Abstract
Both environmental cues and intracellular bioenergetic states profoundly affect intracellular pH (pHi). How a cell responds to pHi changes to maintain bioenergetic homeostasis remains elusive. Here we show that Smad5, a well-characterized downstream component of bone morphogenetic protein (BMP) signaling responds to pHi changes. Cold, basic or hypertonic conditions increase pHi, which in turn dissociates protons from the charged amino acid clusters within the MH1 domain of Smad5, prompting its relocation from the nucleus to the cytoplasm. On the other hand, heat, acidic or hypotonic conditions decrease pHi, blocking the nuclear export of Smad5, and thus causing its nuclear accumulation. Active nucleocytoplasmic shuttling of Smad5 induced by environmental changes and pHi fluctuation is independent of BMP signaling, carboxyl terminus phosphorylation and Smad4. In addition, ablation of Smad5 causes chronic and irreversible dysregulation of cellular bioenergetic homeostasis and disrupted normal neural developmental processes as identified in a differentiation model of human pluripotent stem cells. Importantly, these metabolic and developmental deficits in Smad5-deficient cells could be rescued only by cytoplasmic Smad5. Cytoplasmic Smad5 physically interacts with hexokinase 1 and accelerates glycolysis. Together, our findings indicate that Smad5 acts as a pHi messenger and maintains the bioenergetic homeostasis of cells by regulating cytoplasmic metabolic machinery.
Keywords: Smad5, intracellular pH, BMP, glycolysis, human pluripotent stem cells
Introduction
Given that most biological reactions are pH dependent, it is therefore not a surprise that intracellular pH (pHi) is stringently controlled within a physiological range. While changes in oxygen or nutrient supply states, extracellular pH (pHe) or osmolarity may directly affect pHi homeostasis, cells have a series of immediate pHi sensing and balancing machineries opposing these fluctuations1,2,3,4. To date, the pH sensitive proteins identified are exclusively membrane located transporters. The Na+-H+ exchangers and Na+-HCO3− co-transporters are the main transporters to alkalinize cytosol, while the most common acid-importing transporters are Cl−-HCO3− anion exchangers which acidify the cells2. In addition, lactate and CO2 generated via metabolic reactions tend to acidify the cytoplasm. Whether there is an intracellular pHi messenger which links pHi fluctuations to these metabolic reactions remains unknown.
BMP and TGFβ signaling are evolutionarily conserved pathways, which play pivotal roles in embryonic development, epithelial and mesenchymal cell states transition, and tumor metastasis5. Upon BMP or TGFβ stimulation, cytosolic R-Smad members, including Smad1, Smad2, Smad3, Smad5 and Smad8, become phosphorylated by the type I receptors at the most C-terminal SSXS motif6,7. These phosphorylated R-Smads then oligomerise with Smad4 and translocate into nucleus for gene transcriptional regulation8. Although amino acid sequences of R-Smads share extremely high homology, Smad1/5/8 favor BMP signaling, while Smad2/3 mainly act downstream of the TGFβ pathway9. Transgenic animal studies have demonstrated that Smad1/5/8 or Smad2/3 cannot fully compensate each other, although the exact mechanisms are unclear10,11. Recently, Smad2 has been identified as playing a non-transcriptional role in regulating mitochondrial dynamics and function. This is facilitated by complexing with Mitofusin 2, Rab and Ras interactor 112, suggesting a noncanonical role of the component within the BMP and TGFβ pathways in regulating metabolic processes. In addition, Smad5 deficiency results in abnormally swollen mitochondria and apoptosis in cardiomyocytes, indicating that it might be involved in energy metabolism13.
Here, we show that Smad5 can actively respond to changes of pHi through nucleocytoplasmic shuttling. The MH1 domain of Smad5 plays a critical role in the pHi response, whereas the nuclear export signals (NESs) located in the linker region and the MH2 domain control the process of nuclear export. Importantly, the novel role of Smad5 in responding to pHi changes is BMP signalling independent and its nucleocytoplasmic shuttling does not require interaction with Smad4. Apart from its canonical nuclear role in transcriptional regulation, cytosolic Smad5 enhances glycolysis by interacting with hexokinase 1 (HK1). Loss of function of Smad5 causes glycolysis defects and irreversible dysregulation of cellular bioenergetics homeostasis. These data demonstrate that Smad5 is an immediate pHi fluctuation messenger and an important regulator of physiological bioenergetic homeostasis.
Results
Smad5 is sensitive to environmental cue fluctuations
Living organisms are imbued with the ability to sense and buffer against environmental perturbations to maintain intracellular homeostasis. Molecular mechanisms underlying the sensing of environmental stresses and maintenance of cellular homeostasis are still the focus of intense study. We unexpectedly found that GFP-Smad5 diffused from nucleus to cytoplasm within several minutes when the cells were placed at 25 °C (Figure 1A, Supplementary information, Figure S1A-S1D, Movie S1). The cytoplasmically distributed GFP-Smad5 repopulated in the nucleus 30 min after the cells were placed back at 37 °C (Supplementary information, Figure S1D). Among all Smad family proteins, Smad2, Smad4 and Smad8 are mostly distributed within the cytoplasm (Supplementary information, Figure S1E and S1F)23. Smad1 and Smad3 are largely distributed in the nucleus, but these proteins did not shuttle upon temperature change (Supplementary information, Figure S1D and S1G). These data strongly suggest that Smad5 differs from other Smads by virtue of its ability to sense ambient temperature fluctuation and change its nucleocytoplasmic distribution dynamically.
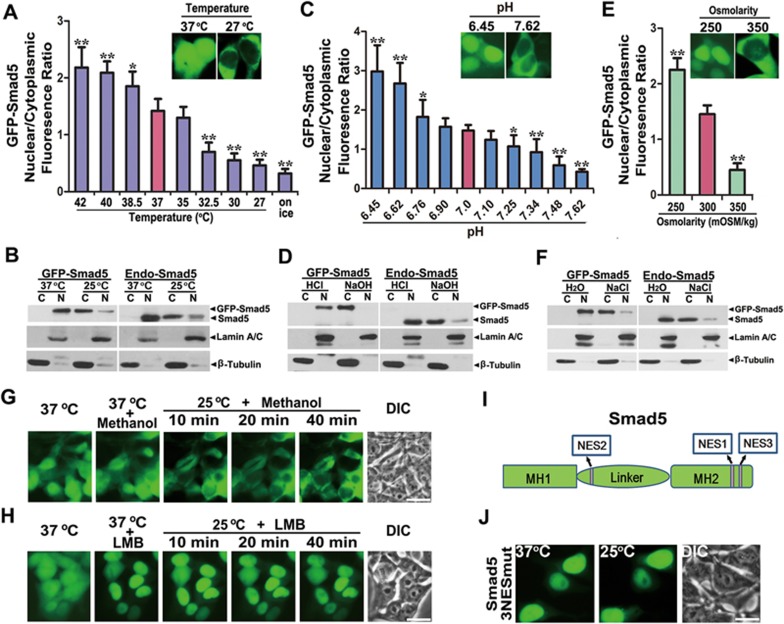
Figure 1.
Temperature, pHe and osmolarity fluctuations dynamically control Smad5 nuclear export. (A) Average fluorescence quantification of nuclear and cytoplasmic localized GFP-Smad5 at various temperatures (n = 30; data are mean ± SEM; *P < 0.05; **P < 0.01). (B) Western blotting shows that endogenous Smad5 also undergoes temperature-sensitive cytoplasmic translocation in HEK293 cells. (C) Average fluorescence quantification of nuclear and cytoplasmic GFP-Smad5 at various extracellular pH values (n = 30; data are mean ± SEM; *P < 0.05; **P < 0.01). (D) Western blotting shows that endogenous Smad5 exhibits pHe-sensitive nucleocytoplasmic shuttling in HEK293 cells. (E) Average fluorescence quantification of nuclear and cytoplasmic localized GFP-Smad5 at various extracellular osmolarity (n = 30; data are mean ± SEM; *P < 0.05; **P < 0.01). (F) Western blotting shows that endogenous Smad5 exhibits extracellular osmolarity-sensitive nucleocytoplasmic shuttling in HEK293 cells. (G) Pretreatment of GFP-Smad5-expressing HEK293 cells with methanol for 1 h at 37 °C does not influence nucleocytoplasmic distribution of Smad5. And in the presence of methanol, low temperature triggers nucleocytoplasmic shuttling of GFP-Smad5. Scale bar, 10 μm. (H) 5 ng/ml LMB treatment for 1 h at 37 °C induces more obvious nuclear accumulation of GFP-Smad5 and completely blocks low temperature-triggered nucleocytoplasmic shuttling. Scale bar, 10 μm. (I) Schematic diagram of the positions of the three Smad5 NESs. (J) Mutation of all three NESs in Smad5 completely abrogates low temperature-driven Smad5 nuclear export even 1 h after shifting the cells to 25 °C. Scale bar, 10 μm.
We then studied the temperature range that affects Smad5 nucleocytoplasmic distribution. At 37 °C, more Smad5 was found in the nucleus than in the cytoplasm (Figure 1A and Supplementary information, Figure S1H). Placing the cells at a temperature lower than 32.5 °C drove most of the nucleus-localized Smad5 to the cytoplasm. Even when the cells were directly placed on ice, Smad5 also quickly accumulated in the cytoplasm. When the environmental temperature was higher than 37 °C, more Smad5 was distributed in the nucleus (Figure 1A and Supplementary information, Figure S1H). These results suggest that Smad5 may be able to sense ambient temperature changes within a wide range. Importantly, western blotting shows that endogenous Smad5 was located largely in the nucleus at 37 °C and translocated into the cytoplasm at 25 °C, in a similar manner to that of GFP-Smad5 (Figure 1B).
The striking temperature-sensitive nucleocytoplasmic redistribution of Smad5 caused us to wonder whether other environmental changes could also induce its redistribution (Supplementary information, Figure S2A-S2C). Among these conditions, we found that changes in external pH (pHe) and osmolarity effectively regulated Smad5 nucleocytoplasmic distributions, in a similar way to that of temperature change (Supplementary information, Movie S2-S5).
GFP-Smad5 was exclusively located in the nucleus when cells were treated with 10 mM hydrochloric acid for 10 min at 37 °C (Supplementary information, Figure S3A and Movie S4). On the other hand, 2 mM sodium hydroxide rapidly induced GFP-Smad5 to translocate into the cytoplasm within 2 min at 37 °C (Supplementary information, Figure S3B and Movie S2). Statistical analysis showed that GFP-Smad5 responded to a subtle range of pHe (Figure 1C). Smad5 was mostly located in the nucleus when pHe was lower than 6.62 (Figure 1C and Supplementary information, Figure S3C), whereas with a pHe higher than 7.62, it was mostly distributed in the cytosol (Figure 1C and Supplementary information, Figure S3D). With an acidic external pH, endogenous Smad5 was again located predominately in the nucleus, whereas alkaline pHe induced its accumulation in the cytoplasm just like GFP-Smad5 (Figure 1D). Therefore, Smad5 may sense delicate pHe changes within a physiological range by nucleocytoplasmic shuttling. In addition, GFP-Smad5 showed obvious nuclear accumulation under hypotonic conditions, whereas it quickly translocated into the cytoplasm under hypertonic conditions (Figure 1E, Supplementary information, Figure S3E and S3F, Movie S3 and S5). Endogenous Smad5 also responded to fluctuations in extracellular osmolarity (Figure 1F). Moreover, these environmental cues did not affect the level of total Smad5 protein (Supplementary information, Figure S3G). Together, these data show that Smad5 is sensitive to fluctuations in temperature, pHe and osmotic pressure.
Dynamic nuclear export of Smad5 is CRM1-dependent
Nuclear export is generally dependent on signaling and accomplished by exportins14. The most versatile nuclear export receptor is CRM1, which was first identified in fission yeast15. Leptomycin B (LMB) binds to CRM1 and inhibits the binding of RanGTP as well as other export substrates16. In the solvent-treated control group, methanol treatment did not cause any change in the nucleocytoplasmic shuttling of Smad5 triggered by low temperature (Figure 1G). GFP-Smad5 accumulated exclusively in the nucleus after LMB treatment at 37 °C for 1 h (Figure 1H). These data suggest that at 37 °C, GFP-Smad5 is highly dynamic and capable of both nuclear import and export. In the presence of LMB, switching the cells to 25 °C for as long as 40 min did not induce any Smad5 cytoplasmic accumulation (Figure 1H). These results indicate that low temperature-triggered Smad5 cytoplasmic accumulation is an event requiring active nuclear export and is CRM1-dependent. To further exclude the possibility that low temperature affects Smad5 nuclear import, we first placed HEK293 cells expressing GFP-Smad5 at 25 °C for 30 min. GFP-Smad5 was found almost completely in the cytoplasm under these conditions (Supplementary information, Figure S4A and S4B). Subsequently, methanol or LMB was added to the cells at 25 °C. Strikingly, after 40 min of LMB treatment, the formerly cytoplasmic GFP-Smad5 now entirely accumulated in the nucleus at 25 °C (Supplementary information, Figure S4B). As a control, methanol treatment did not repopulate Smad5 into the nucleus at 25 °C (Supplementary information, Figure S4A). Thus, Smad5 preserves its ability to be imported into the nucleus at 25 °C but its low temperature-mediated nuclear export results in accelerated cytoplasmic accumulation of Smad5.
Classic NESs are required for CRM1-dependent nuclear export17. We were able to find three such NESs in the Smad5 sequence (Figure 1I). Smad5 mutants with any one of these NES sites mutated showed nuclear accumulation at 37 °C and exhibited cytoplasmic translocation at 25 °C, despite the fact that they all exhibited a slower shuttling rate compared to wild-type (WT) Smad5 (Supplementary information, Figure S4C-S4F). These data indicate that all three NESs take part in low temperature-triggered nucleocytoplasmic shuttling of Smad5 although they are not absolutely indispensable. When we mutated all three NESs in Smad5 (3NESmut), we found that the mutant Smad5 showed exclusive nuclear localization at either 37 °C or 25 °C (Figure 1J). Therefore, the nuclear export of Smad5 triggered by low temperature depends on the three NESs scattered along the linker region and the MH2 domain. In addition, the functional redundancy of the Smad5 NES elements can also account for the immediate cell response to the fluctuation of environmental cues through rapid Smad5 nuclear export.
Nucleocytoplasmic shuttling of Smad5 in response to environmental cues is independent of BMP signaling
To date, the translocation of Smad5 from cytoplasm to nucleus has been exclusively tied to BMP signaling18. Interestingly, we did not detect any changes in Smad5 distribution when BMP signaling was blocked with LDN193189 (LDN) or activated with BMP4; under these conditions, low temperature persistently triggered cytoplasmic accumulation of Smad5 (Supplementary information, Figure S2A and S5A). We therefore generated a deletion mutant (GFP-Smad5 Δ11) of Smad5 by removing the C-terminal 11 amino acids that contain the BMP-responsive SSVS phosphorylation sites. We asked whether the nucleocytoplasmic shuttling of Smad5 in response to temperature is affected by this mutation. We found that GFP-Smad5 Δ11 was largely distributed in the nucleus at 37 °C, similar to WT Smad5, and translocated into the cytoplasm when cells were shifted to 25 °C (Supplementary information, Figure S5B). We then directly mutated the SSVS phosphorylation sites to AAVA (3A) or DDVD (3D) to completely block signaling-mediated phosphorylation or dephosphorylation of Smad5. Similarly, the Smad5 3A or Smad5 3D mutant showed temperature-induced nuclear export comparable to WT Smad5 (Supplementary information, Figure S5B). Even in the absence of Smad4, GFP-Smad5 shuttled from the nucleus to the cytoplasm in response to cold, alkaline pHe or hypertonic conditions, and accumulated in the nucleus under acidic pHe or hypotonic conditions (Supplementary information, Figure S5C-S5E). Collectively, these results suggest that the dynamic nucleocytoplasmic redistribution of Smad5 triggered by environmental cues is independent of BMP signaling, of carboxyl-tail phosphorylation state and of interaction with Smad4.
The temperature, pHe and osmotic pressure response all converge to pHi
Since nucleocytoplasmic shuttling of Smad5 responds to temperature, pHe and osmotic pressure with a very similar bidirectional pattern, we hypothesized that a common factor might link these environmental changes. Low temperature attenuates the cellular metabolic rate and decreases proton production within the mitochondria, subsequently alkalizing the cytosol. It has been demonstrated that pHe levels profoundly affect membrane transporters and lead to intracellular pH (pHi) fluctuation in the same direction2. High-osmotic pressure has also been shown to increase pHi1,2. To test whether temperature, pHe and osmotic pressure indeed converge on pHi, we measured pHi using BCECF-AM19. BCECF fluorescence intensity increased in HEK293 cells shifted from 37 °C to 25 °C, indicating that low temperature induces cytosolic alkalization (Figure 2A and Supplementary information, Figure S6A). The addition of hydrochloric acid (10 mM) caused intracellular acidification and decreased BCECF fluorescence intensity whereas potassium hydroxide (2 mM) treatment rapidly increased pHi (Figure 2B and Supplementary information, Figure S6B). A hypotonic environment also triggered cytosol acidification, whereas hypertonic conditions led to a pHi increase (Figure 2C and Supplementary information, Figure S6C). Taken together, our data confirmed that the response to fluctuation in temperature, pHe and osmotic pressure all converged on a change in pHi.
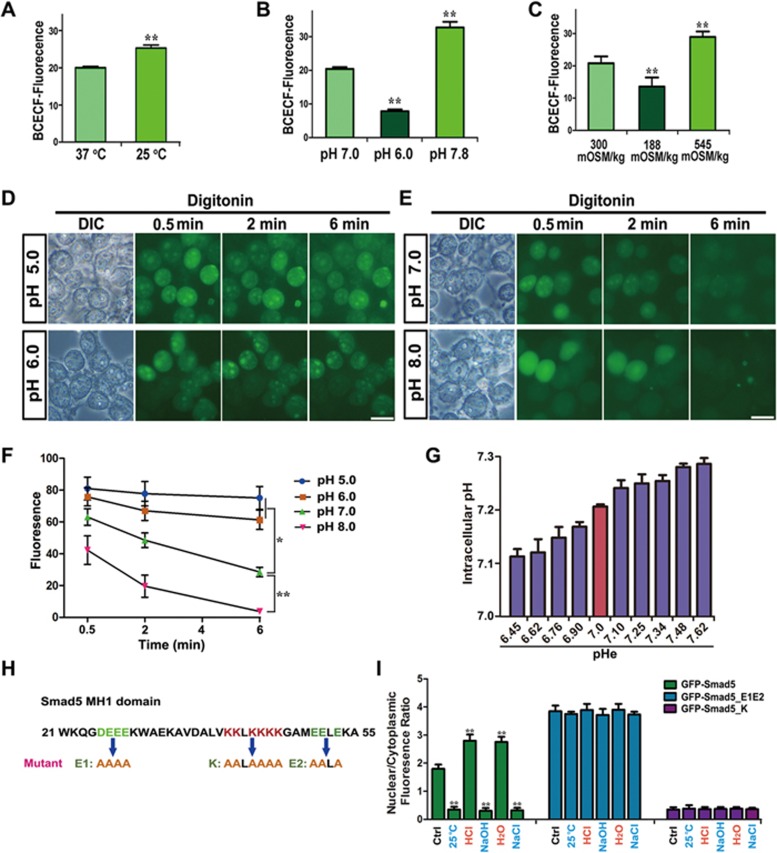
Figure 2.
Smad5 directly senses pHi through charged amino acid clusters. (A-C) Quantifications of intracellular BCECF fluorescence at various temperature (A), pHe (B) and osmolarity (C). Each quantification was made from 10 independent images. Data are presented as mean ± SEM; **P< 0.01. (D, E) Digitonin permeabilization experiments demonstrate that increased pH gradually promotes Smad5 nuclear export. Scale bar, 10 μm. (F) Average fluorescence quantification of nuclear distributed GFP-Smad5 at various pH in D and E (n = 30; data are presented as mean ± SEM; *P < 0.05; **P < 0.01). (G) Measurement of pHi using pHIuorin at different pHe ranges. (H) Diagram showing positions of acidic and basic amino acid clusters. (I) Quantification of nuclear and cytoplasm fluorescence ratio for GFP-Smad5, GFP-Smad5_E1E2 and GFP-Smad5_K at various conditions of temperature, pHe and osmolarity (n = 30; data are presented as mean ± SEM; **P < 0.01).
To further verify that pHi directly regulates Smad5 nucleocytoplasmic shuttling, we treated GFP-Smad5 HEK293 cells with the ionophoric uncoupler of oxidative phosphorylation, FCCP, for 30 min to decrease pHi; this resulted in nuclear accumulation of Smad5 (Supplementary information, Figure S6D and S6E). In addition, we pretreated HEK293 cells carrying the GFP-Smad5 transgene with 10 mM hydrochloric acid for 10 min to trigger nuclear accumulation of Smad5. Cells were then permeabilized by treatment with 160 μg/ml digitonin for 1 min, after which they were soaked in solutions of pH 5.0, 6.0, 7.0 or 8.0. At pH 5.0 or 6.0, nuclear export of Smad5 was almost completely repressed (Figure 2D and 2F). In contrast, GFP-Smad5 was able to exhibit nuclear export at pH 7.0 or 8.0 (Figure 2E and 2F). To determine whether Smad5 responds to pHi changes within a normal physiological range, we measured pHi using pHIuorin20,21 at different pHe as shown in Figure 1C. Statistical analysis showed that pHi increased by 0.17 units when the corresponding pHe changed from 6.45 to 7.62 (Figure 2G), indicating that Smad5 could respond to a subtle physiological range of pHi changes by nucleocytoplasmic shuttling. These data strongly suggest that pHi is a potent factor that regulates the nuclear export rate of Smad5, thus contributing to its dynamic nucleocytoplasmic distribution that is influenced by fluctuations in extracellular cues.
The MH1 domain of Smad5 is essential for its pHi response
Next, we wondered which subdomain of Smad5 responds to pHi changes. To this end we constructed a series of chimeric molecules between Smad1 and Smad5, which are highly homologous but show completely different nucleocytoplasmic translocation properties (Supplementary information, Figure S6F-S6I). Interchanging the MH2 domains between Smad1 and Smad5 did not significantly affect their subcellular distribution at either 37 °C or 25 °C (Supplementary information, Figure S6G). Smad5 maintained its specific properties and could translocate into the cytoplasm at 25 °C upon removal of its entire MH2 domain (Supplementary information, Figure S6H). We then interchanged the MH1 domains of Smad1 and Smad5 and studied their distribution at 37 °C or 25 °C. Interestingly, GFP-Smad5 harboring the MH1 domain of Smad1 behaved similarly to WT GFP-Smad1, and resided in the nucleus at both 37 °C and 25 °C (Supplementary information, Figure S6I). However, GFP-Smad1 harboring the MH1 domain of Smad5 behaved like Smad5 and exhibited nuclear localization at 37 °C but shuttled into the cytoplasm within 10 min at 25 °C (Supplementary information, Figure S6I). Therefore, we conclude that the MH1 domain of Smad5 plays a critical role in responding to pHi changes, while the linker region and MH2 domain contribute to nuclear export at alkaline pHi.
We next asked whether Smad5 directly responds to pHi changes and, if so, what would be the amino acid residues in the MH1 domain responsible for such a role. Because this is a response to pH, we first sought to identify acidic or basic amino acid residues within the MH1 domain that may be essential for the Smad5 pHi response. We identified two acidic and one basic clusters of such charged amino acids in the Smad5 MH1 domain and mutated these residues to alanines (Figure 2H). Compared to the WT, the combined mutant of Smad5's two acidic domains_E1, E2 (E1E2) showed biased nuclear accumulation at 37 °C and exhibited attenuated nuclear export capacity under cold, alkaline pHe or hypertonic conditions (Figure 2I and Supplementary information, Figure S6J). Conversely, the basic domain mutant, Smad5_K, showed cytoplasmic accumulation under normal culture conditions, and acidic pHe or hypotonic environments now failed to promote its nuclear accumulation (Figure 2I and Supplementary information, Figure S6J). Taken together, these data strongly suggest that these three charged amino acid clusters in the MH1 domain of Smad5 are required for the direct sensing of pHi.
Smad5 deficiency causes downregulation of metabolism-related genes
To investigate the biological functions of this highly dynamic and pHi-sensitive protein, we deleted Smad5 in human embryonic stem cells (hESCs) through donor-mediated homologous recombination in combination with transcription activator-like effector nucleases (TALENs)-induced DNA cleavage (Supplementary information, Figure S7A)22,23. Knockout (KO) success was confirmed by genomic PCR and western blotting (Supplementary information, Figure S7B and S7C). A similar strategy was applied to generate HEK293 and HeLa Smad5 KO cell lines (Supplementary information, Figure S7D and S7E).
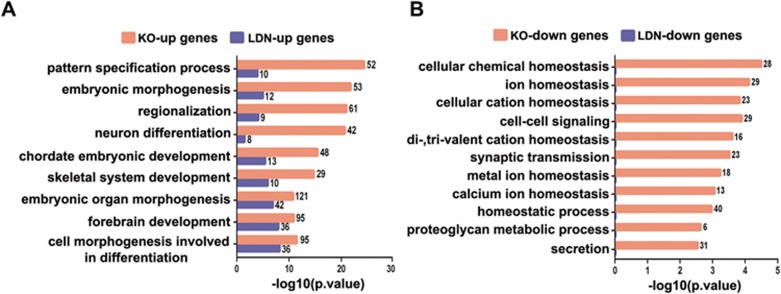
RNA-seq was performed to systematically compare the transcriptomes of WT, Smad5 KO and LDN-treated hES cells. Data retrieved from three independent WT and KO lines revealed ∼468 upregulated and ∼438 downregulated genes upon Smad5 KO (Supplementary information, Figure S7F and S7G). There was a significant overlap between up and downregulated genes among these three WT and KO lines (Supplementary information, Figure S7H). We also treated hES cells with LDN, which prevented Smad1/5/8 SSXS phosphorylation (Supplementary information, Figure S8A) and then analyzed the transcriptome profiles. This showed that LDN treatment induced much fewer changes in gene expression compared to the Smad5 KO (Supplementary information, Figure S8B). Among those upregulated or downregulated genes, only 32 or 1, respectively, overlapped between the Smad5 KO and LDN groups (Supplementary information, Figure S8C). This suggests that disrupting Smad5 results in a different gene expression profile compared to the blocking of BMP signaling. Gene ontology (GO) analyses showed that upregulated genes in the Smad5 KO group were mainly involved in the biological processes of embryonic development and patterning and showing a certain degree of similarity with LDN upregulated genes (Figure 3A). Downregulated gene groups in Smad5 KO cells belonged mainly to the category of cellular hemostasis including cellular chemical homeostasis; ion homeostasis; and proteoglycan metabolic processing. These functions have no overlap with those of LDN downregulated genes (Figure 3B). The expression patterns of the top 10 most downregulated genes were also validated by qPCR in WT, Smad5 KO, LDN-treated and Smad4 KO cells (Supplementary information, Figure S8D).
Figure 3.
Smad5 KO induces specific transcriptional profiling changes related to metabolic homeostasis. (A, B) Functional annotation of up or downregulated gene categories in Smad5 KO and LDN193189-treated hES cells identified by GO.
We then reintroduced Smad5 or GFP-Smad5 into the Smad5 KO hES cell line through lentiviral transduction (Supplementary information, Figure S9A). Surprisingly, neither Smad5 nor GFP-Smad5 restored the expression of those downregulated genes caused by disruption of endogenous Smad5 (Supplementary information, Figure S9B). We therefore carried out pre-knockout rescue, in which Smad5 or corresponding mutants were first overexpressed and then followed by endogenous Smad5 ablation. KO or overexpression efficacy was verified by genomic PCR and western blotting (Supplementary information, Figure S9C and S9D). Under these conditions, downregulation of gene expression induced by the Smad5 KO was significantly rescued by pre-expression of Smad5 (Supplementary information, Figure S9E).
We found that Smad5_E1E2 was mostly present in the nucleus, whereas GFP-Smad5_K was largely distributed in the cytoplasm (Figure 2I and Supplementary information, Figure S6J). A co-immunoprecipitation (Co-IP) experiment showed that BMP4 stimulation significantly triggered the interaction of WT Smad5 with Smad4. In contrast, GFP-Smad5_K showed no significant Smad4 interaction (Supplementary information, Figure S9F). Notably, pre-rescue with Smad5_K, but not Smad5_E1E2, largely prevented Smad5 KO-induced downregulation of gene expression (Supplementary information, Figure S9E). Genome sequencing confirmed the genomic DNA integrity of the transduced Smad5_E1E2 after TALENs targeting (Supplementary information, Figure S9G). The differences between Smad5_K and Smad5_E1E2 in rescuing expression of genes resulting from the Smad5 KO further indicate that it is cytoplasmic Smad5 but not nuclear Smad5 or the BMP/Smad5 signaling pathway, that maintains a normal cellular homeostatic state. Moreover, acidic and basic pH treatment showed that the shuttling of Smad5 into or out of the nucleus for various time periods could not acutely regulate expression of the above characteristic genes (Supplementary information, Figure S10A-S10C), thus indicating a non-transcriptional role of Smad5 in this process.
Disruption of Smad5 reduces glycolytic rate
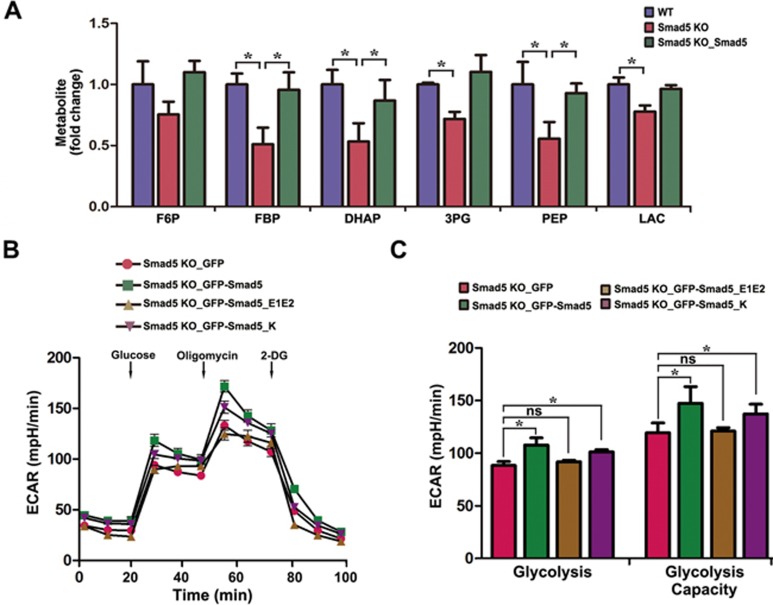
To further delineate the cellular homeostatic deficits in Smad5 KO cells, we profiled metabolites in WT, Smad5 KO and Smad5 KO with Smad5 pre-expression hES cell lines using liquid chromatography tandem mass spectrometry (LC-MS/MS). The Smad5 deficiency resulted in lower levels of glycolytic intermediates. These levels were largely rescued by pre-expression of Smad5 (Figure 4A), indicating that Smad5 is required for maintaining a normal glycolysis rate. In addition, the extracellular acidification rate (ECAR) exhibited that glycolysis was rescued by GFP-Smad5 and GFP-Smad5_K, but not GFP-Smad5_E1E2, suggesting that cytoplasmic Smad5 can promote a higher glycolysis rate (Figure 4B and 4C). In contrast, Smad1 and Smad8 did not affect the glycolysis rate (Supplementary information, Figure S11A and S11B). Together, these results suggest that Smad5 is an important regulator of the rate of glycolysis.
Figure 4.
Loss of function of Smad5 causes glycolysis defects. (A) LC-MS/MS analysis of abundance in glycolytic intermediates in WT, Smad5 KO and Smad5 pre-rescue hES cells. Data are represented as mean ± SEM of three independent experiments. Unpaired two-tailed Student's t-test. *P < 0.05. (B) GFP, GFP-Smad5, GFP-Smad5_E1E2 and GFP-Smad5_K were expressed in Smad5 KO HEK293 cells, and extracellular acidification rate (ECAR) was measured with the Seahorse Analyzer (n = 6). (C) Statistics of glycolysis and glycolysis capacity in B. Data are represented as mean ± SEM of six independent experiments. Unpaired two-tailed Student's t-test. *P < 0.05.
Smad5 KO induces irreversible cellular bioenergetic homeostasis and impairs mitochondrial respiration
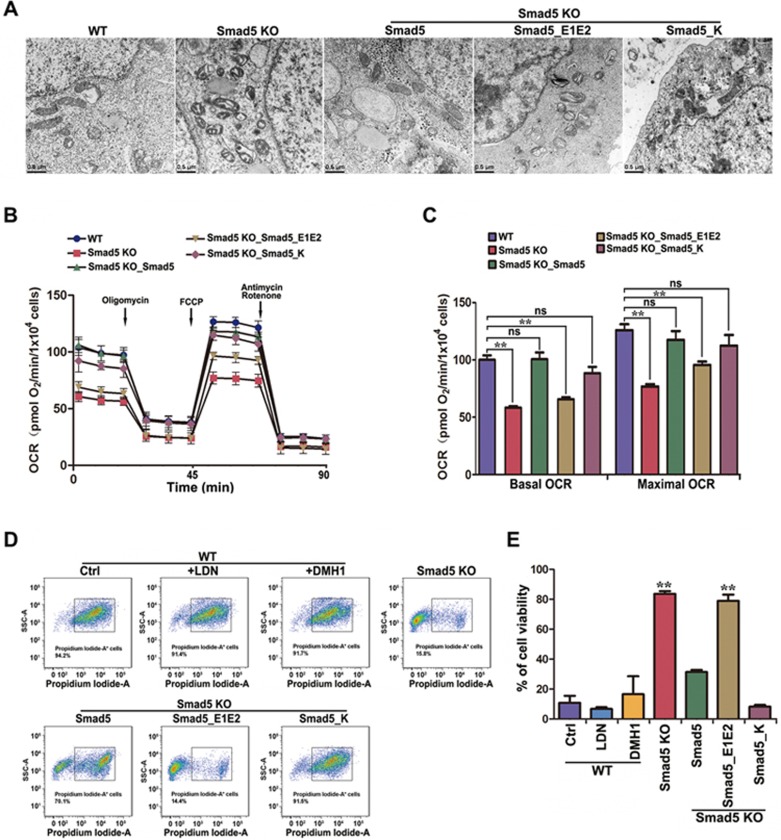
Mitochondria play a central role in cellular respiration and energy production24,25. Transmission electron microscopy showed that the mitochondria of Smad5 KO hES cells mostly exhibited swollen and vacuolated features (Figure 5A), suggesting severe mitochondrial dysfunction. Re-expressing Smad5 in Smad5 KO hES cells through lentiviral transduction could not rescue the abnormal mitochondrial morphology, suggesting that the mitochondrial dysfunction caused by Smad5 KO is irreversible (Supplementary information, Figure S12). The abnormal mitochondrial morphology in Smad5 KO cells can, as predicted, be rescued by pre-expression of ectopic Smad5 or Smad5_K, but not Smad5_E1E2 (Figure 5A), further suggesting a cytoplasmic role of Smad5 in maintaining normal mitochondrial function. We next used a flux analyzer to evaluate the oxygen consumption rate (OCR). Both the basal OCR and maximal OCR decreased in Smad5 KO hES cell lines (Figure 5B and 5C) and this could be rescued by pre-expression of Smad5 and Smad5_K, but not Smad5_E1E2. We also found the same phenomenon in HEK293 cells and HeLa cells (Supplementary information, Figure S13A-S13D). In striking contrast to the Smad5 KO, loss of Smad4 did not profoundly affect the maximal OCR (Supplementary information, Figure S13E and S13F). Although re-introducing GFP-Smad5 into Smad5 KO HEK293 cells increased OCR, overexpression of GFP-Smad1 and GFP-Smad8 had no effect on either basal or maximal OCR (Supplementary information, Figure S13G and S13H).
Figure 5.
Smad5 KO causes deficiency in mitochondrial respiration and malfunction of cellular bioenergetic homeostasis. (A) Electron microscopy images showing mitochondrial morphology in WT, Smad5 KO and pre-rescue hES. Scale bar, 0.5 μm. (B) Oxygen consumption rate (OCR) changes under mitochondrial stress in WT, Smad5 KO and pre-rescue hES as measured using the Seahorse Analyzer (n = 6). (C) Statistics of basal and maximal OCR in B. Data are represented as mean ± SEM of 6 independent experiments. Unpaired two-tailed Student's t-test. **P < 0.01. (D) Flow cytometry sorting of hES cells after propidium iodide staining. (E) Statistics of cell viability following serum withdrawal. Data are presented as mean ± SEM in three independent experiments. **P < 0.01.
Furthermore, considering the obligatory role of Smad5 in maintaining bioenergetic homeostasis, we wondered whether Smad5 KO cells could tolerate acute nutrient stress such as serum deprivation. Surprisingly, we found that WT hES cells were sensitive to nutrient stress and could hardly survive upon removal of serum from the culture. Propidium iodide-A staining followed by flow cytometry analysis revealed massive amounts of cell death 3 days after serum deprivation (Figure 5D and 5E). Remarkably, Smad5 KO hES cells survived well under these extreme nutrient stress conditions (Figure 5D and 5E), suggesting a completely transformed bioenergetic homeostasis state. It should be noted that this abnormal bioenergetic homeostasis state of Smad5 KO cells is not related to deficiencies in BMP signaling, since neither LDN nor DMH1 pretreatment of WT cells could recapitulate the improved survival of Smad5 KO cells under serum deprivation conditions (Figure 5D and 5E). Moreover, pre-expression of ectopic Smad5 or Smad5_K, but not Smad5_E1E2, successfully prevented the transformation of bioenergetic homeostasis (Figure 5D and 5E).
Smad5 KO disrupts neural differentiation of hES cells
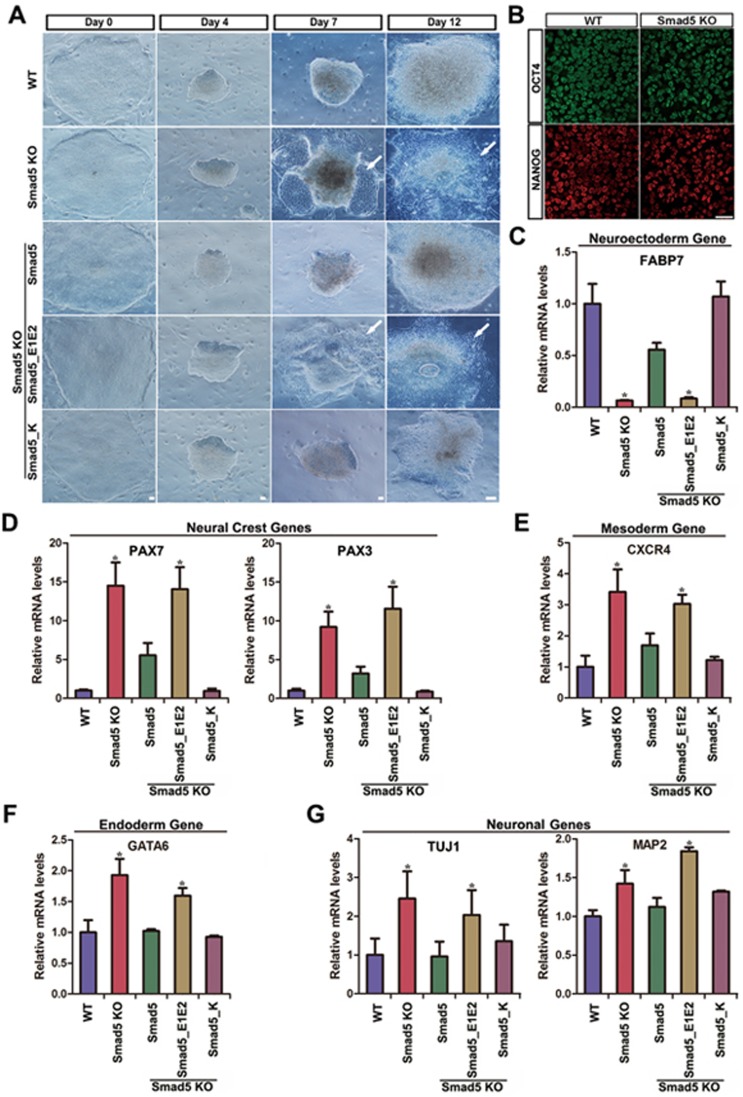
Neural specification is a crucial developmental process which is tightly linked to both BMP signaling and metabolic processes26. Increased pHi is required for embryonic stem cell differentiation27, and more Smad5 was located in the cytosol during early neural differentiation (Supplementary information, Figure S14A and 14B). We therefore explored the function of Smad5 in a neural differentiation model26,28. Smad5 KO hES cells could maintain an ESC-like morphology, and pluripotency transcription factors were equally expressed in WT and Smad5 KO cells (Figure 6A and 6B). However, Smad5 KO cells differentiated toward a neural lineage in an unsynchronized fashion compared to WT cells at day 7 or day 12 (Figure 6A). The abnormal neural morphology in Smad5 KO cells could be rescued by pre-expression of Smad5 and Smad5_K, but not Smad5_E1E2 (Figure 6A). In accord with these morphological findings, qPCR analysis of Smad5 KO cells at day 12 showed lower expression of neuroectodermal genes, but mesendodermal genes, neural crest genes and neuron maturation genes were expressed at higher levels. This further suggests a disrupted neural developmental process (Figure 6C-6G). Again, these abnormal gene expression patterns could be pre-rescued by Smad5 or Smad5_K, but not Smad5_E1E2 (Figure 6C-6G). Together, these results suggest that cytoplasmic Smad5 possesses an important role in guiding neural development and that the bioenergetic state maintained by cytoplasmic Smad5 might be required for normal neural development.
Figure 6.
Smad5 KO hES impairs normal neural development processes. (A) Bright field images show distinct steps of neural induction in WT, Smad5 KO and pre-rescue hES. Arrowhead shows the abnormal neural morphology. Day 0, hES stage; Day 4-7, neuroectoderm; Day 12, neural progenitors. Scale bars, 25 μm. (B) Smad5 KO had no effect on hES pluripotency by criterion of OCT4 and Nanog immunofluorescence staining at Day 0. Scale bar, 25 μm. (C-H) The relative mRNA expression level of neuroectoderm, neural crest, mesoderm, endoderm and neuronal genes in neural progenitor stage at day 12. Data are presented as mean ± SEM of three independent experiments. *P < 0.05.
Smad5 maintains bioenergetic homeostasis by interacting with HK1
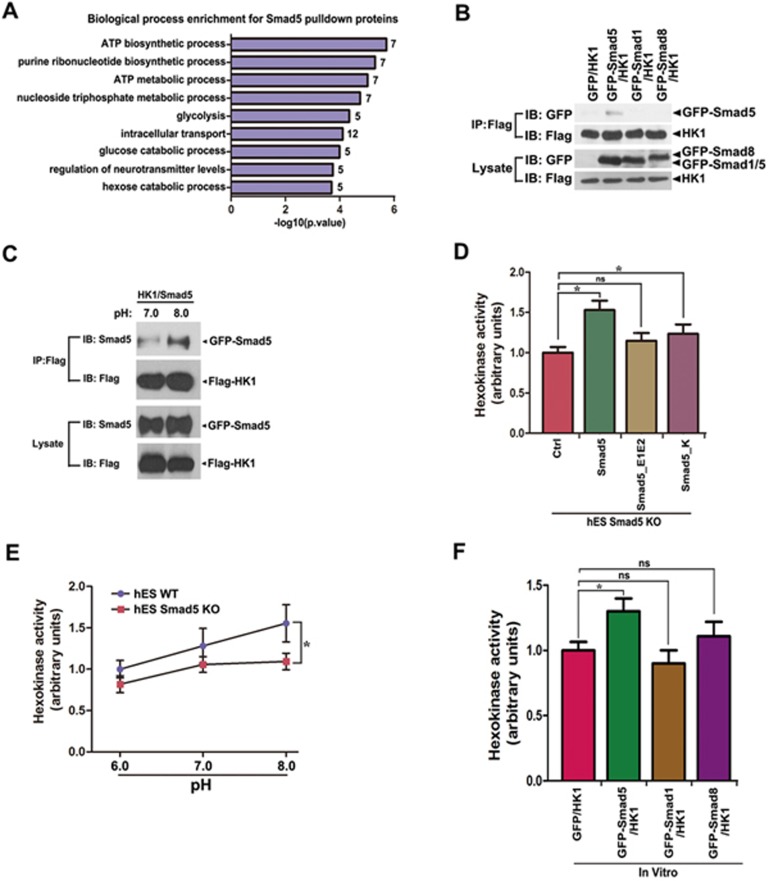
To dissect the underlying molecular mechanisms by which pHi-sensitive Smad5 maintains bioenergetic homeostasis, we performed Co-IP followed by mass spectrometry (IP/MS) to investigate the Smad5 interactome. We identified 67 potential binding partners of GFP-Smad5 (Supplementary information, Table S1). GO analysis of these candidates showed that they were mainly involved in ATP biosynthetic and metabolic processes (Figure 7A), supporting the above conclusion that Smad5 plays an important role in maintaining bioenergetic homeostasis in the cytoplasm. Among these candidates, we identified HK1, a rate-limiting enzyme of glycolysis29,30,31,32. Ectopic expression of GFP-Smad5 and Flag-HK1 in HEK293 cells confirmed their interaction (Figure 7B, Supplementary information, Figure S15A and S15B). Smad1 and Smad8 did not interact with HK1 (Figure 7B). More importantly, when pHe was adjusted from 7.0 to 8.0, the interaction between Smad5 and HK1 was greatly potentiated, coinciding with the cytoplasmic accumulation of Smad5 under the alkaline pHe conditions (Figure 7C).
Figure 7.
Smad5 dynamically regulates HK1 activity. (A) GO analysis of 67 potential Smad5 binding proteins identified by IP/MS. (B) HEK293 cells co-expressing of Flag-HK1 and GFP, GFP-Smad5, GFP-Smad1, GFP-Smad8 were immunoprecipitated with the GFP antibody, and specific interaction of GFP-Smad5 with Flag-HK1 is shown by western blotting. (C) HEK293 cells were cotransfected with GFP-Smad5 and Flag-HK1. Extracellular pH was then adjusted to 7.0 or 8.0 for 30 min followed by immunoprecipitation with the anti-Flag antibody. A larger amount of Smad5 interacts with HK1 at pHe 8.0 as compared with that of pHe 7.0. (D) Hexokinase enzyme activity was detected in Smad5 KO and various pre-rescue hES. Data are presented as mean ± SEM of three independent experiments. *P < 0.05. (E) WT and Smad5 KO hES cells were incubated in pH 6.0, pH 7.0 and pH 8.0 buffer for 30 min, respectively, followed by hexokinase enzyme activity detection. Data are presented as mean ± SEM of three independent experiments. *P < 0.05. (F) GFP-Smad5 binding leads to HK1 activity increase in vitro. HK1 activity is measured in vitro after incubation of GFP, GFP-Smad5, GFP-Smad1 or GFP-Smad8 proteins using hexokinase enzyme activity assay kit. Data are presented as mean ± SEM of three independent experiments. *P < 0.05.
In vitro analysis revealed that HK enzymatic activity decreased in hES, HEK 293 and HeLa cells when the Smad5 gene was deleted (Supplementary information, Figure S15C). HK activity could be pre-rescued by Smad5 or Smad5_K, but not Smad5_E1E2 in Smad5 KO hES or HEK293 cells (Figure 7D and Supplementary information, Figure S15D). Moreover, HK activity gradually increased when pHe was shifted from 6.0 to 8.0 in WT hES, HEK293 and HeLa cells (Figure 7E, Supplementary information, Figure S15E and S15F). However, this tendency of HK activity to increase in response to pHe alkalization was significantly abrogated in Smad5 KO cells (Figure 7E, Supplementary information, Figure S15E and S15F).
We then re-constructed HK1 enzymatic activity in vitro by purifying Flag-HK1 from Smad5 KO HEK293 cells. Strikingly, supplying GFP-Smad5, but not GFP, GFP-Smad1 or GFP-Smad8, led to a significant increase in HK1 activity (Figure 7F), demonstrating a direct role of GFP-Smad5 in HK1 activation. Together, these data suggest that during pHi fluctuation, cytoplasmically localized Smad5 interacts with HK1, thus increasing HK1's enzymatic activity as a glycolytic regulator.
Discussion
Smad5, together with Smad1 and Smad8, act as important substrates for BMP receptors, which play critical roles in embryogenesis and tissue homeostasis33,34. Smad1/5/8 are closely related and share similar sequences. However, our present results show that Smad5 differs from Smad1/8 in several aspects. First, these molecules have different subcellular locations. Smad5 is located both in the cytoplasm and nucleus; Smad1 is largely distributed in the nucleus; and Smad8 is mostly cytoplasmic. Differential subcellular protein localization suggests that their functions may be different. Second, the results presented here demonstrate that Smad5 is sensitive to pHi changes. Basic pHi promotes Smad5 to translocate from nucleus to cytoplasm, whereas it accumulates in the nucleus when pHi decreases. In contrast, Smad1 and Smad8 do not shuttle upon pHi fluctuations. Notably, the charged amino acid clusters in the Smad5 MH1 domain that respond to pHi changes are also found in Smad1 and Smad8, indicating that these amino acid clusters are necessary but not sufficient for the translocation of Smad5. In addition, Smad5 can act in a transcription-independent manner to regulate glycolysis and mitochondrial respiration, thus maintaining cellular bioenergetic states, while Smad1 and Smad8 cannot. Together, these results show that Smad5 possesses unique roles that differ from those of Smad1 and Smad8.
Intracellular pH plays an important role in maintaining the structure and function of proteins, regulating intracellular metabolic reactions and protein interactions. Hence pHi is stringently regulated35. Our findings demonstrate that Smad5 actively senses pHi through nucleocytoplasmic shuttling. Importantly, the pHi changes sensed by Smad5 are within a physiologically relevant range. We show that Smad5 can sense pHi fluctuations and subsequently regulate intracellular metabolic reactions. Therefore, Smad5 plays a key role in pHi sensing and modulation. pHi dynamics also play an essential role in development: increasing pHi is necessary for mouse embryonic stem cell (mES) differentiation, while lower pHi impairs mES differentiation27. Our data also show that pHi increases during neural specification, and that Smad5 KO impairs the differentiation of hES cells towards a neural lineage. Taken together, our findings underscore the importance of Smad5 in pHi sensing and development.
The specific subcellular localization of a protein is crucial for its normal function36,37,38. Transcription factors, protein/RNA chaperones, cell cycle modifiers and various signaling components show precise regulation of their subcellular distribution38,39,40,41. Usually, translocation of a protein is regulated by signaling-mediated phosphorylation/dephosphorylation, protein-protein interactions and ATP/ADP or GTP/GDP binding states8,38,39,40,41. Here we demonstrate that Smad5 is a highly dynamic protein that frequently translocates between the nucleus and the cytoplasm, providing the first example of a protein whose subcellular localization can be dramatically regulated by temperature, pHe, osmolarity as well as pHi. pHi is fundamental for basic cellular biochemistry including enzyme activity, energy metabolism, cell proliferation and migration42,43. Empirically, pHi can be monitored by pH-sensitive dyes, microelectrodes or MRI44. Since Smad5 is highly pHi sensitive, it may also be applied to dynamically monitor pHi within the physiological range in in vitro-cultured cells or in living animals.
In conclusion, our current work demonstrates a noncanonical role of Smad5 in responding to pHi changes and regulating cellular bioenergetic states by controlling the rate of glycolysis. The role of Smad5 in regulating glycolysis by interacting with HK1 may be direct, because the enzymatic activity of HK1 reconstituted in vitro could be modified by adding purified Smad5 and second, the glycolysis rate in Smad5 KO hES cells could be rescued by either post-knockout or pre-knockout re-expression of Smad5 (Supplementary information, Figure S15G and S15H). However, it is also noteworthy that disruption of Smad5 induces an irreversible cellular bioenergetic homeostasis transformation, as evidenced by permanent mitochondrial dysfunction, which could not be reversed by post-knockout re-expression of Smad5 (Supplementary information, Figure S12A). Therefore, Smad5 may play multiple roles in both the immediate balancing and long-term maintenance of cellular bioenergetic states through sensing environmental stresses by acting as a pHi messenger.
Materials and Methods
Plasmids construction
GFP-Smad5, Smad5 and its mutants were cloned into the pLVX-Tight-Puro vector (Clontech). The pLVX-ef1a-rtTA advanced lentiviral vector was used together with the pLVX-Tight-Puro expression vector for making inducible overexpression lines.
Cell culture and generation of stable cell lines
Human Embryonic Kidney (HEK293FT, Invitrogen) cells were regularly cultured in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal bovine serum (GIBCO) in 5% CO2 at 37 °C. Human ES cells (H9, WiCell) were cultured on irradiated mouse embryonic fibroblasts (MEF) as previously described45. Lentiviruses were packaged in HEK293FT cells as previously described45. Viruses were then concentrated by ultracentrifugation and used for making stable lines. Mycoplasma detection tests were conducted in a weekly base to ensure exclusion of any contaminated cells.
Immunofluorescence
HEK293 cell lines stably infected with GFP or GFP-Smad genes were grown on coverslips in 24-well plates. For studying protein distribution at 37 °C, cells were treated with doxycycline for 24 h to induce protein expression and were then immediately fixed for 10 min at 37 °C in 4% paraformaldehyde (PFA)/PBS pre-warmed to 37 °C. Nuclei were stained with Hoechst for 5 min after membrane permeabilization with 0.2% Triton X-100/PBS. To study protein distribution at 25 °C, cells were moved to a 25 °C incubator supplied with 5% CO2 for the indicated time intervals and then fixed for 10 min at 25 °C with 25 °C pre-conditioned 4% PFA/PBS. To study protein distribution at various pH or osmotic pressures, cells were treated with hydrochloric acid, sodium hydroxide, pure water or sodium chloride for 10 min at 37 °C, and then fixed for 10 min at 37 °C with 4% PFA/PBS pre-warmed to 37 °C.
Nuclear and cytoplasmic fractionation and western blotting
HEK293 cells growing in 10 cm dish for 48 h were given indicated treatments for 15 min including temperature shifts (37 °C and 25 °C); extracellular pH changes (10 mM HCl and 2 mM NaOH); or changes in external osmotic pressure (250 mOSM/kg or 350 mOSM/kg adjusted through H2O or NaCl). Cells were then permeabilized using 3 ml of a physiologically isotonic solution (140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM glucose and 10 mM MOPS (pH 6.0 for conditions: 37 °C, extracellular pH 6.0, osmotic pressure 250 mOsm/kg; pH 7.2 for conditions: 25 °C, extracellular pH 7.6, osmotic pressure 350 mOsm/kg)) containing 160 μg/ml digitonin (Sigma) for 3 min. Nuclei were pelleted by centrifugation at 300× g for 5 min and supernatants containing cytoplasmic proteins were collected. Western blotting was performed following electrophoresis on a 10% SDS-polyacrylamide gel. The antibodies used for immunoblotting were rabbit anti-Smad5 (1:1 000, Cell Signaling, 9517), mouse anti-Lamin A/C (1:500, Santa Cruz, SC-7292), mouse anti-β-Tubulin (1:4 000, Sigma, T5201).
Measurement of pHi
The pHi was measured using the pH-sensitive fluorescent probe BCECF-AM [2′,7′-bis-(2-carboxyethyl)-5-carboxyfluorescein-acetoxymethyl ester; Beyotime, China]19. HEK293 cells were incubated for 20 min at 37 °C with 1 μM BCECF-AM in Hank's balanced salt solution. Cells were then rinsed twice for 5 min intervals to fully remove the dye and were treated under differing conditions of: temperature (37 °C and 25 °C); extracellular pH (pH 6.0, pH 7.0, pH 7.8); and external osmotic pressure (188 mOsm/kg, 300 mOsm/kg, 545 mOsm/kg). BCECF fluorescence was imaged using a standard fluorescence filter set19,46.
The cytosolic pH in live cells was measured using pHIuorin20,21, a genetically encoded pH indicator. Cells were seeded in black 96-well flat bottom plates, transfected with the plasmid DNA to express pHIuorin for 30 h. Cells were washed twice with PBS in each well after carefully removing the medium. Readings were recorded after adding 100 μl PBS for 30 min at 37 °C by a Synergy 2 Multi-Mode Microplate Reader (BioTek) with excitation filters of 400 BP 10 nm and 485 BP 20 nm, and emission filter of 528 BP 20 nm (for both excitation wavelengths). Fluorescence values were corrected for background fluorescence by subtracting the intensity of the cell samples not expressing sensors. All samples were run in triplicate.
Derivation of Smad5 and Smad4 knockout cell lines
The Smad5-targeting donor vector comprised a 766 bp 5′ homology arm, puro resistance gene and a 764 bp 3′ homology arm. TALENs were designed and assembled as described47,48. Smad5-targeting sequences were TGTCAAATGACGTCAATGGC for left TALEN, CTAGTCCAGCAGTAAAGCGA for right TALEN. TALEN-mediated homologous recombination in hES cells was performed as previously described48. Genomic DNA PCR primers are as follows: HR-F: AGCATATGGCACTTGTGAAG, Puro-F: TTCCTGGCCACCGTCGGCGTCTC, HR-R: CTTTAGCTCATGATGACTCTGC. Construction of the Smad4 knockout HEK293 cell lines was described as previously23.
Co-immunoprecipitation and western blotting
For co-immunoprecipitation analysis, cells were lysed in IP buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM NaF, 1% Triton X-100, 0.25% sodium deoxycholate and 5% glycerol) supplemented with protease inhibitors and phosphatase inhibitors. Total lysates were incubated in the presence of 1 μg primary GFP antibody (A6455, life technologies) mixed with 30 μl of 50% slurry Protein-G Sepharose beads (Roche) or incubated with anti-Flag M2 beads (Sigma) according to the manufacturer's protocols. Immunoprecipitates and total lysates were subjected to immunoblotting with the indicated antibodies.
Metabolism analysis
Targeted LC-MS/MS-based metabolomics analysis was performed as previously described49. 107 hES cells were harvested in 80% (v/v) methanol at −80 °C. Insoluble material in lysates was centrifuged at 12 000 rpm for 15 min, and the resulting supernatant was evaporated using a refrigerated speed vac.
Seahorse assay
hES cells were cultured on feeder cells in XF96 cell culture microplate (Seahorse Bioscience). HEK293 and Hela cells were seeded onto a Matrigel-coated XF96 cell culture microplate. ECAR and OCR were measured using a standard protocol (Seahorse Bioscience) and were normalized using counted cell numbers.
Electron microscopy
hES cells were fixed overnight at 4 °C in phosphate buffer (PB) containing 2% glutaraldehyde. After five washes in PB buffer (pH 7.4) for 5 min each, cells were post-fixed for 1 h at RT in 1% osmium tetroxide (OSO4). After further washes with PB, cells were dehydrated in 30%, 50%, 70% and 90% acetone (30 min per step), and the cells were then further dehydrated in 100% acetone three times for 11 min each at RT. The samples were then infiltrated in 3:7 (vol/vol) acetone/epon for 2 h, followed by 7:3 acetone/epon overnight at RT, 100% epon for 2 h and finally 100% epon for 48 h at 60 °C for polymerization. The sections were supported on copper grids. The 70 nm sections were post-stained in Uranyl acetate and lead citrate, and the stained sections were imaged using a transmission electron microscope (JEM-1230) operated at 160 kV.
Propidium iodide staining and FACS analysis
To quantify hES cells viability, single-cell suspensions were prepared using trypsin, washed in PBS and then fixed in 70% ethanol. Cells were incubated in 0.1 mg/ml PI (Sigma) for 30 min at room temperature, washed and analyzed by flow cytometry.
Hexokinase activity assay
For HK1 activity measurement in vitro, HEK293 Smad5 KO cells transiently expressing flag-HK1 were lysed in IP buffer and the lysates centrifuged at highest speed. Supernatants were incubated with anti-Flag M2-conjugated agarose beads for 6 h at 4 °C. The M2 beads were then washed three times with wash buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 5% glycerol). The pulled-down flag-HK1 protein was then eluted from the beads with elution buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 150 ng/μl 3×Flag peptide, 1 mM PMSF) for 1 h at 4 °C. For purification of GFP-Smad5, GFP-Smad1, GFP-Smad8 and GFP, HEK293 Smad5 KO cells were transfected with GFP, GFP-Smad5, GFP-Smad1 or GFP-Smad8 plasmids. Cells were lysed in IP buffer, after centrifugation, supernatants were mixed with 1 μg primary GFP antibody (A6455, life technologies) and 30 μl of 50% slurry Protein-G Sepharose beads (Roche) for 6 h at 4 °C. The beads were then washed three times with high salt buffer (50 mM Tris-HCl pH 7.4, 300 mM NaCl, 1 mM EDTA, 1% Triton X-100, 5% glycerol). The purified GFP, GFP-Smad5, GFP-Smad1 and GFP-Smad8 proteins were then incubated with eluted flag-HK1 protein in HK Assay Buffer for 1 h at 4 °C. HK1 activity was measured with Hexokinase Colorimetric Assay Kit (Biovision).
The HK activity in HEK293, Hela and hES were measured with Hexokinase Colorimetric Assay (Biovision), according to the manufacturer's instructions.
Statistical analysis
Data are presented as mean ± SEM. Unpaired two-tailed Student's t-test for two groups. One-way ANOVA was used for multiple comparisons. Statistical significance is considered at P-values below 0.05. *P < 0.05; **P < 0.005.
Data availability
All high-throughput data have been deposited at the Gene Expression Omnibus under accession number GSE99687.
Author Contributions
ZX, LL, XB, YY, ZQ and JG conceived the study. FY performed most of the experiments. LZ, CZ, XX, WY, ZY, FB, LY, SL, HY, ZX, ZB, FH, ML and ZQ helped to setup the gene targeting system, culture hES and part of the molecular study. XM performed the MS/MS analysis and HK activity assay. ZX, DY and JC performed the bioinformatics analysis. FY, XB, LL, DT-K and XZ wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Xinhua Feng for offering Smad cDNAs. We thank Zhengliang Gao, Andong Qiu and Ying Cao for helpful discussions. We also thank Margaret Ho for proof reading of the manuscript. This study was supported by the National Basic Research Program of China (2013CB967600), Science and Technology Commission of Shanghai Municipality (15JC1400202, 15XD1503800), National Natural Science Foundation of China (31471040, 31400934, 31225008, 31671484), Ministry of Education (IRT1168) and Fundamental Research Funds for the Central Universities.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Smad5 accumulates in cytoplasm at low temperature.
Oxidative stress, membrane potential and signaling modifiers could not obviously affect Smad5, Smad1 or Smad8 subcellular distribution under normal or low temperature culture conditions.
Smad5 responses to pHe and osmolarity fluctuations.
Smad5 subjects to accelerated CRM1-dependent nuclear export at low temperature and three NESs are involved in its rapid nuclear export.
Environmental cues triggered Smad5 translocation is BMP signaling-independent.
Temperature, pHe and osmolarity fluctuations can induce pHi changes and Smad5 MH1 domain is responsible for pHi sensing.
Transcriptome analysis of WT and Smad5 KO hES.
Transcriptome analysis of WT and LDN treated hES.
Chronic metabolic adaptation of Smad5 KO hES.
Smad5 shuttling could not regulate metabolism-related gene expression.
Smad5 KO reduces glycolysis.
Smad5 KO induces irreversible mitochondrial morphology changes.
Smad5 KO impaired mitochondrial respiration.
Increased pHi during neural differentiation.
Smad5 interacts with HK1.
Timelapse video recording of nuclear export of GFP-Smad5 at 25°C. GFP-Smad5 HEK293 cells were cultured at 37°C, then the cells were placed at 25°C to record the video.
Timelapse video recording of nuclear export of GFP-Smad5 at basic extracellular pH. GFP-Smad5 HEK293 cells were cultured at 37°C, then the cells were placed at basic extracellular pH conditions to record the video.
Timelapse video recording of nuclear export of GFP-Smad5 at hypertonic conditions. GFP-Smad5 HEK293 cells were cultured at 37°C, then the cells were placed at hypertonic conditions to record the video.
Timelapse video recording of cytosol to nuclear translocation of GFP-Smad5 at acidic extracellular pH. GFP-Smad5 HEK293 cells were cultured at 37°C, then the cells were placed at acidic extracellular pH conditions to record the video.
Timelapse video recording of cytosol to nuclear translocation of GFP-Smad5 at hypotonic conditions. GFP-Smad5 HEK293 cells were cultured at 37°C, then the cells were placed at hypotonic conditions to record the video.
Smad5 (cytosolic) pull down proteins
References
- Salvi A, Quillan JM, Sadee W. Monitoring intracellular pH changes in response to osmotic stress and membrane transport activity using 5-chloromethylfluorescein. AAPS PharmSci 2002; 4:E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol 2010; 11:50–61. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 2011; 13:453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement MV, Hirpara JL, Pervaiz S. Decrease in intracellular superoxide sensitizes Bcl-2-overexpressing tumor cells to receptor and drug-induced apoptosis independent of the mitochondria. Cell Death Differ 2003; 10:1273–1285. [DOI] [PubMed] [Google Scholar]
- Schmierer B, Hill CS. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat Rev Mol Cell Biol 2007; 8:970–982. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang Y, Feng XH. Smads: transcriptional activators of TGF-beta responses. Cell 1998; 95:737–740. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev 2005; 19:2783–2810. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003; 113:685–700. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci 2001; 114:4359–4369. [DOI] [PubMed] [Google Scholar]
- Chang H, Lau AL, Matzuk MM. Studying TGF-beta superfamily signaling by knockouts and knockins. Mol Cell Endocrinol 2001; 180:39–46. [DOI] [PubMed] [Google Scholar]
- Kaivo-oja N, Jeffery LA, Ritvos O, Mottershead DG. Smad signalling in the ovary. Reprod Biol Endocrinol 2006; 4:21.16611366 [Google Scholar]
- Kumar S, Pan CC, Shah N, et al. Activation of Mitofusin2 by Smad2-RIN1 complex during mitochondrial fusion. Mol Cell 2016; 62:520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Zhou J, Liao X, et al. Disruption of Smad5 gene induces mitochondria-dependent apoptosis in cardiomyocytes. Exp Cell Res 2005; 306:85–93. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 1997; 90:1041–1050. [DOI] [PubMed] [Google Scholar]
- Adachi Y, Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene crm1+ which encodes a 115-kD protein preferentially localized in the nucleus and its periphery. J Cell Biol 1989; 108:1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 1997; 390:308–311. [DOI] [PubMed] [Google Scholar]
- Kutay U, Guttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol 2005; 15:121–124. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Signal transduction by the TGF-beta superfamily. Science 2002; 296:1646–1647. [DOI] [PubMed] [Google Scholar]
- Miyauchi K, Marin M, Melikyan GB. Visualization of retrovirus uptake and delivery into acidic endosomes. Biochem J 2011; 434:559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998; 394:192–195. [DOI] [PubMed] [Google Scholar]
- Morimoto YV, Kojima S, Namba K, Minamino T. M153R mutation in a pH-sensitive green fluorescent protein stabilizes its fusion proteins. PLoS One 2011; 6:e19598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi L, Fan B, Feng D, et al. The dorsoventral patterning of human forebrain follows an activation/transformation model. Cereb Cortex 2017; 27:2941–2954. [DOI] [PubMed] [Google Scholar]
- Liu L, Liu X, Ren X, et al. Smad2 and Smad3 have differential sensitivity in relaying TGFbeta signaling and inversely regulate early lineage specification. Sci Rep 2016; 6:21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer. Nat Rev Cancer 2012; 12:685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell 2012; 148:1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 2009; 27:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmschneider B, Grillo-Hill BK, Benitez M, Azimova DR, Barber DL, Nystul TG. Increased intracellular pH is necessary for adult epithelial and embryonic stem cell differentiation. J Cell Biol 2016; 215:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi L, Fan B, Zhang K, et al. Targeted differentiation of regional ventral neuroprogenitors and related neuronal subtypes from human pluripotent stem cells. Stem Cell Rep 2016; 7:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante E, Morris HP, Pedersen PL. Energy metabolism of tumor cells. Requirement for a form of hexokinase with a propensity for mitochondrial binding. J Biol Chem 1981; 256:8699–8704. [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011; 11:85–95. [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, et al. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 2003; 300:332–336. [DOI] [PubMed] [Google Scholar]
- Greiner EF, Guppy M, Brand K. Glucose is essential for proliferation and the glycolytic enzyme induction that provokes a transition to glycolytic energy production. J Biol Chem 1994; 269:31484–31490. [PubMed] [Google Scholar]
- Harland R. Neural induction. Curr Opin Genet Dev 2000; 10:357–362. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors 2004; 22:233–241. [DOI] [PubMed] [Google Scholar]
- Parks SK, Chiche J, Pouyssegur J. pH control mechanisms of tumor survival and growth. J Cell Physiol 2011; 226:299–308. [DOI] [PubMed] [Google Scholar]
- Faust M, Montenarh M. Subcellular localization of protein kinase CK2. A key to its function? Cell Tissue Res 2000; 301:329–340. [DOI] [PubMed] [Google Scholar]
- Reguly T, Wrana JL. In or out? The dynamics of Smad nucleocytoplasmic shuttling. Trends Cell Biol 2003; 13:216–220. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999; 96:857–868. [DOI] [PubMed] [Google Scholar]
- Agarraberes FA, Dice JF. Protein translocation across membranes. Biochim Biophys Acta 2001; 1513:1–24. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 2007; 450:663–669. [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Hebert DN. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell 2003; 112:491–505. [DOI] [PubMed] [Google Scholar]
- Pouyssegur J, Franchi A, L'Allemain G, Paris S. Cytoplasmic pH, a key determinant of growth factor-induced DNA synthesis in quiescent fibroblasts. FEBS Lett 1985; 190:115–119. [DOI] [PubMed] [Google Scholar]
- Brooks C, Ketsawatsomkron P, Sui Y, et al. Acidic pH inhibits ATP depletion-induced tubular cell apoptosis by blocking caspase-9 activation in apoptosome. Am J Physiol Renal Physiol 2005; 289:F410–419. [DOI] [PubMed] [Google Scholar]
- Loiselle FB, Casey JR. Measurement of intracellular pH. Methods Mol Biol 2010; 637:311–331. [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang CT, Chen J, et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 2010; 7:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Huang SL, Zhang XQ, et al. Reversal effects of pantoprazole on multidrug resistance in human gastric adenocarcinoma cells by down-regulating the V-ATPases/mTOR/HIF-1alpha/P-gp and MRP1 signaling pathway in vitro and in vivo. J Cell Biochem 2012; 113:2474–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc 2012; 7:171–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 2011; 29:731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 2011; 43:869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Smad5 accumulates in cytoplasm at low temperature.
Oxidative stress, membrane potential and signaling modifiers could not obviously affect Smad5, Smad1 or Smad8 subcellular distribution under normal or low temperature culture conditions.
Smad5 responses to pHe and osmolarity fluctuations.
Smad5 subjects to accelerated CRM1-dependent nuclear export at low temperature and three NESs are involved in its rapid nuclear export.
Environmental cues triggered Smad5 translocation is BMP signaling-independent.
Temperature, pHe and osmolarity fluctuations can induce pHi changes and Smad5 MH1 domain is responsible for pHi sensing.
Transcriptome analysis of WT and Smad5 KO hES.
Transcriptome analysis of WT and LDN treated hES.
Chronic metabolic adaptation of Smad5 KO hES.
Smad5 shuttling could not regulate metabolism-related gene expression.
Smad5 KO reduces glycolysis.
Smad5 KO induces irreversible mitochondrial morphology changes.
Smad5 KO impaired mitochondrial respiration.
Increased pHi during neural differentiation.
Smad5 interacts with HK1.
Timelapse video recording of nuclear export of GFP-Smad5 at 25°C. GFP-Smad5 HEK293 cells were cultured at 37°C, then the cells were placed at 25°C to record the video.
Timelapse video recording of nuclear export of GFP-Smad5 at basic extracellular pH. GFP-Smad5 HEK293 cells were cultured at 37°C, then the cells were placed at basic extracellular pH conditions to record the video.
Timelapse video recording of nuclear export of GFP-Smad5 at hypertonic conditions. GFP-Smad5 HEK293 cells were cultured at 37°C, then the cells were placed at hypertonic conditions to record the video.
Timelapse video recording of cytosol to nuclear translocation of GFP-Smad5 at acidic extracellular pH. GFP-Smad5 HEK293 cells were cultured at 37°C, then the cells were placed at acidic extracellular pH conditions to record the video.
Timelapse video recording of cytosol to nuclear translocation of GFP-Smad5 at hypotonic conditions. GFP-Smad5 HEK293 cells were cultured at 37°C, then the cells were placed at hypotonic conditions to record the video.
Smad5 (cytosolic) pull down proteins
Data Availability Statement
All high-throughput data have been deposited at the Gene Expression Omnibus under accession number GSE99687.