotoxic stress, such as multiple SSBs, the tumor suppressor p53 reduces the expression level of APE1. This protects the genome from additional single stranded breaks, giving the cell time to properly repair the damage. In the greater than 50% of human cancers that contain p53 mutations abnormally high levels of APE1 could contribute to increased mutagenesis and tumor progression (156, 159). The post translational modification acetylation, which is thought to increase APE1’s activity, is elevated in primary tumor tissues, suggesting that minor changes in function without changes in expression can modify tumorigenesis (160). SNPs within the APE1 gene are also associated with cancer and can cause cytotoxicity by altering substrate binding affinity and catalytic efficiency. D148E is one of the most extensively studied and frequently observed APE1 polymorphisms, at an allele frequency of 0.38 (161, 162). According to one meta-analysis, D148E is associated with colorectal cancer (163). A second APE1 polymorphism, L104R, has diminished endonuclease activity and interferes with APE1 protein-protein interactions, including XRCC1 and pol β (162). A third common APE1 polymorphism, R237C, displays a near 60% reduction in exonuclease activity and also has an impaired association with XRCC1 and pol β (162, 164). These alterations to APE1 activity and correlation with cancer progression again display the advantage of a balanced BER system to maintain genome stability.
Strikingly, approximately 30% of all tumors, regardless of tissue type, exhibit a mutated variant of pol β (165). Mutations in pol β have been particularly associated with human colorectal cancer (166). One explanation for this high correlation is that when the pol β protein contains a mutation that decreases its specificity, more genomic mutations can be created via DNA polymerase misinsertions to promote cancer progression. In a study expressing a single pol β mutation (G231D) near the protein:DNA interface, genomic instability was observed. Additionally, the variant K289M has been shown to enhance mutagenesis within the genome by reducing the ability of pol β to insert the correct nucleotide (i.e. fidelity) (167). The pol β P242R variant displays poor enzyme catalysis, which increases chromosome aberrations and induces cellular transformation (168). During times of high oxidative stress, it is important to also consider that nucleotide damage does not only occur in genomic nucleotides, but also within the free nucleotide triphosphate pools. Mutations that reduce the fidelity of pol β’s nucleotidyl transferase reaction stand to render the entire BER pathway futile; for example, the insertion of the nucleotide 8-oxodGTP instead of a guanine will require another round of DNA repair to remove the DNA polymerase generated DNA damage. Overall, dysfunctions in pol β are a potential source of further genomic instability and can lead to additional complications and futility in the BER pathway during situations with high oxidative stress.
The scaffolding protein XRCC1 does not have an enzymatic function, but still has been identified in epidemiological studies as being associated with cancer common SNPs in XRCC1 can promote genomic instability by altering the ability of XRCC1 to function as a scaffolding protein during repair. The R399Q variant of XRCC1 has been shown to be associated with both breast and pancreatic cancer (169, 170). The XRCC1 polymorphism R194W, also associated with breast cancer (171), results in decreased repair of oxidative lesions like 8-oxoG due to its weakened ability to associate with glycosylase OGG1 (172). Importantly, this lack of repair produces a buildup of oxidative lesions, causing further damage to the cell. These mutations are only a sample of what is known; with the development of third generation sequencing, many more problematic mutations within BER enzymes that mediate biological function may be illuminated in the future.
Although it may initially seem counterintuitive to inhibit DNA repair pathways in patients already suffering from cancer, recent experimental efforts that target BER as a means to further sensitize cancer cells to therapeutic agents have been encouraging. This is because a vast amount of treatments that target rapidly dividing cancer cells act through increasing the amount of DNA damage. BER serves as a key player in repairing the damage generated by therapeutic drugs classified as monofunctional alkylators and antimetabolites (including 5-fluorouracil, thiopurines, and folate analogs) (86). PARP-1 activation, such as after a DNA damaging cancer treatment, depletes the nucleus and cytoplasm of NAD+, effectively blocking glycolysis, and leaves mitochondrial metabolism intact (173). This cascade activates cellular necrotic pathways to induce cell death in cancer cells exhibiting dependence on glycolysis due to the Warburg effect. Aside from reducing BER in combination with DNA damaging therapies, other therapies not specifically targeting DNA, such as ultrasound therapies, show an increase in efficacy when BER proteins are reduced (174). Because so many cancer treatments rely on inducing a lethal amount of mutations, reducing the capacity for repair in conjunction with well-established cancer therapies stands to impact not only the efficacy of treatment, but also the dose of the treatment. This combination approach to improve cancer therapy shows promise in both improving efficacy of treatment and improving quality of life for the patients undergoing treatment.
Aside from inhibiting BER in combination with traditional cancer therapeutics, other DNA-repair targets have recently come to light that would work as stand-alone medications in cases with specific genetic backgrounds. PARP-1 has been shown to be a pivotal protein target in cancers with BRCA (BReast CAncer gene) mutations, leading to the recent approval of the PARP inhibitor, Olaparib, as a cancer therapeutic (175). This is intriguing considering PARP-1 is non-essential in normal cells, but the loss of PARP-1 in BRCA-functional dependence between BRCA and PARP-1 is termed as synthetically lethal (176). The FDA approval of Olaparib was the first instance of a synthetic-lethality-based drug achieving success in the clinic (177–179). In the case of cancers containing BRCA mutations, PARP-1 inhibition causes chromosomal instability, cell cycle arrest, and subsequent apoptosis. Mechanistically, BRCA mutations lead to deficiencies in homologous repair and the inhibition PARP-1 stalls the BER pathway, causing cancer-specific repair (180). Other cancers with upregulated PARP-1, which is thought to occur to facilitate the increased need for repair in an oxidative tumor environment, may also be susceptible to PARP-1 targeting, especially as a combined therapy. As more information about the mechanisms of BER mediated DNA repair are understood, new therapies based on synthetic lethality can be developed, leading to better treatment efficacy and quality of life for patients.
7. PERSPECTIVE
Nucleotides, the building blocks of the genome, must maintain a balance between reactivity and stability. Without reactivity, replication would be energetically unfavorable and repair could not occur. Without stability, an abundance of mutations would quickly render translated proteins dysfunctional and essential genetic content would not persevere from generation to generation. Oxidative stress disrupts this delicate equilibrium, inducing the generation of mutagenic lesions with a wide variety of shapes and sizes, and consequently reducing vital genomic stability (Figure 1). To counterbalance this disruption, DNA repair pathways such as BER mitigate the dangers of oxidative stress by scanning the genome for damage, removing mutagenic lesions, and replacing the gaps left behind with the proper sequence.
Advances in structural biology have complemented standard biochemical and cellular biology techniques to elucidate many of the individual mechanisms of key BER enzymes. To this end, atomic resolution crystal structures of almost every BER enzyme have been solved, with many of these structures being protein:DNA complexes, providing previously elusive details to the enzymes’ mechanisms of handling DNA damage and/or DNA-repair intermediates. Expanding on these methodologies, such as time-lapse X-ray crystallography, to additional DNA-repair enzymes and types of oxidative DNA damage stands to add even further clarity to these complex mechanisms. As technology continues to improve, other approaches such as in singulo studies capitalizing on fluorescence based approaches and atomic force microscopy will illuminate implications of the BER pathway within the context of visualizing protein-protein interactions in transient BER multi-protein co-complexes. These types of studies will be particularly relevant for expanding upon the understanding of how cytotoxic BER intermediates are protected as they are passed from step to step during a BER cycle (Figure 2). Additionally, the generation of increasingly high-resolution protein co-complex structures with techniques such as cryo-electron microscopy and free-electron laser crystallography (where traditionally sized crystals are not needed) could finally provide the advances to solve elusive BER enzyme structures such as NTHL1 and larger multi-protein co-complex structures.
Filling knowledge gaps from a mechanistic perspective has led to an improved understanding of the many diseases that are mediated by oxidative stress and BER, as well as bringing to light new treatments that stand to have higher efficacy and better quality of life for patients. Already, inhibitors of DNA repair, such as Olaparib, are being utilized as a way to sensitize treatment-resistant cancer cells to other DNA damaging therapeutics. In the near future, other DNA repair inhibitors may be developed that target binding pockets and key protein interactions that will be better understood through structural advances, allowing for alternate treatment strategies for patients with cancers that would not have responded to traditional therapeutics. For cases with dysfunctional BER, therapies based on altering signaling pathways that manage BER protein level could be on the horizon, restoring the equilibrium to a functional state. With the power to alter BER activity, many of the diseases discussed in this review stand to be ameliorated.
Acknowledgments
Dr. Amy M. Whitaker and Mathew A. Schaich are co-authors and equally contributed to this paper. This work was supported by the National Institutes of Environmental Health Sciences of the National Institutes of Health under award number R00ES024431. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11(19):3610–8. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 2.Cadet J, Wagner JR. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol. 2013;5(2) doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jovanovic SV, Simic MG. One-Electron Redox Potentials of Purines and Pyrimidines. Journal of Physical Chemistry. 1986;90(5):974–978. doi: 10.1021/j100277a053. [DOI] [Google Scholar]
- 4.Zhang Y, Yuan F, Wu X, Taylor JS, Wang Z. Response of human DNA polymerase iota to DNA lesions. Nucleic Acids Res. 2001;29(4):928–35. doi: 10.1093/nar/29.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oda Y, Uesugi S, Ikehara M, Nishimura S, Kawase Y, Ishikawa H, Inoue H, Ohtsuka E. NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 1991;19(7):1407–12. doi: 10.1093/nar/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19(4):491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 7.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4(1):41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34(8):2305–15. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg MM. The form amidopyrimidines: purine lesions formed in competition with 8-oxopurines from oxidative stress. Acc Chem Res. 2012;45(4):588–97. doi: 10.1021/ar2002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergeron F, Auvre F, Radicella JP, Ravanat JL. HO* radicals induce an unexpected high proportion of tandem base lesions refractory to repair by DNA glycosylases. Proc Natl Acad Sci U S A. 2010;107(12):5528–33. doi: 10.1.0.7.3/pnas.1000193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olinski R, Zastawny T, Budzbon J, Skokowski J, Zegarski W, Dizdaroglu M. DNA base modifications in chromatin of human cancerous tissues. FEBS Lett. 1992;309(2):193–8. doi: 10.1016/0014-5793(92)81093-2. [DOI] [PubMed] [Google Scholar]
- 12.Chetsanga CJ, Grigorian C. A dose-response study on opening of imidazole ring of adenine in DNA by ionizing radiation. Int J Radiat Biol Relat Stud Phys Chem Med. 1983;44(4):321–31. doi: 10.1080/09553008314551261. [DOI] [PubMed] [Google Scholar]
- 13.Delaney MO, Wiederholt CJ, Greenberg MM. Fapy.dA induces nucleotide misincorporation translesionally by a DNA polymerase. Angew Chem Int Ed Engl. 2002;41(5):771–3. doi: 10.1002/1521-3773(20020301)41:5<771::AID-ANIE771>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 14.Kamiya H, Miura H, Murata-Kamiya N, Ishikawa H, Sakaguchi T, Inoue H, Sasaki T, Masutani C, Hanaoka F, Nishimura S, et al. 8-Hydroxyadenine (7,8-dihydro-8-oxoadenine) induces misincorporation in in vitro DNA synthesis and mutations in NIH 3T3 cells. Nucleic Acids Res. 1995;23(15):2893–9. doi: 10.1093/nar/23.15.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan X, Grollman AP, Shibutani S. Comparison of the mutagenic properties of 8-oxo-7,8-dihydro-2′-deoxyadenosine and 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA lesions in mammalian cells. Carcinogenesis. 1999;20(12):2287–92. doi: 10.1093/carcin/20.12.2287. [DOI] [PubMed] [Google Scholar]
- 16.Satou K, Harashima H, Kamiya H. Mutagenic effects of 2-hydroxy-dATP on replication in a HeLa extract: induction of substitution and deletion mutations. Nucleic Acids Res. 2003;31(10):2570–5. doi: 10.1093/nar/gkg368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531(1–2):37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281(33):23445–55. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 19.Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry. 2002;41(19):6090–9. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 20.Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. Human DNA polymerase kappa bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J Biol Chem. 2002;277(40):37604–11. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- 21.Belousova EA, Maga G, Fan Y, Kubareva EA, Romanova EA, Lebedeva NA, Oretskaya TS, Lavrik OI. DNA polymerases beta and lambda bypass thymine glycol in gapped DNA structures. Biochemistry. 2010;49(22):4695–704. doi: 10.1021/bi901792c. [DOI] [PubMed] [Google Scholar]
- 22.van Loon B, Markkanen E, Hubscher U. Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst) 2010;9(6):604–16. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 23.Maki H, Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355(6357):273–5. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 24.Fujikawa K, Kamiya H, Yakushiji H, Nakabeppu Y, Kasai H. Human MTH1 protein hydrolyzes the oxidized ribonucleotide, 2-hydroxy-ATP. Nucleic Acids Res. 2001;29(2):449–54. doi: 10.1093/nar/29.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) EMBO J. 1997;16(11):3341–8. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aravind L, Koonin EV. The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biology. 2000;1(4):research0007.1–research0007.8. doi: 10.1186/gb-2000-1-4-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caulfield JL, Wishnok JS, Tannenbaum SR. Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J Biol Chem. 1998;273(21):12689–95. doi: 10.1074/jbc.273.21.12689. [DOI] [PubMed] [Google Scholar]
- 28.Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol. 2003;20(10):1603–11. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121(1):1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman JI, Stivers JT. Detection of Damaged DNA Bases by DNA Glycosylase Enzymes. Biochemistry. 2010;49(24):4957–4967. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace SS. DNA glycosylases search for and remove oxidized DNA bases. Environ Mol Mutagen. 2013;54(9):691–704. doi: 10.1002/em.21820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buechner CN, Maiti A, Drohat AC, Tessmer I. Lesion search and recognition by thymine DNA glycosylase revealed by single molecule imaging. Nucleic Acids Res. 2015;43(5):2716–29. doi: 10.1093/nar/gkv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403(6772):859–66. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 34.Boal AK, Genereux JC, Sontz PA, Gralnick JA, Newman DK, Barton JK. Redox signaling between DNA repair proteins for efficient lesion detection. Proc Natl Acad Sci U S A. 2009;106(36):15237–42. doi: 10.1073/pnas.0908059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burrows CJ, Muller JG. Oxidative Nucleobase Modifications Leading to Strand Scission. Chem Rev. 1998;98(3):1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 36.Gorodetsky AA, Boal AK, Barton JK. Direct electrochemistry of endonuclease III in the presence and absence of DNA. J Am Chem Soc. 2006;128(37):12082–3. doi: 10.1021/ja064784d. [DOI] [PubMed] [Google Scholar]
- 37.Eley DD, Spivey DI. Semiconductivity of organic substances. Part 9.-Nucleic acid in the dry state. Transactions of the Faraday Society. 1962;58(0):411–415. doi: 10.1039/TF9625800411. [DOI] [Google Scholar]
- 38.Arnold AR, Grodick MA, Barton JK. DNA Charge Transport: from Chemical of the Principles to the Cell. Cell Chem Biol. 2016;23(1):183–97. doi: 10.1016/j.chembiol.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stivers JT, Jiang YL. A mechanistic perspective on the chemistry of DNA repair glycosylases. Chem Rev. 2003;103(7):2729–59. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 40.Coey CT, Malik SS, Pidugu LS, Varney KM, Pozharski E, Drohat AC. Structural basis of damage recognition by thymine DNA glycosylase: Key roles for N-terminal residues. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang YL, Kwon K, Stivers JT. Turning On uracil-DNA glycosylase using a pyrene nucleotide switch. J Biol Chem. 2001;276(45):42347–54. doi: 10.1074/jbc.M106594200. [DOI] [PubMed] [Google Scholar]
- 42.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci U S A. 2000;97(25):13573–8. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S, Verdine GL. Atomic substitution reveals the structural basis for substrate adenine recognition and removal by adenine DNA glycosylase. Proc Natl Acad Sci U S A. 2009;106(44):18497–502. doi: 10.1073/pnas.0902908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maiti A, Morgan MT, Pozharski E, Drohat AC. Crystal structure of human thymine DNA glycosylase bound to DNA elucidates sequence-specific mismatch recognition. Proc Natl Acad Sci U S A. 2008;105(26):8890–5. doi: 10.1073/pnas.0711061105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parikh SS, Walcher G, Jones GD, Slupphaug G, Krokan HE, Blackburn GM, Tainer JA. Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc Natl Acad Sci U S A. 2000;97(10):5083–8. doi: 10.1073/pnas.97.10.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH. Structure and vertebrate anti-mutator specificity uracil-DNA glycosylase SMUG1. Mol Cell. 2003;11(6):1647–59. doi: 10.1016/S1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 47.Zhu C, Lu L, Zhang J, Yue Z, Song J, Zong S, Liu M, Stovicek O, Gao YQ, Yi C. Tautomerization-dependent recognition and excision of oxidation damage in base-excision DNA repair. Proc Natl Acad Sci U S A. 2016;113(28):7792–7. doi: 10.1073/pnas.1604591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullins EA, Shi R, Parsons ZD, Yuen PK, David SS, Igarashi Y, Eichman BF. The DNA glycosylase AlkD uses a non-base-flipping mechanism to excise bulky lesions. Nature. 2015;527(7577):254–8. doi: 10.1038/nature15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernards AS, Miller JK, Bao KK, Wong I. Flipping duplex DNA inside out: a double base-flipping reaction mechanism by Escherichia coli MutY adenine glycosylase. J Biol Chem. 2002;277(23):20960–4. doi: 10.1074/jbc.C200181200. [DOI] [PubMed] [Google Scholar]
- 50.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–30. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 51.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–53. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 52.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc Natl Acad Sci U S A. 2010;107(52):22475–80. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Viswesh V, Gates K, Sun D. Characterization of DNA damage induced by a natural product antitumor antibiotic leinamycin in human cancer cells. Chem Res Toxicol. 2010;23(1):99–107. doi: 10.1021/tx900301r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNeill DR, Lam W, DeWeese TL, Cheng YC, Wilson DM., 3rd Impairment of APE1 function enhances cellular sensitivity to clinically relevant alkylators and antimetabolites. Mol Cancer Res. 2009;7(6):897–906. doi: 10.1158/1541-7786.MCR-08-0519. of Ape1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson DM, 3rd, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res. 2001;485(4):283–307. doi: 10.1016/S0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 56.Maher RL, Bloom LB. Pre-steady-state kinetic characterization of the AP endonuclease activity of human AP endonuclease 1. J Biol Chem. 2007;282(42):30577–85. doi: 10.1074/jbc.M704341200. [DOI] [PubMed] [Google Scholar]
- 57.Fritz G. Human APE/Ref-1 protein. Int J Biochem Cell Biol. 2000;32(9):925–9. doi: 10.1016/S1357-2725(00)00045-5. [DOI] [PubMed] [Google Scholar]
- 58.Freudenthal BD, Beard WA, Cuneo MJ, Dyrkheeva NS, Wilson SH. Capturing snapshots of APE1 processing DNA damage. Nat Struct Mol Biol. 2015;22(11):924–31. doi: 10.1038/nsmb.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination (corrected) Nature. 2000;403(6768):451–6. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 60.Dlakic M. Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem Sci. 2000;25(6):272–3. doi: 10.1016/S0968-0004(00)01582-6. [DOI] [PubMed] [Google Scholar]
- 61.Strauss PR, Beard WA, Patterson TA, Wilson SH. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J Biol Chem. 1997;272(2):1302–7. doi: 10.1074/jbc.272.2.1302. [DOI] [PubMed] [Google Scholar]
- 62.Beloglazova NG, Kirpota OO, Starostin KV, Ishchenko AA, Yamkovoy VI, Zharkov DO, Douglas KT, Nevinsky GA. Thermodynamic, kinetic and structural basis for recognition and repair of abasic sites in DNA by apurinic/apyrimidinic endonuclease from human placenta. Nucleic Acids Res. 2004;32(17):5134–46. doi: 10.1093/nar/gkh846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hadi MZ, Ginalski K, Nguyen LH, Wilson DM., 3rd Determinants in nuclease and Ape2, specificity human homologues of Escherichia coli exonuclease III. J Mol Biol. 2002;316(3):853–66. doi: 10.1006/jmbi.2001.5382. [DOI] [PubMed] [Google Scholar]
- 64.Shen JC, Loeb LA. Mutations in the alpha8 loop of human APE1 alter binding and cleavage of DNA containing an abasic site. J Biol Chem. 2003;278(47):46994–7001. doi: 10.1074/jbc.M309362200. [DOI] [PubMed] [Google Scholar]
- 65.Andres SN, Schellenberg MJ, Wallace BD, Tumbale P, Williams RS. Recognition and Repair of Chemically Heterogeneous Structures at DNA Ends. Environmental and Molecular Mutagenesis. 2015;56(1):1–21. doi: 10.1002/em.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parsons JL, Dianova, Dianov GL. APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res. 2004;32(12):3531–6. doi: 10.1093/nar/gkh676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Izumi T, Hazra TK, Boldogh I, Tomkinson AE, Park MS, Ikeda S, Mitra S. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis. 2000;21(7):1329–34. doi: 10.1093/carcin/21.7.1329. [DOI] [PubMed] [Google Scholar]
- 68.Parsons JL, Dianova, Dianov GL. APE1-dependent repair of DNA single-strand breaks containing 3′-end 8-oxoguanine. Nucleic Acids Res. 2005;33(7):2204–9. doi: 10.1093/nar/gki518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mazouzi A, Vigouroux A, Aikeshev B, Brooks PJ, Saparbaev MK, Morera S, Ishchenko AA. Insight into mechanisms of 3′-5′ exonuclease activity and removal of bulky 8,5′-cyclopurine adducts by apurinic/apyrimidinic endonucleases. Proc Natl Acad Sci U S A. 2013;110(33):E3071–80. doi: 10.1073/pnas.1305281110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chou KM, Cheng YC. An exonucleolytic activity of human apurinic/apyrimidinic endonuclease on 3′ mispaired DNA. Nature. 2002;415(6872):655–9. doi: 10.1038/415655a. [DOI] [PubMed] [Google Scholar]
- 71.Ishchenko AA, Yang X, Ramotar D, Saparbaev M. The 3′->5′ exonuclease of Apn1 provides an alternative pathway to repair 7,8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25(15):6380–90. doi: 10.1128/MCB.25.15.6380-6390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269(5224):699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 73.Piersen CE, Prasad R, Wilson SH, Lloyd RS. Evidence for an imino intermediate in the DNA polymerase beta deoxyribose phosphate excision reaction. J Biol Chem. 1996;271(30):17811–5. doi: 10.1074/jbc.271.30.17811. [DOI] [PubMed] [Google Scholar]
- 74.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36(37):11205–15. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 75.Prasad R, Beard WA, Chyan JY, Maciejewski MW, Mullen GP, Wilson SH. Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase beta as revealed by site-directed mutagenesis. DNA binding and 5′-deoxyribose phosphate lyase activities. J Biol Chem. 1998;273(18):11121–6. doi: 10.1074/jbc.273.18.11121. [DOI] [PubMed] [Google Scholar]
- 76.Deterding LJ, Prasad R, Mullen GP, Wilson SH, Tomer KB. Mapping of the 5′-2-deoxyribose-5-phosphate lyase active site in DNA polymerase beta by mass spectrometry. J Biol Chem. 2000;275(14):10463–71. doi: 10.1074/jbc.275.14.10463. [DOI] [PubMed] [Google Scholar]
- 77.Prasad R, Batra VK, Yang XP, Krahn JM, Pedersen LC, Beard WA, Wilson SH. Structural insight into the DNA polymerase beta deoxyribose phosphate lyase mechanism. DNA Repair (Amst) 2005;4(12):1347–57. doi: 10.1016/j.dnarep.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 78.Caglayan M, Batra VK, Sassa A, Prasad R, Wilson SH. Role of polymerase beta in complementing aprataxin deficiency during abasic-site base excision repair. Nat Struct Mol Biol. 2014;21(5):497–9. doi: 10.1038/nsmb.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beard WA, Wilson SH. Structure and mechanism of DNA polymerase Beta. Chem Rev. 2006;106(2):361–82. doi: 10.1021/cr0404904. [DOI] [PubMed] [Google Scholar]
- 80.Freudenthal BD, Beard WA, Shock DD, Wilson SH. Observing a DNA polymerase choose right from wrong. Cell. 2013;154(1):157–68. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Perera L, Freudenthal BD, Beard WA, Shock DD, Pedersen LG, Wilson SH. Requirement for transient metal ions revealed through computational analysis for DNA polymerase going in reverse. Proc Natl Acad Sci U S A. 2015;112(38):E5228–36. doi: 10.1073/pnas.1511207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freudenthal BD, Beard WA, Perera L, Shock DD, Kim T, Schlick T, Wilson SH. Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature. 2015;517(7536):635–9. doi: 10.1.0.3.8/nature13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Batra VK, Shock DD, Beard WA, McKenna CE, Wilson SH. Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion. Proc Natl Acad Sci U S A. 2012;109(1):113–8. doi: 10.1073/pnas.1112235108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Freudenthal BD, Beard WA, Wilson SH. DNA polymerase minor groove interactions modulate mutagenic bypass of a templating 8-oxoguanine lesion. Nucleic Acids Res. 2013;41(3):1848–58. doi: 10.1093/nar/gks1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fouquerel E, Parikh D, Opresko P. DNA damage processing at telomeres: The ends justify the means. DNA Repair. 2016;44:159–168. doi: 10.1016/j.dnarep.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8(3):193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 87.Rudd SG, Valerie NC, Helleday T. Pathways controlling dNTP pools to maintain genome stability. DNA Repair (Amst) 2016;44:193–204. doi: 10.1016/j.dnarep.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 88.Caglayan M, Wilson SH. Oxidant and environmental toxicant-induced effects compromise DNA ligation during base excision DNA repair. DNA Repair (Amst) 2015;35:85–9. doi: 10.1016/j.dnarep.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rass U, Ahel I, West SC. Actions of aprataxin in multiple DNA repair pathways. J Biol Chem. 2007;282(13):9469–74. doi: 10.1074/jbc.M611489200. [DOI] [PubMed] [Google Scholar]
- 90.Reynolds JJ, El-Khamisy SF, Katyal S, Clements P, McKinnon PJ, Caldecott KW. Defective DNA ligation during short-patch single-strand break repair in ataxia oculomotor apraxia 1. Mol Cell Biol. 2009;29(5):1354–62. doi: 10.1128/MCB.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scott Williams R. 108 Aprataxin and the threat of RNA contamination in DNA. J Biomol Struct Dyn. 2015;33(Suppl 1):68. doi: 10.1080/07391102.2015.1032740. [DOI] [Google Scholar]
- 92.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci. 2011;36(5):262–71. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Doherty AJ, Jackson SP. DNA repair: how Ku makes ends meet. Curr Biol. 2001;11(22):R920–4. doi: 10.1016/S0960-9822(01)00555-3. [DOI] [PubMed] [Google Scholar]
- 94.El-Khamisy SF, Caldecott KW. TDP1-dependent DNA single-strand break repair and neurodegeneration. Mutagenesis. 2006;21(4):219–24. doi: 10.1093/mutage/gel024. [DOI] [PubMed] [Google Scholar]
- 95.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–68. doi: 10.1042/bj3420249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dantzer F, de la Rubia G, Murcia JMD, Hostomsky Z, de Murcia G, Schreiber V. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry. 2000;39(25):7559–7569. doi: 10.1021/bi0003442. [DOI] [PubMed] [Google Scholar]
- 97.Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. Journal of Biological Chemistry. 2002;277(25):23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 98.Lavrik OI, Prasad R, Sobol RW, Horton JK, Ackermann EJ, Wilson SH. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate - Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. Journal of Biological Chemistry. 2001;276(27):25541–25548. doi: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- 99.Sukhanova MV, Khodyreva SN, Lebedeva NA, Prasad R, Wilson SH, Lavrik OI. Human base excision repair enzymes apurinic/apyrimidinic endonuclease1 (APE1), DNA polymerase beta and poly(ADP-ribose) polymerase 1: interplay between strand-displacement DNA synthesis and proofreading exonuclease activity. Nucleic Acids Research. 2005;33(4):1222–1229. doi: 10.1093/nar/gki266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kutuzov MM, Khodyreva SN, Ame JC, Ilina ES, Sukhanova MV, Schreiber V, Lavrik OI. Interaction of PARP-2 with DNA structures mimicking DNA repair intermediates and consequences on activity of base excision repair proteins. Biochimie. 2013;95(6):1208–1215. doi: 10.1016/j.biochi.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 101.Realini CA, Althaus FR. Histone shuttling by poly(ADP-ribosylation) J Biol Chem. 1992;267(26):18858–65. [PubMed] [Google Scholar]
- 102.Ogata N, Ueda K, Kawaichi M, Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J Biol Chem. 1981;256(9):4135–7. [PubMed] [Google Scholar]
- 103.Huletsky A, de Murcia G, Muller S, Hengartner M, Menard L, Lamarre D, Poirier GG. The effect of poly(ADP-ribosyl) ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem. 1989;264(15):8878–86. [PubMed] [Google Scholar]
- 104.Prasad R, Dyrkheeva N, Williams J, Wilson SH. Mammalian Base Excision Repair: Functional Partnership between PARP-1 and APE1 in AP-Site Repair. Plos One. 2015;10(5) doi: 10.1371/journal.pone.0124269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Reynolds P, Cooper S, Lomax M, O’Neill P. Disruption of PARP1 function inhibits base excision repair of a sub-set of DNA lesions. Nucleic Acids Res. 2015;43(8):4028–38. doi: 10.1093/nar/gkv250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9(8):619–31. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 107.Beernink PT, Hwang M, Ramirez M, Murphy MB, Doyle SA, Thelen MP. interactions mediated Specificity by BRCT domains of the XRCC1 DNA repair protein. J Biol Chem. 2005;280(34):30206–13. doi: 10.1074/jbc.M502155200. [DOI] [PubMed] [Google Scholar]
- 108.Hanssen-Bauer A, Solvang-Garten K, Gilljam KM, Torseth K, Wilson DM, Akbari M, Otterlei M. The region of XRCC1 which harbours the three most common nonsynonymous polymorphic variants, is essential for the scaffolding function of XRCC1. DNA Repair. 2012;11(4):357–366. doi: 10.1016/j.dnarep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marintchev A, Mullen MA, Maciejewski MW, Pan B, Gryk MR, Mullen GP. Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nat Struct Biol. 1999;6(9):884–93. doi: 10.1038/12347. [DOI] [PubMed] [Google Scholar]
- 110.Cuneo MJ, London RE. Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase beta binding affinity. Proc Natl Acad Sci U S A. 2010;107(15):6805–10. doi: 10.1073/pnas.0914077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Taylor RM, Wickstead B, Cronin S, Caldecott KW. Role of a BRCT domain in the interaction of DNA ligase III-alpha with the DNA repair protein XRCC1. Curr Biol. 1998;8(15):877–80. doi: 10.1016/S0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- 112.Cuneo MJ, Gabel SA, Krahn JM, Ricker MA, London RE. The structural basis for partitioning of the XRCC1/DNA ligase III-alpha BRCT-mediated dimer complexes. Nucleic Acids Research. 2011;39(17):7816–7827. doi: 10.1093/nar/gkr419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wilson SH. Mammalian base excision repair and DNA polymerase beta. Mutat Res. 1998;407(3):203–15. doi: 10.1016/S0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 114.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7(3):176–8. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 115.Prasad R, Beard WA, Batra VK, Liu Y, Shock DD, Wilson SH. A review of recent experiments on step-to-step “hand-off” of the DNA intermediates in mammalian base excision repair pathways. Mol Biol (Mosk) 2011;45(4):586–600. doi: 10.1134/S0026893311040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: passing the baton. J Biol Chem. 2010;285(52):40479–88. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang H, Clendenin WM, Wong D, Demple B, Slupska MM, Chiang JH, Miller JH. Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch. Nucleic Acids Res. 2001;29(3):743–52. doi: 10.1093/nar/29.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Luncsford PJ, Manvilla BA, Patterson DN, Malik SS, Jin J, Hwang BJ, Gunther R, Kalvakolanu S, Lipinski LJ, Yuan W, Lu W, Drohat AC, Lu AL, Toth EA. Coordination of MYH DNA glycosylase and APE1 endonuclease activities via physical interactions. DNA Repair (Amst) 2013;12(12):1043–52. doi: 10.1016/j.dnarep.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kuznetsova AA, Kuznetsov NA, Ishchenko AA, Saparbaev MK, Fedorova OS. Pre-steady-state fluorescence analysis of damaged DNA transfer from human DNA glycosylases to AP endonuclease APE1. Biochim Biophys Acta. 2014;1840(10):3042–51. doi: 10.1016/j.bbagen.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 120.Moor NA, Vasil’eva IA, Anarbaev RO, Antson AA, Lavrik OI. Quantitative characterization of protein-protein complexes involved in base excision DNA repair. Nucleic Acids Res. 2015;43(12):6009–22. doi: 10.1093/nar/gkv569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brenerman BM, Illuzzi JL, Wilson DM., 3rd Base excision repair capacity in informing healthspan. Carcinogenesis. 2014;35(12):2643–52. doi: 10.1093/carcin/bgu225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A, Schar P. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470(7334):419–23. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 123.Li Z, Gu T-P, Weber AR, Shen J-Z, Li B-Z, Xie Z-G, Yin R, Guo F, Liu X, Tang F, Wang H, Schar P, Xu G-L. Gadd45a promotes DNA demethylation through TDG. Nucleic Acids Research. 2015;43(8):3986–3997. doi: 10.1093/nar/gkv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Meira LB, Bugni JM, Green SL, Lee C-W, Pang B, Borenshtein D, Rickman BH, Rogers AB, Moroski-Erkul CA, McFaline JL, Schauer DB, Dedon PC, Fox JG, Samson LD. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. The Journal of Clinical Investigation. 2008;118(7):2516–2525. doi: 10.1172/jci35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Russo MT, De Luca G, Degan P, Parlanti E, Dogliotti E, Barnes DE, Lindahl T, Yang H, Miller JH, Bignami M. Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Ogg1 DNA glycosylases. Cancer Res. 2004;64(13):4411–4. doi: 10.1158/0008-5472.CAN-04-0355. [DOI] [PubMed] [Google Scholar]
- 126.Meira LB, Devaraj S, Kisby GE, Burns DK, Daniel RL, Hammer RE, Grundy S, Jialal I, Friedberg EC. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001;61(14):5552–7. [PubMed] [Google Scholar]
- 127.Unnikrishnan A, Raffoul JJ, Patel HV, Prychitko TM, Anyangwe N, Meira LB, Friedberg EC, Cabelof DC, Heydari AR. Oxidative stress alters base excision repair pathway and increases apoptotic response in apurinic/apyrimidinic endonuclease 1/redox factor-1 haploinsufficient mice. Free Radic Biol Med. 2009;46(11):1488–99. doi: 10.1016/j.freeradbiomed.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Senejani AG, Dalal S, Liu Y, Nottoli TP, McGrath JM, Clairmont CS, Sweasy JB. Y265C DNA polymerase beta knockin mice survive past birth and accumulate base excision repair intermediate substrates. Proc Natl Acad Sci U S A. 2012;109(17):6632–7. doi: 10.1073/pnas.1200800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nemec AA, Donigan KA, Murphy DL, Jaeger J, Sweasy JB. Colon cancer-associated DNA polymerase beta variant induces genomic instability and cellular transformation. J Biol Chem. 2012;287(28):23840–9. doi: 10.1074/jbc.M112.362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208(2):513–29. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 131.McNeill DR, Lin PC, Miller MG, Pistell PJ, de Souza-Pinto NC, Fishbein KW, Spencer RG, Liu Y, Pettan-Brewer C, Ladiges WC, Wilson DM., 3rd XRCC1 haploinsufficiency in mice has little effect on aging, but adversely modifies exposure-dependent susceptibility. Nucleic Acids Res. 2011;39(18):7992–8004. doi: 10.1093/nar/gkr280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Belanger M, Allaman I, Magistretti PJ. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011;14(6):724–38. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 133.Sliwinska A, Kwiatkowski D, Czarny P, Toma M, Wigner P, Drzewoski J, Fabianowska-Majewska K, Szemraj J, Maes M, Galecki P, Sliwinski T. The levels of 7,8-dihydrodeoxyguanosine (8-oxoG) and 8-oxoguanine DNA glycosylase 1 (OGG1) - A potential diagnostic biomarkers of Alzheimer’s disease. J Neurol Sci. 2016;368:155–9. doi: 10.1016/j.jns.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 134.Shao C, Xiong S, Li GM, Gu L, Mao G, Markesbery WR, Lovell MA. Altered 8-oxoguanine glycosylase in mild cognitive impairment and late-stage Alzheimer’s disease brain. Free Radic Biol Med. 2008;45(6):813–9. doi: 10.1016/j.freeradbiomed.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Weissman L, Jo DG, Sorensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA. Defective DNA base excision repair in brain from individuals with Alzheimer’s disease and amnestic mild cognitive impairment. Nucleic Acids Res. 2007;35(16):5545–55. doi: 10.1093/nar/gkm605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Davydov V, Hansen LA, Shackelford DA. Is DNA repair compromised in Alzheimer’s disease? Neurobiol Aging. 2003;24(7):953–68. doi: 10.1016/S0197-4580(02)00229-4. [DOI] [PubMed] [Google Scholar]
- 137.Sykora P, Misiak M, Wang Y, Ghosh S, Leandro GS, Liu D, Tian J, Baptiste BA, Cong WN, Brenerman BM, Fang E, Becker KG, Hamilton RJ, Chigurupati S, Zhang Y, Egan JM, Croteau DL, Wilson DM, 3rd, Mattson MP, Bohr VA. DNA polymerase beta deficiency leads to neurodegeneration and exacerbates Alzheimer disease phenotypes. Nucleic Acids Res. 2015;43(2):943–59. doi: 10.1093/nar/gku1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Martire S, Mosca L, d’Erme M. PARP-1 involvement in neurodegeneration: A focus on Alzheimer’s and Parkinson’s diseases. Mech Ageing Dev. 2015;146–148:53–64. doi: 10.1016/j.mad.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 139.Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, Weston R, Williams JG, Rossi MN, Galehdari H, Krahn J, Wan A, Trembath RC, Crosby AH, Ahel D, Hay R, Ladurner AG, Timinszky G, Williams RS, Ahel I. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. Embo j. 2013;32(9):1225–37. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kauppinen TM, Swanson RA. The role of poly(ADP-ribose) polymerase-1 in CNS disease. Neuroscience. 2007;145(4):1267–72. doi: 10.1016/j.neuroscience.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 141.Pacher P, Szabo C. Role of poly(ADP-ribose) polymerase-1 activation in the pathogenesis of diabetic complications: endothelial dysfunction, as a common underlying theme. Antioxid Redox Signal. 2005;7(11–12):1568–80. doi: 10.1089/ars.2005.7.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Peralta-Leal A, Rodriguez-Vargas JM, Aguilar-Quesada R, Rodriguez MI, Linares JL, de Almodovar MR, Oliver FJ. PARP inhibitors: new partners in the therapy of cancer and inflammatory diseases. Free Radic Biol Med. 2009;47(1):13–26. doi: 10.1016/j.freeradbiomed.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 143.Strosznajder R, Gadamski R, Walski M. Inhibition of poly(ADP-ribose) polymerase activity protects hippocampal cells against morphological and ultrastructural alteration evoked by ischemia-reperfusion injury. Folia Neuropathol. 2005;43(3):156–65. [PubMed] [Google Scholar]
- 144.Tempera I, Deng Z, Atanasiu C, Chen CJ, D’Erme M, Lieberman PM. Regulation of Epstein-Barr virus OriP replication by poly(ADP-ribose) polymerase 1. J Virol. 2010;84(10):4988–97. doi: 10.1128/JVI.02333-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PARlaying’ NAD+ into a nuclear signal. Genes Dev. 2005;19(17):1951–67. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 146.Noren Hooten N, Fitzpatrick M, Kompaniez K, Jacob KD, Moore BR, Nagle J, Barnes J, Lohani A, Evans MK. Coordination of DNA repair by NEIL1 and PARP-1: a possible link to aging. Aging (Albany NY) 2012;4(10):674–85. doi: 10.18632/aging.100492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Martire S, Fuso A, Mosca L, Forte E, Correani V, Fontana M, Scarpa S, Maras B, d’Erme M. Bioenergetic Impairment in Animal and Cellular Models of Alzheimer’s Disease: PARP-1 Inhibition Rescues Metabolic Dysfunctions. J Alzheimers Dis. 2016;54(1):307–24. doi: 10.3233/JAD-151040. [DOI] [PubMed] [Google Scholar]
- 148.Helleday T. PARP inhibitor receives FDA breakthrough therapy designation in castration resistant prostate cancer: beyond germline BRCA mutations. Annals of Oncology. 2016;27(5):755–757. doi: 10.1093/annonc/mdw048. [DOI] [PubMed] [Google Scholar]
- 149.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447(7143):447–52. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu Y, Prasad R, Beard WA, Hou EW, Horton JK, McMurray CT, Wilson SH. Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J Biol Chem. 2009;284(41):28352–66. doi: 10.1074/jbc.M109.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhao XN, Usdin K. The Repeat Expansion Diseases: The dark side of DNA repair. DNA Repair (Amst) 2015;32:96–105. doi: 10.1016/j.dnarep.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ba X, Aguilera-Aguirre L, Rashid QT, Bacsi A, Radak Z, Sur S, Hosoki K, Hegde ML, Boldogh I. The role of 8-oxoguanine DNA glycosylase-1 in inflammation. Int J Mol Sci. 2014;15(9):16975–97. doi: 10.3390/ijms150916975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Belanger KK, Ameredes BT, Boldogh I, Aguilera-Aguirre L. The Potential Role of 8-Oxoguanine DNA Glycosylase-Driven DNA Base Excision Repair in Exercise-Induced Asthma. Inflammation Mediatorsof. 2016;2016:3762561. doi: 10.1155/2016/3762561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tsuruya K, Furuichi M, Tominaga Y, Shinozaki M, Tokumoto M, Yoshimitsu T, Fukuda K, Kanai H, Hirakata H, Iida M, Nakabeppu Y. Accumulation of 8-oxoguanine in the cellular DNA and the alteration of the OGG1 expression during ischemia-reperfusion injury in the rat kidney. DNA Repair (Amst) 2003;2(2):211–29. doi: 10.1016/S1568-7864(02)00214-8. [DOI] [PubMed] [Google Scholar]
- 155.Akbari M, Morevati M, Croteau D, Bohr VA. The role of DNA base excision repair in brain homeostasis and disease. DNA Repair (Amst) 2015;32:172–9. doi: 10.1016/j.dnarep.2015.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Poletto M, Legrand AJ, Fletcher SC, Dianov GL. p53 coordinates base excision repair to prevent genomic instability. Nucleic Acids Res. 2016;44(7):3165–75. doi: 10.1093/nar/gkw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wilson DM, 3rd, Kim D, Berquist BR, Sigurdson AJ. Variation in base excision repair capacity. Mutat Res. 2011;711(1–2):100–12. doi: 10.1016/j.mrfmmm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Galick HA, Kathe S, Liu M, Robey-Bond S, Kidane D, Wallace SS, Sweasy JB. Germ-line variant of human NTH1 DNA glycosylase induces genomic instability and cellular transformation. Proc Natl Acad Sci U S A. 2013;110(35):14314–9. doi: 10.1073/pnas.1306752110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ozaki T, Nakagawara A. Role of p53 in Cell Death and Human Cancers. Cancers. 2011;3(1):994–1013. doi: 10.3390/cancers3010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sengupta S, Mantha AK, Song H, Roychoudhury S, Nath S, Ray S, Bhakat KK. Elevated level of acetylation of APE1 in tumor cells modulates DNA damage repair. Oncotarget. 2016 doi: 10.18632/oncotarget.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hadi MZ, Coleman MA, Fidelis K, Mohrenweiser HW, Wilson DM., 3rd Functional characterization of Ape1 variants identified in the human population. Nucleic Acids Res. 2000;28(20):3871–9. doi: 10.1093/nar/28.20.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lirussi L, Antoniali G, D’Ambrosio C, Scaloni A, Nilsen H, Tell G. APE1 polymorphic variants cause persistent genomic stress and affect cancer cell proliferation. Oncotarget. 2016;7(18):26293–306. doi: 10.18632/oncotarget.8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gu D, Wang M, Wang M, Zhang Z, Chen J. The DNA repair gene APE1 T1349G polymorphism and cancer risk: a meta-analysis of 27 case-control studies. Mutagenesis. 2009;24(6):507–12. doi: 10.1093/mutage/gep036. [DOI] [PubMed] [Google Scholar]
- 164.Illuzzi JL, Harris NA, Manvilla BA, Kim D, Li M, Drohat AC, Wilson DM., 3rd Functional assessment of population and tumor-associated APE1 protein variants. PLoS One. 2013;8(6):e65922. doi: 10.1371/journal.pone.0065922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12(12):801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 166.Donigan KA, Sun K-w, Nemec AA, Murphy DL, Cong X, Northrup V, Zelterman D, Sweasy JB. Human POLB Gene Is Mutated in High Percentage of Colorectal Tumors. The Journal of Biological Chemistry. 2012;287(28):23830–23839. doi: 10.1074/jbc.M111.324947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lang T, Maitra M, Starcevic D, Li SX, Sweasy JB. A DNA polymerase beta mutant from colon cancer cells induces mutations. Proc Natl Acad Sci U S A. 2004;101(16):6074–9. doi: 10.1073/pnas.0308571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yamtich J, Nemec AA, Keh A, Sweasy JB. A germline polymorphism of DNA polymerase beta induces genomic instability and cellular transformation. PLoS Genet. 2012;8(11):e1003052. doi: 10.1371/journal.pgen.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Duell EJ, Millikan RC, Pittman GS, Winkel S, Lunn RM, Tse CK, Eaton A, Mohrenweiser HW, Newman B, Bell DA. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomarkers Prev. 2001;10(3):217–22. [PubMed] [Google Scholar]
- 170.Wang LJ, Wang HT, Wang XX. Association of XRCC1 gene polymorphisms and pancreatic cancer risk in a Chinese population. Genet Mol Res. 2016;15(2) doi: 10.4238/gmr.15028080. [DOI] [PubMed] [Google Scholar]
- 171.Sanjari Moghaddam A, Nazarzadeh M, Noroozi R, Darvish H, Mosavi Jarrahi A. XRCC1 and OGG1 Gene Polymorphisms and Breast Cancer: A Systematic Review of Literature. Iran J Cancer Prev. 2016;9(1):e3467. doi: 10.17795/ijcp-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Campalans A, Moritz E, Kortulewski T, Biard D, Epe B, Radicella JP. Interaction with OGG1 is required for efficient recruitment of XRCC1 to base excision repair and maintenance of genetic stability after exposure to oxidative stress. Mol Cell Biol. 2015;35(9):1648–58. doi: 10.1128/MCB.00134-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zong W-X, Ditsworth D, Bauer DE, Wang Z-Q, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes & Development. 2004;18(11):1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Xu T, Nie Y, Bai J, Li L, Yang B, Zheng G, Zhang J, Yu J, Cheng X, Jiao J, Jing H. Suppression of human 8-oxoguanine DNA glycosylase (OGG1) augments ultrasound-induced apoptosis in cervical cancer cells. Ultrasonics. 2016;72:1–14. doi: 10.1016/j.ultras.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 175.Konecny GE, Kristeleit RS. PARP inhibitors for BRCA1/2-mutated and sporadic ovarian cancer: current practice and future directions. Br J Cancer. 2016 doi: 10.1038/bjc.2016.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Curtin NJ. Poly(ADP-ribose) polymerase (PARP) and PARP inhibitors. Drug Discovery Today: Disease Models. 2012;9(2):e51–e58. doi: 10.1016/j.ddmod.2012.01.004. [DOI] [Google Scholar]
- 177.Helleday T. DNA repair as treatment target. Eur J Cancer. 2011;47(Suppl 3):S333–5. doi: 10.1016/S0959-8049(11)70192-7. [DOI] [PubMed] [Google Scholar]
- 178.Brown JS, Kaye SB, Yap TA. PARP inhibitors: the race is on. Br J Cancer. 2016;114(7):713–715. doi: 10.1038/bjc.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Weaver A, Yang E. Beyond DNA Repair: Additional Functions of PARP-1 in Cancer. Frontiers in Oncology. 2013;3(290) doi: 10.3389/fonc.2013.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NMB, Jackson SP, Smith GCM, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 181.Roberts VA, Pique ME, Hsu S, Li S, Slupphaug G, Rambo RP, Jamison JW, Liu T, Lee JH, Tainer JA, Ten Eyck LF, Woods VL., Jr Combining H/D exchange mass spectroscopy and computational docking reveals extended DNA-binding surface on uracil-DNA glycosylase. Nucleic Acids Res. 2012;40(13):6070–81. doi: 10.1093/nar/gks291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Darwanto A, Theruvathu JA, Sowers JL, Rogstad DK, Pascal T, Goddard W, 3rd, Sowers LC. Mechanisms of base selection by human single-stranded selective monofunctional uracil-DNA glycosylase. J Biol Chem. 2009;284(23):15835–46. doi: 10.1074/jbc.M807846200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Maiti A, Morgan MT, Drohat AC. Role of two strictly conserved residues in nucleotide flipping and N-glycosylic bond cleavage by human thymine DNA glycosylase. J Biol Chem. 2009;284(52):36680–8. doi: 10.1074/jbc.M109.062356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Moréra S, Grin I, Vigouroux A, Couvé S, Henriot V, Saparbaev M, Ishchenko AA. Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Research. 2012;40(19):9917–9926. doi: 10.1093/nar/gks714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lukina MV, Popov AV, Koval VV, Vorobjev YN, Fedorova OS, Zharkov DO. DNA Damage Processing by Human 8-Oxoguanine-DNA Glycosylase Mutants with the Occluded Active Site. The Journal of Biological Chemistry. 2013;288(40):28936–28947. doi: 10.1074/jbc.M113.487322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Williams SD, David SS. Evidence that MutY is a monofunctional glycosylase capable of forming a covalent Schiff base intermediate with substrate DNA. Nucleic Acids Res. 1998;26(22):5123–33. doi: 10.1093/nar/26.22.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427(6975):652–6. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 188.Prasad A, Wallace SS, Pederson DS. Initiation of base excision repair of oxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase NTH1. Mol Cell Biol. 2007;27(24):8442–53. doi: 10.1128/MCB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. Embo j. 2003;22(13):3461–71. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(25):13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee CY, Delaney JC, Kartalou M, Lingaraju GM, Maor-Shoshani A, Essigmann JM, Samson LD. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG) Biochemistry. 2009;48(9):1850–61. doi: 10.1021/bi8018898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Krokeide SZ, Laerdahl JK, Salah M, Luna L, Cederkvist FH, Fleming AM, Burrows CJ, Dalhus B, Bjoras M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst) 2013;12(12):1159–64. doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(28):10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(6):3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Prakash A, Eckenroth BE, Averill AM, Imamura K, Wallace SS, Doublie S. Structural Investigation of a Viral Ortholog of Human NEIL2/3 DNA Glycosylases. DNA repair. 2013;12(12) doi: 10.1016/j.dnarep.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Liu M, Imamura K, Averill AM, Wallace SS, Doublié S. Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure (London, England: 1993) 2013;21(2) doi: 10.1.0.1.6/j.str.2012.1.2.0.0.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tagami S, Sekine S, Minakhin L, Esyunina D, Akasaka R, Shirouzu M, Kulbachinskiy A, Severinov K, Yokoyama S. Structural basis for promoter RNA polymerase by specificity a phage factor. Genes Dev. 2014;28(5):521–31. doi: 10.1101/gad.233916.113. [DOI] [PMC free article] [PubMed] [Google Scholar]