Abstract
Screening for differentially expressed genes is a straightforward approach to study the molecular basis of a biological system. In the last 10 years, differential screening technology has evolved rapidly and currently high-throughput tools for genome-wide transcript profiling, such as expressed sequence tags and microarray analysis, are becoming widely available. Here, an overview of this (r)evolution is given with emphasis on the differential display method, which for many years has been the preferred technique of scientists in diverse fields of research. Differential display has also been the method of choice for the identification of genes involved in the symbiotic interaction between Azorhizobium caulinodans and Sesbania rostrata. The advantages with respect to tissue specificity of this particular model system for legume nodulation and the results of a screening for early nodulation-related genes have been considered in the context of transcriptome analyses in other rhizobium–legume interactions.
PLANT FUNCTIONS ARE INVOLVED AT DIFFERENT STAGES OF LEGUME NODULATION
The interaction between rhizobia and legumes that leads to symbiotic nitrogen fixation is a process in which a completely new plant organ, the nodule, develops (1,2). At low frequency and under conditions of nitrogen starvation, non-nitrogen-fixing nodule-like structures arise spontaneously on certain alfalfa cultivars, suggesting that the development of the nodule is controlled by the plant (3). In the presence of rhizobia, the ontogenetic process of the nodule is accompanied by bacterial infection of plant tissues in a tightly controlled way. Plant responses, which are reminiscent of defense reactions to pathogens, restrict bacterial entry and guide the deeper invasion (4). The early signal exchange between the two symbionts involves plant-secreted flavonoids and unknown plant receptors that perceive the rhizobial nodulation (Nod) factors (5–8).
The uniqueness of the de novo organogenesis that accompanies this symbiosis together with the great agricultural potential of biological nitrogen fixation has made the study of the molecular mechanisms underlying nodule development and infection a fast-growing field of research. The search for plant genes involved in this program, referred to as ‘nodulin genes’ (9), started 15 years ago and since then a large number of genes have been described that are more or less specific for nodulation.
Nodulins were first identified by in vitro translation (10). Later, the approaches evolved with the available technology. The productivity of differential screens increased when researchers switched from basic screening tools, such as differential hybridization, to more powerful PCR-based methods including differential display. At present, the output is growing exponentially through the application of genome-wide tools, such as genomic and expressed sequence tags (EST) sequencing and the analysis by microarray technology of numerous clones.
THE QUEST FOR DIFFERENTIALLY EXPRESSED GENES
The basis
Although a gene must not necessarily be up- or down-regulated to play a key role in a certain process, screening for differentially expressed genes is one of the most straightforward approaches to unravel the molecular basis of a biological system. The difficulty of isolating differentially expressed genes, particularly low-abundance ones, has been underestimated by some, albeit experienced by many. To fully recognize this, it is important to take into account the complexity of a cell’s transcriptome. A eukaryotic cell contains ∼15 000–30 000 distinct mRNAs with a prevalence ranging from one to several thousands in a total mass of ∼100 000 mRNAs. About 50% of the transcript population is made up of a relatively small number (some hundreds) of abundant transcripts, representing only 1% of the different mRNA species. The other half contains the ‘rare’ mRNAs (11). Not surprisingly, the difficulty of fishing out a gene responsible for a specialized function in a certain biological program often originates from the fact that the gene is expressed at low levels whereas the bulk of a cell’s mRNA is made up of highly abundant transcripts.
In traditional screening methods, such as differential hybridization, the hybridization pattern of the total content of a cDNA library is compared between two samples (12). The fact that the abundant transcripts are also displayed implies high redundancy of non-relevant clones and thus very low labor efficiency. This problem has been solved partly by normalization and subtraction (13); even then, many interesting low-abundance differentially expressed genes are missed because of the low amplification of the hybridization signal (14). Other drawbacks are the limitation to pairwise comparisons and the fact that the techniques are mainly qualitative because of the relative insensitivity of the hybridization (15).
Differential display
With the availability of PCR, low-abundance transcripts could be amplified. One of the first differential screening methods that used this possibility was the differential display technique described by Liang and Pardee (16). By combining 3′ anchored oligo(dT) primers and short 5′ arbitrary primers, subsets of the transcriptome are amplified, the resulting cDNA fragments are separated on a denaturing polyacrylamide gel and visualized autoradiographically. Original statistics indicated that 80 primer combinations would be sufficient to cover the whole transcript mass (17). The expected advantages were numerous: the method would be fast, producing band patterns in 2 days; it was based on simple, well established and widely accessible techniques, making it easily applicable for most researchers; compared with previous methods the sensitivity had been increased dramatically, resulting in a good detection of low-abundance genes; both induced and repressed genes could be detected and more than two samples could be compared, making it highly versatile; furthermore, only a small amount of starting material was needed (16).
It soon became clear from numerous reports that the differential display technique represented a major contribution to the molecular biology toolbox. At the same time, however, investigators experienced drawbacks and limitations and over the years many variations of the original protocol were published (18–23).
Among the major criticisms, one was the questioned ability of the technique to identify rare mRNAs. Some studies suggested that competition for substrates between the PCR products, particularly dNTPs, would be the limiting factor for amplification, so that only abundant transcripts would be amplified to a detectable level by the time dNTPs were depleted (24). Other experiments, however, gave a more optimistic evaluation of the sensitivity (11). By comparing untreated HeLa cells with cells treated with interferon-γ, the calculated prevalences of the isolated clones ranged from 1/214 to ∼1/200 000 with a median of ∼1/20 000. The apparent contradiction between both reports could be explained, at least partially, by a dramatic change in gene expression in the latter system, with the fraction of differentially expressed genes reaching levels of at least 6.6%. A relatively high total number of (rare) differential transcripts increases the probability of detecting some of the low-abundance ones (21).
Once a candidate band has been eluted from the gel, been reamplified and cloned, gene expression analysis tools, such as northern blot, RT–PCR or RNase protection, are applied to confirm the expression pattern and to attribute it to the correct clone. Indeed, most often, cloning of a band from the gel results in a mixture of cDNAs because it is difficult to avoid contamination from neighboring bands during band excision and because one band may consist of several clones, the observed pattern being the additive result of overlapping expression patterns (25,26). Thus, the downstream part of the differential display procedure reveals a major drawback, i.e. the frequency of false positives, which may be as high as 50–75% of the excised bands (11,17,18).
The most significant source of artefacts, however, might be inherent to the design of the differential display method. The combination of short primers and low annealing temperatures during PCR results in non-specific and inefficient amplification (27). Another factor that may generate false positives is the competition for primers by transcripts of different abundance. Experiments showed that increased amounts of a spiked RNA species resulted in increasing amounts of the corresponding PCR product. Concomitantly, the signal intensity of several unrelated bands decreased, questioning the quantitative aspect of the technique (18). The number of false positives might also increase in some cases because of extrinsic factors, such as the systems that are compared and care that is taken in experimental design (e.g. PCR tubes, internal controls; 23).
When the workload of the downstream processing of differential display candidates is considered, another important issue is redundancy. Decamer primers often mismatch and hybridize to distinct regions of the same cDNA (11,24). Consequently, different positive clones might correspond to the same gene. This observation not only implies a reduced screening efficiency, it also means that a lot more primer combinations than theoretically calculated should be used to cover the complete transcriptome. Depending on the design of both 5′ arbitrary and 3′ anchored primer and the cycling program, the proposed amount of primer combinations varies greatly (11,15,16,22).
Most of the published technical modifications and refinements aim to reduce the amount of artefact bands and improve the sensitivity of the differential display. Alternative primer designs have been proposed, such as longer, more specific arbitrary primers, eventually in combination with an altered cycling program (27–29). By reamplifying a subset of the original fingerprint with nested primers, which carry one or more extra selective nucleotides, more reliable results have been reported (30). Other improvements included use of stronger radioactive labels (31), implementation of automatic sequencing machines (15), use of end-labeled 3′ primers (32), combination with a subtraction procedure (33,34) and many other minor modifications (22). Recently, a multicolor fluorescent differential display protocol has been developed that allows digital analysis and the reduction of false positives by inherent signal proofreading (35).
Considering the rate of false positives and redundancy, differential display seemed much less attractive than originally presented. The downstream verification process is not only labor intensive, it also requires significant amounts of RNA, which compromises one of the major advantages of the method. Many different approaches have been proposed to meet the necessity of large-scale screening of candidate cDNA fragments with low amounts of RNA, such as reverse northerns with Southern-blotted (36), slot (37) or dot-blotted (38) clones, eventually in combination with the use of amplified RNA as a probe (39). Another important factor that adds to the post-display effort is intrinsically linked to the design of the technique. Because the cDNA fragments obtained from differential display are short (typically 100–500 bp) and correspond to the 3′ end of the gene that represents mainly the 3′ untranslated region, they usually do not contain a large portion of the coding region. Unless a model system is studied for which a significant amount of sequence information is available in public databases, labor-intensive full-length cDNA screening is needed before significant sequence homology, informative for gene classification and prediction of function, can be obtained. Recently, this problem has been tackled by a number of authors by integrating the protocol for long-distance PCR into differential display. By combining the use of longer primers, higher dNTP concentrations, hot-start PCR at higher stringency and thermostable enzyme mix, bands of up to 2 kb could be displayed (40). The long-distance differential display-PCR (LDD–PCR; 41) produced similarly band patterns of cDNAs ranging from 150 bp to 2 kb. Furthermore, the alterations to the protocol resulted in lower redundancy, reduced amounts of artefact bands and detection of both abundant and rare transcripts.
Beyond differential display
Almost simultaneously with differential display, a conceptually similar technique was developed and published, called RNA fingerprinting by arbitrarily primed PCR (RAP–PCR). It differs from differential display principally in the use of an arbitrary primer both for first-strand cDNA synthesis and subsequent PCR amplification steps (42). A variant in which cDNA fragments are displayed on and excised from agarose gels was subsequently proposed (43).
After differential display had been introduced, more methods using PCR were developed (reviewed in 44–48). A large number of techniques involved the generation of a gel profile to display differences between different mRNA samples, but, in contrast to differential display, tried to evade the use of arbitrary primers and so circumvent problems that originated from mismatch priming during amplification. Instead, restriction sites were used to generate a subset of cDNA fragments that differed in size. Amplification following restriction enzyme digestion was done with primers that matched previously ligated adaptors. Eventually, one or more selective bases were added to the 3′ ends of the primers to further reduce the subset of cDNA fragments that will be displayed, such as in cDNA-amplified fragment length polymorphism (AFLP; 49). The kinetics of gene expression revealed by cDNA–AFLP were similar to those of northern blot analysis, rendering the displayed expression pattern quantitative (49). Other more sophisticated methods include a selection step to end up with only one restriction fragment per mRNA, hence completely ruling out redundancy. In the restriction enzyme analysis of differentially expressed sequences (READS; 50), this aim was obtained through special adaptor design that only allows amplification of the most 3′ restriction fragments. An extension of this method was the ‘ordered differential display’ that utilized the PCR suppression effect for selection of 3′ end restriction fragments of cDNA (51). In two other methods, the 3′ end was selected by biotinylated oligo(dT) primers. In gene expression fingerprinting (GEF; 52) an additional restriction enzyme digestion was performed after PCR amplification and, in the cDNA–AFLP-based transcript profiling method (53), the second digestion occurred before amplification. Still other variants of techniques that combine restriction digest, PCR and gel display have been reported (54–58).
Other types of techniques improved and refined the traditional technology of cDNA library construction and screening. Representational difference analysis (RDA), for example, merged the advantages of subtractive hybridization with the power of PCR amplification (59). Suppression subtractive hybridization (SSH; 60) combined normalization and subtraction in a single procedure. In both techniques, the final product is an amplified pool of cDNAs, enriched for species that are specific for the tester sample. In addition, in the case of SSH, the abundance of cDNAs is equalized. This pool is usually cloned and differentially screened. Still other similar methods that generate enriched pools of cDNAs include linker capture subtraction (LCS; 61), rapid subtraction hybridization (RaSH; 62), enzymatic degrading subtraction (EDS; 63) and selective amplification via biotin- and restriction-mediated enrichment (SABRE; 64).
A completely different sequence-based approach to identify differentially expressed genes is followed by serial analysis of gene expression (SAGE; 65). In this method, very short (10–14 bp) cDNA tags are generated by restriction digestion, amplified by PCR and ligated, after which the resulting concatemers are sequenced. The tags are long enough to identify the corresponding genes unequivocally and the frequency of the tags is a measure of their expression level. This method is very fast (with an automated sequencer more than 1000 transcripts can be analyzed in one 3 h run) and straightforward because it does not imply selection of mRNAs to create displayable subpopulations, nor does it depend on tricky procedures, such as normalization or subtraction. Although regarded as one of the most cost-effective methods (56), the limitation of SAGE is that the corresponding gene can be identified only for the tags deposited in gene banks, making its efficiency dependent on the complexity of available databases. Recently, variants have been published that circumvent some drawbacks of SAGE [tandem arrayed ligation of expressed sequence tags (TALEST; 66) and massively parallel signature sequencing (MPSS; 67)].
Ten years ago, large collections of ESTs were built by single-pass sequencing of random cDNAs (68). These partial sequences of expressed genes were used to discover new genes at only a fraction of the cost of genomic sequencing and, in addition, they facilitated the identification of coding regions in genomic sequences. The ESTs recovered from a certain cell type indicate what kind of genes are expressed. Although the redundancy of a sequence gives an idea regarding the expression level, this approach (referred to as ‘digital northern’ or ‘electronic subtraction’; 11,69) cannot be used routinely as an expression analysis tool. After a method was found to grid large numbers of cDNA clones at high density, thus permitting their efficient screening by differential hybridization, the usefulness of large-scale sequencing projects has been extended from just a source of new genes to a tool for high-throughput transcriptome analysis. Mega-scale reverse northern approaches are now possible thanks to high-speed robotic printing of cDNAs. Originally, cDNAs were spotted on nylon membranes and hybridized with traditional methods (70). With the current microarray technology thousands of clones are displayed on just a couple of square centimeters of glass support and are hybridized in microvolumes with fluorescently labeled cDNA probes, resulting in improved screening sensitivity (71). Alternatively, the DNA targets can be synthetic oligonucleotides synthesized in situ on silicon (72). These oligomers are highly gene specific, thus greatly reducing cross-hybridization.
The power of this technology is self-evident: data can be collected for large numbers of genes in one experiment and genome-wide expression patterns can be observed. However, major limitations reside in the questionable sensitivity of the probes and in the relatively costly and time-consuming collection of the set of (unique) sequences. The thoroughness of the preceding EST sequencing defines the success, because only genes can be analyzed for which sequences are available. However, a feasible approach for organisms that lack extensive EST collections is to use anonymous clones from cDNA libraries; clones with interesting expression patterns can be sequenced after the expression analysis (73). One has also to bear in mind that a hit in a database for a cDNA of interest does not always yield much information, because at present a function has been assigned only to a minority of the submitted sequences (74).
Technological improvements of DNA sequencing resulted in reduced cost for collecting sequences (75). Upcoming new technologies for producing oligonucleotide-based and DNA fragment microarrays will probably significantly cut the costs, making the technology accessible to most laboratories (76). Therefore, the main challenge may be efficient management and analysis of the immense data-flow generated by these techniques. Also experts recently started to worry about imprudent use of the technology. Although experience with microarrays already goes back a number of years, it should be acknowledged that the technology is still in its infancy (77). There is a serious risk that researchers jump on it without fully recognizing the potential for human error caused by the large number of genes that are handled simultaneously. Recently, high error rates have been found even in commercially supplied ‘sequence-verified’ cDNA clone sets (77).
Although microarrays undoubtedly allow high-throughput screening, the efficiency depends on the method used to collect the arrayed sequences. Except for model organisms such as Araidopsis thaliana, the brute-force approach of simple EST sequencing of genuine cDNA pools, which are unbiased for a certain cell type, seems rather unwise in view of the 100 000 mRNAs present in a cell. Several routes can be taken first to enrich the cDNA pool, in order to efficiently screen differentially expressed genes with microarrays. Combinations of microarrays with RDA, SSH or differential display have proven very successful (35,78–80) and, recently, another method has been described that supplies a normalized pool of so-called ‘open reading frame-expressed sequences tags’ (ORESTES; 81). Large-scale sequencing and microarray analysis will probably take the lead for genome-wide studies of gene expression. However, for tackling more limited biological questions, open-ended techniques, such as differential display and cDNA–AFLP-based transcript profiling, will no doubt remain equally useful in the post-genomic future.
THE INTERACTION BETWEEN AZORHIZOBIUM CAULINODANS AND SESBANIA ROSTRATA AS A MODEL SYSTEM FOR THE ISOLATION OF DIFFERENTIALLY EXPRESSED GENES INVOLVED IN THE EARLY STAGES OF NODULATION
The A.caulinodans–S.rostrata symbiosis is very well suited for identifying genes that are differentially expressed during the early stages of the rhizobium–legume symbiosis. Azorhizobium caulinodans not only triggers the formation of nodules on the roots of the tropical legume S.rostrata, but also on the stem. The stem nodules develop at the sites of pre-existing dormant root primordia, which are abundantly present along the stem. Generally, almost all inoculated primordia react and the nodules develop in a synchronous fashion. Consequently, very specific target tissue can be collected even before any sign of a nodule appears. This specificity increases the power of any screening technique employed. The differential display method has been used previously to identify nodule-specific cDNAs induced or enhanced during the first days of the interaction (82). Here we report on the outcome of a differential display-based screening at much earlier stages of the interaction.
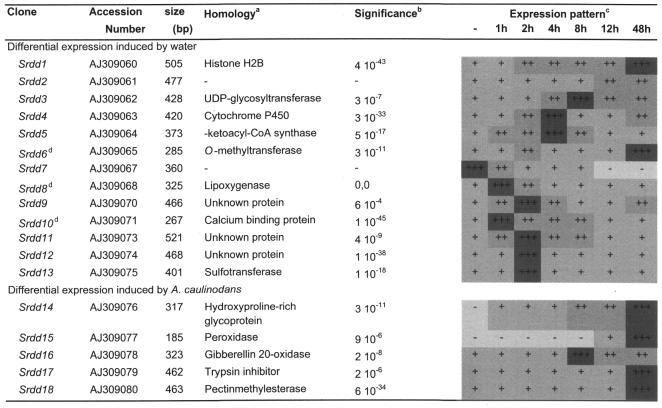
RNA from excised root primordia of 6-week-old S.rostrata plants was compared with RNA from root primordia excised 1, 2, 4, 8, 12 and 48 h after the stems had been inoculated with A.caulinodans ORS571. Twenty 5′ arbitrary decamer primers (Ap-1 to Ap-20) in combination with two different 3′ one-base-anchored primers (dT12MC and dT12MG) were tested (RNAmap, GenHunter, Brookline, MA). After the differential display patterns had been inspected visually, 34 bands that clearly showed differential intensities were selected for further analysis. Thirty-one cDNA fragments could be reamplified after elution from gel and were cloned. For each candidate, the inserts of at least six transformants were sequenced. The cDNA corresponding with the expression pattern of interest was identified by northern blot analyses. For eight candidates the differential display patterns could not be reproduced. Two cDNAs were represented twice in the pool of 31 candidates. Three cDNA clones corresponded with genes previously isolated from S.rostrata stem nodules (82). For the 18 remaining new and unique clones, the differential expression pattern was confirmed by at least two independent northern blot experiments and is presented in Table 1.
Table 1. Sequence homologies and expression profiles of differentially expressed clones.
aResults from a search in the non-redundant protein database with the BLASTX algorithm, except for Srdd14 where BLASTN was used to search the non-redundant DNA sequence database. ‘-’, no significant homology was found (see c).
bE score for the most homologous sequence as given by the BLASTX or BLASTN algorithm; 10–3 was used as the significance limit.
cRelative expression levels in uninfected root primordia (-) and primordia harvested at different timepoints (1, 2, 4, 8, 12 and 48 h) after inoculation with A.caulinodans indicated by signs (– to +++, reflecting undetectable to maximal expression) and shading (the darker the shading, the stronger the expression).
The adventitious root primordia of S.rostrata contain a central vascular bundle and a dormant apical meristem. Upon submergence this meristem is activated, initiating the outgrowth of a lateral root (83). The inoculation procedure used in our experiments implies moistening of the root primordia. Because non-inoculated stems were used as a control sample for the differential display experiment, the observed differential expression might be caused by the brief contact with water rather than by the application of the microsymbiont. Therefore, control experiments were performed in which northern blot analysis was used to follow the expression pattern in root primordia inoculated by water and harvested after inoculation at the same timepoints as in the original differential display experiment (data not shown). The expression of the majority of the isolated cDNAs (13 out of 18) was induced by the water treatment. Although at none of the timepoints tested was any phenotypical sign of root growth initiation visible, it is conceivable that even a relatively short contact with water is enough to trigger differential gene expression. Because initiation of nodule development and adventitious root outgrowth partly involve similar cellular mechanisms, namely cell division, the same genes may play a role in both processes, resulting in overlapping expression patterns in the case of A.caulinodans inoculation. For example, a histone, putatively encoded by clone Srdd1 (Table 1) will be important for cell division both in the root apical meristem and in the nodule meristem in the cortex. Furthermore, the application of water might trigger cellular changes that are essential for nodulation. Thus, the importance for nodulation of the genes that show an enhanced expression pattern upon inoculation with water cannot be excluded on the basis of northern blot analysis. In situ hybridization analysis will highlight whether these genes are induced in nodulation-associated tissues.
Sequence homologies were looked for in DNA and protein databases with the BLASTN and BLASTX algorithm, respectively (84). An overview of the identified clones and their homologies is presented in Table 1. It is premature to hypothesize about the functions these putative proteins could have in nodule formation. Future studies will include isolation of full-length clones, determination of tissue specificity of the expression pattern and spatial expression analysis using in situ hybridization.
TRANSCRIPTOME ANALYSIS AND NODULATION
It is difficult to predict how many genes are expected to be differentially expressed during a process as complex as stem nodulation. Within the timespan covered by the samples analyzed, two plant programs are initiated concurrently by bacterial Nod factor signals (85,86). Cells in the mid-cortex of the root primordium de-differentiate and start to divide, giving rise to a globular nodule primordium that is clearly visible 2 days after inoculation. Meanwhile the bacteria enter the plant via an intercellular infection process during which infection pockets are formed (85,86). Nod factor perception and signal transduction towards the initiation of these events may lead to differential gene expression. At the transcript level, roots and leaves differ by 25 and 30% in tobacco and A.thaliana, respectively (87,88). Reports that compare less different tissue samples indicate 16% differentially expressed genes in a search for light-responsive genes in A.thaliana (89) and ∼200 out of 1800 strawberry genes were enhanced or suppressed during fruit ripening (90). According to the manufacturer (RNAmap, GenHunter), the amount of primer combinations tested here should be sufficient to cover half of the transcriptome. It is clear from our results though that only a very small fraction of the total number of differential mRNAs has been detected. The fact that the differential expression pattern of most clones could be confirmed using northern blot analysis, a rather insensitive technique compared to RT–PCR or the RNase protection assay, indicates that only medium to highly abundant cDNAs have been isolated.
Taking into account the state-of-the-art technology, the results of the differential screening applied to our biological system can be considered as rather poor. Although it has never been the aim to perform a genome-wide screen, the limited output questions the performance of the differential display method. In view of the theoretical considerations discussed above, maybe the limited success should be attributed to too short primers at too low annealing temperature, which made it impossible to amplify efficiently low-abundance cDNAs. However, the weak performance of the technique should not be generalized to the current differential display protocols. As mentioned above, the technique has been the subject of many improvements. In fact, a literature search highlighted that even to date differential display is still a widely used and popular technique.
The clones described here add to the fast-growing list of nodulin genes isolated from a wide number of symbiotic systems. A survey of reported screenings for differentially expressed genes involved in nodulation is presented in Table 2. First, differential hybridization was used, because it was the only transcript screening technique available at the time (91,92). Subtractive hybridization and differential display were soon adopted in nodulation research (82,93). Later, despite the constant evolution and improvement of screening technology, researchers seemed to cling to these methods. This lag either proves the solidity of these traditional methods or indicates the slow penetration of technological (r)evolution from the animal field (from which most of this technology emerges) to the plant world. Two reports successfully applied an improved version of the very simple ‘cold-plaque’ screening procedure, in which non-hybridizing plaques from a differential hybridization are further analyzed (94). Adaption of the protocol included a PCR amplification round of the cold-plaque inserts before subsequent screening by reverse northern (95,96). More recently, the systematic approach of EST sequencing has been initiated in the field of nodulation research (97,98).
Table 2. Survey of differential screening experiments in nodulation research.
| Host plant |
Technique |
Sourcea |
Screenedb |
Candidatesc |
Processedd |
Confirmede |
Newf |
Ref |
| Pea | DH | –; wt 14 d | 600 | – | 6 | 6 | 6 | 91 |
| Soybean | DH | –; wt 10 d | – | 10 | 10 | 3 | >1 | 92 |
| Pea | DH | –; wt 2 d | 3000 | – | – | 3 | 3 | 106 |
| Soybean | SH | –; wt 21 d | 150 000 | 1200 | – | 20 | 9 | 93 |
| –; wt 8 d | 100 000 | 250 | ||||||
| Broad bean | SH | –; wt 30 d | 55 000 | 700 | 700 | 27 | 19 | 104 |
| Alfalfa | DH | –; spontaneous nod. | – | – | – | – | 1 | 107 |
| Pea | SH | –; wt 35 d | 60 000 | 500 | 500 | 19 | – | 108 |
| M.truncatula | SH | –; wt 48, 108 h | – | – | – | – | 1 | 109 |
| S.rostrata | DD | –; wt 2, 3, 4 d | 2800 | 45 | 17 | 5 | 5 | 82 |
| M.truncatula | SH | nodA–; wt 4 d | 30 000 | 473 | 473 | 30 | 29 | 110 |
| Vetch | DD | –; NF 1, 3 h | 27 600 | 34 | 9 | 1 | 1 | 100 |
| L.japonicus | DD | –; wt 7, 11, 13, 21 d | 8000 | 137 | 137 | 19 | 13 | 101 |
| M.truncatula | ESTS | root hair-enriched root | 985 | 899 | – | – | – | 97 |
| M.truncatula | DD | –; NF 2 h | 4000 | 0 | – | – | – | 111 |
| –; NF 24 h | 2400 | 0 | – | – | – | 111 | ||
| –; NF 48 h | 2000 | 4 | 4 | 2 | >1 | 111 | ||
| Alfalfa | CPS | –; spontaneous nod. | 250 | 21 | 21 | 14 | 14 | 95 |
| M.truncatula | ESTS | wt 4–8 d | 408 | 389 | 389 | 117 | 84 | 98 |
| Alfalfa | CPS | Nod–; wt 5 d | 1000 | 38 | 38 | 38 | 22 | 96 |
| Yellow lupine | DD | –; wt 21 d | 3500 | 50 | 14 | 4 | 4 | 112 |
| S.rostrata | DD | –; wt 1, 2, 4, 8, 12, 48 h | 4000 | 57 | 34 | 9 | 6 | this work |
DH, differential hybridization; SH, subtractive hybridization; DD, differential display; ESTS, EST sequencing; CPS, cold-plaque screening.
aSamples compared in the experiment: –, uninoculated; wt, wild-type microsymbiont; nodA– and Nod–, mutant microsymbiont strains; NF, pure Nod factors. For details, see original articles.
bNumber of clones screened as plaques or colonies in the case of differential or subtractive hybridization and cold-plaque screening, bands displayed on gel for differential display, or corresponding to the number of clones sequenced in the case of EST sequencing. For the reports on differential display experiments in which no average amount of bands displayed per gel was cited, 100 was taken as the amount to calculate the approximate number of bands visualized.
cNumber of candidates, putatively differentially expressed, obtained in the differential screening experiment, or the number of readable sequences from EST sequencing.
dNumber of candidates that were further processed to confirm their differential expression pattern.
eNumber of unique clones for which the differential expression pattern could be confirmed.
fNumber of novel genes among the confirmed clones.
The most striking observation when screening reports are overviewed is the low output of novel nodulin genes. This could indicate that the methods applied were unsuitable for large-scale isolation of differentially expressed genes or that the tools for efficient analysis of large amounts of candidate clones were not available. Most researchers limited themselves to the study of just a handful of genes. Only Györgyey et al. (98) analyzed the expression levels of a substantial amount of unique clones, resulting in the identification of 84 genes that were differentially expressed in 4–8-day-old Medicago truncatula nodules when compared to uninoculated roots. On the other hand, the difficulty in identifying novel nodulin genes, particularly low-abundance ones, is probably inherent to the heterogeneity of the plant material. Like other plant organs, symbiotic nodules are complex structures and consist of many distinct tissues and cell types. For most of the nodulin genes, expression is cell type specific and the differential signal is thus present in only a small fraction of the pool of cells that are taken for comparison. Thus, screens for differential gene expression in nodulation will never reach the sensitivity obtained when homogeneous samples are compared, such as cell cultures.
Another remarkable observation is that most reports deal with the later stages of the interaction, presumably as a result of limitations inherent to the biology of root nodulation. There is no strictly defined and easily accessible site of root nodule development. On most legumes root nodule formation is initiated via root hair infection by the rhizobia. The primary target for bacterial invasion is the zone of young, developing root hairs (99). After root hair entry, it takes several days before any phenotypical sign of the developing nodule can be perceived. As a consequence, it is impossible to harvest specific nodulation site samples before this time. When specific material is sampled after nodule primordia are visible, then many early responses will be overlooked. In case whole roots are used to analyze the very early stages, dilution of the material will cause a dramatic decrease in discrimination power. Only in a limited number of reports was plant material enriched for root hairs or for the zone susceptible to rhizobial infection used as source for the screening experiments (97,100,101). Because in the stem nodulation system, nodules develop synchronously at the pre-existing, abundant and easily accessible dormant root primordia, specific nodulation sites can be harvested before nodules become visible and samples can be compared after being collected in the range of hours instead of days after inoculation. The results confirm that the highly specific source of plant material constituted by the S.rostrata root primordia combined with the differential display technique is indeed sensitive enough to detect minor changes in expression levels at the early stages of nodulation. Ten out of the 18 clones described here correspond with genes that are enhanced or repressed within 2 h of inoculation. Although it is not generally recognized as a model legume, the specificity of the stem nodulation sites makes S.rostrata probably one of the best systems for the identification of genes involved in the early steps of nodulation.
What is not immediately noticeable from the overview of differential screening reports is that the more nodulin genes are characterized, the more difficult it will become to add new, previously unknown genes coding for new functions involved in nodulation-related processes. For example, many nodulins isolated until now encode the oxygen-binding leghemoglobins (102). A large number of other nodulins can be classified into the group of the proline-rich proteins that are thought to be involved in cell wall modification correlated with the nodulation process (103). Logically, these easily detectable genes correspond with highly abundant transcripts. Proline-rich proteins are assumed to constitute 50% of all early nodulins (104). Crucial and regulatory genes are presumably expressed at low levels and thus need more exhaustive, genome-wide screening efforts to be picked up. In nodulation research, large-scale EST sequencing in a joined effort between different research groups is the approach currently followed for M.truncatula and Lotus japonicus (105). These legumes were selected as model legumes for their small genome (only three to four times that of A.thaliana), diploid and autogamous nature and rapid generation time. In addition, both species can be readily transformed using Agrobacterium tumefaciens and regenerated into fertile plants. EST sequence data of M.truncatula are collected in the TIGR M.truncatula Gene Index (http://www.tigr.org/tdb/mtgi), which at present already contains more than 30 000 unique sequences.
EST sequencing combined with differential screening using microarray technology is definitely the ultimate approach to identify all nodulation-related variations of the transcriptome. In addition, the approach of systematic EST sequencing has the great advantage that genes also involved in nodulation that do not exhibit a differential expression pattern can be identified. Currently, one of the chief missions in nodulation research is to find the plant receptor(s) that sense(s) the bacterial Nod factor signal. The gene that encodes this key component of the interaction is probably constitutively expressed in the tissue that is prone to react to rhizobial inoculation. Consequently, large-scale sequencing is one of the more straightforward methodologies for detecting putative receptor candidates.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Wilson Ardiles and Caroline Buysschaert for sequencing, Sylvia Herman and Mercé Caturla Goñi for their help with the differential display analysis and Martine De Cock for help in preparing the manuscript. S.L. was a Research Fellow and S.G. is a Postdoctoral fellow of the Fund for Scientific Research (Flanders).
DDBJ/EMBL/GenBank accession nos AJ309060–AJ309080
References
- 1.Schultze M. and Kondorosi,A. (1998) Regulation of symbiotic root nodule development. Annu. Rev. Genet., 32, 33–57. [DOI] [PubMed] [Google Scholar]
- 2.Stougaard J. (2000) Regulators and regulation of legume root nodule development. Plant Physiol., 124, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Truchet G., Barker,D.G., Camut,S., de Billy,F., Vasse,J. and Huguet,T. (1989) Alfalfa nodulation in the absence of Rhizobium. Mol. Gen. Genet., 219, 65–68. [Google Scholar]
- 4.Vasse J., de Billy,F. and Truchet,G. (1993) Abortion of infection during the Rhizobium meliloti–alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J., 4, 555–566. [Google Scholar]
- 5.Bono J.-J., Riond,J., Nicolaou,K.C., Bockovich,N.J., Estevez,V.A., Cullimore,J.V. and Ranjeva,R. (1995) Characterization of a binding site for chemically synthesized lipo-oligosaccharidic NodRm factors in particulate fractions prepared from roots. Plant J., 7, 253–260. [DOI] [PubMed] [Google Scholar]
- 6.Niebel A., Bono,J.-J., Ranjeva,R. and Cullimore,J.V. (1997) Identification of a high affinity binding site for lipo-oligosaccharidic NodRm factors in the microsomal fraction of Medicago cell suspension cultures. Mol. Plant Microbe Interact., 10, 132–134. [Google Scholar]
- 7.Gressent F., Drouillard,S., Mantegazza,N., Samain,E., Geremia,R.A., Canut,H., Niebel,A., Driguez,H., Ranjeva,R., Cullimore,J. and Bono,J.-J. (1999) Ligand specificity of a high-affinity binding site for lipo-chitooligosaccharidic Nod factors in Medicago cell suspension cultures. Proc. Natl Acad. Sci. USA, 96, 4704–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etzler M.E., Kalsi,G., Ewing,N.N., Roberts,N.J., Day,R.B. and Murphy,J.B. (1999) A nod factor binding lectin with apyrase activity from legume roots. Proc. Natl Acad. Sci. USA, 96, 5856–5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Kammen A. (1984) Suggested nomenclature for plant genes involved in nodulation and symbiosis. Plant Mol. Biol. Reporter, 2, 43–45. [Google Scholar]
- 10.Govers F., Gloudemans,T., Moerman,M., van Kammen,A. and Bisseling,T. (1985) Expression of plant genes during the development of pea root nodules. EMBO J., 4, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan J.S., Sharp,S.J., Poirier,G.M.-C., Wagaman,P.C., Chambers,J., Pyati,J., Hom,Y.-L., Galindo,J.E., Huvar,A., Peterson,P.A. et al. (1996) Cloning differentially expressed mRNAs. Nat. Biotechnol., 14, 1685–1691. [DOI] [PubMed] [Google Scholar]
- 12.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Hedrick S.M., Cohen,D.I., Nielsen,E.A. and Davis,M.M. (1984) Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature, 308, 149–153. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z. and Brown,D.D. (1991) A gene expression screen. Proc. Natl Acad. Sci. USA, 88, 11505–11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer D., Müller,H., Reich,J., Riedel,H., Ahrenkiel,V., Warthoe,P. and Strauss,M. (1993) Identification of differentially expressed mRNA species by an improved display technique (DDRT-PCR). Nucleic Acids Res., 21, 4272–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang P. and Pardee,A.B. (1992) Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science, 257, 967–971. [DOI] [PubMed] [Google Scholar]
- 17.Liang P., Averboukh,L. and Pardee,A.B. (1993) Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res., 21, 3269–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Debouck C. (1995) Differential display or differential dismay? Curr. Opin. Biotechnol., 6, 597–599. [Google Scholar]
- 19.McClelland M., Mathieu-Daude,F. and Welsh,J. (1995) RNA fingerprinting and differential display using arbitrarily primed PCR. Trends Genet., 11, 242–246. [DOI] [PubMed] [Google Scholar]
- 20.Liang P. and Pardee,A.B. (1995) Recent advances in differential display. Curr. Opin. Immunol., 7, 274–280. [DOI] [PubMed] [Google Scholar]
- 21.Matz M.V. and Lukyanov,S.A. (1998) Different strategies of differential display: areas of application. Nucleic Acids Res., 26, 5537–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Appel M., Bellstedt,D.U. and Gresshoff,P.M. (1999) Differential display of eukaryotic mRNA: meeting the demands of the new millennium? J. Plant Physiol., 154, 561–570. [Google Scholar]
- 23.Liang P. (1998) Factors ensuring successful use of differential display. Methods, 16, 361–364. [DOI] [PubMed] [Google Scholar]
- 24.Bertioli D.J., Schlichter,U.H.A., Adams,M.J., Burrows,P.R., Steinbiß,H.-H. and Antoniw,J.F. (1995) An analysis of differential display shows a strong bias towards high copy number mRNAs. Nucleic Acids Res., 23, 4520–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li F., Barnathan,E.S. and Karikó,K. (1994) Rapid method for screening and cloning cDNAs generated in differential mRNA display: application of Northern blot for affinity capturing of cDNAs. Nucleic Acids Res., 22, 1764–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Callard D., Lescure,B. and Mazzolini,L. (1994) A method for the elimination of false positives generated by the mRNA differential display technique. Biotechniques, 16, 1096–1103. [PubMed] [Google Scholar]
- 27.Zhao S., Ooi,S.L. and Pardee,A.B. (1995) New primer strategy improves precision of differential display. Biotechniques, 18, 842–850. [PubMed] [Google Scholar]
- 28.Liang P., Zhu,W., Zhang,X., Guo,Z., O’Connell,R.P., Averboukh,L., Wang,F. and Pardee,A.B. (1994) Differential display using one-base anchored oligo-dT primers. Nucleic Acids Res., 22, 5763–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graf D., Fisher,A.G. and Merkenschlager,M. (1997) Rational primer design greatly improves differential display-PCR (DD-PCR). Nucleic Acids Res., 25, 2239–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ralph D., McClelland,M. and Welsh,J. (1993) RNA fingerprinting using arbitrarily primed PCR identifies differentially regulated RNAs in mink lung (My1Lu) cells growth arrested by transforming growth factor β1. Proc. Natl Acad. Sci. USA, 90, 10710–10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tokuyama Y. and Takeda,J. (1995) Use of 33P-labeled primer increases the sensitivity and specificity of mRNA differential display. Biotechniques, 18, 424–425. [PubMed] [Google Scholar]
- 32.Hadman M., Adam,B.-L., Wright,G.L.,Jr and Bos,T.J. (1995) Modification to the differential display technique reduce background and increase sensitivity. Anal. Biochem., 226, 383–386. [DOI] [PubMed] [Google Scholar]
- 33.Fuchs B., Zhang,K., Bolander,M.E. and Sarkar,G. (2000) Identification of differentially expressed genes by mutually subtracted RNA fingerprinting. Anal. Biochem., 286, 91–98. [DOI] [PubMed] [Google Scholar]
- 34.Kang D.-C., LaFrance,R., Su,Z.-Z. and Fisher,P.B. (1998) Reciprocal subtraction differential RNA display: an efficient and rapid procedure for isolating differentially expressed gene sequences. Proc. Natl Acad. Sci. USA, 95, 13788–13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho Y.-j., Meade,J.D., Walden,J.C., Chen,X., Guo,Z. and Liang,P. (2001) Multicolor fluorescent differential display. Biotechniques, 30, 562–571. [DOI] [PubMed] [Google Scholar]
- 36.Consalez G.G., Corradi,A., Ciarmatori,S., Bossolasco,M., Malgaretti,N. and Stayton,C.L. (1996) A new method to screen clones from differential display experiments prior to RNA studies. Trends Genet., 12, 455–456. [DOI] [PubMed] [Google Scholar]
- 37.Vögeli-Lange R., Bürckert,N., Boller,T. and Wiemken,A. (1996) Rapid selection and classification of positive clones generated by mRNA differential display. Nucleic Acids Res., 24, 1385–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mu J.-H., Stains,J.P. and Kao,T.-h. (1994) Characterization of a pollen-expressed gene encoding a putative pectin esterase of Petunia inflata. Plant Mol. Biol., 25, 539–544. [DOI] [PubMed] [Google Scholar]
- 39.Poirier G.M.-C., Pyati,J., Wan,J.S. and Erlander,M.G. (1997) Screening differentially expressed cDNA clones obtained by differential display using amplified RNA. Nucleic Acids Res., 25, 913–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diachenko L.B., Ledesma,J., Chenchik,A.A. and Siebert,P.D. (1996) Combining the technique of RNA fingerprinting and differential display to obtain differentially expressed mRNA. Biochem. Biophys. Res. Commun., 219, 824–828. [DOI] [PubMed] [Google Scholar]
- 41.Jurecic R., Nachtman,R.G., Colicos,S.M. and Belmont,J.W. (1998) Identification and cloning of differentially expressed genes by long-distance differential display. Anal. Biochem., 259, 235–244. [DOI] [PubMed] [Google Scholar]
- 42.Welsh J., Chada,K., Dalai,S.S., Cheng,R., Ralph,D. and McClelland,M. (1992) Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res., 20, 4965–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokolov B.P. and Prockop,D.J. (1994) A rapid and simple PCR-based method for isolation of cDNAs from differentially expressed genes. Nucleic Acids Res., 22, 4009–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozian D.H. and Kirschbaum,B.J. (1999) Comparative gene-expression analysis. Trends Biotechnol., 17, 73–78. [DOI] [PubMed] [Google Scholar]
- 45.Baldwin D., Crane,V. and Rice,D. (1999) A comparison of gel-based, nylon filter and microarray techniques to detect differential RNA expression in plants. Curr. Opin. Plant Biol., 2, 96–103. [DOI] [PubMed] [Google Scholar]
- 46.Lennon G.G. (2000) High-throughput gene expression analysis for drug discovery. Drug Discov. Today, 5, 59–66. [DOI] [PubMed] [Google Scholar]
- 47.Kuhn E. (2001) From library screening to microarray technology: strategies to determine gene expression profiles and to identify differentially regulated genes in plants. Ann. Bot., 87, 139–155. [DOI] [PubMed] [Google Scholar]
- 48.Green C.D., Simons,J.F., Taillon,B.E. and Lewin,D.A. (2001) Open systems: panoramic views of gene expression. J Immunol. Methods, 250, 67–79. [DOI] [PubMed] [Google Scholar]
- 49.Bachem C.W.B., van der Hoeven,R.S., de Bruijn,S.M., Vreugdenhil,D., Zabeau,M. and Visser,R.G.F. (1996) Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J., 9, 745–753. [DOI] [PubMed] [Google Scholar]
- 50.Prashar Y. and Weissman,S.M. (1996) Analysis of differential gene expression by display of 3′ end restriction fragments of cDNAs. Proc. Natl Acad. Sci. USA, 93, 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matz M., Usman,N., Shagin,D., Bogdanova,E. and Lukyanov,S. (1997) Ordered differential display: a simple method for systematic comparison of gene expression profiles. Nucleic Acids Res., 25, 2541–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivanova N.B. and Belyavsky,A.V. (1995) Identification of differentially expressed genes by restriction endonuclease-based gene expression fingerprinting. Nucleic Acids Res., 23, 2954–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breyne P. and Zabeau,M. (2001) Genome-wide expression analysis of plant cell cycle modulated genes. Curr. Opin. Plant Biol., 4, 138–142. [DOI] [PubMed] [Google Scholar]
- 54.Kato K. (1995) Description of the entire mRNA population by a 3′ end cDNA fragment generated by class IIS restriction enzymes. Nucleic Acids Res., 23, 3685–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutcliffe J.G., Foye,P.E., Erlander,M.G., Hilbush,B.S., Bodzin,L.J., Durham,J.T. and Hasel,K.W. (2000) TOGA: an automated parsing technology for analyzing expression of nearly all genes. Proc. Natl Acad. Sci. USA, 97, 1976–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimkets R.A., Lowe,D.G., Tai,J.T.-N., Sehl,P., Jin,H., Yang,R., Predki,P.F., Rothberg,B.E.G., Murtha,M.T., Roth,M.E. et al. (1999) Gene expression analysis by transcript profiling coupled to a gene database query. Nat. Biotechnol., 17, 798–803. [DOI] [PubMed] [Google Scholar]
- 57.Kornmann B., Preitner,N., Rifat,D., Fleury-Olela,F. and Schibler,U. (2001) Analysis of circadian liver gene expression by ADDER, a highly sensitive method for the display of differentially expressed mRNAs. Nucleic Acids Res., 29, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Z.J., Shen,H. and Tew,K.D. (2001) Gene expression profiling using a novel method: amplified differential gene expression (ADGE). Nucleic Acids Res., 29, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lisitsyn N., Lisitsyn,N. and Wigler,M. (1993) Cloning the differences between two complex genomes. Science, 259, 946–951. [DOI] [PubMed] [Google Scholar]
- 60.Diatchenko L., Lau,Y.-F.C., Campbell,A.P., Chenchik,A., Moqadam,F., Huang,B., Lukyanov,S., Lukyanov,K., Gurskaya,N., Sverdlov,E.D. and Siebert,P.D. (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl Acad. Sci. USA, 93, 6025–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang M. and Sytkowski,A.J. (1996) Cloning differentially expressed genes by linker capture subtraction. Anal. Biochem., 237, 109–114. [DOI] [PubMed] [Google Scholar]
- 62.Jiang H., Kang,D.-C., Alexandre,D. and Fisher,P.B. (2000) RaSH, a rapid subtraction hybridization approach for identifying and cloning differentially expressed genes. Proc. Natl Acad. Sci. USA, 97, 12684–12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng J., Gorski,R.A. and Hamer,D. (1994) Differential cDNA cloning by enzymatic degrading subtraction (EDS). Nucleic Acids Res., 22, 4381–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lavery D.J., Lopez-Molina,L., Fleury-Olela,F. and Schibler,U. (1997) Selective amplification via biotin- and restriction-mediated enrichment (SABRE), a novel selective amplification procedure for detection of differentially expressed mRNAs. Proc. Natl Acad. Sci. USA, 94, 6831–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 66.Spinella D.G., Bernardino,A.K., Redding,A.C., Koutz,P., Wei,Y., Pratt,E.K., Myers,K.K., Chappell,G., Gerken,S. and McConnell,S.J. (1999) Tandem arrayed ligation of expressed sequence tags (TALEST): a new method for generating global gene expression profiles. Nucleic Acids Res., 27, e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brenner S., Johnson,M., Bridgham,J., Golda,G., Lloyd,D.H., Johnson,D., Luo,S., McCurdy,S., Foy,M., Ewan,M. et al. (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol., 18, 630–634. [DOI] [PubMed] [Google Scholar]
- 68.Adams M.D., Kelley,J.M., Gocayne,J.D., Dubnick,M., Polymeropoulos,M.H., Xiao,H., Merril,C.R., Wu,A., Olde,B., Moreno,R.F. et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science, 252, 1651–1656. [DOI] [PubMed] [Google Scholar]
- 69.Cushman J.C. and Bohnert,H.J. (2000) Genomic approaches to plant stress tolerance. Curr. Opin. Plant Biol., 3, 117–124. [DOI] [PubMed] [Google Scholar]
- 70.Dunne P.W., Wang,S.-W., Ashizawa,T., Perryman,M.B. and Epstein,H.F. (1992) cDNA surveying of specific tissue expression of human chromosome 19 sequences. Genomics, 14, 263–269. [DOI] [PubMed] [Google Scholar]
- 71.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 72.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C., Kobayashi,M., Horton,H. et al. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 73.Kehoe D.M., Villand,P. and Somerville,S. (1999) DNA microarrays for studies of higher plants and other photosynthetic organisms. Trends Plant Sci., 4, 38–41. [DOI] [PubMed] [Google Scholar]
- 74.Lin X., Kaul,S., Rounsley,S., Shea,T.P., Benito,M.-I., Town,C.D., Fujii,C.Y., Mason,T., Bowman,C.L., Barnstead,M. et al. (1999) Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature, 402, 761–768. [DOI] [PubMed] [Google Scholar]
- 75.Ohlrogge J. and Benning,C. (2000) Unraveling plant metabolism by EST analysis. Curr. Opin. Plant Biol., 3, 224–228. [PubMed] [Google Scholar]
- 76.Richmond T. and Somerville,S. (2000) Chasing the dream: plant EST microarrays. Curr. Opin. Plant Biol., 3, 108–116. [DOI] [PubMed] [Google Scholar]
- 77.Knight J. (2001) When the chips are down. Nature, 410, 860–861. [DOI] [PubMed] [Google Scholar]
- 78.Welford S.M., Gregg,J., Chen,E., Garrison,D., Sorensen,P.H., Denny,C.T. and Nelson,S.F. (1998) Detection of differentially expressed genes in primary tumor tissues using representational differences analysis coupled to microarray hybridization. Nucleic Acids Res., 26, 3059–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang G.P., Ross,D.T., Kuang,W.W., Brown,P.O. and Weigel,R.J. (1999) Combining SSH and cDNA microarrays for rapid identification of differentially expressed genes. Nucleic Acids Res., 27, 1517–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin K.J., Graner,E., Li,Y., Price,L.M., Kritzman,B.M., Fournier,M.V., Rhei,E. and Pardee,A.B. (2001) High-sensitivity array analysis of gene expression for the early detection of disseminated breast tumor cells in peripheral blood. Proc. Natl Acad. Sci. USA, 98, 2646–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dias Neto E., Garcia Correa,R., Verjovski-Almeida,S., Briones,M.R.S., Nagai,M.A., da Silva,W.,Jr, Zago,M.A., Bordin,S., Ferreira Costa,F., Goldman,G.H. et al. (2000) Shotgun sequencing of the human transcriptome with ORF expressed sequence tags. Proc. Natl Acad. Sci. USA, 97, 3491–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goormachtig S., Valerio-Lepiniec,M., Szczyglowski,K., Van Montagu,M., Holsters,M. and de Bruijn,F.J. (1995) Use of differential display to identify novel Sesbania rostrata genes enhanced by Azorhizobium caulinodans infection. Mol. Plant Microbe Interact., 8, 816–824. [DOI] [PubMed] [Google Scholar]
- 83.Duhoux E. and Dreyfus,B.L. (1982) Nature des sites d’infection par le Rhizobium de la tige de la légumineuse Sesbania rostrata Brem. C. R. Hebd. Séances. Acad. Sci. Paris, 294, 407–411.
- 84.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goormachtig S., Alves-Ferreira,M., Van Montagu,M., Engler,G. and Holsters,M. (1997) Expression of cell cycle genes during Sesbania rostrata stem nodule development. Mol. Plant Microbe Interact., 10, 316–325. [DOI] [PubMed] [Google Scholar]
- 86.D’Haeze W., Gao,M., De Rycke,R., Van Montagu,M., Engler,G. and Holsters,M. (1998) Roles for azorhizobial Nod factors and surface polysaccharides in intercellular invasion and nodule penetration, respectively. Mol. Plant Microbe Interact., 11, 999–1008. [Google Scholar]
- 87.Kamalay J.C. and Goldberg,R.B. (1980) Regulation of structural gene expression in tobacco. Cell, 19, 935–946. [DOI] [PubMed] [Google Scholar]
- 88.Ruan Y., Gilmore,J. and Conner,T. (1998) Towards Arabidopsis genome analysis: monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J., 15, 821–833. [DOI] [PubMed] [Google Scholar]
- 89.Desprez T., Amselem,J., Caboche,M. and Höfte,H. (1998) Differential gene expression in Arabidopsis monitored using cDNA arrays. Plant J., 14, 643–652. [DOI] [PubMed] [Google Scholar]
- 90.Lemieux B., Aharoni,A. and Schena,M. (1998) Overview of DNA chip technology. Mol. Breeding, 4, 277–289. [Google Scholar]
- 91.Govers F., Nap,J.-P., Moerman,M., Franssen,H.J., van Kammen,A. and Bisseling,T. (1987) cDNA cloning and developmental expression of pea nodulin genes. Plant Mol. Biol., 8, 425–435. [DOI] [PubMed] [Google Scholar]
- 92.Franssen H.J., Nap,J.-P., Gloudemans,T., Stiekema,W., van Dam,H., Govers,F., Louwerse,J., van Kammen,A. and Bisseling,T. (1987) Characterization of cDNA for nodulin-75 of soybean: a gene product involved in early stages of root nodule development. Proc. Natl Acad. Sci. USA, 84, 4495–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kouchi H. and Hata,S. (1993) Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet., 238, 106–119. [DOI] [PubMed] [Google Scholar]
- 94.Hodge R., Paul,W., Draper,J. and Scott,R. (1992) Cold-plaque screening: a simple technique for the isolation of low abundance, differentially expressed transcripts from conventional cDNA libraries. Plant J., 2, 257–260. [Google Scholar]
- 95.Frugier F., Kondorosi,A. and Crespi,M. (1998) Identification of novel putative regulatory genes induced during alfalfa nodule development with a cold-plaque screening procedure. Mol. Plant Microbe Interact., 11, 358–366. [DOI] [PubMed] [Google Scholar]
- 96.Jiménez-Zurdo J.I., Frugier,F., Crespi,M.D. and Kondorosi,A. (2000) Expression profiles of 22 novel molecular markers for organogenetic pathways acting in alfalfa nodule development. Mol. Plant Microbe Interact., 13, 96–106. [DOI] [PubMed] [Google Scholar]
- 97.Covitz P.A., Smith,L.S. and Long,S.R. (1998) Expressed sequence tags from a root-hair-enriched Medicago truncatula cDNA library. Plant Physiol., 117, 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Györgyey J., Vaubert,D., Jiménez-Zurdo,J.I., Charon,C., Troussard,L., Kondorosi,Á. and Kondorosi,É. (2000) Analysis of Medicago truncatula nodule expressed sequence tags. Mol. Plant Microbe Interact., 13, 62–71. [DOI] [PubMed] [Google Scholar]
- 99.Bhuvaneswari T.V., Turgeon,B.G. and Bauer,W.D. (1980) Early events in the infection of soybean (Glycine max L. Merr) by Rhizobium japonicum. I. Localization of infectible root cells. Plant Physiol., 66, 1027–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heidstra R., Nilsen,G., Martinez-Abarca,F., van Kammen,A. and Bisseling,T. (1997) Nod factor-induced expression of leghemoglobin to study the mechanism of NH4NO3 inhibition on root hair deformation. Mol. Plant Microbe Interact., 10, 215–220. [DOI] [PubMed] [Google Scholar]
- 101.Szczyglowski K., Hamburger,D., Kapranov,P. and de Bruijn,F.J. (1997) Construction of a Lotus japonicus late nodulin expressed sequence tag library and identification of novel nodule-specific genes. Plant Physiol., 114, 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Appleby C.A. (1984) Leghemoglobin and Rhizobium respiration. Annu. Rev. Plant Physiol., 35, 443–478. [Google Scholar]
- 103.Franssen H.J., Scheres,B., Van de Wiel,C. and Bisseling,T. (1988) Characterization of soybean (hydroxy)proline-rich early nodulins. In Palacios,R. and Verma,D.P.S. (eds), Molecular Genetics of Plant-Microbe Interactions. APS Press, St Paul, MN, pp. 321–326.
- 104.Perlick A.M. and Pühler,A. (1993) A survey of transcripts expressed specifically in root nodules of broadbean (Vicia faba L.). Plant Mol. Biol., 22, 957–970. [DOI] [PubMed] [Google Scholar]
- 105.Cook D.R. (1999) Medicago truncatula – a model in the making! Curr. Opin. Plant Biol., 2, 301–304. [DOI] [PubMed] [Google Scholar]
- 106.Scheres B., van Engelen,F., van der Knaap,E., van de Wiel,C., van Kammen,A. and Bisseling,T. (1990) Sequential induction of nodulin gene expression in the developing pea nodule. Plant Cell, 2, 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Crespi M., Jurkevitch,E., Poiret,M., d’Aubenton-Carafa,Y., Petrovics,G., Kondorosi,E. and Kondorosi,A. (1994) enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J., 13, 5099–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Suganuma N., Tamaoki,M. and Kouchi,H. (1995) Expression of nodulin genes in plant-determined ineffective nodules of pea. Plant Mol. Biol., 28, 1027–1038. [DOI] [PubMed] [Google Scholar]
- 109.Cook D., Dreyer,D., Bonnet,D., Howell,M., Nony,E. and VandenBosch,K. (1995) Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell, 7, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gamas P., de Billy,F. and Truchet,G. (1998) Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol. Plant Microbe Interact., 11, 393–403. [DOI] [PubMed] [Google Scholar]
- 111.de Carvalho Niebel F., Lescure,N., Cullimore,J.V. and Gamas,P. (1998) The Medicago truncatula MtAnn1 gene encoding an annexin is induced by Nod factors and during the symbiotic interaction with Rhizobium meliloti. Mol. Plant Microbe Interact., 11, 504–513. [DOI] [PubMed] [Google Scholar]
- 112.Swiderski M.R., Zaborowska,Z. and Legocki,A.B. (2000) Identification of new nodulin cDNAs from yellow lupine by differential display. Plant Sci., 151, 75–83. [Google Scholar]